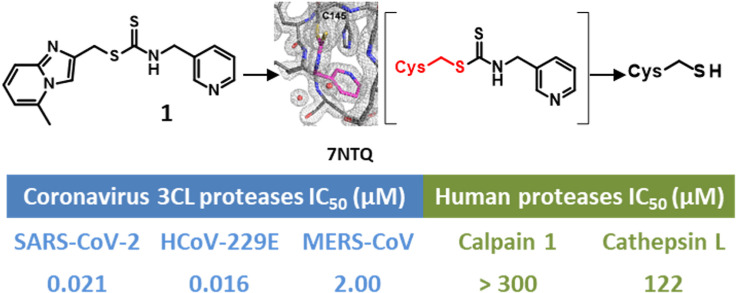
Abstract
Since end of 2019, the global and unprecedented outbreak caused by the coronavirus SARS-CoV-2 led to dramatic numbers of infections and deaths worldwide. SARS-CoV-2 produces two large viral polyproteins which are cleaved by two cysteine proteases encoded by the virus, the 3CL protease (3CLpro) and the papain-like protease, to generate non-structural proteins essential for the virus life cycle. Both proteases are recognized as promising drug targets for the development of anti-coronavirus chemotherapy. Aiming at identifying broad spectrum agents for the treatment of COVID-19 but also to fight emergent coronaviruses, we focused on 3CLpro that is well conserved within this viral family. Here we present a high-throughput screening of more than 89,000 small molecules that led to the identification of a new chemotype, potent inhibitor of the SARS-CoV-2 3CLpro. The mechanism of inhibition, the interaction with the protease using NMR and X-Ray, the specificity against host cysteine proteases and promising antiviral properties in cells are reported.
Keywords: SARS-CoV-2, Coronavirus, 3CL protease Inhibitors, Antiviral
Graphical abstract
1. Introduction
The outbreak of severe acute respiratory syndrome (SARS) [1] in 2002, as well as the Middle-East respiratory syndrome (MERS) [2] in 2012 and COVID-19 [3] since 2019 demonstrates the potential of coronaviruses to cross boundaries between species and highlights the importance of urgently developing efficient antiviral compounds with broad spectrum activity against this virus family.
The newly emerged coronavirus SARS-CoV-2 is the seventh reported coronavirus having the potential to infect human. It is an enveloped, positive single-stranded RNA virus that can infect both humans and animals. Coronaviruses contain the largest known RNA genome encoding, in addition to the structural and accessory proteins, two large viral polyproteins, pp1a and pp1ab. These polyproteins are processed by two viral proteases, the papain-like protease (PLP) and the 3C-like protease (3CLpro, also known as the main protease Mpro) to generate a series of functional non-structural proteins essential for virus replication and transcription.
The viral main protease (3CLpro), a 33.8 kDa cysteine protease with a non-classical Cys-His dyad (Cys145-His41) is active as a homodimer. As an essential component for the formation of the coronavirus replication complex, this protease is an attractive target for the development of anti-coronavirus therapeutics. Moreover, the remarkable degree of conservation of this protease among the viruses of this family (96% sequence identity between the SARS-CoV-2 and SARS-CoV) is a strong asset to design pan-anti-coronavirus drugs to tackle not only the current SARS-CoV-2 but also potential future outbreaks of emergent human coronaviruses [4,5]. Interestingly, 3CLpro cleaves the polyprotein 1 ab at 11 cleavage sites with a common cleavage sequence LQ↓(S/A/G) that is unusual for human proteases [6,7]. This may allow for identifying drug candidates with high specificity for the viral protease reducing unwanted polypharmacology and potential side effects.
Several 3CLpro inhibitors are currently in preclinical and clinical development for the treatment of COVID-19 [[8], [9], [10], [11], [12], [13], [14], [15]]. Amongst the most potent compounds, GC-376, a peptidomimetic broad-spectrum antiviral agent developed for feline coronavirus infections, inhibits the replication of noroviruses, picornaviruses, and coronaviruses [16]. Recently two other peptidomimetic broad-spectrum coronavirus 3CLpro inhibitors with high selectivity over human proteases; PF-07304814, a phosphate prodrug metabolized in vivo to the active moiety PF-00835231 was developed [17] and PF-7321332 (nirmatrelvir), a covalent inhibitor specifically designed to be administered orally, is currently approved [18]. However, the promising clinical results reported by Pfizer for nirmatrelvir are dependent upon coadministration with ritonavir as a pharmacokinetic (PK) enhancer. In addition to peptidomimetic inhibitors that can be challenging in terms of selectivity and PK profile, novel non peptidomimetic small molecule pan-inhibitors of 3CLpro are also highly desirable to fight COVID-19 and potential emerging coronaviruses.
Here we report the high-throughput screening of a large library of more than 89,000 small compounds on the 3CLpro of SARS-CoV-2 and the identification of a novel class of non-peptidomimetic small inhibitors with promising anti-coronavirus activity.
2. Results and discussion
2.1. Development of the enzymatic assay and high-throughput screening
2.1.1. SARS-CoV-2 3CLpro expression
It has been shown that extra amino-acid residues or affinity tags at either the N-terminus or C-terminus significantly decrease 3CLpro enzymatic activity via disturbing its dimerization and/or active site conformation. The native SARS-CoV-2 3CLpro was thus recombinantly expressed in Escherichia coli with a cleavable tag and purified to homogeneity. The N-terminal 6 × His-SUMO tag that was later cleaved using SENP2 protease (His-tagged) to release the native SARS-CoV-2 3CLpro without any extra residue at its extremities. The high yield for expression and purification (∼100 mg/L culture) allowed us to work with a single batch of the protease leading to a high reproducibility of our enzymatic high-throughput screening.
2.1.2. Enzyme characterization, assay development, validation of the assay with known inhibitors
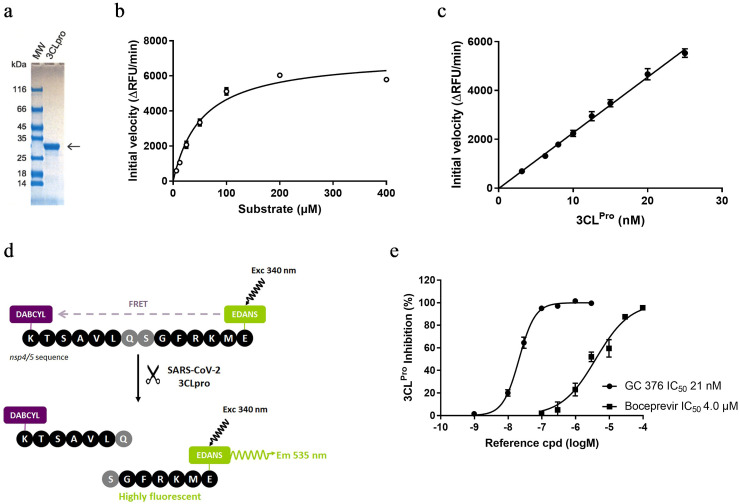
Aiming at rapidly developing a Förster Resonance Energy Transfer (FRET)-based assay and on the basis of the strong identity between SARS-CoV and SARS-CoV-2 main proteases, we have turned to a fluorogenic substrate previously reported to monitor the catalytic activity of the SARS-CoV 3CLpro [19]. The Dabcyl-KTSAVLQ/SGFRKME(Edans) substrate bearing the sequence between polypeptide nsp4-nsp5 junction of SARS-CoV-2 was selected. The fluorescence resulting from the cleavage of this substrate by the protease was measured with a Victor 3V instrument (Perkin-Elmer) using excitation and emission wavelengths of 340(25) nm and 535(25) nm (Fig. 1 d). Using this fluorogenic substrate, proteolytic activity of the 3CLpro was evaluated in a pH 7.4 buffer containing 50 mM HEPES, 0.1 mg/mL BSA, 0.01% Triton X-100 and 2 mM GSH. Due to the presence of a catalytic cysteine in the 3CLpro active site, glutathione (GSH) was added in the assay buffer to both maintain the catalytic cysteine in its active state and discard highly electrophilic compounds from the hit list ([GSH] = 100000 × [3CLpro]). Triton X-100 has also been added to avoid non-specific inhibitory binding of the compounds with the protease by forming aggregate. We characterized the enzymatic activity of the recombinant SARS-CoV-2 3CLpro by determining the Michaelis-Menten constant (Km) value. Fluorogenic substrate concentrations ranging from 6 to 400 μM were used in this kinetic study with a fixed enzyme concentration of 15 nM. The initial velocity was measured and plotted against substrate concentration. Curve fitting with Michaelis-Menten equation gave the best-fit values of Km as 54 μM (Fig. 1b). Previously we had checked that the enzymatic reaction was linear for enzyme concentration between 3.1 nM and 25 nM (Fig. 1c).
Fig. 1.
SARS-CoV-2 3CLpro expression and characterization. (a) SDS-PAGE of 3CLpro; the calculated molecular weight of the 3CLpro is 33,796 Da. (b) Michaelis-Menten plot of 15 nM 3CLpro with various concentrations of FRET substrate (c) Initial velocity plot of enzyme reaction with various concentrations of 3CLpro. (d) Principle of the enzymatic FRET-based assay used to monitor 3CLpro activity. Once the fluorogenic substrate is cleaved by the enzyme, the fluorophore (Edans) and the fluorescence quencher (Dabcyl) are spatially separated, resulting in an increase in fluorescence which is proportional to the enzyme activity. (e) SARS-CoV-2 3CLpro inhibition by reference compounds. 3CLpro was pre-incubated 30 min with various concentration of GC-376 or boceprevir before FRET substrate addition and initial velocity measurement. Data are shown as mean ± SD of duplicates from representative experiments.
As a too high substrate concentration could be detrimental for the purposes of identification of inhibitors that usually compete with substrate for the enzyme active site, we selected the lowest enzyme (15 nM) and substrate concentration (10 μM; 0.2 × Km) that yielded a strong, reliable and reproducible signal in the 384-well plate screening format (0.8 < Z’ < 0.9). Finally, tolerance to DMSO was evaluated and a final maximal DMSO concentration of 1% was used. To validate the enzyme assay, two known inhibitors of the SARS-CoV-2 3CLpro; GC-376 and boceprevir [16] were evaluated in the screening assay. Both compounds dose-dependently inhibited the enzyme activity with IC50 values of 4 μM for boceprevir and 0.02 μM for GC-376, in line with the values reported in the literature (Fig. 1e).
2.1.3. Primary high-throughput screening
A library of 89,193 compounds, selected from commercial vendors or prepared by our chemists using state-of-the-art selection and design criteria, in terms of diversity and “drug/lead-likeness” properties, was screened at 30 μM against the SARS-CoV-2 3CL protease by enzymatic end-point fluorescence intensity assay in 384-well microplate format on a semi-automated system (Fig. 2 ). To allow detection of slow/tight enzyme binders, a 30-min pre-incubation of the compounds with the enzyme was applied before starting the reaction with the substrate. The fluorescence was measured after 30 min. Boceprevir at 4 μM (IC50 value) and 40 μM (10 x IC50) was used as a positive reference compound in each plate. The HTS demonstrated robust performance with an average Z’ factor of 0.85.
Fig. 2.
Left. Overview of the screening. SARS-CoV-2 3CLpro inhibition was reported (%) for each incubate. Green dots: tested compounds at 30 μM, Red dots: positive controls (incubations with vehicle), Blue dots: negative controls (incubations w/o enzyme), Orange dots: reference compound Boceprevir at 40 μM, Light orange dots: Boceprevir at 4 μM. Right. Workflow of the screening campaign.
