Synthetic lethality and mass spectrometry were used to identify novel metabolic vulnerabilities in cancers with IDH1, but not IDH2, mutations that are targetable in the setting of ivosidenib resistance.
Abstract
Isocitrate dehydrogenase 1 and 2 (IDH) are mutated in multiple cancers and drive production of (R)-2-hydroxyglutarate (2HG). We identified a lipid synthesis enzyme [acetyl CoA carboxylase 1 (ACC1)] as a synthetic lethal target in mutant IDH1 (mIDH1), but not mIDH2, cancers. Here, we analyzed the metabolome of primary acute myeloid leukemia (AML) blasts and identified an mIDH1-specific reduction in fatty acids. mIDH1 also induced a switch to b-oxidation indicating reprogramming of metabolism toward a reliance on fatty acids. Compared with mIDH2, mIDH1 AML displayed depletion of NADPH with defective reductive carboxylation that was not rescued by the mIDH1-specific inhibitor ivosidenib. In xenograft models, a lipid-free diet markedly slowed the growth of mIDH1 AML, but not healthy CD34+ hematopoietic stem/progenitor cells or mIDH2 AML. Genetic and pharmacologic targeting of ACC1 resulted in the growth inhibition of mIDH1 cancers not reversible by ivosidenib. Critically, the pharmacologic targeting of ACC1 improved the sensitivity of mIDH1 AML to venetoclax.
Significance:
Oncogenic mutations in both IDH1 and IDH2 produce 2-hydroxyglutarate and are generally considered equivalent in terms of pathogenesis and targeting. Using comprehensive metabolomic analysis, we demonstrate unexpected metabolic differences in fatty acid metabolism between mutant IDH1 and IDH2 in patient samples with targetable metabolic interventions.
See related commentary by Robinson and Levine, p. 266.
This article is highlighted in the In This Issue feature, p. 247
INTRODUCTION
The metabolic enzymes isocitrate dehydrogenase 1 (IDH1) and 2 (IDH2) are mutated in numerous cancers, including acute myeloid leukemia (AML), myelodysplastic syndromes (MDS), glioma, T-cell acute lymphoblastic leukemia, chondrosarcoma, and cholangiocarcinoma (1–3). IDH1 and IDH2 enzymes are present in the cytosol and mitochondria, respectively, and are mutated at high frequency in several of these cancer types (1, 2, 4). Although mutations in both genes occur with similar frequency in AML, IDH1 mutations (mIDH1) are much more common than IDH2 mutations (mIDH2) in glioma, oligodendrocytoma, and chondrosarcoma for unknown reasons (2, 5). Notably, although mIDH1 and mIDH2 do not occur in the same cell, both mutations confer a neomorphic activity for the NADPH-dependent reduction of α-ketoglutarate (αKG) to an oncometabolite (R)-2-hydroxyglutarate (2HG) that competitively inhibits αKG-dependent enzymes involved in epigenetic regulation (3). The mutation-induced altered patterns of DNA methylation and histone modification alter cellular differentiation and contribute to disease pathogenesis (6). Given its central role, 2HG has been used as a disease biomarker for multiple mIDH1/2 cancers.
Small-molecule mIDH1 and mIDH2 inhibitors have been approved for the treatment of mIDH AML, and additional clinical trials are ongoing for other cancers (7–9). In relapsed or refractory AML, these inhibitors, ivosidenib (AG-120) for mIDH1 and enasidenib (AG-221) for mIDH2, produce clinical responses as single agents, but not in all patients (10, 11). Notably, 2HG levels were undetectable in nearly all treated patients, even in those without significant clinical responses (10, 11), which suggests that mIDH may contribute to disease pathogenesis through other mechanisms. Although both mIDH1 and mIDH2 aberrantly produce 2HG, there are potential differences in substrate availability and reaction flux in the cytosol versus mitochondria. The levels of αKG and NADPH may differ between these compartments because of competing forward and reverse reactions, mitochondrial heteroplasmy, and/or differential solute transport (12–14). Thus, other metabolic perturbations in mIDH cancers that are independent of 2HG could be oncogenic. In addition to the canonical forward reaction, which catalyzes the oxidative decarboxylation of isocitrate to αKG and regenerates NADPH (defective in mIDH1 cancers), both wild-type enzymes can also catalyze the reverse anabolic carboxylation reaction, which utilizes αKG and carbon dioxide to produce isocitrate. Cytoplasmic isocitrate can then be shunted into NADPH-dependent pathways used for de novo lipid synthesis that only occurs in the cytoplasm. This reverse reaction, which also consumes rather than regenerating NADPH, appears to be critical for cancer cells under conditions of hypoxia or mitochondrial dysfunction when oxidative phosphorylation linked to the Krebs cycle cannot proceed efficiently. The role of native IDH1 enzyme in reductive carboxylation utilizing glutamine-derived αKG in hypoxia appears to be critical, and gene knockout studies indicate that its loss is incompletely compensated for by IDH2, suggesting mIDH1 cancers may have targetable vulnerabilities that are distinct from IDH2 (14–19). In vitro experiments with the recombinant enzyme have shown mIDH1 cannot efficiently perform reductive carboxylation compared with wild-type (20).
Further nuanced metabolic differences between mIDH1 and mIDH2 have been demonstrated. We recently discovered mutation-specific mutual exclusivity between hypomorphic electron transport chain complex I variants in patients with mIDH1 AML, but not mIDH2, based on germline sequencing (21). Notably, mIDH1 blasts, but not mIDH2 blasts, were highly sensitive to the selective complex I inhibitor IACS-10759, and mIDH1 clones with high variant allele frequencies (VAF) were never found in the same clones as complex I variants. We also recently reported VAF-dependent clinical differences in lactate dehydrogenase in patients with AML with mIDH1 versus mIDH2 (22).
Cancer cells have a fundamental requirement for cellular building blocks, including lipids, to support their continuous proliferation, and metabolic reprogramming for this purpose is a well-established hallmark of cancer (23). Whereas normal cells obtain the bulk of their lipid requirement directly from the circulation, many cancer cells synthesize a significant proportion of lipid de novo for reasons that are not entirely clear (24). De novo fatty acid biosynthesis in mammals is mediated by two related enzymes—ACC1 (the product of ACACA or ACC1 gene) and ACC2 (ACACB or ACC2)—which catalyze the ATP-dependent carboxylation of acetyl-coenzyme A (acetyl CoA) to form malonyl CoA. ACC1 is a dimeric cytoplasmic enzyme that controls fatty acid synthesis and is inhibited by activation of AMP-activated protein kinase (AMPK) in energy-deficient states, whereas ACC2 is tethered to the mitochondrial outer membrane for localized malonyl CoA production, which regulates fatty acid oxidation. The rate-limiting step in de novo lipid synthesis is accepted to be the conversion of acetyl CoA into malonyl CoA by ACC1 in the cytoplasm (25), and in sterol-depleted conditions, IDH1 gene transcription is upregulated by sterol-regulatory element binding proteins, suggesting IDH1 regulates lipogenesis (26). Recently, we identified a synthetic lethal interaction between mIDH1 and ACC1 across human cancers (27), suggesting that de novo lipid synthesis may be altered, constituting a potential therapeutic vulnerability in mIDH1 cancers. Consistent with this, isotope labeling studies using HT-1080 sarcoma cells have demonstrated that an IDH1 R132C mutation can limit lipogenesis by inadequately compensated NADPH consumption (28). Specifically, cytosolic NADPH labeled with deuterium tracer [3–2H], but not NADH (labeled with [4–2H]), was consumed in the production of 2HG in a similar proportion to that utilized for fatty acid production, suggesting both reactions are competing for a limited pool that is incompletely compensated by wild-type IDH2 (28).
Innovative mass spectrometry techniques are currently enabling mutation-specific and cancer type–specific metabolic signatures to be identified, including small polar metabolites and amino acids, with results not always predictable by transcriptomics (29). However, a major challenge is to identify mutation-specific lipidomic alterations, characterizing a large and diverse set of lipid species, spanning a wide range of concentrations in genotyped primary samples (30). Here, we use liquid chromatography–coupled mass spectrometry to characterize lipid profiles in primary AML blasts. Strikingly, we found that nonpolar metabolites in mIDH1 AML are substantially perturbed compared with mIDH2, with specific depletion of single-chain monoglycerides and lysophospholipids in mIDH1, despite both groups having elevated levels of 2HG. mIDH1 cells struggled to grow in lipid-depleted conditions, suggesting an absolute reliance on both the production (via ACC1) and consumption of fatty acids for proliferation. Expression of mIDH1 resulted in decreased reductive carboxylation and NADPH but increased ACC1-dependent fatty acid b-oxidation, consistent with a requirement of IDH1 for ACC1. In human xenograft models, a lipid-free diet markedly slowed the growth of mIDH1, but not mIDH2, AML and prolonged the survival of mice bearing mIDH1, but not wild-type IDH1, sarcomas with ACC1 targeting. Finally, small-molecule ACC1/2 inhibitors or AMPK activators selectively reduced the viability of mIDH1 cancer cells and increased therapeutic targeting in venetoclax-resistant AML in vivo. Our data highlight key metabolic differences between mIDH1 and mIDH2 cancers and pinpoint mIDH1 in having a greater dependence on fatty acid metabolism. These results have implications for dietary and therapeutic approaches for patients with mIDH1 cancers.
RESULTS
ACC1 Is a Synthetic Lethal Metabolic Target for Mutant IDH1, but Not Mutant IDH2
We previously developed a computational algorithm [Mining Synthetic Lethals (MiSL)] that mines pan-cancer human primary tumor data to identify mutation-specific synthetic lethal partners for specific cancers (27). MiSL identified ACACA (ACC1) as a gene that was not deleted when IDH1 mutation was present, and ACC1 mRNA was overexpressed in mIDH1 AML (Fig. 1A and B). ACC1 is a cytoplasmic enzyme controlling fatty acid synthesis downstream of IDH1 but upstream of fatty acid synthase (FASN), which uses malonyl CoA and acetyl CoA to form fatty acids; subsequent elongation and desaturation reactions require fatty acid elongases (ELOVL1–7) and desaturases (SCD, SCD5) to produce a milieu of monounsaturated and saturated fatty acids (Supplementary Fig. S1A). Interestingly, MiSL did not predict ACC1 as a synthetic lethal target in mIDH2 AML. To investigate further, we observed that, unlike mIDH1 AML, ACC1 expression was not consistently increased in mIDH2 AML despite a similar frequency of occurrence in this cancer (Fig. 1B).
Figure 1.
MiSL predicts ACC1 as a metabolic dependency for mutant IDH1, but not mutant IDH2. A, Schematic showing MiSL algorithm and prediction of ACC1 (ACACA) as a potential synthetic lethal partner of mutant IDH1 across pan-cancer. CNA, copy-number alteration. B,ACC1 mRNA expression comparing mutated IDH1 and IDH2 AML vs. wild-type in The Cancer Genome Atlas based on RNA sequencing data. Differences in expression were compared with Student t test with P values as shown. C, Validation of ACC1-specific hairpins on protein expression by Western blot (left) with quantification (right). Experiment was performed 3 independent times. *, P < 0.05. RFU, relative fluorescence unit. D, ACC1-specific hairpins with mRNA quantified by TaqMan qPCR compared with nontargeting shRNA. Experiment was performed in triplicate 2 independent times; ***, P < 0.001; **, P < 0.01; Student t test. E, Knockdown of ACC1 using shRNA#1 (left) or shRNA#2 (right) in THP-1 cells expressing IDH1 R132H (mIDH1) or IDH2 R140Q (mIDH2) under dox-induced promoter in lipid-depleted media. The number of viable RFP+GFP+ double-positive cells at day 10 was enumerated relative to fluorescent counting beads. RFP tracks integrated ACC1-specific shRNA hairpin; eGFP tracks mutant protein after dox induction. This experiment was performed 3 times with a representative graph shown. Bars represent standard deviation. ***, P < 0.001; **, P < 0.01; *, P < 0.05; Student t test performed on 4 sorted biological replicates for each transduction group. F, Similar experiment using wild-type IDH cells. ACC1 was knocked down using shRNA#1 or shRNA#2 in THP-1 cells expressing IDH1 or IDH2 wild-type proteins under dox-induced promoter in lipid-depleted media. As in D, the number of viable RFP+GFP+ double-positive cells at day 10 was enumerated relative to fluorescent counting beads. RFP tracks integrated ACC1-specific shRNA hairpin; eGFP tracks wild-type protein after doxycycline induction. This experiment was performed 3 times with a representative shown. Bars represent standard deviation. NS, nonsignificant; NT, nontargeting shRNA.
To experimentally investigate this computational prediction, we established isogenic IDH1 R132H and IDH2 R140Q doxycycline (dox)-inducible THP-1 AML cell lines, which exhibited similar levels of inducible protein expression (tracked with eGFP and self-cleaving T2A peptide) and 2HG production (Supplementary Fig. S1B–S1D; refs. 27, 31). We validated that 2HG production by IDH1 R132H and IDH2 R140Q lines could be fully inhibited by ivosidenib or enasidenib, respectively (Supplementary Fig. S1E–S1G). We have previously validated that dox did not change the growth characteristics of THP-1 in low serum media (27). Next, we transduced these cell lines with lentivirus encoding nontargeting short hairpin RNA (shRNA) or two independent shRNAs targeting ACC1 (27) and examined their growth in lipid-depleted media. Both shRNAs decreased ACC1 protein (Fig. 1C) and ACC1 transcript (Fig. 1D) compared with nontargeting hairpin, with shRNA #1 having the largest effect. As predicted by MiSL, IDH1 R132H-mutant cells exhibited a marked growth defect in the presence of ACC1 knockdown versus nontargeting control [negative fold change (FC) 7.9, P < 0.0001], whereas the ACC1 knockdown did not cause an equivalent decrease in IDH2 R140Q (negative FC 2.4, P < 0.0001; Fig. 1E). Notably, the ACC1 knockdown had negligible effects on inducible IDH1/2 wild-type expressing cells (Fig. 1F).
