Abstract
Tetracyclines were discovered in the 1940s and exhibited activity against a wide range of microorganisms including gram-positive and gram-negative bacteria, chlamydiae, mycoplasmas, rickettsiae, and protozoan parasites. They are inexpensive antibiotics, which have been used extensively in the prophlylaxis and therapy of human and animal infections and also at subtherapeutic levels in animal feed as growth promoters. The first tetracycline-resistant bacterium, Shigella dysenteriae, was isolated in 1953. Tetracycline resistance now occurs in an increasing number of pathogenic, opportunistic, and commensal bacteria. The presence of tetracycline-resistant pathogens limits the use of these agents in treatment of disease. Tetracycline resistance is often due to the acquisition of new genes, which code for energy-dependent efflux of tetracyclines or for a protein that protects bacterial ribosomes from the action of tetracyclines. Many of these genes are associated with mobile plasmids or transposons and can be distinguished from each other using molecular methods including DNA-DNA hybridization with oligonucleotide probes and DNA sequencing. A limited number of bacteria acquire resistance by mutations, which alter the permeability of the outer membrane porins and/or lipopolysaccharides in the outer membrane, change the regulation of innate efflux systems, or alter the 16S rRNA. New tetracycline derivatives are being examined, although their role in treatment is not clear. Changing the use of tetracyclines in human and animal health as well as in food production is needed if we are to continue to use this class of broad-spectrum antimicrobials through the present century.
The tetracyclines, which were discovered in the 1940s, are a family of antibiotics that inhibit protein synthesis by preventing the attachment of aminoacyl-tRNA to the ribosomal acceptor (A) site. Tetracyclines are broad-spectrum agents, exhibiting activity against a wide range of gram-positive and gram-negative bacteria, atypical organisms such as chlamydiae, mycoplasmas, and rickettsiae, and protozoan parasites. The favorable antimicrobial properties of these agents and the absence of major adverse side effects has led to their extensive use in the therapy of human and animal infections. They are also used prophylactically for the prevention of malaria caused by mefloquine-resistant Plasmodium falciparum. Furthermore, in some countries, including the United States, tetracyclines are added at subtherapeutic levels to animal feeds to act as growth promoters. Although the tetracyclines retain important roles in both human and veterinary medicine, the emergence of microbial resistance has limited their effectiveness. Undoubtedly the use of tetracyclines in clinical practice has been responsible for the selection of resistant organisms. Nevertheless, as we enter the new millennium, the use of tetracyclines and other antibiotics as animal growth promoters is becoming increasingly controversial because of concerns that this practice may be contributing to the emergence of resistance in human pathogens. The increasing incidence of bacterial resistance to tetracyclines has in turn resulted in efforts to establish the mechanisms by which genetic determinants of resistance are transferred between bacteria and the molecular basis of the resistance mechanisms themselves. The improved understanding of tetracycline resistance mechanisms achieved by this work has provided opportunities for the recent discovery of a new generation of tetracyclines, the glycylcyclines (see below). Further research, already under way, is also identifying approaches by which inhibitors of tetracycline resistance mechanisms might be developed for use in conjunction with earlier tetracyclines to restore their antimicrobial activity (185, 186).
The tetracyclines have been extensively reviewed both by the present authors (41, 43, 44, 100, 227–229) and others (56, 73, 263, 275). Nevertheless, in view of continuing interest in this group of antibiotics for both infectious and noninfectious diseases (95), we have decided to write a review that focuses on recent developments in the field.
DISCOVERY AND DEVELOPMENT OF THE TETRACYCLINES
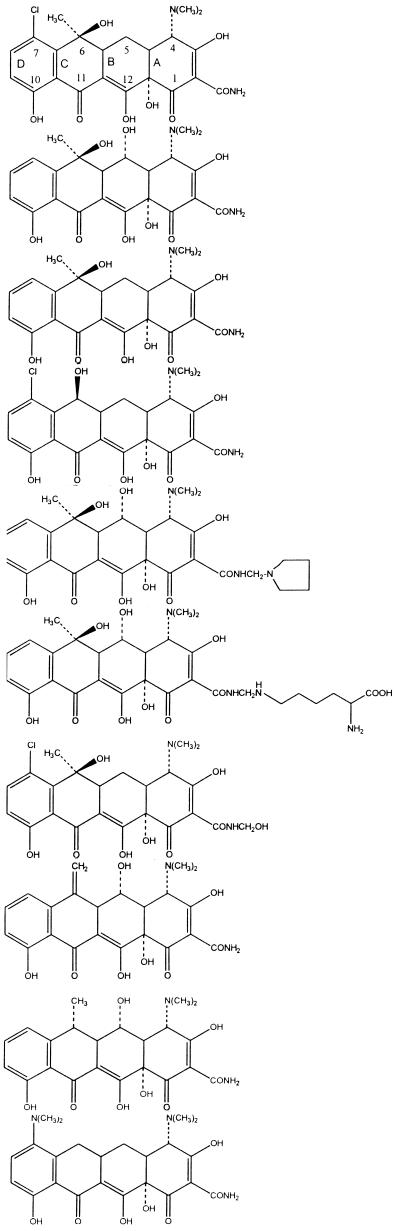
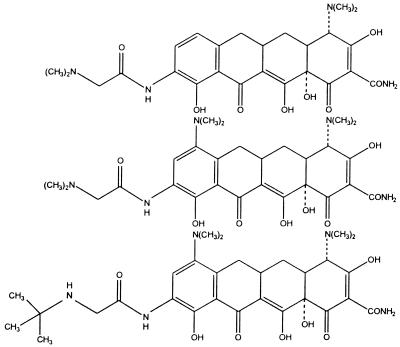
Chlortetracycline and oxytetracycline (Tables 1 and 2), both discovered in the late 1940s, were the first members of the tetracycline group to be described. These molecules were products of Streptomyces aureofaciens and S. rimosus, respectively. Other tetracyclines were identified later, either as naturally occurring molecules, e.g., tetracycline from S. aureofaciens, S. rimosus, and S. viridofaciens and demethylchlortetracycline from S. aureofaciens, or as products of semisynthetic approaches, e.g., methacycline, doxycycline, and minocycline. Despite the success of the early tetracyclines, analogs were sought with improved water solubility either to allow parenteral administration or to enhance oral absorption. These approaches resulted in the development of the semisynthetic compounds rolitetracycline and lymecycline (Tables 1 and 2). The most recently discovered tetracyclines are the semisynthetic group referred to as glycylcyclines, e.g., 9-(N,N-dimethylglycylamido)-6-demethyl-6-deoxytetracycline, 9-(N,N-dimethylglycylamido)-minocycline, and 9-t-(butylglycylamido)-minocycline (Tables 1 and 2). These compounds possess a 9-glycylamido substitutent (Table 2). The antibiotics in Tables 1 and 2 can be referred to as first-generation (1948 to 1963), second-generation (1965 to 1972), and third-generation (glycylcycline) tetracyclines. The 9-t-butylglycylamido derivative of minocycline (tigilcycline; formerly known as GAR-936) commenced phase I studies in October 1999 and is currently undergoing phase II clinical trials (113). Some of the earlier compounds, e.g., clomocycline, are no longer marketed, and others, e.g., rolitetracycline, lymecycline, and chlortetracycline, are not available in all countries (73, 137).
TABLE 1.
Principal members of the tetracycline class
| Chemical name | Generic name | Trade name | Yr of discovery | Status | Therapeutic administration |
|---|---|---|---|---|---|
| 7-Chlortetracycline | Chlortetracycline | Aureomycin | 1948 | Marketed | Oral |
| 5-Hydroxytetracycline | Oxytetracycline | Terramycin | 1948 | Marketed | Oral and parenteral |
| Tetracycline | Tetracycline | Achromycin | 1953 | Marketed | Oral |
| 6-Demethyl-7-chlortetracycline | Demethylchlortetracycline | Declomycin | 1957 | Marketed | Oral |
| 2-N-Pyrrolidinomethyltetracycline | Rolitetracycline | Reverin | 1958 | Marketed | Oral |
| 2-N-Lysinomethyltetracycline | Limecycline | Tetralysal | 1961 | Marketed | Oral and parenteral |
| N-Methylol-7-chlortetracycline | Clomocycline | Megaclor | 1963 | Marketed | Oral |
| 6-Methylene-5-hydroxytetracycline | Methacycline | Rondomycin | 1965 | Marketed | Oral |
| 6-Deoxy-5-hydroxytetracycline | Doxycycline | Vibramycin | 1967 | Marketed | Oral and parenteral |
| 7-Dimethylamino-6-demethyl-6-deoxytetracycline | Minocycline | Minocin | 1972 | Marketed | Oral and parenteral |
| 9-(t-butylglycylamido)-minocycline | Tertiary-butylglycylamidominocycline | Tigilcycline | 1993 | Phase II clinical trials |
TABLE 2.
Structures of the principal members of the tetracycline class
STRUCTURE-ACTIVITY RELATIONSHIPS
The structural features that confer antibacterial activity to the tetracyclines are well established (56, 179, 250) and will only be briefly discussed. More recently, however, new aspects of structure-activity relationships have emerged. This has followed efforts to extend the therapeutic utility of this antibiotic class to encompass bacteria expressing resistance to first- and second-generation compounds through ribosomal protection- and efflux-based mechanisms. This aspect is considered in more detail below.
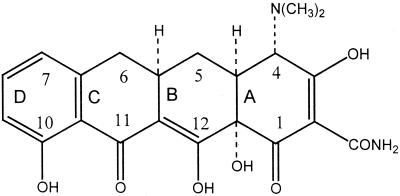
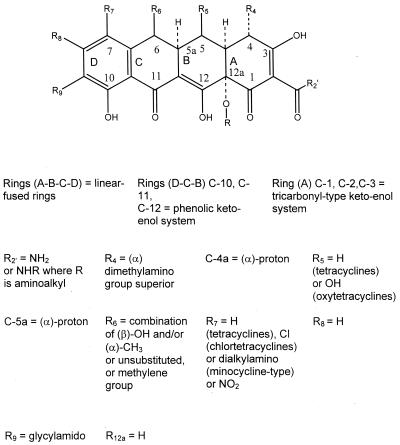
Tetracycline molecules comprise a linear fused tetracyclic nucleus (rings designated A, B, C, and D [Table 2]) to which a variety of functional groups are attached. The simplest tetracycline to display detectable antibacterial activity is 6-deoxy-6-demethyltetracycline (Fig. 1) and so this structure may be regarded as the minimum pharmacophore (179). Features important for antibacterial activity among the tetracyclines are maintenance of the linear fused tetracycle, naturally occurring (α) stereochemical configurations at the 4a, 12a (A-B ring junction), and 4 (dimethylamino group) positions, and conservation of the keto-enol system (positions 11, 12, and 12a) in proximity to the phenolic D ring. The tetracyclines are strong chelating agents (20, 44) and both their antimicrobial and pharmacokinetic properties are influenced by chelation of metal ions (see below). Chelation sites include the β-diketone system (positions 11 and 12) and the enol (positions 1 and 3) and carboxamide (position 2) groups of the A ring (20, 44). The newly discovered glycylcyclines, like other tetracycline derivatives, also form chelation complexes with divalent cations (278). Replacement of the C-2 carboxamide moiety with other groups has generally resulted in analogs with inferior antibacterial activity (179), probably because bacteria accumulate these molecules poorly (42). However, the addition of substituents to the amide nitrogen can impart significant water solubility, as in the case of rolitetracycline and lymecycline (Table 2). Dissociation of the prodrugs in vivo liberates free tetracycline (179, 250). Consistent with the above observations, substitutions at positions 1, 3, 4a, 10, 11, or 12 are invariably detrimental for antibacterial activity (179). Nevertheless, a number of other substitutions at different positions on the B, C, and D rings are tolerated, and molecules possessing these substituents have given rise to the tetracyclines in clinical use today, as well as the new glycylcycline molecules that are currently undergoing clinical trials (Table 2).
FIG. 1.
Structure of 6-deoxy-6-demethyltetracycline, the minimum tetracycline pharmacophore.
The extensive structure-activity studies referred to above revealed that with one exception, each of the rings in the linear fused tetracyclic nucleus must be six membered and purely carbocyclic for the molecules to retain antibacterial activity. For instance, the nortetracyclines, derivatives in which the B ring comprises a five-membered carbocycle, are essentially devoid of antibacterial activity (250). Nevertheless, 6-thiatetracycline, which possesses a sulfur atom at position 6 of the C ring, is an apparent exception to the rule that a purely carbocyclic six-membered ring structure is required for activity, since molecules in this series have potent antibacterial properties (43, 250). Nevertheless, it has now been established that the thiatetracyclines and a number of other tetracycline analogs, referred to collectively as atypical tetracyclines (43, 44), exhibit a different structure-activity relationship from the majority of tetracyclines. These molecules, which also include the anhydrotetracyclines, 4-epi-anhydrotetracyclines, and chelocardin, appear to directly perturb the bacterial cytoplasmic membrane, leading to a bactericidal response (43, 198, 199). This contrasts with the typical tetracyclines, which interact with the ribosome to inhibit bacterial protein synthesis and display a reversible bacteriostatic effect. The membrane-disrupting properties of the atypical tetracyclines are probably related to the relative planarity of the B, C, and D rings so that a lipophilic, nonionized molecule predominates. On interaction with the cell, the atypical tetracyclines are likely to be preferentially trapped in the hydrophobic environment of the cytoplasmic membrane, disrupting its function. These molecules are of no interest as therapeutic candidates because they cause adverse side effects in humans (250), which are probably related to their ability to interact nonspecifically with eukaryotic as well as prokaryotic cell membranes (43). A few other tetracycline molecules have been examined but have not moved on to further study (95, 185, 300).
There has been widespread emergence of efflux- and ribosome-based resistance to first- and second-generation tetracyclines (1, 2, 19, 38, 44, 70, 85, 103, 120, 122, 150, 165, 227–229, 307). To restore the potential of the tetracyclines as a class of useful broad-spectrum agents, a systematic search was undertaken during the early 1990s to discover new analogs that might possess activity against organisms resistant to older members of the class while retaining activity against tetracycline-susceptible organisms (295). This resulted in the discovery of the 9-glycinyltetracyclines (glycylcyclines) (15, 286, 287, 295) (Tables 1 and 2). Previous attempts to introduce substituents at position 9 of the molecule, e.g., 9-nitro, 9-amino, and 9-hydroxy, led to analogs with poor antibacterial activity (179, 250). However, during the 1990s, a team at Lederle Laboratories (now American Home Products) noted that 9-acylamido derivatives of minocycline exhibited antibacterial activities typical of earlier tetracyclines but without activity against tetracycline-resistant organisms (15). Nevertheless, when the acyl group was modified to include an N,N-dialkylamine moiety, e.g., as in the 6-demethyl-6-deoxytetracycline and minocycline derivatives (GAR-936) shown in Table 2, not only was antibacterial activity retained but also the compounds displayed activity against bacteria containing tet genes (Tables 3 to 5) responsible for both efflux of earlier tetracyclines [Tet(A)- to Tet(D) and Tet(K)] and ribosomal protection [Tet(M)] (15, 286, 287, 295). These findings were extended to the 9-t-butylglycylamido derivative of minocycline (Tables 1 and 2) (208). These data suggest that new structure-activity relationships may have been defined for activity against strains expressing efflux or ribosomal protection mechanisms that encompass the previous requirements for activity against tetracycline-susceptible strains but in addition require an N-alkyl glycylamido substitution at the 9 position of the molecule.
TABLE 3.
Mechanisms of resistance for characterized tet and otr genesa
| Genes |
|---|
| Efflux |
| tet(A), tet(B), tet(C), tet(D), tet(E), tet(G), tet(H), tet(I), tet(J), tet(Z), tet(30)btet(31)b |
| tet(K), tet(L) |
| otr(B), tcr3c |
| tetP(A) |
| tet(V) |
| tet(Y)d |
| Ribosomal protection |
| tet(M), tet(O), tet(S), tet(W) |
| tet(Q), tet(T) |
| otr(A), tetP(B),etetc |
| Emzymatic, tet(X) |
| Unknownf |
| tet(U), otr(C) |
TABLE 5.
Distribution of tetracycline resistance genes among gram-positive bacteria, Mycobacterium, Mycoplasma, Nocardia, Streptomyces, and Ureaplasmaa
| One determinant
|
Two determinants
|
Three or more determinants
|
|||
|---|---|---|---|---|---|
| Genus | Gene | Genus | Genes | Genus | Genes |
| Abiotrophia | tet(M) | Actinomyces | tet(L), tet(M) | Eubacteriumb | tet(K), tet(M), tet(Q) |
| Bacterionema | tet(M) | Aerococcus | tet(M), tet(O) | Bacillus | tet(K), tet(L), tet(M) |
| Gemella | tet(M) | Bifidobacteriumb | tet(M), tet(W) | Listeria | tet(K), tet(L), tet(M), tet(S) |
| Mycoplasmac | tet(M) | Gardnerella | tet(M), tet(Q) | Staphylococcus | tet(K), tet(L), tet(M), tet(O) |
| Ureaplasmac | tet(M) | Lactobacillus | tet(O), tet(Q) | Clostridiumb | tet(K), tet(L), tet(M), tet(P), tet(Q) |
| Nocarida | tet(K) | Mobiluncusb | tet(O), tet(Q) | Peptostreptococcusb | tet(K), tet(L), tet(M), tet(O), tet(Q) |
| Corynebacterium | tet(M), tet(Z) | Enterococcus | tet(K), tet(L), tet(M), tet(O), tet(S), tet(U) | ||
| Streptococcus | tet(K), tet(L), tet(M), tet(O), tet(Q), tet(T) | ||||
| Mycobacteriumd | tet(K), tet(L), tet(V), otr(A), otr(B) | ||||
| Streptomycese | tet(K), tet(L), otr(A), otr(B), otr(C), tcr3f, tetf | ||||
Based on information from references 12, 13, 26, 27, 33–38, 45, 47, 48, 62–66, 69, 74–76, 84, 85, 93, 96, 98, 103, 105, 108, 138a, 139, 140, 143, 150–153, 162, 164, 183, 184, 188, 200, 201, 206, 207, 209, 210, 212, 213, 220, 222, 225–230, 235–238, 242–245, 255, 273, 290, 297, and 307; and M. Roberts, unpublished results.
Anaerobic species.
Cell-wall-free bacteria with a gram-positive metabolism.
Acid-fast bacteria.
Multicellular bacteria.
tet and tcr have not been given number designations.
Figure 2 presents a summary of the features that confer optimum antibacterial activity to the tetracycline nucleus.
FIG. 2.
Stereochemical and substitution requirements for optimum antibacterial activity within the tetracycline series.
MODE OF ACTION
It is well established that tetracyclines inhibit bacterial protein synthesis by preventing the association of aminoacyl-tRNA with the bacterial ribosome (44, 263). Therefore, to interact with their targets these molecules need to traverse one or more membrane systems depending on whether the susceptible organism is gram positive or gram negative. Hence, a discussion of the mode of action of tetracyclines requires consideration of uptake and ribosomal binding mechanisms. Also pertinent to this discussion are explanations of the joint antibacterial-antiprotozoal activity of the tetracyclines and the microbial selectivity of the class as a whole. Most of these issues have been considered at length in recent years (44, 67, 78, 263), so the focus here will be on new information.
Tetracyclines traverse the outer membrane of gram-negative enteric bacteria through the OmpF and OmpC porin channels, as positively charged cation (probably magnesium)-tetracycline coordination complexes (44, 263). The cationic metal ion-antibiotic complex is attracted by the Donnan potential across the outer membrane, leading to accumulation in the periplasm, where the metal ion-tetracycline complex probably dissociates to liberate uncharged tetracycline, a weakly lipophilic molecule able to diffuse through the lipid bilayer regions of the inner (cytoplasmic) membrane. Similarly, the electroneutral, lipophilic form is assumed to be the species transferred across the cytoplasmic membrane of gram-positive bacteria. Uptake of tetracyclines across the cytoplasmic membrane is energy dependent and driven by the ΔpH component of the proton motive force (192, 263). Within the cytoplasm, tetracycline molecules are likely to become chelated since the internal pH and divalent metal ion concentrations are higher than those outside the cell (263). Indeed, it is probable that the active drug species which binds to the ribosome is a magnesium-tetracycline complex (44, 144). Association of tetracyclines with the ribosome is reversible, providing an explanation of the bacteriostatic effects of these antibiotics (44).
Several studies have indicated a single, high-affinity binding site for tetracyclines in the ribosomal 30S subunit, with indications through photoaffinity labeling and chemical footprinting studies that protein S7 and 16S rRNA bases G693, A892, U1052, C1054, G1300, and G1338 contribute to the binding pocket (44, 180, 196, 263). However, Schnappinger and Hillen (263) have pointed out that these apparent sites for drug interaction in the ribosome may not necessarily reflect the actual binding site. Indeed, interpretation of the probing studies referred to above is complicated by the observation that binding of tetracycline (which measures approximately 8 by 12 Å) to the ribosome appears to cause wide-ranging structural change in 16 S rRNA (193). Furthermore, photoincorporation methods are subject to the limitation that upon irradiation, tetracycline photoproducts are generated which may react further with the ribosomes (196). Nevertheless, naturally occurring tetracycline-resistant propionibacteria contain a cytosine-to-guanine point mutation at position 1058 in 16S rRNA (251) (see below), which does at least suggest that the neighboring bases U1052 and C1054 identified by chemical footprinting (180) may have functional significance for the binding of tetracyclines to the 30S subunit.
The absence of major antieukaryotic activity explains the selective antimicrobial properties of the tetracyclines. At the molecular level, this results from relatively weak inhibition of protein synthesis supported by 80S ribosomes (302) and poor accumulation of the antibiotics by mammalian cells (78). However, tetracyclines inhibit protein syntheses in mitochondria (221) due to the presence of 70S ribosomes in these organelles. It has been recognized for some time that the spectrum of activity of tetracyclines encompasses various protozoan parasites such as P. falciparum, Entamoeba histolytica, Giardia lamblia, Leishmania major, Trichomonas vaginalis, and Toxoplasma gondii (28, 44, 67, 137, 214). The antiparasitic activity is explained in some cases by the finding that certain organisms, e.g., P. falciparum, contain mitochondria (67). However, a number of other protozoa which lack mitochondria nevertheless remain susceptible to tetracyclines. At present there is no satisfactory molecular explanation for these findings (67).
RESISTANCE TO TETRACYCLINES
Introduction
Prior to the mid-1950s, the majority of commensal and pathogenic bacteria were susceptible to tetracyclines (144), as illustrated by the finding that among 433 different members of the Enterobacteriaceae collected between 1917 and 1954, only 2% were resistant to tetracycline (106). Studies of naturally occurring environmental bacteria, representative of populations existing before the widespread use of tetracyclines by humans (52), also support the view that the emergence of resistance is a relatively modern event that has followed the introduction of these agents for clinical, veterinary, and agricultural use. Indeed, resistance to tetracyclines has now emerged in many commensal and pathogenic bacteria due to genetic acquisition of tet genes. Subsequent parts of this section describe the genetic and biochemical mechanisms of tetracycline resistance, the regulation of resistance gene expression, and the distribution of tet genes in pathogenic and commensal bacteria. The impact of resistance on the use of tetracyclines for human medicine is examined later in the review.
Genetic and Biochemical Mechanisms of Tetracycline Resistance
Nomenclature of resistance determinants.
Mendez et al. (176) in 1980 first examined the genetic heterogeneity of tetracycline resistance determinants from plasmids from members of the Enterobacteriaceae and Pseudomonadaceae. They used restriction enzyme analysis, DNA-DNA hybridization, and expression of resistance to tetracycline and various analogs to categorize the tetracycline-resistant (Tcr) plasmids. Currently, two genes are considered related (i.e., of the same class) and given the same gene designation if they have ≥80% of their amino acid sequences in common. Two genes are considered different from each other if they have ≤79% amino acid sequence identity (150). This comparison can now be done using GenBank sequence information, since with few exceptions [tet(I) and otr(C)], representatives of all tet and otr genes have been sequenced and are available (150, 151) (Table 3).
For most genes, only one representative from each class has been sequenced, making comparisons easier to perform. One exception is the tet(M) gene, which has been sequenced from a number of gram-positive and gram-negative species (228). To determine the distribution of any particular tet or otr gene, it is now customary to prepare specific oligonucleotide probes that hybridize with the specific gene of interest but not to related genes. For example, if one is screening for the presence of the tet(M) gene, the oligonucleotide probes would not hybridize to tet(O) or tet(S) genes, which have approximately 78% sequence identity to the tet(M) gene (228). The number of tet genes has reached the end of the Roman alphabet, and numbers are being assigned to accommodate new tet genes (150). The first two genes with a number designation have now been assigned (Table 4). Stuart Levy has agreed to coordinate the assignment of proposed numbers for a new tetracycline resistance gene to prevent two distinct tet genes from being assigned the same numbers or two related genes (≥80% identity) being assigned different numbers (150).
TABLE 4.
