Abstract
The recent widespread emergence of monkeypox (mpox), a rare and endemic zoonotic disease by monkeypox virus (MPXV), has made global headlines. While transmissibility (R0 ≈ 0.58) and fatality rate (0–3%) are low, as it causes prolonged morbidity, the World Health Organization has declared monkeypox as a public health emergency of international concern. Thus, effective containment and disease management require quick and efficient detection of MPXV. In this bioinformatic overview, we summarize the numerous molecular tests available for MPXV, and discuss the diversity of genes and primers used in the polymerase chain reaction-based detection. Over 90 primer/probe sets are used for the detection of poxviruses. While hemagglutinin and A-type inclusion protein are the most common target genes, tumor necrosis factor receptor and complement binding protein genes are frequently used for distinguishing Clade I and Clade II of MPXV. Problems and possibilities in the detection of MPXV have been discussed.
Supplementary Information
The online version contains supplementary material available at 10.1007/s11262-023-01975-3.
Keywords: Diagnostics, Emerging infectious diseases, Epidemiology and public health, Molecular evolution, Orthopoxvirus, Viral genome surveillance
Introduction
The emergence of new pathogenic viruses is a constant threat to humanity as each year novel viruses are evolving [1]. Unprecedented global changes—population growth, increased trade and travel, and climate change—in the recent decades have made the threat of re-emergence of viruses even more likely [2]. The recent emergence of monkeypox (renamed as mpox by the World Health Organization on 28 Nov 2022) disease by monkeypox virus (MPXV) has made global headlines. Between 13 and 21 May 2022 over 92 monkeypox cases were reported from 12 countries [3]. Although there was a major outbreak of monkeypox in the United States in 2003 via prairie dogs infected from an imported Gambian pouched rat [4, 5], the latest unrelated outbreaks in multiple countries have caused serious concerns since the coronavirus disease (COVID) pandemic and pressed for the need of disease surveillance and viral detection.
Monkeypox is a rare zoonotic disease endemic to the African continent—mainly in the northern and central Democratic Republic of the Congo (DRC). Although the disease has been reported only sporadically in neighbouring Republic of the Congo (ROC) regions, it has emerged several times in west and central Africa between 1970 and 2017 [6]. The MPXV was first identified in 1958 in crab-eating macaque monkeys (Macaca fascicularis), but multiple animal species including rodents can easily transmit the virus [7]. The MPXV is related to variola virus (VARV) that causes smallpox, both belonging to the genus Orthopoxvirus.
Compared to smallpox, monkeypox has a much lower infectivity or human to human transmission with basic reproduction number (R0) < 1 [8, 9], perhaps ≈0.576 [10], and an attack rate (the fraction of at-risk population that contracts the disease in a specified time interval) of ≈50% [11]. While antiviral drug ST-246, a potent Orthopoxvirus egress inhibitor, can protect non-human primates from VARV or MPXV [12], smallpox vaccine (for example, Dryvax) seems 85% effective in humans against monkeypox [13]. The MPXV has two distinct clades [14]. The Clade I causes illness similar to smallpox and has a case fatality rate of up to 10% in unvaccinated populations. The Clade II causes less severe and less inter-human transmissible disease [15, 16].
The disease severity of monkeypox, compared to smallpox, is considerably less with milder rash. However, due to its lengthy incubation period of up to 21 days and prolonged morbidity of two to four weeks, the World Health Organization has declared monkeypox as a public health emergency of international concern. Thus, effective containment and disease management require quick and efficient detection of MPXV. In this bioinformatic overview, we summarize the numerous molecular tests available for MPXV, discuss the diverse genes and primers used in the polymerase chain reaction (PCR)-based detection, and highlight the challenges in detecting MPXV and discriminating different clades and poxviruses.
Methods of detection
There are numerous ways to detect MPXV at the molecular level using appropriate patient specimens. While any type of body fluid might be used, blood in particular, was not found to contain high level of virus. On the other hand, lesions exudate on a swab or crust are considered as the best and least invasive patient samples [17].
Detection of viral particles
Live virus can be grown from patient lesion samples using chorioallantoic membrane or other cell-based viral culture methods. However, apart from the necessity of a fresh/live sample, this approach takes several days. In addition, it requires further characterization for the identification of viral particles. The electron microscopy is used for the conventional physical characterization. However, even if the negative staining reveals a brick-shaped viral particle, as a general form, it cannot be used to distinguish MPXV from other orthopoxviruses [17]. As our emphasis is on molecular detection, this section is not elaborated in depth.
