Abstract
Citrullinated neoepitopes have emerged as key triggers of autoantibodies anti-citrullinated protein antibodies (ACPA) synthesis in rheumatoid arthritis (RA) patients. Apart from their critical role in homeostasis and thrombosis, platelets have a significant contribution to inflammation as well. Although anuclear in nature, platelets have an intricate post-translational modification machinery. Till now, citrullination in platelets and its contribution to trigger autoantibodies ACPA production in RA is an unexplored research direction. Herein, we investigated the expression of peptidylarginine deiminase (PAD) enzymes and citrullinated proteins/peptides in the human platelets and platelet derived microparticles (PDP). Both PAD4 mRNA and protein, but not the other PAD isoforms, are detectable in the human platelets. With a strict filtering criterion,108 citrullination sites present on 76 proteins were identified in the human platelets, and 55 citrullinated modifications present on 37 different proteins were detected in the PDPs. Among them, some are well-known citrullinated autoantigens associated with RA. Citrullinated forms of thrombospondin-1, β-actin, and platelet factor-4 (also known as CXCL4) are highly immunogenic and bound by autoantibodies ACPA. Furthermore, ACPA from RA sera and synovial fluids recognized citrullinated proteins from platelets and significantly activated them as evidenced by P-selectin upregulation and sCD40 L secretion. These results clearly demonstrate the presence of citrullinated autoantigens in platelets and PDPs, thus could serve as potential targets of ACPA in RA.
Keywords: rheumatoid arthritis, citrullination, platelets, anti-citrullinated protein antibodies (ACPA), platelet derived microparticles (PDP)
1. Introduction
Accumulating evidences suggest citrullination is a pre-requisite for triggering autoantibodies ACPA generation in the susceptible rheumatoid arthritis (RA) patients (1–3). The presence of autoantibodies ACPA is a hallmark serological feature for classical RA, and has an association with a more destructive disease course, although their functional role might be both pathogenic and protective depending on their specificity (4). Interestingly, autoantibodies ACPA can be detected decades before the onset of clinical arthritis (5, 6) and it was reported that epitope spreading with an increased recognition of citrullinated antigens occurs before the onset of RA (7). B cells with citrulline specificity are likely to be triggered in response to citrullinated protein targets in vivo but the identity and role of these targets remain unclear.
Previous studies addressed autoantibodies ACPA targets present in the synovium and synovial fluid (8), cartilage (9), lungs (10), and inflammatory cells such as neutrophils and macrophages (11, 12). Herein, we investigated the expression of citrullinated proteins within the platelets and PDPs. Platelets are abundant hematopoietic cells present in the blood circulation. In addition to the well-described homeostasis and thrombosis functions, platelets have important immunologic activities too, by expressing multiple pattern recognition receptors, MHC molecules, and immune co-stimulatory molecules (13). Platelets are the major source of microparticles in the blood circulation, which enhances their communication within the immune system, and also involved in the crosstalk between immune and coagulation systems (14, 15). In RA, platelets and PDPs were detected in both the blood and the synovial fluid samples (16, 17). Depletion of platelets led to reduced vascular leakage in the arthritic joints and attenuated inflammation in the animal models (18, 19). Interestingly, previous studies reported that autoantibodies ACPA contributes to platelet activation via FcγR-dependent pathways (20), platelet-derived microparticals display autoantigen which could be recognition by ACPA, and result in perpetuating formation of inflammatory immune complexes (17).
Although platelets lack nuclei, they have an intricate post-translational modification (PTM) processing machinery. Based on a high resolution proteomic analysis, a diverse set of PTMs such as acetylation/deacetylation (21, 22), phosphorylation, arginylation (23), palmitoylation (24), glycosylation, and methylation (25) were described to fine-tune the biogenesis and function of platelets. Compared to these PTMs, citrullination processes occurring within the platelets have been less explored, including their potential contribution to trigger autoantibodies ACPA production in RA.
Here, for the first time, we provide strong evidences for the presence of citrullination proteome in platelets and PDPs. In addition, we identified certain citrullinated proteins as ACPA targeted autoantigens and in turn, ACPA promoted the activation of platelets. Our findings suggest an important role for citrullinated autoantigens present in the platelets and PDPs in RA pathogenesis.
2. Materials and methods
2.1. Patients and controls
Non-paired blood (n=40 for RA, n=11 for OA) and synovial fluid (SF, n=11 for RA, n=8 for OA) samples were collected from RA and OA patients attending the department of rheumatology, Union hospital, Tongji medical college, university of Wuhan, China. RA patients enrolled met the 1987 ACR/ELAR criteria (26). Blood samples from the age- and gender-matched healthy controls (n=40) were obtained from the blood bank center, University of Wuhan. SF was collected from RA and OA patients requiring arthrocentesis for their affected knees. This study was approved by the local ethics committee of Union hospital, Tongji medical college (No. [2020] IEC-J (130)), and all the study participants gave a written informed consent to participate in this project.
2.2. Isolation of platelets and monocytes from blood
Peripheral blood from the healthy volunteers (n=4) was collected into acid citrate dextrose tube and centrifuged at 200 g for 15 min to collect the platelet-rich plasma, to avoid contamination with other blood cells. Only the upper third of the platelet-rich plasma was used for analysis. Platelets were pelleted at 600 g for 10 min, and washed twice to reduce the contamination of serum proteins using HEPES-buffered Tyrode’s solution (5 mM HEPES, 145 mM NaCl, 5 mM KCl, 0.5 mM Na2HPO4, 1 mM MgSO4, and 5 mM glucose, pH 7.2). The purity of the collected platelets was determined by CD41-staining and subsequent flow cytometry analysis. For monocyte isolation, 5 ml heparinized blood was diluted 1:1 with PBS containing 0.46% sodium citrate and layered over a 5 ml Ficoll cushion, and then centrifuged at 400 g for 15 min. PBMC containing interphase was collected and washed twice with PBS-citrate solution. Thereafter, the PBMCs were cultured in RPMI-1640 medium containing 10% FBS, in six well culture plates for 1h, allowing monocytes to adhere to the plastics. The non-adherent lymphocytes were removed and the adherent monocytes were harvested for further use.
2.3. Identification of PAD isoenzymes expression by RT-PCR
RNA from the platelets and monocytes was isolated using RNAsimple Total RNA Kit (TIANGEN, Beijing, China). The cDNA was synthetized from 1 µg of RNA using HiScript III 1st Strand cDNA Synthesis Kit (Vazyme, Nanjing, China). For PCR, 1 μl of cDNA was added to 25 μl of 2 × Phanta Flash Master Mix (Vazyme, Nanjing, China) containing Phanta Flash Super-Fidelity DNA Polymerase and, 0.5 mM forward and reverse primers. PCR was performed using a ETC811 thermocycler: 30 s at 98°C, 30 cycles of 10 s, 98°C; 5 s, 50-55°C; 5 s, 72°C, and one min elongation time at 72°C. 5 μl of the PCR product was analyzed by electrophoresis on a 1.5% agarose gel. The primers used to amplify the PAD isoforms and β-actin are presented in Table S1 .
2.4. Isolation of the PDPs
For the collection of platelet derived particles (PDPs), platelets were resuspended at 2 × 108/ml and stimulated with 5 μM of ADP (Sigma-Aldrich, Shanghai, China) in platelet aggregometer for 5 min at 37°C. Platelet activation was terminated by placing the tube on ice and then adding ice-cold 0.1% v/v termination buffer containing 10% NP-40, 20 mM PMSF, 200 g/mL trypsin inhibitor, 50 mM N-ethylmaleimide, 10 mM EDTA, 1% SDS and 100 mM benzamidine. Intact platelets and the platelet clots were carefully removed by centrifuging at 800 g for 10 min, sequentially for three times, and the supernatant was harvested each time and pooled before the final spin, which yielded the PDPs.
