Summary
Background
Worldwide, Escherichia coli is the leading cause of neonatal Gram-negative bacterial meningitis, but full understanding of the pathogenesis of this disease is not yet achieved. Moreover, to date, no vaccine is available against bacterial neonatal meningitis.
Methods
Here, we used Transposon Sequencing of saturated banks of mutants (TnSeq) to evaluate E. coli K1 genetic fitness in murine neonatal meningitis. We identified E. coli K1 genes encoding for factors important for systemic dissemination and brain infection, and focused on products with a likely outer-membrane or extra-cellular localization, as these are potential vaccine candidates. We used in vitro and in vivo models to study the efficacy of active and passive immunization.
Results
We selected for further study the conserved surface polysaccharide Poly-β-(1-6)-N-Acetyl Glucosamine (PNAG), as a strong candidate for vaccine development. We found that PNAG was a virulence factor in our animal model. We showed that both passive and active immunization successfully prevented and/or treated meningitis caused by E. coli K1 in neonatal mice. We found an excellent opsonophagocytic killing activity of the antibodies to PNAG and in vitro these antibodies were also able to decrease binding, invasion and crossing of E. coli K1 through two blood brain barrier cell lines. Finally, to reinforce the potential of PNAG as a vaccine candidate in bacterial neonatal meningitis, we demonstrated that Group B Streptococcus, the main cause of neonatal meningitis in developed countries, also produced PNAG and that antibodies to PNAG could protect in vitro and in vivo against this major neonatal pathogen.
Interpretation
Altogether, these results indicate the utility of a high-throughput DNA sequencing method to identify potential immunotherapy targets for a pathogen, including in this study a potential broad-spectrum target for prevention of neonatal bacterial infections.
Fundings
ANR Seq-N-Vaq, Charles Hood Foundation, Hearst Foundation, and Groupe Pasteur Mutualité.
Keywords: Neonatal meningitis, Vaccine, High-throughput sequencing, E. coli K1, PNAG
Research in context.
Evidence before this study
Four factors are involved in this study: (i) Escherichia coli K1, (ii) Group B streptococcus, (iii) the conserved surface polysaccharide Poly-β-(1-6)-N-Acetyl Glucosamine (PNAG) and (IV) the next generation of high-throughput sequencing.
(i–ii) E. coli K1 is a major cause of bacterial neonatal meningitis and is a persistent problem in neonates, despite the application of antibiotics and supportive care. The cross-reactivity of the K1 bacterial capsular polysaccharide with human brain antigens prevents their being used as vaccine candidates. Group B streptococcus is another main cause of neonatal bacterial meningitis. Implementing a vaccine is a promising strategy to prevent neonatal and infant GBS disease and has been identified as a priority by the World Health Organization (WHO). But to date, no vaccine is clinically available (PubMed search using E. coli K1 and Vaccine; then GBS and Vaccine).
(iii) The PNAG surface polysaccharide has been described as a good vaccine candidate and PNAG antibodies have a broad spectrum of activity against both Gram-positive and -negative pathogens. Phase 1 clinical trials using human monoclonal antibodies and vaccines against PNAG were found safe. PNAG has never been studied as an antigen candidate for either neonatal infections nor bacterial meningitis (46 papers found in a PubMed search using PNAG and Vaccine).
(IV) Finally, the use of Transposons (Tn) libraries and high-throughput Sequencing (TnSeq) is an innovative way to evaluate the microbial fitness for E. coli K1 in different settings and provide innovative and novel insights into the host–pathogen interactions at the full genome scale. By contrast, older technologies almost exclusively analysed interactions between a host, a pathogen and a limited set of genes. TnSeq's main advantages come from the rapidity with which comparative genomic data can be generated and analysed and the breadth and scope of the analysis for contributions to fitness that encompasses the entire set of non-essential genes and regulatory factors, including small RNAs. To date only one previous study has used TnSeq in E. coli K1 (PubMed search using E. coli K1, Transposons, Mutants and Sequencing) but while it looked at in vitro growth, gastrointestinal colonizing and survival in serum, it did not look at the final crucial step of the neonatal meningitis: the crossing of blood brain barrier.
Added value of this study
Using TnSeq to study E. coli K1 systemic dissemination and brain infection, we found that PNAG was a virulence factor important for crossing the blood brain barrier and, with an outer membrane localization, constitutes an optimal target for protective antibodies. PNAG was therefore fully explored in a systematic preclinical study using both in vitro and in vivo models. We demonstrated that PNAG was an excellent candidate for both passive and active immunization to either prevent or treat neonatal meningitis caused by E. coli K1. In the same study, we extended this finding about PNAG to neonatal meningitis caused by GBS, once again using both in vitro and in vivo settings. Having a strong, unique antigen candidate, for either passive or active immunization against both E. coli K1 and GBS is a breakthrough in the fight against bacterial neonatal meningitis.
Implications of all the available evidence
High-throughput sequencing can be used as a pipeline to study both bacterial pathogenesis and find antigens of interest. In addition, our finding supports the study of PNAG as a target for immunotherapy in neonatal bacterial meningitis toward the path of a full clinical development.
Introduction
Neonatal bacterial meningitis continues to be a serious disease and even when treated with appropriate antibiotics, mortality rates over 10% still ensue. Up to 40% of survivors exhibit sequelae such as hearing and neurologic impairments and hydrocephalus.1,2 The burden of morbidity and mortality from neonatal bacterial meningitis is substantial in industrialized and developing countries.3,4 Even with the introduction of maternal intrapartum antibiotic prophylaxis that markedly reduced the incidence of infection with Group B Streptococcus (GBS), this organism remains the most common cause of neonatal sepsis and meningitis, responsible for more than 40% of all early-onset infections.5,6 However, worldwide, E. coli strains causing neonatal meningitis (NMEC) are responsible for 15–20% of early-onset infections in term infants and for about 35% in preterm newborns.1 Despite the use of advanced antibiotics, the morbidity and mortality rates associated with E. coli K1 strains (the vastly predominant NMEC) causing meningitis remain unchanged over the last few decades. In addition, due to the emergence of antibiotic resistant E. coli K1 strains, notably those producing the extended spectrum beta-lactamase CTX-M, the mortality rates may further increase significantly in the future.7 Thus, a vaccine preventing E. coli K1 meningitis represents an important medical need. While bacterial capsules are usually promising vaccine targets, the K1 capsule, like that of Neisseria meningitidis group B, are polysialic acid capsules structurally similar to the polysialic acid moiety of the mammalian neural cell adhesion molecule.8 This structural similarity prevents an effective antibody response and attempts to raise antibody to the K1 capsule raise concerns about pathologic cross-reactivity. Therefore, new modes of prevention and treatment are warranted based on an improved understanding of the disease pathophysiology and of E. coli K1 pathogenic factors. Using saturated transposon (Tn) mutagenesis and high-throughput sequencing (TnSeq), a powerful tool to investigate host–pathogen interactions, we fill-in-gaps in our knowledge about the contribution to overall fitness of the multitude of genes in E. coli K1 needed to cause neonatal meningitis. This approach led to the identification of Poly-β-(1-6)-N-Acetyl Glucosamine (PNAG) as a strong candidate vaccine antigen produced by this pathogen as it appears to be both a virulence factor and to have an outer surface localization. PNAG plays an important role in biofilm formation of Gram-positive and -negative bacteria and it is known to participate in immune evasion.9 PNAG has already been described as a good vaccine candidate10 and PNAG antibodies have a broad spectrum of activity against both Gram-positive and -negative pathogens.11,12 Phase 1 clinical trials using human monoclonal antibodies and vaccines against PNAG were found safe.13 We therefore fully explored this candidate in E. coli K1 meningitis, using both in vitro and in vivo studies.
Methods
Ethics statement
The Harvard Medical School and Brigham and Women's Hospital animal management programs are accredited by the Association for the Assessment and Accreditation of Laboratory Animal Care, International (AAALAC), and meet National Institute of Health standards as set forth in the 8th edition of the Guide for the Care and Use of Laboratory Animals (National Research Council “Animal Care and Use Program”. Guide for the Care and Use of Laboratory Animals: Eighth Edition. Washington, DC: The National Academies Press, 2011). The institution also accepts as mandatory the PHS Policy on Humane Care and Use of Laboratory Animals by Awardee Institutions and NIH Principles for the Utilization and Care of Vertebrate Animals used in Testing, Research, and Training.
All animal studies conducted in Boston in this research were approved by the Institutional Animal Care and Use.
All the animal experiments conducted in the University of Paris City were approved by the Ministry of Research.
Two animal protocols were approved with David Skurnik as Principal investigator: APAFIS#26590-2020011519022360 and APAFIS#337319-202201311143868.
Strains
Strains used in this study are listed in Table S1. Primers and plasmids are listed Table S2.
E. coli TnSeq
Illumina libraries were prepared as described.14 Briefly, for the preparation of the library, 100 independent conjugation mixtures were used. The yield was approximately 300,000 individual colonies of E. coli K1 S88 from 100 plates. The pooled library was further incubated for two additional hours in LB with gentamicin. Following centrifugation of the culture, the library was re-suspended in LB containing 20% glycerol and aliquots were frozen at −80 °C. For DNA preparation for high-throughput sequencing, the MmeI type-IIS restriction-modification enzyme was used. MmeI cuts the double-stranded DNA 20 bp away from the recognition site, thus 16 bp outside of the Tn insertion. It produces sticky-end DNA fragments with a two-nucleotide 3′overhang. The transposons inserted into E. coli K1 S88 genome contained MmeI restriction sites on both ends of the Tn, allowing the MmeI enzyme fragments of the genes on both sides of the Tn insertion to be used for sequencing. Moreover, P7 Illumina sequences were also present just adjacent to the MmeI recognition sites on both ends of the Tn. These features allowed the high throughput sequencing to proceed to identify the gene interrupted and place of insertion for each individual Tn insertion. Overall, DNA was extracted, digested with MmeI, and ligated to oligonucleotide adapters, PCR amplified and sequenced using Illumina HiSeq.14,15 Between 20 and 50 million sequencing reads per sample were recovered.
No barcoding was used
All sequences retrieved from the Illumina reactions were trimmed to eliminate those reads with quality scores less than 0.05 and/or sequences with ambiguous nucleotides.14,15 Data were analyzed using the RNA-seq module of the Bioinformatics Workbench software package (CLC) (http://www.clcbio.com/index.php?id=1240). Circos figures in the main text (Fig. 1, Fig. 2, Fig. 3) and in the supporting information figures (Figure S7) were drawn following the instructions provided at http://www.circos.ca/. For each Tn-insert, the significance of the difference in the proportions of the RPKM from one environment to the other was estimated using the statistical test defined by Kal et al.16 All P-values were adjusted for multiple comparisons using Bonferroni corrections. Analysis by operon: for this analysis, we took into account possible polar effects created by the Tn-insertions into genes transcribed from a single promoter and considered that fitness is attributable to the inactivation of operons rather than individual genes.
Fig. 1.
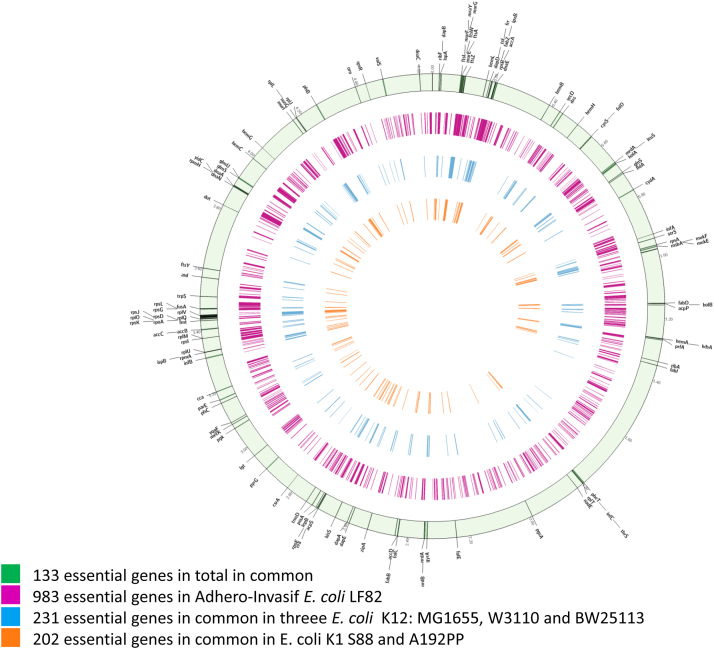
Comparative identification of essential genes in E. coli. Orange circle: common essential genes found in both E. coli K1 S88 and E. coli K1 A192PP. Blue circle: common essential genes found in E. coli K12 MG1655, E. coli K12 W3110 and E. coli K12 BW25113. Purple circle: genes identified as essential in strain E. coli LF82. Green circle: the essential genes found in common for all these strains, annotated and located on E. coli MG1655 chromosome. Table S1 gives a complete listing of the essential genes found for all strains in these conditions.
