Abstract
Cyclic GMP‐AMP synthase (cGAS) is a double‐stranded DNA (dsDNA) sensor that functions in the innate immune system. Upon binding dsDNA, cGAS and dsDNA form phase‐separated condensates in which cGAS catalyzes the synthesis of 2′3′‐cyclic GMP‐AMP that subsequently triggers a STING‐dependent, type I interferon (IFN‐I) response. Here, we show that cytoplasmic RNAs regulate cGAS activity. We discover that RNAs do not activate cGAS but rather promote phase separation of cGAS in vitro. In cells, cGAS colocalizes with RNA and forms complexes with RNA. In the presence of cytoplasmic dsDNA, RNAs colocalize with phase‐separated condensates of cGAS and dsDNA. Further in vitro assays showed that RNAs promote the formation of cGAS‐containing phase separations and enhance cGAS activity when the dsDNA concentration is low. Cotransfection of RNA with a small amount of dsDNA into THP1 cells significantly enhances the production of the downstream signaling molecule interferon beta (IFNB). This enhancement can be blocked by a cGAS‐specific inhibitor. Thus, cytoplasmic RNAs could regulate cGAS activity by modulating the formation of cGAS‐containing condensates.
Keywords: cGAS, innate immunity, phase separation, regulation, RNA
Subject Categories: Immunology, RNA Biology, Signal Transduction
cGAS‐dsDNA phase separation is critical for the dsDNA‐induced activation of cGAS. This study shows that RNA‐modulated phase separation of cGAS plays a role in regulating cGAS activity.
Introduction
Recognition of pathogen‐derived nucleic acids by protein sensors allows the innate immune system to sense infection and initiate host defense mechanisms (Barbalat et al, 2011; Pandey et al, 2014; Wu & Chen, 2014). The cyclic GMP‐AMP synthase (cGAS) is the primary cytosolic double‐stranded DNA (dsDNA) sensor in mammalian cells (Sun et al, 2013; Wu et al, 2013; Li et al, 2013b; Vance, 2016). Upon binding to dsDNA, cGAS undergoes conformational changes that activate its ability to catalyze the synthesis of a noncanonical 2′3′ cyclic GMP‐AMP dinucleotide (2′3′‐cGAMP) that triggers type I interferon production through the endoplasmic reticulum membrane protein STING (also known as TMEM173, MPYS, MITA, and ERIS; Ishikawa & Barber, 2008; Jin et al, 2008; Zhong et al, 2008; Sun et al, 2009; Gao et al, 2013; Zhang et al, 2013). cGAS binds dsDNA in a sequence‐independent manner (Rongvaux et al, 2014; White et al, 2014; Lau et al, 2015; West et al, 2015; Mackenzie et al, 2017; Yang et al, 2017; Wang et al, 2017a). Abnormal activity of cGAS can lead to diseases such as Aicardi‐Goutières syndrome (Gao et al, 2015). cGAS activity is regulated by degradation or modification of the enzyme through ubiquitination, SUMOylation, phosphorylation, or glutamylation (Seo et al, 2015; Hu et al, 2016; Xia et al, 2016; Wang et al, 2017b). A recent report suggests that cGAS activity is also regulated by the formation of phase‐separated condensates upon dsDNA engagement, which confine the activated cGAS to a particular location (Du & Chen, 2018). Previous studies indicated that a variety of parameters, including the concentration and length of the dsDNA, influence the sensitivity of cGAS‐mediated detection of cytosolic DNA (Andreeva et al, 2017; Luecke et al, 2017). However, cellular cGAS activity is not well explained by current structural and biophysical models (Andreeva et al, 2017; Du & Chen, 2018).
Here, we show that cGAS activity can be regulated by RNA‐modulated phase separation. We found that cGAS forms phase‐separated granules with RNA as well as dsDNA. Condensation with cytoplasmic RNAs promotes enzyme activity of cGAS at low concentrations of dsDNA but inhibits cGAS enzyme activity when high concentrations of dsDNA are present. Thus, RNA could play a novel and important role in regulating cGAS activity.
Results and Discussion
DNA‐ and RNA‐induced phase separation of cGAS
We observed the formation of phase‐separated granules soon after mixing the recombinant full‐length human cGAS (FL‐hcGAS) with a 45‐bp double‐stranded interferon stimulatory DNA (ISD; Appendix Fig S1A). Our results are consistent with the published work (Du & Chen, 2018). Previous studies showed that, in addition to dsDNA, cGAS can bind RNAs, single‐stranded DNAs (ssDNAs), and RNA–DNA hybrids (Civril et al, 2013; Kranzusch Philip et al, 2013; Mankan et al, 2014). We also observed that the ssDNA without complementary regions can induce the formation of phase‐separated granules when incubated with FL‐hcGAS (Appendix Fig S1B). However, activation of FL‐hcGAS was not observed when the ssDNAs had no complementary regions (Appendix Fig S1C). Moreover, dsDNAs with less than 20 bp (Fig EV1A) barely form condensates with FL‐cGAS (Figs EV1B and EV2A–C) or activate FL‐hcGAS (Fig EV1C). However, short dsDNAs with one or two ssDNA arms (Fig EV1A) induce phase separations of FL‐hcGAS (Figs EV1B and EV2D–I) and activate the enzyme (Fig EV1C).
Figure EV1. Short dsDNAs with ssDNA arms promote phase separation and activate cGAS.
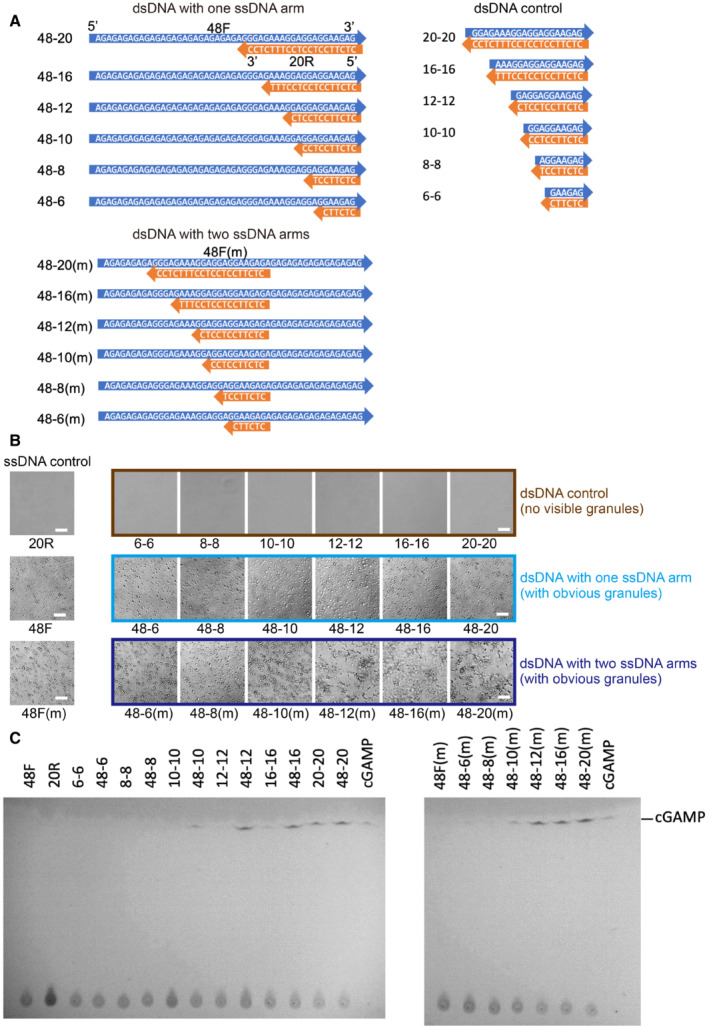
- Diagrams showing sequences of short dsDNAs with ssDNA arms and the short dsDNA controls.
- Photographs of FL‐hcGAS (0.294 mg/ml, 5.0 μM) mixed with indicated DNAs (2.5 μM) showing phase‐separated droplets or condensates induced by short dsDNAs with ssDNA arms but not the short dsDNA controls (n = 2, biological replicates, data from one representative independent biological replicate are shown). The corresponding concentration in mg/ml were 0.009 for 6–6, 0.012 for 8–8, 0.014 for 10–10, 0.018 for 12–12, 0.024 for 16–16, 0.031 for 20–20, 0.043 for 48–6 and 48‐6(m), 0.044 for 48–8 and 48‐8(m), 0.046 for 48–10 and 48–10(m), 0.047 for 48–12 and 48‐12(m), 0.05 for 48–16 and 48–16(m), 0.053 for 48–20 and 48–20(m), 0.015 for 20R, and 0.038 for 48F and 48F(m). The scale bar represents 20 μm.
- TLC analysis for cGAMP, which is indicative of cGAS activation. cGAMP was detected in samples containing short dsDNAs with ssDNA arms but not in samples containing the short dsDNA or ssDNA controls (n = 2, biological replicates, data from one representative independent biological replicate are shown). The FL‐hcGAS and DNA concentrations were (0.294 mg/ml, 5.0 μM) and 2.5 μM, respectively. Samples were prepared in 20 mM HEPES at pH 7.5 and 150 mM NaCl and were incubated at room temperature for 1 h.
Figure EV2. Phase diagrams showing phase separations of FITC‐labeled cGAS induced by short dsDNAs with one or two ssDNA arms.
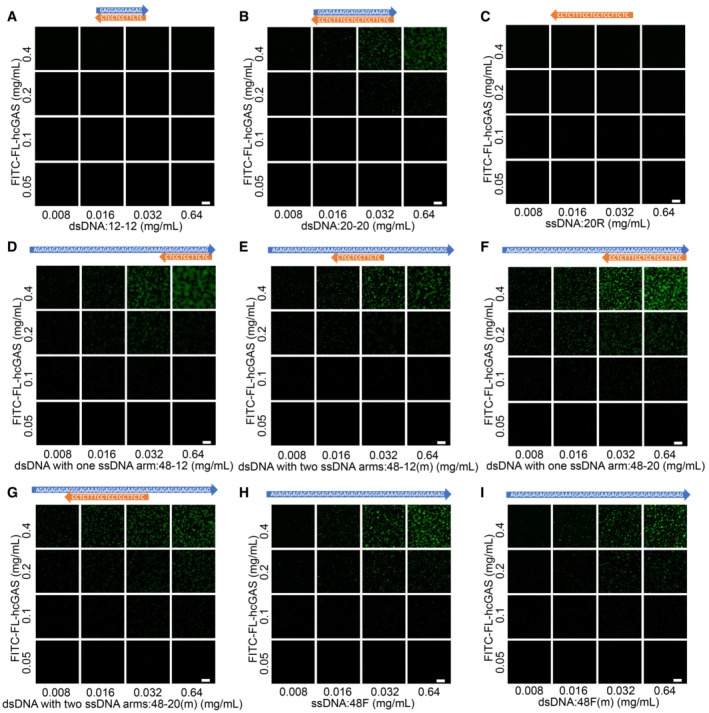
- Phase diagram of cGAS and the 12‐bp dsDNA (12–12) as indicated in Fig EV1A. The scale bar represents 20 μm.
