Ribomethylome profiling of primary acute myeloid leukemia cells uncovered a specific and dynamic rRNA 2′-O-methylation pattern that influences protein translation and thus leukemia stem cell phenotypes and disease progression.
Abstract
The development and regulation of malignant self-renewal remain unresolved issues. Here, we provide biochemical, genetic, and functional evidence that dynamics in ribosomal RNA (rRNA) 2′-O-methylation regulate leukemia stem cell (LSC) activity in vivo. A comprehensive analysis of the rRNA 2′-O-methylation landscape of 94 patients with acute myeloid leukemia (AML) revealed dynamic 2′-O-methylation specifically at exterior sites of ribosomes. The rRNA 2′-O-methylation pattern is closely associated with AML development stage and LSC gene expression signature. Forced expression of the 2′-O-methyltransferase fibrillarin (FBL) induced an AML stem cell phenotype and enabled engraftment of non-LSC leukemia cells in NSG mice. Enhanced 2′-O-methylation redirected the ribosome translation program toward amino acid transporter mRNAs enriched in optimal codons and subsequently increased intracellular amino acid levels. Methylation at the single site 18S-guanosine 1447 was instrumental for LSC activity. Collectively, our work demonstrates that dynamic 2′-O-methylation at specific sites on rRNAs shifts translational preferences and controls AML LSC self-renewal.
Significance:
We establish the complete rRNA 2′-O-methylation landscape in human AML. Plasticity of rRNA 2′-O-methylation shifts protein translation toward an LSC phenotype. This dynamic process constitutes a novel concept of how cancers reprogram cell fate and function.
This article is highlighted in the In This Issue feature, p. 247
INTRODUCTION
Cancers are heterogeneous tissues composed of cells with diverse phenotypes and states. Propagation of many cancers including acute myeloid leukemia (AML) depends on a small subset of self-renewing cells and cancer/leukemia stem cells (LSC; ref. 1). Illustration of the pathways that induce cancer stem cell phenotypes may enable the development of conceptually new therapies. Although self-renewal has been studied primarily and intensively at the level of epigenetics and transcription, the comprehensive repertoire of LSC determinants remains to be fully defined. Recent studies have revealed the crucial role of posttranscriptional mechanisms in the maintenance of normal and malignant stem cells (2, 3). The importance of posttranscriptional regulation is emphasized by the fact that transcription profiles often do not reflect the cellular functional proteome that ultimately defines cell identity and function (4, 5). Importantly, stem cell features are closely linked with fine-tuned protein synthesis (6–9). Protein synthesis determines the switch between quiescent and activated states and defines whether activated stem cells self-renew or differentiate (6–9). Specifically, maintenance of self-renewal requires the coordination of low global protein synthesis with a selective translation of specific proteins crucial for stem cells, including those involved in stress adaptation and energy homeostasis (3, 10, 11). The molecular basis for selective translation in stem cells is largely unknown but might be linked to alterations in the translation apparatus. Mutations that affect the protein translation machinery induce dysfunction of stem cells and increased cancer susceptibility in ribosomopathy patients (12, 13). For example, ribosome protein haploinsufficiency results in the reduced ribosome level and translation of GATA1 mRNA, impairing erythroid lineage commitment in patients with Diamond–Blackfan anemia (12). So far, congenital ribosomal mutations in ribosomopathies and somatic ribosomal mutations in various cancers mainly affect ribosomal proteins. By contrast, cancer-related alterations in ribosomal RNA (rRNA) and the functional consequences of such changes have been much less studied.
One of the key features of rRNA is the high degree of peri- and posttranscriptional chemical modifications during ribosome biogenesis (14). On the molecular level, these modifications stabilize ribosome structure; optimize the interaction of ribosomes with tRNAs, mRNAs, and translation factors; and are supposed to be fundamental to global ribosome topology and functions (14–17). More recently, it was shown that rRNA modifications may represent an important source of ribosome heterogeneity and provide critical variables in protein expression (18, 19). In humans, the most abundant rRNA modification is 2′-O-methylation (2′-O-Me), which is catalyzed by the C/D box snoRNP complex that contains the methyltransferase fibrillarin (FBL); scaffold proteins NOP58, NOP56, and NHP2L1; as well as the guiding C/D box snoRNA (20, 21). Recently, we showed that C/D box snoRNA abundance is associated with LSC frequency (22). Accordingly, C/D box snoRNAs and their directed 2′-O-Me might be involved in leukemic self-renewal (22) and cellular proliferation (23). In addition, the relevance of rRNA 2′-O-Me to human disease has also been identified and described in multiple cancer cell lines and tissues (24–28). Despite these extensive studies, the plasticity, diversity, and dynamics of rRNA 2′-O-Me in human malignancies and, particularly, its role in the acquisition and maintenance of cancer stem cell phenotypes have remained elusive.
AML is an instructive model to address these questions, as cell state diversity is an important disease hallmark of AML, which is composed of a mix of self-renewing leukemic stem cells and leukemic blasts with different stages of differentiation (1, 29). Thus, exploration of rRNA modifications in AML may reveal how protein expression is regulated via ribosome heterogeneity to give rise to the remarkable diversity of cell types and to control unique cell behaviors. In this study, we performed an integrated analysis of the rRNA 2′-O-Me patterns in human primary AMLs, their 3D structural distribution on the ribosome, and the associated gene expression signatures and clinical features. We further evaluated the functional significance based on patient-derived xenograft models and analyzed translation alterations by nascent proteomics and ribosome profiling to dissect the role of dynamic rRNA modifications in the control of tumor cellular hierarchy and leukemic stem cell activity. These analyses demonstrate that enhanced 2′-O-Me on ribosome exterior sites drives AML stem cell phenotypes via preferential translation of amino acid transporter mRNAs in a codon-dependent manner.
RESULTS
A Dynamic rRNA Ribomethylome in Human AML
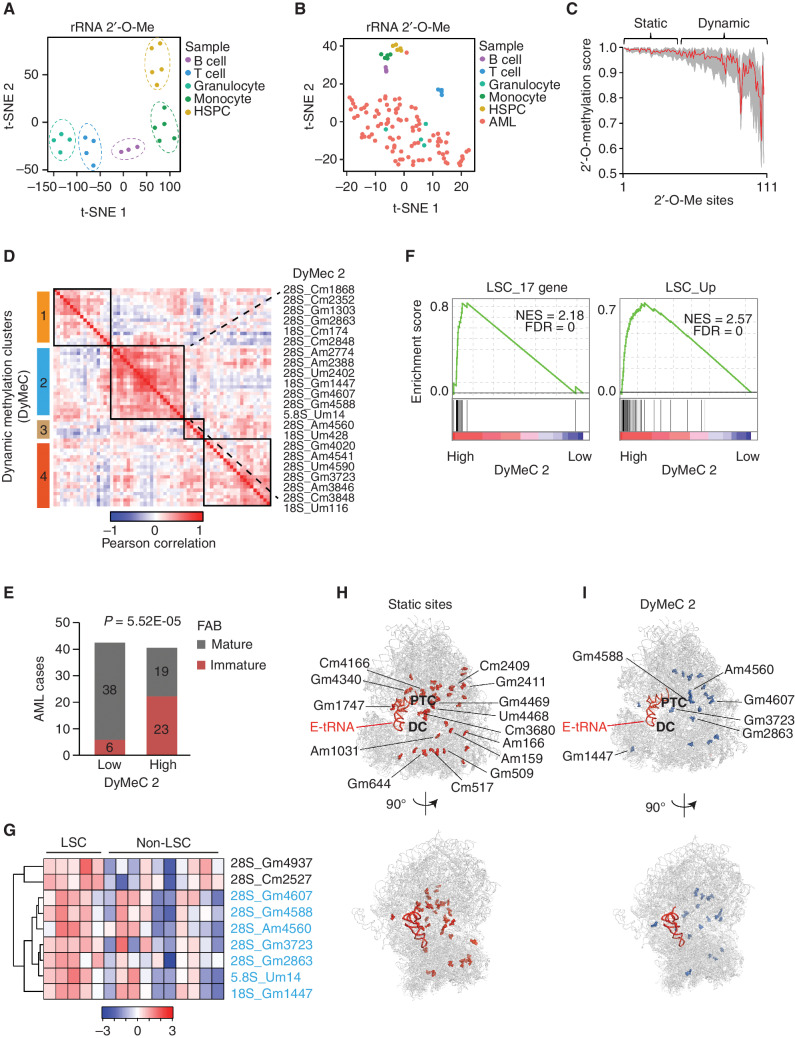
As one of the key steps in ribosome biogenesis, rRNA is posttranscriptionally modified with 2′-O-Me by the methyltransferase FBL. FBL is guided by C/D box snoRNAs for site specificity. In our previously generated LSC proteome dataset, we observed an overrepresentation of FBL and other C/D box snoRNP members in LSC fractions (ref. 30; Supplementary Fig. S1A). Transcriptome analysis in 90 primary AMLs indicated that FBL positively correlated with LSC genes (Supplementary Fig. S1B and S1C) and negatively correlated with hematopoietic differentiation programs (Supplementary Fig. S1D). These findings hinted at a potential association between FBL-induced rRNA 2′-O-Me and AML stem cell phenotypes. Thus, we systematically explored the entire rRNA 2′-O-Me pattern (ribomethylome) for 94 AML specimens, as well as for human CD34+ cord blood cells and peripheral blood B cells, T cells, monocytes, and granulocytes by performing RiboMethSeq (31). In total, 111 sites (69 sites on 28S, 40 on 18S, and 2 on 5.8S rRNA) were found to be 2′-O-methylated in AML blasts and in normal hematopoietic cells (Supplementary Table S1). In healthy cells, the extent of 2′-O-Me of each modification site was rather static, with small fluctuation across the entire rRNA sequence (Supplementary Fig. S1E and S1F). Yet, the overall 2′-O-Me pattern clearly separated each cell type from the others, indicating cell type–specific ribomethylomes (Fig. 1A). The 2′-O-Me patterns in AML blasts showed far higher levels of heterogeneity (Fig. 1B). In patients with AML, only 44 of 111 modification sites were constantly fully methylated (static sites, methylation score > 0.98, variability in methylation score < 0.02), and the other sites exhibited substoichiometric and dynamic methylation (Fig. 1C).
Figure 1.
The dynamic rRNA 2′-O-Me is associated with AML stemness. A, t-Schochastic neighbor embedding (t-SNE) analysis indicating cell type–specific rRNA 2′-O-Me in normal human hematopoietic cells. HSPC, hematopoietic stem and progenitor cells. B, t-SNE analysis indicating distinct and heterogeneous rRNA 2′-O-Me of human primary AMLs (n = 94). C, Variability of 2′-O-Me on each rRNA modification site across human AMLs. The 111 modification sites were ranked according to their 2′-O-Me diversity in AMLs. The red line shows the average 2′-O-Me level of each site in AMLs, and the gray indicates the range of 2′-O-Me. D, Unsupervised hierarchical clustering of pairwise correlation between rRNA dynamic sites based on 2′-O-Me in 94 patients with AML. Each column lists the correlation of one site (in row) with all dynamic sites. Four dynamic clusters (DyMeC, black boxes) with co-occurrence in 2′-O-Me were identified. rRNA sites in DyMeC 2 are labeled. E, Number of AML cases with maturation classification in groups with low and high 2′-O-Me on DyMeC cluster 2. Immature, FAB M0 and M1 subtype; mature, FAB M2 to M6. P = 5.52E−04, Chi-square test. Patients with no FAB subtype information were not included in each group. F, Gene set enrichment analysis plot showing enrichment of LSC gene signatures in patients with AML with higher total 2′-O-Me on DyMeC 2. G, Heat map of 2′-O-Me clusters with increased modification in LSC, supervised by cell types. Samples are in columns; 2′-O-Me sites are in rows. Blue indicates DyMeC 2 sites. NES, normalized enrichment score. H and I, Distribution of static sites (H) and the dynamic DyMeC 2 (I) on cryogenic electron microscopy structure of the human ribosome. Labeled in H are sites located in ribosome-conserved function centers and in I are LSC sites. E-site tRNA is shown in red; rRNA and r-proteins in gray. Protein Data Bank code for the structure is 4UGO. Modification sites from 4UGO are reannotated to the rRNA sequence used for RiboMethSeq. DC, decoding center.
Variation of 2′-O-Me in AML occurred at a subset of apparently coregulated sites. By unsupervised hierarchical clustering based on the Pearson correlation of 2′-O-Me, dynamic sites were categorized into four clusters [dynamic methylation clusters (DyMeC) 1 to 4, Fig. 1D; Supplementary Table S2]. Of note, methylation status on DyMeC 2 correlated with the AML developmental stage. Most patients with high 2′-O-Me on DyMeC 2 (55%, 23 of 42 cases) were diagnosed as AML without maturation [French–American–British classification (FAB) M0 and M1]. This phenotype occurred only in 16% (6 of 44 cases) of patients with low 2′-O-Me (P = 5.52E-05; Fig. 1E). Further, patients with high 2′-O-Me showed higher CD34+ blast counts in the bone marrow and blood (Supplementary Fig. S2A). There was no association between 2′-O-Me and patient's age, gender, bone marrow blast percentage, AML type, cytogenetics, or mutational profile (NPM1, FLT, and CEBPA; Supplementary Fig. S2A). Transcriptome analysis indicated that patients with high rRNA methylation on DyMeC 2 were enriched for LSC signatures and depleted for hematopoietic differentiation programs (Fig. 1F; Supplementary Fig. S2B). Enrichment of LSC signatures in samples with higher 2′-O-Me on DyMeC 2 was confirmed in a second independent patient cohort (n = 18; Supplementary Fig. S2C and S2D). This association was specific for DyMeC 2 and did not occur for the other DyMeC clusters (Supplementary Fig. S2E–S2G).