Fig. 2.
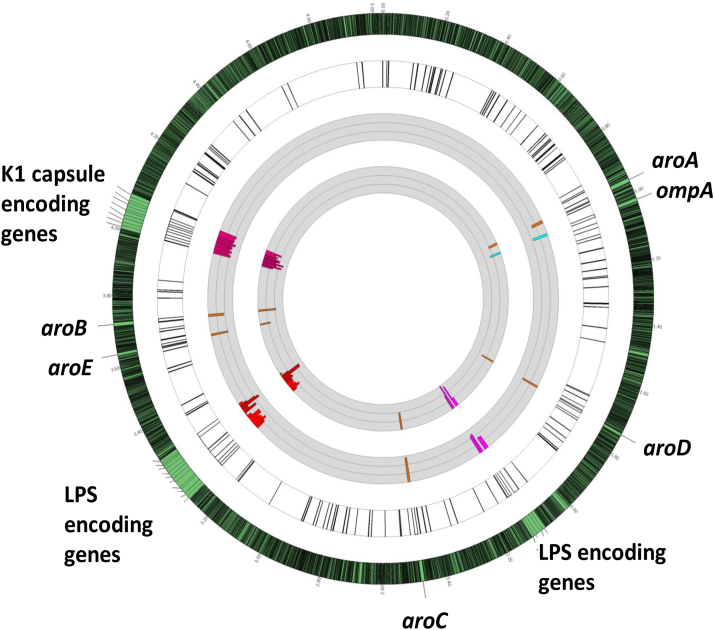
E. coli K1 in vivo (systemic dissemination) genetic fitness analysis using TnSeq localization on E. coli K1 S88 chromosome and quantitative analysis. The outermost circle represents the full E. coli K1 S88 genome with a 30-times magnification of the regions of interest. The first white circle represents the localization on E. coli K1 S88 chromosome of all the genes found important or essential for systemic dissemination (detailed Fig. S7 and Table S1) by TnSeq. Previously described virulence factors of E. coli K1 are highlighted in colour: LPS and K1 capsule encoding genes as examples for gene clusters, OmpA as an example of a unique gene and Aro encoding genes as examples of several individual genes (i.e not in cluster) located on distant parts of the chromosome. The first gray circle represents the fold change of the reads of the Tn insertions from the LB to the Liver after IP challenge. The second gray circle represents the fold change of the reads of the Tn insertions from the LB to the liver after gavage. The thin grey circular lines represent 10-fold-changes (i.e., at log10 scale). The limit of the fold change representation is 1000. Bars represent changes in individual genes. Bars pointing toward the circle's center represent Tn interrupted genes resulting in negative fitness.
Fig. 3.
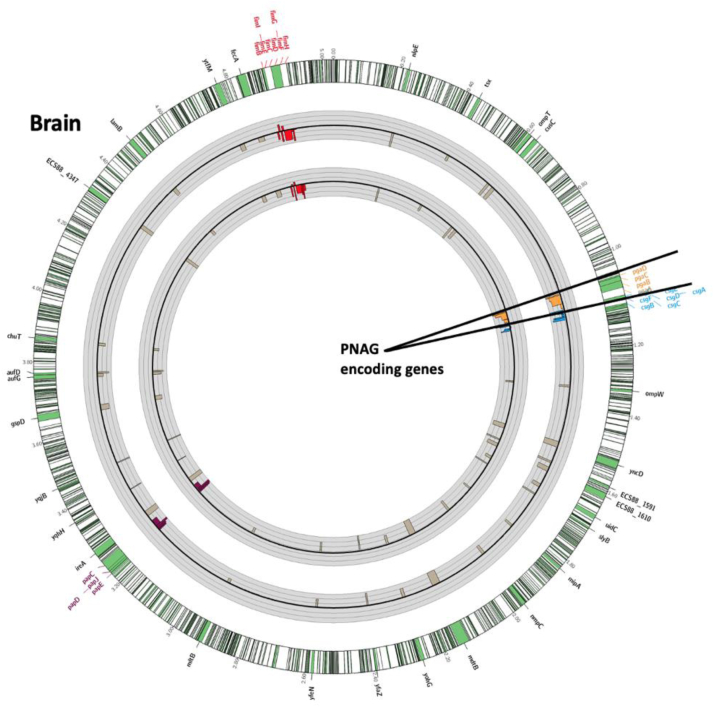
In vivo (brain infection) genetic fitness analysis using TnSeq of E. coli K1 mutants in genes encoding extracellular products or outer-membrane proteins. The outermost circle represents the full E. coli K1 S88 genome with a 30-times magnification of the regions of interest shown as green portions of the chromosome. The first gray circle represents the fold change of the reads of the Tn insertions from the Liver to the Brain after gavage. The second gray circle represents the fold change of the reads of the Tn insertions from the LB to the Brain after gavage. Black circular lines in the middle of the grey circles represent baseline. Thin grey circular lines above and below the central black circular line represent 10-fold-changes (i.e., a log10 scale). The limit of the fold change representation is 1000. Bars represent changes in individual genes. Bars pointing toward the circle's center represent Tn interrupted genes resulting in negative fitness whereas bars pointed outward represent genes whose loss increases fitness. Poly-β-(1-6)-N-Acetyl-Glucosamine (PNAG), Curly, Type 1 Fimbriae and Type 1 pilus encoding genes are highlighted in red, purple, blue and light brown respectively.
Murine models of bacterial neonatal severe infection
Bacterial preparations
The saturated bank of E. coli K1 S88 was grown overnight in lysogeny broth (LB) and then diluted in sodium phosphate buffer, pH 7.6, in an appropriate volume according to the needed bacterial concentration for challenge. E. coli K1 S88, E. coli K1 E11, E. coli K1 E11 Δpga, E. coli K1 E11 Δpga::pga, E. coli K1CTX-M, Group B Streptococcus NEM316 were grown overnight at 37 °C on a blood agar plate. From that plate, a 150 mL flask containing 15 mL of LB was inoculated to reach an Optical Density (O.D) at 650 nm = 0.1 that was incubated at 37 °C for about 2 h, until mid-exponential growth phase (O.D 650 nm = 0.5) was obtained. This suspension was administrated directly or was pelleted and resuspended in sodium phosphate buffer pH 7.6, in an appropriate volume according to the bacterial concentrations needed for the challenge.
E. coli K1 intraperitoneal challenge model
Two or three day-old CD1 mice were separated from their dams and received an intraperitoneal (IP) injection of the infectious bacterial suspension (5 × 106 colony forming units (CFU) of the Tn-mutants bank in 50 μL/mouse). Green food dye was added to visually validate the IP location of the bacterial challenge (Figure S2). The neonates were routinely monitored every 2–6 h and humanely euthanized if they showed signs of severe illness. All surviving animals were euthanized at 24 h after inoculation. The spleens and the livers were harvested and homogenized, serial dilutions of each organ were made and plated on blood agar plates and MacConkey plates to allow the most precise calculation of bacterial load in each organ.
Gavage model of neonatal bacterial meningitis
Two or three day-old CD1 mice were separated from their mothers 2 h before challenge by oral gavage to limit interference from milk in the induction of infection. The neonatal mice also received an IP administration of Ranitidine (20 μg/mouse in 50 μL of saline solution) to decrease the potential for bacterial killing by gastric acidity. For intragastric administration of bacterial suspensions (5 × 106 CFU of the Tn-mutants bank or 105 to 5 × 106 CFU of a specific bacterial strain in 50 μL/animal) to the neonates, anaesthesia using inhaled Isoflurane was performed. Green food dye in the inoculum was used to confirm the intragastric location of the bacterial challenge (Figure S2). Infected neonates were monitored and euthanized as described above, and bacterial burdens in spleens, livers and brains also determined as described above. Litters of ten pups were used both for the IP and the gavage models.
Important genes for systemic dissemination and brain infection
At least 3x106 bacteria were recovered from livers of individual mice in both neonatal infection models and from brains of individual mice following oral gavage leading to meningitis. Genomic DNA was extracted, digested by MmeI and gel-sized fragments ligated to oligonucleotides adapters, PCR amplified and sequenced.
Construction of the E. coli K1 E11 Δpga deletion mutant
Deletion of the pga locus in the E. coli K1 strain E11 was constructed as previously described.17 A kanamycin resistance cassette flanked by FLP recombinase recognition target sites and homology arms to replace the DNA segment of interest were generated by PCR with adequate deletion primers (Table S5). Recombination with the targeted chromosome sequences was mediated by the Red recombinase encoded on pRedET,18 resulting in the replacement of the targeted sequence with a kanamycin-resistant cassette; allele replacement was confirmed by PCR. Subsequently, the kanamycin marker was removed, using the FLP expression plasmid pCP20.
Antibodies
Two fully human IgG1 MAb were obtained by cloning V-regions from human B cells encoding specificity for either Pseudomonas aeruginosa alginate, MAb F429, or PNAG, MAb F598. V-region genes for both were cloned into the TCAE 6.2 expression vector providing them with identical human lambda-light chain and gamma-1 heavy chain constant regions. Both have been described previously.19,20
Animal sera
Goat antiserum was raised to deacetylated PNAG, as previously described,10 using a maleimide-sulfydryl–based conjugation scheme to produce a conjugate vaccine. In brief, purified dPNAG was derivatized with a maleimide group using the chemical linker N- [γ-maleimidobutyryloxy] succinimide ester (GMBS). Free sulfhydryl (SH) groups were introduced into the carrier protein TT by treatment with N-succinimidyl-3-(2-pyridyldithio)-propionate (SPDP). The maleimide-dPNAG and the SH-TT were mixed together for 2 h at room temperature to chemically couple these 2 components together, and the dPNAG-TT conjugate was purified by size exclusion chromatography on a Superose 6 column (GE Healthcare). Goats were immunized as described previously. This serum was replaced during the course of this study by polyclonal goat antiserum raised to a conjugate vaccine comprised of a nonameric β-1-6-linked glucosamine oligosaccharide conjugated to tetanus toxoid (9GlcNH2-TT) that became available after being described in reference 12.
Confocal microscopy and immunochemical detection of PNAG
PNAG expression on different E. coli and GBS strains
Microbial samples from broth were fixed by suspension in 4% paraformaldehyde, then spotted onto microscope slides, air-dried, and covered for about 1 min with ice-cold methanol. After washing, slides were reacted with control MAb F429 or PNAG-specific MAb F598 directly conjugated to Alexa Fluor 488 at 5.2 μg/mL along with a DNA-visualizing dye (4 μM of Syto 83). After 2 h at room temperature or overnight at 4 °C, slides were washed and observed by confocal microscopy. For enzymatic treatments to confirm PNAG identification, samples fixed to slides were incubated in Tris-buffered saline (pH 6.4) containing either 50 μg/mL dispersin B (specifically digests the β-(1-6)-N-acetyl-glucosamine linkages in PNAG) or 50 μg/mL chitinase (no effect on PNAG antigen, digests the β-(1-4)-N-acetyl-glucosamine linkages in chitin) overnight at 37 °C. After washing, cells were treated with the Alexa Fluor 488 directly conjugated to MAb. Samples were washed and mounted for confocal microscopy.11,21
PNAG expression in brains during bacterial meningitis
Sample preparation
We infected a group of 8 baby mice at D3 (three days after birth) IP with the wild-type strain of E. coli K1 E11. The control (uninfected) group received a saline injection. One day after infection, the infected and uninfected mouse pups were sacrificed and the brains were removed and homogenised. For this purpose, the brains were resuspended in ice-cold lysis buffer (50 mM Tris–HCl pH = 7.5, 100 mM NaCl, 5 mM DTT, 1 mM PMSF) and chilled on ice for 10 min. Next, the cell suspension was sonicated with 10 short 10 s bursts, followed by 30 s intervals for cooling. Finally, cell debris was removed by ultracentrifugation at 4 °C for 15 min at 14000 rpm. The supernatant was run on a 10% polyacrylamide gel and analyzed by Western blot.
