Abstract
The aminoacyl-tRNA synthetases (AARSs) and their relationship to the genetic code are examined from the evolutionary perspective. Despite a loose correlation between codon assignments and AARS evolutionary relationships, the code is far too highly structured to have been ordered merely through the evolutionary wanderings of these enzymes. Nevertheless, the AARSs are very informative about the evolutionary process. Examination of the phylogenetic trees for each of the AARSs reveals the following. (i) Their evolutionary relationships mostly conform to established organismal phylogeny: a strong distinction exists between bacterial- and archaeal-type AARSs. (ii) Although the evolutionary profiles of the individual AARSs might be expected to be similar in general respects, they are not. It is argued that these differences in profiles reflect the stages in the evolutionary process when the taxonomic distributions of the individual AARSs became fixed, not the nature of the individual enzymes. (iii) Horizontal transfer of AARS genes between Bacteria and Archaea is asymmetric: transfer of archaeal AARSs to the Bacteria is more prevalent than the reverse, which is seen only for the “gemini group.” (iv) The most far-ranging transfers of AARS genes have tended to occur in the distant evolutionary past, before or during formation of the primary organismal domains. These findings are also used to refine the theory that at the evolutionary stage represented by the root of the universal phylogenetic tree, cells were far more primitive than their modern counterparts and thus exchanged genetic material in far less restricted ways, in effect evolving in a communal sense.
The aminoacyl-tRNA synthetases (AARSs) have long fascinated biologists. They are the linchpin of translation, the link between the worlds of protein and nucleic acid. Their structures and functions, which have both practical and basic significance, are deserving of and have received much attention. However, it is not only the structure-function aspect of these enzymes that has captured the biologist's imagination; it is also the possibility that they could tell us the secrets of the genetic code. To understand these enzymes in standard molecular terms is to add one more piece, a most important one, to the puzzle of what the cell is, how it works. But to understand them in evolutionary terms is to ask what the cell is in a deeper sense, how it evolved, how life came to be—the biologist's ultimate question. Reading the history written into the AARSs was not possible previously for the simple reason that doing so requires molecular sequences from a large number of these molecules, and the necessary body of data was lacking. The progress of genomics in the late 1990s is now providing the needed data, and a picture of AARS evolution is beginning to emerge (5, 7, 15–18, 45, 75). In the present review we examine the still murky image of synthetase evolution from a slightly different perspective and bring forth more of its rich detail and evolutionary depth.
BIOCHEMICAL SKETCH OF AMINOACYL-TRNA SYNTHESIS
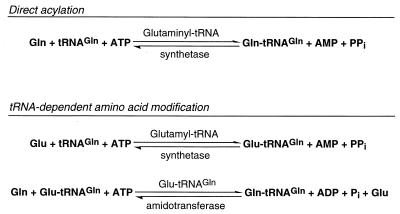
In a departure from the long-accepted view (48) that every cell harbors 20 aminoacyl-tRNA synthetases responsible for the synthesis of the set of 20 canonical aminoacyl-tRNA families, it is now clearly established that there are at least two ways of forming aminoacyl-tRNA (12, 33). The direct acylation of tRNA by aminoacyl-tRNA synthetases is well understood; the ATP-dependent reaction (Fig. 1) is carried out by enzymes which, in general, are exceedingly specific in selecting their substrates, i.e., amino acid and tRNA. They fall into two classes of 10 based on the topology of their ATP binding domain; class I proteins contain a Rossmann fold (characterized by the HIGH and KMSK motifs), while class II enzymes possess an unrelated β-sheet arrangement and are characterized by three degenerate sequence motifs (3, 10, 14, 20). Examples of most of the aminoacyl-tRNA synthetases have been structurally characterized, and it is expected that in the near future the crystal structure of at least one enzyme from all these families will be known (42, 43). There is also an indirect pathway of aminoacyl-tRNA synthesis, tRNA-dependent amino acid modification (Fig. 1). This pathway relies on the acylation of tRNA with a “precursor” amino acid by a nondiscriminating AARS (33). Currently our knowledge of the discriminating versus nondiscriminating AARS is not advanced enough to deduce this property from their amino acid sequence alone. This “precursor” amino acid is then converted, while bound to tRNA, to the correct amino acid (matching the tRNA specificity) by a second, nonsynthetase enzyme, which recognizes only such a mischarged aminoacyl-tRNA species. Our current knowledge about the number and nature of these enzymes is still far from complete, but it is clear that in many organisms this is the essential and only way to form Asn-tRNA and Gln-tRNA (12, 13, 63).
FIG. 1.
Mechanisms of aminoacyl-tRNA formation. Both pathways, direct acylation and tRNA-dependent amino acid modification, are depicted for glutaminyl-tRNA formation. For example, E. coli uses glutaminyl-tRNA synthetase while B. subtilis employs Glu-tRNAGln amidotransferase for this purpose.
EVOLUTIONARY OVERVIEW OF AMINOACYL-TRNA SYNTHETASES
The assumption that translationally produced protein was a part of the very first translation mechanism raises the chicken-and-egg paradox. However, there can be little doubt that once translation did exist, proteins that facilitated tRNA charging would be among the first proteins to evolve, the selective advantage of their specificity being great. Thus, the evolutionary history of the current aminoacyl-tRNA synthetases must go deep into translation's past, to the emergence of the modern genetic code. The central role played by the AARSs in translation would suggest that their histories and that of the genetic code are somehow intertwined. This then raises the question of whether the AARSs in their evolution have contributed to the code's present structure; put another way, are the codon assignments simply reflections of AARS evolutionary wanderings? It is important that conjectures of this sort be examined in detail—and in a genomic era this can be done.
In an evolutionary sense, the most striking thing about the synthetases is the existence of the two distinct classes (3, 10, 14, 20). Common characteristic domain structures and sequence homologies define each class, but the two have nothing in common except the biochemistry of the reactions they catalyze (22): between the two classes, proteins show no structural resemblance, have almost no common motifs (see reference 49 for a possible exception), encounter the tRNA from different angles, and acylate the amino acid to different hydroxyl groups of the terminal ribose of the tRNA (57). This has been widely assumed to suggest that the tRNA-charging function evolved at least twice. In the origin of these two classes of tRNA-charging enzymes lies a clue to one of biology's deepest mysteries (45). Perhaps the two reflect a dichotomous origin of translation itself, in some sort of fusion between two different primitive processes, each associated with its own set of amino acids. Perhaps the two classes are the surviving traces of an ancient evolutionary battle between emerging tRNA-charging mechanisms as biology evolved beyond the RNA world. In any case, the existence of unrelated tRNA-charging systems must be considered a most telling evolutionary relic (45).
The aminoacyl-tRNA synthetases are distributed between the classes according to specific rules. Each class encompasses 10 of the amino acids, and all examples of a given amino acid's synthetase are of the same class, the so-called “class rule.” Within a class, all synthetases associated with a given amino acid are specifically related to one another to the exclusion of the AARSs associated with any other amino acids, the “monophyly rule.” A third, class-independent generalization is that for each organism, all tRNAs assigned to a given amino acid (so-called isoacceptors [50]) can be charged by a single synthetase, a rule that holds even for amino acids such as serine with two distinct sets of codons, UCN and AGY (reviewed in reference 39). Except possibly for the last, these rules have exceptions. The class rule and hence the monophyly rule are violated by lysine; in some organisms its synthetase is class I, while in others it is class II (32, 34, 35). Four more exceptions to the monophyly rule but not the class rule exist, involving glycine, serine, glutamic acid, and aspartic acid. For glycine and serine, each amino acid is associated with two synthetases (for both amino acids they are class II [37, 44]). However, the two enzymes in each case are not specifically related to one another, as the monophyly rule demands. This is most obvious for glycine, where the overall structures of the two enzymes are completely dissimilar, with one being a homodimer and the other being a heterotetramer. For the glutamyl- and aspartyl-tRNA synthetases, the violation of the monophyly rule is of a different nature. In each of these cases, all the synthetases associated with a particular amino acid constitute a related group. However, the synthetase for the amidated form of the amino acid (i.e., glutamine or asparagine) arises from within the same group, which then renders the parent grouping paraphyletic (4, 38, 46, 52, 53). Mention should also be made here of the charging system for cysteine, which breaks the class and monophyly rules in another way. In at least two organisms, the methanogens Methanococcus jannaschii and Methanobacterium thermoautotrophicum, neither a class I nor a class II cysteinyl-tRNA synthetase can be found in the genome, and the exact mechanism of Cys-tRNA formation (direct or indirect) has long remained a mystery. These exceptions to the class and monophyly rules do not rob the rules of their potential evolutionary significance. Erosion of the historical trace is the hallmark of evolution. The exceptions merely restrict what kinds of explanations can be given the rules.
The common general structure and sequence motifs shared by all members of a given synthetase class demand common ancestry and Darwinian descent. The later stages of this descent are captured in the sequence similarities among existing synthetases; importantly, their branching patterns recall structural similarities among the amino acids and patterns in the genetic code. To give examples: the ValRs and IleRS (class I) are impressively similar in sequence; this is not simply a matter of a sequence motif here and there (7). These sequences in turn are somewhat less similar to those of the LeuRS; the MetRSs then join the group at a still lower level of similarity (45). All four corresponding amino acids are nonpolar and aliphatic, and their codons all conform to the general composition NUN. Similarly, the class II enzymes for serine, threonine, proline, histidine, and glycine group phylogenetically and structurally (43). (Only one of the two unrelated forms of the GlyRS shows this specific relatedness, however [17].) The amino acids serine and threonine are obviously related structurally, and both are capable of forming an internal hydrogen-bonded five-membered ring structure that mimics the ring structure of the imino acid proline. However, histidine and glycine appear structurally unrelated to these three (and to one another). In their codons, the first three amino acids are also related; all conform to the general composition NCN, but the codons for histidine (CAY) and glycine (GGN) are not related to the others except, of course, to the CCY codons of proline in the former case.
The third major AARS grouping involves the class II synthetases for lysine, aspartic acid, and asparagine (14, 17, 52), all of which are closely related in structure (4). The amino acids aspartic acid and asparagine are obviously related, but lysine stands apart. In their codons, the three exhibit an overlapping kind of relatedness, with the Asn codons (AAY) being close to both their Asp (GAY) and Lys (AAR) counterparts whereas the last two sets are not closely related.
The close evolutionary relationship between the (class I) synthetases for glutamic acid and glutamine (mentioned above; see also reference 46) is mirrored in the obvious structural relationship between the corresponding amino acids and the relationship between their codons (GAR-Glu versus CAR-Gln). Finally, a pronounced similarity exists between the synthetases for the two aromatic amino acids tyrosine and tryptophan (6, 19), but their codons (UAY and UGG, respectively) are not closely related.
Although the existence of correlations between the genetic code and the evolutionary patterns of the AARSs is clear, their significance is not. Does the fact that the valine, isoleucine, leucine, and methionine enzymes came from a common ancestor mean that this ancestor itself could not distinguish among these amino acids or that the ancestor was able to specifically charge four separate aminoacyl-tRNAs? That seems absurd in the context of modern translation (45). A more acceptable explanation would seem to be that the AARS relationships reflect evolutionary replacement of one tRNA-charging enzyme or acylation system by another. Indeed, what we see here may be only the latest in a series of such evolutionary replacements, a series that traces far back into the code's past and an evolutionary process that still goes on today in a less radical form, involving replacements within the confines of a given amino acid type (see below).
ORDER IN THE GENETIC CODE
The significance of AARS evolution vis-à-vis that of the genetic code cannot be properly assessed without some appreciation of the nature and extent of the code's order. Within the last decade, significant strides have been made in this area. The so-called synonym order in the code, i.e., the degeneracy in codon assignment, which manifests itself almost exclusively in the third codon position, has never been in doubt, except as regards what caused it in the first place. However, such is not the case for the ordering that pertains to related amino acids. Although most biologists accept the existence of such an order, they have disagreed about its exact form, its extent, and its cause. Some have argued that the related amino acid order evolved to ameliorate the phenotypic consequences of mutations, an evolutionary scenario that would produce both synonym and related amino acid orderings (59). An alternative but conceptually related explanation is that the assignments have somehow been adjusted to minimize the consequences of errors in a primitive translation mechanism that was highly inaccurate (66). Seemingly, both error minimization models would lead to a very similar type of order in the code. However, a computer simulation study (26) showed that the assumptions of the first model are unlikely to lead to a synonym order in the code that is almost entirely confined to a single codon position—a type of order that is consistent with, if not predicted by, the second model (which also suggests the third codon position should be the degenerate one [66]). It has also been proposed that the form of the code was predetermined, at least in part, by specific interactions between amino acids and nucleic acids (reference 76 and references therein).
Perhaps the main difficulty in comprehending the code's related amino acid ordering is that amino acid relatedness is context dependent; amino acids that appear similar in one context can be unrelated in another. The amino acid replacement spectra of proteins prove the point: the replacement pattern can differ from position to position in a protein sequence for any amino acid. However, nobody knows what property or properties of the amino acids the code actually reflects.
One important advance in this area was the definition of an amino acid property called the polar requirement, which is a number derived from the paper chromatographic mobility of an amino acid in pyridine-water mixtures of various ratios (71). Simply plotting these numbers on a codon table (Table 1) reveals the existence of a remarkable degree of order, much of which would be unexpected on the basis of amino acid properties as normally understood. For example, codons of the form NUN define a set of five amino acids, all of which have very similar polar requirements. Likewise, the set of amino acids defined by the NCN codons all have nearly the same unique polar requirement. The codon couplets CAY-CAR, AAY-AAR, and GAY-GAR each define a pair of amino acids (histidine-glutamine, asparagine-lysine, and aspartic acid-glutamic acid, respectively) that has a unique polar requirement. Only for the last of these (aspartic and glutamic acids), however, would the two amino acids be judged highly similar by more conventional criteria. Perhaps the most remarkable thing about polar requirement is that although it is only a unidimensional characterization of the amino acids, it still seems to capture the essence of the way in which amino acids, all of which are capable of reacting in varied ways with their surroundings, are related in the context of the genetic code. Also of note is the fact that the context in which polar requirement is defined, i.e., the interaction of amino acids with heterocyclic aromatic compounds in an aqueous environment, is more suggestive of a similarity in the way amino acids might interact with nucleic acids than of any similarity in the way they would behave in a proteinaceous environment (70).
TABLE 1.
| U | C | A | G | ||
|---|---|---|---|---|---|
| Phe 5.0 | Ser 7.5 | Tyr 5.4 | Cys 4.8 | U | |
| U | Phe | Ser | Tyr | Cys | C |
| Leu | Ser | CT | CT | A | |
| Leu | Ser | CT | Trp 5.2 | G | |
| Leu 4.9 | Pro 6.6 | His 8.4 | Arg 9.1 | U | |
| Leu | Pro | His | Arg | C | |
| C | Leu | Pro | Gln 8.6 | Arg | A |
| Leu | Pro | Gln | Arg | G | |
| Ile 4.9 | Thr 6.6 | Asn 10.0 | Ser | U | |
| Ile | Thr | Asn | Ser | C | |
| A | Ile | Thr | Lys 10.1 | Arg | A |
| Met 5.3 | Thr | Lys | Arg | G | |
| Val 5.6 | Ala 7.0 | Asp 13.0 | Gly 7.9 | U | |
| Val | Ala | Asp | Gly | C | |
| G | Val | Ala | Glu 12.5 | Gly | A |
| Val | Ala | Glu | Gly | G |
Taken from reference 71.
The polar requirement is the number following the amino acid.
More recently, computer simulation studies have been used to try to assess the merit of polar requirement as an indicator of the code's related amino acid order is compared to other amino acid properties, how well ordered the code actually is, and the nature of the code's order. An appealingly straightforward approach to the problem was explored by Hurst and his colleagues (23, 28). In summary, they compared the natural code to a series of synthetic codes generated by randomly reassigning the 20 amino acids to the set of synonym codon categories that are defined by the natural code. Each code is then measured for how conservative it is with regard to a given amino acid property under “mutation”; i.e., each codon in a given code is compared to all other codons that are 1 base change removed from it, the numerical difference in that property between the amino acids corresponding to the original and the “mutated” codon is measured, and the squared differences are summed over the code as a whole or over each of the three codon positions individually. For all amino acid properties tested except one, the natural code was not notably superior to the random codes. That exception, polar requirement, revealed a natural code superior to all but 0.01% of the random codes (28). A subsequent, more refined simulation of this sort, which took transition-transversion ratios into account, showed the natural code was “one in a million” (23). There can be no doubt that when viewed in terms of amino acid polar requirements, the genetic code is a highly structured array. It would also seem that it has somehow been optimized to reduce the consequences of translational errors. However, the evolutionary dynamic that shaped the code remains a mystery.
While it must be admitted that the evolutionary relationships among the AARSs bear some resemblance to the related amino acid order of the code, it seems unlikely that they are responsible for that order (45): the evolutionary wanderings of these enzymes alone simply could not produce a code so highly ordered, in both degree and kind, as we now know the genetic code to be. These enzymes could at best be the agents through which other constraints acted to shape the code. However, even in such a capacity they would not be alone: the tRNAs offer a simple and facile alternative mechanism for changing codon assignments (65). It would seem, therefore, that the evolutionary patterns among the aminoacyl-tRNA synthetases do not imply a role for these enzymes in structuring the genetic code (45). The resemblance between their evolutionary patterns and the patterns seen in the code are a loose convergence, forced by the fact that both evolutions independently reflect somewhat similar properties of the amino acids. The evolutionary patterns in the AARSs do seem to represent evolutionary replacements that occurred against the background of an already established, or otherwise fashioned, code (45).
AMINOACYL-TRNA SYNTHETASE EVOLUTION: RECURRING GENE TRANSFER
If the AARSs do not reveal the code's evolution, what do their evolutionary relationships tell us? The answer is clear. Aminoacyl-tRNA synthetase evolution is a superb indicator of the evolutionary dynamic in general.
It should be noted that the AARSs are unique among components of the translation system in their evolutionary behavior. Starting with the rRNAs and continuing through the ribosomal proteins and the translation initiation and elongation factors runs one dominant evolutionary theme—molecules tend to show the same evolutionary history; i.e., their molecular phylogenies are consistent with the accepted overall organismal phylogeny. At the highest level, they tend to yield what we herein call the canonical phylogenetic pattern, which is basically a division of all life into the three primary groupings Bacteria, Archaea, and eukaryotes, with the closest relationship being between the Archaea and eukaryotes (73) (see below). The evolutionary picture painted by the synthetases, however, is a world apart from this canonical pattern. Not only do the phylogenies fail to yield the canonical pattern in a number of cases, but also they typically violate the accepted taxonomic structure within the organismal domains. Furthermore, the molecular phylogenies inferred from the synthetases of different amino acid types tend not to agree with one another—but this is the telling point.
Why should the synthetases show such atypical and disparate evolutionary pictures? The answer again is clear. The AARSs are in essence modular components of the cell; they function in isolation from the rest of the translation apparatus and from the rest of the cell, except for their individual contacts in each case with a small subset of the tRNAs (58). Because of this and because of their universality, the AARSs can function in a wide spectrum of cellular environments, often without disadvantage to the host. In other words, the AARSs are ideal candidates for widespread horizontal gene transfer, and the evidence certainly indicates this, since quite a few examples are known in which two different AARSs for the same amino acid coexist in the same organism. Versions of a given enzyme characteristic of the Archaea can be seen scattered among the bacterial taxa (see below). Versions characteristic of the eukaryotes have been seen in the Bacteria or in the Archaea. Within the Bacteria alone, the different bacterial subtypes of a given enzyme intermix among and within the taxa. There is no set pattern to all this; there is merely evidence consistent with frequent, widespread, indiscriminate horizontal gene transfer.
As suggested above, it is tempting to view the evolution of aminoacyl-tRNA synthesis as a study in horizontal gene transfer from top to bottom: at the deepest level, horizontal replacements involving the ancestors of the two synthetase classes, then replacements that gave rise to the phylogenetic structure within each class, and, finally, the replacements involving the different (modern) synthetases that use the same amino acid.
