Abstract
MMP‐21 is a newly identified member of the matrix metalloproteinase family and has been reported to regulate both embryonic development and tumor progression. However, the roles of MMP‐21 in hemofiltrate C–C chemokine (HCC) remain largely unclear. In this study, we used western blot, qPCR and immunohistochemistry (IHC) to determine the upregulation of MMP‐21 in HCC tissues, and showed that the increase in MMP‐21 was associated with vascular invasion and poor prognosis. Although changing levels of MMP‐21 in HCC cell lines had no significant effect on cell migration or invasion abilities in in vitro transwell tests, both IHC analysis and in vivo mouse models proved that upregulated MMP‐21 promoted metastasis. Functional enrichments of MMP‐21 using The Cancer Genome Atlas (TCGA) data suggested that MMP‐21 might regulate metastasis via macrophages. Further experiments proved that MMP‐21 enhanced macrophage recruitment by increasing CCL‐14 levels and promoted M2‐type polarization of macrophage by elevating the expression of CSF‐1 and FGF‐1. Taken together, this study revealed that MMP‐21 controlled the tumor microenvironment remodeling and functional regulation of macrophages to regulate HCC metastasis.
Keywords: CCL‐14, CSF‐1, FGF‐1, HCC, macrophage, metastasis, MMP‐21
MMP‐21 enhanced macrophage recruitment by upregulating CCL‐14 levels and promoted macrophage M2‐type polarization by increasing the expression of CSF‐1 and FGF‐1, and ultimately facilitated metastasis in HCC.
Abbreviations
- CCL‐14
chemokine (C‐C motif) ligand‐14
- CSF‐1
colony‐stimulating factor‐1
- ECM
extracellular matrix
- FGF‐1
fibroblast growth factor‐1
- HCC
hepatocellular carcinoma
- IHC
immunohistochemistry
- MMP
matrix metalloproteinase
- MMP‐21
matrix metalloproteinase‐21
- qPCR
quantitative PCR
- TAM
tumor‐associated macrophage
1. INTRODUCTION
HCC has a high incidence among malignancies in the world, causing ~750,000 deaths each year. 1 HCC remains a poorly expected tumor due to its high metastasis and recurrence rate. 2 Some molecules have been discovered that assist the prediction of HCC metastasis and prognosis, but the accuracy is limited. 3 , 4 Therefore, more novel biomarkers for HCC are required in diagnosis, prognostic evaluation, and therapeutic approaches.
Matrix metalloproteinases (MMPs) are a family of zinc‐dependent proteases that are engaged in ECM degradation and remolding. 5 MMPs perform considerable roles in tumor progression, 6 including HCC. 7 , 8 , 9 MMP‐21 is a newly identified member of the MMP family. As in other MMPs, it is synthesized as an latent proenzyme and activated as an proteolytic product in the extracellular secretion pathway. 10 MMP‐21 has been illustrated to play essential roles in embryogenesis, inflammation, and stromal remodeling. 10 , 11 , 12 Additional studies on human tumors have illustrated the clinical significance of the MMP‐21 gene. In esophageal squamous cell cancer, upregulated MMP‐21 was associated with tumor aggression and poor prognosis. 13 Elevated MMP‐21 levels were also detected in gastric cancer, 14 colorectal cancer 15 and oral squamous cell carcinoma with lymphatic metastasis. 16 A recent study also showed that MMP‐21 might implement the effect of ADAM17 on HCC invasion and metastasis, 17 indicating that it functions in HCC prognosis. However, the certain role of MMP21 in HCC remains to be explored.
An immune/inflammation response signature is closely related to the HCC progression, 18 while the clearance of senescent hepatocytes in a neoplastic‐prone microenvironment could delay the emergence of HCC. 19 Macrophages are one of the major cell types responsible for the clearance, and therefore in the inhibition of tumor progression. But TAMs often exert opposite effects. In response to distinct inflammatory environmental signals, macrophages are activated to differentiate into type M1 macrophages (proinflammatory and anti‐tumoral) or type M2 macrophages (anti‐inflammatory and protumoral), respectively. 20 TAMs belong to the M2‐type macrophage group, and represent the major compound of infiltrating immune cells in cancer‐related inflammation. 21 TAMs are derived from circulating monocyte precursors and are recruited into the tumor foci by tumor‐associated signals. The recruited macrophages adapt to the local environment and then carry out specific effects. 22 In HCC, TAMs in the peritumor region predominantly participates in fibroblast activation, tissue remodeling, angiogenesis, tumor growth as well as metastasis. 23 , 24 In addition, some cytokines including IL‐3, CSF‐1, and CCL‐2 drive the infiltration of TAMs toward tumor tissues. 25 In addition to regulating macrophage recruitment, 26 some microenvironment factors such as CSF‐1, TGF‐β1, IL‐4, IL‐6, IL‐10, and FGF‐1 also play roles in the polarization of TAMs. 27 , 28 As one of the major parts in tumor microenvironment, MMPs has also been reported to regulate the functions of macrophages. 29 , 30 However, the effects of MMP‐21 on TAMs remain to be studied.
Here, we investigated the expression of MMP‐21 in HCC and explored the potential role of MMP‐21 in HCC progression by in vivo and in vitro experiments. In addition, our study also elaborated how MMP‐21 remodels the tumor microenvironment to promote tumor metastasis.
2. MATERIALS AND METHODS
2.1. Hepatocellular carcinoma patient samples
In total, 104 patients with primary HCC who underwent surgical resection between 2010 and 2012 or between 2014 and 2015, at Department of General Surgery, the First Affiliated Hospital of Wenzhou Medical University (Zhejiang, China) were collected. Clinical tumor stages were classified according to the TNM classification system of International Union against Cancer. Employment of clinicopathological data and human pathological tissues were approved by the research medical ethics committee of Wenzhou Medical University (Zhejiang, China, approval no. 219) and all methods were carried out in accordance with the approved guidelines. The written consent conforming to the ethical guidelines of the Helsinki Declaration were obtained from all patients before the start of the experiment.
2.2. Cell culture
All liver cancer cell lines, human THP1 cells, and mouse RAW264.7 cells were obtained from the Cell Bank of Type Culture Collection of Chinese Academy of Sciences (Shanghai, China). Cells including mouse RAW264.7 cells were cultured in DMEM (Sigma‐Aldrich, St. Louis, MO, USA) supplemented with 10% FBS (Gibco, Grand Island, NY, USA) at 37°C in a humidified incubator containing 5% CO2. Human THP1 cells were maintained in RPMI 1640 medium replenished with 10% FBS. THP1 cells were differentiated using 200 nM phorbol‐12‐myristate‐13‐acetate (PMA; Sigma‐Aldrich) for 3 days.
2.3. Plasmid construction
The plasmid pCMV‐SP‐FLAG‐MMP21 was purchased from Axybio (Hunan, China). The targeting sequence for MMP‐21 siRNA was: 5′‐GUGACAUUCACUUUGACGA‐3′, and the target sequence for CCL‐14 shRNA was 5′‐GCTCCAAGCCCGGAATTGTCT‐3′. Transfections were carried out with Lipofectamine 3000 (Life Technologies, CA, USA) following the manufacturer's instructions. Stable cell lines were generated with G418 (200 μg/ml) in the medium.
2.4. Animal studies
Here, 4‐ to 6‐week‐old male BALB/c nude mice were purchased from the Shanghai Laboratory Animal Center of Chinese Academy Sciences (Shanghai, China) and housed in a separate pathogen‐free room. Animal care and experiments were approved by the research medical ethics committee of Wenzhou Medical University and were performed in strict accordance with the approved guidelines. All the experimental mice were randomized controlled (n = 6 in each group).