We confirmed the rRNA 2′-O-Me patterns in functionally validated LSCs and matched non-LSC counterparts from 5 AML patients (refs. 30, 32; Supplementary Fig. S3A–S3C). Nine rRNA sites showed significantly increased 2′-O-Me modification in LSCs (Fig. 1G). All LSC sites were attributed to DyMeC 2 with the exception of 28S C2527 and G4937 (Fig. 1D and G). Of note, no guide snoRNAs are assigned to 28S C2527 and G4937, suggesting that the two sites might be introduced by distinct mechanisms independent of C/D box snoRNPs (18).
On a 3D ribosome model (33), the static sites mainly located within and surrounding catalytic centers, represented by Gm4166 in the P-tRNA CCA binding pocket, Gm4340 in the E-tRNA binding site, and Um4468 and Gm4469 in the peptidyl transferase center (PTC; Fig. 1H; Supplementary Table S3). In contrast, the DyMeC 2 cluster, including the LSC-associated sites, was mainly located on the ribosome surface distant from catalytic regions (Fig. 1I; Supplementary Fig. S3D), suggestive of a “specialized” function in protein translation control.
The Ribomethylome Regulates Human AML Stemness
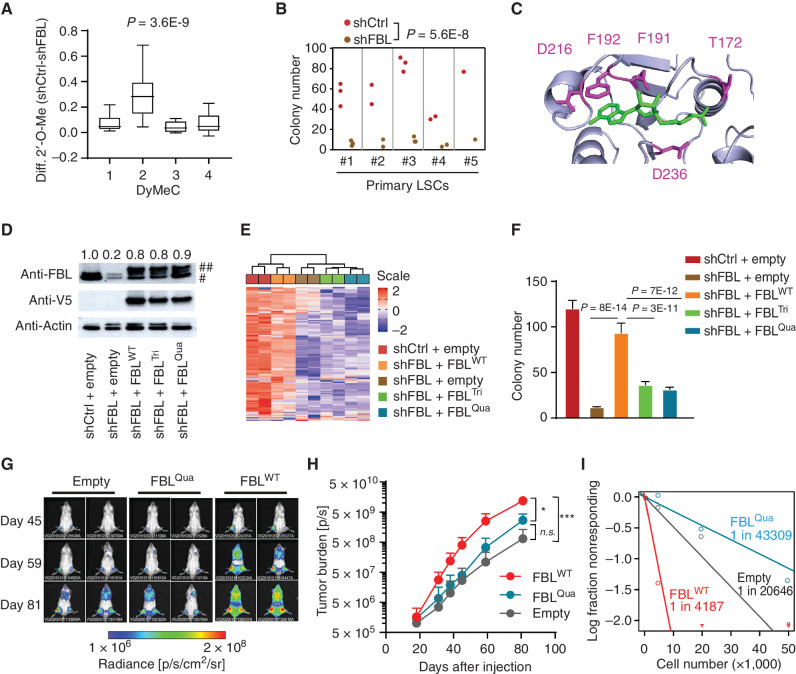
Given the close association of 2′-O-Me, especially that on DyMeC 2, with immature AML phenotypes and LSC gene expression signature, we hypothesized that 2′-O-Me might represent a novel AML stemness regulator. Knockdown of 2′-O-methyltransferase FBL predominantly reduced 2′-O-Me on cluster DyMeC 2, especially on the LSC sites 18S G1447 and 28S G2863, G4560, G4588, and G3723 (Fig. 2A; Supplementary Fig. S4A and S4B). The four static sites in the conserved catalytic centers (28S G4166, G4340, U4468, and G4469) were not affected (Supplementary Fig. S4A and S4C). Decreased FBL expression inhibited colony formation of functionally validated CD34+CD38− leukemic stem cells (Fig. 2B). Next, we aimed to evaluate the relevance of FBL enzymatic activity for leukemogenic functions. Structural analysis of human FBL protein (34) revealed that the amino acids T172, D191, F192, D216, and D236 form a “pocking” domain (Fig. 2C). These sites are predicted to be responsible for substrate binding. We replaced the respective amino acids with alanine to create enzymatically compromised FBL mutants, including mutant FBLT172A, D191A, F192A (referred to as FBLTri) and FBLT172A, D191A, F192A, D216A (referred as FBLQua). Doxycycline-induced target-specific short hairpin RNA (shRNA) achieved an 80% reduction in endogenous FBL protein levels in Kasumi-1 leukemia cells (Fig. 2D). Expression of either wild-type or mutated FBL reinstated total FBL protein levels back to 90% of control (Fig. 2D). The ribomethylome was faithfully reestablished by wild-type but none of the mutant FBL (Fig. 2E; Supplementary Fig. S4D). Functionally, only wild-type FBL rescued AML cell colony formation and proliferation (Fig. 2F; Supplementary Fig. S4E).
Figure 2.
The ribomethylome regulates AML stemness. A, Changes in 2′-O-Me on each dynamic cluster after FBL knockdown in Kasumi-1 AML cells. n = 4 independent experiments. Indicated P value by an unpaired Student t test. B, Colony number formed by control (shCtrl) and FBL knockdown (shFBL) LSC populations. For FBL knockdown, a pool of two different FBL-specific shRNAs was used. Indicated P value by an unpaired Student t test. C, Catalytic center of human FBL protein. Amino acid residues highlighted in purple are substrate binding sites. The substrate S-adenosyl-L-homocysteine (SAH) is highlighted in green. The structure is from Protein Data Bank 5GIO. D, Western blot showing expression of V5-tagged exogenous wild-type (WT) and mutant FBL in FBL knockdown Kasumi-1 cells. Numbers on top indicate the relative expression level of total FBL compared with that in control cells. ##V5-tagged exogenous FBL; #endogenous FBL. E, Unsupervised clustering analysis of rRNA 2′-O-Me in FBL knockdown and rescued cells. The wild-type but not mutant FBL restored rRNA 2′-O-Me. F, Colony formation assay showing the rescue effect of wild-type and mutant FBL. Mean ± SD from n = 9 cultures from three experiments per group is given, and statistical significance was assessed by Student unpaired t test. G, Representative bioluminescence imaging of NSG mice transplanted with AML PDX cells overexpressing empty control, mutant FBL (FBLQua), and wild-type FBL. Images were taken at the indicated time points after transplantation. H, Summary of in vivo proliferation of PDX cells determined by bioluminescence imaging. Note that the signal from FBLWT mice was approaching saturation at day 81. n = 4 for empty control, n = 5 for FBLQua, n = 3 for FBLWT, ***, P = 0.001; *, P = 0.023; n.s., no significance, Student unpaired t test. I, Absolute LSC frequency in the bone marrow of each PDX estimated by in vivo limiting dilution assay. P = 0.0189, FBLWT versus empty vector; P = 0.0006, FBLWT versus FBLQua, Chi-square test.
Engraftment and initiation of human leukemia in immunocompromised mice is the bona fide definition of human LSCs. Thus, we expressed either wild-type FBL or FBLQua mutant in AML patient-derived xenograft (PDX) cells with low leukemogenicity as indicated by long latency. After transplantation into NOD.Prkdcscid.Il2rgnull (NSG) mice, leukemia burden was repetitively monitored by bioluminescence imaging. Wild-type FBL accelerated leukemogenesis, whereas the enzymatically inactive FBLQua mutant failed to do so (Fig. 2G and H). We further observed an increase in the fraction of LSCs (represented by GPR56+CD34+ cells) in PDX cells overexpressing wild-type FBL (Supplementary Fig. S5A and S5B). FBLWT PDX cells formed more colonies upon plating in methylcellulose (Supplementary Fig. S5C). We determined LSC frequencies in these PDX cells by in vivo limiting dilution analysis. FBLWT PDX cells showed nearly a 5-fold and 10-fold increase in LSC frequency compared with empty control and FBLQua PDX cells, respectively (1 in 4,187 vs. 1 in 20,646, P = 0.0189, and vs. 1 in 43,309, P = 0.0006; Fig. 2I; Supplementary Fig. S5D). Forced expression of FBL promoted engraftment of primary AML blast (Supplementary Fig. S5E), which functionally confirmed an FBL-induced LSC phenotype. Moreover, FBL expression transformed non-LSC leukemia cells into functional engrafting LSCs in primary and PDX AML samples. Upon expression of empty vector, CD34+CD38+ populations from two specimens showed very poor engraftment in only 1 of 11 mice (1/7 and 0/4, respectively). Upon enforced FBL expression in these CD34+CD38+ cells, 7 of 8 mice could be engrafted (5/5 and 2/3, respectively; P = 0.0012, Fisher exact test; Supplementary Fig. S5F and S5G). Taken together, these data demonstrated that an FBL-induced ribomethylome induced an LSC phenotype and accelerated leukemia progression.
The Dynamic Ribomethylome Regulates Translation of Amino Acid Transporters
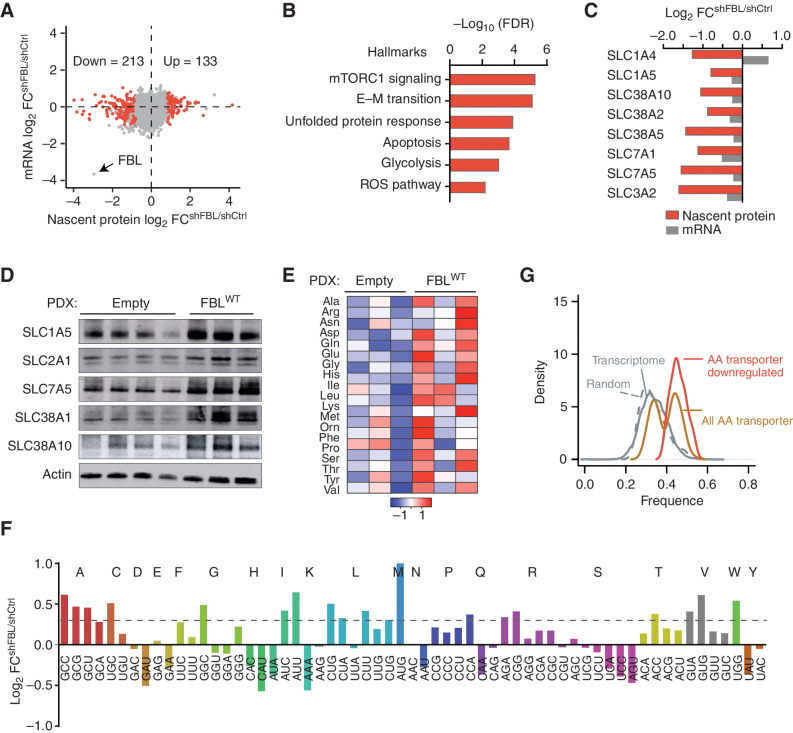
The dynamic ribomethylome might function by redirecting protein translation. We performed nascent proteomics to evaluate active protein synthesis using pulsed azidohomoalanine (pAHA) labeling in combination with pulsed SILAC (pSILAC; ref. 35). Proteins newly synthesized within a 6-hour time window were captured from FBL knockdown or control cells (Supplementary Fig. S6A and S6B), and their abundance was normalized to read density of transcripts identified by mRNA sequencing (mRNA-seq; Supplementary Fig. S6C). This approach enabled us to define genes that were differentially regulated at the translation level. FBL knockdown affected neither the total ribosome level (Supplementary Fig. S6D–S6F) nor global protein translation (Fig. 3A). We obtained a set of 213 proteins with suppressed translation upon decreased FBL levels (Fig. 3A; Supplementary Table S4). The downregulated proteins were enriched for six pathways, with mTORC1 signaling on the top (Fig. 3B). The upregulated proteins were mainly enriched for immune cell development (Supplementary Fig. S6G). The mTORC1 pathway acts as a hub to coordinate protein translation and metabolic demand with the switch of stem cell fate from self-renewal to differentiation (3). Among mTORC1 pathway genes, synthesis of all amino acid transporter proteins captured in our assay was decreased upon suppression of rRNA 2′-O-Me (Fig. 3C). Accordingly, metabolic screening by gas chromatography/mass spectrometry (GC/MS) revealed that amino acids were the metabolites most strongly affected by FBL knockdown (Supplementary Fig. S6H). A targeted metabolite analysis showed that multiple amino acids, especially glutamine, glutamic acid, alanine, and proline, were significantly reduced in FBL knockdown cells (Supplementary Fig. S6I). Amino acid metabolism–related glutathione and α-ketoglutarate were also significantly reduced in knockdown cells (Supplementary Fig. S6J and S6K). Vice versa, overexpression of FBL in PDX cells in vivo increased the amino acid transporter proteins SLC1A5, SLC2A1, SLC7A5, SLC38A1, and SLC38A10, even though the mRNA levels remained unchanged (Fig. 3D; Supplementary Fig. S6L). Consistent with this, we observed increased intracellular levels of glutamine, glutamic acid, glycine, aspartic acid, and histidine in these AML PDX cells (Fig. 3E). Altered amino acid metabolism has been reported as a hallmark feature of leukemic stem cells (36, 37). These data suggested that enhanced amino acid metabolism is an important functional consequence of increased 2′-O-Me in leukemic stem cells.