EVOLUTIONARY PROFILES OF THE INDIVIDUAL AMINOACYL-TRNA SYNTHETASES
We now examine in some detail the evolutionary profiles for each of the 20 aminoacyl-tRNA synthetases, with the principal objective of determining the extent to which each conforms to the canonical phylogenetic pattern (defined below) and asking what, if anything, the exceptions to canonical pattern tell us about these enzymes and about stages in the evolution of the cell.
The organisms mentioned in the figures and tables are listed in Table 2.
TABLE 2.
Organisms listed in figures and tables
| Systematic name | Systematic name | |
|---|---|---|
| Acinetobacter calcoaceticus | Methanosarcina barkeri | |
| Aeropyrum pernix | Methanococcus jannaschii | |
| Aquifex aeolicus | Methanococcus maripaludis | |
| Arabidopsis thaliana | Methanobacterium thermoautotrophicum ΔH | |
| Archaeoglobus fulgidus | Mycoplasma genitalium | |
| Azospirillum brasilense | Mycoplasma pneumoniae | |
| Bacillus subtilis | Mycobacterium tuberculosis | |
| Bartonella bacilliformis | Neisseria gonorrhoeae | |
| Borrelia burgdorferi | Nicotiana tabacum | |
| Bordetella pertussis | Nosema locustae | |
| Caenorhabditis elegans | Oryza sativa | |
| Campylobacter jejuni | Plasmodium falciparum | |
| Caulobacter crescentus | Porphyromonas gingivalis | |
| Cenarchaeum symbiosum | Pseudomonas aeruginosa | |
| Chlorobium tepidum | Pseudomonas fluorescens | |
| Chlamydia trachomatis | Pyrobaculum aerophilum | |
| Clostridium acetobutylicum | Pyrococcus furiosus | |
| Clostridium longisporum | Pyrococcus horikoshii | |
| Corynebacterium glutamicum | Rhizobium meliloti | |
| Coxiella burnetii | Rhodobacter capsulatus | |
| Deinococcus radiodurans | Rickettsia prowazekii | |
| Drosophila melanogaster | Staphylococcus aureus | |
| Escherichia coli | Streptomyces coelicolor | |
| Enterococcus faecalis | Streptococcus pneumoniae | |
| Francisella tularensis | Streptococcus pyogenes | |
| Giardia intestinalis | Sulfolobus solfataricus | |
| Giardia lamblia | Synechocystis sp. strain PCC 6803 | |
| Haemophilus influenzae | Tetrahymena thermophila | |
| Halobacterium salinarum | Thermus aquaticus | |
| Halococcus marismortui | Thermotoga maritima | |
| Helicobacter pylori | Thermus thermophilus | |
| Homo sapiens | Thiobacillus ferrooxidans | |
| Lactobacillus bulgaricus | Treponema pallidum | |
| Lactobacillus delbrueckii | Trichomonas vaginalis | |
| Lactococcus casei | Saccharomyces cerevisiae | |
| Lupinus luteus | Zymomonas mobilis |
The analysis presented is a synthesis of four approaches: (i) conventional phylogenetic trees (see Fig. 2 caption); (ii) visual inspection of alignments to reveal qualitative differences not apparent from the other analyses; (iii) dipeptide similarity matrices (see Table 3 footnotes); and (iv) signature analysis. Signatures are defined in terms of positions in the alignment wherein at least 80% of the members of a given group show a constant composition but one that is found elsewhere in the alignment no more that once within some larger phylogenetic taxonomic context. For example, spirochete signatures would usually be relative to all other bacterial groups but not relative to the more distantly related archaeal and eukaryotic versions of the enzyme.
FIG. 2.
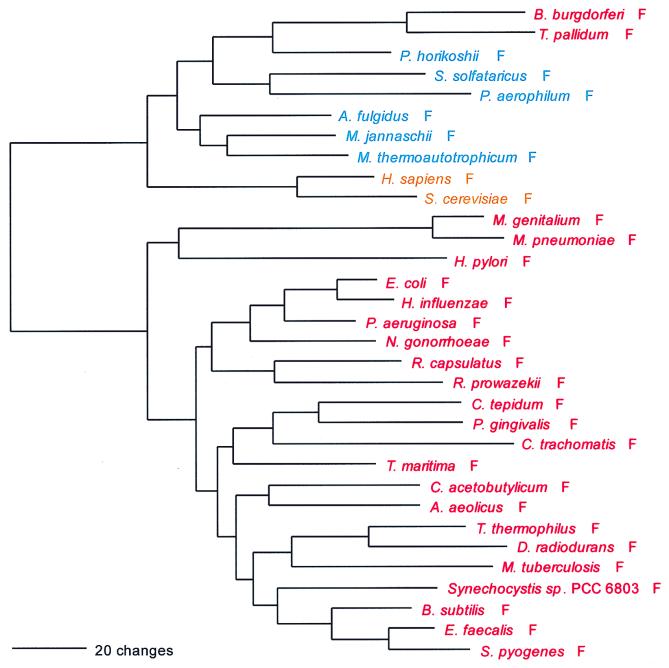
Phylogenetic tree of PheRS sequences. Aligned protein sequences were evaluated for the 1,000 most parsimonious trees (61), using amino acid replacement costs based on the BLOSUM 45 matrix (31, 41). Of these trees, we retained the topology with the maximum likelihood of giving rise to the data under the JTT model in protml version 2.2 of the MOLPHY package (1). Additional optimization was performed by removing sequences, one or more at a time, from the tree and using maximum likelihood to select the best from among the 100 to 500 most parsimonious alternative placements and to assign lengths to the branches. The tree has been rooted between the Bacteria and the Archaea plus Eukarya. The sequence identifiers correspond to an organism defined in Table 2, followed by the one-letter amino acid code (in this case F). Sometimes suffixes 1 and 2 are used; they refer to the fact that the given organism contains more than one specific AARS. Bacteria are shown in red, Archaea in blue, and eukaryotes in yellow.
TABLE 3.
Dipeptide similarity matrix for a representative sampling of taxa from the PheRS sequence alignmenta
| Organism | Similarity to:
|
||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | |
| 1. E. coli | |||||||||||||||
| 2. R. prowazeckii | 15.3 | ||||||||||||||
| 3. B. subtilis | 18.8 | 11.7 | |||||||||||||
| 4. C. tepidum | 13.5 | 10.4 | 15.8 | ||||||||||||
| 5. D. radiodurans | 13.9 | 8.0 | 10.6 | 11.4 | |||||||||||
| 6. T. maritima | 15.6 | 11.4 | 13.0 | 14.4 | 12.4 | ||||||||||
| 7. A. aeolicus | 15.0 | 10.3 | 15.8 | 13.5 | 11.3 | 17.6 | |||||||||
| 8. B. burgdorferi | 3.3 | 3.5 | 4.3 | 3.9 | 4.7 | 5.8 | 3.1 | ||||||||
| 9. P. horikoshii | 5.4 | 5.4 | 4.5 | 5.7 | 2.8 | 5.1 | 4.5 | 14.3 | |||||||
| 10. M. jannaschii | 5.2 | 7.1 | 4.0 | 7.2 | 6.0 | 6.7 | 4.4 | 12.4 | 25.0 | ||||||
| 11. M. thermoautotrophicum | 6.0 | 4.0 | 4.2 | 5.5 | 4.7 | 3.7 | 5.6 | 11.6 | 20.4 | 26.3 | |||||
| 12. A. fulgidus | 5.1 | 4.3 | 5.5 | 6.0 | 5.2 | 6.2 | 6.7 | 12.3 | 19.6 | 20.4 | 24.7 | ||||
| 13. S. solfataricus | 4.7 | 5.2 | 5.1 | 7.7 | 3.5 | 6.7 | 4.4 | 12.7 | 14.2 | 16.7 | 17.1 | 12.6 | |||
| 14. A. pernix | 4.5 | 2.7 | 3.3 | 6.7 | 5.0 | 4.6 | 5.1 | 8.8 | 12.9 | 16.2 | 15.1 | 14.3 | 21.0 | ||
| 15. S. cerevisiae | 2.3 | 2.9 | 2.0 | 2.3 | 2.8 | 2.5 | 2.6 | 7.1 | 6.5 | 9.5 | 8.9 | 8.7 | 9.1 | 7.7 | |
| 16. C. elegans | 2.3 | 3.3 | 2.5 | 3.9 | 3.7 | 4.1 | 4.1 | 7.0 | 9.9 | 11.2 | 11.5 | 12.3 | 12.1 | 10.2 | 27.0 |
A dipeptide similarity matrix resembles a normal amino acid similarity matrix except that adjacent columns in the alignment are considered in pairs (dipeptides), not singly. Only dipeptide identities between sequences are scored. The rationale for using two adjacent columns as the unit measured is that doing so takes the actual sequence of the molecule into account whereas treating the columns singly does not (i.e., shuffling columns does not change the results of the analysis in the latter case but does so in the former).
Because the canonical evolutionary pattern is central to our thesis and because the tRNA-charging enzymes exhibit different partial forms of that pattern, it is necessary to begin by explaining clearly what we mean by the phrase. In its essence (which we will call the basal canonical pattern), the canonical pattern is defined by the relationship between the bacterial and archaeal versions of a given molecule. For the basal canonical pattern to hold, regardless of how many subtypes of a given protein exist, it must be possible to distinguish strongly between characteristic bacterial and archaeal versions of the molecule. This distinction should be a pronounced quantitative one (on the level of sequence similarities) and/or a qualitative one (evident in terms of gross areas in a sequence alignment wherein homology between the two is only weakly evident or nonexistent). In other words, for these two organismal domains, the interdomain differences between the characteristic archaeal and bacterial proteins must far outweigh any intradomain differences: the two must appear to differ in genre. For the full canonical pattern to hold, there must also then exist a characteristic eukaryotic version(s) of the molecule that is distinguishable from both the archaeal and the bacterial versions but which is clearly of the archaeal genre. Tables 3 and 4 are representative dipeptide similarity matrices for two aminoacyl-tRNA synthetases typical of those showing canonical pattern (PheRS and TyrRS), while Tables 5 and 6 are matrices for enzymes (SerRS and CysRS) that do not show canonical pattern.
TABLE 4.
Dipeptide similarity for a representative sampling of taxa from the TyrRS sequence alignmenta
| Organism | Similarity to:
|
|||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | 16 | |
| 1. E. coli | ||||||||||||||||
| 2. B. subtilis 1 | 35.3 | |||||||||||||||
| 3. R. prowazeckii | 16.7 | 16.7 | ||||||||||||||
| 4. B. burgdorferi | 18.1 | 21.5 | 20.9 | |||||||||||||
| 5. H. influenzae | 9.9 | 9.3 | 9.0 | 11.5 | ||||||||||||
| 6. H. pylori | 11.0 | 9.8 | 10.4 | 12.8 | 34.0 | |||||||||||
| 7. A. aeolicus | 11.4 | 11.2 | 10.1 | 11.6 | 35.9 | 36.9 | ||||||||||
| 8. B. subtilis 2 | 9.2 | 8.2 | 8.6 | 11.8 | 29.6 | 24.6 | 25.6 | |||||||||
| 9. D. radiodurans | 9.7 | 7.9 | 6.8 | 9.5 | 33.0 | 24.6 | 27.5 | 22.5 | ||||||||
| 10. M. jannaschii | 2.9 | 3.4 | 3.8 | 3.0 | 8.4 | 6.3 | 8.7 | 6.2 | 6.6 | |||||||
| 11. A. fulgidus | 3.8 | 3.9 | 2.7 | 3.6 | 8.2 | 7.8 | 8.9 | 8.4 | 6.1 | 39.0 | ||||||
| 12. P. horikoshii | 3.0 | 1.5 | 3.9 | 1.9 | 5.6 | 4.1 | 5.1 | 7.1 | 4.4 | 21.9 | 20.8 | |||||
| 13. P. aerophilum | 2.6 | 3.4 | 4.2 | 3.1 | 3.8 | 4.1 | 4.3 | 3.0 | 2.9 | 22.6 | 16.0 | 28.1 | ||||
| 14. S. solfataricus | 1.8 | 3.3 | 5.3 | 3.5 | 8.2 | 6.8 | 9.0 | 8.0 | 6.6 | 23.5 | 21.4 | 22.6 | 20.8 | |||
| 15. N. tabacum | 1.1 | 2.3 | 1.5 | 2.4 | 3.4 | 4.6 | 4.4 | 4.3 | 4.8 | 14.4 | 13.8 | 15.4 | 14.9 | 18.2 | ||
| 16. H. sapiens | 1.7 | 1.4 | 1.8 | 2.2 | 3.5 | 2.8 | 4.7 | 3.8 | 3.3 | 15.8 | 17.1 | 11.9 | 8.1 | 9.9 | 10.0 | |
| 17. S. cerevisiae | 2.1 | 2.1 | 1.4 | 2.2 | 5.8 | 5.5 | 4.9 | 4.3 | 3.7 | 16.9 | 18.8 | 14.7 | 9.3 | 12.3 | 11.3 | 35.4 |
For details, see Table 3 footnote a.
TABLE 5.
Dipeptide similarity for a representative sampling of taxa from the SerRS sequence alignmenta
| Organism | Similarity to:
|
||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | |
| 1. E. coli | |||||||||||||||
| 2. H. pylori | 25.8 | ||||||||||||||
| 3. B. subtilis | 33.2 | 24.9 | |||||||||||||
| 4. P. gingivalis | 19.8 | 17.7 | 22.2 | ||||||||||||
| 5. A. aeolicus | 35.6 | 27.5 | 41.8 | 22.2 | |||||||||||
| 6. B. burgdorferi | 10.1 | 13.2 | 13.9 | 10.2 | 13.4 | ||||||||||
| 7. H. influenzae | 19.0 | 13.1 | 21.0 | 14.0 | 18.8 | 13.3 | |||||||||
| 8. S. cerevisiae 2 | 11.0 | 9.3 | 11.9 | 10.4 | 14.1 | 16.1 | 8.2 | ||||||||
| 9. H. marismortui | 23.3 | 17.5 | 31.2 | 22.2 | 27.4 | 12.3 | 19.2 | 12.9 | |||||||
| 10. A. fulgidus | 14.4 | 10.8 | 20.8 | 12.7 | 18.8 | 19.2 | 14.8 | 14.7 | 13.2 | ||||||
| 11. P. horikoshii | 17.0 | 16.9 | 23.6 | 14.4 | 24.3 | 22.6 | 18.7 | 19.8 | 19.3 | 34.8 | |||||
| 12. P. aerophilum | 13.0 | 11.0 | 19.2 | 9.4 | 19.5 | 14.2 | 11.2 | 10.4 | 10.5 | 29.3 | 24.1 | ||||
| 13. A. pernix | 12.3 | 11.6 | 14.8 | 9.0 | 15.3 | 17.8 | 12.0 | 14.4 | 11.7 | 32.7 | 30.0 | 29.8 | |||
| 14. A. thaliana | 13.5 | 9.4 | 16.5 | 10.6 | 17.8 | 14.6 | 13.8 | 9.5 | 11.9 | 22.6 | 22.6 | 16.1 | 16.4 | ||
| 15. H. sapiens | 13.3 | 9.2 | 12.6 | 8.2 | 13.8 | 13.9 | 9.8 | 9.5 | 9.6 | 18.5 | 18.4 | 13.6 | 18.1 | 33.8 | |
| 16. S. cerevisiae 1 | 10.4 | 7.3 | 10.5 | 9.2 | 15.3 | 12.9 | 8.5 | 8.5 | 9.2 | 17.3 | 18.2 | 13.3 | 12.5 | 34.9 | 29.4 |
For details, see Table 3 footnote a.
TABLE 6.
Dipeptide similarity for a representative sampling of taxa from the CysRS sequence alignmenta
| Organism | Similarity to:
|
|||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | 16 | |
| 1. E. coli | ||||||||||||||||
| 2. B. subtilis | 27.5 | |||||||||||||||
| 3. Synechocystis 6803 | 24.6 | 26.7 | ||||||||||||||
| 4. T. maritima | 25.9 | 27.6 | 22.0 | |||||||||||||
| 5. A. aeolicus | 25.8 | 27.0 | 22.8 | 29.3 | ||||||||||||
| 6. B. burgdorferi | 16.2 | 13.6 | 9.9 | 12.7 | 14.1 | |||||||||||
| 7. P. gingivalis | 22.3 | 21.5 | 17.4 | 19.6 | 22.2 | 15.5 | ||||||||||
| 8. D. radiodurans | 24.3 | 23.7 | 20.9 | 20.9 | 22.4 | 14.1 | 31.4 | |||||||||
| 9. C. trachomatis | 21.2 | 20.5 | 18.9 | 14.7 | 19.0 | 19.7 | 17.1 | 14.5 | ||||||||
| 10. M. barkeri | 21.0 | 21.6 | 19.0 | 18.5 | 18.9 | 15.6 | 15.5 | 18.6 | 22.7 | |||||||
| 11. P. horikoshii | 29.1 | 30.1 | 28.9 | 31.5 | 29.9 | 12.6 | 23.9 | 24.7 | 20.0 | 23.6 | ||||||
| 12. M. maripaludis | 26.1 | 27.8 | 24.1 | 25.8 | 27.8 | 13.9 | 19.5 | 21.6 | 17.9 | 22.2 | 45.7 | |||||
| 13. A. fulgidus | 26.5 | 26.2 | 23.5 | 24.8 | 26.6 | 12.7 | 20.1 | 19.7 | 19.2 | 19.2 | 31.6 | 27.1 | ||||
| 14. C. symbiosum | 23.5 | 24.2 | 22.1 | 23.1 | 25.8 | 11.1 | 17.9 | 21.6 | 19.1 | 22.4 | 29.8 | 28.0 | 29.1 | |||
| 15. P. aerophilum | 22.2 | 20.6 | 20.1 | 25.8 | 27.4 | 11.1 | 17.9 | 17.4 | 15.2 | 18.7 | 26.6 | 26.2 | 23.3 | 21.4 | ||
| 16. H. sapiens | 17.9 | 18.3 | 19.0 | 16.8 | 18.7 | 10.7 | 16.6 | 16.5 | 14.6 | 16.5 | 20.2 | 21.3 | 16.6 | 18.6 | 14.9 | |
| 17. S. cerevisiae | 16.9 | 19.3 | 17.9 | 18.2 | 18.6 | 9.6 | 15.6 | 16.5 | 13.1 | 15.0 | 19.7 | 20.2 | 18.2 | 18.4 | 16.8 | 32.0 |
For details, see Table 3 footnote a.
The aminoacyl-tRNA synthetases are considered individually below in an order defined by their corresponding codons. We have not included most of the mitochondrial data in the analysis, because doing so would add nothing to our conclusions and would needlessly complicate an already complex picture (27, 29).
Synthetases for the NUN-Encoded Amino Acids
Phe; UUY; class II; tetramer of α- and β-subunits.
