Abstract
Filamentous fungi drive carbon and nutrient cycling across our global ecosystems, through its interactions with growing and decaying flora and their constituent microbiomes. The remarkable metabolic diversity, secretion ability, and fiber-like mycelial structure that have evolved in filamentous fungi have been increasingly exploited in commercial operations. The industrial potential of mycelial fermentation ranges from the discovery and bioproduction of enzymes and bioactive compounds, the decarbonization of food and material production, to environmental remediation and enhanced agricultural production. Despite its fundamental impact in ecology and biotechnology, molds and mushrooms have not, to-date, significantly intersected with synthetic biology in ways comparable to other industrial cell factories (e.g. Escherichia coli,Saccharomyces cerevisiae, and Komagataella phaffii). In this review, we summarize a suite of synthetic biology and computational tools for the mining, engineering and optimization of filamentous fungi as a bioproduction chassis. A combination of methods across genetic engineering, mutagenesis, experimental evolution, and computational modeling can be used to address strain development bottlenecks in established and emerging industries. These include slow mycelium growth rate, low production yields, non-optimal growth in alternative feedstocks, and difficulties in downstream purification. In the scope of biomanufacturing, we then detail previous efforts in improving key bottlenecks by targeting protein processing and secretion pathways, hyphae morphogenesis, and transcriptional control. Bringing synthetic biology practices into the hidden world of molds and mushrooms will serve to expand the limited panel of host organisms that allow for commercially-feasible and environmentally-sustainable bioproduction of enzymes, chemicals, therapeutics, foods, and materials of the future.
Keywords: Filamentous fungi, Synthetic biology, Strain optimization, Biomanufacturing, Sustainability, Materials
Graphical abstract
1. Introduction
The largest and oldest known living organism on earth is a filamentous fungus [1]. An individual mushroom-forming Armillaria ostoyae that resides in Oregon spans an estimated 3.5 square miles and is at least 2400 years old [1,2]. An incredible variety of these mushroom-forming fungi colonize terrestrial habitats through the formation of a mycelial network. Unlike yeast, which grow as unicellular organisms, filamentous fungi grow vegetatively as multicellular hyphae filaments, which branch and intermesh into underground mycelial networks (Fig. 1). This not only facilitates the resource foraging and biomass growth of the organism itself, but also mediates interactions between different organisms and ecosystems across space and time [3]. Combined with their mycelial structure, filamentous fungi, in particular white-rot fungi in the class of Agaricomycetes, secrete a diverse portfolio of carbohydrate-active enzymes (CAZymes) at the tips of their growing hyphae to degrade lignocellulose biomass into oligosaccharides, cellobiose and glucose [[4], [5], [6], [7]]. The multifaceted versatility of filamentous fungi also relates to their unique reproductive strategies and spore dispersal traits. Triggered by environmental cues such as the availability of nutrients, Basidiomycetes form large fruiting bodies widely known as mushrooms to generate sexual spores [8,9]. While also able to form fruiting bodies, Ascomycetes predominantly reproduce asexually by dispersible conidia or by fragments [10].
Fig. 1.
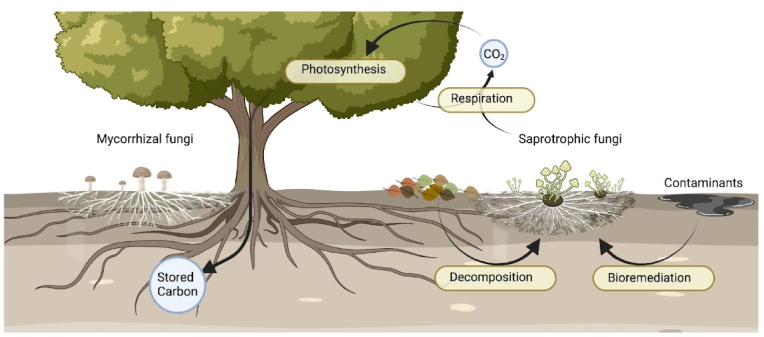
Biochemical, ecological, and structural properties of filamentous fungi. Filamentous fungi drive carbon cycling in terrestrial systems - saprotrophic fungi secrete primary metabolites to decompose organic litters, while mycorrhizal fungi associate with plant roots to store fixed carbon into recalcitrant biomass. Their root-like structure, called mycelium, forms a network of branching, thread-like hyphae that are important crosswalks of interkingdom interactions. These biochemical and ecological roles can be exploited for bioproduction of primary and secondary metabolites, bioremediation of organic contaminants, and carbon sink.
Human interactions with filamentous fungi dates back to the history of food foraging, with the first anthropological record of mushrooms in the human diet predicted to be as early as the Upper Paleolithic Period [11]. The domestication of filamentous fungi as a food source began as mushrooms were cultivated in Asia, first by China in the 7th century, followed by Europe in the 17th century [12]. Meanwhile, mixed cultures of select species of molds, including Aspergillus and Penicillium, by virtue of their metabolic richness, have been exploited in the traditional fermentation of widely appreciated food and beverages such as salami, blue cheese, soy sauce and sake [13].
Since World War II, the isolation of single filamentous fungal strains and their secondary metabolites, such as the beta-lactam antibiotics (e.g. penicillin and cephalosporin) launched a new era of biomanufacturing. In addition, molds’ exceptionally high capacity of expressing and secreting proteins made them ideal workhorses for producing native or recombinant enzymes, accounting for over 50% of the industrial enzyme market [14,15]. Today, interdisciplinary scientists are exploring novel ways to utilize the metabolic, secretory, and structural properties of filamentous fungi that can benefit human societies, animal welfare and ecosystems at large. For example, fungal mycelium can be made into bio-leather materials and fibrous, high-protein meat alternatives [[16], [17], [18]]. Moreover, mycorrhizal fungi that form symbiotic relationships with plants are applied to enhance crop production and climate resilience [19,20]. Filamentous fungi are also being explored for its ability to valorize agricultural and forestry waste [21,22], and biologically remediate ecological hazards [23,24].
Despite their ecological and biotechnological importance, filamentous fungi have been historically overlooked by synthetic biologists [25]. Although there is precedent for engineering mycelial networks (detailed in Section 4), the development of molecular or synthetic biology tools, both to study the biology of filamentous fungi and to genetically improve the strains, has lagged behind compared to yeast and prokaryotes [17]. The increased complexity of their genomes (and their associated metabolic and cellular processes) paired with the lack of molecular tools have greatly impeded the pace of innovation in filamentous fungi engineering. As they are becoming attractive cell factories or biomass products, there is an increasing need to build a fungal synthetic biology toolbox to improve endogenous or heterologous production of proteins and bioactive compounds, and to develop final products with desirable functional and structural characteristics [26,27]. In this review, we will summarize the current applications and technical limitations in the fungal biotechnology space, as well as the genetic engineering, computational, and other synthetic biology tools that are currently applied or can be applied to address these technological challenges. The convergence of synthetic biology and mycobiology may ultimately pave the way for the next revolution in the bioeconomy.
2. Biomanufacturing and sustainability applications
2.1. Organic acids and industrial enzyme bioproduction
Since the early days of biomanufacturing, extracellular primary metabolites derived from filamentous fungi sources have been extracted to support a plethora of industry sectors, from food and pulp and paper to textile, agriculture, animal health, bioenergy and bioremediation [28]. Lacking digestive systems, molds and mushrooms secrete various catabolic enzymes into the surroundings of their mycelia to hydrolyze carbohydrates, proteins, and lipids from other organisms, converting them into smaller molecules before transporting them into the cells [29,30]. For example, white rot fungi secrete several lytic enzymes to attack plant-derived cellulose- and lignin-rich organic matter, cutting high-molecular weight lignocellulose into shorter fragments [31,32]. These hydrolytic and oxidative enzymes play important roles in carbon and nitrogen assimilation and are considered fundamental agents in the generation of primary metabolites (e.g. amino acids, organic acids, nucleosides) vital for the growth of the organisms. Due to high production yields during industrial fermentation, simple cultivation methods and simultaneous biomass production, filamentous-fungi derived enzymes contribute to more than half of the industrial enzyme market, in particular up to 82% of the enzymes in the food and beverage sector [33]. Established industrial workhorse species include Aspergillus niger (citric acid, gluconic acid, CAZymes), Penicillium chrysogenum (penicillin), A. oryzae (amylase, pectinase), Trichoderma reesei (cellulase) (Fig. 2), and more recently Myceliophthora thermophila [33,34].
Fig. 2.
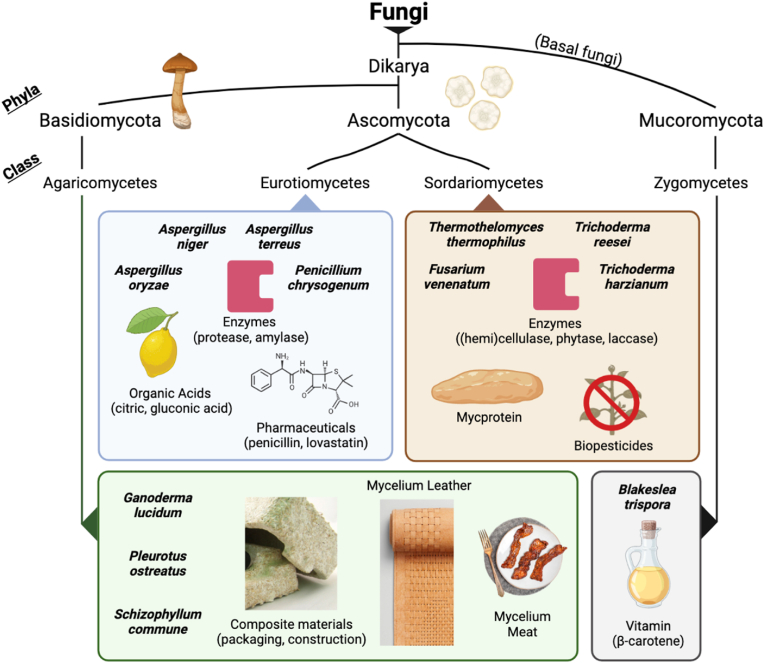
Overview of notable industrial filamentous fungi host species and their associated products, categorized into groups according to taxonomic class (order and family not provided). Production of native enzymes, organic acids, and secondary metabolites (pharmaceuticals) by Ascomycota molds are the predominant applications for filamentous fungi in biomanufacturing. Emerging applications include use of Basidiomycetes organisms for food and materials biomanufacturing, as well as use of established cell factories (e.g. Aspergillus, Penicillium) for heterologous protein production. Composite material shown is Ecovative's Mushroom® packaging, mycelium leather is Bolt Threads Mylo™, and mycelium meat is MyForest Foods' MyBacon. The breadth of organisms used across industries is limited and the rich diversity of the fungal kingdom is highly under-explored.
2.2. Secondary metabolite discovery and therapeutic potential
In nature, the secretion of secondary metabolites, also known as natural products, is critical to fungal development and defense, usually triggered by intra-kingdom and inter-kingdom communications and interactions with other fungi, plants, microorganisms or insects. For example, when encountering the bacterium Ralstonia solanacearum secreting lipopeptide ralsolamycin, Fusarium species form chlamydospore and turn on the expression of bikaverin gene cluster responsible for reducing the bacterium entry [35]. It is estimated that over 40% fungal species exhibit antimicrobial or antifungal activities under natural conditions [36]. This rich source of biologically active secondary metabolites has been harnessed in the past hundred years – the most prominent example is the discovery of penicillin, the first antibiotic compound isolated from the mold Penicillium chrysogenum which revolutionized drug discovery [37]. Many therapeutic agents have since then been derived from fungal secondary metabolites, including fusidic acid and griseofulvin as antimicrobials (isolated from Fusidium coccineum [38] and Penicillium griseofulvum [39], respectively), anidulafungin and caspofungin as antifungals (from Aspergillus rugulovalvus and Saccharopolyspora erythraea, respectively [40]), paclitaxel as an anticancer drug (from the endophyte Taxomyces andreanae [41]), lovastatin and mevastatin as cholesterol-lowering agents (from Aspergillus terreus and Penicillium citrinum, respectively [42]), cyclosporine and mycophenolic acid as immunosuppressants (from Cylindrocarpon lucidum [43] and Penicillium brevicompactum [44], respectively) and ergometrine as vasoconstrictors (from Claviceps purpurea [45]).
The explosion of genomic information and popularization of untargeted metabolomics techniques are unveiling the enormous biosynthetic potential of fungi. Major classes of secondary metabolites encoded in fungal genomes are categorized into polyketides (PKs), terpenes, non-ribosomal peptides (NRPs), indole alkaloids, etc. based on the structural backbones that use acetyl-CoAs or amino acids as building blocks [36,46,47]. These classes of secondary metabolites are mostly encoded by biosynthetic gene clusters (BGCs) whose protein products catalyze and regulate the synthesis and modification of the backbone scaffold of the metabolite. Nowadays, bioinformatics algorithms predict on average 30–70 BGCs in each fungal species rich in secondary metabolites [48]. An estimate of 25,000 BGCs exists just considering the 693 species in the Aspergillus and Penicillium genera alone. The number of BGCs harbored in the fungal kingdom easily exceeds several million under significant underestimation [36]. To explore the pharmaceutical potentials of these uncovered BGCs, synthetic biology methods can be utilized to screen for biochemical activities of synthesized natural products to drug targets [49] and to increase the metabolite production through native or heterologous expression [49].
2.3. Materials and food
Filamentous fungi extend their mycelium to reach organic food sources in the surroundings and incorporate these particles into their hyphal network, forming a composite material [50,51]. In the case of mushroom-forming fungi such as Ganoderma lucidum and Pleurotus ostreatus, their glucan- and chitin-rich hyphae bind plant-derived substrates rich in cellulose and lignin together, conferring high rigidity to the overall interconnected structure [52,53]. After being treated by heat inactivation, these mycelium-based composite materials are mechanically strong enough to serve as packaging materials and structural components in furniture and installations at the architectural scale [[54], [55], [56]]. Several filamentous fungi can use their mycelium as nucleation sites for calcium carbonate biomineralization, further expanding the usability of mycelium composites as structural materials [57,58]. Fungal mycelia also synthesize a wide spectrum of pigments [59,60], form stable communities with engineerable bacteria [61], and are compatible with extrusion-based additive manufacturing [62], making them a popular chassis for engineered living materials [[63], [64], [65], [66], [67]].
Pure mycelium consisting of only fungal biomass can be acquired through the complete digestion of solid feedstock, isolating mycelium pellets from liquid culture, or inducing aerial mycelium formation [68]. After compression and surface treatment, pure mycelium shows texture and flexibility resembling animal hides. Recently, several companies have explored using these materials as sustainable leather alternatives for use in footwear, bags, and apparel [68]. Since the properties of pure mycelium are largely determined by the growth of filamentous fungi, genetically engineering these fungal strains could lead to materials optimization where design parameters are tunable using synthetic biology. For example, allowing genetically modified mycelium to express unique pigments and animal proteins functionalizes the materials as they grow and reduce the environmental impact of sourcing these elements from other organisms.
Beyond leather products, pure mycelium is a prime candidate for animal protein alternatives because of its fibrous texture and high protein content with amino acid profiles similar to meat, with additional health benefits. Mushrooms and food fermented by filamentous fungi have played important roles over millennia in food cultures and technologies worldwide [69]. For example, Tempeh, a traditional Indonesian soy food, is prepared from soybean fermentation using the mold strain Rhizopus oligosporus, whose mycelium binds the beans together and creates a rich smoky flavor [70]. More recently, Fusarium species have become an emergent player in manufacturing mycoproteins in industrial bioreactors, along with others such as A. oryzae, L. edodes, and P. ostreatus [51,71,72]. These pure mycelium materials grown in liquid or solid-state bioreactors can be processed into various forms, including protein powder or mimicking the texture of whole-cut meat when their hyphal filaments are well aligned. The mycelial foam can also serve as a scaffold for growing mammalian cells and cultured meat [73,74]. Growing these versatile fungal foods are highly efficient in land and water use, emerging as an indispensable solution for sustainable protein production.
Besides serving as the structural components of protein-rich food, filamentous fungi are also attractive cell factories for efficient and large-scale recombinant production of animal proteins, due to their cheap feedstock requirements, powerful secretory pathways, and post-translational protein modification ability. A number of animal protein ingredients, mainly milk, dairy and egg proteins, have high market demands because of their flexible functional properties such as gelling, foaming and emulsification [75,76]. Bioproduction by filamentous fungi could enable correct glycosylation of target animal proteins, which plays a role in protein folding and maturation that are essential to replicating the functional properties of the secreted ingredients [29].
For either biomass or precision fermentation, only a limited number of fungal strains have been scaled up for commercial-level manufacturing. Bioprospecting new strains with superior metabolic and physiological properties to identify chassis with better protein quality, less dependency on sugar-based feedstocks, and stable batch-to-batch product yields in large fermenters are key innovation priorities [77]. Alternatively, current fungal strains can be improved with smart breeding tools to select for higher protein and fiber contents, or genetically engineered to increase titers and yields of correctly folded and decorated heterologous proteins [75]. To improve the scalability of the fermentation process and drive down the cost, side streams from food and beverage industry and crop harvesting, such as brewer spent grains and corn residues, can be upcycled by certain fungal species identified through experimental screening or computational prediction of potential feedstock range [78,79].
2.4. Ecological bioremediation
Filamentous fungi, the main decomposers across a wide spectrum of ecosystems, have been gaining scientific interest for its innate abilities for bioremediation applications [31,32]. Compared with bacteria, which are widely deployed for bioremediation and typically require a continuous water phase to grow, filamentous fungi can extend their mycelia across the air-liquid interface into air-filled pores in the soil [80]. Fungal mycelia can thus reach areas inaccessible to bacterial biofilms and form bridges transporting nutrients and chemicals across discontinuous microhabitats [81]. In these environments, fungal cells absorb molecules from enzymatic degradation together with other chemicals, such as heavy metals and metalloids, and convert them into less toxic forms intracellularly.
Many secreted catabolic enzymes have low substrate specificity, allowing filamentous fungi to degrade structurally diverse chemicals, including various organic pollutants and plastics [31,82,83]. Extracellular oxidoreductases, including laccases and various peroxidases, are among the most seen in fungal-secreted enzymes that can degrade a wide spectrum of pollutants like benzene, toluene, ethylbenzene, and xylenes (BTEX) compounds, phenols, endocrine disrupting chemicals (EDCs), synthetic dyes, polycyclic aromatic hydrocarbons (PAHs), explosives, organochlorines, and methyl-tert-butylether (MTBE) [32]. Together with versatile cell-bound enzymes like cytochrome P450 [23], these enzymes make filamentous fungi ideal candidates for removing pollutants in waste streams, topsoil, and water [24,84,85].
Toxic metals, including heavy metals and metalloids, are another important category of pollutants contaminating our environment. Since metals are not degradable, filamentous fungi actively remediate metal pollution by converting them into more harmless species [86]. Mycelia commonly use organic acids like citrate and oxalate to interact with minerals and increase their solubility [87]. The dissolved metals can enter the cells with the help of transporter proteins and translocate away from the site of contamination through the mycelial network [88,89], sometimes to the plants with which they have a symbiotic relationship. Intracellular metals can also interact with proteins in the cytoplasm, forming complexes for storage in different parts of the cell [90]. Some fungal species excrete metal chelators like siderophores and metal-sorbing glycoproteins that facilitate secondary mineral formation near or on the fungal cell wall, immobilizing metals in the mycosphere [[91], [92], [93]].
2.5. Mycorrhiza - symbiosis with plants
In terrestrial ecosystems, fungi and plants often form mutualistic relationships called mycorrhizas [94]. Such symbiosis allows fungal cells to acquire carbon fixed by photosynthesis from plants while providing nitrogen, phosphorus, and metal ions to plants in return [95,96]. Given the large fungal biomass in soil, it is not surprising that mycorrhizas in addition to saprotrophic fungi, play an important role in the biogeochemical cycle of carbon [97]. Specifically, filamentous fungi can form different mycorrhizal types, such as arbuscular mycorrhiza (AM), ectomycorrhiza (EcM), and ericoid mycorrhiza (ErM), depending on the ways mycelia interact with plant roots [98]. Evidence shows that EcM symbionts are major drivers of topsoil carbon accumulation because they produce more recalcitrant biomass and immobilize most of the nitrogen in it, preventing saprotrophic decomposition [99]. The global distribution of EcM plant biomass is positively related to the topsoil and subsoil carbon stocks, showing the potential risk of reduced EcM caused by human activities. However, our understanding of the underground fungal network is still very limited. To address this gap in knowledge, the Society for the Protection of Underground Networks (SPUN) has launched a series of efforts to map and study mycorrhizal fungi across the world [25]. By understanding how specific filamentous fungi help plants grow, we might be able to identify the key fungal species that could promote carbon dioxide removal from the atmosphere and alleviate global warming.
3. Synthetic biology toolkit
3.1. Synthetic biology
Over the past two decades, synthetic biology has grown into a mature engineering field that enables the genetic enhancement of desirable traits in a wide spectrum of organisms, including filamentous fungi, above their native capabilities [[100], [101], [102], [103], [104]]. Synthetic biology brings new genetic and computational tools, such as affordable genome sequencing, gene synthesis, genome editing, and directed evolution technologies, to fungal metabolic engineering, optimizing the production of native and exogenous biomolecules such as proteins and metabolites [[105], [106], [107]] (Fig. 2). However, compared with bacteria and yeasts, the synthetic biology infrastructure is rather underdeveloped in filamentous fungi, especially in the mushroom-forming species. Such gaps in technological development in filamentous fungi result from biological factors, including slow growth rate, low throughput of genetic transformation, and secretion of unwanted enzymes [108]. Despite several filamentous fungal species being established workhorses in biochemical and protein production, the lack of highly engineerable chassis strains limits further optimizing them for manufacturing higher-value biomolecules.