Western blot
For the Western blot, we loaded 15 μl of supernatant onto the gel. The samples were mixed with one time their volume of 2× SDS (Sodium Dodecyl Sulfate) dyes (SDS 25%, Tris–HCl 62.5 mM (pH 6.8), Glycerol 25%, and Bromophenol Blue 0.01%) and denatured for 5 min at 95 °C. They were then plated on a 10% SDS-PAGE gel in 1× Tris-Glycine SDS buffer. Migration was performed at 100 V for approximately 2 h. The gel was removed from the mould and the transfer of the gel contents onto a nitrocellulose membrane (Hybond C super) was carried out by electrophoretic transfer: the gel and the membrane were put into a cassette for transfer onto the membrane. The support to make the sandwich was put in transfer buffer (kept at 4 °C) Tris 20 mM/Glycine 150 mM/Ethanol 20% and the following assembly was made: on the negative side, we put in succession a spontex, 2 Whatman 3 MM papers, the gel, the membrane, 2 Whatmans and a spontex. The holder was closed and placed in the transfer tank containing the transfer buffer. The transfer was performed at 100 V, 250 mA for 2 h at 4 °C. The membrane was removed from the mould and stained with ponceau red to check the transfer and rinsed with 1× PBS/0.1% Tween. The membrane was saturated for 1 h at room temperature in PBS 1×/Tween 0.1%/Milk 5% and rinsed 2 × 5 min in PBS 1×/Tween 0.1%. The membrane was then incubated with 20 μg of Anti-PNAG antibody (F598 mAb) diluted in 20 mL of PBS 1×/Tween 0.1%/Milk 5 overnight at 4 °C and washed 2 × 30 s in PBS 1×/Tween 0.1%, 1 × 15 min in PBS 1×/Tween 0.1% and 3 × 5 min in PBS 1×/Tween 0.1%. It was then incubated with the secondary antibody (Anti-rabbit human IgG coupled to HRP peroxidase, Abcam Ab97160) diluted 1:10000 PBS 1×/Tween 0.1%/Milk 5% for 1 h at room temperature and washed 2 × 30 s in PBS 1×/Tween 0.1%, 1 × 15 min in PBS 1×/Tween 0.1% and 3 × 5 min in PBS 1×/Tween 0.1%. The bands were visualized using the ECL Advance kit and chemiluminescence measured using the Chemidoc system.
Opsonophagocytosis killing assays
The opsonophagocytic killing assays (OPKA) followed published protocols.22,23 Both a polyclonal goat antiserum raised to a conjugate vaccine comprised of a nonameric β-1-6-linked glucosamine oligosaccharide conjugated to tetanus toxoid (9GlcNH2- TT), as well as the fully human IgG1 MAb to PNAG, F598, were used in opsonic killing assays. Normal goat serum (NGS) and MAb F429 to P. aeruginosa alginate were used as controls. Polyclonal sera were first heated at 56 °C for 30 min. To achieve antigenic specificity in the polyclonal sera, all antisera were adsorbed with ∼1010 CFU/mL of E. coli K1 Δpga mutant to remove antibodies reactive with antigens on the surface of the bacteria other than those to PNAG. The actual phagocytic killing assay was performed by mixing 100 μL of the PMN suspension, target bacteria, dilutions of test sera or MAb, and a complement source. The reaction mixture was incubated on a rotor rack at 37 °C for 90 min. The percentage of killing was calculated by determining the ratio of the number of CFU surviving in the tubes with bacteria, PMN, complement, and test antibodies to the number of CFU surviving in tubes containing control antibodies (MAb F429 or NGS), bacteria, complement, and PMN. Sera were classified as positive if, after subtraction of the background killing in control sera using NGS or an irrelevant MAb, more than 30% of the bacterial CFU were killed, and classified as negative if 30% or less killing was obtained. Each OPKA was performed at least two times. The plating for the CFU count was done in triplicate.
Passive protection studies
Groups of five to ten newborn mice (CD1 or Complement-C3 deficient mice) were injected IP with 50 μl of dPNAG-specific goat antiserum to 9GlcNH2-TT or NGS, or with 50 μg of human MAb F598 to PNAG or the irrelevant MAb F429, 24 h before challenge with 105 to 5 × 106 E. coli K1 strains. Twenty-four hours after oral gavage with E. coli K1 E2, E11 or S88 strains, mice were sacrificed and the CFU per brain, liver or spleen determined as described above. In mice passively immunized then challenged with E. coli CTX-M, a model of mortality up to 24 h was used to study the antibodies' efficacy. This same model was used to test the protection conferred by antibodies to PNAG in mice challenged with GBS NEM316.
Passive protection following co-infection with E. coli K1 and S. aureus
Two or three day-old CD1 mice were challenge by gavage with different strains of E. coli K1 (as previously described). At the same time, an IP administration of 105–106 Staphylococcus aureus (either S. aureus USA 300 LAC or S. aureus PS80) was performed. Three hours after the S. aureus IP administration, neonatal mice were injected IP with 50 μl of a dPNAG-specific goat antiserum to 9GlcNH2-TT or NGS, or with 50 μg of human MAb F598 to PNAG or irrelevant human MAb, F429. A mortality model at 24 h following this treatment was used to study the antibodies efficacy. No change in offspring numbers, weight, size were found in pups born from immunized mothers.
Maternal immunization and neonatal protection
Timed-pregnant C3H/HeN female mice (6–8 weeks old) were immunized subcutaneously (s.c.) either with dPNAG-TT or Capsular Polysaccharide 5-TT (CP5) given once a week for 3 weeks after pregnancy confirmed. Immunization efficacy was confirmed by the detection of antibodies to PNAG by ELISA in the blood of the dams after birth. Neonatal mice born to immunized dams were challenged by gavage with E. coli K1. Twenty-four hours after the bacterial challenge, mice were sacrificed and the CFU of E. coli present in the blood and brain were determined as described above. In the second part of the experiments, mice born to CP5-TT immunized dams were placed at birth either in cages with a dPNAG-TT immunized dam or a different CP5-TT immunized dam. One week later, infant mice were challenged by oral gavage with E. coli K1. Twenty-four hours after the bacterial challenge, mice were sacrificed and the CFU of E. coli present in the blood and in the brain were counted.
Potential confounders (such as cage location or order of measurements) were not controlled.
There was no exclusion criteria for the neonates in each litter. All investigators were aware of the group allocation at the different stages of the experiment expect for the co-infections (blinded to all except the principal investigator)
ELISA detection of antibodies to PNAG
Enzyme-linked immunosorbent assay (ELISA) experiments were performed as previously described.10 Briefly, ELISA plates (Maxisorp, Nalge Nunc International, Rochester, NY, USA) were coated with 0.6 μg/ml of purified PNAG diluted in sensitization buffer (0.04 M PO4, pH 7.2) overnight at 4 °C.24,25 The plates were rinsed three times with PBS, blocked with 5% skim milk for 1 h at 37 °C, and rinsed again three times with PBS. Serum samples were then diluted twofold in 5% skim milk with 0.05% Tween 20, incubated for 1 h at 37 °C, washed again, and incubated with appropriate alkaline phosphatase-conjugated secondary antibody (Sigma) for 1 h at 37 °C. After washes, plates were incubated with p-nitrophenyl phosphate substrate, and the optical density at 405 nm (OD405) was determined by an ELISA plate reader (BioTek Instruments, Winoski, Ill.) after a 1-h incubation. Each experiment was done in duplicate in two different times, each time with technical replicates.
E. coli interactions with brain microvascular endothelial cells
Cell cultures
Two different cell lines of human brain microvascular endothelial cells were used in this study. Human brain micro-endothelial cells (HBMEC), immortalized by SV-40 large T antigen transformation, and maintaining the morphological and functional characteristics of the primary endothelium, were obtained from K.S. Kim, MD (Johns Hopkins University, Baltimore).26 HBMEC were grown on collagen-coated tissue culture plates in RPMI medium (Sigma–Aldrich) containing 10% heat inactivated fetal bovine serum, 10% Nu-Serum, 2 mM glutamine, 1 mM pyruvate, 100 U/mL penicillin, 100 μg/mL streptomycin, 1% MEM nonessential amino acids and vitamins at 37 °C in humid atmosphere of 5% CO2 until confluence (about five days). The Human Cerebral Microvascular Endothelial Cell line (hCMEC/D3) (provided by Cedarlane, and developed by P.O Couraud, INSERM, Paris) was prepared from cerebral microvessel endothelial cells by transduction with lentiviral vectors carrying the SV40 T antigen and human telomerase reverse transcriptase.27 hCMEC/D3 were grown on collagen type I coated T75 flasks in EndoGRO-MV Culture MediumTM (provided by EMD Millipore) containing 5% heat inactivated fetal bovine serum, 0.2% EndoGRO-LS Supplement, 5 ng/mL of recombinant human Epidermal Growth Factor, 10 mM of l-glutamine, 1 μg/mL of hydrocortisone hemisuccinate, 0.75 U/mL of heparin sulfate, 50 μg/mL of ascorbic acid, 200 ng/mL of human basic Fibroblast Growth Factor, 100 U/mL penicillin, 100 μg/mL streptomycin, at 37 °C in humid atmosphere of 5% CO2 until confluence (about five days). As for the HBMEC, cells reaching confluence were harvested (using trypsin) and either transferred to a 24-wells plate for further experiments or cryopreserved at −150 °C.
Invasion, binding and translocation assays
Confluent HBMEC or hCMEC/D3 monolayers grown in 24-well plates were infected with bacteria at a multiplicity of infection (MOI) of 100 alone, or with addition of either (i) goat antibody to dPNAG or NGS (diluted 1/10 or 1/100); (ii) with MAb F598 or MAb F429 (100 μL of a 1 mg/mL solution). After 90 min of incubation at 37 °C, the monolayers were washed with the appropriate medium depending on the cell line and then incubated with gentamicin (100 μg/ml) for 1 h to kill extracellular bacteria and washed again. Finally, they were lysed by 0.1% Triton-X100 in PBS and the released intracellular bacteria enumerated. The invasion frequency was calculated by dividing the number of internalized bacteria by the original inoculum. To measure binding, the assay was performed as above except that the gentamicin treatment step and the cell lysis by Triton-X100 were omitted and monolayers were washed at least 8 times by PBS to insure removal of unbound bacteria. For translocation assays, HBMEC or hCMEC/D3 monolayers were established on transwells, then infected with bacteria at a MOI of 100 alone, or with antibodies and translocation levels measured at 0 and 30 min.
All these cell lines experiments were done in duplicate at two different time, each time with technical triplicates.
Trans-endothelial electrical resistance
HBMEC were cultured to confluence then Trans-Epithelial Electrical Resistance was measured using a Millicell-ERS (Millipore, Burlington, MA, USA). The collagen-treated transwell inserts were used to measure the background resistance. In the control monolayer no bacteria was added. Values were expressed as ω × cm2 and were calculated by the formula: [the average resistance of experimental wells − the average resistance of blank wells] × 0.33 (the area of the transwell membrane). The Trans-Epithelial Electrical Resistance values were recorded in real-time every 30 min in triplicates for 90 min.
Group B Streptococcus experiments
Attempts to truncate gene GBS1605 by double crossing over
Deletion of the pgaC-like (gene GBS1605) was attempted in Streptococcus agalactiae NEM316 by allelic exchange with a truncated copy of GBS1605 cloned into the temperature-sensitive shuttle vector, pJRS233, following a procedure previously described.28 Approximately 900 bp fragments upstream and downstream of GBS1605 were amplified by PCR using NEM316 chromosome as the template and primer pairs GBSpgaC1/GBSpgaC2 and GBSpgaC3/GBSpgaC4. The forward primer binding to the 3′-end of pgaC and the reverse primer to the 5′-end were modified to carry a BamH1 restriction site while GBSpgaC1 and GBSpgaC4 primers were modified to carry a pst1 and xho1 site, respectively. Following restriction, ligation and re-amplification using GBSpgaC1 and GBSpgaC4, the resulting fragment carrying the truncated GBS1605 gene (deletion of ca. 50% of the mid-section of the allele) was cloned in xho1/pst1 double digested temperature-sensitive shuttle vector pJRS233 to create plasmid pJRS233pgaC- KO (Table S4). After electroporation and amplification in E. coli DH5-alpha, the recombinant plasmid was recovered (Qiagen plasmid extraction MIDI) and introduced into the chromosome of NEM313 by electro-transformation and homologous recombination followed by excision as described.28 Multiple attempts (confirmation PCR on 1000+ colonies using pgaC flanking primers) yielded the same negative result: the conservation of a wild-type GBS1605 allele in the chromosome and the excision and loss of the truncated allele and the pJRS233 backbone.
