Abstract
Vault RNAs (vtRNAs) are small noncoding RNAs and highly expressed in many eukaryotes. Here, we identified vtRNA2‐1 as a novel regulator of the intestinal barrier via interaction with RNA‐binding protein HuR. Intestinal mucosal tissues from patients with inflammatory bowel diseases and from mice with colitis or sepsis express increased levels of vtRNAs relative to controls. Ectopically expressed vtRNA2‐1 decreases the levels of intercellular junction (IJ) proteins claudin 1, occludin, and E‐cadherin and causes intestinal epithelial barrier dysfunction in vitro, whereas vtRNA2‐1 silencing promotes barrier function. Increased vtRNA2‐1 also decreases IJs in intestinal organoid, inhibits epithelial renewal, and causes Paneth cell defects ex vivo. Elevating the levels of tissue vtRNA2‐1 in the intestinal mucosa increases the vulnerability of the gut barrier to septic stress in mice. vtRNA2‐1 interacts with HuR and prevents HuR binding to claudin 1 and occludin mRNAs, thus decreasing their translation. These results indicate that vtRNA2‐1 impairs intestinal barrier function by repressing HuR‐facilitated translation of claudin 1 and occludin.
Keywords: gut permeability, mucosal defense, posttranscriptional regulation, RNA‐binding proteins, vault RNAs
Subject Categories: Cell Adhesion, Polarity & Cytoskeleton; Molecular Biology of Disease; RNA Biology
Vault RNAs (vtRNAs) are small noncoding RNAs, and their levels increase in intestinal mucosa from patients with IBD. vtRNA2‐1 disrupts intestinal epithelial barrier function by decreasing intercellular junctions via interaction with HuR.

Introduction
Mammalian genomes transcribe a vast number of noncoding RNAs (ncRNAs) with active roles in gene regulation, whereas protein‐coding transcripts account for only a minority (< 2%) of the expressed RNAs (Batista & Chang, 2013). Vault RNAs (vtRNAs) are small (~100 nucleotide long) ncRNAs transcribed by RNA polymerase III and associated with giant cytoplasmic ribonucleoprotein (RNP) particles termed vaults that contain major vault proteins (MVPs; Kedersha & Rome, 1986). vtRNAs are highly conserved across mammalian genomes and expressed in a broad spectrum of eukaryotes. Humans express four vtRNA paralogs (vtRNA1‐1, vtRNA1‐2, vtRNA1‐3, and vtRNA2‐1), while mice only produce one vtRNA (Berger et al, 2009; Stadler et al, 2009). An increasing body of evidence indicates that vtRNAs have important roles outside of vault RNPs and can function independently of MVPs and vault particles (Kickhoefer et al, 1998; Amort et al, 2015; Li et al, 2015; Horos et al, 2019). vtRNAs have been linked to many cellular processes, including mRNA splicing, nuclear transport, drug resistance, synaptogenesis, lysosome function, apoptosis, and tumorigenesis (Kickhoefer et al, 1998; Berger et al, 2009; Kolev et al, 2019; Bracher et al, 2020; Wakatsuki et al, 2021; Wakatsuki & Araki, 2021; Ferro et al, 2022), although only about 5% of the total cellular vtRNAs is incorporated into vaults (Kedersha & Rome, 1986; Kickhoefer et al, 1998). It has been reported that the levels of vtRNAs increase in response to viral infection and a rise in free vtRNA levels favors the replication of the influenza virus via deactivation of RNA‐dependent protein kinase (Li et al, 2015). The free vtRNA1‐1 is shown to regulate selective autophagy by directly interacting with RNA‐binding protein (RBP) p62 (Horos et al, 2019). However, the functions of these abundant small ncRNAs, particularly their involvement in intestinal epithelium homeostasis and pathologies, have been largely unexplored.
The epithelium lines the luminal surface of the intestinal mucosa and functions as a physical and biochemical barrier that protects the subepithelial tissues against a wide array of luminal noxious substances, pathogens, and the microbiome (Camilleri, 2019; Chung et al, 2022). The epithelial barrier is comprised of specialized structures consisting of various intercellular junction (IJ) proteins, including tight junctions (TJs) and adherens junctions (AJs), that surround the subapical region of epithelial cells (Turner, 2009; Peterson & Artis, 2014; Xiao et al, 2021b). The TJ is the most apical element of the junctional complex and establishes a selectively permeable barrier that supports nutrient absorption and prevents even small molecules from leaking between cells (Turner, 2009). TJ complexes are primarily composed of transmembrane proteins such as occludin and one or more members of the claudin family; these proteins also interact with a cytosolic plaque of TJ proteins such as ZO‐1 that associate with the cortical cytoskeleton (Turner, 2009; Peterson & Artis, 2014). Immediately below the TJs are the cadherin‐rich AJs that mediate strong cell‐to‐cell adhesion and play a critical role in forming and modulating the epithelial barrier (Furuse et al, 2014). The AJ E‐cadherin integrates different extracellular and intracellular signals and is crucial for maintaining the stability and effectiveness of the epithelial barrier (Liu et al, 2009; Bhatt et al, 2013). The assembly of TJ and AJ complexes is highly dynamic, and their constituent proteins undergo continuous remodeling and turnover at a rapid pace, which is tightly controlled by different factors and signals at multiple levels (Turner, 2009; Xiao et al, 2021b; Chung et al, 2022). Nonetheless, gut barrier dysfunction occurs commonly in patients with critical illnesses, leading to the translocation of luminal toxic substances and bacteria to the bloodstream (Haussner et al, 2019; Kumar et al, 2020).
Located on chromosome 5q31.1, vtRNA2‐1 (also called nc886) is a 108‐nucleotide long ncRNA that can be incorporated into vault particles or remain free in the cytoplasm (Nandy et al, 2009; Treppendahl et al, 2012). As a highly abundant RNA, vtRNA2‐1 is implicated in different pathological processes and can have either tumor‐suppressive or oncogenic functions, although these observations are still controversial and may be cell type‐dependent (Lee et al, 2011, 2014; Cao et al, 2013; Fort et al, 2020). The expression levels of vtRNA2‐1 are primarily regulated by the extent of promoter methylation, as shown when elevated methylation of the vtRNA2‐1 promoter in maternal blood lowered vtRNA2‐1 content associated with preterm birth (You et al, 2021). In acute myeloid leukemia, patients with hypomethylation of both alleles of the vtRNA2‐1 promoter have a significantly better prognosis, whereas patients with hypermethylation or loss of the second vtRNA2‐1 copy have a poorer outcome (Treppendahl et al, 2012). In this study, we provide evidence that the expression patterns of vtRNAs in the intestinal mucosa change significantly in response to stress and that human intestinal mucosa from patients with inflammatory bowel disease (IBD) exhibited increased levels of vtRNAs. We further discovered that vtRNA2‐1 inhibited IJ expression posttranscriptionally and led to gut barrier dysfunction and that vtRNA2‐1 represses the translation of claudin 1 and occludin by disrupting the interaction of the respective mRNAs with the RBP HuR. These findings support the notion that vtRNA2‐1 is a negative regulator of the intestinal epithelium homeostasis and point to vtRNA2‐1 as a novel therapeutic target for interventions to protect the epithelial barrier function in the clinical setting.
Results
Changes in the levels of vtRNA in intestinal epithelium responding to critical stress
To examine the involvement of vtRNAs in pathologies of the intestinal mucosa, two murine models, dextran sodium sulfate (DSS)‐induced colitis and septic stress induced by cecal ligation and puncture (CLP), were used in this study. As we and others reported previously (Liu et al, 2017; Heidari et al, 2021), administration of 3% DSS in drinking water for 5 days caused damage to the colon epithelium, bloody diarrhea, and granulocyte infiltration in mice, mimicking the injury/erosions and inflammation observed in human ulcerative colitis (UC). As shown in Fig 1A, mucosal vtRNA abundance increased dramatically in the colons of DSS‐treated mice, achieving 6‐fold higher levels than those observed in control animals; the specificity of this response was supported by the observation that DSS treatment did not alter colon mucosal levels of long ncRNA uc.417. As reported previously (Hubbard et al, 2005; Wang et al, 2018), exposure of mice to CLP for 24 h led to acute gut barrier dysfunction, as indicated by an increased mucosal permeability to fluorescein isothiocyanate (FITC)‐dextran. CLP stress for 24 h also resulted in focal villous necrosis and erosions in the small intestinal mucosa. Mucosal levels of vtRNA also increased remarkably in the small intestine of CLP‐mice, without affecting the abundance of tissue uc.417 (Fig 1B), supporting the specificity of the increase in vtRNA in the small intestinal mucosa responding to CLP stress. On the other hand, sham operation did not significantly affect mucosal vtRNA levels in the small intestine compared with mice without any surgical operation.
Figure 1. Mucosal vtRNA expression associated with intestinal pathologies.
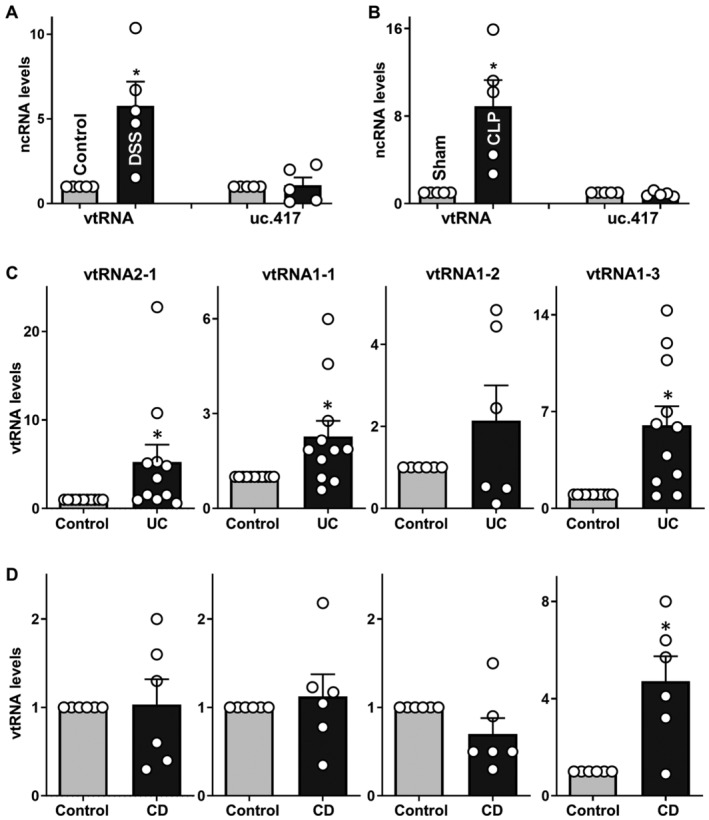
- Levels of mucosal vtRNA and uc.417 in the colons of mice treated with 3% DSS for 5 days, as measured by RT‐Q‐PCR analysis. Values are the means ± SEM (n = 5 mice per group). Unpaired, two‐tailed Student's t‐test was used. *P < 0.05 compared with control mice.
