Abstract
Polyketide natural products show great promise as medicinal agents. Typically the products of microbial secondary biosynthesis, polyketides are synthesized by an evolutionarily related but architecturally diverse family of multifunctional enzymes called polyketide synthases. A principal limitation for fundamental biochemical studies of these modular megasynthases, as well as for their applications in biotechnology, is the challenge associated with manipulating the natural microorganism that produces a polyketide of interest. To ameliorate this limitation, over the past decade several genetically amenable microbes have been developed as heterologous hosts for polyketide biosynthesis. Here we review the state of the art as well as the difficulties associated with heterologous polyketide production. In particular, we focus on two model hosts, Streptomyces coelicolor and Escherichia coli. Future directions for this relatively new but growing technological opportunity are also discussed.
The explosive growth in the number of cloned and sequenced genes has led to an enhanced need for robust heterologous gene expression methods. This need exists, in part, because the proteins encoded by these numerous genes of interest promise to have an important impact in areas ranging from basic biochemical and biophysical research to the practical use of proteins as pharmaceuticals, animal health products, and industrial enzymes. Notwithstanding the enormous efforts that have gone into the development of the heterologous-expression toolbox, the production of a desired protein in a heterologous host remains an empirical and often unpredictable process.
Although there is no universal solution for heterologous protein production, certain generalizations can be made. A few prokaryotic and eukaryotic systems such as Escherichia coli, Saccharomyces cerevisiae, Sf9 insect cells, and Chinese hamster ovary (CHO) cells have emerged as choice hosts due to their simplicity of use, excellent growth characteristics, and a plethora of readily accessible genetic tools. In turn, this has fueled studies aimed at understanding the cellular machinery responsible for protein synthesis, posttranslational modification, protein folding, trafficking, and degradation in these model host cells. Another feature common to good heterologous hosts is the availability of good fermentation protocols that maximize protein productivity, especially those in which cell growth can be decoupled from recombinant gene expression. Finally, it is generally understood that a balance exists between product quality and quantity, regardless of the host used. High levels of gene expression are often accompanied by phenomena such as inclusion body formation, increased amino acid misincorporation, incomplete or inaccurate posttranslational modification, reduced recombinant cell line stability, and a range of other features associated with a greater metabolic burden as a result of heterologous protein production.
In most cases, the ultimate goal of heterologous gene expression is to produce the desired protein in reagent quantities. Over the past 20 years, however, there has been a growing interest in the potential for harnessing the intrinsic metabolic activity of proteins in heterologous hosts. Multiple genes are often coexpressed in metabolic engineering, where the goal is not simply to synthesize large quantities of the target proteins themselves but to optimally interweave their activities among each other as well as among functional protein networks in the host. Examples of metabolic engineering include the bacterial biosynthesis of indigo (19) and the conversion of 3-dehydroshikimic acid, a key intermediate in aromatic amino acid metabolism, into a variety of value-added products such as vanillin (47).
Over the past decade, the study of bioactive natural-product biosynthesis has benefited significantly from the use of heterologous hosts. Notably, the rapid growth in understanding and manipulating polyketide biosynthesis closely parallels developments in the ability to reconstitute these multistep catalytic processes in genetically (and now genomically) friendly heterologous hosts. This review focuses on the development of heterologous expression systems for polyketide synthases (PKSs), and discusses their impact on the field of natural-product biosynthesis and drug development. Reconstitution of polyketide biosynthesis in heterologous hosts demands that large multienzyme assemblies be functionally expressed, their posttranslational modification needs be adequately met, their substrates be available in vivo in reasonable quantities, and the producer cell be protected against the toxicity of the biosynthetic products. Two particular heterologous hosts will be the focus of most of our discussion—Streptomyces coelicolor and Escherichia coli. However, the relative merits of a variety of other heterologous hosts will also be discussed.
WHY POLYKETIDES?
Polyketides, as the name implies, are synthesized from repetitive condensation reactions that link small carbon precursors (typically, 2- and 3-carbon acyl groups derived from coenzyme A (CoA) thioesters) (Fig. 1A) (63). The process is similar in many respects to bacterial and mammalian fatty acid synthesis. In fact, PKSs are classified into type I or II categories based on how closely they mimic the architecture of type I (vertebrate) or type II (bacterial and plant) fatty acid synthases (35). However, unlike fatty acids, the structures of polyketides are far more diverse due to variations in the fatty acid synthesis theme (Fig. 1B) and post-PKS modifications. This structural diversity is also reflected in diversity in their biological modes of action. A number of polyketides have been clinically approved as drugs for treating disorders such as infections, cancer, cardiovascular diseases, and inflammation. Approximately two-thirds of the known bioactive polyketide natural products originate from the actinomycetes. Other major microbial sources of polyketides include the myxobacteria and filamentous fungi. The exact role of polyketides in the life cycles of producing organisms remains unknown, but these secondary metabolites are presumably synthesized to ward off competing microbes during periods of nutrient limitation. Some polyketides are also synthesized as spore pigments (14, 55).
FIG. 1.
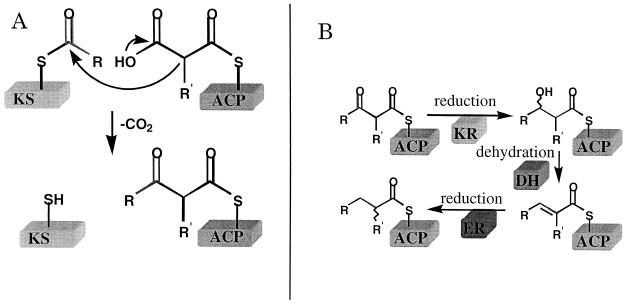
Key reactions and catalysts in polyketide biosynthesis. (A) The decarboxylative condensation reaction that defines a polyketide. The electrophile is attached to the ketosynthase (KS), whereas the nucleophile is attached to an ACP. (The only known exceptions to this rule are the chalcone synthase-like PKSs [see Fig. 3], where the nucleophile remains attached to CoA) (B) In addition to the above-mentioned C—C bond-forming reaction, PKSs catalyze all, some, or none of the following reactions. The β-carbonyl generated on C—C bond formation can be reduced by an NADPH-dependent enzyme called a ketoreductase (KR). The resulting alcohol can be dehydrated by a dehydratase (DH). The resulting olefin can be hydrogenated by another NADPH-dependent enzyme called an enoylreductase (ER). Other, less commonly occurring reactions such as C-methyl transfers (28) (see also Fig. 4) are not shown.
Figure 2 shows examples of familiar polyketides and their representative biological sources. It also highlights differences in the architectures of PKSs that synthesize the carbon skeletons of these natural products. Four broad architectural varieties of PKSs have been isolated thus far from microbes. The first category of PKSs is perhaps the simplest and actually resembles plant PKSs such as the chalcone synthase. An example is shown in Fig. 3 (24). This is the only variety of PKS in which the nucleophilic group involved in each C—C bond-forming reaction is attached to a CoA instead of an acyl carrier protein (ACP). Although the primary amino acid sequences of these PKSs are only weakly related to those of other PKSs, X-ray crystallographic analysis (22) and site-directed mutagenesis (38) of a prototype of this family, a plant chalcone synthase, have clearly established the close evolutionary connection between these PKSs and others. Fungal PKSs represent a second subclass of PKSs that closely resemble vertebrate (type I) fatty acid synthases. The proteins are multidomain and act in an iterative fashion. An example is the lovastatin LNKS, which catalyzes the formation of dihydromonacolin, the polyketide precursor for lovastatin biosynthesis (Fig. 4) (41). Bacterial PKS systems with an architectural relationship to type II fatty acid synthases represent a third PKS category. Also acting in an iterative fashion, these multienzyme systems produce aromatic polyketide products. However, individual active sites occur as distinct polypeptides rather than as domains within a single multifunctional polypeptide. An example is the doxorubicin PKS, which synthesizes the tetracyclic skeleton of this anthracycline antibiotic (Fig. 5) (2). Finally, a fourth category of PKSs, known as modular PKSs, is exemplified by the deoxyerythronolide B synthase (Fig. 6) (13, 17). Here, polyketide biosynthesis proceeds in a processive fashion on a very large megasynthase in which each active site is used once during the overall catalytic cycle.
FIG. 2.
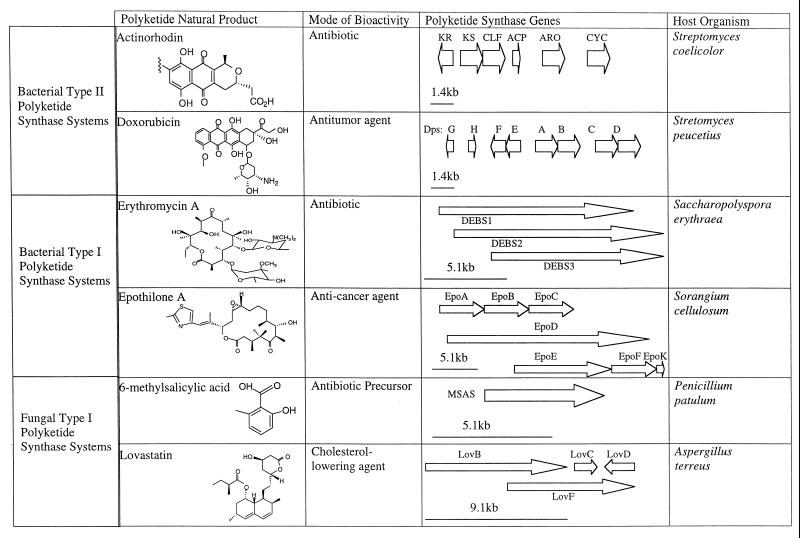
Representative polyketide natural products and PKS systems. This figure illustrates the diversity in polyketide natural products and the PKSs that catalyze their formation. The block arrows represent ORFs encoding different PKS components. The given scales indicate approximate ORF sizes, although individual catalytic domains are not shown. In general, the larger proteins encode several catalytic domains whereas the smaller polypeptides possess a single enzymatic activity. Protein abbreviations: KS, ketosynthase; CLF, chain length factor; KR, ketoreductase; ARO, aromatase; CYC, cyclase; Dps, doxorubicin polyketide synthase; DEBS, deoxyerythronolide B synthase; MSAS, methyl salicylic acid synthase.
