Abstract
Programmed cell death (PCD) in bacteria plays an important role in developmental processes, such as lysis of the mother cell during sporulation of Bacillus subtilis and lysis of vegetative cells in fruiting body formation of Myxococcus xanthus. The signal transduction pathway leading to autolysis of the mother cell includes the terminal sporulation sigma factor EςK, which induces the synthesis of autolysins CwlC and CwlH. An activator of autolysin in this and other PCD processes is yet to be identified. Autolysis plays a role in genetic exchange in Streptococcus pneumoniae, and the gene for the major autolysin, lytA, is located in the same operon with recA. DNA from lysed cells is picked up by their neighbors and recombined into the chromosome by RecA. LytA requires an unknown activator controlled by a sensory kinase, VncS. Deletion of vncS inhibits autolysis and also decreases killing by unrelated antibiotics. This observation suggests that PCD in bacteria serves to eliminate damaged cells, similar to apoptosis of defective cells in metazoa. The presence of genes affecting survival without changing growth sensitivity to antibiotics (vncS, lytA, hipAB, sulA, and mar) indicates that bacteria are able to control their fate. Elimination of defective cells could limit the spread of a viral infection and donate nutrients to healthy kin cells. An altruistic suicide would be challenged by the appearance of asocial mutants without PCD and by the possibility of maladaptive total suicide in response to a uniformly present lethal factor or nutrient depletion. It is proposed that a low rate of mutation serves to decrease the probability that asocial mutants without PCD will take over the population. It is suggested that PCD is disabled in persistors, rare cells that are resistant to killing, to ensure population survival. It is suggested that lack of nutrients leads to the stringent response that suppresses PCD, producing a state of tolerance to antibiotics, allowing cells to discriminate between nutrient deprivation and unrepairable damage. High levels of persistors are apparently responsible for the extraordinary survival properties of bacterial biofilms, and genes affecting persistence appear to be promising targets for development of drugs aimed at eradicating recalcitrant infections. PCD in unicellular eukaryotes is also considered, including aging in Saccharomyces cerevisiae. Apoptosis-like elimination of defective cells in S. cerevisiae and protozoa suggests that all unicellular life forms evolved altruistic programmed death that serves a variety of useful functions.
In animals, apoptosis serves to eliminate cells in the course of development and to eradicate defective cells (61). Cancerous transformation, microbial infection, and lethal factors such as heat, mutagens, oxidants, and chemical toxins all lead to apoptosis. Cell damage is sensed by receptors activating apoptotic signal transduction pathways that converge mainly at the level of caspase proteases.
As in metazoa, programmed cell death (PCD) plays an important role in a number of developmental processes in bacteria, such as lysis of the mother cell in sporulation, lysis of vegetative cells in myxobacterial fruiting body formation, and DNA transformation liberated from cells of streptococci undergoing spontaneous autolysis. A considerable body of data suggests that microorganisms also evolved programmed death of defective cells. A critical examination of this possibility is the focus of the present review.
Perhaps the most common observation of possible PCD in bacteria is autolysis of cells exposed to antibiotics and other harmful conditions. Autolysis is a self-digestion of the cell wall by peptidoglycan hydrolases called autolysins (40, 84). Both synthesis and hydrolysis of peptidoglycan are necessary for building the cell wall, and at least some autolysins are part of this normal cell growth activity. Traditionally, autolysis has been viewed as a maladaptive disbalance in these processes caused by inhibition of cell growth. I will review data suggesting that autolysis represents PCD in bacteria. Other instances of bacterial cell death, not necessarily tied to autolysis, also point to a PCD process and will be considered in this review. The cases I will consider are primarily those in which particular genes affecting cell survival have been identified.
It is not immediately apparent, to put it mildly, that a defective unicellular organism will benefit from committing suicide. This simple consideration has no doubt impeded a serious inquiry into programmed death in bacteria. At the same time, it is becoming increasingly apparent that bacteria live and die in complex communities that in many ways resemble a multicellular organism. Release of pheromones induces bacteria in a population to respond in concert by changing patterns of gene expression, a phenomenon called quorum sensing (30, 38, 75), and reactive oxygen species cause self-aggregation in Escherichia coli (14), a behavior thought to provide mutual protection to the cells. Most bacterial species actually do not live as planktonic suspensions in vivo but form complex biofilms, tightly knit communities of cells (19). From this perspective, programmed death of damaged cells may be beneficial to a multicellular bacterial community. For example, suicide could limit the spread of a viral infection. In the case of serious damage by toxic factors, cells will donate their nutrients to their neighbors instead of draining resources from their kin in a futile attempt to repair themselves. Finally, elimination of cells with damaged DNA would contribute to maintenance of a low mutation rate.
Even if one accepts that programmed death makes sense for a microbial community, the concept of altruistic suicide faces a number of serious challenges. What will happen if a lethal factor such as an antibiotic diffuses throughout the population and damages all the cells? Does the entire population commit suicide? This would clearly be counterproductive, and it is proposed that “persistor” variants are cells in which PCD has been disabled and which are responsible for the survival of the population under these conditions. It appears that persistors are especially important for the extraordinary survival properties of biofilms. What will happen if a nutrient concentration falls? How will a cell distinguish this from unrepairable damage? It is suggested that tolerance—a decreased sensitivity to antibiotics in slow-growing cells—evolved to discriminate between lack of nutrients and unrepairable damage. Finally, mutant egoists can arise and take over a population of altruists. It is suggested that efficient repair and low mutation rate in microorganisms evolved to counter this threat. In the absence of group selection based on social behavior, both cheaters and mutator variants arise in bacterial populations.
This review concludes with an analysis of PCD in eukaryotic unicellular organisms that face similar challenges to altruistic social behavior.
PROGRAMMED DEATH IN BACTERIAL DEVELOPMENT
Sporulation.
Apoptosis in eukaryotic cells was discovered in studies of Caenorhabditis elegans development; the death of certain cells is a necessary stage in ontogenesis (61). Similarly, autolysis is part of the developmental process in a number of bacterial species.
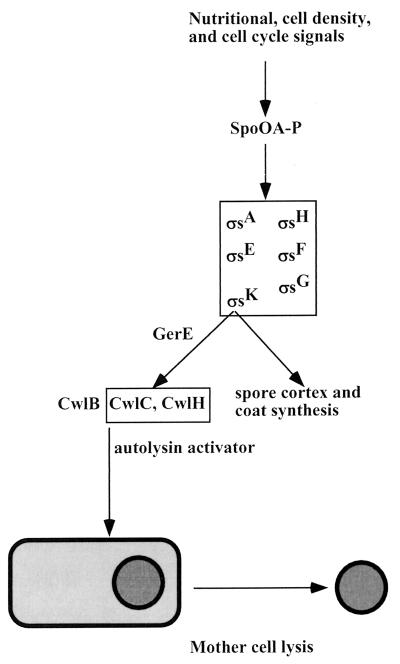
In sporulating Bacillus subtilis, the mother cell is actively lysed prior to release of the spore. Three autolysins have been identified that participate in mother cell lysis. CwlB is the major autolysin produced at the end of the exponential growth phase. A double cwlB cwlC mutant was defective in mother cell lysis (88), but single mutants showed lysis. Similarly, double but not single cwlC cwlH mutants were defective in mother cell lysis (70). CwlC becomes the major autolysin at late sporulation stage. Both CwlC and a minor autolysin, CwlH, are transcribed by sporulation-specific EςK RNA polymerase and require GerE (a coat protein transcriptional activator) for expression (70). Expression of ςK in the process of sporulation has been described in great detail (51, 52, 92). ςK is the final regulator in a complex pathway that begins with Spo0A, a transcriptional factor that integrates signals from nutritional status and cell cycle and in turn activates a cascade of interdependent sigma factors in the forespore and mother cell (Fig. 1). EςK is responsible for spore coat and cortex formation and for mother cell lysis.
FIG. 1.
Programmed death of mother cell in sporulation development of B. subtilis. Environmental as well as internal signals are integrated at the level of Spo0A, which in turn activates a cascade of sporulation sigma factors. The terminal sigma factor ςk controls the final stages of sporulation—formation of the spore cortex, coat synthesis, and expression of autolysins CwlC and CwlH. Activation of autolysins by an unknown factor causes mother cell lysis and liberation of the spore.
It might seem that the knowledge of the signal transduction pathway controlling programmed death of the mother cell is fairly complete. However, the mere presence of autolysin is not sufficient for lysis. Autolysis usually requires an additional, unidentified factor to activate an autolysin. Indeed, an autonomously active CwlB, for example, would exterminate cells at the end of the exponential state of growth. An additional activator released after the spore is formed would allow specific targeting of the mother cell for autolysis.
The obvious function of mother cell autolysis is to eliminate a barrier that could interfere with the outgrowth of a germinating spore. There might be an additional function of autolysis—nutrients released by the mother cell could be used by kin cells, helping them complete the energy-demanding process of sporulation. It is interesting to note that in B. subtilis, rapid autolysis is also observed if cells are transferred into a medium lacking a carbon source. Autolysis is activated by a decrease in proton motive force rather than by a drop in ATP (48). Apparently, sporulation cannot proceed in the complete absence of nutrients. Altruistic lysis under such conditions might provide some of the kin cells with enough of an energy source to sporulate. A simpler explanation, of course, is that B. subtilis cannot cope with a sudden decrease in energy, and cell lysis is a maladaptive consequence of a breakdown in the control of the cell wall synthesis machinery. While one cannot discriminate between these possibilities on the basis of what is currently known, the behavior of a ruminant bacterium, Fibrobacter succinogenes, under similar circumstances provides a telling example, suggesting that bacteria do not leave much to chance. A considerable portion of F. succinogenes cells lyse in a logarithmically growing culture, but if a sugar is withdrawn, autolysis is inhibited by a secreted protease that digests the autolysins (102). This behavior is the direct opposite of what transpires in a B. subtilis population under similar conditions. The ruminant symbiont converts sugar into protein and might be lysing to supply the host with valuable nutrients. When sugar is not provided, as in between the host meals, there is no point for the symbiont to lyse, and lysis is specifically inhibited.
M. xanthus development.
Fruiting body formation and sporulation in Myxococcus bacteria is another example of controlled death aiding development (25, 65, 81). “Autocides,” which include fatty acids as well as glucosamine, induce autolysis in dense cultures of Myxococcus xanthus and are required for normal fruiting body development and sporulation. The pathway leading from these inducers to autolysis is not known, and neither is the exact role of autolysis in development. Most likely, lysis of vegetative cells is altruistic suicide, and the released nutrients feed the fruiting body and developing spores.
Programmed death and sex.
