Abstract
For plants, distinguishing between mutualistic and pathogenic microbes is a matter of survival. All microbes contain microbe-associated molecular patterns (MAMPs) that are perceived by plant pattern recognition receptors (PRRs). Lysin motif receptor-like kinases (LysM-RLKs) are PRRs attuned for binding and triggering a response to specific MAMPs, including chitin oligomers (COs) in fungi, lipo-chitooligosaccharides (LCOs), which are produced by mycorrhizal fungi and nitrogen-fixing rhizobial bacteria, and peptidoglycan in bacteria. The identification and characterization of LysM-RLKs in candidate bioenergy crops including Populus are limited compared to other model plant species, thus inhibiting our ability to both understand and engineer microbe-mediated gains in plant productivity. As such, we performed a sequence analysis of LysM-RLKs in the Populus genome and predicted their function based on phylogenetic analysis with known LysM-RLKs. Then, using predictive models, molecular dynamics simulations, and comparative structural analysis with previously characterized CO and LCO plant receptors, we identified probable ligand-binding sites in Populus LysM-RLKs. Using several machine learning models, we predicted remarkably consistent binding affinity rankings of Populus proteins to CO. In addition, we used a modified Random Walk with Restart network-topology based approach to identify a subset of Populus LysM-RLKs that are functionally related and propose a corresponding signal transduction cascade. Our findings provide the first look into the role of LysM-RLKs in Populus-microbe interactions and establish a crucial jumping-off point for future research efforts to understand specificity and redundancy in microbial perception mechanisms.
Keywords: LysM, Lipo-chitooligosaccharides, Chitin, Populus, Plant-microbe interaction
Graphical Abstract
1. Introduction
Fossil evidence suggests that the successful colonization of land by plants nearly 450 million years ago was likely mediated by arbuscular mycorrhizal fungi (AMF), or a closely related progenitor [1], [2]. This primordial plant-microbe interaction was founded on the fungus providing the host plant with increased access to water and mineral nutrients; in return, the plant provided the fungus with access to fixed carbon [3]. Since then, a plethora of microbes have evolved a variety of mechanisms to directly capture carbon from plants as their primary food source [4]. The mechanisms of carbon capture have been identified in plant mutualistic associations, particularly with mycorrhizal fungi and nitrogen-fixing bacteria, both of which form specialized nutrient-exchange interfaces in the roots of their host plants [5]. However, some microbial pathogens can parasitize the plant and manipulate it to form similar nutrient-exchange interfaces that disproportionately favor the microbe [6]. Thus, the challenge of discriminating mutualistic and pathogenic microbes is vital for plants.
All microbes contain characteristic microbe-associated molecular patterns (MAMPs), including chitin from fungi or peptidoglycan from bacteria. Plants have evolved the specific ability to detect these MAMPs using sophisticated pattern-recognition receptors that activate a preliminary defense response to microbes [7], [8], [9], [10]. Both beneficial and pathogenic microbes produce these MAMPs and, therefore, both activate so-called MAMP-triggered immunity in their host plant [7]. Intriguingly, both beneficial and pathogenic microbes have evolved an arsenal of protein effectors to suppress MAMP-induced host defense responses [11], [12], [13]. In parallel, plants have also evolved the ability to detect effectors using resistance proteins that activate a more robust immune response known as effector-triggered immunity [14]. Given the highly similar response of plants to both pathogenic and beneficial microbes, a prevailing question in the field of plant-microbe interactions is: How do beneficial microbes overcome immune responses and establish a mutualistic association with the host plant? The current theory is that plants use a three-fold approach, which includes: 1) recruiting beneficial microbes by carefully controlling the metabolites they release into their environment, 2) using dual receptor recognition to distinguish between friend and foe, and 3) integrating both intrinsic and extrinsic cues to tune the so-called immune thermostat [15]. The focus of this work is on the second, or dual receptor recognition approach, with a particular emphasis on the role of lysin motif receptor-like kinases (LysM-RLKs) in plant-microbe interactions [16].
In the model plant Arabidopsis thaliana, the LysM-RLK AtCERK1 (Chitin Elicitor Receptor Kinase 1) was identified as a plasma membrane-localized protein required for chitin perception. It contains three extracellular lysin motifs, hereafter referred to as LysM1, LysM2, and LysM3, and an intracellular Ser/Thr kinase domain exhibiting autophosphorylation activity [17]. Direct binding of AtCERK1 to chitin induces homodimerization which is followed by the induction of chitooligosaccharide-responsive genes [18], [19], [20]. However, AtCERK1 does not act alone in chitin perception – in a chitin-dependent manner, it interacts with the LysM-RLK AtLYK5, which has a higher chitin-binding affinity than AtCERK1 [21]. AtLYK5 shares an overlapping function with AtLYK4, a LysM-RLK that also plays a direct role in chitin signaling and plant innate immunity [22]. Like Arabidopsis cerk1 mutants, double lyk4 lyk5 mutants exhibit a total loss of chitin response [21]. Thus, in Arabidopsis, chitin perception is mediated by AtCERK1, AtLYK4, and AtLYK5 [23].
The multi-receptor mechanism of chitin perception in Arabidopsis is conserved in the model legume M. truncatula in which MtLYK9 and MtLYR4 are homologs of AtCERK1 and AtLYK5, respectively [24], [25]. However, in rice (Oryza sativa), the ability of OsCERK1 to serve as an immune receptor for chitin is not dependent on another LysM-RLK, but on the LysM receptor-like protein (RLP) OsCEBiP (Chitin Elicitor Binding Protein), with which it forms a heterodimer [26], [27], [28]. Although three CEBiP homologs exist in Arabidopsis, they do not play any known functional role in chitin signaling [22]. Beyond chitin perception, both AtCERK1 and OsCERK1 also function in the perception of peptidoglycan in concert with LysM-RLPs [29], [30], [31], [32]. Peptidoglycan was recently shown to serve as a dual immunity and symbiosis elicitor [33].
Unlike rice and 72% of all other plant species [34], Arabidopsis cannot associate with AMF because it lacks many components of the common symbiosis signaling pathway (CSSP) [35]. As such, in Arabidopsis, AtCERK1 primarily functions in immune signaling following perception of pathogen-derived chitooligosaccharides (COs), which are often long-chain COs with six to eight N-Acetylglucosamine (GlcNAc) monomers [36]. In contrast, OsCERK1 and MtLYK9 are important not only for immune, but also for symbiotic signaling promoted by short-chain COs that AMF release [24], [25], [27], [28], [32], [37], [38], [39], [40], [41]. OsCERK1 does not act alone in perceiving these signaling molecules. It forms a heterodimer with OsMYR1/OsNFR5, which directly binds tetrameric chitooligosaccharide (CO4) [42]. Furthermore, CO4-bound OsMYR1/OsNFR5 competitively inhibits complex formation between OsCEBiP and OsCERK1, thus providing a species-specific example of a mechanism for host discrimination between COs derived from fungal pathogens and those produced by symbionts [43]. This mechanism strongly supports the hypothesis that dual receptor recognition allows host plants to distinguish between friend and foe [15]. However, the robustness of this hypothesis has recently been challenged, as Chiu and Paszkowski [44] found that the CO4-binding function of OsNFR5/MYR1 may be dispensable for symbiosis.
Beyond short-chain COs, lipo-chitooligosaccharides (LCOs) are also known to serve as symbiotic signals that are produced by rhizobia, AMF, and ectomycorrhizal (ECM) fungi [45], [46], [47]. Recent reports show that LCOs are also produced by most filamentous fungi and play a role not only in symbiosis, but in regulating the development of multiple fungal species [48], [49]. In contrast, LCO production is not widespread among bacteria, but is restricted to rhizobia [50]. In legumes, such as M. truncatula and Lotus japonicus, host plant perception of Nod-LCOs produced by rhizobia is mediated by the LysM-RLKs MtNFP (Nod-Factor Perception)/LjNFR5 (Nod-Factor Receptor 5), which are expressed during the infection thread formation stage of nodule development and are involved in the rhizobial infection process [51], [52], [53]. While required for the perception of Nod-LCOs, MtNFP/LjNFR5 do not exhibit autophosphorylation activity, and therefore, associate with the LysM-RLKs MtLYK3/LjNFR1 [54]. Together, MtNFP/LjNFR5 and MtLYK3/LjNFR1 form a heterodimer receptor complex that directly binds Nod-LCOs [55], [56], [57]. In M. truncatula, the LysM-RLK MtLYR3 is a high-affinity LCO-binding protein that interacts with MtLYK3, which also binds to Nod-LCOs [58], [59], [60]. The third LysM motif in MtLYR3 is crucial for LCO binding and molecular modeling suggests that it harbors amino acid residues forming a hydrophobic tunnel that may be able to accommodate the LCO acyl chain, which is a feature that may facilitate preferential LCO over CO binding [61].
Myc-LCOs are required in combination with COs for the establishment of symbiosis with their host plant ([33], [47], [62], [63]). Yet, despite similar structures among Nod- and Myc-LCOs, NFP/NFR5 and LYK3/NFR1 are not required individually or in combination for AMF colonization in legumes; however, in an M. truncatula nfp lyk9/cerk1 double mutant, AMF root colonization is significantly lower than in a lyk9/cerk1 single mutant, suggesting at least partial overlap in NFP perception of both Nod- and Myc-LCOs ([33], [47], [62], [63]). The tomato (Solanum lycopersicum) NFP/NFR5 homolog, SlLYK10, is also required for AMF root colonization and is a high-affinity LCO receptor [64], [65], further highlighting the potential role of NFP/NFR5 in Myc-LCO perception. Alternatively, the MtNFP homeolog MtLYR1 may serve as a Myc-LCO receptor as it is strongly upregulated during AMF colonization, but is not required for the rhizobia-legume symbiosis [53], [66].
Downstream of the LysM-RLKs described above, Nod-LCOs and both Myc-LCOs and COs activate the CSSP, which is required for host colonization by rhizobia and AMF, and plays a role in the Populus–Laccaria bicolor ECM association [33], [47], [62], [63]. CSSP activation is mediated by DMI2/SYMRK, a leucine-rich repeat receptor-like kinase that serves as an indispensable common co-receptor for both Nod- and Myc-factors [67], [68]. DMI2/SYMRK forms a complex with NFP/NFR5 [69] and possibly with CERK1, but this remains hypothetical [70]. The loss of several components of the CSSP in Arabidopsis, including DMI2, would suggest that Arabidopsis cannot detect and respond to LCOs [35]; but this is not the case. Liang et al. [71] showed that Nod-factors can suppress the plant immune response to the bacterial MAMP flg22. However, in the absence of the LysM-RLK AtLYK3, the Nod-factor-induced suppression is lost, suggesting that AtLYK3 may be involved in or facilitate LCO perception in Arabidopsis [71]. Furthermore, AtLYK3 also plays a role in regulating the cross-talk between immunity signaling and abscisic acid responses [72].
As reviewed above and in greater detail by others [16], the functional characterization of LysM-RLKs is extensive in several crop and model plant species (e.g., A. thaliana, S. lycopersicum, O. sativa, M. truncatula, and L. japonicus) and has facilitated the discovery of both similar and unique molecular mechanisms that plants employ to perceive and respond to many MAMPs and symbiotic signals. This knowledge has the potential to inform targeted engineering of plant LysM-RLKs to allow for the improved establishment of beneficial plant-microbe interactions, which in turn, could drive increased plant yields. However, given the varied role of LysM-RLKs across species, further characterization of LysM-RLKs in candidate biofuel crops—like Populus, a highly geographically distributed woody perennial—is needed to harness microbe-mediated gains in plant biomass production for biofuel feedstocks. As such, in this work we sought to identify LysM-RLKs in the genomes of several biofuel plant species and predict their function based on phylogenetic analysis with LysM-RLKs with known functions. We then focused on Populus to partially validate our prediction by using AlphaFold models [73], molecular dynamics simulations, and comparative structural analysis with previously characterized CO and LCO plant receptors to identify their likely binding sites in Populus LysM-RLK proteins. In addition, several machine learning-based algorithms [74] were used to predict binding affinity rankings of Populus proteins to CO. We also used a network-topology-based approach to identify a subset of Populus LysM-RLKs that are functionally related and propose a corresponding signal transduction cascade.
2. Materials and methods
2.1. Selection of protein amino acid sequences and phylogenetic analysis
We used Phytozome 13 (https://phytozome-next.jgi.doe.gov/) [75] to obtain the full-length amino acid sequences of all putative protein homologs from all listed plant genomes for the five LysM-RLKs from the A. thaliana TAIR10 genome (AtCERK1, AT3G21630; AtLYK2, AT3G01840; AtLYK3, AT1G51940; AtLYK4, AT2G23770; and AtLYK5, AT2G33580) and for MtNFP (Medtr5g019040) from the M. truncatula genome (Mt4.0v1). From all six LysM-RLK gene lists, the gene IDs and associated amino acid sequences for 12 plant species were extracted. These included five monocot (Brachypodium distachyon, Oryza sativa, Panicum virgatum, Setaria italica, and Sorghum bicolor) and four dicot (Eucalyptus grandis, Glycine max, Medicago truncatula, Populus trichocarpa) species that are candidate biofuel crops or associated model species, as well as three species which were included to provide an evolutionary perspective in the phylogenetic analysis (Amborella trichopoda, Physcomitrella patens, and Selaginella moellendorffii). For five gene lists, a threshold of 50% sequence similarity and a bit score> 300 was applied to each list to eliminate unlikely protein homologs; but for the AtLYK2-like list, there were too few gene IDs so a bit score> 170 was used instead. All LysM-RLK gene lists were then compared to one another to identify duplicate listings and gene IDs that were duplicated were retained only in the LysM-RLK list with which they had the highest sequence similarity and bit score.
All phylogenetic analysis steps were performed in MEGA11 [76]. The amino acid sequences from each of the LysM-RLK lists we generated were aligned with MUSCLE [77] using default settings for gap penalties (gap open: −2.90, gap extend: 0.00, hydrophobicity multiplier: 1.20), max iterations (16), and the UPGMA hierarchical clustering method. For each resulting alignment, maximum likelihood fits of 56 different amino acid substitution models were evaluated using neighbor-joining trees with partial deletion at a 95 % cutoff. For most of the LysM-RLK lists, the Jones–Taylor–Thornton (JTT) substitution coupled with gamma distributed with invariant sites (G + I) was the best model, and was, therefore, used for each phylogeny reconstruction coupled with the bootstrap method for calculating confidence intervals based on 1000 replications.
For the Populus proteins that are the focus of this study, we have used a name basis that indicates sequence similarity based on a close ortholog from Arabidopsis, and added the extension “-like” to evoke that experimental evidence of their function is still to be determined. This is consistent with the TAIR Arabidopsis nomenclature guidelines. Accordingly, we do not distinguish putative active and inactive kinases. Our nomenclature leaves the option for someone in the future who completes molecular characterization to change the names according to more precisely determined functions.
