Abstract
Rationale
COPD has been associated repeatedly with single biomarkers of systemic inflammation, ignoring the complexity of inflammatory pathways. This study aimed to cluster patients with COPD based on systemic markers of inflammatory processes and to evaluate differences in their clinical characterisation and examine how these differences may relate to altered biological pathways.
Methods
213 patients with moderate-to-severe COPD in a clinically stable state were recruited and clinically characterised, which included a venous blood sample for analysis of serum biomarkers. Patients were clustered based on the overall similarity in systemic levels of 57 different biomarkers. To determine interactions among the regulated biomarkers, protein networks and biological pathways were examined for each patient cluster.
Results
Four clusters were identified: two clusters with lower biomarker levels (I and II) and two clusters with higher biomarker levels (III and IV), with only a small number of biomarkers with similar trends in expression. Pathway analysis indicated that three of the four clusters were enriched in Rage (receptor for advanced glycation end-products) and Oncostatin M pathway components. Although the degree of airflow limitation was similar, the clinical characterisation of clusters ranged from 1) better functional capacity and health status and fewer comorbidities; 2) more underweight, osteoporosis and static hyperinflation; 3) more metabolically deranged; and 4) older subjects with worse functional capacity and higher comorbidity load.
Conclusions
These new insights may help to understand the functionally relevant inflammatory interactions in the pathophysiology of COPD as a heterogeneous disease.
Short abstract
Biomarker-based clustering in well-characterised COPD patients resulted in four distinct clusters associated with distinct clinical phenotypes. This study provides new insight into relevant inflammatory interactions in the heterogeneous disease COPD. https://bit.ly/3FTI8CB
Introduction
COPD has been shown to be consistently associated with systemic inflammation [1]. Most COPD studies have only focused on one or a limited number of markers of systemic inflammation, revealing the heterogeneity within and between the different systemic inflammatory biomarkers [1–3]. Multiple other inflammatory biomarkers exist; for example, acute phase proteins and the complement system [1, 4, 5], cytokines and chemokines and their receptors [6–10], inhibitors of cytokine signalling [11], adhesion molecules [12, 13], immunoglobulins [14], growth factors [10, 15, 16], proteins involved in tissue remodelling [17, 18], hormones and adipokines [19, 20], coagulation [21, 22] and plasma carrier proteins [23, 24] have all been shown to be associated with COPD. These data suggest that to better understand the complexity of COPD on a systemic level, a variety of serum markers encompassing a wider scope of
Lessons for clinicians
Evidence before the study multiple markers of inflammation, such as acute phase proteins and the complement system, cytokines and chemokines and their receptors, inhibitors of cytokine signalling, adhesion molecules, immunoglobulins, growth factors, proteins involved in tissue remodelling, hormones and adipokines, coagulation and plasma carrier proteins have been shown to be associated with COPD. These data suggest that to better understand the complexity of COPD on a systemic level, a variety of serum markers encompassing the wider scope of inflammatory processes are needed. Previous studies showed that combinations of biomarkers can improve predictive outcome of relevant cross-sectional and longitudinal outcome. Only a limited number of studies have used large-scale proteomics to find different subgroups of well-characterised COPD patients.
Added value of this study Biomarker-based clustering in clinically well-characterised patients with COPD resulted in four distinct clusters. Although a similar degree of airflow limitation was seen among the clusters, they were associated with distinct clinical phenotypes. Pathway analysis revealed components of the RAGE (receptor for advanced glycation end products) pathway to be enriched in three clusters, suggesting a systemic role for this pathway in COPD.
Implications of all the available evidence This study provides new insight into identifying and understanding relevant inflammatory interactions in the heterogeneous disease COPD. Furthermore, the evidence provided by this study may aid in generating new hypotheses into the similarities and differences observed between subgroups of COPD patients.
inflammatory processes are needed. Pinto-Plata and co-workers [25, 26] previously showed a pattern of systemic biomarkers in patients with COPD that can be associated with different clinical variables known to predict disease outcome, including the degree of airflow limitation, lung transfer factor, functional capacity, BODE (body mass index, airflow obstruction, dyspnoea and exercise) index and exacerbation frequency. Additionally, in studies such as COPDGene and Evaluation of Clevidipine in the Perioperative Treatment of Hypertension Assessing Safety Events (ECLIPSE), combinations of biomarkers improved predictive value for relevant cross-sectional and longitudinal COPD outcomes [27].
Only a limited number of studies used large-scale proteomics to find different subgroups of COPD. In a post hoc analysis of the Treatment of Emphysema with a Selective Retinoid Agonist (TESRA) trial, 87 peripheral blood biomarkers in 396 former smokers with emphysema were analysed using multiplex platforms and included in a cluster analysis [28]. A small subgroup of participants with increased inflammatory biomarkers was identified, presented with less emphysema and similar lung function, but worse quality of life compared to the other clusters.
In the present study, we hypothesised that a cluster analysis of a composite panel of biomarkers could express the heterogeneity of different mechanistic pathways in COPD and could to some extent be associated with the phenotypic expressions of COPD, including comorbidities/systemic phenotypes.
Therefore, we aimed to perform a cluster analysis on a broad set of biomarkers, related to inflammatory processes previously associated with COPD. Furthermore, we aimed to compare clinical characteristics among the identified clusters. Finally, we aimed to analyse the pathways enriched in each cluster which may suggest alterations in biologically relevant pathways, thus contributing insight into COPD pathophysiology.
Methods
Study design
This is a secondary analysis of the baseline data from the prospective CIRO comorbidity study [29]. In brief, patients with COPD (forced expiratory volume in 1 s <80% predicted [30]), aged 40–80 years and in a clinically stable state were recruited during the baseline assessment of a comprehensive pulmonary rehabilitation programme at CIRO [31]. During a 3-day assessment, patients were clinically characterised, including multiple comorbidities, as described previously [29].
Venous blood sampling and laboratory analysis
Venous blood was sampled in the fasted state. A total of 57 biomarkers were examined (supplementary table S1). Measurement of leptin, fetuin A (Quantikine; R&D Systems, Minneapolis, MN, USA) and human fibrinogen (Abcam, Cambridge, UK) was performed using ELISA. Measurements of other biomarkers were carried out on multiplex platforms (Meso Scale Discovery, Gaithersburg, MD, USA and Myriad RBM, Austin, TX, USA). If the level of a specific biomarker was below the detection threshold, the value was set to the threshold level. Eight biomarkers with >30% missing values were excluded from the analysis (B-lymphocyte chemoattractant, eotaxin-1, eotaxin-3, interleukin (IL)-1α, IL-12 subunit p70, IL-15, IL-17 and major histocompatibility complex class I chain-related protein-A.) Measurement of leukocytes, haemoglobin, haematocrit, total cholesterol, high-density lipoprotein (HDL), low-density lipoprotein (LDL), triglycerides, bilirubin, creatinine and glucose were determined in the CIRO+ laboratory (validated, custom-made arrays). Glucose, haemoglobin, total cholesterol and creatinine were not evaluated as biomarkers, but as markers of comorbidities: hyperglycaemia, anaemia, dyslipidaemia and renal impairment, respectively.
All markers classified in one of the categories that have shown to be previously linked with the pathophysiology of COPD (see the introduction) were included in the analysis (table 1). Those classified as others were included in the analysis because of availability and previous shown association with COPD, namely ferritin [32], β2-microglobulin [33], myoglobin [34], osteonectin [35], osteopontin [36], bilirubin [37], leukocytes [1], LDL and HDL [38].
TABLE 1.
