Keywords: bronchopulmonary dysplasia, hyperoxia, neonates, pulmonary, single-cell RNA sequencing
Abstract
Hyperoxia disrupts lung development in mice and causes bronchopulmonary dysplasia (BPD) in neonates. To investigate sex-dependent molecular and cellular programming involved in hyperoxia, we surveyed the mouse lung using single cell RNA sequencing (scRNA-seq), and validated our findings in human neonatal lung cells in vitro. Hyperoxia-induced inflammation in alveolar type (AT) 2 cells gave rise to damage-associated transient progenitors (DATPs). It also induced a new subpopulation of AT1 cells with reduced expression of growth factors normally secreted by AT1 cells, but increased mitochondrial gene expression. Female alveolar epithelial cells had less EMT and pulmonary fibrosis signaling in hyperoxia. In the endothelium, expansion of Car4+ EC (Cap2) was seen in hyperoxia along with an emergent subpopulation of Cap2 with repressed VEGF signaling. This regenerative response was increased in females exposed to hyperoxia. Mesenchymal cells had inflammatory signatures in hyperoxia, with a new distal interstitial fibroblast subcluster characterized by repressed lipid biosynthesis and a transcriptomic signature resembling myofibroblasts. Hyperoxia-induced gene expression signatures in human neonatal fibroblasts and alveolar epithelial cells in vitro resembled mouse scRNA-seq data. These findings suggest that neonatal exposure to hyperoxia programs distinct sex-specific stem cell progenitor and cellular reparative responses that underpin lung remodeling in BPD.
INTRODUCTION
Mammalian lung development progresses sequentially through the embryonic, pseudoglandular, canalicular, saccular, and alveolar stages (1). The establishment of the terminal gas exchange unit, the alveolus, lined by alveolar epithelial type 1 cells (AT1s) and juxtaposed with capillary endothelial cells (ECs) is the “summum bonum” of lung development. Lung ontogeny is temporally and spatially regulated by cell-autonomous and nonautonomous signaling networks that specify cellular phenotypes, diversity, maturation, and cell-cell interactions that establish the alveolar-capillary barrier. In recent years, lung single-cell RNA-sequencing (scRNA-seq) has revolutionized our understanding of the complexity and uniqueness of developmental programming of the cells that specify the alveolar niche (2–7). This fundamental knowledge is rapidly allowing us to interrogate deviant programming of cellular phenotypes underlying lung diseases using experimental animal models and human pathology samples.
Bronchopulmonary dysplasia (BPD) is a chronic lung disease that affects newborns born prematurely with canalicular or saccular lungs, not primed for gas exchange (8). Preterm infants are exposed to high concentrations of oxygen (hyperoxia) that causes lung injury and disrupts lung development (8, 9). Pathologically, a reduction in the alveoli with pruning of vascular arborization, along with fibrosis and dysplastic lung development are well described. Signaling cascades related to the VEGF, TGFB, TP53, RhoA, IL1, and fibrosis pathways underlie the pathological changes observed in BPD (10–15). Although disturbances in these signaling pathways are ascribed to arise from alveolar epithelial, endothelial, mesenchymal, and immune populations, transcriptomic networks that program cell-type specific alterations in phenotype specification, development, and function in BPD are incompletely understood. We undertook a single-cell survey in a hyperoxia model of experimental BPD to address these key questions. Strikingly, female preterm neonates are less susceptible to BPD in comparison with gestational age-matched male preterm infants (16, 17). We also explored the mechanisms associated with decreased hyperoxia-induced lung injury in female neonatal mice at single-cell resolution (18–21).
ScRNA-seq studies have defined >35 lung cell types that can be grouped into four major cell lineages: epithelium, endothelium, stromal cells, and immune cells (2, 22). The alveolar epithelium comprises cuboidal alveolar epithelial type 2 (AT2) cells that secrete surfactant, and serve as progenitor cells in the adult lung (23, 24). AT1 cells are thin, flat cells that cover the alveolar surface, and facilitate gas exchange and alveolarization (25, 26). In models of adult lung injury, AT2 cells serving as progenitor cells repopulate the AT1 population (5, 24, 27). Injury-induced AT2 inflammation represses AT2 differentiation to AT1 and leads to alveolar epithelial-to-mesenchymal transition (EMT) (5, 28). TP53 and TGFB inhibit AT2-AT1 intermediate cell maturation to AT1 cells, favoring lung fibrosis (23, 28). Whether hyperoxia-induced lung injury programs changes in AT2 to restore the AT1 population in BPD is unknown. Lung capillary ECs specify into capillary endothelial cell type 2 (Cap2 or aCap) that lie opposed to AT1, participate in gas exchange, and capillary endothelial cell type 1 (Cap1 or gCap), which have distinct vasomotor, immune, and regenerative function (3, 29–32). Car4 expressing Cap2 ECs are known to respond to paracrine signals and might be key to EC and alveolar regeneration (30). However, since Cap2 does not proliferate, it is accepted that Cap1, which retains proliferative and progenitor properties, gives rise to Cap2 during lung injury. The mechanisms by which hyperoxia in the neonatal lung programs cellular and transcriptional networks important for EC recovery is not known. Alveolar myofibroblasts (AMF; myofibroblast 1), ductal myofibroblasts (DMF; myofibroblast 2), distal interstitial fibroblasts (DIF; lipofibroblasts), and proximal interstitial fibroblasts (PIF; matrix fibroblasts) are four major fibroblasts involved in alveolarization and alveolar maintenance (3, 33–35). Excessive proliferation of injury-transformed fibroblasts and extracellular matrix accumulation leads to lung fibrosis (36). The impact of hyperoxia on fibroblast subpopulations and transcriptional networks that contribute to matrix remodeling in human and experimental BPD was elucidated herein.
In this study, we investigated the cell-type specific alterations in transcriptomic networks that program injury, repair, and deviant development in BPD using a model of experimental BPD in which mice are exposed to hyperoxia (85%) from P1 to P14. Combining scRNA-seq data with validation studies in mice and human lung cell lines we show that experimental BPD programs unique phenotypic changes in lung alveolar epithelial cell, capillary EC, fibroblast, and immune cell populations that underpin injury/adaptive responses in hyperoxia. We also investigated sex-specific transcriptomic changes in the injury-responsive cells that might explain the decreased vulnerability or augmented regenerative response to hyperoxia injury seen in female infants.
MATERIALS AND METHODS
Study Approvals
Lab experiments were reviewed and approved under the University of Missouri-Kansas City IBC, protocol number 18–28 and the Children’s Mercy Research Institute IBC, protocol number 025. Animal experiments were reviewed and approved under the University of Missouri-Kansas City IACUC, protocol number 1510-03.
Mice (Mus musculus)
Mice were housed and treated with hyperoxia at the University of Missouri-Kansas City animal facility. Mice were kept in a 12-h light/dark schedule as per the facility animal housing protocol described before (37, 38). WT C57BL/6 (Charles River) mice were used in the study. For hyperoxia, pups with dam were placed in cages in a hyperoxia chamber for 14 days from P1 to P14. We used 3 females and 3 males/experimental condition for scRNA-seq, and 4 females and 4 males for validation studies. The enclosure was flushed with oxygen and the concentration was continuously monitored and controlled with a Pro-ox oxygen controller (BioSpherix) to maintain 85% O2 throughout the study period. Control mice were housed similarly but were not exposed to hyperoxia. Two dams were switched daily between hyperoxia and room air. Pups were euthanized using a 100 mg/kg intraperitoneal injection of pentobarbital, exsanguinated after cessation of heartbeat, and the lungs were harvested as described in Cell Dissociation.
Cell Dissociation
Lungs from experimental C57BL/6 mice were inflated with an enzyme mix consisting of the following: Leibovitz media (Invitrogen) 2 mg/mL of collagenase type 1 and 2 mg/mL of elastase (Worthington Biochemical). Lungs were then chopped with a razor blade into fine chunks and placed in a prewarmed digestion enzyme mix—inflation enzyme mix plus 0.5 mg/mL DNase 1 (Worthington Biochemical). They were placed in an incubator at 37°C and rotated for 15 min; next they were mechanically digested with a 1 mL pipette tip. The digestion mix was placed back into the incubator and rotated for another 15 min, after which the digestion was stopped with 20% FBS and the lysate was filtered through a 70 µm filter (Thermo Fisher Scientific). The lysate was then centrifuged for 5 min at 300 g at 4°C and incubated with 1× red blood cell lysis buffer (Sigma-Aldrich) for 3 min. The sample was centrifuged for 5 min at 300 g at 4°C, and the supernatant was discarded; the sample was then resuspended in Leibovitz media containing 3% FBS and filtered as before. Dissociated cells were counted after centrifugation and used for scRNA-seq.
ScRNA-Seq
Twelve C57BL/6 pups were euthanized using 100 mg/kg intraperitoneal injection of pentobarbital at P14. Fresh lung single-cell digests furnished > 10–20 × 106 cells/mouse with >80% viability. Cells (12,000) were loaded into a 10X Genomics Chromium Single Cell A Chip (10X Genomics; PN-120236), and scRNA-seq was performed using Chromium Single Cell 3' Library & Gel Bead Kit v2 (10X Genomics; PN-120237) following the manufacturer’s protocol (38). Briefly, single-cell gel beads-in-emulsion (GEMs) were produced by running a loaded Chromium Single Cell A Chip on a Chromium Controller instrument (10× Genomics). After GEM-reverse transcription (GEM-RT), GEMs were collected, and cDNA was amplified and cleaned up using SPRISelect (Beckman Coulter; B23318). Indexed libraries were prepared using the Chromium Single Cell 3' Library & Gel Bead Kit v2 (10× Genomics; PN-120237) and sequenced on a NovaSeq 6000 (Illumina) to obtain a sequencing depth of 1.50E + 08 paired-end reads per library. Raw sequencing data were processed through bcl2fastq2 (Illumina), resulting in 1 fastq file per library. Individual 10× libraries were processed through Cell Ranger v3 0.2, and resulting count matrices imported into Seurat v4 1.1 (https://satijalab.org/seurat). Technical variability was reduced using Seurat’s “sctransform,” and quality control metrics were analyzed per sample. The data were then normalized and scaled and the identification of variable genes and PCA were performed, followed by dimensionality reduction. Clusters were analyzed with FindAllMarkers function to identify marker genes for each cell lineage. Each lineage was then subsetted into a new object and reanalyzed. Finally, volcano plots were generated using the library EnhancedVolcano with a Log2 fold-change cutoff of 1, and a P value cutoff at 10e-6 (39). Raw data have been deposited in GEO under the accession number GSE209664.
IPA Analysis
Significance values for differentially expressed genes between groups were defined as P < 0.05, as assessed using unpaired-samples t test. The IPA system (version 73620684, Ingenuity Systems; Qiagen China Co., Ltd.) was used for subsequent bioinformatics analysis, which included canonical pathways, upstream analysis, diseases and functions, and regulator effects. For analyses, the −log (P value) >1.3 (P < 0.05) was taken as the threshold and a Z-score of ± 2 was defined as the threshold of significant activation/repression.
Cell Culture and Immortalized Embryonic Human Lung Fibroblasts Generation
Immortalized embryonic human pulmonary microvascular endothelial cells (HPMEC-Im) were generated from primary HPMEC (fetal 18-wk gestation, female, Cat. No. 3000, ScienCell) and grown as done in previous studies (38, 40). Primary fetal human pulmonary fibroblasts (HPF; male, 20-wk gestation, Cat. No. 3300) and primary fetal pulmonary alveolar epithelial cells (HPAEpiC; female, 20-wk gestation, Cat. No. 3200) were purchased from ScienCell (Carlsbad, CA). We followed the company’s protocol to culture the cells. Immortalized HPF were generated using Lentivirus containing SV40 large T antigens (ABM, Richmond, BC, Canada). The Immortalized HPF (HPF-Im) was verified with fibronectin staining (Supplemental Fig. S3). Cells were cultured in a sealed chamber containing 85% O2 and 5% CO2 for 2-day (for AT cells) or 7-day (HPAEpiC and HPF-Im) hyperoxia treatment. Although we tried to mimic in vivo 85% O2 exposure in our cell culture system, the presence of culture media that is changed every 48 h and covered dishes limit the actual % of O2 the cells are exposed to. Nevertheless, the concentrations of oxygen used in our in vitro models likely exceed physiological oxygen concentrations in these compartments.
Quantitative Reverse Transcription PCR
Total RNA was extracted from mouse lung or cultured cells using the PureLink RNA Mini Kit (Invitrogen) and cDNA was synthesized from 1 µg of RNA using an iScript cDNA synthesis kit (Bio-Rad), according to manufacturer’s instructions. Quantitative reverse transcription PCR (qRT-PCR) was run on a ViiA 7 with SYBR green mastermix (Biotium). 18S was used as the human cell housekeeping gene and Ywhaz was used as the mouse housekeeping gene. The relative gene expression was calculated using the Pflaffl method.
Immunoblotting
Immunoblotting for quantifying changes in protein expression was performed as previously described (38). In brief, mouse lung tissue was homogenized in RIPA lysis buffer containing commercially available protease and phosphatase inhibitors (Sigma) after RA and hyperoxia treatment, with the clarified lysates used for western blotting (WB). Densitometry was performed using ImageJ Software (NIH) and changes were normalized to YWHAZ. Primary antibodies are described in Supplemental Table S1.
