Abstract
How do organisms assess the degree of completion of a large structure, especially an extracellular structure such as a flagellum? Bacteria can do this. Mutants that lack key components needed early in assembly fail to express proteins that would normally be added at later assembly stages. In some cases, the regulatory circuitry is able to sense completion of structures beyond the cell surface, such as completion of the external hook structure. In Salmonella and Escherichia coli, regulation occurs at both transcriptional and posttranscriptional levels. One transcriptional regulatory mechanism involves a regulatory protein, FlgM, that escapes from the cell (and thus can no longer act) through a complete flagellum and is held inside when the structure has not reached a later stage of completion. FlgM prevents late flagellar gene transcription by binding the flagellum-specific transcription factor ς28. FlgM is itself regulated in response to the assembly of an incomplete flagellum known as the hook-basal body intermediate structure. Upon completion of the hook-basal body structure, FlgM is exported through this structure out of the cell. Inhibition of ς28-dependent transcription is relieved, and genes required for the later assembly stages are expressed, allowing completion of the flagellar organelle. Distinct posttranscriptional regulatory mechanisms occur in response to assembly of the flagellar type III secretion apparatus and of ring structures in the peptidoglycan and lipopolysaccharide layers. The entire flagellar regulatory pathway is regulated in response to environmental cues. Cell cycle control and flagellar development are codependent. We discuss how all these levels of regulation ensure efficient assembly of the flagellum in response to environmental stimuli.
INTRODUCTION: THE FLAGELLAR REGULATORY NETWORK
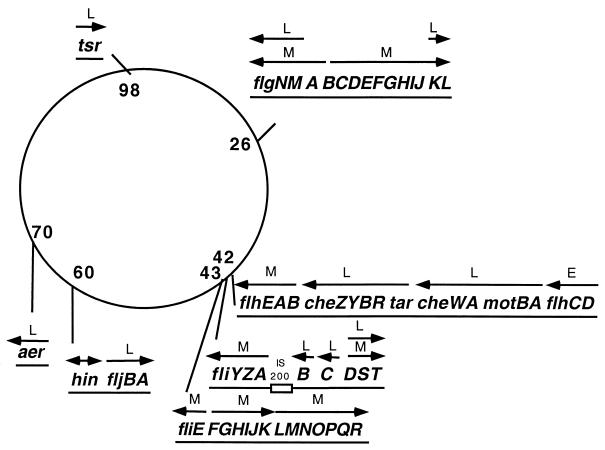
The synthesis and function of the flagellar and chemotaxis system require the expression of more than 50 genes which are divided among at least 17 operons (Fig. 1) that constitute the large, coordinately regulated flagellar regulon (85). The expression of these genes is regulated at a multiplicity of levels in response to environmental cues, in addition to signals that are coupled to the morphological development of the flagellar organelle itself. The elaborate regulation is accomplished via mechanisms that involve a number of transcriptional regulators, some of which are sensitive to one another, and at least one that directly detects the developmental state of the population of flagella expressed by a given cell. Within the regulon, the operons are divided into three temporally regulated, hierarchical transcriptional classes: early, middle, and late. This hierarchical class structure is based on the effect of null mutation in a gene of a given class on the transcription of the remaining genes in the regulon (71). These genes were previously referred to as class 1, class 2, and class 3, but recent findings that many genes are transcribed from multiple promoters added confusion to this nomenclature. The original classification was based upon a requirement for expression of the previous transcriptional class before expression of the next class can occur (71). In this review we will refer to the genes as early, middle, and late and the promoters as class 1, class 2, and class 3.
FIG. 1.
Chromosomal locations of the operons that make up the flagellar regulon of S. enterica serovar Typhimurium. The operons are labeled E, M, or L depending on whether they are expressed early, middle, or late in the temporal induction pathway (see text).
In general, late genes are downregulated in strains defective in early or middle genes, and neither middle nor late genes are expressed in strains defective in early class genes. The three promoter classes that correspond to the transcriptional classification have differences in DNA sequence. The class 1 promoter is a single promoter that transcribes the two early genes in the flhDC operon. As discussed below, the flhD and flhC genes encode transcriptional activators, and transcription of the flhDC operon responds to many environmental cues. The FlhD and FlhC proteins are transcriptional activators for the class 2 promoters upstream of the middle gene operons. The protein products of middle class operons include proteins necessary for the structure and assembly of the hook-basal body, an intermediate structure in flagellar assembly, and the transcriptional regulators FlgM and ς28. The class 3 promoters are specific for ς28 RNA polymerase. With the exception of the hook-associated proteins (HAPs), gene products required at the late assembly stage are transcribed only from class 3 promoters.
The transcriptional classes appeared to correspond to major steps in morphological development of the flagellar structure (49). However, this relatively simple transcriptional hierarchy was complicated by the fact that many of the genes are expressed from more than one promoter class. For example, the flgK, flgL, flgM, flgN, fliD, fliS, and fliT genes are transcribed from both FlhDC-dependent class 2 promoters and ς28-dependent class 3 promoters (32, 66, 72). The coupling of gene expression to flagellar assembly is accomplished by the regulation of ς28 activity by the anti-ς28 factor FlgM (Fig. 2). Regulation of FlgM levels in response to flagellar assembly results in the temporal regulation of ς28-dependent transcription to ensure the efficiency of assembly. In addition to complex transcriptional control, there are layers of posttranscriptional regulation, which are also believed to ensure an efficient temporal assembly process (12, 50, 53).
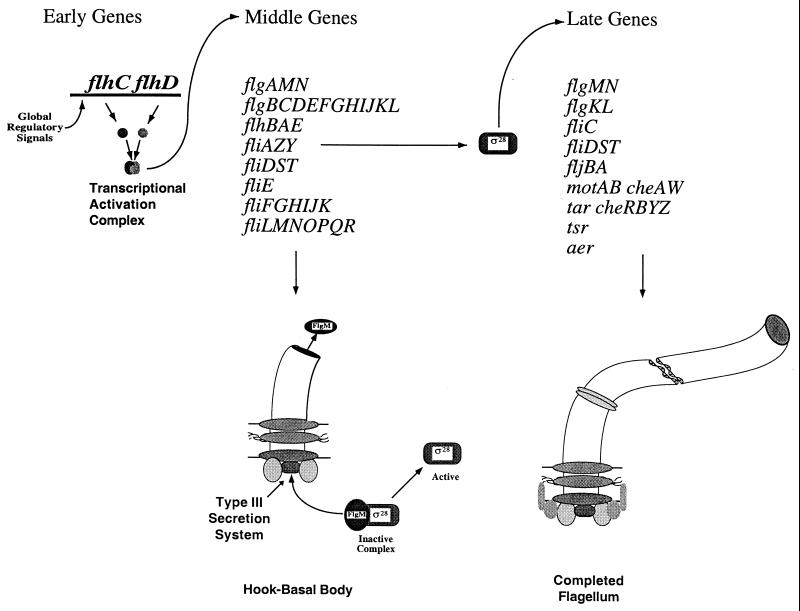
FIG. 2.
Flagellar transcriptional hierarchy coupled to flagellar assembly. There are more than 50 genes in the flagellar and chemotaxis regulon. These genes are transcribed in operons of three temporal classes, early, middle, and late (57, 71). The early genes are included in the master flagellar operon, flhDC. The FlhC and FlhD proteins form a heteromultimeric complex that directs ς70-dependent transcription of class 2 promoters of the middle and some late genes (78). The middle operons encode structural and assembly proteins required for the biosynthesis of the flagellar motor intermediate structure, also known as the hook-basal body (85). In addition to hook-basal body proteins, two competing regulatory proteins, FlgM and FliA (ς28), are also transcribed from class 2 promoters. The fliA gene encodes an alternative transcription factor, ς28, which is specifically required for class 3 promoter transcription. Genes whose products are required late in flagellar assembly, including the external filament (flagellar propeller), are primarily transcribed from class 3 promoters. The FlgM protein binds to ς28 directly to prevent class 3 promoter transcription until after hook-basal body completion. Once the hook-basal body is complete, FlgM is secreted from the cell to free ς28, and class 3 transcription occurs. In this way, the external filament (propeller) is not made until there is a motor (hook-basal body) for it to polymerize onto. A number of late flagellar genes, including flgK, flgL, flgM, flgN, fliD, fliS, and fliT, are expressed in both middle and late operons.
FLAGELLAR PROMOTERS
Class 1 Master Operon
The early genes are transcribed from a class 1 flagellar promoter in a single operon, flhDC, which is sensitive to environmental and cell state sensors such as cyclic AMP (cAMP)-cAMP receptor protein (cAMP-CRP) (119, 133). The class 1 promoter is also known to require a number of other genes outside of the flagellar regulon for its expression (11, 104, 114–116). Six transcriptional start sites have been mapped in the class 1 promoter region of Salmonella enterica serovar Typhimurium (132). It has also been demonstrated that cell cycle and flagellation are interdependent and that the flhDC genes are involved in coupling these processes (96, 103). Likewise, genetic evidence suggests that a growing number of other genes are involved either directly or indirectly in transcriptional regulation of flhDC expression (5, 74, 104, 114–116).
The promoter for the flhDC master operon defines the class 1 flagellar promoter. It is a crucial regulatory point at which the decision to initiate or prevent flagellar biosynthesis can occur. It is not surprising that the class 1 promoter is where a number of global regulatory signals have input on the decision to synthesize flagella. These include cAMP-CRP (119, 133), temperature control (1, 105), heat shock proteins DnaK, DnaJ, and GrpE (116), high concentrations of either inorganic salts, carbohydrates, or alcohols (74, 115), DNA supercoiling (11, 63, 74, 115), growth phase (96), surface-liquid transition (33), phosphatidylethanolamine and phosphatidylglycerol synthesis (91, 114), and cell cycle control (90, 92, 103).
Mutants defective in either adenylate cyclase (cya gene) or CRP (crp gene) failed to produce flagellin, hook, or flagellin mRNA (58, 119, 133). Motile revertants of cya or crp mutants were isolated, and their mutations were found to map to the promoter region of the early flhDC operon (58, 119). These motile revertants were pleiotropic in that they also lost the high-temperature inhibition of flagellar gene expression (119). This suggests that the high-temperature inhibition of flagellum synthesis is an indirect effect on the class 1 promoter through the cAMP-CRP activator complex. A CRP binding site has been identified within the flhDC promoter regions for both E. coli and S. enterica serovar Typhimurium (121, 132).
The presence of six transcriptional start sites in the S. enterica serovar Typhimurium class 1 promoter region indicates sites of transcriptional regulation (132). CRP binding sites have been identified within the flhDC promoter regions for both E. coli and S. enterica serovar Typhimurium (9, 117, 121). One CRP binding site corresponded to a region of near identity 58 to 84 bases upstream of the major S. enterica serovar Typhimurium transcript (P1), which was substantially reduced in a crp mutant background (132). A second was located between the −10 region of the P1 transcript and the GTG start codon. Another transcript identified in S. enterica serovar Typhimurium (P6) was dependent on a functional hns gene for expression (132). The H-NS protein is a bacterial nucleoid-associated protein and a pleiotropic regulator of gene expression (27). In DNase footprinting experiments, purified H-NS protected numerous sites on both sides of the start of the P6 transcript (121). The major site of transcription initiation of the E. coli flhDC transcript was located 17 bp downstream of the S. enterica serovar Typhimurium P1 transcription initiation site, suggesting that some spacing differences exist between the two species (117, 132). Another metabolic intermediate, acetylphosphate, affected expression of the flhDC operon in E. coli, and this effect was dependent on OmpR, another pleiotropic regulatory protein (117). The E. coli transcript was dependent on OmpR protein for expression, and OmpR was shown to footprint large regions upstream and downstream of the transcription initiation site (117). Regulation of flhDC operon expression is further complicated by the fact that it is autogenously regulated and the nature of autogenous regulation is dependent on the intracellular activity of ς28. Under normal growth conditions, FlhDC is an autogenous repressor of flhDC expression, but in the presence of increased ς28 activity, FlhDC is an autogenous activator of flhDC expression (63).
The results presented above suggest a high degree of complexity for the flhDC promoter region that is only beginning to be understood. The interdependence among so many structural and physiological processes and the regulation of flhDC operon expression suggest that motility plays a crucial role in the life cycle of the cell and, as discussed later, serve to couple flagellar biosynthesis to the cell cycle. The production of flagella and resulting motility would represent a significant drain on the cell's resources. However, the ability to reach a food source ahead of competitors or to swim away from substances that adversely affect central metabolic processes would provide a significant survival advantage.