FIG. 3.
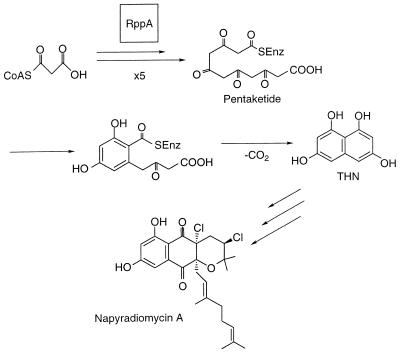
Chalcone synthase-like PKS. The RppA PKS iteratively condenses five malonyl-CoA units, which then cyclize to form 1,3,6,8-tetrahydroxynaphthalene (THN). THN most probably represents an intermediate in the production of antibiotics such as napyradiomycin A. RppA resembles a ketosynthase (KS) domain in other PKS paradigms. However, in this case, RppA condenses malonyl-CoA units directly as opposed to accepting a malonyl unit from an ACP.
FIG. 4.
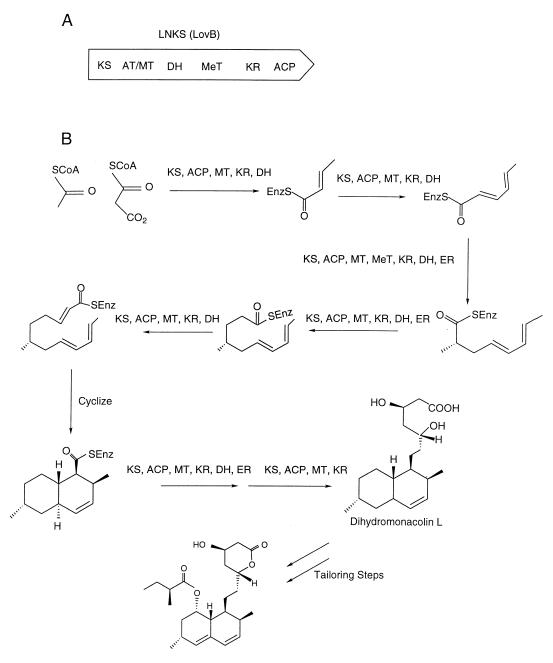
Fungal type I PKS. (A) The lovastatin nonaketide synthase (LNKS; LovB) protein, with its individual enzymatic domains denoted within the PKS. KS, ketosynthase; AT/MT, acetyl/malonyl transferase; DH, dehydratase; MeT, methyltransferase; KR, ketoreductase; ACP, acyl carrier protein. (B) LNKS works in an iterative fashion to produce dihydromonacolin L. The LNKS catalyzes a decarboxylative condensation, and the reaction product (the polyketide chain) then transfers to a new LNKS unit. The reaction arrows indicate condensation reactions between an extender malonyl unit and either a priming acetate unit or the growing polyketide chain (eight condensations in total). The KS domain accepts the starter acetate unit or a growing polyketide chain and catalyzes the condensation with a malonyl unit loaded onto the ACP domain by the AT/MT. The level of reduction applied for a particular iteration is denoted near the arrows. In this case, a separate lovastatin PKS protein (not shown) provides the enoylreductase (ER) activity.
FIG. 5.
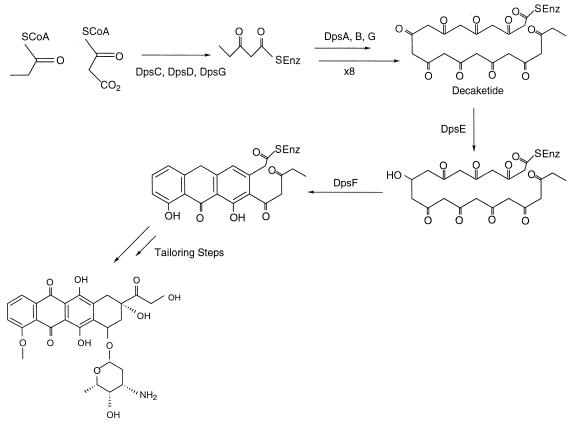
Bacterial type II PKS. The doxorubicin polyketide synthase (Dps) proteins iteratively condense one propionyl-CoA unit and nine malonyl-CoA units. Initially, DpsC, DpsD, and DpsG (analogous to the KS, CLF, and ACP) catalyze a diketide formation between propionyl-CoA and malonyl-CoA. The DpsA, DpsB, and DpsG (again, analogous to the KS, CLF, and ACP) domains catalyze eight more condensation reactions, starting with the previous diketide and adding successive malonyl units to eventually produce a decaketide. The DpsE domain (encoding a KR activity) then reduces the polyketide chain before DpsF cyclizes the chain. DpsH appears to provide another cyclase activity.
FIG. 6.
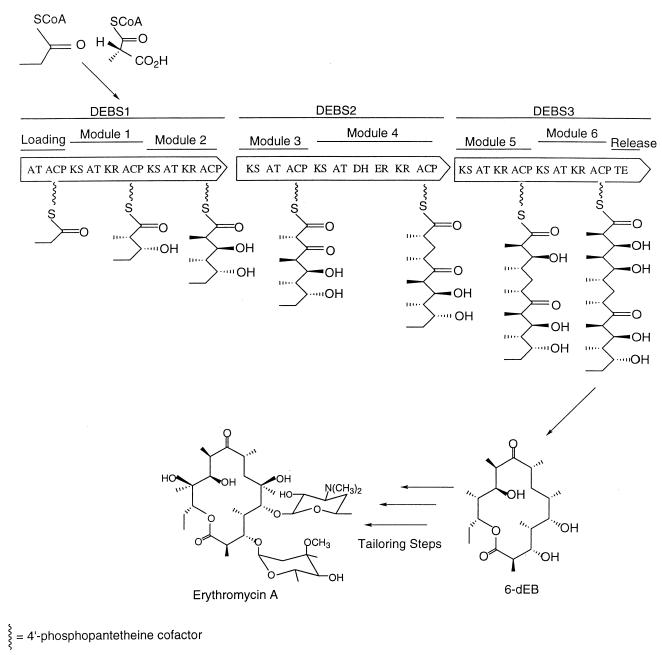
Bacterial type I PKS. The deoxyerythronolide B synthase (DEBS) system catalyzes the formation of the erythromycin derivative 6-deoxyerythronolide B (6-dEB) by processively combining one propionyl-CoA primer unit and six methylmalonyl-CoA extender units. Each DEBS protein contains two modules; modules represent a physical location for a Claisen-like condensation reaction. For example, once loaded onto the KS of module 1 by the loading acyl transferase (AT) and ACP domains, propionyl-CoA condenses with a methylmalonyl unit loaded onto module one ACP by the module 1 AT domain. After the reaction takes place, reductive domains, specific for each module, reduce the resulting ketone group. The chain then passes in a processive manner from module 1 ACP (via the phosphopantetheine arm) to the KS domain of the next module. In this fashion, the polyketide chain grows and diversifies before being released and cyclized by the C-terminal thioesterase (TE) domain.
There are several features associated with microbial polyketide biosynthesis that make these pathways particularly well suited for heterologous expression. First, PKSs are remarkably similar in primary sequences and (by inference) tertiary structures, giving hope that, as improved heuristics for heterologous expression in a given host emerge, they can be applied with greater confidence to newer systems. Second, in all known bacterial and fungal examples, PKS genes have been found to exist as gene clusters (together with transcriptional regulators and self-resistance genes), a feature that greatly simplifies their genetic isolation as well as heterologous expression (at least in bacteria, where multigene operons can be constructed). Third, polyketides are naturally produced as secondary metabolites, suggesting that they can be readily produced in two-stage fermentations in which cell growth can be decoupled from product formation. Fourth, an extraordinary diversity of chemotypes can be synthesized by the PKS paradigm from a relatively small subset of intracellular precursors such as acetyl-CoA, propionyl-CoA, malonyl-CoA, and methylmalonyl-CoA. Moreover, each of these precursors can be derived via multiple metabolic routes from exogenously available carbon sources, thereby presenting a range of options for any potential heterologous host. Finally, as the time lines for converting pharmacologically interesting leads into clinically useful molecules continue to shrink, the earlier paradigm of developing the biology of new microbes for every new polyketide natural product of interest is no longer practical. Lateral transfer of a pathway of interest into a well-developed surrogate host therefore becomes an attractive alternative, both with respect to overproducing the parent natural product itself and for generating novel analogs via biosynthetic engineering. Notwithstanding these advantages, there are several unique characteristics of PKSs that make heterologous expression a definite challenge. Notable PKS characteristics include their relatively large size (100 to 10,000 kDa), the high G+C content of many PKS genes (especially those coming from the actinomycetes, where the G+C content typically exceeds 70%), their requirement for posttranslational modifications, and the relatively rudimentary understanding of the special regulatory and metabolic features associated with the transition from primary to secondary metabolism in any organism.
