Abstract
Osteosarcoma (OS) is the most common primary malignancy of bone. Here, we investigated a possible role of defective osteoblast differentiation in OS tumorigenesis. We found that basal levels of the early osteogenic marker alkaline phosphatase (ALP) activity were low in OS lines. Osteogenic regulators Runx2 and OSX, and the late marker osteopontin (OPN) expressed at low levels in most OS lines, indicating that most OS cells fail to undergo terminal differentiation. Furthermore, OS cells were refractory to osteogenic BMP-induced increases in ALP activity. Osteogenic BMPs were shown to upregulate early target genes, but not late osteogenic markers OPN and osteocalcin (OC). Furthermore, osteogenic BMPs failed to induce bone formation from human OS cells, rather effectively promoted OS tumor growth in an orthotopic OS model. Exogenous expression of early target genes enhanced BMP-stimulated OS tumor growth, whereas osteogenic BMP-promoted OS tumor growth was inhibited by exogenous Runx2 expression. These results suggest that alterations in osteoprogenitors may disrupt osteogenic differentiation pathway. Thus, identifying potential differentiation defects in OS tumors would allow us to reconstruct the tumorigenic events in osteoprogenitors and to develop rational differentiation therapies for clinical OS management.
Keywords: BMPs, bone tumors, mesenchymal stem cells, osteosarcoma, osteogenic differentiation
Osteosarcoma (OS) is the most common nonhematolgic malignant tumor of bone in children and adults.1–3 The peak incidence of OS occurs in the teens.1 OS tumors typically arise around the metaphysis of long bones and are often of high grade with poor prognosis.4 OS is characterized by a high propensity for metastasis, mostly to the lungs, with 10–20% having detectable metastases at diagnosis.5 Only ~10% of OS patients achieve long-term disease-free interval.6 The unifying histologic features of OS are the presence of malignant osteoid produced by neoplastic cells.7
The molecular pathogenesis of OS is poorly understood. Certain genetic or acquired conditions increase the risk of OS.8,9 Patients with hereditary retinoblastoma have a high risk of OS10 and OS may also arise in patients with Paget’s disease of bone.11 We have demonstrated that Wnt/β-catenin and S100A6 are frequently upregulated in human OS tumors.12–14 Cytogenetic studies of OS have documented a variety of genetic alterations, resulting in inactivation of tumor suppressor genes and overexpression of oncogenes.1,2,15,16 However, it is unclear how much these genetic changes contribute to OS development.2,3
Mesenchymal stem cells (MSCs) are adherent bone marrow stromal cells that can differentiate into osteogenic, chondrogenic, adipogenic, and myogenic lineages.17–21 Osteoblastic differentiation of MSCs is an essential process of bone formation,17–20 and is tightly regulated.20,21 BMPs play an important role in osteoblast differentiation.20,22–24 We have analyzed the osteogenic potential of 14 human BMPs and found that BMP-2, 6, 9 are the most potent inducers of osteogenic differentiation both in vitro and in vivo.19,20,24–26 We have further demonstrated that osteogenic BMPs regulate a distinct set of target genes, such as Id HLH factors and connective tissue growth factor (CTGF)/CCN2, during osteogenic differentiation.19,20,27–29
Blockade of stem cell differentiation may lead to tumorigenesis.3,30 The stem cell features of tumor cells have been confirmed by a recent study, in which silencing of Ewing’s sarcoma (EWS)-FLI-1 in Ewing’s tumor cells leads to their recovery of MSC features, such as differentiating along adipogenic and osteogenic lineages,31 whereas expression of the EWS/FLI-1 oncogene in murine MSCs results in EWS-like tumors.32 Human OS tumors usually exhibit osteoblast-like features.3,33–39 We demonstrated that differentiation-promoting agents (eg, PPARγ agonists and 9-cis retinoic acid) can induce osteoblast differentiation and inhibit OS cell proliferation.3,40
Here, we investigate whether the disruption of osteoblast differentiation plays a role in OS development. Using four commonly used OS lines and 12 primary OS lines derived from OS patients, we found that the early stages of osteogenic differentiation were preserved in most OS cells because these OS lines were responsive to osteogenic BMP-upregulated expression of the early target genes, but not late markers. Osteogenic BMPs failed to induce osteogenic differentiation of human OS cells in vivo, but effectively promoted OS tumor growth in an orthotopic model of human OS. Moreover, exogenous Id expression (especially Id2) was shown to enhance BMP-stimulated OS tumor growth, whereas osteogenic BMP-promoted OS tumor growth was inhibited by exogenous expression of Runx2, possibly by promoting osteogenic differentiation of the OS cells. Our findings suggest that genetic and/or molecular changes in osteoprogenitor cells may disrupt osteogenic differentiation pathway, subsequently leading to the OS development.
RESULTS
Early Osteogenic Marker ALP Activity is Low in Most Human OS Tumor Cells
Alkaline phosphatase (ALP) is a well-characterized early osteogenic marker that exhibits an elevated activity in committed preosteoblast cells and a decreased activity in the mature osteoblast cells. We measured the basal ALP activity in four human OS lines, MG63, U2OS, TE85, and SaOS2. As shown in Figure 1a, all OS lines displayed a higher basal ALP activity than that of mouse MSC line C3H10T1/2. The growth conditions (eg, different FCS concentrations) had a limited effect on the basal ALP activity of these OS lines. Among the four OS lines, MG63 exhibited the lowest basal ALP activity, whereas SaOS2 cells had the highest basal ALP activity. In fact, the basal ALP activities were approximately 7, 123, and 4129 times higher in U2OS, TE85, and SaOS2 cells than that in MG63 cells, respectively. The basal ALP activities in these OS lines were also confirmed by histochemical staining (Figure 1b). On the basis of levels of basal ALP activity, we can conclude that SaOS2 is the most differentiated, followed by TE85, whereas MG63 is the least differentiated OS line. These findings are consistent with the in vitro cell proliferative activity and in vivo tumorigenic features of these lines.41
Figure 1.
Basal activity of early osteogenic marker ALP in human osteosarcoma cells. (a) Quantitative analysis of basal ALP activity in four commonly used OS lines and control mouse mesenchymal progenitor line C3H10T1/2. Subconfluent cells were cultured at the indicated serum conditions for 36 h and collected for ALP assay as described in ‘Materials and Methods’. Each assay condition was done in duplicate. (b) Qualitative analysis of basal ALP activity in four OS lines. OS cells were cultured in 1% FCS for 36 h and stained histochemically for ALP activity as described in ‘Materials and Methods’. (c) Basal ALP activity in primary OS lines derived from patients. Twelve primary OS tumor cell lines, primary human marrow stromal cells (hMSC), and immortalized human osteoblast line hFOB were cultured in 1% FCS for 36 h and collected for ALP assays. SaOS2 was included as a control of high basal ALP activity. See ‘Materials and Methods’.
We also analyzed the basal ALP activity in 12 primary OS lines derived from human OS tumors. Isolated normal human bone marrow stromal cells (hMSCs) were used as a control for uncommitted preosteoblasts, whereas the human osteoblast line hFOB1.19 was used as a control for committed osteoblasts.42 As shown in Figure 1c, most primary OS lines exhibited low ALP activity, when compared with that of hFOB1.19 cells. Most primary OS lines also exhibited a much lower basal level of ALP activity than that of SaOS2, suggesting that majority of OS tumor cells may be less differentiated than SaOS2.
