Abstract
The banded krait, Bungarus fasciatus is a widespread elapid snake, likely to comprise several distinct species in different geographic regions of Asia. Therefore, based on molecular phylogenetics and comparative morphology data, we present an overview of the systematic composition of the species to delimit potential biogeographic boundaries. Our phylogenetic analyses, based on four mitochondrial genes, reveal the existence of at least three evolutionary lineages within B. fasciatus, corresponding to Indo-Myanmar, Sundaic and eastern Asian lineages. We are convinced that there are at least three taxonomic entities within the nomen B. fasciatus and restrict the distribution of B. fasciatus sensu stricto to the Indo-Myanmar region. We also provide additional natural history data of the taxon from eastern India. Finally, we advocate further studies to establish the degree of reproductive isolation among these diverging evolutionary lineages and to reassess the systematic status of this species complex especially the Sundaic and eastern Asian lineages.
Subject terms: Herpetology, Phylogenetics, Taxonomy
Introduction
Aside from its taxonomical importance, recognition and ascertainment of independently evolving lineages is crucial for understanding the evolutionary processes affecting the origin of population structure and species diversification1. Because of the growing availability of genetic methods for species delineation2, numerous studies have uncovered cryptic diversity within the widespread vertebrate species including in tropical and sub-tropical Asia; for instance, among fishes3–5, amphibians6–8, birds9–11, and mammals12–14. Moreover, recent phylogeographical and molecular studies have refined our understanding of cryptic speciation across biogeographic boundaries or within biogeographic regions15,16, and even propounded the suitability of reptiles in particular as biogeographic indicators17,18. Recent studies focussing on widespread reptilian species have also established the existence of previously unnoticed cryptic diversity, including in lizards19–22 and snakes23–30.
Bungarus Daudin, 1803, collectively known as kraits, are venomous elapid snakes which inhabit the Asian subcontinent31. Most of the nominal Bungarus species are poorly understood. However, recent study on the diversification and evolution of elapid snakes have highlighted that the diversification of kraits occurred around 30–25 million years ago, and are close relatives of other Australasian elapid genera and sea snakes32. Bungarus fasciatus (Schneider, 1801), commonly known as the banded krait, is a nocturnal and conspicuous krait that grows up to 2,250 mm in total length and is morphologically characterized by its yellow (or cream) and black banded body33. It occurs in various habitat types such as primary forests, agricultural lands as well as domestic gardens up to 2,300 m above sea level33,34. So far, B. fasciatus has been reported from eastern India, Nepal, Bhutan, Bangladesh, and Myanmar, extending southwards through Thailand, Malaysia and Singapore into the Indonesian archipelago, and eastwards through Laos, Vietnam and China35,36. The species is currently listed as a Least Concern (LC) species in the IUCN Red List35. Despite its wide distribution, studies have so far been conducted mainly on its potential medical significance37, ecological importance38,39, or characterization of venom40–45.
Although there are no studies specifically on the molecular systematics of this species, several previous studies have highlighted intra-specific or geographical variability based on genetic barcoding46–48. Accurate species delimitation is crucial in view of the variability in snake venom composition49 and its potential effects on antivenom efficacy50. Most of the existing taxonomic and systematic literature on Bungarus have apparently overlooked the intraspecific diversity of B. fasciatus51–58. Therefore, in this study we fill in the inherent knowledge gaps by providing comparative morphological evidence and molecular phylogeny based on four mitochondrial genes (COI, CYTB, ND4 and 16S rRNA) based on sequences from east and northeast India, Indochina, and the Greater Sunda islands. Moreover, given the minimal knowledge on the natural history, reproductive behaviour, and ecology, which are important for assessing the population status of the species34,59, we also provide natural history data for the populations of B. fasciatus from India.
Materials and methods
Sampling
For this study, we collected both morphological and genetic data for Bungarus fasciatus, which we compared to publicly available or unpublished data. We collected morphological data for the B. fasciatus population represented by 15 specimens from northeastern India between the years 2007–2022. We surveyed during the day and night, collected individuals by hand, and euthanized them with MS-222 following the standard procedure60 in compliance with the American Veterinary Medical Association (AMVA) guidelines and approved by the Institutional Animal Ethics Committee (IAEC) (Permission No. MZU-IAEC/2018/12). We then fixed the specimens in 10% buffered formalin solution overnight, prior to their storage in 70% ethanol. We preserved liver tissue samples for DNA analysis in 95% ethanol, which were stored at −20 °C. Vouchered specimens were deposited at the Departmental Museum of Zoology, Mizoram University (MZMU). Additional blood samples from the caudal sinus were collected from the West Bengal (WB) populations and preserved in EDTA-Tris buffer; these specimens were subsequently released after taking necessary scale counts. Our study is reported in accordance with the ARRIVE 2.0 guidelines (Animal Research: Reporting of In Vivo Experiments)61. The distribution map was prepared using QGIS v3.16.2 and the digital elevation model (DEM) was downloaded from Open Topography (https://opentopography.org/).
DNA extraction, amplification and molecular analyses
Liver tissue or blood was used to extract genomic DNA using DNeasy (Qiagen™) blood and tissue kits following the manufacturer’s instructions. Fragments of four mitochondrial (mt) markers (16S, COI, ND4 and CYTB) were amplified in a 20 μL reaction volume, containing 1X DreamTaq PCR Buffer, 2.5 mM MgCl2, 0.25 mM dNTPs, 0.2 pM of each gene primer pair, approximately 3.0 ng of extracted DNA, and 1 U of Taq polymerase. A negative control with reagent grade water instead of DNA template was always included. Target mt gene sequences were amplified using the thermal profiles and primers given in Supplementary Table S1. PCR products were checked using gel electrophoresis on a 1.5% agarose gel containing ethidium bromide. The PCR products were cleaned using ThermoFisher ExoSAP-IT PCR product cleanup reagent and subsequently sequenced using the Sanger dideoxy method using the ABI 3730xl DNA Analyzer at Barcode BioSciences, Bangalore, India. The generated partial gene sequences were deposited on the NCBI repository (GenBank accession numbers are given in Supplementary Table S2). In this study, a total of one COI, six 16S, six ND4, and nine CYTB sequences were generated and were combined with published sequences of B. fasciatus obtained from the NCBI database; database sequences of B. caeruleus, B. candidus, B. ceylonicus, B. sindanus, and B. multicinctus were used as outgroups. The four mt gene alignments were concatenated in SequenceMatrix62. Using the CYTB dataset, the uncorrected p-distance was estimated in MEGA X using the complete deletion option for the treatment of gaps/missing data63. Prior to the Bayesian analysis, PartitionFinder v2.164 was utilized to search the best partitioning schemes and the best fitting model through Bayesian Information Criterion (BIC) (Supplementary Table S3). Bayesian phylogeny (BI) was reconstructed using the selected models in Mr.Bayes v3.2.565. The MCMC was run with four chains (one cold and three hot chains) for 20 million generations and sampled every 5000 generations. Tracer v1.766 was used to check the convergence of likelihood and the burn-in cut-off. The diagnosis of topological convergence and MCMC and mixing of chains was done in R-Studio67 using the package, R We There Yet (RWTY)68. The BI tree was further illustrated using web-based tree annotator iTOL software v569. The Maximum Likelihood (ML) tree was reconstructed in IQ-TREE70 using 10,000 Ultrafast Bootstrap (UFB)71 based on the dataset partitioned by codon positions with the most appropriate model selected for each partition using ModelFinder72 integrated in IQ-TREE70. The CYTB dataset, partitioned by codon, was utilized for performing BI and ML based Poisson Tree Processes (PTP) species delineation analyses73 implemented in iTaxoTools v0.174. For the input file of PTP, a non-ultrametric tree was produced in IQ-TREE70 with 10,000 UFB replicates71 using the models selected for CYTB partitions. Only the CYTB dataset was selected for the species delimitation analysis as it contains more samples from different geographical regions compared to the other three genes.
Morphology
We obtained morphometric (mensural and meristic) data for species comparisons, and distribution data from examined specimens (Java (JV), Mizoram (MZ) and WB) and published literature54,75–77. We measured the following characters to the nearest millimetre with a Mitutoyo digital caliper and Leica M50 (Leica Microsystems Inc.) dissecting microscopes: eye diameter (ED, horizontal diameter of orbit); eye–nostril length (EN, distance between anteriormost point of eye and middle of nostril); snout length (ES, distance between anteriormost point of eye and snout); head length (HL, distance between posterior edge of mandible and tip of snout); head width (HW, maximum width of head); snout–vent length (SVL, measured from tip of snout to anterior margin of vent); tail length (TaL, measured from anterior margin of vent to tail tip). Meristic characters were taken as follows: supralabials (SL) and infralabials (IL) (first labial scale to last labial scale bordering gape); dorsal scale rows (DSR, counted around the body from one side of ventrals to the other in three positions, on one head length behind neck, at midbody and at one head length prior to cloacal plate); when counting the number of ventral scales (Ve), we scored values according to the method described by Dowling78. We counted subcaudal scales (Sc) from the first subcaudal scale meeting its opposite to the scale before the tip of the tail, the terminal scute is excluded when counting. Sex of the specimens was identified by examining everted hemipenes or by ventral tail dissection. We evaluated the relative size of the nuchal band, the number of the black cross bands of each individual. The number of cross bands on the body (BB) were counted from the first band posterior to the nuchal band on the nape up to the level of cloaca, the count on the tail from the level of cloaca to the tip of tail (BT), and number of vertebral scales covering the nuchal band (NBW). In addition, the number of vertebral scales covering the first cross band is also considered a reliable character for adult individuals. Values for bilateral head characters are given in left/right order. We followed Keogh79 for hemipenial terminology, and the extent of inverted hemipenis in terms of percentage of subcaudal scales (HpR).