Immunostaining
Immunostaining of mouse lung and cultured cells was done as in our previous study (38). Briefly, Lungs were inflated at ∼20 cm H2O pressure until lungs were fully inflated, then held at that pressure for 30 s before tying off the trachea, and then lungs were fixed in 4% formaldehyde and embedded in paraffin. Cultured cells were grown on coverslips and then fixed with 4% formaldehyde. Cells or deparaffined lung sections were incubated with first antibodies after blocking at 4°C overnight and then incubated with corresponding second antibodies at 25°C for 1 h. Several washes with 0.1% Tween in PBS after incubating first and second antibody and then sealed the slides with Vibrance Antifade Mounting Medium with DAPI. Images were taken using ZEISS LSM 510 confocal microscope.
Collagen and Elastic Tissue Staining
Inflated lung sections were stained with Weigert’s Resorcin Fuchsin (without Weigert’s Iron Hematoxylin), and then counter-stained with Van Gieson’s Solution (Rowley Biochemical, Danvers, MA).
Statistical Analysis
The statistical analysis was performed as discussed in our previous study (38). Briefly, data are presented as means ± SD or median with interquartile range. P < 0.05 was considered significant. For cell culture experiments, data are from a minimum of three independent experiments with adequate technical replicates used for quantification. All animal data were obtained in littermate controls. For animal experiments, a minimum of four animals were used for each experimental group with adequate technical replicates used for quantification. For all data, we initially examined whether distribution of data was Gaussian using the D’Agostino-Pearson omnibus normality test. If data were normally distributed, then ANOVA with a post hoc Tukey’s test was used for analysis. If data did not meet Gaussian assumptions, a Mann–Whitney U-test was used for analysis. Comparisons between two groups were made by a one-sample, two-tailed Student’s t test for parametric or nonparametric data. For most analyses, fold changes were calculated related to expression/changes in untreated controls. Statistical analysis and graphs were generated using GraphPad Prism 9.0.
RESULTS
Lung Cellular Composition of the Developing Mouse Lung at P14 after Room Air and Hyperoxia Exposures Revealed by scRNA-Seq
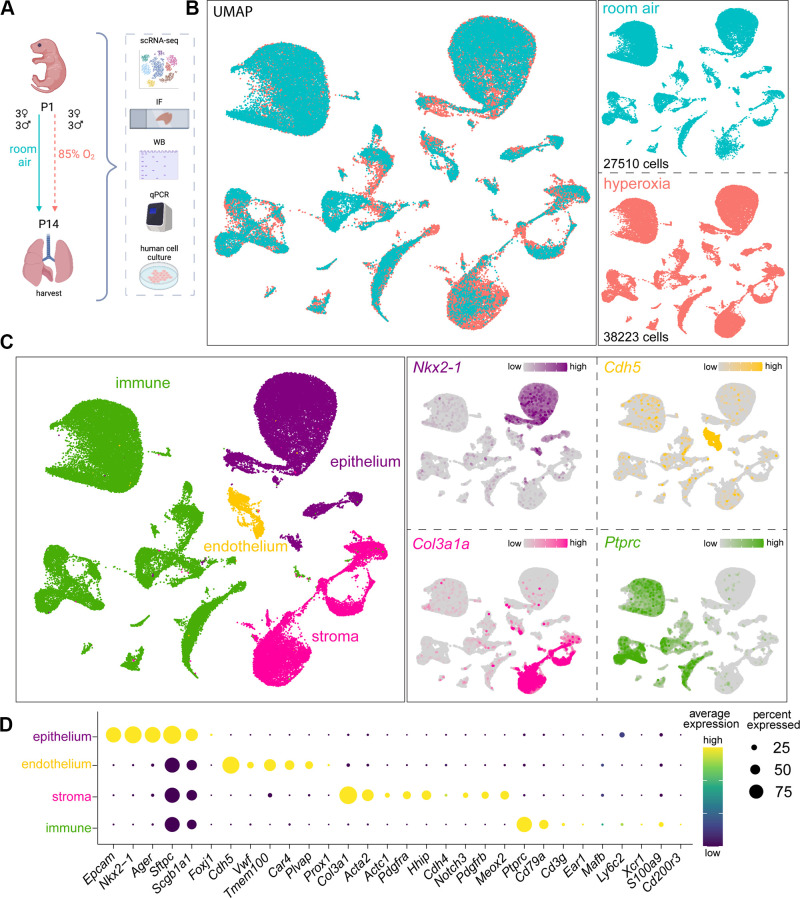
We and others have shown that hyperoxia induces alveolar simplification with arrested vascular and alveolar development (37, 41, 42). To study the effect of hyperoxia on lung cellular lineages and subpopulations, we used a well-established model of experimental BPD. We exposed newborn C57BL/6 mice to room air (RA; 3 males and 3 females) and hyperoxia (3 males and 3 females) from P1 to P14 (Fig. 1A). We used western blotting, immunofluorescence studies, and qRT-PCR in independent mice (4 males and 4 females) and human neonatal lung cells/lines to validate our scRNA-seq findings (Fig. 1A). Hyperoxia reduced radial alveolar counts and increased mean linear intercepts by 40% (Supplemental Fig. S1, A and B). To examine cell type-specific changes in the transcriptome associated with hyperoxia, we performed scRNA-seq (Fig. 1B) and identified four major cell lineages: Nkx2-1+ epithelial cells, Cdh5+ endothelial cells, Col3a1+ stromal cells and Ptprc+ immune cells (3, 29) (Fig. 1C). We identified marker genes for AT1 cells (Aqp5), AT2 cells (Sftpc), ciliated epithelial cells (Foxj1), and club cells (Scgb1a1) in the epithelial cell cluster. We identified lineage markers for Cap1 EC (Plvap), Cap2 EC (Car4), big blood vessel EC (Vwf), and lymphatics marker (Prox1) in EC cluster (Supplemental Table S2). Markers for stromal cells associated with the three axes—vascular, epithelial, and interstitial—were identified, which encompass the pulmonary smooth muscle cells, pericytes, and fibroblast subpopulations (35). Finally, markers for distinct immune cell populations were identified (Fig. 1D and Supplemental Table S2). Because we identified new subclusters within major cell clusters in hyperoxia, a detailed description of subpopulations is described within their respective sections.
Figure 1.
Lung cellular architecture in room air and hyperoxia at P14 by scRNA-seq. A: illustration generated with permission on BioRender of experimental design: mouse pups were exposed to 85% oxygen or RA from P1 to P14, and the lungs were used for single cell RNA-seq (scRNA-seq), immunofluorescence (IF), whole lung lysate qRT-PCR, and western blotting. Human primary cells were used for validation of mouse scRNA-seq data. B: left: scRNA-seq UMAP with merged treatment conditions, RA (blue), and hyperoxia (pink), showing overlapping clusters. Right: UMAPs of cells in each condition show similarities and differences, with cell numbers specified in each case. Three females and 3 males were sequenced per group. C: seurat unbiased clustering grouped all cells into 37 clusters which we attributed to four cell lineages shown in UMAP (left). Epithelial, endothelial, stromal, and immune cell identities were determined by the expression of Nkx2-1, Chd5, Col3a1, and Ptprc, respectively, shown in feature plots (right). D: dot plot depicting expression level (dot color) and percentage of expression (dot size) of markers used to identify subpopulations within each major cell lineage. RA, room air.
Alterations of Canonical Pathways, Upstream Genes and Functions Indicate Upregulation of Oxidative Phosphorylation, NRF2, TP53, and TGFβ1 Signaling in Major Cell Lineages
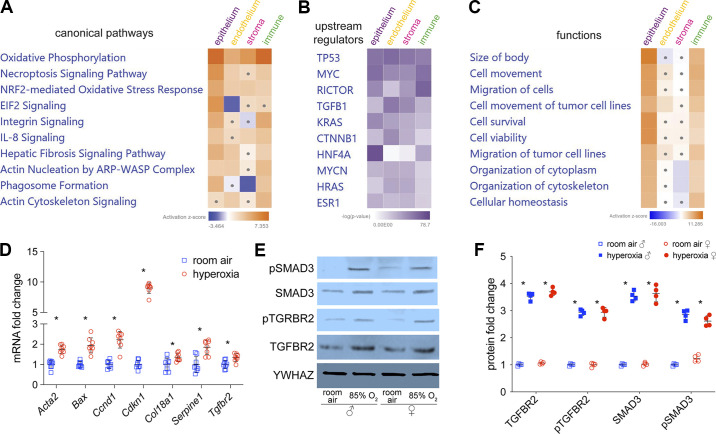
BPD is associated with several changes in developmental, cell injury, fibrosis, and inflammatory pathways (8, 9). We therefore analyzed our scRNA-seq data using Ingenuity Pathway Analysis (IPA; Qiagen) for signaling and functional pathways that were significantly altered in hyperoxia compared with room air (RA) and were shared across major cell lineages (Fig. 2, A–C). Oxidative phosphorylation and NRF2-mediated oxidative stress response were upregulated in all cell lineages (Fig. 2A). Fibrosis signaling was upregulated in all cell lineages except for stromal cells. NRF2-mediated oxidative stress response activates the expression of antioxidant response elements (ARE) that protect the lung from oxidative stress caused by hyperoxia (43, 44). TGFβ signaling is important for lung development but is also a key mediator of fibrosis (45), was altered in all lineages (Fig. 2B). TP53, a regulator of lung injury and fibrosis (13, 46), was also upregulated (Fig. 2B). These changes support the increased fibrosis signaling found in hyperoxia (Fig. 2A). Integrin signaling and actin nucleation by ARP-WASP complex were upregulated in epithelial cells and immune cells, which is consistent with increased cell migration in function analysis (Fig. 2, A and C).
Figure 2.
Alterations of canonical pathways, upstream regulators, and functions in hyperoxia. scRNA-seq differential gene expression data was used to generate IPA heatmaps of top 10 canonical pathways (A), upstream regulators (B), and functions (C) altered by hyperoxia. Dots in heatmaps represent values that had a |Z-score| of <2 (nonsignificant). D: mouse whole lung qRT-PCR shows fibrosis marker genes (Ccnd1, Acta2, Serpine1, and Col18a1), TGFB1 receptor (Tgfbr2) and its downstream genes (Acta2, Tgfbr2, Serpine1, and Cdkn1a) are upregulated by hyperoxia treatment. n = 8/group; *P < 0.05. E and F: mouse whole lung immunoblotting (E) shows expression and phosphorylation of TGFRB2 and SMAD3 are stimulated by hyperoxia in males and females, with densitometry shown graphically in F; n = 4/group; *P < 0.05. IPA, Ingenuity Pathway Analysis.
In independent experiments, we collected whole lung samples from neonatal littermates exposed to hyperoxia or RA from P1 to P14 (n = 8/group, 4 in each sex). We validated genes associated with the signaling of TGFβ and fibrosis. Quantitative reverse transcription PCR (qRT-qPCR) showed that fibrosis marker genes (Ccnd1, Acta2, Serpine1, and Col18a1), TGFB1 downstream genes (Acta2, Tgfbr2, Serpine1, and Cdkn1a), and TP53 signaling genes (Serpine, Bax, and Cdkn1a) in whole mouse lungs with hyperoxia treatment (Fig. 2D) (47–54). Because our gene signatures suggested activation of TGFβ and fibrosis signaling, we performed western blotting (WB) on whole lung lysates to demonstrate that expression and phosphorylation of TGFBR2 and SMAD3, canonical markers of TGFβ activation, were induced in hyperoxia (Fig. 2, E and F). Excessive collagen deposition is a hallmark of lung fibrosis (55). In our model, increased collagen was deposited in alveolar septa, bronchial wall, and blood vessel wall (Supplemental Fig. S1). YWHAZ was used for normalization as β-actin expression changed with hyperoxia. These data show that hyperoxia induces TGFβ and pulmonary fibrosis signaling among several lung cell types, consistent with whole lung data reported before (12, 13, 45, 46).
Damage-Associated Transient Progenitors and Induced AT1 (iAT1) Populations Emerge in Hyperoxia
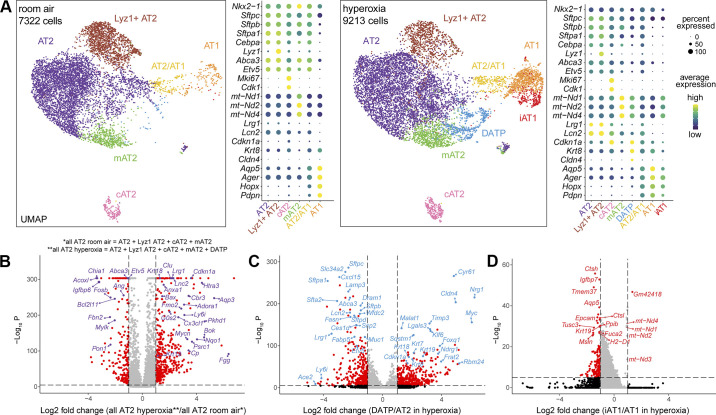
We next analyzed the major cell lineages individually, starting with epithelial cells. In Nkx2-1+/Epcam+/Cdh1+ epithelial cells, we identified 3 main clusters: Etv5−/Pdpn+ AT1 cells, Etv5+/Sftpc+ AT2 cells, and Etv5+/Pdpn+/Sftpc+ AT2/AT1 cells. We also identified minor populations of Foxj1+ bronchiolar ciliated cells and Scgb1a1+ basal cells (data not shown), but we focused on alveolar epithelial cells. Based on gene expression analysis, we categorized AT2 cells into four subclusters in RA: Lyz1+ AT2, AT2, Mki67+ cycling AT2 (cAT2), and mt-Nd1/2/4high mitochondrial AT2 (mAT2; Fig. 3A). cAT2 cells express proliferation-related genes, such as Mki67 and Cdk1 (Fig. 3A) (5). Lyz1+ AT2 cells are enriched with the lysozyme gene Lyz1 (Fig. 3A) and have been previously described (7). Comparing Lyz1+ AT2 to AT2 subcluster, IPA upstream analysis revealed that ETV5 signaling, which controls Lyz1 expression in lung AT2 cells (56), was upregulated in Lyz1+ AT2 cells (Supplemental Fig. S2A). IPA pathway analysis also identified downregulated mTOR, actin cytoskeleton and pulmonary healing signaling pathways, as well as upregulated Hippo signaling in Lyz1+ AT2 (Supplemental Fig. S2B), suggesting less proliferation, migration, and alveologenesis potential. The mAT2 population, which has not been described before, express reduced surfactant genes (Sftpa, Sftpb, and Sftpc) and less AT2 linage marker genes (Etv5, Abca3, and Cebpa) (5, 56, 57) but more mitochondrial genes, such as mt-Nd1/2/4 (Fig. 3A). IPA upstream and canonical pathways analysis demonstrated less ETV5 signaling (Supplemental Fig. S2C) and more mitochondria biogenesis—part of upregulated Sirtuin signaling—in mAT2 compared with AT2 cells (Supplemental Fig. S2D).
