Abstract
Continuous research spanning more than three decades has made the Bacillus bacteriophage φ29 a paradigm for several molecular mechanisms of general biological processes, such as DNA replication, regulation of transcription, phage morphogenesis, and phage DNA packaging. The genome of bacteriophage φ29 consists of a linear double-stranded DNA (dsDNA), which has a terminal protein (TP) covalently linked to its 5′ ends. Initiation of DNA replication, carried out by a protein-primed mechanism, has been studied in detail and is considered to be a model system for the protein-primed DNA replication that is also used by most other linear genomes with a TP linked to their DNA ends, such as other phages, linear plasmids, and adenoviruses. In addition to a continuing progress in unraveling the initiation of DNA replication mechanism and the role of various proteins involved in this process, major advances have been made during the last few years, especially in our understanding of transcription regulation, the head-tail connector protein, and DNA packaging. Recent progress in all these topics is reviewed. In addition to φ29, the genomes of several other Bacillus phages consist of a linear dsDNA with a TP molecule attached to their 5′ ends. These φ29-like phages can be divided into three groups. The first group includes, in addition to φ29, phages PZA, φ15, and BS32. The second group comprises B103, Nf, and M2Y, and the third group contains GA-1 as its sole member. Whereas the DNA sequences of the complete genomes of φ29 (group I) and B103 (group II) are known, only parts of the genome of GA-1 (group III) were sequenced. We have determined the complete DNA sequence of the GA-1 genome, which allowed analysis of differences and homologies between the three groups of φ29-like phages, which is included in this review.
The genus Bacillus incorporates many species of gram-positive, aerobic, endospore-forming bacteria that normally inhabit the soil or decaying plant material. In these habitats, a large variety of phages have been isolated that infect bacilli. All of these phages isolated so far have some common features. First, they all contain double-stranded DNA (dsDNA), and second, the virions have prolate icosohedral heads and are tailed. Modern phage taxonomy is based on properties of the virion and its nucleic acid (see references 74 and 131). The order of tailed phages, named Caudovirales, are classified into three families: Myoviridae (phages with contractile tails), Podoviridae (phages with short tails), and Siphoviridae (phages with long noncontractile tails). For a general review on tailed bacteriophages, see reference 4. In addition to taxonomy based on properties of the virion and its nucleic acid, phages can be divided into three groups based on their infection cycle. The first group contains lytic phages that complete their life cycle within a well-defined period after infection and are unable to lysogenize their host. The second group is formed by the so-called pseudo-temperate phages. These are virulent phages with an extended and irregular latent period. Although this stage mimics lysogeny, it does not involve a stable prophage. The third group contains the temperate phages. The genomes of these phages are able to integrate into the host genome and can be maintained in this lysogenic stage for many generations. Generally, during this stage, the cells are immune to infection with the same phage.
This review specifically focuses on the φ29-like genus of phages, which includes, in addition to φ29, phages PZA, φ15, BS32, B103, M2Y (M2), Nf, and GA-1. They are all lytic phages that belong to the Podoviridae family. Most of these phages infect Bacillus subtilis, but often they also infect other related species such as Bacillus pumilus, Bacillus amyloliquefaciens, and Bacillus licheniformis. Phages of this genus have been subclassified into three groups based on serological properties, DNA physical maps, peptide maps and partial or complete DNA sequences (164, 220, 222). The first group includes phages φ29, PZA, φ15, and BS32; the second group includes B103, Nf, and M2Y; and the third group contains GA-1. Interestingly, the classification of these phages coincides with their geographical distribution. Thus, the phages belonging to group I were isolated in the United States (169), those belonging to group II were isolated in Japan (91, 113, 191), and GA-1 (group III) was isolated in Europe (39).
The genomes of the φ29-like phages consist of a linear dsDNA molecule of about 20 kb that has a phage-encoded protein, named terminal protein (TP), covalently attached at each 5′ DNA end. The DNA sequences of the complete genomes of φ29 and PZA (83, 84, 161, 216, 221, 224) belonging to group I and that of B103 (163) belonging to group II have been determined. However, only parts of the GA-1 sequence, belonging to group III, have been determined so far (78, 86, 111, 114, 222). To gain a comprehensive understanding of the relatedness of the three groups of phages, we have determined the complete DNA sequence of GA-1. The genomes of φ29 and B103 are 19,285 and 18,630 kb, respectively (163, 216). However, the GA-1 genome was reported to be approximately 21.5 kb (220, 223). Thus, an additional incentive to determine the complete DNA sequence of GA-1 was to gain insight into possible additional coding sequences present on the GA-1 genome.
Phage φ29 has been subject to extensive studies, and the results have led to the understanding of several molecular mechanisms of general biological processes, such as DNA replication, regulation of transcription, phage morphogenesis, and phage DNA packaging. These various topics will be discussed in this review, and attention is focused specifically on progress made during the last few years. In general, the views presented are based on results obtained with φ29, since most studies concerned analysis of this phage. In addition, an integrated overview of homologies and differences between the three groups of the φ29 genus based on the complete DNA sequences of φ29 (group I), B103 (group II), and GA-1 (group III) is presented.
GENERAL FEATURES OF PHAGES φ29, B103, AND GA-1
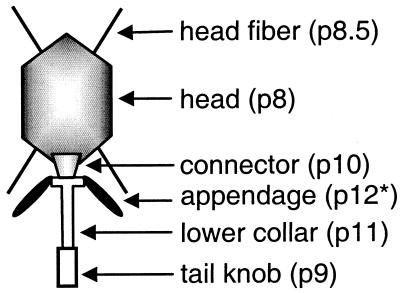
The φ29-like phages are the smallest Bacillus phages isolated so far and are among the smallest known phages containing dsDNA. The sizes of the phage particle of each of the three groups of φ29-like phages are shown in Table 1. Phage φ29 was first isolated by Reilly (169) from garden soil. Phage B103 was first isolated from a nonspecified lysed Bacillus culture (91), and phage GA-1 was first isolated by Bradley (39) from rotting lawn mowings. Electron microscopy analysis showed that the phage particles of φ29, B103, and GA-1 have a sixfold radial symmetry and a short noncontractile tail tube. A schematic representation of a φ29 phage particle in which each protein is indicated is shown in Fig. 1. Analysis of the host range showed that φ29 was able to infect B. subtilis strains 168, 110NA, and Marburg, B. amyloliquefaciens H, and several strains of B. licheniformis and B. pumilus (reviewed in reference 177). The host range of B103 has not been studied, but it is known to infect B. subtilis 9/3 (163). Finally, GA-1 was shown to infect lytically Bacillus species strain G1R (39). Arwert and Venema (10) showed that GA-1 is unable to infect the standard B. subtilis strain 168. Although sequence analysis of G1R 16S rRNA showed that Bacillus strain G1R is most closely related to B. pumilus, GA-1 is unable to infect B. pumilus strains BP1 or B205-L (J. A. Horcajadas, unpublished results). Therefore, the species identification of Bacillus strain G1R, the specific host of GA-1, remains unclear.
TABLE 1.
Structure of the φ29-like phages
| Phage | Size (nm)
|
Reference | |
|---|---|---|---|
| Head | Tail | ||
| φ29 | 41.5 by 31.5 | 32.5 by 6.0 | 6 |
| Nf (B103) | 43.0 by 33.0 | 33.0 by 6.0 | 191 |
| GA-1 | 57.0 by 40.0 | 35.0 by 6.0 | 39 |
FIG. 1.
Schematic representation of a φ29 phage particle. The various proteins are indicated.
SEQUENCE ANALYSIS OF THE GA-1 GENOME
The DNA sequence of the complete genomes of φ29 (group I) (83, 84, 216, 221, 224) and B103 (group II), (163) are known, and they consist of 19,285 and 18,630 bp, respectively. However, only noncontinuous parts of the GA-1 genome have been sequenced before. These include (i) the left (168 bp; GenBank accession number M19512) and right (168 bp; M19519) terminal nucleotide sequences (222); (ii) the central region containing the early promoters A2b and A2c and the late promoter A3 (354 bp; AJ133524) (111); (iii) gene 6 encoding the dsDNA binding protein (DBP) p6 (342 bp; AF148209) (78); (iv) gene 5 encoding the single-stranded DNA (ssDNA) binding protein (SSB) p5 (513 bp; AJ244026) (86); (v) gene 4 encoding the transcriptional regulatory protein p4G (405 bp; AJ133525) (111); (vi) the region spanning genes 3 and 2 encoding the TP (p3) and DNA polymerase (p2), respectively (2,668 bp; X96987) (114); and (vii) a region downstream of gene 2 (549 bp; AJ294726) (A. Bravo and M. Salas, unpublished data). Genes 6 through 2 lie in the same order as the corresponding genes of phages φ29 and B103. Where possible, the individual sequences were integrated in larger contigs and gaps were filled using primers based on the published sequences and purified GA-1 as template DNA. Next, the remaining part (∼17 kbp) of the GA-1 genome sequence was determined of both strands by a primer-walking strategy using purified phage GA-1 DNA as template.
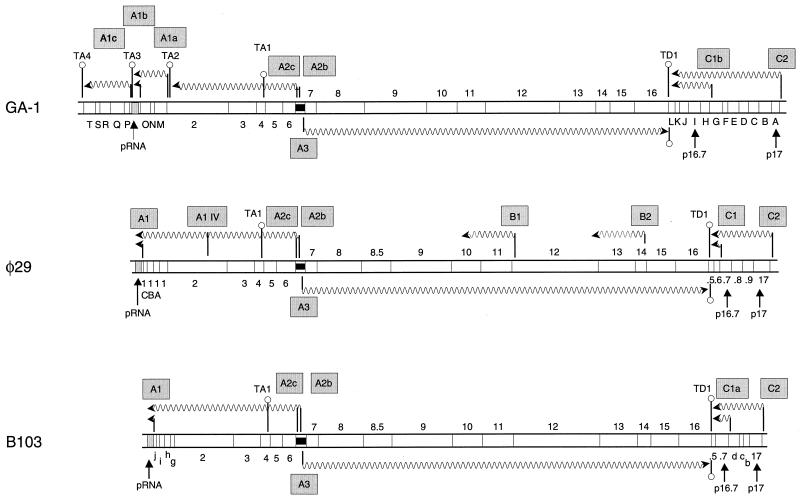
The genome of GA-1 was shown to have a total size of 21,129 bp. The complete nucleotide sequence has been deposited in the EMBL/GenBank/DDBJ nucleotide sequence database and was assigned accession number X96987. Whereas the G+C content of GA-1 is 34.7%, those of φ29 and B103 are 40.0 and 37.7%, respectively. Next, computer-assisted and manual analysis of the DNA sequence were used to identify open reading frames (ORFs), direct and inverted repeats, and putative promoters, ribosomal binding sites, and Rho-independent transcriptional terminators. The deduced amino acid sequences of the various ORFs were compared with protein sequences present in the φ29 and B103 genomes as well as with those present in available databases. In cases where the deduced amino acid sequences of the identified ORFs or genes showed significant homology to those of the φ29 and B103 genes, they were given numbers according to the nomenclature used for these phages. The remaining ORFs were identified with letters. The data obtained were used to construct a putative genetic and transcriptional map of GA-1, which is shown, together with those of φ29 and B103, in Fig. 2. This figure shows that genes 2 to 6, 7 to 16 (with the exception of gene 8.5, which is lacking in GA-1), and 17 and 16.7 are conserved in all three genomes. Characteristics of the proteins synthesized by these GA-1 genes and their levels of similarity to corresponding proteins of φ29 and B103 are given in Table 2, which shows that for all the homologous genes shared by φ29, B103, and GA-1, those of GA-1 are less conserved than those of φ29 and B103. This confirms that within the family of φ29-related phages, GA-1 is the most distantly related one, as suggested previously (164, 220, 222). Features of the putative proteins synthesized by the GA-1 ORFs are given in Table 3.
FIG. 2.
Genetic and transcriptional maps of GA-1 (group III, 21,129 bp), φ29 (group I, 19,285 bp), and B103 (group II, 18,630 bp). The maps are aligned according to the A2b, A2c, A3 promoter region. The direction of transcription and lengths of the transcripts are indicated by wavy arrows. The transcripts of late- and early-expressed operons and the late and early promoters (boxed) are shown below and above the map, respectively. The positions of the various genes and ORFs are indicated between the two DNA strands. Genes are indicated by numbers, ORFs are indicated by letters (capital for GA-1 and φ29, and lowercase for B103). The positions of genes 17 and 16.7 that are conserved in all three phage genomes, located in the right early operon, are indicated. The positions of φ29 ORFs 16.9, 16.8, 16.6, and 16.5 and B103 ORF 16.5, located at the right side of their genomes, are indicated by the numbers .9, .8, .6, and .5. Transcriptional terminators are indicated by hairpin structures. A light grey box indicates the DNA region encoding the pRNA, and a black box indicates the region spanning the early A2b, A2c, and late A3 promoters.
TABLE 2.
Characteristics of GA-1 genes
| Gene | Function of gene product | No. of amino acids | Mol mass (kDa) | Similarity (%)a
|
||
|---|---|---|---|---|---|---|
| GA-1/φ29 | GA-1/B103 | φ29/B103 | ||||
| 2 | DNA polymerase | 578 | 67.1 | 67.3 | 68.0 | 88.5 |
| 3 | Terminal protein | 265 | 30.5 | 51.7 | 53.6 | 74.1 |
| 4 | Transcriptional regulator | 134 | 16.1 | 57.6 | 58.4 | 83.2 |
| 5 | SSB | 170 | 19.2 | 37.9 | 45.1 | 73.8 |
| 6 | DBP, initiation of DNA replication; gene regulation | 93 | 10.2 | 52.7 | 54.8 | 65.0 |
| 7 | Scaffold protein | 107 | 11.9 | 48.0 | 38.6 | 75.5 |
| 8 | Major head protein | 472 | 53.0 | 68.5 | 69.3 | 90.4 |
| 8.5 | Head fiber protein | 63.6 | ||||
| 9 | Tail protein | 612 | 69.2 | 58.3 | 58.5 | 75.3 |
| 10 | Connector (upper collar) | 306 | 35.3 | 63.7 | 63.7 | 84.4 |
| 11 | Lower collar | 287 | 33.7 | 51.6 | 50.9 | 77.8 |
| 12 | Pre-neck appendage protein | 740 | 83.9 | 37.8 | 37.5 | 79.9 |
| 13 | Morphogenesis | 355 | 40.2 | 67.0 | 67.3 | 82.7 |
| 14 | Holin | 133 | 15.3 | 51.9 | 52.3 | 87.1 |
| 15 | Peptidoglycan hydrolase | 239 | 26.5 | 37.2 | 40.6 | 74.8 |
| 16 | DNA encapsidation, ATPase | 336 | 39.0 | 67.5 | 68.8 | 86.3 |
| 16.7 | Distribution of in vivo phage DNA replication | 133 | 15.5 | 48.5 | 47.7 | 68.5 |
| 17 | DNA replication | 87 | 10.3 | 44.8 | 37.9 | 48.1 |
For the calculation of the percent similarity, the following amino acids were considered conservative: L, I, V, A, and M; F, Y, and W; K and R; D and E; Q and N; and S and T.
TABLE 3.
Features of GA-1 ORFs
| ORF | No. of amino acids | Mol mass (kDa) | Putative translation initiation sitesa | Spacingb |
|---|---|---|---|---|
| B | 105 | 12.4 | ATACTTGGAAAGAAGGATGATAACA ATG | 11 |
| C | 94 | 10.7 | GAAACGCAAACTAGGAGGAAGAAAA ATG | 9 |
| D | 101 | 12.1 | TAATGGATAAAAGGGGGTAACAAAT ATG | 10 |
| E | 104 | 12.0 | ACGATAAAGCTTGGTTTGATAAAAA ATG | 14 |
| F | 25 | 2.7 | GAATGGTCTTAAAGGAGGTGATAAA ATG | 9 |
| G | 116 | 14.0 | CTCTAAATTATAAGGAGGAATTAAA ATG | 9 |
| H | 82 | 9.6 | ACATAACAAATAGGAGGAAACAAAA ATG | 10 |
| J | 80 | 9.5 | ACTCTAAGAAAGTTGGTAAATAAAA ATG | 11 |
| K | 48 | 5.8 | AAATTAAAGAATTGAGGTATATTGA ATG | 10 |
| L | 63 | 7.4 | GACATTAGAAAGGAGCTAGACAAAA ATG | 11 |
| M | 130 | 15.3 | GATAGTTAAAAGAGGAGGGTTATAA ATG | 9 |
| N | 33 | 3.5 | TATTAATTTAAAGGAGGAATTTTAA ATG | 10 |
| O | 94 | 11.1 | GTGTTATGATTAGAGGGGGATTCTA ATG | 8 |
| P | 56 | 6.6 | TAAAGATAGAAAGGGAGGGGATAAC ATG | 9 |
| Q | 141 | 16.4 | TTGATCATGTTAGGAGGAGAAAGTA ATG | 10 |
| R | 107 | 12.6 | AAGGGGAAACTAAAGGAGATGTCTA ATG | 8 |
| S | 29 | 3.6 | AAGGGGAAACTAAAGGAGATGTCTA ATG | 8 |
| T | 108 | 12.8 | AGATTCACTGAAAGGAGGTGATAAA ATG | 9 |
The putative initiation codons and the 25 nucleotides upstream are shown; nucleotides complementary to the 3′ end of B. subtilis 16S rRNA (UCUUUCCUCCACUAG) (152) are underlined.
The spacing is calculated as the distance (in bases) from the first base to the right of the 5′-AGAAAGGA-3′ (or the equivalent) to the base adjacent to the initiation codon.
GENETIC AND TRANSCRIPTIONAL ORGANIZATION
Generally, genes with related functions are clustered in phage genomes (4), and Fig. 2 shows that φ29, B103, and GA-1 are no exception to this rule. In addition, Fig. 2 shows that in most aspects, the genomes of φ29, B103, and GA-1 are similarly organized. In all three genomes the genes and ORFs are organized in operons. Depending on the time when they are first expressed during the infection cycle, these can be divided into early and late operons. In all three genomes the early-expressed operons are transcribed leftward and the single late-expressed operon is transcribed rightward. The genes present in the late operon (genes 7 through 16), which is located in the central part of the genome, encode phage structural proteins, proteins involved in phage morphogenesis, and proteins required for lysis of the host. All three genomes contain an early-expressed operon that is divergently transcribed with respect to the late operon (Fig. 2). Genes 6, 5, 3, and 2 of this operon encode the four main proteins required for phage DNA replication. The operon also contains gene 4, which encodes the transcriptional regulator protein. In addition to its role in phage DNA replication, protein p6 also has a role in transcriptional regulation (14, 69, 219). Note that this operon of GA-1 is smaller than the corresponding ones of φ29 and B103. Another early-expressed operon is located at the right side of the phage genomes. However, as described in more detail later, only two genes of this operon, 17 and 16.7, are conserved in all three phage genomes. Finally, another feature shared by all three phages is the presence of a region located in the left part of the genome that encodes an RNA (pRNA) which is required for packaging of phage DNA.
The genome of GA-1 is about 1.8 and 2.5 kb larger than those of φ29 and B103, respectively. Although the structural organization of GA-1 genome is similar to that of φ29 and B103, it contains additional sequences, located at both genome ends, that may encode several proteins, counterparts of which are not present in the genomes of φ29 and B103 (see Fig. 2).
TRANSCRIPTIONAL REGULATION
The (putative) promoters and transcriptional start sites, for these cases already determined, are listed in Table 4. When appropriate, the nomenclature of the GA-1 promoters was adapted to that of φ29 and B103. Expression of most φ29 and GA-1 promoters has been studied. As indicated in Table 4, most of the promoters contain the sequence TG positioned 1 bp upstream of the −10 sequence. This additional sequence is characteristic of the so-called −10 extended promoters first described for Escherichia coli promoters (123, 165). At least in E. coli, the extension of the −10 region is able to compensate for the absence of a good −35 box, helping the sigma 70 RNA polymerase to recognize and bind such promoters (123, 128, 165). The additional TG sequence is also frequently found in ςA-dependent B. subtilis promoters (106, 152). Possible involvement of the TG motif in promoter strength has been recently studied for the φ29 promoters A1, A2c, and A3 (46). In all three promoters, mutation of the TG motif impaired the binding of the ςA-RNA polymerase to the promoter. These and additional results support the view that the TG motif provides contact sites for B. subtilis ςA RNA polymerase that are important for a specific role in the first steps of transcription (46).
TABLE 4.
Promoters of φ29, B103 andGA-1
 |
Promoters are aligned relative to their −35 (except for the A3 promoters) and −10 (except for the A1IV promoter) boxes. Arrows indicate the start sites deduced from primer extension assays of RNAs obtained from infected cells (111, 149; Horcajadas, unpublished results) or, in the case of promoters A1, from pRNA isolated from phage proheads (12). The TG motifs characteristic of the so-called −10 extended promoters are doubly underlined.
Indicated on the right are (i) the length between the −35 and −10 boxes (spacer), (ii) presence of the TG motif 1 bp upstream of the −10 box, and (iii) expression of the promoter during the infection cycle and the genes and/or ORFs expressed from the promoter.
The B. subtilis α-amylase promoters amyP and amyP2 contain the TGTG sequence located 1 bp upstream of its −10 region, called the −16 region. Mutation analysis of the −16 region of these promoters showed that it significantly affected the in vitro promoter strength (217). In addition, a large portion of known gram-positive bacterial promoters contain the −16 TRTG motif (in which R is a purine), suggesting that not only the −10 extended TG motif but also the −16 region is important for promoter strength (217). The −16 region is present in the following phage promoters: A1 and A2b of φ29, A1 of B103, and A1c, A3 and C2 of GA-1 (Table 4). Possible involvement of the −16 region in the activity of these phage promoters has not been studied yet.
