Abstract
Background
Opportunistic infections are a ubiquitous complication in human immunodeficiency virus (HIV)/acquired immunodeficiency syndrome (AIDS) patients. Cryptosporidium spp., Giardia duodenalis, and Enterocytozoon bieneusi are common opportunistic intestinal pathogens in humans. In China, despite the number of HIV/AIDS patients being extremely large, only a few studies have investigated opportunistic infections caused by intestinal pathogens in this patient population. The aims of this study were to elucidate the occurrence and genetic characteristics of Cryptosporidium spp., G. duodenalis, and E. bieneusi in HIV/AIDS patients.
Methods
We collected fecal specimens from 155 HIV/AIDS patients (one from each patient). All of the specimens were examined for the presence of the pathogens by genotyping using polymerase chain reaction and sequencing of the small subunit ribosomal RNA gene for Cryptosporidium spp.; the triosephosphate isomerase, β-giardin and glutamate dehydrogenase genes for G. duodenalis; and the internal transcribed spacer region of the rRNA gene for E. bieneusi. The Cryptosporidium-positive specimens were further subtyped by polymerase chain reacion and sequencing of the 60-kDa glycoprotein gene.
Results
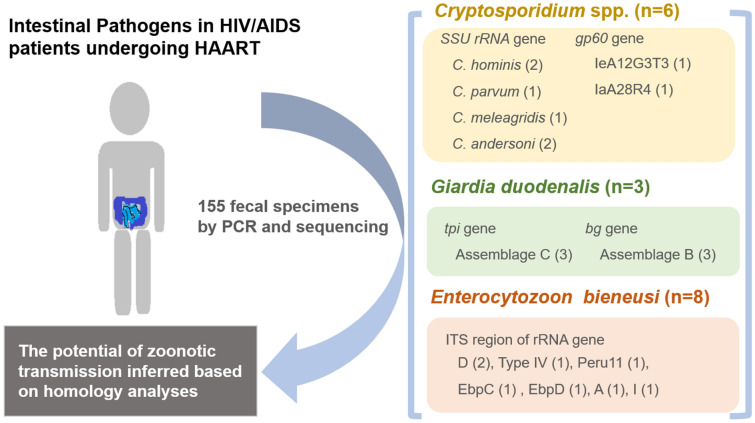
Six (3.9%), three (1.9%), and eight (5.2%) HIV/AIDS patients were positive for Cryptosporidium spp., G. duodenalis, and E. bieneusi, respectively. No statistical differences were observed in occurrence rate between the groups by gender, clinical symptom (diarrhea), and CD4+ cell count. Four Cryptosporidium species were identified: Cryptosporidium hominis (n = 2), Cryptosporidium parvum (n = 1), Cryptosporidium meleagridis (n = 1), and Cryptosporidium andersoni (n = 2). Furthermore, two C. hominis subtypes (IeA12G3T3 and IaA28R4) were detected. Three G. duodenalis-positive specimens were successfully amplified and sequenced at the triosephosphate isomerase and β-giardin loci, which led to the identification of assemblages C and B, respectively. Seven genotypes (D, Type IV, EbpC, Peru11, EbpD, A, and I) were identified in E. bieneusi-positive specimens.
Conclusions
Our findings should increase awareness of AIDS-related opportunistic intestinal pathogens, and indicate the need for routine examination in clinical practice for the detection of Cryptosporidium spp., G. duodenalis, and E. bieneusi. Homology analyses of the three intestinal pathogens at the nucleotide and/or amino acid levels indicated their zoonotic potential.
Graphical Abstract
Keywords: Cryptosporidium species, Giardia duodenalis, Enterocytozoon bieneusi, Human immunodeficiency virus, Genotype, Subtype, Zoonotic transmission
Background
Acquired immunodeficiency syndrome (AIDS) is an important public health concern worldwide, and deprives hundreds of thousands of people of their lives each year. AIDS is the fourth major cause of human death and the leading cause of death due to infectious disease [1]. Serious opportunistic infections (OIs) are the prominent cause of death in patients with advanced AIDS, with many such infections posing a serious threat to the health of human immunodeficiency virus (HIV)/AIDS patients [2]. Cryptosporidium spp., Giardia duodenalis (syn. Giardia intestinalis or Giardia lamblia), and Enterocytozoon bieneusi are common opportunistic intestinal pathogens in humans [3]; their global prevalence in HIV/AIDS patients is 14.0%, 5.0% and 9.2%, respectively [4, 5]. Infections caused by these pathogens are mainly characterized by diarrhea, which can become chronic or life-threatening in immunocompromised individuals, particularly in HIV/AIDS patients [5–7]. The number of patients living with HIV has continued to increase across the globe, from approximately 2 million people in 2016 to approximately 38 million people in 2019 [8, 9]. Early diagnosis of infectious diseases is pivotal to decreasing the occurrence of their severe clinical consequences in this patient population.
Cryptosporidium spp., G. duodenalis and E. bieneusi are found in animals as well as in humans. In humans, they are transmitted via the fecal–oral route, either directly (human-to-human/animal contact) or indirectly (via ingestion of water or food contaminated by human or animal feces) [10, 11]. The role of water and food in the epidemiology of infections caused by these pathogens is now well recognized. Many outbreaks of these infections have been reported worldwide, with waterborne outbreaks being more common (respectively > 524 and > 26 for cryptosporidiosis [12], > 344 and > 38 for giardiasis [12], and one and two for microsporidiosis caused by E. bieneusi [13–15]). Based on their clinical and public health importance, Cryptosporidium spp., G. duodenalis, and E. bieneusi are included in the Environmental Protection Agency Contaminant Candidate List of microbes of concern for waterborne transmission (https://www.epa.gov/ground-water-and-drinking-water/national-primary-drinking-water-regulations). In addition, in 2014, Cryptosporidium spp. and G. duodenalis were ranked, respectively, as the fifth and the 11th most important foodborne parasites worldwide by a joint Food and Agriculture Organization and World Health Organization of the United Nations study [16].
The application of molecular genotyping and subtyping tools has considerably enhanced our ability to trace sources of contamination or infection and to elucidate transmission routes of pathogens. To date, at least 44 Cryptosporidium spp. and 120 genotypes have been identified. Cryptosporidium hominis and C. parvum are highly prevalent (> 90%) in human cases of cryptosporidiosis worldwide [17]. Further, eight assemblages (A–H) of G. duodenalis have been characterized, and assemblages A and B are responsible for the vast majority (99%) of human cases of giardiasis [18]. Enterocytozoon bieneusi is a genetically complex species. To date, at least 685 E. bieneusi genotypes, belonging to 13 distinct genetic groups, have been identified [19]. At least 106 E. bieneusi genotypes are found in humans [20], and genotypes D, EbpC, and Type IV are commonly reported in patients with E. bieneusi infection [21].