- Phase diagram of cGAS and the 20‐bp dsDNA (20–20) as indicated in Fig EV1A. The scale bar represents 20 μm.
- Phase diagram of cGAS and the 20 nt ssDNA (20R) as indicated in Fig EV1A. The scale bar represents 20 μm.
- Phase diagram of cGAS and a 12‐bp dsDNA with one ssDNA arms (48–12) as indicated in Fig EV1A. The scale bar represents 20 μm.
- Phase diagram of cGAS and a 12‐bp dsDNA with two ssDNA arms (48–12(m)) as indicated in Fig EV1A. The scale bar represents 20 μm.
- Phase diagram of cGAS and a 20‐bp dsDNA with one ssDNA arms (48–20) as indicated in Fig EV1A. The scale bar represents 20 μm.
- Phase diagram of cGAS and the 20‐bp dsDNA with two ssDNA arm (48–20(m)) as indicated in Fig EV1A. The scale bar represents 20 μm.
- Phase diagram of cGAS and a 48 nt ssDNA (48F) that has the same sequence as the ssDNA arm strand in “D” and “F.” The scale bar represents 20 μm.
- Phase diagram of cGAS and a 48 nt ssDNA (48F(m)) that has the same sequence as the ssDNA arm strand in “E” and “G.” The scale bar represents 20 μm.
We next performed similar assays using yeast tRNA and total RNA extracted from HeLa cells. tRNAs are abundant in the cytoplasm with an estimated concentration of approximately 1.2–1.8 mg/ml in mammalian cells and up to 20 mg/ml in yeast cells (Frenkel‐Morgenstern et al, 2012; Feijo Delgado et al, 2013; Palazzo & Lee, 2015). Phase‐separated granules were observed when FL‐hcGAS was mixed with yeast tRNA or total RNA from HeLa cells over a wide concentration range of the RNA (Fig 1A and B). No phase‐separated granules were detected when RNAs were mixed with FITC‐labeled C‐terminal domain of mouse cGAS, which does not form phase‐separated granules (Fig 1C and D). No activation of FL‐hcGAS was not detected in the presence of RNA, however (Fig 1E and F). Phase separation of FL‐hcGAS was observed after DNase treatment of the RNA preparation (Appendix Fig S2A) and in the presence of a large amount of BSA/PEG (Appendix Fig S2B and C); the latter mimics the crowded environment in the cytoplasm. The binding of RNAs to FL‐hcGAS was estimated using an electrophoretic mobility shift assay. The results showed that RNA bound cGAS (Fig EV3A and B) with a similar affinity as dsDNA (Fig EV3C and D), which is consistent with previous studies (Civril et al, 2013).
Figure 1. RNA mediates phase separation of cGAS.
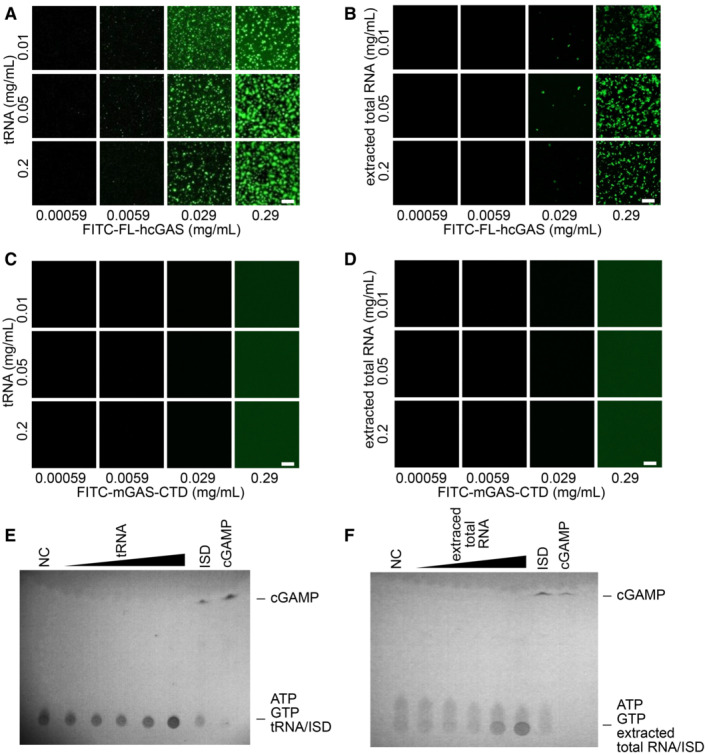
-
A, BFluorescence images of FITC‐labeled full‐length human cGAS (FITC‐FL‐hcGAS) incubated with (A) yeast tRNA or (B) extracted total RNA from HeLa cells at the indicated concentrations (n = 2, biological replicates, data from one representative independent biological replicate are shown). The scale bars represent 20 μm.
-
C, DNegative controls. Fluorescence images of FITC‐labeled C‐terminal domain of mouse cGAS (FITC‐mcGAS‐CTD) incubated with (C) yeast tRNA or (D) extracted total RNA from HeLa cells at the indicated concentrations (n = 2, biological replicates, data from one representative independent biological replicate are shown). mcGAS‐CTD is known not to form phase separation.
-
E, FTLC analysis of cGAMP, which is indicative of cGAS activation, upon incubation with (E) tRNA or (F) total RNA (n = 2, biological replicates, data from one representative independent biological replicate are shown). The FL‐hcGAS concentration was 0.265 mg/ml. The tRNA concentrations from low to high were 0.025, 0.05, 0.1, 0.25, and 0.85 mg/ml. The extracted total RNA concentrations from low to high were 0.025, 0.05, 0.1, 0.25, and 0.5 mg/ml. NC indicates the negative control that has only the FL‐hcGAS. dsDNA ISD at a concentration of 0.05 mg/ml was used as a positive control. All the samples were prepared in 20 mM HEPES at pH 7.5 and 150 mM NaCl.
Figure EV3. Electrophoretic mobility shift analysis of interactions of RNA or DNA with cGAS.
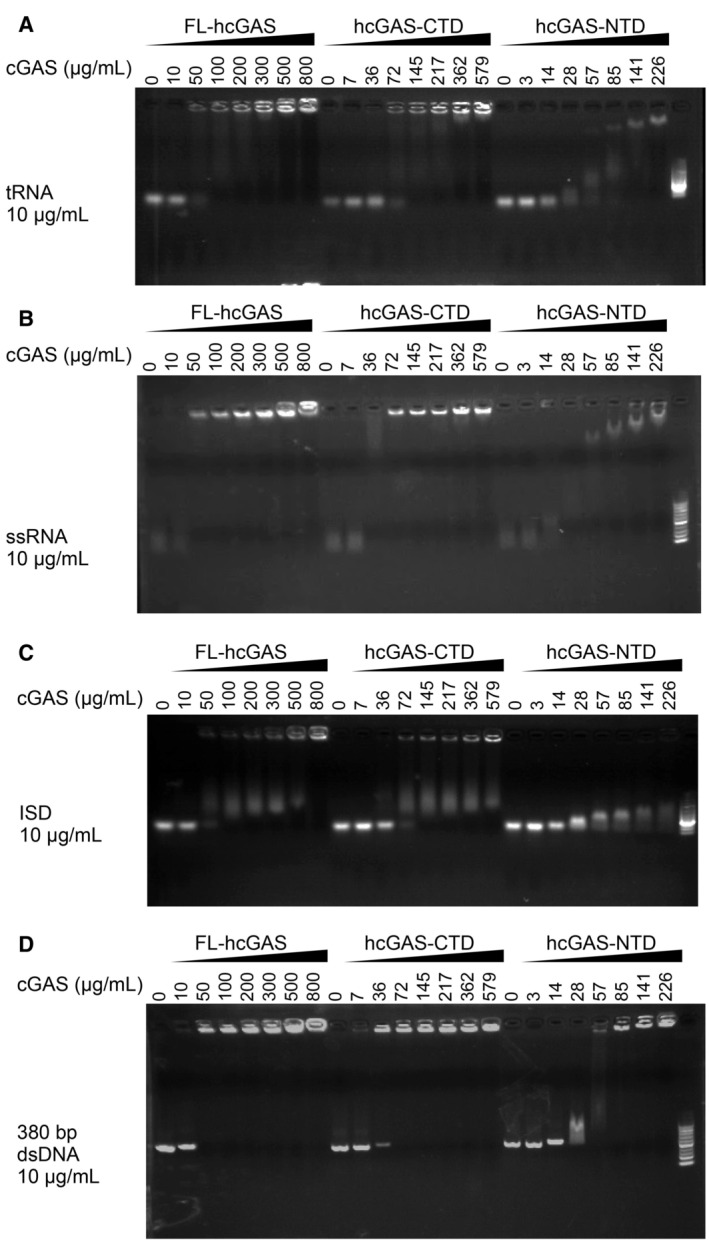
-
A–DElectrophoretic mobility shift analysis (EMSA) of (A) yeast tRNA, (B) ssRNA, (C) ISD, and (D) 380‐bp dsDNA in presence of FL‐hcGAS (n = 2, biological replicates, data from one representative independent biological replicate are shown). The nucleic acid concentration was 10 μg/ml. The FL‐hcGAS concentrations were 0, 10, 50, 100, 200, 300, 500, and 800 μg/ml. The hcGAS‐CTD concentrations were 0, 7, 36, 72, 145, 217, 362, and 579 μg/ml. The hcGAS‐NTD concentrations were 0, 3, 14, 28, 57, 85, 141, and 226 μg/ml.
RNA induces phase separation of cGAS in cells
The concentration of RNAs, including tRNA and mRNA, is much higher than the concentration of DNA in the cytoplasm, probably even under abnormal conditions such as when cells are infected by viruses (Russo & Russo, 2014; Karalyan et al, 2018). To test the hypothesis that cGAS forms phase‐separated granules with RNA in the absence of DNA, we overexpressed hcGAS fused with a C‐terminal GFP tag in the HEK293T cells. The gene for expression of hcGAS‐GFP under control of a doxycycline‐inducible promoter was integrated into the genome of the cell. Confocal microscopy revealed that a portion of the cells with the GFP signals had phase‐separated granules in the cytoplasm (Fig 2A). We then stained the GFP‐positive cells with Hoechst 33342 and pyronin Y, which as previously described stain DNA and RNA, respectively (Darzynkiewicz et al, 2004). Staining with Hoechst 33342 and pyronin Y showed that the hcGAS‐GFP colocalized with cytoplasmic RNAs (Fig 2A). Our in vitro assays showed that dsDNAs and RNAs in the hcGAS‐containing condensates can be differentially stained by combining the two dyes (Appendix Fig S3), confirming that the two dyes can function in condensates. HEK293T cells expressing YFP without hcGAS did not show any phase‐separated granules in the cytoplasm (Fig 2A).