Nisin inducible antisense RNA
For antisense expression, we cloned a 721 bp complementary 3′ end fragment of GBS1605 (primers pgaC-AS1 and pgaC-AS2), including the native transcription start site but avoiding any overlap between adjacent genes, in the antisense orientation downstream of the inducible promoter (PnisA) of pMSP3535.19 Following restriction digestion, the GBS1605 amplicon was cloned into pMSP3535 for expression in the antisense direction under nisin induction. The ligated construct was initially transformed into E. coli DH5-alpha and sequence-verified. Purified recombinant plasmid was then transformed into S. agalactiae NEM316 followed by plating on Todd Hewitt agar supplemented with 10 μg/mL erythromycin. Growth curves were carried out utilizing NEM316 strains with gene-specific antisense fragment or empty vector as follows: Overnight cultures were diluted 1:100 in Todd Hewitt broth supplemented with 5 μg/mL erythromycin. At an approximate O.D. 600 of 0.2, cultures were diluted such that approximately 100 cells were inoculated in triplicate into inducing (0.1–5 μg/mL) or non-inducing Todd Hewitt broth in a microtiter plate. Growth was followed for 24 h by measuring O.D. 600 on a Biochrom WPA spectrophotometer (Biochrom, Holliston, MA).
For qRT-PCR, total S. agalactiae NEM316 RNA was isolated from exponentially growing cells, with or without nisin induction in Todd Hewitt broth, using the RNeasy midi kit (Qiagen) as recommended by the manufacturer. cDNA was synthesized from total RNA (∼1.5 mg) by using the Superscript III First-Strand Synthesis System (Invitrogen) according to the manufacturer's instructions. Using synthesized cDNAs, qRT-PCR was performed using Express SYBR GreenER qPCR Supermix (ThermoFisher, Waltham, MA, USA) and a real-time PCR cycler system (Rotor- Gene Q5-Plex; Qiagen, Hilden, Germany) with the following program: 95 °C for 10 min, and subsequently 40 cycles of 95 °C for 15 s, 55 °C for 1 min. Relative transcript levels were calculated using REST 2009 Software (Qiagen). Expression of elongation factor tuf was used as a housekeeping control gene.
Statistical analysis
Nonparametric data were evaluated by the Mann Whitney U test for 2-group comparisons and multi-group comparisons by the Kruskal Wallis test with Dunn's multiple comparison test for pairwise comparisons. Parametric data were analyzed by t-tests (for 2-group comparisons) or ANOVA with Tukey's multiple comparison test for pair-wise comparisons. Survival analysis utilized the Kaplan–Meier method. All analyses, apart from the Tnseq analyses, used GraphPad Prism 6.0 (GraphPad Software, San Diego, CA).
Role of the funding sources
The funding sources had no role in the study design, in the collection or interpretation of the data and in the decision to submit the paper.
Results
TnSeq and E. coli K1 essential genes
In this study, we initially selected the NMEC sequenced strain E. coli K1 S88, previously reported as an important cause of neonatal meningitis and belonging to the O-serotype 45.29 A bank of 3 × 105 Tn-mutants of E. coli S88 was produced, grown overnight in LB and the DNA recovered and subjected to high-throughput sequencing to identify sites and frequencies of Tn inserts into the genome (TnSeq). As previously described,14 this allowed us to identify genes considered as essential for the growth of E. coli K1 S88 in LB (Figure S1). To further explore these E. coli essential genes, a Tn-bank of 3 × 105 mutants in the genome of the well described commensal strain E. coli K12 MG1655 and a Tn-bank of 3 × 105 inserts in the E. coli LF82 strain,30 associated with inflammatory bowel disease, were also built and analyzed. We then completed the original data generated in the current study with data previously reported in the sequencing of several saturated banks of E. coli grown in LB and selected (i) two other E. coli K12 strains: E. coli K12 W311031 and E. coli K12 BW2511332 and (ii) another E. coli K1 strain: E. coli K1 A192PP.33 This approach allowed us to have a better view of the essential genes in different strains of E. coli (Fig. 1). Overall, we found 133 common essential genes in these six E. coli strains (Table S3).
TnSeq using E. coli K1 in two mouse models of neonatal infection
In order to investigate E. coli K1 pathophysiology, we established two mouse models of neonatal infection (Figure S2). The first in vivo model used 2 to 3 day-old mice given an intraperitoneal (IP) challenge with 5 × 106 Colony Forming Units (CFU)/mouse of the Tn-bank that generated a high level of bacteremia. After 24 h, the neonatal mice were sacrificed, and the livers, spleens, brains and meninges harvested to determine systemic dissemination followed by brain infection (above 108 CFU/organ, Figure S3). The second model was designed to mimic the steps of pathogenesis of neonatal meningitis caused by E. coli K1. Neonatal mice were challenged by oral gavage, as the gastro-intestinal (GI) tract is likely the main site of initial colonization and origin for meningitis pathogens in neonates.34 After 24 h, the neonatal mice were sacrificed and the livers, spleens, brains and meninges harvested (Figure S4). These two models allowed determinations of (i) genes important for systemic dissemination, following either IP infection or after passing of the E. coli through the GI tract (both routes are seen in human infection), and (ii) genes important for brain infection following the route of infection thought to occur in the human disease (from the GI tract to the brain).
Genes important for systemic dissemination using two routes of infection
To identify genes producing factors important for systemic dissemination of E. coli K1, the data generated from these TnSeq experiments were analyzed using the systematic approach we previously reported.14,35 Analysis at the genome scale (Figure S5) revealed that selective pressures in the gavage model were greater than those in the IP model. This result was somewhat expected as the gavage model has an additional selective step compared to the IP model: the passing of E. coli through of the GI tract. While it has been recently reported that this step could lead a bottlenecks through the crossing of the epithelial lining, this barrier is avoided using the IP route of infection.33 Therefore, only mutants with a very strong decrease in fitness (at least a 10-fold decrease in the prevalence of the Tn-mutants between the in vitro and the in vivo conditions) using both routes of infection (gavage and IP) were further analysed. Overall, 179 genes in E. coli K1 S88 were considered important for systemic dissemination (Table S4). As shown, (Fig. 2, Figure S7 and Table S4), among the operons and individual genes having >10-fold changes in the output populations, genes encoding for major E. coli K1 virulence factors such as the K1 capsule,36 and fitness factors such as LPS, and OmpA37,38 were all confirmed to be important for systemic dissemination.
While there is great interest in identifying E. coli K1 genes important for systemic dissemination,33 the main goal of this work studying neonatal bacterial meningitis was to identify what might be the best vaccine candidates in this specific setting. Therefore, to identify the genes likely to be important or essential for brain infection in murine neonatal meningitis, we selected those with Tn-insertions leading to at least a 10-fold reduction in occurrence compared to in vitro growth in LB that also had a greater impact on brain infection compared to systemic dissemination.
Genes important for brain infection
These genes were determined by being at least a 10-fold decreased in representation of the Tn-interrupted genes in the brain compared with the liver. The systematic analysis of the 1203 Tn-mutants with reduced fitness in brain infection under these conditions was undertaken (Table S5), and we decided to focus on those encoding for cell surface factors, outer membrane proteins or extracellular products (Table S6 and Fig. 3). These are strong vaccine candidates due to the potential ability to raise protective antibodies to surface-accessible or extracellular antigens. As highlighted in Fig. 3, four gene clusters were found among the 52 genes (Table S6) encoding for extracellular antigens and with Tn-inserts leading to a >10-fold reduced occurrence in the brains of infected mice. These four clusters were the fim genes, the pap genes, the csg genes, and the pga genes. They encode for the type I Fimbriae, the P Fimbriae, Curli and the PNAG surface polysaccharide, respectively. Finding through our TnSeq experiments and in our animal model, that PNAG encoding genes were among the few operons important or even essential for E. coli K1 neonatal meningitis pathophysiology was considered as an excellent opportunity for vaccine evaluations, as this surface polysaccharide with capsule-like properties has previously been reported as a potential vaccine antigen for other infectious agents.39 Therefore the pga locus, its product, PNAG, and antibodies to PNAG were selected as candidates to develop an efficient immunotherapy (both active and passive vaccination) against neonatal bacterial meningitis caused by E. coli K1.
PNAG is a virulence factor of E. coli K1 and a potential vaccine target
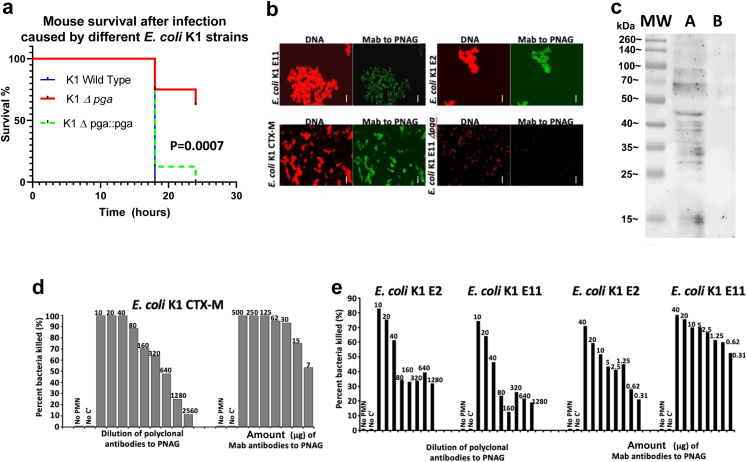
To confirm the data generated using TnSeq, and more precisely the role of PNAG as a fitness factor for E. coli K1 neonatal meningitis, we selected a strain belonging to the major NMEC O-serotype 18, The E. coli K1 clinical isolate E. coli E11. We then deleted the pga genes in strain E11 (E11Δpga) and used E11Δpga, E11 wild type and the complemented strain E11 Δpga::pga for further analysis. Comparing the lethality caused by wild-type and E11Δpga (Fig. 4a), significantly fewer mice died after infection with E. coli E11Δpga in the meningitis model (P = 0.0007, Log–Rank testing). No difference was found between the wild type E11 strain and the complemented strain E11 Δpga::pga, highlighting the specificity of the lower virulence of the Δpga mutant and identifying PNAG as a virulence factor in this model. We next determined that PNAG was expressed on the surface of different strains of E. coli K1, including a multi-drug resistant strain producing the extended spectrum beta-lactamase (ESBL) CTX-M7 (Fig. 4b and Figure S8a–d). The use of an E. coli K11Δpga KO strain (Fig. 4b) confirmed the specificity of the polyclonal antibodies12 and the monoclonal antibody (MAb).19 PNAG production was also detected in vivo by Western blot in infected brains of neonatal mice (Fig. 4c). No PNAG was detected in the brain of uninfected neonates (Fig. 4c), indicating that PNAG was a potential vaccine target for E. coli K1 strains.
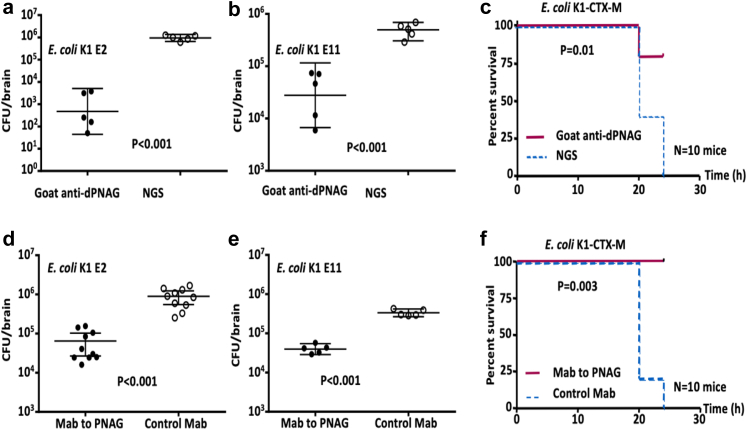
Fig. 4.