FACTORS THAT INFLUENCE HETEROLOGOUS PRODUCTION OF POLYKETIDES
As is now clear from several studies involving reconstituted PKS systems (reviewed in reference 42), PKSs are soluble, cytosolic multienzyme systems that do not require any intracellular substructure or organelle to maintain activity. Individually purified protein components (or in some instances proteins purified as heterodimers) can be mixed in vitro to yield PKS activity. While not completely ruling out the need for additional cellular machinery to enhance PKS activity, these experiments indicate that, if introduced into a foreign cellular host, the DNA encoding PKS genes, when actively expressed, should support polyketide production provided that the required substrates are available. Below we review the important considerations for reconstituting polyketide pathways in a heterologous host.
Posttranslational Modification
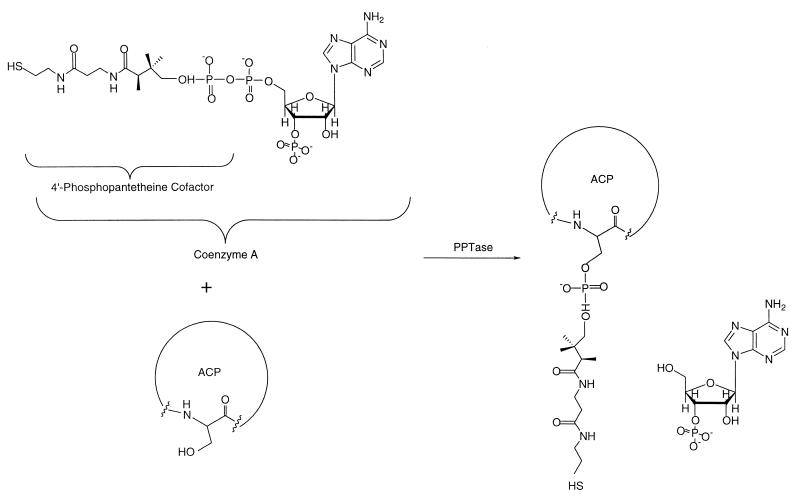
It has long been known that the active site of an ACP involves the thiol moiety of a 4′-phosphopantetheine group that is covalently attached to a conserved serine residue in the polypeptide (79, 83). This pantetheinyl group is posttranslationally derived from intracellular CoASH (Fig. 7) (18). Until recently, very little was known about the enzymatic basis for this modification. Over the past few years, the work of Walsh and coworkers has been instrumental in identifying an evolutionarily related superfamily of enzymes, the phosphopantetheinyl transferases (PPTases), that catalyze this reaction (44, 45). Although these enzymes are known to exist in all organisms except perhaps for the archaea (which do not make fatty acids or polyketides), individual members of this superfamily have significantly distinct configurations and substrate preferences. For example, the E. coli genome contains at least three different PPTase genes (44), each existing as an individual open reading frame (ORF), whereas in Saccharomyces cerevisiae the PPTase that recognizes the cognate fatty acid ACP is a distinct domain within one of the fatty acid synthase subunits (44). Likewise, the E. coli PPTase, which ordinarily pantetheinylates its fatty acid ACP, has relatively tight selectivity for its cognate substrate (44). In contrast, the sfp gene product, which is part of the surfactin biosynthetic gene cluster in Bacillus subtilis, is among the most tolerant PPTase discovered to date and can effectively modify ACPs from all PKS subclasses as well as related peptidyl carrier protein and aryl carrier protein domains from nonribosomal peptide synthetases (NRPSs) (11, 32, 40, 44, 69). (Although a thorough discussion of NRPSs is outside the scope of this review, NRPSs will be occasionally referred to below, given the close relationships between these systems. For further details, the reader is directed to recent reviews on the subject [10, 61].) Therefore, in deciding on a strategy for heterologous PKS gene expression, the choice of a partner PPTase is an important consideration, from both the substrate specificity and the gene regulation perspectives. Depending on the host and expression system chosen, this posttranslational modification may or may not happen.
FIG. 7.
PPTase-catalyzed reaction. The PPTases catalyze the transfer of the 4′-phosphopantetheine arm from free CoASH to a conserved ACP serine. This “swinging arm” facilitates polyketide transfer during biosynthesis.
Substrate Availability
Once functionally expressed and posttranslationally modified within a cellular host, the PKS will require a substrate pool to draw upon for polyketide production. PKSs are known to utilize a broad range of substrates including acetyl-CoA, propionyl-CoA, isobutyryl-CoA, isovaleryl-CoA, malonyl-CoA, methylmalonyl-CoA, ethylmalonyl-CoA, propylmalonyl-CoA, and hydroxymalonyl-CoA (or its methylated counterpart, methoxymalonyl-CoA) (37). When the substrates are chiral, such as methylmalonyl-CoA, the corresponding acyltransferases exhibit strict stereospecificity (52). The α-carboxylated substrates (e.g., malonyl-CoA and methylmalonyl-CoA) are sources of extender units, whereas neutral substrates such as acetyl-CoA are sources of primer units for polyketide chain synthesis. Finally, many “hybrid” natural products are derived from the tandem action of PKSs and NRPSs (9). These multifunctional enzymes utilize an even broader range of substrates, including carboxylic acids such as p-aminobenzoic acid, 3-amino-5-hydroxybenzoic acid, cyclohexenoyl carboxylic acid, and dozens of α- and β-amino acids. Preactivated forms (e.g., CoA thioesters) of these free acids are not required; rather, the acids are activated in situ by ATP-dependent adenylation domains that are intrinsic components of NRPS modules (61).
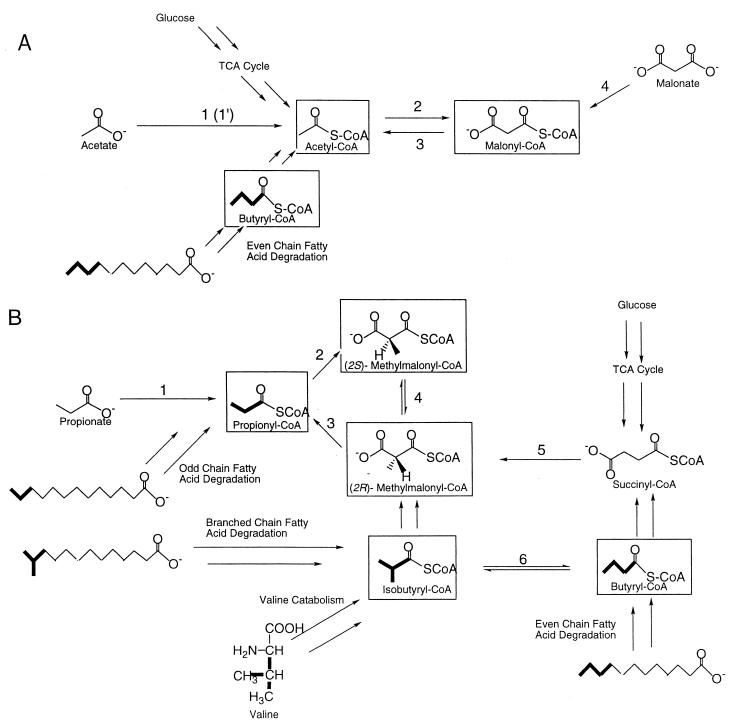
From the above shortlist, it can be appreciated that a good heterologous host must be endowed with the ability to synthesize an impressive range of substrates (in addition, of course, to NADPH and ATP, which are routinely available). Moreover, the supply of these precursors must be coordinately regulated with polyketide biosynthesis so as to ensure availability when required while at the same time avoiding imbalances in tightly controlled metabolite pools (such as those corresponding to various CoA derivatives). Although this might appear to be a formidable problem for the metabolic engineer, several simplifying factors deserve to be noted. (Some of these points are also elaborated below.) First, many polyketide natural products are derived from a subset of acetyl-CoA, propionyl-CoA, malonyl-CoA, and (2S)-methylmalonyl-CoA alone. Therefore, the ability of a candidate host to supply just these four metabolites can make it an attractive environment for heterologous PKS expression. Second, since multiple metabolic routes are known to exist for many of these precursors (Fig. 8), a given host needs to be endowed with only one such pathway in order to facilitate polyketide synthesis. For example, at least four pathways to (2S)-methylmalonyl-CoA are known to exist in bacteria (7, 8, 16, 33, 36, 71, 87). (It should be noted that, from a quantitative viewpoint, not all biosynthetic pathways are energetically or kinetically equivalent.) Third, the broad specificity of certain enzymes that synthesize some of the above metabolites facilitates intracellular production of more than one PKS substrate from a given pathway, depending on the exogenous supply of carboxylic acids. For example, a single enzyme, malonyl-CoA synthetase, can be used to synthesize numerous α-carboxylated CoA thioesters from their corresponding exogenously supplied 1,3-dicarboxylic acids (N. Pohl and C. Khosla, unpublished data). Fourth, the modularity of many PKSs makes it feasible to tailor their substrate preferences to the available intracellular pool of CoA thioesters in a given host (48, 65, 73). A similar approach can also be used to constrain the substrate range of NRPSs, if desired. Fifth, the biosynthetic pathways for relatively uncommon substrates, such as hydroxymalonyl-CoA or 3-amino-5-hydroxybenzoic acid, are often encoded as part of PKS gene clusters that utilize these substrates (1, 75). Therefore, these auxiliary genes can also be harnessed for heterologous expression in very much the same way as the target PKS genes. Finally, since the actinomycetes are the most prolific producers of polyketides, the complete genome sequence of Streptomyces coelicolor (http://www.sanger.ac.uk /Projects/S_coelicolor/), together with its concomitant development as a heterologous host of choice for PKS gene expression, makes the task of the metabolic engineer considerably easier. Orthologs of many known precursor biosynthetic enzymes have already been identified in the genome (unpublished results), and many more can be expected to emerge as functional genomic approaches are applied to investigate the metabolome of this genetically friendly bacterium.