Figure 3.
The ribomethylome regulates translation of amino acid transporters. A, Differentially translated proteins after FBL knockdown in Kasumi-1 AML cells. The difference in newly synthesized proteins (by nascent proteomics) was plotted against changes in mRNA (by mRNA-seq). The red dots highlight differentially translated proteins, defined by log2FC > 0.8 (adjust P < 0.05) on nascent protein without changes on the mRNA level. B, Gene ontology analysis of proteins less translated after FBL knockdown. C, Changes of amino acid transporters on nascent protein and mRNA level. Nascent proteins are shown in red and mRNA expression in gray. D, Western blot showing protein levels of amino acid transporters in PDX cells transduced with empty vector or wild-type FBL. PDX cells were isolated from mice (n = 4 from the empty control group and n = 3 from the FBLWT group) described in Fig. 2H. E, Abundance of cellular amino acid in PDX cells isolated from n = 3 mice per group. F, Comparison of ribosome P-site occupancy on each codon between control and FBL knockdown Kasumi-1 cells. Codons for the same amino acid are shown in the same color. The dashed line indicates log2FC = 0.3. G, Distribution of codons with increased P-site ribosome occupancy in whole-transcriptome, 1,000 random selected transcripts, all amino acid transporter genes, and in amino acid transporter genes with decreased translation after FBL knockdown. The y-axis indicates the proportion of genes within a given range of codons.
Next, we performed ribosome profiling to investigate the interaction of ribosomes with amino acid transporter mRNAs under different rRNA 2′-O-Me conditions (Supplementary Fig. S7A–S7C). Indeed, FBL knockdown altered codon usage (Fig. 3F; Supplementary Fig. S7D and S7E). Increased ribosome occupancy was found at the in-frame P-site at the ATG start codon and codons for alanine (GCC, GCG, GCT), cysteine (TGC), glycine (GGC), isoleucine (ATC, ATT), leucine (CTG, CTA, CTT), proline (CCA), arginine (AGA, CGG), threonine (ACC), valine (GTA, GTG), and tryptophan (TGG; Fig. 3F; Supplementary Fig. S7F), indicating decelerated translation elongation at these codons. Notably, most of these codons belong to optimal codons, which are supposed to increase mRNA stability and translation efficiency (refs. 38–40; Supplementary Fig. S7G). Codon optimality coordinates the expression of functionally related genes—that is, important enzymes involved in glycolysis (41, 42). We analyzed the codon content of amino acid transporter genes. Optimal codons were overrepresented in amino acid transporters, especially in transporters downregulated by 2′-O-Me suppression (Fig. 3G). Accordingly, we observed increased ribosome pausing specifically on amino acid transporter mRNAs (Supplementary Fig. S7H and S7I), consistent with the reduced translation rate. Of note, optimal codons were enriched neither in other genes such as the mTORC1 pathway nor in genes involved in glycolysis shown to be suppressed by FBL knockdown (Supplementary Fig. S7J and S7K). Translational downregulation of such genes might be an in-trans effect of ribosome pausing on the highly expressed amino acid transporters (up to about 20% of actin mRNA; Supplementary Fig. S6L) due to the shared tRNA and translation apparatus (43). Further, we did not observe reduced mTORC1 activity after FBL knockdown, as phosphorylation of the p70 S6 kinase on Thr389 and 4E-BP1 on Thr37/46, the direct downstream targets of mTORC1, was not altered (Supplementary Fig. S7L). Together, these data suggest that FBL-induced rRNA methylation regulates LSC mainly through optimizing the translation of specific amino acid transporters by affecting optimal codon usage, even though FBL may have additional functions.
Methylation of 18S G1447 (Gm1447) Determines LSC Activity
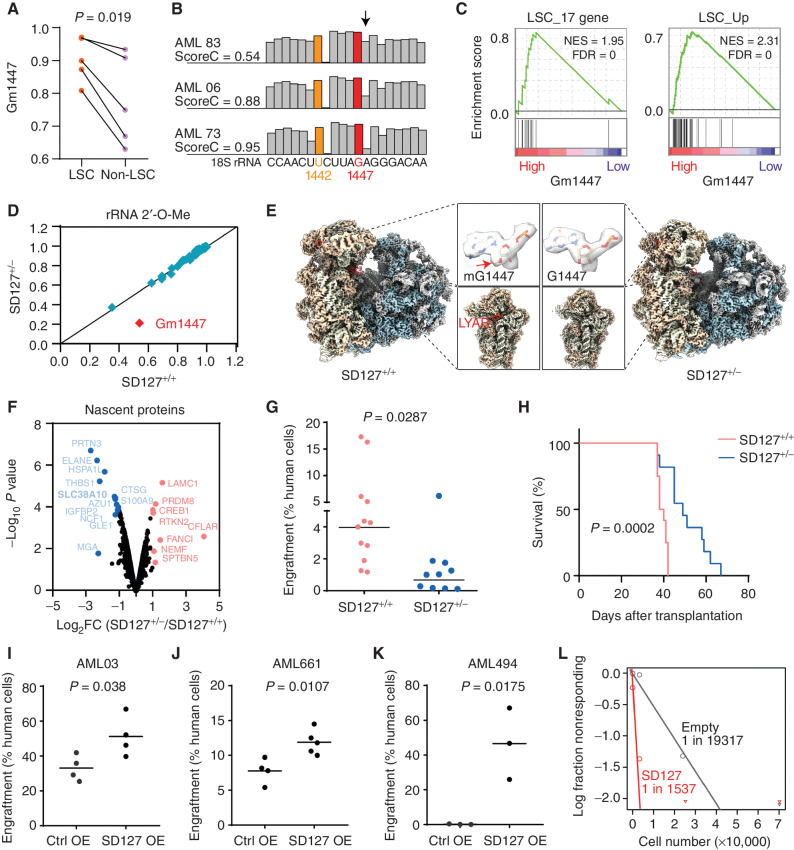
Beyond global rRNA methylation patterns, single-site modification might affect LSC activity. We focused on 18S G1447 (Gm1447), an LSC site in DyMeC 2 that is located on the surface of the small subunit of the human ribosome (Supplementary Fig. S8A). Gm1447 was increased in human LSCs compared with non-LSC counterparts in primary AMLs (Fig. 4A). Gm1447 varied substantially (methylation score ranging from 0.54 to 0.95) across primary AML blasts, whereas the upstream modification site U1442 was constitutively fully methylated (Fig. 4B). Comparing the transcriptomes of Gm1447hi AMLs with Gm1447lo samples revealed a positive enrichment for LSC genes and negative enrichment for hematopoietic differentiation genes in two independent patient cohorts (Fig. 4C; Supplementary Fig. S8B and S8C). These findings suggested an association of Gm1447 with AML stemness.
Figure 4.
Gm1447 determines LSC activity. A, Methylation levels on G1447 in 5 matched LSC and non-LSC fractions as described in Fig. 1G. Indicated P values by Student paired t test. B, Variability of Gm1447 in primary AML samples. Shown are 3 patients with high, medium, and low Gm1447, respectively. G1447 is shown in red, and U1442 with constitutive full methylation in human AMLs is shown in yellow. The arrow indicates +1 position to G1447 that the calculation of the Gm1447 level (score C) is based on (see Methods). C, Gene set enrichment analysis showing that samples with high Gm1447 are enriched for LSC genes. Samples were split into two equal groups at a median of Gm1447. NES, normalized enrichment score. D, Comparison of rRNA 2′-O-Me in SNORD127+/+ and SNORD127+/− Kasumi-1 cells. E, Cryo-EM maps of the 80S ribosome from SNORD127+/+ and SNORD127+/− Kasumi-1 cells. Highlighted are the 2′-O-Me density at G1447 and the decoding center of the 40S ribosomal subunit. The arrow indicates a density bump of 2′-O-Me on G1447 in SNORD127+/+ cells. F, Nascent proteomics from SNORD127+/+ and SNORD127+/− Kasumi-1 cells. Decreased nascent proteins in SNORD127+/− cells are labeled in blue, and increased nascent proteins are in red. G, Engraftment of SNORD127+/+ and SNORD127+/− OCI-AML2 cells in NSG mice (percentages of leukemic cells among bone marrow cells; each dot represents one mouse; n = 12 mice for the SNORD127+/+ group and n = 10 mice for SNORD127+/−). Two different single clones per group were used for transplantation. Short horizontal line indicates the mean; P values are indicated by Student unpaired t test. H, Survival of mice transplanted with SNORD127+/+ and SNORD127+/- OCI-AML2 cells. n = 12 mice for control group and n = 10 mice for SNORD127+/−. Indicated P value by log-rank test. I–K, Engraftment of primary AML cells AML03 (I), AML661 (J), and AML494 (K) transduced with empty vector or SNORD127. Each dot represents the bone marrow engraftment in one mouse. Short horizontal line indicates mean; P values are indicated by Student unpaired t test. OE, overexpression. L, Absolute LSC frequency in primary AML494 cells overexpressing empty vector or SNORD127 estimated by in vivo limiting dilution assay. P = 0.0015, Chi-square test.
The guiding snoRNA for Gm1447 is SNORD127 (Supplementary Fig. S8D), which is expressed from intron 10 of gene PRPF39 on chromosome 14 (44, 45). To suppress Gm1447, we generated SNORD127 knockout in the AML cell lines OCI-AML2 and Kasumi-1 using CRISPR–Cas9 coupled with paired gRNAs flanking SNORD127 sequence (Supplementary Fig. S8E). Of note, no homozygous deletion clone was obtained from 200 screened single clones, which indicated that SNORD127 might be essential at least in leukemic cells. SNORD127 monoallelic deletion (SNORD127+/−) caused an about 60% decrease in SNORD127 levels and in Gm1447 (Fig. 4D; Supplementary Fig. S8F). The loss did not affect other sites and had no effect on host gene expression (Fig. 4D; Supplementary Fig. S8G). To confirm the decrease in Gm1447, we performed cryogenic electron microscopy (cryo-EM) single-particle analysis for 80S ribosome purified from SNORD127+/+ and SNORD127+/− Kasumi-1 cells. We obtained the 80S ribosome structures with high resolution of 2.5 Å and 2.7 Å on average, respectively. Although the 40S head part, where the G1447 locates, displayed decreased local resolution due to its dynamic movement, it was sufficient to identify and compare the 2′-O-Me modification of the rRNA nucleotides. Clearly, the corresponding density bump of the 2′-O-Me modification on G1447 presented in ribosomes in wild-type cells became much weaker in SNORD127+/− cells (Fig. 4E). This finding confirmed decreased 2′-O-Me upon heterozygous SNORD127 deletion. Loss of modification on G1447 did not alter local peptide chain conformation. However, decreased G1448 methylation altered the ribosome interactome. We observed nucleolar protein LYAR on the decoding center of the 40S ribosomal subunit in SNORD127+/+ but not in the SNORD127± cells (Fig. 4E). Loss of LYAR–ribosome association in SNORD127+/− cells was confirmed by sucrose density gradient ribosome fractionation and Western blot analysis (Supplementary Fig. S8H). Of note, LYAR has been implicated in the maintenance of pre-rRNA processing and mouse embryonic stem cell self-renewal (46, 47).
We identified proteins that were differentially translated upon Gm1447 repression. Only 12 genes were found less translated by more than 2-fold in nascent proteomics analyses (Fig. 4F). The SLC38A10 amino acid transporter ranked among the most repressed proteins (Fig. 4F). SLC38A10 mRNA levels were not affected (Supplementary Fig. S8I). Accordingly, SNORD127+/− cells showed reduced intracellular glutamine, glutamic acid, glycine, and aspartic acid levels (Supplementary Fig. S8J). Functionally, the SNORD127 heterozygous deletion reduced the clonogenic capacity of AML cells (Supplementary Fig. S9A and S9B). Lentiviral delivery of SNORD127 in SNORD127+/− cells specifically restored Gm1447 (Supplementary Fig. S9C) and rescued the clonogenic growth defect (Supplementary Fig. S9D). The clonogenic defect was also partially rescued by wild-type but not mutant FBL (Supplementary Fig. S9E), demonstrating the functional importance of Gm1447 for leukemia cells. In in vivo transplantation assays, SNORD127 heterozygous deletion reduced leukemic engraftment (Fig. 4G; Supplementary Fig. S9F) and prolonged the survival of recipient animals (median survival 39 vs. 49 days, P = 0.0002; Fig. 4H). Conversely, enforced expression of SNORD127 in two AML cell lines and three human primary AML cells promoted leukemic engraftment (Fig. 4I–K; Supplementary Fig. S9G and S9H). The enhanced engraftment was in line with the increased LSC pool (CD34+CD38−) in primary AML cells transduced with SNORD127 (Supplementary Fig. S9I and S9J). Finally, the increased LSC frequency upon SNORD127 overexpression was confirmed by in vivo limiting dilution transplantation (1 in 1,537 vs. 1 in 19,317, P = 0.0015; Fig. 4L; Supplementary Fig. S9K).