E. coli K1 and PNAG. (a) Decreased virulence of E. coli K1 Δpga (red line) compared to E. coli K1 wild type (plain dark blue line). The virulence is restored after complementation (E. coli K1 Dpga::pga strain, green dashed line) P values = Log-rank testing. N = 8 mice per group. (b) In vitro detection of PNAG production (green) and bacterial DNA (red) by different strains of E. coli K1 using confocal microscopy to visualize binding of Human monoclonal antibody (MAb) F598 to PNAG. Bacteria in each microscopic field were visualized with a DNA stain, Syto83. White bars = 10 m. (c) Western blots of PNAG from brains of neonatal mice infected with E. coli K1. PNAG was detected using MAb F598 to PNAG. Track (MW) indicates molecular weight standards and their corresponding sizes in kDa. Track (A) mouse brain infected with E. coli K1, track (B) uninfected brain. (d, e) Opsonophagocytosis killing assays: Percent killing of E. coli K1 producing the extended spectrum beta-lactamase CTX-M (d), E. coli K1 E2 and E11 (e) mediated by goat polyclonal antibodies to dPNAG-TT (anti-dPNAG) or a human MAb to PNAG (MAb F598). Killing calculated in comparison to CFU in corresponding tubes that contain normal goat serum or an irrelevant human IgG1 MAb. No bacterial killing is detected in the controls without leukocytes (No PMN) or without complement (No C′). Killing >30% is significant at P < 0.05 using ANOVA and multiple-comparisons test. (d, e): Each OPKA was performed at least two times. The plating for the CFU count was done in triplicate.
In vitro activity of antibodies to PNAG against E. coli K1
An OPKA was used to test the functional activity of antibodies to PNAG against E. coli K1, using both polyclonal antibodies12 and a fully human IgG1 Mab.19 As shown in Fig. 4d–e, both polyclonal antibodies and the MAb to PNAG were able to mediate the killing of different strains of NMEC in the presence of complement and phagocytes in a dose-dependent manner (OPKA).
Prevention of neonatal meningitis caused by E. coli K1 provided by antibodies to PNAG
The prophylactic activity of PNAG antibodies was tested in the mouse model of neonatal meningitis, administering the challenge by oral gavage using four different strains of E. coli K1: S88, E2, E11 and CTX-M (Fig. 5 and Figure S8e). Significant reduction of the bacterial load was found in organs other than the brain (Figure S9). Both polyclonal antibodies and the human MAb to PNAG injected 24 h before infection resulted in a significant decrease in the CFU recovered from the mice infected by S88, E2 and E11 (P < 0.01, P < 0.01, and P < 0.01 respectively, non parametric t-test). When using the E. coli K1 strain producing the ESBL CTX-M, we found this organism led to a moribund/lethal infection in less than 24 h in non-protected controls, but nonetheless injection of either polyclonal antibodies or the human MAb to PNAG resulted in significantly less neonatal deaths in 24 h (P = 0.01 and P < 0.01, respectively, Log–Rank test).
Fig. 5.
In vivo activity of antibodies to PNAGprotect against E. coli K1. Prophylactic (24 h pre-challenge) effect of 50 μl of opsonic goat polyclonal antibodies to PNAG (Goat anti-dPNAG) (a–c) or 50 μg of human MAb F598 to PNAG (d–f) on E. coli K1 levels in the brain (a, b, d, e) or on lethality rate (c, f) 24 h after challenge by gavage. Controls received normal goat serum (NGS) or an irrelevant human IgG1 MAb, F429. Bars indicate medians, errorbars the 95% confidence interval. Challenge doses: 106 CFU/animal (a, b), 105 CFU/animal (c–e) and 104 CFU/animal (f). P values determined by nonparametric t-test (a, b, d, e) and Log-rank test (c, f). Each circle (a, b, d, e) represents one animal.
Treatment of severe neonatal infections caused by E. coli K1, with or without a co-infection caused by Staphylococcus aureus
To try to mimic a clinical scenario in human neonatal meningitis diagnosis and treatment, we next tested the efficacy of antibody to PNAG to impact infection 3 h subsequent to E. coli K1 infection. However, the causative pathogen in a human would take some time to be identified after the onset of signs of infection (whether it is indeed E. coli K1 or another pathogen). This could hamper the potential use of PNAG antibodies after the start of the neonatal infections. However, several other PNAG producing pathogens, including potentially multi-drug resistant (MDR) bacteria such as Staphylococcus epidermidis and more particularly S. aureus, can also cause severe neonatal infections, in particular in premature neonates.40
Therefore, to look for the therapeutic efficacy of antibodies to PNAG in a setting where there could be several different PNAG-producing MDR pathogens causing neonatal meningitis, neonatal mice were treated 3 h after bacterial challenge without knowing if the animals were infected by E. coli K1 (gavage meningitis model), or by S. aureus (bacteremia induced by an intra-peritoneal challenge), or co-infected by both (unlikely clinical event, but tested as proof of concept for polymicrobial infection). We used several strains of E. coli K1 and S. aureus in this challenge setting, including the MDR CTX-M producing E. coli K1, and the methicillin-resistant S. aureus strain USA300 LAC.
As shown in Tables 1 and 2, there was excellent therapeutic efficacy of both polyclonal antibodies and the human MAb to PNAG in reducing mortality and bacterial burdens regardless if the neonatal mice were infected by E. coli alone (E. coli K1 E11 or E. coli K1 CTX-M), S. aureus alone (S. aureus PS80 or S. aureus LAC), or co-infected by E. coli K1 and S. aureus. In some challenges a moribund/mortality outcome was used when the individual strain or combination caused this state to occur in <24 h, but in other challenges when neonates were still alive 24 h after challenge (time point mandated for ending the experiment), CFU from the brain and the liver were counted after euthanasia (indicated by superscript "a" in Tables 1 and 2). In both outcome scenarios, the antibodies to PNAG provided significant protection (P < 0.01, unpaired t-test) (Table 2).
Table 1.
Therapeutic activity of polyclonal and monoclonal antibodies to PNAG 3 h after challenge of neonatal mice with different E. coli K1 strains with and without S. aureus co-infection.
| Strains tested | Survivors/total challenged |
|
|---|---|---|
| Goat polyclonal antibodies to PNAG | Normal goat serum | |
| E. coli CTX-M | 5/5 | 0/5 |
| E. coli CTX-M + S. aureus PS80 | 5/5 | 0/5 |
| E. coli CTX-M + S. aureus LAC | 5/5 | 0/5 |
| E. coli E11 | 5/5 | 5/5a |
| E. coli E11 + S. aureus PS80 | 5/5 | 4/5a |
| E. coli E11 + S. aureus LAC | 4/5 | 0/5 |
| Monoclonal antibody to PNAG (F598) |
Control monoclonal antibody (F429) |
|
| E. coli CTX-M | 4/5 | 0/5 |
| E. coli CTX-M + S. aureus PS80 | 5/10 | 0/10 |
| E. coli CTX-M + S. aureus LAC | 6/10 | 0/10 |
| E. coli E11 | 6/10 | 0/10 |
| E. coli E11 + S. aureus PS80 | 6/10 | 0/10 |
| E. coli E11 + S. aureus LAC | 6/10 | 0/10 |
Significant decrease of CFU in the brains.
Table 2.
Therapeutic activity of polyclonal and monoclonal antibodies to PNAG 1 h after challenge of neonatal mice with two different S. aureus strains.
| Strains tested | Survivors/total challenged |
|
|---|---|---|
| Goat polyclonal antibodies to PNAG | Normal goat serum | |
| S. aureus LAC | 5/5 | 0/5 |
| S. aureus PS80 | 5/5 | 5/5a |
| Monoclonal antibody to PNAG (F598) |
Control monoclonal antibody (F429) |
|
| S. aureus LAC | 5/10 | 0/10 |
| S. aureus PS80 | 6/10 | 0/10 |
Significant decrease of CFU in the livers.
While PNAG was previously described to be expressed by several pathogens,11 no systematic analysis has been performed to date to find the prevalence of the pga genes among the species where these genes have been reported in some isolates. Therefore, we performed a bioinformatic analysis of the presence of the PNAG encoding genes pga A, B, C, and D in the following bacteria that can be either commensal or pathogens depending of the clinical circumstances: E. coli, Klebsiella pneumoniae, Citrobacter freundii, and Enterobacter cloacae.
We used 1294 E. coli genomes published in 41 and all genomes downloaded from NCBI database server that contained metadata about the host and its environment for K. pneumoniae (1000 genomes), C. freundii (347 genomes), and E. cloacae (178 genomes) Table S7. A vast majority of these genomes had the full pga locus reinforcing the potential of PNAG as a vaccine candidate.
Protection of neonates provided by transfer of maternal antibodies to PNAG
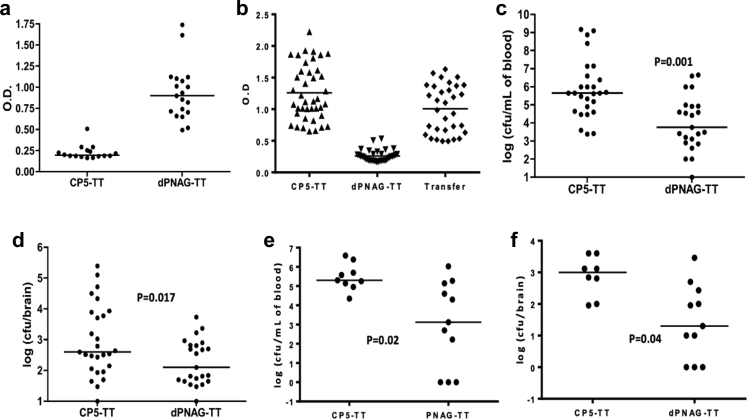
Another approach to prevent neonatal infections is via active vaccination of females, either prior to pregnancy, or in the case of humans, in the third trimester of pregnancy. To assess this means of protection, adult female mice were immunized either with a PNAG vaccine or a control antigen, the capsular polysaccharide 5 (CP5) of S. aureus.42 After immunization with PNAG, specific antibodies were successfully detected in the sera of vaccinated pregnant mice by ELISA (Fig. 6). Then, mice born either from dams immunized with PNAG or CP5 were infected by gavage with E. coli K1. As shown in Fig. 6b–c, significantly less bacteria were recovered from mice born to PNAG immunized dams (P < 0.01, unpaired t-test). Next, we conducted a cross fostering experiment, placing newborn mice from dams immunized with CP5 in a cage with another dam immunized with CP5 or in a cage with a dam immunized with PNAG. Significant protection was observed in neonatal mice nursed by the PNAG immunized mother (P = 0.02, unpaired t-test) compared to the neonatal mice nursed by a CP immunized dam, suggesting that transmission of antibodies to PNAG through the milk could be another source of protective antibodies in this model (Fig. 6d–e). In this regard, it has recently been shown that IgG antibodies in milk can be transferred to the bloodstream of feeding pups via the neonatal Fc receptor.38
Fig. 6.
Protection provided by maternal antibodies to PNAG. (a–b) Antibodies to PNAG were detected by ELISA in the serum of the mothers immunized by dPNAG-TT (a), in the pups born from mothers immunized with dPNAG-TT or transferred to the cage of a mother immunized with dPNAG-TT (b). Dots = individual mice; lines = medians. (a, b): Each experiment was done in duplicate in two different times, each time with technical replicates. (c, d) CFU recovered from the blood taken from the heart (c) or from the brains ((d) of neonates born from dams immunized by either dPNAG-TT or CP5-TT and infected by E. coli K1. P values = unpaired t-test). Bars represent the median. Each circle represents an animal. (e, f) Pups born from CP5-TT immunized mother were placed at birth either into cages with dPNAG-TT immunized dams or CP5-TT immunized dams. Twenty-four hours after infection, CFU were recovered from the blood (e) or from the brains (f). Bars represent the mean. Each circle represents an animal. P values = unpaired t-test.
Antibodies to PNAG and crossing of the blood brain barrier (BBB) by E. coli K1
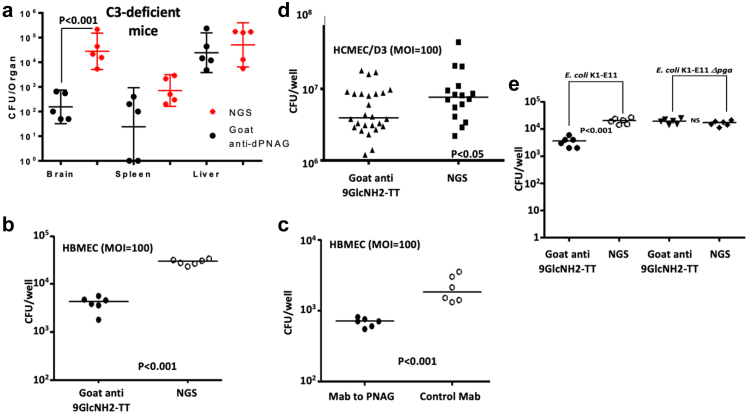
To understand if mechanisms of antibody action other than OPK could also be active in protecting neonatal mice from meningitis, complement-C3 deficient neonatal mice were challenged in the neonatal meningitis murine protocol such that opsonic killing of E. coli by antibody would not be effective. C3-deficient neonatal mice were treated with polyclonal antibodies to PNAG then infected with E. coli K1 via gavage. No differences in the CFU levels were found in the livers and spleens (Fig. 7a) of the C3-deficient mice immunized with antibodies to PNAG compared with those receiving the control antibodies. This result indicates that the OPK activity protects against E. coli K1 systemic dissemination. In contrast, there was a significant reduction in the CFU recovered from the brains of the C3-deficient mice injected with antibodies to PNAG compared to controls (P < 0.01, non-parametric t-test), suggesting an ability of these antibodies to block translocation of E. coli across the BBB.
