Abstract
This manuscript reports Y-chromosomal short tandem repeat (Y-STR) haplotypes for 1032 male U.S. population samples across 30 Y-STR loci characterized by three capillary electrophoresis (CE) length-based kits (PowerPlex Y23 System, Yfiler Plus PCR Amplification Kit, and Investigator Argus Y-28 QS Kit) and one sequence-based kit (ForenSeq DNA Signature Prep Kit): DYF387S1, DYS19, DYS385 a/b, DYS389I, DYS389II, DYS390, DYS391, DYS392, DYS393, DYS437, DYS438, DYS439, DYS448, DYS449, DYS456, DYS458, DYS460, DYS481, DYS505, DYS518, DYS522, DYS533, DYS549, DYS570, DYS576, DYS612, DYS627, DYS635, DYS643, and Y-GATA-H4. The length-based Y-STR haplotypes include six loci that are not reported in the sequence-based kit (DYS393, DYS449, DYS456, DYS458, DYS518, and DYS627), whereas three loci included in the sequence-based kit are not present in length-based kits (DYS505, DYS522, and DYS612). For the latter, a custom multiplex was used to generate CE length-based data, allowing 1032 samples to be evaluated for concordance across the 30 Y-STR loci included in these four commercial Y-STR typing kits. Discordances between typing methods were analyzed further to assess underlying causes such as primer binding site mutations and flanking region insertions/deletions. Allele-level frequency and statistical information is provided for sequenced loci, excluding the multi-copy loci DYF387S1 and DYS385 a/b, for which locus-specific haplotype-level frequencies are provided instead. The resulting data reveals the degree of information gained through sequencing: 88% of sequenced Y-STR loci contain additional sequence-based alleles compared to length-based data, with the DYS389II locus containing the most additional alleles (51) observed by sequencing. Despite these allelic increases, only minimal improvement was observed in haplotype resolution by sequence, with all four commercial kits providing a similar ability to differentiate length-based haplotypes in this sample set. Finally, a subset of 369 male samples were compared to their corresponding additionally sequenced father samples, revealing the sequence basis for the 50 length-based changes observed, and no additional sequence-based mutations. GenBank accession numbers are reported for each unique sequence, and associated records are available in the STRSeq Y-Chromosomal STR Loci National Center for Biotechnology Information (NCBI) BioProject, accession PRJNA380347. Haplotype data is updated in the Y-STR Haplotype Reference Database (YHRD) for the ‘NIST 1032’ data set to now achieve the level of maximal haplotype of YHRD. All supplementary files including revisions to previously published Y-STR data are available in the NIST Public Data Repository: U.S. population data for human identification markers, DOI 10.18434/t4/1500024.
Keywords: Short tandem repeat, Y-STR typing, DNA sequencing, NGS, MPS, Haplotype frequency
1. Introduction
Short Tandem Repeat (STR) markers located on the Y chromosome are useful in forensic mixtures comprised of higher levels of female DNA and lower levels of male DNA, as well as kinship cases involving the patrilineage [1]. Y-STR kits have evolved in a similar fashion as autosomal STR kits, with more loci being added corresponding to advances in capillary electrophoresis (CE) technology, primarily increased resolution of fluorescent dyes and polymerase chain reaction (PCR) fragment distribution range. Early Y-STR kits released between 2003 and 2009 included from 12 to 16 loci: PowerPlex Y12 System, AmpFLSTR Yfiler PCR Amplification Kit, and Investigator Argus Y-12 (note: herein, indistinguishable multi-copy loci are counted once, although this varies in manufacturer product literature). In recent years, expanded versions of these three kits have been introduced: the 22-locus PowerPlex Y23 System released in 2012, the 25-locus Yfiler Plus PCR Amplification Kit released in 2014, and the 26-locus Investigator Argus Y-28 QS Kit released in 2021. The additional loci in these newer kits include Rapidly Mutating Y-STRs (RM Y-STRs), which can be useful in distinguishing close male relatives [2]. In 2015, the ForenSeq DNA Signature Prep Kit was released, which allows for simultaneous sequencing of 24 Y-STR loci along with autosomal STR, X-STR, and single nucleotide polymorphism (SNP) loci. Sequencing Y-STR loci reveals a wider range of allelic variation than can be determined by length-based fragment analysis with CE, thus potentially discriminating between identical length-based haplotypes. See Table 1. for kit manufacturer information and abbreviated kit names used throughout this manuscript.
Table 1.
Y-STR commercial kits included in this study.
| Kit Name | Kit Abbreviation | Vendor |
|---|---|---|
| Investigator Argus Y-12 QS Kit | ArgusY12 | Qiagen, Hilden, Germany |
| Investigator Argus Y-28 QS Kit | ArgusY28 | Qiagen, Hilden, Germany |
| PowerPlex Y12 System | PPY12 | Promega Corp., Madison, WI |
| PowerPlex Y23 System | PPY23 | Promega Corp., Madison, WI |
| Yfiler PCR Amplification Kit | YF | Thermo Fisher, Waltham, MA |
| Yfiler Plus PCR Amplification Kit | YFP | Thermo Fisher, Waltham, MA |
| ForenSeq DNA Signature Prep Kit | ForenSeq | Verogen, San Diego, CA |
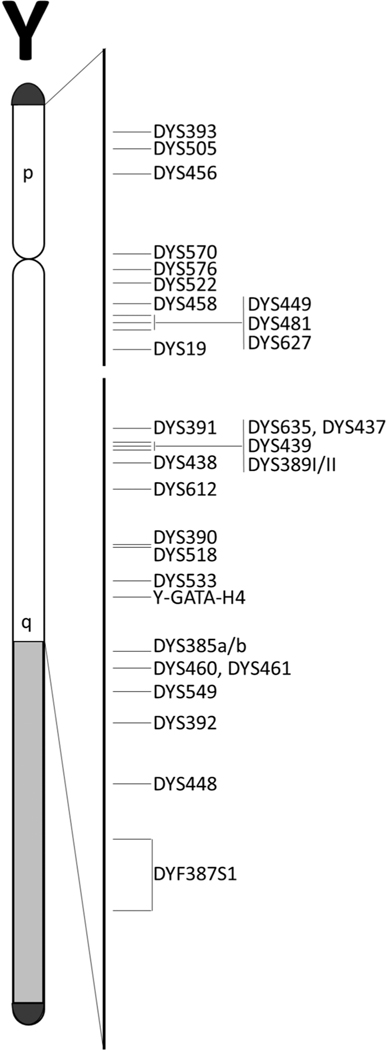
The National Institute of Standards and Technology (NIST) U.S. population sample set consists of 1032 unrelated male individuals (herein referred to as ‘NIST 1032’) with four population groups represented: African American (n = 341), Asian (n = 96), Caucasian (n = 359), and Hispanic (n = 236). Historically, these samples have been genotyped to provide allele frequencies and concordance checks during the development of many STR kits. This manuscript reports the results of an evaluation of ‘NIST 1032’ with the three currently available commercial Y-STR CE kits: PPY23 (previously reported in [3]), YFP and ArgusY28 (data not previously published), and results of ForenSeq Y-STR sequencing (autosomal and X-STR results previously published [4–6]). Haplotypes are reported for this sample set across the 30 Y-STR loci reported from these four kits: DYF387S1, DYS19, DYS385 a/b, DYS389I, DYS389II, DYS390, DYS391, DYS392, DYS393, DYS437, DYS438, DYS439, DYS448, DYS449, DYS456, DYS458, DYS460, DYS481, DYS505, DYS518, DYS522, DYS533, DYS549, DYS570, DYS576, DYS612, DYS627, DYS635, DYS643, and Y-GATA-H4. Table 2. illustrates the Y-STR markers that belong to each of the four commercial kits and includes several characteristics of each marker. Fig. 1. is a schematic of the Y chromosome and the relative positions of each marker in this study determined by their position in the GRCh38 human reference genome.
Table 2.
Y-STR markers analyzed with the ’NIST 1032’ sample set.