3.1.1. Standardized genetic parts and plasmid assembly
Elementary transformations in synthetic biology typically amount to modifications of a gene or its expression logic, relevant for achieving a desired engineering goal. A transcriptional unit is a DNA fragment responsible for producing a protein (or a functional RNA) that consists of a promoter, a coding sequence (CDS), and a terminator. The promoter typically controls the recruitment of RNA polymerases and thus regulates the overall gene expression [109]. Terminators are usually less important but could contribute to unwanted homologous recombination if the same element is used multiple times inside a fungal cell [110]. There are two major categories of promoters: constitutive and inducible (Fig. 3a) [111]. Constitutive promoters are constantly active and drive the expression of genes at various strengths that can be quantified from transcription profile analyses. Most strong constitutive promoters are derived from native promoter sequences in front of highly expressed genes throughout different mycelial growth phases [26]. For example, the Pgpd promoter directing the stable expression of the glyceraldehyde-3-phosphate dehydrogenase (GPD) gene is a commonly used strong constitutive promoter in various filamentous fungi for constant expression of the downstream coding sequences [112]. It is worth noting that using the strongest constitutive promoters does not always guarantee optimal protein production because it might slow down cell growth due to competition for resources. Alternatively, in metabolic engineering, the total yield can be maximized by separating the mycelial growth and biomolecule production phases. Inducible promoters are useful in such cases for turning on gene expression after an ideal cell density is reached [113]. Synthetic inducible promoters based on the Tet system are portable across multiple filamentous fungi and allow transcription activation based on chemical induction [114]. Other inducible promoters that take various optical and chemical inputs have shown great potential in yeast biomanufacturing and could contribute to filamentous fungal strain development after minor modifications [115,116].
Fig. 3.
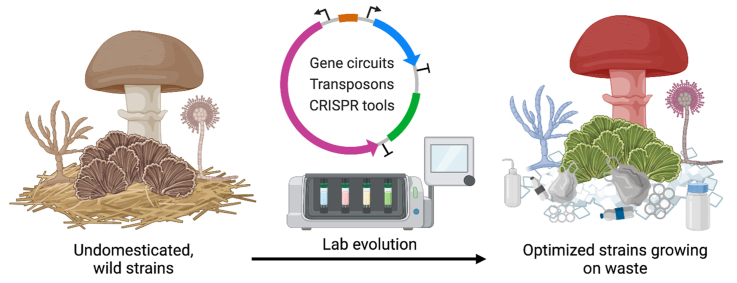
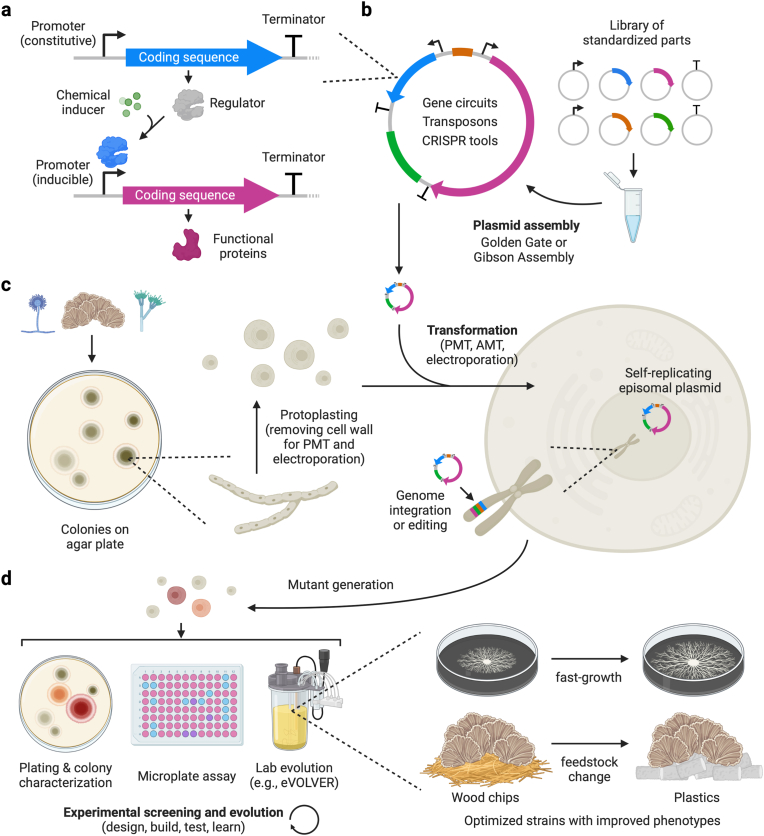
The synthetic biology workflow for strain engineering and optimization. (a) A transcription unit has three components: a promoter, the coding sequence, and a terminator. An inducible promoter can be activated or inactivated by a regulator protein that undergoes conformational changes upon binding with small molecules. (b) Part plasmids hosting promoters, coding sequences, terminators, selection markers, and origin of replication are assembled into a single plasmid using modular cloning strategies. (c) Transformation methods such as PEG-mediated transformation (PMT), Agrobacterium-mediated transformation (AMT), and electroporation transfer the assembled plasmid or linear payload into the fungal cells. The introduced DNA can either self-replicate independently or homologously recombine with the host chromosome. Genetic payloads on an episomal plasmid or integrated into the host genome drive the expression of exogenous proteins or modify the genome, generating mutants with various phenotypes. (d) Researchers often use colony morphology characterization, microplate assay, and continuous evolution techniques to benchmark the mutants. Depending on the selection or evolution methods, the optimized strains could have desirable traits such as fast growth, feedstock change, or higher protein secretion yield.
Multiple transcriptional units can be wired together using the gene products of upstream units to control the expression of downstream units, usually through activating or inactivating their promoters, allowing the expansion of biological functionalities (Fig. 3a) [117]. To build such genetic circuits with multiple transcriptional units, it is necessary to ligate several standardized genetic parts into a single genetic payload, often in the form of a circular plasmid. Modular Cloning (MoClo) based on Golden Gate assembly has become a mainstream language for designing genetic part libraries and assembling them into multiple transcription units on a plasmid [118,119] (Fig. 3b). Recently, a MoClo kit for filamentous fungi containing a collection of promoters, fluorescent reporters, terminators, and selection markers has been constructed [26]. The assembled genetic payload can be integrated into the host genome using a sacrificial plasmid or expressed as an episomal plasmid harboring the AMA1 or UARS element that replicates independently of the chromosomes [120,121]. While genome integration gives more stable gene expression profiles, replicative plasmids are usually better for prototyping because they lead to higher transformation efficiency. However, the number of stable episomal plasmid systems important for rapid prototyping in filamentous fungi is limited, and their portability across species is mostly untested. This is a major bottleneck in mushroom-forming species because no optimized, well-characterized episomal plasmids (other than the Ustilago maydis UARS) are reported. Further exploration and characterization of native plasmids in filamentous fungi are urgently needed to expand the replicative plasmid toolkit.
3.1.2. Transformation methods
For either expressing a foreign protein or modifying the host genome, physically transporting the plasmid or linearized DNA across the cell wall and into the host cytoplasm is an essential step (Fig. 3c). Electroporation, Agrobacterium-mediated transformation (AMT), and protoplast-mediated transformation (PMT) are among the most well-established methods in filamentous fungi [122]. Widely used for introducing DNA into a variety of prokaryotes and eukaryotes, electroporation applies a transient electric field to increase cell membrane permeability that facilitates genetic material transfer [123]. Although several factors, such as electric field profile and buffer solution composition, could drastically change the transformation efficiency across species and require individual fine-tuning [124], electroporation is the simplest and most reliable technique for transforming new filamentous fungal species. AMT is another broad host-range transformation method but usually has higher transformation efficiency. It relies on the native gene transfer mechanism of Agrobacterium to transport and randomly integrate the T-DNA cassette, which harbors the desirable genetic payload, into the host genome as a single copy [125]. However, AMT is incompatible with episomal plasmid transformation and requires tedious preparation steps involving setting up the co-culture system [122]. PMT is the most used method to transfer circular and linear DNA into filamentous fungal cells that are turned into protoplasts through enzymatic cell wall degradation. Crowding agents like polyethylene glycol (PEG) and sorbitol protect the protoplasts and promote their uptake of exogenous DNA [126]. Since the cell wall structure varies between fungal species, each strain requires a specifically optimized enzymatic digest protocol to ensure ideal protoplasting. Nevertheless, PMT is compatible with different cell types, making it a popular, versatile technique in the field.
3.1.3. Genome editing
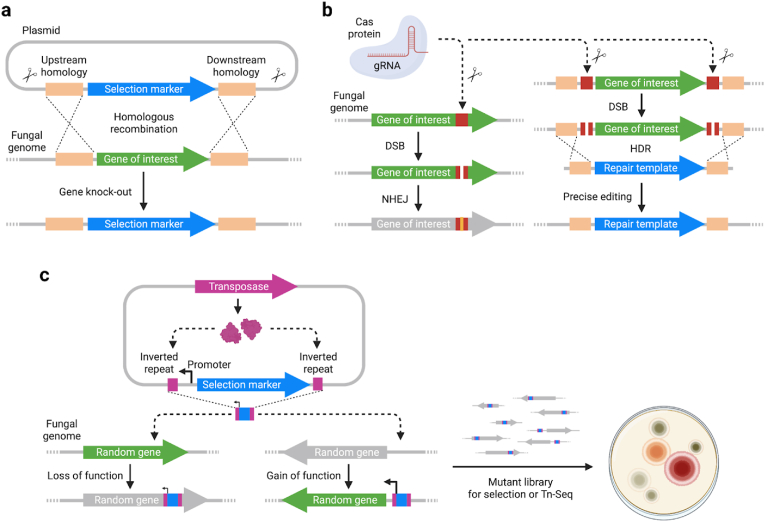
Altering the host genome by inserting, deleting, or substituting genetic elements is crucial for generating stable mutants with heritable modified traits. Traditionally, homologous recombination has been used for generating gene knockouts and integrated genome modifications (Fig. 4a). After a decade of rapid growth and intense development, the CRISPR/Cas system is the dominant genome editing tool in almost all living systems, including molds and mushroom-forming fungi [[127], [128], [129], [130]]. Cas protein, assisted by guide RNA aiming for a specific DNA sequence on the fungal genome, modifies this locus through DNA cleavage, which will later be fixed by DNA repair mechanisms such as the non-homologous end-joining (NHEJ) and homologous directed repair (HDR) (Fig. 4b) [131,132]. NHEJ usually involves the random insertion of DNA base pairs leading to a loss of function mutation, while HDR uses an exogenous DNA template to precisely edit the target genetic element. The functionality of the CRISPR/Cas system can be further expanded by fusing a deactivated Cas protein with proteins such as transcription activator/inactivator for gene activation/inactivation and cytidine deaminase for base editing [133]. In addition to CRISPR/Cas, recombineering based on phage-encoded recombinases has enabled multiplex automated genome engineering in yeast and could potentially be ported into filamentous fungi soon [134,135].
Fig. 4.
Tools for generating mutants and modifying the fungal genome. (a) Homology arms on the genetic payload and the host genome promote homologous recombination. Double crossover of DNA fragments result in swapping of the gene of interest with a selection marker, usually conferring resistance to antibiotics. (b) Cas protein targets a specific DNA sequence on the host genome with the help from guide RNA (gRNA) and generates double-strand break (DSB). DSB repaired by non-homologous end-joining (NHEJ) usually leads to loss of function of the gene. Alternatively, homology directed repair (HDR) uses a repair template, usually a provided linearized DNA, to precisely edit the host genome. (c) Transposon harboring an outward-facing promoter either inactivates a gene when inserted in its coding sequence or activates a previously inactive gene in the neighborhood of insertion, generating a diverse mutant library for downstream selection or Tn-Seq.
3.1.4. Transposon sequencing and mutagenesis
Since the implementation of gene knockouts is extremely important for engineering purposes, but laborious to implement with editing tools at the whole genome-scale, alternative approaches have been devised for high-throughput generation of mutants. Transposon sequencing (Tn-Seq), or transposon-insertion sequencing (TIS), uses transposon insertions to generate many random mutants and characterize their fitness profile under different environmental conditions using next-generation sequencing [136]. In Tn-Seq, transposons cut themselves out from the donor DNA, usually on a non-replicative plasmid, and paste themselves into random genomic locations with low sequence preferences, leading to loss-of-function mutations (Fig. 4c). Transposon mutagenesis can generate a large library with millions of mutants in a single experiment, orders of magnitudes faster than traditional site-specific gene knockout methods. These mutants have a wide spectrum of fitness changes and can be selected under growth conditions like different temperatures and feedstocks. Random barcodes are usually included inside the transposon to facilitate the downstream sequencing workflow [137]. Tn-Seq and transposon mutagenesis are the most popular technologies to determine gene essentiality and have been implemented in yeasts [138] and a few filamentous fungi [[139], [140], [141], [142]]. However, Tn-Seq has not yet been realized in any mushroom-forming fungi and could potentially expand our understanding of these more complex fungal genomes. Moreover, transposon-based mutagenesis is suitable for generating mutants for breeding and directed evolution experiments. Incorporating additional genetic elements enhances the mutagenesis capabilities of transposons beyond inactivating genes [[143], [144], [145], [146]]. For example, adding outward-facing promoters can allow transposons to activate genes in the immediate neighborhood of insertion, leading to greater mutant fitness diversity [144,147].
3.1.5. Experimental screening and evolution
Traditionally, scientists characterize filamentous fungal mutants generated by rational genetic engineering or random mutagenesis using laborious and low-throughput methods like plating on solid media. For example, measuring the colony size from germinating spores after chemical mutagenesis is a common way to screen for fast growers, with no selection or in the presence of antifungals and other chemicals [[148], [149], [150]]. These assays’ high demand for resources often originates from the slow growth of mycelium and its incompatibility in forming a homogeneous liquid culture. New methods involving microplates [151] and droplet microfluidics [152] for mutant separation could slightly accelerate assay turnover but are still far behind the high-throughput methods used in unicellular microorganisms. Several in vivo hypermutation and continuous evolution technologies greatly facilitate the automation of directed evolution experiments in bacteria and yeasts [153]. For instance, the eVOLVER platform is an automated culturing system that allows users to control hundreds of cultures' growth and selection parameters independently [154]. We envision that these examples could soon inspire the development of evolution tools customized for pellet and solid-state mycelium cultures (Fig. 3d). However, besides survival and fast growth, traits shared among the entire population, such as protein secretion, still require the physical separation of mutants and remain challenging to automate.
3.2. Discovery & chassis identification
Currently, more than 1000 fungal genomes, including 634 genomes from Basidiomycota and 1488 from Ascomycota, have been sequenced and shared in the Joint Genome Institute (JGI) Mycocosm database [155]. The increasing availability of fungal genomics and other –omics such as transcriptomics, proteomics, and metabolomics datasets offer a unique opportunity for the biomining of industrially important enzymes, metabolic pathways and bioactive compounds. These -omics information also provide an important basis for the construction of genome-scale metabolic models, to guide the design of fungal systems as cell factories to allow for endogenous or heterologous production of target biomolecules.
3.2.1. In silico bioprospecting
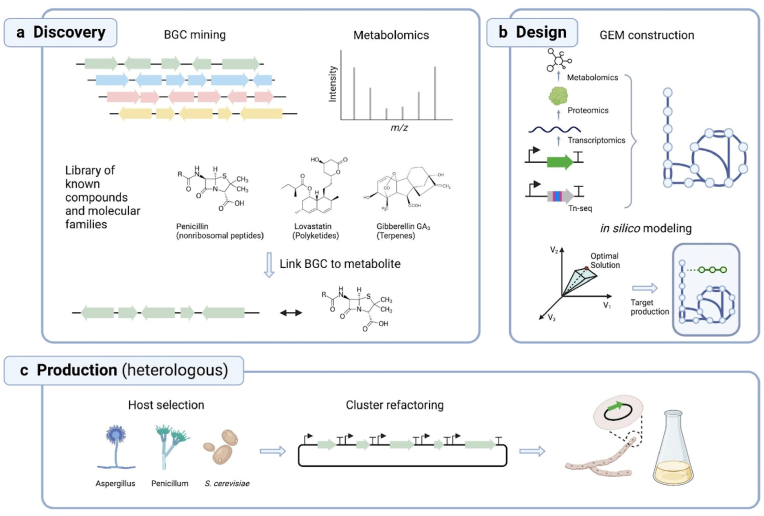
Fungi are underexplored reservoirs of secondary metabolites that are categorized into a number of structural classes such as polyketides (PKs), terpenes, non-ribosome peptides (NRPs), and indole alkaloids [46]. To computationally mine biosynthetic gene clusters (BGCs) from the fungal genomic data, a number of bioinformatics tools, such as antiSMASH [156], SMURF [157], PRISM [158], SMIPS-CASSIS [159], and TOUCAN [160], have been designed to search for conserved motifs of scaffold genes and the clustering patterns. Several strategies exist to link secondary metabolites to BGCs, and vice versa, although the association remains unknown for the majority of secondary metabolites [161,162]. A targeted approach can be taken, if a closely related compound is produced by another organism, where a homology search is performed in the genome of the target organism to identify the candidate BGC [[162], [163], [164]]. The second strategy is based on in silico retro-biosynthesis, to deduce enzymes involved in producing the backbone and decorated chemical groups in the compound structure and subsequently locate putative clusters in the genome responsible for these enzymatic activities [165,166]. With the wealth of genomics and metabolomics data becoming more readily available, newly sequenced BGCs can potentially be mapped to gene cluster families (GCFs) that catalog BGCs based on similarities, some of which are paired to families of molecular scaffolds (Fig. 5a) [167]. Conversely, to infer the direction from putative BGCs to secondary metabolites, target BGCs are either transcriptionally activated if silent, or mutated or deleted if active, to identify the associated secondary metabolites detected or missing [159].
Fig. 5.
Computational and experimental tools for the discovery, design and heterologous production of secondary metabolites. (a) BGC mining in sequenced fungal genomes, combined with untargeted metabolomics approaches, are unlocking the chemical diversity harbored in major fungal lineages. With known libraries of gene cluster families, BGCs and specific metabolites can be linked and verified through downstream genetic approaches. (b) Genome-scale modeling guides rational design of organisms to achieve desirable metabolite flux into the target product. Omics data (e.g. Tn-seq, transcriptomics, proteomics and metabolomics) can be integrated into model construction to improve the mapping accuracy of the core metabolism and constrain the flux at each reaction. (c) Scalable platforms for selecting the optimal host for heterologous production.
Another group of industrially import products secreted by filamentous fungi are carbohydrate-active enzymes (CAZymes) that degrade plant cell wall components such as cellulose, hemicellulose, lignin, pectin, chitin, starch and inulin. These CAZymes are generally cataloged into six major groups, namely glycoside hydrolases, glycosyl transferases, polysaccharide lyases, carbohydrate esterases, auxiliary activity enzymes and carbohydrate-binding modules. Online databases such as CAZy [168] and CAZymes Based Ranking of Fungi (CBRF) [169] exist to organize different classes of enzymes in fungal genomes based on homology search. In addition to comparative genomics studies, transcriptomic and proteomic studies are nowadays more frequently employed to detect dynamic genome-wide transcriptions and synergistic activities among different families of CAZymes in response to culturing on specific lignocellulosic substrates [[170], [171], [172]]. Combined with directed evolution approaches based on random mutagenesis that screens for enhanced enzymatic activities, these discovery and optimization efforts will expand the enzyme repertoire accessible for industrial usage to formulate cocktails of enzymes to ensure maximum synergistic action.
3.2.2. Genome-scale modeling
The rational design of modified organisms for a specific goal can greatly benefit from mathematical and computational tools able to encode and process the large and complex network of biochemical reactions comprising an organism's metabolism. Ideally, such quantitative approaches would quickly translate genomic information into predicted metabolic phenotypes, enabling in silico identification of favorable genetic or environmental modifications. A category of mathematical approaches collectively known as stoichiometric modeling, or constraint-based modeling has emerged as a prominent avenue for this kind of analysis in metabolic engineering [[173], [174], [175]]. These approaches typically necessitate two components: the first is a formal encoding of knowledge about the metabolism of an organism, also known as Genome-scale metabolic model (or GEM); the second is a computational engine that can transform a GEM and environment-specific boundary conditions into prediction of metabolic fluxes for all reaction in the network. GEMs are systems-level representations of the detailed stoichiometry of all known biochemical reactions in a given organism (Fig. 5b). Once mainly constructed through manual curation [176], GEMS are now increasingly produced through automated pipelines, which still require thorough refinement and testing to reach sufficient accuracy [177]. A myriad of computational algorithms can be applied to predict metabolic fluxes from GEMs. For example, flux balance analysis (FBA) [178], one of the most widely used approaches, employs a steady state approximation and linear programming to identify cellular states that maximize a given objective function [179]. This method has been successfully implemented to guide metabolic engineering strategies, such as increasing the final product yield through gene knock-outs [180,181], elevated native pathway production [182], or optimized media input and environmental control [183].
Numerous resources are now available to facilitate the construction and implementation of GEM-base modeling. GEMs are reconstructed from annotated genomes that map functions to genes, using databases such as the KEGG and MetaCyc [184,185]. With the increasing availability of whole-genome sequencing data, and new developments in automated model reconstruction pipelines including ModelSEED, RAVEN, and PathwayTools, GEMs reconstruction of many non-model organisms of filamentous fungi are becoming possible [[186], [187], [188], [189]]. So far, over 30 GEMs have been developed for filamentous fungi, mostly molds and a few well-studied mushroom-forming fungi including Coprinus cinereus and Ganoderma lucidum [[190], [191], [192]]. With the G. lucidum GEM platform prediction, a 38% increase in production of polysaccharide (EPS), one of the main bioactive substances in Ganoderma, was validated when phenylalanine was supplemented in the culture [192]. However, accurate models are challenging to obtain in eukaryotes compared to prokaryotes, due to their large genome sizes, compartmentalization, and the abundance of unannotated or mis-annotated genes [193]. To evaluate and refine the GEMs in eukaryotic models, recent efforts have integrated essential gene analysis acquired from Tn-seq data to confirm predicted roles of many gene products and assign potential functions to uncharacterized genes under one or more incubation conditions [194]. The comparison of essential genes predicted through Tn-seq and in silico modeling using GEMs can be used to evaluate gaps and redundancy in the core metabolic pathways, and refine the gene-protein-reaction (GPR) rules in the GEM, producing a comprehensive understanding of the core cellular metabolism [195]. Other -omics approaches, primarily transcriptomics but also proteomics, metabolomics and fluxomics, can be similarly integrated to generate context-specific metabolic models by constraining the upper and lower bounds of flux across each reaction (Fig. 5b) [[196], [197], [198]].