Early Promoters A2b and A2c and Late Promoter A3: Transcriptional Regulation by Proteins p4 and p6
As described above, the structural organization of the centrally located late operon and the divergently oriented early operon is conserved in the genomes of φ29, B103, and GA-1. In all three phage genomes the promoters that drive the expression of these early and late genes are localized in a short intergenic region between these two operons. The transcriptional regulation of these promoters has been studied extensively for φ29 (for reviews, see references 171 and 182). Two strong promoters named A2c and A2b drive the expression of the early operon of φ29 containing genes 6 to 1. The late φ29 operon is transcribed from a single promoter named A3 (16, 136, 137, 149, 197). The transition from early to late φ29 transcription is controlled by φ29 protein p4, the product of the early gene 4. Protein p4, which is a dimer in solution, binds to its cognate DNA binding sites as a tetramer (142), contacting only one side of the DNA helix (172). The intergenic region comprising promoters A2c, A2b, and A3 contains two p4 binding sites. The center of one of these is located at position −82 relative to the transcription start site of the late promoter A3 (15). Whereas this promoter contains a good consensus sequence at the −10 region for the vegetative B. subtilis ςA RNA polymerase, it lacks a typical −35 box (Table 4). Therefore, the RNA polymerase alone does not bind efficiently to the A3 promoter, which explains why the downstream operon is not expressed during early infection times. Activation of the A3 promoter requires binding of protein p4 to the p4 binding site upstream of the A3 promoter. The main role of protein p4 is to stabilize the binding of RNA polymerase to the A3 promoter as a closed complex, and the protein has little effect on the rest of the steps of the initiation process (157).
The φ29 promoters A2c and A2b drive the expression of the early operon containing genes 6 to 1. Of these, promoter A2b is the one located closest to the oppositely oriented late promoter A3; promoter A2c is located proximal to gene 6. Both early promoters are repressed by protein p4. The p4 binding site that is located upstream of the late A3 promoter and is required for activation of this promoter, as described above, partially overlaps the early A2b promoter. Binding of protein p4 to this site occupies the −35 region of the A2b promoter, preventing the expression of this promoter. Thus, protein p4 activation of the late promoter A3 is accompanied by an efficient repression of the A2b promoter (172). Expression of the other early promoter, A2c, is also repressed by protein p4, but this occurs through a totally different mechanism. In addition to the p4 binding site upstream of the late promoter A3, another p4 binding site is located upstream of promoter A2c (centered at position −72 relative to the transcription start site of A2c). Protein p4 binding to this site is stabilized in the presence of RNA polymerase, indicating that the proteins bind cooperatively to the DNA. In this situation, the RNA polymerase can generate abortive initiation transcripts but is unable to escape from the A2c promoter (150). Thus, repression of the A2c promoter occurs by overstabilization of the RNA polymerase to this promoter (148). Interestingly, both repression at the A2c promoter and activation of the A3 promoter involve interaction between a region of protein p4 containing Arg120 and the C-terminal domain of the RNA polymerase α subunit (140–143, 150, 151, 171).
Recently it was demonstrated that expression of the φ29 A2c, A2b, and A3 promoters is regulated by the viral protein p6 in addition to protein p4 (69). Protein p6 is an abundantly early-expressed dsDNA binding protein that was shown previously to play an important role in initiation of phage DNA replication (see below). Elías-Arnanz and Salas (69) showed that protein p6 promotes p4-mediated repression of the A2b promoter and activation of the A3 promoter by enhancing binding of p4 to its recognition site at promoter A3. In addition, protein p4 promotes p6-mediated repression of the A2c promoter by favoring the formation of a stable p6-nucleoprotein complex that interferes with RNA polymerase binding to promoter A2c.
Although transcriptional regulation of the equivalent promoters of B103 has not been studied, conservation of the main characteristics of this region regarding the A3 and A2b promoters suggests that transcription of these promoters may be regulated in a similar way to those of φ29. Results that at least partially support this assumption may come from the analysis of the corresponding region of phage Nf (147, 158), which belongs to the same group of phages as B103. First, it was shown that activation of the late A3 promoter of Nf requires the Nf-encoded protein gpF (homologue of the φ29 protein p4); (147). Second, Nuez and Salas (158) showed that activation of the Nf A3 promoter is responsive to the φ29 protein p4 in a similar way to that observed for the φ29 A3 promoter.
A first in vivo and in vitro analysis of the transcriptional regulation of the equivalent promoters of GA-1 has been reported recently (111). The in vivo activity of the GA-1 A2b and A2c promoters was shown to diminish 10 min after infection, whereas at this time the expression of the late A3 promoter increased significantly. The GA-1-encoded protein p4 (named p4G, 53% similar to φ29 p4) was purified and used to study its involvement in regulation of these promoters in vitro. As in φ29, a p4G binding site is located upstream of the late A3 promoter that overlaps with the early A2b promoter. As in φ29, binding of p4G to this site prevented the binding of RNA polymerase to the GA-1 early A2b promoter. Surprisingly, however, binding of p4G to this site had no effect on the in vitro expression of the late A3 promoter of GA-1. Both in the absence and in the presence of p4G, promoter A3 was expressed efficiently in vitro. Thus, in contrast to the situation in φ29, p4G is not required in vitro to activate the expression of the GA-1 A3 promoter. Moreover, in contrast to the φ29 protein p4, the GA-1 protein p4G was shown not to interact with the RNA polymerase α subunit (111). Although the A3 promoter of GA-1 was active in the absence of p4G in in vitro assays, it was not active at early infection times in vivo. In addition, in vivo activation of the A3 promoter was completely blocked when protein synthesis was prevented just before infection. Together, these results suggested that the A3 promoter may be repressed in vivo by a host-encoded protein and that protein p4G may function as an antirepressor, permitting A3 expression at late infection times. Finally, it is intriguing that the GA-1 A3 promoter, which, like the A3 promoters of φ29 and B103, lacks a good −35 box, is expressed efficiently in vitro. Studies are under way to unravel the mechanisms that underlie the observed differences in regulation of the φ29 and GA-1 A3 promoters.
At present, it is unknown whether a p4-dependent repression of the A2c promoter, as described for φ29, also applies for the equivalent A2c promoters of Nf/B103 or GA-1. The fact that a typical p4 binding site is lacking upstream of the A2c promoters of B103 (163), Nf (158), and GA-1 (111) may be an indication that p4 is not involved in the repression of these promoters, at least not in a similar way to that in φ29. It is also unknown whether protein p6 of B103/Nf and/or GA-1 plays a role in the regulation of the A2c, A2b, and A3 promoters of these phages.
Early Promoter C2: Transcriptional Regulation by Protein p6
All three phage genomes contain an early-expressed operon located at the right end of their genome, whose expression is under the control of the C2 promoter (Fig. 2). For φ29 it has been demonstrated that the activity of the early promoter C2 decreases rapidly 10 min after infection (110, 122, 149). Protein p6 was shown to be responsible for in vivo and in vitro repression of promoter C2 (14, 219). Thus, the φ29 p6 protein not only plays a role in the regulation of the A3, A2b, and A2c promoters (see above) but also regulates the expression of the C2 promoter. In addition, as described below, it plays an important role in the initiation of φ29 DNA replication. Most probably, binding of p6 to the DNA ends prevents the RNA polymerase to recognize the C2 promoter (A. Camacho and M. Salas, unpublished results). The φ29 mutant sus6(626) contains a suppressible mutation in gene 6, and therefore protein p6 is not synthesized in nonsuppressor cells infected with this mutant phage. When φ29 sus6(626) mutant phage was used for infection, phage DNA replication did occur in suppressor cells but not in nonsuppressor cells (219). However, under these conditions the C2 promoter was not repressed in either nonsuppressor or suppressor cells. It appeared that whereas the amount of p6 protein synthesized under permissive conditions was sufficient to permit in vivo φ29 DNA replication, it was too small to repress the C2 promoter in vivo (47, 219). The observation that a fairly large amount of p6 is required for repression of the C2 promoter in vitro (14, 219) supports this view.
Equivalent C2 promoters are also present in the genomes of B103 and GA-1. Like the C2 promoter of φ29, the GA-1 C2 promoter is expressed almost exclusively during the first 10 min after infection (Horcajadas, unpublished). In vitro expression of the C2 promoter of GA-1 is inhibited in the presence of purified GA-1-encoded protein p6, as well as, although somewhat less efficiently, by protein p6 of φ29. DNase I footprint analysis indicated that DNA binding of protein p6 prevents the RNA polymerase from recognizing the C2 promoter of GA-1 (Horcajadas, unpublished results). Thus, due to protein p6-mediated repression, the φ29 and GA-1 C2 promoters are expressed only during the initial 10 min after infection. Obviously, this repression will limit the amount of proteins encoded by the downstream genes and ORFs.
Early Promoters C1, C1a, and C1b Present in φ29, B103, and GA-1, Respectively
All three phage genomes contain a promoter within the early operon located at the right side of their genome (Fig. 2 and Table 4). The absence of potential transcriptional terminators upstream of these promoters suggests that the last genes or ORFs of these operons may be expressed from two promoters. In φ29 this additional promoter was named C1. It is located within gene 16.7 and may drive the expression of ORFs 16.6 and 16.5. In B103, the promoter is located within ORF d and may drive the expression of gene 16.7 and ORF 16.5. According to the φ29 nomenclature, this promoter of B103 was named C1 (163). Finally, in GA-1 the promoter is located within ORF G and may drive the expression of ORFs H to L. Since these promoters drive the expression of different genes and ORFs, they are not equivalent. Therefore, we named these promoters of B103 and GA-1 Cla and Clb, respectively.
In vitro transcription analysis showed that expression of the φ29 C1 promoter is repressed by protein p6 (14). Although p6 repressed the C2 promoter in the presence of low and high salt concentration, p6 affected C1 expression only at low salt concentrations. This difference may be due to the higher affinity of p6 for the terminal φ29 DNA fragment containing the C2 promoter than for the more internal DNA sequences containing the C1 promoter (14).
Promoter A1, Driving Synthesis of the pRNA
For φ29 it has been demonstrated that packaging of TP DNA into the phage prohead requires a 174-base φ29-encoded RNA (pRNA) (5, 93, 94). This pRNA is produced from promoter A1 (136, 137, 197), which is active throughout the infection cycle (149). Although substantial levels of pRNA were detected at early infection times, a rapid increase in the number of pRNA molecules was detected starting about 15 min after infection, which approximately coincided with the onset of φ29 DNA replication. Therefore, the additional phage DNA templates produced explain this increase of pRNA and suggest a constant transcription rate (149).
Equivalent A1 promoters driving pRNA synthesis of the corresponding phages are present in B103 and GA-1 (Table 4). The pRNA coding sequences of φ29 and B103 are located at the far-left ends of their genomes. Figure 2 shows that the situation is different for GA-1. This genome contains an additional operon downstream of the pRNA-coding region, as well as another operon located between gene 2 and its pRNA-coding region. A promoter is located upstream each of these two unique operons. Thus, whereas the leftmost region of the φ29 and B103 genomes contains only one promoter, this region of GA-1 contains three promoters. To maintain a consistent nomenclature, the GA-1 promoter upstream of ORF M was named A1a, the one driving the expression of the GA-1 pRNA was named A1b, and the one upstream of ORF P was named A1c.
The expression patterns of GA-1 promoter A1b and B103 promoter A1 during the infection cycle have not been studied. Table 4 shows that the −35 and −10 sequences of the A1 promoters of φ29 and B103 and the equivalent A1b promoter of GA-1 are almost identical and very close to the consensus sequence recognized by ςA-containing RNA polymerase. Therefore, it is likely that the A1b promoter of GA-1 and the A1 promoter of B103 behave similarly to the equivalent A1 promoter of φ29.
Other Promoters in the φ29 Genome
In vivo and in vitro experiments revealed two promoters, named B1 and B2, that are located in the φ29 DNA region encoding the late genes (16, 197) (Fig. 2). Transcription from these promoters proceeds leftward. Compared to other φ29 promoters, only minor amounts of RNA were synthesized by the B1 and B2 promoters in vivo (149). No ORF with a reasonable ribosome binding site was found downstream of either of these promoters. Although it has been suggested that the products synthesized by these promoters may function as antisense RNA to modulate the expression of some late genes (16, 136), such a function has not been proven experimentally. The φ29 promoter A1IV, located in the DNA polymerase coding region (Fig. 2), was shown to be weakly expressed in vivo (16) and to contribute to the synthesis of protein p1 (40). The B1, B2, and A1IV promoters are shown in Table 4.
Other Promoters in the GA-1 Genome
The promoters A1c and A1a are unique for GA-1. Primer extension analysis using total RNA isolated at different times after infection showed that these two promoters are active early after infection and that they are progressively downregulated at later infection times (J. A. Horcajadas, unpublished). Therefore, it is likely that promoters A1c and A1a drive the expression of the GA-1 regions containing ORFs P to T and M to O, respectively. At present, the mechanism underlying the in vivo repression of these promoters is unknown. Since the pattern of repression of these promoters is different from that of the abruptly repressed C2 promoter, it is unlikely that these promoters are repressed by protein p6 in a similar way to the C2 promoter.
TRANSCRIPTIONAL TERMINATION
The main early and late in vivo transcription termination sites of φ29 have been determined by S1 nuclease mapping (17). Transcription of the late A3 promoter and that of the early promoters C2 and C1 terminated in the short intergenic region between gene 16 and ORF 16.5 (Fig. 2). This DNA region contains an inverted repeat, and stem-loop structures with calculated free energies of −14.8 and −16.8 kcal could be drawn for the early and late transcripts, respectively. In both directions, a uridine-rich tail follows the stem-loop, indicating that it functions as a Rho-independent bidirectional transcription terminator. This terminator was named TD1. Inverted repeats are located at similar positions in the genomes of B103 and GA-1. As in φ29, uridine-rich tails at either strand follow the stem-loops of B103 and GA-1, indicating that these also constitute bidirectional Rho-independent transcriptional terminators. According to the φ29 nomenclature, these terminators were named TD1. The DNA sequences of the TD1 terminators are shown in Table 5.
TABLE 5.
Transcriptional terminators
| Name | Phage | Sequence (5′-3′)a | Positionsb |
|---|---|---|---|
| → ← | |||
| TD1 | φ29 | AACAATCAAAAGAAAAGCCTATCGTCTGAGGAACGGTAGGCTCTTTTGTAG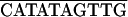 |
17258–17318 |
| →← | |||
| B103 | CGCTTGAAACAAAAACACCTGCTGTTATAATAACGGCAGGCTTTTTAATAGTAATTCATTG | 16974–17034 | |
| →← | |||
| GA-1 | ATGAGAATATACTAAAAGACCGCCTTTAAAGGGCGGTTTTATTTTGTGATCATATATGCTT | 17586–17646 | |
| → ← | |||
| TA4 | GA-1 | AGAAATAGAAATTAAAAGAGTCCTCATTTTGAGGGCTTTTTATTTTGAATTTAAAACTAGT | 140–200 L |
| → ← | |||
| TA3 | GA-1 | ACAAGAAGACAGAGAAAGGGACACTAGAAATAGTGTCTTTTTCTTTTGAAATAATAGTTGT | 1592–1652 L |
| → ← | |||
| TA2 | GA-1 | CATTTTTAGTGAAAAGAAGTTGATAGTTTCTATTGACTTCTTTTTTATTTCATAGTATACT | 2703–2763 L |
| → ← | |||
| TA1 | φ29 | TTGAAGGAATCTGAATACGTGGCATCTAACACCGATGTCACGTTTTTCTTTTCAAGTGAAT | 3859–3919 L |
| → ← | |||
| B103 | TTGAAAGAATCTGAATACGTGGTGTCTAACGGTGATGCCACGTTTTTCTTTTCGAGTGAAT | 3702–3762 L | |
| → ← | |||
| GA-1 | TTGAAAGAATCGAGGTACGTTATCACCTGTGAAAATGGGTTGGTGACGTACTTTTTTTCAT | 5508–5568 L |
Inverted repeats are indicated by arrows. φ29 sequences corresponding to the TD1 and TA1 termination of transcription of the late and early strands are overlined and underlined, respectively.
Positions of the DNA sequences in the phage genomes. L, lower strand.
Another Rho-independent transcriptional terminator, named TA1, was found to be present within gene 4 of φ29 (17). It has been suggested that part of the transcripts initiated at the A2b and A2c promoters terminate at this terminator. This would result in the synthesis of high levels of mRNA coding for proteins p6 (DBP) and p5 (SSB) and lower levels of longer mRNA coding for proteins p6 to p1 (17). Apart from possible differences in translation initiation efficiencies, this explains why p6 and p5 are synthesized in far larger quantities than are proteins p4, p3, p2, and p1 (2, 86, 139). Equivalent TA1 transcriptional terminators are present in the genomes of B103 and GA-1, indicating that a regulatory mechanism similar to that proposed for φ29 exists in B103 and GA-1. In all three genomes, the TA1 transcriptional terminator is located at very similar positions within gene 4. Thus, the mRNAs synthesized up to the TA1 terminators may allow the synthesis of the N-terminal 28 to 30 amino acids of protein p4. Interestingly, this region of the three p4 proteins is far more conserved than the downstream p4 region (Fig. 3), which might imply that the N-terminal 30 amino acids of p4 could have a function on its own.
FIG. 3.
Alignment of the deduced transcriptional regulatory protein sequences encoded by φ29 (p4phi29), B103 (p4B103), and GA-1 (p4GA-1). Black and grey boxes enclose residues that are conserved in all three or two of the three sequences, respectively. The following amino acids were considered conservative: L, I, V, A, and M; F, Y, and W; K and R; D and E; Q and N; and S and T. The position up to which the N-terminal part of the respective gene 4 can be translated, considering the mRNA length terminated at the transcriptional terminator TA1, is indicated by a vertical arrow.
No potential Rho-independent transcriptional terminator is present downstream of the pRNA coding region of φ29, which constitutes the most leftward-reading region of this genome (Fig. 2). This could imply that transcription, starting from the A2b, A2c, and A1 promoters, continues until the left end of the genome is reached. It has indeed been shown that in vivo transcription initiating at these φ29 promoters reaches the very left end of the φ29 DNA molecule as if the RNA polymerase would run off the template (16, 17). The same organization and the absence of a potential Rho-independent terminator downstream of the B103 pRNA-coding region suggests a similar situation for B103.
The situation is different, however, for GA-1. As shown in Fig. 2, three potential Rho-independent terminators are present in the left part of the GA-1 genome. The one located closest to the left DNA end (downstream of ORF T), named TA4, would terminate transcription initiating at the A1c promoter of GA-1. The middle one, named TA3, located downstream of the pRNA coding region, would terminate transcription initiating from the GA-1 promoter A1b and possibly A1a. The third one, named TA2, would terminate transcription initiating from the GA-1 A2c and A2b promoters. Note that in contrast to the situation in φ29 and B103, the GA-1 terminator TA2 is located directly downstream of gene 2. The −35 sequence of the GA-1 promoter A1a is located within this terminator.
PROTEIN-PRIMED MECHANISM OF DNA REPLICATION
The genomes of the φ29-like phages consist of a linear dsDNA molecule of about 20 kb with a phage-encoded protein, TP, covalently attached at each 5′ end. Genomes consisting of a linear dsDNA molecule with a TP covalently linked to their 5′ ends have also been found for (i) other bacteriophages (e.g., the Streptococcus pneumoniae and Escherichia coli phages Cp-1 and PRD1, respectively), (ii) animal viruses (e.g., adenoviruses), (iii) plasmids (e.g., S1 and Kalilo), and (iv) bacteria (e.g., Streptomyces). In most of these cases, initiation of DNA replication occurs via a so-called protein-priming mechanism (for reviews, see references 176, 178, and 181).
The in vitro mechanism of protein-primed DNA replication has been studied in most detail for φ29. The basic features of the protein-primed mechanism of DNA replication, based on the φ29 system, are outlined here. More detailed descriptions of the different steps and the function of the proteins involved are given below. In addition, it should be mentioned that although the main characteristics of protein-primed DNA replication are conserved, some minor differences with respect to the φ29 mechanism have been observed in some cases, especially regarding the sliding-back step (see below). Figure 4 shows a schematic representation of in vitro φ29 DNA replication. Initiation of φ29 DNA replication starts with recognition of the origin of replication, i.e., the TP-containing DNA ends, by a TP-DNA polymerase heterodimer. The virus-encoded protein p6 forms a nucleoprotein complex that would help to open the DNA ends (187), facilitating the formation of a covalent linkage between the first inserted nucleotide (dAMP) and TP, which is catalyzed by the φ29 DNA polymerase (29, 109). The formation of this first TP-dAMP covalent complex is directed by the second nucleotide at the 3′ end of the template; then the TP-dAMP complex slides back 1 nucleotide to recover the information of the terminal nucleotide (144). Next, the φ29 DNA polymerase synthesizes a short elongation product before dissociating from the TP (146). Replication, which starts at both DNA ends, is coupled to strand displacement. This results in the generation of so-called type I replication intermediates consisting of full-length φ29 dsDNA molecules with one or more ssDNA branches of different lengths. The ssDNA stretches generated are bound by the SSB protein (p5). When the two converging DNA polymerases merge, a type I replication intermediate becomes physically separated into two type II replication intermediates. Each of these consists of a full-length φ29 DNA molecule in which a portion of the DNA, starting from one end, is double stranded and the portion spanning to the other end is single -stranded (102, 117). Continuous elongation by the DNA polymerase completes replication of the parental strand.
FIG. 4.
Schematic representation of the mechanism of φ29 protein-primed DNA replication. Primer and parental TP are shown in black and grey, respectively. p6, double-stranded DNA binding protein; p5, single-stranded DNA binding protein (SSB). Only the left DNA end has been drawn, except for type I and type II molecules, where both DNA ends are shown. See the text for details. Adapted with permission from reference 182.