Statistical analyses
The morphological information was obtained from three different populations examined by us: recent and long-term preserved specimens from JV in Indonesia (n = 15), live specimens from WB (n = 8) and live, recent and long-term preserved specimens from MZ (n = 15) states in India. Before performing any further analyses, the meristic data were standardized to zero mean and unit standard deviation to avoid potential bias due to difference in the range of measurement among variables; for mensural data, the combination of characters with the highest R-squared score obtained through linear regression was selected as the best log transformation model to make linear relationship with body size. Since we do not have gender information from the WB population, the meristics of the remaining populations (JV and MZ) were first tested using separate one-way analysis of variance (ANOVA) using sex and locality as factors along with Levene’s test80 to test the homogeneity of variances; if the assumption of homoscedascity was violated, Brown-Forsythe test81 was utilised as an alternative approach. For mensurals (TaL, HL, and HW), a two-way analysis of covariance (ANCOVA) was carried out with snout-vent length (SVL) as a covariate. The meristic variables identified with no sexual dimorphism were utilised for multiple comparison among the three populations by pooling sexes using one-way ANOVA using locality as a factor, and post-hoc was performed with applying Bonferroni correction. In addition, a potential observer difference was screened by repeating measurements on the same specimens and then tested using one-way ANCOVA. The variable characters among lineages identified through the univariate analyses were utilized further for Principal Component Analysis (PCA) to visualize the clustering of the different populations. The correlation matrices between all pairs of the morphological variables, variance explained by each eigenvalue as well as the correlations of each variable to the first two components are explored. Specimens with missing characters were excluded in the multivariate analysis. Statistical analyses were performed using the SPSS v.25.0 statistical package (Armonk, NY: IBM Corp.).
Results
Phylogenetic relationship
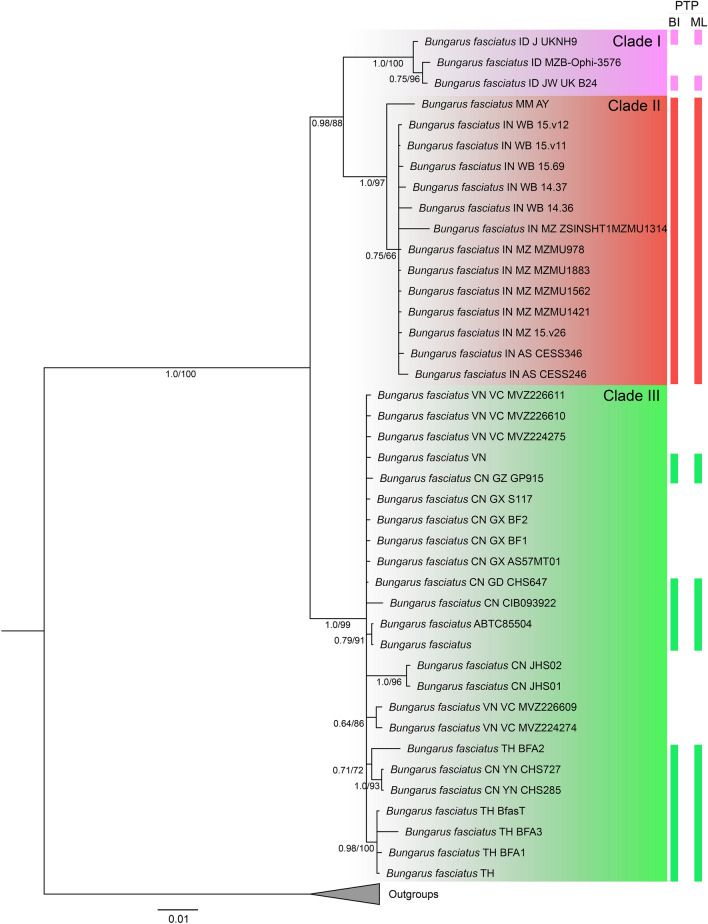
The first 25% of trees from the BI analysis were discarded as burn-in, and the standard deviation of split frequencies were < 0.005 when analyses terminated. The graphs created using RWTY in R-Studio also indicated satisfactory topological mixing. The inferred concatenated trees from BI and ML analyses were congruent with each other. The BI tree, created using Mr.Bayes v3.2.565 and further illustrated using iTOL software v569, is show in Fig. 1, with Bayesian posterior probabilities from the BI analysis and UFB values from the ML analysis. The CYTB dataset consisted of a total of 1047 aligned characters, with 97 variable sites (excluding outgroups).
Figure 1.
Bayesian inference (BI) phylogenetic tree based on concatenated mitochondrial 16S, COI, ND4 and CYTB genes; lineage partitions recovered from CYTB-based PTP analyses are presented besides the BI tree (only the CYTB dataset was utilized for PTP analyses because it contains more representative samples from the three clades compared to the other genes). Values at each node represent Bayesian posterior probabilities (PP) and Ultrafast Bootstrap (UFB) values from the Maximum Likelihood (ML) analysis (PP/UFB). Abbreviations of country and state/province names are: ID Indonesia, JW/J Java, MM Myanmar, AY Ayeyarwady, IN India, WB West Bengal, MZ Mizoram, AS Assam, VN Vietnam, VC Vinh Phuc, CN China, GZ Guizhou, GX Guangxi, GD Guangdong, YN Yunnan, TH Thailand.
Molecular phylogenetic based on the concatenated aligned matrix for four mitochondrial genes (16S, COI, ND4, and CYTB; 2850 bp in length), recovered a monophyletic clade consisting of three lineages within Asia. Both the phylogenetic analyses and the single-locus-based PTP species delineation approach significantly support these three distinct clades which we describe as, (i) B. fasciatus from the Sundaic region, especially from Great Sunda islands which we describe as the Sundaic lineage (Clade I; Fig. 1); (ii) B. fasciatus from Indo-Myanmar (Clade II; Fig. 1), and (iii) B. fasciatus from mainland Sundaland including southern China, here described as east Asian lineage (Clade III; Fig. 1).
The overall mean intra-specific divergence across all lineages of B. fasciatus (uncorrected p-distance) was 3.5%. Furthermore, 0.4% intra-clade genetic divergence was observed within Clade I (between two locations in JV), 0.0%–1.3% within Clade II (between India and Myanmar), and 0.0%–6.5% within Clade III (among China, Vietnam, Thailand, and an unknown locality). The mean inter-clade genetic divergence is 5.0% between Clade I (Sundaic) and Clade II (Indo-Myanmar), 5.3% between Clade II (Indo-Myanmar) and III (east Asia); 5.7% between Clade I (Sundaic) and III (east Asia). Combined B. fasciatus (Clades I + II + III) shows the least inter-specific genetic divergence (19.5%–19.8%) with B. candidus, while inter-specific distances among other species (B. sindanus, B. caeruleus, B. candidus, B. ceylonicus, and B. multicinctus) range from 3.0% (between B. candidus and B. multicinctus) to 19.0% (between these two species and B. ceylonicus) (also see Supplementary Table S4).
Morphometric analysis
In this study, despite limited sampling, morphometric analyses were performed to identify taxonomically informative characters among the examined populations (WB, MZ and JV). Only the mensurals such as TaL (p < 0.001), HW (p < 0.05) and HL (p < 0.05) showed significantly dimorphic characters between males and females within JV and MZ populations. For meristic characters, inter-population differences were statistically significant (p < 0.001) for Ve (MZ vs. JV), BB, BT, and NBW (the latter three characters are tested among three populations), all of which showed a higher number in the MZ population; for mensural characters, inter-population differences were also statistically significant for TaL (p < 0.05) and HL (p < 0.001) (Table 1). Post-hoc tests conducted among the three populations for BB, BT, and NBW showed that, except for BT between MZ and WB populations (p > 0.05), significant differences are seen for all characters: BB (p < 0.001 across all the populations), NBW (p < 0.001 in MZ vs. WB, and JV vs. WB; p < 0.05 in MZ vs. JV), and BT (p < 0.001 in MZ vs. JV; p < 0.01 in JV vs. WB). Comparison was also made based on the identified variable meristic characters among the three populations using a PCA. The correlation matrix showed weak correlations between pairs of variables (r < 0.7); thus, all variables were retained for this analysis. The first two components accounted for 84% of the total variation of the data, with PC1, PC2 and PC3 representing 64%, 20% and 11%, respectively. The loadings of all variables are high on the first axis, while only Ve loads considerably highly on the second axis, with Ve having less effect on PC1 than PC2 (Supplementary Table S5). The representation of the first two components depicts substantial separation of the Javanese and the Indian populations on the first axis (PC1), and marginal separation of the WB and MZ populations on the second axis (PC2) (Fig. 2). Given that the samples from the three populations (WB, MZ and JV) were examined by different recorders, we also tested for potential recorder bias between the East Indian and northeast Indian specimens; however, no significant differences were seen after re-examination of the same specimens (p > 0.05).
Table 1.
Evaluation on the meristic and mensural characters measured for 38 Bungarus fasciatus individuals from Java (JV), Mizoram (MZ), and West Bengal (WB), including mean, standard deviation, minimum and maximum values. Standardized meristic data were utilised for the following tests: Ve of JV and MZ was tested for inter-population difference and sexual dimorphism using separate one-way ANOVA with locality and sex as the factors, respectively; Sc of JV and MZ was tested using two-way ANCOVA using sex and locality as factors; BB and NBW were tested for inter-population difference (among the three populations) and sexual dimorphism (within JV and MZ) using separate one-way ANOVA with locality and sex as the factors, respectively; since BT violated the assumption of homoscedascity, it was tested using the alternative Brown-Forsythe test and was indicated by octothorp (#). For mensurals, two-way ANCOVA was performed for the log transformed TaL, HL, and HW values from JV and MZ by using the log transformed SVL as a covariate, with locality and sex as the factors. The characters with statistically significant variations at the alpha level of 0.05 are shown in boldface. The characters tested for inter-population difference across the three populations are indicated by asterisk (*).