Class 2 and Class 3 Promoters
In addition to autogenous regulation, the FlhDC complex is a positive transcriptional activator of ς70-dependent transcription from the class 2 promoters (71, 78). The FlhDC complex activates class 2 promoter transcription by binding within the class 2 promoter region and interacting with the C-terminal region of the α subunit of RNA polymerase (76). The FlhDC-dependent transcriptional start sites have been mapped for 10 class 2 promoters, which are shown in Fig. 3 (44, 45, 66, 78, 95). Purified FlhDC has been shown to footprint a region between −30 and −80 relative to the transcriptional start site for three of these class 2 promoters (Fig. 3) (78). Sequences termed TTATTCC and GCAATAA elements have been identified in all class 2 promoters in the region where FlhDC binds and, when deleted in the flgB promoter, are defective for transcription (Fig. 3) (44). Operons transcribed from class 2 promoters include genes whose products are required for the morphogenesis of the hook and basal body (49). Included in the genes transcribed from class 2 promoters is the fliA gene, which encodes an alternative sigma subunit of RNA polymerase, ς28 (97).
FIG. 3.
Structure of flagellar class 2 and class 3 promoters. Class 2 promoters are dependent on ς70 and the tetrameric FlhD2C2 transcriptional activation complex for transcription. Class 3 promoters are dependent on the alternative transcription factor ς28 for transcription. The data for class 2 promoters are primarily taken from Ikebe et al. (44). Transcriptional start site mapping was determined as follows: flgAS, flgBS, flhBS, fliES, and fliLS are from reference 44; fliAS is from references 45 and 98; fliDS is from reference 66; flhBE and fliLE are from reference 78; and fliAE is from reference 95. The primary transcriptional start site for the characterized class 2 promoters are underlined and labeled +1. The subscripts E and S after each gene designate whether the promoter corresponds to a flagellar promoter from E. coli or S. enterica serovar Typhimurium, respectively. Sequences that closely match consensus GCAATAA and TTATTCC sequences within known and presumed FlhDC binding regions are labeled by arrows. DNA regions protected by the binding of the FlhDC complex are boxed.
The flagellar alternative sigma factor, ς28, confers specificity for the class 3 flagellar promoters of the late genes (43, 62, 97, 109). DNA sequence analysis of flagellar genes revealed a novel promoter region and suggested that expression of flagellar and chemotaxis genes might occur by an alternative sigma factor (35). A consensus sequence of −35 (TAAAGTTTT) N11 −10 (GCCGATAA) was determined for the ς28-specific class 3 promoter (Fig. 3) (35, 43). Early on, DNA sequence analysis of middle gene promoters revealed that they contained the −10 consensus sequence of the class 3 promoter (9) and suggested a role for ς28 in middle gene expression (34). More recently it was demonstrated that ς28 will transcribe from the promoter regions of the early and middle genes, but the transcriptional start sites differ for FlhDC-dependent class 2 and ς28-dependent class 3 promoters within the middle gene promoter regions (43, 63, 67, 77). A negative regulator of ς28, the FlgM protein (31), is transcribed from a class 2 and a class 3 promoter (32). Among the late genes are those transcribed only from class 3 promoters. These include genes encoding motor torque generator subunits MotA and MotB, chemotaxis proteins, and the flagellin proteins FliC and FljB, which are polymerized outside the cell to form the helical filament (85).
CENTRAL ROLE OF FLGM-ς28 INTERACTION
Positive Regulation by Alternative Sigma Factor ς28
The essential positive regulatory element in the flagellar regulatory cascade is the alternative sigma factor ς28, encoded in the fliA gene. While originally thought to be a transcription factor only of flagellar late assembly genes, there is evidence that ς28 has recognition sequences within the promoter regions of all three flagellar gene classes. A null mutation in the fliA gene resulted in loss of expression of lac operon fusions to the late genes (57, 71). Comparison of the promoter regions for the late gene promoters revealed a promoter consensus sequence distinct from the promoter consensus of the primary sigma factor, ς70, and similar to that of an alternative sigma factor required for flagellar gene expression in Bacillus subtilis, ς28 (35, 43). A thorough analysis of ς28-dependent promoters reveals a revised promoter sequence of TAAAGTTT-N11-GCCGATAA for the −35 and −10 promoter recognition sites (Fig. 3) (43).
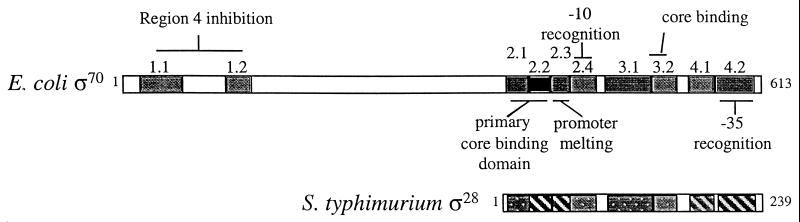
The fliA genes from both S. enterica serovar Typhimurium and E. coli were cloned and sequenced, and their ς28 protein products were purified and characterized (8, 75, 98). One striking feature of the ς28 protein is that it is less than half the size of the primary sigma factor ς70 (Fig. 4). The ς28 protein belongs to a conserved ς70 family of transcription factors (79). There are four main regions of conservation within the ς70 family, regions 1, 2, 3, and 4. DNA sequence comparison revealed that the large N-terminal conserved portion of ς70 (region 1) is not present in ς28 (Fig. 4). Region 1 was found to prevent ς70 from binding promoter sequences when ς70 is not bound to core RNA polymerase (24, 25). Region 1 interacts with region 4 in free ς70. Region 4 contains the −35 promoter-binding domain. Region 1 is not present in ς28 (Fig. 4). Region 4 of ς28 is prevented from binding DNA when free from core RNA polymerase by the interaction with the anti−ς28 factor FlgM (17, 46; M. S. Chadsey and K. T. Hughes, submitted for publication.)
FIG. 4.
Organization of the ς70 and ς28 transcription factors. The ς70 family of transcription factors contain four conserved regions that are subdivided within a given region (79). Region 1 is absent in ς28, but regions 2, 3, and 4 are conserved between the two ς factors. The degree of homology between ς70 and ς28 is indicated within the ς28 structure as follows: hatching, >50% homology; stippling, between 25 and 50% homology; blank, <25% homology. Homologous residue groups are S and T; R and K; N and Q; F, Y, and W; and I, L, V, and M.
The ς28 proteins from both E. coli and S. enterica serovar Typhimurium are 239 amino acids in length and were specifically required for in vitro transcription from the class 3 tar or fliC promoter (75, 98). The fliA gene is located in the fliAZY operon (45, 95). Class 2 and class 3 promoters transcribe the fliAZY and fliAZ transcripts, respectively (45). A separate promoter that is not regulated by the flagellar regulatory system also transcribes the fliY gene (45). Disruption of fliZ caused weak motility (45, 95), while disruption of fliY conferred no motility defect (45). Introduction of a multicopy plasmid carrying the fliZ expressed from vector-derived promoters resulted in a 10-fold increase in expression of a chromosomal fliA-lac operon fusion (68). A null mutation in fliZ resulted in a 10-fold reduction in expression of lac operon fusions to middle genes flgA, flgB, flhB, fliE, fliF, fliL, and fliA but had no effect on expression of an flhD-lac operon fusion (68). This result suggests a positive role for FliZ in class 2 promoter transcription. fliZ null mutants exhibited only a slight defect in motility that could be complemented in trans (45). This is probably why mutations in fliZ had not been identified earlier. Kutsukake et al. (68) pointed out that FliZ protein is predicted to contain a coiled-coil domain, which is believed to be involved in protein-protein interactions (83). They suggest that the role of FliZ is to modulate the binding of the FlhD2C2 complex to class 2 promoters through a direct interaction with the FlhD2C2 complex (68). Another possibility is that FliZ plays a positive role in FlhD2C2 complex formation. Either possibility provides a mechanism by which to amplify the activity of the FlhD2C2 complex on class 2 promoter expression. It also provides the possibility for another checkpoint in the flagellar regulatory cascade. The FlhD2C2 complex activates transcription of the fliAZY operon, and the ς28 (FliA) and FliZ protein products act to increase flagellar gene expression: ς28 will transcribe early, middle, and late genes, and FliZ enhances the function of the FlhD2C2 complex on class 2 promoter transcription.
Feedback Loops for Early and Middle Gene Expression
The ς28 protein is absolutely required for transcription from class 3 promoters. However, overexpression of the fliA gene resulted in increased expression from all middle gene promoters (67). In vitro, Eς28 was shown to transcribe the PfliA, PfliL, and middle gene promoter regions at start sites distinct from those used by Eς70-FlhDC (77). The effect of increased transcription from middle gene promoters by fliA overexpression was dependent on the presence of the early (flhDC) genes (67). This suggested that fliA overexpression led to increased early gene transcription. Indeed, flhDC expression is elevated by increased fliA expression (63). However, this elevation was dependent on the presence of an active flhDC operon. In the absence of fliA, the flhDC operon is negatively autoregulated (63, 132). These results suggested that FlhDC is an autogenous repressor, while ς28 is both an autogenous activator and an activator of the early flhDC operon and other middle operons. Expression of flhDC would lead to induction of class 2 promoter transcription, including the fliA class 2 promoter. Further increase in the expression of flhDC would then depend on the activity of ς28.
Negative Regulation
The coupling of flagellar gene expression to flagellar development by negative regulation was first suggested by Suzuki and Iino from assays of flagellin RNA levels in a variety of flagellar mutant strains (124). Mutations in any one of 11 flagellar genes examined led to a greater than 100-fold reduction in 14C-labeled amino acid incorporated into flagellin compared to incorporation into whole-cell protein. The real breakthroughs in the characterization of the flagellar regulon came with the advent of the Mud-lac fusion technology initially developed by Malcolm Casadaban (14, 15). In the early characterization of a transcriptional fusion of lac to the flagellin structural gene fliC, it was observed that loss of any one of a large number of flagellar genes (23 of the then-known 28) prevented expression of the fliC-lac fusion. Fusions of Mud-lac to all the flagellar genes in both S. enterica serovar Typhimurium and E. coli were obtained, and the effect of other flagellar genes on expression of the different fusions was determined (55, 56, 71). At the same time, many of the functions of the different flagellar genes were determined, so that a correlation between gene regulation and flagellar assembly began to unfold.
The middle genes encoded the structural genes for the hook-basal body intermediate structure, and the late genes encoded proteins needed late in flagellar assembly. Late genes encode hook-associated proteins FlgK, FlgL, and FliD, the filament subunits FliC and FljB, the proton motive force generator subunits MotA and MotB, and the chemosensory system. FlgK and FlgL form the hook-filament junction, and the cap is composed of FliD protein subunits. Expression of individual lac operon fusions to each gene of the flagellar regulatory hierarchy was determined in strains that were individually disrupted in every other gene to determine that the flagellar regulatory hierarchy was divided into the three main classes (Fig. 2) (71). If either flhD or flhC, was disrupted, none of the other genes was expressed. If any of the genes required for hook-basal body formation was disrupted, other hook-basal body genes were still transcribed, as was the flhDC operon, but none of the late genes was transcribed. The FlhD and FlhC proteins were envisioned as transcriptional activators of middle gene promoters (35), and the fliA gene was shown to encode an alternative sigma transcription factor, ς28, which directed transcription of late gene promoters (75, 97). Thus, it was easy to hypothesize that loss of flhDC would prevent expression of both middle and late gene promoters and that loss of fliA would prevent expression of late gene promoters. It was more difficult to hypothesize how loss of any one of the other 34 middle genes could prevent expression of the late genes. Since the majority of middle genes led to hook-basal body formation and the late structural genes were needed after hook-basal body formation, it appeared that gene regulation was coupled to flagellar morphogenesis by negative regulation.
FlgM Acts as an Anti-ς28 Factor
The hypothesis that late flagellar gene expression was coupled to flagellum assembly was demonstrated with the discovery of the negative regulator FlgM (31, 57) and the characterization of its activity as an anti-sigma factor (98). The anti-sigma activity of FlgM was postulated based on the following criteria: (i) purified FlgM inhibited the in vitro transcription of the class 3 fliC promoter using ς28 holoenzyme but had no effect on the in vitro transcription of the tac promoter using ς70 holoenzyme and (ii) a cross-linked product of FlgM and ς28 was obtained both with a mixture of purified proteins and when FlgM and ς28 were cosynthesized in vitro in either minicells or maxicells (98). Later, it was shown that when a fusion of glutathione-S-transferase (GST) to ς28 from either E. coli or S. enterica serovar Typhimurium was isolated from crude extracts on a glutathione column, FlgM was associated with the GST-ς28 fusion without the addition of a cross-linking reagent, suggesting a strong interaction (17, 48). Electrophoresis of a mixture of FlgM and ς28 on a native gel resulted in a novel band that separated into the individual FlgM and ς28 proteins on a denaturing gel (17). Furthermore, nuclear magnetic resonance (NMR) assays and surface plasmon resonance (SPR) assays demonstrated that the interaction between FlgM and ς28 was very tight, with a dissociation constant (Kd) on the order of 2 × 10−10 M (17, 21). This interaction is of the same magnitude as the interaction between ς28 and S. enterica serovar Typhimurium core RNA polymerase (Kd = 8 × 10−10 M) (17).