3.2.3. Host selection
Large scale genomics unveiled a series of genes encoding the synthesis of novel secondary metabolites and enzymes of pharmaceutical and industrial relevance. In nature, fungi adapt their secondary metabolomes and proteomes to environmental stimuli that they encounter, or to certain developmental stages. However, replicating these environments in a laboratory condition is challenging, rendering many gene clusters silent (no observed expression). A silent gene or cluster of genes of interest can either be activated for transcription endogenously in its natural host, or heterologously in a fungal chassis with better genetic tractability. Endogenous activation of silent genes has mostly been achieved in fungi with an established genetic understanding such as the Aspergillus, Fusarium, Penicillium, Trichoderma, and Pestalotiopsis species. Clusters of genes have been activated mainly through promoter refactoring using a constitutive or inducible promoter, regulation of pathway-specific transcription factors [199], epigenetic modifications using small molecules [200], cocultivation of different organisms [48,201,202], and screening in different culturing conditions [203]. However, a large fraction of wildtype filamentous fungi are unculturable in a laboratory condition or not amiable for genetic engineering due to low rate of homologous recombination (HR). Thus, the heterologous expression of fungal enzyme-encoding genes or BGCs for the production of secondary metabolites in strains with accessible genetic manipulation tool boxes becomes a more versatile option (Fig. 5c). Selection of platform strains has been extensively covered by Mózsik et al., 2022 [204], numerating genotypes created through either random mutagenesis, or rational editing to allow HR-mediated targeted gene integration and easy selection. Most widely used heterologous hosts for fungal BGCs activation are Saccharomyces cerevisiae and Aspergillus species [36]. For industrial enzyme production, chassis including Aspergillus niger, Aspergillus oryzae and Trichoderma reesei have been used for decades, and are granted as Generally Recognized As Safe (GRAS) [205]. Strains with low levels of protease activity, such as Aspergillus vadensis, and low background production of unwanted enzymes are also preferable [206]. With the recent strain banking and sequencing efforts from Basecamp Research and SPUN to uncover nature's fungal genetic biodiversity, novel chasses that harbor superior metabolic capacity or demonstrate improved stability and robustness during fermentation processes might emerge, expanding the host repertoire used in biotechnology.
For a given product, prediction of its most production suitable host is still not possible, but rather practically determined by screening through multiple strain backgrounds, expression systems and culturing conditions [33]. A scalable synthetic biology platform has been implemented to systematically refactor groups of pathways derived from ascomycete and basidiomycete genomes in optimized S. cerevisiae strains [207]. Another fungal gene expression platform based on four Aspergillus species, called DIVERSIFY, was invented to search for the best chassis that enable the production of a protein or natural product at a higher titer [208]. These platforms can potentially be expanded to other fungal hosts with transferable synthetic biology tools.
4. Bottlenecks & bioengineering strategies
Many of the challenges encountered in filamentous fungi strain engineering stem from the deep lack of understanding about filamentous fungi physiology, cellular and metabolic processes, regulatory pathways, and its resulting effects on metabolite and enzyme production. This is further complicated by the cultivation context, including fermentation type (e.g. solid-state or submerged fermentation), fermentation vessel, and culture media/feedstock. Thus, forward genetic approaches using large-scale mutagenesis screens have typically been used to identify mutants with phenotypes of interest in set culture conditions. This toolkit includes natural recombination techniques (e.g. sexual reproduction, protoplast fusion), and mutagenesis by physical or chemical mutagens (e.g. UV, ethyl methane sulfonate (EMS)). A key example of its successful application is the development of citric acid hyper producers in A. niger. In one 2004 example, an original yield of 31 g/L citric acid was sequentially improved to 50 g/L by UV-mutagenesis, to 96 g/L by chemical mutagenesis, and then to 114 g/L by culture media modifications [209]. With the explosion in omics data and advances in genetic and metabolic engineering, there is an increasing ability to rationally engineer strains for even higher production yields. There was a recent report as high as 174 g/L citric acid production via the identification and targeted overexpression of glucose transporters in a mutant A. niger background [210]. Despite the evident success in improving production yields of metabolites and enzymes produced natively by the fungal hosts, there remain key scientific mysteries and challenges that yet remain to be solved.
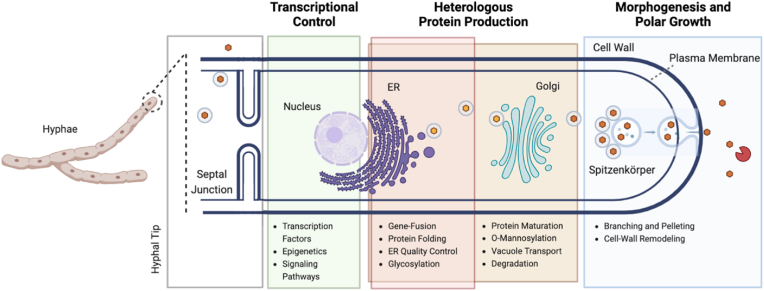
Here we provide three key remaining challenges that hamper the optimization of filamentous fungi strains for bioproduction, and detail the identification/modification of its associated biological targets (Fig. 6). First, heterologous production of mammalian proteins in mycelial cultures has remained prohibitively low. Second, modifying polar hyphal growth and morphogenesis for optimal metabolite/protein production and mycelium material properties has remained elusive. Third, our lack of understanding in regulatory and signaling pathways and its related effects on cellular processes and downstream production is still largely incomplete, preventing targeted transcriptional control. This section will focus on work that has been carried out across the Ascomycota phylum, which hosts industrially-relevant and relatively well-characterized genera (Aspergillus, Trichoderma, Neurospora, and Myceliophthora), and in which the majority of filamentous fungal strain engineering work has been established. If overcome, these bottlenecks may serve as lucrative opportunities to program filamentous fungi as cell factories for biomolecules, or further enhance the product characteristics of future mycelial materials and food production (including alternative leathers, whole-cuts, and bioplastics).
Fig. 6.
Overview of three key technical challenges for filamentous fungi strain engineering in biomanufacturing, categorized by the general milieu of associated bioengineering targets along the polar growth of the hyphal tip. Improvements in heterologous protein production may enable the sustainable production of therapeutics, animal-based proteins, and novel materials. Characterizing and leveraging morphogenic and transcriptional control may enable significant improvements in metabolite/protein production and mycelium material properties. ER: endoplasmic reticulum.
4.1. Improving heterologous protein production
Filamentous fungi have secretion capacities 10–1000 fold higher than that of bacterial, yeast, or mammalian hosts - generally limited to secretion titers lower than 10 g/L [211]. Despite this clear advantage, the production of heterologous proteins by filamentous fungi has faced many challenges that limit secretion titer. This limited production rate is much lower than homologous protein production, ultimately restricting the market penetration and capture of sustainably produced animal-based and therapeutic proteins [212]. These challenges stem from the complex and highly coordinated process of protein production and secretion in fungi, which are covered in more detail in previous reviews [[212], [213], [214]].
For the classical secretory pathway, genes encoding extracellular proteins are transcribed to mRNA in the nucleus, bearing short (15–36 amino acid) signal secretion peptide tags on its N-terminal end [213]. The transcribed mRNA is transported to cytosolic ribosomes, which attach to the membrane of the endoplasmic reticulum (ER) during translation. The growing polypeptide is translocated to the lumen of the ER, where the signal peptide moiety is cleaved and the polypeptide is N-glycosylated and folded. The folded proteins are transferred to the Golgi bodies, where they undergo further maturation and extension of O-mannosylation [215], are then packaged into cytoplasmic vesicles, escorted to the outer membrane at the apical hyphal tip, and released to the extracellular environment by fusion to the cytoplasmic membrane [212,214]. Growing evidence reveals the existence of unconventional protein secretion (UPS) pathways, whereby secreted proteins bypass the Golgi, and sometimes the endoplasmic reticulum, entirely – lacking conventional signal peptides and post-translational modifications [216,217]. Protein secretion has also been shown to take place in intercalary regions (i.e. septal junctions); although the molecular mechanisms are not yet well-understood, there is evidence that certain proteins and stress conditions bias towards these alternative secretion pathways [218,219]. The high variability in regulation, processing, trafficking, and secretion for specific host - heterologous protein pairs must be further explored.
4.1.1. Genetic modifications for improved protein yield
Multiple genetic approaches have been developed to improve protein secretion yields, including the introduction of multiple copies of the target-protein gene, codon optimization, protease depletion, and knockout of native high-yield proteins. The difference in genetic sequences between degenerate codons can have significant effects on mRNA stability and translation efficiency [161], making codon optimization a necessary tool for industrial protein production. For example, the heterologous expression of xylanase in T. reesei only succeeded after significant codon changes were implemented [220].
Provided that filamentous fungi produce a high diversity and quantity of proteases, undesired proteolytic degradation can contribute to the low yields of heterologous proteins. Indeed, early attempts at improving recombinant protein yields targeted proteolytic activity via UV/chemical mutagenesis or direct gene knockout of highly secreted proteases or associated transcription factors [[221], [222], [223]]. For example, deletion of the prtT transcription factor increased extracellular activity of the recombinant cutinase enzyme 36-fold in A. niger; prtT homologs have since been found across the Aspergillus and Penicillium genus and shown to control expression of both intra- and extracellular-proteases [113,224,225]. Improvements in -omics guided approaches, specifically in comparative secretomics – the atlas of secreted proteins for a given strain and growth condition – was recently used to identify and consequently delete 3 proteases in T. reesei, leading to a 78% decrease in protease activity [226]. Despite these attempts, residual levels of protease activity and undesirable reduction in strain capacity can remain detrimental for the scale-up of certain proteins and is an active area of research [29,227].
Disrupting the production of high-expression, native proteins has been explored to increase heterologous expression by optimizing cellular energy use [212] and liberating capacity in the secretory pathways [228]. Indeed, cellobiohydrolase (cbh1/2) in T. reesei and glucoamylase in A. niger account for the majority of their protein production and should theoretically free up production burden in the cell [229,230]. In certain cases, such as the deletion of cellobiohydrolase chb1 in T. reesei, there are significant upticks in heterologous protein production [[231], [232], [233]], but when certain other major secreted proteins are deleted, no effects have been observed [234]. Improved efforts in dynamical flux analysis using genome-scale models may elucidate key additional knockout targets to improve production capacity.
4.1.2. Gene-fusion approaches
Another early attempt to improve protein secretion utilized “carrier-based” gene-fusion approaches, wherein a gene expressing a highly secreted homologous protein is fused in-frame to a heterologous protein. This results in elevated expression of unfolded protein response and protein secretion pathway genes, as well as enhanced proteolytic stability [235]. Since its first reported development in 1990 by Ward et al. [236], the approach has since been refined and used to increase the secretion yields of, to name a few, human granulocyte colony stimulating factor (G-CSF) [237], interleukin 6 [227], bovine chymosin [238], and E. coli -glucuronidase proteins [239]. These efforts have clarified that the carrier-based approach is not always successful [240], the choice of fusion carrier protein is critical for optimal secretion [239], and the activity of the proteolytic cleavage sites (notably KEX-2 type protease cleavage sites) can be highly contextual [212,227,241].
Signal peptides (SP) are another critical engineering target for the high secretion of target heterologous proteins, and thus leveraged in heterologous gene-fusion approaches. These signaling peptide moieties are recognized by signal recognition particles (SRP), which target its entry into the secretory pathway via the ER, where they further assist in the initiation and maintenance of protein folding [242]. Fusion of a heterologous protein with a native SP generally exhibits superior secretion performance compared to fusion with non-native SPs [243,244], with the SP of glucoamylase and cellobiohydrolase commonly used in A. niger and T. reesei, respectively. Even minute differences in the peptide motif sequence can significantly impact the resulting secretion titers, via changes in protein conformation and disruption of SRP recognition [243,245]. By pairing secretomics and advanced bioinformatic analyses, we lend the possibility that signal peptide motifs can be rapidly identified in non-model organisms, diversifying the panel of suitable host chasses in industrial fermentation.
4.1.3. Rewiring the endoplasmic reticulum quality control
The endoplasmic reticulum (ER) serves as the primary site of the protein folding process. This process leverages a plethora of molecular chaperones, lectins, and foldases, including the binding immunoglobulin protein (BiP), calnexin (ClxA), and protein disulfide isomerase (PDI) [212,246,247]. The accumulation of unfolded proteins imposes stress upon the ER, which is managed via three distinct cellular strategies: the Unfolded Protein Response (UPR), Endoplasmic Reticulum-Associated Degradation (ERAD), and Repression Under Secretion Stress (RESS) [[248], [249], [250]]. Upon detection of a high burden of unfolded proteins in the ER, the UPR is activated by a series of signal transduction reactions, culminating in the downstream activity of a spliced Hac1 transcriptional activator that induces the up-regulation of ER proteins that assist in protein folding, including chaperones and foldases [251]. Proteins that cannot be properly folded are transported out of the ER to be degraded by the ubiquitin proteasome system, known as ERAD [212,235]. UPR and ERAD are conserved across eukaryotic systems [252], but the RESS is an additional feedback regulation mechanism in filamentous fungi that inhibits the transcription of secreted proteins [250]. This serves to further reduce stress throughout the secretory pathway.
The multi-faceted, ER-based protein quality control system has been rightfully targeted to enhance the yields of heterologous proteins, by increasing rates of proper folding and decreasing excessive protein clearance. These efforts have been met with highly variable results, demonstrating the importance of host and protein specific design considerations. This is especially clear in reports of co-expression of individual resident ER proteins with heterologous proteins. When PDI was overexpressed in A. awamori, there was a five-fold increase in secretion of plant thaumatin - a heterologous protein with eight disulfide bonds [253]; however, when PDI was overexpressed in a strain of A. niger for the production of hen egg white lysozyme, no increase in secretion yield was observed [254]. In the case of BiP, while overexpression of the ER chaperone in A. awamori (also for thaumatin production) and in T. reesei (for glucose oxidase production) promoted heterologous protein secretion [255,256], “seriously reduced” manganese peroxidase production was observed in A. niger [257]. Constitutive induction of UPR by expression of the activated form of the hacA transcription factor has seen success, leading to significant enhancement of secretion in A. niger [258]. Similar variable results are observed when reducing ERAD activity, with the deletion of a ERAD pathway component gene derA yielding improved protein production in A. niger [259], while detrimentally affecting the growth of A. fumigatus [260]. Careful consideration in ERAD modulation must be made when product fidelity and functionality is required, especially for therapeutics and food. Moreover, the overexpression of one element of the secretion pathway has been shown to simultaneously affect the expression of other elements [255]. Taken together, these results highlight the dearth of understanding behind the complex regulation of the quality control mechanisms in place within the filamentous fungal ER. Specific features of the target product, including post-translational modification sites (e.g. glycosylation, disulfide bonds) and hydrophobic/hydrophilic patches within the protein, as well host-dependent effects need to be considered when tweaking elements of the secretory pathway.
4.1.4. Mirroring mammalian protein glycosylation patterns
Filamentous fungi have garnered industry interest for the production of mammalian proteins due to its superior capability of matching mammalian patterns of glycosylation compared to prokaryotic and yeast systems [227,261]. Nearly 50% of all eukaryotic proteins are decorated with sugars [262], with site-specific glycosylation impacting the folding, functionality, stability, serum half-life, localization, and immunogenicity of a protein [[263], [264], [265]]; incorrectly glycosylated proteins are cleared from the bloodstream, rendering intended human therapeutics obsolete [227]. Additionally, incorrectly or under-glycosylated proteins are recognized by ER quality control systems and may activate the ERAD and RESS response [213,266]. Whereas yeast systems tend towards hyper-glycosylation (of the mannose-type), filamentous fungi are more conservative in hyper-mannosylation [264]. However, filamentous fungi lack the terminal sialylation of the glycans – common and fundamental to functionalized human proteins [228]. An additional challenge is the high level of strain-dependence in patterns of N-glycosylation, which is further altered across cultivation conditions [267,268].
Despite the scarcity of literature regarding the patterns and structures of glycosylation across filamentous fungal-produced proteins, there have been concerted efforts in engineering both the glycosylation sites within the recombinant proteins and the glycosylation systems within the fungal hosts themselves. The introduction of a N-glycosylation site in chymosin yielded an order of magnitude increase in its expression [269]. Leveraging the close relationship between glycosylation and ER quality control, deletion of N-glycosylation sites within the catalytic domain of the natively produced CHB1 protein of T. reesei induced up-regulation of UPR-associated genes [270] – demonstrating a potential for increased secretion of heterologous targets. Concurrent deletion of an ERAD factor, doaA, and overexpression of a glycosylation enzyme, sttC, yielded increased β-glucuronidase secretion in A. niger [271]. Towards humanizing the fungal glycosylation pathway, strides have been made in expressing human glycosylation machinery in Aspergillus and Trichoderma [[272], [273], [274]]. However, given the incredibly complex and poorly understood nature of the glycosylation pathways, targeted engineering of glycosylation sites in the protein remains the preferred strategy.
Towards increasing the yield and functionality of heterologous proteins, reported efforts have clearly demonstrated that the native protein production, secretion, and regulation pathways of filamentous fungi can and have been harnessed. By leveraging traditional mutagenesis and screening, or targeted gene-editing strategies, the protease activity, signaling peptide activity, and ER quality control mechanisms have been successfully modified for higher recombinant protein titers. Looking forward, a systems-level understanding of the mechanisms that underlie the high variability in success across the fungal strain–recombinant protein–engineering target landscape is necessary to realize the vision of sustainable mammalian protein production. Future multidisciplinary efforts that pair the in vitro generation of highly diverse mutational libraries (e.g. transposon mutagenesis) and in silico modeling of cellular protein production (e.g. genome-scale models) will lead us to a new frontier of filamentous fungi-based biomanufacturing.
4.2. Morphogenesis, cell-wall remodeling, and development
4.2.1. Morphogenesis in submerged mycelial fermentation
Filamentous fungi exhibit a diversity of macro-morphologies in both solid and liquid fermentation lifestyles. A 2019 review by Cairns et al. systematically lays out the various forms filamentous fungal cultures can take in submerged fermentation and its impact on downstream biophysical properties and production yields of proteins and metabolites [214]. In short, mycelial growth can range from compact pellets to dispersed hyphae, each with its own advantages and shortcomings respective to the target product to be produced. Although compact pellets exhibit resistance to shear stress and minimize the viscosity of the cultivation media – facilitating downstream compound purification and processing – the mycelia at the inner core of the pellet experience low oxygen and nutrient diffusion and thus low growth and metabolism. Dispersed hyphae do not suffer from local nutrient starvation, but do increase the medium viscosity and are more susceptible to shear stress [[275], [276], [277]]. Controllable fermentation parameters, including media composition, pH, bioreactor-type, agitation, and addition of microparticles can additionally impact the macro-morphology and associated physiology of the fungal cell (e.g. number of hyphal tips) [212]. Improvements in a systems-level understanding of the effects of morphological development on the differential secretion of primary/secondary metabolites and proteins is required to precisely determine optimal production-process parameters. However, generalizable principles have since been learned and leveraged in industrial-scale fermentation.
4.2.2. Hyper-branching for enhanced protein secretion
There exists an intrinsic relationship between polar mycelial growth and protein secretion. Extracellular proteins are generally secreted from the apical hyphal tip, where the hyperbranching phenotype encourages higher yields of protein secretion [212,214,228]. Protein secretion is highest during active periods of growth, during which rapid hyphal extension occurs. A shortened mycelial phenotype is often coincident with hyper-branching mutants, which allows for high-density, low-viscosity cultures – ideal for highly-productive fermentation [278]. The COT-1 pathway plays a key role in cell-wall remodeling, cell differentiation, and polarization in filamentous fungi [279,280]. A key mediator of cell-wall remodeling within the COT-1 pathway, gul1, was disrupted in strains of Neurospora crassa leading to increased protein secretion (25%), increased -glucosidase activity (56%), and markedly reduced medium viscosity [[278], [279], [280]]. Another interesting target is the Rho GTPase RacA, involved in actin polymerization at the mycelial tip. RacA deletion in A. niger significantly increases the number of hyphal tips (20%) and post-Golgi secretory vesicles. In this background, overexpression of glucoamylase using a metabolism-independent gene switch Tet-on yielded a 4-fold increase in protein secretion [219,281]. Growing knowledge of unconventional protein secretion (UPS) pathways also pose possible targets for overexpression of heterologous proteins.
4.2.3. Morphogenesis and metabolite production
Organic acid and secondary metabolite synthesis are tied to specific stages of mycelial growth. Studies in A. niger have demonstrated that the hyper-branched and short-hyphae phenotypes increase citric acid production [282,283] and a shift to phosphate-limited growth induces oxalic and citric synthesis [284]. Moving forward, it would be critical to identify key transporters of these metabolites and determine their dynamic distribution across the mycelial surface (e.g CexA – responsible for citrate production [285]). This will allow us to define optimal morphologies to maximize secretion in production cultures. Fungal secondary metabolites generally seem to be synthesized during periods of very low, or stagnant growth [47]. The production of mycelial pellets with densely-packed cores should limit oxygen and nutrient diffusion at the center – inducing secondary metabolism [214]. This likely explains the increase in product titers observed in production of lovastatin during pelleted fermentation of A. terreus [286]. A heightened understanding of what triggers the transcriptional activation of specific metabolites, as well as which biological factors coordinate morphogenesis is thus critical in developing a microbial-systems awareness of protein and metabolite production, processing, and secretion.
4.2.4. Modifications in cell-wall architecture
Biomass fermentation, unlike precision fermentation, relies on pure mycelial cultures for the production of materials (e.g. mycelial leather) and mycelium whole-cut meat alternatives (e.g. mycelium bacon). The precision modification of cell-wall architecture and resulting material properties is thus an alluring strategy for the generation of novel materials with improved performance or foods with enhanced mouth-feel and taste. The active process of cell wall growth, remodeling, and degradation is a highly coordinated process requiring more than 1000 genes and proteins [287]; this process alone dictates the shape and macro-morphology of the fungus and is well conserved across fungi [288]. A basic filamentous fungi have an inner cell wall composed of rigid polysaccharides (e.g. chitin, -1,3-glucan, and -1,3-glucan) imparting mechanical strength to the mycelial structure, and an outer cell wall composed of polymers and glycosylated proteins [289]. Key performance properties of mycelial materials may be improved by the modification of this intricate structure, including strength and elasticity, hydrophobicity, porosity and permeability [288]. Due to the highly coordinated and complex processes underlying cell-wall remodeling and emergent structure, scalable methods to discern global architecture is required. High-throughput genetic methods (e.g. CRISPR screens) and high-throughput screening methods (e.g. automation or droplet microfluidics) are together required to further characterize and leverage morphology as a toggle for improved biomass and precision fermentation performance. Further research, specifically in host strains such as G. lucidum and P. ostreatus, used in materials and food applications is especially desired.