INITIATION OF DNA REPLICATION
DNA Polymerase-TP Heterodimer Formation
DNA polymerases are unable to initiate de novo DNA synthesis on a DNA template but require the existence of a primer containing a free hydroxyl group to start DNA elongation (126). Generally, RNA primers provide the 3′-hydroxyl (3′-OH) group needed by the DNA polymerase to elongate the DNA chain. However, in most linear genomes containing a TP covalently linked to their 5′ DNA ends, the 3′-OH group of a specific serine, threonine, or tyrosine residue of the TP is used for DNA elongation (reviewed in reference 181). In φ29 DNA polymerase, its TP deoxynucleotidylation activity is responsible for the covalent linkage of 5′-dAMP, via a phosphoester bond, to the hydroxyl group of Ser232 of the TP (24, 29, 109). This reaction requires the formation of a stable heterodimer complex between the TP and the φ29 DNA polymerase (28). Most probably, the active site used for polymerization is also used for the TP deoxynucleotidylation reaction (reviewed in references 31 and 32). This implies that the TP present in the heterodimer complex has to be specifically positioned in order for the DNA polymerase to perform the TP deoxynucleotidylation reaction. Several mutations located in different regions of the φ29 DNA polymerase affect its interaction with the TP (37, 61, 145, 206). In addition, interaction of TP with the purified C-terminal portion of the φ29 DNA polymerase is severely impaired (209). Together, these results suggest that interaction of the TP involves many contacts with different regions of the DNA polymerase.
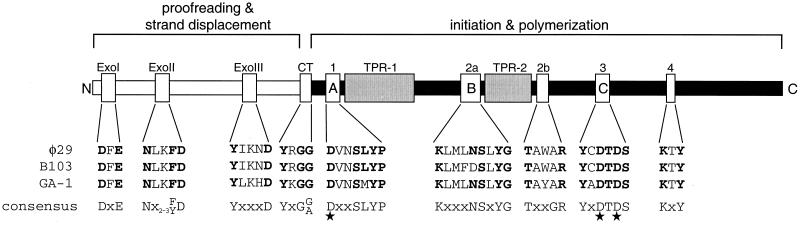
Interestingly, a multiple sequence alignment of DNA polymerases belonging to the B-type family showed that DNA polymerases involved in protein-primed DNA replication contain two regions of amino acids, denoted TPR-1 and TPR-2 (Fig. 5), which are not present in other B-type DNA polymerases (33). Analysis of the φ29 mutant DNA polymerase in which the conserved Asp332 residue of the TPR-1 region was changed into Tyr showed that it was able to form a stable heterodimer with TP and that it had essentially wild-type levels of synthetic activities in DNA primed reactions. However, its activity was drastically affected in φ29 TP-DNA replication, indicating that the mutant DNA polymerase forms a non functional interaction with the TP and hence supporting the view that at least TPR-1 is involved in proper positioning of the TP in the TP-DNA polymerase heterodimer complex (68).
FIG. 5.
Structural and functional map of the DNA polymerase. The N-terminal domain, required for proofreading and strand displacement, and the C-terminal initiation and polymerization domain are indicated by white and black rectangular boxes, respectively (32). The ExoI, ExoII, and ExoIII motifs, as well as the motifs of the C-terminal domain, are indicated: motif 1 (also called motif A), motif 2a (also called motif B), motif 2b, motif 3 (also called motif C), and motif 4. In addition, the position of the YxG(G/A) motif, important for the coordination between 3′-5′ exonuclease and 5′-3′ polymerization, is indicated by CT (cross talk). All these motifs are conserved in B-type proofreading-proficient DNA polymerases. The amino acid sequences of each of these various motifs present in the DNA polymerases of φ29, B103 and GA-1, together with each consensus sequence, are indicated below the map. The three Asp residues that form a metal binding triad required for catalysis at the polymerization active site are indicated by an asterisk. Finally, the position of the TPR-1 and TPR-2 motifs, characteristic of DNA polymerases involved in protein-primed DNA replication, are indicated. See the text for further details.
Sliding-Back Mechanism
Although the TP deoxynucleotidylation reaction can occur in the absence of a DNA template, it is strongly stimulated in the presence of φ29 TP-DNA (24). In the latter case, TP-dAMP is preferentially formed. The DNA ends of φ29 have a short inverted terminal repeat of 6 nucleotides (3′-TTTCAT-5′). The first TP-dAMP is not directed by the terminal nucleotide but by the penultimate nucleotide of the φ29 template strand. Subsequently, the complex slides back 1 nucleotide to recover the information of the 3′-terminal nucleotide (144). Terminal repeats are also present in the genomes of B103 (3′-TTTCAT-5′), GA-1 (3′-TTTATCTT-5′), and all other φ29-related phages analyzed so far. Moreover, this feature is also conserved in other linear genomes containing a TP covalently linked to their DNA ends, such as the E. coli and S. pneumoniae phages PRD1 and Cp-1, respectively, linear plasmids, and the eukaryotic adenovirus. Terminal reiteration is a prerequisite for the sliding-back mechanism. Indeed, the replication initiation site in GA-1 (114), PRD1 (43), Cp-1 (132), and adenovirus (125) corresponds to an internal nucleotide close to the 3′-terminal end, and a sliding-back or similar mechanism has been shown to occur in these cases to recover the information of the terminal nucleotide(s). Probably, the sliding-back mechanism applies to all genomes that replicate via a protein-primed mechanism. Since proofreading does not apply to the TP-dNMP product (72), the sliding-back mechanism would be an alternative way to ensure that the replication origin-containing DNA ends are replicated with high fidelity.
Transition from Protein-Primed to DNA-Primed Replication
After the sliding-back step, the φ29 DNA polymerase and the primer TP do not dissociate immediately. Rather, there is a transition stage in which the DNA polymerase synthesizes a DNA molecule of 5 nucleotides while complexed with the primer TP (initiation mode). During the synthesis of nucleotides 6 to 9 the complex undergoes some structural change (transition mode), and the DNA polymerase finally dissociates from the primer TP when the nucleotide 10 is inserted into the nascent DNA chain (elongation mode) (146). This behavior probably reflects a requirement of the φ29 DNA polymerase for a DNA primer of a minimum length to efficiently carry out DNA-primed elongation. This view is supported by the following data. First, Méndez et al. (146) demonstrated that primer molecules of 6 nucleotides or less are not elongated. This fits well with the observation that φ29 DNA polymerase synthesizes a DNA chain of 5 nucleotides before it changes from the initiation mode to the elongation mode in TP-DNA-primed reactions. Second, abortive replication products consisting of the primer TP linked up to 8 nucleotides were particularly observed under conditions that decrease the strand displacement capacity of φ29 DNA polymerase (146). Finally, de Vega et al. (62) demonstrated that φ29 DNA polymerase covers a DNA region of 10 nucleotides, which may be indicative of the optimum length to carry out polymerization. Interestingly, the φ29 DNA polymerase mutant in which Asp456, belonging to the conserved “YxDTDS” motif at the polymerization domain (see below), has been changed into Gly is unable to proceed further than 5 nucleotides from the initiation complex. This suggested that the φ29 DNA polymerase residue Asp456 is crucial to entry into the transition stage of φ29 DNA replication (185).
A similar transition step has also been demonstrated in replication of adenovirus (124) and probably is a general feature of protein-primed DNA replication.
THE FOUR MAIN PROTEINS REQUIRED FOR IN VITRO DNA REPLICATION
In the φ29, B103, and GA-1 genomes, genes 6, 5, 3 and 2 are located in a single early-expressed operon (Fig. 2). In φ29, these genes are indispensable for in vivo phage DNA replication. Gene 2 encodes the DNA polymerase, gene 3 encodes the TP, gene 5 encodes SSB, and gene 6 encodes DBP. An in vitro φ29 DNA replication system, based on these four purified proteins, has been established (27). The availability of this system has allowed a detailed analysis of the in vitro φ29 DNA replication mechanism and functional analysis of these four main replication proteins. Characteristics of these four proteins are given below.
DNA Polymerase
Gene 2 of φ29, B103, and GA-1 encodes a DNA polymerase. In φ29 and GA-1 the DNA polymerase has been shown to be required for replication of its phage DNA (29, 114). The DNA polymerases encoded by φ29, B103, and GA-1 belong to the B-type superfamily of DNA-dependent DNA polymerases (also referred to as eukaryotic or α-like polymerases). This family includes a large number of prokaryotic and eukaryotic enzymes that are sensitive to certain drugs (aphidicolin and phosphonoacetic acid) and nucleotide analogs (butylanilino-dATP and butylphenyl-dGTP). The DNA polymerase of φ29 has been analyzed in detail (for reviews, see references 31 and 32). The monomeric φ29 DNA polymerase, which has a size of only about 66 kDa, catalyzes both the initiation and elongation stages of DNA synthesis (29, 30). To accomplish this, it is able to carry out two distinguishable synthetic reactions: TP deoxynucleotidylation and DNA polymerization. In addition, it has two degradative activities: pyrophosporolysis and 3′-5′ exonucleolysis. Moreover, it has two intrinsic properties: high processivity and strand displacement ability (25). Due to the φ29 DNA polymerase properties, in vitro φ29 DNA replication does not require accessory proteins and DNA helicases (25).
The enzymatic activities of the φ29 DNA polymerase have been mapped by site-directed mutagenesis. A structural map, given in Fig. 5, shows that the φ29 DNA polymerase has a bimodular organization, with the N-terminal portion constituting the 3′-5′ proofreading domain and the C-terminal portion constituting the domain responsible for its 5′-3′ synthetic activities. The bimodular organization of the φ29 DNA polymerase has been proven experimentally. Analysis of a purified C-terminal deletion derivative of φ29 DNA polymerase containing the 188 N-terminal amino acids showed that it was devoid of any synthetic activity but retained 3′-5′ exonuclease activity (31). Reciprocally, a purified N-terminal deletion derivative containing the C-terminal 388 amino acids had neither 3′-5′ exonuclease nor strand displacement activity but did have synthetic activities (209). Available three-dimensional structures of other DNA polymerases show that the bimodular organization is characteristic of proofreading proficient DNA polymerases (reviewed in reference 121).
C-terminal domain of φ29 DNA polymerase.
The polymerization activity of the φ29 DNA polymerase is confined to the C-terminal domain of the enzyme. This part of the φ29 DNA polymerase has three regions containing motifs that are conserved in other DNA polymerases belonging to family B. These three motifs are Dx2SLYP (motif A, also named motif 1), Kx3NSxYG (motif B, also named motif 2a), and YxDTDS (motif C, also named motif 3). The positions of these and other conserved motifs described below are indicated in Fig. 5, together with the amino acid sequence corresponding to each motif present in the DNA polymerase of φ29, B103, and GA-1. Site-directed mutagenesis at motifs A, B, and C of φ29 DNA polymerase (21, 34–36) showed that these three regions form an evolutionarily conserved polymerization-active site.
Figure 5 shows that of these three motifs, only motif C is fully conserved in the DNA polymerases of B103 and GA-1. Whereas motif A is conserved in the DNA polymerase of B103, a Met residue in the GA-1 polymerase occupies the corresponding position of φ29 Leu253. Analysis of a φ29 DNA polymerase mutant in which Leu253 had been replaced by a Val residue (L253V) showed that whereas it was not affected in template-primer DNA binding, it was strongly affected in reactions involving the use of TP as primer (35). With this result in mind, it would be interesting to study the effects of a φ29 L253M DNA polymerase mutant and relate it to the reciprocal mutation in the GA-1 DNA polymerase (M253L). For motif B, the residue corresponding to Asn387 of φ29 DNA polymerase is occupied by an Asp in the B103 polymerase (Fig. 5). The involvement of φ29 DNA polymerase Asn387 in the correct binding of the primer terminus at the polymerization active site was demonstrated by the analysis of the N387Y mutant (36). Taking into account the protein sequence of the B103 DNA polymerase, it would be interesting to study possible effects of replacing Asn387 by Asp (N387D).
In addition to motifs A, B and C, two other motifs, Tx2GR (motif 2b) and KxY (motif 4), were identified in the C-terminal portion of φ29 DNA polymerase and analyzed by site-directed mutagenesis (37, 145). These two motifs, which are also conserved in the C-terminal portion of B103, GA-1 (Fig. 5), and other B-type DNA polymerases, are involved in primer stabilization at the active site. In addition, motif 2b is involved in TP and metal binding (145). For several DNA polymerases, including the φ29 DNA polymerase, it has been demonstrated that three Asp residues form a metal binding triad required for catalysis at the polymerization active site (reviewed in reference 32). In the φ29 DNA polymerase, the three Asp residues implicated are Asp249, belonging to motif A, and Asp456 and Asp458, both belonging to motif C (21, 35, 185). These three Asp residues are conserved in the DNA polymerases of B103 and GA-1 and in all other known members of the B-type DNA polymerases. Also, Arg438 of motif 2b of φ29 DNA polymerase plays a role in catalysis of the polymerization reaction (145). Moreover, three highly conserved Tyr residues were shown to be involved, directly or indirectly, in interaction with deoxynucleoside triphosphates (dNTPs). These residues, also conserved in the B103 and GA-1 DNA polymerases (Fig. 5), are Tyr254 of motif A (34, 35), Tyr390 of motif B (34, 36), and Tyr454 of motif C (21). Since the φ29 residues Tyr254 (motif A) and Tyr390 (motif B) are also involved in selection of dNTP binding, they play an important role in the fidelity of DNA replication (184). In addition, a single and specific replacement of Tyr254 (motif A) by a Val residue enables the mutant φ29 DNA polymerase to incorporate ribonucleotides without affecting its wild-type affinity for dNTPs (38). This indicates that φ29 Tyr254 is responsible for the discrimination against the 2′-OH group of an incoming ribonucleotide.
In addition, seven residues that are invariant or highly conserved in the C-terminal domain of B-type DNA polymerases were shown to be involved in binding template-primer structures. These residues are Ser252 of motif A (35), Asn387 (see above) and Gly391 of motif B (36), Thr434 and Arg438 of motif 2b (145), and Lys498 and Tyr500 of motif 4 (37).
N-terminal domain of φ29 DNA polymerase.
(i) Proofreading.
The insertion discrimination values of the φ29 DNA polymerase range from 104 to 106 and the efficiency of mismatch elongation is 105- to 106-fold lower compared to a properly paired terminus (72). These values illustrate the high fidelity with which the φ29 DNA polymerase replicates DNA. As with other proofreading-proficient DNA polymerases, the φ29 DNA polymerase owes its high fidelity to its 3′-5′ exonuclease activity (81), which is confined to the N-terminal part of the enzyme. Bernad et al. (20) proposed that three N-terminally located regions, ExoI, ExoII, and ExoIII, form the 3′-5′ exonuclease active site (Fig. 5) and are evolutionarily conserved in prokaryotic and eukaryotic DNA polymerases. This proposal has been proven valid for various DNA polymerases of eukaryotic and prokaryotic origin (for a review, see reference 60). The three Exo domains contain five invariant residues that are involved in metal binding and 3′-5′ exonuclease catalysis. In φ29 DNA polymerase, these residues are Asp12 and Glu14 in ExoI, Asp66 in ExoII, and Tyr165 and Asp169 in ExoIII (20).
The two-metal-ion mechanism, first proposed for the 3′-5′ exonuclease active site of polymerase I (18), was shown also to apply to φ29 DNA polymerase and can be extrapolated to other proofreading-proficient DNA polymerases (73). Another invariant residue, Lys143 of φ29 DNA polymerase, was analyzed and shown to be important for the catalytic efficiency of the 3′-5′ exonuclease activity (63). In addition, other residues in the Exo motifs that are conserved in B103, GA-1, and most other prokaryotic and eukaryotic DNA polymerases were functionally analyzed. Two of these, Thr15 and Asn62, located at the ExoI and ExoII motifs, respectively, were shown to act as single-stranded DNA ligands playing a critical role in the stabilization of the frayed primer terminus at the 3′-5′ exonuclease active site (64). Also, Phe65 of the ExoII motif and residues Ser122 and Leu123, which are part of a newly identified motif [S/T]Lx2h, were shown to be important for (i) stable interaction with ssDNA, (ii) 3′-5′ exonucleolysis of ssDNA substrates, and (iii) proofreading of DNA polymerization errors (65). In addition, these studies showed that the aromatic ring of Phe65 appeared to be critical to orient the ssDNA substrate in a stable conformation to allow 3′-5′ exonucleolytic catalysis. These three residues, Phe65, Ser122, and Leu123, are also conserved in the B103 and GA-1 DNA polymerases.
(ii) Strand displacement.
After the initiation, sliding-back, and transition steps, continuous polymerization, carried out by a single φ29 DNA polymerase molecule, completes the replication of the almost 20-kb DNA strand (30). Using primed M13 DNA as the template, the φ29 DNA polymerase is able to synthesize DNA chains of more than 70 kb (25). This demonstrates the high processivity and strand displacement activity of the φ29 DNA polymerase. Replication of φ29 DNA starts nonsimultaneously from either end of the linear DNA molecule (117), generating so-called type I replication intermediates (Fig. 4). Until the two converging DNA polymerases collide, DNA polymerization is coupled to strand displacement, which makes a helicase unnecessary (25). Various DNA polymerases, but not the one encoded by φ29, are prone to replication slippage. This particular type of error, which results in deletions, is caused when a polymerizing DNA polymerase slips between two short sequence duplications. Recently, evidence has been presented that the high strand displacement activity of the φ29 DNA polymerase prevents replication slippage (48).
Surprisingly, functional analysis of the φ29 DNA polymerases containing mutations in one of the five invariant residues in the Exo motifs critical for 3′-5′ exonuclease activity, Asp12, Glu14, Asp66, Tyr165, or Asp169, showed that they were also strongly affected in their strand displacement activity (73, 194). In addition, mutants corresponding to Lys143, the residue which is conserved in GA-1 and B103 DNA polymerases and was shown to play an auxiliary role in catalysis of the exonuclease reaction, were affected in strand displacement activity (62). These results indicated that the strand displacement activity of φ29 DNA polymerase is located in its N-terminal domain, somehow overlapping with the 3′-5′exonuclease activity.
Mutations of residues Thr15 and Asn62, shown to act as ssDNA ligands but not playing a direct role in the φ29 DNA polymerase 3′-5′ exonuclease catalysis reaction, displayed wild-type levels of strand displacement activity (64). Therefore, it seems that impaired strand displacement activity is restricted to the 3′-5′ exonuclease mutants that act directly as metal ligands or to those that affect the metal binding network. Based on these results, it was proposed that contacts with divalent metal ions assist in interactions with the displaced ssDNA strand (32).
Coordination between synthesis and degradation.
As described above, the φ29 DNA polymerase has a bimodular organization, with its degradative and strand displacement activities present in the N-terminal domain and the synthetic activities present in the C-terminal domain. Effective proofreading of a DNA polymerization error therefore requires that the primer terminus containing the error switch intramolecularly from the polymerization to the 3′-5′ exonuclease active site (62). A conserved motif, YxG[G/A], located between the proofreading and polymerization domains (Fig. 5, indicated by CT for cross talk; note that this motif is also conserved in the DNA polymerases of GA-1 and B103) was recently shown to play an important role in the coordination between DNA synthesis and proofreading (208). Single amino acid substitutions in this motif of φ29 DNA polymerase gave rise to three different mutant phenotypes: (i) favored polymerization, (ii) favored 3′-5′ exonucleolysis, or (iii) favored 3′-5′ exonucleolysis with loss of polymerization. The different phenotypes could be directly related to defects in DNA binding at either active site, thereby stimulating the activity of the other active site. Thus, the YxG[G/A] motif exerts its important role in the coordination between synthesis and degradation primarily through DNA binding.
Terminal Protein p3
The initiation of protein-primed DNA replication requires the formation of a stable heterodimer complex between the TP and the DNA polymerase. Several lines of evidence indicate that the TP occupies the double-stranded DNA binding channel in the DNA polymerase during initiation of replication. First, mutant φ29 DNA polymerases that are affected in interaction with TP also show a reduced capacity to bind a DNA template-primer structure (37, 61, 145, 206). Second, partial proteolysis of the φ29 DNA polymerase-TP heterodimer with endoproteinase LysC resulted in a protection and digestion pattern similar to that obtained with DNA (207). Finally, binding of TP to φ29 DNA polymerase prevents binding and polymerization on DNA template-primer structures (61). Stable heterodimer formation involves many contacts with different regions of the φ29 TP, as shown for the φ29 DNA polymerase. These include (i) an internal region near the N terminus (amino acids 72 to 80) (228), (ii) the C-terminal region (amino acids 242 to 262) (228), and (iii) the RGD motif located at positions 256 to 258 in φ29 TP (116). It should be noted that although the RGD motif is conserved in most other Bacillus phage-encoded TPs (including the one encoded by B103), it is not conserved in the TP of GA-1.
In most linear genomes a TP molecule is covalently linked to the 5′ DNA ends. In φ29 the 5′-terminal dAMP is linked via a phosphoester bond to the hydroxyl group of Ser232 of the TP (109). The observation that a mutant φ29 TP in which Ser232 was replaced by a Thr residue had completely lost its priming activity illustrated the high specificity of Ser232 (82). The Ser232 residue of the φ29 TP is conserved in the TPs encoded by B103 and GA-1. Although experimental evidence is lacking, this suggests that the hydroxyl group of the conserved Ser residue is used for the linkage of the first dAMP in B103 and GA-1 as well.
The DNA ends containing the attached TP molecule constitute the origins of replication. The first step of initiation of DNA replication is the recognition of these origins by a TP-DNA polymerase heterodimer. The TP molecule in the heterodimer functions as primer for the subsequent replication initiation step. To discriminate between the two different functions, the TP molecule linked to the 5′ DNA ends is called parental TP and the TP present in the complex with DNA polymerase is called primer TP.
Blunt-ended DNA fragments containing the left or right φ29 DNA ends, but not internal φ29 DNA fragments, were active as templates in in vitro initiation reactions (80, 103, 104). On one hand, this indicated that specific DNA sequences located at the φ29 DNA ends are involved in origin recognition, and on the other hand it suggested that the TP has DNA binding affinity. The latter feature was indeed demonstrated, and amino acids 13 to 18, 30 to 51 and 56 to 71, all located in the N-terminal portion of the φ29 TP, were shown to be involved in DNA binding (228).