In China, the number of people infected with HIV is high and shows an upward trend. At the end of 2020, there were 1.1 million people living with HIV in China [22]. Epidemiological studies on Cryptosporidium spp., G. duodenalis, and E. bieneusi in this population have been conducted across several provinces and municipalities of China, with reported prevalences of Cryptosporidium spp., G. duodenalis and E. bieneusi of 0.7–60.0% [23], 1.3–16.2% [24, 25], and 5.7–11.6% [26], respectively. However, no associated data are available on infections caused by these opportunistic intestinal pathogens in HIV/AIDS patients in Shanghai (one of the four direct-controlled municipalities). Cryptosporidium spp., G. duodenalis, and E. bieneusi are routinely detected in local patients (adults and children) with diarrhea [27–31]. Studies have also reported the occurrence of these pathogens in animals (e.g., livestock, companion animals, and zoo animals) [32], and the environment (e.g., in source and tap water, combined sewerage systems and wastewater treatment plant effluent) [33–38].
Thus, we investigated the rates of occurrence of the intestinal pathogens Cryptosporidium spp., G. duodenalis, and E. bieneusi in HIV/AIDS patients and determined their genetic characteristics at the genotype and/or subtype levels by using molecular techniques. The sources of contamination or infection by Cryptosporidium spp., G. duodenalis, and E. bieneusi were traced, and the transmission routes of these pathogens assessed by homology analysis of their nucleotide or amino acid sequences.
Methods
Participants
In total, 155 HIV/AIDS patients (of whom only four had diarrhea) were enrolled in the study from July 2013 to March 2017. All of the patients were followed at Shanghai Public Health Clinical Center, and had been on highly active antiretroviral therapy (HAART). All of the patients were adults, aged from 18 to 64 years; 141 (91.0%) were males and 14 (9.0%) were females. At the time of sampling of the fecal specimens, the CD4+ cell count was > 200 cells/μL in 92 of the patients and < 200 cells/μL in 63 of the patients.
Specimen collection and DNA extraction
Fecal specimens were collected from all of the patients (one per patient), immediately transferred to the laboratory in a cooler containing ice packs, and subsequently stored at − 80 °C for molecular detection of Cryptosporidium spp., G. duodenalis, and E. bieneusi. Genomic DNA was directly extracted from an approximately 180- to 200-mg fecal specimen using the QIAamp DNA Stool Mini Kit (QIAgen, Hilden, Germany), in accordance with the manufacturer’s instructions. DNA was eluted in 200 μL AE buffer and stored at − 20 °C until needed.
Polymerase chain reaction amplification
The DNA preparations were screened for the presence of Cryptosporidium spp., G. duodenalis, and E. bieneusi via nested polymerase chain reacion (PCR) and sequence analysis. Cryptosporidium spp. were genotyped and subtyped by amplifying the small subunit ribosomal RNA [SSU rRNA; approximately 830 base pairs (bp)] [39] and 60-kDa glycoprotein (gp60) genes (approximately 800–850 bp) [40], respectively. Giardia duodenalis was genotyped by amplifying the triosephosphate isomerase (tpi) gene (approximately 530 bp) [41], the β-giardin (bg) gene (approximately 510 bp) [42], and the glutamate dehydrogenase gene (approximately 530 bp) [43]. Enterocytozoon bieneusi was identified and genotyped by PCR analysis of an approximately 410-bp region of the rRNA gene covering the entire internal transcribed spacer (ITS) region (243 bp) [44]. PCR was performed with positive controls (chicken-derived Cryptosporidium baileyi DNA for Cryptosporidium spp., cattle-derived assemblage E DNA for G. duodenalis, and deer-derived genotype BEB6 DNA for E. bieneusi) as well as negative controls (DNase-free water). All the secondary PCR products were subjected to electrophoresis in a 1.5% agarose gel, and were visualized by staining the gel with ethidium bromide.
Sequencing and data analyses
All of the PCR amplicons of expected size were purified and then directly sequenced using the corresponding primers on an ABI PRISMTM 3730 DNA Analyzer (Applied Biosystems, Carlsbad, CA) with a BigDye Terminator v3.1 Cycle Sequencing kit (Applied Biosystems). Sequencing data accuracy was validated by sequencing in both directions. Nucleotide sequences were subjected to Basic Local Alignment Search Tool (BLAST) searches (http://www.ncbi.nlm.nih.gov/blast/) and then aligned and analyzed with reference sequences downloaded from GenBank, using Clustal X 1.83 (http://www.clustal.org/) to determine the genotypes and subtypes of Cryptosporidium-, G. duodenalis-, and E. bieneusi-positive specimens. If the obtained nucleotide sequences were different from published sequences, two separate PCR amplicons of the same DNA preparation were sequenced to ensure accuracy.
Statistical analysis
Statistical analysis was performed with SPSS 26.0. Pearson chi-square and Fisher’s exact tests were used to determine statistical significance.
Results
Occurrence of Cryptosporidium spp., G. duodenalis and E. bieneusi
PCR and sequence analyses revealed that, of the 155 HIV/AIDS patients, six, three, and eight patients were positive for Cryptosporidium spp., G. duodenalis, and E. bieneusi, respectively. Enterocytozoon bieneusi (5.2%) was more prevalent than Cryptosporidium spp. (3.9%) and G. duodenalis (1.9%). One patient had a mixed infection with G. duodenalis and E. bieneusi.
Females had a higher rate of infection with Cryptosporidium than males (7.1% vs. 3.6%). Giardia duodenalis and E. bieneusi were only found in males. All Cryptosporidium- and E. bieneusi-positive cases were of non-diarrheal patients. G. duodenalis was identified in both diarrheal and non-diarrheal patients. However, no statistical difference in occurrence rate of diarrhea was found between them (Table 1). Cryptosporidium spp., G. duodenalis, and E. bieneusi were detected in patients with a CD4+ cell count of < 200 cells/μL and in patients with a CD4+ cell count of > 200 cells/μL, and no significant association was found between CD4+ cell count and infections caused by these pathogens.
Table 1.
