Abstract
Introduction
Colorectal cancer is the fourth most common cancer in the UK. There remains a need for improved risk stratification following curative resection. Circulating-tumour DNA (ctDNA) has gained particular interest as a cancer biomarker in recent years. We performed a systematic review to assess the utility of ctDNA in identifying minimal residual disease in colorectal cancer.
Methods
Studies were included if ctDNA was measured following curative surgery and long-term outcomes were assessed. Studies were excluded if the manuscript could not be obtained from the British Library or were not available in English.
Results
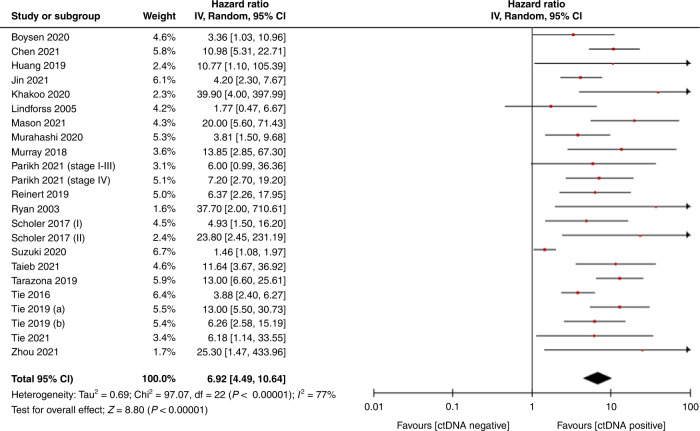
Thirty-seven studies met the inclusion criteria, involving 3002 patients. Hazard ratios (HRs) for progression-free survival (PFS) were available in 21 studies. A meta-analysis using a random effects model demonstrated poorer PFS associated with ctDNA detection at the first liquid biopsy post-surgery [HR: 6.92 CI: 4.49–10.64 p < 0.00001]. This effect was also seen in subgroup analysis by disease extent, adjuvant chemotherapy and assay type.
Discussion
Here we demonstrate that ctDNA detection post-surgery is associated with a greater propensity to disease relapse and is an independent indicator of poor prognosis. Prior to incorporation into clinical practice, consensus around timing of measurements and assay methodology are critical.
Protocol registration
The protocol for this review is registered on PROSPERO (CRD42021261569).
Subject terms: Prognostic markers, Disease-free survival, Tumour biomarkers, Colorectal cancer
Introduction
Colorectal cancer is the fourth most common cancer in the UK. In the last few decades, there has been a steady increase in incidence within developed countries, with the UK now seeing around 35,000 cases a year. Mortality increases with stage, and collectively, colorectal cancer is responsible for 10% of all cancer deaths in the UK [1, 2]. Definitive treatment involves surgical resection, aided by perioperative chemotherapy [3]. Identification of patients who will benefit from adjuvant chemotherapy remains a dilemma, particularly in stage II disease [4].
Minimal residual disease (MRD) is defined as microscopic neoplastic material remaining after curative treatment not detectable clinically [5], and thus holds the potential to precipitate disease relapse. Recently, there has been much interest in the ability of circulating tumour DNA (ctDNA) for detection of MRD and prognostication following curative treatment including surgical resection and radical chemoradiotherapy.
This ctDNA is released from dying cancer cells and is found in varying proportions amongst cell-free DNA (cfDNA) released following the death of normal circulating blood cells. It is released during apoptosis and necrosis and has a half-life of around 2 h [6, 7]. The concept of utilising circulating tumour-derived material to provide diagnostic information on cancer has been coined ‘liquid biopsy’ [8]. The liquid biopsy has many potential advantages over the traditional surgical biopsy. It is minimally invasive and amenable to repeat measurements over time. Liquid biopsies could overcome the spatial limitation of tissue biopsies with variations in genetic profiles seen within the tumour itself and between metastases [9, 10], and could theoretically provide a more complete picture of the molecular profile.
Despite the promise ctDNA holds, there are still a number of limitations. ctDNA comprises only a minor proportion of total cfDNA, thus sensitive methods are required for detection [11]. Clonal haematopoiesis of indeterminate potential (CHIP) are non-tumour derived somatic mutations in haemopoietic cells which can bring the possibility of false positive results [8]. There are two main approaches to ctDNA analysis. Initially measurement relied on PCR-based techniques targeting a few loci. This focused approach is quick and relatively inexpensive. The ability to detect very low variant allele frequencies (VAF) brings high sensitivity, with digital-PCR and BEAMing techniques able to detect VAFs as low as 0.01% [12]. However, PCR-based techniques rely on prior knowledge of the genetic profile of the cancer and have limited capabilities for multiplexing [7]. More recently, the development of next generation sequencing (NGS) has enabled analysis of a much wider panel of target genes and enables screening for unknown variants [7, 13]. There is a growing interest in the characteristics of ctDNA beyond the somatic mutations, including methylation and fragmentation patterns [14].
At present there remains an urgent clinical need for a better post-operative risk stratification paradigm in colorectal cancer, with current tumour markers lacking sensitivity and rising late following disease recurrence [15, 16]. It has been acknowledged that ctDNA holds great potential for this application, evidenced in a number of other primary cancer sites including pancreatic [17], lung [18] and breast [19] cancer, yet there remains little consensus on the validity of this approach in colorectal cancer compounded by a lack of systematic evidence. This systematic review examines the utility of post-surgical ctDNA for detecting MRD following curative surgery in colorectal cancer, and compares study methodologies to facilitate recommendations for optimal study design for future research and integration into clinical practice.
Methods
Search strategy and study selection
An electronic search of MEDLINE, EMBASE and the Cochrane Library was conducted in July 2021. There was no restriction by language and no limits were applied to the search. The search strategy is available in Supplementary Material. The protocol was registered on PROSPERO (CRD42021261569). Study selection, data extraction and quality assessment were performed in duplicate with two authors (LF and LH) working independently. Disagreements were resolved by discussion between authors. All abstracts identified by the search strategy were screened and potentially eligible manuscripts were then reviewed. Study authors were contacted where relevant outcome data was missing from manuscripts.
In order for inclusion, studies had to meet the following prespecified criteria: [1] Participants had to be diagnosed with colorectal cancer and undergoing curative surgical resection. [2] Post-operative ctDNA measurement was performed. [3] Participant follow-up had to be such that long-term outcomes could be assessed.
Surgical procedures on primary colorectal cancer, local recurrences and metastasectomies were included, provided they were carried out with curative intent. The post-operative ctDNA measurement could be carried out at any timepoint post-operatively provided this measurement was then correlated with long-term outcomes. Any length of follow-up were considered provided time to relapse or death were measured during this time. Studies were excluded if the manuscript could not be obtained from the British Library or were not available in English. Unpublished work was not included. We accepted any study design, however case report and reviews were not included. There was no restriction by publication date or sample size.
Data extraction
Data extraction was conducted in accordance with the following criteria: study characteristics (author, date of publication, country); study design (sample size, prospective/retrospective, follow-up time); participant baseline characteristics (age, gender, site, stage, neoadjuvant/adjuvant chemotherapy); ctDNA methodology (timing of samples, assay, gene panel, limit of detection, cut-off value).
At present there is no gold-standard method of detection of MRD, so long-term outcomes were used as surrogate markers, with the hypothesis that those with undetected residual disease will have a higher propensity to relapse. The outcomes collected were the proportion of subjects classified as ctDNA-positive at the first liquid biopsy after surgery, the proportion of participants who relapsed in each group, median progression-free survival (PFS), median overall survival (OS) and the corresponding hazard ratios (HRs) confidence intervals and p values.
Quality assessment
A quality assessment form was designed by considering relevant aspects from each domain in the ROBINS-I risk of bias tool [20]. This generated a ten-point scale. The mapping of each question to the domains of bias according to the ROBINS-I tool are shown in Supplementary Table 1. For each criterion, studies could be graded as ‘low risk’, ‘high risk’ or ‘unsure’. Each study was then scored out of 11, with the final score incorporating study timeline (i.e. prospective/retrospective). We also collected information on centre number, sample size and statistical adjustment.
Both the data extraction form and quality assessment form were pre-piloted and can be found in the supplementary material.
Data synthesis
A meta-analysis was conducted combining the HRs for PFS of ctDNA-positive vs ctDNA-negative groups. HR were pooled by inverse variance using the overall estimated HR and standard error of individual studies, either from data presented in the manuscripts or from a Cox proportional-hazards model from individual participant data available provided as a supplement or obtained directly from the study authors. Heterogeneity was quantified with the I2 statistical test and a random-effect model was used in the presence of significant heterogeneity (p < 0.05 or I2 ≥ 50%). Subgroup analysis was performed according to disease extent (primary resection vs metastasectomy), adjuvant chemotherapy and assay type (NGS vs PCR), as pre-planned. Results were displayed in Forest plots. Publication bias was assessed by Funnel plot to assess for asymmetry.