2.2. Gene expression atlas data
RNA-seq data for P. trichocarpa were downloaded from the JGI Plant Gene Atlas project via Phytozome [75] as previously described [78]. This dataset consists of tissue samples (leaf, stem, root, and bud tissue) taken during a nitrogen source study, and sample descriptions can be accessed online at https://phytozome.jgi.doe.gov/phytomine/aspect.do?name=Expression. Read counts were normalized using the gene length corrected trimmed mean of M-values (GeTMM) method [79] and then transformed as log2 counts per million (CPM) using the EdgeR package [80] in R [81]. Transformed values were visualized as heat maps using the seaborn package [82], [83] in Python (V2.7; http://www.python.org).
2.3. LysM-RLK apo- and holo-structural modeling
The top-ranked AlphaFold models [73] of select Populus proteins (PtCERK1-like1, PtCERK1-like2, PtCERK1-like3, PtLYK3-like, PtLYK4-like1, PtLYK4-like2, PtLYK5-like1, PtLYK5-like2, PtNFP-like1, and PtNFP-like2) were used. An average pLDDT (predicted local distance difference test) above 80 was obtained for these models (Supp. Table S1), indicating high accuracy [84]. For comparative analysis, structural models were also generated for several LysM-RLKs from other species, namely, LYR3–11 from Glycine max (GmLYR-11; Glyma.11G063200.1) and AtLYK3, AtLYK4, and AtLYK5 from A. thaliana. The crystal structures for AtCERK1 solved with a chitin pentamer (PDB id 4EBZ) [19] and MtNFP (PDB id 7AU7) [85] from M. truncatula were used. Models of the CO4-bound Populus LysM ectodomains of PtCERK1-like1, PtCERK1-like2, PtCERK1-like3, PtLYK4-like1, PtLYK4-like2, PtLYK5-like1, and PtLYK5-like2 were obtained by transferring the coordinates of the oligosaccharide from PDB id 4EBZ after using Lovoalign for protein structural alignment [86]. Models of CO4-bound A. thaliana LysM ectodomains of AtLYK3, AtLYK4, and AtLYK5 were built likewise and used, with the crystallographic AtCERK1 complex, as our validation set. These CO-complexes were the inputs in the machine learning models described below to predict relative CO-binding affinity. For that, energy minimization was conducted with steepest descent using GROMACS-2020 [87]. The CHARMM36 force field [88] was used to describe the protein and the CO ligand, and TIP3P water molecules were added for solvation [89]. The Visual Molecular Dynamics (VMD) program was used for visual analysis [90].
We employed a molecular dynamics (MD)-based approach of confirmation selection [91] derived from the strategy of protein structure refinement described in Heo et al. [92] to generate models of GmLYR3–11 bound to LCO-V(C18:1Δ11, S) [61]. These models were used to inform the structural analysis of PtLYK4-like1 and its potential role as an LCO receptor. GmLYR3–11, used as reference in our analysis, effectively binds to a diverse set of LCOs, including Nod and Myc factors. The LCO used in our simulations is appropriate to our goal of fundamentally evaluating the hypothesis of binding of PtLYK4-like1 to LCOs, without deepening the discussion about ligand specificity and its origin. The LCO used is described as a synthetic molecule and is one of the strongest binders among the LCOs experimentally tested [61]. Noticeably, this LCO is very similar to a natural Myc factor described in Maillet et al. [46], namely, LCO-IV(C18:1Δ9Z, S). This Myc-LCO has roughly the same Kd as the molecule we used (20 ± 3.5 nM vs. 18 ± 2.4 nM) [61], indicating that the same key molecular features for binding are present in both molecules.
In this approach, conformational sampling of the protein-ligand complex is accelerated by performing long simulation phases at high temperatures (here up to 360 K) and applying hydrogen mass repartitioning [93], which allows for large integration time steps. To counterbalance the high thermal energy, weak position restraint potentials are applied to selected atoms of the protein and ligand, minimally biasing the system toward its initial configuration. Here, two top-ranked conformations were selected from five independent runs of 256 ns, based on population and energetics criteria. The MD simulations were conducted with GROMACS-2020 [87] on the Summit supercomputer at the Oak Ridge Leadership Computing Facility (OLCF). The MD parameters are described in the Supplementary Information and the simulation phases are summarized in Supp. Table S2.
2.4. LysM-RLK protein-ligand affinity prediction models
The holo structures of LysM-RLKs bound to CO4 were subjected to a range of protein-ligand affinity prediction models, which are thoroughly described previously [74]. In brief, these models are trained on a large, well-curated database known as PDBBind, which consists of protein-ligand X-ray crystal complexes with experimentally measured binding affinities [94], [95], [96]. Affinity prediction models were trained on these targets, using a five-fold cross-validation, combined with validation on an independent data set withheld from the training data that is a subset of PDBBind that forms the CASF-2013 benchmark [97]. Models were trained using regression on the experimental binding affinity targets using either support-vector machines (SVM) [98], random forest (RF) [99], or gradient-boosted trees (BT) [100], [101]. The features were the scores of previously developed models. Either the raw feature values (unscaled) or normalized (scaled) features were used in the training. So-called “reduced” models were trained on a subset of features (scores) that had the highest Spearman correlation with the experimental affinities in the training data. SVM models were trained using a nonlinear kernel (a radial-basis function kernel). Since a SVM model is a function of all the features included in the training, which may be of different orders of magnitude, for numerical stability reasons, the features must be on an equivalent numerical scale, hence necessitating normalization. Therefore, SVM models were only trained with scaled features. The values of the predicted affinities are provided with the resulting rank-order list and are unitless, as they are expressed as -log(Kd). The ML-predicted affinities are used for rankings and the absolute affinities should not be used to compare affinities among different models and with experimental values. This is because the training database consists mostly of proteins bound to relatively small, drug-like ligands. The CO4 is, in general, larger than the ligands found in the PDBbind dataset. Therefore the predicted affinities are not comparable with the binding affinities of the training data. However, the physicochemical space covered by PDBbind is vast, diverse, and representative of that found at the CO4-LysM binding interface, as discussed for other complexes formed with large flexible ligands [91].
2.5. Random walk with restart network analysis
We used Random Walk with Restart (RWR) on multiplex networks to explore the network-topology based associations among LysM-RLKs in Populus. The RandomWalkRestartMH (RWR-MH, version 1.14.0) package in R was used to create multiplex networks and run the RWR process [102]. RWR-MH takes as input a multiplex network and set of genes that will be used as seeds that are the origin nodes for the random walk. We used the following network layers to create a multiplex network using default parameters (delta=0.5): a coexpression network derived from [103], knockout phenotype network derived from [104], predictive expression networks (PEN) from leaf and xylem [105], metabolic pathway network derived from PoplarCyc [106], and protein-protein interactions from STRING-DB [107].
The initial gene set comprised twelve LysM-RLK genes (Table 1). These genes were used as seeds in our RWR-Filter method using default parameters (restart=0.7). Briefly, RWR-Filter begins by using RWR to assign a random-walk score to all genes in the network using network-topology based associations. RWR-Filter then uses the scores to calculate a ‘mutual rank’ among all the genes in the initial gene set; this mutual rank is a measure of how related each gene is to all other genes in the initial gene set. Next, RWR-Filter iteratively rejects each gene from the gene set, starting with the poorest ranking gene, separating the initial gene set into ‘active’ and ‘reject’ sets. At each iteration, RWR-Filter tests the connectivity of the genes in the active set and the reject set by comparing the distribution of ranks to a uniform distribution using the Kolmogorov-Smirnov test. The active set genes were then used as seed genes in the RWR-Lines of Evidence (LOE) method with default parameters (restart=0.7). RWR-LOE also begins with RWR but differs from RWR-Filter in that it attempts to find genes in the network that were functionally related to, but not included in, the given gene set.
Table 1.
Initial hypothesized function of Populus lysin motif receptor-like kinases (LysM-RLKs) based on phylogenetic relatedness with functionally characterized LysM-RLKs from other plant species.
| Poplar LysM-RLK | Phylogenetic Analysis | ||
|---|---|---|---|
| Gene ID | Protein name | Closest homolog(s) | Hypothesized function |
| Potri.002G226600 | PtCERK1-like1 | MtLYK9 | CO perception, dual role in immune and symbiotic signaling[25] |
| Potri.014G156400 | PtCERK1-like2 | MtLYK9 | CO perception, dual role in immune and symbiotic signaling[25] |
| Potri.011G010000 | PtCERK1-like3 | MtLYK9 | CO perception, dual role in immune and symbiotic signaling[25] |
| Potri.001G332800 | PtLYK2-like1 | AtLYK2 | CO perception, elicitor‐induced resistance to pathogens[108] |
| Potri.T115100 | PtLYK-like2 | AtLYK2 | CO perception, elicitor‐induced resistance to pathogens[108] |
| Potri.001G190200 | PtLYK3-like | AtLYK3 | LCO-induced suppression of the innate immune response[71] |
| Potri.007G032100 | PtLYK4-like1 | AtLYK4, MtLYR3 | AtLYK4: CO perception[22]; MtLYR3: LCO perception[59] |
| Potri.005G128200 | PtLYK4-like2 | AtLYK4, MtLYR3 | AtLYK4: CO perception[22]; MtLYR3: LCO perception[59] |
| Potri.005G259600 | PtLYK5-like1 | AtLYK5 | CO perception, interacts with CERK1 during immune signaling[21] |
| Potri.002G001600 | PtLYK5-like2 | AtLYK5 | CO perception, interacts with CERK1 during immune signaling[21] |
| Potri.005G128400 | PtNFP-like1 | MtNFP | LCO perception, direct role in symbiotic signaling[33], [53] |
| Potri.007G032300 | PtNFP-like2 | MtNFP | LCO perception, direct role in symbiotic signaling[33], [53] |
From the 40,815 genes in the multiplex networks, we selected the top 200 (0.5 %) for further analysis. We explored potential common biological processes among these genes by annotating them with both gene ontology (GO) and MapMan ontology and visualizing them in their network-context in Cytoscape [109]. First, we identified the most frequent MapMan terms among the top 200. Then, for the genes annotated with these terms, we selected nodes (genes) with high a degree of connectivity both with the seed genes and with each other. In addition, we performed gene ontology enrichment analysis on these 200 genes using the GO Term Enrichment tool from PlantRegMap (http://plantregmap.gao-lab.org/go.php) [110]. This allowed us to identify GO terms that were significantly represented among the top 200 genes. Furthermore, it helped us identify key genes within significant GO term categories that we could further evaluate for their potential role in signaling events downstream of LysM-RLKs based on functional characterization of gene homologs in other plant species (e.g., Arabidopsis).
3. Results
3.1. Phylogenetic analysis of Populus LysM-RLKs uncovered strong homology with functionally characterized LysM-RLKs in other model plant species
For this study, we generated two classes of phylogenetic trees. The first class included six trees that were restricted by the LysM-RLK used as the query sequence, but included every putative homolog from all 12 plant species considered in this study (Supp. Figs. S1-S6); the second class included just one tree that was species-restricted and only included LysM-RLKs from A. thaliana, M. truncatula, O. sativa, and P. trichocarpa (Fig. 1). Both classes of phylogenetic trees and their accompanying results are described below.
Fig. 1.
Phylogeny and gene atlas expression of Populus lysin-motif receptor-like kinases. A, phylogenetic tree of A. thaliana, M. truncatula, O. sativa, and P. trichocarpa protein homologs for AtCERK1, AtLYK2, AtLYK3, AtLYK4, AtLYK5 and MtNFP. Different phylogenetic LysM-RLK groups are shown in different colors. Protein sequences were aligned using MUSCLE [77] and the phylogenetic tree assembled using the best fit model JTT + G + I in MEGA11 [76]. Confidence intervals were calculated using the bootstrap method with 1000 replications. B, Root tissue expression of 12 Populus LysM-RLK genes putatively likely involved in either lipo- or chitooligosaccharide perception (expression of each of these genes in leaf, stem, and bud tissues is shown in Supp. Fig. S7). The heat map illustrates gene expression from the GeneAtlas data set. LysM-RLK genes are in rows and labeled as "P. trichocarpa gene locus (gene name)." Samples are in columns and labeled as "tissue (experimental condition)." Duplicate sample labels indicate biological replicates. The underlying values are read counts normalized using the gene-normalized trimmed mean of M-values (GeTMM) method and then transformed as log2 counts per million (CPM). The color bar illustrates the color-to-value mapping, where dark blue indicates high values, and light green indicates low values; gray indicates no expression (0 raw counts).
For AtCERK1 (AT3G21630.1) we found that the genomes of all 12 plant species that we queried possessed between one and ten putative protein homologs. Among those, all but one species (P. virgatum) had at least one putative protein ortholog, and several species contained more than one, including: E. grandis (4), G. max (5), M. truncatula (3), P. patens (4), and P. trichocarpa (3) (Supp. Table S3). A phylogenetic tree of the 45 putative AtCERK1-like homologs we identified from all 12 species revealed distinct clustering (Supp. Fig. S1). For example, not only were monocot and dicot AtCERK1 homologs distinctly segregated from one another, but so too were the homologs from the two early diverging plant species (Physcomitrella patens and Selaginella moellendorffii). The three putative Populus homologs of AtCERK1 (PtCERK1-like1, PtCERK1-like2, and PtCERK1-like3) clustered more closely with MtLYK9 than with AtCERK1 (Fig. 1), which suggests that they may play a role in the perception of both long and short-chain COs, including those produced by both pathogenic fungi and AMF [25].
Only seven of the 12 plant species that we queried possessed between one or more protein homologs of AtLYK2 (AT3G01840.1). Among those, only M. truncatula, P. trichocarpa, E. grandis, and A. trichopoda had one or two putative protein orthologs (Supp. Table S4). A phylogenetic tree of the 10 AtLYK2-like homologs revealed that the two Populus homologs (PtLYK2-like1 and PtLYK2-like2) cluster closely with MtLYR10 and AtLYK2 (Supp. Fig. S2), and this was also supported in the species-limited phylogenetic tree (Fig. 1). Therefore, the function of PtLYK2-like1 and PtLYK2-like2 is difficult to predict because AtLYK2, which has been partially functionally characterized, is not required for chitin perception; however, it does contribute to callose deposition induced by chitin [108]. Unfortunately, MtLYR10 has not been functionally characterized, so its function is currently unknown.
All 12 plant species that we queried possessed between one and six protein homologs of AtLYK3 (AT1G51940.1). Among those, only P. virgatum and P. patens lacked putative protein orthologs (Supp. Table S5). The phylogenetic tree we constructed from the 29 AtLYK3-likehomologs essentially consisted of two clusters: one that contained AtLYK3 and another with MtLYK10 (Supp. Fig. S3). One Populus homolog (PtLYK3-like) closely clustered with AtLYK3 (Fig. 1) In contrast, two Populus homologs (Potri.015G082000.1 and Potri.006G252600.2) clustered closely with MtLYK10 (Fig. 1), which is a LysM-RLK involved in exopolysaccharide (EPS) perception [111], [112]. Thus, like the putative role of AtLYK3 in LCO perception, PtLYK3-like may function as an LCO receptor while Potri.015G082000.1 and Potri.006G252600.2 may function as EPS receptors.