4.2.5. Reproductive development
The (a)sexual reproduction of filamentous fungi is linked to enhanced secondary metabolite production, and inhibiting enzyme secretion and biomass growth [46,290,291]. In sporulating zones of A. niger, production of proteins was not observed; deletion of the flbA gene abolishes sporulation and reduces protein secretion heterogeneity across the mycelial colonies [292]. Moreover, flbA affects multiple transcription factor genes – which should be considered for future targets of hyper-secreting mutants [293,294]. Furthermore, several transcription factors (e.g. mpkB, steA, medA) have been identified within the MAPK phosphorylation pathway that have been shown to affect (a)sexual development in the model system A. nidulans [214].
4.3. Transcriptional control of regulatory and signaling pathways
4.3.1. Transcription factors
Transcription factors involved in metabolic processes have been targeted to optimize production. Gene expression can be regulated to optimize downstream production by modulating transcription factor activity [213,295]. As previously described, transcription factors involved in proteolytic digestion and endogenous ER-dependent unfolded protein responses (UPR, ERAD, RESS) were successfully targeted to restructure the protein processing and secretory pathways [212]. Deletion of creA, a Cys2His2-type transcription factor involved in carbon catabolite repression, has been shown to increase expression of endogenous CAZymes [213]. Trans-regulatory elements responsible for alternative nitrogen metabolism (AreA) [213], arabinan degradation (AraR) [295], pectin catabolism (GaaX) [296], and xylan & cellulose degradation (XlnR) [297] have been identified, adding to a list of engineering strategies to optimize cellular productivity. As with many areas of filamentous fungal physiology, the prior limitations of traditional basic science approaches (inherently restricted by lack of scale and breadth across mutational landscapes and screening capacity) leaves scientists with a limited number of putative engineering targets for transcriptional-level control.
4.3.2. Epigenetics
Certain links between epigenetic control and protein and secondary metabolite production are relatively well-established. The genomic integration of heterologous genes into transcriptionally active regions and well-accessible regions within the genome is known to facilitate high transcription and improve mRNA stability [213]. Traditionally, integration at sites of high endogenous production, including the cellobiohydrolase loci in T. reesei (cbhA/B) and the glucoamylase loci in A. niger (glaA/B), have achieved improved heterologous production. Targeted methods (e.g. CRISPR-based or homologous-guided recombination methods) are ideal, if reasonable transformation efficiency can be achieved and appropriate sites are identified. Furthermore, a forward UV-mutagenesis genetics screen revealed that a point mutation in laeA, a putative methyl-transferase-domain protein, c5auses a non-citric acid producing phenotype in A. niger [298]. The heterotrimeric velvet complex in filamentous fungi aids in the coordination of light response, hyphal growth and secondary metabolism [299]. LaeA - a key component of the velvet complex - is hypothesized to reverse formation of transcriptionally silent heterochromatin by H3K9 methylation activity [235]. This has been an attractive target in boosting secondary metabolite activity (e.g. -lactam in Penicillium chrysogenum [300]) and modulating macro-morphology [301], but demonstrated divergent regulatory functions in T. reesei [302]. A panel of epigenetic-regulatory targets will continue to be identified (e.g. StuA, FlbA, BrlA) as transcriptomic analyses shed light on the role of epigenetics in cellular activity [46,303].
4.3.3. Signaling cascades
Numerous studies have supported the hypothesis that a diversity of cell-signaling networks coordinate the polar growth, macro-morphology and development in filamentous fungi [304,305]. These signaling cascades, including the TORC2, CWI, MAPK, PKA/cAMP and calcium/calcineurin signaling, transduce external cues into a wide breadth of intracellular responses; this subject has been previously well-reviewed [214]. The concerted regulation of the rapamycin kinase (TORC2) and cell-wall integrity (CWI) signaling pathways affect sterol, ion transport, amino acid metabolism and protein trafficking [306]. The MAPK pathway senses pheromone and nutrient availability, regulating downstream spore formation, oxidative/osmotic stress responses, and cell wall integrity, via trans-membrane transduction of G-protein coupled receptors (GPCRs) [[307], [308], [309], [310]]. The signal cascades through a phosphorelay system, ultimately affecting the expression of chromatin remodeling proteins, transcription factors, and co-regulatory proteins [304]. The PKA/cAMP pathway senses environmental carbon, regulates vegetative growth, and acts via activation of protein kinase A (PKA) by a GPCR-activated, ATP-derived cAMP [214]. These activated PKA phosphorylates target transcription factors, facilitating entry into the nucleus, modulating gene expression. They have been targeted to affect macro-morphology (e.g. compact colony morphology by PkaC overexpression in A. niger [311]) and secondary metabolism (e.g. elevated production of penicillin and reduced conidiation by fadA overexpression – an -subunit of a G-protein – in A. nidulans [312]). The calcium/calcineurin signaling pathway regulates reproductive development and is well-characterized due to its role in the virulence of human fungal pathogens [313,314], as well as its role in asexual development [315]. Deletion of CrzA, a calcineurin target, caused a hyperbranched phenotype in T. reesei causing concomitant increases in hemicellulose secretion [316]. The complex effects of these signaling cascades confound strain engineering efforts, but may become easier to engineer as broad systems-level understanding is developed.
5. Future remarks and conclusion
Previous efforts in engineering protein secretory pathways and signaling cascades clearly demonstrate the importance of considering specific host and protein features to inform appropriate molecular targets. Efforts to overcome three key bottlenecks in filamentous fungal strain engineering (heterologous protein production, culture morphogenesis and development, and transcriptional control), clarifies the stark lack of understanding across many areas of fungal biology. This is largely due to the limits in scale and specificity inherent to traditional methods, namely the classical UV/chemical mutagenesis and manual colony screening pipeline. The application of high-throughput and single-cell resolution methods, already broadly applied within the synthetic biology community, can ultimately yield an integrated and iterative cycle that accelerates both systems biology characterization and target trait breeding. Upstream of the cycle, better genome-wide mutational strain libraries may now be developed using techniques such as CRISPR screens and transposon mutagenesis, moving beyond simple point mutations and towards large genomic rearrangements and gene-enrichment libraries. Downstream of the cycle, innovations in high-throughput experimental evolution, micro-droplet fluidics, and lab automation platforms greatly improves the power and scale of mutant selection and screening. The greatest challenge that remains to be overcome is the validation, development, and integration of these distinct approaches across a breeding pipeline that enables iterative learning through systems-level characterization and modification. Overcoming these challenges promises previously unprecedented improvements in fermentation performance for industrial biomanufacturing.
Innovations also continue to emerge across cultivation and production-process techniques. This includes a re-emergence in solid-state fermentation research for improved and novel enzyme production [[317], [318], [319]], and improved dynamic modeling of growth in increasingly advanced bioreactors [320]. Microbial mixed cultures are also being explored due to emerging evidence that novel cryptic compounds are only expressed upon co-culture induction [[321], [322], [323]]. Our growing understanding of the complex regulatory and signaling systems in filamentous fungi that coordinate their metabolism, polar growth, protein processing/secretion, morphogenesis, and material properties extend the possibility of commercially-feasible mammalian-protein cell factories, enzymatic workhorses in a circular bioeconomy, and biological agents of carbon capture and remediation [25,51,324,325].
To meet the climate goal set out in the Paris Agreement – constraining global warming to 1.5 °C above pre-industrial levels – the world must take urgent and bold actions to cut 43% of emissions by 2030 and achieve net-zero emissions by 2050 [326]. The fungal biotechnology embodies a myriad of innovations that are clean, versatile and yet functional. Just considering its potential impact on decarbonizing animal protein production alone, fungal-derived proteins chart out an effective way of producing protein-rich biomass and animal functional proteins at cost-parity. A landmark life cycle assessment demonstrated that even a 20% displacement of ruminant meat by mycoproteins can offset its associated annual deforestation and related CO2 emissions in half, as well as significantly lower global methane emissions from animal agriculture [327]. Fungal bio-composite materials, on the other hand, can act as a CO2-sink material with a negative embodied carbon (EC) value, 39.5 kg CO2 eq m−3 [328]. Beyond its potential in decarbonizing the food and material industries, filamentous fungi, specifically mycorrhizal fungi that form mutualistic relationships with plants, naturally sink billions of tons of fixed atmospheric carbon annually in the underground fungal network [329]. It represents a unique, unexplored climate opportunity for creating massive carbon sinks at a global scale. To realize the full potential of filamentous fungi in these novel decarbonization and carbon sequestration avenues, the crosstalks between the synthetic biology community, the mycology community, and the materials science community are needed now more than ever. This review serves as a primer for initiating such collaboration, and calls for holistic solutions that benefit human society, animal welfare and the planetary health at scale.
Credit author statement
Charles Jo, Jing Zhang, Tzu-Chieh Tang wrote the manuscript. Daniel Segrè, Jenny M. Tam, George M. Church, Ahmad S. Khalil reviewed and edited the manuscript.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
The authors thank Dr. Charlie Gilbert (Cultivarium, MA) and Dr. Jinhui Chang (The Hong Kong Polytechnic University) for useful discussions, and Michael Silverstein for reviewing the manuscript. Funding for work at Harvard University was provided by the U.S. Department of Energy under grant no. DE-FG02-02ER63445 and by the National Science Foundation award no. MCB-2037995 (both to G.M.C.). Funding for work at Khalil Lab, Boston University was provided by the National Institute of Health award no. 1R01AI171100-01 (to A.S.K.). A.S.K. acknowledges support from a Department of Defense Vannevar Bush Faculty Fellowship (N00014-20-1-2825). D.S. acknowledges support from the Human Frontier Science Program (HFSP Research Grant RGP0060/2021), the NSF Center for Chemical Currencies of a Microbial Planet, and U.S. Department of Energy, Office of Science, Office of Biological & Environmental Research through Microbial Community Analysis & Functional Evaluation in Soils (m-CAFEs) under contract number DE-AC02-05CH11231 to Lawrence Berkeley National Laboratory). Figures and the graphical abstract were created with BioRender.com.
Contributor Information
Jing Zhang, Email: jillwill00@gmail.com.
Tzu-Chieh Tang, Email: zijaytang@gmail.com.
Data availability
No data was used for the research described in the article.
References
- 1.Maheshwari R. The largest and oldest living organism. Reson. 2005;10:4–9. doi: 10.1007/BF02834644. [DOI] [Google Scholar]
- 2.Sipos G., Prasanna A.N., Walter M.C., O'Connor E., Bálint B., Krizsán K., Kiss B., Hess J., Varga T., Slot J., Riley R., Bóka B., Rigling D., Barry K., Lee J., Mihaltcheva S., LaButti K., Lipzen A., Waldron R., Moloney N.M., Sperisen C., Kredics L., Vágvölgyi C., Patrignani A., Fitzpatrick D., Nagy I., Doyle S., Anderson J.B., Grigoriev I.V., Güldener U., Münsterkötter M., Nagy L.G. Genome expansion and lineage-specific genetic innovations in the forest pathogenic fungi Armillaria. Nat. Ecol. Evol. 2017;1:1931–1941. doi: 10.1038/s41559-017-0347-8. [DOI] [PubMed] [Google Scholar]
- 3.Bahram M., Netherway T. Fungi as mediators linking organisms and ecosystems. FEMS (Fed. Eur. Microbiol. Soc.) Microbiol. Rev. 2022;46 doi: 10.1093/femsre/fuab058. fuab058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Floudas D. In: Advances in Botanical Research. Morel-Rouhier M., Sormani R., editors. Academic Press; 2021. Chapter Two - evolution of lignin decomposition systems in fungi; pp. 37–76. [DOI] [Google Scholar]
- 5.Hage H., Rosso M.-N. Evolution of fungal carbohydrate-active enzyme portfolios and adaptation to plant cell-wall polymers. J. Fungi. 2021;7:185. doi: 10.3390/jof7030185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.van den Brink J., de Vries R.P. Fungal enzyme sets for plant polysaccharide degradation. Appl. Microbiol. Biotechnol. 2011;91:1477–1492. doi: 10.1007/s00253-011-3473-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Klein D.A., Paschke M.W. Filamentous fungi: the indeterminate lifestyle and microbial ecology. Microb. Ecol. 2004;47:224–235. doi: 10.1007/s00248-003-1037-4. [DOI] [PubMed] [Google Scholar]
- 8.Wallen R.M., Perlin M.H. An overview of the function and maintenance of sexual reproduction in dikaryotic fungi. Front. Microbiol. 2018;9 doi: 10.3389/fmicb.2018.00503. https://www.frontiersin.org/articles/10.3389/fmicb.2018.00503 accessed. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pelkmans J.F., Lugones L.G., Wösten H.A.B. In: Growth, Differentiation and Sexuality. Wendland J., editor. Springer International Publishing; Cham: 2016. 15 fruiting body formation in basidiomycetes; pp. 387–405. [DOI] [Google Scholar]
- 10.Nieuwenhuis B.P.S., James T.Y. The frequency of sex in fungi. Phil. Trans. Biol. Sci. 2016;371 doi: 10.1098/rstb.2015.0540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Power R.C., Salazar-García D.C., Straus L.G., González Morales M.R., Henry A.G. Microremains from El Mirón Cave human dental calculus suggest a mixed plant–animal subsistence economy during the Magdalenian in Northern Iberia. J. Archaeol. Sci. 2015;60:39–46. doi: 10.1016/j.jas.2015.04.003. [DOI] [Google Scholar]
- 12.El Sheikha A.F., Hu D.-M. How to trace the geographic origin of mushrooms? Trends Food Sci. Technol. 2018;78:292–303. doi: 10.1016/j.tifs.2018.06.008. [DOI] [Google Scholar]
- 13.Venturini Copetti M. Yeasts and molds in fermented food production: an ancient bioprocess. Curr. Opin. Food Sci. 2019;25:57–61. doi: 10.1016/j.cofs.2019.02.014. [DOI] [Google Scholar]
- 14.Kango N., Jana U.K., Choukade R. In: Advancing Frontiers in Mycology & Mycotechnology: Basic and Applied Aspects of Fungi. Satyanarayana T., Deshmukh S.K., Deshpande M.V., editors. Springer; Singapore: 2019. Fungal enzymes: sources and biotechnological applications; pp. 515–538. [DOI] [Google Scholar]
- 15.Lange L., Agger J.W., Meyer A.S. In: Grand Challenges in Fungal Biotechnology. Nevalainen H., editor. Springer International Publishing; Cham: 2020. Fungal biotechnology: unlocking the full potential of fungi for a more sustainable world; pp. 3–32. [DOI] [Google Scholar]
- 16.Jones M., Gandia A., John S., Bismarck A. Leather-like material biofabrication using fungi. Nat. Sustain. 2021;4:9–16. doi: 10.1038/s41893-020-00606-1. [DOI] [Google Scholar]
- 17.Strong P.J., Self R., Allikian K., Szewczyk E., Speight R., O'Hara I., Harrison M.D. Filamentous fungi for future functional food and feed. Curr. Opin. Biotechnol. 2022;76 doi: 10.1016/j.copbio.2022.102729. [DOI] [PubMed] [Google Scholar]
- 18.Gmoser R., Fristedt R., Larsson K., Undeland I., Taherzadeh M.J., Lennartsson P.R. From stale bread and brewers spent grain to a new food source using edible filamentous fungi. Bioengineered. 2020;11:582–598. doi: 10.1080/21655979.2020.1768694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nath D., Meena V.S. In: Role of Rhizospheric Microbes in Soil: Volume 1: Stress Management and Agricultural Sustainability. Meena V.S., editor. Springer; Singapore: 2018. Mycorrhizae: a potential microorganism and its implication in agriculture; pp. 251–276. [DOI] [Google Scholar]
- 20.Hagh-Doust N., Färkkilä S.M.A., Hosseyni Moghaddam M.S., Tedersoo L. Symbiotic fungi as biotechnological tools: methodological challenges and relative benefits in agriculture and forestry. Fungal Biol. Rev. 2022 doi: 10.1016/j.fbr.2022.06.001. [DOI] [Google Scholar]
- 21.Ragauskas A.J., Beckham G.T., Biddy M.J., Chandra R., Chen F., Davis M.F., Davison B.H., Dixon R.A., Gilna P., Keller M., Langan P., Naskar A.K., Saddler J.N., Tschaplinski T.J., Tuskan G.A., Wyman C.E. Lignin valorization: improving lignin processing in the biorefinery. Science. 2014;344 doi: 10.1126/science.1246843. [DOI] [PubMed] [Google Scholar]
- 22.Ning P., Yang G., Hu L., Sun J., Shi L., Zhou Y., Wang Z., Yang J. Recent advances in the valorization of plant biomass. Biotechnol. Biofuels. 2021;14:102. doi: 10.1186/s13068-021-01949-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lin S., Wei J., Yang B., Zhang M., Zhuo R. Bioremediation of organic pollutants by white rot fungal cytochrome P450: the role and mechanism of CYP450 in biodegradation. Chemosphere. 2022;301 doi: 10.1016/j.chemosphere.2022.134776. [DOI] [PubMed] [Google Scholar]
- 24.Pointing S. Feasibility of bioremediation by white-rot fungi. Appl. Microbiol. Biotechnol. 2001;57:20–33. doi: 10.1007/s002530100745. [DOI] [PubMed] [Google Scholar]
- 25.Popkin G. A fungal safari. Science. 2022;377:142–147. doi: 10.1126/science.add7606. [DOI] [PubMed] [Google Scholar]
- 26.Mózsik L., Pohl C., Meyer V., Bovenberg R.A.L., Nygård Y., Driessen A.J.M. Modular synthetic biology toolkit for filamentous fungi. ACS Synth. Biol. 2021;10:2850–2861. doi: 10.1021/acssynbio.1c00260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang S., Chen H., Tang X., Zhang H., Chen W., Chen Y.Q. Molecular tools for gene manipulation in filamentous fungi. Appl. Microbiol. Biotechnol. 2017;101:8063–8075. doi: 10.1007/s00253-017-8486-z. [DOI] [PubMed] [Google Scholar]
- 28.Dhevagi P., Ramya A., Priyatharshini S., Geetha Thanuja K., Ambreetha S., Nivetha A. In: Recent Trends in Mycological Research: Volume 2: Environmental and Industrial Perspective. Yadav A.N., editor. Springer International Publishing; Cham: 2021. Industrially important fungal enzymes: productions and applications; pp. 263–309. [DOI] [Google Scholar]
- 29.Lübeck M., Lübeck P.S. Fungal cell factories for efficient and sustainable production of proteins and peptides. Microorganisms. 2022;10:753. doi: 10.3390/microorganisms10040753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Barzee T.J., Cao L., Pan Z., Zhang R. Fungi for future foods. J. Fut. Foods. 2021;1:25–37. doi: 10.1016/j.jfutfo.2021.09.002. [DOI] [Google Scholar]
- 31.Ferreira J.A., Varjani S., Taherzadeh M.J. A critical review on the ubiquitous role of filamentous fungi in pollution mitigation. Curr. Pollut. Rep. 2020;6:295–309. doi: 10.1007/s40726-020-00156-2. [DOI] [Google Scholar]
- 32.Harms H., Schlosser D., Wick L.Y. Untapped potential: exploiting fungi in bioremediation of hazardous chemicals. Nat. Rev. Microbiol. 2011;9:177–192. doi: 10.1038/nrmicro2519. [DOI] [PubMed] [Google Scholar]
- 33.Arnau J., Yaver D., Hjort C.M. In: Grand Challenges in Fungal Biotechnology. Nevalainen H., editor. Springer International Publishing; Cham: 2020. Strategies and challenges for the development of industrial enzymes using fungal cell factories; pp. 179–210. [DOI] [Google Scholar]
