Abstract
Tuftelin (TUFT1) is highly expressed in various tumor types and promotes tumor growth and metastasis by activating AKT and other core signaling pathways. However, the effects of post‐translational modifications of TUFT1 on its oncogenic function remain unexplored. In this study, we found that TUFT1 was SUMOylated at K79. SUMOylation deficiency significantly impaired the ability of TUFT1 to promote the proliferation, migration, and invasion of gastric cancer (GC) cells by blocking AKT/mTOR signaling pathway activation. SUMOylation of TUFT1 is mediated by the E3 SUMO ligase tripartite motif‐containing protein 27 (TRIM27), and these two proteins regulate the malignant behavior of GC cells and AKT activation in the same pathway. TUFT1 binds to TRIM27 through its N‐terminus, and decreased binding affinity of TUFT1 to TRIM27 significantly impairs its oncogenic effect. In addition, data collected from GC clinical samples indicated that the combined detection of TUFT1 and TRIM27 expression reflected tumor malignancy and patient survival with higher precision. In addition, we proved that SUMOylated TUFT1 is not only an upstream signal for AKT activation but also directly activates mTOR by forming a complex with Rab GTPase activating protein 1, which further inhibits Rab GTPases and promotes the perinuclear accumulation of mTORC1. Altogether, these data indicate that SUMOylated TUFT1 is the active form that affects GC progression through the AKT/mTOR signaling pathway and might be a promising therapeutic target or biomarker for GC progression.
Keywords: AKT, gastric cancer, SUMOylation, TRIM27, TUFT1
SUMOylated TUFT1 is the active form, which affects GC progression through AKT/mTOR signaling pathway, and might be a promising therapeutic target or biomarker for GC progression. SUMOylated TUFT1 was not only an upstream signal for AKT activation but also directly activated mTOR by forming a complex with RABGAP1, which further inhibited Rab GTPases and promoted the perinuclear accumulation of mTORC1.

1. INTRODUCTION
Gastric cancer (GC) is the fifth most commonly diagnosed cancer and the fourth leading cause of cancer‐related death worldwide, with over 1,000,000 new cases and 769,000 deaths in 2020. 1 The molecular mechanisms underlying GC carcinogenesis and progression are not fully understood, and more in‐depth studies are required to explore new treatment strategies and therapeutic drugs.
SUMOylation is a reversible post‐translational modification involved in various biological processes such as cell cycle regulation, DNA damage repair, and signal transduction. 2 , 3 , 4 SUMOylation follows a conserved catalytic cascade in which small ubiquitin‐like modifier (SUMO) proteins are covalently bound to lysine (K) residues, altering the stability, subcellular localization, or binding affinity of a wide range of proteins. 5 , 6 Several studies have shown that expression of the core SUMOylation machinery (E1 activating enzyme, E2 conjugating enzyme, several E3 ligases, and SUMO1/sentrin specific peptidases) is enhanced in GC, 7 , 8 , 9 , 10 suggesting that SUMOylation is closely related to the survival and malignant behavior of GC cells. However, only a few transcription factors and receptors such as forkhead box protein C2 (FOXC2), N‐myc downstream‐regulated gene 2 (NDRG2), Sp1, CCAAT/enhancer‐binding protein alpha (C/EBPα), insulin‐like growth factor 1 receptor (IGF‐1R), and death domain‐associated protein 6 (DAXX) have been reported to be SUMOylated in GC. 10 , 11 , 12 , 13 , 14 , 15 Uncovering the roles of more SUMOylated proteins in GC will provide a promising perspective for investigating the molecular mechanism of tumorigenesis and progression, finding novel biomarkers with higher sensitivity, and contributing to the early diagnosis and targeted therapies of GC.
Tuftelin (TUFT1) was initially characterized as a protein involved in tooth enamel mineralization in vertebrates and subsequently found in nonmineralized tissues such as lung, stomach, and liver. 16 , 17 Oncological studies indicate that TUFT1 is highly expressed in various tumors, predicting worse clinical outcomes and poorer prognosis. 18 , 19 , 20 , 21 In GC, TUFT1 promotes tumor growth and metastasis by activating AKT and related downstream pathways. 22 Alternatively, TUFT1 directly modulates the activity of mTORC1 by regulating the intracellular lysosome localization and cellular trafficking of Rab GTPases. 19 In other cancers, TUFT1 promotes tumor growth, metastasis, and drug resistance by activating hypoxia inducible factor‐1 (HIF‐1)/Snail and Rab5/Ras‐related C3 botulinum toxin substrate 1 (Rac1)/cadherin‐associated protein, beta 1 (β‐catenin) signaling pathways, or by upregulating long non‐coding RNA differentiation antagonizing non‐protein coding RNA (DANCR). 21 , 23 , 24 , 25 Therefore, TUFT1 is a potential biomarker and therapeutic target for various cancers; however, the effects of post‐translational modification of TUFT1 on the regulation of its biological functions is yet to be explored.
Herein, we demonstrated for the first time that SUMOylation of TUFT1 at K79 is a precondition for its oncogenic effect. Loss of SUMOylation significantly blocked the ability of TUFT1 to promote GC cell proliferation, migration, invasion, and activation of the AKT/mTOR signaling pathway. We further demonstrated that SUMOylation of TUFT1 is mediated by the E3 SUMO ligase tripartite motif‐containing protein 27 (TRIM27), and that the two proteins regulate the malignant behavior and AKT activation of GC cells in the same pathway. In addition, TUFT1 expression is associated with the clinical stage, lymph node metastasis rate, and overall survival (OS) of patients with GC only in the presence of SUMOylation. When TUFT1 was hypo‐SUMOylated, there was no correlation between TUFT1 expression and the clinical parameters mentioned above. Therefore, SUMOylation of TUFT1 is a promising therapeutic target or biomarker for GC progression.
2. MATERIAL AND METHODS
2.1. Cell culture and transfection
GES‐1 and MGC‐803 cells were cultured in RPMI‐1640 medium (BasalMedia) supplemented with 10% FBS (BasalMedia). AGS cells were cultured in Ham's F‐12 medium (BasalMedia) supplemented with 10% FBS. HEK293T and HGC27 cells were cultured in DMEM medium (BasalMedia) supplemented with 10% FBS. All cells were kept at 37°C in a 5% CO2 humidified incubator. JetPRIME® (Polyplus) was used to transfect cells according to the manufacturer's instructions.
2.2. Generation of TUFT1 knockout GC cell lines
All the knockout cell lines were generated using the CRISPR/Cas9 approach. 26 To generate TUFT1 knockout GC cell lines, the target oligo 5′‐CACCGGGAGTCCCATGATGGACATG‐3′ was ligated into a lentiCRISPR v2 vector and transfected into the cells. Puromycin‐resistant cells were selected and manually monocloned. The proliferated clones were subjected to immunoblot analysis and genome sequencing to verify successful knockout of TUFT1.