Low Expression of Osteogenic Regulators Runx2 and Osterix in most Human OS Cells
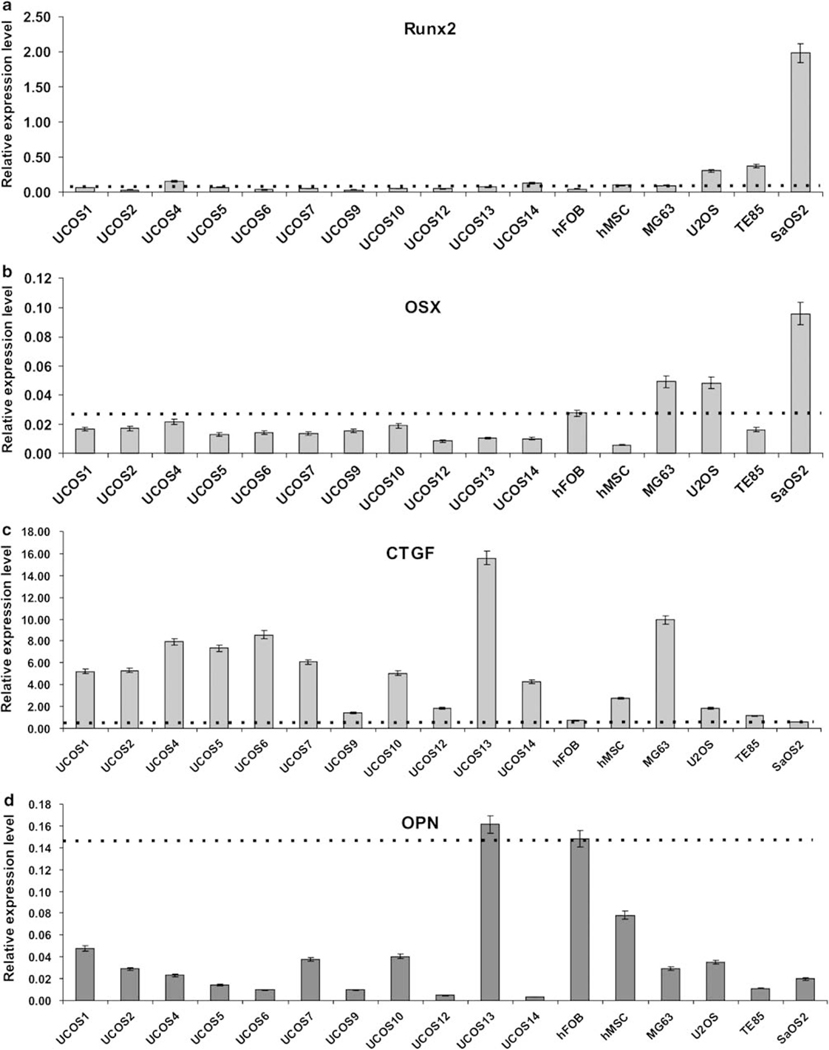
We next examined the endogenous expression levels of Runx2 and Osterix (OSX), both of which are important regulators of osteogenic differentiation,17–21,43 in OS lines. Osteoblast line hFOB1.19 and primary hMSC were used as controls. As shown in Figure 2a and b, most OS lines exhibited low levels of expression of Runx2 and OSX, whereas SaOS2 had the highest expression levels of both genes, consistent with the possibility that SaOS2 is more differentiated. These results indicate that osteogenic regulators Runx2 and/or OSX are expressed at low levels, suggesting that there may be differentiation defects upstream of Runx2 and/or OSX functions in most human OS cells.
Figure 2.
Endogenous expression levels of osteogenic regulators and other osteogenic markers in human osteosarcoma cells. Four OS lines, 11 primary OS lines, human osteoblast line hFOB, and primary human marrow mesenchymal line hMSC were cultured in 1% FCS for 36 h, and were collected for total RNA isolation. The total RNA was subjected to reverse transcription and subsequently quantitative real-time PCR (qPCR) analysis. Gene-specific primers (listed in Supplementary Table 1) were used to determine the expression levels of osteogenic regulators Runx2 (a) and Osterix (OSX) (b), early osteogenic marker CTGF/CCN2 (c), and late osteogenic marker osteopontin (OPN) (d). All qPCR samples were normalized with the expression level of GAPDH. Each condition was done in triplicate. Dotted lines indicate the expression levels of the indicated genes in hFOB, a mature osteoblast line. See ‘Materials and Methods’.
Connective Tissue Growth Factor is Highly Expressed in Uncommitted Preosteoblast Progenitors and in most Human OS Cells
Connective tissue growth factor (a.k.a., CCN2), a member of the CCN family of modulator proteins, is a multifunctional growth factor for osteoblasts, chondrocytes, and vascular endothelial cells.29 We previously demonstrated that CTGF is upregulated at the early stage of osteogenic differentiation.29 We analyzed the basal CTGF expression in human OS cells. As shown in Figure 2c, almost all of the tested primary OS lines exhibited higher expression levels of CTGF than that of hFOB1.19, whereas MG63 (and to a lesser extent, U2OS) also had high basal level of CTGF expression. However, CTGF expression was low in SaOS2 and TE85. These results are consistent with other osteogenic parameters, suggesting that most OS lines may resemble early osteoprogenitors.
Expression Level of Late Osteogenic Marker Osteopontin is Low in most Human OS Cells
Osteopontin (OPN) is a well-characterized late marker of osteogenesis.20,24–26 As expected, mature osteoblast hFOB1.19 exhibited a high expression level of OPN, whereas hMSCs, representing a mix population of progenitors and osteogenic lineages at differentiation stages, had a modestly high expression level of OPN (Figure 2d). However, all but one (ie, UCOS13) OS lines had significantly lower levels of OPN expression than that of hFOB1.19 cells (Figure 2d). These findings indicate that most OS cells fail to undergo terminal osteogenic differentiation.
Human OS Cells are Refractory to Osteogenic BMP-Induced ALP Activity
As shown in Figure 1, most OS lines (except SaOS2, and to a lesser extent, TE85) exhibited low basal ALP activity. We sought to determine whether ALP activity could be induced upon osteogenic BMP stimulation. BMP2, BMP6, BMP7, and BMP9 are the most potent BMPs to induce osteogenic differentiation of MSCs.19,20,24–26 We first analyzed the basal expression of the four BMPs in OS lines. As shown in Figure 3a, BMP7 was relatively highly expressed in the four OS lines tested, whereas BMP2 expression was the lowest in these OS lines. BMP6 and BMP9 expression levels were modest and readily detectable. We further examined the basal expression of Id1, Id2, and Id3 genes, which are important early targets of osteogenic BMP-induced osteogenic differentiation.20,27,28 As shown in Figure 3b, expression of these Id genes was readily detected, although Id3 exhibited much higher expression levels than that of Id1 or Id2 in the tested OS lines.
Figure 3.
Human osteosarcoma cells are refractory to osteogenic BMP-induced ALP activity. (a) Endogenous expression levels of four osteogenic BMPs in human OS lines. Total RNA was isolated from the four OS lines and subjected to reverse transcription and quantitative real-time PCR analysis using primers specific for human BMP2, BMP6, BMP7, and BMP9 (listed in Supplementary Table 1). (b) Endogenous expression levels of early osteogenic genes Id1, Id2, and Id3 in human OS lines. Total RNA and quantitative real-time PCR analysis were carried out essentially similar to that described in (a), except using primers specific for human Id1, Id2, and Id3 (listed in Supplementary Table 1). All qPCR samples were normalized with the expression level of GAPDH. Each condition was done in triplicate. (c) Fold of changes in BMP2- and BMP9-induced ALP activity in the four OS lines. Subconfluent OS cells were infected with AdBMP2, AdBMP9, or AdGFP control. The infected cells were collected for ALP assay at the indicated time points. Each assay condition was done in duplicate. Fold of changes was calculated by dividing the ALP activity from BMP-treated samples with that of GFP-treated samples. The dotted lines indicate that the fold of change is 1.0 (ie, BMP-induced ALP activity is equal to that of GFP control-treated cells). See ‘Materials and Methods’.
