Abstract
Analysis of the bacterial genome sequences shows that many human and animal pathogens encode primary membrane Na+ pumps, Na+-transporting dicarboxylate decarboxylases or Na+-translocating NADH:ubiquinone oxidoreductase, and a number of Na+-dependent permeases. This indicates that these bacteria can utilize Na+ as a coupling ion instead of or in addition to the H+ cycle. This capability to use a Na+ cycle might be an important virulence factor for such pathogens as Vibrio cholerae, Neisseria meningitidis, Salmonella enterica serovar Typhi, and Yersinia pestis. In Treponema pallidum, Chlamydia trachomatis, and Chlamydia pneumoniae, the Na+ gradient may well be the only energy source for secondary transport. A survey of preliminary genome sequences of Porphyromonas gingivalis, Actinobacillus actinomycetemcomitans, and Treponema denticola indicates that these oral pathogens also rely on the Na+ cycle for at least part of their energy metabolism. The possible roles of the Na+ cycling in the energy metabolism and pathogenicity of these organisms are reviewed. The recent discovery of an effective natural antibiotic, korormicin, targeted against the Na+-translocating NADH:ubiquinone oxidoreductase, suggests a potential use of Na+ pumps as drug targets and/or vaccine candidates. The antimicrobial potential of other inhibitors of the Na+ cycle, such as monensin, Li+ and Ag+ ions, and amiloride derivatives, is discussed.
Most bacteria rely on proton motive force as a source of energy for a variety of cellular processes. Usually, an H+ cycle includes generation of the transmembrane electrochemical gradient of H+ (proton motive force) by primary transport systems (H+ pumps) and its use for ATP synthesis, solute transport, motility, reverse electron transport, etc. (reviewed, for example, in references 76, 128, 185, and 186). A substantial body of evidence indicates, however, that certain extremophilic, particularly alkalophilic and thermophilic, bacteria can use Na+ as a coupling ion in an Na+ cycle instead of, or in addition to, the H+ cycle (47, 183–185, 188). As in the H+ cycle, a fully operational Na+ cycle would include a primary Na+ pump that directly couples Na+ translocation to a chemical reaction, an Na+-transporting membrane ATP synthetase, a number of Na+-dependent membrane transporters, and an Na+-dependent flagellar motor. While certain Na+-dependent functions, such as Na+-dependent uptake of melibiose, proline, and glutamate, have been observed in many bacteria, including Escherichia coli and Bacillus subtilis (23, 37, 128, 161, 219), the ion gradients that served as energy sources for these transports have been generated by primary H+ pumps and converted to Na+ gradients by Na+/H+ antiporters (Fig. 1). As a result, until very recently the Na+ cycle has been suspected in many different bacteria but experimentally verified in only precious few of them, such as Vibrio alginolyticus, Propionigenium modestum, and Clostridium fervidus (38, 39, 45, 188). Based on their Na+ requirement for growth and Na+-dependent respiration, Na+ cycling has been proposed in a number of marine bacteria (148, 209; reviewed in reference 109). Here, by analyzing bacterial genomic sequences, including the recently published complete genomes of Vibrio cholerae (83), Pseudomonas aeruginosa (194), and Pasteurella multocida (131), we show that the Na+ cycle may be common among human and animal pathogens and we discuss its potential role in their virulence. Due to its emphasis on genome analysis, this review does not aim to cover detailed biochemical properties of the Na+-dependent systems, which have been extensively reviewed previously (48, 50). Properties of membrane transporters, including Na+-dependent ones, have been reviewed by Saier and coworkers (156, 169–171); recently, an analysis of the distribution of various transporters in the first 18 sequenced microbial genomes has been published (155). An extensive review of the type III protein secretion systems in various bacterial pathogens (93) included brief characterizations of the pathogens involved, some of which are subjects of this review.
FIG. 1.
Proton and sodium ion cycles in bacterial energetics. “Primary pump” indicates any proton or sodium motive force generator (e.g., respiratory ion pump, membrane ATPase, or a Na+-transporting dicarboxylate decarboxylase). “H+ (or Na+) porter” indicates consumers of proton (or sodium) motive force (symporters, flagellar motor, etc.). The actual presence of partial components of both cycles in the membrane of each particular bacterial species may vary, depending on the physiological state of the cell. Na+/H+ antiporters convert proton motive force into sodium motive force and vice versa, playing an important role in cell homeostasis.
PRIMARY NA+ PUMPS
Na+-Transporting Dicarboxylate Decarboxylases
The first evidence of an Na+ cycle in bacteria came from the discovery that decarboxylation of oxaloacetate in the anaerobic bacterium Propionigenium modestum was Na+ dependent and was coupled to the extrusion of Na+ ions from the cytoplasm into the medium (41). In this way, the cells were able to conserve part of the free energy released during the exergonic decarboxylation reaction
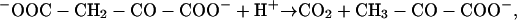 |
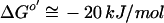 |
in the form of a transmembrane gradient of Na+ ions (50). Further studies of oxaloacetate decarboxylase and similar biotin-dependent membrane-bound decarboxylases have shown that active export of Na+ ions can also be driven by decarboxylation of malonate, methylmalonyl coenzyme A (methylmalonyl-CoA), and glutaconyl-CoA. These energy-conserving “dicarboxylate decarboxylases,” functioning as primary Na+ pumps, have been found in a number of bacteria that grow anaerobically on saturated dicarboxylic acids, such as Klebsiella aerogenes, Veillonella alcalescens, Propionigenium modestum, Malonomonas rubra, Salmonella enterica serovar Typhimurium, and Acidaminococcus fermentans (18, 41; reviewed in references 43, 44, 47, and 48). Na+ gradients, generated by these enzymes, could be used for ATP synthesis and active transport (42, 160). Na+ gradient-driven ATP synthesis, referred to as decarboxylation phosphorylation, is the only ATP-generating mechanism in P. modestum and M. rubra (49, 85). Genetic and enzymological analysis showed that the Na+-transporting oxaloacetate decarboxylase enzyme consists of just three subunits, alpha, beta, and gamma, encoded in the oadGAB operon (51, 120). Malonate, methylmalonyl-CoA, and glutaconyl-CoA decarboxylases have a more complex organization but also contain alpha and beta subunits, homologous to the alpha and beta subunits, respectively, of oxaloacetate decarboxylase (12, 17, 18, 91, 92). Inspection of complete microbial genomes finds conserved oadAB operons (or, in some cases, separate oadA and oadB genes) in a number of phylogenetically distant (Fig. 2) bacteria, from the anaerobic hyperthermophile Thermotoga maritima to such human pathogens as Salmonella enterica serovar Typhi, and Treponema pallidum (Table 1). One could argue whether these data should be interpreted as evidence either of the ancient origin of this enzyme or of its propensity to be horizontally transferred among different bacterial phyla. The latter possibility seems quite plausible, since acquisition of the oadGAB operon would provide the bacterium with the ability to generate membrane potential at the expense of a fairly simple chemical reaction, which should be of selective advantage under some conditions. In any case, Na+ gradient generation by decarboxylase-coupled ion transfer appears to occur in a limited number of (mostly) anaerobic bacteria, making it an exception rather than a rule in microbial world.