FlgM Acts as an Anti-ς28 Holoenzyme Factor
FlgM interacts with ς28 holoenzyme in addition to ς28 alone (17). FlgM prevented ς28 holoenzyme from binding fliC promoter DNA even after formation of Eς28 holoenzyme. Also, in native gel assays, FlgM was found associated with Eς28 in stoichiometric amounts regardless of order of addition (ς28 plus core then FlgM, or ς28 plus FlgM then core), but FlgM did not associate with core RNA polymerase in the absence of ς28. There are at least three ways in which FlgM could prevent the activity of ς28 in transcription: (i) FlgM could act as an anti-ς28 factor and prevent Eς28 formation; (ii) FlgM could act to dissociate Eς28; and (iii) FlgM could bind Eς28 and prevent promoter binding. Using the SPR assay system, it was found that FlgM would actively dissociate core RNA polymerase from Eς28 while having no effect on the dissociation of Eς70 (17). FlgM concentration was estimated at 400 nM, about 1,000-fold higher than the Kd of the FlgM-ς28 complex (200 pM) or the Kd of the Eς28 complex (800 pM). Given that the interactions between ς28 and FlgM and between ς28 and core were within a similar range, there should be no free ς28 in the cell as long as FlgM and core RNA polymerase are in excess to ς28 in the cell. Also, small changes in FlgM concentrations approximately 1,000-fold higher than the Kd of the FlgM-ς28 complex should not affect that interaction, but they may affect the ability of FlgM to destabilize the Eς28 complex.
Genetic and Molecular Analysis of FlgM-ς28 Interaction
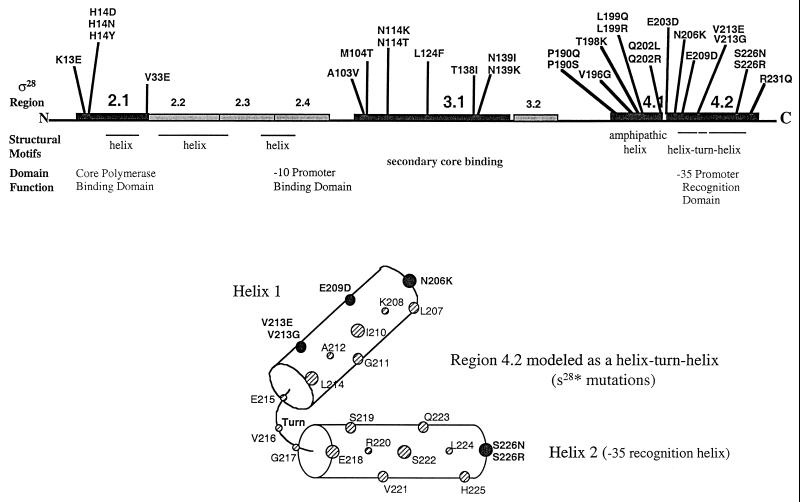
A deletion analysis was carried out on both FlgM and ς28 to assess which portions of these proteins were involved in direct interactions (46, 69). Mutations in ς28 were obtained that express a lac operon fusion to class 3 promoters under FlgM-inhibitory conditions (hook-basal body mutant backgrounds) (69; Chadsey and Hughes, submitted). These mutations resulted in ς28 proteins that bypass negative regulation by FlgM. DNA sequence analysis of the FlgM-insensitive mutations in fliA showed that they were located in three distinct regions (2.1, 3.1, and 4) of the ς28 protein that are conserved among sigma factors (Fig. 5) (69; Chadsey and Hughes, submitted). Regions 2.1 and 3.1 are the regions of sigma factors that interact with core RNA polymerase, while region 4 has been shown to include the −35 promoter DNA-binding region. Expression of C-terminal region 4 alone was sufficient to compete with intact ς28 for binding FlgM in vivo, while expression of N-terminal regions 2 through 3 of ς28 did not compete. SPR assays demonstrated that individuals with mutations in each of the three regions were defective in binding FlgM. This suggested that most of the contacts in ς28 for FlgM were in region 4; however, the V33 position in region 2.1 and the region 3.1 positions were defective in FlgM interaction. These results were consistent with the findings that regions 2, 3, and 4 of the B. subtilis homologue ςF were found to interact with its cognate anti-ςF factor, SpoIIAB (22). Mutants carrying mutations at position H14 of ς28 still bound both FlgM and RNA polymerase with the same affinity as wild-type ς28. It is possible that they represented a direct interaction of regions 2 and 3 with FlgM or an allosteric effect of regions 2 and 3 on region 4. It is not known how the H14 substitution mutations overcome FlgM inhibition. They may affect the ability of FlgM to destabilize the Eς28 complex or the stability of ς28 itself. It is noteworthy that mutations in ς28 region 4.2 that overcome FlgM inhibition do not reside within the DNA recognition helix (see Fig. 5); there are no mutations between positions 213 and 226. This was because ς28 mutants insensitive to FlgM inhibition in vivo were also selected to retain the ability to transcribe class 3 promoters.
FIG. 5.
DNA sequence analysis of FlgM-insensitive mutations in ς28 (69; Chadsey and Hughes, Submitted) (A) Distribution of single amino acid changes in ς28 that allow expression of a fliC-lac transcriptional fusion in a hook-basal body mutant strain (FlgM-inhibitory conditions). The 239-amino-acid ς28 protein is represented by a horizontal bar. Conserved regions of the ς70 family are boxed. Possible structural motifs and putative domain functions based on results with other ς factors are indicated below the protein (79, 86, 112). (B) Conserved helix-turn-helix motif of region 4.2 of ς28. FlgM bypass mutants are indicated by black circles. Note that selection was for FlgM bypass ς28 mutants that still transcribed the fliC promoter. Thus, amino acids within the DNA recognition helix thought to contact the −35 region of the class 3 promoter DNA are not affected.
The C-terminal half of the 97-amino-acid FlgM protein was found to interact directly with ς28 (30, 46). Single and double amino acid changes in FlgM that inhibited anti-ς28 activity in vivo were identified at amino acid positions 57, 58, 63, 66, 67, 76, 77, and 82 (21). Mutant analysis suggested that the anti-ς28 region of FlgM lay between amino acids 42 and 86 (21, 46). NMR analysis showed that FlgM resonances extending from V41 through K97 were all affected by binding to ς28 (21). SPR analysis demonstrated that the single-amino-acid changes L66S and I82T resulted in a fourfold increase in the Kd of FlgM for ς28 (17). These results suggested that amino acids 41 through 97 of FlgM directly contact ς28 in region 4 and possibly in regions 2.1 and 3.1. It remains possible that the defect in the ability of ς28 region 2.1 and 3.1 mutants to bind FlgM may result from an allosteric effect on region 4.
Native FlgM Is an Unfolded Polypeptide
Using multidimensional NMR, Daughdrill et al. made the unexpected discovery that free FlgM protein resided in a mostly unfolded state under physiological conditions (21). It was only after binding ς28 that residues 41 through 97 of FlgM attained a folded conformation, while the N-terminal 40 amino acids remained unstructured in the presence or absence of ς28. These results suggest that the stability of FlgM within the cytoplasm is dependent on the interaction with other proteins. Less FlgM was present in strains lacking ς28, and stability measurements of FlgM in this background indicated that some of this instability is due to an increase in FlgM turnover (50).
The N terminus of FlgM encodes the secretion signal (described below). Deletions of the N-terminal portion resulted in a significant reduction in its cellular levels (20, 46) that may have resulted from increased turnover when the unstructured N terminus was not efficiently bound by another protein. This has yet to be determined.
CHECKPOINT IN FLAGELLAR GENE EXPRESSION
FlgM Secreted in Response to Hook-Basal Body Completion
The remarkable finding that FlgM was regulated by secretion was a clear example of transcriptional regulation in response to cell development as opposed to regulation in response to cellular physiological status. As a result, genes expressed from ς28-dependent class 3 promoters, whose products are required at later assembly stages, are not activated until a morphogenic requirement is fulfilled: the completion of the hook-basal body intermediate structure. It was difficult to understand how the presence of the 30 hook-basal body genes could be sensed by a single protein, given that the combined product of those 30 genes was a structure and not the chemical product of a metabolic pathway. The FlgM protein senses the developmental state of the flagellum by itself being a substrate for secretion through the flagellum-specific type III secretion pathway (42, 64). When the hook-basal body structure is complete and therefore competent for secretion of the hook-associated proteins and flagellin, it also becomes competent for secretion of FlgM (Fig. 2). The onset of FlgM secretion results in a decrease in its cellular concentration and a concomitant derepression of ς28-mediated expression of the class 3 transcripts (50, 53), whose structural products include the HAPs, the flagellins, and motor force generators (49). This finding also challenged the established paradigm that development proceeds in response to gene expression, since in the case of bacterial flagella it is development that cues gene expression (81).
Further proof that FlgM was secreted through the hook-basal body structure came with the ability to artificially induce the flagellar regulon (52). The flhDC operon was placed downstream of the tetracycline-inducible tetA promoter. In this construct, the flagellar transcriptional hierarchy was induced following the addition of tetracycline to the medium. These cells were motile in the presence of tetracycline and nonmotile without inducer present. Following addition of tetracycline, the class 1 (flhDC operon) promoter was immediately transcribed; this was followed by flgM gene transcription at 15 min (transcribed from both class 2 and class 3 promoters); also at 15 min after induction, transcription of the middle class fliA gene was induced; and 30 mins after induction, transcription of the late fliC gene occurred (52). Induction of fliC gene expression 30 min after flhDC induction coincided with the appearance of FlgM anti-sigma factor in the spent growth medium and with the completion of the hook-basal body structure. At 45 min after induction, nascent flagellar filaments were visible under the electron microscope (52). These results support a model in which the flagellar regulatory hierarchy of S. enterica serovar Typhimurium is temporally regulated following induction and both FlgM secretion and class 3 promoter transcription occur upon completion of the hook-basal body intermediate structure.
FlgM Secreted by a Flagellar Type III Pathway
Bacteria translocate proteins by a number of distinct pathways. Translocation systems that secrete proteins to the external environment have been categorized into four main groups, designated types I, II, III, and IV (13, 80), to distinguish them from the signal sequence-dependent, or sec, pathway by which many periplasmic and integral membrane proteins traverse or are inserted into the cytoplasmic membrane, respectively (94).
The type III secretion pathway appears to be most commonly found in pathogenic bacteria (19, 41, 122, 123). The protein components of this type of secretion system have been identified in several species, and in most cases they are both structurally and functionally homologous to components required for flagellar basal body formation (106, 126). These proteins are presumed to form the conduit by which flagellar proteins are secreted (84). Homologous proteins have been shown to form the type III secretion structures for the secretion of virulence determinants (54, 61, 125). Despite the growing number of proteins that have been identified as type III secretory proteins, no clear consensus has emerged for the sequence of the secretion signal. In several examples, the amino-terminal domains of type III secretory proteins have been shown to be sufficient for translocation (46, 73, 87, 110, 120) or to contain information that is essential for secretion (59). However, in some instances, mutations in other regions of type III secretory proteins have also been shown to have an impact on the efficiency of their secretion (38, 73). Paradoxically, short amino-terminal regions of some type III isecretory proteins have been shown to be sufficient to direct secretion (110, 131) but have also been shown to be dispensable for secretion (18). A remarkable finding by Anderson and Schneewind was that a signal for type III-directed secretion could be found in both the 5′-untranslated region (5′-UTR) of the mRNA of the type III secretion substrate and the amino acid sequence of secretion substrates (6, 7). Two distinct amino acid regions have been identified within type III secretion substrates from Yersinia that are required for secretion in addition to mRNA signals (18). This suggested that multiple secretion signals exist for some type III secretory proteins and that some type III secretion substrates may be secreted by more than one type III mechanism (18, 118).
With few exceptions, it is thought that all flagellar proteins, including the transcriptional regulator FlgM, are secreted by a type III pathway that involves passage through the center of the flagellar substructures (84). Following completion of the hook-basal body, secretion of the FlgM anti-ς28 factor occurs by this pathway (52, 108). The work presented below is a characterization of the determinants contained within FlgM that are required for it to be specifically recognized and secreted by this pathway (20).