Fig. 7.
Antibodies to PNAG and crossing of the blood brain barrier. (a) Bacterial levels in the brain, liver and spleen of C3-deficient mice infected by E. coli K1 and immunized with either polyclonal antibodies against PNAG (black dots) or by a normal serum (red dots). Dots = individual mice; lines = medians. P value determined by non-parametric T-test. NGS=Normal goat serum. (b, c) Goat polyclonal antibodies (b) and human monoclonal antibodies (c) to PNAG activity on E. coli K1 adherence to HBMEC cultures. (d) Goat polyclonal antibodies to the synthetic oligosaccharide 9GlcNH2-TT activity on E. coli K1 E11 adherence to HCMEC/D3 cultures. (e) Goat polyclonal antibodies activity on adherence by E. coli K1 E11 Δpga to HBMEC cultures. Symbols are individual wells, lines medians, P values = unpaired t-tests. All cell lines experiments were done in duplicate at two different time, each time with technical triplicates.
Next, we evaluated if this reduction in bacterial burdens in the brains of the C3-deficient mice could be linked to the inhibition of the binding, adhesion or translocation of E. coli K1 through the BBB conferred by the antibodies to PNAG. Two human cell lines were used to test this hypothesis: HBMEC developed by KS Kim and HCMEC/D3 developed by PO Couraud.26,27 As shown in Fig. 7b–d, regardless of the cell line used in the experiments, there was a significant reduction (P < 0.01 and P < 0.05, respectively, unpaired t-tests) in E. coli K1 adhesion to the confluent monolayer cells in the presence of antibodies to PNAG. This result was obtained with polyclonal antibodies and the human MAb and with different strains of E. coli K1 (Figure S10). In addition, a significant reduction of invasion into cells in a monolayer and translocation through cells growing on transwells was also found in the presence of antibodies to PNAG (Figure S10). This decreased translocation of E. coli through the endothelial cells in the presence of anti-PNAG antibodies was not secondary to a change in the monolayer integrity analysed by trans-endothelial electrical resistance, as shown in Figure S11. To study the specificity of the PNAG antibodies in these in vitro experiments, we performed an assay using the parental E. coli K1 strain and the E coli K1 Δpga isogenic mutant. No change in the adhesion to the HBMEC was observed with E. coli K1 Δpga in the presence of antibodies to PNAG, establishing the specificity of the anti-PNAG activity. Taken together, these results suggest that in addition to their OPK activity, antibodies to PNAG may also protect against meningitis caused by E. coli K1 by interfering with the crossing of the BBB.
PNAG antibodies and Group B Streptococci
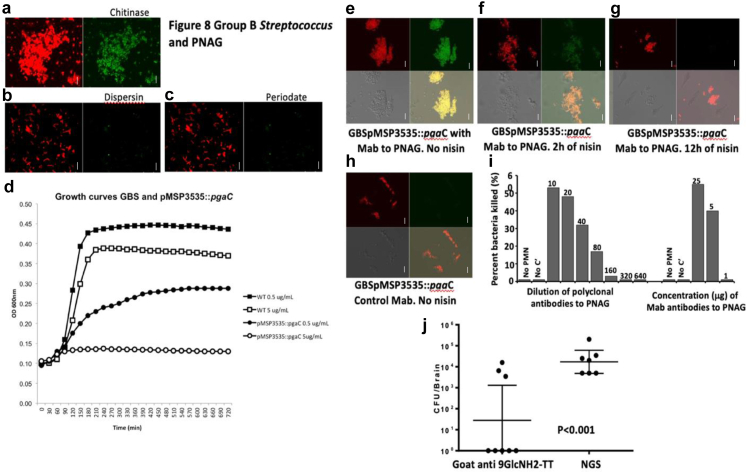
Having found that PNAG antibodies were efficient in our models to treat infections caused by E. coli K1 and other PNAG producing pathogens, including MDR such as MRSA strains, we decided to look at another major cause of bacterial neonatal meningitis: S. agalactiae or Group B streptococcus (GBS).5 Indeed, if PNAG could also target GBS, it would probably significantly increase its potential clinical impact. In this context, we successfully detected PNAG at the surface of GBS as observed by confocal microscopy using the MAb to PNAG F598 and confirmed the properties of the antigen11 by showing that the MAb binding remained after digestion with chitinase but was lost following digestion with the PNAG-degrading enzyme, Dispersin B, as well as following PNAG-degradation with sodium periodate (Fig. 8a–c).
Fig. 8.
Group B streptococcus (GBS) and PNAG. (a–c) Detection of PNAG production (green) and bacterial DNA (red) by GBS using confocal microscopy to visualize binding of MAb F598 to PNAG. MAb binding is resistant to chitinase (a), but susceptible to dispersin B (b) and to periodate treatments (c) White bars represent 10 m. (d) GBS NEM 16 pMSP3535:pgaC growth in the presence of low concentration of nisin compared to the GBS NEM 16 (WT) cultured with low or high concentrations of nisin. A high concentration of nisin completely inhibits GBS pMSP3535:pgaC growth. (e–h) Detection of PNAG production by GBS pMSP3535:pgaC grown in the absence (e) or the presence of nisin (f: 2 h of nisin, g: 12 h) using confocal microscopy. Green colour: detection of the binding to PNAG by the MAb to PNAG (F598). Red colour: bacterial DNA. The MAb F429 to P. aeruginosa alginate was used as control (h). White bars represent 10 µm. The bottom panels represent the visualization with both colours (on the right) or without colour (on the left). (i) Activity on GBS of the polyclonal goat antiserum raised to a a nonameric β-1-6-linked glucosamine oligosaccharide conjugated to tetanus toxoid (9GlcNH2-TT) and of the fully human IgG1 MAb to PNAG, F598. NGS or MAb F429 to P. aeruginosa alginate were used as controls. (j) In vivo activity of the goat polyclonal antibodies to the synthetic oligosaccharide 9GlcNH2-TT against GBS compared to NGS in a murine neonatal meningitis model after gavage. P value = unpaired t-test.
Next, a gene potentially involved in PNAG synthesis was identified in the sequenced GBS strain NEM316, encoding for a protein homologous to E. coli pgaC and designated GBS1605 (Blast P value = 10e-24). Multiple attempts to truncate or delete GBS1605 by double cross overs using a temperature-sensitive shuttle vector28 were unsuccessful, suggesting that GBS1605 was an essential gene for GBS NEM316. Therefore, we used an antisense RNA (asRNA) approach to decrease the expression of GBS1605. A 721 bp asRNA, both complementary and overlapping with the ATG start codon of GBS1605, was cloned under the control of a nisin inducible promoter in the expression vector pMSP3535.43 By qRT-PCR we found that increasing concentrations of nisin caused an increased expression of the asRNA and a corresponding decreased expression of GBS1605 transcripts (Figure S12). Concomitant with decreased GBS1605 mRNA expression, we observed that the growth of strain NEM316 was reduced (0.5 μg/ml nisin induction) or abolished (5 μg/ml nisin) (Fig. 8d). Testing of PNAG production by GBS NEM316 WT carrying either the asRNA for GBS1605 or an empty vector control showed that decreased gene expression led to loss of PNAG production (Fig. 8e–h).
While the reliance of GBS NEM316 on a gene for both growth and PNAG production precluded testing the mutant for changes in virulence, this finding did encourage testing of the efficacy of antibodies to PNAG against GBS for opsonic killing activity and protective efficacy in the neonatal meningitis model. We found that antibodies to PNAG had potent OPK activity against GBS (Fig. 8i). Additionally, significantly less GBS bacteria were recovered from the brains of neonatal mice passively immunized with antibodies to PNAG compared with those given control sera (P < 0.01, unpaired t-test) (Fig. 8j).
Thus, functional antibodies to PNAG have protective potential against the two major causes of neonatal bacterial meningitis (E. coli K1 and GBS) and less frequent cause of infections but more often associated with MDR such as Methicillin resistant S. aureus.
Discussion
New approaches to treat bacterial infections are urgently needed, as even during a viral pandemic such as the COVID-19 crisis44 that started in December 2019, increased use of antibiotics leading to increased selection for antibiotic-resitant microbes persists. In this context, the use of passive45 or active46 immunotherapies against microbial infections are more relevant now than ever. In this work, we were able to show that high-throughput sequencing approaches can be used to identify vaccine targets and accelerate the process of vaccine development for bacterial infections. Prior strategies applied to the development of an effective vaccine against extra-intestinal pathogenic E. coli infections have not yet resulted in an effective vaccine for human use. This is either due to the high antigenic heterogeneity of the major surface polysaccharides, LPS O and capsular K-antigens, that can be targeted for vaccination against many strains of E. coli, or the cross reactivity of potential targets such as the K1 capsule with human oligosaccharides, making it very challenging to design a vaccine when targeting these carbohydrates.47 Using high-throughput sequencing, we determined four operons essential for E. coli K1 meningitis pathophysiology that also encode for surface or extracellular proteins: type I Fimbriae, the P Fimbriae, Curli and PNAG. Type I fimbriae and P fimbriae are well known virulence factors of uropathogenic E. coli (UPEC). They participate in biofilm formation, induce the adhesion of E. coli to kidneys or low urinary tract and increase local inflammation.48, 49, 50 Although they have been characterized in E. coli meningitis, these virulence factors are not expressed in all the clinical strains isolated in newborns diagnosed with E. coli K1 meningitis.51, 52, 53 making them suboptimal vaccine candidates. Notably, Curli fibers were found to be a virulence factor important in E. coli K1 neonatal meningitis and have also been extensively described in the literature for their role in biofilm formation.54, 55, 56 They were found to be highly expressed in E. coli strains isolated from women with upper urinary tract symptoms. However, Curli fibers are less expressed in neonatal meningitis E. coli clinical strains (66.7%).57 Outer membrane protein A was identified as important for systemic dissemination in the mouse model used here (Fig S6). OmpA is already known to be a virulence factor involved in the interaction with the BBB cells58 and to promote the crossing of the BBB by E. coli K1.59 Polymorphisms of OmpA affect E. coli K1 virulence and its ability to invade the CNS.60, 61, 62 While not selected for further investigation, OmpA has potential as a vaccine for neonatal meningitis. The K1 capsule is also known as a virulence factor for E. coli neonatal meningitis and is predominant among E. coli strains causing neonatal meningitis. The K1 capsule is not directly involved in the traversal of the BBB by the bacteria but it confers an increased survival in brain cells.62,63 However, its structural similarity to the polysialic acid moiety of mammalian neural cell adhesion molecules prevent its use as a vaccine candidate.8 Other virulence factors of E. coli neonatal meningitis have been described in the literature. S fimbriae interact with the BBB cells enhancing bacterial adhesion64 but the sfa genes are expressed by less than 40% of newborn meningitis associated E. coli strains.65 aslA,66 Ibes,67,68 TraJ69 and the exotoxin CNF-170,71 have all demonstrated a role in CNS invasion but they have never been studied as vaccine candidates.
In this work, we demonstrated that PNAG was a virulence factor for optimal neonatal meningitis caused by E. coli K1 and that antibodies to PNAG were able to prevent and treat E. coli K1 neonatal meningitis in a mouse model. In addition to their opsonophagocytic activity, these antibodies directly blocked the adhesion and translocation of E. coli across the BBB. A non-specific ability to prevent binding of a bacteria opsonized by antibodies targeting a surface polysaccharide has already been described. Indeed, this mechanism of action has been suggested as responsible for a decreased carriage of S. pneumoniae in patients vaccinated with Prevnar.72 We further found that another frequent bacterial cause of neonatal meningitis, GBS, also produced PNAG and we were able to show protection against GBS in neonatal mice, as well as a more uncommon cause of neonatal meningitis but often associated with antibiotic resistant infections, S. aureus. Two additional points promote the further development of antibodies to PNAG as anti-bacterial reagents. First, an initial conjugate vaccine for PNAG has been tested in humans for safety and immunogenicity73 that elicited broadly opsonic and bactericidal antibodies against a variety of pathogens74 Second, the fully human IgG1 MAb to PNAG, F598, has also been in phase 1 and 2 human trials13,75 and is a potential immunotherapeutic for preventing neonatal meningitis, particularly in infants at high risk for infections. While PNAG is expressed by several commensal bacteria, no impact on the microbiota has been reported in any animal or human.76
These findings support further investigations of PNAG-targeting vaccines in at risk pregnancies and/or MAb immunotherapy for at risk neonates such as the prematurely born. If further testing is successful, it could support human testing of this potential means to prevent serious neurologic infections in neonates.
