Abstract
The review examines the structured organization of interphase nuclei using a range of examples from the plants, animals, and fungi. Nuclear organization is shown to be an important phenomenon in cell differentiation and development. The review commences by examining nuclei in dividing cells and shows that the organization patterns can be dynamic within the time frame of the cell cycle. When cells stop dividing, derived differentiated cells often show quite different nuclear organizations. The developmental fate of nuclei is divided into three categories. (i) The first includes nuclei that undergo one of several forms of polyploidy and can themselves change in structure during the course of development. Possible function roles of polyploidy is given. (ii) The second is nuclear reorganization without polyploidy, where nuclei reorganize their structure to form novel arrangements of proteins and chromosomes. (iii) The third is nuclear disintegration linked to programmed cell death. The role of the nucleus in this process is described. The review demonstrates that recent methods to probe nuclei for nucleic acids and proteins, as well as to examine their intranuclear distribution in vivo, has revealed much about nuclear structure. It is clear that nuclear organization can influence or be influenced by cell activity and development. However, the full functional role of many of the observed phenomena has still to be fully realized.
The study of interphase nuclear organization and the distribution of chromatin has been a subject of interest for over 100 years (23, 24, 131), yet there is little general consensus about the significance of organization patterns, especially concerning the intranuclear locations of individual chromosomes. General questions which were first posed long ago remain pertinent. Does chromosome organization influence genetic function? Do chromosomes occur in reproducible patterns in the nuclei of species? Is chromosome organization important for development? Are chromosomes organized at random with respect to each other? Other factors concerning intranuclear organization are now widely established, perhaps too dogmatically (for example, that chromosomes occur in discrete domains). But is this true of all cell types, and is it a general feature of all organisms?
The major problem for our understanding of the functional significance of nuclear organization is that a huge variety of organisms spanning plants, fungi, and animals have been studied without any particular focus on any one. Furthermore, within the species for which data exist, a range of tissue types have been studied. Few groups, with some notable exceptions, are systematically examining intranuclear organization within well characterized cell types. The time is certainly right to target the nucleus, since probes for nucleic acids and proteins are widely available, as are fluorochromes that can be used to label and investigate living cells. However, few systematic studies are emerging. What is present is a large series of snapshots of different nuclei doing different things in different tissues in different species. This review presents examples that go far beyond the usual model organisms, but strangely, the model organisms add little extra, since in large part the nucleus, as an entity per se, has been ignored in developmental and cell activity studies.
The review commences by describing the nuclear properties of stem cells and dividing cell types (collectively called cycling cells). Following from this, I will show how nuclear structure can change with changing cellular activity in processes that do not involve developmental change, e.g., events that can occur during the cell cycle. The changes that occur are never as dramatic as can occur during development and are perhaps restricted to a certain framework established for the cell type. A fundamental question remains central to our understanding. Does nuclear organization drive changing cell activity, or is it a consequence of that activity? Attention in the review then moves to development. I have divided the developmental processes into a series of possible nuclear outcomes, i.e., polyploidy, nuclear change without polyploidy, and nuclear disintegration associated with apoptosis or programmed cell death (PCD).
There is a clear need to study interphase nuclei more extensively in model organisms. Data from different species are difficult to unify because of widely different genome sizes, chromosome numbers, complexity of cell differentiation, and underlying genetics. Recently, intranuclear organization has attracted increased interest due to in vivo studies using fluorescent markers of chromatin and proteins. It is hoped that this review will enable workers in these fields to put their data in the wider context of nuclei in different cell types and across divergent taxa. The review concentrates on higher-order chromosome organization and, to a limited extent, nucleoli and proteins involved in pre-mRNA splicing. Other important components of the nucleus, particularly nucleolar ultrastructure and proteins involved in ribosomal DNA (rDNA) transcription, rRNA splicing, packaging, and export (for reviews, see references 116 and 141), the nuclear envelope including the nuclear pores (essential for compartmentalizing the nucleus and enabling import into and export from the nucleus [for a review, see reference 62]), and nuclear lamins occurring on the inner face of the nuclear envelope (involved in maintaining nuclear shape and anchoring chromatin at the nuclear periphery in animal nuclei [for a review, see reference 124]) are not covered here.
NUCLEI OF CYCLING CELLS
The nuclei of stem cells and dividing cells, collectively called cycling cells here, are distinctive because they represent a fundamental type of cell from which all differentiated cells are derived (Fig. 1). It follows that the patterns of organization of chromosomes, proteins, and nucleoli in cycling-cell nuclei may also be arranged in a fundamental manner from which novel patterns are formed during changing cell activity and development. Cycling cells are found in all eukaryotes, and it is perhaps in these cells only that unifying nuclear phenomena can be sought. It is only in cycling cells that chromosomes are easily observed, and as a consequence, their nuclei have been the subject of much more intense study than have the nuclei of more derived, differentiated cells. The review commences with a discussion of nuclei of cycling cells because an understanding of these nuclei is essential for a proper understanding of derived cells. However, it must be remembered that cycling cells represent only a small subclass of all cells in a typical eukaryotic organism.
FIG. 1.
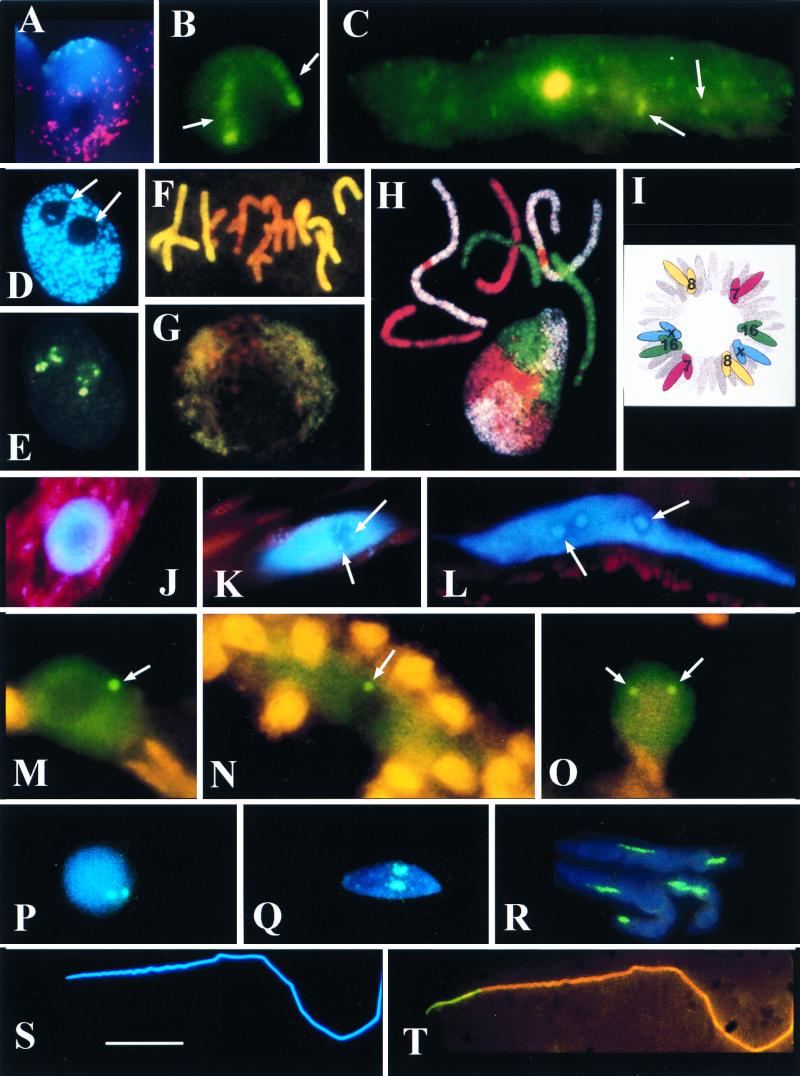
(A and B) Triticum aestivum (wheat) root tip meristematic nuclei. (A) CCS1 labelling for centromeres (digoxigenin-fluorescein isothiocyanate [FITC], cyan fluorescence) at one pole and the telomere consensus sequence (TTTAGGG)n, biotin-Cy3, red fluorescence) at the other. The red dots apparently outside the central nucleus correspond to signal from the telomeric pole of an adjacent nucleus. The nucleus is counterstained for DNA (DAPI stain, blue fluorescence). Photo courtesy of L. Aragon-Alacaide and G. Moore. (B) T. aestivum cv. Beaver carrying two 1Bl/1Rs chromosome arms has 1Rs detected by GISH (digoxigenin-labelled total Secale cereale [rye] DNA-FITC, green fluorescence). Note that the two elongate 1Rs chromosome arm domains (arrows) with the condensed subtelomeric heterochromatin fluoresce more strongly than the remainder of the arm. (C) Protophloem nucleus of T. aestivum cv. Beaver labelled by GISH with total rye DNA (digoxigenin-FITC, green fluorescence). Note that the nucleus is much larger and more elongated than in panel B and is endoreduplicated; the single large 1Rs domain is in the center of the nucleus; there is no evidence of elongate chromosome domains; and fragments of rye signal across the whole volume of the nucleus (arrows). (D and E) Sectioned nucleus from a wheat meristematic cell. (D) The nucleus is counterstained with DAPI (blue fluorescence) for DNA. Note the two spherical nucleoli (arrows). (E) The same nucleus section labelled for rDNA (digoxigenin-labelled pTa71 [59]-FITC, yellow fluorescence). The rDNA signal occurs outside the nucleolus (arrowed in panel D) on a condensed chromatin fiber and inside the nucleolus on chromatin fibers with different levels of condensation (compare with panel D). (F and G) Root tip meristematic metaphase (F) and interphase (G) of the hybrid Hordeum vulgare (barley) × Secale africanum (wild rye) labelled by GISH with total DNA from the wild rye parent (digoxigenin-FITC, yellow fluorescence) and counterstained with propidium iodide for DNA (orange fluorescence). (F) The metaphase plate shows genome separation, with seven chromosomes of wild-rye origin at the periphery and the seven chromosomes of barley origin at the center. (G) Genome separation at interphase with wild-rye chromatin outside the central barley genome. Panels F and G are taken from reference 101. (H) Chromosome painting of a metaphase and interphase nucleus of female fibroblasts of Muntiacus muntjac vaginalis (Indian munjac), chromosome 1 (biotin-Cy5, white), chromosome 2 (FITC–12-dUTP, green), chromosome X +3 (Cy3-dUTP, red) (157). Note that the chromosomes occur in discrete unpaired domains at interphase. Photograph courtesy of F. Yang and M. Ferguson-Smith. Taken from Chromosome Research. (I) Diagram of human fibroblast prometaphase showing the positions of chromosomes 7, 8, 16, and X (from reference 120). Note that a complete set of identified chromosomes are drawn on each side of the prometaphase, suggesting genome separation. Nagele et al. (120) also suggest that there is an order of chromosomes in each genome (i.e. 7, 16, X, 8) in two antiparallel sets. (J to O) Funaria hygrometrica (moss) nuclei from caulonemata (J to N) and from a thallus cell (O). (J to L) DAPI-stained nuclei (blue fluorescence) from an apical cell (J), cell 8 (K), and cell 15 (L) of the caulonemata filament. Note the increasing size and elongation of nucleus and the accumulation of rDNA heterochromatin (arrows). (M to O) Immunocytochemistry to detect D-polypeptide of the spliceosome complex (FITC detection, green fluorescence). In addition to a uniform dispersal of signal across the nucleus but outside the nucleolus, there is one coiled body in the nuclei of cells 4 (M) and 10 (N) of caulonema and two coiled bodies in the nuclei of thallus cells (arrows). All coiled bodies are associated with the nucleolus (O). (P to T) Spermatogenesis in Schistocerca gregaria (locust) stained blue for DNA with DAPI (blue fluorescence) and labelled for rDNA (pTa71, digoxigenin-FITC, green/cyan fluorescence). (P to R) Detection of rDNA in double exposures with DAPI (blue/cyan fluorescence). (P) Early spermatid nucleus with two rDNA loci. (Q and R) Increasingly mature and elongated spermatid nuclei. Note the elongating rDNA loci. (S and T) Fully mature and elongated spermatozoan nucleus, DAPI stained (S) and probed for rDNA (T, yellow fluorescence). Note that all the rDNA signal (yellow) is basal to the nucleus, suggesting intranuclear migration of rDNA. Scale bars, 15 μm for panels A, B, C, F, G, M, N, O, P, Q, R, S, and T; 10 μm for panels D and E; and 20 μm for panels H, J, K, and L.
Rabl Configuration
Rabl (131) described how chromosomes remained in their preceding anaphase configuration as the chromosomes decondensed into interphase following division. Boveri (23) examined Ascaris egg nuclei and showed that some chromosome ends at telophase reappeared at the following prophase in the same position, suggesting that chromosome distribution was fixed during the cell cycle. The distribution of chromosomes, as described by Rabl, results in nuclei with their centromeres toward one pole and their telomeres at the other. Probing interphase nuclei of cereal meristematic cells with a probe against the telomeric consensus sequence (TTTAGGG)n and a probe against the centromeric repeat (CCS1) localizes the two sequences at opposite poles (5, 140) (Fig. 1A). This Rabl configuration is shown diagrammatically in Fig. 2A.
FIG. 2.