FIG. 8.
Polyketide synthase substrate routes. Potential substrates are boxed. (A) Enzymes performing one enzymatic conversion: 1, acetyl-CoA synthetase (alternatively, 1′ represents a two-enzyme pathway, acetate kinase followed by acetylphosphotransferase); 2, acetyl-CoA carboxylase; 3, malonyl-CoA decarboxylase; 4, malonyl-CoA synthetase. (B) Enzymes performing one enzymatic conversion: 1, propionyl-CoA synthetase (1′, propionate kinase followed by propionylphosphotransferase); 2, propionyl-CoA carboxylase; 3, methylmalonyl-CoA decarboxylase; 4, methylmalonyl-CoA epimerase; 5, methylmalonyl-CoA mutase; 6, isobutyryl-CoA mutase.
Other Intracellular Factors
As mentioned above, coincubation of active PKSs and their substrates is adequate for polyketide formation. However, the surrounding cellular environment may contribute to the optimization of this process in many ways. For example, folding and subsequent quaternary assembly of PKSs may benefit from the activity of many known and as yet uncharacterized chaperones. It is unlikely that PKS folding or assembly is absolutely dependent on the presence of dedicated chaperones, since thus far (i) no mutants defective in polyketide biosynthesis have been shown to have mutations that map onto putative chaperone genes, (ii) no chaperone-like genes have been identified within PKS gene clusters, and (c) PKSs have been heterologously expressed in a diverse range of microbial hosts, including E. coli, Streptomyces, Aspergillus, and yeast. However, the search for auxiliary factors that enhance PKS gene expression in vivo has only just begun. It is likely that quantitative aspects of intracellular PKS activity are influenced by factors that affect the stability of >50-kb transcripts, translational processivity of >30-kb ORFs, translational coupling of ORFs encoding subunits that must assemble in 1:1 stoichiometric ratios, cotranslational and posttranslational protein folding, and proteolytic degradation. An understanding of the mechanistic basis for these activities could have important implications for heterologous PKS expression.
Transmembrane Transporters
Given the potential cytotoxicity of most bioactive polyketides, transmembrane proteins are required for their export. Although putative export proteins (often ATP binding cassette transporter homologs) are often found associated with PKS gene clusters, very little is known about their mechanism or selectivity. For example, inactivation of the gene encoding the actinorhodin exporter in S. coelicolor leads to intracellular accumulation of this isochromanequinone antibiotic (20). However, even in a mutant lacking the entire actinorhodin gene cluster, a number of novel polyfunctional aromatic compounds have been produced and are efficiently secreted into the extracellular medium (56). While the possibility of passive diffusion of these compounds cannot be excluded, the lack of observed toxicity due to polyketide production (even in cases where antibacterial activity of the products has been demonstrated) suggests that other relatively tolerant transporter proteins might be encoded by the S. coelicolor genome. Here, too, functional genomic approaches may be useful in providing clues that will lead to the identification of these transporters.
Post-PKS Polyketide Modification
The formation of biologically active polyketides sometimes requires the activity of various “tailoring” enzymes that act on the PKS-derived intermediate to yield the final natural product. Tailoring enzymes are evolutionarily diverse entities that commonly include cyclases, group transferases (e.g., C-, O-, and N-methyltransferases, glycosyltransferases, and acyltransferases), NADP(H)- or FAD(H)-dependent oxidoreductases, and cytochrome P450-type oxygenases. These enzymes are invariably encoded by genes adjacent to PKS genes and can therefore be readily cloned. In most cases, heterologous expression of these monofunctional enzymes is relatively straightforward. Sometimes, however, cosubstrate availability can be an issue. For example, glycosyltransferases associated with polyketide pathways often utilize specialized TDP-deoxysugars, themselves the products of multistep biosynthetic pathways (26, 66, 80). As with PKS genes, the clustering of the genes responsible for TDP-sugar biosynthesis facilitates their expression in heterologous hosts (64).
Self-Resistance
If biologically active polyketides are to be heterologously produced, a final consideration would be the need for a resistance mechanism to inhibit the effect of the natural product on the heterologous host. Fortunately again, self-resistance genes invariably exist within the natural PKS gene cluster (15, 20, 25). However, coexpression of these genes adds another level of complexity to the heterologous production of polyketides and can be particularly challenging in cases where novel polyketides are engineered for which no known resistance mechanisms have been identified.
CANDIDATE HOSTS FOR HETEROLOGOUS POLYKETIDE PRODUCTION
The actual choice of a candidate heterologous host depends on the objective of the exercise. Three general reasons might motivate the transfer of a polyketide pathway from a natural source into a heterologous host. A possible goal could be overproduction of the target natural product. Many useful polyketides are derived from sources that are virtually impossible to ferment on a large scale (e.g., a marine sponge or dinoflagellate, where production is most probably encoded by a symbiotic microbe) or are produced in small quantities by a microbe with unsatisfactory growth characteristics. In such scenarios, successful transfer of the biosynthetic capability into a genetically and physiologically characterized heterologous host can provide an attractive alternative starting point for subsequent strain and process development. A second reason for heterologous expression might be to provide a more efficient platform for combinatorial biosynthesis. Many wild bacterial and fungal strains that produce natural products represent extremely challenging targets for genetic manipulation or biochemical analysis. This is in sharp contrast to the plethora of genetic tools and physiological insights that are available for model organisms such as those described below. Finally, the lateral transfer of polyketide biosynthesis genes might be motivated by the phenotypic implications of synthesizing a bioactive compound (or a library of its derivatives) in a heterologous host. For example, the production of a clonal library of small-molecule ligands and their target receptor within the same cell could facilitate the design of a selectable system for ligand optimization. Likewise, the production of an antifungal or insecticidal agent in a plant of agronomic significance could herald a new paradigm for crop protection. Below, we highlight the characteristics of selected heterologous hosts that make them well suited for polyketide production.
Streptomyces coelicolor
For many reasons, S. coelicolor is an ideal host for the heterologous production of polyketides. As the first few genes encoding polyketide biosynthesis were identified (3, 5, 13, 17, 21, 78), a suitable host was desired for expressing these genes (in either wild-type or altered forms). The extensive knowledge about the biology of S. coelicolor, coupled with its membership of the Streptomyces genus, made this host a desirable choice. S. coelicolor naturally produces at least two known polyketides of its own (Fig. 2), actinorhodin (50) and the whiE spore pigment (14), although the latter is exclusively spore associated and is not produced in liquid culture. To eliminate background polyketide “noise” due to the actinorhodin pathway, a genetically engineered “clean” host strain, CH999, was constructed, in which the entire actinorhodin (act) gene cluster was surgically deleted via homologous recombination and replaced with an ermE marker gene (56). Concomitantly, a low-copy shuttle vector, pRM5, was engineered, which carries the actI-actIII bidirectional promoter together with the actII-ORF4 gene, which encodes an activator for the actI-actIII promoter (56). Introduction of heterologous PKS genes under the control of this expression system allows the production of polyketides in a secondary metabolite-like manner in a wide range of actinomycete strains including S. coelicolor (56), S. lividans (88), S. parvulus (43), and S. erythraea (72). The list of bacterial and fungal polyketides produced using this host-vector system includes products derived from the frenolicin (57), tetracenomycin (56), oxytetracycline (23), R1128 (53), erythromycin (39), picromycin/methymycin (81), oleandomycin (77), megalomicin (84), 6-methylsalicylic acid (4), and epothilone (82) gene clusters. (In some cases, for technical convenience, a “clean” derivative of S. lividans, K4-114, which also lacks the native act gene cluster, has been used as a heterologous host [88].) PKS proteins are produced at ∼1% of the total cellular protein levels (67), and the titers of the resulting polyketide products are typically in the range of 1 to 100 mg/liter of culture.
The introduction of polyketide pathways into S. coelicolor has also influenced the ease with which PKSs can be genetically engineered for the production of “unnatural” natural products. Type II and modular PKSs have been particularly fertile targets of manipulation in this regard. Libraries of novel aromatic polyketides (58, 86) as well as macrolides (59) have been generated by the combinatorial manipulation of PKS domains and subunits. In turn, this has led to the emergence of heuristics for regioselective modification of polyketide structures. While considerable work lies ahead to increase the predictability of these heuristics to the level of the “codon table” for polypeptide biosynthesis, it is reasonable to anticipate that this enhanced biosynthetic capability will draw heavily from advances in heterologous expression technologies for PKS gene clusters.