DISCUSSION
Our study unveils a critical role for dynamic rRNA 2′-O-Me in governing protein translation and cancer stem cell phenotypes. Recent studies illustrated distinct 2′-O-Me changes on rRNA sites in multiple cellular models and tissues (27, 28, 31), highlighting them as a potential source of ribosome diversity for gene expression regulation. But, the mechanisms and the functional outcome remain to be discovered. Here, through profiling rRNA 2′-O-Me in primary AML samples, we show that LSC-enriched FBL and its mediated rRNA 2′-O-Me modulate protein translation, affect LSC phenotypes, and contribute to their propagation.
We established the ribomethylome landscape in human AML and normal hematopoietic cells. This revealed dynamic and cell type–specific rRNA 2′-O-Me patterns. These modification patterns provide an additional layer for ribosome diversity, which, in turn, may confer a strategy to enable cells to co-opt their transcription profile and translation machinery for the formation of a cell type–specific proteome. In AMLs, the dynamic methylation mainly affects ribosome exterior sites but not evolutionarily conserved sites. Our findings complement the concept that a core set of evolutionarily conserved modifications renders the ribosome capable of efficient and accurate translation, whereas the evolutionarily extended modifications might confer regulatory functions to the ribosome (16).
Importantly, we find that LSCs can exploit this regulatory circuit to maintain enhanced self-renewal, providing novel insights into this poorly understood mechanism. FBL and its mediated rRNA 2′-O-Me, as demonstrated by Gm1447, are exquisitely regulated in primary AMLs, with close association with LSC gene expression signature and phenotype. This is consistent with findings that tight control of ribosome biogenesis and protein synthesis is essential to establish and maintain cell identity and function. Accordingly, our study indicates that FBL and rRNA 2′-O-Me preferentially regulate the translation of genes crucial for LSCs. Thus, rRNA 2′-O-Me may present one of the major epitranscriptomic programs that govern protein synthesis to confer AML stemness phenotypes. Of note, the LSCs used for rRNA 2′-O-Me characterization in this study were derived from the CD34+CD38− population. However, because FBL protein abundance was also increased in CD34+CD38+ LSCs (32), it appears that FBL-mediated rRNA 2′-O-Me represents a regulator of LSCs regardless of their immunophenotype. In addition, the rRNA 2′-O-methyltransferase FBL shows high baseline expression in mouse embryonic stem cells and is required for the maintenance of pluripotency (48). Pan-cancer analysis indicates the upregulation of FBL in multiple types of cancer (Supplementary Fig. S9L). Heterogeneity in rRNA 2′-O-Me has also recently been observed in diffuse large B-cell lymphoma and breast cancer (27, 28). Hence, the FBL-mediated dynamic ribomethylome–mediated stemness regulation, similar to those we identify within the context of AML, may therefore extend to other tumor entities, including solid cancer.
A metabolic feature of LSCs is their dependence on elevated amino acid uptake and metabolism for oxidative phosphorylation and survival (36). How LSCs selectively activate proteins involved in amino acid metabolism is not immediately apparent because cancer stem cells are characterized by low global protein translation rates (3, 6, 7). Previous studies showed that selective translation could be steered in stem cells by specific translation initiation factors, mRNA splicing, and ribosome levels (12, 49–51). More recent studies further highlight the importance of dynamic changes in RNA modifications in rewiring the genetic information decoding in stem cells (52). Our study indicates that, through increased 2′-O-Me, LSCs are able to enhance the translation of essential amino acid transporters without changes on mRNA level to meet metabolic requirements. mRNA features in 5′ untranslated region (UTR), upstream ORFs, and 3′ UTR also provide important regulatory elements for translation control during stem cell self-renewal and differentiation (12, 53, 54). We demonstrate that the codon composition of mRNA mediates the selective translation conferred by dynamic rRNA 2′-O-Me. The higher content of optimal codons in amino acid transporter genes confers an advantage to their translation promoted by rRNA 2′-O-Me modification.
Taken together, this study reveals a novel regulatory pathway in cancer pathogenesis. Epitranscriptomic modifications drive cancer stem cell phenotypes by a molecular strategy that integrates dynamic ribosome methylation, selective protein translation, and cellular metabolism.
METHODS
Plasmid Construction and Cell Culture
Human FBL cDNA were purchased from Source BioScience (Clone ID: 3504198). Point mutations of FBL were introduced by custom-designed mutant primers. C-terminal V5 epitope-tagged wild-type and mutant FBL were cloned into lentivirus expression vector PCDH-EF1α-T2A-eGFP between EcoRI/BamHI. shRNAs were cloned into pLKO.1 vector with GFP as the selection marker or doxycycline-inducible Tet-pLKO-puro vector (cat. 21915, Addgene). Targeting sequences of shRNA were GAT TTC GGA AGG AGA TGA CAA for shFBL#2, GCT GTC AGG ATT GCG AGA GAT for shFBL#4, and CGC TGA GTA CTT CGA AAT GTC for shCtrl. For SNORD127 overexpression, the cDNA of SNORD127 was amplified from the genomic DNA of HEK293 cells and was first inserted into pLKO.1-GFP vector between AgeI/EcoRI under U6 promoter. Then, the SNORD127 sequence together with the upstream U6 promoter was subcloned into the PCDH-EF1α-eGFP vector in the ClaI site with the In-Fusion kit (cat. 638909, Takara).
A lentiviral vector encoding a doxycycline-inducible Cas9 was purchased from Addgene (Lenti-iCas9-neo, cat. 85400). Paired single-guide RNAs (sgRNA) for SNORD127 deletion were cloned into the pDECKO-mCherry vector (cat. 78534, Addgene) by In-Fusion cloning. In the pDECKO vector, sgRNA1 (GTG GTC AGT GTA TTT TCA CTG) is driven by the U6 promoter and sgRNA2 (CTG ATT ACT AAG TAG AAC AG) is driven by the H1 promoter.
Cell lines were cultivated at 37°C in a humidified incubator with 5% CO2 (Thermo Scientific. 293T cells were cultured in Iscove's modified Dulbecco's medium (IMDM; cat. 12440061, Thermo Scientific) supplemented with 10% FBS (cat. S0165, BIOCHROM) and 1% penicillin–streptomycin (Sigma). Human leukemia cell lines were cultured in RPMI 1640 medium (cat. R8758, Sigma) with 20% FBS (Kasumi-1 cells) or 10% FBS (OCI-AML2 and HL60 cells), supplemented with 1% penicillin–streptomycin. For cells transduced with Tet (tetracycline)-inducible vectors, doxycycline-free serum (cat. 631105, Clontech) was used. The cell lines have been authenticated and were tested for Mycoplasma contamination.
Human Primary Samples
AML samples for RiboMethSeq and RNA-seq were collected from diagnostic bone marrow aspirations at University Hospital Dresden. The 5 AML samples with determined engrafting capacity for each CD34/CD38 population have been described previously (30, 32). Cord blood samples from normal deliveries were collected from the University Hospital in Heidelberg. Stem and progenitor cells were enriched by positive selection using CD34 UltraPure MicroBead (cat. 130-100-453, Miltenyi Biotec) according to the manufacturer's instructions. Peripheral blood samples from healthy donors were obtained from a blood bank in Heidelberg. Monocytes, granulocytes, B cells, and T cells were isolated from peripheral blood with CD19 MicroBead, CD66abce MicroBead, CD19 MicroBead, and CD3 MicroBead (Miltenyi Biotec). Total RNA was isolated from the primary cells with the miRNeasy Mini Kit (cat. 217004, Qiagen). For sample collection, written informed consent was obtained from all the patients. All experiments involving human samples were conducted in compliance with the Declaration of Helsinki, and all relevant ethical regulations were approved by the ethics committees of the medical faculties of the universities in Heidelberg and Dresden.
Lentivirus Production and Human Leukemia Cell Infection
pLKO.1 (for constitutive FBL knockdown), Tet-pLKO-puro (for inducible FBL knockdown), PCDH vector (for FBL expression), Lenti-iCas9-neo (for inducible Cas9 expression), or pDECKO-mCherry (for paired sgRNA delivery) together either with the secondary generation packaging vectors (pPLP1, pPLP2, and VSV-G) or with the third-generation packaging system (pRSV-Rev, pMDLg/pRRE, pMD2.G for PDX cells; ref. 55) were transfected into 293T cells with Turbofect reagent (Thermo Scientific). The medium containing lentivirus particles was collected 72 hours after transfection, and lentivirus particles were precipitated by centrifugation at 29,000 × g at 4°C for 2 hours. Transductions were conducted by the incubation of virus and cells in the presence of 8 μg/mL polybrene (cat. TR-1003-G, Millipore).
Mouse Experiments
NSG mice were bred and housed under specific pathogen–free conditions in the central animal facility of the Helmholtz Center Munich and the German Cancer Research Center (DKFZ). Animal experiments were approved and performed in accordance with all regulatory guidelines of the official committee (Regierung von Oberbayern and Regierung von Regierungspräsidium Karlsruhe). No statistical methods were used to estimate the sample size.
Generation of AML PDX Models.
Generation of firefly luciferase–expressing AML491 PDX cells was performed as previously described (55, 56). AML491, AML03, AML08, AML661, and AML494 cells were lentivirally transduced to express FBL wild-type, FBL mutant, SNORD127, or empty vectors. Transduced cells were enriched using flow cytometry by gating on GFP expression as previously described (57). For AML491 and AML08, the LSC CD34+CD38− and non-LSC CD34+CD38+ populations were further sorted. Then, sorted AML cells were injected into the tail vein of 10- to 12-week-old NSG mice after irradiation with a dose of 1.75 Gy. The in vivo engrafting experiments of human cell lines, including SNORD127+/+ and SNORD127+/− OCI-AML2 and Kasumi-1 cells, HL60 cells, and OCI-AML2 cells expressing empty vector or SNORD127 were performed with the same procedure as described above.
Assessment of Leukemic Engraftment.
Starting at day 14 after transplantation, tumor burden was regularly assessed using IVIS Lumina II (Caliper) with Living Image version 4.3.1 software (PerkinElmer) for the firefly luciferase–expressing AML491 PDX model. For the other PDX models, 50 μL blood was repetitively collected by tail vein or bone marrow aspiration was performed. Blood samples and bone marrow aspirates were analyzed by flow cytometry after staining for human CD45 and human CD33 as previously described (55). For advanced leukemic disease, mice were sacrificed by exposure to CO2 or cervical dislocation, and PDX cells were reisolated from murine bone marrow for further analyses. In addition, daily monitoring of mice for symptoms of disease (ruffled coat, hunched back, weakness, and reduced mobility) determined the time of sacrificing for injected animals with signs of distress.
In Vivo Limiting Dilution Transplantation Assay.
PDX cells expressing empty vector, wild-type or mutant FBL and DPX cells expressing empty vector or SNORD127 were FACS-sorted from the bone marrow of corresponding animals. To avoid bias from animals, PDX cells from 2 mice of each group were mixed for the limiting dilution transplantation assay. Female NSG mice at the age of 12 weeks were irradiated with 1.75 Gy 1 day before transplantation. Cells were injected into groups of mice at different doses for each animal. Each group contained 4 to 5 mice. Engraftment was assessed 10 weeks after transplantation, and a threshold of 0.1% human PDX cells in bone marrow was used as positive for engraftment.
FBL Rescue Experiments
Rescue of FBL knockdown with wild-type and mutant FBL was performed in Kasumi-1 cells. Kasumi-1 cells were first infected with lentivirus expressing doxycycline-inducible shCtrl and shFBL#4 (targeting 3′-UTR of endogenous FBL mRNA). The infected cells were then selected in a medium with 2 μg/mL puromycin for 10 days. The established inducible knockdown cells were further used for FBL rescue experiments. The empty vector, wild-type, or mutant FBL was delivered to inducible FBL knockdown cells by lentivirus infection. FBL knockdown was induced with 100 ng/mL doxycycline for 3 days.
Colony Formation Assay
For AML LSC populations, cells were transduced with lentivirus expressing shFBL#2, shFBL#4, or shCtrl. Forty-eight hours after transduction, GFP+ cells were FACS-sorted into methylcellulose medium MethoCult H4034 (STEMCELL Technologies) for culture. For AML PDX cells, mCherry+ PDX cells were FACS-sorted from the bone marrow of NSG recipient mice into MethoCult H4034 (STEMCELL Technologies). Each culture contained 20,000 cells in a 0.5 mL methylcellulose medium. For Kasumi-1– and OCI-AML2–derived cells, the MethoCult H4230 methylcellulose medium (STEMCELL Technologies) was used according to the manufacturer's instructions. Each culture contained 300 Kasumi-1 cells or 800 OCI-AML2 cells in 0.5 mL of methylcellulose medium in a 12-well plate. Colonies formed by AML LSCs and leukemia cell lines were evaluated and scored after 14 days in culture, and colonies formed by PDX cells were scanned after 20 days in culture.