| Y-STR Marker | ArgusY28 | PPY23 | YFP | ForenSeq | Position (Mb) GRCh38(Dec 2013) | # Observed Alleles by Length | # Observed Alleles by Sequence | Allele Ranges | Repeat Motif (forward strand)* |
|---|---|---|---|---|---|---|---|---|---|
| DYF387S1 | ✓ | ✓ | 23.8/25.9 | 14 | 61 | 32–43 | [AAAG]n GTAG [GAAG]n [AAAG]n GAAG [AAAG]n [GAAG]n [AAAG]n | ||
| DYS19 | ✓ | ✓ | ✓ | ✓ | 9.7 | 8 | 9 | 12–19 | [TCTA]n ccta [TCTA]n |
| DYS385 a/b | ✓ | ✓ | ✓ | ✓ | 18.6 | 17 | 40 | 8–22 | [TTTC]n [cctt]n |
| DYS389 I | ✓ | ✓ | ✓ | ✓ | 12.5 | 6 | 7 | 9–15 | [TAGA]n [CAGA]n |
| DYS389 II | ✓ | ✓ | ✓ | ✓ | 12.5 | 9 | 60 | 25–34 | [TAGA]n [CAGA]n N48 [TAGA]n [CAGA]n |
| DYS390 | ✓ | ✓ | ✓ | ✓ | 15.2 | 8 | 29 | 20–27 | [TAGA]n CAGA [TAGA]n [CAGA]n |
| DYS391 | ✓ | ✓ | ✓ | ✓ | 11.9 | 7 | 8 | 7–13 | [TCTA]n |
| DYS392 | ✓ | ✓ | ✓ | ✓ | 20.4 | 10 | 11 | 7–16 | [ATA]n |
| DYS393a | ✓ | ✓ | ✓ | 3.3 | 6 | N/A | 11 −16 | [AGAT]n | |
| DYS437 | ✓ | ✓ | ✓ | ✓ | 12.3 | 6 | 20 | 13 −18 | [TCTA]n [TCTG]n [TCTA]n |
| DYS438 | ✓ | ✓ | ✓ | ✓ | 12.8 | 7 | 12 | 8–14 | [TTTTC]n |
| DYS439 | ✓ | ✓ | ✓ | ✓ | 12.4 | 7 | 7 | 9–15 | [GATA]n |
| DYS448 | ✓ | ✓ | ✓ | ✓ | 22.2 | 11 | 28 | 14–23 | [AGAGAT]n N42 [AGAGAT]n |
| DYS449a | ✓ | ✓ | 8.3 | 15 | N/A | 24–37 | [TTCT]n N22 [TTCT]n N12 [TTCT]n | ||
| DYS456b | ✓ | ✓ | ✓ | ✓b | 4.4 | 8 | 9 | 12 −19 | [AGAT]n |
| DYS458a | ✓ | ✓ | ✓ | 7.9 | 15 | N/A | 12–21.2 | [GAAA]n | |
| DYS460 | ✓ | ✓ | ✓ | 18.9 | 6 | 9 | 7–13 | [CTAT]n | |
| DYS461b | ✓b | 18.9 | 7 | 7 | 9–15 | TCTG [TCTA]n | |||
| DYS481 | ✓ | ✓ | ✓ | ✓ | 8.6 | 14 | 30 | 18–31 | [CTT]n |
| DYS505 | ✓ | 3.8 | 9 | 9 | 8–16 | [TCCT]n | |||
| DYS518a | ✓ | ✓ | 15.2 | 17 | N/A | 33–46 | [AAAG]n GAAG [AAAG]n GGAG [AAAG]n gaagag [AAAG]n N27 [AAGG]n | ||
| DYS522 | ✓ | 7.5 | 7 | 10 | 8–14 | [ATAG]n | |||
| DYS533 | ✓ | ✓ | ✓ | ✓ | 16.3 | 6 | 8 | 9–14 | [TATC]n |
| DYS549 | ✓ | ✓ | ✓ | 19.4 | 7 | 9 | 9–15 | [GATA]n | |
| DYS570 | ✓ | ✓ | ✓ | ✓ | 6.9 | 12 | 21 | 12–23 | [TTTC]n |
| DYS576 | ✓ | ✓ | ✓ | ✓ | 7.2 | 9 | 12 | 13–21 | [AAAG]n |
| DYS612 | ✓ | 13.6 | 15 | 37 | 25–41 | [CCT]n CTT [TCT]n CCT [TCT]n | |||
| DYS627a | ✓ | ✓ | 8.8 | 17 | N/A | 14–26 | [AGAA]n N16 [AGAG]n [AAAG]n N82 [AAGG]n | ||
| DYS635 | ✓ | ✓ | ✓ | ✓ | 12.3 | 11 | 28 | 17–27 | [TAGA]n [TACA]n [TAGA]n [TACA]n [TAGA]n [TACA]n [TAGA]n |
| DYS643 | ✓ | ✓ | ✓ | 15.3 | 10 | 11 | 7–15 | [CTTTT]n | |
| Y-GATA-H4 | ✓ | ✓ | ✓ | ✓ | 16.6 | 6 | 8 | 9–14 | [TCTA]n |
- marker present in kit.
- the repeat motifs listed follow the styling of the Forensic STR Sequence Structure Guide (https://strider.online/nomenclature).
Y-STR markers that are only present in length-based capillary electrophoresis (CE) commercial kits.
Y-STR markers that are amplified by the ForenSeq primer mix but not analyzed by the Universal Analysis Software (UAS).
Fig. 1.
Schematic representation of the Y chromosome with the loci analyzed in this study. The relative positions of each marker determined by their position in the GRCh38 human reference genome. The short and long arms of the chromosome are labeled ‘p’ and ‘q’, respectively. The heterochromatic region is represented by grey shading, and the pseudoautosomal regions (PAR) are represented by black caps.
Sequence-based alleles and haplotypes are reported for the 24 Y-STR loci in ForenSeq: DYF387S1, DYS19, DYS385a/b, DYS389I, DYS389II, DYS390, DYS391, DYS392, DYS437, DYS438, DYS439, DYS448, DYS460, DYS481, DYS505, DYS522, DYS533, DYS549, DYS570, DYS576, DYS612, DYS635, DYS643, and Y-GATA-H4. Two additional loci DYS456 and DYS461 are sequenced with ForenSeq but not reported by the associated Universal Analysis Software (UAS, Verogen); therefore, these loci were evaluated individually. The core repeat region as well as flanking region variation was assessed with a customized bioinformatic approach as described previously [6] and supplemented by additional analysis options [7,8]. In addition, the same factors that increase confidence in the NIST autosomal STR, X-STR, and SE33 data sets apply to the Y-STR sequence data described here: minimum coverage requirement, analysis with different bioinformatic pipelines, reporting of high-quality flanking sequence, comparison to CE length-based calls for every sample at every locus, and additional confirmation of null alleles and CE discordant results. The previously reported PPY23 CE data for the ‘NIST 1032’ [3] was compared with the YFP and ArgusY28 CE data generated for the ‘NIST 1032’ in this study, and the combined result was used for concordance comparisons to ForenSeq data. Three loci included in ForenSeq (DYS505, DYS522, and DYS612) are not present in CE-based kits; therefore, a custom multiplex was used to generate length-based calls.
Allele-level and haplotype-level frequencies and relevant forensic genetic parameters are determined for 22 single-copy sequence-based loci. Additionally, for the two multi-copy loci DYF387S1 and DYS385 a/b, copy number variation of the paralogs were detected (up to three distinct alleles) due to gene conversion and structural variations creating deletions and duplications. As this leads to a potentially imprecise determination of the number of alleles, for these two loci, locus-specific haplotype frequencies and a list of observed alleles are reported instead. Finally, a subset of 369 male samples were compared to their corresponding sequenced father samples, revealing the sequence basis for the 50 length-based changes observed, and no additional sequence-based mutations.
In addition to the supplementary files included herein, GenBank records have been submitted for sequences novel to the STRSeq Y-Chromosomal STR Loci NCBI BioProject [9], accession PRJNA380347 and updated haplotypes submitted to the Y-STR Haplotype Reference Database (YHRD) [10]. The information provided in this study will serve to facilitate the application of sequence-based methods to Y-STR profiling in the forensic setting. All supplementary files are available in the NIST Public Data Repository: U.S. population data for human identification markers, DOI 10.18434/t4/1500024.
2. Materials and methods
2.1. Population samples
Anonymous liquid blood samples with self-reported ancestries were purchased from Interstate Blood Bank (Memphis, TN) and Millennium Biotech, Inc. (Ft. Lauderdale, FL) or provided by DNA Diagnostics Center (Fairfield, OH) as buccal swabs from paternity testing samples anonymous to NIST. A total of 1032 unrelated male samples included in this study are a subset of samples previously reported in [11,12], and are divided among four U.S. population sample sets: African American (n = 341), Asian (n = 96), Caucasian (n = 359), and Hispanic (n = 236). The four female samples from the previously published ‘NIST 1036’ were removed: one African American, one Asian, and two Caucasian samples. Previously published Y-STR data [2,3,13,14] using this sample set was verified and revised as part of this study. All work presented in this paper has been reviewed and approved by the NIST Research Protections Office.
Within the ‘NIST 1032’ sample set, a subset of 369 son samples were compared to their corresponding additionally typed father samples. In instances when the son sample was unavailable or data was of insufficient quality, identical haplotypes were confirmed between the sample pairs, and sequence data from the corresponding father sample was used. Replacements were required due to depleted samples, allelic dropout, low fluorescence signal or low coverage of sequencing reads. More details of these replacements are provided in Section 3.1.1 of the Results.
2.2. Capillary electrophoresis testing
CE data was generated using three commercial kits that include PPY23, YFP, and ArgusY28 using manufacturers’ protocols for amplification and CE typing, with the exception of using a 15 s injection time for YFP. PPY23 sample testing was previously described [3]. Samples amplified with YFP and ArgusY28 were typed on a 24-capillary Applied Biosystems Prism 3500xL Genetic Analyzer and the data was analyzed using GeneMapper ID-X Software v1.6 (Thermo Fisher).
Three Y-STR markers which are not included in commercial STR typing CE kits were genotyped in a custom multiplex using previously published primers (DYS505 and DYS522 [13], and DYS612 [2]) and dye labels which are listed in Table S1a. PCR amplification and CE testing with these primers are described in [2,15,16]. Amplification products were diluted in Hi-Di formamide (Thermo Fisher) by adding 1 μL PCR product and 0.5 μL GS600-LIZ, v2.0 internal size standard (Thermo Fisher) to 9.5 μL of Hi-Di formamide. Samples were injected for 15 s at 1.2 kV on the 3500xL Genetic Analyzer without prior denaturation, then separated at 15 kV at a run temperature of 60 °C. Data from the 3500xL were analyzed using GeneMapper ID-X, v1.5 (Thermo Fisher). The bins and panels for the custom multiplex were set in GeneMapper for the Y-STR markers based on positive controls (SRM 2391c and 2800M from Promega) and published data [2].
2.3. Sequencing
2.3.1. Next generation sequencing
Y-STR sequence data was generated for ‘NIST 1032’ using ForenSeq on a MiSeq FGx instrument, as previously described in [4], optimized to obtain a minimum of 30 × sequence coverage per allele. Different parallel bioinformatic analyses were performed: FASTQ files were downloaded and analyzed with custom pipelines using STRait Razor v3.0 [7], methods described previously [6], and FDSTools v1.1.1 [8]. Both pipelines are discussed in more detail below. The Universal Analysis Software (UAS, Verogen) was used for data comparisons. The default output of the UAS is the Sample Level Report (SLR), which includes the repeat region of the loci and occasionally extends into the flanking regions. The UAS Flanking Region Report (FRR) is also available with additional flanking sequences, and this was used to determine the reported range of sequence in this study.
Off-platform analysis of the raw FASTQ files used default and custom anchor sets (see Table S2. for reported sequence ranges) to match or modify the UAS FRR range for analysis with STRait Razor v3.0. After using STRait Razor for all raw FASTQ files, the resulting sample files were run through a custom Perl script to identify the length- and sequenced-based allele calls [6]. For sequences to be considered further, a minimum depth of coverage (DoC) of 10 × was required. For each locus, the sequence with the highest DoC was reported as an allele. Additional sequences were reported as potential alleles if the allele coverage ratio (ACR, defined as the coverage of sequence under consideration divided by coverage of highest DoC sequence) was greater than 20%. Manual evaluation removed high stutter, recovered unbalanced alleles for expected multi-copy loci, and identified discordance that required closer evaluation.