Occurrence of Cryptosporidium spp., Giardia duodenalis and Enterocytozoon bieneusi in human immunodeficiency virus (HIV)/acquired immunodeficiency syndrome (AIDS) patients
| Groups | Examined (n) | Cryptosporidium spp. | G. duodenalis | E. bieneusi | |||
|---|---|---|---|---|---|---|---|
| Positive [n (%)] | χ2/P-value | Positive [n (%)] | χ2/P-value | Positive [n (%)] | χ2/P-value | ||
| Gender | |||||||
| Male | 141 | 5 (3.6) | 0.44/0.51 | 3 (2.1) | 0.57/0.45 | 8 (5.7) | 1.56/0.21 |
| Female | 14 | 1 (7.1) | 0 (0) | 0 (0) | |||
| Diarrhea | |||||||
| Yes | 4 | 0 (0) | 0.32/0.57 | 1 (25.0) | 2.41/0.12 | 0 (0) | 0.43/0.51 |
| No | 151 | 6 (4.0) | 2 (1.3) | 8 (5.3) | |||
| CD4+ T-lymphocyte count (cells/μL) | |||||||
| < 200 | 63 | 4 (6.4) | 1.75/0.19 | 2 (3.2) | 0.86/0.35 | 3 (4.8) | 0.03/0.85 |
| > 200 | 92 | 2 (2.2) | 1 (1.1) | 5 (5.4) | |||
| Total | 155 | 6 (3.9) | 3 (1.9) | 8 (5.2) | |||
Cryptosporidium genotyping and subtyping
Sequence analysis of the SSU rRNA gene revealed the presence of four Cryptosporidium spp.: Cryptosporidium hominis (n = 2), Cryptosporidium parvum (n = 1), Cryptosporidium meleagridis (n = 1), and Cryptosporidium andersoni (n = 2). None of the SSU rRNA gene sequences had been previously described (GenBank accession numbers MT757967–MT757972). Table 2 shows the results of the homology analyses. DNA specimens characterized as originating from C. hominis, C. parvum, or C. meleagridis were further subjected to subtyping by sequence analysis of the gp60 gene. However, only two of these specimens, from C. hominis, were successfully subtyped and identified, namely IaA28R4 (GenBank accession no. OM212052) and IeA12G3T3 (GenBank accession no. OM212051).
Table 2.
Homology analysis of the small subunit ribosomal RNA gene sequences of Cryptosporidium-positive specimens
| Species (n) | Accession nos.a (n) | Accession nos.b (host)/homology |
|---|---|---|
| Cryptosporidium hominis (2) | MT757967 (1) | ON023862 (rhesus monkey)/99.50% |
| MT757968 (1) | MK990042 (human)/97.92% | |
| Cryptosporidium parvum (1) | MT757969 (1) | AY204230 (human)/99.88% |
| Cryptosporidium meleagridis (1) | MT757970 (1) | KT151551 (chicken)/99.88% |
| Cryptosporidium andersoni (2) | MT757971 (1) | KF826309 (human)/94.64% |
| MT757972 (1) | MK982465 (calf)/99.75% |
aAccession nos. indicating the sequences obtained in the present study
bAccession nos. indicating reference sequences deposited in the GenBank database that have the highest similarity with those obtained in the present study
Giardia duodenalis genotyping
Three G. duodenalis-positive specimens were successfully amplified and sequenced at the tpi and bg loci, and all of them showed mixed infections of assemblages C (at the tpi locus) and B (at the bg locus). Three of the tpi gene sequences of assemblage C had not been previously described. However, at the amino acid level, the tpi sequences showed 100% similarity with the sequences of dog-derived G. duodenalis: OM212053 and OM212054 were identical to KY979493, and OM212055 was identical to MW561663. In contrast, three bg gene sequences of assemblage B were identical and shared 100% similarity with the sequence of a squirrel monkey-derived G. duodenalis (GenBank accession no. KJ888974). Table 3 summarizes the results of the homology analyses.
Table 3.
Homology analysis of the tpi and bg genes of Giardia duodenalis-positive specimens at the nucleotide and amino acid levels
| Target gene | Assemblage (n) | GenBank accession nos.a | GenBank accession nos.b (host) | Homology | Codon/amino acid |
|---|---|---|---|---|---|
| Tpi | C (1) | OM212053 | KY979493 (dog); HG970114 (dog) | 100% | |
| C (1) | OM212054 | KY979493 (dog); HG970114 (dog) | 99.39% | G(G → T)G/G → V | |
| C (1) | OM212055 | MW561663 (dog) | 98.80% | G(G → T)G/G → V; G(G → T)G/G → V | |
| Bg | B (3) | OM212056 | KJ888974 (squirrel monkey) | 100% |
aGenBank accession nos. indicating the nucleotide sequences obtained in this study
bGenBank accession nos. indicating the published nucleotide sequences with high similarity to the nucleotide sequences obtained in this study
Enterocytozoon bieneusi genotyping
Based on sequence analysis of the ITS region, seven genotypes of E. bieneusi were identified: D (n = 2), EbpC, TypeIV, Peru11, A, and EbpD (one each) of group 1, and I (n = 1) of group 2.
Discussion
HIV/AIDS patients are at high risk of OIs [45], which are a ubiquitous complication in patients with advanced AIDS. In China, despite the increasing number of patients with HIV/AIDS, little information is available on the occurrence of OIs in these patients, and in particular those OI caused by intestinal pathogens. We report here, to our best knowledge for the first time, the presence of Cryptosporidium spp., G. duodenalis, and E. bieneusi in HIV/AIDS patients in Shanghai, China.
The occurrence rates of Cryptosporidium spp., G. duodenalis, and E. bieneusi were 3.9%, 1.9%, and 5.2% in the HIV/AIDS patients, respectively, which are lower than those reported by most previous studies, particularly those from African countries. For example, the rates of Cryptosporidium spp., G. duodenalis and E. bieneusi in HIV-infected patients were reported to be as high as 79.0% in Nigeria [46], 40.7% in Uganda [47], and 76.9% in Uganda [48], respectively. These differences could be related to the progressive introduction of HAART and the National Free Antiretroviral Therapy Program initiated in 2002 by the Chinese government. In fact, since the introduction of HAART, a marked reduction has been seen in the occurrence of Cryptosporidium spp., G. duodenalis, and E. bieneusi in HIV/AIDS patients. The occurrence ratios between AIDS patients without and with HAART were 15.5 (3.1%/0.2%) for cryptosporidiosis in Australia and 10 European countries [49], 1.2 (6.7%/5.5%) for giardiasis in India, Ethiopia, and Cameroon [5], and 1.5 (13.8%/9.2%) for microsporidiosis caused by E. bieneusi in Guangxi, China [50].
Another reason for the lower occurrence rates of Cryptosporidium spp., G. duodenalis, and E. bieneusi in this study could be that almost all of the patients were non-diarrheal. Diarrhea is the most common clinical symptom associated with intestinal pathogen infections, particularly in immunocompromised individuals. Previous studies have confirmed a significant relationship between infections caused by Cryptosporidium spp., G. duodenalis, and E. bieneusi and the occurrence of diarrhea in HIV/AIDS patients. In India, AIDS patients with diarrhea reportedly showed a higher prevalence of these pathogens than those without diarrhea (Cryptosporidium spp., 46.0% vs. 8.0%; G. duodenalis, 16.0% vs. 8.0%; and microsporidiosis caused by E. bieneusi and Encephalitozoon intestinalis, 10% vs. 0%) [51]. Similar results have been reported for HIV/AIDS patients receiving antiretroviral therapy. In an investigation of diarrhea-associated etiologic agents in 164 HIV/AIDS patients in Kenya, Cryptosporidium spp., G. duodenalis, and E. bieneusi, detected by light microscopy, were found to be more prevalent in patients with diarrhea than in those without diarrhea (Cryptosporidium spp., 16.0% vs. 6.0%; G. duodenalis, 10.0% vs. 3.0%; microsporidia, 10.0% vs. 2.0%) [52]. These findings should raise awareness in clinical practice of OIs caused by intestinal pathogens in HIV/AIDS patients, specifically in those with diarrhea. Early clinical intervention should reduce the occurrence of serious clinical consequences related to OIs.