A principal limitation of S. coelicolor is that its polyketide natural products (actinorhodin and the spore pigment) are exclusively synthesized from malonyl-CoA-derived building blocks. As such, its metabolic apparatus appears to be limited in the supply of other PKS substrates, such as methylmalonyl-CoA, and the productivity of heterologous polyketides derived from these substrates may suffer. Efforts are under way to better understand the pathways employed by S. coelicolor for PKS substrates. For example, genome-sequencing efforts have already led to the identification of its putative methylmalonyl-CoA mutase subunits (GenBank accession number AL138668); understanding the mechanisms by which the activity of this enzyme is regulated at both the transcriptional and posttranscriptional levels could lead to more efficient conversion of glucose into polyketides via succinyl-CoA (a key tricarboxylic acid pathway intermediate). Likewise, at least three different acyl-CoA carboxylase enzymes are encoded within the S. coelicolor genome (71). This apparent redundancy in the biosynthesis of α-carboxylated CoA thioesters might suggest that different alleles have different substrate preferences (e.g., for acetyl-CoA versus propionyl-CoA) and/or regulatory features in response to the transition from primary metabolism (when only malonyl-CoA derived fatty acids are synthesized) to secondary metabolism (when polyketides are synthesized). Finally, the potential exists for the incorporation of entirely new pathways devoted to substrate production. For example, heterologous expression of the genes encoding the malonyl-CoA synthetase and dicarboxylate transporter protein from Rhizobium trifolii into S. coelicolor led to a substantial enhancement in the yield and productivity of methylmalonyl-CoA-derived erythronolide (unpublished data). Importantly, since this synthetase can activate a wide range of 1,3-dicarboxylic acids into the corresponding CoA thioesters (N. L. Pohl, Y. S. Kim, and C. Khosla, submitted for publication), it provides a potentially useful route for the intracellular generation of substrates that are ordinarily unavailable for polyketide biosynthesis.
In addition to optimizing cellular metabolism in S. coelicolor, further developments in understanding and maximizing gene expression (especially in response to the onset of secondary metabolism) are expected to have a significant impact on the utility of this host for the production of heterologous PKSs and polyketides. Of the different promoters that have been investigated thus far (which include the actI [21], tipA [62], and ermE [6] promoters), the actI promoter appears to have the most favorable characteristics with respect to both expression levels and induction at the onset of the stationary phase. However, the exact signals that lead to maximal induction of this promoter system have not yet been elucidated (54), and PKS proteins are produced only during a relatively short (ca. 24-h) window. Moreover, utilization of this expression system on high-copy-number vectors can lead to instability for reasons that are not understood, and the development of vectors that span a range of copy numbers could be valuable in this regard. Together, these features make controlled, high-level expression of PKS genes in S. coelicolor a challenging yet opportune problem.
Escherichia coli
In general, the utility of E. coli as a host for heterologous gene expression as well as metabolic engineering is unquestioned. However, in order to reconstitute polyketide biosynthesis in E. coli, several issues beyond gene expression needed to be addressed. Unlike S. coelicolor, E. coli is a significantly different heterologous host from those that naturally produce polyketide products. The robust E. coli expression systems available can induce PKS proteins to accumulate as inclusion bodies; however, the judicious control of temperature, medium composition, and other induction conditions yields substantial levels (1 to 5% of total cellular protein) of correctly folded protein for PKSs with molecular masses of >200 kDa (unpublished data). Likewise, coexpression of the sfp PPTase gene in a plasmid-borne or chromosomal format leads to stoichiometric pantetheinylation of soluble PKS proteins in E. coli (B. Pfeifer, unpublished results). Also, the high G+C content of actinomycete PKS genes leads to an inappropriate bias in codon usage that is difficult to remedy using synthetic gene approaches due to the large ORF sizes. The use of host strains that contain extra copies of the rare AGG (arginine) and CCC (proline) codons can be helpful in this regard (reference 74 and unpublished data). The availability of PKS substrates in E. coli presents another major challenge, since biosynthesis of malonyl-CoA is under tight control (40) and substrates such as propionyl-CoA and methylmalonyl-CoA are produced under poorly understood conditions (34). Reconstitution of heterologous substrate generation pathways in this host has provided a useful starting point for addressing this problem (unpublished data). Finally, the availability of well-established protocols for two-stage, high-cell-density fermentation protocols, where the growth phase can be decoupled from PKS gene expression and polyketide production, could provide rapid development of processes that yield high volumetric productivities for both PKSs and polyketides (46).
A variety of PKSs have been functionally expressed in E. coli. For example, coexpression of the sfp and 6-methylsalicylic acid synthase genes in E. coli was found to be necessary and sufficient for the intracellular production of 6-methylsalicylic acid, although polyketide production was substantially enhanced under conditions that ordinarily favor higher rates of malonyl-CoA formation (40). E. coli has the means to produce acetyl-, malonyl-, propionyl-, and possibly even methylmalonyl-CoA, and one might expect the ability to increase these levels based on pathway alteration. However, other options also exist. For example, a pathway leading to the formation of malonyl- and (2S)-methylmalonyl-CoA PKS substrates, which has been successfully reconstituted in E. coli, is based on the S. coelicolor acetyl-CoA carboxylase and propionyl-CoA carboxylase enzyme complexes that use biotin as a cofactor (unpublished data). The genes for acetyl-CoA carboxylase and propionyl-CoA carboxylase have recently been cloned and overexpressed in E. coli in an active form and have shown the ability to provide substrates for polyketide formation within E. coli (71). In summary, although E. coli presents a completely new environment for PKS expression and polyketide formation, it represents a promising opportunity for harnessing the biosynthetic capabilities of polyketide pathways. The ability to produce 6-deoxyerythronolide B in E. coli in a robust manner is an example of realizing this promise (unpublished data).
Other Actinomycetes
For reasons similar to those that prompted the utilization of S. coelicolor as a host for polyketide biosynthesis, other polyketide-producing actinomycetes could be considered. For example, as mentioned above, S. lividans has been successfully used to express several PKS systems, due to its close relationship to S. coelicolor and its greater transformation efficiency (which, for example, readily allows the introduction of multiple plasmids [85]). Similarly, S. glaucescens (76) and Saccharopolyspora erythraea (27) have also been tested as heterologous hosts for polyketide production. These hosts have naturally occurring polyketide biosynthetic pathways and may therefore be better suited for the expression of heterologous PKS genes. Finally, of particular interest in the development of heterologous hosts would be the potential utility of highly evolved polyketide-producing strains. For example, through multiple cycles of random mutagenesis and screening, derivatives of Saccharopolyspora erythraea have been selected that produce 8 g erythromycin per liter (which corresponds to a titer that is 50 to 100 times that in the wild-type strain) (60). Recent studies have demonstrated that the genetic basis for overproduction in such hosts is not encoded within the PKS genes themselves but within other loci on the genome (R. McDaniel, unpublished results). Given the multigenic nature of this trait, the development of such organisms as hosts for polyketide production may present an attractive option. Moreover, a systematic dissection of the genetic and physiological basis for overproduction could yield insights that might readily be translated to naive hosts such as S. coelicolor and E. coli.
Other Bacteria
Although the actinomycetes are perhaps the most prolific producers of polyketides, several other bacterial families are known to synthesize structurally diverse polyketides. Chief among these are the myxobacteria, pseudomonads, and mycobacteria. For example, myxobacteria such as Sorangium spp. produce polyketides including soraphen (31) and epothilone (30). Among other compounds, Pseudomonas spp. produce pseudomonic acid (51) and coronatine (70). Mycobacterium ulcerans produces the highly potent immunosuppressive agent mycolactone (29), whereas the genome of Mycobacterium tuberculosis appears to be well endowed with a plethora of as yet uncharacterized PKS pathways (12). Although genetically well-characterized strains representative of these bacterial families, such as Myxococcus xanthus, Pseudomonas putida, and Mycobacterium smegmatis, are known, the credentials of these bacteria as hosts for heterologous polyketide production have not yet been seriously evaluated.
Fungi
Filamentous fungi too are prolific producers of polyketides. The utility of the fungus Aspergillus nidulans as a heterologous host has been successfully demonstrated in the context of lovastatin biosynthesis (41). A major advantage of such hosts would be the ability to splice introns that are frequently present among eukaryotic PKS genes. Moreover, given the well-established fermentation processes for lovastatin and compactin biosynthesis, it is reasonable to assume that highly evolved strains capable of overproducing these metabolites may be available. If so, these systems could present attractive options for reverse engineering into generic hosts for polyketide biosynthesis. Finally, given the premiere status of the yeast Saccharomyces cerevisiae in fungal and eukaryotic genetics and its designation as a GRAS host (Generally Regarded as Safe), its use as a host for polyketide production ought to be seriously investigated. Although yeast has no known polyketide biosynthetic pathway encoded within its genome, it does have a highly active fatty acid biosynthetic pathway. Moreover, recent studies have shown that it is capable of synthesizing polyketides at high levels (40), although several features must be introduced into the yeast genome before this can be generalized.
Plants
Plants offer another option as potential hosts for the heterologous production of polyketides. They are known to produce a number of polyketide products (e.g., chalcones, stilbenes, and coumarins), and their chloroplasts provide a “bacterium-like” environment that appears to be particularly well suited for fatty acid biosynthesis. Over the past two decades, major advances in plant genetic engineering have led to their emergence as viable hosts for heterologous expression and metabolic engineering. In particular, the ability to produce complex polyketides in plants could have a significant impact on strategies for crop protection in transgenic plants.