For the lung metastasis model, transfected Huh7‐Luc cells were resuspended in pre‐cooling PBS (1 × 106 cells/mouse in 100 μl PBS) and were injected into mice through the tail vein. For the subcutaneous xenograft model, transfected Huh7‐Luc cells were resuspended in pre‐cooling PBS (5 × 106 cells/mouse in 100 μl PBS) and were subcutaneously inoculated into the left hind flank regions of 4‐week‐old male nude mice. The mice were sacrificed after 4 weeks, and tumor tissues were harvested for further usage. For the orthotopic transplant model, the harvested tumor was cut into the appropriate size (1 mm in diameter) in PBS. Then the fragments were transplanted into the left lobes of the liver of nude mice. All the mice were monitored for bioluminescence every week with IVIS200 (Xenogen, Caliper, CA) after intraperitoneal administration of 200 μl luciferin (at 15 mg/ml; Promega, WI, USA). The luciferase signal intensity was calculated by region of interest (ROI) analysis.
2.5. Western blot
Samples from liver tissues homogenate and cell lysates were separated by SDS‐PAGE, and transferred onto polyvinylidene fluoride membranes (Millipore, USA). The membranes were incubated with primary antibodies, and then incubated with HRP‐conjugated secondary antibody (Santa Cruz Biotechnology, Dallas, TX, USA). Anti‐MMP21 (55289‐1‐AP), anti‐Flag‐tag (20543‐1‐AP), anti‐CCL14 (14216‐1‐AP), and anti‐GAPDH (60004‐1‐IG) antibodies were purchased from ProteinTech. Anti‐ZEB2 (#97885), anti‐Twist (#69366) and the Epithelial–Mesenchymal Transition (EMT) antibody sampler kit (#9782T) including anti‐vimentin, anti‐Snail, anti‐β‐catenin, anti‐E‐cadherin, anti‐N‐cadherin antibodies were purchased from Cell Signaling Technology (Beverly, MA, USA). The images were visualized using an enhanced chemiluminescence detection kit (Tiangen Biotech, Beijing, China) and the ImageQuant™ LAS‐4000 system (Amersham Biosciences, GE, USA).
2.6. RNA extraction, reverse transcription PCR, and quantitative real‐time PCR
Total RNA was extracted from frozen tissues and cell lines using TRIzol reagent (Invitrogen, Carlsbad, CA, USA). Then the RNA was processed for reverse transcription to cDNA using the PrimeScript RT reagent kit (TaKaRa, Tokyo, Japan). Quantitative real‐time PCR was carried out using SYBR Premix Ex Taq (TaKaRa, Tokyo, Japan) according to the manufacturer's instructions. GAPDH was used as an internal control. The primers used here were as follows: MMP‐21 forward: TGAGGGATCATTTGATACTGCGT and reverse: AATCTCACCATCACCTCTCCAT, ZEB2 forward: GCGATGGTCATGCAGTCAG and reverse: CAGGTGGCAGGTCATTTTCTT, Vimentin forward: CTGCTTCAAGACTCGGTGGAC and reverse: ATCTCCTCCTCGTACAGGTCG, Snail forward: TCGGAAGCCTAACTACAGCGA and reverse: AGATGAGCATTGGCAGCGAG, β‐catenin forward: AAAGCGGCTGTTAGTCACTGG and reverse: CGAGTCATTGCATACTGTCCAT, Twist forward: GTCCGCAGTCTTACGAGGAG and reverse: GCTTGAGGGTCTGAATCTTGCT, E‐cadherin forward: ATTTTTCCCTCGACACCCGAT and reverse: TCCCAGGCGTAGACCAAGA, N‐cadherin forward: AGCCAACCTTAACTGAGGAGT and reverse: GGCAAGTTGATTGGAGGGATG, CCL‐14 forward: GACTGAATCCTCCTCACGGG and reverse: TGGCATCTTCTCTTTATGTCTCTG, CSF‐1 forward: TGGCGAGCAGGAGTATCAC and reverse: AGGTCTCCATCTGACTGTCAAT, FGF‐1 forward: GGGGTTGCTTAGAGCTGTGT and reverse: GGAGCCAAGAATGTCAGCCT. GAPDH forward: GAGTCAACGGATTTGGTCGT and reverse: TTGATTTTGGAGGGATCTCG.
2.7. Immunohistochemistry
Immunohistochemistry (IHC) assay was performed using a Dako REAL EnVision Detection System (Dako, Denmark) in according to the protocol recommended. Anti‐MMP21, anti‐CD68 (CST, #76437), anti‐F4/80(CST, #70076), anti‐CCL14, anti‐CSF1(Abcam, ab233387), anti‐FGF1 (Abcam, ab179455), anti‐CD206 (CST, #24595), anti‐iNOS (ProteinTech, 18985‐1‐AP) and anti‐Arg1 (ProteinTech, 16001‐1‐AP) primary antibodies were used to quantify the protein expression level. The others used in IHC were the same antibodies in used in western blotting. The sections were developed using 3,3′‐diaminobenzidine (DAB), followed by counterstaining with hematoxylin. IHC staining intensity was calculated as 0 for negative, 1 for weak, 2 for moderate, and 3 for strong. The score for the IHC staining area was set as 0 for <5%; 1 for 5–25%; 2 for 26–50%; 3 for 51–75%; and 4 for 76%–100%. The final score was obtained by multiplying two values, and the results ranged from 0 to 12.
2.8. Transwell assays
Transwell assay was carried out in a 12‐well plate (Corning, New York, NY, USA) with 8‐μm transwell filters (Millipore, Billerica, MA, USA). For invasion assays, the bottom of transwell chamber was coated with BD Matrigel Basement Membrane Matrix (BD Biosciences, San Diego, CA, USA). Cells in serum‐free culture medium were added into the upper chamber, and the lower chamber was filled with culture medium with 10% FBS. For macrophage chemotaxis assay, HCC tumor cells were placed into the lower chamber as a chemoattractant. Transwell migration assay was determined 24 h later, invasion of cells and macrophage infiltration was determined 48 h later. Cells on the upper surface of the chamber were wiped off with a cotton swab, and cells on the lower side of the chamber were stained with 0.1% crystal violet. The number of infiltrating cells was counted in five randomly selected fields of view under the microscope.
2.9. The Cancer Genome Atlas and Gene Expression Omnibus data sets
Counts for RNA‐Seq and clinicopathological data were downloaded from The Cancer Genome Atlas (TCGA) database (http://www.cbioportal.org) and Gene Expression Omnibus (GEO) (https://www.ncbi.nlm.nih.gov/geo/). The patient cohorts used were liver cancer (Non‐tumor: 105, Tumor: 319) The difference between groups was assessed using Student's two‐tailed t‐test. The correlations between CCL14, CSF1, FGF1, and MMP‐21 expression were calculated using Pearson's method.
2.10. Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analysis
The genes associated with MMP‐21 in HCC were downloaded from TCGA database. We screened out the genes that were strongly related to MMP21 and performed enrichment analysis on omicsbean (http://www.omicsbean.cn) to obtain the KEGG signaling pathway and protein interaction network.
