PURPOSE
The applicability of FLT3-internal tandem duplications (FLT3-ITD) for assessing measurable residual disease (MRD) in acute myeloid leukemia (AML) in complete remission (CR) has been hampered by patient-specific duplications and potential instability of FLT3-ITD during relapse. Here, we comprehensively investigated the impact of next-generation sequencing (NGS)–based FLT3-ITD MRD detection on treatment outcome in a cohort of patients with newly diagnosed AML in relation to established prognostic factors at diagnosis and other MRD measurements, ie, mutant NPM1 and multiparameter flow cytometry.
METHODS
In 161 patients with de novo FLT3-ITD AML, NGS was performed at diagnosis and in CR after intensive remission induction treatment. FLT3-ITD MRD status was correlated with the cumulative incidence of relapse and overall survival (OS).
RESULTS
NGS-based FLT3-ITD MRD was present in 47 of 161 (29%) patients with AML. Presence of FLT3-ITD MRD was associated with increased risk of relapse (4-year cumulative incidence of relapse, 75% FLT3-ITD MRD v 33% no FLT3-ITD MRD; P < .001) and inferior OS (4-year OS, 31% FLT3-ITD MRD v 57% no FLT3-ITD MRD; P < .001). In multivariate analysis, detection of FLT3-ITD MRD in CR confers independent prognostic significance for relapse (hazard ratio, 3.55; P < .001) and OS (hazard ratio 2.51; P = .002). Strikingly, FLT3-ITD MRD exceeds the prognostic value of most generally accepted clinical and molecular prognostic factors, including the FLT3-ITD allelic ratio at diagnosis and MRD assessment by NGS-based mutant NPM1 detection or multiparameter flow cytometry.
CONCLUSION
NGS-based detection of FLT3-ITD MRD in CR identifies patients with AML with profound risk of relapse and death that outcompetes the significance of most established prognostic factors at diagnosis and during therapy, and furnishes support for FLT3-ITD as a clinically relevant biomarker for dynamic disease risk assessment in AML.
INTRODUCTION
Acute myeloid leukemia (AML) is a heterogeneous disease that arises from the sequential acquisition of specific driver mutations in the leukemic stem cell.1 Internal tandem duplications (ITD) in the FMS-like tyrosine kinase 3 (FLT3) receptor gene are among the most common genetic molecular abnormalities in patients with AML.2,3 In FLT3-ITD AML, the FLT3 kinase is constitutively activated resulting in uncontrolled proliferation of leukemic blasts.4 FLT3-ITDs are generally considered late-event mutations in leukemogenesis and are frequently preceded by the appearance of mutations in DNMT3A and NPM1.2,3 FLT3-ITD has been suggested to characterize an aggressive leukemic phenotype with early relapse and inferior treatment outcome.3-5 Several features present at diagnosis have been postulated to modify the prognostic effect of FLT3-ITD, including the presence of concurrent mutant NPM1 and the FLT3-ITD clone size, ie, allelic ratio.5
CONTEXT
Key Objective
FLT3-internal tandem duplication (ITD) measurable residual disease (MRD) detection in acute myeloid leukemia (AML) has been hampered by the variety in patient-specific duplications. Current guidelines do not recommend FLT3-ITD MRD monitoring because of potential instability at relapse. We comprehensively investigated the impact of FLT3-ITD MRD detection on treatment outcome in AML.
Knowledge Generated
To our knowledge, for the first time, we show that next-generation sequencing–based FLT3-ITD MRD detection identifies patients with AML with profound relapse risk and death that outweighs currently used prognostic factors. FLT3-ITD MRD better identifies patients with AML for relapse compared with multiparameter flow cytometry or next-generation sequencing–based mutant NPM1 MRD alone.
Relevance (C.F. Craddock)
-
Although these results require validation in independent data sets, they present a compelling rationale to incorporate FLT3-ITD MRD monitoring in AML to guide treatment strategies.*
*Relevance section written by JCO Associate Editor Charles F. Craddock, MD.
Increasing evidence indicates that treatment outcome prediction can be improved by assessing the kinetics and depth of response during therapy by detection of measurable residual disease (MRD).6 Currently, MRD detection in FLT3-ITD AML is carried out by a combination of multiparameter flow cytometry (MFC), mutant NPM1 real-time quantitative polymerase chain reaction (RQ-PCR), and next-generation sequencing (NGS).7 In the past, FLT3-ITD MRD detection by RQ-PCR and NGS was hampered by the variety of patient-specific FLT3-ITD, ie, sequence, position, and length. Advances in sequencing technology now enable accurate molecular detection of FLT3-ITD MRD.8-10
Clonal evolution studies revealed that late-event mutations in activated signaling genes, such as those in FLT3, are often subclonal and can be unstable during relapse in up to 25% of patients with AML, which may disqualify these markers for MRD detection.11-16 Systematic studies evaluating the applicability of FLT3-ITD MRD detection as a prognostic biomarker in AML are therefore lacking. Here, we present a comprehensive study investigating the impact of NGS-based FLT3-ITD MRD detection on treatment outcome in a cohort of patients with newly diagnosed AML, enrolled in multicenter prospective phase III HOVON-SAKK clinical trials, in relation to various other established baseline and MRD prognostic markers.