Figure 2. hcGAS interacts with RNA in cells.
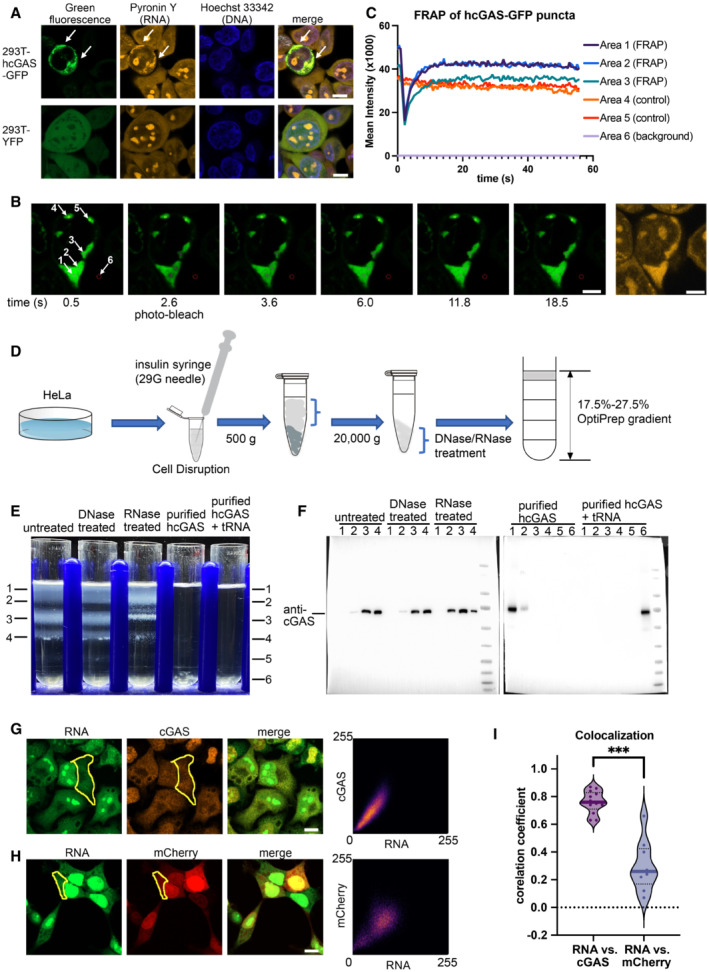
-
AHoechst 33342 and pyronin Y staining of HEK293T cells showing colocalization of the hcGAS‐GFP granules and RNAs (n = 3, biological replicates, data from one representative independent biological replicate are shown). Expression of the hcGAS‐GFP was induced by doxycycline at a concentration of 0.1 μg/ml. HEK293T cells that express YFP were used as the negative controls. The scale bars represent 10 μm.
-
B, CFluorescence recovery after photo‐bleaching (FRAP) assay of hcGAS‐GFP granules in HEK293T cells (n = 4, biological replicates, data from one representative independent biological replicate are shown). (B) Images (left 1–6) of a 293T cell showing the hcGAS‐GFP signal in the cell. The selected areas are indicated with circles and arrows and are numbered in the left‐most image. Areas 1, 2, and 3 were photo‐bleached at the time indicated below images. Areas 4 and 5 were not photo‐bleached and were used as the controls. Signal in area 6 was taken as background. The yellow fluorescence image on the right showing the pyronin Y‐stained RNA signal, which colocalizes with the hcGAS‐GFP signal in the cytoplasm. The scale bars represent 5 μm. (C) Quantification of the fluorescence intensity changes in the 6 selected areas of the FRAP assay indicated in panel “B.”
-
DA schematic diagram showing the HeLa cell cytoplasm extraction and fractionation procedure. To release the cytoplasm without disrupting the nuclear membrane, the cell membranes were disrupted by suspension in a hypotonic buffer and passing the cells through a 29G needle three times. The hypotonic buffer contains 10 mM HEPES at pH 7.5, 5 mM KCl and 3 mM MgCl2.
-
EOptiPrep density gradient analyses of the HeLa cell cytoplasm extracts and purified hcGAS with and without tRNA (n = 3, biological replicates, data from one representative independent biological replicate are shown).
-
FWestern blot analysis of the OptiPrep gradient fractions with or without DNase and RNase treatments.
-
GThe three images on the left showing fluorescence signals of endogenous cGAS and RNA in HeLa cells (n = 13, biological replicates, data from one representative independent biological replicate are shown). The scale bar represents 10 μm. The scatterplot on the right plots the signal intensities of cGAS versus RNA at each pixel in the indicated cytoplasmic area (yellow circle).
-
HThe three images on the left showing the fluorescence signals of mCherry and RNA in HEK293T cells (n = 9, biological replicates, data from one representative independent biological replicate are shown). The scale bar represents 10 μm. The scatterplot on the right plots signal intensities of mCherry versus RNA at each pixel in the indicated cytoplasmic area (yellow circle).
-
IPearson's correlation coefficient (CC) analysis of the fluorescence signals of cGAS vs. RNA (n = 13, biological replicates), and mCherry vs. RNA (n = 9, biological replicates). The median values were calculated and used for the evaluation of significance. ***P < 0.001 (two‐tailed Mann–Whitney test).
To confirm the liquid–liquid phase separation nature of the hcGAS‐GFP‐involved granules, a fluorescence recovery after photo‐bleach (FRAP) assay was performed to measure the fluidity of the granules (Fig 2B and C). The fluorescence in the photo‐bleached region recovered shortly after the bleach (Fig 2B and C) indicative of frequent exchange of the cGAS molecules inside the granules with the cGAS molecules in the environment, typical of liquid‐like phase separation (Brangwynne et al, 2009; Hyman et al, 2014; Alberti et al, 2019). These results demonstrated that the cytoplasmic granules that contain hcGAS‐GFP and RNA are liquid‐like droplets rather than insoluble aggregates.
The interactions between endogenous cGAS and RNAs in a cytoplasmic extract of HeLa cells were further analyzed using an iodixanol‐based (OptiPrep) density gradient ultracentrifugation, in which cytosolic components are separated into layers from top to bottom based on their densities (Fig 2D and E). There were four major bands in the gradient after centrifugation (Fig 2E). Western blot analysis of the bands with a cGAS‐specific antibody showed that endogenous cGAS locates mainly in bands 3 and 4 (Fig 2F). The OptiPrep concentration at band 3 and 4 is between 20 and 25%, which correspond to densities of 1.128–1.152 g/ml (Fig 2E and F). Bands 3 and 4 were sensitive to RNase but not DNase (Fig 2E and F). After the treatment with RNase, the peak fraction of cGAS shifted toward the top of the gradient, indicating that the densities of the bands containing cGAS were reduced by RNase and suggesting that RNA and cGAS most likely form complexes (Fig 2E and F). Sequencing of the RNA in bands 3 and 4 showed high reads for mRNAs, miRNAs, rRNAs, and tRNAs (Appendix Fig S4).
Next, we analyzed the cytoplasmic extract of HEK293T cells that overexpress the hcGAS‐GFP. Similar bands were observed in the OptiPrep gradient (Appendix Fig S5A) to those detected in HeLa cells, and western blot analysis of the bands showed that hcGAS‐GFP is located at similar positions in the gradient as endogenous cGAS (Appendix Fig S5B and C). RNase treatments resulted in a similar shift of hcGAS‐GFP toward the top of the gradient (Appendix Fig S5A–C). These indicate that hcGAS‐GFP‐RNA and endogenous cGAS‐RNA complexes share similar properties. Our FRAP results have shown that hcGAS‐GFP in the cells forms phase‐separated granules with cytoplasmic RNAs (Fig 2A–C). In the control gradient loaded with recombinantly expressed and purified FL‐hcGAS, the FL‐hcGAS signals were detected mainly in fractions that have an OptiPrep concentration between 17.5 and 20%, near the top of the gradient (Fig 2E and F). By contrast, the mixture of FL‐hcGAS and tRNA was detected in fractions that have an OptiPrep concentration of 27.5%, which is located at the bottom of the gradient, and indicate a heavy complex (Fig 2E and F). The difference between the sample prepared in vitro and the sample extracted from the cell reflects complex states of the endogenous cGAS, which probably directly or indirectly binds many other factors as shown in previous studies (Andreeva et al, 2017; Zhao et al, 2022). These results indicate that endogenous hcGAS associates with RNAs in high‐molecular‐weight complexes in cells prior to sensing cytoplasmic dsDNAs, which are usually not present in the cytoplasm.
To further confirm that endogenous cGAS is associated with endogenous RNA in the cytoplasm, we stained endogenous cGAS with a cGAS‐specific antibody in cells in which endogenous RNAs were labeled. The RNAs were labeled using a highly specific labeling method, in which 5′‐ethynyl uridines were incorporated into newly synthesized RNAs in live cells and then the dye Alexa Fluor 488 azide was added to result in a specific click reaction with the ethynyl group of the modified uridine (Jao & Salic, 2008). The Pearson's correlation analysis of the fluorescence signals of endogenous cGAS and RNA in cytoplasm of HeLa cells showed a correlation coefficient of 0.76 (Fig 2G and I), which strongly indicates the colocalization of endogenous cGAS and RNA. As a control, the Pearson's correlation analysis of the fluorescence signals of mCherry and RNA in cytoplasm of HEK293T cells showed a correlation coefficient of 0.26 (Fig 2H and I). These data combined with the OptiPrep density gradient results indicate that endogenous cGAS colocalizes with cytoplasmic RNAs in the form of large complexes. Clear phase‐separated granules of endogenous cGAS and RNA were not observed in the cytoplasm (Fig 2G), probably because the local density of the complex or the ratio between cGAS and RNAs did not reach the threshold required for the formation of observable granules.
dsDNA replaces RNA in preformed phase‐separated granules
It was previously shown that molecules in phase‐separated granules are in a dynamic equilibration with those in solution (Taylor et al, 2019). We observed that when Cy5‐labeled ISD (Cy5‐ISD) was transfected into HEK293T cells, in which phase‐separated granules containing YFP‐hcGAS are present prior to transfection, the Cy5‐ISD was eventually incorporated into the preformed granules (Fig 3A). In addition, Cy5‐ISD colocalized with hcGAS‐GFP and endogenous RNAs (Fig 3B, Appendix Fig S6). As the electrophoretic mobility shift assays showed that the binding affinity of RNAs for hcGAS was comparable to that of dsDNA (Fig EV3), in these cells where phase‐separated granules exist prior to transfection, the transfected Cy5‐ISD may replace RNA molecules in the granules. To verify this hypothesis, we performed in vitro assays with fluorescein‐5‐thiosemicarbazide‐labeled tRNA (FTSC‐tRNA).
Figure 3. Cy5‐ISD colocalizes with both cGAS and RNA in HEK293T cells.