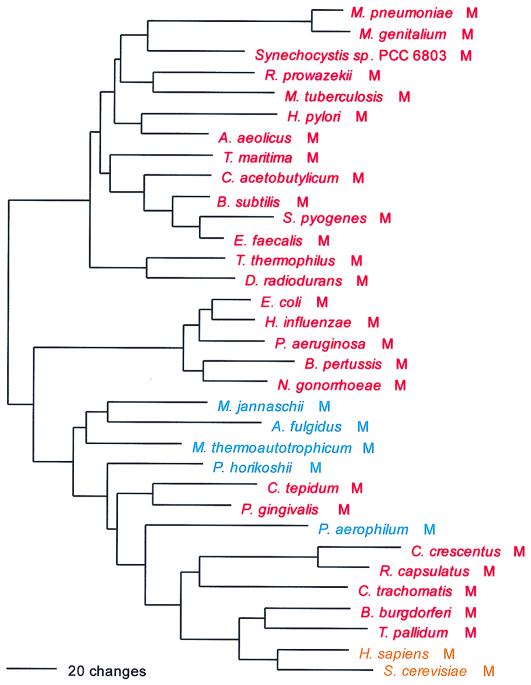
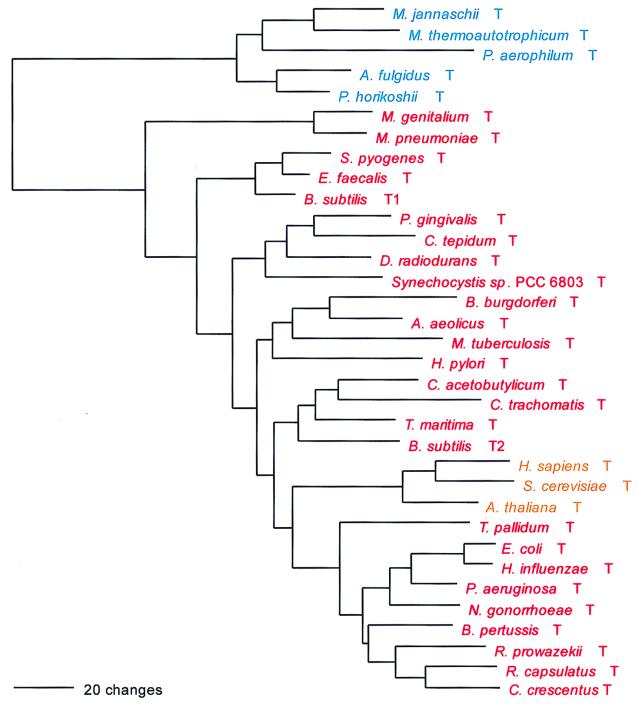
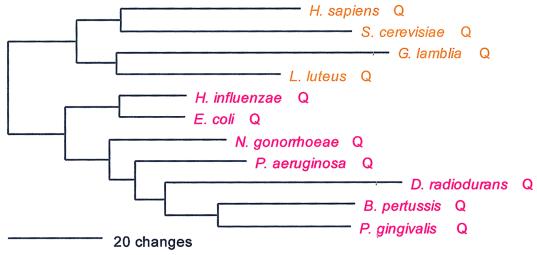
PheRS is the only class II synthetase in the NUN codon group, and it has no close relatives within that class. Not surprisingly, both the α- and β-subunits present the same evolutionary picture; their sequences are combined to produce Fig. 2. PheRS shows the classical full canonical pattern, the only exception being the spirochete PheRSs, which are of the archaeal, not the bacterial genre, and which seem to be specifically related to the Pyrococcus PheRS within that grouping, as sequence signature analysis suggests and Fig. 2 confirms.
For both the α- and β-subunits of PheRS, significant length differences distinguish the bacterial subunits from their archaeal counterpart. The bacterial α-subunit is about 120 amino acids shorter than the archaeal/eukaryotic α-subunit at its N terminus, and the first 90 amino acids of the bacterial sequence show little or no similarity to the archaeal/eukaryotic counterpart. However, for the β-subunit, the bacterial version is the longer, by approximately 250 amino acids. At both termini the bacterial version of the β-subunit extends beyond the archaeal/eukaryotic version by about 100 amino acids; in the N-terminal ∼50 amino acids, the archaeal version of the β-subunit shows no recognizable similarity to its bacterial counterpart. In addition, large sequence gaps distinguish the two genres in the interior of the β-subunit.
Leu; UUR and CUN; class I; monomer.
LeuRS conforms to the full canonical pattern as well, in this case without exception. A striking lack of similarity in various regions of the molecule distinguishes the bacterial and archaeal genres of LeuRS, and a number of sizable insertion and deletion differences distinguish the two genres throughout the alignment. A nearly total lack of sequence similarity between the two is seen in the C-terminal (KMSK) section of the molecule.
Within the Bacteria, however, the accepted phylogenetic relationships are not all preserved—at least two distinct bacterial subtypes of the molecule exist and have obviously migrated horizontally. The best-defined bacterial subtype (by all methods of analysis) is that common to the majority of gram-positive bacteria (and relatives), the spirochetes, chlamydias, and the Cytophaga-Chlorobium grouping (represented by Chlorobium tepidum and Porphyromonas gingivalis). However, this grouping fails to include Clostridium acetobutylicum, a gram-positive species whose LeuRS groups with that of Deinococcus in Fig. 3 (a relationship supported by sequence signature). On the other hand, the proteobacteria (Escherichia coli and relatives) do form a grouping quite consistent with their established phylogeny (Fig. 3).
FIG. 3.
Phylogenetic tree of LeuRS sequences. The tree was rooted using ValRS, IleRS, and MetRS sequences. Other details are as in Fig. 2.
Ile; AUH; class I; monomer.
IleRS also shows the full canonical pattern. As with LeuRS, this fact is obvious upon visual inspection of the alignment, especially its C-terminal section, wherein the bacterial and archaeal genres exhibit very little sequence similarity and show major alignment gaps relative to one another. However, as all methods of analysis clearly show, a sizable minority of bacterial taxa possess an IleRS of the archaeal rather than the bacterial genre (Fig. 4). All of these bacterial examples are specifically related to their eukaryotic counterparts, with the closest relationship being between the eukaryotes and a bacterial subgroup comprising the spirochetes, chlamydias, Mycobacterium, and Rickettsia. Note in Fig. 4 the specific relationship between the IleRS of Mycobacterium and that of Rickettsia, which is strongly suggested by sequence signature as well. Also note the relationship between the C. acetobutylicum IleRS and the plasmid-borne IleRS found in mupirocin-resistant strains of Staphylococcus aureus; this relationship is also supported by sequence signature.
FIG. 4.
Phylogenetic tree of IleRS sequences. The tree has been rooted using ValRS, LeuRS, and MetRS sequences. Other details are as in Fig. 2.
Met; AUG; class I; homodimer.
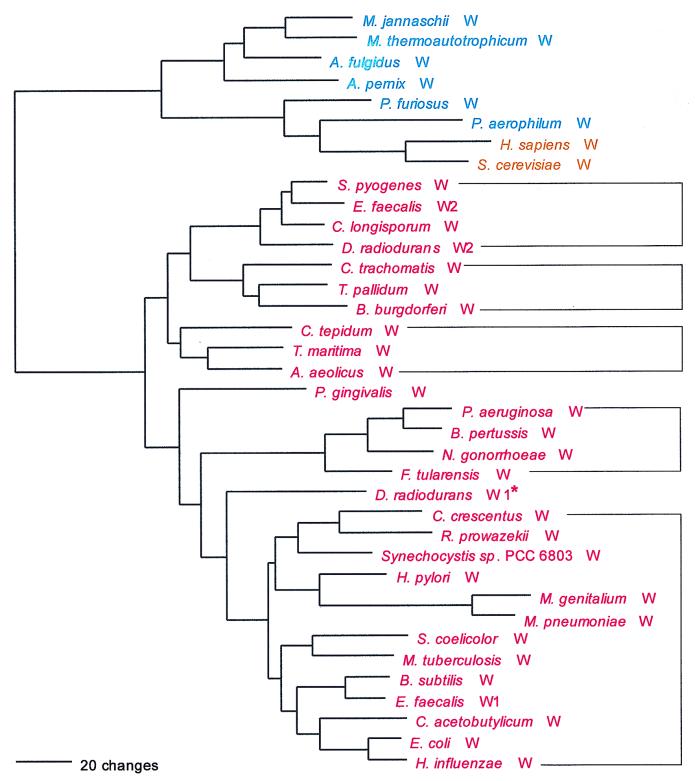
Methionine presents one of the more complex evolutionary profiles among the aminoacyl-tRNA synthetases. The enzyme marginally shows the canonical picture: the majority of bacterial examples—the group represented by Helicobacter in Fig. 5—define a bacterial genre, while the archaea, eukaryotes and a number of bacterial MetRSs constitute the archaeal genre (Fig. 5). However, there is another bacterial grouping, confined to the β and γ proteobacteria, which is of the archaeal genre (Fig. 5). The difference between the bacterial and archaeal genres of MetRS is not as extreme as that seen for the other members of the NUN codon group. However, one large alignment gap (∼25 amino acids) separates the bacterial genre from all others (the latter appear to contain a metal binding region at this point, the consensus sequence of which is CP . C . . . . . a . gD . C . . C . . . . . . . . . . L (where lowercase signifies its presence in only four of the five groupings involved). A strong signature distinguishes the bacterial genre from the others, and its distinctiveness is also evident in a dipeptide similarity matrix.
FIG. 5.
Phylogenetic tree of MetRS sequences. The tree has been rooted using LeuRS, IleRS, and ValRS sequences. Other details are as in Fig. 2.
Relationships within the archaeal genre are themselves complex. The closest relatives of the eukaryotic MetRSs are those of the spirochetes (convincingly demonstrated by sequence signature). The archaea appear paraphyletic; the crenarchaeal examples do not group with their euryarchaeal counterparts to the exclusion of all the bacterial examples (Fig. 5). MetRSs of the bacterial genre (the group represented by Helicobacter) present a mixed phylogenetic picture. The low-G+C gram-positive Bacteria (Bacillus and relatives) cluster well. (Although Fig. 5 does not indicate it, by signature analysis the mycoplasmas do seem to be a part of this grouping.) However, the high-G+C gram-positive representative, Mycobacterium MetRS, falls elsewhere within the tree, and again shows a clear specific relationship to the MetRS of Rickettsia; this is also supported by sequence signature. (Note that the rickettsial MetRS is of a different genre from the MetRSs of the other α proteobacterial representatives.)
The C-terminal domain of MetRS, about 150 amino acids in length, can take one of three forms: (i) it can be covalently linked to the rest of the molecule, as in most bacteria and most archaea; (ii) it can be completely missing, as in a number of bacteria, e.g., cyanobacteria and mycoplasmas; or (iii) it can be present but not covalently linked to the rest of the molecule, as in all eukaryotes (except Caenorhabditis elegans, where it is covalently linked), in Aquifex, and in the Crenarchaeota. In eukaryotes, this separate protein, known as Arc1p, occurs as a part of some higher-order complexes involving eukaryotic synthetases, wherein it is involved in amino acid recognition (54). The C termini of these proteins extend about 60 amino acids beyond the normal C-terminus of MetRS. It is interesting that the spirochete MetRSs (in which the C terminus is a covalently linked part of the molecule) also extend beyond the normal C terminus of MetRS, and in this extension, they show homology to sequences in the Arc1p family, providing further support for a specific relationship between the spirochete and eukaryote enzymes (Fig. 5).
Arc1p-like domains can be seen in a few other aminoacyl-tRNA synthetases as well. Approximately 100 residues of the N terminus of the β-subunit of bacterial PheRS is homologous to a portion of Arc1p. Mammalian TyrRS (only) has appended to its C terminus a more extensive homolog, which is impressively similar to the MetRS extensions just discussed. Also, it has been demonstrated in the mammalian TyrRS case that the extension functions not as Arc1p (i.e., in amino acid recognition) but as a cytokine (64).
Val; GUN; class I; monomer.
The valine-charging enzyme conforms only to the basal canonical pattern; the eukaryotic ValRSs are not archaeal in nature but obviously bacterial (Fig. 6). Also, within the bacterial group a 37-amino-acid insertion in the alignment found only in the eukaryotes and α, β, and γ proteobacteria suggests a specific relationship among them. The distinction between the archaeal and bacterial genres of ValRS is again a strong one and is manifested most strongly in the C-terminal (KMSK) portion of the molecule. The rickettsial ValRS, alone among the bacterial examples, is of the archaeal genre, seemingly specifically related therein to the ValRS of the crenarchaeon Pyrobaculum aerophilum; this relationship is supported by sequence signature.
FIG. 6.
Phylogenetic tree of ValRS sequences. The tree was rooted using IleRS, MetRS, and LeuRS sequences. Other details are as in Fig. 2.
Synthetases for the NCN-Encoded Amino Acids
Serine, threonine, and proline have related structures, codons, and aminoacyl-tRNA synthetases; in this last respect, the group also encompasses histidine and glycine. (However, as mentioned above, only one of the two unrelated GlyRS forms shows the relationship.)
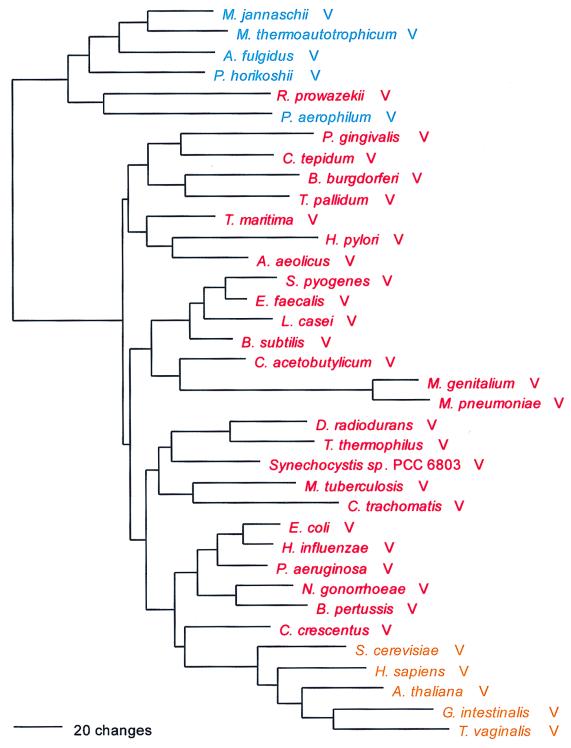
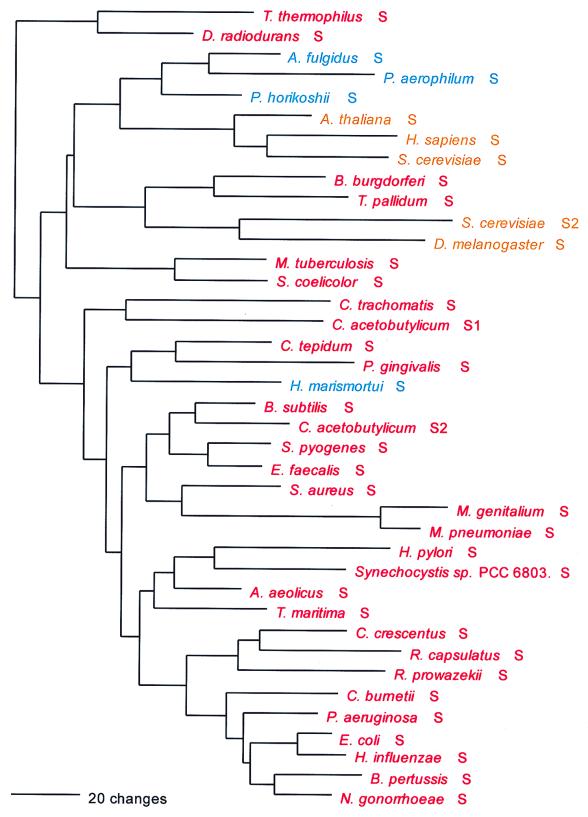
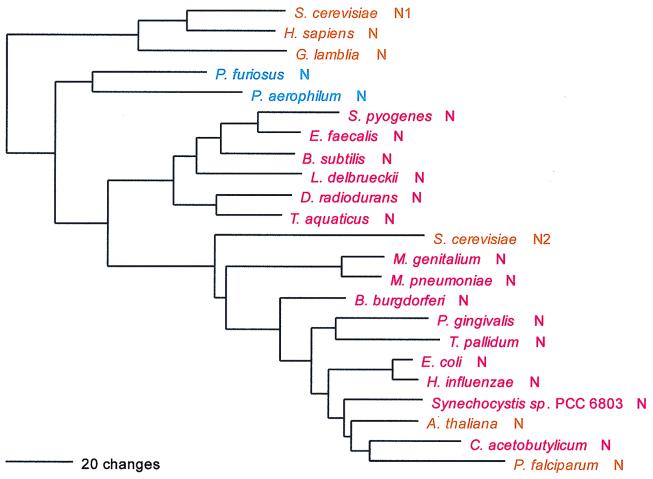
Ser; UCN and AGY; class II; homodimer.
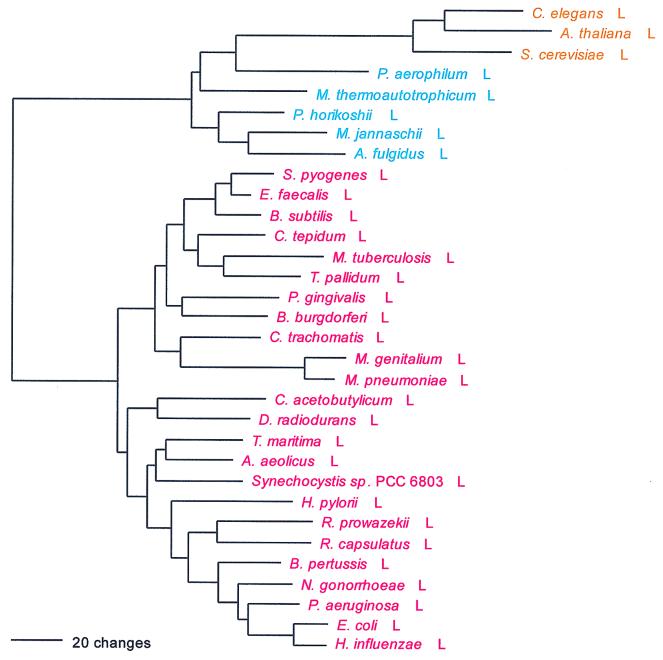
The seryl-tRNA synthetase is of particular interest for two reasons: (i) it clearly fails to conform to the canonical pattern, and (ii) there are two distinct serine-charging enzymes, a very rare form that has been found so far only in M. thermoautotrophicum and the two Methanococcus species examined and a major form that has been found in all other organisms. Although both the major and minor forms of SerRS belong to the above-mentioned Ser-Thr-Pro supercluster, it is unclear whether the two are specifically related to one another therein. (The minor form is not included in Fig. 7.)
FIG. 7.
Phylogenetic tree of SerRS sequences. The tree has been rooted using ThrRS sequences. The unusual SerRSs of the methanogens, which may not be related to other SerRSs, have not been used in constructing the figure. Other details are as in Fig. 2.
Although one can see an archaeal and an eukaryotic grouping in Fig. 7 and the two are specifically related, the true canonical pattern is not exhibited. For example, no alignment gaps separate the archaeal from the bacterial type, and intergroup dipeptide similarities are not strikingly lower than intragroup similarities in general (Table 5). There is considerable evidence suggestive of SerRS horizontal gene transfers. The halobacterial SerRS, for example (62), is not related to other archaeal examples but almost certainly is bacterial in origin, apparently stemming from the group that comprises Porphyromonas and Chlorobium. Two unrelated eukaryotic SerRS groups exist, one of them seemingly related to the main group of archaeal SerRSs and the other (which comprises the Drosophila, plant, and second yeast SerRSs) specifically related to the spirochete SerRSs. Since these latter eukaryotic SerRSs—all clearly related by signature sequence—seem to be mitochondrial, their relationship to spirochetes rather than proteobacteria becomes of interest.
Pro; CCN; class II; homodimer.
ProRS exhibits the full canonical pattern but again with exceptions. The bacterial genre is distinguished from the archaeal by having an insertion of about 180 amino acid residues not seen in the latter at approximate (E. coli) position 190, while the latter extends at the C terminus of the molecule for about 70 residues beyond the former. Dipeptide similarities between the two genre are remarkably low.
The ProRSs of a few bacterial taxa, i.e., the mycoplasmas, Deinococcus, Chlorobium, Porphyromonas, and Borrelia (but not Treponema), are of the archaeal genre, and the eukaryotic enzymes (with the exception of that from Giardia) are included in this phylogenetic grouping; sequence signature analysis shows a sister relationship therein to the genera Borrelia, Chlorobium, and Porphyromonas, which the Fig. 8 tree confirms.
FIG. 8.
Phylogenetic tree of ProRS sequences. The tree was rooted using ThrRS, HisRS, and GlyRS sequences. Other details are as in Fig. 2.
Thr; ACN; class II; homodimer.
Like its valyl counterpart, ThrRS exhibits only the basal canonical pattern, with the eukaryotic versions of the enzyme being bacterial rather than archaeal in nature. The bacterial and archaeal genres are readily distinguished by sizable additions/deletions in the N-terminal ∼250 amino acids or so of the alignment, and evidence of similarity between the two in this portion of the molecule is minimal (Fig. 9). Two of the three available crenarchaeal ThrRSs add a further complication to the picture (see below).
FIG. 9.
Phylogenetic tree of ThrRS sequences. The tree has been rooted using ProRS, GlyRS, and HisRS sequences. Other details are as in Fig. 2.