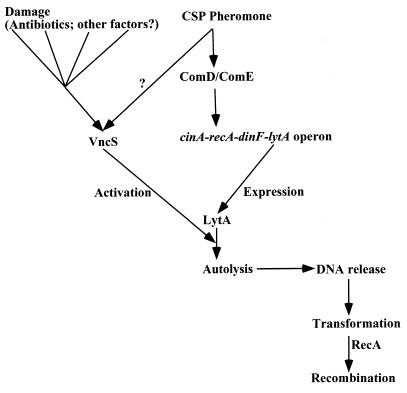
Another example of specialized adaptive autolysis related to development comes from studies of genetic exchange. In a number of species, autolysis is part of a well-controlled mechanism of natural transformation. The released DNA is picked up by cells that did not lyse. Interestingly, the gene for the main autolysin in Streptococcus pneumoniae, lytA, is located in the same operon with recA, which is responsible for homologous recombination with the incoming DNA (62). Both RecA and LytA are induced by the quorum-sensing factor, a peptide pheromone that accumulates at high cell density (Fig. 2).
FIG. 2.
PCD in autolysis of S. pneumoniae. At high cell density, the concentration of a released peptide pheromone (CSP) increases and activates a two-component signal transduction kinase (ComD) and its response regulator (ComE) (2 and references therein). Phosphorylated ComE in turn activates transcription of a competence operon containing genes required for recombination of incoming DNA and the lytA gene, responsible for autolysis. Released DNA is picked up by other cells and recombined with the aid of RecA. LytA requires activation, which is controlled by the sensory kinase VncS. Signals originating from cell damage by antibiotics and possibly by other factors converge prior to or at the level of VncS and activate LytA, causing elimination of defective cells by autolysis.
GENES CONTROLLING CELL DEATH AND SURVIVAL
S. pneumoniae lysis by antibiotics.
The three types of specialized bacterial autolysis participating in development and genetic exchange described above are clearly adaptive and are controlled by programmed death pathways. A number of intriguing observations suggest that programmed death in bacteria also occurs in response to damage, analogously to apoptosis of defective animal cells.
In a recent study of S. pneumoniae, a collection of transposon insertion mutants were screened for resistance to lysis by penicillin (68). Mutations in over 10 genes were obtained, and two of them were identified and characterized. One mutation was in a psa locus coding for an ABC-type Mn translocase (68). The mutation had a pleiotropic phenotype, and its connection to lysis is unclear. Another mutant was more interesting—the affected gene, named vncS, was homologous to VanS, the kinase of the two-component regulatory system controlling induction of vancomycin resistance in Enterococcus faecalis (69). Remarkably, vncS mutants were reported to be resistant to killing by a range of antibiotics hitting unrelated targets (the cell wall, DNA gyrase, and the ribosome, it is not clear if death from antibiotics not targeting the cell wall resulted from lysis). At the same time, growth of the vncS mutant was inhibited by antibiotics as effectively as in the wild type, showing that antibiotics were able to act normally against their targets in vncS cells. In S. pneumoniae, autolysin LytA is responsible for autolysis, and lytA mutants are resistant to killing by cell wall inhibitors like penicillin (95) (antibiotics that are not cell wall inhibitors were not tested with the lytA mutant). LytA was normally expressed in the vncS strain. Apparently, VncS does not control the synthesis of LytA but rather regulates expression of an unknown factor that activates LytA in response to antibiotic action (69) (Fig. 2). vncR, a gene for a putative response regulator, was located in the same operon with vncS. Deletion of vncR caused no obvious phenotype, and the mutant showed normal autolysis. It was suggested that signal transduction from VncS might pass via redundant response regulators, one of which is VncR. It is not known what VncS might be sensing.
What can be concluded from these experiments is that VncS is a component of the autolytic pathway and is required for autolysis induced by antibiotics. But why do antibiotics induce autolysis? One possibility is that the normal pathway of VncS-dependent autolysis in S. pneumoniae is somehow unnaturally activated by different antibiotics (69). Alternatively, elimination of defective cells damaged by antibiotics could represent programmed death.
Persistence—survival of rare cells.
One potentially severe problem with programmed death in bacteria is that suicide could eliminate all cells of a clonal population in response to damage. Indeed, a chemical toxin diffusing into a population can reach and equally damage all cells. A resulting programmed death of the entire population would be counterproductive. However, it appears that bacteria devised an intelligent strategy to avoid such a disastrous outcome. In 1944, Bigger discovered that addition of penicillin to staphylococci produced lysis, but even after prolonged incubation, a small fraction of cells (10−6) remained fully viable (8). These “persistors” are not mutants by two criteria—their growth is completely sensitive to penicillin, and their progeny are not more resistant to lysis by penicillin than the original population. The existence of persistors suggests that a bacterial cell potentially has the choice of whether to live or die, and the real puzzle is not how the rare cells survive, but why the majority of cells choose to be killed by antibiotics. The phenomenon of persistence has been recorded in a wide variety of bacterial species treated with a range of deleterious factors (50). For example, Luria and Laterjet described a “subpopulation” of E. coli with increased resistance to UV (57); we have observed that a small fraction of cells in a Pseudomonas aeruginosa biofilm population are virtually insensitive to killing by antibiotics (11). It seems that bacteria came up with an optimal strategy to respond to external damaging factors. The majority of cells have a program that will determine whether to repair damage or activate suicide, while the suicide program is disabled in a small fraction of persistors in case the damaging factor reaches the entire population.
The nature of persistence is unknown, but it does not seem likely that a special regulatory program is able to assign as few as 10−6 cells to perform a particular function, in this case survival. In bacteria, the number of molecules of a regulator can be very small, for example, about 10 molecules of LacI repressor per E. coli cell. If one or more of the regulators controlling cell survival (like SulA and HipA; see below) were present in small amounts, a random variation in the number of these proteins would produce rare persisting variants. The rate of persistors would then depend on the mean number of molecules of a regulator produced and on the variation of the mean. The evolved setting of this mean and its variation would depend on the need for making persistor cells.
Genes controlling death of E. coli.
Similarly to vncS of S. pneumoniae, a number of regulatory genes were found to dramatically affect cell survival in E. coli without influencing the ability of a cell to grow in the presence of a lethal factor.
A targeted screen for E. coli genes affecting persistence was performed by Moyed and coworkers (63). The rationale was to enrich an ethyl methanesulfonate-mutagenized population with cells surviving penicillin treatment and then screen for colonies producing higher numbers of penicillin persistors. Only mutants whose growth was normally inhibited by penicillin were examined further. This straightforward approach led to the identification of three independent hip (high persistence) loci. All hip mutants produced about 1,000-fold-more persistent cells than the wild type. One of the loci, hipAB, was cloned and sequenced (10, 64). This was the first report of bacterial genes specifically involved in regulation of cell death. A knockout of the wild-type hipA gene did not have an apparent phenotype, while a hipB knockout was not obtained, indicating that a null mutant is nonviable. Deletion of both hipA and hipB was obtained, and this strain and the wild type had similar persistence in the presence of penicillin (9). Biochemical studies have shown that HipB is a transcriptional repressor that binds to the promoter region of the hip operon. HipB is a 10-kDa helix-turn-helix DNA-binding protein that forms a dimer and forms a tight 1-1 complex with HipA. Moderate expression of cloned wild-type HipA produced the same phenotype as the original hipA or hipB mutation—the level of cells persisting through penicillin killing was increased (28). The fact that a null hipB is likely lethal indicates that overexpression of HipA causes death. Indeed, high expression of HipA from a controllable promoter inhibited cell growth, although cell survival was not examined under these conditions (28). Apparently, HipA has the potential to act both as an inhibitor of cell death and as a killing factor. It was suggested that mutants with increased persistence have a decreased affinity of HipA-HipB binding and thus have a higher (and moderate) level of free HipA that protects cells from killing by ampicillin.
Most importantly, the hip mutants showed increased resistance to factors unrelated to ampicillin. Mutated cells had a 1,000-fold-higher survival rate to thymine starvation, which leads to DNA degradation (83), and were more resistant to quinolone antibiotics, which target DNA gyrase and topoisomerase (28). Even more strikingly, the hip mutation protected htpR cells, deficient in induction of heat shock proteins, from killing by increased temperature (83). In this case, hip mutation conferred the highest degree of protection—htpR cells decreased 100-fold after a short incubation at 42°C, while virtually no decrease was observed in htpR hipA cells. Mutations in hipAB did not protect cells from kanamycin, another drug that was tested. Two explanations were offered to account for the high survival of persistors. Koch suggested that survivors of lethal treatments are “dormant” cells present in a population (50). Alternatively, it was suggested that there is a certain stage in the cell cycle that is resistant to penicillin (28, 83). However, it is hard to imagine a distinct developmental stage that occupies a 10−6 part of the cell cycle (cf. the rate of persistors to penicillin), which would mean that it has a duration of ≤30 min/106 = 1.8 (ms). The fact that a mutation can increase the rate of persistors to 100% (as in the case of temperature resistance) suggests that persistors are not dormant and do not represent a special stage of the cell cycle. Rather, the study of hip genes shows that E. coli has a way to dramatically increase survival in the presence of a variety of unrelated lethal factors, but the majority of cells “choose” to die. hip mutants have not been found among natural E. coli isolates, suggesting, paradoxically, that improved survival to lethal factors is a deleterious trait. This paradox suggests that the ability to eliminate defective cells (through programmed death) provides a clonal population with a significant competitive advantage.
hip genes apparently have the potential to strongly influence the “setting” of the cell death pathway, but unfortunately their physiological role (if any) remains unknown. I shall next consider E. coli genes that are good candidates for participation in programmed death and whose function is well understood.
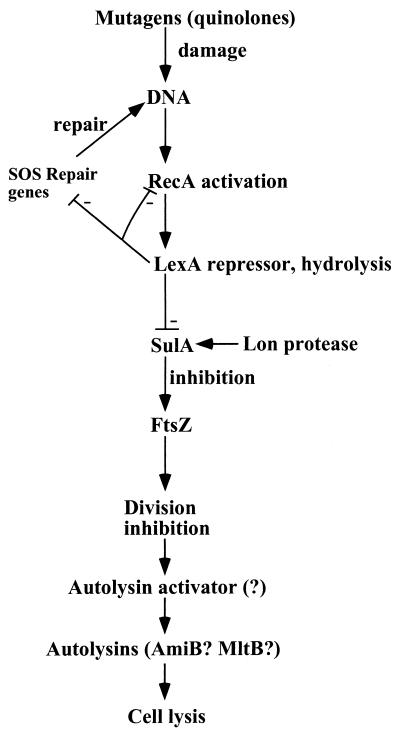
DNA damage by mutagens (which include quinolone antibiotics) is sensed by the RecA protein, which becomes activated and induces hydrolysis and inactivation of the LexA repressor (101). This releases LexA from the promoter regions of a number of lex-box genes and allows the expression of components of the SOS DNA repair response. LexA repressor inactivation also leads to the synthesis of a rapidly hydrolyzed SulA protein that inhibits cell division by binding to FtsZ, the protein that forms the division ring (Fig. 3). SulA therefore acts as a checkpoint—it accumulates after exposure to DNA-damaging agents and inhibits cell division. Subsequently, upon DNA repair, SulA is degraded by the Lon protease, and cell division proceeds. This scenario suggests that without SulA, DNA will not be properly repaired prior to replication, leading to production of nonviable cells. Interestingly, this is not the case—a sulA mutant of an otherwise normal strain (a lon+ background) had a 1,000-fold (!)-higher survival to killing by mutagenic quinolones (77). This experiment strongly suggests that the main role of SulA is not to aid repair but to trigger elimination of cells with serious defects in DNA. sulA mutants have an enormous survival advantage over the wild type, yet the immediate benefit of greater survival of sulA cells is apparently outweighed by the longer-term disadvantage due to the loss of the ability to eliminate defective cells. This analysis indicates just how important programmed death might be in the life of a bacterial population.