Systemic levels of biomarkers of inflammation, chemoattraction, cell activation, tissue destruction and tissue repair per cluster
| Cluster 1: | Cluster 2: | Cluster 3: | Cluster 4: | |
| lower-level cluster I | lower-level cluster II | higher-level cluster I | higher-level cluster II | |
| Patients, n | 64 | 64 | 46 | 39 |
| Acute phase proteins and complement system | ||||
| Complement C3, mg·mL−1 | 1.2±0.2 | 1.1±0.2 | 1.3±0.2 | 1.5±1.0 |
| CRP, ng·mL−1 | 3011±2795 | 1773±1859 | 8214±8153 | 9725±8480 |
| Fibrinogen, μg·mL−1 | 6417±2826 | 6870±4533 | 6465±2374 | 8879±7911 |
| Serum amyloid A, ng·mL−1 | 3059±2705 | 1889±1403 | 6239±7311 | 10 745±7810 |
| Cytokines and chemokines and their receptors | ||||
| Eotaxin-2, pg·mL−1 | 1022±632 | 1398±940 | 1371±954 | 1337±965 |
| IL-1β, pg·mL−1 | 4.8±1.2 | 4.3±1.3 | 4.2±1.1 | 5.0±1.6 |
| IL-12 subunit p40, ng·mL−1 | 0.6±0.2 | 0.5±0.1 | 0.5±0.1 | 0.6±0.1 |
| IL-23, ng·mL−1 | 1.4±0.4 | 1.2±0.3 | 1.2±0.3 | 1.4±0.3 |
| IL-6, pg·mL−1 | 2.9±1.6 | 1.9±1.7 | 3.4±1.7 | 6.3±8.2 |
| IL-8, pg·mL−1 | 12±5 | 13±5 | 14±6 | 14±5 |
| IFN-γ-inducible T-cell α-chemoattractant, pg·mL−1 | 43±22 | 44±24 | 55±25 | 64±47 |
| IFN-γ-inducible protein-10, pg·mL−1 | 93±41 | 73±39 | 90±30 | 142±98 |
| IFN-γ, pg·mL−1 | 0.5±0.3 | 0.4±0.4 | 0.5±0.2 | 1.3±2.6 |
| Macrophage migration inhibitory factor, ng·mL−1 | 0.1±0.1 | 0.1±0.1 | 0.1±0.1 | 0.1±0.1 |
| MCP-1, pg·mL−1 | 511±190 | 469±130 | 606±159 | 513±200 |
| MCP-4, pg·mL−1 | 688±235 | 678±207 | 762±210 | 778±455 |
| T-cell-specific protein RANTES, ng·mL−1 | 19±8 | 23±10 | 31±10 | 25±14 |
| Thymus and activation-regulated chemokine, pg·mL−1 | 524±414 | 636±384 | 784±620 | 770±828 |
| IL-2 receptor-α, pg·mL−1 | 2427±694 | 2020±535 | 2695±927 | 3429±1240 |
| TNF-R2, ng·mL−1 | 6.0±1.6 | 4.9±1.2 | 6.0±1.8 | 9.8±5.0 |
| TNF-R1, pg·mL−1 | 1850±476 | 1531±436 | 1905±588 | 2603±919 |
| Osteoprotegerin, pM | 7.0±1.9 | 6.7±1.1 | 7.2±1.3 | 8.9±2.0 |
| Inhibitors of cytokine signalling | ||||
| IL-1 receptor antagonist, pg·mL−1 | 423±132 | 336±74 | 372±99 | 388±118 |
| Adhesion molecules | ||||
| sICAM-1, ng·mL−1 | 282±76 | 292±76 | 321±102 | 389±113 |
| sVCAM-1, ng·mL−1 | 520±116 | 493±106 | 517±142 | 719±396 |
| Immunoglobulins | ||||
| IgA, mg·mL−1 | 2.9±1.6 | 2.4±1.2 | 3.1±1.4 | 3.6±2.9 |
| IgM, mg·mL−1 | 2.1±1.4 | 1.8±1.9 | 1.7±1.1 | 2.5±4.2 |
| Growth factors | ||||
| Angiopoietin-2, ng·mL−1 | 3.7±1.8 | 3.8±1.3 | 4.4±1.4 | 5.1±1.3 |
| BDNF, ng·mL−1 | 18.1±4.9 | 22±5 | 23±7 | 18±7 |
| Stem cell factor, pg·mL−1 | 397±93 | 330±83 | 354±88 | 473±163 |
| Vascular endothelial growth factor, pg·mL−1 | 242±111 | 258±122 | 354±159 | 290±170 |
| Tissue remodelling | ||||
| α1-Antitrypsim, mg·mL−1 | 1.9±0.3 | 2.0±0.4 | 1.9±0.4 | 2.3±0.6 |
| LAP TGF-β1, ng·mL−1 | 9.4±2.8 | 12±3 | 13±3 | 10±3 |
| MMP-3, ng·mL−1 | 19±14 | 13±7 | 14±7 | 23±16 |
| MMP-9, ng·mL−1 | 147±72 | 130±62 | 153±69 | 159±83 |
| Tissue inhibitor of metalloproteinases-1, ng·mL−1 | 150±23 | 163±27 | 195±30 | 199±65 |
| Hormones and adipokines | ||||
| Adiponectin, μg·mL−1 | 4.4±2.1 | 7.0±3.3 | 5.1±2.7 | 7.4±4.7 |
| Leptin, ng·mL−1 | 14.8±12.3 | 8.2±8.8 | 16.8±14.6 | 13.7±14.8 |
| Erythropoietin, IU·mL−1 | 11.3±5.2 | 8.4±2.6 | 11±7 | 13±4 |
| Osteocalcin (OCN/BGLAP), ng·mL−1 | 117±20 | 127±18 | 116±17 | 135±25 |
| Coagulation | ||||
| α2-Macroglobulin (A2Macro), mg·mL−1 | 1.7±0.4 | 1.7±0.4 | 1.7±0.6 | 1.9±0.7 |
| Factor VII, ng·mL−1 | 447±140 | 406±99 | 403±127 | 422±119 |
| PAI-1, ng·mL−1 | 195±45 | 228±53 | 290±56 | 220±112 |
| Plasma carrier proteins | ||||
| Fetuin A, μg·mL−1 | 574±292 | 706±364 | 603±303 | 652±420 |
| Haptoglobin, mg·mL−1 | 1.7±0.9 | 1.6±1.1 | 2.9±1.4 | 3.5±3.1 |
| IGFBP-1, ng·mL−1 | 2978±2432 | 4368±2771 | 3204±2178 | 4504±2870 |
| IGFBP-2, ng·mL−1 | 79±34 | 110±53 | 80±35 | 132±45 |
| Vitamin D-binding protein, μg·mL−1 | 288±108 | 300±91 | 291±122 | 396±727 |
| Other | ||||
| Ferritin, ng·mL−1 | 180±142 | 151±128 | 190±160 | 183±203 |
| β2-Microglobulin, μg·mL−1 | 2.1±0.5 | 1.8±0.3 | 2.1±0.4 | 2.9±1.0 |
| Myoglobin, ng·mL−1 | 69±37 | 74±111 | 54±28 | 96±62 |
| Osteonectin, ng·mL−1 | 1145±344 | 1394±230 | 1720±322 | 1318±394 |
| Osteopontin, ng·mL−1 | 22±7 | 22±8 | 22±7 | 36±14 |
| Bilirubin, mmol·L−1 | 11.6±4.9 | 14±6 | 10±5 | 12±3 |
| Leukocytes, ×109 cells·L−1 | 6.7±1.4 | 6.9±1.9 | 8.1±1.4 | 8.5±2.4 |
| LDL, mmol·L−1 | 2.7±0.9 | 3.1±0.9 | 3.2±1.2 | 2.9±1.0 |
| HDL, mmol·L−1 | 1.7±0.4 | 1.7±0.5 | 1.7±0.5 | 1.5±0.5 |
Data are presented as mean±sd. Eight biomarkers were measured, but excluded from the statistical analysis, because of >30% missing values (B-lymphocyte chemoattractant, eotaxin-1, eotaxin-3, interleukin (IL)-1α, IL-12 subunit p70, IL-15, IL-17 and major histocompatibility complex (MHC) class I chain-related protein-A). ▪ A significantly higher level compared to the remaining three clusters (p-value <0.01); ▪ a tendency for a significantly higher level compared to the remaining three clusters (p-value between 0.05 and 0.01); ▪ a significantly lower level compared to the remaining three clusters (p<0.01); ▪ a tendency for a significantly lower level compared to the remaining three clusters (p-value between 0.05 and 0.01). CRP: C-reactive protein; IFN: interferon; MCP: monocyte chemotactic protein; TNF-R: tumour necrosis factor receptor; BDNF: brain-derived neurotrophic factor; sICAM: soluble intercellular adhesion molecule; sVCAM: soluble vascular adhesion molecule; LAP TGF-β1: latency-associated peptide of transforming growth factor-β1; MMP: matrix metalloproteinase; PAI: plasminogen activator inhibitor; IGFBP: insulin-like growth factor-binding protein; LDL: low-density lipoprotein; HDL: high-density lipoprotein.