2.11. Statistical analysis
All the analyses were performed with SPSS 22.0 (IBM Corporation, Armonk, NY, USA), ImageJ 1.8.0 (National Institutes of Health, USA) and GraphPad Prism 5.0 (GraphPad software, La Jolla, CA, USA). Results were presented as mean ± standard deviation and each sample was repeated at least three times. The difference between groups was assessed using Student's two‐tailed t‐test. Receiver operating characteristic (ROC) curve analysis was used to determine the cut‐off values for MMP‐21 in IHC. The correlation between clinical factors and MMP‐21 expression was identified using Pearson's chi‐squared test. Kaplan–Meier curves were used to evaluate the overall survival probability and the difference was examined by Log‐rank test. Univariate and multivariate analysis models were established using Cox proportional hazards regression. Statistical significance was set to two‐sided with a p‐value of less than 0.05.
3. RESULTS
3.1. The upregulation of MMP‐21 in HCC is related to poor prognosis
To explore the role of MMP‐21 in HCC, first we determined the expression pattern of MMP‐21 in public HCC data sets. The mRNA expression level of MMP‐21 was significantly upregulated in HCC tissues on data from TCGA database (p = 0.042) and GEO data sets (p = 0.02 for GSE107170, 31 p < 0.001 for GSE78737 32 ) (Figure 1A). Afterward, we examined the mRNA level in 30 pairs of HCC samples, and observed MMP‐21 expression increased in 70% (21/30) cases (Figure 1B). Western blot analysis showed that the MMP‐21 protein level was also markedly elevated in HCC tissues (p = 0.008) (Figure 1C, D). Similar results were found in IHC analysis on 74 pairs of HCC samples at different stages (p < 0.001) (Figure 1E, F).
FIGURE 1.
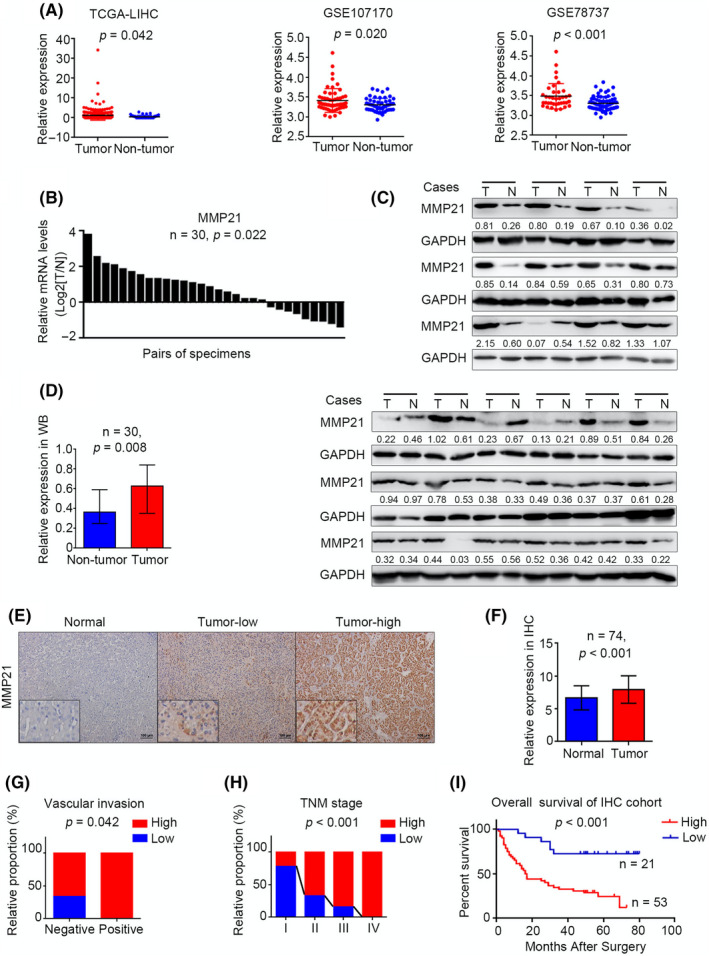
MMP‐21 is upregulated in HCC and related to poor prognosis. (A) Relative mRNA expression of MMP‐21 in HCC tissues and adjacent non‐tumor tissues in public data sets. (B, C) MMP‐21 expression in 30 paired HCC tissues and adjacent non‐tumor tissues. T, HCC tissue, N, adjacent non‐tumor tissue. (D) Statistical data of western blot. (E) Representative IHC staining images in non‐tumor tissue and HCC tissues. (F) Statistical data of the IHC staining score. (G, H) Correlation between MMP‐21 and vascular invasion, TNM stage in IHC cohort. (I) Overall survival of HCC patients with different MMP‐21 expression levels
Next we analyzed the correlation between clinicopathologic features and MMP‐21 expression in HCC patients. Receiver operating characteristic curve analysis divided the patients in the IHC cohort into two groups according to the MMP‐21 expression level, and the representative staining images of MMP‐21low and MMP‐21high are shown, respectively (Figures S1 and 1E). Chi‐squared test showed that the upregulation of MMP‐21 was positively correlated with tumor size (p = 0.046), vascular invasion (p = 0.042), and TNM stage (p < 0.001), and the expression of MMP‐21 was profoundly consistent with disease progression (Figure 1G, H; Table S1). In addition, Kaplan–Meier analysis of the IHC cohort revealed that patients with increased MMP‐21 were predicted poorer overall survival (p < 0.001, Figure 1I). Furthermore, univariate and multivariate Cox analyses were used to evaluate the prognostic significance of MMP‐21 for overall survival (HR, 4.239; 95%CI, 1.785–10.066; p = 0.001 for univariate analysis and HR, 3.094; 95%CI, 1.275–7.507; p = 0.012 for multivariate analysis) (Table S2). We also established a more accurate predictive model for overall survival of HCC patients. Results suggested that the combined TNM stage with MMP‐21 expression system could provide a more powerful approach to cancer evaluation (Figure S2). Taken together, the results demonstrated that the upregulation of MMP‐21 was closely associated with worsening HCC progression.
3.2. MMP‐21 promotes metastasis in vivo whereas confers no significant effects in vitro
As MMPs are known to decompose and remodel ECM, they are generally recognized as EMT‐related protease. 33 Therefore, we focused on this crucial function in our following experiments. First, we investigated MMP‐21 expression levels in liver cancer cells. As shown in Figure S3, the expression of MMP‐21 in liver cancer cell lines was different. Among these cell lines, we chose BEL‐7402 cells with low basal expression to perform MMP‐21 overexpression and SMMC‐7721 cells with high basal expression to process MMP‐21 knockdown. Although qPCR analyses showed that MMP‐21 regulated the mRNA levels of some EMT‐related hallmarks, such as vimentin, Snail, E‐cadherin, and N‐cadherin, very few changes of these hallmarks were observed at the protein level (Figure S4). No significant effects of MMP‐21 on cell migration and invasion could be observed in vitro (Figure S5). In contrast, the positive correlation between MMP‐21 and E‐cadherin as well as N‐cadherin was determined using IHC assay on human HCC tissues (R = 0.301, p < 0.007 for E‐cadherin and R = 0.346, p < 0.002 for N‐cadherin) (Figure 2A, B). Additionally, the metastasis‐promoting effects were proved in vivo. In liver orthotopic transplants models, IHC results of orthotopic xenografts exhibited similar results with those of human HCC tissues (R = 0.598, p = 0.039 for E‐cadherin and R = 0.584, p = 0.046 for N‐cadherin) (Figure 2C–E). MMP‐21 overexpression led to more lung metastasis foci (50% in the Huh7‐Luc‐MMP21 group and 0% in the Huh7‐Luc‐control group) in the lung metastasis model (Figure 2F, G). Furthermore, upregulated MMP‐21 profoundly reduced the survival of tumor‐bearing mice (Figure 2H, I). The contradictory results of in vivo and in vitro experiments indicated that MMP‐21 may play its role by cooperating with other tumor‐associated cells in the microenvironment, and indirectly promote tumor progression.