FIG. 3.
Programmed death of E. coli cells with damaged DNA. Mutagens produce DNA damage, which is sensed by the RecA protein. In turn, RecA induces hydrolysis of a transcriptional repressor (LexA), activating expression of SOS DNA repair proteins (including RecA). Expression of SulA causes inhibition of cell division. If damage is effectively repaired, the level of activated RecA decreases, the level of SulA drops due to the constant hydrolysis by the Lon protease, and cell division resumes. If damage is considerable, prolonged inhibition of division causes cell elongation and production of autolysin activator (of unknown nature), and the cell is lysed.
SulA-dependent death is strikingly similar to the way animal cells respond to mutagens—DNA damage induces repair enzymes and also triggers accumulation of the p53 protein, which may initiate an apoptotic program. Like sulA mutants, cells deficient in p53 survive DNA damage much better than the wild type (87).
While SulA appears to activate death in response to DNA damage, expression of mar genes was found to suppress killing of E. coli by mutagenic quinolones (34). Originally, the global regulator MarA was found to control expression of genes responsible for multiple drug resistance (MDR) to antimicrobials. MarA inhibits the synthesis of the large OmpF porin and simultaneously activates expression of the AcrAB-TolC MDR pump, which results in increased efflux of toxins across the outer membrane (71). In order to compare killing by quinolones of uninduced cells and cells expressing mar genes, each sample was tested with a corresponding minimal concentration of antibiotic completely inhibiting growth (MIC; note that MIC was considerably higher in the case of mar-expressing cells). The probability of survival of cells expressing Mar proteins was increased 100-fold (34). Killing by the other type of factors tested in this study—β-lactam antibiotics—was unaffected by the mar genes. It appears that mar genes have a dual function of inducing antibiotic resistance mechanisms and suppressing a programmed death response triggered by damaged DNA. The fate of cells damaged by mutagens could depend on the balance of SulA and Mar activities.
Another important locus affecting cell survival is relA. It is well established that tolerance to killing by a wide variety of factors (virtually all cidal antibiotics, for example) correlates inversely with growth rate. Slow growth activates the RelA-dependent synthesis of ppGpp, which inhibits anabolic processes in bacterial cells (15). Interestingly, ppGpp suppressed the activity of a major E. coli autolysin, soluble lytic transglycosylase (SLT) (7), which would make the cells more resistant to autolysis and could explain the mechanism of tolerance to antibiotics in slow-growing cells (note that an increase in ppGpp was implicated in cell death controlled by the maz locus in an experimental model [1], as discussed in the Specialized Killer Genes section). A mutation in relA, the gene coding for ppGpp synthase, while not affecting growth rate, made nongrowing cells sensitive to killing by antibiotics that inhibit cell wall synthesis (80). ppGpp inhibits peptidoglycan synthesis, which complicates interpretation of this finding. It would be interesting to learn whether relA mutants also become sensitive to killing by other types of lethal factors that do not target the cell wall. RelA is a potentially very interesting cell death regulator, since homologs of RelA have been found in all bacteria so far, and all species studied become tolerant to killing when growth rate decreases. Suppression of cell death by ppGpp probably allows the cell to avoid mistaking a starvation state for an unrepairable defect.
Cell survival is also modulated by heat shock proteins that function as molecular chaperones in refolding and degradation of damaged polypeptides. Induction of heat shock proteins suppressed autolysis of E. coli by a number of β-lactams that inhibit the synthesis of peptidoglycan (78). This inhibition was also observed in a relA background, indicating that the effect was independent of a possible activation of a stringent response. Interestingly, induction of heat shock specifically inhibits apoptosis of animal cells by Hsp72, preventing activation of c-Jun kinase (103). This regulatory activity of Hsp72 is unrelated to its function as a molecular chaperone. It seems that in bacteria, heat shock proteins can also block the programmed death of damaged cells.
The immediate cause of death in bacteria is often autolysis, and autolytic enzymes are the likely ultimate targets for possible programmed death pathways. In S. pneumoniae, mutation of the LytA autolysin prevents autolysis and causes tolerance to killing by antibiotics inhibiting cell wall synthesis (95). The recent finding of the VncS kinase regulating autolysis in response to antibiotics in S. pneumoniae will undoubtedly facilitate the identification of a pathway linking damage to autolysin. A particular autolysin tied to killing factors has not yet been identified in gram-negative species. Note that unrelated cidal antibiotics (β-lactams, aminoglycosides, and fluoroquinolones) may trigger autolysis in gram-negative bacteria (21, 33, 49, 100), although inhibitors of peptidoglycan synthesis produce the most dramatic and complete hydrolysis of the cell wall. It is important to note that even cell wall inhibitors do not always produce a clear-cut picture of lysis when they kill bacteria (29), including cases when killing does depend on the presence of a functional autolysin (95). It is possible that in some instances lysis is limited and localized; it is also possible that cells might die independently of lysis or autolysins, although this is not known. Several autolysins have been identified in E. coli, and two of them seem to be good candidates for a role in programmed cell death. The SLT autolysin controlled by ppGpp has been mentioned above. AmiB is a particularly interesting E. coli autolysin (98) whose overexpression sensitizes cells to autolysis by cell wall inhibitors. An amiB mutant had no discernible phenotype (tolerance to antibiotics has not been tested). amiB forms a “superoperon” with several genes, including mutL, a mismatch repair protein that protects from mutagens, and hfq, a global regulator that protects cells from stresses, including high temperature and oxidants (apparently through activating expression of the stationary-phase sigma factor). It thus appears that this superoperon harbors elements that can either protect the cell from lethal factors (heat, oxidants, and mutagens) or cause cell death if repair is insufficient to control the damage. The linking of repair and death elements in one operon is suggestive of a program that determines cell fate.
PCD AND OPPORTUNISTIC MUTANTS
In this section, I will consider the dangers posed to a population of altruists by opportunistic mutants. I suggest that a low mutation rate and preventive suicide evolved in response to this threat.
Altruism, cheaters, and mutation rate.
Altruistic suicide such as PCD, and any form of altruism for that matter, is open to subversion by egoists (cheaters) arising within the population of altruists (see reference 55 for a discussion of altruism and its origins). Recently, mutants of M. xanthus were described that are partially defective in sporulation but produce more spores than the wild type when added at a low level to a wild-type culture (99). Conversion to this asocial phenotype occurred spontaneously after culturing cells for 1,000 generations in the absence of conditions for fruiting body formation. Some defined single-locus mutants of M. xanthus deficient in production of A- and C-signals (asgE and asgB) also showed this cheater phenotype—decreased spore formation in pure culture and superior spore production when mixed with the wild type. The ability of these mutants to lyse was not examined, but the simplest possibility seems to be lack of lysis—this would account both for superior spore production in a mixed culture and for decreased sporulation in pure culture.
In S. pneumoniae, rare natural isolates of vncS mutants that are resistant to autolysis have been described (69). Similarly, one may expect egoistic sulA or hip mutants to arise and provide immediate benefit to the cell. The question then arises as to how an altruistic community is maintained. It is clear that colonies of egoists will lose out in competition with altruists who practice mutually beneficial cooperative behavior. The overall outcome of this selection will depend on the relative rate of mutation versus the rate of elimination of egoist colonies through group selection. Thus, a low mutation rate might become the principal factor in defending a population of altruists against a takeover by egoists.
I must note that a low mutation rate alone may not be sufficient to keep the egoists at bay. Let us consider a more complex scenario—two bacterial clones meet and merge. If this mixing occurred frequently, the advantage of benefiting from living among altruists could outweigh the disadvantage of being outcompeted in group selection when the egoists formed their own clones. This parasitism would then severely undermine the advantage of altruistic suicide. A solution to this problem is the emergence of incompatibility strategies. In bacteria, adjacent clones do not usually merge, creating a distinct empty zone between them which is especially apparent on semisolid medium (13). Colonies also cover themselves with a layer of polysaccharide and form biofilms that are hard to penetrate. The next step of this segregation would be the formation of a more permanent clone—a multicellular organism. Permanent isolation effectively prevents a clone from invasion by egoists.
Hostile takeover at stationary state and preventive suicide.
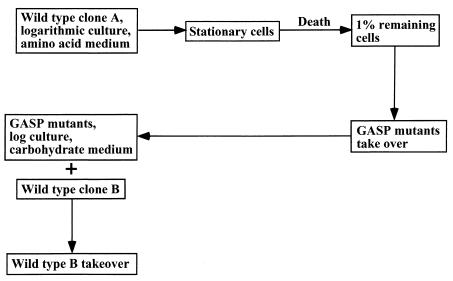
What do bacterial cells die of if they are not damaged by a harsh environment and arrive unscathed at stationary state? When cells reach stationary state, growth practically ceases. In model experiments with E. coli cultured in broth medium, the population declined to about 1% of its original size after several days in stationary state. Thereafter, the size of the population stabilized (97, 105). However, detailed analysis has shown that these populations undergo a fairly complete transformation. Mutant cells that proliferated faster under these conditions increased in numbers and took over the population by day 4 to 5 (105). Continued incubation led to accumulation of multiple mutations in these cells, further increasing their “competitive edge” at stationary state. The first GASP (growth advantage in stationary phase) mutation occurring in these cells usually creates an allele of the stationary-state sigma factor rpoS (rpoS knockout does not produce a survivor phenotype). This mutation leads to, among other things, decreased production of catalase, an enzyme specifically induced under stationary-state conditions. (Note that a reduction in catalase will act as a mutator, further increasing the probability of additional mutations arising in this variant.) The GASP mutants were found to grow faster both in stationary state and in log phase in an amino acid medium (106).
It appears that proliferating mutants arising at stationary state are dead-end variants that lead to a hostile takeover, resulting in the death of the population. Indeed, a GASP mutant is likely to lose out to a wild type once conditions change to low amino acid and high carbohydrate, for example. Similarly, an rpoS mutant unable to upregulate catalase at the onset of stationary state will be at a disadvantage in comparison to the wild type. Abrupt anaerobic-aerobic shift is clearly part of a regular sequence of events experienced by intestinal E. coli (Fig. 4). The GASP mutants are thus analogous to cancer cells—they outcompete the wild type but are likely to ultimately die out without leaving progeny. It is quite possible that the main danger that unicellular organisms face is not competitors, pathogens, or lack of nutrients, but their own kin turning into “unhopeful monsters” and causing the death of the population. Any population surviving to stationary state faces the danger, and it is reasonable to expect that bacteria evolved countermeasures to limit its impact. A low rate of mutations would obviously help to decrease the probability that takeover mutants will appear in a stationary-state population.