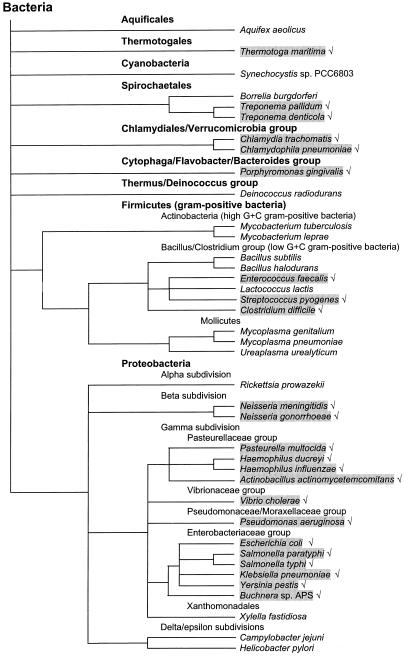
FIG. 2.
Phylogenetic distribution of the bacterial pathogens that use the Na+ cycle. The dendrogram shows the taxonomic positions of the organisms with completely sequenced genomes and several pathogens discussed in the text, according to the NCBI Taxonomy database (http://www.ncbi/nlm.nih.gov/Taxonomy) (218). The branches indicate taxonomic relations only; their lengths do not necessarily reflect evolutionary distances. The main bacterial phyla are shown in boldface. Bacterial species that appear to utilize the Na+ cycle are shaded.
TABLE 1.
Distribution of primary Na+ pumps in bacteria
| Organisma | NQR
|
Oxaloacetate and malonate decarboxylasesd
|
Sequencing center or referencee | ||
|---|---|---|---|---|---|
| Gene orderb | Gene namec | oadGAB gene name | mdcABCDE gene name | ||
| Complete genomes | |||||
| Escherichia coli | nqrEGABCD | ydgLMNOPQ | — | — | 13 |
| Haemophilus influenzae | nqrABCDEF nqrEGABCD | HI0164–HI0171, HI1683–HI1688 | — | — | 59 |
| Neisseria meningitidis | nqrABCDEF | NMB0569–NMB0564 | — | — | 153, 199 |
| Treponema pallidum | — | — | TP0055–TP0057 | — | 217 |
| Chlamydia trachomatis | nqrBCDE, NqrA, NqrF | CT278–CT281, CT634, CT0740 | — | — | 190 |
| Chlamydia pneumoniae | nqrBCDE, nqrA, NqrF | CPn0427–CPn0430 CPn0743, CPn0883 | — | — | 104 |
| Thermotoga maritima | nqrABCDEG | TM0244–TM0249 | TM0128, TM0880 | — | 141 |
| Vibrio cholerae | nqrABCDEF nqrEGABCD | VC2295–VC2290 VC1017–VC1012 | VC0549–VC0551 VC0794–VC0792 | — | 83 |
| Pseudomonas aeruginosa | nqrABCDEF nqrEGABCD | PA2999–PA2994 PA3489–PA3494 | — | PA0208–PA0212 | 194 |
| Pasteurella multocida | nqrABCDEF nqrEGABCD | PM1328–PM1333 PM0387–PM0382 | PM1421–PM1423 | — | 131 |
| Buchnera sp. strain APS | nqrEGABCD | BU113–BU118 | — | — | 179 |
| Unfinished genomes | |||||
| Actinobacillus actinomycetemcomitans | nqrABCDEF | ND | oadGAB | ND | Oklahoma University |
| Clostridium difficile | nqrABCDEG | ND | ND | ND | Sanger Centre |
| Enterococcus faecalis | ND | ND | oadA, oadB | ND | TIGR |
| Haemophilus ducreyi | nqrABCDEF | ND | oadGAB | ND | University of Washington |
| Klebsiella pneumoniae | nqrEGABCD | ND | oadGAB | mdcABCDE | Washington University (120, 176) |
| Neisseria gonorrhoeae | nqrABCDEF | ND | ND | ND | Oklahoma University |
| Porphyromonas gingivalis | nqrABCDEF | ND | oadA, oadB | ND | TIGR, Forsyth Institute |
| Salmonella enterica serovar Paratyphi | nqrEGABCD | ND | oadGAB | ND | Washington University (221) |
| Salmonella enterica serovar Typhi | nqrEGABCD | ND | oadGAB | ND | Sanger Centre |
| Streptococcus pyogenes | ND | ND | oadA, oadB | ND | Oklahoma University Sanger Centre |
| Treponema denticola | nqrABCDE | ND | ND | ND | TIGR |
| Yersinia pestis | nqrEGABCD | ND | ND | ND | Sanger Centre |
Only pathogenic bacteria that encode a primary Na+ pump (NQR or a dicarboxylate decarboxylase) are listed. Other bacteria with completely sequenced genomes, such as Aquifex aeolicus, Bacillus subtilis, B. halodurans, Campylobacter jejuni, Deinococcus radiodurans, Helicobacter pylori, Lactococcus lactis, Mycobacterium tuberculosis, Mycobacterium leprae, Mycoplasma genitalium, Mycoplasma pneumoniae, Rickettsia prowazekii, Synechocystis sp. strain PCC6803, Ureaplasma urealyticum, and Xylella fastidiosa do not appear to encode either of these two primary Na+ pumps and are therefore presumed devoid of a functional Na+ cycle, although most of them encode Na+/H+ antiporters and probably Na+-dependent transporters. Borrelia burgdorferi encodes a homolog of NqrA and NqrB (BB0072) but does not encode other NQR subunits. The list of bacteria with unfinished genomes includes only organisms where a primary Na+ pump could be unequivocally identified from the available sequence data. Both lists are expected to expand as new genomic sequences become available.
