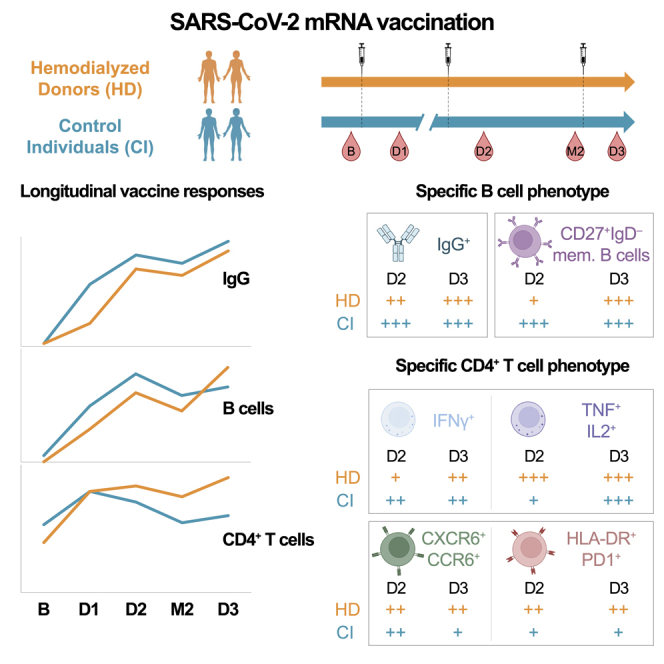
Summary
Cellular immune defects associated with suboptimal responses to severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) mRNA vaccination in people receiving hemodialysis (HD) are poorly understood. We longitudinally analyze antibody, B cell, CD4+, and CD8+ T cell vaccine responses in 27 HD patients and 26 low-risk control individuals (CIs). The first two doses elicit weaker B cell and CD8+ T cell responses in HD than in CI, while CD4+ T cell responses are quantitatively similar. In HD, a third dose robustly boosts B cell responses, leads to convergent CD8+ T cell responses, and enhances comparatively more T helper (TH) immunity. Unsupervised clustering of single-cell features reveals phenotypic and functional shifts over time and between cohorts. The third dose attenuates some features of TH cells in HD (tumor necrosis factor alpha [TNFα]/interleukin [IL]-2 skewing), while others (CCR6, CXCR6, programmed cell death protein 1 [PD-1], and HLA-DR overexpression) persist. Therefore, a third vaccine dose is critical to achieving robust multifaceted immunity in hemodialysis patients, although some distinct TH characteristics endure.
Keywords: COVID-19, SARS-CoV-2, mRNA vaccine, T cell immunity, B cell immunity, Hemodialysis, Vaccine response, Immune cell phenotyping
Graphical abstract
Highlights
-
•
Two doses of SARS-CoV-2 vaccine induce weak B cells in people on hemodialysis (HD)
-
•
People on HD generate less mature B cells after two vaccine doses
-
•
A third immunization in HD robustly boosts B and CD4+ T cell responses
-
•
CD4+ T cell profile in HD is skewed toward CCR6, CXCR6, PD-1, HLA-DR, and TNFα/IL-2
Sannier et al. describe specific humoral, B, and T cell responses in people on hemodialysis (HD) after mRNA vaccination against SARS-CoV-2. Three doses are critical to elicit robust antibody, B cell, and CD8+ T cell responses. CD4+ T cell profile is skewed toward TNF/IL-2, CCR6, CXCR6, PD-1, and HLA-DR expression.
Introduction
Implementation of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) vaccination has led to a sharp decrease in the severity of COVID-19 disease worldwide.1,2 In the general population, mRNA vaccines against SARS-CoV-2 induce robust responses of both humoral3,4,5 and cellular immunity, which is dominated by B cells and T helper (TH) responses with a weaker CD8+ T cell component.6,7 The initial series of mRNA vaccines comprised two doses. A third dose was recommended to offset waning immunity and improve recognition of variants of concern (VOCs), including Omicron.8,9,10 In low-risk populations, substantial protection is conferred by one dose,11 with notable antigen-specific immunity.12,13,14 Some public health agencies delayed the recommended interval between doses to increase population coverage during initial vaccine scarcity.15,16 Studies in low-risk individuals subsequently showed that a longer interval between the first two doses enhanced humoral responses,5,17,18 and increased specific B cell responses and maturation, with little impact on T cells.18,19,20
Patients with end-stage kidney disease receiving hemodialysis (HD) are susceptible to infections and demonstrate suboptimal responses to standard vaccinations against diphtheria, hepatitis B virus (HBV), or influenza.21 They display altered immune functions affecting B and T lymphocytes,22 monocytes,23 dendritic cells, and neutrophils24 due to uremia toxins25 and blood-membrane interactions during the dialysis process.26 However, multiple and/or higher vaccine doses proved to be an effective strategy, e.g., for HBV or influenza vaccination.27
HD patients are vulnerable to SARS-CoV-2 infection, severe COVID-19,28,29 and breakthrough events.30 Therefore, HD are considered a high-priority population for SARS-CoV-2 vaccination. Vaccination in HD generated anti-SARS-CoV-2 antibodies but at lower levels compared with the global population,31,32 and with earlier decline,33 even after three doses.34 While vaccine studies in HD have focused on humoral responses, a better understanding of specific B and T cell immunity is essential to identify underlying defects. CD4+ T cell help plays a critical role in the generation and maintenance of adaptive immunity, particularly of B cell responses,35 and CD8+ T cells may play a direct protective role against the virus. Some studies have shown lower SARS-CoV-2-reactive interferon gamma (IFNγ)-producing T cell frequencies in HD,36,37 strengthening the need to understand this arm of the immune system. While studies suggest that long-interval vaccine regimens are not appropriate for HD, resulting in weaker antibody levels, the impact on cellular immunity remains to be defined.
Herein, we conducted a prospective longitudinal cohort study to define the quantitative and qualitative trajectories of vaccine-induced antibody, B, CD4+, and CD8+ T cell responses in SARS-CoV-2-naive HD patients receiving three mRNA SARS-CoV-2 vaccine doses, compared with antigen-specific responses in low-risk CIs.
Results
Study participants
We assessed cellular and antibody responses in blood samples from three cohorts of SARS-CoV-2-naive participants who received three mRNA vaccine doses (Figure 1A, Table 1): (1) 20 people on HD (HDS cohort) who received the first two doses with a 5-week interval (median, interquartile range [IQ] = 35 [33–35] days); (2) 26 low-risk health care workers with no major kidney disease or immunosuppressive condition (control individuals [CIs]) who received the first two doses at a 16-week interval (median [IQ] = 111 [109–112] days), in agreement with the Quebec Public Health guidelines at the time of the study; (3) seven HD who received a 12-week delayed second dose (median [IQ] = 83 [82–84] days) (HDL long-interval HD cohort). The HDS and CI cohorts were studied in detail, while we performed focused analyses on the HDL cohort.
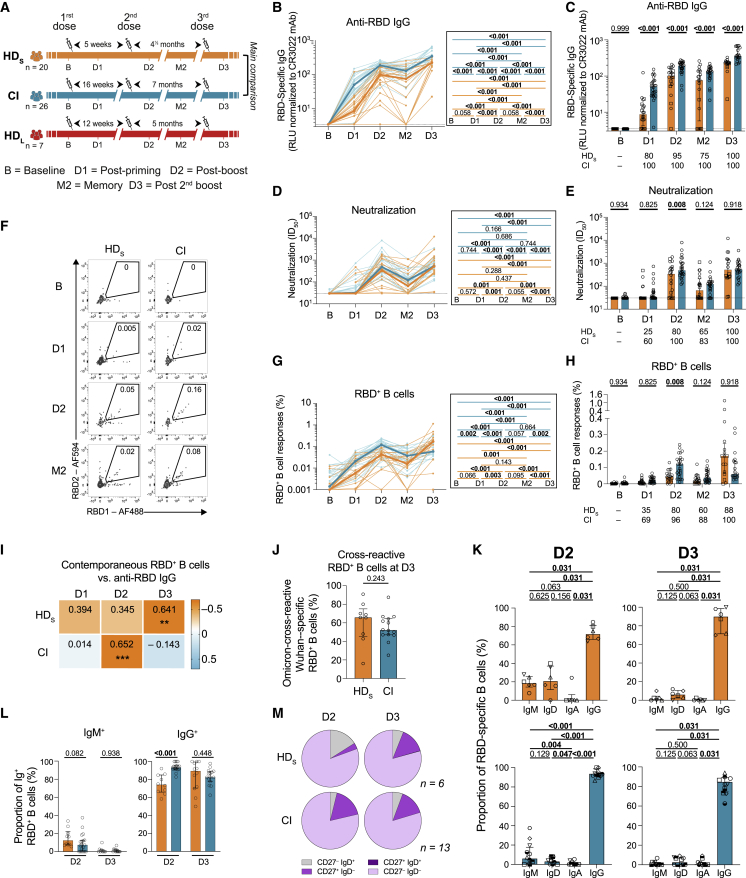
Figure 1.
In HD patients, the initial two mRNA vaccine inoculations elicit poor B cell responses, which are reinvigorated by a third dose
(A) Schematic representation of study design, visits, and vaccine dose administration (indicated by a syringe). Blood samples were collected at five time points: at baseline, B; 3-4 weeks after the first dose, D1 and the second dose, D2; 12–16 weeks after the second dose, M2; and 4 weeks after the third dose, D3. Following provincial vaccination guidelines, 20 HD participants (HDS) received the two doses at 5-week intervals and 26 control individuals (CIs) received the two doses at 16-week intervals. A second group of seven HD participants (HDL) received a delayed second dose with an interval of 12 weeks. Actual times are summarized in Table 1.
(B and C) Kinetics of RBD+ IgG responses in HDS participants (orange) or CI (blue) participants. HDS on immunosuppressive drugs are represented by square symbols, and HDS not on immunosuppressants are represented by circles. (B) Lines connect data points from the same donor. The bold lines represent the median values of each cohort. Right panel: statistical comparisons using a linear mixed model. (C) Comparisons between HDS and CI participants. Bars represent median ± interquartile range. Intercohort statistical comparisons using a linear mixed model are shown. Frequencies of responders are written below the histograms.
(D and E) Kinetics of antibody neutralizing activity measured with plasma from HDS participants (orange) or CI (blue) participants. HDS on immunosuppressive drugs are represented by square symbols; HDS not on immunosuppressants are represented by circles. (D) Lines connect data points from the same donor. The bold lines represent the median values of each cohort. Right panel: statistical comparisons using a linear mixed model. (E) Comparisons between HDS and CI participants. Bars represent medians ± interquartile ranges. Intercohort statistical comparisons using a linear mixed model are shown. Frequencies of responders are written below the histograms.
(F–H) (F) Gating strategy to identify RBD+ B cells. (G and H) Kinetics of RBD+ B cell responses in HDS participants (orange) or CI (blue) participants. HDS on immunosuppressive drugs are represented by square symbols; HDS not on immunosuppressants are represented by circles. (G) Lines connect data points from the same donor. The bold lines represent the median values of each cohort. Right panel: statistical comparisons using a linear mixed model. (H) Comparisons between HDS and CI participants. Bars represent median ± interquartile range. Intercohort statistical comparisons using a linear mixed model are shown. Frequencies of responders are written below the histograms.
(I) Contemporaneous correlations of RBD+ B cells and anti-RBD IgG. Values and colors represent Spearman r, asterisks indicate p values (∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001).
(J) Comparison between HDS and CI of the proportions of Omicron BA.1-RBD+ B cells among wild-type Wuhan-1-specific RBD+ B cells. Mann-Whitney tests are shown.
(K) Proportions of IgD-, IgM-, IgA-, and IgG-positive cells in RBD+ B cells at D2 and D3 in HDS and CI participants, with Wilcoxon tests.
(L) Comparison of IgM+ and IgG+ RBD+ B cells between HDS and CI participants at D2 and D3. Mann-Whitney tests are shown.
(M) Proportion of IgD+/− and CD27+/− populations in RBD+ memory B cells in HDS and CI participants at D2 and D3. In (B–E) and (G–I) n = 20 HDS, n = 26 CI; in (J) n = 9 HDS, n = 14 CI; in (K) and (M) n = 6 HDS, n = 13 CI; in (L) n = 10 HDS, n = 15 CI.
Table 1.