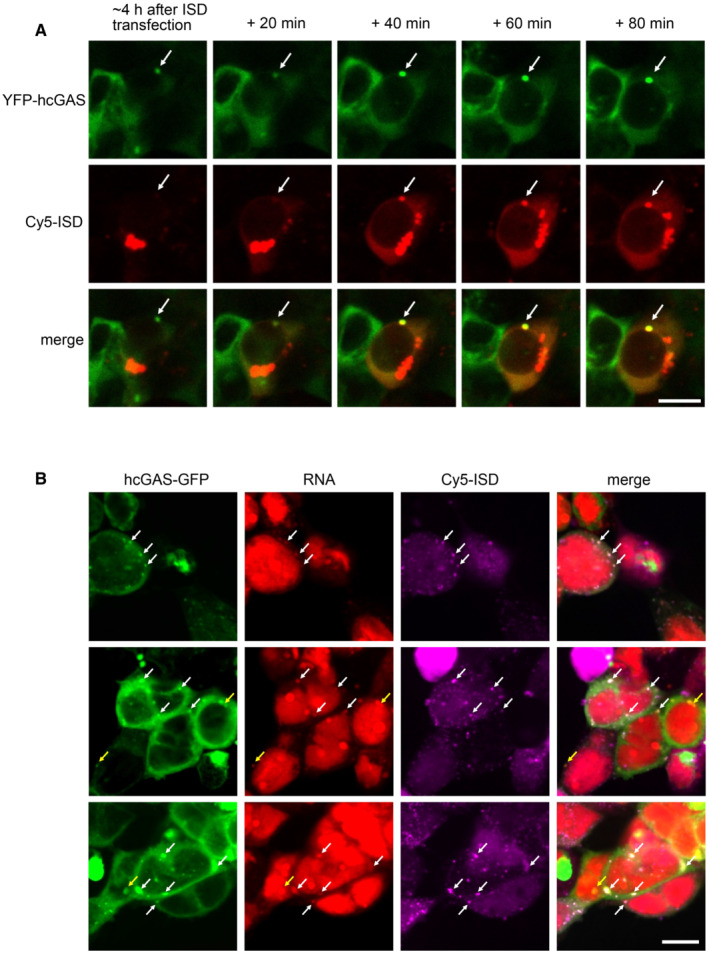
- Real‐time observation showing the gradual incorporation of Cy5‐ISD into a preformed granule of YFP‐hcGAS in a HEK293T cell (n = 2, biological replicates, data from one representative independent biological replicate are shown). The Cy5‐ISD was transfected into cells with Lipofectamine 2000. White arrows indicate the granule. The scale bar represents 10 μm.
- Representative fluorescence images showing colocalization of Cy5‐ISD with both hcGAS‐GFP and endogenous RNA in HEK293T cells (n = 3, biological replicates, data from one representative independent biological replicate are shown). Endogenous RNAs were specifically labeled by using a click reaction‐based method. White arrows indicate the granules with colocalized Cy5‐ISD, hcGAS‐GFP, and endogenous RNA. Yellow arrows indicate the granules with colocalized hcGAS‐GFP and endogenous RNA. The scale bar represents 10 μm.
Different amounts of ISD were added to solutions containing the preformed FTSC‐tRNA‐cGAS granules, the granules were separated from the solution by centrifugation, and the signal due to FTSC‐tRNA was measured in the supernatants (Fig 4A). As the concentration of the dsDNA was increased, the FTSC‐tRNA signal increased in the supernatant until a plateau was reached (Fig 4B). This indicates that dsDNA gradually replaced tRNAs in the granules until a dynamic equilibrium was reached. Similarly, when granules were preformed from ISD and hcGAS, we observed a concentration‐dependent substitution of the dsDNAs by tRNAs (Fig 4C). Analyses by confocal microscopy also demonstrated that the nucleic acid components of the granules are in a dynamic equilibrium (Fig 4D–G).
Figure 4. dsDNA replaces tRNA in cGAS‐tRNA phase separations.
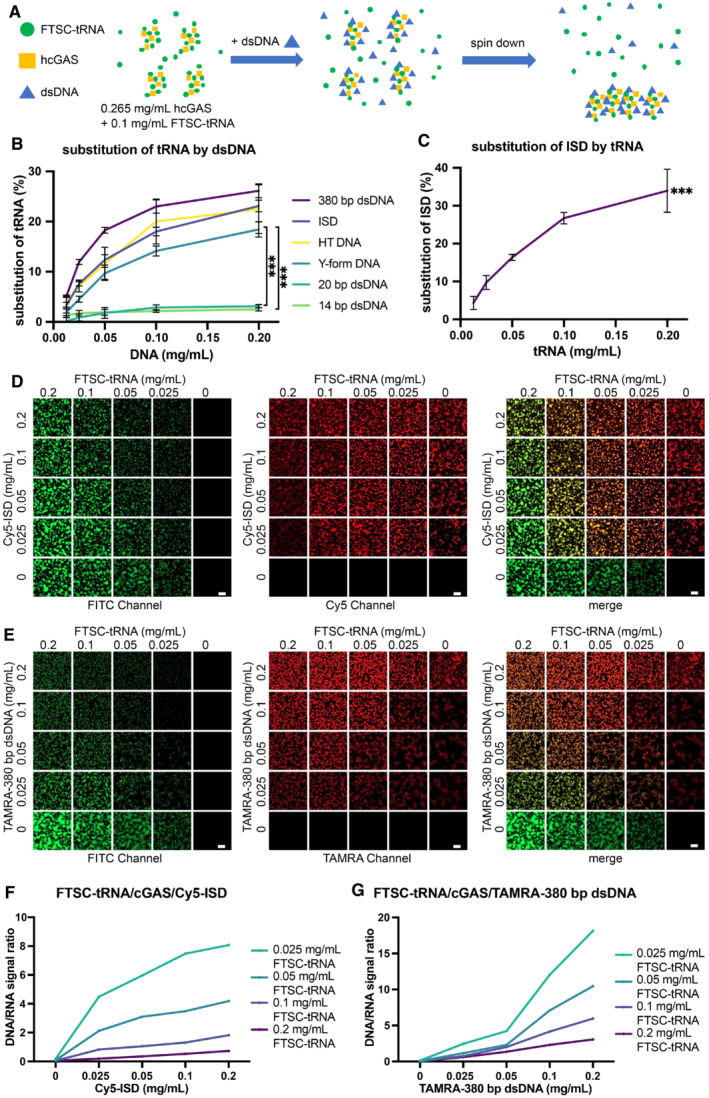
- A schematic diagram of the methods used to quantify tRNA/dsDNA released from the phase‐separated granules. FTSC‐tRNA and FL‐hcGAS were premixed. dsDNA was added, and the sample was centrifuged. The fluorescence signal of FTSC‐tRNA in supernatant was measured.
- The percentage of FTSC‐tRNA released into the supernatant after the addition of the indicated dsDNA as measured by fluorescence spectroscopy (n = 3, biological replicates). The percentage of FTSC‐tRNA released from the condensates was calculated by using the following equation: P = (Fi–F0) * 100/(F–F0). P: percentage of substituted FTSC‐tRNA. Fi: fluorescence of FTSC‐tRNA in the supernatant of each test. F0: fluorescence signal of FTSC‐tRNA in the supernatant when DNA was not added. F: fluorescence signal of FTSC‐tRNA without the addition of DNA and cGAS. The dsDNAs tested were ISD, 14‐bp dsDNA, 20‐bp dsDNA, 380‐bp dsDNA, herring testis DNA (HT DNA) and Y‐form DNA (a 14‐bp dsDNA with unpaired GGG on each end). Data are presented as mean ± SD. Significance was calculated for Y‐form DNA vs. 14‐bp dsDNA and Y‐from DNA vs. 20‐bp dsDNA, ***P < 0.001 (two‐way ANOVA).
- The percentage of Cy5‐ISD released into the supernatant over time after the addition of yeast tRNA as measured by fluorescence spectroscopy (n = 3, biological replicates). The yeast tRNA was used to trigger the release of the ISD from the phase separations of cGAS‐ISD. The percentage of Cy5‐ISD released from the condensates was calculated by using the following equation: P = (Fi–F0) * 100/(F–F0). P: percentage of substituted Cy5‐ISD. Fi: fluorescence signal of Cy5‐ISD in the supernatant of each test. F0: fluorescence signal of Cy5‐ISD in the supernatant when tRNA was not added. F: fluorescence signal of Cy5‐ISD without adding tRNA and cGAS. Data are presented as mean ± SD. ***P < 0.001 (one‐way ANOVA).
- Fluorescence microscopy images showing the replacement of FTSC‐tRNA by Cy5‐ISD at indicated concentrations in preformed granules of hcGAS and FTSC‐tRNA (n = 2, biological replicates, data from one representative independent biological replicate are shown). In each square, a representative area that is 1/16 of the raw image is displayed. The scale bars represent 20 μm.
- Fluorescence microscopy images showing the replacement of FTSC‐tRNA by TAMRA‐380‐bp dsDNA at indicated concentrations in preformed granules of hcGAS and FTSC‐tRNA (n = 2, biological replicates, data from one representative independent biological replicate are shown). In each square, a representative area that is 1/16 of the raw image is displayed. The scale bars represent 20 μm.
- Quantification of the replacement in “D.” For each square, the raw image used was evenly divided into four parts. Fluorescence signal ratios of Cy5‐ISD over FTSC‐tRNA in the four parts were calculated, averaged and plotted.
- Quantification of the replacement in “E.” For each square, the raw image used was evenly divided into four parts. Fluorescence signal ratios of TAMRA‐380‐bp dsDNA over FTSC‐tRNA in the four parts were calculated, averaged and plotted.
Relatively long dsDNAs such as the 45‐bp ISD, a 380‐bp dsDNA, and herring testis DNA displaced the tRNA in the phase‐separated granules even at a low concentration of 0.025 mg/ml (Fig 4B). By contrast, 14‐bp and 20‐bp dsDNAs, which bound to cGAS but barely induce the formation of phase‐separated granules (Appendix Fig S7A–C), did not displace the tRNA (Fig 4B). A Y‐form DNA that has a 14‐bp duplex with unpaired GGG at the termini, a structure previously shown to activate cGAS in cells (Herzner et al, 2015), induced the formation of the phase separations and also displaced tRNA from granules (Appendix Figs S7A, D, and E, and 4B).
tRNA‐induced phase separation affects cGAS activity
Since hcGAS is not activated by binding to RNA, we reasoned that the competitive binding of RNAs to hcGAS should have a negative impact on the dsDNA‐dependent activation of the enzyme. To test this, we measured the hcGAS activity with or without 0.125 mg/ml tRNA and showed that cGAS activity was significantly inhibited by tRNA when DNA concentration exceeded 0.05 mg/ml (Fig 5A). However, tRNA enhanced cGAS activity when the DNA concentration was low and in the range of 0.0034–0.0136 mg/ml (Fig 5A). At a high dsDNA concentration of 0.0544 mg/ml, tRNA inhibited the production of 2′3′‐cGAMP catalyzed by hcGAS in a dose‐dependent manner (Fig 5B). By contrast, at a low dsDNA concentration (0.0068 mg/ml), tRNA promoted the enzyme activity of hcGAS in a dose‐dependent manner (Fig 5C).