| Characters | Sex | Java (n = 15) | Mizoram (n = 15) | West Bengal (n = 8) unsexed | Sexual dimorphism | Inter-population difference | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Mean ± SD | Range | Mean ± SD | Range | Mean ± SD | Range | ||||||
| Ve | Male | 205.44 ± 3.43 | 199–210 | 226 ± 2.10 | 222–228 | 217.63 ± 3.12 | 212–222 | F1,28 = 1.35 | p = 0.256 | F1,28 = 469.80 | p < 0.001 |
| Female | 206.83 ± 1.94 | 205–210 | 229.11 ± 2.15 | 224–231 | |||||||
| Sc | Male | 34.43 ± 0.98 | 33–36 | 35.83 ± 0.75 | 35–37 | 34.63 ± 1.49 | 31–36 | F1,25 = 2.44 | p = 0.131 | F1,25 = 1.30 | p = 0.266 |
| Female | 31.17 ± 1.60 | 30–34 | 33.75 ± 1.28 | 32–36 | |||||||
| BB | Male | 22.67 ± 1.12 | 21–25 | 24.33 ± 1.97 | 22–27 | 28.38 ± 1.73 | 26–31 | F1,28 = 0.44 | p = 0.511 | F2,35 = 39.78* | p < 0.001* |
| Female | 21.83 ± 1.17 | 20–23 | 25.00 ± 1.58 | 23–27 | |||||||
| BT | Male | 3.22 ± 0.67 | 2–4 | 5.00 ± 0.00 | 5 | 5.25 ± 1.09 | 4–7 | F1,21 = 0.12# | p = 0.728# | F2,12 = 17.86*# | p < 0.001*# |
| Female | 3.17 ± 0.41 | 3–4 | 4.22 ± 0.44 | 4–5 | |||||||
| NBW | Male | 19.00 ± 1.00 | 18–20 | 18.20 ± 0.45 | 18–19 | 15.63 ± 1.11 | 14–17 | F1,27 = 0.40 | p = 0.533 | F2,34 = 22.16* | p < 0.001* |
| Female | 19.00 ± 0.63 | 18–20 | 17.67 ± 1.73 | 15–20 | |||||||
| TaL | Male | 120.74 ± 20.01 | 90–145 | 101 ± 38.92 | 47–133 | – | – | F1,24 = 18.96 | p < 0.001 | F1,24 = 6.01 | p = 0.022 |
| Female | 107.86 ± 23.43 | 85–145 | 97.88 ± 15.56 | 76–119 | |||||||
| HL | Male | 35.06 ± 4.97 | 27.10–40.90 | 21.60 ± 5.71 | 12.80–26.60 | – | – | F1,24 = 4.37 | p = 0.047 | F1,24 = 79.38 | p < 0.001 |
| Female | 34.81 ± 6.19 | 25.90–44.50 | 21.03 ± 5.03 | 15.74–29.68 | |||||||
| HW | Male | 20.88 ± 4.03 | 13.80–25.70 | 17.79 ± 5.10 | 12.18–22.46 | – | – | F1,25 = 4.33 | p = 0.048 | F1,25 = 0.97 | p = 0.334 |
| Female | 20.70 ± 3.13 | 16.40–26.20 | 16.12 ± 4.30 | 10.40–22.76 | |||||||
Significant values are in bold.
Figure 2.
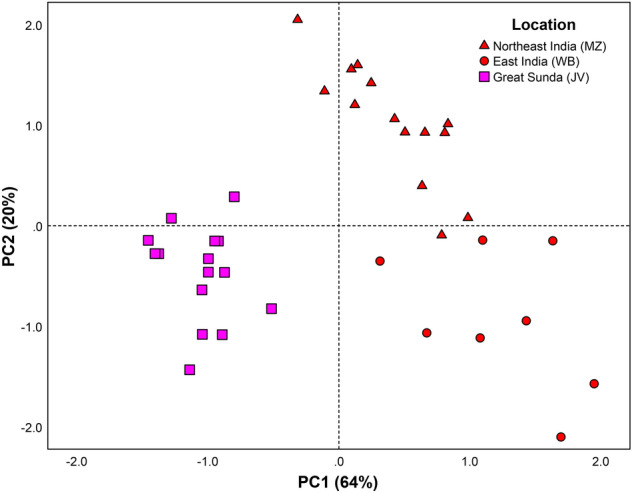
Ordination of Bungarus fasciatus populations from Mizoram (MZ), West Bengal (WB) and Java (JV) along the first two principal components based on a PCA of the characters Ve, BB, BT, and NBW. Total variance associated with PC1 and PC2 are 64% and 20%, respectively.
Systematics
We present diagnostic morphological, morphometric, and meristic data taken for Bungarus fasciatus Clade II from east and northeast India (Supplementary Table S6). The examined specimens of B. fasciatus from India are morphologically distinguishable from the Sundaic population (see Table 2). Based on the present study, we postulate the existence of at least three different taxonomic entities within the nomen B. fasciatus, and also confirm that populations in eastern India (e.g. Odisha, WB, etc.) and northeastern India (e.g. MZ, Assam, etc.) are conspecific. Based on the original description of Pseudoboa fasciata, minimum three specimens were available or referable to Schneider82; hence syntypes. Among these syntypes two specimens (ZMB 2771, 2772) have been deposited at ZMB from the collection of Marcus Bloch (fide Bauer83). In addition, one of syntypes was depicted in Russell84 (page 3, plate 3) as the “Bungarum Pamah”, an adult from “Mansoor Cottah” (now Gobalpur, Odisha (Orissa), India), specimen is now lost (fide Bauer85). So far, the only existing name-bearing type specimens are the two syntypes in the collection of Berlin Zoological Museum (ZMB 2771–72) originating from “Indien” (= India) fide ZMB catalogue36 a detailed taxonomic revision will be published elsewhere (Amarasinghe et al. in preparation). We affirm that the specimen used by Russell84 for his illustration is the same specimen (syntype) housed in the ZMB, thus we adhere with the type locality given by Russell84. Therefore, here we postulate the Indo-Myanmar populations (Clade II) as B. fasciatus sensu stricto, while considering the populations from Sundaic region, especially from Greater Sunda Islands (Clade I) and mainland Sundaland including southern China (Clade III) as B. fasciatus sensu lato. Consequently, we redescribe the B. fasciatus sensu stricto, including hemipenis morphology, based on MZ population, from where a large number of samples are available.
Table 2.
Some comparative morphological data of Bungarus fasciatus sensu lato in each biogeographic region, based on this study and published data.
| Character | Population/clade | ||
|---|---|---|---|
| Indo-Myanmar (n = 23) | East Asia (n = 11) | Greater Sunda (n = 15) | |
| Ventrals | 200–234 | 217–237 | 199–210 |
| Subcaudals | 23–39 | 33–41 | 30–36 |
| Number of dorsal bands on body | 22–31 | 19–21 | 20–25 |
| Number of dorsal bands on tail | 4–7 | ? | 2–4 |
| Nuchal band covered by vertebral scales | 14–20 | ? | 18–20 |
| Background body color | Yellow/cream | Yellow | Yellow/cream |
| Source |
Smith75 This study |
Yang & Rao76; |
This study |
Bungarus fasciatus (Schneider, 1801) sensu stricto
(Tables 1, 2; Figs. 3A–E, 4A,B, 5).
Figure 3.
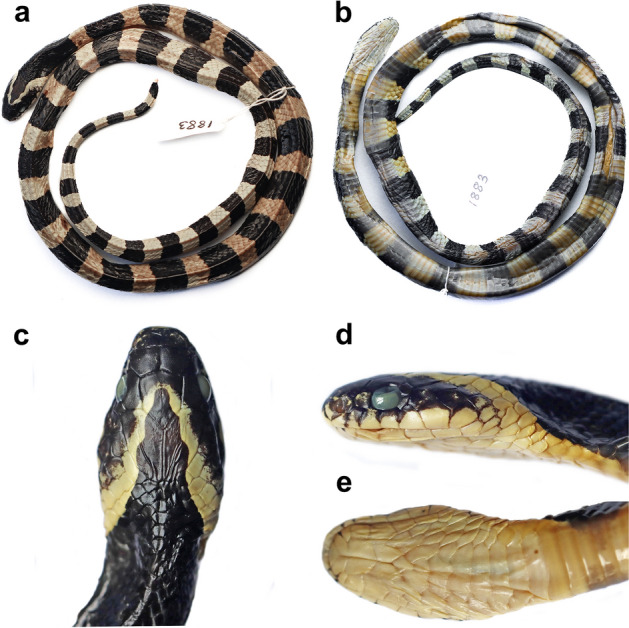
Bungarus fasciatus sensu stricto (MZMU1883) from Northeast India: (a) dorsal view of full body, (b) ventral view of full body, (c) dorsal view of head, (d) lateral view of the left side of head, and (e) ventral view of head.
Figure 4.

Live individuals of Bungarus fasciatus sensu stricto (a) from Keitum village, Mizoram, India (MZMU1421), and (b) a juvenile with creamish dorsum coloration from Saikhawthlir village, Mizoram, India.
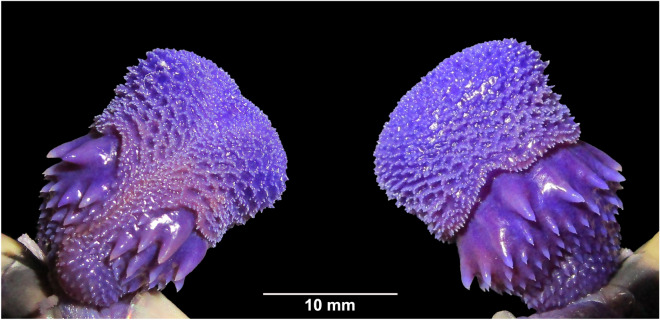
Figure 5.
Sulcal (left) and asulcal (right) views of the right hemipenis of Bungarus fasciatus sensu stricto (MZMU2935) from Mizoram, India.
[English: Banded krait; Bengali: Sankhamuti/Sankhini/Chamorkasa; Mizo: Chawnglei/Tiangsir].
Pseudoboa fasciata Schneider, 1801.
Bungarus annularis Daudin, 1803.
Bungarus fasciatus bifasciatus Mell, 1929.
Bungarus fasciatus insularis Mell, 1930.
Examined materials
Males (n = 7; MZMU 933, 1314, 1320, 1417, 1421, 1883, 2935) and Females (n = 8; MZMU 1319, 1321, 1550, 1562, 1561, 1548, 1572, 2481) collected from MZ, northeast India.