Detection of antigens/antibodies
The easiest way to detect MPXV is to test for the virus-specific antigens in biopsy or other samples. One such method is called specific peptide-based rapid antigen test (RAT) [18]. The peptide-based antigens depend on specific peptides which bind to targets designed using protein–protein interactions. Highly conserved regions of interacting proteins of antigen are used to detect the virus. While RAT might be easy and quick, it might not be specific to MPXV due to high molecular similarities among numerous orthopoxviruses [17]. Despite this, RAT is being utilized for the specific detection of MPXV [19]. Ignoring specificity issues if any, one advantage of RAT is that there might be 100 s or 1000 s of copies of antigens on a virion which greatly increase the possibility of detection compared to a single copy of nucleic acid target for PCR [18]. While RAT might be useful for quick and largescale screening/diagnostics as in COVID-19, it is found to have considerable drawback due to its high false negative rate when the virus load is low [20, 21].
Alternatively, the virus-specific antibodies can also be detected in patient samples. The presence of anti-Orthopoxvirus immunoglobulin M (IgM) antibodies indicate a recent exposure to Orthopoxvirus or it might also be due to smallpox vaccination. However, this can be used as a diagnostic test for Orthopoxvirus infection if prior smallpox vaccination was not so recent, perhaps up to six months [17, 22]. On the other hand, the presence of anti-Orthopoxvirus IgG antibodies indicate a previous (and not so recent) exposure to Orthopoxvirus or smallpox vaccination. Known positive and negative serum samples are always used as assay controls [4].
A number of specific methods such as complement fixation test, hemagglutination inhibition assay, enzyme-linked immunosorbent assay (ELISA), plaque reduction neutralization test, western blot, and electrochemiluminescence (ECL) assay can be used for the detection of antigens or antibodies [4, 7, 22].
The main limitation of antibody-based detection methods is that it is invasive—requires the collection of patient blood/serum samples. In addition, it might also require a cold storage facility for samples/reagents. Further, the assay is not specific to MPXV as it only assesses a previous exposure to any Orthopoxvirus [4] and is affected by prior smallpox vaccination. However, one particular advantage of serological tests is that the polyclonal nature of the immune response permits broader/robust detection compared to the species-specific PCR method that might fail due to mutations in the short genome targets [4].
Mass-spectrometry-based detection
Qualitative and quantitative proteomics using liquid chromatography tandem mass spectrometry (LC–MS/MS) were also used to differentially identify MPXV proteins from other orthopoxviruses. Manes et al. [23] identified numerous MPXV-specific proteins such as J2L (tumor necrosis factor receptor homologue), J3L and D7L (ankyrin repeat containing proteins), and F6R (NF-κB inhibitor), and vaccinia virus-specific proteins such as C22L (tumor necrosis factor receptor homologue), C10L (ankyrin repeat containing protein), and C1L (complement-binding host defence modulator). Similar quantitative proteomics strategy also identified cowpox virus-specific proteins such as host range factor CP77 and secreted chemokine binding protein CPXV-GRI D1, and showed the overrepresentation of A-type inclusion protein in vaccinia virus [24].
Eshoo et al. [25] developed a detection method combining PCR and electrospray ionization-mass spectrometry (PCR/ESI–MS) for the identification of all Orthopoxvirus members based on DNA and RNA helicase and polymerase genes. The method was able to resolve viruses at sub-species level. The PCR/ESI–MS technique was also used to detect MPXV in spiked human blood and aerosol-infected cynomolgus macaque samples, as well as to identify MPXV and vaccinia virus spiked into macaque blood sample at various concentration [26]. The authors claimed that the technique was able to identify all Orthopoxvirus members in a single assay along with quantitative identification of MPXV DNA in clinical specimens, thus eliminating the need for sequencing.
Detection of virus-specific nucleic acid targets
There are numerous variants of PCR-based methods for the detection of Orthopoxvirus/MPXV genomic targets (Table 1 and S1). Over 90 primer/probe sets (see Methods in the Supplemental information) are used to target as many as 38 poxvirus genes (Table S1). Ropp et al. [27] used conventional PCR to amplify near-full-length hemagglutinin (HA, also known as B2R) gene and endonuclease digest electropherograms to distinguish 10 species of orthopoxviruses, including North American orthopoxviruses and MPXV. However, given the close similarities among sequences, primers were not exclusive to species (Table 1 and S1, Fig. S1 and S2) and thus cross-hybridized to multiple members of the genus. Further, the use of multiple endonuclease cleavage profiles overly complicated the species identification. In addition, no attempts were made to distinguish different clades/variants of MPXV.
Table 1.
