Abstract
The claustrum (CLA) is a cluster of neurons located between the insular cortex and striatum. Many studies have shown that the CLA plays an important role in higher brain function. Additionally, growing evidence suggests that CLA dysfunction is associated with neuropsychological symptoms. However, how the CLA is formed during development is not fully understood. In the present study, we analyzed the development of the CLA, especially focusing on the migration profiles of CLA neurons in mice of both sexes. First, we showed that CLA neurons were generated between embryonic day (E) 10.5 and E12.5, but mostly at E11.5. Next, we labeled CLA neurons born at E11.5 using the FlashTag technology and revealed that most neurons reached the brain surface by E13.5 but were distributed deep in the CLA 1 d later at E14.5. Time-lapse imaging of GFP-labeled cells revealed that some CLA neurons first migrated radially outward and then changed their direction inward after reaching the surface. Moreover, we demonstrated that Reelin signal is necessary for the appropriate distribution of CLA neurons. The switch from outward to “reversed” migration of developing CLA neurons is distinct from other migration modes, in which neurons typically migrate in a certain direction, which is simply outward or inward. Future elucidation of the characteristics and precise molecular mechanisms of CLA development may provide insights into the unique cognitive functions of the CLA.
SIGNIFICANCE STATEMENT The claustrum (CLA) plays an important role in higher brain function, and its dysfunction is associated with neuropsychological symptoms. Although psychiatric disorders are increasingly being understood as disorders of neurodevelopment, little is known about CLA development, including its neuronal migration profiles and underlying molecular mechanisms. Here, we investigated the migration profiles of CLA neurons during development and found that they migrated radially outward and then inward after reaching the surface. This switch in the migratory direction from outward to inward may be one of the brain's fundamental mechanisms of nuclear formation. Our findings enable us to investigate the relationship between CLA maldevelopment and dysfunction, which may facilitate understanding of the pathogenesis of some psychiatric disorders.
Keywords: claustrum, development, migration, neuron.
Introduction
The claustrum (CLA) is a thin gray matter structure located beneath the insular cortex and above the striatum. It is reciprocally connected to most cortical areas. The extensive connectivity of the CLA supports the notion that it plays an important role in higher cognitive functions, including multimodal sensory integration, neural correlates of consciousness, sleep regulation, and salience-guided attention (Crick and Koch, 2005; Goll et al., 2015; Benarroch, 2019; Narikiyo et al., 2020; Smith et al., 2020; Chevée et al., 2022). Recently, a growing body of evidence has suggested that CLA dysfunction contributes to neuropsychological symptoms (Sheerin et al., 2004; Goll et al., 2015; Patru and Reser, 2015).
Neuropsychiatric diseases are increasingly understood as disorders of brain development (Kamiya et al., 2005; Tomita et al., 2011; Ishii et al., 2015, 2016; Satterthwaite and Baker, 2015; Niwa et al., 2016; Kubo et al., 2017) and clarifying normal brain development is progressively needed. In particular, the investigation of CLA development may advance our understanding of the pathophysiology of psychiatric disorders. In the past few decades, several studies have investigated the CLA from developmental and evolutionary perspectives. A putative CLA has been described in mammals, birds, and reptiles, indicating the CLA to be an ancient structure in evolutionary terms (Bruguier et al., 2020). Furthermore, developmental origins have been well described by the concept of the pallium's division into four distinct fundamental sectors, each of which produces characteristic derivatives (Puelles et al., 1999, 2000, 2013). The latest pallial model proposed that the CLA originates within the lateral pallium sector together with the insular cortex (Puelles, 2014; Puelles et al., 2016; Bruguier et al., 2020). Moreover, previous studies, solely based on the expression pattern of a CLA marker, have hypothesized that the CLA is transiently formed at the brain surface, and that the insular cortex neurons later migrate radially through the CLA anlage to reach the superficial region, while passively displacing the CLA into the final deeper position (Puelles, 2014; Puelles et al., 2016, 2017; Watson and Puelles, 2017). In addition, CLA development is thought to be closely related to the development of the dorsal endopiriform nucleus (DEn), which lies ventrally to the CLA. One genoarchitectonic observation suggested that the DEn neurons originate jointly with the CLA within the lateral pallium sector, and soon tangentially invade the ventral pallium to take up a final position deep to the piriform cortex (Puelles, 2014; Bruguier et al., 2020).
However, the development of the CLA is not yet fully understood. First, there are different hypotheses regarding the birthdate of CLA neurons. Previous studies have shown that rat CLA neurons are born mainly at E12.5 (Puelles, 2014). In contrast, Fang (2021) and Bayer et al. (1991) reported that the CLA in rats originates at later stages: E14.5-E15.5 and E15-E16, respectively. Second, the migration mode of CLA neurons is unknown. While the migration modes in the cerebral neocortex and the hippocampus during development have been well described, such as “locomotion,” “somal translocation,” “terminal translocation,” “multipolar migration,” and “climbing mode” (Rakic, 1972; Nadarajah et al., 2001; Tabata and Nakajima, 2003; Kitazawa et al., 2014), only a very few migration or lineage studies for CLA (Bruguier et al., 2020) have been conducted; direct lineage tracing methodologies to investigate how CLA neurons migrate to form the nuclear structure are yet to be extensively analyzed. Furthermore, the molecular mechanism underlying CLA development remains unknown.
Here, we determined the birthdates of mouse CLA neurons using BrdU labeling. In addition, we analyzed the migration profiles of CLA neurons by injecting FlashTag or a GFP expression plasmid in combination with time-lapse imaging. We also examined whether Reelin is involved in CLA development.
Materials and Methods
Animals
Pregnant ICR mice were obtained from Japan SLC. To maintain colonies of reeler mice (B6CFe a/a-Relnrl/J) purchased from The Jackson Laboratory, heterozygous females were allowed to mate with homozygous males. The method used to maintain the colonies of Apoer2-KO mice and Vldlr-KO mice has been previously described (Frykman et al., 1995; Hirota et al., 2015). The day females were sperm-positive was designated as E0. Embryos of both sexes were used. All animal experiments were performed using protocols approved by the Keio University Institutional Animal Care and Use Committee in accordance with the Institutional Guidelines on Animal Experimentation at Keio University, the Japanese Government Law Concerning the Protection and Control of Animals, and the Japanese Government Notification of Feeding and Safekeeping of Animals.
In vivo BrdU labeling
BrdU (Sigma) was dissolved in PBS at a concentration of 10 mg/ml. A bolus intraperitoneal injection of BrdU solution was administered at a dose of 50 µg/g body weight.
FlashTag technology
Pregnant mice were deeply anesthetized with isoflurane (1.5 L/min), and their intrauterine embryos were manipulated as described previously (Nakajima et al., 1997). FlashTag was performed as previously described (Telley et al., 2016; Yoshinaga et al., 2021). Briefly, 10 mm 5- or 6-(N-succinimidyloxycarbonyl) fluorescein 3′,6′-diacetate (Cellstain CFSE, C309, Dojindo Molecular Technologies) working stock was prepared by dissolving CFSE in DMSO (Hybri-Max, Sigma-Aldrich). The working solution was further diluted with 1× HEPES-buffered saline to make a 1 mm solution just before surgery. The solution was colored with Fast Green (final concentration 0.01%–0.05%) to monitor the successful injection. Approximately 0.5 µl of the prepared FlashTag solution was injected into the lateral ventricle.
In utero electroporation
Pregnant ICR mice were deeply anesthetized with isoflurane (1.5 L/min), and their intrauterine embryos were surgically manipulated as described previously (Tabata and Nakajima, 2001; Kubo et al., 2010). A modified chicken β-actin promoter with cytomegalovirus-immediate early enhancer (CAG) promoter (provided by Dr. Junichi Miyazaki, Osaka University, Osaka, Japan) (Niwa et al., 1991)-driven GFP expression vector (pEGFP-CAGGS1, a GFP-plasmid) at a concentration of 1.0 mg/ml or a CAG promoter-driven tandem-Tomato fluorescent protein (tdTomato) expression vector (pCAG-tdTomato) (Kanatani et al., 2015) at a concentration of 0.5 mg/ml was injected into a lateral ventricle of the brain of each embryo, and in utero electroporation was performed using a 1-mm-diameter electrode. The angle of inclination of the electrode paddles with respect to the horizontal plane of the brain was zero.
Brain slice preparation
Coronal slices of developing brains were prepared as described previously (Tabata and Nakajima, 2003). In brief, the brains were fixed with 4% PFA and cut into 20-30 µm sections with a cryostat.
Immunohistochemistry
Immunohistochemistry was performed as previously described (Inoue et al., 2019). The primary antibodies used were as follows: anti-nuclear receptor related-1 protein (Nurr1) (goat anti-Nurr1, 1:500, R&D Systems), anti-BrdU (mouse anti-BrdU, 1:50, BD Biosciences), anti-Fluorescein (rabbit anti- Fluorescein, 1:500, Invitrogen), anti-GFP (rabbit anti-GFP, 1:1000, MBL), anti-RFP (rabbit anti-RFP, 1:200, Rockland), anti-Nestin (mouse anti-Nestin, 1:250, BD Pharmingen), anti-Reelin (mouse anti-Reelin, 1:500, Abcam), anti-Ctip2 (rat anti-Ctip2, 1:500, Abcam), and anti-Apolipoprotein E receptor 2 (ApoER2, rabbit anti-ApoER2, 1:500, Abcam). In some sections, nuclei were labeled with DAPI (1:2000, Invitrogen). Green fluorescein images were captured without immunohistochemistry only in Figures 3A–D′, 4C, D and Movies 1, 2, 3. Images were acquired using a confocal laser scanning microscope (FV1000; Olympus).
Figure 3.