Data analysis for the FDSTools pipeline started with initial quality control of the raw FASTQ files as described in [17], including removal of leftover adapters and sequence read-through, low quality base calls, and short reads resulting mainly from excess primer-dimers using Trimmomatic v0.39 [18]. Unique, unequal paired-end read structure of the output from the ForenSeq kit setup generates long first reads (R1s, 351 nucleotides) and very short second reads (R2s, 31 nucleotides). While other methods may omit the latter from analysis, here, to maximize the use of the short R2s, the quality-controlled files were merged using FLASH v1.2.11 [19]. Merging the high quality R2 improves the quality of base calls at the end of R1, creating a short, improved section of the sequence ideal for the placement of locus-specific identifying sequences (referred to as ‘flanks’ in FDSTools) for each of the targeted markers. The quality controlled and merged FASTQ files were used as input files for FDSTools v1.1.1, with library files containing adjusted ‘flanks’ placed to encompass the Y-STR loci amplified in the ForenSeq kit, together with the maximum length and optimal quality of flanking regions possible. Manual evaluation of the results used 30 × DoC for allele calls, and 20% ACR as a general limit for stutter cutoff, higher levels were manually removed, and imbalanced alleles restored. The output files include both sequence strings and bracketed sequences for the called alleles.
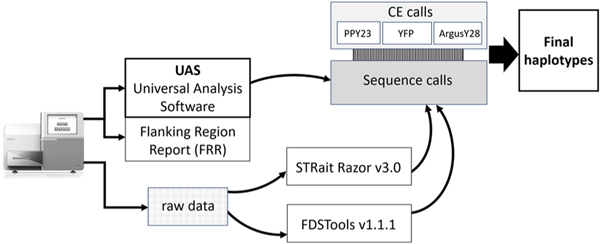
Length-based allele calls from each bioinformatic pipeline were compared to allele calls for the common loci in all three CE kits across all samples. Any differences observed within these evaluations were investigated and arbitrated with maximal information reported in the final data set. An overview of the data analysis pipeline is shown in Fig. 2.
Fig. 2.
Schematic representation of the multipronged data analysis workflow used in this study. On the left a MiSeq FGx represents data generation on this platform. CE kit abbreviations are the same as given in Table 1. Arrows show the progression through the processes to reach the final data set.
Manual review of artifacts which exceeded the pipelines’ DoC and ACR thresholds was performed in the same manner described for autosomal STR data analysis in the NIST Public data repository: U.S. population data for human identification markers, DOI 10.18434/t4/1500024 in the file ‘NIST1036_auSTR_Seq_SuppFile1.pdf’.
2.3.2. Sanger sequencing
Sanger sequencing of the DYF387S1 and DYS385 a/b loci was performed to identify SNPs and insertion/deletions (indels) in the flanking region. The unlabeled primers used for amplification are listed in Table S1b. along with corresponding annealing temperatures (°C). Amplification, purification, Big Dye Terminator v3.1 cycle sequencing (Thermo Fisher) and CE testing on a 3500 8-capillary Genetic Analyzer have all been previously described [20]. Sequences were analyzed using Geneious Prime v2021.1.1 (https://www.geneious.com) (Geneious, Auckland, New Zealand).
2.4. Frequencies and population statistics
A total of 26 Y-STRs are amplified by the ForenSeq kit; from these, 24 are standard loci with their analysis supported by the UAS software (hereafter referred to as the ‘UAS 24’) and two that are unreported by the software. For the UAS 24 loci, allele frequencies were determined using the counting method for the 22 single-copy Y-STRs, while a list of observed alleles and locus-specific haplotype frequencies were given instead for the two multi-copy loci DYF387S1 and DYS385 a/b. The two unreported loci, DYS456 and DYS461, were evaluated individually to determine the possibility of reporting frequency data.
Allele-level forensic statistic parameters were calculated using the webtool STRAF v1.0.5 [21] for the marker sets representing commercial kits from early-stage (PPY, YF, ArgusY12) to current versions (PPY23, YFP, ArgusY28), including the ForenSeq kit (length- and sequence-based calls) as well as the minimal haplotype (MiHT) [10,22]. Multi-copy loci DYF387S1 and DYS385 a/b were considered as locus-specific haplotypes, rather than individual alleles for these calculations. Samples with unexpected copy number variation (e.g., duplications or tri-allelic patterns) in the marker sets considered were omitted from the analysis. Allele-level forensic statistics include Gene Diversity (GD), Polymorphism Information Content (PIC), Match Probability (PM), and Power of Discrimination (PD).
Both allele-level frequencies and statistics were calculated for the ‘NIST 1032’ sample set, as well as for the four population sample sets. Multidimensional scaling (MDS) plot figures were produced from FST matrices using the STRAF output and analyzed in R v4.1.1 [23] with the MASS package [24]. Analysis of MOlecular VAriance (AMOVA) was performed using Arlequin v3.5.2.2 [25].
Haplotype frequencies and haplotype-level forensic parameters are provided for each marker set representing MiHT through early-stage commercial kits (PPY, YF, ArgusY12) to current complexity of commercial kits (PPY23, YFP, ArgusY28), including the ForenSeq sequencing kit (for both length-based and sequence-based calls). Haplotype-level forensic statistics include Haplotype frequency (h), Discrimination capacity (DC), Match probability (MP), Haplotype diversity (HD) and Discrimination potential (DP). The haplotype-level variation was visualized using the Alluvial graph of RawGraphs v2.0 [26].
2.5. Father-son mutations
Father-son pairs were evaluated for the presence of length and sequence-based mutations in the UAS 24 Y-STR loci. Detected mutations were categorized by type (e.g., one-step gain), and mutation rates per locus were calculated.
2.6. Copy number variations
The semi-quantitative method of the relative read-depth ratio test (RRT) [27] [pp. 64–65.] was used to estimate the dosage of allele calls when more than one expected allele was observed. These occurrences were estimated as true duplications or somatic mutations based on read-depth values measured in comparison to similar size loci amplified in the same reaction. This also included correction of DoC values with mean stutter levels relevant to the sequence alleles affected. In this study the father-son pairs were further utilized as confirmations of these two categories, as stable germline duplications are expected to be inherited while somatic mutations should be observed in one generation only.
3. Results
3.1. Quality
3.1.1. Sequence data
Sequencing metrics for the ForenSeq runs reported in this study that include cluster density, percent clusters passing filter, percent phasing and pre-phasing, mean coverage, standard deviation, and coefficient of variance are all previously published [4]. Length-based and sequence-based alleles are reported for the ‘NIST 1032’ at 25 Y-STR loci, which includes the UAS 24 and the unreported locus DYS456. The sequences reported from the UAS FRR and the UAS SLR are illustrated in Table S2. The repeat regions and unique SNPs are highlighted. The base location ranges are defined based on the GRCh38 human reference genome. In all cases except DYS522 the reported sequence is based on the UAS FRR ranges. The DYS522 amplicon has over 200 bp additional 5′ flanking sequence that is not reported in the UAS FRR sequence. Analysis was extended to include a 143 bp 5′ flanking region. This extended sequence facilitates the reporting of a four bp deletion detected in the ‘NIST 1032’ data set, which results in a repeat length change. The UAS FRR DYS19 and DYS392 sequences have no 3′ flanking sequence reported. Longer sequences are reported in the STRSeq records for these loci. No variation from the GRCh38 reference sequence was observed in the 3′ flanking regions for either DYS19 or DYS392.
To improve data quality, different approaches were used: replacement with father samples or alternative data analysis. Across the four kits, 52 father samples replaced son samples, with the majority of these replacements related to the sequencing set (37 samples at 41 allele calls across five loci: DYS385 a/b, DYF387S1, DYS392, DYS448 and DYS612). By population, this replacement affected 20 Asian, 13 African American, one Caucasian and three Hispanic samples. In one case (28H sample pair) the replacement also changed the allele call at the DYS481 locus, where a mutation was observed between the pair. In the alternative data analysis pipeline using FDSTools, an additional QC process was implemented to improve allele calls. This approach achieved more reliable allele calls at multi-copy loci, increasing the mean coverage by 60% (DYS385 a/b) and 40% (DYF387S1) on average across samples.
3.1.2. Sanger sequencing
Sanger sequencing was performed for multi-copy loci DYF387S1 (two samples) and DYS385 a/b (five samples) to further separate and characterize the alleles at these loci to identify the location of flanking region polymorphisms, following previously published methods [28, 29]. Sanger sequencing of the DYS385 ‘a’ and ‘b’ alleles was successful: three observed flanking region polymorphisms were oriented in the flanking region of either the ‘a’ fragment (rs2044536750 and ss5240854442) or the ‘b’ fragment (ss5240854441). The ‘a’ and ‘b’ alleles containing these polymorphisms are differentiated in the ‘Multi--copy loci’ tab of the Table S3., and identified separately in STRSeq DYS385 a/b BioProject PRJNA396120 (e.g., MZ905314.1 is a DYS385 ‘a’ allele with an 8 bp deletion in 3′ flanking region, whereas MZ905312.1 is a DYS385 ‘b’ allele with a SNP in the 5′ flanking region). For DYF387S1, a flanking SNP observed in two samples could not be oriented to a specific genomic location because the Sanger sequencing range did not reach distinguishable sequence regions. In this case, the SNP is indicated at two possible genomic locations (rs1603600992 and rs1603611917).
3.1.3. Capillary electrophoresis typing
3.1.3.1. Revisions to previously published Y-STR length-based data.
Comparing the length-based ForenSeq results to previously published Y-STR data for the ‘NIST 1032’ samples [2,3,14] resulted in detecting 22 erroneous allele calls. Table 3. lists the original and revised allele calls. In addition to these revisions, two data processing errors which affected a large number of samples were identified from allele calls reported in previous publications [13] and [2]. DYS505 and DYS522 data is revised here for n = 190 due to misidentification of samples in [13]. This data processing error also likely affected the data reported for DYS532 and DYS540 from [13]; these loci are not included in currently available Y-STR typing kits. At the DYS570 locus, it appears that a two repeat unit nomenclature shift was inconsistently applied by the study authors, affecting n = 629 of the NIST population sample set in [2]. Moving forward, the data from this manuscript supersedes the data from the previously published manuscripts [2,3,13,14,30] at the overlapping loci in these ‘NIST 1032’ samples.
Table 3.