The bacterial ThrRSs break into subtypes, subtly distinguished from one another yet very evident from the fact that they violate established organismal phylogenies and the fact that B. subtilis possesses two ThrRSs, one of each subtype. Among these taxonomic violations are (i) a grouping of Thermotoga with C. acetobutylicum and one of the two B. subtilis ThrRSs; (ii) a cluster comprising Borrelia, Aquifex, Mycobacterium, and (probably) Helicobacter; and (iii) the clustering of Treponema pallidum with the proteobacteria.
Among the archaea, a close specific relationship is seen between the Pyrococcus and Archaeoglobus ThrRSs, as well as between those of the two methanogens. However, the most striking feature of the archaeal enzymes is that two crenarchaeal examples, Sulfolobus and Aeropyrum, are highly atypical in that for a stretch of about 330 amino acid residues beginning at approximate position 150 (M. jannaschii numbering) these two contain no more than 150 residues in this region, which exhibit no detectable homology to any other sequences in the ThrRS alignment. However, in both cases, a second, unlinked ThrRS-related gene exists that basically covers the region in question (plus a bit more), shows homology therein to other sequences in the alignment, and has by far the highest similarity to the bacterial, not the archaeal, versions. Note, however, that this strange chimeric type of ThrRS is not found in a third crenarchaeon, Pyrobaculum. (The second peptide of the Sulfolobus and Aeropyrum ThrRSs has not been used in the calculations upon which Fig. 9 is based.)
Ala; GCN; class II; homotetramer.
Although a class II enzyme, AlaRS is not a member of the supercluster that contains the other NCN-associated synthetases. The archaeal and bacterial forms of the enzyme are clearly distinguished by dipeptide similarities, sequence signature, and a few small but significant insertions and deletions in the alignment; the N terminus of the archaeal form also begins some 50 amino acids before the bacterial one does (Fig. 10).
FIG. 10.
Phylogenetic tree of AlaRS sequences. The tree has been rooted between the bacterial sequences and the archaeal (plus Giardia) sequences. Other details are as in Fig. 2.
Although the canonical pattern holds for the AlaRS, it is only the basal canonical pattern, since the eukaryotic AlaRSs (except for that of Giardia) cluster with the bacterial AlaRSs; and within that grouping they appear to be specifically related to the Chlorobium-Porphyromonas cluster (Fig. 10), a relationship that is supported by sequence signature. The Giardia AlaRS, however, is of the archaeal genre. This is confirmed by a strong sequence signature, which is also consistent with Giardia's position in Fig. 10 as an outgroup to the archaeal clade. The spirochete AlaRSs, although clearly of the bacterial genre, are highly derived. They both show two characteristic large deletions, one interior and the other C-terminal.
Synthetases for the NAN-Encoded Amino Acids
Tyr; UAY; class I; homodimer.
The TyrRS makes a strong canonical distinction (Fig. 11 and Table 4). In the C-terminal (KMSK) section of the molecule there is very little similarity between the TyrRSs of the bacterial and archaeal genres, and a number of insertion-deletion differences distinguish the two throughout the molecule as well.
FIG. 11.
Phylogenetic tree of TyrRS sequences. The tree has been rooted using TrpRS sequences. Other details are as in Fig. 2.
Two distinct subtypes of bacterial TyrRS can be seen, and these distinguish members of various taxa from one another. Among the enteric-vibrio subgroup of the γ proteobacteria, E. coli, Salmonella, and Yersinia exhibit the first type while Haemophilus, Actinobacillus, and Vibrio exhibit the second. Among the β proteobacteria, Neisseria exhibits the first type while Bordetella and Thiobacillus exhibit the second. Porphyromonas and its relative Chlorobium are phylogenetically split in this way too. B. subtilis and C. acetobutylicum each contain TyrRSs of both subtypes.
Within the archaeal genre, the eukaryotic and archaeal TyrRSs are intermixed. The euryarchaeal enzymes (except for those of the pyrococci) cluster specifically with the animal and fungal TyrRSs, while the three crenarchaeal TyrRSs (and those of the pyrococci) group with the two plant examples (Arabidopsis and tobacco). Sequence signatures strongly support this entire phylogenetic arrangement.
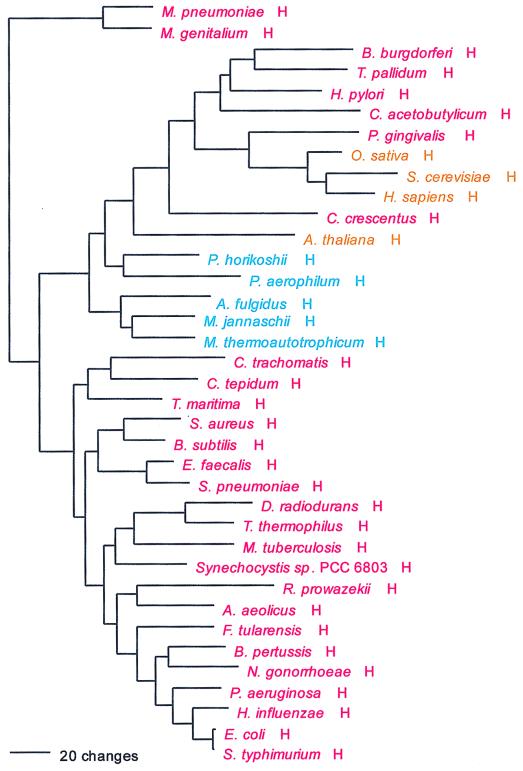
His; CAY; class II; homodimer.
HisRS also shows the full canonical pattern. However, as signature analysis indicates and Fig. 12 confirms, a small group of bacterial taxa—spirochetes, Helicobacter, C. acetobutylicum, Caulobacter, and Porphyromonas—have HisRSs of the archaeal genre. This bacterial grouping in turn encompasses the eukaryotic HisRSs, which shows a specific relationship to Porphyromonas HisRS therein, a relationship supported by sequence signature.
FIG. 12.
Phylogenetic tree of HisRS sequences. The tree has been rooted using ThrRS, ProRS, and GlyRS sequences. Other details are as in Fig. 2.
Gln; CAR; class I; monomer.
It has been convincingly demonstrated that GlnRS stems specifically from the eukaryotic lineage of GluRSs (53). Not only is this evident at the sequence level, but also it has been demonstrated in terms of the overall structure of the molecule (46). In its N-terminal (HIGH) region, the GlnRS sequence is decidedly more similar to eukaryotic than to archaeal GluRSs (and least similar of all to bacterial GluRS). In the C-terminal (KMSK) region, the similarities of GlnRS to the eukaryotic and archaeal versions of GluRS are roughly comparable but sequence similarity to the bacterial GluRS is effectively nonexistent (Fig. 13).
FIG. 13.
Phylogenetic tree of GlnRS sequences. The tree has been rooted using GluRS sequences. Other details are as in Fig. 2.
A GlnRS of a single type seems to occur in all eukaryotes; this generalization is based not only on animals, plants, and fungi but also upon the slime mold Dictyostelium, Trichomonas, and Nosema, with the last two representing deeply branching eukaryotic lineages (8). However, GlnRS is absent from the Archaea, and among bacteria its distribution is very sparse; it is found only in the β and γ subdivisions of the Proteobacteria, the Deinococcus-Thermus division, and Porphyromonas. In other words, among bacteria known not to contain GlnRS are representatives of the α and ɛ subdivisions of the Proteobacteria, the gram-positive bacteria, the cyanobacteria, the spirochetes, the chlamydias, and the genera Aquifex and Thermotoga. The only specific phylogenetic relationship apparent among the bacterial versions of the GlnRS is the proteobacterial grouping, but the proteobacterial representatives are not strongly distinguished from the other bacterial GlnRSs. Indeed, one of the β proteobacteria, Bordetella pertussis, has a GlnRS that appears specifically related to that found in Porphyromonas, a relationship reinforced by a sequence signature.
Asn; AAY; class II; homodimer.
Although they represent different synthetase classes (II and I respectively), in their evolutions the AsnRS and GlnRS families have much in common (Fig. 14). Both arose from within the cluster of the synthetases for their corresponding diacid. For glutamine, it was from the eukaryotic lineage per se that the enzyme arose, while for asparagine, the origin is localized only to the archaeal genre of AspRS in general. In both instances it is in the C-terminal portion of the molecule that the origin of the synthetase for the amidated amino acid is most strikingly seen. As is the case for GluRSs (see below), BLAST sequence similarity searches based upon the C-terminal 40% of the archaeal and eukaryotic AspRS have much higher scores with one another and with the AsnRSs. The root of the AsnRS tree itself separates the eukaryotic AsnRSs from their counterparts (see Fig. 16). The root of a combined phylogenetic tree for the AsnRS and AspRS enzymes (rooted by LysRS) occurs between the bacterial AspRSs and the grouping of archaeal and eukaryotic AspRSs with the AsnRSs.
FIG. 14.
Phylogenetic tree of AsnRS sequences. The tree has been rooted using AspRS sequences. Saccharomyces cerevisiae N1 is the cytoplasmic enzyme, while S. cerevisiae N2 is the mitochondrial enzyme. Other details are as in Fig. 2.
FIG. 16.
Phylogenetic tree of AspRS sequences. The origin of the AsnRS sequences is also shown (designated by the N rather than the D suffixes). The tree was rooted using LysRS sequences. Other details are as in Fig. 2.
The spotty distribution among the organismal taxa characteristic of GlnRS is seen for AsnRS as well, but to a lesser degree. AsnRS appears to be present in all eukaryotes but occurs in only two archaea, Pyrococcus and Pyrobaculum. AsnRS is more widely distributed among the bacteria but still is definitely absent in a number of taxa, i.e., Aquifex, Thermotoga, some proteobacteria (Neisseria, Pseudomonas, and Helicobacter), Mycobacterium, and Chlamydia.
Given their relatively late origins (see above), it is not surprising that the asparagine- and glutamine-charging enzymes show no real evidence of the canonical pattern. There are, for example, no significant areas of deletion-insertion in the alignment that would distinguish an archaeal from a bacterial genre. Dipeptide analysis does not show the pronounced differences between inter- and intra-group similarities, as the canonical pattern requires. Furthermore, no strong signature distinguishes an archaeal from a bacterial version of the enzyme.
The bacterial AsnRSs show two very distinctive subtypes, which are marginally specifically related at best. The first subtype is phylogenetically the more widespread, covering all characterized proteobacterial examples, Porphyromonas, the spirochetes, and the mycoplasmas. The second covers the Bacillus-Lactobacillus area of the gram-positive tree (although not the mycoplasmas) plus the Deinococcus-Thermus division. This second subtype, however, shows more similarity to the AspRSs than does the first subtype, suggesting that the second subtype has retained more ancestral character than have other AsnRSs.
The two known archaeal AsnRSs are specifically related to one another. The eukaryotic AsnRSs, however, fall into two unrelated groupings: the animal and fungal AsnRSs constitute one (distinct from all other AsnRS groups), while the yeast mitochondrial, plant, and Plasmodium enzymes distribute within the first bacterial subtype (and may all be mitochondrial) (Fig. 14).
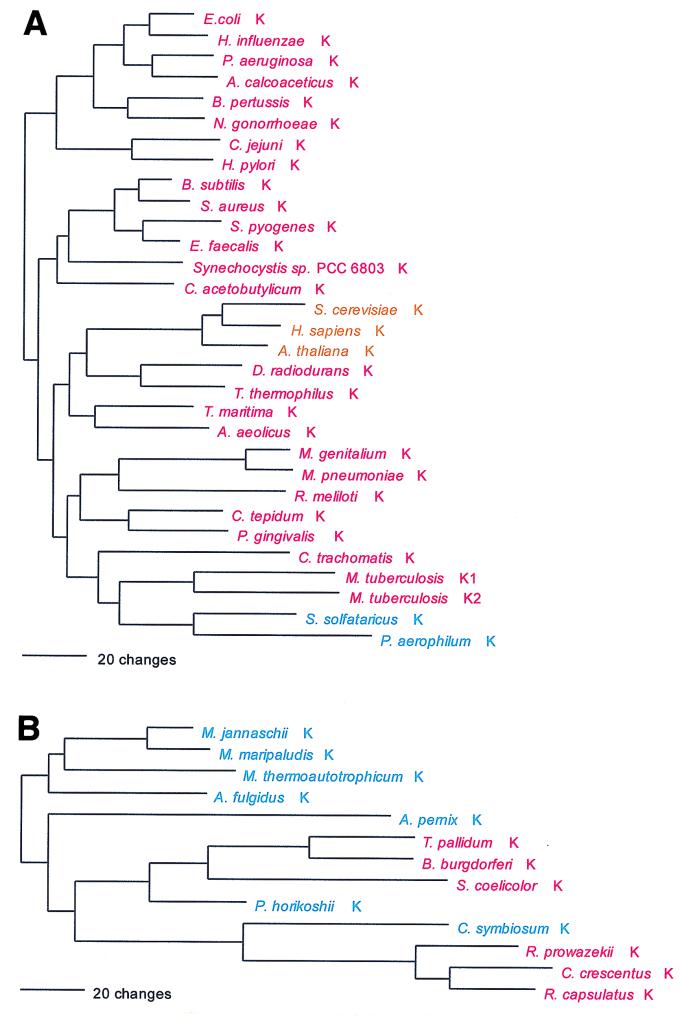
Lys; AAR; class I (monomer) and class II (homodimer).
LysRS represents the only known violation of the class rule: a class II LysRS is found in eukaryotes, most bacteria, and a few archaea (i.e., Sulfolobus and Pyrobaculum) (Fig. 15). However, a class I LysRS is found in the euryarchaeotes, two other members of the Crenarchaeota (Cenarchaeum and Aeropyrum), and a scattering of bacteria (34). The class II LysRSs clearly had a common ancestor with the AspRSs and AsnRSs in the deep past, but the class I enzyme stands essentially alone phylogenetically within its class.
FIG. 15.
Phylogenetic trees of LysRS sequences. (A) The class II synthetases were rooted using AspRS sequences. (B) The class I (archaeal type) sequences were rooted between the crenarchaeal (plus Pyrococcus) and other euryarchaeal sequences. Other details are as in Fig. 2.
The bacterial class II LysRSs are all of a kind and contain the grouping of the two above-mentioned crenarchaeal examples. No specific relationship exists between these archaeal examples and their eukaryotic counterparts, and the canonical pattern is not apparent. The phylogenetic grouping of the class I LysRSs shows the bacterial and archaeal examples to be intermixed. The crenarchaeon Cenarchaeum groups specifically with the known examples of the α proteobacteria, except for Rhizobium meliloti, whose LysRS is class II; however, Aeropyrum, also a crenarchaeon, is not a member of this group. The other known bacterial examples, the spirochete and Streptomyces LysRSs, as a group show a specific relationship to the Pyrococcus enzyme, while the remaining (eury)archaeal examples appear in an outgroup relationship to all those just discussed (Fig. 15).
Asp; GAY; class II; homodimer.
The AspRSs strongly exhibit the full canonical pattern: a single bacterial type exists, which differs dramatically from the AspRSs of the archaeal genre. In the interior of the AspRS sequence alignment, a stretch of about 220 amino acids in the bacterial genre (starting at ca. position 250 in the E. coli sequence) shows almost no similarity to the corresponding (∼100-amino-acid) section in the archaeal genre. Sequence similarity resumes thereafter at ca. bacterial position 470 and continues to the C terminus of the molecule, slightly more than 100 amino acids distant (Fig. 16). Because the AsnRS has arisen from within the grouping of the AspRSs (see above), the latter must be considered paraphyletic, which breaks the monophyly rule.
The arrangement of the AspRSs in all three major groups (bacterial, archaeal, and eukaryotic) does not violate established taxonomy except in minor ways in the bacteria. For example, the Cytophaga-Chlorobium clade (represented by Porphyromonas and Chlorobium) is split by the AspRSs: the Chlorobium version shows remarkably close relationship to the three examples (Rhodobacter, Caulobacter, and Rickettsia) of the α proteobacteria, while the Porphyromonas enzyme shows no clear specific relationships to any other bacterial AspRS.
Glu; GAR; class I; monomer.
Again, the full canonical pattern is strongly evident; the difference between the bacterial genre and its archaeal counterpart is striking. Not only are the bacterial examples about 100 amino acids shorter than the archaeal sequences at the N terminus, but also in the C-terminal (KMSK) section of the molecule the difference between them is extreme: the two show no resemblance, in either sequence or overall structure (46). (BLAST searches based on the archaeal and eukaryotic examples of this region readily detect one another and also the comparable region of all GlnRSs but never detect their bacterial counterparts.) Because the GlnRS has arisen from within the GluRS cluster, the latter breaks the monophyly rule. The bacteria show at least two subtypes of GluRS, which are specifically related to one another (to the exclusion of the GluRSs of the archaeal genre), and a number of bacterial species contain two GluRSs as well (24), all of which makes for a somewhat confusing phylogenetic picture (Fig. 17). It is worth noting that a rather clear grouping emerges that includes the spirochetes, the Cytophaga-Chlorobium group, the Deinococcus-Thermus division, the chlamydias, and two proteobacteria, i.e., Pseudomonas (γ division) and Rhizobium (α division).
FIG. 17.
Phylogenetic tree of GluRS sequences. The tree was rooted between the Bacteria and the Archaea plus Eukarya. The M. thermoautotrophicum ΔH sequence was used. Other details are as in Fig. 2.
Synthetases for the NGN-Encoded Amino Acids
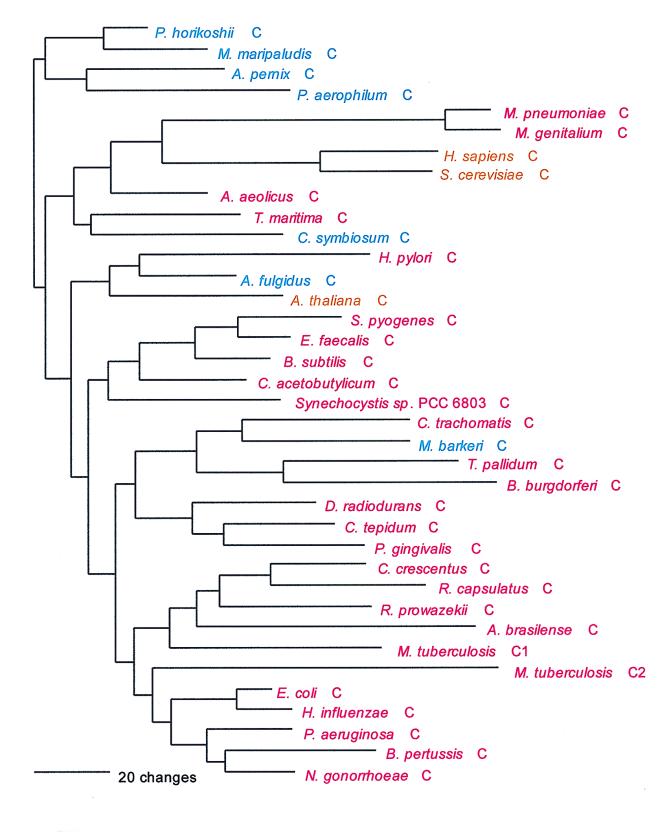
Cys; UGY; class I; monomer.