Figure 5. Regulation of cGAS activity by RNA.
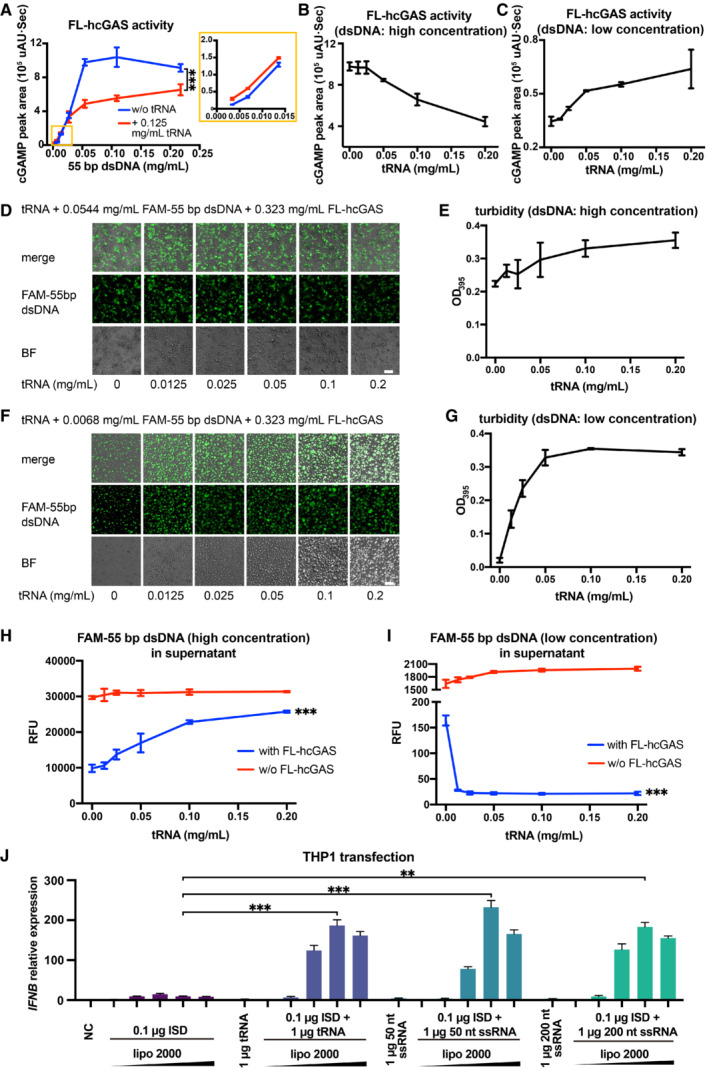
- cGAS activity (as measured by the HPLC peak area of cGAMP) in the presence of a 55‐bp dsDNA at indicated concentrations and in presence or absence of 0.125 mg/ml tRNA (n = 3, biological replicates). The inset is an expanded view of the region boxed in yellow. ***P < 0.001 (two‐way ANOVA).
- cGAS activity in the presence of 0.0544 mg/ml 55‐bp dsDNA and tRNA at indicated concentrations plotted as mean ± SD (n = 3, biological replicates).
- cGAS activity in the presence of 0.0068 mg/ml 55‐bp dsDNA and tRNA at indicated concentrations (n = 3, biological replicates).
- Fluorescence and bright‐field photographs of phase‐separated granules of FL‐hcGAS and 0.0544 mg/ml FAM‐labeled 55‐bp dsDNA in presence of tRNA at indicated concentrations (n = 2, biological replicates, data from one representative independent biological replicate are shown). The scale bar represents 20 μm.
- Turbidities of FL‐hcGAS and 0.0544 mg/ml FAM‐labeled 55‐bp dsDNA in presence of tRNA at indicated concentrations as measured by absorption at 395 nm plotted as mean ± SD (n = 3, biological replicates).
- Fluorescence and bright‐field photographs of phase‐separated granules of FL‐hcGAS and 0.0068 mg/ml FAM‐labeled 55‐bp dsDNA in presence of tRNA at indicated concentrations (n = 2, biological replicates, data from one representative independent biological replicate are shown). The scale bar represents 20 μm.
- Turbidities of FL‐hcGAS and 0.0068 mg/ml FAM‐labeled 55‐bp dsDNA in presence of tRNA at indicated concentrations as measured by absorption at 395 nm plotted as mean ± SD (n = 3, biological replicates).
- Fluorescence signals of the FAM‐labeled 55‐bp dsDNA in the supernatants of FL‐hcGAS and 0.0544 mg/ml FAM‐labeled 55‐bp dsDNA in presence of tRNA at indicated concentrations plotted as mean ± SD (n = 3, biological replicates). ***P < 0.001 (one‐way ANOVA).
- Fluorescence signals of the FAM‐labeled 55‐bp dsDNA in the supernatants of FL‐hcGAS and 0.0068 mg/ml FAM‐labeled 55‐bp dsDNA in presence of tRNA at indicated concentrations (n = 3, biological replicates). ***P < 0.001 (one‐way ANOVA).
- IFNB mRNA levels in THP1 cells induced by co‐transfecting dsDNA (ISD) and different RNAs (tRNA, 50 nt ssRNA and 200 nt ssRNA) plotted as mean ± SEM (n = 4, biological replicates). The volumes of Lipofectamine 2000 from low to high were 1, 2, 4, 6, and 8 μl. Significance was determined by two‐way ANOVA, **P < 0.01, ***P < 0.001.
Robust phase separation of hcGAS was observed at the high dsDNA concentration of 0.0544 mg/ml (Fig 5D and E) but not at the low dsDNA concentration of 0.0068 mg/ml (Fig 5F and G). The phase separation‐related turbidity of the solution did not change significantly as a function of tRNA concentration when the dsDNA concentration was high (0.0544 mg/ml; Fig 5E). However, the fluorescence signal due to FAM‐labeled dsDNA was reduced within the granules upon the addition of higher concentrations of tRNA (Fig 5D and H). The phase separation‐related turbidity of the solution increased significantly as a function of tRNA concentration when the dsDNA concentration was low (0.0068 mg/ml; Fig 5G), and the fluorescence signal due to FAM‐labeled dsDNA in the phase‐separated granules was significantly increased by incorporating tRNA (Fig 5F and I). Thus, tRNA promotes the formation of phase separation when the dsDNA concentration is not high enough to induce phase separation (Fig 5F and I), and this tRNA‐mediated formation of phase separation promotes the activation of cGAS when the dsDNA concentration is low (Fig 5C).
To further investigate the biological relevance of the RNA‐mediated regulation of cGAS activity, we co‐transfected THP1 cells with tRNA (or ssRNA) and ISD and then monitored interferon beta (IFNB) mRNA expression using real‐time quantitative PCR (RT–qPCR). The results showed that both tRNA and ssRNAs in different length significantly enhanced the production of IFNB mRNA with a fold change of approximately 20 when a small amount of ISD (0.1 μg) was transfected (Fig 5J). However, cGAS activity was not influenced significantly by the presence of RNA when a large amount of ISD (1 μg) was transfected (Fig EV4). The RNA‐mediated enhancement of IFNB mRNA production was completely blocked by G150, a specific inhibitor of cGAS (Lama et al, 2019), indicating that this phenotype is dependent on the cGAS‐mediated signaling pathway (Fig EV5A–C).
Figure EV4. RNAs regulate the dsDNA‐induced expression of IFNB in cells.
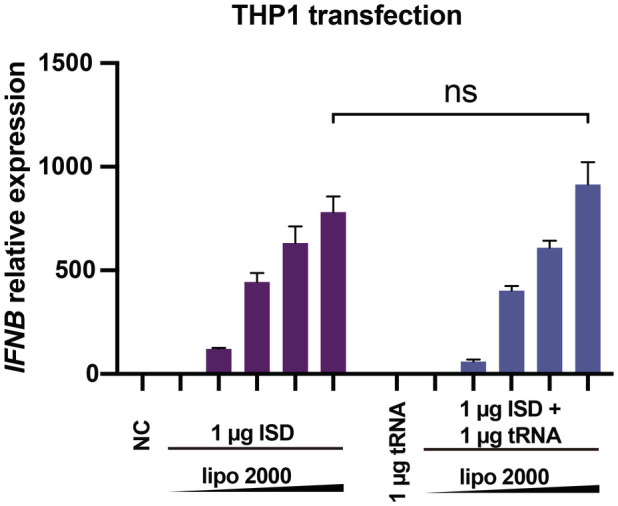
Expression of IFNB mRNA in THP1 cells transfected with 1 μg dsDNA (ISD) and 1 μg tRNA by using different mount of liposomes (n = 4, biological replicates). Data are presented as mean ± SEM, *P < 0.05, **P < 0.01, ***P < 0.001, and ns = not significant (two‐way ANOVA).
Figure EV5. cGAS‐specific inhibitor G150 blocks the RNA‐dsDNA mediated expression of IFNB in cells.
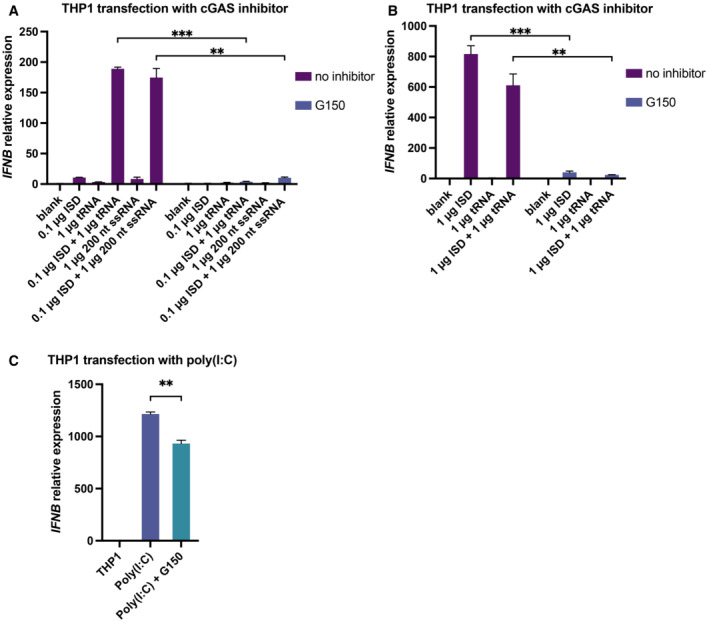
- Expression of IFNB mRNA in THP1 cells transfected with 0.1 μg dsDNA (ISD) and 1 μg tRNA or 200 nt ssRNA with or without the cGAS‐specific inhibitor G150 (n = 4, biological replicates).