Species redescription
Based on the overall examined MZ materials with combined sexes, adults SVL 444.0–1220.0 mm, tail length 47.0–133.0 mm; head elongate (HL 2.0–3.5% of SVL), wide (HW 71.8–92.1% of HL), slightly flattened, indistinct from neck; snout elongate (ES 22.8–40.1% of HL), moderate, flat in dorsal view, rounded in lateral profile, rather depressed. Rostral shield large, flat, slightly visible from above, pointed posteriorly; interorbital width broad; internasals subtriangular; nostrils rather large, nasals large, divided, and elongated, in anterior contact with rostral, and internasal and prefrontal dorsally, 1st and 2nd supralabial ventrally, preocular posteriorly; no loreal; prefrontal rather large, broader than long, and pentagonal; frontal large, hexagonal, short, slightly longer than width; supraoculars narrow, elongate, subrectangular, posteriorly wider; parietals large, elongate, butterfly wing-like in shape, bordered by supraoculars, frontal, upper postocular anteriorly, anterior and upper posterior temporals, and five or six nuchal scales posteriorly; one preocular, vertically slightly elongated, hexagonal, in contact with prefrontal and posterior nasal anteriorly, supraocular dorsally, and 2nd and 3rd supralabials ventrally; eye moderate (ED 10.7–21.7% of HL), round, about half of the size of snout length (ED 41.7–69.9% of ES), pupil rounded; two postoculars, subequal or upper one larger, pentagonal, upper postocular in broad contact with supraocular, parietal and anterior temporal, lower postocular in contact with anterior temporal and 5th supralabials; temporals 1 + 2, large, slightly elongated, subrectangular or pentagonal; anterior temporal larger than posterior temporal, in contact with parietal and both postoculars dorsally, and 5th and 6th supralabial ventrally; lower posterior temporal in contact with 6th and 7th supralabials ventrally. Supralabials seven (on both sides), 5th–7th largest in size; 1st supralabial in contact with rostral anteriorly, nasals dorsally, 2nd with posterior nasal and preocular dorsally, 3rd with preocular and orbit dorsally, 4th with orbit; 5th with orbit, lower postocular, and anterior temporal dorsally, and 6th with anterior and lower posterior temporals dorsally, 7th with lower posterior temporal dorsally and scales of the neck posteriorly.
Mental large, triangular, blunt posteriorly; first infralabial pair larger than mental plate and in broad contact with each other, in contact with anterior chin shields posteriorly; seven infralabials, 1st–4th in contact with anterior chin shields, 4th infralabial largest in size in contact with both anterior and posterior chin shields; 4th–7th infralabials in contact with gular scales; two larger anterior chin shields, and two slightly smaller posterior chin shields; anterior chin shields in broad contact between them; posterior chin shields bordered posteriorly by seven gular scales.
Body robust, elongate and subcylindrical; dorsal scales in 15 midbody rows, all smooth and pointed posteriorly; 222–228 ventrals in males and 224–231 in females; cloacal plate divided. Tail comparatively short, TaL 8.9–10.4% of total length in males and 13.5–17.1% of total length in males, robust and thick; subcaudals 35–37 in males and 32–36 in females, divided.
Coloration
In preservative, dorsum and venter white or yellow; 22–27 black cross bands along the body and 4 or 6 on the tail; cross bands complete laterally, and reaching the ventrals except the nuchal band; the bands on the tail distinct; the nuchal band on the nape anteriorly inverted V-shaped covering 15–20 vertebral scales; nuchal band starts from mid frontal; snout, anterior head, and lateral head black making remaining the white dorsal color an inverted V-shaped marking; first black band on the body covering 6 or 7 vertebral scales; inter-band width covers with 3–5 vertebral scales; lower parts of the supralabials white; ventral head white until the first black band; tail tip black dorsally, white ventrally.
In life (Fig. 4A), same color as in preservative, but the white body color may vary from white, cream, pale yellow to bright yellow. One juvenile with cream and black body bands was encountered in Saikhawthlir, MZ (Fig. 4B), but the snake escaped before recording morphological data.
Variation
Except the anomalous specimen (MZMU1321) which had three postoculars on left and two on right, and temporals 1 + 2 on the left and 2 + 2 on the right, all other meristic and morphometric characters obtained so far did not show any significant variation between the examined populations, and also correspond to the conventional taxonomical characters provided in previously published literature77,86,87.
Hemipenis
Based on MZMU2935, the organ is single and subcylindrical, relatively short, robust, and capitate; inverted hemipenis extends to 4th–7th subcaudal level (i.e. 11.1–20% from the total number of Sc); sulcus spermaticus bifurcate below the crotch, shallow and centripetal; apical lobe less evident with only slight apical flaring; calyculate organ with a complex ornamentation of retiform ridges, papillate flounces, and spines; spines on the upper basal areas enlarged and decreasing the size towards the proximal portion; apical region sharply separated from the basal portion by a well-defined demarcation, so the apex is free and the apical part of the hemipenis is richly capitate (Fig. 5).
Distribution
Within India, B. fasciatus has been reported from Uttar Pradesh (Gorakhpur, fide Masson88; also see Anwar89 and Das et al.90) in the north and central Maharashtra in the west91–93, extending across Telangana (Hyderabad, fide Kinnear94, Andhra Pradesh95, Chhattisgarh96,97, Jharkhand (Koderma, fide Smith86; also see Husain98), Bihar99, Odisha (Mahanadi valley, fide Wall99; also see Boruah et al.100), and northern part of WB101 to northeastern India, including Arunachal Pradesh102,103, Assam99,104,105, Meghalaya106, MZ107,108, Tripura109, Manipur110 and Nagaland111. A few unverified records are available from Madhya Pradesh36, Uttarakhand35, and southern peninsular India in Tamil Nadu, Karnataka and Kerala98.
Here we provide additional distributional records for B. fasciatus sensu stricto based on 44 new localities from MZ, and two from WB, India (Supplementary Table S7). The lowest elevation among these new records is 4 m a.s.l. at Chitrasali in Hooghly District, WB and the highest is 1426 m a.s.l. at Champhai Jailveng in Champhai District, MZ. Based on the previous distribution of the species, the elevation range was between 40 and 2300 m a.s.l.33,34. Moreover, an estimated distribution range of the species was plotted (Fig. 6) following WHO’s range estimation for B. fasciatus112.
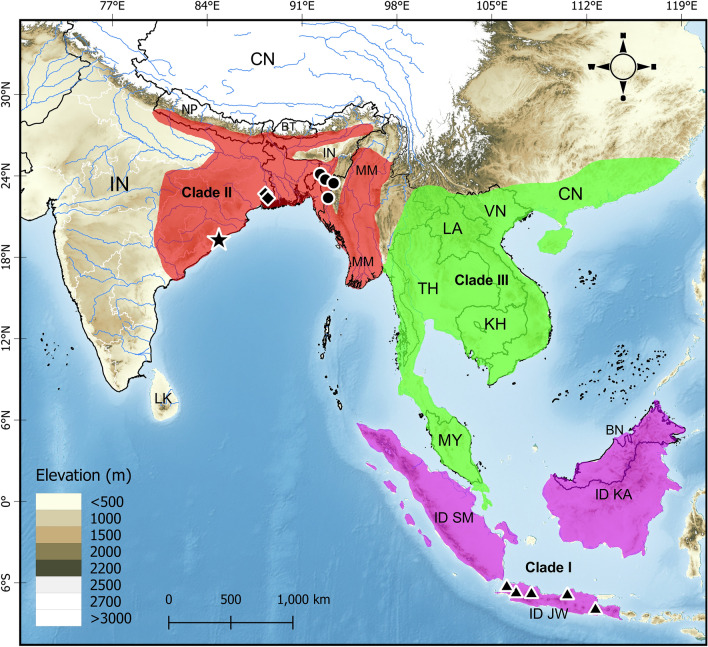
Figure 6.
Map showing the distribution range of Bungarus fasciatus sensu lato, based on the latest species map provided by the World Health Organization (2022); the coloration corresponds to the three distinct evolutionary lineages recovered in the phylogenetic analyses. The type locality of Bungarus fasciatus sensu stricto is indicated by a black star. Localities of specimens used in the morphological analyses are indicated by black filled diamonds (WB), circles (MZ), and triangles (JV). Abbreviations for countries are: IN India, NP Nepal, BT Bhutan, BD Bangladesh, LK Sri Lanka, CN China, MM Myanmar, LA Laos, TH Thailand, VN Vietnam, KH Cambodia, MY Malaysia, BN Brunei Darussalam, ID Indonesia (KA Kalimantan, SM Sumatra, JW Java).
Natural history
Although B. fasciatus is a common species, details on the ecology, habitat, population, and breeding are still sparse and further studies are needed. Therefore, here we provide some natural history data based on two clutches of eggs encountered from two localities in WB State, India:
(i) On 16th May 2020, at ca. 20:00 h, from Chitrasali village, Hooghly, the snake was encountered on the bank of a pond adjacent to a house in the middle of a village. The female was found coiling around a clutch of 19 eggs. The breeding site was located inside a naturally occurring burrow at the base of a dead tree with decayed roots. The burrow was on the bank ca. 6 feet from the pond. The pond had a gentle slope and was surrounded by plentiful vegetation. On the day of the egg collection, the recorded ambient temperature at the natural breeding site was 28–38 °C with average humidity of 78%. The eggs were relocated and incubated in a dedicated herpetoculture room at 27.6 °C using 3 cm thick vermiculite bedding in a perforated box. On 10th June at 20:18 h, the first egg slits were observed, and hatching was completed on 18th June at 05:45 h. The fluctuating room temperature and average humidity from the start of hatching until hatching was completed were 26–35 °C and 81.1%, respectively. Notably, hatchlings crawled out from the pipped eggs on the 12th, 13th, and 14th June. Upon investigation, we found that a total of six eggs failed to hatch, out of which three eggs were unfertilized, two contained partially developed embryos showing deformities, and one egg had a fully developed embryo, possibly unable to cut through the eggshell. On 18th June, we recorded the biometric data of the 13 hatchlings (5 females with average SVL 322.2 mm, TaL 32.4 mm, and body weight 21.2 g; 8 males with average SVL 318.6 mm, TaL 36.5, body weight 19.9 g), and were subsequently released close to where the eggs were collected.