For AtLYK4, five of the 12 plant species that we queried possessed from one to three protein homologs. Among those, only E. grandis, G. max, and P. trichocarpa possessed putative protein orthologs (Supp. Table S6). The phylogenetic tree we constructed from the 13 AtLYK4-like homologs resulted in three clusters: one with AtLYK4 and MtLYR2, another with three G. max putative AtLYK4-like homologs, and a third with two Populus and one E. grandis putative AtLYK4-like homologs (Supp. Fig. S4). However, the species-constrained phylogenetic tree revealed that two Populus homologs (PtLYK4-like1 and PtLYK4-like2) clustered with AtLYK4 and MtLYR3, and two others clustered with MtLYR2 (Potri.005G128300.1 and Potri.014G040000.1; Fig. 1). Based on bit score and sequence similarity, MtLYR3 is included in the AtLYK2-like list (Supp. Table S4), but our phylogenetic analysis suggests that it is more like AtLYK4 (Fig. 1). MtLYR3 functions as an LCO co-receptor with MtLYK3 [59], but AtLYK4 is a CO receptor [22]. As such, this makes it difficult to predict the function of PtLYK4-like1 and PtLYK4-like2 which cluster with both proteins. Similarly, the potential role of Potri.005G128300.1 and Potri.014G040000.1 cannot be predicted since MtLYR2, the protein with which they clustered most closely, has only been partially functionally characterized [59].
Among the 12 plant species that were queried, only S. moellendorfii did not possess one or more protein homologs of AtLYK5. In addition, all but S. bicolor possessed at least one putative protein ortholog (Supp. Table S7). The phylogenetic tree we constructed from the 26 AtLYK5-like homologs divided them into two clades: one with MtLYR5 (unknown function), and a second with the known homologs AtLYK5 and MtLYR4 (Supp. Fig. S5). Two putative Populus homologs (PtLYK5-like1 and PtLYK5-like2) clustered with AtLYK5 and MtLYR4 (Fig. 1). Little is known about MtLYR4 except that it seems to be phosphorylated by MtNFP following addition of LCOs [113]; however, AtLYK5 is well-characterized and binds chitin with high affinity in complex with AtCERK1 [21]. As such, PtLYK5-like1 and PtLYK5-like2 are most likely involved in chitin perception in conjunction with other LysM-RLKs.
Finally, only A. thaliana and P. patens lacked NFP homologs, while the remaining ten species possessed between one and four. All ten of these species possessed at least one putative protein ortholog (Supp. Table S8). The phylogenetic tree we constructed from the 24 MtNFP-like homologs revealed distinct clustering of monocot and dicot gene homologs (Supp. Fig. S6). The monocot species had at least one gene that clustered closely with OsNFR5, but the dicot species clustered more closely with MtNFP and GaNFR5, including the Populus homologs PtNFP-like1 and PtNFP-like2 (Fig. 1). Thus, PtNFP-like1 and PtNFP-like2 most likely function as LCO receptors, and are likely the LysM-RLKs that facilitate Nod-LCO perception in Populus even though rhizobia do not induce nodule development in Populus [114].
In summary, the hypothesized role of each Populus LysM-RLK protein based on their phylogenetic relatedness to functionally characterized LysM-RLKs in other plant species is shown in Table 1.
3.2. The expression of Populus LysM-RLKs varies across tissue types and conditions
To determine which LysM-RLKs should be prioritized for further analysis, we evaluated the expression of each LysM-RLK listed in Table 1 using gene atlas expression data across tissue types and various conditions. All the Populus LysM-RLKs were expressed in nearly all tissue types except for both PtLYK2-like1 and PtLYK2-like2, which exhibited either no expression or extremely low levels of expression in only a few instances (Fig. 1B; Supp. Fig. S7). The LysM-RLKs with the highest expression across tissue types were PtLYK3-like, PtLYK5-like1, PtLYK4-like1, and PtCERK1-like1 (Supp. Fig. S7). In addition to these four, PtCERK1-like3, PtLYK4-like2, PtNFP-like1, and PtNFP-like2 were also moderately expressed in most if not all tissues, but they were most strongly upregulated in root samples under standard conditions, particularly PtNFP-like2 (Fig. 1B). Although the expression of PtCERK1-like2 and PtLYK5-like2 was low, both were consistently expressed across tissue types (Supp. Fig. S7). Based on these expression levels, all the Populus LysM-RLKs except for PtLYK2-like1 and PtLYK2-like2 were advanced for protein modeling analysis.
3.3. Modeling-based analyses uncovered Populus LysM-RLKs putatively involved in CO binding
AlphaFold models generated for PtCERK1-like1, PtCERK1-like2, PtCERK1-like3, PtLYK4-like1, PtLYK4-like2, PtLYK5-like1, PtNFP-like1, and PtNFP-like2 revealed a common trimodular ectodomain formed by the LysM1, LysM2, and LysM3 subdomains (or motifs, Fig. 2A, B). In contrast, models generated for PtLYK3-like and PtLYK5-like2 show a single LysM motif in the extracellular region. Detailed comparative structural analysis between these Populus LysM-RLKs and previously characterized receptors of CO and LCO ligands can provide valuable hints about their function that were not captured with phylogenetic analysis and primary sequence comparison only.
Fig. 2.
Common molecular architecture of Populus lysin motif receptor-like kinases (LysM-RLKs). Structural analysis of the Populus proteins PtCERK1-like1, PtCERK1-like2, PtCERK1-like3, PtLYK3-like, PtLYK4-like1, PtLYK4-like2, PtLYK5-like1, PtLYK5-like2, PtNFP-like1, and PtNFP-like2 was performed. The mature form of all these proteins, except for PtLYK3-like and PtLYK5-like2, includes A, a trimodular LysM ectodomain represented here by the AlphaFold model of the PtCERK1-like2 ectodomain. PtLYK3-like and PtLYK5-like2 possess a single LysM motif in the ectodomain. The putative chitooligosaccharide (CO)- or lipo-chitooligosaccharide (LCO)-binding site is located at the solvent-exposed parallel loops in one of the three subdomains. In PtCERK1-like2, CO is predicted to bind to the groove formed between the beta strands and alpha helices (orange) in LysM2. As shown in B, the LysM ectodomain of each LysM-RLK is anchored to the lipid bilayer via a helical transmembrane region. In addition, each LysM-RLK contains an intracellular kinase domain, that is either active or inactive. In C, the AlphaFold model of the PtCERK1-like2 kinase-like domain is shown, which harbors the ATP-binding P-loop (GxGxF/YG) and Mg-binding loop (DFG). The components of this putatively active Ser/Thr kinase domain are also present in PtCERK1-like1, PtCERK1-like3, and PtLYK3-like; however, they are not present in PtLYK4-like1, PtLYK4-like2, PtLYK5-like1, PtLYK5-like2, PtNFP-like1, and PtNFP-like2, indicating that these LysM-RLK proteins have an inactive kinase-like domain.
The proteins PtCERK1-like1, PtCERK1-like2, and PtCERK1-like3 have similarly high sequence identities to both CO and LCO-specific receptors, such as AtCERK1 and MtLYK3, respectively (Supp. Table S9). As in AtCERK1 and MtLYK3, the putative Ser/Thr kinase domain in these Populus proteins harbors the ATP-binding P-loop (GxGxF/YG) and Mg-binding loop, indicating that they are active kinases [115] (Fig. 2C). In addition, the three Populus proteins harbor a three amino acid sequence in the kinase domain, YAQ, that was identified as an essential feature to induce symbiotic responses [116]. Therefore, these proteins may play a role in symbiosis promoted by short chain COs released by arbuscular mycorrhizal fungi, similarly to orthologs that can trigger both symbiotic and immune responses from short and long-chain COs, respectively, such as OsCERK1 [38]. Deeper analysis of the ectodomain of PtCERK1-like1, PtCERK1-like2, and PtCERK1-like3 revealed that most structural features involved in AtCERK1 CO-binding [19] are present in these proteins, suggesting that they may play an equivalent role in Populus (Fig. 3A). In AtCERK1, several points of interaction with CO are hydrogen bonds (HB) formed with backbone atoms in LysM2, which therefore do not underlie the ligand-specificity of this protein. In contrast, water-mediated HB are formed with the side chains of Gln109 and Asn140. In addition, the hydrophobic pairs Met127/Ala138 and Val107/Ile141 seem to stabilize the acetylamino methyl groups of the chitin residues at the subsites 1 (GlcNAc-1) and 3 (GlcNAc-3), respectively. Despite their highly similar LysM2, Gln109, Met127, and Ala138 are not conserved in any of the three Populus orthologs. However, considering their spatial proximity, Asp139 in PtCERK1-like1 and PtCERK1-like2 and Asn141 in PtCERK1-like3 may play the role of Gln109 in AtCERK1 by interacting with CO via water-mediated HB. In addition, the hydrophobic contacts with the methyl group in GlcNAc-1 may be preserved with the aliphatic portions of Glu134 and Thr123 in PtCERK1-like1 and Glu134, Thr110, and Ile123 in PtCERK1-like2. The crucial role of hydrophobic contacts with the acetyl groups in GlcNAc-1 and GlcNAc-3 for an effective CO-binding has been elegantly demonstrated by [117] by testing the elicitor activity of COs with various acetylation patterns in both wild-type and cerk1 mutant Arabidopsis seedlings.
Fig. 3.
Comparative structural analysis between LysM subdomains in selected Populus trichocarpa lysin motif receptor-like kinases (LysM-RLKs) and known chitooligosaccharide (CO)-binding LysM-RLKs from Arabidopsis thaliana. Structural alignment among key LysM subdomains is shown for: A, AtCERK1 (PDB id 4EBZ) [19] with PtCERK1-like1, PtCERK1-like2, and PtCERK1-like3; B, AtLYK4 with PtLYK4-like1 and PtLYK4-like2; and C, AtLYK5 with PtLYK5-like1. For visual clarity, PtLYK5-like2 LysM2 is not shown superimposed to AtLYK5 because they have more differences than similarities in the region. All structures, except for AtCERK1, were generated with AlphaFold. The crystallographic chito-tetramer from PDB id 4EBZ is depicted as licorice, in violet. The coordinates of this ligand were used to represent the holo structures of AtLYK4 and AtLYK5. Defined (AtCERK1) and inferred (AtLYK4 and AtLYK5) water molecules are shown as red spheres and hydrogen bond interactions are represented as dashed lines. In the aligned structures, only the residues that are discussed in the manuscript or those that differ between the Populus and Arabidopisis proteins are depicted in cyan and yellow licorice, respectively.
The similarity between the LysM2 in the PtCERK1-like proteins and the LysM2 in AtCERK1 is noticeable, but binding to another LysM motif, particularly LysM1, warrants investigation. Bozsoki et al. [58] pointed out highly conserved sequences in the LysM1 of legume CERK proteins that could represent CO-binding motifs. In LjCERK6, these sequences are GSNLTY (“region II”) and KDSVQA (“region IV”). Similar sequences are found in the PtCERK1-like proteins. In region II of PtCERK1-like1, PtCERK1-like2, and PtCERK1-like3 proteins, these are GANLSF, DANLTF, and GSNLTY, respectively, and, in region IV, KDSLPS, KDSLSS, and QDSIRS, respectively.
Unlike AtCERK1, AtLYK4 and AtLYK5 are chitin-binding proteins harboring an inactive kinase-like domain. Similarly, PtLYK4-like1, PtLYK4-like2, PtLYK5-like1, and PtLYK5-like2 lack the functional residues for kinase catalytic activity (Fig. 2B, C), and may therefore work in conjunction with an equivalent protein to AtCERK1 in Populus for CO-triggered immunity [118]. In the ectodomain of AtLYK4 and AtLYK5, the specific location of the CO-binding site remains unclear, and no experimental structure is available for these proteins, making the comparative structural analysis with Populus orthologs more difficult. The point mutations Y128G and S206P introduced in LysM2 and LysM3, respectively, of AtLYK5 were shown to decrease chitin-triggered production of reactive oxygen species [21]; unfortunately, without a resolved crystal structure, it is not clear if these residues are directly interacting with CO or if these non-conservative substitutions indirectly impair CO-binding due to a structural destabilization. However, our structural analysis suggests that the LysM2 domain in Populus orthologs has the most conducive configuration of hydrophobic residues to accommodate the preferable pattern of acetylation described previously [117]. Specifically, PtLYK4-like2 LysM2, which is more similar structurally to AtLYK4 LysM2 than PtLYK4-like1 LysM2, could form hydrophobic contacts with CO residues via Leu128 and Phe129 at subsite 1, and via Val122 and Ile156 at subsite 3 (Fig. 3B). PtLYK5-like1 LysM2 is also remarkably similar to AtLYK5 LysM2 and preserves these key hydrophobic sites, with Phe134 and Met151 at subsite 1, and Leu126 and Leu161 at subsite 3 (Fig. 3C). In contrast, PtLYK5-like2 has lower identity to AtLYK5 (Supp. Table S9), and several differing residues in LysM2 (Fig. 3C). Interestingly, this protein does not carry a signal peptide [119], indicating that it may not effectively be transported via the secretory pathway to the plasma membrane [16].
We used template-based models of LysM2-CO complexes and ten machine learning (ML)-based methods to predict a ranking of CO-binding affinities for the seven potential Populus CO-receptors as well as for the A. thaliana validation set (i.e., CO4-bound AtCERK1, AtLYK3, AtLYK4, and AtLYK5). The result of our meta-prediction of relative binding affinity for the validation set, listed in order of decreasing predicted binding affinity, is AtLYK5 > AtCERK1 > AtLYK3 > AtLYK4 (Supp. Table S10). Encouragingly, eight out of the ten machine learning models correctly predicted AtLYK5 as the strongest CO-binder, and eight predicted AtLYK4 or AtLYK3 as weaker CO-binders. Cao et al. [21] have shown with pull down assays and isothermal titration calorimetry that AtLYK5 has higher affinity to a chitooligosaccharide than AtCERK1. The pull down assays also indicate that AtLYK4 weakly binds to chitooligosaccharides, compared to AtLYK5. AtLYK3 is not involved in chitin signaling. This result indicates that the method applied here is particularly robust in identifying strong binders, which was the focus of its usage in our study. Remarkably, the X-ray structure used as the template to build the other systems, i.e., AtCERK1-CO4, was not predicted to be the strongest binder, which is also encouraging as it indicates that the method is not biased to favor the experimental configuration.
The rankings of the Populus LysM2 proteins for binding CO are listed in Table 2. There is striking consistency in the relative affinity predictions among most of the various models, i.e., PtCERK1-like2 > PtLYK5-like1 > PtCERK1-like3 > PtCERK1-like1 > PtLYK5-like2 > PtLYK4-like2 > PtLYK4-like1. PtCERK1-like2 is predicted by all models to have the highest affinity binding to CO, and all models except one predict PtLYK5-like1 as the second highest in binding affinity. The predicted high affinity of PtLYK5-like1 seems consistent with the relatively strong binding of CO demonstrated for its ortholog AtLYK5 [21]. On the low-affinity end, all models predict PtLYK4-like1 to have the lowest binding affinity for CO, and a majority of models (7 out of 10) predict PtLYK4-like2 to have the second-lowest binding affinity. However, as the results for the validation set indicate that the applied method may have limited predictive power for weaker binders, we gather other lines of evidence, i.e., comparative structural analysis and network analysis, that support the predicted weak binders.