Studies on the determinants for FlgM secretion by the flagellar type III secretion pathway suggest a mechanism that is similar to the results reported for the Yersinia type III secretion system. Evidence suggests that FlgM secretion can occur through both amino acid- and mRNA-dependent pathways. FlgM secretion does not need to be coupled to translation. Replacement of the 5′-UTR of flgM with the 5′-UTR from either the lac operon or the arabinose operon allowed FlgM secretion (20). Evidence also suggests that multiple amino acid regions can serve to direct FlgM secretion. Deletion of amino acids 3 through 11, 17 through 52, or 55 through 92 still allowed secretion even when the 5′-UTR was from the lac operon (20). It is still possible that the mRNA signal lies between amino acids 11 and 17. The efficiency of FlgM secretion was influenced by the 5′-UTR. FlgM translated from the class 2 flgAMN transcript was found primarily in the cytoplasm, whereas FlgM translated from the class 3 flgMN transcript was found primarily in the external environment (50). Thus, during hook-basal body assembly, when the role of FlgM is to inhibit ς28 activity, FlgM is made from a transcript where translation is not efficiently coupled to secretion. After hook-basal body completion, ς28-dependent transcription occurs, but FlgM is also produced from a ς28-dependent class 3 transcript. However, FlgM expressed from this transcript is coupled to its secretion from the cell. Why express FlgM from a class 3 transcript if it is destined to be secreted? Loss of FlgM results in an increase in the number of flagella (67) and an increase in their length (C. Jones and S. Aizawa, unpublished observations), resulting in reduced motility. The coupling of class 3 flgM translation to secretion may provide a mechanism by which to regulate the final length of the flagellum for optimum motility.
FlgN Protein
As mentioned above, evidence exists for independent mRNA and amino acid type III-dependent secretion signals. The N terminus of all late assembly secreted proteins, FlgM, FlgK, FlgL, FliD, FliC, and FljB, is required for secretion, yet there is no homology between the N termini of these proteins. One characteristic common to the late assembly secreted proteins is that their N termini are all disordered in structure (4, 21, 127–129). Perhaps the lack of structure in the N termini of the late assembly secreted proteins provides a polypeptide secretion signal in addition to a possible mRNA signal. Another plausible source for the secretion signal lies outside the secreted proteins themselves in another set of proteins, the type III secretion chaperones. Type III secretion chaperones are known to bind type III secretion substrates, and this binding is thought to be required for secretion of type III secretion substrates and to prevent proteolytic degradation of the secretion substrates in the cytoplasm (10). The FlgN and FliT proteins have been reported to be type III secretion chaperones for the flagellar late assembly secreted proteins (29). However, FlgN and FliT were found to bind the C-terminal region of their target proteins, not the N-terminal secretion signal domain. The flagellum is hollow, and subunits pass through a central channel of 25 to 30 Å (23, 93, 107). The size of the channel requires that the subunits pass through as partially folded monomers. The FlgN protein is known to bind the FlgK and FlgL proteins, while the FliT protein binds FliD (29). Binding of FlgN or FliT to the unstructured N terminus of late assembly secreted proteins might protect them from proteolytic degradation. Also, binding of FlgN or FliT to these proteins suggests that a secretion signal that directs FlgK, FlgL, and FliD (and other late assembly secreted proteins) to the flagellum-specific secretion apparatus could lie in its associated chaperone protein in addition to an amino-terminal secretion signal. Once brought to the secretion apparatus, the unstructured nature of the N termini of the late assembly secreted proteins maintained by chaperone binding could then serve to facilitate passage into the secretion channel.
The flgN gene is transcribed in the middle class flgAMN operon in addition to the ς28-dependent class 3 flgMN transcript (32, 50, 72, 111). Mutations in flgN result in a poorly motile phenotype with only one or two flagella per cell instead of the normal complement of six to ten (72). A closer examination of flagellar substructures revealed that flgN mutant cells possessed hook-basal bodies, but only a few of these had attached filaments. This was consistent with the idea that FlgN binds FlgK and FlgL both to escort these proteins to be secreted and to prevent their intracellular degradation. The defect in motility of flgN mutants can be overcome by loss of FlgM, suggesting that an increase in ς28-dependent expression of late genes can bypass the flgN defect. Overexpression of the flhDC operon also overcomes the motility defect of an flgN mutation (28, 50). The end result of flhDC overexpression is also likely to be an increase in late gene expression. If FlgN served to protect FlgK and FlgL from proteolytic degradation, then overproduction of FlgK and FlgL could lead to increased intracellular levels of FlgK and FlgL and the ability to synthesize normal flagella.
In addition to a role as a type III secretion chaperone, flgN has also been identified as a regulator of FlgM levels in the cell. Evidence suggested that FlgN was a translational regulator of flgM gene expression from the class 3 promoter transcript (50). Loss of flgN also led to increased transcription of the class 3 fliC promoter in a hook-basal body mutant strain (50). Strains defective in flgN show reduced levels of intracellular FlgM levels, which would account for increased class 3 promoter transcription in flgN null strains (50). However, a lacZ translational fusion to flgM was not expressed in an flgN hook-basal body double mutant strain even though this fusion was transcribed at wild-type levels. Finally, FlgM protein produced in strains defective in flgN was still secreted, suggesting that FlgN is not required for FlgM secretion (50). The fact that FlgN acts on class 3 flgMN translation suggests that some ς28-dependent class 3 transcription does occur prior to hook-basal body completion and this transcription is important to the efficient coupling of flagellar development to regulation of gene expression.
The finding that FlgN is a type III chaperone protein for FlgK and FlgL and a translational regulator of flgM transcribed from its class 3 promoters suggested a model for type III secretion that is inclusive of both mRNA and amino acid secretion signals (50). A type III secretion substrate could be targeted to the secretion apparatus by chaperone binding, especially if the chaperone is located at the base of the type III secretion apparatus. In addition, a requirement for type III secretion chaperones for translation of secretion substrates suggests a possible role to keep substrates translated at the base of the apparatus from diffusing away prior to secretion. Perhaps type III secretion chaperones such as FlgN have two roles in secretion. One role is to escort secretion substrates to the secretion apparatus. Another role is to help translate secretion substrates and to keep newly translated substrates from diffusing away from the base of the type III secretion apparatus prior to secretion. After translation, FlgN could bind the C terminus of FlgK and FlgL (and FlgM?) at the flagellar type III secretion apparatus and prevent these proteins from diffusing from the base of the flagellar structure before they can be secreted.
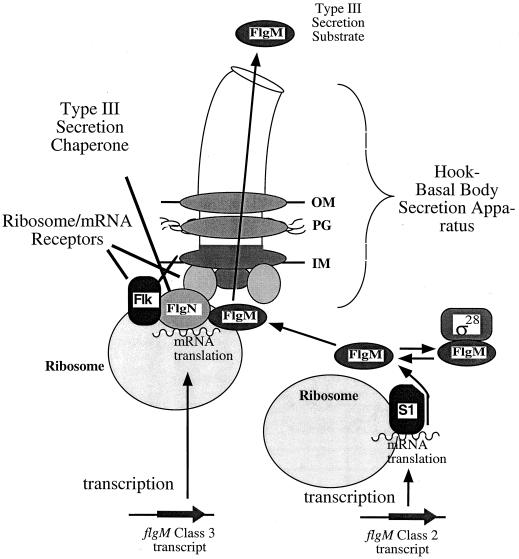
There is likely to be a redundancy in the secretion signal. One is an mRNA secretion signal that favors the coupling of translation to secretion. The second secretion signal would be an amino acid signal in the N terminus of the secreted protein that could be a structural signal (disordered) and/or in the amino acid sequence of the chaperone or translational regulatory proteins recognized by the type III secretion apparatus protein. A model for the coupling of the translation of class 3 transcripts to secretion of the substrate protein by FlgN is presented in Fig. 6. The mRNA transcript is translated at the base of the flagellar secretion apparatus by the action of the chaperone protein and the Flk protein. Flk, discussed below, is currently thought to bind ribosomes to direct the localized translation of specific mRNA transcripts at the cytoplasmic face of the inner membrane (50, 53). In conjunction with FlgN, Flk will localize the translation of the class 3 flgMN transcripts at the base of the flagellar secretion apparatus. Binding of FlgN to the C-terminal regions of the secreted proteins prevents their escape from the base of the secretion apparatus before they can be secreted.
FIG. 6.
Model for the coupling of flgM translation from the class 3 flgMN transcript to secretion by translational regulators Flk and FlgN. This model combines both mRNA and amino acid secretion signals for the FlgM substrate. The hook-basal body is the secretion channel. Some ς28-dependent transcription of class 3 flgMN occurs prior to hook-basal body completion. Secretion of FlgM expressed from the class 3 promoter prior to hook-basal body completion is coupled to mRNA translation by a ribosome/mRNA receptor and a flagellar type III secretion chaperone (right). After hook-basal body completion, secretion can also occur without cotranslation (left). In the case of cotranslation secretion (left), prior to hook-basal body completion, the flgM class 3 transcript (left) is targeted to the secretion channel by the ribosomal protein S1 homologue Flk (or another component of the flagellar type III secretion apparatus), where translation occurs. Both Flk and FlgN facilitate class 3 flgM translation prior to hook-basal body completion. The secretion chaperone FlgN may cooperate with Flk to bind the class 3 flgM transcript. The translated FlgM protein may be held at the secretion channel by a flagellum-specific type III secretion chaperone to keep the nascent secretion substrate from diffusing into the cytoplasm before secretion occurs, using the N-terminal amino acid secretion signal. The FlgN protein is one possible secretion chaperone for this purpose. In the case of translation uncoupled to secretion (right), only the amino acid secretion signal(s) within FlgM targets it to the secretion channel. The class 2 flgM transcript (right) is translated in the cytoplasm using the S1 ribosomal subunit protein. Cytoplasmic FlgM inhibits ς28-dependent transcription. Cytoplasmic FlgM can be targeted to the flagellar secretion channel through amino acid secretion signals. The amino acid signal may include interaction with just the secretion channel or both the chaperone and the secretion channel to facilitate secretion. OM, outer membrane; PG, peptidoglycan; IM, inner membrane.
Hook Completion and Specificity of Type III Secretion
Hook completion is a key checkpoint in flagellar development. It signals FlgM secretion and initiation of flagellin gene expression. FlgM responds to completion of the basal body-hook structure. Still, it remains to be determined what senses completion of the hook and signals the type III secretion apparatus to “stop” secretion of hook subunits and “start” FlgM and late assembly protein secretion. The first clue came with the isolation of mutations in a gene responsible for hook length control, fliK. fliK mutants were identified as having a polyhook phenotype (100). In a wild-type strain, hooks grow to a length of about 55 nm (36), whereas in an fliK null mutant strain, hook length varies from 40 to 900 nm and the mutants are nonmotile (36). Results from several labs support a model in which the switch to late protein secretion is from signals sent to the type III secretion apparatus upon completion of the hook-basal body (36, 70, 88, 130). Because fliK mutants are defective at filament assembly, they are nonmotile. Motile revertants of an fliK null allele have been isolated and include extragenic suppressors that map exclusively to the flhB locus (36, 70). FlhB is a component of the type III secretion apparatus. It has been hypothesized that FliK measures hook length and signals the FlhB component of the type III flagellum-specific secretion apparatus that the hook is complete. The type III secretion machinery switches substrate specificity from secretion of hook-type subunits to secretion of FlgM and late assembly proteins (60, 88).
It was thought that the flhB mutants that allowed secretion of late assembly substrates in the fliK mutant strains (flhB/FliK bypass) were no longer able to distinguish between middle and late class secretion substrates. However, in a strain possessing the flhB/FliK bypass mutation and also defective in the hook (flgE) gene, FlgM was not secreted (65). Kutsukake reported that an additional null mutation in the rflH locus was also required to allow FlgM secretion in the flgE flhB/FliK bypass double mutant background. He proposed that RflH acted as a gate to prevent secretion of late class secretion substrates prior to hook-basal body completion (65). This suggests that the secretion apparatus possesses a physical barrier to secretion of late substrates prior to hook-basal body completion in addition to secretion signal specificity by FliK and FlhB. Null mutations in the rflH locus, called flk, were isolated as causing a defect in flgM translation in strains defective in P- and L-ring assembly (51, 53). Thus, the physical barrier to secretion of FlgM by RflH (Flk) may be associated with a role in cotranslation secretion of FlgM (discussed below).