Clinical characteristics of study participants
| mRNA vaccine |
CIsa |
HDa |
|
|---|---|---|---|
| Long delayb (16 weeks) |
Short delay (HDS)b (5 weeks) |
Long delay (HDL)b (12 weeks) |
|
| Variable | (n = 26) | (n = 20) | (n = 7) |
| Vaccine regimen | |||
| Pfizer BNT162b2 vaccine (3 doses) | n = 25 | n = 19 | n = 7 |
| Heterologous vaccine strategy (Moderna mRNA- 1273 and Pfizer BNT162b2) | n = 1 | n = 1 | n = 0 |
| Age (years)c | 51 (41–56) | 61 (55–64) | 66 (55–77) |
| Gender | |||
| Male | 11 (42%) | 13 (65%) | 4 (57%) |
| Female | 15 (58%) | 7 (35%) | 3 (43%) |
| Vaccine dose spacing | |||
| Days between doses 1 and 2c | 111 (109–112) | 35 (33–35) | 83 (82–84) |
| Days between doses 1 and 3c | 329 (323–334) | 168 (166–168) | 230 (229–231) |
| Days between doses 2 and 3c | 219 (211–222) | 133 (133–133) | 147 (147–147) |
| Visits for immunological profiling | |||
| B, days before first dosec | 1 (0–5) | 12 (7–12) | 1 (01–2) |
| D1, days after first dose | 21 (19–26) | 28 (28–30) | 28 (28–29) |
| D1, days before second dose | 90 (85–92) | 5 (5–7) | 54 (54–56) |
| D1, days before third dosec | 306 (302–310) | 138 (138–139) | 203 (201–203) |
| D2, days after first dosec | 133 (130–139) | 63 (63–63) | 111 (110–112) |
| D2, days after second dose | 21 (20–27) | 28 (28–29) | 28 (28–28) |
| D2, days before third dosec | 196 (193–197) | 105 (103–105) | 119 (119–119) |
| M2, days after first dosec | 224 (222–228) | 119 (117–119) | 167 (167–168) |
| M2, days after second dosec | 112 (110–119) | 84 (84–84) | 84 (84–84) |
| M2, days before third dosec | 104 (101–112) | 49 (49–49) | 63 (63–63) |
| D3, days after first dosec | 362 (355–364) | 198 (198–198) | 265 (264–266) |
| D3, days after second dosec | 249 (245–252) | 163 (163–163) | 182 (182–182) |
| D3, days after third dose | 29 (25–34) | 30 (30–32) | 35 (35–35) |
Values displayed are medians, with interquartile range (IQR) in parentheses for continuous variables, or percentages for categorical variables.
CI, HDS and HDL HD participants cohorts were compared by: Mann-Whitney test for continuous variables, Fisher’s test for categorical variables.
Values statistically different between the CI, HDS, and/or HDL cohorts (p < 0.05).
Blood was sampled at baseline (B) 1–12 days before the first dose; 3–4 weeks after the first dose (D1); 3–4 weeks after the second dose (D2); 3–4 months after the second dose (memory; M2), and 4 weeks after the third dose (D3). Donors with breakthrough COVID-19 events were excluded afterward. There were no significant differences between HDS and HDL in terms of gender, age, or time on HD. HDS and HDL were respectively 10 and 15 years older than the CI. The time of sampling before the first dose (B), between D1 and D2, between D1 and D3, between D2 and D3, and between injection and sampling were significantly different.
Hemodialyzed participants are a heterogeneous population.38 Clinical details for our cohorts are provided in Table S1. The causes of end-stage renal disease (ESRD) were diverse (diabetes mellitus [DM], glomerulonephritis, hypertension, etc.), as were comorbidities other than DM. None were living with HIV. A few participants received immunosuppressive medications (prednisone, cyclosporine, tacrolimus). However, these patients did not behave as outliers in the different immunological analyses. These individuals are represented by different symbols—squares instead of circles—in the figure plots.
In HD patients, the initial two mRNA vaccine inoculations elicit poor B cell responses, which are reinvigorated by a third dose
We measured the levels of immunoglobulin (Ig) G targeting the receptor-binding domain (RBD). This domain is the major target for neutralization39,40 and is associated with vaccine efficacy.41 At baseline, anti-RBD IgG were undetectable in all participants, consistent with their SARS-CoV-2-naive status (Figures 1B and 1C). Both cohorts developed anti-RBD IgG responses after each vaccine dose (D1, D2, and D3) with a small decline at a memory time point (M2) (Figure 1B). However, the antibody levels at D1 were lower in HDS compared with CI, with a median-fold difference of 6. The antibody levels in HDS remained significantly lower through all follow-up time points (Figure 1C). Only 80% (16 out of 20) of HDS seroconverted after the first dose compared with 100% (26 out of 26) of CIs. However, all HDS experienced an increase in anti-RBD IgG responses after the third dose (Figure 1C). In the HDL regimen, the levels of anti-RBD IgG were not significantly different compared with HDS (Figure S1A). We also evaluated the capacity of plasma samples from donors to neutralize pseudoparticles bearing the SARS-CoV-2 spike (S) (D614G) glycoprotein, as previously described.14,42 Neutralizing activity was measured by the neutralization half-maximum inhibitory dilution (ID50). None of the plasma samples collected at baseline was able to neutralize SARS-CoV-2 in HDS and CIs (Figures 1D and 1E). The first dose had only a small impact on the neutralization capacity in most donors (Figures 1D and 1E) with only 25% (5 out of 20) of HDS compared with 60% (15 out of 25) of CI exhibiting a neutralizing activity against the D614G S at D1 (Figure 1E). The second dose increased the neutralization capacity in the HDS cohort but had significantly less activity than in the CI cohort. Strikingly, the third dose abrogated the differences between both groups (Figures 1D and 1E). In line with anti-RBD-IgG levels (Figure S1A), the longer interval between the first two doses in HDL did not lead to higher neutralizing activity against the S glycoprotein (Figure S1B).
We next measured RBD+ B cells using two fluorescently labeled recombinant RBD probes (Figures 1F and S1C).6,19 We observed differences in both magnitude and longitudinal trajectories of B cell responses between cohorts (Figures 1G and 1H). There was a trend for weaker priming of B cell responses (D1) in HDS than in CI, which did not reach statistical significance after correction for multiple comparisons. At D2 and M2, the frequencies of RBD+ B cells in HDS were lower and consistently trailed those in CIs, resulting in almost parallel curves (Figure 1G). In contrast, the responses to the third dose differed, with a more robust expansion of B cell responses in HDS compared with CIs. Consequently, we observed stronger B cell responses at D3 in HDS than in CIs (Figure 1H). Consistent with the antibodies (Figure S1A) and unlike CIs,19 a long interval in HDL did not improve the generation of the RBD+ B cell pool (Figure S1D). The delayed kinetics of anti-RBD IgG responses in HDS compared with CIs is illustrated by contemporaneous associations between B cell and antibody responses: while we observed in CIs a significant positive correlation between RBD+ B cells and anti-RBD IgG at D2, this correlation only appeared at D3 in HDS (Figure 1I).
The rapid worldwide spread of the Omicron variant has decreased vaccine efficacy against infection.43 However, protection against severe diseases remains good and is significantly increased by a third vaccine dose.10 To determine if HD treatment was associated with altered viral cross-recognition by B cells, we tested if HDS immunized with wild-type (WT) Wuhan-1 strain S could elicit cross-reactive B cell responses against Omicron BA.1 RBD (Figure S1E). Among all the WT RBD+ B cells at D3, 65% co-stained for Omicron BA.1 RBD probes, indicating cross-reactivity (Figure 1J). No significant difference was observed between HDS and CIs (Figure 1J).
We next assessed the differentiation of RBD+ B cells following vaccination. To avoid phenotyping bias, we only included donors in whom we detected ≥5 RBD+ B cells at every time point. As the rare RBD+ B cells in HDS at D1 precluded reliable phenotyping, we focused on D2 and D3. We measured the expression of IgM, IgD, IgA, and IgG on RBD+ B cells (Figure S1F). While IgG+ cells were dominant in both cohorts at all time points (Figures 1K, 1L, and S1G), its fraction was lower in HDS at D2, and those of IgM+ and IgD+ cells were higher (Figures 1L and S1H). The profiles converged between cohorts at D3. We next determined the memory differentiation profile of RBD+ B cells using IgD and CD27 co-expression (Figure S1I). CD27 is expressed on memory B cells44 and IgD is mostly found on unswitched B cells.45 In both cohorts, RBD+ B cells were mainly IgD−CD27− (Figure 1M). In HDS, CD27−IgD+ cells represented 15% of RBD+ B cells at D2 and decreased at D3 in favor of mature CD27+IgD− cells. Compared with HDS, we measured in CIs a low fraction of immature CD27−IgD+ RBD+ B cells at D2, in favor of more mature cells. This phenotype was stable at D3. Quantitatively, the magnitude of memory B cell responses increased between M2 and D3 in both groups (Figure S1J).
Our data show that, compared with a CI cohort, HDS elicit low RBD+ and mature B cell responses after two doses, consistent with lower antibody levels. A third immunization in HDS is critical to achieving B cell responses of higher magnitudes than those observed in CIs and leads to convergent differentiation profiles.
Vaccination induces strong CD4+ T cell responses but poor CD8+ T cell immunity in HD participants
SARS-CoV-2-specific CD4+ T cells help is crucial to the development of B cell responses and correlates with long-term humoral responses and CD8+ T cell immunity.6,7,46,47 We measured Spike (S)-specific T cell responses (Figure S2A) using activation-induced marker (AIM)5,6,48,49 and intracellular cytokine staining (ICS) assays.6
S-specific CD4+ and CD8+ T cell responses were assessed by an AND/OR Boolean combination gating strategy of the upregulation of CD69, CD40L, 4-1BB, and OX-40 upon a 15-h stimulation with an overlapping peptide pool spanning the S coding sequence (Figure S2B). This strategy detected S-specific AIM+ CD4+ (Figure S2C) and CD8+ T cell responses (Figure S2D) at all time points. Baseline responses were reported in other studies to result from previous cross-reactive expositions to common coronaviruses,50,51 and possibly to pre-exposition to abortive infection without seroconversion.52 To assess the functionality of the specific T cells, we measured the expression of IFNγ, interleukin (IL)-2, tumor necrosis factor alpha (TNFα), IL-17A, IL-10, and CD107a following a 6-h stimulation with the S peptide pool. We determined total cytokine+ CD4+ T cell responses by a similar OR Boolean combination gating strategy applied to the ICS results (Figure S2E). Specific cytokine+ CD4+ T cell responses were detected at all time points (Figure S2F). In most participants, no significant CD8+ T cell functions were detected and this could not be further assessed (Figure S2G).
AIM+ CD4+ T cell responses in HDS significantly increased after priming, plateaued at D2, waned slightly at M2, and further increased to peak at D3 (Figure 2A). In CIs, the increase at D3 was more muted (Figure 2A). We observed a trend for stronger AIM+ CD4+ T cell responses in HDS than in CIs at M2 and significantly higher magnitudes at D3 (Figure 2B). Cytokine+ CD4+ T cell responses in HDS followed trajectories comparable with the AIM responses (Figures 2C and 2D), with stronger effector CD4+ T cell responses in HDS compared with CI at D3 (Figure 2D). Unlike CD4+ T cells, AIM+ CD8+ T cell responses were lower in HDS than CI at all time points except M2 and D3, at which they converged (Figures 2E and 2F). A trend for increased CD8+ T cell responses in HDS at D3 compared with baseline was observed (Figures 2E and 2F). Of note, the CI cohort was characterized by sizable pre-existing CD8+ T cell responses at baseline, which likely affected the patterns observed (Figures 2E and 2F). Similar to the RBD+ B cell responses, a longer interval in HDL did not show clear benefits for mRNA-vaccine-elicited cellular responses, for either CD4+ (Figure S2H and S2I) or CD8+ (Figure S2J) T cell responses.
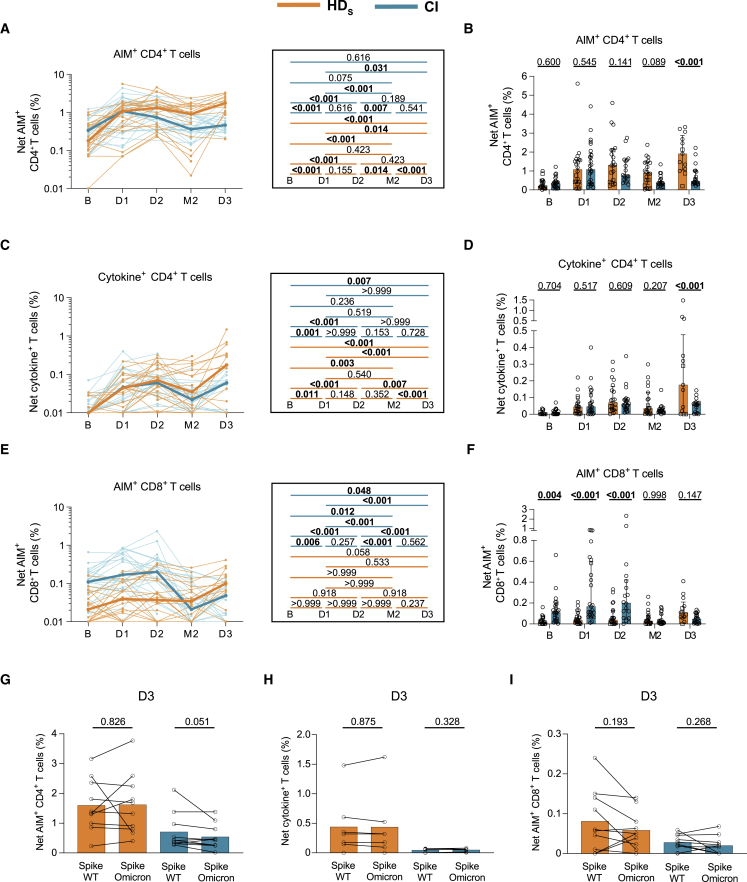
Figure 2.
