Abstract
The H9N2 avian influenza (AI) has become endemic in poultry in many countries since the 1990s, which has caused considerable economic losses in the poultry industry. Considering the long history of the low pathogenicity H9N2 AI in many countries, once H9N2 AI is introduced, it is more difficult to eradicate than high pathogenicity AI. Various preventive measures and strategies, including vaccination and active national surveillance, have been used to control the Y439 lineage of H9N2 AI in South Korea, but it took a long time for the H9N2 virus to disappear from the fields. By contrast, the novel Y280 lineage of H9N2 AI was introduced in June 2020 and has spread nationwide. This study reviews the history, genetic and pathogenic characteristics, and control strategies for Korean H9N2 AI. This review may provide some clues for establishing control strategies for endemic AIV and a newly introduced Y280 lineage of H9N2 AI in South Korea.
Keywords: Avian influenza, H9N2 virus, history, pathogenicity, vaccine
INTRODUCTION
H9N2 avian influenza viruses (AIVs) have spilled over from wild birds, their natural host, to domestic poultry. These viruses have become endemic in poultry in many countries since the 1990s. H9N2 AIVs can be broadly categorized into two major lineages: Eurasian and American. Eurasian H9N2 AIVs, in particular, have circulated in poultry and are classified further into several lineages: G1 (represented by A/quail/Hong Kong/G1/1997), Y280 (represented by A/duck/Hong Kong/Y280/1997; also known as the BJ94 or G9 lineage) and Y439 (represented by A/duck/Hong Kong/Y439/1997; also known as the Korean lineage) lineage [1,2,3].
The Y439 lineage of H9N2 AIV is a group originating from Eurasian wild birds, and it has been reported in many regions, including Europe and Asia [4]. In South Korea, the Y439 lineage of H9N2 AIV was first reported in chicken farms in 1996 and has since spread in poultry and become endemic since 2000s [5,6,7]. Outbreaks of the Y439 lineage of H9N2 AI decreased after vaccinating layer and broiler breeders since 2007. However, even after vaccination, the virus has continued to circulate mainly in Korean native chicken farms and live bird markets (LBM), there have been no reports since it was last detected in 2018 [8,9,10].
The G1 lineage of H9N2 AIV is the most widely distributed H9N2 AIV group in Asia, the Middle East, and Africa [11,12]. The lineage is divided into two sublineages according to the geographical distribution and genetic association: “G1-Eastern” and “G1-Western” [3,13,14]. Among them, the G1-Eastern lineage is endemic to poultry in southern China and neighboring Southeast Asian countries, Vietnam and Cambodia. On the other hand, the G1-Western is distributed across a wide range of regions, from Asia, including Bangladesh and India, to the Middle East and Africa [14].
The Y280 lineage of H9N2 AIV has become the dominant lineage in China since the mid-1990s and has evolved into sublineages (presented as BJ/94, HK/G9, and SH/F98) and various genotypes (A-W, G1-G81) [15,16,17]. This lineage is distributed in Asian countries, such as China, Vietnam, Cambodia, Indonesia, and Myanmar. In Vietnam, which borders China, the Y280 lineage of H9N2 AIV has circulated mainly in poultry since 2012 [4,18]. Recently, it was reported in Japan and eastern Russia, which does not border China [19,20]. In addition, the Y280 lineage of H9N2 AIV was first isolated from LBMs in South Korea in June 2020 and has spread rapidly nationwide [9,10].
Wild birds are the natural host of AIVs, but H9N2 AIV began to be reported in poultry, such as chicken, quail, guinea fowl and partridge in Asia [4], in the mid-1990s and has become endemic in poultry beyond the species barrier without pre-adaptation. The endemicity of Asian H9N2 AI in poultry has promoted the emergence of various novel AIVs and the evolution of H9 AIVs [2]. Infection of H9N2 AIV is an important issue for animal diseases and public health [21]. Previous studies have shown that H9N2 AIVs donated internal gene sets to other human infecting viruses, including H5N1, H5N6, H7N9, and H10N8 [22,23,24,25,26,27].
In South Korea, the Y439 lineage of H9N2 AIV, which occurred for a long period, has not been reported since 2018, but the Y280 lineage of H9N2 AI was newly introduced in 2020. Considering the history of the endemicity of the H9N2 AI in many countries, including South Korea, once H9N2 AI is introduced, it is more difficult to eradicate than high pathogenicity avian influenza (HPAI). This study reviewed the history of Korean H9N2 AI, the genetic and pathogenic characteristics of H9N2 AIVs, and the control strategies, including vaccination in South Korea.
HISTORY AND CURRENT SITUATION OF Y280 LINEAGE OF H9N2 AIV IN ASIA
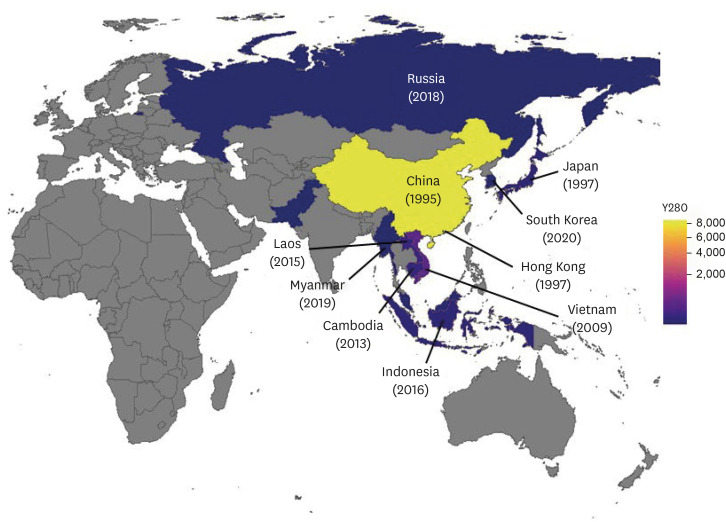
Since the mid-1990s, the Y280 lineage of H9N2 AIV has become the dominant strain and circulated in chickens in China [2,28,29]. From 1995 to June 2022, 8,968 cases of the Y280 lineage of H9N2 AIVs were isolated worldwide (Supplementary Fig. 1, excluding mammal infections, based on the Global Initiative for Sharing All Influenza Data [https://www.gisaid.org/]). Of these, 8,311 cases, approximately 92.7%, were reported in China, where the outbreaks have increased dramatically since 2009. Although vaccination programs for chickens have been in place for a long time in China [17,27,29], the Y280 lineage of H9N2 AIV has been endemic to poultry and has increased the genetic diversity of the virus due to the high proportion of traditional small-scale mixed breeding and the preference for fresh poultry trading through the LBMs [30,31]. According to Gu et al. [27], at least 23 genotypes of Y280 lineage of H9N2 AIV isolated in China from 1994 to 2014 were identified, of which three types were suggested to be major genotypes: A, H, and S. In particular, genotype S is a reassortant of the PB2 and M genes of the G1 lineage of H9N2 AIV based on the gene constellation of the Y280 lineage of H9N2 AIV and has become dominant in China since 2010 [32]. The Y280 lineage of H9N2 AIV, which was almost restricted to China before 2010, has spread to other Asian countries, including Vietnam, Cambodia, Indonesia, and recently South Korea (Fig. 1) [4,9,18,33,34].
Fig. 1. As of July 2022, the global geographical distribution Y280 lineage of H9N2 avian influenza, including the first reporting year by major Asian countries (square) and the number of genetic information of Y280 H9N2 (yellow to deep blue), uploaded to the Global Initiative for Sharing All Influenza Data database (the hemagglutinin gene sequence collected from January 1995 to June 2022, http://gisaid.org/).
In Hong Kong, the Y280 lineage of H9N2 AIV was first isolated in 1997, and some cases subsequently occurred in poultry and humans. Since the early 2000s, cases of H9N2 AI infection have been reported sporadically until recently [1,35,36]. Interestingly, in Japan, the Y280 lineage of H9N2 AIV was first isolated in imported chicken meat products collected in 1997, 2001, and 2002. In addition, in 2015–2016, H9N2 AIVs were isolated in illegally imported poultry products by flight passengers from China and Taiwan into Japan during the quarantine process [19,37].