METHODS
Patients and Samples
In total, 161 treatment-naive patients with de novo FLT3-ITD AML out of 2,274 patients with AML were included (Fig 1). Patients were enrolled in Dutch-Belgian Cooperative Trial Group for Hematology-Oncology (HOVON) or the Swiss Group for Clinical Cancer Research (SAKK) clinical trials HO42A AML, HO102 AML, and HO132 AML (Fig 1). Trial protocols were approved by the ethics committees at each participating site and performed in accordance with the Declaration of Helsinki and after obtaining patient written informed consent. All patients had achieved complete remission (CR; < 5% blast cells in the bone marrow) after two cycles of induction chemotherapy. Treatments protocols and inclusion criteria have been described previously.17-19 Of note, in a subset of patients with AML included in the HOVON 132 trial, the residual disease status by MFC and/or mutant NPM1 RQ-PCR assay was available to the clinical investigator before consolidation therapy to enable subsequent treatment choice.19 Patient samples were taken at diagnosis and during CR after two cycles of standard induction chemotherapy. Details about patient and sample selection are provided in the Data Supplement (online only).
FIG 1.
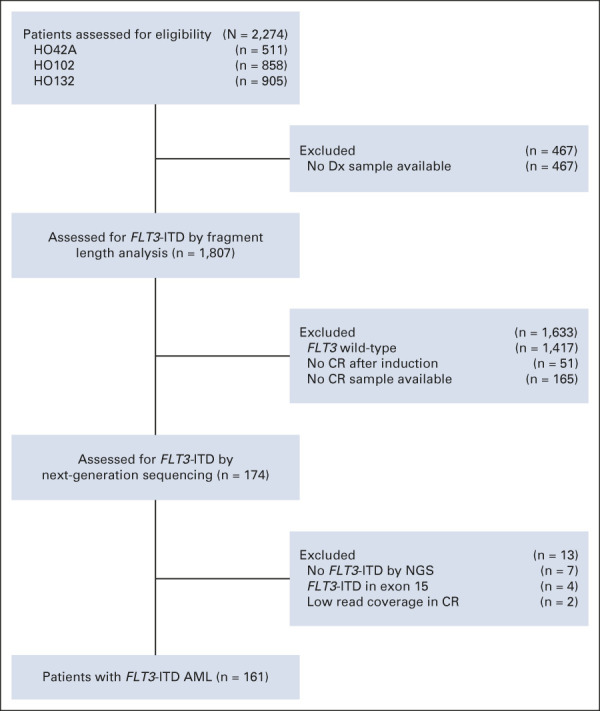
Flow diagram of the FLT3-ITD MRD study. AML, acute myeloid leukemia; CR, complete remission; Dx, diagnosis; ITD, internal tandem duplication; MRD, measurable residual disease; NGS, next-generation sequencing.
FLT3-ITD and Mutant NPM1 Detection by Targeted NGS
The FLT3-ITD status at AML diagnosis was assessed with both capillary fragment length analysis and the NGS TruSight Myeloid Sequencing Panel. Patients were excluded when the presence of FLT3-ITD at diagnosis could not be confirmed by NGS or when the FLT3-ITD was located in exon 15 (Fig 1). High and low allelic FLT3-ITD allelic ratios were determined by capillary fragment length analysis and defined as ≥ 0.5 or < 0.5, respectively.5,20 NPM1 mutations at AML diagnosis were determined by NGS. In CR, FLT3-ITD and mutant NPM1 MRD detection was performed with a single-amplicon NGS library panel, covering exon 14 of FLT3 or exon 12 of NPM1, for targeted deep sequencing analysis. The limit of detection of the FLT3-ITD MRD assay in CR ranges between variant allele frequencies of 0.01% and 0.001% (Data Supplement). In case of insufficient read coverage in CR (< 300,000 reads), the patient samples were excluded (Fig 1). All patients with FLT3-ITD AML were considered for MRD analysis, irrespective of FLT3-ITD ratio and/or number of FLT3-ITDs detectable by NGS at diagnosis. The NGS libraries were paired-end sequenced (2 × 221-bp) on Illumina NGS platforms according to manufacturer's recommendation (Illumina, San Diego, CA). We used our in-house data analysis pipeline for variant calling as previously described.21 Details on the experimental procedures, limit of detection, and bioinformatic analyses are available in the Data Supplement.
Multiparameter Flow Cytometry
Detection of MFC MRD was performed as recommended.7 The residual disease percentage was defined as the number of leukemia-associated immunophenotype cells within the total white blood cell compartment. The threshold between residual disease and no residual disease on the basis of flow cytometry was established at 0.1%. MFC MRD was carried out in 138 out of 161 patients with FLT3-ITD AML.