Vaccination induces strong CD4+ T cell responses but poor CD8+ T cell immunity in HD participants
Frequencies of SARS-CoV-2 S-specific CD4+ and CD8+ T cells in HDS (orange) and CI (blue) who received three vaccine doses. HDS on immunosuppressive drugs are represented by square symbols, HDS not on immunosuppressants are represented by circles. PBMCs were stimulated ex vivo with a pool of overlapping S peptides.
(A and B) Net AIM+ CD4+ T cell responses. (A) Longitudinal analysis of S-specific AIM+ CD4+ T cell responses. Lines connect datapoints from the same donor. The bold lines represent the median values of each cohort. Right panel: statistical comparisons using a linear mixed model. (B) Comparisons between HDS and CI participants. Bars represent median ± interquartile range. Intercohort statistical comparisons using a linear mixed model are shown.
(C and D) Net cytokine+ CD4+ T cell responses measured by ICS. (C) Longitudinal analysis of the magnitude of cytokine+ CD4+ T cell responses. Lines connect datapoints from the same donor. The bold lines represent the median values of each cohort. Right panel: statistical comparisons using a linear mixed model. (D) Comparisons between HDS and CI participants. Bars represent median ± interquartile range. Intercohort statistical comparisons using a linear mixed model are shown.
(E and F) Net AIM+CD8+ T cell responses. (E) Longitudinal analysis of S-specific AIM+CD8+ T cell responses. Lines connect datapoints from the same donor. The bold lines represent the median values of each cohort. Right panel: statistical comparisons using a linear mixed model. (F) Comparisons between HDS and CI participants. Bars represent median ± interquartile range. Intercohort statistical comparisons using a linear mixed model are shown.
(G–I) Comparison of WT Wuhan-1-specific and Omicron BA.1-specific CD4+ and CD8+ T cell responses in HDS (orange) and CI (blue) participants. Mann-Whitney tests are shown. (G) Net S-specific AIM+ CD4+ T cell responses, (H) Net S-specific cytokine+ CD4+ T cell responses, and (I) Net S-specific AIM+ CD8+ T cell responses. In (A–F) and (J) n = 20 HDS, n = 26 CI participants; in (G) n = 10 HDS, n = 10 CI; in (H) n = 7 HDS, n = 7 CI; in (I) n = 9 HDS, n = 10 CI participants.
We next assessed the presence of Omicron-reactive CD4+ and CD8+ T cell responses using an overlapping Omicron BA.1 S peptide pool on D3 samples. We detected Omicron BA.1-reactive AIM+ CD4+ (Figure 2G), cytokine+ CD4+ (Figure 2H), and AIM+ CD8+ (Figure 2I) T cell responses in both cohorts. The magnitude of WT and Omicron BA.1 S-specific CD4+ T cell responses did not differ between groups, suggesting cross-reactivity.
These data demonstrate the emergence of robust SARS-CoV-2-specific CD4+ T cell responses after the initial priming dose in HDS, while a minimum of three doses was required to generate low CD8+ T cell responses.
mRNA vaccines elicit multifaceted AIM+ CD4+ T cell responses with qualitative features in HD distinct from CI participants
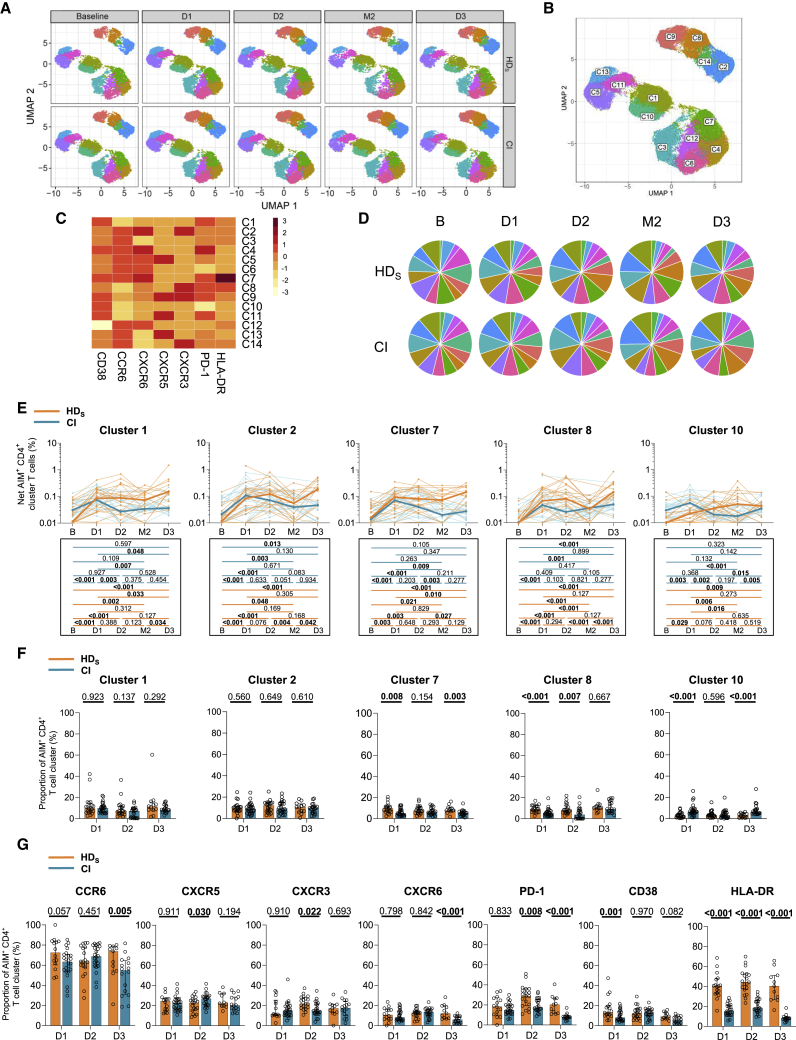
To qualitatively evaluate the S-specific AIM+ CD4+ T cell responses, we applied unsupervised analyses, as described.6 We studied chemokine receptors that are preferentially expressed by some lineages and involved in tissue homing (CXCR5 for TFH; CXCR3 for TH1; CCR6 for TH17/TH22 and mucosal homing; CXCR6 for pulmonary mucosal homing, activation markers [Human Leukocyte Antigen-DR isotype (HLA-DR) and CD38], and an inhibitory checkpoint [programmed cell death protein 1 [PD-1]).
The distribution of clustered populations was represented by the uniform manifold approximation and projection (UMAP) algorithm. Cluster identity was performed using Phenograph, resulting in the identification of 14 clusters (Figures 3A and 3B) based on distinct profiles of relative marker expression (Figures 3C and S3A). The 14 clusters were detectable at all time points (Figures 3A and 3D), most of them following frequency trajectories consistent with those observed for total AIM+ CD4 T cells in both cohorts (Figures 3E and S3B). Some qualitative differences in relative proportions persisted after the third dose, in HDS significant expansion of C4 and C7, two clusters enriched in CXCR6, CCR6, CD38, and PD-1 expression (Figures 3F and S3C). As shown in Figure 3C, C7 differed from C4 only by its enrichment in HLA-DR expression (Figure 3C). In univariate analyses, we observed at D3 in HDS significant expansion of PD-1+, HLA-DR+, CXCR6+ cells, and, to a lesser extent, CD38+ cells both in absolute frequencies and as relative fractions of AIM+ CD4+ T cells (Figures 3G and S3D).
Figure 3.
mRNA vaccines elicit multifaceted AIM+ CD4+ T cell responses with qualitative features in HD distinct from CI participants
(A) Multiparametric UMAP representation of S-specific AIM+ CD4+ T cells based on CD38, HLA-DR, CCR6, CXCR6, CXCR5, CXCR3, and PD-1 expression at each time point, aggregated data for the HDS and CI cohorts. The colors identify 14 populations clustered by unsupervised analysis using Phenograph.
(B) Each cluster is labeled on the global UMAP.
(C) Heatmap summarizing for each cluster the mean fluorescence intensity (MFI) of each loaded parameter.
(D) Pie charts depicting the representation of each identified cluster within total AIM+ CD4+ T cells.
(E) Longitudinal net frequencies of selected AIM+ CD4+ T cell clusters in HDS (orange) and CI (blue) participants for clusters 1, 2, 7, 8, and 10. Lines connect data from the same donor. The bold lines represent the median values of each cohort. Wilcoxon tests for each pairwise comparison are shown below.
(F) Proportions of AIM+ clusters 1, 2, 7, 8, and 10 among AIM+ CD4+ T cells in HDS and CI at D1, D2, and D3. Bars represent median ± interquartile range. Mann-Whitey tests are shown.
(G) Cohort comparison of univariate analyses. Bars represent median ± interquartile range. Statistical comparisons using a linear mixed model are shown. In (A)–(G) n = 20 HDS, n = 26 CI participants.
Therefore, mRNA vaccination elicits in both HDS and CI a multifaceted response already observed after the first dose. After the full course of three vaccinations, TH responses show some qualitative differences between cohorts, with higher expression in HDS of chemokine receptors associated with mucosal immunity (CCR6, CXCR6), immune activation (HLA-DR), and the inhibitory immune checkpoint PD-1.
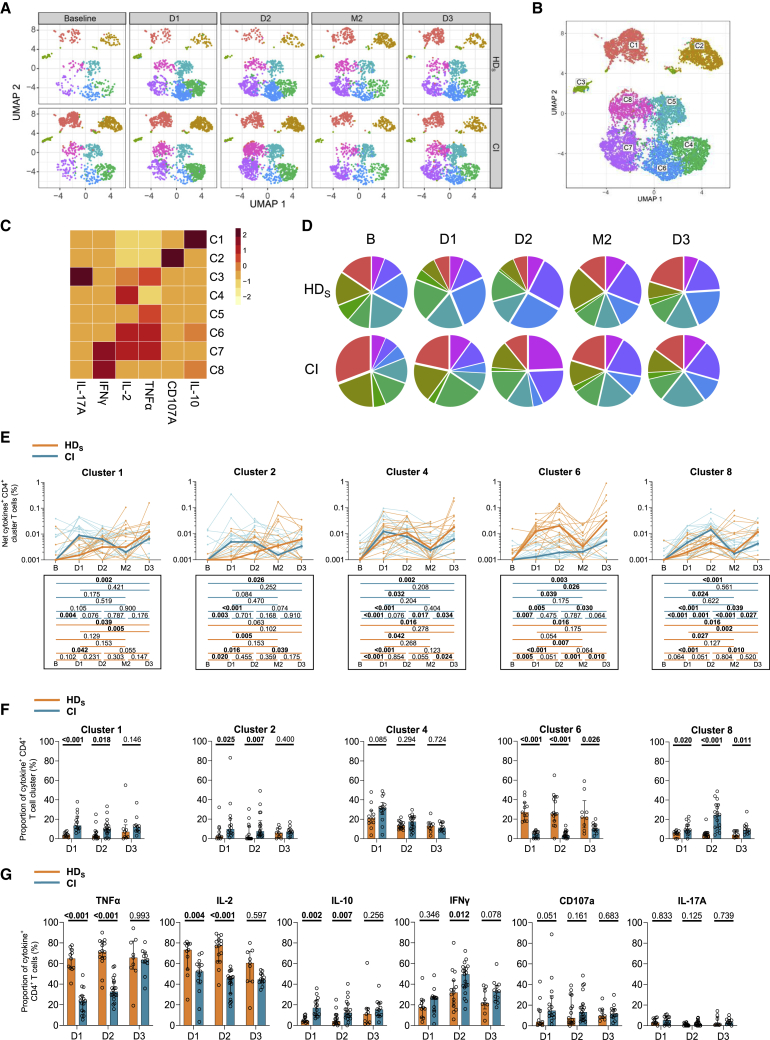
The first two vaccine inoculations elicit in HDS a TNFα/IL-2 skewed TH profile that is attenuated by the third dose
Given the qualitative differences observed with AIM assays, we used the same unsupervised approach to identify differences in CD4+ T cell effector functions. Expression of TNFα, CD107A, IL-10, IFNγ, IL-2, and IL-17A defined eight functional clusters, also detected at all time points (Figures 4A–4C and S4A) in both cohorts (Figure 4D). All clusters increased in magnitude after the first two doses, irrespective of clinical status (Figures 4E and S4B). Several qualitative differences were observed at D1 and D2 between cohorts, with the TNFα/IL-2-expressing C6 enriched in HDS and C1, C2, and C8 overrepresented in CIs (Figure 4F). The third dose led to a partial convergence of the functional profiles, although the differences in C6 and C8 proportions remained significant at D3. In univariate analyses, TNFα and IL-2 expressions were also higher, and IFNγ and IL-10 were lower in HDS compared with CIs during the initial vaccination series. Statistically significant differences were mostly abrogated by the third dose (Figures 4G and S4D).