The cut-off to select hits was set to 50% inhibition of the enzyme activity in order to focus on the most potent inhibitors. 213 compounds were thus cherry-picked from our inventories or repurchased. They were retested in the same assay in dose–response experiments to determine their IC50 values. In parallel the compounds were tested at the highest concentration (100 μM) without enzyme in order to test for fluorescence signal interference. Following the enzyme omission test, 20 compounds were retested in a continuous kinetic assay that allows rate calculation, for confirmation. The DMSO stock solutions of the 176 compounds were controlled for purity and identity by LCMS. From these 176 compounds, 35 were then selected and repurchased based on chemical structures, synthetic access and IP criteria. Finally, 42 confirmed hits were selected using a cut-off of maximal inhibition greater than 50% and IC50 less than 10 μM (Fig. 2).
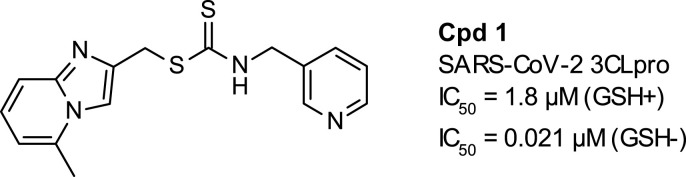
We report here the identification and characterization of a dithiocarbamate 3CLpro inhibitor that represents a novel chemical series with promising anti-coronavirus activity (compound 1, Fig. 3 ).
Fig. 3.
Structure of hit compound 1.
2.2. Hit compound 1 characterization
2.2.1. Slow binding inhibitor
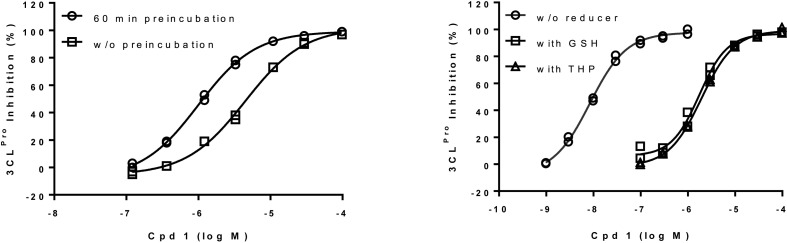
Compound 1 was retested with or without a 60-min pre-incubation step before substrate addition. As can be seen in the concentration-effect curve (Fig. 4 a), the IC50 is lower when the compound is pre-incubated suggesting that the kon is significantly lower than the diffusion rate.
Fig. 4.
(a) SARS-CoV-2 3CLpro inhibition by hit compounds 1. 3CLpro was pre-incubated 60 min or not with various concentration of compound before FRET substrate addition and initial velocity measurement. Inhibitions (%) are shown as duplicate from a representative experiment. (b) SARS-CoV-2 3CLpro inhibition by hit compounds in buffer containing (2 mM) or not GSH and THP. 3CLpro was pre-incubated 60 min with various concentration of compound before FRET substrate addition and initial velocity measurement. Inhibitions (%) are shown as duplicate from a representative experiment.
2.2.2. Effect of reducing agents on inhibitory potency
Enzymatic assays for the SARS-CoV-2 3CLpro inhibition were performed with different reducing conditions classically used with cysteine protease (Fig. 4b). We obtained highly decreased potencies for compound 1 in reducing conditions (IC50 = 0.008 μM without reducing agent, 1.75 μM with 2 mM GSH and 1.94 μM with 2 mM THP (Fig. 4b)). To know if the loss of potency was due to a degradation of the compound by the reducing agents, compound 1 was incubated at 100 μM in the enzymatic assay buffer in presence of 2 mM of GSH. After 1.5 h incubation, the very low disappearance of the compound in both conditions (less than 10%) cannot explain the 2-log decrease in potency observed with the reducing agents suggesting an impact of the reducing condition on the enzyme and rather than on the compound. Others reported this effect but failed to bring mechanistic description [20].
2.2.3. Reversibility of inhibition
To elucidate the binding mode of compound 1 with the 3CLpro, the reversibility of inhibition was evaluated. Jump dilution assay is commonly used to evaluate the reversibility of inhibition. To that aim, the compound was pre-incubated at 10 times its IC50 with 3CLpro in presence of GSH (2 mM) as reducing agent. Then, incubates were quickly diluted to 1/100 with substrate solution before measuring the fluorescence kinetics. Inhibition is compared to control standard incubations at 10 × IC50 and 0.1 × IC50 final concentrations of compounds (Fig. 5 ). Final enzyme and substrate concentrations are 15 nM and 10 μM, respectively. As can be seen in Fig. 5, during the first 10 min after jump dilution, the enzyme was inhibited at around 80%, a value similar to the 10 × IC50 control value, that is consistent with the presence of an electrophilic center (dithiocarbamate function) that may form covalent bond with the nucleophilic Cys145 of the catalytic site. Enzyme activity was progressively recovered and no more inhibition was observed after 2 h.
Fig. 5.

Assessment of the irreversible/reversible mechanism of inhibition with compound 1. SARS-CoV-2 3CLpro was incubated with compound 1 in a jump dilution assay. Initial rates are measured just after the compound dilution with substrate and enzymatic inhibitions (%) are calculated as mean ± SD of triplicate. Data are representative of three other experiments.
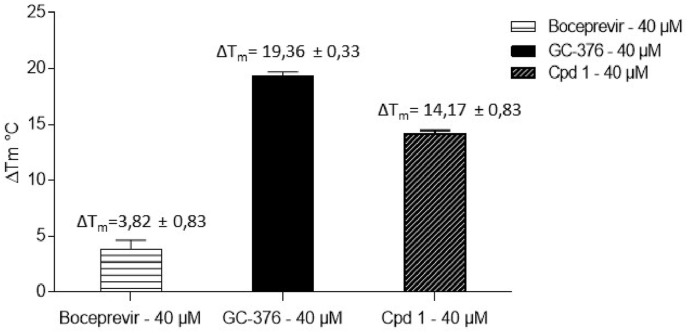
2.2.4. Thermal Shift Assay (TSA)
The direct binding of compound 1 to the 3CL protease was evaluated in the TSA assay. The melting temperature of SARS-CoV-2 3CLpro was shifted by 14.17 °C upon binding of compound 1 supporting the direct binding of compound 1 with the 3CLpro (Fig. 6 ). In agreement with values reported in the literature, Boceprevir and GC-376 shifted the melting curve of 3CLpro by 3.82 and 19.36 °C respectively upon binding [16]. Moreover, the binding of compound 1 to the 3CL protease is maintained when adding GSH 2 mM in the buffer (ΔTm = 11.33 °C).
Fig. 6.
Evaluation of thermal stabilization of SARS-CoV-2 3CLpro by Thermal Shift Assay (TSA) in presence of Boceprevir, GC-376 or Cpd 1 (40 μM). Thermal shift (ΔTm) was calculated by subtracting reference melting temperature of the protease from the Tm in presence of the compound. Values presented are the means of ΔTm ± SD of the eight independent TSA experiments.
2.2.5. Structural data
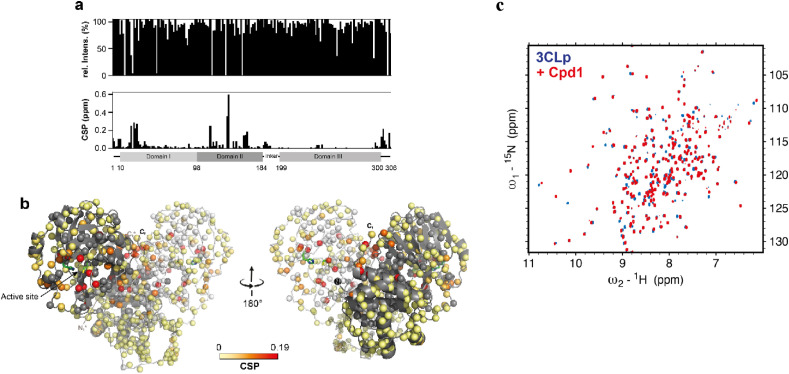
The direct binding of compound 1 to the 3CLpro was also investigated using solution NMR spectroscopy. Using a 2H,15N-labelled 3CLpro we compared the 2D 1H,15N-TROSY-HSQC NMR spectra of the 3CLpro acquired in the absence or the presence of an excess of molecule [21]. Spectral perturbations (chemical shift perturbations and/or signal broadening) were observed on the 3CLpro spectrum upon addition of compound 1 (Fig. 7 ). These chemical shift perturbations (CSP) show the direct binding of this compound in the 3CLpro active site, for which the highest CSP are observed.
Fig. 7.
Interaction of SARS-CoV-2 3CLpro with compound 1 assessed by NMR spectroscopy. a) The variations in NMR resonance intensities (top) and chemical shift perturbations (CSP) (bottom) induced upon cpd 1 addition are displayed along the main protease sequence. b) The CSPs, shown in (a, bottom), have been color coded (from light yellow to red) and are displayed on the structure of the dimeric 3CLpro, with the two protomers shown in dark grey and white, respectively. The side chains of both catalytic His41 and Cys145 are shown in green. c) Overlaid 2D 1H,15N-TROSY-HSQC spectra acquired on 2H,15N-labelled 3CLpro (100 μM) in the absence (in blue) or in the presence (in red) of cpd 1 (target concentration of 2 mM). The final DMSO‑d6 concentration was 3%. The spectra were acquired at 305 K on a 900 MHz NMR spectrometer.
To get additional structural data on the binding mode of compound 1, we solved the crystal structure of the 3CLpro: compound 1 complex. We crystallized the native 3CLpro and obtained the complex with compound 1 using a soaking procedure. The 3CLpro crystals were soaked for 1 h in the crystallization solution containing 10 mM of the molecule (10% DMSO), and then briefly soaked into a cryoprotective solution containing 10% glycerol before flash-freezing in liquid nitrogen. The structure of the complex, solved at 1.49 Å resolution, reveals that the sulfur of active site cysteine 145 undergoes a transthiocarbamoylation by reacting with the electrophilic carbon of the dithiocarbamate function of compound 1, triggering the loss of (5-methylimidazo[1,2-a]pyridin-2-yl)methanethiol (Fig. 8 a). This binding mode is fully consistent with our NMR data where the highest CSP were observed for residues surrounding the S1 pocket. The pyridine ring of the newly formed dithiocarbamate adduct occupies the S1 pocket that usually binds the P1 residue of the substrate. The 3CLpro S1 pocket is defined by the residues Phe140, Leu141, Asn142, Gly143, Ser144, His163, Met165, Glu166, His172. The ligand establishes two hydrogen bonds with the protease, one with His163 and one with Asn142 through a water molecule, as well as many hydrophobic contacts with the surrounding 3CLpro residues (Fig. 8b). The pyridinyl substituent also present in ML188 and calpain inhibitor XII forms the same hydrogen bond with the H163 imidazole in S1 pocket as for compound 1 [22,23]. The His163 residue at the S1 pocket was described as a binding hot spot for 3CLpro inhibitors and the pyridinyl substituent appears as a suitable scaffold to finely fit into the S1 pocket. The 3CLpro S1 pocket is occupied by a cyclobutyl or a glutamine surrogate γ-lactam ring of boceprevir and GC-376 respectively [16,24]. Upon binding of compound 1 the loops (Glu166-His172 and Phe185-Ala194) and the helical segment (Val42-Pro52), forming the walls of the enzymatic cleft, were slightly displaced compared to the structure of the apo-3CLpro, making the cleft slightly wider. These observations are consistent with the location of the observed NMR chemical shift perturbations. In addition, we also observed NMR CSPs for residues that are outside of the active site (Gly2, Phe3, Ser10, Gly11, Ala116, Cys117, Ser121, Ala124, Glu166, Gln299, Ser301, Gly303, Val303 and Phe305) (Fig. 7). This NMR data show that compound 1 while binding at the active site induces structural and/or dynamical perturbations up to the dimerization interface of 3CLpro, where both the N-terminus and C-terminus of the protease, including Ser10, Gly11, R298, have been shown to be essential for its dimerization [25,26]. This path, highlighted by NMR CSPs, clearly shows an allosteric regulation pathway in 3CLpro. The later one could correspond to the conserved communication network (His163, Ser147, Leu115 and Ser10) that have been recently proposed in 3CLpro based on mutational and functional analyses [26]. The formation of the dithiocarbamate function with the Cys145 also confirms the reversible covalent inhibition observed in the jump dilution assay for this compound. Indeed, as the new dithiocarbamate remains electrophilic and hydrolysable, upon dilution, the enzyme recovers slowly its catalytic activity. We have previously observed the ability of compound 1 to react with thiols like GSH derivatives but in the case of 3CLpro, this reactivity is greatly improved as 1 yields complete reaction with 3CLpro and only a low reaction conversion with GSH was observed after 90 min (<10%) despite high GSH concentration (2 mM).