- Levels of mucosal vtRNA and uc.417 in the small intestines of mice exposed to CLP for 24 h. Values are the means ± SEM (n = 5 mice per group). Unpaired, two‐tailed Student's t‐test was used. *P < 0.05 compared with sham mice.
- Levels of mucosal vtRNA2‐1, vtRNA1‐1, vtRNA1‐2, and vtRNA1‐3 in the colon of patients with active ulcerative colitis (UC). Values are the means ± SEM (n = 6 per group). Unpaired, two‐tailed Student's t‐test was used. *P < 0.05 compared with controls.
- Levels of tissue vtRNAs in the ileal mucosa of patients with Crohn's disease (CD). Values are the means ± SEM (n = 6 per group). Unpaired, two‐tailed Student's t‐test was used. *P < 0.05 compared with controls.
To determine the clinical implication of altered vtRNA abundance, we measured the levels of all four human vtRNAs in intestinal mucosal tissues from patients. Colonic and ileal mucosal tissues from patients with active UC and Crohn's disease (CD) were collected to measure vtRNA levels; mucosal samples from healthy individuals without gut mucosal injury/erosions, inflammation, or barrier dysfunction served as controls. As shown in Fig 1C, compared to control tissues, colonic mucosal tissues from patients with active UC exhibited increased levels of vtRNA2‐1, vtRNA1‐1, and vtRNA1‐3 but not vtRNA1‐2. The levels of mucosal vtRNA2‐1, vtRNA1‐1, and vtRNA1‐3 in the colon of UC patients were increased by ~5.1‐, ~2.2‐, and ~5.9‐fold, respectively, compared with controls. In contrast to the findings in UC, only vtRNA1‐3 levels increased significantly in ileal mucosal tissues obtained from CD patients compared to control tissues, while the levels of vtRNA2‐1, vtRNA1‐1, and vtRNA1‐2 in ileal mucosa from CD patients were indistinguishable from those observed in the mucosa from control individuals (Fig 1D). Taken together, these studies conducted in mice and human tissues indicate that the expression levels of vtRNA in the intestinal epithelium increase significantly in response to various critical stresses, strongly supporting the wide implication of vtRNAs in intestinal epithelium homeostasis and pathologies.
vtRNA2‐1 disrupts intestinal epithelial barrier function by lowering TJs and AJ in vitro
Since increased gut permeability occurs commonly in patients with IBD and sepsis and leads to complex pathologies (Turner, 2009; Haussner et al, 2019), we examined the role of altered vtRNAs in the control of intestinal epithelial barrier function. Although the four human vtRNAs are similar and differ slightly in primary and secondary structures, they appear to have distinct biological functions via different targets and mechanisms (Stadler et al, 2009; Horos et al, 2019; Fort et al, 2020). To gain a deeper molecular understanding, we focused specifically on vtRNA2‐1, as this vtRNA changed significantly in the mucosal tissues of UC patients, there is earlier evidence of its possible influence on the epithelial barrier (Lee et al, 2011, 2014; Kong et al, 2015; Fort et al, 2020), and vtRNA2‐1 was a predicted ligand for RBPs including HuR. Two sets of studies were carried out in Caco‐2 cells, a widely accepted cell culture model for intestinal epithelial barrier studies (Guo et al, 2005; Zhang et al, 2020). First, we examined the effect of increasing the levels of vtRNA2‐1 on the expression of TJ and AJ proteins. As shown in Fig 2A, the levels of cellular vtRNA2‐1 were increased by transfection of a plasmid expressing vtRNA2‐1 under control of the pCMV promoter (pcDNA3.1 backbone); 48 h later, vtRNA2‐1 levels increased strongly and specifically, but other vtRNAs such as vtRNA1‐1, vtRNA1‐2, and vtRNA1‐3 did not. Increasing the levels of vtRNA2‐1 specifically inhibited expression of TJs claudin 1 and occludin and AJ E‐cadherin (Fig 2B and Appendix Fig S1A) but failed to alter the expression levels of TJs ZO‐1 or JAM‐A, or the vault protein MVP. Consistent with these findings, immunostaining of claudin 1, occludin, and E‐cadherin, predominantly localized on the plasma membrane of Caco‐2 cells, also decreased significantly after ectopic vtRNA2‐1 overexpression (Fig 2C, top and middle). On the other hand, there were no differences in the intensity of ZO‐1 immunostaining between cells transfected with vtRNA2‐1 and cells transfected with control vector (Fig 2C, bottom). Importantly, elevation of the vtRNA2‐1 levels disrupted the epithelial barrier function, as evidenced by a decrease in transepithelial electrical resistance (TEER) values (Fig 2D, left) and an increase in the levels of paracellular flux of FITC‐dextran (Fig 2D, right). On the other hand, vtRNA2‐1 overexpression did not affect cell viability (Appendix Fig S1B).
Figure 2. Ectopically overexpressed vtRNA2‐1 disrupts intestinal epithelial barrier function in vitro .
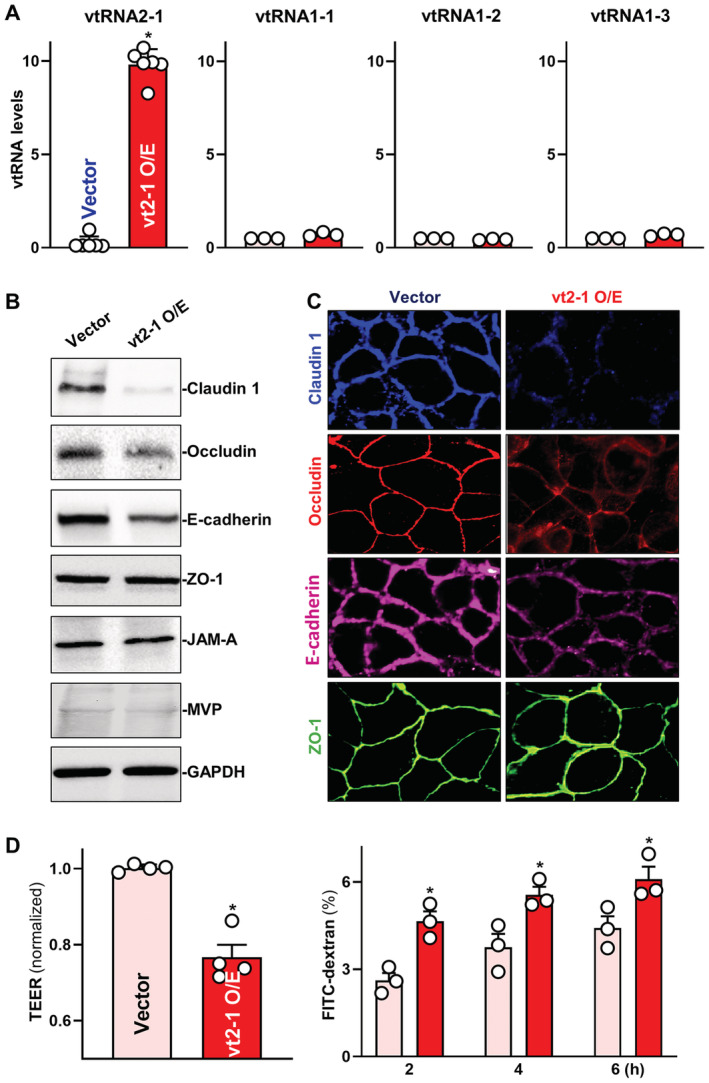
- Levels of vtRNAs in Caco‐2 cells 48 h after transfection with vtRNA2‐1 expression vector (vt2‐1). Values are the means ± SEM (n = 6 or 3 biological replicates). Unpaired, two‐tailed Student's t‐test was used. *P < 0.05 compared with control vector.
- Expression levels of claudin 1, occludin, E‐cadherin, ZO‐1, JAM‐A, and MVP in cells described in (A). Hsc70 immunoblotting was performed as an internal control for equal loading.
- Distribution of claudin 1, occludin, E‐cadherin, and ZO‐1 in cells treated as described in (A). Forty‐eight hours after transfection, cells were fixed, permeabilized, and incubated first with antibodies against different intercellular junction proteins and then with FITC‐conjugated anti‐IgG. Original magnification, ×500. Three separate experiments were performed and showed similar results.
- Changes in TEER (left) and FITC‐dextran paracellular permeability (right) in cells treated as described in (A). TEER assays were performed on 12‐mm Transwell filters; paracellular permeability was assayed by adding the membrane‐impermeable trace molecule FITC‐dextran to the insert medium. Values are the means ± SEM (n = 3 biological replicates). Unpaired, two‐tailed Student's t‐test was used. *P < 0.05 compared with control vector.
Second, we examined the effect of vtRNA2‐1 silencing on TJ/AJ expression and epithelial barrier function. As shown in Fig 3A, the levels of cellular vtRNA2‐1 decreased specifically 48 h after transfection with siRNA targeting vtRNA2‐1 (si‐vt2‐1); there were no significant differences in the levels of vtRNA1‐1, vtRNA1‐2 and vtRNA1‐3 between cells transfected with si‐vt2‐1 and cells transfected with control siRNA (C‐siRNA). This decrease in vtRNA2‐1 levels by si‐vt2‐1 transfection increased the expression of claudin 1 and E‐cadherin, although it slightly reduced ZO‐1 levels and failed to alter the levels of occludin and JAM‐A (Fig 3B and Appendix Fig S1C). Moreover, vtRNA2‐1 silencing enhanced epithelial barrier function, since it increased TEER (Fig 3C, left) and decreased paracellular flux of FITC‐dextran (Fig 3C, right). These results indicate that vtRNA2‐1 inhibits the expression of claudin 1, occludin, and E‐cadherin, thus contributing to dysfunction of the epithelial barrier.
Figure 3. vtRNA2‐1 silencing improves intestinal epithelial barrier function in vitro .
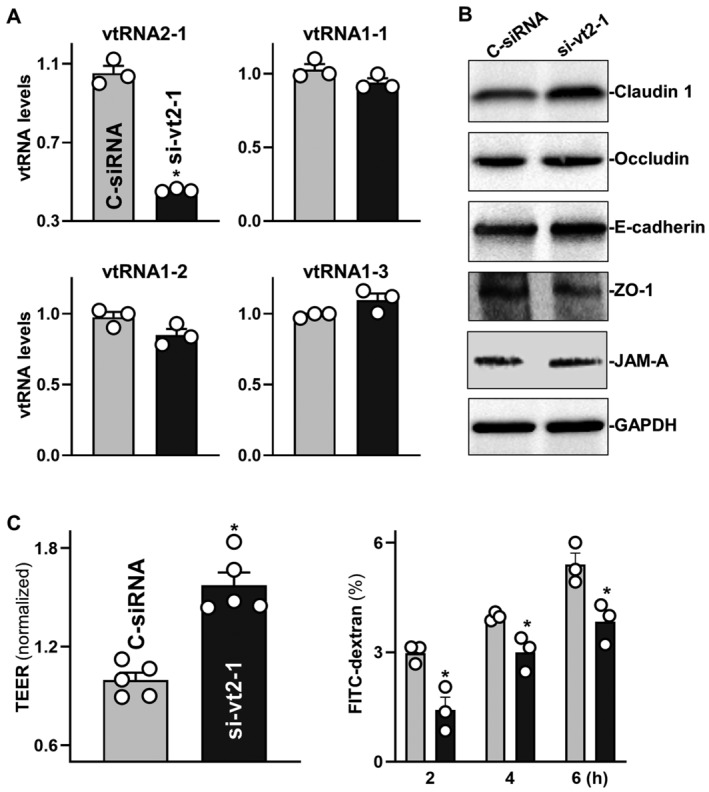
- Levels of vtRNAs in Caco‐2 cells 48 h after transfection with the siRNA targeting vtRNA2‐1 (si‐vt2‐1) or control siRNA (C‐siRNA). Values are the means ± SEM (n = 3 biological replicates). Unpaired, two‐tailed Student's t‐test was used. *P < 0.05 compared with C‐siRNA.