In Vietnam and Cambodia, Y280 H9N2 AI was reported in 2009 and 2013. Since then, the Y280 lineage of H9N2 AIV has become dominant in poultry, mainly in LBMs [38,39,40]. The Y280 lineage of H9N2 AIV of the two countries was genetically closely related to the strain in China. These viruses may have flowed into adjacent countries locally through active poultry trading [19,33,40].
Since the mid-2010s, the Y280 lineage of H9N2 AI has been spreading further in Southeast Asia, and the virus has also been identified in Myanmar, Indonesia, and Laos (Fig. 1, Supplementary Fig. 1). These viruses are genetically closely related to the Y280 lineage of H9N2 AIV in China [41,42,43]. In addition, The H9N2 AIV was first identified in Russia in 2012 but was not defined genetically. Later, in 2018, the Y280 lineage of H9N2 AIV was isolated at a poultry farm in Primorsky Krai, Far East region of Russia, and was found to be genetically related to that isolated in Tajikistan, Central Asia [20].
Long-distance migrating wild birds, as shown in the high pathogenicity H5 AIV, are one of the factors of AIV transmission and spread [44,45,46,47]. In China, there have been several sporadic reports of H9N2 AIV detection in wild birds since 2010 [48,49,50,51,52]. Most H9N2 AIVs in wild birds have been identified as the Eurasian aquatic origin, but some cases were North American and poultry-derived G1 or Y280 lineages. Thus far, there is no direct evidence that the Y280 lineage of H9N2 AIV has been transmitted between countries or continents by wild birds, despite the surveillance programs conducted in several countries [9,53,54,55]. Although the detection of poultry-derived H9N2 AIV in wild birds was limited, the virus can be disseminated by wild migratory birds if this virus acquires more adaptability to wild waterfowl.
Considering the spread of the H9N2 AIV in neighboring countries of China and the detection of H9N2 AIV through the quarantine process in Japan, the Y280 lineage of H9N2 AIV could be transmitted by the movement of contaminated poultry products, people, or goods [56,57]. Another transmission factor, the LBM, is a central point in generating and spreading novel viruses to other species due to the high prevalence and genetic diversity of H9N2 AIV and should be considered a hotspot for surveillance programs [18,30,58].
THE OUTBREAK AND GENOTYPE OF H9N2 AI IN SOUTH KOREA
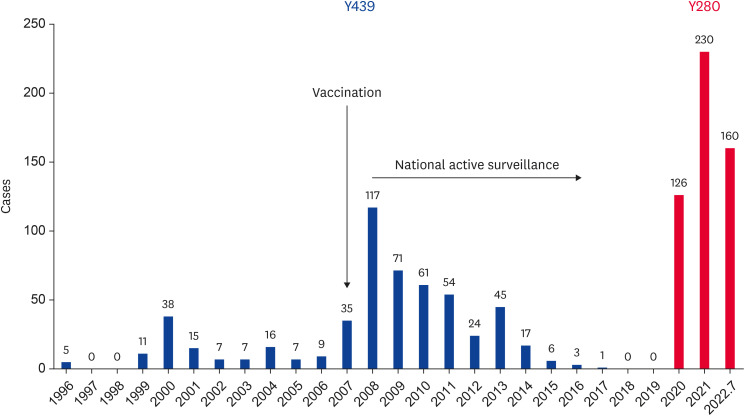
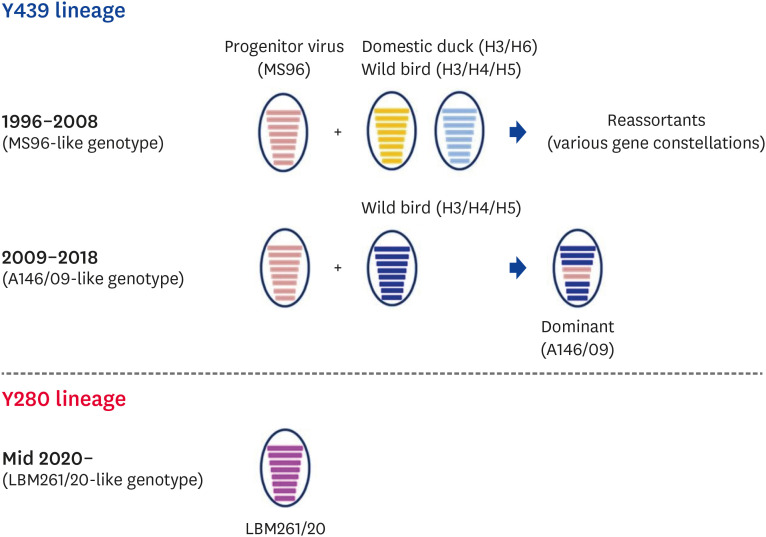
Since the first outbreak of H9N2 in South Korea in 1996, the Y439 lineage of the H9N2 virus has been endemic since the 2000s (Fig. 2). Nationwide outbreaks of H9N2 AI, which have caused considerable economic losses, have led to the use of vaccination programs since 2007 [7]. Since then, the outbreaks of H9N2 AI in poultry farms, such as layers and breeders, have decreased gradually, but the virus was not completely eradicated and was circulated continuously, mainly in Korean native chickens in LBM, until 2018 (data not shown). The Y439 lineage of H9N2 AIV, which has circulated in South Korea for a long time, has continuously evolved by antigenic drift and reassortment with other AIVs from wild birds and domestic ducks in LBMs [5,59,60]. The Y439 lineage of H9N2 AIV in South Korea is divided broadly into two genotypes according to their gene constellation (Fig. 3). The first is the MS96-like genotype, represented by A/chicken/Korea/MS96/1996 (H9N2) and its reassortant viruses with the genes from domestic ducks and wild bird origin, which was distributed in poultry until 2008 (designated as K1, K2, and K3 genotype in Youk et al. [59]). Second, the A146/09-like genotype, represented by A/chicken/Korea/A146/2009 (H9N2), is a reassortant of the hemagglutinin (HA) and nucleoprotein genes of the MS96-like virus with six internal genes originating from wild aquatic birds; this strain has become the dominant strain (designated as K4 genotype in Youk et al. [59]). In South Korea, the national active surveillance program was established for HPAI control, and various measures have been applied, including movement restriction, disinfection, and the culling of infected animals since 2008 (Fig. 4). These preventive measures may play a role in reducing the low pathogenicity avian influenza (LPAI) virus and HPAI virus, particularly in domestic ducks and LBM. Consequently, the emergency of reassortant viruses has been reduced, and finally, the Y439 lineage of H9N2 AIV has disappeared in South Korea since 2018.
Fig. 2. Number of H9N2 avian influenza (Y439 and Y280 lineage) outbreak cases in South Korea from 1996 to July 2022. Before 2007, data was collected from avian disease pathological diagnosis reports in Animal and Plant Quarantine Agency were used, and from 2008, updated from National Animal Disease Statistics in Korea Animal Health Integrated System.
Fig. 3. Illustrative scheme of the gene constellation from Korean representative H9N2 viruses (Y439 and Y280 lineage) depending on their epidemic year. The eight horizontal bars in circle (from top to bottom) represent PB2, PB1, PA, HA, NP, NA, M, and NS genes, respectively.
LBM, live bird market.
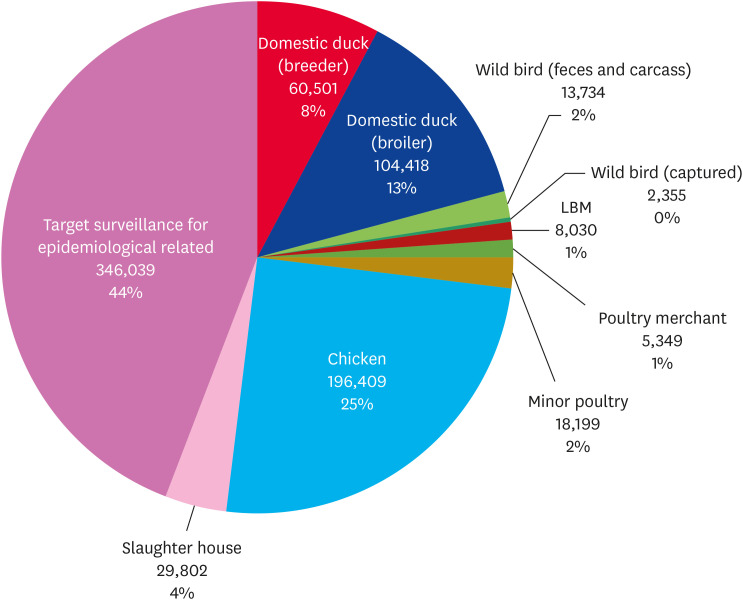
Fig. 4. AI national active surveillance and monitoring results of South Korea in 2021. A total of 784,836 laboratory diagnostic tests were conducted annually in 10 categories, including domestic chickens, ducks, and LBMs (Annual Report on Avian Influenza Surveillance Results for 2021, Animal and Plant Quarantine Agency).