2.5. MS analysis
Platelet pellets and PDPs were lysed in 8 M or 2 M urea solution, respectively. To this lysate, 10 mM HEPES (pH 8) was added and sonicated at 4°C for 20 min. One fraction of platelet sample was separated by gel electrophoresis and subjected to in-gel trypsin digestion as described earlier (27). Briefly, gel slices (20 slices/lane) were excised and the proteins were reduced with 64 mM of dithiothreitol (DTT) at 56°C for 60 min, followed by an alkylation step in the dark for 45 min with 130 mM iodacetamide (IAA). Proteins were digested with trypsin/Lys C mixture (Promega, Wisconsin, USA) overnight at 37°C. The peptides were purified with acetonitrile/0.1% aqueous formic acid 1:1 (v/v), and re-solubilized in 0.1% aqueous formic acid prior to LC-MS/MS analysis. Another fraction of platelet sample as well as PDPs were prepared using a filter-aided sample preparation protocol (FASP) (28). Each sample (100 μg) was transferred to 10 kDa cutoff spin-filter (Millipore, Ireland, UK), reduced with DTT, blocked by IAA and digested with trypsin/Lys C mixture for overnight at 37°C. The peptides were eluted from the spin filters, acidified with 0.1% formic acid, and then desalted with a C18 pipet tip (Millipore, Ireland, UK) for further use.
Peptides were loaded onto a Thermo Acclaim Pepmap precolumn followed by an Acclaim Pepmap Easyspray analytical column. Separation was performed using a Thermo Easy-nLC 1200 at a flow rate of 300 nL/min with a gradient of 2-35% organic phase (0.1% formic acid in acetonitrile) over 200 min. Peptides were analyzed using a high-energy collisional dissociation (HCD) fragmentation mode by data-dependent MS/MS acquisition, with the following settings: MS1-scan resolution 70 ;000, the AGC target was 3e6; MS2-scan resolution 17 ;500, the AGC target was 5e4; and the maximum injection time was 50 ms, NCE stepped set 27. The scan range was set at a resolution of 350-2,000 m/z and the 20 most intense precursors were chosen by data-dependent mode.
2.6. Protein identification and quantification
MaxQuant software (version 1.6.6.0) was used for the identification of peptides and proteins against the human proteome database. Peptide identification was performed with a mass tolerance up to 5 ppm for precursor ion and MS/MS tolerance to 20 ppm. A false discovery rate (FDR) was set to 0.01 at the peptide and protein level. Relative protein quantities were calculated by summing the unique peptide peak areas of each protein in MaxQuant using the LFQ (label-free quantitation) feature (29).
2.7. Identification of citrullination
To ensure the accuracy of identifying citrullination, we applied a strict filtering criterion to verify the citrullinated side chains, as previously described (30, 31). At first, the precursor ions were extracted with a mass tolerance up to 5 ppm to prevent the forced 13C mis-assignment. All the total peptide spectral matches (PSMs) in the C-terminal citrullination site were removed due to inability of trypsin/Lys C catalytic activity to citrulline residues. Next, the assigned citrullinated MS2 spectra were manually inspected to exclude 0.984 Dalton mass increments occurring from deamidation with the neighboring N/Q residues. Only the complete fragment ion series (b, y ions) covering the citrullinated sites were considering as correct assignment. Identification of citrullinated sites was further confirmed by the occurrence of neutral loss on the precursor or fragment ions and a shift in the delayed retention time.
2.8. In vitro citrullination
Human recombinant thrombospondin-1 (TSP-1, Sigma-Aldrich), beta-actin (β-actin, Genway, San Diego, USA), or platelet factor 4 (PF4, Sigma-Aldrich) was incubated with PAD4 enzyme (Sigma-Aldrich) in the siliconized tubes containing 100 mM of Tris pH 7.7, 1 mM of DTT, and 5 mM of CaCl2. After 24 h at 37°C, reactions were stopped by adding 0.1% of trifluoroacetic acid. Citrullinated and non-citrullinated samples were loaded on SDS-PAGE followed by in-gel digestion for further mass spectrometry analysis of citrullination sites, as well as to screen for binding of ACPA by ELISA.
2.9. ELISA for autoantibodies ACPA measurement
The plates were coated with 200~500 ng/well of citrullinated or non-citrullinated forms of TSP-1, β-actin, and PF4 in 0.1 M NaHCO3 (pH 9.6), overnight at 4˚C. The plates were washed with PBS/Tween 0.05%, blocked with 1.5% BSA/PBS for 1 h, at RT, and incubated with RA sera (1:100 dilution) for 2 h, RT. Then the plates were washed and developed with HRP-conjugated rabbit anti-human IgG polyclonal antibodies for 1 h. TMB substrate was added and incubated at 37°C for 5~10 min and the reaction was stopped with 1.0 M sulfuric acid (H2SO4). The absorbance (A450 nm) was read using a microplate reader (Thermo, Shanghai, China). The absorbance value of the pooled healthy sera was set at an arbitrary unit (AU) value of 1 and was used as a standard for all other tests (1 AU/μl).
2.10. Affinity purification of autoantibodies ACPA
Autoantibodies ACPA were purified from selected RA patients’ synovial fluid and serum samples having anti-CCP2 IgG level > 500 AU/ml, using Protein G beads and CCP2-coated resins. CCP2 peptide were purchased from Euro Diagnostica (Malmö, Sweden, obtained from Biolead China), and couple to CNBr-activated Sepharase 4B beads (GE Healthcare) according to the manufacturer’s protocol. SF (5~8 mL/sample) was centrifuged at 4,000 rpm for 10 min, and the supernatant collected was treated with hyaluronidase to decrease the viscosity of the samples. Proteins were precipitated by using a saturated ammonium sulfate solution, and then dissolved in PBS and dialyzed extensively against PBS. While the serum samples were centrifuged and diluted 1:5 in PBS before purification. At first, total IgG was enriched from SF and serum samples using protein G beads. To purify ACPA, 1 mg/ml of CCP2 peptide was coupled to 2 ml of NeutrAvidin Plus UltraLink resin (Pierce Biotechnology/Thermo Scientific) for 1 h at RT. Purified total IgG from SF and serum samples was then applied to the CCP2-coated resin, and washed with PBS. ACPA were eluted by adding 0.1 M glycine-HCl buffer (pH 2.7), neutralized with 1 M Tris (pH 9) and then dialyzed against PBS.
2.11. Platelet activation and flow cytometry analysis
To block the binding of IgG-Fc with CD32, platelets (2×108/ml) were first pre-incubated with 20 μg/ml anti-human CD32 (6C4, eBioscience, Rockford, IL) for 1 h at 37°C and washed twice with PBS, To determine the capability of ACPA mediated platelets activation, platelets were incubated with seleted ACPA high sera, or SF (1:20 dilution) from RA patient or purified autoantibodies ACPA (10 μg/ml in PBS + 0.1% BSA) at 37°C for 90 min. Then supernatants were collected by centrifugation (400×g, 15 min, 20°C) and platelets were resuspended for flow cytometric analysis. The activation status of platelets was determined by flow cytometry by measuring the expression of CD62P and sCD40L level in the collected supernatants. Platelets were incubated with anti-CD62P-FITC (clone AK-4; BD Biosciences, New York, USA) which was diluted in HEPES-buffered Tyrode’s solution, pH 7.2 (1:50 dilution) for 30 minutes at RT, after which cells were fixed using 1% paraformaldehyde and analysed by NovoCyte Flow Cytometer and NovoExpress software (Agilent, Palo Alto, USA). The sCD40L level was determined by using an ELISA Kit (eBioscience, San Diego, USA).
2.12. Statistical analyses
For normally distributed populations, mean ± SD values were used and Student’s t test was applied to evaluate the statistical difference between the groups. Differences in antibody levels were examined using the non-parametric Mann-Whitney U test. Data were analyzed using GraphPad Prism version 9.0. The p values < 0.05 were considered as statistically significant.
