Abstract
Annually, roughly 2.5 billion people are at risk for dengue virus (DENV) infection, and the incidence of infection has increased 30-fold since its discovery in the 1900s. At present, there are no globally licensed antiviral treatments or vaccines that protect against all four of the DENV serotypes. The NIAID Live Attenuated Tetravalent Vaccine (LATV) dengue vaccine candidate is composed of variants of three DENV serotypes attenuated by a 30 nucleotide (Δ30) deletion in the 3′ untranslated region and a fourth component that is a chimeric virus in which the prM and E genes of DENV-2 replace those of DENV-4 on the rDEN4Δ30 backbone. The vaccine candidate encodes the non-structural proteins of DENV-1, DENV-3, and DENV-4, which could be of critical importance in the presentation of DENV-specific epitopes in a manner that facilitates antigen presentation and confers higher protection. Our findings demonstrate that the attenuation mechanism (Δ30) resulted in decreased viral infectivity and replication for each vaccine virus in monocyte-derived dendritic cells but were able to generate a robust innate immune response. When tested as monovalent viruses, DEN-4Δ30 displayed the most immunogenic profile. In addition, we found that the tetravalent DENV formulation induced a significantly greater innate immune response than the trivalent formulation. We demonstrate that the presence of two components with a DENV-4Δ30 backbone is necessary for the induction of RANTES, CD40, IP-10, and Type I IFN by the tetravalent formulation. Finally, we found that the DEN-4Δ30 backbone in the DENV-2 component of the vaccine enhanced its antigenic properties, as evidenced by enhanced ability to induce IP-10 and IFNα2 in monocyte-derived dendritic cells. In sum, our study shows that the Δ30 and Δ30/Δ31 mutations attenuate the DENV vaccine strains in terms of replication and infectivity while still allowing the induction of a robust innate immune response.
Keywords: Dengue, vaccine, innate immunity, vaccine efficacy
Impact statement.
To generate a successful dengue vaccine, the induction of a robust innate immune response is critical for the development of a successful adaptive immune response. In this work, we characterize the differences in innate immune induction between different formulations of what we believe to be the most promising dengue virus (DENV) vaccine candidate currently generated. We investigate the immunogenic contribution of each DENV vaccine serotype and demonstrate a potential role for the DENV non-structural proteins and 3’ untranslated region specific for each serotype in inducing type I IFN responses in primary human cells.
Introduction
Dengue virus (DENV) is the arthropod-borne virus of greatest clinical significance and is transmitted through the bite of an infected Aedes aegypti or Aedes albopictus mosquito. Since its discovery in the early 1900s, DENV has spread globally and put an estimated 3.9 billion people at risk for infection. 1 The WHO reports that the number of dengue cases reported over the course of the last two decades has greatly increased, from 505,430 million in 2000 to 5.2 million in 2019.2,3 Of the 3.9 billion people that are at risk for DENV infection, 96 million or roughly 40% of infected individuals will develop symptoms. Clinical manifestations can range from asymptomatic presentation and mild illness to the development of potentially fatal complications known as severe dengue and dengue shock syndrome.2,4,5 There are four DENV serotypes (DENV1-4), each of which is independently capable of causing the broad spectrum of disease manifestation.1,3,6 Monocytes and dendritic cells (DCs) are the targets during DENV infection, replication, and dissemination.7–13 Upon interaction with a pathogen, immature DCs undergo a maturation process that results in increased antigen presentation and the production of various cytokines, such as TNFα, IL-6 and type I IFNs among others. 14 Mature DCs upregulate chemokine receptors such as CCR7 that result in the migration of the cell to the lymph node where interaction with T-cells can occur. Mature DCs also upregulate costimulatory molecules such as CD40 that play a key role in the activation of antigen-specific T-cells in the lymph nodes.15,16 Human monocyte-derived DCs (MDDCs), generated from peripheral blood mononuclear cells (PBMCs) from healthy blood donors, have been characterized for their ability to support robust infection of DENV and are the primary model system for studying DENV infections ex-vivo.17,18,19
There is currently no globally licensed therapeutics, and the currently licensed vaccine has variable protection against each of the four DENV serotypes. As such, primary measures to reduce the incidence of DENV infections involve management of the mosquito vector and the use of personal protective measures. 20 Dengvaxia by Sanofi Pasteur is a live attenuated tetravalent vaccine (LATV) composed of a yellow fever 17D-204 vaccine backbone with the prM and E proteins of each of the four DENV serotypes represented.6,20 There are currently on-going clinical trials of four dengue vaccines of which Dengvaxia, has been licensed in Mexico, the Philippines, Indonesia, Brazil, El Salvador, Costa Rica, Paraguay, Guatemala, Peru, Thailand, and Singapore, and in 2019, it was licensed by the Food and Drug Administration (FDA) and approved for use by the Center for Disease Control Advisory Committee on Immunization Practices (CDC ACIP) under the measure that all vaccinees had to be shown to be seropositive for dengue prior to administration of the vaccine. 21 Another promising dengue vaccine candidate, TAK-003 (TDV) is being developed by Takeda pharmaceutical company. TAK-003 is based on an attenuated virus and chimeric viruses constructed using recombinant DNA technology. TAK-003 is based on a live attenuated DENV-2 virus that provides the genetic backbone for all four of the vaccine viruses with the prM and E protein of each DENV serotype represented. In a phase III clinical trial in children 4–16 years of age, TAK-003 demonstrated an overall vaccination efficacy of 73.3%. 22 TAK-003 is currently being evaluated for licensure in a number of countries and was granted priority review by the U.S. FDA and is being evaluated for the prevention of dengue disease against all four serotypes of dengue in individuals ages 4-60 years of age. 23
The focus of this study is on the NIAID LATV vaccine candidate that is composed entirely of attenuated variants of the DENV serotypes. The recombinant DENV-1, and DENV-4 vaccine strains (rDEN1Δ30, rDEN4Δ30), components of the vaccine were engineered to have a 30 nucleotide deletion (Δ30) in the A3 region of the 3' UTR.24–26 The rDENV-2 vaccine strain (rDEN2/4Δ30) is an antigenic chimera composed of an rDEN4Δ30 backbone with the precursor pre-membrane (prM) and E genes of DENV-2 replacing those of DENV-4. 27 The rDENV-3 vaccine strain (rDEN3Δ30/31) has a 30-nucleotide deletion (Δ30, nucleotides 173-143) and a 31-nucleotide deletion (Δ31, nucleotides 258-228) in the 3’ UTR.24,28 Data from us and others have shown that the Δ30 deletion in the 3’ UTR of the wild-type (WT) DENV serotypes confer attenuation while inducing a robust immune response in primary cells.26,28
Further, the NIAID LATV candidate encodes the non-structural DENV proteins of three of the four DENV serotypes.3,21 These key differences in the NIAID vaccine candidate are anticipated to result in a more robust innate immune response.25,27,29 The vaccine induced a balanced antibody response for all four viral serotypes in an impressive 90% of vaccinees and did not show significant differences in the incidence of adverse effects exhibited by the vaccine versus the placebo group, with the exception of a mild rash which occurred in ~60% of vaccine recipients. 28 Viral replication and antibody responses to the NIAID DENV vaccine strains have been studied within rhesus macaques and humans but the innate immune response induced by those vaccine candidates at early stages of vaccination have yet to be well characterized.6,20,27–42 However, the development of a safe and efficacious vaccine against DENVs includes a thorough understanding of the innate immune responses and its possible risks as well as the innate immune signatures that are important for shaping a successful adaptive immune response and therefore improving vaccine efficacy.43,44 Little research has been conducted on the innate immune responses induced by the National Institutes of Health/National Institute of Allergy and Infectious Diseases (NIH/NIAID) LATV. Further insight into the modulation of innate immune responses such as type I IFN induction, cytokine responses and immune cell migration in response to vaccination would enhance the current gaps in knowledge with respect to the NIH/NIAID LATV. Here, we provide insights into the innate immune signatures induced by the NIH/NIAID LATV in primary human immune cells ex vivo using state of the art techniques. Our study complements data from pre- and post-NIAID LATV vaccination on innate immune signatures and the induction of a robust adaptive immune response in volunteers receiving the NIAID dengue LATV. We demonstrate that the rDEN4Δ30 plays an immune stimulatory role inducing the highest induction of Type I IFN response that were not seen with the chimeric rDEN-2/4Δ30 virus, supporting our previous findings that DENV-4 confers immune-stimulatory properties while DENV-2 confers immunosuppressant properties. 11 Altogether our findings suggest that the rDENV-4Δ30 vaccine component confers robust immunogenic properties and contributes significantly to vaccine efficacy.