List of commonly used genes, primers, and probes in the detection of MPXV
| Gene | Forward and reverse primers [and probes] | Len | Assay | References |
|---|---|---|---|---|
|
A4L (CP) |
GTCAACGCTGGAAGGAGTG CCAGCAGACAGCCTATCC [CTCCTGTACTAAAACCACGWCAACAAACT] |
217 | rt-PCR | [52] |
|
A27L (ATI) |
AATACAAGGAGGATCT CTTAACTTTTTCTTTCTC |
1549 | PCR | [35] |
|
GAGAGAATCTCTTGATAT ATTCTAGATTGTAATC |
601 | PCR | [36] | |
|
GAGATTAGCAGACTCCAA GATTCAATTTCCAGTTTGTAC [GCAGTCGTTCAACTGTATTTCAAGATCTGAGAT/ CTAGATTGTAATCTCTGTAGCATTTCCACGGC] |
163 | rt-PCR | [37] | |
|
CCGTTACCGTTTTTACAATCGTTAATCAATGCTGATATGGAAAAGAGA ATAGGCTAAAGACTAGAATCAGGGATTCTGATTCATCCTTTGAGAAG |
– | LAMP | [34] | |
|
TACAGTTGAACGACTGCG AGTTCAGTTTTATATGCCGAAT |
221 | |||
|
GATGTCTATCAAGATCCATGATTCT TCTTGAACGATCGCTAGAGA |
115 | |||
|
TGGAGTCTGCTAATCTCTGTAAGATTAGAGAACTAGAGAATAAGTTGACC TGAGTGAATGCCGTGGAAATGCGCAGTCGTTCAACTGTA |
– | |||
|
CACAAGAAGTTGATGCACTG CAGCATTGATTTCATTATTACGT |
235 | |||
|
CGCTCTCGATGCAGTC CAGAGATTACAATCTAGAATCTCAG |
128 | |||
|
A29L (14kd protein) |
CCAGAGATATCATAGCCGCTCTT GAAACTCTCAAACAACGRCTAACT [TAAATAGAACGTCATCATT] |
156/157 | rt-PCR | [53] |
|
B2R (HA) |
ATGACACAATTACCAATAC CTAGACTTTGTTCTCTG |
942 | ORF | [27] |
|
CTGATAATGTAGAAGAC TTGTATTTACGTGGGTG |
406 | PCR | ||
|
GATGATGCAACTCTATCATGTA GTATAATTATCAAAATACAAGACGTC [AGTGCTTGGTATAAGGAG] |
131 | rt-PCR | [13] | |
|
B6R (EEV) |
ATTGGTCATTATTTTTGTCACAGGAACA AATGGCGTTGACAATTATGGGTG [AGAGATTAGAAATA] |
83 | rt-PCR | [48] |
| B7R |
ACGTGTTAAACAATGGGTGATG AACATTTCCATGAATCGTAGTCC [TGAATGAATGCGATACTGTATGTGTGGG] |
99 | rt-PCR | [54] |
| C3L/D14L |
TGGGAGCATTGTAACTTATAGTTGCCCTCCTGAACACATGACA ATCCTCGTATCCGTTATGTCTTCCCACCTATTTGCGAATCTGTT |
- | LAMP | [34] |
|
TGGGTGGATTGGACCATT ATGGTATGGAATCCTGAGG |
199 | |||
|
GATATTCGTTGATTGGTAACTCTGG GTTGGATATAGATGGAGGTGATTGG |
117 | |||
|
TGTCTACCTGGATACAGAAAGCAA GGCATCTCCGTTTAATACATTGAT [CCCATATATGCTAAATGTACCGGTACCGGA] |
100 | rt-PCR | [16] | |
|
E9L (DNA polymerase) |
TCAACTGAAAAGGCCATCTATGA GAGTATAGAGCACTATTTCTAAATCCCA [CCATGCAATATACGTACAAGATAGTAGCCAAC] |
101 | rt-PCR | [48] |
| F3L |
CTCATTGATTTTTCGCGGGATA GACGATACTCCTCCTCGTTGGT [CATCAGAATCTGTAGGCCGT] |
107 | rt-PCR | [7] |
|
CATCTATTATAGCATCAGCATCAGA GATACTCCTCCTCGTTGGTCTAC [TGTAGGCCGTGTATCAGCATCCATT] |
79 | rt-PCR | [55] | |
|
J2R (TNFR) |
CACACCGTCTCTTCCACAGA GATACAGGTTAATTTCCACATCG [AACCCGTCGTAACCAGCAATACATTT] |
82/85 | rt-PCR | [16] |
|
GGAAAATGTAAAGACAACGAATACAG GCTATCACATAATCTGGAAGCGTA [AAGCCGTAATCTATGTTGTCTATCGTGTCC] |
90 | |||
|
AATAAACGGAAGAGATATAGCACCACATGCAC GTGAGATGTAAAGGTATCCGAACCACACG [ACAGAAGCCGTAATCTATGTTGTCTATCGQTFCCTCCGGGAACTTA] |
181 | RPA | [38] | |
| N3R |
AACAACCGTCCTACAATTAAACAACA CGCTATCGAACCATTTTTGTAGTCT [TATAACGGCGAAGAATATACT] |
139 | rt-PCR | [7] |
LAMP loop-mediated isothermal amplification, ORF open reading frame, RPA recombinase polymerase amplification, rt-PCR real-time polymerase chain reaction
Nonetheless, with at least 29 unique primer sets (Table S1), HA gene was the most common target used for the detection of orthopoxviruses [7, 13, 27–33]. Yet, due to its high sequence similarity among different Orthopoxvirus members (Fig. S2), HA gene was not used for the specific detection of MPXV or its clades/variants. For example, an rt-PCR primer and probe set used by Edghill-Smith et al. [13] can detect over a dozen orthopoxviruses. Interestingly, with a unique primer set design, HA gene can be used for the specific detection of MPXV from other orthopoxviruses. For example, the MPXV and cowpox virus have a six-nucleotide deletion at around 431, whereas only cowpox virus has additional six-nucleotide deletions at 323 and 533 (Fig. S2).