Metabolomic Profiling Reveals That mIDH1 Is Specifically Associated with Perturbed Fatty Acid and Phospholipid Biosynthesis
Because ACC1 catalyzes the rate-limiting step for de novo fatty acid synthesis, metabolic profiling was performed in engineered THP-1 lines and primary human AML blasts cultured in lipid-depleted conditions to force the cells to utilize de novo lipid biosynthesis pathways (Supplementary Fig. S2A). We incorporated several mass spectrometry techniques in parallel to maximize metabolomic information including (i) derivatized-labeling of polar carboxylic acid metabolites with o-benzyl hydroxylamine; (ii) derivatized-labeling of polar amine metabolites with dansylation (32); and (iii) nonpolar organic solvent combined with iterative MS/MS data acquisition combined with probability density annotation (Lipid Annotator; ref. 33). Organic solvents enabled sufficient nonpolar metabolite peak resolution from less than 1 × 106 cells, as validated by internal standards (Supplementary Fig. S2B). In positive ion mode, the lipid class with the most distinct measurable metabolites from THP-1 AML cells were the triacylglycerides, followed by various phosphatidylcholine species (Supplementary Fig. S2C and S2D). Fewer distinct lipids were identified in negative ion mode, in which ether-phosphatidylethanolamine species were predominant (Supplementary Fig. S2C and S2D).
Analysis of the nonpolar metabolites in the inducible THP-1 cells revealed that mIDH1 cells (+dox) had a specific metabolic profile, which was distinct from that of mIDH2, wild-type IDH1, and wild-type IDH2 cells, as observed by principal component analysis (Fig. 2A). Compared with endogenous IDH1 wild-type (−dox), mIDH1 cells exhibited a striking decrease in nonpolar monoglyceride [MG(18:0), FC 0.3, dox versus no dox, adjusted P = 0.006; MG(14:0), FC 0.4, P = 0.01] and diglyceride [DG(20:2/20:3), FC 0.1, P = 0.0001] but also phosphatidylcholine [PC(O-16:0/4:0), FC 0.2, P = 1.3 × 10−5; PC(O-12:0/O-2:0), FC 0.3, P = 9.6 × 10−7] and phosphatidylethanolamine species [PE(10:0/11:0), FC 0.1, P = 3.6 × 10−6; PE(12:0/11:0), FC 0.1, P = 4.0 × 10−9; PE(15:0,16:0), FC 0.4, P = 0.0006]. These data are summarized as a heat map of metabolite FC in Fig. 2B. These changes were not observed to the same extent in IDH2 R140Q-mutant cells (Fig. 2B). Although there were notable decreases in specific phosphatidylcholine species in mIDH2[PC(20:0/0:0), FC 0.4, P = 3.2 × 10−5] and monoglycerides [MG(18:0), FC 0.3, P = 0.0009], the total number of distinct and statistically significant decreased nonpolar lipids was only 20 compared with 61 in mIDH1. Thus, despite abundant 2HG production by both mIDH1 and mIDH2, a greater perturbation of multiple lipid classes occurred in mIDH1 cells.
Figure 2.
IDH1-mutant AML associated with decreased single-chain fatty acid metabolites. A, Principal component analysis of extracted nonpolar metabolites identified by mass spectrometry in negative ion mode from THP-1 cells cultured in low serum media following induction of mIDH1 R132H, mIDH2 R140Q, wild-type IDH1, or wild-type IDH2. B, Heat map showing differentially abundant nonpolar lipid species globally decreased in IDH1 R132H THP-1 cells (+dox) compared with IDH2 R140Q (+dox) vs. no-dox controls. C, Schematic showing metabolomics protocol for extracting nonpolar lipid species using organic solvent and derivatization of polar metabolites with O-benzylhydroxylamine and dansyl-tags from primary cell AML extracts. Putative metabolite identifications based on mass/charge ratio and retention time were determined from the Human Metabolome Database, Lipid Annotator software, and a validated in-house lipidomics fragmentation library. HMDB, Human Metabolome Database. D, Heat map showing clustering of primary AML samples based on differentially abundant metabolites (all metabolites, both polar and nonpolar). mIDH1 samples are shown in red. CB, cord blood. E, Volcano plot highlighting significant differentially abundant metabolites in healthy IDH1 wild-type CD34+ progenitors vs. mIDH1 AML. Red dashed line indicates a nonadjusted P value threshold of 0.05. F, Volcano plot highlighting significant differentially abundant metabolites in 6 × mIDH1 vs. 5 × mIDH2 AML samples. Dashed line indicates a nonadjusted P value threshold of 0.05. G, Representative examples of distinct lipid metabolites with decreased abundance in IDH1-mutant vs. IDH2-mutant AML and wild-type AML; x-axis shows metabolite abundance. Bars represent standard error of independent samples. All P values represent comparison with wild-type. LPE, lysophosphatidylethanolamine; MG, monoacylglycerolipids; NS, nonsignificant; WT, wild-type.
We next analyzed primary human genotyped purified AML blasts (immunophenotype CD45midCD33+CD3−CD19−, excluding T cells, mature monocytes, and B cells) with high VAFs of IDH1 or IDH2 mutations and compared these with purified CD34+ cells from cord blood of healthy donors as a consistent negative control and IDH wild-type AML (Fig. 2C; Supplementary Table S1). Strikingly, unsupervised clustering of differentially abundant metabolites, including both polar and nonpolar species, distinguished mIDH1 samples as a distinct metabolic cluster that differed from both mIDH2 AML and normal CD34+ progenitor cells (Fig. 2D). In contrast, analysis of only polar metabolites, which includes 2HG and Krebs cycle substrates, could not distinguish mIDH1 from mIDH2, both of which clustered away from CD34+ cells (Supplementary Fig. S2E). Overall, healthy CD34+ cells had more abundant nonpolar metabolites than either mIDH1 (Fig. 2E) or mIDH2 or wild-type AML (Supplementary Fig. S2F and S2G). Direct comparison of mIDH1 and wild-type AML (n = 6) yielded few statistically significant differences, partly due to increased variability in the wild-type AML group (Supplementary Fig. S2H), with similar results comparing mIDH2 to wild-type AML (Supplementary Fig. S2I).
A direct comparison of the metabolites between mIDH1 and mIDH2 patient samples revealed that there were consistent log-fold decreases in key classes of lipid metabolites in the mIDH1 samples, most notably the single-chain fatty acids: monoacylglycerides and lysophospholipids (Fig. 2F and G). Significantly, these metabolites were not decreased in wild-type AML (Fig. 2G).
Mutant IDH1 Causes Defective Reductive Carboxylation Supplying Carbon to ACC1
These data demonstrate mIDH1-specific differences in nonpolar fatty acid metabolites. We therefore examined whether this was due to a defect in carbon supply induced by mIDH1. Glucose-derived glycerol-3-phosphate is an important precursor for lipid synthesis. To exclude a deficit in glycerol-3-phosphate flux in mIDH1 cells, we traced the metabolism of labeled 13C[1,2] glucose over 16 hours in THP-1 cells, comparing IDH1 wild-type, IDH1 R132C, IDH2 wild-type, and IDH2 R140Q. Significantly, this pathway was highly active in mIDH1 compared with mIDH2 cells, with a high percentage of the M2 glyercol-3-phosphate isotopolog (two out of three carbons labeled) detectable, indicating carbon supply through glucose is not limiting in mIDH1 (Fig. 3A). We also noted no difference in unlabeled M0 isotopolog between mIDH1 and wild-type IDH1 (Supplementary Fig. S3A), with high levels of incorporation into ribose (both M1 and M2) indicative of robust import into the oxidative and nonoxidative branches of the pentose phosphate pathway, respectively (Supplementary Fig. S3B and S3C). Virtually no incorporation of labeled glucose was observed into the tricarboxylic acid cycle (TCA) cycle. Consistent with this, snapshot TCA metabolite abundance studies showed both mIDH1 and mIDH2 AML samples had decreased steady-state levels of isocitrate, fumarate, and αKG TCA intermediates compared with normal CD34+ cells, indicating reduced TCA cycle activity associated with both mIDH1 and mIDH2 as described in other cancers (Supplementary Fig. S3D; refs. 29, 34). As expected, only mIDH1 and mIDH2 AML showed detectable 2HG, with slightly higher levels in mIDH1. Notably, pyruvate levels were higher in mIDH1 AML compared with mIDH2 AML (P = 0.002; Supplementary Fig. S3D), but none of these differences could account for the perturbation in nonpolar metabolites.
Figure 3.
IDH1 mutation is linked to defective reductive carboxylation, increased fatty acid consumption, and decreased NADPH. A, Schematic and graph of flux studies showing the percentage of labeled M2 glycerol-3-phosphate derived from 13C[1,2] labeled glucose (2 of 6 carbons as heavy isotope) across THP-1 cells induced to express mIDH1 vs. IDH1 wild-type compared with mIDH2 vs. IDH2 wild-type. Glucose was added to media in normoxia over 13 hours. Schematic indicates M2 isotopolog of glycerol-3-phosphate de novo synthesis directly from glycolysis utilizing labeled glucose rather than the oxidative pentose phosphate pathway (M1 isotopolog). A two-tailed unpaired Student t test was used to compare differences between groups. This experiment was performed with 6 cell pellets for each sample blinded and randomized on each LC-MS run. DHAP, dihydoxyacetone phosphate; G-6-P, glucose-6-phosphate. B, Decreased reductive carboxylation of mIDH1 compared with IDH1 wild-type and mIDH2 expressed in THP-1 cells measured by the percentage of M5 citrate isotopolog obtained from U-13C5 glutamine labeling in 2% hypoxia over 16 hours, as shown in the schematic. The last bar shows mIDH1 cells cultured in the presence of 10 μmol/L ivosidenib added prior to adding a label. This experiment was performed with 6 cell pellets for each sample blinded and randomized on each LC-MS run. ****, P < 0.0001; Student t test. C, Column graph showing FC increase in acylcarnitine metabolites after induction of mIDH1 (+dox) vs. wild-type (−dox) in comparison with mIDH2 (+dox) vs. IDH2 wild-type (−dox) in THP-1 cells as measured by LC-MS. Student t test is used to compare groups. D, Column graph showing mean fatty acid β-oxidation as measured by the etomoxir (ETO)-sensitive component of oxygen consumption in pmole/min/105 cells in mIDH1 vs. IDH1 wild-type, mIDH2 and IDH2 wild-type in THP-1 cells measured on Seahorse analyzer (3 independent experiments). E, Column graph showing the percentage of b-oxidation of total oxygen consumption (etomoxir-sensitive component) in mIDH1 and mIDH2 before and after the addition of the ACC1 inhibitor 100 nmol/L ND-646. A representative experiment is shown. Statistics represent paired t test with n = 4 replicates. F, Column graph showing percentage of etomoxir-sensitive component in mIDH1 before and after the addition of 10 μmol/L ivosidenib. P = nonsignificant, Student t test, 4 replicates. G, Western blot showing phospho-AMPK (pAMPK) on threonine 172 in THP-1 cells induced with mIDH1, mIDH2, and wild-type counterparts in lipid-replete conditions. Comparison with total AMPK α isoform and Actin is shown in panels below. H, NADPH levels measured in identical 2 × 106 viable cell pellets of mIDH1, mIDH2, or wild-type primary AML blasts vs. normal CD34+ cells grown in culture for 48 hours in normoxia. Symbols indicate individual sample values. Student t test is used to compare groups. **, P < 0.01.I, Schematic summarizing mIDH1-induced mechanisms impacting lipid metabolism. Pathways involved directly in phospholipid synthesis that are perturbed by mIDH1 are shown in red. Thin arrows indicate reduced flux, and thick arrows indicate preserved or increased flux. Black arrows indicate pathways not measurably affected by mIDH1. Mechanistic causes for aberrant lipid metabolism identified in our study include (i) reduced carbon flux arising from defective reductive carboxylation by mutant IDH1 hetero/homodimers exacerbated in relative marrow hypoxia and mitochondrial stress; (ii) cumulative NADP(H) decrease by neomorphic reverse αKG to 2HG reaction; (iii) ongoing depletion of NADPH by residual reductive carboxylation of glutamine; (iv) insufficient NADPH replenishment by impaired forward reaction akin to TCA cycle; (v) increased fatty acid β-oxidation with a concomitant increase in acylcarnitines that is sensitive to ACC1 inhibition, but not ivosidenib; and (vi) decreased AMPK phosphorylation, indicating an AMPK-independent mechanism for enhanced oxidative phosphorylation of fatty acids. In contrast, the flux of glucose to produce glycerol-3-phosphate, the building block for the glycerol component of glycerolipids, is not impaired but rather is upregulated in mIDH1 compared with mIDH2, shown in black. This would suggest de novo synthesis of glycerol from glucose is not the major cause of defective lipid species in mIDH1 AML. Similarly, static levels of citric acid cycle metabolites are decreased to similar levels in both mIDH1 and mIDH2 AML, as shown in Supplementary figures. Blockade of 2HG production by ivosidenib is not sufficient for restoring defective reductive carboxylation or abrogating β-oxidation. NS, nonsignificant; WT, wild-type.
We next examined for a defect in carbon supply derived from glutamine by measuring isotope labeling of αKG derived from labeled glutamine, where every carbon atom is substituted for the heavier isotope. Citrate labeled on 5 out of 6 carbons (M5-citrate) is indicative of direct reductive carboxylation of αKG by the reverse reaction of wild-type IDH1 (35, 36). mIDH1 cells had the lowest rate of reductive carboxylation, as indicated by decreased incorporation of U-13C5 glutamine into M5-citrate isotopolog (Fig. 3B). Importantly, this reductive carboxylation defect was not reversed by preincubation with ivosidenib (Fig. 3B). No differences in TCA cycle–derived M4 citrate or unlabeled M0 citrate were observed (Supplementary Fig. S3E and S3F). mIDH2 cells had lower rates of reductive carboxylation compared with IDH2 wild-type but still higher than mIDH1, consistent with the increased relative dependency of IDH1 for reductive carboxylation in cancer (ref. 15; Fig. 3B). Of note, the mIDH2 deficit in reductive carboxylation was not reversed by enasidenib (Supplementary Fig. S3G).
If mIDH1-dependent alteration in nonpolar metabolites is principally due to carbon deficit, then any growth defect in a lipid-deplete environment should be rescued by the exogenous supply of an alternative carbon source such as Na acetate. In a 4-day growth assay, the proliferation defect observed in mIDH1 cells in lipid-depleted conditions could not be rescued with 1 mmol/L sodium acetate, unlike wild-type cells (Supplementary Fig. S3H), despite equivalent protein expression of acetyl CoA synthetase/ACCS2 (Supplementary Fig. S3I). Notably, ivosdenib treatment did not change the lack of acetate rescue in mIDH1 (Supplementary Fig. S3H). This indicated that limitations to carbon supply are unlikely to account for differences in lipid metabolism in mIDH1.