Figure 3.
scRNA-seq revealed alveolar epithelial diversity and transcriptomic changes in hyperoxia. A: UMAPs showing the distinct alveolar epithelial clusters identified, along with dot plots featuring the marker genes used to characterize them. In RA (right), we found AT2 cells consisted of four subpopulations (AT2, Lyz1+ AT2, mAT2 and cAT2), and found two other clusters including AT2/AT1 and AT1 cells. In hyperoxia (right), two additional subpopulations emerged, a cluster of AT2 cells labeled DATP (blue), and a cluster of AT1 cells labeled iAT1 (red). Volcano plots comparing gene expression between (B) all AT2 cells in hyperoxia vs. AT2 cells in RA, (C) DATP and AT2 in hyperoxia, (D) iAT1 and AT1 in hyperoxia. Down/upregulated genes are shown in red, whereas some genes of biological relevance have been highlighted. RA, room air.
Interestingly, we found major changes in the AT2 transcriptome in hyperoxia (Fig. 3A). The majority of AT2 clusters, including AT2 and Lyz1+ AT2, were enriched for inflammation-related genes (Lcn2 and Lrg1), but expressed less AT2 lineage markers (Etv5, Abca3, and Cebpa; Fig. 3A). A distinct Sftpclow/Krt8+/Cldn4+ AT2 subcluster emerged in hyperoxia that was not evident in normal developing lungs (Fig. 3A). These cells have been labelled as damage-associated transient progenitors (DATPs) in a mouse model of bleomycin-induced fibrosis (5). A comparison between all AT2 cells present in hyperoxia versus RA, showed less AT2 lineage marker genes (Etv5 and Abca3), more AT2 to AT1 differentiation marker genes (Krt18 and Krt19), and more inflammation-related genes (Lcn2 and Lrg1) (5, 56–59) (Fig. 3B). In turn, DATP cluster showed less AT2 lineage markers (Etv5, Abca3, Sftpb, and Sftpc) and more AT2 to AT1 differentiation marker genes (Krt7, Krt8, Krt18, and Krt19) when compared with AT2 in hyperoxia (5, 56–60).
AT2/AT1 cells express AT2 lineage marker genes (Etv5, Abca3, and Cebpa) and, at a lower level, several AT1 cell marker genes (Aqp5, Ager, Hopx, and Pdpn) in RA (Fig. 3A). The AT2/AT1 population had similar signatures in RA versus hyperoxia. We identified one AT1 cell cluster that was enriched with classical AT1 marker genes (Hopx, Pdpn, and Aqp5), and did not express AT2 marker genes (Sftpc, Etv5, and Abca3) in RA or hyperoxia. Interestingly, one new subcluster of AT1 cells, which we labeled “induced” AT1 (iAT1), emerged in hyperoxia (Fig. 3A). Notably, iAT1 had increased expression of mt-Nd1/2/3/4 but less AT1 mature marker genes (Aqp5, Epcam) when compared with AT1 in hyperoxia (Fig. 3D). IPA analysis showed less senescence and lung fibrosis signaling in iAT1 cells compared with AT1 in hyperoxia (Supplemental Fig. S2E). iAT1 cells also express less Vegfa, Pdgfa, Fgf18, and Fgf1 when compared with AT1 in hyperoxia (Supplemental Fig. S2F). Vegfa and Pdgfa play critical roles in alveolarization (61–64), whereas FGF18 regulates key developmental events during pulmonary alveolarization and FGF1 stimulates epithelial proliferation (65, 66). These transcriptional changes in iAT1 cells suggest a decreased ability to contribute to alveolarization.
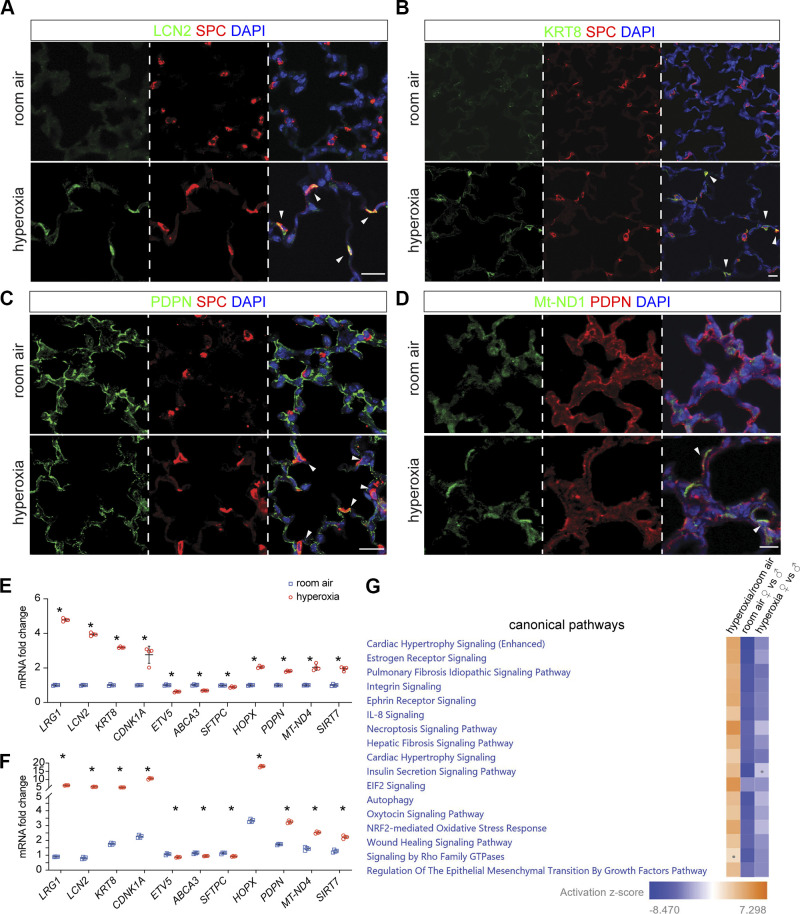
To confirm these interesting findings, we performed immunofluorescence (IF) on lung sections from hyperoxia- and RA-treated pups. We confirmed the general inflamed signature of AT2 cells by demonstrating that LCN2 was increased in AT2 in hyperoxia and colocalized with SPC staining (Fig. 4A). KRT8high/SPC+ staining showed DATP emerged in hyperoxia (Fig. 4B) but was not present in RA. SPC/PDPN staining revealed three clusters of alveolar epithelial cells, which are SFTPC+ AT2 cells, PDPN+ AT1 cells, and PDPN+/SFTPC+ AT2/AT1 cells; the latter were increased in hyperoxia (Fig. 4C). iAT1 cells were visualized with MT-ND1/PDPN staining in hyperoxia (Fig. 4D).
Figure 4.
Mouse in vivo immunostaining and human cell culture confirmed alveolar epithelial diversity and alterations in hyperoxia. A–D: immunofluorescence of mouse lung sections following RA or hyperoxia treatment from P1 to P14; scale bar: 10 µm. A: AT2 cells, labeled with SPC, show coexpression of inflammation marker LCN2 (arrowhead) in hyperoxia. B: emergence of DATP cells is seen in the alveolar region after hyperoxia, where some AT2 cells which express SPC, also express KRT8 (arrowhead). C: PDPN and SPC colocalization (arrowhead) demonstrates increased AT2/AT1 cells in hyperoxia. D: hyperoxia-induced iAT1 cells can be seen by coexpression of Mt-ND1 and AT1 marker PDPN (arrowhead). E: RNA obtained from human pulmonary alveolar epithelial cells (HPAEpiC) exposed to 48 h of RA or hyperoxia (85% O2) was used to quantify LRG1, LCN2, KRT8, CDNK1A, ETV5, ABCA3, SFTPC, HOPX, PDPN, mt-ND4, and SIRT7 with qRT-PCR. n = 4/group; *P < 0.05. F: RNA obtained from 48 h RA and hyperoxia treated HPAEpiC, which were cocultured with immortalized human pulmonary fibroblasts (HPF-Im), was used to quantify LRG1, LCN2, KRT8, CDNK1A, ETV5, ABCA3, SFTPC, HOPX, PDPN, mt-ND4, and SIRT7 with qRT-PCR. n = 4/group; *P < 0.05. G: IPA canonical pathway analysis heatmap showing several upregulated pathways in alln alveolar epithelial cells in hyperoxia; pulmonary fibrosis and wound healing signaling pathways were upregulated, but the differences between female and male becomes less in hyperoxia than in RA. Dots in heatmaps represent values that had a |Z-score| of <2 (nonsignificant). IPA, Ingenuity Pathway Analysis; RA, room air.
IL1β and HIF1α have been shown to induce AT2 differentiation to AT1 via DATP, whereas TGFβ signaling represses AT1 maturation (5, 58). IPA upstream analysis showed that IL1 signaling is activated in Lyz1+ AT2, AT2, mAT2, and AT2/AT1 cells, whereas HIF1α signaling and TGFβ signaling were upregulated in mAT2 and AT2/AT1 cells in hyperoxia (Supplemental Fig. S2G). Upregulated TP53 signaling and HIF1α signaling in DATP were revealed by comparing with AT2 in hyperoxia (Supplemental Fig. S2H). Transcriptomic signatures of IL1R, HIF1α, and TGFβ activation seen in AT2 subpopulations with neonatal hyperoxia resemble those described in adult mouse models of bleomycin-induced fibrosis (5), involved in AT2 reparative responses. These data show that hyperoxia programs AT2 to a transitional “stem-cell-like fate” described in adult models of fibrosis (67).
To validate our mouse data, we exposed neonatal human primary alveolar epithelial cells (HPAEpiC) isolated from the neonatal lung (ScienCell) to hyperoxia or normoxia for 48 h, and then examined marker gene expression of AT2 and AT1 subsets identified in hyperoxia using qRT-PCR. We found that markers of inflammation (LRG1 and LCN2), DATP signature (KRT8), mitochondria biogenesis signature [mt-ND4 and SIRT7 (68)] were upregulated in response to hyperoxia (Fig. 4E). cAT2 and DATP shared marker gene CDNK1A and were also upregulated in hyperoxia (Fig. 4E). AT2 marker genes (ETV5, ABCA3, and SFTPC) were slightly downregulated, and AT1 marker genes (HOPX and PDPN) were upregulated (Fig. 4E). Interestingly, when HPAEpiC were cocultured with human neonatal lung fibroblasts, most of these changes were accentuated (Fig. 4F and Supplemental Fig. S2I). These data indicated that hyperoxia induces signatures of mitochondrial biogenesis and DATP cell autonomously, laying the foundation for the differentiation of AT2 to AT1.
IPA analysis showed that pulmonary fibrosis and epithelial-to-mesenchymal transition (EMT) signaling were upregulated in AT cells in hyperoxia (Fig. 4G). We explored female versus male differences in our scRNA-seq data and found that there was less pulmonary fibrosis and EMT signaling in females in RA and hyperoxia, although differences were less prominent in hyperoxia (Fig. 4G). Less lung fibrosis and EMT signaling could suggest more programming of AT2 to AT1 rather than fibrosis in female mice exposed to hyperoxia.