Figure 4.
The first two vaccine inoculations elicit in HDS a TNFα/IL-2 skewed TH profile that is attenuated by the third dose
(A) Multiparametric UMAP representation of S-specific cytokine+ CD4+ T cells based on TNFα, CD107a, IL-10, IFNγ, IL-2, and IL-17A expression at each time point, aggregated data for the HDS and CI cohorts. The colors identify eight populations clustered by unsupervised analysis using Phenograph.
(B) Each cluster is labeled on the global UMAP.
(C) Heatmap summarizing for each cluster the MFI of each loaded parameter.
(D) Pie charts depicting the representation of each identified cluster within total cytokine+ CD4+ T cells.
(E) Longitudinal frequencies of selected cytokine+ CD4+ T cell clusters 1, 2, 4, 6, and 8 in HDS (orange) and CI (blue) participants. Lines connect data from the same donor. The bold lines represent the median values of each cohort. Wilcoxon tests for each pairwise comparisons are shown below the graphs.
(F) Proportions of cytokine+ clusters 1, 2, 4, 6, and 8 among cytokine+ CD4+ T cells in HDS and CI at D1, D2, and D3. Bars represent median ± interquartile range. Mann-Whitey tests are shown.
(G) Cohort comparison of univariate analyses. Bars represent median ± interquartile range. Statistical comparisons using a linear mixed model are shown. In (A)–(G) n = 20 HDS, n = 26 CI participants.
These analyses show that HDS are associated with a functional skewing upon mRNA vaccination. However, functional responses quantitatively and qualitatively converge between cohorts after the third dose.
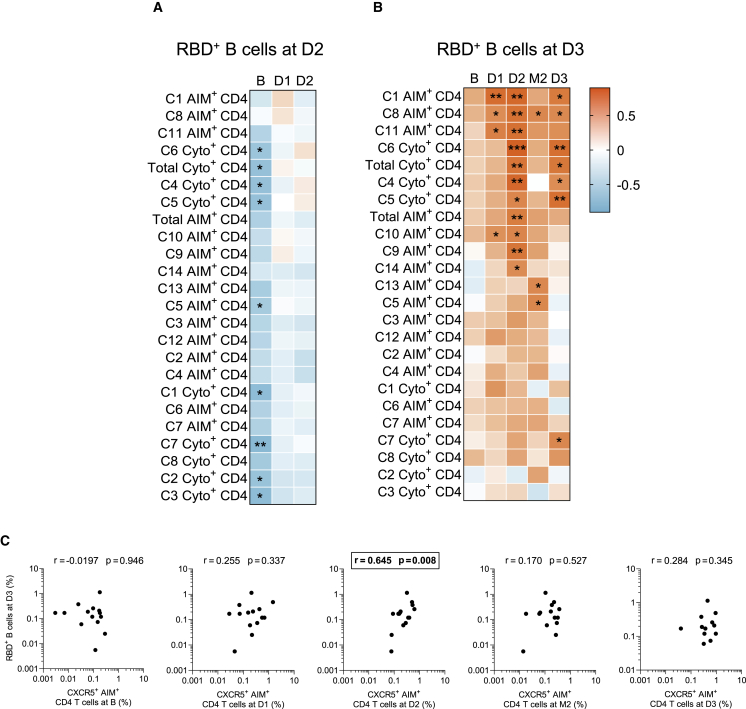
Associations between RBD+ B cell and S-specific CD4+ T cell responses appear late in people on HD
We next examined temporal associations between B and CD4+ T cell responses. The net magnitudes of baseline, D1, and D2 responses for each AIM+ and cytokine+ cluster were correlated with post-boost RBD+ B cell responses at D2 (Figures 5A and S5A) or D3 (Figures 5B and S5B). No positive correlation between the two cellular compartments was found at D2 in HDS (Figure 5A), in contrast to CIs (Figure S5A).6 Instead, several temporal associations were found in HDS between TH and RBD+ B cell responses at D3 (Figure 5B). Among the subsets with significant correlations, we found clusters enriched in CXCR3 and/or CXCR5 (AIM+ C8, C9, C11, and C14), and functional clusters enriched in IL-2 and TNFα (cytokine+ C4, C5, and C6) (Figure 5B).
Figure 5.
Associations between RBD+ B cell and S-specific CD4+ T cell responses appear late in people on HD
Temporal relationships between S-specific-CD4+ T cells and RBD+ B cells in HDS.
(A) Correlation between total CD4+ T cell frequencies at B–D2 and RBD+ B cell frequencies at D2 in HDS (n = 20).
(B) Correlation between total CD4+ T cell frequencies at B–D3 and RBD+ B cell frequencies at D3 in HDS (n = 20). Asterisks indicate statistically significant p values from a Spearman test (∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001). Colors indicate Spearman r.
(C) Correlations between frequencies of AIM+ CXCR5+ CD4+ T cells (for cTFH) at the B–D3 visits and RBD+ B cell frequencies at D3 in HDS. The r and p values from a Spearman test are indicated in each graph.
We previously reported in a low-risk population of vaccinees that S-specific CXCR5+ AIM+ CD4+ T cells (cTFH) after the first vaccine dose were predictive of RBD+ B cell responses after the second dose.6 We observed in HDS an association between the cTFH after the second dose and RBD+ B cell responses after the third dose (Figure 5C). In comparison, we only found a contemporary association between cTFH and the RBD+ B cell responses after the third dose in CIs (Figure S5C). Narrowing our observations to PD-1+ cTFH sub-populations, as they have been more strongly associated with B cell help,53 we only observed temporal and contemporary correlations with the RBD-B+ cells at D3 in HDS but not in CIs (Figure S5D).
These data demonstrate that there are temporal associations between CD4+ T cell help and B cell responses in HDS participants. However, these correlations mostly emerge only after the third dose, consistent with the delayed kinetics of vaccine response in HDS individuals.
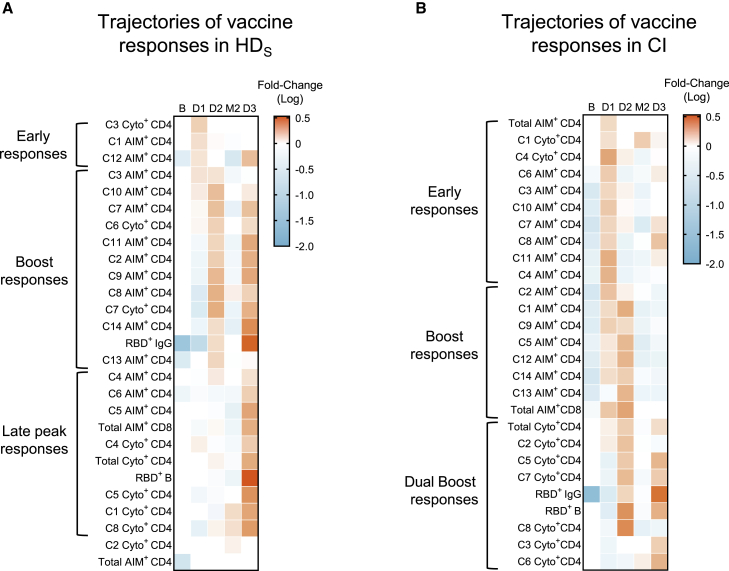
Trajectories of vaccine features highlight the need for multiple boosts in people on HD
Our data reveal multiple immune features whose trajectories differed between cohorts. To compare these trajectories, we used a normalization strategy allowing comparisons between features irrespective of their magnitude. First, we calculated the average response per participant at all time points, for each feature. The ratio of the measured parameter at the time point to its averaged value defined its trajectory. Each ratio was then plotted on a heatmap, and clustered according to the normalized trajectory (Figures 6A and 6B). Three patterns were observed among HDS (Figure 6A): a first group of features peaked early after the priming (AIM+ C1, C12, and cytokine+ C3). A second group showed strong responses after different boosts. This group notably included humoral RBD+ IgG responses, CXCR3+ (C2, C8, C9, and C14) AIM clusters, and the TNFα+ IL-2+-enriched C6 cluster. The third pattern corresponded to late response peaking at D3 and included RBD+ B cells, AIM+ CD8+ T cells, total AIM+ and cytokine+ CD4+ T cells, and several AIM+ and cytokine+ clusters.
Figure 6.
Trajectories of vaccine features highlight the need for multiple boosts in people on HD
(A and B) Trajectories of specific responses to mRNA vaccines in (A) HDS and (B) CI. Trajectories are represented by the fold change of the response at each time point for a given feature, to the mean response of every time point for the same feature. Significant fold change is colored in either orange (if increased) or blue (if decreased) (mixed-effect analysis with Bonferroni correction for multiple comparisons, colors represent adjusted p < 0.05). White color represents fold changes that are not significantly different from the mean response. n = 20 HDS and 26 CI.
Different trajectories were observed in CIs (Figure 6B). Unlike HDS, total CD4 and CD8 responses, along with most AIM+ clusters, were mobilized early at D1 and were boosted at D2. The first boost enhanced total cytokine+ CD4+ T, B cell, and IgG responses and most cytokine+ clusters. The second boost further recalled these responses. In contrast, these immune features were delayed in HDS and mobilized only at D3.
These results highlight the necessity for repeated boosting in HDS to achieve peak immune responses for all immune features. This contrasts with overall earlier peak immune responses in CIs, for whom the immune parameters are robustly generated after the first or the second dose.
Discussion
HD patients are at risk for severe infectious diseases, including COVID-19, and frequently respond poorly to standard vaccinations, including the initial two-dose series of SARS-CoV-2 mRNA vaccines. We show that the administration of a third vaccine dose is pivotal in stimulating B cell expansion and maturation to levels similar to controls. While previous studies reported reduced IFNγ-producing T cell responses, high-dimensional functional assays demonstrate that TH responses in HD are phenotypically and functionally skewed, not quantitatively inferior. Our results on cellular immunity are consistent with vaccination strategies previously proved effective in HD: administration of multiple and/or higher injections can counterbalance their low responses to immunization.27 We show that the optimal vaccine dosing interval is population dependent: in contrast to the general population,18,19,20 increasing the time between the first two doses resulted in weaker humoral and cellular immunity in HD.
The third dose led to partially converging antibody levels between cohorts, although they remained lower in HD than CIs at all time points. This is consistent with studies on HBV, hepatitis C virus (HCV), and influenza vaccines,21 and previous SARS-CoV-2 studies.32,54 Importantly, while the neutralization capacity was significantly lower after the second dose in HDS than in CI participants, the third dose significantly increased the neutralization responses in HDS, bringing them to levels similar to those measured in CIs. The low frequencies of RBD+ B cell responses observed in HD after the first two doses are likely major contributors to these disparities, but their quality may play a role as well. There was a delay in the maturation of B cell responses with the persistence of immature and unswitched IgM+ and IgD+ RBD+ B cells in HD after two mRNA vaccine doses. These features were reported in kidney transplant recipients and dialysis patients55,56 and attributed to chronic inflammation caused by uremia toxins, along with defects of innate and T cell immunity.22,23,25 We also observed incomplete B cell maturation in a cohort of CI vaccinated with the standard 3-week short-interval regimen of mRNA vaccine,19 and thus we cannot univocally delineate such defects in the 5-week-interval regimen applied to the HDS cohort. The longer delay between the two first doses might contribute to this difference in maturation. Some vaccinal studies with a standard 21-day schedule reported that antigen-specific germinal center reactions were maintained after 6 months with an increase in somatic hypermutations over time.57,58
Another key finding is that antigen-specific CD4+ T cell responses in HD were robust. As measured by multiplexed AIM and ICS, their magnitudes were comparable with or greater than those measured in controls, depending on the time point considered. These responses were as highly diverse in phenotype and function in HDS as in CIs, but with qualitative differences that persisted throughout the longitudinal follow-up. We identified a pro-inflammatory/activated skewing of TH responses in HD, with CCR6, CXCR6, and HLA-DR overexpression. Such CD4+ T cell populations have been described as preferentially recruited at sites of inflammation in autoimmune diseases, including inflammatory kidney disease.59,60 The simultaneous overexpression of the inhibitory checkpoint PD-1 by these cells may contribute to suboptimal help to other immune subsets. PD-1 upregulation on both CD4+ and CD8+ T cell populations in HD was previously reported, indicating that this dysregulation is not unique to SARS-CoV-2-specific responses.61 While cTFH cells are heterogeneous,62 with the PD-1+ fraction of cTFH endowed with better helper capacity to B cells in direct ex vivo co-culture experiments than the quiescent PD-1− fraction,53,63,64 the broad expression of PD-1 in HD complicates interpretation of this marker’s upregulation on CXCR5+ CD4+ T cells in HD donors. As we draw a comparison with a control cohort, we therefore mostly used a conservative analysis on total CXCR5+ CD4+ T cells. However, analyses focusing on CXCR5+ PD-1+ cTFH revealed some additional associations between T cell help and RBD+ B cells in the HD cohort.