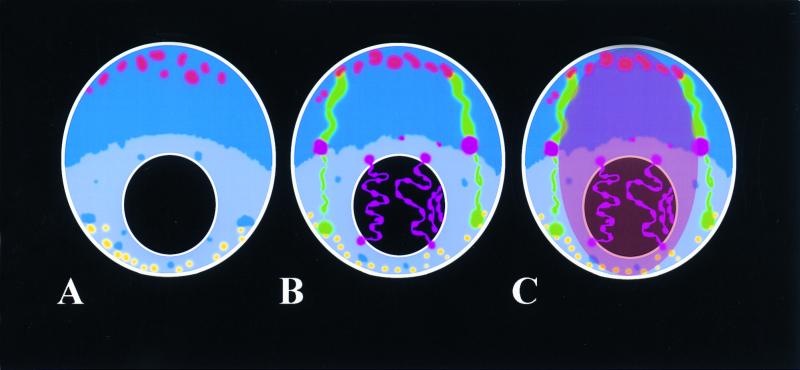
Diagrammatic projections of a root tip meristematic nucleus of a wheat cultivar that carries a 1Bl/1Rs translocation. The intensity of the blue coloration gives an indication of overall DNA condensation levels across the nucleus. The black circle is a nucleolus. (A) The nucleus is drawn in the Rabl configuration, with centromeres clustered at one pole (red) and telomeres at the other pole (yellow). (B) The same nucleus with the inclusion of the 1Rs chromosome arm domains (green), showing different levels of condensation. The rDNA loci (purple) on the 1Rs chromosome arms are drawn condensed and inactive and occupy the region of the nucleus with the highest overall level of DNA condensation. The rDNA loci from wheat chromosomes 6B and 5D are drawn extended through the nucleolus and with varying thickness to illustrate different levels of condensation and activity along each locus. (C) The same nucleus with the inclusion of a central domain to illustrate the possibility of genome separation within these nuclei.
The question whether the Rabl configuration is a feature of dividing cells in all species requires some consideration. It is clearly visible in dividing tissues of several plant species (41), e.g., wheat (2) (Fig. 1A) and field bean (77) root tip meristematic cells. In Chinese hamster cell cultures, localized UV microbeam damage to interphase nuclei, which are then allowed to progress to metaphase, results in damage to one or a few chromosomes in similar regions. Cremer et al. (43, 44) interpret this result to mean that the chromosomes are arranged in a Rabl configuration at interphase. Likewise, a Rabl configuration is predicted at interphase in Indian muntjac lymphocytes in both G1 and G2 nuclei by analysis of chromosome orientations following premature chromatin condensation experiments (147). Funabiki et al. (56) used fluorescent in situ hybridization (FISH) in combination with immunocytochemistry to show that the centromeres of Saccharomyces cerevisiae (budding yeast) were highly clustered and associated with a spindle pole body. More recently, FISH and green fluorescent protein (GFP) labelling of yeast has shown that centromeres are at the nuclear periphery and are clustered in one or a few (up to four) patches at one pole of the nucleus (84, 106). The centromeres are associated with the spindle pole body, which occurs outside the nucleus, an interaction that may be mediated via microtubules (64). The telomeres of yeast are not so strongly clustered at the nuclear envelope and occur at the opposite pole in up to 20 sites (32 telomeres in haploid strains) (84). These data strongly suggest a Rabl configuration throughout the mitotic cell cycle.
Despite these and other examples of Rabl configuration, there are examples of cycling cells where this organization of chromosomes appears to be lacking. Chung et al. (38) used FISH to analyze the positions of telomeres and minichromosomes in Trypanosoma brucei. The minichromosomes clustered together, giving strong polarity to the nucleus, but the telomeres were polarized and peripheral in fewer than 30% of the nuclei. In the remainder, they appeared more scattered across the nucleus. In nuclear spreads of cultured human cells stained for centromere proteins with CREST antibodies, the signal appears over the whole area of the nucleus, with cell-cycle dependent patterns observed (66, 152). There is no poleward clustering of centromeres as there is in, for example, wheat cycling nuclei (Fig. 1A). When lymphocyte nuclei are reconstructed by confocal optical sectioning and probed by FISH for centromeres and whole chromosomes, it is clear that the centromeres are predominately peripheral in G1 nuclei, with telomeres in a more internal location. During G2, the centromeres become more internalized (52). These results certainly show that centromeres can move during the cell cycle. Perhaps this movement causes the loss of the Rabl pattern, which would certainly be present at the end of anaphase.
It is possible that organisms with relatively small genomes and/or chromosomes, as observed in humans and particularly in Trypanosoma, mean that small shifts in centromere and telomere positions disrupt the visualization of an underlying polarity to the nucleus, which is clearly seen in organisms with larger chromosomes and/or genomes. Such an explanation could explain why UV microbeam experiments reveal a Rabl configuration in cultured cells of Chinese hamster cells (43, 44) but not in cultured human cells when probed for centromeres or telomeres (52, 66, 152). Maintenance of a Rabl polarization in the yeast genome, which is very small, may be unusually stabilized by the close association of the centromeres with the spindle pole body.
Chromosome Condensation
The nucleus of dividing cell types includes chromatin fibers at different levels of condensation, and much has been written on the subject (see, e.g., references 40, 110, and 112). Chromatin can occur in a form as condensed as at metaphase to as decondensed as naked DNA fibers. Typically, however, the DNA may be found folded around histones at the level of the nucleosome or “solenoid” (30-nm fiber) or more condensed depending on transcriptional or replication activity and sequence composition. Chromosomes themselves are thought to remain within clearly defined domains, and local decondensation is thought not to cause the domain boundaries between adjacent chromosomes to become heavily intertwined (see “Chromosome domains” below). Manuelidis (110) presented a model showing that small structural changes in the conformation of DNA could occur rapidly and locally to expose nucleosomal DNA for transcription.
The control of DNA condensation is mediated via protein- chromatin interactions and influenced by epigenetic phenomena such as histone acetylation (76) and DNA methylation (89). The abundance of cytosine methylation can vary widely between species, among different sequences in the genome (93), and across blocks of repetitive DNA (55). In species with cytosine methylation, levels of methylation within the genome can be correlated with chromatin condensation. For example, individual units of 18S-5.8S-26S rDNA can have variable methylation levels, with the active units being decondensed and undermethylated compared to inactive units (54, 100). In mammalian cells, hypomethylation is associated with a dramatic inhibition of condensation in the inactive X chromosomes, particularly in the late-replicating regions of the chromosome (67). This might be mediated via the methylation status of the linker DNA between nucleosomes which may regulate H1-dependent chromatin condensation (30).
Cook (40) challenged much conventional thinking on how chromosomes condense and suggested that there are three fundamental levels of organization: nucleosomes, loops of nucleosomal DNA, and transcription factories. He suggested that loops are connected to transcription factories that are fixed on a nuclear skeleton and contain transcription factors, RNA polymerases, and pre-mRNA splicing complexes. He proposed that increased transcriptional activity may increase the number of the factories by counteracting a tendency for them to fuse. In contrast, during mitosis, the nuclear skeleton depolymerizes, transcription ceases, proteins become phosphorylated (e.g., histone H1), and there is increased adhesiveness between factories and nucleosomes. This model suggests little higher-order organization of chromatin compaction and has not been widely accepted. However, the presence of transcription factories in the nucleus is receiving increased interest (see “Interchromosomal domains” below).
The mean level of condensation of chromatin within the interphase nucleus of dividing cells is influenced by the genome size of the organism (the proportion of the genome that is genic) and also the overall activity of the cell (see “Dynamic changes to interphase nuclei in cycling cells” below). The amount of coding DNA is likely to fall somewhere between that of yeast (6,000 genes [61]) and human (85,000 genes [42]). In yeast, coding DNA accounts for ca. 70% of the DNA (61), while in humans it is probably less than 5% (110). At interphase, much of the remaining 95% of the DNA in humans will be condensed to various degrees, up to a condensation state similar to that found at metaphase (110).
Heterochromatin includes tandem repeat sequences and whole or nearly whole chromosomes (109), and it is presumed to be largely inactive (e.g., the Barr body, which is the condensed X chromosome in female mammalian cells [107]). In human cycling cells, heterochromatin is often found either attached to or associated with the nuclear envelope, as well as at internal locations (Fig. 3A). Cavalier-Smith (35) postulated that there is a structural role for this DNA to facilitate chromosome organization at interphase via nuclear envelope interactions. In the human karyotype, housekeeping genes are predominantly clustered in the T bands (136), the majority of which are subtelomeric in location. DNA from these gene-rich bands is likely to make up a substantial component of the decondensed chromatin.
FIG. 3.
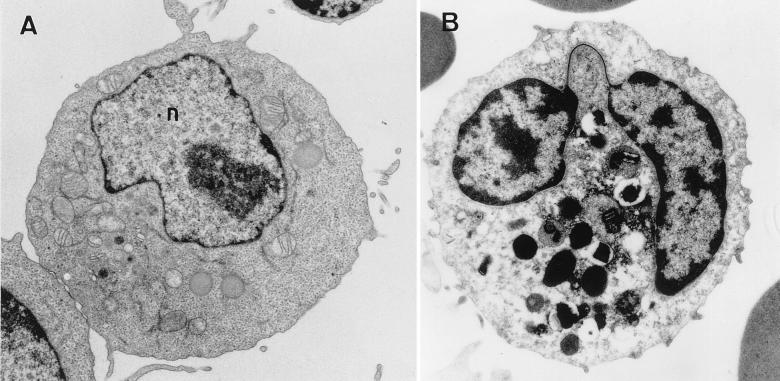
Electron micrograph of a stimulated lymphocyte (A) and a human granulocyte (B). (A) The nucleus (n) has many decondensed chromatin fibers throughout its volume. Condensed fibers are found at the nuclear periphery. (B) The highly lobed nucleus has a thin filament of chromatin connecting the lobes (arrow). Large amounts of condensed chromatin are found close to the nuclear envelope and in clumps internal to the nucleus. Magnification, ×5,000.
In cereal meristematic cell nuclei, the distribution of DNA is nonuniform. The hemisphere of the nucleus that includes the centromeres has more than 70% of the DNA (3). Fig. 1A shows this for a wheat meristematic nucleus that is DAPI (4′,6-diamidino-2-phenylindole) stained (shown diagrammatically in Fig. 2A). Transmission electron microscopy reveals that almost all of the chromatin at the centromeric pole is highly condensed (71). Much of the telomeric hemisphere of the wheat nucleus is occupied by highly decondensed DNA fibers with clumps of subtelomeric heterochromatin often associated with the nuclear envelope (3). Heslop-Harrison et al. (70, 71) speculated that this region may be where most gene expression occurs in cereal dividing cells. Abranches et al. (2) tested the idea of localized transcription within the cereal interphase nucleus by using BrUTP uptake and revealed a few hundred foci or centres, perhaps representing transcription factories, but these occurred across the volume of the nucleus and were not restricted to a particular pole.
Chromatin condensation across the nucleus is not static, and changing patterns do occur through the cell cycle and during cell differentiation, as described below. The mechanism by which changing DNA condensation patterns influence, or are influenced by, phenomena such as DNA methylation and histone acetylation is unknown.
rDNA Distribution
In many organisms rDNA occurs in high copy numbers, with the number of units varying widely between organisms, e.g., 140 rRNA genes in S. cerevisiae (haploid, unreplicated [61]) and 31,000 in Hyacinthus orientalis (80; compare the table of species in reference 4). Species with thousands of copies in the basic genome almost certainly have rDNA units that are redundant and nonfunctional (53).
In species with an apparent excess of the number of rRNA genes to sustain ribosome biosynthesis, many of the inactive gene can occur as condensed chromatin (e.g., in the pea Pisum sativum [135]). In wheat, both decondensed and condensed 18S-5.8S-26S rDNA occurs inside the nucleolus (compare the in situ signal with the DAPI signal in the nucleoli in Fig. 1D and E) while only condensed rDNA is found outside the nucleolus (99, 100) (Fig. 2B). In the root tip, cycling nuclei of wheat cultivars that contain the short arm of rye chromosome 1R (1Rs), the rDNA on this chromosome remains silent and condensed (100) (Fig. 2B). However, in haploid, dividing nuclei of germinating pollen, the 1R rDNA locus in a wheat background is decondensed and active (144), showing that the intranuclear organization and activity of the loci are dependent on the type of cycling cell.
The activity of rDNA at individual loci can also change through the cell cycle. In Petunia hybrida root tip meristematic nuclei, there are four rDNA sites, two are active through most of the interphase stages of the cell cycle. However, after division at telophase/G1, all four rDNA sites show signs of activity, suggesting either cell cycle or ribosome demand regulating gene expression at individual loci (115).
The precise location of actively transcribing genes within the nucleolus is still a matter of controversy, although transcription probably occurs in the dense fibrillar component associated with the fibrillar centers (117). Activity of the genes causes the accumulation of preribosomes as a granular component. In cereal meristematic cells, active rDNA loci tend to become clustered in a domain of the nucleus which appears to favor their transcription (18) (Fig. 2B). In human cell cultures, similar clustering of rDNA-bearing chromosomes is often maintained in the subsequent metaphase (139).