In the present study, sequence analysis of the SSU rRNA gene led to the identification of C. hominis, C. parvum, C. meleagridis, and C. andersoni. Cryptosporidium hominis and C. parvum have the highest prevalences of the genus Cryptosporidium globally, and are responsible for > 90% of all human cases of Cryptosporidium infection/cryptosporidiosis [17]. Cryptosporidium meleagridis is the third most prevalent species of the genus infecting humans [17]. In China, C. hominis (127/265, 47.9%) and C. parvum (44/265, 16.6%) are the most prevalent species of the genus, and are responsible for 64.5% of all human cases of Cryptosporidium infection [23, 53], followed by C. andersoni (59/265, 22.3%) [12]. In a molecular epidemiological investigation of Cryptosporidium in diarrheal patients from southern Assam, India, C. andersoni was reported to be the predominant species, accounting for 79.6% of 98 Cryptosporidium-positive cases [54]. To date, C. andersoni has been found to be responsible for 148 human cases (including those identified in this study) of Cryptosporidium infection/cryptosporidiosis, with the vast majority (96.6%) of cases being reported in developing countries, including China, Myanmar, India, Iran, and Malawi [12]. It is notable that C. andersoni is a common species in ruminants, particularly in adult cattle [17]. However, the extent of its zoonotic transmission remains to be determined because of the absence of multilocus sequence typing (MLST) data for human-derived C. andersoni. MLST subtypes and the population genetic structure of animal-derived C. andersoni, including those from cattle, sheep, horses, golden takins, monkeys, camels, ostriches, and hamsters, have been analyzed in China [23]. Future studies need to employ MLST analysis of human-derived C. andersoni to facilitate elucidation of the sources of infection/contamination with C. andersoni and increase our understanding of its transmission dynamics.
Sequence analysis of the gp60 gene has been widely used to subtype and track the transmission of zoonotic Cryptosporidium spp. and their genotypes [17]. Only two subtypes (IaA28R4 and IeA12G3T3) of C. hominis were identified in the present study, of which subtype IaA28R4 is an emerging C. hominis subtype in humans in China. To date, this subtype has only been identified in human cases of cryptosporidiosis in the USA (62 cases) [55] and Sweden (two cases) [56]. Subtype IeA12G3T3 has been previously reported in HIV/AIDS patients in China [57], and it was also detected in raw wastewater/sewage in wastewater plants in four cities in China, including Shanghai [36, 38]. Moreover, subtype IeA12G3T3 has been found in humans in Vietnam [58], Jamaica [59], Qatar [60], and Slovakia [61]. The data on C. hominis subtypes that are currently available indicate that IaA28R4 and IeA12G3T3 are rarely detected in humans and are restricted to certain countries.
Sequence analysis of the bg gene led to identification of assemblage B in three G. duodenalis-positive specimens. Among the six assemblages (A–F) of G. duodenalis found in humans, A and B are the most common worldwide, and B is responsible for more infections than A. In China, only three assemblages (A–C) have been identified in humans. In Myanmar, 70% (221/287) and 17.4% (50/287) of human cases of G. duodenalis infections were attributable to assemblages A and B, respectively [12]. Meanwhile, sequence analysis of the tpi gene identified assemblage C in three identical G. duodenalis-positive specimens, indicative of mixed infection with assemblages B and C. The increasing use of multilocus genotyping tools in epidemiological studies of G. duodenalis has led to more cases of mixed infections involving different assemblages being found in humans and animals [e.g., assemblages A and B (n = 15) in 54 G. duodenalis-positive asymptomatic immigrants in Qatar [62] and assemblages A and E (n = 4), B and E (n = 1), and C and D (n = 1) in 214 G. duodenalis-positive animal specimens in northeastern China] [63].
The homology analysis indicated that three bg gene sequences of assemblage B were identical. The same bg gene sequence has been previously described in squirrel monkeys from China [64]. Three different tpi gene sequences of assemblage C obtained in the present study have not been previously described. However, at the amino acid level, the tpi sequences showed 100% similarity with those derived from dogs: OM212053 and OM212054 were identical to KY979493 (China), and OM212055 was identical to MW561663 (Thailand). The results of the homology analysis indicated the zoonotic potential of assemblages B and C detected in the present study in patients from Shanghai. Although we identified assemblage C in the present study, it is usually found in canines, and only occasionally in humans. To date, assemblage C has been found in humans in Brazil [65], Egypt [66], Slovakia [67], and China [27]. In China, assemblage C has been found to be responsible for human cases of giardiasis only in Shanghai [27].
Sequence analysis of the ITS region revealed seven genotypes (D, Type IV, EbpC, Peru11, EbpD, A, and I) in the E. bieneusi-positive specimens. Among these, genotypes D, EbpC, and Type IV of group 1 have been most frequently identified in humans and numerous animal species worldwide, and display a low level of host specificity and zoonotic or cross-species transmission potential [11]. In China, the genotypes D, EbpC, and Type IV have been most commonly found in humans (ranked first, second, and fifth, respectively, according to number of cases of E. bieneusi infection) [68], and in at least 22, 12 and seven animal species, respectively [69]. Genotypes Peru11 and EbpD are also frequently detected in humans, and both of these show zoonotic potential; however, their host ranges (five and two animal species for genotypes Peru11 and EbpD, respectively) are smaller than those of genotypes D, EbpC, and Type IV (28, 16, and 9, respectively) [11]. In China, seven cases and one case, respectively, of microsporidiosis have been attributed to genotypes Peru11 and EbpD [68]. Genotype Peru11 has also been found in five non-human primate species and genotype EbpD in black-capped capuchins and pigs [69]. Genotype A is the most prevalent in humans globally [11]. This genotype was previously only found in humans, and was indicative of person-to-person transmission of E. bieneusi [26], but has since been detected in captive baboons in Kenya [70] and in dogs in Spain [71]. In China, besides its detection in HIV/AIDS patients in the present study, this genotype has also been detected in children [72]; however, to date, no studies have reported genotype A in animals. Genotype I of group 2 was initially considered to be cattle-specific due its frequent detection in cattle [73]. E. bieneusi has been found in animal species such as pigs, deer, takins, cats, meerkats, rabbits, and bats, as well as in diarrheal children in China [69, 74]. These findings indicate that genotype I may be of public health concern due to its zoonotic potential.
