Abstract
Despite the need for novel, effective therapeutics for the COVID-19 pandemic, no curative regimen is yet available, therefore patients are forced to rely on supportive and nonspecific therapies. Some SARS-CoV-2 proteins, like the 3 C-like protease (3CLpro) or the major protease (Mpro), have been identified as promising targets for antiviral drugs. The Mpro has major a role in protein processing as well as pathogenesis of the virus, and could be a useful therapeutic target. The antiviral drug nirmatrelvir can keep SARS-CoV-2 from replicating through inhibiting Mpro. Nirmatrelvir was combined with another HIV protease inhibitor, ritonavir, to create Paxlovid (Nirmatrelvir/Ritonavir). The metabolizing enzyme cytochrome P450 3 A is inhibited by ritonavir to lengthen the half-life of nirmatrelvir, so rintonavir acts as a pharmacological enhancer. Nirmatrelvir exhibits potent antiviral activity against current coronavirus variants, despite significant alterations in the SARS-CoV-2 viral genome. Nevertheless, there are still several unanswered questions. This review summarizes the current literature on nirmatrelvir and ritonavir efficacy in treating SARS-CoV-2 infection, and also their safety and possible side effects.
Keywords: Paxlovid, COVID-19, SARS-CoV-2, Nirmatrelvir, Ritonavir, 3 C-like protease
Graphical Abstract
1. Introduction
The culprit agent for coronavirus disease 2019 (COVID-19) pandemic is severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2, formerly 2019-nCoV), which is an enveloped positive-sense single-stranded RNA virus (+ssRNA). Taxonomically speaking, SARS-CoV-2 belongs to the Sarbecovirus subgenus, Betacoronavirus genus, and Coronaviridae family, which includes a variety of species that can cause mild to severe human infections, as well as many animal infections. There were six previously identified human coronaviruses 229E, NL63, OC43, HKU1, middle East respiratory syndrome (MERS)-CoV, and SARS-CoV-1; so SARS-CoV-2 became the seventh coronavirus able to infect humans [1], [2], [3], [4].
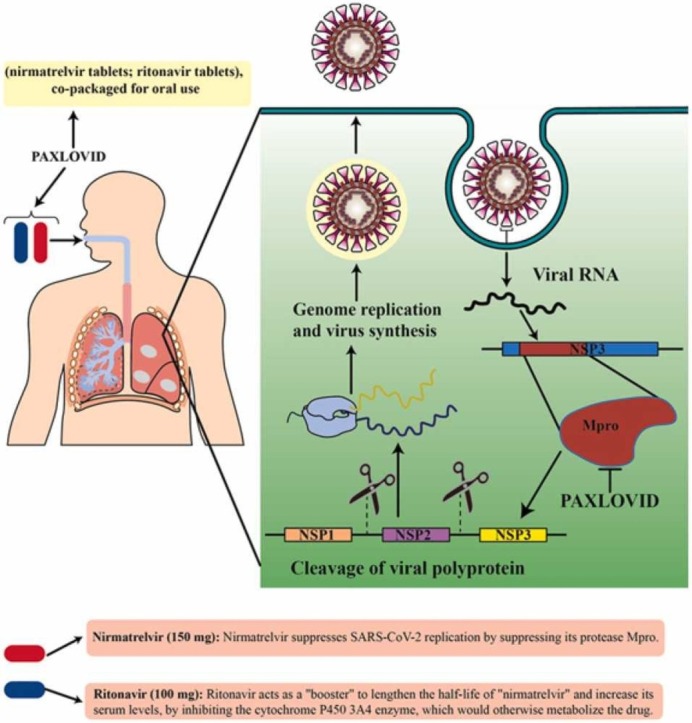
Several treatments for COVID-19 have been proposed, including immunomodulatory drugs, monoclonal antibodies, and various antiviral drugs. However, these treatments have had one or more of these three problems: low efficacy, high price, and toxic side effects, which hampered their use on a large scale [5], [6], [7]. It has been reported that Pfizer's novel antiviral drug called "Paxlovid" exhibited good effectiveness against COVID-19. Paxlovid™ is the brand name for the drug, which is made up of two generic medications—nirmatrelvir and ritonavir. The recommended dosage is 300 mg nirmatrelvir (two 150 mg tablets) with 100 mg ritonavir. (one 100 mg tablet) all taken together orally every 12 h for 5 days. Nirmatrelvir/ritonavir is composed of two antiviral protease inhibitors, i.e. PF-07321332 (also known as nirmatrelvir) and ritonavir. Nirmatrelvir is a new agent that inhibits the 3CLpro enzyme in SARS-CoV-2, while ritonavir is an HIV protease inhibitor. Ritonavir inhibits cytochrome P450 3A4, the enzyme responsible for Nirmatrelvir metabolism, which lengthens Nirmatrelvir presence in the body and boosts its activity [8], [9]. Adults with COVID-19 who were symptomatic but not-hospitalized, had risk factors (at least one) for severe disease, and were seronegative for SARS-CoV-2, were recruited in the EPIC-HR (Evaluation of Protease Inhibition for COVID-19 in High-Risk Patients) trial. On the day 28 of the trial, the nirmatrelvir/ritonavir group showed a reduction of 88% and 75% in the death and hospital admission rate, respectively, in comparison to control [8].