We therefore examined for increased fatty acid consumption. Recent reports have suggested an increased reliance on complex I and oxidative phosphorylation for IDH-mutant tumors with concomitant elevated fatty acid β-oxidation (37), but mIDH1- versus mIDH2-specific differences have not been examined. We first measured acylcarnitine levels by mass spectrometry and noted significant elevation of the long-chain fatty acid transporter cargo octadeanoylcarnitine after induction of mIDH1 in THP-1 cells (FC 11.3, dox vs. no dox, P = 1.5 × 10−5). Similar elevation was observed for palmitoylcarnitine, lauroylcarnitine, and tetradecanoylcarninitine (FC 5.9, 4.7, and 4.0, respectively, all P < 0.0001; Supplementary Fig. S3J). mIDH2 cells also displayed elevated carnitine species (palmitoylcarnitine FC 3.1, dox vs. no dox, P = 5.1 × 10−6; lauroylcarnitine 2.7, P = 0.0003) but at reduced magnitude overall compared with mIDH1 (FC 5.3 vs. 2.9 compared with wild-type, P = 0.02; Fig. 3C). Both mIDH1 and mIDH2 cells showed greater endogenous fatty acid oxidation compared with wild-type counterparts, as shown by an increased fraction of maximal oxidative phosphorylation inhibitable by 4 μmol/L etomoxir, in the absence of exogenous fatty acids or carnitine (Fig. 3D; Supplementary Fig. S3K). Strikingly, the inhibition of mIDH1 cells with the selective ACC1/2 inhibitor ND-646 significantly impaired fatty acid oxidation in mIDH1 cells (P = 0.02), but not mIDH2 cells (P = not significant; Fig. 3E), indicating de novo fatty acid synthesis is necessary for ongoing fatty acid oxidation in mIDH1 in the absence of exogenous lipids. In contrast, treatment with ivosidenib for 24 hours did not reduce fatty acid oxidation in mIDH1 (Fig. 3F).
Fatty acid oxidation is normally regulated by AMPK, which serves as an energy-sensing thermostat that becomes activated on threonine 172 when ATP levels are limiting. Intriguingly, we found complete inactivation of AMPK (both decreased phosphorylation and protein expression), despite the elevation of fatty acid oxidation, in both mIDH1 and mIDH2 cells compared with wild-type, consistent with recent reports (Fig. 3G; ref. 37).
Increased dependence on ACC1 for both ongoing lipid synthesis and β-oxidation may be a consequence of an NADPH deficit (38). Consistent with this, we detected a decreased level of reduced NADPH in mIDH1 primary AML compared with mIDH2 AML, wild-type AML, or normal CD34+ cells (1.9 vs. 4.6 pmol/L per 106 cells, mIDH1 vs. mIDH2, P = 0.004; Fig. 3H). No significant differences in the ratio of reduced NADPH to total NADP(H) were noted (Supplementary Fig. S3L). In contrast, the relative levels of the alternative ROS scavengers (specifically, reduced glutathione and oxidized glutathione), and their overall ratio (reduced:oxidized), were not significantly different in primary mIDH1 versus mIDH2 samples (Supplementary Fig. SM–SO).
Thus, we identified several differences in metabolism between mIDH1 and mIDH2 summarized in Fig. 3I. These include (i) defective reductive carboxylation supplying carbon by mutant IDH1 not rescued by ivosidenib; (ii) a greater NADPH deficit in mIDH1 versus mIDH2; (iii) increased acylcarnitine species (mIDH1 > mIDH2) compared with wild-type, indicating reliance on β-oxidation of fatty acids; and (iv) increased ACC1 dependence on β-oxidation for mIDH1 compared with mIDH2. In contrast, flux of glucose to produce glycerol-3-phosphate, the building block for glycerol and NADH cytoplasm to mitochondria shuttle, was not defective but rather upregulated in mIDH1. Together, our results support a model in which mIDH1 results in increased dependency to maintain both lipid production and lipid oxidation through ACC1, likely due to a longer-term NADPH deficit rather than a limited carbon supply. The NADPH deficit in primary cells and consequent derangements of lipid metabolism is of higher magnitude in mIDH1 compared with mIDH2. We propose that further impairment of de novo lipid synthesis through disruption of ACC1, first identified by MiSL, leads to impaired growth, thereby accounting for the synthetic lethal interaction.
Lipid-Free Diet Impairs Growth of IDH1-Mutant Leukemia In Vivo
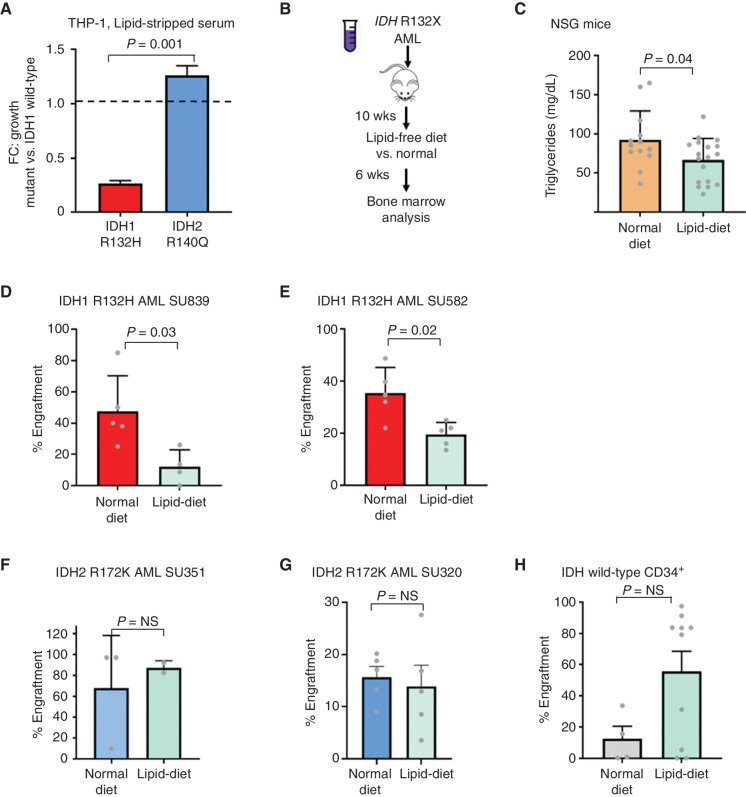
To test for an increased dependency of mIDH1 for exogenous fatty acids, we noted that mIDH1 THP-1 cells (in the absence of ACC1 knockdown) showed a moderate growth impairment in lipid-depleted serum compared with mIDH2 (Fig. 4A). To test whether defective lipid production by IDH1-mutant AML could be exploited for therapeutic advantage, we performed intrafemoral transplantation of primary IDH1-mutant AML samples into immunocompromised (NSG) mice. After confirming engraftment, mice were allocated to two different groups with paired equivalent engraftment levels (baseline median engraftment, nonsignificant, Mann–Whitney U) and were changed to either a strict lipid-free sucrose-rich diet for an additional 6 weeks or continued on a normal diet (Fig. 4B). Consistent with administration of the modified diet, we observed decreased serum triglyceride levels (Fig. 4C) despite steady weight gain (Supplementary Fig. S4A). Notably, with 2 of 2 independent mutant IDH1 samples tested, we detected a significant decrease in engraftment of human CD45+CD33+ leukemic blasts at the end of the 6 weeks (median engraftment 40% vs. 11%, normal vs. lipid-free diet, P = 0.03, mIDH1 SU839; 34% vs. 21%, normal vs. lipid-free, P = 0.02, mIDH1 SU582, Mann–Whitney U; Fig. 4D and E; Supplementary Fig. S4B). In contrast, no statistically significant differences were observed in similar studies with two independent IDH2-mutant primary AML samples (Fig. 4F and G) or normal CD34+ cord blood cells (Fig. 4H).
Figure 4.
Lipid-free diet causes decreased growth of mIDH1 AML. A, Growth of mIDH1 vs. mIDH2 THP-1 cells in lipid-stripped serum over 10 days shown as a relative FC compared with IDH1 wild-type cells. Growth was measured using Presto-Blue cell viability. Bars represent standard deviation of 4 replicates in a representative experiment performed 3 times. B, Schematic showing change in diet to sucrose-rich lipid-free diet at 10 weeks after engraftment of human AML. C, Triglyceride levels measured in NSG mice after 6 weeks of lipid-free dietary supplementation. D–H, Human CD33+ engraftment of mIDH1 AML (D, E), mIDH2 AML (F, G), and normal CD34+ hematopoietic stem/progenitor cells (H) after 6 weeks in mice treated with lipid-free compared with normal rodent diet. Bars represent standard error. Mann–Whitney U test was used to compare engraftments with P values as shown. NS, nonsignificant.
Knockdown of ACC1 Impairs Growth of IDH1-Mutant Solid Tumors
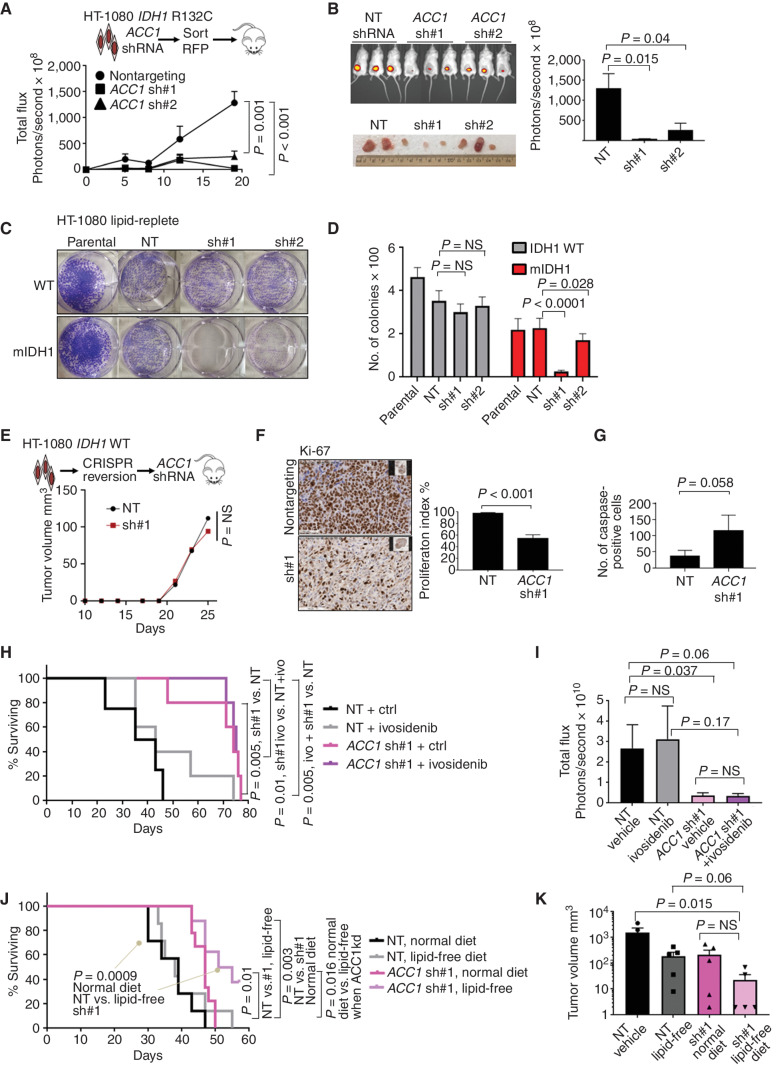
We initially identified the synthetic lethal interaction between ACC1 and mIDH1 in AML. To investigate if this relationship also applies to solid tumors, we utilized the human chondrosarcoma-derived cell line HT-1080 that harbors an endogenous IDH1 R132C mutation (39). Subcutaneous transplantation of ACC1-knockdown HT-1080 cells into immunodeficient mice led to significantly slower tumor growth (Fig. 5A and B). We generated IDH1 wild-type HT-1080 cells by CRISPR-mediated reversion of the R132C mutation as an isogenic control (Supplementary Fig. S5A and S5B). Importantly, knockdown of ACC1 in IDH1 wild-type HT-1080 cells did not result in significant differences in in vitro colony formation in lipid-replete conditions (mean colony number 351 vs. 299, nontargeting vs. ACC1 shRNA#1, P = nonsignificant), unlike mIDH1 parental HT-1080 cells, which showed significant defective growth to both hairpins (mean colony number 225 vs. 25, mIDH1 nontargeting vs. ACC1 shRNA#1, P < 0.0001; mean colony number 225 vs. 176, mIDH1 nontargeting vs. ACC1 shRNA#2, P = 0.028; Fig. 5C and D). Similarly, the knockdown of ACC1 in wild-type HT-1080 cells did not show differences in tumor growth in vivo (Fig. 5E). IDH1 wild-type glioma cells U87 (Supplementary Fig. S5C) and U118 (Supplementary Fig. S5D) did not show in vivo growth defects. ACC1 targeting in mIDH1 HT-1080 resulted in a decreased proliferation index by Ki-67 staining (Fig. 5F; Supplementary Fig. S5E) and a modest increase in the caspase cleavage (Fig. 5G). Significantly, ACC1 knockdown resulted in prolonged overall survival compared with nontargeting shRNA controls (median 74 vs. 39 days, log-rank hazard ratio 0.2, nontargeting control vs. ACC1 shRNA, P = 0.005; Fig. 5H). Importantly, coadministration of ivosidenib (AG-120) at 25 mg/kg daily did not reverse the growth defect or survival advantage for ACC1 knockdown (median survival 74 vs. 75 days, hazard ratio 0.98, ACC1 shRNA vs. ACC1 shRNA + AG-120, P = 0.6). A significant difference remained between nontargeting and ACC1 shRNA tumors treated with AG-120 (median 75 vs. 39 days, hazard ratio 0.2, ACC1 shRNA ivosidenib vs. nontargeting, P = 0.005; Fig. 5H and I). These results provide further evidence that growth impairment by ACC1 targeting is independent of 2HG. Finally, the addition of lipid-free dietary modification further impaired tumor growth and increased overall survival for ACC1 knockdown cells compared with nontargeting control receiving a normal diet (median 53 vs. 39 days, hazard ratio 0.21, ACC1 shRNA + lipid-free vs. nontargeting + normal diet, P = 0.009; Fig. 5J and K).
Figure 5.