Materials and methods
Cell lines
Raji DC-SIGN cells were provided by Viviana Simon (Icahn School of Medicine at Mount Sinai, New York, NY) and originally obtained from the NIAID HIV/AIDS Biorepository. Cells were maintained in culture media (Gibco; RPMI medium with 2 mM L-glutamine (Sigma-Aldrich), 5 mL 100 U/mL Pen/Strep penicillin, 100 μg/mL streptomycin, and 1 mM sodium pyruvate (Thermo Fisher Scientific) supplemented with 500 U/mL human granulocyte-macrophage colony-stimulated factor (GM-CSF) (PeproTech), 1000 U/mL human IL-4 (PeproTech), and 10% (v/v) FBS (Thermo Fisher Scientific) for 5 days at 37°C and 5% CO2
Viruses
Vaccine WT DENV was provided by Steve Whitehead, Laboratory of Viral Diseases (LVD), NIAID/ NIH. Viruses were grown and titrated as previously published. 45
WT viruses include DENV-1 (American genotype; strain Western Pacific/74), DENV-2 (New Guinea C, NGC), DENV-3 (Sleman/78), and DENV-4 (Dominica/81). GenBank accession numbers for WT and vaccine DENV used in this study are: DENV-1 (AY145121.1), DENV-2 (AY744148.1), DENV-3 (AY648961.1), DENV-4 (AY648301), rDEN1Δ30 (AY145123.1), rDEN2Δ30 (AY744149.1), rDEN2/4Δ30 (AY243469.1), rDEN4Δ30 (AF326827.1), and rDEN3Δ30 (AY656170.1).
Generation of MDDCs
MDDCs were generated from buffy coats of healthy human donors (New York Blood Center) using a standard protocol.17–19 Briefly, PBMCs were isolated using Ficoll gradient centrifugation (Histopaque, Sigma-Aldrich). Next, CD14+ cells were isolated from the mononuclear fraction using the MACS CD14+ isolation kit (Miltenyi Biotech) according to the manufacturer’s instructions. CD14 + cells were then differentiated into naive immature DCs by incubation in DC media (RPMI medium with 2 mM L-glutamine, 100 U/mL penicillin (Life Technologies), 100 μg/mL streptomycin (Life Technologies), and 1 mM sodium pyruvate (Life Technologies) supplemented with 500 U/ mL human granulocyte-macrophage colony-stimulating factor (GM-CSF) (PeproTech), 1000 U/mL human IL-4 (PeproTech), and 10% (v/v) Hyclone FBS (Thermo Fisher Scientific) for 5 days at 37°C and 5% CO2.
DENV infection of DCs and Raji DC-SIGN cells
After 5 days in culture, donor-matched naive MDDCs or Raji DC-SIGN cells were mock-infected with DC medium, or with a monovalent DENV(DENV-1 WT (rDEN1 WP), rDEN1Δ30, rDEN2Δ30, DENV-3 WT (rDEN3 Sleman/78), rDEN3Δ30, DENV-4 (rDEN4) WT, rDEN2/4Δ30, rDEN4Δ30 at a multiplicity of infection (MOI) of 1.0 (PFU/cell). Trivalent, tetravalent vaccine formulations and the rDENV-4Δ30×2 vaccine formulation were used at an MOI of 0.25 (PFU/cell) for each DENV serotype represented in the respective vaccine formulation. Infections were performed in DC media containing 10% hyclone FBS (Thermo Fischer Scientific), virus and vaccine inoculum were not removed after infection. After infection, DC media with Hyclone FBS was added to create a total volume of 220 µL per well in a 96-well plate. MDDCs were maintained in culture for the indicated time periods at 37 °C and 5% CO2 at a density of 1×106 cells/mL in DC media.
Multiplex ELISA
The Cytokine Human Magnetic 10-Plex Panel for Luminex Platform (Millipore Milliplex) was used according to the manufacturer’s instructions. The panel was designed to measure the following cytokines and chemokines: TNF-α, IL-1β, IL-6, MIP-1β, RANTES, IL-8, IL-10, MCP-1, IFN-α2, IP-10. The data were acquired on the Luminex 100 System (Millipore Sigma) and were analyzed by a standard curve fit, according to the manufacturer’s protocol. In brief, for each analyte, a seven-point dilution series of the protein standard plus an assay diluent-only background well were run in duplicate. A standard curve was then fitted using a weighted five-parameter logistic function and used to quantify protein concentrations. Data analysis was done with the Milliplex Analyst software version 5.1.
Flow cytometry
After infection, mock or DENV-infected MDDCs were first incubated with 1uL/mL of Human TruStain FcX (Biolegend) for 10 min at 4°C in FACS buffer, followed by incubation using the Live/Dead Fixable Blue Dead Cell Stain Kit (Thermo Fisher Scientific), according to the manufacturer’s instructions. To stain for intracellular DENV E protein a hybridoma was obtained (ATCC cat# D1-4G2-4-15, 4.15 mg/mL) that is referred to as 4G2. The hybridoma was sent to BioXcell for purification of the 4G2 antibody. The DENV anti E-protein antibody was conjugated with Alexa Fluor® 488 Conjugation Kit (Fast)–Lightning-Link® (Abcam, cat: ab236553). A cocktail containing a 1:400 µl ratio of DENV 4G2-AlexaFluor 488 antibody to FACS buffer (500 mL PBS, 2% FBS, 0.2 mL 0.5 M EDTA) was added to cells and incubated overnight at 4°C and subsequently fixed and permeabilized with Cytofix/Cytoperm (BD Pharmingen). To quantify viral infection, a pan-flaviviral E protein was used. Samples were run on a Beckman Coulter Gallios flow cytometer, available through the Microbiology Departments’ Shared Resource Facility. Data analysis was performed with FlowJo (Software version 10.8.1, FlowJo is a trademark of Becton, Dickinson, and Company).
Cytometry by time of flight staining and analysis
All cytometry by time of flight (CyTOF) reagents were provided by the Human Immune Monitoring Core at Mount Sinai and were acquired from Fluidigm Inc. unless otherwise indicated. All antibodies were either purchased pre-conjugated from Fluidigm or conjugated in-house using commercial X8 polymer conjugation kits purchased from Fluidigm at the Human Immune Monitoring Center, Icahn School of Medicine at Mount Sinai, New York. All in-house conjugated antibodies were validated on healthy donor PBMCs or on infected cells for the 4G2 antibody. In brief, the CyTOF protocol is as follows, 4 h before the end of each timepoint, a cocktail of Brefeldin (Thermofisher) and Monensin (Sigma Aldrich) were added to samples. At time of collection cells were centrifuged at 1600 rpm and supernatant was removed. For each viral condition live-cell CyTOF barcoding was performed using anti-B2M antibodies conjugated to unique cadmium isotopes. Rhodium-103 viability (Fluidigm) and Human TruStain FcX (Biolegend) staining was performed simultaneously during this barcoding step. After a 30-min incubation at room temperature, samples were washed twice in CSM and similar timepoints with unique cadmium labels were pooled. After pooling, cells were stained with a cocktail of surface antibodies. Surface-stained cells were then barcoded with CyTOF Cell-ID 20-Plex Palladium Barcoding Kit (Fluidigm) with each pooled time point getting a unique barcode. After washing with Cell staining media (CSM) the timepoints were pooled into a single sample. Cells were then fixed with Cytofix/Cytoperm (BD Biosciences) for 30 min at 4°c. After washing with CSM buffer, the sample was resuspended in heparin and incubated for 5 min. Following the heparin blocking the sample was stained with an antibody cocktail of intracellular antibodies on ice for 30 min. After intracellular staining the sample was fixed in freshly diluted 4% paraformaldehyde containing 125 nM intercalator-Ir (Fluidgm), 2% saponin (Sigma) and 300 nM OsO4 (ACROS Organics). CyTOF was analyzed using Cytobank software version 10 (Cytobank Inc.).
Aurora spectral-flow cytometry
Mock or DENV-infected cells were stained for viability with 1 µL of zombie live/dead (Biolegend) and 1 µL Fc block from (BD biosciences) in 1 mL of FACS buffer. Cells were incubated with a 1:1000 dilution for 20 min at 4°C. Next cells were stained with the following cocktail of antibodies at a concentration of: Brilliant Violet 421™ anti-human HLA-DR (Biolegend cat: 307635), anti-human CD16 Monoclonal antibody-eFlour450 (Thermofisher scientific cat: 48-0168-42), BV510 Rat Anti-Human CXCR5 (BD biosciences, cat: 563105), Brilliant Violet 605™ anti-mouse CD11c (Biolegend, cat:117333), 4G2-AF488 (conjugated as previously described), PerCP/Cyanine5.5 anti-human CD86 (Biolegend cat: 305419), PE anti-human CD80 Antibody (Biolegend, cat:305207), PE-Cy™7 Mouse Anti-Human CD40 (BD biosciences cat:561215), Alexa Fluor 647 anti human CD197 (CCR7) (Biolegend, cat:353217), Human DC-SIGN/CD209 Alexa Fluor® 700-conjugated (RND systems cat: FAB161 N-025), CD14 Monoclonal Antibody (61D3), APC-eFluor 780 (eBioscience, cat: 47-0149-42), Pacific Blue anti-human CD11b antibody (Biolegend, cat:301316), APC anti-human CD83 Antibody (Biolegend, 305311), E1D8-PECy5 DENV NS3 provided by Eva Harris (University of California Berkley, San Francisco, USA) and conjugated with PE/Cy5 conjugation kit (abcam, cat:ab102893). Cells were stained for 1 h at 4°C and were washed and resuspended FACS buffer. Cells were fixed with 200 µL of BD Biosciences perm fixation buffer (BD biosciences cat# 554714) at 4°C for 30 min and subsequently centrifuged at 1800 rpm for 7 min. Post incubation, cells were washed two times with FACS buffer. Data were analyzed using Cytobank software version 10 (Cytobank Inc.) at the Mount Sinai Flow Cytometry facility.
