Abstract
Papillary thyroid carcinoma (PTC) is the most common type of endocrine cancer, with an increasing incidence worldwide. The treatment of PTC is currently the subject of clinical controversy, making it critically important to identify molecular markers that would help improve the risk stratification of PTC patients and optimize the therapeutic approach. The Von Hippel–Lindau (VHL) tumor suppressor gene has been implicated in tumorigenesis of various types of carcinoma and linked with their aggressive biological behavior. The role of VHL in the origin and development of PTC have only recently begun to be revealed. In this narrative review, we attempt to summarize the existing knowledge that implicates VHL in PTC pathogenesis and to outline its potential significance as a candidate molecular biomarker for the grouping of PTC patients into high and low risk groups.
Keywords: Papillary thyroid cancer, papillary thyroid carcinoma (PTC), Von Hippel–Lindau (VHL), biomarker, risk stratification, tumor suppressor
Introduction
Thyroid cancer represents the most common malignancy present in the endocrine organs. Over many decades its incidence has increased worldwide, generating an additional burden on healthcare systems. Papillary thyroid carcinoma (PTC) alone makes up over 80% of all thyroid cancers and for about 95% of the increased incidence worldwide [1, 2].
Molecular biomarker analysis is a significant addition to the traditional pathological evaluation of carcinoma and represents a valuable tool for improving diagnosis and refining clinical management. A number of genetic mutations and other molecular alterations can be detected in fine-needle aspiration biopsies (FNAB) of thyroid nodules and can be of help in diagnosing cancer in a patient and treating patients with thyroid nodules. The American Thyroid Association identified mutations in seven genes and recommended, in its guidelines, that a seven-gene molecular biomarker panel of genetic mutations and rearrangement be set up and tested in FNAB samples [3–5]. This panel consists of BRAFV600E, three isoforms of RAS point mutations and translocations of PAX8/PPARγ and RET/PTC genes. The identification of any of these genetic changes in a thyroid nodule would represent a higher risk of malignancy, which is particularly important for a high number of patients who present with non-specific FNAB cytology. However, there is mounting evidence that the seven-gene MT test shows wide variation, ranging from 44% to 100% [3–6].
Although important advances have been made in the identification of specific genetic alterations and the fundamental role of several signaling pathways in thyroid cancer pathogenesis, 30%–35% of differentiated thyroid carcinomas, including PTC, lack any of these alterations [7]. Therefore, a pressing need to find new and more relevant molecular biomarkers to aid early diagnosis of PTC in order to rule in the malignancy for cytology indeterminate nodules exists.
Risk stratification of diagnosed patients represents another major issue in PTC management. The key outcome to predict is persistent/recurrent disease since, for the majority of PTC cases, the mortality risk is low. PTC tumors are slow growing, so patients usually have an excellent prognosis. However, reports by several groups show that 20%–30% of cases develop recurrence [8–10]. In rare cases, PTC may progress to an undifferentiated thyroid tumor, or the tumor may lose all the differentiation markers and transform into an anaplastic thyroid carcinoma (ATC), a very aggressive form of tumor which is characterized by poor prognosis with very low survival rates [10, 11]. Currently, the prognosis of PTC is essentially based on clinical and pathological factors; among them are: the patient’s age, tumor size, nodal and distant metastases, extrathyroid spread, and histotype [10, 12]. More recent studies suggest that mutational and expressional molecular alterations could be a significant addition to conventional evaluation and a critical addition toward personalized treatment of PTC patients [13–16].
Von Hippel–Lindau (VHL) is a tumor suppressor gene, and loss of its suppressor function is seen in heritable cancers linked with VHL syndrome as well as in some sporadic cancers [17, 18]. A number of reports have shown that VHL protein plays a critical role in oxygen signal transduction, but there is growing evidence to suggest that the function of VHL is likely to extend beyond this and that the loss of its function may result in deregulation of several signaling pathways that have critical roles in biological processes, notably cell proliferation, survival, invasion, and metastasis [19, 20]. In recent years, several lines of evidence, including our two studies, suggest that the VHL gene plays an important role in the development and progression of PTC. This evidence is the focus of the present narrative review.
Application scope and effect of existing molecular markers of PTC
RET-RAS-RAF-MAPK pathway is commonly found activated in PTC. It promotes cell growth, differentiation, proliferation, and survival. The RET/PTC gene rearrangements, RAS-family genes, and point mutations in the BRAF are the most usual genetic changes that activate this pathway in PTC. These genetic mutations are responsible for up to 70% of all PTCs. These genes work independently of each other since each can result in uncontrolled downstream effects. They can therefore be characterized as virtually mutually exclusive [7, 21, 22].
RET gene rearrangements, known as RET/PTC, are identified in about 20% of adults with PTC, 40%–70% of children and adolescents with sporadic PTC, and in 50%–86% of irradiated patients [23]. RET/PTC1 and RET/PTC3 are the most common RET/PTC rearrangements found in PTC. Some pathological features of PTC, e.g., large tumor size and lymph node involvement, are found to correlate with RET/PTC rearrangements [24].
The most common mutation found in PTC in adults is the thymidine to adenine conversion at nucleotide 1799 of exon 15 of the BRAF gene. This has a frequency of 29%–83%, and results in a valine to glutamic acid substitution at amino acid residue 600 (BRAFV600E) [22, 25]. To a lesser degree, BRAFV600E is also detected in poorly differentiated thyroid carcinoma and ATC arising from PTC. This accords with the results in model cells, which suggest that BRAFV600E is involved in dedifferentiation, genomic instability as well as increased invasiveness of cancer [26]. Numerous studies show that BRAF mutation correlates with advanced disease, the incidence in older age, classical papillary as well as poor prognosis and poorer overall survival [27].
Around 11% of PTCs are found to have RAS gene family mutations (0%–11%) [28]. The highest incidence is found in the follicular variant of PTC, 43%. Mutations in the RAS gene generally affect codon 61 of H-RAS and N-RAS and, less often, codons 12 and 13. Mutations in the other codons and in the K-RAS gene are rare [28]. Tumors, which harbor the RAS mutation, are invariably found encapsulated, have a follicular morphology, and show lower rates of nodal disease resulting in a more favorable prognosis. Furthermore, other studies have shown a high rate of RAS mutations in benign tumors, e.g., up to 50% micro follicular adenomas possess RAS mutations. This suggests that these genetic mutations may be the result of an early event in follicular thyroid tumorigenesis [29]. Additionally, RAS mutations are also found in about 50% of poorly differentiated and ATCs and these mutations correlate with poor patient survival [30]. This is highly indicative of the distinct roles that RAS may play in the early and late stages of thyroid cancer.
In one of our earlier studies [31], the above-mentioned genetic alterations were detected in 150 of 266 Serbian PTC patients (56.4%). BRAFV600E was the most abundant mutation noted (84/266, 31.6%). RET/PTC rearrangements were found in 55/266 (20.7%) cases, the RAS mutations were the least frequently seen (11/ 266, 4.1%). We concluded that following radical thyroid surgery followed by radioiodine ablation, BRAFV600E may not be an appropriate measure of poor disease-free survival during the early and middle follow-up period [31].
Other genetic alterations have been identified in PTC, such as PTEN and PIK3CA mutations [7]. However, their prevalence of approximately 1%–2% and lack of specificity limit their biomarker potential in PTC. In the past decade, a significant number of studies were focused on telomerase reverse transcriptase (TERT) promoter mutations in thyroid cancer, as reviewed in [32]. Two TERT promoter mutations, C228T and C250T, have been identified, having a prevalence of 11.3% in PTC. They have been found to be associated with aggressive PTC features, tumor recurrence, and patient mortality. Moreover, in coexistence with BRAFV600E, they show a strong synergistic effect on PTC aggressiveness [32].
A number of gene expression profiles have been identified and proposed for the prediction/prognosis of PTC by various studies [33–36]. However, this is an evolving field and these results need to be reproduced and confirmed by other studies in order to pave their way to clinical practice.
Aside from the gene expression at the mRNA level, expression alterations at the protein level might also have a significant biomarker potential in PTC. A recent systematic review and meta-analysis of the programmed death-ligand 1 (PD-L1) expression level in thyroid carcinoma pointed to the PD-L1 protein expression as a potential biomarker of disease-recurrence in patients with PTC [37].
MicroRNA expression profiles have also been the focus of a plethora of studies investigating their potential as diagnostic/prognostic/predictive biomarkers in PTC and a great number of microRNAs have been found to have deregulated expression [38, 39]. A meta-analysis, including 15 studies involving 807 PTC patients, found that expression levels of miRs-21, -34b, -130b, -135b, -146b, -151, -181b, -199b-5p, -221, -222, -451, -623, -1271, -2861, and let-7e showed significant association with at least one aggressive feature, such as large tumor size, extrathyroidal extension, multifocality, vascular invasion, lymph node metastases, distant metastasis, advanced TNM stage, and presence of the BRAF(V600E) mutation [40]. According to several reports, PTC is most consistently associated with the overexpression of miR-146b, miR-221, and miR-222. Considering that overexpression of these three microRNAs is frequently associated with more aggressive PTC features, their expression profile has been proposed as a potential prognostic biomarker of PTC [39, 41, 42].
VHL tumor suppressor
VHL gene
The VHL gene, located on chromosome 3p25, is 10 kb and comprises of three exons (Figure 1). Distinct isoforms, derived from alternative spliced transcripts have been observed. The best studied is transcript variant 1, which contains all three exons and results in two translation products: a 28- to 30-kDa 213 amino acid protein (pVHL30) translated from the first methionine codon and an 18- to 19-kDA 160 amino acid protein (pVHL19), translated from the second methionine at codon 54. In comparison to pVHL30, the first 53 amino acids are absent from pVHL19 and are less evolutionarily conserved than the rest of the protein [17]. The functional significance of this region is unclear. Both pVHL19 and pVHL30 are biologically active, have equivalent effects in functional assays, and display tumor suppressor activity in in vivo assays [43–47].
Figure 1.