Revisions to previously published Y-STR length-based data.
| Marker | Sample | Originala | Revised |
|---|---|---|---|
| DYS385 a/b | C12A | 19 | 12,19 |
| DYS385 a/b | C26A | 16 | 15,16 |
| DYS385 a/b | C34A | 12 | 12,20 |
| DYS385 a/b | C41A | 10 | 10,19 |
| DYS385 a/b | C42A | 12 | 12,18 |
| DYS385 a/b | C51H | 15 | 12,15 |
| DYS448 | C84A | 16 | del |
| Marker | Sample | Originalb | Revised |
|
| |||
| DYS392 | OT07280 | 11,12 | 11 |
| DYS448 | ZT80358 | 18.4 | 18.5 |
| DYS481 | C96H | 24 | 25 |
| DYS481 | C97H | 23 | 24 |
| DYS481 | C98A | 26 | 27 |
| DYS481 | MT94869 | 22,23 | 22 |
| DYS481 | MT97196 | 22,23 | 22 |
| DYS481 | WT51355 | 22,23 | 22 |
| Marker | Sample | Originalc | Revised |
|
| |||
| DYF387S1 | WA29594 | 36,38 | 35,38 |
| DYF387S1 | WT51556 | 38 | 38,39 |
| DYS449 | JT51467 | 35.2 | 36 |
| DYS518 | TT50700 | 42.1 | 42 |
| DYS570 | GT37913 | 16 | 19 |
| DYS627 | AF78B/C78B | 17.1 | 17.2 |
| DYS627 | MT95104 | 27 | 24 |
| DYS570 | n = 629* | various | various |
| Marker | Sample | Originald | Revised |
|
| |||
| DYS505 | n = 190** | various | various |
| DYS522 | n = 190** | various | various |
|
|
|
|
|
3.2. Concordance
A concordance evaluation was performed between each of the four commercial Y-STR kits that were tested (ArgusY28, PPY23, YFP, and ForenSeq). There were five differences observed between kits listed in Table 4.
Table 4.
Interkit differences from this study.
| Marker | Sample Pair | ArgusY28 |
PPY23 |
YFP |
ForenSeq |
||||
|---|---|---|---|---|---|---|---|---|---|
| son | father | son | father | son | father | son | father | ||
| DYS456 | 77A | null | - | null | null | 16 | 16 | 16.2 | 16.2 |
| DYS460 | 26A | null | - | N/A | N/A | 10 | - | 10 | 10 |
| DYS481 | 69H | - | 24.1 | 25 | - | - | 25 | 25 | 25 |
| DYS533 | 02C | - | 12 | 12 | - | - | 12.1 | 12 | 12 |
|
| |||||||||
| Marker | Sample | ArgusY28 | PPY23 | YFP | ForenSeq | ||||
|
| |||||||||
| DYS481 | GT37420 | 25 | 25.1 | 25 | 25 | ||||
N/A - locus not applicable to the kit.
‘-’ - untested sample.
3.2.1. CE commercial kit concordance
Across the CE kits reported here, 27 Y-STR markers are present within at least two of the three kits (ArgusY28, PPY23, and YFP) except for DYF387S1 which is only present in YFP. From Table 4., all five observed discordances show differences between CE kits. Information regarding the relative PCR primer positions in these commercial Y-STR kits is not available; therefore, potential causes for these differences are described.
There are two discordant results within the DYS481 locus. For one sample pair (69H), the consensus type across kits is an allele 25; however, it appears the ArgusY28 kit captures a 2 bp deletion within the region amplified by the primer set, resulting in a 24.1 allele for this trinucleotide locus. For the other sample (GT37420), the consensus type at the DYS481 locus across kits is also a 25 allele; however, the PPY23 kit types this sample as a 25.1. It is likely the PPY23 amplicon includes a SNP (rs368663163) which has been shown to cause an electrophoretic mobility shift [17,31,32], while the other CE kits do not amplify this flanking SNP. For the DYS533 locus, the consensus type is a 12 allele across the kits except for YFP which types it as a 12.1 allele. This suggests a 1 bp insertion located within the YFP amplicon but outside of the primers of ArgusY28 and PPY23 for this locus. While DYS460 is not present in PPY23, YFP types the affected son sample (26A) as a 10 allele, whereas ArgusY28 exhibits a null allele for this sample at this locus. Finally, there was one discordance at DYS456 in which the sample pair (77A) is typed as a 16 allele with YFP and fails to amplify (i.e., is a null allele) with both ArgusY28 and PPY23 (described further in section 3.2.2).
3.2.2. Sequence vs CE concordance
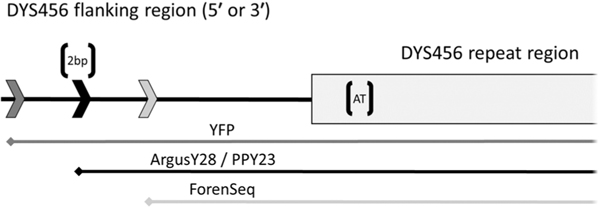
Concordance was also compared between length- and sequence-based allele calls within the loci that overlap between the three commercial CE kits and ForenSeq. There is no corresponding sequence data for five loci that are only included in length-based kits (DYS393, DYS449, DYS458, DYS518, and DYS627). For the three loci (DYS505, DYS522, and DYS612) present in ForenSeq that are not included in the three Y-STR CE kits, a custom multiplex was used to amplify and to confirm length-based concordance. There are no discordant results reported for these three loci. Sequence-based results confirmed the consensus allele calls for four of the five discordant CE results; however, sequence results differed from all CE kits for DYS456 with sample pair 77A (Table 4.). For this sample pair, there are three different results from the four kits compared: ArgusY28 and PPY23 both result in a null allele, YFP genotypes a 16 allele, and ForenSeq sequences a 16.2 allele by length. In the sequenced allele, a 2 bp indel was observed within the repeat region (AGAT AT [AGAT]15). Additionally, these results suggest that there is a 2 bp deletion in the flanking region (either 5′ or 3′) outside of the region defined by the ForenSeq primer, coinciding with the primers of ArgusY28 and PPY23 causing null alleles, while within the amplified region of the YFP kit, as illustrated in Fig. 3.
Fig. 3.
Schematic representation of a (5′ or 3′) flanking region of DYS456 is on the top of the graph. The gray box on the right is the DYS456 repeat region and the attached black line represents the flanking region. The three arrows on the left represent possible primer placements, and the three lines below represent the amplicons of the four kits. These approximate positions are inferred from the allele calls observed from the four commercial kits (Table 4.), suggesting the location of indels ‘[]’ in this sample pair. The ‘[AT]’ represents a sequenced indel inside the repeat region, while the ‘[2 bp]’ represents an expected 2 bp deletion amplified by YFP but not ForenSeq (resulting in 16 and 16.2 allele calls, respectively), and at the primer binding site of ArgusY28 and PPY23 (resulting in null alleles).
3.2.3. UAS vs CE concordance
The UAS data report was also compared to length-based CE data. There were six bioinformatic null alleles (five at DYS385 a/b and one at DYS448) where a CE comparison to UAS output could not be performed (note: the UAS does not report alleles at or below 10 ×). These six alleles were confirmed to be CE concordant using STRait Razor and FDSTools output. Some instances of stutter were observed above the default UAS thresholds. Specifically, for the DYS385 a/b locus, the UAS default stutter threshold of 20% resulted in stutter reported as potential alleles in over 10% of the samples. Additionally, when typing DYS612 with ForenSeq, the resulting allele call reported by the UAS is six repeat units less due to a difference in the nomenclature as described in [2].
3.3. Complete Y-STR data set
Once concordance was established between the four commercial Y-STR kits, the complete length-based allele calls and haplotype data set for the ‘NIST 1032’ for 30 Y-STRs (UAS 24 and one unreported ForenSeq locus plus five CE-only markers) are presented in Table S4. and Table S5. Discordant results were resolved using sequence-based allele calls. The generated haplotype data is accessible through YHRD (from Release 66) through accession numbers YA004438–2 (African American), YA004437–2 (European American), YA003315–3 (Hispanic American), and YA004439–2 (Asian American). This data replaces the previous YHRD submissions (YA004438–1, YA004437–1, YA003315–2, YA004439–1), and supplements them with additional marker data, providing the complete 27-locus YHRD maximal haplotype for all 1032 samples. The DYS505, DYS522 and DYS612 loci are not included in the YHRD database; therefore, the complete 30-locus Y-STR profiles for all 1032 samples are given in Table S4.
3.4. Statistics and population genetics
3.4.1. Allele-level frequencies and forensic statistics
Allele-level frequencies for 22 of the 26 Y-STR loci in ForenSeq are reported in Table S3. ‘Frequency tables’ tab. For the two multi-copy loci (DYF387S1 and DYS385 a/b) locus-specific haplotype frequencies are provided and observed alleles are listed in the ‘Multi-copy loci’ tab of Table S3. For the two UAS unreported loci (DYS456 and DYS461) the list of observed alleles is given in the ‘UAS unreported loci’ tab of Table S3., plus allele-level frequencies are provided for DYS456. Allele frequency data is not provided for the UAS unreported locus DYS461, because allele calls could not be obtained for 13% of samples. Allele-level forensic statistic parameters calculated by STRAF v1.0.5 [21] are summarized in Table S6.