(ii) On 05th May 2021 at 12:30 pm, from a construction site at Ankuni village, Hooghly. A clutch of eight eggs were uncovered under a pile of old bricks at the base of a dead tree with lots of burrows. The breeding site was located on the bank of a pond, and the entire rubble pile was covered in vegetation. However, in this case, the female snake was not found near the eggs, and it is possible that the excavation work might have scared the female away. The eggs were relocated and incubated in the same herpetoculture room using 3 cm thick vermiculite bedding in a perforated box. The room temperature recorded on 5th May fluctuated between 24 and 33 °C, with a relative humidity of 65%. Egg slits were seen on 6th June at ca. 22:00 h. On 8th June at ca. 08:00 h, hatching was completed and all of the juveniles had emerged from the eggs. From the egg relocation until the completed hatching (6th–8th June), the temperature and humidity fluctuated between 24 and 39 °C and 65–75%, respectively. On 8th June, the biometric data of the eight hatchlings were taken (3 females with average SVL 333.3 mm, TaL 38.7 mm, and body weight 21.3 g; 5 males with average SVL 351.0 mm, TaL 43.2, body weight 21.4 g), and they were also released close to the site from which the eggs had been collected.
Discussion
Bungarus fasciatus sensu stricto
Evidence from this study, based on morphology and molecular data, defines three distinct clades of B. fasciatus with non-overlapping distribution clusters. The high genetic divergence among lineages also suggests distinct species-level groups within B. fasciatus as currently conceived. Our morphometric data analysis also provides evidence of their morphological distinctiveness between Clade I and II. Moreover, the lineage from east Asia is basal to the other two lineages but, if these clades were to be accepted as full species, the name-bearing lineage is Clade II. Thus, according to our newly presented evidence, and partly according to Russell84, the distribution range of Bungarus fasciatus sensu stricto (Indo-Myanmar clade) comprises east and northeast India extending towards Myanmar. (Figs. 1, 6).
Systematic challenges
In this study, we elucidate the presence of three independent lineages within B. fasciatus, which is crucial for future nomenclatural revision. In the CYTB gene, while negligible intra-clade genetic divergence was observed within Clade I (0.4%; between two locations in JV) and Clade II (0.0–1.3%; Myanmar, east and northeast India), a wide range of intra-clade genetic divergence (00.0–6.5%) was evident within Clade III (China, Vietnam, Thailand). Consequently, we speculate that there might still be cryptic diversity within the east Asian lineage (Clade III). Moreover, for robust delimitation of the B. fasciatus complex, it is necessary to establish whether these lineages have undergone some degree of extrinsic or intrinsic reproductive isolation to be evolving separately113. For instance, due to the high evolutionary rate of hemipenial traits compared to the other morphological traits114,115, the organ has commonly been used to provide a picture of sexual barrier even among cryptic species116–118.
Although it has been previously stressed that delimiting the taxonomic status of geographically diversified populations of venomous snakes alone cannot necessarily predict patterns of venom variation, it can play a pivotal role in overcoming the consequential variability of venoms119–121. Fry et al.120 further indicated that the medical importance of B. fasciatus has been overestimated. Moreover, the possible existence of undiscovered cryptic species accompanied by more venom diversity with uncharacterized components had been pointed out122. Siqueira-Silva et al.123 observed that more productive environments favour more complex venom, with more toxins in similar proportions. Based on the verbal autopsy we have conducted so far within MZ, there are three cases of fatal envenomation potentially from the bite of banded krait. Therefore, here we highlight the importance of analyzing the venom compositions in different populations in each biogeographically isolated clade.
Further work
The combination of multivariate morphometric analysis and mitochondrial gene-based phylogeography has been applied successfully for species delineation24,124,125 as well as for testing species boundaries126. However, nuclear genes provide an independent test of species boundaries127 as they are capable of measuring the extent of gene flow, and for this reason, recent work has increasingly used a combination of nuclear and mitochondrial genes for phylogeographic analyses and species delineation128. Consequently, we believe that the potentially species-level diversity across different B. fasciatus populations depicted in this study cannot be overlooked, and a thorough comprehension of B. fasciatus systematics is still a fundamental challenge.
Supplementary Information
Acknowledgements
We thank the Chief Wildlife Warden of Environment, Forests and Climate change Department, Government of Mizoram for providing collection permits (No.A.33011/2/99-CWLW/22 and No.B.19060/5/2020-CWLW/20-26). We also thank the Directorate of Forests, Wildlife Wing, Government of West Bengal for continued support to carry out our research and conservation activities. This work is supported by DST-SERB, New Delhi (DST No: EMR/2016/002391), DBT, New Delhi (DBT-NER/AAB/64/2017), National Mission for Himalayan Studies (NMHS), Uttarakhand (GBPNI/NMHS-2017/MG-22/566), DRDO, New Delhi (DGTM/DFTM/GIA/19-20/0422), and DST-SERB, New Delhi (DST No: EEQ/2021/000243). AM would like to acknowledge EU Marie Curie Action PIRSES-GA-2013-612131 (the BITES project); the Bangor University ESRC Impact Acceleration Account, for funding; and Ashok Mallik for his assistance in this work. HTL is grateful to the International Herpetological Symposium (IHS), USA for the award grant. LB is also thankful for the small grant he received from The Rufford Foundation, UK (grant number 36737-1). LB and HTL would like to thank Mathipi Vabeiryureilai, Fanai Malsawmdawngliana, Lal Muansanga, Lalengzuala Tochhawng, Ht. Decemson, Gospel Zothanmawia Hmar, Vanlal Siammawii, H. Laltlanchhuaha; VS would like to thank Biswajit Das, Lakshmi Santra, Aritra Dhara, Pallab Das, Ayan Koley, Rakesh Koley, Rajkumar Chakraborty, Amal Kr Santra, Ananta Katwal, Ishan Santra, Shrabani Santra for their assistance in this study. AATA thanks the Ministry of Environment and Forestry (KLHK) and The Directorate General of Conservation of Natural Resources and Ecosystems (KSDAE) of the Republic of Indonesia for granting research permits; C. Rahmadi, A. Riyanto, A. Hamidy, Syaripudin, and W. Trilaksano (MZB) for their support and facilitating the in-house study of specimens under their care. We are deeply thankful to Patrick David for the insightful taxonomical comments in the draft version of the manuscript.
Author contributions
L.B.: Conceptualization, Data curation, Formal analysis, Investigation, Methodology, Software, Visualization, Writing—original draft. H.T.L.: Conceptualization, Data curation, Funding acquisition, Investigation, Methodology, Project administration, Validation, Visualization, Writing—original draft, Resources, Supervision. V.S.: Investigation, Resources, Writing—review & editing. A.A.T.A.: Data curation, Formal analysis, Software, Validation, Visualization, Investigation, Writing—review & editing. A.D.: Investigation, Resources. M.T.A.: Investigation, Resources. Z.B.M.: Investigation, Resources. S.K.: Investigation, Software, Writing—review & editing. A.M.: Formal analysis, Validation, Writing—review & editing.
Data availability
The generated partial gene sequences were deposited on the NCBI repository (GenBank accession numbers are given in Supplementary Table S2).
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Hmar T. Lalremsanga, Email: htlrsa@yahoo.co.in
A. A. Thasun Amarasinghe, Email: thasun.amarasinghe@ui.ac.id.
Anita Malhotra, Email: a.malhotra@bangor.ac.uk.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-023-28241-8.