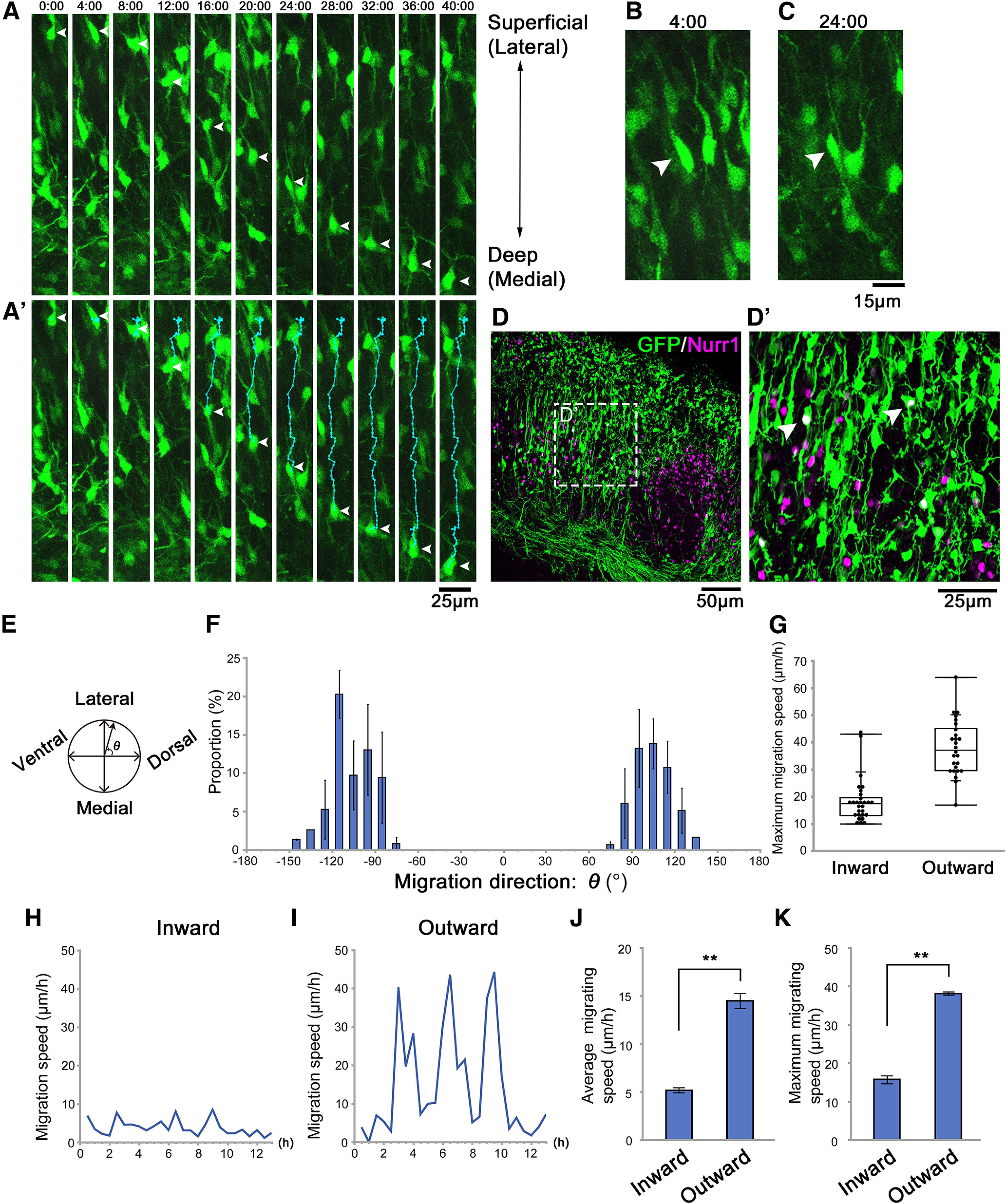
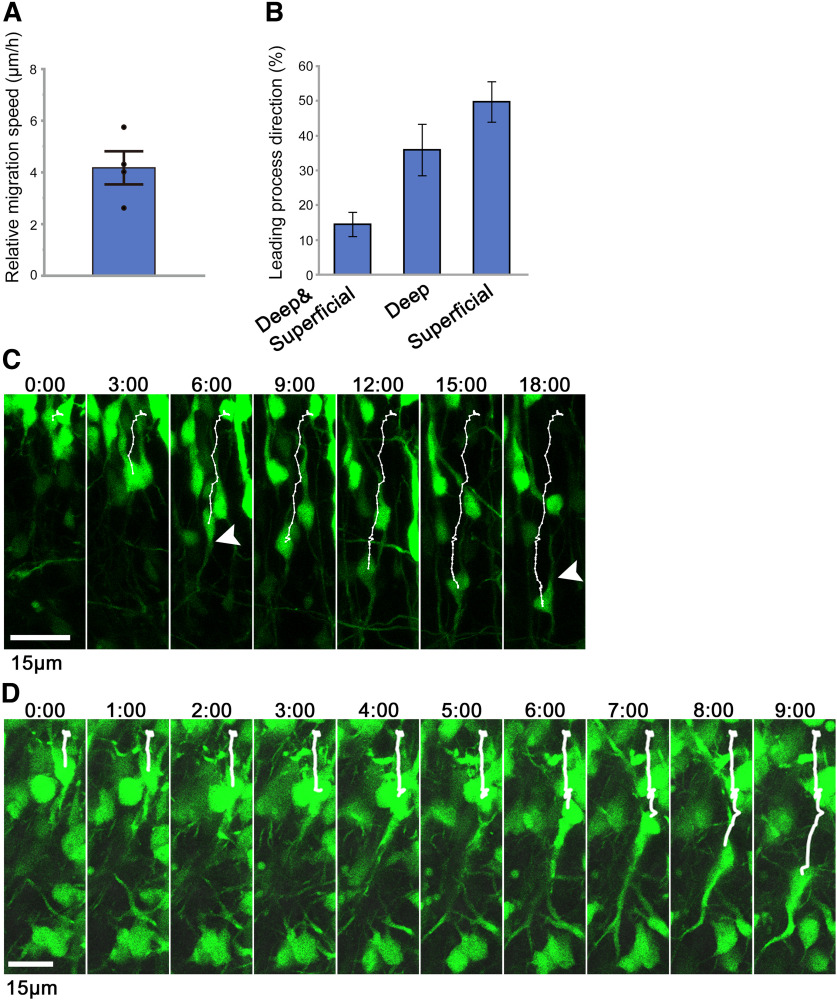
Time-lapse imaging of migrating CLA neurons. A, A′, Time-lapse images were captured from the CLA slices prepared 3 d after electroporation of the GFP-plasmid at E11.5. The time displayed above each panel indicates the elapsed time since the beginning of the observation. The two-way arrow on the right of the panel indicates that the top is the lateral (superficial) side, and the bottom is the medial (deep) side. The orientation is consistent with B–E. The trajectories of representative cells were shown in cyan (A′), and the cell bodies were indicated by white arrowheads (A, A′). Some cells migrated from the superficial region toward the deep region. B, C, Higher magnifications of the cell shown in A′. The representative cell turned its leading process toward the superficial region at first (B, 4:00, arrowhead) and then turned it into the deep direction (C, 24:00, arrowhead). D, Immunohistochemistry for Nurr1 indicated that some GFP-labeled cells were Nurr1-positive. D′, High magnification of the labeled-boxed area in D. E, The angle of migration was defined as θ. The orientation is consistent with A–D′. F, The distribution of migration direction was bimodal: ∼100° (toward the superficial region) and –100° (toward the deep region). G, The maximum migration speeds of cells migrating inward (n = 28) and outward (n = 26) were measured. Each data point represents a different cell in a representative brain. H, I, The trajectories of migrating cells moving inward and outward were traced, and the speeds of the cells in each period were measured. The speed of representative cells migrating inward (H) and outward (I). J, K, Mean of the average migration speeds (J) and mean of the maximum migration speeds (K) of the cells migrating inward and outward. **p < 0.01 (Student's t test). F, J, K, Values are the mean ± SE of four different brains (n = 54, 61, 70, and 77). Scale bars: A-A′, 25 µm; B, C, 15 µm; D, 50 µm; D′, 25 µm.
Figure 4.
Detailed analysis of time-lapse images of migrating CLA neurons. A, The radial speed of representative actively inward-migrating cells relative to the LCS was measured. Error bar indicates mean ± SE (n = 6, 6, 8, and 11 from four different slices). B, The ratios of the number of cells which extended their leading process toward superficial, deep, or both directions to the total number of analyzed GFP-positive cells were measured. Values shown are the mean ± SE (n = 16, 18, 19, and 31, from four different slices). C, D, Time-lapse images were captured from the CLA slices prepared 3 d after electroporation of the GFP-plasmid at E11.5. The time displayed above each panel indicates the elapsed time since the beginning of the observation. Scale bars, 15 µm. Top, Superficial (lateral) side. Bottom, Deep (medial) side.
Time-lapse imaging
Time-lapse imaging was performed, as previously described (Tabata and Nakajima, 2003). Briefly, coronal brain slices (200 µm thick) were placed on a Millicell-CM membrane (pore size, 0.4 µm; Millipore), mounted in low melting temperature agarose, and cultured in Neurobasal medium containing B27 (Invitrogen). The dishes were then mounted in a 40% O2 incubator chamber fitted onto a confocal laser scanning microscope (SP8; Leica). Approximately 10 optical Z-section images were acquired at the indicated interval, and all focal planes (≥ 200 µm thick) were merged.
Analysis of time-lapse imaging of the migrating CLA neurons
The movement of each cell was analyzed using MtrackJ, an ImageJ plugin (Meijering et al., 2012). The leading process directions were classified into three categories: superficial, deep, or both. If cells directed their processes toward both superficial and deep regions at the same time or turned their process toward the superficial first and the deep region later, the cell was judged to direct their leading process toward superficial and deep regions.
Immunoelectron microscopy
Immunoelectron microscopy was performed as described previously (Shin et al., 2019). ICR mice were perfused with a fixative solution (4% PFA, 1% glutaraldehyde, and 0.1% picric acid in 0.1 m PB). The brains were then removed and postfixed for 3-4 h at 4°C in the same fixative solution. After washing with 0.1 m PB, 100-µm-thick coronal brain sections were prepared using a vibratome. The brain sections were incubated with blocking solution (5% normal goat serum, 0.005% saponin in 0.1 m PB) for 30 min at room temperature (RT) and then with an anti-rabbit GFP antibody (FRL-GFP-Rb-AF2020; Frontier Institute) overnight at RT. After washing with 0.005% saponin in 0.1 m PB, the sections were incubated with a Nanogold Fab′ fragment of goat anti-rabbit IgG (N-24916; Fisher Scientific) in blocking solution for 90 min at RT. After washing with 0.1 m PB, the sections were incubated with 0.5% glutaraldehyde in 0.1 m PB for 10 min at RT. After washing with 0.1 m PB, the immunoreactive nanogold particles on the brain sections were enhanced using an HQ silver enhancement kit (2012, Nanoprobes) and washed with distilled water under dark conditions. The sections were then postfixed with 1% osmium tetroxide for 15 min at RT. After dehydration with a series of ethanol solutions and replacement with QY-1, the sections were embedded in an epoxy resin. Ultrathin sections were stained with 2% uranyl acetate for 14 minutes, followed by lead citrate for 12 min at RT. The electron micrographs were obtained using a JEM-1400 microscope (JEOL).
Statistical analysis of cell distribution
To quantitatively analyze the cell distribution of cells in vivo (see Fig. 7), the nuclei of the Nurr1-positive cells were visualized by staining with DAPI. The relative distance of each cell from the pial surface was determined by measuring the distance (X) of the nucleus from the pial surface using the Fiji software (RRID:SCR_002285) (Schindelin et al., 2012) and dividing it (X) by the distance (Y) between the pial surface and the medial border of the cortical migratory stream. The cell distribution was evaluated by dividing the areas into 10 bins and counting the cells in each bin. The most superficial bin was designed as Bin 1, and the deepest bin was Bin 10. The relative distance of each cell from the pial surface (X/Y) was used to assign the cells to each of the 10 bins. For example, cells whose relative distance was > 0.9, but ≤ 1.0 were assigned to Bin 10 and those with distances of 0-0.1 were assigned to Bin 1. The ratio of cells in each bin was calculated by dividing the number of cells in a bin by the total number of cells in all bins.
Figure 7.
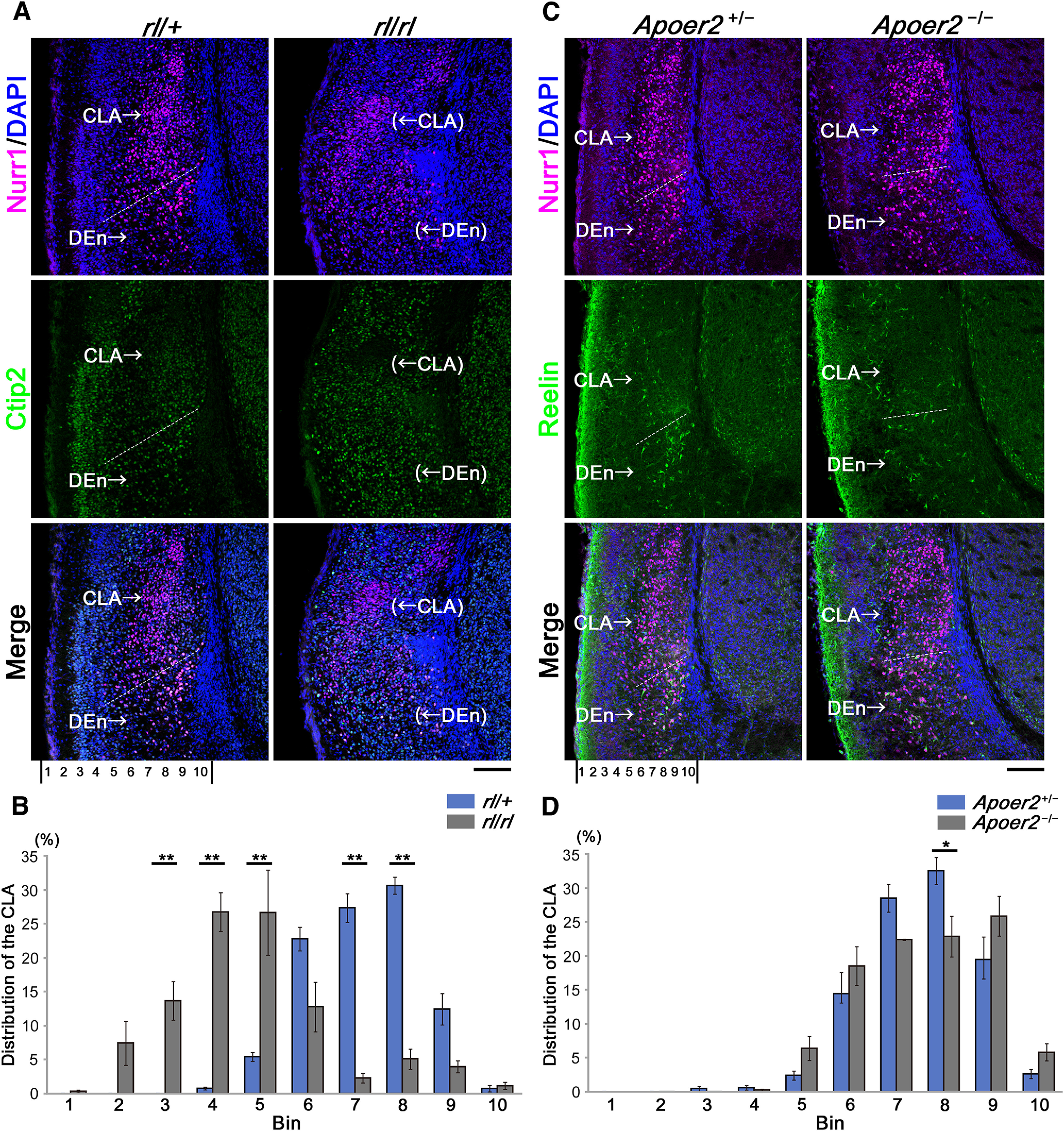
Reelin mutation or KO of Apoer2 resulted in malposition of CLA neurons. A, Immunostaining for Nurr1 (magenta) and Ctip2 (green) was performed in reeler (Reelin-deficient) mice and heterozygous reeler mice at E18.5. C, Immunostaining for Nurr1 (magenta) and Reelin (green) was performed in Apoer2 homozygous and heterozygous KO mice. B, D, A bin analysis was performed to evaluate cell distribution by dividing the space between the pial surface and the medial border of LCS into 10 equal areas (10 bins), and the number of Nurr1-positive cells in each bin was calculated as a percentage of the total number in all 10 bins (B and D correspond to A and C, respectively). Mean ± SE of data obtained from four different brains in rl/+, rl/rl, and Apoer2+/−, and three different brains in Apoer2−/− are shown. The ratio of each corresponding bin was statistically compared between homozygous and heterozygous mutant/KO mice. *p < 0.05; **p < 0.01; Tukey–Kramer test. Scale bars: A, C, 100 µm. Top, Dorsal side. Bottom, Ventral side. Left, Superficial (lateral) side. Right, Deep (medial) side.
