Abstract
Nitroaromatic compounds are xenobiotics that have found multiple applications in the synthesis of foams, pharmaceuticals, pesticides, and explosives. These compounds are toxic and recalcitrant and are degraded relatively slowly in the environment by microorganisms. 2,4,6-Trinitrotoluene (TNT) is the most widely used nitroaromatic compound. Certain strains of Pseudomonas and fungi can use TNT as a nitrogen source through the removal of nitrogen as nitrite from TNT under aerobic conditions and the further reduction of the released nitrite to ammonium, which is incorporated into carbon skeletons. Phanerochaete chrysosporium and other fungi mineralize TNT under ligninolytic conditions by converting it into reduced TNT intermediates, which are excreted to the external milieu, where they are substrates for ligninolytic enzymes. Most if not all aerobic microorganisms reduce TNT to the corresponding amino derivatives via the formation of nitroso and hydroxylamine intermediates. Condensation of the latter compounds yields highly recalcitrant azoxytetranitrotoluenes. Anaerobic microorganisms can also degrade TNT through different pathways. One pathway, found in Desulfovibrio and Clostridium, involves reduction of TNT to triaminotoluene; subsequent steps are still not known. Some Clostridium species may reduce TNT to hydroxylaminodinitrotoluenes, which are then further metabolized. Another pathway has been described in Pseudomonas sp. strain JLR11 and involves nitrite release and further reduction to ammonium, with almost 85% of the N-TNT incorporated as organic N in the cells. It was recently reported that in this strain TNT can serve as a final electron acceptor in respiratory chains and that the reduction of TNT is coupled to ATP synthesis. In this review we also discuss a number of biotechnological applications of bacteria and fungi, including slurry reactors, composting, and land farming, to remove TNT from polluted soils. These treatments have been designed to achieve mineralization or reduction of TNT and immobilization of its amino derivatives on humic material. These approaches are highly efficient in removing TNT, and increasing amounts of research into the potential usefulness of phytoremediation, rhizophytoremediation, and transgenic plants with bacterial genes for TNT removal are being done.
Life on this planet is based on the continuous cycling of elements. In recent years the massive mobilization of natural resources and the industrial synthesis of chemicals have generated a number of environmental problems as a consequence of the limited incorporation of natural and synthesized molecules into ongoing biological cycles. This is particularly true for xenobiotic compounds, which exhibit structural elements or substituents that are rarely found in natural products. The chemical properties of xenobiotic compounds determine their toxicity, their persistence in the environment, and the manner in which they are degraded by microorganisms. Apart from chloramphenicol, nitropyoluteorin (161), oxypyrrolnitrin (101), and phidolopin (232), no other natural nitroaromatic compound has been described to date, which probably explains why nitroaromatic compounds are relatively refractory to biological degradation and why xenobiotic compounds are not easily incorporated into biogenic element cycles, with the consequent impact on living organisms and ecosystems.
A number of nitroaromatic compounds such as nitrobenzenes, o- and p-nitrotoluene, 2,4-dinitrotoluene, and nitrobenzoates are mineralized at a relatively slow pace by microorganisms. These chemicals have multiple applications in the synthesis of polyurethane foams, herbicides, insecticides, pharmaceuticals, and explosives (99), all of which are more recalcitrant than the raw material from which they are synthesized. Comprenhensive reviews of catabolic pathways leading to the degradation of nitroaromatic compounds are available (27, 53, 54, 79, 87, 88, 106, 122, 123, 141, 147, 170, 210–212, 243).
The most widely used nitroaromatic compound is 2,4,6-trinitrotoluene (TNT). TNT is more recalcitrant than mono- and dinitrotoluenes mainly because of the symmetric location of the nitro groups on the aromatic ring, an arrangement that limits attack by classic dioxygenase enzymes involved in the microbial metabolism of aromatic compounds (186). Therefore, the chemical structure of TNT influences its biodegradability. This makes it worth analyzing the mechanism of biodegradation of this molecule in detail.
In this review we first analyze the chemical structure of TNT and then describe how aerobic and anaerobic microorganisms attack it. Certain strains of Pseudomonas and of fungi are able to use TNT as a nitrogen source by removing nitrogen under aerobic conditions (30, 63, 119, 223). This seems to involve, in some cases, the removal of nitro groups from the aromatic ring, which may occur via the formation of an intermediate π-Meisenheimer complex (77), and the further reduction of the released nitrite to ammonium, which is incorporated into carbon skeletons. It has also been suggested that hydroxylaminodinitrotoluenes are key metabolites for the release of nitrogen from the TNT ring by Pseudomonas pseudoalcaligenes (74), although the stage of metabolism when this occurs and the form in which assimilable nitrogen is made available to the cells are unknown.
A number of reports on the mineralization of TNT by Phanerochaete chrysosporium and other fungi that mineralize TNT under ligninolytic conditions are available (47, 107, 129, 151, 164, 183, 194, 196, 219–221, 235–237). Strict anaerobic bacteria of the genera Clostridium and Desulfovibrio (6) can degrade TNT via triaminotoluene, which might subsequently be metabolized to methylphloroglucinol and p-cresol, although conclusive biochemical evidence for this pathway is still needed (54, 60, 64, 80, 142, 180). It has also been suggested that some Clostridium strains reduce TNT to the corresponding hydroxylaminodinitrotoluenes, which, upon Bamberger rearragement, yield aminophenols (111, 113). Another pathway has been described in Pseudomonas sp. strain JLR11 and involves nitrite release and further reduction to ammonium, with almost 85% of the nitrogen of TNT incorporated as organic nitrogen in the cells (68, 69). In this review we intend to highlight the recent discovery of TNT as an alternative electron acceptor in respiratory chains of a facultative Pseudomonas strain (69). We also discuss a number of biotechnological applications of bacteria and fungi, including slurry reactors, composting, and land farming, to remove TNT from polluted soils. These treatments have been designed to achieve either mineralization or reduction of TNT, as well as immobilization of its amino derivatives on humic material. These approaches are highly efficient in removing TNT, and increasing amounts of research into the potential usefulness of phytoremediation, rhizophytoremediation, and transgenic plants with bacterial genes for TNT removal are being done.
CHEMISTRY OF TNT
The π electrons from the aromatic ring of TNT are removed by the electronegative nitro groups, a process that makes the nucleus electrophilic. The nitro group consists of two different elements, N and O, which are both highly electronegative. Oxygen is even more electronegative than the nitrogen atom; hence, the N-O bond is polarized. The partially positive charge of the nitrogen atom, combined with its high electronegativity, makes the nitro group easily reducible (175). Reduction of nitro groups on aromatic rings is widely distributed among living organisms.
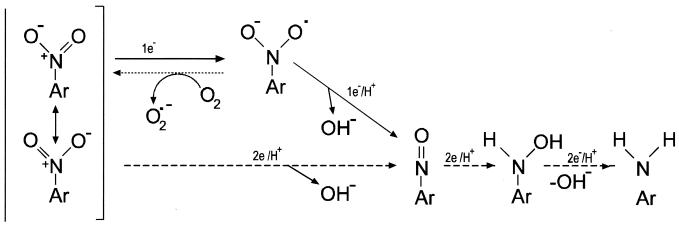
In vitro assays showed that in cell extracts prepared from a number of microorganisms and higher eukaryotes, reduction of the nitro group occurs as a series of two-electron transfers which yield the nitroso, hydroxylamino, and amino derivatives of the parent compound (Fig. 1). This reduction is referred to as oxygen independent (type I), and no radicals are formed (45). Nitroreductase enzymes able to perform this reaction use pairs of electrons donated by reduced pyridine nucleotides (10, 25, 43, 44–46, 86, 98, 130–132, 145, 159, 162, 184, 209, 231, 244, 249, 258, 261–263). Other enzymes that reduce nitroaromatic compounds include aldehyde oxidase (254), dihydrolipic amide dehydrogenase (228), cytochrome b5 reductase (148), diaphorases (126), hydrogenases (58, 149), xanthine oxidase (46), and CO dehydrogenase (111). The nitro group can also be reduced in vitro through single-electron transfers, which form a nitroanion radical (Fig. 1). This radical is a putative intermediate in the reduction of a nitro group to a nitroso group, but it can react with oxygen to produce a superoxide anion and alter the original nitroaromatic compound (Fig. 1). The enzymes that catalyze this reaction are known as oxygen sensitive (type II) nitroreductases and are found in bacteria such as Clostridium (9) and Escherichia coli (171).
FIG. 1.
Mechanisms for the reduction of nitro groups in nitroaromatic compounds. The first step in nitro group reduction can be achieved through one-electron transfer (solid line) or two-electron transfer (dashed line). The first mechanism produces a nitroanion radical that could react with oxygen to form a superoxide radical and the original nitroaromatic compound through a futile cycle (dotted line). If the mechanism occurs via the transfer of two electrons, the nitroso derivative formed is the first putative intermediate; following two consecutive electron transfers, a hydroxylamine and an aromatic amine are produced. The scheme has been adapted from reference 212.
The nitroso and hydroxylamino groups, responsible for the toxicity of nitroaromatic compounds, react with biological molecules (49) and cause chemical mutagenesis and carcinogenesis (16, 20, 36, 51, 85, 110, 178, 189, 213, 224, 226, 234, 255, 260). Complete reduction of the nitro group to an amino group seems to decrease the mutagenic effect of the compound (50, 82, 227). Hydroxylaminodinitrotoluenes also cause hemotoxic symptoms in workers exposed to TNT, since oxyhemoglobin is oxidized by molecular oxygen in the presence of hydroxylaminodinitrotoluenes to yield ferrihemoglobin and nitrosodinitrotoluenes (15, 52, 128, 144, 177). In blood cells this reaction becomes a futile cycle because nitrosodinitrotoluenes are reduced back to hydroxylaminodinitrotoluenes, which again react with oxyhemoglobin.
Abiotic reduction of the nitro groups to the corresponding amines also occurs in sediments, soils, and aquifers (91, 93). Numerous potential electron donors are present in natural systems (e.g., reduced iron species, reduced sulfur species, and natural organic matter) that may reduce nitroaromatic compounds in abiotic reactions (84, 109, 133, 249). TNT also reacts with the siloxane surface of clays to yield covalent complexes (92, 246–248). The roles that living organisms play in these biogeochemical processes are of interest, but this is still a relatively unexplored area of research (108, 109, 192). The behavior of TNT in soil will be examined in depth in “Applications in TNT bioremediation” (below).
In the reduction of the nitro groups of TNT, the reduction of the first nitro group is generally much more rapid than that of the rest. The conversion of nitro to amino groups decreases the electron deficiency of the nitroaromatic ring, and consequently a lower redox potential is required to reduce the rest of the nitro group of the molecule. Because of this, the formation of triaminotoluene requires values below −200 mV, which are found only in anoxic environments (81, 109). Barrows et al. (17) hypothesized that the step that establishes regioselectivity in the abiotic reduction process is the first proton transfer to the radical anion that is created after the initial one-electron reduction and that the nitro group with the greater negative charge localization is likely to be the one most readily protonated. In TNT, the para position is much more easily reduced than the ortho nitro groups.
As Averill (13) pointed out, “the nitro group reduction of nitroaromatic compounds (ArNO2) by bacteria generally proceeds in two-electron processes such as the reduction of nitrate during denitrification”. One might therefore expect the chemical processes in the reduction of NO2− and ArNO2 to be analogous, since both the inorganic and the organic species contain nitrogen in the same oxidation state (+3) and two N-O bonds; however, the arrangement of the valence electron on the nitrogen atom, especially the absence of lone pairs, makes ArNO2 more similar to nitrate (NO3−) than to nitrite (NO2−).
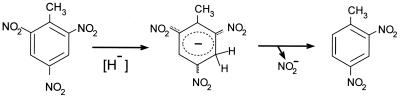
The deficiency of electrons in the aromatic nucleus of TNT makes nucleophilic attack on the ring possible; as a result, a nonaromatic structure such as a Meisenheimer ς complex can be formed (70, 125, 229). These compounds are red-orange and have a negative charge (Fig. 2). Such complexes can be formed in vivo by hydride attack from a reduced pyridine nucleotide. The Meisenheimer complex can be rearomatized after nitrite release, with the formation of dinitrotoluenes. Oxygen is not neccesary for the formation of this kind of compound, and so this process is an alternative for nitroaromatic metabolism when oxygenic removal of the nitro groups is not possible, as in TNT.
FIG. 2.
TNT-Meisenheimer complex formation. The hydride ion can be donated by NAD(P)H, giving rise to a Meisenheimer complex. Aromaticity is restored on the release of a nitro group.
AEROBIC METABOLISM OF TNT BY BACTERIA
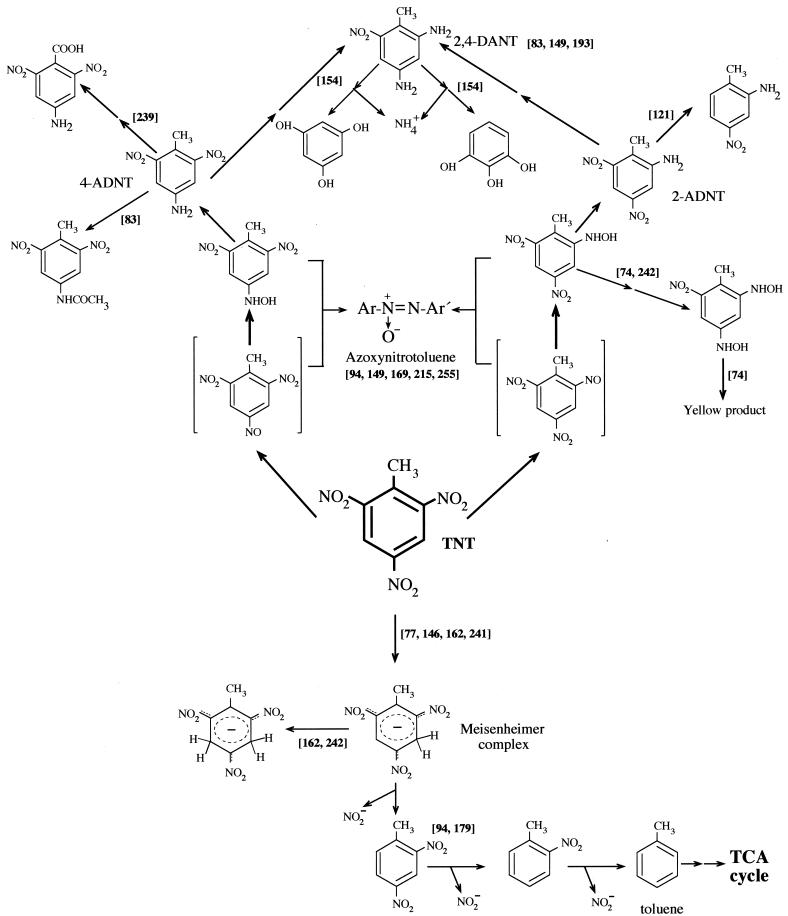
The aim of this section is to review the best-known mechanisms of aerobic metabolism of TNT by bacteria (Fig. 3). Oxygenolytic metabolism of the nitro groups of nitroaromatic compounds is limited to mononitroaromatic and dinitroaromatic compounds (8, 37, 95–97, 105, 155, 156, 158, 182, 191, 211, 214, 217, 225, 261). Oxygenolytic metabolism for aromatic compounds by bacteria does not occur in TNT because of its chemical properties, as described above. Therefore, even in the presence of oxygen, this explosive must be transformed by reductive metabolism.
FIG. 3.
Pathways for the aerobic metabolism of TNT in aerobic microorganisms. The scheme is based on articles cited in the text, which are indicated in the figure in square brackets. When the conversion of a compound to another is believed to occur through a series of intermediates, the steps are indicated by two arrows. TCA, trichloroacetic acid.
In most of the cases described to date, aerobic bacteria tend to transform the TNT molecule by reducing one or two nitro groups to hydroxylamino or amino groups and to produce different isomers of aminonitroaromatic compounds, which in turn usually accumulate in the culture medium without further metabolism. It has also been reported that partially reduced forms of TNT react among themselves in the presence of oxygen to form recalcitrant azoxytetranitrotoluenes (94), which causes a higher rate of mutations than does TNT (82). These transformation reactions remove TNT but yield highly recalcitrant products that are not metabolizable by most of the microorganisms that produce them (16, 36, 110, 118, 215, 226).
Only a few aerobes able to use TNT as a nitrogen or carbon source have been reported, and mineralization of this compound has been described even less frequently (Table 1). Elimination of the nitro group is required to decrease the electrophilic nature of the molecule and allow dioxygenases to use di- or mononitroaromatic compounds as suitable substrates. This fact led researchers to isolate microorganisms able to use TNT as the sole nitrogen source in mineral medium supplemented with additional carbon sources (30, 63, 119, 233). None of these researchers reported TNT mineralization, because the different strains they isolated produced very low levels of 14CO2 from [14C]TNT.
TABLE 1.