References
- 1.Jirsová D, et al. From taxonomic deflation to newly detected cryptic species: Hidden diversity in a widespread African squeaker catfish. Sci. Rep. 2019;9:1–13. doi: 10.1038/s41598-019-52306-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Luo A, Ling C, Ho SY, Zhu CD. Comparison of methods for molecular species delimitation across a range of speciation scenarios. Syst. Biol. 2018;67:830–846. doi: 10.1093/sysbio/syy011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dwivedi AK, et al. Cryptic diversity in the Indian clade of the catfish family Pangasiidae resolved by the description of a new species. Hydrobiologia. 2017;797:351–370. doi: 10.1007/s10750-017-3198-z. [DOI] [Google Scholar]
- 4.Halasan LC, Geraldino PJL, Lin HC. First evidence of cryptic species diversity and population structuring of Selaroides leptolepis in the tropical western Pacific. Front. Mar. Sci. 2021;8:756163. doi: 10.3389/fmars.2021.756163. [DOI] [Google Scholar]
- 5.Matsumoto S, et al. Cryptic diversification of the swamp eel Monopterus albus in East and Southeast Asia, with special reference to the Ryukyuan populations. Ichthyol. Res. 2010;57:71–77. doi: 10.1007/s10228-009-0125-y. [DOI] [Google Scholar]
- 6.Nishikawa K, et al. Molecular phylogeny and biogeography of caecilians from Southeast Asia (Amphibia, Gymnophiona, Ichthyophiidae), with special reference to high cryptic species diversity in Sundaland. Mol. Phylogenet. Evol. 2012;63:714–723. doi: 10.1016/j.ympev.2012.02.017. [DOI] [PubMed] [Google Scholar]
- 7.Ramesh V, Vijayakumar SP, Gopalakrishna T, Jayarajan A, Shanker K. Determining levels of cryptic diversity within the endemic frog genera, Indirana and Walkerana, of the Western Ghats, India. PLoS ONE. 2020;15:e0237431. doi: 10.1371/journal.pone.0237431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stuart BL, Inger RF, Voris HK. High level of cryptic species diversity revealed by sympatric lineages of Southeast Asian forest frogs. Biol. Lett. 2006;2:470–474. doi: 10.1098/rsbl.2006.0505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lohman DJ, et al. Cryptic genetic diversity in “widespread” Southeast Asian bird species suggests that Philippine avian endemism is gravely underestimated. Biol. Conserv. 2010;143:1885–1890. doi: 10.1016/j.biocon.2010.04.042. [DOI] [Google Scholar]
- 10.Outlaw DC, Voelker G. Pliocene climatic change in insular Southeast Asia as an engine of diversification in Ficedula flycatchers. J. Biogeogr. 2008;35:739–752. doi: 10.1111/j.1365-2699.2007.01821.x. [DOI] [Google Scholar]
- 11.Rheindt FE, Wu MY, Movin N, Jønsson KA. Cryptic species-level diversity in Dark-throated Oriole Oriolus xanthonotus. Bull. Br. Ornithol. Club. 2022;142:254–267. doi: 10.25226/bboc.v142i2.2022.a10. [DOI] [Google Scholar]
- 12.Chattopadhyay B, et al. Cryptic diversity of Rhinolophus lepidus in South Asia and differentiation across a biogeographic barrier. Front. Biogeogr. 2021 doi: 10.21425/F5FBG49625. [DOI] [Google Scholar]
- 13.Chen S, et al. Multilocus phylogeny and cryptic diversity of white-toothed shrews (Mammalia, Eulipotyphla, Crocidura) in China. BMC Evol. Biol. 2020;20:1–14. doi: 10.1186/s12862-020-1588-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nater A, et al. Morphometric, behavioral, and genomic evidence for a new orangutan species. Curr. Biol. 2017;27:3487–3498. doi: 10.1016/j.cub.2017.09.047. [DOI] [PubMed] [Google Scholar]
- 15.Pfenninger M, Schwenk K. Cryptic animal species are homogeneously distributed among taxa and biogeographical regions. BMC Evol. Biol. 2007;7:1–6. doi: 10.1186/1471-2148-7-121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vodă R, Dapporto L, Dincă V, Vila R. Cryptic matters: Overlooked species generate most butterfly beta-diversity. Ecography. 2014;38:405–409. doi: 10.1111/ecog.00762. [DOI] [Google Scholar]
- 17.Bauer AM. Reptiles and the biogeographic interpretation of New Caledonia. Tuatara. 1989;30:39–50. [Google Scholar]
- 18.Camargo A, Sinervo B, Sites JW. Lizards as model organisms for linking phylogeographic and speciation studies. Mol. Ecol. 2010;19:3243–3488. doi: 10.1111/j.1365-294x.2010.04722.x. [DOI] [PubMed] [Google Scholar]
- 19.Gowande G, et al. Molecular phylogenetics and taxonomic reassessment of the widespread agamid lizard Calotes versicolor (Daudin, 1802) (Squamata, Agamidae) across South Asia. Vertebr. Zool. 2021;71:669–696. doi: 10.3897/vz.71.e62787. [DOI] [Google Scholar]
- 20.Guo P, et al. Cryptic diversity of green pitvipers in Yunnan, South-west China (Squamata, Viperidae) Amphib. Reptil. 2015;36:265–276. doi: 10.1163/15685381-00003004. [DOI] [Google Scholar]
- 21.Wagner P, et al. Integrative approach to resolve Calotes mystaceus Duméril & Bibron, 1837 species complex (Squamata: Agamidae) Bonn Zool. Bull. 2021;70:141–171. doi: 10.20363/BZB-2021.70.1.141. [DOI] [Google Scholar]
- 22.Zug G, Brown H, Schulte J, Vindum J. Systematics of the Garden Lizards, Calotes versicolor Group (Reptilia, Squamata, Agamidae), in Myanmar: Central Dry Zone Populations. Proc. Calif. Acad. Sci. 2007;57:35–68. [Google Scholar]
- 23.Alfaro ME, Karns DR, Voris HK, Abernathy E, Sellins SL. Phylogeny of Cerberus (Serpentes: Homalopsinae) and phylogeography of Cerberus rynchops: Diversification of a coastal marine snake in Southeast Asia. J. Biogeogr. 2004;31:1277–1292. doi: 10.1111/j.1365-2699.2004.01114.x. [DOI] [Google Scholar]
- 24.Malhotra A, Dawson K, Guo P, Thorpe RS. Phylogenetic structure and species boundaries in the mountain pitviper Ovophis monticola (Serpentes: Viperidae: Crotalinae) in Asia. Mol. Phylogenet. Evol. 2011;59:444–457. doi: 10.1016/j.ympev.2011.02.010. [DOI] [PubMed] [Google Scholar]
- 25.Mallik AK, et al. Disentangling vines: A study of morphological crypsis and genetic divergence in vine snakes (Squamata: Colubridae: Ahaetulla) with the description of five new species from Peninsular India. Zootaxa. 2020;4874:1–62. doi: 10.11646/zootaxa.4874.1.1. [DOI] [PubMed] [Google Scholar]
- 26.Shankar PG, et al. King or royal family? Testing for species boundaries in the King Cobra, Ophiophagus hannah (Cantor, 1836), using morphology and multilocus DNA analyses. Mol. Phylogenet. Evol. 2021;165:107300. doi: 10.1016/j.ympev.2021.107300. [DOI] [PubMed] [Google Scholar]
- 27.Thorpe RS, Pook CE, Malhotra A. Phylogeography of Russell's viper (Daboia russelii) complex in relation to variation in the colour pattern and symptoms of envenoming. Herpetol. J. 2007;10:209–218. [Google Scholar]
- 28.Wüster W. Taxonomic changes and toxinology: Systematic revisions of the Asiatic cobras (Naja naja) species complex. Toxicon. 1996;34:399–406. doi: 10.1016/0041-0101(95)00139-5. [DOI] [PubMed] [Google Scholar]
- 29.Wüster W, Thorpe RS. Naja siamensis, a cryptic species of venomous snake revealed by mtDNA sequencing. Experientia. 1994;50:75–79. doi: 10.1007/BF01992054. [DOI] [PubMed] [Google Scholar]
- 30.Wüster W, Otsuka S, Malhotra A, Thorpe RS. Population systematics of Russell's viper: A multivariate study. Biol. J. Linn. Soc. 1992;47:97–113. doi: 10.1111/j.1095-8312.1992.tb00658.x. [DOI] [Google Scholar]
- 31.Midtgaard, R. Repfocus,aSurveyoftheReptilesoftheWorld. http://repfocus.dk/Bungarus.html (2022).
- 32.Lee MS, Sanders KL, King B, Palci A. Diversification rates and phenotypic evolution in venomous snakes (Elapidae) R. Soc. Open Sci. 2016;3:150277. doi: 10.1098/rsos.150277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ahmed, M. F., Das, A. & Dutta, S. K. AmphibiansandReptilesofNortheastIndia.APhotographicGuide (Aaranyak, 2009).
- 34.Knierim TK, Strine CT, Suwanwaree P, Hill III JG. Spatial ecology study reveals nest attendance and habitat preference of banded kraits (Bungarus fasciatus) Herpetol. Bull. 2019;150:6–13. doi: 10.33256/hb150.613. [DOI] [Google Scholar]
- 35.Stuart, B. et al. Bungarusfasciatus. TheIUCNRedListofThreatenedSpecies2013.https://www.iucnredlist.org/species/192063/2034956 (2013).
- 36.Wallach, V., Williams, K. L. & Boundy, J.SnakesoftheWorld:ACatalogueofLivingandExtinctSpecies (CRC Press, 2014).
- 37.Pe T, et al. Envenoming by Chinese krait (Bungarus multicinctus) and banded krait (B. fasciatus) in Myanmar. Trans. R. Soc. Trop. Med. Hyg. 1997;91:686–688. doi: 10.1016/S0035-9203(97)90524-1. [DOI] [PubMed] [Google Scholar]
- 38.Ahsan MF, Rahman MM. Status, distribution and threats of kraits (Squamata: Elapidae: Bungarus) in Bangladesh. J. Threat. Taxa. 2017;9:9903–9910. doi: 10.11609/jott.2929.9.3.9903-9910. [DOI] [Google Scholar]
- 39.Tongpoo A, et al. Krait envenomation in Thailand. Ther. Clin. Risk Manag. 2018;14:1711–1717. doi: 10.2147/TCRM.S169581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lo, T. B. & Lu, H. S. StudiesonBungarusfasciatusVenom. (Toxins, 1978).