The mechanism of hook length measurement is not known. However, recently it has been shown that the FliK protein is secreted as a hook-type protein (89). Prior to hook completion, FliK was found in the spent growth medium, but in strains with intact hooks, FliK was found in the cytoplasm (89). This result suggested that secretion of FliK itself was dependent on hook length. Once the hook has reached its proper length, FliK was retained in the cytoplasm, where it could interact with FlhB of the type III secretion machinery to change secretion specificity from hook-type secretion substrates to filament-type secretion substrates, including FlgM. It remains to be determined how a completed hook can prevent the secretion of FliK.
flgM mRNA Translation Coupled to Ring Assembly
The flk gene was identified as a positive regulator of the activity of FlgM at an assembly step just prior to hook-basal body completion, at the point of assembly of the P- and L-rings (51). FlgM inhibition of ς28-dependent class 3 flagellar gene transcription was relieved in P- and L-ring assembly mutants (flgA, flgH, or flgI) by introduction of a null mutation in the flk gene (51). Strains defective in hook-basal body synthesis exhibit an increase in the intracellular levels of FlgM, presumably due to the inability of FlgM to be secreted. In P- and L-ring mutant strains, the addition of a recessive mutation in flk resulted in a reduction in intracellular FlgM levels down to those seen in wild-type (Fla+) strains (53). The reduction in intracellular FlgM levels by mutations in the flk gene was concomitant with a significant reduction in flgM-lacZ mRNA translation, expressed from the class 3 flgMN transcript, in P- and L-ring mutant strains, while the translation of flgM-lacZ mRNA from the class 2 flgAMN transcript was unaffected. No reduction in either flgAMN or flgMN mRNA stability was measured in the absence of Flk in Fla+, ring mutant, or hook-basal body deletion strains. This led to the conclusion that the reduction in intracellular FlgM levels by mutation in the flk gene occurs only at the level of flgM mRNA translation (53). The mechanism by which Flk couples flgM translation to P- and L-ring assembly is not understood. Flk is thought to be anchored to the cytoplasmic membrane by a C-terminal 18-amino-acid hydrophobic amino acid segment (53). It has homology to a region of the S1 RNA-binding protein that interacts with the 30S subunit of the ribosome. It is tempting to speculate that Flk can substitute for S1 to bring the ribosome to the cytoplasmic membrane to translate proteins that need to be localized there (Fig. 6). In the case of FlgM, it may facilitate the coupling of FlgM translation to FlgM secretion by the flagellar secretion apparatus.
Both Flk and FlgN were found to regulate translation of flgM expressed from the class 3 flgMN transcript in hook-basal body mutant strains. However, in strains possessing intact hook-basal bodies, loss of either flgN or flk had no effect on translation of flgM from the class 3 transcript (50, 53). This suggests that the effect of Flk and FlgN on translation or cotranslation secretion is redundant to the normal flagellar cotranslation secretion mechanism. It is likely that the flagellar basal structure includes the ability to couple translation to secretion in order to ensure the efficiency of assembly. Such a possibility remains to be addressed.
REGULATION OF NUMBER OF FLAGELLA
The exact regulatory requirements leading to the evolution of the interwoven regulatory circuits of the FlhD-FlhC complex, ς28, and FlgM are not yet completely clear. They may promote ordered and efficient assembly of the flagellum by precisely timing the accumulation of precursor molecules in response to the developmental state of the structure. These regulatory systems may also provide a means of independently regulating the number and length of flagella. Flagellar biogenesis is intimately related to cell division and regulation of flagellar gene expression. The growth of the filament is independent of the cell cycle, and filament length appears to be under a localized control mechanism at the base of each flagellum, whereas the number of filaments (or flagellar basal bodies) is dependent on cell cycle (3). Flagellum number can be increased by increasing either FlhDC levels or ς28 activity. A mutation in flgM has been reported to increase flagellum number by two- to threefold in S. enterica serovar Typhimurium (67). Surface-induced differentiation of S. enterica serovar Typhimurium and E. coli from swimmer cells to swarmer cells also resulted in at least a twofold increase in flagellum number (33). This transition resulted from an increase in flhDC transcription. Since ς28 is now known to activate transcription of the flhDC operon, the number of flagella per cell might ultimately be regulated at the level of ς28 activity.
LINK BETWEEN REGULATION OF EXPRESSION AND EFFICIENCY OF ASSEMBLY
It is known that structural subunits are assembled into the flagella in a specific order. Therefore, if precursors accumulate as cellular pools that are not predisposed to incorporation into any particular growing flagellum, then either ordered protein expression or ordered secretion or both may occur. It is not known how precursor bias is accomplished in flagellar synthesis. The transcriptional hierarchy of the flagellum pathway is one mechanism that leads to the efficient assembly of the flagellum following induction of the pathway. FlgM is secreted following the completion of the hook-basal body intermediate structure (52, 108). This results in the full induction of class 3 promoter transcription. Thus, transcription of the filament genes fliC and fljB does not happen until a flagellar motor is in place onto which the filament can be assembled outside the cell. Given that the filament comprises a significant fraction of the total cell protein and at high concentrations will autopolymerize (82), it makes sense to link filament production to secretion competence.
Efficiency of assembly is further enhanced by the ability to change the protein substrate specificity at the hook-basal body assembly stage. The HAPs FlgK, FlgL, and FliD (the flagellar cap) are transcribed from class 2 promoters (66), yet they are in the late class of secretion substrate (37, 39, 40). During hook-basal body assembly, these proteins are produced in the cytoplasm (66). Once the hook is completed, the secretion apparatus substrate specificity changes to allow secretion and assembly of FlgK, FlgL, and FliD. If the HAPs were transcribed only from a class 3 promoter, as is the case for the filament, then the HAPs and the filament subunits would be produced at the same time and the HAPs might compete with filament subunits for secretion. Any filament subunits secreted prior to assembly of the HAPs would be discarded into the external medium and would lead to inefficient assembly.
In addition to the transcriptional hierarchy and secretion hierarchy, there are additional layers of posttranscriptional control that further increase the efficiency of flagellar assembly. As mentioned above, the Flk protein is hypothesized to regulate flgM mRNA translation in response to completion of the P- and L-rings and the elongation of the hook outside the cell (51, 53). In this way, intracellular FlgM levels may be lowered to allow some filament transcription just prior to hook completion. Thus, Flk may sense that hook-basal body completion is imminent and allow some filament subunits to be produced and ready for secretion as soon as they are needed. Otherwise, if filament transcription occurred only after hook-basal body completion and FlgM secretion, then a time lag would ensue in the assembly process. This time lag would include the time required for filament transcription and translation following FlgM secretion. By reducing the translation of flgM during hook elongation, some class 3 translation could allow a small amount of filament to be ready for secretion as the assembly of the HAPs is completed so that the efficiency of assembly of the entire flagellum is maximized.
Another posttranscriptional regulatory mechanism that enhances the assembly process is at the level of FlgE (hook) expression. Strains defective in basal body assembly showed reduced levels of hook protein present in the cells compared to a strain defective in HAP1 (FlgK) assembly (49). Transcription of flgE was unaffected by basal body mutations (12, 71). Recent studies of hook (FlgE) levels in different basal body-defective strains suggest that accumulation of intact hook protein is dependent upon successful assembly of previously translated flagellar components (12). In strains that were unable to assemble either the MS-ring (fliF), the switch complex (fliG, fliM, fliN), or a functional flagellar type III secretion apparatus (flhA, flhB, fliE, fliH, fliI, fliJ, fliO, fliP, fliQ, and fliR), hook protein was present at levels comparable to those in the wild type (12). However, negligible FlgE protein was detected in strains deficient for any structural or enzymatic component of basal body assembly (flgBCDFGJ) (12). Intermediate levels of FlgE were observed in strains blocked for ring or hook assembly (flgAIHD) (12). Wild-type levels were observed in strains missing the hook-filament junction proteins (flgKL) and in strains that did not have a functional flagellar type III secretion system (flhAB) (12). A threefold increase in FlgE was observed in a strain deleted for the hook length regulator FliK (12). These data suggested that FlgE expression was posttranscriptionally regulated in response to the stage of basal body assembly. FlgE translation or stability was decreased in the absence of assembly of prior hook-basal body components. This regulation of FlgE did not occur until after initiation of the basal body, because wild-type levels of the FlgE protein were present in strains unable to begin flagellar assembly (fliF mutant, MS-ring negative). The C-ring and type III secretion apparatus can be mounted onto the MS-ring once it is embedded into the inner membrane (2). Type III secretion mutant strains had wild-type levels of the FlgE protein, but mutants blocked for assembly of the basal body structural components had negligible levels of the protein (12). This suggests that either the MS-ring or the type III secretion apparatus was required for regulation of FlgE expression or stability until completion of the flagellar basal body. Once the MS-ring and the type III secretion apparatus were complete, FlgE protein levels were negatively regulated until initiation of hook assembly (12). The FlgE protein is the major component of the hook-basal body structure and the last assembled component of the hook-basal body. By preventing the accumulation of FlgE protein from the cell during rod assembly, the FlgE subunits would not compete with rod subunits that are required prior to hook assembly and cotranscribed with the flgE gene. An alternative possibility is that hook subunits and rod subunits are continuously secreted during hook-basal body assembly and that prior to rod formation, hook subunits are proteolyzed in the periplasm. This would alleviate a requirement for different secretion signals that direct rod subunits to be secreted prior to hook subunit secretion.
CELL CYCLE CONTROL
The bacterium Caulobacter crescentus differentiates from a motile swarmer cell into a nonmotile stalked cell (reviewed in references 26, 99, and 113). Synthesis and loss of the flagellum have been clearly demonstrated to be coupled to an asymmetric cell division, and extensive and elegant studies have elucidated many of the mechanisms that couple flagellar biosynthesis to the cell cycle. This has resulted in C. crescentus's being generally regarded as a model developmental system, and the characterization of flagellar regulation has been used to elucidate the mechanism of cell cycle control in Caulobacter (47, 99). Work in E. coli has also shown that flagellar biosynthesis and cell division are coregulated (102, 103), although it has proven more difficult to separate S. enterica serovar Typhimurium and E. coli cells by stage of development than C. crescentus. However, in an elegant study using a membrane filtration technique, Prüss and Matsumura were able to synchronize an E. coli culture (102). The expression of the flhDC operon increases to a maximum during mid-log-phase growth, decreases during late log phase, and then increases to half-maximal levels during stationary phase (102). Expression of a middle flhBAE operon was also growth phase dependent but reached its peak of expression in late log phase (5). Prüss and Matsumura demonstrated that expression of the early flhDC operon, the middle flhBAE operon, and the cells' swimming speed depended on the stage of the cell cycle that the cells were in (102). Expression of flhDC peaked at the middle of a cell cycle, followed by a peak in flhBAE expression half a cycle later (102). This demonstrates the temporal nature of flagellar gene expression in E. coli. Swimming speed was highest at the time of cell division. This suggests that swimming speed is dependent on the formation of new flagella following cell division and that it takes a full cycle to form functional flagella. These results also support earlier findings that expression of flagella decreased in conditional cell division mutants under nonpermissive conditions (96).
In S. enterica serovar Typhimurium, flagellar growth was synchronized by placing the flhDC operon under control of the tetA promoter (52). Placement of the flagellar regulon under control of the inducible tetA promoter allowed the determination of whether multiple flagella can grow simultaneously versus sequentially on a single cell. As described above, the flhDC master operon is influenced by a number of global regulatory signals. Placement of the flhDC structural genes under control of the tetA promoter allowed synchronous flagellar growth following the addition of tetracycline without interference from known or unknown regulatory signals that affect the expression of the flhDC promoter (52). Electron micrograph sampling following induction by tetracycline demonstrated that multiple flagella appeared simultaneously on the cell. However, it took about two cell cycles before the cells exhibited strong swimming behavior following induction of flhDC. It could be that this represents an artifact of using the tetA promoter to induce the flhDC operon, or the increase in swimming speed shown in E. coli was due to induction of flagella from two previous cell cycles.
Prüss and Matsumura also demonstrated that FlhD in the absence of FlhC is a regulator of cell division (103), and this regulation occurs through induction of the acid response gene cadA, encodong lysine decarboxylase (101). They hypothesize that cells degrade serine and synthesize acetylphosphate, which is used to phosphorylate OmpR. Shin and Park showed that acetylphosphate inhibited flhDC expression through OmpR (117). Production of acetylphosphate might serve as an indicator of the metabolic state of the cell or its local environment. FlhD is required to sense serine depletion through acetylphosphate, resulting in a reduction in the rate of cell division. How FlhD regulates cadA expression and how CadA might affect cell division are unclear.
The multiple control mechanisms described for the flagellar regulon appear to make sense only if different motors in the same cell are built simultaneously. These results suggest that normal induction of flagellar biosynthesis would be intimately associated with the cell cycle.
CONCLUSION
Bacterial flagella are complex structures requiring the input of over 50 genes for their assembly and function in response to the presence of attractants or repellents in the environment. Research over the last few years has revealed that regulation of the flagellar regulon is also highly complex. The efficiency at which the flagellar and chemotaxis genes are regulated throughout the assembly process and the sensitivity of the flagellar regulatory complex to numerous global regulatory systems suggest that the efficient coupling of the flagellar regulatory system to flagellum assembly is under enormous selective pressure in the environment.