ACC1 is a potential target in IDH1-mutant solid tumors. A, Growth of HT-1080 tumors over 21 days after knockdown of ACC1 measured by intravital imaging of shRNA-transduced luciferase+ cells. B,In vivo imaging of tumors from the same experiment. C, Colony assays quantified by crystal violet showing decreased colonies in mIDH1 with ACC1 knockdown compared with wild-type in lipid-replete conditions. D, Column graphs showing mean number of colonies after seeding 2,500 cells per well; bars represent standard error of the mean from 2 independent experiments. E,In vivo growth of IDH1 wild-type HT-1080 cells after CRISPR–Cas9 correction. Plots show median tumor growth after 25 days. P = nonsignificant; Mann–Whitney U test. F, Ki-67 IHC stain of explanted HT-1080 (5 tumors × 10 fields of view) after ACC1 knockdown compared with control with Student t test. Right, quantification of the Ki-67 proliferation index in tumors with ACC1 knockdown vs. control. G, Summary bar graph of 5 tumors × 10 fields of view showing increased apoptosis in ACC1 knockdown explants as measured by IHC staining of cleaved caspase-3 in explanted tumors. H, Kaplan–Meier log-rank survival curve for NSG mice transplanted with HT-1080 cells comparing ACC1 knockdown (pink) to nontargeting control (black) ± ivosidenib (ivo) treatment 25 mg/kg/day (gray, purple) or vehicle given by oral gavage for 50 days. Mice eventually succumbed by 72 days in all treatment groups due to increased tumor growth requiring euthanasia. P values represent log-rank Mantel–Cox test between groups as indicated. I, Bar graph summarizing total flux (photons per second) based on intravital imaging at day 21 for the same experiment. Student t test with indicated P values assessed statistical significance. J, Kaplan–Meier log-rank survival curve for NSG mice transplanted with HT-1080 cells and fed lipid-free vs. normal (lipid-replete) rodent diet. As in the previous experiment, mice eventually succumbed by day 70 in all treatment groups due to increased tumor growth. P values represent log-rank Mantel–Cox test between groups as indicated. kd, knockdown. K, Bar graph summarizing tumor size at 21 days after engraftment as measured by calipers. NS, nonsignificant; NT, nontargeting; WT, wild-type.
Pharmacologic Inhibition of ACC1 Has Activity against IDH1 Mutant Cancers
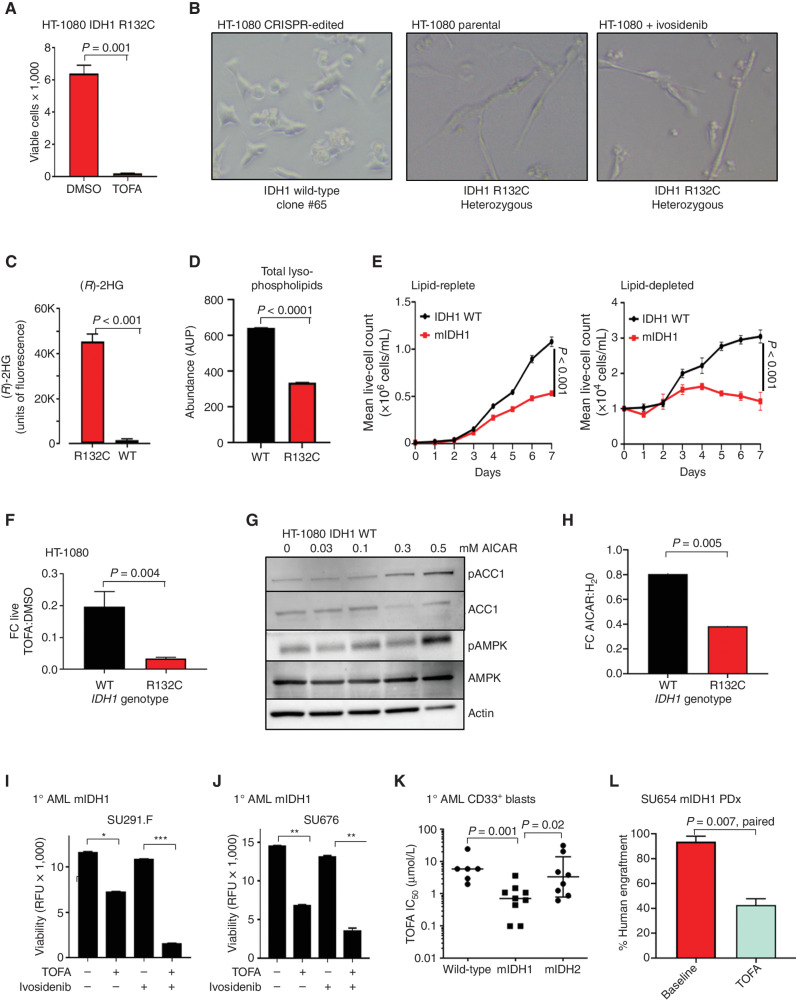
Several small-molecule inhibitors of ACC1 have been described including the agent TOFA (40, 41), which inhibits both ACC1 and ACC2 at low micromolar concentrations. We found that mIDH1 HT-1080 cells showed striking sensitivity to TOFA with half-maximal inhibitory concentration (IC50) of 4 μmol/L in vitro (Fig. 6A). Similarly, a glioblastoma model in which IDH1 wild-type U87 cells were transduced with lentivirus encoding IDH1 R132H demonstrated a 5-fold decrease in TOFA IC50 from 91.8 to 17.9 μmol/L (Supplementary Fig. S6A). To determine whether pharmacologic ACC1 inhibition shows mutation-specific sensitivity, we also tested IDH1 wild-type CRISPR-reversion HT-1080 cells (Supplementary Fig. S5A and S5B). Remarkably, when cultured in lipid-depleted conditions, mIDH1 cells showed an elongated ultrathin spindle-shaped morphology compared with broad triangular and rounded IDH1 wild-type cells (Fig. 6B). Only mIDH1 parental cells made 2HG (Fig. 6C) and, similar to mIDH1 AML, had a profound deficit of single-chain lyso-PE and lyso-PC lipids after mass spectrometry lipidomic analysis (Fig. 6D; Supplementary Fig. S6B and S6C). Consistent with AML, mIDH1 cells had a marked proliferation defect in lipid-depleted serum compared with wild-type revertant cells (Fig. 6E). As expected, mIDH1 cells showed markedly increased sensitivity to ACC1 blockade with TOFA compared with DMSO vehicle and compared with isogenic wild-type cells (Fig. 6F). Likewise, inhibition of ACC1 via AMPK activation with 0.3 mmol/L 5-aminoimidazole-4-carboxamide riboside (AICAR), an AMP mimetic, resulted in upregulation of ACC1 phosphorylation (Fig. 6G) and phenocopied the effect of ACC1 inhibition (Fig. 6H), with mIDH1 showing greater FC death cultured in AICAR compared with wild-type. Conversely, pharmacologic blockade with dorsomorphin or knockdown of AMPK isoform α in mIDH1 cells resulted in improved growth (Supplementary Fig. S6D and S6E). These results imply that mIDH1 cancers are extremely sensitive to any activation of AMPK, preferring to maintain ACC1 in an unphosphorylated and fully active state.
Figure 6.
ACC1 selective inhibitors have activity in mIDH1 cancers. A, Viability of HT-1080 mIDH1 R132C cells after exposure to 4 μmol/L TOFA for 72 hours measured by DAPI-negative cell population. B, Representative images of wild-type IDH1 reversion HT-1080 cells cultured at low density in 4% lipid-depleted serum or IDH1 R132C parental cells or IDH1 R132C treated with 10 μmol/L ivosidenib. Note that cells with IDH1 R132C mutation form ultrathin adherent elongated spindle-like cells. This morphology change was not reversed by coculture with ivosidenib. C, Measurement of 2HG in the supernatant of parental mIHD1 HT-1080 cells but undetectable in wild-type IDH1 reversion HT-1080 cells (clone #65 is shown as representative clone) after 72 hours. D, Total abundance of lysophospholipids (LPC + LPE) measured by LC-MS in mIDH1 vs. WT HT-1080 cultured in lipid-depleted media. AUP, area under the peak. E, Growth curves of mIDH1 vs. wild-type revertant HT-1080 in lipid-replete (left) vs. lipid-depleted (right) serum over 7 days. F, FC decrease in the number of live IDH1 R132C HT-1080 cells compared with IDH1 reversion wild-type HT-1080 cells after 96-hour exposure to the ACC1 inhibitor TOFA compared with DMSO vehicle. Graph shows mean of 3 independent experiments. G, Western blot showing increased phosphorylated ACC1 Serine 79 after treatment of HT-1080 wild-type cells with increasing concentration of AICAR. H, FC decrease in the number of live IDH1 R132C HT-1080 cells after 96-hour exposure to AICAR vs. H2O vehicle in comparison with IDH1 reversion wild-type HT-1080 cells under same conditions. Graph is a summary of 3 independent experiments. I and J, Primary mIDH1 AML cells isolated by flow cytometry from a patient at relapse (I) and a de novo patient (J) are sensitive to 10 μmol/L TOFA over 72 hours, but cytotoxicity is not reversed by 10 μmol/L ivosidenib. ***, P < 0.001;**, P < 0.01; *, P < 0.05. RFU, relative fluorescence unit. K, Summary of TOFA IC50 at 72 hours after in vitro treatment of mIDH1, mIDH2, and IDH wild-type primary AML blasts cultured in low serum media. P value reflects nonparametric two-tailed comparison between groups; bars represent median IC50 in μmol/L. L, Engraftment of mIDH1 AML at baseline and after 30 days of treatment with either vehicle or 50 mg/kg TOFA given by daily intraperitoneal injection. P = 0.007, paired t test, treated vs. baseline. PDx, patient-derived xenograft; WT, wild-type.
We then tested the activity of TOFA in primary AML. TOFA treatment of primary human AML samples also resulted in impaired viability, and the combination with ivosidenib resulted in synergistic IDH1-mutant primary cell death (Fig. 6I and J), similar to results in THP-1 cells (Supplementary Fig. S6F). TOFA treatment of a panel of primary AML samples demonstrated a significant difference in median IC50 between purified mIDH1 and mIDH2 AML blasts (0.7 vs. 2.9 μmol/L, mIDH1 vs. mIDH2, P = 0.02, Mann–Whitney U; Fig. 6K). Notably, mIDH1 samples were also significantly more sensitive to TOFA than wild-type AML (0.7 vs. 6 μmol/L, mIDH1 vs. IDH wild-type, P = 0.001, Mann–Whitney), consistent with our previous observations on a smaller sample set (27). As a pharmacologic agent, TOFA has a suboptimal pharmacokinetic profile (41, 42), and weight loss can be a pharmacodynamic marker of activity. A cohort of NSG mice engrafted with primary human mIDH1 AML was treated with TOFA at 50 mg/kg per day, and mice with a weight less than 22 g at the end of treatment were selected for further analysis (Supplementary Fig. S6G). In this group of low weight mice, TOFA treatment resulted in a decrease in engraftment (P = 0.007, paired t test, TOFA vs. baseline; Fig. 6L).
The Potent and Selective ACC1 Inhibitor ND-646 Has Activity against Venetoclax-Resistant AML
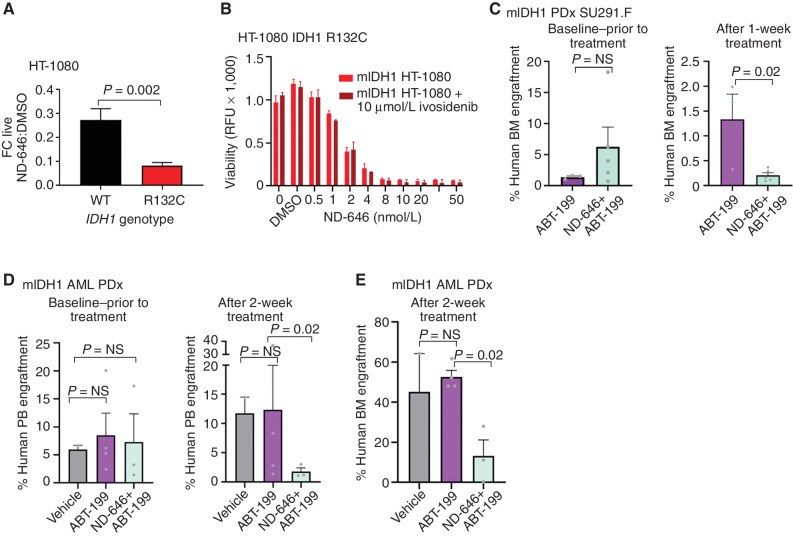
TOFA is a relatively weak inhibitor of ACC1, but several high-potency dual ACC1/ACC2 inhibitors have been developed, including ND-646 (43). After culture in lipid-depleted conditions, mIDH1 HT-1080 cells showed marked sensitivity to ND-646 that was not seen with wild-type reversion cells (Fig. 7A). Importantly, neither the ultrathin morphology in lipid-deplete conditions (Fig. 6B) nor the inhibitory effect of ND-646 was reversed by ivosidenib (Fig. 7B).
Figure 7.
The ACC1-selective inhibitor ND-646 can overcome venetoclax resistance. A, FC decrease in the number of live IDH1 R132C HT-1080 cells compared with IDH1 reversion wild-type (WT) HT-1080 cells after 96 hours exposure to the ACC1 inhibitor ND-646 compared with DMSO vehicle. Graph shows mean of 3 independent experiments. B, Parental IDH1 R132C HT-1080 cells were treated with increasing concentrations of the ACC1/2 inhibitor ND-646 ± 10 μmol/L ivosidenib. Cell viability was measured after 5 days in 4% lipid-stripped serum culture. This experiment was performed 3 times with a representative experiment shown. Bars represent standard deviation of technical replicates. No statistical differences were observed between ivosidenib-treated cells vs. untreated. RFU, relative fluorescence unit. C, Baseline and posttreatment bone marrow (BM) engraftment levels of human CD45+CD33+ mIDH1 AML exposed to ABT-199 50 mg/kg or ABT-199 50 mg/kg in combination with the selective ACC1 inhibitor ND-646 100 mg/kg for 7 days by oral gavage. P values represent the nonparametric Mann–Whitney U test. D, Peripheral blood (PB) engraftment levels of mIDH1 primary AML after treatment for 14 days with either vehicle, ABT-199 50 mg/kg, or ABT-199 in combination with ND-646. E, Bone marrow engraftment levels of the same experiment in D. P values represent the nonparametric Mann–Whitney U test for D and E. NS, nonsignificant; PDx, patient-derived xenograft.