Table 2.
Machine learning-based affinity rankings for the LysM2 domain of select Populus LysM-RLKs bound to CO. “Unscaled” indicates a model trained on raw, unnormalized features. “Reduced” indicates a model trained on a subset of features that had the highest Spearman correlation with the training data (experimental binding affinities of the PDBBind database). For each column, the proteins are listed, from top to bottom, in order of increasing binding affinity. (Abbrev.: BT=boosted trees; RF=random forest; SVM=support-vector machine; PtLYK1-L1 =PtLYK1-like1; PtLYK1-L2 =PtLYK1-like2; PtLYK1-L3 =PtLYK1-like3; PtLYK4-L1 =PtLYK4-like1; PtLYK4-L2 =PtLYK4-like2; PtLYK5-L1 =PtLYK5-like1; PtLYK5-L2 =PtLYK5-like2).
| Machine learning-based affinity rankings | |||||
|---|---|---|---|---|---|
| Reduced | |||||
| Ranking | BT | RF | SVM | BT | RF |
| 1° | PtLYK1-L2 (8.8) | PtLYK5-L1 (9.2) | PtLYK1-L2 (9.0) | PtLYK1-L2 (8.7) | PtLYK5-L1 (9.5) |
| 2° | PtLYK5-L1 (8.2) | PtLYK1-L2 (9.0) | PtLYK5-L1 (8.3) | PtLYK5-L1 (8.2) | PtLYK1-L2 (9.0) |
| 3° | PtLYK1-L3 (7.5) | PtLYK1-L3 (7.8) | PtLYK5-L2 (7.8) | PtLYK1-L3 (7.5) | PtLYK1-L3 (8.4) |
| 4° | PtLYK1-L1 (7.2) | PtLYK1-L1 (7.2) | PtLYK1-L3 (7.5) | PtLYK1-L1 (7.1) | PtLYK1-L1 (7.5) |
| 5° | PtLYK5-L2 (6.6) | PtLYK5-L2 (6.5) | PtLYK1-L1 (7.0) | PtLYK5-L2 (6.6) | PtLYK5-L2 (6.6) |
| 6° | PtLYK4-L2 (4.4) | PtLYK4-L2 (4.7) | PtLYK4-L2 (4.8) | PtLYK4-L2 (4.3) | PtLYK4-L2 (4.6) |
| 7° | PtLYK4-L1 (3.7) | PtLYK4-L1 (3.6) | PtLYK4-L1 (4.2) | PtLYK4-L1 (3.9)) | PtLYK4-L1 (3.8) |
| Unscaled | |||||
| Reduced | |||||
| Ranking | SVM | BT | RF | BT | RF |
| 1° | PtLYK1-L2 (8.5) | PtLYK1-L2 (6.1) | PtLYK1-L2 (6.2) | PtLYK1-L2 (6.0) | PtLYK1-L2 (6.4) |
| 2° | PtLYK5-L1 (7.4) | PtLYK5-L1 (5.8) | PtLYK5-L1 (5.8) | PtLYK5-L1 (5.8) | PtLYK4-L2 (5.9) |
| 3° | PtLYK1-L3 (7.1) | PtLYK1-L3 (5.6) | PtLYK1-L1 (5.6) | PtLYK1-L3 (5.6) | PtLYK5-L1 (5.8) |
| 4° | PtLYK1-L1 (6.9) | PtLYK4-L2 (5.3) | PtLYK4-L2 (5.5) | PtLYK1-L1 (5.4) | PtLYK1-L1 (5.5) |
| 5° | PtLYK5-L2 (6.8) | PtLYK5-L2 (5.2) | PtLYK1-L3 (5.4) | PtLYK5-L2 (5.4) | PtLYK1-L3 (5.4) |
| 6° | PtLYK4-L2 (4.7) | PtLYK1-L1 (5.2) | PtLYK5-L2 (5.1) | PtLYK4-L2 (5.2) | PtLYK5-L2 (5.1) |
| 7° | PtLYK4-L1 (4.0) | PtLYK4-L1 (4.7) | PtLYK4-L1 (4.8) | PtLYK4-L1 (4.3) | PtLYK4-L1 (5.1) |
3.4. Modeling-based analyses identify Populus LysM-RLKs putatively involved in LCO binding
The ability to predict a ranking of LCO-binding affinities among potential receptors in Populus using machine learning models, as performed using AtCERK1 for CO-binding, is limited by the absence of solved structures of homologous LCO-receptors in the holo state that could be used as templates for complex modeling. However, experimental data linked to recently determined apo structures of LCO receptors in model legumes [58], [61], [85] can help to identify strong candidates of LCO receptors in Populus via comparative structural analysis. For instance, PtNFP-like1 and PtNFP-like2 harbor structural features that were demonstrated to be crucial for LCO perception by the LysM2 in MtNFP which appears to selectively bind to Nod factor secreted by Sinorhizobium meliloti [85]. A hydrophobic patch (formed by Leu147 and Leu154 in MtNFP) is found to be essential for LCO perception in the NFP/NFR5 class of receptors [85]. Interestingly, this signature is conserved in the two Populus proteins, i.e., with the pairs of amino acids, Ala153 and Leu146, in PtNFP-like1, and Ala155 and Leu148, in PtNFP-like2. A few non-conservative substitutions relative to MtNFP may be relevant to determine a different LCO-specificity for the Populus proteins. For example, Tyr137 of MtNFP is replaced by a tryptophan in PtNFP-like1 and PtNFP-like2, namely, Trp136 and Trp138, respectively (Fig. 4A). The altered hydropathy at subsite 1 may significantly change the preferred LCO decoration in the reducing end.
Fig. 4.
Comparative structural analysis between selected LysM subdomains in Populus trichocarpa lysin motif receptor-like kinases (LysM-RLKs) and known lipo-chitooligosaccharide (LCO)-binding LysM-RLKs from Medicago truncatula and Glycine max. Structural alignment between key LysM subdomains are shown for: A, MtNFP (PDB id 7AU7) [85] with PtNFP-like1 and PtNFP-like2, and B, GmLYK3–11 with PtLYK4-like1. These structures are rotated 180° in the z-axis relative to the structures in Fig. 3 (axes are identified in the top left). Except for MtNFP, all structures were generated with AlphaFold. In the aligned structures, only the residues that are discussed in the manuscript or differing residues between the subdomains in the Populus and the legume proteins are depicted, in cyan and green licorice, respectively. In C, the two best models generated via a molecular dynamics-based protocol of conformation selection for GmLYR3–11 bound to LCO-V(C18:1Δ11, S) are shown. In the three panels, hydrophobic residues discussed to be key for binding are labeled in bold. These structures are rotated 90° in the x-axis relative to the structures in B.
Like PtCERK1-like1, PtCERK1-like2, and PtCERK1-like3, the Populus proteins PtLYK4-like1 and PtLYK4-like2 have similarly high sequence identities to both CO-receptors and LCO-receptors (Supp. Table S9). In particular, close inspection revealed a remarkably high similarity between the LCO-binding site of LYR3 LysM-RLKs in legumes [61] and the corresponding region of PtLYK4-like1, i.e., the LysM3 subdomain, as demonstrated with the structural alignment with the AlphaFold model of GmLYR3–11 (Fig. 4B). High selectivity towards LCO over CO binding was demonstrated for different LYR3 orthologues in legumes and a tyrosine at the putative subsite was shown to be key for effective binding [61]. In PtLYK4-like1, this tyrosine is conserved (Tyr272) and it harbors Ile264, which may be part of the hydrophobic patch described for NFP/NFR receptors (Fig. 4B).
Relative to CO receptors and MtNFP, there is less structural-functional experimental knowledge available for LYR3 LysM-RLKs in legumes; as such, we used a molecular dynamics-based protocol of conformation selection to generate models of GmLYR3–11 bound to an LCO,l thus allowing us to investigate the potential key interactions that must be present in PtLYK4-like1 for LCO binding. A sulfated LCO molecule was used, LCO-V(C18:1Δ11, S), which was demonstrated to effectively bind to GmLYR3–11 [61]. Two best-fit models were selected based on population and energetics criteria (see Methods, Fig. 4C). These models indicate that Trp192 stabilizes LCO-binding via carbohydrate-π stacking and that Tyr224 makes hydrophobic contacts with the LCO acyl chain. Model 2 suggests that Leu216, as in NFP/NFR receptors, is part of a hydrophobic patch that interacts with the LCO acyl chain. Interestingly, in both models the sulfate substituent interacts with the positive end of the dipole in the ɑ-helix harboring Ser197. These key structural features in GmLYR3 are also present in PtLYK4-like1. This structural information and the predicted low relative binding affinity of PtLYK4-like1 to CO (Table 2) supports the hypothesis that PtLYK4-like1 is most likely not a CO, but rather an LCO receptor.
The PtNFP-like proteins and PtLYK4-like1 are strong candidates for LCO receptors in Populus, but these proteins cannot independently promote signaling since they do not harbor an active kinase domain. Therefore, we investigated if another Populus LysM-RLK could play this complementary role. The single LysM motif in PtLYK3-like is highly conserved relative to AtLYK3 (Supp. Fig. S8), which is known to participate in LCO perception [71]. The discussed structural features that could underlie LCO-binding do not appear evident in these proteins, but it is possible that Ile183, the methyl group in Thr190, and Ala191 suffice to form the hydrophobic patch that anchors the acyl chain of an LCO. Therefore, a function in LCO-signaling of PtLYK3-like and AtLYK3, either by directly binding to it or by an auxiliary role, warrants investigation. In addition, considering the high identity to MtLYK3 (Supp. Table S9), we investigated if PtCERK1-like1, PtCERK1-like2, or PtCERK1-like3 can bind and promote LCO signaling. Among the three proteins PtCERK1-like1 has more conserved residues in LysM1 relative to MtLYK3 LysM1, where LCO binds [58], but, still, this local similarity is very low. These few similar or identical amino acids are shown in Supp. Fig. S8.
3.5. Network analysis links putative CO- but not LCO-binding Populus LysM-RLKs as interacting proteins with other downstream signaling proteins
After evaluating the CO and LCO-binding ability of select Populus LysM-RLKs, we sought to use network analysis to further inform potential interactions among LysM-RLKs and downstream genes possibly involved in signaling pathways. For our analysis, the initial gene set we used was composed of the 12 Populus LysM-RLK genes identified via our phylogenetic analysis (Table 1). These genes were used as seeds for the Random Walk with Restart (RWR)-Filter method applied to a multiplex network we created for Populus as described in the Methods. We found that the genes for four putative CO-binding LysM-RLKs (PtCERK1-like1, PtCERK1-like2, PtLYK4-like2, and PtLYK5-like1) were closely connected based on the RWR-Filter results (Supp. Fig. S9A). These four genes were then used as seed genes in the RWR-Lines of Evidence (LOE) method, from which we derived new gene sets comprising the top 200 ranked genes. We explored potential common biological processes among these top 200 genes plus the four seed genes by annotating them with GO and MapMan terms (Supp. File S1) and visualizing them in their network-context (Supp. File S2). Within the network, we found that the four LysM-RLK genes from RWR-Filter shared the most connections with genes involved in the following processes: protein degradation and ubiquitination (10 genes), MAPK signaling (2 genes), and calcium signaling (4 genes). Shared connections were also present for four transcription factors and one Leucine-rich repeat (LRR) receptor kinase (Fig. 5A). These 21 genes are listed in Supp. File S3 along with their rank among the top 200 genes, closest Arabidopsis gene homolog(s), gene annotation, and potential function based on relevant literature. Their expression levels based on gene atlas expression data are also shown in Supp. Fig. S10.
Fig. 5.
Candidate network genes downstream of the CO-binding LysM-RLK complex and their root expression levels from gene expression atlas data. A, An abbreviated network of select genes that are highly interconnected with each other and with the CO-binding LysM-RLKs. Genes in bold text were annotated with significantly enriched gene ontology terms. B, Expression of the six bolded genes from panel A in root tissue under varying conditions. The heat map illustrates gene expression from the GeneAtlas data set. Genes are in rows and labeled as "P. trichocarpa gene locus (function)." Samples are in columns and labeled as "tissue (experimental condition)." Duplicate sample labels indicate biological replicates. The underlying values are counts normalized using the gene-normalized trimmed mean of M-values (GeTMM) method and then transformed as log2 counts per million (CPM). The color bar illustrates the color-to-value mapping, where dark blue indicates high values, and light green indicates low values; gray indicates no expression (0 raw counts).
Subsequent GO enrichment analysis of the top 200 ranked genes allowed us to determine which GO terms were significantly enriched. Among biological processes, only the GO term ‘protein phosphorylation’ was significantly enriched based on the false-discovery rate-adjusted q-value< 0.05 (Supp. Fig. S11A), and among molecular functions, both the ‘protein serine/threonine kinase activity’ and ‘ATP binding’ GO terms were also significantly enriched (Supp. Fig. S11B). There was substantial overlap in the genes associated with these three GO terms: 16 were listed in all three GO categories, seven were listed in protein phosphorylation and ATP binding, but only five were exclusively listed in ATP binding. These 28 genes are listed in Supp. File S4 along with their rank among the top 200 genes, closest Arabidopsis gene homolog(s), gene annotation, and potential function based on relevant literature. Furthermore, their expression levels based on gene atlas expression data are also shown in Supp. Fig. S12.
Among the 21 manually selected genes and the 28 genes associated with significantly enriched GO terms, six genes were present in both lists, including: PtUBA1/2 (Potri.009G075700), PtPEX4/PtUBC21 (Potri.018G039200), PtMKK5 (Potri.010G249300), PtMAP3KA (Potri.005G062500), PtCCaMK (Potri.010G247400), and a leucine-rich repeat receptor kinase (Potri.014G195100). Gene atlas expression profiles of these six genes indicate that most are expressed at moderate-to-high levels in root (Fig. 5B) and shoot (Supplemental Fig. S10) tissues, which we would expect if they were constitutively expressed as response factors in innate CO-activated signaling pathways.
3.6. Network analysis failed to link putative LCO-binding Populus LysM-RLKs as interacting proteins with other downstream signaling proteins
After identifying a putative gene network associated with CO-binding LysM-RLKs, we hypothesized that a similar network would exist for the LysM-RLKs we identified with putative roles in LCO binding (PtNFP-like1, PtNFP-like2, PtLYK4-like1, and PtLYK3-like). To test this, we manually selected these four genes from the RWR-Filter “reject set” (Supp. Fig. S9A) and used them as seed genes for a second iteration of RWR-LOE (Supp. Fig. S9B). As with the CO-binding LysM-RLK network, the top 200 genes were annotated with MapMan terms (Supp. File S5) and visualized in their network-context using Cytoscape. However, although GO enrichment analysis of the top 200 genes was performed, no significant GO terms were detected (Supp. File S6); as such, no overlapping genes were identified.