This review adheres to the Preferred Reporting Items for Systematic Reviews and Meta-analyses (PRISMA) guidelines [21] and the Meta-analysis of Observational Studies in Epidemiology (MOOSE) guidelines [22]. Statistical analysis was performed on Review Manager (RevMan) Version 5.4, The Cochrane Collaboration (2020).
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Results
Search results
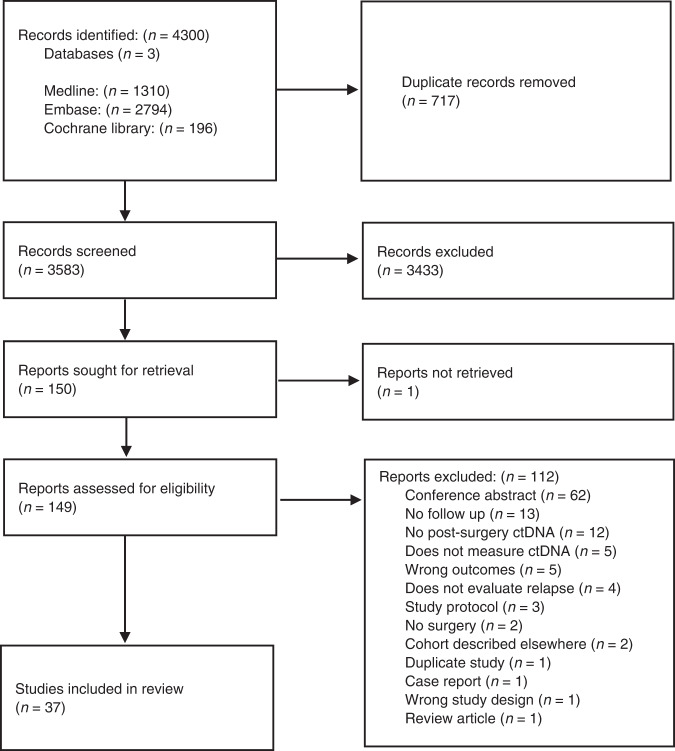
The search identified 3581 papers, after removal of duplicates. Full-text screening was performed for 147 studies, of which 37 studies were included involving 3002 patients (Fig. 1) [23–59]. Details of the key excluded studies can be found in Supplementary Table 2.
Fig. 1. PRISMA flow diagram.
Flow diagram describing the study selection process and number of studies at each stage according to the Preferred Reporting Items for Systematic Reviews and Meta-analyses (PRISMA) guidelines.
Included studies
Included studies incorporated all stages of colorectal cancer (I–IV), with six specific to rectal cancer. On average, 42.2% of patients had rectal cancer and 34.8% exhibited right-sided disease. Articles were published between 1993 and 2021 and were conducted in continents including North America, Europe, Asia and Australasia. Surgical procedures included removal of the primary cancer, local recurrence and metastasectomy. Nine papers addressed metastasectomy alone (liver, peritoneal or lung) with a further two including metastasectomy sub-groups. The median age ranged from 55 to 73 and the proportion of male participants ranged from 33-90%, mean 53.2% (Table 1). Out of 37 papers, only 16 (43.2%) reported the proportion of patients who received neoadjuvant chemotherapy (range = 0–100% participants, mean: 43.6%), and 26 (70.3%) the proportion of patients receiving adjuvant chemotherapy (range: 0–100%, mean: 63.5%). The most common regimen was 5FU-based, either alone or in combination with oxaliplatin. The median follow-up time of the study ranged from 11.7 months to 6.6 years (median 26.2). Post-operative monitoring protocols were described in 23 (62.2%) studies, consisting of physical examination, laboratory tumour markers (CEA, CA19.9) and radiology (Table 1).
Table 1.
Study characteristics.
| Author | Year | Journal | Country | Sample size | Follow-up (months) | Gender (% male) | Age (median) | Cancer stage | Cancer site | Neoadjuvant chemotherapy (%) | Adjuvant chemotherapy (%) | Reference | ||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Median | Range | |||||||||||||
| Allegretti et al. | 2020 | Journal of Experimental and Clinical Research | Italy | 10 | 11.7 | 6–40 | 47.2 | Not stated | Non-metastatic | Colorectal | 0 | Not stated | [23] | |
| Beagan et al. | 2020 | Journal of Clinical Medicine | Netherlands | 24 | Not stated | Max 25 | 58.3 | 66.5 | IV | Metastases (P) | 0 | 37.5 | [24] | |
| Benešová et al. | 2019 | World Journal of Gastroenterology | Czech Republic | 26 | Not stated | Not stated | 48.4 | 63.6a | IV | Metastases (H + L) | Not stated | Not stated | [25] | |
| Boysen et al. | 2020 | Acta Oncologica | Denmark | 35 | 21 | Not stated | 51.4 | 70.5 | IV | Metastases (H + L) | Not stated | 74.3 | [26] | |
| Carpinetti et al. | 2015 | Oncotarget | Brazil | 3 | Not stated | Not stated | 33.3 | Not stated | II–III | Rectal | 100 | 0 | [27] | |
| Chen et al. | 2021 | Journal of Hematology and Oncology | China | 240 | 27.4 | 26.2–28.5 | 56.7 | 60 | II–III | Colon | 0 | 72.5 | [28] | |
| Diehl et al. | 2008 | Nature Medicine | USA | 18 | Not stated | 2–56 days | 44.4 | 59.8a | II–IV | Colon + metastases (H + L) | Not stated | 61.1 | [29] | |
| He et al. | 2020 | Cancer Management and Research | China | 19 | Not stated | 14–18 | 70 | 55a | IV | Metastases (H) | 55 | 80 | [30] | |
| Huang et al. | 2019 | Cancer Biomarkers | China | 41 | 30 | 7–146 weeks | 53.5 | 58 | I–IV | Colorectal | Not stated | 67.4 | [31] | |
| Ji et al. | 2021 | Genomics | China | 32 | Not stated | Not stated | 59.4 | 58.7a | 0–III | Rectal | 100 | Not stated | [32] | |
| Jin et al. | 2021 | PNAS | China | 73 | Not stated | 36–50 months | 63 | 67 | I–IV | Colorectal | 83.6 | 84.9 | [33] | |
| Khakoo et al. | 2020 | Clinical Cancer Research | UK | 23 | 26.4 | (IQR 19.7–31.3) | 56.5 |
50 (+) 59 (−) |
I–III | Rectal | 100 | 91.3 | [34] | |
| Lee et al. | 2021 | Cancers | South Korea | 53 | Not stated | Not stated | 63.8 | 56 | III–IV | Metastases (H/L/P) | 47.2 | Not stated | [35] | |
| Leon Arellano et al. | 2020 | Disease Markers | Spain | 10 | 26 | 25–29 | 30 | 67.5 | II–IV | Colon | 0 | Not stated | [36] | |
| Levy et al. | 2012 | Anticancer Research | Czech Republic | 4 | 22.5 | 12–29 | 50 | 65 | II–IV | Colorectal | 25 | 75 | [37] | |
| Lindforss et al. | 2005 | Anticancer Research | Sweden | 24 | 35 | 5–52 | 37.5 | 72 | I–III | Colorectal | Not stated | Not stated | [38] | |
| López-Rojo et al. | 2020 | Therapeutic Advances in Medical Oncology | Spain | 9 | 28.5 | 8–41 | 36.3 | 56.9a | IV | Metastases (P) | Not stated | 100 | [39] | |
| Mason et al. | 2021 | Journal of the American College of Surgeons | USA | 63 | 30 | 9–53 | 50.8 | 55 | IV | Metastases (H) | 87.3 | 58.7 | [40] | |
| Murahashi et al. | 2020 | British Journal of Cancer | Japan | 59 | Not stated | Not stated | 79.7 | 60 | II–III | Rectal | 100 | Not stated | [41] | |
| Murray et al. | 2018 | Journal of Cancer Research and Clinical Oncology | Australia | 172 | 23.2 | (IQR 14.3–29.5) | 61 | 65.5a | III–IV | Colon + metastases | Not stated | Not stated | [42] | |
| Ng et al. | 2017 | Scientific Reports | Singapore | 10 | 965 days | 786–1253 days | 90 | 65.3 | II–III | Colorectal | Not stated | Not stated | [43] | |
| Parikh et al. | 2021 | Clinical Cancer Research | USA | 70 | 632.5 days | 33–246 days | 60.7 | 60 | I–IV | Colorectal | 45.2 | 54.8 | [44] | |
| Reinert et al. | 2019 | JAMA Oncology | Denmark | 125 | 12.5 | 1.4–38.5 | 56.9 | 69.9 | I–III | Colorectal | Not stated | 61.6 | [45] | |
| Ryan et al. | 2003 | Gut | Netherlands | 85 | 28 | 6–72 | 60.6 | 66 | Dukes A–C | Colorectal | Not stated | 56.4 | [46] | |
| Scøhler et al. | 2017 | Clinical Cancer Research | Denmark |
44 (Cohort 1: 21 Cohort 2:23) |
Not stated | 8 days–36 months | 64.4 | 65 |