Distribution of tet resistance genes among gram-negative bacteriaa
| Efflux
|
Ribosomal protectionb and/or efflux
|
||||||
|---|---|---|---|---|---|---|---|
| One gene
|
Two or more genes
|
One gene
|
Two or more genes
|
||||
| Genus | Gene | Genus | Genes | Genus | Gene | Genus | Genes |
| Actinobacillus | tet(B) | Edwardsiella | tet(A), tet(D) | Eikenella | tet(M) | Butyrivibrioc | tet(O), tet(W) |
| Erwinia | tet(B) | Providencia | tet(B), tet(E), tet(I) | Kingella | tet(M) | Mitsuokellac | tet(Q), tet(W) |
| Moraxella | tet(B) | Plesiomonas | tet(A), tet(B), tet(D) | Neisseria | tet(M) | Selenomonasc | tet(Q), tet(W) |
| Pantoea | tet(B) | Enterobacter | tet(B), tet(C), tet(D) | Campylobacter | tet(O) | Porphyromonasc | tet(Q), tet(W) |
| Treponemad | tet(B) | Mannheimia | tet(B), tet(G), tet(H) | Capnocytophagac | tet(Q) | Bacteroidesc | tet(M), tet(Q), tet(X) |
| Yersinia | tet(D) | Proteus | tet(A), tet(B), tet(C), tet(J) | Prevotellac | tet(Q) | Fusobacteriumc | tet(L), tet(M), tet(W) |
| Alcaligenes | tet(E) | Pseudomonas | tet(A), tet(C), tet(E), tet(G) | Haemophilus | tet(B), tet(K), tet(M) | ||
| Eubacteriumc | tet(K) | Serratia | tet(A), tet(B), tet(C), tet(E) | Veillonellac | tet(L), tet(M), tet(Q) | ||
| Agrobacterium | tet(30)e | Citrobacter | tet(A), tet(B), tet(C), tet(D) | Pasteurella | tet(B), tet(D), tet(H), tet(G), tet(M) | ||
| Klebsiella | tet(A), tet(B), tet(C), tet(D) | ||||||
| Shigella | tet(A), tet(B), tet(C), tet(D) | ||||||
| Salmonella | tet(A), tet(B), tet(C), tet(D), tet(G) | ||||||
| Aeromonas | tet(A), tet(B), tet(D), tet(E), tet(31)e | ||||||
| Vibrio | tet(A), tet(B), tet(C), tet(D), tet(E), tet(G) | ||||||
| Escherichia | tet(A), tet(B), tet(C), tet(D), tet(E), tet(I), tet(Y) | ||||||
Based on information from references 8, 9, 11, 14, 19, 40, 45, 55, 58–61, 81, 94, 97, 104, 111, 115, 116, 120, 122, 132, 139, 143, 149, 151, 152, 155, 163, 168–170, 182, 189, 200, 201, 206, 224, 227, 232, 233, 234, 241, 248, 256, 262, 267, 280, 282, 296, 314, 317, and 319, and M. Roberts, unpublished results).
Ribosomal protection genes have not yet been found in enteric genera, and when these genes are cloned into E. coli the level of resistance conferred is relatively low.
Anaerobic species.
T. denticola anaerobic species, but not all species in the genus are anaerobes.
Beginning of the number designations.
Twenty-nine different tetracycline resistance (tet) genes and three oxytetracycline resistance (otr) genes have been characterized. There is no inherent difference between a tetracycline and an oxytetracycline resistance gene. The oxytetracycline genes were first identified in oxytetracycline-producing organisms, and thus the nomenclature reflects the organisms first shown to carry the particular gene. We have shown that the tet genes are found in the producing Streptomyces spp. and the otr genes are found in the nonproducing Mycobacterium spp. (Table 5). Eighteen of the tet genes and one of the otr genes code for efflux pumps, and seven of the tet genes and one of the otr genes otr(A) code for ribosomal protection proteins (Table 3). The presence of both tet and otr genes with similar efflux or ribosomal protection mechanisms of resistance is consistent with the hypothesis of lateral gene transfer from the tetracycline-producing streptomycetes to other bacteria (16) (Table 5). The tet(P) gene is unusual because it consists of the tetA(P) gene, which encodes a functional efflux protein, linked to the tetB(P) gene, which appears to encode a ribosomal protection protein. tet(P) is counted as one gene in this review, although each component gene is listed in Table 3. tetA(P) has been found without tetB(P), but tetB(P) has not been found alone (164). The otr(C) gene has not been sequenced, while the tet(U) DNA sequence is unrelated to tetracycline efflux, tetracycline ribosomal protection proteins, or enzymatic protein (220, 228). The tet(I) gene has not been sequenced, but phenotypic studies suggest it encodes an efflux pump. It is therefore included in Table 3. An uncharacterized gene has also been described in a gram-negative species (116), and four different ribosomal protection genes, from streptococci, have been cloned using degenerate PCR primers (47). Whether these genes are related to some of the newer tet genes is not clear. The new ribosomal protection genes did not hybridize with tet(M), tet(O), tetP(B), tet(Q), tet(S), or tet(T), suggesting that at least some of the four could be novel, although hybridization with tet(W) was not examined. These five genes have not been characterized in detail and have not been added to Tables 3 to 5. The tet(X) gene encodes an enzyme which modifies and inactivates the tetracycline molecule (281). However, it does not seem to have much clinical relevance since it requires oxygen to function and is found only in strict anaerobes, where oxygen is excluded (281). Thus, it is unlikely that the tet(X) gene functions in its natural host (Bacteroides). No work has been done to determine whether tet(X) is associated with any aerobic species (229). The otr genes were first described in the antibiotic-producing Streptomyces species (63, 66, 218) but more recently have also been found in clinical Mycobacterium spp. (207) and may have a wider distribution among environmental species (Table 5).
There are reports of phenotypically Tcr isolates, which did not hybridize with any of the tet probes examined at the time (59–61, 222, 226, 228, 235). Some of these isolates may carry subsequently characterized tet genes that were not identified at the time of the original work or not thought to be relevant for screening, e.g., determining whether tet genes from gram-positive species were present in gram-negative species. We now know that increasing numbers of gram-negative bacteria carry what have been labeled gram-positive tet genes, such as tet(K), tet(L), tet(O), and tet(M), although these are not often examined when dealing with gram-negative isolates. Similarly, the tet(Q) gene, which was first described in the gram-negative genes Bacteroides, has a low G+C content, can be expressed in both gram-positive and gram-negative species, and is often associated with conjugative transposons. Therefore it should be considered whenever Tcr isolates are examined and not just when gram-negative anaerobes are screened (45, 46, 206) (Table 5). Another possibility is that these Tcr isolates carry novel genes that have yet to be described. Certainly, new tet genes are being identified, e.g., tet(Y) in Escherichia coli, tet(31) in Aeromonas, tet(W) in anaerobic species, and tet(Z) in Corynebacterium (Tables 4 and 5) (14, 150, 267).
Efflux proteins.
The efflux proteins are the best studied of the Tet proteins. The genes encoding then belong to the major facilitator superfamily (MFS), whose products include over 300 individual proteins (205). All the tet efflux genes code for membrane-associated proteins which export tetracycline from the cell. Export of tetracycline reduces the intracellular drug concentration and thus protects the ribosomes within the cell. Efflux genes are found in both gram-positive and gram-negative species (Table 4 and 5). Most of these efflux proteins confer resistance to tetracycline but not to minocycline or glycylcyclines. In contrast, the gram-negative tet(B) gene codes for an efflux protein which confers resistance to both tetracycline and minocycline but not glycylcyclines (44, 295). However, laboratory-derived mutations in tet(A) or tet(B) have led to glycylcycline resistance, suggesting that bacterial resistance to this group of drugs may develop over time and with clinical use (90; M. Tuckman, P. J. Petersen, and S. Projan, Abstr. 38th Intersci. Conf. Antimicrob. Agents Chemother., abstr. C98, p. 97, 1998).
Each of the efflux genes codes for an approximately 46-kDa membrane-bound efflux protein. These proteins have been divided into six groups based on amino acid sequence identity (173). Group 1 contains Tet(A), Tet(B), Tet(C), Tet(D), Tet(E), Tet(G), Tet(H), Tet(Z), and probably Tet(I), Tet(J), and Tet(30) (173, 291). The tetracycline resistance proteins in this group have 41 to 78% amino acid identity. Their tetracycline repressor proteins have 37 to 88% amino acid identity. In this group the proteins have 12 predicted transmembrane α-helices with long central nonconserved cytoplasmic loops connecting transmembrane helices 6 and 7. Among group 1, only Tet(Z) is found in gram-positive species; the others are found only in gram-negative isolates. Tet(Z) is the first gram-positive efflux protein to be described where regulation is controlled by a repressor protein (291). Most of the efflux proteins appear to reside in the lipid bilayer, with the hydrophilic amino acid loops protruding into the periplasmic and cytoplasmic space. The efflux proteins exchange a proton for a tetracycline-cation complex against a concentration gradient (318). The efflux genes from gram-negative bacteria have two functional domains, α and β, which correspond to the N- and C-terminal halves of the protein, respectively (253). Mutations in either half of the protein eliminate resistance, suggesting that residues dispersed across the protein are important for function. More recently, combined mutagenic and labeling approaches have been used to probe topology and structure-function relationships in the gram-negative MFS tetracycline transporter family. Mutations apparently affecting energy coupling have been located in cytoplasmic loops 2–3 and 10–11 of the efflux protein (174). Therefore, within the efflux proteins from gram-negative bacteria, these loops may interact functionally as the proton pump (175). The boundaries of membrane-embedded domains have been defined (126, 134), and the proposed topology of the proteins developed by earlier modeling methods has been confirmed experimentally (127). Furthermore, charge interactions between key residues such as arginine-70 and aspartate-120 in the Tet(B) protein have been identified as a requirement for correct positioning of transmembrane segments in the cytoplasmic membrane (279). Mutation studies have also been used to try to identify the antibiotic binding site within the efflux proteins from gram-negative bacteria. At least part of this site appears to reside in transmembrane helix 4 (110). Binding of the substrate to this region appears to affect the conformation of other regions in the protein, since substrate-induced conformational changes have been detected in transmembrane helices 1 and 11 (129). In a series of studies by Yamaguchi and coworkers, evidence has been obtained for a water-filled transmembrane channel in the Tet(B) efflux protein, flanked by transmembrane helices 2 and 5 and part of helix 4 (110, 128, 129).
Tetracycline efflux proteins have amino acid and protein structure similarities with other efflux proteins involved in multiple-drug resistance, quaternary ammonium resistance, and chloramphenicol and quinolone resistance, including methylenomycin A (MetA) from Streptomyces coelicolor, aminotriazole transport (Atr1) from Saccharomyces, and arabinose transport (AraB) from Escherichia coli (147, 269). Homology between the Tet and other efflux proteins has also been found with a new protein (EfpA) cloned from Mycobacterium tuberculosis (65).
The gram-negative efflux genes are widely distributed and normally associated with large plasmids, most of which are conjugative. They come from a number of different plasmid incompatibility groups (115, 176). These plasmids often carry other antibiotic resistance genes, heavy metal resistance genes, and/or pathogenic factors such as toxins (70). Thus, selection for any of these factors selects for the plasmid. This phenomenon of cross-selection has contributed to the dramatic increase in the number multiple-drug-resistant bacteria over the last 40 years (146, 226).
Group 2 includes Tet(K) and Tet(L), with 58 to 59% amino acid identity; these proteins are found primarily in gram-positive species. Group 2 has 14 predicted transmembrane α-helices. These genes code for proteins which confer resistance to tetracycline and chlortetracycline. Their presence is indicated when gram-positive bacteria are resistant to tetracycline but not to minocycline or glycylcyclines (287, 295). The tet(K) and tet(L) genes are generally found on small transmissible plasmids, which on occasion become integrated into the chromosome of staphylococci (84) or the chromosome of Bacillus subtilis (255) or into larger staphylococcal plasmids (184, 266). Large staphylococcal plasmids carrying the tet(K) genes are relatively uncommon, whereas small plasmids carrying the tet(K) genes are common. The small plasmids represent a family of closely related plasmids, which range in size from 4.4 to 4.7 kb (213). Plasmid pT181 is the prototype of the family and has been completely sequenced (124). The pT181 family of plasmids can carry antibiotic resistance genes other than tet(K) (124).
The large plasmid pJ3358 from Staphylococcus aureus, which codes for mupirocin resistance, carries a complete copy of plasmid pT181 flanked by directly repeating IS257 insertion sequences (184). Chromosomally integrated copies of plasmid pT181 also occur and are also flanked by IS257 elements (84, 252). More recently, we have examined four large staphylococcal plasmids from four species. In each case, a copy of pT181 was found within the large plasmid and was flanked by IS257 sequences (266). In one plasmid, part of the pT181 sequence has been deleted, while in other plasmids the complete pT181 plasmid sequence appears to be present as judged by Southern blot hybridizations. The tet(K) gene is most commonly found in S. aureus but is present in other Staphylococcus species (266). Most staphylococcal species also tend to carry tet(K), with the exception of Staphylococcus intermedius, which preferentially carries tet(M) (266).
A small number of plasmid-borne tet(L) genes have been sequenced and shown to have, in general, 98 to 99% sequence identity (265). One exception is the chromosomal tet(L) gene of B. subtilis (283). This gene has only 81% amino acid sequence identity to the other sequenced tet(L) genes and is just at the limit of what would be considered part of the tet(L) gene. The tet(K) and tet(L) genes can be found together in single isolates of streptococci (27) and C. difficile (243) but cannot be distinguished by their resistance phenotype to different tetracyclines.
Group 3 includes Otr(B) and Tcr3, both found in Streptomyces spp. These proteins have topology similar to group 2 proteins, with 14 predicted transmembrane α-helices. Group 4 includes Tet A(P) from Clostridium spp., with 12 predicted transmembrane α-helices, while group 5 includes Tet(V) from Mycobacterium smegmatis. Group 6 includes unnamed determinants from Corynebacterium striatum (which are not included in Table 3) and includes one protein which is believed to use ATP rather than a proton gradient as the energy source. More information on efflux proteins can be found in the recent chapter by McMurray and Levy (173).
Ribosomal protection proteins.
Nine ribosomal protection proteins are listed in Table 3. These are cytoplasmic proteins that protect the ribosomes from the action of tetracycline and confer resistance to doxycycline and minocycline. They confer a wider spectrum of resistance to tetracyclines than is seen with bacteria that carry tetracycline efflux proteins, with the exception of Tet(B). The ribosomal protection proteins have homology to elongation factors EF-Tu and EF-G (259, 292). The greatest homology is seen at the N-terminal area, which contains the GTP-binding domain. The Tet(M), Tet(O), and OtrA proteins reduce the susceptibility of ribosomes to the action of tetracyclines. The Streptomyces Otr(A) protein has greatest overall amino acid similarity to elongation factors. The mechanism of ribosomal protection works in vivo and in vitro, unlike the action of efflux proteins; which require intact membranes to function. Binding of the Tet(M) protein is not affected by tetracycline but is inhibited by thiostrepton, which also inhibits the binding of the EF-G protein (53). EF-G and the Tet(M) proteins compete for binding on the ribosomes, with Tet(M) having a higher affinity than EF-G. This suggests that these two proteins may have overlapping binding sites and that Tet(M) must be released from the ribosome to allow EF-G to bind (53).
The Tet(M) and Tet(O) proteins are the most extensively characterized of the ribosomal protection group (29–31, 292–294). They have been shown to have ribosome-dependent GTPase activity (173, 298). However, the Tet(M) protein could not replace the function of the EF-G protein in an E. coli isolate with a temperature-sensitive EF-G protein, at the nonpermissive temperature. The Tet(M) protein did not replace either the EF-G or EF-Tu proteins in an in vitro protein synthesis assay (31). The Tet(O) protein binds GDP and GTP. Site-directed mutations in the Tet(O) protein, which reduce the binding of GTP, were correlated with reduction in the susceptibility to tetracycline in isolates. This suggests that the GTP binding is important to the function of the Tet(O) protein (294).
Burdett (31) found that the Tet(M) protein allows the aminoacyl-tRNA to bind to the acceptor site of the ribosome in the presence of tetracycline concentrations that would normally inhibit translation. In the presence of the Tet(M) protein, tetracycline is apparently released from the ribosomes. In the presence of either the Tet(M) or the Tet(O) protein, tetracycline binding to the ribosomes is reduced when GTP but not GDP is present (298). Burdett (31) found that energy from GTP hydrolysis released the tetracycline from the ribosome when a nonhydrolyzabe GTP analog was used. In contrast, in related experiments with the Tet(O) protein, Trieber et al. (298) found that the data were more consistent with a role for GTP hydrolysis in the dissociation of the Tet(O) protein from the ribosomes. It is not clear if true differences between the Tet(M) and Tet(O) proteins exist, as suggested by these experiments from two different laboratories, or whether the results are due to differences in experimental details.
Although only two of the proteins from this group have been extensively examined, it has been assumed that the other proteins in the “ribosomal protection group” [Tet(S), Tet(T), Tet(Q), TetB(P), Tet(W), and Otr(A)] have GTPase activity and interact with tetracycline and the ribosomes in similar ways to those described for the Tet(M) and Tet(O) proteins, because of the similarities at the amino acid sequence level. The ribosomal protection proteins can be divided into groups based on amino acid sequence comparison. The first group includes Tet(M), Tet(O), Tet(S), and the newly described Tet(W) (14). The second group includes the Otr(A) and the TetB(P) proteins, while the third group includes the Tet(Q) and Tet(T) proteins (47, 273).
Like the tet(M) gene, tet(Q) is often associated with a large conjugative transposon which carries the erm(F) gene (encoding an rRNA methylase that confers erythromycin resistance) upstream from the tet(Q) gene (45, 46). Many of the different genera, including gram-positive, gram-negative, aerobic, and anaerobic bacteria, carried both tet(Q) and erm(F) in addition to an open reading frame (ORF) upstream of the erm(F) gene and two genes, rteA and rteB, downstream from the tet(Q) gene (45, 46). The rteA and rteB genes have been thought to play a role in the transfer of the conjugative element in Bacteroides (257). More recently, these genes have been found in a variety of gram-positive and gram-negative genera including Clostridium, Actinobacillus, Prevotella, Selenomonas, and Veillonella (45, 46).
The determinant TetP from Clostridium is unique because its gene consists of two overlapping genes; the first, tetA(P), encodes a classical efflux protein, and the second, tetB(P), encodes a protein which is related to the tetracycline ribosomal protection proteins (Table 5). No other gene with this organization has yet been described. tetA(P) is functional when separated from tetB(P). However, it is not clear whether the tetB(P) gene codes for a functional protein because cloned tetB(P), in both Clostridium perfringens and E. coli, expressed only a low-level resistance to tetracycline (273), which was lower than normally found when other ribosomal protection genes were cloned into these organisms and may not be functional in its natural host (26, 273).
The E. coli miaA gene encodes an enzyme that catalyzes the first step in the modification of A37 on tRNAs that read codons starting with U (173, 294). This is located near the anticodon and with modification decreases the rate of elogation, increases the number of errors at the first position of the codon, and decreases the number of errors at the third position. Mutations in miaA in the presence of Tet(M) reduce the level of tetracycline resistance in E. coli. However, this was not seen when the Tet(O) protein was examined in strains with this mutation (294). Mutations in the rpsL gene, which encodes the S12 ribosomal protein, also decrease tetracycline resistance in the presence of the Tet(M) and Tet(O) proteins.
The current data suggest that the ribosomal protection proteins bind to the ribosome. This causes an alteration in ribosomal conformation which prevents tetracycline from binding to the ribosome, without altering or stopping protein synthesis. The hydrolysis of GTP may provide the energy for the ribosomal conformational change. The ribosomal protection proteins also need to disocciate from the ribosome to allow EF-G to bind, since they have overlapping binding sites on the ribosome. A model for the way chromosomal mutations in the miaA and rpsL genes interfere with the function of the ribosomal protection proteins and reduce resistance to tetracycline can be found in reference 298.
Enzymatic inactivation of tetracycline.
The tet(X) gene (281) encodes the only example of tetracycline resistance due to enzymatic alteration of tetracycline. Two closely related anaerobic Bacteroides transposons containing the tet(X) gene have been described (281). The tet(X) gene was found because it is linked to erm(F), which codes for a rRNA methylase gene. The erm(F) gene was cloned into E. coli, and the clones were found to confer tetracycline resistance in E. coli when grown aerobically. The tet(X) gene product is a 44-kDa cytoplasmic protein that chemically modifies tetracycline in the presence of both oxygen and NADPH. Sequence analysis indicates that this protein has amino acid homology with other NADPH-requiring oxidoreductases and should not be able to function in the natural anaerobic Bacteroides host (281). It has not been found outside Bacteroides. However, to date no surveys have been conducted to assess the distribution of the tet(X) gene. Thus, even though the transposon carrying tet(X) and linked erm(F) is thought to be of gram-positive aerobic or facultative origin, a putative ancestor has not been identified.
Other/unknown mechanisms of resistance.
The tet(U) gene confers low-level tetracycline resistance (220). This gene encodes a 11.8-kDa protein containing 105 amino acids, which is smaller than the efflux proteins (45 kDa) and the ribosomal proteins (72 kDa) (see above). There is 21% similarity over the 105 amino acids between the Tet(U) and Tet(M) proteins, beginning close to the carboxy terminus of the latter. These similarities do not include the consensus GTP-binding sequences, which are thought to play a role in resistance in the Tet(M) and related proteins. However, the sequence is not really similar to either the efflux or ribosomal protection genes, and the mechanism is thus listed as unknown in Table 3.
The mechanism of resistance of the otr(C) gene from Streptomyces has not been determined because it has not yet been sequenced. It has been speculated that the otr(C) gene does not code for either an efflux or ribosomal protection protein. Whether otr(C) encodes an inactivation enzyme, similar to tet(X), or whether it has a novel mechanism of resistance like tet(U) has not yet been determined.
Regulation of Resistance Gene Expression
Efflux genes.
The gram-negative efflux determinants consist of two genes, one coding for an efflux protein and one coding for a repressor protein. Both genes are regulated by tetracycline. The two genes are oriented divergently and share a central regulatory region with overlapping promoters and operators (99). In the absence of tetracycline, the repressor protein occurs as a homodimer, which binds two α-helix–turn–α-helix motifs to the two tandemly orientated tet operators (99, 130). This blocks transcription of the structural genes for both the repressor and the efflux protein. Induction in the system occurs when a tetracycline-Mg2+ complex enters the cell and binds to the repressor protein. Drug binding changes the conformation of the repressor so that it can no longer bind to the DNA operator region. Only nanomolar concentrations of tetracycline are needed for binding to the repressor protein. This system is the most sensitive effector-inducible transcriptional regulation system yet described. After the repressor binds the tetracycline-Mg2+ complex, transcription of the efflux structural and repressor genes occurs. This is a relatively rapid process (99, 144). The tet gene in Tn10 is differentially regulated so that the repressor protein is synthesized before the efflux protein is expressed. The repressor protein will rebind to the DNA only when there is insufficient tetracycline (smaller than nanomolar amounts) present in the cell. This type of regulation most probably occurs with all the gram-negative efflux genes, tet(A), tet(C), tet(D), tet(E), tet(G), and tet(H), and probably also for the tet(I) gene. Crystallography has shown that the three α-helices at the N-terminal region of the repressor protein form the DNA-binding domain in the repressor molecule and that conformational changes in the repressor protein occur in the presence of tetracycline complexed with Mg2+ (130, 203). The tetracycline-binding pocket and the interaction between tetracycline and the repressor protein have also been characterized (99). The structural basis of tet(B) regulation is summarized in reference 203.
Three different strains of Haemophilus parainfluenzae were shown to carry constitutively expressed Tn10 (97). Subsequently it was shown that a truncated nonfunctional repressor protein due to a frameshift mutation in the repressor gene was present (97). This resulted in the constitutive expression of the Tet(B) protein. However, when a functional repressor was added to the cell, the tet(B) gene was inducible and regulated normally. The incidence of defective repressors in nature outside the genus Haemophilus has not been examined.
No repressor proteins have been found in gram-positive tet(K) or tet(L) genes. Upstream of the plasmid tetracycline tet(L) gene is a putative leader peptide with a potential stem-loop mRNA structures with two ribosome-binding sites (RBS), one that overlaps the leader peptide and one downstream that hides the RBS for the structural gene. This suggests that regulation is by translational attenuation when, in the absence of tetracycline, the ribosome binds to the first RBS (RBS1) and a short leader peptide is translated, which ends before the second RBS (RBS2). In the presence of tetracycline, a second stem-loop structure in the mRNA forms which uncovers the RBS2 site and allows the efflux protein to be translated, resulting in the cell becoming phenotypically resistant to tetracycline (265). This is a model similar to that found in regulation of the erm(C) gene, where significant work has been done on the regulation mechanism (inducible versus constitutive). Clinical isolates with either inducible or constitutively regulated erm(C) genes can be isolated. In one isolate, a tandem 26-bp direct repeat was found in the leader sequence, while in a second isolate, a deletion of the 107-bp segment of the leader region was found. In both cases these changes were thought to have converted an inducibly regulated leader region into a nonfunctional leader region, which resulted in constitutive production of the Erm(C) protein (310a).
Induction of the chromosomal tet(L) by tetracycline does not involve unmasking of an RBS and does not occur by the type of translational attenuation described for the plasmid tet(L) gene above. The normal induction from the RBS, for the leader peptide, is more efficient than induction of the RBS from the structural gene. Induction seems to involve an mRNA stem-loop, which overlaps part of the leader peptide sequence just upstream of the structural gene. A model for a tetracycline-promoted stalling of the ribosomes during translation of the early codons of the leader peptide has been proposed (283). This allows stabilization of the larger stem-loop structure, which than guides the ribosomes from the leader sequence to reinitiate translation at the RBS for the structural gene. Tetracycline induction also occurs at the transcriptional level but has not been well studied (173). Naturally occurring constitutive tet(L) plasmid genes have truncated leader peptides (173). This is consistent with both the translational attenuation and reinitiation models. The tet(L) gene also appears to be regulated by elevation in the pH and the presence of Na+ and K+. The sequences upstream of the tet(K) gene suggest regulation of protein synthesis by translational attenuation. Production of the Tet(K) protein is inducibe by tetraycline. However, like the Tet(L) protein, it may also be regulated by other factors (pH, Na+, or K+) as described above (173).
Ribosomal protection.
The expression of both Tet(M) and Tet(O) proteins appears to be regulated. Wang and Taylor (306) have suggested that the 400-bp region directly upstream from the coding region of the tet(O) gene was needed for full expression of the gene; however, the function of the region is not understood (292). Burdett (29) reported that the amount of Tet(M) protein increased when streptococci carrying the determinant were exposed to tetracycline. Similarly, Nesin et al. (188) found that preexposure to subinhibitory concentrations of tetracycline in S. aureus strains carrying the tet(M) gene resulted in both an increase in tetracycline resistance and an increase in the level of mRNA transcripts for tet(M). Su et al. (285) reported a stem-loop structure in the upstream region of the structural gene from Tn916, and both short and long transcripts were found by Northern blot analyses, similar to descriptions for attenuation of mRNA transcription of gram-positive proteins. Based on the DNA sequences from the upstream region in the Ureaplasma urealyticum tet(M) sequence, a similar stem-loop structure to that described for Tn916 would be possible (228). However, we have not looked at the transcripts from this gene.