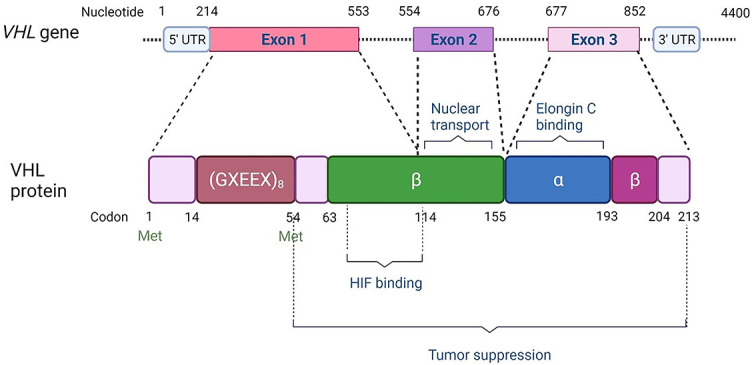
VHL gene and protein structure. The figure is modified under CC BY, based on [133].
VHL disease is a cancer syndrome which is inherited in a dominant manner and its development predisposes to a number of other cancers linked to mutations in the VHL gene. The disease shows marked variation in expression with multifocal and highly vascularized tumors in both mesenchymal and neural crest-derived tissues of multiple organ systems, such as the endocrine system (islet cell tumor), central nervous system (haemangioblastoma—CNS HB), adrenal medulla (pheochromocytoma—PHE), eye (retinal haemangioblastoma—RB), and kidney (clear renal cell carcinoma—cRCC) [17, 18]. Most of the VHL disease cases examined have been shown to exhibit autosomal inherited germline mutations in the VHL gene with over 1000 germline and somatic mutations reported [48]. Within the characterized gene alterations, missense mutations account for approximately 52%, frameshift and nonsense mutations account for 13% and 11%, respectively, inframe indels for about 6%, and deletion of the whole gene for accounts for about 11%. These alterations can be found throughout the coding sequence [48, 49]. Sporadic RCC and CNS have been reported to exhibit somatic mutations in the VHL gene [50] while in other sporadic cancers, such as breast, colon, lung, prostate, and thyroid, they are very rare [51, 52].
VHL protein function and its role in tumor suppression
The VHL protein forms a part of a multiprotein complex. This complex has E3 ubiquitin ligase activity that results in polyubiquitination and proteosomal degradation of particular target proteins. Other members of this complex are elongin B, elongin C, cullin-2 (CUL2), and RING-box1 (RBX1). The main role of VHL in the complex is recognition of the specific protein targets, which are then marked for degradation [53–55]. One particular protein target of VHL, the hypoxia-inducible factor-a (HIF-α), has been the focus of many studies. HIF-α is a transcription factor which plays a pivotal role in the regulation of gene expression by oxygen [55–59] (Figure 2).
Figure 2.
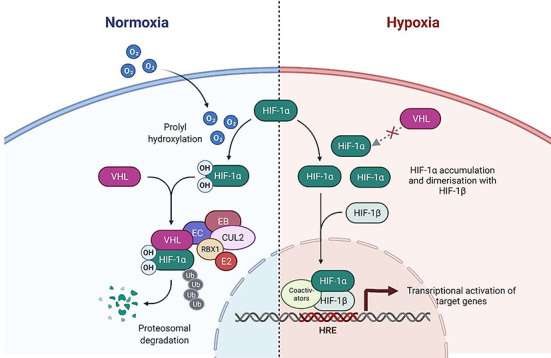
Regulation of the HIF-α by E3 ubiquitin ligase complex in normoxic and hypoxic conditions. Adapted from “HIF Signaling,” by BioRender.com (2021). Retrieved from https://app.biorender.com/biorender-templates.
HIF-α is recognized and marked for degradation under normoxia conditions. In cells exposed to low oxygen levels or which lack functional VHL, HIF-α subunits accumulate and complex with the HIF-β subunit, forming heterodimers. Formation of this heterodimer results in the activation of a number of genes leading to the production of proteins involved in cell adaptation to hypoxia and regulation of angiogenesis [55, 59, 60].
Accumulating evidence suggests that the function of VHL is broader than its established role in oxygen signal transduction. Moreover, the loss of VHL function may affect the regulation of other signaling pathways with important roles in biological processes, such as cell survival, invasion, proliferation, and metastasis [19, 20, 61]. It was found that the VHL protein interacts with a variety of other proteins in the cell, leading to their degradation or inhibition. For example, subsequent to VHL protein interaction, the HIF deubiquitinating enzymes VDU1/2 [62, 63] and Rpb1 subunit of RNA polymerase II are marked for degradation [64]. Studies show that VHL can also inhibit activity in several members of the protein kinase C family [65–67] and the activity of the Sp1 transcription factor [68, 69]. Furthermore, VHL was demonstrated to interact with the ubiquitously expressed Hu family RNA-binding protein HuR [70] that plays a part in mRNA stabilization and, with fibronectin, contributing to the proper assembly of the extracellular matrix [71, 72]. VHL was also found to interact with microtubules and protect them from depolymerization [73]. According to some studies, VHL acts as a positive regulator of the tumor suppressor TP53 (tumor protein p53) by inhibiting its Mdm2-mediated ubiquitination, and by subsequent recruitment of p53-modifying enzymes [74, 75]. On the other hand, there is evidence that VHL negatively regulates p53 activity by controlling the formation of p53 tetramers and reducing the binding of p53 at the promoters of the target genes [76]. A number of other VHL substrates/binding partners and associated signaling pathways have recently been identified, as extensively reviewed elsewhere [61].
There is mounting evidence that VHL performs a wide variety of HIF-α-dependent as well as HIF-α-independent functions affecting thus different cellular processes, some of which have a crucial role in tumorigenesis [50, 77]. It is still unclear, however, to which extent these HIF-α-dependent and HIF-α-independent functions cooperate during the process of tumorigenesis. A summary of the VHL protein functions and their associations with various processes implicated in tumor pathology is given in Tables 1A and 1B.
Table 1A.
HIF-α-dependent functions of VHL protein and their association with cellular processes involved in tumor development and progression. The table is based on data from [50, 55, 61, 118–120]
| Process | HIF-α-dependent functions |
|---|---|
| Cell proliferation and survival | Regulation of TGFα and EGFR |
| Apoptosis | HIF modulation of p53and NF-κB activity, and suppression of BNIP3 |
| Cell cycle progression | Regulation of cyclin D1 |
| Angiogenesis | Regulation of VEGF, PDGF among others |
| Glucose uptake and metabolism | Regulation of GLUT1, GLUT3, HK2, PGK1, LDHA, PFK, and PDH, among others |
| Microtubule stabilization and maintenance of the primary cilium | Primary cilia modulation |
| Chemotaxis | Regulation of SDF1 and CXCR4 |
| Assembly and regulation of the extracellular matrix | Regulation of E-cadherin and MMPs |
| Homeostasis | Regulation of external pH through CAIX |
HIF: Hypoxia-inducible factor; TGFα: Transforming growth factor-α; EGFR: Epidermal growth factor receptor; NF-κB: Nuclear factor-κB; BNIP3: BCL2/adenovirus E1B-interacting protein 3; VEGF: Vascular endothelial growth factor; PDGF: Platelet-derived growth factor; GLUT: Glucose transporter; HK2: Hexokinase 2, PGK1: Phosphoglyceratekinase 1; LDHA: Lactate dehydrogenase A; PFK: Phosphofructokinase; PDH: Pyruvate dehydrogenase; SDF1: Stromal-cell derived factor 1 (encoded by CXCL12); CXCR4: CXC-chemokine receptor 4; MMPs: Matrix metalloproteinases; CAIX: Carbonic anhydrase IX.
Table 1B.
HIF-α-independent functions of VHL protein and their association with cellular processes involved in tumor development and progression. The table is based on data from [50, 55, 61, 118–120]
| Process | HIF-α-independent functions |
|---|---|
| Cell proliferation and survival | Regulation of NDRG3, which accumulates by binding to lactate under hypoxia and further interacts with c-Raf for the activation of the Raf-ERK pathway. Regulation of AKT–VHL binds to hydroxylated AKT induced by EglN1 and inhibits its phosphorylation and kinase activity. |
| Apoptosis | Activation of p53 transcriptional activity, modulation of NF-κB activity and downregulation of JUNB (which is known to blunt neuronal apoptosis during NGF withdrawal). |
| Cell cycle progression | VHL targets B-Myb (MYBL2) for ubiquitination and proteasome degradation |
| Cell senescence | Control of cell senescence through RB and the SWI2/SNF2 chromatin remodeller p400 |
| Transcriptional regulation | Involvement in ubiquitylation of the large subunit of RNA polymerase II in response to oxidative stress, control of influence on HuR, binding to SP1 transcription factor |
| Microtubule stabilization and maintenance of the primary cilium | Association and stabilization of microtubules. Ubiquitination of Aurora kinase A (AURKA) independent of oxygen-dependent PHD activity to regulate formation of the primary cilium in quiescent cells |
| Assembly and regulation of the extracellular matrix | Regulation of fibronectin, collagen IV, adherens, tight junction, integrins and MMPs |
| Homeostasis | Control of cell senescence through RB and the SWI2/SNF2chromatin remodeller p400 |
| Inflammation | VHL functions as an adaptor that promotes the inhibitory phosphorylation of the NF-κB agonist, Card9, by enhancing the interaction between Card9 and CK2 |
| Cell growth and proliferation | Interacts with RAPTOR and increases RAPTOR degradation by ubiquitination, thereby inhibiting mTORC1 signaling |
| Cell growth, apoptosis, cell differentiation, stem-cell self-renewal | Negative regulation of c-Myc transcription |
| Anthracycline cytotoxicity regulation | Transcritional regulation of ALDH2 through interaction with its transcription factor HNF-4α |
NDRG3: N-Myc downstream-regulated gene 3; NF-κB: Nuclear factor-κB; HIF: Hypoxia-inducible factor; HuR: Human antigen R (also known as ELAV1); NGF: Nerve growth factor; RB: Retinoblastoma protein; CARD9: Caspase Recruitment Domain Family Member 9; CK2: Casein kinase II; RAPTOR: Regulatory-associated protein of the mechanistic target of rapamycin complex 1 (mTORC1); ALDH2: Aldehyde dehydrogenase 2; HNF-4α: Hepatocyte nuclear factor 4 alpha.