2.3. Western blotting
Samples were separated using SDS‐PAGE and transferred onto PVDF membranes (Millipore). The membranes were blocked with 4% nonfat milk and then probed with primary antibodies and HRP‐conjugated secondary antibodies (Proteintech, 1:5000). The primary antibodies used are listed in Table S1.
2.4. Immunoprecipitation assays
For immunoprecipitation, cells were lysed in lysis buffer (150 mM NaCl, 50 mM HEPES, 1 mM MgCl2, 1 mM EGTA, 0.5% Triton X‐100, and protease inhibitors; pH 7.4). The supernatant of the cell lysates was mixed with Protein G‐Sepharose beads and antibodies for 2 h at 4°C. The beads were further washed with lysis buffer and boiled at 100°C for 5 min in protein loading buffer.
2.5. Cell proliferation assay
For CCK‐8 assay, 10 μl of CCK‐8 reagent (MedChemExpress) was added to each well and incubated at 37°C for 1 h before measuring absorbance at a wavelength of 450 nm. For the EdU assay, the Cell‐Light™ EdU Apollo488 in vitro kit (RiboBio) was used, according to manufacturer's instructions.
2.6. Transwell assay
For cell migration ability detection, 1 × 105 GC cells were resuspended in 1 ml of serum‐free culture media and 150 μl was seeded onto the upper chamber. For cell invasion ability detection, 100 μl of 300 μg/ml ABW® Matrigengel (ABWbio) was added to the membrane of the upper chamber before cell seeding. A culture medium containing 20% FBS was added to the bottom chamber. After 24 h, cells passing through the membrane were stained with crystal violet and counted under the microscope.
2.7. In vivo growth experiments
All experimental procedures were approved by the Animal Ethics Committee of Central Hospital Affiliated to Shandong First Medical University (#JNCH2021‐20). BALB/c nude mice (male, 5 weeks old) were randomly divided into five groups (n = 6). HGC27 cells (2 × 106; WT, knockout, or stable cell lines) suspended in 100 μl of PBS were subcutaneously injected into right flanks of the mice. Tumor volumes were calculated every 3 days as length × width2 × 0.5. After 24 days, the mice were sacrificed, and the subskin grafts were dissected and weighed.
2.8. Immunohistochemistry staining
Immunohistochemistry was performed as described previously 27 on GC tissue microarrays purchased from Shanghai Outdo Biotech Company (#HstmA160CS01) with anti‐TRIM27 and anti‐TUFT1 antibodies. Two observers independently scored the tissues on a scale of 0–3 according to the percentage of immunoreactive cells and staining intensity.
2.9. Intracellular calcium concentration detection
Cells cultured in 96‐well plates were washed with PBS, and 50 μl of 2 μM Fluo‐4 AM (Beyotime) was added to each well and incubated at 37°C for 30 min. Cells were washed with PBS and the fluorescence intensity of Fluo‐4 was captured under the fluorescence microscope.
2.10. Immunofluorescence
Cells were fixed in 4% paraformaldehyde for 15 min at room temperature and then permeabilized with 0.15% Triton X‐100 for 15 min. Cells were blocked with 3% BSA for 30 min, incubated with primary antibodies (1:100) in 3% BSA at 4°C overnight, and stained with CoraLite488/594‐conjugated secondary antibodies (Proteintech) for 1 h at room temperature. The samples were observed at room temperature using a confocal microscope (Leica).
2.11. Quantification and statistical analysis
The intensity of the immunoblot bands and fluorescent signals was measured using ImageJ software (NIH). Statistical analyses were performed using GraphPad Prism 7 software. Statistical data were analyzed using paired or unpaired two‐tailed Student's t‐test and represented as mean ± SEM. P value <0.05 was considered statistically significant. Asterisks represent statistical significance: ***P < 0.001, **P < 0.01, *P < 0.05. The statistical details of each experiment, including statistical significance and n values, are provided in the figure legends.
3. RESULTS
3.1. TUFT1 is SUMOylated at K79
To identify SUMOylated proteins in GC cells, we performed liquid chromatography tandem mass spectrometry screening following SUMO‐modified protein enrichment of normal gastric mucosa and GC cells, and noticed that TUFT1 was one of the candidates with high confidence. TUFT1 expression was upregulated in GC cells relative to the normal gastric mucosa cells (Figure 1A), which is consistent with the published study. 28 We confirmed SUMOylation of TUFT1 by immunoprecipitation of total SUMOylated proteins from different GC cell line lysates with an anti‐SUMO1 antibody, and this modification was marginally detected in normal gastric mucosa cells (Figure 1A). To further identify the SUMOylation sites of TUFT1, lysine residues were mutated to the non‐SUMOylatable arginine (R) residue, and the SUMOylation level of exogenous TUFT1 was remarkably reduced when K79 was mutated (Figure 1B), indicating that K79 is the main SUMOylation site of TUFT1. Subsequent sequence analysis showed that the amino acid sequences around the SUMOylation site were well‐conserved, with high homology among species (Figure 1C).
FIGURE 1.