- 34.Karnaouri A., Topakas E., Antonopoulou I., Christakopoulos P. Genomic insights into the fungal lignocellulolytic system of Myceliophthora thermophila. Front. Microbiol. 2014:5. doi: 10.3389/fmicb.2014.00281. https://www.frontiersin.org/articles/10.3389/fmicb.2014.00281 accessed. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Spraker J.E., Wiemann P., Baccile J.A., Venkatesh N., Schumacher J., Schroeder F.C., Sanchez L.M., Keller N.P. Conserved responses in a war of small molecules between a plant-pathogenic bacterium and fungi. mBio. 2018;9:e00820. doi: 10.1128/mBio.00820-18. -18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Keller N.P. Fungal secondary metabolism: regulation, function and drug discovery. Nat. Rev. Microbiol. 2019;17:167–180. doi: 10.1038/s41579-018-0121-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Aoki H., Okuhara M. Natural beta-lactam antibiotics. Annu. Rev. Microbiol. 1980;34:159–181. doi: 10.1146/annurev.mi.34.100180.001111. [DOI] [PubMed] [Google Scholar]
- 38.Long J., Ji W., Zhang D., Zhu Y., Bi Y. Bioactivities and structure–activity relationships of fusidic acid derivatives: a review. Front. Pharmacol. 2021:12. doi: 10.3389/fphar.2021.759220. https://www.frontiersin.org/articles/10.3389/fphar.2021.759220 accessed January 19, 2023) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Huber F.M. In: Mechanism of Action. Gottlieb D., Shaw P.D., editors. Springer; Berlin, Heidelberg: 1967. Griseofulvin; pp. 181–189. [DOI] [Google Scholar]
- 40.Butler M.S. The role of natural product chemistry in drug discovery. J. Nat. Prod. 2004;67:2141–2153. doi: 10.1021/np040106y. [DOI] [PubMed] [Google Scholar]
- 41.Stierle A., Strobel G., Stierle D. Taxol and taxane production by Taxomyces andreanae, an endophytic fungus of pacific yew. Science. 1993;260:214–216. doi: 10.1126/science.8097061. [DOI] [PubMed] [Google Scholar]
- 42.Abdel-Razek A.S., El-Naggar M.E., Allam A., Morsy O.M., Othman S.I. Microbial natural products in drug discovery. Processes. 2020;8:470. doi: 10.3390/pr8040470. [DOI] [Google Scholar]
- 43.Wenger R.M. In: Fortschritte Der Chemie Organischer Naturstoffe/Progress in the Chemistry of Organic Natural Products. Inouye H., Jaenicke L., Lounasmaa M., Marner F.-J., Séquin U., Somersalo P., Uesato S., Wenger R.M., Herz W., Grisebach H., Kirby G.W., Tamm Ch, editors. Springer; Vienna: 1986. Cyclosporine and analogues — isolation and synthesis — mechanism of action and structural requirements for pharmacological activity; pp. 123–168. [DOI] [PubMed] [Google Scholar]
- 44.Bentley R. Mycophenolic acid: a one hundred year odyssey from antibiotic to immunosuppressant. Chem. Rev. 2000;100:3801–3826. doi: 10.1021/cr990097b. [DOI] [PubMed] [Google Scholar]
- 45.Douhan G.W., Smith M.E., Huyrn K.L., Westbrook A., Beerli P., Fisher A.J. Multigene analysis suggests ecological speciation in the fungal pathogen Claviceps purpurea. Mol. Ecol. 2008;17:2276–2286. doi: 10.1111/j.1365-294X.2008.03753.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Keller N.P., Turner G., Bennett J.W. Fungal secondary metabolism — from biochemistry to genomics. Nat. Rev. Microbiol. 2005;3:937–947. doi: 10.1038/nrmicro1286. [DOI] [PubMed] [Google Scholar]
- 47.Brakhage A.A. Regulation of fungal secondary metabolism. Nat. Rev. Microbiol. 2013;11:21–32. doi: 10.1038/nrmicro2916. [DOI] [PubMed] [Google Scholar]
- 48.Ninomiya A., Urayama S., Hagiwara D. Antibacterial diphenyl ether production induced by co-culture of Aspergillus nidulans and Aspergillus fumigatus. Appl. Microbiol. Biotechnol. 2022;106:4169–4185. doi: 10.1007/s00253-022-11964-5. [DOI] [PubMed] [Google Scholar]
- 49.Sarkar A., Kim E.Y., Jang T., Hongdusit A., Kim H., Choi J.-M., Fox J.M. Microbially guided discovery and biosynthesis of biologically active natural products. ACS Synth. Biol. 2021;10:1505–1519. doi: 10.1021/acssynbio.1c00074. [DOI] [PubMed] [Google Scholar]
- 50.Jones M., Mautner A., Luenco S., Bismarck A., John S. Engineered mycelium composite construction materials from fungal biorefineries: a critical review. Mater. Des. 2020;187 doi: 10.1016/j.matdes.2019.108397. [DOI] [Google Scholar]
- 51.Meyer V., Basenko E.Y., Benz J.P., Braus G.H., Caddick M.X., Csukai M., de Vries R.P., Endy D., Frisvad J.C., Gunde-Cimerman N., Haarmann T., Hadar Y., Hansen K., Johnson R.I., Keller N.P., Kraševec N., Mortensen U.H., Perez R., Ram A.F.J., Record E., Ross P., Shapaval V., Steiniger C., van den Brink H., van Munster J., Yarden O., Wösten H.A.B. Growing a circular economy with fungal biotechnology: a white paper. Fungal Biol. Biotechnol. 2020;7:5. doi: 10.1186/s40694-020-00095-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Antinori M.E., Ceseracciu L., Mancini G., Heredia-Guerrero J.A., Athanassiou A. Fine-tuning of physicochemical properties and growth dynamics of mycelium-based materials. ACS Appl. Bio Mater. 2020;3:1044–1051. doi: 10.1021/acsabm.9b01031. [DOI] [PubMed] [Google Scholar]
- 53.Haneef M., Ceseracciu L., Canale C., Bayer I.S., Heredia-Guerrero J.A., Athanassiou A. Advanced materials from fungal mycelium: fabrication and tuning of physical properties. Sci. Rep. 2017;7 doi: 10.1038/srep41292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Meyer V., Schmidt B., Freidank-Pohl C., Schmidts C., Pfeiffer S., My-Co S.P.A.C.E. An artistic-scientific vision on how to build with fungi. IOP Conf. Ser. Earth Environ. Sci. 2022;1078 doi: 10.1088/1755-1315/1078/1/012070. [DOI] [Google Scholar]
- 55.Almpani-Lekka D., Pfeiffer S., Schmidts C., Seo S. A review on architecture with fungal biomaterials: the desired and the feasible. Fungal Biol. Biotechnol. 2021;8:17. doi: 10.1186/s40694-021-00124-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Attias N., Danai O., Abitbol T., Tarazi E., Ezov N., Pereman I., Grobman Y.J. Mycelium bio-composites in industrial design and architecture: comparative review and experimental analysis. J. Clean. Prod. 2020;246 doi: 10.1016/j.jclepro.2019.119037. [DOI] [Google Scholar]
- 57.Van Wylick A., Monclaro A.V., Elsacker E., Vandelook S., Rahier H., De Laet L., Cannella D., Peeters E. A review on the potential of filamentous fungi for microbial self-healing of concrete. Fungal Biol. Biotechnol. 2021;8:16. doi: 10.1186/s40694-021-00122-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Menon R.R., Luo J., Chen X., Zhou H., Liu Z., Zhou G., Zhang N., Jin C. Screening of fungi for potential application of self-healing concrete. Sci. Rep. 2019;9:2075. doi: 10.1038/s41598-019-39156-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sharma S., Meyer V. The colors of life: an interdisciplinary artist-in-residence project to research fungal pigments as a gateway to empathy and understanding of microbial life. Fungal Biol. Biotechnol. 2022;9:1. doi: 10.1186/s40694-021-00130-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kalra R., Conlan X.A., Goel M. Fungi as a potential source of pigments: harnessing filamentous fungi. Front. Chem. 2020;8 doi: 10.3389/fchem.2020.00369. https://www.frontiersin.org/articles/10.3389/fchem.2020.00369 accessed. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.McBee R.M., Lucht M., Mukhitov N., Richardson M., Srinivasan T., Meng D., Chen H., Kaufman A., Reitman M., Munck C., Schaak D., Voigt C., Wang H.H. Engineering living and regenerative fungal–bacterial biocomposite structures. Nat. Mater. 2021 doi: 10.1038/s41563-021-01123-y. [DOI] [PubMed] [Google Scholar]
- 62.Elsacker E., Peeters E., De Laet L. Large-scale robotic extrusion-based additive manufacturing with living mycelium materials. Sustain. Fut. 2022;4 doi: 10.1016/j.sftr.2022.100085. [DOI] [Google Scholar]
- 63.Tang T.-C., An B., Huang Y., Vasikaran S., Wang Y., Jiang X., Lu T.K., Zhong C. Materials design by synthetic biology. Nat. Rev. Mater. 2021;6:332–350. doi: 10.1038/s41578-020-00265-w. [DOI] [Google Scholar]
- 64.Rodrigo-Navarro A., Sankaran S., Dalby M.J., del Campo A., Salmeron-Sanchez M. Engineered living biomaterials. Nat. Rev. Mater. 2021;6:1175–1190. doi: 10.1038/s41578-021-00350-8. [DOI] [Google Scholar]
- 65.Nguyen P.Q., Courchesne N.-M.D., Duraj-Thatte A., Praveschotinunt P., Joshi N.S. Engineered living materials: prospects and challenges for using biological systems to direct the assembly of smart materials. Adv. Mater. 2018;30 doi: 10.1002/adma.201704847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Srubar W.V. Engineered living materials: taxonomies and emerging trends. Trends Biotechnol. 2021;39:574–583. doi: 10.1016/j.tibtech.2020.10.009. [DOI] [PubMed] [Google Scholar]
- 67.Gilbert C., Ellis T. Biological engineered living materials: growing functional materials with genetically programmable properties. ACS Synth. Biol. 2019;8:1–15. doi: 10.1021/acssynbio.8b00423. [DOI] [PubMed] [Google Scholar]
- 68.Vandelook S., Elsacker E., Van Wylick A., De Laet L., Peeters E. Current state and future prospects of pure mycelium materials. Fungal Biol. Biotechnol. 2021;8:20. doi: 10.1186/s40694-021-00128-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ghorai S., Banik S.P., Verma D., Chowdhury S., Mukherjee S., Khowala S. Fungal biotechnology in food and feed processing. Food Res. Int. 2009;42:577–587. doi: 10.1016/j.foodres.2009.02.019. [DOI] [Google Scholar]
- 70.Handoyo T., Morita N. Structural and functional properties of fermented soybean (Tempeh) by using Rhizopus oligosporus. Int. J. Food Prop. 2006;9:347–355. doi: 10.1080/10942910500224746. [DOI] [Google Scholar]
- 71.Finnigan T.J.A., Wall B.T., Wilde P.J., Stephens F.B., Taylor S.L., Freedman M.R. Mycoprotein: the future of nutritious nonmeat protein, a symposium review. Curr. Dev. Nutrit. 2019;3:nzz021. doi: 10.1093/cdn/nzz021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.González A., Cruz M., Losoya C., Nobre C., Loredo A., Rodríguez R., Contreras J., Belmares R. Edible mushrooms as a novel protein source for functional foods. Food Funct. 2020;11:7400–7414. doi: 10.1039/D0FO01746A. [DOI] [PubMed] [Google Scholar]
- 73.Antinori M.E., Contardi M., Suarato G., Armirotti A., Bertorelli R., Mancini G., Debellis D., Athanassiou A. Advanced mycelium materials as potential self-growing biomedical scaffolds. Sci. Rep. 2021;11 doi: 10.1038/s41598-021-91572-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Singh S., Yap W.S., Ge X.Y., Min V.L.X., Choudhury D. Cultured meat production fuelled by fermentation. Trends Food Sci. Technol. 2022;120:48–58. doi: 10.1016/j.tifs.2021.12.028. [DOI] [Google Scholar]
- 75.Järviö N., Parviainen T., Maljanen N.-L., Kobayashi Y., Kujanpää L., Ercili-Cura D., Landowski C.P., Ryynänen T., Nordlund E., Tuomisto H.L. Ovalbumin production using Trichoderma reesei culture and low-carbon energy could mitigate the environmental impacts of chicken-egg-derived ovalbumin. Nat. Food. 2021;2:1005–1013. doi: 10.1038/s43016-021-00418-2. [DOI] [PubMed] [Google Scholar]
- 76.Keppler J.K., Heyse A., Scheidler E., Uttinger M.J., Fitzner L., Jandt U., Heyn T.R., Lautenbach V., Loch J.I., Lohr J., Kieserling H., Günther G., Kempf E., Grosch J.H., Lewiński K., Jahn D., Lübbert C., Peukert W., Kulozik U., Drusch S., Krull R., Schwarz K., Biedendieck R. Towards recombinantly produced milk proteins: physicochemical and emulsifying properties of engineered whey protein beta-lactoglobulin variants. Food Hydrocolloids. 2021;110 doi: 10.1016/j.foodhyd.2020.106132. [DOI] [Google Scholar]
- 77.Good Food Institute . GFI; 2022. Fermentation | state of the industry report.https://gfi.org/resource/fermentation-state-of-the-industry-report/ (n.d.) (accessed November 5, [Google Scholar]
- 78.Areniello M., Matassa S., Esposito G., Lens P.N.L. Biowaste upcycling into second-generation microbial protein through mixed-culture fermentation. Trends Biotechnol. 2022 doi: 10.1016/j.tibtech.2022.07.008. [DOI] [PubMed] [Google Scholar]
- 79.Ahlborn J., Stephan A., Meckel T., Maheshwari G., Rühl M., Zorn H. Upcycling of food industry side streams by basidiomycetes for production of a vegan protein source. Int. J. Recycl. Org. Waste Agric. 2019;8:447–455. doi: 10.1007/s40093-019-00317-4. [DOI] [Google Scholar]
- 80.Paul E.A. Academic Press; 2014. Soil Microbiology, Ecology and Biochemistry. [Google Scholar]
- 81.Bornyasz M.A., Graham R.C., Allen M.F. Ectomycorrhizae in a soil-weathered granitic bedrock regolith: linking matrix resources to plants. Geoderma. 2005;126:141–160. doi: 10.1016/j.geoderma.2004.11.023. [DOI] [Google Scholar]
- 82.Srikanth M., Sandeep T.S.R.S., Sucharitha K., Godi S. Biodegradation of plastic polymers by fungi: a brief review. Bioresourc. Bioprocess. 2022;9:42. doi: 10.1186/s40643-022-00532-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Cowan A.R., Costanzo C.M., Benham R., Loveridge E.J., Moody S.C. Fungal bioremediation of polyethylene: challenges and perspectives. J. Appl. Microbiol. 2022;132:78–89. doi: 10.1111/jam.15203. [DOI] [PubMed] [Google Scholar]
- 84.Wikandari R., Hasniah N., Taherzadeh M.J. The role of filamentous fungi in advancing the development of a sustainable circular bioeconomy. Bioresour. Technol. 2022;345 doi: 10.1016/j.biortech.2021.126531. [DOI] [PubMed] [Google Scholar]
- 85.More T.T., Yan S., Tyagi R.D., Surampalli R.Y. Potential use of filamentous fungi for wastewater sludge treatment. Bioresour. Technol. 2010;101:7691–7700. doi: 10.1016/j.biortech.2010.05.033. [DOI] [PubMed] [Google Scholar]
- 86.Burgstaller W., Schinner F. Leaching of metals with fungi. J. Biotechnol. 1993;27:91–116. doi: 10.1016/0168-1656(93)90101-R. [DOI] [Google Scholar]
- 87.Gadd G.M. In: Advances in Microbial Physiology. Poole R.K., editor. Academic Press; 1999. Fungal production of citric and oxalic acid: importance in metal speciation, physiology and biogeochemical processes; pp. 47–92. [DOI] [PubMed] [Google Scholar]
- 88.Darrah P.R., Tlalka M., Ashford A., Watkinson S.C., Fricker M.D. The vacuole system is a significant intracellular pathway for longitudinal solute transport in basidiomycete fungi. Eukaryot. Cell. 2006;5:1111–1125. doi: 10.1128/EC.00026-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Gray S.N. Fungi as potential bioremediation agents in soil contaminated with heavy or radioactive metals. Biochem. Soc. Trans. 1998;26:666–670. doi: 10.1042/bst0260666. [DOI] [PubMed] [Google Scholar]
- 90.Mehra R.K., Winge D.R. Metal ion resistance in fungi: molecular mechanisms and their regulated expression. J. Cell. Biochem. 1991;45:30–40. doi: 10.1002/jcb.240450109. [DOI] [PubMed] [Google Scholar]
- 91.González-Chávez M.C., Carrillo-González R., Wright S.F., Nichols K.A. The role of glomalin, a protein produced by arbuscular mycorrhizal fungi, in sequestering potentially toxic elements. Environ. Pollut. 2004;130:317–323. doi: 10.1016/j.envpol.2004.01.004. [DOI] [PubMed] [Google Scholar]
- 92.Volesky B., Holan Z.R. Biosorption of heavy metals. Biotechnol. Prog. 1995;11:235–250. doi: 10.1021/bp00033a001. [DOI] [PubMed] [Google Scholar]
- 93.Renshaw J.C., Robson G.D., Trinci A.P.J., Wiebe M.G., Livens F.R., Collison D., Taylor R.J. Fungal siderophores: structures, functions and applications. Mycol. Res. 2002;106:1123–1142. doi: 10.1017/S0953756202006548. [DOI] [Google Scholar]
- 94.Read D.J., Perez-Moreno J. Mycorrhizas and nutrient cycling in ecosystems – a journey towards relevance? New Phytol. 2003;157:475–492. doi: 10.1046/j.1469-8137.2003.00704.x. [DOI] [PubMed] [Google Scholar]
- 95.Jansa J., Finlay R., Wallander H., Smith F.A., Smith S.E. In: Phosphorus in Action: Biological Processes in Soil Phosphorus Cycling. Bünemann E., Oberson A., Frossard E., editors. Springer; Berlin, Heidelberg: 2011. Role of mycorrhizal symbioses in phosphorus cycling; pp. 137–168. [DOI] [Google Scholar]
- 96.Govindarajulu M., Pfeffer P.E., Jin H., Abubaker J., Douds D.D., Allen J.W., Bücking H., Lammers P.J., Shachar-Hill Y. Nitrogen transfer in the arbuscular mycorrhizal symbiosis. Nature. 2005;435:819–823. doi: 10.1038/nature03610. [DOI] [PubMed] [Google Scholar]
- 97.Talbot J.M., Allison S.D., Treseder K.K. Decomposers in disguise: mycorrhizal fungi as regulators of soil C dynamics in ecosystems under global change. Funct. Ecol. 2008;22:955–963. doi: 10.1111/j.1365-2435.2008.01402.x. [DOI] [Google Scholar]
- 98.Tedersoo L., Bahram M. Mycorrhizal types differ in ecophysiology and alter plant nutrition and soil processes. Biol. Rev. 2019;94:1857–1880. doi: 10.1111/brv.12538. [DOI] [PubMed] [Google Scholar]
- 99.Soudzilovskaia N.A., van Bodegom P.M., Terrer C., van’t Zelfde M., McCallum I., Luke McCormack M., Fisher J.B., Brundrett M.C., de Sá N.C., Tedersoo L. Global mycorrhizal plant distribution linked to terrestrial carbon stocks. Nat. Commun. 2019;10:5077. doi: 10.1038/s41467-019-13019-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Cameron D.E., Bashor C.J., Collins J.J. A brief history of synthetic biology. Nat. Rev. Microbiol. 2014;12:381–390. doi: 10.1038/nrmicro3239. [DOI] [PubMed] [Google Scholar]
- 101.Meng F., Ellis T. The second decade of synthetic biology: 2010–2020. Nat. Commun. 2020;11:5174. doi: 10.1038/s41467-020-19092-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Voigt C.A. Synthetic biology 2020–2030: six commercially-available products that are changing our world. Nat. Commun. 2020;11:6379. doi: 10.1038/s41467-020-20122-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Park H.-S., Jun S.-C., Han K.-H., Hong S.-B., Yu J.-H. In: Advances in Applied Microbiology. Sariaslani S., Gadd G.M., editors. Academic Press; 2017. Chapter three - diversity, application, and synthetic biology of industrially important Aspergillus fungi; pp. 161–202. [DOI] [PubMed] [Google Scholar]
- 104.Khalil A.S., Collins J.J. Synthetic biology: applications come of age. Nat. Rev. Genet. 2010;11:367–379. doi: 10.1038/nrg2775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Unkles S.E., Valiante V., Mattern D.J., Brakhage A.A. Synthetic biology tools for bioprospecting of natural products in eukaryotes. Chem. Biol. 2014;21:502–508. doi: 10.1016/j.chembiol.2014.02.010. [DOI] [PubMed] [Google Scholar]
- 106.Xiao H., Zhong J.-J. Production of useful terpenoids by higher-fungus cell factory and synthetic biology approaches. Trends Biotechnol. 2016;34:242–255. doi: 10.1016/j.tibtech.2015.12.007. [DOI] [PubMed] [Google Scholar]
- 107.Mattern D.J., Valiante V., Unkles S.E., Brakhage A.A. Synthetic biology of fungal natural products. Front. Microbiol. 2015;6 doi: 10.3389/fmicb.2015.00775. https://www.frontiersin.org/articles/10.3389/fmicb.2015.00775 accessed. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Meyer V., Andersen M.R., Brakhage A.A., Braus G.H., Caddick M.X., Cairns T.C., de Vries R.P., Haarmann T., Hansen K., Hertz-Fowler C., Krappmann S., Mortensen U.H., Peñalva M.A., Ram A.F.J., Head R.M. Current challenges of research on filamentous fungi in relation to human welfare and a sustainable bio-economy: a white paper. Fungal Biol. Biotechnol. 2016;3:6. doi: 10.1186/s40694-016-0024-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Danino Y.M., Even D., Ideses D., Juven-Gershon T. The core promoter: at the heart of gene expression. Biochim. Biophys. Acta (BBA) - Gene Regul. Mech. 2015:1116–1131. doi: 10.1016/j.bbagrm.2015.04.003. 1849. [DOI] [PubMed] [Google Scholar]
- 110.Hossain A., Lopez E., Halper S.M., Cetnar D.P., Reis A.C., Strickland D., Klavins E., Salis H.M. Automated design of thousands of nonrepetitive parts for engineering stable genetic systems. Nat. Biotechnol. 2020;38:1466–1475. doi: 10.1038/s41587-020-0584-2. [DOI] [PubMed] [Google Scholar]
- 111.Nevalainen K.M.H., Te’o V.S.J., Bergquist P.L. Heterologous protein expression in filamentous fungi. Trends Biotechnol. 2005;23:468–474. doi: 10.1016/j.tibtech.2005.06.002. [DOI] [PubMed] [Google Scholar]
- 112.Kolar M., Punt P.J., van den Hondel C.A.M.J.J., Schwab H. Transformation of Penicillium chrysogenum using dominant selection markers and expression of an Escherichia coli lacZ fusion gene. Gene. 1988;62:127–134. doi: 10.1016/0378-1119(88)90586-0. [DOI] [PubMed] [Google Scholar]
- 113.Kluge J., Terfehr D., Kück U. Inducible promoters and functional genomic approaches for the genetic engineering of filamentous fungi. Appl. Microbiol. Biotechnol. 2018;102:6357–6372. doi: 10.1007/s00253-018-9115-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Wanka F., Cairns T., Boecker S., Berens C., Happel A., Zheng X., Sun J., Krappmann S., Meyer V. Tet-on, or Tet-off, that is the question: advanced conditional gene expression in Aspergillus. Fungal Genet. Biol. 2016;89:72–83. doi: 10.1016/j.fgb.2015.11.003. [DOI] [PubMed] [Google Scholar]