VHL expression in PTC
VHL has been shown to be aberrantly expressed in a number of human cancers. These include kidney, colon, breast, gastric cancer, and MEN2-associated medullary thyroid cancer [51, 78, 79]. A few studies have investigated the potential involvement of VHL in PTC development and/or progression. The VHL protein is highly expressed in normal thyroid follicular tissue and is differentially expressed in non-neoplastic and neoplastic thyroid lesions in proportion to the level of tumor differentiation [80–82]. This led to our hypothesis that VHL may be involved in the development of PTC. Consequently, we conducted a study evaluating mutation and methylation status as well as levels of expression of the VHL gene in tumour samples from 264 patients presenting with PTC. We found no somatic mutations or evidence of VHL downregulation via promoter hypermethylation. However, we found strong evidence of deregulated VHL expression at the mRNA level. Moreover, low VHL mRNA levels showed a strong correlation with patients’ older age, advanced clinical stage of the disease, classical PTC histovariant, and tumor multifocality. We also detected a marginal influence of low VHL expression on disease-free interval [83]. Our study was the first to demonstrate the association between VHL levels and clinico-pathological parameters in PTC, providing evidence of the involvement of VHL tumor suppressor in PTC pathology.
Later, in a similar study, Baldini et al. measured the expression levels of the two VHL mRNA splicing variants, VHL-213 (V1) and VHL-172 (V2), in a series of 96 PTC and corresponding normal thyroid tissues. They reported that expression of VHL was deregulated in most of the PTC tissues analyzed, and that the percent of samples with downregulated expression levels of both splicing variants was slightly higher than the percent of samples with upregulated V1 and V2 expression levels [84]. The mechanisms responsible for VHL gene expression regulation were not investigated in this study. In our second study on VHL in PTC [85], we compared the expression levels of VHL mRNA in another tumor series consisting of 42 pairs of PTCs and matched non-tumor thyroid tissues. The results showed that compared to corresponding non-tumor thyroid tissues, the levels of VHL in tumor tissues were either up- or downregulated, which was in line with the results of Baldini et al. [84], despite the opposite trend in the percent of the decreased and increased cases in these two studies. We also evaluated the association between VHL expression levels and clinico-pathological parameters in this patient cohort. Our data showed that lower VHL levels were significantly associated with extrathyroid spread and capsular invasion and there was a trend toward association with the presence of lymph node metastases, which led to the overall conclusion, consistent with our first study of VHL in PTC, that VHL downregulation might be associated with more aggressive tumor features, at least in some PTC cases.
Later on, two other studies addressed the status of VHL in PTC. Zang et al. [86], who evaluated the VHL expression in PTC and corresponding normal thyroid tissues in a group of 52 PTC patients, reported that VHL levels were significantly decreased in PTC. Deregulated VHL gene expression was also found in a recent study conducted on 20 primary tumor and metastatic PTC tissue. Interestingly, lower VHL mRNA levels were found in primary tumors compared to metastatic tissues. In primary tumors, BRAFV600E positive status was associated with higher levels of VHL, while in metastatic tissue, it was associated with lower VHL levels [87].
Summarized results produced from other gene profiling studies showed differences in the expression of over 200 other genes shared between PTC and normal thyroid tissues. The upregulated expression of LGALS3, SERPINA1, MET, KRT19, FN1, and TIMP1 was found within the existing data, as well as downregulated expression of TPO, SLC26A4, DIO1/2, and TFF3 in the well differentiated thyroid carcinomas but there was no evidence of deregulated expression of VHL [42]. This could be attributed to the small sample size of most of the studies or the cut-off values for differential expression being set too high. On the other hand, VHL has been reported to be included in a robust predictive signature for patients with breast cancer. Based on RNA-seq data from The Cancer Genome Atlas and several Gene Expression Omnibus datasets, a 14-gene hypoxia-related signature, which included VHL, was developed and the findings revealed that this signature could serve as a potential prognostic biomarker for breast cancer [88].
VHL expression in other types of cancer
Looking at existing data on the expression of VHL and its correlation with clinicopathological features in other cancer types, Zia et al. [89] reported that in highly aggressive breast cancer cell lines, VHL was either not expressed or was expressed at a low level, affecting cell motility and invasiveness. Zia et al. also found that, in higher grade breast cancer tumors, VHL was expressed at a much lower level compared to its expression in lower grade breast cancer tumors. The downregulated expression of VHL was also seen in tumors from patients with nodal and distant metastasis [89]. A study on ovarian cancer cells also showed that the loss of VHL increased cell aggressiveness [90]. Reduced pVHL expression has also been shown to be correlated with decreased apoptosis and a higher grade of chondrosarcoma [91]. Hoebeeck et al. [92] report that neuroblastoma patients also show a strong correlation between reduced levels of VHL and a poorer outcome in terms of patients’ survival. Similarly, in clear cell renal cell carcinoma, the increase in tumor aggressiveness was found to correlate with reduced expression of VHL identifying VHL downregulation as a risk factor for worse patient overall survival [93]. According to a study of Li et al. [94], although no correlations were observed with patient age, sex, tumor size, lymph node metastasis, or distant metastasis, negative VHL expression associated with a worse prognosis in patients with hepatocellular carcinoma. In a recent study on bladder cancer, differential under-expression of VHL—both mRNA and protein—was found in muscle-invasive bladder cancer in comparison to non-muscle-invasive bladder cancer [95].
Major mechanisms of VHL gene inactivation in cancer
Inactivation of the VHL gene can result from various alterations, such as intragenic mutations, mitotic recombination events, and promoter hypermethylation. VHL gene mutations were found in tumors associated with VHL syndrome as well in some sporadic tumors, such as clear-cell renal carcinomas, hemangioblastomas, and sporadic pheochromocytomaarise harbor VHL gene mutations [96–98]. Somatic VHL mutations on the other hand are rare in histological tumor types not present in VHL disease [51]. The results of our study, which found no evidence for mutations or homozygous deletions of the VHL are consistent with these reports [83]. However, loss of heterozygosity at chromosome 3p, including the VHL gene locus (3p25), was reported in one study [99] where it was found in 29% of PTCs.
The other common mechanism of gene inactivation is the hypermethylation of the promoter region. The VHL gene has been found to be silenced by methylation in 20%–30% of individuals with renal cell carcinoma, acute myeloid leukemia, or multiple myeloma [100–102] while in plasma cell neoplasia methylation of the VHL promoter is a common event [103]. In a recent study on bladder cancer, promoter methylation of the VHL gene was detected in almost 43% of bladder cancer samples, with high methylation being more frequent in muscle-invasive bladder cancer than in non-muscle-invasive bladder cancer [95]. Methylation of the VHL promoter was also detected in different stages of cervical carcinoma [104]. Several groups have reported the presence of epigenetic modifications in thyroid. Promoter hypermethylation was detected in the following tumor suppressors: CDH1, p16INK4A, RASSF1A, SLC5A8, TIMP3, DAPK, MGMT, DNMT1, MLH1, and RARB among others [105–110]. The methylation status of VHL in PTC patients has so far been addressed by only a couple of studies. Migdalska-Sk et al. [111] analyzed the methylation levels of eight tumor suppressor genes, including VHL, in PTC and control, non-cancerous thyroid tissues. According to this study, the highest methylation rate—100%, was found in ARHI, CDH1, p16INK4A, and RASSF1A but the frequency of promoter methylation of the VHL gene was the lowest, both in PTC and noncancerous tissues [111]. Similarly, the analysis of our PTC sample series with reduced VHL levels did not find evidence for VHL gene silencing through methylation. However, since our analysis covered only one part of the VHL promoter we could eliminate the possibility of the presence of methylation in the promoter regions that were not analyzed in our studies [83].
Small non-coding RNAs (microRNAs, miRNAs) have a significant role in gene expression downregulation [112]. This is a class of ~22 nucleotides long non-coding RNAs involved in the posttranscriptional regulation of gene expression. They typically bind to the 3 untranslated regions (UTRs) of target gene mRNAs, which leads to degradation or to translation inhibition of the target mRNA, resulting in expression downregulation of their protein products [113]. Since their discovery, a plethora of studies have demonstrated the importance of miRNAs in cancer biology, with their activity being shown to affect a number of crucial processes in tumorigenesis, such as tumor growth, invasion, angiogenesis, and immune evasion. Depending on their targets, miRNAs can function as oncogenes or tumor suppressors [114]. A number of miRNAs were reported to target VHL directly, downregulating its expression in different cancers, as summarised in Table 2.
Table 2.
MicroRNAs experimentally confirmed to regulate VHL expression in different types of cancer cells
| microRNA | Type of cancer cells | Reference |
|---|---|---|
| miR-17-5p | Renal cell carcinoma cells | [121] |
| miR-21 | Hepatic stellate cells; papillary thyroid carcinoma; pancreatic carcinoma; cervical carcinoma cells | [86, 122–124] |
| miR-23b | Glioma cells | [125] |
| miR-92 | Epithelial ovarian carcinoma; clear cell renal cell carcinoma | [116, 117] |
| miR-101 | Breast carcinoma cells | [126] |
| miR-150 | Glioma cells | [127] |
| miR-155 | Breast carcinoma cells | [78] |
| miR-222 | Retinoblastoma cells | [128] |
| miR-224 | Renal cell carcinoma cells | [121] |
| miR-331-3p | Hepatocellular carcinoma cells | [129] |
| miR-429 | HER2+ breast carcinoma cells | [130] |
| miR-566 | Glioblastoma cells | [131] |
| miR-887 | Hepatocellular carcinoma cells | [132] |
So far, few studies have addressed the regulation of VHL by miRNAs in PTC. Zang et al. [86] showed that VHL can be a potential target of miR-21 in PTC cells. MiR-21 is an oncomiR involved in the tumorogenisis of a number of different cancers [115] and, according to several reports, as summarized in Table 2, it can directly target VHL in different cancers. In one of our studies, we measured the expression levels of VHL and another well documented oncomiR—miR-92a-3p—and explored the correlation between them in PTC and nontumor thyroid tissue. We found that both VHL and miR-92a were deregulated in PTC but a negative correlation between them existed only in a subgroup of PTCs with vascular invasion. Based on these results, we can speculate that VHL, at least at some points during tumor progression, might be regulated by miR-92a-3p in PTC as well, since the possibility of their direct interaction was demonstrated in renal cell carcinoma and epithelial ovarian carcinoma cells [116, 117]. However, more research needs to be done in order to discover the complex interaction network between VHL and functionally related miRNAs in different stages of PTC development and progression, as well as to clarify their roles in disease progression and their prognostic utility.