FUTURE DIRECTIONS
A critical choice in heterologous expression of any protein is the host; this is especially the case when multiple proteins must be coexpressed to effect a phenotypic alteration that is often the target of metabolic engineering. In addition to laboratory convenience, the chosen heterologous hosts must possess (or be engineered to possess) the cellular machinery required for successful protein production and sustained activity. Following in the footsteps of recombinant polypeptide biosynthesis, recombinant polyketide biosynthesis presents a major challenge as well as an opportunity for molecular biology. This article attempts to highlight the unique challenges encountered during the course of heterologous polyketide production. The current state of the art is presented both with respect to PKS gene expression and metabolic engineering. We speculate that, as has occurred with polypeptides, over the next decade a few heterologous systems will emerge as workhorse tools for exploiting the enormous chemical diversity of naturally occurring polyketides. Key criteria that will influence this decision will be the case of manipulating PKS genes in these hosts, metabolic robustness, volumetric productivity (grams of the polyketide produced per liter per hours), and generality of use with different PKS pathways. However, as it becomes increasingly simpler to introduce polyketide pathways into new biological systems, the biologist will be empowered with a unique set of small-molecule-based tools to probe cellular structure and function. This, in turn, should fuel further research into strategies for lateral transfer of polyketides across taxonomic boundaries. After all, this is precisely what nature appears to have accomplished in its attempt to maximize the evolutionary utility of this remarkable library of functional biomolecules.
ACKNOWLEDGMENTS
Research in our laboratory is supported by grants from the National Institutes of Health (CA 66736 and CA 77248) and the National Science Foundation (BES 9806774). B.A.P. is a recipient of a Stanford-NIH Biotechnology Predoctoral Training Fellowship.
REFERENCES
- 1.August P R, Tang L, Yoon Y J, Ning S, Mueller R, Yu T W, Taylor M, Hoffmann D, Kim C G, Zhang X, Hutchinson C R, Floss H G. Biosynthesis of the ansamycin antibiotic rifamycin. Deductions from the molecular analysis of the rif biosynthetic gene cluster of Amycolatopsis mediterranei S699. Chem Biol. 1998;5:69–79. doi: 10.1016/s1074-5521(98)90141-7. [DOI] [PubMed] [Google Scholar]
- 2.Bao W, Sheldon P J, Wendt-Pienkowski E, Hutchinson C R. The Streptomyces peucetius dpsC gene determines the choice of starter unit in biosynthesis of the daunorubicin polyketide. Biochemistry. 1999;38:9752–9758. doi: 10.1128/jb.181.15.4690-4695.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beck J, Ripka S, Siegner A, Schiltz E, Schweizer E. The multifunctional 6-methylsalicylic acid synthase gene of Penicillium patulum: its gene structure relative to that of other polyketide synthases. Eur J Biochem. 1990;192:487–498. doi: 10.1111/j.1432-1033.1990.tb19252.x. [DOI] [PubMed] [Google Scholar]
- 4.Bedford D J, Schweizer E, Hopwood D A, Khosla C. Expression of a functional fungal polyketide synthase in the bacterium Streptomyces coelicolor A3(2) J Bacteriol. 1995;177:4544–4548. doi: 10.1128/jb.177.15.4544-4548.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bibb M J, Bir̀ S, Motamedi H, Collins J F, Hutchinson C R. Analysis of the nucleotide sequence of the Streptomyces glaucescens tcmI genes provides key information about the enzymology of polyketide biosynthesis. EMBO J. 1989;8:2727–2736. doi: 10.1002/j.1460-2075.1989.tb08414.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bibb M J, Janssen G R, Ward J M. Cloning and analysis of the promoter region of the erythromycin resistance gene (ermE) of Streptomyces erythraeus. Gene. 1985;38:215–226. doi: 10.1016/0378-1119(85)90220-3. [DOI] [PubMed] [Google Scholar]
- 7.Birch A, Leiser A, Robinson J. Cloning, sequencing, and expression of the gene encoding methylmalonyl-CoA mutase from Streptomyces cinnamonensis. J Bacteriol. 1993;175:3511–3519. doi: 10.1128/jb.175.11.3511-3519.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bramwell H, Hunter I S, Coggins J R, Nimmo H G. Propionyl-CoA carboxylase from Streptomyces coelicolor A3(2): cloning of the gene encoding the biotin-containing subunit. Microbiology. 1996;142:649–655. doi: 10.1099/13500872-142-3-649. [DOI] [PubMed] [Google Scholar]
- 9.Cane D E, Walsh C T. The parallel and convergent universes of polyketide synthases and nonribosomal peptide synthetases. Chem Biol. 1999;6:R319–R325. doi: 10.1016/s1074-5521(00)80001-0. [DOI] [PubMed] [Google Scholar]
- 10.Cane D E, Walsh C T, Khosla C. Harnessing the biosynthetic code. Combinations, permutations, mutations. Science. 1998;282:63–68. doi: 10.1126/science.282.5386.63. [DOI] [PubMed] [Google Scholar]
- 11.Carreras C W, Gehring A M, Walsh C T, Khosla C. Utilization of enzymatically phosphopantetheinylated acyl carrier proteins and acetyl-acyl carrier proteins by the actinorhodin polyketide synthase. Biochemistry. 1997;36:11757–11761. doi: 10.1021/bi971350+. [DOI] [PubMed] [Google Scholar]
- 12.Cole S T, Brosch R, Parkhill J, Garnier T, Churcher C, Harris D, Gordon S V, Eiglmeier K, Gas S, Barry III C E, Tekaia F, Badcock K, Basham D, Brown D, Chillingworth T, Connor R, Davies R, Devlin K, Feltwell T, Gentles S, Hamlin N, Holroyd S, Hornsby T, Jagels K, Krogh A, McLean J, Moule S, Murphy L, Oliver S, Osborne J, Quail M A, Rajandream M A, Rogers J, Rutter S, Seeger K, Skelton S, Squares S, Sqares R, Sulston J E, Taylor K, Whitehead S, Barrell B G. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature. 1998;393:537–544. doi: 10.1038/31159. [DOI] [PubMed] [Google Scholar]
- 13.Cortes J, Haydock S F, Roberts G A, Bevitt D J, Leadlay P F. An unusually large multifunctional polypeptide in the erythromycin-producing polyketide synthase of Saccharopolyspora erythraea. Nature. 1990;348:176–178. doi: 10.1038/348176a0. [DOI] [PubMed] [Google Scholar]
- 14.Davis N K, Chater K F. Spore colour in Streptomyces coelicolor A3(2) involves the developmentally regulated synthesis of a compound biosynthetically related to polyketide antibiotics. Mol Microbiol. 1990;4:1679–1691. doi: 10.1111/j.1365-2958.1990.tb00545.x. [DOI] [PubMed] [Google Scholar]
- 15.Dhillon N, Hale R S, Cortes J, Leadlay P F. Molecular characterization of a gene from Saccharopolyspora erythraea (Streptomyces erythreus) which is involved in erythromycin biosynthesis. Mol Microbiol. 1989;3:1405–1414. doi: 10.1111/j.1365-2958.1989.tb00123.x. [DOI] [PubMed] [Google Scholar]
- 16.Donadio S, Staver M J, Katz L. Erythromycin production in Saccharopolyspora erythraea does not require a functional propionyl-CoA carboxylase. Mol Microbiol. 1996;19:977–984. doi: 10.1046/j.1365-2958.1996.439969.x. [DOI] [PubMed] [Google Scholar]
- 17.Donadio S, Staver M J, McAlpine J B, Swanson S J, Katz L. Modular organization of genes required for complex polyketide biosynthesis. Science. 1991;252:675–679. doi: 10.1126/science.2024119. [DOI] [PubMed] [Google Scholar]
- 18.Elovson J, Vagelos P R. Acyl carrier protein. X. Acyl carrier protein synthetase. J Biol Chem. 1968;243:3603–3611. [PubMed] [Google Scholar]