Microorganisms reported to degrade or transform TNT under aerobic conditions
| Microorganism | Metabolism | Reference(s) |
|---|---|---|
| Bacillus sp. | Nitrite release from TNT and transformation to 2-amino-4-nitrotoluene | 121 |
| Enterobacter cloacae PB2 | TNT reduction to a Meisenheimer complex with nitrite release | 77 |
| Enterobacter sp. | TNT transformation to unknown polar compounds and 3% mineralization | 14 |
| Mycobacterium vaccae | 50% radioactivity from [14C]TNT incorporated into lipids | 239 |
| Mycobacterium sp. strain HL4NT-1 | Cells grown in 4-nitrotoluene transform TNT to hydride and dihydride Meisenheimer complex | 241, 242 |
| Pseudomonas aeruginosa | Nitrite release from TNT and transformation to 2-amino-4-nitrotoluene | 121 |
| Pseudomonas aeruginosa MA101 | Transforms TNT and ADNTs into highly polar compounds through an O2-dependent process | 7 |
| Pseudomonas fluorescens | An NADPH flavoprotein oxidoreductase catalyzes multiple transformations, generating ArNHOH, H-TNT and H2-TNT | 162 |
| Pseudomonas fluorescens B3468 | TNT degradation to phloroglucinol and pyrogallol; nitrogen of TNT is released as ammonium | 154 |
| Pseudomonas fluorescens | TNT transformation into ADNT, DANTs, and 4-acetamide-2-amino-6-nitrotoluene | 83 |
| Pseudomonas pseudoalcaligenes | Reduction of nitro groups and nitrite release from 2,4-dihydroxylamino-6-nitrotoluene | 74 |
| Pseudomonas savastanoi | Denitration of TNT to release nitrite and reduction of nitro group to amino | 146 |
| Pseudomonas sp. clone A | Strain able to denitrate TNT | 63 |
| Pseudomonas sp. strain CBS3 | Reduces TNT to DANT | 193 |
| Pseudomonas sp. strain FR2 | Reduces TNT to DANT | 149 |
| Pseudomonas sp. strain IIBx | First evidence of [14C]TNT mineralization | 233 |
| Pseudomonas sp. strain 1-2wt | First evidence of [14C]TNT mineralization | 233 |
| Pseudomonas sp. | TNT used as sole nitrogen source; TNT transformation to 2ADNT and 4ADNT | 119 |
| Rhodococcus erythropolis | Cells growing on picric acid catalyze ring hydrogenation, forming hydride and dihydride TNT-Meisenheimer complexes | 242 |
| Serratia marcensens | TNT as sole carbon and energy source in presence of Tween 80 | 153 |
| Staphylococcus sp. | Nitrite release from TNT and transformation to 2-amino-4-nitrotoluene | 121 |
| Stenotrophomonas maltophilia | TNT as sole nitrogen source | 160 |
Duque et al. (63) isolated a Pseudomonas strain from soil around an explosives factory and found that it was able to grow with TNT as the sole nitrogen source. Nitrite accumulated in culture supernatants, and the sequential removal of all three nitro groups was suggested based on the identification of dinitrotoluenes, mononitrotoluenes, and toluene in the culture supernatants. The elimination of TNT by this Pseudomonas strain was proposed to occur via the production of a Meisenheimer complex (94, 179). This kind of complex was also reported by many authors as a metabolite involved in polynitroaromatic metabolism by bacteria (77, 185, 242). Lenke and Knackmuss (139) isolated a Rhodococcus erythropolis strain able to use 2,4,6-trinitrophenol (picric acid) as a nitrogen source after initial reduction of the aromatic ring by a hydride ion (186). Vorbeck et al. (242) suggested that a similar hydrogenation reaction of the aromatic ring of TNT also occurred in resting cells of R. erythropolis, although the strain failed to use TNT as nitrogen source, probably because the corresponding TNT complex undergoes neither nitrite elimination nor rearomatization to 2,4- or 2,6-dinitrotoluene under physiological conditions (138, 242). This route of dihydride complex formation is therefore unproductive for the catabolism of TNT in Rhodococcus spp. In contrast, Haïdour and Ramos (94) reported the formation of 2,4-dinitrotoluene (2,4-DNT) as a result of the in vitro metabolism of the TNT-Meisenheimer complex by Pseudomonas clone A (Fig. 3); however, no stoichiometric analyses were done by these authors, and this reaction could not be confirmed by Vorbeck et al. (242).
Degradation of TNT to 2,4-DNT is desirable, since previous work has shown that other Pseudomonas strains can metabolize this nitroaromatic through dioxygenase attack to produce 4-methyl-5-nitrocatechol and that this is followed by oxidation, release of nitrite, and mineralization (96, 155, 158, 214).
Enterobacter cloacae strain PB2, originally isolated for its ability to use nitrate esters, can also slowly grow with TNT as the sole source of nitrogen. French et al. (77) obtained in vitro evidence for the reduction of TNT with purified pentaerythritol tetranitrate (PETN) reductase and showed that this enzyme reduced TNT to its hydride Meisenheimer complex with concomitant NADPH oxidation and release of nitrite. Because hydroxylaminodinitrotoluene and aminodinitrotoluene were detected, the enzyme was also assumed to have other nitroreductase activities. The gene encoding PETN reductase, designated onr (for “organic nitrate reductase”), has been cloned and overexpressed in E. coli. A recombinant E. coli strain that expressed PETN reductase was also able to release nitrite from TNT. This is the first report of the reduction of the aromatic ring of TNT by a purified enzyme with the concomitant release of nitrite from TNT. A biotechnological application of this enzyme was recently reported (75) and will be discussed below (see “Applications in TNT bioremediation”). Recently, Pak et al. (162) have reported the purification of an NADH-dependent flavoprotein oxidoreductase (XenB) (26) from Pseudomonas fluorescens which catalyzed the reduction of TNT by adding a hydride to the aromatic ring with the concomitant formation of dihydride Meisenheimer complex or catalyzed the reduction of nitro groups.
Other authors also reported denitration reactions in TNT. Martin et al. (146) reported such a reaction in a Pseudomonas savastatoni strain unable to mineralize significant quantities of TNT (less than 1%) but able to transform TNT into 2,4-DNT and nitrite. This reaction seems to be enhanced by removing ammonium and adding nitrite to the growth medium. In contrast, the addition of glucose enhanced 2-amino-4,6-dinitrotoluene (2-ADNT) and 4-amino-2,6-dinitrotoluene (4-ADNT) production and decreased the denitration of TNT.
Recently, Kalafut et al. (121) reported a promising aerobic route for TNT metabolism which led to the production of 2-amino-4-nitrotoluene. This metabolite results from the loss of one of the original nitro groups, leaving two adjacent unsubstituted carbon atoms that can serve as a substrate for oxygenases. This compound was detected in cultures of three different strains of aerobic bacteria (Pseudomonas aeruginosa, Bacillus sp., and Staphylococcus sp.). However, no further details are known.
Bae et al. (14) reported the transformation of TNT to polar products by a strain of Enterobacter. These authors found 3% mineralization, conversion of 80% of the TNT to unidentified polar products, and conversion of 13% to a biomass-associated fraction. Changes in the nature of the TNT molecule leading to increased polarity may indicate hydroxylation or cleavage of the aromatic ring. Such transformations may facilitate further metabolism by other organisms, but characterization of the unknown products is necessary.
Vanderberg et al. (239) isolated a Mycobacterium vaccae strain that was able to metabolize TNT in the presence of propane as the carbon and energy source. Usual TNT reduction metabolites were found, but two novel oxidized metabolites were generated during TNT catabolism: aminodinitrobenzoic acid and diamino-nitrobenzylmethyl ether. The latter compound seems to be a dead-end product, and the authors proposed the o-methylation of the corresponding benzylalcohol as a mechanism of detoxification. Although mineralization was not observed, a substantial portion of the radioactivity from ring-labeled TNT (ca. 50%) was incorporated into the cellular lipid fraction. These results suggest that ring cleavage occurred prior to the incorporation of radiolabeled carbon into polar lipids. Because of its potential interest for biodegradation, further studies of the ring cleavage process in this strain are expected.
The reduction of nitro group compounds to amino compounds has been described as a gratuitous reaction, whereas partial reduction of nitroaromatic compounds to hydroxylamino derivatives has been identified as a key reaction in productive catabolic routes (90, 156). Fiorella and Spain (74) proposed a mechanism of partial reduction of TNT on the basis of their work with cells and cell extracts of nitrobenzene-grown P. pseudoalcaligenes (Fig. 3). These authors identified 2-hydroxylamino-4,6-dinitrotoluene and 2,4-dihydroxy-6-nitrotoluene; the latter seems to be produced by nitrobenzene reductase, and in the presence of oxygen it was converted into an unidentified yellow compound (Fig. 3). The release of nitrogen from the above aromatic compounds has not yet been explained, but the authors suggested that it may occur via the release of nitrite (small amounts were detected). Vorbeck et al. (242) also focused on hydroxylaminoaromatic compounds as key intermediates that make the nitrogen in TNT available for growth. TNT nitrogen users were found to convert TNT to 2-hydroxylamino-4,6-dinitrotoluene and 4-hydroxylamino-2,6-dinitrotoluene, together with a more polar compound that the latter ones. The authors suggested that these strains obtained N for growth from the above-cited hydroxylaminonitrotoluenes, although it is unclear at which stage of metabolism and in which form assimilable nitrogen was made available to the bacterial cells. The removal of N from the TNT ring through hydroxylamine aromatic compounds or via the formation of Meisenheimer complexes seems to explain the aerobic mechanism of TNT degradation by bacteria.
The unspecific reduction of TNT to amino derivatives, which usually accumulate, is a widely distributed reaction among aerobic bacteria (Fig. 3). Only a few reports have examined aerobic metabolism of these aminoaromatic compounds by microorganisms (7, 83, 154). Alvarez et al. (7) reported a strain of P. aeruginosa that aerobically metabolized aminodinitrotoluenes (ADNTs) to highly polar compounds which were not extractable with organic solvents. This transformation was oxygen dependent, and the products were not identified. No mineralization was detected, and the authors suggested that oxidative metabolism of the ADNTs might occur through oxidation of the methyl group and its subsequent decarboxylation or though the action of oxidative deaminases. Naumova et al. (154) cultured a strain of Pseudomonas fluorescens by using the partially reduced metabolites ADNTs as the nitrogen source. Incubation of cell extracts from cultures grown on TNT transformed DANT into phloroglucinol and pyrogallol, and 30% of the DANT nitrogen was released as ammonium. In spite of this activity in vitro, the microorganism was not able to use DANTs as a source of carbon and energy, and the physiological significance of this reaction in vivo remains to be clarified.
ANAEROBIC METABOLISM OF TNT BY BACTERIA
Anaerobic processes have the potential advantages of rapid reduction at low redox potential, which minimize oxidative polymerization of substrates because of the absence of oxygen. Anaerobic systems for TNT degradation have therefore been studied to attain more efficient rates of removal, since azoxynitrotoluene products are not formed (91, 94). Microorganisms reported to anoxically degrade or transform TNT are listed in Table 2.
TABLE 2.
Microorganisms reported to anoxically degrade or transform TNT
| Microorganism | Metabolism | Reference(s) |
|---|---|---|
| Clostridium acetobutylicum | Reduction of TNT to TAT | 64, 127 |
| Clostridium bifermentans CYS-1 | Degrades TNT to alyphatic polar compounds via 4ADNT and 2,4DANT | 205 |
| Clostridium bifermentans LJP-1 | Transforms TNT into TAT and phenolic compounds | 141 |
| Clostridium pasterianum | Reduction of TNT to TAT | 149 |
| Clostridium sordelii | Reduction of TNT to TAT | 64 |
| Clostridium sp. | Bamberger rearrengment of dihydroxylaminodinitrotoluene | 113 |
| Desulfovibrio sp. strain B | TNT as nitrogen source, toluene as a putative intermediate | 29 |
| Desulfovibrio sp. | TNT as the sole nitrogen source; reduction of TNT to TAT | 174 |
| Desulfovibrio sp. | Transforms TNT into TAT and DANT; 42% of radioactivity from [14C]TNT is associated with cell biomass | 60 |
| Escherichia coli | Reduction of TNT to TAT | 64 |
| Lactobacillus sp. | Reduction of TNT to TAT | 64 |
| Methanococcus sp. strain B | Reduction of TNT to DANT | 30 |
| Pseudomonas sp. strain JLR11 | TNT as nitrogen source; TNT as final electron acceptor | 68, 69 |
| Veillonella alkalescens | Reduction of TNT to TAT | 149 |
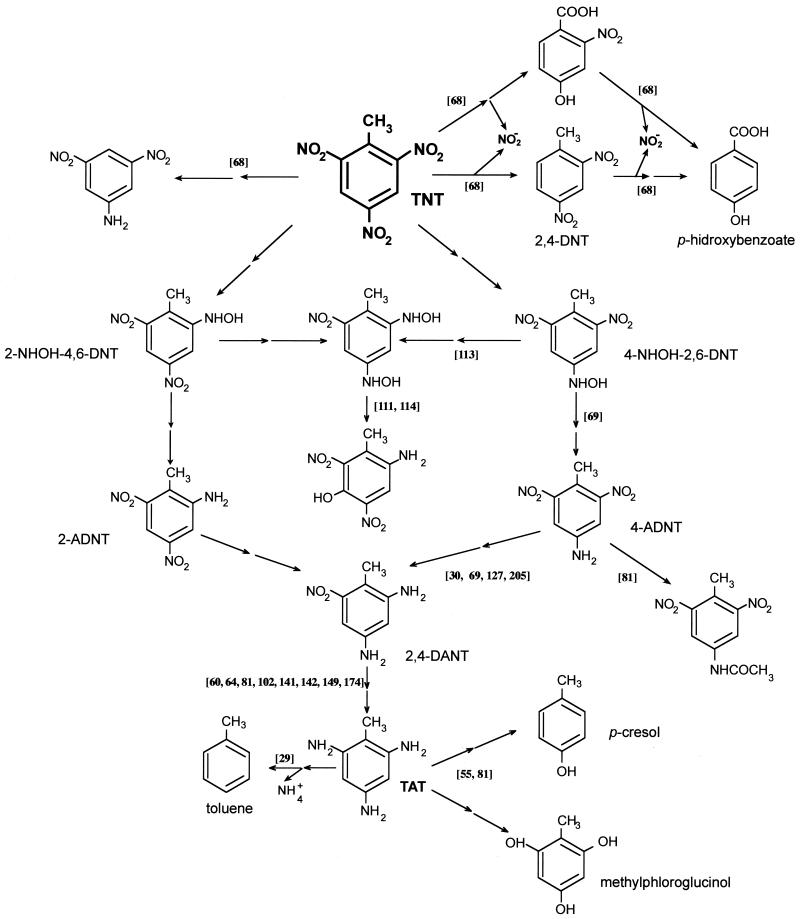
The earliest evidence of the anaerobic metabolism of TNT was reported by McCormick et al. (149), who showed that cell suspensions or crude extracts of Veillonella alkalescens could reduce TNT to triaminotoluene (TAT), with H2 as an electron donor (Fig. 4). In the last 10 years several reports have shown that this reduction is possible under anaerobic conditions because the low potential required for TAT makes this reaction impossible in aerobic microorganisms (60, 64, 80, 174).
FIG. 4.
Proposed mechanisms for anaerobic TNT metabolism in bacteria. The scheme is based on articles cited in the text, which are indicated in the figure in square brackets. When the conversion of a compound to another is believed to occur through a series of intermediates, the steps are indicated by two arrows.
Two genera have been extensively studied because of their anoxic metabolism of TNT: Clostridium and Desulfovibrio. The ability to reduce TNT anaerobically is a general phenomenon among Clostridium species (64). In cell suspension assays with clostridia, reduction of the nitro group could result in the production of TAT and other products, some of which remain unidentified. The question whether the pathways for TNT metabolism are inducible or associated with constitutively expressed metabolic functions in different Clostridium strains remains to be solved (61, 64). Below we describe some in vivo and in vitro reactions of TNT by Clostridium spp. Crawford and colleagues (80, 142, 180) reported cometabolic TNT transformation by Clostridium strains isolated from a long-term bioreactor fed with explosives (TNT, trihydroxytoluene trimethylenetrinitronitramine [RDX], tetramethylenetetranitramine [HMX]), sewage sludge, and soils. TAT was identified in these cultures, and aromatic compounds lacking nitrogen substitution (methylphloroglucinol and p-cresol [Fig. 4]) were detected (54), together with phenolic products from TAT hydrolysis (142). These authors showed that the addition of a cosubstrate that supplied reducing power for nitro group reduction was required for significant TNT degradation (205). Shen et al. (202) have proposed that the p-cresol detected during the anaerobic metabolism of TNT might originate from the metabolism of some aromatic amino acids such as tryptophan rather than from TNT.