We next analyzed the ALP activities in response to osteogenic BMP in OS cells. We previously demonstrated that BMP2 and BMP9 can effectively induce ALP activity in MSCs.19,20,24–26 Our preliminary results indicated that most, if not all, of the TGFβ/BMP types I and II receptors were expressed in the tested OS lines (data not shown). The four OS lines were stimulated with BMP2, BMP9, or GFP control, and their ALP activities were measured at days 5, 7, and 10. As shown in Figure 3c, neither BMP2 nor BMP9 induced any significant increases in ALP activities in the tested four OS lines at all time points. Notably, BMP2 induced a less than onefold increase in ALP activity in MG63 and U2OS cells at day 7, whereas BMP9 induced an approximately 50% increase in ALP activity in TE85 cells at day 10. These results suggest that these OS lines, except SaOS2 cells (and to a lesser extent, TE85) that exhibit high basal ALP activity, may harbor defects that lay upstream of the regulatory circuit of ALP activity at the early stages of osteogenic differentiation.
Osteogenic BMPs Upregulate the Expression of Early Target Genes Id1, Id2, and Id3 in OS Cells, Whereas the Expression of Late Markers OPN and Osteocalcin is not Affected by Osteogenic BMPs
We stimulated the four OS lines and two primary OS lines with BMP2, BMP6, BMP9, or GFP, and analyzed the expression of Id1, Id2, and Id3 by using qPCR. As illustrated in Figure 4a, Id1 expression was readily induced (2- to 9-fold) by all three BMPs in the four OS lines, whereas the induction of Id2 and Id3 expression was apparent but exhibited significant variations among the OS lines and/or three BMPs. Among the three BMPs, BMP2 was shown to be an effective inducer of the Id genes in three of the four OS lines (ie, except MG63). These results may reflect the fact that the basal expression of BMP2 is lower in OS lines than that of BMP6 and BMP9 (Figure 3a). Of the three Id genes, Id3 was shown to be in general the least inducible (Figure 4a), which may be due to the high basal level of Id3 expression (Figure 3b). Similar results about BMP2 and BMP9-induced Id gene expression were obtained in two primary OS lines, UCOS1 and UCOS12 (Figure 4b). These findings suggest that the early stages of osteogenic differentiation may be preserved. We also found that the inhibitory Smad6 and Smad7, which are considered as feedback inhibitors of BMP signaling, were significantly upregulated in these OS cells upon BMP stimulation, indicating that the early stage of the BMP signaling pathway may remain intact in these OS cells (data not shown).
Figure 4.
Osteogenic BMPs effectively induce early osteogenic genes but fail to induce late osteogenic markers. (a) Osteogenic BMPs upregulate Id expression in the four OS lines. Subconfluent OS lines were cultured in 1% FCS and infected with AdBMP2, AdBMP6, AdBMP9, and AdGFP. At 30 h after infection, total RNA was isolated from the infected cells and subjected to reverse transcription and qPCR analysis using primers specific for human Id1, Id2, and Id3. (b) Osteogenic BMPs upregulate Id expression in the patient-derived primary OS lines. Subconfluent cells were cultured in 1% FCS and infected with AdBMP2, AdBMP9, and AdGFP for 30 h. Total RNA and qPCR analysis were carried out essentially the same as that described in panel (a). (c) Osteogenic BMPs exert no significant effect on the expression of late osteogenic marker osteocalcin and osteopontin in human OS lines. Subconfluent OS lines were infected with AdBMP2, AdBMP6, AdBMP9, and AdGFP. At 7 days after infection, total RNA was isolated from the infected cells and subjected to reverse transcription and qPCR analysis using primers specific for human osteocalcin and osteopontin (listed in Supplementary Table 1). Each assay condition was done in triplicate. Fold of changes was calculated by dividing the relative expression level from BMP-treated samples with that of GFP-treated samples. The dotted lines indicate that the fold of change is 1.0 (ie, BMP-induced expression is equal to that of GFP control-treated cells). See text for detail.
We further analyzed the BMP-regulated expression of the late osteogenic markers OPN and osteocalcin (OC) in the OS cells. We previously demonstrated that BMP2, BMP6, and BMP9 can effectively upregulate OPN and OC expression in MSCs.19,20,24–26 As shown in Figure 4c, BMP2, BMP6, and BMP9 failed to induce any significant increases in OC expression in MG63, U2OS, and SaOS2 cells, although there was a small (˂2.5-fold) but consistent induction of OPN expression in TE85 cells. Most OS lines, however, did not exhibit any significant increases in OPN expression upon osteogenic BMP stimulation (although there was a less than twofold increase in MG63 cells; Figure 4c). These findings, together with the results shown in Figure 3c, strongly suggest that these OS lines may contain impairments in the osteogenic pathway, preventing the OS cells from undergoing terminal differentiation.
Osteogenic BMPs Fail to Induce Osteogenic Differentiation of Human OS Cells In Vivo, but Promote Tumor Growth in an Orthotopic Model of Human OS
We next sought to determine the effect of osteogenic BMPs on osteogenic differentiation vs OS tumor growth in an orthotopic model of human OS tumors.41 We used MG63 line and another frequently used OS line 143B for the in vivo studies, as TE85, U2OS, and SaOS2 did not readily form tumors in xenograft models.41 We first analyzed whether BMP2 or BMP9-tranduced OS cells were able to induce bone formation in vivo. We infected MG63 and normal MSC line C3H10T1/2 cells with AdBMP2, AdBMP9, or AdGFP and implanted the transduced cells subcutaneously in athymic mice. At 6 weeks after implantation, animals were killed, and the retrieved masses were subjected to histologic evaluation. It should be noted that GFP-transduced C3H10T1/2 and MG63 cells did not form any detectable masses. As shown in Figure 5a, both BMP2 and BMP9-transduced C3H10T1/2 cells induced robust bone formation (panels i and ii), consistent with our early studies.20,26,44 However, BMP2 or BMP9-transduced MG63 cells did not exhibited any bone formation (Figure 5a, panels iii and iv). Similar results were obtained when we conducted subcutaneous injection with BMP2- or BMP9-transduced other human OS lines (Supplementary Figure 1a). These results further substantiate our in vitro findings, suggesting that most OS cells may harbor differentiation defects and are refractory to osteogenic BMP-induced osteogenic differentiation.
Figure 5.
