Abstract
The ellipsoid body (EB) is a major structure of the central complex of the Drosophila melanogaster brain. Twenty-two subtypes of EB ring neurons have been identified based on anatomic and morphologic characteristics by light-level microscopy and EM connectomics. A few studies have associated ring neurons with the regulation of sleep homeostasis and structure. However, cell type-specific and population interactions in the regulation of sleep remain unclear. Using an unbiased thermogenetic screen of EB drivers using female flies, we found the following: (1) multiple ring neurons are involved in the modulation of amount of sleep and structure in a synergistic manner; (2) analysis of data for ΔP(doze)/ΔP(wake) using a mixed Gaussian model detected 5 clusters of GAL4 drivers which had similar effects on sleep pressure and/or depth: lines driving arousal contained R4m neurons, whereas lines that increased sleep pressure had R3m cells; (3) a GLM analysis correlating ring cell subtype and activity-dependent changes in sleep parameters across all lines identified several cell types significantly associated with specific sleep effects: R3p was daytime sleep-promoting, and R4m was nighttime wake-promoting; and (4) R3d cells present in 5HT7-GAL4 and in GAL4 lines, which exclusively affect sleep structure, were found to contribute to fragmentation of sleep during both day and night. Thus, multiple subtypes of ring neurons distinctively control sleep amount and/or structure. The unique highly interconnected structure of the EB suggests a local-network model worth future investigation; understanding EB subtype interactions may provide insight how sleep circuits in general are structured.
SIGNIFICANCE STATEMENT How multiple brain regions, with many cell types, can coherently regulate sleep remains unclear, but identification of cell type-specific roles can generate opportunities for understanding the principles of integration and cooperation. The ellipsoid body (EB) of the fly brain exhibits a high level of connectivity and functional heterogeneity yet is able to tune multiple behaviors in real-time, including sleep. Leveraging the powerful genetic tools available in Drosophila and recent progress in the characterization of the morphology and connectivity of EB ring neurons, we identify several EB subtypes specifically associated with distinct aspects of sleep. Our findings will aid in revealing the rules of coding and integration in the brain.
Keywords: central complex, Drosophila melanogaster, ellipsoid body, ring neurons, sleep, sleep structure
Introduction
Sleep plays critical roles in many physiological functions. Sleep regulation in the brain is a complex process modulated at the molecular, cellular, circuit, and network levels (John et al., 2016; Scammell et al., 2017; Bringmann, 2018; Herice and Sakata, 2019; D. Liu and Dan, 2019). Previous studies in Drosophila melanogaster have revealed multiple cell types and neural circuits that participate in the regulation of sleep amount, structure, and homeostasis.
The ellipsoid body (EB) contributes to regulation of multiple behaviors, including spatial orientation, navigation, arousal, and sleep (Bausenwein et al., 1994; Lebestky et al., 2009; Ofstad et al., 2011; Seelig and Jayaraman, 2015; Fisher et al., 2019; Kim et al., 2019; Kottler et al., 2019). As one of the central structures on the midline of the fly brain, the EB receives direct input from, and sends output to, many brain regions. This high level of connectivity positions the EB to be a center for integration of multiple information streams, including visual, motor, mechanosensory, and circadian input, allowing it to functionally tune complex behaviors (Franconville et al., 2018).
The organization within the EB also exhibits complexity. With recent progress on morphology and connectivity of the EB, 22 distinct subtypes of ring neurons have been identified (Hulse et al., 2021). Each subtype of ring neuron typically contains a dendritic arborization lateral to the EB, then projects a single axon into the concentric laminated structure within the EB neuropil. The projections from each subtype of ring neuron form distinct layers within the neuropil, terminating in different rings at specific depths along the anterior-posterior axis where they interconnect (Hanesch et al., 1989; Young and Armstrong, 2010; Lin et al., 2013). These connections, between neurons of the same type, provide each ring neuron's strongest inputs (Isaacman-Beck et al., 2020; Hulse et al., 2021) and suggest a structural basis for local communication and synergism for sleep regulation.
Despite the growing understanding of EB connectivity, specific roles for each subtype of ring neuron in sleep are limited. One subtype of R5 neuron (initially referred to as R2) has been shown to drive a persistent sleep on secession of thermoactivation, suggesting a role in sleep drive and homeostasis (Donlea et al., 2014; S. Liu et al., 2016; Pimentel et al., 2016). Another study showed that single R5 neurons get synchronized by circadian input and the power of slow-wave oscillations in R5 neurons has been associated with increased sleep drive (Raccuglia et al., 2019). 5HT7-GAL4+ EB neurons, which consist of several subtypes including R3d, R3p, and R4d and are modulated by serotonergic signaling, can regulate sleep architecture (C. Liu et al., 2019). Despite these important findings, the scope of ring neuron involvement in the regulation of sleep is not clear.
In the present study, we take an unbiased approach, screening 34 drivers that label different combinations of subtypes of ring neurons by thermoactivation using the warmth-sensitive cation channel dTrpA1 (Hamada et al., 2008). Most drivers label multiple ring neurons, and activation of many drivers resulted in significant changes in sleep amount and/or sleep structure. The complexity of the tools and phenotypes necessitated developing computational approaches for assessing the importance of each subtype. Using P(wake) and P(doze) analysis with a mixed Gaussian model, five clusters of drivers were found to regulate sleep depth and pressure during the day and/or at night, respectively. Furthermore, a GLM analysis based on the GAL4 expression pattern and the sleep behavior on 24 h activation suggests several types of ring neuron contribute to sleep regulation consistent with and extending the findings from the Gaussian model. Finally, using genetic suppression of intersected population strategy, we identified a subpopulation of neurons which is sufficient to fragment sleep during both day and night. Although how the ring neurons cooperate to coherently modulate sleep is not yet clear, the identification of roles for specific cell types provides an important piece of the puzzle.
Materials and Methods
Animals
Unless specified, flies were reared on standard cornmeal food (each 1 L H2O: 70 g cornmeal, 50 g sucrose, 10 g soybean powder, 20 g yeast powder, 6 g agar, and 3 g methyl 4-hydroxybenzoate) at 23°C with 60% relative humidity and under a regimen of 12 h light/12 h dark. Flies were allowed to freely mate after eclosion, and mated females aged 2-5 d were used for all experiments. GAL4 lines: R12B01 (RRID:BDSC_48487), R15B07 (RRID:BDSC_48678), R28D01 (RRID:BDSC_47342), R28E01 (RRID:BDSC_49457), R38B06 (RRID:BDSC_49986), R38G08 (RRID:BDSC_50020), R38H02 (RRID:BDSC_47352), R41A08 (RRID:BDSC_50108), R41F05-GAL4 (RRID:BDSC_50133), R47F07 (RRID:BDSC_50320), R48B10 (RRID:BDSC_50352), R49E12 (RRID:BDSC_38693), R53F11 (RRID:BDSC_50443), R53G11 (RRID:BDSC_69747), R54B05 (RRID:BDSC_69148), R56C09 (RRID:BDSC_39145), R64H04 (RRID:BDSC_39323), R70B04 (RRID:BDSC_39513), R70B05 (RRID:BDSC_47721), R73A06 (RRID:BDSC_39805), R73B05 (RRID:BDSC_48312), R81F01 (RRID:BDSC_40120), R84H09 (RRID:BDSC_47803), Aphc507 (RRID:BDSC_30840), C232 (RRID:BDSC_30828), and R44D11-LexA (RRID:BDSC_41264), UAS-dTrpA1 (RRID:BDSC_26263), UAS-mCD8::GFP (RRID:BDSC_5136), UAS-mCD8::RFP, LexAop2-mCD8::GFP (RRID:BDSC_32229), and LexAop-Gal80 (RRID:BDSC_32213) were ordered from the Bloomington Drosophila Stock Center. GAL4 lines: VT012446, VT026841, VT042577, VT042759, VT045108, VT057257, VT038828, VT040539, and VT059775 were ordered from Vienna Drosophila Resource Center originally, but unfortunately not available anymore. 5HT7-GAL4 was provided by Charles Nicols' laboratory. Feb170-GAL4 was generated by Günter Korge's laboratory (Siegmund and Korge, 2001). The WT line wCS was crossed with GAL4 and UAS parental lines as genetic controls. Experimental groups were from the F1 generation of crosses of GAL4 lines to UAS-dTrpA1.
Experimental design for sleep assays and calculation of sleep changes
F1 generation of flies were all maintained on standard food at 23°C. Two- to 5-day-old mated F1 female flies were individually placed into a 65 mm × 5 mm glass tube containing food (2% agar and 5% sucrose). After loading to the DAM2 system (Drosophila Activity Monitor) (Trikinetics; https://www.trikinetics.com/) at 21°C in 12 h:12 h light/dark cycles, flies were entrained for 2-3 d. Then 1 d baseline sleep, 1 d neural activation sleep, as well as 1 d recovery sleep were recorded at 21°C, 30°C, and 21°C, respectively. Total sleep, the number of sleep episodes, and maximum episode length were analyzed for light and dark periods (LP and DP) separately, using MATLAB (RRID:SCR_001622) program (SCAMP2019v2) scripts.
To overview the effects on activation of GAL4+ neurons, all genotypes were arranged in a descending order according to the changes of total sleep during the LP. Sleep changes were calculated by subtracting baseline day sleep of each genotype from its activation day. Since using TRPA1 to activate neurons requires an elevation of ambient temperature (above 25°C), and temperature has been shown to effect sleep (Parisky et al., 2016; Jin et al., 2021; Alpert et al., 2022), it is critical to compare with control groups that have undergone the same temperature shift. With genetic control groups and a subtraction to the baseline day, the temperature effect can be removed and sleep changes because of activation of the neurons can be quantified. For genotypes with significant changes in sleep and/or sleep structure, 3 days' sleep profiles of sleep time in 30 min were plotted. Sleep changes of the recovery day were also calculated. The significant difference was marked when the experimental group is different compared with both genetic controls.
Immunohistochemistry
Brains of adult flies were dissected in 10 mm ice-cold PBS and fixed for 20 min in PBS with 4% PFA at room temperature. Brains were then washed 3 times for 5 min each in PBT (PBS with 0.5% Triton X-100). For GFP and RFP immunostaining, brains were incubated with primary antibodies (1:200, chicken anti-GFP, Abcam, catalog #ab13970, RRID:AB_300798; 1:200, mouse anti-GFP, Roche, catalog #11814460001, RRID:AB_390913; 1:1000, rabbit anti-GFP, Invitrogen, catalog #A-11122, RRID:AB_221569; 1:200, rabbit anti-DsRed, Takara, catalog #632496, RRID:AB_10013483) in 10% NGS in PBT at 4°C for two nights. After 3 times washes for 5 min each with PBT at room temperature, brains were incubated with secondary antibody at 4°C overnight. Second antibodies (488 goat anti-mouse, Invitrogen, catalog #A-11001, RRID:AB_2534069; 488 goat anti-chicken, Invitrogen, catalog #A-11039, RRID:AB_142924; 488 goat anti-rabbit, Invitrogen, catalog #A-11008, RRID:AB_143165; 568 goat anti-rabbit, Fisher Scientific, catalog #A-11011, RRID:AB_143157) were all used in a ratio of 1:200. Samples were then washed 3 times for 5 min each in PBT at room temperature, and mounted on microscope slide in Vectashield mounting medium (Vector Laboratories catalog #H-1000, RRID:AB_2336789). Finally, samples were imaged with Leica TCS SP5/LSM900 confocal microscope (RRID:SCR_002140) and analyzed using the open source of FIJI (ImageJ) software (RRID:SCR_002285).
Probability analysis
The probability of transitioning from a sleep to an awake state (P(wake)), and from a wake state to a sleep state (P(doze)) was used power law distributions analysis as previously described (Wiggin et al., 2020). P(wake) and P(doze) were calculated identically, with calculation of 1 min bin of inactivity and activity reversed. The MATLAB scripts for analysis of P(wake)/P(doze) can be accessed in GitHub at https://github.com/Griffith-Lab/Fly_Sleep_Probability.
Mixed Gaussian model clustering
To figure out different effects of EB drivers on both sleep pressure and depth, we divided all significant subtypes of EB ring neurons into groups with similar distributions of δ P(Wake) and δ P(Doze), using mixed Gaussian model clustering. The clustering analysis was conducted using the scripts of fitgmdist and cluster in MATLAB. Given the small sample size of neuron subtypes (14 and 13 for daytime and nighttime, respectively), the number of cluster k was set to 3, 4, or 5 for both daytime and nighttime. We calculated the silhouette coefficients for each k value using the script of silhouette in MATLAB and chose the final k value whose silhouette coefficient was the closest to one (Lecompte et al., 1986). The size of ellipse for each cluster was decided by the corresponding σ values of its Gaussian mixture distribution.
GLM
To evaluate the effect of a specific anatomic subtype of ring neurons on sleep, the GLM (Generalized linear models) was used to estimate the weights and the corresponding statistical significance of all subtypes for each sleep parameter. The GLM analysis was conducted using the script of glmfit in MATLAB (The MathWorks) to predict each sleep parameter under the combination of all subtypes of neurons. The input variable was defined as 1 or 0 for each subtype of ring neurons (R1, R2, R3d, R3m, R3a, R3p, R3w, R4m, R4d, R5, and R6) when labeled or not labeled by each driver, respectively. And the corresponding output variable was the mean change rate of each sleep parameter of the same driver on the activation to its baseline level (output variable value = (activation – baseline)/baseline). We chose the default parameters for the script of glmfit. According to the weight calculation for each subtype (see Table 6), a positive value represents positive relationship, and a negative value represents negative relationship between the subtype and the sleep parameter, respectively, when the corresponding p value < 0.05.
Table 6.
Statistical table of the weight of the effect of subclasses of ring neurons on the sleep using a GLMa
| Total sleep |
No. of episodes |
Maximum episode length |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| LP |
DP |
LP |
DP |
LP |
DP |
|||||||
| Subtype | Weight | p | Weight | p | Weight | p | Weight | p | Weight | p | Weight | p |
| R1 | −66.963 | 0.806 | 3.71 | 0.789 | −78.081 | 0.218 | −289.982 | 0.025* | −40.711 | 0.918 | 39.093 | 0.058 |
| R2 | −4.43 | 0.986 | 20.206 | 0.117 | −53.171 | 0.351 | −287.956 | 0.015* | 10.987 | 0.976 | 37.288 | 0.047* |
| R3a | 243.881 | 0.394 | −19.084 | 0.192 | 0.156 | 0.998 | 100.72 | 0.43 | 239.529 | 0.562 | −35.538 | 0.095 |
| R3d | −290.368 | 0.202 | 3.727 | 0.742 | 72.222 | 0.166 | −99.637 | 0.323 | −335.803 | 0.306 | −4.78 | 0.769 |
| R3m | 207.085 | 0.45 | 11.389 | 0.413 | −110.33 | 0.086 | 97.25 | 0.428 | 490.865 | 0.222 | 27.874 | 0.168 |
| R3p | 424.718 | 0.038* | −4.8 | 0.628 | 12.879 | 0.772 | −24.945 | 0.775 | 747.409 | 0.014* | −7.29 | 0.608 |
| R3w | −235.362 | 0.422 | 10.664 | 0.472 | −77.07 | 0.253 | −118.224 | 0.367 | −244.843 | 0.563 | 33.605 | 0.122 |
| R4d | −60.809 | 0.768 | −5.332 | 0.61 | −23.994 | 0.611 | −104.199 | 0.264 | −192.1 | 0.522 | 7.413 | 0.621 |
| R4m | 63.512 | 0.793 | −39.476 | 0.003* | −3.416 | 0.951 | 97.465 | 0.371 | 100.607 | 0.774 | −45.005 | 0.016* |
| R5 | −67.36 | 0.751 | 11.467 | 0.292 | −49.769 | 0.31 | −168.218 | 0.086 | −140.881 | 0.648 | 31.922 | 0.048* |
| R6 | 999.282 | 0.069 | −7.214 | 0.788 | 81.007 | 0.505 | 103.387 | 0.663 | 196.977 | 0.798 | −8.583 | 0.823 |
aThe generalized linear model (GLM) analysis was conducted using the script of glmfit in MATLAB with the default parameters setting for total sleep, number of episodes, and maximum episode length. A positive value represents a positive relationship, and a negative value represents a negative relationship between the subtype of ring neurons and the sleep parameter, respectively.
*p < 0.05.
Statistical analysis
Power analysis was conducted using the script of sampsizepwr in MATLAB (The MathWorks) to calculate the power for the sample size in this study. The power analysis was based on the sleep parameters in drivers with significant differences from both control groups presented in the main figures. We selected the mean and SD of control groups under the null hypothesis, and the mean value of experimental groups under the alternative hypothesis during the calculation of power values. Based on current sample size, >80% of the powers of significances of sleep parameters were >0.9 (see Tables 2 and 7).
Table 2.
| Total sleep | Experiment vs GAL4 Control |
Experiment vs UAS Control |
||||||
|---|---|---|---|---|---|---|---|---|
| 30°C |
21°C |
30°C |
21°C |
|||||
| Drivers | LP | DP | LP | DP | LP | DP | LP | DP |
| c232 | 1 | 0.167 | 0.377 | 0.639 | 1 | 0.563 | 0.533 | 0.907 |
| Feb170 | 1 | 1 | 0.946 | 0.209 | 1 | 1 | 0.996 | 0.19 |
| R48B10 | 0.151 | 0.53 | 0.934 | 0.077 | 1 | 0.998 | 0.975 | 0.999 |
| R53F11 | 1 | 0.052 | 0.999 | 0.858 | 0.108 | 0.996 | 0.85 | 0.928 |
| No. of episodes | Experiment vs GAL4 Control |
Experiment vs UAS Control |
||||||
|---|---|---|---|---|---|---|---|---|
| 30°C |
21°C |
30°C |
21°C |
|||||
| Drivers | LP | DP | LP | DP | LP | DP | LP | DP |
| c232 | 0.057 | 0.09 | 0.497 | 0.268 | 0.102 | 0.347 | 0.561 | 0.186 |
| Feb170 | 0.929 | 0.99 | 0.234 | 0.083 | 0.819 | 0.06 | 0.989 | 0.86 |
| R48B10 | 1 | 0.977 | 0.985 | 0.286 | 1 | 0.948 | 0.521 | 0.287 |
| R53F11 | 0.816 | 1 | 0.948 | 0.149 | 0.705 | 0.992 | 0.323 | 0.078 |
| Maximum episode length | Experiment vs GAL4 Control |
Experiment vs UAS Control |
||||||
|---|---|---|---|---|---|---|---|---|
| 30°C |
21°C |
30°C |
21°C |
|||||
| Drivers | LP | DP | LP | DP | LP | DP | LP | DP |
| c232 | 0.999 | 0.182 | 0.075 | 0.05 | 0.975 | 0.46 | 0.159 | 0.057 |
| Feb170 | 1 | 0.795 | 0.053 | 0.05 | 0.993 | 0.859 | 0.673 | 0.121 |
| R48B10 | 0.814 | 0.051 | 0.244 | 0.061 | 1 | 0.21 | 0.502 | 0.086 |
| R53F11 | 0.815 | 0.188 | 0.275 | 0.219 | 0.413 | 0.091 | 0.19 | 0.054 |
aFour GAL4 drivers were included in the analysis: c232, Feb170, R48B10, and R53F11. The experimental group was compared with either GAL4 control group or UAS-dTrpA1 control group for both activation day and recovery day. Total sleep, number of episodes, and maximum episode length for LP and DP were analyzed separately. Power analysis was conducted using the script of sampsizepwr in MATLAB.
Table 7.
| Total sleep | Experiment vs GAL4 Control |
Experiment vs UAS Control |
||||||
|---|---|---|---|---|---|---|---|---|
| 30°C |
21°C |
30°C |
21°C |
|||||
| Drivers | LP | DP | LP | DP | LP | DP | LP | DP |
| R28E01 | 1 | 0.511 | 0.999 | 0.28 | 1 | 0.313 | 0.9 | 0.951 |
| R70B05 | 1 | 1 | 1 | 0.667 | 1 | 1 | 0.994 | 0.985 |
| No. of episodes | Experiment vs GAL4 Control |
Experiment vs UAS Control |
||||||
|---|---|---|---|---|---|---|---|---|
| 30°C |
21°C |
30°C |
21°C |
|||||
| Drivers | LP | DP | LP | DP | LP | DP | LP | DP |
| R28E01 | 0.116 | 0.101 | 0.198 | 0.233 | 0.425 | 0.074 | 0.214 | 0.121 |
| R70B05 | 0.992 | 0.888 | 1 | 0.994 | 0.47 | 0.878 | 1 | 0.997 |
| Maximum episode length | Experiment vs GAL4 Control |
Experiment vs UAS Control |
||||||
|---|---|---|---|---|---|---|---|---|
| 30°C |
21°C |
30°C |
21°C |
|||||
| Drivers | LP | DP | LP | DP | LP | DP | LP | DP |
| R28E01 | 1 | 0.07 | 0.425 | 0.222 | 0.995 | 0.354 | 0.659 | 0.161 |
| R70B05 | 0.947 | 0.652 | 0.981 | 0.873 | 1 | 0.983 | 1 | 1 |
aTwo GAL4 drivers were included in the analysis: R28E01 and R70B05. The experimental group was compared with either GAL4 control group or UAS-dTrpA1 control group for both activation day and recovery day. Total sleep, number of episodes, and maximum episode length for LP and DP were analyzed separately. Power analysis was conducted using the script of sampsizepwr in MATLAB.
Data were performed using GraphPad Prism 8 (RRID:SCR_002798). Group means were compared using one-way ANOVA followed by Bonferroni's multiple comparison test when data were normally distributed, or Kruskal–Wallis test followed by Dunn's multiple comparison test was used when data failed passing normality test (see Tables 1, 3, 4, 8, and 9). All experiments were performed at least 2 replicates, and data presented in the figures were chosen from one representative replicate. To uniform the data presentation, all figures were prepared as mean ± SEM. To visualize all groups in the same figure clearer, error bars were not shown.