FIGURE 2.
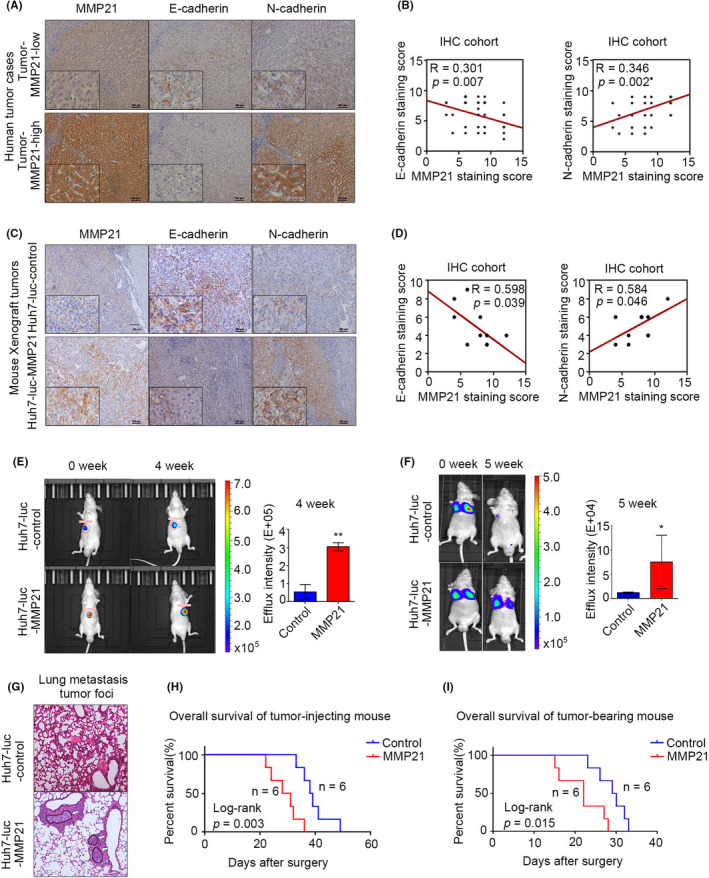
MMP‐21 promotes EMT in vivo, whereas it confers no significant function in vitro. (A, B) Representative IHC staining images of MMP‐21 and E‐cadherin, N‐cadherin in human HCC. (C, D) Representative IHC staining images of MMP‐21 and E‐cadherin, N‐cadherin in liver orthotopic xenografts from mouse. (E) In vivo effect of MMP‐21 in orthotopic liver transplanted mice. (F) In vivo effect of MMP‐21 in mouse lung metastasis models. (G) Representative images of hematoxylin‐eosin staining. The white arrows point to the tumor region. (H) Kaplan–Meier analysis for overall survival of mouse lung metastasis models. (I) Kaplan–Meier analysis for overall survival of orthotopic liver transplanted mice. *p < 0.05, **p < 0.01
3.3. MMP‐21 regulates macrophage recruitment
In HCC, non‐parenchymal cells, especially macrophages, are widely distributed in the tumor microenvironment, and have shown a link with uncontrolled malignant metastasis. 21 , 34 To investigate whether macrophages were engaged in the function of MMP‐21, we examined the number of macrophages in human HCC tissues, and found that upregulation of MMP‐21 significantly raised the number of infiltrated macrophages (R = 0.328, p = 0.004) (Figure 3A, B). Similar IHC results on mouse orthotopic xenografts revealed that overexpressing MMP‐21 in tumor cells increased the number of F4/80‐positive macrophages (p < 0.05) (Figure 3C, D). These data suggested that MMP‐21 was associated with the recruitment of macrophages. To validate the effect of MMP‐21 on macrophage recruitment, we placed liver cancer cells with different MMP‐21 levels in the lower chamber and carried out transwell tests in vitro on mouse RAW264.7 cells and PMA‐differentiated human THP1 cells. The results showed that when we used 7402 cells with overexpressed MMP‐21, the number of macrophages passing through the membrane was significantly increased and MMP‐21 knockdown in 7721 cells conferred the opposite effects (Figure 3E, F). As macrophages and tumor cells were localized in different chambers, we hypothesized that MMP‐21 may promote macrophage recruitment by regulating the secretion of tumor cells. Therefore, we additionally collected conditioned medium from these cancer cells with varying levels of MMP‐21, and then added it in the lower chamber without liver cancer cells. The medium exerted similar functions with their secretory hosts, indicating that MMP‐21 regulates the secretion of tumor cells to facilitate macrophage recruitment (Figure 3G, H).
FIGURE 3.
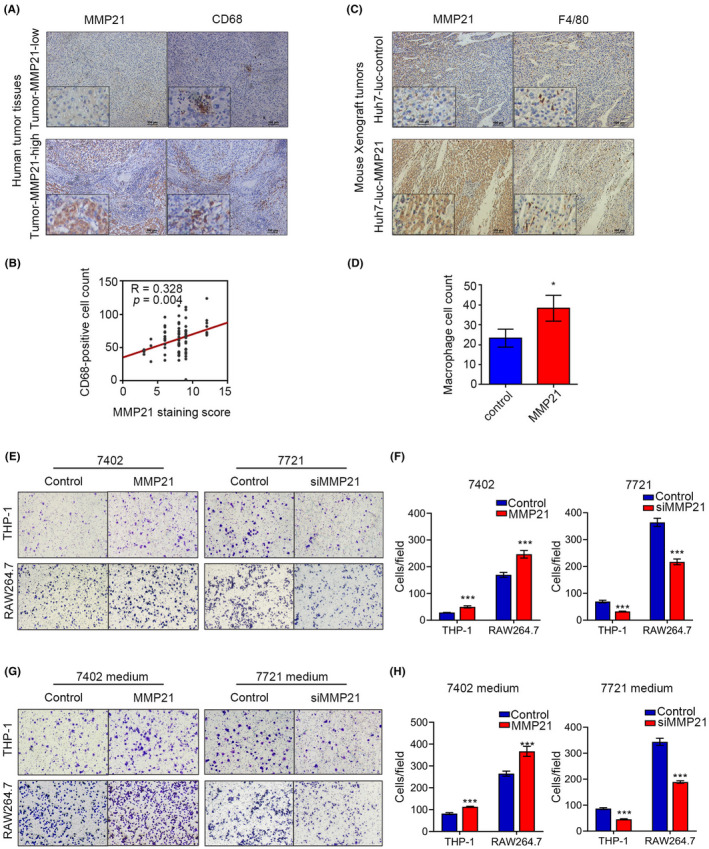
MMP‐21 promotes the recruitment of macrophages. (A) Representative IHC staining images of MMP‐21 and CD68 in human HCC tissues. (B) The correlation between MMP‐21 and CD68‐positive macrophages. (C) Representative IHC staining images of MMP‐21 and F4/80 in liver orthotopic xenografts from tumor‐bearing mouse. (D) Cell count of macrophages. (E, F) Transwell assays to determine macrophage recruitment. (G, H) Transwell assays to determine macrophage recruitment using culture medium from HCC cells. *p < 0.05, **p < 0.01. [Correction added on 21 September 2022, after first online publication: in Figure 3, parts E, F, G and H have been replaced with the correct images].