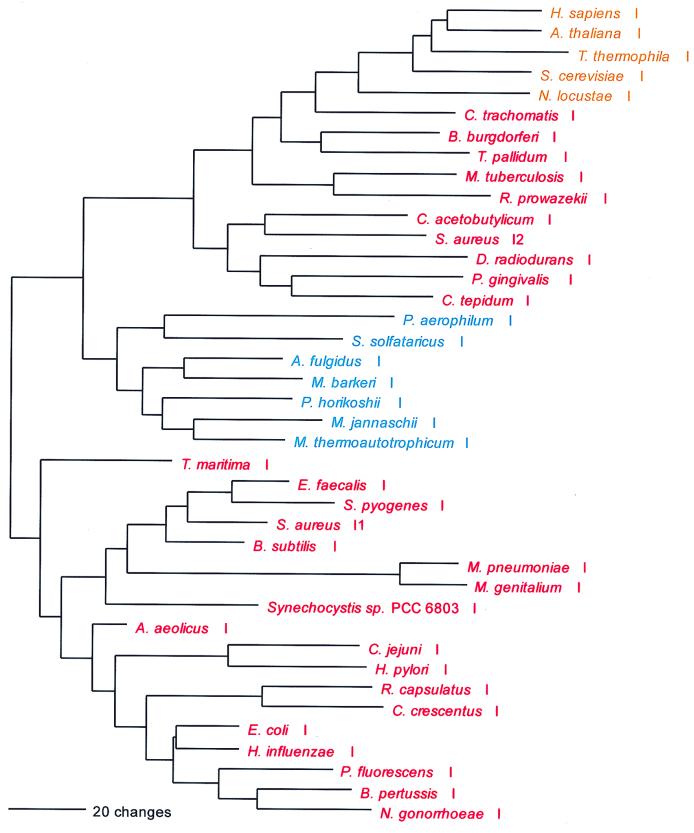
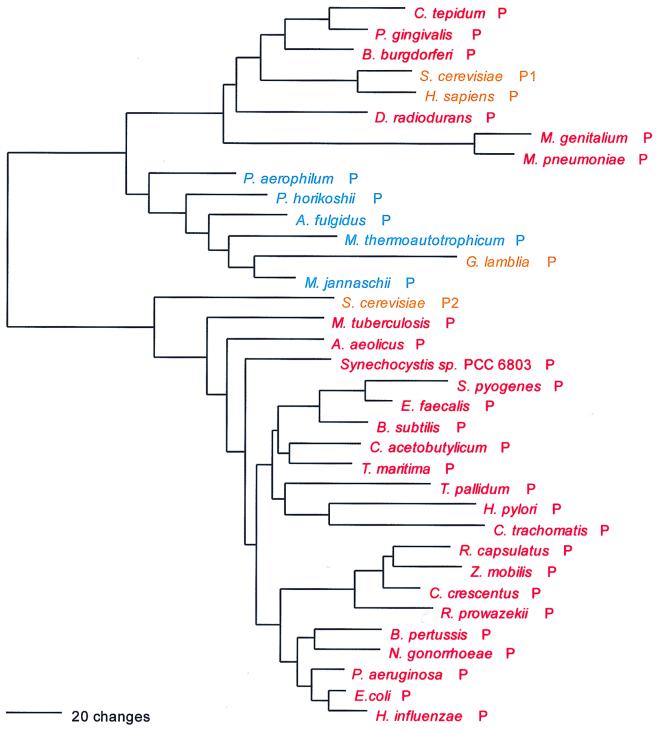
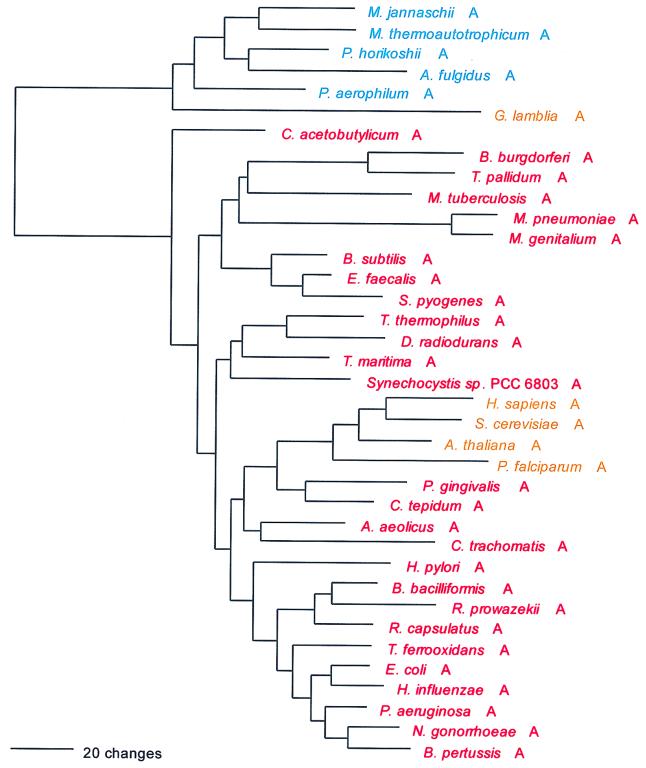
The mechanism of Cys-tRNA formation in M. jannaschii and M. thermoautotrophicum has until now been a mystery. Nothing identifiable as a CysRS was seen in their (complete) genomes. However, a normal functioning CysRS has been identified in Methanococcus maripaludis, a close relative of M. jannaschii (30, 40). Did a third, unrecognized synthetase class exist in these cases, or could the cysteine tRNA be charged indirectly, as in the case of selenocysteinyl-tRNA (11, 33)? The possibility that the highly aberrant SerRS found in M. jannaschii and M. thermoautotrophicum is somehow related to the lack of recognizable CysRS in these organisms was considered (37), the rationale being that such a SerRS might form Ser-tRNACys, which would be a key intermediate in Cys-tRNA formation by a tRNA-mediated amino acid transformation pathway (33). However, in vitro data did not support this view (37). Instead, biochemical and genetic approaches have now revealed that in M. jannaschii and M. thermoautotrophicum, ProRS is able to specifically synthesize both Cys-tRNACys and Pro-tRNAPro (60). This unprecedented dual functionality in an AARS is not reflected in any distinguishing features of these ProRSs at the sequence level. Interpretation of the evolutionary significance of this unexpected versatility among AARSs must now await more detailed biochemical description of its phylogenetic distribution.
As can be inferred from Table 6, the CysRSs do not exhibit the canonical pattern. There is also considerable evidence of interdomain horizontal gene transfer, particularly involving the archaeal CysRSs: In Fig. 18, four of the archaeal CysRSs do cluster. However, the M. maripaludis enzyme (see above) is disturbingly similar to that from Pyrococcus, with pair showing 65% sequence identity (40). Three other archaeal CysRSs, from Methanosarcina, Archaeoglobus, and Cenarchaeum, group among the bacterial examples of the enzyme but show no phylogenetic relationship to one another therein. By contrast, the relationships among the bacterial CysRSs in Fig. 18 are not particularly out of kilter with established bacterial taxonomy, which might suggest that the horizontal gene transfers have been mainly from the Bacteria to the Archaea.
FIG. 18.
Phylogenetic tree of CysRS sequences. The tree was rooted between the largest group of archaeal sequences and all others. Other details are as in Fig. 2.
Trp; UGG; class I; homodimer.
TrpRS is an obvious relative of TyrRS (19), although, as mentioned above, their corresponding codons are not related. The tryptophan enzyme conforms to the full canonical pattern, which can be inferred from Fig. 19, dipeptide similarity matrices, and striking sequence signatures. The TrpRSs of the archaeal genre show a substantial N-terminal extension relative to those of the bacterial genre. Within the bacterial genre, a number of subtypes can be recognized, and two organisms possess two TrpRSs, each of a different bacterial subtype (Fig. 19). By signature analysis, five bacterial subtypes can be identified, designated as such in Fig. 19, with P. gingivalis probably representing a sixth subtype. Within the archaeal genre, neither the archaeal-eukaryotic distinction nor the customary crenarchaeal-euryarchaeal divide holds. Most euryarchaea form a phylogenetic unit, but this unit also includes the crenarchaeon Aeropyrum. The remaining (two) crenarchaeal representatives (Pyrobaculum and Sulfolobus) plus Pyrococcus have a TrpRS of the eukaryotic type (or vice versa). The TyrRSs of Sulfolobus and Pyrococcus are exceptionally close (∼60% sequence similarity). Interestingly, one of the two TrpRSs found in Deinococcus is chimeric. The N-terminal (HIGH) section of the molecule clearly belongs to the bacterial subtype represented by E. coli, while the C-terminal (KMSK) section belongs to the bacterial subtype represented by Aquifex, as shown by sequence signature.
FIG. 19.
Phylogenetic tree of TrpRS sequences. The tree was rooted using TyrRS sequences. Other details are as in Fig. 2. The Deinococcus radiodurans TrpRS denoted by an asterisk is chimeric (see the text). Brackets denote bacterial subtypes defined by sequence signature analysis.
Arg; CGN and AGR; class I; homodimer.
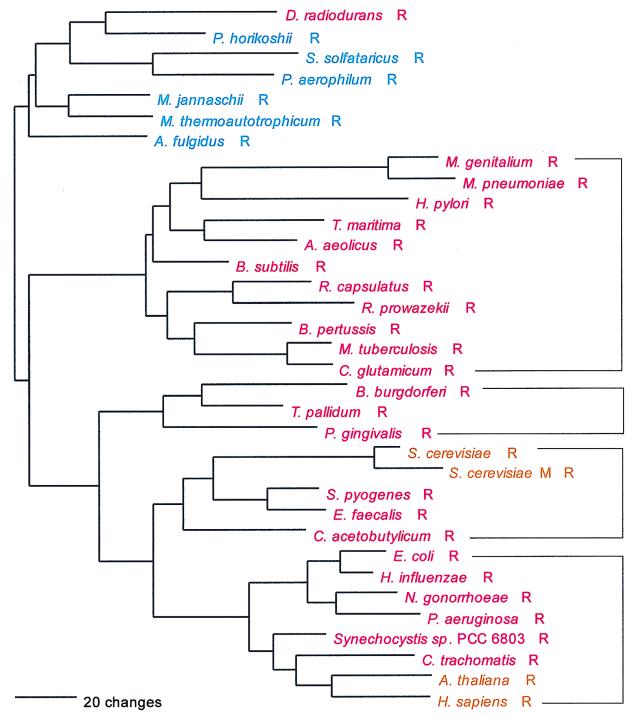
In its evolutionary profile, ArgRS is arguably the most complex of all the aminoacyl-tRNA synthetases: there are at least four bacterial subtypes identifiable by sequence signatures (Fig. 20) and two each for the eukaryotes and archaea. The relationships among them are similarly complex and definitely violate the full canonical pattern. The most distinctive, largest, and most taxonomically diverse of the ArgRS groups is that represented by the mycoplasmas in Fig. 20; it has the strongest sequence signature, and upon gross examination of the alignment it appears the most unusual. It is possible that this group represents a bacterial genre. However, the remaining bacterial subtypes do not appear specifically related to it, and the ArgRS tree cannot be reliably rooted. Given this and its complexity, one cannot confidently state at this point whether ArgRS exhibits any canonical pattern.
FIG. 20.
Phylogenetic tree of ArgRS sequences. The tree was rooted between the archaeal group and the other sequences. The S. cerevisiae M sequence is the mitochondrial enzyme. Other details are as in Fig. 2. Brackets denote bacterial subtypes defined by sequence signature analysis.
As shown in Fig. 20, animal and plant ArgRSs cluster within one of the bacterial subtypes, while the fungal enzyme (both cytoplasmic and mitochondrial) clusters within another. (Note here that although this last cluster contains a mitochondrial ArgRS, none of the bacteria represented therein are proteobacteria, the ancestral source of mitochondria.) The two bacterial groups that contain eukaryotic ArgRSs are specifically related to one another, as the tree in Fig. 20 shows.
While all archaeal ArgRSs appear specifically related to one another, there would seem to be two or three separate subtypes of them: one found in the methanogens and Archaeoglobus, another found in Pyrococcus, and a third involving the crenarchaeal examples. In addition, the Pyrococcus area of the archaeal tree seems to have been the source for the Deinococcus ArgRS, a relationship supported by a strong sequence signature, including a homologous insertion of about 30 amino acids unique to that pair.
Gly; GGN; class II; heterotetramer or homodimer.
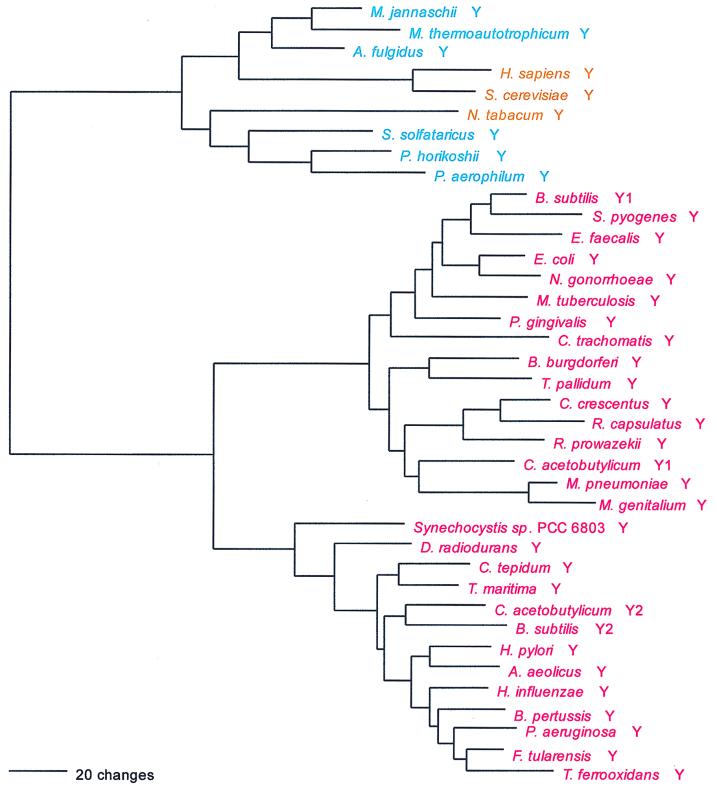
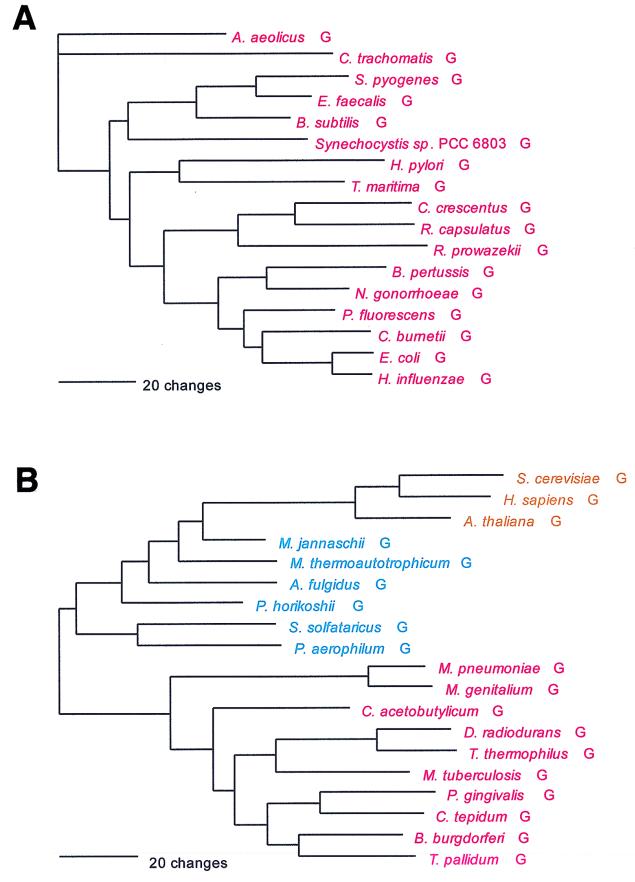
As previously noted, glycine is one of the tRNA-charging enzymes that violates the monophyly rule: two class II GlyRSs exist that are unrelated in both sequence and overall structure. One of the two, confined so far to the bacteria, is a tetramer of two α- and two β-subunits. The other, characteristic of the archaea and eukaryotes but also found in some bacteria, is a homodimer. This latter GlyRS has specific relatives among other members of the class II synthetases (as described above in the section of NCN-associated synthetases), but the former has none. The former cannot follow the canonical pattern, and the latter does not (which is seen clearly by the lack of a strong archaeal-bacterial distinction in a dipeptide similarity matrix) (Fig. 21).
FIG. 21.
Phylogenetic tree of GlyRS sequences. (A) The α2β2 type GlyRS sequences (combining data from both subunits) were rooted on the sequence from Aquifex. (B) The homodimeric GlyRS sequences were rooted using ProRS, HisRS, and ThrRS sequences. Other details are as in Fig. 2.
The first, exclusively bacterial GlyRS type encompasses the majority of bacterial taxa, while its alternative covers the spirochetes, the Deinococcus-Thermus clade, the Cytophaga-Chlorobium clade, and, among the gram-positive bacteria, the mycoplasmas and mycobacteria only. The α-subunit of the strictly bacterial GlyRS is remarkable for its high degree of sequence conservation, far greater than is seen in the β-subunit or other aminoacyl-tRNA synthetases. However, the two subunits, as might be expected, yield the same phylogeny, which (except for those bacteria having the archaeal type of GlyRS) more or less conforms to accepted organismal taxonomy. The so-called archaeal GlyRS type shows three main groupings, i.e., archaeal, eukaryotic, and bacterial. All three groupings appear monophyletic, but the eukaryotic GlyRS cluster lies within the archaeal group in Fig. 21, making the latter paraphyletic.
Recruitment of Aminoacyl-tRNA Synthetases into Other Roles
A number of proteins exist that have obviously derived from particular aminoacyl-tRNA synthetases but are known or suspected for obvious reasons not to function in translation. The pseudo-synthetases in question belong to the glutamate, lysine, histidine, phenylalanine, alanine, and asparagine synthetase families. The glutamate homolog (yadB in E. coli), which is only about 60% the length of a normal GluRS, seems confined to the β and γ proteobacteria and appears marginally specifically related to the normal proteobacterial GluRSs. The class II lysine homolog, which lacks the N-terminal ∼170 amino acids of the normal enzyme, is found in the enteric-vibrio group, Aquifex, and T. pallidum. It bears no specific relationship to any of the normal class II LysRS types. The histidinyl homolog appears to be a normal full-length HisRS. It is found in some gram-positive bacteria, a few proteobacteria, cyanobacteria, and Aquifex. This pseudo-synthetase is marginally of the archaeal HisRS genre, yet it does not occur in the archaea. Recently, it was shown that this gene (hisZ) lacks aminoacylation activity but is a required component of histidine biosynthesis in a number of bacteria in which the hisG gene is truncated (56). In three of the four euryarchaea (Pyrococcus excluded), there is a protein that resembles the α chain of PheRS. The molecule lacks ∼180 amino acids at the N terminus relative to normal archaeal PheA and extends past the normal C terminus by about the same number (63), while other areas exist within the molecule that also show little or no similarity to the normal PheRS. A highly truncated version(s) of the AlaRS exists in some Archaea and a few Bacteria. Finally, Pyrococcus species contain a highly truncated form of their normal AsnRS (e.g., accession no. BAA29342).
Summary of Aminoacyl-tRNA Synthetase Evolutionary Profiles
The aminoacyl-tRNA synthetases have obviously been subject to a great deal of horizontal gene transfer over the evolutionary course covered by the universal phylogenetic tree. Given this, it is somewhat surprising how much of the ancient evolutionary trace these enzymes have retained. The full canonical pattern is shown almost without exception by four of the enzymes (aspartic and glutamic acids, phenylalanine, and leucine) and arguably by two others (tyrosine and tryptophan). In some of these, the bacterial genre contains two or more dramatically dissimilar subtypes, the most extreme example perhaps being TrpRS, where no fewer than six distinct bacterial subtypes can be discerned (Fig. 19). For another four synthetases, i.e., isoleucine, histidine, proline, and probably methionine, the full canonical pattern holds, although a significant minority of bacterial taxa employ a synthetase of the archaeal rather than the bacterial genre. Three more of the amino acids—valine, threonine, and alanine—and perhaps a fourth, arginine, show only the basal canonical pattern; the eukaryotic enzymes in these cases are obviously of the bacterial genre. Almost certainly this pattern results from the horizontal displacement deep in the eukaryotic stem of an original eukaryotic synthetase of the archaeal genre by one of the bacterial genre. Indeed, the Giardia lamblia AlaRS (Fig. 10) lends support to this notion, for that enzyme, unlike its eukaryotic counterparts, is clearly of the archaeal genre (see the alanine section above) but, as expected, is phylogenetically external to the archaeal group per se. The remaining 6 of the 20 amino acids, i.e., cysteine, serine, lysine, glycine, asparagine, and glutamine, do not conform to the canonical pattern. These six we call the “gemini group.”