Nucleoli are dynamic structures in their activity, size, position, and number. These changing parameters must also affect the surrounding chromatin, which does not become incorporated into the nucleolus during changes in nucleolar size and number. The total nucleolar volume is determined predominantly by the transcriptional activity of the rRNA genes and the number of stored preribosomes in the granular component. Jordan et al. (86, 87) showed that the nucleolar volume decreases in cereal meristematic nuclei at high temperatures, probably because increased cytoplasmic demand for ribosomes leads to fewer stored preribosomes as a granular component (98).
Chromosome Domains
Boveri (23) was first to suggest that chromosomes at interphase occurred in territories. More recently, “chromosome painting” of animal chromosomes (105, 128) (Fig. 1H) and genomic in situ hybridization (GISH) of plant chromosomes (73, 118) (Fig. 1B, F, and G) have clearly demonstrated that chromosomes of cycling cells occur in nonintermixed domains.
A detailed structural examination of chromosome domains in interphase nuclei of human cycling cells shows that the chromatin is folded differently both between and within individual domains that reflect different types and/or activities of chromatin (45). Homologues can show different levels of DNA condensation. In cultured cycling female human amniotic cells, the inactive X chromosome domain has a similar volume to the active X domain but the surface area of the active X is significantly higher, suggesting that transcriptional activity is associated with chromatin folding at the domain periphery (49). Kurtz et al. (94) analyzed the positions of three genic sequences in cultured human interphase cells and found that they occurred in characteristic positions at the periphery of chromosome domains; these positions were not altered by transcriptional activity. The surface area of a chromosome domain is important not only because it is probably gene rich but also because it interacts with the nuclear space between chromosome domains, where gene transcription, mRNA splicing, and protein and mRNA transport are thought to occur (160).
Chromosome paints have been made for the three chromosomes in the Indian muntjac (Muntiacus muntjac vaginalis; 2n = 6), and these reveal rounded chromosome domains at interphase (157) (Fig. 1F). Chromosome domains are also visible in wheat carrying the rye chromosome arm 1Rs. This arm can be detected using GISH with total genomic rye DNA as a probe (Fig. 1B; shown diagrammatically in Fig. 2B). The chromosome domains appear more elongated than those seen in mammals (compare Fig. 1B and H). The difference could reflect the larger genome size of wheat (4C = 69.3 pg [Angiosperm DNA C-Values Database, http://www.rbgkew.org.uk/cval/database1.html]), Indian muntjac (4C = 20.4 pg [DNA Mammalian Genome Size Database, http://www.unipv.it /webbio/dbagsdb.htm]), the sequence composition of the chromosomes, or differences in plant and animal chromatin condensation.
Interchromosome Domains
The interchromosomal domain (ICD), occurring between the chromosome domains, is thought to be an important structural and functional compartment of the cycling cell nucleus (160). Protein and mRNA transport is thought to occur here as well as transcription and pre-mRNA splicing, all at the surface of the chromosome domains. Bridger et al. (26) transformed cultured human cells with the Xenopus vimentin gene carrying a nuclear localization signal and showed that the vimentin formed extended, interconnected arrays of filaments that were curvilinear, circular, or branched. The arrays were interpreted to lie within, and be part of, the ICD. Similar ICDs have been revealed in cultured mammalian cells as a reticular network of ribonuclear particles associated with pre-mRNA splicing (146). Nuclei can also have elongated tracts of mRNA in animal nuclei (156), and these too probably occur in the ICD.
The ICD of the nucleus has a complex of proteins and nucleic acids. Perhaps the best localized are the small nuclear RNAs (snRNAs), small nuclear ribonucleoproteins (snRNPs), and non-snRNP splicing factors (31–34) that have been localized in the nucleoplasm of cultured mammalian cells. Proteins involved in pre-mRNA splicing can be dispersed and can occur as speckles and foci with patterns depending on cell activity or type. When foci colocalize with β-coilin, they are termed coiled bodies. Coiled bodies have an intimate relationship with the nucleolus (Fig. 1M and N) and are distinct from intranuclear speckles (31). The role of speckles and foci is still a matter of debate (for a review, see reference 95), but it is likely that the distribution of splicing factors is dynamic, with mRNA being transcribed and processed at the gene locus, and that splicing factors shuttle from speckles in the ICD. Coiled bodies are not found in some cell types (160), and they do not colocalize with pre-mRNA, poly(A) mRNA, the splicing factor SC-35 and DNA. This indicates that they too are probably not directly involved in splicing but may be involved in snRNP storage, maturation, or transport (95).
Chromosome Distribution
The organization of chromosomes in cells at division has been unclear, and many data obtained before the 1990s are contradictory (see, e.g., references 9, 41, 151, and 155). Often this is because data were taken from spread chromosomes, where three-dimensional information has been lost, or mitotic inhibitors were used to accumulate metaphases and these may have perturbed chromosome position (133). For these reasons, and because the data from metaphase spreads is well reviewed (155), this review has concentrated on data derived by probing nuclei for specific proteins, nucleic acids, and chromosomes or by using electron or light microscopy to reconstruct in vivo nuclear organizations.
Labelling of specific chromosomes by in situ hybridization suggests little or no association of homologous chromosomes in dividing human cell types, e.g., amniotic cells (129), fibroblasts, (51), lymphocytes, (37), and HeLa cells (120, 121). Figure 1F shows that the homologues in a muntjac metaphase and interphase cell are also not tightly paired, since six domains are clearly visible. However, it is easy to envisage that in a nucleus like this, which has so few chromosomes, direct contact between almost any chromosome combination could be possible in the active nucleus.
Tight pairing of homologues is a feature of dividing cell types in the diptera, including Drosophila melanogaster (41); this association can be maintained into differentiated cell types (68, 74). Using FISH, Kleckner (90) and Burgess et al. (29) report that homologue association and pairing occurs in diploid cells of S. cerevisiae (yeast), where homologues appear to have multiple interstitial pairing contacts in dividing cells at G1 and G2, although this is disrupted in the S phase of the cell cycle. Burgess and Klechner (29) also demonstrated that interactions of loci on nonhomologous chromosomes can occur, perhaps as a consequence of nonspecific centromere clustering. Kleckner and Weiner (91) speculated that one function of somatic pairing and association of homologues in yeast may be to enable recombinational repair of DNA in G1 and G0 cells in the absence of sister chromatids. Both D. melanogaster males and S. cerevisiae also have achiasmate meiosis, and this may be linked in some way to somatic homologue pairings and associations.
Higher plants have a life cycle that includes an alternation of generations between a haploid stage and a diploid stage, and it is interesting that early land plants are thought to have had a dominant haploid generation. In haploid plants, homologue pairing without nuclear polyploidy is therefore not possible. Labelling of root tip meristematic cells of several plant species shows no pairing of homologous chromosomes (Fig. 1B) (2, 18, 19, 70). However, this may not be the case in all tissues, for example in tapetal cells of the wheat anther (see “Nuclear differentiation and the distribution of chromatin and chromosomes” below). The plant Bryonia dioica may be unusual in that it shows tight associations of homologues in root tip metaphase (46).
It has been proposed that for dividing cell types, heterologous chromosomes may occur in organized patterns with respect to each other in many organisms. Bennett (17) proposed that parental haploid genomes were spatially separate and acted to some extent independently. In hybrid plants, parental genomes can be localized and their positions can be analyzed (Fig. 1F and G). Genome separation has been observed through the cell cycle in the hybrid plant Hordeum vulgare (barley) × Secale africanum (wild rye) (Fig. 1F and G) (101). Here the chromosomes of wild rye can be seen to be spatially separate and to lie outside the central barley chromosomes (Fig. 1F and G). Haploid genome separation has also been observed in mammal cell fusion hybrids (25), early mouse embryos (126), and human fibroblasts and HeLa cells (120) (Fig. 1I). It has also been observed in the wasp Nasonia vitripennis, where it forms the basis of the haplo-diploidy and enables sex determination (154). Here a single chromosome, called the paternal sex ratio chromosome, causes the condensation, peripheralization, and elimination of all paternal chromosomes in the zygote except the paternal sex ratio chromosome to confer maleness to the zygote (154). The mechanisms underlying genome separation in these examples is unknown but may be mediated via genomic imprinting. The model of the wheat interphase nucleus (Fig. 2C) shows how the parental genomes are organized with respect to each other if the organization patterns are as those observed in barley × wild rye hybrids (Fig. 1G).
Bennett (18) suggested that in addition to genome separation, chromosomes can be distributed in predictable patterns within each haploid genome set and that the pattern can be derived from accurate measurements of chromosome arm sizes. Bennett (19) reported that his approach accurately predicts the mean spatial order of centromeres in metaphases of four grass species and can be used to arrange the chromosomes of maize in a manner that is similar to alignments of maize, wheat, and rice chromosomes into syntenic or linkage groups. Nagele et al. (120, 121) have also suggested that in addition to genome separation, chromosomes occur in specific patterns with respect to each other in cultured human fibroblasts and HeLa cells (Fig. 1I). They show that chromosomes organized at prophase are organized in radial arrays (or rosettes), segregated into tandemly linked haploid sets of 23 chromosomes with homologues of chromosomes 7, 8, 11, 16, 21, and 22 on opposite sides of the rosette.
The results of Nagele et al. and Bennett both suggest a fundamental organization of chromosomes in cycling cell types. Bennett (18, 19) suggested that this organization may play important roles in the activity of the nucleus and in maintaining synteny between species. Clearly it would be interesting to apply Bennett's model (18) to the data set of Nagele et al. (120, 121). If there are specific patterns of chromosome organization within the nucleus of cycling cell types, these patterns may not necessarily be maintained in nuclei of cells that have stopped cycling and are differentiated (see below).
These data have demonstrated considerable intranuclear organization, including phenomena such as the Rabl configuration, chromosome domains, and interchromosome domains, in dividing cells. Chromosomes also can be arranged in different patterns in different species, although the full biological significance of this order has still to be realized. Unfortunately, the dispersed nature of the data, derived from disparate organisms with different chromosome numbers, genome sizes, and chromosome morphologies, has hampered our understanding of the functional role of intranuclear chromosome distributions. Furthermore, many data have been obtained from experiments with cells growing under widely different conditions, which potentially make underlying patterns hard to determine. The effects of cell activities on nuclear organization are described below.
DYNAMIC CHANGES TO INTERPHASE NUCLEI IN CYCLING CELLS
This section deals with dynamic nuclear organization within a cell type following changing cellular, metabolic, or transcriptional activity. Changes that occur as a direct consequence of cell differentiation in development are considered later.
Dynamic changes to patterns of chromatin condensation are associated with altered cell activity and the cell cycle. Within the nucleolus, increased rDNA transcription is associated with an altered distribution of rDNA heterochromatin (142) as well as changing size and number of nucleoli (86) and nucleolus ultrastructure (78). Phytohemagglutinin stimulation of mammalian lymphocytes promotes the cell into the cell cycle from G0 and causes the decondensation of chromatin and changing ultrastructure of the nucleolus, particularly a decrease in size and an increase in the number of fibrillar centers (50, 79, 81). In human lymphocytes, these changes are accompanied by a loss of association of chromosome 15 (103) and of nucleolar organizing regions (145) on chromosomes 13, 14, 15, 21, and 22.
Over short timescales there appears to be little chromatin mobility. Abney et al. (1) laser bleached fluorescently labelled living interphase nuclei of mammal cultures (HeLa and Swiss 3T3 cells) and showed that chromatin was immobile over times of around 1 h and over distances greater than 0.4 μm. Similarly, Shelby et al. (143) labelled centromeres of HeLa cells with GFP-cenpB conjugates (cenpB is a centromere-binding protein) and showed that most centromeres remained more or less motionless for up to 2 h.
Over periods equivalent to that of the cell cycle, nuclear changes have been observed. Ferguson and Ward (52) flow sorted human lymphocytes stimulated into G1, S and G2 phase and, using in situ hybridization, observed that in G1, nuclei centromeric regions of the investigated chromosomes were at the nuclear periphery. Progression to G2 was accompanied by a shift in the positions of the centromeres toward the nuclear interior. Similar cell cycle-dependent relocations of centromeres have also been observed using anti-centromere antibodies (66, 152).
In wheat carrying chromosome arm 1Rs, increased transcriptional activity at elevated growing temperatures results in an overall decondensation of 1Rs in the meristematic cells of the root. This decondensation causes the chromosome domains to elongate, which in turn is associated with increased decondensation of rDNA and, an increase in the number and a decrease in size of nucleoli (60, 98).
Structural studies on chromosome domains have been performed in vivo. Zink et al. (159) incorporated Cy3-AR3-dUTP (a fluorescent analogue of dTTP) into replicating DNA and observed shape and positional changes of chromosome domains at interphase after several rounds of the cell cycle. Li et al. (104) and Robinett et al. (134) transformed yeast and a Chinese hamster cell line with multiple copies of the lac operator and observed these sequences in vivo over time with GFP-linked Lac repressor protein. They observed changing condensation patterns of a heterochromatic block through the cell cycle. During DNA replication, the heterochromatin moved to the nuclear interior, decondensed, and replicated to form linear chromatids. These then condensed during G2 interphase and were seen as a compact mass.
In the plant Bryonia dioica, at late S of interphase, condensed chromatin disperses (12). However, this dispersion is not dependent upon DNA synthesis, since inhibition of S phase does not inhibit the dispersion (13). Barlow (13) speculated that DNA conformation is established in late S phase for DNA synthesis without the requirement for DNA synthesis itself.