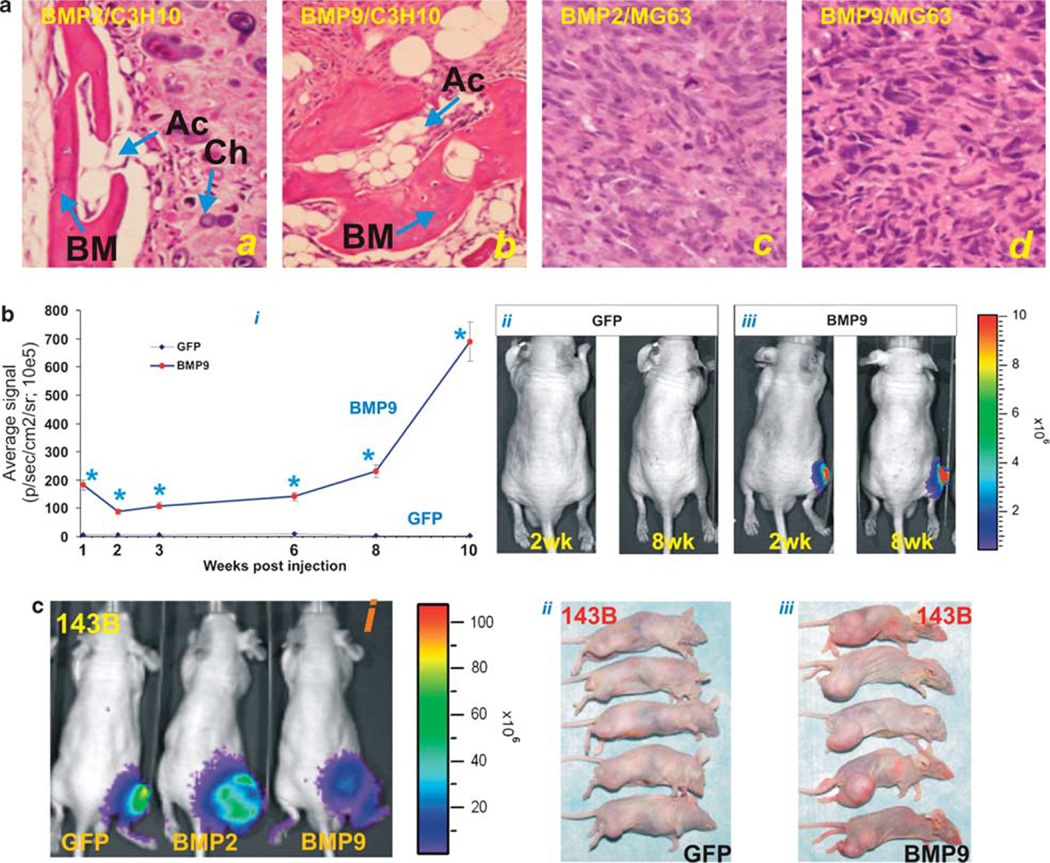
Osteogenic BMPs fail to induce in vivo osteogenic differentiation of OS cells, but effectively promote OS tumor growth. (a) Osteogenic BMPs induce bone formation in normal MSCs but not in OS cells. AdBMP2, AdBMP9, or AdGFP-infected C3H10T1/2 (panels i and ii) and MG63 cells (panels iii and iv) were injected subcutaneously. Animals were killed at 6 weeks after injection, and the retrieved masses were subjected to H&E histologic evaluation. Note that GFP-treated cells did not form any detectable masses at the end point of the experiment. Ac, adipocytes; BM, bone matrix; Ch, chondrocytes. Magnification, × 200. (b) BMP9 promotes tumor growth formed by MG63 cells. Subconfluent MG63-Luc cells were infected with AdBMP9 or AdGFP. At 16 h after infection, cells were collected and injected into athymic mice intratibially (five animals per assay condition), as described in ‘Materials and Methods’. Animals were subjected to Xenogen imaging at the indicated time points. Sizes of tumors (in photons/sec/cm2/steradian) were calculated by using Xenogen’s Living Image software (i). Xenogen optical images of the athymic mice intratibially injected with GFP or BMP9-transduced MG63-Luc cells at weeks 2 and 8 are shown (ii and iii). *P˂0.05 when compared with GFP group at corresponding time points. (c) BMP2 and BMP9 also promote tumor growth formed by human OS line 143B. Athymic mice were intratibially injected with AdBMP2, AdBMP9, or AdGFP-infected 143B-Luc cells. Bioluminescence images taken at 4 weeks after injection are shown (i). Animals were killed after 6 weeks, and the gross images of xenograft tumors formed by intratibially injected with AdGFP- (ii) or AdBMP9- (iii) transduced 143B-Luc OS cells are shown.
We next examined the effect of osteogenic BMPs on tumor growth. Transduced MG63-Luc cells with AdBMP9 or AdGFP were injected into proximal tibiae of athymic mice. The tumor growth was monitored by Xenogen bioluminescence imaging. As shown in Figure 5b, BMP9-promoted tumor growth of MG63 cells in a time-dependent manner (panel i). MG63 cells stimulated with GFP control did not form significant tumors up to 10 weeks after injection, whereas BMP9 was shown to effectively promote tumor growth (Figure 5b, panels ii vs iii). Similar experiments were carried out by using another commonly used human OS line 143B, which is derived from k-Ras transformed TE85 cells and is one of the most tumorigenic OS lines.41 Both BMP2 and BMP9 were shown to effectively enhance the growth of the xenograft tumors formed by transduced 143B-Luc cells (Figure 5c, panel i). The gross appearance of the xenograft tumors is consistent with the bioluminescence imaging results, although Xenogen signal in BMP9-treated group might have underestimated tumor sizes due to the rapid tumor growth and necrosis (Figure 5c, panels ii and iii vs i). The tumor-promoting effect of osteogenic BMPs on OS cells was reproducibly observed when BMP6 or BMP7 were used, or carried out in other OS lines (data not shown). Thus, these in vivo results are consistent with the possibility that OS cells may harbor differentiation defects at the early stages of osteogenesis, rendering the defective osteoprogenitors high proliferative potential. As a result, osteogenic stimuli (such as BMPs) would further promote the proliferation of these OS cells.
Exogenous Id Expression Enhances BMP-Stimulated OS Tumor Growth
It is conceivable that, if the defects are downstream of Id gene regulation, overexpression of Id genes may potentiate BMP-promoted tumor growth. On the basis of our in vitro studies shown in Figure 4a and b, we anticipated that exogenous Id expression could enhance BMPs’ effect on OS tumor growth. To test this possibility, we established MG63 sublines that stably express Id1, Id2, Id3, or vector control. Exogenous expression of Id genes in these pooled stable lines were confirmed (Supplementary Figure 1b). These Id stable lines and vector control line were infected with AdBMP9 or AdGFP and subsequently subjected to intratibial injection as described in Figure 5b. As shown in Figure 6a, Id2 overexpression significantly enhanced BMP9-promoted tumor growth at all time points except at week 10, the decrease in signal at week 10 is likely related to tumor necrosis. Likewise, Id3 expression was also shown to modestly enhance BMP9-promoted tumor growth (Figure 6a, panel i, compared with Figure 5b). Overexpression of Id1 did not exert any significant effect on BMP9-promoted tumor growth, although Id1 expression was shown to slightly enhance tumor growth beyond 8 weeks post-injection. Interestingly, Xenogen signals were in general higher during the first week and ecreased at week 2 prior to sustained increases in BMP9 and/or Id treated groups. The initial signal spikes may be related to the active luciferase expression of the injected cells. Representative bioluminescence images are shown in Figure 6a (panels ii–iv). It is worthy noting that the GFP-treated cells or Id stable lines alone did not form any detectable masses during the course of the study. These results indicate that Id2 (to a much lesser extent, Id3 and Id1) overexpression can enhance OS tumor-promoting effect of BMP9, suggesting that possible differentiation defects may lay downstream of Id genes of the osteogenic pathway.
Figure 6.
Osteogenic BMP-promoted OS tumor growth is enhanced by Id overexpression but is antagonized by Runx2 overexpression. (a) Exogenous expression of Id2 (to a lesser extent, Id3) further enhances BMP9-stimulated MG63 tumor growth. The animal experiments and procedures were carried out at the same time as described in Figure 5b. Briefly, MG63-Id or MG-63-RV cells were infected with AdBMP9, or AdGFP. At 16 h after infection, cells were collected and injected into athymic mice intratibially. Animals were subjected to Xenogen imaging at the indicated time points and tumor sizes were calculated as described in Figure 5b (i). *P˂0.05 when compared with BMP9 only group at corresponding time points. Representative Xenogen images of the athymic mice intratibially injected with BMP9-transduced MG63-Id cells at weeks 2 and 8 are shown (ii and iv). Note that MG63-Id only, and GFP-transduced MG63-Id groups did not form any detectable masses at all time points. (b) Exogenous expression of Runx2 antagonizes BMP9-promoted MG63 tumor growth. The animal experiments and procedures were carried out at the same time as described in Figure 5b. MG63-Luc cells were infected with AdBMP9, AdGFP, AdBMP9 + AdRunx2, or AdGFP + AdRunx2. At 16 h, the infected cells were collected and injected intratibially into athymic mice. Animals were subjected to Xenogen imaging at the indicated time points and tumor sizes were calculated as described in Figure 5b (i). *P˂0.05 when compared with BMP9 only group at corresponding time points. Representative Xenogen images of the athymic mice intratibially injected with BMP9 + Runx2-transduced MG63-Luc cells at weeks 2 and 8 are shown (ii). Note that animals injected with GFP + Runx2-transduced MG63-Luc cells did not form any detectable masses at the injection sites up to 10 weeks. (c) Exogenous expression of Runx2 modestly induces the expression of late osteogenic makers OPN and/or OC in OS lines. Subconfluent OS cells were infected with AdBMP9 or AdGFP. At 5 days after infection, cells were collected for RNA isolation and qPCR analysis using human OPN and OC-specific primers. Each assay condition was done in triplicate. OC, osteocalcin; OPN, osteopontin.