In vitro, ND-646 reduced the growth of primary mIDH1 AML blasts at low micromolar concentrations in 7 out of 8 mIDH1 samples tested (Supplementary Fig. S7A–S7C). We previously reported that IDH1 mutation in AML resulted in a synthetic lethality upon inhibition of BCL2 by venetoclax/ABT-199 (31). We therefore sought to investigate the effect of ACC1 inhibition in comparison with venetoclax against IDH1-mutant AML. Notably, emerging reports indicate that many patients with AML show either intrinsic resistance or can eventually acquire resistance to venetoclax (44, 45), and results with TOFA suggested ACC1 blockade had activity in the setting of venetoclax resistance in mIDH1 THP-1 cells (Supplementary Fig. S7D–S7F). To further examine this, we performed combination experiments in two different settings: (i) an early engraftment model of mIDH1 AML resembling minimal residual disease (Fig. 7C) and (ii) an established model of mIDH1 FLT3-ITD+ AML with high levels of baseline engraftment (Fig. 7D and E). Prior to treatment, engraftment levels in each cohort were not significantly different. In both models, combination treatment of ABT-199 at 50 mg/kg and ND-646 at 100 mg/kg was superior in disease eradication compared with vehicle control or ABT-199 alone, with reductions in blast percentage observed in both peripheral blood (mean hCD33+ % 11.7 vs. 1.7, ABT-199 alone vs. combination, P = 0.02, unpaired t test; Fig. 7D) and bone marrow (mean hCD33+ 1.3 vs. 0.2%, ABT-199 alone vs. combination, P = 0.02, unpaired t test; Fig. 7C; median hCD33+ engraftment 50.2% vs. 11.1%, ABT-199 alone vs. combination, P = 0.02, Mann–Whitney U; Fig. 7E). Importantly, neither significant weight loss nor morbidity was incurred by combination treatment (Supplementary Fig. S7G). Together, these results suggest ACC1 blockade may have utility in the eradication of mIDH1 minimal residual disease after venetoclax treatment and increased efficacy in combination.
DISCUSSION
Computational analysis of primary human tumor data identified a potential synthetic lethal interaction between mIDH1 and ACC1, the rate-limiting enzyme for de novo lipid synthesis (27). A number of reports have demonstrated a critical role for wild-type IDH1 in driving de novo lipogenesis through reductive carboxylation of glutamine-derived αKG in cancer (14, 19, 26). Here we show in primary patient cells that mutant IDH1 markedly perturbs lipid metabolism, characterized by a deficiency of monoacylglycerides and lysophospholipids and increased acylcarnitine species. Our data suggest a number of targetable metabolic differences between mIDH1 and mIDH2, including defective reductive carboxylation, ACC1-dependent β-oxidation, and an NADPH deficit, all of which are likely to contribute to differences in lipid metabolism. Furthermore, genetic and/or pharmacologic inhibition of ACC1 impairs the growth of mIDH1 cancers, which validates the synthetic lethal interaction. Strikingly, these metabolic effects and synthetic lethal interaction are specific to mIDH1, which makes this one of the few functional differences between mIDH1 and mIDH2 cancers and is consistent with our report demonstrating greater sensitivity of mIDH1 to complex I inhibition compared with mIDH2 AML (21). Moreover, some of these effects (increased β-oxidation, defective reductive carboxylation) appear to occur independently of 2HG production, which suggests that these mutations act through additional mechanisms, consistent with recent reports (21, 37).
mIDH1 and mIDH2 are found in a number of different human cancers, but they both increase the production of the oncometabolite 2HG. FDA approval of the mutant-specific inhibitors ivosidenib and enasidenib for the treatment of mIDH1 and mIDH2 AML, respectively, represents a clinical advance for these molecularly defined subgroups. However, the reduction in 2HG that is observed in nearly all patients did not correlate with clinical responses. This also suggests that these mutations act though additional (non-2HG–dependent) pathogenic mechanisms that have yet to be rigorously characterized. Here, we observed a perturbed lipid profile in both primary mIDH1 AML and an mIDH1 chondrosarcoma cell line, consistent with similar results obtained by other groups in which slower growth and decreased de novo lipid synthesis was not reversed by an mIDH1 inhibitor (38). Similar defects in lipid metabolites have been reported in mIDH1 glioblastoma (46, 47) and mIDH1 HCT116 colon cancer (48). Importantly, these derangements were not phenocopied by the exogenous administration of 2HG.
The NADPH deficiency in primary mIDH1 AML cells is notable. This is consistent with data from mIDH1 HT-1080 cells that show a substantial NADPH deficit arising both from de novo lipid synthesis and 2HG production, each of which is equal in magnitude when measured by flux of deuterium-labeled [3–2H]glucose (38). It is therefore likely that multiple compensatory and targetable pathways are co-opted to cope with an ongoing NADPH deficit in mIDH1 cells. Our data suggest mIDH1 cells continue to deplete NADPH via ongoing 2HG production and are unable to replenish effectively via impaired forward reaction in the cytoplasmic equivalent of the canonical TCA cycle but continue to remain committed to reductive carboxylation, an NADPH-consuming reaction, albeit at a lower level of flux (Fig. 3A). Polyunsaturated lipids have a particularly high requirement for reduced equivalents of NADPH, which cannot be compensated directly by NADH (49). Thus, combinatorial therapies inhibiting either the oxidative pentose phosphate pathway or NADPH biosynthetic enzymes in the setting of a lipid-depleted diet may prove effective in the removal of residual mIDH1 leukemia cells. Of note, we did not see differences in pentose pathway flux (glucose → ribose), between mIDH1 and mIDH2 and their wild-type counterparts in both primary cells and cell lines, despite the established role of this pathway in NAPDH regeneration (50, 51).
Our results show that specific microenvironmental conditions or targeted inhibitors may exploit mIDH1-specific vulnerabilities. These vulnerabilities can be (i) 2HG-dependent and are therefore reversible by mIDH1 inhibitors; other vulnerabilities are (ii) secondary to a deficit in NADPH or (iii) caused by a direct defect in reductive carboxylation (14–16), enhanced under conditions of mitochondrial stress. Importantly, many of these metabolic vulnerabilities are druggable. For example, sensitivity to BCL2 inhibitor (31), HIF1α metabolic programming (46), or DNA hypermethylation (6) is predominantly 2HG-dependent and reversible by ivosidenib. Sensitivity to NAMPT inhibitor (52), defective de novo lipid synthesis (38), or ACC1 inhibition, as described here, appears largely 2HG-independent and not reversible with mIDH1 inhibitors. Further research into mIDH-specific metabolic vulnerabilities and their timing, extent, antagonism, and synergy will be vital to the development of novel agents for treatment of mIDH-driven cancers, as well as transcriptional/epigenetic programs induced by mIDH1 that activate fatty acid metabolism.
Metabolomic analysis of primary cancer samples is a field that is accelerating rapidly (53–55). Many of the methods described here are already becoming superseded by more sensitive techniques that do not require derivatization and improved buffer conditions for lipidomic analysis (30). It will be important to apply similar studies, including fluxomics and shotgun metabolomics, to IDH1 mutated preleukemic stem cells and stem cells from healthy persons with clonal hematopoiesis.
Our data suggest that lipid-depleted conditions and potentially a lipid-depleted diet may sensitize mIDH1 AML to other therapies. The tumor microenvironment is generally considered to be nutrient deficient (56) and many tumors upregulate lipogenesis, even when sufficient albumin-bound fatty acids are present in plasma (49, 56), for reasons that are not entirely clear. Emerging data in prostate cancer show metabolic programming in the same tumor for both increased de novo lipid production (57, 58) but also upregulation of β-fatty acid oxidation (59). Our data suggest mIDH1 cancers are also dependent on both but independent of AMPK activation. Future work should investigate other control pathways of β-oxidation in tumors reliant on ACC1 and β-oxidation inhibitors.
For the most part, lipid-deprived diets have not yet proved successful for cancer treatment (49) and may further compound tumor-associated cachexia. Although precision dietetics based on genomic profiling is still in its infancy, it may have utility in certain molecular cancer subgroups (60). Recent improvements in lipidomic mass spectrometry technology applied to cancer patient plasma and heterogeneous tumors is likely to aid in the specific design of diets that (i) maximize NADPH consumption for lipid desaturation and (ii) force local de novo lipid synthesis in mIDH1 tumors. It is notable that in vitro experiments with 10% fatty acid replete serum failed to recapitulate results when using strictly lipid-depleted serum or results seen in vivo. Our data imply that a lipid-deficient diet alone is not likely to be curative but may unmask other mIDH1-induced vulnerabilities that could be used to sensitize tumors to metabolic therapies or for postremission maintenance in patients. Intriguingly, acetate replacement rescued the growth defect in wild-type, but not mIDH1, cells observed in lipid-depleted conditions (Supplementary Fig. S3H and S3I). Future strategies inhibiting fatty acid uptake via CD36 blockade (as shown for prostate cancer; ref. 61) or other SLC27 family members simultaneously with ACC1 inhibitor may increase anticancer efficacy.
In conclusion, we have identified a novel metabolic vulnerability in mIDH1 AML and solid tumors, which arises from impaired de novo lipid synthesis caused by a combination of enhanced β-oxidation, depletion of NADPH, and defective reductive carboxylation. These metabolic changes confer dependency on ACC1, occur primarily independently of 2HG, and are specific for mIDH1 compared with mIDH2. Our results indicate that additional metabolic targets may complement the use of mutation-specific 2HG inhibitors that are currently used in the clinic (7, 10, 62).
METHODS
Cell Lines
THP-1 (RRID:CVCL_0006) cells sourced from ATCC were cultured in RPMI with 10% serum and 50 μmol/L β-mercaptoethanol. HT-1080 (RRID:CVCL_0317), U118 (RRID:CVCL_0633), and U87 (RRID:CVCL_0022) were originally sourced from the ATCC and cultured in DMEM with 10% serum. Confirmation of heterozygous IDH1 R132C mutation was confirmed in HT-1080 cells by Sanger sequencing and (R)-2HG production measured in supernatant after 7 days of growth.
MiSL Algorithm
Details are provided by Sinha and colleagues (27). Briefly, MiSL is a computational pipeline to identify candidate synthetic lethal partners of mutations for subsequent focused experimental validation using high-throughput pan-cancer primary tumor datasets. The starting point is a mutation and a cancer type of interest. For the results here, we utilized 12 cancer datasets from The Cancer Genome Atlas (RRID:SCR_003193). Our underlying assumption is that, across multiple cancers, synthetic lethal partners of a mutation will be amplified more frequently or deleted less frequently in primary tumor samples harboring the mutation with concordant changes in expression.
Chemicals
ND-646, enasidenib, and ivosidenib were purchased from Selleck Chemicals. All other chemicals including dox, AICAR, and TOFA were purchased from Sigma-Aldrich with the exception the following LC-MS solvents: HPLC-grade acetonitrile and water were purchased from Honeywell Burdick and Jackson, and LC-MS–grade formic acid was purchased from Thermo Fisher Scientific.
(R)-2HG Assay
(R)-2HG was measured using Sigma ultrasensitivity fluorescence kit. Briefly, in a total reaction volume of 100 μl, 100 mmol/L HEPES pH 8.0, 100 μmol/L NAD+ (Applichem), 0.1 μg (D)-2-hydroxyglutarate dehydrogenase, 5 μmol/L resazurin (Applichem), and 0.01 U/mL diaphorase (0.01 U/mL, MP Biomedical) were mixed together. Assay solution (75 μL) was added to 25 μL sample volume and incubated at room temperature in the dark for 30 minutes in black, 96-well plates (Thermo Scientific). Fluorometric detection was carried out in triplicate with 25 μL deproteinized sample analyzed in each reaction with excitation at 540 ± 10 nm and emission of 590 ± 10 nm (FLUOstar Omega, BMG Labtech). Cell culture supernatant was deproteinized using a deproteinization kit (Biovision). Briefly, to 100 μL of the sample, 20 μL of the perchloric acid solution was added, neutralized, and incubated on ice for 5 minutes. Samples were centrifuged for 2 minutes at 13,000 × g (Heraeus Fresco 21, Thermo Scientific). Supernatants were collected and stored at −20°C.
Preparation of Lipid-Stripped Serum
Lipids were removed from fetal bovine serum (Sigma) by stirring with 0.8 vol di-isopropyl ether (Sigma-Aldrich) and 0.2 vol n-butanol (Sigma) for 30 minutes at room temperature, followed by centrifugation at 4,000 × g for 15 minutes at 4°C to achieve phase separation. Serum proteins retained in the lower, aqueous phase were carefully collected, leaving behind lipids in the upper phase and interphase. Solvent contaminants were removed by stirring the serum for 2 hours while allowing a slow stream of nitrogen gas to blow over the surface of the serum. Any remaining solvent contaminants were further removed by dialysis for 72 hours using dialysis cassettes (Thermo Scientific).
Construction of ACC1 shRNA Vectors
The human ACACA (GenBank accession code: NM_198834.2) shRNA target sequences were selected using the BLOCK-iT RNAi Designer tool (Life Technologies). The knockdown efficiency of ACACA shRNA constructs was determined by quantitative real-time polymerase chain reaction and Western blotting. A pair of DNA oligonucleotides containing the sense target sequence followed by a loop sequence (5′-TCAAGAG-3′) and the reverse complement of the sense sequence were synthesized and annealed at 50 μmol/L in annealing buffer (10 mmol/L Tris-HCl pH 8.0, 50 mmol/L NaCl, 1 mmol/L EDTA) at 95°C for 10 minutes, followed by a slow cooling over 1 hour to room temperature. The double-stranded DNA template was then cloned into the pRSI9 DECIPHER shRNA expression vector with red fluorescent protein tag under UbiC promoter (Cellecta; RRID:Addgene_28289) digested with BsbI. ACACA sequences targeted by shRNA vectors used in this study were 5′-UGGCAUUGCAGCAGUGAAA-3′ (shRNA #1) and 5′-UGGAAUGAUUGCUGGAGAA-3′ (shRNA #2). Knockdown of ACC1 was validated by Western blotting using ACC1 isoform–specific polyclonal antibody (Cell Signaling Technology, cat. # 4190, RRID:AB_10547752).
Lentiviral Expression Vectors
Lentivirally transduced pools of cells were selected in 1 μg/mL puromycin. IDH1 wild-type, IDH1 R132H, IDH2 wild-type, and IDH2 R140Q proteins were expressed in the pTRIPZ (Open Biosystems) tet-inducible lentiviral vector with GFP encoded in the same open reading frame by T2A peptide. shRNA-transduced cells were marked with RFP. Double-positive DAPI-negative cells after 4 days of dox induction (1 μg/mL) were enumerated with Tru-count beads (BD). Confirmation of 2HG production in supernatants was performed after 72 hours of doxycycline.