- 115.Zhao E.M., Zhang Y., Mehl J., Park H., Lalwani M.A., Toettcher J.E., Avalos J.L. Optogenetic regulation of engineered cellular metabolism for microbial chemical production. Nature. 2018;555:683–687. doi: 10.1038/nature26141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Tan S.Z., Prather K.L. Dynamic pathway regulation: recent advances and methods of construction. Curr. Opin. Chem. Biol. 2017;41:28–35. doi: 10.1016/j.cbpa.2017.10.004. [DOI] [PubMed] [Google Scholar]
- 117.Brophy J.A.N., Voigt C.A. Principles of genetic circuit design. Nat. Methods. 2014;11:508–520. doi: 10.1038/nmeth.2926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Weber E., Engler C., Gruetzner R., Werner S., Marillonnet S. A modular cloning system for standardized assembly of multigene constructs. PLoS One. 2011;6 doi: 10.1371/journal.pone.0016765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Engler C., Kandzia R., Marillonnet S. A one pot, one step, precision cloning method with high throughput capability. PLoS One. 2008;3:e3647. doi: 10.1371/journal.pone.0003647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Aleksenko A., Clutterbuck A.J. Autonomous plasmid replication inAspergillus nidulans:AMA1 and MATE elements. Fungal Genet. Biol. 1997;21:373–387. doi: 10.1006/fgbi.1997.0980. [DOI] [PubMed] [Google Scholar]
- 121.Yu J., Zhang Y., Cui H., Hu P., Yu X., Ye Z. An efficient genetic manipulation protocol for Ustilago esculenta. FEMS (Fed. Eur. Microbiol. Soc.) Microbiol. Lett. 2015;362:fnv087. doi: 10.1093/femsle/fnv087. [DOI] [PubMed] [Google Scholar]
- 122.Li D., Tang Y., Lin J., Cai W. Methods for genetic transformation of filamentous fungi. Microb. Cell Factories. 2017;16:168. doi: 10.1186/s12934-017-0785-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Chen C., Smye S.W., Robinson M.P., Evans J.A. Membrane electroporation theories: a review. Med. Biol. Eng. Comput. 2006;44:5–14. doi: 10.1007/s11517-005-0020-2. [DOI] [PubMed] [Google Scholar]
- 124.Ozeki K., Kyoya F., Fujii, Hizume K., Kanda A., Hamachi M., Nunokawa Y. Transformation of intact Aspergillus Niger by electroporation. Biosci., Biotechnol., Biochem. 1994;58:2224–2227. doi: 10.1271/bbb.58.2224. [DOI] [PubMed] [Google Scholar]
- 125.Michielse C.B., Hooykaas P.J.J., van den Hondel C.A.M.J.J., Ram A.F.J. Agrobacterium-mediated transformation as a tool for functional genomics in fungi. Curr. Genet. 2005;48:1–17. doi: 10.1007/s00294-005-0578-0. [DOI] [PubMed] [Google Scholar]
- 126.Liu Z., Friesen T.L. In: Plant Fungal Pathogens: Methods and Protocols. Bolton M.D., Thomma B.P.H.J., editors. Humana Press; Totowa, NJ: 2012. Polyethylene glycol (PEG)-Mediated transformation in filamentous fungal pathogens; pp. 365–375. [DOI] [PubMed] [Google Scholar]
- 127.Chen B.-X., Wei T., Ye Z.-W., Yun F., Kang L.-Z., Tang H.-B., Guo L.-Q., Lin J.-F. Efficient CRISPR-cas9 gene disruption system in edible-medicinal mushroom cordyceps militaris. Front. Microbiol. 2018;9 doi: 10.3389/fmicb.2018.01157. https://www.frontiersin.org/articles/10.3389/fmicb.2018.01157 accessed. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Wang P.-A., Xiao H., Zhong J.-J. CRISPR-Cas9 assisted functional gene editing in the mushroom Ganoderma lucidum. Appl. Microbiol. Biotechnol. 2020;104:1661–1671. doi: 10.1007/s00253-019-10298-z. [DOI] [PubMed] [Google Scholar]
- 129.Schuster M., Kahmann R. CRISPR-Cas9 genome editing approaches in filamentous fungi and oomycetes. Fungal Genet. Biol. 2019;130:43–53. doi: 10.1016/j.fgb.2019.04.016. [DOI] [PubMed] [Google Scholar]
- 130.Song R., Zhai Q., Sun L., Huang E., Zhang Y., Zhu Y., Guo Q., Tian Y., Zhao B., Lu H. CRISPR/Cas9 genome editing technology in filamentous fungi: progress and perspective. Appl. Microbiol. Biotechnol. 2019;103:6919–6932. doi: 10.1007/s00253-019-10007-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Hille F., Richter H., Wong S.P., Bratovič M., Ressel S., Charpentier E. The biology of CRISPR-cas: backward and forward. Cell. 2018;172:1239–1259. doi: 10.1016/j.cell.2017.11.032. [DOI] [PubMed] [Google Scholar]
- 132.Knott G.J., Doudna J.A. CRISPR-Cas guides the future of genetic engineering. Science. 2018;361:866–869. doi: 10.1126/science.aat5011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Liu G., Lin Q., Jin S., Gao C. The CRISPR-Cas toolbox and gene editing technologies. Mol. Cell. 2022;82:333–347. doi: 10.1016/j.molcel.2021.12.002. [DOI] [PubMed] [Google Scholar]
- 134.Wannier T.M., Ciaccia P.N., Ellington A.D., Filsinger G.T., Isaacs F.J., Javanmardi K., Jones M.A., Kunjapur A.M., Nyerges A., Pal C., Schubert M.G., Church G.M., Recombineering, MAGE Nat. Rev. Methods Primers. 2021;1:1–24. doi: 10.1038/s43586-020-00006-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Barbieri E.M., Muir P., Akhuetie-Oni B.O., Yellman C.M., Isaacs F.J. Precise editing at DNA replication forks enables multiplex genome engineering in eukaryotes. Cell. 2017;171:1453–1467.e13. doi: 10.1016/j.cell.2017.10.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Cain A.K., Barquist L., Goodman A.L., Paulsen I.T., Parkhill J., van Opijnen T. A decade of advances in transposon-insertion sequencing. Nat. Rev. Genet. 2020;21:526–540. doi: 10.1038/s41576-020-0244-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Wetmore K.M., Price M.N., Waters R.J., Lamson J.S., He J., Hoover C.A., Blow M.J., Bristow J., Butland G., Arkin A.P., Deutschbauer A. Rapid quantification of mutant fitness in diverse bacteria by sequencing randomly bar-coded transposons. mBio. 2015;6 doi: 10.1128/mBio.00306-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Schrevens S., Sanglard D. Hijacking transposable elements for saturation mutagenesis in fungi. Front. Fungal Biol. 2021:2. doi: 10.3389/ffunb.2021.633876. https://www.frontiersin.org/articles/10.3389/ffunb.2021.633876 accessed. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Hamer L., Adachi K., Montenegro-Chamorro M.V., Tanzer M.M., Mahanty S.K., Lo C., Tarpey R.W., Skalchunes A.R., Heiniger R.W., Frank S.A., Darveaux B.A., Lampe D.J., Slater T.M., Ramamurthy L., DeZwaan T.M., Nelson G.H., Shuster J.R., Woessner J., Hamer J.E. Gene discovery and gene function assignment in filamentous fungi. Proc. Natl. Acad. Sci. USA. 2001;98:5110–5115. doi: 10.1073/pnas.091094198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Firon A., Villalba F., Beffa R., d'Enfert C. Identification of essential genes in the human fungal pathogen Aspergillus fumigatus by transposon mutagenesis. Eukaryot. Cell. 2003;2:247–255. doi: 10.1128/EC.2.2.247-255.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Hihlal E., Braumann I., van den Berg M., Kempken F. Suitability of vader for transposon-mediated mutagenesis in Aspergillus Niger. Appl. Environ. Microbiol. 2011;77:2332–2336. doi: 10.1128/AEM.02688-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.López-Berges M.S., Di Pietro A., Daboussi M.-J., Wahab H.A., Vasnier C., Roncero M.I.G., Dufresne M., Hera C. Identification of virulence genes in Fusarium oxysporum f. sp. lycopersici by large-scale transposon tagging. Mol. Plant Pathol. 2009;10:95–107. doi: 10.1111/j.1364-3703.2008.00512.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Castilho B.A., Olfson P., Casadaban M.J. Plasmid insertion mutagenesis and lac gene fusion with mini-mu bacteriophage transposons. J. Bacteriol. 1984;158:488–495. doi: 10.1128/jb.158.2.488-495.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Perkins J.B., Youngman P.J. Construction and properties of Tn917-lac, a transposon derivative that mediates transcriptional gene fusions in Bacillus subtilis. Proc. Natl. Acad. Sci. USA. 1986;83:140–144. doi: 10.1073/pnas.83.1.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Calamia J., Manoil C. Lac permease of Escherichia coli: topology and sequence elements promoting membrane insertion. Proc. Natl. Acad. Sci. USA. 1990;87:4937–4941. doi: 10.1073/pnas.87.13.4937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Gregory J.A., Becker E.C., Jung J., Tuwatananurak I., Pogliano K. Transposon assisted gene insertion technology (tagit): a tool for generating fluorescent fusion proteins. PLoS One. 2010;5 doi: 10.1371/journal.pone.0008731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Zagorec M. Steinmetz, Construction of a derivative of Tn917 containing an outward-directed promoter and its use in Bacillus subtilis. Microbiology. 1991;137:107–112. doi: 10.1099/00221287-137-1-107. [DOI] [PubMed] [Google Scholar]
- 148.Teimoori B., Pourianfar H., Jami Moeini M., Janpoor J. Chemically and physically induced mutagenesis in basidiospores ofoyster mushroom Pleurotus ostreatusvar. Florida. 2014;2:915–921. [Google Scholar]
- 149.Leonard C.A., Brown S.D., Hayman J.R. Random mutagenesis of the Aspergillus oryzae genome results in fungal antibacterial activity. Int. J. Microbiol. 2013 doi: 10.1155/2013/901697. 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Nevalainen K.M.H. In: Applied Mycology and Biotechnology. Khachatourians G.G., Arora D.K., editors. Elsevier; 2001. Strain improvement in filamentous fungi-an overview; pp. 289–304. [DOI] [Google Scholar]
- 151.Chadha S., Kale S.p. Simple fluorescence-based high throughput cell viability assay for filamentous fungi. Lett. Appl. Microbiol. 2015;61:238–244. doi: 10.1111/lam.12460. [DOI] [PubMed] [Google Scholar]
- 152.Beneyton T., Wijaya I.P.M., Postros P., Najah M., Leblond P., Couvent A., Mayot E., Griffiths A.D., Drevelle A. High-throughput screening of filamentous fungi using nanoliter-range droplet-based microfluidics. Sci. Rep. 2016;6 doi: 10.1038/srep27223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Molina R.S., Rix G., Mengiste A.A., Álvarez B., Seo D., Chen H., Hurtado J.E., Zhang Q., García-García J.D., Heins Z.J., Almhjell P.J., Arnold F.H., Khalil A.S., Hanson A.D., Dueber J.E., Schaffer D.V., Chen F., Kim S., Fernández L.Á., Shoulders M.D., Liu C.C. In vivo hypermutation and continuous evolution. Nat. Rev. Methods Primers. 2022;2:1–22. doi: 10.1038/s43586-022-00119-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Wong B.G., Mancuso C.P., Kiriakov S., Bashor C.J., Khalil A.S. Precise, automated control of conditions for high-throughput growth of yeast and bacteria with eVOLVER. Nat. Biotechnol. 2018;36:614–623. doi: 10.1038/nbt.4151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Grigoriev I.V., Nikitin R., Haridas S., Kuo A., Ohm R., Otillar R., Riley R., Salamov A., Zhao X., Korzeniewski F., Smirnova T., Nordberg H., Dubchak I., Shabalov I. MycoCosm portal: gearing up for 1000 fungal genomes. Nucleic Acids Res. 2014;42:D699. doi: 10.1093/nar/gkt1183. –D704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Blin K., Shaw S., Steinke K., Villebro R., Ziemert N., Lee S.Y., Medema M.H., Weber T. antiSMASH 5.0: updates to the secondary metabolite genome mining pipeline. Nucleic Acids Res. 2019;47:W81. doi: 10.1093/nar/gkz310. –W87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Khaldi N., Seifuddin F.T., Turner G., Haft D., Nierman W.C., Wolfe K.H., Fedorova N.D. SMURF: genomic mapping of fungal secondary metabolite clusters. Fungal Genet. Biol. 2010;47:736–741. doi: 10.1016/j.fgb.2010.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Skinnider M.A., Dejong C.A., Rees P.N., Johnston C.W., Li H., Webster A.L.H., Wyatt M.A., Magarvey N.A. Genomes to natural products PRediction informatics for secondary metabolomes (PRISM) Nucleic Acids Res. 2015;43:9645–9662. doi: 10.1093/nar/gkv1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Hautbergue T., Jamin E.L., Debrauwer L., Puel O., Oswald I.P. From genomics to metabolomics, moving toward an integrated strategy for the discovery of fungal secondary metabolites. Nat. Prod. Rep. 2018;35:147–173. doi: 10.1039/C7NP00032D. [DOI] [PubMed] [Google Scholar]
- 160.Almeida H., Palys S., Tsang A., Diallo A.B. TOUCAN: a framework for fungal biosynthetic gene cluster discovery. NAR Genom. Bioinfo. 2020;2 doi: 10.1093/nargab/lqaa098. lqaa098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Nielsen J.C., Nielsen J. Development of fungal cell factories for the production of secondary metabolites: linking genomics and metabolism. Synth. Syst. Biotechnol. 2017;2:5–12. doi: 10.1016/j.synbio.2017.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Kjærbølling I., Mortensen U.H., Vesth T., Andersen M.R. Strategies to establish the link between biosynthetic gene clusters and secondary metabolites. Fungal Genet. Biol. 2019;130:107–121. doi: 10.1016/j.fgb.2019.06.001. [DOI] [PubMed] [Google Scholar]
- 163.Kautsar S.A., Blin K., Shaw S., Navarro-Muñoz J.C., Terlouw B.R., van der Hooft J.J.J., van Santen J.A., Tracanna V., Suarez Duran H.G., Pascal Andreu V., Selem-Mojica N., Alanjary M., Robinson S.L., Lund G., Epstein S.C., Sisto A.C., Charkoudian L.K., Collemare J., Linington R.G., Weber T., Medema M.H. MIBiG 2.0: a repository for biosynthetic gene clusters of known function. Nucleic Acids Res. 2020;48:D454. doi: 10.1093/nar/gkz882. –D458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Pi B., Yu D., Dai F., Song X., Zhu C., Li H., Yu Y. A genomics based discovery of secondary metabolite biosynthetic gene clusters in Aspergillus ustus. PLoS One. 2015;10 doi: 10.1371/journal.pone.0116089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Khater S., Anand S., Mohanty D. In silico methods for linking genes and secondary metabolites: the way forward. Synth. Syst. Biotechnol. 2016;1:80–88. doi: 10.1016/j.synbio.2016.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Gao X., Chooi Y.-H., Ames B.D., Wang P., Walsh C.T., Tang Y. Fungal indole alkaloid biosynthesis: genetic and biochemical investigation of the tryptoquialanine pathway in Penicillium aethiopicum. J. Am. Chem. Soc. 2011;133:2729–2741. doi: 10.1021/ja1101085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Robey M.T., Caesar L.K., Drott M.T., Keller N.P., Kelleher N.L. An interpreted atlas of biosynthetic gene clusters from 1,000 fungal genomes. Proc. Natl. Acad. Sci. USA. 2021;118 doi: 10.1073/pnas.2020230118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Lombard V., Golaconda Ramulu H., Drula E., Coutinho P.M., Henrissat B. The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Res. 2014;42:D490. doi: 10.1093/nar/gkt1178. –D495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Kameshwar A.K.S., Ramos L.P., Qin W. CAZymes-based ranking of fungi (CBRF): an interactive web database for identifying fungi with extrinsic plant biomass degrading abilities. Bioresourc. Bioprocess. 2019;6:51. doi: 10.1186/s40643-019-0286-0. [DOI] [Google Scholar]
- 170.Henske J.K., Wilken StE., Solomon K.V., Smallwood C.R., Shutthanandan V., Evans J.E., Theodorou M.K., O'Malley M.A. Metabolic characterization of anaerobic fungi provides a path forward for bioprocessing of crude lignocellulose. Biotechnol. Bioeng. 2018;115:874–884. doi: 10.1002/bit.26515. [DOI] [PubMed] [Google Scholar]
- 171.Miyauchi S., Navarro D., Grisel S., Chevret D., Berrin J.-G., Rosso M.-N. The integrative omics of white-rot fungus Pycnoporus coccineus reveals co-regulated CAZymes for orchestrated lignocellulose breakdown. PLoS One. 2017;12 doi: 10.1371/journal.pone.0175528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Arntzen M.Ø., Bengtsson O., Várnai A., Delogu F., Mathiesen G., Eijsink V.G.H. Quantitative comparison of the biomass-degrading enzyme repertoires of five filamentous fungi. Sci. Rep. 2020;10 doi: 10.1038/s41598-020-75217-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Papoutsakis E.T. Equations and calculations for fermentations of butyric acid bacteria. Biotechnol. Bioeng. 1984;26:174–187. doi: 10.1002/bit.260260210. [DOI] [PubMed] [Google Scholar]
- 174.O'Brien E.J., Monk J.M., Palsson B.O. Using genome-scale models to predict biological capabilities. Cell. 2015;161:971–987. doi: 10.1016/j.cell.2015.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Gottstein W., Olivier B.G., Bruggeman F.J., Teusink B. Constraint-based stoichiometric modelling from single organisms to microbial communities. J. R. Soc. Interface. 2016;13 doi: 10.1098/rsif.2016.0627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Aminian-Dehkordi J., Mousavi S.M., Jafari A., Mijakovic I., Marashi S.-A. Manually curated genome-scale reconstruction of the metabolic network of Bacillus megaterium DSM319. Sci. Rep. 2019;9 doi: 10.1038/s41598-019-55041-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Bernstein D.B., Sulheim S., Almaas E., Segrè D. Addressing uncertainty in genome-scale metabolic model reconstruction and analysis. Genome Biol. 2021;22:64. doi: 10.1186/s13059-021-02289-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Orth J.D., Thiele I., Palsson B.Ø. What is flux balance analysis? Nat. Biotechnol. 2010;28:245–248. doi: 10.1038/nbt.1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Fang X., Lloyd C.J., Palsson B.O. Reconstructing organisms in silico: genome-scale models and their emerging applications. Nat. Rev. Microbiol. 2020;18:731–743. doi: 10.1038/s41579-020-00440-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Simeonidis E., Price N.D. Genome-scale modeling for metabolic engineering. J. Ind. Microbiol. Biotechnol. 2015;42:327–338. doi: 10.1007/s10295-014-1576-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Massaiu I., Pasotti L., Sonnenschein N., Rama E., Cavaletti M., Magni P., Calvio C., Herrgård M.J. Integration of enzymatic data in Bacillus subtilis genome-scale metabolic model improves phenotype predictions and enables in silico design of poly-γ-glutamic acid production strains. Microb. Cell Factories. 2019;18:3. doi: 10.1186/s12934-018-1052-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Mishra P., Lee N.-R., Lakshmanan M., Kim M., Kim B.-G., Lee D.-Y. Genome-scale model-driven strain design for dicarboxylic acid production in Yarrowia lipolytica. BMC Syst. Biol. 2018;12:12. doi: 10.1186/s12918-018-0542-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Banerjee D., Eng T., Lau A.K., Sasaki Y., Wang B., Chen Y., Prahl J.-P., Singan V.R., Herbert R.A., Liu Y., Tanjore D., Petzold C.J., Keasling J.D., Mukhopadhyay A. Genome-scale metabolic rewiring improves titers rates and yields of the non-native product indigoidine at scale. Nat. Commun. 2020;11:5385. doi: 10.1038/s41467-020-19171-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Kanehisa M., Araki M., Goto S., Hattori M., Hirakawa M., Itoh M., Katayama T., Kawashima S., Okuda S., Tokimatsu T., Yamanishi Y. KEGG for linking genomes to life and the environment. Nucleic Acids Res. 2008;36:D480. doi: 10.1093/nar/gkm882. –D484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Caspi R., Billington R., Keseler I.M., Kothari A., Krummenacker M., Midford P.E., Ong W.K., Paley S., Subhraveti P., Karp P.D. The MetaCyc database of metabolic pathways and enzymes - a 2019 update. Nucleic Acids Res. 2020;48:D445. doi: 10.1093/nar/gkz862. –D453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Karlsen E., Schulz C., Almaas E. Automated generation of genome-scale metabolic draft reconstructions based on KEGG. BMC Bioinf. 2018;19:467. doi: 10.1186/s12859-018-2472-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Agren R., Liu L., Shoaie S., Vongsangnak W., Nookaew I., Nielsen J. The RAVEN toolbox and its use for generating a genome-scale metabolic model for Penicillium chrysogenum. PLoS Comput. Biol. 2013;9 doi: 10.1371/journal.pcbi.1002980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Karp P.D., Midford P.E., Billington R., Kothari A., Krummenacker M., Latendresse M., Ong W.K., Subhraveti P., Caspi R., Fulcher C., Keseler I.M., Paley S.M. Pathway Tools version 23.0 update: software for pathway/genome informatics and systems biology. Briefings Bioinf. 2021;22:109–126. doi: 10.1093/bib/bbz104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Seaver S.M.D., Liu F., Zhang Q., Jeffryes J., Faria J.P., Edirisinghe J.N., Mundy M., Chia N., Noor E., Beber M.E., Best A.A., DeJongh M., Kimbrel J.A., D’haeseleer P., McCorkle S.R., Bolton J.R., Pearson E., Canon S., Wood-Charlson E.M., Cottingham R.W., Arkin A.P., Henry C.S. The ModelSEED Biochemistry Database for the integration of metabolic annotations and the reconstruction, comparison and analysis of metabolic models for plants, fungi and microbes. Nucleic Acids Res. 2021;49 doi: 10.1093/nar/gkaa746. D575–D588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Brandl J., Andersen M.R. Current state of genome-scale modeling in filamentous fungi. Biotechnol. Lett. 2015;37:1131–1139. doi: 10.1007/s10529-015-1782-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Pitkänen E., Jouhten P., Hou J., Syed M.F., Blomberg P., Kludas J., Oja M., Holm L., Penttilä M., Rousu J., Arvas M. Comparative genome-scale reconstruction of gapless metabolic networks for present and ancestral species. PLoS Comput. Biol. 2014;10 doi: 10.1371/journal.pcbi.1003465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Ma Z., Ye C., Deng W., Xu M., Wang Q., Liu G., Wang F., Liu L., Xu Z., Shi G., Ding Z. Reconstruction and analysis of a genome-scale metabolic model of Ganoderma lucidum for improved extracellular polysaccharide production. Front. Microbiol. 2018;9 doi: 10.3389/fmicb.2018.03076. https://www.frontiersin.org/articles/10.3389/fmicb.2018.03076 accessed. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Wilken StE., Swift C.L., Podolsky I.A., Lankiewicz T.S., Seppälä S., O'Malley M.A. Linking ‘omics’ to function unlocks the biotech potential of non-model fungi. Curr. Opin. Struct. Biol. 2019;14:9–17. doi: 10.1016/j.coisb.2019.02.001. [DOI] [Google Scholar]