Table 1.
| △ Total sleep | LP (Fig. 1B) |
DP (Fig. 1G) |
||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Nonparametric/parametric test |
Post hoc comparisons |
Nonparametric/parametric test |
Post hoc comparisons |
|||||||||||||
| Driver | Test | DFn, DFd | F | p | n1 | n2 | Mean difference | p | Test | F | p | Mean difference | p | |||
| R47F07 | ANOVA | 2,92 | 118 | <0.0001 | 1 vs 2 | 24 | 39 | 350.6 | <0.0001 | **** | ANOVA | 46.59 | <0.0001 | −152.8 | <0.0001 | **** |
| 1 vs 3 | 24 | 32 | 257.9 | <0.0001 | **** | −249.2 | <0.0001 | **** | ||||||||
| R28E01 | K-W | 3,95 | 54.91 | <0.0001 | 1 vs 2 | 32 | 31 | 51.12 | <0.0001 | **** | K-W | 10.74 | 0.0046 | −15.22 | 0.0569 | NS |
| 1 vs 3 | 32 | 32 | 30.41 | <0.0001 | **** | 7.094 | 0.6067 | NS | ||||||||
| C232 | K-W | 3,91 | 31.94 | <0.0001 | 1 vs 2 | 30 | 31 | 36.11 | <0.0001 | **** | K-W | 10.32 | 0.0057 | −3.126 | >0.9999 | NS |
| 1 vs 3 | 30 | 30 | 29.27 | <0.0001 | **** | 17.13 | 0.024 | * | ||||||||
| R70B04 | K-W | 3,94 | 43.46 | <0.0001 | 1 vs 2 | 30 | 32 | 43.7 | <0.0001 | **** | K-W | 8.177 | 0.0168 | 6.665 | 0.6727 | NS |
| 1 vs 3 | 30 | 32 | 11.9 | 0.2195 | NS | 19.43 | 0.0101 | * | ||||||||
| R53F11 | K-W | 3,95 | 51.75 | <0.0001 | 1 vs 2 | 31 | 32 | 40.65 | <0.0001 | **** | K-W | 23.45 | <0.0001 | 1.31 | >0.9999 | NS |
| 1 vs 3 | 31 | 32 | −4.4 | >0.9999 | NS | 29.62 | <0.0001 | **** | ||||||||
| R56C09 | ANOVA | 2,51 | 5.698 | 0.0058 | 1 vs 2 | 22 | 11 | 125.5 | 0.0077 | ** | ANOVA | 0.579 | 0.5642 | −44.95 | 0.5146 | NS |
| 1 vs 3 | 22 | 21 | 89.39 | 0.0232 | * | −27.91 | 0.6812 | NS | ||||||||
| R54B05 | K-W | 3,89 | 22.79 | <0.0001 | 1 vs 2 | 26 | 31 | 19.65 | 0.0085 | ** | K-W | 12.29 | 0.0021 | −24.07 | 0.0009 | *** |
| 1 vs 3 | 26 | 32 | −11.12 | 0.2059 | NS | −12.41 | 0.1378 | NS | ||||||||
| R38B06 | K-W | 3,86 | 34.64 | <0.0001 | 1 vs 2 | 29 | 29 | 37.19 | <0.0001 | **** | K-W | 2.427 | 0.2972 | −8.966 | 0.3431 | NS |
| 1 vs 3 | 29 | 28 | 9.566 | 0.2964 | NS | −8.772 | 0.3697 | NS | ||||||||
| Aphc507 | K-W | 3,77 | 13.32 | 0.0013 | 1 vs 2 | 28 | 28 | 21.04 | 0.0009 | *** | K-W | 21.59 | <0.0001 | −24.18 | 0.0001 | *** |
| 1 vs 3 | 28 | 21 | 4.952 | 0.8863 | NS | −25.18 | 0.0002 | *** | ||||||||
| R49E12 | ANOVA | 2,93 | 26.93 | <0.0001 | 1 vs 2 | 32 | 32 | 99.88 | 0.0003 | *** | ANOVA | 3.407 | 0.0373 | −47.66 | 0.0236 | * |
| 1 vs 3 | 32 | 32 | −86.66 | 0.0019 | ** | −33.16 | 0.1413 | NS | ||||||||
| R81F01 | K-W | 3,96 | 16.46 | 0.0003 | 1 vs 2 | 32 | 32 | 10.72 | 0.2475 | NS | K-W | 18.51 | <0.0001 | −25.95 | 0.0004 | *** |
| 1 vs 3 | 32 | 32 | −17.28 | 0.0262 | * | −0.01563 | >0.9999 | NS | ||||||||
| R53G11 | K-W | 3,96 | 11.82 | 0.0027 | 1 vs 2 | 32 | 32 | 23.38 | 0.0016 | ** | K-W | 19.26 | <0.0001 | −20.84 | 0.0055 | ** |
| 1 vs 3 | 32 | 32 | 7.188 | 0.604 | NS | 8.938 | 0.3987 | NS | ||||||||
| VT026841 | ANOVA | 2122 | 30.52 | <0.0001 | 1 vs 2 | 31 | 31 | 105.9 | <0.0001 | **** | ANOVA | 3.524 | 0.0325 | 37.39 | 0.1971 | NS |
| 1 vs 3 | 31 | 63 | −39.68 | 0.0634 | NS | 54.29 | 0.0167 | * | ||||||||
| VT059775 | ANOVA | 2114 | 20.89 | <0.0001 | 1 vs 2 | 28 | 26 | 103.2 | 0.0003 | *** | ANOVA | 16.31 | <0.0001 | 85.44 | 0.01 | * |
| 1 vs 3 | 28 | 63 | −43.83 | 0.0888 | NS | 142.6 | <0.0001 | **** | ||||||||
| R73B05 | ANOVA | 2,51 | 3.395 | 0.0413 | 1 vs 2 | 17 | 16 | 72.13 | 0.1285 | NS | ANOVA | 2.099 | 0.133 | −87.99 | 0.0842 | NS |
| 1 vs 3 | 17 | 21 | −23.49 | 0.7478 | NS | −35.22 | 0.5896 | NS | ||||||||
| R38H02 | K-W | 3,86 | 0.235 | 0.889 | 1 vs 2 | 27 | 31 | 0.6565 | >0.9999 | NS | K-W | 1.185 | 0.5529 | −5.4 | 0.8227 | NS |
| 1 vs 3 | 27 | 28 | −2.376 | >0.9999 | NS | −6.99 | 0.5986 | NS | ||||||||
| VT040539 | ANOVA | 2121 | 30.93 | <0.0001 | 1 vs 2 | 29 | 32 | 87.69 | 0.0001 | *** | ANOVA | 2.882 | 0.0599 | −15.65 | 0.7011 | NS |
| 1 vs 3 | 29 | 63 | −53.56 | 0.0093 | ** | 28.5 | 0.2546 | NS | ||||||||
| R64H04 | ANOVA | 2,55 | 3.678 | 0.0317 | 1 vs 2 | 15 | 22 | 48.58 | 0.2599 | NS | ANOVA | 4.723 | 0.0128 | −113.7 | 0.0111 | * |
| 1 vs 3 | 15 | 21 | −35.03 | 0.4794 | NS | −104.1 | 0.0222 | * | ||||||||
| R48B10 | K-W | 3,95 | 24.6 | <0.0001 | 1 vs 2 | 31 | 32 | −3.269 | >0.9999 | NS | K-W | 19.05 | <0.0001 | 15.12 | 0.0591 | NS |
| 1 vs 3 | 31 | 32 | −31.21 | <0.0001 | **** | 30.32 | <0.0001 | **** | ||||||||
| R28D01 | K-W | 3,90 | 32.67 | <0.0001 | 1 vs 2 | 32 | 30 | 31.6 | <0.0001 | **** | K-W | 4.767 | 0.0922 | 0.05104 | >0.9999 | NS |
| 1 vs 3 | 32 | 28 | −3.527 | >0.9999 | NS | −12.96 | 0.1104 | NS | ||||||||
| R41A08 | K-W | 3,88 | 6.072 | 0.048 | 1 vs 2 | 32 | 28 | 8.607 | 0.3858 | NS | K-W | 4.948 | 0.0842 | −13.51 | 0.0819 | NS |
| 1 vs 3 | 32 | 28 | −8.214 | 0.428 | NS | −1.172 | >0.9999 | NS | ||||||||
| VT042759 | ANOVA | 2118 | 21.26 | <0.0001 | 1 vs 2 | 27 | 31 | 80.45 | 0.0069 | ** | ANOVA | 0.487 | 0.6156 | 2.115 | 0.996 | NS |
| 1 vs 3 | 27 | 63 | −65.97 | 0.0117 | * | 21.63 | 0.6054 | NS | ||||||||
| VT045108 | ANOVA | 2120 | 38.59 | <0.0001 | 1 vs 2 | 28 | 32 | 107.2 | <0.0001 | **** | ANOVA | 0.528 | 0.591 | 12.84 | 0.805 | NS |
| 1 vs 3 | 28 | 63 | −67.04 | 0.0031 | ** | −8.04 | 0.8938 | NS | ||||||||
| R12B01 | K-W | 3,78 | 12.95 | 0.0015 | 1 vs 2 | 32 | 25 | 15.31 | 0.0227 | * | K-W | 6.563 | 0.0376 | −14.36 | 0.0352 | * |
| 1 vs 3 | 32 | 21 | −8.075 | 0.4089 | NS | −0.7254 | >0.9999 | NS | ||||||||
| VT057257 | ANOVA | 2123 | 19.46 | <0.0001 | 1 vs 2 | 31 | 32 | 47.47 | 0.0708 | NS | ANOVA | 23.47 | <0.0001 | 98.3 | 0.0004 | *** |
| 1 vs 3 | 31 | 63 | −70.94 | 0.001 | ** | 161.2 | <0.0001 | **** | ||||||||
| VT038828 | K-W | 3,62 | 5.49 | 0.0643 | 1 vs 2 | 26 | 15 | 5.356 | 0.7197 | NS | K-W | 4.588 | 0.1008 | −9.454 | 0.2121 | NS |
| 1 vs 3 | 26 | 21 | −8.482 | 0.2182 | NS | 3.346 | >0.9999 | NS | ||||||||
| R38G08 | K-W | 3,83 | 10.76 | 0.0046 | 1 vs 2 | 26 | 29 | 8.57 | 0.376 | NS | K-W | 6.406 | 0.0406 | −15.45 | 0.0353 | * |
| 1 vs 3 | 26 | 28 | −12.3 | 0.1221 | NS | −13.08 | 0.0928 | NS | ||||||||
| R15B07 | K-W | 3,85 | 14.76 | 0.0006 | 1 vs 2 | 29 | 28 | 11.53 | 0.1559 | NS | K-W | 5.134 | 0.0768 | −3.488 | >0.9999 | NS |
| 1 vs 3 | 29 | 28 | −13.78 | 0.0702 | NS | −14.26 | 0.0585 | NS | ||||||||
| VT042577 | K-W | 3120 | 58.2 | <0.0001 | 1 vs 2 | 27 | 31 | 9.205 | 0.6295 | NS | K-W | 12.25 | 0.0022 | −19.2 | 0.0719 | NS |
| 1 vs 3 | 27 | 62 | −43.13 | <0.0001 | **** | −28.07 | 0.0009 | *** | ||||||||
| R84H09 | K-W | 3,90 | 18.2 | 0.0001 | 1 vs 2 | 32 | 30 | −12.04 | 0.1397 | NS | K-W | 17.35 | 0.0002 | −23.64 | 0.0007 | *** |
| 1 vs 3 | 32 | 28 | −28.79 | <0.0001 | **** | −24.29 | 0.0007 | *** | ||||||||
| VT012446 | ANOVA | 2117 | 75.08 | <0.0001 | 1 vs 2 | 28 | 29 | 30.76 | 0.341 | NS | ANOVA | 7.622 | 0.0008 | −48.21 | 0.1101 | NS |
| 1 vs 3 | 28 | 63 | −191 | <0.0001 | **** | −85.36 | 0.0004 | *** | ||||||||
| R73A06 | K-W | 3120 | 75.21 | <0.0001 | 1 vs 2 | 25 | 32 | 8.963 | 0.6687 | NS | K-W | 1.036 | 0.5958 | −3.546 | >0.9999 | NS |
| 1 vs 3 | 25 | 63 | −49.77 | <0.0001 | **** | −7.989 | 0.6624 | NS | ||||||||
| Feb170 | K-W | 3,80 | 18.24 | 0.0001 | 1 vs 2 | 28 | 24 | −22.63 | 0.0009 | *** | K-W | 51.75 | <0.0001 | −35.42 | <0.0001 | **** |
| 1 vs 3 | 28 | 28 | −23.77 | 0.0003 | *** | −41.73 | <0.0001 | **** | ||||||||
| R70B05 | K-W | 3,92 | 26.7 | <0.0001 | 1 vs 2 | 28 | 32 | −26.62 | 0.0002 | *** | K-W | 53.05 | <0.0001 | −47.15 | <0.0001 | **** |
| 1 vs 3 | 28 | 32 | −34.32 | <0.0001 | **** | −40.02 | <0.0001 | **** | ||||||||
| △ No. of episodes | LP (Fig. 1C) |
DP (Fig. 1H) |
||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Nonparametric/parametric test |
Post hoc comparisons |
Nonparametric/parametric test |
Post hoc comparisons |
|||||||||||||
| Driver | Test | DFn, DFd | F | p | n1 | n2 | Mean difference | p | Test | F | p | Mean difference | p | |||
| R47F07 | K-W | 3,95 | 7.568 | 0.0227 | 1 vs 2 | 24 | 39 | 19.64 | 0.0119 | * | K-W | 18.4 | 0.0001 | 29.67 | <0.0001 | **** |
| 1 vs 3 | 24 | 32 | 12.74 | 0.1729 | NS | 24.76 | 0.0017 | ** | ||||||||
| R28E01 | K-W | 3,95 | 2.654 | 0.2653 | 1 vs 2 | 32 | 31 | −4.62 | >0.9999 | NS | K-W | 0.739 | 0.6911 | 1.878 | >0.9999 | NS |
| 1 vs 3 | 32 | 32 | −11.16 | 0.2099 | NS | −3.953 | >0.9999 | NS | ||||||||
| C232 | K-W | 3,91 | 0.678 | 0.7125 | 1 vs 2 | 30 | 31 | 3.171 | >0.9999 | NS | K-W | 2.079 | 0.3536 | 2.794 | >0.9999 | NS |
| 1 vs 3 | 30 | 30 | −2.367 | >0.9999 | NS | 9.55 | 0.3222 | NS | ||||||||
| R70B04 | K-W | 3,94 | 35.74 | <0.0001 | 1 vs 2 | 30 | 32 | −32.64 | <0.0001 | **** | K-W | 0.779 | 0.6776 | 3.006 | >0.9999 | NS |
| 1 vs 3 | 30 | 32 | −38.64 | <0.0001 | **** | 6.1 | 0.7555 | NS | ||||||||
| R53F11 | K-W | 3,95 | 9.895 | 0.0071 | 1 vs 2 | 31 | 32 | 20.93 | 0.0051 | ** | K-W | 9.558 | 0.0084 | 18.14 | 0.0178 | * |
| 1 vs 3 | 31 | 32 | 15.93 | 0.0432 | * | 19.06 | 0.012 | * | ||||||||
| R56C09 | K-W | 3,54 | 2.258 | 0.3233 | 1 vs 2 | 22 | 11 | 5.568 | 0.6736 | NS | K-W | 6.358 | 0.0416 | −4.023 | 0.9765 | NS |
| 1 vs 3 | 22 | 21 | −3.209 | >0.9999 | NS | 9.295 | 0.1051 | NS | ||||||||
| R54B05 | K-W | 3,89 | 26.26 | <0.0001 | 1 vs 2 | 26 | 31 | 18.94 | 0.0114 | * | K-W | 4.334 | 0.1145 | 10.62 | 0.2432 | NS |
| 1 vs 3 | 26 | 32 | 34.87 | <0.0001 | **** | −2.155 | >0.9999 | ns | ||||||||
| R38B06 | ANOVA | 2,83 | 2.258 | 0.1109 | 1 vs 2 | 29 | 29 | 4.103 | 0.0839 | NS | ANOVA | 3.078 | 0.0514 | 0.7586 | 0.9196 | NS |
| 1 vs 3 | 29 | 28 | 0.899 | 0.87 | NS | 5.187 | 0.0442 | * | ||||||||
| Aphc507 | K-W | 3,77 | 24.3 | <0.0001 | 1 vs 2 | 28 | 28 | 27.54 | <0.0001 | **** | K-W | 7.434 | 0.0243 | 7.679 | 0.3972 | NS |
| 1 vs 3 | 28 | 21 | 23.79 | 0.0005 | *** | 17.59 | 0.0128 | * | ||||||||
| R49E12 | K-W | 3,96 | 1.664 | 0.4351 | 1 vs 2 | 32 | 32 | 8.453 | 0.4482 | NS | K-W | 6.715 | 0.0348 | 13.72 | 0.0972 | NS |
| 1 vs 3 | 32 | 32 | 6.828 | 0.6522 | NS | 16.98 | 0.0292 | * | ||||||||
| R81F01 | K-W | 3,96 | 3.284 | 0.1936 | 1 vs 2 | 32 | 32 | 12.59 | 0.14 | NS | K-W | 9.191 | 0.0101 | 16.44 | 0.0362 | * |
| 1 vs 3 | 32 | 32 | 6.484 | 0.7018 | NS | −3.219 | >0.9999 | NS | ||||||||
| R53G11 | K-W | 3,96 | 22.95 | <0.0001 | 1 vs 2 | 32 | 32 | 32.3 | <0.0001 | **** | K-W | 44.77 | <0.0001 | 44.39 | <0.0001 | **** |
| 1 vs 3 | 32 | 32 | 23.25 | 0.0017 | ** | 34.36 | <0.0001 | **** | ||||||||
| VT026841 | K-W | 3125 | 8.142 | 0.0171 | 1 vs 2 | 31 | 31 | 16.61 | 0.1409 | NS | K-W | 22.53 | <0.0001 | −36.65 | 0.0001 | *** |
| 1 vs 3 | 31 | 63 | −5.966 | 0.9039 | NS | −1.65 | >0.9999 | NS | ||||||||
| VT059775 | K-W | 3117 | 12.24 | 0.0022 | 1 vs 2 | 28 | 26 | 31.59 | 0.0012 | ** | K-W | 0.976 | 0.614 | −8.577 | 0.7052 | NS |
| 1 vs 3 | 28 | 63 | 19.7 | 0.0209 | * | −2.032 | >0.9999 | NS | ||||||||
| R73B05 | K-W | 3,54 | 2.625 | 0.2692 | 1 vs 2 | 17 | 16 | 0.5919 | >0.9999 | NS | K-W | 5.997 | 0.0499 | −6.77 | 0.4324 | NS |
| 1 vs 3 | 17 | 21 | −6.804 | 0.369 | NS | 5.99 | 0.4854 | NS | ||||||||
| R38H02 | K-W | 3,86 | 9.21 | 0.01 | 1 vs 2 | 27 | 31 | 19.9 | 0.0048 | ** | K-W | 5.21 | 0.0739 | −8.433 | 0.398 | NS |
| 1 vs 3 | 27 | 28 | 10.16 | 0.2613 | NS | 6.322 | 0.6947 | NS | ||||||||
| VT040539 | K-W | 3124 | 24.96 | <0.0001 | 1 vs 2 | 29 | 32 | 19.98 | 0.0599 | NS | K-W | 3.677 | 0.159 | −3.832 | >0.9999 | NS |
| 1 vs 3 | 29 | 63 | −18.53 | 0.0428 | * | 10.06 | 0.4235 | NS | ||||||||
| R64H04 | ANOVA | 2,55 | 1.367 | 0.2633 | 1 vs 2 | 15 | 22 | 2.088 | 0.6098 | NS | ANOVA | 4.35 | 0.0176 | −8.248 | 0.0391 | * |
| 1 vs 3 | 15 | 21 | −1.724 | 0.713 | NS | 0.1238 | 0.999 | NS | ||||||||
| R48B10 | K-W | 3,95 | 46.85 | <0.0001 | 1 vs 2 | 31 | 32 | 40.93 | <0.0001 | **** | K-W | 7.392 | 0.0248 | 17.16 | 0.0267 | * |
| 1 vs 3 | 31 | 32 | 41.57 | <0.0001 | **** | 15.45 | 0.0517 | NS | ||||||||
| R28D01 | K-W | 3,90 | 3.352 | 0.1871 | 1 vs 2 | 32 | 30 | −3.553 | >0.9999 | NS | K-W | 9.549 | 0.0084 | −5.297 | 0.8486 | NS |
| 1 vs 3 | 32 | 28 | −12.11 | 0.1453 | NS | 15.17 | 0.0493 | * | ||||||||
| R41A08 | ANOVA | 2,85 | 5.785 | 0.0044 | 1 vs 2 | 32 | 28 | −0.1295 | 0.9951 | NS | ANOVA | 1.715 | 0.1861 | 2.393 | 0.3985 | NS |
| 1 vs 3 | 32 | 28 | −4.772 | 0.006 | ** | 3.679 | 0.1318 | NS | ||||||||
| VT042759 | K-W | 3121 | 21.94 | <0.0001 | 1 vs 2 | 27 | 31 | 20.81 | 0.0481 | * | K-W | 8.514 | 0.0142 | −25.67 | 0.0108 | * |
| 1 vs 3 | 27 | 63 | −15.04 | 0.1239 | NS | −8.14 | 0.6248 | NS | ||||||||
| VT045108 | K-W | 3123 | 8.938 | 0.0115 | 1 vs 2 | 28 | 32 | 9.393 | 0.6156 | NS | K-W | 3.369 | 0.1856 | −15.18 | 0.1992 | NS |
| 1 vs 3 | 28 | 63 | −13.03 | 0.2138 | NS | −2.889 | >0.9999 | NS | ||||||||