Fig. 8.
Crystallographic structure of the SARS-CoV-2 3CLpro bound to the N-(pyridin-3-ylmethyl)thioformamide moiety (in pink) from the compound 1 (PDB ID: 7NTQ) (a). The 2Fo-Fc electron-density map, contoured at 1.5 σ, is shown as light grey mesh. (b) Interaction of the ligand (cpd 1 after Cys145 binding) with the 3CLpro S1 pocket. The analysis of the interaction was made using LigPlot+ (v2.2.4). 3CLpro and ligand bonds are shown in brown and purple, respectively. Water molecules are displayed as cyan spheres. Dashed green lines and dashed red lines represent hydrogen bonds and hydrophobic interactions, respectively. The 3CLpro residues that are involved in hydrogen bonds have their name displayed in green whereas the residues making hydrophobic contact(s) are indicated with their name surrounded by red spikes. Atoms involved in hydrophobic contact(s) are surrounded by red spikes.
2.3. Analogs synthesis of compound 1 and enzymatic activity on 3CLpro
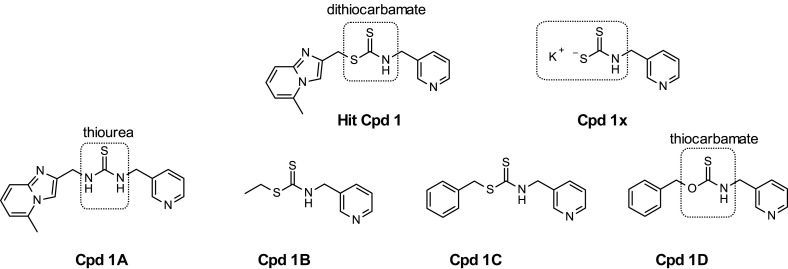
To gain molecular insight into the binding properties of the dithiocarbamate-based inhibitor 1 within the catalytic site, four analogs were synthesized. In compound 1A, the electrophilic warhead dithiocarbamate was replaced by a thiourea moiety. After covalent binding of compound 1 with the catalytic Cys145 of the 3CLpro, only the pyridyl part remains in the catalytic site. To evaluate the importance of the leaving moiety (5-methylimidazo[1,2-a]pyridin-2-yl)methanethiol for the binding of 1, this part was replaced by either an ethyl (1B) or a benzyl (1C) moiety while keeping the dithiocarbamate warhead. Finally, the dithiocarbamate in 1C was replaced by a thiocarbamate in 1D (Fig. 9 ).
Fig. 9.
Structures of analogs 1x and 1A-1D of hit compound 1.
2.3.1. Chemistry
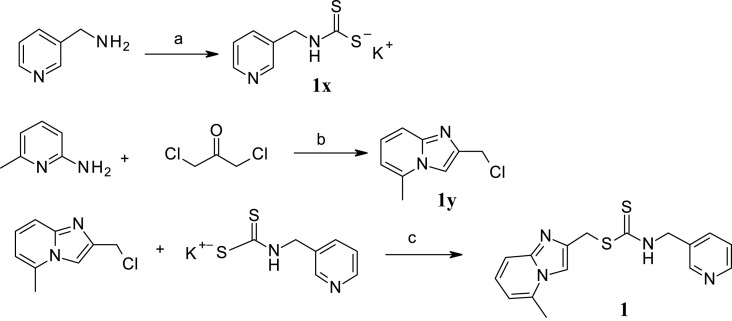
Hit compound 1 was resynthesized in three steps described in Scheme 1 . 3-(aminomethyl)pyridine was reacted with carbon disulfide in presence of potassium hydroxide to obtain intermediate 1x. 1,3-dichloroacetone and 2-amino-6-methylpyridine were refluxed in ethanol to lead to chlorine intermediate 1y. Finally, compound 1 was obtained by nucleophilic substitution between 1x and 1y.
Scheme 1.
Synthetic Route to hit compound 1. Reagents and conditions: (a) CS2, KOH, MeOH, 2 h, 0 °C, 43% (b) EtOH, reflux, overnight, 30% (c) NEt3, MeOH, 3 h, room temp., 85%.
1A-1D were obtained from isothiocyanate 1i (Scheme 2 ) and different nucleophiles (amine, thiol and alcohol). 1i was prepared according a reported procedure using 3-picolylamine and bis(2-pyridyloxy)methanethione (DPT) [27].
Scheme 2.
Synthetic Routes to 1A-1D. Reagents and conditions: (a) NaH (60% dispersion in mineral oil) in dry THF, 0 °C, 1 h then bis(2-pyridyloxy)methanethione, room temp., overnight, 67% (b) (8-methylimidazo[1,2-a]pyridin-2-yl)methanamine dihydrochloride, NEt3, DCM, 1 h, room temp., 96% (c) Ethanethiol or benzylthiol, NEt3, DCM, 1 h, room temp., 90–96% (d) Benzyl alcohol, NaH (60% dispersion in mineral oil), dry THF, 0 °C then 1i, room temp., 1 h, 85%.
2.3.2. SARS-CoV-2 3CLpro inhibition
Enzymatic assays for the SARS-CoV-2 3CLpro inhibition were performed with and without the reducing agent GSH. The activities presented in Table 1 show that the dithiocarbamate function present in 1, 1B and 1C is essential for the inhibitory effect as replacement by thiourea (1A) or thiocarbamate (1D) led to inactive compounds. The replacement of the 5-methylimidazo[1,2-a]pyridin-2-yl moiety of compound 1 by a phenyl in compound 1C led to a similar inhibitory activity while the replacement by an ethyl group in 1B resulted in 8-fold lower potency. Interestingly, the potassium dithiocarbamate intermediate 1x devoid of thiol leaving group is as potent as compound 1. Salts of dithiocarbamate as ditiocarb, the sodium salt of diethyldithiocarbamate, are known to be powerful metal chelating agent. As zinc complexes are known to inhibit 3CLpro [28], we hypothesized that compound 1x could inhibit the 3CLpro as a zinc complex. The compound was thus tested in presence of the zinc chelator EDTA (500 μM) but 1x revealed as potent in presence or in absence of EDTA in contrast to zinc acetate that completely loose its inhibitory activity in presence of EDTA. To get insight into the binding mode of compound 1x, we solved the crystal structure of the 3CLpro: compound 1x complex. As previously for compound 1, we crystallized the native 3CLpro and obtained the complex with compound 1x using a soaking procedure. The structure of the complex reveals that the active site cysteine 145 makes a covalent bound with the electrophilic carbon of the dithiocarbamate function of compound 1x leading to the same fragment bound to the protease as with compound 1 (Fig. S1a). The superimposition of the structures of the SARS-CoV-2 3CLpro bound to the compound 1 and to the compound 1x (Fig. S2b) shows that the N-(pyridin-3-ylmethyl)thioformamide moiety bound to the protease has the same binding mode (Fig. S1b).
Table 1.
SARS-CoV-2 3CLpro inhibition of hit compound 1 and analogs 1A-D, 1x.
| Cpd | Assay without GSH |
Assay with GSH |
||
|---|---|---|---|---|
| IC50 (μM) | pIC50 ± SD | IC50 (μM) | pIC50 ± SD | |
| 1 | 0.021 | 7.67 ± 0.30 | 1.46 | 5.85 ± 0.05 |
| 1A | 35.000 | 4.46 ± 0.19 | >100 | <4 |
| 1B | 0.178 | 6.75 ± 0.63 | 22.40 | 4.65 ± 0.07 |
| 1C | 0.023 | 7.65 ± 0.32 | 9.36 | 5.03 ± 0.10 |
| 1D | >100 | <4 | >100 | <4 |
| 1x | 0.027 | 7.57 ± 0.22 | 1.36 | 5.87 ± 0.04 |
Enzymatic assays were performed with or without GSH 2 mM in the buffer. The enzyme was pre-incubated for 1 hour with compound at increasing concentrations before starting the reaction with the substrate. Initial rates were recorded to calculate % inhibition and IC50 were obtained from concentration-response curves by a nonlinear regression analysis of the data. IC50 values are averages of three independent experiments.
2.3.3. Inhibition of the 3CL protease of different human coronaviruses
Since the catalytic sites of 3CL proteases are highly conserved among coronaviruses family, we hypothesized that compound 1 could act as a pan-inhibitor of coronaviruses 3CL proteases. Consistent with this hypothesis, compound 1 was shown to be inhibitor of the 3CL protease of coronaviruses of alpha (HCoV-229E) and beta (SARS-CoV-2, SARS-CoV, MERS-CoV) groups of Coronaviridae (Table 2 ) and could represent a good starting point to develop a broad-spectrum anti-coronavirus compound to fight emerging coronaviruses. A weaker potency has been obtained for all tested compounds on the 3CLpro of MERS-CoV. Moreover, an increase in enzymatic activity was observed in the presence of low concentrations of compounds while inhibition of enzymatic activity was observed at higher inhibitor concentrations (Fig. S2). The activation of MERS-CoV 3CLpro by ligands at low concentration was previously described as a result of dimerization induced upon partial occupation of the substrate binding pocket. Indeed, MERS-CoV 3CLpro is a weakly associated dimer requiring ligand binding for dimer formation and enzymatic activity [29].
Table 2.