3.4.2. Haplotype-level frequencies and forensic statistics
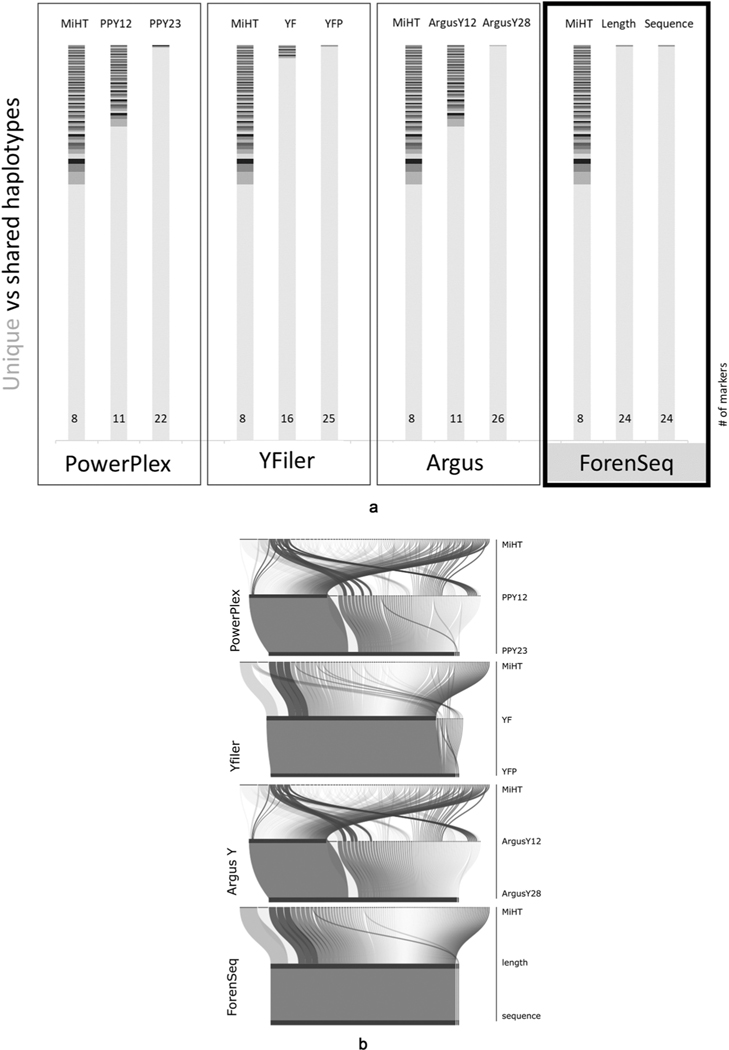
Haplotype frequencies and haplotype-level forensic parameters are provided in Table S5. for each marker set representing from MiHT through early-stage commercial kits (PPY, YF, ArgusY12) to current commercial kits (PPY23, YFP, ArgusY28), including the ForenSeq sequencing kit (for both length- and sequence-based calls). Haplotype resolution across the four commercial kits with progression from MiHT through the earlier versions to current kits are shown in Fig. 4a. and b. These demonstrate that more markers facilitate the increase of haplotype diversity, reaching almost unique haplotype resolution in the ‘NIST 1032’ samples across all current kits. Each kit achieved similar resolution with only three haplotype pairs (PPY23), two haplotype pairs (YFP and ForenSeq) and one haplotype pair (ArgusY28) remaining unresolved. The ForenSeq kit length-based and sequence-based calls resulted in the same two unresolved haplotype pairs.
Fig. 4.
Resolution of haplotypes of the ‘NIST 1032’ sample set through the evolution of commercial kits. Fig. 4a. shows the histograms of haplotypes for four different series of Y-STR kits, starting with MiHT for each series with progression through the earlier to current kit versions. In each histogram unique haplotypes in the set are shown in light grey, and shared haplotypes across samples are colored darker. Number of markers for each set are noted at the bottom of each histogram. The first three boxes of the figure show data for the CE kits, while the last box (with black border) depicts the haplotypes described by the ForenSeq marker set, based on derived length alleles in the middle and sequenced alleles on the right. Maximum haplotype resolution would show a single light grey bar, which was not achieved for this sample set by any of these kits. Fig. 4b. is an alluvial graph representing the break-down (bended connector lines) of the shared haplotypes of the MiHT (excluding the n = 668 haplotypes which were unique by the MiHT) by marker sets of commercial CE kits in three stages: starting from the shared haplotypes of the MiHT, followed by early versions of the kits, and the current commercial versions at the third horizontal line for each series/manufacturer. Below the three CE-based kits is the ForenSeq kit, starting from haplotypes of MiHT, then change via length to sequence-based haplotypes. The thickness of the connectors represents the number of samples within the shared haplotypes. The horizontal black bars represent unique haplotypes and remaining unresolved haplotype pairs in the current kits. Maximum haplotype resolution would show a single horizontal black bar, which was not achieved by any of these kits, leaving one to three unresolved pairs for this sample set.
3.4.2.1. Unresolved haplotype pairs.
Within marker sets for these four commercial kits, a total of five unresolved haplotype pairs remained; however, only one pair was indistinguishable across all four kits. These five pairs were checked with additional distinguishing Y-chromosomal markers from existing data. In a set of 884 Y-SNPs [33] these pairs cannot be further resolved; however, using data from additional RM Y-STRs [2] each of the five pairs can be separated into unique haplotypes, including the one pair which was previously indistinguishable across all four kits. These five pairs and their differentiating markers are summarized in Table S7. Additional in-house data addresses the question of possible relatedness within these pairs: an evaluation of 30 autosomal STR loci in this sample set using the blind search function of Familias software v3.2.8 [34] did not provide evidence of relatedness when tested out to the level of second cousin.
3.4.3. Population-level calculations
The four population sample sets are compared in the form of MDS plots visualizing genetic distances of the populations. These plots are generated from the pairwise FST matrices calculated by STRAF, based on the alleles of the UAS 24 Y-STR loci plus the unreported DYS456 locus of the ForenSeq kit. Graphs of MDS plots for each marker set representing commercial kits including the markers with both the length- and sequence-based alleles of the ForenSeq kit are provided in Fig. S1. AMOVA was also performed to measure the genetic variation between the four population sample sets using the genetic distance among haplotypes of each marker sets and the results are shown in Table S8.
3.5. Mutation rates
3.5.1. Father and son mutations
In total, 369 father-son pairs were evaluated for the UAS 24 Y-STR loci of the ForenSeq kit. The length- and sequence-based allele changes for each locus are illustrated in Table 5. Table S9. lists the number of meiotic transfers that were evaluated at each locus. Fifty changes were observed across 18 Y-STR loci for this data set and were also observed by CE. At two of these occurrences (at DYS439 and at DYS505) the father samples exhibit additional minor alleles, likely somatic mutations, which were not inherited by their sons. Forty-eight of these occurrences were mutations observed between father and son pairs, and six pairs had mutations at two distinct loci. Forty-five of the mutations are one-step, while two-step mutations are observed at DYS612 (two samples) and Y-GATA-H4 (one sample). Seven loci did not exhibit mutations between the pairs. Mutation rates calculated from this set, alongside published mutation rates, are included in Table S9.
Table 5.
Changes observed via ForenSeq sequencing of n = 369 father-son pairs.
| Locus | Father Alleles by Length and Sequence | Son Alleles by Length and Sequence (Mutation Bolded) | Mutation | ||
|---|---|---|---|---|---|
| DYS19 | 17 | [TCTA]14 CCTA [TCTA]3 |
18 | [TCTA]15 CCTA [TCTA]3 |
+ 1 |
|
|
|
|
|
|
|
| DYS385 a/b | 11,15 | [TTTC]11 [CCTT]6, [TTTC]15 [CCTT]6 |
12,15 | [TTTC]12 [CCTT]6, [TTTC]15 [CCTT]6 |
+1 |
|
|
|
|
|
|
|
| DYS389Ia | 14 | [TAGA]11 [CAGA]3 |
13 | [TAGA]10 [CAGA]3 |
−1 |
| 15 | [TAGA]12 [CAGA]3 |
14 | [TAGA]11 [CAGA]3 |
−1 | |
| 14 | [TAGA]11 [CAGA]3 |
15 | [TAGA]12 [CAGA]3 |
+1 | |
| 13 | [TAGA]10 [CAGA]3 |
14 | [TAGA]11 [CAGA]3 |
+1 | |
|
|
|
|
|
|
|
| DYS389II | 30 | [TAGA]9 [CAGA]3 N48 [TAGA]13 [CAGA]5 |
31 | [TAGA]9 [CAGA]3 N48 [TAGA]14 [CAGA]5 |
+1 |
|
|
|
|
|
|
|
| DYS390 | 24 | [TAGA]4 CAGA [TAGA]11 [CAGA]8 |
23 | [TAGA]4 CAGA [TAGA]10 [CAGA]8 |
−1 |
|
|
|
|
|
|
|
| DYS439 | 12, (13) | [GATA]12, ([GATA]13) | 12 | [GATA]12 | 0b |
| 13 | [GATA]13 | 12 | [GATA]12 | −1 | |
| 13 | [GATA]13 | 12 | [GATA]12 | −1 | |
| 13 | [GATA]13 | 12 | [GATA]12 | −1 | |
| 12 | [GATA]12 | 13 | [GATA]13 | +1 | |
| 14 | [GATA]14 | 15 | [GATA]15 | +1 | |
|
|
|
|
|
|
|
| DYS456 | 15 | [AGAT]15 | 16 | [AGAT]16 | +1 |
|
|
|
|
|
|
|
| DYS460 | 11 | [CTAT]11 | 10 | [CTAT]10 | −1 |
| 11 | [CTAT]11 | 10 | [CTAT]10 | −1 | |
| 12 | [CTAT]12 | 11 | [CTAT]11 | −1 | |
| 10 | [CTAT]10 | 11 | [CTAT]11 | +1 | |
|
|
|
|
|
|
|
| DYS481 | 24 | [CTT]24 | 25 | [CTT]25 | +1 |
| 25 | [CTT]25 | 26 | [CTT]26 | +1 | |
|
|
|
|
|
|
|
| DYS505 | 13, (14) | [TCCT]13, ([TCCT]14) | 13 | [TCCT]13 | 0b |
| 12 | [TCCT]12 | 11 | [TCCT]11 | −1 | |
| 14 | [TCCT]14 | 13 | [TCCT]13 | −1 | |
| 14 | [TCCT]14 | 13 | [TCCT]13 | −1 | |
|
|
|
|
|
|
|
| DYS522 | 12 | [ATAG]12 | 11 | [ATAG]11 | −1 |
|
|
|
|
|
|
|
| DYS549 | 13 | [GATA]13 | 12 | [GATA]12 | −1 |
| 13 | [GATA]13 | 14 | [GATA]14 | +1 | |
|
|
|
|
|
|
|
| DYS570c | 17 | [TTTC]17 | 16 | [TTTC]16 | −1 |
| 21 | [TTTC]21 | 20 | [TTTC]20 | −1 | |
| 17 | [TTTC]17 | 18 | [TTTC]18 | +1 | |
| 17 | [TTTC]17 | 18 | [TTTC]18 | +1 | |
| 17 | [TTTC]17 | 18 | [TTTC]18 | +1 | |
|
|
|
|
|
|
|
| DYS576c | 19 | [AAAG]19 | 18 | [AAAG]18 | −1 |
| 21 | [AAAG]21 | 20 | [AAAG]20 | −1 | |
| 16 | [AAAG]16 | 17 | [AAAG]17 | +1 | |
| 19 | [AAAG]19 | 20 | [AAAG]20 | +1 | |
|
|
|
|
|
|
|
| DYS612c | 34 | [CCT]5 CTT [TCT]4 CCT [TCT]23 |
32 | [CCT]5 CTT [TCT]4 CCT [TCT]21 |
−2 |
| 35 | [CCT]5 CTT [TCT]4 CCT [TCT]24 |
34 | [CCT]5 CTT [TCT]4 CCT [TCT]23 |
−1 | |
| 37 | [CCT]5 CTT [TCT]4 CCT [TCT]26 |
36 | [CCT]5 CTT [TCT]4 CCT [TCT]25 |
−1 | |
| 39 | [CCT]5 CTT [TCT]4 CCT [TCT]28 |
40 | [CCT]5 CTT [TCT]4 CCT [TCT]29 |
−1 | |
| 37 | [CCT]5 CTT [TCT]4 CCT [TCT]26 |
38 | [CCT]5 CTT [TCT]4 CCT [TCT]27 |
+1 | |
| 38 | [CCT]5 CTT [TCT]4 CCT [TCT]27 |
39 | [CCT]5 CTT [TCT]4 CCT [TCT]28 |
+1 | |
| 38 | [CCT]5 CTT [TCT]4 CCT [TCT]27 |
40 | [CCT]5 CTT [TCT]4 CCT [TCT]29 |
+2 | |
|
|
|
|
|
|
|
| DYS635 | 23 | [TAGA]13 [TACA]2 [TAGA]2 [TACA]2 [TAGA]4 |
22 | [TAGA]12 [TACA]2 [TAGA]2 [TACA]2 [TAGA]4 |
−1 |
| 22 | [TAGA]12 [TACA]2 [TAGA]2 [TACA]2 [TAGA]4 |
23 | [TAGA]13 [TACA]2 [TAGA]2 [TACA]2 [TAGA]4 |
+1 | |
|
|
|
|
|
|
|
| DYS643 | 14 | [CTTTT]14 | 15 | [CTTTT]15 | +1 |
|
|
|
|
|
|
|
| Y-GATA-H4 | 11 | [TCTA]11 | 9 | [TCTA]9 | −2 |
| 12 | [TCTA]12 | 11 | [TCTA]11 | −1 | |
| 12 | [TCTA]12 | 13 | [TCTA]13 | + 1 | |
mutations reported at locus DYS389I were also observed at the encompassing DYS389II locus.