Although blunt-ended DNA fragments comprising the left or right φ29 DNA ends were active in in vitro initiation reactions, their activity was far lower than that of DNA ends containing a parental TP (103, 104). These results showed that the parental TP is the major signal in the template for origin recognition and strongly suggested that the TP-DNA polymerase heterodimer is recruited to the origin through interaction with the parental TP. The observation that parental TP molecules can interact with each other (159, 179) suggests that recruitment of the heterodimer to the origin is brought about by protein-protein contacts between the parental and primer TP. The φ29 TP residues Asn80 and Tyr82, which are conserved in other Bacillus phage-encoded TPs including those of B103 and GA-1, were shown to be specifically involved in this recruitment (115). Interestingly, these residues are located just before a region, spanning amino acids 84 to 118, that has a high probability of forming an amphipathic α-helix, which might provide an appropriate surface for the interaction between parental and primer TP through the formation of a coiled coil. This putative amphipathic α-helix is conserved in all other TPs encoded by φ29-like phages. Recently, evidence has been presented that this putative coiled-coil domain of φ29 TP is indeed an important element for origin recognition (186). In addition to interactions between primer and parental TP, it has been shown that origin recognition also involves interactions between the DNA polymerase and the parental TP (87).
DBP Protein p6
The DBP protein p6, which has been described as a histone-like protein (187), is able to bind in vitro to the whole φ29 DNA, and a role in genome organization has been proposed (100). Its specific role in regulating the expression of the φ29 promoters A2b, A2c, A3, and C2 is described above. The high intracellular abundance of p6 in infected cells suggests that it also plays a role in φ29 genome organization in vivo (2, 187). Although the large amounts of p6 synthesized in infected cells are sufficient to bind the whole φ29 DNA, it binds preferentially at the DNA ends (167, 189) through the minor groove (77, 190). The preferred protein p6 binding sites are located at nucleotides 46 to 68 and 62 to 125 at the left and right φ29 DNA ends, respectively. Whereas these regions do not show sequence similarity, they do contain DNA sequences that are predicted to be bendable every 12 bp (189), and this feature was indeed shown to be the major determinant for protein p6 recognition (188). Interestingly, the φ29 DNA region containing promoters A2b and A3 has intrinsic curvature (143, 173), which may be important for protein p6 binding and thus for its role in regulation of these promoters.
Protein p6 was shown to be essential for in vivo φ29 DNA replication (50, 70). Binding of protein p6 to the DNA ends activates initiation of φ29 DNA replication (26, 180). When protein p6 binds to circular DNA, it restrains positive supercoiling, supporting a model in which a right-handed superhelix of DNA wraps tightly around a multimeric protein p6 core (190). It was indeed demonstrated that the φ29 DNA ends adopt a right-handed toroidal conformation that wraps around a multimeric p6 core (188). On protein p6 binding, the DNA is compacted 4.2-fold. The parameters that define the path taken by the DNA in the protein p6 complex have been defined: one superhelical turn has 63 bp with a pitch of 5.1 nm and a diameter of 6.6 nm. Consequently, the DNA should be strongly bent (66° every 12 bp) and underwound (11.5 bp/turn) (188). The specific conformation of this nucleoprotein complex would help to open the DNA ends (180, 188), facilitating the formation of the covalent linkage between dAMP and the primer TP catalyzed by φ29 DNA polymerase.
A deletion derivative lacking the N-terminal 13 amino acids of φ29 protein p6 is unable to bind DNA, indicating that this region of p6 is involved in DNA binding (160). Secondary-structure predictions indicated the existence of a putative amphipathic α-helix in this region. To test the possibility that the positively charged residues of this putative helix are involved in DNA interaction, the Lys2 and Arg6 residues were changed into Ala. The observation that both purified mutant p6 proteins were affected in DNA binding supported this hypothesis (77). Also the N-terminal regions of the p6 proteins encoded by B103 and GA-1 are predicted to form an α-helix with an amphipathic character. In addition, the two positively charged residues of the φ29 protein p6 that are important for DNA binding are conserved in the p6 proteins encoded by B103 and GA-1. These conserved sequence features in the N-terminal part of the various p6 proteins may reflect a conserved function for this region, i.e., its involvement in DNA binding.
The φ29-protein p6 was shown to form dimers in solution (162). Self-association of φ29 protein p6 was studied in more detail using analytical ultracentrifugation analysis (2). In these studies it was shown that in the absence of DNA and in a concentration range between 1 and 100 μM, protein p6 is in a monomer-dimer equilibrium, in agreement with earlier studies. However, at concentrations around 1 mM, which corresponds approximately to the intracellular concentration of p6 in infected cells, protein p6 associates into oligomers (2). These results further supported the view that protein p6 can form a scaffold for DNA binding. In subsequent studies the structure of the oligomers were analyzed by transmission electron microscopy. This revealed that protein p6 aggregates into crooked oligomers, compatible with a helical structure (1).
Residues of φ29 protein p6 that were critical for self-association of the protein were identified by random mutagenesis (3). The mutations found were mainly clustered in two regions, one located at the N terminus and the other located in the central part of the protein. Two φ29 p6 single mutants, A44V and I8T, were further analyzed. These mutants showed, in addition to impaired dimer formation ability, reduced DNA binding affinity; consequently, they were also affected in activation of in vitro initiation of φ29 DNA replication. These two residues of the φ29 protein p6 are conserved in the p6 proteins of B103 and GA-1. Dimer formation capacity was enhanced in protein φ29 p6 mutants containing C-terminal deletions. In these latter mutants, a highly acidic region, which is conserved in the p6 proteins of GA-1 and B103, was removed, and hence it was proposed that this C-terminal acidic region modulates φ29 protein p6 self-association (3).
Protein p6 binds with a precise phase to the replication origins of φ29 DNA, which is crucial for the activation of DNA replication (190). Thus, protein p6 did not activate a DNA fragment with a 4-bp insertion between the nucleation and the initiation site because the p6 binding was out of phase with respect to the replication origin. Restoring the p6 binding phase by the insertion of 24 bp resulted in recovery of the replication initiation activity (190). The phasing of protein p6 encoded by GA-1 and Nf (the latter belonging to the B103 group of φ29-related phages) on homologous and heterologous DNAs was studied by hydroxyl radical and DNase I footprinting analyses (78). The p6 proteins of Nf and B103 are 95% identical. When tested in the presence of homologous DNA, the periodicity of protection of the p6 proteins of φ29 and Nf was 12 bp (corresponding to one monomer) and that of GA-1 was 11 bp. Similar protection assays using heterologous DNA showed that the periodicity and positioning of the Nf protein p6 monomers on φ29 DNA was almost the same as for those formed by φ29 protein p6 on its homologous DNA. However, in the reciprocal situation the protection of φ29 protein p6 bound to Nf DNA was shifted 4 bp with respect to the homologous Nf situation, indicating that Nf and φ29 p6 proteins recognized different signals in the Nf DNA. Thus, although the phasing was not conserved, protein p6 of φ29 and Nf are able to bind to the respective heterologous DNA. A different outcome was observed for protein p6 of GA-1. In this case, no clearly structured complexes of GA-1 protein p6 were formed on φ29 or Nf DNA, although these were formed with GA-1 DNA. Moreover, Freire et al. (78) showed that correct phasing of p6 with respect to the origins of replication was required for activation of GA-1 and Nf DNA replication, as observed for φ29. These studies nicely illustrated, on one hand, that the protein p6 mechanism of DNA replication activation is conserved for φ29, Nf, and GA-1 and, on the other hand, that each of the three p6 proteins forms highly specific structures with their respective DNA origins that are required for activation of initiation of DNA replication.
SSB Protein p5
Due to its symmetrical mode of DNA replication, coupled to strand displacement, the replication intermediates generated during φ29 DNA replication contain large stretches of ssDNA. Gene 5 of phages φ29, B103, and GA-1 encodes an SSB that is highly abundant in infected cells. The SSB protein p5 is essential for elongation of replication in vivo (139). Direct binding of φ29 protein p5 to φ29 DNA replication intermediates has been demonstrated (101). This binding protects the ssDNA branches against nuclease degradation and greatly stimulates dNTP incorporation during φ29 DNA replication in vitro (133). Because binding of protein p5 prevents nonproductive binding of the DNA polymerase to ssDNA (101), this probably explains the strong enhancement of DNA replication by SSB. In addition, binding of p5 to ssDNA increases the DNA elongation velocity about fivefold in in vitro φ29 DNA replication using φ29 DNA polymerase mutants that are defective in strand displacement. This effect is most probably due to the helix-destabilizing properties of φ29 SSB (196). Protein p5 of φ29 binds ssDNA in a cooperative way (Keff = 105 M−1, ω = 50 to 70), covering 3.4 nucleotides per p5 monomer (85, 195).
The importance of SSB in replication is further illustrated in in vitro φ29 DNA amplification assays. Omission of protein p5 from the reaction mixtures results in the generation and amplification of short φ29 DNA products (27). Esteban et al. (71) have shown that such small φ29 DNA products have a palindromic nature and that they are caused by a DNA polymerase template-switching event. The presence of sufficient protein p5 prevents the DNA polymerase from switching template by either one or both of the following two mechanisms. First, binding of protein p5 to ssDNA may preclude the displaced ssDNA strand from being an alternative template for the DNA polymerase. Second, binding of SSB will prevent the formation of secondary structures in the displaced ssDNA strand, which otherwise impedes efficient progression of the DNA polymerase and which probably stimulates the DNA polymerase to switch template (71).
Recently, comparative studies of the structural complexes formed by the φ29, Nf, and GA-1 SSB with DNA have been performed (85). The sequences of the Nf and B103 SSB are 96% identical. Whereas the SSB of φ29 and Nf formed stable monomers in solution under the conditions used, the SSB of GA-1 formed hexamers. In addition, the GA-1 SSB occluded a larger binding site (51 nucleotides/hexamer) than did the φ29 and Nf SSBs (3.4 and 4.7 nucleotides/monomer, respectively). Moreover, on binding, the GA-1 SSB compacted the ssDNA far more than the φ29 SSB did. Whereas the length of the ssDNA was reduced about sixfold on binding of GA-1 SSB, only a twofold reduction was obtained on binding of φ29 SSB (85, 101). In addition to the structural complexes formed between the SSB of φ29, Nf, and GA-1, their functional behavior has been analyzed recently (86). In agreement with the structural analysis, far less GA-1 SSB than Nf or φ29 SSB was required to display a helix-destabilizing effect and to stimulate dNTP incorporation in in vitro DNA replication assays. In summary, the SSB of GA-1 behaved structurally and functionally quite differently from the SSBs of Nf and φ29. Comparison of the three SSB sequences showed that the SSB of GA-1 has an N-terminal extension of about 40 amino acids. Possibly, this additional protein domain is involved in the different characteristics of the GA-1 SSB.
OTHER GENES AND OPEN READING FRAMES DOWNSTREAM OF GENE 2 IN φ29 AND B103
Figure 2 shows that a transcriptional terminator (TA2) is present in the GA-1 genome downstream of gene 2, strongly indicating that genes 6 to 2 constitute one operon that is expressed by the GA-1 promoters A2b and A2c. In contrast, the corresponding operon of φ29 contains, in addition to genes 6 to 2, gene 1 (86 codons) as well as three ORFs, named ORF 1A, ORF 1B and ORF 1C, that may encode proteins of 56, 58, and 47 amino acids, respectively. Also, the corresponding operon of B103 contains four additional ORFs downstream of its gene 2. These ORFs, which we named ORF g, ORF h, ORF i, and ORF j, may encode proteins of 36, 67, 56, and 22 amino acids, respectively. The function of the putative small proteins encoded by φ29 ORFs 1A to 1C and B103 ORFs g to j is yet unknown. It should be mentioned, however, that the deduced protein sequence of B103 ORF h shows significant similarity to the 46 C-terminal amino acids of φ29 protein p1 (50 and 62.5% identity and similarity, respectively). In addition, the deduced protein sequences of φ29 ORF 1A and B103 ORF i, both 56 amino acids long, have a high level of similarity (64 and 75% identity and similarity, respectively). Finally, the deduced protein sequence of B103 ORF j (22 amino acids) is similar to the N-terminal part of φ29 ORF 1B (50 and 59% identity and similarity, respectively). These similarities suggest that the putative proteins encoded by these ORFs may have similar functions.
Gene 1 of φ29
The φ29 sus1(629) mutant contains a point mutation which changes codon 7 of gene 1 (CAA) into a nonsense TAA codon (166). Compared to wild-type φ29-infected cells, two proteins with molecular masses of about 8.5 and 4.5 kDa were not detected when nonsuppressor cells were infected with sus1(629) mutant phage (8, 50). The 8.5-kDa protein is most probably the product of gene 1. The absence of the other protein may be due to either a polar effect or the presence of a second mutation. Phage φ29 DNA replication was severely affected in sus1(629) phage-infected nonsuppressor cells when these were grown at 37°C (41, 42, 166). However, when the infected cells were grown at 30°C, the rate of sus1(629) DNA synthesis was only slightly lower than in cells infected with wild-type phage (42). These results showed that in vivo DNA synthesis of sus1(629) is affected in a temperature-dependent manner. Protein p1 was found to be associated with the cell membranes, and the 43 C-terminal amino acid residues are required for membrane association (41). In addition, purified protein p1 lacking its 33 N-terminal amino acid residues assembled into long protofilaments that associated in a highly ordered, parallel array forming large two-dimensional sheets (42). Together with other data (41), these results suggested that protein p1 is a component of a virus-encoded membrane-associated structure which would provide an anchoring site for the viral DNA replication machinery (41, 42). The observation that an excess of p1 interferes with the in vitro φ29 DNA replication initiation reaction and that p1 interacts with the TP, as assessed by in vitro cross-linking studies, supports this view (40).
Another function has been attributed to φ29 protein p1 by Take-Uchi et al. (199, 200). They found that larger amounts of φ29 DNA polymerase were produced in nonsuppressor cells infected with sus1(629) than in those infected with wild-type φ29 phage (200). This indicated that protein p1 might down-regulate the synthesis of the DNA polymerase. Experiments performed with E. coli showed that in addition to the DNA polymerase, the synthesis of TP (gene 3) and the transcriptional regulatory protein (gene 4) was diminished by protein p1. Because similar levels of mRNA encoding genes 4-1 were synthesized in the absence or presence of functional gene 1, protein p1 appeared not to affect the transcription of these genes. The observation, however, that protein p1 was able to bind mRNA of genes 1 to 4 suggested that the observed repression might be at the translational level (199).
GA-1 OPERONS CONTAINING OPEN READING FRAMES M-O AND P-T
Surprisingly, a gene 1 homologue is not present in the genome of GA-1. As described above, the GA-1 promoters A1c and A1a are expressed early after infection. Therefore, it is likely that proteins are synthesized from ORFs M to O and P to T. The deduced protein sequences of none of these ORFs showed significant homology to proteins present in available databases. Therefore, the functions of the putative products encoded by these ORFs are unknown. Since these ORFs are not conserved in the genomes of φ29 or B103, it seems unlikely that the putative products will play an essential role. We consider it possible that they play a role in interaction with the infected host. Note that whereas φ29 and B103 are able to infect B. subtilis and various other Bacillus species, GA-1 seems to infect only a specific, still undefined, Bacillus species (see above). Clearly, additional studies are required to attribute a function to these ORFs.
EARLY OPERON LOCATED AT THE RIGHT SIDE OF THE PHAGE GENOMES
All three phages contain an early-expressed operon located at the right side of their genome (Fig. 2). Genes 17 and 16.7, which are the only ones conserved in all three phage genomes, are discussed below. In φ29, this operon also contains genes 16.9 and 16.8, which, together with genes 17 and 16.7, are transcribed from promoter C2. The φ29 ORFs 16.6 and 16.5 may be transcribed from promoters C2 and C1. By Western blot analysis, the products of φ29 genes 16.9 and 16.8 have been detected in infected cells (our unpublished results); their function, however, is still unknown. B103 and GA-1 do not contain homologues of these two genes. ORF 16.5, but not ORF 16.6, is conserved in B103. Neither of these two ORFs is conserved in GA-1.
In B103, gene 17 and ORFs b to d may be transcribed from promoter C2 and gene 16.7 and ORF 16.5 may be transcribed from promoters C2 and C1a. The deduced protein sequences of ORFs b to d do not show significant homology to those of φ29 or GA-1 or those of proteins present in available databases, and their function is unknown.
Compared to those of φ29 and B103, the corresponding operon of GA-1 is considerably larger (Fig. 2). In this case, gene 17 and ORFs B to G may be transcribed from promoter C2 and gene 16.7, ORF H, and ORFs J to L may be transcribed from promoters C2 and C1b. The deduced protein sequences of ORFs B to H and those of ORFs J to L do not have significant similarity to protein sequences present in available databases, and their function is unknown.
Gene 17
The φ29 mutant sus17(112) contains a point mutation that changes the fifth CAA codon of gene 17 into a TAA stop codon (19). Nonsuppressor cells infected with this mutant phage produced a reduced phage yield and viral DNA synthesis (50, 120, 153). In addition, infection with the sus17(112) φ29 mutant in solid media gave only very tiny plaques (138). Crucitti et al. (59) showed that the presence of protein p17 is required for efficient viral DNA synthesis when cells were infected at a low multiplicity of infection but not at high multiplicities of infection. In addition, a moderate stimulatory effect of p17 on in vitro φ29 DNA amplification was observed when the reaction mixtures contained small amounts of template DNA and protein p6. Together, these results indicate that protein p17 stimulates DNA replication under conditions of limiting amounts of template DNA and DNA replication proteins, a situation that is expected to occur at early infection times in cells infected at a low multiplicity.
Phages PZA and φ15 are closely related to φ29 (164). Nevertheless, compared with φ29, the central part of the PZA gene 17 is reorganized and portions of it have been deleted (161). In addition, comparison of genes 17 of φ29 and φ15 showed that the latter contains a deletion of 63 bp near the 3′ end (19). This showed that, in particular, the central and C-terminal parts of genes 17 of φ29, φ15, and PZA have diverged considerably during evolution of these closely related phages. Compared to p17 of φ29, the homologues of B103 and GA-1 are 37 and 79 amino acids smaller, respectively. The C-terminal 43 amino acids of the φ29 protein p17 are absent in both the B103 and GA-1 p17 protein. Moreover, compared to that of φ29, these proteins contain additional deletions clustered in the N-terminal and central parts of the protein. Finally, analysis of the GA-1 p17 protein sequence revealed the presence of a putative transmembrane-spanning domain located in the N-terminal part of the protein (amino acids 15 to 36), suggesting that it may be an integral membrane protein (W. J. J. Meijer, unpublished results). Transmembrane-spanning domains are not predicted for the other p17 proteins. In fact, the p17 protein encoded by φ29 is known to be a soluble protein (59). A comparison of the gene 17 products of φ29, BS32, PZA, φ15, Nf, B103, and GA-1 has been presented previously (163).
Gene 16.7
Computer-assisted analysis of the deduced protein sequence of φ29 p16.7 revealed some interesting data. First, the N-terminal region (amino acids 1 to 22) of p16.7 has a very hydrophobic character and may constitute a transmembrane-spanning domain. Membrane topology predictions indicated that p16.7 may be an integral membrane protein with its N and C regions at the outside and inside of the cell, respectively. Second, the region spanning amino acids 19 to 60 has a high potential to form an α-helical coiled-coil structure, suggesting that this region may function as a protein multimerization domain. Third, the C-terminal part of the p16.7 protein sequence (amino acids 70 to 130) shows some similarity to DNA binding proteins. Thus, these analyses suggested that p16.7 might be an integral dimeric or multimeric DNA binding protein. Over the last decades, data have accumulated showing that prokaryotic DNA replication, including that of phage DNA, occurs at the cell membrane (for reviews, see references 75, 76, 130, and 192). The first evidence that φ29 DNA replication occurs at the membrane of the infected cell was obtained by Ivarie and Pène (118). They recovered parental and replicating φ29 DNA molecules together with membranes in a rapidly sedimenting complex in linear density gradients. Interestingly, the formation of these complexes required an early-expressed viral protein(s) (118). The general view is that the DNA replication factors, probably present in an organized structure, are recruited to one or more sites on the membrane. Despite many studies, our current knowledge about factors involved in bacterial DNA-membrane association is rather poor. As described above, the φ29 protein p1 probably plays a role in this process. The predicted features of protein p16.7 suggested that this protein may also be involved in membrane-associated φ29 DNA replication. We showed that protein p16.7 is abundantly expressed at early times after infection (135). In addition, p16.7 appeared to be a membrane protein and the N-terminally located transmembrane-spanning domain was shown to be required for its membrane localization. A derivative, p16.7A, in which the N-terminal membrane anchor was replaced by a histidine tag, was purified and characterized. Purified p16.7A was shown to form dimers in solution and to have nonspecific affinity for DNA. To study a possible role of p16.7 in in vivo phage DNA replication, a φ29 mutant containing a suppressible mutation in gene 16.7 was constructed. In vivo phage DNA replication was affected in the absence of p16.7, especially at early infection times, supporting the view that p16.7 is involved in in vivo φ29 DNA replication (135). Further insight in the role of p16.7 in in vivo φ29 DNA replication was obtained by comparing the localization of φ29 DNA replication in infected cells by immunofluorescence techniques in the presence or absence of p16.7 protein (134). In wild-type-infected cells, initiation of phage DNA replication was localized to a single focus within the cell, nearly always toward one end of the host cell nucleoid. Already a few minutes later, phage replication had redistributed to multiple sites around the periphery of the nucleoid, just under the cell membrane. These results showed that φ29 DNA replication occurs at various sites at the membrane of the infected cell. As in wild-type-infected cells, initiation of phage DNA replication also localized to a single focus toward one end of the host cell nucleoid in the absence of protein p16.7. However, in this case the redistribution of replicating phage DNA from the initial replication site to various sites surrounding the nucleoid was strongly delayed. Thus, although p16.7 is not essential for in vivo φ29 DNA replication under laboratory conditions, it ensures optimal phage DNA replication by spatially redistributing replicating phage DNA in infected cells.