Conclusion
The VHL tumor suppressor has been implicated in the development of a dominantly inherited cancer syndrome known as VHL disease, as well as a number of sporadic cancers. By regulation of the availability of HIF-α in the cell, the VHL has important effects on the tumorigenesis of the cell. VHL protein negatively controls angiogenesis, a critical factor in the progression of cancer. Accumulating evidence strongly indicates that VHL is also involved both through HIF-α-dependent as well as HIF-α-independent actions in several other processes, such as cell proliferation and survival, cell cycle progression, apoptosis, extracellular matrix regulation, inflammation, etc. Moreover, the latest evidence suggests that aside from a tumor suppressor function, VHL may also demonstrate pro-tumor function in some circumstances. In this context, VHL has definitively been shown to be a strong potential candidate as a biomarker and/or a therapeutic target in cancer. However, more research needs to be done since the complexity of its role in the cell, both in normal and pathological conditions, has only recently started to be revealed. So far, just a few studies have investigated VHL in papillary thyroid cancer, and all reported it to be deregulated. The significance of this deregulation, as well as its potential as a diagnostic/prognostic biomarker has yet to be clarified. In this review, we summarized the existing knowledge about the implication of VHL in PTC pathogenesis with the aim to bring attention to it and emphasize its potential utility as an expression biomarker for the stratification of PTC patients into high and low risk groups for recurrent disease.
Acknowledgments
Figures in this article were created with BioRender.com
Conflicts of interest: The authors declare no conflicts of interest.
Funding: This work was funded by the Ministry of Education, Science and Technological Development of The Republic of Serbia, grant number 451-03-9/2021-14/ 200017 and Cell and Gene Therapy group, King’s College London, The Rayne Institute, 123 Cold harbour Lane, London SE59NU, UK.
References
- 1. Cancer stat facts: thyroid cancer. [cited 2021 September 20]. Available from: https://seer.cancer.gov/statfacts/html/thyro.html.
- 2. ECIS - European cancer information system. 2021 [cited 2021 September 20]. Available from: https://ecis.jrc.ec.europa.eu/.
- 3.Nikiforov YE, Ohori NP, Hodak SP, Carty SE, Lebeau SO, Ferris RL, et al. Impact of mutational testing on the diagnosis and management of patients with cytologically indeterminate thyroid nodules: a prospective analysis of 1056 FNA samples. J Clin Endocrinol Metab. 2011;96(11):3390–7. doi: 10.1210/jc.2011-1469. https://doi.org/10.1210/jc.2011-1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yip L, Ferris RL. Clinical application of molecular testing of fine-needle aspiration specimens in thyroid nodules. Otolaryngol Clin North Am. 2014;47(4):557–71. doi: 10.1016/j.otc.2014.04.003. https://doi.org/10.1016/j.otc.2014.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nikiforov YE, Steward DL, Robinson-Smith TM, Haugen BR, Klopper JP, Zhu Z, et al. Molecular testing for mutations in improving the fine-needle aspiration diagnosis of thyroid nodules. J Clin Endocrinol Metab. 2009;94(6):2092–8. doi: 10.1210/jc.2009-0247. https://doi.org/10.1210/jc.2009-0247.0. [DOI] [PubMed] [Google Scholar]
- 6.Haugen BR, Alexander EK, Bible KC, Doherty GM, Mandel SJ, Nikiforov YE, et al. 2015 American Thyroid Association Management Guidelines for adult patients with thyroid nodules and differentiated thyroid cancer: The American Thyroid Association Guidelines Task Force on thyroid nodules and differentiated thyroid cancer. Thyroid. 2016;26(1):1–133. doi: 10.1089/thy.2015.0020. https://doi.org/10.1089/thy.2015.0020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xing M. Molecular pathogenesis and mechanisms of thyroid cancer. Nat Rev Cancer. 2013;13(3):184–99. doi: 10.1038/nrc3431. https://doi.org/10.1038/nrc3431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chan S, Karamali K, Kolodziejczyk A, Oikonomou G, Watkinson J, Paleri V, et al. Systematic review of recurrence rate after hemithyroidectomy for low-risk well-differentiated thyroid cancer. Eur Thyroid J. 2020;9(2):73–84. doi: 10.1159/000504961. https://doi.org/10.1159/000504961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Grogan RH, Kaplan SP, Cao H, Weiss RE, Degroot LJ, Simon CA, et al. A study of recurrence and death from papillary thyroid cancer with 27 years of median follow-up. Surgery. 2013;154(6):1436–47. doi: 10.1016/j.surg.2013.07.008. https://doi.org/10.1016/j.surg.2013.07.008. [DOI] [PubMed] [Google Scholar]
- 10.Tuttle RM, Ball DW, Byrd D, Dilawari RA, Doherty GM, Duh QY, et al. Thyroid carcinoma. J Natl Compr Canc Netw. 2010;8(11):1228–74. doi: 10.6004/jnccn.2010.0093. https://doi.org/10.6004/jnccn.2010.0093. [DOI] [PubMed] [Google Scholar]
- 11.Omur O, Baran Y. An update on molecular biology of thyroid cancers. Crit Rev Oncol Hematol. 2014;90(3):233–52. doi: 10.1016/j.critrevonc.2013.12.007. https://doi.org/10.1016/j.critrevonc.2013.12.007. [DOI] [PubMed] [Google Scholar]
- 12.Mcleod DSA. Progress towards improving initial risk stratification for patients with papillary thyroid cancer. Clin Endocrinol (Oxf) 2020;92(4):282–3. doi: 10.1111/cen.14163. https://doi.org/10.1111/cen.14163. [DOI] [PubMed] [Google Scholar]
- 13.Gomes-Lima CJ, Auh S, Thakur S, Zemskova M, Cochran C, Merkel R, et al. A novel risk stratification system for thyroid nodules with indeterminate cytology—a pilot cohort study. Front Endocrinol (Lausanne) 2020;11:53. doi: 10.3389/fendo.2020.00053. https://doi.org/10.3389/fendo.2020.00053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.He J, Tian Z, Yao X, Yao B, Liu Y, Yang J. A novel RNA sequencing-based risk score model to predict papillary thyroid carcinoma recurrence. Clin Exp Metastasis. 2020;37(2):257–67. doi: 10.1007/s10585-019-10011-4. https://doi.org/10.1007/s10585-019-10011-4. [DOI] [PubMed] [Google Scholar]
- 15.Hosseinkhan N, Honardoost M, Blighe K, Moore CBT, Khamseh ME. Comprehensive transcriptomic analysis of papillary thyroid cancer: potential biomarkers associated with tumor progression. J Endocrinol Invest. 2020;43(7):911–23. doi: 10.1007/s40618-019-01175-7. https://doi.org/10.1007/s40618-019-01175-7. [DOI] [PubMed] [Google Scholar]
- 16.Lin AJ, Samson P, Dewees T, Henke L, Baranski T, Schwarz J, et al. A molecular approach combined with American Thyroid Association classification better stratifies recurrence risk of classic histology papillary thyroid cancer. Cancer Med. 2019;8(1):437–46. doi: 10.1002/cam4.1857. https://doi.org/10.1002/cam4.1857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Maher ER, Kaelin WG., Jr von Hippel-Lindau disease. Medicine (Baltimore) 1997;76(6):381–1. doi: 10.1097/00005792-199711000-00001. https://doi.org/10.1097/00005792-199711000-00001. [DOI] [PubMed] [Google Scholar]
- 18.Lonser RR, Glenn GM, Walther M, Chew EY, Libutti SK, Linehan WM, et al. von Hippel-Lindau disease. Lancet. 2003;361(9374):2059–67. doi: 10.1016/S0140-6736(03)13643-4. https://doi.org/10.1016/S0140-6736(03)13643-4. [DOI] [PubMed] [Google Scholar]
- 19.Barry RE, Krek W. The von Hippel-Lindau tumour suppressor: a multi-faceted inhibitor of tumourigenesis. Trends Mol Med. 2004;10(9):466–72. doi: 10.1016/j.molmed.2004.07.008. https://doi.org/10.1016/j.molmed.2004.07.008. [DOI] [PubMed] [Google Scholar]
- 20.Czyzyk-Krzeska MF, Meller J. von Hippel-Lindau tumor suppressor: not only HIF’s executioner. Trends Mol Med. 2004;10(4):146–9. doi: 10.1016/j.molmed.2004.02.004. https://doi.org/10.1016/j.molmed.2004.02.004. [DOI] [PubMed] [Google Scholar]
- 21.Haroon Al Rasheed MR, Xu B. Molecular alterations in thyroid carcinoma. Surg Pathol Clin. 2019;12(4):921–30. doi: 10.1016/j.path.2019.08.002. https://doi.org/10.1016/j.path.2019.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Prete A, Borges De Souza P, Censi S, Muzza M, Nucci N, Sponziello M. Update on fundamental mechanisms of thyroid cancer. Front Endocrinol (Lausanne) 2020;11:102. doi: 10.3389/fendo.2020.00102. https://doi.org/10.3389/fendo.2020.00102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Santoro M, Melillo RM, Carlomagno F, Fusco A, Vecchio G. Molecular mechanisms of RET activation in human cancer. Ann NY Acad Sci. 2002;963:116–21. doi: 10.1111/j.1749-6632.2002.tb04102.x. https://doi.org/10.1111/j.1749-6632.2002.tb04102.x. [DOI] [PubMed] [Google Scholar]