CD4+ T cell responses in HD also presented functional skewing, with an overrepresentation of TNFα+ and IL-2+, at the expense of IL-10+ and IFNγ+ cells. These patterns raise questions about the impact of these cytokines in the establishment of vaccine responses. It has been shown that high levels of TNFα in COVID-19 could induce downstream activation of TH1 cells and block the final step of cTFH differentiation.65 This skewing may contribute to the delay observed in B cell responses to SARS-CoV-2 vaccination due to insufficient feedback inhibition of pro-inflammatory cytokines.66 The third dose was characterized in HD by the normalization of the effector function profile compared with the CI. Therefore, differences in the assays used (e.g., high-dimensional flow cytometry versus IFNγ Elispot) likely explain discrepancies between our data, in which we found robust CD4+ T cell responses in HD, and studies showing weaker T cell responses in this population.36,37
CD8+ T cell responses tend to be generated in HD only after the third dose of vaccine, in light of previous results showing that people with ESRD have more exhausted and anergic CD8+ T cells than CIs.61 In both cohorts, CD8+ T cell responses remain low compared with their CD4+ T cell counterparts, consistent with the TH-biased profile of responses elicited by SARS-CoV-2 mRNA vaccines.67
Our results show that a long (12-week) interval between the first two doses is not beneficial for people receiving HD: both B cell and antibody responses in HDL after the second dose tend to be weaker than those observed in the HDS cohort. The optimal dosing interval in people receiving HD remains uncertain; another study suggests that a slightly longer interval (up to 45 days, compared with 35 days in our study) was associated with stronger humoral responses.68
VOCs are an evolving challenge. After the third dose, HD developed B and CD4+ T cell responses specific to SARS-CoV-2 cross-reactive to Omicron BA.1 and at levels similar to CI. These findings complement previous reports showing that a third vaccine dose in HD enhanced neutralizing capacity of antibody responses against VOCs.9 Therefore, they might have protection against VOCs such as Omicron similar to CIs.8,9,10
The global immune profiles observed longitudinally are consistent with a model in which HD respond more slowly to vaccination, with a third dose required to achieve B and T cell responses quantitatively and qualitatively close to those generated after two doses in CIs. Temporal associations between SARS-CoV-2-specific CD4+ T cell and RBD+ B cell responses are shifted by one dose in HD, with a delayed link between the two features compared with CIs. As cellular immune responses are comparatively less affected by the third dose in CIs than HD individuals, responses from both cohorts globally converged after the full vaccination course. Some studies have highlighted such convergence for anti-RBD IgG responses between HD and CIs after three mRNA vaccine doses.69,70 We believe that the finding that low cellular immunity responsiveness in HD can be overcome by repeat dosing is a major positive conclusion of our study and provides an immunological basis for previous findings on the antibody responses elicited by a third dose in this vulnerable population.
Determining whether the qualitative skewing of CD4+ T cell responses observed in HD can alter protection against breakthrough infection, how long cellular responses persist after the third dose, and how additional booster doses can further modulate the immune profiles identified will warrant further study.
Limitations of the study
Our study identified several alterations of adaptive immunity elicited by SARS-CoV-2 vaccines in HD patients, with a spectrum of responsiveness in this population. Further studies are needed to better understand what individual factors may contribute to this heterogeneity. Mechanistic studies of related immune defects are very challenging, as no animal model for long-term chronic HD exists.
HD individuals are known to have frequent comorbidities and in our study were older than CIs. These factors might affect immune responses independently of HD. However, while the cohorts studied are too small to conduct multivariate analyses, chronic diseases are highly prevalent in HD patients, therefore distinguishing individual factors would have limited practical impact.
This study focuses on SARS-CoV-2 naive individuals. Additional studies are required to evaluate how prior infection shapes hybrid immunity in HD upon vaccination.
The size of the long-interval hemodialyzed cohort is small. It was not possible to recruit more suitable participants as the standard of care shifted to a short-interval regimen soon after the vaccination campaign began. The time points between D1 and D3, and D2 and D3, were not matched between cohorts. Therefore, this difference in timing might lead to potential quantitative differences in anti-SARS-CoV-2 antibodies due to the higher rate of RBD-B cell maturation in the short-delay cohort.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| UCHT1 (BUV395) [Human anti-CD3] | BD Biosciences | Cat#563546; Lot:9,058,566; RRID:AB_2744387 |
| 1B5 (BUV 395) [Human anti-CCR10] | BD Biosciences | Cat# 565322; Lot:1,198,884; RRID:AB_2739181 |
| IA6-2 (BUV 563) [Human anti-IgD] | BD Biosciences | Cat# 741394; Lot:2,048,494; RRID:AB_2870889 |
| MI15 (BUV 661) [Human anti-CD138] | BD Biosciences | Cat# 749873; Lot:1,140,733; RRID:AB_2874113 |
| UCH-B1 (BUV 737) [Human anti-IgM] | BD Biosciences | Cat# 748928; Lot:1,154,015; RRID:AB_2873331 |
| ML5 (BUV 805) [Human anti-CD24] | BD Biosciences | Cat# 742010; Lot:1,154,017; RRID:AB_2871308 |
| G18-145 (BV421) [Human anti-IgG] | BD Biosciences | Cat# 562581; Lot:1,033,053; RRID:AB_2737665 |
| SJ25C1 (BV650) [Human anti-CD19] | Biolegend | Cat# 363026; Lot:B328293; RRID:AB_2564255 |
| 2H7 (BV711) [Human anti-CD20] | BD Biosciences | Cat# 563126; Lot:2,032,072; RRID:AB_2313579 |
| B-LY4 (BV786) [Human anti-CD21] | BD Biosciences | Cat# 740969; Lot:1,167,364; RRID:AB_2740594 |
| G46-6 (BB700) [Human anti-HLADR] | BD Biosciences | Cat# 566480; Lot:1,053,189; RRID:AB_2744477 |
| HIT2 (BB790) [Human anti-CD38] | BD Biosciences | Cat#624296; Lot:9,119,974; CUSTOM |
| IS11-8E10 (PE) [Human anti-IgA] | Miltenyi | Cat# 130-113-476; Lot:5,210,405,486; RRID:AB_2733861 |
| M-T271 (APC-R700) [Human anti-CD27] | BD Biosciences | Cat# 565116; Lot:0,262,146; RRID:AB_2739074 |
| UCHT1 (BUV496) [Human anti-CD3] | BD Biosciences | Cat#612941; Lot:1,022,424; RRID:AB_2870222 |
| L200 (BV711) [Human anti-CD4] | BD Biosciences | Cat#563913; Lot:03,000,025; RRID:AB_2738484 |
| SK3 (BB630) [Human anti-CD4] | BD Biosciences | Cat#624294; Lot:0,289,566; CUSTOM |
| RPA-T8 (BV570) [Human anti-CD8] | Biolegend | Cat#301037; Lot:B281322; RRID:AB_10933259 |
| M5E2 (BUV805) [Human anti-CD14] | BD Biosciences | Cat#612902; Lot:0,262,150; RRID:AB_2870189 |
| M5E2 (BV480) [Human anti-CD14] | BD Biosciences | Cat#746304; Lot:9,133,961; RRID:AB_2743629 |
| 3G8 (BV650) [Human anti-CD16] | Biolegend | Cat#302042; Lot:B323847; RRID:AB_2563801 |
| HIB19 (APC-eFluor780) [Human anti-CD19] | Thermo Fisher Scientific | Cat#47-0199; Lot:2,145,095; RRID:AB_1582231 |
| HIB19 (BV480) [Human anti-CD19] | BD Biosciences | Cat#746457; Lot:1,021,649; RRID:AB_2743759 |
| HI100 (PerCP Cy5.5) [Human anti-CD45RA] | BD Biosciences | Cat#563429; Lot:8,332,746; RRID:AB_2738199 |
| NCAM16.2 (BUV737) [Human anti-CD56] | BD Biosciences | Cat#564448; Lot:8,288,818; RRID:AB_2744432 |
| FN50 (PerCP-eFluor710) [Human anti-CD69] | Thermo Fisher Scientific | Cat#46-0699-42; Lot:1,920,361; RRID:AB_2573694 |
| FN50 (BV650) [Human anti-CD69] | Biolegend | Cat# 310934; Lot:B303462; RRID:AB_2563158 |
| H4A3 (BV786) [Human anti-CD107A] | BD Biosciences | Cat#563869; Lot:8,144,866; RRID:AB_2738458 |
| ACT35 (APC) [Human anti-CD134 (OX40)] | BD Biosciences | Cat#563473; Lot:1,015,537; RRID:AB_2738230 |
| 4B4-1 (PE-Dazzle 594) [Human anti-CD137 (4-1BB)] | Biolegend | Cat# 309826; Lot:B253152; RRID:AB_2566260 |
| TRAP1 (BV421) [Human anti-CD154 (CD40L)] | BD Biosciences | Cat#563886; Lot:9,037,850; RRID:AB_2738466 |
| TRAP1 (PE) [Human anti-CD154 (CD40L)] | BD Biosciences | Cat#555700; Lot:7,086,896; RRID:AB_396050 |
| J25D4 (BV421) [Human anti-CD185 (CXCR5)] | Biolegend | Cat# 356920; Lot:B325837; RRID:AB_2562303 |
| B27 (PECy7) [Human anti-IFN-γ] | BD Biosciences | Cat#557643; Lot:8,256,597; RRID:AB_396760 |
| MQ1-17H12 (PE-Dazzle594) [Human anti-IL-2] | Biolegend | Cat#500344; Lot:B2261476; RRID:AB_2564091 |
| JES3-9D7 (PE) [Human anti-IL-10] | BD Biosciences | Cat#554498; Lot:8,198,773; RRID:AB_395434 |
| eBio64CAP17 (eFluor660) [Human anti-IL-17A] | Thermo Fisher Scientific | Cat#50-7179-42; Lot:2,151,998; RRID:AB_11149126 |
| Mab11 (Alexa Fluor 488) [Human anti-TNF-α] | Biolegend | Cat#502915; Lot:B285221; RRID:AB_493121 |
| LIVE/DEAD Fixable dead cell | Thermo Fisher Scientific | L34960 |
| Cell lines | ||
| HEK293T cells | ATCC | Cat#CRL-3216 RRID:CVCL_0063 |
| 293T-ACE2 cells | Ref. 42 | N/A |
| Chemicals, peptides, recombinant proteins, and DNA | ||
| RBD1 probe (Alexa Fluor 488) | In house | N/A |
| RBD2 probe (Alexa Fluor 594) | In house | N/A |
| RBD Omicron (Alexa Fluor 647) | In house | N/A |
| PepMix™ SARS-CoV-2 (Spike Glycoprotein) | JPT | Cat#PM-WCPV-S-1 |
| Staphylococcal Enterotoxin B (SEB) | Toxin technology | Cat#BT202 |
| PepMix™ SARS-CoV-2 (Spike B.1.1.529/Omicron Glycoprotein) | JPT | Cat#PM-SARS2-SMUT08-1 |
| SARS-CoV-2 Spike RBD, His Tag (B.1.1.529/Omicron) | Acrobiosystems | Cat#SPD-C522e-100ug |
| pNL4.3 R-E− Luc | NIH AIDS Reagent program | Cat#3418 |
| pCG1-SARS-CoV-2 Spike D614G | Ref. 71 | N/A |
| Software and algorithms | ||
| Flow Jo v10.8.0 | Flow Jo | https://www.flowjo.com |
| GraphPad Prism v8.4.1 | GraphPad | https://www.graphpad.com |
| R studio v4.1.0 | R studio | https://rstudio.com |
| R codes scripted | Github | https://github.com/otastet/Nayrac_et_al |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Daniel E. Kaufmann (daniel.kaufmann@chuv.ch or daniel.kaufmann@umontreal.ca).
Materials availability
All unique reagents generated during this study are available from the lead contact upon a material transfer agreement (MTA).
Data and code availability
-
•
The published article includes all datasets generated and analyzed for this study. Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact Author (daniel.kaufmann@chuv.ch).
-
•
We adapted previously submitted6 R codes scripted to perform unsupervised analyzes on B and T cells from hemodialyzed donors and controls individuals. All original codes have been deposited on Github and are publicly available as of the date of publication. URL link is listed in the key resources table.
Experimental model and subject details
Institutional permissions and oversight
All work was conducted in accordance with the Declaration of Helsinki and approved by the institutional boards of the participating institutions (Quebec Renal Network31 multicentric protocol MP-02-2021-9006; and CHUM protocols 19.381 and 20.065).
Informed consent
All work was conducted in accordance with the Declaration of Helsinki and written informed consent obtained before enrollment into the study.