Activity of compounds against 3CLpro of different human coronaviruses.
| Cpd | hCoV-229E (Alpha-CoV) 3CLpro IC50 μM (pIC50 ± SD) | SARS-CoV-2 (Beta-CoV) 3CLpro IC50 μM (pIC50 ± SD) | SARS-CoV (Beta-CoV) 3CLpro IC50 μM (pIC50 ± SD) | MERS-CoV (Beta-CoV) 3CLpro IC50 μM (pIC50 ± SD) |
|---|---|---|---|---|
| 1 | 0.016 (7.79 ± 0.14) | 0.021 (7.67 ± 0.30) | 0.383 (6.42 ± 0.18) | 2.00 (5.70 ± 0.10) |
| 1x | 0.019 (7.73 ± 0.10) | 0.027 (7.57 ± 0.22) | 0.165 (6.78 ± 0.32) | 1.36 (5.87 ± 0.21) |
| GC-376 | 0.028 (7.56 ± 0.09) | 0.012 (7.92 ± 0.02) | 0.161a (6.79 ± 0.43) | 0.43 (6.37 ± 0.06) |
Enzymatic assays were performed without GSH in the buffer. The enzymes were pre-incubated for 1 h with compound at increasing concentrations before starting the reaction with the substrate. Initial rates were used to calculate % inhibitions and IC50 were obtained from concentration-response curves by a nonlinear regression analysis of the data. IC50 values are averages of at least three independent experiments.
n = 2.
2.3.4. Selectivity against human cysteine proteases (Human Calpain 1, Cathepsin L)
Compound 1 and 1x were tested on two host-cell proteases, the human cysteine proteases calpain 1 and cathepsin L. Both compounds showed no inhibitory activity against Calpain 1 (IC50 > 300 μM) and an 80 and 30-fold decrease of potency on Cathepsin L compared to SARS-CoV-2 3CLpro was obtained respectively for compound 1 and 1x, suggesting promising selectivity for coronavirus 3CL proteases. In contrast, GC-376 is highly potent on SARS-CoV-2 3CLpro, human Calpain 1 and Cathepsin L (Table 3 ).
Table 3.
Selectivity profile of compounds against human cysteine proteases.
| Cpd | 3CLpro SARS-CoV-2 IC50 μM (pIC50 ± SD)a | Human Calpain 1 IC50 μM (pIC50 ± SD)b | Cathepsin L IC50 μM (pIC50 ± SD)b |
|---|---|---|---|
| 1 | 1.46 (5.85 ± 0.05) | >300 (>3.52) | 122 (3.91 ± 0.19) |
| 1x | 1.36 (5.87 ± 0.04) | >300 (>3.52) | 42.8 (4.37 ± 0.10) |
| GC-376 | 0.02 (7.64 ± 0.27) | 0.016 (7.79 ± 0.05) | 0.0007 (9.16 ± 0.11) |
Enzymatic assays were performed with GSH (2 mM) as reducing agent. The enzymes were pre-incubated with compound at increasing concentrations before starting the reaction with the substrate. Initial rates were used to calculate % inhibitions and IC50 were obtained from concentration-response curves by a nonlinear regression analysis of the data.
IC50 values are averages of three independent experiments; enzyme and compound pre-incubation of 60 min.
IC50 values are averages of two independent experiments; enzyme and compound pre-incubation of 30 min.
2.3.5. Antiviral activity of 3CLpro inhibitor 1 in SARS-CoV-2 live virus assay
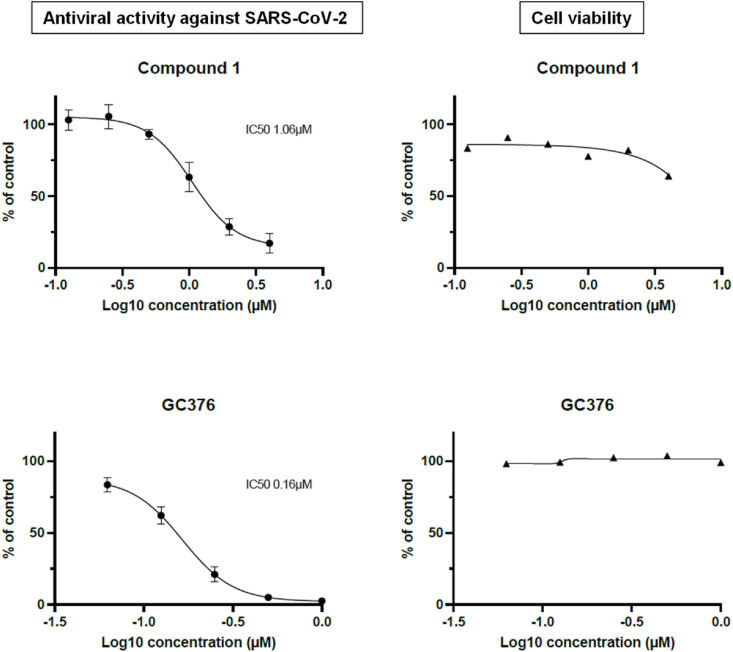
To evaluate the antiviral activity of our 3CLpro inhibitors against SARS-CoV-2, compounds 1 and 1x were tested in a cellular assay in Vero-81 cells stably expressing a fluorescent reporter probe to detect SARS-CoV-2 infection (F1G cells) [30]. As Vero cells are known to express high levels of the efflux transporter P-glycoprotein (P-gp), the assay was performed in presence of the P-gp inhibitor CP-100356 (0.5 μM) that had no antiviral or cytotoxic activity at the concentration used. As can be seen in Fig. 10 , compound 1 demonstrated promising antiviral with micromolar potency (IC50 = 1.06 μM) without cytoxicity at the active doses. For compound 1x, it was unfortunately toxic at the doses that gave antiviral activity.
Fig. 10.
F1G cells were infected with SARS-CoV-2 in presence of P-gp inhibitor (CP-100356) and increasing concentrations of compound 1 (top) or GC-376 (bottom). 16 h later, cells were fixed in presence of Hoechst 33342. Infected cells (GFP positive nuclei) and the total number of cells (number of nuclei, Hoechst staining) were determined. Results are presented as the percentage of the control and are the average of four independent experiments. Error bars represent the SEM.
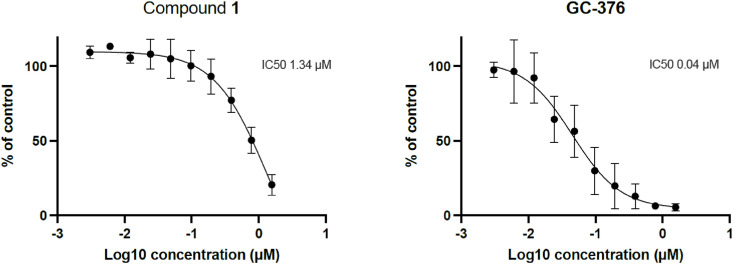
2.3.6. Antiviral activity of 3CLpro inhibitor 1 in HCoV-229E live virus assay
As compound 1 is a potent inhibitor of the 3CLpro of the human coronavirus 229E, its antiviral activity in cellulo on this coronavirus was also evaluated. Compound toxicity was first evaluated in a high-content screening apoptosis assay to determine the range of concentrations devoid of toxicity. This assay allows the rapid detection and quantification of apoptotic Huh-7 cells by detection of Caspase-3/7 activity in real-time imaging using NucView™ 488 substrate containing peptide sequence DEVD attached to a nucleic acid dye (data not shown). Then, in cellulo (Huh-7 cells) antiviral activity on the human coronavirus 229E was assessed using recombinant HCoV-229E expressing the Renilla luciferase (Rluc) reporter gene. Rluc activity was measured using the Renilla-Glo luciferase assay system (Promega). As can be seen in Fig. 11 compound 1 inhibits viral replication of this coronavirus with a micromolar potency showing that compound 1 is a good starting point to develop a new broad spectrum anti-coronavirus candidate.
Fig. 11.
Antiviral activity on the human coronavirus 229E assessed using recombinant HCoV-229E expressing the Renilla luciferase (Rluc) reporter gene for compounds 1 (left) and GC-376 (right). Results are the mean from at least 3 independent experiments. Error bars represent the SEM.
3. Conclusion
Compound 1 bearing a dithiocarbamate warhead represents a novel class of covalent, 3CLpro inhibitor with potency in the nanomolar range on the 3CLpro of SARS-CoV-2 (IC50 = 21 nM). This compound also inhibited the 3CL protease of three other coronaviruses; HCoV-229E (Alpha-CoV), MERS-CoV and SARS-CoV (Beta-CoV). The selectivity over host proteases, often challenging with electrophilic covalent inhibitors, was evaluated on two human cysteine proteases and revealed a more than 80-fold selectivity. We also showed that inhibition of the enzyme, performed through the formation of a labile dithiocarbamate with the sulfur atom of the catalytic cysteine, is reversible within 2 h, a blockade time likely sufficient to cover the protein turnover time in infected cells. Indeed, a robust cellular antiviral activity in the low micromolar range on both SARS-CoV-2 and HCoV-229E was obtained. Therefore, these results collectively support compound 1, a selective 3CL protease inhibitor with antiviral activity, as a promising starting point for further development of this series as broad-spectrum anti-coronavirus agents.
4. Experimental section
4.1. Biology
3CLpro substrate {Dabcyl}-KTSAVLQSGFRKM-{Glu(Edans)} was purchased from LifeTein (purity >95%). Boceprevir was purchased from MedChemExpress, GC-376 from Carbosynth (BG167367), compound 1 from enamine (T5466440). SARS-CoV 3CL protease was purchased from Boston Biochem.
4.1.1. Expression and purification of the 3CLpro of SARS-CoV-2, MERS-CoV and hCoV-229E
The gene coding for the SARS-CoV-2 main protease (3CLpro) was synthesized, with codon optimization, and inserted into an in-house modified pET24a plasmid in order to produce the 3CLpro fused to a N-terminal 6 × His-SUMO tag in Escherichia coli BL21(DE3) (pHis-SUMO-3CLpro). The bacteria were grown at 37 °C in Luria-Bertani (LB) medium supplemented with Kanamycin (25 μg/mL). When A600nm reached ∼0.9, the temperature was lowered to 21 °C and induction was carried out with 0.3 mM isopropyl-β-d-galactopyranoside for 12 h. Harvested cells were lysed using homogenizer (Emulsiflex C-3) in lysis buffer (50 mM Tris.Cl pH 8.0, 300 mM NaCl) supplemented with both DNAseI and RNAseA. The fusion protein was first purified using a HisTrap HP column (Cytiva) and eluted with an elution buffer containing 400 mM imidazole. The 6xHis-SUMO-3CLpro fractions were selected from SDS-PAGE analysis, then pooled and dialyzed (cut-off 6–8 kDa) 2 h at 4 °C against 2 L of cleavage buffer (40 mM Tris-Cl pH 7.5, 100 mM NaCl, 5 mM β-mercaptoethanol). The 6xHis-SUMO tag was cleaved by adding SENP2 protease (His tagged) into the dialysis bag and by dialyzing the sample over-night at 4 °C against 2 L of fresh cleavage buffer. The sample was then passed through a HisTrap HP column (Cytiva) to eliminate the SENP2 protease and the 6xHis-SUMO tag. The flow-through containing the native 3CLpro was dialyzed (cut-off 6–8 kDa) at 4 °C against 2 times 3 L of storage buffer (50 mM Tris-Cl pH 7.5, 20 mM NaCl, 1 mM EDTA, 1 mM DTT). The 3CLpro was concentrated using a stirred cell with (Amicon) a 10 kDa membrane. A 15.8 mg/mL batch was kept at 4 °C whereas a 12.6 mg/ml batch containing 20% glycerol was flash frozen in liquid nitrogen and then stored at −80 °C until used. The final yield was about 100 mg per L of culture. The native sequence of the purified 3CLpro was checked by MALDI-tof analysis (Axima Assurance, Shimadzu).