2. Nirmatrelvir and SARS-CoV-2 variants
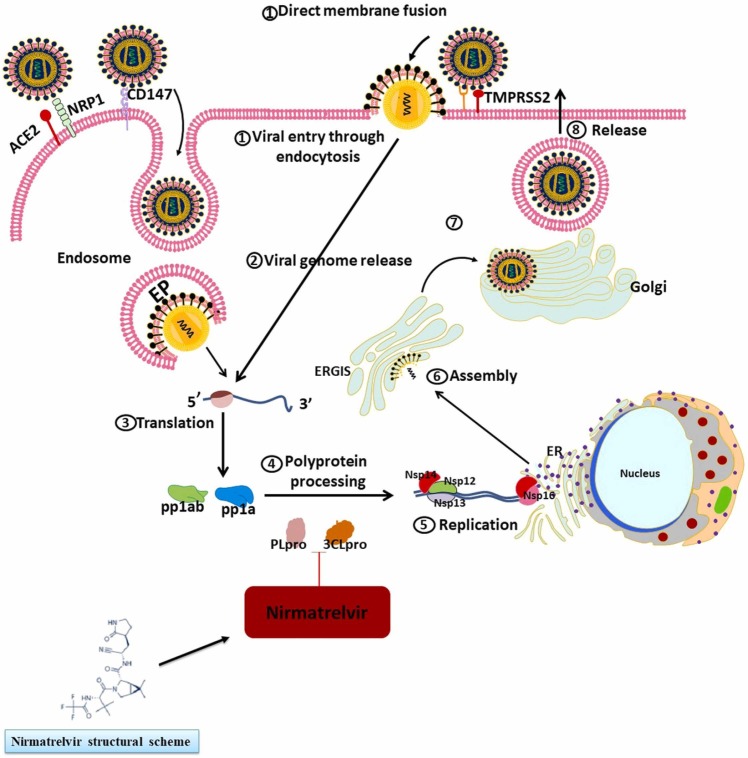
The papain-like protease (PLpro) as well as the 3CLpro (aka Mpro) are two promising molecules to target for designing medical agents against SARS-CoV-2. Both of these cysteine proteases play important roles for the SARS-CoV-2 to keep living. Antiviral protease inhibitors may be relatively safe drugs in humans because mammalian protease enzymes have different substrate specificity. In particular, the enzyme 3CLpro (cysteine protease; EC 3.4.22.69) is needed for the cleavage and maturation of proteins to make them functional for the viral replication processes [10], [11]. Various medicinal chemistry strategies have focused on the enzyme that catalyzes the process of protein cleavage, which is characterized by a catalytic dyad of Cys145 and His41 [12]. Nirmatrelvir was discovered in 2021 in an attempt to produce agents that inhibit SARS-CoV-2 3CLpro, and functions as a protease inhibitor and can be taken orally; it binds from its nitrile site to the cited dyad of Mpro [13]. Ritonavir is a tripeptide structure which inhibits HIV protease by binding to its functional site. According to the literature, the half-life of nirmatrelvir was 5.1 h in rats and 0.8 h in monkeys. When taken by os, monkeys show relatively poor bioavailability for Nirmatrelvir (8.5%), primarily because this species carries out oxidative metabolism throughout the gastrointestinal tract, while the bioavailability is moderate in rats (34–50%). Overall, nirmatrelvir showed moderate plasma protein binding, with 0.310–0.478 being the average proportion of its free plasma level in rats, monkeys, and humans [14]. In hepatocytes of these species, nirmatrelvir was similarly metabolized with the cytochrome P450 (CYP450) being the main enzyme that oxidized different functional groups of this drug. CYP3A4 was the main mediator of Nirmatrelvir oxidation in humans (fraction metabolized = 0.99). Sandwich-cultured human and animal hepatocytes (between two collagen sheets) showed there was likely to be minor elimination of unchanged nirmatrelvir by renal and biliary excretion. In vitro studies demonstrated that nirmatrelvir reversibly inhibited Mpro in a time-dependent manner and could also promote CYP3A function. The first study in humans showed increasing concentrations of nirmatrelvir when coadministered with ritonavir which inhibited CYP3A4 as the main metabolizer of nirmatrelvir [14].
Moreover, nirmatrelvir can potently inhibit the Mpro proteolytic activity in all seven types of human coronavirus, which include alpha-coronaviruses (HCoV-NL63 and HCoV-229E) as well as beta-coronaviruses (MERS-CoV, SARS-CoV-1, SARS-CoV-2, HCoV-OC43, and HCoV-HKU1) [13], [15]. At the highest tested dose of nirmatrelvir (100 μM), no inhibitory activity was observed against several mammalian proteases, including aspartyl, serine, and cysteine proteases, as well as the HIV protease, which is a retroviral aspartyl protease (retropepsin). In a study by Owen et al. [13], A549 and dNHBE cell lines (two cell lines with human respiratory epithelium origin) were infected by human coronaviruses and nirmatrelvir was administered to assess this drug activity against these species. Nirmatrelvir showed significant antiviral activity against SARS-CoV-2, MERS-CoV, HCoV-229E, and SARS-CoV-1; Moreover, cytotoxic effects were not observed at doses lower than 3 μM. They also infected a murine model with SARS-CoV-2 to assess nirmatrelvir efficacy in vivo. Nirmatrelvir significantly lowered weight loss as well as death rate in comparison to control. Furthermore, viral titer levels measured in the lungs of euthanized mice were significantly lower in the nirmatrelvir group [13].
High risk COVID-19 patients at emergent conditions were prescribed nirmatrelvir after it was approved by FDA on December 2021. Nirmatrelvir may be started after the disease has lasted for 5 days, and should be continued for 5 consecutive days [9]. The EPIC-HR trial assessed nirmatrelvir efficacy in adults with precited criteria [8]. This research was performed at the time of delta variant pandemic; therefore, it lacks data about nirmatrelvir effects on the Omicron variant and also adults with previous exposure to SARS-CoV-2. Arbel et al. recently evaluated nirmatrelvir effects in adults with previous exposure to SARS-CoV-2 and at the time of Omicron pandemic [16]. They included 3902 individuals who met the inclusion criteria and then administered nirmatrelvir. They observed that for patients ≥ 65 years, nirmatrelvir induced a significant reduction in admission as well as death rate, compared to the control. However, for younger cases, nirmatrelvir did not notably alter the course of the disease [16]. Furthermore, in a study by Li et al. nirmatrelvir effects on Omicron-infected Calu-3 cells was evaluated [17]. The researchers found that even low concentrations of nirmatrelvir were able to suppress Omicron variant replication. To model the effect of nirmatrelvir in COVID-19–vaccinated patients, Calu-3 cells were treated with the serum of vaccinated individuals and then exposed to either wild-type (WT) or Omicron form. Wild-type SARS-CoV-2 was unable to replicate in the serum-incubated cells while Omicron variant did replicate, albeit at low levels. However, nirmatrelvir treatment was able to successfully suppress the residual replication [17]. Nirmatrelvir antiviral activity was evaluated in another investigation against several different SARS-CoV-2 variants of concern (VOCs) (alpha, beta, gamma, delta, and Omicron) by Vangeel et al. [18]. They showed that nirmatrelvir was effective against all currently known VOCs, including Omicron. They used ORF1ab software to show that there were two known amino acid alterations in the 3CLpro sequence (K90R at position 3353 in Beta, and P132H at position 3395 in Omicron), which did not involve the active site of the 3CLpro, making them unlikely to affect the sensitivity to nirmatrelvir. Similarly, they predicted that nirmatrelvir would remain effective against the alpha, beta, gamma, and delta forms [18]; furthermore, an in silico study of P132H mutation by Dawood showed similar results [19].
It has been shown that some missense point mutations in SARS-CoV-1 3CLpro (with 96% similarity to SARS-CoV-2 3CLpro in the order of amino acids) may affect the protease activity [20]. Catalytic activity may be somewhat higher (S284, T285, I286) or significantly decreased (F140, R298, N28, G11, N214, S139, E166) as a result of these known mutations [21], [22], [23], [24]. Lineage comparison [25] of the SARS-CoV-2 genomic material showed three missense mutations with > 20% frequency at the Mpro portion of the ORF1a/b gene [26]. In addition, it has been reported that some of these mutations and SARS-CoV-2 variants may be associated with each other. For example, Lambda (or C.37 in PANGO nomenclature) has a G15S mutation that is > 85% common. Beta (B.1.351) carries a K90R mutation that is > 95% common, and Omicron (B.1.1.529) carries a P132H mutation that is > 95% common [20], [25], [27], [28]. However, the Delta (B.1.617.2) had no association with any specific mutations [25]. This suggests the similarity of Mpro of Delta and WT virus. Recently, Ullrich et al. examined whether nirmatrelvir efficacy alters with Mpro mutations [20]; and therefore, recruited six SARS-CoV-2 variants: Lambda, Omicron, Beta, P.2 Zeta, B.1.1.318, and B.1.2, each of them carrying a specific mutation. The results suggested that nirmatrelvir would be just as effective against the variations as it is against the WT virus, and that the current COVID-19 variations would not impact the effectiveness of nirmatrelvir [20].