The gemini group derives its name from the fact that for each of its six members (and only these six), two unrelated charging systems exist. For glutamine and asparagine, the difference between the two systems is of one kind: one system is a normal direct charging, while the other is indirect, with a “precursor” amino acid being placed on the tRNA and then biochemically converted in situ to the appropriate amino acid by amidation (see above) (12, 33). For cysteine, the alternative mode of charging, which involves ProRS (60), is completely novel (see the cysteine section above).
The other three members of the gemini group, i.e., serine, lysine, and glycine, each have two normal but not (specifically) related charging systems, thereby violating the monophyly rule. The second (minor) charging enzyme for serine, which so far has been found only in two genera of methanogens, belongs to the same class II supercluster as does the dominant (normal) SerRS but is probably not specifically related to the dominant SerRS therein (see the serine section above). The two LysRS enzymes belong to different structural classes; the class I enzyme occurs in most archaea and a small number of bacterial taxa, while its class II counterpart dominates the bacteria and is seen in a few archaea and all of the eukaryotes so far examined. The heterotetrameric form of GlyRS is confined to and dominates the bacteria, while its unrelated homodimeric counterpart occurs in all characterized archaea and eukaryotes as well as in a minority of bacterial taxa.
We do not consider it coincidental that the members of the gemini group share these two characteristics, i.e., lack of canonical pattern and twin charging systems, because in our opinion it reflects an evolutionary circumstance unique to this group—one that may well shed light on the meaning of the canonical pattern and the long-term dynamic of horizontal gene transfer (see the discussion below).
PHYLOGENETIC PICTURE EMERGING FROM THE AMINOACYL-TRNA SYNTHETASES
Much has been said and written about aminoacyl-tRNA synthetases contradicting established organismal phylogenies. From the above analysis, one can see that in many cases they do so—sometimes in spectacular ways. In at least as many other cases, however, they do not. In this section, we discuss the question of phylogenetic congruence between the aminoacyl-tRNA synthetases and established organismal phylogenies. The main framework for the discussion is Table 7, a tabulation of the extents to which the various aminoacyl-tRNA synthetase relationships confirm certain established major taxonomic groupings. As we shall see, none of the established organismal relationships are disproven, most are confirmed (by a majority of the synthetases), and the horizontal gene transfer characteristics of these enzymes suggests other evolutionary relationships that were not detected by the phylogenetic analyses of vertically inherited genes.
TABLE 7.
Support or lack thereof from the individual aminoacyl-tRNA synthetases for various major bacterial taxonomic groupings
| Taxonomic groupa | Support for presence of aminoacyl-tRNA synthetaseb:
|
|||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| F | L | I | M | V | S | P | T | A | Y | H | K | D | E | C | W | R | G | |
| Proteobacteria | ||||||||||||||||||
| β,γ | tsd | tsd | tsd | tsd | tsd | tsd | tsd | tsd | tsd | ∗ | tsd | tsd | tsd | tsd | tsd | ∗ | tsd | ts |
| α,β,γ | t d | ts | ts | − | s | tsd | tsd | tsd | tsd | ∗ | ∗ | − | tsd | sd | ∗ | − | s | |
| α,β,γ,ɛ | d | ts | ts | − | s | − | ts | ∗ | − | − | ts | s | ∗ | − | s | |||
| β,γ,ɛ | t | s | ||||||||||||||||
| Bacillus-Streptococcus | ||||||||||||||||||
| BES | tsd | tsd | tsd | tsd | tsd | ts | tsd | tsd | tsd | ∗ | tsd | tsd | tsd | tsd | t d | ∗ | − | t |
| BES,Cl | d | − | − | tsd | sd | tsd | s | ∗ | − | sd | d | − | t d | ∗ | − | − | ||
| BES,My | sd | s | tsd | s | s | td | − | sd | sd | ∗ | s | sd | tsd | ∗ | − | − | ||
| BES,Mb | d | − | − | s | − | ∗ | − | − | ||||||||||
| BES,Cl,My | s | − | − | s | ts | − | − | ∗ | − | − | ||||||||
| Spirochetes | tsd | s | tsd | tsd | ts | ts | − | − | tsd | tsd | ts | tsd | tsd | tsd | ts | ts | tsd | tsd |
| Cytophaga-Chlorobium | ts | s | tsd | tsd | ts | ts | tsd | ts | tsd | − | − | tsd | − | tsd | ts | − | − | tsd |
| Deinococcus-Thermus | tsd | ND | + | tsd | tsd | ts | ND | + | tsd | + | tsd | tsd | tsd | tsd | + | + | + | tsd |
Taxonomic abbreviations are more or less evident: α, β, etc., are the various divisions of the Proteobacteria. BES, Bacillus-Enterococcus-Streptococcus taxon; Cl, Clostridium acetobutylicum; My, mycoplasmas; Mb, mycobacteria.
The amino acids are given their one-letter codes. t, monophyletic (orthophyletic) unit in corresponding phylogenetic tree; s, signature analysis supports the unit; d, dipeptide similarity supports the unit; ∗, relationships too complex to be displayed in this tabular form; −, relationship definitely ruled out; + (in Deinococcus row), specific relationship can be detected using BLAST scores for Thermus provided by the Göttingen Website (www.g2l.bio.uni-goettingen.de); ND (in Deinococcus row), no definitive data available.
The aminoacyl-tRNA synthetases most strikingly confirm the profound evolutionary divide that separates the Bacteria from the Archaea and Eucarya. Over and above this, these enzymes support most of the accepted major taxa (kingdoms, divisions, etc.) within each of the three domains (in cases where a sufficient number of species have been characterized to permit making reliable assessments). The aminoacyl-tRNA synthetases do not confirm the branching orders among the major taxa within each domain. Here they present only a confused, and so unreliable, picture.
Spirochetes
The spirochetes constitute a major bacterial grouping that by multicellular eukaryotic standards would be considered at least a division or phylum (47, 68). The two sequenced spirochete genomes—from Borrelia burgdorferi and Treponema pallidum—do not cover the full phylogenetic breadth of the division, but they are well separated phylogenetically within it. The two spirochetes possess 19 of the aminoacyl-tRNA synthetases (they have no GlnRS). In only two cases, i.e., threonine, and proline, are the spirochete enzymes clearly not specifically related to one another. In the first case, the Treponema ThrRS groups with those of the proteobacteria, quite distant from the grouping that encompasses the Borrelia ThrRS (Fig. 9). The T. pallidum ProRS clusters within the main bacterial group, while the B. burgdorferi ProRS is of the archaeal genre and is specifically related to the eukaryotic ProRS (Fig. 8). In a few other trees (Fig. 3 and 14), the two spirochetes do not appear to be specifically related but nevertheless are clustered by sequence signature analysis. Moreover, in these trees the two genera lie close enough that the conclusion that they are not specifically related is by no means certain.
The spirochetes are notable for the number of cases in which their aminoacyl-tRNA synthetases are of the archaeal and eukaryotic genre: 7 of their 19 synthetases are so (Table 8). In four of these, i.e., isoleucine, methionine, proline, and histidine, the group to which a spirochete enzyme belongs not only lies within the general archaeal and eukaryotic grouping but therein is a sister lineage to the eukaryotic synthetases. (For proline, only the Borrelia enzyme is of this nature.) These four examples cover all those in which a bacterial subtype is of the archaeal and eukaryotic genre and specifically related to its eukaryotic counterparts (Table 8).
TABLE 8.
Type of aminoacyl-tRNA synthetase represented in a given taxon
| Taxonomic groupa | Aminoacyl-tRNA synthetase typeb
|
|||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| F | L | I | M | V | S | P | T | A | Y | H | Q | N | K | D | E | C | W | R | G | |
| Proteobacteria | ||||||||||||||||||||
| β | B | B | B | A | B | B | B | B | B | B | B | B | B | B | ? | B | ||||
| γ | B | B | B | A | B | B | B | B | B | B | B | B | B | B | ? | B | ||||
| α | B | B | B/A | A/B | B/A | B | B | B | B | A/B | A | B | B | B | B | B | ||||
| ɛ | B | B | B | B | B | B | B | B | B | A | B | B | B | B | B | B | ||||
| Bacillus-Streptococcus | ||||||||||||||||||||
| BES | B | B | B | B | B | B | B | B | B | B | B | B | B | B | ? | B | ||||
| C. acetobutylicum | B | B | A | B | B | B | B | B | B | A | B | B | B | B | ? | A | ||||
| Mycoplasma | B | B | B | B | B | A | B | B | B | B | B | B | B | B | B | A | ||||
| Mycobacteria | B | B | A | B | B | B | B | B | B | B | B/A | B | B | B | B | A | ||||
| Cyanobacteria | B | B | B | B | B | B | B | B | B | B | B | B | B | B | ? | B | ||||
| Aquifex | B | B | B | B | B | B | B | B | B | B | B | B | B | B | B | B | ||||
| Thermotoga | B | B | B | B | B | B | B | B | B | B | B | B | B | B | B | B | ||||
| Chlamydia | B | B | A | A | B | B | B | B | B | B | B | B | B | B | ? | B | ||||
| Spirochetes | A | B | A | A | B | A/B | B | B | B | A | A | B | B | B | ? | A | ||||
| Cytophaga-Chlorobium | B | B | A | A | B | A | B | B | B | A/B | B | B | B | B | ? | A | ||||
| Deinococcus-Thermus | B | B | A | B | B | A | B | B | B | B | B | B | B | B | A | A | ||||
| Archaea | M | MC | MC | mC | MC | P | MC | MC | MC | m | MC | P | MC | M | P | m | MC | mC | ||
| Eukaryotes | A | A | b2,1 | b1 | B3 | b1,4 | B2 | B4 | A | b4 | B | P | B | A | A | A | B2 | A | ||
Abbreviations are defined in Table 7 footnote a.
Abbreviations: A, synthetase of the archaeal genre; B, synthetase of the bacterial genre. In the Archaea and Eukaryotes section of table: A, synthetase of the archaeal genre; B, synthetase of the bacterial genre; b, eukaryotic synthetase of archaeal genre with closest relatives being bacterial; M, Archaea are monophyletic and exclude the eukaryotes; m, Archaea are monophyletic but include the eukaryotes; C, Archaea comprise euryarchaeal and crenarchaeal groups, and a signature exists for them; P, polyphyletic. Closest relatives: 1, spirochetes; 2, Chlamydia; 3, Proteobacteria; 4, Porphyromonas. ?, uncertain.
For two others of the seven spirochete exceptions (phenylalanine and lysine), the synthetase is precisely of the archaeal type. The spirochete PheRS is the only bacterial PheRS of the archaeal genre and is most similar to its Pyrococcus counterpart (Fig. 2) (see the phenylalanine section above). The spirochetes are one of three bacterial taxa known to use the class I LysRSs. While the spirochete enzyme again appears Pyrococcus-like, the other bacterial class I LysRSs (found in the α proteobacteria and streptomycetes) resemble the class I LysRSs of the crenarchaeal branch of the Archaea. The final (seventh) example of a “nonbacterial” synthetase in the spirochetes is GlyRS. Here the spirochete enzyme belongs to a minor but significant clade of bacterial GlyRSs that are of the archaeal and eukaryotic type and almost certainly of archaeal origin (see the glycine section above).
ArgRS, the most evolutionarily complex of the 20 synthetases, should also be mentioned here. The spirochete ArgRS is unusual in that it (and the Porphyromonas enzyme) forms a phylogenetically coherent grouping peripherally related to the two bacterial groupings in which the eukaryotic ArgRSs reside—predicted by sequence signature (Fig. 20). This entire combined unit appears not to be specifically related to the “main” bacterial type (genre) and can at best be only marginally related to the archaeal genre (see the arginine section above).
Cytophaga-Chlorobium Kingdom
While the two divisions that comprise the Cytophaga-Chlorobium kingdom were each solidly defined by 16S rRNA analysis (47, 68), their sister relationship was only weakly suggested thereby. The addition of 23S rRNA data made the case for the kingdom convincing (74). Porphyromonas, on the bacteroides branch of the Cytophaga division, and Chlorobium tepidum together span the full phylogenetic breadth of this kingdom. The aminoacyl-tRNA synthetases from these two organisms further reinforce the specific relationship between the Cytophaga and Chlorobium (green sulfur) divisions.
As Table 7 shows, for 13 of the amino acids, the Porphyromonas and Chlorobium synthetases are of the same bacterial subtype and specifically related to one another. (In one of these, leucine, the relationship is seen only in sequence signatures, not in the phylogenetic trees. However, the tree in Fig. 3 does put the two taxa in close enough proximity that the branching order shown cannot be considered definitive.) For two of the seven (confirmed) exceptions to the specific relationship, i.e., asparagine and glutamine, no synthetase has been detected in the currently available Chlorobium sequence data. For the remaining five exceptions, i.e., aspartic acid, arginine, histidine, tryptophan, and tyrosine, the Porphyromonas and Chlorobium enzymes clearly belong to different bacterial subtypes (Fig. 11, 12, 16, 19, and 20). Table 8 shows that a total of four of the synthetases in both taxa, i.e., isoleucine, methionine, proline and glycine, are of the archaeal genre, while a fifth (histidine) is so in Porphyromonas only.
Deinococcus-Thermus Division
The Deinococcus-Thermus bacterial division was recognized initially through 16S rRNA analysis. (The kingdom-level unit to which it belongs includes a second division, represented by Chloroflexus and Thermomicrobium, which has yet to be characterized in genomic sequence terms [47, 68].) Although no extensive genomic data are publically available for Thermus species, sequences exist in the public databases for 11 different Thermus aminoacyl-tRNA synthetases. In all 11 cases, the Thermus enzyme is specifically related to its Deinococcus counterpart. (Using the BLAST score server provided by the Göttingen Genomics Laboratory [www.g2l.bio.uni-goettingen.de], one can infer that another seven Thermus enzymes at least, i.e., CysRS, GlnRS, IleRS, ThrRS, TyrRS, TrpRS, and ArgRS, also are specifically related to their Deinococcus counterparts.) These 18 examples give solid support to the predicted specific relationship between the genera Deinococcus and Thermus. For 2 of the 20 synthetases, i.e., isoleucine and proline, the Deinococcus (and Thermus) enzymes are known to cluster with their spirochete and Cytophaga-Chlorobium counterparts in a unit that also contains the eukaryotic synthetases (Fig. 4 and 8). In all, 4 of the 20 Deinococcus-Thermus synthetases are of archaeal character (Table 8).
The extent to which the three major bacterial divisions or kingdoms just discussed, i.e., the spirochetes, the Cytophaga-Chlorobium kingdom, and the Deinococcus-Thermus division, show the same minor bacterial synthetase type is noteworthy. For the amino acids isoleucine, proline, glutamic acid, cysteine, and glycine (Fig. 4, 8, 17, 18, and 21), all three bacterial taxa are represented in the same minor bacterial synthetase group and, except for glutamic acid and cysteine, that synthetase is of archaeal character. For another two or three amino acids, i.e., methionine, histidine, and possibly serine, two of the three taxa are represented in the same minor bacterial synthetase group (Tables 7 and 8). One wonders whether this correlation is suggestive of some evolutionary relationship among these three divisions and kingdoms that is too subtle to have been detected by analyses of rRNA or other vertically inherited genes.
Proteobacteria (Purple Bacteria)
As defined by rRNA analysis, the kingdom Proteobacteria comprises five divisions (originally designated subdivisions), α through ɛ (47, 68). The β division is actually a highly divergent branch within γ, and these two together show a sister relationship to α. The remaining two divisions, δ and ɛ, appear specifically related to one another and together specifically relate to the α-β-γ grouping (47, 68). Except for δ, all the proteobacterial divisions have genomically characterized representatives.
For over half of the 20 amino acids, the aminoacyl-tRNA synthetases of the major bacterial subtype encompass all four characterized proteobacterial divisions. Otherwise, the divisions, or various taxa within them, show different subtypes, usually bacterial. For asparagine and glutamine (as in many other bacterial taxa), some proteobacteria possess no synthetases for one or both of these amino acids. The phylogenetic unit that comprises the α, β, and γ divisions is seen in at least nine of the synthetases; the higher-level relationship that then includes the ɛ division as well can be seen in about eight cases (Table 7). Only one of the proteobacterial synthetases, MetRS, is of the archaeal genre, with the exception of four cases in which some or all α proteobacteria are so (Table 8).
Gram-Positive Bacteria
The gram-positive bacteria as defined by rRNA comprise a number of major lineages, not merely the two conventionally recognized high- and low-G+C types (47, 68). The kingdom is phylogenetically sufficiently disperse that its coherence is only minimally suggested even by rRNA analyses. It is not unexpected, therefore, that the aminoacyl-tRNA synthetases do not give evidence supporting the phylogenetic coherence of the kingdom as a whole. At this point in time, three of the gram-positive lineages are represented in genomic terms, the high-G+C gram-positive lineage (mycobacteria and some streptomycetes), the Bacillus-Lactobacillus lineage (genomes of Bacillus, Staphylococcus, Streptococcus, Enterococcus, and two mycoplasmas), and a third lineage represented by C. acetobutylicum.
The aminoacyl-tRNA synthetases of B. subtilis are never of the archaeal genre, and almost without exception the organism has a synthetase of the main bacterial subtype. For two of the amino acids, i.e., threonine and tyrosine, B. subtilis carries two synthetases, each of a different bacterial subtype. Almost without exception, the synthetases of Streptococcus and Enterococcus are of the same bacterial subtype as those of B. subtilis and are specifically related therein to their B. subtilis counterpart. The corresponding mycoplasma synthetases are somewhat more idiosyncratic (discussed below).
The synthetases of C. acetobutylicum (representing a distinct gram-positive division) are of the same bacterial subtype as those of Bacillus in the majority of cases, at least six of which show a specific relationship to the Bacillus “family” (Table 7).
The synthetases representing the high-G+C division of the gram-positive bacteria give no definitive indication that this division is related to the others in the gram-positive kingdom. In most cases, however, the mycobacterial version of a synthetase belongs to the same bacterial subtype as does its Bacillus counterpart. Although the representative mycobacterial synthetases do not confirm the organism's relationship to other gram-positive bacteria, neither do they contradict it, since the mycobacterial enzymes show no consistent specific relationship to synthetases from other taxa.
Bacterial taxa represented by a single genome only are not discussed in this context, since the data are too few to draw meaningful conclusions and little in the way of solid relationships to other taxa are seen (however, see Table 7).
Rickettsias, Mycoplasmas, and Certain Other Bacteria
Paleontologists have known for the better part of this century that different lineages evolve at different rates or tempos (55). Molecular evolutionists encountered this phenomenon early in the comparative study of molecular sequences. What paleontologists also understood was that this quantitative measure, tempo, was associated with a qualitative one, “mode,” i.e., the nature of the evolutionary outcomes (55). The question then became whether evolutionary tempo at the molecular level also has some kind of mode associated with it. Comparative analysis of rRNA sequences showed that this was indeed the case, in turn suggesting that the tempo-mode relationship was a fundamental characteristic of the evolutionary process (68).
Molecular mode can be explained as follows. If changes (residue replacements and the like) at the sequence level were purely a matter of tempo, the frequency distribution for residue replacements (as a function of position in a given sequence) would have the same relative shape for rapidly and slowly evolving lineages. However, it has been observed that the shape of this distribution is different in the two cases: rapidly evolving lineages show a disproportionate increase in the replacement rate for the more highly conserved residues in the molecule (68). This in principle then allows the detection of rapidly (or slowly) evolving lineages without resort to outgroup rooting of a tree (68).
Three good examples of rapidly evolving lineages defined by rRNA criteria are represented by full genomic sequences: those of the mycoplasmas, Rickettsia prowazekii, and Helicobacter pylori (47, 68). Mycoplasmas are considered the most rapidly evolving lineage of all (68). The mycoplasmas have been shown by rRNA and other analyses to form a phylogenetic grouping that is specifically related to the major gram-positive taxon built around the genera Bacillus, Lactobacillus, Streptococcus, and relatives (47, 68); a specific relationship may even exist between the mycoplasmas and streptococci. In the aminoacyl-tRNA synthetase trees, the mycoplasmas cluster specifically with Bacillus and the others in only two cases, isoleucine and glutamic acid (Table 7). In two others, the mycoplasmas have a synthetase of the archaeal genre (which Bacillus never has). In the remaining cases, the mycoplasmas belong to the same bacterial synthetase subtype as does Bacillus, but they do not appear specifically related to the latter group. Indeed, in the majority of trees the mycoplasmas (alone or in combination with a small number of other bacterial taxa) appear to form the deepest-branching lineage in their bacterial synthetase subtype grouping. In contrast, a weak to strong sequence signature suggests specific relationship between the mycoplasmas and Bacillus and relatives in at least 10 cases (Table 7).