The A-type inclusion (ATI, also known as A27L) protein/gene was the second most common target [34–37]. The MPXV ATI gene has 72 to 95.3% identity with other Orthopoxvirus species such as variola, vaccinia, cowpox, ectromelia, and camelpox viruses. By designing a primer to the region containing 8-bp deletion in MPXV, Neubauer et al. [36] differentiated 19 strains of MPXV from five Orthopoxvirus species. While Saijo et al. [37] used rt-PCR, Iizuka et al. [34] used loop-mediated isothermal amplification (LAMP) method to detect MPXV (Table 1). Both the sets of primers were designed such that the non-coding region downstream of ATI gene was also included. Further, given the large difference in this stretch of genome (Fig. S1) due to multiple long indels, the primers/methods could uniquely detect Clade I and Clade II of MPXV.
The reverse transcription polymerase chain reaction (rt-PCR) is a standard method for the universal detection of poxviruses [11]. Numerous variants of the method—for example—multiplexed PCR, assay based on dried PCR reagent, etc. have been tried and tested. Combination of primers and/or probes were also used. For example, only variola virus was specifically detected by a FAM (6-carboxyfluorescein)-labelled probe while camelpox, cowpox, monkeypox, and vaccinia viruses were detected by a TET (6-carboxytetramethylrhodamine)-labelled probe in a single PCR reaction [29]. Davi et al. [38] used recombinase polymerase amplification (RPA) assay targeting the G2R (= J2R) gene for the specific and rapid detection of MPXV within 3 to 10 min. The RPA-assay was claimed to be highly sensitive with a limit of detection of 16 DNA molecules/μl.
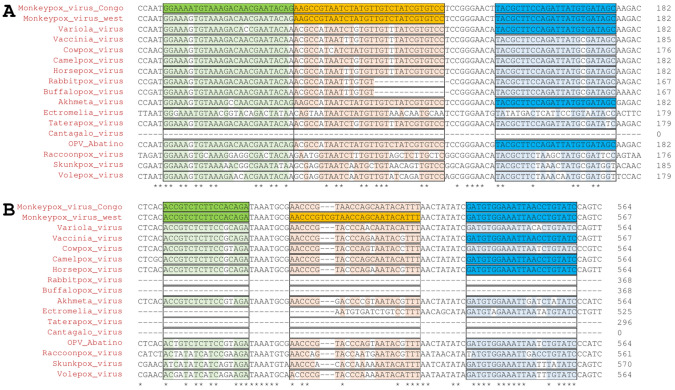
The tumor necrosis factor receptor (TNFR, also known as J2R) gene in particular seems to be a good target for the specific detection of MPXV and its two clades—Clade I and Clade II. For example, Li et al. [16] used two sets of rt-PCR primers and probes. While the first set of primers/probes would potentially detect MPXV over other PXVs (Fig. 1A), a second set of primers/probes seemed specific for Clade II of MPXV due to a three-nucleotide insertion in the sequence (Fig. 1B).
Fig. 1.
Detection of MPXV using TNFR (J2R) gene. Li et al. [16] proposed two sets of primers and probes for the specific rt-PCR detection of MPXV and its two clades—Clade I and Clade II. A While the primer GGAAAATGTAAAGACAACGAATACAG seems specific for Clade I, it might bind non-specifically to Clade II MPXV and other PXVs. However, due to multiple mismatches with probe, other PXV sequences might unlikely to be detected. On the other hand, B the probe AACCCGTCGTAACCAGCAATACATTT seems to be specific for Clade II MPXV due to a three-nucleotide insertion in the sequence. Forward (green) and reverse (blue) primers and probes (orange) are shaded
C3L/D14L gene was another target used in either LAMP [34] or rt-PCR method [16]. It seems that C3L/D14L is completely missing in the Clade II of MPXV. However, as many orthopoxvirus (OPV) members such as vaccinia virus (VACV) and VARV have a highly similar C3L/D14L sequence, it would be very misleading to use C3L/D14L as a distinguishing target for the detection of the Clade I of MPXV.