- Expression of IFNB mRNA in THP1 cells transfected with 1 μg dsDNA (ISD) and 1 μg tRNA with or without the cGAS‐specific inhibitor G150 (n = 4, biological replicates).
- Expression of IFNB mRNA in THP1 cells transfected with 1 μg poly(I:C) with or without cGAS‐specific inhibitor G150 (n = 3, biological replicates).
Data information: The concentration of G150 used was 10 μM. The transfection reagent used was Lipofectamine 2000 and the amount of transfection reagent used per well in a 6‐well plate was 6 μl. Data are presented as mean ± SEM, *P < 0.05, **P < 0.01, ***P < 0.001, and ns = not significant (two‐way ANOVA for A and B, unpaired two‐tailed t‐test for C).
In summary, by combining biochemical and cellular assays, we have unequivocally established that cGAS forms phase‐separated granules with RNAs in vitro and in the cytoplasm of human cells that overexpress cGAS. Endogenous cGAS is associated with, and most likely forms complexes with, endogenous RNAs. Our study did not reveal the physical nature of the complexes formed by endogenous cGAS and RNAs, which might be nanoscopic condensates, and this needs further investigation. Nucleic acid‐binding proteins in cytoplasm such as G3BP1 might also contribute to the formation of the cGAS‐containing granules or complexes (Zhao et al, 2022). Our data suggest that RNAs might regulate the sensitivity of cGAS activity to cytosolic dsDNA. We envision that at a low dsDNA concentration that is not enough to induce the formation of phase separation, preformed complexes of cGAS and cytoplasmic RNAs, especially mRNAs and tRNAs, provide platforms that promote the formation of condensates of cGAS and dsDNA, which is required for cGAS activation. When the concentration of dsDNA is high enough to replace RNAs in the preformed complexes of cGAS‐RNA and to induce strong phase separation, dsDNA and RNAs are in a dynamic equilibrium, through which RNAs compete with dsDNA and inhibit cGAS activity, presumably to limit overactivation of the enzyme. Given the high RNA concentration in the cytoplasm, RNAs are likely the dominant regulators of cGAS activity. Our observations suggest a reasonable mechanism by which cGAS sensitively detects cytosolic dsDNA but is modulated to ensure an appropriate immune response to cytosolic dsDNA. Our finding that short dsDNAs, such as 14‐ and 20‐bp duplexes, do not efficiently displace RNAs from phase‐separated granules could provide an explanation for why short dsDNAs do not activate cGAS in cells, although they can activate cGAS in vitro (Li et al, 2013a; Herzner et al, 2015). The abundances of tRNAs and mRNAs are altered under stress conditions (Torrent et al, 2018), suggesting that RNAs could have complex functions in regulating cGAS‐mediated innate immune responses.
Materials and Methods
Cell lines
HEK293T cells (cat# CRL‐3216) and HeLa cells (cat# CRM‐CCL‐2) were purchased from American Type Culture Collection. THP1 cells were purchased from National Infrastructure of Cell Line Resource of China (cat# 1101HUM‐PUMC000057). STR authentication was performed by the vendors. All cell lines were negative in the tests for mycoplasma contamination in our laboratory.
Protein expression and purification
The coding sequence of hcGAS was optimized for E. coli expression using the GeneOptimizer algorithm (Thermo Fisher). The synthetic gene (Qinglan) was cloned into pETDuet‐1 (Novagen) at a site designed to fuse a 10× His tag and a sumo tag at the N terminus of the protein. The pETDuet‐hcGAS plasmid was transformed into E. coli BL21 Star (DE3) competent cells. The transformed cells were cultured in six 800‐ml aliquots of LB at 37°C until the absorbance at 600 nm reached ~ 0.6. The cells and medium were cooled to 16°C, and 1 mM IPTG was added to induce protein expression. The cells were harvested 16 h after the induction and were resuspended in 100 ml PBS buffer at pH 7.0 with 300 mM NaCl. The resuspended cells were homogenized, and the cell lysate was centrifugated at 20,000 g for 15 min. The supernatant was applied to 4‐ml Talon Metal Affinity Resin (Clontech, cat# 635503). After washing with 30 ml wash buffer containing 20 mM HEPES at pH 7.5, 150 mM NaCl, and 10 mM imidazole, the resin was resuspended in 11 ml of buffer containing 20 mM HEPES, pH 7.5, 150 mM NaCl, and 0.1 mg/ml ULP1 and incubated at 4°C for 12 h to remove the sumo tag. The hcGAS released from the resin was purified over a heparin column (GE Healthcare) to remove any dsDNA contamination and was further purified over a Superdex 75 size‐exclusion column (GE Healthcare) using a running buffer containing 20 mM HEPES, pH 7.5, and 150 mM NaCl.
Fluorescence labeling of the tRNA, dsDNA, ssDNA, cGAS
Yeast tRNAs (Solarbio, cat# T8630) were labeled with FTSC as described by Qiu et al (2015). In brief, a solution containing 20 μl tRNA (10 mg/ml) was mixed with 200 μl of 0.25 M sodium acetate. The mixture was diluted with ddH2O to a final volume of 700 μl, and 50 μl of 1 mM NaIO4 was added to oxidize the tRNAs. After incubation for 90 min in the dark at room temperature, the oxidization reaction was stopped by adding 40 μl of 2.5 mM Na2SO3 to the solution, followed by incubation for 15 min at room temperature. Labeling of the tRNAs was performed by adding 60 μl of 2.5 mM FTSC in DMF, and the solution was incubated for 3 h in the dark at room temperature. Excess FTSC was removed using a 10‐kDa cutoff Ultra Centrifugal Filter (Millipore) using 20 mM HEPES, pH 7.5, and 150 mM NaCl as the buffer.
Interferon stimulatory DNA and the 55‐bp dsDNA were labeled by primer extension with the forward primer conjugated to FAM or Cy5 on the 5′ terminus (Sangon). The ISD sequence is 5′‐TACAGATCTACTAGTGATCTATGACTGATCTGTACATGATCTACA‐3′. The 55‐bp dsDNA sequence is 5′‐TCGATACAGATCTACTAGTGATCTATGACTGATCTGTACATGATCTACAATCACT‐3′. The 380‐bp dsDNA was PCR amplified from a plasmid that contains part of the SARS‐CoV cDNA genome using a forward primer with a 5′ TAMRA label (Sangon). The primer sequences are forward 5′‐TAATACGACTCACTATAGGGATGTCTGATAATGG‐3′ and reverse 5′‐ AGCTTCTGGGCCAGTTCCTAG‐3′.
The cGAS protein was labeled with FITC (Thermo Fisher, cat# 46424). The protein in 20 mM HEPES, pH 7.5, and 150 mM NaCl at a concentration of 1–5 mg/ml and FITC were mixed at a molar ratio of 1:1 to 1:1.5. After incubation for 5 min in the dark at room temperature, 2 mM Tris–HCl, pH 8.0, was added to the solution to stop the reaction. Protein aggregates were removed by centrifugation at 20,000 g for 5 min. Excess FITC was removed using a 30‐kDa cutoff Ultra Centrifugal Filter (Millipore) with the 20 mM HEPES, pH 7.5, and 150 mM NaCl as buffer.
Observation of RNA‐induced phase separation of cGAS in vitro
For the phase separations induced by the total RNA from HeLa cells or yeast tRNA, RNAs and FITC‐labeled cGAS were mixed at the indicated concentrations and were transferred into a 384‐well plate. Total RNA was extracted from HeLa cells with an RNA purification kit (Thermo, cat# K0731). The samples were prepared in 20 mM HEPES, pH 7.5, and 150 mM NaCl. Phase separations of the RNAs and FITC‐labeled cGAS were observed using laser scanning confocal microscopy with excitation at 488 nm and emission at 499–641 nm.
For substitution of FTSC‐tRNA in cGAS‐associated phase‐separated granules by dsDNAs, FTSC‐tRNA and cGAS were mixed first, and dsDNAs (Cy5‐ISD or TAMRA‐labeled 380‐bp dsDNA) were then added at the indicated concentrations. The samples were prepared in 20 mM HEPES, pH 7.5, 150 mM NaCl. The samples were transferred into a 384‐well plate and observed using laser scanning confocal microscopy. For each fluorophore, excitation lasers and emission filters were as follows: FTSC excitation, 488 nm; FTSC emission, 491–535 nm; Cy5 excitation, 633 nm; Cy5 emission, 638–759 nm; TAMRA excitation, 543 nm; and TAMRA emission, 558–682 nm.
To observe the influence of phase separation on cGAS activity, FAM‐labeled 55‐bp dsDNA, tRNA and cGAS were mixed at the same concentrations in the cGAS activity assays. The samples were prepared in 25 mM Tris–HCl, pH 8.0, 20 mM NaCl, 5 mM MgCl2, 1 mM ATP, and 1 mM GTP. Phase separation was observed using laser scanning confocal microscopy with excitation at 488 nm and emission at 499–641 nm.
Stable cell line generation
To generate the cell line that stably expresses doxycycline‐inducible YFP‐hcGAS or hcGAS‐GFP, the sequence encoding YFP‐hcGAS or hcGAS‐GFP was cloned into the lentiviral vector pLVX‐TetOne‐Puro (Takara). A mixture of 2 μg pLVX‐TetOne‐Puro construct, 1 μg pMD2.G, and 1 μg psPAX2 was transfected into the HEK293T cells using Lipofectamine 2000 (Thermo Fisher, cat# 11668019) when cells were at ~ 70% confluence. The cells were cultured in a 6‐well plate with the DMEM medium (Thermo Fisher) and 10% FBS (Gibco, cat# 10091148) to generate the lentivirus. The medium containing lentiviruses was collected 60 h after the transfection, and the dead cell debris was removed by centrifugation. Polybrene was added to the solution at a final concentration of 0.8 μg/ml to enhance the lentiviral infection. The lentivirus was added to HEK293T cells cultured in a 6‐well plate when cells were at 50–60% confluence. The virus was replaced 12 h after infection with fresh DMEM medium containing 10% FBS. The cells were cultured for another 12 h, and then puromycin was added to the medium at a final concentration of 2.5 μg/ml. After passaging the cells three times in medium containing 2.5 μg/ml puromycin, the cells were maintained in the medium containing 2.5 μg/ml puromycin.
Cell imaging
For RNA/DNA differential staining and imaging, hcGAS‐GFP HEK293T cells were seeded on a coverslip. After 24 h, expression of hcGAS‐GFP was induced by adding 0.1 μg/ml doxycycline, and cells were allowed to grow for 9–20 h. Cells were fixed with 4% paraformaldehyde in PBS and were washed with PBS after 15 min. The cells were permeabilized with 0.5% Triton X‐100 in PBS for 15 min, and then the cells were washed three times with PBS. PBS containing 2 μg/ml Hoechst 33342 and 4 μg/ml pyronin Y (Amresco, cat# 0207) was added to the cells to stain DNA and RNA, respectively. After 15 min, coverslips were mounted on glass slides with Prolong Glass Antifade Mountant (Thermo Fisher). After 2 h or longer, the slides were observed by confocal microscopy (Zeiss LSM 880). For each dye, excitation lasers and emission filters were as follows: Hoechst 33342 excitation, 405 nm; Hoechst 33342 emission, 410–489 nm; YFP excitation, 514 nm; YFP emission, 525–588 nm; pyronin Y excitation, 561 nm; and pyronin Y emission, 625–758 nm. Phase‐separated granules of cGAS and the 55‐bp dsDNA, total RNA, or tRNA were observed as controls (Appendix Fig S3).