This study has some limitations. The number of analyzed specimens was small, and the number of positive specimens was low. In accordance with the One World—One Health Manhattan Principles, we believe that future molecular studies on the epidemiology of Cryptosporidium spp., G. duodenalis, and E. bieneusi need to be conducted using a sufficient number of human specimens, a greater variety of animal hosts and environmental specimens, to assess and validate the zoonotic potential of these pathogens. The types of data thus collected should facilitate the development of efficient control strategies for the management and prevention of OIs in HIV/AIDS patients.
Conclusions
Herein we report the occurrence and genetic characteristics of Cryptosporidium spp., G. duodenalis, and E. bieneusi in HIV/AIDS patients in Shanghai, China. Our findings should increase awareness of opportunistic intestinal pathogens in AIDS patients, and indicate the importance in clinical practice of routine examination of patients for the detection of the common pathogens discussed here. The high diversity of Cryptosporidium spp. and the rare gp60 subtypes of C. hominis identified in this study possibly reflect the genetic characteristics of Cryptosporidium endemic to China. Mixed infections with different assemblages were indicated by identification of assemblages B and C in three identical G. duodenalis-positive specimens. Seven zoonotic genotypes of E. bieneusi, including genotypes D, EbpC, Type IV (high frequency), Peru 11, EbpD, and A (commonly found), and I (rarely found), were also identified. Homology analysis of nucleotide and/or amino acid sequences of these pathogens revealed their zoonotic potential.
Acknowledgements
We thank the staff at Shanghai Public Health Clinical Center for their assistance with sample collection and case investigation.
Abbreviations
- AIDS
Acquired immunodeficiency syndrome
- bg
β-giardin
- BLAST
Basic Local Alignment Search Tool
- HAART
Highly active antiretroviral therapy
- HIV
Human immunodeficiency virus
- ITS
Internal transcribed spacer
- OI
Opportunistic infection
- SSU rRNA
Small subunit ribosomal RNA
- tpi
Triosephosphate isomerase
Author contributions
YS and JC designed the study. YJ, LL and ZY performed the experiments. YJ, AL and ZY analyzed the data. YS and JC contributed reagents/materials. YJ and LL wrote the first draft of the manuscript and prepared the tables and the figures. YS and JC undertook the final revision of the manuscript. All authors read and approved the submitted version of the manuscript.
Funding
This research was partially supported by the National Key Research and Development Program of China (no. 2021YFC2300902), the National Science and Technology Major Program of China (no. 2018ZX10713001-004), the Fifth Round of the Three-Year Public Health Action Plan of Shanghai (no. GWV-10.1-XK13), and the National Nature Science Foundation of China (no. 82273693). The funders had no role in the study design, data collection or analysis, decision to publish or preparation of the manuscript.
Availability of data and materials
The representative nucleotide sequences obtained in the present study were deposited in GenBank database under the following accession nos.: MT757967–MT757972 (Cryptosporidium), OM212053–OM212056 (G. duodenalis).
Declarations
Ethics approval and consent to participate
The research protocol was reviewed and approved by the Ethics Committee of the National Institute of Parasitic Diseases, Chinese Centre for Disease Control and Prevention, China (approval no. 2012-12). All the study participants were HIV/AIDS patients undergoing HAART. They were informed of the aims of this study and of the procedures that would be used in it at the time of enrollment, and permission was obtained from each of them before collection of the fecal specimens. A structured questionnaire was used to collect demographic data and clinical signs and symptoms of each participant.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Yanyan Jiang and Li Liu contributed equally to this work
Contributor Information
Yanyan Jiang, Email: jiangyy@nipd.chinacdc.cn.
Li Liu, Email: liuli@shaphc.org.
Zhongying Yuan, Email: yuanzy@nipd.chinacdc.cn.
Aiqin Liu, Email: liuaiqin1128@126.com.
Jianping Cao, Email: caojp@chinacdc.cn.
Yujuan Shen, Email: shenyj@nipd.chinacdc.cn.
References
- 1.Sohn AH, Lumbiganon P, Kurniati N, Lapphra K, Law M, Do VC, et al. Determining standardized causes of death of infants, children, and adolescents living with HIV in Asia. AIDS. 2020;34:1527–1537. doi: 10.1097/QAD.0000000000002583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Betancourth S, Archaga O, Moncada W, Rodríguez V, Fontecha G. First molecular characterization of Cryptosporidium spp. in patients living with HIV in Honduras. Pathogens. 2021;10:336. doi: 10.3390/pathogens10030336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Maikai BV, Umoh JU, Lawal IA, Kudi AC, Ejembi CL, Xiao L. Molecular characterizations of Cryptosporidium, Giardia, and Enterocytozoon in humans in Kaduna State, Nigeria. Exp Parasitol. 2012;131:452–456. doi: 10.1016/j.exppara.2012.05.011. [DOI] [PubMed] [Google Scholar]
- 4.Wang ZD, Liu Q, Liu HH, Li S, Zhang L, Zhao YK, et al. Prevalence of Cryptosporidium, microsporidia and Isospora infection in HIV-infected people: a global systematic review and meta-analysis. Parasit Vectors. 2018;11:28. doi: 10.1186/s13071-017-2558-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mahdavi F, Shams M, Sadrebazzaz A, Shamsi L, Omidian M, Asghari A, et al. Global prevalence and associated risk factors of diarrheagenic Giardia duodenalis in HIV/AIDS patients: a systematic review and meta-analysis. Microb Pathog. 2021;160:105202. doi: 10.1016/j.micpath.2021.105202. [DOI] [PubMed] [Google Scholar]
- 6.Wang RJ, Li JQ, Chen YC, Zhang LX, Xiao LH. Widespread occurrence of Cryptosporidium infections in patients with HIV/AIDS: epidemiology, clinical feature, diagnosis, and therapy. Acta Trop. 2018;187:257–263. doi: 10.1016/j.actatropica.2018.08.018. [DOI] [PubMed] [Google Scholar]
- 7.Qiu L, Xia W, Li W, Ping J, Ding S, Liu H. The prevalence of microsporidia in China: a systematic review and meta-analysis. Sci Rep. 2019;9:3174. doi: 10.1038/s41598-019-39290-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dabis F, Bekker LG. We still need to beat HIV. Science. 2017;20:eaao4197. doi: 10.1126/science.aao4197. [DOI] [PubMed] [Google Scholar]