- 24.Romei C, Ciampi R, Elisei R. A comprehensive overview of the role of the RET proto-oncogene in thyroid carcinoma. Nat Rev Endocrinol. 2016;12(4):192–202. doi: 10.1038/nrendo.2016.11. https://doi.org/10.1038/nrendo.2016.11. [DOI] [PubMed] [Google Scholar]
- 25.Acquaviva G, Visani M, Repaci A, Rhoden KJ, De Biase D, Pession A, et al. Molecular pathology of thyroid tumours of follicular cells: a review of genetic alterations and their clinicopathological relevance. Histopathology. 2018;72(1):6–31. doi: 10.1111/his.13380. https://doi.org/10.1111/his.13380. [DOI] [PubMed] [Google Scholar]
- 26.Mitsutake N, Knauf JA, Mitsutake S, Mesa C, Jr, Zhang L, Fagin JA. Conditional BRAFV600E expression induces DNA synthesis, apoptosis, dedifferentiation, and chromosomal instability in thyroid PCCL3 cells. Cancer Res. 2005;65(6):2465–73. doi: 10.1158/0008-5472.CAN-04-3314. https://doi.org/10.1158/0008-5472.CAN-04-3314. [DOI] [PubMed] [Google Scholar]
- 27.Vuong HG, Duong UN, Altibi AM, Ngo HT, Pham TQ, Tran HM, et al. A meta-analysis of prognostic roles of molecular markers in papillary thyroid carcinoma. Endocr Connect. 2017;6(3):R8–17. doi: 10.1530/EC-17-0010. https://doi.org/10.1530/EC-17-0010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Howell GM, Hodak SP, Yip L. RAS mutations in thyroid cancer. Oncologist. 2013;18(8):926–32. doi: 10.1634/theoncologist.2013-0072. https://doi.org/10.1634/theoncologist.2013-0072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vasko V, Ferrand M, Di Cristofaro J, Carayon P, Henry JF, De Micco C. Specific pattern of RAS oncogene mutations in follicular thyroid tumors. J Clin Endocrinol Metab. 2003;88(6):2745–52. doi: 10.1210/jc.2002-021186. https://doi.org/10.1210/jc.2002-021186. [DOI] [PubMed] [Google Scholar]
- 30.Garcia-Rostan G, Zhao H, Camp RL, Pollan M, Herrero A, Pardo J, et al. ras mutations are associated with aggressive tumor phenotypes and poor prognosis in thyroid cancer. J Clin Oncol. 2003;21(17):3226–35. doi: 10.1200/JCO.2003.10.130. https://doi.org/10.1200/JCO.2003.10.130. [DOI] [PubMed] [Google Scholar]
- 31.Stanojevic B, Dzodic R, Saenko V, Milovanovic Z, Pupic G, Zivkovic O, et al. Mutational and clinico-pathological analysis of papillary thyroid carcinoma in Serbia. Endocr J. 2011;58(5):381–93. doi: 10.1507/endocrj.k11e-054. https://doi.org/10.1507/endocrj.k11e-054. [DOI] [PubMed] [Google Scholar]
- 32.Liu R, Xing M. TERT promoter mutations in thyroid cancer. Endocr Relat Cancer. 2016;23(3):R143–55. doi: 10.1530/ERC-15-0533. https://doi.org/10.1530/ERC-15-0533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Saftencu M, Braicu C, Cojocneanu R, Buse M, Irimie A, Piciu D, et al. Gene expression patterns unveil new insights in papillary thyroid cancer. Medicina (Kaunas) 2019;55(8):500. doi: 10.3390/medicina55080500. https://doi.org/10.3390/medicina55080500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Arora C, Kaur D, Naorem LD, Raghava GPS. Prognostic biomarkers for predicting papillary thyroid carcinoma patients at high risk using nine genes of apoptotic pathway. PLoS One. 2021;16(11):e0259534. doi: 10.1371/journal.pone.0259534. https://doi.org/10.1371/journal.pone.0259534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shibata M, Inaishi T, Ichikawa T, Shimizu D, Soeda I, Takano Y, et al. Identifying the tumor-progressive gene expression profile in high-risk papillary thyroid cancer. Surg Today. 2021;51(10):1703–12. doi: 10.1007/s00595-021-02262-0. https://doi.org/10.1007/s00595-021-02262-0. [DOI] [PubMed] [Google Scholar]
- 36.Wang Y, Mckelvey BA, Liu Z, Rooper L, Cope LM, Zeiger MA, et al. Retrospective analysis of cancer-specific gene expression panel for thyroid fine needle aspiration specimens. J Cancer Res Clin Oncol. 2021;147(10):2983–91. doi: 10.1007/s00432-021-03706-3. https://doi.org/10.1007/s00432-021-03706-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Girolami I, Pantanowitz L, Mete O, Brunelli M, Marletta S, Colato C, et al. Programmed death-ligand 1 (PD-L1) is a potential biomarker of disease-free survival in papillary thyroid carcinoma: a systematic review and meta-analysis of pd-l1 immunoexpression in follicular epithelial derived thyroid carcinoma. Endocr Pathol. 2020;31(3):291–300. doi: 10.1007/s12022-020-09630-5. https://doi.org/10.1007/s12022-020-09630-5. [DOI] [PubMed] [Google Scholar]
- 38.Hitu L, Gabora K, Bonci EA, Piciu A, Hitu AC, Stefan PA, et al. MicroRNA in papillary thyroid carcinoma: a systematic review from 2018 to June 2020. Cancers (Basel) 2020;12(11):3118. doi: 10.3390/cancers12113118. https://doi.org/10.3390/cancers12113118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Papaioannou M, Chorti AG, Chatzikyriakidou A, Giannoulis K, Bakkar S, Papavramidis TS. MicroRNAs in papillary thyroid cancer: what is new in diagnosis and treatment. Front Oncol. 2021;11:755097. doi: 10.3389/fonc.2021.755097. https://doi.org/10.3389/fonc.2021.755097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Aragon Han P, Weng CH, Khawaja HT, Nagarajan N, Schneider EB, Umbricht CB, et al. MicroRNA expression and association with clinicopathologic features in papillary thyroid cancer: a systematic review. Thyroid. 2015;25(12):1322–9. doi: 10.1089/thy.2015.0193. https://doi.org/10.1089/thy.2015.0193. [DOI] [PubMed] [Google Scholar]
- 41.Celano M, Rosignolo F, Maggisano V, Pecce V, Iannone M, Russo D, et al. MicroRNAs as biomarkers in thyroid carcinoma. Int J Genomics. 2017;2017:6496570. doi: 10.1155/2017/6496570. https://doi.org/10.1155/2017/6496570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Abdullah MI, Junit SM, Ng KL, Jayapalan JJ, Karikalan B, Hashim OH. Papillary thyroid cancer: genetic alterations and molecular biomarker investigations. Int J Med Sci. 2019;16(3):450–460. doi: 10.7150/ijms.29935. https://doi.org/10.7150/ijms.29935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chou A, Toon C, Pickett J, Gill AJ. von Hippel-Lindau syndrome. Front Horm Res. 2013;41:30–49. doi: 10.1159/000345668. https://doi.org/10.1159/000345668. [DOI] [PubMed] [Google Scholar]
- 44.Woodward ER, Buchberger A, Clifford SC, Hurst LD, Affara NA, Maher ER. Comparative sequence analysis of the VHL tumor suppressor gene. Genomics. 2000;65(3):253–65. doi: 10.1006/geno.2000.6144. https://doi.org/10.1006/geno.2000.6144. [DOI] [PubMed] [Google Scholar]
- 45.Schoenfeld A, Davidowitz EJ, Burk RD. A second major native von Hippel-Lindau gene product, initiated from an internal translation start site, functions as a tumor suppressor. Proc Natl Acad Sci USA. 1998;95(15):8817–22. doi: 10.1073/pnas.95.15.8817. https://doi.org/10.1073/pnas.95.15.8817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Iliopoulos O, Ohh M, Kaelin WG., Jr pVHL19 is a biologically active product of the von Hippel-Lindau gene arising from internal translation initiation. Proc Natl Acad Sci USA. 1998;95(20):11661–6. doi: 10.1073/pnas.95.20.11661. https://doi.org/10.1073/pnas.95.20.11661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Decker J, Brauch H. Von Hippel-Lindau tumor suppressor gene, in Encyclopedia of cancer. Schwab M, editor. Berlin (Germany): Springer; 2011. pp. 3929–36. [Google Scholar]
- 48.Tabaro F, Minervini G, Sundus F, Quaglia F, Leonardi E, Piovesan D, et al. VHLdb: a database of von Hippel-Lindau protein interactors and mutations. Sci Rep. 2016;6:31128. doi: 10.1038/srep31128. https://doi.org/10.1038/srep31128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nordstrom-O’brien M, Van Der Luijt RB, Van Rooijen E, Van Den Ouweland AM, Majoor-Krakauer DF, Lolkema MP, et al. Genetic analysis of von Hippel-Lindau disease. Hum Mutat. 2010;31(5):521–37. doi: 10.1002/humu.21219. https://doi.org/10.1002/humu.21219. [DOI] [PubMed] [Google Scholar]
- 50.Gossage L, Eisen T, Maher ER. VHL, the story of a tumour suppressor gene. Nat Rev Cancer. 2015;15(1):55–64. doi: 10.1038/nrc3844. https://doi.org/10.1038/nrc3844. [DOI] [PubMed] [Google Scholar]
- 51.Kim WY, Kaelin WG. Role of VHL gene mutation in human cancer. J Clin Oncol. 2004;22(24):4991–5004. doi: 10.1200/JCO.2004.05.061. https://doi.org/10.1200/JCO.2004.05.061. [DOI] [PubMed] [Google Scholar]