Real-time quantitative PCR
Evaluation of the relative levels of RNA expression of human cytokines from different cell types and viral RNA was carried out using iQ SYBR green Supermix (Bio-Rad) according to the manufacturer’s instructions. The PCR temperature profile was 95°C for 10 min, followed by 40 cycles of 95°C for 10s and elongation at 60°C for 60s for 40 cycles. Real-time quantitative PCR (RT-qPCR) was carried out using the BioRad 1000 C Thermal Cycler. Quantification of gene expression levels was performed based on Ct values of a given normalized to one housekeeping gene ribosomal protein S11 (RPS11). To quantify viral RNA levels, a set of primers for a conserved region of the DENV 3'UTR was used: F-AAGGACTAGAGGTTAGAGGAGACCC, R-GGCGTCCTGTGCCTGGAATGATG. The following primers were used to quantify genomic RIG-I induction: F-AAAGCCTTGGCATGTTACAC, R-GGCTTGGGATGTGGTCTACT.
Statistical analysis
The statistical analyses were performed using two-way analysis of variance (ANOVA), with multiple comparisons and Bonferroni correction. P values were calculated using Graphpad Prism 9 (GraphPad Software, LLC). Data considered significant had P values ⩽0.05. No statistical methods were used to predetermine the sample size.
Results
NIAID LATV TV003 vaccine strains have a lower replication and infection efficiency than their wild-type virus counterparts in MDDCs
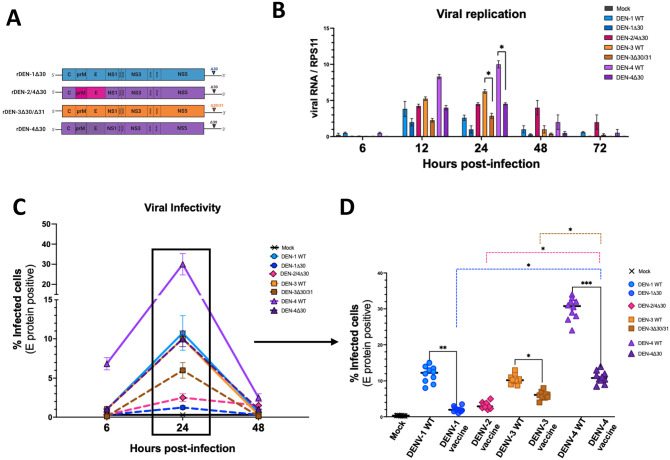
In a first step, we aimed to investigate the phenotypic differences in infection profiles of the WT DENVs and vaccine strains. To do so, we infected human donor-matched MDDCs with WT DENV-1 [rDEN1 WP], DENV-3 [rDEN3 Sleman/78], DENV-4 [rDEN4] and rDEN1Δ30, rDEN2/4Δ30, rDEN3Δ30/31 and rDEN4Δ30 included in the NIAID TV003 dengue vaccine candidate (Figure 1(A)). We quantified viral replication and measured viral RNA using a RT-qPCR approach to detect a region in the 3' UTR that is conserved in all DENV strains, including the vaccine strains. We found that each of the vaccine strains replicated to lower levels relative to their WT DENV counterparts (Figure 1(B)). The DENV-4 WT [rDEN4] replicated to highest levels, followed by the rDEN-4Δ30 strain (Figure 1(B)). In contrast, DENV-1 WT, [rDEN1 WP], and rDENV-1Δ30 showed the lowest level of replication (Figure 1(B)). Interestingly, while the replication of DENV-3 [rDEN3 Sleman/78] and DENV-4 [rDEN4 Dominica 81] and vaccine strains peak at 24 h post-infection (hpi), the DENV-1 WT [rDEN1 WP] does so already at 12 hpi in human MDDCs (Figure 1(B)). To compare viral replication with the production of viral proteins in MDDCs we quantified the levels of DENV E protein by flow cytometry. Our data show that each of the DENV vaccine strain produced less DENV E protein in MDDCs when compared to their WT DENV counterparts (Figure 1(C)). Notably, the rDEN-4Δ30 infects MDDCs to the highest level when compared to all other vaccine strains (Figure 1(D)). In sum, our data show that the Δ30 deletions in the 3'UTR region present in the vaccine strains result in reduced viral replication and production of viral proteins in MDDCs.
Figure 1.
Viral infectivity and replication of the NAID LATV TV003 vaccine viruses and their wild-type counterparts. (A) The DENV-1 vaccine component was generated via the introduction of a 30-nucleotide (Δ30) deletion into the 3' untranslated region (UTR) of a DENV-1 Western Pacific (WP) strain. The DENV-2 vaccine strain was generated via a chimerization strategy in which the prM and E proteins of a DENV-2 New Guinea C strain were introduced into the backbone of the DENV-4 vaccine virus. The DENV-3 vaccine strain was generated similarly to the DENV-1 vaccine strain, with the introduction of the Δ30 deletion in the 3' UTR of the DENV-3 Sleman/78 strain. To sufficiently attenuate the DENV-3 virus an additional 31 nucleotide deletion was introduced into the 3' UTR of a Sleman/78 isolate. The DENV-4 vaccine virus was generated via the introduction of the Δ30 deletion in the 3' UTR of a DENV-4 Dominica isolate. (B) Viral replication was quantified via RT-qPCR using a primer designed to capture a conserved region of the DENV 3' UTR that is not affected by the presence of the Δ30 deletion. Monocyte-derived dendritic cells (MDDC)s were infected with either DENV-1 WT, rDEN-1Δ30, rDEN-2/4Δ30, DENV-3 WT, rDEN-3Δ30/31, DENV-4 WT or rDEN-4Δ30 at an MOI of 1.0. Cells were lysed and collected at 6, 12, 24, 48, and 72 hpi. Replication levels were standardized to the RPS11 housekeeping gene and relative levels were determined using the 2^-ΔΔCt method. (C) Flow cytometry was performed on MDDCs infected with either mock, DENV-1 WT, rDEN-1Δ30, rDEN-2/4Δ30, DENV-3 WT, rDEN-3Δ30/31, DENV-4 WT, or rDEN-4Δ30 viruses at an MOI of 1.0 for 6, 24, and 48 hpi. Infectivity was measured via the binding and fluorescence of the E protein binding, pan-flaviviral 4G2 antibody. (D) Detailed analysis of viral infectivity at 24 hpi indicating significant differences in infectivity between the DENV-1 WT and vaccine strains, the DENV-3 WT and vaccine strains, the DENV-4 WT and vaccine strains. Hashed colored lines represent significant differences between the DENV-1, rDEN-2/4Δ30, and rDEN-3Δ30/31vaccine strains compared to the rDEN-4Δ30 vaccine strain. Statistical significance between the four vaccine viruses was calculated using two-way analysis of variance (ANOVA) with multiple comparisons and Bonferroni correction (*P ⩽ 0.05, **P ⩽ 0.01, **P ⩽ 0.001). (A color version of this figure is available in the online journal.)
The DENV-4 (rDEN-4Δ30) vaccine component induces the highest secretion levels of pro-inflammatory cyto- and chemokines by MDDCs
Viral infections activate the innate immune response and therefore the secretion of pro-inflammatory cyto- and chemokines that regulate the recruitment and activation of immune cells. 27 Among them, there are cells involved in inflammation that are known to play a pivotal role in enhancing the innate immune response and the induction of the adaptive immunity. 46 To compare the cyto- and chemokine profile of DENV vaccine strains, we utilized Multiplex ELISAs analyzed a panel of cyto- and chemokines (IFNα2, IL-10, IL-1β, Il-6, IL-8, IP-10, MCP-1, MIP-1β, RANTES, TNFα) that have been characterized to play an important role in DENV infection.47–51 We did not observe differences in the secretion between the vaccine viruses for IL-8, MIP-1β, TNFα, and RANTES (Supplementary Figure 1, A-E). However, the rDEN1Δ30 and rDEN4Δ30 vaccine strains showed significant induction of IFNα2 at 48 hpi (Figure 2(A)). Interestingly, only MDDCs infected with rDEN-4Δ30 showed high secretion of the pro-inflammatory cytokine IP-10 at 24 hpi (Figure 2(B)). These data reveal that the rDEN-4Δ30 induces the highest secretion levels of IFNα2 and IP-10 in MDDCs compared to the other vaccine strains.
Figure 2.