The Pan American Health Organization (PAHO) interim guidance on laboratory testing for monkeypox virus mentions that while commercial PCR kits for detecting OPV and MPXV in particular are under development, no validated PCR kits are available in the market at present. PAHO/WHO currently follow two protocols for OPV and MPXV detection [39]. The first one involves subjecting the samples (lesion swabs, vesicular fluids, and crusts) from suspected cases to OPV rt-PCR and the positive samples are subjected to MPXV-specific rt-PCR. In the second one, the samples are directly subjected to MPXV-specific rt-PCR followed by differentiation of Clade I and Clade II. The PAHO/WHO uses primers/probes specific for G2R (= J2R) and C3L/D14L genes that have several issues as discussed in detail in this review.
Detection of MPXV: challenges and opportunities
The 196,858-bp MPXV genome has at least 190 open reading frames of ≥ 60 amino acid residues each [40]. However, it seems that just 11 genes have been used as likely targets (Table 1). The choice of target for the specific detection of MPXV is important but tricky. The sequence in the central region of the MPXV genome, which encodes essential enzymes and structural proteins, is 96.3% identical to VARV. On the contrary, MPXV and VARV are said to have considerable differences in the regions encoding virulence and host-range factors near the ends of the genome [41].
While highly similar sequences lead to nonspecific primer/probe binding and lead to ambiguous detection of OPVs, highly divergent or variable sequence regions pose considerable challenge to detection. As in other viruses [42], comparison among OPVs such as VACV and VARV revealed that indels of 3–25 bp are common events in poxviruses [43]. The A27L (ATI) gene is one such hypervariable target containing variations including truncations, deletions, insertions, and base changes in the two clades of MPXV [35]. The C3L/D14L gene is completely missing in Clade II [16]. While these targets are often used to differentiate Clade I and Clade II of MPXV [37], given the inherent issues in the molecular detection, a negative result should not be taken as a confirmation. For, poxviruses can undergo rapid changes as VACV was shown to acquire 7–10% increase in genome size via K3L gene amplification [44] and there are considerable differences among MPXV clades [45].
Further, like in all viruses, the evolution of the genome due to ongoing point-mutations is leading to additional challenges in detection. Compared to RNA viruses such as Poliovirus-1 that has a very high rate of 0.01 substitutions per site per year, the MPXV has a far lower rate of 7 × 10–7 [46]. Yet, different MPXV/OPV sequences contain or can acquire a considerable number of substitutions (Fig. 1, S2, and S3), which may result in undesirable outcomes. For example, primers with one or more mismatches can bind to templates and effect PCR amplification by < 1.5 to > 7 cycle threshold [47]. The substitutions are also problematic in restriction fragment length polymorphism-based detection [27]. Thus, it is advisable to use multiple targets and primer/probe sets for the unambiguous detection of MPXV [7, 48].
While rt-PCR is the state-of-the-art method, it is a costly, highly sophisticated and laborious/multistep process, needing fine instrumentation and technical expertise. The development of LAMP and RPA overcomes some of these issues, even as MPXV detection urgently requires a validated test kit [39].
Conclusions and future outlook
Since its recent emergence, at least 84,639 cases of MPXV and 80 deaths have been reported from 110 countries [49, 50]. At present it is not a serious concern per se. However, given that MPXV has multiple natural hosts—mainly rodents—that find expanding conducive geographical range, pet trade, intercontinental travel, etc. all point to the possibility that monkeypox infections could continue to intensify or re-emerge in the future [51]. Although smallpox vaccination or infection has given some immunity, MPXV is a persistent threat. Thus, for effective disease surveillance, easy, quick, and effective viral detection is required. As explored in this work, over 90 primer/probe sets are used for the detection of poxviruses. HA and ATI are the common target genes, whereas TNFR and C3L are frequently used for distinguishing Clade I and Clade II of MPXV. There are multiple issues in the primer/probe sets and/or targets used in the detection of MPXV that require considerable attention while developing a validated test kit [39].
Supplementary Information
Below is the link to the electronic supplementary material.
Acknowledgements
This work did not receive any specific funding.
Author contributions
SDG and RSPR planned and performed the work, and wrote the manuscript. All authors contributed intellectually, and reviewed the manuscript.
Data availability
The data used in this work were collected/curated from the literature/NCBI. Relevant data are given in the supplemental information section.
Declarations
Conflict of interest
The authors declare that there is no conflict of interest.