The flagellar genes are transcriptionally regulated through an elaborate combination of mechanisms that create a regulon sensitive to environmental conditions, the physiology of the cell, and the developmental state of the motility and chemotaxis systems. Continued study is likely to unveil additional mechanisms, possibly including posttranscriptional mechanisms, in addition to the secretion-dependent regulation of FlgM. These may include regulation of protein stability or the efficiency of translation of individual cistrons. The numerous flagellar genes with nonstructural and unknown functions are candidates for regulators mediating nontranscriptional effects.
The exact regulatory requirements leading to the evolution of the interwoven regulatory circuits of the FlhD-FlhC complex, ς28, and FlgM are not yet completely clear. They may promote ordered and efficient assembly of the flagellum by precisely timing the accumulation of precursor molecules in response to the developmental state of the structure. These regulatory systems may also provide a means of independently regulating the number and length of flagella. It is certain that flagellation is a significant biosynthetic undertaking for the cell and that the more rapidly these complicated organelles can be synthesized, the faster an individual cell can swim toward a more favorable growth environment. This seems to have placed speed and efficiency at the forefront of selective pressures that resulted in such an intricate and versatile regulatory system. Given the importance of a large extracellular organelle system that constitutes a significant fraction of the total cell protein of flagellated cells, it is not surprising that novel regulatory mechanisms have been discovered during the study of the flagellar regulons of bacteria.
ACKNOWLEDGMENTS
We thank Shin-Ichi Aizawa for comments and suggestions.
This work was supported by Public Health Service grant GM56141 from the National Institutes of Health to K.T.H. G.S.C. is a recipient of a PHS National Research Award (G32 T GM07270).
REFERENCES
- 1.Adler J, Templeton B. The effect of environmental conditions on the motility of Escherichia coli. J Gen Microbiol. 1967;46:175–184. doi: 10.1099/00221287-46-2-175. [DOI] [PubMed] [Google Scholar]
- 2.Aizawa S-I. Flagellar assembly in Salmonella typhimurium. Mol Microbiol. 1996;20:1–4. doi: 10.1046/j.1365-2958.1996.344874.x. [DOI] [PubMed] [Google Scholar]
- 3.Aizawa S-I, Kubori T. Bacterial flagellation and cell division. Genes Cells. 1998;3:625–634. doi: 10.1046/j.1365-2443.1998.00219.x. [DOI] [PubMed] [Google Scholar]
- 4.Aizawa S-I, Vonderviszt F, Ishima R, Akasaka K. Termini of Salmonella flagellin are disordered and become organized upon polymerization into flagellar filament. J Mol Biol. 1990;211:673–677. doi: 10.1016/0022-2836(90)90064-S. [DOI] [PubMed] [Google Scholar]
- 5.Amsler C D, Cho M, Matsumura P. Multiple factors underlying the maximum motility of Escherichia coli as cultures enter post-exponential growth. J Bacteriol. 1993;175:6238–6244. doi: 10.1128/jb.175.19.6238-6244.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Anderson D, Schneewind O. A mRNA signal for the type III secretion of Yop proteins by Yersinia enterocolitica. Science. 1997;278:1140–1143. doi: 10.1126/science.278.5340.1140. [DOI] [PubMed] [Google Scholar]
- 7.Anderson D M, Schneewind O. Yersinia enterocolitica type III secretion: an mRNA signal that couples translation and secretion of YopQ. Mol Microbiol. 1999;31:1139–1148. doi: 10.1046/j.1365-2958.1999.01254.x. [DOI] [PubMed] [Google Scholar]
- 8.Arnosti D N, Chamberlin M J. Secondary ς factor controls transcription of flagellar and chemotaxis genes in Escherichia coli. Proc Natl Acad Sci USA. 1989;86:830–834. doi: 10.1073/pnas.86.3.830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bartlett D H, Frantz B B, Matsumura P. Flagellar transcriptional activators FlbB and FlaI: gene sequences and 5′ consensus sequences of operons under FlbB and FlaI control. J Bacteriol. 1988;170:1575–1581. doi: 10.1128/jb.170.4.1575-1581.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bennett J C, Hughes C. From flagellum assembly to virulence: the extended family of type III export chaperones. Trends Microbiol. 2000;8:202–204. doi: 10.1016/s0966-842x(00)01751-0. [DOI] [PubMed] [Google Scholar]
- 11.Bertin P, Terao E, Lee E H, Lejeune P, Colson C, Danchin A, Collatz E. The H-NS protein is involved in the biogenesis of flagella in Escherichia coli. J Bacteriol. 1994;176:5537–5540. doi: 10.1128/jb.176.17.5537-5540.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bonifield H R, Yamaguchi S, Hughes K T. The flagellar hook protein, FlgE, of Salmonella enterica serovar Typhimurium is posttranscriptionally regulated in response to the stage of flagellar assembly. J Bacteriol. 2000;182:4044–4050. doi: 10.1128/jb.182.14.4044-4050.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Burns D L. Biochemistry of type IV secretion. Curr Opin Microbiol. 1999;2:25–29. doi: 10.1016/s1369-5274(99)80004-6. [DOI] [PubMed] [Google Scholar]
- 14.Casadaban M J, Chou J. In vivo formation of gene fusions encoding hybrid beta-galactosidase proteins in one step with a transposable Mu-lac transducing phage. Proc Natl Acad Sci USA. 1984;81:535–539. doi: 10.1073/pnas.81.2.535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Casadaban M J, Cohen S N. Lactose genes fused to exogenous promoters in one step using a Mu-lac bacteriophage: in vivo probe for transcriptional control sequences. Proc Natl Acad Sci USA. 1979;76:4530–4533. doi: 10.1073/pnas.76.9.4530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Reference deleted.
- 17.Chadsey M S, Karlinsey J E, Hughes K T. The flagellar anti-sigma factor FlgM actively dissociates Salmonella typhimurium ς28 RNA polymerase holoenzyme. EMBO J. 1998;17:3123–3136. doi: 10.1101/gad.12.19.3123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cheng L W, Anderson D M, Schneewind O. Two independent type III secretion mechanisms for YopE in Yersinia enterocolitica. Mol Microbiol. 1997;24:757–765. doi: 10.1046/j.1365-2958.1997.3831750.x. [DOI] [PubMed] [Google Scholar]
- 19.Cheng L W, Schneewind O. Type III machines of Gram-negative bacteria: delivering the goods. Trends Microbiol. 2000;8:214–220. doi: 10.1016/s0966-842x(99)01665-0. [DOI] [PubMed] [Google Scholar]
- 20.Chilcott G S, Hughes K T. The type III secretion determinants of the flagellar anti-transcription factor, FlgM, extend from the amino-terminus into the anti-ς28 domain. Mol Microbiol. 1998;30:1029–1040. doi: 10.1046/j.1365-2958.1998.01131.x. [DOI] [PubMed] [Google Scholar]
- 21.Daughdrill G W, Chadsey M S, Karlinsey J E, Hughes K T, Dahlquist F W. The C-terminal half of the anti-sigma factor, FlgM, becomes structured when bound to its target ς28. Nat Struct Biol. 1997;4:285–291. doi: 10.1038/nsb0497-285. [DOI] [PubMed] [Google Scholar]
- 22.Decatur A, Losick R. Three sites of contact between the Bacillus subtilis transcription factor ςF and its antisigma factor SpoIIAB. Genes Dev. 1996;10:2348–2358. doi: 10.1101/gad.10.18.2348. [DOI] [PubMed] [Google Scholar]
- 23.DeRosier D J. The turn of the screw: the bacterial flagellar motor. Cell. 1998;93:17–20. doi: 10.1016/s0092-8674(00)81141-1. [DOI] [PubMed] [Google Scholar]
- 24.Dombroski A J, Walter W A, Gross C A. Amino-terminal amino acids modulated sigma-factor DNA-binding activity. Genes Dev. 1993;7:2446–2455. doi: 10.1101/gad.7.12a.2446. [DOI] [PubMed] [Google Scholar]
- 25.Dombroski A J, Walter W A, Record M T J, Siegele D A, Gross C A. Polypeptides containing highly conserved regions of transcription initiation factor sigma 70 exhibit specificity of binding to promoter DNA. Cell. 1992;70:501–512. doi: 10.1016/0092-8674(92)90174-b. [DOI] [PubMed] [Google Scholar]
- 26.Domian I J, Quon K C, Shapiro L. The control of temporal and spatial organization during the Caulobacter cell cycle. Curr Cpin Genet Dev. 1996;6:538–544. doi: 10.1016/s0959-437x(96)80081-5. [DOI] [PubMed] [Google Scholar]
- 27.Dorman C J, Hinton J C, Free A. Domain organization and oligomerization among H-NS-like nucleoid-associated proteins in bacteria. Trends Microbiol. 1999;7:124–128. doi: 10.1016/s0966-842x(99)01455-9. [DOI] [PubMed] [Google Scholar]
- 28.Dufour A, Furness R B, Hughes C. Novel genes that upregulate the Proteus mirabilis flhDC master operon controlling flagellar biogenesis and swarming. Mol Microbiol. 1998;29:741–751. doi: 10.1046/j.1365-2958.1998.00967.x. [DOI] [PubMed] [Google Scholar]
- 29.Fraser G M, Bennett J C, Hughes C. Substrate-specific binding of hook-associated proteins by FlgN and FliT, putative chaperones for flagellum assembly. Mol Microbiol. 1999;32:569–580. doi: 10.1046/j.1365-2958.1999.01372.x. [DOI] [PubMed] [Google Scholar]
- 30.Gillen K L, Hughes K T. Molecular characterization of flgM, a gene encoding a negative regulator of flagellin synthesis in Salmonella typhimurium. J Bacteriol. 1991;173:6453–6459. doi: 10.1128/jb.173.20.6453-6459.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gillen K L, Hughes K T. Negative regulatory loci coupling flagellin synthesis to flagellar assembly in Salmonella typhimurium. J Bacteriol. 1991;173:2301–2310. doi: 10.1128/jb.173.7.2301-2310.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gillen K L, Hughes K T. Transcription from two promoters and autoregulation contribute to the control of expression of the Salmonella typhimurium flagellar regulatory gene flgM. J Bacteriol. 1993;175:7006–7015. doi: 10.1128/jb.175.21.7006-7015.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Harshey R M, Matsuyama T. Dimorphic transition in Escherichia coli and Salmonella typhimurium: surface-induced differentiation into hyperflagellate swarmer cells. Proc Natl Acad Sci USA. 1994;91:8631–8635. doi: 10.1073/pnas.91.18.8631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Helmann J D. Alternative sigma factors and the regulation of flagellar gene expression. Mol Microbiol. 1991;5:2875–2882. doi: 10.1111/j.1365-2958.1991.tb01847.x. [DOI] [PubMed] [Google Scholar]
- 35.Helmann J D, Chamberlin M J. DNA sequence analysis suggests that expression of flagellar and chemotaxis genes in Escherichia coli and Salmonella typhimurium is controlled by an alternative sigma factor. Proc Natl Acad Sci USA. 1987;84:6422–6424. doi: 10.1073/pnas.84.18.6422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hirano T, Yamaguchi S, Oosawa K, Aizawa S-I. Roles of FliK and FlhB in determination of flagellar hook length in Salmonella typhimurium. J Bacteriol. 1994;176:5439–5449. doi: 10.1128/jb.176.17.5439-5449.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Homma M, Fujita H, Yamaguchi S, Iino T. Excretion of unassembled flagellin by Salmonella typhimurium mutants deficient in hook-associated proteins. J Bacteriol. 1964;159:1056–1059. doi: 10.1128/jb.159.3.1056-1059.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Homma M, Fujita H, Yamaguchi S, Iino T. Regions of Salmonella typhimurium flagellin essential for its polymerization and excretion. J Bacteriol. 1987;169:291–296. doi: 10.1128/jb.169.1.291-296.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Homma M, Iino T. Locations of hook-associated proteins in flagellar structures of Salmonella typhimurium. J Bacteriol. 1985;162:183–189. doi: 10.1128/jb.162.1.183-189.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Homma M, Kutsukake K, Iino T, Yamaguchi S. Hook-associated proteins essential for flagellar filament formation in Salmonella typhimurium: J. Bacteriol. 1964;157:100–108. doi: 10.1128/jb.157.1.100-108.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hueck C. Type III protein secretion systems in the bacterial pathogens of animals and plants. Microbiol Mol Biol Rev. 1998;62:379–433. doi: 10.1128/mmbr.62.2.379-433.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hughes K T, Gillen K L, Semon M J, Karlinsey J E. Sensing structural intermediates in bacterial flagellar assembly by export of a negative regulator. Science. 1993;262:1277–1280. doi: 10.1126/science.8235660. [DOI] [PubMed] [Google Scholar]