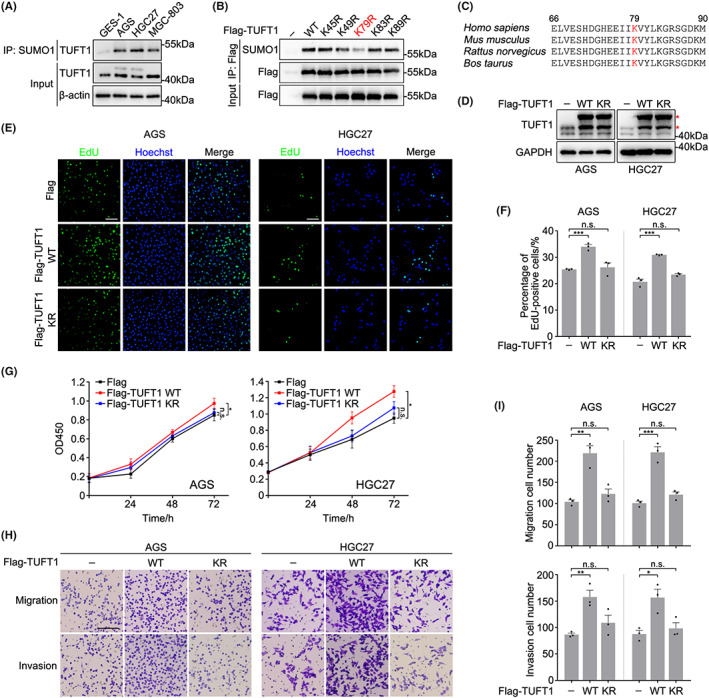
SUMOylation of TUFT1 at K79 is essential for promoting the proliferation, migration, and invasion of gastric cancer cells. (A) Lysates of gastric cancer cells (AGS, HGC27, MGC‐803) and gastric mucosa cells (GES‐1) were subjected to immunoprecipitation (IP) with anti‐SUMO1 antibody and immunoblotted with anti‐TUFT1 antibody. β‐actin was used as a loading control. (B) HEK293T cells overexpressing TUFT1‐WT or point mutants were subjected to immunoprecipitation with anti‐Flag antibody and immunoblotted with anti‐SUMO1 antibody. (C) Comparison of TUFT1 amino acid sequences around the SUMOylation site (shown in red) among species. (D) Lysates of AGS and HGC27 cells overexpressing TUFT1 were immunoblotted with anti‐TUFT1 antibody. Asterisks indicate exogenous TUFT1. GAPDH was used as a loading control. (E, F) EdU assays were performed in AGS and HGC27 cells overexpressing TUFT1‐WT or TUFT1‐KR to evaluate cell proliferation. DNA was stained with Hoechst (blue). Scale bar, 50 μm. Quantification is shown in (F) (n = 3, five randomly selected fields). (G) CCK‐8 assays were performed in AGS and HGC27 cells overexpressing TUFT1‐WT or TUFT1‐KR to evaluate cell proliferation (n = 3). (H, I) Transwell assays were performed in AGS and HGC27 cells overexpressing TUFT1‐WT or TUFT1‐KR to evaluate cell migration and invasion. Scale bar, 100 μm. Quantification is shown in (I) (n = 3, five randomly selected fields). Data are presented as means ± SEM. P values were determined by unpaired (F, I) or paired two‐tailed Student's t‐test (G). ***P < 0.001, **P < 0.01, *p < 0.05; n.s., not significant.
3.2. SUMOylation of TUFT1 is essential for promoting proliferation, migration, and invasion of GC cells
Since TUFT1 promotes the malignant behavior of various cancer cells, we investigated whether SUMOylation influences the biological functions of TUFT1. EdU and CCK‐8 assays indicated that overexpression of TUFT1‐WT significantly increased the proliferation of AGS and HGC27 cells (Figure 1D–G). However, overexpression of the non‐SUMOylatable mutant TUFT1 (TUFT1‐KR) did not promote GC cell proliferation (Figure 1D–G). Accordingly, the results of the Transwell assays showed that TUFT1‐WT overexpression remarkably enhanced the migration and invasion of GC cells, which was significantly impaired in the TUFT1‐KR transfection groups (Figure 1H,I). These results indicate that SUMOylation of TUFT1 at K79 is a prerequisite for its oncogenic effect in GC.
To further confirm the importance of TUFT1 SUMOylation, we generated TUFT1 knockout AGS and HGC27 cell lines by a CRISPR‐Cas9 approach (Figure S1). TUFT1 knockout remarkably suppressed the proliferation, migration, and invasion of GC cells, and this phenotype could only be rescued by transient re‐introduction of TUFT1‐WT but not TUFT1‐KR (Figure 2A–E). A similar phenotype was observed in AGS cells transfected with TUFT1 siRNA, and this phenotype could only be rescued by siRNA‐resistant TUFT1‐WT transfection (Figure S2A–E). Overall, these results suggest that SUMOylation is essential for TUFT1 to promote the malignant behavior of GC cells.
FIGURE 2.

The oncogenic effect of TUFT1 is impaired in the absence of SUMOylation. (A, B) EdU assays were performed in WT or TUFT1 knockout (KO) AGS and HGC27 cells rescued by exogenous TUFT1‐WT or TUFT1‐KR. DNA was stained with Hoechst (blue). Scale bar, 50 μm. Quantification is shown in (B) (n = 3, five randomly selected fields). (C) CCK‐8 assays were performed in WT or TUFT1 KO AGS and HGC27 cells rescued by exogenous TUFT1‐WT or TUFT1‐KR (n = 3). (D, E) Transwell assays were performed in WT or TUFT1 KO AGS and HGC27 cells rescued by exogenous TUFT1‐WT or TUFT1‐KR. Scale bar, 100 μm. Quantification is shown in (E) (n = 3, five randomly selected fields). Data are presented as means ± SEM. P values were determined by unpaired (B, E) or paired two‐tailed Student's t‐test (C). ***P < 0.001, **P < 0.01, *P < 0.05; n.s., not significant.
3.3. TUFT1 SUMOylation is necessary for AKT/mTOR signaling pathway activation
Previous studies have indicated that the AKT/mTOR signaling pathway is one of the key pathways activated by TUFT1 to promote tumor growth and metastasis. 18 , 20 , 22 Based on these results, we detected whether AKT activation is influenced by TUFT1 SUMOylation. Consistent with our hypothesis, the ability of TUFT1‐KR to activate AKT was notably suppressed compared with that of TUFT1‐WT (Figure 3A,B). Meanwhile, the phosphorylation state of 70‐kDa ribosomal S6 kinase (S6K1), a downstream effector of mTORC1, was not elevated as much (Figure 3A). Accordingly, TUFT1‐KR could not restore the blockage of AKT/mTOR pathway activation caused by TUFT1 knockout or knockdown (Figures 3C,D and S2F,G). These results indicate that SUMOylation of TUFT1 promotes the proliferation, migration, and invasion of GC cells by activating the AKT/mTOR signaling pathway.
FIGURE 3.

TUFT1 SUMOylation is necessary for AKT/mTOR activation. (A, B) Evaluating the effects of TUFT1 overexpression on AKT/mTOR signaling activation in AGS and HGC27 cells by immunoblotting. Phospho‐AKT (p‐AKT) indicates AKT activation and 70‐kDa ribosomal S6 kinase (S6K1) is a downstream effector of mTOR. Quantification is shown in (B) (n = 3). (C, D) Evaluating the effects of TUFT1 KO on AKT/mTOR signaling activation in AGS and HGC27 cells by immunoblotting (n = 3). Exogenous TUFT1‐WT or TUFT1‐KR were transiently transfected into the KO cells. Asterisks indicate exogenous TUFT1. Quantification is shown in (D) (n = 3). Data are presented as means ± SEM. P values were determined by unpaired two‐tailed Student's t‐test. ***P < 0.001, **P < 0.01, *P < 0.05; n.s., not significant.