Ethical approval
The work is in compliance with ethical standards. No ethical clearance was necessary.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Sudeep D. Ghate, Email: sudeep1129@gmail.com
R. Shyama Prasad Rao, Email: drrsprao@gmail.com.
References
- 1.Woolhouse M, Scott F, Hudson Z, et al. Human viruses: discovery and emergence. Philos Trans R Soc B: Biol Sci. 2012;367:2864–2871. doi: 10.1098/rstb.2011.0354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baker RE, Mahmud AS, Miller IF, et al. Infectious disease in an era of global change. Nat Rev Microbiol. 2022;20:193–205. doi: 10.1038/s41579-021-00639-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.WHO (2022a) Multi-country monkeypox outbreak in non-endemic countries. https://www.who.int/emergencies/disease-outbreak-news/item/2022-DON385. Accessed 12 Jan 2023.
- 4.Karem KL, Reynolds M, Braden Z, et al. Characterization of acute-phase humoral immunity to monkeypox: use of immunoglobulin M enzyme-linked immunosorbent assay for detection of monkeypox infection during the 2003 north American outbreak. Clin Diagn Lab Immunol. 2005;12:867–872. doi: 10.1128/CDLI.12.7.867-872.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Reed KD, Melski JW, Graham MB, et al. The detection of monkeypox in humans in the western hemisphere. N Engl J Med. 2004;350:342–350. doi: 10.1056/NEJMoa032299. [DOI] [PubMed] [Google Scholar]
- 6.Durski KN, McCollum AM, Nakazawa Y, et al. Emergence of monkeypox—west and central Africa, 1970–2017. MMWR Morb Mortal Wkly Rep. 2018;67:306–310. doi: 10.15585/mmwr.mm6710a5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kulesh DA, Loveless BM, Norwood D, et al. Monkeypox virus detection in rodents using real-time 3′-minor groove binder TaqMan® assays on the Roche LightCycler. Lab Invest. 2004;84:1200–1208. doi: 10.1038/labinvest.3700143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Learned LA, Reynolds MG, Wassa DW, et al. Extended interhuman transmission of monkeypox in a hospital community in the Republic of the Congo, 2003. Am J Trop Med Hyg. 2005;73:428–434. doi: 10.4269/ajtmh.2005.73.428. [DOI] [PubMed] [Google Scholar]
- 9.Sklenovská N, Van Ranst M. Emergence of monkeypox as the most important orthopoxvirus infection in humans. Front Public Health. 2018;6:241. doi: 10.3389/fpubh.2018.00241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.McMullen CL (2015) A one health perspective on disease dynamics: human monkeypox transmission in Sankuru district, Democratic Republic of Congo. Duke University ProQuest Dissertations Publishing 1587691.
- 11.Luciani L, Inchauste L, Ferraris O, et al. A novel and sensitive real-time PCR system for universal detection of poxviruses. Sci Rep. 2021;11:1798. doi: 10.1038/s41598-021-81376-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huggins J, Goff A, Hensley L, et al. Nonhuman primates are protected from smallpox virus or monkeypox virus challenges by the antiviral drug ST-246. Antimicrob Agents Chemother. 2009;53:2620–2625. doi: 10.1128/AAC.00021-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Edghill-Smith Y, Golding H, Manischewitz J, et al. Smallpox vaccine–induced antibodies are necessary and sufficient for protection against monkeypox virus. Nat Med. 2005;11:740–747. doi: 10.1038/nm1261. [DOI] [PubMed] [Google Scholar]
- 14.Likos AM, Sammons SA, Olson VA, et al. A tale of two clades: monkeypox viruses. J Gen Virol. 2005;86:2661–2672. doi: 10.1099/vir.0.81215-0. [DOI] [PubMed] [Google Scholar]
- 15.Chen N, Li G, Liszewski MK, et al. Virulence differences between monkeypox virus isolates from west Africa and the Congo basin. Virology. 2005;340:46–63. doi: 10.1016/j.virol.2005.05.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li Y, Zhao H, Wilkins K, et al. Real-time PCR assays for the specific detection of monkeypox virus West African and Congo Basin strain DNA. J Virol Methods. 2010;169:223–227. doi: 10.1016/j.jviromet.2010.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McCollum AM, Damon IK. Human monkeypox. Clin Infect Dis. 2014;58:260–267. doi: 10.1093/cid/cit703. [DOI] [PubMed] [Google Scholar]
- 18.Vangala RK (2022) Peptides and conjugates thereof as ACE-2 and S1 subunit mimics against Severe Acute Respiratory Syndrome Coronavirus-2 (SARS-CoV2) infection. PCT/IN2021/051030. https://ipindiaservices.gov.in/PublicSearch/PublicationSearch/PatentDetails. Accessed 6 July 2022.
- 19.JOYSBIO (2022) Monkeypox rapid test kit. https://en.joysbio.com/monkeypox-rapid-test-kit/. Accessed 12 Jan 2023.