| R12B01 | K-W | 3,78 | 3.412 | 0.1816 | 1 vs 2 | 32 | 25 | 9.155 | 0.2589 | NS | K-W | 12.76 | 0.0017 | 6.425 | 0.5756 | NS |
| 1 vs 3 | 32 | 21 | −2.077 | >0.9999 | NS | 22.53 | 0.0008 | *** | ||||||||
| VT057257 | K-W | 3126 | 22.64 | <0.0001 | 1 vs 2 | 31 | 32 | 41.51 | <0.0001 | **** | K-W | 15.57 | 0.0004 | −12.24 | 0.3661 | NS |
| 1 vs 3 | 31 | 63 | 11.37 | 0.3106 | NS | 17.93 | 0.0502 | NS | ||||||||
| VT038828 | ANOVA | 2,59 | 1.74 | 0.1845 | 1 vs 2 | 26 | 15 | 4.382 | 0.2177 | NS | ANOVA | 5.011 | 0.0098 | 9.069 | 0.0096 | ** |
| 1 vs 3 | 26 | 21 | −0.7418 | 0.9423 | NS | 6.46 | 0.0475 | * | ||||||||
| R38G08 | ANOVA | 2,80 | 1.988 | 0.1436 | 1 vs 2 | 26 | 29 | −2.434 | 0.3897 | NS | ANOVA | 2.548 | 0.0846 | 0.3448 | 0.9817 | NS |
| 1 vs 3 | 26 | 28 | −4.121 | 0.0904 | NS | 4.429 | 0.0875 | NS | ||||||||
| R15B07 | K-W | 3,85 | 10.93 | 0.0042 | 1 vs 2 | 29 | 28 | 17.1 | 0.0176 | * | K-W | 14.44 | 0.0007 | 6.181 | 0.6876 | NS |
| 1 vs 3 | 29 | 28 | −3.076 | >0.9999 | NS | 23.97 | 0.0005 | *** | ||||||||
| VT042577 | K-W | 3120 | 19.98 | <0.0001 | 1 vs 2 | 27 | 31 | 16.35 | 0.1474 | NS | K-W | 14.08 | 0.0009 | −31.35 | 0.0012 | ** |
| 1 vs 3 | 27 | 62 | −17.24 | 0.0625 | NS | −7.047 | 0.7584 | NS | ||||||||
| R84H09 | K-W | 3,90 | 20.78 | <0.0001 | 1 vs 2 | 32 | 30 | 29.04 | <0.0001 | **** | K-W | 11.58 | 0.0031 | −1.535 | >0.9999 | NS |
| 1 vs 3 | 32 | 28 | 6.507 | 0.6706 | NS | 19.42 | 0.008 | ** | ||||||||
| VT012446 | K-W | 3120 | 18.28 | 0.0001 | 1 vs 2 | 28 | 29 | 38.62 | <0.0001 | **** | K-W | 21.18 | <0.0001 | −29.3 | 0.0029 | ** |
| 1 vs 3 | 28 | 63 | 14.5 | 0.1319 | NS | 6.21 | 0.8629 | NS | ||||||||
| R73A06 | K-W | 3120 | 9.067 | 0.0107 | 1 vs 2 | 25 | 32 | 3.286 | >0.9999 | NS | K-W | 4.374 | 0.1123 | −8.631 | 0.7041 | NS |
| 1 vs 3 | 25 | 63 | −17.14 | 0.0736 | NS | 7.051 | 0.7813 | NS | ||||||||
| Feb170 | ANOVA | 2,77 | 3.437 | 0.0372 | 1 vs 2 | 28 | 24 | 6.054 | 0.0387 | * | ANOVA | 11.1 | <0.0001 | −10.18 | 0.0001 | *** |
| 1 vs 3 | 28 | 28 | 5.25 | 0.0666 | NS | −0.75 | 0.9254 | NS | ||||||||
| R70B05 | K-W | 3,92 | 8.747 | 0.0126 | 1 vs 2 | 28 | 32 | 18.15 | 0.0171 | * | K-W | 2.965 | 0.2271 | 9.442 | 0.3428 | NS |
| 1 vs 3 | 28 | 32 | 1.772 | >0.9999 | NS | 11.15 | 0.2129 | NS | ||||||||
| △ Maximum episode length | LP (Fig. 1D) |
DP (Fig. 1I) |
||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Nonparametric/parametric test |
Post hoc comparisons |
Nonparametric/parametric test |
Post hoc comparisons |
|||||||||||||
| Driver | Test | DFn, DFd | F | p | n1 | n2 | Mean difference | p | Test | F | p | Mean difference | p | |||
| R47F07 | K-W | 3,95 | 22.94 | <0.0001 | 1 vs 2 | 24 | 39 | 33.99 | <0.0001 | **** | K-W | 8.847 | 0.012 | −19.7 | 0.0117 | * |
| 1 vs 3 | 24 | 32 | 17.46 | 0.038 | * | −18.91 | 0.0221 | * | ||||||||
| R28E01 | K-W | 3,95 | 37.95 | <0.0001 | 1 vs 2 | 32 | 31 | 42.57 | <0.0001 | **** | K-W | 4.146 | 0.1258 | −1.289 | >0.9999 | NS |
| 1 vs 3 | 32 | 32 | 17.2 | 0.0251 | * | 11.5 | 0.1904 | NS | ||||||||
| C232 | K-W | 3,91 | 13.18 | 0.0014 | 1 vs 2 | 30 | 31 | 21.43 | 0.0031 | ** | K-W | 2.027 | 0.3629 | 5.958 | 0.7569 | NS |
| 1 vs 3 | 30 | 30 | 21.33 | 0.0035 | ** | 9.617 | 0.317 | NS | ||||||||
| R70B04 | K-W | 3,94 | 21.76 | <0.0001 | 1 vs 2 | 30 | 32 | 32.33 | <0.0001 | **** | K-W | 3.214 | 0.2005 | 12.38 | 0.1483 | NS |
| 1 vs 3 | 30 | 32 | 16.28 | 0.0376 | * | 5.442 | 0.865 | NS | ||||||||
| R53F11 | K-W | 3,95 | 18.06 | 0.0001 | 1 vs 2 | 31 | 32 | 11.71 | 0.184 | NS | K-W | 0.858 | 0.6513 | −6.345 | 0.7222 | NS |
| 1 vs 3 | 31 | 32 | −17.4 | 0.0245 | * | −4.142 | >0.9999 | NS | ||||||||
| R56C09 | K-W | 3,54 | 3.97 | 0.1374 | 1 vs 2 | 22 | 11 | 6.636 | 0.5066 | NS | K-W | 1.94 | 0.379 | 0.04545 | >0.9999 | NS |
| 1 vs 3 | 22 | 21 | 9.381 | 0.1013 | NS | −6.102 | 0.4072 | NS | ||||||||
| R54B05 | K-W | 3,89 | 25.17 | <0.0001 | 1 vs 2 | 26 | 31 | −12.84 | 0.1231 | NS | K-W | 6.834 | 0.0328 | −8.372 | 0.446 | NS |
| 1 vs 3 | 26 | 32 | −33.55 | <0.0001 | **** | 8.645 | 0.41 | NS | ||||||||
| R38B06 | K-W | 3,86 | 5.417 | 0.0667 | 1 vs 2 | 29 | 29 | 15.22 | 0.0405 | * | K-W | 2.062 | 0.3566 | −1.224 | >0.9999 | NS |
| 1 vs 3 | 29 | 28 | 6.685 | 0.6245 | NS | −8.794 | 0.3675 | NS | ||||||||
| Aphc507 | K-W | 3,77 | 8.111 | 0.0173 | 1 vs 2 | 28 | 28 | −4 | >0.9999 | NS | K-W | 15.37 | 0.0005 | −17.68 | 0.0062 | ** |
| 1 vs 3 | 28 | 21 | −17.85 | 0.0114 | * | −23.57 | 0.0005 | *** | ||||||||
| R49E12 | K-W | 3,96 | 21.38 | <0.0001 | 1 vs 2 | 32 | 32 | 13.92 | 0.0912 | NS | K-W | 0.052 | 0.9745 | −1.578 | >0.9999 | NS |
| 1 vs 3 | 32 | 32 | −18.19 | 0.018 | * | −0.6719 | >0.9999 | NS | ||||||||
| R81F01 | K-W | 3,96 | 15.93 | 0.0003 | 1 vs 2 | 32 | 32 | 0.1719 | >0.9999 | NS | K-W | 8.752 | 0.0126 | −10.81 | 0.241 | NS |
| 1 vs 3 | 32 | 32 | −23.98 | 0.0011 | ** | 9.781 | 0.3203 | NS | ||||||||
| R53G11 | K-W | 3,96 | 14.58 | 0.0007 | 1 vs 2 | 32 | 32 | −26.19 | 0.0003 | *** | K-W | 16.56 | 0.0003 | −27.7 | 0.0001 | *** |
| 1 vs 3 | 32 | 32 | −17.08 | 0.0284 | * | −19.03 | 0.0126 | * | ||||||||
| VT026841 | K-W | 3125 | 11.13 | 0.0038 | 1 vs 2 | 31 | 31 | 13.68 | 0.2743 | NS | K-W | 12.01 | 0.0025 | 31.89 | 0.0011 | ** |
| 1 vs 3 | 31 | 63 | −12.52 | 0.2302 | NS | 16.09 | 0.0859 | NS | ||||||||
| VT059775 | K-W | 3117 | 18.86 | <0.0001 | 1 vs 2 | 28 | 26 | −9.118 | 0.6472 | NS | K-W | 12.9 | 0.0016 | 18.48 | 0.0909 | NS |
| 1 vs 3 | 28 | 63 | −30.99 | 0.0001 | *** | 27.66 | 0.0007 | *** | ||||||||
| R73B05 | K-W | 3,54 | 2.125 | 0.3456 | 1 vs 2 | 17 | 16 | 7.397 | 0.3541 | NS | K-W | 3.092 | 0.2131 | −6.57 | 0.4611 | NS |
| 1 vs 3 | 17 | 21 | 1.171 | >0.9999 | NS | −8.835 | 0.1704 | NS | ||||||||
| R38H02 | K-W | 3,86 | 4.33 | 0.1147 | 1 vs 2 | 27 | 31 | −13.12 | 0.092 | NS | K-W | 8.219 | 0.0164 | 6.246 | 0.684 | NS |
| 1 vs 3 | 27 | 28 | −3.622 | >0.9999 | NS | −12.2 | 0.1399 | NS | ||||||||
| VT040539 | K-W | 3124 | 1.149 | 0.5629 | 1 vs 2 | 29 | 32 | −3.626 | >0.9999 | NS | K-W | 3.704 | 0.1569 | 11.61 | 0.4152 | NS |
| 1 vs 3 | 29 | 63 | −8.339 | 0.6022 | NS | 15.48 | 0.1098 | NS | ||||||||
| R64H04 | K-W | 3,58 | 2.046 | 0.3595 | 1 vs 2 | 15 | 22 | −7.117 | 0.4163 | NS | K-W | 5.632 | 0.0598 | 9.37 | 0.195 | NS |
| 1 vs 3 | 15 | 21 | −7.367 | 0.3938 | NS | −2.267 | >0.9999 | NS | ||||||||
| R48B10 | K-W | 3,95 | 43.17 | <0.0001 | 1 vs 2 | 31 | 32 | −21.71 | 0.0036 | ** | K-W | 2.044 | 0.3598 | 1.821 | >0.9999 | NS |
| 1 vs 3 | 31 | 32 | −45.61 | <0.0001 | **** | 9.336 | 0.358 | NS | ||||||||
| R28D01 | K-W | 3,90 | 8.714 | 0.0128 | 1 vs 2 | 32 | 30 | 19.28 | 0.0074 | ** | K-W | 7.384 | 0.0249 | 7.048 | 0.5768 | NS |
| 1 vs 3 | 32 | 28 | 6.165 | 0.7236 | NS | −11.47 | 0.1796 | NS | ||||||||
| R41A08 | K-W | 3,88 | 7.201 | 0.0273 | 1 vs 2 | 32 | 28 | 10.37 | 0.2333 | NS | K-W | 1.163 | 0.5592 | −3.431 | >0.9999 | NS |
| 1 vs 3 | 32 | 28 | 17.57 | 0.0157 | * | −7.127 | 0.562 | NS | ||||||||
| VT042759 | K-W | 3121 | 1.694 | 0.4288 | 1 vs 2 | 27 | 31 | 3.568 | >0.9999 | NS | K-W | 2.454 | 0.2932 | 14.21 | 0.2475 | NS |
| 1 vs 3 | 27 | 63 | −6.024 | 0.9105 | NS | 5.741 | 0.9534 | NS | ||||||||
| VT045108 | K-W | 3123 | 11.3 | 0.0035 | 1 vs 2 | 28 | 32 | 8.79 | 0.6813 | NS | K-W | 0.231 | 0.8908 | −0.9107 | >0.9999 | NS |
| 1 vs 3 | 28 | 63 | −16.04 | 0.0952 | NS | −3.512 | >0.9999 | NS | ||||||||
| R12B01 | K-W | 3,78 | 2.066 | 0.3559 | 1 vs 2 | 32 | 25 | −4.789 | 0.857 | NS | K-W | 6.516 | 0.0385 | −8.618 | 0.3084 | NS |
| 1 vs 3 | 32 | 21 | −9.04 | 0.3109 | NS | −16.03 | 0.0235 | * | ||||||||
| VT057257 | K-W | 3126 | 10.88 | 0.0043 | 1 vs 2 | 31 | 32 | −21.74 | 0.0364 | * | K-W | 3.119 | 0.2102 | 12.92 | 0.3204 | NS |
| 1 vs 3 | 31 | 63 | −26.02 | 0.0023 | ** | 13.53 | 0.1824 | NS | ||||||||
| VT038828 | K-W | 3,62 | 1.464 | 0.4808 | 1 vs 2 | 26 | 15 | −3.295 | >0.9999 | NS | K-W | 4.462 | 0.1074 | −12.33 | 0.0701 | NS |
| 1 vs 3 | 26 | 21 | −6.39 | 0.4546 | NS | −5.159 | 0.6594 | NS | ||||||||
| R38G08 | K-W | 3,83 | 1.082 | 0.5821 | 1 vs 2 | 26 | 29 | 6.631 | 0.6168 | NS | K-W | 2.933 | 0.2307 | −6.452 | 0.6432 | NS |
| 1 vs 3 | 26 | 28 | 2.31 | >0.9999 | NS | −11.22 | 0.175 | NS | ||||||||
| R15B07 | K-W | 3,85 | 8.564 | 0.0138 | 1 vs 2 | 29 | 28 | −18.1 | 0.0113 | * | K-W | 7.204 | 0.0273 | −9.044 | 0.3332 | NS |
| 1 vs 3 | 29 | 28 | −14.3 | 0.0576 | NS | −17.54 | 0.0146 | * | ||||||||
| VT042577 | K-W | 3120 | 13.2 | 0.0014 | 1 vs 2 | 27 | 31 | −3.256 | >0.9999 | NS | K-W | 7.474 | 0.0238 | 21.56 | 0.0371 | * |
| 1 vs 3 | 27 | 62 | −24.72 | 0.0041 | ** | 2.697 | >0.9999 | NS | ||||||||
| R84H09 | K-W | 3,90 | 8.832 | 0.0121 | 1 vs 2 | 32 | 30 | −17.27 | 0.0186 | * | K-W | 5.947 | 0.0511 | −0.7583 | >0.9999 | NS |
| 1 vs 3 | 32 | 28 | −16.91 | 0.0248 | * | −14.86 | 0.0559 | NS | ||||||||
| VT012446 | K-W | 3120 | 45.35 | <0.0001 | 1 vs 2 | 28 | 29 | −7.68 | 0.8093 | NS | K-W | 4.805 | 0.0905 | 12.13 | 0.3761 | NS |
| 1 vs 3 | 28 | 63 | −46.4 | <0.0001 | **** | −4.972 | >0.9999 | NS | ||||||||
| R73A06 | K-W | 3120 | 45.9 | <0.0001 | 1 vs 2 | 25 | 32 | −4.716 | >0.9999 | NS | K-W | 1.651 | 0.438 | 11.93 | 0.3978 | NS |
| 1 vs 3 | 25 | 63 | −45.6 | <0.0001 | **** | 6.513 | 0.8566 | NS | ||||||||
| Feb170 | K-W | 3,80 | 26.9 | <0.0001 | 1 vs 2 | 28 | 24 | −29.38 | <0.0001 | **** | K-W | 18.19 | 0.0001 | −16.12 | 0.0253 | * |
| 1 vs 3 | 28 | 28 | −27.16 | <0.0001 | **** | −26.29 | <0.0001 | **** | ||||||||
| R70B05 | K-W | 3,92 | 26.38 | <0.0001 | 1 vs 2 | 28 | 32 | −31.9 | <0.0001 | **** | K-W | 12.54 | 0.0019 | −15.88 | 0.043 | * |
| 1 vs 3 | 28 | 32 | −30.17 | <0.0001 | **** | −24.21 | 0.0009 | *** | ||||||||
| △ P(doze) | LP (Fig. 1E) |
DP (Fig. 1J) |
||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Nonparametric/parametric test |
Post hoc comparisons |
Nonparametric/parametric test |
Post hoc comparisons |
|||||||||||||
| Driver | Test | DFn, DFd | F | p | n1 | n2 | Mean difference | p | Test | F | p | Mean difference | p | |||
| R47F07 | K-W | 3,95 | 45.66 | <0.0001 | 1 vs 2 | 32 | 31 | 46.4 | <0.0001 | **** | K-W | 23.39 | <0.0001 | 25.36 | 0.0005 | *** |
| 1 vs 3 | 32 | 32 | 16.66 | 0.0313 | * | 31.47 | <0.0001 | **** | ||||||||
| R28E01 | K-W | 3,96 | 15.34 | 0.0005 | 1 vs 2 | 32 | 32 | 26.91 | 0.0002 | *** | K-W | 5.817 | 0.0545 | 10.84 | 0.2389 | NS |
| 1 vs 3 | 32 | 32 | 9.563 | 0.3394 | NS | 16.53 | 0.0352 | * | ||||||||
| C232 | K-W | 3,92 | 12.34 | 0.0021 | 1 vs 2 | 31 | 31 | 17.03 | 0.0241 | * | K-W | 8.184 | 0.0167 | −2.323 | >0.9999 | NS |
| 1 vs 3 | 31 | 30 | 23.11 | 0.0015 | ** | 15.71 | 0.0433 | * | ||||||||
| R70B04 | K-W | 3,90 | 21.41 | <0.0001 | 1 vs 2 | 27 | 31 | 30.78 | <0.0001 | **** | K-W | 13.43 | 0.0012 | 22.41 | 0.0022 | ** |
| 1 vs 3 | 27 | 32 | 9.712 | 0.3097 | NS | 21.63 | 0.0031 | ** | ||||||||
| R53F11 | K-W | 3,94 | 62.65 | <0.0001 | 1 vs 2 | 30 | 32 | 54.01 | <0.0001 | **** | K-W | 38.71 | <0.0001 | 34.1 | <0.0001 | **** |
| 1 vs 3 | 30 | 32 | 36.17 | <0.0001 | **** | 40.22 | <0.0001 | **** | ||||||||
| R56C09 | K-W | 3,55 | 7.995 | 0.0184 | 1 vs 2 | 23 | 11 | 15.14 | 0.0199 | * | K-W | 1.245 | 0.5367 | −4.585 | 0.87 | NS |
| 1 vs 3 | 23 | 21 | 10.06 | 0.075 | NS | 2.06 | >0.9999 | NS | ||||||||
| R54B05 | K-W | 3,87 | 16.31 | 0.0003 | 1 vs 2 | 24 | 31 | 27.73 | 0.0001 | *** | K-W | 1.889 | 0.3889 | 2.621 | >0.9999 | NS |
| 1 vs 3 | 24 | 32 | 15.73 | 0.0422 | * | −5.938 | 0.768 | NS | ||||||||
| R38B06 | K-W | 3,85 | 20.07 | <0.0001 | 1 vs 2 | 29 | 29 | 26.55 | <0.0001 | **** | K-W | 4.584 | 0.1011 | 9.897 | 0.2536 | NS |
| 1 vs 3 | 29 | 27 | 2.854 | >0.9999 | NS | 13.58 | 0.0793 | NS | ||||||||
| Aphc507 | K-W | 3,78 | 48.18 | <0.0001 | 1 vs 2 | 28 | 29 | 35.39 | <0.0001 | **** | K-W | 40.34 | <0.0001 | 31.56 | <0.0001 | **** |
| 1 vs 3 | 28 | 21 | 39.21 | <0.0001 | **** | 36.68 | <0.0001 | **** | ||||||||
| R49E12 | K-W | 3,94 | 23.26 | <0.0001 | 1 vs 2 | 31 | 31 | 24.03 | 0.001 | ** | K-W | 0.85 | 0.6536 | −6.226 | 0.7378 | NS |
| 1 vs 3 | 31 | 32 | −7.883 | 0.503 | NS | −4.345 | >0.9999 | NS | ||||||||
| R81F01 | ANOVA | 2,93 | 12.57 | <0.0001 | 1 vs 2 | 32 | 32 | 0.08838 | <0.0001 | **** | ANOVA | 0.488 | 0.6154 | 0.03071 | 0.6904 | NS |
| 1 vs 3 | 32 | 32 | 0.02067 | 0.5304 | NS | 0.02308 | 0.9553 | NS | ||||||||
| R53G11 | ANOVA | 2,91 | 140.8 | <0.0001 | 1 vs 2 | 31 | 31 | 0.4689 | <0.0001 | **** | ANOVA | 83.57 | <0.0001 | 0.4356 | <0.0001 | **** |
| 1 vs 3 | 31 | 32 | 0.4239 | <0.0001 | **** | 0.4201 | <0.0001 | **** | ||||||||