- 9.Alemayehu E, Gedefie A, Adamu A, Mohammed J, Kassanew B, Kebede B, et al. Intestinal parasitic infections among HIV-infected patients on antiretroviral therapy attending Debretabor General Hospital, northern Ethiopia: a cross-sectional study. HIV AIDS (Auckl) 2020;12:647–655. doi: 10.2147/HIV.S275358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ryan UM, Feng Y, Fayer R, Xiao L. Taxonomy and molecular epidemiology of Cryptosporidium and Giardia—a 50 year perspective (1971–2021) Int J Parasitol. 2021;51:1099–1119. doi: 10.1016/j.ijpara.2021.08.007. [DOI] [PubMed] [Google Scholar]
- 11.Li W, Feng Y, Santin M. Host specificity of Enterocytozoon bieneusi and public health implications. Trends Parasitol. 2019;35:436–451. doi: 10.1016/j.pt.2019.04.004. [DOI] [PubMed] [Google Scholar]
- 12.Wu Y, Gong B, Liu X, Jiang Y, Cao J, Yao L, et al. Identification of uncommon Cryptosporidium viatorum (a novel subtype XVcA2G1c) and Cryptosporidium andersoni as well as common Giardia duodenalis assemblages A and B in humans in Myanmar. Front Cell Infect Microbiol. 2020;10:614053. doi: 10.3389/fcimb.2020.614053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cotte L, Rabodonirina M, Chapuis F, Bailly F, Bissuel F, Raynal C, et al. Waterborne outbreak of intestinal microsporidiosis in persons with and without human immunodeficiency virus infection. J Infect Dis. 1999;180:2003–2008. doi: 10.1086/315112. [DOI] [PubMed] [Google Scholar]
- 14.Michlmayr D, Alves de Sousa L, Müller L, Jokelainen P, Ethelberg S, Vestergaard LS, et al. Incubation period, spore shedding duration, and symptoms of Enterocytozoon bieneusi genotype C infection in a foodborne outbreak in Denmark, 2020. Clin Infect Dis. 2022;75:468–475. doi: 10.1093/cid/ciab949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Decraene V, Lebbad M, Botero-Kleiven S, Gustavsson AM, Löfdahl M. First reported foodborne outbreak associated with microsporidia, Sweden, October 2009. Epidemiol Infect. 2012;140:519–527. doi: 10.1017/S095026881100077X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Plutzer J, Lassen B, Jokelainen P, Djurković-Djaković O, Kucsera I, Dorbek-Kolin E, et al. Review of Cryptosporidium and Giardia in the eastern part of Europe, 2016. Euro Surveill. 2018;23:16–00825. doi: 10.2807/1560-7917.ES.2018.23.4.16-00825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ryan U, Zahedi A, Feng Y, Xiao L. An update on zoonotic Cryptosporidium species and genotypes in humans. Animals (Basel) 2021;11:3307. doi: 10.3390/ani11113307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sprong H, Cacciò SM, van der Giessen JW. ZOOPNET network and partners. Identification of zoonotic genotypes of Giardia duodenalis. PLoS Negl Trop Dis. 2009;3:e558. doi: 10.1371/journal.pntd.0000558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang T, Ren G, Zhou H, Qiang Y, Li J, Zhang Y, et al. Molecular prevalence and genetic diversity analysis of Enterocytozoon bieneusi in humans in Hainan Province, China: high diversity and unique endemic genetic characteristics. Front Public Health. 2022;10:1007130. doi: 10.3389/fpubh.2022.1007130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shen Y, Gong B, Liu X, Wu Y, Yang F, Xu J, et al. First identification and genotyping of Enterocytozoon bieneusi in humans in Myanmar. BMC Microbiol. 2020;20:10. doi: 10.1186/s12866-019-1694-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li W, Xiao L. Ecological and public health significance of Enterocytozoon bieneusi. One Health. 2020;12:100209. doi: 10.1016/j.onehlt.2020.100209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.He N. Research progress in the epidemiology of HIV/AIDS in China. China CDC Wkly. 2021;3:1022–1030. doi: 10.46234/ccdcw2021.249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu A, Gong B, Liu X, Shen Y, Wu Y, Zhang W, et al. A retrospective epidemiological analysis of human Cryptosporidium infection in China during the past three decades (1987–2018) PLoS Negl Trop Dis. 2020;14:e0008146. doi: 10.1371/journal.pntd.0008146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tian LG, Chen JX, Wang TP, Cheng GJ, Steinmann P, Wang FF, et al. Co-infection of HIV and intestinal parasites in a rural area of China. Parasit Vectors. 2012;5:36. doi: 10.1186/1756-3305-5-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pang XL, Chen SY, Gao K, Mai HX, Han ZG, Xu HF, et al. Serum epidemiological analysis of opportunistic infection of pathogenic protozoa in HIV/AIDS. J Trop Med. 2015;15:1425–1428. [Google Scholar]
- 26.Gong B, Yang Y, Liu X, Cao J, Xu M, Xu N, et al. First survey of Enterocytozoon bieneusi and dominant genotype Peru6 among ethnic minority groups in southwestern China's Yunnan Province and assessment of risk factors. PLoS Negl Trop Dis. 2019;13:e0007356. doi: 10.1371/journal.pntd.0007356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu H, Shen Y, Yin J, Yuan Z, Jiang Y, Xu Y, et al. Prevalence and genetic characterization of Cryptosporidium, Enterocytozoon, Giardia and Cyclospora in diarrheal outpatients in China. BMC Infect Dis. 2014;14:25. doi: 10.1186/1471-2334-14-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jiang Y, Yuan Z, Liu H, Yin J, Qin Y, Jiang X, et al. Intestinal protozoan infections in patients with diarrhea—Shanghai Municipality, Zhenjiang City, and Danyang City, China, 2011–2015 and 2019–2021. China CDC Wkly. 2022;4:143–147. doi: 10.46234/ccdcw2022.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Feng Y, Wang L, Duan L, Gomez-Puerta LA, Zhang L, Zhao X, et al. Extended outbreak of cryptosporidiosis in a pediatric hospital. China Emerg Infect Dis. 2012;18:312–314. doi: 10.3201/eid1802.110666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang L, Xiao L, Duan L, Ye J, Guo Y, Guo M, et al. Concurrent infections of Giardia duodenalis, Enterocytozoon bieneusi, and Clostridium difficile in children during a cryptosporidiosis outbreak in a pediatric hospital in China. PLoS Negl Trop Dis. 2013;7:e2437. doi: 10.1371/journal.pntd.0002437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang Y, Li N, Guo Y, Wang L, Wang R, Feng Y, et al. Persistent occurrence of Cryptosporidium hominis and Giardia duodenalis subtypes in a welfare institute. Front Microbiol. 2018;9:2830. doi: 10.3389/fmicb.2018.02830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yin JH, Shen YJ, Cao JP. Burden of Cryptosporidium infections in the Yangtze River Delta in China in the 21st century: a One Health perspective. Zoonoses. 2022;7:2. [Google Scholar]