All these data suggest that changes in the distribution of chromatin and levels of chromatin condensation are associated with changing cell activity. The time frame for these changes is typically many hours within the timescale of the cell cycle. However, in most of these cases the changes are subtle and occur within a framework defined by the cell type being investigated. This is unlike many of the events associated with cell differentiation, where the nuclear framework changes completely. Gross reorganization of the nucleus is probably associated with a changing cellular role, as occurs during cell differentiation and development, the subject of the following sections of the review.
NUCLEI OF NONCYCLING CELLS
This section details the many ways in which a nucleus can reorganize in association with development. Nuclei do change in development. Nuclei of differentiated cells are frequently distinct from those of cycling cells. In brief, the process of cell differentiation can alter, sometimes profoundly, the way the nucleus is organized; its volume and shape; the overall condensation and distribution of the chromatin; the number, size, and distribution of nucleoli; and the nuclear protein content. Differentiation of the cell is often mirrored by changes to the nucleus, although which comes first and whether changes to the nucleus drive or are driven by cell differentiation are not known.
Cell differentiation is divided into three categories depending on the fate of the nucleus: (i) cell differentiation associated with polyploidy, including endoreduplication, polyteny, and endomitosis; (ii) cell differentiation associated with reorganization of some, many, or all components of the nucleus without changing the nuclear ploidy; and (iii) cell differentiation associated with nuclear disintegration, which may or may not be followed by cell death.
Nuclear Differentiation with Polyploidy
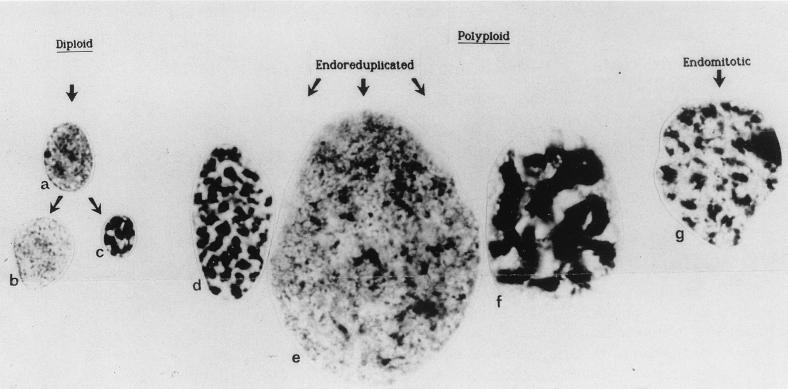
A common way for a nucleus to differentiate in development is by undergoing many rounds of DNA synthesis without an accompanied cell division, i.e., a process of nuclear polyploidization. Nuclear polyploidy is commonly encountered in eukaryotic tissues, and its occurrence in plants and animals is well reviewed (27, 122). There can be several types of polyploid nucleus even within a single organism, although the functional distinction of each is not understood. Therman (149) demonstrated this very nicely and showed a range of polyploid nuclear types in human (Fig. 4). This section of the review details some of the range of polyploid nuclei and attempts to define some of their properties, including how intranuclear components are organized. It concludes by summarizing potential roles of nuclear polyploidy, which is assumed to go beyond just the amplification of genes.
FIG. 4.
(a to e and g) Variation in the morphology and ploidy levels interphase nuclei from human female cell types. (f) Nucleus from a pea root. Taken from reference 149.
General types of polyploidy.
Nuclear polyploidy can hugely increase the DNA content of a cell. Nagl (122) reviewed maximum levels of polyploidy and reported values as high as 8,192C in the suspensor cells of the plant Phaseolus coccineus and 524,288C (i.e., 219C) in silk glands of the insect Bombyx mori. Therman (149) reviewed several mechanisms that give rise to polyploidy. (i) The first is endoreduplication, where genomes replicate without cell division. This is common. In many organisms the chromatids remain tightly associated, forming polytene chromosomes, and these have been found in a diverse range of tissues and taxa (see the table in reference 122). In Drosophila, many tissues of the embryo contain cells with polytene chromosomes (74), and in vascular plants, they regularly occur in synergid and tapetal cells (122). (ii) Another mechanism is endomitosis, where replicated chromosomes condense as if entering mitosis but then do not segregate; instead, they remain together within a single nucleus. Endomitosis has been elegantly filmed in vivo in colchicine-treated plant endosperm cells. The sister chromatids at metaphase can be seen to separate but not to segregate, leading to chromosomes in “ski pairs” (11). A process like this is thought to be important in plant evolution, where increases in chromosome number in germ cell lineages give rise to polyploid gametes. Up to 70% of angiosperms and 95% of pteridophyte species are thought to have undergone polyploidization in their evolution (102).
Differential amplification, where there is nonuniform amplification of particular sequences, chromosomes, or subsets of the chromosomes is also encountered in plants (149) (see below) and animals (122). In D. melanogaster, this is associated with underrepresentation of heterochromatin (57, 69). Polyploidy may also be accompanied by genome reorganization via DNA splicing, as occurs in the ciliate Oxytricha (122), and by polytene chromosome breakage, as occurs in some cells of D. melanogaster (74).
Polyploid nucleus differentiation.
It is often difficult to monitor the fate of nuclei during cell differentiation, because events occur within complex three-dimensional tissues. Perhaps for this reason, few polyploid cell lineages have been examined during development. However, during development polyploid nuclei can themselves differentiate. In ciliates, this is associated with a complete restructuring of the genome (130). In Delia antiqua (Diptera), polytene chromosomes develop in nurse cells that nourish the oocyte. During maturation of the oocyte, the polytene chromosomes condense and then fall apart as a large number of separate, highly condensed metaphase-like chromosomes before decondensing into an apparently “ordinary” polyploid nucleus (68). The reasons for these transitions are unknown.
Moss caulonemata lend themselves to developmental studies of the nucleus because cell and nuclear differentiation can be observed over time in vivo (92). Caulonemata are filaments of cells growing by cell division of an apical cell. Nuclei of the apical cell are haploid and spherical with a large central nucleolus. Mitotic divisions of this cell cut off a linear series of cells behind. Nuclei of these cells undergo polyploidy up to 8C (Fig. 1J to L), increase their volume, and acquire long polar extensions (Fig. 1L). The increase in DNA amount does not occur by DNA doubling as is usual for polyploid cells (1C, 2C, 4C, 8C, e.g., as can occur in polyteny [122]) but by the addition of single genomes (1C, 2C, 3C, … 8C). This suggests differentiation between the haploid genomes such that only one genome at a time replicates (92). Such specialization of component genomes may be mediated through DNA methylation or other genomic imprinting mechanism.
In the development of roots of higher plants, the cell lineages giving rise to protophloem cells become polyploid (47, 48). In wheat, development starts at the root apex in a series of well-defined steps in which changing patterns of nuclear morphology are observed. Initially there are formative divisions. At completion, the nuclei endoreduplicate and the chromatin becomes condensed and peripheralized but the nuclear volume does not decline. Thereafter, the nucleus becomes lobed and fragmented, and small dense chromatin masses lie at the periphery of the cell, often surrounded by membranes that connect with the endoplasmic reticulum (16, 47). Thus, in the protophloem lineage, nuclei enlarge and endoreduplicate, the DNA condenses, and the nucleus fragments.
These examples show that nuclear polyploidy is involved in development and that during the course of cell differentiation, the nucleus can enlarge its DNA content in different ways. Furthermore, once DNA replication is complete, the nucleus can continue to change and undergo further differentiation. The reasons for these complexities are unknown.
Chromosome distribution in polyploid nuclei.
Apart from a few examples, very little is known about the distribution of chromosomes in polyploid nuclei. The best understood are the chromosomes in polytene nuclei of the diptera. Here, homologues are intimately paired as well as endoreduplicated (122), and in some cell types they are organized in a Rabl configuration that is thought to have been established in early diploid cells (63). Hochstrasser and Sedat (74) showed that in D. melanogaster polytene nuclei of prothoracic and salivary gland cells, heterologous chromosomes can also be associated. Chromosome arm 2L is regularly next to 2R and 3L is next to 3R. In some species of mosquitoes, there is such extensive pairing of heterologues that mapping of polytenes using polytene cell spreads is almost impossible (21). These examples of heterologue interaction may well play a role in the regulation of nuclear activity, perhaps through the interactions of gene regulators on genes of adjacent chromosomes.
In wheat cultivars that carry the rye chromosome arm 1Rs, the distribution of 1Rs during the development of vascular tissues can be examined. Meristematic nuclei show two elongate 1Rs domains arranged in a Rabl configuration (Fig. 1B). After endoreduplication, this pattern of nuclear organization is lost, the Rabl configuration disappears, and, instead, some nuclei show one large region of rye chromatin (as identified using GISH [Fig. 1C]). Endoreduplication is associated here with an entirely different organization from that found in meristematic cells.
Significance of polyploid nuclei.
It is generally assumed that polyploidy occurs to amplify genes without the energetically demanding process of cell division. Thus, many secretory cell types are polyploid (e.g., D. melanogaster salivary gland cells). Polyploid nuclei can be induced following some form of stimulus; for example, when resistant cultivars of barley are challenged with powdery mildew, there is a detectable increase in ploidy levels after only 2 h (10). This may occur to generate sufficient gene product to elicit the resistance response.
A gene amplification-expression argument to explain polyploidy does not explain the several types of polyploidy found in a single organism (Fig. 4) or why polyploid nuclei themselves undergo developmental changes. In the formation of the polyploid macronucleus of ciliates, development is associated with the elimination of nonfunctional DNA, the restructuring of the DNA, and the massive amplification of functional genes (130). In D. melanogaster larvae, polytene chromosomes of the gut can appear broken, with autosome arms apparently separated across the nucleus (74). Perhaps polyploidy is associated with chromatin fragmentation and genome restructuring occurs more commonly than is generally thought, but there are too few data to confirm this. There are other potential roles for polyploidy. In polyploid nuclei, chromosome arms might be able to associate in a manner that is impossible without multiple copies of each chromosome. Such interactions might be important for chromosome trans-sensing (148). Alternatively, nuclear polyploidy could amplify the genetic component of a cell which is destined to be long-lived and perhaps vulnerable to mutation. In so doing, nuclear polyploidy might extend the duration of cell viability.
This section has shown that a range of polyploid nuclear types can be found in a single organism and that polyploid nuclei can themselves differentiate during the course of development. Little is known about the intranuclear structure of polyploid nuclei, except perhaps in cells of D. melanogaster with polytene chromosomes (74, 75). Both homologue and heterologue associations can be found in polyploid nuclei along with differential amplification and chromosome fragmentation. The full significance of polyploidy is unknown. However, it could play roles in gene amplification, genome restructuring, chromosome interactions, and cell longevity.
The following section addresses the fate of nuclei during cell differentiation, but in this case without associated polyploidy. There are similarities to polyploid nuclei, including associations of homologous and heterologous chromosomes and substantial structural reorganization of the nucleus from that found in cycling cells.
Nuclear Differentiation without Polyploidy
Nuclei can reorganize in development without undergoing polyploidy, although how and why the nuclear changes occur is largely guesswork. More work is needed to relate changing cell activity and the expression of genes in development to specific organizational properties of the nucleus, including the intranuclear distribution of genes and chromosomes. Only then will the full functional significance of nuclear organization in differentiated cells be properly understood. This section illustrates the different organizational properties of differentiated nuclei that are not polyploid and demonstrates how these organization patterns differ from those previously encountered in cycling cell types. Unfortunately, it is not clearly understood why nuclei differentiate at all. However, nuclear differentiation is a real phenomenon. It is clearly important to consider cell type in any analysis of intranuclear organization, since with changing cell type, different intranuclear organizations are likely to be encountered.
Noncycling, differentiated cells that are diploid (or haploid, as in lower-plant thalli) can have much more nuclear variation than the cycling cells from which they were derived. Nuclei can differ in shape; volume (18); organization of chromatin and chromosomes (111); size, number, and distribution of nucleoli (127); and distribution of nuclear proteins (160). One of the most obvious examples of nuclear differentiation without polyploidy is found in mammalian blood cell types, where nuclei can be highly lobed (Fig. 3B), and in spermatogenesis, where nuclei may take on extremely elongated morphologies (Fig. 1S).
Nuclear differentiation and nuclear proteins.
Electron microscopy of nuclei from differentiated, noncycling cells that have not undergone polyploidy shows that some cell types can accumulate intranuclear proteins and that these can appear as crystals, crystalline bodies, coiled bodies, or electron-dense structures (153). An example of how nuclear proteins can change during development can be seen in the moss Funaria hygrometrica. In apical haploid cells of the caulonemata, a component of the pre-mRNA splicing machinery (the D polypeptide) occurs dispersed across the volume of the nucleus but outside the nucleolus. In derived cells behind the apex, which do not divide, a single coiled body is additionally observed. The coiled body is associated with the nucleolus and is found in all remaining cells of the caulonemata irrespective of ploidy level (92, Fig. 1M and N). However in differentiated haploid cells of the thallus, all nuclei have two coiled bodies associated with the nucleolus (Fig. 1O). Therefore, haploid cells of different F. hygrometrica tissues regularly have no, one, or two coiled bodies depending on the particular cell type examined.