Cohort 1: I–III Cohort 2: IV |
Colorectal + metastases (H) | Not stated | 34.1 | [47] | |
| Suzuki et al. | 2020 | Oncotarget | Japan | 44 | 366 days | Not stated | 58.4 | 71 | II–III | Colorectal | Not stated | 36.4 | [48] | |
| Taieb et al. | 2021 | Clinical Cancer Research | France | 1017 | 6.6 years | (95% CI 6.5–6.8) | 56.6 | 64.4 | III | Colon | Not stated | 100 | [49] | |
| Tanaka et al. | 2021 | Scientific reports | Japan | 11 | 22 | 15.4–23.6 | 36.4 | 69 | I–III | Colorectal | Not stated | 45.5 | [50] | |
| Tarazona et al. | 2019 | Annals of Oncology | Spain | 69 | 24.7 | 1–45.2 | 64.9 | 71 | I–III | Colon | 0 | 37.3 | [51] | |
| Tie et al. | 2016 | Science Translational Medicine | Australia | 167 | 27 | 2–52 | 57 | 65 | II | Colon | 0 | 23 | [47] | |
| Tie et al. (a) | 2019 | JAMA Oncology | Australia | 96 | 28.9 | 11.6–46.4 | 51 | 64 | III | Colon | Not stated | 99 | [37] | |
| Tie et al. (b) | 2019 | Gut | Australia | 159 | 24 | 1–55 | 67.3 | 62 | II–III | Rectal | 100 | 64.2 | [52] | |
| Tie et al. | 2021 | PLoS Medicine | Australia | 38 | 50.5 | 5–82 | 71.4 | 64 | IV | Metastases (H) | 42.6 | 77.8 | [31] | |
| Yamada et al. | 2016 | Cancer Science | Japan | 7 | Not stated | Not stated | 71.4 | 73 | III–IV | Metastases (H + L) | 14.3 | Not stated | [53] | |
| Zhou et al. | 2016 | PLoS ONE | China | 5 | 46 | 32–47 | 33.3 | 64 | I–III | Colorectal | 0 | 66.7 | [54] | |
| Zhou et al. | 2021 | Clinical Cancer Research | China | 89 | 18.8 | 3.1–21.3 | 65.2 | 60 | II–III | Rectal | 99 | 80.6 | [55] | |
| Zou et al. | 2020 | Carcinogenesis | New Zealand | 28 | 510.5 days | 98–692 days | 48.3 | 65 | II–IV | Colorectal + metastases (H + L) | Not stated | Not stated | [56] | |
Table shows study characteristics and participant characteristics of the included studies.
IQR interquartile range, metastases site: H hepatic, L lung, P peritoneal.
aAge presented as mean.
Timing of the first post-operative ctDNA measurement varied from the day of surgery to 13 months post-surgery. PCR-based methods were used in 19 (51%) studies and 15 (31%) used NGS, with 3 studies monitoring epigenetic changes. Fourteen (38%) reported a limit of detection (LoD) of the assay and 31 (86%) specified a cut-off level to establish ctDNA positivity. There was little consensus on the gene panel breadth, with the number of genes evaluated ranging from 1 to 1021. In 16 studies, the mutations evaluated in ctDNA were based on those previously identified in tissue (15) or plasma (1). Within these, the size of the gene panel evaluated in the tumour ranged from 4 genes to whole-genome sequencing (WGS). ctDNA was also measured pre-operatively in 32 (86%) of the studies (Table 2).
Table 2.
Methodology.
| Author, Year | Timing of post-op liquid biopsy | Limit of detection (VAF) | Cut-off level | Method of detection | Gene panel | Number of genes |
|---|---|---|---|---|---|---|
| Allegretti, 2020 | 3 months | ≥0.2% | Any mutations detected | NextSeq Digital PCR plus validation with dPCR | TruSight Tumour panel | 15 |
| Beagan, 2020 | 2 weeks–3 months | Not stated | Not stated | ddPCR | Variants in metastases VAF ≥ 3% | (48) |
| Benešová, 2019 | 1 week | 0.03–1% | Not stated | PCR and DCE | KRAS, TP53, APC, PIK3CA, BRAF, CTNNB1 | 6 |
| Boysen, 2020 | 2 weeks | 0.10% | Any mutations detected | ddPCR and MassARRAY | UltraSEEK MA Colon Panel | 5 |
| Carpinetti, 2015 | Not stated | 1 amplifiable copy/mL plasma | 3 positive droplets from 10 to 15,000 droplets | Taqman assays | Chromosomal rearrangements from WGS of tumour | (WGS) |
| Chen, 2021 | 3–7 days | Not stated | >5% of total tracking variants | Geneseeq Prime | Geneseeq Prime™ 425-gene panel | 425 |
| Diehl, 2008 | 1 day | Not stated | Fraction of beads bound to mutant fragments higher than the negative control, mean mutant DNA fragments plus one standard deviation >1.0 | BEAMing | Mutations detected in tumour FFPE sequencing | (4) |
| He, 2020 | Within 7 days | Not stated | Not stated | Capture-based targeted deep sequencing | ColonCore panel NextSeq 500 system (Illumina, Inc.) | 41 |
| Huang, 2019 | 1 month | Not stated | >4 mutant reads in plasma with >1 read on each strand | Illumina NextSeq 500 | 85 genes | 85 |
| Ji, 2021 | 1 day | Not stated | TMB > 10/ctDNA—any mutations/change in TMB | Illumina HiSeq X-Ten | 30 mutation signatures | 30 |
| Jin, 2021 | 1–14 days | 0.05% tumour DNA | mqMSP assay: ΔCq value > −1. SEPT9 assay: at least one out of three qPCR replicates had a Ct value <45 | Methylation-specific quantitative PCR assay (mqMSP) | Septin 9 (SEPT9) gene hypermethylated | NA |
| Khakoo, 2020 | 4–12 weeks | not stated | Two mutant-positive droplets present for at least one variant | ddPCR | 1–3 variants with highest VAF in tumour | 1–3 (6) |
| Lee, 2021 | 3–4 weeks | 1% | VAF ≥ 1% | Ultra-deep targeted sequencing | Somatic variants identified from primary and metastatic tumour | (50) |
| Leon Arellano, 2020 | 3 months | Valid when the ACTB Ct was ≤32.1 | SEPT9 Ct <45 cycles | Duplex quantitative PCR, Fast Real-Time PCR | Septin 9 (SEPT9) hypermethylated | 1* |
| Levy, 2012 | <1 week | Not stated | 5% of mutated alleles | Fluorescently labelled PCR and DCE | Somatic mutations previously found in tumour | (5) |
| Lindforss, 2005 | 3 days | Not stated | Not stated | PCR | KRAS | 1 |
| López-Rojo, 2020 | 48 h | Not stated | Concentration compared between samples and wild-type controls using a Z test, p < 0.05 used for positivity | ddPCR | KRAS | 1 |
| Mason, 2021 | Median 13 months (range 1–45 months) | 0.30% | Any mutations detected | Guardant360 CDx | 70 genes | 70 |
| Murahashi, 2020 | 12 weeks | Not stated | VAF 0.15% | Amplicon-based deep sequencing | 14 genes | 14 |
| Murray, 2018 | Within 12 months | Not stated | At least one PCR replicate positive for methylation | Triplex real-time qPCR assay | BCAT1 and IKZF1 methylation | 2* |
| Ng, 2017 | Within 5 days | 0.05% | Positive on a one-tailed exact conditional test of the ratio of two Poisson rates to distinguish from negative controls | Multiplex-PCR amplicon sequencing | Somatic variants identified from the primary tumour | 1–14 (799) |
| Parikh, 2021 | 11–148 days | Not stated | Any mutations detected | Guardant Reveal test | Not stated | Not stated |