- Immunoblots of claudin 1, occludin, E‐cadherin, ZO‐1, and JAM‐A in cells treated as described in (A). Hsc70 immunoblotting served as an internal control for equal loading. Three separate experiments were performed and showed similar results.
- Changes in TEER (left) and FITC‐dextran paracellular permeability (right) in cells treated as described in (A). Values are the means ± SEM (n = 3 biological replicates). Unpaired, two‐tailed Student's t‐test was used. *P < 0.05 compared with C‐siRNA.
vtRNA2‐1 reduces IJs and impairs epithelial renewal and Paneth cells ex vivo
In an ex vivo model consisting of the primary culture of intestinal organoids derived from mouse small intestinal mucosa, elevation of the levels of the human vtRNA2‐1 ortholog also decreased claudin 1, occludin, and E‐cadherin without the effect on ZO‐1 content, as measured by immunostaining assays. As shown in Fig 4A, transfection of intestinal organoids with the vtRNA2‐1 expression vector increased the levels of vtRNA2‐1, although it did not alter the levels of endogenous mouse vtRNA or U6 RNA (Appendix Fig S2A). In the control group, claudin 1 and occludin clearly lined the basolateral and apical membrane regions, with slight cytoplasmic staining present in some organoids (Fig 4B, left). E‐cadherin predominantly lined the basolateral membrane areas and was noticeably absent from the cytoplasm and apical region of cells, while ZO‐1 was limited primarily at or near the apical membrane (Fig 4B, right). Consistent with the observations in Caco‐2 cells, ectopically expressed vtRNA2‐1 also inhibited expression of claudin 1, occludin, and E‐cadherin, as shown by a significant decrease in the levels of immunostaining intensity of claudin 1, occludin, and E‐cadherin in organoids transfected with vtRNA2‐1 expression vector relative to organoids transfected with control vector (Fig 4B, bottom). In contrast, vtRNA2‐1 overexpression did not alter ZO‐1 levels or its subcellular distribution in intestinal organoids.
Figure 4. Ectopically overexpressed vtRNA2‐1 decreases intercellular junction levels and lowers epithelial renewal and Paneth cell function ex vivo .
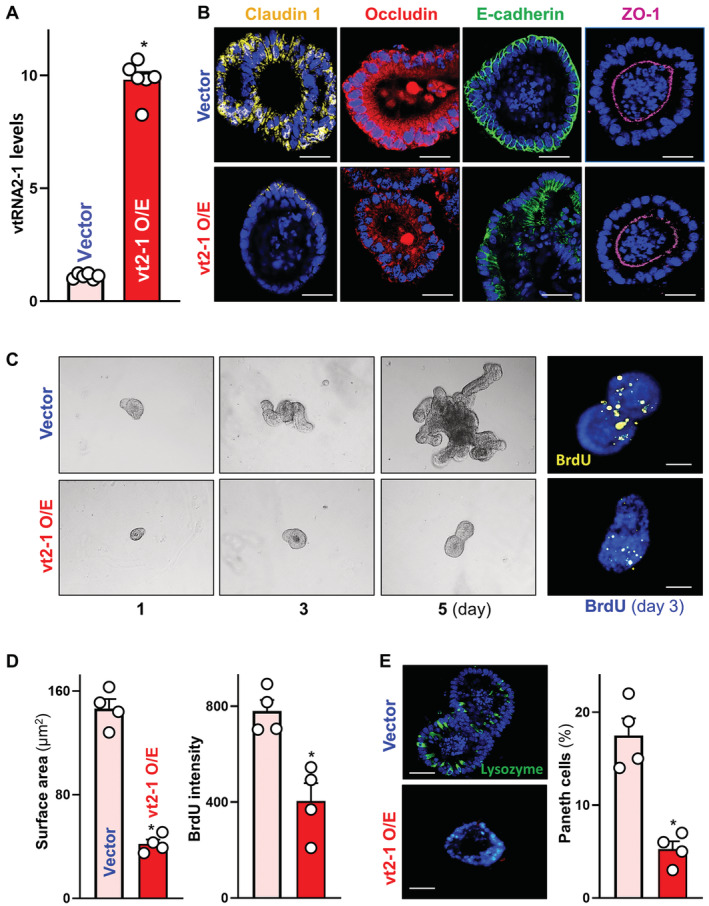
- Levels of vtRNA2‐1 in primarily cultured intestinal organoids 48 h after transfection with vtRNA2‐1 expression vector (vt2‐1). Organoids were derived from proximal small intestine and grown in the indicated conditions. Values are the means ± SEM (n = 6 biological replicates). Unpaired, two‐tailed Student's t‐test was used. *P < 0.05 compared with control vector.
- Images of immunofluorescence staining of claudin 1, occludin, E‐cadherin, and ZO‐1 in intestinal organoids treated as described in (A). Scale bars, 100 μm. Three separate experiments showed similar results.
- Sizes of intestinal organoids at different times after vtRNA2‐1 overexpression. Left, bright field microscopy analysis of growth of organoids; right, confocal analysis of BrdU (yellow) and DAPI (blue) on day 3 after culture. Scale bars, 100 μm.
- Quantitative data of surface area of organoids (left) and BrdU (right) in organoids treated as described in (C). Values are means ± SEM (n = 4 biological replicates). Unpaired, two‐tailed Student's t‐test was used. *P < 0.05 compared with control vector.
- Immunostaining (left) of Paneth (lysozyme‐positive) cells and quantitative data (right) in intestinal organoids treated as described in (A). Lysozyme: green. Scale bars, 100 μm. Values are means ± SEM (n = 4 biological replicates). Unpaired, two‐tailed Student's t‐test was used. *P < 0.05 compared with control vector.
In addition, elevation of the vtRNA2‐1 levels inhibited the growth of intestinal organoids as shown in Fig 4C. Intestinal organoids were initially grown from tiny proliferating crypts, but after 5 days in culture, the organoids consisted of multiple cells and buds in both control and vtRNA2‐1‐transfected organoids. However, ectopically expressing vtRNA2‐1 markedly reduced the sizes of organoids and inhibited DNA synthesis in multiple cells, as indicated by a reduction in their surface areas (Fig 4D, left) and a decrease in BrdU incorporation (Fig 4D, right) in the organoids transfected with vtRNA2‐1 expression vector, compared with organoids transfected with control vector. Moreover, vtRNA2‐1 overexpression led to Paneth cell defects in intestinal organoids (Fig 4E), as determined by using lysozyme‐immunostaining assays. Lysozyme‐positive cells in control organoids were highly enriched, but they decreased dramatically and were almost undetectable in the organoids transfected with vtRNA2‐1 expression vector. These results indicate that increased vtRNA2‐1 inhibits TJ/AJ expression and lowers organoid growth and Paneth cell function ex vivo.
Induced vtRNA2‐1 increases the vulnerability of the gut barrier to septic stress in mice
In an effort to define the in vivo importance of vtRNA2‐1 in regulating intestinal barrier function, we increased the levels of the human vtRNA2‐1 ortholog by infecting mice with a lentiviral vector expressing vtRNA2‐1 (lenti‐vtRNA2‐1) under the control of the suCMV‐promoter as described (Xiao et al, 2021a). By 5 days after infection with lenti‐vtRNA2‐1, there was a sustained increase (~2.5‐fold) in the levels of vtRNA2‐1 in the intestinal mucosa (Fig 5A) compared with those from animals infected with the control lentiviral vector. Consistent with the findings obtained from cultured Caco‐2 cells and organoids, ectopic expression of vtRNA2‐1 also decreased the levels of claudin 1 and occludin in the small intestinal mucosa, but it failed to alter mucosal abundances of JAM‐A, ZO‐1, and E‐cadherin (Fig 5B and Appendix Fig S2B). In contrast to the results observed in culture findings observed in vitro, increasing the levels of vtRNA2‐1 in the mucosa did not affect the basal level of gut permeability in sham mice (Fig 5C, left), as examined by FITC‐dextran flux assay.
Figure 5. Elevation of mucosal vtRNA2‐1 enhances gut barrier dysfunction in mice exposed to septic stress.
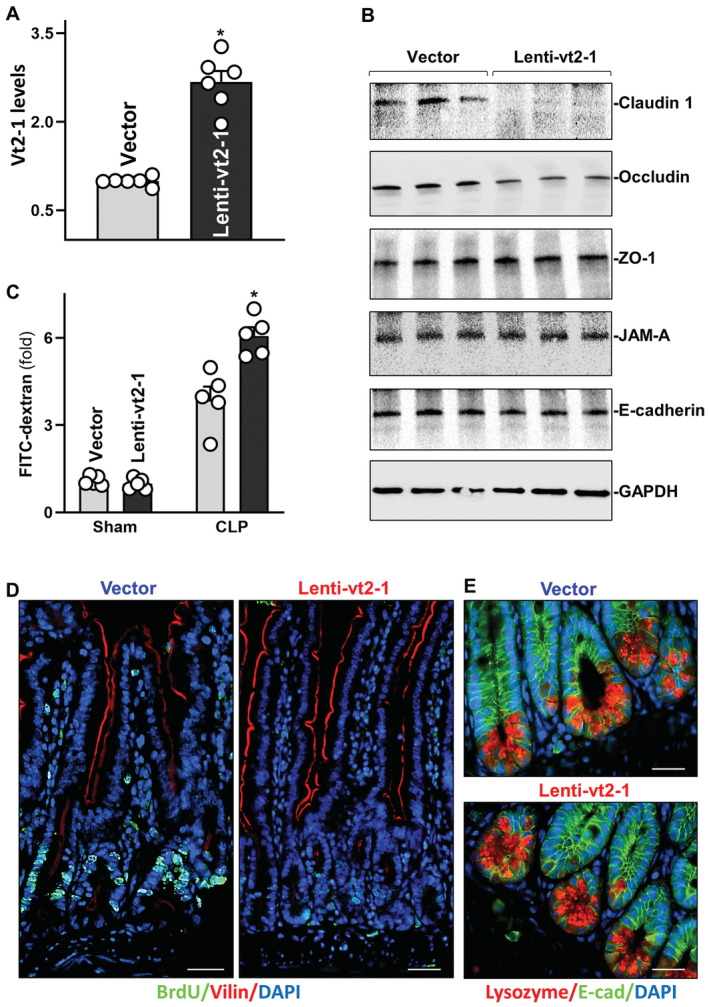
- Levels of vtRNA2‐1 in the small intestinal mucosa of mice on day 5 after intraperitoneal injection with a recombinant vtRNA2‐1 lentiviral expression vector (Lenti‐vt2‐1) or empty control lentiviral vector (vector). Values are means ± SEM (n = 6 biological replicates). Unpaired, two‐tailed Student's t‐test was used. *P < 0.05 compared with vector.