The expression of native main protease from both 229E and MERS-CoV was performed following the same strategy as above. The proteins were stored at −80 °C, in storage buffer (20 mM Tris-Cl pH 7.5, 50 mM NaCl, 1 mM EDTA, 2 mM DTT) at 6 mg/mL and 12.6 mg/mL, respectively. The final yields were about 80 mg and 35 mg per L of culture, respectively.
4.1.2. SARS-CoV-2 3CLpro enzymatic assays
For HTS, the FRET-based assay was optimized and miniaturized in Corning 384-wells plates (dark, low-binding, low volume). The assay was conducted at room temperature using a reaction volume of 20 μL and a buffer containing 50 mM HEPES, 0.1 mg/mL BSA, 0.01% Triton and 2 mM GSH at pH 7.5. Compounds stored as 10 mM stock solutions in DMSO were dispensed by acoustic nanodispensing with an Echo Liquid Handler (Labcyte). 10 μL 3CLpro was added to the tested compounds and preincubated for 30 min before addition of 10 μL FRET-based substrate {Dabcyl}-KTSAVLQSGFRKM-{Glu(Edans)}. Final concentrations were 15 nM, 10 μM and 30 μM, for enzyme, substrate and compound library respectively. Final concentration of DMSO did not exceed 1%. After 30 min, the fluorescence intensity was monitored with a Victor 3V instrument (Perkin-Elmer) using excitation and emission wavelengths of 340(25) nm and 535(25) nm. Boceprevir was used as a positive reference compound at 4 μM (close to the IC50 value) and 40 μM. Minimal and maximal fluorescence values were obtained in each test plates by incubating the substrate alone (negative control) or with the 3CLpro enzyme (positive control), respectively. Data were normalized intra-plate with these controls: the average of the negative control values was subtracted from all raw data and percentages of inhibition were calculated by the following equation:
Z’ factors were calculated according to Zhang et al. [31] using mean and standard deviations from positive and negative controls. Plates were validated if their respective Z’ factors were ≥0.5 and if enzyme inhibition with 40 μM or 4 μM boceprevir were ≥80% and ≥40%, respectively.
For determination of Km or compound IC50, the reaction progress was monitored for 30 min to measure the initial velocities used in calculations. Km was obtained by curve fitting with Michaelis-Menton equation using GraphPad Prism 7. IC50 values were obtained from concentration-response curves by a nonlinear regression analysis of the data using an equation at four parameters (using XL fit™ 5.2.0.0. from IDBS (Guilford, United Kingdom) or GraphPad Prism 7 (San Diego, USA).
A, minimum y value; B, maximum y value; C, LogIC50 value; D, slope factor.
A jump dilution assay was used to evaluate inhibition reversibility. Compounds are pre-incubated for 1 h at 10 times their IC50 with 3CLpro (100X). Then, the incubates are quickly diluted to 100-fold with substrate solution before measuring the fluorescence kinetics. Final concentrations after dilution are 0.1 × IC50, 15 nM and 10 μM for compound, enzyme and substrate, respectively. In the same experiment, control pre-incubations of 3CLpro with compounds are performed (60 min) with just, as usual, a two-fold dilution with the substrate to obtain 10 × IC50 and 0.1 × IC50 final concentrations of compounds. Enzyme and substrate are at 15 nM and 10 μM respectively. All incubates are performed in triplicate.
4.1.3. hCov-229E and MERS-CoV 3CLpro Enzymatic assays
Incubations are performed similarly to the SARS-CoV-2 3CLpro assay, using the same substrate and the same buffer. Enzyme is at 15 nM, 400 nM and 50 nM for hCov-229E, MERS-CoV and SARS-CoV 3CLpro assay, respectively. Substrate is at 30 μM (0.2 × Km), 60 μM (0.2 × Km) and 20 μM (0.2 × Km), for hCoV-229E, MERS-CoV and SARS-CoV 3CLpro assay, respectively. The reaction progress was monitored for 30 min to measure the initial velocities and inhibition data expressed as % inhibition, in the same way that for the SARS-CoV-2 3CLpro enzymatic assay.
4.1.4. Expression and purification of 2H,15N labelled SARS-CoV-2 3CLpro
The protocol is similar to the one for unlabeled 3CLpro (see above) but with the following modifications. Bacteria were grown in a M9-based semi-rich medium (M9 medium supplemented with 15NH4Cl (1 g/L), d-glucose-C-d7 (3 g/L), Isogro N,D-powder growth medium (0.5 g/L; Sigma-Aldrich), and kanamycin (25 μg/mL). The final storage buffer was (50 mM NaPi pH 6.8, 40 mM NaCl, 0.2 mM EDTA, 3 mM THP). The 2H,15N-labelled 3CLpro was concentrated up to 8.37 mg/mL, flash frozen in liquid nitrogen and then stored at −80 °C until used.
4.1.5. NMR analysis of SARS-CoV-2 3CLpro with ligands
All NMR experiments were performed at 305 K using Bruker Neo 900 MHz NMR spectrometer equipped with a cryogenic triple resonance probe (Bruker, Karlsruhe, Germany). The proton chemical shifts were referenced using the methyl signal of TMSP (sodium 3-trimethylsillyl-[2,2,3,3-d4]-propionate) at 0 ppm. Spectra were processed with the Bruker TopSpin software package 4.0.6. Data analysis was done with Sparky software [32].
NMR data were acquired on 200 μL samples in 3 mm tubes containing 100 μM of 2H,15N-doubly labelled 3CLpro sample in NMR buffer (50 mM NaPi pH 6.8, 40 mM NaCl, 3 mM THP, 3% DMSO‑d 6, 5% D2O) and 2 mM of the ligands. 2D 1H,15N-TROSY-HSQC spectra were acquired with 64 scans and 2048 and 128 complex points in the 1H and 15N dimensions respectively.
4.1.6. Crystallization of SARS-CoV-2 3CLpro
A 3CLpro sample at 5 mg/mL in storage buffer (50 mM Tris-Cl pH 7.5, 20 mM NaCl, 1 mM EDTA, 1 mM DTT) was used for crystallogenesis. Crystals with flower-shape were obtained in 0.2 M sodium formate, 20% PEG 3350 at room temperature. These crystals were crushed with a micro-tool to make a seed stock and new crystals were grown in the same condition using the microseeding technique with a cat whisker.
For the complexes with compounds 1 and 1x, the 3CLpro crystals were soaked for 1 h in a solution containing 10 mM compound 1, 10% DMSO, 0.2 M sodium formate, 20% PEG 3350 and then briefly soaked into 0.2 M sodium formate, 20% PEG 3350, 10% glycerol before freezing.
4.1.7. X-ray data collection and processing
X-Ray data were collected on the Proxima1 or Proxima2a beamlines [33] of the SOLEIL synchrotron facility (Paris, France). The data collection was done remotely using the MXCuBE2 [34] software and the crystals were handled by a Staubli sample changer. The data were collected at 100 K using an Eiger-X 16 M or 9 M (Dectris) detector. The data were processed with XDS [35] (xdsme scripts from the synchrotron facility, https://github.com/legrandp/xdsme). The molecular replacement (using the PDB entry 7K3T, https://doi.org/10.2210/pdb7K3T/pdb) and the refinement steps were done using the CCP4i2 interface [36] of the CCP4 program suite [37]. The statistics for data collection and refinement are summarized in the Table 1 in supporting Information.
The final models and the structure factors corresponding to the 3CLpro bound to N-(pyridin-3-ylmethyl)thioformamide from compounds 1 and 1x have been deposited in the Protein Data Bank as entries 7NTQ and 8AEB respectively.
The Fig. 8a has been generated using Pymol (The PyMOL Molecular Graphics System, Version 2.0 Schrödinger, LLC.
4.1.8. Thermal Shift Assay
Fluorescent dye SYPRO® Orange Protein Gel Stain was produced by Sigma-aldrich (S692-500UL). Two compounds have been used as positive shifter of the 3CLpro melting temperature protein: Boceprevir (HY-10237, Medchemexpress), GC-376 (BG167367, Biosynth Carbosynth®). The assay was performed in 50 mM Tris/HCl, pH 7.5, 150 mM NaCl, 2 mM MgCl2 and 1% DMSO in LightCycler® 480 white Multiwell Plate 96 (Roche, 04729692001). First, 5 μL of reference and compound 1 have been transferred in assay microplates. Then, 10 μL of a 3CLpro of SARS-Cov2 and Sypro Orange Dye mix were loaded to achieve final concentrations of 40 or 100 μM for tested and reference compounds, 5X for Sypro orange dye and 2.5 μM for 3CLpro. After 30 min of incubation at room temperature, assay microplate was launched in LightCycler480 device (Roche) for thermal denaturation of the protein under a temperature gradient range from 37 to 95 °C with 0.05 °C/s incremental step. The melting temperature (Tm) was calculated as the mid log of the transition phase from the native to the unfolded protein using a Boltzmann model in LightCycler_Thermal_Shif_Analysis software v2.0. ΔTm was obtained by subtracting reference Tm of proteins in the presence of DMSO from the Tm in the presence of compounds.
4.1.9. Calpain assay
Effect on human Calpain 1 activity was measured using the InnoZyme™ Calpain 1/2 Activity kit purchased by Calbiochem®. Assay was performed in Corning 96-wells plates according to the kit instructions with 1/400 diluted enzyme and 0.5 mM (DABCYL)-TPLKSPPPSPR-(EDANS) substrate in the presence of calcium ions and reducing agent (GSH 2 mM). Compounds were pre-incubated for 30 min at room temperature with Calpain 1. The fluorescence (excitation at 320 nm and emission at 480 nm) was measured for 30 min at room temperature with a Victor 3V (Perkin-Elmer) and initial velocities used for inhibition (%) calculations.
4.1.10. Cathepsin assay
Effect on human Cathepsin L activity was measured using the SensoLyte® 520 Cathepsin L Assay Kit purchased by Anaspec®. Assay was performed in 96-wells plates according to the kit instructions with 1/1000 diluted enzyme and QXL™ 520/HiLyte Fluor™ 488 substrate. GSH (2 mM) was used as reducing agent. Compounds were pre-incubated for 60 min at room temperature with Cathepsin L. The fluorescence (excitation at 490 nm and emission at 520 nm) was measured for 30 min at room temperature with a Victor 3V (Perkin-Elmer) and initial velocities used for inhibition (%) calculations.
4.1.11. Cellular antiviral activity
Cells and viruses. Vero-81 cells stably expressing a fluorescent reporter probe to detect SARS-CoV-2 infection (F1G cells) and Huh-7 cells were grown at 37 °C with 5% CO2 in Dulbecco's modified eagle medium (DMEM, Gibco) containing Glutamax and supplemented with 10% FBS (Life technologies).
SARS-CoV-2 virus (hCoV-19_IPL_France strain; NCBI MW575140) was propagated on Vero-81 cells expressing TMPRSS2 at 37 °C and recombinant HCoV-229E expressing the Renilla luciferase (kindly provided by Dr Volker Thiel, University of Bern, Switzerland) was propagated on Huh-7 cells at 32 °C. After complete lysis of the cells, supernatants containing the viruses were centrifuged, aliquoted and stored at −80 °C.