| Reinert, 2019 | 30 days | Not stated | At least 2 variants detected | HiSeq 2500 system | 16 somatic single-nucleotide variants and short indels based on WES of tumour | 16 (WES) |
| Ryan, 2003 | 1 week | Not stated | Not stated | PCR | KRAS | 1 |
| Scøhler, 2017 | 8 days | 0.50% | Any mutations detected | ddPCR | Mutations identified on WES of tumour | Mean 4.2 (WES) |
| Schou, 2018 | 3 months | 170 ng/mL | Above 75th quartile | Direct fluorescent assay | Not applicable | not applicable |
| Suzuki, 2020 | At the end of hospitalisation | 0.02% |
NGS: at least one mutated ctDNA PCR: one copy of mutated ctDNA |
Pre op: Oncomine Pan Cancer Cell Free Assay. Post op: ddPCR | Mutations identified pre-surgery in plasma by NGS | ((52)) |
| Taieb, 2021 | After surgery, before adjuvant chemotherapy | Above limit of blank | Above limit of blank | Multiplex droplet-based digital PCR (ddPCR) and NGS | WIF1 and NPY gene hypermethylation (AmpliSeq Colon and Lung Cancer Panel V2 performed in a subset of patients) | 2*+22 |
| Tanaka, 2021 | 1 day | 0.10% | VAF 0.15% | dPCR Taqman assays | BRAF | 1 |
| Tarazona, 2019 | 6–8 weeks | Not stated | VAF 5% | Orthogonal droplet digital PCR | Two mutations with the highest VAF on NGS of tumour | 2 (29) |
| Tie, 2016 | 4–10 weeks | Not stated | Permutation test comparing mutation frequency between samples and controls | Illumina MiSeq | Somatic mutation with highest VAF in tumour FFPE | 1 (15) |
| Tie, 2019 (1) | 4–10 weeks | Not stated | Permutation test comparing mutation frequency between samples and controls | Illumina MiSeq | Mutation with the highest VAF in tissue from surgery | 1 (15) |
| Tie, 2019 (2) | 4–10 weeks | Not stated | Permutation test comparing mutation frequency between samples and controls | Safe-SeqS and Illumina MiSeq | Somatic mutation with the highest VAF in tumour tissue | 1 (15) |
| Tie, 2021 | 4–10 weeks | Not stated | Permutation test comparing mutation frequency between samples and controls | Safe-SeqS | Mutation with the highest VAF in tumour tissue | 1 (15) |
| Yamada, 2016 | Within 1 month | <1.00%. | Ratio of 0.1% mutant to 99.9% wild type | Invader Plus assay with peptide nucleic acid clamping method and digital PCR | KRAS | 1 |
| Zhou, 2016 | 1 month | Not stated | VAF > 0 | Illumina HiSeq 2500 | 545 genes | 545 |
| Zhou, 2021 | Within 1 month | Not stated | At least one mutation in ctDNA also detected in tissue | HiSeq 3000 Sequencing System (Illumina) | 1021 genes | 1021 |
| Zou, 2020 | 12 weeks | Not stated | VAF 1% | ddPCR | Somatic mutations from targeted sequencing of FFPE slides | 2 (71) |
Table shows analysis methods of measurement of ctDNA in post-operative blood samples. *Gene methylation; () genes evaluated in tumour tissue; (()) genes evaluated in pre-op blood samples.
DCE denaturing capillary electrophoresis, VAF variant allele frequency, ddPCR digital droplet PCR, NGS next-generation sequencing, PCR polymerase chain reaction, qPCR quantitative PCR, VAF variant allele frequency, WES whole-exome sequencing, WGS whole-genome sequencing.
Association of ctDNA with PFS
The proportion of participants classified as ctDNA-positive at the first liquid biopsy after surgery ranged from 0 to 90.9% (median 20%). In 3 studies, no patients had detectable ctDNA at the first liquid biopsy after surgery [23–25]. The proportion of patients who relapsed during follow-up was consistently higher in ctDNA positive participants concurrent with shorter median PFS (Table 3). Time-to-event analysis for PFS according to post-operative ctDNA was available for 21 studies including 2645 participants. This included outcomes calculated from data available in the supplementary material [26] and data sent by the study authors [27]. Multivariate analysis had been performed in 15 studies and OS was assessed in 12 (Supplementary Table 3). A shorter PFS associated with ctDNA-positivity was consistently observed, with HRs varying between 1.36 and 39.9. This was statistically significant in 19 studies via univariate analysis and in all multivariate analysis (Table 3).
Table 3.
Disease relapse.
| Study | ctDNA positive n (%) | Relapse (%) | Median PFS (months) | Univariate | Multivariate | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ctDNA positive | ctDNA negative | ctDNA positive | ctDNA negative | Hazard ratio | Confidence interval | p value | Hazard ratio | Confidence interval | p value | |||||
| Allegretti, 2020 | 3 (30%) | 100 | 0 | 3 | Not reached | – | – | – | – | – | – | |||
| Beagan, 2020 | 1 (4.2%) | 100 | 56.5 | 7 | Not reached | – | – | – | – | – | – | |||
| Benešová, 2019 | 2 (7.1%) | 100 | 58.3 | 6 | Not stated | – | – | – | – | – | – | |||
| Boysen, 2020 | 5 (14.3%) | 80 | 37.1 | 9 | Not reached | 3.36 | 1.03–10.94 | 0.03 | 7.48 | 1.47–38.36 | 0.02 | |||
| Carpinetti, 2015 | 1 (33.3%) | 100 | 0 | 10 | Not reached | – | – | – | – | – | – | |||
| Chen, 2021 | 20 (8.3%) | 60 | 0.5 | Not stated | Not stated | 10.98 | 5.31–22.72 | <0.001 | 8.02 | 3.59–17.92 | <0.001 | |||
| 9.99** | 4.40–22.69** | <0.001** | – | – | – | |||||||||
| Diehl, 2008 | 10 (55.6%) | 100 | 33.3 | Not stated | Not stated | – | – | – | – | – | – | |||
| He, 2020 | 4 (20%) | Not stated | Not stated | 5 | Not reached | – | – | – | – | – | – | |||
| Huang, 2019 | 9 (23.1%) | 33.3 | 3.3 | Not stated | Not stated | 10.767 | 1.1–103.8 | 0.04 | ||||||
| Ji, 2021 | 19 (59.4%) | 38.5 | 10.5 | Not stated | Not stated | – | – | – | – | – | – | |||
| Jin, 2021 | 21 (28.8%) | 52.4 | 17.3 | Not stated | Not stated | 4.2 | 2.3–18.73 | 0.0005 | – | – | – | |||
| 4.08* | 1.26–75.05* | 0.037* | – | – | – | |||||||||
| 5.16** | 2.31–29.78** | 0.001** | – | – | – | |||||||||
| Localised | – | – | – | – | – | 4.04 | 1.98–23.07 | 0.001 | – | – | – | |||
| Khakoo, 2020 | 3 (13%) | 100 | 0 | Not stated | Not stated | 39.9 | 4.0–399.5 | 0.002 | ||||||
| Lee, 2021 | 0 | NA | 52.8 | NA | 23.6 | – | – | – | – | – | – | |||
| Leon Arellano, 2020 | 4 (40%) | 100 | 0 | Not stated | Not stated | – | – | – | – | – | – | |||
| Levy, 2012 | 0 | NA | 25 | NA | 23 | – | – | – | – | – | – | |||
| Lindforss, 2005 | 8 (33.3%) | 50 | 23.5 | 12 | Not reached | 1.77 | 0.47–6.62 | 0.396 | ||||||