We have sequenced the 4.9-kb Hincll fragment containing the tet(M) gene from U. urealyticum and Tn916 has also been completely sequenced (75). We have compared the upstream regions of seven tet(M) genes using the U. urealyticum gene as the standard. We found that the seven upstream regions examined had between 96 and 100% sequence identity to the U. urealyticum upstream DNA sequences (228). In contrast, when downstream regions were compared, sequence identity was more variable and ranged from 81 to 99%, with the S. aureus sequences being the most divergent (188). Finding little variability in the upstream sequences and more variability in the downstream regions is consistent with the hypothesis that the upstream regions are important for regulation, while no role has been described for sequences directly downstream of the structural gene. The sequences of both the upstream and downstream regions, as well as the structural genes, have G+C contents of <40%. This is despite the fact the genes were isolated from bacteria which have chromosomal G+C contents that varied from a low of 28% (U. urealyticum) to a high of 50% (Neisseria spp.). This is consistent with the hypothesis that the tet(M) genes have come from gram-positive bacteria and illustrates the spread of gram-positive genes into gram-negative species (227). This information, along with that of Wang and Taylor (306), is consistent with the hypothesis that tet(O) and tet(S) genes may also be regulated. However, the regulation of tet(Q) expression is unclear since most of the work has been done to determine the regulation of self-transfer and mobilization of both coresident plasmids and unlinked integrated elements, rather than expression of the Tet(Q) protein (152, 256, 257). There are also no direct data for regulation in tet(S). There have not been adequate downstream sequences (<100 bp) available in GenBank, and thus we could not compare downstream regions of tet(O), tet(S), and tet(Q) with the tet(M) downstream sequences.
Incidence of Tetracycline Resistance
Overview for pathogenic and opportunistic organisms.
Most work on bacterial resistance has either been conducted with pathogenic bacteria, which usually cause disease when present, or opportunistic bacteria, which occasionally cause disease (145, 146, 157, 160, 223, 229, 230, 232–244). Opportunistic bacteria are often part of the host's normal flora and can cause disease when they leave their normal sites. The majority of tet genes in bacteria have been associated with mobile plasmids, transposons, conjugative transposons, and integrons (gene cassettes) (176, 216, 224–228, 230, 232). These mobile units have enabled the tet genes to move from species to species and into a wide range of genera by conjugation (Tables 4 and 5). The gram-negative tet genes, first described in the Enterobacteriaceae and Pseudomonadaceae, are now also found in Neisseria, Haemophilus, Mannheimia, Treponema, and Vibrio (Table 4) (121, 132, 144, 149, 168, 182, 232, 233, 271). The tet(B) gene has the widest host range of the gram-negative tet genes and has been identified in 20 gram-negative genera (Table 4), while tet(M) is found in 26 genera including gram-negative and gram-positive bacteria (Tables 4 and 5). We have speculated that some genes, such as tet(E), may have a more limited host range because they are located on nonmobile plasmids, which reduces opportunities for transfer to other species and genera (61, 280). Some tet genes in some species or genera may confer low levels of resistance, which would be unlikely to protect the bacterial cell exposed to tetracyclines in clinical or environmental settings.
In 1953, the first tetracycline-resistant bacterium, Shigella dysenteriae, was isolated (70, 307). The first multiple-drug resistant Shigella was isolated in 1955. This later isolate was resistant to tetracycline, streptomycin, and chloramphenicol (4, 70, 155) and represented 0.02% of the isolates tested. By 1960, multiple-drug-resistant Shigella represented almost 10% of the strains tested in Japan (4, 70, 155), a dramatic increase in 5 years. The increase in multiple-drug resistant Shigella strains has continued to the present. The study by Lima et al. (155) showed that over 60% of the S. flexneri strains isolated between 1988 and 1993 were resistant to tetracycline, streptomycin, and chloramphenicol, which is the same combination of antibiotic resistance determinants found in the 1953 S. dysenteriae isolate. It was demonstrated that these antibiotic-resistant bacteria could transfer all their antibiotic-resistant phenotypes to susceptible isolates by cocultivation. This transfer was dependent on direct contact of the viable growing bacteria (308). We now know that the Japanese studies were the first reports of tetracycline resistance genes carried on conjugative R-plasmids. These tetracycline resistance genes conferred efflux of tetracycline from the cell and encoded the first of the three different types of tetracycline resistance mechanisms to be found in bacteria (228).
Isolation of Salmonella enterica serovar Typhimurium DT104 has become more common in recent years in both human and animal sources, and the isolates have been genetically characterized. Many of the isolates are multiple-drug resistant and carry a class 1 integron containing a variety of different antibiotic resistance genes including those encoding resistance to tetracycline (104, 189, 296). In a recent Canadian study (189), 10 human and 8 nonhuman isolates carried the tet(G) gene, along with genes conferring resistance to one or more other antibiotics including ampicillin, chloramphenicol, streptomycin, spectinomycin, and sulfonamide. One human isolate carried the tet(A) gene, and three carried tet(B) in place of the tet(G) gene. Multiple-drug resistance, which includes Tcr, has been identified in an increasing number of gram-negative pathogens and opportunistic bacteria.
Gram-positive species have also acquired Tcr, especially those that are multiple-drug resistant. A 1994 study (86) found that approximately 90% of the methicillin-resistant Staphylococcus aureus, 70% of Streptococcus agalactiae, 70% of multiple-drug resistant Enterococcus faecalis, and 60% of the multiple-drug resistant Streptococcus pneumoniae strains were now Tcr.
“Gram-negative tet genes” are those which have been found only in gram-negative bacteria. These genes have higher G+C contents (>40%) than those of gram-positive origin. All of the gram-negative tet genes encode efflux proteins and do not express well if moved into gram-positive hosts. Most of the gram-negative tet genes are regulated by a repressor, which is transcribed in the opposite direction from the structural gene. “Gram-positive tet genes” are those which are usually found in gram-positive species but, more importantly, have relatively low G+C contents (<35%). These genes are found in an increasing number of gram-negative species, including anaerobes (Table 4). This is especially true with the tet(M) gene, which has been identified in clinical isolates from 8 gram-negative genera and 18 gram-positive genera (Tables 4 and 5). The tet(M) gene has been conjugally transferred in the laboratory to an even larger group of species and genera than has been found in natural isolates (18, 35, 48, 74, 209, 274).
Previous work by Levy (146) found that long-term use of tetracycline selects not only for tetracycline-resistant gram-negative bacteria but also for multiple-drug-resistant gram-negative species. Tcr genes in both gram-positive and gram-negative species are often found on the same units (plasmids, transposons, or integrons) as other antibiotic resistance genes. For example, all chloramphenicol-resistant (Cmr) Haemophilus influenzae strains isolated in the 1970s and 1980s were also Tcr (111, 168, 282, 304). One hypothesis was that the first H. influenzae strains to obtain Cmr had acquired the Tcr Cmr transposon, which was then passed between strains and species. In contrast, some H. influenzae strains obtained tetracycline resistance transposons without the chloramphenicol gene and thus passed on only the Tcr. One plasmid had the tetracycline transposon inserted within one of the two inverted repeats of the chloramphenicol transposon (111). In another, the ampicillin transposon was integrated into the inverted repeats of the tetracycline transposon (111). Unfortunately, little work on antibiotic resistance plasmids from H. influenzae, or other Haemophilus spp., has been done recently.
Clewell and coworkers hypothesized that the transposon carrying the tet(M) gene, as typified by Tn916, was the original gram-positive conjugative transposon (75). It is suggested that over time other antibiotic resistance genes were inserted directly into this family of transposons, creating larger units carrying two to four different antibiotic resistance genes (35, 48, 108). This could be one explanation why the tet(M) gene is often linked to the erm(B) gene, which codes for an rRNA methylase and confers resistance to macrolides, lincosamides, and B streptogramins (MLSB). The combination of tet(M) and erm(B) genes is common in gram-positive streptococci, staphylococci, and enterococci (48). Similarly, a chloramphenicol acetyltransferase gene and an aminoglycoside phosphotransferase-encoding kanamycin resistance (Knr) gene, aph A-3, are often linked to tet(M) in the same transposon (35). The presence of these genes in common transposons may explain why Cmr and/or Knr Streptococcus pneumoniae strains have been isolated in North America recently, even though the use of chloramphenicol and kanamycin has essentially been discontinued in much of the industrialized world, including North America (64, 164). In addition, multiple conjugative transposons, which have one complete transposon inserted within another transposon, have been described in some of the cocci. These can transfer as a single unit, or the inserted transposon can be transferred separately, giving flexibility for transfer of antibiotic resistance genes (48, 140). Selection for any antibiotic on these multiple-drug-resistant units normally selects for the entire unit and may explain why the isolation of Tcr S. pneumoniae in children occurs even though tetracycline is not used in this age group (162, 289).
Obligatory intracellular pathogens such as Chlamydia and Rickettsia have not yet acquired tetracycline resistance. Since these bacteria grow only inside cells, it would require that cells be infected with two genera to allow gene exchange into the obligate intercellular pathogen. Mutations to increased tetracycline resistance would be more likely to occur in such intracellular bacteria. A few reports have described “heterotypic” tetracycline resistance in C. trachomatis when grown at high density (>5 × 104 IFU/ml), but there was no clear correlation of the phenotype to isolates from patients who did not respond to tetracycline therapy (277; R. B. Jones, B. Van der Pol, and B. F. Batteiger, Program Abstr. 30th Intersci. Conf. Antimicrob. Agents Chemother., abstr. 679, p. 199, 1990). On further examination, the phenotype was not stably transferred and was most probably an artifact of the growth conditions (Jones et al., 30th ICAAC). Lefevre et al. (142) described a C. trachomatis strain for which the tetracyline MIC was >64 μg/ml, but <1% of the population showed this resistance. Unfortunately, this isolate has not been further examined, and it is unclear whether the apparent resistance is also due to growth conditions or a permanent change, such as a mutation. More recently, Somani et al. (277) described what appear to be stable multidrug-resistant urogenital solates of C. trachomatis that included doxycycline-resistant strains expressing levels of resistance up to 4 μg of doxycycline/ml. These strains were responsible for treatment failures with antibiotics and caused relapsing or persistent infections. This appears to be the first report of clinically significant infection with C. trachomatis resistant to a member of the tetracycline class.
Overview for commensal microorganisms.
The commensal flora consists of microorganisms which are present in and on surfaces of a host and are not thought to cause disease. These organisms are often beneficial to the host, providing nutrients and inhibiting the growth of potential pathogens by preventing them from becoming established (10). Not surprisingly, these bacteria have the same tet genes, plasmids, transposons, conjugative transposons, and integrons as their disease-producing counterparts among the opportunistic and pathogenic bacteria. Many oral viridans streptococci (74, 93, 201) have acquired tet(M), tet(O), tet(L), or tet(K), as have the pathogenic streptococcal species S. pneumoniae and S. pyogenes. Most people today carry Tcr viridans streptococci in their mouth regardless of use of tetracycline therapy or age, while Tcr S. pneumoniae and S. pyogenes are significantly less common in most populations (162). This differs from isolates recovered before the introduction of tetracycline therapy, when the majority of bacteria were susceptible to tetracycline (12).
Another observation is that over time the gram-positive commensal bacteria have converted from carrying single tet genes to carrying multiple tet genes (13, 226, 236). The different tet genes can have either the same mode of action (efflux or ribosomal protection), or different modes of action (efflux and ribosomal protection), just like the pathogenic and opportunistic species do (230). The carriage of multiple tet genes of different classes is commonly found in individual gram-positive isolates (37, 236, 266, 267, 307) and in Mycobacterium spp. and Streptomyces spp. (207) but is uncommon in facultative gram-negative bacteria, especially enteric species (8, 60, 61, 81, 115, 168–170, 176). The reason for this is unknown, but a similar situation exists for the carriage of other antibiotic resistance genes (155). Differences in carriage of antibiotic resistance genes are found among different age groups within the same population and different areas of the world. This is why surveillance is needed for antibiotic resistance in commensal bacteria. In genera such as Neisseria, the commensal bacteria often carry more antibiotic resistance genes and acquired them earlier than the pathogenic N. gonorrhoeae and N. meningitidis (223, 234).
To stimulate the study of commensal bacteria, a new group recently came together to focus on antibiotic-resistant commensal bacteria and was named the Reservoirs of Antibiotic Resistance or (ROAR) project. The purpose of the group is to promote the study of carriage of antibiotic-resistant bacteria in humans, during food production and agricultural processes, and in the environment. The ROAR project is providing a source of information on resistance in commensal bacteria and can be found at. http://www.roar.antibiotic.org. Another Web site, which can be accessed from the ROAR site or directly, is The Alliance for the Prudent Use of Antibiotics (APUA) (http://www.APUA.org) (231). This is a nonprofit organization whose purpose is to promote proper antibiotic use and curb antibiotic resistance worldwide.
In general, it has been found that most disease-producing species of gram-negative genera carry the same tetracycline resistance genes as do the commensal species within the same genus. Many examples exist in the literature for the gram-negative genera Haemophilus (168), Neisseria (132), and Bacteriodes (45, 57, 143) and for gram-positive genera such as Streptococcus, which all carry the same tet genes as their pathogenic and opportunistic related species (45, 74, 140) Commensal Neisseria species carry an incomplete tet(M) transposon integrated into their chromosomal DNA while the pathogenic N. gonorrhoeae and N. meningitidis carry an incomplete tet(M) transposon integrated on a 25.2-MDa plasmid of gonococcal origin (132, 223). However, in the laboratory the 25.2-MDa plasmid can be transferred and maintained in the commensal Neisseria spp. (223). This is an exception, since in most other genera examined the tet genes are located in the same place (plasmid or chromosome) and often carry the same or related plasmids, transposons, or conjugative transposons (222, 230). In view of the above observations, it has been proposed that commensal bacteria act as a reservoir for tet and other antibiotic resistance genes found in, human pathogens and are thus very important in our understanding of how antibiotic resistance genes are maintained and spread through bacterial populations (222, 230).
Environmental bacteria such as Bacillus subtilis carry the same tet genes as clinical gram-positive bacteria (255, 283). Similarly, gram-negative bacteria isolated from aquaculture, including bacteria from fish (both suface and intestinal floras), water, marine sediment, and plants, all have the same tet genes as do clinical isolates (8, 59–61). Clinical isolates of streptomycetes, which may or may not be producing disease, have the same otr genes as do industrial strains (207).
Distribution and Mobility of tet Genes
Overview.
The tet genes are found in a variety of bacteria isolated from humans, animals, and the environment (Tables 4 and 5). The majority of the tet genes are associated with either conjugative or mobilizable elements, which may partially explain their wide distribution among bacterial species (115, 176, 216). The gram-negative tet efflux genes are found on transposons inserted into a diverse group of plasmids from a variety of incompatibility groups (115, 176). Gram-positive efflux genes are associated with small plasmids (124, 265, 266). The ribosomal protection genes tet(S) and tet(O) can be found on conjugative plasmids, or in the chromosome, where they are not self-mobile (36, 37, 162). The tet(M) and tet(Q) genes are generally associated with conjugative chromosomal elements, which code for their own transfer (48, 152, 256, 257). These conjugative transposons transfer mobilizable plasmids to other isolates and species and even unlinked genomic DNA (26, 48, 152, 166, 184, 272). Genes in the tet(Q) operon have been identified which mediate excision and circularization of discrete nonadjacent segments of chromosomal DNA in Bacteroides. The transfer origin (oriT) region of one of the Bacteroides conjugative transposons has been located near the middle of the conjugative transposon (152). Bacteroides conjugative transposons range from 65 to over 150 kb; most elements carry both tet(Q) and erm(F) and belong to a family of elements with the prototype being Tcr Emr DOT (257). A 16-kb region of the transposon is required and sufficient for conjugal transfer of the element and for mobilization of both coresident plasmids and unlinked integrated elements. DNA transfer is tetracycline regulated and mediated by at least three regulatory genes including a putative sensor (rteA), a putative regulator (rteB), and a third gene, rteC, which seems to stimulate transfer in an unknown fashion (152). This differs from the mechanism proposed for tetracycline regulation of conjugation of the Tn916 family of elements, where Manganelli et al. (166) suggest that tetracycline increases the number of circular intermediates present in the cell, which leads to more transconjugants. A gene can be induced to increase conjugal transfer by the presence of low doses of tetracycline. This has been illustrated in vitro with tet(M) and gram-positive cocci and rods (69, 222, 272) and in the gram-positive Listeria spp. (69). Transfer of the tet(Q) gene is also inducible by tetracyclines in Bacteriodes spp. (256, 257).
Movement of the Tn916-like and Bacteroides conjugative elements is hypothesized to involve a Rec-independent excision event which produces a nonreplicative circular intermediate that can insert at a different site within the cell or transfer to a new host by a conjugative plasmid-like process (48, 268). Integration of the Bacteroides conjugative transposon into a new host is relatively site specific, while the Tn916-like transposons can be relatively site specific or more randomly integrated into the host chromosome, depending on the host (121, 268). Integration into plasmids can occur (224), as can integration within conjugative transposons to create composite elements (48). These composite gram-positive elements are ≥50 kb and have been found in streptococci and enterococci. The prototype of the composite element is Tn3701, first described in S. pyogenes (140). In some of these composite elements, the central Tn916-like element can be removed and the nonhomologous segment can undergo conjugative transposition independently, as occurs in Tn5253 from S. pneumoniae (48). The structural organization of these composite elements varies. Both the composite and Tn916 families of elements can carry antibiotic resistance genes which confer resistance to chloramphenicol, erythromycin, and kanamycin, in addition to the tet(M) gene, which confers resistance to both tetracycline and minocycline (48, 140).
The Bacteroides conjugative transposons can mobilize resident plasmids either in trans or in cis. In trans the transposon provides all the proteins needed for mating and the plasmid provided the proteins that nick the plasmid and initiate plasmid transfer. In cis, the transposon provides the proteins needed for transfer (257). The Tn916 family can mobilize plasmids in trans (48, 183, 297). Low levels of tetracycline increase the transfer of these genes and also increase the ability of the bacterial host to spread antibiotic resistance genes to other isolates, species, and genera (48, 69, 297). With their transfer ability, one might hypothesize that the ribosomal protection genes would be found in virtually all tetracycline-resistant genera examined; however, this is not the case (Tables 4 and 5).
A few isolates of Haemophilus spp. and two clones of high-level tetracycline-resistant Moraxella catarrhalis carry the tet(B) gene in their chromosome (246, 247). The tet(B) gene is not conjugative in these isolates but can be moved by transformation using chromosomal DNA. More recently, we have found the tet(B) gene in Treponema denticola, an anaerobic spirochete thought to play an important role in periodontal disease (233). In the T. denticola isolates the tet(B) gene was nonmobile and not associated with a plasmid, suggesting that it was most likely to be located on the chromosome (233). This is the first description of a gram-negative efflux gene in a strictly anaerobic species. When a PCR product from the T. denticola tet(B) gene was sequenced, there was 90% DNA sequence identity between the gene from T. denticola and the tet(B) gene from Tn10. However, it has been difficult to assess whether the tet(B) gene confers tetracycline resistance in T. denticola, because of the organism's growth requirements. In contrast to the tet(B) gene, which is nonmobile, T. denticola does carry conjugative erm genes (233).
The tet(E) gene differs from the tet(A), tet(B), tet(C), and tet(D) genes because it is associated with large plasmids which are neither mobile nor conjugative (61, 280). This may explain its limited distribution and predominance in aquatic environments and its prevalence in polluted marine sediment (8) (Table 4). The tet(E) gene has also recently been associated with the chromosome (141). Jones et al. (115) found a correlation between the plasmid incompatibility group and the particular tet genes carried by the plasmid. They suggested that some of the tet genes may have become genetically linked to specific incompatibility and/or replication genes and thus the distribution of these genes could reflect the occurrence of particular incompatibility groups in particular genera or species. This hypothesis has not been thoroughly examined, and this relationship was not shown in an earlier study by Mendez et al. (176).
Gram-negative bacteria.
Currently 39 genera of gram-negative bacteria (Table 4) and 23 genera of gram-positive bacteria and related genera (Table 5) have been described in which the mechanism of tetracycline resistance has been determined. Other uncharacterized tet genes probably exist since studies have identified tetracycline-resistant isolates which do not carry any of the known tet genes (58, 61, 115, 225, 235, 244). New tet genes are continuing to be described (14, 47, 62, 150, 220, 267), and new genera have been identified which carry acquired tet genes (9, 45, 206, 207).
The tet(A), tet(B), tet(C), tet(D), tet(E), tet(G), tet(H), tet(I), tet(J), tet(Y), tet(30), and tet(31) genes are found exclusively in gram-negative genera (Table 4). Most of these genera belong to the facultative enteric group. The tet(B) gene has the widest host range among gram-negative species and has been identified in such diverse species as Actinobacillus actinomycetemcomitans, H. influenzae, Moraxella catarrhalis, and T. denticola (Table 4). Actinobacillus, Haemophilus, Moraxella, and Treponema species are all found in the oral area and the respiratory tract. The tet(B) gene is found on conjugative plasmids in Actinobacillus and Haemophilus (224, 248), and we have recently been able to show transfer of the tet(B)-carrying plasmid from A. actinomycetemcomitans to H. influenzae (248). The tet(B) gene is not mobile in the small number of Moraxella (246) and Treponema (233) isolates examined, but it would be interesting to determine whether either A. actinomycetemcomitans or Haemophilus spp. could transfer their tet(B) genes into either of these genera and if the transconjugants could then transfer the tet(B) gene.
Of the species which carry the tet(M) gene, some have complete conjugative elements, like Veilbonella parvula and Fusobacterium nucleatum, which are mobile, while another species, H. ducreyi, has a complete conjugative element integrated into a conjugative plasmid (224, 241). Other species carry nonconjugative incomplete elements in their chromosome, like Neisseria spp. and most Listeria innocua and Gardnarella vaginalis spp. (69, 105), or incomplete elements on conjugative plasmids, like N. gonorrhoeae (223, 239). In N. gonorrhoeae, the incomplete transposons are associated with 25.2-MDa conjugative plasmids (223). The 25.2-MDa conjugative plasmids have one of two different deletions of the tet(M) transposon (223, 317). One plasmid has a deletion downstream of a HincII site and is found in plasmids isolated from N. gonorrhoeae, Kingella denitrificans, and Eikenella corrodens (317). The second plasmid type has a deletion of over 800 bp upstream of the HincII site and is found in N. gonorrhoeae, N. meningitidis, and K. denitrificans isolates (317). Both types of plasmid confer resistance to ≥16 μg of tetracycline/ml and can transfer to other N. gonorrhoeae strains at frequencies ranging from 10−1 to 10−9 per recipient. Even though N. meningitidis has been found naturally with only one of the two plasmid types, both are readily transferred by conjugation into N. meningitidis (223). The 25.2-MDa plasmids can be stably transferred into a number of the commensal Neisseria spp. in the laboratory, but to date only two species, N. perflava/sicca and N. mucosa, have been found that naturally carry the tet(M) gene (223, 240). In all these isolates, an incomplete Tet(M) transposon located in the chromosome rather than on the 25.2-MDa plasmid has been found. This illustrates that plasmids or genes can be moved in the laboratory, resulting in stable transconjugants, but similar strains are not found in natural isolates.
Both 25.2-MDa plasmids can be transferred in the laboratory into Haemophilus spp., K. denitrificans, and E. corrodens but not into Moraxella catarrhalis, which carries nonmobile tet(B) rather than tet(M) (132, 240). Once the Neisseria 25.2-MDa plasmid has been transferred into Haemophilus spp., it is easily transferred among species within the genus Haemophilus but not back to Neisseria spp. (239). Similarly, an H. ducreyi plasmid which carries the complete Tet(M) transposon can be transferred among Haemophilus spp. but not outside the genus (224). This type of plasmid incompatibility may influence the spread of particular tet genes into new genera.
The tet(Q) gene was first described in human colonic Bacteroides spp. and is normally associated with conjugative elements (152, 256, 257). This gene is now found in a number of Prevotella spp. from the oral cavity and in other oral gram-negative genera such as Capnocytophaga, Mitsuokella, Selenomonas, and Veillonella (200, 201, 230). From other sites we found tet(Q) in gram-positive Clostridium, Gardnerella, and Peptostreptococcus spp. (45). The tet(Q) gene has been found in seven gram-positive genera (Table 5) and six gram-negative genera (Table 4); it is possible that the host range of the tet(Q) gene might be similar to that found for the tet(M) gene.
The number of different tet genes found in a particular gram-negative genus varies from one tet gene carrying either tet(B) (efflux), tet(M), or tet(Q) (both ribosomal protection), to a maximum of seven different tet efflux genes found in the genus Escherichia (Table 4). However, Jones et al. (115) and Mendez et al. (176) found that no E. coli plasmids carried more than one type of tet gene. In contrast, Streptomyces and gram-positive genera often have individual isolates that carry multiple genes coding for the same antimicrobial resistance including tetracycline (207, 307). The tet(G) (11, 319), tet(H) (94), and tet(I) genes (116) have not been extensively examined for their distribution in gram-negative bacteria. It is clear that many environmental (8, 52) and food and animal (14, 40, 46, 55, 60, 61, 101, 102, 133, 135, 141, 266, 276, 280) isolates are tetracycline resistant. However, these bacteria have not yet been as extensively examined as those associated with human disease. Little work has been done to elucidate why some genera carry only a single tet gene, while members of other genera can carry a variety of different tet genes. More work is required to better understand the factors, which influence not only whether a particular genus or species of bacteria will carry tetracycline resistance genes but also whether they will encode efflux, ribosomal protection, or both.