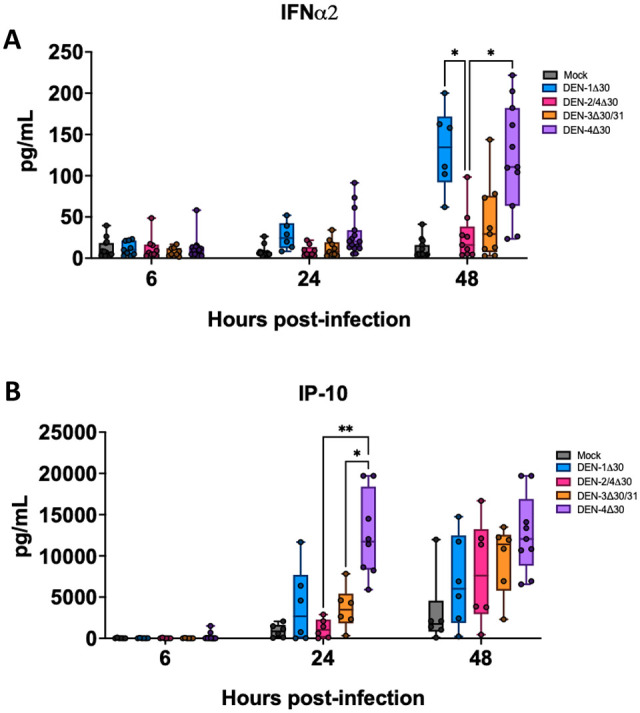
Cytokine induction of the NAID LATV TV003 vaccine viruses. (A, B) Monocyte-derived dendritic cells (MDDCs) were infected with either mock, rDEN-1Δ30, rDEN-2/4Δ30, rDEN-3Δ30/31, or c vaccine viruses at a multiplicity of infection (MOI) of 1.0 for 6, 24, and 48 hpi. Multiplex ELISAs were performed to evaluate extracellular cytokine secretion. Data shown represent the levels of secreted IFNα2, IP-10, and IL-6 in cell supernatant. Black asterisks represent statistical significance by the two-way analysis of variance (ANOVA), with adjustment for multiple comparisons and the Bonferroni correction (*P ⩽ 0.05, **P ⩽ 0.01). (A color version of this figure is available in the online journal.)
The DENV-4 (rDEN-4Δ30) non-structural proteins and 3' UTR confer unique immunogenic properties and are sufficient to induce a type I IFN response
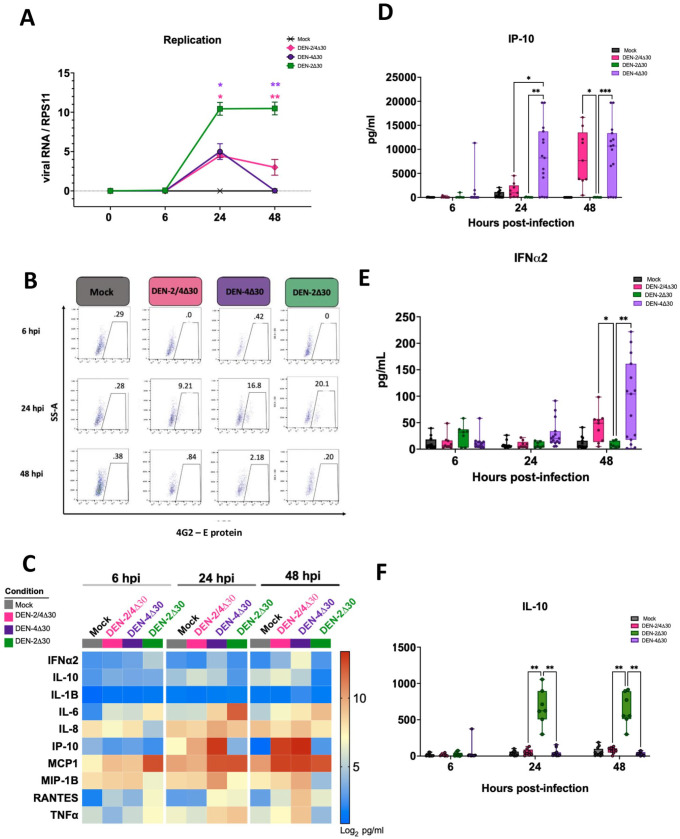
To further explore the contribution of the rDEN4Δ30 vaccine backbone to the immune response elicited by the tetravalent vaccine (rDEN1Δ30, rDEN3Δ30, rDEN4Δ30), we infected MDDCs with rDEN2Δ30, which was generated via the introduction of the Δ30 deletion into the 3' UTR of WT DENV-2 Tonga/74. This rDEN2Δ30 was used in clinical trials (CIR299 and CIR300) to challenge participants vaccinated with the tetravalent or trivalent DENV vaccines. rDENV-2Δ30 was given to participants 180 days post initial vaccination to assess the protective efficacy of the vaccines. 52 Utilizing flow cytometry to quantify levels of viral protein production and subsequently infection efficiency, we found that the rDEN2Δ30 challenge virus infected MDDCs to roughly double the levels of the rDENV-2/4Δ30 vaccine virus at 24 hpi (Figure 3(B)), suggesting that the chimerization strategy employed to generate the DENV-2 [DEN2/4Δ30] vaccine virus results in additional attenuation properties. As a following step, we measured levels of cyto- and chemokine secretion at early time points post-infection (Figure 3(C)). Interestingly, our results demonstrate that despite being significantly less infective relative to the rDEN2Δ30 challenge virus, the infection with the rDEN2/4Δ30 induces significantly higher secretion levels of the pro-inflammatory cytokines IP-10 and IFNα2 (Figure 3(D) and E)). This suggests that the presence of the DENV-4Δ30 backbone in the rDEN2/4Δ30 is sufficient to induce type-I IFN responses. Strikingly, when we measured secretion levels of the anti-inflammatory cytokine IL-10, we found that the rDEN2Δ30 challenge virus induced significantly higher levels of IL-10 secretion when compared to the rDEN2/4Δ30 and rDENV-4Δ30. IL-10 is known for its inhibition of DENV-specific T-cell responses during acute DENV infections. Therefore, our data strongly suggest that the non-structural proteins of DENV-4 included in the chimeric rDENV-2/4Δ30 contribute to its immunogenic properties, but that we cannot exclude that the structural proteins of DENV-2 may have some immunogenic as well.
Figure 3.
DENV-2 vaccine and DENV-4 vaccine viruses induce robust levels of IP-10 and IFNα2 but the DENV-2 challenge strain does not. (A) Viral replication was quantified via real-time quantitative PCR (RT-qPCR) using the previously described 3' untranslated region (UTR) primer. Monocyte-derived dendritic cells (MDDCs) were infected at 0 (1 h after infection), 6, 24, and 48 hpi. Viral RNA was quantified as previously described. (B) MDDCs were infected with either Mock, rDEN-2/4Δ30, rDEN-4Δ30 vaccine, or rDEN-2Δ30 challenge strains and cells were stained by flow cytometry (Data shown from one representative donor). The percentages of infected cells were quantified by measuring E protein levels at 6, 24, and 48 hpi. (C) Heat map comparing specific cytokine production levels by MDDCs after mock, trivalent, tetravalent, or DEN-4Δ30 × 2 infection. Calculations were made using data acquired from 10-plex ELISA. The cytokine levels were calculated as the log2 values obtained in pg/mL (nine DC donors are represented. Red indicates greater expression levels. (D to F) Data are reflective of values represented in panel B. Black asterisks represent statistical significance by the two-way analysis of variance (ANOVA), with adjustment for multiple comparisons and the Bonferroni correction (*P ⩽ 0.05, **P ⩽ 0.01, **P ⩽ 0.001). (A color version of this figure is available in the online journal.)