Hughes et al. (113), working with cell extracts of Clostridium strains, detected the formation of 2-amino-4-hydroxylamino-5-hydroxyl-6-nitrotoluene and 2-hydroxylamino-4-amino-5-hydroxyl-6-nitrotoluene (Fig. 4), which may have arisen from Bamberger rearrangement of dihydroxydinitrotoluene. Recently Huang et al. (111) have suggested that the inital reaction could be mediated by CO dehydrogenase and that this enzyme may transform TNT into 2-hydroxylamine-4,6-dinitrotoluene, 4-hydroxylamine-2,6-dinitrotoluene, and 2,4-dihydroxylamine-6-nitrotoluene as reduction products. Via Bamberger rearrangement, the last compound may subsequently yield 2-amino-4-hydroxylamino-5-hydroxyl-6-nitrotoluene and hydroxylamine-4-amino-5-hydroxyl-6-nitrotoluene. Subsequently, Ahmad and Hughes (6) suggested that the incorporation of an aerobic stage to cleave aminophenols formed in clostridial systems could help to mineralize nitroaromatic contaminants. In some Clostridium strains, dihydroxylamino intermediates seem to play an important role not just in TNT but also in DNT metabolism (115). However, further metabolism of dihydroxylaminotoluenes resulted in the formation of arylamines and not in aminophenols as in the dihydroxylaminonitrotoluene rearrangement (113).
Desulfovibrio strains have been also extensively studied because of their ability to metabolize TNT under anoxic conditions. Boopathy and Kulpa (29) isolated a strain, called Desulfovibrio sp. strain B, that used TNT as the sole nitrogen source and accumulated toluene in the culture medium (Fig. 4). The authors hypothesized that the nitro group was reduced to the corresponding amines to form TAT, which then underwent reductive elimination of the amino groups by a mechanism analogous to that described by Schnell and Schink (198) for aniline. The proposal was preliminary, since the authors failed to detect TAT or any evidence of deamination.
Preuss et al. (174) isolated a pure culture of Desulfovibrio which was selected by using TNT as the sole nitrogen source and pyruvate as the carbon and energy source. This strain was able to catalyze the complete reduction of TNT to TAT in cell suspensions in the absence of any reducing agent. The authors suggested that the reduction of DANTs to TAT was the rate-determining step in the overall process. No reductively deaminating reaction was found, and no toluene was detected in cell suspensions of the isolate. The authors also carried out biochemical studies of the microbial enzymes that catalyze TNT reduction. The kinetics of DANT reduction by Desulfovibrio spp. depended on the electron donor. With pyruvate and H2, DANTs was converted to TAT; in contrast, the addition of carbon monoxide (CO) inhibited the production of TAT, and 2,4-diamino-6-hydroxylaminotoluene accumulated in the medium. These observations led the authors to believe that sulfite reductase was responsible for the reduction of 2,4-diamino-6-hydroxylaminotoluene to TAT because this enzyme is known to be inhibited by CO. However, assays with purified enzymes have not yet been done.
Drzyzga et al. (60) described biotransformation assays with [14C]TNT in Desulfovibrio strains and found that although the explosive was not mineralized, 42% of the initial radiolabel was associated with the cell biomass. TAT and both DANT isomers were detected in liquid supernatants.
When TNT degradation by Clostridium and Desulfovibrio spp. seems to proceed via TAT, the mineralization of TNT under anaerobic conditions must involve the elimination of the amino groups from TAT. The biological transformation of TAT is possible only at neutral pH and under anoxic conditions; otherwise the molecule is unstable (175). However, the role of TAT as the key metabolite in the anaerobic metabolism of TNT has been questioned by Hawari et al. (103), who suggested that this compound is a dead-end product which prevents mineralization of the nitroaromatic. The chemical properties of TAT in an anaerobic atmosphere were used by Rieger and Knackmuss (185) to design a bioremediation technique (see “Applications in TNT bioremediation” below) which consists of full reduction of the explosive to TAT using anaerobic incubations with sewage sludge cultures.
The anaerobic removal of TNT under different electron-accepting conditions (nitrate, sulfate, and carbon dioxide) has been reported. Nitrate respiraton conditions gave the best degradation rate with a consortium of soil bacteria (33), whereas methanogenesis seemed to be optimal with slurries of aquifer sediment (135).
Ederer et al. (64) tested anaerobic degradation of TNT by enterobacteria (E. coli and Salmonella enterica serovar Typhimurium) and lactobacilli (Lactobacillus acidophilus, L. casei, and L. latis) and found that these species were able to reduce TNT but not DANT.
More recently, Ramos and colleagues described a different mechanism for the anoxic degradation of TNT (68, 69). They isolated a facultative Pseudomonas sp., designated strain JLR11, which was able to grow on TNT as the sole nitrogen source with glucose as a cosubstrate. Mass balance assays with TNT revealed that about 85% of the total nitrogen in the nitroaromatic compound was incorporated as cell biomass. Analysis of the culture supernatants detected pathway intermediates lacking nitro groups, e.g., 2-nitro-4-hydroxybenzoic acid, 4-hydroxybenzaldehyde, and 4-hydroxybenzoic acid (Fig. 4). The conversion of TNT to these compounds requires not only the removal of nitro groups but also oxidation of the lateral methyl group of TNT to the corresponding aldehyde and carboxylic acid. In assays with [14C]TNT, only 1% of the radioactivity was detected as 14CO2 but 45% was associated to trichloroacetic acid-precipitable cell material. The productive initial attack on the nitroaromatic compound seemed to involve nitrite release, since nitrite accumulated transitorily with TNT consumption. Cell extracts of Pseudomonas sp. strain JLR11 grown anoxically in TNT showed nitrite reductase activity (69). A mutant selected after mini-Tn5-tellurite mutagenesis as being unable to grow on TNT as the sole nitrogen source had also lost the capacity to grow on nitrite as the sole nitrogen source (67). Cell suspension assays with the mutant in a TNT medium resulted in the release of nitrite, which accumulated with time. This accumulation was not observed in identical experiments using the wild-type strain.
Strain JLR11 reduced a fraction of the total TNT to ADNTs, and these products accumulated with time and were not used by the strain as a nitrogen source. The authors determined that the reduced forms of TNT are produced by Pseudomonas sp. strain JLR11, because TNT acted as a final electron acceptor. To verify this, strain JLR11 was cultured with TNT and acetate as a cosubstrate; under these conditions the oxidation of acetate under anoxic conditions required an electron acceptor, a role that only TNT could play in this assay. Proton translocation, together with the reduction of TNT, was detected in anoxic Pseudomonas sp. strain JLR11 cell suspensions cultured in medium containing TNT, but this extrusion did not occur when nitrite- or nitrate-grown cells were incubated with TNT.
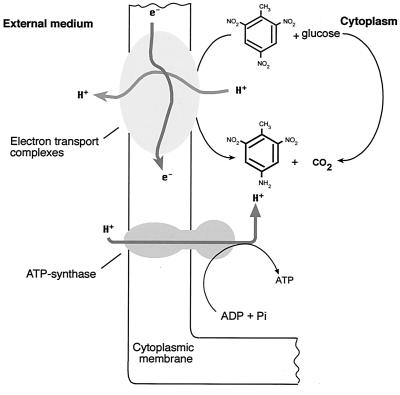
Esteve-Núñez et al. (69) also found synthesis of ATP coupled to H2-TNT oxidation-reduction in membrane vesicles prepared from JLR11 cells cultured with TNT as the final electron acceptor (Fig. 5). When nitrate or nitrite replaced TNT as the final electron acceptor, the rates of ATP synthesis were on the order of 10 and 35%, respectively, of the rate in the presence of TNT. The role of TNT as an electron acceptor was suggested previously by Boopathy and Kulpa (29), although Esteve-Núñez et al. (69) demonstrated experimentally that the reduction of TNT in Pseudomonas sp. strain JLR11 was linked to proton extrusion, which may contribute to a transmembrane electrochemical proton gradient of sufficient magnitude to drive ATP synthesis. The physiological role of TNT respiration raises the possibility of interesting environmental applications in anaerobic environments polluted with TNT, in which the pollutant might be used not only as a nitrogen source but also as a terminal electron acceptor.
FIG. 5.
Scheme showing the coupling of electron donor compounds, TNT oxidoreduction, and ATP synthesis. Pi, inorganic phosphate. The scheme has been adapted from reference (69).
FUNGAL METABOLISM OF TNT
The use of fungi for the bioremediation of TNT has generated considerable interest in the last decade. The wood white rot fungus Phanerochaete chrysosporium has been the subject of intensive study, and more recently other white rot fungi and litter decay fungi have been investigated for their ability to transform TNT.
The interest in white rot fungi arises from their capacity to degrade a diverse group of environmentally persistent and toxic chemicals. Early observations revealed TNT metabolism by P. chrysosporium under nitrogen-limiting conditions (47, 72, 102). Later it was shown that different nutrient starvation conditions (carbon, nitrogen, or sulfur limitation) favoured TNT and other xenobiotic attack and that these reactions were carried out by secreted enzymes of the lignin-degrading system. This system contains lignin peroxidase, manganese peroxidase (MnP), oxidases, reductases, hydrogen peroxidase, veratryl alcohol, oxalate, and quinol oxidases (221).
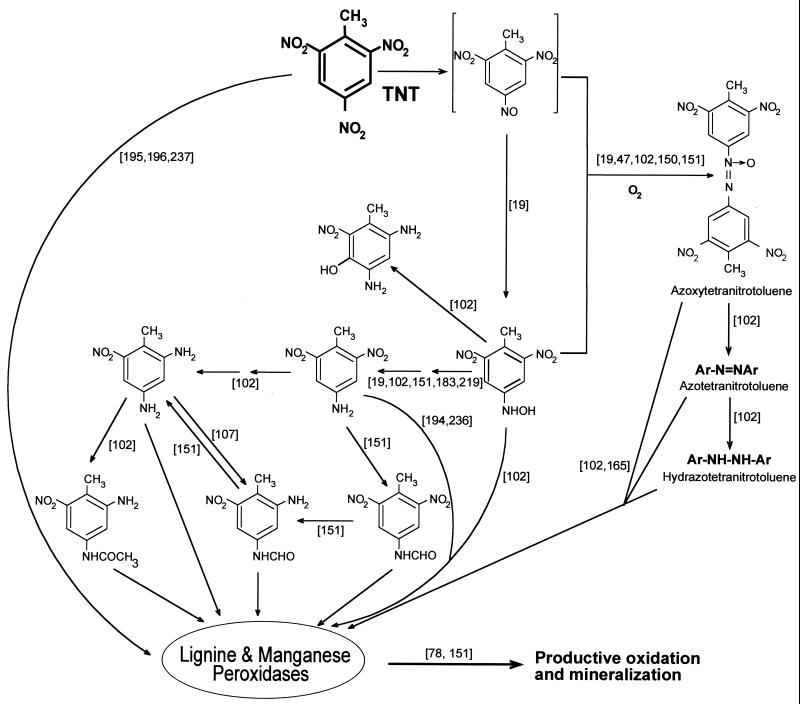
As in other organisms, the initial steps in the fungal degradation of TNT involve the reduction of nitro groups (164). Mycelia of P. chrysosporium reduce TNT to a mixture of 4-ADNT, 2-ADNT, 4-hydroxylamino-2,6-dinitrotoluene, and azoxytetranitrotoluenes (Fig. 6). Under ligninolytic conditions the aromatic compounds and azoxytetranitrotoluenes disappear, and mineralization can be fairly extensive.
FIG. 6.
Mechanisms for fungal degradation of TNT. The scheme is based on articles cited in the text, which are indicated in the figure in s brackets.
Stahl and Aust (219–221) provided evidence that TNT is reduced by a plasma membrane redox system in P. chrysosporium that requires live and intact mycelia. Any conditions that disrupt the integrity of the plasma membrane destroy the reductase activity. The presence of compounds known to inhibit the membrane redox systems also inhibits TNT reductase. The authors suggested that reduction is coupled to the proton export system used by the fungus to maintain the extracellular physiological pH at approximately 4.5. In contrast, Rieble el al. (183) reported a membrane-associated TNT reductase activity that required NADPH as a cosubstrate and the absence of molecular oxygen. Because activity was detected in the detergent-solubilized form, where a membrane potential could not be maintained, a membrane potential redox system was assumed to be inessential for this reaction. On the other hand, NAD(P)H-dependent intracellular TNT reductase activity has also been described (151).
Regardless of the site where of TNT is reduced, further degradation of these compounds and eventual mineralization of TNT by P. chrysosporium occur only when cultures are ligninolytic, implying that lignin peroxidase, MnP, and/or other enzymes of the lignolytic system further transforms the reduced products of TNT. The mechanism by which the fungi mineralize the explosive is not known, but preliminary information about the process has been reported (107, 151). When ligininolytic cultures of P. chrysosporium were incubated with 4-ADNT, a compound identified as 4-formamide-2,6-DNT was detected (Fig. 6). This intermediate was transformed into 2-amino-4-formamide-6-nitrotoluene, a compound that disappeared rapidly under ligninolytic conditions but not under nonligninolytic ones. In nonligninolytic environments the ADNT metabolites were slowly reduced to DANTs and the concentration of azoxytetranitrotoluenes increased (Fig. 6).
The hydroxylaminodinitrotoluene products of TNT reduction inhibit the veratryl alcohol oxidase activity of lignin peroxidase (47, 150). This activity protects lignin peroxidase from H2O2 inactivation, and explains why the presence of the hydroxylaminodinitrotoluenes makes ADNT more readily mineralizable than TNT (221). The design of bioremediation processes for TNT-contaminated soils with P. chrysosporium therefore requires conditions under which hydroxylaminodinitrotoluenes do not accumulate.
The degradation of azoxytetranitrotoluenes by P. chrysosporium is not unexpected, because the reduction of azoxy compounds to azo compounds is chemically simple and the mineralization of various azo dyes by this fungus has been demonstrated (165). The catabolic potential of P. chrysosporium for TNT mineralization is limited because growth of this fungus is inhibited at relatively low concentrations of TNT and because reduction of this polynitroaromatic compound by immobilized P. chrysosporium mycelia is inhibited above 20 ppm. The low tolerance of P. chrysosporium to TNT prevents this fungus from being a good candidate for bioremediation of TNT-contaminated sites, which usually show a high concentration of this nitroaromatic compound.
Other fungi also exhibit potential for TNT degradation (129). In this regard it was reported that the MnP of several white rot fungus attacks TNT (194–196). Scheibner and Hofrichter (194) showed that cell-free preparations of MnP from the white rot basidiomycete Nematoloma frowardii and from the litter-decaying basidiomycete Stropharia rugosoannulata were able to mineralize a mixture of reduction products from [14C]TNT as well as 2-amino-4,6-[14C]dinitrotoluene (195, 196). More recently, Van Aken et al. (237, 238) showed that a concentrated preparation of MnP from a different white rot fungus, Phlebia radiata, was able to completely transform TNT (22% mineralization) and 2-amino-4,6-dinitrotoluene (76% mineralization). In these in vitro assays, reduced glutathione and cysteine in small amounts were shown to strongly enhance the transformation and mineralization of TNT, probably through the generation of the highly reactive radical and because the activity of the concentrated MnP enzymatic system was much higher than that of the ligninolytic extracellular fluid (235, 237). This enzyme could play an important role in TNT degradation because its in vivo production is not regulated by carbon sources (78).
Recently, several fungal species have been reported to tolerate high TNT concentrations (19). Cladosporium resinae and Cunninghamella echinulata var. elegans transform TNT to reduced products, mainly azoxytetranitrotoluenes. Although these fungi convert significant amounts of 14C to polar metabolites (nonextractable after organic extraction), no mineralization was detected. Bayman and Radkar (19) consequently proposed a two-step process in which TNT is initially reduced to ADNTs by a soil fungus such as C. resinae and then mineralized by P. chrysosporium. This process may both speed up the rate of mineralization of the explosive and protect P. chrysosporium from TNT toxicity caused by hydroxylaminodinitrotoluene.
Apart from white rot fungi, other fungal strains belonging to wood- and litter-decaying basidiomycetes have been tested for their ability to metabolize and mineralize TNT (196, 236). Significant mineralization was detected in Clitocybula dusenii TMB12 (42%) and Stropharia rugosoanulata DSM11372 (36%) under lignolytic conditions, which suggests an important role for ligninolytic systems in the mineralization process by these fungi. A disadvantage of this system is that these fungi are native to wood rather than soil and may not compete well with native soil fungi in field remediation situations (19, 73).
APPLICATIONS IN TNT BIOREMEDIATION
The manufacture, processing, and packaging of TNT at ammunitions plants for several decades have resulted in high concentrations of contaminants in soil (168, 199, 257), and eventually this pollutant reaches the groundwater (18, 169, 206). A number of physicochemical treatments have been proposed, and some of them are in use (190). Currently, incineration is the most effective remediation alternative, but it is expensive for use with polluted soils because of the costs of soil excavation, transport, and energy for incineration. For the treatment of TNT in liquid wastes, several technologies have been proposed, e.g., chemical reduction (5, 11, 12, 66, 137), thermal decomposition, subcritical water (104), photocatalytic degradation (197), electrohydraulic discharge (250), granular iron (59), and ozonolysis (136, 216, 222). Several biologically based technologies have also been developed for the remediation of TNT-contaminated soils; of these approaches, the most thoroughly investigated are soil slurry reactors, composting, and land farming. Each approach has advantages and disadvantages, and the main applications are summarized below. Many groups have developed bench-scale assays, but only a few have tried full-scale assays (80, 140).