| VT026841 | K-W | 3125 | 35.75 | <0.0001 | 1 vs 2 | 31 | 31 | 13.48 | 0.2857 | NS | K-W | 6.354 | 0.0417 | −14.97 | 0.2077 | NS |
| 1 vs 3 | 31 | 63 | −30.83 | 0.0002 | *** | 4.997 | >0.9999 | NS | ||||||||
| VT059775 | ANOVA | 2110 | 37.94 | <0.0001 | 1 vs 2 | 25 | 25 | 0.2231 | <0.0001 | **** | ANOVA | 7.847 | 0.0007 | −0.00487 | >0.9999 | NS |
| 1 vs 3 | 25 | 63 | 0.22 | <0.0001 | **** | 0.08162 | 0.0052 | ** | ||||||||
| R73B05 | K-W | 3,54 | 0.229 | 0.8917 | 1 vs 2 | 17 | 16 | −1.706 | >0.9999 | NS | K-W | 4.328 | 0.1148 | −3.813 | 0.9732 | NS |
| 1 vs 3 | 17 | 21 | 0.7703 | >0.9999 | NS | 6.762 | 0.3754 | NS | ||||||||
| R38H02 | K-W | 3,85 | 26.52 | <0.0001 | 1 vs 2 | 27 | 31 | 32.8 | <0.0001 | **** | K-W | 8.025 | 0.0181 | −15.33 | 0.0365 | * |
| 1 vs 3 | 27 | 27 | 11.67 | 0.1649 | NS | 0.8148 | >0.9999 | NS | ||||||||
| VT040539 | K-W | 3124 | 6.565 | 0.0375 | 1 vs 2 | 29 | 32 | 14.85 | 0.214 | NS | K-W | 4.511 | 0.1048 | 12.19 | 0.3718 | NS |
| 1 vs 3 | 29 | 63 | −5.067 | >0.9999 | NS | 17.12 | 0.0675 | NS | ||||||||
| R64H04 | K-W | 3,59 | 4.236 | 0.1203 | 1 vs 2 | 15 | 23 | 11.35 | 0.093 | NS | K-W | 8.076 | 0.0176 | −15.03 | 0.0168 | * |
| 1 vs 3 | 15 | 21 | 4.429 | 0.8913 | NS | −4.143 | 0.9511 | NS | ||||||||
| R48B10 | K-W | 3,93 | 33.55 | <0.0001 | 1 vs 2 | 30 | 31 | 38.6 | <0.0001 | **** | K-W | 36.21 | <0.0001 | 35.53 | <0.0001 | **** |
| 1 vs 3 | 30 | 32 | 28.67 | <0.0001 | **** | 36.49 | <0.0001 | **** | ||||||||
| R28D01 | K-W | 3,88 | 11.65 | 0.0029 | 1 vs 2 | 31 | 30 | 10.33 | 0.2287 | NS | K-W | 3.733 | 0.1546 | −0.2624 | >0.9999 | NS |
| 1 vs 3 | 31 | 27 | −12.79 | 0.1143 | NS | 11.28 | 0.1871 | NS | ||||||||
| R41A08 | K-W | 3,86 | 15.5 | 0.0004 | 1 vs 2 | 31 | 28 | 15.05 | 0.0415 | * | K-W | 3.114 | 0.2108 | −4.499 | 0.9791 | NS |
| 1 vs 3 | 31 | 27 | −11.35 | 0.1686 | NS | 7.286 | 0.5354 | NS | ||||||||
| VT042759 | K-W | 3119 | 25.96 | <0.0001 | 1 vs 2 | 26 | 30 | 29.2 | 0.0032 | ** | K-W | 2.782 | 0.2489 | −10.97 | 0.4702 | NS |
| 1 vs 3 | 26 | 63 | −9.69 | 0.4564 | NS | −13.3 | 0.1963 | NS | ||||||||
| VT045108 | K-W | 3123 | 9.84 | 0.0073 | 1 vs 2 | 28 | 32 | 16.79 | 0.1377 | NS | K-W | 6.609 | 0.0367 | −20.93 | 0.0466 | * |
| 1 vs 3 | 28 | 63 | −7.48 | 0.7112 | NS | −3.385 | >0.9999 | NS | ||||||||
| R12B01 | K-W | 3,77 | 12.13 | 0.0023 | 1 vs 2 | 31 | 25 | 18.97 | 0.0032 | ** | K-W | 2.735 | 0.2547 | −5 | 0.8115 | NS |
| 1 vs 3 | 31 | 21 | 16.92 | 0.0149 | * | 5.952 | 0.693 | NS | ||||||||
| VT057257 | K-W | 3126 | 17.41 | 0.0002 | 1 vs 2 | 31 | 32 | 35.45 | 0.0002 | *** | K-W | 25.56 | <0.0001 | −8.33 | 0.7308 | NS |
| 1 vs 3 | 31 | 63 | 7.576 | 0.6886 | NS | 28.13 | 0.0009 | *** | ||||||||
| VT038828 | K-W | 3,62 | 5.289 | 0.071 | 1 vs 2 | 19 | 22 | 12.44 | 0.0553 | NS | K-W | 0.959 | 0.619 | 5.335 | 0.6902 | NS |
| 1 vs 3 | 19 | 21 | 9.882 | 0.1673 | NS | 4.123 | 0.9409 | NS | ||||||||
| R38G08 | K-W | 3,81 | 4.512 | 0.1047 | 1 vs 2 | 25 | 29 | 3.23 | >0.9999 | NS | K-W | 1.653 | 0.4377 | −7.363 | 0.503 | NS |
| 1 vs 3 | 25 | 27 | −9.71 | 0.2741 | NS | −0.7319 | >0.9999 | NS | ||||||||
| R15B07 | K-W | 3,84 | 12.6 | 0.0018 | 1 vs 2 | 29 | 28 | −0.351 | >0.9999 | NS | K-W | 1.506 | 0.4709 | −7.621 | 0.4767 | NS |
| 1 vs 3 | 29 | 27 | −20.39 | 0.0035 | ** | −1.806 | >0.9999 | NS | ||||||||
| VT042577 | K-W | 3123 | 25.99 | <0.0001 | 1 vs 2 | 28 | 32 | 21.61 | 0.0384 | * | K-W | 4.099 | 0.1288 | −9.924 | 0.5641 | NS |
| 1 vs 3 | 28 | 63 | −17.6 | 0.0595 | NS | 5.738 | 0.9571 | NS | ||||||||
| R84H09 | K-W | 3,88 | 39.79 | <0.0001 | 1 vs 2 | 31 | 30 | 38.04 | <0.0001 | **** | K-W | 1.079 | 0.5831 | 6.716 | 0.6093 | NS |
| 1 vs 3 | 31 | 27 | 4.256 | >0.9999 | NS | 2.368 | >0.9999 | NS | ||||||||
| VT012446 | K-W | 3120 | 21.99 | <0.0001 | 1 vs 2 | 28 | 29 | 43.22 | <0.0001 | **** | K-W | 18.98 | <0.0001 | −17.35 | 0.1195 | NS |
| 1 vs 3 | 28 | 63 | 21.6 | 0.0125 | * | 16.15 | 0.0819 | NS | ||||||||
| R73A06 | K-W | 3120 | 1.409 | 0.4943 | 1 vs 2 | 25 | 32 | −6.146 | >0.9999 | NS | K-W | 1.733 | 0.4204 | −10.97 | 0.4748 | NS |
| 1 vs 3 | 25 | 63 | −9.716 | 0.4747 | NS | −2.466 | >0.9999 | NS | ||||||||
| Feb170 | K-W | 3,81 | 21.58 | <0.0001 | 1 vs 2 | 29 | 25 | 28.27 | <0.0001 | **** | K-W | 11.39 | 0.0034 | −10.35 | 0.2139 | NS |
| 1 vs 3 | 29 | 27 | 21.31 | 0.0014 | ** | 11.65 | 0.128 | NS | ||||||||
| R70B05 | ANOVA | 2,86 | 23.94 | <0.0001 | 1 vs 2 | 27 | 30 | 0.2709 | <0.0001 | **** | ANOVA | 69.04 | <0.0001 | 0.4665 | <0.0001 | **** |
| 1 vs 3 | 27 | 32 | 0.1498 | 0.0004 | *** | 0.4365 | <0.0001 | **** | ||||||||
| △ P(wake) | LP (Fig. 1F) |
DP (Fig. 1K) |
||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Nonparametric/parametric test |
Post hoc comparisons |
Nonparametric/parametric test |
Post hoc comparisons |
|||||||||||||
| Driver | Test | DFn, DFd | F | p | n1 | n2 | Mean difference | p | Test | F | p | Mean difference | p | |||
| R47F07 | K-W | 3,95 | 20.67 | <0.0001 | 1 vs 2 | 32 | 31 | −22.98 | 0.0019 | ** | K-W | 33.22 | <0.0001 | 21.38 | 0.0042 | ** |
| 1 vs 3 | 32 | 32 | −29.97 | <0.0001 | **** | 39.69 | <0.0001 | **** | ||||||||
| R28E01 | K-W | 3,96 | 21.46 | <0.0001 | 1 vs 2 | 32 | 32 | −31.44 | <0.0001 | **** | K-W | 9.154 | 0.0103 | 20.5 | 0.0065 | ** |
| 1 vs 3 | 32 | 32 | −9.438 | 0.3507 | NS | 6.031 | 0.7729 | NS | ||||||||
| C232 | K-W | 3,92 | 5.439 | 0.0659 | 1 vs 2 | 31 | 31 | −10.77 | 0.2243 | NS | K-W | 0.371 | 0.8309 | −0.1935 | >0.9999 | NS |
| 1 vs 3 | 31 | 30 | −15.53 | 0.0464 | * | −3.708 | >0.9999 | NS | ||||||||
| R70B04 | K-W | 3,90 | 24.58 | <0.0001 | 1 vs 2 | 27 | 31 | −32.25 | <0.0001 | **** | K-W | 1.492 | 0.4741 | 7.388 | 0.5653 | NS |
| 1 vs 3 | 27 | 32 | −7.976 | 0.4854 | NS | 0.603 | >0.9999 | NS | ||||||||
| R53F11 | K-W | 3,94 | 31.47 | <0.0001 | 1 vs 2 | 30 | 32 | −18.44 | 0.0156 | * | K-W | 5.14 | 0.0765 | 15.13 | 0.0582 | NS |
| 1 vs 3 | 30 | 32 | 19.81 | 0.0085 | ** | 4.16 | >0.9999 | NS | ||||||||
| R56C09 | K-W | 3,55 | 0.134 | 0.9352 | 1 vs 2 | 23 | 11 | 1.277 | >0.9999 | NS | K-W | 1.029 | 0.5978 | 5.387 | 0.718 | NS |
| 1 vs 3 | 23 | 21 | −0.8965 | >0.9999 | NS | 3.669 | 0.896 | NS | ||||||||
| R54B05 | K-W | 3,87 | 3.497 | 0.174 | 1 vs 2 | 24 | 31 | −11.23 | 0.2039 | NS | K-W | 16.74 | 0.0002 | 27.92 | <0.0001 | **** |
| 1 vs 3 | 24 | 32 | −1.24 | >0.9999 | NS | 13.17 | 0.1071 | NS | ||||||||
| R38B06 | K-W | 3,85 | 22.1 | <0.0001 | 1 vs 2 | 29 | 29 | −29.45 | <0.0001 | **** | K-W | 3.352 | 0.1871 | 5.931 | 0.7203 | NS |
| 1 vs 3 | 29 | 27 | −7.777 | 0.4775 | NS | 12.08 | 0.1343 | NS | ||||||||
| Aphc507 | K-W | 3,78 | 7.019 | 0.0299 | 1 vs 2 | 28 | 29 | −7.612 | 0.4097 | NS | K-W | 35.78 | <0.0001 | 32.48 | <0.0001 | **** |
| 1 vs 3 | 28 | 21 | 9.583 | 0.2858 | NS | 31.29 | <0.0001 | **** | ||||||||
| R49E12 | K-W | 3,94 | 13.47 | 0.0012 | 1 vs 2 | 31 | 31 | −12.16 | 0.1585 | NS | K-W | 10.84 | 0.0044 | 18.94 | 0.0126 | * |
| 1 vs 3 | 31 | 32 | 13.06 | 0.1149 | NS | 20.36 | 0.0061 | ** | ||||||||
| R81F01 | K-W | 3,96 | 11.28 | 0.0036 | 1 vs 2 | 32 | 32 | 0.5 | >0.9999 | NS | K-W | 13.12 | 0.0014 | 22.13 | 0.003 | ** |
| 1 vs 3 | 32 | 32 | 20.5 | 0.0065 | ** | 0.5625 | >0.9999 | NS | ||||||||
| R53G11 | K-W | 3,94 | 10.85 | 0.0044 | 1 vs 2 | 31 | 31 | 4.968 | 0.9468 | NS | K-W | 25.01 | <0.0001 | 34.29 | <0.0001 | **** |
| 1 vs 3 | 31 | 32 | 21.58 | 0.0034 | ** | 12.88 | 0.1219 | NS | ||||||||
| VT026841 | K-W | 3125 | 12.64 | 0.0018 | 1 vs 2 | 31 | 31 | −27.65 | 0.0053 | ** | K-W | 1.709 | 0.4254 | −10.32 | 0.5239 | NS |
| 1 vs 3 | 31 | 63 | −1.502 | >0.9999 | NS | −0.809 | >0.9999 | NS | ||||||||
| VT059775 | K-W | 3113 | 29.41 | <0.0001 | 1 vs 2 | 25 | 25 | 7 | 0.9001 | NS | K-W | 2.853 | 0.2402 | −9.72 | 0.5885 | NS |
| 1 vs 3 | 25 | 63 | 36.83 | <0.0001 | **** | −13.07 | 0.1827 | NS | ||||||||
| R73B05 | K-W | 3,54 | 4.271 | 0.1182 | 1 vs 2 | 17 | 16 | −5.555 | 0.6214 | NS | K-W | 5.594 | 0.061 | 12.86 | 0.0379 | * |
| 1 vs 3 | 17 | 21 | 5.216 | 0.6191 | NS | 7.521 | 0.2857 | NS | ||||||||
| R38H02 | K-W | 3,85 | 2.746 | 0.2533 | 1 vs 2 | 27 | 31 | 9.661 | 0.274 | NS | K-W | 2.862 | 0.2391 | 0.5317 | >0.9999 | NS |
| 1 vs 3 | 27 | 27 | 9.37 | 0.3261 | NS | 10 | 0.2731 | NS | ||||||||
| VT040539 | K-W | 3124 | 33.57 | <0.0001 | 1 vs 2 | 29 | 32 | −13.52 | 0.2846 | NS | K-W | 4.112 | 0.128 | 18.66 | 0.0858 | NS |
| 1 vs 3 | 29 | 63 | 29.09 | 0.0006 | *** | 10.51 | 0.3848 | NS | ||||||||
| R64H04 | K-W | 3,59 | 5.863 | 0.0533 | 1 vs 2 | 15 | 23 | −4.896 | 0.7808 | NS | K-W | 4.294 | 0.1169 | 10.76 | 0.1182 | NS |
| 1 vs 3 | 15 | 21 | 7.61 | 0.38 | NS | 10.5 | 0.1408 | NS | ||||||||
| R48B10 | K-W | 3,93 | 19.43 | 19.43 | 1 vs 2 | 30 | 31 | 16.61 | 0.0325 | * | K-W | 1.99 | 0.3698 | 2.994 | >0.9999 | NS |
| 1 vs 3 | 30 | 32 | 30.21 | <0.0001 | **** | −6.388 | 0.7035 | NS | ||||||||
| R28D01 | K-W | 3,88 | 16.8 | 0.0002 | 1 vs 2 | 31 | 30 | −20.47 | 0.0035 | ** | K-W | 5.059 | 0.0797 | 5.797 | 0.7513 | NS |
| 1 vs 3 | 31 | 27 | 5.559 | 0.8169 | NS | 15.06 | 0.0503 | NS | ||||||||
| R41A08 | K-W | 3,86 | 3.179 | 0.204 | 1 vs 2 | 31 | 28 | 6.783 | 0.5948 | NS | K-W | 5.348 | 0.069 | 14.63 | 0.0492 | * |
| 1 vs 3 | 31 | 27 | 11.61 | 0.1545 | NS | 10.1 | 0.2486 | NS | ||||||||
| VT042759 | K-W | 3119 | 8.507 | 0.0142 | 1 vs 2 | 26 | 30 | −1.505 | >0.9999 | NS | K-W | 0.097 | 0.9526 | −1.413 | >0.9999 | NS |
| 1 vs 3 | 26 | 63 | 17.64 | 0.0564 | NS | 0.9634 | >0.9999 | NS | ||||||||
| VT045108 | K-W | 3123 | 20.09 | <0.0001 | 1 vs 2 | 28 | 32 | −6.754 | 0.9282 | NS | K-W | 8.545 | 0.014 | 11.15 | 0.4535 | NS |
| 1 vs 3 | 28 | 63 | 24.84 | 0.0043 | ** | 23.06 | 0.0088 | ** | ||||||||
| R12B01 | K-W | 3,77 | 12.28 | 0.0022 | 1 vs 2 | 31 | 25 | −3.947 | >0.9999 | NS | K-W | 2.253 | 0.3241 | 7.408 | 0.436 | NS |
| 1 vs 3 | 31 | 21 | 17.95 | 0.0091 | ** | −1.604 | >0.9999 | NS | ||||||||
| VT057257 | K-W | 3126 | 15.43 | 0.0004 | 1 vs 2 | 31 | 32 | −2.342 | >0.9999 | NS | K-W | 12.62 | 0.0018 | −27.62 | 0.0054 | ** |
| 1 vs 3 | 31 | 63 | 24.32 | 0.0048 | ** | −26.39 | 0.002 | ** | ||||||||
| VT038828 | K-W | 3,62 | 3.449 | 0.1783 | 1 vs 2 | 19 | 22 | −1.543 | >0.9999 | NS | K-W | 3.602 | 0.1651 | 9.1 | 0.2145 | NS |
| 1 vs 3 | 19 | 21 | 8.065 | 0.316 | NS | 0.02256 | >0.9999 | NS | ||||||||
| R38G08 | K-W | 3,81 | 5.615 | 0.0604 | 1 vs 2 | 25 | 29 | −7.476 | 0.4886 | NS | K-W | 5.318 | 0.07 | 10.32 | 0.2157 | NS |
| 1 vs 3 | 25 | 27 | 7.43 | 0.5104 | NS | 14.71 | 0.0485 | * | ||||||||
| R15B07 | K-W | 3,84 | 5.756 | 0.0562 | 1 vs 2 | 29 | 28 | −5.112 | 0.8579 | NS | K-W | 9.158 | 0.0103 | 1.052 | >0.9999 | NS |
| 1 vs 3 | 29 | 27 | 10.4 | 0.2219 | NS | 17.74 | 0.0131 | * | ||||||||
| VT042577 | K-W | 3123 | 23.35 | <0.0001 | 1 vs 2 | 28 | 32 | −7.221 | 0.8676 | NS | K-W | 31.86 | <0.0001 | 28.4 | 0.0042 | ** |
| 1 vs 3 | 28 | 63 | 26.82 | 0.0019 | ** | 45.57 | <0.0001 | **** | ||||||||
| R84H09 | K-W | 3,88 | 19.71 | <0.0001 | 1 vs 2 | 31 | 30 | 21.25 | 0.0023 | ** | K-W | 27.23 | <0.0001 | 28.34 | <0.0001 | **** |
| 1 vs 3 | 31 | 27 | 28.32 | <0.0001 | **** | 31.12 | <0.0001 | **** | ||||||||
| VT012446 | K-W | 3120 | 47.36 | <0.0001 | 1 vs 2 | 28 | 29 | 2.543 | >0.9999 | NS | K-W | 39.32 | <0.0001 | 27.13 | 0.0065 | ** |
| 1 vs 3 | 28 | 63 | 45.02 | <0.0001 | **** | 49.01 | <0.0001 | **** | ||||||||
| R73A06 | K-W | 3120 | 77.72 | <0.0001 | 1 vs 2 | 25 | 32 | −4.016 | >0.9999 | NS | K-W | 15.11 | 0.0005 | 10.21 | 0.5429 | NS |
| 1 vs 3 | 25 | 63 | 53.74 | <0.0001 | **** | 29.44 | 0.0007 | *** | ||||||||
| Feb170 | K-W | 3,81 | 21.73 | <0.0001 | 1 vs 2 | 29 | 25 | 26.48 | <0.0001 | **** | K-W | 52.79 | <0.0001 | 33.97 | <0.0001 | **** |
| 1 vs 3 | 29 | 27 | 24.31 | 0.0002 | *** | 43.34 | <0.0001 | **** | ||||||||
| R70B05 | K-W | 3,89 | 29.88 | <0.0001 | 1 vs 2 | 27 | 30 | 30.76 | <0.0001 | **** | K-W | 53.64 | <0.0001 | 45.1 | <0.0001 | **** |
| 1 vs 3 | 27 | 32 | 34 | <0.0001 | **** | 42.08 | <0.0001 | **** | ||||||||
aChange in sleep parameters for total sleep, number of episodes, maximum episode length, P(doze), and P(wake) on the activation day (30°C) were analyzed for day (LP) and night (DP) separately. Datasets that had a normal distribution, one-way ANOVA followed by Bonferroni test was applied. For datasets that did not pass the normality test, Kruskal–Wallis (K-W) followed by Dunn's test was applied. Post hoc tests were applied between the experimental group (F1 generation of the cross of GAL4 lines to UAS-dTrpA1)(1) and the genetic control groups (F1 generation of the crosses of either GAL4 lines to wCS or UAS-dTrpA1 to wCS)(2 or 3).
*p < 0.05.