Gene and operon assignments were made on the basis of BLAST searches (1) against finished and unfinished microbial genome databases at NCBI (http://www.ncbi.nlm.nih.gov/Microb_blast/unfinishedgenome.html) and TIGR (http://www.tigr.org/cgi-bin/BlastSearch/blast.cgi?) using the sequences of protein products of the nqrABCDEF operon from V. alginolyticus (138), oadGAB operon from K. pneumoniae (120, 176), and mdcABCDE operon from K. pneumoniae (88), respectively, as queries. Gene assignments for the complete genomes were also compared against the COG database (197). Two nqr operons were detected in H. influenzae, V. cholerae, P. aeruginosa, and P. multocida, and two oadGAB operons were found in V. cholerae; the VC0793 gene is apparently disrupted by a frameshift mutation (83). The dash and ND (not detected) indicate the absence of the corresponding gene(s) in complete and unfinished genomes, respectively.
The gene names are those from the complete genome sequences; they can be used to retrieve corresponding DNA sequences from GenBank or the deduced proteins from the NCBI protein database.
Due to the high level of sequence similarity between the corresponding subunits of oxaloacetate, malonate, methylmalonyl-CoA, and glutaconyl-CoA decarboxylases (12, 91, 120, 221), gene assignments were made based solely on the operon structure; the exact substrate specificity of each of these enzymes remains to be determined.
The WWW sites of the unfinished genome sequencing projects, listed in the table, are as follows: A. actinomycetemcomitans, N. gonorrhoeae, and S. pyogenes, http://www.genome.ou.edu; C. difficile, S. enterica serovar Typhi, S. pyogenes, and Y. pestis, http://www.sanger.ac.uk/Projects/Microbes; E. faecalis and T. denticola, http://www.tigr.org/tdb/mdb/mdb.html; H. ducreyi, http://www.htsc.washington.edu/hducreyi/info/index.cfm; K. pneumoniae and S. enterica serovar Paratyphi, http://genome.wustl.edu/gsc/Projects/bacteria.shtml; and P. gingivalis, http://www.pgingivalis.org. See http://www.niaid.nih.gov/factsheets/seqmicrobes.htm for more details.
Na+-Transporting NADH Dehydrogenase
Shortly after the discovery of the Na+-transporting oxaloacetate decarboxylase, a respiratory Na+ pump, the Na+-translocating NADH:ubiquinone oxidoreductase (NQR), was reported in a marine bacterium, Vibrio alginolyticus (202, 204). Similar Na+-transporting respiratory pumps have since been found in other Vibrio spp., Alcaligenes spp., Bacillus spp., and even Escherichia coli (8, 110, 206). In contrast to dicarboxylate decarboxylases, which appear to function mostly in anaerobes, NQR is the dedicated Na+ pump in aerobic bacteria (see references 44 and 48 for reviews). After the genes encoding the V. alginolyticus pump were cloned and sequenced (10, 79, 80), homologous nqrABCDEF operons were found in Haemophilus influenzae, Vibrio cholerae, and Vibrio harveyi (77, 82, 228). The availability of complete microbial genome sequences allowed the identification of homologous genes encoding the NQR in a wide variety of bacteria, from E. coli to Chlamydia trachomatis (190, 192, 228) (Table 1). Remarkably, this enzyme is encoded even in the genome of the aphid symbiont Buchnera sp. strain APS, the second smallest of all known bacterial genomes (179). Our analysis of unfinished genome sequences, available through the web sites of the Sanger Centre (http://www.sanger.ac.uk). The Institute for Genomic Research (TIGR) (http://www.tigr.org), and the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov/BLAST), showed that the nqr operon is widely distributed in bacteria, including such important pathogens as Neisseria gonorrhoeae, Pasteurella multocida, Porphyromonas gingivalis, and Yersinia pestis (Table 1). The deduced protein products encoded by these operons display a significant degree of sequence conservation (typically, more than 25% identity). Among the sequenced genomes, there is no deviation from the V. alginolyticus gene order in V. cholerae, H. influenzae, Neisseria meningitidis, and Pseudomonas aeruginosa (Table 1). In Thermotoga maritima, the gene order is the same, but the nqrF gene, encoding the beta subunit of the enzyme, is replaced by a gene encoding a different Fe-S center-containing protein (referred to as nqrG in Table 1). In E. coli and Buchnera sp., nqrE and nqrG precede the other four genes instead of following them. This gene order is the same as in the previously described mfABCDEF (Rhodobacter nitrogen fixation) operon from Rhodobacter capsulatus (173). Indeed, sequence comparisons show that the mfA gene is homologous to nqrE, the mfCDEF genes are homologous, to nqrABCD, and mfB corresponds to nqrG. This observation shows that the terminal acceptor of electrons from quinone reduction, catalyzed by NQR, does not necessarily need to be oxygen; it can be nitrate or even 2,4-dinitrophenol (168). In addition, NQR can catalyze reverse electron transport from quinol to NAD+ (see below). This would explain the presence of the NQR in Thermotoga maritima, an obligate anaerobe (141). Similar nqrEGABCD operons (in addition to nqrABCDEF operons) are present in the genomes of V. cholerae, H. influenzae, and P. aeruginosa (Table 1); the reason why these organisms should have two copies of the nqr genes is unclear. Finally, in Chlamydia trachomatis and C. pneumoniae, the nqrA and nqrF genes are located separately from the nqrBCDE operon (Table 1).