3.4. TUFT1 SUMOylation is regulated by TRIM27
Since SUMOylation is of great importance in regulating the biological function of TUFT1, we aimed to determine the E3 SUMO ligase that mediates TUFT1 SUMOylation. By searching related protein interaction databases, we noticed that TRIM27, a zinc finger protein with SUMO transferase activity, 29 may interact with TUFT1. Interactions between TUFT1 and TRIM27 were detected by co‐immunoprecipitation assays (Figure 4A,B). Recombinant TUFT1 and TRIM27 expressed in and purified from bacteria bound to each other in vitro (Figure S3A), further suggesting their direct interactions. To investigate whether TUFT1 is a substrate for TRIM27, we transfected AGS cells with TRIM27 siRNA and found that SUMOylation of TUFT1 was remarkably decreased after TRIM27 knockdown (Figure 4C). We further co‐overexpressed TRIM27 and TUFT1 (WT or KR), and the immunoblot results showed that overexpression of TRIM27 significantly increased SUMOylation of TUFT1‐WT but not TUFT1‐KR (Figure 4D), indicating that the K79 residue of TUFT1 is SUMOylated by TRIM27. Since TRIM27 possesses both ubiquitin ligase activity and SUMO transferase activity, 29 , 30 we examined whether TRIM27 participates in the ubiquitin‐proteasome‐mediated degradation of TUFT1. The mutation at K79 did not block the ubiquitination of TUFT1 (Figure S3B). TRIM27 overexpression or knockdown did not alter the ubiquitination level of TUFT1 (Figure S3C,D), indicating that TRIM27 only participates in TUFT1 SUMOylation.
FIGURE 4.
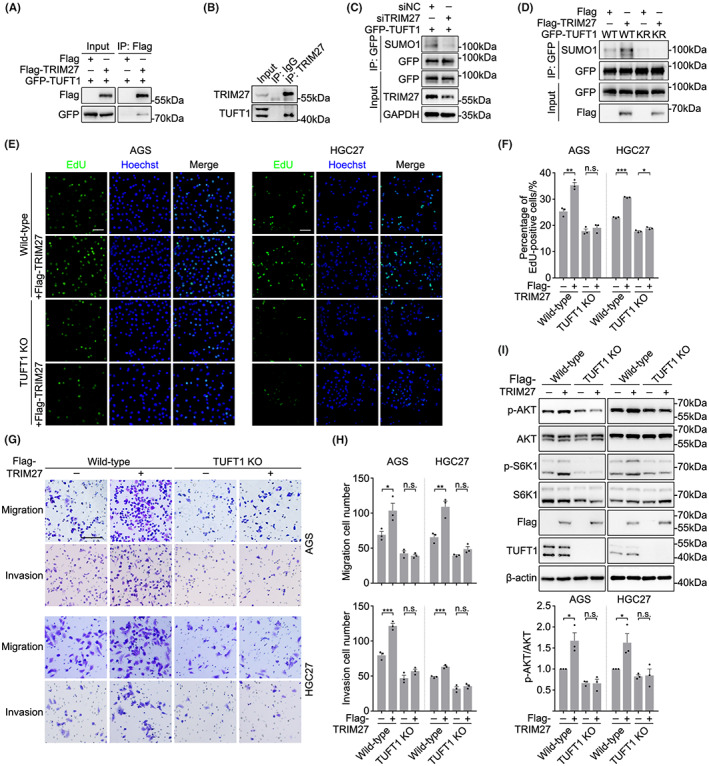
TUFT1 SUMOylation is regulated by TRIM27. (A) HEK293T cells overexpressing Flag‐TRIM27 and GFP‐TUFT1 were subjected to immunoprecipitation with an anti‐Flag antibody and immunoblotted with anti‐Flag and anti‐GFP antibodies. (B) TRIM27 was co‐immunoprecipitated with TUFT1 in AGS cells. The samples were immunoblotted with anti‐TRIM27 and anti‐TUFT1 antibodies. (C) Negative control (siNC) or TRIM27 knockdown (siTRIM27) AGS cells overexpressing GFP‐TUFT1 were subjected to immunoprecipitation with anti‐GFP antibody and immunoblotted with anti‐SUMO1 antibody. GAPDH was used as a loading control. (D) AGS cells overexpressing GFP‐TUFT1 (WT or KR) and Flag‐TRIM27 were subjected to immunoprecipitation with anti‐GFP antibody and immunoblotted with anti‐SUMO1 antibody. (E, F) EdU assays were performed in WT or TUFT1 KO AGS and HGC27 cells overexpressing TRIM27. DNA was stained with Hoechst (blue). Scale bar, 50 μm. Quantification is shown in (F) (n = 3, five randomly selected fields). (G, H) Transwell assays were performed in WT or TUFT1 KO AGS and HGC27 cells overexpressing TRIM27. Scale bar, 100 μm. Quantification is shown in (H) (n = 3, five randomly selected fields). (I) Evaluating the effects of TRIM27 overexpression on AKT/mTOR activation in WT and TUFT1 KO AGS and HGC27 cells by immunoblotting. Quantification is shown below (n = 3). Data are presented as means ± SEM. P values were determined by unpaired two‐tailed Student's t‐test. ***P < 0.001, **P < 0.01, *P < 0.05; n.s., not significant.
Since several studies have demonstrated that TRIM27 promotes esophageal and colorectal cancer progression by activating the AKT signaling pathway, 31 , 32 we next investigated whether TRIM27 promoted GC progression and AKT activation by SUMOylating TUFT1. Indeed, TRIM27 overexpression remarkably enhanced the proliferation, migration, and invasion of WT GC cells, and this phenotype was almost abolished in TUFT1 knockout or knockdown groups (Figures 4E–H and S3E–I). Accordingly, overexpression of TRIM27 activated the AKT signaling pathway in WT but not TUFT1‐deficient GC cells (Figures 4I and S3J,K), suggesting that TRIM27 and TUFT1 promote the malignant behavior and AKT activation in GC cells via the same pathway.
3.5. TUFT1 interact with TRIM27 through its N‐terminus
Having confirmed the physical and regulatory relationships between TUFT1 and TRIM27, it was essential to determine the binding region of TUFT1 to TRIM27. We constructed a series of truncation mutants based on the functional domains of TUFT1, and co‐immunoprecipitation assays revealed that the first 59 amino acids of TUFT1 were indispensable for its binding affinity to TRIM27 (Figures S4 and 5A). Consistent with this conclusion, the SUMOylation level of the TUFT1 ΔN mutant (60–390 amino acids; TUFT1‐ΔN) was remarkably decreased (Figure 5B), although the SUMOylation site (K79) was incorporated. Thus, TUFT1‐ΔN did not promote proliferation, migration, invasion, or AKT activation in GC cells (Figure 5C–H), similar to the phenotype observed in the TUFT1‐KR mutant groups.
FIGURE 5.