Statistics
All statistical analyses were performed using Viscovery SOMine 7.1 (Viscovery Software; www.viscovery.net). Self-organising maps (SOMs, also known as Kohonen maps) were used to create an ordered representation of the data [39]. The SOM method can be viewed as a nonparametric regression technique that converts multidimensional data spaces into lower dimensional abstractions. A SOM generates a nonlinear representation of the data distribution and allows the user to visually identify homogenous data groups.
Patients were ordered by their overall similarity regarding their systemic levels of biomarkers. Based on the created SOM model, clusters were generated using the SOM-Ward cluster algorithm of Viscovery, a hybrid algorithm that applies the classical hierarchical method of Ward on top of the SOM topology. The method begins by defining each individual node as a separate cluster. In each step of the algorithm, two clusters with minimal distance according to the SOM-Ward distance measure are merged. This measure heeds the Ward distances as well as the positioning of two clusters in the map picture by defining that the distance of nonadjacent clusters is always infinite, limiting merging to topologically neighbouring clusters. A detailed description of SOMs can be found in the supplementary methods.
Summary variables on the systemic levels of biomarkers and clinical characteristics for the study sample and for each cluster are presented as mean±sd for quantitative variables and percentage for discrete variables. Viscovery automatically identified for each cluster the attributes that significantly differ from the average of the whole study sample of 213 patients using the integrated two-sided t-test with a confidence of 95% and 99%.
Pathway analysis
To determine interactions between similarly regulated biomarkers, protein networks were made for each cluster using String [40], including the serum markers for which an official gene symbol could be identified (supplementary table S1). The minimum required interaction score was set to high confidence (0.7) and all possible active interactions were allowed. The edges of the network display predicted molecular modes of action and unconnected nodes were not shown. Gene ontology of biological processes as well as pathway enrichment were analysed in each cluster (supplementary tables S2–S9) [40, 41].
Results
General patient characteristics
213 out of 255 patients admitted to CIRO were eligible for the study. These patients had moderate to very-severe COPD, with a substantial smoking history, moderately impaired diffusion capacity, increased static lung volumes and multimorbidity (table 2).
TABLE 2.
General characteristics of study subjects
| Subjects, n | 213 |
| Age, years | 63.6±7.0 |
| Male | 59 |
| BMI, kg·m−2 | 26.2±5.1 |
| FFMI, kg·m−2 | 17.0±2.4 |
| mMRC dyspnoea grade | 2.1±1.1 |
| Current smoker | 28 |
| Smoking pack-years | 46±26 |
| Long-term oxygen therapy | 17 |
| FEV1, L | 1.40±0.54 |
| FEV1, % predicted | 51.2±16.9 |
| FEV1/FVC | 0.40±0.11 |
| ITGV, % predicted | 148±33 |
| DLCO, % predicted | 56±17 |
| 6MWD, m | 470±106 |
| SGRQ, total score | 51.3±17.5 |
| Updated BODE score | 2.9±2.5 |
| Framingham 10-year risk, % | 9.4±6.7 |
Data are presented as mean±sd or %, unless otherwise stated. BMI: body mass index; FFMI: fat-free mass index; mMRC: modified Medical Research Council; FEV1: forced expiratory volume in 1 s; FVC: forced vital capacity; ITGV: intrathoracic gas volume; DLCO: diffusion capacity of the lung for carbon monoxide; 6MWD: 6-min walk distance; SGRQ: St George's Respiratory Questionnaire; BODE: body mass index, obstruction, dyspnoea, exercise capacity index.
Clustering based on the systemic levels of the biomarkers
57 serum biomarkers were used to cluster the 213 patients with COPD, which resulted in four clusters with distinct biomarker profiles. Table 1 describes how these clusters are constituted and which biomarkers have significantly higher or lower levels compared to the mean values of the whole sample. 15 biomarkers were not significantly different between the four clusters at the 99% confidence level.
In cluster 1 (“lower-level cluster I”), the levels of 17 biomarkers were significantly lower compared to the mean values of the whole sample, while two biomarkers were significantly higher. In cluster 2 (“lower-level cluster II”), the systemic levels of 19 biomarkers were significantly lower compared to the mean values of the whole sample, while three biomarkers were significantly higher. Only three biomarkers were significantly lower in both clusters. In cluster 3 (“higher-level cluster I”), the systemic levels of three biomarkers were significantly lower compared to the mean values of the whole sample, while 12 biomarkers were significantly higher. In cluster 4 (“higher-level cluster II”), the systemic level of only one biomarker was significantly lower compared to the mean values of the whole sample, while the levels of 27 biomarkers were significantly higher. Only four biomarkers were found to be significantly higher in both clusters 3 and 4.
Clinical characteristics and objectively identified comorbidities related to the different clusters
Tables 3 and 4 summarise the clinical characteristics and the comorbidities of the four clusters, respectively. Spirometry results were similar among different clusters as were the mean pack years, mean score on the modified Medical Research Council (mMRC) dyspnoea scale and the Charlson Comorbidity Index. Additionally, no difference in exacerbations in the past 12 months was observed among clusters.
TABLE 3.