Gene 16.7 homologues are present in the genomes of both B103 and GA-1. Whereas the deduced p16.7 protein sequences of φ29 and B103 have 68% similarity, the GA-1 p16.7 sequence is 48% similar to those of φ29 and B103. A comparison of the p16.7 protein sequences of φ29, B103 and GA-1 is shown in Fig. 6. Whereas the N-terminal 80 amino acids of the φ29 and B103 p16.7 sequences, encompassing the transmembrane-spanning and coiled-coil domain, have high similarity, this region is far less conserved in p16.7 of GA-1. Nevertheless, this region is predicted to share the same features. Thus, the region spanning amino acids 5 to 27 of GA-1 p16.7 is predicted to constitute a transmembrane-spanning domain and the region encompassing amino acids 33 to 67 is predicted to form a coiled-coil structure, probably involved in protein dimerisation (Meijer, unpublished). The observation that the features of this region are conserved despite considerable deviation at the protein level suggests that they are especially important for the localization and structure of the protein. On the other hand, the predicted DNA binding domain of φ29 and B103 proteins p16.7, located in the C-terminal half of the protein, is rather well conserved in the p16.7 sequence of GA-1 (the 56 C-terminal amino acid residues have 60% similarity). This may further support the view that this region constitutes the functional domain of the p16.7 protein.
FIG. 6.
Comparison of the deduced p16.7 protein sequences encoded by φ29 (group I), B103 (group II), and GA-1 (group III). Amino acid residues are given on the right. Black and grey boxes enclose residues that are conserved in all three or in two of the three sequences, respectively. The following amino acids were considered conservative: L, I, V, M, and A; F, Y, and W; K and R; D and E; Q and N; and S and T. The transmembrane, coiled-coil, and putative DNA binding domain are indicated above the protein sequence. Note that the p16.7 sequences of φ29 and B103 have a high level of homology throughout their entire protein sequences but that only the putative DNA binding domain is conserved in all three p16.7 protein sequences. An alignment of the deduced p16.7 protein sequences from φ29 (group I) and B103 (group II), together with those of the group I phages φ15, PZA, and BS32, has been published previously (135). The percent identities between the deduced p16.7 protein sequence of φ29 and those of φ15, PZA, and BS32 are 91, 89, and 87%, respectively.
LATE OPERON
Gene 8.5, Encoding the Head Fiber Protein
In all three phage genomes, the late genes encoding the structural phage proteins, proteins involved in phage morphogenesis, and proteins required for lysis of the host are present in a single operon located in the center of the phage genome. The levels of similarity between the various homologous proteins are presented in Table 2. A major difference between these operons is that GA-1 does not contain a homologue of φ29 and B103 gene 8.5, encoding the head fiber protein. GA-1 is not the first phage of the group of φ29-related phages known to lack the gene encoding the head fiber protein. Phage M2Y, belonging to the B103 group of φ29 phages, was also reported to lack this gene (220). In fact, apart from a small deletion in the M2Y genome, the restriction maps of M2Y and Nf are identical, suggesting that M2Y may have arisen from Nf by deletion of gene 8.5 (220). These observations show that head fibers are not essential for infection or morphology of phages GA-1 and M2Y. In agreement with that, Salas et al. (183) showed that φ29 phage particles from which the head fibers were released by treatment with 50% dimethyl sulfoxide retained their infectivity. In addition, the plating efficiency of a set of φ29 mutants that were unable to synthesize the head fiber protein was shown to be similar to that of wild-type φ29 phage (170). Together, these results show that the head fibers are not essential for phage morphogenesis or infection. It has been proposed that the head fibers play a role in stabilizing the capsid (201).
Structural Phage Proteins and φ29 Phage Morphogenesis
Prohead formation.
Although phage morphogenesis and DNA packaging have not yet been analyzed for B103 or GA-1, they have been studied extensively for φ29 (for a review, see reference 9), and a highly efficient in vitro φ29 DNA-packaging system has been established (23, 96). Moreover, the three-dimensional structures of the φ29 virion and its empty prohead precursor, obtained by reconstruction of cryoelectron microscopy (cryo-EM) images, have been determined recently (201). Mutant infections or in vitro assembly have been used to study the morphogenesis pathway of φ29. The prolate icosohedral proheads of φ29 consist of 235 copies of the major capsid protein (p8), approximately 180 copies of the scaffolding protein (p7), 55 dimers of the head fiber protein (p8.5), 12 copies of the head-tail connector (p10), 5 or 6 copies of the 174-base pRNA, and 5 or 6 copies of the ATPase protein (p16). The major capsid proteins are organized as hexameric structures at each of the threefold axes and as pentameric structures at each of the fivefold axes. The head fibers attach as dimers to the p8 subunits at quasi-threefold axes that relate one pentamer to a pair of hexamers. The pentameric opening in the prohead, occupied by the structure of the 12 connector molecules, is too small to accommodate insertion of this rigid connector structure after prohead shell assembly. Therefore, it is believed that the connector structure is the origin from which shell assembly is initiated (95). The observation that isometric particles are formed in gene 7 mutant-infected cells suggests an interaction between p7 and connector protein (9). The structure formed by the head-tail connector protein and the pRNA molecules forms an extremely efficient DNA-translocating motor that, together with the aid of the DNA packaging protein p16 and ATP, actively pumps the φ29 DNA into the prohead (for reviews, see references 53, 108, and 213).
DNA translocating/packaging machine.
(i) Connector.
The head-tail connector is a preformed oligomer with 12-fold symmetry (49; for reviews, see references 53 and 213). Its structure has been studied by atomic force microscopy (154), cryo-EM of two-dimensional arrays (212), immunoelectron microscopy (211), and X-ray crystallography (11, 92, 193). In addition, the topology of the connector and other components of the packaging machinery has been studied (112). The 12 connector molecules assemble into a toroidal structure with a total height of 75 Å. A channel runs along the longitudinal axis, which also has a tronco-conical or bottle-neck shape (diameter of about 36 Å at the narrow end, increasing to 60 Å at the wide end). The connector structure can be divided into three main regions: the narrow end, the central part, and the wide end. Whereas the wide end has a 12-fold symmetry, the narrow end has an apparent 6-fold symmetry. The connector is positioned such that only the narrow end protrudes from the portal vertex of the prohead; the wide end of the connector is buried inside the prohead (112, 201, 212). The structure of the connector resembles a propeller. It uses a fitting mechanism that maintains the connector at its place in the prohead vertex but allows rotation with respect to the prohead. As discussed below, encapsidation of φ29 DNA probably involves rotation of the connector structure. It is worth noting that the φ29 connector has a number of important similarities to other toroidal translocases such as several helicases, processivity factors, and exonucleases, suggesting that the mechanism of DNA movement coupled to ATP hydrolysis used by these various multiprotein complexes may be based on similar principles (reviewed in reference 53).
(ii) pRNA ring.
Surprisingly, it was shown that in vivo and in vitro packaging of φ29 DNA required a specific RNA molecule that is encoded by φ29 (93, 94, 97, 205). This RNA molecule, named pRNA, is required for both the translocation and the selection of the φ29 DNA to be packaged (5, 214). The φ29 pRNA is a small molecule of 174 nucleotides, although a C-terminally truncated form of 120 nucleotides has the full activity of the 174-nucleotide form. In contrast to all the other components of the prohead and mature phage particle, the pRNA is not encoded by the late operon but is constitutively expressed from the A1 promoter (see above), which is located in the left part of the genome (Fig. 2). The pRNA, as the connector, has received considerable attention, and major advances in determination of its structure and function have been made during the last few years. Mg2+ induces a conformational change in the pRNA, which leads to its binding to the connector (55). The secondary pRNA structure has a high helical content, containing seven bulges and three loops (12, 57, 204). Recently, it has been shown that intermolecular pairing between bases of two loops, bases 45 to 48 (AACC) and bases 85 to 82 (UUGG), leads to a ring-like structure with a hole in the middle (99, 229). Using functional analyses, evidence has been provided that the active form of the pRNA ring is composed of 6 pRNA molecules (99, 229). Nevertheless, cryo-EM and image reconstruction analyses of φ29 proheads indicate a pentameric pRNA ring (193).
Three-dimensional modeling of the pRNA hexamer showed that it had a diameter similar to that of the accessible end of the connector and that maintenance of these dimensions in binding would result in superposition of the cyclic pRNA hexamer on the connector, forming a double-ring structure (229). Together, this complex forms the essence of the DNA-packaging machine; it plays a key role in recognition of the φ29 DNA and probably is responsible for the specificity of the packaging of φ29 DNA from its left end (22). The view that the cyclic pRNA structure interacts with the accessible end of the connector, made up of amino acids 1 to 94, is experimentally supported by analysis of site-directed mutants with mutations in this region of the connector, protein p10 (66). The RNA binding domain of p10 would include amino acids 21 to 94. In addition, the positively charged N terminus of p10 was shown to be absolutely required for DNA packaging as well as for efficient DNA binding (67).
In addition to that of φ29, the sequence and predicted secondary structure of the GA-1-encoded pRNA have been described (12). An updated version that also includes the sequence and predicted secondary structure of the B103-encoded pRNA has been recently published (57). Despite a very low level of sequence identity among these pRNAs, their predicted secondary structures show a high level of similarity (12, 57). In addition, similar secondary structures were predicted for pRNAs of other φ29-related phages such as PZA (belonging to group I φ29 phages) and M2 and Nf (belonging to group II φ29 phages). In all of these pRNAs, the loops involved in intermolecular pRNA interactions, as demonstrated for the φ29 pRNA (see above), contain complementary sequences. The bases involved are 5′-AACC/UUGG-5′ for φ29, 5′-UAUC/AUAG-5′ for B103, and 5′-CC/GG-5′ for GA-1. The requirement of at least one GC pair was demonstrated for φ29 pRNA (57). The observation that the paired sequences of the two loops of all pRNAs analyzed contain at least one GC pair is therefore, in agreement with the experimental data. In addition, Zhang et al. (229) demonstrated that base pairing of φ29 residues (5′-CC/GG-5′) was sufficient for biological activity. This conclusion may be further supported by the finding that this is the only paired loop sequence present in GA-1 pRNA.
(iii) ATPase protein p16.
Several observations suggest that φ29 protein p16 binds to the pRNA. First, protein p16, as well as the connector protein p10, contains an RNA recognition motif characteristic of a number of RNA-associated proteins (89). Second, binding of p16 to the prohead protects the pRNA from RNase (89). Finally, cryo-EM images of partially DNA filled proheads, but not empty proheads, revealed additional density associated with each of the pRNA molecules. This additional density was attributed to protein p16 (193). Protein p16 of φ29 was shown to possess a DNA- and prohead-dependent ATPase activity (97). After binding of p16 to the prohead, it binds to φ29 DNA, which generates a conformational change in p16 that allows the binding of ATP and its hydrolysis during DNA packaging (98). Later it was shown that φ29 p16 activity depends on pRNA (89). Two typical ATP binding sites were detected in the φ29 p16 sequence (98). A Walker A motif and a Walker B motif are located in the N-terminal (amino acids 24 to 39) and C-terminal (amino acids 248 to 256) part of the φ29 p16 protein, respectively (98). Both motifs are conserved in the p16 proteins encoded by B103 and GA-1 (our unpublished data), strongly suggesting that these conserved motifs are required for ATP hydrolysis, which generate the energy for DNA encapsidation. Five or six copies of φ29 protein p16 are required for φ29 DNA packaging (89, 193). Thus, the very efficient φ29 packaging motor involves a structure that is built from 12 connector molecules, 5 or 6 pRNA molecules, and probably 5 or 6 p16 molecules. Probably, this situation is similar for B103 and GA-1.
Putative mechanism of φ29 DNA packaging.
Several models for phage DNA packaging in general and φ29 DNA packaging in particular have been proposed. These have been recently reviewed by Hendrix (108). Packaging of φ29 DNA is a very energy-consuming process since it takes approximately 1 ATP molecule to package 2 bp of φ29 DNA (98). In an attractive model, whose first version was proposed more than two decades ago (107), it is envisioned that the ATP consumption is used to drive rotation of the connector protein with respect to the rest of the prohead. In particular, the mismatch between the 5-fold symmetry of the prohead and the 12-fold symmetry of the wide end of the connector was rationalized to be crucial, because it would allow rotation of the connector structure by reducing the energy barrier. In addition, the exterior of the connector has no significant regions of charge accumulation, implying an oily, smooth external surface, which would further facilitate its rotation (193). The observation that pRNA causes a fourfold increase in the ATPase activity of protein p16 (89) suggests that the pRNA structure may play a role in the energetics of the DNA translocation. Chen and Guo (56) showed that the six pRNA molecules act sequentially, which favors the rotation hypothesis. These authors envision that the sequential action of the six pRNAs would result in turning of the connector. In this case, the inter-pRNA interactions may serve as a link to pass a signal to adjacent pRNAs. Further support for this is the observation that the pRNAs most probably remain associated with the prohead during the entire packaging process and thus are not needed only for the initiation of DNA packaging (56). In a slightly different version, it is suggested that the prohead, the pRNA ring, and the protein p16 ATPase together act as a stator delivering the energy for rotation of the connector (193). In this latter model, each monomer of the pRNA ring would interact with the head and the orientation of the pRNA would be determined by its interaction with the head rather than with the connector.
Basically, two models have been proposed to explain how rotation of the connector would translocate the phage DNA into the prohead. In the first model it is assumed that supercoiled DNA wraps around the outside of the connector protein and that rotation of the connector allows the DNA to pass into the prohead via the outside of the connector (210). The observation that protein p16 is able to introduce supercoils in TP-containing φ29 DNA after binding to it (90) and predictions of the energy consumption may support this “wrapping” model (56). In the second model, it is assumed that the DNA is translocated through the axial hole of the connector protein (107). Recently, results have been obtained that strongly favor this latter model (193).
Phage maturation.
During φ29 DNA packaging, the prohead becomes more angular and rigid (22), the surface charge decreases, and the scaffolding p7 protein molecules are expelled (201). After packaging, the ATPase protein p16 and the pRNA molecules are also released from the prohead (94) by an unknown mechanism. Next, 6 copies of the lower collar (p11), 3 or 4 copies of the tail knob (p9), and 12 appendages (dimers of protein p12* cleaved from p12 precursor molecules) are assembled sequentially onto a stable, DNA-filled head (45, 79, 120, 168; for a review, see reference 9) (Fig. 1). EM analysis of purified neck crystals without appendages (p12*) and the tail knob (p9) protein revealed a hexagonal array of the necks composed of the connector (p10) and protein p11, which had a hole in the middle. The diameter of the inner, sixfold region of the neck is about 7 nm. In addition, the connector, also called upper collar, and the lower collar (p11) were found to be tightly bound (52, 54). Thus, the lower collar has, as the distal end of the connector, a sixfold symmetry, which may explain, at least in part, the high stability of this neck complex and also may be involved in releasing the less stable bound pRNA from its place. In the absence of the lower collar protein (p11), the tail knob protein (p9) is unable to assemble to the phage particle (45). This shows that first the lower collar and then the tail knob is assembled. Removing the tail knobs from the phage results in release of the encapsidated DNA of the particles, indicating that it functions directly or indirectly as a mechanical stop for DNA exit (45).
Interestingly, although protein p13 is not present in mature phage heads, the number of DNA-containing particles produced in cells infected with gene 13 mutant φ29 phage was very small. Analysis of the empty viral heads produced under these conditions showed that they had characteristics typical of DNA-filled particles. This indicated that the DNA was lost after packaging and therefore suggested that protein p13 somehow interacts with p9 and/or p11 to generate stable mature phage particles (105, 120, 127). Using complementation assays, García et al. (79) demonstrated that protein p13 is required in vivo for the production of functional protein p9. Finally, the appendages (dimers of p12*), which are involved in the adsorption of the phage to bacteria (215), are assembled to complete the phage particle (44, 105, 155, 203). Gene 12 encodes a precursor protein with a calculated molecular mass of 92 kDa. Processing of the precursor results in the active form of the appendage protein, whose monomers have an apparent molecular mass of 75 to 80 kDa (7, 51, 203). The proteolytic processing does not require interaction of the precursor protein with a maturing neck structure (203). So far, the protease responsible for processing and the processing site(s) have not been determined. The sites for assembly of the appendages are most probably created by the interaction of p10 and p11 (52). Final stable assembly of the appendages is obtained only when, in addition to the connector (p10) and the lower-collar (p11) proteins, the tail knob (p9) is also present. Therefore, it was assumed that p9 somehow stabilizes the assembly sites at the connector–lower-collar connection (45).
Lysis cassette.
In the three phage genomes, genes 14 and 15 encode a holin and a peptidoglycan hydrolase, respectively. Both of these proteins are required for efficient lysis of the infected host to release the progeny phages. Lysis is delayed when cells are infected with gene 14 mutant φ29 phages, resulting in larger burst sizes (50). This mutation has no effect on either DNA replication or morphogenesis of the phage, and this mutant phage is often used for in vivo studies, since it allows analysis of phage features at late infection times without interference of lysis. Holins, which are small membrane proteins, introduce pores in the cell membrane, allowing the peptidoglycan hydrolase to exit the cytoplasm and attack the cell wall. Thus, holins control the timing of lysis. Reviews describing structural, functional, and evolutionary aspects of phage-encoded holins and peptidoglycan hydrolases have been published recently (175, 218, 225–227).
(i) Holin-encoding genes of φ29, B103, and GA-1.
Steiner et al. (198) demonstrated that φ29 gene 14 encodes a holin. Analysis of the deduced protein sequence of φ29 gene 14 showed that it contains two or possibly three potential transmembrane domains (198). Based on the number and size of potential transmembrane domains, most holins can be divided in two classes. Whereas class I holins may have two or three transmembrane domains, class II holins are limited to two transmembrane domains and are smaller than the class I holins. Thus, the holin of φ29 belongs to class I. Gene 14 of φ29 contains two potential start codons (position 1 and 3), each with a properly spaced potential ribosomal binding site, which would allow synthesis of two products, of 131 and 129 amino acids. Indeed, this was shown to be the case (202). Many other holin-encoding genes use this so-called dual start motif. Prototypes of holins with a dual start motif are encoded by the S genes of lambda (class I) and phage 21 (class II). In both cases and probably in all other cases with a dual start motif, despite the nearly identical sequences, the two proteins have opposing functions, with the longer product acting as an inhibitor of the shorter product, which is called the lysis effector (references 13 and 88 and references therein). Cooperative action of the inhibitor and effector would result in proper scheduling of cell lysis (“lysis clock”). Major progress in understanding the molecular mechanism underlying regulation of lysis by the dual start motif has been reported recently for the lambda encoded holin (88).
Gene 14 of B103 has similar features to that of φ29, even though the second amino acid residue of the predicted inhibitor species is an Asp instead of a Lys. Analysis of the GA-1 p14 gene shows that it does not contain the typical dual start motif. In fact, alignment of the φ29, B103, and GA-1 holin sequences aligns the Met start residue of the GA-1 holin to the second Met residue of the predicted lysis effector species of φ29 and B103 holins (Fig. 7). The second Met residue in the GA-1 holin is located at position 9 (Fig. 7). However, no potential ribosomal binding site is found at the appropriate location upstream of this second Met codon. These observations suggest that GA-1 would encode only the lysis effector.
FIG. 7.
Alignment of the deduced holin (p14) sequences of φ29 (p14phi29; group I), B103 (p14B103; group II) and GA-1 (p14GA-1; group III). Amino acid residues are given on the right. Black and grey boxes enclose residues that are conserved in all three or in two of the three sequences, respectively. The following amino acids were considered conservative: L, I, V, M, and A; F, Y, and W; K and R; D and E; Q and N; and S and T. Note that the holin sequence of GA-1 lacks the dual start motif.
(ii) Peptidoglycan hydrolase-encoding genes of φ29, B103, and GA-1.
Gene 15 of φ29, B103, and GA-1 encodes a peptidoglycan hydrolase that, at the end of the infection cycle, attacks the cell wall, resulting in lysis of the infected cell and release of the phage progeny. Peptidoglycan hydrolases of tailed phages can be classified into the following groups: muramidases, amidases, peptidases, and transglycosylases (4). The peptidoglycan hydrolases encoded by gene 15 of φ29 and B103 have a high level of homology and both belong to the group of muramidases (83, 163, 174). Independently of their muralytic activity, the peptidoglycan hydrolases of phages T4, T7, lambda, and φ29 have also been named lysozyme. The peptidoglycan hydrolase encoded by gene 15 of GA-1 shows only moderate homology to those of φ29 and B103 (Table 2). Comparison of this protein sequence to those in available databases showed that the GA-1 peptidoglycan hydrolase is most closely related to several Bacillus-encoded autolysins (all belonging to the group of amidases), such as CwlC of B. subtilis, B. lichenoformis, and Bacillus halodurans (about 55% similarity), and the peptidoglycan hydrolase gene of phage SPP1 (data not shown). As described by Ackermann (4), there are many lines of evidence that phage peptidoglycan hydrolases are spread by horizontal gene transfer, e.g., exchange of these genes with their hosts. The strong homology of the GA-1 gene to the Bacillus autolysin genes suggests that GA-1 has “borrowed” its amidase from its host.
CONCLUSIONS AND FUTURE PERSPECTIVES
Here we have described features of the φ29-related phages φ29 (group I), B103 (group II), and GA-1 (group III), the result of more than 30 years of ongoing research. We have attempted to provide a comprehensive overview of DNA replication, regulation of transcription, DNA packaging, phage morphogenesis, and host cell lysis. The fundamental studies, which were focused mainly on φ29, turned out to reveal various molecular mechanisms of general biological processes. One example is the discovery of the interesting dual role of protein p6 in activation of DNA replication and transcriptional regulation.