LBM, live bird market.
In June 2020, the Y280 lineage of H9N2 AIV was first isolated from Korean native chickens in LBM by active surveillance programs, and has since spread nationwide (Fig. 2). A/chicken/Korea/LBM261/2020 (H9N2), which was the virus of the index case in South Korea, was closely related to the A/chicken/Shandong/1844/2019 (H9N2) virus of China. The Korean Y280 lineage of H9N2 AIV is designated as the LBM261/20-like genotype, which belongs to a subgroup of genotype S in China (Fig. 3) [9]. Five hundred sixteen cases of the Korean Y280 lineage of H9N2 AI have been detected nationwide in various breeds, such as Korean native chickens, layer and broiler chickens by active surveillance of domestic poultry from June 2020 to July 2022 (Fig. 2). Although the route of introduction of the novel H9N2 AIV into South Korea remains unclear, the likelihood of introduction by wild migratory birds is considered low. This is because the poultry-derived Y280 lineage of H9N2 AIV in wild birds is rarely reported even in China [48,49,50,51,52], and there is no virus isolation in wild birds, including feces, captive birds, carcass through intensive active surveillance in South Korea. Therefore, the virus is likely to be introduced through contaminated poultry products or human activities, as shown in the periodical AIV detection cases in the quarantine process in Japan [9].
PATHOGENIC CHARACTERISTICS OF H9N2 AIV IN CHICKENS AND DUCKS
Although H9N2 AIV is classified as a low pathogenicity virus in poultry, it is causing economic damage to the poultry industry by the decrease in spawning and some mortality rates in commercial chickens. Most chickens infected with H9N2 AIV at farms showed typical signs of influenza, such as respiratory symptoms, egg drop, and mortality (0% to 40%) (summarized in Table 1) [2,5,61,62]. On the other hand, experimental infections in specific-pathogen-free chickens showed no mortality and only mild symptoms, such as depression and decreased feed intake [5,19,63,64,65,66]. This disparity between laboratory and field infections with H9N2 AIV suggested that the pathogenicity of H9N2 AIV can vary depending on ages, breeds, the level of immunity, and another secondary opportunistic pathogen infection [5,64,67,68,69].
Table 1. Summary of clinical signs of H9N2 viruses from the Y439 and Y280 lineage in farms and animal experiments, respectively.
| Cases | Species | Y439 lineage | Y280 lineage | ||
|---|---|---|---|---|---|
| Clinical signs | Reference | Clinical signs | Reference | ||
| Field (farm) | Chicken (commercial) | Egg drop, respiratory sign, depression, diarrhea, weight loss, decreased feed intake, mortality (0%–30%) | [2,5,61,63] | Egg drop, respiratory signs (coughing, sneezing, gasping), mortality (10%–40%) | [1,2,62] |
| Animal experiment | Chicken (SPF) | No mortality, depression | [5,63,65,66] | No mortality, depression, diarrhea, decreased feed intake | [16,17,19,64,66,71,89,97,102] |
| Viral shedding: higher titer via CL route | Viral shedding: higher titer via OP route | ||||
| Mice | Mostly no clinical signs and mortality, weight loss, inappetence | [5,92,107] | Inappetence, huddling, ruffled fur, labored breathing, hunched posture, respiratory distress, weight loss, mortality (0%–30%) | [2,36,52,82,83,89,91,92,93] | |
Bold: observed major clinical symptoms.
SPF, specific-pathogen-free; OP, oropharyngeal; CL, cloacal.
Previous studies have shown that similar clinical signs were observed in infection between the Y439 and Y280 lineage of H9N2 AIVs (Table 1). In the viral shedding, however, there was a significant difference in the preferential replication between the two viruses. The Y280 lineage of H9N2 AIV was replicated more efficiently in the respiratory tract, while the Y439 lineage of H9N2 AIV was replicated more efficiently in the intestinal tract [5,19,65,66]. Thus, the Y280 lineage of H9N2 AIV can be transmitted airborne more efficiently via the oral-to-oral pathway than the Y439 lineage of H9N2 AIV. This feature can cause a more efficiently spread virus between poultry in the same space. It can be a risk factor that increases the chance of viral infection even between species in contact with infected poultry [2,70,71].
Domestic ducks are intermediate species between poultry and wild waterfowl and have susceptibility and resistance to AIVs [72,73,74]. Experiments with H9N2 AIV infections in domestic ducks are limited, but the results show that most infected ducks were asymptomatic [2,66,75,76]. In addition, viral replication was not detected in most infected ducks and was identified as low titers in oropharyngeal (OP) and cloacal (CL) swabs from a few infected ducks. According to Wang et al. [76], it was confirmed that the genotype S of the Y280 lineage of H9N2 AIV could replicate with relatively high titers in the respiratory tract of the Muscovy duck. Despite the limited cases, some experimental results have shown viral replication of H9N2 AIV in ducks. If the chicken-adapted H9N2 AIV replicates more efficiently in ducks, it can be a potential risk factor in AIV transmission by domestic ducks and wild migratory ducks.
HUMAN INFECTION BY H9N2 AIV
Human infection by the H9N2 AIV was first reported in Hong Kong in 1998 [4]. Since then, sporadic cases have been reported continuously in various countries, mainly China. As of June 2022, 112 cases have been identified in eight countries, including China, Egypt, Bangladesh, and Cambodia. Cases of infection have been reported mainly in people in close contact with infected poultry and meats or exposed to contaminated environments [77,78]. Children under the age of 10 were most infected with H9N2 AIV but developed mild symptoms [79]. On the other hand, the H9N2 AIV is closely involved in other fatal human infections as well as direct infections. The high pathogenicity H5N1 AIV in Hong Kong in 1997 was found to have reassorted from six internal genes of the G1 lineage of H9N2 AIV, excluding HA and neuraminidase [22,80]. In addition, the internal genes of H7N9 AIV, which has 1,568 human infections, including 616 fatal cases (case fatality rate, 39%) in China since 2013, originated from the Y280 lineage of H9N2 AI [23].
Poultry-adapted AIVs exhibit asymptomatic or weak signs and can evolve as potential infection sources in mammals through circulation in poultry [81,82,83]. The HA protein of AIV is determined in the host range by binding with sialic acid on the surface of the host cell. In general, AIV has the highest binding affinity with the α2,3-linked sialic acid of birds, but mutations on the receptor binding sites for high affinity with α2,6-linked sialic acid have been found to increase infectivity in mammals [84,85]. Previous studies reported that leucine (L) in position 226 of the HA proteins plays an important role in the binding affinity to sialic acid as a representative mammalian affinity marker [86,87]. Thus far, the human infection cases by H9N2 AIV were only reported in Y280 and G1 lineages, most of which have a Q226L substitution on the HA protein. In addition, the genotype S of the Y280 lineage, which has been dominant in poultry in China since 2010, has acquired various mammalian affinity markers: H183N, T190V, and Q226L in the HA protein; A588V in the PB2 protein; K356R and S409N in the PA protein; V15I in the M1 protein; I28V and L55F in M2 protein [4,65,88,89,90,91,92]. Newly introduced H9N2 AIV into South Korea in 2020 belonged to genotype S of the Y280 lineage of H9N2 AIV, which has similar genetic characteristics [9].