- 19.Ensley B D, Ratzkin B J, Osslund T D, Simon M J, Wackett L P, Gibson D T. Expression of naphthalene oxidation genes in Escherichia coli results in the biosynthesis of indigo. Science. 1983;222:167–169. doi: 10.1126/science.6353574. [DOI] [PubMed] [Google Scholar]
- 20.Fernǹdez-Moreno M A, Caballero J L, Hopwood D A, Malpartida F. The act cluster contains regulatory and antibiotic export genes, direct targets for translational control by the bldA tRNA gene of Streptomyces. Cell. 1991;66:769–780. doi: 10.1016/0092-8674(91)90120-n. [DOI] [PubMed] [Google Scholar]
- 21.Fernǹdez-Moreno M A, Martǹez E, Boto L, Hopwood D A, Malpartida F. Nucleotide sequence and deduced functions of a set of cotranscribed genes of Streptomyces coelicolor A3(2) including the polyketide synthase for the antibiotic actinorhodin. J Biol Chem. 1992;267:19278–19290. [PubMed] [Google Scholar]
- 22.Ferrer J L, Jez J M, Bowman M E, Dixon R A, Noel J P. Structure of chalcone synthase and the molecular basis of plant polyketide biosynthesis. Nat Struct Biol. 1999;6:775–784. doi: 10.1038/11553. [DOI] [PubMed] [Google Scholar]
- 23.Fu H, Ebert-Khosla S, Hopwood D A, Khosla C. Relaxed specificity of the oxytetracycline polyketide synthase for an acetate primer in the absence of a malonamyl primer. J Am Chem Soc. 1994;116:6443–6444. [Google Scholar]
- 24.Funa N, Ohnishi Y, Shibuya M, Ebizuka Y, S H. A new pathway for polyketide synthesis in microorganisms. Nature. 1999;400:897–899. doi: 10.1038/23748. [DOI] [PubMed] [Google Scholar]
- 25.Furuya K, Hutchinson C R. The DrrC protein of Streptomyces peucetius, a UvrA like protein, is a DNA binding protein whose gene is induced by daunorubicin. FEMS Microbiol Lett. 1998;168:243–249. doi: 10.1111/j.1574-6968.1998.tb13280.x. [DOI] [PubMed] [Google Scholar]
- 26.Gaisser S, Bohm G A, Doumith M, Raynal M C, Dhillon N, Cortes J, Leadlay P F. Analysis of eryBI, eryBIII and eryBVII from the erythromycin biosynthetic gene cluster in Saccharopolyspora erythraea. Mol Gen Genet. 1998;258:78–88. doi: 10.1007/s004380050709. [DOI] [PubMed] [Google Scholar]
- 27.Gaisser S, Reather J, Wirtz G, Kellenberger L, Staunton J, Leadlay P F. A defined system for hybrid macrolide biosynthesis in Saccharopolyspora erythraea. Mol Microbiol. 2000;36:391–401. doi: 10.1046/j.1365-2958.2000.01856.x. [DOI] [PubMed] [Google Scholar]
- 28.Gehring A M, DeMoll E, Fetherston J D, Mori I, Mayhew G F, Blattner F R, Walsh C T, Perry R D. Iron acquisition in plague: modular logic in enzymatic biogenesis of yersiniabactin by Yersinia pestis. Chem Biol. 1998;5:573–586. doi: 10.1016/s1074-5521(98)90115-6. [DOI] [PubMed] [Google Scholar]
- 29.George K M, Chatterjee D, Gunawardana G, Welty D, Hayman J, Lee R, Small P L. Mycolactone: a polyketide toxin from Mycobacterium ulcerans required for virulence. Science. 1999;283:854–857. doi: 10.1126/science.283.5403.854. [DOI] [PubMed] [Google Scholar]
- 30.Gerth K, Bedorf N, Hofle G, Irschik H, Reichenbach H. Epothilons A and B: antifungal and cytotoxic compounds from Sorangium cellulosum (Myxobacteria). Production, physico-chemical and biological properties. J Antibiot. 1996;49:560–563. doi: 10.7164/antibiotics.49.560. [DOI] [PubMed] [Google Scholar]
- 31.Gerth K, Bedorf N, Irschik H, Hofle G, Reichenbach H. The soraphens: a family of novel antifungal compounds from Sorangium cellulosum (Myxobacteria). I. Soraphen A1 alpha: fermentation, isolation, biological properties. J Antibiot. 1994;47:23–31. doi: 10.7164/antibiotics.47.23. [DOI] [PubMed] [Google Scholar]
- 32.Gokhale R S, Tsuji S Y, Cane D E, Khosla C. Dissecting and exploiting intermodular communication in polyketide synthases. Science. 1999;284:482–485. doi: 10.1126/science.284.5413.482. [DOI] [PubMed] [Google Scholar]
- 33.Gottschalk G. Bacterial metabolism. 2nd ed. New York, N.Y: Springer-Verlag; 1986. [Google Scholar]
- 34.Haller T, Buckel T, Retey J, Gerlt J A. Discovering new enzymes and metabolic pathways: conversion of succinate to propionate by Escherichia coli. Biochemistry. 2000;39:4622–4629. doi: 10.1021/bi992888d. [DOI] [PubMed] [Google Scholar]
- 35.Hopwood D A, Sherman D H. Molecular genetics of polyketides and its comparison to fatty acid biosynthesis. Annu Rev Genet. 1990;24:37–66. doi: 10.1146/annurev.ge.24.120190.000345. [DOI] [PubMed] [Google Scholar]
- 36.Hunaiti A R, Kolattukudy P E. Isolation and characterization of an acyl-coenzyme A carboxylase from an erythromycin-producing Streptomyces erythyreus. Arch Biochem Biophys. 1982;216:362–371. doi: 10.1016/0003-9861(82)90222-3. [DOI] [PubMed] [Google Scholar]
- 37.Hutchinson C R, Decker H, Madduri K, Otten S L, Tang L. Genetic control of polyketide biosynthesis in the genus Streptomyces. Antonie Leeuwenhoek. 1993;64:165–176. doi: 10.1007/BF00873025. [DOI] [PubMed] [Google Scholar]
- 38.Jez J M, Ferrer J L, Bowman M E, Dixon R A, Noel J P. Dissection of malonyl-coenzyme A decarboxylation from polyketide formation in the reaction mechanism of a plant polyketide synthase. Biochemistry. 2000;39:890–902. doi: 10.1021/bi991489f. [DOI] [PubMed] [Google Scholar]
- 39.Kao C M, Katz L, Khosla C. Engineered biosynthesis of a complete macrolactone in a heterologous host. Science. 1994;265:509–512. doi: 10.1126/science.8036492. [DOI] [PubMed] [Google Scholar]
- 40.Kealey J, Liu L, Santi D V, Betlach M, Barr P J. Production of a polyketide natural product in non-polyketide producing prokaryotic and eukaryotic hosts. Proc Natl Acad Sci USA. 1998;95:505–509. doi: 10.1073/pnas.95.2.505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kennedy J, Auclair K, Kendrew S G, Park C, Vederas J C, Hutchinson C R. Modulation of polyketide synthase activity by accessory proteins during lovastatin biosynthesis. Science. 1999;284:1368–1372. doi: 10.1126/science.284.5418.1368. [DOI] [PubMed] [Google Scholar]
- 42.Khosla C, Gokhale R, Jacobsen J R, Cane D E. Tolerance and specificity of polyketide synthases. Annu Rev Biochem. 1999;68:219–253. doi: 10.1146/annurev.biochem.68.1.219. [DOI] [PubMed] [Google Scholar]
- 43.Kim E S, Cramer K D, Sherve A L, Sherman D H. Heterologous expression of an engineered biosynthetic pathway: functional dissection of type II polyketide synthase components in Streptomyces species. J Bacteriol. 1995;177:1202–1207. doi: 10.1128/jb.177.5.1202-1207.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lambalot R H, Gehring A M, Flugel R S, Zuber P, LaCelle M, Marahiel M A, Reid R, Khosla C, Walsh C T. A new enzyme superfamily—the phosphopantetheinyl transferases. Chem Biol. 1996;3:923–936. doi: 10.1016/s1074-5521(96)90181-7. [DOI] [PubMed] [Google Scholar]
- 45.Lambalot R H, Walsh C T. Cloning, overproduction, and characterization of the Escherichia coli holo-acyl carrier protein synthase. J Biol Chem. 1995;270:24658–24661. doi: 10.1074/jbc.270.42.24658. [DOI] [PubMed] [Google Scholar]
- 46.Lee S Y. High cell-density culture of Escherichia coli. Trends Biotechnol. 1996;14:98–105. doi: 10.1016/0167-7799(96)80930-9. [DOI] [PubMed] [Google Scholar]
- 47.Li K, Frost J W. Synthesis of vanillin from glucose. J Am Chem Soc. 1998;120:10545–10546. [Google Scholar]
- 48.Liu L, Thamchaipenet A, Fu H, Betlach M, Ashley G. Biosynthesis of 2-nor-6-deoxyerythronolide B by rationally designed domain substitution. J Am Chem Soc. 1997;119:10553–10554. [Google Scholar]
- 49.Reference deleted.