The majority of gram-negative isolates described in the literature carry a single type of tet gene, although it may occur on multiple plasmids. This was evident from the earliest studies of the distribution of tet genes, when it was found that only 3.5% of the lactose-fermenting coliforms carried two different tet genes (169). Similar results have been found in bacteria isolated from catfish and their environment (59–61), in Shigella spp. isolated in Mexico (170), and in S. enterica serovar Typhimurium isolated in Africa (120). In fact, the only study where large numbers of gram-negative species were reported to carry more than a single tet gene was the recent study of polluted marine sediments from Norway (8). In that study, 26% of tetracycline-resistant isolates carried both tet(D) and tet(E) genes.
The incompatibility groups of gram-negative plasmids, which determines their bacterial host range, has been studied (115, 176). Plasmid host ranges vary from very restrictive, such as with the large conjugative Haemophilus R-plasmids (224, 247), which do not readily survive outside their own genus, to broad, which allow the plasmid to survive in diverse host backgrounds (2). These differences in plasmid host range may influence the spread of particular tet genes associated with them.
Gram-negative anaerobic genera and some nonenteric gram-negative genera like Neisseria, Eikenella, and Kingella most commonly or exclusively carry ribosomal protection genes. One exception is the genus Haemophilus, especially H. influenzae and H. parainfluenzae, where all of the early isolates (from the 1970s) carried the tet(B) efflux gene (168). Unfortunately, with the use of the H. influenzae b vaccine, interest in this organism has faded and no recent studies have been performed to see whether current isolates also contain only tet(B). A few strains of H. aphrophilus, isolated from periodontal patients in the 1990s, carry the tet(K) gene (206), while H. ducreyi can carry either tet(B) or tet(M) (168, 224) (Table 4). A few isolates of V. parvula have been found which carry either tet(L) or tet(Q); however, most of the isolates examined carry the tet(M) gene (45, 206). Different ribosomal protection genes have been found in tetracycline-resistant Bacteroides spp. (45, 57, 152), in addition to the tet(X) gene (281). A variety of different tet genes have been found within the genus Pasteurella (40, 94) (Table 4).
Eight gram-negative genera currently carry tet(M) genes, seven carry tet(Q), five carry tet(W) genes, two carry tet(O), two carry tet(K), and one each carries tet(H), tet(I), tet(J), tet (Y), tet(30), and tet(31) (Table 4). Based on their low G+C content and their regulation (see “Regulation of resistance gene expression” above), tet(K), tet(L), tet(M), tet(O), tet(P), tet(S), tet(T), tet(Q), tet(W), and maybe tet(Z) are thought to be gram-positive origin. This strengthens the hypothesis that antibiotic resistance genes from gram-positive species, especially the ribosomal protection genes, have been exchanged throughout the bacterial population without regard to species or genus and can be successfully integrated and expressed in a variety of bacterial host backgrounds (18, 24, 229).
Gram-positive bacteria.
Seventeen Gram-positive genera, two cell wall-free genera (Mycoplasma and Ureaplasma), Actinomyces, Nocadia, Mycobacterium, and Streptomyces all carry known tetracycline resistance genes (Table 5). However, as shown above with gram-negative isolates, not all tetracycline-resistant gram-positive bacteria have been correlated with possession of specific known tet genes (244). A total of 18 genera carry the tet(M) gene, 11 carry tet(K), 10 carry tet(L), 7 carry tet(Q), 5 carry tet(O), 2 carry tet(S), and 1 carries tet(P). Recently, Bifidobacterium spp. have been shown to carry the tet(W) gene (267) and Corynebacterium carries either tet(M) or the newly described tet(Z) (150). The new tet(U) gene is found in Enterococcus (220), the tet(T) gene is found in Streptococcus (47), and the tet(V) gene is found in Mycobacterium smegmatis (62) (Table 5).
The tet(K) and tet(L) genes are widely distributed among gram-positive species associated with humans, animals, and the soil (Table 5) and have been found in rapidly growing Mycobacterium, Norcardia, and Streptomyces spp. (65, 207) isolated from patients (Table 5). This is the first time that acquisition of an antibiotic-resistance determinant has been documented in Mycobacterium and Nocardia spp., and it suggests that gene exchange between tetracycline-resistant gram-positive bacteria, Mycobacterium spp., Nocardia spp., and Streptomyces spp. has occurred. The hypothesis that Streptomyces spp. exchange antibiotic resistance genes with other genera is strengthened by the finding of the otr genes, from industrial Streptomyces spp., in clinical isolates of Mycobacterium and Streptomyces spp. (207). This is consistent with the hypothesis that some of the tet genes could be ancestrally related to genes found in the antibiotic-producing Streptomyces spp. (16). Work is still needed to determine the host range of the otr genes.
The tet(M) gene is often associated with a conjugative element of the Tn916-Tn1545 family (48). This group of elements form nonreplicating circular intermediates, which are essential for both intracellular transposition and intercellular conjugative transfer (75). Both types of movements occur by an excision-integration process, where excision and formation of a covalently closed circular molecule precedes movement of the conjugative element. In both Tn916 and Tn1545, imperfect inverted repeats (20 of 26 bp) are present at the ends of the transposon and integration occurs without duplication of the target DNA sequence (48, 268). More recently, Manganelli et al. (166) have shown that the number of circular intermediates varies in different Enterococcus faecalis strains. Their work indicates that the number of circular intermediates influences the frequency of conjugation between 5.1 × 10−8 and 2.8 × 10−6, while Rice et al. (219) demonstrated that the number of circular intermediates increased when the strains were grown in the presence of tetracycline.
Clewell et al. (48) determined that the Tn916 family of elements was found naturally or could be transferred in the laboratory into over 50 different species representing 24 bacterial genera. However, the host range of the Tn916 family is even greater (Tables 4 and 5). The tet(M) gene can also be introduced into a significant number of other genera including gram-negative and gram-positive organisms and species lacking cell walls (18, 24, 69, 237, 274, 297). In a majority of gram-positive species, the tet(M) gene is found in the chromosome, most often on conjugative elements (48, 140, 226). Little has been done with the tet(S) gene other than to show transfer between L. monocytogenes and E. faecalis at frequencies ranging from 10−4 to 10−9 (37). The tet(O) gene is not associated with conjugative elements, is mobile only when found on conjugative plasmids, and can be transferred among streptococci and Campylobacter (27, 293). The tet(Q) genes have been identified in gram-positive species (Table 5), and we have recently shown that they are located on conjugative elements in these species, where they are often linked with the erm(F) gene (45, 46).
Distribution of Other Resistance Determinants
Efflux systems.
Bacteria have a number of innate chromosomally encoded proteins, which transport molecules in and out of the cell. These have been divided into groups which include the MFS, resistance-nodulation-cell division (RND) family, the small multidrug resistance (SMR) family, and the ATP-binding cassette transport family (21, 204). Some, but not all of these efflux pumps confer resistance to tetracycline (190, 191). An external layer of peptidoglycan surrounds gram-positive bacterial cytoplasmic membranes, while gram-negative bacteria have both peptidoglycan and an outer membrane outside the cytoplasmic membrane. Consequently, exported molecules need to go through these layers. It is hypothesized that a membrane fusion protein (MFP) is connected to the outer membrane protein channel, enabling transport of small molecules across the outer membrane of gram-negative bacteria. The pumps of the RND, MFS, and SMR families use the proton motive force as the driving force for efflux. In contrast, the ATP-binding cassette transporters use ATP hydrolysis (205). Interest in these efflux pumps has increased dramatically over the last few years, and there are a number of recent reviews dealing with them (21, 190, 191, 205).
A number of the RND pumps have tetracycline as a substrate (191). RND efflux pumps have 12 predicted transmembrane segments and are unrelated to the MFS family, of which the above-described tet efflux pumps are members. The RND pumps are found mainly in gram-negative bacteria, and most are involved in pumping multiple ligands (antibiotics, toxic metal ions, etc.) out of the bacterial cell (191). Many of the RND efflux pumps are associated with a linker protein and an outer membrane channel. RND pumps which transport tetracycline comprise the Acr system found in E. coli, the multiple Mex systems found in Pseudomonas aeruginosa, the Mtr system found in N. gonorrhoeae, and the Mtr-like system recently found in Stenotrophomonas maltophilia (6). P. aeruginosa has a number of different RND efflux pump systems including MexA-MexB-OprM. Strains which overproduce the efflux operon are more resistant to tetracycline, chloramphenicol, and quinolones, while mutants with deletions in this operon are hypersensitive to these antibiotics. Another operon, MexC-MexD-OprM, also confers resistance to tetracycline, chloramphenicol, and quinolones (21, 87, 153).
A related MexA-MexB-OprM operon has been isolated from multiple-drug-resistant Burkholderia cepacia (32), while a similar system has been identified in multiple-drug resistant Campylobacter jejuni (39). An analogous system (the mtr system) has been found in N. gonorrhoeae (161). Production of the MtrCDE efflux proteins is controlled by transcription with both cis- and trans-acting factors involving the mtrR gene. The MtrR protein has amino acid similarities to the tetracycline repressors regulating the gram-negative efflux genes tet(A) to tet(I). Missense or deletion mutations within the mtrR coding region result in increased transcription of the mtrCDE genes, protein production, and increased resistance to antibiotics and other agents (161). The N. gonorrhoeae mtr system makes these isolates clinically resistant to a variety of antibiotics, and its prevalence has increased in N. gonorrhoeae populations since its discovery in the 1970s.
A chromosomal tetracycline efflux system associated with multiple-antibiotic resistance (the mar locus) has been described in E. coli (147). The mutation in the marR region of the chromosome enhances intrinsic resistance to a large group of antibiotics including tetracycline. The level of tetracycline resistance increases as the cells are grown continually in the presence of tetracycline. MarA is a transcriptional activator of a common group of promoters (5, 147). The mar locus appears to be widely established in bacteria (5, 147). Mutants expressing high levels of MarA show decreased accumulation of tetracycline (147). The efflux is dependent on the expression of marA. Inactivation of the region creates a susceptible phenotype (147). Weak acids, uncouplers, and antibiotics such as tetracycline induce expression of the mar operon. The regulator MarR negatively controls the expression of the mar operon. Overexpression of MarA causes decreased expression of the OmpF porin and increased expression of the multiple-drug efflux pump AcrAB, another operon in E. coli that affects antibiotic resistance (197). Spontaneous multiple-drug-resistant mutants of K. pneumoniae, which have increased resistance to a range of unrelated antibiotics including tetracycline, have also been described (82). Similar mutants have been found in Serratia marcescens, Enterobacter spp., and Campylobacter (39). These mutants exhibited efflux of tetracycline, similar to the E. coli Mar mutants. An acrAB homolog coding for a functional multiple-drug efflux pump has been described in H. influenzae (258), although disruption of the region did not produce detectable changes in tetracycline susceptibility. However, only one strain, with disruption in two different genes, was examined (258). Currently, none of the RND systems has been associated with natural transposable elements. The effect of this has been to limit the spread of these systems and hence, in general, to limit their importance in clinical tetracycline resistance. However, whether any of these will, or could, be picked up by integron-like elements is unknown. If this does happen, these new elements would allow for transfer between strains and species and more rapid spread through the bacterial population of these mutant genes.
The SMR family codes for proton-dependent efflux pumps with a multiple-drug proton antiport mechanism. The E. coli emrE gene produces a protein which confers resistance to monovalent cations and antibiotics including tetracycline (205).
Point mutations.
Recently 15 Tcr clinical isolates (MICs, 2 to 64 μg of tetracycline per ml, 1 to 32 μg of doxycycline per ml) of cutaneous propionibacteria were found to have a cytosine instead of guanine at position 1058 in the 16S rRNA (251). This change was associated with increased tetracycline resistance. This region of the 16S rRNA, known as helix 34, is important for peptide chain termination and translational accuracy. The clinical significance of these strains is not clear at present. Mutations which alter the permeability of the outer membrane porins and/or lipopolysaccharides can also affect bacterial susceptibility to tetracycline and other agents (21, 154). How often these mutations occur and whether they are of clinical importance has not been established.
APPLICATIONS OF TETRACYCLINES
Administration and Pharmacokinetic Behavior in Humans
Tetracyclines are usually administered orally, although some are also available as parenteral products (Table 1) (73, 137). Rolitetracycline is available only as a parenteral product. The ability to use either oral or parenteral formulations of doxycycline has been used advantageously to permit switching programs from intravenous to oral administration (51).
The dosing regimens and pharmacokinetic properties of the tetracyclines have been extensively reviewed in previous publications (73, 137, 313). Therefore only a brief summary is provided here. Absorption following oral administration occurs largely in the stomach and proximal small intestine and is influenced by the presence of food, milk, or divalent cations, particularly, calcium, with which tetracyclines form nonabsorbable chelates. Levels achieved in serum after normal oral dosing are in the range of 2 to 5 μg/ml, and most tetracyclines have to be given four times daily to maintain therapeutic concentrations in the serum. However, the long elimination half-lives of doxycycline and minocycline permit once- or twice-daily dosing. Tetracyclines generally penetrate moderately well into body fluids and tissues and are excreted in the urine. For instance, levels in sputum about 20% of those in serum, can be achieved, which explains why the tetracyclines have a role in the treatment of respiratory tract infections. Tetracyclines also penetrate into the sebum and are excreted in perspiration, properties which contribute to their usefulness in the management of acne.
Human Therapy and Prophylactics
Although 9-(t-butylglycylamido)-minocycline, a third-generation compound, is currently undergoing clinical trials, it is now nearly 30 years since the last tetracycline, minocycline, was introduced (Table 1). During this period, as already discussed, there have been increases in the incidence of bacterial resistance to the tetracyclines and in availability of more active and better tolerated agents from different antimicrobial classes. Consequently, in recent years the clinical use of tetracyclines has significantly declined in most countries since they are no longer drugs of choice in many instances (44, 73, 79, 137). However, in other cases new applications have been identified. For instance, tetracycline has been used as part of a triple therapy for management of gastritis and peptic ulcer disease associated with Helicobacter pylori. Although omeprazole, clarithromycin, and amoxicillin (or metronidazole) are standard therapy, the role of tetracycline may increase as more clarithromycin- and methronidazole-resistant H. pylori isolates are encountered (303). Tetracyclines are active against malaria, and this has unexpectedly become important for prophlylaxis following the rapid increase of mefloquine-resistant P. falciparum strains (28, 211, 264).
Table 6 provides a summary of current anti-infective applications of the tetracyclines in humans. These antibiotics have also been evaluated for their potential in other situations. However, such applications may not necessarily gain widespread acceptance or become components of standard therapeutic regimens. For instance, on the basis of limited clinical evaluation, Ji et al. (119) consider that minocycline, in combination with ofloxacin, may have a role in the treatment of leprosy. Although larger clinical trials would be needed to establish benefit, it seems likely that a combination of minocycline and ofloxacin could provide the opportunity for supervised monthly administration of these antibiotics, thereby greatly improving patient compliance.
TABLE 6.
Current applications of the tetracyclines for therapy and prophylaxis of human infectionsa
| Infection for which tetracyclines are:
| |
|---|---|
| First choice | Acceptable alternative to other agentsb |
| Respiratory | |
| Atypical pneumonia due to Mycoplasma pneumoniae, Chlamydia pneumoniae, C. psittaci | Community-acquired pneumoniac |
| Infective exacerbations of chronic bronchitisc | |
| Legionellosis (doxycycline) | |
| Bowel | |
| Cholera | |
| Prophylaxis of traveler's diarrhea | |
| Genital | |
| Nongonococcal urethritis | Syphilis |
| Cervicitis | Epididymitis |
| Lymphogranuloma venereum | Prostatitis |
| Pelvic inflammatory disease | |
| Granuloma inguinale | |
| Local and systemic | |
| Rocky mountain spotted fever | MRSA |
| Endemic and epidemic typhus | MRSE (minocycline) when vancomycin or other agents inappropriate |
| Q fever | Plague |
| Brucellosis (in combination with rifampin or streptomycin) | Tularerira |
| Lyme disease | Bartonellosis |
| Relapsing fever | Leptospirosis |
| Periodontal infection (topical therapy with tetracycline or minocycline) | Whipple's disease Cutaneous infections caused by Mycobacterium marinum and in multiple-drug regimens for ocular infections caused by M. cheloni |
| Acne vulgaris (topical and systemic treatment) | |
| Gastritis caused by Helicobacter pylori (tetracycline in multiple-drug regimens). | |
| Prophylaxis of mefloquine-resistant Plasmodium falciparum malaria | |
Based on information from references 3, 73, 79, 88, 107, 118, 131, 171, 275, 301, 303, 305, and 313.
MRSA, methicillin-resistant Staphylococcus aureus; MRSE; methicillin-resistant Staphylococcus epidermidis.
Except in situations where there is a high rate of resistance among pneumococci and/or H. influenzae.
Veterinary Medicine
The tetracyclines have applications for the treatment of infections in poultry, cattle, sheep, and swine. In some cases, e.g., for therapeutic treatment of large numbers of poultry reared on commercial farms, the antibiotics are added directly to feed or water or can be administered in aerosols. The use of tetracyclines in the rearing of farm animals has been reviewed in recent years (44, 91) and readers are referred to these earlier papers for details. Tetracyclines are also used for treatment of infections in domestic pets (135, 146).
Animal Growth Promoters
Antibiotics represent one of the few classes of drugs that can be used in food animals both therapeutically to treat disease and subtherapeutically, usually over long periods, to improve their rate of growth and feed conversion efficiency. The practice of adding low concentrations of antibiotics, defined in the United States as < 200 g/ton of feed (50, 83, 109), to animals to improve growth and feed efficiency is referred to as growth promotion or growth enhancement (44, 50, 83, 91, 109, 113a). An obvious outcome of this practice is that animals need less food to reach marketable weight. The mechanisms responsible for growth promotion have not been fully elucidated but appear to include enhancement of vitamin production by gastrointestinal microorganisms, elimination of subclinical populations of pathogenic organisms, and increased intestinal absorption of nutrients (50).
The growth-promoting properties of tetracyclines were discovered in 1949, when it was observed that low levels of chlortetracycline in livestock rations beneficially affected the rate of growth and feed utilization by young chickens (284). The initial observations in chickens were confirmed and soon extended to swine and cattle, leading to the development of both chlortetracycline and oxytetracycline as animal growth promoters (91). In the United States these antibiotics were approved by Food and Drug Administration as feed additives in 1951 (chlortetracycline) and 1953 (oxytetracycline) (109). Increasing concerns about growth promoters followed the publication in the United Kingdom of the Swann report in 1969 (288). This report, suggesting that subtherapeutic application of tetracyclines and other antibiotics to farm animals might contribute to the development of resistant human isolates, led to a ban of the use of tetracyclines for growth promotion in Europe in the early 1970s (EC directive 70/524; see http://europa.eu.int/comm/dg24/health/sc/index_en.html). Although issues surrounding the use of growth-promoting antibiotics have been widely discussed in other countries, particularly the United States (50, 83, 91, 136, 146), and Australia (113a), no such ban has been imposed on the use of tetracyclines for growth promotion in these and many other countries.
Other Uses
Tetracyclines are used in aquaculture to control infections in salmon, catfish, and lobsters (59, 109, 146). They are also sprayed onto fruit trees and other plants to treat infection by Erwinia amylovara, injected into palm trees to treat mycoplasma infections (lethal yellow), and used to control infection of seeds by Xanthomonas campestis (black rot) (146; http://europa.eu.int/comm/dg24/health/sc/index_en.html). They also have applications in the treatment of insects of commercial value; e.g., oxytetracycline is used to treat foulbrood disease of the honeybee, which is caused by either Bacillus larvae or Streptococcus pluton (146).
QUANTITIES OF TETRACYCLINES USED
Introduction
Tetracyclines are one of the cheapest classes of antibiotic available, and their cost in real terms is declining due to improved manufacturing technology (159). The pricing structure makes them particularly attractive for use in developing nations (73). Furthermore, the HIV Meeting 2000 suggested the use of tetracyclines to reduce bacterial sexually transmitted diseases in the developing world. Nevertheless, the emergence of bacterial resistance to tetracyclines, the development of alternative agents that are better tolerated and more potent, and the introduction in some countries of legislation to prevent the use of tetracyclines as animal growth promoters are factors influencing the usage of tetracyclines. Although reliable data on production and consumption of antibiotics, including tetracyclines, are notoriously difficult to obtain (49, 109, 112), such data where available can assist analyses of trends in relation to the clinical and legislative factors mentioned above (109, 159). Furthermore, such data provide an indication of the extent to which continuing selection pressure for the emergence of tetracycline resistance is being imposed on human and veterinary pathogens and commensal bacteria (109). The following sections present data on human and animal consumption of tetracyclines. It has not been possible to obtain data relating to quantities used in aquaculture and agriculture.
Current data on the total quantities of tetracyclines used in human therapy and prophylaxis have, with the exception of Australia, New Zealand, and Norway (Table 7), been difficult to acquire. As discussed above, tetracyclines are of value primarily in the prophylaxis and treatment of community-acquired infections, with a more minor role for nosocomial infections. Therefore it is expected that the use of tetracyclines in humans (Table 7) primarily represents community use. A recent report on the use of antibiotics in Dutch hospitals certainly supports this view (112). The use of tetracyclines for community medicine is declining in many countries, with reduced prescription rates recently recorded for tetracyclines in Spain (23), the United States (172), and the United Kingdom (54). In the United Kingdom this followed an earlier period of decline between 1967 and 1984 (44). A similar downward trend in Norway has occurred, where annual consumption of tetracyclines for therapeutic use in humans fell from 3,185 kg in 1992 to 2,191 kg in 1996 (89).
TABLE 7.
Estimates of human and animal consumption of tetracyclines in a number of countries during the mid-1990sa
| Region | Tetracycline consumption (kg/yr) by:
|
|
|---|---|---|
| Human | Farm animals | |
| United States | NAb | 3,488,000c |
| Canada | NA | 398,000c |
| Europe | NA | 2,294,000d |
| Norway | 2,191 | NA |
| Australia | 12,677 | 77,619c |
| New Zealand | 1,343 | 2,311c |
Based on information from references 89 and 113a, http://europa.eu.int/comm/dg24/health/sc/index_en.html, and http://www.maf.govt.nz/ACVM/.
NA, not available.
Does not distinguish between therapeutic and subtherapeutic levels.
Therapeutic use only.
Veterinary Medicine and Animal Growth Promotion
As discussed above, tetracyclines have applications both in veterinary medicine and as animal growth promoters. Current data on their use in farm animals are presented in Table 7. These data do not distinguish between therapeutic and subtherapeutic uses. However, in countries where use of tetracyclines at subtherapeutic levels in animal feed is still permitted, it can be estimated from earlier data (109) that approximately 90% of tetracyclines administered to cattle and swine are used at subtherapeutic concentrations whereas only 15% of usage in poultry reflects subtherapeutic administration. On the basis of animal consumption data, the use of tetracyclines in farm animals appears to be increasing in the United States, since approximately 2.6 × 106 kg was consumed in 1985 (109), rising to current levels of 3.5 × 106 kg (Table 7). This probably reflects an increase in the numbers of farm animals being raised following a switch from grain farms to pig and cattle farms due to the low prices of cereal crops.
RESISTANCE DEVELOPMENT AND ITS IMPLICATIONS
Introduction
Although bacterial resistance to tetracyclines has emerged in plant and fish pathogens as a consequence of using these antibiotics to control disease (144), it is the development of resistance in the context of human and animal use that has raised the greatest levels of concern. Since the tetracyclines have been used in humans and animals for some 50 years, it is not surprising that the selection pressure resulting from their use has resulted in the emergence of resistant bacterial variants, particularly those containing the tet genes described above. The emergence of resistance in human and veterinary pathogens has already had consequences for the use of tetracyclines as therapeutic agents. However, one of the biggest issues to emerge in recent years concerns the use of tetracyclines as animal growth promoters and the implications of this practice for human health. These topics are addressed in the following sections.
Resistance Following Human Therapy and Prophylaxis
The reported rates of bacterial resistance to tetracyclines have varied widely on the basis of geographical locale and year of isolation (137). However, by the mid-1970s resistance to tetracycline was common among the Enterobacteriaceae, staphylococci, streptococci, and bacteroides (144). In some locations resistance rates have been very high. For instance, in a Boston Hospital in 1969, 38% of S. aureus, 61% of E. coli, 62% of Klebsiella spp., 58% of Enterobacter spp., 91% of Proteus spp., and 97% of Serratia spp. were resistant to tetracyclines (254). Comparable, high rates of resistance were also recorded in Bacteroides fragilis and H. influenzae in the early 1980s, both in the United States and in Europe (137). The emergence of resistance to the tetracyclines in human clinical isolates has severely limited the further utility of these drugs and has undoubtedly been an important factor in the declining use of these antibiotics, in most countries, for the therapy of human infections (44, 73, 137). Fortunately, resistance has not yet become a problem for most situations (Table 6) where tetracyclines are still the drugs of choice, e.g., as demonstrated by the apparent absence of resistance in Brucella melitensis (3) and Coxiella burnetii (171) and low rates or only sporadic reports of resistance in periodontal bacteria (139), in Helicobacter pylori (58, 138, 177), and possibly Chlamydia trachomatis (142, 277). However, this situation may change in light of the recommendations for tetracyclines as first-line agents for acne (Table 6), since resistance rates as high as 25% have recently been reported for cutaneous propionibacteria (114). However, resistance in the propionibacteria is due to mutations rather than acquisition of tet genes (251).
For other antibiotic classes there is some relationship between use and the emergence of resistance in human isolates (261, 309; W. L. Nelson, J. N. Kuritsky, D. L. Kennedy, and C. S. Lao, Program Abstr. 27th Intersci. Conf. Antimicrob. Agents Chemother., abstr. 455, p. 175, 1987). Since tetracyclines are still used quite extensively in certain developing countries, is there evidence that the incidence of resistance is higher in these countries than in regions where use has declined? Recent extensive data on the global antibiotic resistance patterns of community-acquired lower respiratory tract pathogens (71) permit such an analysis. In centers in Brazil, Mexico, and South Africa the incidence of resistance in S. pneumoniae for 1996 and 1997 is relatively high, ranging between 7.2 and 27.5%. Nevertheless, similar ranges in resistance rates, with possibly higher upper levels (5.3 to 39.4%) were recorded in the United States and many European countries (71), where sales of tetracyclines have been diminishing both during the last decade and in earlier periods. Comparison of data for global resistance rates in H. influenzae (71) also provides no indication of resistance trends in relation to tetracycline usage.