Although the Y439 lineage of H9N2 AIV had circulated for a long period (1996–2018), there have been no human infection cases in South Korea (Fig. 2). The Korean Y439 lineage of H9N2 AIV had retained poultry affinity markers rather than mammals [9,86]. In mouse experiments, the Y280 lineage of H9N2 AIV replicated well in the respiratory tract of infected mice without adaptation and showed various clinical signs, body weight loss, and mortality, whereas the Y439 lineage of H9N2 AIV showed mostly no clinical signs or mild symptoms, such as inappetence and weight loss (Table 1) [5,89,90,91,92,93]. These results show that the Y439 lineage of H9N2 AIV is at least less lethal in mammalian infections than the Y280 lineage of H9N2 AIV.
CONTROL STRATEGIES OF H9N2 AI IN SOUTH KOREA
National active surveillance for AI has been conducted since 2008 to monitor HPAI in South Korea. Although there is a slight difference annually, 784,836 laboratory diagnostic tests were conducted in 2021 (Fig. 4). The main targets of active surveillance were domestic chickens (approximately 25%), domestic ducks (approximately 21%), wild birds (approximately 2%), LBMs and poultry traders (approximately 2%), and the epidemiological-related places with HPAI outbreaks (approximately 44%). Surveillance has been applied to wild birds for an early warning of HPAI introduction, including fecal samples, captive wild birds, and carcasses. High pathogenicity H5Nx AIVs have been detected in wild birds at the early time of migration before poultry outbreaks [94,95]. Domestic ducks are considered an important target of active surveillance because they can be a potential viral transmission factor, despite not showing clinical symptoms when infected with HPAIV [72]. LBM, which has a high risk of viral transmission by live bird trading, is one of the main targets of surveillance [18,30,58]. For effective control of AIV, surveillance has also been conducted in the place of poultry merchants and farms related to LBMs. Through the surveillance of LBM, the introduction of the Y280 lineage of H9N2 AIV into South Korea was also found [9]. Overall, intensive national active surveillance and followed control measures, such as disinfection, restriction of movement, ban of poultry trading, and stamping out of HPAI-infected birds, have gradually reduced LPAI as well as HPAI in South Korea. Therefore, active surveillance programs are essential to monitor the emergence of new viruses and to control the spread of the viruses in the early stages after detection.
As a preventive measure, vaccination has been used to control H9N2 AI in many countries, particularly in endemic regions. China has implemented vaccination programs for H9N2 AI on chicken farms since 1998 [65,96]. On the other hand, the H9N2 AI still has a high prevalence in China (Supplementary Fig. 1). Moreover, the long-term circulation of the H9N2 AIV in a vaccinated population has caused many virus mutations [17,97,98,99,100,101,102]. This is considered to have been compositely caused by factors, such as inefficient application of vaccines, low doses, low vaccination coverage, and limited updates of vaccine strains [98,100,103]. At least 20 commercial vaccines have been used in China to cope with various viruses, which need to be updated regularly [97,98,101].
The H9N2 AIV has been prevalent nationwide in South Korea since 2000 but officially reported outbreaks were limited (Fig. 2) [7]. Therefore, since 2007, Korean animal health authorities have permitted the use of H9N2 vaccines, which use a single vaccine strain (A/chicken/Korea/01310/2001) of the Y439 lineage of H9N2 AI in layer and breeder chicken to prevent damage to the poultry industry [104,105]. Although outbreaks of the Y439 lineage of H9N2 AI have decreased since the vaccine program, it took more than a decade to disappear from the field (Fig. 2). The H9N2 AIV had remained especially in LBMs and small-scale Korean native chicken farms for a long time. This fact suggests a limit to controlling the H9N2 AI with vaccination alone.
Another factor to consider in vaccination strategy is the possibility of virus mutations and the need to update the vaccine strain. Immune pressure by long-term vaccination may cause genetic and antigenic changes, as shown in China and South Korea [8,28,59,65,101,106,107]. This leads to a gradual decrease in the suitability of vaccine strain and vaccine efficacy in the field. Although the vaccine strain for the Y439 lineage of H9N2 AIV has never been updated in South Korea, but depending on the situation in which the Y439 lineage of H9N2 AI is circulated in poultry again, it will be necessary to update the vaccine strain by the genetic and antigenic characteristics of the field virus.
Unfortunately, as the Y280 lineage of H9N2 AIV was newly introduced into South Korea in 2020, previously authorized vaccines against the Y439 lineage of H9N2 AIV may not be an appropriate option to control the current Y280 lineage of H9N2 AIV because of the difference in the genetic and antigenic features (81.8% nucleotide similarity) [108]. In animal experiments, the Y439 lineage of the vaccine showed only limited efficacy to heterogeneous Y280 lineage of H9N2 AIV (Y439 lineage of the vaccine reduced the replication of the Y280 lineage of H9N2 AIV in the cecal tonsils by 37.5%, and also partially inhibits viral shedding in respiratory and intestinal tracts) (Fig. 5). By contrast, the rgHS314 virus (derived from A/chicken/Korea/H314/2020), which was newly developed as an autogenous vaccine for the current epizootic Y280 lineage of H9N2 AIV, can reduce viral replication significantly with 100% inhibition of virus recovery in the cecal tonsil and no viral shedding in OP and CL swabs (Fig. 5) [108]. New commercial vaccines using the Y280 lineage of the H9N2 vaccine seed strain may be available in the field in the first half of 2023. However, active surveillance and enhanced biosecurity levels must be combined with vaccination to control the H9N2 AI effectively [86,103].
Fig. 5. Assessment of the protective efficacy of the commercial Y439 vaccine and newly developed Y280 vaccine (used homologous strain, A/chicken/Korea/H314/2020) when challenged with the Y280 H9N2 virus. In an animal experiment, the commercial Y439 vaccine has been found only partially to inhibit viral replication and shedding and has been shown to provide incomplete protection against the Y280 H9N2 virus [108].
AIV, avian influenza virus.
CONCLUSION
This report provides an overview of the history of outbreaks and the control strategies for H9N2 AI in South Korea. Unlike many endemic countries, including China, where new variants of H9N2 AIV are emerging by genetic mutations, in South Korea, the Y439 lineage of H9N2 AI has disappeared by effective control measures, such as continued large-scale national surveillance, improved levels of biosecurity, appropriate vaccination, and culling of poultry in the case of HPAI. Therefore, in order to control the new Korean Y280 lineage of H9N2 AI, measures such as updating vaccine strain, organizing surveillance based on the potential risks of H9N2 AI (breeds and prevalence rate, etc.) and strengthening follow-up monitoring of LBM's supply farms and distribution networks are urgently needed. These intensive measures and strategies will help control the Y280 lineage of H9N2 AI as soon as possible. This review paper is expected to assist in establishing control strategies and provide insight for low pathogenicity H9N2 AI in endemic countries.
Footnotes
Funding: This work was supported by the Animal and Plant Quarantine Agency, Republic of Korea (grant numbers B-1543418-2022-24-01).
Conflict of Interest: The authors declare no conflicts of interest.
- Conceptualization: Lee YJ.
- Data curation: Sagong M, Lee KN, Lee EK, Kang H.
- Formal analysis: Sagong M.
- Investigation: Sagong M, Lee KN, Lee EK, Kang H.
- Methodology: Sagong M.
- Project administration: Lee YJ.
- Resources: Sagong M, Lee KN, Lee EK, Kang H.
- Supervision: Choi YK, Lee YJ.
- Validation: Lee YJ.
- Visualization: Sagong M.
- Writing - original draft: Sagong M.
- Writing - review & editing: Choi YK, Lee YJ.
SUPPLEMENTARY MATERIAL
Y280 lineage H9 subtype virus detection graph by country and year (total 8,967 cases based on the GISAID database).