Soil slurry technologies consist of reactors filled with a mixture of soil and water to which cosubstrates and nutrients can be added as necessary. These systems are designed to optimize mass transfer of nutrients and electron acceptors by using mechanical mixing and/or aeration. All work with this technology has used anoxic incubation to avoid the polymerization reactions that take place in the presence of oxygen; sometimes an additional aerobic treatment is required. The metabolism of TNT in anoxic processes requires an additional cosubstrate (e.g., starch, glucose, sucrose, or molasses), which plays two main roles: oxygen removal by growing aerobes, and electron donation for nitro group reduction of TNT. There are two different approaches to TNT bioremediation by slurry reactors: mineralization of the explosive as the main target (31, 32, 80, 81) and irreversible binding of TNT metabolites to the soil matrix (140, 178, 185). The method of Funk et al. (80, 81), developed, patented, and licensed at the University of Idaho, appears to be useful for the biological remediation of soils contaminated with nitroaromatic compounds and explosives. This procedure was previously used for the strictly anaerobic microbial bioremediation of nitroaromatic-herbicide-contaminated soils (188). The process, designated SABRE (sequencial anaerobic biological remediation ex situ), consists of a consortium of facultative anaerobic organisms that transform explosives such as TNT to nontoxic, nonaromatic, and aerobically mineralizable products. The consortium contained strains of the genus Clostridium, and spores of these strains are expected to be useful as inoculants for bioremediation (200).
The accumulation of starchy material in treated soil produces a high oxygen demand, which may be detrimental in agricultural soils because of the rapid development of anaerobic conditions when the soil is wetted, such as after irrigation or rainfall. Roberts et al. (187) proposed an aerobic treatment after the anoxic stage to remove excess external carbon and obtain treated soil with a lower oxygen demand. The process they reported required glucose as a cosubstrate and the presence in the soil of a population of nitroaromatic-degrading anaerobes (80, 81). Nishino et al. (157) reported that the addition of specific nitroaromatic-mineralizing bacteria seemed to enhance the mineralization of dinitrotoluenes in soil slurries whereas native bacteria did not transform the compound during the incubation time required for the mineralization of DNTs.
The U.S. Army Environmental Center has developed a bioslurry system to provide a nonproprietary process for the treatment of explosives-contaminated soil (117). The process is similar to the SABRE and was conducted in a trench reactor. The results of analytical and toxicity tests support the potential usefulness of land applications of the processed water, although an additional aerobic treatment seems to be necessary to bring down the biological oxygen demand of the treated waters.
Boopathy et al. (28) did a laboratory study using a soil slurry reactor with a dissolved-oxygen profile which made the reactor to function as an aerobic-anaerobic system. TAT was removed in the batch mode during the first 2 weeks; however, reduction metabolites persisted and semicontinuous operation over a period of 3 months was recommended by the authors. TNT degradation was demonstrated by observing mineralization of [14C]TNT (23% of total 14C evolved as 14CO2) and by finding 27% of the radioactivity as trichloroacetic acid-precipitable material.
The soil slurry reactor technology has also been used to enhance the anaerobic fixation of xenobiotic compounds to the soil organic matrix, a process mediated by microbial cometabolic activities (185, 223). Bench-scale assays have been developed, and all of them showed complete TNT removal and incorporation of 84 to 99% radioactivity ([14C]TNT) to the soil matrix (56, 60, 61) after anoxic-aerobic treatment. This biologically induced immobilization of TNT was tested on a technical scale (140) in a Terranox reactor with 18 m3 of TNT-contaminated soil (314 mg of TNT/g of soil) mixed with 10 m3 of water supplied with sucrose and ammonium chloride. The process led to removal of 98% of the TNT, DNT, and RDX in 30 weeks. The decrease in the concentrations of the explosives was slower than in the laboratory assay, possibly because of the lower temperature and mass transfer as a result of less intense mixing. The extraction of bioremediated soils with various chemicals did not lead to the release of TNT or its metabolites (1). Binding seems to increase with the number of amino groups in the aromatic ring, making TNT undetectable in the supernatants. The results of the ecotoxicological evaluation of the supernatant after anaerobic incubation showed toxic effects, but no toxicity was detectable after the subsequent aerobic treatment. Terrestrial tests performed at the end of the bioremediation process showed no mortality for earthworms.
Composting is considered a feasible technology for the remediation of munitions-contaminated soils. Research thus far indicates that TNT can be removed from contaminated soils (35, 42, 61, 62, 124, 167, 253). In composting, the soil must be mixed with a bulk agent and additional organic substrates which are usually by-products of agricultural processes (straw, alfalfa, wood chips, sugarbeet stems, and leaves). Windrow composting and anaerobic-aerobic composting systems are highly efficient for treatment of TNT-contaminated soils (42). Under these conditions the explosive was reduced to amino- and diaminonitrotoluenes during the anoxic phase, and the subsequent aerated phase eliminated most of the transformation products, possibly by covalent binding to the soil (39, 40, 134). In tests of the toxic effects of these compounds on human monocytes, leachates of bioremediated soil produced a response similar to that of compost-free of nitroaromatic compounds (41). Tan et al. (227) reported that composting of TNT markedly reduced mutagenicity in the contaminated soil, and this was confirmed by others (89, 116). The main criticisms of the compost technique are the long incubation time, the cost of setting up and maintaining the system, and the lack of knowledge about the bacteria and fungi involved in the process.
Land farming is a solid-phase treatment process in which contaminated soil is mixed with nutrients and moisture, using periodical mechanical turning of the soil mixture to increase aeration (71, 117, 250). Widrig et al. (250) investigated an innovative approach to land farming for the bioremediation of TNT-contaminated soil using molasses as a cosubstrate and shredded grass to improve soil management properties. Degradation of TNT was demonstrated by mineralization of [14C]TNT and the presence of 14C in the cell biomass as trichloroacetic acid-precipitable material. The results of this bench-scale study are promising with regard to transferring the process to full-scale applications. Due to the solubility of TNT and its derivatives in water, hydraulic control of infiltrated water may be required in the design of any on-site or in situ method.
The ubiquitous reductive metabolism of TNT present in all treatments describe above yields large amounts of hydroxylaminonitrotoluenes and ADNTs. Most of the reduction products bind strongly to the mineral (56, 109) and organic (1–4, 38, 56, 57, 60–62, 65, 143) fractions of the soil. After treatment, the reduction metabolites cannot be desorbed from soil by alkaline or acidic hydrolysis or by methanolic extraction, so the authors concluded that metabolites become unavailable for further microbial degradation and mineralization. The covalent binding of reduced metabolites of [15N]TNT to soil organic matter was analyzed by 15N nuclear magnetic resonance (NMR) spectroscopy (1, 134, 173); the NMR shifts showed that nitrogen was covalently bound to humic acid as substituted amines and amides, whereas the NMR spectra of silylated humin suggested the formation of azoxy compounds and imine linkages. This indicates immobilization of the aminoaromatic compounds to the soil matrix and can be considered a detoxification process, although information about the long-term fate of these bound metabolites is not available. Recently, Sheremata and Hawari (204) reported finding a natural surfactant able to desorb TNT, aminonitrotoluenes, and diaminonitrotoluenes from polluted soils.
Phytoremediation seems to be another alternative for the in situ treatment of explosives-contaminated soils. Recent reports have described assays with a variety of plant systems such as yellow nutsedge (163), bush bean (100), switchgrass (172), aquatic and wetland species (21, 22, 23, 48, 112, 166, 240), axenic root cultures (112), and hybrid poplar (230). In most studies, TNT transformation by plants was accompanied by the appearance of its monoamino derivatives 2- and 4-ADNT. Little is known about the product profile of TNT biocatalysis of plants beyond these early products. Some plants seem to sequester or integrate TNT metabolites into biomass as conjugates (201, 245). The formation of such conjugates of organic xenobiotic compounds is an important protective phase in plant detoxification metabolism. Some of these conjugates are compartmentalized into plant organelles such as the vacuoles. The primary reduction products of TNT seem to be involved in such conjugation processes (24).
Plant-bacterium combinations to phytoremediate contaminated soil are also being developed (207) with a Pseudomonas strain capable of transforming TNT to its monodinitrotoluene and diaminonitrotoluene metabolites. Inoculation of meadow bromegrass (Bromus erectus) with this strain increased the portion of the rhizosphere community involved in nitroaromatic metabolism and led to a reduction in soil TNT levels.
More recently, transgenic plants that express microbial degradative enzymes for TNT bioremediation have been developed (75). Transgenic tobacco plants that carry the onr gene, which encodes the pentaerythritol tetranitrate reductase from Enterobacter cloacae (76, 77), showed detectable expression of the reductase enzyme in leaf and root tissue. Transgenic plants that express the explosives-degrading enzyme are able to germinate and grow in the presence of explosives concentrations inhibitory to wild-type plants. Further research is required to determine whether transgenic plants are capable of contributing to the degradation of TNT.
PERSPECTIVES IN TNT DEGRADATION AND UNEXPLORED RESEARCH
TNT degradation has attracted the attention of many scientists in the last decade, probably because a number of Environmental Protection Agencies have declared it a pollutant whose removal is a priority. The symmetrical positioning of the nitro groups prevents the attack of classical dioxygenases which generate hydroxylated aromatic rings that subsequently become the substrates of ring cleavage enzymes. In contrast, reduction of the nitro groups seems to be a widely distributed reaction among aerobic and anerobic bacteria and fungi. However, there are not many details about the biochemical properties of these TNT reductases or the genes that encode them, and we expect many important contributions in this research area. Many anaerobic bacteria and fungi are able to reduce TNT. The derivatives of this reduction are futher metabolized and eventually mineralized. In contrast, when these products are produced by aerobic bacteria, they usually constitute dead-end products. The productive steps leading to the mineralization of reduced TNT derivatives by anaerobic bacteria and fungi are unknown, and advances are envisaged by using microorganisms susceptible to the combination genetic and biochemical analyses. These studies will be illuminating not only in terms of understanding the catabolic pathways themselves but also in attempts to gain insights into the evolution of these activities in bacteria and fungi. The wide distribution of TNT-degrading microbes and the abundance of catabolic pathways that can lead to mineralization of TNT lead us to envisage that interest in the field will be focused on the optimization of the catabolic properties of these microbes rather than on the development of recombinant strains, as suggested previously (179), as an alternative to TNT degradation.
A number of laboratories have shown that TNT denitration can yield many different products depending on the microorganism under scrutiny. Except for the reaction catalyzed by pentaerythritol tetranitrate reductase on TNT, little, if anything, is known about these denitrase activities. We have recently cloned the denitrase cluster of Pseudomonas sp. strain JLR11 as a means of giving Burkholderia cepacia R34 the ability to grow on TNT. An in-depth analysis of this gene cluster may shed light on the mechanism underlying the reaction. In turn, gene probes can be useful for studying the distribution of this set of genes in nitroaromatic-polluted soils. The use of gene probes will be useful in identifying niches in which these kinds of genes prevail and the conditions under which the population of microbes bearing these genes increases (251). This may also be useful in the design of biological treatments for sites polluted with TNT. Rhizoremediation of TNT by microbes capable of colonizing the rhizospheres of plants will provide a cheap, fast, and efficient process for the removal of this pollutant from the upper layers of the soil.
ACKNOWLEDGMENTS
Work in our laboratory was supported by grant BIO2000-0964 from the Comisión Interministerial de Ciencia y Tecnologia, and by grant QLRT2001-00345 from the European Commission.
REFERENCES
- 1.Achtnich C, Fernandes E, Bollag J-M, Knackmuss H-J, Lenke H. Covalent binding of reduced metabolites of 15N-TNT to soil organic matter during a bioremediation process analyzed by 15N NMR spectroscopy. Environ Sci Technol. 1999;33:4448–4456. [Google Scholar]
- 2.Achtnich C, Lenke H. Stability of immobilized 2,4,6-trinitrotoluene metabolites in soil under long-term leaching conditions. Environ Toxicol Chem. 2001;20:280–283. doi: 10.1897/1551-5028(2001)020<0280:soitmi>2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 3.Achtnich C, Lenke H, Klaus U, Spiteller M, Knackmuss H-J. Stability of immobilized TNT derivatives in soil as a function of nitro group reduction. Environ Sci Technol. 2000;34:3698–3704. [Google Scholar]
- 4.Achtnich C, Pfortner P, Weller M G, Niessner R, Lenke H, Knackmuss H-J. Reductive transformation of bound trinitrophenyl residues and free TNT during a bioremediation process analyzed by immunoassay. Environ Sci Technol. 1999;33:3421–3426. [Google Scholar]
- 5.Agrawal A, Tratnyek P G. Reduction of nitroaromatic compounds by zero-valent iron metal. Environ Sci Technol. 1996;30:153–160. [Google Scholar]
- 6.Ahmad F, Hughes J B. Anaerobic transformation of TNT by Clostridium. In: Spain J C, Hughes J B, Knackmuss H-J, editors. Biodegradation of nitroaromatic compounds and explosives. Boca Raton, Fla: Lewis-Publishers; 2000. pp. 185–212. [Google Scholar]