The results obtained after various regimens of nirmatrelvir therapy were documented by Peluso et al. in four consecutive cases from a post-COVID cohort analysis [29]. Despite timely antiviral treatment, the first patient suffered a clinical rebound and ultimately acquired long COVID-19. In the two subsequent patients, who took nirmatrelvir 25 and 60 days after their COVID-19 symptoms first appeared, both participants showed an improvement in their condition. The last patient, who had suspected long COVID for two years with a new superimposed COVID, experienced significant relief from chronic symptoms after taking nirmatrelvir. In conclusion, further research is required to elucidate the effects of timely nirmatrelvir therapy on progression of acute COVID to long COVID and also its effects on long COVID itself. [29].
As a surface protein, the S (spike) protein of SARS-CoV-2 functions as a major antigen for the host immune system [30]. This intense selection pressure against the S protein leads to the appearance of variant mutations, thus decreasing the effectiveness of vaccines based on this protein [31], [32], [33], [34]. Thus, other proteins with fewer variations may be better targets for facing this virus; Mpro as well as RNA-dependent RNA polymerase (RdRp) are two of these proteins mainly focused on, in the studies [35], which are also exposed to possible mutations. Iketani and colleagues recently examined possible mechanisms of resistance to nirmatrelvir [36]. They used different SARS-CoV-2 variants for cellular infection and nirmatrelvir in different doses to evaluate resistance development. They reported that several resistant viruses appeared, and analysis of their genomes showed that they included several 3CL protease mutations. Their large-scale experiment found mutations in 23 different amino acid residues in 53 distinct viral lineages. T21I, P252L, or T304I were found in most viral lineages as precursor mutations which only conferred mild resistance, but the accumulation of further mutations could lead to increased resistance. The most significant resistance was found with the E166V mutation, which reduced the replicative competence of the virus, but this was restored by compensating mutations T21I and L50F. In conclusion, unlike the study of Ullrich et al. [20], their results showed that 3CLpro mutations may cause resistance to nirmatrelvir in SARS-CoV-2 variants, a finding that needs further evaluation [36]. ( Fig. 1).
Fig. 1.
Nirmatrelvir can inhibit 3CLpro, an enzyme involved in maturation of proteins, in different variants of SARS-CoV-2, and therefore, suppress their replication.
3. Ritonavir implications in COVID-19 treatment
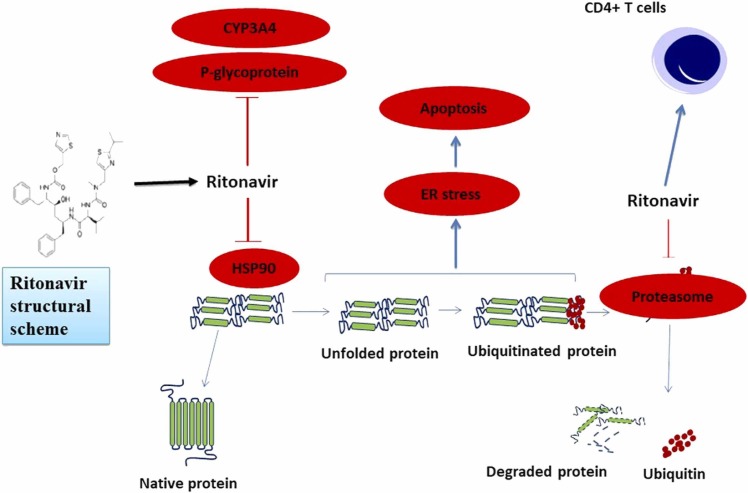
After saquinavir, ritonavir was the second agent with protease inhibition property which was confirmed by FDA to be used in AIDS treatment. Studies have shown that, in addition to inhibiting the HIV protease, ritonavir can also inhibit CYP450–3A4 ( Fig. 2). It is now used primarily to enhance the bioavailability of co-administered antiretroviral drugs (ARVs); its combination with standard anti-HIV triple therapy, resulted in increased CD4 + counts and lower HIV RNA levels in treatment-naive individuals [37]. Ritonavir has been found to increase the effectiveness of various ARVs. For example, the combination of lopinavir plus ritonavir significantly reduced viral load and improved immunological parameters in HIV-infected patients who were both previously treated and treatment-naïve [38], [39]. Because of its mechanisms of action, ritonavir is being studied in combination with antineoplastic drugs for treating cancer. In addition, its combination with ombitasvir, paritaprevir, as well as dasabuvir has received FDA approval for HCV genotype 1 treatment. Moreover, the FDA approved lopinavir/ritonavir for HIV treatment in the early 2000 s [38].
Fig. 2.
Cellular processes affected by Ritonavir. Ritonavir may inhibit proteasomes, HSP90 (heat shock protein 90), CYP3A4, as well as P-glycoprotein; it may also modulate the function of immune cells [54].
Pharmacologic enhancement is used to increase the tolerability and effectiveness of protease inhibitors. Ritonavir inhibits two essential metabolic processes, making it the ideal pharmacologic enhancer. Ritonavir inhibits first-pass metabolism, which takes place during absorption. Both P-glycoprotein, a drug efflux transporter that pumps drugs out of the gut wall and back into the intestinal lumen, as well as CYP3A4, an important cytochrome P450 isoenzyme involved in drug metabolism, are found in the enterocytes that line the intestine [40]. Firstly, since ritonavir inhibits P-glycoprotein it may increase the Cmax of the ARV drug. Secondly, ritonavir lengthens the plasma half-life of the drug by inhibiting CYP3A4. The P-glycoprotein in CD4 + cells may also be inhibited by ritonavir to prolong the intracellular half-life of the ARV drug [40]. Although ritonavir-mediated inhibition of CYP3A4 is clinically significant, its precise mechanism of action is not yet established. Still, ritonavir stands out as a powerful mechanism-based enhancer because of its ability to irreversibly inhibit CYP3A4. This irreversible inhibition is thought to occur by four distinct mechanisms: (a) ritonavir metabolites bind to the heme group; (b) unmodified ritonavir strongly binding to the heme iron; (c) damage to the heme molecule; (d) covalent attachment of a reactive ritonavir intermediate to the CYP3A4 apoprotein [41]. Ritonavir can inhibit CYP3A4 and CYP3A5 with almost equivalent potency, and since it has been used in patients from different racial origins, these results are significant. None of the suggested hypothesized mechanism are likely to be completely erroneous, but it is hard to establish what the principal mechanism of action in vivo actually is. In addition, its function through different mechanisms may enhance its inhibitory effectiveness [41].
Patients with HIV who have failed all available treatments have complex mutations that make them resistant to or unable to tolerate nucleoside reverse transcriptase inhibitors, and are therefore eligible for double-boost protease inhibitor combinations such as lopinavir/ritonavir [42], [43]; a strategy that may also be used against SARS-CoV-2. It is necessary to mention that although it has been suggested that ritonavir is of the ability to inhibit SARS-CoV-2 protease, ritonavir alone has not shown in vitro activity against this virus [44]. Talking about combination of ritonavir and lopinavir in COVID-19 treatment, lopinavir can also inhibit Mpro of coronavirus, and ritonavir delays its clearance from the body [45], [46]. The use of lopinavir-ritonavir in the treatment of a marmoset model infected with MERS-CoV resulted in positive clinical, radiological, as well as pathological results, and also reduced viral load [47]. Lopinavir-ritonavir decreased clinical symptoms in ferrets infected with SARS-CoV-2, but had no impact on viral load [48]. In a human study of COVID-19, the combination of lopinavir-ritonavir and ribavirin resulted in more favorable prognosis as well as decreased virus titer [49]. Lopinavir-ritonavir shortened the duration of viral shedding [50], and was successful in controlling fever [51] in some COVID-19 cases. In another clinical study of COVID-19, lopinavir-ritonavir made no alterations in the course of disease or the outcomes [52]; there was similar lack of efficacy in the study of Horby et al. [53].