- 41.Lu J, et al. A novel serine protease inhibitor from Bungarus fasciatus venom. Peptides. 2008;29:369–374. doi: 10.1016/j.peptides.2007.11.013. [DOI] [PubMed] [Google Scholar]
- 42.Rusmili MRA, Yee TT, Mustafa MR, Hodgson WC, Othman I. Proteomic characterization and comparison of Malaysian Bungarus candidus and Bungarus fasciatus venoms. J. Proteom. 2014;110:129–144. doi: 10.1016/j.jprot.2014.08.001. [DOI] [PubMed] [Google Scholar]
- 43.Tan NH, Ponnudurai G. A comparative study of the biological properties of krait (genus Bungarus) venoms. Comp. Biochem. Physiol. C. Toxicol. Pharmacol. 1990;95:105–109. doi: 10.1016/0742-8413(90)90089-r. [DOI] [PubMed] [Google Scholar]
- 44.Tsai IH, Tsai HY, Saha A, Gomes A. Sequences, geographic variations and molecular phylogeny of venom phospholipases and threefinger toxins of eastern India Bungarus fasciatus and kinetic analyses of its Pro31 phospholipases A2. FEBS J. 2007;274:512–525. doi: 10.1111/j.1742-4658.2006.05598.x. [DOI] [PubMed] [Google Scholar]
- 45.Ziganshin RH, et al. Quantitative proteomic analysis of Vietnamese krait venoms: Neurotoxins are the major components in Bungarus multicinctus and phospholipases A2 in Bungarus fasciatus. Toxicon. 2015;107:197–209. doi: 10.1016/j.toxicon.2015.08.026. [DOI] [PubMed] [Google Scholar]
- 46.Kundu S, et al. Mitochondrial DNA discriminates distinct population of two deadly snakes (Reptilia: Elapidae) in Northeast India. Mitochondrial DNA B Resour. 2020;5:1530–1534. doi: 10.1080/23802359.2020.1742210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Laopichienpong N, et al. Assessment of snake DNA barcodes based on mitochondrial COI and Cytb genes revealed multiple putative cryptic species in Thailand. Gene. 2016;594:238–247. doi: 10.1016/j.gene.2016.09.017. [DOI] [PubMed] [Google Scholar]
- 48.Supikamolseni A, Ngaoburanawit N, Sumontha M, Chanhome L, Suntrarachun S, Peyachoknagul S, Srikulnath K. Molecular barcoding of venomous snakes and species-specific multiplex PCR assay to identify snake groups for which antivenom is available in Thailand. Genet. Mol. Res. 2015;14:13981–13997. doi: 10.4238/2015.october.29.18. [DOI] [PubMed] [Google Scholar]
- 49.Chippaux JP, Williams V, White J. Snake venom variability: Methods of study, results and interpretation. Toxicon. 1991;29:1279–1303. doi: 10.1016/0041-0101(91)90116-9. [DOI] [PubMed] [Google Scholar]
- 50.Harrison RA, Wüster W, Theakston RDG. The conserved structure of snake venom toxins confers extensive immunological cross-reactivity to toxin-specific antibody. Toxicon. 2003;41:441–449. doi: 10.1016/s0041-0101(02)00360-4. [DOI] [PubMed] [Google Scholar]
- 51.Abtin E, Nilson G, Mobaraki A, Hosseini AA, Dehgannejhad M. A new species of krait, Bungarus (Reptilia, Elapidae, Bungarinae) and the first record of that genus in Iran. Russ. J. Herpetol. 2014;21:243–250. doi: 10.30906/1026-2296-2014-21-4-243-250. [DOI] [Google Scholar]
- 52.Ashraf MR, et al. Phylogenetic analysis of the Common Krait (Bungarus caeruleus) in Pakistan based on mitochondrial and nuclear protein coding genes. Amphib. Reptile Conserv. 2019;13:203–211. [Google Scholar]
- 53.Biakzuala L, Purkayastha J, Rathee YS, Lalremsanga HT. New data on the distribution, morphology, and molecular systematics of two venomous snakes, Bungarus niger and Bungarus lividus (Serpentes: Elapidae), from north-east India. Salamandra. 2021;57:219–228. [Google Scholar]
- 54.Chen ZN, Shi SC, Vogel G, Ding L, Shi JS. Multiple lines of evidence reveal a new species of Krait (Squamata, Elapidae, Bungarus) from Southwestern China and Northern Myanmar. Zookeys. 2021;1025:35–71. doi: 10.3897/zookeys.1025.62305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Keogh JS. Molecular phylogeny of elapid snakes and a consideration of their biogeographic history. Biol. J. Linn. Soc. 1998;63:177–203. doi: 10.1111/J.1095-8312.1998.TB01513.X. [DOI] [Google Scholar]
- 56.Kuch U, et al. A new species of krait (Squamata: Elapidae) from the Red River system of northern Vietnam. Copeia. 2005;818–833:2005. doi: 10.1643/0045-8511(2005)005[0818:ANSOKS]2.0.CO;2. [DOI] [Google Scholar]
- 57.Slowinski JB. A phylogenetic analysis of Bungarus (Elapidae) based on morphological characters. J. Herpetol. 1994;28:440–446. doi: 10.2307/1564956. [DOI] [Google Scholar]
- 58.Slowinski JB, Keogh JS. Phylogenetic relationships of elapid snakes based on cytochrome b mtDNA sequences. Mol. Phylogenet. Evol. 2000;15:157–164. doi: 10.1006/mpev.1999.0725. [DOI] [PubMed] [Google Scholar]
- 59.Maritz B, Penner J, Martins M, Crnobrnja-Isailović J, Spear S, Alencar LR, Sigala-Rodriguez J, Messenger K, Clark RW, Soorae P, Luiselli L. Identifying global priorities for the conservation of vipers. Biol. Conserv. 2016;204:94–102. doi: 10.1016/j.biocon.2016.05.004. [DOI] [Google Scholar]
- 60.Conroy CJ, Papenfuss T, Parker J, Hahn NE. Use of tricainemethanesulfonate (MS222) for euthanasia of reptiles. J. Am. Assoc. Lab. Anim. Sci. 2009;48:28–32. [PMC free article] [PubMed] [Google Scholar]
- 61.Percie du Sert, N. etal. The ARRIVE guidelines 2.0: Updated guidelines for reporting animal research. J.Cereb.BloodFlowMetab.40, 1769–1777 (2020). [DOI] [PMC free article] [PubMed]
- 62.Vaidya G, Lohman DJ, Meier R. SequenceMatrix: Concatenation software for the fast assembly of multi-gene datasets with character set and codon information. Cladistics. 2011;27:1716–2180. doi: 10.1111/j.1096-0031.2010.00329.x. [DOI] [PubMed] [Google Scholar]
- 63.Kumar S, Stecher G, Li M, Knyaz C, Tamura K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018;35:1547–1549. doi: 10.1093/molbev/msy096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lanfear R, Frandsen PB, Wright AM, Senfeld T, Calcott B. PartitionFinder 2: New methods for selecting partitioned models of evolution for molecular and morphological phylogenetic analyses. Mol. Biol. Evol. 2017;34:772–773. doi: 10.1093/molbev/msw260. [DOI] [PubMed] [Google Scholar]
- 65.Ronquist F, et al. MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 2012;61:539–542. doi: 10.1093/sysbio/sys029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Rambaut A, Drummond AJ, Xie D, Baele G, Suchard MA. Posterior summarization in Bayesian phylogenetics using Tracer 1.7. Syst. Biol. 2018;67:901–904. doi: 10.1093/sysbio/syy032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.R Core Team. R:ALanguageandEnvironmentforStatisticalComputinghttps://www.R-project.org/ (R Foundation for Statistical Computing, 2020).
- 68.Warren DL, Geneva AJ, Lanfear R. RWTY (R We There Yet): An R package for examining convergence of Bayesian phylogenetic analyses. Mol. Biol. Evol. 2017;34:1016–1020. doi: 10.1093/molbev/msw279. [DOI] [PubMed] [Google Scholar]
- 69.Letunic I, Bork P. Interactive Tree Of Life (iTOL) v5: An online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 2021;49:W293–W296. doi: 10.1093/nar/gkab301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Nguyen LT, Schmidt HA, von Haeseler A, Minh BQ. IQ-TREE: A fast and effective stochastic algorithm for estimating maximum likelihood phylogenies. Mol. Biol. Evol. 2015;32:268–274. doi: 10.1093/molbev/msu300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Minh BQ, Nguyen MAT, von Haeseler A. Ultrafast approximation for phylogenetic bootstrap. Mol. Biol. Evol. 2013;30:1188–1195. doi: 10.1093/molbev/mst024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kalyaanamoorthy S, Minh BQ, Wong TK, Von Haeseler A, Jermiin LS. ModelFinder: fast model selection for accurate phylogenetic estimates. Nat. Methods. 2017;14:587–589. doi: 10.1038/nmeth.4285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zhang J, Kapli P, Pavlidis P, Stamatakis A. A general species delimitation method with applications to phylogenetic placements. Bioinform. 2013;29:2869–2876. doi: 10.1093/bioinformatics/btt499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Vences M, et al. iTaxoTools 0.1: Kickstarting a specimen-based software toolkit for taxonomists. Megataxa. 2021;6:77–92. doi: 10.11646/megataxa.6.2.1. [DOI] [Google Scholar]
- 75.Smith, M. A. TheFaunaofBritishIndia,CeylonandBurma.ReptiliaandAmphibia. Vol. 3. Serpentes (Taylor & Francis, 1943)
- 76.Yang DT, Rao DQ. Amphibia and Reptilia of Yunnan. Yunnan Science and Technology Press; 2008. [Google Scholar]
- 77.Leviton AE, et al. The dangerously venomous snakes of Myanmar illustrated checklist with keys. Proc. Cal. Acad. Sci. 2003;54:407–462. [Google Scholar]
- 78.Dowling HG. A proposed standard system of counting ventrals in snakes. Br. J. Herpetol. 1951;1:97–99. [Google Scholar]
- 79.Keogh JS. Evolutionary implications of hemipenial morphology in the terrestrial Australian elapid snakes. Zool. J. Linn. Soc. 1999;125:239–278. doi: 10.1111/j.1096-3642.1999.tb00592.x. [DOI] [Google Scholar]
- 80.Levene, H. RobustTestsforEqualityofVariances.ContributionstoProbabilityandStatistics.EssaysinHonorofHaroldHotelling (eds. Olkin, I. et al.). 279–292 (Stanford University Press, 1961).
- 81.Brown MB, Forsythe AB. Robust tests for the equality of variances. J. Am. Stat. Assoc. 1974;69:364–367. doi: 10.1080/01621459.1974.10482955. [DOI] [Google Scholar]
- 82.Schneider, J. G. HistoriaeAmphibiorumNaturalisetLiterariae.FasciculusSecundusContinensCrocodilos,Scincos,Chamaesauras,Boas.Pseudoboas,Elapes,Angues.AmphisbaenasetCaecilias. (Frommanni, 1801).
- 83.Bauer, A. M. & Lavilla, E. O. (eds.). J.G.Schneider’sHistoriaeAmphibiorum:HerpetologyattheDawnofthe19thCentury. (SSAR, 2021).
- 84.Russell, P. AnAccountofIndianSerpents,CollectedontheCoastofCoromandel:ContainingDescriptionsandDrawingsofEachSpecies;TogetherwithExperimentsandRemarksonTheirSeveralPoisons (W. Bulmer and Co., 1796).