- 43.Ide N, Ikebe T, Kutsukake K. Reevaluation of the promoter structure of the class 3 flagellar operons of Escherichia coli and Salmonella. Genes Genet Syst. 1999;74:113–116. doi: 10.1266/ggs.74.113. [DOI] [PubMed] [Google Scholar]
- 44.Ikebe T, Iyoda S, Kutsukake K. Promoter analysis of the class 2 flagellar operons of Salmonella. Genes Genet Syst. 1999;74:179–183. doi: 10.1266/ggs.74.179. [DOI] [PubMed] [Google Scholar]
- 45.Ikebe T, Iyoda S, Kutsukake K. Structure and expression of the fliA operon of Salmonella typhimurium. Microbiology. 1999;145:1389–1396. doi: 10.1099/13500872-145-6-1389. [DOI] [PubMed] [Google Scholar]
- 46.Iyoda S, Kutsukake K. Molecular dissection of the flagellum-specific anti-sigma factor, FlgM, of Salmonella typhimurium. Mol Gen Genet. 1995;249:417–424. doi: 10.1007/BF00287103. [DOI] [PubMed] [Google Scholar]
- 47.Jacobs C, Domian I J, Maddock J R, Shapiro L. Cell cycle-dependent polar localization of an essential bacterial histidine kinase that controls DNA replication and cell division. Cell. 1999;97:111–120. doi: 10.1016/s0092-8674(00)80719-9. [DOI] [PubMed] [Google Scholar]
- 48.Jishage M, Ishihama A. A stationary phase protein in Escherichia coli with binding activity to the major sigma subunit of RNA polymerase. Proc Natl Acad Sci USA. 1998;95:4953–4958. doi: 10.1073/pnas.95.9.4953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jones C J, Macnab R M. Flagellar assembly in Salmonella typhimurium: analysis with temperature-sensitive mutants. J Bacteriol. 1990;172:1327–1339. doi: 10.1128/jb.172.3.1327-1339.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Karlinsey J E, Lonner J, Brown K L, Hughes K T. Translation secretion coupling by type III secretion systems. Cell. 2000;102:487–497. doi: 10.1016/s0092-8674(00)00053-2. [DOI] [PubMed] [Google Scholar]
- 51.Karlinsey J E, Pease A J, Winkler M E, Bailey J L, Hughes K T. The flk gene of Salmonella typhimurium couples flagellar P- and L-ring assembly to flagellar morphogenesis. J Bacteriol. 1997;179:2389–2400. doi: 10.1128/jb.179.7.2389-2400.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Karlinsey J E, Shugo Tanaka S, Bettenworth V, Yamaguchi S, Boos W, Aizawa S I, Hughes K T. Completion of the hook-basal body of the Salmonella typhimurium flagellum is coupled to FlgM secretion and fliC transcription. Mol Microbiol. 2000;37:1220–1231. doi: 10.1046/j.1365-2958.2000.02081.x. [DOI] [PubMed] [Google Scholar]
- 53.Karlinsey J E, Tsui H-C T, Winkler M E, Hughes K T. Flk couples flgM translation to flagellar ring assembly in Salmonella typhimurium. J Bacteriol. 1998;180:5384–5397. doi: 10.1128/jb.180.20.5384-5397.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kimbrough T G, Miller S I. Contribution of Salmonella typhimurium type III secretion components to needle complex formation. Proc Natl Acad Sci USA. 2000;97:11008–11013. doi: 10.1073/pnas.200209497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Komeda Y. Fusions of flagellar operons to lactose genes on a Mu lac bacteriophage. J Bacteriol. 1982;150:16–26. doi: 10.1128/jb.150.1.16-26.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Komeda Y. Isolation of fla-lacZ fusions in Escherichia coli K-12: most fusions result in soluble β-galactosidase. J Bacteriol. 1988;170:1980–1983. doi: 10.1128/jb.170.4.1980-1983.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Komeda Y. Transcriptional control of flagellar genes in Escherichia coli K-12. J Bacteriol. 1986;168:1315–1318. doi: 10.1128/jb.168.3.1315-1318.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Komeda Y, Suzuki H, Ishidsu J I, Iino T. The role of cAMP in flagellation of Salmonella typhimurium. Mol Gen Genet. 1975;142:289–298. doi: 10.1007/BF00271253. [DOI] [PubMed] [Google Scholar]
- 59.Kornacker M G, Newton A. Information essential for cell-cycle-dependent secretion of the 591-residue Caulobacter hook protein is confined to a 21-amino-acid sequence near the N-terminus. Mol Microbiol. 1994;14:73–85. doi: 10.1111/j.1365-2958.1994.tb01268.x. [DOI] [PubMed] [Google Scholar]
- 60.Koroyasu S, Yamazato M, Hirano T, Aizawa S-I. Kinetic analysis of the growth rate of the flagellar hook in Salmonella typhimurium by the population balance method. Biophys J. 1998;74:436–443. doi: 10.1016/S0006-3495(98)77801-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kubori T, Matsushima Y, Nakamura D, Uralil J, Lara-Tejero M, Sukhan A, Galan J E, Aizawa S I. Supramolecular structure of the Salmonella typhimurium type III protein secretion system. Science. 1998;280:602–605. doi: 10.1126/science.280.5363.602. [DOI] [PubMed] [Google Scholar]
- 62.Kundu T K, Kusano S, Ishihama A. Promoter selectivity of Escherichia coli RNA polymerase ςF holoenzyme involved in transcription of flagellar and chemotaxis genes. J Bacteriol. 1997;179:4264–4269. doi: 10.1128/jb.179.13.4264-4269.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kutsukake K. Autogenous and global control of the flagellar master operon, flhD, in Salmonella typhimurium. Mol Gen Genet. 1997;254:440–448. doi: 10.1007/s004380050437. [DOI] [PubMed] [Google Scholar]
- 64.Kutsukake K. Excretion of the anti-sigma factor through a flagellar substructure couples flagellar gene expression with flagellar assembly in Salmonella typhimurium. Mol Gen Genet. 1994;243:605–612. doi: 10.1007/BF00279569. [DOI] [PubMed] [Google Scholar]
- 65.Kutsukake K. Hook-length control of the export-switching machinery involves a double-locked gate in Salmonella typhimurium flagellar morphogenesis. J Bacteriol. 1997;179:1268–1273. doi: 10.1128/jb.179.4.1268-1273.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kutsukake K, Ide N. Transcriptional analysis of the flgK and fliD operons of Salmonella typhimurium which encode flagellar hook-associated proteins. Mol Gen Genet. 1995;247:275–281. doi: 10.1007/BF00293195. [DOI] [PubMed] [Google Scholar]
- 67.Kutsukake K, Iino T. Role of the FliA-FlgM regulatory system on the transcriptional control of the flagellar regulon and flagellar formation in Salmonella typhimurium. J Bacteriol. 1994;176:3598–3605. doi: 10.1128/jb.176.12.3598-3605.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kutsukake K, Ikebe T, Yamamoto S. Two novel regulatory genes, fliT and fliZ, in the flagellar regulon of Salmonella. Genes Genet Syst. 1999;74:287–292. doi: 10.1266/ggs.74.287. [DOI] [PubMed] [Google Scholar]
- 69.Kutsukake K, Iyoda S, Ohnishi K, Iino T. Genetic and molecular analyses of the interaction between the flagellum-specific sigma and its anti-sigma factors in Salmonella typhimurium. EMBO J. 1994;13:4568–4576. doi: 10.1002/j.1460-2075.1994.tb06778.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kutsukake K, Minamino T, Yokoseki T. Isolation and characterization of FliK-independent flagellation mutants from Salmonella typhimurium. J Bacteriol. 1994;176:7625–7629. doi: 10.1128/jb.176.24.7625-7629.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kutsukake K, Ohya Y, Iino T. Transcriptional analysis of the flagellar regulon of Salmonella typhimurium. J Bacteriol. 1990;172:741–747. doi: 10.1128/jb.172.2.741-747.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kutsukake K, Okada T, Yokoseki T, Iino T. Sequence analysis of the flgA gene and its adjacent region in Salmonella typhimurium, and identification of another flagellar gene, flgN. Gene. 1994;143:49–54. doi: 10.1016/0378-1119(94)90603-3. [DOI] [PubMed] [Google Scholar]
- 73.Kuwajima G, Kawagishi I, Homma M, Asaka J, Kondo E, Macnab R M. Export of an N-terminal fragment of Escherichia coli flagellin by a flagellum-specific pathway. Proc Natl Acad Sci USA. 1989;86:4953–4957. doi: 10.1073/pnas.86.13.4953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Li C, Louise C J, Shi W, Adler J. Adverse conditions which cause lack of flagella in Escherichia coli. J Bacteriol. 1993;175:2229–2235. doi: 10.1128/jb.175.8.2229-2235.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Liu X, Matsumura P. An alternative sigma factor controls transcription of flagellar class-III operons in Escherichia coli: gene sequence, overproduction, purification and characterization. Gene. 1995;164:81–84. doi: 10.1016/0378-1119(95)00480-t. [DOI] [PubMed] [Google Scholar]
- 76.Liu X, Matsumura P. The C-terminal region of the alpha subunit of Escherichia coli RNA polymerase is required for transcriptional activation of the flagellar level II operons by the FlhD/FlhC complex. J Bacteriol. 1995;177:5186–5188. doi: 10.1128/jb.177.17.5186-5188.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Liu X, Matsumura P. Differential regulation of multiple overlapping promoters in flagellar class II operons in Escherichia coli. Mol Microbiol. 1996;21:613–620. doi: 10.1111/j.1365-2958.1996.tb02569.x. [DOI] [PubMed] [Google Scholar]
- 78.Liu X, Matsumura P. The FlhD/FlhC complex, a transcriptional activator of the Escherichia coli flagellar class II operons. J Bacteriol. 1994;176:7345–7351. doi: 10.1128/jb.176.23.7345-7351.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Lonetto M, Gribskow M, Gross C A. The ς70 family: sequence conservation and evolutionary relationships. J Bacteriol. 1992;174:3843–3849. doi: 10.1128/jb.174.12.3843-3849.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Lory S. Secretion of proteins and assembly of bacterial surface organelles: shared pathways of extracellular protein targeting. Curr Opin Microbiol. 1998;1:27–35. doi: 10.1016/s1369-5274(98)80139-2. [DOI] [PubMed] [Google Scholar]
- 81.Losick R, Shapiro L. Checkpoints that couple gene expression to morphogenesis. Science. 1993;262:1227–1228. doi: 10.1126/science.8235653. [DOI] [PubMed] [Google Scholar]
- 82.Lowry J, McDonough M W. Structure of filaments produced by re-aggregation of Salmonella flagellin. Nature. 1964;204:125–127. doi: 10.1038/204125a0. [DOI] [PubMed] [Google Scholar]
- 83.Lupas A. Coiled coils: new structures and new functions. Trends Biochem Sci. 1996;21:375–382. [PubMed] [Google Scholar]
- 84.Macnab R M. The bacterial flagellum: reversible rotary propellor and type III export apparatus. J Bacteriol. 1999;181:149–153. doi: 10.1128/jb.181.23.7149-7153.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Macnab R M. Flagella and motility. In: Neidhart F C, et al., editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C.: ASM Press; 1996. pp. 123–145. [Google Scholar]
- 86.Malhotra A, Severinova E, Darst S A. Crystal structure of a sigma 70 subunit fragment from E. coli RNA polymerase. Cell. 1996;87:127–136. doi: 10.1016/s0092-8674(00)81329-x. [DOI] [PubMed] [Google Scholar]
- 87.Michiels T, Cornelis G R. Secretion of hybrid proteins by the Yersinia Yop export system. J Bacteriol. 1991;173:1677–1685. doi: 10.1128/jb.173.5.1677-1685.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Minamino T, Doi H, Kutsukake K. Substrate specificity switching of the flagellum-specific export apparatus during flagellar morphogenesis in Salmonella typhimurium. Biosci Biotechnol Biochem. 1999;63:1301–1303. doi: 10.1271/bbb.63.1301. [DOI] [PubMed] [Google Scholar]