Cell-type-specific distributions of snRNPs involved in pre-mRNA splicing have also been observed in mammals. In this case the examples come from cell cultures from different tissues. Zirbel et al. (160) used immunocytochemistry to examine the distribution of snRNPs in 10 different primary mammalian cell cultures and found cell type differences with signal in a dispersed, patchy, or speckled distribution in some nuclei, while others had additional larger foci at the surface of chromosome domains. These differences presumably represent differing specific or total activities of the different cell types. Sahlas et al. (137) showed that the distributions of snRNP, the non-snRNP splicing factor SC-35, and nuclear actin were different in cultured neuronal and nonneuronal cell types. Once again, the differences probably reflect the different total activities of the cell types. However, in all these cases the underlying reasons for the different distribution patterns are unknown, since all the cells have a requirement for pre-mRNA splicing.
Nuclear differentiation and DNA condensation.
The amount and distribution of condensed chromatin can vary without polyploidy in different nuclei of the same organisms. Examination of electron microscopic ultrastructure in a range of cell types of the same organism will reveal much variation (compare Fig. 3A and B). At its extreme, chromatin can be completely condensed, as occurs in sperm nuclei of locust (Fig. 1S). Rae and Franke (132) localized heterochromatin in interphase nuclei by in situ hybridization to sections of different mouse tissues. They observed cell type patterns of chromatin condensation in Sertoli cells and spermatids of the testis. Later, Chandley and Speed (36) showed that during puberty and the onset of spermatogenesis in mice, the condensation of the Y chromosome changes. In prepubertal Sertoli cells, which are a cycling cell type, the Y chromosome is condensed, while in primordial germ cells, it is more decondensed. Later in development, and coinciding with the first appearance of spermatids, the prepubertal Sertoli cells mature and cease to divide. This is associated with a decondensation of the Y chromosome. Thus, condensation patterns of individual chromosomes are associated with changing cell activity associated with development.
Nuclear differentiation and the distribution of chromatin and chromosomes.
Chromatin and chromosomes can redistribute in the absence of nuclear polyploidy during cell differentiation. During spermatogenesis, the nuclei of some animals and lower plants can undergo dramatic reorganization associated with substantial nuclear elongation. In vertebrate spermatogenesis, CREST sera raised against centromeres in haploid developing sperm show that development is associated with the pairing of at least some heterologous centromeres (65). In the locust Schizocerca gregaria, probes against rDNA reveal two loci in early spermatogonia (Fig. 1P and Q). With maturation, the nuclei elongate enormously and, as they do, the rDNA elongates and then migrates to the base of the nucleus (Fig. 1R to T). Similar observations have also been made during spermatogenesis in planarians (85). The factor that drives these chromatin relocations is unknown.
In the formation of blood cell types, some extremely differentiated forms of nuclei can be observed. For example, mammalian neutrophils have nuclear lobes separated by regions that are highly constricted and contain very little chromatin (as in the granulocyte [Fig. 3B]). In human females, the inactive X chromosome occurs in a minor lobe described as a drumstick (114). Likewise, in males, the Y chromosome may be found in a club-shaped minor lobe (96). These observations led Sanchez et al. (138) to question whether the major lobes of the neutrophil nuclei also had characteristic chromosome contents. They investigated the distribution of the sex chromosomes and autosomes 2 and 18 by using chromosome paints and found the distribution of chromosomes in the major lobes to be variable, although there were significant biases toward cosegregation of the two X chromosomes in four lobed female neutrophils and of both homologues in the same lobe.
Close homologue pairing is a feature of the diptera at metaphase (68) and in polytene nuclei (122), and it can be assumed to occur in other differentiated cells on the basis of analysis of gene expression patterns. Pairing may be responsible for the trans-sensing (transvection) effects described by Tartof and Henikoff (148), whereby the expression of a gene is influenced by the gene sensing its homologue. Tartof and Henikoff (148) viewed the trans-sensing phenomenon as a pathway of interactions whose final physiological result was appropriate gene expression. This may be mediated via the diffusion of gene products or transcription precursors between closely situated chromosomes.
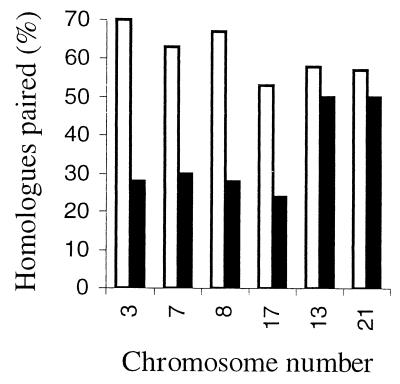
Leitch et al. (97) speculated that a reorganization of chromosomes giving rise to homologue pairing was associated with cell differentiation in human cells. No marked association of homologues was observed in cycling cells (see above). However, several differentiated noncycling cell types do show homologue pairing. In cerebellar cell nuclei, chromosomes 1 and 17 are associated (7, 8). In human Sertoli cells, the homologues of chromosomes 3, 7, 8, and 17 were associated in 53 to 70% of cells as opposed to only 24 to 30% in phytohemagglutinin-stimulated lymphocytes, which are cycling cells (Fig. 5). Interestingly, chromosomes 13 and 21, which are nucleolus-organizing chromosomes, are similarly distributed in Sertoli cells and stimulated lymphocytes, and this is probably because these chromosomes have a tendency to cluster owing to their activity in the formation of the nucleolus (37).
FIG. 5.
Graph to show the association of homologous chromosomes in nondividing adult human Sertoli cells (open) and stimulated lymphocytes (solid). The distribution of chromosomes is significantly different between Sertoli cells and stimulated lymphocytes for chromosomes 3, 7, 8, and 17. Only for the acrocentric chromosomes 13 and 21, which form nucleoli in the center of the nucleus in both cell types, are there similar distributions. Chandley et al. (37) suggested that homologue pairing was a feature associated with cell differentiation.
In cereals, root tip cycling cells do not show pairing of homologous chromosomes (2). However, this might not be the case in derived differentiated cell types. Aragon-Alcaide et al. (6) used GISH with total genomic DNA from barley and observed homologue pairing of two barley chromosomes in premeiotic anther cells of a wheat cultivar carrying a barley substitution. Interestingly, when homologue pairing becomes apparent in the developing meiocytes, the tapetal cells that surround them, which do not themselves undergo meiosis, also show homologous chromosome associations. This suggests that homologue pairing and meiosis are independent processes. Perhaps there is also a diffusable factor from the meiocytes or the tapetal cells which promotes the homologue associations.
Other chromosome interactions may also be important, such as those predicted in the models of Bennett (18, 19) and Nagele et al. (120, 121). However, distribution patterns can change with cellular activity. Borden and Manuelidis (22) examined the distribution of the X chromosome in human neurons. They showed that in individuals with epilepsy, relocation of an X chromosome centromere to a more internal nuclear position was associated with the different cellular activities of the neurons in epileptic individuals.
This section has demonstrated that nuclei can reorganize substantially without nuclear polyploidy during development. Indeed, almost any aspect of nuclear organization examined will reveal cell-type-specific patterns. However, data sets show clearly that nuclear organization is not identical in every cell of a type. It is only possible to speculate about the reasons for the variability. Perhaps the reasons relate to different activities of individual cells being examined or some lack of uniformity in the cell types being investigated.
The review now considers nuclear differentiation associated with apoptosis (PCD). Here, too, the nucleus is intimately involved. Interestingly, PCD is first observed in the nucleus, many of the defining features of PCD centre on nuclear morphology, and the nucleus is the last structure to disappear with the death of the cell.
Nuclear Disintegration
PCD is crucial in development. It is now the subject of intense interest to developmental biologists (see, for example, reference 113). Many of the characters which define PCD and distinguish it from necrosis relate to the nucleus. PCD characteristics include the marginalization and condensation of chromatin in the nucleus, internucleosomal DNA fragmentation, nuclear shrinkage, membrane blebbing associated with membrane retention, and sequestration of the cellular components into intact membrane-bound vesicles termed apoptopic bodies (88). The nuclear envelope survives until quite late in the process, being one of the last structures to disappear with autolysis of the cell (15). The fragmentation of nuclear DNA, typically into ∼50-kb fragments (39), is one of the best-defined biochemical events of PCD. These fragments can be detected in vitro by electrophoresis and in vivo by terminal deoxynucleotidyl transferase-mediated dUTP nick end labelling (TUNEL) of DNA (58).
PCD is intimately associated with a family of intracellular cysteine proteases, called caspases, which are believed to bring about many of the morphological changes observed in the cell (82, 119, 123). For example, nuclear structure is thought to be lost in mammalian cells when caspases disassemble the nuclear lamina involved in maintaining nuclear integrity (150). Caspases have been found in organisms ranging from the nematode Caenorhabditis elegans to humans.
Cells undergoing PCD have been investigated by cell biologists for many years, and elegant studies have been conducted (although most papers are not couched in the modern terminology associated with PCD). For example, in Zea mays root cap development, the nuclei undergo many changes over a few hundred micrometers of root tissue. The cells replicate from 2C up to 8C, a process accompanied by increased nuclear volume and increased condensed chromatin. Then, apoptosing nuclei become highly heterochromatinized and pycnotic; this is followed by nuclear degradation and DNA loss. Nevertheless, these dying cells still take up [3H]uridine, showing that RNA synthesis is still occurring during nuclear degradation and apoptosis (14).
Although the nucleus is one of the first structures where the initiation of PCD can be detected, only one work (as far as I am aware) addresses nuclear disintegration systematically using in situ probes and chromosome paints (A. Jausch, C. Lengauer, B. Schoell, H. H. Holtgreve-Grez, and T. Cremer, Cytogenet. Cell Genet. 77:22, 1997 [abstract]). This subject will surely be a fruitful area for considerable study. In the intriguing situations where the nucleus disintegrates but the cell continues to survive, for example in the formation of erythrocytes in mammals and sieve elements in plant, it is likely that only part of the PCD pathway has been activated. Comparing nuclear disintegration in these tissues with that in cells undergoing complete PCD is likely to shed much light on the nuclear processes involved and on PCD in general.
CONCLUDING STATEMENT AND FUTURE DIRECTIONS
Current data suggest that the nucleus of each cell type has a structured framework within which local changes in the organization and distribution of proteins and nucleic acids can occur with changing cell activity. The structured framework appears to change fundamentally during cell differentiation, establishing new patterns that are characteristic of that cell type. It is thus of absolute importance to define cell type in performing experiments to analyze nuclear structure.
The many results described here suggest that chromatin, chromosome, and nuclear organization are important to the control of development and differentiation; the central question is, what is that role? At present it is only possible to speculate on its identity. (i) One possible function is to bring individual genes or clusters of genes required for the particular cell type into transcriptionally active regions of the nucleus. Immunocytochemical methods have already shown that transcription is highly compartmentalized within the nucleus (33, 34). Thus, genes or gene clusters may have to be brought into regions of the nucleus which are transcriptionally or DNA replication active. In cells where a particular activity is not required, chromatin may be “moved” away into sectors of the nucleus to become inactive. (ii) A second possible function is to bring individual genes or banks of genes on chromosome arms into close proximity to their homologous partner or to gene regulators and promoters on heterologous chromosomes. Clearly, chromosomal interactions need not necessarily occur only between homologous chromosomes, and, as already described, heterologous associations have been reported, for example in the distribution of centromeres in vertebrate spermatogonia (65), the organization of chromosomes in human fibroblasts (120), and the distribution of D. melanogaster polytene chromosomes (74).
More well-targeted experiments and developmental analyses of nuclear reorganization are needed to more fully understand the role of nuclear reorganization in development. Time course experiments are powerful, as so elegantly shown in Bajer's (11) cine films of mitosis. Later, Oakley (125) studied meiosis in vivo in the nematode Mesostoma ehrenbergii and showed shuffling of chromosomes into haploid sets, giving hints about some cellular recognition system for chromosome identity. More recently, Abney et al. (1), Zink et al. (159), and Li et al. (104) used fluorochromes to analyze chromatin mobility over time, and these studies together are starting to give us insight into the dynamics of interphases. More of these experiments are needed, in particular to monitor individual nuclei in cell lineages over time during development. As Therman (149) rightly says, “the field is ripe for exploration.”
ACKNOWLEDGMENTS
I thank the BBSRC for support.
I thank M. C. P. Glyn, T. Kalynyak, K. Kingham, and K. Y. Lim for assistance with the data in this paper. I also thank Drs P. Barlow, L. Aragon-Alcaide, A. Levy, and T. Hartman for helpful comments on the manuscript.