Statistical Analysis
The primary end point of the study was the cumulative incidence of relapse (CIR). Relapse and survival time were calculated from the date when CR bone marrow samples were taken until the date of the event of interest or censoring set at the date last known alive. Competing-risk analysis was performed for relapse with adjustment for nonrelapse mortality according to the method of Gray22 and the Fine and Gray23 model. The secondary end point was overall survival (OS), defined by death from any cause. Survival curves were estimated using the Kaplan-Meier method and differences in survival were assessed with the log-rank test. Multivariable modeling was performed with the Cox proportional hazards model. The proportional hazards assumption was tested by including the interaction with time-varying coefficients. Differences in patient or molecular characteristics were tested using the Fisher's exact test for categorical variables and Mann-Whitney U test for continuous variables. All statistical tests were two-sided, and P values < .05 were considered statistically significant. Statistical analyses were executed with Stata Statistics and Data Science software, Release 17.0 (Stata, College Station, TX).
RESULTS
FLT3-ITD Detection at Diagnosis, Relapse, and in Complete Remission
NGS-based FLT3-ITD detection was carried out in a cohort of 161 patients with AML, with a median survival time of 29.6 months (Table 1 and Fig 1). At diagnosis, 74 out of 161 (46%) patients had FLT3-ITD low allelic ratios and 87 (54%) patients had high allelic ratios. Patients with FLT3-ITD AML carried a variety of concurrent gene mutations (Table 1 and Data Supplement). The most frequent coexisting mutations were found in NPM1 (57%) and DNMT3A (47%). At relapse, identical FLT3-ITDs were observed in 88% of patients with AML (n = 25). Two patients with AML had gained mutations in different genes at relapse that had been wild-type at diagnosis (Data Supplement).
TABLE 1.
Patient Characteristics of FLT3-ITD Acute Myeloid Leukemia by Detection of FLT3-ITD MRD
FLT3-ITD MRD was detected in 47 out of 161 (29%) patients with AML, with a median variant allele frequency of 0.008% (range, 0.00031%-3.10%; Table 1 and Data Supplement). All FLT3-ITDs found in CR were identical (ie, sequence, position, and length) compared with diagnosis and multiple FLT3-ITD MRD clones were detected in five patients with AML. FLT3-ITD MRD was significantly associated with aberrant cytogenetics at diagnosis (35% FLT3-ITD MRD v 19% no FLT3-ITD MRD; P = .040) and patients with NPM1 wild-type AML (72% FLT3-ITD MRD v 32% no FLT3-ITD MRD; P < .001; Table 1). Interestingly, FLT3-ITD MRD was observed in 22% of patients with triple-mutant AML (DNMT3A/NPM1/FLT3-ITD), whereas none of the patients with double–mutant AML (NPM1/FLT3-ITD) showed FLT3-ITD MRD. Although significantly higher FLT3-ITD ratios were observed in patients with FLT3-ITD MRD AML (P = .035), the association of FLT3-ITD MRD with the FLT3-ITD allelic ratio status at diagnosis was not significant (P = .121). Detection of FLT3-ITD MRD was significantly more frequent in patients with AML who needed two cycles rather than one induction cycle to attain CR (45% FLT3-ITD MRD v 11% no FLT3-ITD MRD; P < .001; Table 1).
FLT3-ITD Residual Disease and Outcome
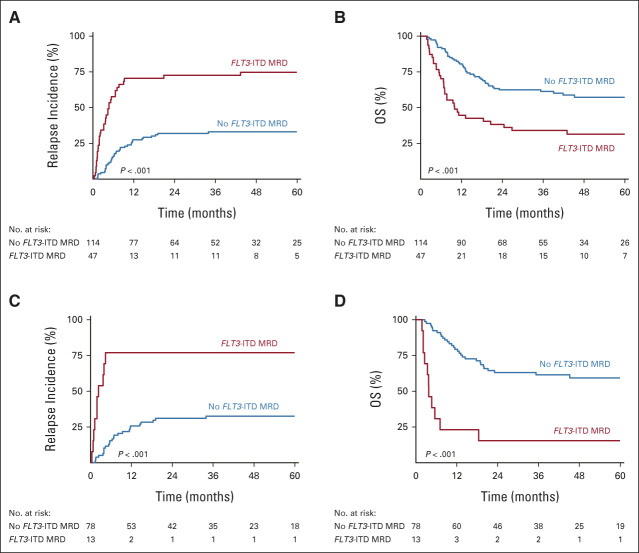
We next assessed whether FLT3-ITD MRD was prognostic for relapse and OS. FLT3-ITD MRD was associated with an increased risk of relapse (4-year CIR, 75% FLT3-ITD MRD v 33% no FLT3-ITD MRD; hazard ratio [HR], 3.70; 95% CI, 2.31 to 5.94; P < .001) and reduced OS (4-year OS, 31% FLT3-ITD MRD v 57% no FLT3-ITD MRD; HR, 2.47; 95% CI, 1.59 to 3.84; P < .001; Figs 2A and 2B). To increase the sensitivity of the FLT3-ITD MRD assay, we compared 100 ng and 500 ng DNA input in a selected subset of patients with AML for whom sufficient DNA was available (n = 122). Although the number of patients with FLT3-ITD MRD AML increased (n = 13) at very low levels (< 0.01%), the association with relapse did not improve (Data Supplement). The FLT3-ITD MRD clone size, as indicated by the variant allele frequency in remission, was directly correlated with the risk of relapse (Data Supplement). The number of persisting FLT3-ITD MRD clones did not associate with relapse (Data Supplement). Within the mutant NPM1 FLT3-ITD AML subset, the prognostic value for relapse (4-year CIR, 77% FLT3-ITD MRD v 33% no FLT3-ITD MRD; HR, 4.87; 95% CI, 1.92 to 12.3; P < .001) and survival (4-year OS, 15% FLT3-ITD MRD v 59% no FLT3-ITD MRD; HR, 5.36; 95% CI, 2.65 to 10.8; P < .001) was preserved (Figs 2C and 2D).