Quantification and statistical analysis
Statistical parameters, including the number of cells (n) and brains (N), are reported in Figures 1–5, and 7 and the figure legends. Data are presented as mean ± SEM. All statistical analyses were performed using JMP statistical software version 16.0.0 (SAS Institute). For comparison of two groups of data, a two-tailed, unpaired Student's t test was used (see Fig. 3J,K). Some data were analyzed by one-way ANOVA, followed by Turkey post hoc comparisons (see Fig. 2F,7B,7D). Differences were considered statistically significant at p < 0.05.
Figure 1.
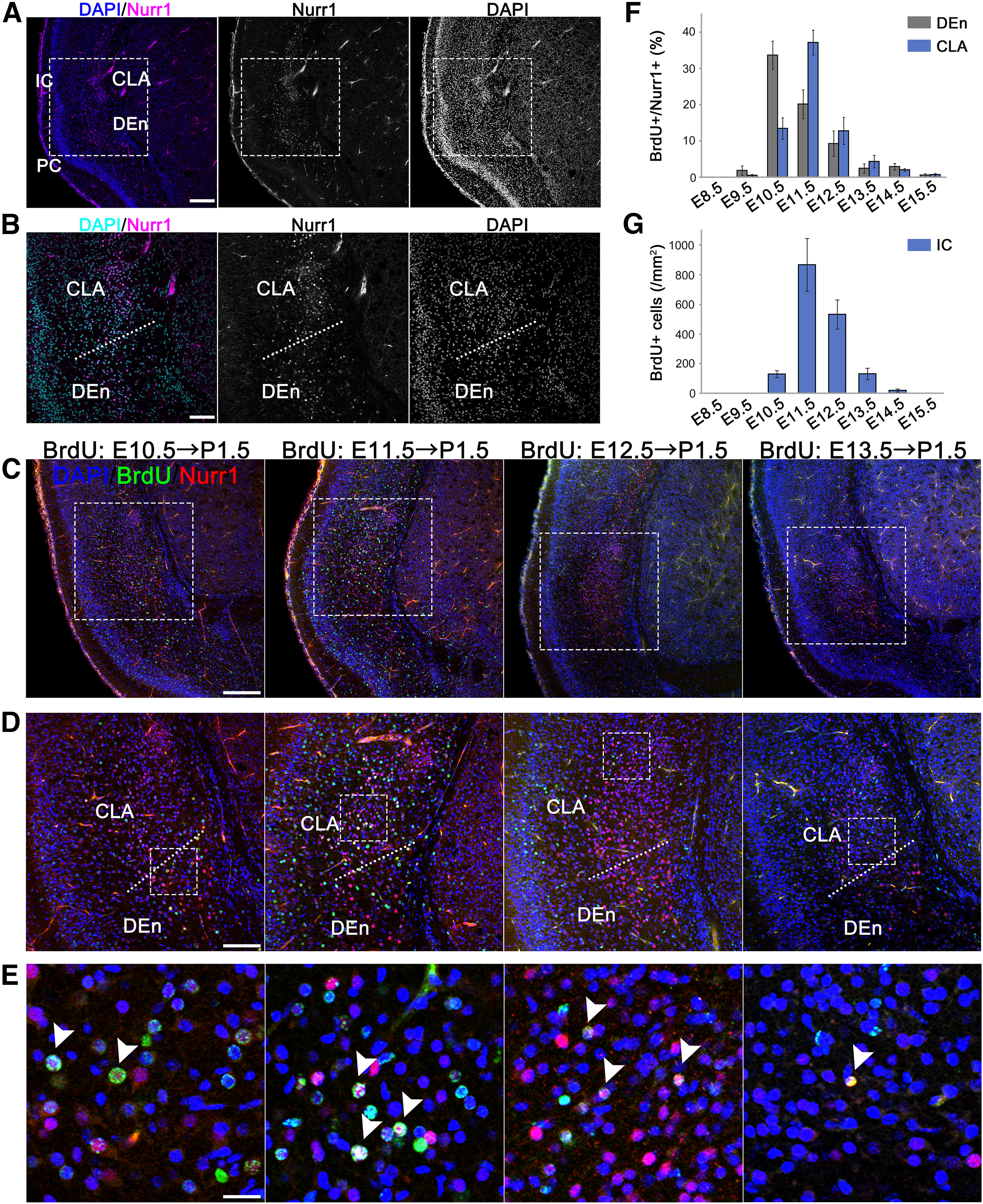
Mapping of the CLA and DEn cell birthdate analyzed by BrdU incorporation. BrdU was injected into pregnant mice at E8.5, 9.5, 10.5, 11.5, 12.5, 13.5, 14.5, or 15.5. The nuclei were labeled with DAPI (blue in A, C–E; cyan in B). A, A coronal section at the medial level. In order to identify the CLA and DEn, we performed immunohistochemistry for Nurr1 (magenta), a molecular marker of the CLA and DEn. IC, Insular cortex; PC, piriform cortex. B, Higher magnification of A. C, Representative images of the CLA and DEn slices showing BrdU-labeled cells (green) generated between E10.5 and E13.5 and analyzed at P1.5. The CLA and DEn were labeled with Nurr1 (red). D, E, Higher magnification of the labeled-boxed areas in C and D, respectively. BrdU-labeled cells of the CLA or DEn were indicated (white arrowheads). F, Ratios of BrdU-labeled Nurr1-positive cells among total Nurr1-positive cells in the CLA and DEn at P1.5. CLA was considered to be Nurr1-positive cells above the CLA/DEn dividing line and included both the ventral and dorsal CLA. Total numbers of cells counted: 9265. Data are mean ± SE of the data obtained in seven different brains from three litters (E8.5, E9.5, E13.5, E14.5, and E15.5) or nine brains derived from four litters (E10.5, E11.5, and E12.5). G, Cell density of BrdU-positive cells in the insular cortex at P1.5. Total number of cells counted: 870. Data are mean ± SE of the data obtained from the seven different brains derived from three litters. Scale bars: A, 250 µm; B, 125 µm; C, 200 µm; D, 100 µm; E, 20 µm. Top, Dorsal side. Bottom, Ventral side. Left, Superficial (lateral) side. Right, Deep (medial) side.
Figure 5.
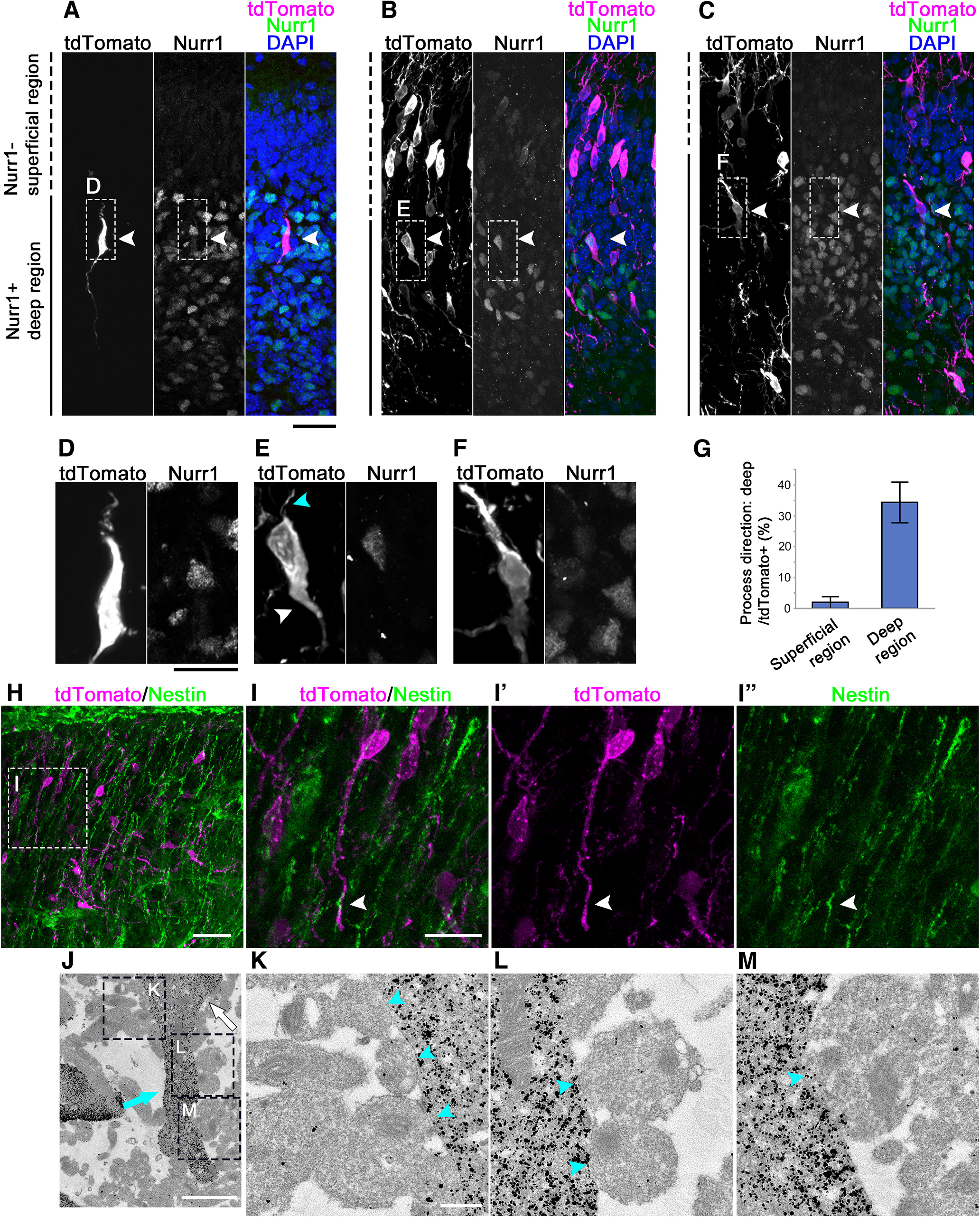
Sparse-cell labeling of migrating neurons. tdTomato plasmid was transfected into the embryos at E11.5, and the brains were fixed 3.5 d later. A–C, Immunostaining for RFP (magenta) and Nurr1 (green). Representative cells with the major process directed toward the superficial region (A), the deep region (B), or both directions (C) are shown (white arrowheads). The nuclei were labeled with DAPI (blue). Top, Superficial side. Right, Dorsal side. D–F, Higher magnifications of the labeled-boxed areas in A–C are shown, respectively. Some tdTomato-labeled cells directed their leading process toward the deep region (a white arrowhead), and they also oriented a thin process toward the superficial region (a cyan arrowhead). G, The mean ± SE of cells extending their process toward the deep region as a proportion of the tdTomato-labeled cells in the Nurr1-negative superficial area and the Nurr1-positive deep area in four different brains (n = 19, 24, 44, and 45) were calculated. H, Immunostaining for RFP (magenta) and Nestin (green). Top, Superficial side. Right, Ventral side. I–I′′, Higher magnifications of the boxed area in H are shown. J, Immunoelectron microscopic analysis of an inward-migrating neuron. The embryos at E11.5 were transfected with GFP-expressing plasmid, fixed 3.5 d later, and observed through an immunoelectron microscope. Migrating neurons were immunolabeled with an anti-GFP antibody (visualized with nanogold, black spots). A representative immunostained cell that turned its major process toward the deep region (cyan arrow) was shown. Its cell body was indicated by a white arrow. K–M, Higher magnifications of the labeled-boxed areas in J. The cell had contact with multiple fibers (cyan arrowheads). Top, Superficial side. Bottom, Deep side. Scale bars: A–C, 50 µm; D–F, 25 µm; H, 100 µm; I–I′′, 50 µm; J, 2 µm; K–M, 500 nm.