| López-Rojo, 2020 | 5 (55.6%) | 100 | 0 | 8.3 | 35.4 | – | – | – | – | – | – | |||
| Mason, 2021 | 42 (66.7%) | 26.2 | Not stated | Not stated | Not stated | – | – | – | – | – | – | |||
| Murahashi, 2020 | 21 (35.6%) | 23.8 | 18.8 | Not stated | Not stated | 20 | 5.6–72 | <0.0001 | 7.7 | 1.6–42 | 0.0127 | |||
| Murray, 2018 | 28 (16.3%) | 25 | 11.1 | Not stated | Not stated | – | – | – | 3.81 | 1.5–9.5 | 0.004 | |||
| Ng, 2017 | 0 | NA | 10 | NA | Not stated | – | – | – | – | – | – | |||
| Parikh, 2021 | 17 (24.2%) | 88.2 | 24.5 | 5.3 | Not reached | 11.2 | Not stated | <0.0001 | – | – | – | |||
| – | – | – | – | – | 12.03* | 1.77–81.7* | <0.0001* | – | – | – | ||||
| – | – | – | – | – | 7.35** | 1.72–31.42** | <0.0001** | – | – | – | ||||
| Reinert, 2019 | 10 (10.6%) | 70 | 11.9 | Not stated | Not stated | – | – | – | – | – | – | |||
| – | – | – | 4.5* | 1.6–12.8* | 0.004* | |||||||||
| Post-surgery | 7.2** | 2.7–19.0** | <0.001** | |||||||||||
| Post-ACT | 17.5** | 5.4–56.5** | <0.001** | – | – | – | ||||||||
| Ryan, 2003 | 15 (17.6%) | 60 | 11.4 | Not stated | Not stated | – | – | – | 6.37 | 2.26–18.0 | 0 | |||
| Scøhler, 2017 | Primary resection | 6 (28.6%) | 100 | 27 | Not stated | Not stated | 37.7 | 2–335.5 | <0.001 | – | – | – | ||
| Metastasectomy | 7 (30.4%) | 100 | 50 | Not stated | Not stated | 4.93 | 1.5–15.7 | 0.007 | – | – | – | |||
| Suzuki, 2020 | 6 (13.6%) | 50 | 2.6 | Not stated | Not stated | 23.8 | 2.45–250 | 0.006 | – | – | – | |||
| Taieb, 2021 | 139 (13.7%) | 35 | 27 | (66.4% at 3 years) | (76.7% at 3 years) | 1.46** | 1.08–1.97** | 0.015** | 1.55 | 1.13–2.12 | 0.006 | |||
| Tanaka, 2021 | 10 (90.9%) | 40 | 0 | 15.4 | Not reached | – | – | – | – | – | – | |||
| Tarazona, 2019 | 14 (20.3%) | 57.1 | 13 | Not stated | Not stated | 6.96 | Not stated | 0.0001 | 11.64 | 3.67–36.88 | <0.001 | |||
| Tie, 2016 | All | 13 | 6.6–27 | <0.001 | 14 | 6.8–28 | <0.001 | |||||||
| No chemotherapy | 14 (7.9%) | 78.6 | 9.8 |
9.9 (0% at 3 years) |
Not reached (90% at 3 years) |
18* | 7.9–40* | <0.001* | 28* | 11–68* | <0.001* | |||
| Adjuvant chemo | 6 (11.5%) | Not stated | Not stated | Not stated | Not stated | 11** | 1.8–68** | 0.001** | – | – | – | |||
| Tie, 2019 (a) | Post-surgery | 20 (20.8%) | 45 | 14.5 |
20.6 (47% at 3 years) |
Not reached (76% at 3 years) |
3.8** | 2.4–21** | <0.001** | 7.5** | 3.5–16.1** | <0.001** | ||
| Post-chemo | 15 (17%) | 66.7 | 17.8 | (30% at 3 years) | (77% at 3 years) | 6.8** | 11–157** | <0.001** | – | – | – | |||
| Tie, 2019 (b) | All | 19 (11.9%) | 58 | 8.60 | (33% at 3 years) | (87% at 3 years) | 13 | 5.5–31 | <0.001 | 6.0 | 2.2–16 | <0.001 | ||
| No chemo | 22* | 4.2–110* | <0.001* | – | – | – | ||||||||
| Adjuvant chemo | 11 (10.8%) | 100 | 17.3 | (50% at 3 years) | (85% at 3 years) | 10** | 3.4–29** | <0.001** | – | – | – | |||
| Tie, 2021 | Post-op | 21 (24.5%) | 83.3 | 30 | (69.3% at 5 years) | (16.7% at 5 years) | 6.26 | 2.58–15.2 | <0.001 | 3.13 | 1–9.82 | 0.05 | ||
| Post-chemo | 14.94 | 4.94–44.7 | <0.001 | – | – | – | ||||||||
| Yamada, 2016 | 2 (28.6%) | 0 | 60 | Not stated | 7.5 | – | – | – | – | – | – | |||
| Zhou, 2016 | 1 (20%) | 25 | 0 | Not stated | Not stated | – | – | – | – | – | – | |||
| Zhou, 2021 | 6 (6.7%) | 100 | 6 | Not stated | Not stated | 25.3 | 1.475–434 | <0.001 | 1.267 | Not stated | <0.001 | |||
| Zou, 2020 | 2 (7.14%) | 100 | 0 | 11.3 | 62.7 | – | – | – | – | – | – | |||
Table shows the proportion of patients who were identified as ctDNA positive from the first liquid biopsy after surgery, disease relapse rate, median PFS and time-to-event analysis for PFS according to ctDNA results. Percentage PFS from Kaplan–Meier estimates.
CI confidence interval, HR hazard ratio, PFS progression-free survival, NA not applicable.
aSample did not receive adjuvant chemotherapy.
bSample received adjuvant chemotherapy.
Meta-analysis of PFS according to ctDNA
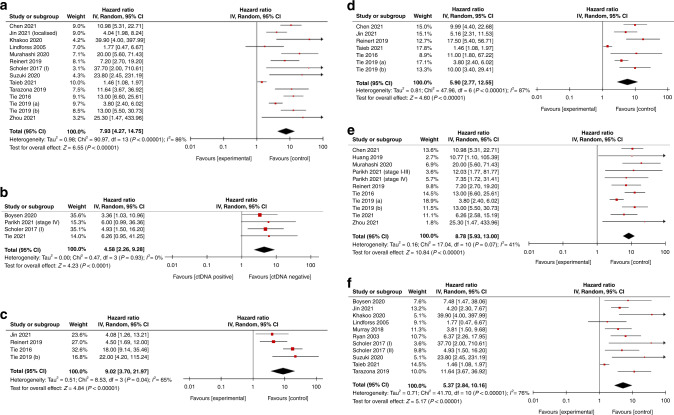
A meta-analysis confirmed poor prognosis associated with ctDNA detection post-operatively, which was found to be statistically significant [HR 6.92, CI 4.49–10.64, p < 0.00001] (Fig. 2). This effect was also seen in subgroup analysis according to adjuvant chemotherapy use [adjuvant chemotherapy HR 6.01, CI 2.96–12.21, p < 0.00001, no adjuvant chemotherapy HR 10.3, CI 6.46–16.45, p < 0.00001], disease extent [primary resection HR 7.93, CI 4.27–14.75, p < 0.00001 metastasectomy HR 5.08, CI 2.85–9.05, p < 0.00001] and assay type [NGS: HR 8.87, CI 5.93–13, p < 0.00001; PCR: HR 5.37, CI 2.84–10.16, p < 0.00001] (Fig. 3). A meta-analysis was also performed where multivariate analysis was available [HR 5.73, CI 3.34–9.84, p < 0.00001] (Supplementary Fig. 1). Statistical testing demonstrated significant heterogeneity (p < 0.00001) with an I2 value of 77%, hence a random effects model was used. The funnel plots of effect size (HR) plotted against standard error showed asymmetry suggestive of publication bias (Fig. 4).
Fig. 2. Forest plot showing meta-analysis for PFS according to post-operative ctDNA following surgery for colorectal cancer.
Data displayed as HR with 95% confidence intervals on a logarithmic scale. HR hazard ratio, PFS progression-free survival, SE standard error.
Fig. 3. Subgroup analysis.
Forest plot showing subgroup meta-analysis for PFS according to post-operative ctDNA according to disease extent, adjuvant chemotherapy and assay type: a resection of primary disease; b metastasectomy, c did not receive adjuvant chemotherapy; d received adjuvant chemotherapy; e NGS; f PCR data displayed as HR with 95% confidence intervals on a logarithmic scale. HR hazard ratio, NGS next-generation sequencing, PCR polymerase chain reaction, PFS progression-free survival.
Fig. 4. Funnel plot.
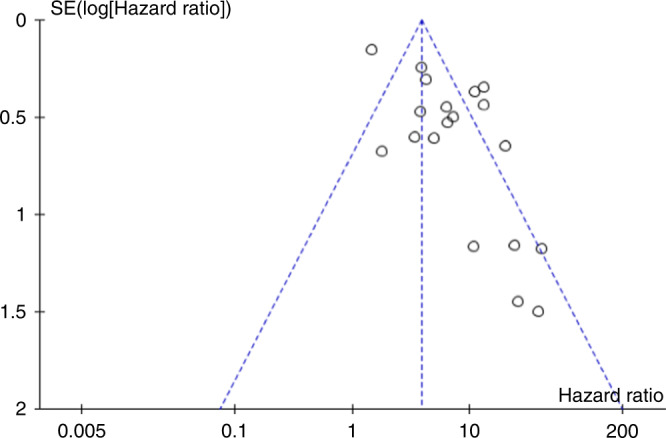
Funnel plot to show effect size against standard error for HR of PFS according to ctDNA status. HR hazard ratio, PFS progression-free survival, SE standard error.
Association of ctDNA with OS
Hazard ratios comparing overall survival were available in five papers [28–32]. An association of poor prognosis with post-operative ctDNA detection was also seen on meta-analysis when comparing overall survival [HR 3.64, CI 1.63-8.12, p = 0.002] (Supplementary Table 3 and Supplementary Fig. 2).