3. Results
3.1. PAD4 expression was detectable in the human platelets
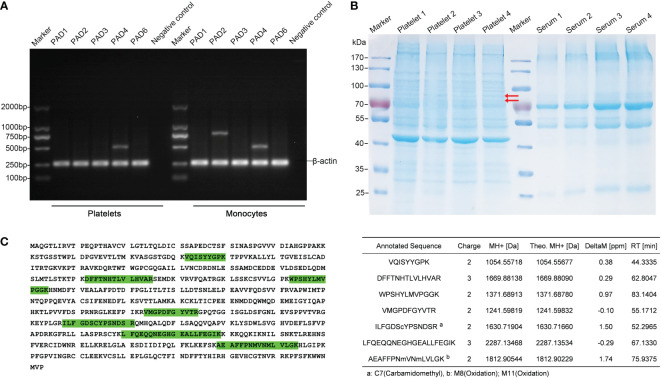
To date, five Ca2+-dependent PAD isozymes designated as PAD1-4, and PAD6, which catalyze the conversion of arginine to citrulline have been identified, each with a unique tissue and cell distribution pattern (32). To examine the presence of PAD isoforms in human platelets, we first investigated the mRNA expression of all the five PAD isozymes by RT-PCR. Platelets (> 98% CD41+ cells by flow cytometry analysis, data not shown) and paired monocytes were isolated from the peripheral blood of four healthy volunteers. Total RNA was isolated from the platelets and monocytes, and each isotype of PAD gene was amplified by RT-PCR. Surprisingly, a substantial number of PAD4 mRNA was detectable in all the four platelet samples, but not the mRNAs of PAD2 or PAD1, 3 and 6 isozymes, the other isoforms described to be present in the platelets. On the other hand, we detected both PAD2 and PAD4 mRNAs in the monocytes ( Figure 1A ).
Figure 1.
Identification of PAD4 expression in the human platelets. (A) Agarose gel electrophoresis of RT-PCR products after amplifying the mRNA of PAD isoforms in platelets and monocytes. The β-actin expression was used as an internal control. PCR without the template served as a negative control. (B) SDS-PAGE depicting the proteins from platelets. Lanes 2-5 are corresponding to platelet samples divided into 20 slices from top to bottom, followed by an in gel-digestion and LC-MS/MS analysis. Arrow indicates PAD4 expression in the gel by MS analysis. Lanes 7-10, show the paired serum samples indicating the limitation present in the carry-through of plasma proteins into platelet proteomics. (C) Detected PAD4 peptide sequence coverage. Left section: the identified tryptic peptides are indicated in green shadow. Right section: list of detected and mapped peptides.
We then analyzed the expression of PAD proteins by mass spectrometry (MS). To make the platelet samples less complex to achieve a higher resolution of PADs, platelet proteins were fractioned by gel electrophoresis into 20 gel slices prior to MS analysis ( Figure 1B ). Clearly, unique peptides from PAD4 could be detected in two gel slices between 70 to 100 KDa bands in all the four platelet samples, which correspond to the expected molecular mass of PAD4 enzyme. BLAST search in the UniProt confirmed the aa sequences of these peptides to be unique for human PAD4 isoform, with an amino acid sequence coverage of 14.5%. In contrast, peptides of PAD2, or PAD1, 3, and 6 isoforms were not detectable in the platelet protein extracts ( Figure 1C ). Taken together, the identified PAD4 mRNA and protein expressed in the platelets suggests the importance and critical contribution of PAD4 isoform in citrullinating the proteins/peptides within the platelets in humans.
3.2. Identification of citrullinated proteins in human platelets
Next, we identified the proteins and specific arginine residues that were citrullinated in the platelets. To facilitate the peptide recovery, we performed a filter-aided sample preparation (FASP) method in the four platelet samples. MS analysis identified more than 44, 639 unique peptides and 3, 972 proteins with an abundance covering 10 orders of magnitude. The 100 most abundant proteins are well-known cytoskeleton, α-granule, and cytoskeletal-linked signaling proteins ( Table S2 and Figure S1 ).
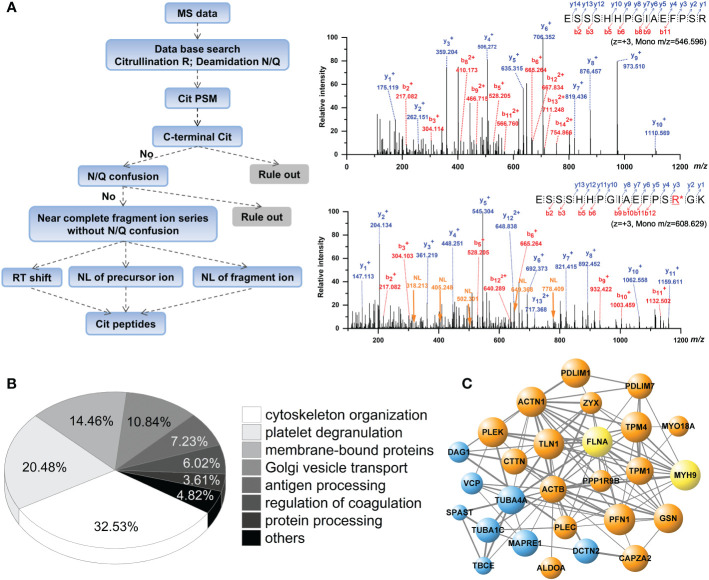
When peptides were analyzed for citrullinated modifications, a strict filtering criterion was applied to eliminate the interferences from N/Q deamidation and 13C isotopes ( Figure 2A ). Each MS/MS spectrum was manually verified by comparing it with the theoretically fragmented ions. The FASP short-gun MS and SDS-pre-fraction LC-MS/MS analysis revealed 108 citrullination sites on 76 different proteins ( Table 1 ). These 76 citrullinated proteins included previously characterized substrates of PAD enzymes, such as actin, alpha-enolase, fibrinogen α/β, filamin-A, transgelin-2, and tubulin α (11, 30). In addition, some proteins with more specific roles in platelet functions were observed to be modified by citrullination, including integrin α-IIb, plasminogen, platelet glycoprotein Ib α, platelet glycoprotein IX, TSP-1, and von Willebrand factor. We noted that proteins with a broad range of expression levels were modified by citrullination, including highly and less abundant proteins in human platelets ( Table 1 ).
Figure 2.
Analysis of citrullinated peptides present in the human platelets by mass spectrometry. (A) Decision tree for the identification of citrullination is shown in the left panel. Representative dimer-isotopic MS2 spectrum of non-citrullinated and citrullinated 560E–575K peptide of human fibrinogen α is shown in the right panel. Purple arrow indicates the neutral loss fragment (NL, -43.0058 Da) corresponding to y3+~y7+ ions. (B) Pie-chart showing the classification of identified citrullinated proteins in the platelets, according to functional categories. (C) Protein interaction network of citrullinated proteins. Orange bobbles show the actin-myosin filaments; blue bobbles show the microtubules; yellow bobbles show the actin-myosin filaments and microtubules.
Table 1.
Citrullination modifications present in human platelets.