Contributors
SP and DR performed in vitro and in vivo experiments, analysed data and worked on the manuscript. EF analysed data and edited the manuscript, YS and CG performed in vivo experiments on E. coli K1 and edited the manuscript, FL worked on the genetics of GBS and edited the manuscript, AK performed in vivo experiments on E. coli K1 and edited the manuscript, OD performed high-throughput sequencing experiments and edited the manuscript, LS, SC, AM, AF LH, BS, HA and HS analysed the data, CCB performed in vitro experiments on PNAG, JJM analysed data and edited the manuscript, TG performed in vitro experiments on E. coli K1, analysed the data and edited the manuscript, GBP analysed the data, designed experiments, worked on the manuscript, D.S. developed the study concept, supervised the project, designed and performed in vitro and in vivo experiments on GBS, S. aureus and E. coli K1, analysed data and wrote the manuscript. All authors read and approved the final version of the manuscript.
Data sharing statement
All data are available in the main text or the supplementary materials. All RPKM recovered after sequencing for all in vitro and in vivo experiments are available upon request.
Declaration of interests
GBP is an inventor of intellectual properties [human monoclonal antibody to PNAG and PNAG vaccines] that are licensed by Brigham and Women's Hospital to Alopexx, Inc. in which GBP also holds equity. As an inventor of intellectual properties, GBP also has the right to receive a share of licensing-related income (royalties, fees) through Brigham and Women's Hospital from Alopexx, Inc. GBP's interests were reviewed and are managed by the Brigham and Women's Hospital and MGB Healthcare in accordance with their conflict of interest policies. CCB and DS are inventors of intellectual properties [use of human monoclonal antibody to PNAG and use of PNAG vaccines] that are licensed by Brigham and Women's Hospital to Alopexx Inc. As inventors of intellectual properties, they also have the right to receive a share of licensing-related income (royalties, fees) through Brigham and Women's Hospital from Alopexx Inc. All other authors declare they have no competing interests.
Acknowledgments
Funding: SP received an award from the Groupe Pasteur Mutualité, DR received an award from the Hearst Foundation, OD was supported by AI-026289 from National Institute of Allergy and Infectious Diseases, CCB received an unrestricted gift from Alopexx Vaccines, DS received an award from the ANR (AAPG2020-Seq-N-Vaq, France), DS received an award from the Charles H. Hood Foundation.
Footnotes
Supplementary data related to this article can be found at https://doi.org/10.1016/j.ebiom.2023.104439.
Appendix A. Supplementary data
References
- 1.Ouchenir L., Renaud C., Khan S., et al. The epidemiology, management, and outcomes of bacterial meningitis in infants. Pediatrics. 2017;140(1):e20170476. doi: 10.1542/peds.2017-0476. [DOI] [PubMed] [Google Scholar]
- 2.Kohli-Lynch M., Russell N.J., Seale A.C., et al. Neurodevelopmental impairment in children after Group B Streptococcal disease worldwide: systematic review and meta-analyses. Clin Infect Dis. 2017;65(suppl_2):S190–S199. doi: 10.1093/cid/cix663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Okike I.O., Johnson A.P., Henderson K.L., et al. Incidence, etiology, and outcome of bacterial meningitis in infants aged <90 days in the United Kingdom and Republic of Ireland: prospective, enhanced, national population-based surveillance. Clin Infect Dis. 2014;59(10):e150–e157. doi: 10.1093/cid/ciu514. [DOI] [PubMed] [Google Scholar]
- 4.Tan J., Kan J., Qiu G., et al. Clinical prognosis in neonatal bacterial meningitis: the role of cerebrospinal fluid protein. PLoS One. 2015;10(10) doi: 10.1371/journal.pone.0141620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Joubrel C., Tazi A., Six A., et al. Group B streptococcus neonatal invasive infections, France 2007-2012. Clin Microbiol Infect. 2015;21(10):910–916. doi: 10.1016/j.cmi.2015.05.039. [DOI] [PubMed] [Google Scholar]
- 6.Verani J.R., McGee L., Schrag S.J., Division of Bacterial Diseases NCfI, Respiratory Diseases CfDC, Prevention Prevention of perinatal group B streptococcal disease--revised guidelines from CDC, 2010. MMWR Recomm Rep. 2010;59(RR-10):1–36. [PubMed] [Google Scholar]
- 7.Dubois D., Prasadarao N.V., Mittal R., Bret L., Roujou-Gris M., Bonnet R. CTX-M beta-lactamase production and virulence of Escherichia coli K1. Emerg Infect Dis. 2009;15(12):1988–1990. doi: 10.3201/eid1512.090928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Finne J., Leinonen M., Mäkelä P.H. Antigenic similarities between brain components and bacteria causing meningitis. Implications for vaccine development and pathogenesis. Lancet. 1983;2(8346):355–357. doi: 10.1016/s0140-6736(83)90340-9. [DOI] [PubMed] [Google Scholar]
- 9.Cerca N., Jefferson K.K., Oliveira R., Pier G.B., Azeredo J. Comparative antibody-mediated phagocytosis of Staphylococcus epidermidis cells grown in a biofilm or in the planktonic state. Infect Immun. 2006;74(8):4849–4855. doi: 10.1128/IAI.00230-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Maira-Litran T., Kropec A., Goldmann D.A., Pier G.B. Comparative opsonic and protective activities of staphylococcus aureus conjugate vaccines containing native or deacetylated staphylococcal poly-N-acetyl- -(1-6)-glucosamine. Infect Immun. 2005;73(11):7789. doi: 10.1128/IAI.73.10.6752-6762.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cywes-Bentley C., Skurnik D., Zaidi T., et al. Antibody to a conserved antigenic target is protective against diverse prokaryotic and eukaryotic pathogens. Proc Natl Acad Sci U S A. 2013;110(24):E2209–E2218. doi: 10.1073/pnas.1303573110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gening M.L., Maira-Litrán T., Kropec A., et al. Synthetic {beta}-(1->6)-linked N-acetylated and nonacetylated oligoglucosamines used to produce conjugate vaccines for bacterial pathogens. Infect Immun. 2010;78(2):764–772. doi: 10.1128/IAI.01093-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vlock D, Lee JC, Kropec-Huebner A, Pier GB. Pre-clinical and initial phase i evaluations of a fully human monoclonal antibody directed against the PNAG surface polysaccharide on Staphylococcus aureus. Abstracts of the 50th ICAAC 2010; Abstract G1-1654/329. 2010.
- 14.Skurnik D., Roux D., Aschard H., et al. A comprehensive analysis of in vitro and in vivo genetic fitness of Pseudomonas aeruginosa using high-throughput sequencing of transposon libraries. PLoS Pathog. 2013;9(9) doi: 10.1371/journal.ppat.1003582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Skurnik D., Roux D., Cattoir V., et al. Enhanced in vivo fitness of carbapenem-resistant oprD mutants of Pseudomonas aeruginosa revealed through high-throughput sequencing. Proc Natl Acad Sci U S A. 2013;110(51):20747–20752. doi: 10.1073/pnas.1221552110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kal A.J., van Zonneveld A.J., Benes V., et al. Dynamics of gene expression revealed by comparison of serial analysis of gene expression transcript profiles from yeast grown on two different carbon sources. Mol Biol Cell. 1999;10(6):1859–1872. doi: 10.1091/mbc.10.6.1859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lu X., Skurnik D., Pozzi C., et al. A Poly-N-acetylglucosamine-Shiga toxin broad-spectrum conjugate vaccine for Shiga toxin-producing Escherichia coli. mBio. 2014;5(2) doi: 10.1128/mBio.00974-14. 009744-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yu D., Ellis H.M., Lee E.C., Jenkins N.A., Copeland N.G., Court D.L. An efficient recombination system for chromosome engineering in Escherichia coli. Proc Natl Acad Sci U S A. 2000;97(11):5978–5983. doi: 10.1073/pnas.100127597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kelly-Quintos C., Cavacini L.A., Posner M.R., Goldmann D., Pier G.B. Characterization of the opsonic and protective activity against Staphylococcus aureus of fully human monoclonal antibodies specific for the bacterial surface polysaccharide poly-N-acetylglucosamine. Infect Immun. 2006;74(5):2742–2750. doi: 10.1128/IAI.74.5.2742-2750.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pier G.B., Boyer D., Preston M., et al. Human monoclonal antibodies to Pseudomonas aeruginosa alginate that protect against infection by both mucoid and nonmucoid strains. J Immunol. 2004;173(9):5671–5678. doi: 10.4049/jimmunol.173.9.5671. [DOI] [PubMed] [Google Scholar]
- 21.Skurnik D., Roux D., Pons S., et al. Extended-spectrum antibodies protective against carbapenemase-producing Enterobacteriaceae. J Antimicrob Chemother. 2016;71(4):927–935. doi: 10.1093/jac/dkv448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kropec A., Maira-Litran T., Jefferson K.K., et al. Poly-N-acetylglucosamine production in Staphylococcus aureus is essential for virulence in murine models of systemic infection. Infect Immun. 2005;73(10):6868–6876. doi: 10.1128/IAI.73.10.6868-6876.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Skurnik D., Merighi M., Grout M., et al. Animal and human antibodies to distinct Staphylococcus aureus antigens mutually neutralize opsonic killing and protection in mice. J Clin Invest. 2010;120(9):3220–3233. doi: 10.1172/JCI42748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Maira-Litrán T., Kropec A., Abeygunawardana C., et al. Immunochemical properties of the staphylococcal poly-N-acetylglucosamine surface polysaccharide. Infect Immun. 2002;70(8):4433–4440. doi: 10.1128/IAI.70.8.4433-4440.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cywes-Bentley C., Rocha J.N., Bordin A.I., et al. Antibody to Poly-N-acetyl glucosamine provides protection against intracellular pathogens: mechanism of action and validation in horse foals challenged with Rhodococcus equi. PLoS Pathog. 2018;14(7) doi: 10.1371/journal.ppat.1007160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stins M.F., Gilles F., Kim K.S. Selective expression of adhesion molecules on human brain microvascular endothelial cells. J Neuroimmunol. 1997;76(1–2):81–90. doi: 10.1016/s0165-5728(97)00036-2. [DOI] [PubMed] [Google Scholar]
- 27.Weksler B., Romero I.A., Couraud P.-O. The hCMEC/D3 cell line as a model of the human blood brain barrier. Fluids Barriers CNS. 2013;10(1):16. doi: 10.1186/2045-8118-10-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Perez-Casal J., Price J.A., Maguin E., Scott J.R. An M protein with a single C repeat prevents phagocytosis of Streptococcus pyogenes: use of a temperature-sensitive shuttle vector to deliver homologous sequences to the chromosome of S. pyogenes. Mol Microbiol. 1993;8(5):809–819. doi: 10.1111/j.1365-2958.1993.tb01628.x. [DOI] [PubMed] [Google Scholar]
- 29.Bonacorsi S., Clermont O., Houdouin V., et al. Molecular analysis and experimental virulence of French and North American Escherichia coli neonatal meningitis isolates: identification of a new virulent clone. J Infect Dis. 2003;187(12):1895–1906. doi: 10.1086/375347. [DOI] [PubMed] [Google Scholar]