Quality assessment
The total quality assessment score of included studies ranged from 7 to 11 out of 11 (Supplementary Table 4). Patient baseline characteristics and ctDNA methodologies were generally well described. Most studies were conducted in single centres [32] and sample size calculations were rarely performed [4]. There were a number of studies with small sample sizes and inclusion of only a few participants; however, of those included in the meta-analysis the minimum sample size was 24 owing to the need for sufficient data for meaningful survival analysis in these studies.
Discussion
In this review, we demonstrate that ctDNA detection after curative surgery in colorectal cancer is associated with shorter time to disease relapse. This relationship was consistently demonstrated across multiple studies, and here we demonstrate for the first time that this effect is statistically significant when combined through a meta-analysis. The role of ctDNA as a marker of prognosis has previously been explored in Stage IV disease; a systematic review included four studies looking at resectable disease incorporating 123 patients. They report a ‘lead time’ with ctDNA appearance and disease relapse compared to detection by imaging, but did not find a significant relationship between pre-surgery ctDNA and overall survival [33]. As far as we are aware this is the first meta-analysis combining survival analysis between ctDNA detection and long-term outcomes and is the first review examining this effect in resectable disease across all disease stages. Despite the large volume of research on this topic, there remains a lack of consensus on a number of practical aspects. This resulted in considerable variability between studies, introducing heterogeneity into the analysis and was the main limitation of this review.
Post-operative ctDNA measurement could influence clinical management at a number of points. Recognition of patients at low-risk of relapse would enable identification of individuals in whom adjuvant therapy was unnecessary, whereas ctDNA measurement after completion of adjuvant treatment could be used to determining the need for further treatment [34, 35]. ctDNA could also be incorporated into ‘watch and wait’ protocol in rectal cancer following complete response to neoadjuvant chemotherapy. Liquid biopsy could also be incorporated into the assessment of response to other modalities of curative treatments including radical radiotherapy. Additionally, ctDNA could be used to guide post-treatment surveillance through identification of patients in whom more intensive monitoring is warranted.
There was little consensus across studies regarding timing of ctDNA sampling. Three studies measured ctDNA both post-surgery and after completion of adjuvant chemotherapy, demonstrating the post-chemotherapy time-point to be a stronger predictor of prognosis [31, 36, 37]. In order to be of clinical utility, detection of MRD should be performed at a time when it is possible to influence disease management. Delay in commencing adjuvant chemotherapy beyond eight weeks is associated with worse long-term outcomes [38], meaning that post-surgical ctDNA timings will be a critical consideration when being incorporated into treatment pathways. Analysis should be performed once ctDNA from the primary tumour has been cleared from the circulation. Clearance of ctDNA following surgery was investigated by Chen et al. through serial measurement in the immediate post-operative period following resection of lung cancer; they showed that ctDNA continues to decrease until three days post-surgery and that detection past this time point correlated better with prognosis [39]. Another important consideration in assay timing is that cfDNA rises with physiological stresses, including surgery. Henriksen et al. recently investigated the sequence of cfDNA and ctDNA post-operatively in colorectal and bladder cancer; they found that short cfDNA rose and remained significantly elevated for four weeks following surgery and recommend repeat ctDNA analysis at four weeks for any patients in whom ctDNA is not detected immediately post-op [40].
Gene panel selection remains a challenge in many aspects of precision oncology. There was a wide variation in the breadth of gene panels in this review as a result of the combination of PCR and NGS-based techniques. More comprehensive gene/mutation panels will enable detection of rarer mutations [41], but bring the possibility of false positives from CHIP [8]. Some of the studies in this review investigated presence of germline mutations either by sequencing DNA from peripheral blood leucocytes or based on the ctDNA VAF.
A tumour-informed approach was adopted by 16 studies, tracking previously identified mutations. This personalised approach brings the advantage of improved specificity whilst also achieving a high sensitivity using PCR-based assays [42]. However, the need for individualised assay development will be more logistically difficult to incorporate into routine care.
An alternative approach to identifying somatic mutations is to assess epigenetic changes. Although technically more challenging to measure, methylation changes are more consistent across a cancer type and occur early in the cancer pathophysiology. Four papers in this review assessed gene methylation [29, 30, 43, 44]. Parikh et al. investigated both genetic and epigenetic changes in NGS analysis of 103 patients undergoing curative surgery for stage I–IV colorectal cancer and concluded that integrating both genetic and epigenetic changes increases sensitivity for MRD detection [44].
Assay sensitivity is of significance in the setting of MRD, where disease bulk is low. Of our included studies, Suzuki et al. report the lowest LoD of 0.02% using ddPCR [27] (Table 2). In three studies, none of the cohort had detectable ctDNA after surgery [23–25] (Table 3), yet in all three studies, a subset of patients went on to relapse which may have represented ctDNA levels below the sensitivity of these assays. The majority of studies in this review measured pre-surgical ctDNA. In three studies, detection of ctDNA pre-surgery was a requirement for inclusion in the post-operative analysis [27, 45, 46], which may serve to remove ‘non-shedders’ or ‘low shedders’, a subset of patients whose tumour does not release ctDNA.
Statistical testing showed significant heterogeneity between studies, which is likely to affect the repeatability and external validity of this review. This remains the main limitation of this review and of application to clinical practice. Clinical heterogeneity will have arisen from differences in study design. Differences in the approach to removal of CHIP and requirement for ctDNA detection pre-operatively will have affected the pre-test probability of post-operative ctDNA detection. This review will also have been subject to methodological heterogeneity due to the range of assays used for ctDNA analysis. Subgroup analysis was performed to partially overcome this. There remained significant heterogeneity in subgroup analysis, probably as a result of the large number of contributing variables. Of note, statistical testing demonstrated no appreciable heterogeneity within the metastasectomy subgroup, confirming disease stage to be one of the sources of heterogeneity.
Many of the studies in this review were small and exploratory in nature. There was no minimum sample size for inclusion, resulting in the inclusion of a few studies with small numbers of patients. However, for inclusion in the meta-analysis there had to be sufficient participants for survival analysis calculation to be performed. Quality assessment looked at the likelihood of bias due to differences in the management of ctDNA-positive and -negative groups. For a ‘low bias’ score the treating clinicians had to be blinded to the ctDNA results, which was the case in 15 studies. A further significant source of bias would be confounding due to the effects of adjuvant chemotherapy with only 12 studies outlining the proportion of participants who received adjuvant chemotherapy. Overall, it was felt that bias due to the classification of interventions and measurement outcomes was low.
Funnel plot asymmetry was observed, suggestive of publication bias. This is likely due to inclusion of a number of smaller studies and was partly overcome by obtaining individual participant data where possible to calculate HRs. Whilst this might exaggerate the magnitude of effect, the fact that the association was consistently observed across the studies suggests a true relationship. In addition, sample size calculations were performed in four of the included studies, demonstrating that shorter PFS associated with ctDNA detection reaches statistical significance when suitably powered [29, 31, 37, 47]. Large scale observational trials are already underway to establish the prognostic implications of ctDNA detection following surgery. Preliminary results from the GALAXY trial demonstrated a significantly shorter PFS with ctDNA detection at both 4 and 12 weeks post-op, and a higher rate of ctDNA clearance with adjuvant chemotherapy [48]. Interventional trials are also underway investigating the effectiveness of ctDNA in directing adjuvant chemotherapy use [49] and recent results from the DYNAMIC trial demonstrated non-inferiority with ctDNA guided selection to adjuvant chemotherapy [50].
A further limitation of this review was the inclusion of participants with incomplete surgical resections within some of the studies, which would preclude the analysis of MRD. Inclusion of studies that did not test for matched germline mutations may have resulted in false positives due to CHIP. Patients who had undergone curative treatment by other modalities such as chemoradiotherapy were not included, as this was outside the scope of this review.
Conclusions
To conclude, ctDNA detection after curative surgery for colorectal cancer is a marker of poor prognosis. Here we demonstrate for the first time via meta-analysis that ctDNA detection post-operatively is associated with a significantly shorter PFS. Despite this wide body of evidence, there remains no consensus on many logistical aspects, most notably in the timing and method of analysis resulting in the considerable heterogeneity of this review and remains the greatest limitation to the clinical utility of this phenomenon.
Supplementary information
Author contributions
The review was designed by LGF with input from LMH and ALT. Database searching was carried out by CP. Processes in the systematic review were carried out by LGF and LMH. Manuscript was written by LGF with input from LMH, J.A.S. and AT. All authors approved the final manuscript.