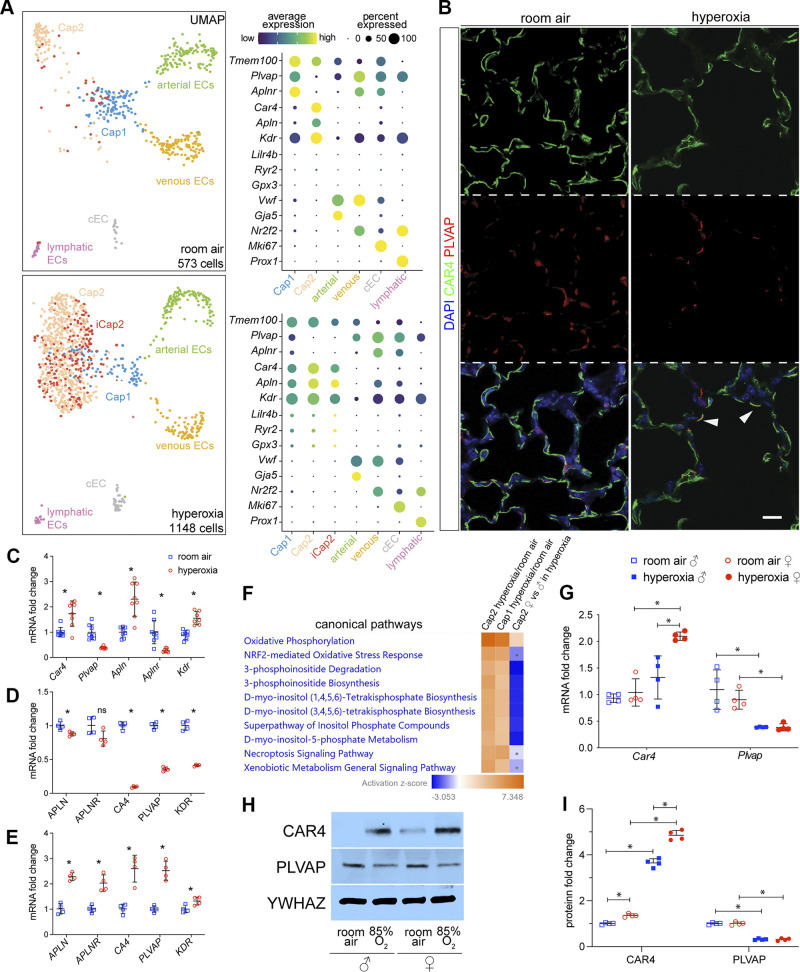
New Subpopulation of Car4+ Cap2 Endothelial Cells Emerges in Hyperoxia
We analyzed EC in RA and identified six previously known clusters: Cdh5+/Plvap+/Aplnr+/Vwf- Cap1, Cdh5+/Car4+/Apln+/Plvap- Cap2, Cdh5+/Vwf+/Nr2f2+ venous EC, Cdh5+/VWF+/Gja5+ arterial EC, Cdh5+/Prox1+ lymphatic EC, and Cdh5+/Mki67+ cycling EC (cEC; Fig. 5A) (3, 29–32). The proportion of capillary EC identified as Cap1 decreased in hyperoxia, whereas the Cap2 proportion increased (Fig. 5A). Dot plots showed that Car4 and Apln expression was upregulated, whereas Aplnr and Plvap were downregulated in Cap1 in hyperoxia (Fig. 5A), which may indicate a skewing of capillary transcriptomic signature from Cap1 to Cap2 in hyperoxia. Interestingly, in adult mouse models of influenza-induced lung injury, Cap1 cells are considered to be progenitor cells, giving rise to Cap2 (CAR4+) cells after infection-induced injury (32). This is also known to be the case during normal lung ontogeny (30, 31). Unlike in RA, the Cap2 population in hyperoxia appeared heterogenous with an immature Cap2 (iCap2) subcluster (Fig. 5A). These cells were specifically enriched for Lilr4b, Ryr2, and Gpx3 (Fig. 5A), had less Car4 expression but more Apln expression compared with regular Cap2, and did not express the Cap1 markers Plvap and Aplnr (Fig. 5A). GPx3, a major scavenger of reactive oxygen species (ROS) in plasma, acts as a redox signal modulator. More GPx3 expression may indicate a protective response to hyperoxia that limits oxidative damage in iCap2. Interestingly, the comparison between iCap2 and Cap2 in hyperoxia showed significant downregulation of Icam2, a marker of lumenized vessels, further indicating these cells are immature and may be undergoing vascular remodeling (Supplemental Fig. S3A) (69). IPA analysis revealed that VEGF signaling, which is important for Cap2 differentiation (30), was repressed in iCap2 compared with Cap2 (Supplemental Fig. S3B).
Figure 5.
Hyperoxia induces a new subpopulation of Car4+ Cap2 cells and causes sex-specific transcriptomic changes in the endothelium. A: UMAPs of endothelial cell (EC) clusters in RA (top) and hyperoxia (bottom). In RA, six clusters were identified using the marker genes shown in dot plots: Plvap+ Cap1, Car4+ Cap2, Gja5+ arterial EC, Nr2f2+ venous EC, Mki67+ cEC, and Prox1+ lymphatic EC. In hyperoxia, an additional subpopulation of Cap2 emerged which we named iCap2 (red). B: immunostaining of mouse lung sections showing a general reduction in vessel density in hyperoxia compared with hyperoxia. Coexpression of CAR4 and PLVAP was also observed in hyperoxia (arrowhead). Scale bar: 10 µm. C: mouse whole lung qRT-PCR depicts upregulated expression of Cap2 EC marker genes (Car4, Apln, and Kdr) and downregulated expression of Cap1 EC marker genes (Plvap and Aplnr) in hyperoxia compared with RA. n = 8/group; *P < 0.005. D: RNA obtained from immortalized human pulmonary microvascular endothelial cells (HPMEC-Im) exposed to 7 days of RA or hyperoxia (85% O2) was used to quantify APLN, APLNR, CA4, PLVAP, and KDR with qRT-PCR. n = 4/group; *P < 0.05. E: RNAs obtained from HPMEC-Im exposed to 7 days of RA or hyperoxia (85% O2) that were cocultured with immortalized human pulmonary fibroblasts (HPF-IM) were used to quantify APLN, APLNR, CA4, PLVAP, and KDR with qRT-PCR. n = 4/group; P < 0.05. F: heatmap of top 10 hyperoxia-altered canonical pathways analyzed with IPA showing that female Cap2 EC had increased oxidative phosphorylation compared with their male counterpart, whereas other pathways were downregulated. The dots in heatmaps represent values that had a |Z-score| of <2 (nonsignificant). G: mouse whole lung qRT-PCR showing decreased Plvap mRNA expression in hyperoxia samples in both females and males, but higher Car4 mRNA expression in females only. n = 4/group; *P < 0.05. H and I: mouse whole lung immunoblotting (H) reveals that hyperoxia represses PLVAP expression and stimulates CAR4 expression in both females and males but stimulates more CAR4 expression in females vs. males, with densitometry shown graphically in I; n = 4/groups; *P < 0.05. IPA, Ingenuity Pathway Analysis; RA, room air.
Next, we proceeded to validate our scRNA-seq findings in independent RA- and hyperoxia-treated mouse samples with IF. CAR4+ EC (Cap2) covers most of the alveolar surface abutting the air interface, whereas PLVAP+ EC (Cap1) localizes to the opposite side of the alveolar-capillary interface (30, 31) (Fig. 5B). In addition to a general reduction in vessel density, we observed more colocalization between CAR4 and PLVAP staining in hyperoxia (Fig. 5B), which would suggest a greater proportion of Cap1 cells may be transitioning to Cap2 in hyperoxia. Car4, Apln, and Kdr were enriched in Cap2, whereas Plvap and Aplnr were enriched in Cap1, as expected (Fig. 5A). Whole lung RNA expression by qRT-PCR demonstrated increased expression of Car4, Apln, and Kdr, but decreased expression of Aplnr and Plvap (Fig. 5C) in hyperoxia, which was also confirmed by our immunoblotting data (Fig. 5, H and I) showing increased CAR4 and decreased PLVAP in hyperoxia. These results suggest an increase in the proportion of Cap2 and decreased Cap1 as noted in our scRNA-seq UMAP (Fig. 5A).
To validate our mouse data in human lung EC, we used immortalized fetal human pulmonary microvascular endothelial cells (HPMEC-Im) that we had generated before (38). Some cells expressed the Cap1 marker (APLNR), whereas others the Cap2 marker (CA4; Supplemental Fig. S4C) (30, 70). We exposed HPMEC-Im to hyperoxia (85%) or normoxia for 7 days in the presence or absence of immortalized fetal human pulmonary fibroblasts (HPF-Im) to test cell-autonomous changes in gene expression in hyperoxia. Surprisingly, qRT-PCR demonstrated that hyperoxia suppressed Cap2 and Cap1 markers [APLN, APLNR, CA4 (orthologous to mouse Car4), PLVAP, and KDR] in the absence of fibroblasts (Fig. 5D). However, when cocultured with HPF-Im for seven days, expression of both Cap2 and Cap1 markers was increased in hyperoxia (Fig. 5E). These data suggest fibroblasts are required for the induction or maintenance of CAR4 expression in hyperoxia.
We then did IPA analysis on gene expression profiles in Cap2 and Cap1 in hyperoxia versus RA. The top differentially expressed canonical pathways in hyperoxia included oxidative phosphorylation, NRF2-oxidative stress response genes, and inositol biosynthesis signaling (Fig. 5F). Comparing female versus male Cap2 population showed that female Cap2 cells had increased oxidative phosphorylation compared with male Cap2 cells, whereas other pathways were suppressed (Fig. 5F). We did not have enough Cap1 cells in hyperoxia to do a male versus female comparison.
Whole lung Car4 mRNA expression was upregulated by hyperoxia in females, but not in males, and its expression was higher in females than males in hyperoxia (Fig. 5G). Plvap mRNA expression was downregulated by hyperoxia in both males and females (Fig. 5G). Western blotting showed that CAR4 protein was upregulated by hyperoxia in both females and males, and its expression was higher in female than males in hyperoxia, whereas PLVAP protein expression was downregulated by hyperoxia in both males and females (Fig. 5, H and I). This suggests a more robust reparative EC response with increased transitioning of Cap1 to Cap2.
Hyperoxia Induces Inflammation Signature in Stromal Cells while Specifying a Fibrotic Fibroblast Subpopulation
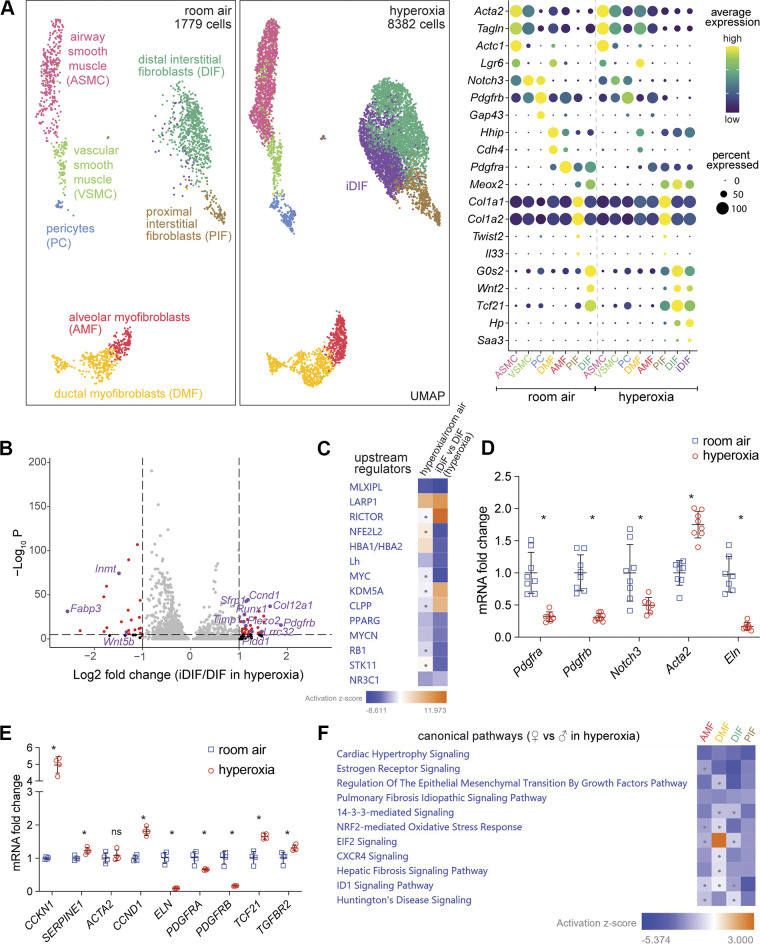
In RA, we classified Col3a1+ stromal mesenchymal cells into three groups based on their proximity to vascular or epithelial structures nearby or “axes” according to terminology published recently (35). The vascular axis consists of Notch3high/Gap43+/Pdgfrbhigh pericytes (PC) and Notch3high/Gap43−/Acta2+ vascular smooth muscle cells (VSMC); the epithelial axis is formed by Pdgfrahigh/Lgr6−/Hhip+/Cdh4+ alveolar myofibroblasts (AMF), Pdgfra+/Lgr6+/Hhiphigh/Cdh4high ductal myofibroblasts (DMF), and Notch3+/Actc1+/Lgr6+ airway smooth muscle cells (ASMC); the interstitial axis includes Pdgfra+/Twist2+/Il33+/Col1a1high/Col1a2high proximal interstitial fibroblasts (PIF), and Pdgfra+/Wnt2+/Tcf-21high/G0s2high distal interstitial fibroblasts (DIF) (3, 29, 34, 35) (Fig. 6A).
Figure 6.
New population of distal interstitial fibroblasts emerges in hyperoxia and shows sex-specific characteristics. A: UMAP of stromal cell showing seven clusters in RA (left) identified as DIF, PIF, AMF, DMF, PC, VSMC, and ASM. In hyperoxia (middle), a new cluster emerged labeled as iDIF (purple). Dot plot showing marker genes used for identification of each population (right). B: volcano plot shows gene expression differences between iDIF and DIF in hyperoxia. Significantly affected genes are shown in red, with some biological relevant genes highlighted in purple. C: IPA analysis revealing downregulation of MLXIPL and PPARG signaling in hyperoxia, whereas these two upstream regulators are even more repressed in iDIF vs. DIF. The dots in heat maps represent values that had a |Z-score| of <2 (nonsignificant). D: mouse whole lung qRT-PCR reveals downregulated expression of different stromal cluster marker genes, such as Pdgfra, pdgfrb, Notch3, and Eln, and upregulated Acta expression in hyperoxia. n = 8/group; *P < 0.02. E: immortalized human pulmonary fibroblasts (HPF-IM) in culture were exposed to 7 days of RA or hyperoxia (85% O2) and used to quantify CCKN1, SERPINE1, ACTA2, CCND1, ELN, PDGFRA, PDGFRB, TCF21, and TGFBR2 RNA expression by qRT-PCR. n = 4; *P < 0.05. F: IPA analysis indicating less pulmonary fibrosis signaling in female fibroblasts compared with male fibroblasts in hyperoxia. The dots in heatmaps represent values that had a |Z-score| of <2 (nonsignificant). IPA, Ingenuity Pathway Analysis; RA, room air.