FIG 2.
Survival outcome of FLT3-ITD MRD. (A) Relapse incidence and (B) OS of FLT3-ITD AML according to FLT3-ITD MRD in complete remission (n = 161). (C) Relapse incidence and (D) OS according to FLT3-ITD MRD in patients with mutant NPM1 FLT3-ITD AML (n = 91). AML, acute myeloid leukemia; ITD, internal tandem duplication; MRD, measurable residual disease; OS, overall survival.
To assess whether the detection of FLT3-ITD MRD serves as an independent prognostic factor, we performed univariate analysis and multivariate analysis. In univariate analysis, significantly increased risk of relapse and reduced OS were observed among patients with a high white blood cell count at diagnosis (> 100 × 109/L), late CR (ie, achieved after two cycles of induction chemotherapy), and patients with a high FLT3-ITD allelic ratio at diagnosis (Data Supplement) and within the ELN intermediate and adverse risk classification (Data Supplement). In multivariable modeling, FLT3-ITD MRD confers profound independent prognostic significance with respect to the relapse rate (HR, 3.55; 95% CI, 1.92 to 6.56; P < .001) and OS (HR, 2.51; 95% CI, 1.42 to 4.43; P = .002; Table 2). Besides FLT3-ITD MRD, only a high white blood cell count and late CR appeared to be independently associated with relapse and OS. Remarkably, the NPM1 mutation status and the FLT3-ITD allelic ratio at diagnosis lost their prognostic value for relapse and survival when FLT3-ITD MRD was taken into account (Table 2). In sensitivity analysis, no significant clinically relevant or treatment-related interactions were observed. The prognostic value of FLT3-ITD MRD was unaffected in a correction for variation in sampling time (Data Supplement).
TABLE 2.
Multivariate Analysis of Prognostic Factors for Relapse and OS
FLT3-ITD Residual Disease and Allogeneic Transplantation
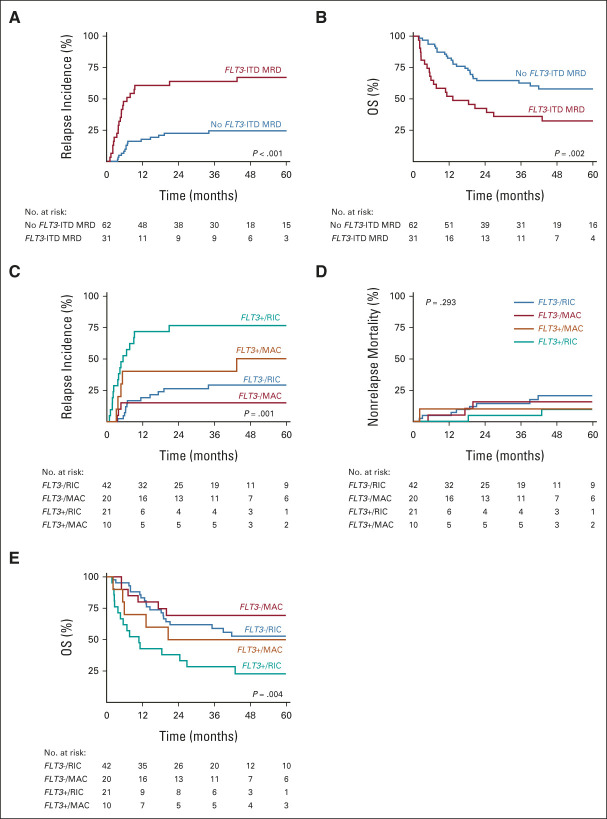
Regarding the poor risk features of FLT3-ITD AML, many patients will undergo allogeneic transplantation. Therefore, we explored the prognostic value of FLT3-ITD MRD in the 93 patients who underwent allogeneic transplantation. In total, 30 patients with AML received myeloablative conditioning (MAC) and 63 patients received reduced-intensity conditioning (RIC). Although the overall risk of relapse is reduced in the transplanted FLT3-ITD AML patients, we demonstrate increased relapse incidence and inferior outcome of patients with FLT3-ITD MRD AML (Fig 3). The prognostic value of FLT3-ITD MRD is comparable in both MAC and RIC groups (P = .858). However, the risk of relapse is lower in patients with FLT3-ITD MRD AML who had received MAC conditioning. No significant differences for nonrelapse mortality between the RIC and MAC groups were observed, resulting in improved OS for patients with AML with residual disease after MAC conditioning (Fig 3). In a time-dependent correction for allogeneic hematopoietic stem-cell transplant, the prognostic significance of FLT3-ITD MRD is maintained (Data Supplement).