Na+-Transporting ATPases
There appear to be two major classes of ATP-dependent primary Na+ pumps that are capable of generating Na+ gradients at the expense of ATP hydrolysis. One of them is an ABC (ATP binding cassette)-type transporter, NatAB, recently characterized in Bacillus subtilis (26). Similar Na+-transporting ATPases are encoded in B. firmus (215) and in the genomes of Deinococcus radiodurans, Thermotoga maritima, Clostridium difficile, and Legionella pneumophila. However, the primary (if not the only) function of this Na+ pump appears to be in prevention of Na+ toxicity, that is, accumulation of Na+ in the cytoplasm at the levels that would impair the normal cell functions (26, 215). Short of that, ATP expenditure for Na+ export would be just too costly for cellular metabolism. Indeed, since the intracellular concentration of H+ ions is approximately 106-fold lower than the concentration of Na+ ions (10−7 to 10−8 M versus 10−1 to 10−2 M), it takes many fewer ATP molecules to create a 103-fold gradient of H+ ions than of Na+ ions, even taking into account the buffering capacity of the cytoplasm (182). As a result, for ATP-dependent uptake of nutrients, bacterial cells use ABC-type transporters rather than mediating it by ATP-dependent generation of the Na+ gradient (155).
Na+-transporting ATPases of the second class are simply F0F1-type and archaeal/vacuolar-type (V-type) ATP synthetases working in the reverse direction. Surprisingly, Na+-transporting F0F1-type ATP synthetases are remarkably similar to the H+-transporting ones (48). In fact, the cation specificity of an F0F1-type ATP synthetase can be switched just by several amino acid changes in the a or c subunits of its membrane component (101, 227). The same appears to be true for V-type ATPases that are also found in Na+-transporting and H+-transporting variants (89). It is clear that these enzymes are capable of Na+ extrusion (89, 102, 103, 142). However, due to the huge energy costs of this process (see above), it would seem unlikely to be their function under natural conditions. Indeed, expression of the Na+-transporting V-type ATPase of Enterococcus hirae is induced only by high pH or by high levels of intracellular Na+ (94, 136). It therefore appears that bacterial cells spend ATP on Na+ excretion only under extreme conditions, when it is necessary for their survival.
Recently, a P-type Na+-transporting ATPase has been found in the facultatively anaerobic alkalophilic gram-positive bacterium Exiguobacterium aurantiacum (207). Previously, P-type ATPases in bacteria were not known to transport Na+, as opposed to the eukaryotic Na+/K+ ATPase. If true, this would be an interesting example of the diversity of Na+-transporting ATPases in bacteria. A mutation in the P-type K+-transporting ATPase KdpFABC of E. coli has recently been shown to result in low-level Na+ transport (174).
Na+-Transporting Terminal Oxidases
There have been reports that the cytochrome d terminal oxidase of E. coli is not an H+ pump (162) but a Na+ pump (6, 8, 15, 135). An Na+-transporting terminal oxidase has also been found in a representative of the genus Bacillus (8, 115, 178), later identified as Bacillus halodurans (71). Those reports still remain unconfirmed, even though cytochrome d-type terminal oxidases are encoded in genomes of many organisms, including such Na+ cycle-dependent ones as E. coli, P. aeruginosa, V. cholerae, H. influenzae, C. trachomatis, and C. pneumoniae (Table 2). On the other hand, very similar cytochrome d-type oxidases are encoded in the genomes of B. subtilis, Synechocystis spp., Campylobacter jejuni, and Rickettsia prowazekii, which do not seem to encode any (other) Na+ pumps (Table 1) or require Na+ for growth. It is possible also that cytochrome d-type oxidases do not pump either Na+ or H+ and charge the membrane solely by consuming H+ ions from the cytoplasm to produce H2O (162). Due to this uncertainty, we do not count cytochrome d-type enzymes as primary Na+ pumps (Table 1) but, rather, tentatively consider them to be H+ pumps (Table 2).
TABLE 2.
Pathogenic bacteria that utilize the Na+ cycle
| Organisma | Disease(s) caused | Primary Na+-pump typesa | Primary H+ pump typesb | Na+/H+ antiporter typesc | Na+-dependent transportsc | Comments |
|---|---|---|---|---|---|---|
| Treponema pallidum | Syphilis | DD | None | None | Ala, Pi | T. pallidum appears to rely exclusively on the Na+ cycle, while T. denticola is more flexible |
| Treponema denticola | Necrotizing gingivitis | NQR | TH | NhaC | Ala, Glu, Pi, citrate | |
| Chlamydia trachomatis | Trachoma, vaginitis | NQR | CYD | NhaD | Ala | The presence of a single Na+ pump and just one Na+/H+ antiporter suggests that Na+ circulation is critical for chlamydial physiology |
| Chlamydia pneumoniae | Bronchitis, pneumonia | NQR | CYD | NhaD | Ala, Pro | |
| Porphyromonas gingivalis | Adult periodontitis | NQR, DD | TH, CYD | NhaA NhaP | Pro | Na+ gradient may help P. gingivalis to survive pH swings and high Ca2+ levels |
| Enterococcus faecalis | Endocarditis; wound and urinary tract infections | DD | CYD | NhaC NhaP | Ser, citrate | oadB and oadA genes form an operon with citCDEFG, indicating a possibility of the Na+/citrate cycle in both E. faecalis and S. pyogenes |
| Streptococcus pyogenes | Pharyngitis, rheumatic fever | DD | ND | ND | Ala, Ser, Pi, citrate | |
| Clostridium difficile | Pseudomembranous colitis | NQR | ND | NhaC | Ala, Glu, Pro, Pi | |
| Neisseria meningitidis | Meningitidis | NQR | NDH, TH, CBB | NhaC | Ala, Glu, Pro, Ser | Both species of Neisseria have versatile membrane energetics; the Na+ cycle is unlikely to be crucial for their survival |