Funding
LGF has received funding support from the National Institute of Health Research [ACF-2019-11-008]. LMH is supported by Cancer Research UK in conjunction with the UK Department of Health on an Experimental Cancer Medicine Centre grant [C10604/A25151].
Data availability
All data supporting the findings of this study are available within the article and Supplementary Files.
Competing interests
The authors declare no competing interests.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version contains supplementary material available at 10.1038/s41416-022-02017-9.
References
- 1.Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71:209–49. doi: 10.3322/caac.21660. [DOI] [PubMed] [Google Scholar]
- 2.CRUK. Bowel cancer incidence statistics. Cancer Research UK. 2017. https://www.cancerresearchuk.org/health-professional/cancer-statistics/statistics-by-cancer-type/bowel-cancer/incidence%0Ahttps://www.cancerresearchuk.org/health-professional/cancer-statistics/statistics-by-cancer-type/bowel-cancer/incidence#heading-One%250.
- 3.Dekker E, Tanis PJ, Vleugels JLA, Kasi PM, Wallace MB. Colorectal cancer. Lancet. 2019;394:1467–80. doi: 10.1016/S0140-6736(19)32319-0. [DOI] [PubMed] [Google Scholar]
- 4.Benson AB, Schrag D, Somerfield MR, Cohen AM, Figueredo AT, Flynn PJ, et al. American Society of Clinical Oncology recommendations on adjuvant chemotherapy for stage II colon cancer. J Clin Oncol. 2004;22:3408–19. doi: 10.1200/JCO.2004.05.063. [DOI] [PubMed] [Google Scholar]
- 5.Pantel K, Alix-Panabières C. Liquid biopsy and minimal residual disease - latest advances and implications for cure. Nat Rev Clin Oncol. 2019;16:409–24. doi: 10.1038/s41571-019-0187-3. [DOI] [PubMed] [Google Scholar]
- 6.Cheng F, Su L, Qian C. Circulating tumor DNA: a promising biomarker in the liquid biopsy of cancer. Oncotarget. 2016;7:48832–41. doi: 10.18632/oncotarget.9453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wan JCM, Massie C, Garcia-Corbacho J, Mouliere F, Brenton JD, Caldas C, et al. Liquid biopsies come of age: towards implementation of circulating tumour DNA. Nat Rev Cancer. 2017;17:223–38. doi: 10.1038/nrc.2017.7. [DOI] [PubMed] [Google Scholar]
- 8.Merker JD, Oxnard GR, Compton C, Diehn M, Hurley P, Lazar AJ, et al. Circulating tumor DNA analysis in patients with cancer: American society of clinical oncology and college of American pathologists joint review. J Clin Oncol. 2018;36:1631–41. doi: 10.1200/JCO.2017.76.8671. [DOI] [PubMed] [Google Scholar]
- 9.Kim M-J, Lee HS, Kim JH, Kim YJ, Kwon JH, Lee J-O, et al. Different metastatic pattern according to the KRAS mutational status and site-specific discordance of KRAS status in patients with colorectal cancer. BMC Cancer. 2012;12:347. doi: 10.1186/1471-2407-12-347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kogita A, Yoshioka Y, Sakai K, Togashi Y, Sogabe S, Nakai T, et al. Inter- and intra-tumor profiling of multi-regional colon cancer and metastasis. Biochem Biophys Res Commun. 2015;458:52–6. doi: 10.1016/j.bbrc.2015.01.064. [DOI] [PubMed] [Google Scholar]
- 11.Heitzer E, Haque IS, Roberts CES, Speicher MR. Current and future perspectives of liquid biopsies in genomics-driven oncology. Nat Rev Genet. 2019;20:71–88. doi: 10.1038/s41576-018-0071-5. [DOI] [PubMed] [Google Scholar]
- 12.Denis JA, Guillerm E, Coulet F, Larsen AK, Lacorte J-M. The role of BEAMing and digital PCR for multiplexed analysis in molecular oncology in the era of next-generation sequencing. Mol Diagn Ther. 2017;21:587–600. doi: 10.1007/s40291-017-0287-7. [DOI] [PubMed] [Google Scholar]
- 13.Chen M, Zhao H. Next-generation sequencing in liquid biopsy: cancer screening and early detection. Hum Genomics. 2019;13:34. doi: 10.1186/s40246-019-0220-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Keller L, Belloum Y, Wikman H, Pantel K. Clinical relevance of blood-based ctDNA analysis: mutation detection and beyond. Br J Cancer. 2021;124:345–58. doi: 10.1038/s41416-020-01047-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nicholson BD, Shinkins B, Pathiraja I, Roberts NW, James TJ, Mallett S, et al. Blood CEA levels for detecting recurrent colorectal cancer. Cochrane Database Syst Rev. 2015;10a:CD011134.. doi: 10.1002/14651858.CD011134.pub2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shinkins B, Nicholson BD, Primrose J, Perera R, James T, Pugh S, et al. The diagnostic accuracy of a single CEA blood test in detecting colorectal cancer recurrence: results from the FACS trial. PLoS ONE. 2017;12:e0171810. doi: 10.1371/journal.pone.0171810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lee JS, Rhee TM, Pietrasz D, Bachet JB, Laurent-Puig P, Kong SY, et al. Circulating tumor DNA as a prognostic indicator in resectable pancreatic ductal adenocarcinoma: a systematic review and meta-analysis. Sci Rep. 2019;9:1–7. doi: 10.1038/s41598-019-53271-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chae YK, Oh MS. Detection of minimal residual disease using ctDNA in lung cancer: current evidence and future directions. J Thorac Oncol. 2019;14:16–24. doi: 10.1016/j.jtho.2018.09.022. [DOI] [PubMed] [Google Scholar]
- 19.Cullinane C, Fleming C, O’Leary DP, Hassan F, Kelly L, O’Sullivan MJ, et al. Association of circulating tumor DNA with disease-free survival in breast cancer: a systematic review and meta-analysis. JAMA Netw Open. 2020;3:1–10. doi: 10.1001/jamanetworkopen.2020.26921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sterne JA, Hernán MA, Reeves BC, Savović J, Berkman ND, Viswanathan M, et al. ROBINS-I: a tool for assessing risk of bias in non-randomised studies of interventions. BMJ. 2016;355:i4919. doi: 10.1136/bmj.i4919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Page MJ, McKenzie JE, Bossuyt PM, Boutron I, Hoffmann TC, Mulrow CD, et al. The PRISMA 2020 statement: an updated guideline for reporting systematic reviews. BMJ. 2021;372:n71. doi: 10.1136/bmj.n71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stroup DF, Berlin JA, Morton SC, Olkin I, Williamson GD, Rennie D, et al. Meta-analysis of observational studies in epidemiology: a proposal for reporting. Meta-analysis Of Observational Studies in Epidemiology (MOOSE) group. JAMA. 2000;283:2008–12. doi: 10.1001/jama.283.15.2008. [DOI] [PubMed] [Google Scholar]
- 23.Levy M, Benesova L, Lipska L, Belsanova B, Minarikova P, Veprekova G, et al. Utility of cell-free tumour DNA for post-surgical follow-up of colorectal cancer patients. Anticancer Res. 2012;32:1621–6. [PubMed] [Google Scholar]
- 24.Lee S, Park Y-S, Chang W-J, Choi JY, Lim A, Kim B, et al. Clinical implication of liquid biopsy in colorectal cancer patients treated with metastasectomy. Cancers. 2021;13:2231. [DOI] [PMC free article] [PubMed]
- 25.Ng SB, Chua C, Ng M, Gan A, Poon PS, Teo M, et al. Individualised multiplexed circulating tumour DNA assays for monitoring of tumour presence in patients after colorectal cancer surgery. Sci Rep. 2017;7:40737. doi: 10.1038/srep40737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lindforss U, Zetterquist H, Papadogiannakis N, Olivecrona H. Persistence of K-ras mutations in plasma after colorectal tumor resection. Anticancer Res. 2005;25:657–61. [PubMed] [Google Scholar]
- 27.Suzuki T, Suzuki T, Yoshimura Y, Yahata M, Yew PY, Nakamura T, et al. Detection of circulating tumor DNA in patients of operative colorectal and gastric cancers. Oncotarget. 2020;11:3198–207. doi: 10.18632/oncotarget.27682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Khakoo S, Carter PD, Brown G, Valeri N, Picchia S, Bali MA, et al. MRI tumor regression grade and circulating tumor DNA as complementary tools to assess response and guide therapy adaptation in rectal cancer. Clin Cancer Res. 2020;26:183–92. doi: 10.1158/1078-0432.CCR-19-1996. [DOI] [PubMed] [Google Scholar]