**p < 0.01.
***p < 0.001.
****p < 0.0001.
Table 3.
| LP |
DP |
|||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Driver | Nonparametric/parametric test |
Post hoc comparisons |
Nonparametric/parametric test |
Post hoc comparisons |
||||||||||||
| D3-D1 21°C | Test | DFn, DFd | F | p | n1 | n2 | Mean difference | p | Test | F | p | Mean difference | p | |||
| C232 | ||||||||||||||||
| △ Total sleep | K-W | 3,91 | 4.103 | 0.1285 | 1 vs 2 | 30 | 31 | 8.725 | 0.3943 | NS | K-W | 16.77 | 0.0002 | −21.16 | 0.0035 | ** |
| 1 vs 3 | 30 | 30 | 13.63 | 0.0912 | NS | −26.32 | 0.0002 | *** | ||||||||
| △ No. of episodes | K-W | 3,91 | 4.451 | 0.108 | 1 vs 2 | 30 | 31 | 8.845 | 0.3803 | NS | K-W | 2.186 | 0.3352 | 9.984 | 0.2786 | NS |
| 1 vs 3 | 30 | 30 | 14.22 | 0.0735 | NS | 4.9 | 0.9435 | NS | ||||||||
| △ Maximum episode length | K-W | 3,91 | 1.39 | 0.4991 | 1 vs 2 | 30 | 31 | −2.339 | >0.9999 | NS | K-W | 0.326 | 0.8494 | −1.673 | >0.9999 | NS |
| 1 vs 3 | 30 | 30 | 5.45 | 0.8483 | NS | 2.183 | >0.9999 | NS | ||||||||
| △ P(doze) | K-W | 3,92 | 28.85 | <0.0001 | 1 vs 2 | 31 | 31 | −30.23 | <0.0001 | **** | K-W | 4.809 | 0.0903 | −13.87 | 0.0817 | NS |
| 1 vs 3 | 31 | 30 | 2.694 | >0.9999 | NS | −2.237 | >0.9999 | NS | ||||||||
| △ P(wake) | K-W | 3,92 | 5.069 | 0.0793 | 1 vs 2 | 31 | 31 | −13.42 | 0.0957 | NS | K-W | 10.1 | 0.0064 | 17.19 | 0.0225 | * |
| 1 vs 3 | 31 | 30 | −13.09 | 0.1112 | NS | 19.97 | 0.007 | ** | ||||||||
| Feb170 | ||||||||||||||||
| △ Total sleep | K-W | 3,80 | 27.64 | <0.0001 | 1 vs 2 | 28 | 24 | 15.15 | 0.0382 | * | K-W | 3.219 | 0.2 | −10.73 | 0.1939 | NS |
| 1 vs 3 | 28 | 28 | 32.63 | <0.0001 | **** | −8.661 | 0.3263 | NS | ||||||||
| △ No. of episodes | ANOVA | 2,77 | 9.999 | 0.0001 | 1 vs 2 | 28 | 24 | 2.488 | 0.1756 | NS | ANOVA | 4.401 | 0.0155 | 1.405 | 0.7464 | NS |
| 1 vs 3 | 28 | 28 | 7.679 | <0.0001 | **** | 5.964 | 0.0108 | * | ||||||||
| △ Maximum episode length | K-W | 3,80 | 8.277 | 0.0159 | 1 vs 2 | 28 | 24 | 0.6994 | >0.9999 | NS | K-W | 0.452 | 0.7979 | −0.7917 | >0.9999 | NS |
| 1 vs 3 | 28 | 28 | 15.98 | 0.0201 | * | −3.964 | >0.9999 | NS | ||||||||
| △ P(doze) | K-W | 3,81 | 44.43 | <0.0001 | 1 vs 2 | 29 | 25 | 18.76 | 0.007 | ** | K-W | 18.67 | <0.0001 | 11.07 | 0.1695 | NS |
| 1 vs 3 | 29 | 27 | 41.91 | <0.0001 | **** | 27.1 | <0.0001 | **** | ||||||||
| △ P(wake) | K-W | 3,81 | 11.13 | 0.0038 | 1 vs 2 | 29 | 25 | 0.06207 | >0.9999 | NS | K-W | 10.96 | 0.0042 | 19.4 | 0.005 | ** |
| 1 vs 3 | 29 | 27 | −18.47 | 0.0067 | ** | 16.48 | 0.0176 | * | ||||||||
| R48B10 | ||||||||||||||||
| △ Total sleep | K-W | 3,95 | 17.59 | 0.0002 | 1 vs 2 | 31 | 32 | 23.65 | 0.0013 | ** | K-W | 0.76 | 0.684 | −5.828 | 0.803 | NS |
| 1 vs 3 | 31 | 32 | 26.67 | 0.0002 | *** | −1.546 | >0.9999 | NS | ||||||||
| △ No. of episodes | K-W | 3,95 | 11.89 | 0.0026 | 1 vs 2 | 31 | 32 | 23.91 | 0.0011 | ** | K-W | 4.2 | 0.1224 | 11.79 | 0.1781 | NS |
| 1 vs 3 | 31 | 32 | 12 | 0.167 | NS | 12.82 | 0.1289 | NS | ||||||||
| △ Maximum episode length | K-W | 3,95 | 2.567 | 0.2771 | 1 vs 2 | 31 | 32 | 8.717 | 0.4192 | NS | K-W | 1.002 | 0.606 | −3.074 | >0.9999 | NS |
| 1 vs 3 | 31 | 32 | 10.39 | 0.2696 | NS | −6.933 | 0.6365 | NS | ||||||||
| △ P(doze) | ANOVA | 2,90 | 3.153 | 0.0475 | 1 vs 2 | 30 | 31 | 0.02628 | 0.4415 | NS | ANOVA | 13.58 | <0.0001 | 0.1613 | 0.0003 | *** |
| 1 vs 3 | 30 | 32 | 0.05309 | 0.0277 | * | 0.2017 | <0.0001 | **** | ||||||||
| △ P(wake) | K-W | 3,93 | 8.553 | 0.0139 | 1 vs 2 | 30 | 31 | −16.77 | 0.0305 | * | K-W | 2.183 | 0.3357 | 10.15 | 0.2844 | NS |
| 1 vs 3 | 30 | 32 | −18.15 | 0.0163 | * | 6.156 | 0.7389 | NS | ||||||||
| R53F11 | ||||||||||||||||
| △ Total sleep | K-W | 3,95 | 15.56 | 0.0004 | 1 vs 2 | 31 | 32 | 23.22 | 0.0017 | ** | K-W | 9.302 | 0.0096 | 20.46 | 0.0064 | ** |
| 1 vs 3 | 31 | 32 | 24.33 | 0.0009 | *** | 5.681 | 0.8266 | NS | ||||||||
| △ No. of episodes | K-W | 3,95 | 7.201 | 0.0273 | 1 vs 2 | 31 | 32 | 18.6 | 0.0147 | * | K-W | 1.607 | 0.4479 | −7.84 | 0.5069 | NS |
| 1 vs 3 | 31 | 32 | 10.08 | 0.2921 | NS | −0.7308 | >0.9999 | NS | ||||||||
| △ Maximum episode length | K-W | 3,95 | 1.743 | 0.4183 | 1 vs 2 | 31 | 32 | −8.788 | 0.4117 | NS | K-W | 2.279 | 0.3201 | 8.549 | 0.4369 | NS |
| 1 vs 3 | 31 | 32 | −6.726 | 0.6659 | NS | −0.8881 | >0.9999 | NS | ||||||||
| △ P(doze) | ANOVA | 2,95 | 1.461 | 0.2371 | 1 vs 2 | 30 | 34 | 0.01617 | 0.8543 | NS | K-W | 9.719 | 0.0078 | 9.725 | 0.3443 | NS |
| 1 vs 3 | 30 | 34 | 0.03458 | 0.183 | NS | 22.08 | 0.0039 | ** | ||||||||
| △ P(wake) | K-W | 3,98 | 9.365 | 0.0093 | 1 vs 2 | 30 | 34 | −19.12 | 0.0146 | * | K-W | 5.342 | 0.0692 | −15.8 | 0.0531 | NS |
| 1 vs 3 | 30 | 34 | −19.03 | 0.0151 | * | −4.475 | >0.9999 | NS | ||||||||
aChange in sleep parameters, total sleep, number of episodes, maximum episode length, P(doze), and P(wake) on the recovery day (21°C) were analyzed for day (LP) and night (DP) separately. One-way ANOVA followed by Bonferroni test or Kruskal–Wallis (K-W) followed by Dunn's test was applied based on distribution of the datasets.
*p < 0.05.
**p < 0.01.
***p < 0.001.
****p < 0.0001.
Table 4.
| LP |
DP |
|||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Driver | Nonparametric/parametric test |
Post hoc comparisons |
Nonparametric/parametric test |
Post hoc comparisons |
||||||||||||
| D3-D1 21°C | Test | DFn, DFd | F | p | n1 | n2 | Mean difference | p | Test | F | p | Mean difference | p | |||
| R64H04 | ||||||||||||||||
| △ Total sleep | ANOVA | 2,55 | 5.451 | 0.0069 | 1 vs 2 | 15 | 22 | −121.1 | 0.0035 | ** | ANOVA | 2.027 | 0.1415 | −29.82 | 0.6373 | NS |
| 1 vs 3 | 15 | 21 | −85.58 | 0.0464 | * | −75.3 | 0.0959 | NS | ||||||||
| △ No. of episodes | ANOVA | 2,55 | 0.0772 | 0.9258 | 1 vs 2 | 15 | 22 | −0.5091 | 0.9676 | NS | ANOVA | 0.098 | 0.9072 | −1.006 | 0.9253 | NS |
| 1 vs 3 | 15 | 21 | −0.981 | 0.8884 | NS | −1.4 | 0.8643 | NS | ||||||||
| △ Maximum episode length | K-W | 3,58 | 8.367 | 0.0152 | 1 vs 2 | 15 | 22 | −16.35 | 0.0077 | ** | K-W | 2.913 | 0.2331 | −9.277 | 0.2017 | NS |
| 1 vs 3 | 15 | 21 | −9.386 | 0.2003 | NS | −7.681 | 0.3569 | NS | ||||||||
| △ P(doze) | ANOVA | 2,56 | 3.591 | 0.0341 | 1 vs 2 | 15 | 23 | −0.02696 | 0.7393 | NS | K-W | 1.256 | 0.2927 | −0.07932 | 0.2415 | NS |
| 1 vs 3 | 15 | 21 | 0.04529 | 0.2829 | NS | −0.05507 | 0.5749 | NS | ||||||||
| △ P(wake) | ANOVA | 2,56 | 5.536 | 0.0064 | 1 vs 2 | 15 | 23 | 0.1066 | 0.016 | * | K-W | 3.68 | 0.1588 | 0.5768 | >0.9999 | NS |
| 1 vs 3 | 15 | 21 | 0.124 | 0.0054 | ** | 9.295 | 0.2188 | NS | ||||||||
| R47F07 | ||||||||||||||||
| △ Total sleep | ANOVA | 2,92 | 7.799 | 0.0007 | 1 vs 2 | 24 | 39 | −107.8 | 0.001 | ** | ANOVA | 12.99 | <0.0001 | −54.83 | 0.0075 | ** |
| 1 vs 3 | 24 | 32 | −25.5 | 0.6125 | NS | 30.61 | 0.1952 | NS | ||||||||
| △ No. of episodes | K-W | 3,95 | 9.574 | 0.0083 | 1 vs 2 | 24 | 39 | 12.56 | 0.1576 | NS | K-W | 2.283 | 0.3194 | −10.53 | 0.2796 | NS |
| 1 vs 3 | 24 | 32 | −7.448 | 0.633 | NS | −4.609 | >0.9999 | NS | ||||||||
| △ Maximum episode length | K-W | 3,95 | 13.04 | 0.0015 | 1 vs 2 | 24 | 39 | −20.25 | 0.0093 | ** | K-W | 0.042 | 0.9793 | −0.00641 | >0.9999 | NS |
| 1 vs 3 | 24 | 32 | 0.8698 | >0.9999 | NS | −1.229 | >0.9999 | NS | ||||||||
| △ P(doze) | K-W | 3,95 | 3.265 | 0.1954 | 1 vs 2 | 32 | 31 | 12.13 | 0.1615 | NS | K-W | 5.942 | 0.0513 | −5.134 | 0.9198 | NS |
| 1 vs 3 | 32 | 32 | 8.75 | 0.4085 | NS | 11.38 | 0.1977 | NS | ||||||||
| △ P(wake) | ANOVA | 2,92 | 2.319 | 0.1041 | 1 vs 2 | 32 | 31 | 0.07036 | 0.1217 | NS | K-W | 9.721 | 0.0077 | 6.953 | 0.6339 | NS |
| 1 vs 3 | 32 | 32 | −0.00178 | 0.9983 | NS | −14.25 | 0.0774 | NS | ||||||||
| R84H09 | ||||||||||||||||
| △ Total sleep | K-W | 3,90 | 0.3417 | 0.8429 | 1 vs 2 | 32 | 30 | −2.221 | >0.9999 | NS | K-W | 6.194 | 0.0452 | 10.71 | 0.2137 | NS |
| 1 vs 3 | 32 | 28 | 1.777 | >0.9999 | NS | −6.096 | 0.7344 | NS | ||||||||
| △ No. of episodes | K-W | 3,90 | 0.934 | 0.6269 | 1 vs 2 | 32 | 30 | −0.725 | >0.9999 | NS | K-W | 1.779 | 0.4108 | 1.976 | >0.9999 | NS |
| 1 vs 3 | 32 | 28 | −6.054 | 0.7396 | NS | 8.681 | 0.3972 | NS | ||||||||
| △ Maximum episode length | K-W | 3,90 | 1.147 | 0.5634 | 1 vs 2 | 32 | 30 | −2.349 | >0.9999 | NS | K-W | 1.838 | 0.399 | 1.258 | >0.9999 | NS |
| 1 vs 3 | 32 | 28 | 4.877 | 0.9412 | NS | −7.375 | 0.5506 | NS | ||||||||
| △ P(doze) | K-W | 3,88 | 8.562 | 0.0138 | 1 vs 2 | 31 | 30 | 12.25 | 0.1223 | NS | K-W | 2.214 | 0.3305 | −1.055 | >0.9999 | NS |
| 1 vs 3 | 31 | 27 | −7.252 | 0.5617 | NS | 8.217 | 0.4435 | NS | ||||||||
| △ P(wake) | K-W | 3,88 | 0.9652 | 0.6172 | 1 vs 2 | 31 | 30 | 6.324 | 0.6676 | NS | K-W | 3.736 | 0.1545 | −9.147 | 0.3242 | NS |
| 1 vs 3 | 31 | 27 | 2.068 | >0.9999 | NS | 3.382 | >0.9999 | NS | ||||||||
aChange in sleep parameters, total sleep, number of episodes, maximum episode length, P(doze), and P(wake) on the recovery day (21°C) were analyzed for day (LP) and night (DP) separately. One-way ANOVA followed by Bonferroni test or Kruskal–Wallis (K-W) followed by Dunn's test was applied based on distribution of the datasets.
*p < 0.05.
**p < 0.01.
Table 8.
| LP |
DP |
|||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Driver | Nonparametric/parametric test |
Post hoc comparisons |
Nonparametric/parametric test |
Post hoc comparisons |
||||||||||||
| D3-D1 21°C | Test | DFn, DFd | F | p | n1 | n2 | Mean difference | p | Test | F | p | Mean difference | p | |||
| R28E01 | ||||||||||||||||
| △ Total sleep | K-W | 3,95 | 22 | <0.0001 | 1 vs 2 | 32 | 31 | −31.06 | <0.0001 | **** | K-W | 4.966 | 0.0835 | 5.503 | 0.8565 | NS |
| 1 vs 3 | 32 | 32 | −23.77 | 0.0011 | ** | 15.17 | 0.0554 | NS | ||||||||
| △ Maximum episode length | K-W | 3,95 | 7.5 | 0.0235 | 1 vs 2 | 32 | 31 | −15.71 | 0.0474 | * | K-W | 1.406 | 0.4951 | −7.219 | 0.5974 | NS |
| 1 vs 3 | 32 | 32 | −16.97 | 0.0276 | * | −6.969 | 0.6239 | NS | ||||||||
| △ P(doze) | K-W | 3,96 | 4.376 | 0.1121 | 1 vs 2 | 32 | 32 | −7.656 | 0.5432 | NS | K-W | 8 | 0.0183 | 9.281 | 0.3653 | NS |
| 1 vs 3 | 32 | 32 | −14.56 | 0.073 | NS | 19.69 | 0.0094 | ** | ||||||||
| △ P(wake) | K-W | 3,96 | 12.76 | 0.0017 | 1 vs 2 | 32 | 32 | 22.16 | 0.0029 | ** | K-W | 2.335 | 0.3111 | −2.031 | >0.9999 | NS |
| 1 vs 3 | 32 | 32 | 20.88 | 0.0054 | ** | −10.06 | 0.297 | NS | ||||||||
| R70B05 | ||||||||||||||||
| △ Total sleep | K-W | 3,92 | 16.31 | 0.0003 | 1 vs 2 | 28 | 32 | −26.52 | 0.0002 | *** | K-W | 13.3 | 0.0013 | −7.692 | 0.5311 | NS |
| 1 vs 3 | 28 | 32 | −21.48 | 0.0038 | ** | −24.4 | 0.0008 | *** | ||||||||
| △ Maximum episode length | K-W | 3,92 | 27.48 | <0.0001 | 1 vs 2 | 28 | 32 | −35.09 | <0.0001 | **** | K-W | 20.64 | <0.0001 | −23.01 | 0.0017 | ** |
| 1 vs 3 | 28 | 32 | −26.31 | 0.0003 | *** | −30.33 | <0.0001 | **** | ||||||||
| △ P(doze) | ANOVA | 2,86 | 14.35 | <0.0001 | 1 vs 2 | 27 | 30 | 0.1319 | <0.0001 | **** | ANOVA | 10.82 | <0.0001 | 0.2261 | <0.0001 | **** |
| 1 vs 3 | 27 | 32 | 0.1144 | <0.0001 | **** | 0.1582 | 0.0035 | ** | ||||||||
| △ P(wake) | K-W | 3,89 | 18.94 | <0.0001 | 1 vs 2 | 27 | 30 | 28.97 | <0.0001 | **** | K-W | 14.74 | 0.0006 | 15.24 | 0.0523 | NS |
| 1 vs 3 | 27 | 32 | 21.15 | 0.0035 | ** | 25.88 | 0.0003 | *** | ||||||||
aChange in sleep parameters, total sleep, number of episodes, maximum episode length, P(doze), and P(wake) on the recovery day (21°C) were analyzed for day (LP) and night (DP) separately. One-way ANOVA followed by Bonferroni test or Kruskal–Wallis (K-W) followed by Dunn's test was applied based on distribution of the datasets.
*p < 0.05.
**p < 0.01.
***p < 0.001.
****p < 0.0001.
Table 9.
| LP |
DP |
|||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Nonparametric/parametric test |
Post hoc comparisons |
Nonparametric/parametric test |
Post hoc comparisons |
|||||||||||||
| D3-D1 21 °C | Test | DFn, DFd | F | p | n1 | n2 | Mean difference | p | Test | F | p | Mean difference | p | |||
| Figure 7C | ||||||||||||||||
| △ Total sleep | K-W | 3,93 | 2.44 | 0.2952 | 1 vs 2 | 31 | 32 | 0.2535 | >0.9999 | NS | K-W | 4.394 | 0.1111 | −10.63 | 0.2363 | NS |
| 1 vs 3 | 31 | 30 | −9.22 | 0.3644 | NS | −13.76 | 0.0929 | NS | ||||||||
| △ No. of episodes | K-W | 3,93 | 0.7446 | 0.6892 | 1 vs 2 | 31 | 32 | −1.765 | >0.9999 | NS | K-W | 7.075 | 0.0291 | 17.92 | 0.0167 | * |
| 1 vs 3 | 31 | 30 | −5.817 | 0.799 | NS | 7.035 | 0.6165 | NS | ||||||||
| △ Maximum episode length | K-W | 3,93 | 0.1451 | 0.93 | 1 vs 2 | 31 | 32 | −2.587 | >0.9999 | NS | ANOVA | 1.287 | 0.281 | −72.87 | 0.202 | NS |
| 1 vs 3 | 31 | 30 | −1.191 | >0.9999 | NS | −26.9 | 0.7888 | NS | ||||||||
| Figure 7E | ||||||||||||||||
| △ Total sleep | ANOVA | 2,93 | 0.5201 | 0.5962 | 1 vs 2 | 40 | 30 | 25.21 | 0.5593 | NS | K-W | 1.289 | 0.525 | 7.621 | 0.5146 | NS |
| 1 vs 3 | 40 | 26 | 0.2596 | >0.9999 | NS | 2.791 | >0.9999 | NS | ||||||||
| △ No. of episodes | K-W | 3,96 | 27.92 | <0.0001 | 1 vs 2 | 40 | 30 | 24.85 | 0.0004 | *** | ANOVA | 21.09 | <0.0001 | 11.26 | <0.0001 | **** |
| 1 vs 3 | 40 | 26 | 34.78 | <0.0001 | **** | 12.6 | <0.0001 | **** | ||||||||
| △ Maximum episode length | K-W | 3,96 | 1.574 | 0.4551 | 1 vs 2 | 40 | 30 | −5.592 | 0.8118 | NS | K-W | 20.92 | <0.0001 | −25.46 | 0.0003 | *** |
| 1 vs 3 | 40 | 26 | −8.41 | 0.4615 | NS | −27.34 | 0.0002 | *** | ||||||||
aChange in sleep parameters, total sleep, number of episodes, and maximum episode length on the activation day (30°C) were analyzed for LP and DP separately. One-way ANOVA followed by Bonferroni test or Kruskal–Wallis (K-W) followed by Dunn's test was applied based on distribution of the datasets.
*p < 0.05.