| Neisseria gonorrhoeae | Gonorrhea | NQR | NDH, TH, CBB | NhaC NhaP | Ala, Glu, Pro, Ser | |
| Pasteurella multocida | Fowl cholera in poultry | NQR, DD | TH, CYD | NhaA NhaB NhaC NhaP | Ala, Pro, drugs | |
| Haemophilus ducreyi | Chancroid | NQR, DD | TH, CYD | NhaA NhaB | Ala, Glu, Pro, Ser, drugs | |
| Haemophilus influenzae | Pneumonia, otitis media | NQR | TH, CYD | NhaA NhaB NhaC | Ala, Glu, Pro, Ser, drugs | |
| Actinobacillus actinomycetemcomitans | Juvenile periodontitis | NQR | TH, CYD | NhaB NhaC | Glu, Pro, drugs | |
| Vibrio cholerae | Cholera | NQR, DD | TH, CBB, CYD | NhaA NhaB NhaC NhaD NhaP Mnh | Ala, Glu, Pro, Ser, Pi, citrate, drugs | Na+ gradient plays an important role in the motility and virulence of V. cholerae |
| Pseudomonas aeruginosa | Lung and skin infection | NQR, DD | NDH, CYO, CBB, TH, CYD | NhaB NhaP | Ala, Glu, Pro, Ser, Pi, drugs | |
| Escherichia coli | Enteritidis, urinary tract infections | NQR | NDH, CYO, TH, CYD | NhaA NhaB NhaP | Ala, Glu, Pro, Ser, Pi, drugs | E. coli might use the Na+ cycle for survival at alkaline pH |
| Salmonella enterica serovar Paratyphi | Paratyphoid fever | NQR, DD | NDH, CYO, TH, CYD | NhaA NhaB NhaC NhaP | Ala, Glu, Pro, Ser, Pi, citrate, drugs | K. pneumoniae, S. enterica serovar Typhi and Paratyphi are extremely versatile pathogens; they might use the Na+ cycle for energy buffering and for survival at high pH; they are all capable of Na+-dependent citrate fermentation that uses NQR for NAD+ reduction |
| Salmonella enterica serovar Typhi | Typhoid fever | NQR, DD | NDH, CYO, TH, CYD | NhaA NhaB NhaC NhaP | Ala, Glu, Pro, Ser, Pi, citrate, drugs | |
| Klebsiella pneumoniae | Pneumonia | NQR, DD | NDH, TH, CYD | NhaA NhaB NhaP | Ala, Glu, Pro, Ser, Pi, citrate, drugs | |
| Yersinia pestis | Plague | NQR | NDH, CYO, TH, CYD | NhaA NhaB NhaC NhaP | Glu, Pro, Ser, drugs |
Only pathogenic bacteria that have a primary Na+ pump of the NQR or DD (dicarboxylate decarboxylase) type (see Table 1) are listed. The list is ordered according to the position of the respective microorganism on the 16S rRNA-based phylogenetic tree (Fig. 2).
NDH, H+-translocating NADH:ubiquinone oxidoreductase; CYO, cytochrome o-type terminal oxidase; CBB, cbb3-type terminal oxidase; TH, pyrimidine nucleotide transhydrogenase; CYD, cytochrome bd-type terminal oxidase (see the text for a discussion of the cation specificity of cytochrome o-type and cytochrome d-type terminal oxidases). The presence of each type of terminal oxidase has been determined on the basis of TBLASTN (1) searches against the databases of complete and unfinished genomes, maintained at NCBI and TIGR, using the following E. coli proteins as queries: NDH, NuoA, NuoB, and NuoCD (216); CYO, CyoA, CyoB, and CyoC (27); TH, PntA and PntB (28); CYD, CydA and CydB (70). For CBB, the CcoN and CcoO proteins of Rhodobacter capsulatus (201) were used as queries. Functional assignments of the database hits obtained in these searches were checked by comparing each of them to the COG database, as described earlier (140, 197). ND, not detected (in the unfinished genome).
The distribution of the Na+/H+ antiporters and Na+ symporters has been established on the basis of TBLASTN (1) searches, using the following proteins as queries: NhaA, NHAA_ECOLI (106); NhaB, NHAB_ECOLI (158); NhaC, NHAC_BACFI (97); NhaD, gil3123728 (146); NhaP, gil3327262 (210); Mnh, MnhABCDEFG, GenBank accession number AB015981 (87); Ala, ALCP_BACP3 (105); Glu, GLTS_ECOLI (34); Pro, PUTP_ECOLI (137); Pi, NPT2_HUMAN (127); Ser, P42602 (147); citrate, CitS of Klebsiella pneumoniae (211); drugs, NorM of Vibrio parahaemolyticus (134). Functional assignments of the database hits obtained in these searches were checked by comparing each of them to the COG database, as described previously (140, 197). The list of (predicted) Na+-dependent transporters is not meant to be complete; other Na+-dependent transporters are likely to be encoded in the genomes of these bacteria.
Because the cytochrome bo-type type terminal oxidase of E. coli has been directly demonstrated to be an H+ pump (162, 163), similar enzymes in other bacteria are generally assumed to be specific to H+ ions. However, in Vitreoscilla, a beta-subdivision proteobacterium that belongs to the Neisseriaceae family, cytochrome o has been repeatedly shown to function as a primary Na+ pump (55, 56, 108, 152). Because the sequence of this Na+-transporting cytochrome o is still not available, it is difficult to judge how unusual it is and whether other bacteria might also be able to utilize cytochrome o complexes as Na+ pumps. It is remarkable that Neisseria gonorrhoeae and N. meningitidis, closely related to Vitreoscilla spp., do not encode cytochrome o complexes; instead, their terminal oxidases are of the cbb3 type (Table 2).
Cytochrome c oxidases of the cbb3- type are found primarily in microaerophiles, such as Neisseria spp., Helicobacter pylori, and C. jejuni (129, 187). This enzyme complex translocates H+ ions across the membrane (33, 164, 205). The possibility that this complex could (also) pump Na+ ions has not been investigated.
Na+-Transporting Methyltransferase
Yet another type of primary Na+ pump, found in methanogenic archaea, couples Na+ export to methyl group transfer from tetrahydromethanopterin to CoM (see reference 35 for a recent review). No such enzyme has been reported in any bacteria.
UTILIZATION OF NA+ GRADIENTS
Once the chemical energy is transformed into the electrochemical energy of the Na+ gradient, it can be used to drive ATP synthesis, Na+-dependent transports, and Na+-dependent motility.