- 7.Alvarez M A, Kitts C L, Botsford J L, Unkefer P J. Pseudomonas aeruginosa strain MA01 aerobically metabolizes the aminodinitrotoluenes produced by 2,4,6-trinitrotoluene nitro group reduction. Can J Microbiol. 1995;41:984–991. doi: 10.1139/m95-137. [DOI] [PubMed] [Google Scholar]
- 8.An D, Gibson D T, Spain J C. Oxidative release of nitrite from 2-nitrotoluene by a three component enzyme system from Pseudomonas sp. strain JS42. J Bacteriol. 1994;176:7462–7467. doi: 10.1128/jb.176.24.7462-7467.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Angermaier L, Simon H. On nitroaryl reductase activities in several Clostridia. Hoppe-Seylers Z Physiol Chem. 1983;364:1653–1663. doi: 10.1515/bchm2.1983.364.2.1653. [DOI] [PubMed] [Google Scholar]
- 10.Anlezark G M, Melton R G, Sherwood R F, Coles B, Friedlos F, Knox R. The bioactivation of 5-(aziridin-1-yl)-2,4-dinitrobenzamide (CB1954)-1. Purification and properties of a nitroreductase enzyme from Escherichia coli—a pontential enzyme for antibody-directed enzyme prodrug therapy (ADEPT) Biochem Pharmacol. 1992;44:2289–2295. doi: 10.1016/0006-2952(92)90671-5. [DOI] [PubMed] [Google Scholar]
- 11.Arienzo M. Degradation of 2,4,6-trinitrotoluene in water and soil slurry utilizing a calcium peroxide compound. Chemosphere. 2000;40:331–337. doi: 10.1016/s0045-6535(99)00212-x. [DOI] [PubMed] [Google Scholar]
- 12.Arienzo M. Use of abiotic oxidative-reductive technologies for remediation of munition contaminated soil in a bioslurry reactor. Chemosphere. 2000;40:441–448. doi: 10.1016/s0045-6535(99)00326-4. [DOI] [PubMed] [Google Scholar]
- 13.Averill B A. Transformation of inorganic N-oxides by denitrifying and nitrifying bacteria: Pathways, mechanisms, and relevance to biotransformation of nitroaromatic compounds. In: Spain J C, editor. Biodegradation of nitroaromatic compounds. New York, N.Y: Plenum Press; 1995. pp. 183–197. [Google Scholar]
- 14.Bae B, Autenrieth R L, Bonner J S. Aerobic biotransformation and mineralization of 2,4,6-trinitrotoluene. In: Hinchee R E, Hoeppel R E, Anderson D B, editors. Bioremediation of recalcitrant organics. Columbus, Ohio: Battelle Press; 1995. pp. 231–238. [Google Scholar]
- 15.Bakhtiar R, Leung K H. Covalent binding of 2,4,6-trinitrotoluene to human hemoglobin. Evidence for protein adducts probed by electrospray ionization mass spectrometry. Rapid Commun Mass Spectrom. 1997;11:1935–1937. doi: 10.1002/(SICI)1097-0231(199711)11:17<1935::AID-RCM98>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 16.Banerjee H N, Verma M, Hou L H, Ashraf M, Dutta S K. Cytotoxicity of TNT and its metabolites. Yale J Biol Med. 1999;72:1–4. [PMC free article] [PubMed] [Google Scholar]
- 17.Barrows S E, Cramer C J, Truhlar D G, Elovitz M S, Weber E J. Factors controlling regioselectivity in the reduction of polynitroaromatics in aqueous solution. Environ Sci Technol. 1996;30:3028–3038. [Google Scholar]
- 18.Bart J C, Judd L L, Hoffman K E, Wilkins A M, Kusterbeck A W. Application of a portable immunosensor to detect the explosives TNT and RDX in groundwater samples. Environ Sci Technol. 1997;31:1505–1511. [Google Scholar]
- 19.Bayman P, Radkar G V. Transformation and tolerance of TNT (2,4,6-trinitrotoluene) by fungi. Int Biodeterior Biodegrad. 1997;39:45–53. [Google Scholar]
- 20.Berthe-Corti L, Jacobi H, Kleihauer S, White I. Cytotoxicity and mutagenicity of a 2,4,6-trinitrotoluene (TNT) and hexogen contaminated soil in Salmonella typhimurium and mammalian cells. Chemosphere. 1998;37:209–218. doi: 10.1016/s0045-6535(98)00039-3. [DOI] [PubMed] [Google Scholar]
- 21.Best E P, Sprecher S L, Larson S L, Fredrickson H L, Bader D F. Environmental behavior of explosives in groundwater from the Milan army ammunition plant in aquatic and wetland plant treatments. Removal, mass balances and fate in groundwater of TNT and RDX. Chemosphere. 1999;38:3383–3396. doi: 10.1016/s0045-6535(98)00550-5. [DOI] [PubMed] [Google Scholar]
- 22.Best E P, Sprecher S L, Larson S L, Fredrickson H L, Bader D F. Environmental behavior of explosives in groundwater from the Milan army ammunition plant in aquatic and wetland plant treatments. Uptake and fate of TNT and RDX in plants. Chemosphere. 1999;39:2057–2072. doi: 10.1016/s0045-6535(99)00117-4. [DOI] [PubMed] [Google Scholar]
- 23.Bhadra R, Spanggord R J, Wayment D G, Hughes J B, Shanks J V. Characterization of oxidation products of TNT metabolism in aquatic phytoremediation systems of Myriophyllum aquaticum. Environ Sci Technol. 1999;33:3354–3361. [Google Scholar]
- 24.Bhadra R, Wayment D G, Hughes J B, Shanks J V. Confirmation of conjugation processes during TNT metabolism by axenic plant roots. Environ Sci Technol. 1999;33:446–452. [Google Scholar]
- 25.Blasco R, Castillo F. Characterization of a nitrophenol reductase from the phototrophic bacterium Rhodobacter capsulatus E1F1. Appl Environ Microbiol. 1993;59:1774–1778. doi: 10.1128/aem.59.6.1774-1778.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Blehert D S, Fox B G, Chambliss G H. Cloning and sequence analysis of two Pseudomonas flavoprotein xenobiotic reductases. J Bacteriol. 1999;181:6254–6263. doi: 10.1128/jb.181.20.6254-6263.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Blotevogel K-H, Gorontzy T. Microbial degradation of compounds with nitro functions. In: Klein J, editor. Biotechnology. 11b. Environmental processes II. Weinheim, Germany: Wiley-VCH; 2000. pp. 274–302. [Google Scholar]
- 28.Boopathy R, Gurgas M, Ullian J, Manning J F. Metabolism of explosive compounds by sulfate-reducing bacteria. Curr Microbiol. 1998;37:127–131. doi: 10.1007/s002849900350. [DOI] [PubMed] [Google Scholar]
- 29.Boopathy R, Kulpa C F. Trinitrotoluene as a sole nitrogen source for a sulfate-reducing bacterium Desulfovibrio sp. (B strain) isolated from an anaerobic digester. Curr Microbiol. 1992;25:235–241. doi: 10.1007/BF01570724. [DOI] [PubMed] [Google Scholar]
- 30.Boopathy R, Kulpa C F. Biotransformation of 2,4,6-trinitrotoluene (TNT) by a Methanococcus sp. (strain B) isolated from a lake sediment. Can J Microbiol. 1994;40:273–278. doi: 10.1139/m94-044. [DOI] [PubMed] [Google Scholar]
- 31.Boopathy R, Manning J, Kulpa C F. Optimization of environmental factors for the biological treatment of trinitrotoluene-contaminated soil. Arch Environ Contam Toxicol. 1997;32:94–98. doi: 10.1007/s002449900159. [DOI] [PubMed] [Google Scholar]
- 32.Boopathy R, Manning J, Kulpa C F. A laboratory study of the bioremediation of 2,4,6-trinitrotoluene-contaminated soil using aerobic/anoxic soil slurry reactor. Water Environ Res. 1998;70:80–86. [Google Scholar]
- 33.Boopathy R, Wilson M, Kulpa C F. Anaerobic removal of 2,4,6-TNT under different electron accepting conditions: laboratory study. Water Environ Res. 1993;65:271–275. [Google Scholar]
- 34.Bradley P M, Chapelle F H. Factors affecting microbial 2,4,6-TNT mineralization in contaminated soil Environ. Sci Technol. 1995;29:802–806. doi: 10.1021/es00003a031. [DOI] [PubMed] [Google Scholar]
- 35.Breitung J. Bioremediation of 2,4,6-trinitrotoluene-contaminated soils by two different aerated compost systems. Appl Microbiol Biotechnol. 1996;44:795–800. doi: 10.1007/BF00178621. [DOI] [PubMed] [Google Scholar]
- 36.Brooks L R, Jacobson R W, Warren S H, Kohan M J, Donnelly K C, George S E. Mutagenicity of HPLC-fractionated urinary metabolites from 2,4,6-trinitrotoluene-treated Fischer 344 rats. Environ Mol Mutagen. 1997;30:298–302. [PubMed] [Google Scholar]
- 37.Bruhn C, Lenke H, Knackmuss H J. Nitrosubstituted aromatic compounds as nitrogen source for bacteria. Appl Environ Microbiol. 1987;53:208–210. doi: 10.1128/aem.53.1.208-210.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bruns-Nagel D, Bretung J, von Löw E, Steinbach K, Borontzy T, Kahl M, Blotevogel K-H, Gemsa D. Microbial transformation of 2,4,6-trinitrotoluene in aerobic soil columns. Appl Environ Microbiol. 1996;62:2651–26562. doi: 10.1128/aem.62.7.2651-2656.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bruns-Nagel D, Drzyzga O, Steinbach K, Schmidt C, von Löw E, Gorontzy T, Blotevogel K-H, Gemsa D. Anaerobic/aerobic composting of 2,4,6-trinitrotoluene-contaminated soil in a reactor system. Environ Sci Technol. 1998;32:1676–1679. [Google Scholar]
- 40.Bruns-Nagel D, Knicker H, Drzyzga O, von Löw E, Steinbach K. Characterization of 15N-TNT residues after an anaerobic/aerobic treatment of soil/molasses mixtures by solid state 15N-NMR spectroscopy. II. Systematic investigation of whole soil and different humic fractions. Environ Sci Technol. 2000;34:1549–1556. [Google Scholar]
- 41.Bruns-Nagel D, Scheffer S, Casper B, Garn H, Drzyzga O, von Löw E, Gemsa D. Effect of 2,4,6-trinitrotoluene and its metabolites on human monocytes. Environ Sci Technol. 1999;33:2566–2570. [Google Scholar]
- 42.Bruns-Nagel D, Steinbach K, Gemsa D, von Löw E. Composting (humification) of nitroaromatic compounds. In: Spain J, Hughes J B, Knackmuss H-J, editors. Biodegradation of nitroaromatic compounds and explosives. Boca Raton, Fla: Lewis Publishers; 2000. pp. 357–393. [Google Scholar]
- 43.Bryant C, Hubbard L, McElroy W D. Cloning, nucleotide sequence, and expression of the nitroreductase gene from Enterobacter cloacae. J Biol Chem. 1991;266:4126–4130. [PubMed] [Google Scholar]
- 44.Bryant C, DeLuca M. Purification and characterization of an oxygen-insensitive NAD(P)H nitroreductase from Enterobacter cloacae. J Biol Chem. 1991;266:4119–4125. [PubMed] [Google Scholar]
- 45.Bryant D W, McCalla D R, Leelsma M, Laneuville P. Type I nitroreductases of Escherichia coli. Can J Microbiol. 1981;27:81–86. doi: 10.1139/m81-013. [DOI] [PubMed] [Google Scholar]
- 46.Bueding E, Jollife N. Metabolism of trinitrotoluene (TNT) in vitro. J Pharmacol Exp Ther. 1946;88:300–312. [PubMed] [Google Scholar]
- 47.Bumpus J A, Tatarko M. Biodegradation of 2,4,6-trinitrotoluene by Phanerochaete chrysosporium: identification of initial degradation products and the discovery of a TNT metabolite that inhibits lignin peroxidase. Curr Microbiol. 1994;28:185–190. [Google Scholar]
- 48.Burken J G, Shanks J V, Thompson P L. Phytoremediation and plant metabolism of explosives and nitroaromatic compounds. In: Spain J, Hughes J B, Knackmuss H-J, editors. Biodegradation of nitroaromatic compounds and explosives. Boca Raton, Fla: Lewis Publishers; 2000. pp. 239–275. [Google Scholar]
- 49.Carpenter D F, McCormick N G, Cornell J H, Kaplan A. Microbial transformation of 14C-labeled 2,4,6-TNT in an activated-sludge system. Appl Environ Microbiol. 1978;35:949–954. doi: 10.1128/aem.35.5.949-954.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cash G G. Prediction of chemical toxicity to aquatic microorganism: ECOSAR vs. Microtox assay. Environ Toxicol Water Qual. 1998;132:211–216. [Google Scholar]
- 51.Channon H J, Mills G T, Williams R T. The metabolism of 2,4,6-trinitrotoluene (α-T.N.T.) Biochem J. 1944;38:70–85. doi: 10.1042/bj0380070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Coombs M, Schillack V. Determination of trinitrotoluene and metabolites in urine by means of gas-chromatography with mass detection. Int Arch Occup Environ Health. 1998;71:S22–S25. [PubMed] [Google Scholar]
- 53.Crawford R L. Biotreatment of nitroaromatic compounds. Trends Biotechnol. 1993;11:411–412. [Google Scholar]
- 54.Crawford R L. The microbiology and treatment of nitroaromatic compounds. Curr Opin Biotechnol. 1995;6:329–336. [Google Scholar]
- 55.Crawford R L. Biodegradation of nitrated munition compounds and herbicides by obligately anaerobic bacteria. In: Spain J C, editor. Biodegradation of nitroaromatic compounds. New York, N.Y: Plenum Press; 1995. pp. 87–98. [Google Scholar]
- 56.Daun G, Lenke H, Reuss M, Knackmuss H-J. Biological treatment of TNT-contaminated soil. 1. Anaerobic cometabolic reduction and interaction of TNT and metabolites with soil components. Environ Sci Technol. 1998;32:1956–1963. [Google Scholar]
- 57.Dawel G, Kastner M, Michels J, Poppitz W, Günther W, Fritsche W. Structure of a laccase-mediated product of coupling of 2,4-diamino-6-nitrotoluene to guaiacol, a model for coupling of 2,4,6-trinitrotoluene metabolites to a humic organic soil matrix. Appl Environ Microbiol. 1997;63:2560–2565. doi: 10.1128/aem.63.7.2560-2565.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Del Campo F F, Ramirez J M, Paneque A, Losada M. Ferredoxin and the dark and light reduction of dinitrophenol. Biochem Biophys Res Commun. 1966;22:547–553. doi: 10.1016/0006-291x(66)90309-3. [DOI] [PubMed] [Google Scholar]
- 59.Devlin J F, Klausen J, Schwarzenbach R P. Kinetics of nitroaromatic reduction on granular iron in recirculating batch experiments. Environ Sci Technol. 1998;32:1941–1947. [Google Scholar]
- 60.Drzyzga O, Bruns-Nagel D, Gorontzy T, Blotevogel K-H, Gemsa D. Mass balance studies with 14C-labeled 2,4,6-trinitrotoluene (TNT) mediated by an anerobic Desulfovibrio species and an aerobic Serratia species. Curr Microbiol. 1998;37:380–386. doi: 10.1007/s002849900397. [DOI] [PubMed] [Google Scholar]
- 61.Drzyzga O, Bruns-Nagel D, Gorontzy T, Blotevogel K-H, Gemsa D, von Low E. Incorporation of 14C-labeled 2,4,6-trinitrotoluene metabolites into different soil fractions after anaerobic and anaerobic-aerobic treatment of soil/molasses mixtures. Environ Sci Technol. 1998;32:3529–3535. [Google Scholar]
- 62.Drzyzga O, Bruns-Nagel D, Gorontzy T, Blotevogel K-H, von Löw E. Anaerobic incorporation of the radiolabeled explosive TNT and metabolites into the organic soil matrix of contaminated soil after different treatment procedures. Chemosphere. 1999;38:2081–2095. doi: 10.1016/s0045-6535(98)00426-3. [DOI] [PubMed] [Google Scholar]
- 63.Duque E, Haïdour A, Godoy F, Ramos J L. Construction of a Pseudomonas hybrid strain that mineralizes 2,4,6-trinitrotoluene. J Bacteriol. 1993;175:2278–2283. doi: 10.1128/jb.175.8.2278-2283.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ederer M M, Lewis T A, Crawford R L. 2,4,6-Trinitrotoluene (TNT) transformation by Clostridia isolated from a munition-fed bioreactor: comparison with non-adapted bacteria. J Ind Microbiol Biotechnol. 1997;18:82–88. doi: 10.1038/sj.jim.2900257. [DOI] [PubMed] [Google Scholar]
- 65.Elovitz M S, Weber E J. Sediment-mediated reduction of 2,4,6-trinitrotoluene and fate of the resulting aromatic (poly)amines. Environ Sci Technol. 1999;33:2617–2625. [Google Scholar]
- 66.Emmrich M. Kinetics of the alkaline hydrolysis of 2,4,6-trinitrotoluene in aqueous solution and highly contaminated soils. Environ Sci Technol. 1999;33:3802–3805. doi: 10.1021/es0014990. [DOI] [PubMed] [Google Scholar]
- 67.Esteve-Núñez A. Metabolismo anaerobio del explosivo 2,4,6-trinitrotolueno (TNT) por bacterias de género Pseudomonas. Ph.D. thesis. Granada, Spain: University of Granada; 2000. [Google Scholar]
- 68.Esteve-Núñez A, Ramos J L. Metabolism of 2,4,6-trinitrotoluene by Pseudomonas sp. JLR11. Environ Sci Technol. 1998;32:3802–3808. [Google Scholar]
- 69.Esteve-Núñez A, Luchessi G, Philipps B, Schink B, Ramos J L. Respiration of 2,4,6-trinitrotoluene by Pseudomonas sp. strain JLR11. J Bacteriol. 2000;182:1352–1355. doi: 10.1128/jb.182.5.1352-1355.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Fant F, de Sloovere A, Matthijse K, Marle C, el Fantroussi S, Werstraete W. The use of amino compounds for binding 2,4,6-trinitrotoluene in water. Environ Pollut. 2001;111:503–507. doi: 10.1016/s0269-7491(00)00077-4. [DOI] [PubMed] [Google Scholar]