Western Blot Analysis
Cells were washed two times with ice-cold PBS and lysed in RIPA buffer (cat. 89900, Thermo Scientific) with a proteinase inhibitor cocktail (cat. 11873580001, Roche). After incubation on ice for 10 minutes, cell lysates were centrifuged at 12,000 × g for 10 minutes. Supernatants were collected and heated at 70°C for 10 minutes after adding NuPAGE LDS Sample Buffer (Cat. NP0007, Thermo Scientific). Protein lysates were resolved on NuPAGE 4% to 12% Bis-Tris gels (cat. NP0321BOX, Thermo Scientific) with NuPAGE MES SDS running buffer (cat. NP0002, Thermo Scientific) and blotted on Amersham Protran Western blotting membranes, nitrocellulose (cat. GE10600001, Sigma). Membranes were blocked overnight at 4°C in PBS containing 0.5% (v/v) Tween-20 with 5% milk powder. Primary antibodies against anti-FBL (cat. ab5821, Abcam), anti-V5 tag (cat. ab9116, Abcam), anti–β-Actin (cat. ab6276, Abcam), anti-PRPF39 (cat. GTX104949, Genetex), anti-SLC38A10 (cat. ab121830, Abcam), anti-LAT1/SLC7A5 (cat. 5347S, Cell Signaling Technology), anti-ASCT2/SLC1A5 (cat. 8057S, Cell Signaling Technology), anti-Glut1/SLC2A1 (cat. 12939S, Cell Signaling Technology), anti-SNAT17SLC38A1 (cat. 36057S, Cell Signaling Technology), anti-LYAR (cat. ab182138, Abcam), anti-RPL23A (cat. Ab157110, Abcam), anti–4E-BP1 (cat. 9644T, Cell Signaling Technology), anti–phospho-4E-BP1 (Thr37/46; cat. 2588T, Cell Signaling Technology), anti–p70 S6 kinase (cat. 2708T, Cell Signaling Technology), and anti–phospho-p70 S6 kinase (Thr389; cat. 9234T, Cell Signaling Technology) were incubated 2 hours at room temperature in blocking solution. Secondary antibodies (horseradish peroxidase-linked anti-rabbit-IgG or anti-mouse-IgG antibody, Dianova) were incubated for 90 minutes at room temperature in a blocking solution. Immunocomplexes were detected using an ECL Prime reagent (cat. RPN2232, GE HealthCare) and acquired with an Amersham Imager 600 system (GE HealthCare).
Quantitative RT-PCR
The total RNA was extracted with the miRNeasy Mini Kit (Qiagen) combined with RNase-free DNase (Qiagen) treatment according to the manufacturer's instructions. cDNA of mRNA was synthesized using a SuperScript IV Reverse Transcriptase (cat. 18090050, Thermo Scientific) according to the manufacturer's protocol. Real-time PCR was performed on a CFX96 Touch System (Bio-Rad) using ssoAdvanced Universal SYBR Green SuperMix (cat. 1725272, Bio-Rad). The following primers were used: SLC7A5 (forward: 5′-GTG TAC GTG CTG ACC AAC CT; reverse: 5′-TGA CGC CCA GGT GAT AGT TC), SLC1A5 (forward: 5′-TTA CTC TTT GCC CGC CTT GG; reverse: 5′-TAG GGG TTT TTG CGG GTG AA), SLC2A1 (forward: 5′-CTG GCA TCA ACG CTG TCT TC; reverse: 5′-AAC AGC GAC ACG ACA GTG AA), SLC38A1 (forward: 5′-TCC CTG CAT TGT TCC AGA GC; reverse: 5′-TGA CGG GTG GCA AAC AAA TG), SLC1A5 (forward: 5′-CTC GAG ACT CCA AGG GGC T; reverse: 5′-CCG GGA ACC GCA GTA GC), Actin (forward: 5′-ACA GAG CCT CGC CTT TGC; reverse: 5′-CGC GGC GAT ATC ATC ATC CA), and GAPDH (forward: 5′-ACT GCC AAC GTG TCA GTG G; reverse: 5′-CAC CCT GTT GCT GTA GCC A).
cDNA for snoRNA were synthesized with the Mir-X miRNA First-Strand Synthesis Kit (cat. 638313, Clontech). SnoRNAs were detected with gene-specific forward primer and universal reverse primer. The forward primer for SNORD127 is 5′-GGC AAC TGT GAT GAA AGA TTT GGT.
Flow Cytometry
LSC and non-LSC fractions were FACS-sorted from primary AML samples. Frozen vials of AML samples were thawed in a 37°C water bath in IMDM with 10% FBS and 100 μg/mL DNase I (Sigma). After filtering through the 40-μm cell strainer, cells were stained with the following antibodies: Alexa Fluor 488 anti-human CD34 (cat. 343518, BioLegend); PE anti-human CD38 antibody (cat. 303506, BioLegend); and a lineage cocktail consisting of CD4-APC (cat. 300514, BioLegend), APC anti-human CD20 (cat. 302310, BioLegend), APC anti-human CD8a (cat. 301014, BioLegend), and APC anti-human CD19 (cat. 302212, BioLegend). Before sorting, 1 μL of 10 mg/mL DAPI (cat. 422801, BioLegend) was added to the cell suspension. Cells were sorted into four quadrants according to CD34 and CD38 expression within the DAPI-negative and lineage-negative gate.
To check the purity of isolated healthy hematopoietic cells, the following antibodies were used: CD34-BV421 (cat. 562577, BD Biosciences), CD14-PE (cat. 301850, BioLegend), CD15-PE-Cy7 (cat. 560827, BD Biosciences), CD3-BV421 (cat. 300434, BioLegend), and CD19-PE-Cy7 (cat. 302216, BioLegend).
For PDX cells, the total bone marrow of NSG recipient mice was stained with APC anti-human CD34 (cat. 340441, BD) and PE anti-human GPR56 (cat. 358203, BioLegend). Before flow cytometry analysis, 1 μL of DAPI (10 μg/mL) was added to the cell suspension to gate out the dead cells.
RiboMethSeq
RiboMethSeq was performed as previously described with minor modifications (31). Briefly, 100 ng total RNA was hydrolyzed in an alkaline buffer (50 mmol/L bicarbonate, pH 9.2) at 95°C for 12 minutes to achieve an average fragment size of about 30 nucleotides. The rRNA fragment was purified by ethanol precipitation using 3 M Na-OAc pH 5.2 and glycogen as a carrier. RNA fragments were proceeded to end repair with 5 U of Antarctic Phosphatase (NEB) for 30 minutes at 37°C for 3′-end dephosphorylated. After heat inactivation of the phosphatase, the 5′-end of RNA fragments were phosphorylated using T4 PNK and 1 mmol/L ATP for 1 hour at 37°C. Libraries were prepared using the NEBNext Small RNA library prep kit, followed by single-end sequencing on an Illumina NextSeq 500. Reads trimming was performed with cutadapt to remove the adapter sequence, and reads below 15 nucleotides were discarded. The filtered reads were mapped to rDNA sequences with the bowtie2 reference sequence file containing rDNA sequences of U18, U28, and U5.8S. Methylation was calculated by ScoreC as described before (31) based on the reads on 5′-end of the fragment. Annotation of modification sites, as well as rRNA sequence, was downloaded from https://www-snorna.biotoul.fr.
Ribosome Profiling Library Preparation
Cells were grown in culture medium (RPMI 1640 with 20% FBS) with 100 μg/mL cycloheximide (CHX; cat. 4859, Sigma) at 37°C for 10 minutes. Cells were collected and washed with PBS (with 100 μg/mL CHX) twice. Afterward, cell lysis was performed on ice for 10 minutes in lysis buffer [20 mmol/L Tris-HCl pH 7.4 (cat. AM9850G and cat. AM9855G, Thermo Scientific), 150 mmol/L NaCl (cat. AM9760G, Thermo Scientific), 5 mmol/L MgCl2 (cat. AM9530G, Thermo Scientific), 1 mmol/L dithiothreitol (DTT; cat. 43816, Sigma), 1% Triton X-100 (cat. T8787, Sigma), and 25 U/mL of Turbo DNase I (cat. AM2238, Thermo Fisher Scientific)] containing 100 μg/mL of CHX. Lysates were further sheared by passing through a 26-G needle four times. Nuclei and debris were removed by centrifugation at 12,000 × g for 10 minutes, and the RNA concentration of the cell lysate was measured with a Qubit kit (Thermo Fisher Scientific). To generate ribosome-protected fragments (RPF), 20 μg of total RNA were diluted with polysome buffer (20 mmol/L Tris-Cl pH 7.4, 150 mmol/L NaCl, 5 mmol/L MgCl2, 1 mmol/L DTT, and 100 μg/mL of CHX) to 200 μL and treated with 7.5 μL of RNase I (100 U/μL, cat. AM2294, Invitrogen) for 45 minutes at room temperature with gentle mixing. Digestion was blocked by adding 10 μL of SUPERase·In RNase inhibitor. The reaction was cleaned up with MicroSpin S-400 Columns (cat. GE27-5140-01, Sigma), followed by purification using the RNA Clean and Concentrator-25 kit (cat. R1017, Zymo Research). The eluted RNA was pelleted by ethanol precipitation. Size selection of RPF with a length of 26 to 34 nucleotides was performed on 15% polyacrylamide TBE-urea gel (cat. EC6885BOX, Thermo Fisher Scientific). End repair of RPFs was performed with 10 U T4 PNK treatment (cat. M0201S, NEB) at 37°C for 1 hour. The libraries were prepared with NEBNext Small RNA Library Prep Set for Illumina. The library DNA was purified with AMPure beads (cat. 63881, Beckman Coulter) at the ratio of 1.8:1. The library size was measured on Bioanalyzer (Agilent). All samples were multiplexed and sequenced on an Illumina NextSeq 500.
Ribosome Profiling Data Processing and Analysis
Reads were processed by removing 5′ adapters, discarding reads shorter than 20 nucleotides, and trimming the first nucleotide of the remaining reads. Reads mapping on human rRNAs (obtained from https://www.ncbi.nlm.nih.gov/nuccore/555853) and tRNAs (obtained from GtRNAdb at http://gtrnadb.ucsc.edu/genomes/eukaryota/Hsapi19/hg19-tRNAs.fa) were removed. The remaining reads were mapped onto the human transcriptome. Antisense and duplicate reads were removed. All alignments were performed with Bowtie 2 (v.2.2.6) using the default settings.
Data analysis was performed with the riboWaltz R package, RiboToolkit, and diricore (58–60). Briefly, BAM files were loaded into the riboWaltz R package. Reads were regarded as duplicates and removed by the function “duplicates_filter” when they were mapped on the same transcript and shared both the 5′ extremity and the 3′ extremity. After annotation, the function “psite offset” was executed to the identification of the P-site position within the reads. Then “region_psite” was used to calculate the percentage of P-sites falling in the three transcript regions (5′ UTR, coding sequences, and 3′ UTR). “rlength_distr” was performed to calculate the distribution of read lengths. “frame_psite” was used to calculate the percentage of P-sites falling in the three possible translation reading frames. The function “metaprofile_psite” was used to generate the metaprofiles based on the P-site position and to visualize trinucleotide periodicity along coding sequences. After the above quality control steps, “codon_usage_psite” was used to compare the codon usage index between shCtrl and shFBL samples. “codon_coverage” was used to compute the read footprints or P-sites mapping on each triplet of annotated coding sequences and UTRs. The changes in codon usage identified by riboWaltz were further confirmed with RiboToolkit and diricore. To calculate the pause score, we normalized the read count at each codon of a gene by dividing it by the mean read count at all codons for the gene.
RNA-seq
A total amount of 1 μg RNA per sample was used as input material for the RNA sample preparations. Sequencing libraries were generated using NEBNext Ultra RNA Library Prep Kit for lllumina (NEB) following the manufacturer's recommendations. The generated libraries were applied to the Illumina platform for paired-end sequencing. Raw reads of FASTQ format were first processed through fastp. Clean reads were obtained by removing reads containing adapter and poly-N sequences and reads with low quality from raw data. Paired-end clean reads were mapped to the human genome using HISAT2 software. FeatureCounts was used to count the read numbers mapped of each gene, including known and novel genes. Then fragments per kilobase of exon per million mapped fragments (FPKM) of each gene were calculated based on the length of the gene and read count mapped to this gene. Differential expression analysis between two conditions/groups was performed using the DESeq2 R package. The resulting P values were adjusted using the Benjamini and Hochberg approach for controlling the FDR. Genes with an adjusted P < 0.05 found by DESeq2 were assigned as differentially expressed.
Nascent Proteomics and Data Analysis
Nascent proteomics was performed as previously described (20). The cells were washed twice with PBS and cultured in amino acid-depletion medium [RPMI formulation without methionine, arginine, and lysine (Gibco), with 20% dialyzed FBS (Gibco) and 200 mg/mL L-proline] for 45 minutes. The cells were pelleted and cultured in intermediate (depletion medium supplemented with 2H4-lysine, 13C6-arginine, and AHA) or heavy (depletion medium supplemented with 15N213C6-lysine, 15N413C6-arginine, and AHA) labeling medium for 3 hours. Cells were pelleted and lysed in lysis buffer (8 M urea, 0.3 M HEPES, 0.75 M NaCl, and 6.2% CHAPS detergent) including protease inhibitor cocktail without EDTA (Roche) and sonicated on ice using a probe sonicator (Branson). Equal amounts of precleared heavy and intermediate SILAC-labeled lysates were merged.