fathers exhibit likely somatic mutations at these loci which are not inherited to the next generation.
RM Y-STRs.
3.5.2. Copy number variations
A total of 24 supernumerary alleles were observed instead of the expected single allele call across 11 Y-STR loci. The relative read-depth ratio test [27] [pp. 64–65] was used to estimate these occurrences as true duplications or somatic mutations. Fourteen of these are labeled as more likely to be duplications and ten of these are more likely to be somatic mutations. From the father-son pairs, four were present for each of these categories and confirmed the expected; for duplication the relevant fathers also exhibit the extra allele, while for somatic mutations the fathers only have the expected single allele call. These are distinguished in Table S10. Deletions generated six null alleles at five Y-STR loci, three of which are found in one sample (33B) at neighboring markers (DYS389I/II and DYS439). The other null alleles are at DYS19 (one sample) and DYS448 (two samples). There are 11 length-based tri-allelic combinations observed within three loci: DYF387S1 (nine samples), DYS448 (one sample) and DYS19 (one sample). There are an additional four sequence-based tri-allelic combinations which arose from isoalleles at DYF387S1. Alleles with copy number variations were removed from the allele frequency tables Table S3. ‘Allele frequencies’ tab.
3.6. Benefits observed from sequencing
3.6.1. Y-STR allelic and gene diversity increase
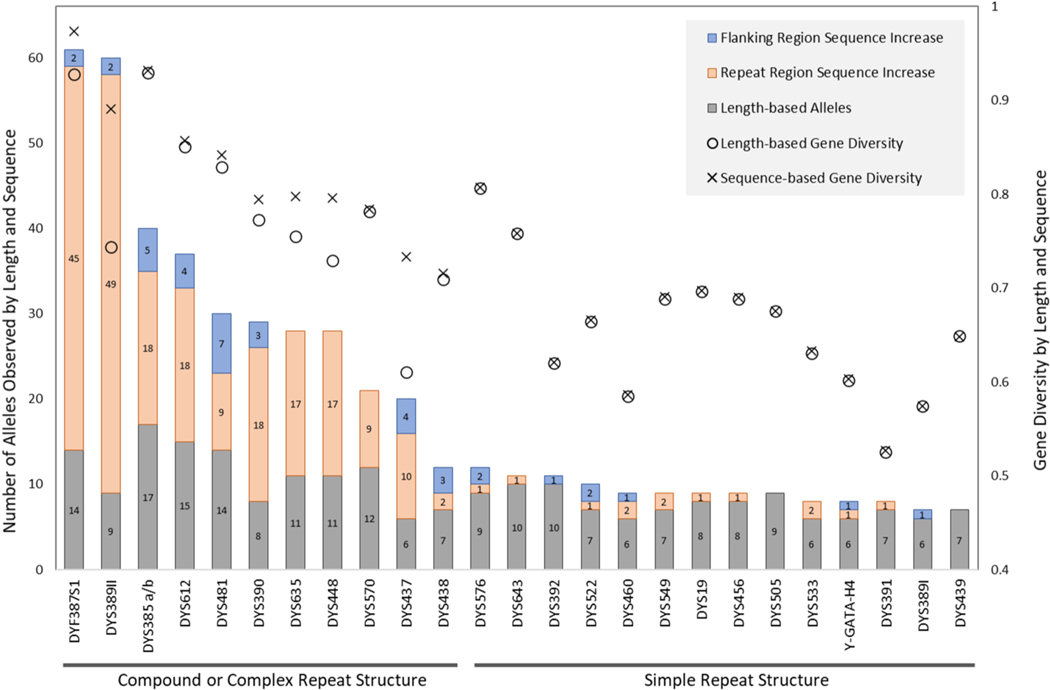
The degree of information gained through sequencing 25 Y-STR loci of the ForenSeq kit increased the number of alleles observed as well as improved gene diversity (Tables S6. and S11.). The unreported locus DYS456 is included in these calculations because the sequence data quality was sufficient for allele calls to be obtained for the complete set of ‘NIST 1032’ samples. The unreported locus DYS461 is omitted from this summary due to incomplete results, as previously described. The majority of the loci exhibit at least one additional allele from sequencing as compared to length-based data; only two Y-STR loci (DYS439 and DYS505) did not show an increase in number of alleles by sequence (Fig. 5.). Markers benefiting the most from sequencing are DYS389II, which has the highest number of additional alleles (51), followed by DYF387S1, for which 47 additional sequence-based alleles were observed. In total, there are 263 sequence-based alleles for all loci in this study of 1032 male samples, which represents an increase of 114% from 230 length-based alleles from the same set (Table S11.). Calculated gene diversity values showed an increase for a subset of loci, with substantial increases observed for DYS389II and DYS437; however, most loci had minimal or no increase in gene diversity from sequencing compared to length-based values.
Fig. 5.
Allele counts and gene diversity values for both length-based and sequence-based alleles for 25 Y-STRs in the ForenSeq kit (UAS 24 plus the unreported DYS456) for the ‘NIST 1032’ set. The ‘x’ axis lists the Y-STRs, the left ‘y’ axis is the number of alleles, and the right ‘y’ axis is the gene diversity. For each locus, the bars of the histogram consist of up to three sections: grey = number of length-based alleles, orange = number of new alleles resulting from repeat region sequence variations and blue = additional new alleles from flanking region variants. The number inside of each section indicates the count of unique alleles. Gene diversity values are plotted for length- (◯) and sequence-based alleles (×). Gene diversity values for the two multi-copy loci (DYF387S1 and DYS385 a/b) are calculated based on locus-specific haplotype frequencies.
3.6.2. Flanking region polymorphisms
Evaluating sequence data beyond the repeat region allows characterization of variation within the flanking regions of the STR markers. In this data set using the ForenSeq sequencing kit and the reporting range indicated in Table S2., we observed 22 different flanking region polymorphisms across 14 loci (Fig. 5.). These flanking region variations are described in Table S12. by their unique identifiers (rs numbers cataloged by the dbSNP database) and by the changes in Human Genome Variation Society (HGVS) nomenclature on the human reference genome GRCh38. Only five of these 22 variants are included in the UAS SLR range, the rest can be identified by extended analysis of the flanking region. In the current Y-STR sequencing multiplex kits, the fragments of the multi-copy loci DYS385 and DYF387S1 cannot be oriented to specific genomic locations; therefore, Sanger sequencing was attempted to orient the fragments for the samples exhibiting flanking region variation at these loci (results given in Section 3.1.2). All of these flanking region variants are also illustrated and cataloged by their respective Y-STRs in Table S2. with their genomic position highlighted in the flanking region in relation to the repeat region, and the different reporting ranges.
4. Discussion
4.1. Concordance
The ‘NIST 1032’ set was typed by three different currently available length-based Y-STR CE kits with results compared to previously published Y-STR data (revisions are outlined in Table 3.). Differences between proprietary primer designs of different manufacturers can generate discrepant allele calls from the same samples due to different primer placement in the flanking region of the targeted STRs combined with sequence variation (SNPs, indels) of the flanking regions or in primer binding sites. Sequence variation in a primer binding region may interfere with successful amplification and result in less PCR product or even the absence of amplification (i.e., null alleles). To detect this expected phenomenon, length-based allele calls from all four kits (ArgusY28, PPY23, YFP, and ForenSeq) were compared. From this comparison, five discordant calls (see Table 4.) were identified across these kits. Alleles were in agreement from three of the four kits in each case, except one occurrence where the four kits called a DYS456 allele with three different results. The haplotype frequency data reported in Table S5. are kit specific and differ across kits for these five samples, while the length-based allele calls in Table S4. are reported from ForenSeq data for all discrepant loci.
Discordances were also considered when comparing the default data output from the UAS against the consensus CE calls, and six discrepancies were found. Because these ‘bioinformatic null alleles’ can result from sequences present in the raw data which are unrecognized by a particular data analysis method, in this study, a multipronged approach was used with two different software beyond the default UAS. If bioinformatic null alleles are not considered, incomplete data could be generated and easily dismissed as homozygotes in diploid markers or reported as null alleles in haploid markers. Sequences for all bioinformatic null alleles omitted by the UAS are recognized and reported by both additional data analysis methods (STRait Razor and FDSTools). This highlights that performing additional data analysis is beneficial to report complete and concordant allele calls.
4.2. Father–son pairs
A subset of the ‘NIST 1032’ set included 369 male samples with their additionally typed father samples. Fifty-one of these father samples were used as replacements when their respective son samples were depleted, or their typing was challenged by drop-outs. This substitution allowed the reporting of a complete set of 1032 samples for all the 30 Y-STRs.