Na+-Dependent ATP Synthesis
In contrast to ATP-dependent Na+ transport, which has been demonstrated in a number of organisms (40, 118), Na+-dependent ATP synthesis requires a relatively large Na+ gradient and probably occurs only in a few bacteria. Such a reaction was first reported in Propionigenium modestum, one of the few organisms that appear to be exclusively dependent on the Na+ cycle (46, 85). Shortly thereafter, ATP synthesis in response to an artificially imposed Na+ gradient was demonstrated in V. alginolyticus and E. coli (7, 39). In each of these bacteria, Na+-dependent ATP synthesis was catalyzed by a typical F0F1-type ATPase originally thought to be an exclusively H+-transporting enzyme. It transpired that the cation specificity of this enzyme was not absolute and could be changed by mutations (100, 119, 227). As a result, there is currently no clear way to predict the cation specificity of a given F0F1-type ATPase from its sequence, even taking into account the latest data identifying some potentially important residues (53, 100, 101). One could safely assume, though, that under conditions of low proton motive force and high sodium motive force a normally H+-transporting F0F1-type ATPase may function as an Na+-transporting ATP synthetase. Hence, the ability to generate an Na+ gradient through any primary Na+ pump could be considered an important trait, helping the organism to synthesize ATP and ultimately survive under certain unfavorable conditions.
An analysis of ATP synthesis in microorganisms that encode an archaeal/vacuolar type (V-type) ATPase is similarly unable to determine the selectivity of the enzyme toward Na+ and H+ ions. It is particularly interesting to compare the two spirochetes Treponema pallidum and Borrelia burgdorferi, the causative agents of syphilis and Lyme disease, respectively, which both have V-type ATPases encoded by similarly organized operons (66). While T. pallidum has a primary Na+ pump of the oxaloacetate decarboxylase type (Tables 1 and 2) and conceivably could utilize an Na+ gradient for ATP synthesis, B. burgdorferi does not encode any known Na+ pumps and appears to rely solely on the H+ cycle.
Na+-Dependent Symports
Obligately parasitic bacteria generally have smaller genomes than free-living ones and may depend on their hosts for essential nutrients such as amino acids, nucleobases, and cofactors (vitamins) (66, 113, 114). These nutrients are transported into the cell at the expense of energy that comes in the form of either ATP (ABC-type transport systems) or proton (and/or sodium) motive force (secondary transport). The diversity of bacterial transport systems encoded in complete microbial genomes varies widely but generally correlates with the genome size (155, 156). It has long been known that Na+ symports are the principal, and sometimes the only, form of secondary transporters in alkalophilic and thermophilic bacteria (84, 189). The presence of primary Na+ pumps in many pathogens (Table 1) indicates that they, too, could use sodium motive force for solute uptake. Indeed, most of them encode Na+-dependent symporters for alanine, proline, and several other amino acids (Table 2).
While substrate specificity of many permeases encoded in microbial genomes is not known, it can often be predicted, at least in general terms, using the protein family assignment based on the recently developed transporter classification (TC) (see reference 170 for a review). Certain transporter families include both Na+-dependent and H+-dependent members. For example, the branched-chain amino acid/cation symporter family (LIVCS; TC 2.A.26) includes both an Na+-dependent transporter, BraB, from Pseudomonas aeruginosa and an H+-dependent transporter, BrnQ, from Lactobacillus delbruckii (90, 195). However, analysis of permeases that belong to conserved families of transporters shows that in most cases, the cation specificity stays the same throughout the family (166). Reizer et al. have identified 11 conserved protein families of (mostly) Na+/solute symporters that comprised the sodium solute symporter superfamily (166). So, although the exact substrate specificity of most permeases encoded in microbial genomes is still obscure, the pathogens that utilize the Na+ cycle (Table 2) seem to encode a significant share of permeases that belong to Na+-dependent transporter families (155).
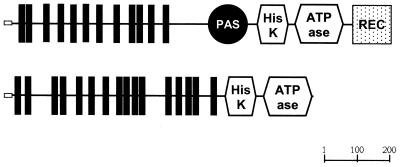
One such conserved family, the solute/sodium symporter family (SSS; TC 2.A.21) unifies the experimentally characterized Na+/proline and Na+/pantothenate permeases PutP and PanF from E. coli (165) with the Na+/glucose symporter SglT from V. parahaemolyticus (170). Homologous transporters are encoded in the genomes of P. gingivalis, C. pneumoniae, C. difficile, N. meningitidis, N. gonorrhoeae, P. multocida, H. influenzae, H. ducreyi, A. actinomycetemcomitans, K. pneumoniae, P. aeruginosa, S. enterica serovars Typhi and Paratyphi, V. cholerae, and Y. pestis (Table 2). All these proteins are likely to function as Na+-dependent symporters, although some of them contain additional domains similar to sensory kinase components of signal transduction systems and may be involved in signal transduction. For instance, in Rickettsia prowazekii protein RP465, V. cholerae protein VC0303, and P. aeruginosa proteins PA3271 and PA4725, putative members of the solute/sodium symporter family are fused to PhoR-like sensory kinase domains (Fig. 3). The signal transduced by these proteins, if any, has not been identified.
FIG. 3.
Domain organization of a new type of sensor histidine kinases. SMART (175) diagrams of the common domain structure V. cholerae protein VC0303 and P. aeruginosa proteins PA3271 and PA4725 (A) and R. prowazekii protein RP465 (B). (A) The small open box on the left indicates the likely signal peptide, predicted by the SignalP program (144). The vertical boxes indicate 12 transmembrane helices of the solute/sodium symporter family (TC 2.A.21) transporter, predicted by the TopPred program (29). The circle indicates the PAS domain (198); the two hexagons indicate the phosphoacceptor and ATPase domains, respectively, of the histidine kinase; and the dotted square on the right indicates the CheY-type receiver domain (193). (B) R. prowazekii protein RP465 contains 16 predicted transmembrane helices and both domains of a histidine kinase but lacks PAS and CheY-type domains. The ruler indicates the length of the protein in amino acid residues.
Another conserved family of Na+-dependent transporters, identified by Reizer et al. (166), includes experimentally characterized alanine and glycine transporters from the marine bacterium Alteromonas haloplanktis and the thermophilic bacterium PS3 (105). Proteins belonging to this family (AGCS; TC 2.A.25) and probably involved in the uptake of alanine and/or glycine are found in many bacteria, including most human pathogens (Table 2).