TUFT1 interacts with TRIM27 through its N‐terminus. (A) HEK293T cells co‐overexpressing GFP‐TRIM27 and Flag‐TUFT1 (WT or ΔN) were subjected to immunoprecipitation with anti‐Flag antibody and immunoblotted with anti‐GFP and anti‐Flag antibodies. (B) AGS cells overexpressing Flag‐TUFT1 (WT or ΔN) were subjected to immunoprecipitation with anti‐Flag antibody and immunoblotted with anti‐SUMO1 antibody. (C) CCK‐8 assays were performed in WT or TUFT1 KO AGS and HGC27 cells rescued by exogenous TUFT1‐WT or TUFT1‐ΔN (n = 3). (D, E) EdU assays were performed in WT or TUFT1 KO AGS and HGC27 cells rescued by exogenous TUFT1‐WT or TUFT1‐ΔN. DNA was stained with Hoechst (blue). Scale bar, 50 μm. Quantification is shown in (E) (n = 3, five randomly selected fields). (F, G) Transwell assays were performed in WT or TUFT1 KO AGS and HGC27 cells rescued by exogenous TUFT1‐WT or TUFT1‐ΔN. Scale bar, 100 μm. Quantification is shown in (G) (n = 3, five randomly selected fields). (H) Evaluating the effects of TUFT1 KO on AKT/mTOR activation in AGS and HGC27 cells by immunoblotting. Exogenous TUFT1‐WT or TUFT1‐ΔN were transiently transfected into the KO cells. Asterisks indicate exogenous TUFT1. Quantification is shown below (n = 3). Data are presented as means ± SEM. P values were determined by unpaired (E–G, H) or paired two‐tailed Student's t‐test (C). ***P < 0.001, **P < 0.01, *P < 0.05; n.s., not significant.
To confirm that TUFT1 SUMOylation deficiency impaired tumor growth in vivo, TUFT1 knockout HGC27 cells stably expressing GFP‐TUFT1 (WT, KR, or ΔN) and control cells were subcutaneously injected into nude mice to induce ectopic tumor formation. Compared with WT cells, the volume and weight of tumor tissues remarkably decreased in mice injected with TUFT1 knockout cells (Figure 6A–C). In addition, TUFT1‐KR and TUFT1‐ΔN insignificantly restored tumor growth compared to that with TUFT1‐WT (Figure 6A–C), further confirming that TRIM27‐mediated SUMOylation is crucial for the oncogenic effect of TUFT1.
FIGURE 6.

Upregulation of the TRIM27/TUFT1 axis is associated with poor outcomes in GC. (A–C) WT, TUFT1 knockout, and stable rescue HGC27 cells were injected subcutaneously into the right flanks of nude mice (n = 6). Representative photographs of excised tumors are shown in (A). The tumor volume was measured every 3 days (B). The tumor weight was measured at the end of experiment (C). (D) Representative images of immunohistochemistry staining of TRIM27 and TUFT1 in GC tissue microarrays. Arrows indicate the cytoplasmic staining. Scale bar, 100 μm. (E, F) Relationship between TUFT1 expression and clinical stages (E) or lymph node metastasis rate (F) in TRIM27 low and high expression subgroups, using the GC patient cohort in (D). (G) Survival analysis of TRIM27/TUFT1 expression in GC. Data are presented as means ± SEM. P values were determined by unpaired two‐tailed Student's t‐test (B, C–F) or two‐sided log‐rank test (G). ***P < 0.001, *P < 0.05; n.s., not significant.
3.6. Upregulation of the TRIM27/TUFT1 axis is associated with poor outcomes in GC
We further investigated the potential effects of TUFT1 SUMOylation on the prognosis of patients with GC. Since there was no available antibody specific for SUMOylated TUFT1, we assessed the correlation between TRIM27/TUFT1 expression in GC samples and patient outcomes instead. We performed immunohistochemical staining on commercial tissue microarrays of GC samples with TUFT1 and TRIM27 antibodies, and scored every sample on a scale of 0–3 based on the percentage of immunoreactive cells and the staining intensity. Scores of 0–1 were marked as low, while scores of 2–3 were marked as high (Figure 6D and Table S2). Patients with high TRIM27 and TUFT1 expression tended to develop more advanced clinical stages accompanied by higher rates of lymph node metastasis (Figure 6E,F). Meanwhile, there was no correlation between TUFT1 expression and GC malignancy in patients with low TRIM27 expression (Figure 6E,F), indicating that the expression of TRIM27 is a precondition for assessing the oncogenic function of TUFT1 in GC progression. We assessed the correlation between TRIM27/TUFT1 expression and patient survival. Cohorts of patients with GC collected from published studies 33 , 34 were divided into two groups according to the protein level of TRIM27. Consistent with the above results, TUFT1 expression was negatively correlated with OS only in the TRIM27 high expression group (Figure 6G). In summary, the TRIM27/TUFT1 axis well reflects outcomes and survival of patients with GC and might be a promising biomarker for GC diagnosis.
3.7. SUMOylation of TUFT1 affects the perinuclear accumulation of lysosomes and mTORC1
Having confirmed the importance of TUFT1 SUMOylation in GC progression, we investigated the molecular mechanism of TUFT1 SUMOylation‐induced AKT activation in GC. It is reported that TUFT1 activates the AKT signaling pathway by elevating the concentration of intracellular calcium in hepatocellular carcinoma cells. 20 However, the ability of the TUFT1‐KR mutant to elevate the concentration of intracellular calcium in AGS cells was not impaired compared with that of TUFT1‐WT (Figure 7A), indicating that TUFT1 SUMOylation deficiency blocks AKT activation downstream of the intracellular calcium signal. On the other hand, a recent study reported that TUFT1 directly activated mTOR, bypassing AKT phosphorylation. 19 Mechanically, TUFT1 forms a complex with Rab GTPase‐activating protein 1 (RABGAP1) and inhibits Rab GTPases. Inhibition of Rab GTPases promotes the perinuclear accumulation of mTORC1, which is indispensable for mTORC1 activation. 19 , 35 Co‐immunoprecipitation assays showed that the binding affinity of TUFT1‐KR or TUFT1‐ΔN to RABGAP1 compared to that with TUFT1‐WT was significantly decreased (Figure 7B). Accordingly, we observed diffuse localization of lysosomes and mTOR in TUFT1 knockout AGS cells, and only TUFT1‐WT restored perinuclear accumulation (Figure 7C). Together, TRIM27‐mediated SUMOylation is of great importance for TUFT1 to promote the malignant behavior of GC cells by activating the AKT/mTOR signaling pathway. On the one hand, TUFT1 SUMOylation is a necessary upstream signal for AKT activation. On the other hand, SUMOylated TUFT1 forms a complex with RABGAP1 and directly activates mTOR by promoting its perinuclear accumulation (Figure 7D).