REFERENCES
- 1.Abney J R, Cutler B, Fillbacj M L, Axelrod D, Scalettar B A. Chromatin dynamics in interphase nuclei and its implications for nuclear structure. J Cell Biol. 1997;137:1459–1468. doi: 10.1083/jcb.137.7.1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Abranches R, Bevan A F, Aragon-Alcaide L, Shaw P J. Transcription sites are not correlated with chromosome territories in wheat nuclei. J Cell Biol. 1998;143:5–12. doi: 10.1083/jcb.143.1.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Anamthawat-Jönsson K, Heslop-Harrison J S. Centromeres, telomeres and chromatin in the interphase nucleus of cereals. Caryologia. 1990;43:205–213. [Google Scholar]
- 4.Appels R, Honeycutt R L. rDNA: evolution over a billion years. In: Datta S K, editor. DNA systematics. II. Plants. Boca Raton, Fla: CRC Press Inc.; 1986. pp. 81–135. [Google Scholar]
- 5.Aragon-Alcaide L, Miller T, Schwarzacher T, Reader S, Moore G. A cereal centromeric sequence. Chromosoma. 1996;105:261–268. doi: 10.1007/BF02524643. [DOI] [PubMed] [Google Scholar]
- 6.Aragon-Alcaide, L., S. Reader, A. Bevan, P. Shaw, T. Miller, and G. Moore. Association of homologous chromosomes during floral development. Curr. Biol. 7:905–908. [DOI] [PubMed]
- 7.Arnoldus E P J, Noordermeyer I A, Peters A C B, Raap A K, Van der Ploeg M. Interphase cytogenetics reveals somatic pairing of chromosome 17 centromeres in normal human brain tissue, but no trisomy 7 or sex chromosome loss. Cytogenet Cell Genet. 1991;56:214–216. doi: 10.1159/000133092. [DOI] [PubMed] [Google Scholar]
- 8.Arnoldus E P J, Peters A C B, Bots G T A M, Raap A K, Van der Ploeg M. Somatic pairing of chromosome 1 centromeres in interphase nuclei of human cerebellum. Hum Genet. 1989;83:231–234. doi: 10.1007/BF00285162. [DOI] [PubMed] [Google Scholar]
- 9.Avivi L, Feldman M. Arrangement of chromosomes in the interphase nucleus of plants. Hum Genet. 1980;55:281–295. doi: 10.1007/BF00290206. [DOI] [PubMed] [Google Scholar]
- 10.Baluska F, Bacigalova K, Oud J L, Hauskrecht M, Kubica S. Rapid reorganisation of microtubular cytoskeleton accompanies early changes in nuclear ploidy and chromatin structure in post-mitotic cells of barley leaves infected with powdery mildew. Protoplasma. 1995;185:140–151. [Google Scholar]
- 11.Bajer A. Cine-micrographic studies on mitosis in endosperm 1. Acta Soc Bot Pol. 1954;23:383–412. [Google Scholar]
- 12.Barlow P W. Changes in chromatin structure during the cell cycle. Protoplasma. 1977;91:207–211. doi: 10.1007/BF01276735. [DOI] [PubMed] [Google Scholar]
- 13.Barlow P W. The dispersion of chromocentres in plant nuclei and its relation to DNA synthesis. Caryologia. 1984;37:167–176. [Google Scholar]
- 14.Barlow P W. Nuclear chromatin structure in relation to cell differentiation and cell activation in the cap and quiescent centre of Zea mays L. J Exp Bot. 1985;36:1492–1503. [Google Scholar]
- 15.Barinaga M. Stroke-damaged neurons may commit cellular suicide. Science. 1998;281:302–303. doi: 10.1126/science.281.5381.1302. [DOI] [PubMed] [Google Scholar]
- 16.Behnke H D, Sjolund R D. Sieve elements. Berlin, Germany: Springer-Verlag KG; 1990. [Google Scholar]
- 17.Bennett M D. Nucleotypic basis of the spatial ordering of chromosomes in eukaryotes and the implication of order for genome evolution and phenotypic variation. In: Dover G A, Flavell R B, editors. Genome evolution. London, United Kingdom: Academic Press, Ltd.; 1982. pp. 239–261. [Google Scholar]
- 18.Bennett M D. Nuclear architecture and its manipulation. In: Gustafson J P, editor. Gene manipulation in plant improvement. New York, N.Y: Plenum Publishing Corp.; 1984. pp. 469–502. [Google Scholar]
- 19.Bennett M D. The nucleotype, the natural karyotype and the ancestral genome. In: Heslop-Harrison J S, editor. Unifying plant genomes. Society for Experimental Biology symposium 50. Cambridge, United Kingdom: The Company of Biologists; 1996. pp. 45–52. [PubMed] [Google Scholar]
- 20.Reference deleted.
- 21.Besansky N J, Collins F H. The mosquito genome: organisation and manipulation. Parasitol Today. 1992;8:186–192. doi: 10.1016/0169-4758(92)90262-z. [DOI] [PubMed] [Google Scholar]
- 22.Borden J, Manuelidis L. Movement of the X chromosome in epilepsy. Science. 1988;242:1687–1691. doi: 10.1126/science.3201257. [DOI] [PubMed] [Google Scholar]
- 23.Boveri T. Die Befruchtung und Teilung des Eies von Ascaris megalocephala. In: Fischer G, editor. Zellen-Studien, H. 2. 1888. pp. 1–189. Jena, Germany. [Google Scholar]
- 24.Boveri T. Die Blastomerenkerne von Ascaris megalocephala und die Theorie der Chromosomenindividualität. Arch Zellforsch. 1909;3:181–286. [Google Scholar]
- 25.Brandriff B F, Gordon L A, Segraves R, Pinkel D. The male-derived genome after sperm-egg fusion: spatial distribution of chromosomal DNA and paternal-maternal genomic association. Chromosoma. 1991;100:262–266. doi: 10.1007/BF00344160. [DOI] [PubMed] [Google Scholar]
- 26.Bridger J M, Hermann H, Münkel C, Lichter P. Identification of an interchromosomal compartment by polymerization of nuclear-targeted vimentin. J Cell Sci. 1998;111:1241–1253. doi: 10.1242/jcs.111.9.1241. [DOI] [PubMed] [Google Scholar]
- 27.Brodsky V Y, Uryvaeva I V. Genome multiplication in growth and development: biology of polyploid and polytene cells. Dev. Cell Biol. Ser. Vol. 15. Cambridge, United Kingdom: Cambridge University Press; 1985. [Google Scholar]
- 28.Burgess S M, Kleckner N. Collisions between yeast chromosomal loci in vivo are governed by three layers of organisation. Genes Dev. 1999;15:1871–1973. doi: 10.1101/gad.13.14.1871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Burgess S M, Kleckner N, Weiner B M. Somatic pairing of homologs in budding yeast: existence and modulation. Genes Dev. 1999;15:1627–1642. doi: 10.1101/gad.13.12.1627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Caiafa P, Reale A, Santoro R, Marenzi S, Zardo G, Strom R. Does hypomethyation of linker DNA play a role in chromatin condensation? Gene. 1995;157:247–251. doi: 10.1016/0378-1119(95)00115-m. [DOI] [PubMed] [Google Scholar]
- 31.Carmo-Fonseca M, Pepperkok R, Carvalho M, Lamond A. Transcription-dependent colocalisation of the U1, U2, U4, and U5 snRNPs in coiled bodies. J Cell Biol. 1992;117:1–14. doi: 10.1083/jcb.117.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Carmo-Fonseca M, Tollervey D, Pepperkok R, Barzbino S M L, Merdes A, Brunner C, Zamore P D, Green M R, Hurt E, Lamond A. Mammalian nuclei contain foci which are highly enriched in components of the pre-mRNA splicing machinery. EMBO J. 1991;10:195–206. doi: 10.1002/j.1460-2075.1991.tb07936.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Carter K C, Bowman D, Carrington W, Fogarty K, McNeil J A, Fay F S, Lawrence J B. A three-dimensional view of precursor messenger RNA metabolism within the mammalian nucleus. Science. 1993;259:1330–1334. doi: 10.1126/science.8446902. [DOI] [PubMed] [Google Scholar]
- 34.Carter K C, Taneja K L, Lawrence J B. Discrete nuclear domains of Poly(A)RNA and their relationship top the functional organisation of the nucleus. J Cell Biol. 1993;115:1191–1202. doi: 10.1083/jcb.115.5.1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cavalier-Smith T. Nuclear volume control by nucleoskeletal DNA, selection for cell volume and cell growth, and the solution of the DNA C-value paradox. J Cell Sci. 1978;34:247–278. doi: 10.1242/jcs.34.1.247. [DOI] [PubMed] [Google Scholar]
- 36.Chandley A C, Speed R M. A reassessment of the Y chromosomal behaviour in germ cells and Sertoli cells of the mouse revealed by in situ hybridisation. Chromosoma. 1995;104:282–286. doi: 10.1007/BF00352259. [DOI] [PubMed] [Google Scholar]
- 37.Chandley A C, Speed R M, Leitch A R. Different distributions of homologous chromosomes in adult Sertoli cells and in lymphocytes signify nuclear differentiation. J Cell Sci. 1996;109:773–776. doi: 10.1242/jcs.109.4.773. [DOI] [PubMed] [Google Scholar]
- 38.Chung H-M M, Shea C, Fields S, Taub R N, Van der Ploeg LHT. Architectural organization in the interphase nucleus of the protozoan Trypanosoma brucei: location of telomeres and mini-chromosomes. EMBO J. 1990;9:2611–2619. doi: 10.1002/j.1460-2075.1990.tb07443.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cohen J J. Apoptosis. Immunol Today. 1993;14:126–130. doi: 10.1016/0167-5699(93)90214-6. [DOI] [PubMed] [Google Scholar]
- 40.Cook P R. A chromomeric model for nuclear and chromosomal structure. J Cell Sci. 1995;108:2927–2935. doi: 10.1242/jcs.108.9.2927. [DOI] [PubMed] [Google Scholar]
- 41.Comings D E. Arrangement of chromatin in the nucleus. Hum Genet. 1980;53:131–143. doi: 10.1007/BF00273484. [DOI] [PubMed] [Google Scholar]
- 42.Craig J M, Bickmore W A. Chromosome bands—flavours to savour. Bioessays. 1993;15:349–354. doi: 10.1002/bies.950150510. [DOI] [PubMed] [Google Scholar]
- 43.Cremer T, Cremer C, Baumann H, Luedtke E-K, Sperling K, Teuber V, Zorn C. Rabl's model of interphase chromosome arrangement tested in chinese hamster cells by premature chromatin condensation and laser UV microbeam analysis. Hum Genet. 1982;60:46–56. doi: 10.1007/BF00281263. [DOI] [PubMed] [Google Scholar]
- 44.Cremer T, Cremer C, Schneider T, Baumann H, Hens L, Kirsch-Volders M. Analysis of chromosome positions in the interphase nucleus of chinese hamster cells by laser-UV-microirradiation experiments. Hum Genet. 1982;62:201–209. doi: 10.1007/BF00333519. [DOI] [PubMed] [Google Scholar]
- 45.Dietzel S, Jauch A, Kienle D, Qu G, Holtgreve H, Eils R, Münkel C, Bittner M, Meltzer P S, Trent J M, Cremer T. Separate and variable shaped chromosome arm domains are disclosed by chromosome arm painting in human cell nuclei. Chromosome Res. 1998;6:25–33. doi: 10.1023/a:1009262223693. [DOI] [PubMed] [Google Scholar]
- 46.Duriau-Frankignoul M. Evolution interphasique des prochromosomes chez le Bryonia dioica Jacq. Rev Cytol Biol Veg. 1972;35:295–302. [Google Scholar]
- 47.Eleftheriou E P. Ultrastructural studies on protophloem sieve elements in Triticum aestivum L. nuclear disintegration. J Ultrastruct Mol Struct Res. 1986;95:47–60. [Google Scholar]
- 48.Eleftheriou E P. Developmental features of protophloem sieve elements in roots of wheat (Triticum aestivum L.) Protoplasma. 1996;193:204–212. [Google Scholar]
- 49.Eils R, Dietzel S, Bertin E, Schröck E, Speicher M R, Reid T, Robert-Nicoud M, Cremer C, Cremer T. Three-dimensional reconstruction of painted human interphase chromosomes: active and inactive X chromosome territories have similar volumes but differ in shape and surface structure. J Cell Biol. 1996;135:1427–1440. doi: 10.1083/jcb.135.6.1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Elves M W, Gough J, Chapman J A, Israels M C G. Electron microscopic studies of lymphocytes. Transformation under the influence of phytohaemagglutinin. Lancet. 1964;ii:306–308. [Google Scholar]
- 51.Emmerich P, Loos P, Jauch A, Hopman A H N, Wiegant J, Higgins M J, White B N, van der Ploeg M, Cremer C, Cremer T. Double in situ hybridization in combination with digital image analysis: a new approach to study interphase chromosome tomography. Exp Cell Res. 1989;181:126–140. doi: 10.1016/0014-4827(89)90188-2. [DOI] [PubMed] [Google Scholar]
- 52.Ferguson M, Ward D C. Cell cycle dependent chromosomal movement in pre-mitotic human T-lymphocyte nuclei. Chromosoma. 1992;101:557–565. doi: 10.1007/BF00660315. [DOI] [PubMed] [Google Scholar]
- 53.Flavell R B. The structure and control of expression of ribosomal RNA genes. Oxford Surv Plant Mol Cell Biol. 1986;3:252–274. [Google Scholar]
- 54.Flavell R B, O'Dell M, Thompson W F. Regulation of cytosine methylation in ribosomal DNA and nucleolus organizer expression in wheat. J Mol Biol. 1988;204:523–534. doi: 10.1016/0022-2836(88)90352-x. [DOI] [PubMed] [Google Scholar]