FIG 3.
Survival outcome of FLT3-ITD MRD and allogeneic transplantation. (A) Relapse incidence and (B) OS of FLT3-ITD MRD in patients with AML who received allogeneic transplantation (n = 93). (C) Relapse incidence, (D) nonrelapse mortality, and (E) OS of FLT3-ITD MRD in patients with AML stratified by RIC or MAC regimens. AML, acute myeloid leukemia; ITD, internal tandem duplication; MAC, myeloablative conditioning; MRD, measurable residual disease; OS, overall survival; RIC, reduced-intensity conditioning.
Residual Disease Detection by Mutant NPM1 and MFC
MFC MRD and mutant NPM1 MRD are recommended methods for relapse prediction in patients with AML in CR.7 We compared the assessment of FLT3-ITD MRD with available NGS-based mutant NPM1 and MFC residual disease measurements in 91 and 138 patients with AML, respectively.
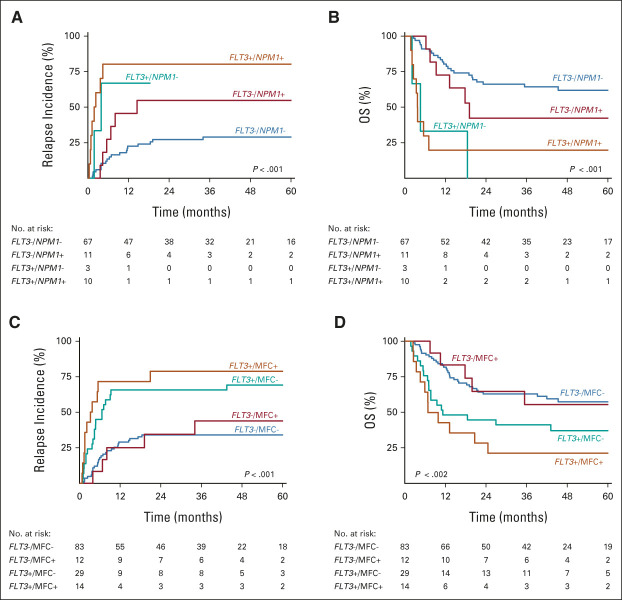
Interestingly, FLT3-ITD MRD significantly associated with high relapse risk and adverse OS irrespective of the mutant NPM1 MRD or MFC MRD status (Fig 4). Increased risk of relapse and inferior survival was observed in patients with AML without FLT3-ITD MRD but with persistent mutant NPM1 MRD; however, the association was not significant (P = .081 and P = .236). By contrast, MFC MRD without FLT3-ITD MRD appeared to confer limited prognostic value for relapse and survival (Figs 4C and 4D).
FIG 4.
Survival outcome of FLT3-ITD MRD and NGS-based mutant NPM1 and MFC MRD. (A) Relapse incidence and (B) OS of FLT3-ITD MRD and mutant NPM1 MRD combined (n = 91). (C and D) Relapse incidence and OS of both FLT3-ITD MRD and MFC MRD (n = 138). ITD, internal tandem duplication; MFC, multiparameter flow cytometry; MRD, measurable residual disease; NGS, next-generation sequencing; OS, overall survival.
DISCUSSION
Current risk classification and management of FLT3-ITD AML relies on the assessment of gene mutations at diagnosis and dynamic residual disease response measurements during therapy.5 However, the prognostic significance of FLT3-ITD has been subject of ongoing debate as it may depend on allelic burden and the presence of other gene mutations, in particular mutant NPM1.5 Here, we performed a comprehensive study in FLT3-ITD AML investigating the impact of persistence of FLT3-ITD in CR after induction chemotherapy and treatment outcome. Our results reveal FLT3-ITD MRD as a strong independent prognostic factor that identifies patients with AML with profound risk of relapse and death. Furthermore, FLT3-ITD MRD outcompetes the impact of other currently established prognostic factors in FLT3-ITD AML, including the NPM1 mutation status and FLT3-ITD allelic ratio at diagnosis and residual disease measurements in CR by NGS-based mutant NPM1 detection or MFC.