- 89.Minamino T, Gonzalez-Pedrajo B, Yamaguchi K, Aizawa S-I, Macnab R M. FliK, the protein responsible for flagellar hook length control in Salmonella, is exported during hook assembly. Mol Microbiol. 1999;34:295–304. doi: 10.1046/j.1365-2958.1999.01597.x. [DOI] [PubMed] [Google Scholar]
- 90.Mizushima T, Koyanagi R, Katayama T, Miki T, Sekimizu K. Decrease in expression of the master operon of flagellin synthesis in a dnaA46 mutant of Escherichia coli. Biol Pharm Bull. 1997;20:327–331. doi: 10.1248/bpb.20.327. [DOI] [PubMed] [Google Scholar]
- 91.Mizushima T, Koyanagi R, Suzuki E, Tomura A, Kutsukake K, Miki T T, Sekimizu K. Control by phosphotidylglycerol of expression of the flhD gene of Escherichia coli. Biochim Biophys Acta. 1995;1245:397–401. doi: 10.1016/0304-4165(95)00114-x. [DOI] [PubMed] [Google Scholar]
- 92.Mizushima T, Tomura A, Shinpuku T, Miki T, Sekimizu K. Loss of flagellation in dnaA mutants of Escherichia coli. J Bacteriol. 1994;176:5544–5546. doi: 10.1128/jb.176.17.5544-5546.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Morgan D G, Owen C, Melanson L A, DeRosier D J. Structure of bacterial flagellar filaments at 11 Å resolution: packing of the alpha-helices. J Mol Biol. 1995;249:88–110. doi: 10.1006/jmbi.1995.0282. [DOI] [PubMed] [Google Scholar]
- 94.Murphy C K, Beckwith J. Export of proteins to the cell envelope in Escherichia coli. In: Neidhart F C, et al., editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C.: ASM Press; 1996. pp. 967–978. [Google Scholar]
- 95.Mytelka D S, Chamberlin M J. Escherichia coli fliAZY operon. J Bacteriol. 1996;178:24–34. doi: 10.1128/jb.178.1.24-34.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Nishimura A, Hirota Y. A cell division regulatory mechanism controls the flagellar regulon in Escherichia coli. Mol Gen Genet. 1989;216:340–346. doi: 10.1007/BF00334374. [DOI] [PubMed] [Google Scholar]
- 97.Ohnishi K, Kutsukake K, Suzuki H, Iino T. Gene fliA encodes an alternative sigma factor specific for flagellar operons in Salmonella typhimurium. Mol Gen Genet. 1990;221:139–147. doi: 10.1007/BF00261713. [DOI] [PubMed] [Google Scholar]
- 98.Ohnishi K, Kutsukake K, Suzuki H, Iino T. A novel transcriptional regulatory mechanism in the flagellar regulon of Salmonella typhimurium: an anti sigma factor inhibits the activity of the flagellum-specific sigma factor, ςF. Mol Microbiol. 1992;6:3149–3157. doi: 10.1111/j.1365-2958.1992.tb01771.x. [DOI] [PubMed] [Google Scholar]
- 99.Osteras M, Jenal U. Regulatory circuits in Caulobacter. Curr Opin Microbiol. 2000;3:171–176. doi: 10.1016/s1369-5274(00)00071-0. [DOI] [PubMed] [Google Scholar]
- 100.Patterson-Delafield J, Martinez R J, Stocker B A, Yamaguchi S. A new fla gene in Salmonella typhimurium—flaR—and its mutant phenotype—superhooks. Arch Mikrobiol. 1973;90:107–120. doi: 10.1007/BF00414513. [DOI] [PubMed] [Google Scholar]
- 101.Prüss B M, Markovic D, Matsumura P. The Escherichia coli flagellar transcriptional activator FlhD regulates cell division through induction of the acid response gene cadA. J Bacteriol. 1997;179:3818–3821. doi: 10.1128/jb.179.11.3818-3821.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Prüss B M, Matsumura P. Cell cycle regulation of flagellar genes. J Bacteriol. 1997;179:5602–5604. doi: 10.1128/jb.179.17.5602-5604.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Prüss B M, Matsumura P. A regulator of the flagellar regulon of Escherichia coli, flhD, also affects cell division. J Bacteriol. 1996;178:668–674. doi: 10.1128/jb.178.3.668-674.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Prüss B M, Wolfe A J. Regulation of acetyl phosphate synthesis and degradation, and the control of flagellar expression in Escherichia coli. Mol Microbiol. 1994;12:973–984. doi: 10.1111/j.1365-2958.1994.tb01085.x. [DOI] [PubMed] [Google Scholar]
- 105.Qualding C, Stocker B A D. An environmentally-induced transition from the flagellated to the non-flagellated state in Salmonella typhimurium: the fate of parental flagella at cell division. J Gen Microbiol. 1962;28:257–270. doi: 10.1099/00221287-28-2-257. [DOI] [PubMed] [Google Scholar]
- 106.Rosqvist R, Hakansson S, Forsberg A, Wolf-Watz H. Functional conservation of the secretion and translocation machinery for virulence proteins of yersiniae, salmonellae and shigellae. EMBO J. 1995;14:4187–4195. doi: 10.1002/j.1460-2075.1995.tb00092.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Ruiz T, Francis N R, Morgan D G, DeRosier D J. Size of the export channel in the flagellar filament of Salmonella typhimurium. Ultramicroscopy. 1993;49:417–425. doi: 10.1016/0304-3991(93)90247-u. [DOI] [PubMed] [Google Scholar]
- 108.Saito T, Ueno T, Kubori T, Yamaguchi S, Iino T, Aizawa S-I. Flagellar filament elongation can be impaired by mutations in the hook protein FlgE of Salmonella typhimurium: a possible role of the hook as a passage for the anti-sigma factor FlgM. Mol Microbiol. 1998;27:1129–1139. doi: 10.1046/j.1365-2958.1998.00738.x. [DOI] [PubMed] [Google Scholar]
- 109.Schaubach O L, Dombroski A J. Transcription initiation at the flagellin promoter by RNA polymerase carrying sigma28 from Salmonella typhimurium. J Biol Chem. 1999;274:8757–8763. doi: 10.1074/jbc.274.13.8757. [DOI] [PubMed] [Google Scholar]
- 110.Schesser K, Frithz-Lindsten E, Wolf-Watz H. Delineation and mutational analysis of the Yersinia pseudotuberculosis YopE domains which mediate translocation across bacterial and eukaryotic cellular membranes. J Bacteriol. 1996;178:7227–7233. doi: 10.1128/jb.178.24.7227-7233.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Schmitt C K, Darnell S C, Tesh V L, Stocker B A, O'Brien A D. Mutation of flgM attenuates virulence of Salmonella typhimurium, and mutation of fliA represses the attenuated phenotype. J Bacteriol. 1994;176:368–377. doi: 10.1128/jb.176.2.368-377.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Severinova E, Severinov K, Fenyo D, Marr M, Brody E N, Roberts J W, Chait B T, Darst S A. Domain organization of the Escherichia coli RNA polymerase sigma 70 subunit. J Mol Biol. 1996;263:637–647. doi: 10.1006/jmbi.1996.0604. [DOI] [PubMed] [Google Scholar]
- 113.Shapiro L, Losick R. Dynamic spatial regulation in the bacterial cell. Cell. 2000;100:89–98. doi: 10.1016/s0092-8674(00)81686-4. [DOI] [PubMed] [Google Scholar]
- 114.Shi W, Bogdanov M, Dowhan W, Zusman D R. The pss and psd genes are required for motility and chemotaxis in Escherichia coli. J Bacteriol. 1993;175:7711–7714. doi: 10.1128/jb.175.23.7711-7714.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Shi W, Li C, Louise C J, Adler J. Mechanism of adverse conditions causing lack of flagella in Escherichia coli. J Bacteriol. 1993;175:2236–2240. doi: 10.1128/jb.175.8.2236-2240.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Shi W, Zhou Y, Wild J, Adler J, Gross C A. DnaK, DnaJ, and GrpE are required for flagellum synthesis in Escherichia coli. J Bacteriol. 1992;174:6256–6263. doi: 10.1128/jb.174.19.6256-6263.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Shin S, Park C. Modulation of flagellar expression in Escherichia coli by acetyl phosphate and the osmoregulator OmpR. J Bacteriol. 1995;177:4696–4702. doi: 10.1128/jb.177.16.4696-4702.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Silhavy T J. Death by lethal injection. Science. 1997;278:1085–1086. doi: 10.1126/science.278.5340.1085. [DOI] [PubMed] [Google Scholar]
- 119.Silverman M, Simon M. Characterization of Escherichia coli flagellar mutants that are insensitive to catabolite repression. J Bacteriol. 1974;120:1196–1203. doi: 10.1128/jb.120.3.1196-1203.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Sory M P, Boland A, Lambermont I, Cornelis G R. Identification of the YopE and YopH domains required for secretion and internalization into the cytosol of macrophages, using the cyaA gene fusion approach. Proc Natl Acad Sci USA. 1995;92:11998–12002. doi: 10.1073/pnas.92.26.11998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Soutourina O, Kolb A, Krin E, Laurent-Winter C, Rimsky S, Danchin A, Bertin P. Multiple control of flagellum biosynthesis in Escherichia coli: role of H-NS protein and the cyclic AMP-catabolite activator protein complex in transcription of the flhDC master operon. J Bacteriol. 1999;181:7500–7508. doi: 10.1128/jb.181.24.7500-7508.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Stephens C, Shapiro L. Bacterial protein secretion—a target for new antibiotics? Chem Biol. 1997;4:637–641. doi: 10.1016/s1074-5521(97)90217-9. [DOI] [PubMed] [Google Scholar]
- 123.Stephens C, Shapiro L. Delivering the payload: bacterial pathogenesis. Curr Biol. 1996;6:927–930. doi: 10.1016/s0960-9822(02)00628-0. [DOI] [PubMed] [Google Scholar]
- 124.Suzuki H, Iino T. Absence of messenger ribonucleic acid specific for flagellin in non-flagellate mutants of Salmonella. J Mol Biol. 1975;95:549–556. doi: 10.1016/0022-2836(75)90316-2. [DOI] [PubMed] [Google Scholar]
- 125.Tamano K, Aizawa S I, Katayama E, Nonaka T, Imajoh-Ohmi S, Kuwae A, Nagai S, Sasakawa C. Supramolecular structure of the Shigella type III secretion machinery: the needle part is changeable in length and essential for delivery of effectors. EMBO J. 2000;19:3876–3887. doi: 10.1093/emboj/19.15.3876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Van Gijsegem F, Gough C, Zischek C, Niqueux E, Arlat M, Genin S, Barberis P, German S, Castello P, Boucher C. The hrp gene locus of Pseudomonas solanacearum, which controls the production of a type III secretion system, encodes eight proteins related to components of the bacterial flagellar biogenesis complex. Mol Microbiol. 1995;15:1095–1114. doi: 10.1111/j.1365-2958.1995.tb02284.x. [DOI] [PubMed] [Google Scholar]
- 127.Vonderviszt F, Aizawa S, Namba K. Role of the disordered terminal regions of flagellin in filament formation and stability. J Mol Biol. 1991;221:1461–1474. doi: 10.1016/0022-2836(91)90946-4. [DOI] [PubMed] [Google Scholar]
- 128.Vonderviszt F, Ishima R, Akasaka K, Aizawa S. Terminal disorder: a common structural feature of the axial proteins of bacterial flagellum. J Mol Biol. 1992;226:575–579. doi: 10.1016/0022-2836(92)90616-r. [DOI] [PubMed] [Google Scholar]
- 129.Vonderviszt F, Kanto S, Aizawa S, Namba K. Terminal regions of flagellin are disordered in solution. J Mol Biol. 1989;209:127–133. doi: 10.1016/0022-2836(89)90176-9. [DOI] [PubMed] [Google Scholar]
- 130.Williams A W, Yamaguchi S, Togashi F, Aizawa S I, Kawagishi I, Macnab R M. Mutations in fliK and flhB affecting flagellar hook and filament assembly in Salmonella typhimurium. J Bacteriol. 1996;178:2960–2970. doi: 10.1128/jb.178.10.2960-2970.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Woestyn S, Sory M P, Boland A, Lequenne O, Cornelis G R. The cytosolic SycE and SycH chaperones of Yersinia protect the region of YopE and YopH involved in translocation across eukaryotic cell membranes. Mol Microbiol. 1996;20:1261–1271. doi: 10.1111/j.1365-2958.1996.tb02645.x. [DOI] [PubMed] [Google Scholar]
- 132.Yanagihara S, I;-da S, Ohnishi K, Iino T, Kutsukake K. Structure and transcriptional control of the flagellar master operon of Salmonella typhimurium. Genes Genet Syst. 1999;74:105–111. doi: 10.1266/ggs.74.105. [DOI] [PubMed] [Google Scholar]
- 133.Yokota T, Gots J S. Requirement of adenosine 3′,5′-cyclic phosphate for flagellum formation in Escherichia coli and Salmonella typhimurium. J Bacteriol. 1970;103:513–516. doi: 10.1128/jb.103.2.513-516.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]