- 55.Fulnecek J, Matyášek R, Kovarík A, Bezdek M. Methylation of tobacco 5SrRNA genes determined by genomic sequencing. Mol Gen Genet. 1998;259:133–141. doi: 10.1007/s004380050798. [DOI] [PubMed] [Google Scholar]
- 56.Funabiki H, Hagan I, Uzawa S, Yanigida M. Cell cycle-dependent specific positioning and clustering of centromeres and telomeres in fission yeast. J Cell Biol. 1993;121:961–976. doi: 10.1083/jcb.121.5.961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gall J G, Cohen E H, Polan M L. Repetitive DNA sequences in Drosophila. Chromosoma. 1971;33:319–344. doi: 10.1007/BF00284948. [DOI] [PubMed] [Google Scholar]
- 58.Gavrieli Y, Sherman Y, Ben-Sasson S A. Identification of programmed cell death in situ via specific labelling of nuclear DNA fragmentation. J Cell Biol. 1992;119:493–501. doi: 10.1083/jcb.119.3.493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gerlach W L, Bedbrook J R. Cloning and characterisation of ribosomal RNA genes from wheat and barley. Nucleic Acids Res. 1979;7:1869–1885. doi: 10.1093/nar/7.7.1869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Glyn M C P, Leitch A R. The distribution of a spliceosome protein in cereal (Triticeae) interphase nuclei from cells with different metabolic activities and through the cell cycle. Plant J. 1995;8:531–540. doi: 10.1046/j.1365-313x.1995.8040531.x. [DOI] [PubMed] [Google Scholar]
- 61.Goffeau A, Barell B G, Bussey H, Davis R W, Dujon B, Feldman H, Gailbert F, Homeisel J D, Jacq C, Johnston M, Lewis E J, Mewes H W, Murakami Y, Philippsen P, Tettelin H, Oliver S G. Life with 6,000 genes. Science. 1996;274:546–567. doi: 10.1126/science.274.5287.546. [DOI] [PubMed] [Google Scholar]
- 62.Grant T M, Goldberg M W, Allen T D. Nuclear envelope and nuclear pore assembly: analysis of assembly intermediates by electron microscopy. Curr Opin Cell Biol. 1998;10:409–415. doi: 10.1016/s0955-0674(98)80018-5. [DOI] [PubMed] [Google Scholar]
- 63.Gruenbaum Y, Hochstrasser M, Mathog D, Saumweber H, Agard D A, Sedat J W. Spatial organization of the Drosophila nucleus: A three-dimensional cytogenetic study. J Cell Sci Suppl. 1984;1:223–234. doi: 10.1242/jcs.1984.supplement_1.14. [DOI] [PubMed] [Google Scholar]
- 64.Guacci V, Hogan E, Koshland D. Centromere position in budding yeast: evidence for anaphase A. Mol Biol Cell. 1997;8:957–972. doi: 10.1091/mbc.8.6.957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Haaf T, Grunenberg H, Schmid M. Paired arrangement of non-homologous centromeres during spermatogenesis. Exp Cell Res. 1990;187:157–161. doi: 10.1016/0014-4827(90)90130-3. [DOI] [PubMed] [Google Scholar]
- 66.Haaf T, Schmid M. Centromeric association and non-random distribution of centromeres in human tumour cells. Hum Genet. 1989;81:137–143. doi: 10.1007/BF00293889. [DOI] [PubMed] [Google Scholar]
- 67.Haaf T, Werner P, Schmid M. 5-azacytidine distinguishes between active and inactive X-chromosome condensation. Cytogenet Cell Genet. 1993;63:160–168. doi: 10.1159/000133526. [DOI] [PubMed] [Google Scholar]
- 68.Hartman T P V, Southern D I. Genome re-organization from polytene to polyploid in the nurse cells found in onion fly (Delia antiqua) and cabbage root fly (Delia radicum) ovaries (Diptera, Anthomyiidae) Chromosome Res. 1995;3:271–280. doi: 10.1007/BF00713064. [DOI] [PubMed] [Google Scholar]
- 69.Heitz E. Über α- und β-Heterochromatin sowie Konstanz und Bau der Chromomeren bei Drosophila. Biol Zentbl. 1971;54:588–609. [Google Scholar]
- 70.Heslop-Harrison J S, Smith J B, Bennett M D. The absence of the somatic association of centromeres of homologous chromosomes in grass mitotic metaphases. Chromosoma. 1988;96:119–131. [Google Scholar]
- 71.Heslop-Harrison J S, Huelskamp M, Wendroth S, Atkinson M D, Leitch A R, Bennett M D. Chromatin and centromeric structures in interphase nuclei. In: Brandham P E, editor. Kew Chromosome Conference III. London, United Kingdom: Allen & Unwin; 1988. pp. 209–271. [Google Scholar]
- 72.Heslop-Harrison J S, Leitch A R, Schwarzacher T. The physical organisation of interphase nuclei. In: Heslop-Harrison J S, Flavell R, editors. The chromosome. Oxford, United Kingdom: Bios Scientific Publishers; 1993. pp. 221–230. [Google Scholar]
- 73.Heslop-Harrison J S, Leitch A R, Schwarzacher T, Anamthawat-Jönsson K. Detection and characterisation of 1B/1R translocations in hexaploid wheat. Heredity. 1990;65:385–392. [Google Scholar]
- 74.Hochstrasser M, Sedat J W. Three-dimensional organisation of Drosophila melanogaster interphase nuclei. I. Tissue-specific aspects of polytene architecture. J Cell Biol. 1987;104:1455–1470. doi: 10.1083/jcb.104.6.1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Hochstrasser M, Sedat J W. Three-dimensional organisation of Drosophila melanogaster interphase nuclei. II. Chromosome spatial organisation and gene regulation. J Cell Biol. 1987;104:1471–1483. doi: 10.1083/jcb.104.6.1471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Houben A, Belyaev N D, Turner B M, Schubert I. Differential immunostaining of plant chromosomes by antibodies, recognising acetylated histone H4 variants. Chromosome Res. 1996;4:191–195. doi: 10.1007/BF02254958. [DOI] [PubMed] [Google Scholar]
- 77.Houben A, Guttenbach M, Kress W, Pich U, Schubert I, Schmid M. Immunostaining and interphase arrangement of field bean kinetochores. Chromosome Res. 1995;3:27–31. doi: 10.1007/BF00711158. [DOI] [PubMed] [Google Scholar]
- 78.Hozak P, Zatsepina O, Vasilyeva I, Chentsov Y. An electron microscopic study of the nucleolus-organising regions at some stages of the cell cycle (G0 period, G2 period, mitosis) Biol Cell. 1986;57:197–206. doi: 10.1111/j.1768-322x.1986.tb00476.x. [DOI] [PubMed] [Google Scholar]
- 79.Hozak P, Novak J T, Smetana K. Three-dimensional reconstructions of nucleolus-organizing regions in PHA-stimulated human lymphocytes. Biol Cell. 1989;66:225–233. doi: 10.1111/j.1768-322x.1989.tb00839.x. [DOI] [PubMed] [Google Scholar]
- 80.Ingle J, Timmis J N, Sinclair J. The relationship between satellite deoxyribonucleic acid, ribosomal RNA gene redundancy, and genome size in plants. Plant Physiol. 1975;55:496–501. doi: 10.1104/pp.55.3.496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Inman D R, Cooper E H. Electron microscopy of human lymphocytes stimulated with phytohemagglutinin. J Cell Biol. 1963;19:441–445. doi: 10.1083/jcb.19.2.441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Jacobson M D, Weil M, Raff M C. Programmed cell death in animal development. Cell. 1997;88:347–354. doi: 10.1016/s0092-8674(00)81873-5. [DOI] [PubMed] [Google Scholar]
- 83.Reference deleted.
- 84.Jin Q, Trelles-Stricken E, Scherthan H, Loidl J. Yeast nuclei display prominent centromere clustering that is reduced in nondividing cells in meiotic prophase. J Cell Biol. 1998;141:21–29. doi: 10.1083/jcb.141.1.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Joffe B I, Solovei I V, MacGregor H C. Ordered arrangement and rearrangement of chromosomes during spermatogenesis in two species of planarians (Plathelminthes) Chromosoma. 1998;107:173–183. doi: 10.1007/s004120050294. [DOI] [PubMed] [Google Scholar]
- 86.Jordan E G, Martini G, Bennett M D, Flavell R B. Nucleolar fusion in wheat. J Cell Sci. 1982;56:485–495. [Google Scholar]
- 87.Jordan E G, Cooper P J, Martini G, Bennett M D, Flavell R B. The effects of temperature and ageing on root apical meristems during seedling growth of Triticum aestivum L.: a specific effect on nucleoli. Plant Cell Environ. 1985;8:325–331. [Google Scholar]
- 88.Kerr J F R, Wyllie A H, Currie A R. Apoptosis: a basic biological phenomenon with wide ranging implications in tissue kinetics. Br J Cancer. 1972;26:239–257. doi: 10.1038/bjc.1972.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Keshet I, Yisraeli J, Cedar H. DNA methylation affects the formation of active chromatin. Cell. 1986;44:535–543. doi: 10.1016/0092-8674(86)90263-1. [DOI] [PubMed] [Google Scholar]
- 90.Kleckner N. Interactions between and along meiotic chromosomes. Cytogenet Cell Genet. 1998;81:100. [Google Scholar]
- 91.Klechner N, Weiner B M. Potential advantages of unstable interactions for pairing of chromosomes in meiotic, somatic, and premeiotic cells. Cold Spring Harbor Symp Quant Biol. 1993;58:553–565. doi: 10.1101/sqb.1993.058.01.062. [DOI] [PubMed] [Google Scholar]
- 92.Kingham K I, Duckett J G, Glyn M C P, Leitch A R. Nuclear differentiation in the filamentous caulonema of the moss Funaria hygrometrica Hedw. New Phytol. 1995;131:543–556. doi: 10.1111/j.1469-8137.1995.tb03090.x. [DOI] [PubMed] [Google Scholar]
- 93.Kovarik A, Matyášek R, Leitch A, Gazdová B, Fulnecek J, Bezdek M. Variability in CNG methylation in higher plant genomes. Gene. 1997;204:25–33. doi: 10.1016/s0378-1119(97)00503-9. [DOI] [PubMed] [Google Scholar]
- 94.Kurtz A, Lampel S, Nickolenko J E, Bradl J, Benner A, Zirbel R M, Cremer T, Lichter P. Active and inactive genes localize preferentially in the periphery of chromosome territories. J Cell Biol. 1996;135:1195–1206. doi: 10.1083/jcb.135.5.1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Lamond A I, Earnshaw W C. Structure and function in the nucleus. Science. 1998;280:547–553. doi: 10.1126/science.280.5363.547. [DOI] [PubMed] [Google Scholar]
- 96.Lamborot-Mazur M, Tishler P V, Atkins L. Fluorescent drumsticks in male polymorphs. Lancet. 1971;i:973–974. doi: 10.1016/s0140-6736(71)91480-2. [DOI] [PubMed] [Google Scholar]
- 97.Leitch A R, Brown J K M, Mosgöller W, Schwarzacher T, Heslop-Harrison J S. The spatial localization of homologous chromosomes in human fibroblasts at mitosis. Hum Genet. 1994;93:275–280. doi: 10.1007/BF00212022. [DOI] [PubMed] [Google Scholar]
- 98.Leitch A R, Glyn M, Kingham K, Quinn M, Aragon-Alcaide L F, Somasekaram A, Duckett J G. The dynamic organisation of interphase nuclei during cell differentiation and with changing cell activity. In: Brandham P E, Bennett M D, editors. Kew Chromosome Conference IV. London, United Kingdom: Her Majesty's Stationery Office; 1995. pp. 83–95. [Google Scholar]
- 99.Leitch A R, Heslop-Harrison J S. Ribosomal RNA gene expression and localization in cereals. Chromosomes Today. 1993;11:91–100. [Google Scholar]
- 100.Leitch A R, Mosgöller W, Shi M, Heslop-Harrison J S. Different patterns of rDNA organisation in nuclei of wheat and rye. J Cell Sci. 1992;101:751–757. doi: 10.1242/jcs.101.4.751. [DOI] [PubMed] [Google Scholar]
- 101.Leitch A R, Schwarzacher T, Mosgöller W, Bennett M D, Heslop-Harrison J S. Parental genomes are separated throughout the cell cycle in a plant hybrid. Chromosoma. 1991;101:206–213. [Google Scholar]
- 102.Leitch I J, Bennett M D. Polyploidy in angiosperms. Trends Plant Sci. 1997;12:470–476. [Google Scholar]
- 103.Lewis J P, Tanke H J, Raap A K, Beverstock G C, Kluin-Nelemans H C. Somatic pairing of centromeres and short arms of chromosome 15 in the hematopoitic and lymphoid system. Hum Genet. 1993;92:577–582. doi: 10.1007/BF00420942. [DOI] [PubMed] [Google Scholar]
- 104.Li G, Sudlow G, Belmont A S. Interphase cell dynamics of a late replicating, heterochromatic homogenously staining region: precise choreography of condensation/decondensation and nuclear positioning. J Cell Biol. 1998;140:975–989. doi: 10.1083/jcb.140.5.975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Lichter P, Cremer T, Borden J, Manuelidis L, Ward D C. Delineation of individual human chromosomes in metaphase and interphase cells by in situ suppression hybridisation using recombinant DNA libraries. Hum Genet. 1988;80:224–234. doi: 10.1007/BF01790090. [DOI] [PubMed] [Google Scholar]
- 106.Loidl J. General principles of meiosis derived from yeast and other model system. Cytogenet Cell Genet. 1999;85:113. [Google Scholar]
- 107.Lyon M F. Gene action in the X chromosome of mouse (Mus musculus L.) Nature. 1961;190:372–373. doi: 10.1038/190372a0. [DOI] [PubMed] [Google Scholar]
- 108.Reference deleted.