In hyperoxia, Pdgfra expression decreased profoundly in all fibroblasts, especially AMF (Fig. 6A and Supplemental Fig. S4A). PDGFRA signaling in AMF plays a critical role in secondary septation and alveolarization (64, 71). ELN, which forms looping elastic bundles to surround alveolar ducts and forms a continuous sheet or network in blood vessel walls (72), is highly expressed in AMF and VSMC in RA, and its expression in these cells dramatically decreased in hyperoxia (Supplemental Fig. S4A). ACTA2 expression in myofibroblasts is considered to be a key determinant of remodeling and disease progression in lung fibrosis (48, 73). Acta2 expression was upregulated in DMF in hyperoxia (Fig. 6A and Supplemental Fig. S4A). Notch3 and Pdgfrb were highly expressed in smooth muscle (like) cells in RA, and their expression was repressed in hyperoxia (Fig. 6A and Supplemental Fig. S4A). Notch3 signaling in mural cells is important for cell proliferation, differentiation, and survival and vascular integrity (74–76). These patterns of changes in stromal cells could contribute to the interrupted secondary septation and alveolarization and damaged integrity of the vasculature in hyperoxia, which are consistent with the reported increased fibrosis in human BPD (77).
Interestingly, a hyperoxia-induced DIF (iDIF) subcluster emerged in hyperoxia (Fig. 6A). Dot plot shows Haptoglobin (HP) and serum amyloid A3 (SAA3) were specific markers for this population whereas DIF marker genes, such as Wnt2, Tcf21, and G0S2, were less expressed in iDIF (Fig. 6A). Volcano plot comparing iDIF to DIF in hyperoxia shows significantly increased expression of TGFB signaling and its downstream genes (Pdgfrb, Ccnd1, Col12a1, Runx1, Timp1) (78–82), and less expression of Inmt, a gene enriched in DIF (Fig. 6B) (83). DIF, as lipid-storing fibroblasts, provide lipids to AT2 cells for surfactant production (84). MLX interacting protein-like (MLXIPL) signaling induces lipogenesis in adipocytes and the liver, and PPARG signaling is involved in adipocyte lipid metabolism and inhibits the conversion of lipofibroblasts to myofibroblasts (85–87). IPA analysis revealed that both MLXIPL and PPARG upstream regulators were repressed in hyperoxia and even more repressed in the iDIF subcluster (Fig. 6C). Consistent with this finding, we noted upregulation of Runx1, a transcription factor recently described to regulate the differentiation of fibroblasts into myofibroblasts, which produce matrix proteins, thus contributing to fibrosis (83).
We validated scRNA-seq data in independent mouse lung samples exposed to hyperoxia or RA (n = 4/group). qRT-PCR confirmed increased expression of Acta2 and decreased expression of Pdgfra, Pdgfrb, Notch3, and Eln in the whole lung in hyperoxia (Fig. 6D). We wanted to determine whether changes in mouse lung fibroblasts can be recapitulated in human fibroblasts in a cell-autonomous manner. We obtained human lung fibroblasts and immortalized them. We confirmed fibronectin expression in immortalized fetal human pulmonary fibroblasts (HPF-Im) using immunofluorescence staining (Supplemental Fig. S4B). HPF-Im were exposed to 85% oxygen (hyperoxia) or normoxia for 7 days, and the cells were used to perform qRT-PCR, which demonstrated that hyperoxia repressed the expression of PDGFRA, PDGFRB, and ELN, while upregulating TCF21 expression (Fig. 6E). TP53 target genes (SERPINE1 and CDKN1A), TGFβ signaling marker genes (TGFBR2, SERPINE1 and CDKN1A), and fibrosis signaling marker genes (CCND1 and SERPINE1) were upregulated in hyperoxia, but their shared downstream gene ACTA2 did not change (Fig. 6E). Our HPF-Im data is mostly consistent with our mouse lung data in hyperoxia. The increase in TCF21 with decrease in ACTA2 in HPF-Im with hyperoxia is likely representative of a shift toward a Tcf21+ DIF signature, which is consistent with an increase in this population with hyperoxia in the mouse lung (Fig. 6A).
Next, we compared signaling pathways between female and male mouse lung fibroblasts after hyperoxia treatment using the gene expression data from scRNA-seq. IPA canonical pathway analysis showed less pulmonary fibrosis signaling pathway in four types of fibroblasts in female in hyperoxia (Fig. 6F). Less pulmonary fibrosis signaling in female fibroblasts may lead to reduced lung fibrosis in females in hyperoxia, though this will have to be experimentally proven.
Immune Cells Show Sex-Specific Changes in Their Transcriptome following Hyperoxia
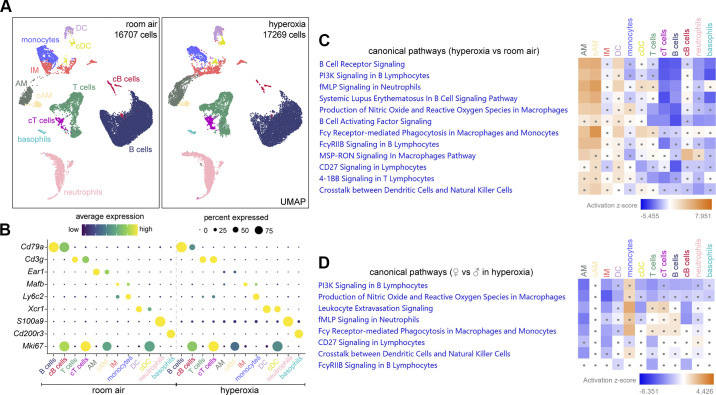
Based on Ptprc and individual marker genes, 12 major clusters of Ptprc+ immune cells were identified (Fig. 7A): Cd79a+ B cells, Cd79a+/Mki67+ cycling B (cB) cells, Cd3g+ T cells, Cd3g+/Mki67+ cycling T (cT) cells, Ear1+ alveolar macrophages (AM), Ear1+/Mki67+ cycling AM (cAM), Mafbhigh interstitial macrophages (IM), Ly6c2high monocytes, Xcr1+ dendritic cells (DC), Xcr1+/Mki67+ cycling dendritic cell (cDC), S100a9+ neutrophils, and Cd200r3+ basophils (Fig. 7B). We used IPA canonical pathways analysis to identify major transcriptomic changes filtered by the different cell types (Fig. 7C). It revealed less B-cell receptor signaling, PI3K signaling, B cells activating factor signaling, CD27 signaling, and leukocyte extravasation signaling in B cells. It also showed less 4-1BB signaling in T cells. Interestingly, IPA analysis demonstrated more leukocyte extravasation signaling, production of nitric oxide (NO) and reactive oxygen species (ROS), Fcy receptor-mediated phagocytosis, and MSP-RON signaling in AM in hyperoxia (Fig. 7C).
Figure 7.
Hyperoxia induces sex-dependent changes in immune cells. A: UMAPs of immune cells showing distinct clusters of B cells, cycling (c)B cells, T cells, cTcells, AM, cAM, IM, monocytes, DC, cDC, neutrophils, and basophils in RA and hyperoxia. B: dot plot showing marker genes used to identify each immune cell cluster. C: IPA analysis revealing less B-cell receptor signaling, B cells activating factor signaling, CD27 signaling and leukocyte extravasation signaling in B cells, but upregulated production of NO and ROS, Fcy receptor-mediated phagocytosis, and MSP-RON signaling in AM in hyperoxia. D: IPA analysis indicating less upregulated leukocyte extravasation signaling, production of NO and ROS and Fcy receptor-mediated phagocytosis in female AM compared with male AM in hyperoxia. Dots in heatmaps (C and D) represent values that had a |Z-score| of <2 (nonsignificant). AM, alveolar macrophages; DC, dendritic cells; IM, interstitial macrophages; IPA, Ingenuity Pathway Analysis; RA, room air; ROS, reactive oxygen species.
Next, we compared canonical pathways between females and males in hyperoxia. IPA analysis revealed less PI3K signaling in B cells and less CD27 signaling in T cells. Remarkably, it also revealed female AM had less upregulation of leukocyte extravasation signaling, production of NO and ROS, and Fcy receptor-mediated phagocytosis in hyperoxia (Fig. 7D). NADPH oxidase, a major source of ROS, is constituted by CYBA, CYBB, NCF1, NCF2, and NCF4. ScRNAseq revealed that their expression was upregulated 2.1-, 2.3-, 1.9-, 1.3-, and 1.4-folds respectively significantly in AM in hyperoxia. NO and ROS produced by activated macrophages protect hosts against a broad spectrum of pathogens and stimulation (88, 89), whereas Fcy receptor-mediated phagocytosis plays a key role in clearing pathogens (90). AM are the first line of defense in the lung against airborne pathogens, clearing injured cells, and debris (91, 92), but they also contribute to pathogenesis and pulmonary fibrosis by releasing excessive cytokines, oxygen radicals and proteases (6, 93–95). Hyperoxia induced more macrophage infiltration in male mice compared with female mice (18). Excessive activated macrophages could cause tissue damage in lung injury (96). Less activation of ROS pathways in female mice in hyperoxia is also consistent with less macrophage activation and may contribute to less BPD susceptibility.
DISCUSSION
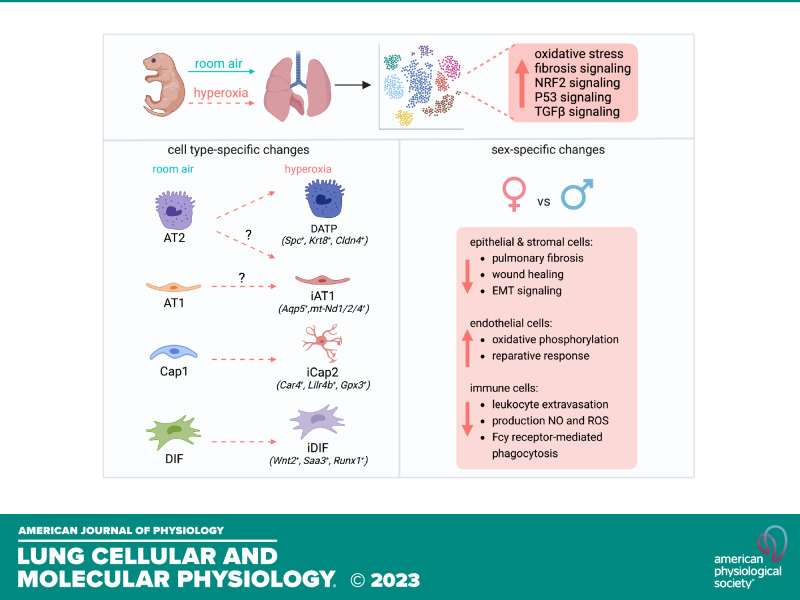
To investigate how hyperoxia programs transcriptional networks regulating cellular identity, plasticity, injury, and repair in the neonatal lung and the impact of sex on transcriptomic signatures, we combined scRNA-seq with validation studies in independent mouse lung samples and neonatal human lung cell lines. Hyperoxia acutely induces AT1 injury, and it is postulated that AT2 cells serve as the progenitor cells that repopulate AT1 cells on injury (97). In the developing mouse lung, we identified Lyz1+ AT2, AT2, cAT2 cells, and another AT2 cluster, which we labeled mAT2. The Lyz1+ AT2 cluster had previously been noted by Hurskainen et al. (7) in their study describing cellular populations after hyperoxia. However, mAT2 cells have not been described in the neonatal lung before. This cell population showed enrichment of mitochondrial biogenesis genes such as mt-ND1 and mtND2 while having decreased expression of Sftpc, Etv5, and Abca3. In hyperoxia, we first noted and then confirmed by IF that AT2 cells assumed an “inflamed” phenotype with increased Lcn2 expression, an observation Hurskainen et al. also noted in their study. The striking finding among AT2 was the emergence of DATP population (Sftpclow/Krt8+/Cldn4+) in hyperoxia, which was not present in RA. This population has been described as a transient stem cell progenitor state induced by bleomycin in the adult lung (5) but it had not been described before in hyperoxia models. IPA analysis revealed activation of IL1β and HIF1α pathway signaling in hyperoxia-exposed AT2 cells, as reported by Choi et al. (5) and Nold et al. (98), which revealed that these pathways program AT2 toward an AT1 phenotype. The switch from AT2 to inflamed AT2 to DATP in the neonatal lung with hyperoxia, suggests that AT2 have similar adaptive/regenerative mechanisms to various noxious stimuli. In primary neonatal human alveolar epithelial cells in vitro, we showed that hyperoxia cell-autonomously induces expression of the inflamed AT2 and DATP signatures, suggesting a conserved response across species. Scafa et al. (99) used a brief exposure to hyperoxia (P1–P3) to study AT1 and AT2 populations using scRNA-seq, and although they identified several AT clusters, they did not comment on the populations we identified. Whether this response is able to reconstitute the AT1 pool, and the key signaling mechanisms mediating it, are topics for future research.
In AT1 cells, we noted a distinct cluster emerge in hyperoxia marked by increased mitochondrial biogenesis genes, which we labeled as iAT1. These iAT1 cells had decreased expression of the traditional AT1 markers and AT1-derived growth factors. We also confirmed their emergence in vivo by colocalization of mt-ND1 and PDPN in the mouse lung after hyperoxia. Whether these cells represent an injured state of AT1 or an immature population of newly formed AT1 cells, and the specific mechanisms governing their emergence—whether from DAPT, AT2/AT1, or from AT2 cells—needs to be further investigated. We compared alveolar epithelial cells between females and males in hyperoxia to determine transcriptomic changes regulating the resilience of female pups to hyperoxia, and the analysis suggests less EMT, pulmonary fibrosis, and necroptosis signaling in females.