4. Discussion
The results described in this study provide a foundational understanding of the likely role of LysM-RLKs in the bioenergy crop Populus. They further provide a clear trajectory for future research efforts in characterizing both Populus LysM-RLKs and putatively associated genes involved in downstream signaling. Overall, our phylogenetically predicted function of Populus LysM-RLKs (Fig. 1A, Table 1) aligned well with our comparative protein modeling (Fig. 2, Fig. 3, Fig. 4) and binding affinity predictions (Table 2), which suggested that PtCERK1-like1, PtCERK1-like2, PtCERK1-like3, PtLYK4-like2, and PtLYK5-like1 are most likely CO-binding LysM-RLKs (Fig. 3), whereas PtLYK3-like, PtLYK4-like1, PtNFP-like1, and PtNFP-like2 are most likely LCO-binding LysM-RLKs (Fig. 4). Furthermore, our network analysis highlighted distinct connections between Populus LysM-RLKs putatively involved in chitin signaling and downstream components with known functions in immune and symbiotic signaling, as well as other genes which are less well studied (Fig. 5).
4.1. The putative Populus LysM-RLK chitooligosaccharide-binding complex resembles the one found in other eudicots
Our phylogenetic results revealed that the framework for a LysM-RLK complex capable of binding COs exists in Populus trichocarpa that is similar to Arabidopsis and M. truncatula (Fig. 1; Supp. Fig. 1, Fig. 4, Fig. 5; Table 1). Protein modeling of the seven putative CO-binding LysM-RLKs in Populus provided key insights into their predicted function and machine-learning models predicted that PtCERK1-like2 has the highest CO-binding affinity (Table 2). We hypothesize that the presence of Phe135 in LysM2 of PtCERK1-like2 contributes to this predicted strong binding as it allows for the formation of a stacking geometry with the chitin residue at subsite 1 (Fig. 3 A). Indeed, carbohydrate-π interactions are a common attribute of carbohydrate-binding proteins [120]. Preceding PtCERK1-like1 in the predicted ranking there is PtCERK1-like3, for which we speculate that Ile136 is key for the additional stabilization of the acetyl methyl group at subsite 1. Like AtCERK1, PtCERK1-like1 and PtCERK1-like2 possess aliphatic residue moieties capable of forming hydrophobic contacts with the acetyl group in GlcNAc-1 and GlcNAc-3. Gubaeva et al. [117] showed that these hydrophobic contacts in AtCERK1 are crucial for effective CO-binding. They proposed that optimum CO-binding occurs by a ‘slipped sandwich’ arrangement of two LysM-containing proteins that can accommodate the acetyl groups at subsites 1 and 3 in the LysM motifs. This model is supported by the necessity of AtCERK1 to interact with AtLYK5, which has a higher chitin-binding affinity, for kinase activation [21].
Similar to AtCERK1 and AtLYK5, our results indicate that PtCERK1-like2 and PtLYK5-like1 are the best candidates to form a heteromer for chitin perception in Populus (Fig. 6). In contrast to the Arabidopsis counterparts, our data suggest that PtLYK5-like1 may not bind to CO with higher affinity than PtCERK1-like2; however, the predicted PtLYK5-like1 CO-binding affinity did consistently exceed that of PtCERK1-like3 and PtCERK1-like1 (Table 2). Comparative structural analysis indicates that the PtCERK1-like proteins may be functionally redundant. Indeed, the three PtCERK1-like proteins are highly ranked in our prediction of binding affinity to CO, together with PtLYK5-like1. In addition, network analysis suggests a close relationship between PtCERK1-like1 and PtCERK1-like2 and analysis of expression levels indicates higher expression of PtCERK1-like1 across plant tissues than PtCERK1-like2 and PtCERK1-like3. Future binding assays are crucial to evaluate these predictions and to determine the magnitude of the difference in CO-binding affinity between PtCERK1-like2 and PtLYK5-like1.
Fig. 6.
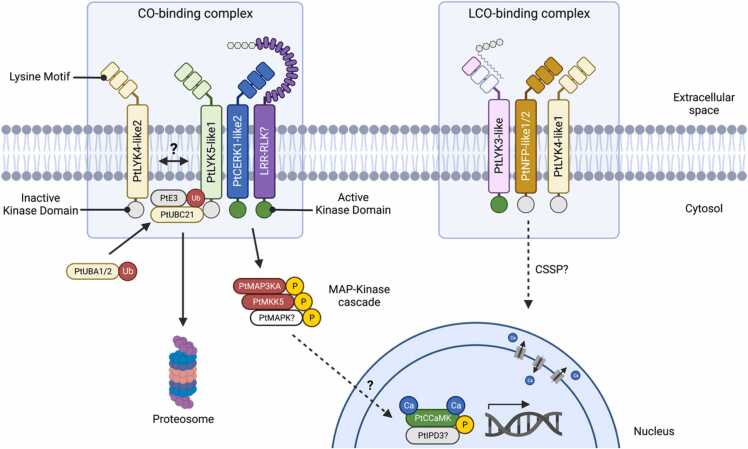
Hypothesized model of CO- and LCO-binding LysM-RLK complexes in Populus and putative downstream signaling components. CO-binding complex: Protein modeling and machine-learning-based methods predicted that PtCERK1-like2 and PtLYK5-like1 have the highest CO-binding affinities and are therefore the strongest candidates for the CO-binding LysM-RLK complex; however, PtCERK1-like2 may be interchangeable with PtCERK1-like1 or PtCERK1-like3, and PtLYK5-like1 may be interchangeable with PtLYK4-like2. Network analysis with RWR-Filter revealed strong connectivity among PtCERK1-like1, PtCERK1-like2, PtLYK4-like2, and PtLYK5-like1. Subsequent RWR-LOE network analysis using these four LysM-RLKs as seed genes uncovered strong connectivity with six genes that likely function as follows. CO-binding by the putative LysM-RLK complex may be facilitated by the leucine-rich repeat (LRR)-RLK encoded by Potri.014G195100. Following stable CO-binding, the kinase domain of one of the PtLYK1 proteins likely initiates the phosphorylation of PtMAP3KA and PtMKK5 in a mitogen-activated protein (MAP) kinase cascade that ultimately triggers transcriptional changes in response to CO perception by the LysM-RLK complex. PtCCaMK, a calcium- and calmodulin-dependent protein kinase, may play a key role in the activation of said transcriptional changes. Activation of this putative signaling cascade is likely halted by the activity of PtUBA1/2 and PtUBC21, which are ubiquitin-activating and ubiquitin conjugating enzymes, respectively. Both are associated with the ubiquitination pathway and could therefore play a role in tagging CO-bound LysM-RLKs for protein turnover via proteasome degradation, thus halting the cellular response to CO binding by the LysM-RLK complex. LCO-binding complex: Protein modeling showed that PtNFP-like1 and PtNFP-like2 possess key hydrophobic patches that are conserved in known LCO-binding LysM-RLKs like MtNFP. Similarly, protein modeling and molecular dynamics revealed that PtLYK4-like1 has conserved amino acid residues with GmLYR3–11 that are also essential for LCO binding. Finally, protein modeling also suggests that PtLYK3-like has sufficient similarities with MtLYK3 to qualify as candidate LCO-binding LysM-RLKs. Although we represent a single putative LCO-binding complex, other configurations are possible, for example, the formation of distinct complexes, PtLYK3-like—PtNFP-like1/2 and PtLYK3-like—PtLYK4-like1, which could display different LCO specificities. Among the top 200 genes identified by targeted RWR-LOE network analysis with the strongest LCO-binding LysM-RLK candidates (i.e., PtNFP-like1/2, PtLYK4-like1, and PtLYK3-like), no gene ontology terms were significantly enriched making it difficult to confidently predict significant downstream signaling components. Regardless, the genome of Populus encodes key components of the common symbiosis signaling pathway (CSSP), some of which are already known to play a key role in the transduction of LCO perception (e.g., the nuclear membrane-localized calcium-regulated calcium channels CASTOR and POLLUX). While this diagram summarizes the key findings and predictions from this study, it is not intended to portray the degree of protein oligomerization depicted among the LysM-RLKs. Significant knowledge gaps in the literature on specific CO- and LCO-binding LysM-RLKs and downstream signaling components are indicated using a “?”.
Even though the predicted CO-binding affinity of the two PtLYK4-like proteins was lower than for the PtCERK1-like and PtLYK5-like LysM-RLKs (Table 2), the similarity to AtLYK4, particularly of PtLYK4-like2, suggests that it may play a redundant or perhaps additive role with PtLYK5-like1 in CO binding. The predicted weak CO-binding of PtLYK4-like2 is consistent with the weak binding inhibition of AtLYK4 to chitin-magnetic beads by chitoheptaose and chitooctaose relative to AtLYK5 [21].
4.2. A putative Populus LysM-RLK lipo-chitooligosaccharide-binding complex resembles well-described complexes in legumes
Our phylogenetic results also predicted that Populus possesses LysM-RLKs that resemble the components of the LCO-binding complex in M. truncatula, even though it does not engage in the root-nodule symbiosis with rhizobia and therefore, in theory, does not need to perceive Nod-LCOs. However, LCOs with similar compositions may be found in rhizobia and in mycorrhizal fungi [46], [61]. Using protein modeling, we evaluated these candidate LysM-RLKs for their ability to potentially bind LCOs. We found that PtNFP-like1 and PtNFP-like2 harbor a hydrophobic patch comparable to one in MtNFP that is crucial for LCO perception by the LysM2 [85] (Fig. 4A), and that a non-conservative substitution of tyrosine at the reducing end subsite may be relevant for determining variations in LCO specificity, as shown for other species [45], [46], [121], [122]. The study of the specificity of Populus receptors towards different types of LCOs (e.g., Myc or Nod factors) would probably require enzymatic assays, which is beyond the scope of this work, but recent work has shown that both purified and rhizobia-derived sLCO and nsLCO can activate the “common symbiosis signaling pathway” (CSSP) in Populus leading to nuclear calcium spiking [47], [114], suggesting more versatile LCO-binding sites or the presence of receptors with different specificities. Both hypotheses remain to be tested.
All machine learning models predicted that PtLYK4-like1 had the lowest CO-binding affinity (Table 2), and it clustered with the known LCO receptor MtLYR3 (Fig. 1A). As such, we evaluated its ability to function as an LCO receptor. Remarkably, we observed high similarity between the LCO-binding site (LysM3 subdomain) of LYR3 LysM-RLKs in legumes [61] and the corresponding region in PtLYK4-like1 (Fig. 4B). Via molecular dynamics-based predictions of GmLYR3–11 bound to a sulphated LCO, we identified potential key residues underlying this interaction and found that these are highly conserved in PtLYK4-like1. This provides strong evidence that PtLYK4-like1 is an LCO-binding LysM-RLK, and not a CO-binding LysM-RLK like PtLYK4-like2.
The putative LCO-binding LysM-RLKs PtNFP-like1/2 and PtLYK4-like1 do not harbor an active kinase domain; therefore, their association with an additional LysM-RLK that does is essential for inducing downstream signaling. For example, in M. truncatula, MtLYK3 fulfills this role in the MtLYK3/MtNFP LCO-binding complex [54]. Our analyses suggest that PtLYK3-like, which possesses a putatively active kinase domain (Fig. 2C), can have a similar function in Populus. It is structurally similar to and is clustered with AtLYK3 (Fig. 1A), which plays a role in LCO perception in Arabidopsis [71].
Collectively, from comparative protein structural analysis, molecular dynamics simulations, and inference from our binding affinity predictions, we have strong evidence that the NFP-like LysM-RLKs and PtLYK4-like1 are candidate LCO receptors in Populus, with PtNFP-like1 and PtNFP-like2 likely playing a redundant functional role. Furthermore, some evidence exists suggesting that PtLYK3-like may serve as the active kinase domain-bearing receptor in this complex (Fig. 6); however, the structural elements underlying LCO-binding are not as clearly identified for PtLYK3-like as it is in the other putative LCO receptors and further investigation is needed to test this hypothesis.
4.3. Potential role of Populus LysM-RLKs beyond chitin binding
The significantly suppressed expression levels of both PtLYK2-like homologs (Fig. 1B) suggests that they do not play a critical role in innate CO signaling. This is further supported by the rejection of these proteins by RWR-Filter (Supp. Fig. S9). However, both PtLYK2-like proteins are clustered with AtLYK2 (Fig. 1A), which has been partially functionally characterized and, although it is not required for chitin perception, it contributes to callose deposition induced by chitin [108]. Therefore, PtLYK2-like homologs could possibly play a similar role in Populus.
Two Populus homologs (Potri.015G082000.1 and Potri.006G252600.2) clustered closely with MtLYK10 (Fig. 1A), which is a LysM-RLK involved in exopolysaccharide EPS perception [111], [112]. The focus in this study was the identification of LysM-RLKs capable of binding either COs or LCOs; however, work in this area is warranted because EPS plays an important role not only for immune signaling in response to bacterial pathogens, but also for symbiotic signaling in response to mutualistic bacteria [123], [124]. Thus, identifying EPS-binding LysM-RLKs in Populus could improve our understanding of how Populus perceives and responds to bacterial pathogens and mutualists.
4.4. Network analysis revealed a CO-binding LysM-RLK network that includes known immune signaling components
In strong support of our protein modeling-based hypothesis that PtCERK1-like2, PtLYK5-like1, and potentially PtLYK4-like2 form the core CO-binding LysM-RLK complex in Populus, our network analysis using RWR-Filter retained these three proteins plus PtCERK1-like1 as the active gene set (Supp. Fig. S9). Together, these four LysM-RLKs shared multiple connections both with each other and with several genes that, based on homology with characterized Arabidopsis genes, are putatively involved in protein degradation and ubiquitination, MAPK signaling, and calcium signaling, as well as one that encodes a leucine-rich repeat (LRR)-RLK (Fig. 5).
We focused on six of these genes for further investigation as they were among the list of genes associated with significantly enriched GO terms (Supp. Fig. S11; Supp. File S3). These six genes included: PtUBA1/2, which plays a role in the ubiquitination pathway during the activation and downstream signaling of several R-proteins [125]; PtUBC21/PtPEX4, which is involved in regulating peroxisomal protein import [126]; Potri.014G195100, which is an LRR-RLK involved in chitin perception and respiratory burst associated with defense responses [127]; PtMAP3KA, which is required for melatonin-mediated induction of an innate immune response [128]; PtMKK5, which is a central component of MAPK cascade that, when activated, confers resistance to both bacterial and fungal pathogens [129]; and PtCCaMK (Fig. 6). While no homolog of PtCCaMK exists in Arabidopsis [35], in M. truncatula, it plays a crucial role in symbiosis signaling [130]. Recently, it was also found to play an important role in mediating the Populus association with the ectomycorrhizal fungus L. bicolor [47]. However, in tomato, knocked-down expression of SlCCaMK led to reduced resistance to the fungal pathogen Sclerotinia sclerotiorum and the bacterial pathogen Pseudomonas syringae pv. tomato DC3000, thus demonstrating a potentially wider and less explored role in pathogen detection, and therefore immune signaling[131].