- 20.Peeling RW, Olliaro PL, Boeras DI, Fongwen N. Scaling up COVID-19 rapid antigen tests: promises and challenges. Lancet Infect Dis. 2021;21:e290–e295. doi: 10.1016/S1473-3099(21)00048-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Scohy A, Anantharajah A, Bodéus M, et al. Low performance of rapid antigen detection test as frontline testing for COVID-19 diagnosis. J Clin Virol. 2020;129:104455. doi: 10.1016/j.jcv.2020.104455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Usme-Ciro JA, Paredes A, Walteros DM, et al. Detection and molecular characterization of zoonotic poxviruses circulating in the Amazon region of Colombia, 2014. Emerg Infect Dis. 2017;23:649–653. doi: 10.3201/eid2304.161041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Manes NP, Estep RD, Mottaz HM, et al. Comparative proteomics of human monkeypox and vaccinia intracellular mature and extracellular enveloped virions. J Proteome Res. 2008;7:960–968. doi: 10.1021/pr070432+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Doellinger J, Schaade L, Nitsche A. Comparison of the cowpox virus and vaccinia virus mature virion proteome: analysis of the species-and strain-specific proteome. PLoS ONE. 2015;10:e0141527. doi: 10.1371/journal.pone.0141527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Eshoo MW, Whitehouse CA, Nalca A, et al. Rapid and high-throughput pan-Orthopoxvirus detection and identification using PCR and mass spectrometry. PLoS ONE. 2009;4:e6342. doi: 10.1371/journal.pone.0006342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Grant RJ, Baldwin CD, Nalca A, et al. Application of the Ibis-T5000 pan-orthopoxvirus assay to quantitatively detect monkeypox viral loads in clinical specimens from macaques experimentally infected with aerosolized monkeypox virus. Am J Trop Med Hyg. 2010;82:318–323. doi: 10.4269/ajtmh.2010.09-0361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ropp SL, Jin Q, Knight JC, et al. PCR strategy for identification and differentiation of small pox and other orthopoxviruses. J Clin Microbiol. 1995;33:2069–2076. doi: 10.1128/jcm.33.8.2069-2076.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Aitichou M, Javorschi S, Ibrahim MS. Two-color multiplex assay for the identification of orthopox viruses with real-time LUX-PCR. Mol Cell Probes. 2005;19:323–328. doi: 10.1016/j.mcp.2005.05.003. [DOI] [PubMed] [Google Scholar]
- 29.Aitichou M, Saleh S, Kyusungb P, et al. Dual-probe real-time PCR assay for detection of variola or other orthopoxviruses with dried reagents. J Virol Methods. 2008;153:190–195. doi: 10.1016/j.jviromet.2008.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Damaso CR, Esposito JJ, Condit RC, et al. An emergent poxvirus from humans and cattle in Rio de Janeiro State: Cantagalo virus may derive from Brazilian smallpox vaccine. Virology. 2000;277:439–449. doi: 10.1006/viro.2000.0603. [DOI] [PubMed] [Google Scholar]
- 31.Espy MJ, Cockerill FR, III, Meyer RF, et al. Detection of smallpox virus DNA by LightCycler PCR. J Clin Microbiol. 2002;40:1985–1988. doi: 10.1128/JCM.40.6.1985-1988.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ibrahim SM, Kulesh DA, Saleh SS, et al. Real-time PCR assay to detect smallpox virus. J Clin Microbiol. 2003;41:3835–3839. doi: 10.1128/JCM.41.8.3835-3839.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Putkuri N, Piiparinen H, Vaheri A, et al. Detection of human orthopoxvirus infections and differentiation of smallpox virus with real-time PCR. J Med Virol. 2009;81:146–152. doi: 10.1002/jmv.21385. [DOI] [PubMed] [Google Scholar]
- 34.Iizuka I, Saijo M, Shiota T, et al. Loop-mediated isothermal amplification-based diagnostic assay for monkeypox virus infections. J Med Virol. 2009;81:1102–1108. doi: 10.1002/jmv.21494. [DOI] [PubMed] [Google Scholar]
- 35.Meyer H, Ropp SL, Esposito JJ. Gene for A-type inclusion body protein is useful for a polymerase chain reaction assay to differentiate orthopoxviruses. J Virol Methods. 1997;64:217–221. doi: 10.1016/S0166-0934(96)02155-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Neubauer H, Reischl U, Ropp S, et al. Specific detection of monkeypox virus by polymerase chain reaction. J Virol Methods. 1998;74:201–207. doi: 10.1016/S0166-0934(98)00099-8. [DOI] [PubMed] [Google Scholar]
- 37.Saijo M, Ami Y, Suzaki Y, et al. Diagnosis and assessment of monkeypox virus (MPXV) infection by quantitative PCR assay: differentiation of Congo Basin and West African MPXV strains. Jpn J Infect Dis. 2008;61:140–142. doi: 10.7883/yoken.JJID.2008.140. [DOI] [PubMed] [Google Scholar]
- 38.Davi SD, Kissenkötter J, Faye M, et al. Recombinase polymerase amplification assay for rapid detection of monkeypox virus. Diagn Microbiol Infect Dis. 2019;95:41–45. doi: 10.1016/j.diagmicrobio.2019.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.PAHO/WHO (2022) Laboratory guidelines for the detection and diagnosis of monkeypox virus infection. https://www.paho.org/en/documents/laboratory-guidelines-detection-and-diagnosis-monkeypox-virus-infection. Accessed 12 Jan 2023.