We identified two populations of capillary EC as reported by other groups (3, 29–31). Cap2 appear to be specialized for gas exchange but do not proliferate, whereas Cap1 function as a progenitor cell for Cap2 during lung development and injury recovery (31, 32). In our study, Car4+ Cap2 and Car4 expression in Cap1 increased after hyperoxia, indicating a reparative capillary response. Hurskainen et al. (7) also found a significant increase in the Cap2 population in hyperoxia, which is consistent with our data, but they did not address sex-specific endothelial signatures. We also noted the emergence of iCap2, a new subpopulation of Cap2 cells not described before. These cells expressed Car4 and Apln to a lesser extent than bona fide Cap2, but not Plvap or Aplnr, the Cap1 markers. Other distinguishing features of this population were decreased Kdr and Nrp1, which are key mediators of VEGF signaling (100). Whether iCap2 cells are injured or newly formed Cap2 remains to be deciphered, but a decrease in lumenization marker Icam2 would suggest they represent an immature state. ScRNA-seq showed more Car4 expression in female Cap1 than male Cap1 cells in hyperoxia. Validation studies further confirmed higher Car4 mRNA and protein expression in female neonatal mice. We did not have the tools to isolate Cap2 from Cap1 to validate increased CAR4 in the Cap1 population, but this sex-dependent difference was so pronounced that we could detect it in whole lung lysates. Altogether, these data might indicate a more robust EC regenerative response in females (32). Expression of both Cap2 and Cap1 markers, such as APLN, CA4, APLNR, and PLVAP, along with KDR was repressed by hyperoxia in primary canalicular stage HPMEC, whereas the addition of human fetal lung fibroblasts in a coculture system upregulated gene expression of both Cap1 and Cap2 markers. These data suggest that the EC recovery from hyperoxia requires paracrine input from fibroblasts and AT1. We speculate that AT1-derived VEGFA might be required for selective Cap2 expansion in hyperoxia, as it is a known key factor for the emergence of Car4+ Cap2 during development (30). Moreover, although scRNA-seq shows an increase in Car4+ Cap2 cells, IF shows a decrease in CAR4 staining. This could be explained by the formation of new Cap2 cells in hyperoxia, which would potentially have shorter extensions as they start to develop, visually leading to a less dense CAR4 network. Both Cap2 EC and AM express Car4 under normal conditions. However, we believe that the main source of increased Car4 expression in mouse whole lung experiments in hyperoxia comes from the expansion of Cap2 vessels since Car4 seems unchanged in scRNA-seq data comparing AM between both conditions.
We defined stromal mesenchymal cells as previously described in a recent paper (35). Among these subpopulations, Pdgfra+/Wnt2+/Tcf-21high/G0s2high DIF, Pdgfrahigh/Lgr6-/Hhip+/Cdh4+ AMF, and Pdgfra+/Twist2+/Il33+/Col1a1high/Col1a2high PIF were similarly defined as Col13a1+ fibroblasts, myofibroblasts, and Col14a1+ fibroblasts, respectively, in another neonatal hyperoxia study (7). SAA3 was enriched in interstitial fibroblasts in hyperoxia, which is consistent with Hurskainen study (7). Pdgfra expression was repressed in all fibroblasts in hyperoxia, which is also consistent with scRNA-seq data from Hurskainen et al. (7). We also identified a new subcluster of DIF in hyperoxia, iDIF, in which expression of DIF marker genes was reduced, but TGFB downstream genes were upregulated. Incidentally, the upregulation of Runx1 and suppression of Pparg in iDiF also suggest these cells might differentiate into collagen-synthesizing myofibroblasts and contribute to the fibrotic response seen in hyperoxia and BPD (101). EC induces differentiation and maturation of mural cells via NOTCH3 (102, 103), whose expression was decreased in mural cells in hyperoxia. PDGFRA signaling in AMF plays a critical role in alveolar septation, and Pdgfra−/− mice exhibit alveolar simplification (64). Repressed expression of Pdgfra and Eln in AMF could be a key cause of disrupted alveolar development in hyperoxia. We revealed that Pdgfra and Eln mRNA expression was repressed in hyperoxia, whereas TGFBR2 and SMAD3 protein expression and phosphorylation were upregulated in the whole lung. We further confirmed this in neonatal human fibroblasts in vitro. Hyperoxia repressed PDGFRA and ELN expression, which are critical to secondary septation, whereas hyperoxia upregulated the expression of TGFBR2 and ACTA2, key determinants of remodeling and disease progression in lung fibrosis (48, 73). IPA analysis showed less pulmonary fibrosis signaling in fibroblast clusters in female mice exposed to hyperoxia, which might explain decreased BPD vulnerability in female infants.
Although our focus was on epithelial, endothelial, and mesenchymal cells that directly contribute to alveolarization, we also examined the immune cell population. Our results did not identify any new populations in hyperoxia, but IPA analysis revealed hyperoxia stimulated important proinflammatory signaling pathways in AM. The expansion of activated AM population in hyperoxia could represent monocyte-derived alternatively activated macrophages (104). Interestingly, hyperoxia-induced leukocyte extravasation signaling, production of NO and ROS, and Fcy receptor-mediated phagocytosis were decreased in females compared with males, which may be relevant to decreased BPD incidence in female infants, as AM activation is known to interrupt lung development (6).
In conclusion, our study shows that hyperoxia programs distinct cellular and transcriptomic changes that signal injury and reparative responses in the neonatal lung. Although hyperoxia-induced alveolar simplification does not recapitulate the complex human BPD phenotype, it is a well-established model to study the detrimental effects of hyperoxia on lung development (105). The emergence of DATP and iCap2 most likely represents reparative responses, although whether iAT1 and iDIF populations represent injury states or newly generated immature populations remains to be discerned. We speculate that continuing hyperoxia exposure in our model restricts reparative responses resulting in incomplete/inadequate repair. Female mice showed signatures of decreased fibrosis signaling, a more robust EC iCap2 response, and decreased macrophage activation, all of which could contribute to the decreased vulnerability of females to hyperoxia injury. Interestingly, neonatal human alveolar epithelial cells and fibroblasts exhibit cell-autonomous reparative gene expression signatures in hyperoxia that resemble our mouse data, whereas cell-autonomous EC responses mimic an injury response, which is rescued with fibroblast coculture. Although we report several novel and interesting findings, a lack of insight into the transcriptional mechanisms that program these distinct cell-type specific changes remains a limitation. However, our findings lay a foundation for future mechanistic studies that can target the cell types and signaling pathways here identified.
DATA AVAILABILITY
Raw data have been deposited in GEO under the accession number GSE209664.
SUPPLEMENTAL DATA
Supplemental Tables S1 and S2 and Supplemental Figs. S1–S4: https://doi.org/10.6084/m9.figshare.20435898.
GRANTS
H.M., S.X., S.M.M., and V.S. were supported by 1R01HL128374-05 (V.S.) and 1R01HL162937-01 (V.S.). L.V.E. was supported by 5K99HL155845; J.C. was supported by R01HL153511 and R01HL130129.
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the authors.
AUTHOR CONTRIBUTIONS
S.X., J.C., and V.S. conceived and designed research; S.X., H.M., S.M.M., A.V., and D.L. performed experiments; S.X., L.V.E., K.W., H.M., S.M.M., M.G., E.G., J.C., and V.S. analyzed data; S.X., L.V.E., and V.S. interpreted results of experiments; S.X., L.V.E., H.M., and V.S. prepared figures; S.X., H.M., and V.S. drafted manuscript; S.X., L.V.E., and V.S. edited and revised manuscript; J.C., L.V.E., and V.S. approved final version of manuscript.
ACKNOWLEDGMENTS
The authors thank Aparna Venkataraman from Children Mercy Hospital, Kansas City for critical input and Wei Yu, PhD from Children Mercy Hospital, Kansas City for producing immortalized cell lines. We also thank Qiagen for permission to use images generated with IPA software. Figure 1 and graphical abstract image created with BioRender.com and published with permission. Preprint is available at https://doi.org/10.1101/2022.07.12.499826.
REFERENCES
- 1. Schittny JC. Development of the lung. Cell Tissue Res 367: 427–444, 2017. doi: 10.1007/s00441-016-2545-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Travaglini KJ, Nabhan AN, Penland L, Sinha R, Gillich A, Sit RV, Chang S, Conley SD, Mori Y, Seita J, Berry GJ, Shrager JB, Metzger RJ, Kuo CS, Neff N, Weissman IL, Quake SR, Krasnow MA. A molecular cell atlas of the human lung from single-cell RNA sequencing. Nature 587: 619–625, 2020. doi: 10.1038/s41586-020-2922-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Guo M, Du Y, Gokey JJ, Ray S, Bell SM, Adam M, Sudha P, Perl AK, Deshmukh H, Potter SS, Whitsett JA, Xu Y. Single cell RNA analysis identifies cellular heterogeneity and adaptive responses of the lung at birth. Nat Commun 10: 37, 2019. doi: 10.1038/s41467-018-07770-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Reyfman PA, Walter JM, Joshi N, Anekalla KR, McQuattie-Pimentel AC, Chiu S, et al.. Single-cell transcriptomic analysis of human lung provides insights into the pathobiology of pulmonary fibrosis. Am J Respir Crit Care Med 199: 1517–1536, 2019. doi: 10.1164/rccm.201712-2410OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Choi J, Park J-E, Tsagkogeorga G, Yanagita M, Koo B-K, Han N, Lee J-H. Inflammatory signals induce AT2 cell-derived damage-associated transient progenitors that mediate alveolar regeneration. Cell Stem Cell 27: 366–382.e7, 2020. doi: 10.1016/j.stem.2020.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Sahoo D, Zaramela LS, Hernandez GE, Mai U, Taheri S, Dang D, Stouch AN, Medal RM, McCoy AM, Aschner JL, Blackwell TS, Zengler K, Prince LS. Transcriptional profiling of lung macrophages identifies a predictive signature for inflammatory lung disease in preterm infants. Commun Biol 3: 259, 2020. doi: 10.1038/s42003-020-0985-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Hurskainen M, Mižíková I, Cook DP, Andersson N, Cyr-Depauw C, Lesage F, Helle E, Renesme L, Jankov RP, Heikinheimo M, Vanderhyden BC, Thébaud B. Single cell transcriptomic analysis of murine lung development on hyperoxia-induced damage. Nat Commun 12: 1565, 2021. doi: 10.1038/s41467-021-21865-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Morty RE. Recent advances in the pathogenesis of BPD. Semin Perinatol 42: 404–412, 2018. doi: 10.1053/j.semperi.2018.09.001. [DOI] [PubMed] [Google Scholar]
- 9. Lignelli E, Palumbo F, Myti D, Morty RE. Recent advances in our understanding of the mechanisms of lung alveolarization and bronchopulmonary dysplasia. Am J Physiol Lung Cell Mol Physiol 317: L832–L887, 2019. doi: 10.1152/ajplung.00369.2019. [DOI] [PubMed] [Google Scholar]
- 10. Meller S, Bhandari V. VEGF levels in humans and animal models with RDS and BPD: temporal relationships. Exp Lung Res 38: 192–203, 2012. doi: 10.3109/01902148.2012.663454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Stark A, Dammann C, Nielsen HC, Volpe MV. A pathogenic relationship of bronchopulmonary dysplasia and retinopathy of prematurity? A review of angiogenic mediators in both diseases. Front Pediatr 6: 125, 2018. doi: 10.3389/fped.2018.00125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Alejandre-Alcázar MA, Kwapiszewska G, Reiss I, Amarie OV, Marsh LM, Sevilla-Pérez J, Wygrecka M, Eul B, Köbrich S, Hesse M, Schermuly RT, Seeger W, Eickelberg O, Morty RE. Hyperoxia modulates TGF-β/BMP signaling in a mouse model of bronchopulmonary dysplasia. Am J Physiol Lung Cell Mol Physiol 292: L537–L549, 2007. doi: 10.1152/ajplung.00050.2006. [DOI] [PubMed] [Google Scholar]
- 13. Das KC, Ravi D, Holland W. Increased apoptosis and expression of p21 and p53 in premature infant baboon model of bronchopulmonary dysplasia. Antioxid Redox Signal 6: 109–116, 2004. doi: 10.1089/152308604771978417. [DOI] [PubMed] [Google Scholar]
- 14. Ni J, Dong Z, Han W, Kondrikov D, Su Y. The role of RhoA and cytoskeleton in myofibroblast transformation in hyperoxic lung fibrosis. Free Radic Biol Med 61: 26–39, 2013. doi: 10.1016/j.freeradbiomed.2013.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Rindfleisch MS, Hasday JD, Taciak V, Broderick K, Viscardi RM. Potential role of interleukin-1 in the development of bronchopulmonary dysplasia. J Interferon Cytokine Res 16: 365–373, 1996. doi: 10.1089/jir.1996.16.365. [DOI] [PubMed] [Google Scholar]