The recent comparative studies of the DNA polymerase, TP, transcriptional regulator, SSB (p5), and DBP (p6) proteins of φ29, B103/Nf, and GA-1 have shown that whereas the main characteristics of these proteins are conserved, interesting differences are also found. In particular, the observed differences will be important tools in further unraveling the molecular mechanisms by which these proteins act and in understand in more detail the evolutionary adaptation of these proteins with respect to their function for their specific phage. Another important discovery is the involvement of the pRNA in phage DNA packaging. In particular, the finding of a mechanistic role of a joint action of several identical units of RNA is exciting and has widened the activities of RNA molecules. Based on the available knowledge and the effort displayed by various laboratories, it may be expected that the mechanism of phage DNA packaging and the exact role of the pRNA molecules in this process will be resolved in the near future.
The idea that DNA replication is attached to an underlying structure is gaining a wide audience in the scientific community. During the last few years, results have been obtained indicating that replication of eukaryotic DNA, either chromosomal or viral, takes place at specific sites (for reviews, see references 58, 119, and 156). In addition to the DNA polymerases, other necessary replication factors are localized to these sites (156). Together, these results led to the notion that eukaryotic DNA replication takes place in so-called replication factories, each of which contains a relative high concentration of most, if not all, of the DNA replication factors. The replication factories remain at relatively fixed positions, and it is believed that the DNA is threaded through them (58). The static position of the replication factories suggests that they are attached to a substructure. Recently, the concept of a static replication factory has also been demonstrated for the B. subtilis chromosome (129). For prokaryotes it is well known that the membrane is the cellular substructure to which DNA replication is attached. However, very little is known about factors involved in substructure attachment and organization of in vivo DNA replication in replication factories in general. The detailed knowledge of the in vitro mechanism of φ29 DNA replication forms a sound basis for a study of the fundamental process of in vivo DNA replication. In fact, our recent results strongly indicate that at least proteins p16.7 and p1 are involved in this process. A further characterization of these proteins may lead to a more detailed insight in the in vivo organization of DNA replication in general and that of φ29 DNA replication in particular.
Finally, it will be interesting to determine the functions of the putative proteins, encoded by the ORFs that are unique for GA-1. In summary, although the studies performed on these phages have resulted in a wealth of specific and general information on fundamental biological processes, there are still exciting questions to be resolved.
ACKNOWLEDGMENTS
We are indebted to Encarna Madueño for excellent assistance in sequencing.
This investigation was supported by research grants 2R01 GM27242-21 from the National Institutes of Health, PB98-0645 from the Dirección General de Investigación Científica y Técnica, and ERBFMX CT97 0125 from the European Economic Community and an Institutional grant from Fundación Ramón Areces. W.J.J.M. and J.A.H. were supported by postdoctoral fellowships from the Spanish Ministry of Education and Culture and Fundación Raul González-Salas, respectively.
REFERENCES
- 1.Abril A M, Marco S, Carrascosa J L, Salas M, Hermoso J M. Oligomeric structures of the phage φ29 histone-like protein p6. J Mol Biol. 1999;292:581–588. doi: 10.1006/jmbi.1999.3078. [DOI] [PubMed] [Google Scholar]
- 2.Abril A M, Salas M, Andreu J M, Hermoso J M, Rivas G. Phage φ29 protein p6 is in a monomer-dimer equilibrium that shifts to higher association states at the millimolar concentrations found in vivo. Biochemistry. 1997;36:11901–11908. doi: 10.1021/bi970994e. [DOI] [PubMed] [Google Scholar]
- 3.Abril A M, Salas M, Hermoso J M. Identification of residues within two regions involved in self-association of viral histone-like protein p6 from phage φ29. J Biol Chem. 2000;275:26404–26410. doi: 10.1074/jbc.M002739200. [DOI] [PubMed] [Google Scholar]
- 4.Ackermann H-W. Tailed bacteriophages: the order Caudovirales. Adv Virus Res. 1998;51:135–201. doi: 10.1016/S0065-3527(08)60785-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Anderson D L, Bodley J W. Role of RNA in bacteriophage φ29 DNA packaging. J Struct Biol. 1990;104:70–74. doi: 10.1016/1047-8477(90)90059-l. [DOI] [PubMed] [Google Scholar]
- 6.Anderson D L, Hickman D D, Reilly B E. Structure of Bacillus subtilis bacteriophage φ29 and the length of φ29 deoxyribonucleic acid. J Bacteriol. 1966;91:2081–2089. doi: 10.1128/jb.91.5.2081-2089.1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Anderson D L, Reilly B E. Analysis of bacteriophage φ29 gene function: protein synthesis in suppressor-sensitive mutant infection of Bacillus subtilis. J Virol. 1974;13:211–221. doi: 10.1128/jvi.13.1.211-221.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Anderson D L, Reilly B E. Analysis of gene function in Bacillus subtilis bacteriophage φ29. In: Schlessinger D, editor. Microbiology—1976. Washington, D.C.: American Society for Microbiology; 1976. pp. 254–274. [Google Scholar]
- 9.Anderson D L, Reilly B E. Morphogenesis of bacteriophage φ29. In: Sonenshein A L, Hoch J A, Losick R, editors. Bacillus subtilis and other gram-positive bacteria: biochemistry, physiology, and molecular genetics. Washington, D.C.: American Society for Microbiology; 1993. pp. 859–867. [Google Scholar]
- 10.Arwert F, Venema G. Protease-sensitive transfection of Bacillus subtilis with bacteriophage GA-1 DNA: a probable case of heterologous transfection. J Virol. 1974;13:584–589. doi: 10.1128/jvi.13.3.584-589.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Badasso M O, Leiman P G, Tao Y, He Y, Ohlendorf D H, Rossmann M G, Anderson D. Purification, crystallization and initial X-ray analysis of the head-tail connector of bacteriophage φ29. Acta Crystallogr Sar D. 2000;56:1187–1190. doi: 10.1107/s0907444900009239. [DOI] [PubMed] [Google Scholar]
- 12.Bailey S, Wichitwechkarn J, Johnson D, Reilly B E, Anderson D L, Bodley J W. Phylogenetic analysis and secondary structure of the Bacillus subtilis bacteriophage RNA required for DNA packaging. J Biol Chem. 1990;265:22365–22370. [PubMed] [Google Scholar]
- 13.Barenboim M, Chang C-Y, dib Hajj F, Young R. Characterization of the dual start motif of a class II holin gene. Mol Microbiol. 1999;32:715–727. doi: 10.1046/j.1365-2958.1999.01385.x. [DOI] [PubMed] [Google Scholar]
- 14.Barthelemy I, Mellado R P, Salas M. In vitro transcription of bacteriophage φ29 DNA: inhibition of early promoters by the viral replication protein p6. J Virol. 1989;63:460–462. doi: 10.1128/jvi.63.1.460-462.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Barthelemy I, Salas M. Characterization of a new prokaryotic transcriptional activator and its DNA recognition site. J Mol Biol. 1989;208:225–232. doi: 10.1016/0022-2836(89)90384-7. [DOI] [PubMed] [Google Scholar]
- 16.Barthelemy I, Salas M, Mellado R P. In vivo transcription of bacteriophage φ29 DNA: transcription initiation sites. J Virol. 1986;60:874–879. doi: 10.1128/jvi.60.3.874-879.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Barthelemy I, Salas M, Mellado R P. In vivo transcription of bacteriophage φ29 DNA: transcription termination. J Virol. 1987;61:1751–1755. doi: 10.1128/jvi.61.5.1751-1755.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Beese L S, Steitz T A. Structural basis for the 3′-5′ exonuclease activity of Escherichia coli DNA polymerase I: a two metal ion mechanism. EMBO J. 1991;10:25–33. doi: 10.1002/j.1460-2075.1991.tb07917.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Benes V, Arnold L, Smrt J, Paces V. Nucleotide sequence of the right early region of Bacillus phage φ15 and comparison with related phages: reorganization of gene 17 during evolution. Gene. 1989;75:341–347. doi: 10.1016/0378-1119(89)90281-3. [DOI] [PubMed] [Google Scholar]
- 20.Bernad A, Blanco L, Lázaro J M, Martín G, Salas M. A conserved 3′-5′ exonuclease active site in prokaryotic and eukaryotic DNA polymerases. Cell. 1989;59:219–228. doi: 10.1016/0092-8674(89)90883-0. [DOI] [PubMed] [Google Scholar]
- 21.Bernad A, Lázaro J M, Salas M, Blanco L. The highly conserved amino acid sequence Tyr-Gly-Asp-Thr-Asp-Ser in α-like DNA polymerases is required by phage φ29 DNA polymerase for protein-primed initiation and polymerization. Proc Natl Acad Sci USA. 1990;87:4610–4614. doi: 10.1073/pnas.87.12.4610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bjornsti M A, Reilly B E, Anderson D. Morphogenesis of bacteriophage φ29 of Bacillus subtilis: oriented and quantized in vitro packaging of DNA protein gp3. J Virol. 1983;45:383–396. doi: 10.1128/jvi.45.1.383-396.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bjornsti M A, Reilly B E, Anderson D L. In vitro assembly of the Bacillus subtilis bacteriophage φ29. Proc Natl Acad Sci USA. 1981;78:5861–5865. doi: 10.1073/pnas.78.9.5861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Blanco L, Bernad A, Esteban J A, Salas M. DNA-independent deoxynucleotidylation of the φ29 terminal protein by the φ29 DNA polymerase. J Biol Chem. 1992;267:1225–1230. [PubMed] [Google Scholar]
- 25.Blanco L, Bernad A, Lázaro J M, Martín G, Garmendia C, Salas M. Highly efficient DNA synthesis by the phage φ29 DNA polymerase. Symmetrical mode of DNA replication. J Biol Chem. 1989;264:8935–8940. [PubMed] [Google Scholar]
- 26.Blanco L, Gutiérrez J, Lázaro J M, Bernad A, Salas M. Replication of phage φ29 DNA in vitro: role of the viral protein p6 in initiation and elongation. Nucleic Acids Res. 1986;14:4923–4937. doi: 10.1093/nar/14.12.4923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Blanco L, Lázaro J M, De Vega M, Bonnin A, Salas M. Terminal protein-primed DNA amplification. Proc Natl Acad Sci USA. 1994;91:12198–12202. doi: 10.1073/pnas.91.25.12198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Blanco L, Prieto I, Gutiérrez J, Bernad A, Lázaro J M, Hermoso J M, Salas M. Effect of NH4+ ions on φ29 DNA-protein p3 replication: formation of a complex between the terminal protein and the DNA polymerase. J Virol. 1987;61:3983–3991. doi: 10.1128/jvi.61.12.3983-3991.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Blanco L, Salas M. Characterization and purification of a phage φ29-encoded DNA polymerase required for the initiation of replication. Proc Natl Acad Sci USA. 1984;81:5325–5329. doi: 10.1073/pnas.81.17.5325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Blanco L, Salas M. Replication of phage φ29 DNA with purified terminal protein and DNA polymerase: synthesis of full length DNA. Proc Natl Acad Sci USA. 1985;82:6404–6408. doi: 10.1073/pnas.82.19.6404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Blanco L, Salas M. Bacteriophage φ29 DNA polymerase. In: Eckstein F, Lilley D M J, editors. Nucleic acids and molecular biology. Berlin, Germany: Springer-Verlag KG; 1995. pp. 328–341. [Google Scholar]
- 32.Blanco L, Salas M. Relating structure to function in φ29 DNA polymerase. J Biol Chem. 1996;271:8509–8512. doi: 10.1074/jbc.271.15.8509. [DOI] [PubMed] [Google Scholar]
- 33.Blasco M A, Blanco L, Parés E, Salas M, Bernad A. Structural and functional analysis of temperature-sensitive mutants of the phage φ29 DNA polymerase. Nucleic Acids Res. 1990;18:4763–4770. [PMC free article] [PubMed] [Google Scholar]
- 34.Blasco M A, Lázaro J M, Bernad A, Blanco L, Salas M. φ29 DNA polymerase active site: mutants in conserved residues Tyr254 and Tyr390 are affected in dNTP binding. J Biol Chem. 1992;267:19427–19434. [PubMed] [Google Scholar]
- 35.Blasco M A, Lázaro J M, Blanco L, Salas M. φ29 DNA polymerase active site. Residue Asp249 of conserved amino acid motif DX2SLYP is critical for synthetic activities. J Biol Chem. 1993a;268:24106–24113. [PubMed] [Google Scholar]
- 36.Blasco M A, Lázaro J M, Blanco L, Salas M. φ29 DNA polymerase active site. The conserved amino acid motif “Kx3NSxYG” is involved in template-primer binding and dNTP selection. J Biol Chem. 1993b;268:16763–16770. [PubMed] [Google Scholar]
- 37.Blasco M A, Méndez J, Lázaro J M, Blanco L, Salas M. Primer-terminus stabilization at the φ29 DNA polymerase active site: mutational analysis of conserved motif KxY. J Biol Chem. 1995;270:2735–2740. doi: 10.1074/jbc.270.6.2735. [DOI] [PubMed] [Google Scholar]
- 38.Bonnin A, Lázaro J M, Blanco L, Salas M. A single tyrosine prevents insertion of ribonucleotides in the eukaryotic-type φ29 DNA polymerase. J Mol Biol. 1999;290:241–251. doi: 10.1006/jmbi.1999.2900. [DOI] [PubMed] [Google Scholar]
- 39.Bradley D E. The isolation and morphology of some new bacteriophages specific for Bacillus and Acetobacter species. J Gen Microbiol. 1965;41:233–241. doi: 10.1099/00221287-41-2-233. [DOI] [PubMed] [Google Scholar]
- 40.Bravo A, Illana B, Salas M. Compartmentalization of phage φ29 DNA replication: interaction between the primer terminal protein and the membrane-associated protein p1. EMBO J. 2000;19:5575–5584. doi: 10.1093/emboj/19.20.5575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bravo A, Salas M. Initiation of bacteriophage φ29 DNA replication in vivo: assembly of a membrane-associated multiprotein complex. J Mol Biol. 1997;269:102–112. doi: 10.1006/jmbi.1997.1032. [DOI] [PubMed] [Google Scholar]
- 42.Bravo A, Salas M. Polymerization of bacteriophage φ29 replication protein p1 into protofilament sheets. EMBO J. 1998;17:6096–6105. doi: 10.1093/emboj/17.20.6096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Caldentey J, Blanco L, Bamford D H, Salas M. In vitro replication of bacteriophage PRD1 DNA. Characterization of the protein-primed initiation site. Nucleic Acids Res. 1993;21:3725–3730. doi: 10.1093/nar/21.16.3725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Camacho A, Jiménez F, de la Torre J, Carrascosa J L, Mellado R P, Vásquez C, Viñuela E, Salas M. Assembly of Bacillus subtilis phage φ29. I. Mutants in the cistrons coding for the structural proteins. Eur J Biochem. 1977;73:39–55. doi: 10.1111/j.1432-1033.1977.tb11290.x. [DOI] [PubMed] [Google Scholar]
- 45.Camacho A, Jiménez F, Viñuela E, Salas M. Order of assembly of the lower collar and the tail proteins of Bacillus subtilis bacteriophage φ29. J Virol. 1979;29:540–545. doi: 10.1128/jvi.29.2.540-545.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Camacho A, Salas M. Effect of mutations in the “extended −10” motif of three Bacillus subtilis ςA-RNA polymerase-dependent promoters. J Mol Biol. 1999;286:683–693. doi: 10.1006/jmbi.1998.2526. [DOI] [PubMed] [Google Scholar]
- 47.Camacho A, Salas M. Pleiotropic effect of protein p6 on the viral cycle of bacteriophage φ29. J Bacteriol. 2000;182:6927–6932. doi: 10.1128/jb.182.24.6927-6932.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Canceill D, Vigueras E, Ehrlich S D. Replication slippage of different DNA polymerases is inversely related to their strand displacement efficiency. J Biol Chem. 1999;274:27481–27490. doi: 10.1074/jbc.274.39.27481. [DOI] [PubMed] [Google Scholar]
- 49.Carazo J M, Donate L E, Herranz L, Secilla J P, Carrascosa J L. Three-dimensional reconstruction of the connector of bacteriophage φ29 at 1.8 nm resolution. J Mol Biol. 1986;192:853–867. doi: 10.1016/0022-2836(86)90033-1. [DOI] [PubMed] [Google Scholar]
- 50.Carrascosa J L, Camacho A, Moreno F, Jiménez F, Mellado R P, Viñuela E, Salas M. Bacillus subtilis phage φ29; characterization of gene products and functions. Eur J Biochem. 1976;66:229–241. doi: 10.1111/j.1432-1033.1976.tb10512.x. [DOI] [PubMed] [Google Scholar]
- 51.Carrascosa J L, Camacho A, Viñuela E, Salas M. A precursor of the neck appendage protein of B. subtilis phage φ29. FEBS Lett. 1974;44:317–321. [PubMed] [Google Scholar]
- 52.Carrascosa J L, Carazo J M, García N. Structural localization of the proteins of the head to tail connecting region of bacteriophage φ29. Virology. 1983;124:133–144. [PubMed] [Google Scholar]
- 53.Carrascosa J L, Valpuesta J M. Bacteriophage connectors: structural features of a DNA translocating motor. Recent Res Dev Virol. 1999;1:449–465. [Google Scholar]
- 54.Carrascosa J L, Viñuela E, García N, Santisteban A. Structure of the head-tail connector of bacteriophage φ29. J Mol Biol. 1982;154:311–324. doi: 10.1016/0022-2836(82)90066-3. [DOI] [PubMed] [Google Scholar]
- 55.Chen C, Guo P. Mg2+-induced conformational change of packaging RNA for procapsid recognition and binding during phage φ29 DNA encapsidation. J Virol. 1997;71:495–500. doi: 10.1128/jvi.71.1.495-500.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chen C, Guo P. Sequential action of six virus-encoded DNA-packaging RNAs during phage φ29 genomic DNA translocation. J Virol. 1997;71:3864–3871. doi: 10.1128/jvi.71.5.3864-3871.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chen C, Zhang C, Guo P. Sequence requirements for hand-in hand interaction in formation of RNA dimers and hexamers to gear φ29 DNA translocation motor. RNA. 1999;5:805–818. doi: 10.1017/s1355838299990350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cook P R. The organization of replication and transcription. Science. 1999;284:1790–1795. doi: 10.1126/science.284.5421.1790. [DOI] [PubMed] [Google Scholar]
- 59.Crucitti P, Lázaro J M, Benes V, Salas M. Bacteriophage φ29 early protein p17 is conditionally required for the first rounds of viral DNA replication. Gene. 1998;223:135–142. doi: 10.1016/s0378-1119(98)00167-x. [DOI] [PubMed] [Google Scholar]
- 60.Derbyshire V, Pinsonneault J K, Joyce C M. Structure-function analysis of 3′→5′-exonuclease of DNA polymerases. Methods Enzymol. 1995;262:363–385. doi: 10.1016/0076-6879(95)62030-3. [DOI] [PubMed] [Google Scholar]
- 61.De Vega M, Blanco L, Salas M. φ29 DNA polymerase residue Ser122, a single-stranded DNA ligand for 3′-5′ exonucleolysis, is required to interact with the terminal protein. J Biol Chem. 1998;273:28966–28977. doi: 10.1074/jbc.273.44.28966. [DOI] [PubMed] [Google Scholar]
- 62.De Vega M, Blanco L, Salas M. Processive proofreading and the spatial relationship between polymerase and exonuclease active sites of bacteriophage φ29 DNA polymerase. J Mol Biol. 1999;292:39–51. doi: 10.1006/jmbi.1999.3052. [DOI] [PubMed] [Google Scholar]
- 63.De Vega M, Ilyina T, Lázaro J M, Salas M, Blanco L. An invariant lysine residue is involved in catalysis at the 3′-5′ exonuclease active site of eukaryotic-type DNA polymerases. J Mol Biol. 1997;270:65–78. doi: 10.1006/jmbi.1997.1093. [DOI] [PubMed] [Google Scholar]
- 64.De Vega M, Lázaro J M, Salas M, Blanco L. Primer-terminus stabilization at the 3′-5′ exonuclease active site of φ29 DNA polymerase. Involvement of two amino acid residues highly conserved in proofreading DNA polymerases. EMBO J, 1996;15:1182–1192. [PMC free article] [PubMed] [Google Scholar]
- 65.De Vega M, Lázaro J M, Salas M, Blanco L. Mutational analysis of φ29 DNA polymerase residues acting as ssDNA ligands for 3′-5′ exonucleolysis. J Mol Biol. 1998;279:807–822. doi: 10.1006/jmbi.1998.1805. [DOI] [PubMed] [Google Scholar]
- 66.Donate L E, Valpuesta J M, Mier C, Rojo F, Carrascosa J L. Characterization of an RNA-binding domain in the bacteriophage φ29 connector. J Biol Chem. 1993;268:20198–20204. [PubMed] [Google Scholar]