- 71.Ferguson R. Pilot-scale bioremediation of soils containing TNT at the former Raritan arsenal using sequential strong reducing conditions and aerobic degradation. Second International Symposium on Biodegradation of Nitroaromatic Compounds and Explosives. 1999. [Google Scholar]
- 72.Fernando T, Bumpus J A, Aust S D. Biodegradation of TNT (2,4,6-trinitrotoluene) by Phanerochaete chrysosporium. Appl Environ Microbiol. 1990;56:1666–1671. doi: 10.1128/aem.56.6.1666-1671.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Field J A, De Jong E, Feijoo-Costa G, de Bont J A M. Screening for ligninolytic fungi applicable to the biodegradation of xenobiotics. Trends Biotechnol. 1993;11:44–49. [Google Scholar]
- 74.Fiorella P D, Spain J C. Transformation of 2,4,6-trinitrotoluene by Pseudomonas pseudoalcaligenes JS52. Appl Environ Microbiol. 1997;63:2007–2015. doi: 10.1128/aem.63.5.2007-2015.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.French C E, Hosser S J, Davies G J, Nicklin S, Bruce N C. Biodegradation of explosives by transgenic plants expressing pentaerythritol tetranitrate reductase. Nat Biotechnol. 1999;17:491–494. doi: 10.1038/8673. [DOI] [PubMed] [Google Scholar]
- 76.French C E, Nicklin S, Bruce N. Sequence and properties of pentaerythritol tetranitrate reductase from Enterobacter cloacae PB2. J Bacteriol. 1995;178:6623–6627. doi: 10.1128/jb.178.22.6623-6627.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.French C E, Nicklin S, Bruce N C. Aerobic degradation of 2,4,6-trinitrotoluene by Enterobacter cloacae PB2 and by pentaerythritol tetranitrate reductase. Appl Environ Microbiol. 1998;64:2864–2868. doi: 10.1128/aem.64.8.2864-2868.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Fritsche W, Scheibner K, Herre A, Hofrichter M. Fungal degradation of explosives: TNT and related nitroaromatic compounds. In: Spain J C, Hughes J B, Knackmuss H-J, editors. Biodegradation of nitroaromatic compounds and explosives. Boca Raton, Fla: Lewis Publishers; 2000. pp. 213–238. [Google Scholar]
- 79.Fuller M E, Manning J., Jr Aerobic gram-positive and gram-negative bacteria exhibit differential sensitivity to and transformation of 2,4,6-trinitrotoluene (TNT) Curr Microbiol. 1997;35:77–83. doi: 10.1007/s002849900216. [DOI] [PubMed] [Google Scholar]
- 80.Funk S B, Crawford D L, Crawford R L, Mead G, Davis-Hooker W. Full-scale anaerobic bioremediation of trinitrotoluene contaminated soil. Appl Biochem Biotechnol. 1995;51:625–633. [Google Scholar]
- 81.Funk S B, Roberts D J, Crawford D L, Crawford R L. Initial-phase optimization for bioremediation of munition compound-contaminated soils. Appl Environ Microbiol. 1993;59:2171–2177. doi: 10.1128/aem.59.7.2171-2177.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.George S E, Huggins-Clark G, Brooks L R. Use of a Salmonella microsuspension bioassay to detect the mutagenicity of munitions compounds at low concentrations. Mutat Res. 2001;490:45–56. doi: 10.1016/s1383-5718(00)00150-9. [DOI] [PubMed] [Google Scholar]
- 83.Gilcrease C P, Murphy V G. Bioconversion of 2,4-diamino-6-nitrotoluene to a novel metabolite under anoxic and aerobic conditions. Appl Environ Microbiol. 1995;61:4209–4214. doi: 10.1128/aem.61.12.4209-4214.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Glaus M A, Heijman C G, Schwarzenbach R P, Zeyer J. Reduction of nitroaromatic compounds mediated by Streptomyces sp. exudates. Appl Environ Microbiol. 1992;58:1945–1951. doi: 10.1128/aem.58.6.1945-1951.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Gong P, Wilke B, Fleischmann S. Soil-based phytotoxicity of 2,4,6-trinitrotoluene (TNT) to terrestrial higher plants. Arch Environ Contam Toxicol. 1999;36:152–157. doi: 10.1007/s002449900455. [DOI] [PubMed] [Google Scholar]
- 86.Goodwin A, Kersulyte D, Sisson G, Veldhuyzen van Zanten S J O, Berg D E, Hoffman P S. Metronidazole resistance in Helicobacter pylori is due to null mutations in a gene (rdxA) that encodes an oxygen-insensitive NADPH nitroreductase. Mol Microbiol. 1998;28:383–393. doi: 10.1046/j.1365-2958.1998.00806.x. [DOI] [PubMed] [Google Scholar]
- 87.Gorontzy T, Drzyzga O, Kahl M W, Bruns-Nagel D, Breitung J, von Löw E, Blotevogel K-H. Microbial degradation of explosives and related compounds. Crit Rev Microbiol. 1994;20:265–284. doi: 10.3109/10408419409113559. [DOI] [PubMed] [Google Scholar]
- 88.Gorontzy T, Küver J, Blotevogel K. Microbial transformation of nitroaromatic compounds under anaerobic conditions. J Gen Microbiol. 1993;139:1331–1336. doi: 10.1099/00221287-139-6-1331. [DOI] [PubMed] [Google Scholar]
- 89.Griest W H, Tyndall R L, Stewart A J, Caton J E, Vass A A, Ho C-H, Caldwell W M. Chemical characterization and toxicological testing of windrow composts from explosives-contaminated sediments. Environ Toxicol Chem. 1995;14:51–59. [Google Scholar]
- 90.Groenewegen P E J, de Bont J M A. Degradation of 4-nitrobenzoate via 4-hydroxylaminobenzoate and 3,4-dihydroxybenzoate in Comamonas acidovorans NBA-10. Arch Microbiol. 1992;158:381–386. [Google Scholar]
- 91.Haderlein S B, Schwarzenbach R P. Environmental processes influencing the rate of abiotic reduction of nitroaromatic compounds in the subsurface. In: Spain J C, editor. Biodegradation of nitroaromatic compounds. New York, N.Y: Plenum Press; 1995. pp. 199–225. [Google Scholar]
- 92.Haderlein S B, Weissmahr K W, Schwarzenbach R P. Specific adsorption of nitroaromatic explosives and pesticides to clay minerals. Environ Sci Technol. 1996;30:612–622. [Google Scholar]
- 93.Haderlein S, Hofstetter B, Schwarzenbach R. Subsurface chemistry of nitroaromatic compounds. In: Spain J C, Hughes J B, Knackmuss H-J, editors. Biodegradation of nitroaromatic compounds and explosives. Boca Raton, Fla: Lewis Publishers; 2000. pp. 311–356. [Google Scholar]
- 94.Haïdour A, Ramos J L. Identification of products resulting from the biological reduction of 2,4,6-trinitrotoluene, 2,4-dinitrotoluene and 2,6-dinitrotoluene by Pseudomonas sp. Environ Sci Technol. 1996;30:2365–2370. [Google Scholar]
- 95.Haigler B E, Spain J C. Biodegradation of 4-nitrotoluene by Pseudomonas sp. strain 4NT. Appl Environ Microrbiol. 1993;59:2239–2343. doi: 10.1128/aem.59.7.2239-2243.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Haigler B E, Wallace W H, Spain J C. Biodegradation of 2-nitrotoluene by Pseudomonas sp. strain JS42. Appl Environ Microbiol. 1994;6077:3466–3469. doi: 10.1128/aem.60.9.3466-3469.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Hanne L F, Kirk L L, Appel S M, Narayan A D, Bains K K. Degradation and induction specificity in actinomycetes that degrade p-nitrophenol. Appl Environ Microbiol. 1993;59:3505–3508. doi: 10.1128/aem.59.10.3505-3508.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Harade N, Omura T. Participation of cytocrome P-450 in the reduction of nitro compounds by rat liver microsomes. J Biochem. 1980;87:1539–1554. doi: 10.1093/oxfordjournals.jbchem.a132895. [DOI] [PubMed] [Google Scholar]
- 99.Hartter D R. The use and importance of nitroaromatic compounds in the chemical industry. In: Rickert D E, editor. Chemical Institute of Toxicology Series. Toxicity of nitroaromatic compounds. Washington, D.C.: Hemisphere Publishing; 1985. pp. 1–14. [Google Scholar]
- 100.Harvey S D, Fellows R J, Cataldo D A, Bean R M. Analysis of 2,4,6-trinitrotoluene and its transformation products in soils and plant tissues by high-performance liquid chromatography. J Chromatogr. 1990;518:361–374. [Google Scholar]
- 101.Hattor, K., M. Hashimoto, and M. Ohashi. June 1970. Oxypyrrolnitrin: a new antibiotic. Japanese patent 45029432.
- 102.Hawari J, Halasz A, Beaudet S, Paquet L, Ampleman G, Thiboutot S. Biotransformation of 2,4,6-trinitrotoluene with Phanerochaete chrysosporium in agitated cultures at pH 4.5. Appl Environ Microbiol. 1999;65:2977–2986. doi: 10.1128/aem.65.7.2977-2986.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Hawari J, Halasz A, Paquet L, Zhou E, Spencer B, Ampleman G, Thiboutot S. Characterization of metabolites in the biotransformacion of 2,4,6-trinitrotoluene with anaerobic sludge: role of triaminotoluene. Appl Environ Microbiol. 1998;64:2200–2206. doi: 10.1128/aem.64.6.2200-2206.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Hawthorne S B, Lagadec A J M, Kalderis D, Lilke A V, Miller D J. Pilot-scale destruction of TNT, RDX and HMX on contaminated soils using subcritical water. Environ Sci Technol. 2000;34:3224–3228. [Google Scholar]
- 105.Hess T F, Schmidt S K, Silverstein J, Howe B. Supplemental substrate enhancement of 2,4-dinitrophenol mineralization by a bacterial consortium. Appl Environ Microbiol. 1990;56:1551–1558. doi: 10.1128/aem.56.6.1551-1558.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Higson F K. Microbial degradation of nitroaromatic compounds. Adv Appl Microbiol. 1992;37:1–19. doi: 10.1016/s0065-2164(08)70250-8. [DOI] [PubMed] [Google Scholar]
- 107.Hodgson J, Rho D, Guiot S R, Ampleman G, Thiboutot S, Hawari J. Tween 80 enhanced TNT mineralization by Phanerochaete chrysosporium. Can J Microbiol. 2000;46:110–118. doi: 10.1139/w99-126. [DOI] [PubMed] [Google Scholar]
- 108.Hofstetter T B. Reduction of polynitroaromatic compounds by reduced iron species. Coupling biogeochemical processes with pollutant transformation. Ph.D. thesis. Zürich, Switzerland: Swiss Federal Institute of Technology (EHT); 1999. [Google Scholar]
- 109.Hofstetter T B, Heijman C G, Haderlein S B, Holliger C, Schwarzenbach R P. Complete reduction of TNT and other (poly)nitroaromatic compounds under iron-reducing subsurface conditions. Environ Sci Technol. 1999;33:1479–1487. [Google Scholar]
- 110.Honeycutt M E, Jarvis A S, McFarland V A. Cytotoxicity and mutagenicity of 2,4,6-trinitrotoluene and its metabolites. Ecotoxicol Environ Saf. 1996;35:282–287. doi: 10.1006/eesa.1996.0112. [DOI] [PubMed] [Google Scholar]
- 111.Huang S, Lindahl P A, Wang C, Bennett G N, Rudolph F B, Hughes J B. 2,4,6-Trinitrotoluene reduction by carbon monoxide dehydrogenase from Clostridium thermoaceticum. Appl Environ Microbiol. 2000;66:1474–1478. doi: 10.1128/aem.66.4.1474-1478.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Hughes J B, Shanks J, Vanderford M, Lauritzen J, Bhadra R. Transformation of TNT by aquatic plants and plant tissue cultures. Environ Sci Technol. 1997;31:266–271. [Google Scholar]
- 113.Hughes J B, Wang C, Yesland K, Bhadra R, Richardson A, Bennet G, Rudolph F. Reduction of 2,4,6-trinitrotoluene by Clostridium acetobutylicum through hydroxylamino intermediates. Environ Toxicol Chem. 1998;17:343–348. [Google Scholar]
- 114.Hughes J B, Wang C, Yesland K, Richardson A, Bhadra R, Bennet G, Rudolph F. Bamberger rearrangement during TNT metabolism by Clostridium acetobutylicum. Environ Sci Technol. 1998;32:494–500. [Google Scholar]
- 115.Hughes J B, Wang C Y, Zhang C. Anaerobic biotransformation of 2,4-dinitrotoluene and 2,6-dinitrotoluene by Clostridium acetobutylicum: a pathway through dihydroxylamino intermediates. Environ Sci Technol. 1999;33:1065–1070. [Google Scholar]
- 116.Jarvis A S, McFarland V A, Honeycutt M E. Assesment of the effectiviness of composting for the reduction of toxicity and mutagenicity of explosives-contaminated soil. Ecotoxicol Environ Saf. 1998;39:131–135. doi: 10.1006/eesa.1997.1617. [DOI] [PubMed] [Google Scholar]
- 117.Jerger D E, Woodhull P M. Applications and costs for biological treatment of explosives-contaminated soils in the U.S. In: Spain J, Hughes J B, Knackmuss H-J, editors. Biodegradation of nitroaromatic compounds and explosives. BocaRaton, Fla: Lewis Publishers; 2000. pp. 395–423. [Google Scholar]
- 118.Johnson L R, Davenport R, Balbachand H, Schaeffer D J. Phototoxicity 3. Comparative toxicity of trinitrotoluene and aminodinitrotoluenes to Daphnia magna, Dugesi dorotocephala, and sheep erythrocytes. Ecotoxicol Environ Saf. 1994;27:34–39. doi: 10.1006/eesa.1994.1005. [DOI] [PubMed] [Google Scholar]
- 119.Jones A M, Greer C W, Ampleman G, Thiboutot S, Lavigne J, Hawari J. Biodegradability of selected highly energetic pollutants under aerobic conditions. In: Hinchee E, Hoeppel R E, Anderson D B, editors. Bioremediation of recalcitrant organics. Columbus, Ohio: Battelle Press; 1995. pp. 251–257. [Google Scholar]
- 120.Kaake R H, Crawford D L, Crawford R L. Biodegradation of the nitroaromatic herbicide dinoseb (2-sec-butyl-4,6-dinitrophenol) under reducing conditions. Biodegradation. 1995;6:329–337. doi: 10.1007/BF00695263. [DOI] [PubMed] [Google Scholar]
- 121.Kalafut T, Wales M E, Rastogi V K, Naumova R P, Zaripova S K, Wild J R. Biotransformation patterns of 2,4,6-trinitrotoluene by aerobic bacteria. Curr Microbiol. 1998;36:45–54. doi: 10.1007/s002849900278. [DOI] [PubMed] [Google Scholar]
- 122.Kaplan D L. Biotransformation pathways of hazardous energetic organo-nitro compounds. Adv Appl Biotechnol Ser. 1989;4:157–181. [Google Scholar]
- 123.Kaplan D L. Biological degradation of explosives and chemical agents. Curr Opin Biotechnol. 1992;3:253–260. [Google Scholar]
- 124.Kaplan D L, Kaplan A M. Thermophilic biotransformation of 2,4,6-trinitrotoluene under simulated composting conditions. Appl Environ Microbiol. 1982;44:757–760. doi: 10.1128/aem.44.3.757-760.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Kaplan L A, Siedle A R. Studies in boron hydrides. IV. Stable hydride Meisenheimer adducts. J Org Chem. 1971;36:937–939. [Google Scholar]
- 126.Kato R, Oshima T, Kakanak A. Studies of the mechanism of nitro reduction by rat liver. Mol Pharmacol. 1969;5:487–498. [PubMed] [Google Scholar]
- 127.Khan T A, Bhadra R, Hughes J. Anaerobic transformation of 2,4,6-TNT and related nitroaromatic compounds by Clostridium acetobutylicum. J Ind Microbiol Biotechnol. 1997;18:198–203. [Google Scholar]
- 128.Kiese M, Tager K. The fate of phenylhydroxylamine in human red cells. Naunyn Schmiedebergs Arch Pharmacol. 1976;292:59–66. doi: 10.1007/BF00506490. [DOI] [PubMed] [Google Scholar]
- 129.Kim H Y, Song H G. Comparison of 2,4,6-trinitrotoluene degradation by seven strains of white rot fungi. Curr Microbiol. 2001;41:317–320. doi: 10.1007/s002840010142. [DOI] [PubMed] [Google Scholar]
- 130.Kinouchi T, Manabe Y, Wakisaki K, Ohnishi Y. Biotransformation of 1-nitropyrene in intestinal anaerobic bacteria. Microbiol Immunol. 1982;26:992–1005. doi: 10.1111/j.1348-0421.1982.tb00249.x. [DOI] [PubMed] [Google Scholar]
- 131.Kitts C L, Green C E, Otley R A, Alvarez M A, Unkefer P J. Type I nitroreductases in soil enterobacteria reduce TNT (2,4,6-trinitrotoluene) and RDX (hexahydro-1,3,5-trinitro-1,3,5-triazine) Can J Microbiol. 2000;46:278–282. doi: 10.1139/w99-134. [DOI] [PubMed] [Google Scholar]
- 132.Kobori T, Hiroshi S, Lee W C, Zenno S, Saigo K, Murphy M E P, Tanokura M. Structure and site-directed mutagenesis of a flavoprotein from Escherichia coli that reduces nitro compounds. J Biol Chem. 2001;276:2816–2823. doi: 10.1074/jbc.M002617200. [DOI] [PubMed] [Google Scholar]
- 133.Klausen J, Tröber S P, Haderlein S B, Schwarzenbach R P. Reduction of substituted nitrobenzenes by Fe(II) in aqueous mineral suspensions. Environ Sci Technol. 1995;29:2396–2404. doi: 10.1021/es00009a036. [DOI] [PubMed] [Google Scholar]