FIGURE 7.

SUMOylation of TUFT1 affects the perinuclear accumulation of lysosomes and mTORC1. (A) The concentration of intracellular calcium was detected in AGS cells overexpressing TUFT1. Data are presented as means ± SEM. P values were determined by unpaired two‐tailed Student's t‐test. ***P < 0.001; n.s., not significant. (B) HEK293T cells overexpressing Flag‐TUFT1 (WT, KR, or ΔN) were subjected to immunoprecipitation with anti‐Flag antibody and immunoblotted with anti‐RABGAP1 and anti‐Flag antibodies. (C) Immunostaining of LAMP1 (red) and mTOR (green) in WT or TUFT1 KO AGS cells. DNA was stained with DAPI (blue). Scale bar, 5 μm. (D) Schematic of TRIM27/TUFT1 axis in promoting GC progression. TRIM27‐mediated TUFT1 SUMOylation is a necessary upstream signal for AKT activation. In addition, SUMOylated TUFT1 forms a complex with RABGAP1 and directly activates mTOR by promoting its perinuclear accumulation.
4. DISCUSSION
TUFT1 has been proven to play a critical role in tumor growth, metastasis, and drug resistance in various cancers and is associated with worse outcomes and poorer prognosis. 18 , 21 , 25 , 36 , 37 However, the effects of post‐translational modifications of TUFT1 on its oncogenic function remain unexplored. In this study, we demonstrated, for the first time, that TUFT1 is SUMOylated at K79. SUMOylation deficiency significantly impairs the ability of TUFT1 to promote the proliferation, migration, and invasion of GC cells by blocking the activation of the AKT/mTOR signaling pathway, indicating that SUMOylation determines the oncogenic effect of TUFT1.
Elevated TUFT1 or TRIM27 expression has been proven to be associated with unfavorable clinical features and poor prognosis in GC (Figure S5A,B). 19 , 38 Through analysis of clinical samples from tissue microarrays and public databases, we further uncovered that TUFT1 expression only correlates with tumor malignancy and OS of patients with high TRIM27 expression (Figures 6D–G and S5C), indicating that the joint detection of TRIM27 and TUFT1 expression reflects GC progression more accurately. In fact, we also found that, in addition to GC, the TRIM27/TUFT1 axis reflects patient OS in triple‐negative breast cancer, ovarian cancer, and lung adenocarcinoma (Figure S6), 39 , 40 , 41 , 42 further confirming that SUMOylation of TUFT1 might be a promising therapeutic target or biomarker for tumor progression.
The AKT/mTOR signaling pathway is one of the core pathways activated by TUFT1 to promote tumor progression. 18 , 20 , 43 Herein, we demonstrated that SUMOylated TUFT1 regulates this pathway through two distinct mechanisms. First, SUMOylated TUFT1 is an upstream signal for AKT activation. In future studies, we will investigate how SUMOylated TUFT1 activates AKT in GC cells. Second, SUMOylated TUFT1 directly activates mTOR by forming a complex with RABGAP1, which further inhibits Rab GTPases and promotes perinuclear accumulation of mTORC1. Loss of SUMOylation remarkably decreased the binding affinity of TUFT1 to RABGAP1 (Figure 7B). In addition to the AKT/mTOR signaling pathway, TUFT1 promotes tumor growth, metastasis, and drug resistance through other pathways such as HIF‐1/Snail and Rab5/Rac1/β‐catenin. 21 , 23 , 25 Moreover, TUFT1 upregulates the expression of LncRNA DANCR, promoting the malignancy and invasiveness of triple‐negative breast cancer. 24 Whether SUMOylation of TUFT1 influences these downstream effectors in GC needs further investigation.
TRIM27 plays an oncogenic role in various types of cancer, and its expression positively correlates with tumor malignancy and anticancer drug resistance. 32 , 44 , 45 , 46 In colorectal, esophageal, and ovarian serous carcinoma cells, TRIM27 promotes tumor growth and metastasis by activating AKT. 31 , 32 , 47 Herein, we found that TUFT1 is a new intermediate between the TRIM27 and AKT signaling pathways in GC. TRIM27 interacted with TUFT1 and promotes AKT/mTOR signaling activation by SUMOylating TUFT1 (Figure 4A–D,I). Consistent with this conclusion, the ability of TRIM27 to promote cell proliferation, migration, and invasion was almost abolished in TUFT1‐deficient GC cells (Figures 4E–H and S3E–I). On the other hand, it was found that TRIM27 regulates GC cell proliferation and 5‐fluorouracil resistance through the Hippo‐baculoviral IAP repeat‐containing protein 5 (BIRC5) pathway 38 ; whether there is any crosstalk between the two pathways requires further studies.
Based on previous studies and our results, SUMOylation is closely related to tumor growth, survival, metastasis, and drug resistance as it regulates the function of core signaling transduction proteins or transcription factors in GC. We believe that a comprehensive quantitative SUMOylome mass spectrometry analysis of GC clinical samples or cell lines is of great necessity to investigate the mechanism of tumorigenesis and progression from a new perspective, contributing to the exploration of early diagnosis and targeted therapies of GC.
AUTHOR CONTRIBUTIONS
T.W. performed most of the molecular biology experiments, cell biology experiments, clinical sample analysis, and data analysis. L.M., Y.G., and M.Z. conducted some molecular biology and cell biology experiments. S.F. and H.W. performed some clinical sample analysis. Y.Z. and Y.W. are the senior authors who conceived and designed the project. T.W., Y.Z., and Y.W. wrote the manuscript.
FUNDING INFORMATION
This work was supported by the Natural Science Foundation of Shandong Province (ZR2021QH005, ZR2021QH304) and the China Postdoctoral Science Foundation (2021M701406, 2021M701407).
DISCLOSURE
The authors declare no potential conflicts of interest.
ETHICAL APPROVAL
Approval of the research protocol by an Institutional Reviewer Board: The research protocol was approved by the Ethics Committee of Central Hospital Affiliated to Shandong First Medical University (#SZR2021‐017‐01) and the Ethics Committee of Shanghai Outdo Biotech Company (#YBM‐05‐02).
Informed consent: All samples were obtained with written informed consent from participants.
Registry and the registration
No of the study/trial: None.
Animal studies
All experimental procedures were approved by Animal Ethics Committee of Central Hospital Affiliated to Shandong First Medical University (#JNCH2021‐20).
Supporting information
Appendix S1.
ACKNOWLEDGMENT
The authors thank all members of the Wang laboratory for helpful comments and suggestions.
Wang T, Min L, Gao Y, et al. SUMOylation of TUFT1 is essential for gastric cancer progression through AKT/mTOR signaling pathway activation. Cancer Sci. 2023;114:533‐545. doi: 10.1111/cas.15618
Contributor Information
Yunshan Wang, Email: zxsyswys@126.com.