Figure 2.
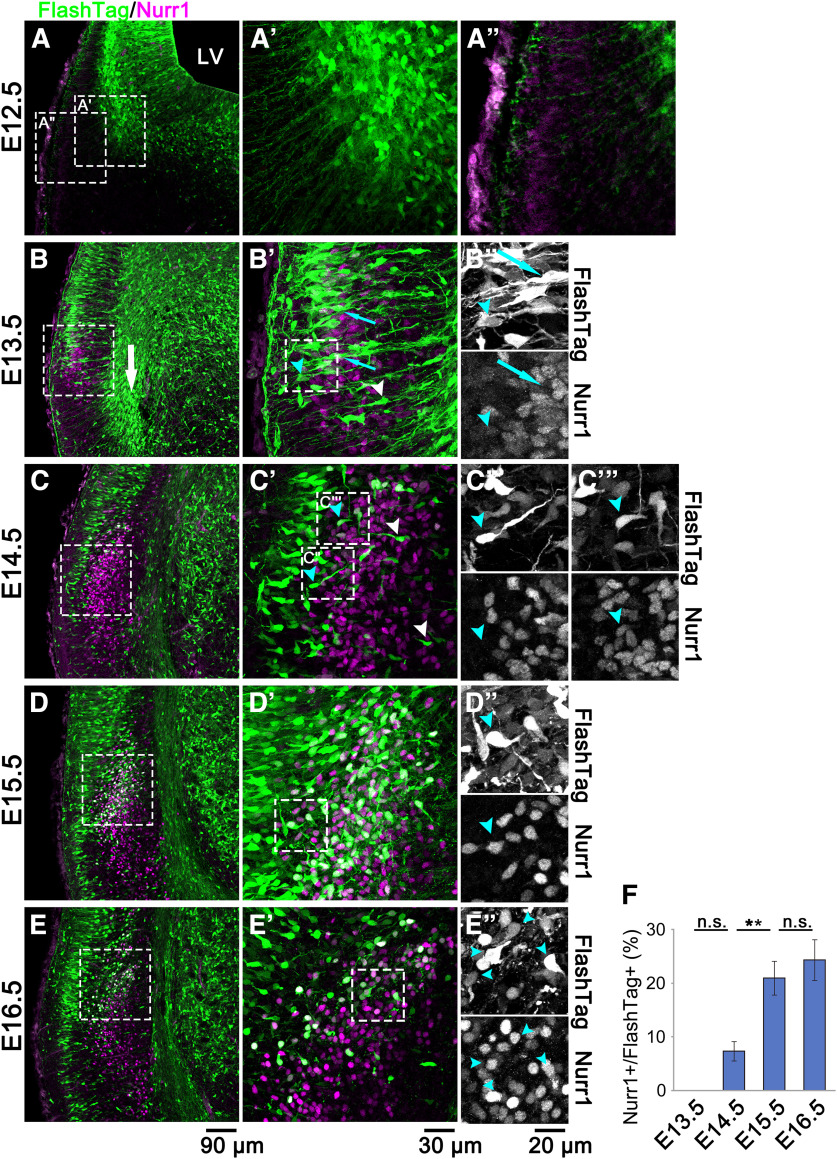
Migration profile of the CLA neurons at each stage of development. A–E, FlashTag was injected at E11.5, and the brains were fixed at E12.5 (A), E13.5 (B), E14.5 (C), E15.5 (D), and E16.5 (E). B, White arrow indicates cortical migratory stream. A′–E′, A′′, Higher magnifications of the boxed areas in A–E. B–E′′, C′′′, Higher magnifications of the labeled-boxed areas in B′–E′. FlashTag-labeled Nurr1-positive cells were rarely observed at E13.5, but they became visible clearly at E15.5 (cyan arrowheads in D′′). F, The percentages of Nurr1-positive cells among FlashTag-positive cells in the presumptive CLA and insular cortex were measured. Values shown are the mean ± SE (from four different slices at E13.5-E15.5 or three different slices at E16.5). Total number of cells counted: 1169. **p < 0.01 (Tukey–Kramer test). Scale bars: A–D, 90 µm; A′-D′, 30 µm; A′′-D′′, C′′′, 20 µm. Left, Superficial (lateral) side. Right, Deep (medial) side.
Results
Analysis of birthdates of the CLA neurons
The CLA lies dorsally to the endopiriform nucleus (En) and beneath the insular cortex, whereas the En is deep to the piriform cortex (Smith et al., 2019). It is known that both the CLA and DEn express Nurr1 (also known as Nuclear Receptor Subfamily 4 Group A Member 2 or Nr4a2), which is a nuclear orphan receptor (Puelles, 2014; Puelles et al., 2016; Bruguier et al., 2020). To identify the CLA and the DEn, we performed immunohistochemistry for Nurr1 on coronal sections at the level of the medial telencephalon (Fig. 1A,B). Two populations of Nurr1-positive cells were distinguished from each other by differences in their cell densities. The population located dorsally and deep to the insular cortex had a higher cell density, while the population positioned ventral and deep to the piriform cortex had a lower cell density. Based on the previous findings (Smith et al., 2019), we concluded that the dorsal and dense population of Nurr1-positive cells was the CLA and that the ventral and sparse population was the DEn; thus, the former (the CLA) would include both the ventral and dorsal CLA as explicated by a previous study (Fang et al., 2021).
Next, to determine the birthdates of the CLA and DEn neurons, we intraperitoneally injected BrdU, which would label newly born neurons (Kubo et al., 2010), into pregnant mice at E8.5, E9.5, E10.5, E11.5, E12.5, E13.5, E14.5, or E15.5. On postnatal day (P) 1.5, the majority of the Nurr1-positive cells in the CLA were positive for BrdU when BrdU was injected at E11.5, suggesting that CLA neurons were generated mainly at E11.5 (Fig. 1C–E). When the ratio of BrdU-positive cells among the Nurr1-positive cells in the CLA and DEn was calculated, a peak was at E11.5 and E10.5, respectively (Fig. 1F), indicating that most CLA neurons were generated during E10.5-E12.5, especially around E11.5. In addition, Nurr1-positive cells had a gradient along the ventrodorsal axis of the CLA and DEn, which is, earlier-born and later-born Nurr1-positive cells tended to be distributed ventrally and dorsally, respectively (Fig. 1C–E). Thus, neurons in the DEn were born earlier, mainly at E10.5, whereas neurons in the CLA were born later, mainly at E11.5. Moreover, BrdU-positive cells in the insular cortex were particularly abundant in the embryos injected with BrdU at E11.5 and E12.5, indicating that cells in the insular cortex were mainly born between E11.5 and E12.5 (Fig. 1G).
Migration profiles of the CLA neurons were elucidated by FlashTag labeling
Next, to examine the process of CLA formation, we used FlashTag labeling, which is a fluorescence birthdating technique to label and track isochronic cohorts of newborn cells in the embryonic CNS in vivo (Govindan et al., 2018; Yoshinaga et al., 2021). To perform FlashTag labeling, we injected carboxyfluorescein esters (CFSEs) into the lateral ventricle at E11.5, fixed the brains at E12.5, E13.5, E14.5, E15.5, or E16.5, and analyzed them at the level of the foramen of Monro.
At E12.5, 1 d after the injection of FlashTag, most FlashTag-positive cells had already left the ventricular surface and were distributed in the middle of the brain surface and ventricle. FlashTag-positive cells showed multipolar morphologies with multiple and complex processes (Fig. 2A,A′). At this stage, weakly Nurr1-positive cells were aligned near the brain surface to form a cellular band of ∼30 µm thickness (Fig. 2A,A′′). Radially oriented thin processes, which presumably extended from the FlashTag-labeled radial glial cells in the ventricular zone, ran between the weakly Nurr1-positive cells toward the brain surface. These radially oriented thin processes are thought to function as scaffolds for the migration of FlashTag-positive cells at later stages.
At E13.5, 2 d after the injection of FlashTag, a small cluster of Nurr1-positive cells was formed near the brain surface (Fig. 2B,B′). At this stage, the insular cortex was not formed because the majority of cells in the insular cortex were negative for BrdU when it was injected at E10.5 and earlier (Fig. 1C–E). Nurr1-positive cells near the brain surface were rarely labeled with FlashTag (Fig. 2B′′) and were considered CLA or DEn neurons born earlier than E11.5. In contrast, we observed many FlashTag-positive cells in the cortical migratory stream (Fig. 2B, white arrow), which is consistent with the previous finding that CLA neurons first migrate in the cortical migratory stream (Puelles, 2014). Several FlashTag-labeled cells left the stream and oriented their leading processes radially outward (Fig. 2B′, white arrowhead). Some FlashTag-positive cells arrived at the outermost region and were located superficially to the cluster of Nurr1-positive cells (Fig. 2B′, cyan arrowhead), whereas other FlashTag-positive cells were aligned between the Nurr1-positive cells (Fig. 2B′,B′′, cyan arrows). The latter cells are thought to migrate radially outward.
At E14.5, 3 d after injection of FlashTag, the cluster of Nurr1-positive cells became larger. Most FlashTag-labeled cells were located superficially to the Nurr1-positive cells, indicating that they had already arrived at the surface, whereas a few FlashTag-labeled cells with a major process directed outward were observed between the Nurr1-positive cells (Fig. 2C′, white arrowheads). Interestingly, we also found some FlashTag-positive cells that oriented their processes inward and were negative (Fig. 2C′′, cyan arrowhead) or positive for Nurr1 (Fig. 2C′′′, cyan arrowhead), suggesting the possibility that they migrated inward.
At E15.5, 4 d after injection of FlashTag, a substantial number of neurons labeled with FlashTag at E11.5 reached near the brain surface, settling in the insular cortex that lay superficially to the CLA. In addition, a large number of FlashTag-labeled cells were located deep in the presumptive CLA region and were positive for Nurr1. Some of them directed their process towards the deep regions (Fig. 2D–D′′, cyan arrowheads). These results suggest that it took 4 d for CLA neurons labeled with FlashTag at E11.5 to reach their final destination (the CLA region) and become positive for Nurr1. At E16.5, 5 d after injection of FlashTag, FlashTag-positive cells in the CLA region developed variously directed processes, suggesting that most CLA neurons had completed their migration by this stage (Fig. 2E-E′′, cyan arrowheads). The percentage of Nurr1-positive cells among FlashTag-positive cells in the presumptive CLA and insular cortex significantly increased from E14.5 to E15.5 but stabilized at E15.5 and E16.5, suggesting that CLA neurons expressed Nurr1 at the end of migration (Fig. 2F; N = 4 at E13.5-E15.5, and N = 3 at E16.5; E13.5 vs E14.5, p = 0.184; E14.5 vs E15.5, p = 0.008; E15.5 vs E16.5, p = 0.786; Tukey's test).
Together, these findings raise the possibility that at least some CLA neurons move superficially past the presumptive CLA region, turn near the brain surface, and then move in the reverse direction, inwardly, toward the CLA region. Another possibility is that, although less likely, some Nurr1-positive future CLA neurons might halt their outward migration when they reach the presumptive CLA region and turn their process toward the deep regions.
Time-lapse imaging revealed that some CLA neurons migrate inward before they settle
To distinguish between these two possibilities and elucidate how neurons migrate toward the presumptive CLA region, we introduced a GFP-expressing plasmid at E11.5 with in utero electroporation at the approximate locus of the ventral and lateral pallium, to successfully label CLA and insular neurons, as was done in previous studies (Rueda-Alaña et al., 2018; Bruguier et al., 2020). Thereon, we prepared coronal slices containing the presumptive CLA region 3 d after electroporation and acquired time-lapse images every 30 min for 48 h (Fig. 3A, only data every 4 h are shown; Movie 1).
Time-lapse imaging of migrating cells labeled with GFP by in utero electroporation. Images were captured every 30 min. The video clip corresponds to Figure 3A,A′. Top, Superficial (lateral) side. Bottom, Deep (medial) side.
After the GFP-labeled cells migrated in the cortical migratory stream, some cells changed their direction radially outward. They moved toward the brain surface (presumptive insular cortex) radially passing through the presumptive CLA region. A portion of the neurons that had reached the brain surface migrated inward toward the deep region (Fig. 3A, arrowheads; Fig. 3A′, trajectory shown by cyan lines; Movie 1). In addition, some outward-migrating neurons initially directed their leading process toward the brain surface (Fig. 3A, 4:00; Fig. 3B); then they changed the direction of their processes toward the deep region and migrated inward (Fig. 3A, 16:00 and 24:00; Fig. 3C; the arrowheads of Fig. 3B and 3C indicate the same neuron). Other examples of cells that showed a reversal of the direction of their leading processes and migrated inward are shown in Movies 1, 2, 3 (cells tracked with lines).