4. Nirmatrelvir/ritonavir and COVID-19 therapy
At the time of Omicron pandemic and to assess nirmatrelvir/ritonavir efficacy against SARS-CoV-2, Najjar-Debbiny et al. recruited adults (≥18 years) who were experiencing COVID-19 for the first time and had risk factors for progression into severe disease and followed them for 28 days [55]. They observed that COVID-19 patients receiving nirmatrelvir/ritonavir had significant reduction in the death rate as well as the risk of progression into severe disease in comparison to control; furthermore, the cited effects were more pronounced in the conditions of immunosuppression and also neurological or cardiovascular comorbidities. In conclusion, their findings is in favor nirmatrelvir/ritonavir ability to alter the course of the disease and also death rate [55].
After FDA approved nirmatrelvir/ritonavir in December 2021 to be used for non-severe COVID-19 cases but in risk of progressing into severe disease, Malden et al. decided to evaluate its efficacy in lowering the risk of the concerned progression [56]. Therefore, they gathered the data of 5287 patients who received nirmatrelvir/ritonavir between December 2021 and May 2022, and explored whether they needed presence to health care system between 5 and 15 days after nirmatrelvir/ritonavir course had finished. They observed that of these cases only 45 presented to the health care system of whom six were hospitalized; furthermore, approximately half of these 45 cases had either the risk factor of age (≥65) or the presence of medical comorbidity. Taken together, the low rate of further requirement to medical care observed in their study is again in favor of nirmatrelvir/ritonavir ability to alter the course of the diseases [56].
In a similar retrospective work, Shah et al. analyzed the data of a population of 699,848 Americans who were diagnosed with COVID-19 in the spring and summer of 2022, and evaluated the proportion of them who received nirmatrelvir/ritonavir, and also how nirmatrelvir/ritonavir altered the course of disease [57]. They observed that nirmatrelvir/ritonavir, in general as well as with various demographic adjustments, was of the ability to significantly reduce the rate of admission to hospital, which could be interpreted into its ability to inhibit the progression toward severe COVID-19; regarding their findings, they insisted on a more pervasive use of this medication in outpatient settings [57].
Regarding examining nirmatrelvir/ritonavir use in pregnant women, Loza et al. performed a short term follow up on seven pregnant women of different gestational ages who received nirmatrelvir/ritonavir with a diagnosis of non-severe COVID-19 [58]. Of these seven patients, four with smaller gestational ages did not come to delivery in the time of follow up, and neither showed any specific maternal or fetal adverse events. Of the three remaining patients who came to delivery in the time of follow up, one did not complete the five day course of treatment, and the remaining two did not show any specific complications attributable to nirmatrelvir/ritonavir. In conclusion, their study had various pitfalls, the most important one being lack of long term follow up, and therefore, not much could be concluded from it regarding the safety of nirmatrelvir/ritonavir use in pregnancy [58].( Table 1).
Table 1.
Summary of clinical trials and preclinical studies using nirmatrelvir/ritonavir and nirmatrelvir.
| Drug | Model | Cell line, species, number of patients | Administrative way | Does | Time | Findings | Follow up | Phase | Ref |
|---|---|---|---|---|---|---|---|---|---|
| Nirmatrelvir/ritonavir | Human | Overall: 2246 | Oral | nirmatrelvir (300 mg) + ritonavir (100 mg) | Every 12 h for 5 days | Reduced development of severe cases | 28 days | 2–3 | [8] |
| Nirmatrelvir/ritonavir | Human | Overall: 180,351 Paxlovid group: 4737 |
- | - | - | Reduced death rate as well as number of severe cases | 28 days | - | [55] |
| Nirmatrelvir/ritonavir | Human | - | Oral | Single‐ascending dose (SAD): nirmatrelvir (150–1500 mg) and next nirmatrelvir (250–750 mg) +ritonavir (100 mg) and final dose: nirmatrelvir (250 mg) + ritonavir (100 mg). Multiple‐ascending dose (MAD): nirmatrelvir dose (75, 250, and 500 mg)+ ritonavir (100 mg). |
SAD: − 12, 0, and 12 h relative to nirmatrelvir MAD: twice daily for 10 days |
Reduced development of severe cases | 28 days | 1 | [59] |
| Nirmatrelvir/ritonavir | Human | 14 | Oral | - | 6.5-days | - | 14 days | - | [60] |
| Nirmatrelvir/ritonavir | Human | 30322 | Oral | - | - | Reduced hospitalization, better effects in unvaccinated and obese patients | 28 days | - | [61] |
| Nirmatrelvir/ritonavir | Human | 25 (solid organ transplant recipients) | Oral | - | 5 days | It is better to use a lower dose in transplant recipients | 30 days | - | [62] |
| Nirmatrelvir | In vitro | HeLa, S3–ENT1, S3-ENT2 | - | - | - | Human ENT1 as well as ENT2 are not affected by Nirmatrelvir | - | - | [63] |
| Nirmatrelvir/ritonavir | Human | mild (n = 8), moderate (n = 8), and severe renal impairment (n = 8) | Oral | 100–mg nirmatrelvir with 100 mg ritonavir | 5 day (ritonavir received 12 h before, together with and 12 and 24 h after the nirmatrelvi) | Renal insufficiency may implicate dose adjustment for nirmatrelvir/ritonavir (150/100 mg nirmatrelvir/ritonavir) | 28–35 days | 1 | [64] |
| Nirmatrelvir/ritonavir | Human | 5287 | Oral | nirmatrelvir (300 mg) + ritonavir (100 mg) | Every 12 h for 5 days | Reduced hospitalization and Emergency Department visits | 5–15 days | - | [56] |
| Nirmatrelvir/ritonavir | Human | 482 | Oral | nirmatrelvir (300 mg) + ritonavir (100 mg) | Every 12 h for 5 days | Lowered number of the shed viruses and therefore, hampered ability of the virus to infect new cases | 7–21 days | - | [65] |
| Nirmatrelvir/ritonavir | Human | 7 | Oral | - | - | Improvement in symptoms as well as elevated rate of negative rapid tests | 17 days | - | [66] |
| Nirmatrelvir, Nirmatrelvir/ritonavir | In vitro | BL21 (DE3) | - | - | - | Ritonavir significantly lengthens the half-life of nirmatrelvir | - | - | [67] |
| Nirmatrelvir, Nirmatrelvir/ritonavir | In vivo | Rats and monkeys | - | - | - | No significant adverse effects were observed for the drug | 2-week, 1-month |
Cardiovascular, pharmacokinetics, recovery phase | [68] |
| Nirmatrelvir/ritonavir | Human | 2260 | Oral | nirmatrelvir (300 mg) + ritonavir (100 mg) | Every 12 h for 5 days | Reduced death rate as well as number of severe cases | 30 days | - | [69] |
| Nirmatrelvir/ritonavir | Human | 1279 (469 patients (36.7%) received Paxlovid) | Oral | nirmatrelvir (300 mg) + ritonavir (100 mg) | Every 12 h for 5 days | Reduced viral load and mortality | - | - | [70] |
| Nirmatrelvir | In vitro | VeroE6 | - | - | - | The L50F, E166A, and L167F mutation are resistance to nirmatrelvir. | - | - | [71] |
5. Safety and tolerability of nirmatrelvir/ritonavir
Although nirmatrelvir/ritonavir seems potent in combating COVID-19, its interactions with transplant drugs should be taken into account [72]. Transplant patients often suffer from immunosuppression and vaccination failure, which along with other concurrent conditions make them vulnerable to development of severe COVID-19 and death [73], [74]. Because ritonavir inhibits CYP3A, it has been reported that the concentrations of CYP3A-metabolized drugs can rise by 1.8 up to 20 fold [75]. The rapid increase in plasma levels of tacrolimus, cyclosporine, calcineurin inhibitors (CNIs), or mTOR inhibitors: everolimus and sirolimus in patients exposed to ritonavir is due to the dependence of these drugs on CYP3A metabolism [76]. The intestinal cytochrome CYP3A enzymes break down the tacrolimus molecule, and a sudden rise in its concentration in the bloodstream could cause posterior reversible encephalopathy syndrome, seizures, kidney damage, and even death [76], [77]. Recently, Prikis and Cameron reported that nirmatrelvir/ritonavir therapy in a kidney transplant patient receiving tacrolimus was associated with acute renal damage, and a suddenly higher tacrolimus level, which required treatment discontinuation [78]. Their results suggested that elevated concentration of tacrolimus and its metabolites could produce harmful effects, including acute renal damage, resulting from the inhibition of its metabolism by nirmatrelvir/ritonavir; therefore, alternative immunosuppressive drugs or a decrease in daily tacrolimus dosage should be considered throughout the COVID-19 treatment [78].