***p < 0.001.
****p < 0.0001.
Results
Thermoactivation of ring neurons changes sleep amount
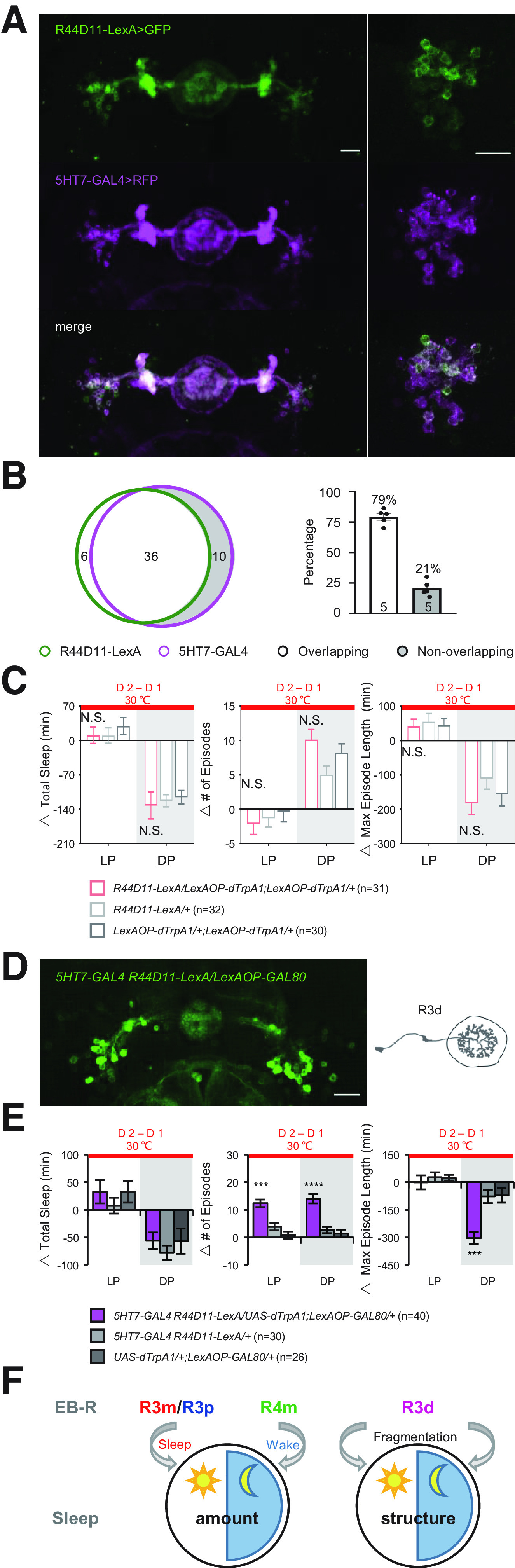
To investigate the roles of ring neuron types, we collected 34 GAL4 drivers that label different populations of ring neurons and used them to drive the thermogenetic tool dTrpA1, allowing the use of elevated temperature to drive neuronal firing (Hamada et al., 2008). Animals were placed in DAM2 system tubes and entrained at 21°C in a 12 h:12 h light/dark cycle. Sleep was then recorded for 3 d: 1 d of baseline sleep at 21°C, 1 d of neural activation sleep at 30°C, then 1 d of recovery sleep at 21°C (Fig. 1A). Changes in sleep parameters for each genotype on the activation and recovery days were calculated by subtracting the baseline day value (Fig. 1A). Changes were only considered significant when the experimental group was different from both genetic controls. Changes in total daytime sleep of the 34 drivers on the activation day are arranged in descending order (Fig. 1B), and changes of total nighttime sleep (Fig. 1G) as well as changes in the number of episodes (Fig. 1C,H), maximum episode duration (Fig. 1D,I), P(doze) (Fig. 1E,J), and P(wake) (Fig. 1F,K) are displayed in the same order as the daytime sleep data to allow assessment of all parameter changes for each genotype. The color-coding of the histogram bars corresponds to the Gaussian clusters shown in Figure 5 and is also used to identify lines in Figures 2, 3, 6, and 7 as part of particular clusters.
Figure 1.
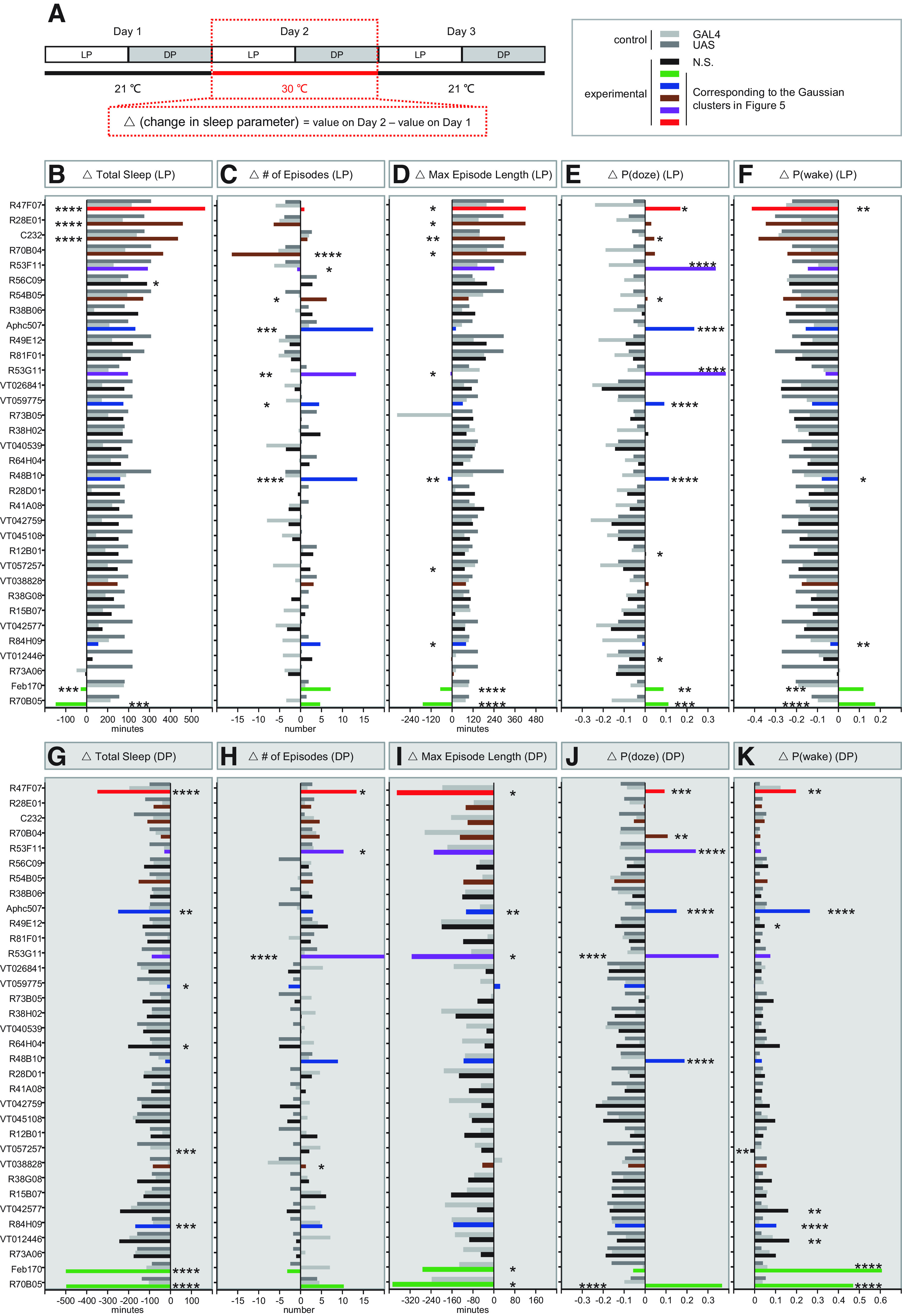
Sleep changes with activation of subtypes of ring neurons. A, Design of the experiments and calculation of sleep parameters on the activation day (red dashed box). B, G, Changes in sleep amount during day (LP) and night (DP). C, H, Changes in number of sleep episodes during daytime and night. D, I, Changes in maximum sleep episodes during daytime and night. E, J, Changes in P(doze) during daytime and night. F, K, Changes in P(wake) during daytime and night. Colored and black bars represent the experimental groups. Color codes are consistent through all of the figures and are based on the daytime cluster analysis in Figure 5. Gray and dark gray bars represent GAL4 control and UAS control, respectively. One-way ANOVA analysis and Dunn's multiple comparisons test were used. Significance, only when the experimental group is significantly different from both GAL4 and UAS controls: *p < 0.05. **p < 0.01. ***p < 0.001. ****p < 0.0001. Data are mean.
Figure 5.
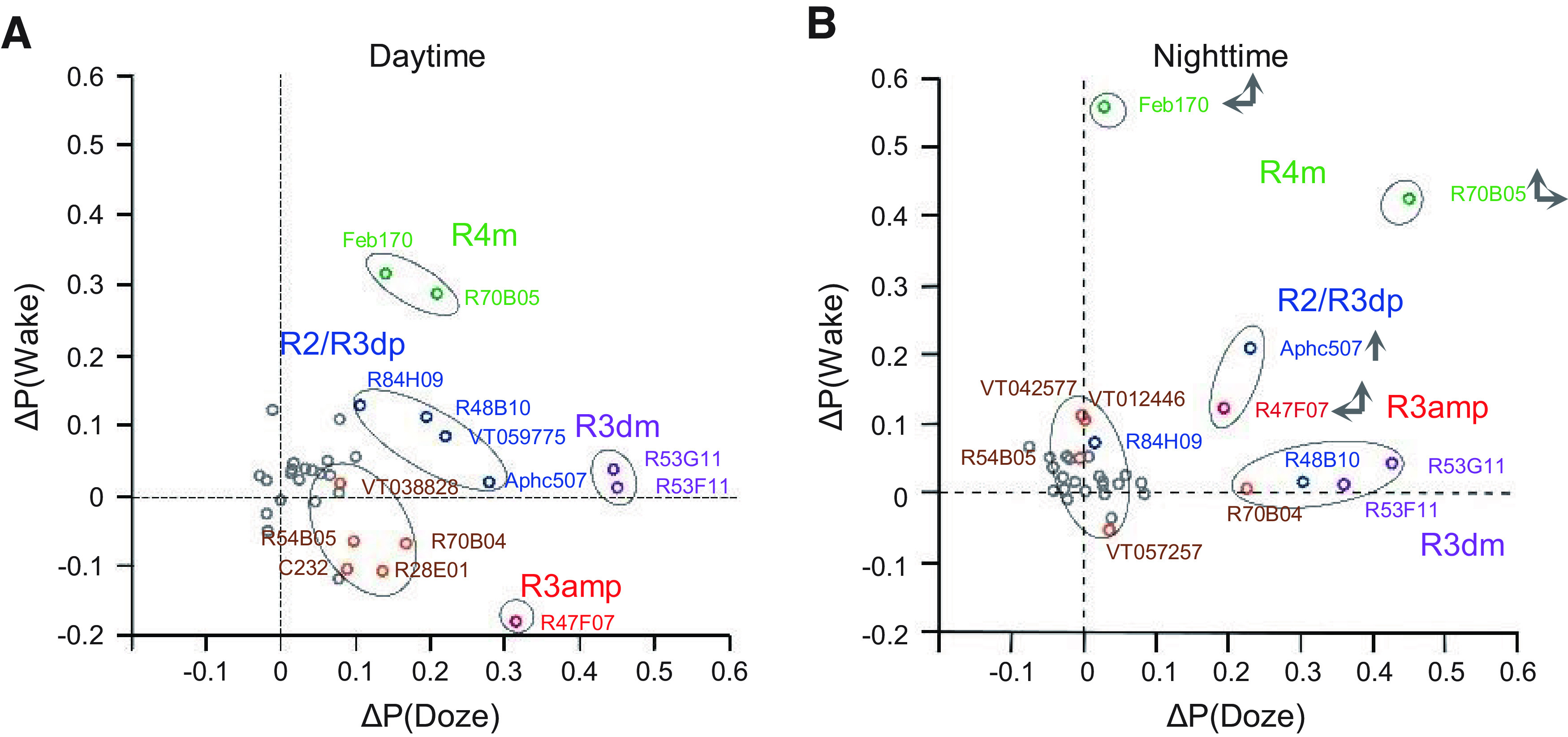
Association of changes in arousal and sleep drive with GAL4+ groups of ring neurons. Mixed Gaussian model cluster analysis for drivers have similar patterns during the daytime (A) and at night (B). Gray dots represent activation did not show significance in P(wake)/P(doze) analysis. Green dots represent increase in both P(wake) and P(doze). Blue dots represent mild increase in both P(wake) and P(doze). Brown dots represent weak increase in both P(wake) and P(doze). Purple dots represent increase only in P(doze). Red dots represent increase in P(doze) and decrease in P(wake). Vertical and horizontal arrows in nighttime panel represent shifts in location of P(doze) and P(wake) compared with the daytime.
Figure 2.
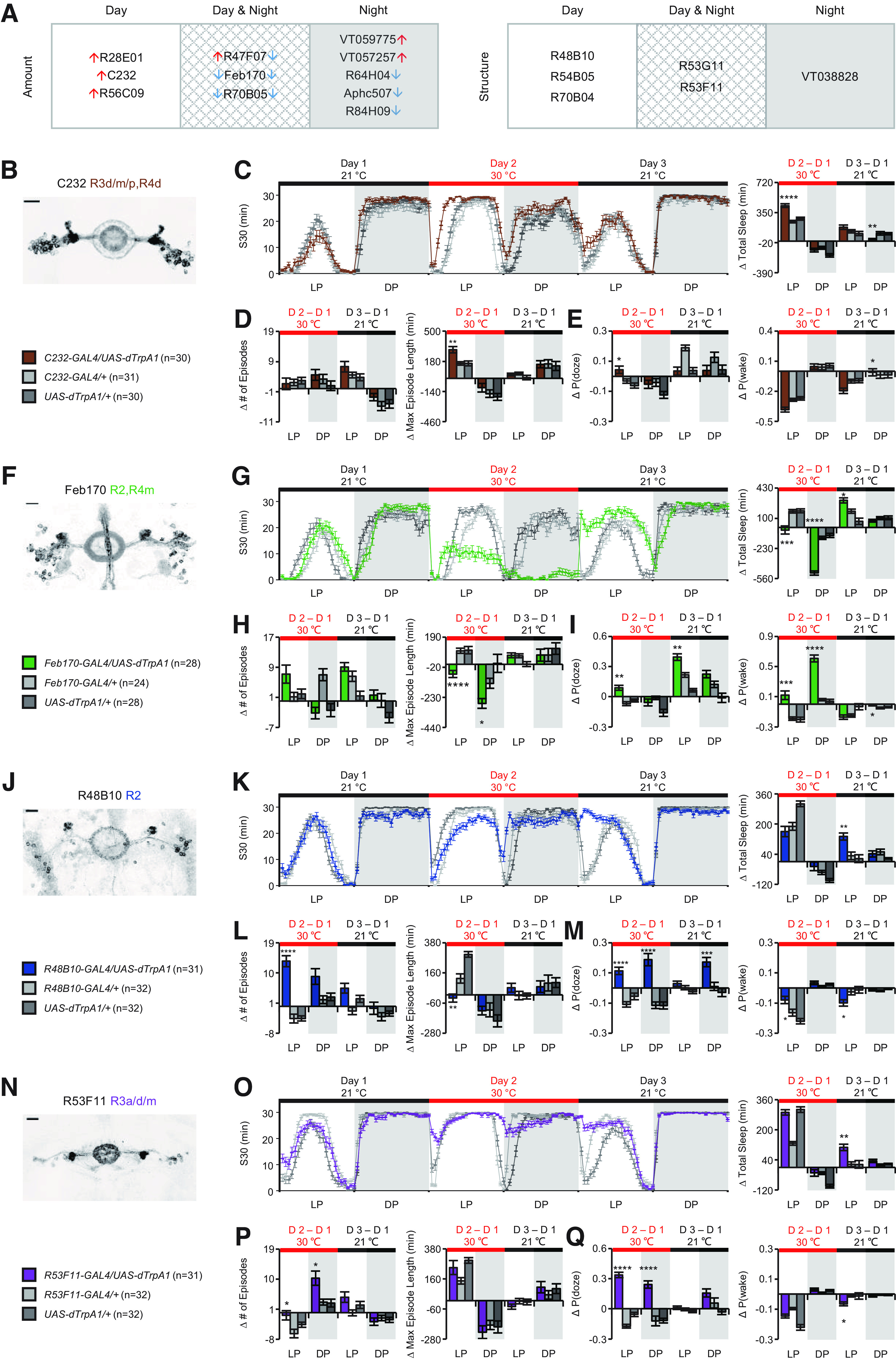
Complex effects on sleep homeostasis with thermoactivation of ring neurons. A, Summary of the drivers exhibited significant changes in total amount of sleep and sleep structure, respectively. Arrows on the left and right represent changes during the day and night, respectively. Up arrows represent increased total amount of sleep. Down arrows represent decreased total amount of sleep. Clusters represent the phenotypes observed day only, night only, or both day and night. Expression patterns of c232-GAL4 (B), Feb170-GAL4 (F), R48B10-GAL4 (J), and R53F11-GAL4 (N). Sleep profiles with quantification of changes of sleep parameters of each driver: total sleep (C,G,K,O), the number of episodes and maximum episode length (D,H,L,P), and P(doze) and P(wake) (E,I,M,Q). Scale bar, 20 μm. *p < 0.05. **p < 0.01. ***p < 0.001. ****p < 0.0001.
Figure 3.
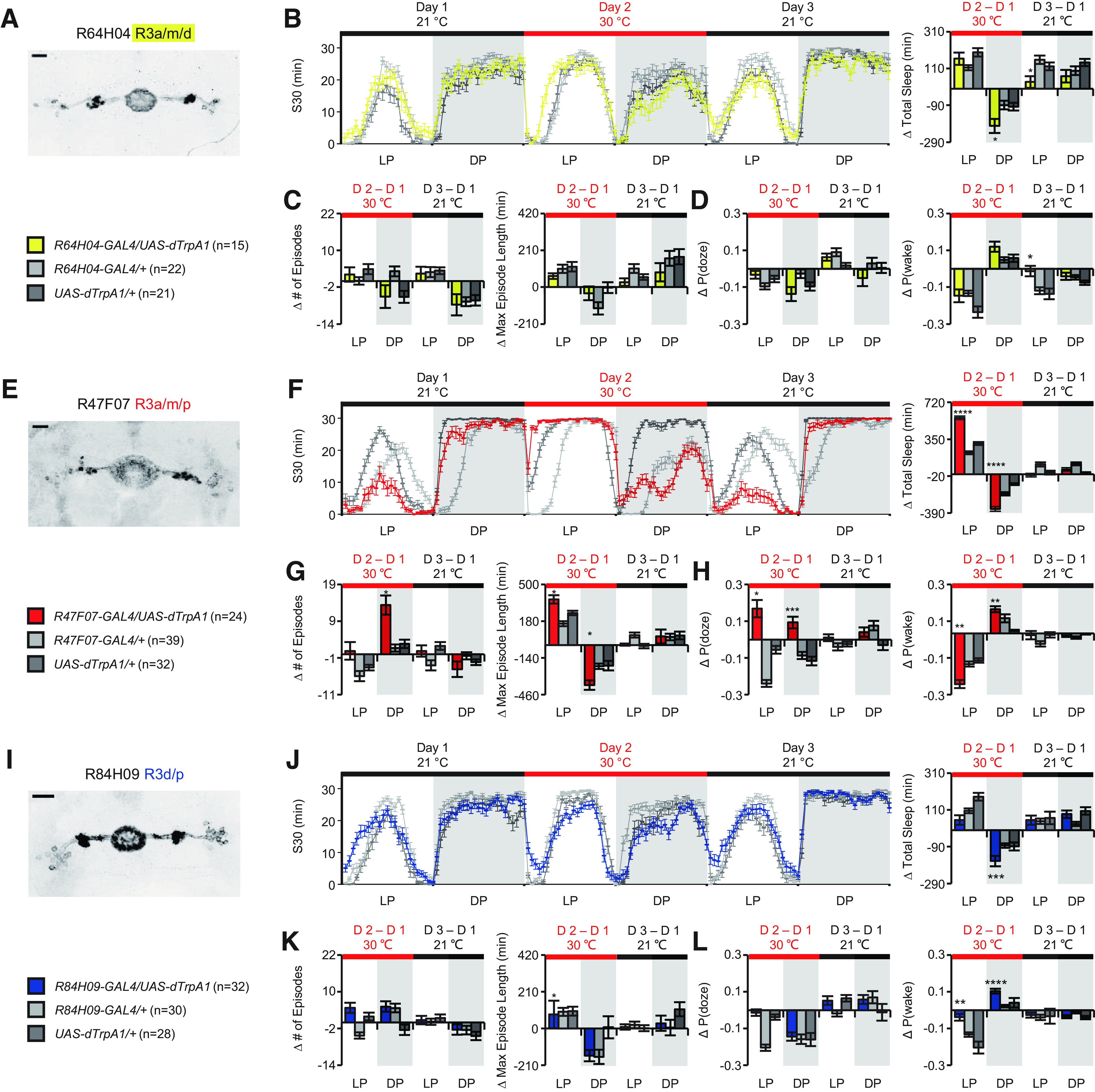
Complex effects on sleep homeostasis with thermoactivation of ring neurons. A–D, Decreased nighttime sleep often fails to induce rebound sleep on cessation of thermoactivation of ring neurons. A, Expression pattern of R64H04-GAL4 which labels R3a, R3m, and R3d neurons. B, Sleep profile and quantification of total sleep before, during, and after activation. Activation of R64H04-GAL4+ neurons reduced total sleep at night, and a persisting reduced sleep on cessation of activation. C, No significant change was observed in the number of episodes and maximum episode length. D, No change of P(doze) was observed, and significantly higher change in P(wake) than controls was found on cessation of activation. E–L, Drivers involved in the regulation of sleep amount and/or structure do not exhibit homeostatic rebound on cessation of thermoactivation. Expression pattern, sleep profile, quantification of sleep amount, sleep structure, and sleep drive/arousal threshold of each driver were presented. E–H, R47F07-GAL4. I–L, R84H09-GAL4. *p < 0.05. **p < 0.01. ***p < 0.001. ****p < 0.0001. Data are mean ± SEM. Scale bar: 20 μm.