References
- 1.Guan Y, Shortridge KF, Krauss S, Webster RG. Molecular characterization of H9N2 influenza viruses: were they the donors of the “internal” genes of H5N1 viruses in Hong Kong? Proc Natl Acad Sci U S A. 1999;96(16):9363–9367. doi: 10.1073/pnas.96.16.9363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Guo YJ, Krauss S, Senne DA, Mo IP, Lo KS, Xiong XP, et al. Characterization of the pathogenicity of members of the newly established H9N2 influenza virus lineages in Asia. Virology. 2000;267(2):279–288. doi: 10.1006/viro.1999.0115. [DOI] [PubMed] [Google Scholar]
- 3.Dong G, Luo J, Zhang H, Wang C, Duan M, Deliberto TJ, et al. Phylogenetic diversity and genotypical complexity of H9N2 influenza A viruses revealed by genomic sequence analysis. PLoS One. 2011;6(2):e17212. doi: 10.1371/journal.pone.0017212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Peacock TH, James J, Sealy JE, Iqbal M. A global perspective on H9N2 avian influenza virus. Viruses. 2019;11(7):620. doi: 10.3390/v11070620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lee YJ, Shin JY, Song MS, Lee YM, Choi JG, Lee EK, et al. Continuing evolution of H9 influenza viruses in Korean poultry. Virology. 2007;359(2):313–323. doi: 10.1016/j.virol.2006.09.025. [DOI] [PubMed] [Google Scholar]
- 6.Lee DH, Song CS. H9N2 avian influenza virus in Korea: evolution and vaccination. Clin Exp Vaccine Res. 2013;2(1):26–33. doi: 10.7774/cevr.2013.2.1.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mo IP, Bae YJ, Lee SB, Mo JS, Oh KH, Shin JH, et al. Review of avian influenza outbreaks in South Korea from 1996 to 2014. Avian Dis. 2016;60(1) Suppl:172–177. doi: 10.1637/11095-041715-Review. [DOI] [PubMed] [Google Scholar]
- 8.Lee DH, Fusaro A, Song CS, Suarez DL, Swayne DE. Poultry vaccination directed evolution of H9N2 low pathogenicity avian influenza viruses in Korea. Virology. 2016;488:225–231. doi: 10.1016/j.virol.2015.11.023. [DOI] [PubMed] [Google Scholar]
- 9.Heo GB, Kye SJ, Sagong M, Lee EK, Lee KN, Lee YN, et al. Genetic characterization of H9N2 avian influenza virus previously unrecognized in Korea. J Vet Sci. 2021;22(2):e21. doi: 10.4142/jvs.2021.22.e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Youk S, Cho AY, Lee DH, Jeong S, Kim YJ, Lee S, et al. Detection of newly introduced Y280-lineage H9N2 avian influenza viruses in live bird markets in Korea. Transbound Emerg Dis. 2022;69(2):881–885. doi: 10.1111/tbed.14014. [DOI] [PubMed] [Google Scholar]
- 11.Davidson I, Fusaro A, Heidari A, Monne I, Cattoli G. Molecular evolution of H9N2 avian influenza viruses in Israel. Virus Genes. 2014;48(3):457–463. doi: 10.1007/s11262-014-1037-0. [DOI] [PubMed] [Google Scholar]
- 12.El Houadfi M, Fellahi S, Nassik S, Guérin JL, Ducatez MF. First outbreaks and phylogenetic analyses of avian influenza H9N2 viruses isolated from poultry flocks in Morocco. Virol J. 2016;13(1):140. doi: 10.1186/s12985-016-0596-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nagy A, Mettenleiter TC, Abdelwhab EM. A brief summary of the epidemiology and genetic relatedness of avian influenza H9N2 virus in birds and mammals in the Middle East and North Africa. Epidemiol Infect. 2017;145(16):3320–3333. doi: 10.1017/S0950268817002576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Carnaccini S, Perez DR. H9 influenza viruses: an emerging challenge. Cold Spring Harb Perspect Med. 2020;10(6):a038588. doi: 10.1101/cshperspect.a038588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gu M, Xu L, Wang X, Liu X. Current situation of H9N2 subtype avian influenza in China. Vet Res. 2017;48(1):49. doi: 10.1186/s13567-017-0453-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Choi YK, Ozaki H, Webby RJ, Webster RG, Peiris JS, Poon L, et al. Continuing evolution of H9N2 influenza viruses in Southeastern China. J Virol. 2004;78(16):8609–8614. doi: 10.1128/JVI.78.16.8609-8614.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li C, Yu K, Tian G, Yu D, Liu L, Jing B, et al. Evolution of H9N2 influenza viruses from domestic poultry in Mainland China. Virology. 2005;340(1):70–83. doi: 10.1016/j.virol.2005.06.025. [DOI] [PubMed] [Google Scholar]
- 18.Sealy JE, Fournie G, Trang PH, Dang NH, Sadeyen JR, Thanh TL, et al. Poultry trading behaviours in Vietnamese live bird markets as risk factors for avian influenza infection in chickens. Transbound Emerg Dis. 2019;66(6):2507–2516. doi: 10.1111/tbed.13308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shibata A, Hiono T, Fukuhara H, Sumiyoshi R, Ohkawara A, Matsuno K, et al. Isolation and characterization of avian influenza viruses from raw poultry products illegally imported to Japan by international flight passengers. Transbound Emerg Dis. 2018;65(2):465–475. doi: 10.1111/tbed.12726. [DOI] [PubMed] [Google Scholar]
- 20.Zinyakov NG, Sosipatorova VY, Andriyasov AV, Ovchinnikova EV, Nikonova ZB, Kozlov AA, et al. Genetic analysis of genotype G57 H9N2 avian influenza virus isolate A/chicken/Tajikistan/2379/2018 recovered in Central Asia. Arch Virol. 2021;166(6):1591–1597. doi: 10.1007/s00705-021-05011-3. [DOI] [PubMed] [Google Scholar]
- 21.World Health Organization. Avian Influenza Weekly Update Number 853 -15 July 2022. Geneva: World Health Organization; 2022. [Google Scholar]
- 22.Guan Y, Shortridge KF, Krauss S, Chin PS, Dyrting KC, Ellis TM, et al. H9N2 influenza viruses possessing H5N1-like internal genomes continue to circulate in poultry in southeastern China. J Virol. 2000;74(20):9372–9380. doi: 10.1128/jvi.74.20.9372-9380.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lam TT, Wang J, Shen Y, Zhou B, Duan L, Cheung CL, et al. The genesis and source of the H7N9 influenza viruses causing human infections in China. Nature. 2013;502(7470):241–244. doi: 10.1038/nature12515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen H, Yuan H, Gao R, Zhang J, Wang D, Xiong Y, et al. Clinical and epidemiological characteristics of a fatal case of avian influenza A H10N8 virus infection: a descriptive study. Lancet. 2014;383(9918):714–721. doi: 10.1016/S0140-6736(14)60111-2. [DOI] [PubMed] [Google Scholar]
- 25.Shen YY, Ke CW, Li Q, Yuan RY, Xiang D, Jia WX, et al. Novel Reassortant Avian Influenza A(H5N6) Viruses in Humans, Guangdong, China, 2015. Emerg Infect Dis. 2016;22(8):1507–1509. doi: 10.3201/eid2208.160146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang M, Zhang X, Xu K, Teng Q, Liu Q, Li X, et al. Characterization of the pathogenesis of H10N3, H10N7, and H10N8 subtype avian influenza viruses circulating in ducks. Sci Rep. 2016;6(1):34489. doi: 10.1038/srep34489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gu M, Xu L, Wang X, Liu X. Current situation of H9N2 subtype avian influenza in China. Vet Res. 2017;48(1):49. doi: 10.1186/s13567-017-0453-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xu KM, Smith GJ, Bahl J, Duan L, Tai H, Vijaykrishna D, et al. The genesis and evolution of H9N2 influenza viruses in poultry from southern China, 2000 to 2005. J Virol. 2007;81(19):10389–10401. doi: 10.1128/JVI.00979-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sun Y, Pu J, Jiang Z, Guan T, Xia Y, Xu Q, et al. Genotypic evolution and antigenic drift of H9N2 influenza viruses in China from 1994 to 2008. Vet Microbiol. 2010;146(3-4):215–225. doi: 10.1016/j.vetmic.2010.05.010. [DOI] [PubMed] [Google Scholar]