3.4. CCL‐14 is identified as the key secreted molecule from liver cancer cells in MMP21‐mediated macrophage recruitment
To find out key factors in MMP21‐associated secretion, we established the functional enrichment analysis of KEGG pathways about all the genes strongly associated with MMP‐21 (Pearson |R|≥0.3 and Spearman |R|≥0.1) and mapped some pathways related to the communication between cells, the top pathways are shown in Figure 4A and Table S3. We found that CCL‐14 was involved in most of these pathways, especially chemotaxis. To verify the relationship between CCL‐14 and MMP‐21, we first analyzed the correlation between the expression of them in TCGA data sets and observed a positive result (Figure 4B). Furthermore, western blot, qPCR and IHC (R = 0.305, p < 0.001) on human tumor tissues also revealed similar results (Figure 4C–E). IHC results showed that the degree of macrophage infiltration in HCC tissues was related to the expression level of CCL‐14 (Figure 4E). In vitro data also indicated that MMP‐21 positively regulated CCL‐14 expression in 7402 cells and 7721 cells at both protein and mRNA levels (Figure 4F, G). In addition, knocking down CCL‐14 in the 7402 cell line could reduce the number of migrated macrophages (Figure 4H, I), whereas overexpressing MMP‐21 in the CCL14‐knock‐down 7402 stable cell line could not increase the number significantly (Figure 4J, K), indicating that knocking down CCL‐14 could attenuate the effects of MMP‐21 on macrophage recruitment. The efficiency of CCL‐14 knockdown was detected using western blot and qPCR (Figure 4H). Overall, MMP‐21 promoted macrophage recruitment via upregulating CCL‐14 levels.
FIGURE 4.
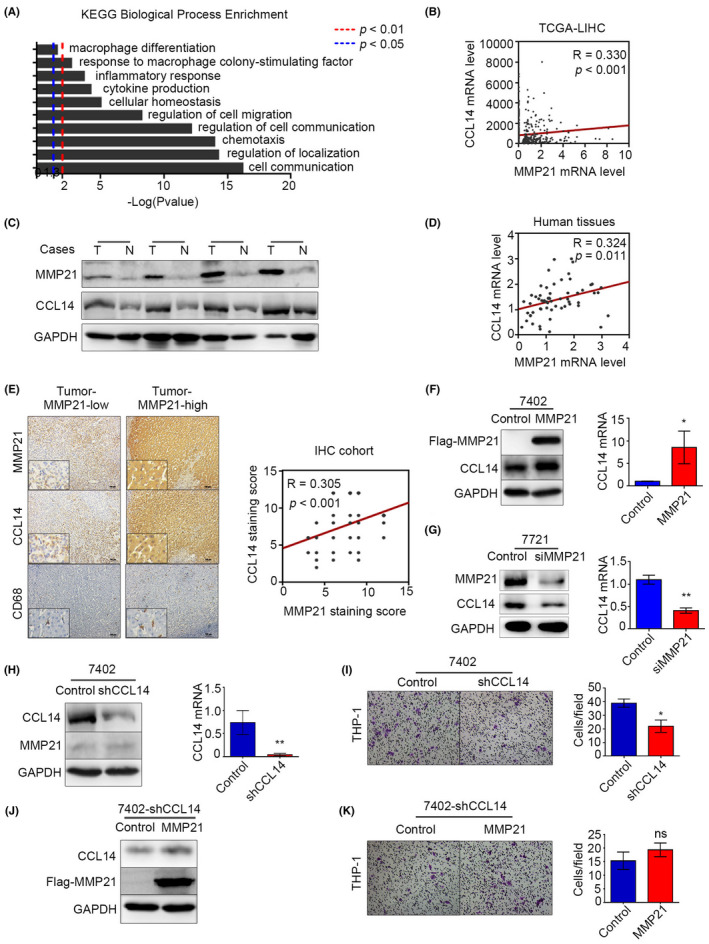
CCL‐14 is the key secreted molecule from HCC cells in MMP21‐mediated macrophage recruitment. (A) Functional KEGG pathway enrichment from a TCGA‐LIHC data set. (B) The correlation between MMP‐21 and CCL‐14 from a TCGA‐LIHC data set. (C) Protein levels of MMP‐21 and CCL‐14 in HCC tissues. (D) Correlation between MMP‐21 and CCL‐14 for mRNA levels in HCC tissues. (E) IHC analysis of MMP‐21, CCL‐14, and CD68 in HCC tissues. (F) CCL‐14 expression in 7402 cells with increasing MMP‐21. (G) CCL‐14 expression in 7721 cells with decreasing MMP‐21. (H) Knockdown efficiency of CCL‐14 in 7402 cells. (I) Transwell test to determine the macrophage recruitment using 7402 cells with CCL‐14 knockdown. (J) Overexpression efficiency of MMP21 in a CCL14‐knockdown 7402 stable cell line. (K) Macrophage recruitment assessment using the CCL14‐knockdown 7402 stable cell line with MMP‐21 overexpression. *p < 0.05, **p < 0.01; ns, no significance. [Correction added on 21 September 2022, after first online publication: in Figure 4, part G has been replaced with the correct image].
3.5. MMP‐21 regulates the transcription of some cytokines to promote TAM polarization
Aberrant expression of some genes such as CSF‐1 and FGF‐1 has been reported to promote malignant differentiation of TAMs. 28 , 35 In MMP‐21‐associated KEGG functional enrichment results, we found that CSF‐1 and FGF‐1 were involved in macrophage differentiation, so we speculated that MMP‐21 might affect the polarization of macrophages via these cytokines (Figure 5A). Analysis of TCGA data sets displayed the potential connections between MMP‐21 and these two molecules (R = 0.343, p < 0.001 for CSF‐1 and R = 0.614, p < 0.001 for FGF‐1) (Figure 5B). In vitro data showed that the overexpression of MMP‐21 triggered the expression of CSF‐1 and FGF‐1, and their transcriptional levels were reduced by MMP‐21 knockdown (Figure 5C). As CSF‐1 and FGF‐1 are secretory factors, we used conditioned medium from liver cancer cell lines with different MMP‐21 levels to incubate PMA‐induced THP1 cells, and detect the M1‐ and M2‐related markers on the treated cells. We found that M2‐related markers IL‐1RA and Arg‐1 were increased in MMP21‐overexpressing groups and reduced in MMP‐21‐knockdown groups, whereas M1‐related markers IL‐6 and TNF‐α had no significant changes (Figure 5D). Conversely, we performed IHC in human HCC tissues. The results showed that the staining level of Arg‐1 increased in liver cancer tissues with high expression of MMP‐21, while the staining level of iNOS was attenuated (Figure S6). Similar results were observed in IHC analysis in that MMP‐21 had a positive relationship with CSF‐1 and FGF‐1 at the protein levels (R = 0.279, p = 0.002 for CSF‐1 and R = 0.303, p < 0.001 for FGF‐1) (Figure 5E, F). Furthermore, CD206‐positive TAMs, which represented the M2‐type macrophages, were more widely distributed in HCC tissues with higher MMP‐21 expression (R = 0.293, p = 0.011) (Figure 5E, F). These results indicated that MMP‐21 promoted the polarization of TAMs by regulating the transcription of CSF‐1 and FGF‐1 in HCC tumor cells.
FIGURE 5.