FIG. 4.
Model of the fate of a bacterial population. Upon entering stationary state, the majority of cells die, which decreases the mutant load. Eventually, existing or arising GASP mutants carrying defects in global regulator (rpoS) that can proliferate under conditions of this particular stationary culture take over. When conditions improve, such as by the appearance of carbohydrate nutrients, the population consisting of GASP mutants will be outcompeted by an unrelated wild-type clone B. The likely end result is the elimination of clone A due to the emergence of GASP mutants. A low mutation rate, preventive suicide upon entering the stationary state, and death of GASP mutants due to decreased survival (loss of tolerance) of dividing cells in the presence of deleterious factors will contribute to delaying a takeover by GASP mutants.
As in animal cells, a low mutation rate alone can only postpone, not prevent, the ultimate takeover of a stationary population by deleterious mutants. Apoptosis of rapidly proliferating mutants is an additional mechanism preventing cancer in animal cells, and it is possible that programmed death also helps eliminate bacterial mutants that rapidly divide at stationary state. Cells in stationary state find themselves under unfavorable conditions, where the presence of toxic compounds produced by self and other species is increased. Virtually nongrowing wild-type cells will become tolerant to killing by lethal factors such as antibiotics produced by neighboring species (recall that tolerance correlates inversely with growth rate), while faster-growing takeover mutants will have an active death program and will be susceptible to killing.
It is possible that, unlike multicellular organisms, bacteria came up with an additional unique strategy to prevent takeover of a population by opportunistic mutants. We have discussed the case of S. pneumoniae cells that undergo lysis, which enables genetic exchange. This is an extravagantly wasteful approach to sex—conjugation might have produced the same result without sacrificing cells. It is especially puzzling that competence for DNA transformation is a transient property of late-log-phase cells that precedes the massive autolysis occurring at stationary state (2). It seems that this massive suicide per se could actually be the main function of spontaneous autolysis. Once the population density becomes dangerously high and likely to produce takeover mutants, a quorum-sensing mechanism triggers lysis. This strategy is essentially preventive suicide. Paradoxically, a smaller, mutant-free population entering stationary state might have a far better chance of ultimately surviving (i.e., producing viable progeny) than a much larger population burdened by mutant cells. For example, a typical clone of 108 cells could be reduced by autolysis to about 103 cells, and given a mutation rate of 10−7, this small population entering stationary state is likely to be free of takeover mutants and unlikely to create them. DNA exchange probably evolved as a secondary function of lysis, building upon an existing process of preventive suicide. Indeed, decrease in viability upon entering stationary state is common among bacteria, but spontaneous DNA transformation is not. As noted above, an E. coli population rapidly diminishes to 1% in size shortly after entering stationary state (it is not clear whether this death is due to autolysis). E. coli does not have spontaneous DNA transformation. This 100-fold decrease in numbers might represent preventive suicide as well.
Mutation rates in asocial populations.
Is deterrence of opportunistic mutants a driving force for a low mutation rate that evolved in microorganisms? Such a causative link clearly exists in a very different system—a low mutation rate allows animals to avoid cancer for years and even decades. We must note that a role for the very low mutation rate observed in bacteria has not been determined (common explanations such as that a low mutation rate is important to maintain the integrity of the genome are not particularly illuminating). The mutation rate in E. coli is around 10−7 per gene per cell per generation. Systems like proofreading, DNA repair, and programmed death of cells with DNA defects, as discussed above, ensure that the mutation rate is kept at this extremely low level. As a result, a population of 107 cells of a species with 5,000 genes will only produce about 5,000 mutants, each having approximately only one mutation per genome. A low mutation rate produces the immediate benefit of preventing the formation of defective cells, and thus a population with a lower mutation rate might grow faster. However, given the very low probability of mutations, it does not seem like a population would gain a useful selective advantage from keeping the death rate from mutations at ≤0.05%. Predation, host attack, disease (phage), and lethal environmental factors are likely to claim significantly more than 0.05% of a bacterial population in vivo. A replication and DNA maintenance system that worked faster and cheaper and produced more mutants would seem like a better option. It appears therefore that there must be an important reason for a low mutation rate, and decreasing the probability that egotistic cells will arise is a strong contender.
If low mutation rates evolved to check the appearance of asocial mutants, be it cheaters that do not exhibit PCD or takeover mutants that lead to the death of a stationary-state population, one would expect that growth under asocial conditions would lead to an increased rate of mutations. This is indeed the case—among 12 clones of E. coli reinoculated daily for 10,000 generations, three populations evolved into mutator strains with defects in mismatch repair (89). The mutation rate of the mutator strains increased by 100- to 1,000-fold. Note that a very simple minimal glucose medium was used in the experiment, and it is possible that a more complex or variable environment would result in mutators evolving in all populations. When the environment gave mutants a distinct advantage, reversion to a Lac+ phenotype on a lactose medium, the frequency of mutator generation by an E. coli lac population was increased 500-fold (59). This experiment was also carried out in the absence of a prolonged stationary state, or conditions in which PCD would be advantageous to the population. It appears that without the need for selection against asocial mutants, a higher mutation rate gives cells a distinct advantage.
DEATH AND SURVIVAL IN BIOFILMS
In this section, I will consider the possibility of a higher rate of persistor generation and decreased PCD being responsible for biofilm survival.
One of the most dramatic documented cases of bacterial survival is observed in biofilms. It is important to stress that biofilm cells are not necessarily more resistant to antimicrobial agents, if resistance is defined in a generally accepted way as the ability to grow in the presence of a test substance. What biofilms are good at is survival in the presence of extremely high concentrations of bactericidal antimicrobials. The genetics of biofilm development is an area of intense research (23, 73, 79), but little is known about the mechanism of biofilm survival.
A number of factors have been considered for the resilience of biofilm cells—the presence of a diffusion barrier to antimicrobials formed by the biofilm glycocalix, slow rate of cell growth, and perhaps expression of certain resistance genes (19). Some antibiotics, like vancomycin and fluoroquinolones, were shown to freely diffuse into biofilms (22, 67, 91). Slower growth will result in “tolerance,” a decreased susceptibility to killing by antibiotics, possibly through a relA-dependent suppression of PCD, as described in previous sections. This will definitely contribute to survival of biofilm cells, but the observed resistance of biofilms to killing can be higher than in nongrowing planktonic cell cultures (11). The expression of resistance genes in a biofilm is an open question.
We recently examined dose-response killing of P. aeruginosa biofilms and found that fluoroquinolones produce a distinctly biphasic killing—at low concentrations, most of the population dies, and the remaining fraction (a decrease of 1,000- to 100,000-fold) is completely resistant to further increases in antibiotic (11). The planktonic population did not produce noticeable numbers of persistors under these conditions. The surviving biofilm cells are not mutants, but rather persistor variants of the wild type. Surprisingly, the role of persistors in biofilm survival has not been considered, yet several papers describe experiments that show killing patterns very similar to our findings of persistor cells in P. aeruginosa biofilms. In E. coli, increasing concentrations of ciprofloxacin or imipenem caused an initial 100- to 1,000-fold decrease in live cells of a biofilm, while the remaining small population was essentially insensitive to further increases in drug concentration (4). This pattern was also observed with amoxicillin and clindamycin in Lactobacillus acidophilus and with erythromycin and metronidazole in the case of Gardnera vaginalis biofilms, where initial rapid killing was followed by a plateau of resistant cells (66).
It appears that biofilm survival can be largely explained by the increased production of persistor cells. In planktonic cells treated with an antibiotic, a small residual population of persistors will be eliminated by the immune system and does not present a clinical problem (it can be a problem in immunocompromised individuals). Elimination of remaining persistors by the immune system has not been specifically studied, but we know very well that the immune system is capable of eliminating an entire population of pathogens that would otherwise persist in the presence of a bacteriostatic antibiotic. The capability of the immune system is the reason for the recommended MBC to be defined as a concentration of antibiotic that causes a ≥99.9% drop in cell numbers. Unlike planktonic cells, biofilm cells are protected from the immune system by a polysaccharide matrix (41), and remaining persistors will be responsible for biofilm regrowth when the antibiotic concentration drops or when the treatment is discontinued based on the apparent absence of infection. This explains why it is so hard to eradicate a biofilm. At the same time, it is clear that the ability of biofilms to produce increased numbers of persistors predates our use of antibiotics (it is also possible that in some cases, biofilms do not produce more persistors than a planktonic population and will still be more resistant to antibiotic treatment in vivo because of protection from the immune system). Perhaps bacteria have two main life styles—planktonic cells for rapid proliferation and spread into new territories and biofilms, slower-growing populations focused on perseverance, that use increased production of persistors as the main mechanism of survival.
The role of persistors in both planktonic and biofilm populations might be very significant. According to the currently accepted view, survival of (nonsporulating) bacterial populations is explained by two types of mechanisms—induction of stress responses (SOS, heat shock, oxidation stress, etc.) and genetic mechanisms, such as the appearance of resistant mutants or acquisition of foreign genes carrying resistance elements. However, a sudden challenge by a lethal factor will not allow the expression of a stress response. For example, a sudden rise in temperature will not allow expression of heat shock proteins, and the majority of cells will die. Persistors do not need time to develop resistance and can survive a sudden challenge by a killing factor. Persistors arise at a considerably higher rate (10- to 10,000-fold) than mutants. It seems likely that the process of nonheritable variation that produces persistors plays a significant role in survival of bacterial populations encountering damaging factors. Antibiotic-resistant strains have become a major clinical problem (54), and persistors, especially those found in biofilm cells, may be important intermediates in the development of resistance.
SPECIALIZED KILLER GENES
The focus of this review is on programmed death that is potentially beneficial to a population of kin cells. However, suicide of a cell can also be directed by parasitic DNA. The demarcation between the two types of suicide may be blurred, and I will briefly consider the case of specialized killer genes (see references 26 and 32 for detailed reviews).
A number of bacterial plasmids harbor toxin-antitoxin coding genes that are part of the plasmid maintenance mechanism. These genes and proteins belong to several classes, but the overall design of the suicide strategy is similar (32). A well-studied example is maintenance of the E. coli F plasmid (39). The ccdB gene codes for a cidal toxin that inhibits DNA gyrase, while ccdA, located in the same operon, codes for an antitoxin. CcdA is rapidly degraded by the Lon protease, and if a daughter cell does not receive a copy of the F plasmid, it will be killed by the stable CcdB toxin.