The nascent proteins were enriched with alkyne agarose resin slurry (Click-iT Protein Enrichment Kit, Thermo Fisher Scientific). On-beads digestion was performed by overnight digestion with 5 μg of trypsin at 37°C in digestion buffer (100 mmol/L ammonium bicarbonate in ULCMS water) in a shaking incubator. The samples were centrifuged for 5 minutes (1,000 × g, RT), and the supernatant with digested peptides was collected. Peptides were desalted using an Oasis PRiME HKB mElution Plate according to the manufacturer's instructions. The samples were vacuum-dried and dissolved in 0.1% formic acid. High pH reversed-phase fractionation was done on an Infinity 1260 LC system (Agilent) with Gemini 3 μm C18 column (Phenomonex). Separation occurred along a 60-minute linear gradient of buffer A (20 mmol/L ammonium formate pH 10) and buffer B (100% acetonitrile). Samples were dried and dissolved in 0.1% formic acid.
MS was performed on Q-Exactive HF or Orbitrap Fusion mass spectrophotometers connected to an EASY-nLC 1,200 (Thermo Scientific). Peptides were separated with a C18 ultra-performance liquid chromatography (UPLC) column (Thermo Scientific). The Q-Exactive HF mass spectrometer was operated in data-dependent mode, acquiring one survey MS scan in the Orbitrap followed by up to 20 fragmentation scans (TOP20) of the most abundant ions analyzed in the MS by HCD fragmentation. Orbitrap fusion was operated using Orbitrap as an MS1 analyzer and Ion Trap as an MS2 analyzer.
The raw MS data were processed with MaxQuant. A minimum of 2 peptides were required for protein identification, and the FDR at peptide and protein levels was set to 0.01. R/Bioconductor was used to calculate the percentages of SILAC MS/MS spectra as a percentage of the total MS/MS per protein group. SILAC ratios below the threshold of 0.1 were removed. The SILAC ratios were inverted to account for SILAC label swap between replicate experiments. Subsequently, ratios were log2 transformed and corrected by the median of the log2 ratios to obtain filtered and normalized ratios for further analysis. Proteins quantified in both replicates were subjected to differential expression analysis by the Limma R package. Multiple test correction was applied according to the Benjamini–Hochberg procedure. Adjusted P values equal to or lower than 0.05 pointed to statistically significant protein expression change.
SNORD127+/− Generation
SNORD127 heterozygous knockout (SNORD127+/−) lines were generated either by transducing inducible Cas9-expressing cells with lentiviral constructs expressing paired sgRNA specific to SNORD127 locus (for Kasumi-1) or by transfecting Cas9 protein and synthetic paired sgRNA (Synthego; for OCI-AML2). To generate doxycycline-inducible Cas9-expressing cells, Kasumi-1 cells were transduced with Lenti-iCas9-neo and selected with 1 mg/mL G418 for 2 weeks. Cells were treated with 100 ng/mL doxycycline to induce Cas9-P2A-GFP expression, and single cells with high Cas9-P2A-GFP expression were selected and expanded as parent cells to make SNORD127 knockout. Kasumi-1 iCas9 cells were infected with pDECKO lentivirus expressing paired sgRNA and FACS-sorted for mCherry-positive cells. To induce SNORD127 knockout in the selected population, Cas9 expression was induced by treating cells with 100 ng/mL doxycycline for 4 days. Single clones were picked and expanded to screen for SNORD127 deletion.
For SNORD127 knockout in OCI-AML2 cells, Cas9-Cy3 protein (cat. CP06-100, PNA Bio) and synthetic SNOR127 paired sgRNA (Synthego) were electroporated into OCI-AML2 cells with the Neon transfection system (Thermo Scientific). Briefly, 1 μg of sgRNA was incubated with 1 μg Cas9-Cy3 protein for 15 minutes at room temperature in 2 μL PBS without Ca2+ and Mg2+. The cells were washed with PBS (without Ca2+ and Mg2+) and resuspended in resuspension buffer R to a density of 2 × 107 cells/mL. A total of 13 μL cell suspension was added to the premixed 2 μL of the Cas9 protein/sgRNA RNP. A volume of 10 μL cell/RNP mixture was used for electroporation on the Neon system. Afterward, cells were cultured in 500 μL medium (RPMI 1640 with 10% FBS) without antibiotics. Forty-eight hours later, single cells were sorted into a 96-well plate from Cy3-positive populations. Genomic DNA was extracted from the single clones, and the knockout clones were screened by genotyping PCR with forward primer CTG TCA AAC TAG CCC GGC ATC and reverse primer GGC AAC GCC TAT GCT TTT GTT T. All the knockouts were further confirmed by Sanger sequencing. Single clones without any mutations in SNORD127 loci were used as wild-type control clones.
Gene Set Enrichment Analysis
Gene set enrichment analysis (GSEA) was conducted using the GSEA desktop application from the Broad Institute. The LSC_17 gene set contains a list of 17 genes generated in (61). The LSC_Up gene set was composed of 43 highly expressed genes (reads per kilobase of exon per million reads mapped > 1), which showed at least a 2-fold increase in LSChi versus LSClo AMLs (62). The HEMATOPOIETIC_CELL_LINEAGE and BROWN_MYELOID_CELL_DEVELOPMENT_UP gene set was downloaded from the GSEA Molecular Signatures Database (MSigDB). For FBL mRNA expression–associated signatures (Supplementary Fig. S1B-S1D), we used our mRNA-seq data from AML patient samples and calculated the Pearson correlation between FBL mRNA with each of the other transcripts. The transcripts were then ranked from high to low correlation score (from positive to negative correlation, shown “corr” and “anticorr” in figures). The preranked gene list was applied to GSEA with the indicated gene sets. For rRNA 2′-O-Me–associated signatures, patients with AML were divided into two groups (high and low) according to the total 2′-O-Me score on the relevant clusters or sites. We then compared the mRNA-seq data of these two groups to calculate fold change for each gene with their mean value in each group. The genes were ranked based on fold change and applied to GSEA with the indicated gene set.
Gene Ontology Analysis
Gene ontology analysis was performed with ShinyGo (http://bioinformatics.sdstate.edu/go/).
Metabolite Screening by GC/MS
Semitargeted metabolite screening was performed with GC/MS analysis. Briefly, frozen pellets were extracted in 180 μL of 100% MeOH for 15 minutes at 70°C with vigorous shaking. As an internal standard, 10 μL ribitol (0.2 mg/mL) was added to each sample. After the addition of 100 μL chloroform, samples were shaken at 37°C for 5 minutes. To separate polar and organic phases, 200 μL water was added and samples were centrifuged for 10 minutes at 11,000 × g. For derivatization, 300 μL of the polar (upper) phase was transferred to a fresh tube and dried in a speed-vac (vacuum concentrator) without heating. GC/MS-QP2010 Plus (Shimadzu) fitted with a Zebron ZB 5MS column (Phenomenex) was used for GC/MS analysis. The GC was operated with an injection temperature of 230°C, and 2 μL sample was injected with split mode. The MS was operated with ion source and interface temperatures of 250°C, a solvent cut time of 6.3 minutes, and a scan range (m/z) of 40 to 1,000 with an event time of 0.3 seconds. The “GCMS solution” software (Shimadzu) was used for data processing. Metabolite set enrichment analysis was performed with MetaboAnalyst5.0.
Targeted Metabolites via UPLC
Target analysis for amino acids, thiols, and metabolites in the tricarboxylic acid cycle was conducted by UPLC. Free amino acids and thiols were extracted from frozen cell pellets with 0.3 mL of 0.1 M HCl in an ultrasonic ice bath for 10 minutes. The resulting extracts were centrifuged twice for 10 minutes at 4°C and 16,400 × g to remove cell debris. Amino acids were derivatized with AccQ-Tag reagent (Waters) and determined as described in Weger and colleagues (63). Total glutathione was quantified by reducing disulfides with DTT followed by thiol derivatization with the fluorescent dye monobromobimane (Thiolyte, Calbiochem). For quantification of GSSG, free thiols were first blocked by NEM, followed by DTT reduction and monobromobimane derivatization. GSH equivalents were calculated by subtracting GSSG from total glutathione levels. Derivatization was performed as described by Wirtz and colleagues (64). UPLC-FLR analysis was carried out using an Acquity H-class UPLC system. Separation was achieved with a binary gradient of buffer A (100 mmol/L potassium acetate pH 5.3) and solvent B (acetonitrile). The column (Acquity BEH Shield RP18 column, Waters) was maintained at 45°C, and the sample temperature was kept constant at 14°C. Monobromobimane conjugates were detected by fluorescence at 480 nm after excitation at 380 nm and quantified using ultrapure standards (Sigma). Determination of organic acids was adapted from Uran and colleagues (65). In brief, cell pellets were extracted in 0.2 mL ice-cold methanol with sonication on ice. A volume of 50 μL extract was mixed with 25 μL 140 mmol/L 3-nitrophenylhydrazine hydrochloride (Sigma-Aldrich), 25 μL methanol, and 100 μL 50 mmol/L ethyl-3-(3-dimethylaminopropyl) carbodiimide hydrochloride (Sigma-Aldrich) and incubated for 20 minutes at 60°C. Separation was carried out on the above-described UPLC system coupled to a QDa mass detector (Waters) using an Acquity HSS T3 column (Waters). Separation of derivates was achieved by increasing the concentration of 0.1% formic acid in acetonitrile (B) in 0.1% formic acid in water (A) at 0.55 mL/minutes. Mass signals for the following compounds were detected in single ion record (SIR) mode using negative detector polarity and 0.8 kV capillary voltage: lactate (224.3 m/z; 25 V CV), malate (403.3 m/z; 25 V CV), succinate (387.3 m/z; 25 CV), fumarate (385.3 m/z; 30 V), citrate (443.3 m/z; 10 V), pyruvate (357.3 m/z; 15 V), and ketoglutarate (550.2 m/z; 25 CV). Data acquisition and processing were performed with the Empower3 software suite (Waters).
Ribosome Purification by Sucrose Gradient
Purification of 80S ribosomes of the SNORD127+/+ and SNORD127+/− cells was conducted by sucrose gradient centrifugation. In brief, cells were resuspended in lysis buffer (20 mmol/L HEPES pH 7.4, 100 mmol/L KOAc, 7.5 mmol/L Mg(OAc)2, 1 mmol/L DTT, 0.5% NP-40) with protease inhibitor cocktail (cat.11873580001, Roche). Cell lysates were incubated on ice for 10 minutes and pushed through a 26-G needle. Cell debris was discarded by centrifugation at 12,000 × g for 15 minutes at 4°C, and the cytoplasmic fraction without any lipids was loaded onto 1M sucrose cushion (50 mmol/L HEPES pH 7.4, 100 mmol/L KOAc, 7.5 mmol/L Mg(OAc)2, 5 mmol/L β-mercaptoethanol, 1 M sucrose, 0.1% NP-40). The ribosome-enriched pellet was obtained by centrifugation at 100,000 rpm at 4°C for 45 minutes in a TLA110 rotor. The pellet was resuspended in ribosome storage buffer (20 mmol/L HEPES pH 7.4, 100 mmol/L KOAc, 5 mmol/L Mg(OAc)2, 2 mmol/L DTT). Ribosome particles were purified on a 10%-40% sucrose gradient using an SW40 rotor at 16,700 rpm for 18 hours at 4°C. Fractions containing 80S ribosomes were collected into chilled 2 mL tubes on ice. The 80S ribosomes were concentrated at 2,000 rpm in 15 mL Pierce Protein Concentrator, 100K MWCO (cat. 88533, Thermo Scientific) to 500 μL, diluted with ribosome storage buffer to 5 mL and concentrated again to 500 μL. The concentration–dilution was repeated 4 times to remove sucrose traces. The ribosomal pellet was resuspended in a ribosome storage buffer and stored at −80°C until further use.
Cryo-EM Structure Analysis of Ribosome
Electron Microscopy and Image Processing.
After ribosome purification by sucrose gradient, a final concentration of 0.05% NP-40 was added to the samples. Purified 3.5 μL of the sample was applied to precoated (2 nm) R3/3 holey-carbon–supported copper grids (Quantifoil), blotted for 2 to 3 seconds at 4°C, and plunge-frozen in liquid ethane using an FEI Vitrobot Mark IV (FEI Company). Data were collected on a Titan Krios operated at 300 keV. All data were collected with a pixel size of 1.059 Å/pixel and within a defocus range of −0.8 to −2.5 μm using a K2 Summit direct electron detector under low-dose conditions with a total dose of 44 e-/Å2. Original image stacks were dose weighted, aligned, summed, and drift-corrected using MotionCor2 (66). Contrast-transfer function (CTF) parameters and resolutions were estimated for each micrograph using CTFFIND4 and GCTF, respectively (67, 68). Micrographs with an estimated resolution better than 5 Å and astigmatism below 5% were manually screened for contamination or carbon rupture.
Reference-free particle picking was carried out using Gautomatch. In total, 582,101 particles were picked from 12,443 good micrographs of the SNORD127+/+ cell dataset and 538,797 particles were picked from 7,533 good micrographs of the SNORD127+/− cell dataset. After 2D classification, 188,471 particles and 394,983 particles of the SNORD127+/+ and SNORD127+/− cell dataset, respectively, were selected for 3D classification and 3D refinement in Relion 3.1 (69). The final class that contained the most particle number and display higher resolution was picked because we aimed for a high-resolution map to see the 2′-O methylation. To improve the overall resolution and local resolution, CTF refinement and multibody refinement were applied for the picked classes. In general, the 80S ribosome was divided into three bodies: the 60S ribosome, the 40S ribosome body, and the 40S ribosome head. To better illustrate the map, a local resolution filter was carried out for all three bodies.