The above examples illustrate how little we know about the precise factors that lead to selection and maintenance of tetracycline-resistant organisms in bacterial populations. A factor contributing to this lack of knowledge is the absence of studies designed to evaluate changes in antibiotic resistance rates over time, with an emphasis in many cases on point prevalence studies (http://europa.eu.int/comm/dg24/health/sc/index_en.html). However, the observation that tetracycline resistance rates in S. pneumoniae are still high in countries where use of these drugs has declined illustrates the potential for stabilization of tet genes in bacterial populations even when selection pressure is reduced. Indeed, the persistence of resistance in pathogens such as S. pneumoniae might relate to a general ability of gram-positive pathogens to acquire and maintain multiple tet genes and the spread of multiple-drug-resistant clones. In addition, many of the transposons carry multiple-antibiotic resistance genes, any one of which can be selected, thereby maintaining all genes (see above).
In view of the extensive dissemination of tet genes into a wide range of bacteria and their apparent stabilization in these species, it is not surprising that tetracyclines are no longer regarded as reliable for use as initial therapy in many infections (195). Consequently, even if a reversal in the rates of tetracycline resistance could be achieved through decreased usage, there is unlikely to be renewed interest in the older tetracyclines by clinicians.
Resistance Following Applications in Veterinary Medicine and Use as Animal Growth Promoters
There is a substantial body of evidence that the use of antibiotics for veterinary therapy, prophylaxis, and animal growth promotion results in the selection of resistant animal pathogens and commensals (http://europa.eu.int/comm/dg24/health /sc/index_en.html; http://www.fda.gov/cvm/fda/mappgs/narms.html) (83, 125, 144, 146, 148, 158, 167, 316). The primary impact of resistance to antibacterials in terms of veterinary medicine is failure of empiric therapy of bacterial infection, which causes an increase in morbidity and mortality and hence prolonged suffering of infected animals; untreatable infections do occur but are rare and are not yet a significant problem in veterinary medicine (http://europa.eu.int/comm/dg24/health/sc/index_en.html). Antibiotic resistance in animals arises in a number of zoonotic pathogens such as Salmonella serovars, Campylobacter spp., and Yersinia spp and in commensals such as E. coli and enterococci, which exist in both human and animal ecosystems. Examples exist showing direct linkage between use of antibiotics in food animals and development of resistant infections in humans, indicating that transfer of antibiotic-resistant bacteria from animals to humans can occur (http://www.fda.gov/cvm/fda/mappgs/narms.html) (72, 83, 109, 146, 181, 316). Thereafter, these organisms might themselves cause infections that are difficult to treat, as in the case of food-borne diseases, or might transfer their resistance genes to other organisms that affect humans. These issues have been the subject of intense and continuous debate since the 1960s, when the Swann Committee in the United Kingdom concluded that antibiotics used in human chemotherapy or those that promote cross-resistance to other therapeutically valuable agents should not be used as growth promoters in animals (288). The antibiotics used as growth promoters in animals are considered to be associated with the greatest risk for selection of resistance because of the continuous subtherapeutic levels used in growth promotion regimens compared to the higher, short-term, therapeutic levels used to treat animal infections (50, 133, 217). Even though there is only circumstantial evidence that resistance in humans is exacerbated by the use of antibiotics in animal feed (http://europa.eu.int/comm/dg24/health/sc/index_en.html)(50, 83, 109), there are increasing concerns, and growing public awareness, that use of antibiotics in animal feed could constitute a human health hazard. In the European Union this has led to application of the precautionary principle whereby the European Commission banned, in animal feed, the use of bacitracin, spiromycin, virginiamycin, and tylosin on 1 July 1999 (Council Directive 2821/98) followed by olaquindox and carbadox on 1 October 1999 (Council Directive 2788/98). These decisions followed earlier ones banning the subtherapeutic use of tetracyclines in 1970 and avoparcin in 1997. Furthermore, the World Health Organization favors the elimination of antibiotics used to treat human diseases as growth promoters in food animals (www.who.ch/programmes/emc/zoo/oct97.pdf). Recently, the World Health Organization has placed on the internet, for public comment, their recommendations Prudent Use of Antimicrobials in Food-Producing Animals (http://www.who.int/emc/diseases/zoo/edg/draft.html.). Key factors of this proposal include proactive approaches by governments to reduce the need for antibiotics in animals, consideration of safety issues regarding the human health impact of resistance development in food animals, and surveillance of antimicrobial consumption by food animals. If such recommendations can be adopted, they will go a long way toward dealing with the agricultural issues of antibiotic use.
CONCLUSIONS
The tetracyclines are a class of antibiotics discovered more than 50 years ago. They are relatively inexpensive drugs with a broad spectrum of activity. Consequently, they have been extensively used in the prophylaxis and therapy of human and animal infections and as animal growth promoters. The selection pressures exerted by the use of tetracyclines in these various environments have resulted in the emergence of resistant organisms. The first tetracycline resistance R-factors were identified over 40 years ago in Japan (70, 146, 308). Since then, tetracycline resistance genes have spread in both gram-negative and gram-positive genera, primarily by conjugal transfer of plasmids and/or transposons. The dramatic increase in the number of species and genera that have acquired tetracycline resistance since the 1950s has led to a reduction in the efficacy and use of current tetracycline therapy for many diseases.
The gram-negative efflux genes have a common genetic organization and show ancestral relationships both in the efflux and regulation proteins. Genetic relationships are found with the gram-positive efflux proteins [Tet(K) and Tet(L)] and among the different ribosomal protection proteins [Tet(M), Tet(O), and Tet(S)]. Both efflux and ribosomal protection proteins are found in antibiotic-producing streptomycetes (33, 63, 206, 218).
The facultative gram-negative efflux genes are associated primarily with classic transposons and integrons, which are located on a diverse group of plasmids, as well as in the chromosome (19, 115, 176, 216). The gram-negative tet(B) gene has the widest host range of the efflux genes and is the only efflux gene which confers resistance to both tetracycline and minocycline. Escherichia spp., Vibrio spp., and Streptomyces spp. host the largest number of different tet genes (Table 4). Many of the Vibrio spp. are associated with fish diseases and/or water and have been subjected to extensive exposure to antibiotics because of fish aquaculture (8, 11, 59–61, 146, 280, 319). E. coli is also found in this environment (60, 61). Whether the environment can or does influence the ability of bacteria to acquire different tet genes or other antibiotic resistance genes is unknown. However, it is likely that an environment where there are large numbers of bacteria with many different species each carrying different tet genes provides an excellent climate for gene exchange. Why various gram-negative enteric genera carry only some of the gram-negative efflux genes is also not clear. We have speculated that the reduced host range of tet(E) may be due to its association with nonmobile and nonconjugative plasmids (61, 280). Perhaps there is a linkage between tet genes and particular incompatible-group plasmids, as suggested by Jones et al. (115). It is also possible that the ribosomal protection genes have not yet been found in enteric species because they confer relatively low-level tetracycline resistance in these hosts. However, given time and enough tetracycline exposure, this barrier may be breached in the future, as have so many in the past (7).
Gram-positive efflux genes [tet(K) and tet(L)] have recently been found in isolates of Haemophilus spp. and Veillonella spp., both of which can also carry the ribosomal protection tet(M) gene. More work must be done to look for gram-positive tet genes in natural gram-negative isolates. The ability to acquire tet genes from both gram-positive and gram-negative bacteria gives gram-positive species more options for acquiring different tet genes. The ribosomal protection genes, especially tet(M), are found in a number of gram-positive and gram-negative species, suggesting that they may have an advantage when it comes to being distributed among the different genera.
The same tet genes are found in bacterial pathogens, opportunistic microbes, and members of the normal flora. Significant numbers of normal flora bacteria from humans and animals, as well as bacteria isolated from food and the environment, are resistant to tetracycline (8, 69, 141, 146, 178, 266). The normal human flora may act as a reservoir for antibiotic resistance genes in general and for tet genes in particular (45, 46, 143, 225, 230, 236, 249). Similarly, the normal floras in animals, plants, and bacteria in the environment may also play import roles as reservoirs for antibiotic resistance genes (8, 46, 125, 230, 266, 319).
The widespread distribution of specific tet genes like tet(B) or tet(M) supports the hypothesis that the tet genes are exchanged by bacteria from many different ecosystems and between humans and both pet and food animals. Thus, bacteria exposed to antibiotics in the environment or in animals can ultimately influence antibiotic resistance in bacteria of human origin (125, 316). The presence of gram-positive tet genes in gram-negative species supports the hypothesis that gram-positive genes are being introduced and maintained in gram-negative species in nature and is not simply an in vitro artifact. It is very likely that this trend will continue, with more gram-positive genes becoming stably maintained in gram-negative hosts.
The use of tetracyclines to combat active infection in individual animals and herds should continue, not only because it reduces animal suffering but also because animals of considerable economic value can be saved (50; Miller, Abstract, 1999). However, the use of tetracyclines as animal growth promoters is a more contentious issue. The use of tetracyclines in food production is considerable and contributes to the worldwide exposure of bacteria to tetracyclines. There is no doubt that this practice results in the selection of resistant organisms and that in some cases these can be transmitted to humans. Furthermore, since tet genes can be located in integrons, the use of tetracyclines as growth promoters could result in selection and transfer to humans of resistance to unrelated antibiotics whose resistance gene cassettes are also incorporated within the integron. Although there is only poor scientific evidence that the use of growth-promoting antibiotics, including tetracyclines, poses a significant hazard to public health, the use of tetracyclines and other antibiotics as animal growth promoters has been banned in the European Union. Eliminating the use of tetracyclines and other antibiotics at subtherapeutic levels in animal feeds in other countries should also be a priority for feed manufacturers and policy makers, in the hope that these changes will help to reduce the incidence of resistant bacteria in the environment. It could be argued that further transmission of tetracycline resistance from animals to humans is of minor consequence because of the declining importance of the older tetracyclines in human medicine. Nevertheless, these agents still have valuable roles in the first-line treatment of certain infections, and for some applications improved formulations are being developed to further enhance their efficacy (260). These important medical uses of tetracyclines could be eroded if the human pathogens concerned acquired tet genes from the animal environment. In addition, the consequences of continued selection pressure with older tetracyclines in the animal setting for the success of the new glycylcyclines in human medicine are unknown. The observation that tet genes with the capacity to confer resistance to glycylcyclines apparently already exist in veterinary Salmonella isolates (M. Tuckman, P. J. Petersen, and S. Projan, Program Abstr. 38th Intersci. Conf. Antimicrob. Agents Chemother., abstr. C98, p. 97, 1998) is clearly a matter for concern.
FUTURE DIRECTION
The emergence of bacterial resistance to tetracyclines mirrors the situation with most antimicrobials currently in use. New derivatives of the tetracyclines, such as the glycylcyclines, dactylocycline, and other analogs with large hydrophobic groups, are all under examination for potential introduction as clinical agents to circumvent existing tetracycline resistance mechanisms (173, 300). There is also interest in discovering tetracycline efflux pump inhibitors that could be used in conjunction with older tetracyclines to restore their activity (185, 186). Among these agents, the glycycline analog tigilcycline (formerly known as GAR-936) is closest to introduction as a therapeutic agent since it is currently in phase II clinical trials (113).
Glycylcyclines display activity against strains expressing a variety of different tet genes, including those that encode ribosomal protection and efflux mechanisms. The glycylcyclines inhibit protein synthesis catalyzed by translation systems prepared from cells expressing Tet(M) and Tet(O) proteins (17, 215). Glycylcyclines compete with tetracycline for ribosomal binding but have a higher binding affinity for ribosomes than earlier tetracyclines (17). This is the most likely reason that ribosomal protection proteins are unable to confer resistance to glycylcyclines. The molecular basis of the activity of glycylcyclines against strains containing tet genes that encode efflux has also been examined. Activity could be due to the failure of the Tet efflux proteins to recognize glycylcyclines per se or to the inability of the Tet transporters to translocate glycylcyclines across the cytoplasmic membrane even though these proteins may recognize and bind the new analogs. The result of either mechanism would be failure to remove glycylcyclines from the bacterial cytoplasm so that inhibitor concentrations necessary to prevent protein synthesis would be maintained. Using everted membrane vesicles prepared from E. coli expressing the Tet(B) efflux transporter, Someya et al. (278) obtained data to suggest that 9-(N,N-dimethylglycylamido)-6-demethyl-6-deoxytetracycline (Table 2) is not recognized and hence not bound by the efflux protein. Structural studies with Tet(B) repressor protein, albeit from another class of tetracycline-interactive (water-soluble) proteins, demonstrate that the repressor does interact with glycylcyclines (202). Compared to tetracycline, the chelating nucleus of 9-(N,N-dimethylglycylamido)-6-demethyl-6-deoxytetracycline was unable to reach the normal inducer binding position within the repressor, as a result of a weakened hydrogen-bonding pattern within and around the tetracycline binding site.
Glycylcyclines possess the basic structural features associated with the broad spectrum of activity of the class, and, as already discussed, they are also active against strains containing tet determinants. A number of studies have been conducted on clinical isolates taken from patients with respiratory tract infections, skin and soft tissue infections, urinary tract infections, enterococcal infections, anaerobic infections, gonorrhoea, and cutaneous infections caused by rapidly growing mycobacteria, and they show promise for the glycylcyclines in these infections (25, 68, 77, 86, 92, 123, 194, 208, 270, 287, 295, 310–312, 315). In addition, 9-(t-butylglycylamido)-minocycline (tigilcycline; GAR-936) (Tables 1 and 2) could have therapeutic potential in cystic fibrosis patients infected with P. aeruginosa (113). More recently, 1,730 clinical isolates were examined in two studies with tigilcycline and older tetracyclines (22, 80). In general, the glycylcycline was 2- to 4-fold more active than minocycline and 2- to 16-fold more active than tetracycline against the Enterobacteriaceae. An exception was found with Proteus mirabilis and indole-positive Proteus spp. where the tigilcycline MIC against 90% of isolates was ≥8 μg/ml (80). Tigilcycline was active against virtually all gram-positive clinical isolates including those resistant to earlier tetracyclines (22). Surveillance studies have not so far identified any naturally occurring high-level glycylcycline-resistant strains among human clinical isolates (113). However, resistance to the investigational glycylcyclines 9-(N,N-dimethylglycylamido)-6-demethyl-6-deoxytetracycline and 9-(N,N-dimethylglycylamido)minocycline has been reported in two veterinary isolates, one of S. enterica serovar Typhimurium and one of serovar Choleraesuis (Tuckman et al. 38th ICAAC). Resistance was also found in African multiple-drug-resistant Salmonella and Shigella isolates (M. Roberts, unpublished results).
The outcome of the current phase II clinical trials with tigicycline will indicate whether this member of the latest class of tetracyclines has the potential for introduction as a chemotherapeutic agent. The observation that tet genes with the capacity to confer resistance to some glycylcyclines apparently already exist in veterinary Salmonella and Shigella isolates is clearly a potential issue.
Unfortunately, since resistance to new tetracycline deriviates may occur quickly, other strategies must also be considered to combat bacterial resistance, such as the development of new classes of drugs with unique antibacterial mechanisms. We also require a better understanding of the ecology of resistance to all antibiotics, including tetracyclines, that could lead to the design of new intervention strategies. We need to better understand the fate of tetracyclines in the environment. This is important because uneaten tetracycline-supplemented animal feeds go directly into the soil and water environment and it is not clear how long tetracyclines remain active in these environments. We need to develop methods to contain or detoxify antimicrobials, including tetracyclines. Antibacterial substances, such as triclosan, are being added to household products such as soap, children's toys, and clothing. How these substances affect the development of bacterial antibiotic resistance, including tetracycline resistance, is not clear. The experience gained with tetracycline highlights the need for a better understanding of bacterial resistance at the global level. We need a variety of approaches to reduce the amounts of antimicrobials being used worldwide in human and animal medicine and food production. This is the only way we will be able to keep tetracyclines and other antibiotics as resources for the next generation.
ACKNOWLEDGMENTS
We thank F. J. Piraces and V. J. Lee for valuable discussions.
REFERENCES
- 1.Acar J F. Consequences of bacterial resistance to antibiotics in medical practice. Clin Infect Dis. 1997;24:S17–S18. doi: 10.1093/clinids/24.supplement_1.s17. [DOI] [PubMed] [Google Scholar]
- 2.Acar J F, Bouanchaud D H, Chabbert Y A. Evolutionary aspects of plasmid mediated resistance in a hospital environment. In: Drews J, Hogerauers G, editors. Topics in infectious diseases. R-factors: their properties and possible control. Vol. 2. Vienna, Austria: Springer-Verlag; 1977. pp. 5–23. [Google Scholar]
- 3.Agalar C, Usubutun S, Turkyilmaz R. Ciprofloxacin and rifampicin versus doxycycline and rifampicin in the treatment of brucellosis. Eur J Clin Microbiol Infect Dis. 1999;18:535–538. doi: 10.1007/s100960050344. [DOI] [PubMed] [Google Scholar]
- 4.Akiba T, Koyama K, Ishiki Y, Kimura S, Fukushima T. On the mechanism of the development of multiple-drug-resistant clones of Shigella. Jpn J Microbiol. 1960;4:219. doi: 10.1111/j.1348-0421.1960.tb00170.x. [DOI] [PubMed] [Google Scholar]
- 5.Alekshum M N, Levy S B. Regulation of chromosomally mediated multiple antibiotic resistance: the mar regulation. Antimicrob Agents Chemother. 1997;41:2067–2075. doi: 10.1128/aac.41.10.2067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Alonso A, Marinez J L. Multiple antibiotic resistance in Stenotrophomonas maltophilia. Antimicrob Agents Chemother. 1997;41:2119–2121. doi: 10.1128/aac.41.5.1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.American Society for Microbiology. Report of the ASM task force on antibiotic resistance. Antimicrob Agents Chemother. 1995;1995(Suppl.):1–23. [PubMed] [Google Scholar]
- 8.Andersen S R, Sandaa R A. Distribution of tetracycline resistance determinants among gram-negative bacteria isolated from polluted and unpolluted marine sediments. Appl Environ Microbiol. 1994;60:908–912. doi: 10.1128/aem.60.3.908-912.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Andres M T, Chung W O, Roberts M C, Fierro J F. Antimicrobial susceptibilities of Porphyromonas gingivalis, Prevotella intermedia, and Prevotella nigrescens isolated in Spain. Antimicrob Agents Chemother. 1998;42:3022–3023. doi: 10.1128/aac.42.11.3022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Antonio M A D, Hawes S E, Hillier S L. The identification of vaginal Lactobacillus species and the demographic and microbiologic characteristics of women colonized by these species. J Infect Dis. 1999;180:1950–1956. doi: 10.1086/315109. [DOI] [PubMed] [Google Scholar]
- 11.Aoki T, Satoh T, Kitao T. New tetracycline resistance determinant on R-plasmids from Vibrio anguillarum. Antimicrob Agents Chemother. 1987;31:1446–1449. doi: 10.1128/aac.31.9.1446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Atkinson B A, Abu-Al-Jaibat A, LeBlanc D J. Antibiotic resistance among enterococci isolated from clinical specimens between 1953 and 1954. Antimicrob Agents Chemother. 1997;41:1598–1600. doi: 10.1128/aac.41.7.1598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Banai M, LeBlanc D J. Genetic, molecular, and functional analysis of Streptococcus faecalis plasmid pJH1. J Bacteriol. 1983;155:1094–1104. doi: 10.1128/jb.155.3.1094-1104.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Barbosa T M, Scott K P, Flint H J. Evidence for recent intergeneric transfer of new tetracycline resistance gene, tet(W), isolated from Butyrivibrio fibrisolens, and the occurrence of tet(O) in ruminal bacteria. Environ Microbiol. 1999;1:53–64. doi: 10.1046/j.1462-2920.1999.00004.x. [DOI] [PubMed] [Google Scholar]
- 15.Barden T C, Buckwalter B L, Testa R T, Petersen P J, Lee V J. “Glycylcyclines.” 3. 9-Aminodoxycyclinecarboxamides. J Med Chem. 1994;37:3205–3211. doi: 10.1021/jm00046a003. [DOI] [PubMed] [Google Scholar]
- 16.Benveniste R, Davies J. Aminoglycoside antibiotic-inactivation enzymes in actinomycetes similar to those present in clinical isolates of antibiotic resistant bacteria. Proc Natl Acad Sci USA. 1973;172:3628–3632. doi: 10.1073/pnas.70.8.2276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bergeron J, Ammirati M, Danley D, James L, Norcia M, Retsema J, Strick C A, Su W G, Sutcliffe J, Wondrack L. Glycylcyclines bind to the high-affinity tetracycline ribosomal binding site and evade Tet(M)- and Tet(O)-mediated ribosomal protection. Antimicrob Agents Chemother. 1996;40:2226–2228. doi: 10.1128/aac.40.9.2226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bertram J, Stratz M, Durre P. Natural transfer of conjugative transposon Tn916 between gram-positive and gram-negative bacteria. J Bacteriol. 1991;173:443–448. doi: 10.1128/jb.173.2.443-448.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Biggs C E, Fratamico P M. Molecular characterization of an antibiotic resistance gene cluster of Salmonella typhimurium DT104. Antimicrob Agents Chemother. 1999;43:846–849. doi: 10.1128/aac.43.4.846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Blackwood R K. Structure determination and total synthesis of the tetracyclines. In: Hlavka J J, Boothe J H, editors. Handbook of experimental pharmacology. Vol. 78. Berlin, Germany: Springer-Verlag KG; 1985. pp. 59–136. [Google Scholar]