Growth in Lipid-Depleted Serum
Lentivirally transduced cells were plated at 5 × 104 per mL grown in 0.5% charcoal-stripped serum (Sigma) over 10 days after dox induction. Cell growth was measured by Presto-Blue fluorescence (Thermo Fisher).
Metabolite Extraction
Cell pellets were homogenized in 75% methanol with a homogenizer Precellys 24 (Bertin Technologies). The homogenized mixture was then extracted with 3 volumes of the cold acetonitrile:acetone:isopropanol (1:1:1 v:v:v:) mixture. The extracted mixtures were centrifuged at 13,000 rpm for 15 minutes and supernatants were isolated. For lipid analysis, the supernatant was injected directly onto a hypersil C18 column (Thermo Fisher) for analysis. For polar compound analysis, aliquots of samples were taken and dried in a Speedvac (Thermo Scientific). The dried extract was then resuspended in 50% acetonitrile/water.
Semitargeted Metabolomic Analysis Methods
Dansylation of cellular metabolites was performed using previously described methods (32, 63–67). In brief, a volume of the extracted metabolites was combined with ½ volume of 0.1 mol/L sodium tetraborate buffer and then combined with an equal volume of 50 mmol/L dansyl chloride and vortexed. The mixture was incubated at room temperature for 30 minutes before the addition of an equal volume of 0.5% formic acid to stop the reaction. The supernatant of the reaction mixture was then placed in an autosampler vial.
O-benzylhydroxylamine (OBHA) labeling of cellular metabolites was performed according to Tan and colleagues (68). In brief, a volume of the cell extract 50 μL was combined with ½ volume of 0.5 mol/L OBHA and one volume of 1M EDC [1-ethyl-3-(3-dimethylaminopropyl) carbodiimide] in 8.6% pyridine at 25°C for 1 hour. Then, 300 μL ethyl acetate was added. After mixing, the organic layer was removed to a clean tube and dried down. The material was then resuspended in 50 μL of 20% acetonitrile and 0.1% formic acid buffer and placed in an autosampler vial.
All samples were analyzed on an accurate mass Q-TOF 6520 (Agilent Technologies) coupled with an Agilent UHPLC infinity 1290 system. For polar compounds, chromatography runs were carried out on a Phenomenex Kinetex reversed-phase C18 column (dimension 2.1 mm × 100 mm, 2.6 mm particles, 100A° pore size). Solvent A was HPLC water with 0.1% formic acid, and solvent B was acetonitrile with 0.1% formic acid. A 30-minute gradient at 0.5 mL/minute was as follows: t = 0.5 minutes, 5% solvent B; t = 20.5 minutes, 60% solvent B; t = 25 minutes, 95% solvent B; and t = 30 minutes, 95% solvent B. The column was equilibrated at 5% solvent B for 5 minutes. All data were acquired by positive electrospray ionization (ESI) using Masshunter acquisition software (Agilent Technologies, RRID:SCR_013575). For lipid analysis, chromatography runs were carried out on a BDS hypersil C8 column (Thermo Fisher Scientific; dimension 2.1 mm × 100 mm, 2.4 mm particles). Solvent A was 20% acetonitrile in water with 0.16% acetic acid, and solvent B was isopropanol and acetonitrile (9:1). Data were acquired with both positive and negative ESI.
Analysis of Mass Spectrometry Data
Molecular feature extraction for all data was carried out using Masshunter Profinder (Agilent Technologies) software. For lipids, ion spectra were matched against in silico–generated databases that use a combination of Bayesian scoring, a probability density algorithm, and nonnegative least squares fit to search the theoretical lipid library (LipidBlast; ref. 69). This approach minimized over-annotation of lipid entities by only providing structural information that was confidently informed by MS/MS spectra with distinct lipid classes detected in both positive and negative ion mode.
All metabolites that were present in more than one sample were analyzed. We noted that some metabolites were either not detected or had a very low intensity, and the resulting inaccurate abundance measurements could lead to an overestimation of the FC between samples. Therefore, the minimum threshold abundance was empirically set to 1,000. If a metabolite was not detected, or if it had an intensity value below this threshold in a sample, its abundance value in that sample was assumed to be the threshold value. Metabolic features were identified as differentially abundant between two conditions if (i) the FC was ≥2 (either direction); (ii) the FDR-adjusted (using the Benjamini–Hochberg method; ref. 70) P value (two-sample t test on the log scale) was less than 0.05; and (iii) the average abundance was >5,000 in at least one of the two conditions. The latter condition was to ensure that the observed difference was not due to artifacts generated by measurements made near the lower detection limit.
Patient Sample Data
Primary bone marrow and peripheral blood AML samples were obtained with written informed consent according to institutional guidelines (Stanford University Institutional Review Board No. 6453 and No. 18329 and Central Adelaide Local Health Network Human Ethics Committee Approval #12986 for samples obtained from Royal Adelaide Hospital) in accordance with the Declaration of Helsinki. Inclusion criteria were adults with a confirmed diagnosis of AML (blast count >20%) and somatic mutation profiling performed. Exclusion criteria were lymphoid leukemias and prior treatment. Mononuclear cells from each sample were isolated by Ficoll separation and cryopreserved in liquid nitrogen. All of the primary samples were tested for mutations in FLT3, NPM1, IDH1, and IDH2 either by the Stanford Anatomic Pathology and Clinical Laboratories or SA Pathology. IDH1/2 mutations were detected using SNaPshot methodology (Life Technologies). For TOFA and ND-646 studies, AML blasts were cultured at low density (3 × 104 cells in 200 μL 96-well plates) after T- and B-cell depletion and dead-cell removal in IMDM with 0.5% lipid-stripped serum; Plasmocin antibiotic (InvivoGen); and 20 ng/mL each of IL3, GM-CSF, G-CSF, IL6, SCF, FLT3L, and TPO (PeproTech). For NADP(H) quantification, purified blasts were cultured at high density (106/mL) in IMDM with 20% serum; 50 μmol/L b-mercaptoethanol; and 20 ng/mL each of IL3, GM-CSF, G-CSF, IL6, SCF, FLT3L, and TPO (PeproTech).
Metabolic Flux Analysis
THP-1 (RRID:CVCL_0006) cells were cultured for 12 hours in glucose-free RPMI (Gibco) with 2 g/L D-glucose-1,2–13C2 (Sigma-Aldrich) or U-13C5 4 mmol/L glutamine in glutamine-free IMDM and 10% lipid-stripped serum. Metabolites were extracted by applying 80% cold methanol on cells that were washed and collected on a 0.45-μm nylon filter membrane using a vacuum filter. The metabolite extracts were lyophilized and sent for analysis at Agilent and the UCLA Metabolomics Center where chromatographic separation was performed on a Vanquish Flex (Thermo Scientific). Metabolites were detected with a Thermo Scientific Q Exactive mass spectrometer and quantified by TraceFinder 4.1 (Thermo Fisher Scientific, RRID:SCR_008452). All results were corrected for the 1.1% natural abundance of 13C.
NADPH Analysis of Primary Cells
Prior to measurement, primary AML blast cells were cultured for 48 hours following thawing and assessed for the viability of >75%. A minimum of 2 million live cells were used to measure NADPH using the High Sensitivity NADPH Quantitation Fluorometric Assay Kit (Sigma Aldrich, MAK216) following the manufacturer's protocol.
Lipidomics in HT-1080 Cells
Aliquots of HT-1080 (RRID:CVCL_0317) cell pellets were homogenized using a Precellys 24 Tissue homogenizer (Bertin Technologies SAS). Total protein (10 μg) of each sample was combined with 145 μL of extraction buffer (173:173:2 v/v/v acetonitrile:propan-2-ol:EquiSPLASH lipid standard; Avanti Polar Lipids) and then stored at −20°C overnight allowing the protein to precipitate. Following centrifugation, 130 μL of protein-free supernatant was transferred to KX+ LC-MS 1.2 mL clear screw top vials (Kinesis). A quality control sample was generated by combining a 10 μL volume from each sample. Reversed-phase separation and mass spectrometric analysis was performed as described in White and colleagues (71), with the difference that data were acquired in continuum mode rather than centroid mode. The lipid database was expanded to include ether lipids, cholesterol esters, triacylglycerides, and sphingolipids. Data were processed using Progenesis QI (v2.4.6911.27652). Import thresholds for low energy data and elevated energy, respectively, were set to 250 counts and 0.01% of base peak. Databank search parameters were better than 5 ppm mass accuracy (average 0.8 ppm) and better than 0.15-minute retention time accuracy (average observed 0.06 minutes).
AMPK Pathway Experiments
Dorsomorphin was purchased from Selleck. shRNA constructs specific for PRKAA1 (shRNA#2 GAAGGTTGTAAACCCATATTA and shRNA#5 TGATTGATGATGAAGCCTTAA) were obtained from Vector Builder and validated with AMPKα1 isoform–specific polyclonal antibody (Cell Signaling Technology; cat. # 2795, RRID:AB_560856) and phospho-AMPK antibody (Cell Signaling Technology; cat. # 2535, RRID: RRID:AB_331250).
Measurement of Fatty Acid Oxidation
THP-1 pTRIPZ cells were cultured overnight in a substrate-limited medium consisting of XF RPMI base medium pH 7.4 (Seahorse Bioscience, cat. # 103576-100) supplemented with 2 mmol/L glucose, 1 mmol/L glutamine, and 1% serum. On the day of assay, cells were seeded at 1 × 105 cells/well in XF RPMI base medium supplemented with 2 mmol/L glucose and incubated in a non-CO2 incubator for 60 minutes. Fifteen minutes prior to starting the assay, 4 μmol/L etomoxir or vehicle was added to appropriate wells. Respiration was measured using the Seahorse XFe96 Extracellular Flux Analyzer with XFe96 FluxPaks (Seahorse Bioscience, cat. # 102416-100) and XF Cell Mito Stress Test Kit (Seahorse Bioscience, cat. # 103015-100).
Animal Care
All mouse experiments were conducted according to an Institutional Animal Care and Use Committee–approved protocols (Stanford Administrative Panel on Laboratory Animal Care no. 22264; South Australian Bioresources Animal Ethics Protocol SAM-20-037v2) and in adherence to the NIH's Guide for the Care and Use of Laboratory Animals and the Australian code for the care and use of animals for scientific purposes. At the end of each study, mice deeply anesthetized by isofluorane were euthanized by carbon dioxide inhalation, followed by cervical dislocation.
Xenograft of Primary AML
Freshly thawed primary AML samples from patients with mIDH1 were transplanted via intrafemoral injection into 6- to 12-week-old NSG mice (The Jackson Laboratory, RRID:IMSR_JAX:005557) conditioned with 180 rad of irradiation. Both female and male mice were used. All mouse experiments were conducted according to an Institutional Animal Care and Use Committee–approved protocol (Stanford Administrative Panel on Laboratory Animal Care no. 22264) and in adherence to the NIH's Guide for the Care and Use of Laboratory Animals. Up to 5 mice were transplanted for each treatment group if a sufficient amount of primary patient material was available, giving enough power to see a statistical difference of >30% Mann–Whitney U test. The sample size was based on estimations by power analysis with a level of significance of 0.05 and a power of 0.8. A smaller number of mice were used if the sample was limiting. Blinding could not be performed due to obvious differences in drug suspension appearance. Randomization was performed for all treatment groups matched for baseline engraftment levels. ND-646 was administered by daily oral gavage in 0.9% NaCl, 1% Tween-80, and 0.5% methylcellulose. ABT-199 was administered by daily oral gavage suspended in 60% Phosal-50PG, 30% PEG-400, and 10% ethanol vehicle.
Lipid-Free Rodent Diet
Teklad custom diet TD.03314 (Envigo Bioproducts Inc.) comprising 62% sucrose and no fat was used to feed mice after 6 weeks of engraftment. Diet consisted of casein 219 g/kg, DL methionine 2.5 g/kg, sucrose 620 g/kg, cellulose 109.5 g/kg, calcium carbonate 4 g/kg plus vitamin and mineral mix 45 g/kg.
Bone Marrow Engraftment Analysis
Bone marrow cells were collected by aspiration of the femur using a 27-gauge needle and stained for 30 minutes at 4°C with the following fluorophore-conjugated monoclonal antibodies: mTER199-PE-Cy5 (dilution 1:100; clone TER-199, Thermo Fisher Scientific; cat. #15-5921-82, RRID:AB_468810), mCD45-PE-Cy7 (dilution 1:50; clone A20, Thermo Fisher Scientific; cat. #25-0453-82, RRID:AB_469629), hCD45-V450 (dilution 1:25; clone HI30, BD Biosciences; cat. #560367, RRID:AB_1645573), and hCD33-APC (dilution 1:25, clone WM53, BD Biosciences; cat. #551378, RRID:AB_398502). Viable cells were identified by propidium iodide exclusion. The human leukemic population was identified as mTER199−, mCD45−, hCD45+, and hCD33+. Cells stably transduced with shRNA were identified as RFP+.
Statistical Data Analysis
Unless otherwise stated, P values comparing two means were calculated using the two-tailed unpaired Student t test in Prism version 6 (GraphPad Prism; RRID:SCR_002798). For in vivo engraftment data, nonparametric Mann–Whitney U test was used. A P value less than 0.05 was considered statistically significant. IC50 values were determined using the dose–response (inhibition) function in Prism version 6.0. The data were normalized and fitted using a variable Hill Slope model.
Data Availability
Mass spectrometric retention time and m/z data can be made available at request from the corresponding author.
Supplementary Material
Table S1 showing AML primary cell details for metabolomics
Supplementary Figure file showing Figures S1-S7
Acknowledgments
We acknowledge Feifei Zhao for lab management and Subarna Sinha and David Dill for the computational development of the MiSL algorithm. D. Thomas is supported by a National Cancer Institute K99 Pathway to Independence Award (CA2077301), a Commonwealth Serum Laboratories Centenary Fellowship, The Hospital Research Foundation, the Australian Medical Research Future Fund, and National Health and Medical Research Council Ideas Grants 1182564 and 1184485. This work was supported by NIH grant 1R01CA25133 and the Stanford Ludwig Center for Cancer Stem Cell Research and Medicine. R. Majeti is a recipient of The Leukemia & Lymphoma Society Scholar Award. D. Thomas and R. Majeti are supported by The Leukemia & Lymphoma Society (www.lls.org), the Snowdome Foundation (https://www.snowdome.org.au/), and the Leukaemia Foundation (www.leukaemia.org.au) grants 6619-21 and 6650-23. G. Peltz., M. Zheng, and M. Wu were supported by NIH/National Institute on Drug Abuse (NIDA) Award 1U01DA04439901. Y. Nakauchi was supported by the Nakayama Foundation for Human Science and a Stanford University School of Medicine Dean's Postdoctoral Fellowship. G. Peltz was supported by NIH/NIDA 5U01DA04439902.