- 194.Burger B.T., Imam S., Scarborough M.J., Noguera D.R., Donohue T.J. Combining genome-scale experimental and computational methods to identify essential genes in rhodobacter sphaeroides. mSystems. 2017;2:e00015–e00017. doi: 10.1128/mSystems.00015-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.diCenzo G.C., Mengoni A., Fondi M. Tn-core: a toolbox for integrating tn-seq gene essentiality data and constraint-based metabolic modeling. ACS Synth. Biol. 2019;8:158–169. doi: 10.1021/acssynbio.8b00432. [DOI] [PubMed] [Google Scholar]
- 196.Blazier A., Papin J. Integration of expression data in genome-scale metabolic network reconstructions. Front. Physiol. 2012:3. doi: 10.3389/fphys.2012.00299. https://www.frontiersin.org/articles/10.3389/fphys.2012.00299 accessed. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Zampieri G., Vijayakumar S., Yaneske E., Angione C. Machine and deep learning meet genome-scale metabolic modeling. PLoS Comput. Biol. 2019;15 doi: 10.1371/journal.pcbi.1007084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Yizhak K., Benyamini T., Liebermeister W., Ruppin E., Shlomi T. Integrating quantitative proteomics and metabolomics with a genome-scale metabolic network model. Bioinformatics. 2010;26 doi: 10.1093/bioinformatics/btq183. i255–i260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Pillay L.C., Nekati L., Makhwitine P.J., Ndlovu S.I. Epigenetic activation of silent biosynthetic gene clusters in endophytic fungi using small molecular modifiers. Front. Microbiol. 2022;13 doi: 10.3389/fmicb.2022.815008. https://www.frontiersin.org/articles/10.3389/fmicb.2022.815008 accessed. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Nützmann H.-W., Fischer J., Scherlach K., Hertweck C., Brakhage A.A. Distinct amino acids of histone H3 control secondary metabolism in Aspergillus nidulans. Appl. Environ. Microbiol. 2013;79:6102–6109. doi: 10.1128/AEM.01578-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Netzker T., Fischer J., Weber J., Mattern D.J., König C.C., Valiante V., Schroeckh V., Brakhage A.A. Microbial communication leading to the activation of silent fungal secondary metabolite gene clusters. Front. Microbiol. 2015:6. doi: 10.3389/fmicb.2015.00299. https://www.frontiersin.org/articles/10.3389/fmicb.2015.00299 accessed. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Oh D.-C., Kauffman C.A., Jensen P.R., Fenical W. Induced production of emericellamides A and B from the marine-derived fungus Emericella sp. in competing co-culture. J. Nat. Prod. 2007;70:515–520. doi: 10.1021/np060381f. [DOI] [PubMed] [Google Scholar]
- 203.Bode H.B., Bethe B., Höfs R., Zeeck A. Big effects from small changes: possible ways to explore nature's chemical diversity. Chembiochem. 2002;3:619–627. doi: 10.1002/1439-7633(20020703)3:7<619::AID-CBIC619>3.0.CO. 2-9. [DOI] [PubMed] [Google Scholar]
- 204.Mózsik L., Iacovelli R., Bovenberg R.A.L., Driessen A.J.M. Transcriptional activation of biosynthetic gene clusters in filamentous fungi. Front. Bioeng. Biotechnol. 2022:10. doi: 10.3389/fbioe.2022.901037. https://www.frontiersin.org/articles/10.3389/fbioe.2022.901037 accessed. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Lubertozzi D., Keasling J.D. Developing Aspergillus as a host for heterologous expression. Biotechnol. Adv. 2009;27:53–75. doi: 10.1016/j.biotechadv.2008.09.001. [DOI] [PubMed] [Google Scholar]
- 206.de Vries R.P., Burgers K., van de Vondervoort P.J.I., Frisvad J.C., Samson R.A., Visser J. A new black Aspergillus species, A. vadensis, is a promising host for homologous and heterologous protein production. Appl. Environ. Microbiol. 2004;70:3954–3959. doi: 10.1128/AEM.70.7.3954-3959.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Harvey C.J.B., Tang M., Schlecht U., Horecka J., Fischer C.R., Lin H.-C., Li J., Naughton B., Cherry J., Miranda M., Li Y.F., Chu A.M., Hennessy J.R., Vandova G.A., Inglis D., Aiyar R.S., Steinmetz L.M., Davis R.W., Medema M.H., Sattely E., Khosla C., Onge R.P. St, Tang Y., Hillenmeyer M.E. HEx: a heterologous expression platform for the discovery of fungal natural products. Sci. Adv. 2018;4 doi: 10.1126/sciadv.aar5459. eaar5459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Jarczynska Z.D., Rendsvig J.K.H., Pagels N., Viana V.R., Nødvig C.S., Kirchner F.H., Strucko T., Nielsen M.L., Mortensen U.H. DIVERSIFY: a fungal multispecies gene expression platform. ACS Synth. Biol. 2021 doi: 10.1021/acssynbio.0c00587. [DOI] [PubMed] [Google Scholar]
- 209.Ikram-ul H., Ali S., Qadeer M.A., Iqbal J. Citric acid production by selected mutants of Aspergillus Niger from cane molasses. Bioresour. Technol. 2004;93:125–130. doi: 10.1016/j.biortech.2003.10.018. [DOI] [PubMed] [Google Scholar]
- 210.Xue X., Bi F., Liu B., Li J., Zhang L., Zhang J., Gao Q., Wang D. Improving citric acid production of an industrial Aspergillus Niger CGMCC 10142: identification and overexpression of a high-affinity glucose transporter with different promoters. Microb. Cell Factories. 2021;20:168. doi: 10.1186/s12934-021-01659-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Meyer V. Metabolic Engineering. John Wiley & Sons; 2021. Metabolic engineering of filamentous fungi; pp. 765–801. [DOI] [Google Scholar]
- 212.Sun X., Su X. Harnessing the knowledge of protein secretion for enhanced protein production in filamentous fungi. World J. Microbiol. Biotechnol. 2019;35:54. doi: 10.1007/s11274-019-2630-0. [DOI] [PubMed] [Google Scholar]
- 213.Sakekar A.A., Gaikwad S.R., Punekar N.S. Protein expression and secretion by filamentous fungi. J. Biosci. 2021;46:5. doi: 10.1007/s12038-020-00120-8. [DOI] [PubMed] [Google Scholar]
- 214.Cairns T.C., Zheng X., Zheng P., Sun J., Meyer V. Moulding the mould: understanding and reprogramming filamentous fungal growth and morphogenesis for next generation cell factories. Biotechnol. Biofuels. 2019;12:77. doi: 10.1186/s13068-019-1400-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Lommel M., Strahl S. Protein O-mannosylation: conserved from bacteria to humans. Glycobiology. 2009;19:816–828. doi: 10.1093/glycob/cwp066. [DOI] [PubMed] [Google Scholar]
- 216.Rabouille C. Pathways of unconventional protein secretion. Trends Cell Biol. 2017;27:230–240. doi: 10.1016/j.tcb.2016.11.007. [DOI] [PubMed] [Google Scholar]
- 217.Shoji J., Kikuma T., Kitamoto K. Vesicle trafficking, organelle functions, and unconventional secretion in fungal physiology and pathogenicity. Curr. Opin. Microbiol. 2014;20:1–9. doi: 10.1016/j.mib.2014.03.002. [DOI] [PubMed] [Google Scholar]
- 218.Hayakawa Y., Ishikawa E., Shoji J., Nakano H., Kitamoto K. Septum-directed secretion in the filamentous fungus Aspergillus oryzae. Mol. Microbiol. 2011;81:40–55. doi: 10.1111/j.1365-2958.2011.07700.x. [DOI] [PubMed] [Google Scholar]
- 219.Fiedler M.R.M., Barthel L., Kubisch C., Nai C., Meyer V. Construction of an improved Aspergillus Niger platform for enhanced glucoamylase secretion. Microb. Cell Factories. 2018;17:95. doi: 10.1186/s12934-018-0941-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Te’o V.S.J., Cziferszky A.E., Bergquist P.L., Nevalainen K.M.H. Codon optimization of xylanase gene xynB from the thermophilic bacterium Dictyoglomus thermophilum for expression in the filamentous fungus Trichoderma reesei. FEMS (Fed. Eur. Microbiol. Soc.) Microbiol. Lett. 2000;190:13–19. doi: 10.1111/j.1574-6968.2000.tb09255.x. [DOI] [PubMed] [Google Scholar]
- 221.van den Hombergh J.P.T.W., van de Vondervoort P.J.I., Fraissinet-Tachet L., Visser J. Aspergillus as a host for heterologous protein production: the problem of proteases. Trends Biotechnol. 1997;15:256–263. doi: 10.1016/S0167-7799(97)01020-2. [DOI] [PubMed] [Google Scholar]
- 222.Mattern I.E., van Noort J.M., van den Berg P., Archer D.B., Roberts I.N., van den Hondel C.A.M.J.J. Isolation and characterization of mutants of Aspergillus Niger deficient in extracellular proteases. Mol. Gen. Genet. 1992;234:332–336. doi: 10.1007/BF00283855. [DOI] [PubMed] [Google Scholar]
- 223.Reilly M.C., Kim J., Lynn J., Simmons B.A., Gladden J.M., Magnuson J.K., Baker S.E. Forward genetics screen coupled with whole-genome resequencing identifies novel gene targets for improving heterologous enzyme production in Aspergillus Niger. Appl. Microbiol. Biotechnol. 2018;102:1797–1807. doi: 10.1007/s00253-017-8717-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Kamaruddin N., Storms R., Mahadi N.M., Illias R.Md, Bakar F.D.A., Murad A.M.A. Reduction of extracellular proteases increased activity and stability of heterologous protein in $${ Aspergillus}$$$${ Niger. Arabian J. Sci. Eng. 2018;43:3327–3338. doi: 10.1007/s13369-017-2914-3. [DOI] [Google Scholar]
- 225.Chen L., Zou G., Zhang L., de Vries R.P., Yan X., Zhang J., Liu R., Wang C., Qu Y., Zhou Z. The distinctive regulatory roles of PrtT in the cell metabolism of Penicillium oxalicum. Fungal Genet. Biol. 2014;63:42–54. doi: 10.1016/j.fgb.2013.12.001. [DOI] [PubMed] [Google Scholar]
- 226.Wei H., Wu M., Fan A., Su H. Recombinant protein production in the filamentous fungus Trichoderma. Chin. J. Chem. Eng. 2021;30:74–81. doi: 10.1016/j.cjche.2020.11.006. [DOI] [Google Scholar]
- 227.Punt P.J., van Biezen N., Conesa A., Albers A., Mangnus J., van den Hondel C. Filamentous fungi as cell factories for heterologous protein production. Trends Biotechnol. 2002;20:200–206. doi: 10.1016/S0167-7799(02)01933-9. [DOI] [PubMed] [Google Scholar]
- 228.Nevalainen H., Peterson R. Making recombinant proteins in filamentous fungi- are we expecting too much? Front. Microbiol. 2014:5. doi: 10.3389/fmicb.2014.00075. https://www.frontiersin.org/articles/10.3389/fmicb.2014.00075 accessed. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Ward O.P. Production of recombinant proteins by filamentous fungi. Biotechnol. Adv. 2012;30:1119–1139. doi: 10.1016/j.biotechadv.2011.09.012. [DOI] [PubMed] [Google Scholar]
- 230.Nummi M., Niku-Paavola M.L., Lappalainen A., Enari T.M., Raunio V. Cellobiohydrolase from Trichoderma reesei. Biochem. J. 1983;215:677–683. doi: 10.1042/bj2150677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Karhunen T., Mäntylä A., Nevalainen K.M.H., Suominen P.L. High frequency one-step gene replacement in Trichoderma reesei. I. Endoglucanase I overproduction. Mol. Gen. Genet. 1993;241:515–522. doi: 10.1007/BF00279893. [DOI] [PubMed] [Google Scholar]
- 232.Joutsjoki V.V. Construction by one-step gene replacement of Trichoderma reesei strains that produce the glucoamylase P of Hormoconis resinae. Curr. Genet. 1994;26:422–429. doi: 10.1007/BF00309929. [DOI] [PubMed] [Google Scholar]
- 233.Miettinen-Oinonen A., Suominen P. Enhanced production of Trichoderma reesei endoglucanases and use of the new cellulase preparations in producing the stonewashed effect on denim fabric. Appl. Environ. Microbiol. 2002;68:3956–3964. doi: 10.1128/AEM.68.8.3956-3964.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Miettinen-Oinonen A., Torkkeli T., Paloheimo M., Nevalainen Helena. Overexpression of the Aspergillus Niger pH 2.5 acid phosphatase gene in a heterologous host Trichoderma reesei. J. Biotechnol. 1997;58:13–20. doi: 10.1016/S0168-1656(97)00121-1. [DOI] [PubMed] [Google Scholar]
- 235.Wang Q., Zhong C., Xiao H. Genetic engineering of filamentous fungi for efficient protein expression and secretion. Front. Bioeng. Biotechnol. 2020;8 doi: 10.3389/fbioe.2020.00293. https://www.frontiersin.org/articles/10.3389/fbioe.2020.00293 accessed. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Ward M., Wilson L.J., Kodama K.H., Rey M.W., Berka R.M. Improved production of chymosin in Aspergillus by expression as a glucoamylase-chymosin fusion. Nat. Biotechnol. 1990;8:435–440. doi: 10.1038/nbt0590-435. [DOI] [PubMed] [Google Scholar]
- 237.Krasevec N., Milunović T., Lasnik M.A., Lukančič I., Komel R., Porekar V.G. Human granulocyte colony stimulating factor (G-CSF) produced in the filamentous fungus Aspergillus Niger. Acta Chim. Slov. 2014;61:709–717. [PubMed] [Google Scholar]
- 238.Ohno A., Maruyama J., Nemoto T., Arioka M., Kitamoto K. A carrier fusion significantly induces unfolded protein response in heterologous protein production by Aspergillus oryzae. Appl. Microbiol. Biotechnol. 2011;92:1197–1206. doi: 10.1007/s00253-011-3487-9. [DOI] [PubMed] [Google Scholar]
- 239.Alcocer M.J.C., Furniss C.S.M., Kroon P.A., Campbell M., Archer D.B. Comparison of modular and non-modular xylanases as carrier proteins for the efficient secretion of heterologous proteins from Penicillium funiculosum. Appl. Microbiol. Biotechnol. 2003;60:726–732. doi: 10.1007/s00253-002-1184-4. [DOI] [PubMed] [Google Scholar]
- 240.Conesa A., van den Hondel C.A.M.J.J., Punt P.J. Studies on the production of fungal peroxidases inAspergillus Niger. Appl. Environ. Microbiol. 2000;66:3016–3023. doi: 10.1128/AEM.66.7.3016-3023.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Koseki T., Otsuka M., Mizuno T., Shiono Y. Mutational analysis of Kex2 recognition sites and a disulfide bond in tannase from Aspergillus oryzae. Biochem. Biophys. Res. Commun. 2017;482:1165–1169. doi: 10.1016/j.bbrc.2016.12.006. [DOI] [PubMed] [Google Scholar]
- 242.Zoglowek M., Lübeck P.S., Ahring B.K., Lübeck M. Heterologous expression of cellobiohydrolases in filamentous fungi – an update on the current challenges, achievements and perspectives. Process Biochem. 2015;50:211–220. doi: 10.1016/j.procbio.2014.12.018. [DOI] [Google Scholar]
- 243.Adney W.S., Chou Y.-C., Decker S.R., Ding S.-Y., Baker J.O., Kunkel G., Vinzant T.B., Himmel M.E. Applications of Enzymes to Lignocellulosics. American Chemical Society; 2003. Heterologous expression of Trichoderma reesei 1,4-β-D-Glucan cellobiohydrolase (cel 7A) pp. 403–437. [DOI] [Google Scholar]
- 244.Chou Y.-C., Adney W.S., Decker S.R., Baker J.O., Kunkel G., Templeton D.W., Himmel M.E. Lignocellulose Biodegradation. American Chemical Society; 2004. Cloning and heterologous expression of the gene encoding a family 7 glycosyl hydrolase from Penicillium funiculosum; pp. 170–193. [DOI] [Google Scholar]
- 245.Matoba S., Ogrydziak D.M. Another factor besides hydrophobicity can affect signal peptide interaction with signal recognition particle. J. Biol. Chem. 1998;273:18841–18847. doi: 10.1074/jbc.273.30.18841. [DOI] [PubMed] [Google Scholar]
- 246.Gasser B., Saloheimo M., Rinas U., Dragosits M., Rodríguez-Carmona E., Baumann K., Giuliani M., Parrilli E., Branduardi P., Lang C., Porro D., Ferrer P., Tutino M.L., Mattanovich D., Villaverde A. Protein folding and conformational stress in microbial cells producing recombinant proteins: a host comparative overview. Microb. Cell Factories. 2008;7:11. doi: 10.1186/1475-2859-7-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Almanza A., Carlesso A., Chintha C., Creedican S., Doultsinos D., Leuzzi B., Luís A., McCarthy N., Montibeller L., More S., Papaioannou A., Püschel F., Sassano M.L., Skoko J., Agostinis P., de Belleroche J., Eriksson L.A., Fulda S., Gorman A.M., Healy S., Kozlov A., Muñoz-Pinedo C., Rehm M., Chevet E., Samali A. Endoplasmic reticulum stress signalling – from basic mechanisms to clinical applications. FEBS J. 2019;286:241–278. doi: 10.1111/febs.14608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Bernasconi R., Molinari M. ERAD and ERAD tuning: disposal of cargo and of ERAD regulators from the mammalian ER. Curr. Opin. Cell Biol. 2011;23:176–183. doi: 10.1016/j.ceb.2010.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.Wang G., Zhang D., Chen S. Effect of earlier unfolded protein response and efficient protein disposal system on cellulase production in Rut C30. World J. Microbiol. Biotechnol. 2014;30:2587–2595. doi: 10.1007/s11274-014-1682-4. [DOI] [PubMed] [Google Scholar]
- 250.Pakula T.M., Laxell M., Huuskonen A., Uusitalo J., Saloheimo M., Penttilä M. The effects of drugs inhibiting protein secretion in the filamentous fungus Trichoderma reesei: evidence for down-regulation of genes that encode secreted proteins in the stressed cells. J. Biol. Chem. 2003;278:45011–45020. doi: 10.1074/jbc.M302372200. [DOI] [PubMed] [Google Scholar]
- 251.Saloheimo M., Valkonen M., Penttilä M. Activation mechanisms of the HACI-mediated unfolded protein response in filamentous fungi. Mol. Microbiol. 2003;47:1149–1161. doi: 10.1046/j.1365-2958.2003.03363.x. [DOI] [PubMed] [Google Scholar]
- 252.Bravo R., Parra V., Gatica D., Rodriguez A.E., Torrealba N., Paredes F., Wang Z.V., Zorzano A., Hill J.A., Jaimovich E., Quest A.F.G., Lavandero S. In: International Review of Cell and Molecular Biology. Jeon K.W., editor. Academic Press; 2013. Chapter five - endoplasmic reticulum and the unfolded protein response: dynamics and metabolic integration; pp. 215–290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Moralejo F., Watson A., Jeenes D., Archer D., Martín J. A defined level of protein disulfide isomerase expression is required for optimal secretion of thaumatin by Aspergillus awamori. Mol. Genet. Genom. 2001;266:246–253. doi: 10.1007/s004380100550. [DOI] [PubMed] [Google Scholar]
- 254.Ngiam C., Jeenes D.J., Punt P.J., Van Den Hondel C.A.M.J.J., Archer D.B. Characterization of a foldase, protein disulfide isomerase A, in the protein secretory pathway ofAspergillus Niger. Appl. Environ. Microbiol. 2000;66:775–782. doi: 10.1128/AEM.66.2.775-782.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Wu Y., Sun X., Xue X., Luo H., Yao B., Xie X., Su X. Overexpressing key component genes of the secretion pathway for enhanced secretion of an Aspergillus Niger glucose oxidase in Trichoderma reesei. Enzym. Microb. Technol. 2017;106:83–87. doi: 10.1016/j.enzmictec.2017.07.007. [DOI] [PubMed] [Google Scholar]
- 256.Lombraña M., Moralejo F.J., Pinto R., Martín J.F. Modulation of Aspergillus awamori thaumatin secretion by modification of bipA gene expression. Appl. Environ. Microbiol. 2004;70:5145–5152. doi: 10.1128/AEM.70.9.5145-5152.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257.Conesa A., Jeenes D., Archer D.B., van den Hondel C.A.M.J.J., Punt P.J. Calnexin overexpression increases manganese peroxidase production in Aspergillus Niger. Appl. Environ. Microbiol. 2002;68:846–851. doi: 10.1128/AEM.68.2.846-851.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.Valkonen M., Ward M., Wang H., Penttilä M., Saloheimo M. Improvement of foreign-protein production in Aspergillus Niger var. awamori by constitutive induction of the unfolded-protein response. Appl. Environ. Microbiol. 2003;69:6979–6986. doi: 10.1128/AEM.69.12.6979-6986.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259.Carvalho N.D.S.P., Arentshorst M., Kooistra R., Stam H., Sagt C.M., van den Hondel C.A.M.J.J., Ram A.F.J. Effects of a defective ERAD pathway on growth and heterologous protein production in Aspergillus Niger. Appl. Microbiol. Biotechnol. 2011;89:357–373. doi: 10.1007/s00253-010-2916-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.Richie D.L., Feng X., Hartl L., Aimanianda V., Krishnan K., Powers-Fletcher M.V., Watson D.S., Galande A.K., White S.M., Willett T., Latgé J.-P., Rhodes J.C., Askew D.S. The virulence of the opportunistic fungal pathogen Aspergillus fumigatus requires cooperation between the endoplasmic reticulum-associated degradation pathway (ERAD) and the unfolded protein response (UPR) Virulence. 2011;2:12–21. doi: 10.4161/viru.2.1.13345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261.Karnaukhova E., Ophir Y., Trinh L., Dalal N., Punt P.J., Golding B., Shiloach J. Expression of human α1-proteinase inhibitor in Aspergillus Niger. Microb. Cell Factories. 2007;6:34. doi: 10.1186/1475-2859-6-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262.Apweiler R., Hermjakob H., Sharon N. On the frequency of protein glycosylation, as deduced from analysis of the SWISS-PROT database11Dedicated to Prof. Akira Kobata and Prof. Harry Schachter on the occasion of their 65th birthdays. Biochim. Biophys. Acta Gen. Subj. 1999;1473:4–8. doi: 10.1016/S0304-4165(99)00165-8. [DOI] [PubMed] [Google Scholar]