Glutamate, aspartate, serine, and threonine also can be taken up by bacterial cells by an Na+ symport mechanism. The Na+/glutamate symporter GltS from E. coli remains the only characterized member of the glutamate/sodium symporter family (ESS; TC 2.A.27). Members of this family in other bacteria can be assumed to have the same narrow substrate specificity (Table 2). The Na+/serine-threonine symporter, SstT, from E. coli belongs to the diverse family of transporters (DAACS; TC 2.A.23) that also includes Na+- and H+-dependent symporters for glutamate and dicarboxylic intermediates of the Krebs cycle. Members of this family are widely represented in bacterial genomes (156). Unfortunately, their exact substrate and cation specificity is still difficult to establish based solely on sequence comparisons.
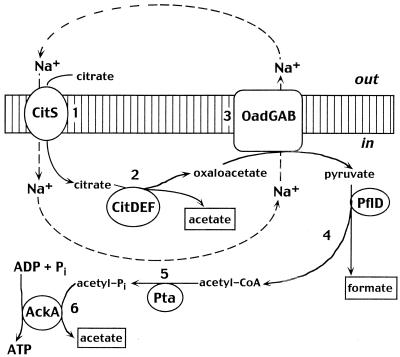
Members of the citrate/cation symporter family (CCS; TC 2.A.24) are involved in Na+-dependent uptake of such 2-hydroxycarboxylates as citrate, malate, and lactate (211, 212). Some of these permeases reportedly can also transport citrate or malate in symport with H+ ions (130). Actually, the true substrate of the Na+/citrate symporter CitS, best studied in Klebsiella pneumoniae, appears to be the protonated (divalent) form of citrate, transported in symport with two Na+ ions. Thus, technically, CitS is an H+/2Na+/citrate symporter (52, 211). In K. pneumoniae, S. enterica serovar Typhimurium, and several other bacteria, CitS catalyzes the first stage of anaerobic citrate fermentation pathway. In this remarkable Na+-dependent pathway, citrate is first transported into the cell at the expense of the Na+ gradient (Fig. 4) and then split into acetate and oxaloacetate by citrate lyase (16). Decarboxylation of oxaloacetate into pyruvate by the Na+-transporting oxaloacetate decarboxylase restores the Na+ gradient and produces pyruvate, which is further metabolized into acetate with acetyl-CoA and acetyl phosphate as intermediates (16). The last stage of this pathway, catalyzed by acetate kinase, yields ATP and thus results in energy conservation. Based on the presence of genes encoding both CitS-type carrier and oxaloacetate decarboxylase (Table 2), such a pathway can be assumed to function in Treponema denticola, V. cholerae, S. enterica serovars Typhi and Paratyphi, and, possibly, Streptococcus pyogenes. Not surprisingly, in K. pneumoniae, V. cholerae, and S. enterica serovar Typhi, the genes for the Na+/citrate symporter and oxaloacetate decarboxylase are located within a single operon (16) (Table 1).
FIG. 4.
Scheme of the Na+-dependent citrate fermentation pathway. Citrate is transported into the cell in symport with Na+ ions by CitS (step 1) and split into acetate and oxaloacetate by citrate lyase CitDEF (step 2). Decarboxylation of oxaloacetate into pyruvate by Na+-transporting oxaloacetate decarboxylase, OadGAB (step 3), restores the Na+ gradient and produces pyruvate. Pyruvate-formate lyase PflD splits pyruvate into formate and acetyl-CoA (step 4), which is further converted into acetylphosphate by phosphotransacetylase Pta (step 5). Dephosphorylation of acetylphosphate by acetate kinase AckA (step 6) yields ATP, resulting in energy conservation. The enzymes are indicated by their standard gene names, where available. The fermentation end products are boxed. See reference 16 for more details.
Na+-Dependent Drug Efflux
Drug resistance of human pathogens is a growing problem, threatening to negate the success of the antibacterial efforts of the last 50 years and make humans once again vulnerable to a siew of infectious diseases. Comparative genomics is being widely used for identification of potential drug targets (2, 65, 172). The recurring problem in drug design, however, is the presence of multidrug efflux pumps that excrete a wide variety of compounds, decreasing their cellular concentrations below the required MICs (122, 123, 154). Indeed, preventing drug efflux significantly increases the efficacy of even standard antibiotics, such as streptomycin and tetracycline (121, 191). Most of the known multidrug efflux pumps belong to one of three groups of secondary transporters, typically energized by the protonmotive force: the major facilitator superfamily (MFS; TC 2.A.1), the small multidrug resistance family (SMR; TC 2.A.7.1), and the resistance/nodulation/cell division family (RND; TC 2.A.6) (154). Several other groups of multidrug transporters belong to the ATP-dependent ABC superfamily (ABC; TC 3.A.1). Recently, a multidrug transporter, NorM, from V. parahaemolyticus that caused Na+-dependent efflux of norfloxacin has been described (133, 134). NorM turned out to be a representative of a new large family of multidrug efflux pumps (20, 134), termed the multiantimicrobial extrusion family (MATE; TC 2.A.66). Members of this family are encoded in almost every sequenced genome, including nearly identical NorM proteins in V. cholerae, E. coli, H. influenzae, and P. aeruginosa. Close homologs of NorM are also found in S. enterica serovars Typhi and Paratyphi, K. pneumoniae, A. actinomycetemcomitans, P. multocida, H. ducreyi, and Y. pestis (Table 2). While it is still too early to speculate, which members of this family of transporters are Na+ dependent and which, if any, are H+ dependent, there is little doubt that the sodium motive force can serve as an energy source for drug efflux pumps in a number of bacterial pathogens.
Reverse Electron Transport
The presence of nqr genes in the genomes of anaerobic microorganisms, including the obligately anaerobic hyperthermophile Thermotoga maritima (Table 1), indicates that NQR can also work in the reverse direction, using the energy of the Na+ gradient to reduce NAD+ to NADH. Indeed, Na+-dependent NAD+ reduction by formate has been experimentally demonstrated in K. pneumoniae grown anaerobically on citrate (157). This reaction was sensitive to the quinone analog 2-heptyl-4-hydroxyquinoline N-oxide (HQNO), a well-characterized inhibitor of NQR. For K. pneumoniae, Na+-dependent reverse electron transport could help to produce reducing equivalents, needed for assimilation of the formate formed in the anaerobic citrate fermentation pathway (see above) (Fig. 4).