- 16. Silveyra P, Fuentes N, Rodriguez Bauza DE. Sex and gender differences in lung disease. Adv Exp Med Biol 1304: 227–258, 2021. doi: 10.1007/978-3-030-68748-9_14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Grimm SL, Dong X, Zhang Y, Carisey AF, Arnold AP, Moorthy B, Coarfa C, Lingappan K. Effect of sex chromosomes versus hormones in neonatal lung injury. JCI Insight 6: e146863, 2021. doi: 10.1172/jci.insight.146863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Lingappan K, Jiang W, Wang L, Moorthy B. Sex-specific differences in neonatal hyperoxic lung injury. Am J Physiol Lung Cell Mol Physiol 311: L481–L493, 2016. doi: 10.1152/ajplung.00047.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Bajar BT, Wang ES, Zhang S, Lin MZ, Chu J. A guide to fluorescent protein FRET pairs. Sensors (Basel) 16: 1488, 2016. doi: 10.3390/s16091488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Zhang Y, Dong X, Shirazi J, Gleghorn JP, Lingappan K. Pulmonary endothelial cells exhibit sexual dimorphism in their response to hyperoxia. Am J Physiol Heart Circ Physiol 315: H1287–H1292, 2018. doi: 10.1152/ajpheart.00416.2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Pozarska A, Rodríguez-Castillo JA, Surate Solaligue DE, Ntokou A, Rath P, Mižíková I, Madurga A, Mayer K, Vadász I, Herold S, Ahlbrecht K, Seeger W, Morty RE. Stereological monitoring of mouse lung alveolarization from the early postnatal period to adulthood. Am J Physiol Lung Cell Mol Physiol 312: L882–L895, 2017. doi: 10.1152/ajplung.00492.2016. [DOI] [PubMed] [Google Scholar]
- 22. Franks TJ, Colby TV, Travis WD, Tuder RM, Reynolds HY, Brody AR, Cardoso WV, Crystal RG, Drake CJ, Engelhardt J, Frid M, Herzog E, Mason R, Phan SH, Randell SH, Rose MC, Stevens T, Serge J, Sunday ME, Voynow JA, Weinstein BM, Whitsett J, Williams MC. Resident cellular components of the human lung current knowledge and goals for research on cell phenotyping and function. Proc Am Thorac Soc 5: 763–766, 2008. doi: 10.1513/pats.200803-025HR. [DOI] [PubMed] [Google Scholar]
- 23. Wu H, Tang N. Stem cells in pulmonary alveolar regeneration. Development 148: dev193458, 2021. doi: 10.1242/dev.193458. [DOI] [PubMed] [Google Scholar]
- 24. Olajuyin AM, Zhang X, Ji HL. Alveolar type 2 progenitor cells for lung injury repair. Cell Death Discov 5: 63, 2019. doi: 10.1038/s41420-019-0147-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Yang J, Hernandez BJ, Martinez Alanis D, Narvaez del Pilar O, Vila-Ellis L, Akiyama H, Evans SE, Ostrin EJ, Chen J. The development and plasticity of alveolar type 1 cells. Development 143: 54–65, 2016. doi: 10.1242/dev.130005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Wang Y, Tang Z, Huang H, Li J, Wang Z, Yu Y, Zhang C, Li J, Dai H, Wang F, Cai T, Tang N. Pulmonary alveolar type I cell population consists of two distinct subtypes that differ in cell fate. Proc Natl Acad Sci USA 115: 2407–2412, 2018. doi: 10.1073/pnas.1719474115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Desai TJ, Brownfield DG, Krasnow MA. Alveolar progenitor and stem cells in lung development, renewal and cancer. Nature 507: 190–194, 2014. doi: 10.1038/nature12930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Vyas-Read S, Wang W, Kato S, Colvocoresses-Dodds J, Fifadara NH, Gauthier TW, Helms MN, Carlton DP, Brown LAS. Hyperoxia induces alveolar epithelial-to-mesenchymal cell transition. Am J Physiol - Lung Cell Mol Physiol 306: L326–L340, 2014. doi: 10.1152/ajplung.00074.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Angelidis I, Simon LM, Fernandez IE, Strunz M, Mayr CH, Greiffo FR, Tsitsiridis G, Ansari M, Graf E, Strom TM, Nagendran M, Desai T, Eickelberg O, Mann M, Theis FJ, Schiller HB. An atlas of the aging lung mapped by single cell transcriptomics and deep tissue proteomics. Nat Commun 10: 963, 2019. doi: 10.1038/s41467-019-08831-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Vila Ellis L, Cain MP, Hutchison V, Flodby P, Crandall ED, Borok Z, Zhou B, Ostrin EJ, Wythe JD, Chen J. Epithelial Vegfa specifies a distinct endothelial population in the mouse lung. Dev Cell 52: 617–630.e6, 2020. doi: 10.1016/j.devcel.2020.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Gillich A, Zhang F, Farmer CG, Travaglini KJ, Tan SY, Gu M, Zhou B, Feinstein JA, Krasnow MA, Metzger RJ. Capillary cell-type specialization in the alveolus. Nature 586: 785–789, 2020. doi: 10.1038/s41586-020-2822-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Niethamer TK, Stabler CT, Leach JP, Zepp JA, Morley MP, Babu A, Zhou S, Morrisey EE. Defining the role of pulmonary endothelial cell heterogeneity in the response to acute lung injury. eLife 9: e53072, 2020. doi: 10.7554/eLife.53072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Rodríguez-Castillo JA, Pérez DB, Ntokou A, Seeger W, Morty RE. Understanding alveolarization to induce lung regeneration. Respir Res 19: 148, 2018. doi: 10.1186/s12931-018-0837-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Xie T, Wang Y, Deng N, Huang G, Taghavifar F, Geng Y, Liu N, Kulur V, Yao C, Chen P, Liu Z, Stripp B, Tang J, Liang J, Noble PW, Jiang D. Single-cell deconvolution of fibroblast heterogeneity in mouse pulmonary fibrosis. Cell Rep 22: 3625–3640, 2018. doi: 10.1016/j.celrep.2018.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Narvaez Del Pilar O, Gacha Garay MJ, Chen J. Three-axis classification of mouse lung mesenchymal cells reveals two populations of myofibroblasts. Development 149: dev200081, 2022. doi: 10.1242/dev.200081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Kendall RT, Feghali-Bostwick CA. Fibroblasts in fibrosis: novel roles and mediators. Front Pharmacol 5: 123, 2014. doi: 10.3389/fphar.2014.00123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Nyp MF, Mabry SM, Navarro A, Menden H, Perez RE, Sampath V, Ekekezie II. Lung epithelial-specific TRIP-1 overexpression maintains epithelial integrity during hyperoxia exposure. Physiol Rep 6: e13585, 2018. doi: 10.14814/phy2.13585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Xia S, Menden HL, Townley N, Mabry SM, Johnston J, Nyp MF, Heruth DP, Korfhagen T, Sampath V. Delta-like 4 is required for pulmonary vascular arborization and alveolarization in the developing lung. JCI Insight 6: e134170, 2021. doi: 10.1172/jci.insight.134170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Blighe K, Rana S, Lewis M. EnhancedVolcano: publication-ready volcano plots with enhanced colouring and labeling (Online). https://github.com/kevinblighe/EnhancedVolcano.
- 40. Nitkin CR, Xia S, Menden H, Yu W, Xiong M, Heruth DP, Ye SQ, Sampath V. FOSL1 is a novel mediator of endotoxin/lipopolysaccharide-induced pulmonary angiogenic signaling. Sci Rep 10: 13143, 2020. doi: 10.1038/s41598-020-69735-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Ambalavanan N, Morty RE. Searching for better animal models of BPD: a perspective. Am J Physiol Lung Cell Mol Physiol 311: L924–L927, 2016. doi: 10.1152/ajplung.00355.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Garcia D, Carr JF, Chan F, Peterson AL, Ellis KA, Scaffa A, Ghio AJ, Yao H, Dennery PA. Short exposure to hyperoxia causes cultured lung epithelial cell mitochondrial dysregulation and alveolar simplification in mice. Pediatr Res 90: 58–65, 2021. doi: 10.1038/s41390-020-01224-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Cho H-Y, Jedlicka AE, Reddy SPM, Kensler TW, Yamamoto M, Zhang L-Y, Kleeberger SR. Role of NRF2 in protection against hyperoxic lung injury in mice. Am J Respir Cell Mol Biol 26: 175–182, 2002. doi: 10.1165/ajrcmb.26.2.4501. [DOI] [PubMed] [Google Scholar]
- 44. Sampath V, Garland JS, Helbling D, Dimmock D, Mulrooney NP, Simpson PM, Murray JC, Dagle JM. Antioxidant response genes sequence variants and BPD susceptibility in VLBW infants. Pediatr Res 77: 477–483, 2015. doi: 10.1038/pr.2014.200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Biernacka A, Dobaczewski M, Frangogiannis NG. TGF-β signaling in fibrosis. Growth Factors 29: 196–202, 2011. doi: 10.3109/08977194.2011.595714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Bhandary YP, Shetty SK, Marudamuthu AS, Ji HL, Neuenschwander PF, Boggaram V, Morris GF, Fu J, Idell S, Shetty S. Regulation of lung injury and fibrosis by p53-mediated changes in urokinase and plasminogen activator inhibitor-1. Am J Pathol 183: 131–143, 2013. doi: 10.1016/j.ajpath.2013.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Watts KL, Cottrell E, Hoban PR, Spiteri MA. RhoA signaling modulates cyclin D1 expression in human lung fibroblasts; implications for idiopathic pulmonary fibrosis. Respir Res 7: 88, 2006. doi: 10.1186/1465-9921-7-88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Peyser R, MacDonnell S, Gao Y, Cheng L, Kim Y, Kaplan T, Ruan Q, Wei Y, Ni M, Adler C, Zhang W, Devalaraja-Narashimha K, Grindley J, Halasz G, Morton L. Defining the activated fibroblast population in lung fibrosis using single-cell sequencing. Am J Respir Cell Mol Biol 61: 74–85, 2019. doi: 10.1165/rcmb.2018-0313OC. [DOI] [PubMed] [Google Scholar]
- 49. Ghosh AK, Vaughan DE. PAI-1 in tissue fibrosis. J Cell Physiol 227: 493–507, 2012. doi: 10.1002/jcp.22783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Ghandikota S, Sharma M, Ediga HH, Madala SK, Jegga AG. Consensus gene co-expression network analysis identifies novel genes associated with severity of fibrotic lung disease. Int J Mol Sci 23: 5447, 2022. doi: 10.3390/ijms23105447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Park C, Kim TM, Malik AB. Transcriptional regulation of endothelial cell and vascular development. Circ Res 112: 1380–1400, 2013. doi: 10.1161/CIRCRESAHA.113.301078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Xu Y, Wang Y, Besnard V, Ikegami M, Wert SE, Heffner C, Murray SA, Donahue LR, Whitsett JA. Transcriptional programs controlling perinatal lung maturation. PLoS One 7: e37046, 2012. doi: 10.1371/journal.pone.0037046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Macleod KF, Sherry N, Hannon G, Beach D, Tokino T, Kinzler K, Vogelstein B, Jacks T. p53-dependent and independent expression of p21 during cell growth, differentiation, and DNA damage. Genes Dev 9: 935–944, 1995. doi: 10.1101/gad.9.8.935. [DOI] [PubMed] [Google Scholar]
- 54. Kortlever RM, Higgins PJ, Bernards R. Plasminogen activator inhibitor-1 is a critical downstream target of p53 in the induction of replicative senescence. Nat Cell Biol 8: 877–884, 2006. doi: 10.1038/ncb1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Zhang K, Rekhter MD, Gordon D, Phan SH. Myofibroblasts and their role in lung collagen gene expression during pulmonary fibrosis. A combined immunohistochemical and in situ hybridization study. Am J Pathol 145: 114–125, 1994. [PMC free article] [PubMed] [Google Scholar]
- 56. Zhang Z, Newton K, Kummerfeld SK, Webster J, Kirkpatrick DS, Phu L, Eastham-Anderson J, Liu J, Lee WP, Wu J, Li H, Junttila MR, Dixit VM. Transcription factor Etv5 is essential for the maintenance of alveolar type II cells. Proc Natl Acad Sci USA 114: 3903–3908, 2017. doi: 10.1073/pnas.1621177114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Tomer Y, Wambach J, Knudsen L, Zhao M, Rodriguez LR, Murthy A, White FV, Venosa A, Katzen J, Ochs M, Hamvas A, Beers MF, Mulugeta S. The common ABCA3(E292V) variant disrupts AT2 cell quality control and increases susceptibility to lung injury and aberrant remodeling. Am J Physiol Lung Cell Mol Physiol 321: L291–L307, 2021. doi: 10.1152/ajplung.00400.2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Russell REK, Sc B, Ph D. Ineffectual type 2 to type 1 alveolar epithelial cell differentiation in idiopathic pulmonary fibrosis: persistence of the KRT8hi transitional state. Am J Respir Crit Care Med 201: 1443–1447, 2020. doi: 10.1164/rccm.201909-1726LE. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Kobayashi Y, Tata A, Konkimalla A, Katsura H, Lee RF, Ou J, Banovich NE, Kropski JA, Tata PR. Persistence of a regeneration-associated, transitional alveolar epithelial cell state in pulmonary fibrosis. Nat Cell Biol 22: 934–946, 2020. doi: 10.1038/s41556-020-0542-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Auyeung VC, Sheppard D. Stuck in a moment: does abnormal persistence of epithelial progenitors drive pulmonary fibrosis? Am J Respir Crit Care Med 203: 667–669, 2021. doi: 10.1164/rccm.202010-3898ED. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Kunig AM, Balasubramaniam V, Markham NE, Morgan D, Montgomery G, Grover TR, Abman SH. Recombinant human VEGF treatment enhances alveolarization after hyperoxic lung injury in neonatal rats. Am J Physiol Lung Cell Mol Physiol 289: 529–535, 2005. doi: 10.1152/ajplung.00336.2004. [DOI] [PubMed] [Google Scholar]