- 263.De Pourcq K., De Schutter K., Callewaert N. Engineering of glycosylation in yeast and other fungi: current state and perspectives. Appl. Microbiol. Biotechnol. 2010;87:1617–1631. doi: 10.1007/s00253-010-2721-1. [DOI] [PubMed] [Google Scholar]
- 264.Deshpande N., Wilkins M.R., Packer N., Nevalainen H. Protein glycosylation pathways in filamentous fungi. Glycobiology. 2008;18:626–637. doi: 10.1093/glycob/cwn044. [DOI] [PubMed] [Google Scholar]
- 265.Brooks S.A. Appropriate glycosylation of recombinant proteins for human use. Mol. Biotechnol. 2004;28:241–255. doi: 10.1385/MB:28:3:241. [DOI] [PubMed] [Google Scholar]
- 266.Kruszewska J.S., Perlińska-Lenart U., Górka-Nieć W., Orłowski J., Zembek P., Palamarczyk G. Alterations in protein secretion caused by metabolic engineering of glycosylation pathways in fungi. Acta Biochim. Pol. 2008;55:447–456. doi: 10.18388/abp.2008_3050. [DOI] [PubMed] [Google Scholar]
- 267.Bergquist P.L., Te’o V.S.J., Gibbs M.D., Curach N.C., Nevalainen K.M.H. Recombinant enzymes from thermophilic micro-organisms expressed in fungal hosts. Biochem. Soc. Trans. 2004;32:293–297. doi: 10.1042/bst0320293. [DOI] [PubMed] [Google Scholar]
- 268.Stals I., Sandra K., Geysens S., Contreras R., Van Beeumen J., Claeyssens M. Factors influencing glycosylation of Trichoderma reesei cellulases. I: postsecretorial changes of the O- and N-glycosylation pattern of Cel7A. Glycobiology. 2004;14:713–724. doi: 10.1093/glycob/cwh080. [DOI] [PubMed] [Google Scholar]
- 269.Berka R.M., Kodama K.H., Rey M.W., Wilson L.J., Ward M. The development of Aspergillus Niger var. awamori as a host for the expression and secretion of heterologous gene products. Biochem. Soc. Trans. 1991;19:681–685. doi: 10.1042/bst0190681. [DOI] [PubMed] [Google Scholar]
- 270.Qi F., Zhang W., Zhang F., Chen G., Liu W. Deciphering the effect of the different N-glycosylation sites on the secretion, activity, and stability of cellobiohydrolase I from Trichoderma reesei. Appl. Environ. Microbiol. 2014;80:3962–3971. doi: 10.1128/AEM.00261-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271.Jacobs D.I., Olsthoorn M.M.A., Maillet I., Akeroyd M., Breestraat S., Donkers S., van der Hoeven R.A.M., van den Hondel C.A.M.J.J., Kooistra R., Lapointe T., Menke H., Meulenberg R., Misset M., Müller W.H., van Peij N.N.M.E., Ram A., Rodriguez S., Roelofs M.S., Roubos J.A., van Tilborg M.W.E.M., Verkleij A.J., Pel H.J., Stam H., Sagt C.M.J. Effective lead selection for improved protein production in Aspergillus Niger based on integrated genomics. Fungal Genet. Biol. 2009;46 doi: 10.1016/j.fgb.2008.08.012. S141–S152. [DOI] [PubMed] [Google Scholar]
- 272.Zhong Y., Liu X., Xiao P., Wei S., Wang T. Expression and secretion of the human erythropoietin using an optimized cbh1 promoter and the native CBH I signal sequence in the industrial fungus Trichoderma reesei. Appl. Biochem. Biotechnol. 2011;165:1169–1177. doi: 10.1007/s12010-011-9334-8. [DOI] [PubMed] [Google Scholar]
- 273.Kainz E., Gallmetzer A., Hatzl C., Nett J.H., Li H., Schinko T., Pachlinger R., Berger H., Reyes-Dominguez Y., Bernreiter A., Gerngross T., Wildt S., Strauss J. N-glycan modification in Aspergillus species. Appl. Environ. Microbiol. 2008;74:1076–1086. doi: 10.1128/AEM.01058-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 274.Gerngross T.U. Advances in the production of human therapeutic proteins in yeasts and filamentous fungi. Nat. Biotechnol. 2004;22:1409–1414. doi: 10.1038/nbt1028. [DOI] [PubMed] [Google Scholar]
- 275.Gibbs P.A., Seviour R.J., Schmid F. Growth of filamentous fungi in submerged culture: problems and possible solutions. Crit. Rev. Biotechnol. 2000;20:17–48. doi: 10.1080/07388550091144177. [DOI] [PubMed] [Google Scholar]
- 276.Driouch H., Hänsch R., Wucherpfennig T., Krull R., Wittmann C. Improved enzyme production by bio-pellets of Aspergillus Niger: targeted morphology engineering using titanate microparticles. Biotechnol. Bioeng. 2012;109:462–471. doi: 10.1002/bit.23313. [DOI] [PubMed] [Google Scholar]
- 277.Kurt T., Marbà-Ardébol A.-M., Turan Z., Neubauer P., Junne S., Meyer V. Rocking Aspergillus: morphology-controlled cultivation of Aspergillus Niger in a wave-mixed bioreactor for the production of secondary metabolites. Microb. Cell Factories. 2018;17:128. doi: 10.1186/s12934-018-0975-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 278.Lin L., Sun Z., Li J., Chen Y., Liu Q., Sun W., Tian C. Disruption of gul-1 decreased the culture viscosity and improved protein secretion in the filamentous fungus Neurospora crassa. Microb. Cell Factories. 2018;17:96. doi: 10.1186/s12934-018-0944-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 279.Herold I., Yarden O. Regulation of Neurospora crassa cell wall remodeling via the cot-1 pathway is mediated by gul-1. Curr. Genet. 2017;63:145–159. doi: 10.1007/s00294-016-0625-z. [DOI] [PubMed] [Google Scholar]
- 280.Herold I., Zolti A., Garduño-Rosales M., Wang Z., López-Giráldez F., Mouriño-Pérez R.R., Townsend J.P., Ulitsky I., Yarden O. The GUL-1 protein binds multiple RNAs involved in cell wall remodeling and affects the MAK-1 pathway in Neurospora crassa. Front. Fungal Biol. 2021:2. doi: 10.3389/ffunb.2021.672696. https://www.frontiersin.org/articles/10.3389/ffunb.2021.672696 accessed. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 281.Meyer V., Wanka F., van Gent J., Arentshorst M., van den Hondel C.A.M.J.J., Ram A.F.J. Fungal gene expression on demand: an inducible, tunable, and metabolism-independent expression system for Aspergillus Niger. Appl. Environ. Microbiol. 2011;77:2975–2983. doi: 10.1128/AEM.02740-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 282.Papagianni M., Mattey M. Morphological development of Aspergillus Niger in submerged citric acid fermentation as a function of the spore inoculum level. Application of neural network and cluster analysis for characterization of mycelial morphology. Microb. Cell Factories. 2006;5:3. doi: 10.1186/1475-2859-5-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 283.Yin X., Shin H., Li J., Du G., Liu L., Chen J. Comparative genomics and transcriptome analysis of Aspergillus Niger and metabolic engineering for citrate production. Sci. Rep. 2017;7 doi: 10.1038/srep41040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 284.Upton D.J., McQueen-Mason S.J., Wood A.J. An accurate description of Aspergillus Niger organic acid batch fermentation through dynamic metabolic modelling. Biotechnol. Biofuels. 2017;10:258. doi: 10.1186/s13068-017-0950-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 285.Steiger M.G., Rassinger A., Mattanovich D., Sauer M. Engineering of the citrate exporter protein enables high citric acid production in Aspergillus Niger. Metab. Eng. 2019;52:224–231. doi: 10.1016/j.ymben.2018.12.004. [DOI] [PubMed] [Google Scholar]
- 286.Gonciarz J., Bizukojc M. Adding talc microparticles to Aspergillus terreus ATCC 20542 preculture decreases fungal pellet size and improves lovastatin production. Eng. Life Sci. 2014;14:190–200. doi: 10.1002/elsc.201300055. [DOI] [Google Scholar]
- 287.de Groot P.W., Ruiz C., Vázquez de Aldana C.R., Duenas E., Cid V.J., Del Rey F., Rodríquez-Peña J.M., Pérez P., Andel A., Caubín J., Arroyo J., García J.C., Gil C., Molina M., García L.J., Nombela C., Klis F.M. A genomic approach for the identification and classification of genes involved in cell wall formation and its regulation in Saccharomyces cerevisiae. Comp. Funct. Genom. 2001;2:124–142. doi: 10.1002/cfg.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 288.Gow N.A.R., Lenardon M.D. Architecture of the dynamic fungal cell wall. Nat. Rev. Microbiol. 2022:1–12. doi: 10.1038/s41579-022-00796-9. [DOI] [PubMed] [Google Scholar]
- 289.Gow N.A.R., Latge J.-P., Munro C.A. The fungal cell wall: structure, biosynthesis, and function. Microbiol. Spectr. 2017;5 doi: 10.1128/microbiolspec.FUNK-0035-2016. [DOI] [PubMed] [Google Scholar]
- 290.Pelkmans J.F., Patil M.B., Gehrmann T., Reinders M.J.T., Wösten H.A.B., Lugones L.G. Transcription factors of Schizophyllum commune involved in mushroom formation and modulation of vegetative growth. Sci. Rep. 2017;7:310. doi: 10.1038/s41598-017-00483-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 291.Wösten H.A.B., van Veluw G.J., de Bekker C., Krijgsheld P. Heterogeneity in the mycelium: implications for the use of fungi as cell factories. Biotechnol. Lett. 2013;35:1155–1164. doi: 10.1007/s10529-013-1210-x. [DOI] [PubMed] [Google Scholar]
- 292.Krijgsheld P., Nitsche B.M., Post H., Levin A.M., Müller W.H., Heck A.J.R., Ram A.F.J., Altelaar A.F.M., Wösten H.A.B. Deletion of flbA results in increased secretome complexity and reduced secretion heterogeneity in colonies of Aspergillus Niger. J. Proteome Res. 2013;12:1808–1819. doi: 10.1021/pr301154w. [DOI] [PubMed] [Google Scholar]
- 293.Aerts D. Utrecht University; 2018. Wosten, Han, University Utrecht, Sub Molecular Microbiology, Regulators Controlled by the Sporulation Gene flbA of Aspergillus niger.https://dspace.library.uu.nl/handle/1874/363348 accessed. [Google Scholar]
- 294.Krijgsheld P., Wosten H. Transcriptome analysis of zones of colonies of the ΔflbA strain of Aspergillus Niger. Fungal Genom. Biol. 2013;3:109. doi: 10.4172/2165-8056.1000109. [DOI] [Google Scholar]
- 295.Alazi E., Ram A.F.J. Modulating transcriptional regulation of plant biomass degrading enzyme networks for rational design of industrial fungal strains. Front. Bioeng. Biotechnol. 2018:6. doi: 10.3389/fbioe.2018.00133. https://www.frontiersin.org/articles/10.3389/fbioe.2018.00133 accessed. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 296.Niu J., Alazi E., Reid I.D., Arentshorst M., Punt P.J., Visser J., Tsang A., Ram A.F.J. An evolutionarily conserved transcriptional activator-repressor module controls expression of genes for D-galacturonic acid utilization in Aspergillus Niger. Genetics. 2017;205:169–183. doi: 10.1534/genetics.116.194050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 297.Hasper A.A., Visser J., De Graaff L.H. The Aspergillus Niger transcriptional activator XlnR, which is involved in the degradation of the polysaccharides xylan and cellulose, also regulates d-xylose reductase gene expression. Mol. Microbiol. 2000;36:193–200. doi: 10.1046/j.1365-2958.2000.01843.x. [DOI] [PubMed] [Google Scholar]
- 298.Bok J.W., Keller N.P. LaeA, a regulator of secondary metabolism in Aspergillus spp. Eukaryot. Cell. 2004;3:527–535. doi: 10.1128/EC.3.2.527-535.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 299.Bayram Ö., Krappmann S., Ni M., Bok J.W., Helmstaedt K., Valerius O., Braus-Stromeyer S., Kwon N.-J., Keller N.P., Yu J.-H., Braus G.H. VelB/VeA/LaeA complex coordinates light signal with fungal development and secondary metabolism. Science. 2008;320:1504–1506. doi: 10.1126/science.1155888. [DOI] [PubMed] [Google Scholar]
- 300.Salo O.V., Ries M., Medema M.H., Lankhorst P.P., Vreeken R.J., Bovenberg R.A.L., Driessen A.J.M. Genomic mutational analysis of the impact of the classical strain improvement program on β–lactam producing Penicillium chrysogenum. BMC Genom. 2015;16:937. doi: 10.1186/s12864-015-2154-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 301.Niu J., Arentshorst M., Nair P.D.S., Dai Z., Baker S.E., Frisvad J.C., Nielsen K.F., Punt P.J., Ram A.F.J. Identification of a classical mutant in the industrial host Aspergillus Niger by systems genetics: LaeA is required for citric acid production and regulates the formation of some secondary metabolites. G3 (Bethesda) 2015;6:193–204. doi: 10.1534/g3.115.024067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 302.Karimi-Aghcheh R., Bok J.W., Phatale P.A., Smith K.M., Baker S.E., Lichius A., Omann M., Zeilinger S., Seiboth B., Rhee C., Keller N.P., Freitag M., Kubicek C.P. Functional analyses of Trichoderma reesei LAE1 reveal conserved and contrasting roles of this regulator. G3 (Bethesda) 2013;3:369–378. doi: 10.1534/g3.112.005140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 303.Hu P., Wang Y., Zhou J., Pan Y., Liu G. AcstuA, which encodes an APSES transcription regulator, is involved in conidiation, cephalosporin biosynthesis and cell wall integrity of Acremonium chrysogenum. Fungal Genet. Biol. 2015;83:26–40. doi: 10.1016/j.fgb.2015.08.003. [DOI] [PubMed] [Google Scholar]
- 304.Kwon M.J., Jørgensen T.R., Nitsche B.M., Arentshorst M., Park J., Ram A.F., Meyer V. The transcriptomic fingerprint of glucoamylase over-expression in Aspergillus Niger. BMC Genom. 2012;13:701. doi: 10.1186/1471-2164-13-701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 305.Meyer V., Arentshorst M., Flitter S.J., Nitsche B.M., Kwon M.J., Reynaga-Peña C.G., Bartnicki-Garcia S., van den Hondel C.A.M.J.J., Ram A.F.J. Reconstruction of signaling networks regulating fungal morphogenesis by transcriptomics. Eukaryot. Cell. 2009;8:1677–1691. doi: 10.1128/EC.00050-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 306.Kwon M.J., Nitsche B.M., Arentshorst M., Jørgensen T.R., Ram A.F.J., Meyer V. The transcriptomic signature of RacA activation and inactivation provides new insights into the morphogenetic network of Aspergillus Niger. PLoS One. 2013;8 doi: 10.1371/journal.pone.0068946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 307.Wang M., Zhang M., Li L., Dong Y., Jiang Y., Liu K., Zhang R., Jiang B., Niu K., Fang X. Role of Trichoderma reesei mitogen-activated protein kinases (MAPKs) in cellulase formation. Biotechnol. Biofuels. 2017;10:99. doi: 10.1186/s13068-017-0789-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 308.Frawley D., Bayram Ö. The pheromone response module, a mitogen-activated protein kinase pathway implicated in the regulation of fungal development, secondary metabolism and pathogenicity. Fungal Genet. Biol. 2020;144 doi: 10.1016/j.fgb.2020.103469. [DOI] [PubMed] [Google Scholar]
- 309.Bayram Ö., Bayram Ö.S., Ahmed Y.L., Maruyama J., Valerius O., Rizzoli S.O., Ficner R., Irniger S., Braus G.H. The Aspergillus nidulans MAPK module AnSte11-ste50-ste7-fus3 controls development and secondary metabolism. PLoS Genet. 2012;8 doi: 10.1371/journal.pgen.1002816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 310.Futagami T., Nakao S., Kido Y., Oka T., Kajiwara Y., Takashita H., Omori T., Furukawa K., Goto M. Putative stress sensors WscA and WscB are involved in hypo-osmotic and acidic pH stress tolerance in Aspergillus nidulans. Eukaryot. Cell. 2011;10:1504–1515. doi: 10.1128/EC.05080-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 311.Bencina M., Panneman H., Ruijter G.J.G., Legiša M. Visser, Characterization and overexpression of the Aspergillus Niger gene encoding the cAMP-dependent protein kinase catalytic subunit. Microbiology. 1997;143(n):1211–1220. doi: 10.1099/00221287-143-4-1211. [DOI] [PubMed] [Google Scholar]
- 312.Tag A., Hicks J., Garifullina G., Ake C., Jr., Phillips T.D., Beremand M., Keller N. G-protein signalling mediates differential production of toxic secondary metabolites. Mol. Microbiol. 2000;38:658–665. doi: 10.1046/j.1365-2958.2000.02166.x. [DOI] [PubMed] [Google Scholar]
- 313.Liu S., Hou Y., Liu W., Lu C., Wang W., Sun S. Components of the calcium-calcineurin signaling pathway in fungal cells and their potential as antifungal targets. Eukaryot. Cell. 2015;14:324–334. doi: 10.1128/EC.00271-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 314.Steinbach W.J., Cramer R.A., Perfect B.Z., Asfaw Y.G., Sauer T.C., Najvar L.K., Kirkpatrick W.R., Patterson T.F., Benjamin D.K., Heitman J., Perfect J.R. Calcineurin controls growth, morphology, and pathogenicity in Aspergillus fumigatus. Eukaryot. Cell. 2006;5:1091–1103. doi: 10.1128/EC.00139-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 315.Dinamarco T.M., Freitas F.Z., Almeida R.S., Brown N.A., dos Reis T.F., Ramalho L.N.Z., Savoldi M., Goldman M.H.S., Bertolini M.C., Goldman G.H. Functional characterization of an Aspergillus fumigatus calcium transporter (PmcA) that is essential for fungal infection. PLoS One. 2012;7 doi: 10.1371/journal.pone.0037591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 316.Chen L., Zou G., Wang J., Wang J., Liu R., Jiang Y., Zhao G., Zhou Z. Characterization of the Ca2+-responsive signaling pathway in regulating the expression and secretion of cellulases in Trichoderma reesei Rut-C30. Mol. Microbiol. 2016;100:560–575. doi: 10.1111/mmi.13334. [DOI] [PubMed] [Google Scholar]
- 317.Leite P., Sousa D., Fernandes H., Ferreira M., Costa A.R., Filipe D., Gonçalves M., Peres H., Belo I., Salgado J.M. Recent advances in production of lignocellulolytic enzymes by solid-state fermentation of agro-industrial wastes. Curr. Opin. Green Sustain. Chem. 2021;27 doi: 10.1016/j.cogsc.2020.100407. [DOI] [Google Scholar]
- 318.Steudler S., Werner A., Cheng J.J., editors. Solid State Fermentation: Research and Industrial Applications. Springer International Publishing; Cham: 2019. [DOI] [Google Scholar]
- 319.de Castro R.J.S., Sato H.H. Enzyme production by solid state fermentation: general aspects and an analysis of the physicochemical characteristics of substrates for agro-industrial wastes valorization. Waste Biomass Valor. 2015;6:1085–1093. doi: 10.1007/s12649-015-9396-x. [DOI] [Google Scholar]
- 320.Meyer V., Cairns T., Barthel L., King R., Kunz P., Schmideder S., Müller H., Briesen H., Dinius A., Krull R. Understanding and controlling filamentous growth of fungal cell factories: novel tools and opportunities for targeted morphology engineering. Fungal Biol. Biotechnol. 2021;8:8. doi: 10.1186/s40694-021-00115-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 321.Nai C., Meyer V. From axenic to mixed cultures: technological advances accelerating a paradigm shift in microbiology. Trends Microbiol. 2018;26:538–554. doi: 10.1016/j.tim.2017.11.004. [DOI] [PubMed] [Google Scholar]
- 322.Zhuang L., Zhang H. Utilizing cross-species co-cultures for discovery of novel natural products. Curr. Opin. Biotechnol. 2021;69:252–262. doi: 10.1016/j.copbio.2021.01.023. [DOI] [PubMed] [Google Scholar]
- 323.Knowles S.L., Raja H.A., Roberts C.D., Oberlies N.H. Fungal–fungal co-culture: a primer for generating chemical diversity. Nat. Prod. Rep. 2022;39:1557–1573. doi: 10.1039/D1NP00070E. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 324.Wösten H.A.B. Filamentous fungi for the production of enzymes, chemicals and materials. Curr. Opin. Biotechnol. 2019;59:65–70. doi: 10.1016/j.copbio.2019.02.010. [DOI] [PubMed] [Google Scholar]
- 325.Malyan S.K., Kumar A., Baram S., Kumar J., Singh S., Kumar S.S., Yadav A.N. In: Recent Advancement in White Biotechnology through Fungi: Volume 3: Perspective for Sustainable Environments. Yadav A.N., Singh S., Mishra S., Gupta A., editors. Springer International Publishing; Cham: 2019. Role of fungi in climate change abatement through carbon sequestration; pp. 283–295. [DOI] [Google Scholar]
- 326.Martin, Sustainable development goals report, Unit. Nat. Sustain. Dev.. (n.d.). https://www.un.org/sustainabledevelopment/progress-report/(accessed November 6, 2022).
- 327.Humpenöder F., Bodirsky B.L., Weindl I., Lotze-Campen H., Linder T., Popp A. Projected environmental benefits of replacing beef with microbial protein. Nature. 2022;605:90–96. doi: 10.1038/s41586-022-04629-w. [DOI] [PubMed] [Google Scholar]
- 328.Livne A., Wösten H.A.B., Pearlmutter D., Gal E. Fungal mycelium bio-composite acts as a CO2-sink building material with low embodied energy. ACS Sustainable Chem. Eng. 2022;10:12099–12106. doi: 10.1021/acssuschemeng.2c01314. [DOI] [Google Scholar]
- 329.Dodge E. In: Transforming Climate Finance and Green Investment with Blockchains. Marke A., editor. Academic Press; 2018. Chapter 16 - carbon deposits—using soil and blockchains to achieve net-zero emissions; pp. 217–228. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
No data was used for the research described in the article.