Mutations in activated signaling genes, such as FLT3-ITD, generally represent late events in AML development and can be lost or gained at relapse.11-16 Therefore, residual disease detection of these late mutations have previously been considered to be of limited prognostic value, in contrast to some of the more stable leukemic driver mutations, such as mutant NPM1.7,16 The results of the current study, however, indicate stability of FLT3-ITD during relapse in the majority of patients with AML and substantiate residual FLT3-ITD in CR as a clinically useful indicator for relapse. We showed that leukemic clones with residual FLT3-ITDs that were present at baseline carry high impact on the risk of relapse, whereas AML clones with persistent mutant NPM1 without FLT3-ITD had limited prognostic value for relapse. This implies that, within FLT3-ITD AML with concurrent mutant NPM1, residual disease characterized by FLT3-ITD better identifies patients with AML for relapse than mutant NPM1 MRD detection alone. Future studies exploring the comparison of DNA- and RNA-based mutant NPM1 and FLT3-ITD MRD testing as well as using peripheral blood may further improve MRD monitoring of these genetically defined AML subtypes.
FLT3-ITD MRD failed to identify all cases of AML relapse. Although NGS-based sequencing was carried out at substantial depth, FLT3-ITDs at ultralow levels (< 0.001%) may have been missed. The FLT3-ITD assay may even further improve prediction of relapse by the ability to sequence at even higher depth or by increased DNA sample input, as well as the capability to sequence larger FLT3-ITDs and those in exon 15 of the FLT3 gene. Of note, we studied FLT3-ITD MRD at a single time point. With respect to the correlation of FLT3-ITD MRD clone size with relapse, it will be of interest to examine how longitudinal monitoring may support relapse prediction or guide different treatment strategies and allogeneic conditioning regimens for FLT3-ITD AML.
We have studied FLT3-ITD MRD in patients treated with intensive chemotherapy only. It will be interesting to investigate the prognostic value of FLT3-ITD MRD in the context of treatments that include the use of FLT3 inhibitors, since these patients have been shown to acquire novel FLT3-ITD mutations or mutations in other activated signaling genes during relapse.24-26 From this perspective, it will be important to expand NGS-based MRD detection to additional relevant late-event mutations in activated signaling genes and investigate the efficiency of novel and more specific FLT3 inhibitors on the eradication of residual FLT3-ITD cells and the prevention of impending relapse in AML.
In conclusion, NGS-based detection of FLT3-ITD MRD in CR identifies patients with AML with profound relapse risk and death, and outweighs the prognostic factors that are currently used in AML risk stratification. Therefore, we propose to incorporate FLT3-ITD MRD in AML treatment protocols.
ACKNOWLEDGMENT
We thank all the participating centers of the Dutch–Belgian Cooperative Trial Group for Hematology–Oncology (HOVON) and Swiss Group for Clinical Cancer Research (SAKK), where the clinical trials that formed the basis for this study were conducted; H. Berna Beverloo for performing cytogenetic analyses, Remco Hoogenboezem, Elodie Stoetman and Jolinda Konijnenburg for assisting with bioinformatics, and Egied Simons for assisting with preparation of the figures (Erasmus University Medical Center).
Jacqueline Cloos
Consulting or Advisory Role: Novartis (Inst)
Speakers' Bureau: Astellas Pharma (Inst)
Research Funding: Novartis (Inst), Merus (Inst), Helsinn Therapeutics (Inst), Genentech (Inst)
Patents, Royalties, Other Intellectual Property: Royalties for our designed leukemia stem cell flow cytometry tube (Inst), License for our validated MRD assay to be used in clincal trials (Inst)
Yngvar Fløisand
Honoraria: Arog Pharmaceuticals
Consulting or Advisory Role: Arog Pharmaceuticals
Gert J. Ossenkoppele
Consulting or Advisory Role: Celgene, Novartis, Amgen, Pfizer, Roche, Bristol Myers Squibb, AGIOS, AbbVie/Genentech, Astellas Pharma, Otsuka, Daiichi Sankyo, Gilead Sciences, Servier, Jazz Pharmaceuticals
Research Funding: Janssen (Inst), Celgene (Inst), Novartis (Inst)
Travel, Accommodations, Expenses: Roche
Violaine Havelange
Consulting or Advisory Role: Novartis, Incyte, Celgene/Bristol Myers Squibb, AbbVie
Bob Löwenberg
Stock and Other Ownership Interests: Frame Therapeutics, CureVac
Consulting or Advisory Role: Astex Pharmaceuticals, Clear Creek Bio, Agios, Celgene, AbbVie, Roche, GEMoaB, AIMM Therapeutics, Oxford Biomedica, Bristol Myers Squibb, Servier, Catamaran Bio, Astellas Pharma, Kronos Bio, CureVac
Peter J.M. Valk
Honoraria: BMS GmbH & Co, KG, Astellas Scientific and Medical Affairs Inc, Jazz Pharmaceuticals
Travel, Accommodations, Expenses: Seattle Genetics, Gilead Sciences
No other potential conflicts of interest were reported.