Furthermore, comparing sequenced Y-STR alleles in these father-son pairs allowed the calculation of mutation rates at the ForenSeq loci, using length- and sequence-based alleles that reveal not just the magnitude and direction of changes by length but the basis of these changes in the sequence of the repeats as well. In this study, there were no changes affecting only the underlying sequence; therefore, all 48 observed mutations between father-son pairs were also confirmed by CE. The RM Y-STR loci (DYS570, DYS576, and DYS612) have high mutation rates in this sample set, including DYS612 with the highest mutation rate (1.96 × 10−2) from seven distinct events, comparable to previously published rates [35]. The only outlier from the expected mutation rate is RM Y-STR DYF387S1, which did not present any mutations in this set, possibly resulting from the limited number of populations sampled here. Most of the mutations observed in this data set were one-step mutations followed by a small number of two-step mutations, similar to previous observations [35], while three or more step mutations were not observed. Two-step mutations occur less often and do not follow the stepwise mutation model (SMM) [36,37]. Adding more samples could minimally affect the calculated mutation rates and it may account for additional mutations across the loci with no changes observed in this set; however, when checked against the combined mutation rates calculated by the YHRD database, these were comparable (Table S9.).
Eight of the available father samples were also utilized to confirm the results of the dosage estimation from the semi-quantitative RRT tests (Table S10.), and whether the respective son samples typed with supernumerary alleles were germline duplications or somatic mutations. As expected, stable germline duplications were present in both, while somatic mutations were only detected in the son samples. This method is not suitable to estimate dosage from tri-allelic combinations, which were observed in this data set both by length and sequence.
4.3. Sequence-based allele diversity
For forensic autosomal STR markers, sequencing increases the allelic and gene diversity values. This increase mostly correlates with the complexity of the repeat structure, where more complex loci exhibit more new sequence-based alleles [4]. This thoroughly characterized ‘NIST 1032’ set was an ideal subject for evaluating the usefulness of Y-STR sequence variants and the overall benefit of sequencing these markers. For Y-STR markers, an unequal improvement of allelic diversity was observed across loci, with less than half of the analyzed loci showing more than three additional alleles by sequence compared to length-based methods. Allelic increases were concentrated in Y-STRs with a compound or complex repeat structure, where the new alleles arose mainly from repeat pattern variations and, to a lesser extent, SNPs/indels present in the sequence (Fig. 5. and Table S12.).
Interestingly, DYS481 clusters with these more complex repeats (Fig. 5.) despite being historically characterized as a simple CTT repeat. The increase in alleles by sequence at this locus owes to a minimally variable CTG = 0/1/2 in the adjacent 5′ flanking region: [CTG]0–2 [CTT]n. The most commonly observed variant in this data set (also found in the GRCh38 reference sequence) contains a single CTG (CTG = 1) which is reported as part of the flanking region. When CTG = 0, it is interpreted as a flanking SNP (CTG > CTT; rs370750300), whereas, when CTG = 2, it is reported as a nucleotide change (CTT > CTG) inside of the CTT repeat region. Such variations can be considered together as repeat pattern variations of the CTG, an uncounted repeat preceding the main CTT repeat [17]. Alternative nomenclatures for the locus DYS481 are summarized in Table S13., which compares two alternatives plus the leftmost interpretation of the locus, changing the main repeat from CTT to TTC. Similarly, DYS385 is traditionally considered a simple TTTC repeat; however, posterior to the 3′ end of the core repeat there is a variable number of repeat units of CCTT = 5/6/7/8/9: [TTTC]n [CCTT] 5–9. In GRCh38, CCTT = 6, and this is also the most commonly observed variant in ‘NIST 1032’ (87.5% of alleles). Its compound structure contributes to the increased number of sequence-based variants observed at this locus (Fig. 5.). This nomenclature including both the core and the flanking repeats is used across the manuscript and supplementary files, specifically: visualized in Table S2., reported in Table S3., and listed in Table 5.
In contrast to the compound and complex repeat structures, Y-STRs that were minimally improved by sequencing had a simple repeat structure, with the main source of the observed new variant alleles being an occasional SNP/indel in the repeat or the flanking regions (Fig. 5. and Table S12.). Two Y-STRs did not yield additional alleles in this data set with sequence analysis (Fig. 5.; DYS505 and DYS439).
When comparing gene diversity (plotted in Fig. 5.) and other forensic statistics (PIC, PM, PD in Table S6.) from length- to sequence-based methods, not all of the Y-STRs benefit equally from the introduction of sequencing. For example, with DYS389II and DYS437 these statistics improved the most, but loci such as DYS385 and DYS612 showed similar statistics by length and sequence, despite a more than doubling of alleles by sequence (Fig. 5.).
4.4. Haplotype-level diversity
Y-STR markers are analyzed individually but considered in haplotypes: the string of markers inherited together on the male-specific region of the Y chromosome (MSY). MSY is not affected by recombination; therefore, the haplotypes are shared across paternal-line relatives except for mutations occurring between generations [38]. When reporting haplotypes, the sequence-level variation introduced for each locus may not collectively increase the diversity in haplotypes because the high number of length-based markers already differentiates the haplotypes to a nearly individual-specific level [39]. By analyzing the extended range of the UAS FRR, 22 different flanking region variants were identified in this sample set; however, only five of these variants are included when the smaller range of the UAS SLR (the default UAS software output) is considered. Using the UAS SLR, therefore, could create discordance at the allele level, but not necessarily at the haplotype level. In this data set, the number of unique and unresolved haplotypes remained unchanged between reporting at the UAS FRR and the smaller UAS SLR sequence ranges. This is not surprising, as the rarely occurring flanking variants are less differentiating than the combinations of the faster changing repeat arrays. The length- and sequence-based haplotype frequencies show a very high degree of haplotype resolution (HD = 0.999996), and other haplotype-level forensic parameters are not improved by sequence either, as listed in Table S5. Haplotypes were compared across marker sets from the lowest number of markers, MiHT, and representing commercial kits including the early versions and the currently available, larger multiplexes. This comparison highlights that the number of Y-STRs included in these kits is the main factor for reaching maximum haplotype diversity, rather than the typing methodology (CE compared to sequencing). All of the larger kits currently available performed equally well, providing nearly the same level of haplotype resolution for this sample set. The few remaining unresolved haplotype pairs in this study (one to three; Table S7.) were distinguished only by typing additional RM Y-STRs [2] which are not included in any of these kits. Haplotype-level resolution is visually compared in Fig. 4a. and b. and the individual haplotypes for all considered marker sets are reported in Table S5., including haplotype-level statistics that show improvement correlating with the increasing number of markers.
4.5. Population-level diversity
The increase in sequence-based alleles between populations is only observed in certain Y-STRs. For example, the loci with the most increase in gene diversity and other forensic statistics (DYS389II and DYS437) show a skewed gain in sequence-based alleles in the African American population sample set compared to the other three, especially the Caucasian population samples, which shows the least improvement (Table S6.). The gain seen in the African American population sample set is in line with high genetic variation observed in African-origin populations; however, these samples do not show any new alleles by sequence at the RM Y-STR DYS612, which is unexpected. Another RM Y-STR, DYS570, showed a biased increase in the Hispanic population sample set as compared to the others (Table S6.). Finally, DYS635 showed the most new alleles in the Caucasian population samples, which correlates to the high number of new alleles generated by an insertion of repeat units in the general structure of this locus. This change is observed on the phylogeny, originating from the super-haplogroup P and its descendants [17], in which most Caucasian samples belong. Samples of this population, however, do not exhibit higher increase in gene diversity at this locus, compared to samples in the other three populations. Using the marker sets representing the MiHT, each CE commercial kit and the length- and sequence-based ForenSeq kit, the pairwise genetic distances of the four populations are differentiated on MDS plots, even with the least number of markers (MiHT). As opposed to the increasing haplotype diversity, the genetic distances are minimally affected by additional markers. The introduction of sequencing does not increase haplotype diversity or genetic distance (Fig. S1.). AMOVA results also showed no obvious difference between the marker sets when exploring the source of genetic variance among and within the four population sample sets (Table S8.). These results indicate significant but limited genetic differentiation among the four population sample sets, regardless of the marker sets used for the calculation. This proportion of genetic variance is accounted for by allele frequency differences among the four populations.
5. Conclusions
This study used the well-established ‘NIST 1036’ sample set from four main U.S. populations which was previously characterized for other forensic markers [4–6,11,12,40] as well as for the Y-STRs in CE-based studies in previous publications [2,3,13,14,41]. To adhere to the high standards of data quality associated with this sample set, here a complete set of Y-STR haplotypes for 30 Y-STR loci is given, including revisions of Y-STR calls published earlier, which are now further confirmed by sequencing, as well as the currently available commercial Y-STR CE kits. Data from the haplotypes published here therefore supersedes previously published data sets for these samples.
The final set of Y-STR haplotypes generated by this study is provided for the community in the form of openly available data via the NIST Public Data Repository: U.S. population data for human identification markers, DOI 10.18434/t4/1500024. Furthermore, all observed Y-STR allele variants are reported with unique GenBank accession numbers, and associated records are available in the STRSeq NCBI BioProject, accession PRJNA380127 under the Y-Chromosomal STR Loci subproject (PRJNA380347). New flanking region polymorphisms were also submitted to dbSNP. The revised Y-STR haplotypes were used to update the population sets in YHRD (YA004438–2, YA004437–2, YA003315–3, and YA004439–2) extended to the YHRD maximal haplotype for all 1032 samples, where these haplotype frequencies contribute to population level haplotype frequencies. For matching haplotypes, these population level haplotype frequencies can be retrieved from the database to add statistical weight to matches on the paternal line reported by practitioners.
Those interested in using the ‘NIST 1032’ sequence-level Y-STR data should consider the sequence range provided, which is equivalent to the UAS FRR, except for one locus (DYS522) where a larger range of sequence is provided. It is possible to truncate this sequence-based data set to a smaller range, collapsing frequencies when flanking region polymorphisms are outside of the region of interest. As mentioned in the discussion, reducing the sequence range from the UAS FRR to the UAS SLR does not result in different haplotype frequencies in this data set, but this would need to be evaluated for any similar study. If this sequence-based data is of a smaller range than a different sequencing multiplex, then the ranges need to be matched with this data, or a different data set should be used.