To confirm that the inward-migrating neurons that we observed were CLA neurons, we fixed the slices after time-lapse imaging and performed immunohistochemistry for Nurr1. Cells with a deeply oriented process were considered inward-migrating neurons, as shown by time-lapse imaging. The results revealed that some GFP-positive cells with a major process directed inward expressed Nurr1, indicating that some neurons migrating inward were CLA neurons (Fig. 3D,D′, white arrowheads). These results demonstrate that at least some CLA neurons initially migrate outward and then move inward, namely, “reversely.”
We then quantitatively analyzed cell movement by tracing the trajectories of GFP-positive cells and measuring their speed and angle of migration during each observation period. The angle of migration (Fig. 3E) had a roughly symmetrical bimodal distribution, ∼100° (toward the lateral/superficial regions) and –100° (toward the medial/deep regions), indicating that some neurons migrated outward toward the brain surface, whereas others migrated in the opposite direction toward deep regions. The former was thought to be a migration toward the insular cortex and the latter toward the presumptive CLA region (Fig. 3F). Analyses of inward and outward migration speeds showed that both migrations alternated fast and slow movements. However, the peak of fast movement of neurons migrating outward was higher than that of neurons migrating inward (Fig. 3H,I). In addition, the overall migration speed of the two populations was different from each other: those migrating inward (5.2 ± 0.6 µm/h, N = 4) were significantly slower than those migrating outward (14.5 ± 0.6 µm/h, N = 4, p < 0.001, Student's t test; Fig. 3J). Similarly, the maximum migrating speed of the neurons that move inward (15.7 ± 1.0 µm/h, N = 4) was significantly slower than that of those migrating outward (38.1 ± 0.4 µm/h, N = 4, p < 0.001, Student's t test; Fig. 3K). Although the maximum migration speed of the inward-migrating neurons was relatively slow (∼16 µm/h), some inward-migrating neurons moved at a speed of >40 µm/h, which was as fast as outward migration (Fig. 3G).
Furthermore, to exclude the possibility that the inward migration of CLA neurons was caused by brain expansion, we measured the relative distance from the inward-migrating neurons to the outermost margin of the cortical migratory stream in the radial direction. The quantitative analysis indicated that the distance was shortened at 4.2 µm/h (95% CI, 2.1–6.2 µm/h; Fig. 4A), which was equivalent to the above-mentioned average migration speed of the neurons migrating inward (Fig. 3J), supporting that the CLA neurons indeed migrated toward the deep regions.
Next, we focused on neurons migrating medially inward and quantified the orientation of their major processes during time-lapse imaging (Fig. 4B, N = 4). The results showed that 49.7 ± 5.8% of the neurons directed their major process toward the brain surface, while 35.9 ± 7.4% of the neurons directed their major process toward the deep regions. Interestingly, 14.4 ± 3.5% of the neurons directed their major process toward the brain surface at first, whereas they extended a new major process toward the deep regions later during the time-lapse observations.
To visualize the cell morphology and movement of CLA neurons in more detail, time-lapse images were acquired at higher frequencies and magnifications (Fig. 4C,D; Movies 2, 3). The result (Movie 2) showed that the representative migrating neuron directed its leading (major) process toward the brain surface at first (Fig. 4C, 3:00), then extended a novel leading (major) process toward the deep regions (Fig. 4C, 6:00, white arrowhead), while the original leading process remained thinner (Fig. 4C, 18:00, white arrowhead). Another time-lapse observation (Movie 3) confirmed that the migrating neurons oriented their leading processes toward deep regions at least transiently. The representative neuron initially moved slowly (Fig. 4D, 0:00–6:00). However, at the end of the observation period, the cell body moved rapidly while the tip of its leading process seemed to remain mostly at the same position (Fig. 4D, 7:00-9:00).
Time-lapse imaging of migrating cells labeled with GFP by in utero electroporation. Images were captured every 25 min. The video clip corresponds to Figure 4C. Top, Superficial (lateral) side. Bottom, Deep (medial) side.
Time-lapse imaging of migrating cells labeled with GFP by in utero electroporation. Images were captured every 8 min. The video clip corresponds to Figure 4D. Top, Superficial (lateral) side. Bottom, Deep (medial) side.
Neurons migrating inward had contacts with radial glial fibers
To analyze the morphology of the migrating CLA neurons in detail, we transfected a tdTomato-expressing plasmid into the embryos at E11.5 using in utero electroporation and fixed them 3.5 d later. The results showed that tdTomato-labeled neurons were distributed both in the Nurr1-positive region and the superficial Nurr1-negative region, the latter corresponding to the insular cortex (Fig. 5A–C). Most tdTomato-positive neurons directed their major and thin processes to superficial and deep regions, respectively (Fig. 5A, arrowhead; Fig. 5D). In contrast, some tdTomato-labeled neurons directed their major processes toward the deep regions, most of which were Nurr1-positive (Fig. 5B, arrowhead; Fig. 5E, white arrowhead) and had a thin process oriented toward the superficial regions, resembling a trailing process (Fig. 5E, cyan arrowhead). Interestingly, a small number of neurons had two major processes oriented toward both deep and superficial regions (Fig. 5C,F), which may correspond to neurons that extended a new process directed toward deep regions during time-lapse imaging (Movies 1, 2, 3). The quantitative analysis indicated that 1.9 ± 1.9% of tdTomato-positive cells in the superficial Nurr1-negative region and 34.3 ± 6.6% of tdTomato-positive cells in the deep Nurr1-positive region directed their major process toward the deep/medial regions (Fig. 5G, N = 4, respectively, for the superficial and deep regions). These results confirmed that a substantial number of neurons in the deep Nurr1-positive region directed their processes toward the deep regions, suggesting that these neurons had migrated inward.
In the embryonic neocortex, newly generated neurons use radial glial fibers as a scaffold for migration (Rakic et al., 1974). However, it remains unknown whether the radial glial fibers also function as a scaffold for the migration of CLA neurons. To examine this issue, we performed immunohistochemistry for nestin, a marker of radial glial fibers, in addition to the cell labeling described above. The results showed that some neurons had their processes directed toward the deeper regions that were in line with the radial glial fibers. Interestingly, the tip of the process occasionally seemed to have contact with a radial glial fiber different from the one with which the rest of the process and the cell body had contact (Fig. 5H,I–I′′).
To further investigate the interaction between CLA neurons and radial glial fibers, we performed immune electron microscopy analysis. We transfected a GFP-expressing plasmid into the embryos at E11.5, fixed the mice 3.5 d later, and observed them using an anti-GFP antibody with an electron microscope. The results indicated that the deeply/inward-directed process had direct contact with fibers (Fig. 5J–M), suggesting that radial glial fibers act as a scaffold for CLA migration.
The CLA expresses ApoER2 during development
Reelin is an essential molecule for the development of the cerebral neocortex. Previous studies have proposed that Reelin has several functions (Hirota and Nakajima, 2017), such as inducing neuronal aggregation (Kubo et al., 2010), stopping signals for migration (Sekine et al., 2014; Hirota et al., 2018; Hirota and Nakajima, 2020), regulation of the multipolar-bipolar transition (Franco et al., 2011; Jossin and Cooper 2011; Sekine et al., 2011, 2012; Gil-Sanz et al., 2013), terminal translocation (Sekine et al., 2011, 2012), and radial migration in the intermediate zone (Hack et al., 2007; Britto et al., 2011; Hirota et al., 2018). Reelin binds to two receptors: ApoER2 and very-low-density lipoprotein receptor (VLDLR). We hypothesized that Reelin may also play a role in the development of CLA. To test this hypothesis, we performed immunohistochemistry for ApoER2 and Reelin at E13.5, E14.5, and E15.5.
We observed strong signals for ApoER2 in the cortical migratory stream (Fig. 6A–C, arrows), indicating that cells migrating in the stream or fibers running through the stream strongly expressed ApoER2. In contrast, although the signals in the future CLA regions were weak to moderate at E14.5, we observed a small cluster of ApoER2-positive cells in the presumptive CLA region (Fig. 6B,B′). At E15.5, the number of cells that had reached the presumptive CLA region increased and moderately expressed ApoER2 (Fig. 6C,C′).
Figure 6.
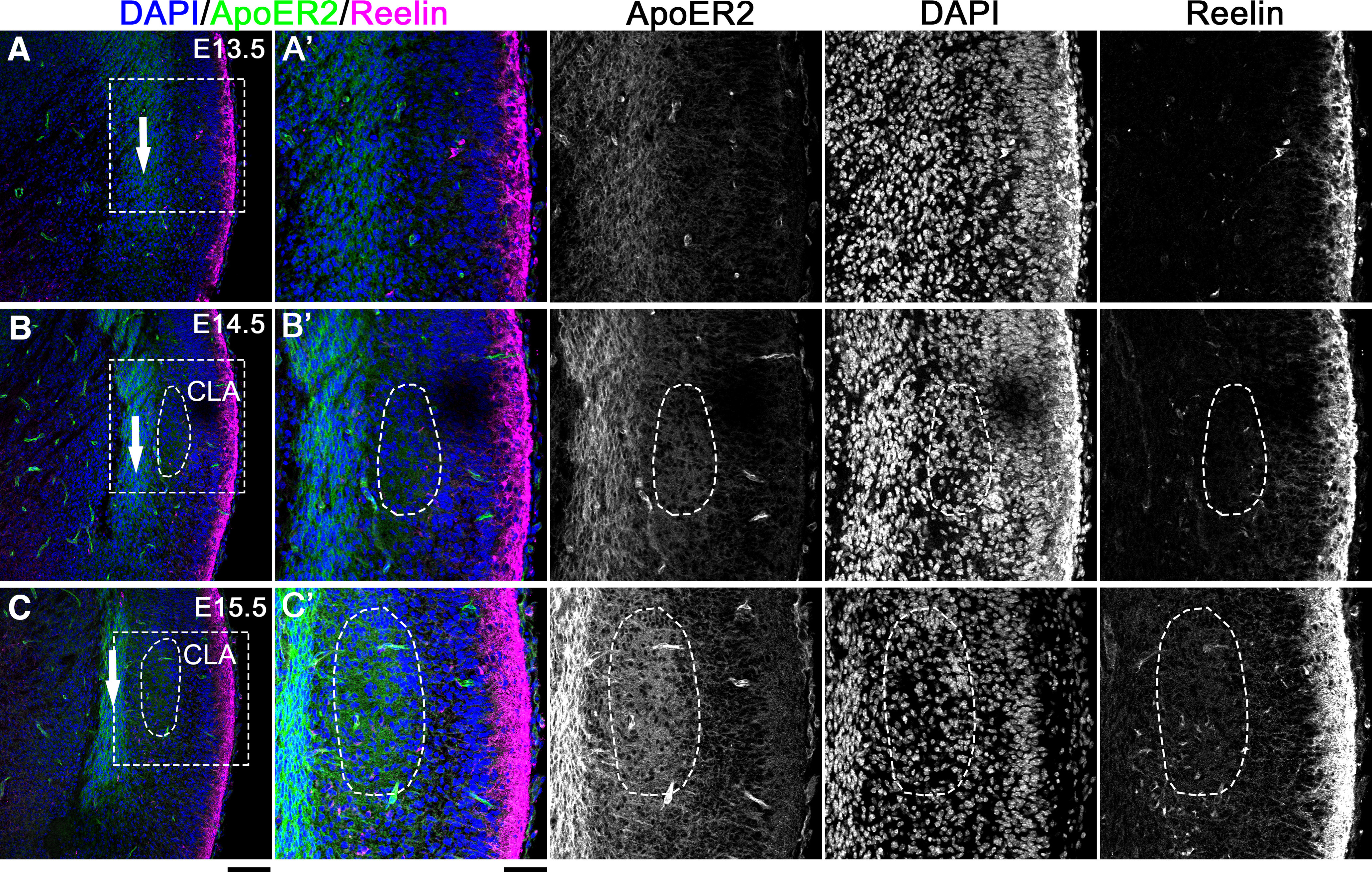
The CLA expressed ApoER2 during development. A–C, Brains were immunostained with anti-ApoER2 (green) and anti-Reelin (magenta) antibodies at E13.5 (A), E14.5 (B), and E15.5 (C). White arrows indicate cortical migratory stream. The nuclei were labeled with DAPI (blue). A′–C′, Higher magnifications of the labeled-boxed area in A–C, respectively. Scale bars: A–C, 200 µm; A′–C′, 100 µm. Left, Deep (medial) side. Right, Superficial (lateral) side. Top, Dorsal side. Bottom, Ventral side.