Two toxin-antitoxin loci, chpA (mazEF) and chpB, homologous to parD and pem of plasmids R1 and R100, respectively, were identified in the E. coli chromosome (60). The mazEF genes were studied in detail, and when expressed, recombinant MazF appeared to be toxic in the absence of MazE (1, 27). The mazEF genes are located immediately downstream of relA. It was shown that a sudden increase in ppGpp triggered by overexpression of recombinant RelA from a controllable promoter results in cell death that is partially dependent upon the presence of a mazEF locus (1). The physiological role of this locus remains unclear. Another E. coli locus, relB, originally described as a possible stringent response control element (mutations in relB relax stringent control [15]), was found to code for relBE genes that are homologous to a ccd-type toxin-antitoxin system. RelE acts as a toxin which is neutralized by RelB (35). It is not known if RelB has a physiological function. It would be interesting to learn, for example, whether a strain with deletions in both relB and relE was affected in stringent response or had any apparent phenotype. Close homologs of RelE and RelB were found in E. coli plasmid P307, where they are responsible for plasmid maintenance. Homologs of RelB and RelE were also found to be widely spread among bacteria, including gram-positive and gram-negative species, and archaea. The role of chromosomal RelBE elements remains unclear. Recently, it was reported that E. coli K-12 carries five distinct loci belonging to the hok class of toxin-antitoxins (74). All five are inactivated by insertional elements or by mutations.
The E. coli hipAB locus reviewed here resembles a toxin-antitoxin system, although it does not have apparent homologs in GenBank. Unlike typical toxin-antitoxin genes, the hipAB locus has the potential of both killing the cell and improving survival from lethal factors. It seems that the cell could make use of its arsenal of toxin-antitoxin proteins, but the example of nonfunctional hok genes makes this possibility uncertain.
While suicide directed by plasmids is clearly detrimental to the host and beneficial to the parasite, suicide directed by some prophages does appear to benefit the host as well (90). Prophages present in the chromosome of some E. coli strains activate expression of a toxin if the host is infected by another phage. For example, a Lit protein of E. coli prophage e14 interacts with the product of the gol gene of invading phage T4, resulting in a toxin that cleaves elongation factor Tu, leading to cell death and exclusion of the phage (6, 104). Programmed suicide in this case seems to have originated in the e14 prophage and represents the result of a competition between two types of parasitic DNA, with the host as a battlefield. The prophage-coded toxin limits the spread of the phage infection that will benefit both the parasitic prophage DNA and the host it permanently inhabits.
APOPTOSIS IN UNICELLULAR EUKARYOTES
Unicellular life presents unrelated organisms with similar challenges, which could lead to the development of comparable adaptations. A number of studies indicate that PCD resembling metazoan apoptosis is found in unicellular eukaryotes. PCD eliminates cells in developing nematodes, causes autolysis of the mother cell in Bacillus sporulation, and is required for the formation of the stalk of dead cells in the single-cell slime mold Dictyostelium discoideum development (18, 72). These observations are significant but not surprising, given the apparent functional role of PCD in development. But does PCD have a role in eliminating defective cells in unicellular organisms, as in metazoa?
Yeast programmed death and aging.
In a recent study by Frohlich and colleagues, morphological changes in Saccharomyces cerevisiae in response to a lethal dose of H2O2 were found to closely resemble changes in apoptotic metazoan cells (58). These included condensation of chromatin along the nuclear membrane, permeabilization of the cytoplasmic membrane, and fragmentation of DNA. Inhibition of protein synthesis by cycloheximide strongly protected S. cerevisiae from the lethal action of H2O2 and prevented the development of apoptotic morphology. Protein synthesis is required for apoptosis in mammalian cells, and cycloheximide acts as an antiapoptotic substance. The authors suggested that yeast cells have an “altruistic apoptosis” that eliminates defective cells that would otherwise compete with healthy cells for nutrients. Indeed, without invoking apoptosis, it is difficult to imagine why a protein synthesis inhibitor should rescue cells encountering this common deleterious factor instead of exacerbating killing (by blocking the expression of antioxidant enzymes). This situation closely resembles the action of the bacteriostatic agent chloramphenicol, which inhibits protein synthesis in bacteria, preventing death caused by penicillin (96). It seems that in both cases, suppression of a death program provides a satisfactory explanation for the paradoxical action of protein synthesis inhibitors.
In spite of substantial morphological similarities between apoptotic yeast and metazoan cells, homologs of specialized apoptotic proteins are not found in the yeast genome. At the same time, several mammalian proapoptotic proteins (Bak, Bax, and Ced-4) caused cell death and produced a characteristic apoptotic morphology when expressed in yeast cells (36, 37, 42, 45, 82), and death of a superoxide dismutase-deficient mutant of S. cerevisiae was suppressed by the human apoptotic inhibitor Bcl-2 (56). It has been suggested that the action of apoptotic proteins in yeast cells derives from the known ability of proapoptotic proteins to induce the formation of reactive oxygen species (ROS), while Bcl-2 suppresses ROS (58). ROS are important inducers and signaling intermediates in metazoan apoptosis and could play a similar role in yeast cells. However, the presence in S. cerevisiae of an apoptosis morphologically similar to animal apoptosis in the apparent absence of homologous apoptotic proteins is puzzling. This could mean that the components of an ancient eukaryotic-style apoptosis are no longer recognized as homologs by our BLAST comparisons of proteins from yeasts and metazoan animals. Perhaps unicellular protozoans would be better candidates for the search for ancestral apoptotic proteins of animals. The sequencing of several protozoan genomes will soon be complete, and we will learn whether they carry homologs of apoptotic proteins.
As in bacteria, PCD in unicellular yeasts will present a serious problem to a population uniformly challenged by a lethal factor such as hydrogen peroxide. Programmed death of all yeast cells would be counterproductive. We can expect that as with bacteria, part of the yeast population consists of persistors with a disabled death program. Another challenge that altruistic yeast cells will face is takeover by egoistic mutants. As in the case of bacteria, a low mutation rate in eukaryotic yeast cells might have evolved in response to this threat. In bacteria, the mutation rate increases dramatically if the population evolves in the absence of a need for a social behavior like PCD or survival at stationary state, as discussed above. I am not aware of similar experiments with yeast cells, but analyzing what we know about yeast mutation rates suggests that the function of high-fidelity repair of chromosomal DNA requires an explanation. In S. cerevisiae, mutations that lead to mitochondria with defective oxidative phosphorylation are found in approximately 1% of cells growing on a medium with sugars (16, 44, 47). These are essentially lethal mutations in vivo, since yeast strains lacking oxidative phosphorylation have not been isolated from the environment. Given a chromosomal mutation rate of ≈10−7 per gene per cell per generation and a genome size on the order of 104 bases, a yeast population of 107 cells will produce 104 mutants. The loss of life from chromosomal mutations is then ≤0.1% (not all mutations are lethal), and lethal mitochondrial mutations will account for 1% of deaths. The obvious question is why the chromosomal mutation rate should be kept this low in the face of substantial death from mitochondrial mutations. It seems that a need to decrease the probability of producing egoists provides a satisfactory explanation to the yeast mutation rate paradox.
Apart from PCD in response to a lethal factor, programmed death has been proposed as a mechanism of eliminating aged yeast cells (86). It was reported that after producing buds approximately 24 times, the mother cell can no longer reproduce and apparently dies. Note that “aging” in these studies has been defined as the number of divisions rather than the duration of survival of a single cell. Genes affecting the “setting” of senescence, both increasing and decreasing the mean number of divisions, have been described (31, 46, 85). The big debate in the field of aging is whether death is a result of random accumulation of mistakes, and organisms die essentially as automobiles, or whether death is triggered by a specialized program, providing surviving kin and progeny with some adaptive advantage. The maximal number of yeast divisions was found to be narrowly distributed and to vary among different strains. This has led to a suggestion that yeast cells provide a precedent for programmed death, with a molecular clock controlling accumulation of a senescence factor that eventually overwhelms old cells (86).
Let us calculate the possible advantage of a putative longevity-limiting program to a population of kin yeast cells. After 24 generations, a cell will produce around 107 progeny, of which 105 will die of mitochondrial mutations. It does not seem that the fate of one cell is of much relevance to a population where death from old age occurs with a probability that is 100,000 times lower than that of death from mitochondrial mutations. Perhaps programmed death is aimed at eliminating old cells which, if unchecked, will have a higher probability of producing dangerous egotistic mutants? This too does not seem realistic. The probability of mutations in the old cell is only 24 times higher than in a young cell, or equivalent to the mutation rate of a population made of 24 cells. This is 400,000 times lower than the total mutation probability of the 107 progeny cells. This analysis suggests, by default, that aged yeast cells stop reproducing and most likely die due to the accumulation of mistakes. The appearance of this process as programmed death is misleading. Genes that affect the number of permissible divisions likely affect the accumulation of mistakes, making their study both interesting and relevant to the aging of multicellular organisms, where mistakes, especially from oxidative damage, are considered a major cause of aging (5). Yeast cells seem to provide the first clear example of aging resulting from accumulated mistakes, since the case for an adaptive programmed death of old cells in this organism is hardly realistic.
Protozoa.
In Tetrahymena, elimination of old macronuclei in the course of development involves condensation and degradation of DNA into oligonucleosome-sized fragments, similar to apoptotic changes in DNA of animal cells (24). A possible link between a lethal factor and PCD was reported in a study of staurosporin action in Tetrahymena. Staurosporin is a protein kinase C inhibitor and causes cell death. The morphology of cells treated with staurosporin resembled apoptotic animal cells, and the RNA polymerase inhibitor actinomycin D prevented killing by staurosporin, suggesting the involvement of programmed death (17). DNA fragmentation in response to a lethal factor was reported in another protozoan, Plasmodium falciparum. The characteristic oligonucleosomal DNA fragmentation was observed after treatment with the antimalarial cloroquine (76).
The study of PCD in unicellular eukaryotes has only begun, but the evidence suggests the involvement of programmed death both in development and in elimination of defective cells. Possible reasons for apoptosis in protozoans would be the same as for yeasts and bacteria—ridding a population of defective cells, obtaining nutrients from neighbors committing altruistic suicide, stemming an infection by pathogens, decreasing the mutation rate, and lowering the probability that takeover mutants will arise.
Bacteria, mitochondria, and eukaryotic PCD—in search of homologies.