- 62. Thébaud B, Ladha F, Michelakis ED, Sawicka M, Thurston G, Eaton F, Hashimoto K, Harry G, Haromy A, Korbutt G, Archer SL. Vascular endothelial growth factor gene therapy increases survival, promotes lung angiogenesis, and prevents alveolar damage in hyperoxia-induced lung injury: evidence that angiogenesis participates in alveolarization. Circulation 112: 2477–2486, 2005. doi: 10.1161/CIRCULATIONAHA.105.541524. [DOI] [PubMed] [Google Scholar]
- 63. Gouveia L, Kraut S, Hadzic S, Vazquéz-Liébanas E, Kojonazarov B, Wu CY, Veith C, He L, Mermelekas G, Schermuly RT, Weissmann N, Betsholtz C, Andrae J. Lung developmental arrest caused by PDGF-A deletion: consequences for the adult mouse lung. Am J Physiol Lung Cell Mol Physiol 318: L831–L843, 2020. doi: 10.1152/AJPLUNG.00295.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Li R, Bernau K, Sandbo N, Gu J, Preissl S, Sun X. Pdgfra marks a cellular lineage with distinct contributions to myofibroblasts in lung maturation and injury response. eLife 7: e36865, 2018. doi: 10.7554/eLife.36865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Franco-Montoya ML, Boucherat O, Thibault C, Chailley-Heu B, Incitti R, Delacourt C, Bourbon JR. Profiling target genes of FGF18 in the postnatal mouse lung: possible relevance for alveolar development. Physiol Genomics 43: 1226–1240, 2011. doi: 10.1152/physiolgenomics.00034.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Cardoso WV, Itoh A, Nogawa H, Mason I, Brody JS. FGF-1 and FGF-7 induce distinct patterns of growth and differentiation in embryonic lung epithelium. Dev Dyn 208: 398–405, 1997. doi:. [DOI] [PubMed] [Google Scholar]
- 67. Strunz M, Simon LM, Ansari M, Kathiriya JJ, Angelidis I, Mayr CH, Tsidiridis G, Lange M, Mattner LF, Yee M, Ogar P, Sengupta A, Kukhtevich I, Schneider R, Zhao Z, Voss C, Stoeger T, Neumann JHL, Hilgendorff A, Behr J, O’Reilly M, Lehmann M, Burgstaller G, Königshoff M, Chapman HA, Theis FJ, Schiller HB. Alveolar regeneration through a Krt8+ transitional stem cell state that persists in human lung fibrosis. Nat Commun 11: 3559, 2020. doi: 10.1038/s41467-020-17358-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Yan W-W, Liang Y-L, Zhang Q-X, Wang D, Lei M-Z, Qu J, He X-H, Lei Q-Y, Wang Y-P. Arginine methylation of SIRT7 couples glucose sensing with mitochondria biogenesis. EMBO Rep 19: e46377, 2018. doi: 10.15252/embr.201846377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Franco CA, Jones ML, Bernabeu MO, Geudens I, Mathivet T, Rosa A, Lopes FM, Lima AP, Ragab A, Collins RT, Phng L-K, Coveney PV, Gerhardt H. Dynamic endothelial cell rearrangements drive developmental vessel regression. PLoS Biol 13: e1002125, 2015. [Erratum in PLoS Biol 13: e1002163, 2015]. doi: 10.1371/journal.pbio.1002125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Fina L, Molgaard HV, Robertson D, Bradley NJ, Monaghan P, Delia D, Sutherland DR, Baker MA, Greaves MF. Expression of the CD34 gene in vascular endothelial cells. Blood 75: 2417–2426, 1990. doi: 10.1182/blood.V75.12.2417.2417. [DOI] [PubMed] [Google Scholar]
- 71. Gouveia L, Betsholtz C, Andrae J. PDGF-A signaling is required for secondary alveolar septation and controls epithelial proliferation in the developing lung. Dev 145: dev161976, 2018. doi: 10.1242/dev.161976. [DOI] [PubMed] [Google Scholar]
- 72. Mecham RP. Elastin in lung development and disease pathogenesis. Matrix Biol 73: 6–20, 2018. doi: 10.1016/j.matbio.2018.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Phan SH. Genesis of the myofibroblast in lung injury and fibrosis. Proc Am Thorac Soc 9: 148–152, 2012. doi: 10.1513/pats.201201-011AW. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Wang YY, Pan LY, Moens CB, Appel B. Notch3 establishes brain vascular integrity by regulating pericyte number. Development 141: 307–317, 2014. doi: 10.1242/dev.096107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Liu H, Zhang W, Lilly B. Evaluation of Notch3 deficiency in diabetes-induced pericyte loss in the Retina. J Vasc Res 55: 308–318, 2018. doi: 10.1159/000493151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Monickaraj F, Rangasamy S, Cabrera A, McGuire P, Das A. Role of Notch3 signaling in pericyte survival: a novel pathway of blood retinal barrier alteration in diabetic retinopathy (Abstract). Invest Ophthalmol Vis Sci 61: 724, 2020. https://iovs.arvojournals.org/article.aspx?articleid=2766695. [Google Scholar]
- 77. Bonikos DS, Bensch KG, Northway WHJ, Edwards DK. Bronchopulmonary dysplasia: the pulmonary pathologic sequel of necrotizing bronchiolitis and pulmonary fibrosis. Hum Pathol 7: 643–666, 1976. doi: 10.1016/s0046-8177(76)80077-9. [DOI] [PubMed] [Google Scholar]
- 78. Peng Y, Yan S, Chen D, Cui X, Jiao K. Pdgfrb is a direct regulatory target of TGFβ signaling in atrioventricular cushion mesenchymal cells. PLoS One 12: e0175791, 2017. doi: 10.1371/journal.pone.0175791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Dai M, Al-Odaini AA, Fils-Aimé N, Villatoro MA, Guo J, Arakelian A, Rabbani SA, Ali S, Lebrun J. Cyclin D1 cooperates with p21 to regulate TGFβ-mediated breast cancer cell migration and tumor local invasion. Breast Cancer Res 15: R49, 2013. [Erratum in Breast Cancer Res 19: 43, 2017]. doi: 10.1186/bcr3441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Castro NE, Kato M, Park JT, Natarajan R. Transforming growth factor β1 (TGF-β1) enhances expression of profibrotic genes through a novel signaling cascade and microRNAs in renal mesangial cells. J Biol Chem 289: 29001–29013, 2014. doi: 10.1074/jbc.M114.600783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Ito Y, Miyazono K. RUNX transcription factors as key targets of TGF-β superfamily signaling. Curr Opin Genet Dev 13: 43–47, 2003. doi: 10.1016/s0959-437x(03)00007-8. [DOI] [PubMed] [Google Scholar]
- 82. Kwak H-J, Park M-J, Cho H, Park C-M, Moon S-I, Lee H-C, Park I-C, Kim M-S, Rhee CH, Hong S-I. Transforming growth factor-β1 induces tissue inhibitor of metalloproteinase-1 expression via activation of extracellular signal-regulated kinase and Sp1 in human fibrosarcoma cells. Mol Cancer Res 4: 209–220, 2006. doi: 10.1158/1541-7786.MCR-05-0140. [DOI] [PubMed] [Google Scholar]
- 83. Liu X, Rowan S, Liang CJ, Yao C, Deng N, Huang G, Xie T, Wang Y, Stripp BR, Noble PW, Jiang D. Definition and signatures of lung fibroblast populations in development and fibrosis in mice and men. Am J Respir Crit Care Med 201: A4025, 2020. doi: 10.1164/ajrccm-conference.2020.201.1_meetingabstracts.a4025. [DOI] [Google Scholar]
- 84. Schultz CJ, Torres E, Londos C, Torday JS. Role of adipocyte differentiation-related protein in surfactant phospholipid synthesis by type II cells. Am J Physiol Lung Cell Mol Physiol 283: L288–L296, 2002. doi: 10.1152/ajplung.00204.2001. [DOI] [PubMed] [Google Scholar]
- 85. Czech MP, Tencerova M, Pedersen DJ, Aouadi M. Insulin signalling mechanisms for triacylglycerol storage. Diabetologia 56: 949–964, 2013. doi: 10.1007/s00125-013-2869-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Xu X, So J-S, Park J-G, Lee A-H. Transcriptional control of hepatic lipid metabolism by SREBP and ChREBP. Semin Liver Dis 33: 301–311, 2013. doi: 10.1055/s-0033-1358523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Schoonjans K, Staels B, Auwerx J. The peroxisome proliferator activated receptors (PPARS) and their effects on lipid metabolism and adipocyte differentiation. Biochim Biophys Acta 1302: 93–109, 1996. doi: 10.1016/0005-2760(96)00066-5. [DOI] [PubMed] [Google Scholar]
- 88. Forman HJ, Torres M. Redox signaling in macrophages. Mol Aspects Med 22: 189–216, 2001. doi: 10.1016/s0098-2997(01)00010-3. [DOI] [PubMed] [Google Scholar]
- 89. Bedard K, Krause KH. The NOX family of ROS-generating NADPH oxidases: physiology and pathophysiology. Physiol Rev 87: 245–313, 2007. doi: 10.1152/physrev.00044.2005. [DOI] [PubMed] [Google Scholar]
- 90. Aderem A, Underhill DM. Mechanisms of phagocytosis in macrophages. Annu Rev Immunol 17: 593–623, 1999. doi: 10.1146/annurev.immunol.17.1.593. [DOI] [PubMed] [Google Scholar]
- 91. Tan SYS, Krasnow MA. Developmental origin of lung macrophage diversity. Development 143: 1318–1327, 2016. doi: 10.1242/dev.129122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Sibille Y, Reynolds HY. Macrophages and polymorphonuclear neutrophils in lung defense and injury. Am Rev Respir Dis 141: 471–501, 1990. doi: 10.1164/ajrccm/141.2.471. [DOI] [PubMed] [Google Scholar]
- 93. Oberdörster G, Ferin J, Gelein R, Soderholm SC, Finkelstein J. Role of the alveolar macrophage in lung injury: Studies with ultrafine particles. Environ Health Perspect 97: 193–199, 1992. doi: 10.1289/ehp.97-1519541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Cheng P, Li S, Chen H. Macrophages in lung injury, repair and fibrosis. Cells 10: 436, 2021. doi: 10.3390/cells10020436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Clement A, Chadelat K, Sardet A, Grimfeld A, Tournier G. Alveolar macrophage status in bronchopulmonary dysplasia. Pediatr Res 23: 470–473, 1988. doi: 10.1203/00006450-198805000-00007. [DOI] [PubMed] [Google Scholar]
- 96. Lu H-L, Huang X-Y, Luo Y-F, Tan W-P, Chen P-F, Guo Y-B. Activation of M1 macrophages plays a critical role in the initiation of acute lung injury. Biosci Rep 38: BSR20171555, 2018. doi: 10.1042/BSR20171555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Yee M, Vitiello PF, Roper JM, Staversky RJ, Wright TW, McGrath-Morrow SA, Maniscalco WM, Finkelstein JN, O’Reilly MA. Type II epithelial cells are critical target for hyperoxia-mediated impairment of postnatal lung development. Am J Physiol Lung Cell Mol Physiol 291: L1101–L1111, 2006. doi: 10.1152/ajplung.00126.2006. [DOI] [PubMed] [Google Scholar]
- 98. Nold MF, Mangan NE, Rudloff I, Cho SX, Shariatian N, Samarasinghe TD, Skuza EM, Pedersen J, Veldman A, Berger PJ, Nold-Petry CA. Interleukin-1 receptor antagonist prevents murine bronchopulmonary dysplasia induced by perinatal inflammation and hyperoxia. Proc Natl Acad Sci USA 110: 14384–14389, 2013. doi: 10.1073/pnas.1306859110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Scaffa A, Yao H, Oulhen N, Wallace J, Peterson AL, Rizal S, Ragavendran A, Wessel G, De Paepe ME, Dennery PA. Single-cell transcriptomics reveals lasting changes in the lung cellular landscape into adulthood after neonatal hyperoxic exposure. Redox Biol 48: 102091, 2021. doi: 10.1016/j.redox.2021.102091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Herzog B, Pellet-Many C, Britton G, Hartzoulakis B, Zachary IC. VEGF binding to NRP1 is essential for VEGF stimulation of endothelial cell migration, complex formation between NRP1 and VEGFR2, and signaling via FAK Tyr407 phosphorylation. Mol Biol Cell 22: 2766–2776, 2011. doi: 10.1091/mbc.E09-12-1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Dubey S, Dubey PK, Umeshappa CS, Ghebre YT, Krishnamurthy P. Inhibition of RUNX1 blocks the differentiation of lung fibroblasts to myofibroblasts. J Cell Physiol 237: 2169–2182, 2022. doi: 10.1002/jcp.30684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Liu H, Kennard S, Lilly B. NOTCH3 expression is induced in mural cells through an autoregulatory loop that requires endothelial-expressed JAGGED1. Circ Res 104: 466–475, 2009. doi: 10.1161/CIRCRESAHA.108.184846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Fouillade C, Monet-Leprêtre M, Baron-Menguy C, Joutel A. Notch signalling in smooth muscle cells during development and disease. Cardiovasc Res 95: 138–146, 2012. doi: 10.1093/cvr/cvs019. [DOI] [PubMed] [Google Scholar]
- 104. Gundra UM, Girgis NM, Ruckerl D, Jenkins S, Ward LN, Kurtz ZD, Wiens KE, Tang MS, Basu-Roy U, Mansukhani A, Allen JE, Loke P. Alternatively activated macrophages derived from monocytes and tissue macrophages are phenotypically and functionally distinct. Blood 123: e110-22–e122, 2014. doi: 10.1182/blood-2013-08-520619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Nardiello C, Mižíková I, Silva DM, Ruiz-Camp J, Mayer K, Vadász I, Herold S, Seeger W, Morty RE. Standardisation of oxygen exposure in the development of mouse models for bronchopulmonary dysplasia. Dis Model Mech 10: 185–196, 2017. doi: 10.1242/dmm.027086. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Tables S1 and S2 and Supplemental Figs. S1–S4: https://doi.org/10.6084/m9.figshare.20435898.
Data Availability Statement
Raw data have been deposited in GEO under the accession number GSE209664.