- Immunoblots of claudin 1, occludin, JAM‐A, ZO‐1, and E‐cadherin in the mucosa from mice treated as described in (A). GAPDH immunoblotting served as an internal control for equal loading.
- Changes in gut permeability in sham‐ and CLP (septic stress)‐mice. CLP was induced on day 5 after injection with lenti‐vt2‐1 or control vector. FITC‐dextran was given orally 20 h after CLP, and blood samples were collected 4 h later. Values are means ± SEM (n = 5 biological replicates). Unpaired, two‐tailed Student's t‐test was used. *P < 0.05 compared with vector.
- Proliferating cells as measured by BrdU labeling assays in the mucosa from mice treated as described in (A). BrdU (1 h post‐injection, S phase): green. Scale bars, 25 μm. Experiments were repeated in four mice injected with either lenti‐vt2‐1 or control vectors and showed similar results.
- Paneth cells (lysozyme‐positive cells) in the small intestinal mucosa from mice treated as described in (A). Lysozyme: red. Scale bars, 25 μm.
To determine if induction in the levels of mucosal vtRNA2‐1 affected the vulnerability of the gut barrier under critical pathological conditions, we subjected the mice to septic stress using CLP model as described (Hubbard et al, 2005). Exposure of mice to CLP for 24 h led to an acute gut barrier dysfunction in both the lenti‐vtRNA2‐1‐infected mice and mice infected with the control lentiviral vector, as indicated by an increase in gut mucosal permeability to FITC‐dextran. Importantly, however, the increase in gut permeability in mice infected with lenti‐vtRNA2‐1 was much higher than that observed in mice infected with control vector after exposure to CLP (Fig 5C, right). These results reveal that elevating the levels of tissue vtRNA2‐1 increases the vulnerability of the gut barrier to septic stress in mice.
In agreement with the results from intestinal organoids, vtRNA2‐1 overexpression in mice also inhibited growth of the small intestinal mucosa, as demonstrated by a significant decrease in the proliferating crypt cell population (marked by BrdU) in lenti‐vtRNA2‐1‐infected mice compared with animals infected with control vector (Fig 5D). The numbers of BrdU‐positive cells in crypts of the intestinal mucosa decreased by ~85% in lenti‐vtRNA2‐1‐infected mice relative to controls. Unexpectedly, increasing the levels of vtRNA2‐1 did not affect Paneth cell function in the small intestine of mice (Fig 5E). There were no significant changes in the numbers of lysozyme‐positive cells in the mucosa between lentivtRNA2‐1‐infected mice and mice infected with control vector. These results strongly suggest that vtRNA2‐1 is a potent inhibitor of intestinal epithelial renewal but plays a smaller role in the regulation of Paneth cell function.
vtRNA2‐1 inhibits expression of claudin 1, occludin, and E‐cadherin posttranscriptionally
To investigate the mechanisms underlying the repression of claudin 1, occludin, and E‐cadherin by vtRNA2‐1, we examined the levels of mRNAs encoding these IJ proteins in cells overexpressing vtRNA2‐1. Ectopically expressed vtRNA2‐1 did not alter the levels and stability of claudin 1, occludin, and JAM‐A (Fig 6A and B, and Appendix Fig S3A), but it decreased the levels of E‐cadherin mRNA by enhancing its degradation, since ectopic vtRNA2‐1 overexpression selectively lowered the half‐life of E‐cadherin mRNA. To test the possibility that vtRNA2‐1 regulates expression of claudin 1 and occludin at the translational level, we used the Click‐iT technology (Xiao et al, 2019). The levels of newly synthesized claudin 1 and occludin proteins decreased significantly in cells overexpressing vtRNA2‐1 relative to cells transfected with control vector (Fig 6C). The translational rates of claudin 1 and occludin in vtRNA2‐1‐transfected cells decreased by ~85 and ~50%, respectively, compared with those from cells transfected control vector (Fig 6D). The repression of claudin 1 and occludin protein synthesis by vtRNA2‐1 was specific since there were no changes in the levels of nascent GAPDH synthesis after vtRNA2‐1 overexpression.
Figure 6. vtRNA2‐1 inhibits translation of claudin 1 and occludin and enhances decay of E‐cadherin mRNA.
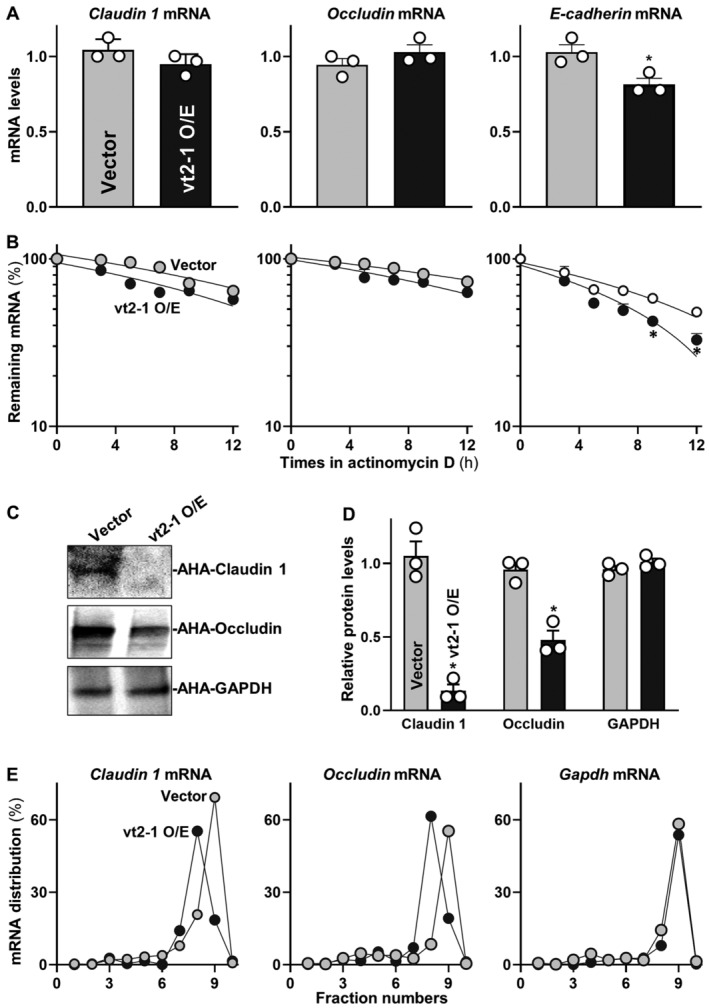
- Levels of claudin‐1, occludin, and E‐cadherin mRNAs in Caco‐2 cells 48 h after transfection with vtRNA2‐1 expression vector. Values are means ± SEM (n = 3 biological replicates). Unpaired, two‐tailed Student's t‐test was used. *P < 0.05 compared with control vector.
- Stability of the mRNAs in cells treated as described in (A). The mRNA levels were examined at different times after administration with actinomycin D. Values are means ± SEM (n = 3 biological replicates). Unpaired, two‐tailed Student's t‐test was used. *P < 0.05 compared with control vector.
- Newly synthesized claudin 1 and occludin proteins in cells treated as described in (A). After cells were exposed to L‐azidohomoalaine (AHA), cell lysates were incubated with the reaction buffer containing biotin/alkyne reagent; the biotin‐alkyne‐azide‐modified protein complex was pulled down by paramagnetic streptavidin‐conjugated dynabeads.
- Quantitative data of immunoblots shown in (C). Values are means ± SEM (n = 3 biological replicates). Unpaired, two‐tailed Student's t‐test was used. *P < 0.05 compared with control vector.
- Distribution of claudin 1 (left), occludin (center), and GAPDH (right) mRNAs in each gradient fraction of polysomal profiles prepared from cells treated as described in (A).
To further investigate the role of vtRNA2‐1 in the regulation of claudin 1 and occludin translation, we measured the relative distribution of claudin 1 and occludin mRNAs in individual fractions from polyribosome gradients after the ectopic overexpression of vtRNA2‐1 as described (Zhang et al, 2017). Although increasing the levels of vtRNA2‐1 did not alter global polysomal profiles (Appendix Fig S3B), the association of claudin 1 and occludin mRNAs with actively translating fractions (fractions 8, 9) decreased in cells overexpressing vtRNA2‐1, along with an induction in the levels of claudin 1 and occludin mRNAs in low‐translating fractions (fractions 6, 7; Fig 6E, left and middle). In contrast, GAPDH mRNA, encoding a housekeeping protein, distributed similarly in both groups (Fig 6E, right). Together, these data indicate that vtRNA2‐1 destabilizes E‐cadherin mRNA, but it inhibits the translation of claudin 1 and occludin without affecting the stability of their respective mRNAs.
vtRNA2‐1 represses translation of claudin 1 and occludin by interacting with HuR
Since HuR was found to directly interact with claudin 1 and occludin mRNAs via the 3′‐untranslated regions (UTRs) and regulates their translation (Yu et al, 2011; Xiao et al, 2016), we tested the possibility that vtRNA2‐1 represses translation of claudin 1 and occludin by altering HuR association with claudin 1 and occludin mRNAs. First, we synthesized biotin‐labeled vtRNA2‐1 and examined the direct interaction of vtRNA2‐1 with HuR in an in vitro system. Cytoplasmic lysates isolated from Caco‐2 cells were initially incubated with biotinylated vtRNA2‐1 as described (Xiao et al, 2016), and then, the levels of HuR and other proteins in the pull‐down material were assessed by Western blot analysis. As shown in Fig 7A, biotinylated vtRNA2‐1 specifically bound to HuR but did not bind AU‐binding factor 1 (AUF1), claudin 1, occludin, and E‐cadherin, as determined by biotin pull‐down analysis. Like vtRNA1‐1 (Horos et al, 2019), vtRNA2‐1 also bound to p62. To further examine the association of endogenous vtRNA2‐1 with endogenous HuR, we performed RNP immunoprecipitation (RIP) assays using anti‐HuR and control IgG antibodies, followed by isolation of bound RNA in both RIP reactions. After reverse transcription, Q‐PCR analysis was used to measure the levels of vtRNA enrichment in the HuR IP relative to IgG IP, as described (Zou et al, 2006). Results in Fig 7B (top) showed that vtRNA2‐1 was highly enriched in HuR IP compared to control IgG IP. In contrast, there were no significant changes in the levels of vtRNA1‐1 in HuR IP samples (Fig 7B, bottom).