The publication costs of this article were defrayed in part by the payment of publication fees. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Footnotes
NoteSupplementary data for this article are available at Cancer Discovery Online (http://cancerdiscovery.aacrjournals.org/).
Authors’ Disclosures
T. Köhnke reports grants from The Leukemia & Lymphoma Society and other support from TenSixteen Bio outside the submitted work. R. Majeti reports grants and other support from Gilead, other support from Kodikaz Therapeutic Solutions, Pheast Therapeutics, MyeloGene, and Orbital Therapeutics, and personal fees from Syros Pharmaceuticals, TenSixteen Bio, Roche, and Cullgen outside the submitted work. No disclosures were reported by the other authors.
Authors’ Contributions
D. Thomas: Conceptualization, funding acquisition, methodology, writing–review and editing. M. Wu: Conceptualization, formal analysis, supervision, investigation, writing–original draft, writing–review and editing. Y. Nakauchi: Resources, formal analysis, investigation, writing–review and editing. M. Zheng: Resources, formal analysis, investigation, writing–review and editing. C.A.L. Thompson-Peach: Investigation. K. Lim: Investigation. N. Landberg: Data curation, formal analysis, investigation. T. Köhnke: Formal analysis, investigation. N. Robinson: Validation, investigation, writing–review and editing. S. Kaur: Resources, formal analysis, investigation, writing–review and editing. M. Kutyna: Conceptualization, formal analysis, supervision, validation, investigation, writing–original draft, writing–review and editing. M. Stafford: Formal analysis, investigation. D. Hiwase: Writing–review and editing. A. Reinisch: Validation, investigation, writing–review and editing. G. Peltz: Resources, data curation, formal analysis, investigation, writing–review and editing. R. Majeti: Conceptualization, formal analysis, supervision, validation, investigation, writing–original draft, writing–review and editing.
References
- 1. Chou W, Hou H, Chen C, Tang J, Yao M, Tsay W, et al. Distinct clinical and biologic characteristics in adult acute myeloid leukemia bearing the isocitrate dehydrogenase 1 mutation. Blood 2010;115:2749–54. [DOI] [PubMed] [Google Scholar]
- 2. Dang L, Jin S, Su SM. IDH mutations in glioma and acute myeloid leukemia. Trends Mol Med 2010;16:387–97. [DOI] [PubMed] [Google Scholar]
- 3. Dang L, White DW, Gross S, Bennett BD, Bittinger MA, Driggers EM, et al. Cancer-associated IDH1 mutations produce 2-hydroxyglutarate. Nature 2009;462:739–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. DiNardo CD, Ravandi F, Agresta S, Konopleva M, Takahashi K, Kadia T, et al. Characteristics, clinical outcome, and prognostic significance of IDH mutations in AML. Am J Hematol 2015;90:732–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Saha SK, Parachoniak CA, Bardeesy N. IDH mutations in liver cell plasticity and biliary cancer. Cell Cycle 2014;13:3176–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Figueroa ME, Abdel-Wahab O, Lu C, Ward PS, Patel J, Shih A, et al. Leukemic IDH1 and IDH2 mutations result in a hypermethylation phenotype, disrupt TET2 function, and impair hematopoietic differentiation. Cancer Cell 2010;18:553–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Fan B, Mellinghoff IK, Wen PY, Lowery MA, Goyal L, Tap WD, et al. Clinical pharmacokinetics and pharmacodynamics of ivosidenib, an oral, targeted inhibitor of mutant IDH1, in patients with advanced solid tumors. Invest New Drugs 2020;38:433–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kats LM, Vervoort SJ, Cole R, Rogers AJ, Gregory GP, Vidacs E, et al. A pharmacogenomic approach validates AG-221 as an effective and on-target therapy in IDH2 mutant AML. Leukemia 2017;31:1466–70. [DOI] [PubMed] [Google Scholar]
- 9. Lowery MA, Burris HA 3rd, Janku F, Shroff RT, Cleary JM, Azad NS, et al. Safety and activity of ivosidenib in patients with IDH1-mutant advanced cholangiocarcinoma: a phase 1 study. Lancet Gastroenterol Hepatol 2019;4:711–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. DiNardo CD, Stein EM, de Botton S, Roboz GJ, Altman JK, Mims AS, et al. Durable remissions with ivosidenib in IDH1-mutated relapsed or refractory AML. N Engl J Med 2018;378:2386–98. [DOI] [PubMed] [Google Scholar]
- 11. Stein EM, DiNardo CD, Pollyea DA, Fathi AT, Roboz GJ, Altman JK, et al. Enasidenib in mutant IDH2 relapsed or refractory acute myeloid leukemia. Blood 2017;130:722–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Gaude E, Schmidt C, Gammage PA, Dugourd A, Blacker T, Chew SP, et al. NADH shuttling couples cytosolic reductive carboxylation of glutamine with glycolysis in cells with mitochondrial dysfunction. Mol Cell 2018;69:581–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Dexter JP, Ward PS, Dasgupta T, Hosios AM, Gunawardena J, Vander Heiden MG. Lack of evidence for substrate channeling or flux between wildtype and mutant isocitrate dehydrogenase to produce the oncometabolite 2-hydroxyglutarate. J Biol Chem 2018;293:20051–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Metallo CM, Gameiro PA, Bell EL, Mattaini KR, Yang J, Hiller K, et al. Reductive glutamine metabolism by IDH1 mediates lipogenesis under hypoxia. Nature 2011;481:380–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Grassian AR, Parker SJ, Davidson SM, Divakaruni AS, Green CR, Zhang X, et al. IDH1 mutations alter citric acid cycle metabolism and increase dependence on oxidative mitochondrial metabolism. Cancer Res 2014;74:3317–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Jiang L, Shestov AA, Swain P, Yang C, Parker SJ, Wang QA, et al. Reductive carboxylation supports redox homeostasis during anchorage-independent growth. Nature 2016;532:255–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Mullen AR, Hu Z, Shi X, Jiang L, Boroughs LK, Kovacs Z, et al. Oxidation of alpha-ketoglutarate is required for reductive carboxylation in cancer cells with mitochondrial defects. Cell Rep 2014;7:1679–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Mullen AR, Wheaton WW, Jin ES, Chen PH, Sullivan LB, Cheng T, et al. Reductive carboxylation supports growth in tumour cells with defective mitochondria. Nature 2011;481:385–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Fan J, Kamphorst JJ, Rabinowitz JD, Shlomi T. Fatty acid labeling from glutamine in hypoxia can be explained by isotope exchange without net reductive isocitrate dehydrogenase (IDH) flux. J Biol Chem 2013;288:31363–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Leonardi R, Subramanian C, Jackowski S, Rock CO. Cancer-associated isocitrate dehydrogenase mutations inactivate NADPH-dependent reductive carboxylation. J Biol Chem 2012;287:14615–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Bassal MA, Samaraweera SE, Lim K, Benard BA, Bailey S, Kaur S, et al. Germline mutations in mitochondrial complex I reveal genetic and targetable vulnerability in IDH1-mutant acute myeloid leukaemia. Nat Commun 2022;13:2614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Benard BA, Leak LB, Azizi A, Thomas D, Gentles AJ, Majeti R. Clonal architecture predicts clinical outcomes and drug sensitivity in acute myeloid leukemia. Nat Commun 2021;12:7244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ward PS, Thompson CB. Metabolic reprogramming: a cancer hallmark even Warburg did not anticipate. Cancer Cell 2012;21:297–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ookhtens M, Kannan R, Lyon I, Baker N. Liver and adipose tissue contributions to newly formed fatty acids in an ascites tumor. Am J Physiol 1984;247:R146–153. [DOI] [PubMed] [Google Scholar]
- 25. Baenke F, Peck B, Miess H, Schulze A. Hooked on fat: the role of lipid synthesis in cancer metabolism and tumour development. Dis Model Mech 2013;6:1353–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Shechter I, Dai P, Huo L, Guan G. IDH1 gene transcription is sterol regulated and activated by SREBP-1a and SREBP-2 in human hepatoma HepG2 cells: evidence that IDH1 may regulate lipogenesis in hepatic cells. J Lipid Res 2003;44:2169–80. [DOI] [PubMed] [Google Scholar]
- 27. Sinha S, Thomas D, Chan S, Gao Y, Brunen D, Torabi D, et al. Systematic discovery of mutation-specific synthetic lethals by mining pan-cancer human primary tumor data. Nat Commun 2017;8:15580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Badur MG, Muthusamy T, Parker SJ, Ma S, McBrayer SK, Cordes T, et al. Oncogenic R132 IDH1 mutations limit NADPH for de novo lipogenesis through (D)2-hydroxyglutarate production in fibrosarcoma cells. Cell Rep 2018;25:1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Biedermann J, Preussler M, Conde M, Peitzsch M, Richter S, Wiedemuth R, et al. Mutant IDH1 differently affects redox state and metabolism in glial cells of normal and tumor origin. Cancers 2019;11:2028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Murphy RC. Lipid mass spectrometry: a path traveled for 50 years. J Mass Spectrom 2020;55:e4492. [DOI] [PubMed] [Google Scholar]
- 31. Chan SM, Thomas D, Corces-Zimmerman MR, Xavy S, Rastogi S, Hong WJ, et al. Isocitrate dehydrogenase 1 and 2 mutations induce BCL-2 dependence in acute myeloid leukemia. Nat Med 2015;21:178–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Tapuhi Y, Schmidt DE, Lindner W, Karger BL. Dansylation of amino acids for high-performance liquid chromatography analysis. Anal Biochem 1981;115:123–9. [DOI] [PubMed] [Google Scholar]
- 33. Koelmel JP, Li X, Stow SM, Sartain MJ, Murali A, Kemperman R, et al. Lipid annotator: towards accurate annotation in non-targeted liquid chromatography high-resolution tandem mass spectrometry (LC-HRMS/MS) lipidomics using a rapid and user-friendly software. Metabolites 2020;10:101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Molenaar RJ, Maciejewski JP, Wilmink JW, van Noorden CJF. Wild-type and mutated IDH1/2 enzymes and therapy responses. Oncogene 2018;37:1949–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Jang C, Chen L, Rabinowitz JD. Metabolomics and isotope tracing. Cell 2018;173:822–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Yoo H, Antoniewicz MR, Stephanopoulos G, Kelleher JK. Quantifying reductive carboxylation flux of glutamine to lipid in a brown adipocyte cell line. J Biol Chem 2008;283:20621–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Stuani L, Sabatier M, Saland E, Cognet G, Poupin N, Bosc C, et al. Mitochondrial metabolism supports resistance to IDH mutant inhibitors in acute myeloid leukemia. J Exp Med 2021;218:e20200924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Badur MG, Muthusamy T, Parker SJ, Ma S, McBrayer SK, Cordes T, et al. Oncogenic R132 IDH1 mutations limit NADPH for de novo lipogenesis through (D)2-hydroxyglutarate production in fibrosarcoma sells. Cell Rep 2018;25:1018–1026e1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Li L, Paz AC, Wilky BA, Johnson B, Galoian K, Rosenberg A, et al. Treatment with a small molecule mutant IDH1 inhibitor suppresses tumorigenic activity and decreases production of the oncometabolite 2-hydroxyglutarate in human chondrosarcoma cells. PLoS One 2015;10:e0133813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Li S, Qiu L, Wu B, Shen H, Zhu J, Zhou L, et al. TOFA suppresses ovarian cancer cell growth in vitro and in vivo. Mol Med Rep 2013;8:373–8. [DOI] [PubMed] [Google Scholar]