- 30.Chen LJ, Lin XD, Guo WP, Tian JH, Wang W, Ying XH, et al. Diversity and evolution of avian influenza viruses in live poultry markets, free-range poultry and wild wetland birds in China. J Gen Virol. 2016;97(4):844–854. doi: 10.1099/jgv.0.000399. [DOI] [PubMed] [Google Scholar]
- 31.Chen LJ, Lin XD, Tian JH, Liao Y, Ying XH, Shao JW, et al. Diversity, evolution and population dynamics of avian influenza viruses circulating in the live poultry markets in China. Virology. 2017;505:33–41. doi: 10.1016/j.virol.2017.02.009. [DOI] [PubMed] [Google Scholar]
- 32.Gu M, Chen H, Li Q, Huang J, Zhao M, Gu X, et al. Enzootic genotype S of H9N2 avian influenza viruses donates internal genes to emerging zoonotic influenza viruses in China. Vet Microbiol. 2014;174(3-4):309–315. doi: 10.1016/j.vetmic.2014.09.029. [DOI] [PubMed] [Google Scholar]
- 33.Jonas M, Sahesti A, Murwijati T, Lestariningsih CL, Irine I, Ayesda CS, et al. Identification of avian influenza virus subtype H9N2 in chicken farms in Indonesia. Prev Vet Med. 2018;159:99–105. doi: 10.1016/j.prevetmed.2018.09.003. [DOI] [PubMed] [Google Scholar]
- 34.Karlsson EA, Horm SV, Tok S, Tum S, Kalpravidh W, Claes F, et al. Avian influenza virus detection, temporality and co-infection in poultry in Cambodian border provinces, 2017-2018. Emerg Microbes Infect. 2019;8(1):637–639. doi: 10.1080/22221751.2019.1604085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Saito T, Lim W, Suzuki T, Suzuki Y, Kida H, Nishimura SI, et al. Characterization of a human H9N2 influenza virus isolated in Hong Kong. Vaccine. 2001;20(1-2):125–133. doi: 10.1016/s0264-410x(01)00279-1. [DOI] [PubMed] [Google Scholar]
- 36.Choi YK, Ozaki H, Webby RJ, Webster RG, Peiris JS, Poon L, et al. Continuing evolution of H9N2 influenza viruses in Southeastern China. J Virol. 2004;78(16):8609–8614. doi: 10.1128/JVI.78.16.8609-8614.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mase M, Eto M, Imai K, Tsukamoto K, Yamaguchi S. Characterization of H9N2 influenza A viruses isolated from chicken products imported into Japan from China. Epidemiol Infect. 2007;135(3):386–391. doi: 10.1017/S0950268806006728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nomura N, Sakoda Y, Endo M, Yoshida H, Yamamoto N, Okamatsu M, et al. Characterization of avian influenza viruses isolated from domestic ducks in Vietnam in 2009 and 2010. Arch Virol. 2012;157(2):247–257. doi: 10.1007/s00705-011-1152-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Thuy DM, Peacock TP, Bich VT, Fabrizio T, Hoang DN, Tho ND, et al. Prevalence and diversity of H9N2 avian influenza in chickens of Northern Vietnam, 2014. Infect Genet Evol. 2016;44:530–540. doi: 10.1016/j.meegid.2016.06.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Suttie A, Tok S, Yann S, Keo P, Horm SV, Roe M, et al. The evolution and genetic diversity of avian influenza A(H9N2) viruses in Cambodia, 2015 - 2016. PLoS One. 2019;14(12):e0225428. doi: 10.1371/journal.pone.0225428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lin TN, Nonthabenjawan N, Chaiyawong S, Bunpapong N, Boonyapisitsopa S, Janetanakit T, et al. Influenza A(H9N2) virus, Myanmar, 2014-2015. Emerg Infect Dis. 2017;23(6):1041–1043. doi: 10.3201/eid2306.161902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Novianti AN, Rahardjo K, Prasetya RR, Nastri AM, Dewantari JR, Rahardjo AP, et al. Whole-genome sequence of an avian influenza A/H9N2 virus isolated from an apparently healthy chicken at a live-poultry market in Indonesia. Microbiol Resour Announc. 2019;8(17):e01671-18. doi: 10.1128/MRA.01671-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nugroho CM, Silaen OS, Kurnia RS, Soejoedono RD, Poetri ON, Soebandrio A. Isolation and molecular characterization of the hemagglutinin gene of H9N2 avian influenza viruses from poultry in Java, Indonesia. J Adv Vet Anim Res. 2021;8(3):423–434. doi: 10.5455/javar.2021.h530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Capua I, Alexander DJ. Avian influenza infections in birds--a moving target. Influenza Other Respi Viruses. 2007;1(1):11–18. doi: 10.1111/j.1750-2659.2006.00004.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Capua I, Alexander DJ. Avian influenza infection in birds: a challenge and opportunity for the poultry veterinarian. Poult Sci. 2009;88(4):842–846. doi: 10.3382/ps.2008-00289. [DOI] [PubMed] [Google Scholar]
- 46.Sakoda Y, Sugar S, Batchluun D, Erdene-Ochir TO, Okamatsu M, Isoda N, et al. Characterization of H5N1 highly pathogenic avian influenza virus strains isolated from migratory waterfowl in Mongolia on the way back from the southern Asia to their northern territory. Virology. 2010;406(1):88–94. doi: 10.1016/j.virol.2010.07.007. [DOI] [PubMed] [Google Scholar]
- 47.Jeong J, Kang HM, Lee EK, Song BM, Kwon YK, Kim HR, et al. Highly pathogenic avian influenza virus (H5N8) in domestic poultry and its relationship with migratory birds in South Korea during 2014. Vet Microbiol. 2014;173(3-4):249–257. doi: 10.1016/j.vetmic.2014.08.002. [DOI] [PubMed] [Google Scholar]
- 48.Wang B, Chen Q, Chen Z. Complete genome sequence of an H9N2 avian influenza virus isolated from egret in Lake Dongting wetland. J Virol. 2012;86(21):11939. doi: 10.1128/JVI.02042-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhu G, Wang R, Xuan F, Daszak P, Anthony SJ, Zhang S, et al. Characterization of recombinant H9N2 influenza viruses isolated from wild ducks in China. Vet Microbiol. 2013;166(3-4):327–336. doi: 10.1016/j.vetmic.2013.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang H, Zhang Z, Chen Z, Zhang Y, Lv Q, An X, et al. High genetic diversity and frequent genetic reassortment of avian influenza A(H9N2) viruses along the East Asian-Australian migratory flyway. Infect Genet Evol. 2016;39:325–329. doi: 10.1016/j.meegid.2016.02.013. [DOI] [PubMed] [Google Scholar]
- 51.Li X, Sun J, Lv X, Wang Y, Li Y, Li M, et al. Novel reassortant avian influenza A(H9N2) virus isolate in migratory waterfowl in Hubei Province, China. Front Microbiol. 2020;11:220. doi: 10.3389/fmicb.2020.00220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhang X, Li Y, Jin S, Wang T, Sun W, Zhang Y, et al. H9N2 influenza virus spillover into wild birds from poultry in China bind to human-type receptors and transmit in mammals via respiratory droplets. Transbound Emerg Dis. 2022;69(2):669–684. doi: 10.1111/tbed.14033. [DOI] [PubMed] [Google Scholar]
- 53.Liu JH, Okazaki K, Shi WM, Kida H. Phylogenetic analysis of hemagglutinin and neuraminidase genes of H9N2 viruses isolated from migratory ducks. Virus Genes. 2003;27(3):291–296. doi: 10.1023/a:1026304117797. [DOI] [PubMed] [Google Scholar]
- 54.Jackwood MW, Stallknecht DE. Molecular epidemiologic studies on North American H9 avian influenza virus isolates from waterfowl and shorebirds. Avian Dis. 2007;51(1) Suppl:448–450. doi: 10.1637/7536-032706R.1. [DOI] [PubMed] [Google Scholar]
- 55.Lee DH, Park JK, Yuk SS, Erdene-Ochir TO, Kwon JH, Lee JB, et al. Complete genome sequence of a natural reassortant H9N2 avian influenza virus found in bean goose (Anser fabalis): direct evidence for virus exchange between Korea and China via wild birds. Infect Genet Evol. 2014;26:250–254. doi: 10.1016/j.meegid.2014.06.007. [DOI] [PubMed] [Google Scholar]