- 134.Knicker H, Bruns-Nagel D, Drzyzga O, von Löw E, Steinbach K. Characterization of 15N-TNT residues after an anaerobic/aerobic treatment of soil/molasses mixtures by solid-state 15N NMR spectroscopy. 1. Determination and optimization of relevant NMR spectroscopy parameters. Environ Sci Technol. 1999;33:343–349. [Google Scholar]
- 135.Krumholz L R, Clarkson W W, Wilber G G, Suflita J M. Transformations of TNT and related aminotoluenes in groundwater aquifer slurries under different electron-accepting conditions. J Ind Microbiol Biotechnol. 1997;18:161–169. doi: 10.1038/sj.jim.2900317. [DOI] [PubMed] [Google Scholar]
- 136.Lang P S, Ching W-K, Willberg D M, Hoffmann M R. Oxidative degradation of 2,4,6-trinitrotoluene by ozone in an electrohydraulic discharge reactor. Environ Sci Technol. 1998;32:3142–3148. [Google Scholar]
- 137.Larson R A, Miller P L, Crowley T O. Borohydride photoreduction of nitroaromatic compounds related to military ordenance constituents. Environ Sci Technol. 1996;30:1192–1197. [Google Scholar]
- 138.Lenke H, Achtnich C, Knackmuss H-J. Perspectives of bioelimination of polynitroaromatic compounds. In: Spain J C, Hughes J B, Knackmuss H-J, editors. Biodegradation of nitroaromatic compounds and explosives. Boca Raton, Fla: Lewis Publishers; 2000. pp. 91–126. [Google Scholar]
- 139.Lenke H, Knackmuss H-J. Initial hydrogenation during catabolism of picric acid by Rhodococcus erythropolis HL 24-2. Appl Environ Microbiol. 1992;58:2933–2937. doi: 10.1128/aem.58.9.2933-2937.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Lenke H, Warrelmann J, Daun G, Hund K, Sieglen U, Knackmuss H-J. Biological treatment of TNT-contaminated soil. 2. Biologically induced immobilization of the contaminants and full-scale application. Environ Sci Technol. 1998;32:1964–1971. [Google Scholar]
- 141.Lewis T A, Ederer M M, Crawford R L, Crawford D L. Microbial transformation of 2,4,6-trinitrotoluene. J Ind Microbiol Biotechnol. 1997;18:89–96. doi: 10.1038/sj.jim.2900257. [DOI] [PubMed] [Google Scholar]
- 142.Lewis T A, Goszczynski S, Crawford R L, Korus R A, Admassu W. Products of anaerobic 2,4,6-trinitrotoluene (TNT) transformation by Clostridium bifermentans. Appl Environ Microbiol. 1996;62:4669–4674. doi: 10.1128/aem.62.12.4669-4674.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Li A Z, Marx K A, Walker J, Kaplan D L. Trinitrotoluene and metabolites binding to humic acid. Environ Sci Technol. 1997;31:584–589. [Google Scholar]
- 144.Liu Y Y, Yao M, Fang J L, Wang Y W. Monitoring human risk and exposure to trinitrotoluene (TNT) using haemoglobin adducts as biomarkers. Toxicol Lett. 1995;77:281–287. doi: 10.1016/0378-4274(95)03308-4. [DOI] [PubMed] [Google Scholar]
- 145.Lovering A L, Hyde E I, Searle P F, White S A. The structure of Escherichia coli nitroreductase complexed with nicotinic acid. Three crystal forms at 1.7 Å, 1.8 Å and 2.4 Å resolution. J Mol Biol. 2001;309:203–213. doi: 10.1006/jmbi.2001.4653. [DOI] [PubMed] [Google Scholar]
- 146.Martin J L, Comfort S D, Shea P J, Kokjohn T A, Drijber R A. Denitration of 2,4,6-trinitrotoluene by Pseudomonas savastanoi. Can J Microbiol. 1997;43:447–455. doi: 10.1139/m97-063. [DOI] [PubMed] [Google Scholar]
- 147.Marvin-Sikkema F D, de Bont J A M. Degradation of nitroaromatic compounds by microorganisms. Appl Microbiol Biotechnol. 1994;42:499–507. doi: 10.1007/BF00173912. [DOI] [PubMed] [Google Scholar]
- 148.Mason R P, Holtzmann J L. The role of catalytic superoxide formation in the O2 inhibition of nitroreductase. Biochem Biophys Res Commun. 1975;67:1267–1274. doi: 10.1016/0006-291x(75)90163-1. [DOI] [PubMed] [Google Scholar]
- 149.McCormick N G, Feeherry F E, Levinson H S. Microbial transformation of 2,4,6-TNT and other nitroaromatic compounds. Appl Environ Microbiol. 1976;31:949–958. doi: 10.1128/aem.31.6.949-958.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Michels J, Gottschalk G. Inhibition of the lignin peroxidase of Phanerochaete chrysosporium by hydroxylamino-dinitrotoluene, an early intermediate in the degradation of 2,4,6-TNT. Appl Environ Microbiol. 1994;60:187–194. doi: 10.1128/aem.60.1.187-194.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Michels J, Gottschalk G. Pathway of 2,4,6-trinitrotoluene (TNT) degradation by Phanerochaete chrysosporium. In: Spain J C, editor. Biodegradation of nitroaromatic compounds. New York, N.Y: Plenum Press; 1995. pp. 135–149. [Google Scholar]
- 152.Mondecar M, Bender J, Ross J, George W, Preslan J. Removal of 2,4,6-TNT from contaminated water with microbial mats. In: Hinchee E, editor. Applied biotechnology for site remediation. Columbus, Ohio: Battelle Press; 1997. pp. 342–345. [Google Scholar]
- 153.Montpas S, Samson J, Langlois E, Lei J, Piche Y, Chêvenert R. Degradation of 2,4,6-trinitrotoluene by Serratia marcescens. Biotechnol Lett. 1997;19:291–294. [Google Scholar]
- 154.Naumova R P, Selivanovskaya S L U, Mingatina F A. Possibilities for the deep bacterial destruction of 2,4,6-trinitrotoluene. Mikrobiologia. 1988;57:218–222. [PubMed] [Google Scholar]
- 155.Nishino S F, Paoli G C, Spain J C. Aerobic degradation of dinitrotoluenes and pathway for bacterial degradation of 2,6-dinitrotoluene. Appl Environ Microbiol. 2000;66:2139–2147. doi: 10.1128/aem.66.5.2139-2147.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Nishino S F, Spain J C. Degradation of nitrobenzene by Pseudomonas pseudoalcaligenes. Appl Environ Microbiol. 1993;59:2520–2525. doi: 10.1128/aem.59.8.2520-2525.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Nishino S F, Spain J C, Lenke H, Knackumuss H-J. Mineralization of 2,4-dinitrotoluene and 2,6-dinitrotoluene in soil slurries. Environ Sci Technol. 1999;33:1060–1064. [Google Scholar]
- 158.Nishino S F, Spain J C, He Z. Strategies for aerobic degradation of nitroaromatic compounds by bacteria: process discovery to field application. In: Spain J, Hughes J B, Knackmuss H-J, editors. Biodegradation of nitroaromatic compounds and explosives. Boca Raton, Fla: Lewis Publishers; 2000. pp. 7–61. [Google Scholar]
- 159.Oh B-T, Sarath G, Shea P J. TNT nitroreductase from a Pseudomonas aeruginosa strain isolated from TNT-contaminated soil. Soil Biol Biochem. 2001;33:875–881. [Google Scholar]
- 160.Oh K, Kim Y. Degradation of explosive 2,4,6-trinitrotoluene by s-triazine degrading bacterium isolated from contaminated soil. Bull Environ Contam Toxicol. 1998;61:702–708. doi: 10.1007/s001289900818. [DOI] [PubMed] [Google Scholar]
- 161.Ohmori T, Hagiwara S, Ueda A, Minoda Y, Yamada K. Production of pyoluteorin and its derivatives from n-paraffin by Pseudomonas aeruginosa S10B2. Agric Biol Chem. 1978;42:2031–2036. [Google Scholar]
- 162.Pak J W, Knoke K L, Noguera D R, Fox B G, Chambliss G H. Transformation of 2,4,6-trinitrotoluene by purified xenobiotic reductase B from Pseudomonas fluorescens I-C. Appl Environ Microbiol. 2000;66:4742–4750. doi: 10.1128/aem.66.11.4742-4750.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Palazzo A J, Leggett D C. Effect and disposition of TNT in a terrestrial plant. J Environ Qual. 1986;15:49–52. [Google Scholar]
- 164.Parrish F W. Fungal transformation of 2,4-dinitrotoluene and 2,4,6-trinitrotoluene (TNT) Appl Environ Microbiol. 1977;34:232–233. doi: 10.1128/aem.34.2.232-233.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Pasti-Grigsby M B, Lewis T A, Crawford D L, Crawford R L. Transformation of 2,4,6-trinitrotoluene (TNT) by Actinomycetes isolated from TNT-contaminated and uncontaminated environments. Appl Environ Microbiol. 1996;62:1120–1123. doi: 10.1128/aem.62.3.1120-1123.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Pavlostathis S G, Comstock K G, Jacobson M E, Saunders F M. Transformation of 2,4,6-trinitrotoluene by the aquatic plant Myriophyllum spicatum. Environ Toxicol Chem. 1998;17:2266–2273. [Google Scholar]
- 167.Pennington J, Hayes C A, Myers K F, Ochman M, Gunnison D, Felt D R, McCormick E F. Fate of 2,4,6-trinitrotoluene in a stimulated compost system. Chemosphere. 1995;30:429–438. [Google Scholar]
- 168.Pennington J C, Patrick W H., Jr Adsorption and desorption of 2,4,6-trinitrotoluene by soils. J Environ Qual. 1990;19:559–567. [Google Scholar]
- 169.Pereira W E, Short D L, Manigold D B, Roscio P K. Isolation and characterization of TNT and its metabolites in groundwater by gas chromatograph-mass spectrometer-computer techniques. Bull Environ Contam Toxicol. 1979;21:554–562. doi: 10.1007/BF01685469. [DOI] [PubMed] [Google Scholar]
- 170.Peres C M, Agathos S N. Biodegradation of nitroaromatic pollutants: from pathways to remediation. Biotechnol Annu Rev. 2000;6:197–220. doi: 10.1016/s1387-2656(00)06023-3. [DOI] [PubMed] [Google Scholar]
- 171.Peterson F J, Mason R P, Horspian J, Holtzman J L. Oxygen-sensitive and insensitive nitroreduction by Escherichia coli and rat hepatic microcosomes. J Biol Chem. 1979;254:4009–4014. [PubMed] [Google Scholar]
- 172.Peterson M M, Horst G L, Shea P J, Comfort S D. Germination and seedling development of switchgrass and smooth bromegrass exposed to 2,4,6-trinitrotoluene. Environ Pollut. 1998;99:53–59. doi: 10.1016/s0269-7491(97)00175-9. [DOI] [PubMed] [Google Scholar]
- 173.Pfortner P, Weller M G, Niessner R. Immunological method for detection of nitroaromatic residues bound to humic acids. Fresenius J Anal Chem. 1998;360:192–198. [Google Scholar]
- 174.Preuss A, Fimpel J, Dickert G. Anaerobic transformation of 2,4,6-trinitrotoluene (TNT) Arch Microbiol. 1993;159:345–353. doi: 10.1007/BF00290917. [DOI] [PubMed] [Google Scholar]
- 175.Preuss A, Rieger P G. Anaerobic transformation of 2,4,6-TNT and other nitroaromatic compounds. In: Spain J C, editor. Biodegradation of nitroaromatic compounds. New York, N.Y: Plenum Press; 1995. pp. 69–85. [Google Scholar]
- 176.Price C B, Brannon J M, Hayes C A. Effect of redox potential and pH on TNT transformation in soil-water slurries. J Environ Eng. 1997;123:988–992. [Google Scholar]
- 177.Rafii F, Franklin W, Heflich R H, Cerniglia C. Reduction of nitroaromatic compounds by anaerobic bacteria isolated from the human gastrointestinal tract. Appl Environ Microbiol. 1991;57:962–968. doi: 10.1128/aem.57.4.962-968.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Rafii F, Selby A L, Newton R K, Cerniglia C E. Reduction and mutagenic activation of nitroaromatic compounds by a Mycobacterium sp. Appl Environ Microbiol. 1994;60:4263–4267. doi: 10.1128/aem.60.12.4263-4267.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Ramos J L, Haïdour A, Delgado A, Duque E, Fandila M D, Gil M, Piñar G. Potential of toluene-degrading systems for the construction of hybrid pathways for nitrotoluene metabolism. In: Spain J C, editor. Biodegradation of nitroaromatic compounds. New York, N.Y: Plenum Press; 1995. pp. 53–67. [Google Scholar]
- 180.Regan K M, Crawford R L. Characterization of Clostridium bifermentans and its biotransformation of 2,4,6-trinitrotoluene (TNT) and 1,3,5-triaza-1,3,5-trinitrocyclohexane (RDX) Biotechnol Lett. 1994;16:1081–1086. [Google Scholar]
- 181.Rho D, Hodgson J, Thiboutot S, Ampleman G, Hawari J. Transformation of 2,4,6-trinitrotoluene (TNT) by immobilized Phanerochaete chrysosporium under fed-batch and continuous TNT feeding conditions. Biotechnol Bioeng. 2001;73:271–281. doi: 10.1002/bit.1060. [DOI] [PubMed] [Google Scholar]
- 182.Rhys-Williams W, Taylor S C, Williams P A. A novel pathway for the catabolism of 4-nitrotoluene by Pseudomonas. J Gen Microbiol. 1993;139:1967–1972. doi: 10.1099/00221287-139-9-1967. [DOI] [PubMed] [Google Scholar]
- 183.Rieble S, Joshi D K, Gold M. Aromatic nitroreductase from the basidiomycete Phanerochaete chrysospurium. Biochem Biophys Res Commun. 1994;205:298–304. doi: 10.1006/bbrc.1994.2664. [DOI] [PubMed] [Google Scholar]
- 184.Riefler R G, Smets B F. Enzymatic reduction of 2,4,6-trinitrotoluene and related nitroarenes: kinetics linked to one-electron redox potentials. Environ Sci Technol. 2000;34:3900–3906. [Google Scholar]
- 185.Rieger P, Knackmuss H-J. Basic knowledge and perspectives on biodegradation of 2,4,6-TNT and related nitroaromatic compounds in contaminated soil. In: Spain J C, editor. Biodegradation of nitroaromatic compounds. New York, N.Y: Plenum Press; 1995. pp. 1–8. [Google Scholar]
- 186.Rieger P-G, Sinnwell V, Preuss A, Franke W, Knackmuss H-J. Hydride-Meisenheimer complex formation and protonation as key reactions of 2,4,6-trinitrophenol biodegradation by Rhodococcus erythropolis. J Bacteriol. 1999;181:1189–1195. doi: 10.1128/jb.181.4.1189-1195.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Roberts D J, Ahmad F, Pendharkar S. Optimization of an aerobic polishing stage to complete the anaerobic treatment of munitions-contaminated soils. Environ Sci Technol. 1996;30:2021–2026. [Google Scholar]
- 188.Roberts D J, Kaake R H, Funk S B, Crawford D L, Crawford R L. Field-scale anaerobic bioremediation of dinoseb-contaminated soils. In: Gealt M, Levin M, editors. Biotreatment of industrial and hazardous wastes. New York, N.Y: McGraw Hill Book Co.; 1993. pp. 219–244. [Google Scholar]
- 189.Robidoux P Y, Hawari J, Thiboutot S, Ampleman G, Sunahara G I. Acute toxicity of 2,4,6-trinitrotoluene in earthworm (Eisenia andrei) Ecotoxicol Environ Saf. 1999;44:311–321. doi: 10.1006/eesa.1999.1839. [DOI] [PubMed] [Google Scholar]
- 190.Rodgers J D, Bunce N J. Treatment methods for the remediation of nitroaromatic explosives. Water Res. 2001;35:2101–2111. doi: 10.1016/s0043-1354(00)00505-4. [DOI] [PubMed] [Google Scholar]
- 191.Roldán M D, Blasco R, Caballero F J, Castillo F. Degradation of p-nitrophenol by the phototrophic bacterium Rhodobacter capsulatus. Arch Microbiol. 1998;169:36–42. doi: 10.1007/s002030050538. [DOI] [PubMed] [Google Scholar]
- 192.Rügge K, Hofstetter T B, Haderlein S B, Bjerg P L, Knudsen S, Zraunig C, Mosbaek H, Christensen T H. Characterization of predominant reductants in an anaerobic leachate-contaminated aquifer by nitroaromatic probe compounds. Environ Sci Technol. 1998;32:23–31. [Google Scholar]
- 193.Schackmann A, Müller R. Reduction of nitroaromatic compounds by different Pseudomonas species under aerobic conditions. Appl Microbiol Biotechnol. 1991;34:809–813. [Google Scholar]
- 194.Scheibner K, Hofrichter M. Conversion of aminonitrotoluenes by fungal manganese peroxidase. J Basic Microbiol. 1998;38:51–59. [PubMed] [Google Scholar]
- 195.Scheibner K, Hofrichter M, Fritsche W. Mineralization of 2-amino-4,6-dinitrotoluene by manganese peroxidase of the white-rot fungus Nematoloma frowardii. Biotechnol Lett. 1997;19:835–839. [Google Scholar]
- 196.Scheibner K, Hofrichter M, Herre A, Michels J, Fritsche W. Screening for fungi intensevely mineralizing 2,4,6-trinitrotoluene. Appl Microbiol Biotechnol. 1997;47:452–457. doi: 10.1007/s002530050955. [DOI] [PubMed] [Google Scholar]
- 197.Schmelling D C, Gray K A, Kamat P V. Role of reduction in the photocatalytic degradation of TNT. Environ Sci Technol. 1996;30:2547–2555. [Google Scholar]
- 198.Schnell S, Schinck B. Anaerobic aniline degradation via reductive deamination of 4-aminobenzoyl-CoA in Desulfobacterium anilini. Arch Microbiol. 1991;155:183–190. [Google Scholar]
- 199.Selim H M, Xue S K, Iskandar I K. Transport of 2,4,6-trinitrotoluene and hexahydro-1,3,5-trinitro-1,3,5-triazine in soils. Soil Sci. 1995;160:328–339. [Google Scholar]