The DENV-4 (rDEN-4Δ30) vaccine component drives the activation of the innate immune response in the tetravalent vaccine
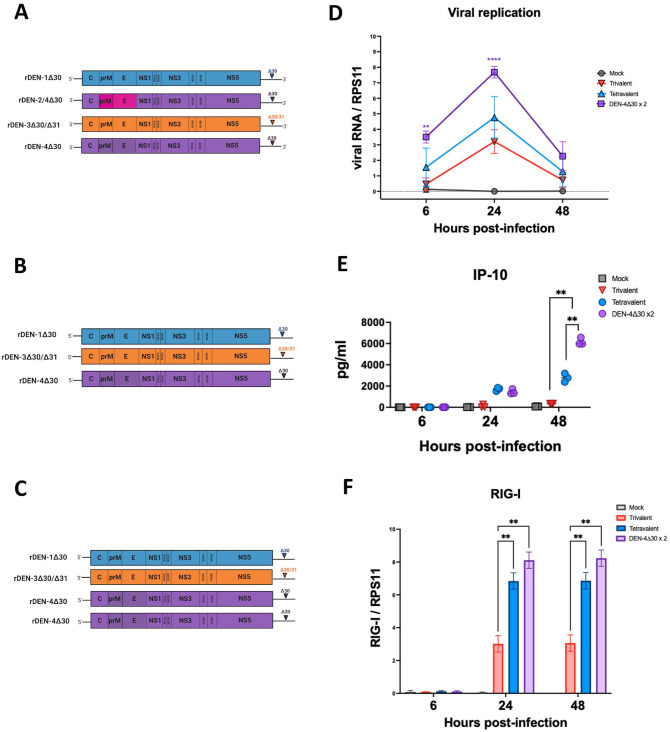
The tetravalent formulation is composed of a full length DENV-1 Western Pacific strain with a 30 nucleotide deletion In the 3' UTR, a DENV-2 component that is a chimera composed of the prM and E proteins of a DENV-2 New Guinea C strain Introduced into a full length DENV-4 (Dominica/81) strain with a 30 nucleotide deletion in the 3' UTR; this is due to the chimerization strategy employed to generate the rDEN-2/4 Δ30 component in which the prM and E proteins of DENV-2 New Guinea C strain were introduced into the backbone of the rDEN4Δ30 vaccine strain, a DENV-2 (Sleman/78) strain with the 30 nucleotide deletion and an additional 31 nucleotide deletion in the 3' UTR and a DENV-4 component of DENV-4 (Dominica/81) with a 30 nucleotide deletion in the 3' UTR, To further assess the contribution of the two DEN-4 Δ30 backbones to the immunogenicity of the tetravalent vaccine, we generated an admixture (DEN-4Δ30 × 2) composed of one component of rDEN-1Δ30, one component of rDEN-3Δ30 and two components of rDEN-4Δ30 (Figure 4(A)). In addition, to assess the contribution of the rDEN2/4Δ30 component, we also infected the MDDCs with a trivalent formulation including only rDEN-1Δ30, rDEN-3Δ30, and rDEN-4Δ30, and an MOI of 0.25 each (Figure 4(B)). We compared replication kinetics in MDDCs at 6, 24, and 48 hpi using RT-qPCR (Figure 4(C)). Despite infecting MDDCs with the same amount of plaque forming units per cell, the DENV-4Δ30×2 replicated to significantly higher levels than the original tetravalent vaccine formulation (Figure 4(C)). In a next step, we measured cyto- and chemokine production and found that the DENV-4Δ30×2 admixture induced significantly higher secretion levels of IP-10 when compared to the tetravalent and trivalent formulations (Figure 4(D)). To assess the effects of the vaccine formulations on immune sensing we quantified levels of RIG-I expression in Raji DC-SIGN cells infected with either the trivalent, tetravalent, or DENV-4Δ30×2 formulations. Our data show that both the tetravalent and DENV-4Δ30×2 formulations induced significantly higher levels of RIG-I expression at 24 and 48 hpi when compared to the trivalent formulation (Figure 4(E)). Therefore, our results demonstrate that the rDENV-4Δ30 vaccine component drives the innate immune response in MDDCs.
Figure 4.
The addition of a second DENV-4 component to the trivalent dengue vaccine results in an enhanced viral replication and IP-10 in dendritic cells (DCs). (A) The tetravalent admixture is composed of one component of the rDEN-1Δ30, rDEN-2/4Δ30, rDEN-3Δ30/31, and rDEN-4Δ30 vaccine viruses and was used to infect human monocyte-derived dendritic cells (MDDCs) at a multiplicity of infection (MOI) of 0.25 for each vaccine virus present. (B) The DEN-4Δ30 × 2 admixture is composed of one component of the DENV-1 and DENV-3 vaccines and two components of the DENV-4 vaccine virus and was used to infect human MDDCs at an MOI of 0.25 for each vaccine virus present. (C) The trivalent admixture is composed of one component of the rDEN-1Δ and rDEN-3Δ30/31 vaccines and one component of the rDEN-4Δ30 vaccine virus at an MOI of 0.25 for each vaccine virus present. (D) Real-time quantitative PCR (RT-qPCR) quantifying levels of viral replication in MDDCs infected with the indicated dengue vaccine admixtures. (E) Multiplex ELISA was performed as in previous assays to quantify the presence of IP-10 in the supernatant of MDDC cultures treated with the indicated DENV vaccine admixtures. (F) RT-qPCR capturing the induction of RIG-I. Expression levels were quantified in Raji DC-Sign cells infected with mock, trivalent vaccine, tetravalent vaccine or DEN-4Δ30 × 2 formulation. Statistical significance was calculated using Two-way analysis of variance (ANOVA) with multiple comparisons and Bonferroni correction (*P ⩽ 0.05, **P ⩽ 0.01). (A color version of this figure is available in the online journal.)
An additional the DENV-4 (rDEN-4Δ30) vaccine component favors migration of innate immune cells critical for the development of an adaptive immune response
In the last step, we aimed to examine how the vaccine formulation influences the migration of antigen-presenting cells like DCs, an important process in vivo where DCs have to migrate to lymph nodes in order to present the antigen to T cells for T cell priming. 53 We compared the tetravalent, trivalent and DEN-4Δ30×2 vaccine formulations in their ability to induce the cyto- and chemokine secretion by MDDCs. Interestingly, only the DEN-4Δ30×2 vaccine formulation induced significant induction of RANTES at 48 hpi (Figure 5(A)). As previously stated, RANTES is a key chemokine that plays an important role in the homing and migration of T-cells and influences the activation of the adaptive immune response. Next, we used CyTOF to capture single-cell data and expression profiles of specific immune markers. Treatment of MDDCs with all three vaccine formulations resulted in the upregulation of DC migration marker CCR7 indicating MDDCs migration to lymph nodes (Figure 5(C)). However, the tetravalent and DENV-4Δ30x2 formulations induced a significantly higher expression levels of CCR7 with respect to the trivalent formulation with DENV-4Δ30x2 formulation inducing the highest levels of CCR7 expression (Figure 5(C)). Similarly, the DC costimulatory marker CD40 shows the highest upregulation in MDDCs infected with the DEN-4Δ30×2 formulation when compared to the tetravalent and trivalent formulations (Figure 5(D)). Altogether these data indicated that the presence of an additional rDENV-4Δ30 component elicits the most MDDC activation and migration.
Figure 5.
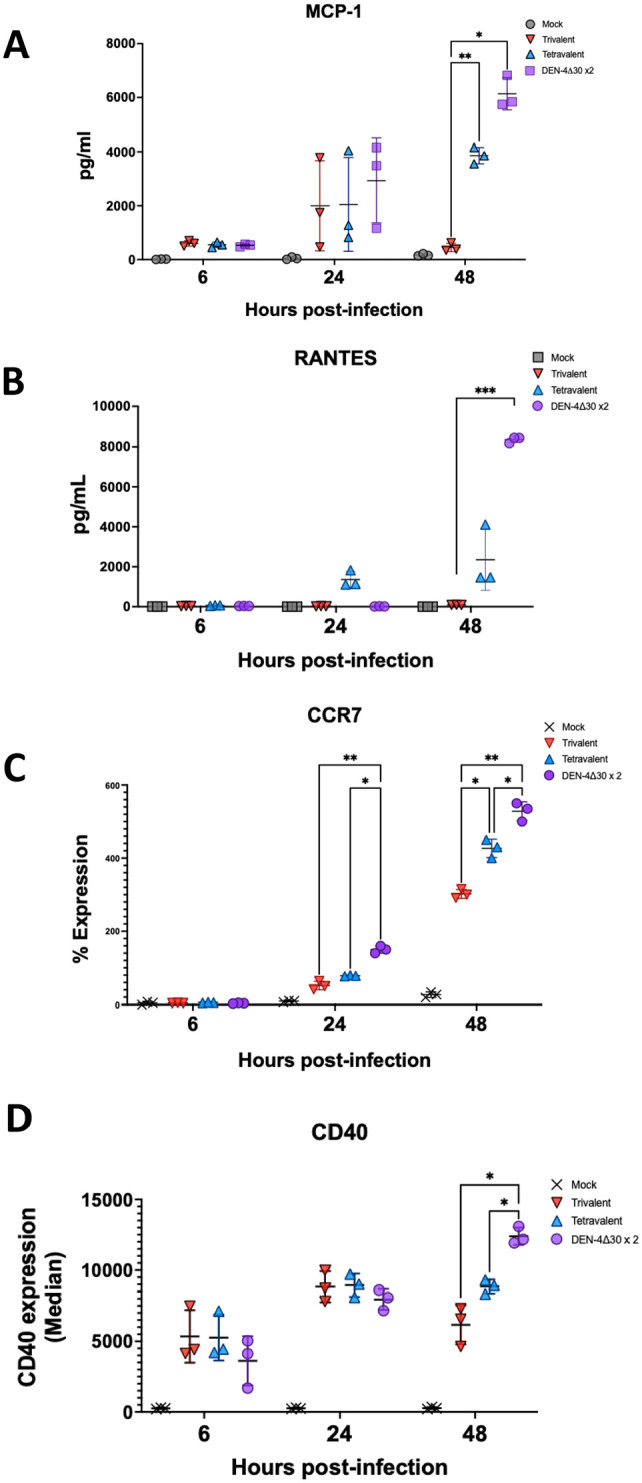
The presence of a second DENV-4 component confers induction of markers that are key in dendritic cell (DC) migration. (A, B) Monocyte-derived DCs (MDDCs) were infected with mock, trivalent vaccine, tetravalent vaccine or DEN-4Δ30 × 2 formulation. Multiplex ELISA was performed as in previous experiments to quantify the levels of MCP-1 and RANTES secretion, respectively, to the culture supernatant at 6, 24, and 48 hpi. (C) Using mass cytometry by time-of-flight (CyTOF) analysis for each mock, trivalent vaccine, tetravalent vaccine, or DEN-4Δ30 × 2 formulation sample. The percent of cells expressing CCR7 was quantified. (D) CyTOF depicting the median expression of CD40 of total cell populations is shown. Black asterisks represent statistical significance by the two-way analysis of variance (ANOVA), with adjustment for multiple comparisons and the Bonferroni correction (*P ⩽ 0.05, **P ⩽ 0.01, **P ⩽ 0.001). (A color version of this figure is available in the online journal.)