- 56.Hu M, Jin Y, Zhou J, Huang Z, Li B, Zhou W, et al. Genetic characteristic and global transmission of influenza a H9N2 virus. Front Microbiol. 2017;8:2611. doi: 10.3389/fmicb.2017.02611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yang J, Xie D, Nie Z, Xu B, Drummond AJ. Inferring host roles in bayesian phylodynamics of global avian influenza A virus H9N2. Virology. 2019;538:86–96. doi: 10.1016/j.virol.2019.09.011. [DOI] [PubMed] [Google Scholar]
- 58.Ge FF, Zhou JP, Liu J, Wang J, Zhang WY, Sheng LP, et al. Genetic evolution of H9 subtype influenza viruses from live poultry markets in Shanghai, China. J Clin Microbiol. 2009;47(10):3294–3300. doi: 10.1128/JCM.00355-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Youk SS, Lee DH, Jeong JH, Pantin-Jackwood MJ, Song CS, Swayne DE. Live bird markets as evolutionary epicentres of H9N2 low pathogenicity avian influenza viruses in Korea. Emerg Microbes Infect. 2020;9(1):616–627. doi: 10.1080/22221751.2020.1738903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lee HJ, Kwon JS, Lee DH, Lee YN, Youn HN, Lee YJ, et al. Continuing evolution and interspecies transmission of influenza viruses in live bird markets in Korea. Avian Dis. 2010;54(1) Suppl:738–748. doi: 10.1637/8785-040109-ResNote.1. [DOI] [PubMed] [Google Scholar]
- 61.Mo IP, Song CS, Kim KS, Rhee JC. An occurrence of non-highly pathogenic avian influenza in Korea. Avian Dis. 2003;47(Special Issue):379–383. [Google Scholar]
- 62.Lai VD, Kim JW, Choi YY, Kim JJ, So HH, Mo J. First report of field cases of Y280-like LPAI H9N2 strains in South Korean poultry farms: pathological findings and genetic characterization. Avian Pathol. 2021;50(4):327–338. doi: 10.1080/03079457.2021.1929833. [DOI] [PubMed] [Google Scholar]
- 63.Lee CW, Song CS, Lee YJ, Mo IP, Garcia M, Suarez DL, et al. Sequence analysis of the hemagglutinin gene of H9N2 Korean avian influenza viruses and assessment of the pathogenic potential of isolate MS96. Avian Dis. 2000;44(3):527–535. [PubMed] [Google Scholar]
- 64.Kishida N, Sakoda Y, Eto M, Sunaga Y, Kida H. Co-infection of Staphylococcus aureus or Haemophilus paragallinarum exacerbates H9N2 influenza A virus infection in chickens. Arch Virol. 2004;149(11):2095–2104. doi: 10.1007/s00705-004-0372-1. [DOI] [PubMed] [Google Scholar]
- 65.Kim HR, Park CK, Oem JK, Bae YC, Choi JG, Lee OS, et al. Characterization of H5N2 influenza viruses isolated in South Korea and their influence on the emergence of a novel H9N2 influenza virus. J Gen Virol. 2010;91(Pt 8):1978–1983. doi: 10.1099/vir.0.021238-0. [DOI] [PubMed] [Google Scholar]
- 66.Kye SJ, Park MJ, Kim NY, Lee YN, Heo GB, Baek YK, et al. Pathogenicity of H9N2 low pathogenic avian influenza viruses of different lineages isolated from live bird markets tested in three animal models: SPF chickens, Korean native chickens, and ducks. Poult Sci. 2021;100(9):101318. doi: 10.1016/j.psj.2021.101318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bano S, Naeem K, Malik SA. Evaluation of pathogenic potential of avian influenza virus serotype H9N2 in chickens. Avian Dis. 2003;47(3) Suppl:817–822. doi: 10.1637/0005-2086-47.s3.817. [DOI] [PubMed] [Google Scholar]
- 68.Kwon JS, Lee HJ, Lee DH, Lee YJ, Mo IP, Nahm SS, et al. Immune responses and pathogenesis in immunocompromised chickens in response to infection with the H9N2 low pathogenic avian influenza virus. Virus Res. 2008;133(2):187–194. doi: 10.1016/j.virusres.2007.12.019. [DOI] [PubMed] [Google Scholar]
- 69.Xing Z, Cardona CJ, Li J, Dao N, Tran T, Andrada J. Modulation of the immune responses in chickens by low-pathogenicity avian influenza virus H9N2. J Gen Virol. 2008;89(Pt 5):1288–1299. doi: 10.1099/vir.0.83362-0. [DOI] [PubMed] [Google Scholar]
- 70.Shi H, Ashraf S, Gao S, Lu J, Liu X. Evaluation of transmission route and replication efficiency of H9N2 avian influenza virus. Avian Dis. 2010;54(1):22–27. doi: 10.1637/8937-052809-Reg.1. [DOI] [PubMed] [Google Scholar]
- 71.Yao M, Lv J, Huang R, Yang Y, Chai T. Determination of infective dose of H9N2 Avian Influenza virus in different routes: aerosol, intranasal, and gastrointestinal. Intervirology. 2014;57(6):369–374. doi: 10.1159/000365925. [DOI] [PubMed] [Google Scholar]
- 72.Chen H, Deng G, Li Z, Tian G, Li Y, Jiao P, et al. The evolution of H5N1 influenza viruses in ducks in southern China. Proc Natl Acad Sci U S A. 2004;101(28):10452–10457. doi: 10.1073/pnas.0403212101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Sturm-Ramirez KM, Hulse-Post DJ, Govorkova EA, Humberd J, Seiler P, Puthavathana P, et al. Are ducks contributing to the endemicity of highly pathogenic H5N1 influenza virus in Asia? J Virol. 2005;79(17):11269–11279. doi: 10.1128/JVI.79.17.11269-11279.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Bi Y, Chen J, Zhang Z, Li M, Cai T, Sharshov K, et al. Highly pathogenic avian influenza H5N1 Clade 2.3.2.1c virus in migratory birds, 2014-2015. Virol Sin. 2016;31(4):300–305. doi: 10.1007/s12250-016-3750-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Teng Q, Shen W, Liu Q, Rong G, Chen L, Li X, et al. Protective efficacy of an inactivated vaccine against H9N2 avian influenza virus in ducks. Virol J. 2015;12(1):143. doi: 10.1186/s12985-015-0372-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wang C, Wang Z, Ren X, Wang L, Li C, Sun Y, et al. Infection of chicken H9N2 influenza viruses in different species of domestic ducks. Vet Microbiol. 2019;233:1–4. doi: 10.1016/j.vetmic.2019.04.018. [DOI] [PubMed] [Google Scholar]
- 77.Huang R, Wang AR, Liu ZH, Liang W, Li XX, Tang YJ, et al. Seroprevalence of avian influenza H9N2 among poultry workers in Shandong Province, China. Eur J Clin Microbiol Infect Dis. 2013;32(10):1347–1351. doi: 10.1007/s10096-013-1888-7. [DOI] [PubMed] [Google Scholar]
- 78.Ma MJ, Zhao T, Chen SH, Xia X, Yang XX, Wang GL, et al. Avian influenza A virus infection among workers at live poultry markets, China, 2013-2016. Emerg Infect Dis. 2018;24(7):1246–1256. doi: 10.3201/eid2407.172059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.European Food Safety Authority. European Centre for Disease Prevention and Control. European Union Reference Laboratory for Avian Influenza. Adlhoch C, Fusaro A, Gonzales JL, et al. Avian influenza overview March - June 2022. EFSA J. 2022;20(8):e07415. doi: 10.2903/j.efsa.2022.7415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Suarez DL, Perdue ML, Cox N, Rowe T, Bender C, Huang J, et al. Comparisons of highly virulent H5N1 influenza A viruses isolated from humans and chickens from Hong Kong. J Virol. 1998;72(8):6678–6688. doi: 10.1128/jvi.72.8.6678-6688.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Jegede A, Fu Q, Berhane Y, Lin M, Kumar A, Guan J. H9N2 avian influenza virus retained low pathogenicity after serial passage in chickens. Can J Vet Res. 2018;82(2):131–138. [PMC free article] [PubMed] [Google Scholar]
- 82.Ren W, Zhang CH, Li G, Liu G, Shan H, Li J. Two genetically similar H9N2 influenza viruses isolated from different species show similar virulence in minks but different virulence in mice. Acta Virol. 2020;64(1):67–77. doi: 10.4149/av_2020_109. [DOI] [PubMed] [Google Scholar]