Reelin is necessary for the development of the CLA
Next, to investigate whether Reelin signaling is necessary for the development of the CLA, we performed immunohistochemical analyses on E18.5 reeler (Reelin-deficient) brains at the level of the foramen of Monro. Immunohistochemistry for Nurr1 revealed that the distribution of Nurr1-positive cells in heterozygous mice was similar to that in the WT mice. In contrast, immunohistochemistry for Nurr1 in reeler mouse brains showed that the distribution of Nurr1-positive cells at the level of the CLA and DEn in the dorsoventral axis extended more superficially (and widely) in reeler mice than in heterozygous mice (Fig. 7A). Quantitative bin analysis showed that a significantly larger percentage of Nurr1-positive cells in the CLA was distributed in the superficial bins (Bins 3-5) in reeler mice than in heterozygous mice (see Fig. 7B, N = 4, respectively; Bin 3, p = 0.007; Bin 4, p < 0.001; Bin 5, p < 0.001; Bin 7, p < 0.001; Bin 8, p < 0.001; Tukey's test). The abnormal distribution of Nurr1-positive cells in reeler mice indicated that Reelin is necessary for the normal development of the CLA and DEn.
Reelin signaling pathway through ApoER2 is involved in the development of the CLA
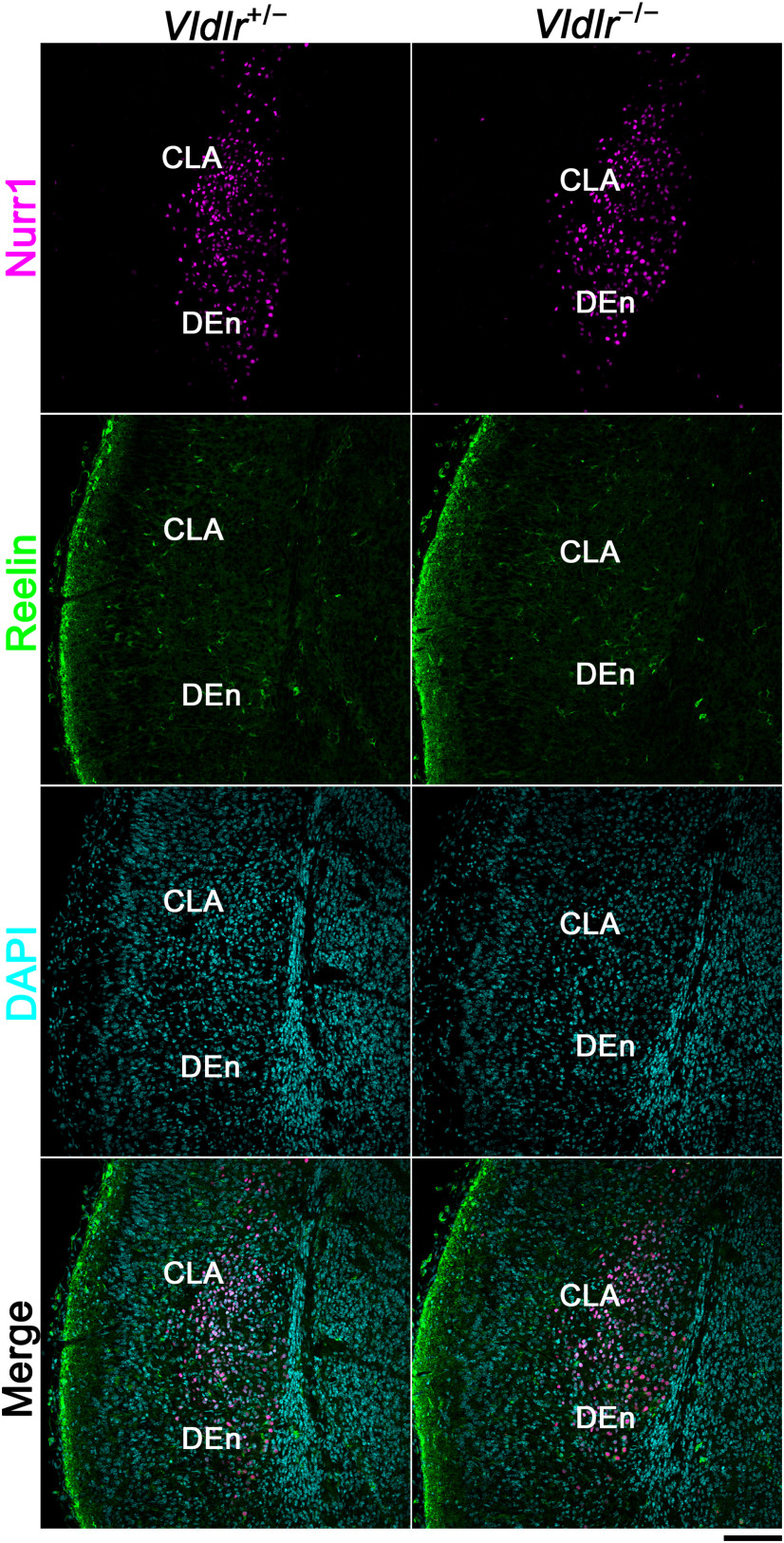
Next, to further investigate whether Reelin signaling is required for CLA development, we examined the brains of Apoer2 or Vldlr KO mice at E18.5. Immunohistochemistry for Nurr1, which is a marker for the CLA and DEn, and immunohistochemistry for Reelin, which itself could label the DEn at stages later than E15.5, showed that the Nurr1-positive cells in the CLA in Apoer2 KO mice were separated into two parts: the superficial and deep parts (Fig. 7C). The distribution of Nurr1-positive cells in the CLA was evaluated by quantitative bin analysis. Although the distribution of Nurr1-positive cells in Apoer2 heterozygous mice showed a peak in Bin 8, a significantly smaller percentage of the Nurr1-positive cells was distributed in Bin 8 in Apoer2 KO mice (p = 0.017; Tukey's test), indicating that Nurr1-positive cells in Apoer2 KO mice were more evenly distributed than those in heterozygous mice (Fig. 7D; Apoer2−/−, N = 3; Apoer2+/−, N = 4). In addition, the Nurr1-positive cells in the DEn seemed to be sparsely distributed, which was also observed in reeler mice. Thus, in Apoer2 homozygous KO mice, some Nurr1-positive cells in the CLA might be located more superficially, like those in the reeler, and others might be located deeper compared with those in Apoer2 heterozygous mice, resulting in a more flattened distribution of the Nurr1-positive cells in the Apoer2 KO mice.
In contrast, immunohistochemistry for Nurr1 and Reelin showed no remarkable difference between Vldlr KO mice and heterozygous mice (Fig. 8). Therefore, ApoER2 is thought to play a more important role than very-low-density lipoprotein receptor (VLDLR) in the CLA and DEn formation.
Figure 8.
Vldlr KO resulted in no remarkable difference in the alignment of CLA neurons. Immunostaining for Nurr1 (magenta) and Reelin (green) was performed in Vlder homozygous and heterozygous KO mice at E18.5. The nuclei were labeled with DAPI (cyan). Scale bars, 100 µm. Top, Dorsal side. Bottom, Ventral side. Left, Superficial (lateral) side. Right, Deep (medial) side.
Discussion
In the present study, BrdU analysis revealed that CLA and DEn neurons were mainly born between E10.5 and E12.5 (Fig. 1F). It was previously reported that rat CLA neurons were generated between E15 and E16, and the DEn neurons were generated between E14 and E15, when the day the females became sperm positive was designated as El (Bayer and Altman, 1991). Another rat study showed the similar results: CLA neurons were produced at E14.5-E15.5 and DEn neurons were produced at E13.5-E14.5 (Fang et al., 2021). Other studies have reported that CLA neurons were born earlier, between E11 and E12 in mice (Smart and Smart, 1977, 1982), and mainly at E12.5, in rats (Bisconte and Marty, 1975). Considering the 1-3 d variation in embryonic brain development between rats and mice (Clancy et al., 2001), our data are considerably consistent with these previous studies. In addition, our results suggested that early-born neurons in the insular cortex were mainly generated at the same time as CLA neurons (Fig. 1G), which is also consistent with the previous findings in rats (Fang et al., 2021). In a previous study, both CLA and insular neurons were labeled when in utero electroporation of EGFP was performed at E11 and E12 (Bruguier et al., 2020), supporting the idea that the birthdates of CLA and insular neurons overlap.
Next, we showed that the Nurr1-positive cluster was originally formed beneath the surface (Fig. 2B), suggesting that the CLA neurons transiently reached the surface but then shifted to a deeper position. In addition, at E13.5, 2 d after injection of FlashTag, we observed that most FlashTag-labeled cells were distributed superficially to the Nurr1-positive cluster (more mature CLA neurons), also supporting that the migrating CLA neurons once reached the surface. At E14.5, 1 d later, we found a few FlashTag-positive cells that oriented their processes inward and were positive for Nurr1 (Fig. 2C,C′,C′′′), further supporting the possibility that some CLA neurons moved superficially past the presumptive CLA region and migrated inward to their final destination.
To test this possibility, we performed time-lapse imaging and observed that some cells moved inward from the surface toward the deeper positions. This change in the migratory direction from outward to “inward” is a unique type of neuronal migration mode as opposed to conventional ones, such as “locomotion,” “somal translocation,” “terminal translocation,” “multipolar migration,” and “climbing mode” (Rakic, 1972; Nadarajah et al., 2001; Tabata and Nakajima, 2003; Noctor et al., 2004; Cooper, 2008; Tabata et al., 2009; Sekine et al., 2011, 2012; Yoshinaga et al., 2012; Kitazawa et al., 2014). Interestingly, although the CLA and insular cortex are close to each other in terms of location and birthdate, the migration profiles of CLA neurons were distinct from those of neurons in the insular cortex, which seemed to migrate only outward (Movie 1; Fig. 9). However, in time-lapse imaging studies, we observed that some cells born at E11.5 left the cortical migratory stream and wandered around the presumptive CLA region, which suggests that a certain population of CLA neurons might migrate radially outward and “directly join” the CLA without reversed migration. Further studies are necessary to uncover what proportions of the CLA neurons are supplied through the radial “reversed” migration or only the outward migration.
Figure 9.
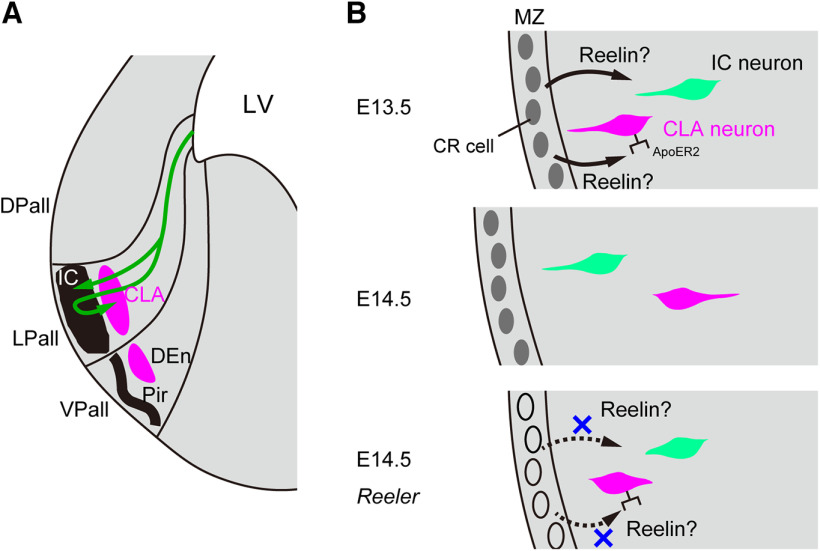
A schematic representation of the migratory behavior of neurons in the insular cortex and the CLA. A, They are generated in the ventricular zone and migrate tangentially via cortical migratory stream at first. Next, they migrate radially outward to reach the surface. Cortical neurons in the insular cortex complete the migration after they reach the superficial region. However, some CLA neurons migrate inward to take a position in the deep region after they reach the superficial region. B, The CLA neuron (magenta cell) reaches brain surface around E13.5 and moves inward probably via Reelin signaling secreted from Cajal-Retzius cells. The insular neuron (green cell) migrates toward the brain surface presumably via Reelin signaling. However, in reeler mice, those migrations are disrupted and CLA neurons are located in the superficial region. IC, Insular cortex; Pir, piriform cortex; LV, lateral ventricle; DPall, dorsal pallium; LPall, lateral pallium; VPall, ventral pallium; MZ, marginal zone; CR cell, Cajal-Retzius cell.