Figure 7. vtRNA2‐1 inhibits translation of claudin 1 and occludin via interaction with HuR.
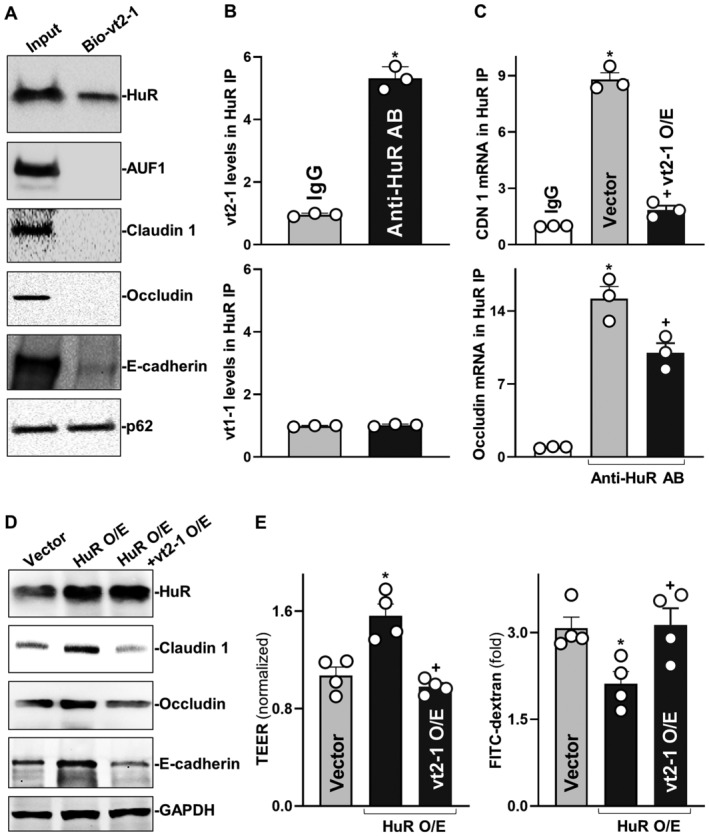
- Association of vtRNA2‐1 with HuR in Caco‐2 cells as measured by biotin‐labeled RNA pull‐down assays. After cytoplasmic lysates were incubated with biotinylated vtRNA2‐1, levels of HuR and other proteins in the pull‐down material were assessed. Three separate experiments showed similar results.
- Association of endogenous HuR with endogenous vtRNA2‐1 (top) and vtRNA1‐1 (bottom). After immunoprecipitation (IP) of RNA‐protein complexes from cell lysates using anti‐HuR antibody (AB) or control IgG, RNAs were isolated and measured by RT‐Q‐PCR analysis. Values are means ± SEM (n = 3 biological replicates). Unpaired, two‐tailed Student's t‐test was used. *P < 0.05 compared with IgG.
- Effect of increasing the levels of vtRNA2‐1 on HuR binding to claudin 1 (CDN 1; top) and occludin (bottom) mRNAs. Twenty‐four hours after cells were transfected with vtRNA2‐1 expression vector, the levels of CLDN1 and occludin mRNAs in the materials pulled down by anti‐HuR AB were examined. Values are means ± SEM (n = 3 biological replicates). One‐way ANOVA was utilized with Tukey's post hoc test. *, + P < 0.05 compared with IgG or cells transfected with control vector, respectively.
- Changes in the expression levels of claudin‐1, occludin, and E‐cadherin proteins 48 h after cells were transfected with HuR expression vector alone or co‐transfected with HuR and vtRNA2‐1 expression vectors.
- Changes in TEER (left) and FITC‐dextran paracellular permeability (right) in cells treated as described in (D). Values are the means ± SEM (n = 4 biological replicates). One‐way ANOVA was utilized with Tukey's post hoc test. *, + P < 0.05 compared with cells transfected with control vector or cells transfected with HuR expression vector alone, respectively.
Second, we determined if increasing the level of cellular vtRNA2‐1 prevented HuR interaction with claudin 1 and occludin mRNAs. The levels of claudin 1 and occludin mRNAs were highly enriched in HuR IP samples, whereas ectopically expressed vtRNA2‐1 by transfecting cells with vtRNA2‐1 expression vector almost totally blocked HuR binding to claudin 1 mRNA (Fig 7C, top) and significantly decreased the levels of HuR/occludin mRNA complex (Fig 7C, bottom). vtRNA2‐1 overexpression did not affect total HuR abundance or its subcellular distribution. As shown in Fig 7D and Appendix Fig S3C, ectopically overexpressed HuR increased the levels of claudin 1 and occludin proteins but this induction was abolished by increasing vtRNA2‐1 in cells overexpressing HuR. The levels of claudin 1 and occludin proteins in cells co‐transfected with HuR and vtRNA2‐1 expression vectors were similar to those observed in cells transfected with control vector. Interestingly, increasing the levels of vtRNA2‐1 also prevented HuR binding to the E‐cadherin mRNA (Appendix Fig S4A) and blocked HuR‐induced expression of E‐cadherin (Fig 7D and Appendix Fig S4B).
Finally, we examined if the interaction between vtRNA2‐1 and HuR played a role in regulating epithelial barrier function in vitro. As anticipated, ectopically expressed HuR promoted the epithelial barrier function, as shown by increased TEER (Fig 7E, left) and decreased paracellular flux of FITC‐dextran (Fig 7E, right) in HuR‐transfected cells relative to cells transfected with control vector. However, this stimulation of epithelial barrier function by HuR was prevented by vtRNA2‐1 overexpression since there were no significant differences in the levels of TEER and paracellular flux of FITC‐dextran between cells co‐transfected with HuR and vtRNA2‐1 expression vectors and cells transfected with control vector. Because HuR is a potent and biological enhancer of the epithelial barrier function and its binding to target mRNAs is tightly regulated (Yu et al, 2011, 2013; Zhuang et al, 2013; Chung et al, 2018), these findings strongly suggest that increased vtRNA2‐1 disrupts the intestinal barrier function at least partially by preventing HuR association with claudin 1, occludin, and E‐cadherin mRNAs, thereby inhibiting their expression.
Discussion
Gut barrier dysfunction causes complex pathologies, especially in critically ill patients, leading to leaky gut and structural abnormalities of the epithelium, and, in some extreme cases, resulting in multiple organ dysfunction syndrome and death (Carter et al, 2013; Payen, 2020). Effective therapies to preserve gut barrier integrity are limited, because of poorly understood mechanisms of acute gut barrier dysfunction in various pathological conditions (Camilleri, 2019; Chung et al, 2022). Identifying the underlying causes and successful medical treatment has been a major challenge, and finding effective therapeutic targets to protect the barrier function is extremely important. Here we identified a small ncRNA, vtRNA2‐1, as a novel regulator of intestinal epithelial barrier by altering IJ expression at a posttranscriptional level. Increased levels of vtRNA2‐1 in the intestinal epithelium not only caused barrier dysfunction in culture, but it also increased the vulnerability of the gut barrier to septic stress in mice as a result of inhibition of IJ expression. Our results further revealed that vtRNA2‐1 represses the translation of claudin 1 and occludin by reducing the binding of HuR to claudin 1 and occludin mRNAs. These results represent a significant conceptual advance by linking vtRNAs with IJ expression and subsequent gut permeability and demonstrate the important impact of altering the relative levels of vtRNA2‐1 and HuR on the pathogenesis of gut barrier dysfunction in patients with critical illnesses.
The results reported here indicate that human intestinal mucosa epithelium expressed all four vtRNAs (vtRNA1‐1, vtRNA1‐2, vtRNA1‐3, and vtRNA2‐1) and that their levels increased dramatically in the mucosal tissues from patients with IBD, particularly in UC. Intestinal mucosal samples from mice with DSS‐induced colitis or CLP‐induced sepsis also exhibited increased levels of vtRNA, compared with the tissues from control animals. Overexpression and silencing experiments demonstrate that vtRNA2‐1 negatively affects the expression of claudin 1, occludin, and E‐cadherin in the intestinal epithelium. Our further results obtained from studies in cultured cells, intestinal organoids, and CLP in mice strongly support the notion that vtRNA2‐1 is a biological repressor of the intestinal epithelium homeostasis and that elevation of mucosal vtRNA2‐1 levels plays an important role in the pathogenesis of gut barrier dysfunction. Since ectopically expressed vtRNA2‐1 failed to alter the levels of MVP in cultured IECs, we propose that regulation of the intestinal epithelial barrier function by vtRNA2‐1 is independent of vault complex formation. In support of our findings, several studies have shown a critical role of vtRNA2‐1 in various cellular and disease processes (Treppendahl et al, 2012; Cao et al, 2013; Silver et al, 2015). Decreased levels of vtRNA2‐1 by elevated methylation of its promoter were associated with preterm birth (You et al, 2021) and poor outcomes in patients with acute myeloid leukemia and small lung cancer (Lee et al, 2011; Treppendahl et al, 2012). Inhibition of vtRNA2‐1 also enhanced apoptosis by increasing Bax expression in cervical cancer cells (Kong et al, 2015).
Our results also indicate that vtRNA2‐1 inhibits growth of the intestinal epithelium, although vtRNA2‐1 can act as a tumor suppressor or an oncogene depending on the particular cell type (Lee et al, 2011, 2014; Cao et al, 2013; Silver et al, 2015; Fort et al, 2020). There was a significant decrease in the proliferating crypt cell population in primary cultures of intestinal organoids and in small intestinal mucosa of mice after vtRNA2‐1 overexpression. This repression in intestinal mucosal growth may also contribute to vtRNA2‐1‐mediated epithelial barrier dysfunction observed in vitro and in vivo systems, since rapid and continuous intestinal epithelial renewal is crucial for maintaining the epithelial integrity that is essential for normal gut barrier structure and function (Xiao et al, 2016, 2019). Although ectopically overexpressed vtRNA2‐1 led to Paneth cell defects in intestinal organoids ex vivo, it did not affect Paneth cell function in mouse small intestinal mucosa in vivo. Unlike most IECs that are quickly turned over within a few days, Paneth cells are specialized IECs that are located at the base of small intestinal crypts and can persist for months in healthy individuals after maturation (Beumer & Clevers, 2021; Chung et al, 2021). On the other hand, Paneth cells develop rapidly with growth of intestinal organoids within days after primary culture (Lueschow & McElroy, 2020). The differences in the response of Paneth cells to vtRNA2‐1 overexpression between ex vivo and in vivo model systems suggest that vtRNA2‐1 inhibits the development of Paneth cells in the small intestinal mucosa but has minor role in regulating the activity of differentiated Paneth cells.