Functional characterization of these candidate genes in Populus will likely uncover whether they play a role in CO-induced signaling responses downstream of the putative CO-binding LysM-RLK complex. If they do, this will demonstrate conservation of CO-induced signaling mechanisms in plant species beyond Arabidopsis. Furthermore, functionally characterizing these genes will also allow for greater characterization of potentially common signaling mechanisms in response to COs from both fungal pathogens and fungal mutualists, like mycorrhizal fungi.
4.5. Signaling components downstream of LCO-binding LysM-RLKs in Populus are poorly characterized
The network analysis we performed with four putative LCO-binding LysM-RLKs failed to yield as informative of results as in the network analysis with CO-binding LysM-RLKs, specifically because no significantly enriched GO terms were identified among the top 200 genes. This outcome is not entirely unexpected because, in plants, GO enrichment analysis relies heavily on the annotation of genes from the highly studied model plant Arabidopsis to determine putative gene function in other plant species based on gene orthology. However, as already discussed, in stark contrast to the ability of Arabidopsis to perceive COs, its ability to perceive LCOs has only been weakly demonstrated [71]; as such, the number of characterized genes and associated pathways in Arabidopsis that exist downstream of LCO perception are extremely limited. Thus, confidently detecting putative signaling components downstream of Populus LCO-binding LysM-RLKs proved difficult using the methods described herein.
Alternatively, there is strong evidence in the literature that signaling components downstream of LCO perception do exist in Populus. Several genes and pathways that have been characterized as essential for LCO perception in non-Arabidopsis model plant species also appear to be present in the genome of P. trichocarpa [132]. For example, Garcia et al. reported that the core components of the CSSP are present in Populus [133]. Subsequently, Cope et al. demonstrated that Populus both perceives and responds to a variety of LCOs in a CSSP-dependent manner [47], [114]. More specifically, they demonstrated that RNA interference-mediated knock-down of the CSSP genes CASTOR and POLLUX – which encode calcium-regulated calcium channels [134] – abolished LCO-induced nuclear calcium spiking [47]. Given the critical role of these Populus proteins in signal transduction downstream of LCO perception, it is very likely that many, if not all, components of the CSSP [63] also play an important role in LCO signaling in Populus (Fig. 6). Thus, functional characterization of additional CSSP genes in Populus is warranted.
4.6. Future directions for further characterizing Populus LysM-RLKs
One of the limiting factors affecting genetic and molecular studies in Populus when compared to other plant species is the long-life cycle and the inability to obtain seeds from mutants in a timely manner. Nevertheless, techniques have been developed to circumvent this and other roadblocks. For example, Populus can be transformed using Agrobacterium [135]. In addition, the release of the P. trichocarpa genome [132] preceding the advent of CRISPR/Cas9 gene-editing technology has allowed for the recent development of efficient multi-site gene editing techniques in Populus [136]. Thus, these tools provide the framework for functionally characterizing the Populus LysM-RLKs described in this study. For example, site-directed mutagenesis, base-editing and binding assays with Populus LysM-RLKs would further resolve specific amino acid residues that are responsible for CO and LCO binding, and single, double, or triple knock-out CRISPR/Cas9-mutants could be developed to test the hypotheses raised here based on multiple lines of evidence. Furthermore, complementation of Arabidopsis and M. truncatula LysM-RLK mutants with Populus LysM-RLKs could also facilitate the validation of evolutionarily conserved roles of Populus LysM-RLKs in CO and LCO signaling. Co-immunoprecipitation could be performed to test the LysM-RLK protein-protein complexes suggested in this study. Finally, domain swapping techniques could facilitate the engineering of Populus LysM-RLKs for optimizing perception of specific mutualistic microbes, thereby potentially maximizing microbial-mediated nutrient uptake which could directly lead to biomass gains.
5. Conclusions
In summary, LysM-RLKs play a pivotal role in both pathogenic and mutualistic plant–microbe interactions [16]. Similar to legumes, genome duplication events have expanded the suite of LysM-RLKs within Populus, thus opening the door for neofunctionalization of LysM-RLKs with specific roles potentially not found in other plant species, like Arabidopsis [137] combined use of protein modeling, molecular dynamics, and network analysis in this study proved highly effective in formulating strong hypotheses for the proposed function of several Populus LysM-RLKs. Experimental validation of these hypotheses and functionally characterizing these proteins will improve our ability to harness Populus–microbe interactions for maximizing Populus biomass as feedstock for biofuel production and enhancing carbon sequestration in soil.
CRediT authorship contribution statement
Kevin R Cope: Conceptualization, Methodology, Data Curation, Formal analysis, Investigation, Visualization, Validation, Writing- original draft, review & Editing. Erica T. Prates: Conceptualization, Methodology, Data Curation, Formal analysis, Investigation, Visualization, Writing- original draft, review & Editing. John I Miller: Data Curation, Software, Methodology, Formal analysis, Investigation, Visualization, Writing- original draft, review & Editing. Omar N A Demerdash: Data Curation, Formal analysis, Validation, Writing- original draft, review & Editing. Manesh Shah: Methodology, Data Curation, Formal analysis, Visualization, Writing- original draft, review & editing. David Kainer: Data Curation, Software, Resources, Methodology. Ashley Cliff: Data Curation, Formal analysis, Software, Methodology. Kyle Sullivan: Software, Methodology. Mikaela Cashman: Software, Methodology. Matthew Lane: Software, Methodology. Anna Matthiadis: Data Curation, Resources, Writing- original draft. Jesse Labbe: Funding acquisition, Conceptualization, Writing- review & editing. Timothy J Tschaplinski: Funding acquisition, Project administration, Resources, Writing- review & editing. Daniel A Jacobson: Supervision, Funding acquisition, Conceptualization, Methodology, Resources, Writing - review & editing. Udaya C Kalluri: Supervision, Project administration, Funding acquisition, Conceptualization, Methodology, Resources, Writing- original draft, review & Editing.
Declaration of Competing Interest
None. The funding agency [DOE BER] had no involvement on the study design, data collection and analysis or interpretation of results reported here.
Acknowledgments
This research was sponsored by the Genomic Science Program, U.S. Department of Energy, Office of Science, Biological and Environmental Research, as part of the Plant Microbe Interfaces Scientific Focus Area at Oak Ridge National Laboratory (http://pmi.ornl.gov). Oak Ridge National Laboratory is managed by UT-Battelle, LLC, for the U.S. Department of Energy under contract DE-AC05–00OR22725.
Footnotes
This manuscript has been authored by UT-Battelle, LLC under Contract No. DE-AC05–00OR22725 with the U.S. Department of Energy. The United States Government retains and the publisher, by accepting the article for publication, acknowledges that the United States Government retains a non-exclusive, paid-up, irrevocable, world-wide license to publish or reproduce the published form of this manuscript, or allow others to do so, for United States Government purposes. The Department of Energy will provide public access to these results of federally sponsored research in accordance with the DOE Public Access Plan (http://energy.gov/downloads/doe-public-access-plan).
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.csbj.2022.12.052.
Appendix A. Supplementary material
Supplementary material.
.
Supplementary material.
.
Supplementary material.
.
References
- 1.Remy W., Taylor T.N., Hass H., Kerp H. Four hundred-million-year-old vesicular arbuscular mycorrhizae. Proc Natl Acad Sci USA. 1994;91:11841–11843. doi: 10.1073/pnas.91.25.11841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Redecker D., Kodner R., Graham L.E. Glomalean fungi from the Ordovician. Science. 2000;289:1920–1921. doi: 10.1126/science.289.5486.1920. [DOI] [PubMed] [Google Scholar]
- 3.Rich M.K., Vigneron N., Libourel C., Keller J., Xue L., Hajheidari M., et al. Lipid exchanges drove the evolution of mutualism during plant terrestrialization. Science. 2021;372:864–868. doi: 10.1126/science.abg0929. [DOI] [PubMed] [Google Scholar]
- 4.Lambers H., Mougel C., Jaillard B., Hinsinger P. Plant-microbe-soil interactions in the rhizosphere: an evolutionary perspective. Plant Soil. 2009;321:83–115. [Google Scholar]
- 5.Banasiak J., Jamruszka T., Murray J.D., Jasiński M. A roadmap of plant membrane transporters in arbuscular mycorrhizal and legume-rhizobium symbioses. Plant Physiol. 2021;187:2071–2091. doi: 10.1093/plphys/kiab280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sun Y., Wang M., Mur L.A.J., Shen Q., Guo S. The cross-kingdom roles of mineral nutrient transporters in plant-microbe relations. Physiol Plant. 2021;171:771–784. doi: 10.1111/ppl.13318. [DOI] [PubMed] [Google Scholar]
- 7.Newman M.-A., Sundelin T., Nielsen J.T., Erbs G. MAMP (microbe-associated molecular pattern) triggered immunity in plants. Front Plant Sci. 2013;4:139. doi: 10.3389/fpls.2013.00139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Böhm H., Albert I., Fan L., Reinhard A., Nürnberger T. Immune receptor complexes at the plant cell surface. Curr Opin Plant Biol. 2014;20:47–54. doi: 10.1016/j.pbi.2014.04.007. [DOI] [PubMed] [Google Scholar]
- 9.Zipfel C., Oldroyd G.E.D. Plant signalling in symbiosis and immunity. Nature. 2017;543:328–336. doi: 10.1038/nature22009. [DOI] [PubMed] [Google Scholar]
- 10.Saijo Y., Loo E.P.-I., Yasuda S. Pattern recognition receptors and signaling in plant-microbe interactions. Plant J. 2018;93:592–613. doi: 10.1111/tpj.13808. [DOI] [PubMed] [Google Scholar]
- 11.Sędzielewska Toro K., Brachmann A. The effector candidate repertoire of the arbuscular mycorrhizal fungus Rhizophagus clarus. BMC Genomics. 2016;17:101. doi: 10.1186/s12864-016-2422-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Miwa H., Okazaki S. How effectors promote beneficial interactions. Curr Opin Plant Biol. 2017;38:148–154. doi: 10.1016/j.pbi.2017.05.011. [DOI] [PubMed] [Google Scholar]
- 13.Khan M., Seto D., Subramaniam R., Oh Desveaux D. The places they’ll go! A survey of phytopathogen effectors and their host targets. Plant J. 2018;93:651–663. doi: 10.1111/tpj.13780. [DOI] [PubMed] [Google Scholar]
- 14.Peng Y., van Wersch R., Zhang Y. Convergent and divergent signaling in PAMP-triggered immunity and effector-triggered immunity. Mol Plant Microbe Interact. 2018;31:403–409. doi: 10.1094/MPMI-06-17-0145-CR. [DOI] [PubMed] [Google Scholar]
- 15.Thoms D., Liang Y., Haney C.H. Maintaining symbiotic homeostasis: how do plants engage with beneficial microorganisms while at the same time restricting pathogens? Mol Plant Microbe Interact. 2021;34:462–469. doi: 10.1094/MPMI-11-20-0318-FI. [DOI] [PubMed] [Google Scholar]
- 16.Buendia L., Girardin A., Wang T., Cottret L., Lefebvre B. LysM receptor-like kinase and LysM receptor-like protein families: an update on phylogeny and functional characterization. Front Plant Sci. 2018;9:1531. doi: 10.3389/fpls.2018.01531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Miya A., Albert P., Shinya T., Desaki Y., Ichimura K., Shirasu K., et al. CERK1, a LysM receptor kinase, is essential for chitin elicitor signaling in Arabidopsis. Proc Natl Acad Sci USA. 2007;104:19613–19618. doi: 10.1073/pnas.0705147104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Iizasa E. ichi, Mitsutomi M., Nagano Y. Direct binding of a plant lysM receptor-like kinase, LysM RLK1/CERK1, to chitin in vitro*. J Biol Chem. 2010;285:2996–3004. doi: 10.1074/jbc.M109.027540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu T., Liu Z., Song C., Hu Y., Han Z., She J., et al. Chitin-induced dimerization activates a plant immune receptor. Science. 2012;336:1160–1164. doi: 10.1126/science.1218867. [DOI] [PubMed] [Google Scholar]
- 20.Wan J., Zhang X.-C., Neece D., Ramonell K.M., Clough S., Kim S.-Y., et al. A LysM receptor-like kinase plays a critical role in chitin signaling and fungal resistance in Arabidopsis. Plant Cell. 2008;20:471–481. doi: 10.1105/tpc.107.056754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cao Y., Liang Y., Tanaka K., Nguyen C.T., Jedrzejczak R.P., Joachimiak A., et al. The kinase LYK5 is a major chitin receptor in Arabidopsis and forms a chitin-induced complex with related kinase CERK1. eLife. 2014;3 doi: 10.7554/eLife.03766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wan J., Tanaka K., Zhang X.-C., Son G.H., Brechenmacher L., Nguyen T.H.N., et al. LYK4, a lysin motif receptor-like kinase, is important for chitin signaling and plant innate immunity in Arabidopsis. Plant Physiol. 2012;160:396–406. doi: 10.1104/pp.112.201699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shinya T., Motoyama N., Ikeda A., Wada M., Kamiya K., Hayafune M., et al. Functional characterization of CEBiP and CERK1 homologs in arabidopsis and rice reveals the presence of different chitin receptor systems in plants. Plant Cell Physiol. 2012;53:1696–1706. doi: 10.1093/pcp/pcs113. [DOI] [PubMed] [Google Scholar]
- 24.Bozsoki Z., Cheng J., Feng F., Gysel K., Vinther M., Andersen K.R., et al. Receptor-mediated chitin perception in legume roots is functionally separable from Nod factor perception. Proc Natl Acad Sci USA. 2017;114:E8118–E8127. doi: 10.1073/pnas.1706795114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gibelin-Viala C., Amblard E., Puech-Pages V., Bonhomme M., Garcia M., Bascaules-Bedin A., et al. The Medicago truncatula LysM receptor-like kinase LYK9 plays a dual role in immunity and the arbuscular mycorrhizal symbiosis. New Phytol. 2019;223:1516–1529. doi: 10.1111/nph.15891. [DOI] [PubMed] [Google Scholar]