- 67.Donate L E, Valpuesta J M, Rocher A, Mendez E, Rojo F, Salas M, Carrascosa J L. Role of the amino-terminal domain of bacteriophage φ29 connector in DNA binding and packaging. J Biol Chem. 1992;267:10919–10924. [PubMed] [Google Scholar]
- 68.Dufour E, Méndez J, Lázaro J M, De Vega M, Blanco L, Salas M. An aspartic acid residue in TPR-1, a specific region of protein-priming DNA polymerases, is required for the functional interaction with primer terminal protein. J Mol Biol. 2000;304:289–300. doi: 10.1006/jmbi.2000.4216. [DOI] [PubMed] [Google Scholar]
- 69.Elías-Arnanz M, Salas M. Functional interactions between a phage histone-like protein and a transcriptional factor in regulation of φ29 early-late transcriptional switch. Genes Dev. 1999;13:2502–2513. doi: 10.1101/gad.13.19.2502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Escarmís C, Guirao D, Salas M. Replication of recombinant φ29 DNA molecules in Bacillus subtilis protoplasts. Virology. 1989;169:152–160. doi: 10.1016/0042-6822(89)90051-2. [DOI] [PubMed] [Google Scholar]
- 71.Esteban J A, Blanco L, Villar L, Salas M. In vitro evolution of terminal protein-containing genomes. Proc Natl Acad Sci USA. 1997;94:2921–2926. doi: 10.1073/pnas.94.7.2921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Esteban J A, Salas M, Blanco L. Fidelity of φ29 DNA polymerase. Comparison between protein-primed initiation and DNA polymerization. J Biol Chem. 1993;268:2719–2726. [PubMed] [Google Scholar]
- 73.Esteban J A, Soengas M S, Salas M, Blanco L. 3′→5′ exonuclease active site of φ29 DNA polymerase. Evidence favoring a metal ion-assisted reaction mechanism. J Biol Chem. 1994;269:31946–31954. [PubMed] [Google Scholar]
- 74.Fauquet C M. Taxonomy, classification and nomenclature of viruses. In: Granoff A, Webster R G, editors. Encyclopaedia of virology. 2nd ed. London, United Kingdom: Academic Press, Ltd; 1999. pp. 1730–1756. [Google Scholar]
- 75.Firshein W. Role of the DNA/membrane complex in prokaryotic DNA replication. Annu Rev Microbiol. 1989;43:89–120. doi: 10.1146/annurev.mi.43.100189.000513. [DOI] [PubMed] [Google Scholar]
- 76.Firshein W, Kim P. Plasmid replication and partition in Escherichia coli: is the cell membrane the key? Mol Microbiol. 1997;23:1–10. doi: 10.1046/j.1365-2958.1997.2061569.x. [DOI] [PubMed] [Google Scholar]
- 77.Freire R, Salas M, Hermoso J M. A new protein domain for binding to DNA through the minor groove. EMBO J. 1994;13:4353–4360. doi: 10.1002/j.1460-2075.1994.tb06755.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Freire R, Serrano M, Salas M, Hermoso J M. Activation of replication origins in φ29-related phages requires the recognition of initiation proteins to specific nucleoprotein complexes. J Biol Chem. 1996;271:31000–31007. doi: 10.1074/jbc.271.48.31000. [DOI] [PubMed] [Google Scholar]
- 79.García J A, Carrascosa J L, Salas M. Assembly of the tail protein of the Bacillus subtilis phage φ29. Virology. 1983;125:18–30. doi: 10.1016/0042-6822(83)90060-0. [DOI] [PubMed] [Google Scholar]
- 80.García J A, Peñalva M A, Blanco L, Salas M. Template requirements for initiation of phage φ29 DNA replication in vitro. Proc Natl Acad Sci USA. 1984;81:80–84. doi: 10.1073/pnas.81.1.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Garmendia C, Bernad A, Esteban J A, Blanco L, Salas M. The bacteriophage φ29 DNA polymerase, a proofreading enzyme. J Biol Chem. 1992;267:2594–2599. [PubMed] [Google Scholar]
- 82.Garmendia C, Salas M, Hermoso J M. Site-directed mutagenesis in the DNA linking site of bacteriophage φ29 terminal protein: isolation and characterization of a Ser232Thr mutant. Nucleic Acids Res. 1988;16:5727–5740. doi: 10.1093/nar/16.13.5727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Garvey K J, Saedi M S, Ito J. Nucleotide sequence of Bacillus phage φ29 genes 14 and 15: homology of gene 15 with other phage lysozymes. Nucleic Acids Res. 1986;14:10001–10008. doi: 10.1093/nar/14.24.10001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Garvey K J, Yoshikawa H, Ito J. The complete sequence of the Bacillus phage φ29 right early region. Gene. 1985;40:301–309. doi: 10.1016/0378-1119(85)90053-8. [DOI] [PubMed] [Google Scholar]
- 85.Gascón I, Gutiérrez C, Salas M. Structural and functional comparative study of the complexes formed by viral φ29, Nf and GA-1 SSB proteins with DNA. J Mol Biol. 2000;296:989–999. doi: 10.1006/jmbi.2000.3521. [DOI] [PubMed] [Google Scholar]
- 86.Gascón I, Lázaro J M, Salas M. Differential functional behavior of viral φ29, Nf, and GA-1 SSB proteins. Nucleic Acids Res. 2000;28:2034–2042. doi: 10.1093/nar/28.10.2034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.González-Huici V, Lázaro J M, Salas M, Hermoso J M. Specific recognition of parental terminal protein by DNA polymerase for initiation of protein-primed DNA replication. J Biol Chem. 2000;275:14678–14683. doi: 10.1074/jbc.m910058199. [DOI] [PubMed] [Google Scholar]
- 88.Graschopf A, Bläsi U. Molecular function of the dual-start motif in the λ S holin. Mol Microbiol. 1999;33:569–582. doi: 10.1046/j.1365-2958.1999.01501.x. [DOI] [PubMed] [Google Scholar]
- 89.Grimes S, Anderson D. RNA dependence of the bacteriophage φ29 DNA packaging ATPase. J Mol Biol. 1990;215:559–566. doi: 10.1016/s0022-2836(05)80168-8. [DOI] [PubMed] [Google Scholar]
- 90.Grimes S, Anderson D. The bacteriophage φ29 packaging proteins supercoil the DNA ends. J Mol Biol. 1997;266:901–914. doi: 10.1006/jmbi.1996.0843. [DOI] [PubMed] [Google Scholar]
- 91.Grunow R, Schonherr M, Taubeneck U, Zimmermann I. Zur Struktur von Bakteriophagen aus Enzym-produzierenden Bacillus Kulturen. Acta Biotechnol. 1981;1:17–19. [Google Scholar]
- 92.Guasch A, Pous J, Párraga A, Valpuesta J M, Carrascosa J L, Coll M. Crystallographic analysis reveals the 12-fold symmetry of the bacteriophage φ29 connector particles. J Mol Biol. 1998;281:219–225. doi: 10.1006/jmbi.1998.1928. [DOI] [PubMed] [Google Scholar]
- 93.Guo P, Bailey S, Bodley J W, Anderson D. Characterization of the small RNA of the bacteriophage φ29 DNA packaging machine. Nucleic Acids Res. 1987;15:7081–7090. doi: 10.1093/nar/15.17.7081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Guo P, Erickson S, Anderson D. A small viral RNA is required for in vitro packaging of bacteriophage φ29 DNA. Science. 1987;236:690–694. doi: 10.1126/science.3107124. [DOI] [PubMed] [Google Scholar]
- 95.Guo P, Erickson S, Xu W, Olson N, Baker T S, Anderson D. Regulation of the phage φ29 prohead shape and size by the portal vertex. Virology. 1991;183:366–373. doi: 10.1016/0042-6822(91)90149-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Guo P, Grimes S, Anderson D. A defined system for in vitro packaging of DNA-gp3 of the Bacillus subtilis bacteriophage φ29. Proc Natl Acad Sci USA. 1986;83:3505–3509. doi: 10.1073/pnas.83.10.3505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Guo P, Peterson C, Anderson D. Initiation events in in vitro packaging of bacteriophage φ29 DNA-p3. J Mol Biol. 1987;197:219–228. doi: 10.1016/0022-2836(87)90120-3. [DOI] [PubMed] [Google Scholar]
- 98.Guo P, Peterson C, Anderson D. Prohead and DNA-gp3-dependent ATPase activity of the DNA packaging protein gp16 of bacteriophage φ29. J Mol Biol. 1987;197:229–236. doi: 10.1016/0022-2836(87)90121-5. [DOI] [PubMed] [Google Scholar]
- 99.Guo P, Zhang C, Chen C, Garver K, Trottier M. Inter-RNA interaction of phage φ29 pRNA to form a hexameric complex for viral DNA transportation. Mol Cell. 1998;2:149–155. doi: 10.1016/s1097-2765(00)80124-0. [DOI] [PubMed] [Google Scholar]
- 100.Gutiérrez C, Freire R, Salas M, Hermoso J M. Assembly of phage φ29 genome with viral protein p6 into a compact complex. EMBO J. 1994;13:269–276. doi: 10.1002/j.1460-2075.1994.tb06257.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Gutiérrez C, Martín G, Sogo J M, Salas M. Mechanism of stimulation of DNA replication by bacteriophage φ29 single-stranded DNA-binding protein p5. J Biol Chem. 1991;266:2104–2111. [PubMed] [Google Scholar]
- 102.Gutiérrez C, Sogo J M, Salas M. Analysis of replicative intermediates produced during bacteriophage φ29 DNA replication in vitro. J Mol Biol. 1991;222:983–994. doi: 10.1016/0022-2836(91)90589-x. [DOI] [PubMed] [Google Scholar]
- 103.Gutiérrez J, García J A, Blanco L, Salas M. Cloning and template activity of the origins of replication of phage φ29 DNA. Gene. 1986;43:1–11. doi: 10.1016/0378-1119(86)90002-8. [DOI] [PubMed] [Google Scholar]
- 104.Gutiérrez J, Vinós J, Prieto I, Méndez E, Hermoso J M, Salas M. Signals in the φ29 DNA-terminal protein template for the initiation of phage φ29 DNA replication. Virology. 1986;155:474–483. doi: 10.1016/0042-6822(86)90209-6. [DOI] [PubMed] [Google Scholar]
- 105.Hagen E W, Reilly B E, Tosi M E, Anderson D L. Analysis of gene function of bacteriophage φ29 of Bacillus subtilis: identification of cistrons essential for viral assembly. J Virol. 1976;19:501–517. doi: 10.1128/jvi.19.2.501-517.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Helmann J D. Compilation and analysis of Bacillus subtilis ςA-dependent promoter sequences: evidence for extended contact between RNA polymerase and upstream promoter DNA. Nucleic Acids Res. 1995;23:2351–2360. doi: 10.1093/nar/23.13.2351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Hendrix R W. Symmetry mismatch and DNA packaging in large bacteriophages. Proc Natl Acad Sci USA. 1978;75:4779–4783. doi: 10.1073/pnas.75.10.4779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Hendrix R W. Bacteriophage DNA packaging: RNA gears in a DNA transport machine. Cell. 1998;94:147–150. doi: 10.1016/s0092-8674(00)81413-0. [DOI] [PubMed] [Google Scholar]
- 109.Hermoso J M, Méndez E, Soriano F, Salas M. Location of the serine residue involved in the linkage between the terminal protein and the DNA of φ29. Nucleic Acids Res. 1985;13:7715–7728. doi: 10.1093/nar/13.21.7715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Holder R D, Whiteley H R. In vitro synthesis of late bacteriophage φ29 RNA. J Virol. 1983;46:690–702. doi: 10.1128/jvi.46.3.690-702.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Horcajadas J A, Monsalve M, Rojo F, Salas M. The switch from early to late transcription in phage GA-1: characterization of the regulatory protein p4G. J Mol Biol. 1999;290:917–928. doi: 10.1006/jmbi.1999.2932. [DOI] [PubMed] [Google Scholar]
- 112.Ibarra B, Carrascosa J L, Llorca O, Valle M, Valpuesta J M. Topology of the components of the DNA packaging machinery in the phage φ29 prohead. J Mol Biol. 2000;298:807–815. doi: 10.1006/jmbi.2000.3712. [DOI] [PubMed] [Google Scholar]
- 113.Ikeda Y, Saito H, Miura K, Takagi J, Aoki H. DNA base composition, susceptibility to bacteriophages, and interspecific transformation as criteria for classification in the genus Bacillus. J Gen Appl Microbiol. 1965;11:181–190. [PubMed] [Google Scholar]
- 114.Illana B, Blanco L, Salas M. Functional characterization of the genes coding for the terminal protein and DNA polymerase from bacteriophage GA-1. Evidence for a sliding-back mechanism during protein-primed GA-1 DNA replication. J Mol Biol. 1996;264:453–464. doi: 10.1006/jmbi.1996.0653. [DOI] [PubMed] [Google Scholar]
- 115.Illana B, Lázaro J M, Gutiérrez C, Meijer W J J, Blanco L, Salas M. Phage φ29 terminal protein residues Asn80 and Tyr82 are recognition elements of the replication origins. J Biol Chem. 1999;274:15073–15079. doi: 10.1074/jbc.274.21.15073. [DOI] [PubMed] [Google Scholar]
- 116.Illana B, Zaballos A, Blanco L, Salas M. The RGD sequence in phage φ29 terminal protein is required for interaction with φ29 DNA polymerase. Virology. 1998;248:12–19. doi: 10.1006/viro.1998.9276. [DOI] [PubMed] [Google Scholar]
- 117.Inciarte M R, Salas M, Sogo J M. The structure of replicating DNA molecules of Bacillus subtilis phage φ29. J Virol. 1980;34:187–190. doi: 10.1128/jvi.34.1.187-199.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Ivarie R D, Pène J J. DNA replication in bacteriophage φ29: the requirement of a viral-specific product for association of φ29 DNA with the cell membrane of Bacillus amyloliquefaciens. Virology. 1973;52:351–362. doi: 10.1016/0042-6822(73)90330-9. [DOI] [PubMed] [Google Scholar]
- 119.Jackson D A, Cooper C S. The structural basis of nuclear function. Int Rev Cytol. 1995;162:125–149. doi: 10.1016/s0074-7696(08)61230-9. [DOI] [PubMed] [Google Scholar]
- 120.Jiménez F, Camacho A, de la Torre J, Viñuela E, Salas M. Assembly of Bacillus subtilis phage φ29. 2. Mutants in the cistrons coding for the non-structural proteins. Eur J Biochem. 1977;73:57–72. doi: 10.1111/j.1432-1033.1977.tb11291.x. [DOI] [PubMed] [Google Scholar]
- 121.Joyce C M, Steitz T A. Function and structure relationships in DNA polymerases. Annu Rev Biochem. 1994;63:777–822. doi: 10.1146/annurev.bi.63.070194.004021. [DOI] [PubMed] [Google Scholar]
- 122.Kawamura F, Ito J. Transcription of the genome of bacteriophage φ29: isolation and mapping of the major early mRNA synthesized in vivo and in vitro. J Virol. 1977;23:562–577. doi: 10.1128/jvi.23.3.562-577.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Keilty S, Rosenberg M. Constitutive function of a positively regulated promoter reveals new sequences essential for activity. J Biol Chem. 1987;262:6389–6395. [PubMed] [Google Scholar]
- 124.King A J, Teertstra W R, van der Vliet P C. Dissociation of the protein primer and DNA polymerase after initiation of adenovirus DNA replication. J Biol Chem. 1997;272:24617–24623. doi: 10.1074/jbc.272.39.24617. [DOI] [PubMed] [Google Scholar]
- 125.King A J, van der Vliet P C. A precursor terminal protein-trinucleotide intermediate during initiation of adenovirus DNA replication: regeneration of molecular ends in vitro by a jumping back mechanism. EMBO J. 1994;13:5786–5792. doi: 10.1002/j.1460-2075.1994.tb06917.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Kornberg A, Baker T A. DNA replication. W. H. New York, N.Y: Freeman & Co.; 1992. [Google Scholar]
- 127.Krawiec S, Jiménez F, García J A, Villanueva N, Sogo J M, Salas M. The orderly, in vitro emergence of DNA from bacteriophage φ29 particles. Virology. 1981;111:440–454. doi: 10.1016/0042-6822(81)90347-0. [DOI] [PubMed] [Google Scholar]
- 128.Kumar A, Malloch R A, Fujita N, Smillie D A, Ishihama A, Hayward R S. The minus 35 recognition region of Escherichia coli sigma 70 is inessential for initiation of transcription at the “extended minus 10” promoters. J Mol Biol. 1993;232:406–418. doi: 10.1006/jmbi.1993.1400. [DOI] [PubMed] [Google Scholar]
- 129.Lemon K P, Grossman A D. Localization of bacterial DNA polymerase: evidence for a factory model of replication. Science. 1998;282:1516–1519. doi: 10.1126/science.282.5393.1516. [DOI] [PubMed] [Google Scholar]
- 130.Liebowitz P J, Schaechter M. The attachment of the bacterial chromosome to the membrane. Int Rev Cytol. 1975;41:1–28. doi: 10.1016/s0074-7696(08)60964-x. [DOI] [PubMed] [Google Scholar]
- 131.Maniloff J, Ackermann H-W, Jarvis A. Phage taxonomy and classification. In: Granoff A, Webster R G, editors. Encyclopaedia of virology. 2nd ed. London, United Kingdom: Academic Press, Ltd.; 1999. pp. 1221–1228. [Google Scholar]
- 132.Martín A C, Blanco L, García P, Salas M, Méndez J. In vitro initiation of pneumococcal phage Cp-1 DNA replication occurs at the third 3′ nucleotide of the linear template: a stepwise sliding-back mechanism. J Mol Biol. 1996;260:369–377. doi: 10.1006/jmbi.1996.0407. [DOI] [PubMed] [Google Scholar]
- 133.Martín G, Lázaro J M, Méndez E, Salas M. Characterization of the phage φ29 protein p5 as a single-stranded DNA binding protein. Function in φ29 DNA-protein p3 replication. Nucleic Acids Res. 1989;17:3663–3672. doi: 10.1093/nar/17.10.3663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Meijer W J J, Lewis P J, Errington J, Salas M. Dynamic relocalization of phage φ29 DNA during replication and the role of the viral protein p16.7. EMBO J. 2000;19:4182–4190. doi: 10.1093/emboj/19.15.4182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Meijer W J J, Serna-Rico A, Salas M. Characterization of the bacteriophage φ29-encoded protein p16.7: a membrane protein involved in phage DNA replication. Mol Microbiol. 2001;39:731–746. doi: 10.1046/j.1365-2958.2001.02260.x. [DOI] [PubMed] [Google Scholar]
- 136.Mellado R P, Barthelemy I, Salas M. In vitro transcription of bacteriophage φ29 DNA. Correlation between in vitro and in vivo promoters. Nucleic Acids Res. 1986a;14:4731–4741. doi: 10.1093/nar/14.12.4731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Mellado R P, Barthelemy I, Salas M. In vivo transcription of bacteriophage φ29 DNA early and late promoter sequences. J Mol Biol. 1986b;191:191–197. doi: 10.1016/0022-2836(86)90256-1. [DOI] [PubMed] [Google Scholar]
- 138.Mellado R P, Moreno F, Viñuela E, Salas M, Reilly B E, Anderson D L. Genetic analysis of bacteriophage φ29 of Bacillus subtilis: integration and mapping of reference mutants of two collections. J Virol. 1976;19:495–500. doi: 10.1128/jvi.19.2.495-500.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Mellado R P, Peñalva M A, Inciarte M R, Salas M. The protein covalently linked to the 5′ termini of the DNA of Bacillus subtilis phage φ29 is involved in the initiation of DNA replication. Virology. 1980;104:84–96. doi: 10.1016/0042-6822(80)90367-0. [DOI] [PubMed] [Google Scholar]
- 140.Mencía M, Monsalve M, Rojo F, Salas M. Transcription activation by phage φ29 protein p4 is mediated by interaction with the α subunit of Bacillus subtilis RNA polymerase. Proc Natl Acad Sci USA. 1996;93:6616–6620. doi: 10.1073/pnas.93.13.6616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Mencía M, Monsalve M, Rojo F, Salas M. Substitution of the C-terminal domain of the Escherichia coli RNA polymerase α subunit by that from Bacillus subtilis makes the enzyme responsive to a Bacillus subtilis transcriptional activator. J Mol Biol. 1998;275:177–185. doi: 10.1006/jmbi.1997.1463. [DOI] [PubMed] [Google Scholar]
- 142.Mencía M, Monsalve M, Salas M, Rojo F. Transcriptional activator of phage φ29 late promoter: mapping of residues involved in interaction with RNA polymerase and in DNA bending. Mol Microbiol. 1996;20:273–282. doi: 10.1111/j.1365-2958.1996.tb02616.x. [DOI] [PubMed] [Google Scholar]
- 143.Mencía M, Salas M, Rojo F. Residues of the Bacillus subtilis phage φ29 transcriptional activator required both to interact with RNA polymerase and to activate transcription. J Mol Biol. 1993;233:695–704. doi: 10.1006/jmbi.1993.1546. [DOI] [PubMed] [Google Scholar]
- 144.Méndez J, Blanco L, Esteban J A, Bernad A, Salas M. Initiation of φ29 DNA replication occurs at the second 3′ nucleotide of the linear template: a sliding-back mechanism for protein-primed DNA replication. Proc Natl Acad Sci USA. 1992;89:9579–9583. doi: 10.1073/pnas.89.20.9579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Méndez J, Blanco L, Lázaro J M, Salas M. Primer-terminus stabilization at the φ29 DNA polymerase active site. Mutational analysis of conserved motif TX2GR. J Biol Chem. 1994;269:30030–30038. [PubMed] [Google Scholar]
- 146.Méndez J, Blanco L, Salas M. Protein-primed DNA replication: a transition between two modes of priming by a unique DNA polymerase. EMBO J. 1997;16:2519–2527. doi: 10.1093/emboj/16.9.2519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Miyagi T. Studies on phage structures coded by bacteriophage M2. Ph.D. thesis. Tokyo, Japan: Graduate School of Biological Science. Sophia University; 1985. [Google Scholar]
- 148.Monsalve M, Calles B, Mencía M, Salas M, Rojo F. Transcription activation or repression by phage φ29 protein p4 depends on the strength of the RNA polymerase-promoter interactions. Mol Cell. 1997;1:99–107. doi: 10.1016/s1097-2765(00)80011-8. [DOI] [PubMed] [Google Scholar]