- 52.Costa V, Esposito R, Ziviello C, Sepe R, Bim LV, Cacciola NA, et al. New somatic mutations and WNK1-B4GALNT3 gene fusion in papillary thyroid carcinoma. Oncotarget. 2015;6(13):11242–51. doi: 10.18632/oncotarget.3593. https://doi.org/10.18632/oncotarget.3593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Iwai K, Yamanaka K, Kamura T, Minato N, Conaway RC, Conaway JW, et al. Identification of the von Hippel-Lindau tumor-suppressor protein as part of an active E3 ubiquitin ligase complex. Proc Natl Acad Sci USA. 1999;96(22):12436–41. doi: 10.1073/pnas.96.22.12436. https://doi.org/10.1073/pnas.96.22.12436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lisztwan J, Imbert G, Wirbelauer C, Gstaiger M, Krek W. The von Hippel-Lindau tumor suppressor protein is a component of an E3 ubiquitin-protein ligase activity. Genes Dev. 1999;13(14):1822–33. doi: 10.1101/gad.13.14.1822. https://doi.org/10.1101/gad.13.14.1822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zurlo G, Guo J, Takada M, Wei W, Zhang Q. New insights into protein hydroxylation and its important role in human diseases. Biochim Biophys Acta. 2016;1866(2):208–20. doi: 10.1016/j.bbcan.2016.09.004. https://doi.org/10.1016/j.bbcan.2016.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Maxwell PH, Wiesener MS, Chang GW, Clifford SC, Vaux EC, Cockman ME, et al. The tumour suppressor protein VHL targets hypoxia-inducible factors for oxygen-dependent proteolysis. Nature. 1999;399(6733):271–5. doi: 10.1038/20459. https://doi.org/10.1038/20459. [DOI] [PubMed] [Google Scholar]
- 57.Ohh M, Park CW, Ivan M, Hoffman MA, Kim TY, Huang LE, et al. Ubiquitination of hypoxia-inducible factor requires direct binding to the beta-domain of the von Hippel-Lindau protein. Nat Cell Biol. 2000;2(7):423–7. doi: 10.1038/35017054. https://doi.org/10.1038/35017054. [DOI] [PubMed] [Google Scholar]
- 58.Kamura T, Sato S, Iwai K, Czyzyk-Krzeska M, Conaway RC, Conaway JW. Activation of HIF1alpha ubiquitination by a reconstituted von Hippel-Lindau (VHL) tumor suppressor complex. Proc Natl Acad Sci USA. 2000;97(19):10430–5. doi: 10.1073/pnas.190332597. https://doi.org/10.1073/pnas.190332597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tanimoto K, Makino Y, Pereira T, Poellinger L. Mechanism of regulation of the hypoxia-inducible factor-1 alpha by the von Hippel-Lindau tumor suppressor protein. EMBO J. 2000;19(16):4298–309. doi: 10.1093/emboj/19.16.4298. https://doi.org/10.1093/emboj/19.16.4298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kaelin WG. Von Hippel-Lindau disease. Annu Rev Pathol. 2007;2:145–73. doi: 10.1146/annurev.pathol.2.010506.092049. https://doi.org/10.1146/annurev.pathol.2.010506.092049. [DOI] [PubMed] [Google Scholar]
- 61.Zhang J, Zhang Q. VHL and hypoxia signaling: beyond HIF in cancer. Biomedicines. 2018;6(1):35. doi: 10.3390/biomedicines6010035. https://doi.org/10.3390/biomedicines6010035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Li Z, Na X, Wang D, Schoen SR, Messing EM, Wu G. Ubiquitination of a novel deubiquitinating enzyme requires direct binding to von Hippel-Lindau tumor suppressor protein. J Biol Chem. 2002;277(7):4656–62. doi: 10.1074/jbc.M108269200. https://doi.org/10.1074/jbc.M108269200. [DOI] [PubMed] [Google Scholar]
- 63.Li Z, Wang D, Messing EM, Wu G. VHL protein-interacting deubiquitinating enzyme 2 deubiquitinates and stabilizes HIF-1alpha. EMBO Rep. 2005;6(4):373–8. doi: 10.1038/sj.embor.7400377. https://doi.org/10.1038/sj.embor.7400377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kuznetsova AV, Meller J, Schnell PO, Nash JA, Ignacak ML, Sanchez Y, et al. von Hippel-Lindau protein binds hyperphosphorylated large subunit of RNA polymerase II through a proline hydroxylation motif and targets it for ubiquitination. Proc Natl Acad Sci USA. 2003;100(5):2706–11. doi: 10.1073/pnas.0436037100. https://doi.org/10.1073/pnas.0436037100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Okuda H, Hirai S, Takaki Y, Kamada M, Baba M, Sakai N, et al. Direct interaction of the beta-domain of VHL tumor suppressor protein with the regulatory domain of atypical PKC isotypes. Biochem Biophys Res Commun. 1999;263(2):491–7. doi: 10.1006/bbrc.1999.1347. https://doi.org/10.1006/bbrc.1999.1347. [DOI] [PubMed] [Google Scholar]
- 66.Pal S, Claffey KP, Dvorak HF, Mukhopadhyay D. The von Hippel-Lindau gene product inhibits vascular permeability factor/vascular endothelial growth factor expression in renal cell carcinoma by blocking protein kinase C pathways. J Biol Chem. 1997;272(44):27509–12. doi: 10.1074/jbc.272.44.27509. https://doi.org/10.1074/jbc.272.44.27509. [DOI] [PubMed] [Google Scholar]
- 67.Datta K, Nambudripad R, Pal S, Zhou M, Cohen HT, Mukhopadhyay D. Inhibition of insulin-like growth factor-I-mediated cell signaling by the von Hippel-Lindau gene product in renal cancer. J Biol Chem. 2000;275(27):20700–6. doi: 10.1074/jbc.M909970199. https://doi.org/10.1074/jbc.M909970199. [DOI] [PubMed] [Google Scholar]
- 68.Cohen HT, Zhou M, Welsh AM, Zarghamee S, Scholz H, Mukhopadhyay D, et al. An important von Hippel-Lindau tumor suppressor domain mediates Sp1-binding and self-association. Biochem Biophys Res Commun. 1999;266(1):43–50. doi: 10.1006/bbrc.1999.1767. https://doi.org/10.1006/bbrc.1999.1767. [DOI] [PubMed] [Google Scholar]
- 69.Mukhopadhyay D, Knebelmann B, Cohen HT, Ananth S, Sukhatme VP. The von Hippel-Lindau tumor suppressor gene product interacts with Sp1 to repress vascular endothelial growth factor promoter activity. Mol Cell Biol. 1997;17(9):5629–39. doi: 10.1128/mcb.17.9.5629. https://doi.org/10.1128/MCB.17.9.5629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Datta K, Mondal S, Sinha S, Li J, Wang E, Knebelmann B, et al. Role of elongin-binding domain of von Hippel Lindau gene product on HuR-mediated VPF/VEGF mRNA stability in renal cell carcinoma. Oncogene. 2005;24(53):7850–8. doi: 10.1038/sj.onc.1208912. https://doi.org/10.1038/sj.onc.1208912. [DOI] [PubMed] [Google Scholar]
- 71.Ohh M, Yauch RL, Lonergan KM, Whaley JM, Stemmer-Rachamimov AO, Louis DN, et al. The von Hippel-Lindau tumor suppressor protein is required for proper assembly of an extracellular fibronectin matrix. Mol Cell. 1998;1(7):959–68. doi: 10.1016/s1097-2765(00)80096-9. https://doi.org/10.1016/s1097-2765(00)80096-9. [DOI] [PubMed] [Google Scholar]
- 72.Kurban G, Hudon V, Duplan E, Ohh M, Pause A. Characterization of a von Hippel Lindau pathway involved in extracellular matrix remodeling, cell invasion, and angiogenesis. Cancer Res. 2006;66(3):1313–9. doi: 10.1158/0008-5472.CAN-05-2560. https://doi.org/10.1158/0008-5472.CAN-05-2560. [DOI] [PubMed] [Google Scholar]
- 73.Hergovich A, Lisztwan J, Barry R, Ballschmieter P, Krek W. Regulation of microtubule stability by the von Hippel-Lindau tumour suppressor protein pVHL. Nat Cell Biol. 2003;5(1):64–70. doi: 10.1038/ncb899. https://doi.org/10.1038/ncb899. [DOI] [PubMed] [Google Scholar]
- 74.Roe JS, Youn HD. The positive regulation of p53 by the tumor suppressor VHL. Cell Cycle. 2006;5(18):2054–6. doi: 10.4161/cc.5.18.3247. https://doi.org/10.4161/cc.5.18.3247. [DOI] [PubMed] [Google Scholar]
- 75.Semenza GL. VHL and p53: tumor suppressors team up to prevent cancer. Mol Cell. 2006;22(4):437–9. doi: 10.1016/j.molcel.2006.05.001. https://doi.org/10.1016/j.molcel.2006.05.001. [DOI] [PubMed] [Google Scholar]
- 76.Kinnaird A, Boukouris AE, Saleme B, Dromparis P, Zervopoulos SD, Gurtu V, et al. Interaction with p53 explains a pro-proliferative function for VHL in cancer. J Mol Med (Berl) 2020;98(9):1269–78. doi: 10.1007/s00109-020-01951-6. https://doi.org/10.1007/s00109-020-01951-6. [DOI] [PubMed] [Google Scholar]
- 77.Frew IJ, Krek W. Multitasking by pVHL in tumour suppression. Curr Opin Cell Biol. 2007;19(6):685–90. doi: 10.1016/j.ceb.2007.10.001. https://doi.org/10.1016/j.ceb.2007.10.001. [DOI] [PubMed] [Google Scholar]
- 78.Kong W, He L, Richards EJ, Challa S, Xu CX, Permuth-Wey J, et al. Upregulation of miRNA-155 promotes tumour angiogenesis by targeting VHL and is associated with poor prognosis and triple-negative breast cancer. Oncogene. 2014;33(6):679–89. doi: 10.1038/onc.2012.636. https://doi.org/10.1038/onc.2012.636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Koch CA, Brouwers FM, Vortmeyer AO, Tannapfel A, Libutti SK, Zhuang Z, et al. Somatic VHL gene alterations in MEN2-associated medullary thyroid carcinoma. BMC Cancer. 2006;6:131. doi: 10.1186/1471-2407-6-131. https://doi.org/10.1186/1471-2407-6-131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Corless CL, Kibel AS, Iliopoulos O, Kaelin WG., Jr Immunostaining of the von Hippel-Lindau gene product in normal and neoplastic human tissues. Hum Pathol. 1997;28(4):459–64. doi: 10.1016/s0046-8177(97)90035-6. https://doi.org/10.1016/s0046-8177(97)90035-6. [DOI] [PubMed] [Google Scholar]