- 21.Bolhuis H, van Veen H W, Poolman B, Driessen A J M, Konings W N. Mechanisms of multidrug transporters. FEMS Microbiol Rev. 1997;21:55–84. doi: 10.1111/j.1574-6976.1997.tb00345.x. [DOI] [PubMed] [Google Scholar]
- 22.Boucher H W, Wennersten C B, Eliopoulos G M. In vitro activities of the glycylcycline GAR-936 against gram-positive bacteria. Antimicrob Agents Chemother. 2000;44:2225–2229. doi: 10.1128/aac.44.8.2225-2229.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bremon A R, Ruiz-Tovar M, Gorricho B P, de Torres P D, Rodgriguez R L. Non-hospital consumption of antibiotics in Spain: 1987–1997. J Antimicrob Chemother. 2000;45:395–400. doi: 10.1093/jac/45.3.395. [DOI] [PubMed] [Google Scholar]
- 24.Brisson-Noel A, Arthur M, Courvalin P. Evidence for natural gene transfer from gram-positive cocci to Escherichia coli. J Bacteriol. 1988;170:1739–1745. doi: 10.1128/jb.170.4.1739-1745.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Brown B A, Wallace R J, Onyi G. Activities of the glycylcyclines N,N-dimethylglycylamido-minocycline and N,N-dimethylglycylamido-6-demethyl-6-deoxytetracycline against Nocardia spp. and tetracycline-resistant isolates of rapidly growing mycobacteria. Antimicrob Agents Chemother. 1996;40:874–878. doi: 10.1128/aac.40.4.874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Brown J T, Roberts M C. Cloning and characterization of tetM from a Ureaplasma urealyticum strain. Antimicrob Agents Chemother. 1987;31:1852–1854. doi: 10.1128/aac.31.11.1852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Brown M B, Roberts M C. Tetracycline resistance determinants in streptococcal species isolated from the bovine mammary gland. Vet Microb. 1991;29:173–180. doi: 10.1016/0378-1135(91)90124-x. , 1991. [DOI] [PubMed] [Google Scholar]
- 28.Bunnag D, Karbwang J, Na-Bangchang K, Thanavibul A, Chittamas S, Harinasuta T. Quinine-tetracycline for multidrug resistant falciparum malaria. Southeast Asia J Trop Med Public Health. 1996;27:15–18. [PubMed] [Google Scholar]
- 29.Burdett V. Purification and characterization of Tet(M), a protein that renders ribosomes resistant to tetracycline. J Biol Chem. 1991;266:2872–2877. [PubMed] [Google Scholar]
- 30.Burdett V. tRNA modification activity is necessary for Tet(M)-mediated tetracycline resistance. J Bacteriol. 1993;175:7209–7215. doi: 10.1128/jb.175.22.7209-7215.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Burdett V. Tet(M)-promoted release of tetracycline from ribosomes is GTP dependent. J Bacteriol. 1996;178:3246–3251. doi: 10.1128/jb.178.11.3246-3251.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Burns J L, Wadsworthk C D, Barry J J, Goodall C P. Nucleotide sequence analysis of a gene from Burkholderia (Pseudomonas) cepacia encoding an outer membrane lipoprotein involved in multiple antibiotic resistance. Antimicrob Agents Chemother. 1996;40:307–313. doi: 10.1128/aac.40.2.307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Butler M J, Friend E J, Hunter I S, Kaczmarek F S, Sugden D A, Warren M. Molecular cloning of resistance genes and architecture of a linked gene cluster involved in biosynthesis of oxytetracycline by Streptomyces rimosus. Mol Gen Genet. 1989;215:231–238. doi: 10.1007/BF00339722. [DOI] [PubMed] [Google Scholar]
- 34.Buu-Hoi A, Le Bouguenec C, Horaud T. Genetic basis of antibiotic resistance in Aerococcus viridans. Antimicrob Agents Chemother. 1989;33:529–534. doi: 10.1128/aac.33.4.529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Caillaud F, Carlier C, Courvalin P. Physical analysis of the conjugative shuttle transposon Tn1545. Plasmid. 1987;17:58–60. doi: 10.1016/0147-619x(87)90009-6. [DOI] [PubMed] [Google Scholar]
- 36.Charpentier E, Gerbaud G, Courvalin P. Characterization of a new class of tetracycline-resistance gene tet(S) in Listeria monocytogenes BM4210. Gene. 1993;131:27–34. doi: 10.1016/0378-1119(93)90665-p. [DOI] [PubMed] [Google Scholar]
- 37.Charpentier E, Gerbaud G, Courvalin P. Presence of the Listeria tetracycline resistance gene tet(S) in Enterococcus faecalis. Antimicrob Agents Chemother. 1994;38:2330–2335. doi: 10.1128/aac.38.10.2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Charpentier E, Courvalin P. Antibiotic resistance in Listeria spp. Antimicrob Agents Chemother. 1999;43:2103–2108. doi: 10.1128/aac.43.9.2103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Charvalos E, Tselentis Y, Hamzehpour M M, Kohler T, Pechere J-C. Evidence for an efflux pump in multidrug-resistant Campylobacter jejuni. Antimicrob Agents Chemother. 1995;39:2019–2022. doi: 10.1128/aac.39.9.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chaslus-Dancla E, Lesage-Descauses M-C, Leroy-Setrin S, Marel J-L, Lafont J-P. Tetracycline resistance determinants, Tet B and Tet M detected in Pasteurella haemolytica and Pasteurella multocida from bovine herds. J Antimicrob Chemother. 1995;36:815–819. doi: 10.1093/jac/36.5.815. [DOI] [PubMed] [Google Scholar]
- 41.Chopra I. Mode of action of the tetracyclines and the nature of bacterial resistance to them. In: Hlavka J J, Boothe J H, editors. Handbook of experimental pharmacology. Vol. 78. Berlin, Germany: Springer-Verlag KG; 1985. pp. 315–392. [Google Scholar]
- 42.Chopra I. Transport of tetracyclines into Escherichia coli requires a carboxamide group at the C2 position of the molecule. J Antimicrob Chemother. 1986;18:661–666. doi: 10.1093/jac/18.6.661. [DOI] [PubMed] [Google Scholar]
- 43.Chopra I. Tetracycline analogs whose primary target is not the bacterial ribosome. Antimicrob Agents Chemother. 1994;38:637–640. doi: 10.1128/aac.38.4.637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chopra I, Hawkey P M, Hinton M. Tetracyclines, molecular and clinical aspects. J Antimicrob Chemother. 1992;29:245–277. doi: 10.1093/jac/29.3.245. [DOI] [PubMed] [Google Scholar]
- 45.Chung W O, Young K, Leng Z, Roberts M C. Mobile elements carrying ermF and tetQ genes in Gram-positive and Gram-negative bacteria. J Antimicrob Chemother. 1999;44:329–335. doi: 10.1093/jac/44.3.329. [DOI] [PubMed] [Google Scholar]
- 46.Chung W O, Werckenthin C, Schwarz S, Roberts M C. Host range of the ermF rRNA methylase gene in bacteria of human and animal origin. J Antimicrob Chemother. 1999;43:5–14. doi: 10.1093/jac/43.1.5. [DOI] [PubMed] [Google Scholar]
- 47.Clermont D, Chesneau O, De Cespedes G, Horaud T. New tetracycline resistance determinants coding for ribosomal protection in streptococci and nucleotide sequence of tet(T) isolated from Streptococcus pyogenes A498. Antimicrob Agents Chemother. 1997;41:112–116. doi: 10.1128/aac.41.1.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Clewell D B, Flannagan S E, Jaworski D D. Unconstrained bacterial promiscuity: the Tn916-Tn1545 family of conjugative transposons. Trends Microbiol. 1995;3:229–236. doi: 10.1016/s0966-842x(00)88930-1. [DOI] [PubMed] [Google Scholar]
- 49.Col N F, O'Connor R W. Estimating worldwide current antibiotic usage: report of task force 1. Rev Infect Dis. 1987;9:S232–S243. doi: 10.1093/clinids/9.supplement_3.s232. [DOI] [PubMed] [Google Scholar]
- 50.Committee on Drug Use in Food Animals. The use of drugs in food animals, benefits and risks. Washington, D.C.: National Academy Press; 1999. [Google Scholar]
- 51.Cunha B A. Doxycycline re-visited. Arch Intern Med. 1999;159:1006–1007. doi: 10.1001/archinte.159.9.1006. [DOI] [PubMed] [Google Scholar]
- 52.Dancer S J, Shears P, Platt D J. Isolation and characterization of coliforms from glacial ice and water in Canada's high arctic. J Appl Microbiol. 1997;82:597–609. doi: 10.1111/j.1365-2672.1997.tb03590.x. [DOI] [PubMed] [Google Scholar]
- 53.Dantley K A, Dannelly H K, Burdett V. Binding interaction between Tet(M) and the ribosome: requirements for binding. J Bacteriol. 1998;180:4089–4092. doi: 10.1128/jb.180.16.4089-4092.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Davey P G, Bax R P, Newey J, Reeves D, Rutherford D, Slack R, Warren R E, Watt B, Wilson J. Growth in the use of antibiotics in the community in England and Scotland in 1980–93. B Med J. 1996;312:613. doi: 10.1136/bmj.312.7031.613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Davis M A, Hancock D D, Besser T E, Rice D H, Gay J M, Gay C, Gearhart L, DiGiacomo R. Changes in antimicrobial resistance among Salmonella enterica serovar typhimurium isolates from humans and cattle in the northwestern United States, 1982–1997. Emerg Infect Dis. 1999;5:802–806. doi: 10.3201/eid0506.990610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Dax S L. Antibacterial chemotherapeutic agents. London, United Kingdom: Blackie Academic and Professional; 1997. [Google Scholar]
- 57.de Barbeyrac B, Dutilh B, Quentin C, Renaudin H, Bebear C. Susceptibility of Bacteroides ureolyticus to antimicrobial agents and identification of a tetracycline resistance determinant related to tetM. J Antimicrob Chemother. 1991;27:721–731. doi: 10.1093/jac/27.6.721. [DOI] [PubMed] [Google Scholar]
- 58.Debets-Ossenkopp Y J, Herscheid A J, Pot R G J, Kuipers E J, Kusters J G, Vandenbroucke-Grauls C M J E. Prevalence of Helicobacter pylori resistance to metronidazole, clarithromycin, amoxycillin, tetracycline and trovafloxacin in the Netherlands. J Antimicrob Chemother. 1999;43:511–515. doi: 10.1093/jac/43.4.511. [DOI] [PubMed] [Google Scholar]
- 59.DePaola A, Flynn P A, McPherarson R M, Levy S B. Phenotypic and genotypic characterization of tetracycline- and oxytetracycline-resistant Aeromonas hydrophila from cultured channel catfish (Ictalurus punctatus) and their environments. Appl Environ Microbiol. 1988;54:1861–1863. doi: 10.1128/aem.54.7.1861-1863.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.DePaola A, Hill W E, Harrell F M. Oligonucleotide probe determination of tetracycline-resistant bacteria isolated from catfish ponds. Mol Cell Probes. 1993;7:345–348. doi: 10.1006/mcpr.1993.1051. [DOI] [PubMed] [Google Scholar]
- 61.DePaola A, Roberts M C. Class D and E tetracycline resistance determinants in gram-negative catfish pond bacteria. Mol Cell Probes. 1995;9:311–313. doi: 10.1016/s0890-8508(95)91572-9. [DOI] [PubMed] [Google Scholar]
- 62.De Rossi E, Blokpoel M C J, Cantoni R, Branzoni M, Riccardi G, Young D B, De Set K A L, Ciferri O. Molecular cloning and functional analysis of a novel tetracycline resistance determinant, tet(V), from Mycobacterium smegmatis. Antimicrob Agents Chemother. 1998;42:1931–1937. doi: 10.1128/aac.42.8.1931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Dittrich W, Schrempf H. The unstable tetracycline resistance gene of Streptomyces lividans 1326 encodes a putative protein with similarities to translational elongation factors and Tet (M) and Tet (O) proteins. Antimicrob Agents Chemother. 1992;36:1119–1124. doi: 10.1128/aac.36.5.1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Doern G V, Brueggemann A B, Huynh H, Wingert E, Rhombert P. Antimicrobial resistance with Streptococcus pneumoniae in the United States, 1997–98. Emerg Infect Dis. 1999;5:757–765. doi: 10.3201/eid0506.990603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Doran J L, Pang Y, Mdluli K E, Moran A J, Victor T C, Stokes R W, van Helden E, Roberts M C, Nano F E. Mycobacterium tuberculosis efpA encodes an efflux protein of the QacA transporter family. Clin Diagn Lab Immunol. 1997;4:23–32. doi: 10.1128/cdli.4.1.23-32.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Doyle D, McDowall K J, Bulter M J, Hunter I S. Characterization of an oxytetracycline-resistance gene, otrA, of Streptomyces rimosus. Mol Microbiol. 1991;5:2923–2933. doi: 10.1111/j.1365-2958.1991.tb01852.x. [DOI] [PubMed] [Google Scholar]
- 67.Edlind T D. Protein synthesis as a target for antiprotozoal drugs. In: Coombs G, North M, editors. Biochemical protozoology. London, United Kingdom: Taylor & Francis; 1991. pp. 569–586. [Google Scholar]
- 68.Eliopoulos G M, Wennersten C B, Cole G, Moellering R C. In vitro activities of two glycylcyclines against gram-positive bacteria. Antimicrob Agents Chemother. 1994;38:534–541. doi: 10.1128/aac.38.3.534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Facinelli B, Roberts M C, Giovanetti E, Casolari C, Fabio U, Varaldo P E. Genetic basis of tetracycline resistance in food borne isolates of Listeria innocua. Appl Environ Microbiol. 1993;59:614–616. doi: 10.1128/aem.59.2.614-616.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Falkow S. Infectious multiple drug resistance. London, United Kingdom: Pion Ltd.; 1975. [Google Scholar]
- 71.Felmingham D, Gruneberg R N the Alexander Project Group. The Alexander Project 1996–1997: latest susceptibility data from this international study of bacterial pathogens from community-acquired lower respiratory tract infections. J Antimicrob Chemother. 2000;45:191–203. doi: 10.1093/jac/45.2.191. [DOI] [PubMed] [Google Scholar]
- 72.Fey P D, Safranek T J, Rupp M E, Dunne E F, Ribot E, Iwen P C, Bradford P A, Angulo F J, Hinrichs S H. Ceftriaxone-resistant salmonella infection acquired by a child from cattle. N Engl J Med. 2000;342:1242–1249. doi: 10.1056/NEJM200004273421703. [DOI] [PubMed] [Google Scholar]
- 73.Finch R G. Tetracyclines. In: O'Grady F, Lambert H P, Finch R G, Greenwood D, editors. Antibiotic and chemotherapy. 7th ed. New York, N.Y: Churchill Livingstone Ltd.; 1997. pp. 469–484. [Google Scholar]
- 74.Fitzgerald G F, Clewell D B. A conjugative transposon (Tn919) in Streptococcus sanguis. Infect Immun. 1985;47:415–420. doi: 10.1128/iai.47.2.415-420.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Flannagan S E, Zitzow L A, Su Y A, Clewell D B. Nucleotide sequence of the 18-kb conjugative transposon Tn916 from Enterococcus faecalis. Plasmid. 1994;32:350–354. doi: 10.1006/plas.1994.1077. [DOI] [PubMed] [Google Scholar]
- 76.Fletcher H M, Daneo-Moore L. A truncated Tn916-like element in a clinical isolates of Enterococcus faecium. Plasmid. 1992;27:155–160. doi: 10.1016/0147-619x(92)90015-3. [DOI] [PubMed] [Google Scholar]
- 77.Fraise A P, Brenwald N, Andrews J M, Wise R. In-vitro activity of two glycylcyclines against enterococci resistant to other agents. J Antimicrob Chemother. 1995;35:877–881. doi: 10.1093/jac/35.6.877. [DOI] [PubMed] [Google Scholar]
- 78.Franklin T J. Mode of action of the tetracyclines. In: Newton B A, Reynolds P E, editors. Biochemical studies of antimicrobial drugs. Sixteenth Symposium of the Society for General Microbiology. Cambridge, United Kingdom: Cambridge University Press; 1966. pp. 192–212. [Google Scholar]
- 79.Freeman C D, Nightingale C H, Quintiliani R. Minocycline: old and new therapeutic uses. Int J Antimicrob Agents. 1994;4:325–335. doi: 10.1016/0924-8579(94)90034-5. [DOI] [PubMed] [Google Scholar]
- 80.Gales A C, Jones R N. Antimicrobial activity and spectrum of the new glycylcycline, GAR-936 tested against 1,203 recent clinical bacterial isolates. Diagn Microbiol Infect Dis. 2000;36:19–36. doi: 10.1016/s0732-8893(99)00092-9. [DOI] [PubMed] [Google Scholar]
- 81.Galimand M, Guiyoule A, Gerbaud G, Rasoamanana B, Chanteau S, Carniel E, Courvalin P. Multidrug resistance in Yersinia pestis by a transferable plasmid. N Engl J Med. 1997;337:677–680. doi: 10.1056/NEJM199709043371004. [DOI] [PubMed] [Google Scholar]
- 82.George A M, Hall R M, Stokes H W. Multidrug resistance in Klebsiella pneumoniae: a novel gene, ramA, confers a multidrug resistance phenotype in Escherichia coli. Microbiology. 1995;141:1909–1920. doi: 10.1099/13500872-141-8-1909. [DOI] [PubMed] [Google Scholar]
- 83.Georgetown University Center for Food and Nutrition Policy. Antibiotic use in animals: food safety and risk assessment. Conference proceedings. Washington, D.C.: Georgetown University; 1999. [Google Scholar]
- 84.Gillespie M T, Lyon B R, Loo L S L, Mathhews P R, Stewart P R, Skurray R A. Homologous direct repeat sequences associated with mercury, methicillin, tetracycline and trimethoprim resistance in Staphylococcus aureus. FEMS Microbiol Lett. 1987;43:165–171. [Google Scholar]
- 85.Gillespie M T, May J W, Skurray R. Detection of an integrated tetracycline resistance plasmid in the chromosome of methicillin-resistant Staphylococcus aureus. J Gen Microbiol. 1986;132:1723–1728. doi: 10.1099/00221287-132-6-1723. [DOI] [PubMed] [Google Scholar]
- 86.Goldstein F W, Kitzis M D, Acar J F. N,N-Dimethylglycyl-amido derivative of minocycline and 6-demethyl-6-deoxytetracycline, two new glycylcyclines highly effective against tetracycline-resistant gram-positive cocci. Antimicrob Agents Chemother. 1994;38:2218–2220. doi: 10.1128/aac.38.9.2218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Gotoh N, Tsujimoto H, Poole K, Yamagishi J-I, Nishino T. The outer membrane protein OprM of Pseudomonas aeruginosa is encoded by oprK of the mexA-mexB-oprK multidrug resistance operon. Antimicrob Agents Chemother. 1995;39:2567–2569. doi: 10.1128/aac.39.11.2567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Graeme K A, Pollack C V. Antibiotic use in the emergency department. II. The aminoglycosides, macrolides, tetracyclines. Sulfa drugs, and urinary antiseptics. J Emerg Med. 1996;14:361–371. doi: 10.1016/0736-4679(96)00035-2. [DOI] [PubMed] [Google Scholar]
- 89.Grave K, Lingaas E, Bangen M, Ronning M. Surveillance of the overall consumption of antibacterial drugs in humans, domestic animals and farmed fish in Norway in 1992 and 1996. J Antimicrob Chemother. 1999;43:243–252. doi: 10.1093/jac/43.2.243. [DOI] [PubMed] [Google Scholar]
- 90.Guay G G, Tuckman M, Rothstein D M. Mutations in the tetA(B) gene that cause a change in substrate specificity of the tetracycline efflux pump. Antimicrob Agents Chemother. 1994;38:857–860. doi: 10.1128/aac.38.4.857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Gustafson R H, Kiser J S. Nonmedical uses of the tetracyclines. In: Hlavka J J, Boothe J H, editors. Handbook of experimental pharmacology. Vol. 78. Berlin, Germany: Springer-Verlag KG; 1985. pp. 405–446. [Google Scholar]
- 92.Hamilton-Miller J M T, Shah S. Activity of glycylcyclines CL 329998 and CL 331002 against minocycline-resistant and other strains of methicillin-resistant Staphylococcus aureus. J Antimicrob Chemother. 1996;37:1171–1175. doi: 10.1093/jac/37.6.1171. [DOI] [PubMed] [Google Scholar]
- 93.Hartley D L, Hones K R, Tobian J A, LeBlanc D J, Macrina F L. Disseminated tetracycline resistance in oral streptococci: implication of a conjugative transposon. Infect Immun. 1984;45:13–17. doi: 10.1128/iai.45.1.13-17.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Hansen L M, McMurry L M, Levy S B, Hirsch D C. A new tetracycline resistance determinant, Tet H, from Pasteurella multocida specifying active efflux of tetracycline. Antimicrob Agents Chemother. 1993;37:2699–2705. doi: 10.1128/aac.37.12.2699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Hatsu M, Sasaki T, Gomi S, Kodama Y, Sezaki M, Inouye S, Kondo S. A new tetracycline with antitumor activity: II the structural elucidation of SF2575. J Antibiot. 1992;45:325–330. doi: 10.7164/antibiotics.45.325. [DOI] [PubMed] [Google Scholar]
- 96.Hawley R J, Lee L N, LeBlanc D J. Effects of tetracycline on the streptococcal flora of periodontal pockets. Antimicrob Agents Chemother. 1980;17:372–378. doi: 10.1128/aac.17.3.372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Heur C, Hickman R K, Curiale M S, Hillen W, Levy S B. Constitutive expression of tetracycline resistance mediated by a Tn10-like element in Haemophilus parainfluenzae results from a mutation in the repressor gene. J Bacteriol. 1987;169:990–994. doi: 10.1128/jb.169.3.990-994.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Higlander S K, Novick R P. Mutational and physiological analyses of plasmid pT181 functions expressing incompatibility. Plasmid. 1990;23:1–15. doi: 10.1016/0147-619x(90)90039-f. [DOI] [PubMed] [Google Scholar]
- 99.Hillen W, Berens C. Mechanisms underlying expression of Tn10-encoded tetracycline resistance. Annu Rev Microbiol. 1994;48:345–369. doi: 10.1146/annurev.mi.48.100194.002021. [DOI] [PubMed] [Google Scholar]
- 100.Hlavka J J, Ellestad G A, Chopra I. Tetracyclines. In: Kroschwitz J I, Howe-Grant M, editors. Kirk-Othmer encyclopedia of chemical technology. 4th ed. Vol. 3. New York, N.Y: John Wiley & Sons, Inc.; 1992. pp. 331–346. [Google Scholar]
- 101.Holmberg S D, Osterholm M T, Senger K A, Cohen M L. Drug-resistant Salmonella from animals fed antimicrobials. N Engl J Med. 1984;311:617. doi: 10.1056/NEJM198409063111001. [DOI] [PubMed] [Google Scholar]
- 102.Holmberg S D, Wells J G, Cohen M L. Animal-to-man transmissions of antimicrobial-resistant Salmonella: investigation of US outbreaks, 1971–1983. Science. 1984;225:833–835. doi: 10.1126/science.6382605. [DOI] [PubMed] [Google Scholar]
- 103.Horaud T, de Cerspedes G, Clermont D, David F, Delbos F. Variability of chromosomal genetic elements in streptococci. In: Dunny G M, Cleary P P, McKay L L, editors. Genetics and molecular biology of streptococci, lactococci and enterococci. Washington, D.C.: American Society for Microbiology; 1991. pp. 16–20. [Google Scholar]
- 104.Hosek G, Leschinksy D D, Irons S, Safranek T J. Multidrug-resistant Salmonella serotype Typhimurium—United States, 1996. Morb Mortal Wkly Rep. 1997;46:308–310. [PubMed] [Google Scholar]
- 105.Huang R, Gascoyne-Binzi D M, Hawkey P M, Yu M, Heritage J, Eley A. Molecular evolution of the tet(M) gene in Gardnerella vaginalis. J Antimicrob Chemother. 1997;40:561–565. doi: 10.1093/jac/40.4.561. [DOI] [PubMed] [Google Scholar]
- 106.Hughes V M, Datta N. Conjugative plasmids in bacteria of the ‘pre-antibiotic’ era. Nature. 1983;302:725–726. doi: 10.1038/302725a0. [DOI] [PubMed] [Google Scholar]
- 107.Humbert P, Treffel P, Chapuis J F, Buchet S, Derancourt C, Agache P. The tetracyclines in dermatology. J Am Acad Dermatol. 1991;25:691–697. doi: 10.1016/0190-9622(91)70255-z. [DOI] [PubMed] [Google Scholar]
- 108.Inamine J M, Burdett V. Structural organization of a 67-kilobase streptococcal conjugative element mediating multiple antibiotic resistance. J Bacteriol. 1985;161:620–626. doi: 10.1128/jb.161.2.620-626.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Institute of Medicine, Division of Health Promotion and Disease Prevention. Report of a study. Human health risks with the subtherapeutic use of penicillin or tetracyclines in animal feed. Washington, D.C.: National Academy Press; 1998. [Google Scholar]
- 110.Iwaki, S., N. Tamura, T. Kimura-Someya, S. Nada, and A. Yamaguchi. Cysteine-scanning mutagenesis of transmembrane segments 4 and 5 of the Tn10-encoded metal-tetracycline/H+ antiporter reveals a permeability barrier in the middle of a transmembrane water-filled channel. J. Biol. Chem. 275:22704–22712. [DOI] [PubMed]
- 111.Jahn G, Laufs R, Kaulfers P-M, Kolenda H. Molecular nature of two Haemophilus influenzae R factors containing resistance and the multiple integration of drug resistance transposons. J Bacteriol. 1979;138:584–597. doi: 10.1128/jb.138.2.584-597.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Janknegt R, Lashof A O, Gould I M, van der Meer J W M. Antibiotic use in Dutch hospitals 1991–1996. J Antimicrob Chemother. 2000;45:251–256. doi: 10.1093/jac/45.2.251. [DOI] [PubMed] [Google Scholar]
- 113.Johnson A P. GAR-936. Curr Opin Anti-infect Investig Drugs. 2000;2:164–170. [Google Scholar]
- 113a.Joint expert advisory committee on antibiotic resistance (JETACAR) The use of antibiotics in food-producing animals: antibiotic-resistant bacteria in animals and humans. Canberra, Australia: Commonwealth Department of Health and Aged Care and Commonwealth Department of Agriculture, Fisheries and Forestry; 1999. [Google Scholar]
- 114.Jones C E, Vyakrnam S, Eady E A, Cove J H, Cunliffe W J. Antibiotic resistant propionibacteria and acne: crisis or conundrum? J Investig Dermatol. 1996;108:381. [Google Scholar]
- 115.Jones C S, Osborne D J, Stanley J. Enterobacterial tetracycline resistance in relation to plasmid incompatibility. Mol Cell Probes. 1992;6:313–317. doi: 10.1016/0890-8508(92)90007-k. [DOI] [PubMed] [Google Scholar]
- 116.Jones C S, Osborne D J, Stanley J. Cloning of a probe for a previously undescribed enterobacterial tetracycline resistance gene. Lett Appl Microbiol. 1992;15:106–108. doi: 10.1111/j.1472-765x.1992.tb00738.x. [DOI] [PubMed] [Google Scholar]
- 117.Reference deleted.