- 30.Hoffmann T.W., Pham H.-P., Bridonneau C., et al. Microorganisms linked to inflammatory bowel disease-associated dysbiosis differentially impact host physiology in gnotobiotic mice. ISME J. 2016;10(2):460–477. doi: 10.1038/ismej.2015.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Goodall E.C.A., Robinson A., Johnston I.G., et al. The essential genome of Escherichia coli K-12. mBio. 2018;9(1) doi: 10.1128/mBio.02096-17. e02096-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Baba T., Ara T., Hasegawa M., et al. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol Syst Biol. 2006;2 doi: 10.1038/msb4100050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McCarthy A.J., Stabler R.A., Taylor P.W. Genome-wide identification by transposon insertion sequencing of Escherichia coli K1 genes essential for in vitro growth, gastrointestinal colonizing capacity, and survival in serum. J Bacteriol. 2018;200(7) doi: 10.1128/JB.00698-17. e00698-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Witcomb L.A., Collins J.W., McCarthy A.J., Frankel G., Taylor P.W. Bioluminescent imaging reveals novel patterns of colonization and invasion in systemic Escherichia coli K1 experimental infection in the neonatal rat. Infect Immun. 2015;83(12):4528–4540. doi: 10.1128/IAI.00953-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Roux D., Danilchanka O., Guillard T., et al. Fitness cost of antibiotic susceptibility during bacterial infection. Sci Transl Med. 2015;7(297) doi: 10.1126/scitranslmed.aab1621. [DOI] [PubMed] [Google Scholar]
- 36.Kim K.S., Itabashi H., Gemski P., Sadoff J., Warren R.L., Cross A.S. The K1 capsule is the critical determinant in the development of Escherichia coli meningitis in the rat. J Clin Invest. 1992;90(3):897–905. doi: 10.1172/JCI115965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Khan N.A., Shin S., Chung J.W., et al. Outer membrane protein A and cytotoxic necrotizing factor-1 use diverse signaling mechanisms for Escherichia coli K1 invasion of human brain microvascular endothelial cells. Microb Pathog. 2003;35(1):35–42. doi: 10.1016/s0882-4010(03)00090-1. [DOI] [PubMed] [Google Scholar]
- 38.Zheng W., Zhao W., Wu M., et al. Microbiota-targeted maternal antibodies protect neonates from enteric infection. Nature. 2020;577(7791):543–548. doi: 10.1038/s41586-019-1898-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Roux D., Pier G.B., Skurnik D. Magic bullets for the 21st century: the reemergence of immunotherapy for multi- and pan-resistant microbes. J Antimicrob Chemother. 2012;67(12):2785–2787. doi: 10.1093/jac/dks335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Giannoni E., Agyeman P.K.A., Stocker M., et al. Neonatal sepsis of early onset, and hospital-acquired and community-acquired late onset: a prospective population-based cohort study. J Pediatr. 2018;201:106–114.e4. doi: 10.1016/j.jpeds.2018.05.048. [DOI] [PubMed] [Google Scholar]
- 41.Touchon M., Perrin A., de Sousa J.A.M., et al. Phylogenetic background and habitat drive the genetic diversification of Escherichia coli. PLoS Genet. 2020;16(6) doi: 10.1371/journal.pgen.1008866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Skurnik D., Kropec A., Roux D., Theilacker C., Huebner J., Pier G.B. Natural antibodies in normal human serum inhibit Staphylococcus aureus capsular polysaccharide vaccine efficacy. Clin Infect Dis. 2012;55(9):1188–1197. doi: 10.1093/cid/cis624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bryan E.M., Bae T., Kleerebezem M., Dunny G.M. Improved vectors for nisin-controlled expression in gram-positive bacteria. Plasmid. 2000;44(2):183–190. doi: 10.1006/plas.2000.1484. [DOI] [PubMed] [Google Scholar]
- 44.Vaughn V.M., Gandhi T., Petty L.A., et al. Empiric antibacterial therapy and community-onset bacterial co-infection in patients hospitalized with COVID-19: a multi-hospital cohort study. Clin Infect Dis. 2021;72(10):e533–e541. doi: 10.1093/cid/ciaa1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Baum A., Fulton B.O., Wloga E., et al. Antibody cocktail to SARS-CoV-2 spike protein prevents rapid mutational escape seen with individual antibodies. Science. 2020;369(6506):1014–1018. doi: 10.1126/science.abd0831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Corbett K.S., Flynn B., Foulds K.E., et al. Evaluation of the mRNA-1273 vaccine against SARS-CoV-2 in nonhuman primates. N Engl J Med. 2020;383(16):1544–1555. doi: 10.1056/NEJMoa2024671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nesta B., Pizza M. In: Escherichia coli, a versatile pathogen. 416. Frankel G., Ron E.Z., editors. Springer International Publishing; Cham: 2018. Vaccines against Escherichia coli; pp. 213–242. [Google Scholar]
- 48.Melican K., Sandoval R.M., Kader A., et al. Uropathogenic Escherichia coli P and type 1 fimbriae act in synergy in a living host to facilitate renal colonization leading to nephron obstruction. PLoS Pathog. 2011;7(2) doi: 10.1371/journal.ppat.1001298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lane M.C., Mobley H.L.T. Role of P-fimbrial-mediated adherence in pyelonephritis and persistence of uropathogenic Escherichia coli (UPEC) in the mammalian kidney. Kidney Int. 2007;72(1):19–25. doi: 10.1038/sj.ki.5002230. [DOI] [PubMed] [Google Scholar]
- 50.Stærk K., Khandige S., Kolmos H.J., Møller-Jensen J., Andersen T.E. Uropathogenic Escherichia coli express type 1 fimbriae only in surface adherent populations under physiological growth conditions. J Infect Dis. 2016;213(3):386–394. doi: 10.1093/infdis/jiv422. [DOI] [PubMed] [Google Scholar]
- 51.Yao Y., Xie Y., Kim K.S. Genomic comparison of Escherichia coli K1 strains isolated from the cerebrospinal fluid of patients with meningitis. Infect Immun. 2006;74(4):2196–2206. doi: 10.1128/IAI.74.4.2196-2206.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Basmaci R., Bonacorsi S., Bidet P., et al. Escherichia coli meningitis features in 325 children from 2001 to 2013 in France. Clin Infect Dis. 2015;61(5):779–786. doi: 10.1093/cid/civ367. [DOI] [PubMed] [Google Scholar]
- 53.Cole B.K., Ilikj M., McCloskey C.B., Chavez-Bueno S. Antibiotic resistance and molecular characterization of bacteremia Escherichia coli isolates from newborns in the United States. PLoS One. 2019;14(7) doi: 10.1371/journal.pone.0219352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hung C., Marschall J., Burnham C.-A.D., Byun A.S., Henderson J.P. The bacterial amyloid curli is associated with urinary source bloodstream infection. PLoS One. 2014;9(1) doi: 10.1371/journal.pone.0086009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nhu N.T.K., Phan M.-D., Peters K.M., et al. Discovery of new genes involved in curli production by a uropathogenic Escherichia coli strain from the highly virulent O45:K1:H7 lineage. mBio. 2018;9(4) doi: 10.1128/mBio.01462-18. 014622-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Luna-Pineda V.M., Moreno-Fierros L., Cázares-Domínguez V., et al. Curli of uropathogenic Escherichia coli enhance urinary tract colonization as a fitness factor. Front Microbiol. 2019;10:2063. doi: 10.3389/fmicb.2019.02063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Fakruddin M., Mannan K.S.B., Mazumdar R.M. Correlation between in vitro biofilm formation and virulence properties of extra-intestinal pathogenic Escherichia coli (Expec) Online J Biol Sci. 2014;14(4):261–270. [Google Scholar]
- 58.Prasadarao N.V. Identification of Escherichia coli outer membrane protein A receptor on human brain microvascular endothelial cells. Infect Immun. 2002;70(8):4556–4563. doi: 10.1128/IAI.70.8.4556-4563.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Prasadarao N.V., Wass C.A., Kim K.S. Endothelial cell GlcNAc beta 1-4GlcNAc epitopes for outer membrane protein A enhance traversal of Escherichia coli across the blood-brain barrier. Infect Immun. 1996;64(1):154–160. doi: 10.1128/iai.64.1.154-160.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mittal R., Krishnan S., Gonzalez-Gomez I., Prasadarao N.V. Deciphering the roles of outer membrane protein A extracellular loops in the pathogenesis of Escherichia coli K1 meningitis. J Biol Chem. 2011;286(3):2183–2193. doi: 10.1074/jbc.M110.178236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Nielsen D.W., Ricker N., Barbieri N.L., Allen H.K., Nolan L.K., Logue C.M. Outer membrane protein A (OmpA) of extraintestinal pathogenic Escherichia coli. BMC Res Notes. 2020;13(1):51. doi: 10.1186/s13104-020-4917-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hoffman J.A., Wass C., Stins M.F., Kim K.S. The capsule supports survival but not traversal of Escherichia coli K1 across the blood-brain barrier. Infect Immun. 1999;67(7):3566–3570. doi: 10.1128/iai.67.7.3566-3570.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kim K.J., Elliott S.J., Di Cello F., Stins M.F., Kim K.S. The K1 capsule modulates trafficking of E. coli-containing vacuoles and enhances intracellular bacterial survival in human brain microvascular endothelial cells. Cell Microbiol. 2003;5(4):245–252. doi: 10.1046/j.1462-5822.2003.t01-1-00271.x. [DOI] [PubMed] [Google Scholar]
- 64.Stins M.F., Prasadarao N.V., Ibric L., Wass C.A., Luckett P., Kim K.S. Binding characteristics of S fimbriated Escherichia coli to isolated brain microvascular endothelial cells. Am J Pathol. 1994;145(5):1228–1236. [PMC free article] [PubMed] [Google Scholar]
- 65.Kaczmarek A., Budzynska A., Gospodarek E. Prevalence of genes encoding virulence factors among Escherichia coli with K1 antigen and non-K1 E. coli strains. J Med Microbiol. 2012;61(Pt 10):1360–1365. doi: 10.1099/jmm.0.044263-0. [DOI] [PubMed] [Google Scholar]
- 66.Hoffman J.A., Badger J.L., Zhang Y., Huang S.H., Kim K.S. Escherichia coli K1 aslA contributes to invasion of brain microvascular endothelial cells in vitro and in vivo. Infect Immun. 2000;68(9):5062–5067. doi: 10.1128/iai.68.9.5062-5067.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Huang S.H., Chen Y.H., Fu Q., et al. Identification and characterization of an Escherichia coli invasion gene locus, ibeB, required for penetration of brain microvascular endothelial cells. Infect Immun. 1999;67(5):2103–2109. doi: 10.1128/iai.67.5.2103-2109.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wang Y., Kim K.S. Role of OmpA and IbeB in Escherichia coli K1 invasion of brain microvascular endothelial cells in vitro and in vivo. Pediatr Res. 2002;51(5):559–563. doi: 10.1203/00006450-200205000-00003. [DOI] [PubMed] [Google Scholar]
- 69.Hill V.T., Townsend S.M., Arias R.S., et al. TraJ-dependent Escherichia coli K1 interactions with professional phagocytes are important for early systemic dissemination of infection in the neonatal rat. Infect Immun. 2004;72(1):478–488. doi: 10.1128/IAI.72.1.478-488.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Khan N.A., Wang Y., Kim K.J., Chung J.W., Wass C.A., Kim K.S. Cytotoxic necrotizing factor-1 contributes to Escherichia coli K1 invasion of the central nervous system. J Biol Chem. 2002;277(18):15607–15612. doi: 10.1074/jbc.M112224200. [DOI] [PubMed] [Google Scholar]
- 71.Doye A., Mettouchi A., Bossis G., et al. CNF1 exploits the ubiquitin-proteasome machinery to restrict Rho GTPase activation for bacterial host cell invasion. Cell. 2002;111(4):553–564. doi: 10.1016/s0092-8674(02)01132-7. [DOI] [PubMed] [Google Scholar]
- 72.Jochems S.P., Weiser J.N., Malley R., Ferreira D.M. The immunological mechanisms that control pneumococcal carriage. PLoS Pathog. 2017;13(12) doi: 10.1371/journal.ppat.1006665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.A study of AV0328 administered to healthy adult volunteers. ClinicalTrials.gov identifier: NCT02853617.
- 74.Cywes-Bentley C, Vinacur M, Roberts C, et al. Av0328-A synthetic oligosaccharide-tetanus toxoid conjugate targeting the broadly expressed microbial surface polysaccharide PNAG is safe and immunogenic in humans, eliciting high titers of functional antibody to multiple pathogens. Abstracts from Microbe. 2018;Session 243:Abstract number 4861.
- 75.Trial to assess the efficacy of F598 in preventing an experimental urethral infection with N. Gonorrhoeae in healthy males ClinicalTrials.gov identifier: NCT03222401.
- 76.Hülsdünker J., Thomas O.S., Haring E., et al. Immunization against poly- N -acetylglucosamine reduces neutrophil activation and GVHD while sparing microbial diversity. Proc Natl Acad Sci U S A. 2019;116(41):20700–20706. doi: 10.1073/pnas.1908549116. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.