| Accession | Description | Site | iBAQ | Accession | Description | Site | iBAQ |
|---|---|---|---|---|---|---|---|
| P12814 | Alpha-actinin-1 | R350 | 2 | P35579 | Myosin-9 | R1912 | 23 |
| R760 | 22 | R1923 | |||||
| Q08043 | Alpha-actinin-3 | R74 | P23368 | ME2 | R413 | 422 | |
| P06733 | Alpha-enolase | R429 | 3660 | Q96SB3 | Neurabin-2 | R810 | 1518 |
| P08758 | Annexin A5 | R18 | 55 | O00151 | PDLIM1 | R177 | 51 |
| P60709 | Beta-Actin | R256 | 444 | Q9NR12 | PDLIM7 | R137 | 304 |
| Q9BXS5 | AP1M1 | R304 | 580 | R158 | |||
| O95810 | CAVIN2 | R141 | 128 | P08567 | Pleckstrin | R307 | 18 |
| P14209 | CD99 antigen | R170 | 432 | Q15149 | Plectin | R2767 | 626 |
| Q96G23 | Ceramide synthase | R363 | 1038 | R3337 | |||
| Q00610 | Clathrin | R574 | 190 | P07737 | Profilin-1 | R75 | 4 |
| Q6PJW8 | Consortin | R250 | 384 | P02760 | Protein AMBP | R185 | 1177 |
| Q9H7D0 | DOCK5 | R1721 | 2063 | Q9H0Q0 | Protein FAM49A | R147 | 1042 |
| Q13561 | Dynactin subunit | R212 | 351 | Q15404 | RSU1 | R165 | 33 |
| O00429 | DNM1L | R108 | 218 | Q9H0U4 | RAB1B | R183 | 186 |
| Q14118 | Dystroglycan | R689 | 1437 | Q9NS28 | RGS18 | R205 | 516 |
| Q9NZN3 | EHD3 | R404 | 98 | O94804 | STK10 | R451 | 1577 |
| P11021 | HSPA5 | R49 | 109 | P02768 | Serum albumin | R281 | 11 |
| Q9BSJ8 | ESYT1 | R1003 | 366 | R509 | |||
| P47755 | CAPZA2 | R109 | 245 | O75368 | SH3BGRL | R58 | 198 |
| P02671 | Fibrinogen alpha | R123 | 32 | Q9UJC5 | SH3BGRL2 | R74 | 165 |
| R271 | Q9UBP0 | Spastin | R488 | 3193 | |||
| R287 | Q14247 | CTTN | R59 | 259 | |||
| R510 | R434 | ||||||
| R573 | Q7Z422 | SZRD1 | R65 | 2288 | |||
| R591 | O00161 | SNAP23 | R148 | 140 | |||
| P02675 | Fibrinogen beta | R60 | 28 | Q9Y490 | Talin-1 | R35 | 27 |
| P21333 | Filamin-A | R2001 | 15 | R181 | |||
| R2003 | R606 | ||||||
| R2334 | R2154 | ||||||
| R2484 | P50991 | CCT4 | R153 | 393 | |||
| P04075 | ALDOA | R331 | 48 | O43396 | TXNL1 | R211 | 447 |
| O15117 | FYB1 | R19 | 288 | P07996 | Thrombospondin-1 | R314 | 25 |
| Q3ZCW2 | LGALSL | R106 | 122 | R412 | |||
| P06396 | Gelsolin | R397 | 39 | R751 | |||
| Q02153 | GUCY1B1 | R197 | 979 | R767 | |||
| P0DMV9 | HSPA1B | R357 | 234 | R926 | |||
| P10316 | HLA-A | R86 | 3871 | Q01664 | TFAP4 | R293 | 3887 |
| Q9H2U2 | PPA2 | R84 | 959 | P37802 | Transgelin-2 | R160 | 20 |
| P23229 | Integrin alpha-6 | R635 | 292 | P55072 | VCP | R210 | 134 |
| P08514 | Integrin alpha-IIb | R104 | 42 | P09493 | TPM1 | R55 | 84 |
| R334 | P67936 | TPM4 | R27 | 10 | |||
| R399 | Q9BQE3 | Tubulin alpha-1C | R422 | 371 | |||
| R433 | P68366 | Tubulin alpha-4A | R79 | 80 | |||
| R799 | Q15813 | TBCE | R13 | 2743 | |||
| P05106 | Integrin beta-3 | R505 | 35 | O43399 | TPD52L2 | R182 | 382 |
| R662 | Q92614 | MYO18A | R1302 | 1524 | |||
| P13645 | KRT10 | R15 | 967 | P46459 | NSF | R533 | 1014 |
| P35908 | KRT2A | R524 | 1917 | P18206 | Vinculin | R547 | 49 |
| Q15691 | MAPRE1 | R162 | 171 | R622 | |||
| Q13201 | Multimerin-1 | R450 | 160 | R874 | |||
| P35579 | Myosin-9 | R165 | 23 | P04275 | von Willebrand factor | R1136 | 214 |
| R930 | R1597 | ||||||
| R1342 | Q15942 | Zyxin | R517 | 70 |
String and GO analysis showed 32.53% (n = 27 proteins) of the identified citrullinated proteins are involved in the organization of cytoskeleton (p = 3.21×10−11) related to actin-myosin filaments and microtubules in platelets ( Figures 2B, C ). Protein interactome analysis indicated that a substantial portion of the citrullinated proteins was enriched after platelet degranulation (20.48%, p = 5.80×10−16; Figure 2B ). Previous studies using brain, gut mucosa, and myocardial tissues showed a more preferential targeting of extracellular- and membrane-bound proteins by citrullination (33–35). Similarly, in our analysis, proteins annotated as membrane-bound proteins involved in cell-cell and cell-substrate adhesions were over-represented in the human platelet citrullinome (14.46%, p = 8.57×10−10), especially in the processes involved in integrin α-IIb and fibrinogen, as well as in the interactions related to the cytoskeletal remodeling and platelet degranulation.
3.3. Citrullinated proteins are released in the platelet derived microparticles
Since human platelets expressed PAD4 enzyme and abundant citrullinated proteins were identified, we next addressed whether platelets could release the citrullinated proteins outside the cells. To investigate the citrullinated protein contents of PDPs, platelets from 12 volunteers were stimulated with ADP to induce the release of PDPs, and then the PDPs were harvested and subjected to MS analysis. We identified 5, 476 peptides of 687 proteins at FDR 0.01 ( Table S3 ). As expected, many PDPs were found to be citrullinated, with the identification of 55 citrullinated peptides from 37 different proteins ( Table 2 ). As described above, some proteins having more specific roles in platelet functions were observed to be citrullinated in the PDPs as well, such as integrin α-IIb, multimerin-1, pleckstrin, TSP-1, and von Willebrand factor. However, the majority of proteins identified to be in the citrullinated form were present within a single sample. This may be due to technical reasons, such as varying sampling depth or low abundance of the modification, or individual differences in the citrullination patterns.
Table 2.
Citrullination modifications detected in platelet delivery PRMs.
| Accession | Description | Site | iBAQ | Accession | Description | Site | iBAQ |
|---|---|---|---|---|---|---|---|
| P35609 | Alpha-actinin-3 | R067 | 620 | P02776 | Platelet factor 4 | R080 | 1 |
| P02749 | Beta-2-glycoprotein 1 | R210 | 122 | P07359 | PG-Ib-alpha | R080 | 74 |
| P60709 | β-actin | R256 | 4 | P01127 | PDGFB | R142 | 110 |
| P27797 | Calreticulin | R177 | 176 | P08567 | Pleckstrin | R307 | 21 |
| O95810 | CAVIN2 | R141 | 220 | P07737 | Profilin-1 | R075 | 13 |
| P10909 | Clusterin | R194 | 19 | A6NCN2 | KRT87P | R072 | 559 |
| P01024 | Complement C3 | R1591 | 69 | P02787 | Serotransferrin | R239 | 25 |
| P29279 | CCN2 | R240 | 161 | R651 | |||
| Q86UX7 | FERMT3 | R098 | 48 | R682 | |||
| P02671 | Fibrinogen alpha | R218 | 15 | P02768 | Serum albumin | R122 | 3 |
| P02675 | Fibrinogen beta | R410 | 14 | R141 | |||
| P21333 | Filamin-A | R2003 | 64 | R281 | |||
| P06396 | Gelsolin | R048 | 23 | R469 | |||
| R295 | R509 | ||||||
| R397 | Q9H299 | SH3BGRL3 | R047 | 164 | |||
| P04406 | GAPDH | R080 | 32 | P09486 | SPARC | R181 | 5 |
| P01857 | IGHG1 | R299 | 7 | Q9Y490 | Talin-1 | R606 | 80 |
| P08514 | Integrin alpha-IIb | R312 | 136 | P07996 | Thrombospondin-1 | R314 | 6 |
| P35527 | KRT9 | R327 | 153 | R412 | |||
| Q14766 | LTBP1 | R1445 | 85 | R460 | |||
| R1508 | R751 | ||||||
| R1538 | R926 | ||||||
| Q13201 | Multimerin-1 | R450 | 65 | P01137 | TGF beta-1 | R255 | 67 |
| R589 | P67936 | Tropomyosin alpha-4 | R065 | 17 | |||
| R890 | P07437 | Tubulin beta | R086 | 44 | |||
| P14543 | Nidogen-1 | R1017 | 54 | P04275 | von Willebrand factor | R647 | 102 |
| P02775 | Platelet basic protein | R062 | 2 | R1597 | |||
| P02776 | Platelet factor 4 | R051 | 1 |
3.4. Autoantibodies ACPA bind to citrullinated TSP-1, β-actin, and PF4
To address the citrullination activities by PAD4 on platelet proteins, we chose TSP-1, β-actin, and PF4 as testing substrates, because of their identified citrullinated forms present in the platelets and PDPs. After the treatment with PAD4 in vitro, in total 18 arginine residues of rTSP-1 were citrullinated, which included the five in vivo citrullination sites of TSP-1 observed in the platelets and PDPs, as well as additional 12 citrullinated residues ( Table 3 ). For β-actin samples treated with the PAD4 enzyme, 12 citrullination sites were identified, including the 10 citrullination sites that had previously been reported by another group (36) ( Table 3 ). Human PF4 (also named as CXCL4) contained 4 Arg residues, including Arg51 and Arg80 that could be citrullinated in vitro ( Table 3 ). Over all, PAD4 was efficient in citrullinating the platelet proteins TSP-1, β-actin, and PF4.