Subject characteristics
Subject characteristics are summarized in Table 1. Patients with end-stage renal disease receiving hemodialysis (HD) were enrolled into the Quebec Renal Network (QRN) COVID-19 Study as previously described31 and followed every 2-3 days at the Center Universitaire de Santé McGill (CUSM), the Center Hospitalier de l’Université de Montréal (CHUM), and the Hôpital du Sacré-Coeur de Montréal (HSCM). Participants from this cohort were followed and sampled before and after vaccination. Blood draws were performed at baseline (B) 12 days before the first dose of vaccine with mRNA vaccine, 4 weeks after the first dose (D1), 4 weeks post second dose (D2), 12 weeks after the second dose (M2) and 4-5 weeks post third dose (D3). Hemodialyzed participants were divided into two cohorts: a short interval cohort for which the first two vaccine doses were administered 5 weeks apart (HDS, n = 20); and a long interval cohort (HDL, n = 7) for which the first two doses were given 12 weeks apart when vaccine scarcity was limiting.
The cohort of control individuals (CI, n = 26) consisted of healthcare workers who did not have a major medical precondition qualifying for a short interval schedule (e.g, immunosuppression) and who received the first two vaccine doses 16 weeks apart per Quebec public health guidelines early in the vaccination campaign in Canada. The third inoculation was given 7 months after the second dose. Blood draws were performed at baseline (B) 1 day before the first dose of mRNA vaccine, 3 weeks after the first dose (D1), 3 weeks following the second dose (D2), 16 weeks after the second dose (M2), and 4 weeks after the third dose (D3).
Median age and interquartile range for the HDS cohort was 61 [55-64], and 13 individuals were males (65%). Median age and interquartile range for the CI cohort were younger (median = 51 [41-56], p < 0.001), and 11 individuals were males (42%). The HDL cohort was not significantly older (median = 66 [55-77]) and 4 individuals were males (57%). Time on hemodialysis between each cohort was comparable (See Table 1).
Method details
PBMCs and plasma isolation
Peripheral blood mononuclear cells (PBMCs) were isolated from blood samples by Ficoll density gradient centrifugation and cryopreserved in liquid nitrogen until use. Plasma was stored at −80°C. For antibody assays, plasma was heat-inactivated for 1 h at 56°C prior to experiments. Plasma from uninfected donors collected before the pandemic were used as negative controls and used to calculate the seropositivity threshold in our ELISA assay.
Enzyme-linked immunosorbent assay
The SARS-CoV-2 RBD ELISA assay used was previously described.42 Briefly, recombinant SARS-CoV-2 RBD proteins or BSA (2.5 μg/mL) as negative control were prepared in PBS and adsorbed to plates overnight at 4°C. Coated wells were subsequently blocked with blocking buffer and then washed. CR3022 mAb (50 ng/ml) or a ½50 dilution of plasma from HD, or CI donors were prepared in a diluted solution of blocking buffer and incubated with the RBD-coated wells. Plates were washed followed by incubation with the respective secondary Abs. The binding of CR3022 IgG was quantified with HRP-conjugated antibodies specific for the Fc region of human IgG and used to normalize the RLU from each plate. HRP enzyme activity was determined after the addition of a 1:1 mix of Western Lightning oxidizing and luminol reagents (Perkin Elmer Life Sciences). Light emission was measured with an LB942 Tri-Star luminometer (Berthold Technologies). Signal obtained with BSA was subtracted for each plasma and was then normalized to the signal obtained with CR3022 present in each plate. The seropositivity threshold was established using the following formula: mean of pre-pandemic SARS-CoV-2 negative plasma + (3 SD of the mean of pre-pandemic SARS-CoV-2 negative plasma).
Virus neutralization assay
The SARS-CoV-2 virus neutralization assay was used previously.42 Briefly, 293T cells were transfected with the lentiviral vector pNL4.3 R-E− Luc plasmid (NIH AIDS Reagent Program) and a plasmid encoding for the full-length SARS-CoV-2 Spike D614G glycoprotein42,71 at a ratio of 10:1. Two days post-transfection, cell supernatants were harvested and stored at −80°C until use. Pseudoviral particles were incubated with the indicated plasma dilutions (1/50; ½50; 1/1250; 1/6250; ⅓1/31,250) for 1h at 37°C and were then added to the 293T-ACE2 target cells followed by incubation for 48 h at 37°C. Then, cells were lysed and followed by one freeze-thaw cycle. An LB942 Tri-Star luminometer (Berthold Technologies) was used to measure the luciferase activity. The neutralization half-maximal inhibitory dilution (ID50) represents the plasma dilution to inhibit 50% of the infection of 293T-ACE2 cells by SARS-CoV-2 pseudoviruses.
RBD-specific B cell staining
PBMCs were resuspended at 4x106 cells/mL in RPMI (Gibco by Life Technologies) supplemented with penicillin/streptomycin (Gibco by Life Technologies), 10% heat inactivated FCS and incubated at 37°C, 5% CO2 for 2hrs in the presence of fluorescently labeled CCR10 antibody.
For surface stain, PBMCs were first stained for viability dye (Aquavivid, Thermofisher, 20min, 4°C) next with a mix containing a brilliant stain buffer (BD Biosciences), the surface markers for B cells detection (CD19, CD20, CD21, IgM and IgD), B cells memory phenotype (CD24, CD27, IgG and IgA), plasmablasts and plasma cells (CD38 and CD138) phenotypes, T-cells and monocytes exclusion (CD3, CD56, CD14, and CD16) (30min, 4°C) (see Table S2 for antibodies), as well as fluorescently-labeled probes for RBD+ B cells detection targeting two different epitopes of the RBD (RBD1-AF488 and RBD2 AF594). Omicron BA.1-RBD peptide (Accrobiosystem) was labeled, and the Omicron BA.1-RBD probe was also added into the mix where appropriate (RBD Omicron BA.1 AF647). Cells were fixed with 1% paraformaldehyde (Sigma-Aldritch) for 15 min at room temperature before filtration for acquisition on a FACSymphony A5 Cell Analyzer (BD Biosciences) and analyzed using FlowJo (BD, v10.6.2).
Activation-induced markers (AIM) assay
PBMCs were plated in a 96-wells flat bottom plate, at 10x106 cells/mL RPMI (Gibco by Life Technologies) supplemented with penicillin/streptomycin (Gibco by Life Technologies), 10% heat inactivated FCS and incubated at 37°C, 5% CO2. After a rest of 3hrs, a CD40 blocking antibody (Miltenyi) was added to the culture to prevent the interaction of CD40L with CD40 and its subsequent downregulation. In addition, antibodies for chemokine receptors CXCR6, CXCR3, CXCR5, and CCR6 were added in culture. After 15min incubation at 37°C, 5% CO2, cells were stimulated with 0.5 μg/mL staphylococcal enterotoxin B (SEB) or 0.5 μg/mL of overlapping peptide pools for Wuhan-1 or Omicron BA.1 variants SARS-CoV-2 Spike (JPT) for 15 hrs at 37°C, 5% CO2. An unstimulated condition with 0.4μL of DMSO served as a negative control.
Cells were stained for viability dye (Aquavivid, Thermofisher, 20min, 4°C), surface markers (30min, 4°C) (see Table S3 for antibodies) and fixed using 2% paraformaldehyde (Sigma-Aldritch, 15min, RT) before filtration for acquisition on the flow cytometer (FACSymphony A5 Cell Analyzer; BD Biosciences) and analyzed using FlowJo (BD, v10.6.2). For phenotypic analysis of antigen-specific CD4+ T cells, only responses that were >2-fold over unstimulated condition were included to limit the impact of background staining. In contrast, for analysis of antigen-specific CD4+ T cell subsets as percentage of total CD4+ T cells, background-subtracted net values were used, which did not require excluding responses.
Intracellular cytokines staining (ICS) assay
PBMCs were resuspended at 10x106 cells/mL RPMI (Gibco by Life Technologies) supplemented with penicillin/streptomycin (Gibco by Life Technologies), 10% heat inactivated FCS, and incubated at 37°C, 5% CO2. After a rest of 2hrs, cells were stimulated with 0.5 μg/mL staphylococcal enterotoxin B (SEB) or 0.5 μg/mL of overlapping peptide pools for Wuhan-1 or Omicron BA.1 variants SARS-CoV-2 Spike (JPT) for 6 hrs at 37°C, 5% CO2. An unstimulated condition with 0.4μL of DMSO served as a negative control. Brefeldin A (BD Biosciences), Monensin-1 (BD Biosciences), and a fluorescently labeled CD107a antibody were added for the remaining 5hrs.
Cells were stained for viability dye (Aquavivid, Thermofisher, 20min, 4°C), surface markers (30min, 4°C), and intracellularly for cytokines (30min, room temperature) using the IC Fixation/Permeabilization kit (eBioscience) (see Table S4 for antibodies) and filtrated before acquisition on the flow cytometer (FACSymphony A5 Cell Analyzer, BD Biosciences) and analyzed using FlowJo (BD, v10.6.2).
Quantification and statistical analysis
Statistics
Symbols represent biologically independent samples from HD and CI donors. Lines connect data from the same donor. Thick lines represent median values. Linear mixed models fitting cell frequencies in terms of cohort, time point, and their interaction were run using R and the package ‘‘nlme’’. Model diagnostics were performed, checking for heteroscedasticity and normality among residuals. All retained models used a square-root transform on the response variable, which helped in reducing the impact of outliers. Post-hoc contrasts across all pairwise comparisons of factor levels were obtained with the package ‘‘emmeans’’, correcting the p values by the method of Holm-Bonferroni where applicable. An important caveat of the square-root transform is that the reported contrast estimates and their confidence intervals remain on this scale, making their interpretation tricky. This was not deemed too great an obstacle, as qualitative statements on significant contrasts could be made based on p values. Thirty-five linear mixed models were retained, those being anti-RBD IgG, RBD B, AIM CD4, ICS CD4, AIM CD8, CXCR3, CXCR5, CXCR6, CCR6, PD-1, CD38, HLA-DR, IFNγ, IL-2, TNFα, IL-10, CD107a and IL-17A being compared between HDS, CI and HDL cohorts. There were also comparisons of HDS, HDL, and CI for anti-RBD IgG, RBD B, AIM CD4, ICS CD4, and AIM CD8. Models without satisfactory diagnostics were abandoned in favor of non-parametric methods. Differences in responses for the same patient before and after vaccination were performed using Wilcoxon matched pair tests. Differences in responses between HDS and CI were measured by Mann-Whitney tests. Wilcoxon and Mann-Whitney tests were generated using GraphPad Prism (version 9.2.0). p values < 0.05 were considered significant. p values are indicated for each comparison assessed. For descriptive correlations, Spearman’s R correlation coefficient was applied. For graphical representation on a log scale (but not for statistical tests), null values were arbitrarily set at the minimal values for each assay.
Software scripts and visualization
Graphics and pie charts were generated using GraphPad PRISM (v9.2.0) and ggplot2 (v3.3.3) in R (v4.1.0). Heat maps were generated in R (v4.1.0) using the pheatmap package (v1.0.12). Principal component analyses were performed with the prcomp function (R). Uniform manifold approximation and projection (UMAP) was performed using package M3C (v1.14.0) on gated FCS files loaded through the flowCore package (v2.4.0). Samples were downsampled to a comparable number of events (300 cells for AIM, 100 cells for ICS). Scaling and logical transformation of the flow cytometry data were applied using the FlowSOM72 R package (v2.0.0). All samples at all time points were loaded. Clustering was achieved using Phenograph (v0.99.1) with the hyperparameter k (number of nearest neighbors) set to 150). We previously provided all R codes scripted for this paper in another study.6 We obtained an initial 14 AIM+ and 8 ICS+ clusters. For B and CD4+ T cell phenotyping, only participants with ≥5 events across all depicted time points were analyzed.
Acknowledgments
The authors are grateful to the study participants. We thank the CRCHUM BSL3 and Flow Cytometry platforms for technical assistance and Dr. Johanne Poudrier for advice and discussions. This work was supported by an FRQS Merit Research Scholar award #268471 (D.E.K.), the Réseau rénal Québécois, the Fondation du CHUM, le Ministère de l’Économie et de l’Innovation du Québec, Programme de soutien aux organismes de recherche et d’innovation (A.F.), a Canadian Institutes of Health Research (CIHR) operating grant #178344 (D.E.K. and A.F.), a CIHR Rapid Research COVID-19 funding opportunity grant #447760 (R.S.S.), a foundation grant #352417 (A.F.), a CIHR operating Pandemic and Health Emergencies Research grant #177958 (A.F.), and an Exceptional Fund COVID-19 from the Canada Foundation for Innovation (CFI) #41027 to A.F. and D.E.K. The Symphony flow cytometer was funded by a John R. Evans Leaders Fund from the Canada Foundation for Innovation (#37521 to D.E.K.) and the Fondation Sclérodermie Québec. A.F. is the recipient of Canada Research Chair on Retroviral Entry no. RCHS0235 950-232424. A.P. holds a Canada Research Chair in Multiple Sclerosis and the Power Corporation of Canada Chair of Université de Montréal. C.T. holds the Pfizer/Université de Montréal Chair on HIV translational research. V.M.L. is supported by an FRQS Junior 1 salary award. G.S. is supported by an FRQS doctoral fellowship and by a scholarship from the Department of Microbiology, Infectious Disease, and Immunology of the University of Montreal. M.B. was supported by a CIHR fellowship. A.T. was supported by a Mitacs Accélération postdoctoral fellowship. The funders had no role in the study design, data collection, analysis, decision to publish, or preparation of the manuscript.