Osteogenic BMP-Promoted OS Tumor Growth is Inhibited by Exogenous Expression of Runx2
We further examined whether overexpression of Runx2 would exert any effect on BMP-promoted OS tumor growth. MG63-Luc cells transduced with AdBMP9, AdGFP, and/or AdRunx2 were injected intratibially as described in Figure 5b. Exogenous expression of Runx2 was shown to inhibit BMP9-stimulated tumor growth at all time points (Figure 6b, panel i). Representative Xenogen images are shown in Figure 6b, panel ii. Moreover, we overexpressed Runx2 in MG63, U2OS, and TE85, and determined the expression levels of Runx2 target OC and late osteogenic marker OPN. As shown in Figure 6c, OC expression was induced in MG63 and U2OS cells, but not in TE85 cells, whereas OPN expression was upregulated in all three tested OS lines. These results suggest that most, if not all steps of the differentiation pathway downstream Runx2 may be preserved in OS cells. Runx2 expression seemingly decreased the proliferative activity in these OS cells, as demonstrated by Ki-67 immunohistochemical staining (Supplementary Figure 2), although the detailed mechanism requires further investigations. These in vivo and in vitro findings suggest that some of the OS lines may harbor differentiation defects upstream Runx2 functionality and may become more differentiated when induced with Runx2.
Osteogenic BMPs Promote Cell Proliferation in Early Progenitors and in OS Cells
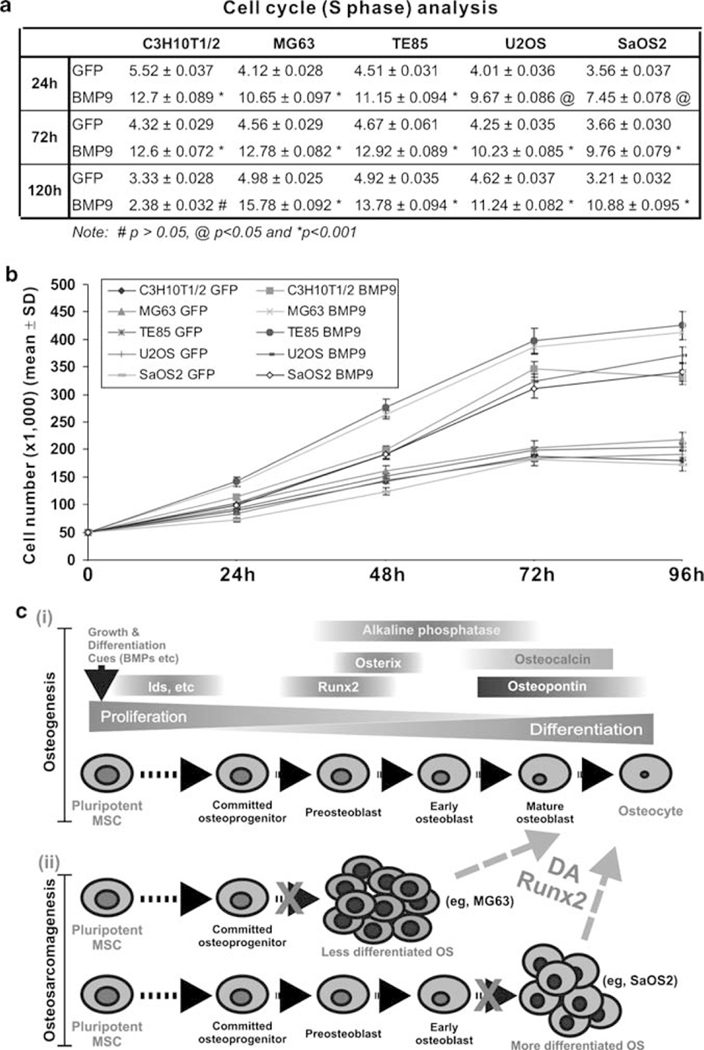
Lastly, we examined whether osteogenic BMPs exert any effects on cell proliferation of MSCs and OS cells. C3H10T1/2 cells were infected with AdGFP or AdBMP9 for 24, 72, and 120 h post-infection, and subjected to flow cytometry. As shown in Figure 7a, AdBMP9-infected C3H10T1/2 cells exhibited much higher percentages of cells in S phase than that of control (P˂0.01), consistent with the notion that BMP9 may promote progenitor cell expansion at the early stage of differentiation at 24 and 72 h, whereas cell numbers in S phase were not different in both BMP9 and GFP-transduced cells at 120 h (P˃0.05). However, in AdBMP9-infected OS cells the percentages of cells in S phase is much higher than that of AdGFP-infected cells at all three points (P˂0.05) (Figure 7a), consistent with the in vivo results in which BMPs were shown to promote tumor growth without inducing terminal differentiation. The BMP9-promoted cell proliferation of OS cells was also confirmed by counting viable cells at different time points. As shown in Figure 7b, the proliferation rates of AdBMP9-infected OS cells were significantly higher than that of the respective AdGFP-infected OS cells at 48, 72, and 96 h (P˂0.01).
Figure 7.
Osteogenic BMPs promote cell proliferation in early progenitors and in OS cells. (a) C3H10T1/2 cells and human OS cells were infected with GFP or BMP9 adenovirus. At 24 h, 72 h, and 120 h post-infection, cells were collected, fixed, incubated with the propidium iodide (PI)/RNase staining buffer, and subjected to flow cytometry. Percentage of cells in S phase was statistically analyzed. Each assay condition was done in triplicate. (b) C3H10T1/2 cells and human OS cells were infected with AdBMP9 or AdGFP. At the indicated time points, cells were collected. Viable cells were counted. Each assay condition was done in triplicate. (c) Relationship between osteogenic differentiation and bone tumorigenesis. Osteogenic differentiation is a well-coordinated process including the repopulation of early progenitors and terminal differentiation of committed osteoblast cells (i). Any impairment in the early stages of osteogenic differentiation may prevent osteoprogenitor cells from undergoing terminal differentiation, hence leading to the development of bone tumors (ii). Reintroduction of Runx2 or treatment of differentiation agents (DA) may increase differentiation potential of OS cells and convert those cells into more differentiated osteoblasts or osteocytes. See text for detail.
DISCUSSION
Here, we investigated whether disruption of osteoblast terminal differentiation plays a role in OS development. Using four commonly used OS lines and 12 primary OS lines derived from OS tumors, we found that the basal levels of early osteogenic marker ALP activity were low in most OS cells, suggesting that majority of these OS tumor cells may be less differentiated. Similarly, low basal expression levels of the osteogenic regulators Runx2 and OSX were detected in most OS cells, suggesting that there may be differentiation defects upstream of Runx2 and/or OSX functions in OS cells. Early osteogenic factor CTGF was found highly expressed in uncommitted preosteoblast progenitors and in most OS cells, whereas expression levels of late osteogenic marker OPN were lower in most human OS tumor cells than that of mature osteoblastic cells, indicating that most OS cells fail to undergo terminal osteogenic differentiation. OS cells were refractory to osteogenic BMP-induced increases in ALP activity. Further, osteogenic BMPs failed to induce bone formation from human OS cells, rather promoted OS tumor growth in an orthotopic OS model. Exogenous Id expression (especially Id2) was shown to enhance BMP-stimulated OS tumor growth, whereas osteogenic BMP-promoted OS tumor growth was inhibited by exogenous Runx2 expression, possibly by promoting terminal differentiation of the OS cells.