In addition to the cited immunosuppressive drugs, other drugs like statins, calcium channel blockers, and warfarin must also be taken into account, when administering nirmatrelvir/ritonavir. The FDA European Union Authorization document has a more comprehensive list of possible drug interactions [72].
Considering the novelty of nirmatrelvir/ritonavir, not much is known about its long-term efficacy or any possible adverse effects. However, some studies have reported that nirmatrelvir/ritonavir could induce common side effects like headache, emesis, loose stool, as well as a disturbed sense of taste. Infrequently, muscular aches and elevated blood pressure were also reported [79].
Specifically talking about calcium channel blockers, a case of verapamil toxicity induced by nirmatrelvir/ritonavir was reported, which resulted in decreased level of consciousness as well as bradycardia [80]. The bradycardia prompted the use of an internal pacemaker, besides discontinuation of both verapamil and nirmatrelvir/ritonavir. Fortunately, no serious complications occurred, and the pacemaker was removed after four days [80]. Furthermore, the potential of nirmatrelvir/ritonavir to interfere with normal sinoatrial node function has also been reported [81].
Sathish et al. used rats and monkeys to evaluate the safety profile of nirmatrelvir [68]. The duration of oral nirmatrelvir administration was one month for both species, and the max dose was 1000 mg/kg.day for rats and 600 mg/kg.day for the monkeys. In rats, locomotion, respiratory rate, and bleeding time were observed to be increased. In monkeys, the max dose of nirmatrelvir caused the blood pressure to transiently elevate, the pulse rate to drop, and the transaminase levels to increase, but there were no arrhythmic changes. Taken together, they concluded that nirmatrelvir seems reasonably safe for use in human cases [68].
To assess the efficacy as well as the side effects of nirmatrelvir/ritonavir, Zheng et al. performed a meta-analysis on 13 studies [82]. Their final statistical results denied the nirmatrelvir/ritonavir efficacy in reducing the number rebound cases of COVID-19, but showed that the side effects attributed to nirmatrelvir/ritonavir were not significant [82].
6. Conclusion
Nirmatrelvir, alone or in combination with ritonavir (Paxlovid), seems to be a potent antiviral drug to treat COVID-19, but there are some unanswered questions. First, the full findings of extensive clinical studies are yet to be released. Second, keeping a careful eye on the efficacy of nirmatrelvir against new COVID-19 strains in the years to come is essential. Selection pressure on the virus can cause additional mutations to arise in the protease protein, resulting in a decline in nirmatrelvir/ritonavir effectiveness. Third, regarding the inhibitory effect of ritonavir on CYP3A4, the interactions with other drugs should be kept in mind. Despite these facts, the existing evidence from randomized trials has shown that nirmatrelvir/ritonavir is effective in treating COVID-19 with a reasonable safety profile, and with the most prominent effects of this drug being reduced chance of progression to severe disease and also death rate. Finally, further evaluation is needed to confirm nirmatrelvir/ritonavir efficacy in treatment of COVID-19 and it low manufacturing cost and easy administration make it a valuable tool in fighting COVID-19, especially for countries with a low vaccination rate.
CRediT authorship contribution statement
Hamed Mirzaei involved in conception, design, and drafting of the manuscript. Seyed Mohammad Reza Hashemian, Amirhossein Sheida, Mohammad Taghizadieh, Mohammad Yousef Memar, Michael R Hamblin, Hossein Bannazadeh Baghi, Javid Sadri Nahand and Zatollah Asemi contributed in data collection and manuscript drafting. All authors approved the final version for submission.
Conflict of interest statement
The authors have no relevant financial or non-financial interests to disclose.
References
- 1.Cui J., Li F., Shi Z.L. Origin and evolution of pathogenic coronaviruses. Nat. Rev. Microbiol. 2019;17(3):181–192. doi: 10.1038/s41579-018-0118-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Khailany R.A., Safdar M., Ozaslan M. Genomic characterization of a novel SARS-CoV-2. Gene Rep. 2020;19 doi: 10.1016/j.genrep.2020.100682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhu N., Zhang D., Wang W., Li X., Yang B., Song J., et al. A novel coronavirus from patients with pneumonia in China, 2019. N. Engl. J. Med. 2020;382(8):727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Su S., Wong G., Shi W., Liu J., Lai A.C.K., Zhou J., et al. Epidemiology, genetic recombination, and pathogenesis of coronaviruses. Trends Microbiol. 2016;24(6):490–502. doi: 10.1016/j.tim.2016.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Drożdżal S., Rosik J., Lechowicz K., Machaj F., Szostak B., Przybyciński J., et al. An update on drugs with therapeutic potential for SARS-CoV-2 (COVID-19) treatment. Drug Resist Updat. 2021;59 doi: 10.1016/j.drup.2021.100794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Parums D.V. Editorial: current status of oral antiviral drug treatments for SARS-CoV-2 infection in non-hospitalized patients. Med Sci. Monit. 2022;28 doi: 10.12659/MSM.935952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Weinreich D.M., Sivapalasingam S., Norton T., Ali S., Gao H., Bhore R., et al. REGN-COV2, a neutralizing antibody cocktail, in outpatients with covid-19. N. Engl. J. Med. 2021;384(3):238–251. doi: 10.1056/NEJMoa2035002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hammond J., Leister-Tebbe H., Gardner A., Abreu P., Bao W., Wisemandle W., et al. Oral nirmatrelvir for high-risk, nonhospitalized adults with covid-19. N. Engl. J. Med. 2022;386(15):1397–1408. doi: 10.1056/NEJMoa2118542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.JA OS. EUA 105 Pfizer Paxlovid LOA (12222021). In: Administration USFaD, editor. 2021.