Furthermore, labeling of the migrating CLA neurons showed that some migrating CLA neurons had one main leading process as seen during neuronal migration by the “locomotion” mode in the neocortex. This process was directed toward the deep region (Fig. 5B,E), suggesting that the neurons were moving toward the deep region. Additionally, most neurons with a deeply directed process were located in the Nurr1-positive deep region, supporting the possibility that neurons that had reached the presumptive CLA region elongated a deeply oriented process during their inward migrations. Moreover, some CLA neurons contacted the radial glial fibers via a deeply oriented process. Interestingly, the tip of the process seemed to come into contact with a different fiber from the one that the cell body had contact with, raising the hypothesis that the CLA neurons depend on multiple radial glial fibers during inward migration, which is similar to the hippocampal neurons during their migration using the “climbing mode” (Kitazawa et al., 2014).
In addition, we found that Reelin was necessary for the migration of CLA neurons. However, the role of Reelin in CLA development remains unclear. We propose the following three hypotheses: First, Reelin plays a role rather in the migration of the insular neurons. Late-born insular neurons migrate radially through the developing CLA region and reach the superficial region. If the insular neurons were not able to migrate past the developing CLA region in the reeler, they might have been stuck beneath the CLA region and pushed it up to shift them more superficially. However, this non–cell-autonomous mechanism does not seem to be consistent with the extended distribution of the insular cortex of reeler mice. Ctip2-positive insular cortical neurons were not trapped on the deeper side of the CLA (Fig. 7A), reducing the possibility that Reelin acts only on the outward migration of insular neurons, although a possible secondary effect of Reelin deficiency on CLA distribution cannot be ruled out. The second hypothesis is that defects in Reelin signaling alter the adhesion properties of CLA neurons, causing migrating insular neurons to adhere to CLA neurons. In the cerebral neocortex, a sensu lato non–cell-autonomous function of Reelin downstream signaling has been proposed. A previous study suggested that Dab1−/− deep layer neurons, when present in high cell density below the subplate, became impermissible to the migration of Dab1−/− superficial layer neurons, which otherwise had the potential to migrate past the subplate (Yoshinaga et al., 2022). Similarly, it is likely that Reelin acts on the interaction between the CLA and insular neurons. On the other hand, the third hypothesis is that the Reelin signaling is necessary for cell-autonomous inward migration of CLA neurons. The superficially distributed population in reeler and Apoer2 KO mice might be CLA neurons that could not completely migrate inward. To test these hypotheses, it is necessary to disrupt the Reelin signaling pathway exclusively in cells that migrate inward in a future study.
The reversed migration mode of CLA neurons has not been reported previously. A study by [3H]thymidine injection demonstrated that CLA neurons left the cortical migratory stream and migrated outward to settle in the final position (Bayer and Altman, 1991), whereas the migration direction could not be determined by [3H]thymidine autoradiography. Another study using the CLA marker Nurr1 suggested that the insular neurons migrated radially across the developing CLA region and passively displaced it into its final deep position (in an inside-out pattern) (Puelles, 2014), which is partly consistent with our observation that the FlashTag-positive cells migrate through the Nurr1-positive cells in the presumptive CLA region (Fig. 2). The results of the present study indicate that CLA neurons may actively migrate inward and take a deep position. We labeled CLA neurons and analyzed their migratory profiles of CLA neurons with in utero electroporation or FlashTag technology. These approaches revealed the distinct migratory behaviors of CLA neurons, such as migrating inward relatively slowly and using multiple radial glial fibers as a scaffold for inward migration. Time-lapse imaging of migrating neurons supports the hypothesis of inward CLA migration. Apoer2 KO also suggested that CLA neurons migrated toward the deep region because the superficial distribution of Nurr1-positive cells in Apoer2 KO mice is consistent with the hypothesis of inward migration of CLA neurons. The spindle shape with one main leading process of migrating CLA neurons is similar to that of cortical neurons migrating by the “locomotion” mode. However, reversed migration is distinct from the conventional migration modes of neocortical neurons in terms of migration direction.
Proper neuronal migration is a critical step in the formation of a highly organized mammalian brain structure and is essential for normal brain function. Disruption of this developmental process causes cytoarchitectural deficits, including mildly altered cell positioning and is related to altered axonal projections and pathogenesis of some neuropsychiatric disorders (Ishii et al., 2016; Kubo et al., 2017). For instance, previous studies have suggested that mispositioning of serotonergic neurons in the dorsal raphe nucleus is associated with reduced serotonergic projections to rostral brain areas (Tikker et al., 2020). Moreover, abnormal neuronal alignments have been reported in neuropsychiatric disorders associated with cognitive impairment, such as schizophrenia (Casanova et al., 2008) and autism (Bailey et al., 1998). Therefore, migration defects in CLA neurons are likely to be associated with disruption of their extensive connectivity with cortical areas and their unique cognitive functions. Future elucidation of the characteristics and precise molecular mechanisms of CLA neuronal migration may provide insights into the pathogenesis of neuropsychiatric symptoms.
Footnotes
This work was supported by Ministry of Education, Culture, Sports, Science, and Technology/Japan Society for the Promotion of Science Grants-in-Aid for Scientific Research JP20H05688, JP16H06482, JP21H02853, JP20H03649, JP19H05227, and JP19H01152; Keio Gijuku Academic Development Funds; Keio Gijuku Fukuzawa Memorial Fund for the Advancement of Education and Research; Takeda Science Foundation; Uehara Memorial Foundation; PRIME; AMED JP19gm6310004, JP20gm6310004, and JP21gm6310004; SENSHIN Medical Research Foundation; and the Jikei University Exploratory Collaboration Research Fund. We thank the members of our laboratories for their valuable discussions; and Dr. J. Miyazaki (pCAGGS vector) for permission to use the CAG promoter.
The authors declare no competing financial interests.
References
- Bailey A, Luthert P, Dean A, Harding B, Janota I, Montgomery M, Rutter M, Lantos P (1998) A clinicopathological study of autism. Brain 121:889–905. 10.1093/brain/121.5.889 [DOI] [PubMed] [Google Scholar]
- Bayer SA, Altman J (1991) Development of the endopiriform nucleus and the claustrum in the rat brain. Neuroscience 45:391–412. 10.1016/0306-4522(91)90236-h [DOI] [PubMed] [Google Scholar]
- Benarroch EE (2019) Insular cortex: functional complexity and clinical correlations. Neurology 93:932–938. 10.1212/WNL.0000000000008525 [DOI] [PubMed] [Google Scholar]
- Bisconte JC, Marty R (1975) Chronoarchitectonic analysis of the rat brain using radioautography: I. Histogenesis of the telencephalon. J Hirnforsch 16:55–74. [PubMed] [Google Scholar]
- Britto JM, Tait KJ, Johnston LA, Hammond VE, Kalloniatis M, Tan SS (2011) Altered speeds and trajectories of neurons migrating in the ventricular and subventricular zones of the reeler neocortex. Cereb Cortex 21:1018–1027. 10.1093/cercor/bhq168 [DOI] [PubMed] [Google Scholar]
- Bruguier H, Suarez R, Manger P, Hoerder-Suabedissen A, Shelton AM, Oliver DK, Packer AM, Ferran JL, García-Moreno F, Puelles L, Molnár Z (2020) In search of common developmental and evolutionary origin of the claustrum and subplate. J Comp Neurol 528:2956–2977. 10.1002/cne.24922 [DOI] [PubMed] [Google Scholar]
- Casanova MF, Kreczmanski P, Trippe J 2nd, Switala A, Heinsen H, Steinbusch HW, Schmitz C (2008) Neuronal distribution in the neocortex of schizophrenic patients. Psychiatry Res 158:267–277. 10.1016/j.psychres.2006.12.009 [DOI] [PubMed] [Google Scholar]
- Chevée M, Finkel EA, Kim SJ, O'Connor DH, Brown SP (2022) Neural activity in the mouse claustrum in a cross-modal sensory selection task. Neuron 110:486–501.e7. 10.1016/j.neuron.2021.11.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clancy B, Darlington RB, Finlay BL (2001) Translating developmental time across mammalian species. Neuroscience 105:7–17. 10.1016/s0306-4522(01)00171-3 [DOI] [PubMed] [Google Scholar]
- Cooper JA (2008) A mechanism for inside-out lamination in the neocortex. Trends Neurosci 31:113–119. 10.1016/j.tins.2007.12.003 [DOI] [PubMed] [Google Scholar]
- Crick FC, Koch C (2005) What is the function of the claustrum? Philos Trans R Soc Lond B Biol Sci 360:1271–1279. 10.1098/rstb.2005.1661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang C, Wang H, Naumann RK (2021) Developmental patterning and neurogenetic gradients of nurr1 positive neurons in the rat claustrum and lateral cortex. Front Neuroanat 15:786329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franco SJ, Martinez-Garay I, Gil-Sanz C, Harkins-Perry SR, Müller U (2011) Reelin regulates cadherin function via Dab1/Rap1 to control neuronal migration and lamination in the neocortex. Neuron 69:482–497. 10.1016/j.neuron.2011.01.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frykman PK, Brown MS, Yamamoto T, Goldstein JL, Herz J (1995) Normal plasma lipoproteins and fertility in gene-targeted mice homozygous for a disruption in the gene encoding very low density lipoprotein receptor. Proc Natl Acad Sci USA 92:8453–8457. 10.1073/pnas.92.18.8453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gil-Sanz C, Franco SJ, Martinez-Garay I, Espinosa A, Harkins-Perry S, Müller U (2013) Cajal-Retzius cells instruct neuronal migration by coincidence signaling between secreted and contact-dependent guidance cues. Neuron 79:461–477. 10.1016/j.neuron.2013.06.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goll Y, Atlan G, Citri A (2015) Attention: the claustrum. Trends Neurosci 38:486–495. 10.1016/j.tins.2015.05.006 [DOI] [PubMed] [Google Scholar]
- Govindan S, Oberst P, Jabaudon D (2018) In vivo pulse labeling of isochronic cohorts of cells in the central nervous system using FlashTag. Nat Protoc 13:2297–2311. 10.1038/s41596-018-0038-1 [DOI] [PubMed] [Google Scholar]
- Hack I, Hellwig S, Junghans D, Brunne B, Bock HH, Zhao S, Frotscher M (2007) Divergent roles of ApoER2 and Vldlr in the migration of cortical neurons. Development 134:3883–3891. 10.1242/dev.005447 [DOI] [PubMed] [Google Scholar]
- Hirota Y, Nakajima K (2017) Control of neuronal migration and aggregation by reelin signaling in the developing cerebral cortex. Front Cell Dev Biol 5:40. 10.3389/fcell.2017.00040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirota Y, Kubo K, Katayama K, Honda T, Fujino T, Yamamoto TT, Nakajima K (2015) Reelin receptors ApoER2 and VLDLR are expressed in distinct spatiotemporal patterns in developing mouse cerebral cortex. J Comp Neurol 523:463–478. 10.1002/cne.23691 [DOI] [PubMed] [Google Scholar]
- Hirota Y, Kubo K, Fujino T, Yamamoto TT, Nakajima K (2018) ApoER2 controls not only neuronal migration in the intermediate zone but also termination of migration in the developing cerebral cortex. Cereb Cortex 28:223–235. 10.1093/cercor/bhw369 [DOI] [PubMed] [Google Scholar]
- Hirota Y, Nakajima K (2020) VLDLR is not essential for reelin-induced neuronal aggregation but suppresses neuronal invasion into the marginal zone. Development 147:dev189936. 10.1242/dev.189936 [DOI] [PubMed] [Google Scholar]