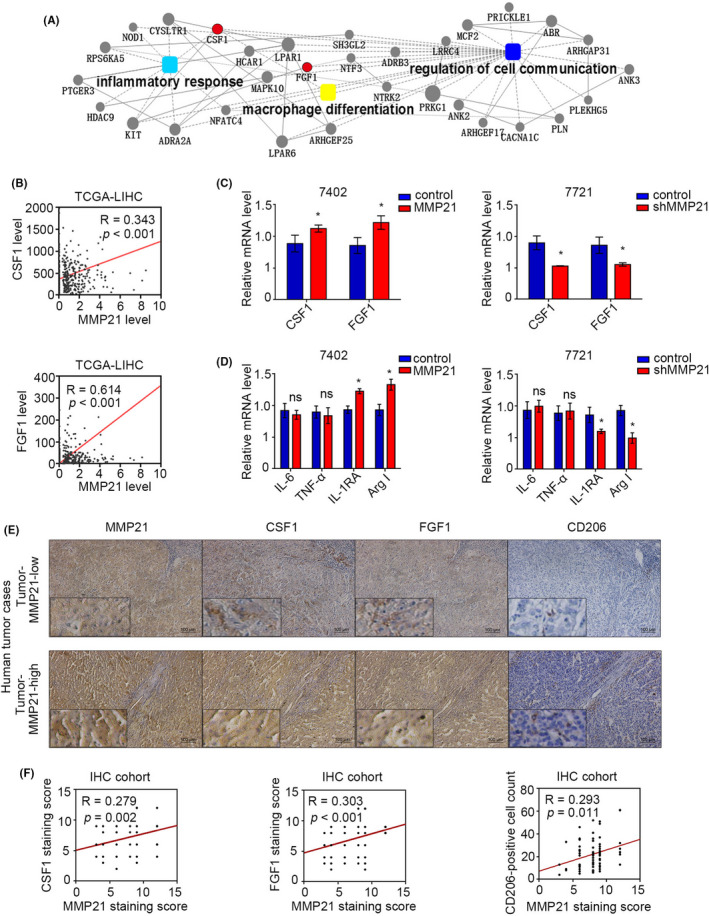
MMP‐21 regulates the expression of FGF‐1 and CSF‐1 to promote macrophage polarization toward M2‐type. (A) Genes strongly correlated with MMP‐21 in the three pathways (R ≥ 0.3). (B) Correlations of CSF‐1, FGF‐1, and MMP‐21 from the TCGA‐LIHC data set. (C) Relative mRNA expression of CSF‐1 and FGF‐1 in HCC cell lines with overexpression or knockdown of MMP‐21. (D) Relative mRNA expression of M1 and M2 markers in THP1 cells incubated with culture medium from HCC cell lines. (E) IHC analysis of CSF‐1, FGF‐1 and CD206 in human HCC tissues with different MMP‐21 levels. (F) Statistical data for IHC staining score and the cell count of CD206‐positive macrophages in human HCC tissues. *p < 0.05; ns, no significance
4. DISCUSSION
In this study, our data revealed that the upregulation of MMP‐21 was closely associated with tumor vascular invasion and poor prognosis. Mechanistically, we demonstrated that MMP‐21 increased the expression of CCL‐14, CSF‐1, and FGF‐1 to regulate the recruitment and polarization of TAMs. Ultimately, the recruited macrophages might cooperate with MMP‐21 to facilitate tumor metastasis (Figure 6).
FIGURE 6.
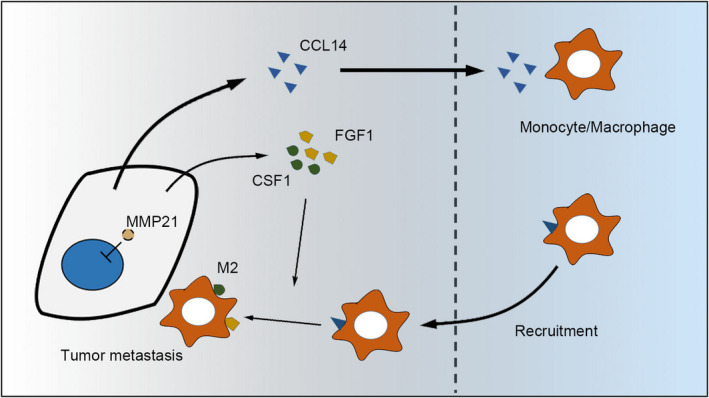
Schematic describing how MMP‐21 functions in HCC
HCC is obviously an example among the inflammation‐related cancers. 36 , 37 In our study, due to the contradictory effects of MMP‐21 between in vitro and in vivo experiments, we hypothesized that some components of the tumor microenvironment might be involved in the MMP21‐mediated promoting effects on tumor metastasis. The most familiar one among these potential effective ingredients is TAMs. 21 TAMs in tumors are mostly polarized from PMBCs, for which monocyte recruitment is important. 38 Monocyte recruitment relies on a series of cytokines, which present a wide range of interactions. 39 , 40 Among these molecules, CCL‐14 is mostly homologous to the MIP‐1α, which is identified to work in macrophage recruitment, and experiments on cloned chemokine receptors showed that CCL‐14 specifically activated CCR‐1, the same receptor as MIP‐1α activating. 41 CCL‐14 is not a chemoattractant for leukocytes, 42 but induces chemotaxis of purified human monocytes, THP1 cells, and CCR1‐transfected cells. 41 In our study, MMP‐21 elevated the expression of CCL‐14 to recruit macrophages and this phenomenon was verified in in vitro both on human THP1 cells and mouse RAW264.7 cells.
Tumor‐derived CSF‐1 has been found to be correlated with faster tumor growth, more frequent metastasis and poor prognosis in cancers including HCC. 34 , 43 , 44 , 45 A previous study demonstrated that HCC patients with high CSF‐1 expression levels in peritumoral liver tissue had a shorter survival. 46 In addition, CSF‐1 also works in macrophage polarization. It could stimulate macrophages to exhibit M2‐type activation. 47 Similar to CSF‐1, FGF‐1 is a potent growth factor for hepatocytes that is involved in inflammation, tissue repair, angiogenesis, and carcinogenesis. 48 , 49 Existing studies implied that FGF‐1 induced IL‐4 expression to cause a higher arginase I (Arg1)‐positive M2 macrophage response. 28 Recombinant FGF1‐treated mice exerted anti‐inflammatory properties in the liver. 50 Consistent with these findings, our data describe that MMP‐21 could induce macrophages to M2‐type transformation by increasing CSF‐1 and FGF‐1 expression. Although CSF‐1 is also a chemoattractant of macrophages, CCL‐14 is more markedly increased at the expression level. Therefore, we emphasize that CCL‐14 played a leading role in MMP21‐mediated macrophage recruitment, CSF‐1 and FGF‐1 are involved in the M2 polarization of macrophage.
Targeting TAMs is increasingly becoming a promising therapy in cancer treatment. A growing number of studies have suggested that suppressing TAM recruitment or M2‐type programming via targeting cytokines may be effective in inhibiting tumor progression. 51 , 52 Therefore, the identification of essential inflammation‐related cytokines leading to chronic liver damage in primary HCC and even secondary distant metastasis will portray a new blueprint of predictive targets to analyze and treat patients with liver disease. 53 Clinical studies have flooded us with a considerable amount of inflammatory mediators implicated in HCC. 37 , 54 , 55 Here, we provide a potential approach to treat HCC via inhibiting MMP‐21, which might dampen the recruitment and activation of TAMs at the same time.