- 26.Kaku H., Nishizawa Y., Ishii-Minami N., Akimoto-Tomiyama C., Dohmae N., Takio K., et al. Plant cells recognize chitin fragments for defense signaling through a plasma membrane receptor. Proc Natl Acad Sci USA. 2006;103:11086–11091. doi: 10.1073/pnas.0508882103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shimizu T., Nakano T., Takamizawa D., Desaki Y., Ishii-Minami N., Nishizawa Y., et al. Two LysM receptor molecules, CEBiP and OsCERK1, cooperatively regulate chitin elicitor signaling in rice. Plant J. 2010;64:204–214. doi: 10.1111/j.1365-313X.2010.04324.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kouzai Y., Nakajima K., Hayafune M., Ozawa K., Kaku H., Shibuya N., et al. CEBiP is the major chitin oligomer-binding protein in rice and plays a main role in the perception of chitin oligomers. Plant Mol Biol. 2014;84:519–528. doi: 10.1007/s11103-013-0149-6. [DOI] [PubMed] [Google Scholar]
- 29.Gimenez-Ibanez S., Ntoukakis V., Rathjen J.P. The LysM receptor kinase CERK1 mediates bacterial perception in Arabidopsis. Plant Signal Behav. 2009;4:539–541. doi: 10.4161/psb.4.6.8697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Willmann R., Lajunen H.M., Erbs G., Newman M.-A., Kolb D., Tsuda K., et al. Arabidopsis lysin-motif proteins LYM1 LYM3 CERK1 mediate bacterial peptidoglycan sensing and immunity to bacterial infection. Proc Natl Acad Sci USA. 2011;108:19824–19829. doi: 10.1073/pnas.1112862108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu B., Li J.-F., Ao Y., Qu J., Li Z., Su J., et al. Lysin motif-containing proteins LYP4 and LYP6 play dual roles in peptidoglycan and chitin perception in rice innate immunity. Plant Cell. 2012;24:3406–3419. doi: 10.1105/tpc.112.102475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ao Y., Li Z., Feng D., Xiong F., Liu J., Li J.-F., et al. OsCERK1 and OsRLCK176 play important roles in peptidoglycan and chitin signaling in rice innate immunity. Plant J. 2014;80:1072–1084. doi: 10.1111/tpj.12710. [DOI] [PubMed] [Google Scholar]
- 33.Feng F., Sun J., Radhakrishnan G.V., Lee T., Bozsóki Z., Fort S., et al. A combination of chitooligosaccharide and lipochitooligosaccharide recognition promotes arbuscular mycorrhizal associations in Medicago truncatula. Nat Commun. 2019;10:5047. doi: 10.1038/s41467-019-12999-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Brundrett M.C., Tedersoo L. Evolutionary history of mycorrhizal symbioses and global host plant diversity. New Phytol. 2018;220:1108–1115. doi: 10.1111/nph.14976. [DOI] [PubMed] [Google Scholar]
- 35.Delaux P.-M., Varala K., Edger P.P., Coruzzi G.M., Pires J.C., Ané J.-M. Comparative phylogenomics uncovers the impact of symbiotic associations on host genome evolution. PLOS Genet. 2014;10 doi: 10.1371/journal.pgen.1004487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cao Y., Halane M.K., Gassmann W., Stacey G. The Role of Plant Innate Immunity in the Legume-Rhizobium Symbiosis. Annu Rev Plant Biol. 2017;68:535–561. doi: 10.1146/annurev-arplant-042916-041030. [DOI] [PubMed] [Google Scholar]
- 37.Desaki Y., Kouzai Y., Ninomiya Y., Iwase R., Shimizu Y., Seko K., et al. OsCERK1 plays a crucial role in the lipopolysaccharide-induced immune response of rice. New Phytol. 2018;217:1042–1049. doi: 10.1111/nph.14941. [DOI] [PubMed] [Google Scholar]
- 38.Miyata K., Kozaki T., Kouzai Y., Ozawa K., Ishii K., Asamizu E., et al. The bifunctional plant receptor, OsCERK1, regulates both chitin-triggered immunity and arbuscular mycorrhizal symbiosis in rice. Plant Cell Physiol. 2014;55:1864–1872. doi: 10.1093/pcp/pcu129. [DOI] [PubMed] [Google Scholar]
- 39.Zhang X., Dong W., Sun J., Feng F., Deng Y., He Z., et al. The receptor kinase CERK1 has dual functions in symbiosis and immunity signalling. Plant J. 2015;81:258–267. doi: 10.1111/tpj.12723. [DOI] [PubMed] [Google Scholar]
- 40.Genre A., Chabaud M., Balzergue C., Puech-Pagès V., Novero M., Rey T., et al. Short-chain chitin oligomers from arbuscular mycorrhizal fungi trigger nuclear Ca2+ spiking in Medicago truncatula roots and their production is enhanced by strigolactone. New Phytol. 2013;198:190–202. doi: 10.1111/nph.12146. [DOI] [PubMed] [Google Scholar]
- 41.Carotenuto G., Chabaud M., Miyata K., Capozzi M., Takeda N., Kaku H., et al. The rice LysM receptor-like kinase OsCERK1 is required for the perception of short-chain chitin oligomers in arbuscular mycorrhizal signaling. New Phytol. 2017;214:1440–1446. doi: 10.1111/nph.14539. [DOI] [PubMed] [Google Scholar]
- 42.He J., Zhang C., Dai H., Liu H., Zhang X., Yang J., et al. A LysM receptor heteromer mediates perception of arbuscular mycorrhizal symbiotic signal in rice. Mol Plant. 2019;12:1561–1576. doi: 10.1016/j.molp.2019.10.015. [DOI] [PubMed] [Google Scholar]
- 43.Zhang C., He J., Dai H., Wang G., Zhang X., Wang C., et al. Discriminating symbiosis and immunity signals by receptor competition in rice. Proc Natl Acad Sci USA. 2021:118. doi: 10.1073/pnas.2023738118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chiu C.H., Paszkowski U. Receptor-like kinases sustain symbiotic scrutiny. Plant Physiol. 2020;182:1597–1612. doi: 10.1104/pp.19.01341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lerouge P., Roche P., Faucher C., Maillet F., Truchet G., Promé J.C., et al. Symbiotic host-specificity of Rhizobium meliloti is determined by a sulphated and acylated glucosamine oligosaccharide signal. Nature. 1990;344:781–784. doi: 10.1038/344781a0. [DOI] [PubMed] [Google Scholar]
- 46.Maillet F., Poinsot V., André O., Puech-Pagès V., Haouy A., Gueunier M., et al. Fungal lipochitooligosaccharide symbiotic signals in arbuscular mycorrhiza. Nature. 2011;469:58–63. doi: 10.1038/nature09622. [DOI] [PubMed] [Google Scholar]
- 47.Cope K.R., Bascaules A., Irving T.B., Venkateshwaran M., Maeda J., Garcia K., et al. The ectomycorrhizal fungus laccaria bicolor produces lipochitooligosaccharides and uses the common symbiosis pathway to colonize populus roots. Plant Cell. 2019;31:2386–23410. doi: 10.1105/tpc.18.00676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rush T.A., Puech-Pagès V., Bascaules A., Jargeat P., Maillet F., Haouy A., et al. Lipo-chitooligosaccharides as regulatory signals of fungal growth and development. Nat Commun. 2020;11:3897. doi: 10.1038/s41467-020-17615-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Villalobos Solis M.I., Engle N.L., Spangler M.K., Cottaz S., Fort S., Maeda J., et al. Expanding the biological role of lipo-chitooligosaccharides and chitooligosaccharides in Laccaria bicolor growth and development. Front Fungal Biol. 2022;3 doi: 10.3389/ffunb.2022.808578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dénarié J., Debellé F., Promé J.C. Rhizobium lipo-chitooligosaccharide nodulation factors: signaling molecules mediating recognition and morphogenesis. Annu Rev Biochem. 1996;65:503–535. doi: 10.1146/annurev.bi.65.070196.002443. [DOI] [PubMed] [Google Scholar]
- 51.Amor B.B., Shaw S.L., Oldroyd G.E.D., Maillet F., Penmetsa R.V., Cook D., et al. The NFP locus of Medicago truncatula controls an early step of Nod factor signal transduction upstream of a rapid calcium flux and root hair deformation. Plant J. 2003;34:495–506. doi: 10.1046/j.1365-313x.2003.01743.x. [DOI] [PubMed] [Google Scholar]
- 52.Radutoiu S., Madsen L.H., Madsen E.B., Felle H.H., Umehara Y., Grønlund M., et al. Plant recognition of symbiotic bacteria requires two LysM receptor-like kinases. Nature. 2003;425:585–592. doi: 10.1038/nature02039. [DOI] [PubMed] [Google Scholar]
- 53.Arrighi J.-F., Barre A., Ben Amor B., Bersoult A., Soriano L.C., Mirabella R., et al. The Medicago truncatula lysin [corrected] motif-receptor-like kinase gene family includes NFP and new nodule-expressed genes. Plant Physiol. 2006;142:265–279. doi: 10.1104/pp.106.084657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Smit P., Limpens E., Geurts R., Fedorova E., Dolgikh E., Gough C., et al. Medicago LYK3, an entry receptor in rhizobial nodulation factor signaling. Plant Physiol. 2007;145:183–191. doi: 10.1104/pp.107.100495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Radutoiu S., Madsen L.H., Madsen E.B., Jurkiewicz A., Fukai E., Quistgaard E.M.H., et al. LysM domains mediate lipochitin-oligosaccharide recognition and Nfr genes extend the symbiotic host range. EMBO J. 2007;26:3923–3935. doi: 10.1038/sj.emboj.7601826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Moling S., Pietraszewska-Bogiel A., Postma M., Fedorova E., Hink M.A., Limpens E., et al. Nod factor receptors form heteromeric complexes and are essential for intracellular infection in medicago nodules. Plant Cell. 2014;26:4188–4199. doi: 10.1105/tpc.114.129502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Broghammer A., Krusell L., Blaise M., Sauer J., Sullivan J.T., Maolanon N., et al. Legume receptors perceive the rhizobial lipochitin oligosaccharide signal molecules by direct binding. Proc Natl Acad Sci USA. 2012;109:13859–13864. doi: 10.1073/pnas.1205171109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bozsoki Z., Gysel K., Hansen S.B., Lironi D., Krönauer C., Feng F., et al. Ligand-recognizing motifs in plant LysM receptors are major determinants of specificity. Science. 2020;369:663–670. doi: 10.1126/science.abb3377. [DOI] [PubMed] [Google Scholar]
- 59.Fliegmann J., Jauneau A., Pichereaux C., Rosenberg C., Gasciolli V., Timmers A.C.J., et al. LYR3, a high-affinity LCO-binding protein of Medicago truncatula, interacts with LYK3, a key symbiotic receptor. FEBS Lett. 2016;590:1477–1487. doi: 10.1002/1873-3468.12191. [DOI] [PubMed] [Google Scholar]
- 60.Fliegmann J., Canova S., Lachaud C., Uhlenbroich S., Gasciolli V., Pichereaux C., et al. Lipo-chitooligosaccharidic symbiotic signals are recognized by LysM receptor-like kinase LYR3 in the legume Medicago truncatula. ACS Chem Biol. 2013;8:1900–1906. doi: 10.1021/cb400369u. [DOI] [PubMed] [Google Scholar]
- 61.Malkov N., Fliegmann J., Rosenberg C., Gasciolli V., Timmers A.C.J., Nurisso A., et al. Molecular basis of lipo-chitooligosaccharide recognition by the lysin motif receptor-like kinase LYR3 in legumes. Biochem J. 2016;473:1369–1378. doi: 10.1042/BCJ20160073. [DOI] [PubMed] [Google Scholar]
- 62.Sun J., Miller J.B., Granqvist E., Wiley-Kalil A., Gobbato E., Maillet F., et al. Activation of symbiosis signaling by arbuscular mycorrhizal fungi in legumes and rice. Plant Cell. 2015;27:823–838. doi: 10.1105/tpc.114.131326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Oldroyd G.E.D. Speak, friend, and enter: signalling systems that promote beneficial symbiotic associations in plants. Nat Rev Microbiol. 2013;11:252–263. doi: 10.1038/nrmicro2990. [DOI] [PubMed] [Google Scholar]
- 64.Buendia L., Wang T., Girardin A., Lefebvre B. The LysM receptor-like kinase SlLYK10 regulates the arbuscular mycorrhizal symbiosis in tomato. New Phytol. 2016;210:184–195. doi: 10.1111/nph.13753. [DOI] [PubMed] [Google Scholar]
- 65.Girardin A., Wang T., Ding Y., Keller J., Buendia L., Gaston M., et al. LCO receptors involved in arbuscular mycorrhiza are functional for rhizobia perception in legumes. Curr Biol. 2019;29(4249–59) doi: 10.1016/j.cub.2019.11.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gomez S.K., Javot H., Deewatthanawong P., Torres-Jerez I., Tang Y., Blancaflor E.B., et al. Medicago truncatula and Glomus intraradices gene expression in cortical cells harboring arbuscules in the arbuscular mycorrhizal symbiosis. BMC Plant Biol. 2009;9:10. doi: 10.1186/1471-2229-9-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Stracke S., Kistner C., Yoshida S., Mulder L., Sato S., Kaneko T., et al. A plant receptor-like kinase required for both bacterial and fungal symbiosis. Nature. 2002;417:959–962. doi: 10.1038/nature00841. [DOI] [PubMed] [Google Scholar]
- 68.Endre G., Kereszt A., Kevei Z., Mihacea S., Kaló P., Kiss G.B. A receptor kinase gene regulating symbiotic nodule development. Nature. 2002;417:962–966. doi: 10.1038/nature00842. [DOI] [PubMed] [Google Scholar]
- 69.Antolín-Llovera M., Ried M.K., Parniske M. Cleavage of the symbiosis receptor-like kinase ectodomain promotes complex formation with Nod factor receptor 5. Curr Biol. 2014;24:422–427. doi: 10.1016/j.cub.2013.12.053. [DOI] [PubMed] [Google Scholar]
- 70.Genre A., Bonfante P. A rice receptor for mycorrhizal fungal signals opens new opportunities for the development of sustainable agricultural practices. Mol Plant. 2020;13:181–183. doi: 10.1016/j.molp.2020.01.009. [DOI] [PubMed] [Google Scholar]
- 71.Liang Y., Cao Y., Tanaka K., Thibivilliers S., Wan J., Choi J., et al. Nonlegumes respond to rhizobial Nod factors by suppressing the innate immune response. Science. 2013;341:1384–1387. doi: 10.1126/science.1242736. [DOI] [PubMed] [Google Scholar]
- 72.Paparella C., Savatin D.V., Marti L., De Lorenzo G., Ferrari S. The Arabidopsis lysin motif-containing receptor-like kinase3 regulates the cross talk between immunity and abscisic acid responses. Plant Physiol. 2014;165:262–276. doi: 10.1104/pp.113.233759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Jumper J., Evans R., Pritzel A., Green T., Figurnov M., Ronneberger O., et al. Highly accurate protein structure prediction with AlphaFold. Nature. 2021;596:583–589. doi: 10.1038/s41586-021-03819-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Demerdash O.N.A. Using diverse potentials and scoring functions for the development of improved machine-learned models for protein-ligand affinity and docking pose prediction. J Comput Aided Mol Des. 2021;35:1095–1123. doi: 10.1007/s10822-021-00423-4. [DOI] [PubMed] [Google Scholar]
- 75.Goodstein D.M., Shu S., Howson R., Neupane R., Hayes R.D., Fazo J., et al. Phytozome: a comparative platform for green plant genomics. Nucleic Acids Res. 2012;40:D1178–D1186. doi: 10.1093/nar/gkr944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Tamura K., Stecher G., Kumar S. MEGA11: molecular evolutionary genetics analysis version 11. Mol Biol Evol. 2021;38:3022–3027. doi: 10.1093/molbev/msab120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Edgar R.C. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Weighill D., Jones P., Shah M., Ranjan P., Muchero W., Schmutz J., et al. Pleiotropic and epistatic network-based discovery: integrated networks for target gene discovery. Front Energy Res. 2018:6. doi: 10.3389/fenrg.2018.00030. [DOI] [Google Scholar]
- 79.Smid M., Coebergh van den Braak R.R.J., van de Werken H.J.G., van Riet J., van Galen A., de Weerd V., et al. Gene length corrected trimmed mean of M-values (GeTMM) processing of RNA-seq data performs similarly in intersample analyses while improving intrasample comparisons. BMC Bioinform. 2018;19:236. doi: 10.1186/s12859-018-2246-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Chen Y., Lun A.T.L., Smyth G.K. From reads to genes to pathways: differential expression analysis of RNA-Seq experiments using Rsubread and the edgeR quasi-likelihood pipeline. F1000Res. 2016;5:1438. doi: 10.12688/f1000research.8987.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.R Core Team (2013). R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. URL http://www.R-project.org/.