- 33.Huang C, Hu Y, Wang L, Wang Y, Li N, Guo Y, et al. Environmental transport of emerging human-pathogenic Cryptosporidium species and subtypes through combined sewer overflow and wastewater. Appl Environ Microbiol. 2017;83:e00682–e717. doi: 10.1128/AEM.00682-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ma J, Feng Y, Hu Y, Villegas EN, Xiao L. Human infective potential of Cryptosporidium spp., Giardia duodenalis and Enterocytozoon bieneusi in urban wastewater treatment plant effluents. J Water Health. 2016;14:411–423. doi: 10.2166/wh.2016.192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hu Y, Feng Y, Huang C, Xiao L. Occurrence, source, and human infection potential of Cryptosporidium and Enterocytozoon bieneusi in drinking source water in Shanghai, China, during a pig carcass disposal incident. Environ Sci Technol. 2014;48:14219–14227. doi: 10.1021/es504464t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li N, Xiao L, Wang L, Zhao S, Zhao X, Duan L, et al. Molecular surveillance of Cryptosporidium spp., Giardia duodenalis, and Enterocytozoon bieneusi by genotyping and subtyping parasites in wastewater. PLoS Negl Trop Dis. 2012;6:e1809. doi: 10.1371/journal.pntd.0001809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Feng Y, Zhao X, Chen J, Jin W, Zhou X, Li N, et al. Occurrence, source, and human infection potential of Cryptosporidium and Giardia spp. in source and tap water in Shanghai, China. Appl Environ Microbiol. 2011;77:3609–3616. doi: 10.1128/AEM.00146-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Feng Y, Li N, Duan L, Xiao L. Cryptosporidium genotype and subtype distribution in raw wastewater in Shanghai, China: evidence for possible unique Cryptosporidium hominis transmission. J Clin Microbiol. 2009;47:153–157. doi: 10.1128/JCM.01777-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xiao L, Singh A, Limor J, Graczyk TK, Gradus S, Lal A. Molecular characterization of Cryptosporidium oocysts in samples of raw surface water and wastewater. Appl Environ Microbiol. 2001;67:1097–1101. doi: 10.1128/AEM.67.3.1097-1101.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stensvold CR, Elwin K, Winiecka-Krusnell J, Chalmers RM, Xiao L, Lebbad M. Development and application of a gp60-based typing assay for Cryptosporidium viatorum. J Clin Microbiol. 2015;53:1891–1897. doi: 10.1128/JCM.00313-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sulaiman IM, Fayer R, Bern C, Gilman RH, Trout JM, Schantz PM, et al. Triosephosphate isomerase gene characterization and potential zoonotic transmission of Giardia duodenalis. Emerg Infect Dis. 2003;9:1444–1452. doi: 10.3201/eid0911.030084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lalle M, Pozio E, Capelli G, Bruschi F, Crotti D, Cacciò SM. Genetic heterogeneity at the beta-giardin locus among human and animal isolates of Giardia duodenalis and identification of potentially zoonotic subgenotypes. Int J Parasitol. 2005;35:207–213. doi: 10.1016/j.ijpara.2004.10.022. [DOI] [PubMed] [Google Scholar]
- 43.Cacciò SM, Beck R, Lalle M, Marinculic A, Pozio E. Multilocus genotyping of Giardia duodenalis reveals striking differences between assemblages A and B. Int J Parasitol. 2008;38:1523–1531. doi: 10.1016/j.ijpara.2008.04.008. [DOI] [PubMed] [Google Scholar]
- 44.Buckholt MA, Lee JH, Tzipori S. Prevalence of Enterocytozoon bieneusi in swine: an 18-month survey at a slaughterhouse in Massachusetts. Appl Environ Microbiol. 2002;68:2595–2599. doi: 10.1128/AEM.68.5.2595-2599.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pang W, Shang P, Li Q, Xu J, Bi L, Zhong J, et al. Prevalence of opportunistic infections and causes of death among hospitalized HIV-infected patients in Sichuan, China. Tohoku J Exp Med. 2018;244:231–242. doi: 10.1620/tjem.244.231. [DOI] [PubMed] [Google Scholar]
- 46.Adesiji YO, Lawal RO, Taiwo SS, Fayemiwo SA, Adeyeba OA. Cryptosporidiosis in HIV infected patients with diarrhoea in Osun State, southwestern Nigeria. Eur J Gen Med. 2007;4:119–122. [Google Scholar]
- 47.Johnston AR, Gillespie TR, Rwego IB, McLachlan TL, Kent AD, Goldberg TL. Molecular epidemiology of cross-species Giardia duodenalis transmission in western Uganda. PLoS Negl Trop Dis. 2010;4:e683. doi: 10.1371/journal.pntd.0000683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tumwine JK, Kekitiinwa A, Bakeera-Kitaka S, Ndeezi G, Downing R, Feng X, et al. Cryptosporidiosis and microsporidiosis in Ugandan children with persistent diarrhea with and without concurrent infection with the human immunodeficiency virus. Am J Trop Med Hyg. 2005;73:921–925. [PubMed] [Google Scholar]
- 49.Babiker A, Darbyshire J, Pezzotti P, Porter K, Rezza G, Walker SA, et al. Changes over calendar time in the risk of specific first AIDS-defining events following HIV seroconversion, adjusting for competing risks. Int J Epidemiol. 2002;31:951–958. doi: 10.1093/ije/31.5.951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Liu H, Jiang Z, Yuan Z, Yin J, Wang Z, Yu B, et al. Infection by and genotype characteristics of Enterocytozoon bieneusi in HIV/AIDS patients from Guangxi Zhuang Autonomous Region, China. BMC Infect Dis. 2017;17:684. doi: 10.1186/s12879-017-2787-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dwivedi KK, Prasad G, Saini S, Mahajan S, Lal S, Baveja UK. Enteric opportunistic parasites among HIV infected individuals: associated risk factors and immune status. Jpn J Infect Dis. 2007;60:76–81. [PubMed] [Google Scholar]