- 41. Wang C, Xu C, Sun M, Luo D, Liao DF, Cao D. Acetyl-CoA carboxylase-alpha inhibitor TOFA induces human cancer cell apoptosis. Biochem Biophys Res Commun 2009;385:302–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Thupari JN, Pinn ML, Kuhajda FP. Fatty acid synthase inhibition in human breast cancer cells leads to malonyl-CoA-induced inhibition of fatty acid oxidation and cytotoxicity. Biochem Biophys Res Commun 2001;285:217–23. [DOI] [PubMed] [Google Scholar]
- 43. Svensson RU, Parker SJ, Eichner LJ, Kolar MJ, Wallace M, Brun SN, et al. Inhibition of acetyl-CoA carboxylase suppresses fatty acid synthesis and tumor growth of non-small-cell lung cancer in preclinical models. Nat Med 2016;22:1108–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Konopleva M, Pollyea DA, Potluri J, Chyla B, Hogdal L, Busman T, et al. Efficacy and biological correlates of response in a phase II study of venetoclax monotherapy in patients with acute myelogenous leukemia. Cancer Discov 2016;6:1106–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Pollyea DA, Stevens BM, Jones CL, Winters A, Pei S, Minhajuddin M, et al. Venetoclax with azacitidine disrupts energy metabolism and targets leukemia stem cells in patients with acute myeloid leukemia. Nat Med 2018;24:1859–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Zhao S, Lin Y, Xu W, Jiang W, Zha Z, Wang P, et al. Glioma-derived mutations in IDH1 dominantly inhibit IDH1 catalytic activity and induce HIF-1alpha. Science 2009;324:261–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Fack F, Tardito S, Hochart G, Oudin A, Zheng L, Fritah S, et al. Altered metabolic landscape in IDH-mutant gliomas affects phospholipid, energy, and oxidative stress pathways. EMBO Mol Med 2017;9:1681–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Gelman SJ, Naser F, Mahieu NG, McKenzie LD, Dunn GP, Chheda MG, et al. Consumption of NADPH for 2-HG synthesis increases pentose phosphate pathway flux and sensitizes cells to oxidative stress. Cell Rep 2018;22:512–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Butler LM, Perone Y, Dehairs J, Lupien LE, de Laat V, Talebi A, et al. Lipids and cancer: emerging roles in pathogenesis, diagnosis and therapeutic intervention. Adv Drug Deliv Rev 2020;159:245–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Chen L, Zhang Z, Hoshino A, Zheng HD, Morley M, Arany Z, et al. NADPH production by the oxidative pentose-phosphate pathway supports folate metabolism. Nat Metab 2019;1:404–15. [PMC free article] [PubMed] [Google Scholar]
- 51. Fan J, Ye J, Kamphorst JJ, Shlomi T, Thompson CB, Rabinowitz JD. Quantitative flux analysis reveals folate-dependent NADPH production. Nature 2014;510:298–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Tateishi K, Wakimoto H, Iafrate AJ, Tanaka S, Loebel F, Lelic N, et al. Extreme vulnerability of IDH1 mutant cancers to NAD+ depletion. Cancer Cell 2015;28:773–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Kumar A, Misra BB. Challenges and opportunities in cancer metabolomics. Proteomics 2019;19:e1900042. [DOI] [PubMed] [Google Scholar]
- 54. Rinschen MM, Ivanisevic J, Giera M, Siuzdak G. Identification of bioactive metabolites using activity metabolomics. Nat Rev Mol Cell Biol 2019;20:353–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Wishart DS. Metabolomics for investigating physiological and pathophysiological processes. Physiol Rev 2019;99:1819–75. [DOI] [PubMed] [Google Scholar]
- 56. Ackerman D, Simon MC. Hypoxia, lipids, and cancer: surviving the harsh tumor microenvironment. Trends Cell Biol 2014;24:472–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Zadra G, Photopoulos C, Loda M. The fat side of prostate cancer. Biochim Biophys Acta 2013;1831:1518–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Suburu J, Gu Z, Chen H, Chen W, Zhang H, Chen YQ. Fatty acid metabolism: implications for diet, genetic variation, and disease. Food Bioscience 2013;4:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Ma Y, Zha J, Yang X, Li Q, Zhang Q, Yin A, et al. Long-chain fatty acyl-CoA synthetase 1 promotes prostate cancer progression by elevation of lipogenesis and fatty acid beta-oxidation. Oncogene 2021;40:1806–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Camp KM, Trujillo E. Position of the academy of nutrition and dietetics: nutritional genomics. J Acad Nutr Diet 2014;114:299–312. [DOI] [PubMed] [Google Scholar]
- 61. Watt MJ, Clark AK, Selth LA, Haynes VR, Lister N, Rebello R, et al. Suppressing fatty acid uptake has therapeutic effects in preclinical models of prostate cancer. Sci Transl Med 2019;11:eaau5758. [DOI] [PubMed] [Google Scholar]
- 62. Birendra KC, DiNardo CD. Evidence for clinical differentiation and differentiation syndrome in patients with acute myeloid leukemia and IDH1 mutations treated with the targeted mutant IDH1 inhibitor, AG-120. Clin Lymphoma Myeloma Leuk 2016;16:460–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Park W, Wu M, Bowen R, Zheng M, Fitch WL, Pai RK, et al. Metabolomic markers differentiate mucinous and non-mucinous pancreatic cysts. Gastrointest Endosc 2013;78:295–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Wu M, Fitch WL, Zheng M, Merritt RE, Shrager JB, Weiruo Z, et al. Increased dipeptide abundance in non-small cell lung cancer rapid commun. Mass Spectrom 2013;27:2091–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Wu M, Sahbaie P, Zheng M, Lobato R, Boison D, David Clark J, et al. Opiate-induced changes in brain adenosine levels and narcotic drug responses. Neuroscience 2013;228:235–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Wu M, Zheng M, Zhang W, Suresh S, Schlecht U, Fitch WL, et al. Identification of drug targets by chemogenomic and metabolomic profiling in yeast. Pharmacogenet Genomics 2012;22:877–86. [DOI] [PubMed] [Google Scholar]
- 67. Tang Y, Li L. Differential isotope dansylation labeling combined with liquid chromatography mass spectrometry for quantification of intact and N-terminal truncated proteins. Anal Chim Acta 2013;792:79–85. [DOI] [PubMed] [Google Scholar]
- 68. Tan B, Lu Z, Dong S, Zhao G, Kuo MS. Derivatization of the tricarboxylic acid intermediates with O-benzylhydroxylamine for liquid chromatography-tandem mass spectrometry detection. Anal Biochem 2014;465:134–47. [DOI] [PubMed] [Google Scholar]
- 69. Kind T, Liu KH, Lee DY, DeFelice B, Meissen JK, Fiehn O. LipidBlast in silico tandem mass spectrometry database for lipid identification. Nat Methods 2013;10:755–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J Roy Stat Soc B 1995;57:289–300. [Google Scholar]
- 71. White JB, Trim PJ, Salagaras T, Long A, Psaltis PJ, Verjans JW, et al. Equivalent carbon number and interclass retention time conversion enhance lipid identification in untargeted clinical lipidomics. Anal Chem 2022;94:3476–84. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1 showing AML primary cell details for metabolomics
Supplementary Figure file showing Figures S1-S7
Data Availability Statement
Mass spectrometric retention time and m/z data can be made available at request from the corresponding author.



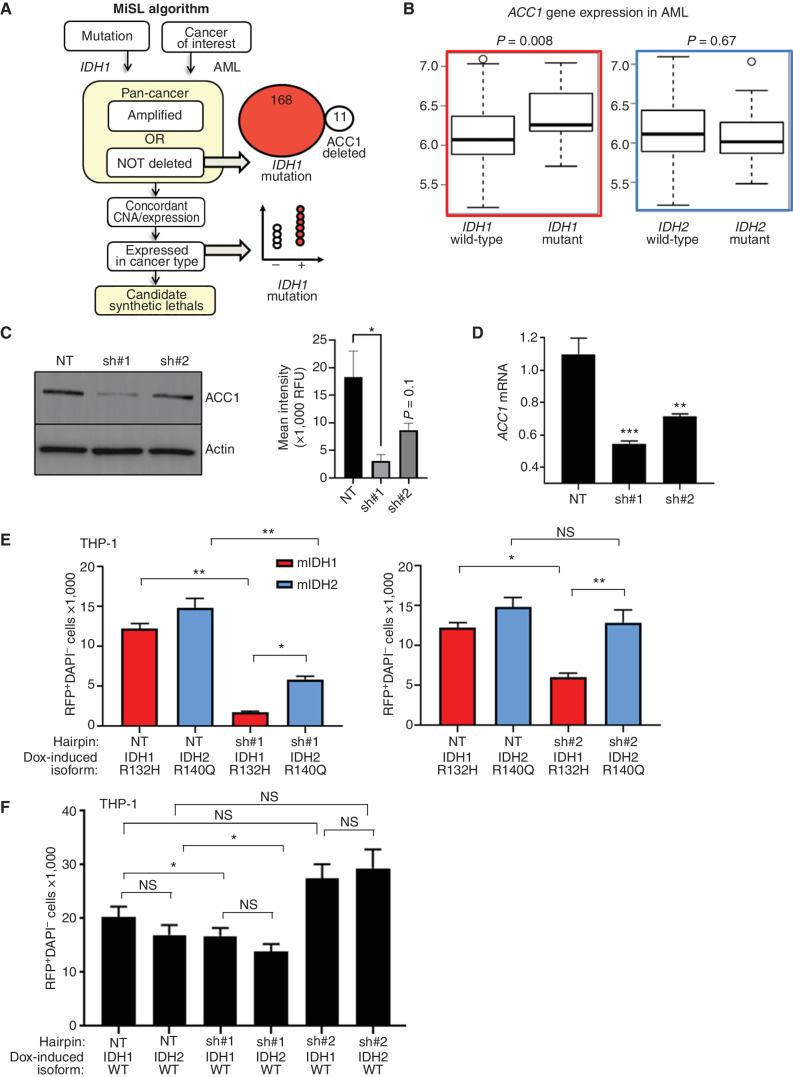
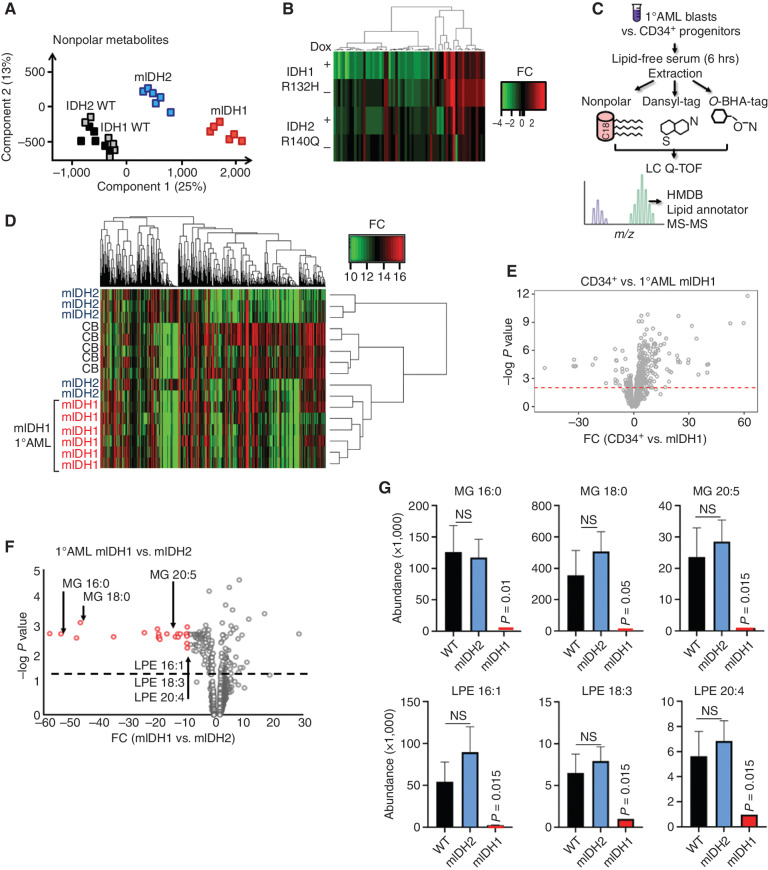
![Figure 3. IDH1 mutation is linked to defective reductive carboxylation, increased fatty acid consumption, and decreased NADPH. A, Schematic and graph of flux studies showing the percentage of labeled M2 glycerol-3-phosphate derived from 13C[1,2] labeled glucose (2 of 6 carbons as heavy isotope) across THP-1 cells induced to express mIDH1 vs. IDH1 wild-type compared with mIDH2 vs. IDH2 wild-type. Glucose was added to media in normoxia over 13 hours. Schematic indicates M2 isotopolog of glycerol-3-phosphate de novo synthesis directly from glycolysis utilizing labeled glucose rather than the oxidative pentose phosphate pathway (M1 isotopolog). A two-tailed unpaired Student t test was used to compare differences between groups. This experiment was performed with 6 cell pellets for each sample blinded and randomized on each LC-MS run. B, Decreased reductive carboxylation of mIDH1 compared with IDH1 wild-type and mIDH2 expressed in THP-1 cells measured by the percentage of M5 citrate isotopolog obtained from U-13C5 glutamine labeling in 2% hypoxia over 16 hours as shown in the schematic. The last bar shows mIDH1 cells cultured in the presence of 10 μmol/L ivosidenib added prior to adding a label. This experiment was performed with 6 cell pellets for each sample blinded and randomized on each LC-MS run; ***, P < 0.001; **, P < 0.01, Student t test. C, Column graph showing fold change increase in acylcarnitine metabolites after induction of mIDH1 (+dox) vs. wild-type (−dox) in comparison with mIDH2 (+dox) vs. IDH2 wild-type (−dox) in THP-1 cells as measured by LC-MS. Student t test is used to compare groups. D, Column graph showing mean fatty acid β-oxidation as measured by the etomoxir-sensitive component of oxygen consumption in pmole/min/105 cells in mIDH1 vs. IDH1 wild-type, mIDH2 and IDH2 wild-type in THP-1 cells measured on Seahorse analyzer, 3 independent experiments. E, Column graph showing the percentage of beta-oxidation of total oxygen consumption (etomoxir-sensitive component) in mIDH1 and mIDH2 before and after the addition of the ACC1 inhibitor 100 nmol/L ND-646. A representative experiment is shown. Statistics represent paired t test with n = 4 replicates. F, Column graph showing % etomoxir-sensitive component in mIDH1 before and after the addition of 10 μmol/L ivosidenib. P = nonsignificant, Student t test, 4 replicates. G, Western blot showing phospho-AMPK on threonine 172 in THP-1 cells induced with mIDH1, mIDH2 and wild-type counterparts in lipid-replete conditions. Comparison with total AMPK alpha isoform and actin is shown in panels below. H, NADPH levels measured in identical 2 × 106 viable cell pellets of mIDH1, mIDH2, or wild-type primary AML blasts vs. normal CD34+ cells grown in culture for 48 hours in normoxia. Symbols indicate individual sample values. Student t test is used to compare groups. I, Schematic summarizing mIDH1-induced mechanisms impacting lipid metabolism. Pathways involved directly in phospholipid synthesis that are perturbed by mIDH1 are shown in red. Thin arrows indicate reduced flux, and thick arrows indicate preserved or increased flux. Black arrows indicate pathways not measurably affected by mIDH1. Mechanistic causes for aberrant lipid metabolism identified in our study include: (i) reduced carbon flux arising from defective reductive carboxylation by mutant IDH1 hetero/homodimers exacerbated in relative marrow hypoxia and mitochondrial stress, (ii) cumulative NADP(H) decrease by neomorphic reverse αKG to 2HG reaction, (iii) ongoing depletion of NADPH by residual reductive carboxylation of glutamine, (iv) insufficient NADPH replenishment by impaired forward reaction akin to TCA cycle, (v) increased fatty acid β-oxidation with a concomitant increase in acylcarnitines that is sensitive to ACC1 inhibition but not ivosidenib, and (vi) decreased AMPK phosphorylation, indicating an AMPK-independent mechanism for enhanced oxidative phosphorylation of fatty acids. In contrast, the flux of glucose to produce glycerol-3-phosphate, the building block for the glycerol component of glycerolipids, is not impaired and is rather upregulated in mIDH1 compared with mIDH2, shown in black. This would suggest de novo synthesis of glycerol from glucose is not the major cause of defective lipid species in mIDH1 AML. Similarly, static levels of citric acid cycle metabolites are decreased to similar in both mIDH1 and mIDH2 AML, as shown in Supplementary figures. Blockade of 2HG production by ivosidenib is not sufficient for restoring defective reductive carboxylation nor abrogating β-oxidation.](https://cdn.ncbi.nlm.nih.gov/pmc/blobs/8f73/9900324/036f12733f94/496fig3.jpg)