Yan Zheng, Email: zhengdr@hotmail.com.
REFERENCES
- 1. Sung H, Ferlay J, Siegel RL, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71:209‐249. doi: 10.3322/caac.21660 [DOI] [PubMed] [Google Scholar]
- 2. Qin G, Tu X, Li H, et al. Long noncoding RNA p53‐stabilizing and activating RNA promotes p53 signaling by inhibiting heterogeneous nuclear ribonucleoprotein K deSUMOylation and suppresses hepatocellular carcinoma. Hepatology. 2020;71:112‐129. doi: 10.1002/hep.30793 [DOI] [PubMed] [Google Scholar]
- 3. Dou H, Huang C, van Nguyen T, Lu LS, Yeh ET. SUMOylation and de‐SUMOylation in response to DNA damage. FEBS Lett. 2011;585:2891‐2896. doi: 10.1016/j.febslet.2011.04.002 [DOI] [PubMed] [Google Scholar]
- 4. Schimmel J, Eifler K, Sigurðsson JO, et al. Uncovering SUMOylation dynamics during cell‐cycle progression reveals FoxM1 as a key mitotic SUMO target protein. Mol Cell. 2014;53:1053‐1066. doi: 10.1016/j.molcel.2014.02.001 [DOI] [PubMed] [Google Scholar]
- 5. Rabellino A, Andreani C, Scaglioni PP. The role of PIAS SUMO E3‐ligases in cancer. Cancer Res. 2017;77:1542‐1547. doi: 10.1158/0008-5472.CAN-16-2958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Seeler JS, Dejean A. SUMO and the robustness of cancer. Nat Rev Cancer. 2017;17:184‐197. doi: 10.1038/nrc.2016.143 [DOI] [PubMed] [Google Scholar]
- 7. Wei J, Costa C, Shen J, et al. Differential effect of MMSET mRNA levels on survival to first‐line FOLFOX and second‐line docetaxel in gastric cancer. Br J Cancer. 2014;110:2662‐2668. doi: 10.1038/bjc.2014.231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Shao DF, Wang XH, Li ZY, et al. High‐level SAE2 promotes malignant phenotype and predicts outcome in gastric cancer. Am J Cancer Res. 2015;5:140‐154. [PMC free article] [PubMed] [Google Scholar]
- 9. Wei J, Costa C, Ding Y, et al. mRNA expression of BRCA1, PIAS1, and PIAS4 and survival after second‐line docetaxel in advanced gastric cancer. J Natl Cancer Inst. 2011;103:1552‐1556. doi: 10.1093/jnci/djr326 [DOI] [PubMed] [Google Scholar]
- 10. Ren YH, Liu KJ, Wang M, et al. De‐SUMOylation of FOXC2 by SENP3 promotes the epithelial‐mesenchymal transition in gastric cancer cells. Oncotarget. 2014;5:7093‐7104. doi: 10.18632/oncotarget.2197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Wang M, Sang J, Ren Y, et al. SENP3 regulates the global protein turnover and the Sp1 level via antagonizing SUMO2/3‐targeted ubiquitination and degradation. Protein Cell. 2016;7:63‐77. doi: 10.1007/s13238-015-0216-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hu XY, Liu Z, Zhang KL, et al. SUMO‐specific protease 2‐mediated deSUMOylation is required for NDRG2 stabilization in gastric cancer cells. Cancer Biomark. 2017;21:195‐201. doi: 10.3233/cbm-170651 [DOI] [PubMed] [Google Scholar]
- 13. Liu J, Zhang Q, Ruan B, et al. MORC2 regulates C/EBPalpha‐mediated cell differentiation via sumoylation. Cell Death Differ. 2019;26:1905‐1917. doi: 10.1038/s41418-018-0259-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Liu D, Li Z, Yang Z, Ma J, Mai S. Ginkgoic acid impedes gastric cancer cell proliferation, migration and EMT through inhibiting the SUMOylation of IGF‐1R. Chem Biol Interact. 2021;337:109394. doi: 10.1016/j.cbi.2021.109394 [DOI] [PubMed] [Google Scholar]
- 15. Chen C, Sun X, Xie W, et al. Opposing biological functions of the cytoplasm and nucleus DAXX modified by SUMO‐2/3 in gastric cancer. Cell Death Dis. 2020;11:514. doi: 10.1038/s41419-020-2718-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Deutsch D, Leiser Y, Shay B, et al. The human tuftelin gene and the expression of tuftelin in mineralizing and nonmineralizing tissues. Connect Tissue Res. 2002;43:425‐434. doi: 10.1080/03008200290001186 [DOI] [PubMed] [Google Scholar]
- 17. Leiser Y, Blumenfeld A, Haze A, et al. Localization, quantification, and characterization of tuftelin in soft tissues. Anat Rec. 2007;290:449‐454. doi: 10.1002/ar.20512 [DOI] [PubMed] [Google Scholar]
- 18. Liu H, Zhu J, Mao Z, Zhang G, Hu X, Chen F. Tuft1 promotes thyroid carcinoma cell invasion and proliferation and suppresses apoptosis through the Akt‐mTOR/GSK3beta signaling pathway. Am J Transl Res. 2018;10:4376‐4384. [PMC free article] [PubMed] [Google Scholar]
- 19. Kawasaki N, Isogaya K, Dan S, et al. TUFT1 interacts with RABGAP1 and regulates mTORC1 signaling. Cell Discov. 2018;4:1. doi: 10.1038/s41421-017-0001-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Dou C, Zhou Z, Xu Q, et al. Hypoxia‐induced TUFT1 promotes the growth and metastasis of hepatocellular carcinoma by activating the Ca(2+)/PI3K/AKT pathway. Oncogene. 2019;38:1239‐1255. doi: 10.1038/s41388-018-0505-8 [DOI] [PubMed] [Google Scholar]
- 21. Zhou B, Zhan H, Tin L, et al. TUFT1 regulates metastasis of pancreatic cancer through HIF1‐Snail pathway induced epithelial‐mesenchymal transition. Cancer Lett. 2016;382:11‐20. doi: 10.1016/j.canlet.2016.08.017 [DOI] [PubMed] [Google Scholar]
- 22. Mu J, Sun X, Zhao Z, Sun H, Sun P. BRD9 inhibition promotes PUMA‐dependent apoptosis and augments the effect of imatinib in gastrointestinal stromal tumors. Cell Death Dis. 2021;12:962. doi: 10.1038/s41419-021-04186-6 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 23. Liu W, Han J, Shi S, Dai Y, He J. TUFT1 promotes metastasis and chemoresistance in triple negative breast cancer through the TUFT1/Rab5/Rac1 pathway. Cancer Cell Int. 2019;19:242. doi: 10.1186/s12935-019-0961-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Wu G, Zhou H, Li D, et al. LncRNA DANCR upregulation induced by TUFT1 promotes malignant progression in triple negative breast cancer via miR‐874‐3p‐SOX2 axis. Exp Cell Res. 2020;396:112331. doi: 10.1016/j.yexcr.2020.112331 [DOI] [PubMed] [Google Scholar]