For RNA‐specific staining, HEK293T cells that express hcGAS‐GFP or HeLa cells were seeded on a coverslip in a 24‐well plate. The expression of hcGAS‐GFP was induced by adding 0.1 μg/ml doxycycline. The culture medium was supplemented with 1 mM 5‐ethynyl uridine. After 18 h, HeLa cells were fixed by the addition of 4% paraformaldehyde in PBS. HEK293T‐hcGAS‐GFP cells were transfected with 0.5 μg Cy5‐ISD and 1.5 μg PEI max. After 20 h, 293T‐hcGAS‐GFP cells were fixed with 4% paraformaldehyde in PBS. The fixed cells were washed with PBS, permeabilized by the addition of 0.5% TritonX‐100 in PBS and washed with PBS again. RNAs containing 5‐ethynyl uridine were stained by the addition of a buffer containing 100 mM Tris, pH 8.5, 1 mM CuSO4, 25 μM Alexa Fluor 488‐azide (Thermo Fisher, C10329) or azide‐fluor 545 (Sigma, 760757). After 30 min, the staining buffer was removed, and cells were washed with PBS containing 0.02% sodium azide. HEK293T‐hcGAS‐GFP cells were washed by PBST three times and mounted onto a slide. HeLa cells were further stained with a cGAS‐specific rabbit antibody (CST, cat# 79978) for 1 h at room temperature, washed with PBST three times, stained with Alexa Fluor plus 555‐conjugated goat anti‐rabbit antibody (Thermo Fisher, cat# A32732), washed with PBST five times, and mounted onto a slide. The slides were observed using a confocal microscope (Zeiss LSM 980) or a spinning disk confocal microscope (Olympus IXplore SpinSR).
For imaging YFP‐hcGAS in HEK293T cells transfected with Cy5‐ISD, cells were cultured in a four‐chamber dish with a glass bottom (Cellvis, cat# D35C4‐20‐1.5‐N). Approximately 6–9 h after the induction of expression with 0.1 μg/ml doxycycline, 0.5 μg of Cy5‐ISD was transfected into the cells in one chamber using Lipofectamine 2000. Cells were observed using a confocal microscope (Zeiss LSM 710).
tRNA displacement assays
FTSC‐labeled tRNA and cGAS were mixed to allow tRNA‐cGAS condensates to form, and then DNA was added. After a short incubation of 5 min at room temperature, cGAS‐nucleic acid condensates were spun down at 20,000 g for 5 min. The fluorescence signal of the free FTSC‐tRNA in the supernatant was measured using a NanoDrop 3300 (Thermo Scientific). Release of the FAM‐labeled ISD from the condensates was measured similarly. The FTSC‐tRNA and the FAM‐dsDNA concentrations had linear relationships with fluorescent signals in the range of 0.0125–0.2 mg/ml (Appendix Fig S8). The percentage of FTSC‐tRNA displaced was calculated by the following equation: P = (Fi–F0) * 100/(F–F0). P: percentage of substituted FTSC‐tRNA. Fi: fluorescence signal of FTSC‐tRNA in the supernatant of each test. F0: fluorescence signal of FTSC‐tRNA in the supernatant when DNA was not added. F: fluorescence signal of FTSC‐tRNA without adding DNA or cGAS.
Extraction of cytoplasmic cGAS and density gradient centrifugation
Extraction of cytoplasmic cGAS from HeLa cells was previously described (Du & Chen, 2018). The cytoplasm was extracted by hypotonic treatment (10 mM HEPES, pH 7.5, 5 mM KCl, 3 mM MgCl2) and homogenized through a 29G needle. The homogenized cells were centrifuged at 500 g to remove cell debris and nuclei. The supernatant was collected and centrifuged at 20,000 g to obtain the supernatant fraction S20 and the pellet fraction (P20). P20 was resuspended in an isotonic buffer (20 mM HEPES, pH 7.5, 250 mM sucrose, 25 mM KCl, 5 mM MgCl2) containing 17.5% OptiPrep (Sigma, cat# D1556). The resuspended P20 (400 μl) was further fractionated over an OptiPrep density gradient after treatment with RNase (20 μl of 10 mg/ml RNase A, 5 μl of 10 U/μl RNase I), DNase (25 μl of 1 U/μl DNase I), or buffer for 30 min at 37°C. After further fractionation over an OptiPrep density gradient (isotonic buffer containing 27.5, 25, 22.5, and 20% OptiPrep, from bottom to top, 900 μl per layer, with resuspended P20 on the top of the gradient).
In vitro analysis of cGAS activity
In vitro enzymatic activities of cGAS were measured by monitoring the formation of the product 2′3′‐cGAMP by using an HPLC system (SHIMADZU, LC‐10A) equipped with LC‐10AT pumps and an SPD‐10AV ultraviolet detector. The strands of the 55‐bp dsDNA were chemically synthesized: 5′‐TCGATACAGATCTACTAGTGATCTATGACTGATCTGTACATGATCTACAATCACT‐3′ and 5′‐AGTGATTGTAGATCATGTACAGATCAGTCATAGATCACTAGTAGA CTGTATCGA‐3′. The reaction mixture contained 5.5 μM (0.323 mg/ml) FL‐hcGAS, 25 mM Tris–HCl, pH 8.0, 20 mM NaCl, 5 mM MgCl2, 1 mM ATP, 1 mM GTP, the 55‐bp dsDNA at various concentrations (0.0034–0.2166 mg/ml) with or without 5 μM (0.125 mg/ml) tRNA. Reactions were also performed by varying tRNA concentrations (0.5–8 μM, 0.005–0.200 mg/ml) at 0.0068 or 0.0544 mg/ml 55‐bp dsDNA with all other components the same as described above. Each reaction in 40 μl total volume was incubated at 25°C for 5 min and terminated by heating at 85°C for 15 min. After centrifuging at 16,000 g for 10 min, the supernatant was analyzed using an YMC‐pack pro‐C18 reverse phase column (4.6 × 250 mm, 5 μm). Analytes were monitored using 254 nm light. Buffer A contained 5 mM ammonium acetate (pH 5.0) in water, and phase B was 100% acetonitrile. The samples were eluted using a linear gradient from 2% B to 15% B at a flow rate of 1 ml/min over 15 min. The product 2′3′‐cGAMP was quantified by measuring the peak area (μAU·sec) of 2′3′‐cGAMP visualized with 254 nm light. Each experiment was performed in triplicate.
IFNB mRNA quantification in THP1 cells
The ssRNA used for transfection was prepared using an in vitro transcription kit (Thermo Fisher, cat# AM1334), treated with CIAP (calf intestinal alkaline phosphatase, Thermo Fisher, cat# 18009019), and purified with a clean‐up kit (Thermo Fisher, cat# AM1908) according to the instructions provided by the vendor. THP1 cells were seeded in 6‐well plates. RNA and DNA were transfected with Lipofectamine 2000 (Thermo Fisher). For cGAS inhibitor G150 treated groups, 10 μM G150 (MCE, cat# HY‐128583) was added to the medium 1 h before transfection. After 6 h, cells were harvested and RNA was extracted using the GeneJet RNA purification kit (Thermo Fisher). The cDNAs were produced by reverse transcriptions using the PrimeScript™ RT reagent kit with gDNA Eraser (Takara). RT‐qPCR was performed by using the iTaq Universal SYBR Green Supermix on a CFX96 RT–qPCR machine (Bio‐Rad). PPIB was chosen as the reference gene. The primers for the RT–qPCRs were designed by using the PrimerBank server (http://pga.mgh.harvard.edu/primerbank/). Primer sequences are as follows: PPIB: forward 5′‐GTCCGTCTTCTTCCTGCTG‐3′, reverse 5′‐CATCTTCATCTCCAATTCGTAGG‐3′. IFNB: forward 5′‐ACGCCGCATTGACCATCTAT‐3′, reverse 5′‐TGGCCTTCAGGTAATGCAGA‐3′. The RT–qPCR results were analyzed with the Bio‐Rad software CFX manager, and the graphs were drawn by using the software GraphPad Prism.
Author contributions
Silian Chen: Data curation; investigation; writing – review and editing. Miao Rong: Data curation. Yun Lv: Data curation; writing – review and editing. Deyu Zhu: Data curation; funding acquisition; methodology; writing – review and editing. Ye Xiang: Conceptualization; supervision; funding acquisition; writing – original draft; project administration; writing – review and editing.
Disclosure and competing interests statement
The authors declare that they have no conflict of interest.
Supporting information
Appendix S1
Expanded View Figures PDF
PDF+
Acknowledgments
We thank the Core Facility and the Cell Biology Facility of the Center of Biomedical Analysis, Tsinghua University, and the imaging core facility of China National Center for Protein Sciences (Beijing)/Technology Center for Protein Sciences, Tsinghua University, for assistance with imaging. We thank Dr. Jiaying Yang for providing the extracted total RNA from HeLa cells. This work was supported by funds from the Ministry of Science and Technology of China (grant number: 2021YFA1300204), the Tsinghua University Vanke Special Fund for Public Health and Health Discipline Development (NO. 2022Z82WKJ013), the National Natural Science Foundation of China (grant numbers: 31925023, 21827810, 31861143027, & 31470721), the Spring Breeze Fund of Tsinghua University, the Beijing Frontier Research Center for Biological Structure, and the Beijing Advanced Innovation Center for Structure Biology to YX and the Key R&D Project of Shandong Province, China (grant number: 2019GSF108212) to DZ.
EMBO reports (2023) 24: e51800
Contributor Information
Deyu Zhu, Email: zhudeyu@sdu.edu.cn.
Ye Xiang, Email: yxiang@mail.tsinghua.edu.cn.
Data availability
No large primary datasets have been generated and deposited.