Failure of the mycoplasmas to tree reliably is a common experience. Mycoplasma proteins are generally quite idiosyncratic in sequence (with a number of positions therein that break with what otherwise is the universal composition). Because of their highly derived nature (manifested also in impressively low dipeptide similarities), the parsimony treeing algorithm at least tends to place them improperly (21). The conclusion of nonrelationship of the mycoplasma synthetases to their Bacillus group counterparts implied by the above phylogenetic trees is best held in abeyance until sequence data are available from closer relatives of the mycoplasmas, such as their walled relatives (68); as might be expected, experience has shown that the sequences from rapidly evolving species are “better behaved” when treed in the presence of specific close relatives that are not so rapidly evolving (68).
Helicobacter is another case in which aminoacyl-tRNA synthetase sequences appear somewhat highly derived. Although in the above trees the organism groups with the other proteobacteria in some cases, in others it tends to wander among the taxa, often branching near the bottom of a given major bacterial group and sometimes “specifically” with the mycoplasmas, both of which are signs of rapidly evolving, highly derived sequences. As with the mycoplasmas, the treeing of Helicobacter sequences should benefit from sequence data from more closely related but slower-evolving relatives, for example, members of the δ division of the Proteobacteria.
The R. prowazekii aminoacyl-tRNA synthetase sequences appear less highly derived than those of the two taxa just discussed. The above trees show that 14 of the 18 rickettsial aminoacyl-tRNA synthetases cluster with their α proteobacterial counterparts, as do rickettsial proteins in general (2). In the four remaining cases, the rickettsial synthetases are of a different genre from their α proteobacterial counterparts. It is too early to draw firm conclusions about rapidly evolving lineages from these three organisms (mycoplasmas, rickettsias, and Helicobacter), but the data appear to suggest that at least some types of rapidly evolving lineages might be more prone to horizontal gene transfers than are their more slowly evolving relatives.
Archaea
Too few archaeal aminoacyl-tRNA synthetases have been sequenced to permit detailed evolutionary conclusions to be drawn. However, some loose general conclusions can be made at this point. (i) The archaeal synthetases usually form a monophyletic (though sometimes paraphyletic) grouping. (ii) The fundamental euryarchaeal versus crenarchaeal divide within the Archaea is respected in most but not in all cases; it is seen in 11 cases (Table 8) and in several more in which the crenarchaeal cluster is joined by a Pyrococcus synthetase. (iii) Horizontal gene transfers of aminoacyl-tRNA synthetases have occurred both within the archaea and between the archaea and eukaryotes. (The TyrRSs and TrpRS give especially clear indications of this [see the tyrosine and tryptophan sections above].) (iv) While synthetases of the archaeal genre seem often to have migrated into the bacteria, the opposite seems not to occur, with the exception of the gemini group of synthetases (discussed above), which show a unique evolutionary dynamic.
EVOLUTIONARY SYNTHESIS
Understanding in an evolutionary context is not like understanding in a general (modern) biological context. It is nature, not the biologist, that does the evolutionary experiment, and essentially all such experiments have been done in the distant past and certainly cannot be repeated in the laboratory. Such limitations require that evolutionary understanding draw heavily upon inference and inspired conjecture. Therefore, if we are to reconstruct the history of life on this planet and understand the process by which it arose, theories capable of relating and interpreting the available facts in broad, meaningful ways, capable of defining and focusing scientific thinking on particular ideas, will have to be developed. In this concluding section, we try to do exactly this, i.e., to use the aminoacyl-tRNA synthetases to help construct a tentative picture of what the primary lineages are and what the evolutionary process that produced them is. The goal is to develop a clearer concept of how modern cells evolved and to identify various stages in that process.
Because of their modular nature (see above) (51), the aminoacyl-tRNA synthetases readily undergo horizontal gene transfer. Also, because their functions are ancient and universal, these transfers are expected to be broad in scope (which they are), to occur throughout the recorded evolutionary course (which they appear to do), and to be largely selectively neutral in character (which is certainly the simplest explanation, for example, for the fact that the same organism can harbor genes for two very different but functionally equivalent subtypes of a given synthetase). Because the 20 charging enzymes are in essence alike in function (and presumably alike in their relationships to the cell as a whole), one would expect the horizontal gene transfer profiles of the 20 synthetases all to be similar in some general respect (although in their details, of course, each would be unique). However, this is clearly not the case: the horizontal gene transfer profiles of the different synthetases can be qualitatively different. Nevertheless, these profiles seem to be of several general types, which are crudely distinguished by the extent to which and ways in which they manifest the canonical pattern.
Evolutionary Significance of the Canonical Pattern
What is the significance of the canonical pattern; at what stage in the evolution of the cell did the canonical pattern arise? The horizontal gene transfer patterns of the aminoacyl-tRNA synthetases begin to provide answers here, but far too few data now exist to treat these answers as anything but theoretical conjectures requiring more thorough testing in the future.
It is essentially self-evident that, other factors being equal, most horizontal gene transfers will be taxonomically local (the donor and recipient would be close neighbors on the phylogenetic tree), for the simple reason that in such cases, horizontal displacement of an indigenous gene by a foreign equivalent would be minimally perturbing of cellular function, in that the indigenous and foreign genes are relatively similar in sequence. With the caveat that the more taxonomically local the horizontal gene replacement, the harder it is to recognize (the more data are needed), we nevertheless think that the data available at present suggest that horizontal gene transfers of the aminoacyl-tRNA synthetases are not predominantly local. The horizontal displacements that have occurred deep in the phylogenetic tree, involving transfers from one major taxon to another major taxon, appear to be unexpectedly prevalent.
The majority of transfers that introduce synthetases of the archaeal genre into the bacteria appear to involve a gene that arose deep in the common archaeal/eukaryotic lineage and transferred to the ancestor of some major bacterial taxon. These can only have been very ancient evolutionary events. Also, the fact that they appear to dominate the landscape of aminoacyl-tRNA relationships suggests that the dynamic of horizontal gene transfer has not remained constant over the evolutionary course covered by the universal phylogenetic tree.
We assume that the primary determinant of the nature of horizontal transfer is the nature of the recipient cell. If this is so, a dynamic of horizontal gene transfer that changes over the evolutionary course means that the nature of cells has changed over that course. Our theory, then, is that the deeper branching in the universal phylogenetic tree corresponds to evolutionary stages when the evolution of the cell was not yet complete, when the modern cell(s) had yet to emerge. In other words, the primary branchings in the phylogenetic tree (the canonical pattern) and the deep branchings in each of the domains involved primitive entities that were in the process of evolving to become modern cells. This would easily explain why the sequences of bacterial and archaeal aminoacyl-tRNA synthetases (different genres) differ so dramatically from one another while the sequences within each of the domains differ in relatively trivial ways (reference 68 offers a more detailed discussion of this matter).
The complex, highly refined cells of today obviously came from primitive counterparts that were far simpler and more rudimentary. Not only were the subsystems of the cell—the translation and transcription mechanisms, the genome and its replication apparatus, and so on—simpler and less accurate in their functions than today's versions thereof, but also the primitive cell as a whole was more loosely and less hierarchically organized and its states were fewer and less well defined (72). Such simple systems, loosely and simply organized and defined, can undergo changes of a more dramatic sort than would be permissible in complex, highly integrated modern cells. Horizontal transfer of genes would be less disruptive of these primitive cells and their subsystems, and so the process of horizontal gene transfer would be of a very different nature, i.e., more pervasive, widespread, and spectacular, than it now is. Indeed, at very early times, horizontal gene transfer could have encompassed all aspects of the cell, all its genes (67). The process would dominate the primitive evolutionary dynamic; most of the evolutionary innovation would be horizontally acquired. At such early evolutionary stages, life (no matter how varied at the level of the individual organism) can be looked at as communal in the evolutionary sense, for it is only through horizontally shared innovation that the evolution leading to modern life was possible; i.e., the community evolved as a whole, not as individual organismal lineages (69; see also discussions of earlier forms of the idea in references 36 and 67).
Gradually, then, as the subsystems of the cell over time became increasingly complex and refined and as they became more intricately interwoven into the evolving fabric of the cell, horizontal gene transfers would become increasingly restricted: foreign parts tend not to be compatible with complex, precisely defined machines. Of course, the first systems in the cell to become so refined, which probably were the information-processing mechanisms—in particular translation (72)—would be the first to become refractory to horizontal gene transfer.
What the basal canonical pattern then represents is an early stage when the first (the more complex) subsystems of the cell became more or less refractory to horizontal gene transfer and the universal ancestor had differentiated into the communities that would spawn the primary organismal groupings. At that early time, the eukaryotic and archaeal lineages were still communally unified (at least in terms of the information processing systems) and all aspects of the cell had long evolutionary developments ahead of them.
The aminoacyl-tRNA synthetases that exhibit the canonical phylogenetic pattern are therefore those whose organismal taxonomic distributions became fixed at this early stage in cellular evolution. In their evolutionary profiles, these enzymes retain some record of the evolutions of the three basic cell types. Conversely, the aminoacyl-tRNA synthetases whose evolutionary profiles show little or no canonical pattern were still in evolutionary flux at this early stage; their organismal distributions would not stabilize until much later in the evolutionary course, after modern cells had evolved in some cases. The last synthetases to achieve their present taxonomic distribution, we think, are the members of the gemini group, to which we now return our attention.
The gemini group exemplifies the evolution of the amino acid charging systems. Multiple ways of associating amino acids with their tRNAs have always existed—as the existence of the two main synthetase structural classes implies. A considerable body of experimental data now shows that structures derived from RNA alone can both chemically discriminate between amino acids and catalyze aminoacylation with a high degree of substrate specificity (77). This ability of RNA to mimic some of the essential characteristics of contemporary aminoacyl-tRNA synthetases strongly supports older ideas that direct interactions between nucleic acids and amino acids (or peptides) contributed to the origin of translation (25, 71).
The evolutionary field is strewn with the relics of apparent takeover (replacement) battles among the tRNA-charging systems. Long before the universal ancestor gave rise to the primary organismal groupings, the ancestor of the LeuRS, IleRS, and ValRS, say, spawned some variant that came to displace earlier versions of the leucine, isoleucine, and valine-charging systems. We know these to be early events because the LeuRS, IleRS, and ValRS evolutionary profiles retain semblances of the canonical pattern. At that early time or before, the two unrelated enzymes of the glycine-charging system as well as the two lysine-charging enzyme also existed, but from their evolutionary profiles (i.e., from the lack of canonical pattern shown), it would appear that the takeover battles in these cases continued to some extent into the modern evolutionary era.
Serine and cysteine represent takeovers in which one of the two systems has achieved almost complete dominance. There seems to have been an archaeal type for both systems, quite different from the corresponding bacterial type. However, these archaeal types have now largely been displaced by their bacterial counterparts. The lack of canonical pattern in these cases is prima facie evidence that the spread of the bacterial types occurred late in the evolutionary course, probably after the major branchings in each of the primary organismal groupings had begun to form.
The final two aminoacyl-tRNA synthetases in the gemini group, glutamine and asparagine, add a solid time point to the developing picture of the evolutionary course. For each of these amino acids, the aminoacyl-tRNA synthetase has arisen from within the cluster defined by the synthetases of the corresponding dicarboxylic acid, and for both, the ancestral source has been a GluRS or AspRS of the archaeal/eukaryotic genre. (As seen above, the GluRS and AspRS enzymes exhibit strongly canonical pattern in their own right; i.e., they are ancient in both origin and taxonomic distribution.)
GlnRS arises specifically from the eukaryotic GluRS lineage (see the glutamine section above). Although AsnRS shows clear affinity with the archaeal and eukaryotic AspRSs (to the exclusion of bacterial AsnRSs), no convincing specific affinity to either the eukaryotic or archaeal AspRS is evident. This could be explained by the AsnRS arising somewhat prior to the GlnRS, which is also consistent with the wider taxonomic distribution of the former. Given its ancestry, the modern GlnRS, unlike its parental GluRS, had to have come into being after the three organismal domains themselves arose; the enzyme achieved its taxonomic distribution subsequently. We think that the lack of canonical pattern shown by all synthetases in the gemini group has a similar explanation, i.e., that all their taxonomic distributions, like that of GlnRS, were established relatively late in the evolutionary course, well after the domains themselves had arisen and their major branchings had begun to coalesce.
Some General Evolutionary Matters
It is one thing to note that certain bacterial taxa contain one or more aminoacyl-tRNA synthetases of the archaeal genre or that there are two very dissimilar types of a bacterial synthetase that are nevertheless of the same genre; e.g., TyrRS. It is another to understand how these came to be. In some cases, the source of the enzyme can be satisfactorily localized. Several examples exist (see above) in which the bacterial enzyme is not only of the archaeal genre but distinctly Pyrococcus-like. Such localizations will refine with the sequencing of additional archaeal genomes, just as will the precise source of the (plasmid-borne) mupirocin-resistant IleRS recently acquired by some Staphylococcus strains (9) (currently localized phylogenetically only to the general vicinity of the major gram-positive bacterial taxon represented by C. acetobutylicum [see the isoleucine section above]).
However, in a number of cases a bacterial synthetase is obviously of the general archaeal genre but resembles neither the archaeal nor the eukaryotic version specifically. This immediately raises a nontrivial question about the source of the synthetase. The same general question arises in the context of the bacteria alone when two very disparate versions of a synthetase exist, both bacterial in genre. The reflex answer to such questions, of course, is that undiscovered deeply branching lineages in either the Archaea or the Bacteria are the sources. Such explanations serve only to trivialize the questions. What are these hypothetical lineages? In the archaeal cases, some of them would need to branch not from the archaeal stem itself but from the common stem shared by the archaea and eukaryotes, in which case they would represent undiscovered organismal domains! Obviously, microbiologists have yet to uncover a great deal of the diversity in the microbial world, but the idea that in this day and age they have missed entire domains stretches credulity somewhat.
A more satisfying explanation may lie in the classical evolutionists' concept of evolutionary radiations, which again involves the tempo-mode relationship (55). The fossil record holds key evidence that otherwise would not be available, i.e., evidence of extinct lineages. In the world of macroscopic organisms, an evolution radiation (which is a period of rapid evolution) has three important characteristics: (i) accelerated evolutionary tempo, (ii) remarkable and remarkably diverse evolutionary invention, and (iii) creation of new lineages, most of which are short-lived (on the evolutionary timescale).
This last characteristic offers a conceptually challenging explanation for the point under discussion: the evolutions that underlay the development of the Bacteria and the Archaea (and Eucarya) were not the simple straightforward courses one might naively infer from the ancestral stems on phylogenetic trees. Rather, these ostensibly bare linear stems actually correspond to periods of great evolutionary turmoil, invention, and radiation. However, the evidence for almost all of this is gone (especially on the organismal level); the lineages are extinct. However, what the aminoacyl-tRNA synthetases may be telling us is that not all traces of extinct lineages are necessarily erased. Horizontal gene transfer makes it possible for some of the genes, some of the evolutionary inventions that occurred in these extinct lineages, to survive today, preserved by their transfer to lineages that have persisted.
As mentioned above, the horizontal transfer of aminoacyl-tRNA synthetase genes between Archaea and Bacteria is asymmetric in the sense that transfers of archaeal-type synthetases into the bacteria are common, both early on and after the organismal domains coalesced (and their major taxa had begun to emerge). Transfers of bacterial synthetases into the Archaea appear not to have occurred (at least early on [see the discussion of the gemini group above]). This asymmetry has no satisfactory explanation right now. It is not because archaea are refractory to the transfer of aminoacyl-tRNA synthetase (or any other) genes. We have seen ample evidence above that intra-archaeal exchanges occur. The explanation could be that the archaeal cell type and the bacterial cell type are of different natures, the one more permissive as regards accepting foreign genetic material than the other. Alternatively, the archaeal and bacterial cell types may not have matured evolutionarily (become modern cells) at the same time, so that transfer of genes occurred from a relatively mature archaeal type of cell to a relatively immature bacterial type of cell, which would therefore be more receptive to foreign genetic material.
It is not intended that the reader accept the above hypothetical scheme as truth; nor do we wish him or her to view it as idle and so useless speculation. The ideas we have put forth in this section are simply part of our attempt to paint a picture of the course of cellular evolution and to generate a theory that has the consistency, explanatory power, and conceptual power to inspire and inform studies of deep evolutionary questions.
CONCLUSION
It is unlikely that the aminoacyl-tRNA synthetases played any specific role in the evolution of the genetic code; their evolutions did not shape the codon assignments. Far from diminishing the evolutionary significance of the aminoacyl-tRNA synthetases, this realization, in a sense, enhances it. The separation of the two problems allows biologists to see the evolution of the code for the profound problem that it is and to focus on what the evolutions of these synthetases do tell us. Aminoacyl-tRNA synthetase genes are among the more frequently exchanged of the functionally important molecules in the cell. However, this does not diminish, but enhances, the value of the AARSs as evolutionary indicators. Admittedly, horizontal transfer introduces noise into organismal genealogical records, but evolution has dimensions, qualities, that are not captured in vertically inherited genes; it is precisely because the AARSs are horizontally transferred that they provide information about some of these otherwise inaccessible dimensions. The aminoacyl-tRNA synthetases appear to tell of the existence of deeply branching lineages in both the Bacteria and Archaea that do not have known (or extant) representatives. In that the evolutionary dynamic of horizontal gene transfer is in the first instance a function of the nature of the recipient cell, the fact that this dynamic appears increasingly to change (becoming more extreme in nature) the further back in time evolution is traced implies that even after the phylogenetic structure of the domains began to form, cells may not have been as they are today; they had yet to evolve, to mature into modern cells.
In all this is the suggestion that the evolutionary process is not what it might seem when attention is confined to the genome sequence level, i.e., a more or less continuous progression leading from very simple primitive forms to the modern ones. Rather, evolution may proceed through a series of discontinuous stages, with the quality of the evolving systems changing dramatically from stage to stage, and these stages can be defined by the type of horizontal gene transfer each exhibits. If we can infer the dynamics of horizontal gene transfer finely enough at early enough evolutionary times, the stages should reveal themselves. In the present review, we have used the AARSs merely for what they can tell about the later evolutionary stages, those associated with the emergence of the primary lines of descent. It is obvious that the synthetases collectively carry a record of far earlier times, earlier and more genetically “violent” evolutionary stages. These were stages that preceded those represented by the root of the universal (rRNA) phylogenetic tree, and they can be glimpsed (and so identified) by the evolutionary relationships among aminoacyl-tRNA synthetases of different amino acid types, in other words, by synthetase takeovers that are not confined within a given amino acid type.
Individually, the synthetases represent universal, constant, and ancient functions, which are defined mainly by the tRNAs—by all accounts among the most ancient and structurally constant of molecules in the cell. The selective pressures against the horizontal transfer of these enzymes seem minimal and general. In that the 20 aminoacyl-tRNA synthetases come as close as possible to being identical in evolutionary respects, their evolutions collectively serve to “interpret” one another; comparisons among them tend to sort out the trivial, and the idiosyncratic in each of their evolutionary records from the telling aspects of their histories. This is what makes the aminoacyl-tRNA synthetases so valuable a tool in attempts to reconstruct the history of life on this planet.
ACKNOWLEDGMENTS
Work in our laboratories on this subject was supported by grants from NASA and DOE (to C.R.W. and G.J.O.) and the National Institute of General Medical Sciences (to D.S.). M.I. is the recipient of an Investigator award from the Alfred Benzon Foundation. The computational work used resources at the W. M. Keck Center for Comparative and Functional Genomics.
We are extremely grateful to S. T. Fitz-Gibbon, W. Metcalf, and M. L. Sogin, who have provided data prior to publication. We thank D. Graham for help with the manuscript.