- 29.Murray DH, Symonds EL, Young GP, Byrne S, Rabbitt P, Roy A, et al. Relationship between post-surgery detection of methylated circulating tumor DNA with risk of residual disease and recurrence-free survival. J Cancer Res Clin Oncol. 2018;144:1741–50. doi: 10.1007/s00432-018-2701-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Taieb J, Taly V, Henriques J, Bourreau C, Mineur L, Bennouna J, et al. Prognostic value and relation with adjuvant treatment duration of ctDNA in stage III colon cancer: a post-hoc analysis of the PRODIGE-GERCOR IDEA-France trial. Clin Cancer Res. 2021;27:5638–46. [DOI] [PubMed]
- 31.Tie J, Wang Y, Cohen J, Li L, Hong W, Christie M, et al. Circulating tumor DNA dynamics and recurrence risk in patients undergoing curative intent resection of colorectal cancer liver metastases: a prospective cohort study. PLoS Med. 2021;18:1–16. doi: 10.1371/journal.pmed.1003620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schøler LV, Reinert T, Ørntoft MBW, Kassentoft CG, Arnadøttir SS, Vang S, et al. Clinical implications of monitoring circulating Tumor DNA in patients with colorectal cancer. Clin Cancer Res. 2017;23:5437–45. doi: 10.1158/1078-0432.CCR-17-0510. [DOI] [PubMed] [Google Scholar]
- 33.Jones RP, Pugh SA, Graham J, Primrose JN, Barriuso J. Circulating tumour DNA as a biomarker in resectable and irresectable stage IV colorectal cancer; a systematic review and meta-analysis. Eur J Cancer. 2021;144:368–81. doi: 10.1016/j.ejca.2020.11.025. [DOI] [PubMed] [Google Scholar]
- 34.Coakley M, Garcia-Murillas I, Turner NC. Molecular residual disease and adjuvant trial design in solid tumors. Clin Cancer Res. 2019;25:6026–34. doi: 10.1158/1078-0432.CCR-19-0152. [DOI] [PubMed] [Google Scholar]
- 35.Dasari A, Grothey A, Kopetz S. Circulating tumor DNA-defined minimal residual disease in solid tumors: opportunities to accelerate the development of adjuvant therapies. J Clin Oncol. 2018. 10.1200/JCO.2018.78.9032. [DOI] [PMC free article] [PubMed]
- 36.Reinert T, Henriksen TV, Christensen E, Sharma S, Salari R, Sethi H, et al. Analysis of plasma cell-free DNA by ultradeep sequencing in patients with stages I to III colorectal cancer. JAMA Oncol. 2019;5:1124–31. doi: 10.1001/jamaoncol.2019.0528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tie J, Cohen JD, Wang Y, Christie M, Simons K, Lee M, et al. Circulating tumor DNA analyses as markers of recurrence risk and benefit of adjuvant therapy for stage III colon cancer. JAMA Oncol. 2019;5:1710–7. doi: 10.1001/jamaoncol.2019.3616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Petrelli F, Zaniboni A, Ghidini A, Ghidini M, Turati L, Pizzo C, et al. Timing of adjuvant chemotherapy and survival in colorectal, gastric, and pancreatic cancer. A systematic review and meta-analysis. Cancers. 2019;11:1–11. doi: 10.3390/cancers11040550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chen K, Zhao H, Shi Y, Yang F, Wang LT, Kang G, et al. Perioperative dynamic changes in circulating tumor DNA in patients with lung cancer (Dynamic) Clin Cancer Res. 2019;25:7058–67. doi: 10.1158/1078-0432.CCR-19-1213. [DOI] [PubMed] [Google Scholar]
- 40.Henriksen TV, Reinert T, Christensen E, Sethi H, Birkenkamp-Demtroder K, Gogenur M, et al. The effect of surgical trauma on circulating free DNA levels in cancer patients-implications for studies of circulating tumor DNA. Mol Oncol. 2020;14:1670–9. doi: 10.1002/1878-0261.12729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kinde I, Wu J, Papadopoulos N, Kinzler KW, Vogelstein B. Detection and quantification of rare mutations with massively parallel sequencing. Proc Natl Acad Sci USA. 2011;108:9530–5. doi: 10.1073/pnas.1105422108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Killock D. Personalized MRD assays and therapy? Nat Rev Clin Oncol. 2019;16:593. doi: 10.1038/s41571-019-0269-2. [DOI] [PubMed] [Google Scholar]
- 43.Leon Arellano M, García-Arranz M, Ruiz R, Olivera R, Magallares S, Olmedillas-Lopez S, et al. A first step to a biomarker of curative surgery in colorectal cancer by liquid biopsy of methylated septin 9 gene. Dis Markers. 2020;2020:9761406. [DOI] [PMC free article] [PubMed]
- 44.Parikh AR, Van Seventer EE, Siravegna G, Hartwig AV, Jaimovich A, He Y, et al. Minimal residual disease detection using a plasma-only circulating tumor DNA assay in colorectal cancer patients. Clin Cancer Res. 2021;27:5586–94. [DOI] [PMC free article] [PubMed]
- 45.Jin S, Zhu D, Shao F, Chen S, Guo Y, Li K, et al. Efficient detection and post-surgical monitoring of colon cancer with a multi-marker DNA methylation liquid biopsy. Proc Natl Acad Sci USA. 2021;118:e2017421118. [DOI] [PMC free article] [PubMed]
- 46.Benešová L, Hálková T, Ptáčková R, Semyakina A, Menclová K, Pudil J, et al. Significance of postoperative follow-up of patients with metastatic colorectal cancer using circulating tumor DNA. World J Gastroenterol. 2019;25:6939–48. doi: 10.3748/wjg.v25.i48.6939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tie J, Wang Y, Tomasetti C, Li L, Springer S, Kinde I, et al. Circulating tumor DNA analysis detects minimal residual disease and predicts recurrence in patients with stage II colon cancer. Sci Transl Med. 2016;8:346ra92. doi: 10.1126/scitranslmed.aaf6219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kotaka M, Shirasu H, Watanabe J, Yamazaki K, Hirata K, Akazawa N, et al. Association of circulating tumor DNA dynamics with clinical outcomes in the adjuvant setting for patients with colorectal cancer from an observational GALAXY study in CIRCULATE-Japan. J Clin Oncol. 2022;40:9. [Google Scholar]
- 49.Taniguchi H, Nakamura Y, Kotani D, Yukami H, Mishima S, Sawada K, et al. CIRCULATE-Japan: circulating tumor DNA-guided adaptive platform trials to refine adjuvant therapy for colorectal cancer. Cancer Sci. 2021;112:2915–20. doi: 10.1111/cas.14926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tie J, Cohen JD, Lahouel K, Lo SN, Wang Y, Kosmider S, et al. Circulating tumor DNA analysis guiding adjuvant therapy in stage II colon cancer. N Engl J Med. 2022;386:2261–72. doi: 10.1056/NEJMoa2200075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tarazona N, Gimeno-Valiente F, Gambardella V, Zuñiga S, Rentero-Garrido P, Huerta M, et al. Targeted next-generation sequencing of circulating-tumor DNA for tracking minimal residual disease in localized colon cancer. Ann. Oncol. 2019;30:1804–12. [DOI] [PubMed]
- 52.Tie J, Cohen JD, Wang Y, Li L, Christie M, Simons K, et al. Serial circulating tumour DNA analysis during multimodality treatment of locally advanced rectal cancer: a prospective biomarker study. Gut 2019;68:663–71. [DOI] [PMC free article] [PubMed]
- 53.Yamada T, Iwai T, Takahashi G, Kan H, Koizumi M, Matsuda A, et al. Utility of KRAS mutation detection using circulating cell-free DNA from patients with colorectal cancer. Cancer Sci. 2016;107:936–43. [DOI] [PMC free article] [PubMed]
- 54.Zhou J, Chang L, Guan Y, Yang L, Xia X, Cui L, et al. Application of Circulating Tumor DNA as a Non-Invasive Tool for Monitoring the Progression of Colorectal Cancer. PLoS One 2016;11:e0159708. [DOI] [PMC free article] [PubMed]
- 55.Zhou J, Wang C, Lin G, Xiao Y, Jia W, Xiao G et al. Serial Circulating Tumor DNA in Predicting and Monitoring the Effect of Neoadjuvant Chemoradiotherapy in Patients with Rectal Cancer: A Prospective Multicenter Study. Clin Cancer Res. 2021;27:301–10. [DOI] [PubMed]
- 56.Zou D, Day R, Cocadiz JA, Parackal S, Mitchell W, Black MA, et al. Circulating tumor DNA is a sensitive marker for routine monitoring of treatment response in advanced colorectal cancer. Carcinogenesis 2020;41:1507–17. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data supporting the findings of this study are available within the article and Supplementary Files.