- 85.Bauer AM. Patrick Russell’s snakes and their role as type specimens. Hamadryad. 2015;37:18–65. [Google Scholar]
- 86.Smith OA. Large common and banded krait. J. Bombay Nat. Hist. Soc. 1911;21:283–284. [Google Scholar]
- 87.Whitaker R, Captain A. Snakes of India: The Field Guide. Draco Books; 2008. [Google Scholar]
- 88.Masson J. The distribution of the Banded Krait (Bungarus fasciatus) J. Bombay Nat. Hist. Soc. 1930;34:256–257. [Google Scholar]
- 89.Anwar M. First record of banded krait (Bungarus fasciatus) from Pilibhit District, Uttar Pradesh-India. Taprobanica. 2012;3:102–103. doi: 10.4038/tapro.v3i2.3967. [DOI] [Google Scholar]
- 90.Das A, Basu D, Converse L, Suresh CC. Herpetofauna of Katerniaghat Wildlife Sanctuary, Uttar Pradesh, India. J. Threat. Taxa. 2012;4:2553–2568. doi: 10.11609/JoTT.o2587.2553-68. [DOI] [Google Scholar]
- 91.Bhandarkar WR, Paliwal GT, Bhandarkar SV, Kali AA. Herpetofaunal diversity at navegaon national park, Distt. Gondia Maharashtra. Int. J. Environ. Rehabil. Conserv. 2012;3:42–49. [Google Scholar]
- 92.Deshmukh RV, Deshmukh SA, Badhekar SA, Naitame RY. Snakes of Bhandara District, Maharashtra, Central India with notes on natural history. Reptil. Amphib. 2020;27:10–17. doi: 10.17161/randa.v27i1.14438. [DOI] [Google Scholar]
- 93.Joshi PS, Charjan AP, Tantarpale VT. A herpetofaunal inventory of Vidarbha region, Maharashtra. India. Bio. Disc. 2017;8:582–587. [Google Scholar]
- 94.Kinnear NB. Banded Krait (Bungarus fasciatus) in Hyderabad State. J. Bombay Nat. Hist. Soc. 1913;22:635–636. [Google Scholar]
- 95.Srinivasulu C, Venkateshwarlu D, Seetharamaraju M. Rediscovery of the Banded Krait Bungarus fasciatus (Schneider 1801) (Serpentes: Elapidae) from Warangal District, Andhra Pradesh, India. J. Threat. Taxa. 2009;1:353–354. doi: 10.11609/JoTT.o1986.353-4. [DOI] [Google Scholar]
- 96.Chandra K, Raha A, Majumder A, Parida A, Sarsavan A. First Record of Banded Krait, Bungarus fasciatus (Schneider, 1801), (Reptilia: Elapidae), from Guru Ghasidas National Park, Koriya District, Chhattisgarh. India. Rec. Zool. Surv. India. 2013;113:77–80. [Google Scholar]
- 97.Ingle M. Herpetofauna of Naglok Region, Jashpur District. Chhattisgarh. Rec. Zool. Surv. India. 2011;111:99–109. [Google Scholar]
- 98.Hussain A. New record of banded krait Bungarus fasciatus (Schneider, 1801) from Ranchi (Jharkhand) with its preying on checkered keel-back snake. Biol. Forum. 2020;12:29–32. [Google Scholar]
- 99.Wall F. A popular treatise on the common Indian snakes. Part 15. Bungarus fasciatus and Lycodon striatus. J. Bombay Nat. Hist. Soc. 1912;20:933–953. [Google Scholar]
- 100.Boruah B, et al. Diversity of herpetofauna and their conservation in and around North Orissa University Campus, Odisha, India. NeBIO. 2016;7:138–145. [Google Scholar]
- 101.Sharma, R. C. TheFaunaofIndiaandtheAdjacentCountries. Vol. 3. Reptilia(Serpentes). (Zoological Survey of India, 2007).
- 102.Borang A, Bhatt BB, Chaudhury SB, Borkotoki A, Bhutia PT. Checklist of the snakes of Arunachal Pradesh, northeast India. J. Bombay Nat. Hist. Soc. 2005;102:19–26. [Google Scholar]
- 103.Das A. Notes on Snakes of the Genus Bungarus (Serpentes: Elapidae) from Northeast India. Indian Hotspots. Springer; 2018. [Google Scholar]
- 104.Mathew R. On a collection of snakes from North-east India (Reptilia: Serpentes) Rec. Zool. Surv. India. 1983;80:449–458. doi: 10.26515/rzsi/v80/i3-4/1982/161206. [DOI] [Google Scholar]
- 105.Purkayastha J, Das M, Sengupta S. Urban herpetofauna: A case study in Guwahati City of Assam. India. Herpetol. Notes. 2011;4:195–202. [Google Scholar]
- 106.Mathew, R. StateFaunaSeries4:FaunaofMeghalaya,PartI;Reptilia (ed. Director). 379–454 (Zoological Survey of India, 1995).
- 107.Lalremsanga HT, Sailo S, Chinliansiama H. Diversity of snakes (Reptilia: Squamata) and role of environmental factors in their distribution in Mizoram, Northeast India. Proc. Adv. Environ. Chem. 2011;64:265–269. [Google Scholar]
- 108.Pawar S, Birand A. A Survey of Amphibians, Reptiles, and Birds in Northeast India. Centre for Ecological Research and Conservation; 2001. [Google Scholar]
- 109.Majumder J, Bhattacharjee PP, Majumdar K, Debnath C, Agarwala BK. Documentation of herpetofaunal species richness in Tripura, northeast India. NeBio. 2012;3:60–70. [Google Scholar]
- 110.Singh S. On a collection of reptiles and amphibians of Manipur. Geobios New Rep. 1995;14:135–145. [Google Scholar]
- 111.Dasgupta, G. & Raha, S. FaunaofNagaland,StateFaunaSeries12;Reptilia (ed. Director). 433–460 (Zoological Survey of India, 2006).
- 112.World Health Organization. SnakebiteInformationandDataPlatform. https://www.who.int/teams/control-of-neglected-tropical-diseases/snakebiteenvenoming/snakebite-information-and-data-platform/overview#tab=tab_1 (2022).
- 113.Hillis DM. Species delimitation in herpetology. J. Herpetol. 2019;53:3–12. doi: 10.1670/18-123. [DOI] [Google Scholar]
- 114.Gilman CA, Corl A, Sinervo B, Irschick DJ. Genital morphology associated with mating strategy in the polymorphic lizard, Uta stansburiana. J. Morphol. 2019;280:184–192. doi: 10.1002/jmor.20930. [DOI] [PubMed] [Google Scholar]
- 115.Klaczko J, Ingram T, Losos J. Genitals evolve faster than other traits in Anolis lizards. J. Zool. 2015;295:44–48. doi: 10.1111/jzo.12178. [DOI] [Google Scholar]
- 116.Arnold EN. Why copulatory organs provide so many useful taxonomic characters: the origin and maintenance of hemipenial differences in lacertid lizards (Reptilia: Lacertidae) Biol. J. Linn. Soc. 1986;29:263–281. doi: 10.1111/j.1095-8312.1986.tb00279.x. [DOI] [Google Scholar]
- 117.Myers CW, McDowell SB. New taxa and cryptic species of neotropical snakes (Xenodontinae), with commentary on hemipenes as generic and specific characters. Bull. Am. Museum Nat. Hist. 2014;385:1–112. doi: 10.1206/862.1. [DOI] [Google Scholar]
- 118.Nunes PMS, Fouquet A, Curcio FF, Kok PJR, Rodrigues MT. Cryptic species in Iphisa elegans Gray, 1851 (Squamata: Gymnophthalmidae) revealed by hemipenial morphology and molecular data. Zool. J. Linn. Soc. 2012;166:361–376. doi: 10.1111/j.1096-3642.2012.00846.x. [DOI] [Google Scholar]
- 119.Daltry JC, Wüster W, Thorpe RS. Diet and snake venom evolution. Nature. 1996;379:537–540. doi: 10.1038/379537a0. [DOI] [PubMed] [Google Scholar]
- 120.Fry BG, Winkel KD, Wickramaratna JC, Hodgson WC, Wüster W. Effectiveness of snake antivenom: Species and regional venom variation and its clinical impact. J. Toxicol. Toxin Rev. 2003;22:23–34. doi: 10.1081/TXR-120019018. [DOI] [Google Scholar]
- 121.Williams HF, et al. The urgent need to develop novel strategies for the diagnosis and treatment of snakebites. Toxins. 2019;11:363. doi: 10.3390/toxins11060363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Chatrath ST, et al. Identification of novel proteins from the venom of a cryptic snake Drysdalia coronoides by a combined transcriptomics and proteomics approach. J. Proteome Res. 2011;10:739–750. doi: 10.1021/pr1008916. [DOI] [PubMed] [Google Scholar]
- 123.Siqueira-Silva T, et al. Ecological and biogeographic processes drive the proteome evolution of snake venom. Glob. Ecol. Biogeogr. 2021;30:1978–1989. doi: 10.1111/geb.13359. [DOI] [Google Scholar]
- 124.Wüster W, Broadley DG. A new species of spitting cobra from northeastern Africa (Serpentes: Elapidae: Naja) J. Zool. 2003;259:345–359. doi: 10.1017/S0952836902003333. [DOI] [Google Scholar]
- 125.Wüster W, Broadley DG. Get an eyeful of this: a new species of giant spitting cobra from eastern and north-eastern Africa (Squamata: Serpentes: Elapidae: Naja) Zootaxa. 2007;1532:51–68. doi: 10.11646/zootaxa.1532.1.4. [DOI] [Google Scholar]
- 126.Puorto G, et al. Combining mitochondrial DNA sequences and morphological data to infer species boundaries: phylogeography of lanceheaded pitvipers in the Brazilian Atlantic forest, and the status of Bothrops pradoi (Squamata: Serpentes: Viperidae) J. Evol. Biol. 2001;14:527–538. doi: 10.1046/j.1420-9101.2001.00313.x. [DOI] [Google Scholar]
- 127.Hare MP. Prospects for nuclear gene phylogeography. Trends Ecol. Evol. 2001;16:700–706. doi: 10.1016/S0169-5347(01)02326-6. [DOI] [Google Scholar]
- 128.Wüster W, et al. Integration of nuclear and mitochondrial gene sequences and morphology reveals unexpected diversity in the forest cobra (Naja melanoleuca) species complex in Central and West Africa (Serpentes: Elapidae) Zootaxa. 2018;4455:68–98. doi: 10.11646/zootaxa.4455.1.3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The generated partial gene sequences were deposited on the NCBI repository (GenBank accession numbers are given in Supplementary Table S2).