Table 3.
Citrullination sites present in TSP-1, β-actin, and PF4 by PAD4 catalysis.
| protein | site | Annotated Sequence a | In vivo |
|---|---|---|---|
| TSP-1 (18) | R83 | FQDLVDAVRAEK | |
| R101 | TRGTLLALERK | ||
| R196 | DLASIARLRIAK | ||
| R216 | GGVNDNFQGVLQNVRFVFGTTPEDILR | ||
| R285 | DLQAICGISCDELSSMVLELRGLR | ||
| R288 | GLRTIVTTLQDSIR | ||
| R314 | RPPLCYHNGVQYR | R314 | |
| R404 | GRSCDSLNNR | ||
| R412 | SCDSLNNRCEGSSVQTR | R412 | |
| R421 | CEGSSVQTRTCHIQECDK | ||
| R458 | QDGGWSHWSPWSSCSVTCGDGVITRIR | ||
| R460 | IRLCNSPSPQMNGK | ||
| R517 | RSRLCNNPTPQFGGK | ||
| R607 | EVPDACFNHNGEHRCENTDPGYNCLPCPPR | ||
| R675 | CNYLGHYSDPMYRCECK | ||
| R751 | IPDDRDNCPFHYNPAQYDYDR | R751 | |
| R767 | DNCPFHYNPAQYDYDRDDVGDR | R767 | |
| R926 | DSDGDGRGDACKDDFDHDSVPDIDDICPENVDISETDFR | R926 | |
| β-Actin (10) | R62 | RGILTLK | |
| R95 | IWHHTFYNELRVAPEEHPVLLTEAPLNPK | ||
| R183 | LDLAGRDLTDYLMK | ||
| R196 | ILTERGYSFTTTAER | ||
| R206 | GYSFTTTAEREIVR | ||
| R210 | REIVRDIK | ||
| R254 | SYELPDGQVITIGNERFR | ||
| R256 | FRCPEALFQPSFLGMESCGIHETTFNSIMK | R256 | |
| R312 | DLYANTVLSGGTTMYPGIADRMQK | ||
| R372 | QEYDESGPSIVHRK | ||
| PF4 (2) | R51 | TTSQVRPR | R51 |
| R80 | AGPHCPTAQLIATLKNGR*K | R80 |
red color indicates citrullinated arginine residue.
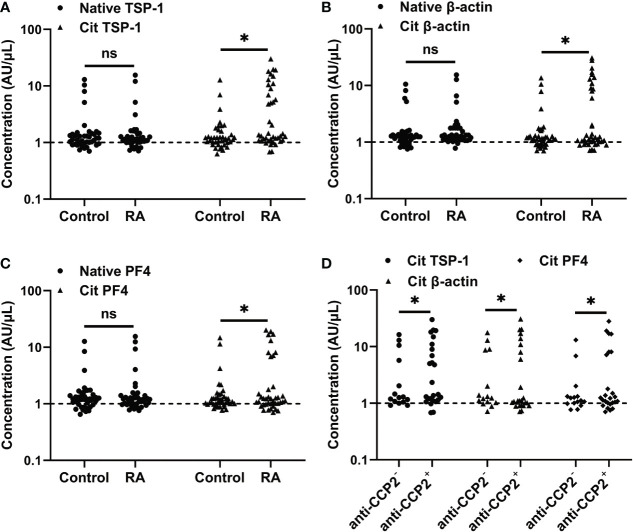
We then investigated whether citrullinated TSP-1, β-actin, and PF4 could serve as autoantigens in RA. The reactivity of RA serum with native and citrullinated forms of TSP-1, β-actin, and PF4 was assessed by ELISA. RA serum showed a low reactivity towards native forms of TSP-1, β-actin, and PF4, similar to healthy controls ( Figures 3A–C ). In comparison, an increased frequency of RA sera showed reactivity to citrullinated forms of TSP-1 (37.5%), β-actin (30.0%), and PF4 (22.5%) ( Figures 3A–C ), while the control serum had no such increased reactivity. As expected, the majority of the citrullinated TSP-1, β-actin, and PF4 positive RA cases were confined to the anti-CCP2+ RA group ( Figure 3D ).
Figure 3.
Measurement of citrullinated TSP-1, β-actin, and PF4 - specific antibodies in the RA sera. (A-C) Determination of citrullinated TSP-1, β-actin, and PF4-specific antibodies in the sera (1:100 dilution) of healthy controls and RA patients. (D) Distribution of citrullinated TSP-1, β-actin, and PF4-specific antibodies in the anti-CCP2+ and anti-CCP2- RA sera. Broken lines indicate the cutoff values. Cit, denotes citrullination, *p < 0.05, ns = not significant.
3.5. Autoantibodies ACPA bind to citrullinated platelet proteins and enhance the activation of platelets.
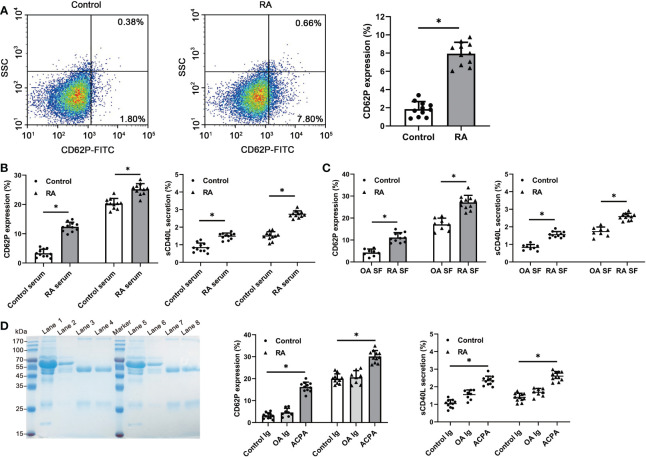
Since citrullinated proteins are present in the human platelets, we hypothesized that autoantibodies ACPA in RA patients may react to the citrullinated platelet proteins and interact with the activation of platelets. To investigate the ACPA reactivity, we used RA plasma and SF having a high titer of anti-CCP2 antibodies (> 500 AU/ml) in the stimulation experiments. A higher basal activation status was detected in platelets from high anti-CCP2+ RA patients, as indicated by an increased expression of P-selectin (RA patients 7.94 ± 1.25% vs control 1.86 ± 0.82; p < 0.05) ( Figure 4A ). Upon stimulation, both the serum and SF from high anti-CCP2+ RA patients have potently induced platelets collected from healthy and RA patients, when compared to the healthy serum or OA SF, and also increased the production of sCD40L ( Figures 4B, C ). We further purified the ACPA from RA patients’ SF and serum samples. Pooled ACPA samples have significantly increased the expression of both P-selectin and the production of sCD40L in the healthy and RA neutrophils, when compared to control IgG or IgG from OA SF ( Figure 4D ). These observations demonstrate ACPA reactivity to the citrullinated proteins present in the platelets and their subsequent activation of platelets.