As to why MMP21 is abnormally upregulated in HCC, previous studies have reported that there are multiple transcription factor binding motifs in the promoter of the MMP21 gene, such as Tcf‐4, Pax and RBP‐Jκ (Notch), which indicates that the expression of MMP21 may be affected by a variety of transcription factors, and under the influence of different tumor microenvironment, the level of MMP21 will change accordingly. 10 Conversely, MMP21 is activated as an proteolytic product in the extracellular secretion pathway, and the prodomain of MMP21 contains a peptide sequence similar to that of TNF‐α. A recent paper has reported that ADAM17 may cleave the precursor of MMP21 and ultimately increase the invasive ability of HCC, which is known as a TNF‐α converting enzyme. 56 This suggests that some enzymes identified for processing TNF‐α to sTNF (secreted TNF‐α) may activate MMP21 and positively regulate the secretion of MMP21. The specific mechanism needs further research.
CONFLICT OF INTEREST
The authors declare that they have no competing interests.
Supporting information
Fig S1
Fig S2
Fig S3
Fig S4
Fig S5
Fig S6
Table S1
Table S2
Table S3
ACKNOWLEDGEMENTS
We are indebted to the School of Laboratory Medicine and Life Sciences, Wenzhou Medical University and the First Affiliated Hospital of Wenzhou Medical University, for their support and understanding throughout the years of our working. And we thank all staff members and patients for their trust and involvement in the study.
Zhou J, Liu L, Hu X, et al. Matrix metalloproteinase‐21 promotes metastasis via increasing the recruitment and M2 polarization of macrophages in HCC. Cancer Sci. 2023;114:423–435. doi: 10.1111/cas.15368
Funding information
This research did not receive any specific grant from funding agencies in the public, commercial, or not‐for‐profit sectors
Contributor Information
Wei Li, Email: liweiwzmc@163.com.
Yunfeng Shan, Email: shanyf@yahoo.com.
Jing Jin, Email: jinjing@wmu.edu.cn.
REFERENCES
- 1. Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394‐424. [DOI] [PubMed] [Google Scholar]
- 2. Govaere O, Roskams T. Pathogenesis and prognosis of hepatocellular carcinoma at the cellular and molecular levels. Clin Liver Dis. 2015;19(2):261‐276. [DOI] [PubMed] [Google Scholar]
- 3. Malaguarnera G, Giordano M, Paladina I, Berretta M, Cappellani A, Malaguarnera M. Serum markers of hepatocellular carcinoma. Dig Dis Sci. 2010;55(10):2744‐2755. [DOI] [PubMed] [Google Scholar]
- 4. Chen T, Dai X, Dai J, et al. AFP promotes HCC progression by suppressing the HuR‐mediated Fas/FADD apoptotic pathway. Cell Death Dis. 2020;11(10):822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Visse R, Nagase H. Matrix metalloproteinases and tissue inhibitors of metalloproteinases: structure, function, and biochemistry. Circ Res. 2003;92(8):827‐839. [DOI] [PubMed] [Google Scholar]
- 6. Stetlerstevenson WG. The role of matrix metalloproteinases in tumor invasion, metastasis, and angiogenesis. Surg Oncol Clin N Am. 2001;10(2):383‐392. [PubMed] [Google Scholar]
- 7. Li T, Zhu Y, Han L, Ren W, Liu H, Qin C. VEGFR‐1 activation‐induced MMP‐9‐dependent invasion in hepatocellular carcinoma. Future Oncol. 2015;11(23):3143‐3157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ng TP, Qi X, Kong KL, et al. Overexpression of matrix metalloproteinase‐12 (MMP‐12) correlates with poor prognosis of hepatocellular carcinoma. Eur J Cancer. 2011;47(15):2299‐2305. [DOI] [PubMed] [Google Scholar]
- 9. Wu J, Hao ZW, Zhao YX, et al. Full‐length soluble CD147 promotes MMP‐2 expression and is a potential serological marker in detection of hepatocellular carcinoma. J Transl Med. 2014;12(1):190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Marchenko GN, Marchenko ND, Strongin AY. The structure and regulation of the human and mouse matrix metalloproteinase‐21 gene and protein. Biochem J. 2003;372(Pt 2):503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ahokas K, Lohi J, Lohi H, et al. Matrix metalloproteinase‐21, the human orthologue for XMMP, is expressed during fetal development and in cancer. Gene. 2002;301(1):31‐41. [DOI] [PubMed] [Google Scholar]
- 12. Ahokas K, Lohi J, Illman SA, et al. Matrix metalloproteinase‐21 is expressed epithelially during development and in cancer and is up‐regulated by transforming growth factor‐beta1 in keratinocytes. Laboratory investigation. J Tech Method Pathol. 2003;83(12):1887. [DOI] [PubMed] [Google Scholar]
- 13. Zhao Z, Yan L, Li S, Sun H, Zhou Y, Li X. Increased MMP‐21 expression in esophageal squamous cell carcinoma is associated with progression and prognosis. Med Oncol. 2014;31(8):1‐7. [DOI] [PubMed] [Google Scholar]
- 14. Wu T, Li Y, Lu J, et al. Increased MMP‐21 expression is associated with poor overall survival of patients with gastric cancer. Med Oncol. 2013;30(1):323. [DOI] [PubMed] [Google Scholar]
- 15. Huang Y, Li W, Chu D, et al. Overexpression of matrix metalloproteinase‐21 is associated with poor overall survival of patients with colorectal cancer. J Gastrointest Surg. 2011;15(7):1188‐1194. [DOI] [PubMed] [Google Scholar]
- 16. Pu Y, Wang L, Wu H, Feng Z, Wang Y, Guo C. High MMP‐21 expression in metastatic lymph nodes predicts unfavorable overall survival for oral squamous cell carcinoma patients with lymphatic metastasis. Oncol Rep. 2014;31(6):2644‐2650. [DOI] [PubMed] [Google Scholar]
- 17. Xiang Y, Liu L, Wang Y, Li B, Peng J, Feng D. ADAM17 promotes the invasion of hepatocellular carcinoma via upregulation MMP21. Cancer Cell Int. 2020;20:516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Budhu A, Forgues M, Ye QH, et al. Prediction of venous metastases, recurrence, and prognosis in hepatocellular carcinoma based on a unique immune response signature of the liver microenvironment. Cancer Cell. 2006;10(2):99‐111. [DOI] [PubMed] [Google Scholar]
- 19. Marongiu F, Serra MP, Sini M, Angius F, Laconi E. Clearance of senescent hepatocytes in a neoplastic‐prone microenvironment delays the emergence of hepatocellular carcinoma. Aging. 2014;6(1):26‐34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Mantovani A, Sica A. Macrophages, innate immunity and cancer: balance, tolerance, and diversity. Curr Opin Immunol. 2010;22(2):231‐237. [DOI] [PubMed] [Google Scholar]
- 21. Solinas G, Germano G, Mantovani A, Allavena P. Tumor‐associated macrophages (TAM) as major players of the cancer‐related inflammation. J Leukoc Biol. 2009;86(5):1065‐1073. [DOI] [PubMed] [Google Scholar]
- 22. Qian BZ, Pollard JW. Macrophage diversity enhances tumor progression and metastasis. Cell. 2010;141(1):39‐51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kuang DM, Zhao Q, Peng C, et al. Activated monocytes in peritumoral stroma of hepatocellular carcinoma foster immune privilege and disease progression through PD‐L1. J Exp Med. 2009;206(6):1327‐1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Fujita N, Nishie A, Aishima S, et al. Role of tumor‐associated macrophages in the angiogenesis of well‐differentiated hepatocellular carcinoma: pathological‐radiological correlation. Oncol Rep. 2014;31(6):2499‐2505. [DOI] [PubMed] [Google Scholar]