On the basis of these findings, we propose a model that depicts the relationship between osteogenic differentiation and OS tumorigenesis (Figure 7c). Osteogenic differentiation of MSCs is a multistep process that requires a balanced regulation of proliferation and differentiation of osteoprogenitors17–20,43 (Figure 7c, panel i). Disruptions of osteogenic differentiation may lead to the development of OS.3 It is conceivable that osteoprogenitor cells harboring defects at the early stage of osteogenic pathway may lead to the development of less differentiated and more aggressive OS tumors, whereas OS tumors may be more differentiated if they are caused by defects occurring at later stages of osteogenic pathway (Figure 7c, panel ii). Some of the OS lines may harbor differentiation defects upstream Runx2 functionality and may become more differentiated when induced with Runx2 or other differentiation agents.3,40
This model is supported by the following facts. First, OS cells exhibit the characteristics of undifferentiated osteoblasts.3,33–39 Second, we demonstrated that differentiation-promoting agents (eg, PPARγ agonists and 9-cis retinoic acid) can induce OS differentiation and inhibit OS proliferation.3,40 Third, a recent study that showed Runx2 and p27KIP1-mediated osteoblast terminal differentiation is disrupted in OS.39 Fourth, in EWS, the second most common malignant pediatric bone tumor caused by oncogenes EWS/FLI-1 or EWS/ETS, EWS/ETS fusion proteins block differentiation along osteogenic and adipogenic lineages of MSCs.45 Fifth, EWS-FLI1-silenced Ewing’s cell lines can differentiate along adipogenic lineage, or osteogenic lineage when stimulated with appropriate differentiation cocktails, suggesting that the inhibition of EWS-FLI1 may allow Ewing’s cells to recover the phenotype of their MSC progenitors.31 Lastly, expression of EWS/FLI-1 oncogene in murine primary MSCs results in EWS/FLI-1-dependent, Ewing’s sarcoma-like tumors.32
Numerous cytogenetic studies of OS have described a variety of genetic alterations resulting in inactivation of tumor suppressor genes and overexpression of oncogenes.1,2,15,16 In particular, many OS tumors harbor genetic lesions that inactivate Rb and/or p53 pathways.1,15,46 Conversely, several oncogenes, such as FOS, MYC, MET, SAS, GLI, MDM2, CDK4, and ERBB2, are reportedly overexpressed or activated in some OS.1,16 We and others have reported that Wnt signaling pathway is also deregulated in OS tumors.12,47,48 Currently, it is unclear how much these genetic and molecular changes contribute to the development of human OS.2,3 However, it is conceivable that some of the genetic and/or molecular changes in osteoprogenitors may disrupt osteogenic differentiation pathway, subsequently leading to the development of human OS tumors.
It has been reported that Rb functions as a direct transcriptional co-activator promoting osteoblast differentiation.49 Loss of pRb has been shown to block late osteoblast differentiation, and pRb physically interacts with Runx2, resulting in synergistic transactivation of an osteoblast-specific reporter.49 It has been reported that osteoblast differentiation is regulated by MDM2-p53 signaling pathway.50 Osteoprogenitor cells deleted for MDM2 have elevated p53 activity, reduced level of Runx2 and decreased osteogenic differentiation. Conversely, the p53-null osteoprogenitor cells have increased Runx2 expression, increased osteoblast maturation, and yet increased tumorigenic potential as mice specifically deleted p53 in osteoblasts develop OS tumors.50 Future investigations should be directed to identify the genetic and molecular alterations that may disrupt osteogenic differentiation.
In summary, our results suggest that in most OS cells the early stages of osteogenic differentiation may be preserved, but these OS lines may have defects in osteogenic terminal differentiation. As a result, osteogenic stimuli (such as BMPs) were unable to induce bone formation from OS cells, and rather promoted OS tumor growth because the OS cells were locked in the early proliferative phase of osteogenic pathway. We have shown that differentiation agents, such as PPARγ agonists and retinoid acid receptor ligands, are able to inhibit OS proliferation and induce osteogenic differentiation.3,40 Interestingly, BMPs and/or their receptors are readily expressed in OS cells.51–56 Thus, understanding the genetic and molecular underpinning of the differentiation defects in human OS tumors should allow us to reconstruct the tumorigenic events in osteoprogenitors and to develop rational differentiation therapeutics as an integral part of the clinical management of human OS.
MATERIALS AND METHODS
Cell Culture and Chemicals
Human OS lines MG63, TE85, U2OS, SaOS2, and 143B, as well as HEK293 and C3H10T1/2, were from ATCC. The hFOB1.19 was kindly provided by Dr Thomas C Spelsberg. Primary OS cells were isolated from resected OS specimens according to the approved by the Institutional Review Board. The cell lines were maintained in the conditions as described in Supplementary Methods.
Recombinant Adenoviruses Expressing GFP, BMPs, and Runx2
Adenoviruses expressing BMP2, BMP6, BMP9, and Runx2 were generated using the AdEasy technology as described.25–27,57,58 An analogous adenovirus expressing only GFP (AdGFP) was used as a control.57,59,60
ALP Assays
Alkaline phosphatase activity was assessed by colorimetric assay and/or histochemical staining assay as described.25–29,61
Establishment of OS Lines Stably Expressing Luciferase
MG63-Luc and 143B-Luc were generated by using a retroviral vector expressing firefly luciferase. Luciferase activity of the pooled stable cells was assessed by using Promega’s Luciferase Assay system (see Supplementary Methods).
Establishment of Id-Expressing Stable Lines
Retroviral viruses expressing mouse Id1, Id2, and Id3 were used to infect MG63-Luc cells under hygromycin selection.62 The resultant stable pools were designated as MG63-Id1, MG63-Id2, MG-Id3, and control MG63-RV. Overexpression of Id genes was verified by quantitative real-time PCR (qPCR; see Supplementary Methods).
RNA Isolation and Quantitative Real-Time PCR Analysis
Total RNA was isolated using Trizol Reagents (Invitrogen). qPCR was carried out as described.28,29,61 qPCR primers (Supplementary Table 1) were 18-mers, designed by using the Primer3 program to amplify the gene of interest (approximately 120 bp). All samples were normalized by the expression level of GAPDH (see Supplementary Methods).
Subcutaneous Implantation of MSCs and OS Cells
C3H10T1/2 and MG63 cells infected with AdBMP2, AdBMP9, or AdGFP for 16 h were collected for subcutaneous injection (5 × 106 cells per injection, 4 injections per group) into the flanks of athymic mice. At 6 weeks, animals were killed and the implantation sites were retrieved for histologic evaluation (see Supplementary Methods).
Intratibial Tumor Injection
MG63-Luc and 143B-Luc cells were infected with adenoviruses (BMPs, GFP, and/or Runx2). Cells were harvested, and resuspended in PBS to a final density of 2 × 107 cells per ml. Cells (1 × 106) in 50 μl of PBS were injected into the proximal tibiae of athymic mice as described.41 Animals injected with MG63-Luc were killed after 10 weeks, whereas 143B-Luc injected animals were killed after 6 weeks (see Supplementary Methods).
Xenogen Bioluminescence Imaging
Animal were anesthetized with isoflurane attached to a nosecone mask within Xenogen IVIS 200 imaging system. For bioluminescence imaging, animals were injected (i.p.) with D-Luciferin sodium salt (Gold BioTechnology) at 100 mg/kg in 0.1 ml sterile saline. The pseudoimages were obtained by superimposing the emitted light over the gray-scale photographs of the animal. Quantitative analysis was done with Xenogen’s Living Image V2.50.1 software (see Supplementary Methods).
Histological Evaluation
Retrieved tissues were fixed in 10% formalin overnight (decalcified if necessary) and embedded in paraffin. Serial sections of the embedded specimens were stained with hematoxylin and eosin, and other staining.63
Cell-Cycle Analysis
Cells were infected with GFP or BMP9 adenovirus. At indicated time points, cells were collected, fixed, and then incubated with the propidium iodide (PI)/RNase staining buffer (BD Biosciences Pharmingen), followed by flow cytometry assay (see Supplementary Methods).