Model Building and Refinement.
The human 80S ribosome structure [Protein Data Bank (PDB) ID: 6ZMI] was used as the initial reference to do rigid body fit, followed by manual adjustment in Coot (such as removing the NSP1 protein; refs. 70, 71). In the case of the model from SNORD127+/− cell, E-site tRNA and LYAR were removed. Because most of the region has sufficient resolution to distinguish 2′-O methylation, all the modified bases were manually built. In the center of the structure, magnesium and water molecules were visible; thus, we also manually built them. The final models were real-space refined with secondary structure restraints using the PHENIX suite (72). Final model evaluation was performed with MolProbity (73). Maps and models were visualized, and figures were created with ChimeraX (74).
Statistical Analysis
No statistical method was used to predetermine the sample size. The experiments were not randomized. Animals with the same gender and age were used in paired groups. The investigators were not blinded to allocation during experiments and outcome assessment. To test the statistical significance between samples from two different groups, two-tailed Student t tests were used. When comparing samples from the same patient, paired t tests were applied. Data are represented as means ± SD unless otherwise indicated. Sample sizes and significance are shown in the corresponding figure legends. The relative importance of individual covariates in multivariate logistic regression models was estimated by examining the Pearson chi-squared test. The survival curves were constructed according to Kaplan–Meier with the log-rank test. Bioinformatics statistical analysis was performed using R.
Data Availability
The sequencing data have been deposited in the Gene Expression Omnibus SuperSeries under accession numbers GSE184721, GSE184722, GSE184724, GSE184727, and GSE185489. Ribosome cryo-EM structures from SNORD127+/+ and SNORD127+/− leukemia cells have been deposited in the Electron Microscopy Data Bank (EMD) and PDB under the accession number EMD_33329, PDB7XNX, EMD_33330, and PDB7XNY. The ribosome structure to visualize the distribution of 2′-O-Me sites is from PDB 4UG0. Previously published human FBL structure is from PDB 5GIO. The MS proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD029527.
Code Availability
All codes and scripts used in the paper are available upon reasonable request.
Supplementary Material
Supplementary Figure S1 shows enrichment of FBL in LSC, correlation of FBL with LSC signature and rRNA 2'-O-Me in healthy hematopoietic cells. Supplementary Figure S2 shows the association of rRNA 2’-O-Me with LSC and hematopoietic differentiation signatures. Supplementary Figure S3 shows strategy for LSC sorting and distribution of LSC methylation sites on ribosomes. Supplementary Figure S4 shows the effect of FBL knockdown on rRNA 2’-O-methylation and in vitro proliferation of leukemia cells. Supplementary Figure S5 show the effect of FBL overexpression on in vivo engraftment of primary AML cells. Supplementary Figure S6 shows the effect of FBL knockdown on nascent proteome and metabolism of AML cells. Supplementary Figure S7 shows the ribosome footprinting analysis after FBL knockdown. Supplementary Figure S8 shows the association of Gm1447 with LSC signature and the effect of Gm1447 supression on cellular amino acid levels. Supplementary Figure S9 shows regulatory effect of Gm1447 on in vivo leukemic self-renewal.
Supplementary table S1 shows all rRNA 2’-O-methylation sites identified in human AMLs. Supplementary table S2 shows dynamic rRNA 2’-O-methylation sites in each DyMeC cluster. Supplementary table S3 shows the location of rRNA 2’-O-methylation static sites in ribosome regions. Supplementary table S4 shows proteins with altered translation after FBL knockdown.
Acknowledgments
We thank Stefanie Hofmann and Dr. Volker Eckstein for flow cytometry cell sorting. We thank the Metabolomics Core Technology Platform of the Excellence Cluster CellNetworks for support with metabolite quantification. We thank K. Reifenberg, P. Prückl, M. Durst, A. Rathgeb, and animal caretakers of the DKFZ Central Animal Laboratory for excellent animal welfare and husbandry; members of the DKFZ Core Facility Flow Cytometry for technical assistance; and Dr. Darja Karpova and Prof. Halvard B. Bönig (IHBT, Frankfurt am Main) for providing the AML patient samples. This work was supported by Deutsche Forschungsgemeinschaft (DFG) ZH 831/1-1 (F. Zhou), DFG YU 406/1-1 (H. Yun), DFG MU 1328/15-1, MU 1328/9-2, MU 1328/18-1, MU 1328/23-1 (C. Müller-Tidow), DFG RA 3166/2-1 (S. Raffel), DFG SPP2036, FOR2674, and SFB873 (A. Trumpp); Deutsche Krebshilfe 70113908, 111286, 70112974 (C. Müller-Tidow), and 70115215 (F. Zhou); Deutsche José-Carreras-Leukämie-Stiftung DJCLS 04R/2022, DJCLS 22R/2017, and DJCLS R 13/04 (C. Müller-Tidow); Sander Stiftung Nr. 2017.004.1 and 2021.145.1 (C. Müller-Tidow); BMBF 01ZX1911D (S. Raffel); the European Research Council Advanced Grant SHATTER-AML 101055270 (A. Trumpp); DKTK joint funding project “RiskY-AML” (A. Trumpp); and the Dietmar Hopp Foundation (A. Trumpp).
The publication costs of this article were defrayed in part by the payment of publication fees. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Footnotes
Note: Supplementary data for this article are available at Cancer Discovery Online (http://cancerdiscovery.aacrjournals.org/).
Authors’ Disclosures
T. Oellerich reports grants and personal fees from Merck KGaA, grants from GILEAD, and personal fees from Roche and Kronos Bio outside the submitted work. C. Röllig reports personal fees from Amgen, Astellas, Bristol Myers Squibb, Roche, and Servier and grants and personal fees from AbbVie, Pfizer, and Novartis outside the submitted work. M. Bornhäuser reports personal fees from Jazz Pharmaceuticals, MSD, and Gilead outside the submitted work. C. Thiede reports other support from AgenDix GmbH, grants and personal fees from Novartis, and personal fees from Jazz, Astellas, and Janssen outside the submitted work. M. Frye reports grants from Merck outside the submitted work. C. Müller-Tidow reports other support from Janssen-Cilag GmbH and Pfizer Pharma GmbH, and personal fees from Wilhelm Sander Foundation, Exeter Pharma Consultancy, MorphoSys AG, and The Kay Kendall Leukaemia Fund outside the submitted work. No disclosures were reported by the other authors.
Authors’ Contributions
F. Zhou: Conceptualization, data curation, formal analysis, supervision, validation, investigation, visualization, methodology, writing–original draft. N. Aroua: Formal analysis, investigation, methodology. Y. Liu: Investigation, visualization, methodology. C. Rohde: Software, formal analysis, investigation, visualization, methodology. J. Cheng: Investigation, visualization, methodology. A.-K. Wirth: Data curation, investigation, methodology. D. Fijalkowska: Data curation, investigation, methodology. S. Göllner: Writing–review and editing. M. Lotze: Investigation. H. Yun: Formal analysis, methodology. X. Yu: Investigation. C. Pabst: Resources. T. Sauer: Methodology. T. Oellerich: Investigation. H. Serve: Writing–review and editing. C. Röllig: Resources. M. Bornhäuser: Resources. C. Thiede: Resources. C. Baldus: Writing–review and editing. M. Frye: Writing–review and editing. S. Raffel: Resources, data curation, investigation. J. Krijgsveld: Data curation, investigation, methodology, writing–review and editing. I. Jeremias: Investigation, methodology, writing–review and editing. R. Beckmann: Investigation, writing–review and editing. A. Trumpp: Conceptualization, data curation, supervision, investigation, methodology, writing–review and editing. C. Müller-Tidow: Conceptualization, resources, supervision, funding acquisition, investigation, writing–original draft, project administration.
References
- 1. Bonnet D, Dick JE. Human acute myeloid leukemia is organized as a hierarchy that originates from a primitive hematopoietic cell. Nat Med 1997;3:730–7. [DOI] [PubMed] [Google Scholar]
- 2. Chua BA, Van Der Werf I, Jamieson C, Signer RAJ. Post-transcriptional regulation of homeostatic, stressed, and malignant stem cells. Cell Stem Cell 2020;26:138–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Saba JA, Liakath-Ali K, Green R, Watt FM. Translational control of stem cell function. Nat Rev Mol Cell Biol 2021;22:671–90. [DOI] [PubMed] [Google Scholar]
- 4. Schwanhausser B, Busse D, Li N, Dittmar G, Schuchhardt J, Wolf J, et al. Global quantification of mammalian gene expression control. Nature 2011;473:337–42. [DOI] [PubMed] [Google Scholar]
- 5. Khan Z, Ford MJ, Cusanovich DA, Mitrano A, Pritchard JK, Gilad Y. Primate transcript and protein expression levels evolve under compensatory selection pressures. Science 2013;342:1100–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Signer RA, Magee JA, Salic A, Morrison SJ. Haematopoietic stem cells require a highly regulated protein synthesis rate. Nature 2014;509:49–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Blanco S, Bandiera R, Popis M, Hussain S, Lombard P, Aleksic J, et al. Stem cell function and stress response are controlled by protein synthesis. Nature 2016;534:335–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Sanchez CG, Teixeira FK, Czech B, Preall JB, Zamparini AL, Seifert JR, et al. Regulation of ribosome biogenesis and protein synthesis controls germline stem cell differentiation. Cell Stem Cell 2016;18:276–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Zismanov V, Chichkov V, Colangelo V, Jamet S, Wang S, Syme A, et al. Phosphorylation of eIF2alpha is a translational control mechanism regulating muscle stem cell quiescence and self-renewal. Cell Stem Cell 2016;18:79–90. [DOI] [PubMed] [Google Scholar]
- 10. Harding HP, Novoa I, Zhang Y, Zeng H, Wek R, Schapira M, et al. Regulated translation initiation controls stress-induced gene expression in mammalian cells. Mol Cell 2000;6:1099–108. [DOI] [PubMed] [Google Scholar]
- 11. Teslaa T, Teitell MA. Pluripotent stem cell energy metabolism: an update. EMBO J 2015;34:138–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Khajuria RK, Munschauer M, Ulirsch JC, Fiorini C, Ludwig LS, McFarland SK, et al. Ribosome levels selectively regulate translation and lineage commitment in human hematopoiesis. Cell 2018;173:90–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ruggero D, Grisendi S, Piazza F, Rego E, Mari F, Rao PH, et al. Dyskeratosis congenita and cancer in mice deficient in ribosomal RNA modification. Science 2003;299:259–62. [DOI] [PubMed] [Google Scholar]
- 14. Decatur WA, Fournier MJ. rRNA modifications and ribosome function. Trends Biochem Sci 2002;27:344–51. [DOI] [PubMed] [Google Scholar]
- 15. Polikanov YS, Melnikov SV, Soll D, Steitz TA. Structural insights into the role of rRNA modifications in protein synthesis and ribosome assembly. Nat Struct Mol Biol 2015;22:342–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Natchiar SK, Myasnikov AG, Kratzat H, Hazemann I, Klaholz BP. Visualization of chemical modifications in the human 80S ribosome structure. Nature 2017;551:472–7. [DOI] [PubMed] [Google Scholar]
- 17. Burakovsky DE, Prokhorova IV, Sergiev PV, Milon P, Sergeeva OV, Bogdanov AA, et al. Impact of methylations of m2G966/m5C967 in 16S rRNA on bacterial fitness and translation initiation. Nucleic Acids Res 2012;40:7885–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Sloan KE, Warda AS, Sharma S, Entian KD, Lafontaine DLJ, Bohnsack MT. Tuning the ribosome: The influence of rRNA modification on eukaryotic ribosome biogenesis and function. RNA biology 2017;14:1138–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Sharma S, Lafontaine DLJ. ‘ View from a bridge’: a new perspective on eukaryotic rRNA base modification. Trends Biochem Sci 2015;40:560–75. [DOI] [PubMed] [Google Scholar]
- 20. Kiss-Laszlo Z, Henry Y, Bachellerie JP, Caizergues-Ferrer M, Kiss T. Site-specific ribose methylation of preribosomal RNA: a novel function for small nucleolar RNAs. Cell 1996;85:1077–88. [DOI] [PubMed] [Google Scholar]
- 21. Cavaille J, Nicoloso M, Bachellerie JP. Targeted ribose methylation of RNA in vivo directed by tailored antisense RNA guides. Nature 1996;383:732–5. [DOI] [PubMed] [Google Scholar]
- 22. Zhou F, Liu Y, Rohde C, Pauli C, Gerloff D, Kohn M, et al. AML1-ETO requires enhanced C/D box snoRNA/RNP formation to induce self-renewal and leukaemia. Nat Cell Biol 2017;19:844–55. [DOI] [PubMed] [Google Scholar]
- 23. Pauli C, Liu Y, Rohde C, Cui C, Fijalkowska D, Gerloff D, et al. Site-specific methylation of 18S ribosomal RNA by SNORD42A is required for acute myeloid leukemia cell proliferation. Blood 2020;135:2059–70. [DOI] [PubMed] [Google Scholar]
- 24. Jansson MD, Hafner SJ, Altinel K, Tehler D, Krogh N, Jakobsen E, et al. Regulation of translation by site-specific ribosomal RNA methylation. Nat Struct Mol Biol 2021;28:889–99. [DOI] [PubMed] [Google Scholar]