Author contributions
Conceptualization, G.S., A.N., M.D., A.F., R.S.S., and D.E.K.; methodology, G.S., A.N., M.D., M.N., J.N., A.F., and D.E.K.; software, O.T. and G.S.; formal analysis, J.B. and R.L.B.; investigation, G.S., A.N., L.M., M.N., M.L., D.C., A.T., M.B., S.Y.G., C.B., G.G.L., and H.M.; resources, M.L., R.C., A.M.S.F., C.B., G.G.L., H.M., N.B., G.G.D., L.G., C.M., P.A., C.T., V.M.L., G.G., and SD; writing – original draft, G.S., A.N., M.D., and D.E.K.; writing – review & editing, G.S., A.N., M.D., and D.E.K.; supervision, D.E.K., A.F., and R.S.S.; funding acquisition, D.E.K., A.F., and R.S.S.
Declaration of interests
C.T. serves as a consultant for Merck, Gilead, GSK, AstraZeneca, and Medicago.
Published: March 21, 2023
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.xcrm.2023.100955.
Contributor Information
Andrés Finzi, Email: andres.finzi@umontreal.ca.
Rita S. Suri, Email: rita.suri@mcgill.ca.
Daniel E. Kaufmann, Email: daniel.kaufmann@chuv.ch.
Supplemental information
References
- 1.Haas E.J., Angulo F.J., McLaughlin J.M., Anis E., Singer S.R., Khan F., Brooks N., Smaja M., Mircus G., Pan K., et al. Impact and effectiveness of mRNA BNT162b2 vaccine against SARS-CoV-2 infections and COVID-19 cases, hospitalisations, and deaths following a nationwide vaccination campaign in Israel: an observational study using national surveillance data. Lancet. 2021;397:1819–1829. doi: 10.1016/S0140-6736(21)00947-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tenforde M.W., Self W.H., Adams K., Gaglani M., Ginde A.A., McNeal T., Ghamande S., Douin D.J., Talbot H.K., Casey J.D., et al. Association between mRNA vaccination and COVID-19 hospitalization and disease severity. JAMA. 2021;326:2043–2054. doi: 10.1001/jama.2021.19499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Earle K.A., Ambrosino D.M., Fiore-Gartland A., Goldblatt D., Gilbert P.B., Siber G.R., Dull P., Plotkin S.A. Evidence for antibody as a protective correlate for COVID-19 vaccines. Vaccine. 2021;39:4423–4428. doi: 10.1016/j.vaccine.2021.05.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gilbert P.B., Montefiori D.C., McDermott A.B., Fong Y., Benkeser D., Deng W., Zhou H., Houchens C.R., Martins K., Jayashankar L., et al. Immune correlates analysis of the mRNA-1273 COVID-19 vaccine efficacy clinical trial. Science. 2022;375:43–50. doi: 10.1126/science.abm3425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tauzin A., Gong S.Y., Beaudoin-Bussières G., Vézina D., Gasser R., Nault L., Marchitto L., Benlarbi M., Chatterjee D., Nayrac M., et al. Strong humoral immune responses against SARS-CoV-2 Spike after BNT162b2 mRNA vaccination with a 16-week interval between doses. Cell Host Microbe. 2022;30:97–109.e5. doi: 10.1016/j.chom.2021.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nayrac M., Dubé M., Sannier G., Nicolas A., Marchitto L., Tastet O., Tauzin A., Brassard N., Lima-Barbosa R., Beaudoin-Bussières G., et al. Temporal associations of B and T cell immunity with robust vaccine responsiveness in a 16-week interval BNT162b2 regimen. Cell Rep. 2022;39:111013. doi: 10.1016/j.celrep.2022.111013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Painter M.M., Mathew D., Goel R.R., Apostolidis S.A., Pattekar A., Kuthuru O., Baxter A.E., Herati R.S., Oldridge D.A., Gouma S., et al. Rapid induction of antigen-specific CD4(+) T cells is associated with coordinated humoral and cellular immunity to SARS-CoV-2 mRNA vaccination. Immunity. 2021;54:2133–2142.e3. doi: 10.1016/j.immuni.2021.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu J., Chandrashekar A., Sellers D., Barrett J., Jacob-Dolan C., Lifton M., McMahan K., Sciacca M., VanWyk H., Wu C., et al. Vaccines elicit highly conserved cellular immunity to SARS-CoV-2 Omicron. Nature. 2022;603:493–496. doi: 10.1038/s41586-022-04465-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schmidt F., Muecksch F., Weisblum Y., Da Silva J., Bednarski E., Cho A., Wang Z., Gaebler C., Caskey M., Nussenzweig M.C., et al. Plasma neutralization of the SARS-CoV-2 Omicron variant. N. Engl. J. Med. 2022;386:599–601. doi: 10.1056/NEJMc2119641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tauzin A., Gong S.Y., Chatterjee D., Ding S., Painter M.M., Goel R.R., Beaudoin-Bussières G., Marchitto L., Boutin M., Laumaea A., et al. A boost with SARS-CoV-2 BNT162b2 mRNA vaccine elicits strong humoral responses independently of the interval between the first two doses. Cell Rep. 2022;41:111554. doi: 10.1016/j.celrep.2022.111554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Polack F.P., Thomas S.J., Kitchin N., Absalon J., Gurtman A., Lockhart S., Perez J.L., Pérez Marc G., Moreira E.D., Zerbini C., et al. Safety and efficacy of the BNT162b2 mRNA covid-19 vaccine. N. Engl. J. Med. 2020;383:2603–2615. doi: 10.1056/NEJMoa2034577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Baden L.R., El Sahly H.M., Essink B., Kotloff K., Frey S., Novak R., Diemert D., Spector S.A., Rouphael N., Creech C.B., et al. Efficacy and safety of the mRNA-1273 SARS-CoV-2 vaccine. N. Engl. J. Med. Overseas. Ed. 2021;384:403–416. doi: 10.1056/NEJMoa2035389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Absalon J., Koury K., Gruber W.C. Safety and efficacy of the BNT162b2 mRNA covid-19 vaccine. N. Engl. J. Med. 2021;384 doi: 10.1056/NEJMc2036242. 1578-1577. [DOI] [PubMed] [Google Scholar]
- 14.Tauzin A., Nayrac M., Benlarbi M., Gong S.Y., Gasser R., Beaudoin-Bussières G., Brassard N., Laumaea A., Vézina D., Prévost J., et al. A single dose of the SARS-CoV-2 vaccine BNT162b2 elicits Fc-mediated antibody effector functions and T cell responses. Cell Host Microbe. 2021;29:1137–1150.e6. doi: 10.1016/j.chom.2021.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Paltiel A.D., Zheng A., Schwartz J.L. Speed versus efficacy: quantifying potential tradeoffs in COVID-19 vaccine deployment. Ann. Intern. Med. 2021;174:568–570. doi: 10.7326/M20-7866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tuite A.R., Zhu L., Fisman D.N., Salomon J.A. Alternative dose allocation strategies to increase benefits from constrained COVID-19 vaccine supply. Ann. Intern. Med. 2021;174:570–572. doi: 10.7326/M20-8137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Grunau B., Goldfarb D.M., Asamoah-Boaheng M., Golding L., Kirkham T.L., Demers P.A., Lavoie P.M. Immunogenicity of extended mRNA SARS-CoV-2 vaccine dosing intervals. JAMA. 2022;327:279–281. doi: 10.1001/jama.2021.21921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hall V.G., Ferreira V.H., Wood H., Ierullo M., Majchrzak-Kita B., Manguiat K., Robinson A., Kulasingam V., Humar A., Kumar D. Delayed-interval BNT162b2 mRNA COVID-19 vaccination enhances humoral immunity and induces robust T cell responses. Nat. Immunol. 2022;23:380–385. doi: 10.1038/s41590-021-01126-6. [DOI] [PubMed] [Google Scholar]
- 19.Nicolas A., Sannier G., Dubé M., Nayrac M., Tauzin A., Painter M.M., Goel R.R., Laporte M., Gendron-Lepage G., Medjahed H., et al. An extended SARS-CoV-2 mRNA vaccine prime-boost interval enhances B cell immunity with limited impact on T cells. iScience. 2023;26:105904. doi: 10.1016/j.isci.2022.105904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Payne R.P., Longet S., Austin J.A., Skelly D.T., Dejnirattisai W., Adele S., Meardon N., Faustini S., Al-Taei S., Moore S.C., et al. Immunogenicity of standard and extended dosing intervals of BNT162b2 mRNA vaccine. Cell. 2021;184:5699–5714.e11. doi: 10.1016/j.cell.2021.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zimmermann P., Curtis N. Factors that influence the immune response to vaccination. Clin. Microbiol. Rev. 2019;32:000844-18. doi: 10.1128/CMR.00084-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Betjes M.G. Uremia-associated ageing of the thymus and adaptive immune responses. Toxins. 2020;12:224. doi: 10.3390/toxins12040224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Girndt M., Trojanowicz B., Ulrich C. Monocytes in uremia. Toxins. 2020;12:340. doi: 10.3390/toxins12050340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kim J.K., Hong C.W., Park M.J., Song Y.R., Kim H.J., Kim S.G. Increased neutrophil extracellular trap formation in uremia is associated with chronic inflammation and prevalent coronary artery disease. J. Immunol. Res. 2017;2017:8415179. doi: 10.1155/2017/8415179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cohen G. Immune dysfunction in uremia 2020. Toxins. 2020;12:439. doi: 10.3390/toxins12070439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Abdelrasoul A., Westphalen H., Saadati S., Shoker A. Hemodialysis biocompatibility mathematical models to predict the inflammatory biomarkers released in dialysis patients based on hemodialysis membrane characteristics and clinical practices. Sci. Rep. 2021;11:23080. doi: 10.1038/s41598-021-01660-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Krueger K.M., Ison M.G., Ghossein C. Practical guide to vaccination in all stages of CKD, including patients treated by dialysis or kidney transplantation. Am. J. Kidney Dis. 2020;75:417–425. doi: 10.1053/j.ajkd.2019.06.014. [DOI] [PubMed] [Google Scholar]
- 28.Clark A., Jit M., Warren-Gash C., Guthrie B., Wang H.H.X., Mercer S.W., Sanderson C., McKee M., Troeger C., Ong K.L., et al. Global, regional, and national estimates of the population at increased risk of severe COVID-19 due to underlying health conditions in 2020: a modelling study. Lancet Global Health. 2020;8:e1003–e1017. doi: 10.1016/S2214-109X(20)30264-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hsu C.M., Weiner D.E., Aweh G., Miskulin D.C., Manley H.J., Stewart C., Ladik V., Hosford J., Lacson E.C., Johnson D.S., Lacson E., Jr. COVID-19 among US dialysis patients: risk factors and outcomes from a national dialysis provider. Am. J. Kidney Dis. 2021;77:748–756.e1. doi: 10.1053/j.ajkd.2021.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Anand S., Montez-Rath M.E., Han J., Garcia P., Cadden L., Hunsader P., Morgan C., Kerschmann R., Beyer P., Dittrich M., et al. SARS-CoV-2 vaccine antibody response and breakthrough infection in patients receiving dialysis. Ann. Intern. Med. 2021;175:371–378. doi: 10.7326/M21-4176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Goupil R., Benlarbi M., Beaubien-Souligny W., Nadeau-Fredette A.C., Chatterjee D., Goyette G., Gunaratnam L., Lamarche C., Tom A., Finzi A., et al. Short-term antibody response after 1 dose of BNT162b2 vaccine in patients receiving hemodialysis. CMAJ (Can. Med. Assoc. J.) 2021;193:E793–E800. doi: 10.1503/cmaj.210673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Grupper A., Sharon N., Finn T., Cohen R., Israel M., Agbaria A., Rechavi Y., Schwartz I.F., Schwartz D., Lellouch Y., Shashar M. Humoral response to the pfizer BNT162b2 vaccine in patients undergoing maintenance hemodialysis. Clin. J. Am. Soc. Nephrol. 2021;16:1037–1042. doi: 10.2215/CJN.03500321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Alcázar-Arroyo R., Portolés J., López-Sánchez P., Zalamea F., Furaz K., Méndez Á., Nieto L., Sánchez-Hernández R., Pizarro S., García A., et al. Rapid decline of anti-SARS-CoV-2 antibodies in patients on haemodialysis: the COVID-FRIAT study. Clin. Kidney J. 2021;14:1835–1844. doi: 10.1093/ckj/sfab048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ducloux D., Colladant M., Chabannes M., Yannaraki M., Courivaud C. Humoral response after 3 doses of the BNT162b2 mRNA COVID-19 vaccine in patients on hemodialysis. Kidney Int. 2021;100:702–704. doi: 10.1016/j.kint.2021.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Crotty S. T follicular helper cell biology: a decade of discovery and diseases. Immunity. 2019;50:1132–1148. doi: 10.1016/j.immuni.2019.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Broseta J.J., Rodríguez-Espinosa D., Rodríguez N., Mosquera M.D.M., Marcos M.Á., Egri N., Pascal M., Soruco E., Bedini J.L., Bayés B., Maduell F. Humoral and cellular responses to mRNA-1273 and BNT162b2 SARS-CoV-2 vaccines administered to hemodialysis patients. Am. J. Kidney Dis. 2021;78:571–581. doi: 10.1053/j.ajkd.2021.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Melin J., Svensson M.K., Albinsson B., Winqvist O., Pauksens K. Humoral and cellular response to SARS-CoV-2 BNT162b2 mRNA vaccine in hemodialysis patients. BMC Immunol. 2021;22:70. doi: 10.1186/s12865-021-00458-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Webster A.C., Nagler E.V., Morton R.L., Masson P. Chronic kidney disease. Lancet. 2017;389:1238–1252. doi: 10.1016/S0140-6736(16)32064-5. [DOI] [PubMed] [Google Scholar]