Discussion
The human immune responses induced in response to DENV infection and other flaviviruses are complex and characterized by high-level induction of pro-inflammatory chemokines and cytokines. 54 Despite there being multiple DENV vaccines in development, there is no bonafide correlate of protection that can be applied across the different types of DENV vaccines. With respect to the vaccines that are either licensed or in late clinical stages of development, the induction of neutralizing antibodies has been used as the benchmark for protective efficacy. However, neutralizing antibody was not predictive of protection in the Phase IIb efficacy trial of Dengvaxia. 21 In this report, we provide a comprehensive, detailed analysis of the infection kinetics and innate immune responses to the individual strains in the NIAID LATV TV003 vaccine. We demonstrate that a tetravalent formulation, which includes rDEN-4Δ30 induces enhanced immunogenicity. Individual DENV serotypes confer unique innate immune signatures; however, the rDEN-4Δ30 component is absolutely necessary to induce significant host innate immune reaction.
These results correlate with previous findings from collaborators that demonstrated that the tetravalent, but not trivalent vaccine formulation was protective against a DEN-2 challenge (rDEN-2Δ30). 49 To investigate if the immunogenicity elicited by the rDEN-4Δ30 was due to the role of the NS proteins of the virus, we compared the rDEN-2/4Δ30 and rDEN-4Δ30 vaccine strains with the DENV-2 challenge strain in MDDCs and evaluated differences in viral replication, infectivity and cyto- and chemokine secretion. We found that the DENV-2 challenge strain replicates to significantly higher levels when compared to the rDEN-2/4Δ30 and rDEN-4Δ30. Despite this, the rDEN-2Δ30 challenge virus induced significantly lower levels of pro-inflammatory cytokines that are key mediators of Type 1 IFN responses. However, the rDEN2Δ30 induced significantly higher levels of the anti-inflammatory marker IL-10. Altogether this supports previous findings from our group that DENV-4 strains confer immuno-stimulatory properties while DENV-2 Tonga shows a less immunogenic profile than other serotypes tested.
To further investigate which innate immune signatures are underlying this protective advantage witnessed in vaccinees receiving the tetravalent vaccine formulation, we compared infectivity profiles, infection kinetics and cytokine and chemokine profiles between the tetravalent and trivalent vaccines. Our data indicate that the tetravalent vaccine induces higher levels of monocyte migration markers when compared to the trivalent formulation (Figure 4(C) and (D)). We also tested a rDENV-4Δ30×2 vaccine formulation that would contain the same amount of virus as the tetravalent admixture. We found that the DEN-4Δ30×2 formulation replicated to significantly higher levels than the tetravalent and trivalent formulations. Given that the tetravalent vaccine and the DEN-4Δ30×2 formulation both contain two components of the rDEN-4Δ30 vaccine strain, one could anticipate that the tetravalent vaccine and the DEN-4Δ30×2 formulation would behave similarly in terms of replication. One possibility for the replicative differences seen is that the chimerization strategy employed to generate the DENV-2 vaccine may have introduced an additional source of attenuation. The DENV-4 backbone and non-structural proteins may not efficiently replicate the DENV-2 prM and E proteins. Another explanation could be the differences in binding efficiency between the prM and E proteins of DENV-2 and DENV-4. Previous data from our group demonstrated that a DENV-2 strain infects MDDCs to lower levels compared to a DENV-4 strain, but the cells that are infected with DENV-2 express higher levels of viral proteins. 55 This suggests that the E protein of DENV-4 strains may display enhanced capacity for binding immune cells, resulting in higher infection levels. Despite having the same amount of total virus, the DEN-4Δ30x2 vaccine formulation induced significantly higher levels of IP-10, suggesting that the addition of a DENV-4 vaccine component confers immunogenic properties. These data are consistent with our previous findings, in which DENV-4 was found to induce strong innate immune response compared to DENV-2 in human MDDCs and showed different infection kinetics in human DCs. 55
In addition to comparing the infectivity and replicative levels of the vaccines and the DENV-4Δ30×2 vaccine formulation, we assessed the phenotypic differences in immune markers that regulate the DC activation to induce cellular responses. 1 Given that there are serotype-specific differences in DENV NS proteins that antagonize host innate immune responses 56 and therefore may affect the magnitude and profile of the innate immune reaction including DCs activation. The tetravalent vaccine formulation induces higher magnitude of IP-10 and type I IFN which may drive the ability of DCs toward a TH1 response in the lymph node which is important for the stimulation of B cells to produce IgM and IgG 57 (Figure 5(C)). The results of this study support previous data from our group showing that WT DENV-4 induces enhanced production of IFNα, IP-10 and IL-12 which when compared to WT DENV-2 that may skew the adaptive immune response toward a Th1 response. 55 Interestingly, we found that expression of CCR7 and CD40, two markers that mediate the migration of DCs to the lymph node was significantly upregulated in the DEN-4Δ30×2 vaccine formulation relative to the tetravalent and trivalent formulations further supporting the significant role of the rDEN-4Δ30 component for vaccine efficacy. In hand with this, our data show that RANTES, a key chemokine that plays an important role in the homing and migration of T-cells25,26,29 was significantly upregulated in MDDCs infected with the DEN-4Δ30×2 vaccine formulation. Altogether these findings suggest that the presence of an additional rDEN-4Δ30 vaccine component confers robust immunogenic properties and contributes significantly to vaccine efficacy regardless of viral replication level.
In conclusion, our study highlights the importance of the rDEN-4Δ30 vaccine component for a robust host innate immune response. This response is characterized by an early and high secretion of IP-10 and IFNα2 and the upregulation of DCs activation and migration markers. Therefore, the results of this study highlight the innate immune profile underlying high vaccine protection and the importance to consider the innate immune profile of the different DENV vaccine strains for future vaccine design.
Supplemental Material
Supplemental material, sj-pdf-1-ebm-10.1177_15353702231151241 for The dengue virus 4 component of NIAID’s tetravalent TV003 vaccine drives its innate immune signature by Jessica Pintado Silva, Rafael Fenutria, Dabeiba Bernal-Rubio, Irene Sanchez-Martin, Annika Hunziker, Eva Chebishev, Jeury Veloz, Geoffrey Kelly, Seunghee Kim-Schulze, Steve Whitehead, Anna Durbin, Irene Ramos and Ana Fernandez-Sesma in Experimental Biology and Medicine
Acknowledgments
We would like to thank Michael Schotsaert for his support with the Luminex reader and the Gallios flow cytometer. We thank Mayte Suarez-Fariñas, Ruixue Hou, Lewis Tomalin, Sandra Bos, Adeeb Rahman and Eun-young Kim for insightful discussion. We thank Eva Harris and Viviana Simon for providing reagents. In addition, we would like to thank Jordi Ochando from the Mount Sinai Flow cytometry core for his support with the Aurora Spectral-flow cytometry. Finally, we would like to thank every member of the DHIPC consortium that was not explicitly recognized for the intellectual contribution.
Footnotes
Authors’ Contributions: All authors participated in the design, interpretation of the studies, and analysis of the data and review of the manuscript. Specific contribution; JPS and RF conducted the experiments, DB generated the MDDCs, IRS contributed to the analysis of Aurora-flow cytometry, EC contributed cell lines, IR contributed to the data analysis and experimental design, GK and SKS performed CyTOF experiments, SW provided the viruses used in the study, AD provided input in the experimental design and AFS procured funding, designed the study, edited the manuscript, and supervised the experiments and data analyses.
The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This work was funded partially by the NIAID Human Immunology Project Consortia (HIPC) AI118610 (DHIPC) to AFS and AD and the diversity supplement 3 U19 AI118610-04 S1 to JPS. Reagents provided by SW were funded by NIAID intramural funds.
ORCID iD: Jessica Pintado Silva  https://orcid.org/0000-0002-3405-6702
https://orcid.org/0000-0002-3405-6702
Supplemental Material: Supplemental material for this article is available online.