Dose-response Experiments. For SARS-CoV-2 infection assay, F1G cells were plated on coverslips in 24 well-plates. The next day, cells were infected at an MOI of 0.1 in presence of 0.5 μM CP-100356 and increasing concentrations of GC376 or compound 1. Sixteen hours later, cells were fixed with 4% PFA for 30 min containing 10 μg/ml Hoechst 33342 (life technologies). Cells were rinsed with PBS and mounted on glass slides in Mowiol 4–88 containing medium. Images acquisitions were performed with an EVOS M5000 imaging system (Thermo Fischer Scientific) equipped with a 10X objective and light cubes for DAPI and GFP. The total number of cells was determined by counting the number of nuclei and the number of infected cells was determined by counting the number of GFP positive nuclei. The experiment was performed four times.
For HCoV-229E infection assays, Huh-7 cells were plated in 96-well plates and infected 24 h later in presence of increasing concentration of GC376 or compound 1. Cells were incubated for 6h and lysed. Luciferase activity was measured by using the renilla luciferase assay system (Promega) and a Berthold luminometer.
4.2. Chemistry
All commercial reagents and solvents were used without further purification. Flash chromatography was performed using a Puriflash®430 with prepacked silica columns. UV detection was used to collect the desired product.
NMR spectra were recorded on a Bruker DRX-300 spectrometer. The assignments were made using one-dimensional (1D) 1H and 13C spectra and two-dimensional (2D) HSQC, HMBC and COSY spectra. Chemical shifts are in parts per million (ppm).
LC-MS Waters system was equipped with a 2747 sample manager, a 2695 separations module, a 2996 photodiode array detector (200–400 nm) and a Micromass ZQ2000 detector. XBridge C18 column (50 mm × 4.6 mm, 3.5 mm, Waters) was used. The injection volume was 20 μL. A mixture of water and acetonitrile was used as mobile phase in gradient-elution. The pH of the mobile phase was adjusted with HCOOH and NH4OH to form a buffer solution at pH 3.8. The analysis time was 5 min (at a flow rate at 2 mL/min). Purity (%) was determined by reversed phase HPLC using UV detection (215 nm), and all isolated compounds showed purity greater than 95%. HRMS analysis was performed on a LC-MS system equipped with a LCT Premier XE mass spectrometer (Waters), using a XBridge C18 column (50 mm_ 4.6 mm, 3.5 mm,Waters). A gradient starting from 98% H2O 5 mM Ammonium Formate pH 3.8 and reaching 100% CH3CN 5 mM Ammonium Formate pH 3.8 within 3 min at a flow rate of 1 mL/min was used.
4.2.1. Potassium 3-pyridylmethylimino(thioxo)methanethiolate (1x)
To a solution of 3-(aminomethyl) pyridine (400 mg, 3.70 mmol) in methanol (15 ml) was added CS2 (1.55 ml, 25.09 mmol, 7.0 equiv) and KOH (137 mg, 3.70 mmol, 1.0 equiv) at 0 °C. The reaction mixture was stirred for 2 h at 0 °C. Then the reaction mixture was concentrated under reduced pressure and recrystallized in ethanol to afford the desired compound as white amorphous solid (350 mg, 43%). HRMS (ESI): [M+H]+ C7H9N2S2: calcd. 185.0207 found 185.0192. 1H NMR (300 MHz, CD3OD): δ (ppm) 8.55–8.51 (m, 1H), 8.41–8.36 (m, 1H), 7.88–7.82 (m, 1H), 7.37 (ddd, 1H, J = 0.6, 4.8, 7.8 Hz), 4.88 (s, 2H). 13C NMR (75 MHz, CD3OD): δ (ppm) 216.3, 149.4, 148.3, 137.6, 137.0, 125.0, 49.1.
4.2.2. 2-(chloromethyl)-5-methylimidazo[1,2-a]pyridine (1y)
A solution of 1,3-dichloroacetone (593 mg, 4.67 mmol) and 2-amino-6- methylpyridine (500 mg, 4.62 mmol) in ethanol (5.0 mL) was heated to reflux overnight. Then, the reaction mixture was cooled to room temperature and concentrated under reduced pressure. The residue was diluted with aqueous saturated solution of NaHCO3 (5.0 mL) and extracted with ethyl acetate (3 x 5.0 mL). The combined organic layers were dried over MgSO4 and concentrated under reduced pressure. The crude product was purified on silica gel column chromatography eluting by (Hexane/EtOAc 5/5) to give 2- (chloromethyl)-5-methylimidazo[1,2-a]pyridine (250 mg, 30%). LC-MS: tR = 1.75 min. MS (ESI): [M+H]+ 181.00. 1H NMR (300 MHz, CDCl3): δ (ppm) 7.53–7.47 (m, 2H), 7.16 (dd, 1H, J = 6.9, 9.0 Hz), 6.66–6.60 (m, 1H), 4.81–4.78 (m, 2H), 2.57 (s, 3H). 13C NMR (75 MHz, CDCl3): δ (ppm) 145.9, 143.1, 134.8, 125.5, 115.2, 111.9, 108.2, 39.9, 18.8.
4.2.3. (5-methylimidazo[1,2-a]pyridin-2-yl)methyl (pyridin-3-ylmethyl)carbamodithioate (1)
To a solution of potassium N-(3-pyridylmethyl)carbamodithioate (100 mg, 0.45 mmol, 1.0 equiv) and 2-(chloromethyl)-5-methyl-imidazo[1,2-a]pyridine (81 mg, 0.45 mmol, 1.0 equiv) in MeOH (2.0 mL) was added Et3N (0.01 mL, 0.67 mmol, 1.5 equiv). The reaction was stirred at room temperature for 3 h. Then, the solvent was evaporated under reduced pressure and the residue was diluted with saturated solution of NaHCO3 and EtOAc. The organic layer was separated and the aqueous layer was extracted for 3 times with EtOAc. The combined organic layers were dried over MgSO4 and concentrated under reduced pressure to give the crude product which was purified though column chromatography eluting by (EtOAc/MeOH 100/0 to 100/10) to give the desired product as yellow solid which upon washing with Et2O gave white solid (130 mg, 85%). LC-MS: tR = 2.08 min. MS (ESI) m/z = 329.02 [M+H]+. HRMS (ESI): [M+H]+ C16H17N4S2: calcd. 329.0895 found 329.0896. 1H NMR (300 MHz, CDCl3): δ (ppm) 11.82 (br, 1H). 8.66 (d, J = 1.7 Hz, 1H), 8.49 (dd, 1H, J = 1.5, 4.8 Hz), 7.78–7.72 (m, 1H), 7.33 (s, 1H), 7.22 (dd, 1H, J = 4.8, 13 Hz), 7.13–7.06 (m, 1H), 7.04–7.98 (m, 1H), 6.61–6.56 (m, 1H), 4.97 (d, 2H, J = 4.8 Hz), 4.27 (s, 2H), 2.50 (s, 3H). 13C NMR (75 MHz, CDCl3): δ (ppm) 197.0, 149.8, 148.9, 144.8, 142.8, 136.4, 134.8, 132.4, 126.0, 123.5, 113.8, 112.1, 107.1, 48.3, 33.3, 18.7.
4.2.4. 3-(isothiocyanatomethyl)pyridine (1i)
To a suspension of NaH (60% dispersion in mineral oil) (163 mg, 4.07 mmol, 1.1 equiv.) in dry THF (10.0 mL), 3-Picolylamine (400 mg, 3.70 mmol) dissolved in dry THF (15 mL) was added dropwise at 0 °C. After 1 h, bis(2-pyridyloxy)methanethione (DPT) (868 mg, 3.74 mmol, 1.0 equiv.) was added and the mixture gradually allowed to reach to room temperature and stirred overnight. Then, the reaction was quenched with drops of water and the volatiles were removed under reduced pressure. The residue was diluted with EtOAc and washed with water for 3 times. The combined organic layers were dried over MgSO4 and concentrated under reduced pressure to give dark yellow crude product. The crude product was purified through column chromatography eluting by (Hexane/EtOAc 50/50) to give the desired product as pale-yellow oil (370 mg, 67%). LC-MS: tR = 2.22 min. MS (ESI+): m/z = 151 [M+ H]+. 1H NMR (300 MHz, CDCl3): δ (ppm) 8.56 (dd, 1H, J = 1.3, 4.8 Hz), 8.53 (d, 1H, J = 1.8 Hz), 7.68–7.61 (m, 1H), 7.30 (dd, 1H, J = 4.8, 7.8 Hz), 4.71 (s, 2H). 13C NMR (75 MHz, CDCl3): δ (ppm) 149.8, 148.3, 134.6, 134.0, 130.2, 123.8, 46.4.
4.2.5. General procedure A for the synthesis of 1A-1C
To a solution of 3-(isothiocyanatomethyl)pyridine 1i and thiol or primary amine (1.10 equiv.) in DCM was added Et3N (1.2 equiv.). The reaction mixture was stirred at room temperature for 1 h. Then, the volatiles were removed under reduced pressure and the residue was diluted by saturated solution of NaHCO3 and EtOAc. The organic layer was separated and the aqueous layer was extracted with EtOAc for more 3 times. The combined organic layers were dried over MgSO4 and concentrated under reduced pressure. The crude product was purified through column chromatography.
4.2.5.1. 1-((5-methylimidazo[1,2-a]pyridin-2-yl)methyl)-3-(pyridin-3-ylmethyl)thiourea (1A)
The titled compound was synthesized according to the general procedure A using 3-(isothiocyanatomethyl)pyridine (70 mg, 0.46 mmol), (8-methylimidazo[1,2-a]pyridin-2-yl)methanamine dihydrochloride (120 mg, 0.51 mmol, 1.10 equiv.) and Et3N (156 mg, 1.54 mmol, 3.30 equiv.) in DCM (5.0 mL). The crude product was purified through column chromatography eluting by (DCM/MeOH 95/5) to give 1A as a white solid (140 mg, 96%). LC-MS: tR = 1.70 min. HRMS (ESI): [M+H]+ C16H18N5S: calcd. 312.1283 found 312.1294. 1H NMR (300 MHz, CDCl3): δ (ppm) 8.45 (s, 1H), 8.38 (d, 1H, J = 4.2 Hz), 7.63–7.55 (m, 1H), 7.39 (s, 1H), 7.21–7.14 (m, 1H), 7.13–7.02 (m, 2H), 6.58 (d, 1H, J = 6.9 Hz), 4.89–4.67 (m, 4H), 3.10 (br, 2H), 2.49 (s, 3H). 13C NMR (75 MHz, CDCl3): δ (ppm) 183.7, 149.2, 148.4, 145.2, 142.8, 135.8, 135.0, 134.0, 125.8, 123.4, 113.7, 112.1, 107.8, 46.1, 42.1, 18.8.
4.2.5.2. Ethyl (pyridin-3-ylmethyl)carbamodithioate (1B)
The titled compound was synthesized according the general procedure A using 3-(isothiocyanatomethyl)pyridine (70 mg, 0.46 mmol), ethanethiol (31.9 mg, 0.513 mmol, 1.10 equiv.) and Et3N (56.6 mg, 0.55 mmol, 1.20 equiv.) in DCM (5.0 mL) then. The crude product was purified through column chromatography eluting by (EtOAc 100%) to give the 1B as a white solid (95 mg, 96%). LC-MS: tR = 2.28 min. HRMS (ESI): [M+H]+ C9H13N2S2: calcd. 213.0520 found 213.0516. 1H NMR (300 MHz, CDCl3): δ (ppm) 8.50 (d, 2H, J = 1.6 Hz), 8.42 (dd, 1H, J = 1.5, 4.9 Hz), 7.84–7.77 (m, 1H), 7.38 (dd, 1H, J = 4.9, 7.7 Hz), 4.93 (s, 2H), 3.23 (q, 2H, J = 12.0 Hz), 1.29 (t, 3H, J = 12.5 Hz). 13C NMR (75 MHz, CDCl3): δ (ppm) 200.5, 149.6, 148.8, 137.9, 135.6, 125.1, 48.0, 30.0, 14.8.