- 25. Pollard JW. Trophic macrophages in development and disease. Nat Rev Immunol. 2009;9(4):259‐270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Chai ZT, Zhu XD, Ao JY, et al. microRNA‐26a suppresses recruitment of macrophages by down‐regulating macrophage colony‐stimulating factor expression through the PI3K/Akt pathway in hepatocellular carcinoma. J Hematol Oncol. 2015;8(1):56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Sica A, Schioppa T, Mantovani A, Allavena P. Tumour‐associated macrophages are a distinct M2 polarised population promoting tumour progression: potential targets of anti‐cancer therapy. Eur J Cancer. 2006;42(6):717‐727. [DOI] [PubMed] [Google Scholar]
- 28. Kuo HS, Tsai MJ, Huang MC, et al. Acid fibroblast growth factor and peripheral nerve grafts regulate Th2 cytokine expression, macrophage activation, polyamine synthesis, and neurotrophin expression in transected rat spinal cords. J Neurosci. 2011;31(11):4137‐4147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Tan TK, Zheng G, Hsu TT, et al. Matrix metalloproteinase‐9 of tubular and macrophage origin contributes to the pathogenesis of renal fibrosis via macrophage recruitment through osteopontin cleavage. Lab Invest. 2013;93(4):434‐449. [DOI] [PubMed] [Google Scholar]
- 30. Gharib SA, Johnston LK, Huizar I, et al. MMP28 promotes macrophage polarization toward M2 cells and augments pulmonary fibrosis. J Leukoc Biol. 2014;95(1):9‐18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Diaz G, Engle RE, Tice A, et al. Molecular Signature and Mechanisms of Hepatitis D Virus‐Associated Hepatocellular Carcinoma. Molecular Cancer Research Mcr. 2018;16(9):1406‐1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Sekhar V, Pollicino T, Diaz G, et al. Infection with hepatitis C virus depends on TACSTD2, a regulator of claudin‐1 and occludin highly downregulated in hepatocellular carcinoma. PLoS Pathog. 2018;14(3):e1006916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Egeblad M, Werb Z. New functions for the matrix metalloproteinases in cancer progression. Nat Rev Cancer. 2002;2(3):161‐174. [DOI] [PubMed] [Google Scholar]
- 34. Capece D, Fischietti M, Verzella D, et al. The inflammatory microenvironment in hepatocellular carcinoma: a pivotal role for tumor‐associated macrophages. Biomed Res Int. 2013;2012:187204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Osman A, Bhuyan F, Hashimoto M, Nasser H, Maekawa T, Suzu S. M‐CSF inhibits anti‐HIV‐1 activity of IL‐32, but they enhance M2‐like phenotypes of macrophages. J Immunol. 2014;192(11):5083‐5089. [DOI] [PubMed] [Google Scholar]
- 36. Mantovani A, Allavena P, Sica A, Balkwill F. Cancer‐related inflammation. Nature. 2008;454(7203):436‐444. [DOI] [PubMed] [Google Scholar]
- 37. Nakagawa H, Maeda S. Inflammation‐and stress‐related signaling pathways in hepatocarcinogenesis. World J Gastroenterol. 2012;18(31):4071‐4081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Shirabe K, Mano Y, Muto J, et al. Role of tumor‐associated macrophages in the progression of hepatocellular carcinoma. Surg Today. 2012;42(1):1. [DOI] [PubMed] [Google Scholar]
- 39. Coussens LM, Werb Z. Inflammation and cancer. Nature. 2002;420(6917):860‐867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Murdoch C, Giannoudis A, Lewis C. Mechanisms regulating the recruitment of macrophages into hypoxic areas of tumors and other ischemic tissues. Blood. 2004;104(8):2224‐2234. [DOI] [PubMed] [Google Scholar]
- 41. Tsou CL, Gladue RP, Carroll LA, et al. Identification of C‐C chemokine receptor 1 (CCR1) as the monocyte hemofiltrate C‐C chemokine (HCC)‐1 receptor. J Exp Med. 1998;188(3):603‐608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Schulz‐Knappe P, Mägert HJ, Dewald B, et al. HCC‐1, a novel chemokine from human plasma. J Exp Med. 1996;183(1):295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Scholl SM, Pallud C, Beuvon F, et al. Anti‐colony‐stimulating factor‐1 antibody staining in primary breast adenocarcinomas correlates with marked inflammatory cell infiltrates and prognosis. J Natl Cancer Inst. 1994;86(2):120. [DOI] [PubMed] [Google Scholar]
- 44. Kawamura K, Komohara Y, Takaishi K, Katabuchi H, Takeya M. Detection of M2 macrophages and colony‐stimulating factor 1 expression in serous and mucinous ovarian epithelial tumors. Pathol Int. 2009;59(5):300‐305. [DOI] [PubMed] [Google Scholar]
- 45. Behnes CL, Bremmer F, Hemmerlein B, Strauss A, Ströbel P, Radzun HJ. Tumor‐associated macrophages are involved in tumor progression in papillary renal cell carcinoma. Virchows Arch. 2014;464(2):191‐196. [DOI] [PubMed] [Google Scholar]
- 46. Zhu XD, Zhang JB, Zhuang PY, et al. High expression of macrophage colony‐stimulating factor in peritumoral liver tissue is associated with poor survival after curative resection of hepatocellular carcinoma. J Clin Oncol. 2008;26(16):2707‐2716. [DOI] [PubMed] [Google Scholar]
- 47. Fleetwood A, Lawrence T, Hamilton J, Cook A. Granulocyte‐macrophage colony‐stimulating factor (CSF) and macrophage CSF‐dependent macrophage phenotypes display differences in cytokine profiles and transcription factor activities: implications for CSF blockade in inflammation. J Immunol. 2007;178(8):5245‐5252. [DOI] [PubMed] [Google Scholar]
- 48. Genin M, Clement F, Fattaccioli A, Raes M, Michiels C. M1 and M2 macrophages derived from THP‐1 cells differentially modulate the response of cancer cells to etoposide. BMC Cancer. 2015;15(1):577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Korc M, Friesel RE. The role of fibroblast growth factors in tumor growth. Curr Cancer Drug Targets. 2009;9(5):639‐651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Kirov A, Duarte M, Guay J, et al. Transgenic expression of nonclassically secreted FGF suppresses kidney repair. PLoS One. 2012;7(5):e36485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Mantovani A, Marchesi F, Malesci A, Laghi L, Allavena P. Tumour‐associated macrophages as treatment targets in oncology. Nat Rev Cancer Clin Oncol. 2017;7(7):399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Ngambenjawong C, Gustafson HH, Pun SH. Progress in tumor‐associated macrophage (TAM)‐targeted therapeutics. Adv Drug Deliv Rev. 2017;114:206‐221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Weber A, Boege Y, Reisinger F, Heikenwälder M. Chronic liver inflammation and hepatocellular carcinoma: persistence matters. Swiss Med Wkly. 2011;141(18):w13197. [DOI] [PubMed] [Google Scholar]
- 54. Szabo G, Lippai D. Molecular Hepatic Carcinogenesis: Impact of Inflammation. Dig Dis. 2012;30(3):243‐248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Wai PY, Kuo PC. Intersecting pathways in inflammation and cancer: Hepatocellular carcinoma as a paradigm. World J Clin Oncol. 2012;3(2):15‐23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Xiang Y, Liu L, Wang Y, Li B, Feng D. ADAM17 promotes the invasion of hepatocellular carcinoma via upregulation MMP21; 2020. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig S1
Fig S2
Fig S3
Fig S4
Fig S5
Fig S6
Table S1
Table S2
Table S3