Our present results also suggest that vtRNA2‐1 regulates the translation of claudin 1 and occludin and the stability of E‐cadherin mRNA by interacting with HuR. HuR has three RNA recognition motifs through which it interacts with numerous mRNAs to modulate their translation and/or stability (Dixon et al, 2001; Li et al, 2020; Si et al, 2021). Our previous studies have revealed that HuR functions as a master regulator of TJ expression in the intestinal epithelium and that inhibition of HuR expression and disruption of its subcellular distribution or binding affinity for target mRNAs cause the epithelial barrier dysfunction in vitro as well as in vivo (Yu et al, 2011, 2013; Xiao et al, 2016). Intestinal epithelial tissue‐specific deletion of HuR in mice compromised gut mucosal regeneration (Liu et al, 2014), delayed wound healing (Liu et al, 2017), and impaired epithelium‐host defense by lowering Paneth cell function (Xiao et al, 2019). HuR also interacts with microRNAs such as miR‐195 (Zhuang et al, 2013; Kwon et al, 2021), long ncRNAs including H19 and SPRY4‐IT1 (Xiao et al, 2016; Zou et al, 2016), and circular RNA circPABPN1 (Li et al, 2020) to jointly regulate target transcripts in the intestinal epithelium antagonistically or synergistically. Although ectopically overexpressed vtRNA2‐1 failed to alter HuR levels or subcellular localization, it prevented HuR binding to the claudin 1, occludin, and E‐cadherin mRNAs and thus abolished HuR‐mediated stimulation of these IJ expression. In addition, HuR also regulates TJ expression via interaction with other RBPs such as CUG‐binding protein 1 (CUGBP1) and AU‐binding protein 1 (AUF1; Zou et al, 2010; Yu et al, 2013), but the effect of HuR interaction with CUGBP1 or AUF1 on vtRNA2‐1‐regulated IJ expression remains unknown. Because HuR is highly expressed in the intestinal epithelium and its binding affinity for target mRNAs is tightly regulated by ncRNAs in response to stress (Peterson & Artis, 2014; Xiao et al, 2021b), it is likely that vtRNA2‐1 affects gene regulatory programs governing IJ expression and epithelial barrier function at least partially by its association with HuR or other RBPs.
Our results have potential clinical relevance, because human intestinal mucosa with gut barrier dysfunction, injury/erosions, and inflammation from patients with IBD exhibited increased levels of vtRNAs including vtRNA2‐1. As reported (Lee et al, 2011; Treppendahl et al, 2012; Li et al, 2015; Horos et al, 2019; Wakatsuki et al, 2021), deregulated expression of vtRNAs occurs commonly in patients with different diseases including various cancers, viral infection, drug resistance, and preterm birth. Abnormalities in the abundance of tissue vtRNAs can alter many biological processes including autophagy, proliferation, and apoptosis and also influences cancer growth, viral replication, and host defense. In this study, we identified a novel role for vtRNA2‐1 in regulating intestinal epithelial barrier function. Increasing the levels of cellular vtRNA2‐1 inhibited IJ expression posttranscriptionally and increased the vulnerability of gut barrier to CLP‐induced septic stress. vtRNA2‐1 inhibits translation of claudin 1 and occludin and destabilizes E‐cadherim mRNA via its direct interaction with HuR, and this regulation of protein function by a ncRNA could represent a widespread general principle of biological control, complementing well‐established forms of regulation such as protein–protein interactions and posttranslational modification. In sum, our results indicate that vtRNA2‐1 regulates intestinal epithelial barrier function mainly by interacting with HuR. These findings help us to understand how vtRNAs and their interactions with RBPs, as seen for vtRNA2‐1/HuR, control gut permeability under various stressful environments and uncover novel therapeutic targets to protect the barrier integrity in pathologic conditions.
Materials and Methods
Chemicals and cell culture
Caco‐2 cells were purchased from the American Type Culture Collection (Manassas, VA) and were maintained under standard culture conditions (Zou et al, 2010; Liu et al, 2015). The culture medium and fetal bovine serum were purchased from Invitrogen (Carlsbad, CA), and biochemicals were from Sigma (St. Louis, MO). Antibodies recognizing claudin 1 (sc‐166338), occludin (sc‐133256), JAM‐A (sc‐56323), ZO‐1(sc‐33725), E‐cadherin (sc‐8426), lysozyme (sc‐518012), HuR (sc‐5261), AUF1 (07‐260), p62 (sc‐28359), HSC70 (MABE1120), and GAPDH (G9545) were obtained from Santa Cruz Biotechnology and BD Biosciences (Sparks, MD), while the antibody against MVP (MA513871) was from Invitrogen. The secondary antibody conjugated to horseradish peroxidase was purchased from Sigma. All antibodies utilized in this study were validated for species specificity. Antibody dilutions used for Western blots of claudin 1, occludin, JAM‐A, ZO‐1, E‐cadherin, HuR, AUF1, p62, HSP70, and GAPDH were 1:800 or 1,000 (first Ab) and 1:2,000 (second AB), respectively, whereas antibody dilutions for immunostaining were 1:200 (first) and 1:2,000 (second). Relative protein levels were analyzed by using Biorad Chemidoc and XRS system equipped with Image lab software (version 4.1). We also utilized “Quantity tool” to determine the band intensity volume; the values were normalized with internal loading control GAPDH.
Studies in murine and human tissues
Wild‐type C57BL/6J mice (male and female, 6–9 weeks old) were purchased from The Jackson Laboratory and housed in a pathogen‐free animal facility at the Baltimore VA Medical Center. All animal experiments were conducted in accordance with NIH guidelines and were approved by the Institutional Animal Care and Use Committee of University Maryland School of Medicine and Baltimore VA hospital. Acute colitis was induced in mice by adding 3% dextran sodium sulfate (DSS, MP Biomedicals) to the drinking water for 5 consecutive days as reported (Heidari et al, 2021). The colonic mucosal tissues were harvested for various chemical and histological analyses. To generate the model of cecal ligation and puncture (CLP)‐induced injury, mice were anesthetized by Nembutal, and CLP was performed as described (Hubbard et al, 2005). The distal portion of the cecum (1 cm) was ligated with 5‐0 silk suture. The ligated cecum was then punctured with a 25‐gauge needle and slightly compressed with an applicator until a small amount of stool appeared. In sham‐operated animals, the cecum was manipulated but without ligation and puncture and was placed back in the peritoneum. Twenty‐four hours after CLP, two 4‐cm segments taken from the middle of the small intestine were removed in each animal as described previously (Wang et al, 2018).
Human tissue samples were obtained from surplus discarded tissue from Department of Surgery, University of Maryland Health Science Center and commercial tissue banks. The study was approved by the University Maryland Institutional Review Board. All ileal and colonic mucosal tissue samples were divided into two portions, one for extraction of RNA and the other for histological examination. The procedures for RNA extraction and histological analysis were carried out according to the method described (Xiao et al, 2019).
Intestinal organoid culture
Isolation and culture of primary enterocytes were conducted following the method described previously (Lindemans et al, 2015; Xiao et al, 2018). Briefly, primary crypts were released from the small intestinal mucosa in mice; isolated crypts were mixed with matrigel (Corning 356231) and cultured in mouse IntestiCult™ organoid growth medium (Stemcell Technology 06005). The levels of DNA synthesis were measured by BrdU incorporation, and the growth of organoids was examined by measuring surface area of organoid horizontal cross sections using NIS‐Elements AR4.30.02 program.
Plasmid construction and RNA interference
Lenti‐vtRNA2‐1 was custom‐made by AMSBIO Inc., in which vtRNA2‐1 cDNA/GFP was subcloned into expression lentiviral vector via Eco cloning technology and expressed under the control of the suCMV‐promoter. Lenti‐vtRNA2‐1 and control lentiviral were packaged in lentiviral production cells, concentrated by ultracentrifugation, resuspended in PBS, and used at the dose of 2.5 × 107 IFU/25‐g body weight to increase vtRNA2‐1 in vivo as described (Xiao et al, 2016, 2021b). An expression vector containing vtRNA2‐1 cDNA under control of pCMV‐promoter was constructed and used to increase vtRNA2‐1 in Caco‐2 cells, whereas a vector containing a scrambled sequence of vtRNA2‐1 was used as control.
Expression of vtRNA2‐1 was silenced by transfection with specific small interfering RNA targeting vtRNA2‐1 (si‐vt2‐1) as described (Xiao et al, 2018). The si‐vt2‐1 and C‐siRNA (a scrambled version of si‐vt2‐1) were purchased from Santa Cruz Biotechnologies. For each 60‐mm cell culture dish, 15 μl of the 20 μM stock duplex si‐vt2‐1 or C‐siRNA was used. Forty‐eight hours after transfection using LipofectAMINE (Invitrogen 116668019), cells were harvested for analysis.
Q‐PCR and immunoblotting analyses
Total RNA was isolated by using the RNeasy mini kit (Qiagen, Valencia, CA) and used in reverse transcription (RT) and PCR amplification reactions as described (Zhang et al, 2020). Q‐PCR analysis was performed using Step‐one‐plus Systems with specific primers, probes, and software (Applied Biosystems, Foster City, CA). All primers used for Q‐PCR analysis were purchased from Thermo Fisher Scientific (Waltham, MA). The levels of Gapdh mRNA were assessed to monitor the evenness in RNA input in Q‐PCR analysis.
To examine protein levels, whole‐cell lysates were prepared using 2% SDS, sonicated, and centrifuged. The supernatants were boiled and size‐fractionated by SDS–PAGE. After transferring proteins onto nitrocellulose filters, the blots were incubated with primary antibody, following incubations with secondary antibody.
Biotin‐labeled vtRNA2‐1 pull‐down and RNP‐IP assays
Biotinylated‐RNA pull‐down assays were conducted as described previously (Xiao et al, 2016). After biotin‐labeled vtRNA2‐1 was incubated with cytoplasmic proteins at room temperature for 1 h, the mixture was mixed with Streptavidin‐Dynal beads (Invitrogen 35136) and incubated at 4°C on a rotator overnight. The beads were washed thoroughly, and the beads‐bound RNA was isolated and subjected to RT followed by Q‐PCR analysis. To examine association of vtRNA2‐1 with RBPs, levels of HuR and other proteins in the pull‐down material were assessed after cytoplasmic lysates were incubated with biotinylated vtRNA2‐1.
Immunoprecipitation (IP) of RNP complexes was carried out to assess the association of endogenous HuR with endogenous mRNAs encoding claudin 1 and occludin as described (Yu et al, 2020). Twenty million cells were collected per sample, and lysates were used for IP for 4 h at room temperature in the presence of excess (30 μg) IP antibody (IgG, anti‐HuR). RNA in IP materials was used in RT reactions followed by Q‐PCR analysis. The amplification of GAPDH mRNA, found in all samples as low‐level contaminating housekeeping transcripts (not HuR target), served to monitor the evenness of sample input, as reported previously (Yu et al, 2011).
Assays of newly translated protein and polysome analysis
Nascent claudin 1 and occludin proteins were detected by Click‐iT protein analysis detection kit (Life Technologies, C33372) and conducted following the company's manual. Briefly, cells were incubated in methionine‐free medium and then exposed to L‐azidohomoalanine (AHA). After mixing cell lysates with the reaction buffer for 20 min, the biotin‐alkyne/azide‐modified protein complex was pulled down using paramagnetic Streptavidin‐conjugated Dynabeads. The pull‐down material was resolved by 10% SDS–PAGE and analyzed by Western immunoblotting analysis using antibodies against claudin 1, occludin, or GAPDH.