4.2.5.3. Benzyl (pyridin-3-ylmethyl)carbamodithioate (1C)
The titled compound was synthesized according the general procedure A using 3-(isothiocyanatomethyl)pyridine (70 mg, 0.46 mmol), benzylthiol (63.7 mg, 0.513 mmol, 1.10 equiv.) and Et3N (56.6 mg, 0.55 mmol, 1.20 equiv.) in DCM (5.0 mL). The crude product was purified through column chromatography eluting by (EtOAc 100%) to give 1C as a white solid (115 mg, 90%). LC-MS: tR = 2.73 min. HRMS (ESI): [M+H]+ C14H15N2S2: calcd. 275.0677 found 275.0667. 1H NMR (300 MHz, CD3OD): δ (ppm) 8.50 (d, 1H, J = 1.6 Hz), 8.41 (dd, 1H, J = 1.5, 4.9 Hz), 7.80–7.74 (m, 1H), 7.40–7.31 (m, 3H), 7.30–7.17 (m, 3H), 4.93 (s, 2H), 4.54 (s, 2H). 13C NMR (75 MHz, CD3OD): δ (ppm) 199.9, 149.6, 148.4, 138.4, 137.9, 135.4, 130.1, 129.5, 128.3, 125.1, 48.3, 40.3.
4.2.6. O-benzyl(pyridin-3-ylmethyl)carbamothioate (1D)
To a suspension of NaH (60% dispersion in mineral oil) (15.9 mg, 0.41 mmol, 1.25equiv.) in dry THF (2.0 mL), benzyl alcohol (39.6 mg, 0.36 mmol, 1.10 equiv.) dissolved in dry THF (1.0 mL) was added dropwise at 0 °C. Then, 3-(isothiocyanatomethyl)pyridine (50 mg, 0.33 mmol) was added and the mixture gradually allowed to reach to room temperature and stirred for further 1 h. The reaction was quenched with drops of water and the volatiles were removed under reduced pressure. The residue was diluted by saturated solution of NaHCO3 and EtOAc. The organic layer was separated and the aqueous layer was extracted with EtOAc for more 3 times. The combined organic layers were dried over MgSO4 and concentrated under reduced pressure. The crude product was purified through column chromatography eluting by (Hexane/EtOAc 50/50) to give 1D as a white solid (73 mg, 85%). LC-MS: tR = 2.60 min. HRMS (ESI): [M+H]+ C14H15N2OS: calcd. 259.0905 found 259.0898. 1H NMR (300 MHz, CDCl3): δ (ppm) 8.53–8.38 (m, 2H), 7.88–7.67 (m, 1H), 7.57–7.47 (m, 1H), 7.41–7.17 (m, 6H), 5.55–5.48 (m, 2H), 4.80–4.37 (m, 2H). 13C NMR (75 MHz, CDCl3): δ (ppm) 190.8, 189.7, 149.1, 149.0, 148.9, 136.0, 135.7, 135.5, 135.2, 132.9, 132.5, 128.6, 128.5, 128.4, 123.7, 73.5, 72.4, 46.6, 44.6 (mixture of rotamers).
Accession codes
Coordinates for the crystal structures have been deposited in the Protein Data Bank with ID 7NTQ (compound 1) and 8AEB (compound 1x).
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
We are grateful to the institutions that support our laboratory: INSERM, Université de Lille, Institut Pasteur de Lille and CNRS. This work was supported by the Institut Pasteur de Lille (to JeD, XH, and BD), the Centre National de la Recherche Scientifique (CNRS: COVID and ViroCrib programs to JeD) and the I-SITE ULNE Foundation (I-Site_Covid20_ANTI-SARS2 to JeD, 3CLPRO-SCREEN-NMR to XH) and fondation Rotary (to BD), Vinted (to BD), Crédit Mutuel Nord Europe (to BD), Entreprises et Cités (to BD), AG2R (to BD), DSD Système (to BD), M comme Mutuelle (to BD), Protecthoms (to BD), RBL Plastiques (to BD), Saverglass (to BD), Brasserie 3 Monts (to BD), Coron Art (to BD). The platform used in this work was supported by the European Union (ERC-STG INTRACELLTB grant 260901), the ANR (ANR-10-EQPX-04-01), the “Fonds Européen de Développement Régional” (FEDER-ERDF) (12001407 [D-AL] EquipEx ImagInEx BioMed), CPER-CTRL (Centre Transdisciplinaire de Recherche sur la Longévité) and the Région Hauts-de-France (convention 12000080). European Union – ERDF REACT-EU funds, Union response to Coronavirus pandemic (convention 22003061). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. We thank the infrastructure ChemBioFrance (http://www.chembiofrance.org/) and particularly the platform ARIADNE-criblage to provide access to the facilities and for their methodological support during library screening. We thank also ChemBioFrance for financial support and to provide the Essential Chemical Library (1040 compounds) from the Chimiothèque Nationale. We thank the company Life Chemicals that provided 640 compounds of their anti-coronavirus screening libraries. L.B. is a recipient of a PhD fellowship from the University of Lille and Région Hauts-de-France. We thank Valentin Guillaume for helpful discussions and Fanny Bourgeois, Mathilde Malessan, Clémence Wattebled, Chloe Ribes for technical assistance. We thank Dr V. Villeret for his help for crystallogenesis and data processing. We acknowledge SOLEIL for the provision of synchrotron-radiation facilities. We would like to thank Tatiana Isabet and Serena Sirigu for their valuable support during data collection at beamlines PX1 and PX2A at the SOLEIL synchrotron facility (Paris, France). Financial support from the Infranalytics (NMR division) FR 2054 CNRS for conducting the research is gratefully acknowledged. The NMR facilities were funded by the Nord Region Council, CNRS, European Union (FEDER- ERDF), French Research Ministry and Univ. Lille.
Handling Editor: Dr. Z Liu
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.ejmech.2023.115186.
Abbreviations
- DCM
dichloromethane
- DMSO
dimethylsulfoxide;
- DPT
bis(2-pyridyloxy)methanethione
- MERS
Middle East respiratory syndrome
- SARS
severe acute respiratory Syndrome
- TCEP
Tris(2-carboxyethyl)phosphine;
- THF
tetrahydrofuran
- 3CLpro
chymotrypsin-like cysteine protease
- Mpro
main protease
- HPLC
high performance liquid chromatography
- HCoV-229E
human coronavirus 229E
Appendix A. Supplementary data
The following is the Supplementary data to this article:
Additional experimental details and materials, including Crystallography data collection and refinement statistics; crystallographic structure of the SARS-CoV-2 3CLpro after binding to 1x, dose-response curves in the enzymatic assay on the MERS-CoV 3CLpro; 1H NMR and 13C NMR spectra of synthetic compounds (PDF).
Data availability
Data is available upon reasonable request.
References
- 1.Zhong N.S., Zheng B.J., Li Y.M., Xie Z.H., Chan K.H., Li P.H., Tan S.Y., Chang Q., Xie J.P., Liu X.Q., Xu J., Li D.X., Yuen K.Y., Peiris J.S.M., Guan Y. Epidemiology and cause of severe acute respiratory syndrome (SARS) in Guangdong, people's Republic of China, in February, 2003. Lancet. 2003;362:1353–1358. doi: 10.1016/S0140-6736(03)14630-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zaki A.M., van Boheemen S., Bestebroer T.M., Osterhaus A.D.M.E., Fouchier R.A.M. Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N. Engl. J. Med. 2012;367:1814–1820. doi: 10.1056/NEJMoa1211721. [DOI] [PubMed] [Google Scholar]
- 3.Zhu N., Zhang D., Wang W., Li X., Yang B., Song J., Zhao X., Huang B., Shi W., Lu R., Niu P., Zhan F., Ma X., Wang D., Xu W., Wu G., Gao G.F., Tan W. A novel coronavirus from patients with pneumonia in China, 2019. N. Engl. J. Med. 2020;382:727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shitrit A., Zaidman D., Kalid O., Bloch I., Doron D., Yarnizky T., Buch I., Segev I., Ben-Zeev E., Segev E., Kobiler O. Conserved interactions required for inhibition of the main protease of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) Sci. Rep. 2020;10 doi: 10.1038/s41598-020-77794-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang H., He S., Deng W., Zhang Y., Li G., Sun J., Zhao W., Guo Y., Yin Z., Li D., Shang L. Comprehensive insights into the catalytic mechanism of Middle East respiratory syndrome 3C-like protease and severe acute respiratory syndrome 3C-like protease. ACS Catal. 2020;10:5871–5890. doi: 10.1021/acscatal.0c00110. [DOI] [PubMed] [Google Scholar]
- 6.Fan K., Ma L., Han X., Liang H., Wei P., Liu Y., Lai L. The substrate specificity of SARS coronavirus 3C-like proteinase. Biochem. Biophys. Res. Commun. 2005;329:934–940. doi: 10.1016/j.bbrc.2005.02.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Anand K., Ziebuhr J., Wadhwani P., Mesters J.R., Hilgenfeld R. Coronavirus main proteinase (3CLpro) structure: basis for design of anti-SARS drugs. Science. 2003;300:1763–1767. doi: 10.1126/science.1085658. [DOI] [PubMed] [Google Scholar]
- 8.Pillaiyar T., Manickam M., Namasivayam V., Hayashi Y., Jung S.-H. An overview of severe acute respiratory syndrome coronavirus (SARS-CoV) 3CL protease inhibitors: peptidomimetics and small molecule chemotherapy. J. Med. Chem. 2016;59:6595–6628. doi: 10.1021/acs.jmedchem.5b01461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cannalire R., Cerchia C., Beccari A.R., Di Leva F.S., Summa V. Targeting SARS-CoV-2 proteases and polymerase for COVID-19 treatment: state of the art and future opportunities. J. Med. Chem. 2022;65:2716–2746. doi: 10.1021/acs.jmedchem.0c01140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Konwar M., Sarma D. Advances in developing small molecule SARS 3CLpro inhibitors as potential remedy for corona virus infection. Tetrahedron. 2021;77 doi: 10.1016/j.tet.2020.131761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu Y., Liang C., Xin L., Ren X., Tian L., Ju X., Li H., Wang Y., Zhao Q., Liu H., Cao W., Xie X., Zhang D., Wang Y., Jian Y. The development of Coronavirus 3C-Like protease (3CLpro) inhibitors from 2010 to 2020. Eur. J. Med. Chem. 2020;206 doi: 10.1016/j.ejmech.2020.112711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gao K., Wang R., Chen J., Tepe J.J., Huang F., Wei G.-W. Perspectives on SARS-CoV-2 main protease inhibitors. J. Med. Chem. 2021;64:16922–16955. doi: 10.1021/acs.jmedchem.1c00409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kitamura N., Sacco M.D., Ma C., Hu Y., Townsend J.A., Meng X., Zhang F., Zhang X., Ba M., Szeto T., Kukuljac A., Marty M.T., Schultz D., Cherry S., Xiang Y., Chen Y., Wang J. Expedited approach toward the rational design of noncovalent SARS-CoV-2 main protease inhibitors. J. Med. Chem. 2022;65:2848–2865. doi: 10.1021/acs.jmedchem.1c00509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Han S.H., Goins C.M., Arya T., Shin W.