- 109.Manuelidis L. Different central nervous system cell types display distinct and non-random arrangements of satellite DNA sequences. Proc Natl Acad Sci USA. 1984;81:3123–3127. doi: 10.1073/pnas.81.10.3123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Manuelidis L. Interphase chromosome positions and structure during silencing, transcription and replication. In: Van Driel R, Otte A P, editors. Nuclear organisation, chromatin structure, and gene expression. New York, N.Y: Oxford University Press; 1997. pp. 145–168. [Google Scholar]
- 111.Manuelidis L, Borden J. Reproducible compartmentalization of individual chromosome domains in human CNS cells revealed by in situ hybridisation and three-dimensional reconstruction. Chromosoma. 1988;96:397–410. doi: 10.1007/BF00303033. [DOI] [PubMed] [Google Scholar]
- 112.Manuelidis L, Chen T L. A unified model of eukaryotic chromosomes. Cytometry. 1990;11:8–25. doi: 10.1002/cyto.990110104. [DOI] [PubMed] [Google Scholar]
- 113.Miller L J, Marx J. Apoptosis. Science. 1998;281:1301. [Google Scholar]
- 114.Mittwoch U. Frequency of drumsticks in normal women and in patients with chromosomal abnormalities. Nature. 1964;201:317–319. doi: 10.1038/201317a0. [DOI] [PubMed] [Google Scholar]
- 115.Montijn M B, ten Hoopen R, Fransz P F, Oud J L, Nanninga N. Characterisation of the nucleolar organising regions during the cell cycle in two varieties of Petunia hybrida as visualised by fluorescence in situ hybridisation and silver staining. Chromosoma. 1998;107:80–86. doi: 10.1007/s004120050283. [DOI] [PubMed] [Google Scholar]
- 116.Mosgöller W, Jordan E G, Hernandez-Verdun D. News and views on the nucleolus in 1996. Report on the Colloquium on the Nucleolus, a meeting at Paris-Grignon, July 18–20, 1996. Biol Cell. 1996;88:1–4. doi: 10.1016/s0248-4900(97)86824-4. [DOI] [PubMed] [Google Scholar]
- 117.Mosgöller W, Schofer C, Wesierska-Gadek J, Steiner M, Muller M, Wachtler F. Ribosomal gene transcription is organised in foci within nucleolar components. Histochem Cell Biol. 1998;109:111–118. doi: 10.1007/s004180050208. [DOI] [PubMed] [Google Scholar]
- 118.Mukai Y, Gill B S. Detection of barley chromatin added to wheat by genomic in situ hybridisation. Genome. 1991;34:448–452. [Google Scholar]
- 119.Nagata S. Apoptosis by death factor. Cell. 1997;88:355–365. doi: 10.1016/s0092-8674(00)81874-7. [DOI] [PubMed] [Google Scholar]
- 120.Nagele R, Freeman T, McMorrow L, Lee H-Y. Precise positioning of chromosomes during prometaphase: evidence for chromosomal order. Science. 1995;270:1831–1835. doi: 10.1126/science.270.5243.1831. [DOI] [PubMed] [Google Scholar]
- 121.Nagele R G, Freeman T, Fazekas J, Lee K-M, Thompson Z, Lee H-Y. Chromosome spatial order in human cells: evidence for early origin and faithful propagation. Chromosoma. 1998;107:330–338. doi: 10.1007/s004120050315. [DOI] [PubMed] [Google Scholar]
- 122.Nagl W. Endopolyploidy and polyteny in differentiation and evolution. Amsterdam, The Netherlands: North-Holland Publishing Co./Elsevier; 1978. [Google Scholar]
- 123.Nicholson D W, Thornberry N A. Caspases: killer proteases. Trends Biochem Sci. 1997;22:299–306. doi: 10.1016/s0968-0004(97)01085-2. [DOI] [PubMed] [Google Scholar]
- 124.Nigg E A. The nuclear envelope. Curr Opin Cell Biol. 1989;1:435–440. doi: 10.1016/0955-0674(89)90002-1. [DOI] [PubMed] [Google Scholar]
- 125.Oakley H A. Male meiosis in Mesostoma ehrenbergii ehrenbergii. In: Brandham P E, Bennett M D, editors. Kew Chromosome Conference II. London, United Kingdom: George Allen & Unwin; 1982. pp. 195–199. [Google Scholar]
- 126.Odartchenko N, Keneklis T. Localization of paternal DNA in interphase nuclei of mouse eggs during early cleavage. Nature. 1973;241:528–529. doi: 10.1038/241528a0. [DOI] [PubMed] [Google Scholar]
- 127.Park P C, De Boni U. Dynamics of nucleolar fusion in neuronal interphases nuclei in vitro: association with nuclear rotation. Exp Cell Res. 1991;197:213–221. doi: 10.1016/0014-4827(91)90425-t. [DOI] [PubMed] [Google Scholar]
- 128.Pinkel D, Landegent J, Collins C, Fuscoe J, Segraves R, Lucas J, Gray J. Fluorescence in situ hybridisation with human chromosome-specific libraries: detection of trisomy 21 and translocations of chromosome 4. Proc Natl Acad Sci USA. 1988;85:9138–9142. doi: 10.1073/pnas.85.23.9138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Popp S, Scholl H P, Loos P, Jauch A, Stelzer E, Cremer C, Cremer T. Distribution of chromosome 18 and X centric heterochromatin in the interphase nucleus of cultured human cells. Exp Cell Res. 1990;189:1–12. doi: 10.1016/0014-4827(90)90249-a. [DOI] [PubMed] [Google Scholar]
- 130.Prescott D M. The unusual organization and processing of genomic DNA in hypotrichous ciliates. Trends Genet. 1992;8:439–445. doi: 10.1016/0168-9525(92)90328-2. [DOI] [PubMed] [Google Scholar]
- 131.Rabl C. Über Zelltheilung. Morphol Jahrb. 1885;10:214–330. [Google Scholar]
- 132.Rae P M M, Franke W W. The interphase distribution of satellite DNA-containing heterochromatin in mouse nuclei. Chromosoma. 1972;39:443–456. doi: 10.1007/BF00326177. [DOI] [PubMed] [Google Scholar]
- 133.Rohlf F J, Rodman T C, Flehinger B J. The use of nonmetric multidimensional scaling for the analysis of chromosomal association. Comp Biomed Res. 1980;13:19–35. doi: 10.1016/0010-4809(80)90003-8. [DOI] [PubMed] [Google Scholar]
- 134.Robinett C C, Straight A, Li G, Willhelm C, Sudlow G, Murray A, Belmont A S. In vivo localization of DNA sequences and visualization of large scale chromatin organisation using lac operator/repressor recognition. J Cell Biol. 1996;135:1685–1700. doi: 10.1083/jcb.135.6.1685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Rawlins D J, Shaw P J. Three dimensional organization of ribosomal DNA in interphase nuclei of Pisum sativum by in situ hybridization and optical tomography. Chromosoma. 1990;99:143–151. [Google Scholar]
- 136.Saccone S, De Sario A, Valle G D, Berbardi G. The highest gene concentrations in the human genome are in the telomeric bands of metaphase chromosomes. Proc Natl Acad Sci USA. 1992;89:4913–4917. doi: 10.1073/pnas.89.11.4913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Sahlas D J, Milankov K, Park P C, De Boni U. Distribution of snRNPs, splicing factor SC-35 and actin in interphase nuclei: immunocytochemical evidence for differential distribution during changes in functional states. J Cell Sci. 1993;105:347–357. doi: 10.1242/jcs.105.2.347. [DOI] [PubMed] [Google Scholar]
- 138.Sanchez J A, Karni R J, Wangh L J. Fluorescent in situ hybridisation (FISH) analysis of the relationship between chromosomal location and nuclear morphology in human neutrophils. Chromosoma. 1997;106:168–177. doi: 10.1007/s004120050236. [DOI] [PubMed] [Google Scholar]
- 139.Schwarzacher H G. Chromosomes in mitosis and interphase. Berlin, Germany: Springer-Verlag KG; 1976. [Google Scholar]
- 140.Schwarzacher T, Heslop-Harrison J S. In situ hybridisation to plant telomeres using synthetic oligomeres. Genome. 1991;34:317–324. [Google Scholar]
- 141.Shaw P J, Jordan E G. The nucleolus. Annu Rev Cell Dev Biol. 1995;11:93–121. doi: 10.1146/annurev.cb.11.110195.000521. [DOI] [PubMed] [Google Scholar]
- 142.Shaw P, Rawlins D, Highett M. Nuclear and nucleolar structure. In: Heslop-Harrison J S, Flavell R B, editors. The chromosome. Oxford, United Kingdom: Bios Scientific Publishers; 1993. pp. 161–171. [Google Scholar]
- 143.Shelby R D, Hahn K M, Sullivan K F. Dynamic elastic behavior of α-satellite DNA domains visualised in situ in living human cells. J Cell Biol. 1996;135:545–557. doi: 10.1083/jcb.135.3.545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Silva M, Queiroz A, Neves N, Barao A, Castilho A, Morais-Cecilio L, Viegas W. Reprogramming of rye rDNA in triticale during microsporogenesis. Chromosome Res. 1995;3:492–496. doi: 10.1007/BF00713964. [DOI] [PubMed] [Google Scholar]
- 145.Sigmund J, Schwarzacher H G, Mikelsaar A-V. Satellite frequency and number of nucleoli depend in cell cycle duration and NOR-activity. Hum Genet. 1979;50:81–91. doi: 10.1007/BF00295594. [DOI] [PubMed] [Google Scholar]
- 146.Spector D L. Higher order nuclear organization: three-dimensional distribution of small nuclear ribonucleoprotein particles. Proc Natl Acad Sci USA. 1990;87:147–151. doi: 10.1073/pnas.87.1.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Sperling K, Luedeke E-K. Arrangement of prematurely condensed chromosomes in cultured cells and lymphocytes of the Indian Muntjac. Chromosoma. 1981;83:541–553. doi: 10.1007/BF00328278. [DOI] [PubMed] [Google Scholar]
- 148.Tartof K D, Henikoff S. Trans-sensing effects from Drosophila to humans. Cell. 1991;65:201–203. doi: 10.1016/0092-8674(91)90153-p. [DOI] [PubMed] [Google Scholar]
- 149.Therman E. Chromosome behavior in cell differentiation: a field ripe for exploration. Genetics. 1995;141:799–804. doi: 10.1093/genetics/141.3.799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Thornberry N A, Lazebnik Y. Caspases: enemies within. Science. 1998;281:1312–1316. doi: 10.1126/science.281.5381.1312. [DOI] [PubMed] [Google Scholar]
- 151.Vogel F, Krüger J. Is there a general relationship between estimated chromosome distances in interphase and location of genes with related functions? Hum Genet. 1983;63:362–368. doi: 10.1007/BF00274763. [DOI] [PubMed] [Google Scholar]
- 152.Weimer R, Haaf T, Kruger J, Poot M, Schmid M. Characterisation of centromere arrangements and test for random distribution in G0, G1, S, G2, G1 and early S' phase in human lymphocytes. Hum Genet. 1992;88:673–682. doi: 10.1007/BF02265296. [DOI] [PubMed] [Google Scholar]
- 153.Wergin W P, Gruber P J, Newcomb E H. Fine structural investigation of nuclear inclusions in plants. J Ultrastruct Res. 1970;30:533–557. doi: 10.1016/s0022-5320(70)90051-1. [DOI] [PubMed] [Google Scholar]
- 154.Werren J H, Nur U, Charnov E L. An extrachromosomal factor causing loss of paternal chromosomes. Nature. 1987;327:75–76. doi: 10.1038/327075a0. [DOI] [PubMed] [Google Scholar]
- 155.Wollenberg C, Kiefaber M P, Zang K D. Quantitative studies on the arrangement of human metaphase chromosomes. IX. Arrangement of chromosomes with and without spindle apparatus. Hum Genet. 1982;62:310–315. doi: 10.1007/BF00304545. [DOI] [PubMed] [Google Scholar]
- 156.Xing Y, Johnson C V, Dobner P R, Lawrence J B. Higher level organisation of individual gene transcription and RNA splicing. Science. 1993;259:1326–1330. doi: 10.1126/science.8446901. [DOI] [PubMed] [Google Scholar]
- 157.Yang F, O'Brien P C M, Weinberg J, Ferguson-Smith M A. A reappraisal of the tandem fusion theory of karyotype evolution in the Indian muntjac using chromosome painting. Chromosome Res. 1997;5:109–117. doi: 10.1023/a:1018466107822. [DOI] [PubMed] [Google Scholar]
- 158.Reference deleted.
- 159.Zink D, Cremer T, Saffrich R, Fisher R, Trendelenberg M F, Ansorge W, Stelzer E H K. Structure and dynamics of human interphase chromosome territories in vivo. Hum Genet. 1998;102:241–251. doi: 10.1007/s004390050686. [DOI] [PubMed] [Google Scholar]
- 160.Zirbel R M, Mathieu U R, Kurtz A, Cremer T, Lichter P. Evidence for a nuclear compartment of transcription and splicing located at chromosome domain boundaries. Chromosome Res. 1993;1:93–106. doi: 10.1007/BF00710032. [DOI] [PubMed] [Google Scholar]