Polysome analysis was carried out as described previously (Xiao et al, 2019). Briefly, cells at ~70% confluence were incubated in 0.1 mg/ml cycloheximide, then lifted by scraping in PEB lysis buffer. Nuclei were pelleted, and the resulting supernatant was centrifuged through a 10–50% linear sucrose gradient to fractionate cytoplasmic components according to their molecular weights. The eluted fractions were prepared with a fraction collector (Brandel, Gaithersburg, MD), and their quality was monitored at 254 nm using a UV‐6 detector (ISCO, Louisville, KY). After RNA in each fraction was extracted, the levels of each individual mRNA were quantified by Q‐PCR analysis in each of the fractions.
Immunofluorescence staining
The immunofluorescence staining procedure of Caco‐2 cells and intestinal organoids was carried out according to the method described in our previous publications (Xiao et al, 2021a). Slides were fixed in 3.7% formaldehyde in phosphate‐buffered saline and rehydrated. All slides were incubated with the primary antibody against different proteins in the blocking buffer at concentration of 1:200 or 1:300 dilution at 4°C overnight and then incubated with secondary antibody conjugated with Alexa Fluor‐594 (Invitrogen A20185) or Alexa Fluor‐488 (Invitrogen A11054) for 2 h at room temperature. After rinsing three times, the slides were incubated with 1 μM DAPI (Invitrogen D3571) for 10 min to stain cell nuclei. Finally, the slides were mounted and viewed through a Zeiss confocal microscope (model LSM710). Slides were examined in a blinded fashion by coding and decoded only after examination was completed. Images were processed using Photoshop software (Adobe, San Jose, CA).
Measurements of gut epithelial barrier function
The epithelial barrier function in vitro was examined by using the 12‐mm Transwell plate as described (Yu et al, 2013). FITC‐dextran (70 kDa; Sigma‐Aldrich), a membrane‐impermeable molecule, served as the paracellular tracer and was added to a final concentration of 0.25 mM to the apical bathing wells that contained 0.5 ml of medium. The basal bathing well had no added tracers and contained 1.5 ml of the same flux assay medium as in the apical compartment. All flux assays were performed at 37°C, and the basal medium was collected at different times after addition of the FITC‐dextran. The concentration of the FITC‐dextran in the basal medium was determined using a fluorescence plate reader with an excitation wavelength at 490 nm and an emission wavelength of 530 nm. Transepithelial electrical resistance (TEER) was measured with an epithelial voltmeter under open‐circuit conditions (WPI, Sarasota, FL) as described (Zou et al, 2016), and the TEER of all monolayers was normalized to that of control monolayers in the same experiment.
Gut permeability in vivo was determined by examining the appearance in blood of FITC‐dextran administered by gavage as described (Yu et al, 2011). Briefly, mice were gavaged with FITC‐dextran at a dose of 60 mg/100‐g wt 4 h before harvest. Blood sample was collected by cardiac puncture. The serum concentration of the FITC‐dextran was determined using a fluorescence plate reader as described above.
Statistical analysis
All values were expressed as the means ± SEM. Unpaired, two‐tailed Student's t‐test was used when indicated with P < 0.05 considered significant. When assessing multiple groups, one‐way ANOVA was utilized with Tukey's post hoc test (Harter, 1960). The statistical software used was GraphPad InStat Prism 9.0 (San Diego, CA). For non‐parametric analysis rank comparison, the Kruskal–Wallis test was conducted.
Author contributions
Xiang‐Xue Ma: Conceptualization; data curation; formal analysis; validation; visualization; methodology. Lan Xiao: Conceptualization; data curation; software; formal analysis; supervision; validation; visualization; methodology. Susan J Wen: Data curation; validation; methodology. Ting‐Xi Yu: Data curation; validation; visualization; methodology. Shweta Sharma: Data curation; formal analysis; validation; methodology. Hee K Chung: Data curation; validation; investigation; methodology. Bridgette Warner: Data curation; validation; methodology. Caroline G Mallard: Data curation; validation; methodology. Jaladanki N Rao: Conceptualization; data curation; validation; investigation; methodology. Myriam Gorospe: Conceptualization; investigation; writing – review and editing. Jian‐Ying Wang: Conceptualization; resources; data curation; software; formal analysis; supervision; funding acquisition; validation; investigation; visualization; methodology; writing – original draft; project administration; writing – review and editing.
Disclosure and competing interests statement
This author discloses the following: J‐YW is a Senior Research Career Scientist at the Biomedical Laboratory Research and Development Service (US Department of Veterans Affairs). The remaining authors declare that they have no conflict of interest.
Supporting information
Appendix
Acknowledgements
This work was supported by Merit Review Awards (to J‐YW; JNR) from US Department of Veterans Affairs; grants from National Institutes of Health (NIH; DK57819, DK61972, DK68491 to J‐YW); and funding from the National Institute on Aging‐Intramural Research Program, NIH (to MG).
EMBO reports (2023) 24: e54925
Data availability
No primary datasets have been generated and deposited.
References
- Amort M, Nachbauer B, Tuzlak S, Kieser A, Schepers A, Villunger A, Polacek N (2015) Expression of the vault RNA protects cells from undergoing apoptosis. Nat Commun 6: 7030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batista PJ, Chang HY (2013) Long noncoding RNAs: cellular address codes in development and disease. Cell 152: 1298–1307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger W, Steiner E, Grusch M, Elbling L, Micksche M (2009) Vaults and the major vault protein: novel roles in signal pathway regulation and immunity. Cell Mol Life Sci 66: 43–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beumer J, Clevers H (2021) Cell fate specification and differentiation in the adult mammalian intestine. Nat Rev Mol Cell Biol 22: 39–53 [DOI] [PubMed] [Google Scholar]
- Bhatt T, Rizvi A, Batta SP, Kataria S, Jamora C (2013) Signaling and mechanical roles of E‐cadherin. Cell Commun Adhes 20: 189–199 [DOI] [PubMed] [Google Scholar]
- Bracher L, Ferro I, Pulido‐Quetglas C, Ruepp MD, Johnson R, Polacek N (2020) Human vtRNA1‐1 levels modulate signaling pathways and regulate apoptosis in human cancer cells. Biomolecules 10: 614–651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camilleri M (2019) Leaky gut: mechanisms, measurement and clinical implications in humans. Gut 68: 1516–1526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao J, Song Y, Bi N, Shen J, Liu W, Fan J, Sun G, Tong T, He J, Shi Y et al (2013) DNA methylation‐mediated repression of miR‐886‐3p predicts poor outcome of human small cell lung cancer. Cancer Res 73: 3326–3335 [DOI] [PubMed] [Google Scholar]
- Carter SR, Zahs A, Palmer JL, Wang L, Ramirez L, Gamelli RL, Kovacs EJ (2013) Intestinal barrier disruption as a cause of mortality in combined radiation and burn injury. Shock 40: 281–289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung HK, Wang SR, Xiao L, Rathor N, Turner DJ, Yang P, Gorospe M, Rao JN, Wang JY (2018) α4 coordinates small intestinal epithelium homeostasis by regulating stability of HuR. Mol Cell Biol 38: e00631‐17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung HK, Xiao L, Jaladanki KC, Wang JY (2021) Regulation of Paneth cell function by RNA‐binding proteins and noncoding RNAs. Cells 10: 2107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung HK, Rao JN, Wang JY (2022) Regulation of gut barrier function by RNA‐binding proteins and noncoding RNAs. In Comprehensive pharmacology, Kenakin T (ed), Vol. 5, pp 194–213. Amsterdam: Elsevier; [Google Scholar]
- Dixon DA, Tolley ND, King PH, Nabors LB, McIntyre TM, Zimmerman GA, Prescott SM (2001) Altered expression of the mRNA stability factor HuR promotes cyclooxygenase‐2 expression in colon cancer cells. J Clin Invest 108: 1657–1665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferro I, Gavini J, Gallo S, Bracher L, Landolfo M, Candinas D, Stroka DM, Polacek N (2022) The human vault RNA enhances tumorigenesis and chemoresistance through the lysosome in hepatocellular carcinoma. Autophagy 18: 191–203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fort RS, Garat B, Sotelo‐Silveira JR, Duhagon MA (2020) vtRNA2‐1/nc886 produces a small RNA that contributes to its tumor suppression action through the microRNA pathway in prostate cancer. Noncoding RNA 6: 7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuse M, Izumi Y, Oda Y, Higashi T, Iwamoto N (2014) Molecular organization of tricellular tight junctions. Tissue Barriers 2: e28960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo X, Rao JN, Liu L, Zou T, Keledjian KM, Boneva D, Marasa BS, Wang JY (2005) Polyamines are necessary for synthesis and stability of occludin protein in intestinal epithelial cells. Am J Physiol Gastrointest Liver Physiol 288: G1159–G1169 [DOI] [PubMed] [Google Scholar]
- Harter HL (1960) Critical values for Duncan's new multiple range test. Biometrics 16: 671–685 [Google Scholar]
- Haussner F, Chakraborty S, Halbgebauer R, Huber‐Lang M (2019) Challenge to the intestinal mucosa during sepsis. Front Immunol 10: 891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heidari N, Abbasi‐Kenarsari H, Namaki S, Baghaei K, Zali MR, Ghaffari Khaligh S, Hashemi SM (2021) Adipose‐derived mesenchymal stem cell‐secreted exosome alleviates dextran sulfate sodium‐induced acute colitis by Treg cell induction and inflammatory cytokine reduction. J Cell Physiol 236: 5906–5920 [DOI] [PubMed] [Google Scholar]
- Horos R, Buscher M, Kleinendorst R, Alleaume AM, Tarafder AK, Schwarzl T, Dziuba D, Tischer C, Zielonka EM, Adak A et al (2019) The small non‐coding vault RNA1‐1 acts as a riboregulator of autophagy. Cell 176: 1054–1067 [DOI] [PubMed] [Google Scholar]
- Hubbard WJ, Choudhry M, Schwacha MG, Kerby JD, Rue LW III, Bland KI, Chaudry IH (2005) Cecal ligation and puncture. Shock 24: 52–57 [DOI] [PubMed] [Google Scholar]
- Kedersha NL, Rome LH (1986) Isolation and characterization of a novel ribonucleoprotein particle: Large structures contain a single species of small RNA. J Cell Biol 103: 699–709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kickhoefer VA, Rajavel KS, Scheffer GL, Dalton WS, Scheper RJ, Rome LH (1998) Vaults are up‐regulated in multidrug‐resistant cancer cell lines. J Biol Chem 273: 8971–8974 [DOI] [PubMed] [Google Scholar]