The clinical characteristics of each cluster
|
Cluster 1:
lower-level cluster I |
Cluster 2:
lower-level cluster II |
Cluster 3:
higher-level cluster I |
Cluster 4:
higher-level cluster II |
|
| Female | 34 | 47 | 48 | 33 |
| Age, years | 65±7 | 61±6 | 62±6 | 69±6 |
| Lung function | ||||
| FEV1, L | 1.5±0.5 | 1.4±0.5 | 1.4±0.5 | 1.3±0.6 |
| FEV1, % predicted | 54±16 | 51±19 | 50±15 | 48±16 |
| FEV1/FVC, % | 41±12 | 40±11 | 40±11 | 38±9 |
| DLCO, % predicted | 60±17 | 54±16 | 58±17 | 48±14 |
| ITGV, % predicted | 139±32 | 156±35 | 147±34 | 152±29 |
| RV, % predicted | 153±45 | 175±54 | 162±44 | 165±39 |
| TLC, % predicted | 116±17 | 123±16 | 119±17 | 116±16 |
| PaCO2, kPa | 5.3±0.6 | 5.2±0.5 | 5.5±0.7 | 5.4±0.6 |
| PaO2, kPa | 9.6±1.1 | 9.6±1.2 | 9.3±0.9 | 9.2±0.9 |
| SaO2, % | 95.1±1.9 | 95.2±1.7 | 94.7±2.0 | 94.4±1.8 |
| PImax, % predicted | 85±24 | 77±25 | 78±22 | 75±18 |
| PEmax, % predicted | 64±20 | 62±22 | 60±17 | 56±17 |
| COPD-specific characteristics | ||||
| mMRC dyspnoea grade | 2.0±1.1 | 2.0±1.1 | 2.1±0.9 | 2.4±1.3 |
| Long-term oxygen therapy | 11 | 14 | 17 | 31 |
| Smoking pack-years | 44±21 | 47±28 | 48±30 | 47±23 |
| Current smoker | 14 | 31 | 37 | 36 |
| Hospital admissions for COPD in past 12 months, n | 0.51±1.17 | 0.42±1.17 | 0.49±0.84 | 0.69±1.28 |
| Steroid/antibiotic courses for COPD in past 12 months, n | 1.53±1.95 | 1.42±1.61 | 1.01±1.32 | 1.92±1.92 |
| Exacerbations in past 12 months, n | 2.05±2.60 | 1.85±2.41 | 1.54±1.81 | 2.55±2.18 |
| GOLD group A/B/C/D | 19/30/20/30 | 18/26/15/40 | 11/40/7/40 | 15/18/15/51 |
| Physical fitness | ||||
| 6MWD, m | 493±91 | 497±101 | 461±91 | 402±126 |
| Peak work rate, W | 86±30 | 76±23 | 75±27 | 58±26 |
| Peak work rate, % predicted | 63±25 | 62±25 | 61±29 | 47±22 |
| Constant work-rate test, s | 409±264 | 408±329 | 296±191 | 234±187 |
| Health status | ||||
| SGRQ symptoms domain | 54±19 | 53±21 | 57±18 | 57±25 |
| SGRQ activity domain | 65±22 | 68±22 | 73±17 | 69±27 |
| SGRQ impact domain | 35±19 | 38±18 | 47±16 | 43±26 |
| SGRQ total score | 48±19 | 50±17 | 57±12 | 53±23 |
| Prognostic indices | ||||
| Updated BODE index | 2.2±1.8 | 2.7±2.3 | 2.6±2.0 | 4.6±3.7 |
| Framingham risk score | 9.7±6.1 | 7.5±5.9 | 9.3±7.2 | 12.3±7.2 |
Data are presented as mean±sd or %, unless otherwise stated. ▪ A significantly higher level compared to the remaining three clusters (p<0.01); ▪ a tendency for a significantly higher level compared to the remaining three clusters (p-value between 0.05 and 0.01); ▪ a significantly lower level compared to the remaining three clusters (p<0.01); ▪ a tendency for a significantly lower level compared to the remaining three clusters (p-value between 0.05 and 0.01). FEV1: forced expiratory volume in 1 s; FVC: forced vital capacity; DLCO: diffusion capacity of the lung for carbon monoxide; ITGV: intrathoracic gas volume; RV: residual volume; TLC: total lung capacity; PaCO2: arterial carbon dioxide partial pressure; PaO2: arterial oxygen partial pressure; SaO2: arterial haemoglobin oxygen saturation; PImax: maximal inspiratory mouth pressure; PEmax: maximal expiratory mouth pressure; mMRC: modified Medical Research Council; GOLD: Global Initiative for Chronic Obstructive Lung Disease; 6MWD: 6-min walk distance; SGRQ: St George's Respiratory Questionnaire; BODE: body mass index, obstruction, dyspnoea, exercise capacity index.
TABLE 4.
Comorbidities per biomarker-based cluster
| Cluster 1: lower-level cluster I | Cluster 2: lower-level cluster II | Cluster 3: higher-level cluster I | Cluster 4: higher-level cluster II | |
| Comorbidities, n | 3.5±1.5 | 3.4±1.5 | 3.7±1.7 | 4.3±1.8 |
| Charlson Comorbidity Index | 1.6±0.7 | 1.5±0.9 | 1.6±0.8 | 1.8±1.1 |
| eGFR, mL·min−1 | 78±21 | 83±23 | 87±25 | 66±20 |
| Creatinine, μmol·L−1 | 93±20 | 78±15 | 84±14 | 99±32 |
| Renal impairment | 23 | 14 | 15 | 41 |
| Haemoglobin, mmol·L−1 | 9.0±0.6 | 9.1±0.7 | 9.1±0.9 | 8.7±0.7 |
| Haematocrit | 44±4 | 44±4 | 44±5 | 42±4 |
| Anaemia | 2 | 2 | 11 | 10 |
| Systolic blood pressure, mmHg | 140±17 | 136±25 | 138±15 | 145±28 |
| Diastolic blood pressure, mmHg | 83±8 | 82±11 | 83±9 | 83±12 |
| Hypertension | 45 | 44 | 48 | 62 |
| BMI, kg·m−2 | 27.4±4.8 | 24.2±4.9 | 28.1±5.4 | 25.2±4.2 |
| Obese | 28 | 16 | 33 | 18 |
| Underweight | 9 | 25 | 4 | 15 |
| FFMI, kg·m−2 | 17.6±2.0 | 16.1±2.5 | 17.5±2.8 | 16.9±2.0 |
| Muscle wasting | 14 | 50 | 20 | 26 |
| Glucose, mmol·L−1 | 5.8±1.0 | 5.7±0.8 | 5.9±1.0 | 5.6±0.9 |
| Hyperglycaemia | 56 | 55 | 61 | 44 |
| Triglycerides, mmol·L−1 | 1.6±0.7 | 1.3±0.5 | 2.0±1.3 | 1.4±0.7 |
| Cholesterol, mmol·L−1 | 5.0±1.0 | 5.4±1.0 | 5.8±1.5 | 5.1±1.1 |
| Cholesterol/HDL ratio | 3.2±1.0 | 3.3±0.9 | 3.8±1.3 | 3.6±1.1 |
| Dyslipidaemia | 41 | 22 | 52 | 33 |
| Thrombocytes, ×109 cells·L−1 | 235±57 | 253±65 | 334±138 | 260±72 |
| HADS Anxiety | 5.5±3. | 6.7±4.3 | 7.0±3.8 | 6.0±3.7 |
| Anxiety | 16 | 23 | 25 | 22 |
| HADS Depression | 5.1±3.5 | 5.5±3.9 | 6.7±3.3 | 5.7±3.2 |
| Depression | 16 | 15 | 20 | 14 |
| c-IMT, mm | 1.0±0.2 | 0.9±0.2 | 1.0±0.2 | 1.0±0.2 |
| Atherosclerosis | 61 | 35 | 57 | 62 |
| Cardiac Infarction Injury Score | 10.4±7.2 | 11.2±5.4 | 11.3±5.7 | 13.3±7.0 |
| T-score total body | −0.6±1.1 | −1.4±1.3 | −0.8±1.2 | −1.4±1.5 |
| Osteoporosis | 22 | 41 | 22 | 41 |
Data are presented as mean±sd or %, unless otherwise stated. ▪ A significantly higher level compared to the remaining three clusters (p<0.01); ▪ a tendency for a significantly higher level compared to the remaining three clusters (p-value between 0.05 and 0.01); ▪ a significantly lower level compared to the remaining three clusters (p<0.01); ▪ a tendency for a significantly lower level compared to the remaining three clusters (p-value between 0.05 and 0.01). eGFR: estimated glomerular filtration rate; BMI: body mass index; FFMI: fat-free mass index; HDL: high-density lipoprotein; HADS: Hospital Anxiety and Depression Score; c-IMT: carotid intima-media thickness.
While patients in cluster 1 had a similar level of airway obstruction and diffusion capacity, these patients exhibited less static hyperinflation, better mouth pressures, a higher peak work rate, a longer 6-min walk distance and were less often current smokers. Subjects in cluster 1 had lower prevalence of low muscle mass and a better bone mineral density compared to the whole study population. In addition, subjects had better renal function, a higher body mass index (BMI), lower blood cholesterol and lower St George's Respiratory Questionnaire (SGRQ) levels.
Cluster 2 had a lower mean age, BMI and fat-free mass index and were more likely to be underweight. In contrast to cluster 1, cluster 2 had more hyperinflation. Bone mineral density was worse, with a corresponding higher prevalence of osteoporosis. Conversely, atherosclerosis and dyslipidaemia were less prevalent, with a lower Framingham cardiovascular risk score.
Cluster 3 had a higher SGRQ score and higher scores on the depression score of the Hospital Anxiety and Depression Score. Cluster 3 had a higher BMI, triglycerides and cholesterol, but worse renal function and more anaemia.