- 149.Monsalve M, Mencía M, Rojo F, Salas M. Transcription regulation in Bacillus subtilis phage φ29: expression of the viral promoters throughout the infection cycle. Virology. 1995;207:23–31. doi: 10.1006/viro.1995.1048. [DOI] [PubMed] [Google Scholar]
- 150.Monsalve M, Mencía M, Rojo F, Salas M. Activation and repression of transcription at two different phage φ29 promoters are mediated by interaction of the same residues of regulatory protein p4 with RNA polymerase. EMBO J. 1996;15:383–391. [PMC free article] [PubMed] [Google Scholar]
- 151.Monsalve M, Mencía M, Salas M, Rojo F. Protein p4 represses phage φ29 A2c promoter by interacting with the α subunit of Bacillus subtilis RNA polymerase. Proc Natl Acad Sci USA. 1996;93:8913–8918. doi: 10.1073/pnas.93.17.8913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Moran C P, Lang N, LeGrice S F J, Lee G, Stephans M, Sonenshein A L, Pero J, Losick R. Nucleotide sequence that signal the initiation of transcription and translation in Bacillus subtilis. Mol Gen Genet. 1982;186:339–346. doi: 10.1007/BF00729452. [DOI] [PubMed] [Google Scholar]
- 153.Moreno F, Camacho A, Viñuela E, Salas M. Suppressor-sensitive mutants and genetic map of Bacillus subtilis bacteriophage φ29. Virology. 1974;62:1–16. doi: 10.1016/0042-6822(74)90298-0. [DOI] [PubMed] [Google Scholar]
- 154.Müller D J, Engel A, Carrascosa J L, Vélez M. The bacteriophage φ29 head-tail connector imaged at high resolution with the atomic force microscope in buffer solution. EMBO J. 1997;16:2547–2553. doi: 10.1093/emboj/16.10.2547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Nelson R A, Reilly B E, Anderson D L. Morphogenesis of bacteriophage φ29 of Bacillus subtilis: preliminary isolation and characterization of intermediate particles of the assembly pathway. J Virol. 1976;19:518–532. doi: 10.1128/jvi.19.2.518-532.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Newport J, Yan H. Organization of DNA into foci during replication. Curr Opin Cell Biol. 1996;8:365–368. doi: 10.1016/s0955-0674(96)80011-1. [DOI] [PubMed] [Google Scholar]
- 157.Nuez B, Rojo F, Salas M. Phage φ29 regulatory protein p4 stabilizes the binding of the RNA polymerase to the late promoter in a process involving direct protein-protein contacts. Proc Natl Acad Sci USA. 1992;89:11401–11405. doi: 10.1073/pnas.89.23.11401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Nuez B, Salas M. Bacteriophage Nf DNA region controlling late transcription: structural and functional homology with bacteriophage φ29. Nucleic Acids Res. 1993;21:2861–2865. doi: 10.1093/nar/21.12.2861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Ortín J, Viñuela E, Salas M, Vásquez C. DNA-protein complex in circular DNA from phage φ29. Nature (London) New Biol. 1971;234:275–277. doi: 10.1038/newbio234275a0. [DOI] [PubMed] [Google Scholar]
- 160.Otero M J, Lázaro J M, Salas M. Deletions at the N-terminus of bacteriophage φ29 protein p6: DNA binding and activity in φ 29 DNA replication. Gene. 1990;95:25–30. doi: 10.1016/0378-1119(90)90409-k. [DOI] [PubMed] [Google Scholar]
- 161.Paces V, Vlcek C, Urbanek P, Hostomsky Z. Nucleotide sequence of the right early region of Bacillus subtilis phage PZA completes the 19366-bp sequence of PZA genome. Comparison with the homologous sequence of phage φ29. Gene. 1986;44:115–120. doi: 10.1016/0378-1119(86)90049-1. [DOI] [PubMed] [Google Scholar]
- 162.Pastrana R, Lázaro J M, Blanco L, Garcia J A, Méndez E, Salas M. Overproduction and purification of protein p6 of Bacillus subtilis phage φ29: role in the initiation of DNA replication. Nucleic Acids Res. 1985;13:3083–3100. doi: 10.1093/nar/13.9.3083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Pecenkova T, Benes V, Paces J, Vlcek C, Paces V. Bacteriophage B103: complete DNA sequence of its genome and relationship to other Bacillus phages. Gene. 1997;199:157–163. doi: 10.1016/s0378-1119(97)00363-6. [DOI] [PubMed] [Google Scholar]
- 164.Pecenkova T, Paces V. Molecular phylogeny of φ29-like phages and their evolutionary relatedness to other protein-primed replicating phages and other phages hosted by Gram-positive bacteria. J Mol Evol. 1999;48:197–208. doi: 10.1007/pl00006458. [DOI] [PubMed] [Google Scholar]
- 165.Ponnambalan S, Webster C, Bingham A, Busby S. Transcription initiation at the Escherichia coli galactose operon promoters in the absence of the normal −35 region. J Biol Chem. 1986;261:16043–16048. [PubMed] [Google Scholar]
- 166.Prieto I, Méndez E, Salas M. Characterization, overproduction and purification of the product of gene 1 of Bacillus subtilis phage φ29. Gene. 1989;77:195–204. doi: 10.1016/0378-1119(89)90067-x. [DOI] [PubMed] [Google Scholar]
- 167.Prieto I, Serrano M, Lázaro J M, Salas M, Hermoso J M. Interaction of the bacteriophage φ29 protein p6 with double-stranded DNA. Proc Natl Acad Sci USA. 1988;85:314–318. doi: 10.1073/pnas.85.2.314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Ramirez G, Méndez E, Salas M, Viñuela E. Head-neck connecting protein in phage φ29. Virology. 1972;48:263–265. doi: 10.1016/0042-6822(72)90134-1. [DOI] [PubMed] [Google Scholar]
- 169.Reilly B E. A study of the bacteriophages of Bacillus subtilis and their infectious nucleic acids. Ph.D. thesis. Detroit, Mich: Case Western Reserve University; 1965. [Google Scholar]
- 170.Reilly B E, Nelson R A, Anderson D L. Morphogenesis of bacteriophage φ29 of Bacillus subtilis: mapping and functional analysis of the head fiber gene. J Virol. 1977;24:363–377. doi: 10.1128/jvi.24.1.363-377.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Rojo F, Mencía M, Monsalve M, Salas M. Transcription activation and repression by interaction of a regulator with the α subunit of RNA polymerase: the model of phage φ29 protein p4. Prog Nucleic Acid Res Mol Biol. 1998;60:29–46. doi: 10.1016/s0079-6603(08)60888-0. [DOI] [PubMed] [Google Scholar]
- 172.Rojo F, Salas M. A DNA curvature can substitute phage φ29 regulatory protein p4 when acting as a transcriptional repressor. EMBO J. 1991;10:3429–3438. doi: 10.1002/j.1460-2075.1991.tb04907.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Rojo F, Zaballos A, Salas M. Bend induced by phage φ29 transcriptional activator in the viral late promoter is required for activation. J Mol Biol. 1990;211:713–725. doi: 10.1016/0022-2836(90)90072-t. [DOI] [PubMed] [Google Scholar]
- 174.Saedi M S, Garvey K J, Ito J. Cloning and purification of a unique lysozyme produced by Bacillus phage φ29. Proc Natl Acad Sci USA. 1987;84:955–958. doi: 10.1073/pnas.84.4.955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Saier M H. Families of proteins forming transmembrane channels. J Membr Biol. 2000;175:165–180. doi: 10.1007/s00232001065. [DOI] [PubMed] [Google Scholar]
- 176.Salas M. Protein-priming of DNA replication. Annu Rev Biochem. 1991;60:39–71. doi: 10.1146/annurev.bi.60.070191.000351. [DOI] [PubMed] [Google Scholar]
- 177.Salas M. Bacillus phage φ29. In: Webster R G, Granoff A, editors. Encyclopaedia of virology. 2nd ed. London, United Kingdom: Academic Press, Ltd.; 1999. pp. 119–130. [Google Scholar]
- 178.Salas M. Mechanism of initiation of linear DNA replication in prokaryotes. Genet Eng. 1999;21:159–171. doi: 10.1007/978-1-4615-4707-5_8. [DOI] [PubMed] [Google Scholar]
- 179.Salas M, Mellado R P, Viñuela E, Sogo J M. Characterization of a protein covalently linked to the 5′ termini of the DNA of Bacillus subtilis phage φ29. J Mol Biol. 1978;119:269–291. doi: 10.1016/0022-2836(78)90438-2. [DOI] [PubMed] [Google Scholar]
- 180.Salas M, Méndez J, Esteban J A, Serrano M, Gutiérrez C, Hermoso J M, Bravo A, Soengas M S, Lázaro J M, Blasco M A, Freire R, Bernad A, Sogo J M, Blanco L. Terminal protein priming of DNA replication: bacteriophage φ29 as a model system. In: Doerfler W, Böhm P, editors. Virus strategies. Molecular biology and pathogenesis. Weinheim, Germany: VCH; 1993. pp. 3–19. [Google Scholar]
- 181.Salas M, Miller J T, Leis J, DePamphilis M L. Mechanisms for priming DNA synthesis. In: DePamphilis M L, editor. DNA replication in eukaryotic cells. New York, N.Y: Cold Spring Harbor Laboratory Press; 1996. pp. 131–176. [Google Scholar]
- 182.Salas M, Rojo F. Replication and transcription of bacteriophage φ29. In: Sonenshein A L, Hoch J A, Losick R, editors. Bacillus subtilis and other gram-positive bacteria: biochemistry, physiology, and molecular genetics. Washington, D.C.: American Society for Microbiology; 1993. pp. 843–858. [Google Scholar]
- 183.Salas M, Vásquez C, Méndez E, Viñuela E. Head fibers of bacteriophage φ29. Virology. 1972;50:180–188. doi: 10.1016/0042-6822(72)90358-3. [DOI] [PubMed] [Google Scholar]
- 184.Saturno J, Blanco L, Salas M, Esteban J A. A novel kinetic analysis to calculate nucleotide affinity of proofreading DNA polymerases. Application to φ29 DNA polymerase fidelity mutants. J Biol Chem. 1995;270:31235–31243. doi: 10.1074/jbc.270.52.31235. [DOI] [PubMed] [Google Scholar]
- 185.Saturno J, Lázaro J M, Blanco L, Salas M. Role of the first aspartate residue of the “YxDTDS” motif of φ29 DNA polymerase as a metal ligand during both TP-primed and DNA-primed DNA synthesis. J Mol Biol. 1998;283:633–642. doi: 10.1006/jmbi.1998.2121. [DOI] [PubMed] [Google Scholar]
- 186.Serna-Rico A, Illana B, Salas M, Meijer W J J. The putative coiled coil domain of the φ29 terminal protein is a major determinant involved in recognition of the origin of replication. J Biol Chem. 2000;275:40529–40538. doi: 10.1074/jbc.M007855200. [DOI] [PubMed] [Google Scholar]
- 187.Serrano M, Gutiérrez C, Freire R, Bravo A, Salas M, Hermoso J M. Phage φ29 protein p6: a viral histone-like protein. Biochimie. 1994;76:981–991. doi: 10.1016/0300-9084(94)90023-x. [DOI] [PubMed] [Google Scholar]
- 188.Serrano M, Gutiérrez C, Salas M, Hermoso J M. Superhelical path of the DNA in the nucleoprotein complex that activates the initiation of phage φ29 DNA replication. J Mol Biol. 1993;230:248–259. doi: 10.1006/jmbi.1993.1140. [DOI] [PubMed] [Google Scholar]
- 189.Serrano M, Gutiérrez J, Prieto I, Hermoso J M, Salas M. Signals at the bacteriophage φ29 DNA replication origins required for protein p6 binding and activity. EMBO J. 1989;8:1879–1885. doi: 10.1002/j.1460-2075.1989.tb03584.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Serrano M, Salas M, Hermoso J M. A novel nucleoprotein complex at a replication origin. Science. 1990;248:1012–1016. doi: 10.1126/science.2111580. [DOI] [PubMed] [Google Scholar]
- 191.Shimizu N, Miura K, Aoki H. Characterization of Bacillus subtilis bacteriophage. II. Isolation and morphology of phage Nf and properties of its DNA. J Biochem Tokyo. 1970;68:277–286. doi: 10.1093/oxfordjournals.jbchem.a129357. [DOI] [PubMed] [Google Scholar]
- 192.Siegel P J, Schaechter M. The role of the host cell membrane in the replication and morphogenesis of bacteriophages. Annu Rev Microbiol. 1973;27:261–281. doi: 10.1146/annurev.mi.27.100173.001401. [DOI] [PubMed] [Google Scholar]
- 193.Simpson A A, Tao Y, Leiman P G, Badasso M O, He Y, Jardine P J, Olson N H, Morais M C, Grimes S, Anderson D L, Baker T S, Rossmann M G. Structure of the bacteriophage φ29 DNA packaging motor. Nature. 2000;408:745–750. doi: 10.1038/35047129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Soengas M S, Esteban J A, Lázaro J M, Bernad A, Blasco M A, Salas M, Blanco L. Site-directed mutagenesis at the Exo III motif of φ29 DNA polymerase: overlapping structural domains for the 3′-5′ exonuclease and strand-displacement activities. EMBO J. 1992;11:4227–4237. doi: 10.1002/j.1460-2075.1992.tb05517.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Soengas M S, Esteban J A, Salas M, Gutiérrez C. Complex formation between phage φ29 single-stranded DNA binding protein and DNA. J Mol Biol. 1994;239:213–226. doi: 10.1006/jmbi.1994.1364. [DOI] [PubMed] [Google Scholar]
- 196.Soengas M S, Gutiérrez C, Salas M. Helix-destabilizing activity of φ29 single-stranded DNA binding protein: effect on the elongation rate during strand displacement DNA replication. J Mol Biol. 1995;253:517–529. doi: 10.1006/jmbi.1995.0570. [DOI] [PubMed] [Google Scholar]
- 197.Sogo J M, Inciarte M R, Corral J, Viñuela E, Salas M. RNA polymerase binding sites and transcription map of the DNA of Bacillus subtilis phage φ29. J Mol Biol. 1979;127:411–436. doi: 10.1016/0022-2836(79)90230-4. [DOI] [PubMed] [Google Scholar]
- 198.Steiner M, Lubitz W, Bläsi U. The missing link in phage lysis of Gram-positive bacteria: gene 14 of Bacillus subtilis phage φ29 encodes the functional homolog of lambda S protein. J Bacteriol. 1993;175:1038–1042. doi: 10.1128/jb.175.4.1038-1042.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Take-Uchi A, Makino O, Hirokawa H. Gene 1, one of the dna genes of bacteriophage φ29, represses other dna genes through binding to mRNA. Biochem Biophys Res Commun. 1998;243:566–571. doi: 10.1006/bbrc.1998.8139. [DOI] [PubMed] [Google Scholar]
- 200.Take-Uchi A, Makino O, Matsumoto K, Hirokawa H. Inhibition of protein synthesis from DNA replication genes by the product of gene 1, a dna gene of Bacillus subtilis phage φ29. J Gen Appl Microbiol. 1995;41:507–516. [Google Scholar]
- 201.Tao Y, Olson N H, Xu W, Anderson D L, Rossmann M G, Baker T S. Assembly of a tailed bacterial virus and its genome release studied in three dimensions. Cell. 1998;95:431–437. doi: 10.1016/s0092-8674(00)81773-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Tedin K, Resch A, Steiner M, Bläsi U. Dual translational start motif evolutionary conserved in the holin gene of Bacillus subtilis phage φ29. Virology. 1995;206:479–484. doi: 10.1016/s0042-6822(95)80063-8. [DOI] [PubMed] [Google Scholar]
- 203.Tosi M E, Reilly B E, Anderson D L. Morphogenesis of bacteriophage φ29 of Bacillus subtilis: cleavage and assembly of the neck appendage protein. J Virol. 1975;16:1282–1295. doi: 10.1128/jvi.16.5.1282-1295.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Trottier M, Mat-Arip Y, Zhang C, Chen C, Sheng S, Shao Z, Guo P. Probing the structure of monomers and dimers of the bacterial virus φ29 hexamer RNA complex by chemical modification. RNA. 2000;6:1257–1266. doi: 10.1017/s1355838200992501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Trottier M, Zhang C L, Guo P. Complete inhibition of virion assembly in vivo with mutant pRNA essential for phage φ29 DNA packaging. J Virol. 1996;70:55–61. doi: 10.1128/jvi.70.1.55-61.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Truniger V, Blanco L, Salas M. Role of the “YxGG/A” motif of φ29 DNA polymerase in protein-primed replication. J Mol Biol. 1999;286:57–69. doi: 10.1006/jmbi.1998.2477. [DOI] [PubMed] [Google Scholar]
- 207.Truniger V, Blanco L, Salas M. Analysis of φ29 DNA polymerase by partial proteolysis: binding of terminal protein in the double-stranded DNA channel. J Mol Biol. 2000;295:441–453. doi: 10.1006/jmbi.1999.3370. [DOI] [PubMed] [Google Scholar]
- 208.Truniger V, Lázaro J M, Salas M, Blanco L. A DNA binding motif coordinating synthesis and degradation in proofreading DNA polymerases. EMBO J. 1996;15:3430–3441. [PMC free article] [PubMed] [Google Scholar]
- 209.Truniger V, Lázaro J M, Salas M, Blanco L. φ29 DNA polymerase requires the N-terminal domain to bind terminal protein and DNA primer substrates. J Mol Biol. 1998;278:741–755. doi: 10.1006/jmbi.1998.1724. [DOI] [PubMed] [Google Scholar]
- 210.Turnquist S, Simon M, Egelman E, Anderson D. Supercoiled DNA wraps around the bacteriophage φ29 head-tail connector. Proc Natl Acad Sci USA. 1992;89:10479–10483. doi: 10.1073/pnas.89.21.10479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Valle M, Kremer L, Martinez-A C, Roncal F, Valpuesta J M, Albar J P, Carrascosa J L. Domain architecture of the bacteriophage φ29 connector protein. J Mol Biol. 1999;288:899–909. doi: 10.1006/jmbi.1999.2731. [DOI] [PubMed] [Google Scholar]
- 212.Valpuesta J M, Carazo J M, Carrascosa J L. The three-dimensional structure of a DNA translocating machine at 10 Å resolution. Structure. 1999;7:289–296. doi: 10.1016/s0969-2126(99)80039-2. [DOI] [PubMed] [Google Scholar]
- 213.Valpuesta J M, Carrascosa J L. Structure of viral connectors and their function in bacteriophage assembly and DNA packaging. Q Rev Biophys. 1994;27:107–155. doi: 10.1017/s0033583500004510. [DOI] [PubMed] [Google Scholar]
- 214.Valpuesta J M, Donate L E, Mier C, Herranz L, Carrascosa J L. RNA-mediated specificity of DNA packaging into hybrid proheads. EMBO J. 1993;12:4453–4459. doi: 10.1002/j.1460-2075.1993.tb06131.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Villanueva N, Salas M. Adsorption of bacteriophage φ29 to Bacillus subtilis through the neck appendages of the viral particle. J Virol. 1981;38:15–19. doi: 10.1128/jvi.38.1.15-19.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Vlcek C, Paces V. Nucleotide sequence of the late region of Bacillus phage φ29 completes the 19285-bp sequence of φ29. Comparison with the homologous sequence of phage PZA. Gene. 1986;46:215–225. doi: 10.1016/0378-1119(86)90406-3. [DOI] [PubMed] [Google Scholar]
- 217.Vosknil M I, Chambliss G H. The −16 region of Bacillus subtilis and other gram-positive bacterial promoters. Nucleic Acids Res. 1998;26:3584–3590. doi: 10.1093/nar/26.15.3584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Wang I N, Smith D L, Young R. HOLINS: the protein clocks of bacteriophage infections. Annu Rev Microbiol. 2000;54:799–825. doi: 10.1146/annurev.micro.54.1.799. [DOI] [PubMed] [Google Scholar]
- 219.Whiteley H R, Ramey W D, Spiegelman G B, Holder R D. Modulation of in vivo and in vitro transcription of bacteriophage φ29 early genes. Virology. 1986;155:392–401. doi: 10.1016/0042-6822(86)90202-3. [DOI] [PubMed] [Google Scholar]
- 220.Yoshikawa H, Elder J H, Ito J. Comparative studies on the small Bacillus bacteriophages. J Gen Appl Microbiol. 1986;33:39–49. [Google Scholar]
- 221.Yoshikawa H, Friedmann T, Ito J. Nucleotide sequences at the termini of φ29 DNA. Proc Natl Acad Sci USA. 1981;78:1336–1340. doi: 10.1073/pnas.78.3.1336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Yoshikawa H, Garvey K J, Ito J. Nucleotide sequence analysis of DNA replication origins of the small bacteriophages: evolutionary relationships. Gene. 1985;37:125–130. doi: 10.1016/0378-1119(85)90264-1. [DOI] [PubMed] [Google Scholar]
- 223.Yoshikawa H, Ito J. Terminal proteins and short inverted terminal repeats of the small Bacillus bacteriophage genomes. Proc Natl Acad Sci USA. 1981;78:2596–2600. doi: 10.1073/pnas.78.4.2596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Yoshikawa H, Ito J. Nucleotide sequence of the major early region of bacteriophage φ29. Gene. 1982;17:323–335. doi: 10.1016/0378-1119(82)90149-4. [DOI] [PubMed] [Google Scholar]
- 225.Young I, Wang I, Roof W D. Phages will out: strategies of host cell lysis. Trends Microbiol. 2000;8:120–128. doi: 10.1016/s0966-842x(00)01705-4. [DOI] [PubMed] [Google Scholar]
- 226.Young R. Bacteriophage lysis: mechanism and regulation. Microbiol Rev. 1992;56:430–481. doi: 10.1128/mr.56.3.430-481.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Young R, Bläsi U. Holins: form and function in bacteriophage lysis. FEMS Microbiol Rev. 1995;17:191–205. doi: 10.1111/j.1574-6976.1995.tb00202.x. [DOI] [PubMed] [Google Scholar]
- 228.Zaballos A, Salas M. Functional domains in the bacteriophage φ29 terminal protein for interaction with the φ29 DNA polymerase and with DNA. Nucleic Acids Res. 1989;17:10353–10366. doi: 10.1093/nar/17.24.10353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Zhang F, Lemieux S, Wu X, St.-Arnaud D, McMurray T C, Major F, Anderson D. Function of hexameric RNA in packaging of bacteriophage φ29 DNA in vitro. Mol Cell. 1998;2:141–147. doi: 10.1016/s1097-2765(00)80123-9. [DOI] [PubMed] [Google Scholar]