Figure 4.
Detection of platelet activation after RA serum, SF or purified ACPA stimulation. (A) Flow cytometric quantification of CD62P expression in the platelets from healthy controls and RA patients. Data are representative of six independent experiments. (B, C) Both the serum and SF from RA induced significantly a higher level of CD62P expression and sCD40L secretion in RA and healthy platelets, compared to the healthy serum or OA SF (RA serum and SF: n=11, healthy control serum: n=11, OA SF: n=8). (D) Purified ACPA from RA SF and serum samples has significantly enhanced the CD62P expression and sCD40L secretion in RA and healthy platelets. SDS-PAGE showing the purified ACPA from SF of 2 RA patients by protein G plus beads and CCP2-peptide coated resin. Lane1/lane5: SF; Lane2/lane6: flow through from protein G column; Lane3/lane7: eluted IgG from protein G column; Lane4/lane8: eluted ACPA from CCP2-coated resin. Data are presented as mean ± SD, *p < 0.05.
4. Discussion
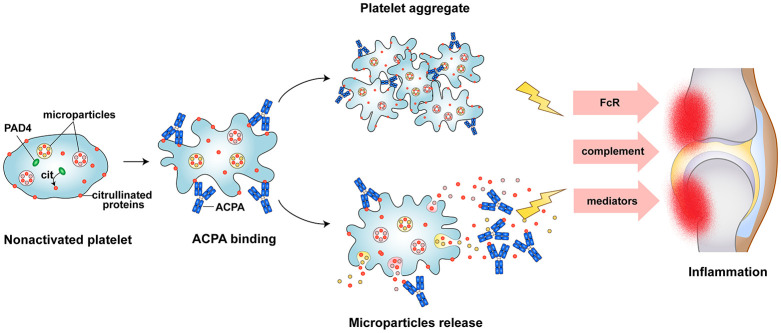
In this study for the first time, we have demonstrated the expression of PAD4 enzyme and citrullinated proteins present within the human platelets and PDPs. In addition, we also found platelet activation mediated by ACPA from RA patients. In RA, platelet aggregates are often observed in the blood and the joint, microparticles releasing as consequence of platelet activation accumulate in the joints of patients. While the mechanistic events that lead to platelet activation in RA have not been extensively characterised. We hypothesized that citrullinated proteins from platelet and PDP may play prominence role in stimulating platelet activation in RA. Here we shown that plenty of citrullinated protein present in platelets and PDPs can be recognized by autoantibodies ACPA. and ACPA can stimulate platelet activation lead to releasing active inflammatory molecules and citrullinated autoantigens that may sustain inflammatory responses in RA joint ( Figure 5 ).
Figure 5.
Schematic picture of platelet as a source of autoantigens and persistent inflammation in RA. Autoantibodies ACPA recognized citrullinated proteins in platelet, result in platelet activation, aggregation, and microparticles releasing. This will further lead to citrullinated proteins exposure to the immune cell, triggering of ACPA production and contribute to the persistent inflammation.
The most noticeable finding is the unique expression of PAD4 in platelets both at mRNA and protein levels, which citrullinated platelet proteins/peptides. Recently, PAD4 was identified to be inextricably associated with RA than other PAD isoforms. PAD4 enzymes have been founded in the rheumatoid synovial tissue and fluid (37, 38). In addition, PAD4 gene and certain SNPs were detected within a susceptibility locus associated with RA (39). PAD4 is unique because only this PAD isoform contains a canonical nuclear localization signal (NLS) found predominantly in the nucleus (40). Many in vitro experiments demonstrated that both PAD4 and PAD2 isoforms are efficient in generating citrullinated target sites for autoantibodies ACPA, while ACPA preferentially bind to histone H3 and fibrinogen citrullinated by PAD4 than PAD2 (41, 42).
Our data uncovered a list of previously unreported citrullinated proteins present in the platelets and PDPs. We also identified citrullinated forms of fibrinogen α/β, α-enolase, β-actin, and α/β-tubulin, which are all well-known ACPA auto-antigens in the platelets and PDPs (8, 41, 43). In addition, our MS data uncovered a list of citrullinated proteins including numerous cytoskeletal and cell-motility proteins such as myosin-9, caveolae-associated protein 2, tubulin, and actin. Platelets are highly specialized cells and their activation involves a series of rapid rearrangements in the cytoskeleton. In this context, we hypothesized that the citrullination may modify the protein’s functional properties regulating their actin dynamics, similar to the functions of other PTMs present in the actin cytoskeleton like acetylation, phosphorylation, tyrosination and polyglutamylation (22, 24, 44). Furthermore, upon stimulation by various dysregulated molecules in the synovium and circulation, platelet could quick release immunostimulatory microparticals and intracellular citrullinated proteins in their environment, the exposed citrullinated proteins recognition by ACPA leading to formation of inflammatory immune complexes. Thus, activated platelet, immunostimulatory microparticals, and inflammatory immune complexes perpetuate pathogenic mechanisms in RA disease ( Figure 5 ).
Intriguingly, we detected citrullination on some cytokines in PDPs, such as PF-4 and TGFβ-1. Previous studies have found that citrullination of cytokines could regulate their biochemical functions. For example, citrullinated chemokines CXCL10, CXCL11, CXCL8 and CXCL12 have reduced chemoattracting capacity, and their ability to signal through the chemokine receptors (45–47). Thus, it is important to underscore that the citrullination process should no longer be considered as a rare modification, because a large number of citrullinated proteins are being identified in RA (8, 10, 34) and in many other diseases as well. Our proteomic analysis of platelets and PDPs has shown the presence of abundant citrullinated proteins/peptides that overlap with ACPA autoantigens. Citrullination of proteins, even among the important targets of autoantibodies ACPA, is a physiological event, not specific for RA, although the production of ACPA is more specific for RA.
Citrullination of TSP-1, β-actin, and PF4 led to exposure of epitopes recognized by ACPA may facilitate our understanding of the role of ACPA in RA development by providing evidences for previous observations involving platelet activation by ACPA in RA (20), the elevated levels of PDPs associated with disease activity (16, 48), and possibly the increased risk of cardiovascular events observed in RA patients (49). Although ACPA and tested platelet used in this study are non-paired sample, we speculate in matched samples, ACPA could reactive with self-generation platelet derived citrullination protein and lead to self-platelet activation, since both citrullinated protein and ACPA are heterogeneity, PTM as citrullinated modification could break of autoreactive B cell tolerance (50).
This study is the first comprehensive description of the platelet citrullination and expands our understanding of the scope of citrullination in platelets. We propose PAD4 enzyme could be responsible for eliciting protein citrullination in human platelets and thus suitable as a target protein for future drug development. In addition, our results may help for better understanding of the origin and consequences of citrullination processes as well as the citrullination-associated autoimmune mechanisms involved in RA.
Data availability statement
The mass spectrometry proteomics data was deposited in the ProteomeXchange with a dataset identifier PXD037743.
Ethics statement
The study was approved by the local ethics committee of Union Hospital, Tongji Medical College (No. [2020] IEC-J(130)). The patients/participants provided their written informed consent to participate in this study.
Author contributions
All authors were involved in drafting the article or revising it critically for important intellectual content, and all authors approved the final version to be published. HG had full access to all of the data in the study and take responsibility for the integrity of the data and the accuracy of the data analysis. All authors contributed to the article and approved the submitted version.
Funding Statement
This work was supported by the Natural Science Foundation of China (32170906), Fundamental Research Funds for the Central University of Central China Normal University (CCNU20TS02112), and the Swedish Research Council (2019-01209).
Abbreviations
ACPA, anti-citrullinated protein antibodies; RA, rheumatoid arthritis; OA, osteoarthritis; SF, synovial fluid; Cit, citrullination; ELISA, enzyme-linked immunosorbent assay; FASP, filter-aided sample preparation protocol; MHC, Major histocompatibility complex; PAD, peptidylarginine deiminase; PDP, platelet derived microparticles; PTM, post-translational modification; TSP-1, thrombospondin-1; β-actin, beta actin; PF4, platelet factor 4.