Supplementary Material
ACKNOWLEDGEMENTS
We are grateful for Dr Michael Simon and Dr Terrance Peabody of The University of Chicago Medical Center for their encouragement of this work and assistance on collecting human OS samples. We thank Dr Thomas C Spelsberg of Mayo Clinic College of Medicine, Rochester, MN, for generous provision of the hFOB1.19 osteoblast cell line. This work was supported in part by research grants from China Natural Science Foundation (JL, ZLD, and QK), the China Ministry of Science and Technology (JL, ZLD, BCH, QK, LC, and TCH), the American Cancer Society (TCH and HHL). The Brinson Foundation (TCH), the National Institutes of Health (RCH, TCH and HHL), and the Orthopaedic Research and Education Foundation (HHL). The Xenogen bioluminescence imaging performed at the Optical Imaging Core Facility was supported by a pilot grant from The University of Chicago Cancer Research Center.
Footnotes
Supplementary Information accompanies the paper on the Laboratory Investigation website (http://www.laboratoryinvestigation.org)
References
- 1.Sandberg AA, Bridge JA. Updates on the cytogenetics and molecular genetics of bone and soft tissue tumors: osteosarcoma and related tumors. Cancer Genet Cytogenet 2003;145:1–30. [PubMed] [Google Scholar]
- 2.Helman LJ, Meltzer P. Mechanisms of sarcoma development. Nat Rev Cancer 2003;3:685–694. [DOI] [PubMed] [Google Scholar]
- 3.Haydon RC, Luu HH, He TC. Osteosarcoma and osteoblastic differentiation: a new perspective on oncogenesis. Clin Orthop Relat Res 2007;454:237–246. [DOI] [PubMed] [Google Scholar]
- 4.Unni KK, Dahlin DC. Osteosarcoma: pathology and classification. Semin Roentgenol 1989;24:143–152. [DOI] [PubMed] [Google Scholar]
- 5.Bruland OS, Pihl A. On the current management of osteosarcoma. A critical evaluation and a proposal for a modified treatment strategy. Eur J Cancer 1997;33:1725–1731. [DOI] [PubMed] [Google Scholar]
- 6.Meyers PA, Gorlick R. Osteosarcoma. Pediatr Clin North Am 1997;44:973–989. [DOI] [PubMed] [Google Scholar]
- 7.Dorfman HD, Weiss SW. Borderline osteoblastic tumors: problems in the differential diagnosis of aggressive osteoblastoma and low-grade osteosarcoma. Semin Diagn Pathol 1984;1:215–234. [PubMed] [Google Scholar]
- 8.Keel SB, Jaffe KA, Petur Nielsen G, et al. Orthopaedic implant-related sarcoma: a study of twelve cases. Mod Pathol 2001;14:969–977. [DOI] [PubMed] [Google Scholar]
- 9.Mark RJ, Poen J, Tran LM, et al. Postirradiation sarcomas. A single-institution study and review of the literature. Cancer 1994;73: 2653–2662. [DOI] [PubMed] [Google Scholar]
- 10.Draper GJ, Sanders BM, Kingston JE. Second primary neoplasms in patients with retinoblastoma. Br J Cancer 1986;53:661–671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Arndt CA, Crist WM. Common musculoskeletal tumors of childhood and adolescence. N Engl J Med 1999;341:342–352. [DOI] [PubMed] [Google Scholar]
- 12.Haydon RC, Deyrup A, Ishikawa A, et al. Cytoplasmic and/or nuclear accumulation of the beta-catenin protein is a frequent event in human osteosarcoma. Int J Cancer 2002;102:338–342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Luu HH, Zhang R, Haydon RC, et al. Wnt/beta-catenin signaling pathway as a novel cancer drug target. Curr Cancer Drug Targets 2004;4:653–671. [DOI] [PubMed] [Google Scholar]
- 14.Luo J, Chen J, Deng ZL, et al. Wnt signaling and human diseases: what are the therapeutic implications? Lab Invest 2007;87:97–103. [DOI] [PubMed] [Google Scholar]
- 15.Horowitz JM, Park SH, Bogenmann E, et al. Frequent inactivation of the retinoblastoma anti-oncogene is restricted to a subset of human tumor cells. Proc Natl Acad Sci USA 1990;87:2775–2779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gorlick R, Anderson P, Andrulis I, et al. Biology of childhood osteogenic sarcoma and potential targets for therapeutic development: meeting summary. Clin Cancer Res 2003;9:5442–5453. [PubMed] [Google Scholar]
- 17.Aubin JE. Regulation of osteoblast formation and function. Rev Endocr Metab Disord 2001;2:81–94. [DOI] [PubMed] [Google Scholar]
- 18.Ducy P, Schinke T, Karsenty G. The osteoblast: a sophisticated fibroblast under central surveillance. Science 2000;289:1501–1504. [DOI] [PubMed] [Google Scholar]
- 19.He TC. Distinct osteogenic activity of BMPs and their orthopaedic applications. J Musculoskelet Neuronal Interact 2005;5:363–366. [PubMed] [Google Scholar]
- 20.Luu HH, Song WX, Luo X, et al. Distinct roles of bone morphogenetic proteins in osteogenic differentiation of mesenchymal stem cells. J Orthop Res 2007;25:665–677. [DOI] [PubMed] [Google Scholar]
- 21.Deng ZL, Sharff KA, Tang N, et al. Regulation of osteogenic differentiation during skeletal development. Front Biosci 2008;13:2001–2021. [DOI] [PubMed] [Google Scholar]
- 22.Urist MR. Bone: formation by autoinduction. Science 1965;150:893–899. [DOI] [PubMed] [Google Scholar]
- 23.Wozney JM, Rosen V, Celeste AJ, et al. Novel regulators of bone formation: molecular clones and activities. Science 1988;242: 1528–1534. [DOI] [PubMed] [Google Scholar]
- 24.Luo J, Sun MH, Kang Q, et al. Gene therapy for bone regeneration. Curr Gene Ther 2005;5:167–179. [DOI] [PubMed] [Google Scholar]
- 25.Cheng H, Jiang W, Phillips FM, et al. Osteogenic activity of the fourteen types of human bone morphogenetic proteins (BMPs). J Bone Joint Surg Am 2003;85-A:1544–1552. [DOI] [PubMed]
- 26.Kang Q, Sun MH, Cheng H, et al. Characterization of the distinct orthotopic bone-forming activity of 14 BMPs using recombinant adenovirus-mediated gene delivery. Gene Therapy 2004;11:1312–1320. [DOI] [PubMed] [Google Scholar]
- 27.Peng Y, Kang Q, Cheng H, et al. Transcriptional characterization of bone morphogenetic proteins (BMPs)-mediated osteogenic signaling. J Cell Biochem 2003;90:1149–1165. [DOI] [PubMed] [Google Scholar]
- 28.Peng Y, Kang Q, Luo Q, et al. Inhibitor of DNA binding/differentiation helix-loop-helix proteins mediate bone morphogenetic protein-induced osteoblast differentiation of mesenchymal stem cells. J Biol Chem 2004;279:32941–32949. [DOI] [PubMed] [Google Scholar]
- 29.Luo Q, Kang Q, Si W, et al. Connective tissue growth factor (CTGF) is regulated by Wnt and bone morphogenetic proteins signaling in osteoblast differentiation of mesenchymal stem cells. J Biol Chem 2004;279:55958–55968. [DOI] [PubMed] [Google Scholar]
- 30.Reya T, Morrison SJ, Clarke MF, et al. Stem cells, cancer, and cancer stem cells. Nature 2001;414:105–111. [DOI] [PubMed] [Google Scholar]
- 31.Tirode F, Laud-Duval K, Prieur A, et al. Mesenchymal stem cell features of Ewing tumors. Cancer Cell 2007;11:421–429. [DOI] [PubMed] [Google Scholar]