- 81.Hinze R, Boltze C, Meye A, Holzhausen HJ, Dralle H, Rath FW. Expression of the von Hippel-Lindau tumor suppressor gene in nonneoplastic and neoplastic lesions of the thyroid. Endocr Pathol. 2000;11(2):145–55. doi: 10.1385/ep:11:2:145. https://doi.org/10.1385/ep:11:2:145. [DOI] [PubMed] [Google Scholar]
- 82.Sakashita N, Takeya M, Kishida T, Stackhouse TM, Zbar B, Takahashi K. Expression of von Hippel-Lindau protein in normal and pathological human tissues. Histochem J. 1999;31(2):133–44. doi: 10.1023/a:1003554712386. https://doi.org/10.1023/a:1003554712386. [DOI] [PubMed] [Google Scholar]
- 83.Stanojevic B, Saenko V, Todorovic L, Petrovic N, Nikolic D, Zivaljevic V, et al. Low VHL mRNA expression is associated with more aggressive tumor features of papillary thyroid carcinoma. PLoS One. 2014;9(12):e114511. doi: 10.1371/journal.pone.0114511. https://doi.org/10.1371/journal.pone.0114511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Baldini E, Tuccilli C, Arlot-Bonnemains Y, Chesnel F, Sorrenti S, De Vito C, et al. Deregulated expression of VHL mRNA variants in papillary thyroid cancer. Mol Cell Endocrinol. 2017;443:121–7. doi: 10.1016/j.mce.2017.01.019. https://doi.org/10.1016/j.mce.2017.01.019. [DOI] [PubMed] [Google Scholar]
- 85.Todorovic L, Stanojevic B, Mandusic V, Petrovic N, Zivaljevic V, Paunovic I, et al. Expression of VHL tumor suppressor mRNA and miR-92a in papillary thyroid carcinoma and their correlation with clinical and pathological parameters. Med Oncol. 2018;35(2):17. doi: 10.1007/s12032-017-1066-3. https://doi.org/10.1007/s12032-017-1066-3. [DOI] [PubMed] [Google Scholar]
- 86.Zang C, Sun J, Liu W, Chu C, Jiang L, Ge R. miRNA-21 promotes cell proliferation and invasion via VHL/PI3K/AKT in papillary thyroid carcinoma. Hum Cell. 2019;32(4):428–36. doi: 10.1007/s13577-019-00254-4. https://doi.org/10.1007/s13577-019-00254-4. [DOI] [PubMed] [Google Scholar]
- 87.Spirina LV, Chizhevskaya SY, Kovaleva IV, Kondakova IV. The association of the BRAF-V600E mutation with the expression of the molecular markers in the primary tumor and metastatic tissue in papillary thyroid cancer. Asian Pac J Cancer Prev. 2021;22(7):2017–24. doi: 10.31557/APJCP.2021.22.7.2017. https://doi.org/10.31557/APJCP.2021.22.7.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Wang J, Wang Y, Xing P, Liu Q, Zhang C, Sui Y, et al. Development and validation of a hypoxia-related prognostic signature for breast cancer. Oncol Lett. 2020;20(2):1906–14. doi: 10.3892/ol.2020.11733. https://doi.org/10.3892/ol.2020.11733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Zia MK, Rmali KA, Watkins G, Mansel RE, Jiang WG. The expression of the von Hippel-Lindau gene product and its impact on invasiveness of human breast cancer cells. Int J Mol Med. 2007;20(4):605–11. https://doi.org/10.3892/ijmm.20.4.605. [PubMed] [Google Scholar]
- 90.Liu T, Zhao L, Chen W, Li Z, Hou H, Ding L, et al. Inactivation of von Hippel-Lindau increases ovarian cancer cell aggressiveness through the HIF1alpha/miR-210/VMP1 signaling pathway. Int J Mol Med. 2014;33(5):1236–42. doi: 10.3892/ijmm.2014.1661. https://doi.org/10.3892/ijmm.2014.1661. [DOI] [PubMed] [Google Scholar]
- 91.Chen C, Zhou H, Liu X, Liu Z, Ma Q. Reduced expression of von Hippel-Lindau protein correlates with decreased apoptosis and high chondrosarcoma grade. J Bone Joint Surg Am. 2011;93(19):1833–40. doi: 10.2106/JBJS.I.01553. https://doi.org/10.2106/JBJS.I.01553. [DOI] [PubMed] [Google Scholar]
- 92.Hoebeeck J, Vandesompele J, Nilsson H, De Preter K, Van Roy N, De Smet E, et al. The von Hippel-Lindau tumor suppressor gene expression level has prognostic value in neuroblastoma. Int J Cancer. 2006;119(3):624–9. doi: 10.1002/ijc.21888. https://doi.org/10.1002/ijc.21888. [DOI] [PubMed] [Google Scholar]
- 93.Hogner A, Krause H, Jandrig B, Kasim M, Fuller TF, Schostak M, et al. PBRM1 and VHL expression correlate in human clear cell renal cell carcinoma with differential association with patient’s overall survival. Urol Oncol. 2018;36(3):94 e91–94 e14. doi: 10.1016/j.urolonc.2017.10.027. https://doi.org/10.1016/j.urolonc.2017.10.027. [DOI] [PubMed] [Google Scholar]
- 94.Li G, Shen Y, Wang F, Hong S, Cai M. Correlation between von Hippel-Lindau gene expression and tumor SUVmax and survival prognosis in hepatocellular carcinoma. Med Sci Monit. 2020;26:e920473. doi: 10.12659/MSM.920473. https://doi.org/10.12659/MSM.920473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Basu M, Chatterjee A, Chakraborty B, Chatterjee E, Ghosh S, Samadder S, et al. High nuclear expression of HIF1alpha, synergizing with inactivation of LIMD1 and VHL, portray worst prognosis among the bladder cancer patients: association with arsenic prevalence. J Cancer Res Clin Oncol. 2021;147(8):2309–22. doi: 10.1007/s00432-021-03661-z. https://doi.org/10.1007/s00432-021-03661-z. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 96.Sgambati MT, Stolle C, Choyke PL, Walther MM, Zbar B, Linehan WM, et al. Mosaicism in von Hippel-Lindau disease: lessons from kindreds with germline mutations identified in offspring with mosaic parents. Am J Hum Genet. 2000;66(1):84–91. doi: 10.1086/302726. https://doi.org/10.1086/302726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Chen F, Kishida T, Yao M, Hustad T, Glavac D, Dean M, et al. Germline mutations in the von Hippel-Lindau disease tumor suppressor gene: correlations with phenotype. Hum Mutat. 1995;5(1):66–75. doi: 10.1002/humu.1380050109. https://doi.org/10.1002/humu.1380050109. [DOI] [PubMed] [Google Scholar]
- 98.Zbar B, Kishida T, Chen F, Schmidt L, Maher ER, Richards FM, et al. Germline mutations in the Von Hippel-Lindau disease (VHL) gene in families from North America, Europe, and Japan. Hum Mutat. 1996;8(4):348–57. doi: 10.1002/(SICI)1098-1004(1996)8:4<348::AID-HUMU8>3.0.CO;2-3. . [DOI] [PubMed] [Google Scholar]
- 99.Grebe SK, Mciver B, Hay ID, Wu PS, Maciel LM, Drabkin HA, et al. Frequent loss of heterozygosity on chromosomes 3p and 17p without VHL or p53 mutations suggests involvement of unidentified tumor suppressor genes in follicular thyroid carcinoma. J Clin Endocrinol Metab. 1997;82(11):3684–91. doi: 10.1210/jcem.82.11.4352. https://doi.org/10.1210/jcem.82.11.4352. [DOI] [PubMed] [Google Scholar]
- 100.Herman JG, Latif F, Weng Y, Lerman MI, Zbar B, Liu S, et al. Silencing of the VHL tumor-suppressor gene by DNA methylation in renal carcinoma. Proc Natl Acad Sci USA. 1994;91(21):9700–4. doi: 10.1073/pnas.91.21.9700. https://doi.org/10.1073/pnas.91.21.9700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Dulaimi E, Ibanez De Caceres I, Uzzo RG, Al-Saleem T, Greenberg RE, Polascik TJ, et al. Promoter hypermethylation profile of kidney cancer. Clin Cancer Res. 2004;10(12 Pt 1):3972–9. doi: 10.1158/1078-0432.CCR-04-0175. https://doi.org/10.1158/1078-0432.CCR-04-0175. [DOI] [PubMed] [Google Scholar]
- 102.Benetatos L, Dasoula A, Syed N, Hatzimichael E, Crook T, Bourantas KL. Methylation analysis of the von Hippel-Lindau gene in acute myeloid leukaemia and myelodysplastic syndromes. Leukemia. 2008;22(6):1293–5. doi: 10.1038/sj.leu.2405053. https://doi.org/10.1038/sj.leu.2405053. [DOI] [PubMed] [Google Scholar]
- 103.Hatzimichael E, Dranitsaris G, Dasoula A, Benetatos L, Stebbing J, Crook T, et al. Von Hippel-Lindau methylation status in patients with multiple myeloma: a potential predictive factor for the development of bone disease. Clin Lymphoma Myeloma. 2009;9(3):239–42. doi: 10.3816/CLM.2009.n.047. https://doi.org/10.3816/CLM.2009.n.047. [DOI] [PubMed] [Google Scholar]
- 104.Chakraborty C, Mitra S, Roychowdhury A, Samadder S, Dutta S, Roy A, et al. Deregulation of LIMD1-VHL-HIF-1alpha-VEGF pathway is associated with different stages of cervical cancer. Biochem J. 2018;475(10):1793–806. doi: 10.1042/BCJ20170649. https://doi.org/10.1042/BCJ20170649. [DOI] [PubMed] [Google Scholar]
- 105.Hoque MO, Rosenbaum E, Westra WH, Xing M, Ladenson P, Zeiger MA, et al. Quantitative assessment of promoter methylation profiles in thyroid neoplasms. J Clin Endocrinol Metab. 2005;90(7):4011–8. doi: 10.1210/jc.2005-0313. https://doi.org/10.1210/jc.2005-0313. [DOI] [PubMed] [Google Scholar]