- 10.Thiel V., Ivanov K.A., Putics Á., Hertzig T., Schelle B., Bayer S., et al. Mechanisms and enzymes involved in SARS coronavirus genome expression. J. Gen. Virol. 2003;84(Pt 9):2305–2315. doi: 10.1099/vir.0.19424-0. [DOI] [PubMed] [Google Scholar]
- 11.Zhu W., Xu M., Chen C.Z., Guo H., Shen M., Hu X., et al. Identification of SARS-CoV-2 3CL protease inhibitors by a quantitative high-throughput screening. ACS Pharm. Transl. Sci. 2020;3(5):1008–1016. doi: 10.1021/acsptsci.0c00108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dai W., Zhang B., Jiang X.M., Su H., Li J., Zhao Y., et al. Structure-based design of antiviral drug candidates targeting the SARS-CoV-2 main protease. Science. 2020;368(6497):1331–1335. doi: 10.1126/science.abb4489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Owen D.R., Allerton C.M.N., Anderson A.S., Aschenbrenner L., Avery M., Berritt S., et al. An oral SARS-CoV-2 M(pro) inhibitor clinical candidate for the treatment of COVID-19. Science. 2021;374(6575):1586–1593. doi: 10.1126/science.abl4784. [DOI] [PubMed] [Google Scholar]
- 14.Eng H., Dantonio A.L., Kadar E.P., Obach R.S., Di L., Lin J., et al. Disposition of nirmatrelvir, an orally bioavailable inhibitor of SARS-CoV-2 3C-Like Protease, across Animals and Humans. Drug Metab. Dispos. 2022;50(5):576–590. doi: 10.1124/dmd.121.000801. [DOI] [PubMed] [Google Scholar]
- 15.Zarei A., Fardood S.T., Moradnia F., Ramazani A. A review on coronavirus family persistency and considerations of novel type, COVID-19 features. Eurasia Chem. Commun. 2020:798–811. [Google Scholar]
- 16.Arbel R., Wolff Sagy Y., Hoshen M., Battat E., Lavie G., Sergienko R., et al. Nirmatrelvir use and severe covid-19 outcomes during the omicron surge. N. Engl. J. Med. 2022;387(9):790–798. doi: 10.1056/NEJMoa2204919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li P., Wang Y., Lavrijsen M., Lamers M.M., de Vries A.C., Rottier R.J., et al. SARS-CoV-2 Omicron variant is highly sensitive to molnupiravir, nirmatrelvir, and the combination. Cell Res. 2022;32(3):322–324. doi: 10.1038/s41422-022-00618-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vangeel L., Chiu W., De Jonghe S., Maes P., Slechten B., Raymenants J., et al. Remdesivir, Molnupiravir and Nirmatrelvir remain active against SARS-CoV-2 Omicron and other variants of concern. Antivir. Res. 2022;198 doi: 10.1016/j.antiviral.2022.105252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dawood A.A. The efficacy of Paxlovid against COVID-19 is the result of the tight molecular docking between M(pro) and antiviral drugs (nirmatrelvir and ritonavir) Adv. Med Sci. 2022;68(1):1–9. doi: 10.1016/j.advms.2022.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ullrich S., Nitsche C. The SARS-CoV-2 main protease as drug target. Bioorg. Med. Chem. Lett. 2020;30(17) doi: 10.1016/j.bmcl.2020.127377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cheng S.C., Chang G.G., Chou C.Y. Mutation of Glu-166 blocks the substrate-induced dimerization of SARS coronavirus main protease. Biophys. J. 2010;98(7):1327–1336. doi: 10.1016/j.bpj.2009.12.4272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen S., Hu T., Zhang J., Chen J., Chen K., Ding J., et al. Mutation of Gly-11 on the dimer interface results in the complete crystallographic dimer dissociation of severe acute respiratory syndrome coronavirus 3C-like protease: crystal structure with molecular dynamics simulations. J. Biol. Chem. 2008;283(1):554–564. doi: 10.1074/jbc.M705240200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shi J., Sivaraman J., Song J. Mechanism for controlling the dimer-monomer switch and coupling dimerization to catalysis of the severe acute respiratory syndrome coronavirus 3C-like protease. J. Virol. 2008;82(9):4620–4629. doi: 10.1128/JVI.02680-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shi J., Song J. The catalysis of the SARS 3C-like protease is under extensive regulation by its extra domain. Febs J. 2006;273(5):1035–1045. doi: 10.1111/j.1742-4658.2006.05130.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Abdel Latif A., Mullen J., Alkuzweny M., Tsueng G., Cano M., Haag E., et al. Outbreak. info-lineage comparison. 2021. 2021.