References
- 1. Weiskopf D, Angelo MA, Bangs DJ, Sidney J, Paul S, Peters B, de Silva AD, Lindow JC, Diehl SA, Whitehead S, Durbin A, Kirkpatrick B, Sette A. The human CD8+ T cell responses induced by a live attenuated tetravalent dengue vaccine are directed against highly conserved epitopes. J Virol 2015;89(1):120–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Centers for Disease Control and Prevention. Areas with risk of dengue, 2021, https://www.cdc.gov/dengue/areaswithrisk/index.html
- 3. WHO. Dengue and severe dengue, 2022, https://www.who.int/news-room/fact-sheets/detail/dengue-and-severe-dengue
- 4. Weiskopf D, Sette A. T-cell immunity to infection with dengue virus in humans. Front Immunol 2014;5:93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Yauch LE, Zellweger RM, Kotturi MF, Qutubuddin A, Sidney J, Peters B, Prestwood TR, Sette A, Shresta S. A protective role for dengue virus-specific CD8+ T cells. J Immunol 2009;182:4865–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Guy B, Barrere B, Malinowski C, Saville M, Teyssou R, Lang J. From research to phase III: preclinical, industrial and clinical development of the Sanofi Pasteur tetravalent dengue vaccine. Vaccine 2011;29:7229–41 [DOI] [PubMed] [Google Scholar]
- 7. Durbin AP, Vargas MJ, Wanionek K, Hammond SN, Gordon A, Rocha C, Balmaseda A, Harris E. Phenotyping of peripheral blood mononuclear cells during acute dengue illness demonstrates infection and increased activation of monocytes in severe cases compared to classic dengue fever. Virology 2008;376:429–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ho LJ, Wang JJ, Shaio MF, Kao CL, Chang DM, Han SW, Lai JH. Infection of human dendritic cells by dengue virus causes cell maturation and cytokine production. J Immunol 2001;166:1499–506 [DOI] [PubMed] [Google Scholar]
- 9. Kyle JL, Beatty PR, Harris E. Dengue virus infects macrophages and dendritic cells in a mouse model of infection. J Infect Dis 2007;195:1808–17 [DOI] [PubMed] [Google Scholar]
- 10. Libraty DH, Pichyangkul S, Ajariyakhajorn C, Endy TP, Ennis FA. Human dendritic cells are activated by dengue virus infection: enhancement by gamma interferon and implications for disease pathogenesis. J Virol 2001;75(8):3501–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Marovich M, Grouard-Vogel G, Louder M, Eller M, Sun W, Wu SJ, Putvatana R, Murphy G, Tassaneetrithep B, Burgess T, Birx D, Hayes C, Schlesinger-Frankel S, Mascola J. Human dendritic cells as targets of dengue virus infection. J Investig Dermatol Symp Proc 2001;6(3):219–24 [DOI] [PubMed] [Google Scholar]
- 12. Pham AM, Langlois RA, TenOever BR. Replication in cells of hematopoietic origin is necessary for dengue virus dissemination. PLoS Pathog 2012;8(1):e1002465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Wu SJ, Grouard-Vogel G, Sun W, Mascola JR, Brachtel E, Putvatana R, Louder MK, Filgueira L, Marovich MA, Wong HK, Blauvelt A, Murphy GS, Robb ML, Innes BL, Birx DL, Hayes CG, Frankel SS. Human skin Langerhans cells are targets of dengue virus infection. Nat Med 2000;6(7):816–20 [DOI] [PubMed] [Google Scholar]
- 14. Fujii S, Liu K, Smith C, Bonito AJ, Steinman RM. The linkage of innate to adaptive immunity via maturing dendritic cells in vivo requires CD40 ligation in addition to antigen presentation and CD80/86 costimulation. J Exp Med 2004;199:1607–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Banchereau J, Steinman RM. Dendritic cells and the control of immunity. Nature 1998;392:245–52 [DOI] [PubMed] [Google Scholar]
- 16. Lee AW, Truong T, Bickham K, Fonteneau JF, Larsson M, Da Silva I, Somersan S, Thomas EK, Bhardwaj N. A clinical grade cocktail of cytokines and PGE2 results in uniform maturation of human monocyte-derived dendritic cells: implications for immunotherapy. Vaccine 2002;20:A8–22 [DOI] [PubMed] [Google Scholar]
- 17. Rodriguez-Madoz JR, Belicha-Villanueva A, Bernal-Rubio D, Ashour J, Ayllon J, Fernandez-Sesma A. Inhibition of the type I interferon response in human dendritic cells by dengue virus infection requires a catalytically active NS2B3 complex. J Virol 2010;84(19):9760–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Rodriguez-Madoz JR, Bernal-Rubio D, Kaminski D, Boyd K, Fernandez-Sesma A. Dengue virus inhibits the production of type I interferon in primary human dendritic cells. J Virol 2010;84(9):4845–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Aguirre S, Maestre AM, Pagni S, Patel JR, Savage T, Gutman D, Maringer K, Bernal-Rubio D, Shabman RS, Simon V, Rodriguez-Madoz JR, Mulder LC, Barber GN, Fernandez-Sesma A. DENV inhibits type I IFN production in infected cells by cleaving human STING. PLoS Pathog 2012;8(10):e1002934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Guy B, Saville M, Lang J. Development of Sanofi Pasteur tetravalent dengue vaccine. Hum Vaccin 2010;6:696–705 [DOI] [PubMed] [Google Scholar]
- 21. Sabchareon A, Wallace D, Sirivichayakul C, Limkittikul K, Chanthavanich P, Suvannadabba S, Jiwariyavej V, Dulyachai W, Pengsaa K, Wartel TA, Moureau A, Saville M, Bouckenooghe A, Viviani S, Tornieporth NG, Lang J. Protective efficacy of the recombinant, live-attenuated, CYD tetravalent dengue vaccine in Thai schoolchildren: a randomised, controlled phase 2b trial. Lancet 2012;380: 1559–67 [DOI] [PubMed] [Google Scholar]
- 22. Biswal S, Borja-Tabora C, Martinez Vargas L, Velasquez H, Theresa Alera M, Sierra V, Johana Rodriguez-Arenales E, Yu D, Wickramasinghe VP, Duarte Moreira E, Jr, Fernando AD, Gunasekera D, Kosalaraksa P, Espinoza F, Lopez-Medina E, Bravo L, Tuboi S, Hutagalung Y, Garbes P, Escudero I, Rauscher M, Bizjajeva S, LeFevre I, Borkowski A, Saez-Llorens X, Wallace D, TIDES study group. Efficacy of a tetravalent dengue vaccine in healthy children aged 4–16 years: a randomised, placebo-controlled, phase 3 trial. Lancet 2020;395:1423–33 [DOI] [PubMed] [Google Scholar]
- 23. Takeda. Takeda’s dengue vaccine candidate provides continued protection against dengue fever through 4.5 years in pivotal clinical trial. Takeda, 9 June 2022, https://www.takeda.com/newsroom/newsreleases/2022/takedas-dengue-vaccine-candidate-provides-continued-protection-against-dengue-fever-through-4.5-years-in-pivotal-clinical-trial/
- 24. Durbin AP, Kirkpatrick BD, Pierce KK, Elwood D, Larsson CJ, Lindow JC, Tibery C, Sabundayo BP, Shaffer D, Talaat KR, Hynes NA, Wanionek K, Carmolli MP, Luke CJ, Murphy BR, Subbarao K, Whitehead SS. A single dose of any of four different live attenuated tetravalent dengue vaccines is safe and immunogenic in flavivirus-naive adults: a randomized, double-blind clinical trial. J Infect Dis 2013;207:957–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Alvarez DE, De Lella Ezcurra AL, Fucito S, Gamarnik AV. Role of RNA structures present at the 3’UTR of dengue virus on translation, RNA synthesis, and viral replication. Virology 2005;339:200–12 [DOI] [PubMed] [Google Scholar]
- 26. Durbin A. Evaluating the safety and immune response to a dengue virus vaccine in healthy adults, 2015, https://www.clinicaltrials.gov/ct2/show/NCT01931176
- 27. Durbin AP, McArthur JH, Marron JA, Blaney JE, Thumar B, Wanionek K, Murphy BR, Whitehead SS. rDEN2/4Delta30(ME), a live attenuated chimeric dengue serotype 2 vaccine is safe and highly immunogenic in healthy dengue-naive adults. Hum Vaccin 2006;2:255–60 [DOI] [PubMed] [Google Scholar]
- 28. Blaney JE, Jr, Durbin AP, Murphy BR, Whitehead SS. Development of a live attenuated dengue virus vaccine using reverse genetics. Viral Immunol 2006;19(1):10–32 [DOI] [PubMed] [Google Scholar]
- 29. Durbin AP, Whitehead SS, McArthur J, Perreault JR, Blaney JE, Jr, Thumar B, Murphy BR, Karron RA. rDEN4delta30, a live attenuated dengue virus type 4 vaccine candidate, is safe, immunogenic, and highly infectious in healthy adult volunteers. J Infect Dis 2005;191:710–8 [DOI] [PubMed] [Google Scholar]
- 30. Villar L, Dayan GH, Arredondo-Garcia JL, Rivera DM, Cunha R, Deseda C, Reynales H, Costa MS, Morales-Ramirez JO, Carrasquilla G, Rey LC, Dietze R, Luz K, Rivas E, Miranda Montoya MC, Cortes Supelano M, Zambrano B, Langevin E, Boaz M, Tornieporth N, Saville M, Noriega F, Group CYDS. Efficacy of a tetravalent dengue vaccine in children in Latin America. N Engl J Med 2015;372:113–23 [DOI] [PubMed] [Google Scholar]