References
- Alberti S, Gladfelter A, Mittag T (2019) Considerations and challenges in studying liquid‐liquid phase separation and biomolecular condensates. Cell 176: 419–434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreeva L, Hiller B, Kostrewa D, Lassig C, de Oliveira Mann CC, Jan Drexler D, Maiser A, Gaidt M, Leonhardt H, Hornung V et al (2017) cGAS senses long and HMGB/TFAM‐bound U‐turn DNA by forming protein‐DNA ladders. Nature 549: 394–398 [DOI] [PubMed] [Google Scholar]
- Barbalat R, Ewald SE, Mouchess ML, Barton GM (2011) Nucleic acid recognition by the innate immune system. Annu Rev Immunol 29: 185–214 [DOI] [PubMed] [Google Scholar]
- Brangwynne CP, Eckmann CR, Courson DS, Rybarska A, Hoege C, Gharakhani J, Julicher F, Hyman AA (2009) Germline P granules are liquid droplets that localize by controlled dissolution/condensation. Science 324: 1729–1732 [DOI] [PubMed] [Google Scholar]
- Civril F, Deimling T, de Oliveira Mann CC, Ablasser A, Moldt M, Witte G, Hornung V, Hopfner KP (2013) Structural mechanism of cytosolic DNA sensing by cGAS. Nature 498: 332–337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darzynkiewicz Z, Juan G, Srour EF (2004) Differential staining of DNA and RNA. Curr Protoc Cytom Chapter 7: Unit 7.3 [DOI] [PubMed] [Google Scholar]
- Du M, Chen ZJ (2018) DNA‐induced liquid phase condensation of cGAS activates innate immune signaling. Science 361: 704–709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feijo Delgado F, Cermak N, Hecht VC, Son S, Li Y, Knudsen SM, Olcum S, Higgins JM, Chen J, Grover WH et al (2013) Intracellular water exchange for measuring the dry mass, water mass and changes in chemical composition of living cells. PLoS One 8: e67590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frenkel‐Morgenstern M, Danon T, Christian T, Igarashi T, Cohen L, Hou YM, Jensen LJ (2012) Genes adopt non‐optimal codon usage to generate cell cycle‐dependent oscillations in protein levels. Mol Syst Biol 8: 572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao P, Ascano M, Zillinger T, Wang W, Dai P, Serganov AA, Gaffney BL, Shuman S, Jones RA, Deng L et al (2013) Structure‐function analysis of STING activation by c[G(2′,5′)pA(3′,5′)p] and targeting by antiviral DMXAA. Cell 154: 748–762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao D, Li T, Li X‐D, Chen X, Li Q‐Z, Wight‐Carter M, Chen ZJ (2015) Activation of cyclic GMP‐AMP synthase by self‐DNA causes autoimmune diseases. Proc Natl Acad Sci U S A 112: E5699–E5705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herzner A‐M, Hagmann CA, Goldeck M, Wolter S, Kübler K, Wittmann S, Gramberg T, Andreeva L, Hopfner K‐P, Mertens C et al (2015) Sequence‐specific activation of the DNA sensor cGAS by Y‐form DNA structures as found in primary HIV‐1 cDNA. Nat Immunol 16: 1025–1033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu MM, Yang Q, Xie XQ, Liao CY, Lin H, Liu TT, Yin L, Shu HB (2016) Sumoylation promotes the stability of the DNA sensor cGAS and the adaptor STING to regulate the kinetics of response to DNA virus. Immunity 45: 555–569 [DOI] [PubMed] [Google Scholar]
- Hyman AA, Weber CA, Julicher F (2014) Liquid‐liquid phase separation in biology. Annu Rev Cell Dev Biol 30: 39–58 [DOI] [PubMed] [Google Scholar]
- Ishikawa H, Barber GN (2008) STING is an endoplasmic reticulum adaptor that facilitates innate immune signalling. Nature 455: 674–678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jao CY, Salic A (2008) Exploring RNA transcription and turnover in vivo by using click chemistry. Proc Natl Acad Sci U S A 105: 15779–15784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin L, Waterman PM, Jonscher KR, Short CM, Reisdorph NA, Cambier JC (2008) MPYS, a novel membrane tetraspanner, is associated with major histocompatibility complex class II and mediates transduction of apoptotic signals. Mol Cell Biol 28: 5014–5026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karalyan ZA, Izmailyan RA, Abroyan LO, Avetisyan AS, Hakobyan LA, Zakaryan HS, Karalova EM (2018) Evaluation of viral genome copies within viral factories on different DNA viruses. J Histochem Cytochem 66: 359–365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kranzusch Philip J, Lee Amy S‐Y, Berger James M, Doudna Jennifer A (2013) Structure of human cGAS reveals a conserved family of second‐messenger enzymes in innate immunity. Cell Rep 3: 1362–1368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lama L, Adura C, Xie W, Tomita D, Kamei T, Kuryavyi V, Gogakos T, Steinberg JI, Miller M, Ramos‐Espiritu L et al (2019) Development of human cGAS‐specific small‐molecule inhibitors for repression of dsDNA‐triggered interferon expression. Nat Commun 10: 2261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau L, Gray EE, Brunette RL, Stetson DB (2015) DNA tumor virus oncogenes antagonize the cGAS‐STING DNA‐sensing pathway. Science 350: 568–571 [DOI] [PubMed] [Google Scholar]
- Li X, Shu C, Yi G, Chaton Catherine T, Shelton Catherine L, Diao J, Zuo X, Kao CC, Herr Andrew B, Li P (2013a) Cyclic GMP‐AMP synthase is activated by double‐stranded DNA‐induced Oligomerization. Immunity 39: 1019–1031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li XD, Wu J, Gao D, Wang H, Sun L, Chen ZJ (2013b) Pivotal roles of cGAS‐cGAMP signaling in antiviral defense and immune adjuvant effects. Science 341: 1390–1394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luecke S, Holleufer A, Christensen MH, Jonsson KL, Boni GA, Sorensen LK, Johannsen M, Jakobsen MR, Hartmann R, Paludan SR (2017) cGAS is activated by DNA in a length‐dependent manner. EMBO Rep 18: 1707–1715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackenzie KJ, Carroll P, Martin CA, Murina O, Fluteau A, Simpson DJ, Olova N, Sutcliffe H, Rainger JK, Leitch A et al (2017) cGAS surveillance of micronuclei links genome instability to innate immunity. Nature 548: 461–465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mankan AK, Schmidt T, Chauhan D, Goldeck M, Honing K, Gaidt M, Kubarenko AV, Andreeva L, Hopfner KP, Hornung V (2014) Cytosolic RNA:DNA hybrids activate the cGAS‐STING axis. EMBO J 33: 2937–2946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palazzo AF, Lee ES (2015) Non‐coding RNA: what is functional and what is junk? Front Genet 6: 2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey S, Kawai T, Akira S (2014) Microbial sensing by toll‐like receptors and intracellular nucleic acid sensors. Cold Spring Harb Perspect Biol 7: a016246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu C, Liu WY, Xu YZ (2015) Fluorescence labeling of short RNA by oxidation at the 3′‐end. Methods Mol Biol 1297: 113–120 [DOI] [PubMed] [Google Scholar]
- Rongvaux A, Jackson R, Harman CC, Li T, West AP, de Zoete MR, Wu Y, Yordy B, Lakhani SA, Kuan CY et al (2014) Apoptotic caspases prevent the induction of type I interferons by mitochondrial DNA. Cell 159: 1563–1577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russo J, Russo IH (2014) Techniques and methodological approaches in breast cancer research. New York: Springer; [Google Scholar]
- Seo GJ, Yang A, Tan B, Kim S, Liang Q, Choi Y, Yuan W, Feng P, Park HS, Jung JU (2015) Akt kinase‐mediated checkpoint of cGAS DNA sensing pathway. Cell Rep 13: 440–449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun W, Li Y, Chen L, Chen H, You F, Zhou X, Zhou Y, Zhai Z, Chen D, Jiang Z (2009) ERIS, an endoplasmic reticulum IFN stimulator, activates innate immune signaling through dimerization. Proc Natl Acad Sci U S A 106: 8653–8658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun L, Wu J, Du F, Chen X, Chen ZJ (2013) Cyclic GMP‐AMP synthase is a cytosolic DNA sensor that activates the type I interferon pathway. Science 339: 786–791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor NO, Wei MT, Stone HA, Brangwynne CP (2019) Quantifying dynamics in phase‐separated condensates using fluorescence recovery after photobleaching. Biophys J 117: 1285–1300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torrent M, Chalancon G, de Groot NS, Wuster A, Babu MM (2018) Cells alter their tRNA abundance to selectively regulate protein synthesis during stress conditions. Sci Signal 11: eaat6409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vance RE (2016) Cytosolic DNA sensing: the field narrows. Immunity 45: 227–228 [DOI] [PubMed] [Google Scholar]
- Wang H, Hu S, Chen X, Shi H, Chen C, Sun L, Chen ZJ (2017a) cGAS is essential for the antitumor effect of immune checkpoint blockade. Proc Natl Acad Sci U S A 114: 1637–1642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Huang L, Hong Z, Lv Z, Mao Z, Tang Y, Kong X, Li S, Cui Y, Liu H et al (2017b) The E3 ubiquitin ligase RNF185 facilitates the cGAS‐mediated innate immune response. PLoS Pathog 13: e1006264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- West AP, Khoury‐Hanold W, Staron M, Tal MC, Pineda CM, Lang SM, Bestwick M, Duguay BA, Raimundo N, MacDuff DA et al (2015) Mitochondrial DNA stress primes the antiviral innate immune response. Nature 520: 553–557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- White MJ, McArthur K, Metcalf D, Lane RM, Cambier JC, Herold MJ, van Delft MF, Bedoui S, Lessene G, Ritchie ME et al (2014) Apoptotic caspases suppress mtDNA‐induced STING‐mediated type I IFN production. Cell 159: 1549–1562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, Chen ZJ (2014) Innate immune sensing and signaling of cytosolic nucleic acids. Annu Rev Immunol 32: 461–488 [DOI] [PubMed] [Google Scholar]
- Wu J, Sun L, Chen X, Du F, Shi H, Chen C, Chen ZJ (2013) Cyclic GMP‐AMP is an endogenous second messenger in innate immune signaling by cytosolic DNA. Science 339: 826–830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia P, Ye B, Wang S, Zhu X, Du Y, Xiong Z, Tian Y, Fan Z (2016) Glutamylation of the DNA sensor cGAS regulates its binding and synthase activity in antiviral immunity. Nat Immunol 17: 369–378 [DOI] [PubMed] [Google Scholar]
- Yang H, Wang H, Ren J, Chen Q, Chen ZJ (2017) cGAS is essential for cellular senescence. Proc Natl Acad Sci U S A 114: E4612–E4620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Shi H, Wu J, Zhang X, Sun L, Chen C, Chen ZJ (2013) Cyclic GMP‐AMP containing mixed phosphodiester linkages is an endogenous high‐affinity ligand for STING. Mol Cell 51: 226–235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao M, Xia T, Xing JQ, Yin LH, Li XW, Pan J, Liu JY, Sun LM, Wang M, Li T et al (2022) The stress granule protein G3BP1 promotes pre‐condensation of cGAS to allow rapid responses to DNA. EMBO Rep 23: e53166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong B, Yang Y, Li S, Wang YY, Li Y, Diao F, Lei C, He X, Zhang L, Tien P et al (2008) The adaptor protein MITA links virus‐sensing receptors to IRF3 transcription factor activation. Immunity 29: 538–550 [DOI] [PubMed] [Google Scholar]