Bacteria are ancestral to mitochondria, which play an important role in the apoptotic pathway of animal cells (reviewed in references 12, 20, 43, and 94). It is not known whether mitochondria have a role in apoptosis of unicellular eukaryotes, but is there a similarity between the apoptotic processes observed in mitochondria and the death of their bacterial ancestors? It is known that ROS activate the opening of the large permeability transition pore (PTP) in the inner mitochondrial membrane, which causes swelling, rupture of the outer membrane, and the release into the cytoplasm of several intermembrane proteins—cytochrome c, procaspase 9, and Aif-1. Cytochrome c forms a complex with cytosolic Apaf-1 and procaspase 9, producing active caspase 9. This in turn activates procaspase 3, which is one of the terminal caspases of apoptosis. Aif-1 goes directly to the nucleus and activates DNA degradation. The pro- and antiapoptotic proteins Bax and Bcl-2, respectively, mentioned above, are mitochondrial proteins that apparently modify the activity of PTP. In the process of releasing apoptotic proteins, mitochondria are destroyed. This destruction does not appear to be related to autolysis of bacterial cells. At the same time, bacteria harbor large stretch-activated channels in the cytoplasmic membrane that open in response to a sudden decrease in osmolarity of the environment (53, 93). The opened channels allow solutes to escape rapidly from the cell, relieving osmotic pressure. Known bacterial channels are not homologous to the mitochondrial PTP. It is conceivable that the channels also play a role in programmed death of bacterial cells, but there are currently no data to support this.
Several eukaryotic apoptotic proteins have homologs in bacteria (3). Whole-genome searching shows that an “apoptotic” Ap-ATP domain found in several apoptotic proteins, including Apaf-1, is present in B. subtilis and actinomycetes. A TIR domain responsible for interaction of some apoptotic proteins shows up in Streptomyces, B. subtilis, Synechocystis sp., and Rhizobium spp., and caspase homologs are found in Streptomyces spp. In the absence of functional data, it is not possible to decide whether these proteins are involved in PCD in bacteria or do other useful things. However, the presence of apoptotic protein homologs in only several bacteria would argue against the conservation of a homologous PCD among pro- and eukaryotes.
Little is known about death in Archaea, whose physiology and life style closely resemble those of eubacteria. As I have already mentioned, RelBE-type toxin-antitoxin specialized suicide genes are found in the genomes of archaea (74), but their role is unknown. Whether organisms from all domains of life have programmed elimination of defective cells remains an intriguing open question.
CONCLUDING REMARKS
PCD may serve a number of important functions in bacteria—aid development, facilitate genetic exchange, eliminate defective cells from a population, decrease the load of mutants that can potentially take over, and lead to the death of a population.
The strongest argument for programmed death of defective cells comes from the finding of persistor cells and genes that dramatically affect survival to lethal factors without changing growth susceptibility (such as hip, vncS, mar, and sulA). The only apparent cost attached to high persistence seems to be the loss of an ability to eliminate defective cells from a population. It appears that the cost is too high to pay. At the same time, it seems prudent to introduce a note of caution—we do not know for certain the physiological roles of genes affecting cell death. It is conceivable, for example, that the better survival of sulA mutants observed in the presence of quinolones might change to a decreased survival in a different environment. However, we can only base our interpretations on what is known, and the current body of knowledge points to PCD as the minimal hypothesis explaining the role of genes controlling cell death.
The concept of programmed death in bacteria prompts us to reexamine a broad range of important yet poorly understood phenomena in the life of microbial cells, such as the mechanism of killing by antibiotics, the role of a low mutation rate, death and survival at stationary state, the nature of persistence, and the related issues of population survival and biofilm resilience. The practical implications of understanding programmed death can be significant as well. Components that make up the signal transduction program of PCD should be prime candidates for the development of drugs that will activate programmed death and aid the elimination of pathogens by antibiotics. This might be especially important in the case of slow-growing cells and biofilms, where tolerance and persistence often produce infections that are untreatable with current antimicrobial therapies.
ACKNOWLEDGMENTS
V. P. Skulachev, M. Sherman, S. Epstein, and E. Lozovskaya are gratefully acknowledged for critical reading of the manuscript and helpful discussions.
Work in my laboratory that was described in this review was supported by NIH grant R01GM54412.
REFERENCES
- 1.Aizenman E, Engelberg-Kulka H, Glaser G. An Escherichia coli chromosomal “addiction module” regulated by guanosine 3′,5′-bispyrophosphate: a model for programmed bacterial cell death. Proc Natl Acad Sci USA. 1996;93:6059–6063. doi: 10.1073/pnas.93.12.6059. . (Erratum, 93:9991.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alloing G, Martin B, Granadel C, Claverys J P. Development of competence in Streptococcus pneumoniae pheromone autoinduction and control of quorum sensing by the oligopeptide permease. Mol Microbiol. 1998;29:75–83. doi: 10.1046/j.1365-2958.1998.00904.x. [DOI] [PubMed] [Google Scholar]
- 3.Aravind L, Dixit V M, Koonin E V. The domains of death: evolution of the apoptosis machinery. Trends Biochem Sci. 1999;24:47–53. doi: 10.1016/s0968-0004(98)01341-3. [DOI] [PubMed] [Google Scholar]
- 4.Ashby M J, Neale J E, Knott S J, Critchley I A. Effect of antibiotics on non-growing planktonic cells and biofilms of Escherichia coli. J Antimicrob Chemother. 1994;33:443–452. doi: 10.1093/jac/33.3.443. [DOI] [PubMed] [Google Scholar]
- 5.Beckman K B, Ames B N. The free radical theory of aging matures. Physiol Rev. 1998;78:547–581. doi: 10.1152/physrev.1998.78.2.547. [DOI] [PubMed] [Google Scholar]
- 6.Bergsland K J, Kao C, Yu Y T, Gulati R, Snyder L. A site in the T4 bacteriophage major head protein gene that can promote the inhibition of all translation in Escherichia coli. J Mol Biol. 1990;213:477–494. doi: 10.1016/S0022-2836(05)80209-8. [DOI] [PubMed] [Google Scholar]
- 7.Betzner A S, Ferreira L C, Holtje J V, Keck W. Control of the activity of the soluble lytic transglycosylase by the stringent response in Escherichia coli. FEMS Microbiol Lett. 1990;55:161–164. doi: 10.1016/0378-1097(90)90187-u. [DOI] [PubMed] [Google Scholar]
- 8.Bigger J W. Treatment of staphylococcal infections with penicillin. Lancet. 1944;ii:497–500. [Google Scholar]
- 9.Black D S, Irwin B, Moyed H S. Autoregulation of hip, an operon that affects lethality due to inhibition of peptidoglycan or DNA synthesis. J Bacteriol. 1994;176:4081–4091. doi: 10.1128/jb.176.13.4081-4091.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Black D S, Kelly A J, Mardis M J, Moyed H S. Structure and organization of hip, an operon that affects lethality due to inhibition of peptidoglycan or DNA synthesis. J Bacteriol. 1991;173:5732–5739. doi: 10.1128/jb.173.18.5732-5739.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brooun A, Liu S, Lewis K. A dose-response study of antibiotic resistance in Pseudomonas aeruginosa biofilms. Antimicrob Agents Chemother. 2000;44:640–646. doi: 10.1128/aac.44.3.640-646.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Budihardjo I, Oliver H, Lutter M, Luo X, Wang X. Biochemical pathways of caspase activation during apoptosis. Annu Rev Cell Dev Biol. 1999;15:269–290. doi: 10.1146/annurev.cellbio.15.1.269. [DOI] [PubMed] [Google Scholar]
- 13.Budrene E O. Formation of space-ordered structures in colonies of motile bacteria on agar. Dokl Akad Nauk SSSR. 1985;283:470–473. [PubMed] [Google Scholar]
- 14.Budrene E O, Berg H C. Complex patterns formed by motile cells of Escherichia coli. Nature. 1991;349:630–633. doi: 10.1038/349630a0. [DOI] [PubMed] [Google Scholar]
- 15.Cashel M, Gentry D R, Hernandez V J, Vinella D. The stringent response. In: Neidhardt F C, Curtiss R I, Ingraham J L, Lin C C L, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. Washington, D.C.: ASM Press; 1996. pp. 1400–1416. [Google Scholar]
- 16.Cherry J R, Denis C L. Overexpression of the yeast transcriptional activator ADR1 induces mutation of the mitochondrial genome. Curr Genet. 1989;15:311–317. doi: 10.1007/BF00419910. [DOI] [PubMed] [Google Scholar]
- 17.Christensen S T, Chemnitz J, Straarup E M, Kristiansen K, Wheatley D N, Rasmussen L. Staurosporine-induced cell death in Tetrahymena thermophila has mixed characteristics of both apoptotic and autophagic degeneration. Cell Biol Int. 1998;22:591–598. doi: 10.1006/cbir.1998.0320. [DOI] [PubMed] [Google Scholar]
- 18.Cornillon S, Foa C, Davoust J, Buonavista N, Gross J D, Golstein P. Programmed cell death in Dictyostelium. J Cell Sci. 1994;107:2691–2704. doi: 10.1242/jcs.107.10.2691. [DOI] [PubMed] [Google Scholar]
- 19.Costerton J W, Stewart P S, Greenberg E P. Bacterial biofilms: a common cause of persistent infections. Science. 1999;284:1318–1322. doi: 10.1126/science.284.5418.1318. [DOI] [PubMed] [Google Scholar]
- 20.Crompton M. The mitochondrial permeability transition pore and its role in cell death. Biochem J. 1999;341:233–249. [PMC free article] [PubMed] [Google Scholar]
- 21.Crosby H A, Bion J F, Penn C W, Elliott T S. Antibiotic-induced release of endotoxin from bacteria in vitro. J Med Microbiol. 1994;40:23–30. doi: 10.1099/00222615-40-1-23. [DOI] [PubMed] [Google Scholar]