- 39.Piccoli L., Park Y.J., Tortorici M.A., Czudnochowski N., Walls A.C., Beltramello M., Silacci-Fregni C., Pinto D., Rosen L.E., Bowen J.E., et al. Mapping neutralizing and immunodominant sites on the SARS-CoV-2 spike receptor-binding domain by structure-guided high-resolution serology. Cell. 2020;183:1024–1042.e21. doi: 10.1016/j.cell.2020.09.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shi R., Shan C., Duan X., Chen Z., Liu P., Song J., Song T., Bi X., Han C., Wu L., et al. A human neutralizing antibody targets the receptor-binding site of SARS-CoV-2. Nature. 2020;584:120–124. doi: 10.1038/s41586-020-2381-y. [DOI] [PubMed] [Google Scholar]
- 41.Barin B., Kasap U., Selçuk F., Volkan E., Uluçkan Ö. Comparison of SARS-CoV-2 anti-spike receptor binding domain IgG antibody responses after CoronaVac, BNT162b2, ChAdOx1 COVID-19 vaccines, and a single booster dose: a prospective, longitudinal population-based study. Lancet. Microbe. 2022;3:e274–e283. doi: 10.1016/S2666-5247(21)00305-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Prévost J., Gasser R., Beaudoin-Bussières G., Richard J., Duerr R., Laumaea A., Anand S.P., Goyette G., Benlarbi M., Ding S., et al. Cross-sectional evaluation of humoral responses against SARS-CoV-2 spike. Cell Rep. Med. 2020;1:100126. doi: 10.1016/j.xcrm.2020.100126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hacisuleyman E., Hale C., Saito Y., Blachere N.E., Bergh M., Conlon E.G., Schaefer-Babajew D.J., DaSilva J., Muecksch F., Gaebler C., et al. Vaccine breakthrough infections with SARS-CoV-2 variants. N. Engl. J. Med. 2021;384:2212–2218. doi: 10.1056/NEJMoa2105000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Macallan D.C., Wallace D.L., Zhang Y., Ghattas H., Asquith B., de Lara C., Worth A., Panayiotakopoulos G., Griffin G.E., Tough D.F., Beverley P.C.L. B-cell kinetics in humans: rapid turnover of peripheral blood memory cells. Blood. 2005;105:3633–3640. doi: 10.1182/blood-2004-09-3740. [DOI] [PubMed] [Google Scholar]
- 45.Kaminski D.A., Wei C., Qian Y., Rosenberg A.F., Sanz I. Advances in human B cell phenotypic profiling. Front. Immunol. 2012;3:302. doi: 10.3389/fimmu.2012.00302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Goel R.R., Apostolidis S.A., Painter M.M., Mathew D., Pattekar A., Kuthuru O., Gouma S., Hicks P., Meng W., Rosenfeld A.M., et al. Distinct antibody and memory B cell responses in SARS-CoV-2 naive and recovered individuals following mRNA vaccination. Sci. Immunol. 2021;6:eabi6950. doi: 10.1126/sciimmunol.abi6950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sette A., Crotty S. Adaptive immunity to SARS-CoV-2 and COVID-19. Cell. 2021;184:861–880. doi: 10.1016/j.cell.2021.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Morou A., Brunet-Ratnasingham E., Dubé M., Charlebois R., Mercier E., Darko S., Brassard N., Nganou-Makamdop K., Arumugam S., Gendron-Lepage G., et al. Altered differentiation is central to HIV-specific CD4(+) T cell dysfunction in progressive disease. Nat. Immunol. 2019;20:1059–1070. doi: 10.1038/s41590-019-0418-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Niessl J., Baxter A.E., Morou A., Brunet-Ratnasingham E., Sannier G., Gendron-Lepage G., Richard J., Delgado G.G., Brassard N., Turcotte I., et al. Persistent expansion and Th1-like skewing of HIV-specific circulating T follicular helper cells during antiretroviral therapy. EBioMedicine. 2020;54:102727. doi: 10.1016/j.ebiom.2020.102727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Braun J., Loyal L., Frentsch M., Wendisch D., Georg P., Kurth F., Hippenstiel S., Dingeldey M., Kruse B., Fauchere F., et al. SARS-CoV-2-reactive T cells in healthy donors and patients with COVID-19. Nature. 2020;587:270–274. doi: 10.1038/s41586-020-2598-9. [DOI] [PubMed] [Google Scholar]
- 51.Grifoni A., Weiskopf D., Ramirez S.I., Mateus J., Dan J.M., Moderbacher C.R., Rawlings S.A., Sutherland A., Premkumar L., Jadi R.S., et al. Targets of T Cell responses to SARS-CoV-2 coronavirus in humans with COVID-19 disease and unexposed individuals. Cell. 2020;181:1489–1501.e15. doi: 10.1016/j.cell.2020.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Swadling L., Diniz M.O., Schmidt N.M., Amin O.E., Chandran A., Shaw E., Pade C., Gibbons J.M., Le Bert N., Tan A.T., et al. Pre-existing polymerase-specific T cells expand in abortive seronegative SARS-CoV-2. Nature. 2022;601:110–117. doi: 10.1038/s41586-021-04186-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Locci M., Havenar-Daughton C., Landais E., Wu J., Kroenke M.A., Arlehamn C.L., Su L.F., Cubas R., Davis M.M., Sette A., et al. Human circulating PD-1+CXCR3-CXCR5+ memory Tfh cells are highly functional and correlate with broadly neutralizing HIV antibody responses. Immunity. 2013;39:758–769. doi: 10.1016/j.immuni.2013.08.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Stumpf J., Siepmann T., Lindner T., Karger C., Schwöbel J., Anders L., Faulhaber-Walter R., Schewe J., Martin H., Schirutschke H., et al. Humoral and cellular immunity to SARS-CoV-2 vaccination in renal transplant versus dialysis patients: a prospective, multicenter observational study using mRNA-1273 or BNT162b2 mRNA vaccine. Lancet Reg. Health. Eur. 2021;9:100178. doi: 10.1016/j.lanepe.2021.100178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rincon-Arevalo H., Choi M., Stefanski A.L., Halleck F., Weber U., Szelinski F., Jahrsdörfer B., Schrezenmeier H., Ludwig C., Sattler A., et al. Impaired humoral immunity to SARS-CoV-2 BNT162b2 vaccine in kidney transplant recipients and dialysis patients. Sci. Immunol. 2021;6:eabj1031. doi: 10.1126/sciimmunol.abj1031. [DOI] [PubMed] [Google Scholar]
- 56.Lederer K., Bettini E., Parvathaneni K., Painter M.M., Agarwal D., Lundgreen K.A., Weirick M., Muralidharan K., Castaño D., Goel R.R., et al. Germinal center responses to SARS-CoV-2 mRNA vaccines in healthy and immunocompromised individuals. Cell. 2022;185:1008–1024.e15. doi: 10.1016/j.cell.2022.01.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Cho A., Muecksch F., Wang Z., Ben Tanfous T., DaSilva J., Raspe R., Johnson B., Bednarski E., Ramos V., Schaefer-Babajew D., et al. Antibody evolution to SARS-CoV-2 after single-dose Ad26.COV2.S vaccine in humans. J. Exp. Med. 2022;219:e20220732. doi: 10.1084/jem.20220732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kim W., Zhou J.Q., Horvath S.C., Schmitz A.J., Sturtz A.J., Lei T., Liu Z., Kalaidina E., Thapa M., Alsoussi W.B., et al. Germinal centre-driven maturation of B cell response to mRNA vaccination. Nature. 2022;604:141–145. doi: 10.1038/s41586-022-04527-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hou L., Yuki K. CCR6 and CXCR6 identify the Th17 cells with cytotoxicity in experimental autoimmune encephalomyelitis. Front. Immunol. 2022;13:819224. doi: 10.3389/fimmu.2022.819224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Linke A., Tiegs G., Neumann K. Pathogenic T-cell responses in immune-mediated glomerulonephritis. Cells. 2022;11:1625. doi: 10.3390/cells11101625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hartzell S., Bin S., Cantarelli C., Haverly M., Manrique J., Angeletti A., Manna G.L., Murphy B., Zhang W., Levitsky J., et al. Kidney failure associates with T cell exhaustion and imbalanced follicular helper T cells. Front. Immunol. 2020;11:583702. doi: 10.3389/fimmu.2020.583702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Schmitt N., Ueno H. Blood Tfh cells come with colors. Immunity. 2013;39:629–630. doi: 10.1016/j.immuni.2013.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Crotty S. T follicular helper cell differentiation, function, and roles in disease. Immunity. 2014;41:529–542. doi: 10.1016/j.immuni.2014.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.He J., Tsai L.M., Leong Y.A., Hu X., Ma C.S., Chevalier N., Sun X., Vandenberg K., Rockman S., Ding Y., et al. Circulating precursor CCR7(lo)PD-1(hi) CXCR5(+) CD4(+) T cells indicate Tfh cell activity and promote antibody responses upon antigen reexposure. Immunity. 2013;39:770–781. doi: 10.1016/j.immuni.2013.09.007. [DOI] [PubMed] [Google Scholar]
- 65.Kaneko N., Kuo H.H., Boucau J., Farmer J.R., Allard-Chamard H., Mahajan V.S., Piechocka-Trocha A., Lefteri K., Osborn M., Bals J., et al. Loss of bcl-6-expressing T follicular helper cells and germinal centers in COVID-19. Cell. 2020;183:143–157.e13. doi: 10.1016/j.cell.2020.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ulrich C., Wilke A., Schleicher N., Girndt M., Fiedler R. Hypervolemia-induced immune disturbances do not involve IL-1ss but IL-6 and IL-10 activation in haemodialysis patients. Toxins. 2020;12:159. doi: 10.3390/toxins12030159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Corbett K.S., Edwards D.K., Leist S.R., Abiona O.M., Boyoglu-Barnum S., Gillespie R.A., Himansu S., Schäfer A., Ziwawo C.T., DiPiazza A.T., et al. SARS-CoV-2 mRNA vaccine design enabled by prototype pathogen preparedness. Nature. 2020;586:567–571. doi: 10.1038/s41586-020-2622-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Haarhaus M., Duhanes M., Leševic N., Matei B., Ramsauer B., Da Silva Rodrigues R., Su J., Haase M., Santos-Araújo C., Macario F. Improved immunologic response to COVID-19 vaccine with prolonged dosing interval in haemodialysis patients. Scand. J. Immunol. 2022;95:e13152. doi: 10.1111/sji.13152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Shashar M., Nacasch N., Grupper A., Benchetrit S., Halperin T., Erez D., Rozenberg I., Shitrit P., Sela Y., Wand O., Cohen-Hagai K. Humoral response to pfizer BNT162b2 vaccine booster in maintenance hemodialysis patients. Am. J. Nephrol. 2022;53:207–214. doi: 10.1159/000521676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Verdier J.F., Boyer S., Chalmin F., Jeribi A., Egasse C., Maggi M.F., Auvray P., Yalaoui T. Response to three doses of the Pfizer/BioNTech BNT162b2 COVID-19 vaccine: a retrospective study of a cohort of haemodialysis patients in France. BMC Nephrol. 2022;23:189. doi: 10.1186/s12882-022-02751-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Beaudoin-Bussières G., Laumaea A., Anand S.P., Prévost J., Gasser R., Goyette G., Medjahed H., Perreault J., Tremblay T., Lewin A., et al. Decline of humoral responses against SARS-CoV-2 spike in convalescent individuals. mBio. 2020;11:025900-20. doi: 10.1128/mBio.02590-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Quintelier K., Couckuyt A., Emmaneel A., Aerts J., Saeys Y., Van Gassen S. Analyzing high-dimensional cytometry data using FlowSOM. Nat. Protoc. 2021;16:3775–3801. doi: 10.1038/s41596-021-00550-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
The published article includes all datasets generated and analyzed for this study. Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact Author (daniel.kaufmann@chuv.ch).
-
•
We adapted previously submitted6 R codes scripted to perform unsupervised analyzes on B and T cells from hemodialyzed donors and controls individuals. All original codes have been deposited on Github and are publicly available as of the date of publication. URL link is listed in the key resources table.