REFERENCES
- 1.Adduce J, Hoosegow M. MOLPHY: programs for maximum likelihood inference of protein phylogeny. Computer science monographs, no. 27. Tokyo, Japan: Institute of Statistical Mathematics; 1992. [Google Scholar]
- 2.Andersson S G, Zomorodipour A, Andersson J O, Sicheritz-Ponten T, Alsmark U C, Podowski R M, Naslund A K, Eriksson A S, Winkler H H, Kurland C G. The genome sequence of Rickettsia prowazekii and the origin of mitochondria. Nature. 1998;396:133–140. doi: 10.1038/24094. [DOI] [PubMed] [Google Scholar]
- 3.Arnez J G, Moras D. Structural and functional considerations of the aminoacylation reaction. Trends Biochem Sci. 1997;22:211–216. doi: 10.1016/s0968-0004(97)01052-9. [DOI] [PubMed] [Google Scholar]
- 4.Berthet-Colominas C, Seignovert L, Härtlein M, Grotli M, Cusack S, Leberman R. The crystal structure of asparaginyl-tRNA synthetase from Thermus thermophilus and its complexes with ATP and asparaginyl-adenylate: the mechanism of discrimination between asparagine and aspartic acid. EMBO J. 1998;17:2947–2960. doi: 10.1093/emboj/17.10.2947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brown J R. Aminoacyl-tRNA synthetases: evolution of a troubled family. In: Wiegel J, Adams M H W, editors. Thermophiles: the keys to molecular evolution and the origin of life? London, United Kingdom: Taylor & Francis; 1998. pp. 217–230. [Google Scholar]
- 6.Brown J R, Robb F T, Weiss R, Doolittle W F. Evidence for the early divergence of tryptophanyl- and tyrosyl-tRNA synthetases. J Mol Evol. 1997;45:9–16. doi: 10.1007/pl00006206. [DOI] [PubMed] [Google Scholar]
- 7.Brown J R, Doolittle W F. Root of the universal tree of life based on ancient aminoacyl-tRNA synthetase gene duplications. Proc Natl Acad Sci USA. 1995;92:2441–2445. doi: 10.1073/pnas.92.7.2441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brown J R, Doolittle W F. Gene descent, duplication, and horizontal transfer in the evolution of glutamyl- and glutaminyl-tRNA synthetases. J Mol Evol. 1999;49:485–495. doi: 10.1007/pl00006571. [DOI] [PubMed] [Google Scholar]
- 9.Brown J R, Zhang J, Hodgson J E. A bacterial antibiotic resistance gene with eukaryotic origins. Curr Biol. 1998;8:R365–R367. doi: 10.1016/s0960-9822(98)70238-6. [DOI] [PubMed] [Google Scholar]
- 10.Burbaum J J, Schimmel P. Structural relationships and the classification of aminoacyl-tRNA synthetases. J Biol Chem. 1991;266:16965–16968. [PubMed] [Google Scholar]
- 11.Commans S, Böck A. Selenocysteine inserting tRNAs: an overview. FEMS Microbiol Rev. 1999;23:335–351. doi: 10.1111/j.1574-6976.1999.tb00403.x. [DOI] [PubMed] [Google Scholar]
- 12.Curnow A W, Hong K-W, Yuan R, Kim S-I, Martins O, Winkler W, Henkin T M, Söll D. Glu-tRNAGln amidotransferase: a novel heterotrimeric enzyme required for correct decoding of glutamine codons during translation. Proc Natl Acad Sci USA. 1997;94:11819–11826. doi: 10.1073/pnas.94.22.11819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Curnow A W, Tumbula D L, Pelaschier J T, Min B, Söll D. Glutamyl-tRNAGln amidotransferase in Deinococcus radiodurans may be confined to asparagine biosynthesis. Proc Natl Acad Sci USA. 1998;95:12838–12843. doi: 10.1073/pnas.95.22.12838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cusack S. Aminoacyl-tRNA synthetases. Curr Opin Struct Biol. 1997;7:881–889. doi: 10.1016/s0959-440x(97)80161-3. [DOI] [PubMed] [Google Scholar]
- 15.Dennis P P. Ancient ciphers: translation in Archaea. Cell. 1997;89:1007–1010. doi: 10.1016/s0092-8674(00)80288-3. [DOI] [PubMed] [Google Scholar]
- 16.Diaz-Lazcoz Y, Aude J C, Nitschke P, Chiapello H, Landes-Devauchelle C. Evolution of genes, evolution of species: the case of aminoacyl-tRNA synthetases. Mol Biol Evol. 1998;15:1548–1561. doi: 10.1093/oxfordjournals.molbev.a025882. [DOI] [PubMed] [Google Scholar]
- 17.Doolittle R F, Handy J. Evolutionary anomalies among the aminoacyl-tRNA synthetases. Curr Opin Genet Dev. 1998;8:630–636. doi: 10.1016/s0959-437x(98)80030-0. [DOI] [PubMed] [Google Scholar]
- 18.Doolittle W F. Phylogenetic classification and the universal tree. Science. 1999;284:2124–2129. doi: 10.1126/science.284.5423.2124. [DOI] [PubMed] [Google Scholar]
- 19.Doublié S, Bricogne G, Gilmore C, Carter C W., Jr Tryptophanyl-tRNA synthetase crystal structure reveals an unexpected homology to tyrosyl-tRNA synthetase. Structure. 1995;3:17–31. doi: 10.1016/s0969-2126(01)00132-0. [DOI] [PubMed] [Google Scholar]
- 20.Eriani G, Delarue M, Poch O, Gangloff J, Moras D. Partition of tRNA synthetases into two classes based on mutually exclusive sets of sequence motifs. Nature. 1990;347:203–206. doi: 10.1038/347203a0. [DOI] [PubMed] [Google Scholar]
- 21.Felsenstein J. Cases in which parsimony or compatibility methods will be positively misleading. Syst Zool. 1978;27:401–410. [Google Scholar]
- 22.First E A. Catalysis of aminoacylation by class I and class II aminoacyl-tRNA synthetases. In: Sinnott M L, editor. Comprehensive biological catalysis. London, United Kingdom: Academic Press, Ltd.; 1998. pp. 573–607. [Google Scholar]
- 23.Freeland S J, Hurst L D. The genetic code is one in a million. J Mol Evol. 1998;47:238–248. doi: 10.1007/pl00006381. [DOI] [PubMed] [Google Scholar]
- 24.Gagnon Y, Lacoste L, Champagne N, Lapointe J. Widespread use of the Glu-tRNAGln transamidation pathway among bacteria. A member of the α purple bacteria lacks glutaminyl-tRNA synthetase. J Biol Chem. 1996;271:14856–14863. doi: 10.1074/jbc.271.25.14856. [DOI] [PubMed] [Google Scholar]
- 25.Gamow G A. Possible relation between deoxyribonucleic acid and protein structures. Nature. 1954;173:318. [Google Scholar]
- 26.Goldman N. Further results on error minimization in the genetic code. J Mol Evol. 1993;37:662–664. doi: 10.1007/BF00182752. [DOI] [PubMed] [Google Scholar]
- 27.Gray M W, Lang B F, Cedergren R, Golding G B, Lemieux C, Sankoff D, Turmel M, Brossard N, Delage E, Littlejohn T G, Plante I, Rioux P, Saint-Louis D, Zhu Y, Burger G. Genome structure and gene content in protist mitochondrial DNAs. Nucleic Acids Res. 1998;26:865–878. doi: 10.1093/nar/26.4.865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Haig D, Hurst L D. A quantitative measure of error minimization in the genetic code. J Mol Evol. 1991;33:412–417. doi: 10.1007/BF02103132. [DOI] [PubMed] [Google Scholar]
- 29.Hashimoto T, Sanchez L B, Shirakura T, Muller M, Hasegawa M. Secondary absence of mitochondria in Giardia lamblia and Trichomonas vaginalis revealed by valyl-tRNA synthetase phylogeny. Proc Natl Acad Sci USA. 1998;95:6860–6865. doi: 10.1073/pnas.95.12.6860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hamann C S, Sowers K R, Lipman R S, Hou Y M. An archaeal aminoacyl-tRNA synthetase missing from genomic analysis. J Bacteriol. 1999;181:5880–5884. doi: 10.1128/jb.181.18.5880-5884.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Henikoff S, Henikoff J G. Amino acid substitution matrices from protein blocks. Proc Natl Acad Sci USA. 1992;89:10915–10919. doi: 10.1073/pnas.89.22.10915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ibba M, Bono J L, Rosa P A, Söll D. Archaeal-type lysyl-tRNA synthetase in the Lyme disease spirochete, Borrelia burgdorferi. Proc Natl Acad Sci USA. 1997;94:14383–14388. doi: 10.1073/pnas.94.26.14383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ibba M, Curnow A W, Söll D. Aminoacyl-tRNA synthesis: divergent routes to a common goal. Trends Biochem Sci. 1997;22:39–42. doi: 10.1016/s0968-0004(96)20033-7. [DOI] [PubMed] [Google Scholar]
- 34.Ibba M, Losey C H, Kawarabayasi Y, Kikuchi H, Bunjun S, Söll D. Substrate recognition by class I lysyl-tRNA synthetases: a molecular basis for gene displacement. Proc Natl Acad Sci USA. 1999;96:418–423. doi: 10.1073/pnas.96.2.418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ibba M, Morgan S, Curnow A W, Pridmore D R, Vothknecht U C, Gardner W, Lin W, Woese C R, Söll D. A euryarchaeal lysyl-tRNA synthetase: resemblance to class I synthetases. Science. 1997;278:1119–1122. doi: 10.1126/science.278.5340.1119. [DOI] [PubMed] [Google Scholar]
- 36.Kandler O. Cell wall biochemistry in Archaea and its phylogenetic implications. J Biol Phys. 1994;20:165–169. [Google Scholar]
- 37.Kim H S, Vothknecht U C, Hedderich R, Celic I, Söll D. Sequence divergence of seryl-tRNA synthetases in Archaea. J Bacteriol. 1998;180:6446–6449. doi: 10.1128/jb.180.24.6446-6449.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lamour V, Quevillon S, Diriong S, N'Guyen V C, Lipinski M, Mirande M. Evolution of the Glx-tRNA synthetase family: the glutaminyl enzyme as a case of horizontal gene transfer. Proc Natl Acad Sci USA. 1994;91:8670–8674. doi: 10.1073/pnas.91.18.8670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lenhard B, Orellana O, Ibba M, Weygand-Durasevic I. tRNA recognition and evolution of determinants in seryl-tRNA synthesis. Nucleic Acids Res. 1999;27:721–729. doi: 10.1093/nar/27.3.721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li T, Graham D, Stathopoulos C, Haney P, Kim H-S, Vothknecht U, Kitabatake M, Hong K-W, Eggertsson G, Curnow A W, Lin W, Celic I, Whitman W, Söll D. Cysteinyl-tRNA formation: the last puzzle of aminoacyl-tRNA synthesis. FEBS Lett. 1999;462:302–306. doi: 10.1016/s0014-5793(99)01550-1. [DOI] [PubMed] [Google Scholar]
- 41.Marsh T L, Reich C I, Whitelock R B, Olsen G J. Transcription factor IID in the Archaea: Sequences in the Thermococcus celer genome would encode a product closely related to the TATA-binding protein of eukaryotes. Proc Natl Acad Sci USA. 1994;81:4180–4184. doi: 10.1073/pnas.91.10.4180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Martinis S A, Plateau P, Cavarelli J, Florentz C. Aminoacyl-tRNA synthetases: a family of expanding functions. Mittelwihr, France, October 10–15. EMBO J. 1999;18:4591–4596. doi: 10.1093/emboj/18.17.4591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Martinis S A, Plateau P, Cavarelli J, Florentz C. Aminoacyl-tRNA synthetases: a new image for a classical family. Biochimie. 1999;81:683–700. doi: 10.1016/s0300-9084(99)80126-6. [DOI] [PubMed] [Google Scholar]
- 44.Mazauric M H, Keith G, Logan D, Kreutzer R, Giegé R, Kern D. Glycyl-tRNA synthetase from Thermus thermophilus-wide structural divergence with other prokaryotic glycyl-tRNA synthetases and functional inter-relation with prokaryotic and eukaryotic glycylation systems. Eur J Biochem. 1998;251:744–757. doi: 10.1046/j.1432-1327.1998.2510744.x. [DOI] [PubMed] [Google Scholar]
- 45.Nagel G M, Doolittle R F. Phylogenetic analysis of the aminoacyl-tRNA synthetases. J Mol Evol. 1995;40:487–498. doi: 10.1007/BF00166617. [DOI] [PubMed] [Google Scholar]
- 46.Nureki O, Vassylyev D G, Katayanagi K, Shimizu T, Sekine S, Kigawa T, Miyazawa T, Yokoyama S, Morikawa K. Architectures of class-defining and specific domains of glutamyl-tRNA synthetase. Science. 1995;267:1958–1965. doi: 10.1126/science.7701318. [DOI] [PubMed] [Google Scholar]
- 47.Olsen G J, Woese C R, Overbeek R. The winds of (evolutionary) change: breathing new life into microbiology. J Bacteriol. 1994;176:1–6. doi: 10.1128/jb.176.1.1-6.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.RajBhandary U L. Once there were twenty. Proc Natl Acad Sci USA. 1997;94:11761–11763. doi: 10.1073/pnas.94.22.11761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ribas de Pouplana L, Buechter D, Sardesai N Y, Schimmel P. Functional analysis of peptide motif for RNA microhelix binding suggests new family of RNA-binding domains. EMBO J. 1998;17:5449–5457. doi: 10.1093/emboj/17.18.5449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Saks M E, Sampson J R, Abelson J. Evolution of a transfer RNA gene through a point mutation in the anticodon. Science. 1998;279:1665–1670. doi: 10.1126/science.279.5357.1665. [DOI] [PubMed] [Google Scholar]
- 51.Schimmel P, Ripmaster T. Modular design of components of the operational RNA code for alanine in evolution. Trends Biochem Sci. 1995;20:333–334. doi: 10.1016/s0968-0004(00)89067-2. [DOI] [PubMed] [Google Scholar]
- 52.Shiba K, Motegi H, Yoshida M, Noda T. Human asparaginyl-tRNA synthetase: molecular cloning and the inference of the evolutionary history of Asx-tRNA synthetase family. Nucleic Acids Res. 1998;26:5045–5051. doi: 10.1093/nar/26.22.5045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Siatecka M, Rozek M, Barciszewski J, Mirande M. Modular evolution of the Glx-tRNA synthetase family—rooting of the evolutionary tree between the bacteria and archaea/eukarya branches. Eur J Biochem. 1998;256:80–87. doi: 10.1046/j.1432-1327.1998.2560080.x. [DOI] [PubMed] [Google Scholar]
- 54.Simos G, Sauer A, Fasiolo F, Hurt E C. A conserved domain within Arc1p delivers tRNA to aminoacyl-tRNA synthetases. Mol Cell. 1998;1:235–242. doi: 10.1016/s1097-2765(00)80024-6. [DOI] [PubMed] [Google Scholar]
- 55.Simpson G G. Tempo and mode in evolution. New York, N.Y: Columbia University Press; 1944. [Google Scholar]
- 56.Sissler M, Delorme C, Bond J, Elrlich S D, Renault P, Francklyn C. An aminoacyl-tRNA synthetase paralog with a catalytic role in histidine biosynthesis. Proc Natl Acad Sci USA. 1999;96:8985–8990. doi: 10.1073/pnas.96.16.8985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Söll, D., and M. Ibba. Aminoacyl-tRNA synthesis. Annu. Rev. Biochem., in press. [DOI] [PubMed]
- 58.Söll D, RajBhandary U L. tRNA: structure, biosynthesis and function. Washington, D.C.: ASM Press; 1995. [Google Scholar]
- 59.Sonneborn T M. Degeneracy of the genetic code: extent, nature and genetic implications. In: Bryson V, Vogel J H, editors. Evolving genes and proteins. New York, N.Y: Academic Press, Inc.; 1965. pp. 377–397. [Google Scholar]
- 60.Stathopoulos C, Li T, Longman R, Vothknecht U C, Becker H, Ibba M, Söll D. One polypeptide with two aminoacyl-tRNA synthetase activities. Science. 2000;287:479–482. doi: 10.1126/science.287.5452.479. [DOI] [PubMed] [Google Scholar]
- 61.Swofford D L. PAUP 4.0: phylogenetic analysis using parsimony (and other methods). Sunderland, Mass: Sinauer Associates; 1999. [Google Scholar]
- 62.Taupin C M, Leberman R. Archaebacterial seryl-tRNA synthetases: adaptation to extreme environments and evolutionary analysis. J Mol Evol. 1999;48:408–420. doi: 10.1007/pl00006485. [DOI] [PubMed] [Google Scholar]
- 63.Tumbula D, Vothknecht U C, Kim H S, Ibba M, Min B, Li T, Pelaschier J T, Stathopoulos C, Becker H, Söll D. Archaeal aminoacyl-tRNA synthesis: diversity replaces dogma. Genetics. 1999;152:1269–1276. doi: 10.1093/genetics/152.4.1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wakasugi K, Schimmel P. Two distinct cytokines released from a human aminoacyl-tRNA synthetase. Science. 1999;284:147–151. doi: 10.1126/science.284.5411.147. [DOI] [PubMed] [Google Scholar]
- 65.Watanabe K, Osawa S. In: tRNA: structure, biosynthesis, and function. Söll D, RajBhandary U, editors. Washington, D.C.: ASM Press; 1995. [Google Scholar]
- 66.Woese C R. On the evolution of the genetic code. Proc Natl Acad Sci USA. 1965;54:1546–1552. doi: 10.1073/pnas.54.6.1546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Woese C R. Archaebacteria and cellular origins: an overview. Zentbl Bakteriol Mikrobiol Hyg I Abt Orig C. 1982;3:1–17. [Google Scholar]
- 68.Woese C R. Bacterial evolution. Microbiol Rev. 1987;51:221–271. doi: 10.1128/mr.51.2.221-271.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Woese C R. The universal ancestor. Proc Natl Acad Sci USA. 1998;95:6854–6859. doi: 10.1073/pnas.95.12.6854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Woese C R, Dugre D H, Dugre S A, Kondo M, Saxinger W C. On the fundamental nature and evolution of the genetic code. Cold Spring Harbor Symp Quant Biol. 1966;31:723–736. doi: 10.1101/sqb.1966.031.01.093. [DOI] [PubMed] [Google Scholar]
- 71.Woese C R, Dugre D H, Saxinger W C, Dugre S A. The molecular basis for the genetic code. Proc Natl Acad Sci USA. 1966;55:966–974. doi: 10.1073/pnas.55.4.966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Woese C R, Fox G E. The concept of cellular evolution. J Mol Evol. 1977;10:1–6. doi: 10.1007/BF01796132. [DOI] [PubMed] [Google Scholar]
- 73.Woese C R, Kandler O, Wheelis M L. Towards a natural system of organisms: proposal for the domains Archaea, Bacteria, and Eucarya. Proc Natl Acad Sci USA. 1990;87:4576–4579. doi: 10.1073/pnas.87.12.4576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Woese C R, Mandelco L, Yang D, Gherna R, Madigan M T. The case for relationship of the flavobacteria and their relatives to the green sulfur bacteria. Syst Appl Microbiol. 1990;13:258–262. doi: 10.1016/s0723-2020(11)80196-7. [DOI] [PubMed] [Google Scholar]
- 75.Wolf Y I, Aravind L, Grishin N V, Koonin E V. Evolution of aminoacyl-tRNA synthetases-analysis of unique domain architectures and phylogenetic trees reveals a complex history of horizontal gene transfer events. Genome Res. 1999;9:689–710. [PubMed] [Google Scholar]
- 76.Yarus M. Amino acids as RNA ligands: a direct-RNA-template theory for the code's origin. J Mol Evol. 1998;47:109–117. doi: 10.1007/pl00006357. [DOI] [PubMed] [Google Scholar]
- 77.Yarus M, Illangasekare M. Aminoacyl-tRNA synthetases and self acylating ribozymes. In: Gesteland R F, Cech T R, Atkins J F, editors. The RNA world. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1999. pp. 183–196. [Google Scholar]