- 26.Zhukova A., Blassel L., Lemoine F., Morel M., Voznica J., Gascuel O. Origin, evolution and global spread of SARS-CoV-2. C. R. Biol. 2020 doi: 10.5802/crbiol.29. [DOI] [PubMed] [Google Scholar]
- 27.Hughes L., Gangavarapu K., Latif A.A., Mullen J., Alkuzweny M., Hufbauer E., et al. Outbreak.info genomic reports: scalable and dynamic surveillance of SARS-CoV-2 variants and mutations. Res Sq. 2022 doi: 10.1038/s41592-023-01769-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rambaut A., Holmes E.C., O'Toole Á., Hill V., McCrone J.T., Ruis C., et al. A dynamic nomenclature proposal for SARS-CoV-2 lineages to assist genomic epidemiology. Nat. Microbiol. 2020;5(11):1403–1407. doi: 10.1038/s41564-020-0770-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Peluso M.J., Anglin K., Durstenfeld M.S., Martin J.N., Kelly J.D., Hsue P.Y., et al. Effect of oral nirmatrelvir on long COVID symptoms: 4 cases and rationale for systematic studies. Pathog. Immun. 2022;7(1):95–103. doi: 10.20411/pai.v7i1.518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Maleki Dana P., Sadoughi F., Hallajzadeh J., Asemi Z., Mansournia M.A., Yousefi B., et al. An insight into the sex differences in COVID-19 patients: what are the possible causes? Prehosp. Disaster Med. 2020;35(4):438–441. doi: 10.1017/S1049023X20000837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.VanBlargan L.A., Errico J.M., Halfmann P.J., Zost S.J., Crowe J.E., Jr., Purcell L.A., et al. An infectious SARS-CoV-2 B.1.1.529 Omicron virus escapes neutralization by therapeutic monoclonal antibodies. Nat. Med. 2022;28(3):490–495. doi: 10.1038/s41591-021-01678-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Planas D., Saunders N., Maes P., Guivel-Benhassine F., Planchais C., Buchrieser J., et al. Considerable escape of SARS-CoV-2 Omicron to antibody neutralization. Nature. 2022;602(7898):671–675. doi: 10.1038/s41586-021-04389-z. [DOI] [PubMed] [Google Scholar]
- 33.Weisblum Y., Schmidt F., Zhang F., DaSilva J., Poston D., Lorenzi J.C.C., et al. Escape from neutralizing antibodies by SARS-CoV-2 spike protein variants. bioRxiv. 2020 doi: 10.7554/eLife.61312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Harvey W.T., Carabelli A.M., Jackson B., Gupta R.K., Thomson E.C., Harrison E.M., et al. SARS-CoV-2 variants, spike mutations and immune escape. Nat. Rev. Microbiol. 2021;19(7):409–424. doi: 10.1038/s41579-021-00573-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Beheshtirouy S., Khani E., Khiali S., Entezari-Maleki T. Investigational antiviral drugs for the treatment of COVID-19 patients. Arch. Virol. 2022;167(3):751–805. doi: 10.1007/s00705-022-05368-z. [DOI] [PubMed] [Google Scholar]
- 36.Iketani S., Mohri H., Culbertson B., Hong S.J., Duan Y., Luck M.I., et al. Multiple pathways for SARS-CoV-2 resistance to nirmatrelvir. Nature. 2022 doi: 10.1038/s41586-022-05514-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lea A.P., Faulds D.Ritonavir. Drugs. 1996;52(4):7–8. doi: 10.2165/00003495-199652040-00007. 541-6; discussion. [DOI] [PubMed] [Google Scholar]
- 38.Talha B., Dhamoon A.S.Ritonavir. StatPearls Publishing LLC,; 2022. StatPearls. Treasure Island (FL) StatPearls Publishing Copyright © 2022. [Google Scholar]
- 39.Croxtall J.D., Perry C.M. Lopinavir/Ritonavir: a review of its use in the management of HIV-1 infection. Drugs. 2010;70(14):1885–1915. doi: 10.2165/11204950-000000000-00000. [DOI] [PubMed] [Google Scholar]
- 40.Boffito M. From concept to care: pharmacokinetic boosting of protease inhibitors. PRN Noteb. 2004;9(4):15–18. [Google Scholar]
- 41.Loos N.H.C., Beijnen J.H., Schinkel A.H. The mechanism-based inactivation of CYP3A4 by ritonavir: what mechanism? Int J. Mol. Sci. 2022;23(17) doi: 10.3390/ijms23179866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wensing A.M., van Maarseveen N.M., Nijhuis M. Fifteen years of HIV protease inhibitors: raising the barrier to resistance. Antivir. Res. 2010;85(1):59–74. doi: 10.1016/j.antiviral.2009.10.003. [DOI] [PubMed] [Google Scholar]
- 43.Stebbing J., Scourfield A., Koh G., Taylor C., Taylor S., Wilkins E., et al. A multicentre cohort experience with double-boosted protease inhibitors. J. Antimicrob. Chemother. 2009;64(2):434–435. doi: 10.1093/jac/dkp192. [DOI] [PubMed] [Google Scholar]
- 44.Patel T.K., Patel P.B., Barvaliya M., Saurabh M.K., Bhalla H.L., Khosla P.P. Efficacy and safety of lopinavir-ritonavir in COVID-19: A systematic review of randomized controlled trials. J. Infect. Public Health. 2021;14(6):740–748. doi: 10.1016/j.jiph.2021.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liu X., Wang X.J. Potential inhibitors against 2019-nCoV coronavirus M protease from clinically approved medicines. J. Genet Genom. 2020;47(2):119–121. doi: 10.1016/j.jgg.2020.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nukoolkarn V., Lee V.S., Malaisree M., Aruksakulwong O., Hannongbua S. Molecular dynamic simulations analysis of ritonavir and lopinavir as SARS-CoV 3CL(pro) inhibitors. J. Theor. Biol. 2008;254(4):861–867. doi: 10.1016/j.jtbi.2008.07.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chan J.F., Yao Y., Yeung M.L., Deng W., Bao L., Jia L., et al. Treatment with lopinavir/ritonavir or interferon-β1b improves outcome of MERS-CoV infection in a nonhuman primate model of common marmoset. J. Infect. Dis. 2015;212(12):1904–1913. doi: 10.1093/infdis/jiv392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Park S.J., Yu K.M., Kim Y.I., Kim S.M., Kim E.H., Kim S.G., et al. Antiviral efficacies of FDA-approved drugs against SARS-CoV-2 infection in ferrets. mBio. 2020;11(3) doi: 10.1128/mBio.01114-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chu C.M., Cheng V.C., Hung I.F., Wong M.M., Chan K.H., Chan K.S., et al. Role of lopinavir/ritonavir in the treatment of SARS: initial virological and clinical findings. Thorax. 2004;59(3):252–256. doi: 10.1136/thorax.2003.012658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yan D., Liu X.Y., Zhu Y.N., Huang L., Dan B.T., Zhang G.J., et al. Factors associated with prolonged viral shedding and impact of lopinavir/ritonavir treatment in hospitalised non-critically ill patients with SARS-CoV-2 infection. Eur. Respir. J. 2020;56(1) doi: 10.1183/13993003.00799-2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ye X.T., Luo Y.L., Xia S.C., Sun Q.F., Ding J.G., Zhou Y., et al. Clinical efficacy of lopinavir/ritonavir in the treatment of Coronavirus disease 2019. Eur. Rev. Med Pharm. Sci. 2020;24(6):3390–3396. doi: 10.26355/eurrev_202003_20706. [DOI] [PubMed] [Google Scholar]
- 52.Cao B., Wang Y., Wen D., Liu W., Wang J., Fan G., et al. A trial of lopinavir-ritonavir in adults hospitalized with severe covid-19. N. Engl. J. Med. 2020;382(19):1787–1799. doi: 10.1056/NEJMoa2001282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lopinavir-ritonavir in patients admitted to hospital with COVID-19 (RECOVERY): a randomised, controlled, open-label, platform trial. Lancet. 2020;396(10259):1345–1352. doi: 10.1016/S0140-6736(20)32013-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sato A. The human immunodeficiency virus protease inhibitor ritonavir is potentially active against urological malignancies. Onco Targets Ther. 2015;8:761–768. doi: 10.2147/OTT.S79776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Najjar-Debbiny R., Gronich N., Weber G., Khoury J., Amar M., Stein N., et al. Effectiveness of Paxlovid in Reducing Severe COVID-19 and Mortality in High Risk Patients. Clin. Infect. Dis. 2022 doi: 10.1093/cid/ciac443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Malden D.E., Hong V., Lewin B.J., Ackerson B.K., Lipsitch M., Lewnard J.A., et al. Hospitalization and emergency department encounters for COVID-19 after paxlovid treatment - California, December 2021-May 2022. MMWR Morb. Mortal. Wkly Rep. 2022;71(25):830–833. doi: 10.15585/mmwr.mm7125e2. [DOI] [PubMed] [Google Scholar]