PRIOR PRESENTATION
Presented in part at the European Hematology Association 2022 congress, Vienna, Austria, June 10, 2022; and at the Society of Hematologic Oncology 10th annual meeting, Houston, TX, September 28-October 1, 2022.
SUPPORT
Supported by grants from the Queen Wilhelmina Fund Foundation of the Dutch Cancer Society (EMCR 2019-12507).
AUTHOR CONTRIBUTIONS
Conception and design: Tim Grob, Christian M. Vonk, Bob Löwenberg, Mojca Jongen-Lavrencic, Peter J.M. Valk
Financial support: Peter J.M. Valk
Provision of study materials or patients: Yngvar Fløisand, Marinus van Marwijk Kooy, Markus G. Manz, Gert J. Ossenkoppele, Lidwine W. Tick, Violaine Havelange, Bob Löwenberg, Mojca Jongen-Lavrencic
Collection and assembly of data: Tim Grob, Mathijs A. Sanders, Christian M. Vonk, Franҫois G. Kavelaars, Melissa Rijken, Diana W. Hanekamp, Jacqueline Cloos, Yngvar Fløisand, Marinus van Marwijk Kooy, Markus G. Manz, Gert J. Ossenkoppele, Lidwine W. Tick, Violaine Havelange, Bob Löwenberg, Mojca Jongen-Lavrencic, Peter J.M. Valk
Data analysis and interpretation: Tim Grob, Mathijs A. Sanders, Christian M. Vonk, Franҫois G. Kavelaars, Melissa Rijken, Patrycja L. Gradowska, Bob Löwenberg, Mojca Jongen-Lavrencic, Peter J.M. Valk
Manuscript writing: All authors
Final approval of manuscript: All authors
Accountable for all aspects of the work: All authors
AUTHORS' DISCLOSURES OF POTENTIAL CONFLICTS OF INTEREST
Prognostic Value of FLT3-Internal Tandem Duplication Residual Disease in Acute Myeloid Leukemia
The following represents disclosure information provided by authors of this manuscript. All relationships are considered compensated unless otherwise noted. Relationships are self-held unless noted. I = Immediate Family Member, Inst = My Institution. Relationships may not relate to the subject matter of this manuscript. For more information about ASCO's conflict of interest policy, please refer to www.asco.org/rwc or ascopubs.org/jco/authors/author-center.
Open Payments is a public database containing information reported by companies about payments made to US-licensed physicians (Open Payments).
Jacqueline Cloos
Consulting or Advisory Role: Novartis (Inst)
Speakers' Bureau: Astellas Pharma (Inst)
Research Funding: Novartis (Inst), Merus (Inst), Helsinn Therapeutics (Inst), Genentech (Inst)
Patents, Royalties, Other Intellectual Property: Royalties for our designed leukemia stem cell flow cytometry tube (Inst), License for our validated MRD assay to be used in clincal trials (Inst)
Yngvar Fløisand
Honoraria: Arog Pharmaceuticals
Consulting or Advisory Role: Arog Pharmaceuticals
Gert J. Ossenkoppele
Consulting or Advisory Role: Celgene, Novartis, Amgen, Pfizer, Roche, Bristol Myers Squibb, AGIOS, AbbVie/Genentech, Astellas Pharma, Otsuka, Daiichi Sankyo, Gilead Sciences, Servier, Jazz Pharmaceuticals
Research Funding: Janssen (Inst), Celgene (Inst), Novartis (Inst)
Travel, Accommodations, Expenses: Roche
Violaine Havelange
Consulting or Advisory Role: Novartis, Incyte, Celgene/Bristol Myers Squibb, AbbVie
Bob Löwenberg
Stock and Other Ownership Interests: Frame Therapeutics, CureVac
Consulting or Advisory Role: Astex Pharmaceuticals, Clear Creek Bio, Agios, Celgene, AbbVie, Roche, GEMoaB, AIMM Therapeutics, Oxford Biomedica, Bristol Myers Squibb, Servier, Catamaran Bio, Astellas Pharma, Kronos Bio, CureVac
Peter J.M. Valk
Honoraria: BMS GmbH & Co, KG, Astellas Scientific and Medical Affairs Inc, Jazz Pharmaceuticals
Travel, Accommodations, Expenses: Seattle Genetics, Gilead Sciences
No other potential conflicts of interest were reported.