- 22.Darouiche R O, Dhir A, Miller A J, Landon G C, Raad I I, Musher D M. Vancomycin penetration into biofilm covering infected prostheses and effect on bacteria. J Infect Dis. 1994;170:720–723. doi: 10.1093/infdis/170.3.720. [DOI] [PubMed] [Google Scholar]
- 23.Davies G D, Parsek M R, Pearson J P, Iglewski B H, Costerton J W, Greenberg E P. The involvement of cell-to-cell signals in the development of a bacterial biofilm. Science. 1998;280:295–298. doi: 10.1126/science.280.5361.295. [DOI] [PubMed] [Google Scholar]
- 24.Davis M C, Ward J G, Herrick G, Allis C D. Programmed nuclear death: apoptotic-like degradation of specific nuclei in conjugating Tetrahymena. Dev Biol. 1992;154:419–432. doi: 10.1016/0012-1606(92)90080-z. [DOI] [PubMed] [Google Scholar]
- 25.Downard J, Toal D. Branched-chain fatty acids: the case for a novel form of cell-cell signalling during Myxococcus xanthus development. Mol Microbiol. 1995;16:171–175. doi: 10.1111/j.1365-2958.1995.tb02290.x. [DOI] [PubMed] [Google Scholar]
- 26.Engelberg-Kulka H, Glaser G. Addiction modules and programmed cell death and antideath in bacterial cultures. Annu Rev Microbiol. 1999;53:43–70. doi: 10.1146/annurev.micro.53.1.43. [DOI] [PubMed] [Google Scholar]
- 27.Engelberg-Kulka H, Reches M, Narasimhan S, Schoulaker-Schwarz R, Klemes Y, Aizenman E, Glaser G. rexB of bacteriophage lambda is an anti-cell death gene. Proc Natl Acad Sci USA. 1998;95:15481–15486. doi: 10.1073/pnas.95.26.15481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Falla T J, Chopra I. Joint tolerance to beta-lactam and fluoroquinolone antibiotics in Escherichia coli results from overexpression of hipA. Antimicrob Agents Chemother. 1998;42:3282–3284. doi: 10.1128/aac.42.12.3282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fujimoto D F, Bayles K W. Opposing roles of the Staphylococcus aureus virulence regulators, Agr and Sar, in Triton X-100- and penicillin-induced autolysis. J Bacteriol. 1998;180:3724–3726. doi: 10.1128/jb.180.14.3724-3726.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fuqua C, Greenberg E P. Self perception in bacteria: quorum sensing with acylated homoserine lactones. Curr Opin Microbiol. 1998;1:183–189. doi: 10.1016/s1369-5274(98)80009-x. [DOI] [PubMed] [Google Scholar]
- 31.Gartenberg M R. The Sir proteins of Saccharomyces cerevisiae: mediators of transcriptional silencing and much more. Curr Opin Microbiol. 2000;3:132–137. doi: 10.1016/s1369-5274(00)00064-3. [DOI] [PubMed] [Google Scholar]
- 32.Gerdes K. Toxin-antitoxin modules may regulate synthesis of macromolecules during nutritional stress. J Bacteriol. 2000;182:561–572. doi: 10.1128/jb.182.3.561-572.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gilleland L B, Gilleland H E, Gibson J A, Champlin F R. Adaptive resistance to aminoglycoside antibiotics in Pseudomonas aeruginosa. J Med Microbiol. 1989;29:41–50. doi: 10.1099/00222615-29-1-41. [DOI] [PubMed] [Google Scholar]
- 34.Goldman J D, White D G, Levy S B. Multiple antibiotic resistance (mar) locus protects Escherichia coli from rapid cell killing by fluoroquinolones. Antimicrob Agents Chemother. 1996;40:1266–1269. doi: 10.1128/aac.40.5.1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gotfredsen M, Gerdes K. The Escherichia coli relBE genes belong to a new toxin-antitoxin gene family. Mol Microbiol. 1998;29:1065–1076. doi: 10.1046/j.1365-2958.1998.00993.x. [DOI] [PubMed] [Google Scholar]
- 36.Greenhalf W, Stephan C, Chaudhuri B. Role of mitochondria and C-terminal membrane anchor of Bcl-2 in Bax induced growth arrest and mortality in Saccharomyces cerevisiae. FEBS Lett. 1996;380:169–175. doi: 10.1016/0014-5793(96)00044-0. [DOI] [PubMed] [Google Scholar]
- 37.Hanada M, Aime-Sempe C, Sato T, Reed J C. Structure-function analysis of Bcl-2 protein: identification of conserved domains important for homodimerization with Bcl-2 and heterodimerization with Bax. J Biol Chem. 1995;270:11962–11969. doi: 10.1074/jbc.270.20.11962. [DOI] [PubMed] [Google Scholar]
- 38.Hardman A M, Stewart G S, Williams P. Quorum sensing and the cell-cell communication dependent regulation of gene expression in pathogenic and non-pathogenic bacteria. Antonie Van Leeuwenhoek J Microbiol Serol. 1998;74:199–210. doi: 10.1023/a:1001178702503. [DOI] [PubMed] [Google Scholar]
- 39.Hiraga S, Jaffe A, Ogura T, Mori H, Takahashi H. F plasmid ccd mechanism in Escherichia coli. J Bacteriol. 1986;166:100–104. doi: 10.1128/jb.166.1.100-104.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Holtje J V. From growth to autolysis: the murein hydrolases in Escherichia coli. Arch Microbiol. 1995;164:243–254. doi: 10.1007/BF02529958. [DOI] [PubMed] [Google Scholar]
- 41.Hoyle B D, Jass J, Costerton J W. The biofilm glycocalyx as a resistance factor. J Antimicrob Chemother. 1990;26:1–5. doi: 10.1093/jac/26.1.1. [DOI] [PubMed] [Google Scholar]
- 42.Ink B, Zornig M, Baum B, Hajibagheri N, James C, Chittenden T, Evan G. Human Bak induces cell death in Schizosaccharomyces pombe with morphological changes similar to those with apoptosis in mammalian cells. Mol Cell Biol. 1997;17:2468–2474. doi: 10.1128/mcb.17.5.2468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jacotot E, Costantini P, Laboureau E, Zamzami N, Susin S A, Kroemer G. Mitochondrial membrane permeabilization during the apoptotic process. Ann NY Acad Sci. 1999;887:18–30. doi: 10.1111/j.1749-6632.1999.tb07919.x. [DOI] [PubMed] [Google Scholar]
- 44.James A P, Johnson B F, Inhaber E R, Gridgeman N T. A kinetic analysis of spontaneous rho-mutations in yeast. Mutat Res. 1975;30:199–208. [PubMed] [Google Scholar]
- 45.James C, Gschmeissner S, Fraser A, Evan G I. CED-4 induces chromatin condensation in Schizosaccharomyces pombe and is inhibited by direct physical association with CED-9. Curr Biol. 1997;7:246–252. doi: 10.1016/s0960-9822(06)00120-5. [DOI] [PubMed] [Google Scholar]
- 46.Jazwinski S M. Molecular mechanisms of yeast longevity. Trends Microbiol. 1999;7:247–252. doi: 10.1016/s0966-842x(99)01509-7. [DOI] [PubMed] [Google Scholar]
- 47.Jimenez J, Benitez T. Yeast cell viability under conditions of high temperature and ethanol concentrations depends on the mitochondrial genome. Curr Genet. 1988;13:461–469. doi: 10.1007/BF02427751. [DOI] [PubMed] [Google Scholar]
- 48.Jolliffe L K, Doyle R J, Streips U N. The energized membrane and cellular autolysis in Bacillus subtilis. Cell. 1981;25:753–763. doi: 10.1016/0092-8674(81)90183-5. [DOI] [PubMed] [Google Scholar]
- 49.Kadurugamuwa J L, Beveridge T J. Natural release of virulence factors in membrane vesicles by Pseudomonas aeruginosa and the effect of aminoglycoside antibiotics on their release. J Antimicrob Chemother. 1997;40:615–621. doi: 10.1093/jac/40.5.615. [DOI] [PubMed] [Google Scholar]
- 50.Koch A L. Similarities and differences of individual bacteria within a clone. In: Neidhardt F C, Curtiss R I, Ingraham J L, Lin C C L, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. Washington, D.C.: ASM Press; 1996. pp. 1640–1651. [Google Scholar]
- 51.Kroos L, Zhang B, Ichikawa H, Yu Y T. Control of sigma factor activity during Bacillus subtilis sporulation. Mol Microbiol. 1999;31:1285–1294. doi: 10.1046/j.1365-2958.1999.01214.x. [DOI] [PubMed] [Google Scholar]
- 52.Levin P A, Grossman A D. Cell cycle and sporulation in Bacillus subtilis. Curr Opin Microbiol. 1998;1:630–635. doi: 10.1016/s1369-5274(98)80107-0. [DOI] [PubMed] [Google Scholar]
- 53.Levina N, Totemeyer S, Stokes N R, Louis P, Jones M A, Booth I R. Protection of Escherichia coli cells against extreme turgor by activation of MscS and MscL mechanosensitive channels: identification of genes required for MscS activity. EMBO J. 1999;18:1730–1737. doi: 10.1093/emboj/18.7.1730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Levy S B. The challenge of antibiotic resistance. Sci Am. 1998;278:46–53. doi: 10.1038/scientificamerican0398-46. [DOI] [PubMed] [Google Scholar]
- 55.Lewis K. Pathogen resistance as the origin of kin altruism. J Theor Biol. 1998;193:359–363. doi: 10.1006/jtbi.1998.0725. [DOI] [PubMed] [Google Scholar]
- 56.Longo V D, Ellerby L M, Bredesen D E, Valentine J S, Gralla E B. Human Bcl-2 reverses survival defects in yeast lacking superoxide dismutase and delays death of wild-type yeast. J Cell Biol. 1997;137:1581–1588. doi: 10.1083/jcb.137.7.1581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Luria S E, Latarjet R. Ultraviolet irradiation of bacteriophage during intracellular growth. J Bacteriol. 1947;53:149–163. doi: 10.1128/jb.53.2.149-163.1947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Madeo F, Frohlich E, Ligr M, Grey M, Sigrist S J, Wolf D H, Frohlich K U. Oxygen stress: a regulator of apoptosis in yeast. J Cell Biol. 1999;145:757–767. doi: 10.1083/jcb.145.4.757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mao E F, Lane L, Lee J, Miller J H. Proliferation of mutators in a cell population. J Bacteriol. 1997;179:417–422. doi: 10.1128/jb.179.2.417-422.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Masuda Y, Miyakawa K, Nishimura Y, Ohtsubo E. chpA and chpB, Escherichia coli chromosomal homologs of the pem locus responsible for stable maintenance of plasmid R100. J Bacteriol. 1993;175:6850–6856. doi: 10.1128/jb.175.21.6850-6856.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Metzstein M M, Stanfield G M, Horvitz H R. Genetics of programmed cell death in C. elegans: past, present and future. Trends Genet. 1998;14:410–416. doi: 10.1016/s0168-9525(98)01573-x. [DOI] [PubMed] [Google Scholar]
- 62.Mortier-Barriere I, de Saizieu A, Claverys J P, Martin B. Competence-specific induction of recA is required for full recombination proficiency during transformation in Streptococcus pneumoniae. Mol Microbiol. 1998;27:159–170. doi: 10.1046/j.1365-2958.1998.00668.x. [DOI] [PubMed] [Google Scholar]