- 32.Castillero-Trejo Y, Eliazer S, Xiang L, et al. Expression of the EWS/FLI-1 oncogene in murine primary bone-derived cells Results in EWS/FLI-1-dependent, ewing sarcoma-like tumors. Cancer Res 2005;65:8698–8705. [DOI] [PubMed] [Google Scholar]
- 33.Nozaki K, Kadosawa T, Nishimura R, et al. 1,25-Dihydroxyvitamin D3, recombinant human transforming growth factor-beta 1, and recombinant human bone morphogenetic protein-2 induce in vitro differentiation of canine osteosarcoma cells. J Vet Med Sci 1999;61:649–656. [DOI] [PubMed] [Google Scholar]
- 34.Hong SH, Kadosawa T, Nozaki K, et al. In vitro retinoid-induced growth inhibition and morphologic differentiation of canine osteosarcoma cells. Am J Vet Res 2000;61:69–73. [DOI] [PubMed] [Google Scholar]
- 35.Zenmyo M, Komiya S, Hamada T, et al. Transcriptional activation of p21 by vitamin D(3) or vitamin K(2) leads to differentiation of p53-deficient MG-63 osteosarcoma cells. Hum Pathol 2001;32:410–416. [DOI] [PubMed] [Google Scholar]
- 36.Carpio L, Gladu J, Goltzman D, et al. Induction of osteoblast differentiation indexes by PTHrP in MG-63 cells involves multiple signaling pathways. Am J Physiol Endocrinol Metab 2001;281:E489–E499. [DOI] [PubMed] [Google Scholar]
- 37.Siggelkow H, Schenck M, Rohde M, et al. Prolonged culture of HOS 58 human osteosarcoma cells with 1,25-(OH)2-D3, TGF-beta, and dexamethasone reveals physiological regulation of alkaline phosphatase, dissociated osteocalcin gene expression, and protein synthesis and lack of mineralization. J Cell Biochem 2002;85:279–294. [DOI] [PubMed] [Google Scholar]
- 38.Postiglione L, Domenico GD, Montagnani S, et al. Granulocyte-macrophage colony-stimulating factor (GM-CSF) induces the osteoblastic differentiation of the human osteosarcoma cell line SaOS-2. Calcif Tissue Int 2003;72:85–97. [DOI] [PubMed] [Google Scholar]
- 39.Thomas DM, Johnson SA, Sims NA, et al. Terminal osteoblast differentiation, mediated by runx2 and p27KIP1, is disrupted in osteosarcoma. J Cell Biol 2004;167:925–934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Haydon RC, Zhou L, Feng T, et al. Nuclear receptor agonists as potential differentiation therapy agents for human osteosarcoma. Clin Cancer Res 2002;8:1288–1294. [PMC free article] [PubMed] [Google Scholar]
- 41.Luu HH, Kang Q, Park JK, et al. An orthotopic model of human osteosarcoma growth and spontaneous pulmonary metastasis. Clin Exp Metastasis 2005;22:319–329. [DOI] [PubMed] [Google Scholar]
- 42.Harris SA, Enger RJ, Riggs BL, et al. Development and characterization of a conditionally immortalized human fetal osteoblastic cell line. J Bone Miner Res 1995;10:178–186. [DOI] [PubMed] [Google Scholar]
- 43.Caplan AI, Bruder SP. Mesenchymal stem cells: building blocks for molecular medicine in the 21st century. Trends Mol Med 2001;7:259–264. [DOI] [PubMed] [Google Scholar]
- 44.Tseng Y-H, He T-C. Bone morphogenetic proteins and adipocyte differentiation. Cell Sci Rev 2007;3:324–360. [Google Scholar]
- 45.Torchia EC, Jaishankar S, Baker SJ. Ewing tumor fusion proteins block the differentiation of pluripotent marrow stromal cells. Cancer Res 2003;63:3464–3468. [PubMed] [Google Scholar]
- 46.Abramson DH, Ellsworth RM, Kitchin FD, et al. Second nonocular tumors in retinoblastoma survivors. Are they radiation-induced? Ophthalmology 1984;91:1351–1355. [DOI] [PubMed] [Google Scholar]
- 47.Zhou L, An N, Haydon RC, et al. Tyrosine kinase inhibitor STI-571/Gleevec down-regulates the beta-catenin signaling activity. Cancer Lett 2003;193:161–170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hoang BH, Kubo T, Healey JH, et al. Dickkopf 3 inhibits invasion and motility of Saos-2 osteosarcoma cells by modulating the Wnt-beta-catenin pathway. Cancer Res 2004;64:2734–2739. [DOI] [PubMed] [Google Scholar]
- 49.Thomas DM, Carty SA, Piscopo DM, et al. The retinoblastoma protein acts as a transcriptional coactivator required for osteogenic differentiation. Mol Cell 2001;8:303–316. [DOI] [PubMed] [Google Scholar]
- 50.Lengner CJ, Steinman HA, Gagnon J, et al. Osteoblast differentiation and skeletal development are regulated by Mdm2-p53 signaling. J Cell Biol 2006;172:909–921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yoshikawa H, Takaoka K, Masuhara K, et al. Prognostic significance of bone morphogenetic activity in osteosarcoma tissue. Cancer 1988;61:569–573. [DOI] [PubMed] [Google Scholar]
- 52.Yoshikawa H, Rettig WJ, Lane JM, et al. Immunohistochemical detection of bone morphogenetic proteins in bone and soft-tissue sarcomas. Cancer 1994;74:842–847. [DOI] [PubMed] [Google Scholar]
- 53.Guo W, Gorlick R, Ladanyi M, et al. Expression of bone morphogenetic proteins and receptors in sarcomas. Clin Orthop Relat Res 1999;365:175–183. [DOI] [PubMed] [Google Scholar]
- 54.Gobbi G, Sangiorgi L, Lenzi L, et al. Seven BMPs and all their receptors are simultaneously expressed in osteosarcoma cells. Int J Oncol 2002;20:143–147. [PubMed] [Google Scholar]
- 55.Sulzbacher I, Birner P, Trieb K, et al. The expression of bone morphogenetic proteins in osteosarcoma and its relevance as a prognostic parameter. J Clin Pathol 2002;55:381–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yoshikawa H, Nakase T, Myoui A, et al. Bone morphogenetic proteins in bone tumors. J Orthop Sci 2004;9:334–340. [DOI] [PubMed] [Google Scholar]
- 57.He TC, Zhou S, da Costa LT, et al. A simplified system for generating recombinant adenoviruses. Proc Natl Acad Sci USA 1998;95: 2509–2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Luo J, Deng ZL, Luo X, et al. A protocol for rapid generation of recombinant adenoviruses using the AdEasy system. Nat Protoc 2007;2:1236–1247. [DOI] [PubMed] [Google Scholar]
- 59.He TC, Sparks AB, Rago C, et al. Identification of c-MYC as a target of the APC pathway. Science 1998;281:1509–1512. [DOI] [PubMed] [Google Scholar]
- 60.He TC, Chan TA, Vogelstein B, et al. PPARdelta is an APC-regulated target of nonsteroidal anti-inflammatory drugs. Cell 1999;99:335–345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Si W, Kang Q, Luu HH, et al. CCN1/Cyr61 is regulated by the canonical Wnt signal and plays an important role in Wnt3A-induced osteoblast differentiation of mesenchymal stem cells. Mol Cell Biol 2006;26:2955–2964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Luo Q, Kang Q, Song WX, et al. Selection and validation of optimal siRNA target sites for RNAi-mediated gene silencing. Gene 2007;395:160–169. [DOI] [PubMed] [Google Scholar]
- 63.Luu HH, Zhou L, Haydon RC, et al. Increased expression of S100A6 is associated with decreased metastasis and inhibition of cell migration and anchorage independent growth in human osteosarcoma. Cancer Lett 2005;229:135–148. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.