- 118.Joshi N, Miller D Q. Doxycycline revisited. Arch Intern Med. 1997;157:1421–1428. [PubMed] [Google Scholar]
- 119.Ji B, Sow S, Perani E, Lienhardt C, Diderot V, Grosset J. Bactericidal activity of a single-dose combination of ofloxacin plus minocycline, with or without rifampin, against Mycobacterium leprae in mice and in lepromatous patients. Antimicrob Agents Chemother. 1998;42:1115–1120. doi: 10.1128/aac.42.5.1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Kariuki S, Mirza N B, Wasteson Y, Senerwa D, Gathuma J M, Olsvik O. Tetracycline resistance genes in Kenyan hospital isolates of Salmonella typhimurium. APMIS. 1992;100:629–634. doi: 10.1111/j.1699-0463.1992.tb03977.x. [DOI] [PubMed] [Google Scholar]
- 121.Kauc L, Goodgal S H. Introduction of transposon Tn916 DNA into Haemophilus influenzae and Haemophilus parainfluenzae. J Bacteriol. 1989;171:6625–6628. doi: 10.1128/jb.171.12.6625-6628.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Kehrenberg C, Werckenthin C, Schwarz S. Tn5706, a transposon-like element from Pasteurella multocida mediating tetracycline resistance. Antimicrob Agents Chemother. 1998;42:2116–2118. doi: 10.1128/aac.42.8.2116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Kenny G E, Cartwright F D. Susceptibilities of Mycoplasma hominis, Mycoplasma pneumoniae, and Ureaplasma urealyticum to new glycylcyclines in comparison with those to older tetracyclines. Antimicrob Agents Chemother. 1994;38:2628–2632. doi: 10.1128/aac.38.11.2628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Khan S A, Novick R P. Complete nucleotide sequence of pT181, a tetracycline resistance plasmid from Staphylococcus aureus. Plasmid. 1983;30:163–166. doi: 10.1016/0147-619x(83)90039-2. [DOI] [PubMed] [Google Scholar]
- 125.Khachatourians G G. Agricultural use of antibiotics and the evolution and transfer of antibiotic-resistant bacteria. Can Med Assoc J. 1998;159:1129–1136. [PMC free article] [PubMed] [Google Scholar]
- 126.Kimura T, Suzuki M, Sawai T, Yamaguchi A. Determination of a transmembrane segment using cysteine-scanning mutants of transposon Tn10-encoded metal-tetracycline/H+ antiporter. Biochemistry. 1996;35:15896–15899. doi: 10.1021/bi961568g. [DOI] [PubMed] [Google Scholar]
- 127.Kimura T, Ohnuma M, Sawai T, Yamaguchi A. Membrane topology of the transposon 10-encoded metal-tetracycline/H+ antiporter as studied by site-directed chemical labeling. J Biol Chem. 1997;272:580–585. doi: 10.1074/jbc.272.1.580. [DOI] [PubMed] [Google Scholar]
- 128.Kimura-Someya T, Iwaki S, Yamaguchi A. Site-directed chemical modification of cysteine-scanning mutants as to transmembrane segment II and its flanking regions of the Tn10-encoded metal-tetracycline/H+ antiporter reveals a transmembrane water-filled channel. J Biol Chem. 1998;273:32806–32811. doi: 10.1074/jbc.273.49.32806. [DOI] [PubMed] [Google Scholar]
- 129.Kimura-Someya T, Iwaki S, Konishi S, Tamura N, Kubo Y, Yamaguchi A. Cysteine-scanning mutagenesis around transmembrane segments 1 and 11 and their flanking loop regions of Tn10-encoded metal-tetracycline/H+ antiporter. J Biol Chem. 2000;275:18692–18697. doi: 10.1074/jbc.M000354200. [DOI] [PubMed] [Google Scholar]
- 130.Kisker C, Hinrichs W, Tovar K, Hillen W, Saenger W. The complex formed between Tet repressor and tetracycline-Mg2+ reveals mechanism of antibiotic resistance. J Mol Biol. 1995;247:260–280. doi: 10.1006/jmbi.1994.0138. [DOI] [PubMed] [Google Scholar]
- 131.Klein N C, Cunha B A. Tetracyclines. Med Clin North Am. 1995;79:789–801. doi: 10.1016/s0025-7125(16)30039-6. [DOI] [PubMed] [Google Scholar]
- 132.Knapp J S, Johnson S R, Zenilman J M, Roberts M C, Morse S A. High-level tetracycline resistance resulting from TetM in strains of Neisseria species, Kingella denitrificans, and Eikenella corrodens. Antimicrob Agents Chemother. 1988;32:765–767. doi: 10.1128/aac.32.5.765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Kobland J D, Gale G O, Gustafson R H, Simkins K L. Comparison of therapeutic versus subtherapeutic levels of chlortetracycline in the diet for selection of resistant Salmonella in experimentally challenged chickens. Poult Sci. 1987;66:1129–1137. doi: 10.3382/ps.0661129. [DOI] [PubMed] [Google Scholar]
- 134.Konishi S, Iwaki S, Kimura-Someya T, Yamaguchi A. Cysteine-scanning mutagenesis around transmembrane segment VI of Tn10-encoded metal-tetracycline/H+ antiporter. FEBS Lett. 1999;461:315–318. doi: 10.1016/s0014-5793(99)01490-8. [DOI] [PubMed] [Google Scholar]
- 135.Kordick D L, Papich M G, Breitschwerdt E B. Efficacy of enrofloxacin or doxycycline for treatment of Bartonella henselae or Bartonella clarridgeiae infection in cats. Antimicrob Agents Chemother. 1997;41:2448–2455. doi: 10.1128/aac.41.11.2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Kiser J S. A perspective on the use of antibiotics in animal feeds. J Anim Sci. 1976;42:1058–1072. doi: 10.2527/jas1976.4241058x. [DOI] [PubMed] [Google Scholar]
- 137.Kucers A, McK. Bennett N. The use of antibiotics. 4th ed. Oxford, United Kingdom: Heinemann Medical; 1987. [Google Scholar]
- 138.Kwon D H, Kim J J, Lee M, Yamaoka Y, Kato M, Osato M S, El-Zaatari F A K, Graham D Y. Isolation and characterization of tetracycline-resistant clinical isolates of Helicobacter pylori. Antimicrob Agents Chemother. 2000;44:3203–3205. doi: 10.1128/aac.44.11.3203-3205.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138a.Lacks S, Lopez A P, Greenberg B, Espinosa M. Identification and analysis of genes for tetracycline resistance and replication functions in the broad-host-range plasmid pSL1. J Mol Biol. 1986;192:753–765. doi: 10.1016/0022-2836(86)90026-4. [DOI] [PubMed] [Google Scholar]
- 139.Lacroix J-M, Walker C B. Detection and incidence of the tetracycline resistance determinant tet(M) in the microflora associated with adult periodontitis. J Periodontol. 1993;66:102–108. doi: 10.1902/jop.1995.66.2.102. [DOI] [PubMed] [Google Scholar]
- 140.Le Bouguenec C, de Cespedes G, Horaud T. Presence of chromosomal elements resembling the composite structure Tn3701 in streptococci. J Bacteriol. 1988;172:727–734. doi: 10.1128/jb.172.2.727-734.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Lee C, Langlois B E, Dawson K L. Detection of tetracycline resistance determinants in pig isolates from three herds with different histories of antimicrobial agent exposure. Appl Environ Microbiol. 1993;59:1467–1472. doi: 10.1128/aem.59.5.1467-1472.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Lefevre F C, Lepargneur J P, Guion D, Bei S. Tetracycline-resistant Chlamydia trachomatis in Toulouse, France. Pathol Biol. 1997;45:376–378. [PubMed] [Google Scholar]
- 143.Leng Z, Riley D E, Berger R E, Krieger J N, Roberts M C. Distribution and mobility of the tetracycline resistant determinant Tet Q. J Antimicrob Chemother. 1997;40:551–559. doi: 10.1093/jac/40.4.551. [DOI] [PubMed] [Google Scholar]
- 144.Levy S B. Resistance to the tetracyclines. In: Bryan L E, editor. Antimicrobial drug resistance. Orlando, Fla: Academic Press; 1984. pp. 191–240. [Google Scholar]
- 145.Levy S B. Tetracycline resistance determinants are widespread. ASM News. 1988;54:418–421. [Google Scholar]
- 146.Levy S B. The antibiotic paradox: how miracle drugs are destroying the miracle. New York, N.Y: Plenum Press; 1992. [Google Scholar]
- 147.Levy S B. Active efflux mechanisms for antimicrobial resistance. Antimicrob Agents Chemother. 1992;36:695–703. doi: 10.1128/aac.36.4.695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Levy S B, Fitzgerald G B, Macone A B. Changes in intestinal flora of farm personnel after introduction of a tetracycline-supplemented feed on a farm. N Engl J Med. 1976;295:583–588. doi: 10.1056/NEJM197609092951103. [DOI] [PubMed] [Google Scholar]
- 149.Levy S B, Buu-Hoi A, Marshall B. Transposon Tn10-like tetracycline resistance determinants in Haemophilus parainfluenzae. J Bacteriol. 1984;160:87–94. doi: 10.1128/jb.160.1.87-94.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Levy S B, McMurry L M, Barbosa T M, Burdett V, Courvalin P, Hillen W, Roberts M C, Rood J I, Taylor D E. Nomenclature for new tetracycline resistance determinants. Antimicrob Agents Chemother. 1999;43:1523–1524. doi: 10.1128/aac.43.6.1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Levy S B, McMurry L M, Burdett V, Courvalin P, Hillen W, Roberts M C, Taylor D E. Nomenclature for tetracycline resistance determinants. Antimicrob Agents Chemother. 1989;33:1373–1374. doi: 10.1128/aac.33.8.1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Li L-Y, Shoemaker N B, Salyers A A. Location and characteristics of the transfer region of Bacteriodes conjugative transposon and regulation of transfer genes. J Bacteriol. 1995;177:4992–4999. doi: 10.1128/jb.177.17.4992-4999.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Li X-Z, Livermore D M, Nikaido H. Role of efflux pumps in intrinsic resistance of Pseudomonas aeruginosa: resistance to tetracycline, chloramphenicol and norfloxacin. Antimicrob Agents Chemother. 1994;38:1732–1741. doi: 10.1128/aac.38.8.1732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Li X-Z, Nikaido H, Poole K. Role of mexA-mexB-oprM in antibiotic efflux in Pseudomonas aeruginosa. Antimicrob Agents Chemother. 1995;39:1948–1953. doi: 10.1128/aac.39.9.1948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Lima A A M, Lima N L, Pinho M C, Barros E A, Jr, Teixeria M J, Marins M C V, Guerrant R I. High frequency of strains multiply resistant to ampicillin, trimethoprim-sulfamethoxazole, streptomycin, chloramphenicol, and tetracycline isolated from patients with shigellosis in northeastern Brazil during the period 1988 to 1993. Antimicrob Agents Chemother. 1995;39:256–259. doi: 10.1128/aac.39.1.256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Reference deleted.
- 157.Lina G, Quaglia A, Reverdy M-E, LeClercq R, Vandenesch F, Etienne J. Distribution of genes encoding resistance to macrolides, lincosamides, and streptogramins among staphylococci. Antimicrob Agents Chemother. 1999;43:1062–1066. doi: 10.1128/aac.43.5.1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Linton A H. Antibiotic-resistant bacteria in animal husbandry. Br Med Bull. 1984;40:91–95. doi: 10.1093/oxfordjournals.bmb.a071953. [DOI] [PubMed] [Google Scholar]
- 159.Liss R H, Batchelor F R. Economic evaluations of antibiotic use and resistance—a perspective: report of task force 6. Rev Infect Dis. 1987;9(Suppl. 3):S297–S312. doi: 10.1093/clinids/9.supplement_3.s297. [DOI] [PubMed] [Google Scholar]
- 160.Louie M, Louie L, Papia G, Talbot J, Lovgren M, Simor A E. Molecular analysis of the genetic variation among penicillin-susceptible and penicillin-resistant Streptococcus pneumoniae serotypes in Canada. J Infect Dis. 1999;179:892–900. doi: 10.1086/314664. [DOI] [PubMed] [Google Scholar]
- 161.Lucas C E, Balthazar J T, Hagman K E, Shager W M. The MtrR repressor binds the DNA sequence between the mtrR and the mtrC genes in Neisseria gonorrhoeae. J Bacteriol. 1997;179:4123–4128. doi: 10.1128/jb.179.13.4123-4128.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Luna V A, Roberts M C. The presence of the tetO gene in a variety of tetracycline resistant Streptococcus pneumoniae serotypes from Washington State. J Antimicrob Chemother. 1998;42:613–619. doi: 10.1093/jac/42.5.613. [DOI] [PubMed] [Google Scholar]
- 163.Luo Z-O, Farrand S K. Cloning and characterization of a tetracycline resistance determinant present in Agrobacterium tumefaciens C58. J Bacteriol. 1999;181:618–626. doi: 10.1128/jb.181.2.618-626.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Lyras D, Rood J I. Genetic organization and distribution of tetracycline resistance determinants in Clostridium perfringens. Antimicrob Agents Chemother. 1996;40:2500–2504. doi: 10.1128/aac.40.11.2500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Manavathu E K, Fernandez C L, Cooperman B S, Taylor D E. Molecular studies on the mechanism of tetracycline resistance mediated by Tet(O) Antimicrob Agents Chemother. 1990;34:71–77. doi: 10.1128/aac.34.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Manganelli R, Romano L, Ricci S, Zazzi M, Pozzi G. Dosage of Tn916 circular intermediates in Enterococcus faecalis. Plasmid. 1995;34:48–57. doi: 10.1006/plas.1995.1032. [DOI] [PubMed] [Google Scholar]
- 167.Manie T, Khan S, Brozel V S, Veith W J, Gouws P A. Antimicrobial resistance of bacteria isolated from slaughtered and retail chickens in South Africa. Lett Appl Microbiol. 1998;26:253–258. doi: 10.1046/j.1472-765x.1998.00312.x. [DOI] [PubMed] [Google Scholar]
- 168.Marshall B, Roberts M, Smith A, Levy S B. Homogeneity of tetracycline-resistance determinants in Haemophilus species. J Infect Dis. 1984;149:1028–1029. doi: 10.1093/infdis/149.6.1028. [DOI] [PubMed] [Google Scholar]
- 169.Marshall B, Tachibana C, Levy S B. Frequency of tetracycline resistance determinant classes among lactose-fermenting coliforms. Antimicrob Agents Chemother. 1983;24:835–840. doi: 10.1128/aac.24.6.835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Martinez-Salazar J M, Alvarez G, Gomez-Eichelmann M C. Frequency of four classes of tetracycline resistance determinants in Salmonella and Shigella spp. clinical isolates. Antimicrob Agents Chemother. 1986;30:630–631. doi: 10.1128/aac.30.4.630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Maurin M, Raoult D. Q fever. Clin Microbiol Rev. 1999;12:518–553. doi: 10.1128/cmr.12.4.518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.McCaig L F, Hughes J M. Trends in antimicrobial drug prescribing among office-based physicians in the United States. JAMA. 1995;273:214–219. [PubMed] [Google Scholar]
- 173.McMurry L M, Levy S B. Tetracycline resistance in gram-positive bacteria. In: Fischetti V A, Novick R P, Ferretti J J, Portnoy D A, Rood J I, editors. Gram-positive pathogens. Washington, D.C.: American Society for Microbiology; 2000. pp. 660–677. [Google Scholar]
- 174.McNicholas P, Chopra I, Rothstein D M. Genetic analysis of the TetA(C) gene on plasmid pBR322. J Bacteriol. 1992;174:7926–7933. doi: 10.1128/jb.174.24.7926-7933.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.McNicholas P, McGlynn M, Guay G G, Rothstein D M. Genetic analysis suggests functional interactions between the N- and C-terminal domains of the TetA(C) efflux pump encoded by pBR322. J Bacteriol. 1995;177:5355–5357. doi: 10.1128/jb.177.18.5355-5357.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Mendez B, Tachibana C, Levy S B. Heterogeneity of tetracycline resistance determinants. Plasmid. 1980;3:99–108. doi: 10.1016/0147-619x(80)90101-8. [DOI] [PubMed] [Google Scholar]
- 177.Midolo P D, Korman M G, Turnidge J D, Lambert J R. Helicobacter pylori resistance to tetracycline. Lancet. 1996;347:1194–1195. [PubMed] [Google Scholar]
- 178.Reference deleted.
- 179.Mitscher L A. The chemistry of the tetracycline antibiotics. New York, N.Y: Marcel Dekker, Inc; 1978. [Google Scholar]
- 180.Moazed D, Noller H F. Interaction of antibiotics with functional sites in 16S ribosomal RNA. Nature. 1987;327:389–394. doi: 10.1038/327389a0. [DOI] [PubMed] [Google Scholar]
- 181.Molbak K, Baggesen D L, Aarestrup F M, Ebbesen J M, Engberg J, Frydendahl K, Gerner-Smidt P, Petersen A M, Wegener H C. An outbreak of multidrug-resistant, quinolone-resistant Salmonella enterica serotype Typhimurum DT104 N. Engl J Med. 1999;341:1420–1425. doi: 10.1056/NEJM199911043411902. [DOI] [PubMed] [Google Scholar]
- 182.Morse S A, Johnson S J, Biddle J W, Roberts M C. High-level tetracycline resistance in Neisseria gonorrhoeae due to the acquisition of the tetM determinant. Antimicrob Agents Chemother. 1986;30:664–670. doi: 10.1128/aac.30.5.664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Naglich J G, Andrews R E., Jr Tn916-dependent conjugal transfer of pC194 and pUB110 from Bacillus subtillis into Bacillus thuringiensis subsp. israelensis. Plasmid. 1988;20:113–126. doi: 10.1016/0147-619x(88)90014-5. [DOI] [PubMed] [Google Scholar]
- 184.Needham C, Rahman M, Dyke K G H, Noble W C. An investigation of plasmids from Staphylococcus aureus that mediate resistance to mupirocin and tetracycline. Microbiology. 1994;140:2577–2583. doi: 10.1099/00221287-140-10-2577. [DOI] [PubMed] [Google Scholar]
- 185.Nelson M L, Park B H, Andrew J S, Georgian V A, Thomas B C, Levy S B. Inhibition of the tetracycline efflux antiport protein by 13-thio-substituted 5-hydroxy-6-deoxytetracyclines. J Med Chem. 1993;36:370–377. doi: 10.1021/jm00055a008. [DOI] [PubMed] [Google Scholar]
- 186.Nelson M L, Levy S B. Reversal of tetracycline resistance mediated by different bacterial tetracycline resistance determinants by an inhibitor of the Tet(B) antiport protein. Antimicrob Agents Chemother. 1999;43:1719–1724. doi: 10.1128/aac.43.7.1719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Reference deleted.
- 188.Nesin M, Svec P, Lupski J R, Godson G N, Kreiswirth B, Kornblum J, Projan S J. Cloning and nucleotide sequence of a chromosomally encoded tetracycline resistance determinant, tetA(M), from a pathogenic, methicillin-resistant strain of Staphylococcus aureus. Antimicrob Agents Chemother. 1990;34:2273–2276. doi: 10.1128/aac.34.11.2273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Ng L-K, Mulvery M R, Martin I, Peters G A, Johnson W. Genetic characterization of antimicrobial resistance in Canadian isolates of Salmonella serovar Typhimurium DT104. Antimicrob Agents Chemother. 1999;43:3018–3021. doi: 10.1128/aac.43.12.3018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Nikaido H. Multidrug efflux pumps of gram-negative bacteria. J Bacteriol. 1998;178:5853–5859. doi: 10.1128/jb.178.20.5853-5859.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Nikaido H. Multiple antibiotic resistance and efflux. Curr Opin Microbiol. 1998;1:516–523. doi: 10.1016/s1369-5274(98)80083-0. [DOI] [PubMed] [Google Scholar]
- 192.Nikaido H, Thanassi D G. Penetration of lipophilic agents with multiple protonation sites into bacterial cells: tetracyclines and fluroquinolones as examples. Antimicrob Agents Chemother. 1993;37:1393–1399. doi: 10.1128/aac.37.7.1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Noah J W, Dolan M A, Babin P, Wollenzien P. Effects of tetracycline and spectinomycin on the tertiary structure of ribosomal RNA in the Escherichia coli 30S ribosomal subunit. J Biol Chem. 1999;274:16576–16581. doi: 10.1074/jbc.274.23.16576. [DOI] [PubMed] [Google Scholar]
- 194.Nord C E, Lindmark A, Persson I. In vitro activity of DMG-Mino and DMG-DM Dot, two new glycylcyclines, against anaerobic bacteria. Eur J Clin Microbiol Infect Dis. 1993;12:784–786. doi: 10.1007/BF02098471. [DOI] [PubMed] [Google Scholar]
- 195.O'Brien T F the Members of Task Force 2. Resistance of bacteria to antibacterial agents: report of task force 2. Rev Infect Dis. 1987;9:S244–S260. doi: 10.1093/clinids/9.supplement_3.s244. [DOI] [PubMed] [Google Scholar]
- 196.Oehler R, Polacek N, Steiner G, Barta A. Interaction of tetracycline with RNA: photoincorporation into ribosomal RNA of Escherichia coli. Nucleic Acids Res. 1997;25:1219–1224. doi: 10.1093/nar/25.6.1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Oethinger M, Kern W V, Jellen-Ritter A S, McMurry L M, Levy S B. Ineffectiveness of topoisomerase mutations in mediating clinically significant fluoroquinolone resistance in Escherichia coli in the absence of the AcrAB efflux pump. Antimicrob Agents Chemother. 2000;44:10–13. doi: 10.1128/aac.44.1.10-13.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Oliva B, Chopra I. Tet determinants provide poor protection against some tetracyclines: further evidence for division of tetracyclines into two classes. Antimicrob Agents Chemother. 1992;36:876–878. doi: 10.1128/aac.36.4.876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Oliva B, Gordon G, McNicholas P, Ellestad G, Chopra I. Evidence that tetracycline analogs whose primary target is not the bacterial ribosome cause lysis of Escherichia coli. Antimicrob Agents Chemother. 1992;36:913–919. doi: 10.1128/aac.36.5.913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Olsvik B, Olsen I, Tenover F C. Tet tetQ gene in bacteria isolated from patients with refractory periodontal disease. Oral Microbiol Immunol. 1994;9:251–255. doi: 10.1111/j.1399-302x.1994.tb00067.x. [DOI] [PubMed] [Google Scholar]
- 201.Olsvik B, Olsen I, Tenover F C. Detection of tet(M) and tet(O) using the polymerase chain reaction in bacteria isolated from patients with periodontal disease. Oral Microbiol Immun. 1995;10:87–92. doi: 10.1111/j.1399-302x.1995.tb00124.x. [DOI] [PubMed] [Google Scholar]
- 202.Orth P, Schnappinger D, Sum P-E, Ellestad G A, Hillen W, Saenger W, Hinrichs W. Crystal structure of the Tet repressor in complex with a novel tetracycline, 9-(N,N-dimethylglycylamido)-6-demethyl-6-deoxytetracycline. J Mol Biol. 1999;285:455–461. doi: 10.1006/jmbi.1998.2290. [DOI] [PubMed] [Google Scholar]
- 203.Orth P, Schnappinger D, Hillen W, Saenger W, Hinrichs W. Structural basis of gene regulation by the tetracycline inducible Tet repressor-operator system. Nat Struct Biol. 2000;7:215–219. doi: 10.1038/73324. [DOI] [PubMed] [Google Scholar]
- 204.Ouellette M, Kundig C. Microbial multidrug resistance. Int J Antimicrob Agents. 1997;8:179–187. doi: 10.1016/s0924-8579(96)00370-6. [DOI] [PubMed] [Google Scholar]
- 205.Paulsen I T, Brown M H, Skurray R A. Proton-dependent multidrug efflux systems. Microbiol Rev. 1996;60:575–608. doi: 10.1128/mr.60.4.575-608.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Pang Y, Bosch T, Roberts M C. Single polymerase chain reaction for the detection of tetracycline resistant determinants Tet K and Tet L. Mol Cell Probes. 1994;8:417–422. doi: 10.1006/mcpr.1994.1059. [DOI] [PubMed] [Google Scholar]
- 207.Pang Y, Brown B A, Steingrube V A, Wallace R J, Jr, Roberts M C. Acquisition of gram-positive tetracycline resistance genes in Mycobacterium and Streptomyces species. Antimicrob Agents Chemother. 1994;38:1408–1412. doi: 10.1128/aac.38.6.1408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Petersen P J, Jacobus N V, Weiss W J, Sum P E, Testa R T. In vitro and in vivo antibacterial activities of a novel glycylcycline, the 9-t-butylglycylamido derivative of minocycline (GAR-936) Antimicrob Agents Chemother. 1999;43:738–744. doi: 10.1128/aac.43.4.738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Poyart C, Celli J, Trieu-Cuot P. Conjugative transposition of Tn916-related elements from Enterococcus faecalis to Escherichia coli and Pseudomonas fluorescens. Antimicrob Agents Chemother. 1995;39:500–506. doi: 10.1128/aac.39.2.500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Poyart C, Quesne G, Acar P, Berche P, Trieu-Cuot P. Characterization of the Tn916-like transposon Tn3872 on a strain of Abiotrophia defectiva (Streptococcus defectivus) causing sequential episodes of endocarditis in a child. Antimicrob Agents Chemother. 2000;44:790–793. doi: 10.1128/aac.44.3.790-793.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Pradines B, Spiegel A, Rogier C, Tall A, Mosnier J, Fusai T, Trape J F, Parzy D. Antibiotics for prophylaxis of Plasmodium falciparum infections: in vitro activity of doxycycline against Senegalese isolates. Am J Trop Med Hyg. 2000;62:82–85. doi: 10.4269/ajtmh.2000.62.82. [DOI] [PubMed] [Google Scholar]
- 212.Projan S J, Monod M, Narayanan C S, Dubnau D. Replication properties of pIM13, a naturally occurring plasmid found in Bacillus subtilis, and of its close relative pE5, a plasmid native to Staphylococcus aureus. J Bacteriol. 1987;169:5131–5139. doi: 10.1128/jb.169.11.5131-5139.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Projan S J, Novick R. Comparative analysis of five related staphylococcal plasmids. Plasmid. 1988;19:203–221. doi: 10.1016/0147-619x(88)90039-x. [DOI] [PubMed] [Google Scholar]
- 214.Pukrittayakamee S, Chantra A, Vanijanonta S, Clemens R, Looareesuwan S, White N J. Therapeutic responses to quinine and clindamycin in multidrug-resistant falciparum malaria. Antimicrob Agents Chemother. 2000;44:2395–2398. doi: 10.1128/aac.44.9.2395-2398.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Rasmussen B A, Gluzman Y, Tally F P. Inhibition of protein synthesis occurring on tetracycline-resistant, TetM-protected ribosomes by a novel class of tetracyclines, the glycylcyclines. Antimicrob Agents Chemother. 1994;38:1658–1660. doi: 10.1128/aac.38.7.1658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Recchia G D, Hall R M. Gene cassettes: a new class of mobile element. Microbiology. 1995;141:3015–3027. doi: 10.1099/13500872-141-12-3015. [DOI] [PubMed] [Google Scholar]
- 217.Reference deleted.