Figure 6.
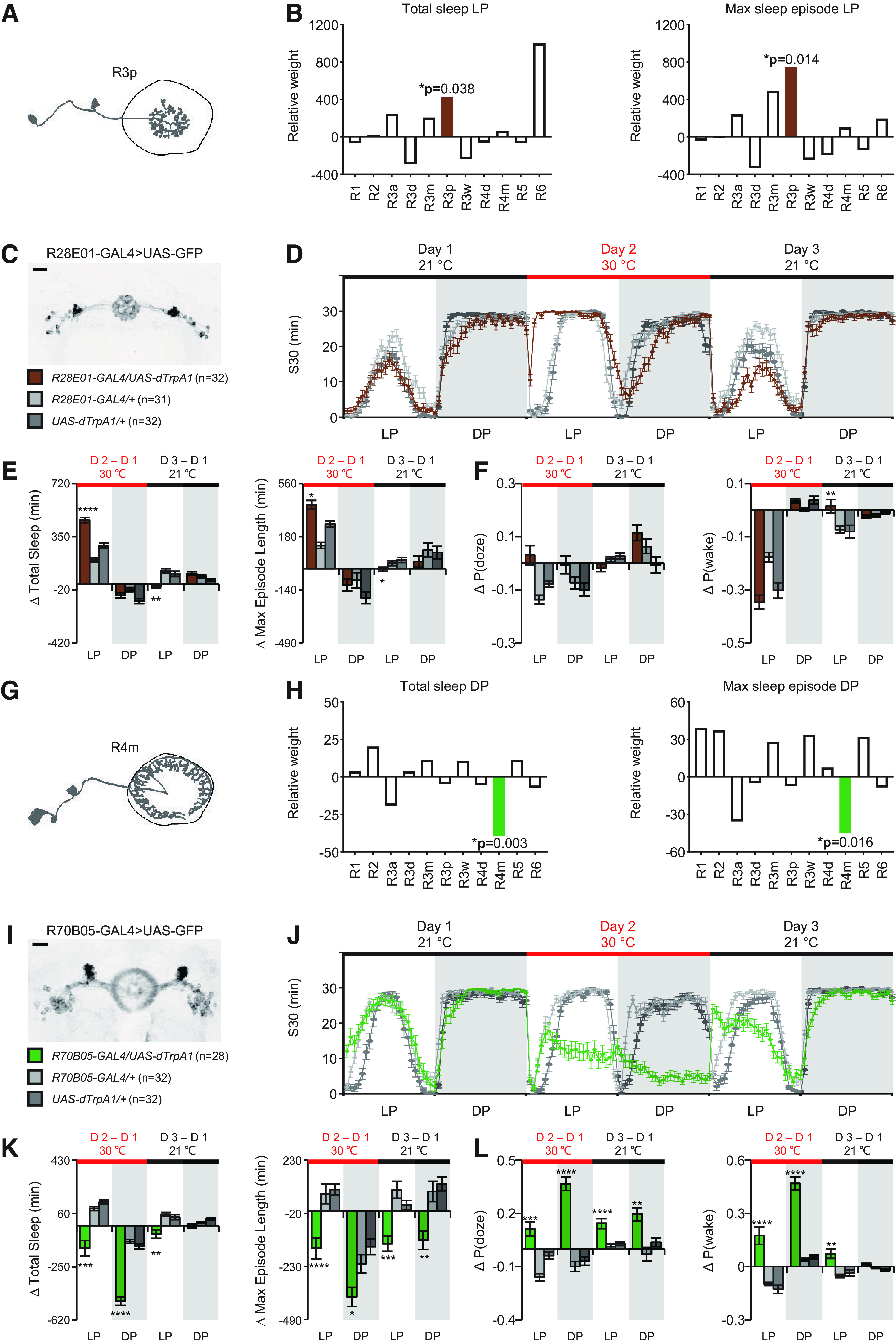
Two subtypes of ring neurons identified by GLM that significantly contribute in the regulation of total sleep and episode length. A, G, Schematic morphologic pattern of a single R3p neuron and R4m neuron, respectively. B, H, R3p neuron and R4m neuron are highly correlated to regulate daytime sleep and nighttime sleep, respectively. The weight of each subclass was analyzed with a GLM. C, I, Expression pattern of R28E01-GAL4 and R70B05-GAL4 as representative for R3p and R4m. D, J, Sleep profiles of total sleep before, during and after activation of R28E01-GAL4+ and R70B05-GAL4+ neurons with two controls. E, K, Changes in total amount of sleep and maximum episodes on the activation day and the recovery day. R70B05-GAL4+ neurons not only significantly reduced nighttime sleep but also exhibited strong impact on reducing daytime sleep. F, No detectable changes in P(doze) on and after activation of R28E01-GAL4+ neurons. Weak elevation of P(wake) on the recovery day was found. L, Strong increase in P(doze) was found when R70B05-GAL4+ neurons were activated, and this effect lasted with cessation of activation. Significant increase in P(wake) was also observed on and after activation. *p < 0.05. **p < 0.01. ***p < 0.001. ****p < 0.0001. Data are mean ± SEM. Scale bar: 20 μm.
Figure 7.
R3d neurons contribute to sleep fragmentation. A, Expression pattern of R44D11-LexA+ neurons and 5HT7-GAL4+ neurons labeled by GFP and RFP, respectively; 79% of the R44D11-LexA+ neurons (green) overlap with 5HT7-GAL4+ neurons (magenta). B, Venn diagram represents the overlapping and nonoverlapping cells between 5HT7-GAL4+ and R44D11-LexA+. Bar graph represents the quantified ratio of nonoverlapping neurons. C, Activation of R44D11-LexA+ neurons do not change sleep or sleep structure during both day and night. D, R3d populations are labeled by suppressing the overlapped neurons of R44D11-LexA+ and 5HT7-GAL4+ by using LexAOP-GAL80. Scale bar, 20 μm. E, Activation of the nonoverlapping R3d neurons fragments sleep without significant effect on total amount during LP and DP. F, Schematic of sleep/structure regulation by multiple subtypes of ring neurons. ***p < 0.001. ****p < 0.0001. Data are mean ± SEM.
Activation of GAL4+ neurons produced many different patterns of change in the amount of sleep. During the daytime, a significant increase in total sleep was found when R47F07-GAL4+, R28E01-GAL4+, C232-GAL4+, and R56C09-GAL4+ neurons were activated (Fig. 1B; Table 1). Since change in total sleep is often associated with change in sleep structure (C. Liu et al., 2019; Wiggin et al., 2020), we also evaluated the number of sleep episodes, episode length, and the behavioral transition probabilities, P(doze) and P(wake) (Wiggin et al., 2020) to further understand the changes in sleep drive and arousal threshold. The increased sleep observed in the above three drivers was accompanied by a significant increase in maximum episode length but no change in the number of episodes compared with their genetic controls (Fig. 1C,D). These flies had increased P(doze) and decreased P(wake), suggesting that these neurons possibly contribute to increased sleep pressure and sleep depth (Fig. 1E,F).
We also found cell groups which, when activated, induced a significant reduction in total sleep: Feb170-GAL4+ and R70B05-GAL4+ neurons (Fig. 1B; Table 1). Sleep reduction was associated with significant decreases in maximum episode length with no change in the number of episodes compared with their genetic controls (Fig. 1C,D; Table 1). The reduced sleep amount and episode length were possibly because of the increased P(doze) and P(wake) (Fig. 1E,F; Table 1), suggesting neurons labeled by these two drivers are involved in upregulation of sleep pressure and downregulation of sleep depth during the daytime.
Nighttime effects of thermogenetic neuron activation are more complex to interpret. Data have to be viewed in the context of the sleep-suppressing effects of elevated temperature on normal WT animal sleep (Parisky et al., 2016; Jin et al., 2021). This temperature effect can be visualized in the continuous sleep plots for most of the GAL4 and UAS control lines in Figures 2, 3, and 6. VT059775-GAL4+ and VT057257-GAL4+ neuron activation led to almost no change of total sleep compared with their own baseline, but this reflects a significant difference from genetic controls, which respond to heat with at large reduction in sleep. These lines also had only small reductions in P(wake) compared with controls, implying that these neurons may be involved in sleep promotion by changing sleep depth (Fig. 1G,K; Table 1).
We also found a number of GAL4 drivers, including R47F07, Aphc507, R64H04, R84H09, Feb170, and R70B05 which significantly reduced nighttime sleep amount compared with their controls, suggesting they contribute to promoting wakefulness (Fig. 1G; Table 1). These reductions in total sleep were accompanied by changes in sleep structure, featured as fragmentation where the number of episodes significantly increased and/or episode length reduced (Fig. 1H,I; Table 1). Many drivers exhibited increased P(doze) and P(wake) (Fig. 1J,K; Table 1), suggesting sleep pressure and sleep depth play important roles in nighttime sleep.
Thermoactivation of ring neurons can change sleep structure independent of sleep amount
We also found cases where sleep structure was changed without alterations in total sleep, supporting the idea that structure can be regulated independently (C. Liu et al., 2019). Activation of neurons from several GAL4 drivers, including R70B04, R53F11, R54B05, R53G11, R48B10, and VT038828, resulted in significant change only in sleep structure. Except for R70B04, which induced consolidated daytime sleep with a decrease in the number of episodes and an increase in the episode length, all drivers mentioned above exhibited fragmented sleep either during the day or at night (Fig. 1C,D,H,I; Table 1). Fragmentation was accompanied by a robust increase in P(doze) for the majority drivers (Fig. 1E,J; Table 1). P(doze) is believed to correlate with sleep pressure (Wiggin et al., 2020), suggesting the fragmentation reflects an increase in the probability of switching from wake to sleep (i.e., high sleep drive rather than from an inability to maintain the sleep state).
The circadian period during which fragmentation occurred varied with GAL4 line. Daytime fragmentation was observed when R54B05-GAL4+ and R48B10-GAL4+ neurons were activated (Fig. 1C; Table 1), and nighttime fragmentation was seen when VT038828-GAL4+ neurons were activated (Fig. 1H; Table 1). Fragmentation of both day and night was found when R53G11-GAL4+ and R53F11-GAL4+ neurons were activated (Fig. 1C,D,H,I; Table 1).
The structural parameters that were altered were also variable. Three GAL4 drivers, R53F11, R54B05, and VT038828, only exhibited a significant increase in the number of episodes. R53G11 and R48B10 only showed reduced episode length. All of these changes contributed to increases in P(doze) with little or weak P(wake) effects, especially during the day (Fig. 1E,F,J,K; Table 1). Interestingly, R12B01-GAL4 did not exhibit detectable changes in the number of episodes or episode length, but had a significant increase in P(doze) compared with both controls (Fig. 1E; Table 1), suggesting a potential specific contribution of R28E01-GAL4+ neurons to control of sleep pressure. Together, changes in sleep structure are highly associated with P(doze), but when sleep structure changes are accompanied by changes in total sleep amount, P(wake) becomes an important component of the regulation.
Thermoactivation of ring neurons has complex effects on sleep homeostasis
We summarized drivers with significant changes of total amount of sleep or sleep structure during the day, at night, or both (Fig. 2A). We plotted sleep and changes in parameters over 3 d to provide a more nuanced picture of the lasting effects of activation of these neurons and present the lines ordered from largest to smallest rebound sleep on the recovery day (Figs. 2B–Q, 3; Tables 2–4). For some of the lines, the changes in total sleep appeared to activate homeostatic changes that were evident during the recovery day. Activation of C232-GAL4+ neurons, which increases sleep on the activation day, leads to a negative rebound (decrease in sleep) on cessation of activation (Fig. 2C; Tables 2 and 3). Activation of Feb170-GAL4+ neurons decreased sleep both in the day and night, and this was followed by a homeostatic rebound increase in sleep (Fig. 2G; Tables 2 and 3). Activation of R48B10-GAL4+ or R53F11-GAL4+ neurons led to fragmentation during either the day or both in the day and night, and a robust homeostatic rebound increase occurred (Figs. 2J–Q; Tables 2 and 3). Interestingly, some drivers exhibited decreased sleep without a rebound change in sleep afterward (e.g., R64H04, R47F07, and R84H09; Fig. 3; Table 4), suggesting that, for these lines, sleep loss was either not able to be compensated for or was not “counted” by the homeostat. These may represent cell types that are not integrated into the homeostat (Seidner et al., 2015).
Association of changes in arousal and sleep drive with GAL4+ groups of ring neurons
The majority of the GAL4 lines we screened contained more than one subtype of ring neuron (Fig. 4A), and exhibited expression outside the EB in other areas of the central brain (Table 5). To examine the linkage between ring neuron types and distinct aspects of sleep amount and/or sleep structure, we first separated drivers into two groups (Fig. 2A): (1) those that exhibited changes in sleep amount and (2) those that exhibited no change in sleep amount but had changes in sleep structure. Based on the time of day when the phenotype was observed (day only, night only, or both day and night), we classed those drivers into three clusters. For lines that changed total sleep, we noted their effects in Figure 2A as increasing or decreasing. The second type of information we layered into the analysis was the identification of the subtypes of ring neurons in each line according to anatomic features and recent nomenclature (Omoto et al., 2018; Hulse et al., 2021) (Fig. 4A). Based on this primary classification, many subtypes of ring neurons, including R1, R2, R4m, R4d, R5, and many R3 subtypes (R3a, R3m, R3d, and R3p), may participate in the regulation of sleep amount (Fig. 4B). Because of the multiplicity of ring neurons in these EB drivers, it was hard to a priori link a single subtype of EB neuron with a specific function in the regulation of sleep amount/structure. Thus, we used statistical models to try to identify links between ring subtypes and phenotypes.
Figure 4.
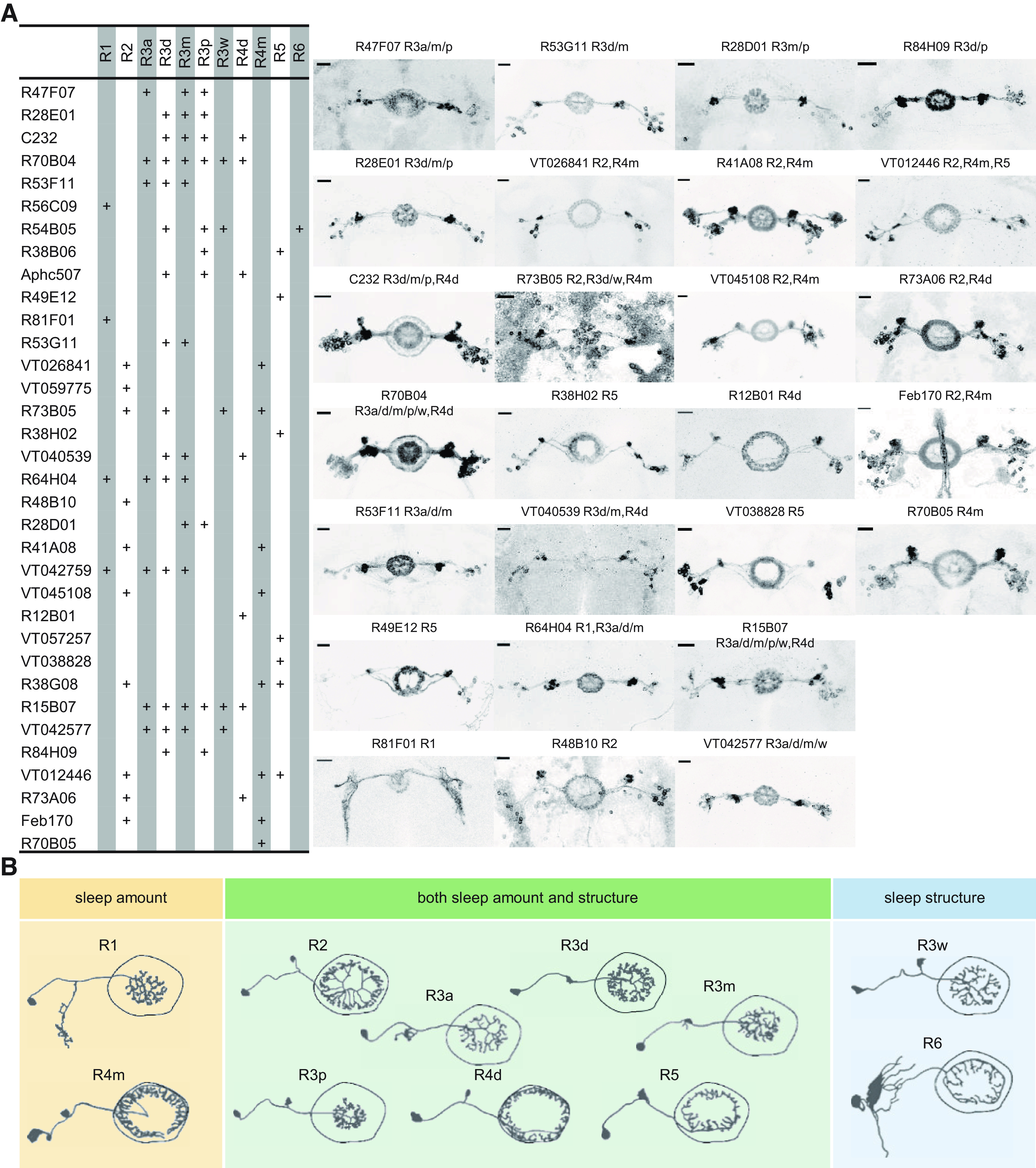
Expression patterns of EB drivers in the screen. A, Distinct subtypes of ring neurons were labeled by the 34 drivers. Expression patterns of all the publicly available drivers (26) are shown. B, Single subtype of ring neurons that are involved in the regulation of sleep amount (yellow), structure (blue), and both amount and structure (green). “+” in columns indicates where the driver has expression. Scale bar: 20 μm.
Table 5.
Expression patterns of 34 drivers outside the EB in the central braina
| Sleep Amount | Sleep Structure | AL | AMMC | AOTU | ATL | AVLP | CL | FB | GA | GNG | ICL | LH | LO | LOP | MB | ME | NO | OL | PI | PRW | SAD | SCL | SEZ | SIP | SLP | SMP | WED | Ventrolateral protocerebrum | Adult pheromone projection PPN1 neuron | Large field neuron | Source | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| R47F07 | √ | √ | + | + | + | + | + | VF | ||||||||||||||||||||||||
| R28E01 | √ | FB | ||||||||||||||||||||||||||||||
| C232 | √ | + | FB | |||||||||||||||||||||||||||||
| R70B04 | √ | FB | ||||||||||||||||||||||||||||||
| R53F11 | √ | FB | ||||||||||||||||||||||||||||||
| R56C09 | √ | + | FB | |||||||||||||||||||||||||||||
| R54B05 | √ | + | + | + | + | + | + | + | VF | |||||||||||||||||||||||
| R38B06 | + | + | + | FB | ||||||||||||||||||||||||||||
| Aphc507 | √ | √ | + | + | FB | |||||||||||||||||||||||||||
| R49E12 | FB | |||||||||||||||||||||||||||||||
| R81F01 | VF | |||||||||||||||||||||||||||||||
| R53G11 | √ | + | + | + | + | + | + | + | + | + | + | + | + | + | + | VF | ||||||||||||||||
| VT026841 | + | + | + | + | + | + | + | + | + | FB | ||||||||||||||||||||||
| VT059775 | √ | √ | + | + | + | + | + | + | + | + | + | + | + | + | + | FB | ||||||||||||||||
| R73B05 | + | + | VF | |||||||||||||||||||||||||||||
| R38H02 | + | + | FB | |||||||||||||||||||||||||||||
| VT040539 | + | + | + | + | + | + | + | FB | ||||||||||||||||||||||||
| R64H04 | √ | FB | ||||||||||||||||||||||||||||||
| R48B10 | √ | + | + | + | FB | |||||||||||||||||||||||||||
| R28D01 | FB | |||||||||||||||||||||||||||||||
| R41A08 | + | FB | ||||||||||||||||||||||||||||||
| VT042759 | + | + | + | + | + | + | + | FB | ||||||||||||||||||||||||
| VT045108 | + | + | + | + | + | + | + | + | FB | |||||||||||||||||||||||
| R12B01 | FB | |||||||||||||||||||||||||||||||
| VT057257 | √ | + | + | + | + | + | + | + | + | + | + | FB | ||||||||||||||||||||
| VT038828 | √ | + | + | + | + | + | + | FB | ||||||||||||||||||||||||
| R38G08 | FB | |||||||||||||||||||||||||||||||
| R15B07 | FB | |||||||||||||||||||||||||||||||
| VT042577 | + | + | + | + | + | + | + | + | FB | |||||||||||||||||||||||
| R84H09 | √ | + | + | + | VF | |||||||||||||||||||||||||||
| VT012446 | + | + | + | + | + | + | + | + | + | + | + | FB | ||||||||||||||||||||
| R73A06 | FB | |||||||||||||||||||||||||||||||
| Feb170 | √ | + | + | FB | ||||||||||||||||||||||||||||
| R70B05 | √ | + | + | + | + | + | + | + | VF |
aDrivers are listed in the first column. √ (in the second or third column) indicates whether they had a phenotype for sleep amount and/or sleep structure. + (in subsequent columns) indicates where the driver has expression. The regions of the central brain were in abbreviation based on description of FlyBase and Virtual FlyBrain in alphabet order. AL, Antennal lobe; AMMC, antennal mechanosensory and motor center; AOTU, anterior optic tubercle; ATL, antler; AVLP, anterior ventrolateral protocerebrum; CL, clamp; GA, gall; GNG, gnathal ganglion; ICL, inferior clamp; LH, lateral horn; LO, lobula; LOP, lobula plate; ME, medulla; NO, nodulus; OL, optic lobe; PI, pars intercerebralis; PRW, prow; SAD, saddle; SCL, superior clamp; SEZ, subesophageal zone; SIP, superior intermediate protocerebrum; SLP, superior lateral protocerebrum; SMP, superior medial protocerebrum; WED, wedge. Source of images for expression analysis is listed in the last column. VF, virtual fly brain; FB, FlyBase. Non-EB expression was not predictive of either total sleep or sleep structure phenotypes.