Cluster 4 had the highest mean age. The mean score on the updated BODE index and for the Framingham risk score within 10 years were increased in cluster 4, suggesting an overall more severe disease status. Although the degree of airflow obstruction was similar compared to the other clusters, diffusion capacity was lower. Exercise capacity was significantly less compared to the whole study population and there was a higher mMRC, a lower haemoglobin oxygen saturation and a higher proportion of patients on long-term oxygen therapy. Cluster 4 had a higher number of comorbidities, including a significantly higher degree of renal impairment, lower bone mineral density than in other clusters and a significantly increased Framingham cardiovascular risk and a higher cardiac infarction injury score.
A pathway analysis based on the identified clusters of biomarkers
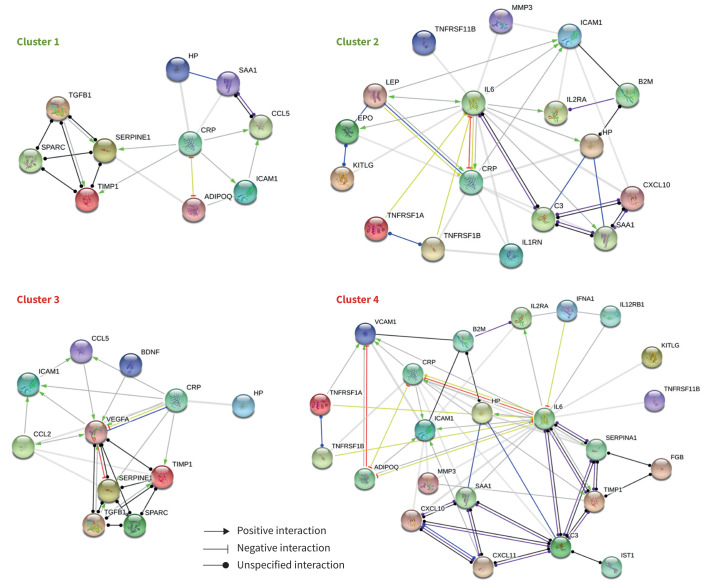
Protein levels that were altered in the same direction (significantly upregulated or downregulated) per cluster were used to generate networks and predict enriched pathways (figure 1). In all clusters there were significant degrees of protein–protein interactions (p<3.5×10−11; figure 1). Interestingly, when we enriched for pathways associated with these protein networks, the most significant were common between clusters 1, 3 and 4, namely the Oncostatin M (OSM) and receptor for advanced glycation end products (RAGE) pathways (supplementary tables S2, S4 and S5). In cluster 2, cytokine–cytokine receptor interactions and IL-1 regulation of extracellular matrix were the two most significantly enriched pathways (supplementary table S3).
FIGURE 1.
Network interactions of predicted upregulated or downregulated proteins by cluster. Proteins that were similarly and significantly upregulated (clusters 3 and 4) or downregulated (clusters 1 and 2) were entered into String [35], all possible interaction sources were allowed and subjected to a minimum required high-confidence interaction score (0.7). Disconnected nodes are not shown. All clusters had a p-value of ≥3.53×10−11. The mode of interaction is depicted as an arrow (positive interaction), inhibition (negative interaction) or ball (unspecified interaction). Gene names, with corresponding biomarker names in parenthesis, used in this figure: ADIPOQ (adiponectin), BDNF (brain-derived neurotrophic factor), B2M (β2-microglobulin), C3 (complement C3), CCL2 (monocyte chemotactic protein-1), CCL5 (RANTES), CRP (C-reactive protein), CXCL10 (interferon (IFN)-γ-induced protein-10), CXCL11 (IFN-γ-inducible T-cell α chemoattractant), EPO (erythropoietin), HP (haptoglobin), ICAM1 (intercellular adhesion molecule-1), IFNG (IFN-γ), IL1RN (interleukin (IL)-1ra), IL2RA (IL-2 receptor-α), IL6 (IL-6), IL12RB1 (IL12p40), BGLAP (osteocalcin), KITLG (stem cell factor), LEP (leptin), MMP3 (matrix metalloproteinase-3), SAA1 (serum amyloid A), SERPINA1 (α1-antitrypsin), SERPINE1 (plasminogen activator inhibitor-1), SPARC (osteonectin), TGFB1 (latency-associated peptide of transforming growth factor-β1), TIMP1 (tissue inhibitor of metalloproteinases-1), TNFRSF11B (osteoprotegerin), TNFRSF1A (tumour necrosis factor receptor (TNFR)I), TNFRSF1B (TNFR2), VEGFA (vascular endothelial growth factor), VCAM1 (vascular cell adhesion molecule-1).
Discussion
This is the first study to cluster a sample of well-characterised patients with COPD based on a set of biomarkers. This resulted in four clusters with distinct biomarker patterns. Although the clusters had a similar degree of airflow limitation, distinct patient profiles could, to a certain extent, be attributed to the different biomarker clusters. Additionally, those clusters appeared connected and enriched for numerous signalling pathways.
A large heterogeneity in the levels of the different biomarkers was observed in this study, which corroborates previous findings [3, 25–27]. The current clustering resulted in two lower-level clusters (1 and 2) and two higher-level clusters (3 and 4). Interestingly, only three biomarkers in clusters 1 and 2 and only four biomarkers in clusters 3 and 4 overlapped in the same direction. In contrast, four biomarkers in the low-level and three in the high-level clusters exhibited opposing expression patterns. These findings illustrate that different biological processes can be involved in patients with a high inflammatory state, as well as in patients with a low inflammatory state. 15 biomarkers were not significantly altered in any of the four clusters; however, multiple biomarkers have been found to be altered in COPD and/or shown to play a role in the pathogenesis, and could thus point to common pathogenic mechanisms. (e.g. α2-macroglobulin [42], factor VII [21], ferritin [32, 43, 44], fetuin A [23], IL-1β [6], IL-8 [1], monocyte chemotactic protein-4 [7], macrophage migration inhibitory factor [8], matrix metalloproteinase-9 [17], thymus and activation-regulated chemokine [9] and vitamin D-binding protein [24]).
The degree of airflow limitation was comparable between clusters, indicating the limited value of the degree of airflow limitation in predicting systemic levels of the biomarkers and vice versa. Interestingly, there were some significant differences in clinical characteristics between the clusters. Overall, the lower-level biomarker clusters have less disease manifestations compared to the high-level clusters. This is in line with Pinto-Plata et al. [26], who reported that patients in the highest quartile of a broader set of inflammatory biomarkers were more clinically compromised and had higher mortality. Cluster 1 could be considered as metabolically healthier with fewer comorbidities compared to the whole study population. Moreover, this cluster showed one of the smallest protein interaction networks, suggesting limited systemic alterations. Clinically, cluster 2 relates in some way to the cachectic or implosive cluster or phenotype [29]. This finding might be in contrast with previous data suggesting that cachectic patients have increased levels of systemic inflammation [45], as the majority of the systemic biomarkers were decreased.
The clear differences in body composition between clusters 1 and 2 may help to explain some biomarker differences between the groups, such as the low leptin, high adiponectin and brain-derived neurotrophic factor (BDNF) levels in cluster 2. The higher levels of the selective competitive inhibitor of IL-1, IL-1Ra, in cluster 1 confirms a robust low inflammatory status. Indeed, the lack of upregulation of IL-1Ra in cluster 2 may be underlying the implosive phenotype rather than upregulation of other pro-inflammatory mediators as seen in the high clusters. Conversely, cluster 2 exhibited decreased levels of IL-23, which is involved in the proliferation and maintenance of T-helper 17 cells and, thus, proper mucosal defences via, for example, the induction and production of secretory immunoglobulin (sIg)A. Importantly, sIgA levels were also lower in cluster 2, which could indicate a specific defect in mucosal immunity [46].
Cluster 4 is clearly the most inflamed cluster in terms of the number of significantly altered biomarkers (27 out of 57 increased). Perhaps this high level of inflammation is unsurprising due to the clinical characteristics of this cluster (table 2), as age, smoking and cardiovascular risk have indeed been repeatedly associated with increased inflammatory status [1].