- 218.Reynes J P, Calmels T, Drocourt D, Tiragy G. Cloning, expression in Escherichia coli and nucleotide sequence of a tetracycline-resistance gene from Streptomyces rimosus. J Gen Microbiol. 1988;134:585–598. doi: 10.1099/00221287-134-3-585. [DOI] [PubMed] [Google Scholar]
- 219.Rice L B, Marshall S H, Carias L L. Tn5381, a conjugative transposon identifiable as a circular form in Enterococcus faecalis. J Bacteriol. 1992;174:7380–7315. doi: 10.1128/jb.174.22.7308-7315.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Ridenhour M B, Fletcher H M, Mortensen J E, Daneo-Moore L. A novel tetracycline-resistant determinant, tet(U), is encoded on the plasmid pKQ10 in Enterococcus faecium. Plasmid. 1996;35:71–80. doi: 10.1006/plas.1996.0009. [DOI] [PubMed] [Google Scholar]
- 221.Riesbeck K, Bredberg A, Forsgren A. Ciprofloxacin does not inhibit mitochondrial functions but other antibiotics do. Antimicrob Agents Chemother. 1990;34:167–169. doi: 10.1128/aac.34.1.167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Roberts M C. Gene transfer in the urogenital and respiratory tract. In: Levy S, Miller R V, editors. Gene transfer in the environment. New York, N.Y: McGraw-Hill Publishing Co.; 1989. pp. 347–375. [Google Scholar]
- 223.Roberts M C. Plasmids of Neisseria gonorrhoeae and other Neisseria species. Rev Clin Microbiol. 1989;2:S18–S23. doi: 10.1128/cmr.2.suppl.s18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Roberts M C. Plasmid-mediated Tet M in Haemophilus ducreyi. Antimicrob Agents Chemother. 1989;33:1611–1613. doi: 10.1128/aac.33.9.1611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Roberts M C. Characterization of the Tet M determinant in urogenital and respiratory bacteria. Antimicrob Agents Chemother. 1990;34:476–478. doi: 10.1128/aac.34.3.476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Roberts M C. Tetracycline resistance in Peptostreptococcus species. Antimicrob Agents Chemother. 1991;35:1682–1684. doi: 10.1128/aac.35.8.1682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Roberts M C. Epidemiology of tetracycline resistance determinants. Trends Microbiol. 1994;2:353–357. doi: 10.1016/0966-842x(94)90610-6. [DOI] [PubMed] [Google Scholar]
- 228.Roberts M C. Tetracycline resistant determinants: mechanisms of action, regulation of expression, genetic mobility and distribution. FEMS Microbiol Rev. 1996;19:1–24. doi: 10.1111/j.1574-6976.1996.tb00251.x. [DOI] [PubMed] [Google Scholar]
- 229.Roberts M C. Genetic mobility and distribution of tetracycline resistance determinants. Ciba Found Symp. 1997;207:206–218. [PubMed] [Google Scholar]
- 230.Roberts M C. Oral bacteria: reservoirs for antibiotic resistance traits. APUA Newsl. 1997;15:1–6. [Google Scholar]
- 231.Roberts M C. APUA—Alliance for the prudent use of antibiotics. ASM News. 2000;66:108. [Google Scholar]
- 232.Roberts M C, Actis L A, Crosa J H. Molecular characterization of chloramphenicol resistant Haemophilus parainfluenzae and Haemophilus ducreyi. Antimicrob Agents Chemother. 1985;28:176–180. doi: 10.1128/aac.28.2.176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Roberts M C, Chung W, Roe D E. Characterization of tetracycline and erythromycin determinants in Treponema denticola. Antimicrob Agents Chemother. 1996;40:1690–1694. doi: 10.1128/aac.40.7.1690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Roberts M C, Chung W, Roe D E, Xia M, Marquez C, Borthagaray G, Whittington W L, Holmes K K. Erythromycin-resistant Neisseria gonorrhoeae and oral commensal Neisseria spp. carry known rRNA methylase genes. Antimicrob Agents Chemother. 1999;43:1367–1372. doi: 10.1128/aac.43.6.1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Roberts M C, Hillier S L. Genetic basis of tetracycline resistance in urogenital bacteria. Antimicrob Agents Chemother. 1990;34:261–264. doi: 10.1128/aac.34.2.261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Roberts M C, Hillier S L, Hale J, Holmes K K, Kenny G E. Tetracycline resistance and tetM in pathogenic urogenital bacteria. Antimicrob Agents Chemother. 1986;30:810–812. doi: 10.1128/aac.30.5.810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Roberts M C, Kenny G E. Dissemination of the tetM tetracycline resistance determinant to Ureaplasma urealyticum. Antimicrob Agents Chemother. 1986;29:350–352. doi: 10.1128/aac.29.2.350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Roberts M C, Kenny G E. Conjugal transfer of transposon Tn916 from Streptococcus faecalis to Mycoplasma hominis. J Bacteriol. 1987;169:3836–3839. doi: 10.1128/jb.169.8.3836-3839.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Roberts M C, Knapp J S. Host range of the conjugative 25.2 Mdal tetracycline resistance plasmid from Neisseria gonorrhoeae. Antimicrob Agents Chemother. 1988;32:488–491. doi: 10.1128/aac.32.4.488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Roberts M C, Knapp J S. Transfer frequency of various 25.2 Mdal TetM-containing plasmids in Neisseria gonorrhoeae. Sex Transm Dis. 1989;16:91–94. doi: 10.1097/00007435-198904000-00010. [DOI] [PubMed] [Google Scholar]
- 241.Roberts M C, Lansciardi J. Transferable TetM in Fusobacterium nucleatum. Antimicrob Agents Chemother. 1990;34:1836–1838. doi: 10.1128/aac.34.9.1836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Roberts M C, Leonard R B, Briselden A M, Schoenknecht F D, Coyle M B. Characterization of antibiotic resistant Corynebacterium striatum strains. J Antimicrob Chemother. 1992;30:463–474. doi: 10.1093/jac/30.4.463. [DOI] [PubMed] [Google Scholar]
- 243.Roberts M C, McFarland L V, Mullany P, Mulligan M E. Characterization of the genetic basis of antibiotic resistance in Clostridium difficile. J Antimicrob Chemother. 1994;33:419–429. doi: 10.1093/jac/33.3.419. [DOI] [PubMed] [Google Scholar]
- 244.Roberts M C, Moncla B J, Hillier S L. Characterization of unusual tetracycline-resistant gram-positive bacteria. Antimicrob Agents Chemother. 1991;35:2655–2657. doi: 10.1128/aac.35.12.2655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Roberts M C, Pang Y, Riley D E, Hillier S L, Berger R, Krieger J N. Detection of Tet M and Tet O tetracycline resistance genes by polymerase chain reaction. Mol Cell Probes. 1993;7:387–393. doi: 10.1006/mcpr.1993.1057. [DOI] [PubMed] [Google Scholar]
- 246.Roberts M C, Pang Y, Spencer R C, Winstanley T G, Brown B A, Wallace R J., Jr Tetracycline resistance in Moraxella (Branhamella) catarrhalis—demonstration of two clonal outbreaks using pulsed-field gel electrophoresis. Antimicrob Agents Chemother. 1991;35:2453–2455. doi: 10.1128/aac.35.11.2453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Roberts M C, Smith A L. Molecular characterization of “plasmid-free” antibiotic resistant Haemophilus influenzae. J Bacteriol. 1980;144:476–479. doi: 10.1128/jb.144.1.476-479.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Roe D E, Braham P, Weinberg A, Roberts M C. Characterization of tetracycline resistance in Actinobacillus actinomycetemcomitans. Oral Microbiol Immunol. 1995;10:227–232. doi: 10.1111/j.1399-302x.1995.tb00147.x. [DOI] [PubMed] [Google Scholar]
- 249.Roe D E, Weinberg A, Roberts M C. Characterization of erythromycin resistance in Campylobacter (Wolinella) rectus. Clin Infect Dis. 1995;20:S370–S371. doi: 10.1093/clinids/20.supplement_2.s370. [DOI] [PubMed] [Google Scholar]
- 250.Rogalski W. Chemical modification of the tetracyclines. In: Hlavka J J, Boothe J H, editors. Handbook of experimental pharmacology. Vol. 78. Berlin, Germany: Springer-Verlag KG; 1985. pp. 179–316. [Google Scholar]
- 251.Ross J I, Eady E A, Cove J H, Cunliffe W J. 16S rRNA mutation associated with tetracycline resistance in a gram-positive bacterium. Antimicrob Agents Chemother. 1998;42:1702–1705. doi: 10.1128/aac.42.7.1702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Rouch D A, Skurray R A. IS257 from Staphylococcus aureus: a member of an insertion sequence superfamily prevalent among Gram-positive and Gram-negative bacteria. Gene. 1989;76:195–205. doi: 10.1016/0378-1119(89)90160-1. [DOI] [PubMed] [Google Scholar]
- 253.Rubin R A, Levy S B. Interdomain hybrid Tet proteins confer tetracycline resistance only when they are derived from closely related members of the tet gene family. J Bacteriol. 1991;173:4503–4509. doi: 10.1128/jb.172.5.2303-2312.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.Sabath L D. Current concepts: drug resistance of bacteria. N Engl J Med. 1969;280:91–94. doi: 10.1056/NEJM196901092800208. [DOI] [PubMed] [Google Scholar]
- 255.Sakaguchi R, Shishido K. Molecular cloning of a tetracycline-resistance determinant from Bacillus subtilis chromosomal DNA and its expression in Escherichia coli and B. subtilis. Biochim Biophys Acta. 1988;949:9–57. doi: 10.1016/0167-4781(88)90053-x. [DOI] [PubMed] [Google Scholar]
- 256.Salyers A A, Shoemaker N B, Li L-Y. In the Driver's seat: the Bacteroides conjugative transposons and the elements they mobilize. J Bacteriol. 1995;177:5727–5731. doi: 10.1128/jb.177.20.5727-5731.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257.Salyers A A, Shoemaker N B, Stevens A M, Li L-Y. Conjugative transposons: an unusual and diverse set of integrated gene transfer elements. Microbiol Rev. 1995;59:579–590. doi: 10.1128/mr.59.4.579-590.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.Sanchez L, Pan W, Vinas M, Nikaido H. The acrAB homology of Haemophilus influenzae codes for a functional multidrug efflux pump. J Bacteriol. 1997;179:6855–6857. doi: 10.1128/jb.179.21.6855-6857.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259.Sanchez-Pescador R, Brown J T, Roberts M, Urdea M S. Homology of the TetM with translational elongation factors: implications for potential modes of tetM conferred tetracycline resistance. Nucleic Acids Res. 1988;16:1218. doi: 10.1093/nar/16.3.1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.Sangare L, Morrisset R, Ravaoarinoro M. In-vitro anti-chlamydial activities of free and liposomal tetracycline and doxycycline. J Med Microbiol. 1999;48:689–693. doi: 10.1099/00222615-48-7-689. [DOI] [PubMed] [Google Scholar]
- 261.Seppala H, Klaukka T, Vuopio-Varkila J, Muotiala A, Helenius H, Lager K, Huovinen P. The effect of changes in the consumption of macrolide antibiotics on erythromycin resistance in group A streptococci in Finland. N Engl J Med. 1997;337:441–446. doi: 10.1056/NEJM199708143370701. [DOI] [PubMed] [Google Scholar]
- 262.Schnabel E L, Jones A L. Distribution of tetracycline resistance genes and transposons among phylloplane bacteria in Michigan apple orchards. Appl Environ Microbiol. 1999;65:4898–4907. doi: 10.1128/aem.65.11.4898-4907.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Schnappinger D, Hillen W. Tetracyclines: antibiotic action, uptake, and resistance mechanisms. Arch Microbiol. 1996;165:359–369. doi: 10.1007/s002030050339. [DOI] [PubMed] [Google Scholar]
- 264.Schwarz E, Regev-Yochay G. Primaquine as prophylaxis for malaria for nonimmune travelers: a comparison with mefloquine and doxycycline. Clin Infect Dis. 1999;29:1502–1506. doi: 10.1086/313527. [DOI] [PubMed] [Google Scholar]
- 265.Schwarz S, Cardoso M, Wegener H C. Nucleotide sequence and phylogeny of tet(L) tetracycline resistance determinant encoded by plasmid pSTE1 from Staphylococcus hyicus. Antimicrob Agents Chemother. 1992;36:580–588. doi: 10.1128/aac.36.3.580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266.Schwarz S, Roberts M C, Werckenthin C, Pang Y, Lange C. Tetracycline resistance in Staphylococcus spp. from domestic and pet animals. Vet Microbiol. 1998;63:217–228. doi: 10.1016/s0378-1135(98)00234-x. [DOI] [PubMed] [Google Scholar]
- 267.Scott K P, Melville C M, Barbosa T M, Flint H J. Occurrence of the new tetracycline resistance gene tet(W) in bacteria from the human gut. Antimicrob Agents Chemother. 2000;44:775–777. doi: 10.1128/aac.44.3.775-777.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268.Scott J R. Sex and the single circle: conjugative transposition. J Bacteriol. 1992;174:6005–6010. doi: 10.1128/jb.174.19.6005-6010.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269.Sheridan R P, Chopra I. Origin of tetracycline efflux proteins: conclusions from nucleotide sequence analysis. Mol Microbiol. 1991;5:895–900. doi: 10.1111/j.1365-2958.1991.tb00763.x. [DOI] [PubMed] [Google Scholar]
- 270.Shonekan D, Handwerger S, Mildvan D. Comparative in-vitro activities of RP59500 (quinupristin/dalfopristin), CL 329,998, CL 331,002, trovafloxacin, clinafloxacin, teicoplanin and vancomycin against Gram-positive bacteria. J Antimicrob Chemother. 1997;39:405–409. doi: 10.1093/jac/39.3.405. [DOI] [PubMed] [Google Scholar]
- 271.Shaw W V, Bouanchaud D H, Goldstein F W. Mechanism of transferable resistance to chloramphenicol in Haemophilus parainfluenzae. Antimicrob Agents Chemother. 1978;13:326–330. doi: 10.1128/aac.13.2.326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 272.Showsh S A, Andrews R E., Jr Tetracycline enhances Tn916-mediated conjugal transfer. Plasmid. 1992;28:213–224. doi: 10.1016/0147-619x(92)90053-d. [DOI] [PubMed] [Google Scholar]
- 273.Sloan J, McMurry L M, Lyras D, Levy S B, Rood J I. The Clostridium perfringens Tet P determinant comprises two overlapping genes: tetA(P), which mediates active tetracycline efflux, and tetB(P), which is related to the ribosomal protection family of tetracycline-resistance determinants. Mol Microbiol. 1994;11:403–415. doi: 10.1111/j.1365-2958.1994.tb00320.x. [DOI] [PubMed] [Google Scholar]
- 274.Smidt H, Song D, van der Oost J, de Vos W M. Random transposition by Tn916 in Desulfitobacterium dehalogenans allows for isolation and characterization of halorespiration-deficient mutants. J Bacteriol. 1999;181:6882–6888. doi: 10.1128/jb.181.22.6882-6888.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 275.Smilack J D. The tetracyclines. Mayo Clin Proc. 1999;74:727–729. doi: 10.4065/74.7.727. [DOI] [PubMed] [Google Scholar]
- 276.Smith H W, Crabb W E. The effect of the continuous administration of diets containing low levels of tetracyclines on the incidence of drug-resistant Bacterium coli in the faeces of pigs and chickens: the sensitivity of the Bacterium coli to other chemotherapeutic agents. Vet Rec. 1976;69:24–30. [Google Scholar]
- 277.Somani J, Bhullar V B, Workowski K A, Farshy C E, Black C M. Multiple drug-resistant Chlamydia trachomatis associated with clinical treatment failure. J Infect Dis. 2000;181:1420–1427. doi: 10.1086/315372. [DOI] [PubMed] [Google Scholar]
- 278.Someya Y, Yamaguchi A, Sawai T. A novel glycylcycline, 9-(N,N-dimethylglycylamido)-6-demethyl-6-deoxytetracycline, is neither transported nor recognized by the transposon Tn10-encoded metal-tetracycline/H+ antiporter. Antimicrob Agents Chemother. 1995;39:247–249. doi: 10.1128/aac.39.1.247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 279.Someya Y, Kimura-Someya T, Yamaguchi A. Role of the charge interaction between Arg(70) and Asp(120) in the Tn10-encoded metal-tetracycline/H+ antiporter of Escherichia coli. J Biol Chem. 2000;275:210–214. doi: 10.1074/jbc.275.1.210. [DOI] [PubMed] [Google Scholar]
- 280.Sorum H, Roberts M C, Crosa J H. Identification and cloning of a tetracycline resistance gene from the fish pathogen Vibrio salmonicida. Antimicrob Agents Chemother. 1992;36:611–615. doi: 10.1128/aac.36.3.611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 281.Speer B S, Bedzyk L, Salyers A A. Evidence that a novel tetracycline resistance gene found on two Bacteroides transposons encodes an NADP-requiring oxidoreductase. J Bacteriol. 1991;173:176–183. doi: 10.1128/jb.173.1.176-183.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 282.Spies T, Laufs R, Riess F-C. Amplification of resistance genes in Haemophilus influenzae plasmids. J Bacteriol. 1983;155:839–846. doi: 10.1128/jb.155.2.839-846.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 283.Stasinopoulos S J, Farr G A, Bechhofer D H. Bacillus subtilis tetA(L) gene expression: evidence for regulation by translational reinitiation. Mol Microbiol. 1998;30:923–932. doi: 10.1046/j.1365-2958.1998.01119.x. [DOI] [PubMed] [Google Scholar]
- 284.Stockstad E L R, Jukes T H, Pierce J, Page A C, Franklin A L. The multiple nature of the animal protein factor. J Biol Chem. 1949;180:647–654. [PubMed] [Google Scholar]
- 285.Su Y A, He P, Clewell D B. Characterization of the tet(M) determinant of Tn916: evidence for regulation by transcription attenuation. Antimicrob Agents Chemother. 1992;36:769–778. doi: 10.1128/aac.36.4.769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 286.Sum P-E, Lee V J, Testa R T, Hlavka J J, Ellestad G A, Bloom J D, Gluzman Y, Tally F P. Glycylcyclines. 1. A new generation of potent antibacterial agents through modification of 9-aminotetracyclines. J Med Chem. 1994;37:184–188. doi: 10.1021/jm00027a023. [DOI] [PubMed] [Google Scholar]
- 287.Sum P E, Petersen P. Synthesis and structure-activity relationship of novel glycylcycline derivatives leading to the discovery of GAR-936. Bioorg Med Chem Lett. 1999;9:1459–1462. doi: 10.1016/s0960-894x(99)00216-4. [DOI] [PubMed] [Google Scholar]
- 288.Swann M M. Report of joint committee on the use of antibiotics in animal husbandry and veterinary medicine. Cmnd. 4190. London, United Kingdom: Her Majesty's Stationery Office; 1969. [Google Scholar]
- 289.Tamayo M, Sa-Leao R, Sanches I S, Castaneda E, de Lencastre H. Dissemination of a chloramphenicol-and tetracycline-resistant but penicillin-susceptible invasive clone of serotype 5 Streptococcus pneumoniae in Colombia. J Clin Microbiol. 1999;7:2337–2342. doi: 10.1128/jcm.37.7.2337-2342.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 290.Tannock G W, Luchansky J B, Miller L, Connell H, Thode-Andersen S, Mercer A A, Kaenhammer T R. Molecular characterization of a plasmid-borne (pGT633) erythromycin resistance determinant (ermGT) from Lactobacillus reuteri 100–63. Plasmid. 1994;31:60–71. doi: 10.1006/plas.1994.1007. [DOI] [PubMed] [Google Scholar]
- 291.Tauch A, Puhler A, Kalinowski J, Thierbach G. TetZ, a new tetracycline resistance determinant discovered in gram-positive bacteria, shows high homology to gram-negative regulated efflux systems. Plasmid. 2000;44:285–291. doi: 10.1006/plas.2000.1489. [DOI] [PubMed] [Google Scholar]
- 292.Taylor D E, Chau A. Tetracycline resistance mediated by ribosomal protection. Antimicrob Agents Chemother. 1996;40:1–5. doi: 10.1128/aac.40.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 293.Taylor D E, Courvalin P. Mechanisms of antibiotic resistance in Campylobacter species. Antimicrob Agents Chemother. 1988;32:1107–1112. doi: 10.1128/aac.32.8.1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 294.Taylor D E, Trieber C A, Trescher G, Bekkering M. Host mutations (miaA and rpsL) reduce tetracycline resistance mediated by Tet(O) and Tet(M) Antimicrob Agents Chemother. 1998;42:59–64. doi: 10.1128/aac.42.1.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 295.Testa R T, Petersen P J, Jacobus N L, Sum P-E, Lee V J, Tally F P. In vitro and in vivo antibacterial activities of the glycylcyclines, a new class of semisynthetic tetracyclines. Antimicrob Agents Chemother. 1993;37:2270–2277. doi: 10.1128/aac.37.11.2270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 296.Threlfall E J, Frost J A, Ward L R, Rowe B. Epidemic in cattle and humans of Salmonella typhimurium DT104 with chromsomally integrated multiple drug resistance. Vet Rec. 1994;134:577. doi: 10.1136/vr.134.22.577. [DOI] [PubMed] [Google Scholar]
- 297.Torres O R, Korman R Z, Zahler S A, Dunny G M. The conjugative transposon Tn925: enhancement of conjugal transfer by tetracycline in Enterococcus faecalis and mobilization of chromosomal genes in Bacillus subtilis and E. faecalis. Mol Gen Genet. 1991;225:395–400. doi: 10.1007/BF00261679. [DOI] [PubMed] [Google Scholar]
- 298.Trieber C A, Burkhardt N, Nierhaus K H, Taylor D E. Ribosomal protection from tetracycline mediated by Tet(O): Tet(O) interaction with ribosomes is GTP-dependent. Biol Chem. 1998;379:847–855. doi: 10.1515/bchm.1998.379.7.847. [DOI] [PubMed] [Google Scholar]
- 299.Reference deleted.
- 300.Tymiak A A, Aklonis C, Bolgar M S, Kahle A D, Kirsch D R, O'Sullivan J, Porubcan M A, Principe P, Trejo W H, Ax H A, Wells J S, Andersen H H, Devasthale P V, Telikepalli H, Vander Velde D, Zou J-Y, Mitscher L A. Novel tetracycline glycosides active against tetracycline-resistant bacteria. J Org Chem. 1993;58:535–537. [Google Scholar]
- 301.Urquhart E, Addy M. Topical antimicrobials: new horizons for management of periodontal disease in general practice? Dent Update. 1995;1995(Apr.):104–111. [PubMed] [Google Scholar]
- 302.van den Bogert C, Kroon A M. Tissue distribution and effects on mitochondrial protein synthesis of tetracyclines after prolonged continuous intravenous administration to rats. Biochem Pharmacol. 1981;30:1706–1709. doi: 10.1016/0006-2952(81)90403-2. [DOI] [PubMed] [Google Scholar]
- 303.van der Hulst R W M, Keller J J, Rauws E A J, Tytgat G N J. Treatment of Helicobacter pylori infection: a review of the world literature. Helicobacter. 1996;1:6–19. doi: 10.1111/j.1523-5378.1996.tb00003.x. [DOI] [PubMed] [Google Scholar]
- 304.van Klingeren B, van Embden J D A, Dessens-Kroon M. Plasmid-mediated chloramphenicol resistance in Haemophilus influenzae. Antimicrob Agents Chemother. 1977;11:383–387. doi: 10.1128/aac.11.3.383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 305.van Steenberghe D, Rosling B, Soder P-O, Landry R G, van der Velden U, Timmerman M F T, McCarthy E F, Vandenhoven G, Wouters C, Wilson M, Matthews J, Newman H N. A 15-month evaluation of the effects of repeated subgingival minocycline in chronic adult periodontitis. J Periodontol. 1999;70:657–667. doi: 10.1902/jop.1999.70.6.657. [DOI] [PubMed] [Google Scholar]
- 306.Wang Y, Taylor D E. A DNA sequence upstream of the tet(O) gene is required for full expression of tetracycline resistance. Antimicrob Agents Chemother. 1991;35:2020–2025. doi: 10.1128/aac.35.10.2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 307.Wasteson Y, Hoie S, Roberts M C. Characterization of antibiotic resistance in Streptococcus suis. Vet Microbiol. 1994;41:41–49. doi: 10.1016/0378-1135(94)90134-1. [DOI] [PubMed] [Google Scholar]
- 308.Watanabe T. Infectious heredity of multiple drug resistance in bacteria. Bacteriol Rev. 1963;27:87–115. doi: 10.1128/br.27.1.87-115.1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 309.Weinstein R A. Endemic emergence of cephalosporin-resistant enterobacter: relation to prior therapy. Infect Control. 1986;7:120–123. doi: 10.1017/s0195941700065632. [DOI] [PubMed] [Google Scholar]
- 310.Weiss W J, Jacobus N V, Petersen P J, Testa R T. Susceptibility of enterococci, methicillin-resistant Staphylococcus aureus and Streptococcus pneumoniae to the glycylcyclines. J Antimicrob Chemother. 1995;36:225–230. doi: 10.1093/jac/36.1.225. [DOI] [PubMed] [Google Scholar]
- 310a.Werckenthin C, Schwarz S, Westh H. Structural alterations in the translational attenuator of constitutively expressed ermC genes. Antimicrob Agents Chemother. 1999;43:1681–1685. doi: 10.1128/aac.43.7.1681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 311.Wexler H M, Molitoris E, Finegold S M. In vitro activities of two new glycylcyclines, N,N-dimethylglycylamido derivatives of minocycline and 6-demethyl-6-deoxytetracycline, against 339 strains of anaerobic bacteria. Antimicrob Agents Chemother. 1994;38:2513–2515. doi: 10.1128/aac.38.10.2513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 312.Whittington W L, Roberts M C, Hale J, Holmes K K. Susceptibilities of Neisseria gonorrhoeae to the glycylcyclines. Antimicrob Agents Chemother. 1995;39:1864–1865. doi: 10.1128/aac.39.8.1864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 313.Williams D N. Tetracyclines. In: Gorbach S L, Bartlett J G, Blacklow N R, editors. Infectious diseases. Philadelphia, Pa: The W. B. Saunders Co.; 1992. pp. 211–214. [Google Scholar]
- 314.Winterscheid K K, Whittington W L, Roberts M C, Schwebke J R, Holmes K K. Decreased susceptibility to penicillin G and Tet M plasmids in genital and anorectal isolates of Neisseria meningitidis. Antimicrob Agents Chemother. 1994;38:1661–1663. doi: 10.1128/aac.38.7.1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 315.Wise R, Andrews J M. In vitro activities of two glycylcyclines. Antimicrob Agents Chemother. 1994;38:1096–1102. doi: 10.1128/aac.38.5.1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 316.Witte W. Medical consequences of antibiotic use in agriculture. Science. 1998;279:996–997. doi: 10.1126/science.279.5353.996. [DOI] [PubMed] [Google Scholar]
- 317.Xia M, Pang Y, Roberts M C. Detection of two groups of 25.2 Mda Tet M plasmids by polymerase chain reaction of the downstream region. Mol Cell Probes. 1995;9:327–332. doi: 10.1016/s0890-8508(95)91620-2. , 1995. [DOI] [PubMed] [Google Scholar]
- 318.Yamaguchi A, Ono N, Akasaka T, Noumi T, Sawai T. Metal-tetracycline/H+ antiporter of Escherichia coli encoded by a transposon, Tn10. J Biol Chem. 1990;265:15525–15530. [PubMed] [Google Scholar]
- 319.Zhao J, Aoki T. Nucleotide sequence analysis of the class G tetracycline resistance determinant from Vibrio anguillarum. Microbiol Immunol. 1992;36:1051–1060. doi: 10.1111/j.1348-0421.1992.tb02109.x. [DOI] [PubMed] [Google Scholar]