- 50.Malpartida F, Hopwood D A. Molecular cloning of the whole biosynthetic pathway of a Streptomyces antibiotic and its expression in a heterologous host. Nature. 1984;309:462–464. doi: 10.1038/309462a0. [DOI] [PubMed] [Google Scholar]
- 51.Mantle P G, Macgeorge K M. The role of monic acid A in pseudomonic acid A biosynthesis in Pseudomonas fluorescens. Appl Microbiol Biotechnol. 1990;33:709–711. doi: 10.1007/BF00604943. [DOI] [PubMed] [Google Scholar]
- 52.Marsden A F A, Caffrey P, Aparicio J F, Loughran M S, Staunton J, Leadlay P F. Stereospecific acyl transfers on the erythromycin-producing polyketide synthase. Science. 1994;263:378–380. doi: 10.1126/science.8278811. [DOI] [PubMed] [Google Scholar]
- 53.Marti T, Hu Z, Pohl N L, Shah A N, Khosla C. Cloning, nucleotide sequence, and heterologous expression of the biosynthetic gene cluster for R1128, a non-steroidal estrogen receptor antagonist. Insights into an unusual priming mechanism. J Biol Chem. 2000;275:33443–33448. doi: 10.1074/jbc.M006766200. [DOI] [PubMed] [Google Scholar]
- 54.Martinez-Costa O H, Martin-Triana A J, Martinez E, Fernandez-Moreno M A, Malpartida F. An additional regulatory gene for actinorhodin production in Streptomyces lividans involves a LysR-type transcriptional regulator. J Bacteriol. 1999;181:4353–4364. doi: 10.1128/jb.181.14.4353-4364.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mayorga M E, Timberlake W E. The developmentally regulated Aspergillus nidulans wA gene encodes a polypeptide homologous to polyketide and fatty acid synthases. Mol Gen Genet. 1992;235:205–212. doi: 10.1007/BF00279362. [DOI] [PubMed] [Google Scholar]
- 56.McDaniel R, Ebert-Khosla S, Hopwood D A, Khosla C. Engineered biosynthesis of novel polyketides. Science. 1993;262:1546–1550. doi: 10.1126/science.8248802. [DOI] [PubMed] [Google Scholar]
- 57.McDaniel R, Ebert-Khosla S, Hopwood D A, Khosla C. Engineered biosynthesis of novel polyketides: manipulation and analysis of an aromatic polyketide synthase with unproven catalytic specificities. J Am Chem Soc. 1993;115:11671–11675. [Google Scholar]
- 58.McDaniel R, Ebert-Khosla S, Hopwood D A, Khosla C. Rational design of aromatic polyketide natural products by recombinant assembly of enzymatic subunits. Nature. 1995;375:549–554. doi: 10.1038/375549a0. [DOI] [PubMed] [Google Scholar]
- 59.McDaniel R, Thamchaipenet A, Gustafsson C, Fu H, Betlach M, Ashley G. Multiple genetic modifications of the erythromycin polyketide synthase to produce a library of novel “unnatural” natural products. Proc Natl Acad Sci USA. 1999;96:1846–1851. doi: 10.1073/pnas.96.5.1846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Minas W, Brunker P, Kallio P T, Bailey J E. Improved erythromycin production in a genetically engineered industrial strain of Saccharopolyspora erythraea. Biotechnol Prog. 1998;14:561–566. doi: 10.1021/bp980055t. [DOI] [PubMed] [Google Scholar]
- 61.Mootz H D, Marahiel M A. Biosynthetic systems for nonribosomal peptide antibiotic assembly. Curr Opin Chem Biol. 1997;1:543–551. doi: 10.1016/s1367-5931(97)80051-8. [DOI] [PubMed] [Google Scholar]
- 62.Murukami T, Holt T G, Thompson C J. Thiostrepton-induced gene expression in Streptomyces lividans. J Bacteriol. 1989;171:1459–1466. doi: 10.1128/jb.171.3.1459-1466.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.O'Hagan D. The polyketide metabolites. Chichester, United Kingdom: Ellis Horwood; 1991. [Google Scholar]
- 64.Olano C, Lomovskaya N, Fonstein L, Roll J T, Hutchinson C R. A two-plasmid system for the glycosylation of polyketide antibiotics: bioconversion of epsilon-rhodomycinone to rhodomycin D. Chem Biol. 1999;6:845–855. doi: 10.1016/s1074-5521(00)80004-6. [DOI] [PubMed] [Google Scholar]
- 65.Oliynyk M, Brown M J B, Cortes J, Staunton J, Leadlay P F. A hybrid modular polyketide synthase obtained by domain swapping. Chem Biol. 1996;3:833–839. doi: 10.1016/s1074-5521(96)90069-1. [DOI] [PubMed] [Google Scholar]
- 66.Otten S L, Gallo M A, Madduri K, Liu X, Hutchinson C R. Cloning and characterization of the Streptomyces peucetius dnmZUV genes encoding three enzymes required for biosynthesis of the daunorubicin precursor thymidine diphopho-l-daunosamine. J Bacteriol. 1997;179:4446–4450. doi: 10.1128/jb.179.13.4446-4450.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Pieper R, Luo G, Cane D E, Khosla C. Cell-free biosynthesis of polyketides by recombinant erythromycin polyketide synthases. Nature. 1995;378:263–266. doi: 10.1038/378263a0. [DOI] [PubMed] [Google Scholar]
- 68.Reference deleted.
- 69.Quadri L E, Weinreb P H, Lei M, Nakano M M, Zuber P, Walsh C T. Characterization of Sfp, a Bacillus subtilis phosphopantetheinyl transferase for peptidyl carrier protein domains in peptide synthetases. Biochemistry. 1998;37:1585–1595. doi: 10.1021/bi9719861. [DOI] [PubMed] [Google Scholar]
- 70.Rangaswamy V, Jiralerspong S, Parry R, Bender C L. Biosynthesis of the Pseudomonas polyketide coronafacic acid requires monofunctional and multifunctional polyketide synthase proteins. Proc Natl Acad Sci USA. 1998;95:15469–15474. doi: 10.1073/pnas.95.26.15469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Rodriguez E, Gramajo H. Genetic and biochemical characterization of the alpha and beta components of a propionyl-CoA carboxylase complex of Streptomyces coelicolor A3(2) Microbiology. 1999;145:3109–3119. doi: 10.1099/00221287-145-11-3109. [DOI] [PubMed] [Google Scholar]
- 72.Rowe C J, Cortes J, Gaisser S, Staunton J, Leadlay P F. Construction of new vectors for high-level expression in actinomycetes. Gene. 1998;216:215–223. doi: 10.1016/s0378-1119(98)00327-8. [DOI] [PubMed] [Google Scholar]
- 73.Ruan X, Pereda A, Stassi D L, Zeidner D, Summers R G, Jackson M, Shivakumar A, Kakavas S, Staver M J, Donadio S, Katz L. Acyltransferase domain substitutions in erythromycin polyketide synthase yield novel erythromycin derivatives. J Bacteriol. 1997;179:6416–6425. doi: 10.1128/jb.179.20.6416-6425.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Schenk P M, Baumann S, Mattes R, Steinbiss H. Improved high level expression systems for eukaryotic genes in Escherichia coli using T7 RNA polymerase and rare ArgtRNAs. BioTechniques. 1995;19:196–200. [PubMed] [Google Scholar]
- 75.Schweke T, Aparicio J F, Molnar I, Konig A, Khaw L E, Haydock S F, Oliynyk M, Caffrey P, Cortes J, Lester J B, Bohm G, Staunton J, Leadlay P F. The biosynthetic gene cluster for the polyketide immunosuppressant rapamycin. Proc Natl Acad Sci USA. 1995;92:7839–7843. doi: 10.1073/pnas.92.17.7839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Seow K T, Meurer G, Gerlitz M, Wendt-Pienkowski E, Hutchinson C R, Davies J. A study of iterative type II polyketide synthases, using bacterial genes cloned from soil DNA: a means to access and use genes from uncultured microorganisms. J Bacteriol. 1997;179:7360–7368. doi: 10.1128/jb.179.23.7360-7368.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Shah S, Xue Q, Tang L, Carney J R, Betlach M, McDaniel R. Cloning, characterization, and heterologous expression of a polyketide synthase and P-450 oxidase involved in the biosynthesis of the antibiotic oleandomycin. J Antibiot. 2000;53:502–508. doi: 10.7164/antibiotics.53.502. [DOI] [PubMed] [Google Scholar]
- 78.Sherman D H, Malpartida F, Bibb M J, Kieser H M, Bibb M J, Hopwood D A. Structure and deduced function of the granaticin-producing polyketide synthase gene cluster of Streptomyces violaceoruber Tü22. EMBO J. 1989;8:2717–2725. doi: 10.1002/j.1460-2075.1989.tb08413.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Simoni R D, Criddle R S, Stumpf P K. Fat metabolism in higher plants. XXXI. Purification and properties of plant and bacterial acyl carrier proteins. J Biol Chem. 1967;242:573–581. [PubMed] [Google Scholar]
- 80.Summers R G, Donadio S, Staver M J, Wendt-Pienkowski E, Hutchinson C R, Katz L. Sequencing and mutagenesis of genes from the erythromycin biosynthetic gene cluster of Saccharopolyspora erythraea that are involved in l-mycarose and d-desosamine production. Microbiology. 1997;143:3251–3262. doi: 10.1099/00221287-143-10-3251. [DOI] [PubMed] [Google Scholar]
- 81.Tang L, Fu H, Betlach M, McDaniel R. Elucidating the mechanism of chain termination switching in the picromycin/methymycin polyketide synthase. Chem Biol. 1999;6:553–558. doi: 10.1016/S1074-5521(99)80087-8. [DOI] [PubMed] [Google Scholar]
- 82.Tang L, Shah S, Chung L, Carney J, Katz L, Khosla C, Julien B. Cloning and heterologous expression of the epothilone gene cluster. Science. 2000;287:640–642. doi: 10.1126/science.287.5453.640. [DOI] [PubMed] [Google Scholar]
- 83.Vanaman T C, Wakil S J, Hill R L. The complete amino acid sequence of the acyl carrier protein of Escherichia coli. J Biol Chem. 1968;243:6420–6431. [PubMed] [Google Scholar]
- 84.Volchegurski Y, Hu Z, Katz L, McDaniel R. Biosynthesis of the anti-parasite agent megalomicin: transformation of erythromycin to megalomicin in Saccharopolyspora erythraea. Mol Microbiol. 2000;37:752–762. doi: 10.1046/j.1365-2958.2000.02059.x. [DOI] [PubMed] [Google Scholar]
- 85.Xue Q, Ashley G, Hutchinson C R, Santi D V. A multiplasmid approach to preparing large libraries of polyketides. Proc Natl Acad Sci USA. 1999;96:11740–11745. doi: 10.1073/pnas.96.21.11740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Yu T W, Shen Y, McDaniel R, Floss H G, Khosla C, Hopwood D A, Moore B S. Engineered biosynthesis of novel polyketides from Streptomyces spore pigment polyketide synthases. J Am Chem Soc. 1998;120:7749–7759. [Google Scholar]
- 87.Zerbe-Burkhardt K, Ratnatilleke A, Phillipon N, Birch A, Leiser A, Vrijbloed J, Hess D, Hunziker P, Robinson J. Cloning, sequencing, expression, and insertional inactivation of the gene for the large subunit of the coenzyme B12-dependent isobutyryl-CoA mutase from Streptomyces cinnamonensis. J Biol Chem. 1998;273:6508–6517. doi: 10.1074/jbc.273.11.6508. [DOI] [PubMed] [Google Scholar]
- 88.Ziermann R, Betlach M C. Recombinant polyketide synthesis in Streptomyces: engineering of improved host strains. BioTechniques. 1999;26:106–110. doi: 10.2144/99261st05. [DOI] [PubMed] [Google Scholar]