Cluster 3, the other “high-level” biomarker cluster is clearly different from cluster 4 in terms of number (12 versus 27) and type of increased markers. On the clinical level, this cluster was mainly characterised by metabolic dysregulation. It is likely that the biomarker pattern in this cluster is partly driven by increased fat mass. The adipokine leptin was slightly increased in this cluster. The adipokines BDNF and insulin-like growth factor-binding protein 2, which have been implicated in the pathogenesis of metabolic syndrome [47], were bidirectionally different between clusters 3 and 4. Increased levels of IL12p40 have been found in the serum of obese adolescents compared to normal-weight controls [48]. Additionally, plasminogen activator inhibitor (PAI)-1, a major inhibitor of fibrinolysis, which was elevated in cluster 3, but not in cluster 4, has been shown to significantly correlate with components of metabolic syndrome [22].
Through examination of protein–protein interactions and pathway analysis we can begin to shed light on the underlying molecular mechanisms that may be partially responsible for aspects of disease in the different clusters. Interestingly, we found that three out of the four clusters were enriched in the OSM and RAGE pathways. Ample evidence has already implicated the RAGE pathway in the pathogenesis of COPD. This evidence includes protection from experimental emphysema in RAGE-deficient mice and the identification of the AGER, the gene encoding RAGE protein, as a susceptibility gene in genome-wide association studies [49]. Due to the direction of alterations of key molecules associated with the RAGE pathway (supplementary tables S2, S4 and S5), we would predict that the RAGE pathway is downregulated in cluster 1 and upregulated in clusters 3 and 4, which fits the role of RAGE as a major contributor to inflammation and the data obtained in animal studies. Additionally, RAGE-deficient mice were not prone to weight gain, even when fed a high-fat diet [50]. We see that our COPD patients in cluster 1 show clinical characteristics of lower cholesterol and normal body weight, suggesting that these clinical manifestations may be due to systemic downregulation of the RAGE pathway. Conversely, we find that where the RAGE pathway is predicted to be increased, significantly higher body weight and higher cholesterol are observed (cluster 3). Furthermore, an increase in RAGE is reported to be associated with platelet activation and, in agreement, we find a significant increase in thrombocytes and PAI-1 in cluster 3 and a significant decrease in cluster 1 [51]. The enrichment of the RAGE pathway in cluster 4 only partially overlaps with cluster 3, with additional effects on inflammatory mediators such as IL-6 and soluble vascular adhesion molecule 1. Important to note is that our dataset included mostly inflammatory mediators downstream of RAGE, whereas ligands activating the pathway are underrepresented. It is likely that levels of the various ligands identified for RAGE also differ between clusters, which could be a reason for the observed discriminatory downstream effects between clusters 3 and 4 in particular. As the RAGE signalling pathway is furthermore inhibited by soluble RAGE (of which levels are in general lower in COPD patients), this is yet another unexplored mechanism through which the pathway could be differentially regulated between clusters and contributing to the pathogenesis and clinical characteristics between patients and clusters. Due to the prevalence of RAGE components and enrichment of the pathway, it would be interesting to determine these additional aspects, as well as genetic variations of AGER in our identified COPD clusters in future studies.
OSM pathways were highly significant in the same direction and in the same clusters as RAGE. This is not entirely surprising, since a number of biomarkers are indeed shared between both pathways (supplementary tables S1, S2 and S4). OSM is a pleiotropic cytokine of the IL-6 family of cytokines, which like RAGE, contains multiple positive feedback loops to enhance its pro-inflammatory actions. Moreover, it has previously been shown to be increased in COPD and is involved in airway remodelling [52, 53]. Interestingly, it was also demonstrated using RAGE-deficient mice that RAGE mediates hypomethylation in the promoter of OSM and that the resulting elevated expression of OSM is likely to contribute to the observed effects of RAGE in cigarette-induced airway inflammation and emphysema development [54].
Previously, a subanalysis of the TESRA study was the first to use proteomics to subtype stable ex-smoking COPD patients [28]. The authors included 87 biomarkers measured in 396 COPD patients and identified three different clusters. They report that 18 biomarkers were different between the three clusters, but do not report the distribution of markers among the three clusters. Similar to our results, BDNF, fibrinogen, osteoprotegerin, stem cell factor and vascular endothelial growth factor had discriminating power in differentiating the clusters. The authors specifically pointed out a smaller cluster of participants with increased inflammatory biomarkers, impaired quality of life yet less emphysema, which could be reminiscent of cluster 4 in the current study, as this cluster was also characterised by increased markers of α1-antitrypsin, C-reactive protein, haptoglobin and tumour necrosis factor receptor I. Although diffusion capacity was lower in cluster 4, which commonly relates to the degree of emphysema. In TESRA, subjects were characterised with less detail; for example, no information on comorbidities and functional performance is provided. Most COPD patients have multiple chronic conditions which are strongly interconnected. Previously, five different comorbidity clusters have been identified in COPD patients of the CIROCO cohort [29]. The present study highlights the complexity of the pathophysiology, as the clinical characteristics of patients in the four observed clusters of circulating biomarkers did not seem to correspond well with the previously described comorbidity clusters [29]. This probably reflects the heterogeneity of patients with COPD, where different clinical traits and diseases coexist to varying degrees within the same individual, and there is an inability to delineate mutually exclusive clusters of (mechanisms of) disease(s) [55].
The current results are hypothesis-generating rather than definitive and there are several limitations to the current study. Indeed, our findings need to be corroborated, using additional cohorts of individuals to determine the robustness of our clustering approach. We have thus far only examined patients with moderate-to-severe COPD, but not those in early stages of the disease. Including early-stage individuals and following them over time might gain additional insight into systemic changes in inflammation and how they may mirror the development of clinical characteristics. Although a large set of biomarkers was used, this still does not reflect the complexity of biological interactions in the human body. Moreover, there is considerable debate over how reflective systemic inflammation is of local inflammation in the airways. However, using a relatively noninvasive sampling method, such as whole-blood sampling, numerous biomarkers can be examined relatively easily without causing unnecessary stress to the patient. Recognising the fact that the discussion is valid only for the biomarkers we have explored, our findings may help to shed light on the underlying pathogenic processes involved in COPD.
Biomarker-based clustering in patients with COPD resulted in four clusters, illustrating the heterogeneity of biomarkers in patients with COPD. A dichotomy in clusters was seen based on mainly lower/higher levels of biomarkers, which in turn resulted in a unique biomarker composition for each cluster. Although the degree of airflow limitation was similar among the clusters, differences in clinical characterisation were seen. Pathway analysis revealed components of the RAGE pathway to be enriched in three of the four clusters, suggesting a systemic role for this pathway in COPD. These new insights may help to understand the functionally relevant inflammatory interactions in the pathophysiology of COPD as a heterogeneous disease.
Supplementary material
Please note: supplementary material is not edited by the Editorial Office, and is uploaded as it has been supplied by the author.
Supplement on data analysis and statistical methods 00301-2022.supplementary_methods (175.9KB, pdf)
Tables S1-S9 00301-2022.supplementary_tables (381.8KB, pdf)
Footnotes
Provenance: Submitted article, peer reviewed.
Author contributions: L.E.G.W. Vanfleteren, E.F.M. Wouters and M.A. Spruit contributed to conception and design; L.E.G.W. Vanfleteren, J. Weidner, F.M.E. Franssen, S. Gaffron, N.L. Reynaert and M.A. Spruit contributed to acquisition, analysis or interpretation of the data; L.E.G.W. Vanfleteren, J. Weidner and M.A. Spruit drafted the manuscript; all authors critically revised the manuscript, gave final approval, and agree to be accountable for all aspects of work ensuring integrity and accuracy.
Conflict of interest: L.E.G.W. Vanfleteren has received grants and personal fees from AstraZeneca, and personal fees from Novartis, GSK, Resmed, Boehringer and Verona Pharma; and is an associate editor of this journal.