- 25. Liu W, Chen G, Sun L, et al. TUFT1 promotes triple negative breast cancer metastasis, stemness, and chemoresistance by up‐regulating the Rac1/beta‐catenin pathway. Front Oncol. 2019;9:617. doi: 10.3389/fonc.2019.00617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ran FA, Hsu PD, Wright J, Agarwala V, Scott DA, Zhang F. Genome engineering using the CRISPR‐Cas9 system. Nat Protoc. 2013;8:2281‐2308. doi: 10.1038/nprot.2013.143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Li P, Jia YF, Ma XL, et al. DEC2 suppresses tumor proliferation and metastasis by regulating ERK/NF‐kappaB pathway in gastric cancer. Am J Cancer Res. 2016;6:1741‐1757. [PMC free article] [PubMed] [Google Scholar]
- 28. Meng P, Xia R, Shao Z. MicroRNA‐145 regulates the proliferation of the human gastric cancer cells by targeting tuftelin 1 (TUFT1). Acta Biochim Pol. 2022;69:357‐362. doi: 10.18388/abp.2020_5798 [DOI] [PubMed] [Google Scholar]
- 29. Chu Y, Yang X. SUMO E3 ligase activity of TRIM proteins. Oncogene. 2011;30:1108‐1116. doi: 10.1038/onc.2010.462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Jia X, Zhao C, Zhao W. Emerging roles of MHC class I region‐encoded E3 ubiquitin ligases in innate immunity. Front Immunol. 2021;12:687102. doi: 10.3389/fimmu.2021.687102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ma L, Yao N, Chen P, Zhuang Z. TRIM27 promotes the development of esophagus cancer via regulating PTEN/AKT signaling pathway. Cancer Cell Int. 2019;19:283. doi: 10.1186/s12935-019-0998-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Zhang Y, Feng Y, Ji D, et al. TRIM27 functions as an oncogene by activating epithelial‐mesenchymal transition and p‐AKT in colorectal cancer. Int J Oncol. 2018;53:620‐632. doi: 10.3892/ijo.2018.4408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Ge S, Xia X, Ding C, et al. A proteomic landscape of diffuse‐type gastric cancer. Nat Commun. 2018;9:1012. doi: 10.1038/s41467-018-03121-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Ni X, Tan Z, Ding C, et al. A region‐resolved mucosa proteome of the human stomach. Nat Commun. 2019;10:39. doi: 10.1038/s41467-018-07960-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Sancak Y, Bar‐Peled L, Zoncu R, Markhard AL, Nada S, Sabatini DM. Ragulator‐Rag complex targets mTORC1 to the lysosomal surface and is necessary for its activation by amino acids. Cell. 2010;141:290‐303. doi: 10.1016/j.cell.2010.02.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Xiao Z, Liu Y, Zhao J, et al. Long noncoding RNA LINC01123 promotes the proliferation and invasion of hepatocellular carcinoma cells by modulating the miR‐34a‐5p/TUFT1 axis. Int J Biol Sci. 2020;16:2296‐2305. doi: 10.7150/ijbs.45457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Luo X, Wei J, Yang FL, et al. Exosomal lncRNA HNF1A‐AS1 affects cisplatin resistance in cervical cancer cells through regulating microRNA‐34b/TUFT1 axis. Cancer Cell Int. 2019;19:323. doi: 10.1186/s12935-019-1042-4 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 38. Yao Y, Liu Z, Cao Y, et al. Downregulation of TRIM27 suppresses gastric cancer cell proliferation via inhibition of the Hippo‐BIRC5 pathway. Pathol Res Pract. 2020;216:153048. doi: 10.1016/j.prp.2020.153048 [DOI] [PubMed] [Google Scholar]
- 39. Szász AM, Lánczky A, Nagy A, et al. Cross‐validation of survival associated biomarkers in gastric cancer using transcriptomic data of 1,065 patients. Oncotarget. 2016;7:49322‐49333. doi: 10.18632/oncotarget.10337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Gyorffy B, Surowiak P, Budczies J, Lánczky A. Online survival analysis software to assess the prognostic value of biomarkers using transcriptomic data in non‐small‐cell lung cancer. PLoS One. 2013;8:e82241. doi: 10.1371/journal.pone.0082241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Gyorffy B, Lanczky A, Szallasi Z. Implementing an online tool for genome‐wide validation of survival‐associated biomarkers in ovarian‐cancer using microarray data from 1287 patients. Endocr Relat Cancer. 2012;19:197‐208. doi: 10.1530/ERC-11-0329 [DOI] [PubMed] [Google Scholar]
- 42. Gyorffy B. Survival analysis across the entire transcriptome identifies biomarkers with the highest prognostic power in breast cancer. Comput Struct Biotechnol J. 2021;19:4101‐4109. doi: 10.1016/j.csbj.2021.07.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Dou C, Sun L, Wang L, et al. Bromodomain‐containing protein 9 promotes the growth and metastasis of human hepatocellular carcinoma by activating the TUFT1/AKT pathway. Cell Death Dis. 2020;11:730. doi: 10.1038/s41419-020-02943-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Xing L, Tang X, Wu K, Huang X, Yi Y, Huan J. TRIM27 functions as a novel oncogene in non‐triple‐negative breast cancer by blocking cellular senescence through p21 ubiquitination. Mol Ther Nucleic Acids. 2020;22:910‐923. doi: 10.1016/j.omtn.2020.10.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Zhang HX, Xu ZS, Lin H, et al. TRIM27 mediates STAT3 activation at retromer‐positive structures to promote colitis and colitis‐associated carcinogenesis. Nat Commun. 2018;9:3441. doi: 10.1038/s41467-018-05796-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Liu S, Tian Y, Zheng Y, et al. TRIM27 acts as an oncogene and regulates cell proliferation and metastasis in non‐small cell lung cancer through SIX3‐beta‐catenin signaling. Aging. 2020;12:25564‐25580. doi: 10.18632/aging.104163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Ma Y, Wei Z, Bast RC, et al. Downregulation of TRIM27 expression inhibits the proliferation of ovarian cancer cells in vitro and in vivo. Lab Invest. 2016;96:37‐48. doi: 10.1038/labinvest.2015.132 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix S1.