- 82.Waskom M. seaborn: statistical data visualization. J Open Source Softw. 2021;6:3021. [Google Scholar]
- 83.Hunter J.D. Matplotlib: A 2D Graphics Environment. Comput Sci Eng. 2007;9:90–95. [Google Scholar]
- 84.Mariani V., Biasini M., Barbato A., Schwede T. lDDT: a local superposition-free score for comparing protein structures and models using distance difference tests. Bioinformatics. 2013;29:2722–2728. doi: 10.1093/bioinformatics/btt473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Gysel K., Laursen M., Thygesen M.B., Lironi D., Bozsóki Z., Hjuler C.T., et al. Kinetic proofreading of lipochitooligosaccharides determines signal activation of symbiotic plant receptors. Proc Natl Acad Sci USA. 2021:118. doi: 10.1073/pnas.2111031118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Martínez L., Andreani R., Martínez J.M. Convergent algorithms for protein structural alignment. BMC Bioinform. 2007;8:306. doi: 10.1186/1471-2105-8-306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Abraham M.J., Murtola T., Schulz R., Páll S., Smith J.C., Hess B., et al. GROMACS: High performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX. 2015;1–2:19–25. [Google Scholar]
- 88.Huang J., MacKerell A.D., Jr. CHARMM36 all-atom additive protein force field: validation based on comparison to NMR data. J Comput Chem. 2013;34:2135–2145. doi: 10.1002/jcc.23354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Berendsen H.J.C., Grigera J.R., Straatsma T.P. The missing term in effective pair potentials. J Phys Chem. 1987;91:6269–6271. [Google Scholar]
- 90.Humphrey W., Dalke A., Schulten K. VMD: visual molecular dynamics. J Mol Graph. 1996;14(33–8):27–28. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 91.Hameedi M.A., T Prates E., Garvin M.R., Mathews, Amos B.K., Demerdash O., et al. Structural and functional characterization of NEMO cleavage by SARS-CoV-2 3CLpro. Nat Commun. 2022;13:5285. doi: 10.1038/s41467-022-32922-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Heo L., Arbour C.F., Janson G., Feig M. Improved sampling strategies for protein model refinement based on molecular dynamics simulation. J Chem Theory Comput. 2021;17:1931–1943. doi: 10.1021/acs.jctc.0c01238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Hopkins C.W., Le Grand S., Walker R.C., Roitberg A.E. Long-time-step molecular dynamics through hydrogen mass repartitioning. J Chem Theory Comput. 2015;11:1864–1874. doi: 10.1021/ct5010406. [DOI] [PubMed] [Google Scholar]
- 94.Wang R., Fang X., Lu Y., Wang S. The PDBbind database: collection of binding affinities for protein-ligand complexes with known three-dimensional structures. J Med Chem. 2004;47:2977–2980. doi: 10.1021/jm030580l. [DOI] [PubMed] [Google Scholar]
- 95.Wang R., Fang X., Lu Y., Yang C.-Y., Wang S. The PDBbind database: methodologies and updates. J Med Chem. 2005;48:4111–4119. doi: 10.1021/jm048957q. [DOI] [PubMed] [Google Scholar]
- 96.Liu Z., Li Y., Han L., Li J., Liu J., Zhao Z., et al. PDB-wide collection of binding data: current status of the PDBbind database. Bioinformatics. 2015;31:405–412. doi: 10.1093/bioinformatics/btu626. [DOI] [PubMed] [Google Scholar]
- 97.Li Y., Liu Z., Li J., Han L., Liu J., Zhao Z., et al. Comparative assessment of scoring functions on an updated benchmark: 1. Compilation of the test set. J Chem Inf Model. 2014;54:1700–1716. doi: 10.1021/ci500080q. [DOI] [PubMed] [Google Scholar]
- 98.Cortes C., Vapnik V. Support-vector networks. Mach Learn. 1995;20:273–297. [Google Scholar]
- 99.Breiman L. Random forests. Mach Learn. 2001;45:5–32. [Google Scholar]
- 100.Friedman J.H. Greedy function approximation: a gradient boosting machine. Aos. 2001;29:1189–1232. [Google Scholar]
- 101.Freund Y., Schapire R.E. A decision-theoretic generalization of on-line learning and an application to boosting. J Comput System Sci. 1997;55:119–139. [Google Scholar]
- 102.Valdeolivas A., Tichit L., Navarro C., Perrin S., Odelin G., Levy N., et al. Random walk with restart on multiplex and heterogeneous biological networks. Bioinformatics. 2019;35:497–505. doi: 10.1093/bioinformatics/bty637. [DOI] [PubMed] [Google Scholar]
- 103.Zhang J., Yang Y., Zheng K., Xie M., Feng K., Jawdy S.S., et al. Genome-wide association studies and expression-based quantitative trait loci analyses reveal roles of HCT2 in caffeoylquinic acid biosynthesis and its regulation by defense-responsive transcription factors in Populus. New Phytol. 2018;220:502–516. doi: 10.1111/nph.15297. [DOI] [PubMed] [Google Scholar]
- 104.Oellrich A., Walls R.L., Cannon E.K., Cannon S.B., Cooper L., Gardiner J., et al. An ontology approach to comparative phenomics in plants. Plant Methods. 2015;11:10. doi: 10.1186/s13007-015-0053-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Cliff A., Romero J., Kainer D., Walker A., Furches A., Jacobson D. A high-performance computing implementation of iterative random forest for the creation of predictive expression networks. Genes. 2019;10 doi: 10.3390/genes10120996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Zhang P., Dreher K., Karthikeyan A., Chi A., Pujar A., Caspi R., et al. Creation of a genome-wide metabolic pathway database for Populus trichocarpa using a new approach for reconstruction and curation of metabolic pathways for plants. Plant Physiol. 2010;153:1479–1491. doi: 10.1104/pp.110.157396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Szklarczyk D., Gable A.L., Lyon D., Junge A., Wyder S., Huerta-Cepas J., et al. STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019;47:D607–D613. doi: 10.1093/nar/gky1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Giovannoni M., Lironi D., Marti L., Paparella C., Vecchi V., Gust A.A., et al. The arabidopsis thaliana LysM-containing receptor-like kinase 2 is required for elicitor-induced resistance to pathogens. Plant Cell Environ. 2021;44:3545–3562. doi: 10.1111/pce.14192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Shannon P., Markiel A., Ozier O., Baliga N.S., Wang J.T., Ramage D., et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Tian F., Yang D.-C., Meng Y.-Q., Jin J., Gao G. PlantRegMap: charting functional regulatory maps in plants. Nucleic Acids Res. 2020;48:D1104–D1113. doi: 10.1093/nar/gkz1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Kawaharada Y., Kelly S., Nielsen M.W., Hjuler C.T., Gysel K., Muszyński A., et al. Receptor-mediated exopolysaccharide perception controls bacterial infection. Nature. 2015;523:308–312. doi: 10.1038/nature14611. [DOI] [PubMed] [Google Scholar]
- 112.Maillet F., Fournier J., Mendis H.C., Tadege M., Wen J., Ratet P., et al. Sinorhizobium meliloti succinylated high-molecular-weight succinoglycan and the Medicago truncatula LysM receptor-like kinase MtLYK10 participate independently in symbiotic infection. Plant J. 2020;102:311–326. doi: 10.1111/tpj.14625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Rose C.M., Venkateshwaran M., Volkening J.D., Grimsrud P.A., Maeda J., Bailey D.J., et al. Rapid phosphoproteomic and transcriptomic changes in the rhizobia-legume symbiosis. Mol Cell Proteomics. 2012;11:724–744. doi: 10.1074/mcp.M112.019208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.R Cope K., B Irving T., Chakraborty S., Ané J.-M. Perception of lipo-chitooligosaccharides by the bioenergy crop Populus. Plant Signal Behav. 2021;16 doi: 10.1080/15592324.2021.1903758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Tanaka K., Nguyen C.T., Liang Y., Cao Y., Stacey G. Role of LysM receptors in chitin-triggered plant innate immunity. Plant Signal Behav. 2013;8 doi: 10.4161/psb.22598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Nakagawa T., Kaku H., Shimoda Y., Sugiyama A., Shimamura M., Takanashi K., et al. From defense to symbiosis: limited alterations in the kinase domain of LysM receptor-like kinases are crucial for evolution of legume-Rhizobium symbiosis. Plant J. 2011;65:169–180. doi: 10.1111/j.1365-313X.2010.04411.x. [DOI] [PubMed] [Google Scholar]
- 117.Gubaeva E., Gubaev A., Melcher R.L.J., Cord-Landwehr S., Singh R., El Gueddari N.E., et al. “Slipped sandwich” model for chitin and chitosan perception in arabidopsis. Mol Plant Microbe Interact. 2018;31:1145–1153. doi: 10.1094/MPMI-04-18-0098-R. [DOI] [PubMed] [Google Scholar]
- 118.Li C.-L., Xue D.-X., Wang Y.-H., Xie Z.-P., Staehelin C. A method for functional testing constitutive and ligand-induced interactions of lysin motif receptor proteins. Plant Methods. 2020;16:3. doi: 10.1186/s13007-020-0551-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Teufel F., Almagro Armenteros J.J., Johansen A.R., Gíslason M.H., Pihl S.I., Tsirigos K.D., et al. SignalP 6.0 predicts all five types of signal peptides using protein language models. Nat Biotechnol. 2022;40:1023–1025. doi: 10.1038/s41587-021-01156-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Spiwok V. CH/π interactions in carbohydrate recognition. Molecules. 2017;22 doi: 10.3390/molecules22071038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Walker L., Lagunas B., Gifford M.L. Determinants of host range specificity in legume-rhizobia symbiosis. Front Microbiol. 2020;11 doi: 10.3389/fmicb.2020.585749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Truchet G., Roche P., Lerouge P., Vasse J., Camut S., de Billy F., et al. Sulphated lipo-oligosaccharide signals of Rhizobium meliloti elicit root nodule organogenesis in alfalfa. Nature. 1991;351:670–673. [Google Scholar]
- 123.Marczak M., Mazur A., Koper P., Żebracki K., Skorupska A. Synthesis of rhizobial exopolysaccharides and their importance for symbiosis with legume plants. Genes. 2017;8 doi: 10.3390/genes8120360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Nishad R., Ahmed T., Rahman V.J., Kareem A. Modulation of plant defense system in response to microbial interactions. Front Microbiol. 2020;11:1298. doi: 10.3389/fmicb.2020.01298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Goritschnig S., Zhang Y., Li X. The ubiquitin pathway is required for innate immunity in Arabidopsis. Plant J. 2007;49:540–551. doi: 10.1111/j.1365-313X.2006.02978.x. [DOI] [PubMed] [Google Scholar]
- 126.Nito K., Kamigaki A., Kondo M., Hayashi M., Nishimura M. Functional classification of Arabidopsis peroxisome biogenesis factors proposed from analyses of knockdown mutants. Plant Cell Physiol. 2007;48:763–774. doi: 10.1093/pcp/pcm053. [DOI] [PubMed] [Google Scholar]
- 127.Mata-Pérez C., Sánchez-Calvo B., Begara-Morales J.C., Luque F., Jiménez-Ruiz J., Padilla M.N., et al. Transcriptomic profiling of linolenic acid-responsive genes in ROS signaling from RNA-seq data in Arabidopsis. Frontiers in Plant Science. 2015;6 doi: 10.3389/fpls.2015.00122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Lee H.Y., Back K. Melatonin is required for H2 O2 - and NO-mediated defense signaling through MAPKKK3 and OXI1 in Arabidopsis thaliana. J Pineal Res. 2017:62. doi: 10.1111/jpi.12379. [DOI] [PubMed] [Google Scholar]
- 129.Asai T., Tena G., Plotnikova J., Willmann M.R., Chiu W.-L., Gomez-Gomez L., et al. MAP kinase signalling cascade in Arabidopsis innate immunity. Nature. 2002;415:977–983. doi: 10.1038/415977a. [DOI] [PubMed] [Google Scholar]
- 130.Lévy J., Bres C., Geurts R., Chalhoub B., Kulikova O., Duc G., et al. A putative Ca2+ and calmodulin-dependent protein kinase required for bacterial and fungal symbioses. Science. 2004;303:1361–1364. doi: 10.1126/science.1093038. [DOI] [PubMed] [Google Scholar]
- 131.Wang J.-P., Munyampundu J.-P., Xu Y.-P., Cai X.-Z. Phylogeny of plant calcium and calmodulin-dependent protein kinases (CCaMKs) and functional analyses of tomato CCaMK in disease resistance. Front Plant Sci. 2015;6:1075. doi: 10.3389/fpls.2015.01075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Tuskan G.A., Difazio S., Jansson S., Bohlmann J., Grigoriev I., Hellsten U., et al. The genome of black cottonwood, Populus trichocarpa (Torr. & Gray) Science. 2006;313:1596–1604. doi: 10.1126/science.1128691. [DOI] [PubMed] [Google Scholar]
- 133.Garcia K., Delaux P.-M., Cope K.R., Ané J.-M. Molecular signals required for the establishment and maintenance of ectomycorrhizal symbioses. New Phytol. 2015;208:79–87. doi: 10.1111/nph.13423. [DOI] [PubMed] [Google Scholar]
- 134.Kim S., Zeng W., Bernard S., Liao J., Venkateshwaran M., Ane J.-M., et al. Ca2+-regulated Ca2+ channels with an RCK gating ring control plant symbiotic associations. Nat Commun. 2019;10:3703. doi: 10.1038/s41467-019-11698-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Parsons T.J., Sinkar V.P., Stettler R.F., Nester E.W., Gordon M.P. Transformation of poplar by agrobacterium tumefaciens. Biotechnology. 1986;4:533–536. [Google Scholar]
- 136.Triozzi P.M., Schmidt H.W., Dervinis C., Kirst M., Conde D. Simple, efficient and open-source CRISPR/Cas9 strategy for multi-site genome editing in Populus tremula × alba. Tree Physiol. 2021;41:2216–2227. doi: 10.1093/treephys/tpab066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Gough C., Cottret L., Lefebvre B., Bono J.-J. Evolutionary history of plant LysM receptor proteins related to root endosymbiosis. Front Plant Sci. 2018;9:923. doi: 10.3389/fpls.2018.00923. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material.
Supplementary material.
Supplementary material.