- 40.Shchelkunov SN, Totmenin AV, Safronov PF, et al. Analysis of the monkeypox virus genome. Virology. 2002;297:172–194. doi: 10.1006/viro.2002.1446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shchelkunov SN, Totmenin AV, Babkin IV, et al. Human monkeypox and smallpox viruses: genomic comparison. FEBS Lett. 2001;509:66–70. doi: 10.1016/S0014-5793(01)03144-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rao RSP, Ahsan N, Xu C, et al. Evolutionary dynamics of indels in SARS-CoV-2 spike glycoprotein. Evol Bioinform. 2021;17:1064616. doi: 10.1177/11769343211064616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Coulson D, Upton C. Characterization of indels in poxvirus genomes. Virus Genes. 2011;42:171–177. doi: 10.1007/s11262-010-0560-x. [DOI] [PubMed] [Google Scholar]
- 44.Elde NC, Child SJ, Eickbush MT, et al. Poxviruses deploy genomic accordions to adapt rapidly against host antiviral defenses. Cell. 2012;150:831–841. doi: 10.1016/j.cell.2012.05.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kugelman JR, Johnston SC, Mulembakani PM, et al. Genomic variability of monkeypox virus among humans, Democratic Republic of the Congo. Emerg Infect Dis. 2014;20:232–239. doi: 10.3201/eid2002.130118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Stern A, Andino R (2016) Viral evolution—it is all about mutations. In Katze MG, et al. (eds) Viral pathogenesis—from basics to systems biology, pp 233–240. 10.1016/B978-0-12-800964-2.00017-3.
- 47.Stadhouders R, Pas SD, Anber J, et al. The effect of primer-template mismatches on the detection and quantification of nucleic acids using the 5' nuclease assay. J Mol Diagn. 2010;12:109–117. doi: 10.2353/jmoldx.2010.090035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li Y, Olson VA, Laue T, et al. Detection of monkeypox virus with real-time PCR assays. J Clin Virol. 2006;36:194–203. doi: 10.1016/j.jcv.2006.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.OWID (2022) Monkeypox data explorer. https://ourworldindata.org/explorers/monkeypox?tab=table&facet=none&Metric=Confirmed+cases&Frequency=Cumulative&Shown+by=Date+of+confirmation&country=~OWID_WRL. Accessed 13 Jan 2023.
- 50.WHO (2022b) Multi-country monkeypox outbreak: situation update. https://www.who.int/emergencies/disease-outbreak-news/item/2022-DON396#:~:text=Since%201%20January%20and%20as,new%20countries%20have%20reported%20cases. Accessed 12 Jan 2023.
- 51.Parker S, Nuara A, Buller RML, Schultz DA. Human monkeypox: an emerging zoonotic disease. Future Microbiol. 2007;2:17–34. doi: 10.2217/17460913.2.1.17. [DOI] [PubMed] [Google Scholar]
- 52.Kuo S-C, Wang Y-M. Identification of pan-orthopoxvirus, monkeypox-specific and smallpox-specific DNAs by real-time PCR assay. J Med Sci. 2013;33:293–303. [Google Scholar]
- 53.Dumont C, Irenge LM, Magazani EK, et al. Simple technique for in field samples collection in the cases of skin rash illness and subsequent PCR detection of orthopoxviruses and varicella zoster virus. PLoS ONE. 2014;9:e96930. doi: 10.1371/journal.pone.0096930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shchelkunov SN, Shcherbakov DN, Maksyutov RA, et al. Species-specific identification of variola, monkeypox, cowpox, and vaccinia viruses by multiplex real-time PCR assay. J Virol Methods. 2011;175:163–169. doi: 10.1016/j.jviromet.2011.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Maksyutov RA, Gavrilova EV, Meyer H, et al. Real-time PCR assay for specific detection of cowpox virus. J Virol Methods. 2015;211:8–11. doi: 10.1016/j.jviromet.2014.10.004. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data used in this work were collected/curated from the literature/NCBI. Relevant data are given in the supplemental information section.