- Kolev NG, Rajan KS, Tycowski KT, Toh JY, Shi H, Lei Y, Michaeli S, Tschudi C (2019) The vault RNA of Trypanosoma brucei plays a role in the production of trans‐spliced mRNA. J Biol Chem 294: 15559–15574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong L, Hao Q, Wang Y, Zhou P, Zou B, Zhang YX (2015) Regulation of p53 expression and apoptosis by vault RNA2‐1‐5p in cervical cancer cells. Oncotarget 6: 28371–28388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar M, Leon Coria A, Cornick S, Petri B, Mayengbam S, Jijon HB, Moreau F, Shearer J, Chadee K (2020) Increased intestinal permeability exacerbates sepsis through reduced hepatic SCD‐1 activity and dysregulated iron recycling. Nat Commun 11: 483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon MS, Chung HK, Xiao L, Yu TX, Wang SR, Piao JJ, Rao JN, Gorospe M, Wang JY (2021) MicroRNA‐195 regulates tuft cell function in the intestinal epithelium by altering translation of DCLK1. Am J Physiol Cell Physiol 320: C1042–C1054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee K, Kunkeaw N, Jeon SH, Lee I, Johnson BH, Kang GY, Bang JY, Park HS, Leelayuwat C, Lee YS (2011) Precursor miR‐886, a novel noncoding RNA repressed in cancer, associates with PKR and modulates its activity. RNA 17: 1076–1089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee HS, Lee K, Jang HJ, Lee GK, Park JL, Kim SY, Kim SB, Johnson BH, Zo JI, Lee JS et al (2014) Epigenetic silencing of the non‐coding RNA nc886 provokes oncogenes during human esophageal tumorigenesis. Oncotarget 5: 3472–3481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F, Chen Y, Zhang Z, Ouyang J, Wang Y, Yan R, Huang S, Gao GF, Guo G, Chen JL (2015) Robust expression of vault RNAs induced by influenza a virus plays a critical role in suppression of PKR‐mediated innate immunity. Nucleic Acids Res 43: 10321–10337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li XX, Xiao L, Chung HK, Ma XX, Liu X, Song JL, Jin CZ, Rao JN, Gorospe M, Wang JY (2020) Interaction between HuR and circPABPN1 modulates autophagy in the intestinal epithelium by altering ATG16L1 translation. Mol Cell Biol 40: e00492‐19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindemans CA, Calafiore M, Mertelsmann AM, O'Connor MH, Dudakov JA, Jenq RR, Velardi E, Young LF, Smith OM, Lawrence G et al (2015) Interleukin‐22 promotes intestinal‐stem‐cell‐mediated epithelial regeneration. Nature 528: 560–564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L, Guo X, Rao JN, Zou T, Xiao L, Yu T, Timmons JA, Turner DJ, Wang JY (2009) Polyamines regulate E‐cadherin transcription through c‐Myc modulating intestinal epithelial barrier function. Am J Physiol Cell Physiol 296: C801–C810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L, Christodoulou‐Vafeiadou E, Rao JN, Zou T, Xiao L, Chung HK, Yang H, Gorospe M, Kontoyiannis D, Wang JY (2014) RNA‐binding protein HuR promotes growth of small intestinal mucosa by activating the Wnt signaling pathway. Mol Biol Cell 25: 3308–3318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L, Ouyang M, Rao JN, Zou T, Xiao L, Chung HK, Wu J, Donahue JM, Gorospe M, Wang JY (2015) Competition between RNA‐binding proteins CELF1 and HuR modulates MYC translation and intestinal epithelium renewal. Mol Biol Cell 26: 1797–1810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L, Zhuang R, Xiao L, Chung HK, Luo J, Turner DJ, Rao JN, Gorospe M, Wang JY (2017) HuR enhances early restitution of the intestinal epithelium by increasing Cdc42 translation. Mol Cell Biol 37: e00574‐16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lueschow SR, McElroy SJ (2020) The Paneth cell: the curator and defender of the immature small intestine. Front Immunol 11: 587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nandy C, Mrazek J, Stoiber H, Grasser FA, Huttenhofer A, Polacek N (2009) Epstein‐barr virus‐induced expression of a novel human vault RNA. J Mol Biol 388: 776–784 [DOI] [PubMed] [Google Scholar]
- Payen D (2020) The gut as a hidden source of sepsis. Minerva Anestesiol 86: 662–669 [DOI] [PubMed] [Google Scholar]
- Peterson LW, Artis D (2014) Intestinal epithelial cells: regulators of barrier function and immune homeostasis. Nat Rev Immunol 14: 141–153 [DOI] [PubMed] [Google Scholar]
- Si R, Cabrera JTO, Tsuji‐Hosokawa A, Guo R, Watanabe M, Gao L, Lee YS, Moon JS, Scott BT, Wang J et al (2021) HuR/Cx40 downregulation causes coronary microvascular dysfunction in type 2 diabetes. JCI Insight 6: e147982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silver MJ, Kessler NJ, Hennig BJ, Dominguez‐Salas P, Laritsky E, Baker MS, Coarfa C, Hernandez‐Vargas H, Castelino JM, Routledge MN et al (2015) Independent genomewide screens identify the tumor suppressor VTRNA2‐1 as a human epiallele responsive to periconceptional environment. Genome Biol 16: 118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadler PF, Chen JJ, Hackermuller J, Hoffmann S, Horn F, Khaitovich P, Kretzschmar AK, Mosig A, Prohaska SJ, Qi X et al (2009) Evolution of vault RNAs. Mol Biol Evol 26: 1975–1991 [DOI] [PubMed] [Google Scholar]
- Treppendahl MB, Qiu X, Sogaard A, Yang X, Nandrup‐Bus C, Hother C, Andersen MK, Kjeldsen L, Mollgard L, Hellstrom‐Lindberg E et al (2012) Allelic methylation levels of the noncoding VTRNA2‐1 located on chromosome 5q31.1 predict outcome in AML. Blood 119: 206–216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner JR (2009) Intestinal mucosal barrier function in health and disease. Nat Rev Immunol 9: 799–809 [DOI] [PubMed] [Google Scholar]
- Wakatsuki S, Araki T (2021) Novel molecular basis for synapse formation: small non‐coding vault RNA functions as a riboregulator of MEK1 to modulate synaptogenesis. Front Mol Neurosci 14: 748721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakatsuki S, Ohno M, Araki T (2021) Human vault RNA1‐1, but not vault RNA2‐1, modulates synaptogenesis. Commun Integr Biol 14: 61–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang JY, Cui YH, Xiao L, Chung HK, Zhang Y, Rao JN, Gorospe M, Wang JY (2018) Regulation of intestinal epithelial barrier function by long noncoding RNA uc.173 through interaction with microRNA 29b. Mol Cell Biol 38: e00010‐18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao L, Rao JN, Cao S, Liu L, Chung HK, Zhang Y, Zhang J, Liu Y, Gorospe M, Wang JY (2016) Long noncoding RNA SPRY4‐IT1 regulates intestinal epithelial barrier function by modulating the expression levels of tight junction proteins. Mol Biol Cell 27: 617–626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao L, Wu J, Wang JY, Chung HK, Kalakonda S, Rao JN, Gorospe M, Wang JY (2018) Long noncoding RNA uc.173 promotes renewal of the intestinal mucosa by inducing degradation of microRNA 195. Gastroenterology 154: 599–611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao L, Li XX, Chung HK, Kalakonda S, Cai JZ, Cao S, Chen N, Liu Y, Rao JN, Wang HY et al (2019) RNA‐binding protein HuR regulates Paneth cell function by altering membrane localization of TLR2 via post‐transcriptional control of CNPY3. Gastroenterology 157: 731–743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao L, Ma XX, Luo J, Chung HK, Kwon MS, Yu TX, Rao JN, Kozar R, Gorospe M, Wang JY (2021a) Circular RNA circHIPK3 promotes homeostasis of the intestinal epithelium by reducing microRNA 29b function. Gastroenterology 161: 1303–1317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao L, Rao JN, Wang JY (2021b) RNA‐binding proteins and long noncoding RNAs in intestinal epithelial autophagy and barrier function. Tissue Barriers 9: 1895648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- You YA, Kwon EJ, Hwang HS, Choi SJ, Choi SK, Kim YJ (2021) Elevated methylation of the vault RNA2‐1 promoter in maternal blood is associated with preterm birth. BMC Genomics 22: 528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu TX, Wang PY, Rao JN, Zou T, Liu L, Xiao L, Gorospe M, Wang JY (2011) Chk2‐dependent HuR phosphorylation regulates occludin mRNA translation and epithelial barrier function. Nucleic Acids Res 39: 8472–8487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu TX, Rao JN, Zou T, Liu L, Xiao L, Ouyang M, Cao S, Gorospe M, Wang JY (2013) Competitive binding of CUGBP1 and HuR to occludin mRNA controls its translation and modulates epithelial barrier function. Mol Biol Cell 24: 85–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu TX, Chung HK, Xiao L, Piao JJ, Lan S, Jaladanki SK, Turner DJ, Raufman JP, Gorospe M, Wang JY (2020) Long noncoding RNA H19 impairs the intestinal barrier by suppressing autophagy and lowering Paneth and goblet cell function. Cell Mol Gastroenterol Hepatol 9: 611–625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Zhang Y, Xiao L, Yu TX, Li JZ, Rao JN, Turner DJ, Gorospe M, Wang JY (2017) Cooperative repression of insulin‐like growth factor type 2 receptor translation by microRNA 195 and RNA‐binding protein CUGBP1. Mol Cell Biol 37: e00225‐17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang T, Liu Y, Yan JK, Cai W (2020) Early downregulation of P‐glycoprotein facilitates bacterial attachment to intestinal epithelial cells and thereby triggers barrier dysfunction in a rodent model of total parenteral nutrition. FASEB J 34: 4670–4683 [DOI] [PubMed] [Google Scholar]
- Zhuang R, Rao JN, Zou T, Liu L, Xiao L, Cao S, Hansraj NZ, Gorospe M, Wang JY (2013) miR‐195 competes with HuR to modulate stim1 mRNA stability and regulate cell migration. Nucleic Acids Res 41: 7905–7919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou T, Mazan‐Mamczarz K, Rao JN, Liu L, Marasa BS, Zhang AH, Xiao L, Pullmann R, Gorospe M, Wang JY (2006) Polyamine depletion increases cytoplasmic levels of RNA‐binding protein HuR leading to stabilization of nucleophosmin and p53 mRNAs. J Biol Chem 281: 19387–19394 [DOI] [PubMed] [Google Scholar]
- Zou T, Rao JN, Liu L, Xiao L, Yu TX, Jiang P, Gorospe M, Wang JY (2010) Polyamines regulate the stability of JunD mRNA by modulating the competitive binding of its 3′ untranslated region to HuR and AUF1. Mol Cell Biol 30: 5021–5032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou T, Jaladanki SK, Liu L, Xiao L, Chung HK, Wang JY, Xu Y, Gorospe M, Wang JY (2016) H19 long noncoding RNA regulates intestinal epithelial barrier function via microRNA 675 by interacting with RNA‐binding protein HuR. Mol Cell Biol 36: 1332–1341 [DOI] [PMC free article] [PubMed] [Google Scholar]