Conflict of interest
The authors declare that this research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The reviewer SC declared a shared parent affiliation with the author RD at the time of review.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2023.1084283/full#supplementary-material
References
- 1. Schellekens GA, De Jong BA, Van Den Hoogen FH, Van De Putte LB, Van Venrooij WJ. Citrulline is an essential constituent of antigenic determinants recognized by rheumatoid arthritis-specific autoantibodies. J Clin Invest (1998) 101:273–81. doi: 10.1172/JCI1316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Hill JA, Bell DA, Brintnell W, Yue D, Wehrli B, Jevnikar AM, et al. Arthritis induced by posttranslationally modified (citrullinated) fibrinogen in DR4-IE transgenic mice. J Exp Med (2008) 205:967–79. doi: 10.1084/jem.20072051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Trouw LA, Rispens T, Toes REM. Beyond citrullination: Other post-translational protein modifications in rheumatoid arthritis. Nat Rev Rheumatol (2017) 13:331–9. doi: 10.1038/nrrheum.2017.15 [DOI] [PubMed] [Google Scholar]
- 4. Ge C, Holmdahl R. The structure, specificity and function of anti-citrullinated protein antibodies. Nat Rev Rheumatol (2019) 15:503–8. doi: 10.1038/s41584-019-0244-4 [DOI] [PubMed] [Google Scholar]
- 5. Rantapää-Dahlqvist S, De Jong BA, Berglin E, Hallmans G, Wadell G, Stenlund H, et al. Antibodies against cyclic citrullinated peptide and IgA rheumatoid factor predict the development of rheumatoid arthritis. Arthritis Rheum (2003) 48:2741–9. doi: 10.1002/art.11223 [DOI] [PubMed] [Google Scholar]
- 6. Johansson L, Pratesi F, Brink M, Ärlestig L, D'amato C, Bartaloni D, et al. Antibodies directed against endogenous and exogenous citrullinated antigens pre-date the onset of rheumatoid arthritis. Arthritis Res Ther (2016) 18:127. doi: 10.1186/s13075-016-1031-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. van der Woude D, Rantapää-Dahlqvist S, Ioan-Facsinay A, Onnekink C, Schwarte CM, Verpoort KN, et al. Epitope spreading of the anti-citrullinated protein antibody response occurs before disease onset and is associated with the disease course of early arthritis. Ann Rheum Dis (2010) 69:1554–61. doi: 10.1136/ard.2009.124537 [DOI] [PubMed] [Google Scholar]
- 8. van Beers JJ, Schwarte CM, Stammen-Vogelzangs J, Oosterink E, Božič B, Pruijn GJ. The rheumatoid arthritis synovial fluid citrullinome reveals novel citrullinated epitopes in apolipoprotein e, myeloid nuclear differentiation antigen, and β-actin. Arthritis Rheum (2013) 65:69–80. doi: 10.1002/art.37720 [DOI] [PubMed] [Google Scholar]
- 9. Haag S, Schneider N, Mason DE, Tuncel J, Andersson IE, Peters EC, et al. Identification of new citrulline-specific autoantibodies, which bind to human arthritic cartilage, by mass spectrometric analysis of citrullinated type II collagen. Arthritis Rheumatol (2014) 66:1440–9. doi: 10.1002/art.38383 [DOI] [PubMed] [Google Scholar]
- 10. Ytterberg AJ, Joshua V, Reynisdottir G, Tarasova NK, Rutishauser D, Ossipova E, et al. Shared immunological targets in the lungs and joints of patients with rheumatoid arthritis: identification and validation. Ann Rheum Dis (2015) 74:1772–7. doi: 10.1136/annrheumdis-2013-204912 [DOI] [PubMed] [Google Scholar]
- 11. Tutturen AE, Fleckenstein B, De Souza GA. Assessing the citrullinome in rheumatoid arthritis synovial fluid with and without enrichment of citrullinated peptides. J Proteome Res (2014) 13:2867–73. doi: 10.1021/pr500030x [DOI] [PubMed] [Google Scholar]
- 12. Spengler J, Lugonja B, Ytterberg AJ, Zubarev RA, Creese AJ, Pearson MJ, et al. Release of active peptidyl arginine deiminases by neutrophils can explain production of extracellular citrullinated autoantigens in rheumatoid arthritis synovial fluid. Arthritis Rheumatol (2015) 67:3135–45. doi: 10.1002/art.39313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Maouia A, Rebetz J, Kapur R, Semple JW. The immune nature of platelets revisited. Transfus Med Rev (2020) 34:209–20. doi: 10.1016/j.tmrv.2020.09.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Gaertner F, Ahmad Z, Rosenberger G, Fan S, Nicolai L, Busch B, et al. Migrating platelets are mechano-scavengers that collect and bundle bacteria. Cell (2017) 171:1368–1382.e1323. doi: 10.1016/j.cell.2017.11.001 [DOI] [PubMed] [Google Scholar]
- 15. Banerjee M, Huang Y, Joshi S, Popa GJ, Mendenhall MD, Wang QJ, et al. Platelets endocytose viral particles and are activated via TLR (Toll-like receptor) signaling. Arterioscler Thromb Vasc Biol (2020) 40:1635–50. doi: 10.1161/ATVBAHA.120.314180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Boilard E, Nigrovic PA, Larabee K, Watts GF, Coblyn JS, Weinblatt ME, et al. Platelets amplify inflammation in arthritis via collagen-dependent microparticle production. Science (2010) 327:580–3. doi: 10.1126/science.1181928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Cloutier N, Tan S, Boudreau LH, Cramb C, Subbaiah R, Lahey L, et al. The exposure of autoantigens by microparticles underlies the formation of potent inflammatory components: The microparticle-associated immune complexes. EMBO Mol Med (2013) 5:235–49. doi: 10.1002/emmm.201201846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Cloutier N, Paré A, Farndale RW, Schumacher HR, Nigrovic PA, Lacroix S, et al. Platelets can enhance vascular permeability. Blood (2012) 120:1334–43. doi: 10.1182/blood-2012-02-413047 [DOI] [PubMed] [Google Scholar]
- 19. Mott PJ, Lazarus AH. CD44 antibodies and immune thrombocytopenia in the amelioration of murine inflammatory arthritis. PloS One (2013) 8:e65805. doi: 10.1371/journal.pone.0065805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Habets KL, Trouw LA, Levarht EW, Korporaal SJ, Habets PA, De Groot P, et al. Anti-citrullinated protein antibodies contribute to platelet activation in rheumatoid arthritis. Arthritis Res Ther (2015) 17:209. doi: 10.1186/s13075-015-0665-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Sadoul K, Wang J, Diagouraga B, Vitte AL, Buchou T, Rossini T, et al. HDAC6 controls the kinetics of platelet activation. Blood (2012) 120:4215–8. doi: 10.1182/blood-2012-05-428011 [DOI] [PubMed] [Google Scholar]
- 22. Aslan JE, Rigg RA, Nowak MS, Loren CP, Baker-Groberg SM, Pang J, et al. Lysine acetyltransfer supports platelet function. J Thromb Haemost (2015) 13:1908–17. doi: 10.1111/jth.13070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lian L, Suzuki A, Hayes V, Saha S, Han X, Xu T, et al. Loss of ATE1-mediated arginylation leads to impaired platelet myosin phosphorylation, clot retraction, and in vivo thrombosis formation. Haematologica (2014) 99:554–60. doi: 10.3324/haematol.2013.093047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Dowal L, Yang W, Freeman MR, Steen H, Flaumenhaft R. Proteomic analysis of palmitoylated platelet proteins. Blood (2011) 118:e62–73. doi: 10.1182/blood-2011-05-353078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Rocheleau AD, Melrose AR, Cunliffe JM, Klimek J, Babur Ö, Tassi Yunga S, et al. Identification, quantification, and system analysis of protein n-ϵ lysine methylation in anucleate blood platelets. Proteomics (2019) 19:e1900001. doi: 10.1002/pmic.201900001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Arnett FC, Edworthy SM, Bloch DA, Mcshane DJ, Fries JF, Cooper NS, et al. The American rheumatism association 1987 revised criteria for the classification of rheumatoid arthritis. Arthritis Rheum (1988) 31:315–24. doi: 10.1002/art.1780310302 [DOI] [PubMed] [Google Scholar]