REFERENCES
- 1.Welch JS, Ley TJ, Link DC, et al. : The origin and evolution of mutations in acute myeloid leukemia. Cell 150:264-278, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cancer Genome Atlas Research N, Ley TJ, Miller C, et al. : Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N Engl J Med 368:2059-2074, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Papaemmanuil E, Gerstung M, Bullinger L, et al. : Genomic classification and prognosis in acute myeloid leukemia. N Engl J Med 374:2209-2221, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Daver N, Schlenk RF, Russell NH, Levis MJ: Targeting FLT3 mutations in AML: Review of current knowledge and evidence. Leukemia 33:299-312, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dohner H, Estey E, Grimwade D, et al. : Diagnosis and management of AML in adults: 2017 ELN recommendations from an international expert panel. Blood 129:424-447, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vonk CM, Al Hinai ASA, Hanekamp D, Valk PJM: Molecular minimal residual disease detection in acute myeloid leukemia. Cancers (Basel) 13:5431, 2021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Heuser M, Freeman SD, Ossenkoppele GJ, et al. : 2021 update on MRD in acute myeloid leukemia: A consensus document from the European LeukemiaNet MRD Working Party. Blood 138:2753-2767, 2021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Thol F, Kolking B, Damm F, et al. : Next-generation sequencing for minimal residual disease monitoring in acute myeloid leukemia patients with FLT3-ITD or NPM1 mutations. Genes Chromosomes Cancer 51:689-695, 2012 [DOI] [PubMed] [Google Scholar]
- 9.Levis MJ, Perl AE, Altman JK, et al. : A next-generation sequencing-based assay for minimal residual disease assessment in AML patients with FLT3-ITD mutations. Blood Adv 2:825-831, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Blätte TJ, Schmalbrock LK, Skambraks S, et al. : getITD for FLT3-ITD-based MRD monitoring in AML. Leukemia 33:2535-2539, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kottaridis PD, Gale RE, Langabeer SE, et al. : Studies of FLT3 mutations in paired presentation and relapse samples from patients with acute myeloid leukemia: Implications for the role of FLT3 mutations in leukemogenesis, minimal residual disease detection, and possible therapy with FLT3 inhibitors. Blood 100:2393-2398, 2002 [DOI] [PubMed] [Google Scholar]
- 12.Jan M, Snyder TM, Corces-Zimmerman MR, et al. : Clonal evolution of preleukemic hematopoietic stem cells precedes human acute myeloid leukemia. Sci Transl Med 4:149ra118, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Corces-Zimmerman MR, Hong WJ, Weissman IL, et al. : Preleukemic mutations in human acute myeloid leukemia affect epigenetic regulators and persist in remission. Proc Natl Acad Sci USA 111:2548-2553, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Garg M, Nagata Y, Kanojia D, et al. : Profiling of somatic mutations in acute myeloid leukemia with FLT3-ITD at diagnosis and relapse. Blood 126:2491-2501, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shih LY, Huang CF, Wu JH, et al. : Internal tandem duplication of FLT3 in relapsed acute myeloid leukemia: A comparative analysis of bone marrow samples from 108 adult patients at diagnosis and relapse. Blood 100:2387-2392, 2002 [DOI] [PubMed] [Google Scholar]
- 16.Kronke J, Bullinger L, Teleanu V, et al. : Clonal evolution in relapsed NPM1-mutated acute myeloid leukemia. Blood 122:100-108, 2013 [DOI] [PubMed] [Google Scholar]
- 17.Pabst T, Vellenga E, van Putten W, et al. : Favorable effect of priming with granulocyte colony-stimulating factor in remission induction of acute myeloid leukemia restricted to dose escalation of cytarabine. Blood 119:5367-5373, 2012 [DOI] [PubMed] [Google Scholar]
- 18.Lowenberg B, Pabst T, Maertens J, et al. : Therapeutic value of clofarabine in younger and middle-aged (18-65 years) adults with newly diagnosed AML. Blood 129:1636-1645, 2017 [DOI] [PubMed] [Google Scholar]
- 19.Löwenberg B, Pabst T, Maertens J, et al. : Addition of lenalidomide to intensive treatment in younger and middle-aged adults with newly diagnosed AML: The HOVON-SAKK-132 trial. Blood Adv 5:1110-1121, 2021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stone RM, Mandrekar SJ, Sanford BL, et al. : Midostaurin plus chemotherapy for acute myeloid leukemia with a FLT3 mutation. N Engl J Med 377:454-464, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jongen-Lavrencic M, Grob T, Hanekamp D, et al. : Molecular minimal residual disease in acute myeloid leukemia. N Engl J Med 378:1189-1199, 2018 [DOI] [PubMed] [Google Scholar]
- 22.Gray RJ: A class of K-sample tests for comparing the cumulative incidence of a competing risk. Ann Stat 16:1141-1154, 1988 [Google Scholar]
- 23.Fine JP, Gray RJ: A proportional hazards model for the subdistribution of a competing risk. J Am Stat Assoc 94:496-509, 1999 [Google Scholar]
- 24.Piloto O, Wright M, Brown P, et al. : Prolonged exposure to FLT3 inhibitors leads to resistance via activation of parallel signaling pathways. Blood 109:1643-1652, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schmalbrock LK, Dolnik A, Cocciardi S, et al. : Clonal evolution of acute myeloid leukemia with FLT3-ITD mutation under treatment with midostaurin. Blood 137:3093-3104, 2021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McMahon CM, Ferng T, Canaani J, et al. : Clonal selection with RAS pathway activation mediates secondary clinical resistance to selective FLT3 inhibition in acute myeloid leukemia. Cancer Discov 9:1050-1063, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]