Conflict of interest: J. Weidner is an employee of AstraZeneca.
Conflict of interest: F.M.E. Franssen has received grants and personal fees from AstraZeneca, and personal fees from Boehringer Ingelheim, Chiesi, GSK and Novartis.
Conflict of interest: S. Gaffron has nothing to disclose.
Conflict of interest: N.L. Reynaert has nothing to disclose.
Conflict of interest: E.F.M. Wouters has nothing to disclose.
Conflict of interest: M.A. Spruit received a grant for the present study from AstraZeneca, paid to Ciro.
Support statement: This study was supported by AstraZeneca. Funding information for this article has been deposited with the Crossref Funder Registry.
References
- 1.Agusti A, Edwards LD, Rennard SI, et al. Persistent systemic inflammation is associated with poor clinical outcomes in COPD: a novel phenotype. PLoS One 2012; 7: e37483. doi: 10.1371/journal.pone.0037483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Núñez B, Sauleda J, Garcia-Aymerich J, et al. Lack of correlation between pulmonary and systemic inflammation markers in patients with chronic obstructive pulmonary disease: a simultaneous, two-compartmental analysis. Arch Bronconeumol 2016; 52: 361–367. doi: 10.1016/j.arbres.2016.01.003 [DOI] [PubMed] [Google Scholar]
- 3.Arellano-Orden E, Calero-Acuña C, Cordero JA, et al. Specific networks of plasma acute phase reactants are associated with the severity of chronic obstructive pulmonary disease: a case-control study. Int J Med Sci 2017; 14: 67–74. doi: 10.7150/ijms.16907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Westwood JP, Mackay AJ, Donaldson G, et al. The role of complement activation in COPD exacerbation recovery. ERJ Open Res 2016; 2: 00027-2016. doi: 10.1183/23120541.00027-2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhao D, Abbasi A, Rossiter HB, et al. Serum amyloid A in stable COPD patients is associated with the frequent exacerbator phenotype. Int J Chron Obstruct Pulmon Dis 2020; 15: 2379–2388. doi: 10.2147/COPD.S266844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pauwels NS, Bracke KR, Dupont LL, et al. Role of IL-1α and the Nlrp3/caspase-1/IL-1β axis in cigarette smoke-induced pulmonary inflammation and COPD. Eur Respir J 2011; 38: 1019–1028. doi: 10.1183/09031936.00158110 [DOI] [PubMed] [Google Scholar]
- 7.Eagan TM, Ueland T, Wagner PD, et al. Systemic inflammatory markers in COPD: results from the Bergen COPD Cohort Study. Eur Respir J 2010; 35: 540–548. doi: 10.1183/09031936.00088209 [DOI] [PubMed] [Google Scholar]
- 8.Husebø GR, Bakke PS, Grønseth R, et al. Macrophage migration inhibitory factor, a role in COPD. Am J Physiol Lung Cell Mol Physiol 2016; 311: L1–L7. doi: 10.1152/ajplung.00461.2015 [DOI] [PubMed] [Google Scholar]
- 9.Machida H, Inoue S, Shibata Y, et al. Thymus and activation-regulated chemokine (TARC/CCL17) predicts decline of pulmonary function in patients with chronic obstructive pulmonary disease. Allergol Int 2021; 70: 81–88. doi: 10.1016/j.alit.2020.04.004 [DOI] [PubMed] [Google Scholar]
- 10.Bade G, Khan MA, Srivastava AK, et al. Serum cytokine profiling and enrichment analysis reveal the involvement of immunological and inflammatory pathways in stable patients with chronic obstructive pulmonary disease. Int J Chron Obstruct Pulmon Dis 2014; 9: 759–773. doi: 10.2147/COPD.S61347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sapey E, Ahmad A, Bayley D, et al. Imbalances between interleukin-1 and tumor necrosis factor agonists and antagonists in stable COPD. J Clin Immunol 2009; 29: 508–516. doi: 10.1007/s10875-009-9286-8 [DOI] [PubMed] [Google Scholar]
- 12.Blidberg K, Palmberg L, James A, et al. Adhesion molecules in subjects with COPD and healthy non-smokers: a cross sectional parallel group study. Respir Res 2013; 14: 47. doi: 10.1186/1465-9921-14-47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Aaron CP, Schwartz JE, Bielinski SJ, et al. Intercellular adhesion molecule 1 and progression of percent emphysema: the MESA Lung Study. Respir Med 2015; 109: 255–264. doi: 10.1016/j.rmed.2014.10.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ladjemi MZ, Lecocq M, Weynand B, et al. Increased IgA production by B-cells in COPD via lung epithelial interleukin-6 and TACI pathways. Eur Respir J 2015; 45: 980–993. doi: 10.1183/09031936.00063914 [DOI] [PubMed] [Google Scholar]
- 15.Stoll P, Wuertemberger U, Bratke K, et al. Stage-dependent association of BDNF and TGF-β1 with lung function in stable COPD. Respir Res 2012; 13: 116. doi: 10.1186/1465-9921-13-116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kranenburg AR, de Boer WI, Alagappan VK, et al. Enhanced bronchial expression of vascular endothelial growth factor and receptors (Flk-1 and Flt-1) in patients with chronic obstructive pulmonary disease. Thorax 2005; 60: 106–113. doi: 10.1136/thx.2004.023986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wells JM, Parker MM, Oster RA, et al. Elevated circulating MMP-9 is linked to increased COPD exacerbation risk in SPIROMICS and COPDGene. JCI Insight 2018; 3: e123614. doi: 10.1172/jci.insight.123614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Atkinson JJ, Lutey BA, Suzuki Y, et al. The role of matrix metalloproteinase-9 in cigarette smoke-induced emphysema. Am J Respir Crit Care Med 2011; 183: 876–884. doi: 10.1164/rccm.201005-0718OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Carolan BJ, Kim YI, Williams AA, et al. The association of adiponectin with computed tomography phenotypes in chronic obstructive pulmonary disease. Am J Respir Crit Care Med 2013; 188: 561–566. doi: 10.1164/rccm.201212-2299OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Oh YM, Jeong BH, Woo SY, et al. Association of plasma adipokines with chronic obstructive pulmonary disease severity and progression. Ann Am Thorac Soc 2015; 12: 1005–1012. doi: 10.1513/AnnalsATS.201501-005OC [DOI] [PubMed] [Google Scholar]
- 21.Undas A, Jankowski M, Kaczmarek P, et al. Thrombin generation in chronic obstructive pulmonary disease: dependence on plasma factor composition. Thromb Res 2011; 128: e24–e28. doi: 10.1016/j.thromres.2011.05.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Waschki B, Watz H, Holz O, et al. Plasminogen activator inhibitor-1 is elevated in patients with COPD independent of metabolic and cardiovascular function. Int J Chron Obstruct Pulmon Dis 2017; 12: 981–987. doi: 10.2147/COPD.S128689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Minas M, Mystridou P, Georgoulias P, et al. Fetuin-A is associated with disease severity and exacerbation frequency in patients with COPD. COPD 2013; 10: 28–34. doi: 10.3109/15412555.2012.727922 [DOI] [PubMed] [Google Scholar]