-J., Maw J., Hooper A., Sonawane D.P., Porter M.R., Bannister B.E., Crouch R.D., Lindsey A.A., Lakatos G., Martinez S.R., Alvarado J., Akers W.S., Wang N.S., Jung J.U., Macdonald J.D., Stauffer S.R. Structure-based optimization of ML300-derived, noncovalent inhibitors targeting the severe acute respiratory syndrome coronavirus 3CL protease (SARS-CoV-2 3CLpro) J. Med. Chem. 2022;65:2880–2904. doi: 10.1021/acs.jmedchem.1c00598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang C.-H., Spasov K.A., Reilly R.A., Hollander K., Stone E.A., Ippolito J.A., Liosi M.-E., Deshmukh M.G., Tirado-Rives J., Zhang S., Liang Z., Miller S.J., Isaacs F., Lindenbach B.D., Anderson K.S., Jorgensen W.L. Optimization of triarylpyridinone inhibitors of the main protease of SARS-CoV-2 to low-nanomolar antiviral potency. ACS Med. Chem. Lett. 2021;12:1325–1332. doi: 10.1021/acsmedchemlett.1c00326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ma C.A.-O., Sacco M.D., Hurst B., Townsend J.A., Hu Y., Szeto T., Zhang X., Tarbet B., Marty M.T., Chen Y., Wang J. Boceprevir, GC-376, and calpain inhibitors II, XII inhibit SARS-CoV-2 viral replication by targeting the viral main protease. Cell Res. 2020;30:678–692. doi: 10.1038/s41422-020-0356-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hoffman R.L., Kania R.S., Brothers M.A., Davies J.F., Ferre R.A., Gajiwala K.S., He M., Hogan R.J., Kozminski K., Li L.Y., Lockner J.W., Lou J., Marra M.T., Mitchell L.J., Murray B.W., Nieman J.A., Noell S., Planken S.P., Rowe T., Ryan K., Smith G.J., Solowiej J.E., Steppan C.M., Taggart B. Discovery of ketone-based covalent inhibitors of coronavirus 3CL proteases for the potential therapeutic treatment of COVID-19. J. Med. Chem. 2020;63:12725–12747. doi: 10.1021/acs.jmedchem.0c01063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hammond J., Leister-Tebbe H., Gardner A., Abreu P., Bao W., Wisemandle W., Baniecki M., Hendrick V.M., Damle B., Simón-Campos A., Pypstra R., Rusnak J.M., EPIC-HR Investigators Oral nirmatrelvir for high-risk, nonhospitalized adults with covid-19. N. Engl. J. Med. 2022;386:1397–1408. doi: 10.1056/NEJMoa2118542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee H., Mittal A., Patel K., Gatuz J.L., Truong L., Torres J., Mulhearn D.C., Johnson M.E. Identification of novel drug scaffolds for inhibition of SARS-CoV 3-Chymotrypsin-like protease using virtual and high-throughput screenings. Bioorg. Med. Chem. 2014;22:167–177. doi: 10.1016/j.bmc.2013.11.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee H., Torres J., Truong L., Chaudhuri R., Mittal A., Johnson M.E. Reducing agents affect inhibitory activities of compounds: results from multiple drug targets. Anal. Biochem. 2012;423:46–53. doi: 10.1016/j.ab.2012.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cantrelle F.X., Boll E., Brier L., Moschidi D., Belouzard S., Landry V., Leroux F., Dewitte F., Landrieu I., Dubuisson J., Deprez B., Charton J., Hanoulle X. NMR spectroscopy of the main protease of SARS-CoV-2 and fragment-based screening identify three protein hotspots and an antiviral fragment. Angew. Chem., Int. Ed. Engl. 2021;60:25428–25435. doi: 10.1002/anie.202109965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sacco M.D., Ma C., Lagarias P., Gao A., Townsend J.A., Meng X., Dube P., Zhang X., Hu Y., Kitamura N., Hurst B., Tarbet B., Marty M.T., Kolocouris A., Xiang Y., Chen Y., Wang J. Structure and inhibition of the SARS-CoV-2 main protease reveal strategy for developing dual inhibitors against M(pro) and cathepsin L. Sci. Adv. 2020;6 doi: 10.1126/sciadv.abe0751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jacobs J., Grum-Tokars V., Zhou Y., Turlington M., Saldanha S.A., Chase P., Eggler A., Dawson E.S., Baez-Santos Y.M., Tomar S., Mielech A.M., Baker S.C., Lindsley C.W., Hodder P., Mesecar A., Stauffer S.R. Discovery, synthesis, and structure-based optimization of a series of N-(tert-butyl)-2-(Narylamido)-2-(pyridin-3-yl) acetamides (ML188) as potent noncovalent small molecule inhibitors of the severe acute respiratory syndrome coronavirus (SARS-CoV) 3CL protease. J. Med. Chem. 2013;56:534–546. doi: 10.1021/jm301580n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fu L., Ye F., Feng Y., Yu F., Wang Q., Wu Y., Zhao C., Sun H., Huang B., Niu P., Song H., Shi Y., Li X., Tan W., Qi J., Gao G.F. Both boceprevir and GC376 efficaciously inhibit SARS-CoV-2 by targeting its main protease. Nat. Commun. 2020;11:4417. doi: 10.1038/s41467-020-18233-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen S., Zhang J., Hu T., Chen K., Jiang H., Shen X. Residues on the dimer interface of SARS coronavirus 3C-like protease: dimer stability characterization and enzyme catalytic activity analysis. J. Biochem. 2008;143:525–536. doi: 10.1093/jb/mvm246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Flynn J.M., Samant N., Schneider-Nachum G., Barkan D.T., Yilmaz N.K., Schiffer C.A., Moquin S.A., Dovala D., Bolon D.N. Comprehensive fitness landscape of SARS-CoV-2 Mpro reveals insights into viral resistance mechanisms. Elife. 2022;11 doi: 10.7554/eLife.77433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Maingot L., Elbakali J., Dumont J., Bosc D., Cousaert N., Urban A., Deglane G., Villoutreix B., Nagase H., Sperandio O., Leroux F., Deprez B., Deprez-Poulain R. Aggrecanase-2 inhibitors based on the acylthiosemicarbazide zinc-binding group. Eur. J. Med. Chem. 2013;69:244–261. doi: 10.1016/j.ejmech.2013.08.027. [DOI] [PubMed] [Google Scholar]
- 28.Lee C.C., Kuo C.J., Hsu M.F., Liang P.H., Fang J.M., Shie J.J., Wang A.H. Structural basis of mercury- and zinc-conjugated complexes as SARS-CoV 3C-like protease inhibitors. FEBS Lett. 2007;581:5454–5458. doi: 10.1016/j.febslet.2007.10.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tomar S., Johnston M.L., St John S.E., Osswald H.L., Nyalapatla P.R., Paul L.N., Ghosh A.K., Denison M.R., Mesecar A.D. Ligand-induced Dimerization of Middle East Respiratory Syndrome (MERS) Coronavirus nsp5 Protease (3CLpro): implications for nsp5 regulation and the development of antivirals. J. Biol. Chem. 2015;290:19403–19422. doi: 10.1074/jbc.M115.651463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Desmarets L., Callens Nr, Hoffmann E., Danneels A., Lavie M., Couturier C., Dubuisson J., Belouzard S., Rouillé Y. A reporter cell line for the automated quantification of SARS-CoV-2 infection in living cells. Front. Microbiol. 2022;13 doi: 10.3389/fmicb.2022.1031204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang J.H., Chung T.D., Oldenburg K.R. A simple statistical parameter for use in evaluation and validation of high throughput screening assays. J. Biomol. Screen. 1999;4:67–73. doi: 10.1177/108705719900400206. [DOI] [PubMed] [Google Scholar]
- 32.Lee W., Tonelli M., Markley J.L. NMRFAM-SPARKY: enhanced software for biomolecular NMR spectroscopy. Bioinformatics. 2015;31:1325–1327. doi: 10.1093/bioinformatics/btu830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Coati A., Chavas L.M.G., Fontaine P., Foos N., Guimaraes B., Gourhant P., Legrand P., Itie J.-P., Fertey P., Shepard W., Isabet T., Sirigu S., Solari P.-L., Thiaudiere D., Thompson A. Status of the Crystallography beamlines at synchrotron SOLEIL. Eur. Phys. J. Plus. 2017;132:174. doi: 10.1140/epjp/i2017-11403-3. [DOI] [Google Scholar]
- 34.Oscarsson M., Beteva A., Flot D., Gordon E., Guijarro M., Leonard G., McSweeney S., Monaco S., Mueller-Dieckmann C., Nanao M., Nurizzo D., Popov A., von Stetten D., Svensson O., Rey-Bakaikoa V., Chado I., Chavas L., Gadea L., Gourhant P., Isabet T., Legrand P., Savko M., Sirigu S., Shepard W., Thompson A., Mueller U., Nan J., Eguiraun M., Bolmsten F., Nardella A., Milan-Otero A., Thunnissen M., Hellmig M., Kastner A., Schmuckermaier L., Gerlach M., Feiler C., Weiss M.S., Bowler M.W., Gobbo A., Papp G., Sinoir J., McCarthy A., Karpics I., Nikolova M., Bourenkov G., Schneider T., Andreu J., Cuní G., Juanhuix J., Boer R., Fogh R., Keller P., Flensburg C., Paciorek W., Vonrhein C., Bricogne G., de Sanctis D. MXCuBE2: the dawn of MXCuBE collaboration. J. Synchrotron Radiat. 2019;26:393–405. doi: 10.1107/S1600577519001267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kabsch W.X.D.S. Acta Crystallogr. D Biol. Crystallogr. 2010;66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Potterton L., Agirre J., Ballard C., Cowtan K., Dodson E., Evans P.R., Jenkins H.T., Keegan R., Krissinel E., Stevenson K., Lebedev A., McNicholas S.J., Nicholls R.A., Noble M., Pannu N.S., Roth C., Sheldrick G., Skubak P., Turkenburg J., Uski V., von Delft F., Waterman D., Wilson K., Winn M., Wojdyr M. CCP4i2: the new graphical user interface to the CCP4 program suite. Acta Crystallogr. D Struct Biol. 2018;74:68–84. doi: 10.1107/S2059798317016035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Winn M.D., Ballard C.C., Cowtan K.D., Dodson E.J., Emsley P., Evans P.R., Keegan R.M., Krissinel E.B., Leslie A.G.W., McCoy A., McNicholas S.J., Murshudov G.N., Pannu N.S., Potterton E.A., Powell H.R., Read R.J., Vagin A., Wilson K.S. Overview of the CCP4 suite and current developments. Acta Crystallogr. D Biol. Crystallogr. 2011;67:235–242. doi: 10.1107/S0907444910045749. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional experimental details and materials, including Crystallography data collection and refinement statistics; crystallographic structure of the SARS-CoV-2 3CLpro after binding to 1x, dose-response curves in the enzymatic assay on the MERS-CoV 3CLpro; 1H NMR and 13C NMR spectra of synthetic compounds (PDF).
Data Availability Statement
Data is available upon reasonable request.