- 200.Sembries S, Crawford R L. Production of Clostridium bifermentans spores as inoculum for bioremediation of nitroaromatic contaminants. Appl Environ Microbiol. 1997;63:2100–2104. doi: 10.1128/aem.63.5.2100-2104.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Sens C, Sheidemann P, Werner D. The distribution of 14C-TNT in different biochemical compartments of the monocotyledoneous Triticum aestivum. Environ Pollut. 1999;104:113–119. [Google Scholar]
- 202.Shen C F, Hawari J A, Ampleman G, Thiboutot S, Guiot S R. Origin of p-cresol in the anaerobic degradation of trinitrotoluene. Can J Microbiol. 2000;46:119–124. doi: 10.1139/w99-124. [DOI] [PubMed] [Google Scholar]
- 203.Sheremata T W, Thiboutot S, Ampleman G, Paquet L, Halasz A, Hawari J. Fate of 2,4,6-trinitrotoluene and its metabolites in natural and model soil systems. Environ Sci Technol. 1999;33:4002–4008. [Google Scholar]
- 204.Sheremata T W, Hawari J. Cyclodextrins for desorption and solubilization of 2,4,6-trinitrotoluene and its metabolites from soil. Environ Sci Technol. 2000;34:3462–3468. [Google Scholar]
- 205.Shim C Y, Crawford D L. Biodegradation of trinitrotoluene (TNT) by a strain of Clostridium bifermentans. In: Hinchee R E, Fredrickson J, Alleman B C, editors. Bioaugmentation for site remediation. Columbus, Ohio: Battelle Press; 1995. pp. 57–69. [Google Scholar]
- 206.Shriver-Lake L C, Donner B L, Ligler F S. On-site detection of TNT with a portable fiber optic biosensor. Environ Sci Technol. 1997;31:837–841. [Google Scholar]
- 207.Siciliano S D, Roy R, Greer C W. Reduction in denitrification activity in field soils exposed to long term contamination by 2,4,6-trinitrotoluene (TNT) FEMS Microbiol Ecol. 2000;32:61–68. doi: 10.1111/j.1574-6941.2000.tb00699.x. [DOI] [PubMed] [Google Scholar]
- 208.Smock L A, Stoneburner D L, Clark J R. The toxic effects of trinitrotoluene (TNT) and its primary degradation products on two species of algae and the fathead minnow. Water Res. 1976;10:537. [Google Scholar]
- 209.Somerville C C, Nishino S F, Spain J C. Purification and characterization of nitrobenzene nitroreductase from Pseudomonas pseudoalcaligenes JS45. J Bacteriol. 1995;177:3837–3842. doi: 10.1128/jb.177.13.3837-3842.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Spain J. Bacterial degradation of nitroaromatic compounds under aerobic conditions. In: Spain J, editor. Biodegradation of nitroaromatic compounds. New York, N.Y: Plenum Press; 1995. pp. 19–35. [Google Scholar]
- 211.Spain J, Hughes J B, Knackmuss H-J, editors. Biodegradation of nitroaromatic compounds and explosives. Boca Raton, Fla: Lewis Publishers; 2000. [Google Scholar]
- 212.Spain J C. Biodegradation of nitroaromatic compounds. Annu Rev Microbiol. 1995;49:523–555. doi: 10.1146/annurev.mi.49.100195.002515. [DOI] [PubMed] [Google Scholar]
- 213.Spanggord R J, Mortelmans K E, Griffin A F, Simmon V F. Mutagenicity in Salmonella typhimurium and structure-activity relationships of wastewater components emanating from the manufacture of trinitrotoluene. Environ Mutagen. 1982;4:163–179. doi: 10.1002/em.2860040207. [DOI] [PubMed] [Google Scholar]
- 214.Spanggord R J, Spain J C, Nishino S F, Mortelmans K E. Biodegradation of 2,4-dinitrotoluene by a Pseudomonas sp. Appl Environ Microbiol. 1991;57:3200–3205. doi: 10.1128/aem.57.11.3200-3205.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Spanggord R J, Stewart K R, Riccio E S. Mutagenicity of tetranitroazoxytoluenes: a preliminary screening in Salmonella typhimurium strains TA100 and TA100NR. Mutat Res. 1995;335:207–211. doi: 10.1016/0165-1161(95)00022-4. [DOI] [PubMed] [Google Scholar]
- 216.Spanggord R J, Yao C D, Mill T. Oxidation of aminodinitrotoluenes with ozone: products and pathways. Environ Sci Technol. 2000;34:497–504. [Google Scholar]
- 217.Spiess T, Desiere F, Fischer P, Spain J C, Knackmuss H-J, Lenke H. A new 4-nitrotoluene degradation pathway in a Mycobacterium strain. Appl Environ Microbiol. 1998;64:446–452. doi: 10.1128/aem.64.2.446-452.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Spiker J K, Crawford D L, Crawford R L. Influence of 2,4,6-trinitrotoluene (TNT) concentration on the degradation of TNT in explosive-contaminated soils by the white rot fungus Phanerochaete chrysosporium. Appl Environ Microbiol. 1992;58:3199–3202. doi: 10.1128/aem.58.9.3199-3202.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Stahl J D, Aust S D. Plasma membrane dependent reduction of 2,4,6-TNT by Phanerochaete chrysosporium. Biochem Biophys Res Commun. 1993;192:471–476. doi: 10.1006/bbrc.1993.1439. [DOI] [PubMed] [Google Scholar]
- 220.Stahl J D, Aust S D. Metabolism and detoxification of TNT by Phanerochaete chrysosporium. Biochem Biophys Res Comm. 1993;192:477–482. doi: 10.1006/bbrc.1993.1440. [DOI] [PubMed] [Google Scholar]
- 221.Stahl J D, Aust S D. Biodegradation of 2,4,6-trinitrotoluene by the white rot fungus Phanerochaete chrysosporium. In: Spain J C, editor. Biodegradation of nitroaromatic compounds. New York, NY: Plenum Press; 1995. pp. 117–134. [Google Scholar]
- 222.Stockinger H, Heinzle E, Kut O M. Removal of chloro- and nitroaromatic wastewater pollutants by ozonation and biotreatment. Environ Sci Technol. 1998;29:2016–2022. doi: 10.1021/es00008a021. [DOI] [PubMed] [Google Scholar]
- 223.Stolpmann H, Lenke H, Warrelmann J, Heuermann E, Fruechtnicht A, Daun G, Knackmuss H-J. Bioremediation of TNT-contaminated soil by the TERRANOX® system. In: Hindee R E, Skeen R S, Sayles G P, editors. Biological unit processes for hazardous waste treatment. Columbus, Ohio: Batelle Press; 1999. pp. 283–288. [Google Scholar]
- 224.Styles J A, Cross M F. Activity of 2,4,6-TNT in an in vitro mammalian gene mutation assay. Cancer Lett. 1983;20:103–108. doi: 10.1016/0304-3835(83)90194-5. [DOI] [PubMed] [Google Scholar]
- 225.Suen W, Haigler B E, Spain J C. 2,4-Dinitrotoluene dioxygenase from Burkholderia sp. strain DNT: similarity to naphthalene dioxygenase. J Bacteriol. 1996;178:4926–4934. doi: 10.1128/jb.178.16.4926-4934.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Tadros M G, Crawford A, Mateo-Sullivan A, Zhang C, Hughes J B. Toxic effects of hydroxylamino intermediates from microbial transformation of trinitrotoluene and dinitrotoluenes on algae Selenastrum capricornutum. Bull Environ Contam Toxicol. 2000;64:579–585. doi: 10.1007/s001280000042. [DOI] [PubMed] [Google Scholar]
- 227.Tan E L, Ho C H, Griest W H, Tyndall R L. Mutagenicity of trinitrotoluene and its metabolites formed during composting. J Toxicol Environ Health. 1992;36:165–175. doi: 10.1080/15287399209531632. [DOI] [PubMed] [Google Scholar]
- 228.Tatsumi K, Koga N, Tamura S K, Yoshimura H, Wardman P, Kato Y. Enzymatic cis/trans isomerization of nitrofuran derivatives-isomerizing activity of xanthine oxidase lipoyl dehydrogenase, DT-diaphorase and liver microsomes. Biochim Biophys Acta. 1979;567:75–87. doi: 10.1016/0005-2744(79)90174-8. [DOI] [PubMed] [Google Scholar]
- 229.Taylor R P. Novel Meisenheimer complexes: the preparation of tetramethylammonium 1,1-dihydro-2,4,6-trinitrocyclohexadienate. J Chem Soc Chem Comm. 1970;1970:1463–1463. [Google Scholar]
- 230.Thompson P L, Ramer L A, Schnoor J L. Uptake and transformation of TNT by hybrid poplar trees. Environ Sci Technol. 1998;32:975–980. [Google Scholar]
- 231.Thornton-Manning J R, Campbell W L, Hass B S, Chen J J, Fu P P, Cerniglia C E, Heflich R H. The role of nitro reduction in the synergistic mutational response induced by mixtures of 1- and 3-nitrobenzo[a]pyrene in Salmonella typhymurium. Environ Mol Mutagen. 1989;13:203–210. doi: 10.1002/em.2850130303. [DOI] [PubMed] [Google Scholar]
- 232.Tischler M, Ayer S W, Andersen R J. Nitrophenols from northeast Pacific bryozoans. Comp Biochem Physiol Ser B. 1986;84:43–45. [Google Scholar]
- 233.Traxler R W, Wood E, Delaney J M. Bacterial degradation of alpha-TNT. Dev Ind Microbiol. 1974;16:71–76. [Google Scholar]
- 234.Vaatanen A R. Spectrum of spontaneous and 2,4,6-trinitrotoluene (TNT)-induced mutations in Salmonella typhymurium strains with different nitroreductase and o-acetyltransferase activities. Mutat Res. 1997;379:185–190. doi: 10.1016/s0027-5107(97)00136-x. [DOI] [PubMed] [Google Scholar]
- 235.Van Aken B, Cameron M D, Stahl J D, Plumat A, Naveau H, Aust S I, Agathos S N. Glutathione-mediated mineralization of 14C-labeled 2-amin-dinitrotoluene by Mn-dependent peroxidase HS from white-rot fungus Phanerochaete chrysosporiun. Appl Microbiol Biotechnol. 2000;54:659–664. doi: 10.1007/s002530000436. [DOI] [PubMed] [Google Scholar]
- 236.Van Aken B, Godefroid L M, Peres C M, Naveau H, Agathos S N. Mineralization of 14C-ring labeled 4-hydroxylamino-2,6-dinitrotoluene by manganese-dependent peroxidase of the white-rot basidiomycete Phlebia radiata. J Biotechnol. 1999;68:159–169. [Google Scholar]
- 237.Van Aken B, Hofrichter M, Scheibner K, Hatakka A I, Naveau H, Agathos S N. Transformation and mineralization of 2,4,6-trinitrotoluene (TNT) by manganese peroxidase from the white-rot basidiomycete Phlebia radiata. Biodegradation. 1999;10:83–91. doi: 10.1023/a:1008371209913. [DOI] [PubMed] [Google Scholar]
- 238.Van Aken B, Skubusz K, Naveau H, Agathos S N. Biodegradation of 2,4,6-trinitrotoluene by the white-rot basidiomycete Phlebia radiata. Biotechnol Lett. 1997;19:813–817. doi: 10.1023/a:1008371209913. [DOI] [PubMed] [Google Scholar]
- 239.Vanderberg L A, Perry J J, Unkefer P J. Catabolism of 2,4,6-trinitrotoluene by Mycobacterium vaccae. Appl Microbiol Biotechnol. 1995;43:937–945. doi: 10.1007/BF02431931. [DOI] [PubMed] [Google Scholar]
- 240.Vanderford M, Shanks J V, Hughes J B. Phytotransformation of trinitrotoluene (TNT) and distribution of metabolic products in Myriophyllum aquaticum. Biotechnol Lett. 1997;3:277–280. [Google Scholar]
- 241.Vorbeck C, Lenke H, Fischer P, Knackmuss H-J. Identification of a hydride-Meisenheimer complex as a metabolite of 2,4,6-trinitrotoluene by a Mycobacterium strain. J Bacteriol. 1994;176:932–934. doi: 10.1128/jb.176.3.932-934.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Vorbeck C, Lenke H, Fischer P, Spain J C, Knackmuss H-J. Initial reductive reactions in aerobic microbial metabolism of 2,4,6-trinitrotoluene. Appl Environ Microbiol. 1998;64:246–252. doi: 10.1128/aem.64.1.246-252.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Walker J E, Kaplan D L. Biological degradation of explosives and chemical agents. Biodegradation. 1992;3:369–385. [Google Scholar]
- 244.Watanabe M, Nishino T, Takio K, Sofuni T, Nohmi T. Purification and characterization of wild-type and mutant “classical” nitroreductases of Salmonella typhimurium. J Biol Chem. 1998;273:23922–23928. doi: 10.1074/jbc.273.37.23922. [DOI] [PubMed] [Google Scholar]
- 245.Wayment D G, Bhadra R, Lauritzen J, Hughes J, Shanks J V. A transient study of formation of conjugates during TNT metabolism by plant tissues. Int J Phytoremed. 1999;1:227–239. [Google Scholar]
- 246.Weissmahr K W, Haderlein S B, Schwarzenbach R P. In situ spectroscopic investigations of adsorption mechanisms of nitroaromatic compounds at clay minerals. Environ Sci Technol. 1997;31:240–247. [Google Scholar]
- 247.Weissmahr K W, Haderlein S B, Schwarzenbach R P. Complex formation of soil minerals with nitroaromatic explosives and other p-acceptors. Soil Sci Soc Am J. 1998;62:369–378. [Google Scholar]
- 248.Weissmahr K W, Hildenbrand M, Schwarzenbach R P, Haderlein S B. Laboratory and field scale evaluation of geochemical controls on groundwater transport of nitroaromatic ammunition residues. Environ Sci Technol. 1999;33:2593–2600. [Google Scholar]
- 249.Whiteway J, Koziarz P, Veall J, Sandhu N, Kumar P, Hoecher B, Lambert I B. Oxygen-insensitive nitroreductases: analysis of the roles of nfsA and nfsB in development of resistance to 5-nitrofuran derivatives in Escherichia coli. J Bacteriol. 1998;180:5529–5539. doi: 10.1128/jb.180.21.5529-5539.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Widrig D L, Boopathy R, Manning J F., Jr Bioremediation of TNT-contaminated soil: a laboratory study. Environ Toxicol Chem. 1997;16:1141–1148. [Google Scholar]
- 251.Wikstrom P, Andersson A C, Nygren Y, Sjostrom J, Forsman M. Influence of TNT transformation on microbial community structure in four different lake microcosms. J Appl Microbiol. 2000;89:302–308. doi: 10.1046/j.1365-2672.2000.01111.x. [DOI] [PubMed] [Google Scholar]
- 252.Willberg D M, Lang P S, Höchemer R H, Kratel A, Hoffmann M R. Degradation of 4-chlorophenol, 3,4-dichloroaniline and 2,4,6-trinitrotoluene in an electrohydraulic discharge reactor. Environ Sci Technol. 1996;30:2526–2534. [Google Scholar]
- 253.Williams R T, Ziegenfuss P S, Sisk W E. Composting of explosives and propellant contaminated soils under thermophilic and mesophilic conditions. J Ind Microbiol. 1992;9:137–144. [Google Scholar]
- 254.Wolper M K, Althans J R, Johns D G. Nitroreductase activity of mammaliam liver aldehyde oxidase. J Pharmacol Exp Ther. 1973;185:202–213. [PubMed] [Google Scholar]
- 255.Won W D, Disalvo L H, Ng J. Toxicity and mutagenicity of 2,4,6-TNT and its microbial metabolites. Appl Environ Microbiol. 1976;31:576–580. doi: 10.1128/aem.31.4.576-580.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Won W D, Heckly R J, Glover D J, Hoffsommer J C. Metabolic disposition of 2,4,6-trinitrotoluene. Appl Microbiol. 1974;27:513–516. doi: 10.1128/am.27.3.513-516.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257.Xue S K, Iskandar I K, Selim H M. Adsorption-desorption of 2,4,6-trinitrotoluene and hexahidro-1,3,5-trinitro-1,3,5-triazine in soils. Soil Sci. 1995;160:317–327. [Google Scholar]
- 258.Yamashina I, Shikata S, Egami F. Enzymatic reduction of aromatic nitro, nitroso and hydroxylamino compounds. Bull Chem Soc Jpn. 1953;27:44–45. [Google Scholar]
- 259.Yu S Y, Bailey G W. Reduction of nitrobenzene by four sulfide minerals: kinetics, products and solubility. J Environ Qual. 1992;21:86–94. [Google Scholar]
- 260.Zachariah P K, Juchau M R. The role of gut flora in the reduction of aromatic nitro-groups. Drug Metab Dispos. 1974;2:74–78. [PubMed] [Google Scholar]
- 261.Zenno S, Kobori T, Tanokura M, Saigo K. Conversion of NfsA, the major Escherichia coli nitroreductase, to a flavin reductase with an activity similar to that of Frp, a flavin reductase in Vibrio harveyi, by a single amino acid substitution. J Bacteriol. 1998;180:422–425. doi: 10.1128/jb.180.2.422-425.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262.Zenno S, Koike H, Kumar A N, Jarayaman R, Tanokura M, Saigo K. Biochemical characterization of NfsA, the Escherichia coli major nitroreductase exhibiting a high amino acid sequence homology to Frp, a Vibrio harveyi flavin oxidoreductase. J Bacteriol. 1996;178:4508–4514. doi: 10.1128/jb.178.15.4508-4514.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Zenno S, Koike H, Tanokura M, Saigo K. Conversion of NfsB, a minor Escherichia coli nitroreductase, to a flavin reductase similar in biochemical properties to FRase I, the major flavin reductase in Vibrio fischeri, by a single amino acid substitution. J Bacteriol. 1996;178:4731–4733. doi: 10.1128/jb.178.15.4731-4733.1996. . J. C. [DOI] [PMC free article] [PubMed] [Google Scholar]