- 83.Niu X, Wang H, Zhao L, Lian P, Bai Y, Li J, et al. All-trans retinoic acid increases the pathogenicity of the H9N2 influenza virus in mice. Virol J. 2022;19(1):113. doi: 10.1186/s12985-022-01809-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Butt AM, Siddique S, Idrees M, Tong Y. Avian influenza A (H9N2): computational molecular analysis and phylogenetic characterization of viral surface proteins isolated between 1997 and 2009 from the human population. Virol J. 2010;7(1):319. doi: 10.1186/1743-422X-7-319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Kimble B, Nieto GR, Perez DR. Characterization of influenza virus sialic acid receptors in minor poultry species. Virol J. 2010;7(1):365. doi: 10.1186/1743-422X-7-365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Pusch EA, Suarez DL. The multifaceted zoonotic risk of H9N2 avian influenza. Vet Sci. 2018;5(4):82. doi: 10.3390/vetsci5040082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Sun X, Belser JA, Maines TR. Adaptation of H9N2 influenza viruses to mammalian hosts: a review of molecular markers. Viruses. 2020;12(5):541. doi: 10.3390/v12050541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Huang Y, Li X, Zhang H, Chen B, Jiang Y, Yang L, et al. Human infection with an avian influenza A (H9N2) virus in the middle region of China. J Med Virol. 2015;87(10):1641–1648. doi: 10.1002/jmv.24231. [DOI] [PubMed] [Google Scholar]
- 89.Song Y, Zhang Y, Chen L, Zhang B, Zhang M, Wang J, et al. Genetic characteristics and pathogenicity analysis in chickens and mice of three H9N2 avian influenza viruses. Viruses. 2019;11(12):1127. doi: 10.3390/v11121127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Zhang RR, Yang X, Shi CW, Yu LJ, Lian YB, Huang HB, et al. Improved pathogenicity of H9N2 subtype of avian influenza virus induced by mutations occurred after serial adaptations in mice. Microb Pathog. 2021;160:105204. doi: 10.1016/j.micpath.2021.105204. [DOI] [PubMed] [Google Scholar]
- 91.Murakami J, Shibata A, Neumann G, Imai M, Watanabe T, Kawaoka Y. Characterization of H9N2 avian influenza viruses isolated from poultry products in a mouse model. Viruses. 2022;14(4):728. doi: 10.3390/v14040728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Park SJ, Kang YM, Cho HK, Kim DY, Kim S, Bae Y, et al. Cross-protective efficacy of inactivated whole influenza vaccines against Korean Y280 and Y439 lineage H9N2 viruses in mice. Vaccine. 2021;39(42):6213–6220. doi: 10.1016/j.vaccine.2021.09.028. [DOI] [PubMed] [Google Scholar]
- 93.Lin Z, Xu C, Liu B, Ji Y, Fu Y, Guo J, et al. Analysis of the phylogeny of Chinese H9N2 avian influenza viruses and their pathogenicity in mice. Arch Virol. 2014;159(10):2575–2586. doi: 10.1007/s00705-014-2110-7. [DOI] [PubMed] [Google Scholar]
- 94.Baek YG, Lee YN, Lee DH, Shin JI, Lee JH, Chung DH, et al. Multiple reassortants of H5N8 clade 2.3.4.4b highly pathogenic avian influenza viruses detected in South Korea during the winter of 2020-2021. Viruses. 2021;13(3):490. doi: 10.3390/v13030490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Sagong M, Lee YN, Song S, Cha RM, Lee EK, Kang YM, et al. Emergence of clade 2.3.4.4b novel reassortant H5N1 high pathogenicity avian influenza virus in South Korea during late 2021. Transbound Emerg Dis. 2022;69(5):e3255–e3260. doi: 10.1111/tbed.14551. [DOI] [PubMed] [Google Scholar]
- 96.Sun Y, Liu J. H9N2 influenza virus in China: a cause of concern. Protein Cell. 2015;6(1):18–25. doi: 10.1007/s13238-014-0111-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Zhang P, Tang Y, Liu X, Peng D, Liu W, Liu H, et al. Characterization of H9N2 influenza viruses isolated from vaccinated flocks in an integrated broiler chicken operation in eastern China during a 5 year period (1998-2002) J Gen Virol. 2008;89(Pt 12):3102–3112. doi: 10.1099/vir.0.2008/005652-0. [DOI] [PubMed] [Google Scholar]
- 98.Sun Y, Pu J, Fan L, Sun H, Wang J, Zhang Y, et al. Evaluation of the protective efficacy of a commercial vaccine against different antigenic groups of H9N2 influenza viruses in chickens. Vet Microbiol. 2012;156(1-2):193–199. doi: 10.1016/j.vetmic.2011.10.003. [DOI] [PubMed] [Google Scholar]
- 99.Liu Y, Li S, Sun H, Pan L, Cui X, Zhu X, et al. Variation and molecular basis for enhancement of receptor binding of H9N2 avian influenza viruses in China isolates. Front Microbiol. 2020;11:602124. doi: 10.3389/fmicb.2020.602124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Cui H, de Jong MC, Beerens N, van Oers MM, Teng Q, Li L, et al. Vaccination with inactivated virus against low pathogenic avian influenza subtype H9N2 does not prevent virus transmission in chickens. J Virus Erad. 2021;7(3):100055. doi: 10.1016/j.jve.2021.100055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Zhang Y, Yin Y, Bi Y, Wang S, Xu S, Wang J, et al. Molecular and antigenic characterization of H9N2 avian influenza virus isolates from chicken flocks between 1998 and 2007 in China. Vet Microbiol. 2012;156(3-4):285–293. doi: 10.1016/j.vetmic.2011.11.014. [DOI] [PubMed] [Google Scholar]
- 102.Bi J, Deng G, Dong J, Kong F, Li X, Xu Q, et al. Phylogenetic and molecular characterization of H9N2 influenza isolates from chickens in Northern China from 2007-2009. PLoS One. 2010;5(9):e13063. doi: 10.1371/journal.pone.0013063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Liu S, Zhuang Q, Wang S, Jiang W, Jin J, Peng C, et al. Control of avian influenza in China: strategies and lessons. Transbound Emerg Dis. 2020;67(4):1463–1471. doi: 10.1111/tbed.13515. [DOI] [PubMed] [Google Scholar]
- 104.Choi JG, Lee YJ, Kim YJ, Lee EK, Jeong OM, Sung HW, et al. An inactivated vaccine to control the current H9N2 low pathogenic avian influenza in Korea. J Vet Sci. 2008;9(1):67–74. doi: 10.4142/jvs.2008.9.1.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Cho HK, Kang YM, Kim HM, Lee CH, Kim DY, Choi SH, et al. Sales and immunogenicity of commercial vaccines to H9N2 low pathogenic avian influenza virus in Korea from 2007 to 2017. Vaccine. 2020;38(16):3191–3195. doi: 10.1016/j.vaccine.2020.02.083. [DOI] [PubMed] [Google Scholar]
- 106.Su H, Zhao Y, Zheng L, Wang S, Shi H, Liu X. Effect of the selection pressure of vaccine antibodies on evolution of H9N2 avian influenza virus in chickens. AMB Express. 2020;10(1):98. doi: 10.1186/s13568-020-01036-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Park KJ, Song MS, Kim EH, Kwon HI, Baek YH, Choi EH, et al. Molecular characterization of mammalian-adapted Korean-type avian H9N2 virus and evaluation of its virulence in mice. J Microbiol. 2015;53(8):570–577. doi: 10.1007/s12275-015-5329-4. [DOI] [PubMed] [Google Scholar]
- 108.Kim DY, Kang YM, Cho HK, Park SJ, Lee MH, Lee YJ, et al. Development of a recombinant H9N2 influenza vaccine candidate against the Y280 lineage field virus and its protective efficacy. Vaccine. 2021;39(42):6201–6205. doi: 10.1016/j.vaccine.2021.08.089. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Y280 lineage H9 subtype virus detection graph by country and year (total 8,967 cases based on the GISAID database).