- 25. Krogh N, Jansson MD, Hafner SJ, Tehler D, Birkedal U, Christensen-Dalsgaard M, et al. Profiling of 2’-O-Me in human rRNA reveals a subset of fractionally modified positions and provides evidence for ribosome heterogeneity. Nucleic Acids Res 2016;44:7884–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Erales J, Marchand V, Panthu B, Gillot S, Belin S, Ghayad SE, et al. Evidence for rRNA 2’-O-methylation plasticity: control of intrinsic translational capabilities of human ribosomes. Proc Natl Acad Sci U S A 2017;114:12934–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Krogh N, Asmar F, Come C, Munch-Petersen HF, Gronbaek K, Nielsen H. Profiling of ribose methylations in ribosomal RNA from diffuse large B-cell lymphoma patients for evaluation of ribosomes as drug targets. NAR Cancer 2020;2:zcaa035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Marcel V, Kielbassa J, Marchand V, Natchiar KS, Paraqindes H, Nguyen Van Long F, et al. Ribosomal RNA 2'O-methylation as a novel layer of inter-tumour heterogeneity in breast cancer. NAR cancer 2020;2:zcaa036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Dick JE. Stem cell concepts renew cancer research. Blood 2008;112:4793–807. [DOI] [PubMed] [Google Scholar]
- 30. Raffel S, Falcone M, Kneisel N, Hansson J, Wang W, Lutz C, et al. BCAT1 restricts alphaKG levels in AML stem cells leading to IDHmut-like DNA hypermethylation. Nature 2017;551:384–8. [DOI] [PubMed] [Google Scholar]
- 31. Marchand V, Blanloeil-Oillo F, Helm M, Motorin Y. Illumina-based RiboMethSeq approach for mapping of 2’-O-Me residues in RNA. Nucleic Acids Res 2016;44:e135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Raffel S, Klimmeck D, Falcone M, Demir A, Pouya A, Zeisberger P, et al. Quantitative proteomics reveals specific metabolic features of acute myeloid leukemia stem cells. Blood 2020;136:1507–19. [DOI] [PubMed] [Google Scholar]
- 33. Khatter H, Myasnikov AG, Natchiar SK, Klaholz BP. Structure of the human 80S ribosome. Nature 2015;520:640–5. [DOI] [PubMed] [Google Scholar]
- 34. Yang Z, Lin J, Ye K. Box C/D guide RNAs recognize a maximum of 10 nt of substrates. Proc Natl Acad Sci U S A 2016;113:10878–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Eichelbaum K, Winter M, Berriel Diaz M, Herzig S, Krijgsveld J. Selective enrichment of newly synthesized proteins for quantitative secretome analysis. Nat Biotechnol 2012;30:984–90. [DOI] [PubMed] [Google Scholar]
- 36. Jones CL, Stevens BM, D'Alessandro A, Reisz JA, Culp-Hill R, Nemkov T, et al. Inhibition of amino acid metabolism selectively targets human leukemia stem cells. Cancer Cell 2018;34:724–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Jones CL, Stevens BM, D'Alessandro A, Culp-Hill R, Reisz JA, Pei S, et al. Cysteine depletion targets leukemia stem cells through inhibition of electron transport complex II. Blood 2019;134:389–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Presnyak V, Alhusaini N, Chen YH, Martin S, Morris N, Kline N, et al. Codon optimality is a major determinant of mRNA stability. Cell 2015;160:1111–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Hanson G, Coller J. Codon optimality, bias and usage in translation and mRNA decay. Nat Rev Mol Cell Biol 2018;19:20–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Wu Q, Medina SG, Kushawah G, DeVore ML, Castellano LA, Hand JM, et al. Translation affects mRNA stability in a codon-dependent manner in human cells. eLife 2019;8:e45396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Najafabadi HS, Goodarzi H, Salavati R. Universal function-specificity of codon usage. Nucleic Acids Res 2009;37:7014–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Akashi H. Translational selection and yeast proteome evolution. Genetics 2003;164:1291–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Frumkin I, Lajoie MJ, Gregg CJ, Hornung G, Church GM, Pilpel Y. Codon usage of highly expressed genes affects proteome-wide translation efficiency. Proc Natl Acad Sci U S A 2018;115:E4940–E9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Yang JH, Zhang XC, Huang ZP, Zhou H, Huang MB, Zhang S, et al. snoSeeker: an advanced computational package for screening of guide and orphan snoRNA genes in the human genome. Nucleic Acids Res 2006;34:5112–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Maden BE, Salim M. The methylated nucleotide sequences in HELA cell ribosomal RNA and its precursors. J Mol Biol 1974;88:133–52. [DOI] [PubMed] [Google Scholar]
- 46. Miyazawa N, Yoshikawa H, Magae S, Ishikawa H, Izumikawa K, Terukina G, et al. Human cell growth regulator Ly-1 antibody reactive homologue accelerates processing of preribosomal RNA. Genes Cells 2014;19:273–86. [DOI] [PubMed] [Google Scholar]
- 47. Li H, Wang B, Yang A, Lu R, Wang W, Zhou Y, et al. Ly-1 antibody reactive clone is an important nucleolar protein for control of self-renewal and differentiation in embryonic stem cells. Stem Cells 2009;27:1244–54. [DOI] [PubMed] [Google Scholar]
- 48. Watanabe-Susaki K, Takada H, Enomoto K, Miwata K, Ishimine H, Intoh A, et al. Biosynthesis of ribosomal RNA in nucleoli regulates pluripotency and differentiation ability of pluripotent stem cells. Stem Cells 2014;32:3099–111. [DOI] [PubMed] [Google Scholar]
- 49. Thoreen CC, Chantranupong L, Keys HR, Wang T, Gray NS, Sabatini DM. A unifying model for mTORC1-mediated regulation of mRNA translation. Nature 2012;485:109–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Hsieh AC, Liu Y, Edlind MP, Ingolia NT, Janes MR, Sher A, et al. The translational landscape of mTOR signalling steers cancer initiation and metastasis. Nature 2012;485:55–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Tahmasebi S, Jafarnejad SM, Tam IS, Gonatopoulos-Pournatzis T, Matta-Camacho E, Tsukumo Y, et al. Control of embryonic stem cell self-renewal and differentiation via coordinated alternative splicing and translation of YY2. Proc Nat Acad Sci USA 2016;113:12360–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Roundtree IA, Evans ME, Pan T, He C. Dynamic RNA modifications in gene expression regulation. Cell 2017;169:1187–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Blair JD, Hockemeyer D, Doudna JA, Bateup HS, Floor SN. Widespread translational remodeling during human neuronal differentiation. Cell Rep 2017;21:2005–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Ingolia NT, Lareau LF, Weissman JS. Ribosome profiling of mouse embryonic stem cells reveals the complexity and dynamics of mammalian proteomes. Cell 2011;147:789–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Vick B, Rothenberg M, Sandhofer N, Carlet M, Finkenzeller C, Krupka C, et al. An advanced preclinical mouse model for acute myeloid leukemia using patients’ cells of various genetic subgroups and in vivo bioluminescence imaging. PLoS One 2015;10:e0120925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Ebinger S, Zeller C, Carlet M, Senft D, Bagnoli JW, Liu WH, et al. Plasticity in growth behavior of patients’ acute myeloid leukemia stem cells growing in mice. Haematologica 2020;105:2855–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Terziyska N, Castro Alves C, Groiss V, Schneider K, Farkasova K, Ogris M, et al. In vivo imaging enables high-resolution preclinical trials on patients’ leukemia cells growing in mice. PLoS One 2012;7:e52798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Lauria F, Tebaldi T, Bernabo P, Groen EJN, Gillingwater TH, Viero G. riboWaltz.: Optimization of ribosome P-site positioning in ribosome profiling data. PLoS Comput Biol 2018;14:e1006169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Loayza-Puch F, Rooijers K, Buil LC, Zijlstra J, Oude Vrielink JF, Lopes R, et al. Tumour-specific proline vulnerability uncovered by differential ribosome codon reading. Nature 2016;530:490–4. [DOI] [PubMed] [Google Scholar]
- 60. Liu Q, Shvarts T, Sliz P, Gregory RI. RiboToolkit: an integrated platform for analysis and annotation of ribosome profiling data to decode mRNA translation at codon resolution. Nucleic Acids Res 2020;48:W218–W29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Ng SW, Mitchell A, Kennedy JA, Chen WC, McLeod J, Ibrahimova N, et al. A 17-gene stemness score for rapid determination of risk in acute leukaemia. Nature 2016;540:433–7. [DOI] [PubMed] [Google Scholar]
- 62. Pabst C, Bergeron A, Lavallee VP, Yeh J, Gendron P, Norddahl GL, et al. GPR56 identifies primary human acute myeloid leukemia cells with high repopulating potential in vivo. Blood 2016;127:2018–27. [DOI] [PubMed] [Google Scholar]
- 63. Weger BD, Weger M, Gorling B, Schink A, Gobet C, Keime C, et al. Extensive regulation of diurnal transcription and metabolism by glucocorticoids. PLos Genet 2016;12:e1006512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Wirtz M, Droux M, Hell R. O-acetylserine (thiol) lyase: an enigmatic enzyme of plant cysteine biosynthesis revisited in Arabidopsis thaliana. J Exp Bot 2004;55:1785–98. [DOI] [PubMed] [Google Scholar]
- 65. Uran S, Landmark KE, Hjellum G, Skotland T. Quantification of 13C pyruvate and 13C lactate in dog blood by reversed-phase liquid chromatography-electrospray ionization mass spectrometry after derivatization with 3-nitrophenylhydrazine. J Pharm Biomed Anal 2007;44:947–54. [DOI] [PubMed] [Google Scholar]
- 66. Zheng SQ, Palovcak E, Armache JP, Verba KA, Cheng Y, Agard DA. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat Methods 2017;14:331–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Rohou A, Grigorieff N. CTFFIND4: Fast and accurate defocus estimation from electron micrographs. J Struct Biol 2015;192:216–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Zhang Ki. Gctf: Real-time CTF determination and correction. J Struct Biol 2016;193:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Zivanov J, Nakane T, Forsberg BO, Kimanius D, Hagen WJ, Lindahl E, et al. New tools for automated high-resolution cryo-EM structure determination in RELION-3. eLife 2018;7:e42166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Thoms M, Buschauer R, Ameismeier M, Koepke L, Denk T, Hirschenberger M, et al. Structural basis for translational shutdown and immune evasion by the Nsp1 protein of SARS-CoV-2. Science 2020;369:1249–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr 2004;60:2126–32. [DOI] [PubMed] [Google Scholar]
- 72. Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr 2010;66:213–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Chen VB, Arendall WB 3rd, Headd JJ, Keedy DA, Immormino RM, Kapral GJ, et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr D Biol Crystallogr 2010;66:12–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Goddard TD, Huang CC, Meng EC, Pettersen EF, Couch GS, Morris JH, et al. UCSF ChimeraX: Meeting modern challenges in visualization and analysis. Protein Sci 2018;27:14–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure S1 shows enrichment of FBL in LSC, correlation of FBL with LSC signature and rRNA 2'-O-Me in healthy hematopoietic cells. Supplementary Figure S2 shows the association of rRNA 2’-O-Me with LSC and hematopoietic differentiation signatures. Supplementary Figure S3 shows strategy for LSC sorting and distribution of LSC methylation sites on ribosomes. Supplementary Figure S4 shows the effect of FBL knockdown on rRNA 2’-O-methylation and in vitro proliferation of leukemia cells. Supplementary Figure S5 show the effect of FBL overexpression on in vivo engraftment of primary AML cells. Supplementary Figure S6 shows the effect of FBL knockdown on nascent proteome and metabolism of AML cells. Supplementary Figure S7 shows the ribosome footprinting analysis after FBL knockdown. Supplementary Figure S8 shows the association of Gm1447 with LSC signature and the effect of Gm1447 supression on cellular amino acid levels. Supplementary Figure S9 shows regulatory effect of Gm1447 on in vivo leukemic self-renewal.
Supplementary table S1 shows all rRNA 2’-O-methylation sites identified in human AMLs. Supplementary table S2 shows dynamic rRNA 2’-O-methylation sites in each DyMeC cluster. Supplementary table S3 shows the location of rRNA 2’-O-methylation static sites in ribosome regions. Supplementary table S4 shows proteins with altered translation after FBL knockdown.
Data Availability Statement
The sequencing data have been deposited in the Gene Expression Omnibus SuperSeries under accession numbers GSE184721, GSE184722, GSE184724, GSE184727, and GSE185489. Ribosome cryo-EM structures from SNORD127+/+ and SNORD127+/− leukemia cells have been deposited in the Electron Microscopy Data Bank (EMD) and PDB under the accession number EMD_33329, PDB7XNX, EMD_33330, and PDB7XNY. The ribosome structure to visualize the distribution of 2′-O-Me sites is from PDB 4UG0. Previously published human FBL structure is from PDB 5GIO. The MS proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD029527.