In this study, sequencing data detects at least one new allele for 88% of the sequenced loci; however, sequencing Y-STRs did not provide additional resolution of shared haplotypes. The increase in loci for CE kit iterations was impactful enough in haplotype resolution, as the current Y-STR multiplexes of 22 to 26 loci are able to individualize nearly all haplotypes with length-based alleles in this sample set. Sequencing Y-STR markers can be beneficial for other reasons. As these markers can be included in the same workflow/multiplex as autosomal STRs, X-STRs and SNPs, sequencing approaches can maximize obtained information when sample quantity is limited. Also, there may be other populations or particular cases where sequence information is impactful.
Using the ‘NIST 1032’ set for this multi-dimensional study to evaluate concordance across assays, platforms, and bioinformatic pipelines continually adds to the extensive forensic marker information available for these samples and maintains their relevance to the forensic community in facilitating technology transition.
Supplementary Material
Acknowledgments
The authors wish to thank John M. Butler for his review of the manuscript and Kevin M. Kiesler for collating Y-SNP data on the five shared haplotypes and for his related laboratory work. The authors are grateful to QIAGEN for providing the Investigator Argus Y-28 QS kits used in this study.
Funding and disclaimer
This work was funded in part by the Federal Bureau of Investigation (FBI) interagency agreements DJF-15-1200-0000174 and DJF-16-1200-0000246: “Forensic DNA Typing as a Biometric Tool”. T.I.H. was funded by the NIST Special Programs Office (Forensic Genetics Focus Area). Points of view in this document are those of the authors and do not necessarily represent the official position or policies of the U.S. Department of Commerce or the Department of Justice. Certain commercial equipment, instruments, and materials are identified in order to specify experimental procedures as completely as possible. In no case does such identification imply a recommendation or endorsement by NIST, nor does it imply that any of the materials, instruments, or equipment identified are necessarily the best available for the purpose. All work presented has been reviewed and approved by the NIST Research Protections Office.
The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Footnotes
Appendix A. Supporting information
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.fsigen.2021.102655.
Conflict of interest
The authors declare no conflict of interest.
References
- [1].Kayser M, Forensic use of Y-chromosome DNA: a general overview, Hum. Genet. 136 (5) (2017) 621–635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Ballantyne KN, et al. , Toward male individualization with rapidly mutating y-chromosomal short tandem repeats, Hum. Mutat. 35 (8) (2014) 1021–1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Coble MD, Hill CR, Butler JM, Haplotype data for 23 Y-chromosome markers in four U.S. population groups, Forensic Sci. Int. Genet 7 (3) (2013) e66–e68. [DOI] [PubMed] [Google Scholar]
- [4].Gettings KB, et al. , Sequence-based U.S. Population Data for 27 Autosomal STR loci, 37, Forensic Sci. Int., Genet., 2018, pp. 106–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Borsuk LA, et al. , Sequence-based U.S. population data for the SE33 locus, Electrophoresis 0 (2018) 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Borsuk LA, Steffen CR, Keisler KM, Vallone PM, Gettings KB, Sequence-based U.S. population data for 7 X-STR loci, Forensic Sci. Int. Rep. (2020) 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Woerner AE, King JL, Budowle B, Fast STR Allele Identification with STRait Razor 3.0, Forensic Sci. Int., Genet., 2017. [DOI] [PubMed]
- [8].Hoogenboom J, et al. , FDSTools: A software package for analysis of massively parallel sequencing data with the ability to recognise and correct STR stutter and other PCR or sequencing noise, Forensic Sci. Int. Genet. 27 (2017) 27–40. [DOI] [PubMed] [Google Scholar]
- [9].Gettings KB, et al. , STRSeq: A catalog of sequence diversity at human identification Short Tandem Repeat loci. Forensic Sci. Int. Genet. 31 (2017) 111–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Willuweit S, Roewer L, The new Y chromosome haplotype reference database, Forensic Sci. Int. Genet. 15 (2015) 43–48. [DOI] [PubMed] [Google Scholar]
- [11].Hill CR, et al. , U.S. population data for 29 autosomal STR loci, Forensic Sci. Int. Genet. 7 (3) (2013) e82–e83. [DOI] [PubMed] [Google Scholar]
- [12].Steffen CR, et al. , Corrigendum to ‘U.S. Population Data for 29 Autosomal STR Loci’ [Forensic Sci. Int. Genet. 7 (2013) e82-e83], Forensic Sci. Int. Genet. 31 (2017) e36–e40. [DOI] [PubMed] [Google Scholar]
- [13].Butler JM, et al. , Allele frequencies for 27 Y-STR loci with U.S. Caucasian, African American, and Hispanic samples, Forensic Sci. Int. 156 (2–3) (2006) 250–260. [DOI] [PubMed] [Google Scholar]
- [14].Coble MD, Butler JM, Characterization of new miniSTR loci to aid analysis of degraded DNA, J. Forensic Sci. 50 (1) (2005) 43–53. [PubMed] [Google Scholar]
- [15].Hill CR, et al. , Characterization of 26 miniSTR loci for improved analysis of degraded DNA samples, J. Forensic Sci. 53 (1) (2008) 73–80. [DOI] [PubMed] [Google Scholar]
- [16].Hill CR, Butler JM, Vallone PM, A 26plex autosomal STR assay to aid human identity testing*, J. Forensic Sci. (2009). [DOI] [PubMed]
- [17].Huszar TI, Jobling MA, Wetton JH, A phylogenetic framework facilitates Y-STR variant discovery and classification via massively parallel sequencing, Forensic Sci. Int. Genet. 35 (2018) 97–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Bolger AM, Lohse M, Usadel B, Trimmomatic: a flexible trimmer for Illumina sequence data, Bioinformatics 30 (15) (2014) 2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Magoc T, Salzberg SL, FLASH: fast length adjustment of short reads to improve genome assemblies, Bioinformatics 27 (21) (2011) 2957–2963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Kline MC, et al. , STR sequence analysis for characterizing normal, variant, and null alleles, Forensic Sci. Int. Genet. 5 (4) (2011) 329–332. [DOI] [PubMed] [Google Scholar]
- [21].Gouy A, Zieger M, STRAF-A convenient online tool for STR data evaluation in forensic genetics, Forensic Sci. Int. Genet. 30 (2017) 148–151. [DOI] [PubMed] [Google Scholar]
- [22].Kayser M, et al. , Evaluation of Y-chromosomal STRs: a multicenter study, Int. J. Leg. Med. 110 (3) (1997) 125–133. [DOI] [PubMed] [Google Scholar]
- [23].RCoreTeam. R: A language and environment for statistical computing. 2014. Available from: 〈https://www.R-project.org/〉.
- [24].Venables WN, Ripley BD Modern Applied Statistics with S, Fourth edition ed.,, Springer New York, 2002. [Google Scholar]
- [25].Excoffier L, Lischer HE, Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and Windows, Mol. Ecol. Resour. 10 (3) (2010) 564–567. [DOI] [PubMed] [Google Scholar]
- [26].Mauri M, Elli T, Caviglia G, Uboldi G, Azzi M, RAWGraphs: A Visualisation Platform to Create Open Outputs, in CHITALY ‘17: Proceedings of the 12th Biannual Conference on Italian SIGCHI Chapter, 2017., Italy. p. 1–5. [Google Scholar]
- [27].Huszar TI, Massively parallel sequencing of forensic markers: sequence variation and forensic application. Genetics and Genome Biology, University of Leicester,, Leicester, UK., 2020. [Google Scholar]
- [28].Niederstatter H, et al. , Separate analysis of DYS385a and b versus conventional DYS385 typing: is there forensic relevance? Int. J. Leg. Med 119 (1) (2005) 1–9. [DOI] [PubMed] [Google Scholar]
- [29].Watahiki H, et al. , Polymorphisms and microvariant sequences in the Japanese population for 25 Y-STR markers and their relationships to Y-chromosome haplogroups, Forensic Sci. Int. Genet. 41 (2019) e1–e7. [DOI] [PubMed] [Google Scholar]
- [30].Decker AE, et al. , Analysis of mutations in father-son pairs with 17 Y-STR loci, Forensic Sci. Int. Genet. 2 (2008) e31–e35. [DOI] [PubMed] [Google Scholar]
- [31].Lee EY, Lee HY, Shin KJ, Off-ladder alleles due to a single nucleotide polymorphism in the flanking region at DYS481 detected by the PowerPlex((R)) Y23 System, Forensic Sci. Int. Genet. 24 (2016) e7–e8. [DOI] [PubMed] [Google Scholar]
- [32].Aliferi A, et al. , UK and Irish Y-STR population data-A catalogue of variant alleles, Forensic Sci. Int. Genet. 34 (2018) e1–e6. [DOI] [PubMed] [Google Scholar]
- [33].Ralf A, et al. , Forensic Y-SNP analysis beyond SNaPshot: High-resolution Y-chromosomal haplogrouping from low quality and quantity DNA using Ion AmpliSeq and targeted massively parallel sequencing, Forensic Sci. Int. Genet 41 (2019) 93–106. [DOI] [PubMed] [Google Scholar]
- [34].Kling D, Tillmar AO, Egeland T, Familias 3 - Extensions and new functionality, Forensic Sci. Int. Genet. 13 (2014) 121–127. [DOI] [PubMed] [Google Scholar]
- [35].Ballantyne KN, et al. , Mutability of Y-chromosomal microsatellites: rates, characteristics, molecular bases, and forensic implications, Am. J. Hum. Genet. 87 (3) (2010) 341–353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Ohta T, Kimura M, A model of mutation appropriate to estimate the number of electrophoretically detectable alleles in a finite population, Genet. Res. 22 (2) (1973) 201–204. [DOI] [PubMed] [Google Scholar]
- [37].Di Rienzo A, et al. , Mutational processes of simple-sequence repeat loci in human populations, Proc. Natl. Acad. Sci. USA 91 (8) (1994) 3166–3170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Jobling MA, Tyler-Smith C, The human Y chromosome: an evolutionary marker comes of age, Nat. Rev. Genet. 4 (8) (2003) 598–612. [DOI] [PubMed] [Google Scholar]
- [39].Beleza S, et al. , Extending STR markers in Y chromosome haplotypes, Int. J. Leg. Med. 117 (1) (2003) 27–33. [DOI] [PubMed] [Google Scholar]
- [40].Taylor CR, et al. , Platinum-quality mitogenome haplotypes from United States populations, Genes (Basel) 11 (11) (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Decker AE, et al. , The impact of additional Y-STR loci on resolving common haplotypes and closely related individuals, Forensic Sci. Int. Genet. 1 (2007) 215–217. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.