- 27. Su YL, Wang B, Hu MD, Cui ZW, Wan J, Bai H, et al. Site-specific n-glycan characterization of grass carp serum IgM. Front Immunol (2018) 9:2645. doi: 10.3389/fimmu.2018.02645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Wiśniewski JR, Zougman A, Nagaraj N, Mann M. Universal sample preparation method for proteome analysis. Nat Methods (2009) 6:359–62. doi: 10.1038/nmeth.1322 [DOI] [PubMed] [Google Scholar]
- 29. Tyanova S, Temu T, Cox J. The MaxQuant computational platform for mass spectrometry-based shotgun proteomics. Nat Protoc (2016) 11:2301–19. doi: 10.1038/nprot.2016.136 [DOI] [PubMed] [Google Scholar]
- 30. Lee CY, Wang D, Wilhelm M, Zolg DP, Schmidt T, Schnatbaum K, et al. Mining the human tissue proteome for protein citrullination. Mol Cell Proteomics (2018) 17:1378–91. doi: 10.1074/mcp.RA118.000696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Maurais AJ, Salinger AJ, Tobin M, Shaffer SA, Weerapana E, Thompson PR. A streamlined data analysis pipeline for the identification of sites of citrullination. Biochemistry (2021) 60:2902–14. doi: 10.1021/acs.biochem.1c00369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Mondal S, Thompson PR. Protein arginine deiminases (PADs): Biochemistry and chemical biology of protein citrullination. Acc Chem Res (2019) 52:818–32. doi: 10.1021/acs.accounts.9b00024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Fert-Bober J, Giles JT, Holewinski RJ, Kirk JA, Uhrigshardt H, Crowgey EL, et al. Citrullination of myofilament proteins in heart failure. Cardiovasc Res (2015) 108:232–42. doi: 10.1093/cvr/cvv185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Bennike TB, Ellingsen T, Glerup H, Bonderup OK, Carlsen TG, Meyer MK, et al. Proteome analysis of rheumatoid arthritis gut mucosa. J Proteome Res (2017) 16:346–54. doi: 10.1021/acs.jproteome.6b00598 [DOI] [PubMed] [Google Scholar]
- 35. Faigle W, Cruciani C, Wolski W, Roschitzki B, Puthenparampil M, Tomas-Ojer P, et al. Brain citrullination patterns and T cell reactivity of cerebrospinal fluid-derived CD4(+) T cells in multiple sclerosis. Front Immunol (2019) 10:540. doi: 10.3389/fimmu.2019.00540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Darrah E, Rosen A, Giles JT, Andrade F. Peptidylarginine deiminase 2, 3 and 4 have distinct specificities against cellular substrates: novel insights into autoantigen selection in rheumatoid arthritis. Ann Rheum Dis (2012) 71:92–8. doi: 10.1136/ard.2011.151712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Chang X, Yamada R, Suzuki A, Sawada T, Yoshino S, Tokuhiro S, et al. Localization of peptidylarginine deiminase 4 (PADI4) and citrullinated protein in synovial tissue of rheumatoid arthritis. Rheumatol (Oxford) (2005) 44:40–50. doi: 10.1093/rheumatology/keh414 [DOI] [PubMed] [Google Scholar]
- 38. Kinloch A, Lundberg K, Wait R, Wegner N, Lim NH, Zendman AJ, et al. Synovial fluid is a site of citrullination of autoantigens in inflammatory arthritis. Arthritis Rheum (2008) 58:2287–95. doi: 10.1002/art.23618 [DOI] [PubMed] [Google Scholar]
- 39. Suzuki A, Yamada R, Chang X, Tokuhiro S, Sawada T, Suzuki M, et al. Functional haplotypes of PADI4, encoding citrullinating enzyme peptidylarginine deiminase 4, are associated with rheumatoid arthritis. Nat Genet (2003) 34:395–402. doi: 10.1038/ng1206 [DOI] [PubMed] [Google Scholar]
- 40. Nakashima K, Hagiwara T, Yamada M. Nuclear localization of peptidylarginine deiminase V and histone deimination in granulocytes. J Biol Chem (2002) 277:49562–8. doi: 10.1074/jbc.M208795200 [DOI] [PubMed] [Google Scholar]
- 41. Blachère NE, Parveen S, Frank MO, Dill BD, Molina H, Orange DE. High-titer rheumatoid arthritis antibodies preferentially bind fibrinogen citrullinated by peptidylarginine deiminase 4. Arthritis Rheumatol (2017) 69:986–95. doi: 10.1002/art.40035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Damgaard D, Bawadekar M, Senolt L, Stensballe A, Shelef MA, Nielsen CH. Relative efficiencies of peptidylarginine deiminase 2 and 4 in generating target sites for anti-citrullinated protein antibodies in fibrinogen, alpha-enolase and histone H3. PloS One (2018) 13:e0203214. doi: 10.1371/journal.pone.0203214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Kinloch A, Tatzer V, Wait R, Peston D, Lundberg K, Donatien P, et al. Identification of citrullinated alpha-enolase as a candidate autoantigen in rheumatoid arthritis. Arthritis Res Ther (2005) 7:R1421–1429. doi: 10.1186/ar1845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Shevchuk O, Begonja AJ, Gambaryan S, Totzeck M, Rassaf T, Huber TB, et al. Proteomics: A tool to study platelet function. Int J Mol Sci (2021) 22(9):4776. doi: 10.3390/ijms22094776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Loos T, Mortier A, Gouwy M, Ronsse I, Put W, Lenaerts JP, et al. Citrullination of CXCL10 and CXCL11 by peptidylarginine deiminase: A naturally occurring posttranslational modification of chemokines and new dimension of immunoregulation. Blood (2008) 112:2648–56. doi: 10.1182/blood-2008-04-149039 [DOI] [PubMed] [Google Scholar]
- 46. Proost P, Loos T, Mortier A, Schutyser E, Gouwy M, Noppen S, et al. Citrullination of CXCL8 by peptidylarginine deiminase alters receptor usage, prevents proteolysis, and dampens tissue inflammation. J Exp Med (2008) 205:2085–97. doi: 10.1084/jem.20080305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Struyf S, Noppen S, Loos T, Mortier A, Gouwy M, Verbeke H, et al. Citrullination of CXCL12 differentially reduces CXCR4 and CXCR7 binding with loss of inflammatory and anti-HIV-1 activity via CXCR4. J Immunol (2009) 182:666–74. doi: 10.4049/jimmunol.182.1.666 [DOI] [PubMed] [Google Scholar]
- 48. Knijff-Dutmer EA, Koerts J, Nieuwland R, Kalsbeek-Batenburg EM, Van De Laar MA. Elevated levels of platelet microparticles are associated with disease activity in rheumatoid arthritis. Arthritis Rheum (2002) 46:1498–503. doi: 10.1002/art.10312 [DOI] [PubMed] [Google Scholar]
- 49. Avina-Zubieta JA, Thomas J, Sadatsafavi M, Lehman AJ, Lacaille D. Risk of incident cardiovascular events in patients with rheumatoid arthritis: A meta-analysis of observational studies. Ann Rheum Dis (2012) 71:1524–9. doi: 10.1136/annrheumdis-2011-200726 [DOI] [PubMed] [Google Scholar]
- 50. Dekkers JS, Verheul MK, Stoop JN, Liu B, Ioan-Facsinay A, Van Veelen PA, et al. Breach of autoreactive b cell tolerance by post-translationally modified proteins. Ann Rheum Dis (2017) 76:1449–57. doi: 10.1136/annrheumdis-2016-210772 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The mass spectrometry proteomics data was deposited in the ProteomeXchange with a dataset identifier PXD037743.