The first approach we used was aimed at determining the effects of the GAL4 lines (each of which has a different mixture of ring neuron subtypes) in regulating sleep. We used a mixed Gaussian model for changes in P(wake) or P(doze) on the activation day compared with the baseline day (Fig. 5A,B). We chose to use these transition probabilities since they capture some of the more complex aspects of sleep: P(wake) correlates with arousal state/sleep depth, while P(doze) is a measure of sleep drive (Wiggin et al., 2020). A single value of ΔP(wake) and ΔP(doze) for each line was calculated by subtracting the average of the genetic controls for that driver (experimental ΔP – (UAS ΔP + GAL4 ΔP)/2)). These values were then plotted in ΔP(wake)–ΔP(doze) space and clustered with the model to find groups with similar effects on sleep depth and pressure. We identified five clusters of GAL4 lines for day and night, respectively (Fig. 5A,B). These clusters define the color codes used in Figures 1, 2, 3, 5, 6 and Figure 7.
Using our anatomic analysis of these lines, we found that the lines within each cluster shared a common ring neuron subtype. During the daytime (Fig. 5A), R4m (and perhaps R2 neurons) emerged as strong candidates for the regulation of sleep depth/arousal since they are present in lines that have high ΔP(wake) values. R3dm cells appeared to increase sleep drive (i.e., increase the probability of falling asleep); consistent with this, lines with these cells had high P(doze). R2, R3d, and R3p neurons were present in several clusters and did not appear to have unique functionality with regard to sleep depth and drive, but a role in facilitation of the effects of R2 and R3m neurons, or in more specialized functions in sleep structure, cannot be ruled out. We also observed that many drivers play different roles during the day and night (Fig. 5B). For example, R70B05 exhibits relative strong P(wake) but weak P(doze) effect during the day, but at night increases its influence on P(doze); R47F07 has little effect on P(wake) in the day but becomes much more wake-promoting at night.
Association of specific ring neuron subtypes with changes in sleep parameters
Since the variable analyzed using Gaussian clustering was the GAL4 line, which is most often a collection of different ring neuron subtypes, the effects we saw could also be the result of particular combinations of subtypes rather than the result of one dominant subtype alone. To try to isolate effects specific to subtypes, and to look at more specific sleep parameters, we used a second method to extract the contributions of each ring neuron subtype to functional outcomes. Using a GLM with ring neuron subtype as the variable allowed us to calculate the weights of the potential contribution of each subtype of ring neuron to all the sleep parameters for daytime and nighttime, respectively (Table 6). R3p exhibited a significantly positive effect on daytime sleep amount which was associated with its positive weight in episode length (Fig. 6A,B; Table 7). As an example, activation of R28E01-GAL4+ neurons, which include the R3p subtype, elevated daytime sleep and maximum episode length (Fig. 6C–E; Table 8). But the R3p subtype had little effect on P(doze) or P(wake) (Fig. 6F; Table 8). We also found that R4m had a significantly inhibiting effect on total sleep at night and a negative effect on episode length (Fig. 6G,H; Table 7), consistent with the results of Gaussian clustering. R70B05-GAL4+ neurons include the R4m subtype, and activation of neurons labeled by this driver caused a dramatic reduction of sleep in both day and night, which is likely because of the shortened episode length (Fig. 6I–K; Table 8). The effects of activation of these R70B05-GAL4+ neurons persisted into the recovery day, with flies exhibiting significantly elevated sleep pressure and lightened sleep depth (Fig. 6L; Table 8).
Ring neuron synergy is important for sculpting sleep
Interestingly, there were effects uncovered in the GLM analysis that were not seen with GAL4 drivers that labeled only that specific subtype. R1 and R2 neurons exhibit a significantly negative weight in the number of episodes at night, suggesting that these neurons may contribute to consolidation of sleep structure (Table 6). However, we failed to observe consolidation after activation of R1- or R2-specific GAL4 drivers; R56C09 and R81F01 had little significant effect on sleep structure (Fig. 1; Table 1), while activation of R48B10 produced a moderately strong increase in P(doze/wake) (Fig. 2M; Table 3). This suggests that the sleep consolidation effects of activating these neurons uncovered by the GLM requires coactivation of other subtypes.
Supporting the complexity of ring neuron subtype interactions, we observed that activation of the R47F07-GAL4 driver, which labels R3a, R3m, and R3p ring neurons, induced increased daytime sleep but reduced nighttime sleep (Figs. 3E,F, 5; Table 4). Increased daytime sleep was associated with an increase of episode length, explained by elevated sleep pressure and “deeper” sleep depth (Figs. 3G,H, 5; Table 4). Opposite to the daytime change, reduced nighttime sleep was accompanied by fragmentation, resulting in increased sleep pressure and/or light sleep depth (Figs. 3G,H, 5; Table 4). How these three subtypes of ring neurons coordinate to segregate, and effect a sign change on, day and night sleep still needs to be determined but may provide insight into coordination of the EB circuit.
Regulation of sleep fragmentation by a specific ring neuron subset
One of the interesting findings of this screen was that there appeared to be circuits that regulate sleep structure independent of sleep amount. These data were consistent with our previous studies, which identified 5HT in EB as a modulator of sleep structure; activation of 5HT7-GAL4+ neurons fragmented sleep without changing the amount of sleep (C. Liu et al., 2019). 5HT7-GAL4+ neurons include R3d, R3p, and R4d subtypes (Hulse et al., 2021). To examine whether sleep structure regulation could be attributed to a specific subtype, we identified a driver R44D11-LexA that had an expression pattern similar to 5HT7-GAL4 (Fig. 7A). LexA+ neurons overlapped nearly 79% with 5HT7-GAL4+ neurons (Fig. 7B), but activation of R44D11-LexA+ neurons does not induce sleep/structure changes on activation (Fig. 7C; Table 9). To test the hypothesis that sleep fragmentation might be induced by the nonoverlapping population of 5HT7-GAL4+ neurons, we introduced LexAop-GAL80 to suppress the overlapping neurons between R44D11-LexA+ and 5HT7-GAL4+ neurons (Fig. 7D). We found that activation of the nonoverlapping 5HT7-GAL4+ neurons increased the number of episodes and reduced episode length (Fig. 7E; Table 9), suggesting that the nonoverlapping neurons play a critical role in sleep fragmentation. Interestingly, the nonoverlapping neurons morphologically are R3d subtypes (Fig. 7D). This subtype of ring neuron was present in 4 of 6 of the lines we identified in this screen as affecting structure only (R70B04, R53F11, R54B05, R53G11), and there were also R3d neurons in some lines that fragmented sleep in addition to changing its amount (Aphc507, R84H09). The fact that not all lines that contain this ring neuron subtype fragment sleep may be because of interactions with other ring neuron types or heterogeneity within the R3d population.
Discussion
Sleep is crucial for survival and overall health across animal kingdoms. Fly sleep exhibits the majority of the highly conserved features of vertebrate sleep, and the tractability of Drosophila as an experimental model has produced a growing number of studies, which contribute to our knowledge of sleep mechanisms and circuits. In addition to the importance in learning and memory of the mushroom body (MB), multiple subtypes of intrinsic MB Kenyon cells (KCs) have been identified as influencing sleep (Joiner et al., 2006; Sitaraman et al., 2015; Artiushin and Sehgal, 2017; Bringmann, 2018). For example, α′β′ and γm KCs contribute to wake promotion, and γd KCs contribute to sleep promotion (Sitaraman et al., 2015). A pair of GABAergic and serotonergic dorsal paired medial neurons, which are MB extrinsic projecting neurons and play a role in memory consolidation (Keene et al., 2004, 2006; Zhang et al., 2013), were shown to be involved in promoting sleep (Haynes et al., 2015). Dopaminergic PPL1 and PPM3 neurons that project to different layers of fan-shaped body (FB) have been shown to have specific roles in wake, via suppression of the FB, which is thought as a sleep-induction center (Q. Liu et al., 2012; Ueno et al., 2012; Pimentel et al., 2016). In addition to these central neurons, peripheral neurons, such as ppk+ neurons that project to the central brain, have been shown to have a role in the regulation of sleep homeostasis (Satterfield et al., 2022).
Many of these brain structures have been implicated in multiple behaviors. Like the MB and FB mentioned above, the EB has been shown to integrate sensory inputs to formulate locomotor output commands, but our understanding of its role in sleep is still limited. In the present study, we identified subtypes of ring neurons that regulate sleep/structure by the following: (1) screening a small collection of EB drivers using thermogenetic activation; and (2) specifying the roles of several single subtypes in different sleep components using two models and intersection strategies. We found that R3m/R3p neurons contribute to daytime sleep, R4m neurons to wakefulness, and R3d neurons fragment sleep structure (Fig. 7F).
The role of these neurons in sleep may be intimately involved with their other functions. Previous studies found that R2, R3, R4d, and R4m subtypes appear to be tuned to visual stimuli (Shiozaki and Kazama, 2017; Fisher et al., 2019; Kim et al., 2019; Hardcastle et al., 2021). This sensory input may be an important cue to change sleep/wake status, and is likely influenced by the circadian system. Previous studies showed that the R5 subtype is linked to the control of sleep homeostasis and stabilization of sleep structure (S. Liu et al., 2016; C. Liu et al., 2019), and our analysis supports these findings. A recent study released on bioRxiv identified two subtypes: sleep-promoting R3m neurons and wake-promoting R3d neurons (Aleman et al., 2021). Consistently, we also observed that R3m contributes both sleep amount and sleep structure. 5HT7-GAL4+ neurons play an important role in sleep maintenance, when they are activated, sleep became fragmented (C. Liu et al., 2019). According to a recent anatomic analysis (Hulse et al., 2021), 5HT7-GAL4+ neurons include R3d, R3p, and R4d subtypes, and we narrowed the fragmentation effect down to a specific subtype (R3d) in the present study. However, more efforts are still needed to understand how a certain subtype of ring neuron responds to sensory inputs and how neuronal activity patterns form in the network. Future work examining the neural activity of each subtype of ring neurons that control distinct sleep components and the interaction with other behaviors may reveal fundamental information about the rules of the coding and integration of the brain.
Footnotes
This work was supported by National Natural Science Foundation of China 32071009 to C.L.; Guangdong Basic and Applied Basic Research Foundation 2020A1515011055 to C.L.; CAS Key Laboratory of Brain Connectome and Manipulation (2019DP173024); Shenzhen Fundamental Research Program JCYJ20210324103014037 to H.L.; and National Institutes of Health Grant R01MH67284 to L.C.G.
The authors declare no competing financial interests.
References
- Aleman A, Omoto JJ, Singh P, Nguyen BC, Kandimalla P, Hartenstein V, Donlea JM (2021) Opposing subclasses of Drosophila ellipsoid body neurons promote and suppress sleep. bioRxiv 464469. 10.1101/2021.10.19.464469. [DOI] [Google Scholar]
- Alpert MH, Gil H, Para A, Gallio M (2022) A thermometer circuit for hot temperature adjusts Drosophila behavior to persistent heat. Curr Biol 32:4079–4087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Artiushin G, Sehgal A (2017) The Drosophila circuitry of sleep-wake regulation. Curr Opin Neurobiol 44:243–250. 10.1016/j.conb.2017.03.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bausenwein B, Muller NR, Heisenberg M (1994) Behavior-dependent activity labeling in the central complex of Drosophila during controlled visual stimulation. J Comp Neurol 340:255–268. 10.1002/cne.903400210 [DOI] [PubMed] [Google Scholar]
- Bringmann H (2018) Sleep-active neurons: conserved motors of sleep. Genetics 208:1279–1289. 10.1534/genetics.117.300521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donlea JM, Pimentel D, Miesenbock G (2014) Neuronal machinery of sleep homeostasis in Drosophila. Neuron 81:1442. 10.1016/j.neuron.2014.03.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher YE, Lu J, D'Alessandro I, Wilson RI (2019) Sensorimotor experience remaps visual input to a heading-direction network. Nature 576:121–125. 10.1038/s41586-019-1772-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franconville R, Beron C, Jayaraman V (2018) Building a functional connectome of the Drosophila central complex. Elife 7:e37017. 10.7554/eLife.37017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamada FN, Rosenzweig M, Kang K, Pulver SR, Ghezzi A, Jegla TJ, Garrity PA (2008) An internal thermal sensor controlling temperature preference in Drosophila. Nature 454:217–220. 10.1038/nature07001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanesch U, Fischbach KF, Heisenberg M (1989) Neuronal architecture of the central complex in Drosophila melanogaster. Cell Tissue Res 257:343–366. 10.1007/BF00261838 [DOI] [Google Scholar]
- Hardcastle BJ, Omoto JJ, Kandimalla P, Nguyen BM, Keles MF, Boyd NK, Hartenstein V, Frye MA (2021) A visual pathway for skylight polarization processing in Drosophila. Elife 10:e63225. 10.7554/eLife.63225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haynes PR, Christmann BL, Griffith LC (2015) A single pair of neurons links sleep to memory consolidation in Drosophila melanogaster. Elife 4:e03868. 10.7554/eLife.03868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herice C, Sakata S (2019) Pathway-dependent regulation of sleep dynamics in a network model of the sleep-wake cycle. Front Neurosci 13:1380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hulse BK, Haberkern H, Franconville R, Turner-Evans DB, Takemura SY, Wolff T, Noorman M, Dreher M, Dan C, Parekh R, Hermundstad AM, Rubin GM, Jayaraman V (2021) A connectome of the Drosophila central complex reveals network motifs suitable for flexible navigation and context-dependent action selection. Elife 10:e66039. 10.7554/eLife.66039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isaacman-Beck J, Paik KC, Wienecke CF, Yang HH, Fisher YE, Wang IE, Ishida IG, Maimon G, Wilson RI, Clandinin TR (2020) SPARC enables genetic manipulation of precise proportions of cells. Nat Neurosci 23:1168–1175. 10.1038/s41593-020-0668-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin X, Tian Y, Zhang ZC, Gu P, Liu C, Han J (2021) A subset of DN1p neurons integrates thermosensory inputs to promote wakefulness via CNMa signaling. Curr Biol 31:2075–2087.e6. 10.1016/j.cub.2021.02.048 [DOI] [PubMed] [Google Scholar]
- John B, Bellipady SS, Bhat SU (2016) Sleep promotion program for improving sleep behaviors in adolescents: a randomized controlled pilot study. Scientifica (Cairo) 2016:8013431. 10.1155/2016/8013431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joiner WJ, Crocker A, White BH, Sehgal A (2006) Sleep in Drosophila is regulated by adult mushroom bodies. Nature 441:757–760. 10.1038/nature04811 [DOI] [PubMed] [Google Scholar]
- Keene AC, Stratmann M, Keller A, Perrat PN, Vosshall LB, Waddell S (2004) Diverse odor-conditioned memories require uniquely timed dorsal paired medial neuron output. Neuron 44:521–533. 10.1016/j.neuron.2004.10.006 [DOI] [PubMed] [Google Scholar]
- Keene AC, Krashes MJ, Leung B, Bernard JA, Waddell S (2006) Drosophila dorsal paired medial neurons provide a general mechanism for memory consolidation. Curr Biol 16:1524–1530. 10.1016/j.cub.2006.06.022 [DOI] [PubMed] [Google Scholar]
- Kim SS, Hermundstad AM, Romani S, Abbott LF, Jayaraman V (2019) Generation of stable heading representations in diverse visual scenes. Nature 576:126–131. 10.1038/s41586-019-1767-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kottler B, Faville R, Bridi JC, Hirth F (2019) Inverse control of turning behavior by dopamine D1 receptor signaling in columnar and ring neurons of the central complex in Drosophila. Curr Biol 29:567–577.e566. 10.1016/j.cub.2019.01.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebestky T, Chang JS, Dankert H, Zelnik L, Kim YC, Han KA, Wolf FW, Perona P, Anderson DJ (2009) Two different forms of arousal in Drosophila are oppositely regulated by the dopamine D1 receptor ortholog DopR via distinct neural circuits. Neuron 64:522–536. 10.1016/j.neuron.2009.09.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lecompte D, Kaufman L, Rousseeuw P (1986) Hierarchical cluster analysis of emotional concerns and personality characteristics in a freshman population. Acta Psychiatr Belg 86:324–333. [PubMed] [Google Scholar]
- Lin CY, Chuang CC, Hua TE, Chen CC, Dickson BJ, Greenspan RJ, Chiang AS (2013) A comprehensive wiring diagram of the protocerebral bridge for visual information processing in the Drosophila brain. Cell Rep 3:1739–1753. 10.1016/j.celrep.2013.04.022 [DOI] [PubMed] [Google Scholar]
- Liu C, Meng Z, Wiggin TD, Yu J, Reed ML, Guo F, Zhang Y, Rosbash M, Griffith LC (2019) A serotonin-modulated circuit controls sleep architecture to regulate cognitive function independent of total sleep in Drosophila. Curr Biol 29:3635–3646.e3635. 10.1016/j.cub.2019.08.079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu D, Dan Y (2019) A motor theory of sleep-wake control: arousal-action circuit. Annu Rev Neurosci 42:27–46. 10.1146/annurev-neuro-080317-061813 [DOI] [PubMed] [Google Scholar]
- Liu Q, Liu S, Kodama L, Driscoll MR, Wu MN (2012) Two dopaminergic neurons signal to the dorsal fan-shaped body to promote wakefulness in Drosophila. Curr Biol 22:2114–2123. 10.1016/j.cub.2012.09.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S, Liu Q, Tabuchi M, Wu MN (2016) Sleep drive is encoded by neural plastic changes in a dedicated circuit. Cell 165:1347–1360. 10.1016/j.cell.2016.04.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ofstad TA, Zuker CS, Reiser MB (2011) Visual place learning in Drosophila melanogaster. Nature 474:204–207. 10.1038/nature10131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omoto JJ, Nguyen BM, Kandimalla P, Lovick JK, Donlea JM, Hartenstein V (2018) Neuronal constituents and putative interactions within the Drosophila ellipsoid body neuropil. Front Neural Circuits 12:103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parisky KM, Agosto Rivera JL, Donelson NC, Kotecha S, Griffith LC (2016) Reorganization of sleep by temperature in Drosophila requires light, the homeostat, and the circadian clock. Curr Biol 26:882–892. 10.1016/j.cub.2016.02.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pimentel D, Donlea JM, Talbot CB, Song SM, Thurston AJ, Miesenbock G (2016) Operation of a homeostatic sleep switch. Nature 536:333–337. 10.1038/nature19055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raccuglia D, Huang S, Ender A, Heim MM, Laber D, Suarez-Grimalt R, Liotta A, Sigrist SJ, Geiger JR, Owald D (2019) Network-specific synchronization of electrical slow-wave oscillations regulates sleep drive in Drosophila. Curr Biol 29:3611–3621.e3613. 10.1016/j.cub.2019.08.070 [DOI] [PubMed] [Google Scholar]
- Satterfield LK, De J, Wu M, Qiu T, Joiner WJ (2022) Inputs to the sleep homeostat originate outside the brain. J Neurosci 42:5695–5704. 10.1523/JNEUROSCI.2113-21.2022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scammell TE, Arrigoni E, Lipton JO (2017) Neural circuitry of wakefulness and sleep. Neuron 93:747–765. 10.1016/j.neuron.2017.01.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seelig JD, Jayaraman V (2015) Neural dynamics for landmark orientation and angular path integration. Nature 521:186–191. 10.1038/nature14446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seidner G, Robinson JE, Wu M, Worden K, Masek P, Roberts SW, Keene AC, Joiner WJ (2015) Identification of neurons with a privileged role in sleep homeostasis in Drosophila melanogaster. Curr Biol 25:2928–2938. 10.1016/j.cub.2015.10.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiozaki HM, Kazama H (2017) Parallel encoding of recent visual experience and self-motion during navigation in Drosophila. Nat Neurosci 20:1395–1403. 10.1038/nn.4628 [DOI] [PubMed] [Google Scholar]
- Siegmund T, Korge G (2001) Innervation of the ring gland of Drosophila melanogaster. J Comp Neurol 431:481–491. [DOI] [PubMed] [Google Scholar]
- Sitaraman D, Aso Y, Jin X, Chen N, Felix M, Rubin GM, Nitabach MN (2015) Propagation of homeostatic sleep signals by segregated synaptic microcircuits of the Drosophila mushroom body. Curr Biol 25:2915–2927. 10.1016/j.cub.2015.09.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueno T, Tomita J, Tanimoto H, Endo K, Ito K, Kume S, Kume K (2012) Identification of a dopamine pathway that regulates sleep and arousal in Drosophila. Nat Neurosci 15:1516–1523. 10.1038/nn.3238 [DOI] [PubMed] [Google Scholar]
- Wiggin TD, Goodwin PR, Donelson NC, Liu C, Trinh K, Sanyal S, Griffith LC (2020) Covert sleep-related biological processes are revealed by probabilistic analysis in Drosophila. Proc Natl Acad Sci USA 117:10024–10034. 10.1073/pnas.1917573117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young JM, Armstrong JD (2010) Building the central complex in Drosophila: the generation and development of distinct neural subsets. J Comp Neurol 518:1525–1541. 10.1002/cne.22285 [DOI] [PubMed] [Google Scholar]
- Zhang Z, Li X, Guo J, Li Y, Guo A (2013) Two clusters of GABAergic ellipsoid body neurons modulate olfactory labile memory in Drosophila. J Neurosci 33:5175–5181. 10.1523/JNEUROSCI.5365-12.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]