- 63.Moyed H S, Bertrand K P. hipA, a newly recognized gene of Escherichia coli K-12 that affects frequency of persistence after inhibition of murein synthesis. J Bacteriol. 1983;155:768–775. doi: 10.1128/jb.155.2.768-775.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Moyed H S, Broderick S H. Molecular cloning and expression of hipA, a gene of Escherichia coli K-12 that affects frequency of persistence after inhibition of murein synthesis. J Bacteriol. 1986;166:399–403. doi: 10.1128/jb.166.2.399-403.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Mueller C, Dworkin M. Effects of glucosamine on lysis, glycerol formation, and sporulation in Myxococcus xanthus. J Bacteriol. 1991;173:7164–7175. doi: 10.1128/jb.173.22.7164-7175.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Muli F W, Struthers J K. The growth of Gardnerella vaginalis and Lactobacillus acidophilus in Sorbarod biofilms. J Med Microbiol. 1998;47:401–405. doi: 10.1099/00222615-47-5-401. [DOI] [PubMed] [Google Scholar]
- 67.Nichols W W, Evans M J, Slack M P E, Walsmsley H L. The penetration of antibiotics into aggregates of mucoid and non-mucoid Pseudomonas aeruginosa. J Gen Microbiol. 1989;135:1291–1303. doi: 10.1099/00221287-135-5-1291. [DOI] [PubMed] [Google Scholar]
- 68.Novak R, Braun J S, Charpentier E, Tuomanen E. Penicillin tolerance genes of Streptococcus pneumoniae: the ABC-type manganese permease complex Psa. Mol Microbiol. 1998;29:1285–1296. doi: 10.1046/j.1365-2958.1998.01016.x. [DOI] [PubMed] [Google Scholar]
- 69.Novak R, Henriques B, Charpentier E, Normark S, Tuomanen E. Emergence of vancomycin tolerance in Streptococcus pneumoniae. Nature. 1999;399:590–593. doi: 10.1038/21202. [DOI] [PubMed] [Google Scholar]
- 70.Nugroho F A, Yamamoto H, Kobayashi Y, Sekiguchi J. Characterization of a new sigma K-dependent peptidoglycan hydrolase gene that plays a role in Bacillus subtilis mother cell lysis. J Bacteriol. 1999;181:6230–6237. doi: 10.1128/jb.181.20.6230-6237.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Okusu H, Ma D, Nikaido H. AcrAB efflux pump plays a major role in the antibiotic resistance phenotype of Escherichia coli multiple-antibiotic-resistance (Mar) mutants. J Bacteriol. 1996;178:306–308. doi: 10.1128/jb.178.1.306-308.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Olie R A, Durrieu F, Cornillon S, Loughran G, Gross J, Earnshaw W C, Golstein P. Apparent caspase independence of programmed cell death in Dictyostelium. Curr Biol. 1998;8:955–958. doi: 10.1016/s0960-9822(98)70395-1. [DOI] [PubMed] [Google Scholar]
- 73.O'Toole G A, Pratt L A, Watnick P I, Newman D K, Weaver V B, Kolter R. Genetic approaches to study of biofilms. Methods Enzymol. 1999;310:91–109. doi: 10.1016/s0076-6879(99)10008-9. [DOI] [PubMed] [Google Scholar]
- 74.Pedersen K, Gerdes K. Multiple hok genes on the chromosome of Escherichia coli. Mol Microbiol. 1999;32:1090–1102. doi: 10.1046/j.1365-2958.1999.01431.x. [DOI] [PubMed] [Google Scholar]
- 75.Pesci E C, Iglewski B H. The chain of command in Pseudomonas quorum sensing. Trends Microbiol. 1997;5:132–134. doi: 10.1016/S0966-842X(97)01008-1. ; discussion, 134–135. [DOI] [PubMed] [Google Scholar]
- 76.Picot S, Burnod J, Bracchi V, Chumpitazi B F, Ambroise-Thomas P. Apoptosis related to chloroquine sensitivity of the human malaria parasite Plasmodium falciparum. Trans R Soc Trop Med Hyg. 1997;91:590–591. doi: 10.1016/s0035-9203(97)90039-0. [DOI] [PubMed] [Google Scholar]
- 77.Piddock L J, Walters R N. Bactericidal activities of five quinolones for Escherichia coli strains with mutations in genes encoding the SOS response or cell division. Antimicrob Agents Chemother. 1992;36:819–825. doi: 10.1128/aac.36.4.819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Powell J K, Young K D. Lysis of Escherichia coli by beta-lactams which bind penicillin-binding proteins 1a and 1b: inhibition by heat shock proteins. J Bacteriol. 1991;173:4021–4026. doi: 10.1128/jb.173.13.4021-4026.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Pratt L A, Kolter R. Genetic analyses of bacterial biofilm formation. Curr Opin Microbiol. 1999;2:598–603. doi: 10.1016/s1369-5274(99)00028-4. [DOI] [PubMed] [Google Scholar]
- 80.Rodionov D G, Ishiguro E E. Direct correlation between overproduction of guanosine 3′,5′-bispyrophosphate (ppGpp) and penicillin tolerance in Escherichia coli. J Bacteriol. 1995;177:4224–4229. doi: 10.1128/jb.177.15.4224-4229.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Rosenbluh A, Rosenberg E. Role of autocide AMI in development of Myxococcus xanthus. J Bacteriol. 1990;172:4307–4314. doi: 10.1128/jb.172.8.4307-4314.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Sato T, Hanada M, Bodrug S, Irie S, Iwama N, Boise L H, Thompson C B, Golemis E, Fong L, Wang H G, et al. Interactions among members of the Bcl-2 protein family analyzed with a yeast two-hybrid system. Proc Natl Acad Sci USA. 1994;91:9238–9242. doi: 10.1073/pnas.91.20.9238. . (Erratum, 92:2016, 1995.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Scherrer R, Moyed H S. Conditional impairment of cell division and altered lethality in hipA mutants of Escherichia coli K-12. J Bacteriol. 1988;170:3321–3326. doi: 10.1128/jb.170.8.3321-3326.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Shockman G D, Daneo-Moore L, Kariyama R, Massidda O. Bacterial walls, peptidoglycan hydrolases, autolysins, and autolysis. Microb Drug Resist. 1996;2:95–98. doi: 10.1089/mdr.1996.2.95. [DOI] [PubMed] [Google Scholar]
- 85.Sinclair D, Mills K, Guarente L. Aging in Saccharomyces cerevisiae. Annu Rev Microbiol. 1998;52:533–560. doi: 10.1146/annurev.micro.52.1.533. [DOI] [PubMed] [Google Scholar]
- 86.Sinclair D A, Mills K, Guarente L. Molecular mechanisms of yeast aging. Trends Biochem Sci. 1998;23:131–134. doi: 10.1016/s0968-0004(98)01188-8. [DOI] [PubMed] [Google Scholar]
- 87.Sionov R V, Haupt Y. The cellular response to p53: the decision between life and death. Oncogene. 1999;18:6145–6157. doi: 10.1038/sj.onc.1203130. [DOI] [PubMed] [Google Scholar]
- 88.Smith T J, Foster S J. Characterization of the involvement of two compensatory autolysins in mother cell lysis during sporulation of Bacillus subtilis 168. J Bacteriol. 1995;177:3855–3862. doi: 10.1128/jb.177.13.3855-3862.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Sniegowski P D, Gerrish P J, Lenski R E. Evolution of high mutation rates in experimental populations of E. coli. Nature. 1997;387:703–705. doi: 10.1038/42701. [DOI] [PubMed] [Google Scholar]
- 90.Snyder L. Phage-exclusion enzymes: a bonanza of biochemical and cell biology reagents? Mol Microbiol. 1995;15:415–420. doi: 10.1111/j.1365-2958.1995.tb02255.x. [DOI] [PubMed] [Google Scholar]
- 91.Stewart P S. Theoretical aspects of antibiotic diffusion into microbial biofilms. Antimicrob Agents Chemother. 1996;40:2517–2522. doi: 10.1128/aac.40.11.2517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Stragier P, Losick R. Molecular genetics of sporulation in Bacillus subtilis. Annu Rev Genet. 1996;30:297–241. doi: 10.1146/annurev.genet.30.1.297. [DOI] [PubMed] [Google Scholar]
- 93.Sukharev S I, Blount P, Martinac B, Blattner F R, Kung C. A large-conductance mechanosensitive channel in E. coli encoded by mscL alone. Nature. 1994;368:265–268. doi: 10.1038/368265a0. [DOI] [PubMed] [Google Scholar]
- 94.Thress K, Kornbluth S, Smith J J. Mitochondria at the crossroad of apoptotic cell death. J Bioenerg Biomembr. 1999;31:321–326. doi: 10.1023/a:1005471701441. [DOI] [PubMed] [Google Scholar]
- 95.Tomasz A, Albino A, Zanati E. Multiple antibiotic resistance in a bacterium with suppressed autolytic system. Nature. 1970;227:138–140. doi: 10.1038/227138a0. [DOI] [PubMed] [Google Scholar]
- 96.Tomasz A, Waks S. Mechanism of action of penicillin: triggering of the pneumococcal autolytic enzyme by inhibitors of cell wall synthesis. Proc Natl Acad Sci USA. 1975;72:4162–4166. doi: 10.1073/pnas.72.10.4162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Tormo A, Almiron M, Kolter R. surA, an Escherichia coli gene essential for survival in stationary phase. J Bacteriol. 1990;172:4339–4347. doi: 10.1128/jb.172.8.4339-4347.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Tsui H C, Feng G, Winkler M E. Transcription of the mutL repair, miaA tRNA modification, hfq pleiotropic regulator, and hflA region protease genes of Escherichia coli K-12 from clustered Esigma32-specific promoters during heat shock. J Bacteriol. 1996;178:5719–5731. doi: 10.1128/jb.178.19.5719-5731.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Velicer G J, Kroos L, Lenski R E. Developmental cheating in the social bacterium Myxococcus xanthus. Nature. 2000;404:598–601. doi: 10.1038/35007066. [DOI] [PubMed] [Google Scholar]
- 100.Vincent S, Glauner B, Gutmann L. Lytic effect of two fluoroquinolones, ofloxacin and pefloxacin, on Escherichia coli W7 and its consequences on peptidoglycan composition. Antimicrob Agents Chemother. 1991;35:1381–1385. doi: 10.1128/aac.35.7.1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Walker G C. The SOS response of Escherichia coli. In: Neidhardt F C, Curtiss R I, Ingraham J L, Lin C C L, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. Washington, D.C.: ASM Press; 1996. pp. 1400–1416. [Google Scholar]
- 102.Wells J E, Russell J B. Why do many ruminal bacteria die and lyse so quickly? J Dairy Sci. 1996;79:1487–1495. doi: 10.3168/jds.S0022-0302(96)76508-6. [DOI] [PubMed] [Google Scholar]
- 103.Yaglom J A, Gabai V L, Meriin A B, Mosser D D, Sherman M Y. The function of HSP72 in suppression of c-Jun N-terminal kinase activation can be dissociated from its role in prevention of protein damage. J Biol Chem. 1999;274:20223–20228. doi: 10.1074/jbc.274.29.20223. [DOI] [PubMed] [Google Scholar]
- 104.Yu Y T, Snyder L. Translation elongation factor Tu cleaved by a phage-exclusion system. Proc Natl Acad Sci USA. 1994;91:802–806. doi: 10.1073/pnas.91.2.802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Zambrano M M, Siegele D A, Almiron M, Tormo A, Kolter R. Microbial competition: Escherichia coli mutants that take over stationary phase cultures. Science. 1993;259:1757–1760. doi: 10.1126/science.7681219. [DOI] [PubMed] [Google Scholar]
- 106.Zinser E R, Kolter R. Mutations enhancing amino acid catabolism confer a growth advantage in stationary phase. J Bacteriol. 1999;181:5800–5807. doi: 10.1128/jb.181.18.5800-5807.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]