- 31. Durbin AP, McArthur J, Marron JA, Blaney JE, Jr, Thumar B, Wanionek K, Murphy BR, Whitehead SS. The live attenuated dengue serotype 1 vaccine rDEN1Delta30 is safe and highly immunogenic in healthy adult volunteers. Hum Vaccin 2006;2(4):167–73 [DOI] [PubMed] [Google Scholar]
- 32. Blaney JE, Jr, Hanson CT, Firestone CY, Hanley KA, Murphy BR, Whitehead SS. Genetically modified, live attenuated dengue virus type 3 vaccine candidates. Am J Trop Med Hyg 2004;71(6):811–21 [PubMed] [Google Scholar]
- 33. Blaney JE, Jr, Matro JM, Murphy BR, Whitehead SS. Recombinant, live-attenuated tetravalent dengue virus vaccine formulations induce a balanced, broad, and protective neutralizing antibody response against each of the four serotypes in rhesus monkeys. J Virol 2005;79(9):5516–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Durbin AP, Karron RA, Sun W, Vaughn DW, Reynolds MJ, Perreault JR, Thumar B, Men R, Lai CJ, Elkins WR, Chanock RM, Murphy BR, Whitehead SS. Attenuation and immunogenicity in humans of a live dengue virus type-4 vaccine candidate with a 30 nucleotide deletion in its 3’-untranslated region. Am J Trop Med Hyg 2001;65(5):405–13 [DOI] [PubMed] [Google Scholar]
- 35. Durbin AP, Kirkpatrick BD, Pierce KK, Schmidt AC, Whitehead SS. Development and clinical evaluation of multiple investigational monovalent DENV vaccines to identify components for inclusion in a live attenuated tetravalent DENV vaccine. Vaccine 2011;29:7242–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Durbin AP, Schmidt A, Elwood D, Wanionek KA, Lovchik J, Thumar B, Murphy BR, Whitehead SS. Heterotypic dengue infection with live attenuated monotypic dengue virus vaccines: implications for vaccination of populations in areas where dengue is endemic. J Infect Dis 2011;203:327–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Hanley KA, Manlucu LR, Manipon GG, Hanson CT, Whitehead SS, Murphy BR, Blaney JE., Jr. Introduction of mutations into the non-structural genes or 3’ untranslated region of an attenuated dengue virus type 4 vaccine candidate further decreases replication in rhesus monkeys while retaining protective immunity. Vaccine 2004;22:3440–8 [DOI] [PubMed] [Google Scholar]
- 38. Lindow JC, Durbin AP, Whitehead SS, Pierce KK, Carmolli MP, Kirkpatrick BD. Vaccination of volunteers with low-dose, live-attenuated, dengue viruses leads to serotype-specific immunologic and virologic profiles. Vaccine 2013;31:3347–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. McArthur JH, Durbin AP, Marron JA, Wanionek KA, Thumar B, Pierro DJ, Schmidt AC, Blaney JE, Jr, Murphy BR, Whitehead SS. Phase I clinical evaluation of rDEN4Delta30-200,201: a live attenuated dengue 4 vaccine candidate designed for decreased hepatotoxicity. Am J Trop Med Hyg 2008;79(5):678–84 [PMC free article] [PubMed] [Google Scholar]
- 40. Troyer JM, Hanley KA, Whitehead SS, Strickman D, Karron RA, Durbin AP, Murphy BR. A live attenuated recombinant dengue-4 virus vaccine candidate with restricted capacity for dissemination in mosquitoes and lack of transmission from vaccinees to mosquitoes. Am J Trop Med Hyg 2001;65(5):414–9 [DOI] [PubMed] [Google Scholar]
- 41. Whitehead SS, Falgout B, Hanley KA, Blaney JE, Jr, Markoff L, Murphy BR, Markoff L, Murphy BR. A live, attenuated dengue virus type 1 vaccine candidate with a 30-nucleotide deletion in the 3’ untranslated region is highly attenuated and immunogenic in monkeys. J Virol 2003;77(2):1653–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Wright PF, Durbin AP, Whitehead SS, Ikizler MR, Henderson S, Blaney JE, Thumar B, Ankrah S, Rock MT, McKinney BA, Murphy BR, Schmidt AC. Phase 1 trial of the dengue virus type 4 vaccine candidate rDEN4{Delta}30-4995 in healthy adult volunteers. Am J Trop Med Hyg 2009;81(5):834–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Kasturi SP, Skountzou I, Albrecht RA, Koutsonanos D, Hua T, Nakaya HI, Ravindran R, Stewart S, Alam M, Kwissa M, Villinger F, Murthy N, Steel J, Jacob J, Hogan RJ, Garcia-Sastre A, Compans R, Pulendran B. Programming the magnitude and persistence of antibody responses with innate immunity. Nature 2011;470:543–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Pulendran B. Learning immunology from the yellow fever vaccine: innate immunity to systems vaccinology. Nat Rev Immunol 2009;9(10):741–7 [DOI] [PubMed] [Google Scholar]
- 45. Bustos-Arriaga J, Gromowski GD, Tsetsarkin KA, Firestone CY, Castro-Jimenez T, Pletnev AG, Cedillo-Barron L, Whitehead SS. Decreased accumulation of subgenomic RNA in human cells infected with vaccine candidate DEN4Delta30 increases viral susceptibility to type I interferon. Vaccine 2018;36:3460–7 [DOI] [PubMed] [Google Scholar]
- 46. Imad HA, Phumratanaprapin W, Phonrat B, Chotivanich K, Charunwatthana P, Muangnoicharoen S, Khusmith S, Tantawichien T, Phadungsombat J, Nakayama E, Konishi E, Shioda T. Cytokine expression in dengue fever and dengue hemorrhagic fever patients with bleeding and severe hepatitis. Am J Trop Med Hyg 2020;102(5):943–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Bozza FA, Cruz OG, Zagne SM, Azeredo EL, Nogueira RM, Assis EF, Bozza PT, Kubelka CF. Multiplex cytokine profile from dengue patients: MIP-1beta and IFN-gamma as predictive factors for severity. BMC Infect Dis 2008;8:86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Chen JP, Lu HL, Lai SL, Campanella GS, Sung JM, Lu MY, Wu-Hsieh BA, Lin YL, Lane TE, Luster AD, Liao F. Dengue virus induces expression of CXC chemokine ligand 10/IFN-gamma-inducible protein 10, which competitively inhibits viral binding to cell surface heparan sulfate. J Immunol 2006;177:3185–92 [DOI] [PubMed] [Google Scholar]
- 49. Hou R, Tomalin LE, Silva JP, Kim-Schulze S, Whitehead SS, Fernandez-Sesma A, Durbin AP, Suarez-Farinas M. The innate immune response following multivalent dengue vaccination and implications for protection against dengue challenge. JCI Insight 2022;7:e157811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Islam M, Kalita T, Saikia AK, Begum A, Baruah V, Singh N, Borkotoky R, Bose S. Significance of RANTES-CCR5 axis and linked downstream immunomodulation in dengue pathogenesis: a study from Guwahati, India. J Med Virol 2019;91(12):2066–73 [DOI] [PubMed] [Google Scholar]
- 51. Kwissa M, Nakaya HI, Onlamoon N, Wrammert J, Villinger F, Perng GC, Yoksan S, Pattanapanyasat K, Chokephaibulkit K, Ahmed R, Pulendran B. Dengue virus infection induces expansion of a CD14(+)CD16(+) monocyte population that stimulates plasmablast differentiation. Cell Host Microbe 2014;16:115–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Anna Durbin KP. Evaluating the safety and protective efficacy of a single dose of a trivalent live attenuated dengue vaccine to protect against infection with DENV-2, 2018, https://clinicaltrials.gov/ct2/show/NCT02433652
- 53. Yewdall AW, Drutman SB, Jinwala F, Bahjat KS, Bhardwaj N. CD8+ T cell priming by dendritic cell vaccines requires antigen transfer to endogenous antigen presenting cells. PLoS ONE 2010;5:e11144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Lee YH, Leong WY, Wilder-Smith A. Markers of dengue severity: a systematic review of cytokines and chemokines. J Gen Virol 2016;97(12): 3103–19 [DOI] [PubMed] [Google Scholar]
- 55. Hamlin RE, Rahman A, Pak TR, Maringer K, Mena I, Bernal-Rubio D, Potla U, Maestre AM, Fredericks AC, Amir ED, Kasarskis A, Ramos I, Merad M, Fernandez-Sesma A. High-dimensional CyTOF analysis of dengue virus-infected human DCs reveals distinct viral signatures. JCI Insight 2017;2:e92424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Hannemann H, Sung P-Y, Chiu H-C, Yousuf A, Bird J, Lim SP, Davidson AD. Serotype-specific differences in dengue virus non-structural protein 5 nuclear localization. J Biol Chem 2013;288:22621–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Tian Y, Grifoni A, Sette A, Weiskopf D. Human T cell response to dengue virus infection. Front Immunol 2019;10:2125. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material, sj-pdf-1-ebm-10.1177_15353702231151241 for The dengue virus 4 component of NIAID’s tetravalent TV003 vaccine drives its innate immune signature by Jessica Pintado Silva, Rafael Fenutria, Dabeiba Bernal-Rubio, Irene Sanchez-Martin, Annika Hunziker, Eva Chebishev, Jeury Veloz, Geoffrey Kelly, Seunghee Kim-Schulze, Steve Whitehead, Anna Durbin, Irene Ramos and Ana Fernandez-Sesma in Experimental Biology and Medicine