- 106.Hu S, Ewertz M, Tufano RP, Brait M, Carvalho AL, Liu D, et al. Detection of serum deoxyribonucleic acid methylation markers: a novel diagnostic tool for thyroid cancer. J Clin Endocrinol Metab. 2006;91(1):98–104. doi: 10.1210/jc.2005-1810. https://doi.org/10.1210/jc.2005-1810. [DOI] [PubMed] [Google Scholar]
- 107.Xing M. Gene methylation in thyroid tumorigenesis. Endocrinology. 2007;148(3):948–53. doi: 10.1210/en.2006-0927. https://doi.org/10.1210/en.2006-0927. [DOI] [PubMed] [Google Scholar]
- 108.Hu S, Liu D, Tufano RP, Carson KA, Rosenbaum E, Cohen Y, et al. Association of aberrant methylation of tumor suppressor genes with tumor aggressiveness and BRAF mutation in papillary thyroid cancer. Int J Cancer. 2006;119(10):2322–9. doi: 10.1002/ijc.22110. https://doi.org/10.1002/ijc.22110. [DOI] [PubMed] [Google Scholar]
- 109.Khatami F, Larijani B, Heshmat R, Nasiri S, Saffar H, Shafiee G, et al. Promoter methylation of four tumor suppressor genes in human papillary thyroid carcinoma. Iran J Pathol. 2019;14(4):290–8. doi: 10.30699/ijp.2019.94401.1922. https://doi.org/10.30699/ijp.2019.94401.1922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Botezatu A, Iancu IV, Plesa A, Manda D, Popa O, Bostan M, et al. Methylation of tumour suppressor genes associated with thyroid cancer. Cancer Biomark. 2019;25(1):53–65. doi: 10.3233/CBM-182265. https://doi.org/10.3233/CBM-182265. [DOI] [PubMed] [Google Scholar]
- 111.Migdalska-Sk M, Pastuszak-Lewandoska D, Czarnecka K, Nawrot E, Domańska D, Brzeziński J, et al. Methylation profile of selected TSGs in non-cancerous thyroid tissue adjacent to primary PTC. Contemporary Oncology. 2011;15(4):191–7. https://doi.org/10.5114/wo.2011.24312. [Google Scholar]
- 112.Acunzo M, Romano G, Wernicke D, Croce CM. MicroRNA and cancer—a brief overview. Adv Biol Regul. 2015;57:1–9. doi: 10.1016/j.jbior.2014.09.013. https://doi.org/10.1016/j.jbior.2014.09.013. [DOI] [PubMed] [Google Scholar]
- 113.Tetreault N, De Guire V. miRNAs: their discovery, biogenesis and mechanism of action. Clin Biochem. 2013;46(10–11):842–5. doi: 10.1016/j.clinbiochem.2013.02.009. https://doi.org/10.1016/j.clinbiochem.2013.02.009. [DOI] [PubMed] [Google Scholar]
- 114.Hayes J, Peruzzi PP, Lawler S. MicroRNAs in cancer: biomarkers, functions and therapy. Trends Mol Med. 2014;20(8):460–9. doi: 10.1016/j.molmed.2014.06.005. https://doi.org/10.1016/j.molmed.2014.06.005. [DOI] [PubMed] [Google Scholar]
- 115.Pfeffer SR, Yang CH, Pfeffer LM. The role of miR-21 in cancer. Drug Dev Res. 2015;76(6):270–7. doi: 10.1002/ddr.21257. https://doi.org/10.1002/ddr.21257. [DOI] [PubMed] [Google Scholar]
- 116.Guo FJ, Shao YP, Wang YP, Jin YM, Liu SS, Wang QY. MIR-92 stimulates VEGF by inhibiting von Hippel-Lindau gene product in epithelial ovarian cancer. J Biol Regul Homeost Agents. 2017;31(3):615–24. https://www.biolifesas.org/EN/Y2017/V31/I3/615. [PubMed] [Google Scholar]
- 117.Valera VA, Walter BA, Linehan WM, Merino MJ. Regulatory effects of microRNA-92 (miR-92) on VHL gene expression and the hypoxic activation of miR-210 in clear cell renal cell carcinoma. J Cancer. 2011;2:515–26. doi: 10.7150/jca.2.515. https://doi.org/10.7150/jca.2.515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Ganner A, Gehrke C, Klein M, Thegtmeier L, Matulenski T, Wingendorf L, et al. VHL suppresses RAPTOR and inhibits mTORC1 signaling in clear cell renal cell carcinoma. Sci Rep. 2021;11(1):14827. doi: 10.1038/s41598-021-94132-5. https://doi.org/10.1038/s41598-021-94132-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Hwang IY, Roe JS, Seol JH, Kim HR, Cho EJ, Youn HD. pVHL-mediated transcriptional repression of c-Myc by recruitment of histone deacetylases. Mol Cells. 2012;33(2):195–201. doi: 10.1007/s10059-012-2268-3. https://doi.org/10.1007/s10059-012-2268-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Gao YH, Wu ZX, Xie LQ, Li CX, Mao YQ, Duan YT, et al. VHL deficiency augments anthracycline sensitivity of clear cell renal cell carcinomas by down-regulating ALDH2. Nat Commun. 2017;8:15337. doi: 10.1038/ncomms15337. https://doi.org/10.1038/ncomms15337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Lichner Z, Mejia-Guerrero S, Ignacak M, Krizova A, Bao TT, Girgis AH, et al. Pleiotropic action of renal cell carcinoma-dysregulated miRNAs on hypoxia-related signaling pathways. Am J Pathol. 2012;180(4):1675–87. doi: 10.1016/j.ajpath.2011.12.030. https://doi.org/10.1016/j.ajpath.2011.12.030. [DOI] [PubMed] [Google Scholar]
- 122.Wu N, Mcdaniel K, Zhou T, Ramos-Lorenzo S, Wu C, Huang L, et al. Knockout of microRNA-21 attenuates alcoholic hepatitis through the VHL/NF-kappaB signaling pathway in hepatic stellate cells. Am J Physiol Gastrointest Liver Physiol. 2018;315(3):G385–98. doi: 10.1152/ajpgi.00111.2018. https://doi.org/10.1152/ajpgi.00111.2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Sun J, Jiang Z, Li Y, Wang K, Chen X, Liu G. Downregulation of miR-21 inhibits the malignant phenotype of pancreatic cancer cells by targeting VHL. Onco Targets Ther. 2019;12:7215–26. doi: 10.2147/OTT.S211535. https://doi.org/10.2147/OTT.S211535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Cai L, Wang W, Li X, Dong T, Zhang Q, Zhu B, et al. MicroRNA-21-5p induces the metastatic phenotype of human cervical carcinoma cells in vitro by targeting the von Hippel-Lindau tumor suppressor. Oncol Lett. 2018;15(4):5213–9. doi: 10.3892/ol.2018.7937. https://doi.org/10.3892/ol.2018.7937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Chen L, Han L, Zhang K, Shi Z, Zhang J, Zhang A, et al. VHL regulates the effects of miR-23b on glioma survival and invasion via suppression of HIF-1alpha/VEGF and beta-catenin/Tcf-4 signaling. Neuro Oncol. 2012;14(8):1026–36. doi: 10.1093/neuonc/nos122. https://doi.org/10.1093/neuonc/nos122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Liu N, Xia WY, Liu SS, Chen HY, Sun L, Liu MY, et al. MicroRNA-101 targets von Hippel-Lindau tumor suppressor (VHL) to induce HIF1alpha mediated apoptosis and cell cycle arrest in normoxia condition. Sci Rep. 2016;6:20489. doi: 10.1038/srep20489. https://doi.org/10.1038/srep20489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Li SJ, Liu HL, Tang SL, Li XJ, Wang XY. MicroRNA-150 regulates glycolysis by targeting von Hippel-Lindau in glioma cells. Am J Transl Res. 2017;9(3):1058–66. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC5375998/ https://e-century.us/files/ajtr/9/3/ajtr0036923.pdf. [PMC free article] [PubMed] [Google Scholar]
- 128.Li C, Zhao J, Sun W. microRNA-222-mediated VHL downregulation facilitates retinoblastoma chemoresistance by increasing HIF1alpha expression. Invest Ophthalmol Vis Sci. 2020;61(10):9. doi: 10.1167/iovs.61.10.9. https://doi.org/10.1167/iovs.61.10.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Cao Y, Zhang J, Xiong D, Wang D, Wu T, Huang A, et al. Hsa-miR-331-3p inhibits VHL expression by directly targeting its mRNA 3’-UTR in HCC cell lines. Acta Biochim Pol. 2015;62(1):77–82. doi: 10.18388/abp.2014_779. https://doi.org/10.18388/abp.2014_779. [DOI] [PubMed] [Google Scholar]
- 130.Cava C, Novello C, Martelli C, Lodico A, Ottobrini L, Piccotti F, et al. Theranostic application of miR-429 in HER2+ breast cancer. Theranostics. 2020;10(1):50–61. doi: 10.7150/thno.36274. https://doi.org/10.7150/thno.36274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Xiao B, Zhou X, Ye M, Lv S, Wu M, Liao C, et al. MicroRNA-566 modulates vascular endothelial growth factor by targeting Von Hippel-Landau in human glioblastoma in vitro and in vivo. Mol Med Rep. 2016;13(1):379–85. doi: 10.3892/mmr.2015.4537. https://doi.org/10.3892/mmr.2015.4537. [DOI] [PubMed] [Google Scholar]
- 132.Zou W, Cheng J. MiR-887 promotes the progression of hepatocellular carcinoma via targeting VHL. Technol Cancer Res Treat. 2020;19:1533033820940425. doi: 10.1177/1533033820940425. https://doi.org/10.1177/1533033820940425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Peng S, Zhang J, Tan X, Huang Y, Xu J, Silk N, et al. The VHL/HIF axis in the development and treatment of pheochromocytoma/paraganglioma. Front Endocrinol. 2020;11(903):586857. doi: 10.3389/fendo.2020.586857. https://doi.org/10.3389/fendo.2020.586857. [DOI] [PMC free article] [PubMed] [Google Scholar]