- 57.Shah M.M., Joyce B., Plumb I.D., Sahakian S., Feldstein L.R., Barkley E., et al. Paxlovid associated with decreased hospitalization rate among adults with COVID-19—United States, April–September 2022. Am. J. Transplant. 2023;23(1):150–155. doi: 10.1016/j.ajt.2022.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Loza A., Farias R., Gavin N., Wagner R., Hammer E., Shields A. Short-term pregnancy outcomes after nirmatrelvir-ritonavir treatment for mild-to-moderate coronavirus disease 2019 (COVID-19) Obstet. Gynecol. 2022;140(3):447–449. doi: 10.1097/AOG.0000000000004900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Singh R.S.P., Toussi S.S., Hackman F., Chan P.L., Rao R., Allen R., et al. Innovative randomized phase i study and dosing regimen selection to accelerate and inform pivotal COVID-19 trial of nirmatrelvir. Clin. Pharm. Ther. 2022;112(1) doi: 10.1002/cpt.2603. 101-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Epling B.P., Rocco J.M., Boswell K.L., Laidlaw E., Galindo F., Kellogg A., et al. COVID-19 redux: clinical, virologic, and immunologic evaluation of clinical rebound after nirmatrelvir/ritonavir. medRxiv. 2022 [Google Scholar]
- 61.Dryden-Peterson S., Kim A., Kim A.Y., Caniglia E.C., Lennes I., Patel R., et al. Nirmatrelvir plus ritonavir for early COVID-19 and hospitalization in a large US health system. medRxiv. 2022 doi: 10.7326/M22-2141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Salerno D.M., Jennings D.L., Lange N.W., Kovac D.B., Shertel T., Chen J.K., et al. Early clinical experience with nirmatrelvir/ritonavir for the treatment of COVID-19 in solid organ transplant recipients. Am. J. Transpl. 2022;22(8):2083–2088. doi: 10.1111/ajt.17027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hau R.K., Wright S.H., Cherrington N.J. PF-07321332 (Nirmatrelvir) does not interact with human ENT1 or ENT2: Implications for COVID-19 patients. Clin. Transl. Sci. 2022;15(7):1599–1605. doi: 10.1111/cts.13292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Toussi S.S., Neutel J.M., Navarro J., Preston R.A., Shi H., Kavetska O., et al. Pharmacokinetics of Oral Nirmatrelvir/Ritonavir, a Protease Inhibitor for Treatment of COVID-19, in Subjects With Renal Impairment. Clin. Pharm. Ther. 2022;112(4):892–900. doi: 10.1002/cpt.2688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Li H., Gao M., You H., Zhang P., Pan Y., Li N., et al. Association of nirmatrelvir/ritonavir treatment on upper respiratory SARS-CoV-2 RT-PCR negative conversion rates among high-risk patients with COVID-19. Clin. Infect. Dis. 2022 doi: 10.1093/cid/ciac600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Boucau J., Uddin R., Marino C., Regan J., Flynn J.P., Choudhary M.C., et al. Characterization of virologic rebound following nirmatrelvir-ritonavir treatment for COVID-19. Clin. Infect. Dis. 2022 doi: 10.1093/cid/ciac512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Greasley S.E., Noell S., Plotnikova O., Ferre R., Liu W., Bolanos B., et al. Structural basis for the in vitro efficacy of nirmatrelvir against SARS-CoV-2 variants. J. Biol. Chem. 2022;298(6) doi: 10.1016/j.jbc.2022.101972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sathish J.G., Bhatt S., DaSilva J.K., Flynn D., Jenkinson S., Kalgutkar A.S., et al. Comprehensive Nonclinical Safety Assessment of Nirmatrelvir Supporting Timely Development of the SARS-COV-2 Antiviral Therapeutic, Paxlovid™. Int J. Toxicol. 2022;41(4):276–290. doi: 10.1177/10915818221095489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ganatra S., Dani S.S., Ahmad J., Kumar A., Shah J., Abraham G.M., et al. Oral Nirmatrelvir and Ritonavir in Non-hospitalized Vaccinated Patients with Covid-19. Clin. Infect. Dis. 2022 doi: 10.1093/cid/ciac673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Cai H., Yan J., Li P., Ding L., Liu S., Zhan Y., et al. Paxlovid for Treating Hospitalized COVID-19 Patients with Kidney Injury.
- 71.Jochmans D., Liu C., Donckers K., Stoycheva A., Boland S., Stevens S.K., et al. The Substitutions L50F, E166A, and L167F in SARS-CoV-2 3CLpro Are Selected by a Protease Inhibitor In Vitro and Confer Resistance To Nirmatrelvir. Mbio. 2023 doi: 10.1128/mbio.02815-22. e02815-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Fishbane S., Hirsch J.S., Nair V. Special considerations for paxlovid treatment among transplant recipients with SARS-CoV-2 infection. Am. J. Kidney Dis. 2022;79(4):480–482. doi: 10.1053/j.ajkd.2022.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Stock P.G., Henrich T.J., Segev D.L., Werbel W.A. Interpreting and addressing suboptimal immune responses after COVID-19 vaccination in solid-organ transplant recipients. J. Clin. Invest. 2021;131:14. doi: 10.1172/JCI151178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Nair V., Jandovitz N., Hirsch J.S., Nair G., Abate M., Bhaskaran M., et al. COVID-19 in kidney transplant recipients. Am. J. Transpl. 2020;20(7):1819–1825. doi: 10.1111/ajt.15967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Hsu A., Granneman G.R., Bertz R.J. Ritonavir. Clinical pharmacokinetics and interactions with other anti-HIV agents. Clin. Pharm. 1998;35(4):275–291. doi: 10.2165/00003088-199835040-00002. [DOI] [PubMed] [Google Scholar]
- 76.Mertz D., Battegay M., Marzolini C., Mayr M. Drug-drug interaction in a kidney transplant recipient receiving HIV salvage therapy and tacrolimus. Am. J. Kidney Dis. 2009;54(1):e1–e4. doi: 10.1053/j.ajkd.2009.01.268. [DOI] [PubMed] [Google Scholar]
- 77.Luo X., Zhu L.J., Cai N.F., Zheng L.Y., Cheng Z.N. Prediction of tacrolimus metabolism and dosage requirements based on CYP3A4 phenotype and CYP3A5(*)3 genotype in Chinese renal transplant recipients. Acta Pharm. Sin. 2016;37(4):555–560. doi: 10.1038/aps.2015.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Prikis M., Cameron A. Paxlovid (Nirmatelvir/Ritonavir) and tacrolimus drug-drug interaction in a kidney transplant patient with SARS-2-CoV infection: a case report. Transpl. Proc. 2022;54(6):1557–1560. doi: 10.1016/j.transproceed.2022.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Duffy E. Treat. COVID-19 Paxlovid Prim. care. 2022 [Google Scholar]
- 80.Haque O.I., Mahar S., Hussain S., Sloane P. Pharmacokinetic interaction between verapamil and ritonavir-boosted nirmatrelvir: implications for the management of COVID-19 in patients with hypertension. BMJ Case Rep. Cp. 2023;16(1) doi: 10.1136/bcr-2022-252677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Ganipisetti V.M., Bollimunta P., Maringanti S. Paxlovid-induced symptomatic bradycardia and syncope. Cureus. 2023;15:1. doi: 10.7759/cureus.33831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Zheng Q., Ma P., Wang M., Cheng Y., Zhou M., Ye L., et al. Efficacy and safety of paxlovid for COVID-19:a meta-analysis. J. Infect. 2023;86(1):66–117. doi: 10.1016/j.jinf.2022.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]