- Inoue S, Hayashi K, Fujita K, Tagawa K, Okazawa H, Kubo K, Nakajima K (2019) Drebrin-like (Dbnl) controls neuronal migration via regulating N-cadherin expression in the developing cerebral cortex. J Neurosci 39:678–691. 10.1523/JNEUROSCI.1634-18.2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishii K, Kubo K, Endo T, Yoshida K, Benner S, Ito Y, Aizawa H, Aramaki M, Yamanaka A, Tanaka K, Takata N, Tanaka KF, Mimura M, Tohyama C, Kakeyama M, Nakajima K (2015) Neuronal heterotopias affect the activities of distant brain areas and lead to behavioral deficits. J Neurosci 35:12432–12445. 10.1523/JNEUROSCI.3648-14.2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishii K, Kubo K, Nakajima K (2016) Reelin and neuropsychiatric disorders. Front Cell Neurosci 10:229. 10.3389/fncel.2016.00229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jossin Y, Cooper JA (2011) Reelin, Rap1 and N-cadherin orient the migration of multipolar neurons in the developing neocortex. Nat Neurosci 14:697–703. 10.1038/nn.2816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamiya A, Kubo K, Tomoda T, Takaki M, Youn R, Ozeki Y, Sawamura N, Park U, Kudo C, Okawa M, Ross CA, Hatten ME, Nakajima K, Sawa A (2005) A schizophrenia-associated mutation of DISC1 perturbs cerebral cortex development. Nat Cell Biol 7:1167–1178. 10.1038/ncb1328 [DOI] [PubMed] [Google Scholar]
- Kanatani S, Honda T, Aramaki M, Hayashi K, Kubo K, Ishida M, Tanaka DH, Kawauchi T, Sekine K, Kusuzawa S, Kawasaki T, Hirata T, Tabata H, Uhlén P, Nakajima K (2015) The COUP-TFII/Neuropilin-2 is a molecular switch steering diencephalon-derived GABAergic neurons in the developing mouse brain. Proc Natl Acad Sci USA 112:E4985–E4994. 10.1073/pnas.1420701112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitazawa A, Kubo K, Hayashi K, Matsunaga Y, Ishii K, Nakajima K (2014) Hippocampal pyramidal neurons switch from a multipolar migration mode to a novel 'climbing' migration mode during development. J Neurosci 34:1115–1126. 10.1523/JNEUROSCI.2254-13.2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubo K, Honda T, Tomita K, Sekine K, Ishii K, Uto A, Kobayashi K, Tabata H, Nakajima K (2010) Ectopic Reelin induces neuronal aggregation with a normal birthdate-dependent 'inside-out' alignment in the developing neocortex. J Neurosci 30:10953–10966. 10.1523/JNEUROSCI.0486-10.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubo K, et al. (2017) Association of impaired neuronal migration with cognitive deficits in extremely preterm infants. JCI Insight 2:e88609. 10.1172/jci.insight.88609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meijering E, Dzyubachyk O, Smal I (2012) Methods for cell and particle tracking. Methods Enzymol 504:183–200. 10.1016/B978-0-12-391857-4.00009-4 [DOI] [PubMed] [Google Scholar]
- Nadarajah B, Brunstrom JE, Grutzendler J, Wong RO, Pearlman AL (2001) Two modes of radial migration in early development of the cerebral cortex. Nat Neurosci 4:143–150. 10.1038/83967 [DOI] [PubMed] [Google Scholar]
- Nakajima K, Mikoshiba K, Miyata T, Kudo C, Ogawa M (1997) Disruption of hippocampal development in vivo by CR-50 mAb against reelin. Proc Natl Acad Sci USA 94:8196–8201. 10.1073/pnas.94.15.8196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narikiyo K, Mizuguchi R, Ajima A, Shiozaki M, Hamanaka H, Johansen JP, Mori K, Yoshihara Y (2020) The claustrum coordinates cortical slow-wave activity. Nat Neurosci 23:741–753. 10.1038/s41593-020-0625-7 [DOI] [PubMed] [Google Scholar]
- Niwa H, Yamamura K, Miyazaki J (1991) Efficient selection for high-expression transfectants with a novel eukaryotic vector. Gene 108:193–199. 10.1016/0378-1119(91)90434-d [DOI] [PubMed] [Google Scholar]
- Niwa M, Cash-Padgett T, Kubo K, Saito A, Ishii K, Sumitomo A, Taniguchi Y, Ishizuka K, Jaaro-Peled H, Tomoda T, Nakajima K, Sawa A, Kamiya A (2016) DISC1 a key molecular lead in psychiatry and neurodevelopment: No-More Disrupted-in-Schizophrenia 1. Mol Psychiatry 21:1488–1489. 10.1038/mp.2016.154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noctor SC, Martínez-Cerdeño V, Ivic L, Kriegstein AR (2004) Cortical neurons arise in symmetric and asymmetric division zones and migrate through specific phases. Nat Neurosci 7:136–144. 10.1038/nn1172 [DOI] [PubMed] [Google Scholar]
- Patru MC, Reser DH (2015) A new perspective on delusional states: evidence for claustrum involvement. Front Psychiatry 6:158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puelles L, Kuwana E, Puelles E, Rubenstein JL (1999) Comparison of the mammalian and avian telencephalon from the perspective of gene expression data. Eur J Morphol 37:139–150. 10.1076/ejom.37.2.139.4756 [DOI] [PubMed] [Google Scholar]
- Puelles L, Kuwana E, Puelles E, Bulfone A, Shimamura K, Keleher J, Smiga S, Rubenstein JL (2000) Pallial and subpallial derivatives in the embryonic chick and mouse telencephalon, traced by the expression of the genes Dlx-2, Emx-1, Nkx-2.1, Pax-6, and Tbr-1. J Comp Neurol 424:409–438. [DOI] [PubMed] [Google Scholar]
- Puelles L (2014) Development and evolution of the claustrum. In: The claustrum, pp 119–176. Amsterdam: Elsevier. [Google Scholar]
- Puelles L (2017) Comments on the updated tetrapartite pallium model in the mouse and chick, featuring a homologous claustro-insular complex. Brain Behav Evol 90:171–189. 10.1159/000479782 [DOI] [PubMed] [Google Scholar]
- Puelles L, Harrison M, Paxinos G, Watson C (2013) A developmental ontology for the mammalian brain based on the prosomeric model. Trends Neurosci 36:570–578. 10.1016/j.tins.2013.06.004 [DOI] [PubMed] [Google Scholar]
- Puelles L, Ayad A, Alonso A, Sandoval JE, MartÍnez-de-la-Torre M, Medina L, Ferran JL (2016) Selective early expression of the orphan nuclear receptor Nr4a2 identifies the claustrum homolog in the avian mesopallium: impact on sauropsidian/mammalian pallium comparisons. J Comp Neurol 524:665–703. 10.1002/cne.23902 [DOI] [PubMed] [Google Scholar]
- Rakic P (1972) Mode of cell migration to the superficial layers of fetal monkey neocortex. J Comp Neurol 145:61–83. 10.1002/cne.901450105 [DOI] [PubMed] [Google Scholar]
- Rakic P, Stensas LJ, Sayre E, Sidman RL (1974) Computer-aided three-dimensional reconstruction and quantitative analysis of cells from serial electron microscopic montages of foetal monkey brain. Nature 250:31–34. 10.1038/250031a0 [DOI] [PubMed] [Google Scholar]
- Rueda-Alaña E, Martínez-Garay I, Encinas JM, Molnár Z, García-Moreno F (2018) Dbx1-derived pyramidal neurons are generated locally in the developing murine neocortex. Front Neurosci 12:792. 10.3389/fnins.2018.00792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satterthwaite TD, Baker JT (2015) How can studies of resting-state functional connectivity help us understand psychosis as a disorder of brain development? Curr Opin Neurobiol 30:85–91. 10.1016/j.conb.2014.10.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, Tinevez JY, White DJ, Hartenstein V, Eliceiri K, Tomancak P, Cardona A (2012) Fiji: an open-source platform for biological-image analysis. Nat Methods 9:676–682. 10.1038/nmeth.2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekine K, Honda T, Kawauchi T, Kubo K, Nakajima K (2011) The outermost region of the developing cortical plate is crucial for both the switch of the radial migration mode and the Dab1-dependent 'inside-out' lamination in the neocortex. J Neurosci 31:9426–9439. 10.1523/JNEUROSCI.0650-11.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekine K, Kawauchi T, Kubo K, Honda T, Herz J, Hattori M, Kinashi T, Nakajima K (2012) Reelin controls neuronal positioning by promoting cell-matrix adhesion via inside-out activation of integrin α5β1. Neuron 76:353–369. 10.1016/j.neuron.2012.07.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekine K, Kubo K, Nakajima K (2014) How does Reelin control neuronal migration and layer formation in the developing mammalian neocortex? Neurosci Res 86:50–58. 10.1016/j.neures.2014.06.004 [DOI] [PubMed] [Google Scholar]
- Sheerin AH, Nylen K, Zhang X, Saucier DM, Corcoran ME (2004) Further evidence for a role of the anterior claustrum in epileptogenesis. Neuroscience 125:57–62. 10.1016/j.neuroscience.2004.01.043 [DOI] [PubMed] [Google Scholar]
- Shin M, Kitazawa A, Yoshinaga S, Hayashi K, Hirata Y, Dehay C, Kubo K, Nakajima K (2019) Both excitatory and inhibitory neurons transiently form clusters at the outermost region of the developing mammalian cerebral neocortex. J Comp Neurol 527:1577–1597. 10.1002/cne.24634 [DOI] [PubMed] [Google Scholar]
- Smart IH, Smart M (1977) The location of nuclei of different labelling intensities in autoradiographs of the anterior forebrain of postnatial mice injected with [3H]thymidine on the eleventh and twelfth days post-conception. J Anat 123:515–525. [PMC free article] [PubMed] [Google Scholar]
- Smart IH, Smart M (1982) Growth patterns in the lateral wall of the mouse telencephalon: I. Autoradiographic studies of the histogenesis of the isocortex and adjacent areas. J Anat 134:273–298. [PMC free article] [PubMed] [Google Scholar]
- Smith JB, Alloway KD, Hof PR, Orman R, Reser DH, Watakabe A, Watson GD (2019) The relationship between the claustrum and endopiriform nucleus: a perspective towards consensus on cross-species homology. J Comp Neurol 527:476–499. 10.1002/cne.24537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith JB, Lee AK, Jackson J (2020) The claustrum. Curr Biol 30:R1401–R1406. 10.1016/j.cub.2020.09.069 [DOI] [PubMed] [Google Scholar]
- Tabata H, Nakajima K (2001) Efficient in utero gene transfer system to the developing mouse brain using electroporation: visualization of neuronal migration in the developing cortex. Neuroscience 103:865–872. 10.1016/S0306-4522(01)00016-1 [DOI] [PubMed] [Google Scholar]
- Tabata H, Nakajima K (2003) Multipolar migration: the third mode of radial neuronal migration in the developing cerebral cortex. J Neurosci 23:9996–10001. 10.1523/JNEUROSCI.23-31-09996.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabata H, Kanatani S, Nakajima K (2009) Differences of migratory behavior between direct progeny of apical progenitors and basal progenitors in the developing cerebral cortex. Cereb Cortex 19:2092–2105. 10.1093/cercor/bhn227 [DOI] [PubMed] [Google Scholar]
- Telley L, Govindan S, Prados J, Stevant I, Nef S, Dermitzakis E, Dayer A, Jabaudon D (2016) Sequential transcriptional waves direct the differentiation of newborn neurons in the mouse neocortex. Science 351:1443–1446. 10.1126/science.aad8361 [DOI] [PubMed] [Google Scholar]
- Tikker L, Casarotto P, Singh P, Biojone C, Piepponen TP, Estartús N, Seelbach A, Sridharan R, Laukkanen L, Castrén E, Partanen J (2020) Inactivation of the GATA cofactor ZFPM1 results in abnormal development of dorsal raphe serotonergic neuron subtypes and increased anxiety-like behavior. J Neurosci 40:8669–8682. 10.1523/JNEUROSCI.2252-19.2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomita K, Kubo K, Ishii K, Nakajima K (2011) Disrupted-in-Schizophrenia-1 (Disc1) is necessary for migration of the pyramidal neurons during mouse hippocampal development. Hum Mol Genet 20:2834–2845. 10.1093/hmg/ddr194 [DOI] [PubMed] [Google Scholar]
- Watson C, Puelles L (2017) Developmental gene expression in the mouse clarifies the organization of the claustrum and related endopiriform nuclei. J Comp Neurol 525:1499–1508. 10.1002/cne.24034 [DOI] [PubMed] [Google Scholar]
- Yoshinaga S, Ohkubo T, Sasaki S, Nuriya M, Ogawa Y, Yasui M, Tabata H, Nakajima K (2012) A phosphatidylinositol lipids system, lamellipodin, and Ena/VASP regulate dynamic morphology of multipolar migrating cells in the developing cerebral cortex. J Neurosci 32:11643–11656. 10.1523/JNEUROSCI.0738-12.2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshinaga S, Shin M, Kitazawa A, Ishii K, Tanuma M, Kasai A, Hashimoto H, Kubo K, Nakajima K (2021) Comprehensive characterization of migration profiles of murine cerebral cortical neurons during development using FlashTag labeling. iScience 24:102277. 10.1016/j.isci.2021.102277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshinaga S, Honda T, Kubo K, Nakajima K (2022) Dab1-deficient deep layer neurons prevent Dab1-deficient superficial layer neurons from entering the cortical plate. Neurosci Res 180:23–35. 10.1016/j.neures.2022.03.011 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Time-lapse imaging of migrating cells labeled with GFP by in utero electroporation. Images were captured every 30 min. The video clip corresponds to Figure 3A,A′. Top, Superficial (lateral) side. Bottom, Deep (medial) side.
Time-lapse imaging of migrating cells labeled with GFP by in utero electroporation. Images were captured every 25 min. The video clip corresponds to Figure 4C. Top, Superficial (lateral) side. Bottom, Deep (medial) side.
Time-lapse imaging of migrating cells labeled with GFP by in utero electroporation. Images were captured every 8 min. The video clip corresponds to Figure 4D. Top, Superficial (lateral) side. Bottom, Deep (medial) side.