- 52.Wanyiri JW, Kanyi H, Maina S, Wang DE, Ngugi P, O'Connor R, et al. Infectious diarrhoea in antiretroviral therapy-naive HIV/AIDS patients in Kenya. Trans R Soc Trop Med Hyg. 2013;107:631–638. doi: 10.1093/trstmh/trt078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Xu N, Liu H, Jiang Y, Yin J, Yuan Z, Shen Y, et al. First report of Cryptosporidium viatorum and Cryptosporidium occultus in humans in China, and of the unique novel C. viatorum subtype XVaA3h. BMC Infect Dis. 2020;20:16. doi: 10.1186/s12879-019-4693-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hussain G, Roychoudhury S, Singha B, Paul J. Incidence of Cryptosporidium andersoni in diarrheal patients from southern Assam, India: a molecular approach. Eur J Clin Microbiol Infect Dis. 2017;36:1023–1032. doi: 10.1007/s10096-016-2887-2. [DOI] [PubMed] [Google Scholar]
- 55.Feng Y, Tiao N, Li N, Hlavsa M, Xiao L. Multilocus sequence typing of an emerging Cryptosporidium hominis subtype in the United States. J Clin Microbiol. 2014;52:524–530. doi: 10.1128/JCM.02973-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lebbad M, Winiecka-Krusnell J, Stensvold CR, Beser J. High diversity of Cryptosporidium species and subtypes identified in cryptosporidiosis acquired in Sweden and abroad. Pathogens. 2021;10:523. doi: 10.3390/pathogens10050523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wang L, Zhang H, Zhao X, Zhang L, Zhang G, Guo M, et al. Zoonotic Cryptosporidium species and Enterocytozoon bieneusi genotypes in HIV-positive patients on antiretroviral therapy. J Clin Microbiol. 2013;51:557–563. doi: 10.1128/JCM.02758-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Iwashita H, Takemura T, Tokizawa A, Sugamoto T, Thiem VD, Nguyen TH, et al. Molecular epidemiology of Cryptosporidium spp. in an agricultural area of northern Vietnam: a community survey. Parasitol Int. 2021;83:102341. doi: 10.1016/j.parint.2021.102341. [DOI] [PubMed] [Google Scholar]
- 59.Gatei W, Barrett D, Lindo JF, Eldemire-Shearer D, Cama V, Xiao L. Unique Cryptosporidium population in HIV-infected persons, Jamaica. Emerg Infect Dis. 2008;14:841–843. doi: 10.3201/eid1405.071277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Boughattas S, Behnke JM, Al-Sadeq D, Ismail A, Abu-Madi M. Cryptosporidium spp., prevalence, molecular characterisation and socio-demographic risk factors among immigrants in Qatar. PLoS Negl Trop Dis. 2019;13:e0007750. doi: 10.1371/journal.pntd.0007750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hatalová E, Valenčáková A, Luptáková L, Špalková M, Kalinová J, Halánová M, et al. The first report of animal genotypes of Cryptosporidium parvum in immunosuppressed and immunocompetent humans in Slovakia. Emerg Dis. 2019;66:243–249. doi: 10.1111/tbed.13009. [DOI] [PubMed] [Google Scholar]
- 62.Chourabi M, Boughattas S, Abdallah AM, Ismail A, Behnke JM, Al-Mekhlafi HM, et al. Genetic diversity and prevalence of Giardia duodenalis in Qatar. Front Cell Infect Microbiol. 2021;11:652946. doi: 10.3389/fcimb.2021.652946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wu Y, Yao L, Chen H, Zhang W, Jiang Y, Yang F, et al. Giardia duodenalis in patients with diarrhea and various animals in northeastern China: prevalence and multilocus genetic characterization. Parasit Vectors. 2022;15:165. doi: 10.1186/s13071-022-05269-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Karim MR, Wang R, Yu F, Li T, Dong H, Li D, et al. Multi-locus analysis of Giardia duodenalis from nonhuman primates kept in zoos in China: geographical segregation and host-adaptation of assemblage B isolates. Infect Genet Evol. 2015;30:82–88. doi: 10.1016/j.meegid.2014.12.013. [DOI] [PubMed] [Google Scholar]
- 65.Durigan M, Abreu AG, Zucchi MI, Franco RM, de Souza AP. Genetic diversity of Giardia duodenalis: multilocus genotyping reveals zoonotic potential between clinical and environmental sources in a metropolitan region of Brazil. PLoS ONE. 2014;9:e115489. doi: 10.1371/journal.pone.0115489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Soliman RH, Fuentes I, Rubio JM. Identification of a novel assemblage B subgenotype and a zoonotic assemblage C in human isolates of Giardia intestinalis in Egypt. Parasitol Int. 2011;60:507–511. doi: 10.1016/j.parint.2011.09.006. [DOI] [PubMed] [Google Scholar]
- 67.Štrkolcová G, Maďar M, Hinney B, Goldová M, Mojžišová J, Halánová M. Dog's genotype of Giardia duodenalis in human: first evidence in Europe. Acta Parasitol. 2015;60:796–799. doi: 10.1515/ap-2015-0113. [DOI] [PubMed] [Google Scholar]
- 68.Zhou K, Liu M, Wu Y, Zhang R, Wang R, Xu H, et al. Enterocytozoon bieneusi in patients with diarrhea and in animals in the northeastern Chinese city of Yichun: genotyping and assessment of potential zoonotic transmission. Parasite. 2022;29:40. doi: 10.1051/parasite/2022041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wang SS, Wang RJ, Fan XC, Liu TL, Zhang LX, Zhao GH. Prevalence and genotypes of Enterocytozoon bieneusi in China. Acta Trop. 2018;183:142–152. doi: 10.1016/j.actatropica.2018.04.017. [DOI] [PubMed] [Google Scholar]
- 70.Li W, Kiulia NM, Mwenda JM, Nyachieo A, Taylor MB, Zhang X, Xiao L. Cyclospora papionis, Cryptosporidium hominis, and human-pathogenic Enterocytozoon bieneusi in captive baboons in Kenya. J Clin Microbiol. 2011;49:4326–4329. doi: 10.1128/JCM.05051-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Galván-Díaz AL, Magnet A, Fenoy S, Henriques-Gil N, Haro M, Gordo FP, et al. Microsporidia detection and genotyping study of human pathogenic E. bieneusi in animals from Spain. PLoS ONE. 2014;9:e92289. doi: 10.1371/journal.pone.0092289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Qi M, Yu F, Zhao A, Zhang Y, Wei Z, Li D, et al. Unusual dominant genotype NIA1 of Enterocytozoon bieneusi in children in southern Xinjiang, China. PLoS Negl Trop Dis. 2020;14:e0008293. doi: 10.1371/journal.pntd.0008293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Santín M, Dargatz D, Fayer R. Prevalence and genotypes of Enterocytozoon bieneusi in weaned beef calves on cow-calf operations in the USA. Parasitol Res. 2012;110:2033–2041. doi: 10.1007/s00436-011-2732-6. [DOI] [PubMed] [Google Scholar]
- 74.Zhang X, Wang Z, Su Y, Liang X, Sun X, Peng S, et al. Identification and genotyping of Enterocytozoon bieneusi in China. J Clin Microbiol. 2011;49:2006–2008. doi: 10.1128/JCM.00372-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The representative nucleotide sequences obtained in the present study were deposited in GenBank database under the following accession nos.: MT757967–MT757972 (Cryptosporidium), OM212053–OM212056 (G. duodenalis).