- 24.Gao J, Törölä T, Li CX, et al. Sputum vitamin D binding protein (VDBP) GC1S/1S genotype predicts airway obstruction: a prospective study in smokers with COPD. Int J Chron Obstruct Pulmon Dis 2020; 15: 1049–1059. doi: 10.2147/COPD.S234464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pinto-Plata V, Toso J, Lee K, et al. Profiling serum biomarkers in patients with COPD: associations with clinical parameters. Thorax 2007; 62: 595–601. doi: 10.1136/thx.2006.064428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pinto-Plata V, Casanova C, Müllerova H, et al. Inflammatory and repair serum biomarker pattern: association to clinical outcomes in COPD. Respir Res 2012; 13: 71. doi: 10.1186/1465-9921-13-71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zemans RL, Jacobson S, Keene J, et al. Multiple biomarkers predict disease severity, progression and mortality in COPD. Respir Res 2017; 18: 117. doi: 10.1186/s12931-017-0597-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zarei S, Mirtar A, Morrow JD, et al. Subtyping chronic obstructive pulmonary disease using peripheral blood proteomics. Chronic Obstr Pulm Dis 2017; 4: 97–108. doi: 10.15326/jcopdf.4.2.2016.0147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vanfleteren LE, Spruit MA, Groenen M, et al. Clusters of comorbidities based on validated objective measurements and systemic inflammation in patients with chronic obstructive pulmonary disease. Am J Respir Crit Care Med 2013; 187: 728–735. doi: 10.1164/rccm.201209-1665OC [DOI] [PubMed] [Google Scholar]
- 30.Vestbo J, Hurd SS, Agustí AG, et al. Global strategy for the diagnosis, management and prevention of chronic obstructive pulmonary disease: GOLD executive summary. Am J Respir Crit Care Med 2013; 187: 347–365. doi: 10.1164/rccm.201204-0596PP [DOI] [PubMed] [Google Scholar]
- 31.Spruit MA, Vanderhoven-Augustin I, Janssen PP, et al. Integration of pulmonary rehabilitation in COPD. Lancet 2008; 371: 12–13. doi: 10.1016/S0140-6736(08)60048-3 [DOI] [PubMed] [Google Scholar]
- 32.Cloonan SM, Mumby S, Adcock IM, et al. The ‘iron’-y of iron overload and iron deficiency in chronic obstructive pulmonary disease. Am J Respir Crit Care Med 2017; 196: 1103–1112. doi: 10.1164/rccm.201702-0311PP [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gao N, Wang Y, Zheng CM, et al. β2-Microglobulin participates in development of lung emphysema by inducing lung epithelial cell senescence. Am J Physiol Lung Cell Mol Physiol 2017; 312: L669–L677. doi: 10.1152/ajplung.00516.2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Loza MJ, Watt R, Baribaud F, et al. Systemic inflammatory profile and response to anti-tumor necrosis factor therapy in chronic obstructive pulmonary disease. Respir Res 2012; 13: 12. doi: 10.1186/1465-9921-13-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Delgado-Eckert E, James A, Meier-Girard D, et al. Lung function fluctuation patterns unveil asthma and COPD phenotypes unrelated to type 2 inflammation. J Allergy Clin Immunol 2021; 148: 407–419. doi: 10.1016/j.jaci.2020.12.652 [DOI] [PubMed] [Google Scholar]
- 36.Papaporfyriou A, Loukides S, Kostikas K, et al. Increased levels of osteopontin in sputum supernatant in patients with COPD. Chest 2014; 146: 951–958. doi: 10.1378/chest.13-2440 [DOI] [PubMed] [Google Scholar]
- 37.Apperley S, Park HY, Holmes DT, et al. Serum bilirubin and disease progression in mild COPD. Chest 2015; 148: 169–175. doi: 10.1378/chest.14-2150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xuan L, Han F, Gong L, et al. Association between chronic obstructive pulmonary disease and serum lipid levels: a meta-analysis. Lipids Health Dis 2018; 17: 263. doi: 10.1186/s12944-018-0904-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kohonen T. Essentials of the self-organizing map. Neural Netw 2013; 37: 52–65. doi: 10.1016/j.neunet.2012.09.018 [DOI] [PubMed] [Google Scholar]
- 40.Szklarczyk D, Gable AL, Lyon D, et al. STRING v11: protein–protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res 2019; 47: D607–D613. doi: 10.1093/nar/gky1131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kuleshov MV, Jones MR, Rouillard AD, et al. Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res 2016; 44: W90–W97. doi: 10.1093/nar/gkw377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Verrills NM, Irwin JA, He XY, et al. Identification of novel diagnostic biomarkers for asthma and chronic obstructive pulmonary disease. Am J Respir Crit Care Med 2011; 183: 1633–1643. doi: 10.1164/rccm.201010-1623OC [DOI] [PubMed] [Google Scholar]
- 43.Zhang YH, Hoopmann MR, Castaldi PJ, et al. Lung proteomic biomarkers associated with chronic obstructive pulmonary disease. Am J Physiol Lung Cell Mol Physiol 2021; 321: L1119–L1130. doi: 10.1152/ajplung.00198.2021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ghio AJ, Hilborn ED, Stonehuerner JG, et al. Particulate matter in cigarette smoke alters iron homeostasis to produce a biological effect. Am J Respir Crit Care Med 2008; 178: 1130–1138. doi: 10.1164/rccm.200802-334OC [DOI] [PubMed] [Google Scholar]
- 45.Remels AH, Gosker HR, Langen RC, et al. The mechanisms of cachexia underlying muscle dysfunction in COPD. J Appl Physiol 2013; 114: 1253–1262. doi: 10.1152/japplphysiol.00790.2012 [DOI] [PubMed] [Google Scholar]
- 46.Jaffar Z, Ferrini ME, Herritt LA, et al. Cutting edge: lung mucosal Th17-mediated responses induce polymeric Ig receptor expression by the airway epithelium and elevate secretory IgA levels. J Immunol 2009; 182: 4507–4511. doi: 10.4049/jimmunol.0900237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Motamedi S, Karimi I, Jafari F. The interrelationship of metabolic syndrome and neurodegenerative diseases with focus on brain-derived neurotrophic factor (BDNF): kill two birds with one stone. Metab Brain Dis 2017; 32: 651–665. doi: 10.1007/s11011-017-9997-0 [DOI] [PubMed] [Google Scholar]
- 48.Lichtenauer M, Franz M, Fritzenwanger M, et al. Elevated plasma levels of interleukin-12p40 and interleukin-16 in overweight adolescents. Biomed Res Int 2015; 2015: 940910. doi: 10.1155/2015/940910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Reynaert NL, Gopal P, Rutten EPA, et al. Advanced glycation end products and their receptor in age-related, non-communicable chronic inflammatory diseases; overview of clinical evidence and potential contributions to disease. Int J Biochem Cell Biol 2016; 81: 403–418. doi: 10.1016/j.biocel.2016.06.016 [DOI] [PubMed] [Google Scholar]
- 50.Song F, Hurtado del Pozo C, Rosario R, et al. RAGE regulates the metabolic and inflammatory response to high-fat feeding in mice. Diabetes 2014; 63: 1948–1965. doi: 10.2337/db13-1636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Fuentes E, Rojas A, Palomo I. Role of multiligand/RAGE axis in platelet activation. Thromb Res 2014; 133: 308–314. doi: 10.1016/j.thromres.2013.11.007 [DOI] [PubMed] [Google Scholar]
- 52.Botelho FM, Rodrigues R, Guerette J, et al. Extracellular matrix and fibrocyte accumulation in BALB/c mouse lung upon transient overexpression of oncostatin M. Cells 2019; 8: 126. doi: 10.3390/cells8020126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Baines KJ, Simpson JL, Gibson PG. Innate immune responses are increased in chronic obstructive pulmonary disease. PLoS One 2011; 6: e18426. doi: 10.1371/journal.pone.0018426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Li P, Wang T, Chen M, et al. RAGE-mediated functional DNA methylated modification contributes to cigarette smoke-induced airway inflammation in mice. Biosci Rep 2021; 41: BSR20210308. doi: 10.1042/BSR20210308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Castaldi PJ, Benet M, Petersen H, et al. Do COPD subtypes really exist? COPD heterogeneity and clustering in 10 independent cohorts. Thorax 2017; 72: 998–1006. doi: 10.1136/thoraxjnl-2016-209846 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Please note: supplementary material is not edited by the Editorial Office, and is uploaded as it has been supplied by the author.
Supplement on data analysis and statistical methods 00301-2022.supplementary_methods (175.9KB, pdf)
Tables S1-S9 00301-2022.supplementary_tables (381.8KB, pdf)