Na+-Dependent Motility
Motility is widely believed to be an important virulence factor in bacterial pathogens, since it allows the bacterium to penetrate different host tissues and/or helps it to attach to the surface of epithelial cells (86, 107, 132, 222). Indeed, loss of motility has been reported to correlate with significant decrease of virulence in host-parasite models for H. pylori, V. cholerae, Y. enterocolitica, and other bacteria (72, 98, 150, 225).
Many bacterial pathogens that depend on an Na+ cycle (Table 2) are motile. Their genomes contain the full set of ca. 30 genes that are required for the formation of a functional flagellum (126). For several of them, motility has been directly shown to be Na+ dependent (5, 14, 78, 111). In V. parahaemolyticus and V. alginolyticus, lateral and polar flagella are reportedly powered by proton motive force and sodium motive force, respectively (5, 14, 111). Some components of the Na+-dependent motor can be functionally replaced by homologous components of the H+-dependent motor (3, 68). Uncovering the details of the organization of the Na+-dependent motors of V. cholerae and related bacteria is quite important, because it could clarify the contribution of motility to their pathogenicity.
Is the Na+ Gradient Involved in Toxin Export?
The contribution of the flagellar genes to virulence is not limited to the colonization stage of infection. The flagellar export apparatus, which is responsible for the secretion and assembly of a functional organelle, participates in the secretion of a variety of virulence factors, including certain bacterial toxins (reviewed in references 30, 62, and 93). This toxin export apparatus is referred to as a type III protein secretion system and participates, for example, in secretion of extracellular proteins by Y. enterocolitica, including the virulence-associated phospholipase YplA (226). Because the flagellar export system traverses the cytoplasmic membrane, the peptidoglycan layer, and the outer membrane, proteins exported via this system can be delivered directly to the exterior and may even penetrate the cytoplasm of the host cell (93). The energy for this complex process is apparently supplied in the form of ATP and is used by the ATPase FliI, which is homologous to the catalytic beta subunit of the F0F1-type H+-ATPase (58, 93). Genome comparisons show that genes encoding type III protein secretion systems have a wider phylogenetic distribution than the rest of the flagellar genes; they can be found even in such nonmotile organisms as C. trachomatis and C. pneumoniae (143, 190). It is important to note here that in a motile cell, a significant part of the flagellar apparatus (“rotor”) rotates together with the flagellar filament relative to the remainder of the flagellar machinery (“stator”) and the rest of the cell, providing torque that propels the bacterium (11, 126). Remarkably, homologs of the FliF protein that forms the two membrane rings (M and S) of the rotor, i.e., the flagellar motor switch protein FliG, located at the interface between the rotor and the stator, and the FliN protein, which forms the inner (cytoplasmic) ring of the rotor (149, 200, 208), appear to be involved in the functioning of the export machinery in several bacterial pathogens (93). In Y. enterocolitica and Y. pestis, these homologs of FliF, FliG, and FliN, referred to as YscJ, YscD, and YscQ, respectively (SctJ, SctD, and SctQ according to the new nomenclature suggested by Hueck), are encoded on large virulence plasmids that encode both components of the export machinery and secreted virulence proteins. These observations suggest that SctD, SctQ, and particularly SctJ might be able to rotate in the membrane. Unfortunately, the relation, if any, between the proper functioning of the flagellar export machinery and its rotation remains unknown. Early work on the role of proton motive force in the elongation of the flagellar filament suggested that flagellar rotation might be needed for flagellin export in E. coli (63). It is tempting to speculate that Na+-dependent rotation of the basal body might be related to the secretion of virulence proteins, either promoting or impeding it. This idea, however, is far from having any experimental support.
INTERPLAY OF NA+ AND H+ CYCLES IN BACTERIAL PATHOGENS
The presence of genes encoding primary Na+ pumps in a number of important human pathogens (Table 1) indicates that these bacteria rely on the Na+ cycle for at least part of their energy metabolism. However, in addition to a primary Na+ pump, it appears that most of them encode primary H+ pumps (Table 2). Although one cannot be sure that every primary Na+ pump and H+ pump has been accounted for, we can judge whether a particular microorganism uses the Na+ cycle based on the presence of any of the two proven primary Na+ pumps, NQR and dicarboxylate decarboxylases. In any case, most genomes encode multiple Na+/H+ antiporters (Table 2), which should allow the generation of an H+ gradient at the expense of an Na+ gradient and vice versa (see Fig. 1). This (at least partial) interchangeability of proton motive force and sodium motive force is a striking feature of the bioenergetics of nearly all bacteria studied to date. In the following section we consider the possible role(s) of Na+ cycle in the energy metabolism of several potentially Na+-dependent bacterial pathogens in more detail.
Treponema pallidum and T. denticola
T. pallidum, the causative agent of syphilis, has remained quite an enigmatic organism even after its genome was completely sequenced (61, 217). It still cannot be continuously cultivated in vitro, and a syphilis vaccine remains elusive (181). An analysis of the genome sequence of T. pallidum showed that this organism encodes a very limited number of biosynthetic pathways for amino acids, nucleotides, and cofactors, which partly explains its complex growth requirements (61). A recent analysis of the protein set of T. pallidum revealed a number of organism-specific gene products whose exact biological role remains unclear (196, 217). An analysis of the energy metabolism of T. pallidum (Table 2) reveals a stunning picture. This organism does not appear to encode any primary H+ pumps or Na+/H+ antiporters, and oxalate decarboxylase seems to be the only ionic pump encoded in its genome. The apparent absence of respiratory ionic pumps is quite unexpected, since T. pallidum is a microaerophile rather than an obligate anaerobe and should be routinely attacked by superoxide radicals generated by the host defense systems; it even has a dedicated superoxide reductase (125, 217). It seems likely that “decarboxylation phosphorylation,” i.e., ATP synthesis at the expense of the Na+ gradient generated by oxalate decarboxylation (48, 50), serves as a major energy source for T. pallidum. The only peculiarity of this process in T. pallidum is that its ATP synthetase is of the archaeal/vacuolar type, very similar to the Na+-transporting V-type ATPase of Enterococcus hirae (66). As was noted above, solute uptake, energized by ion gradients that have been generated at the expense of ATP, is energetically costly. Indeed, T. pallidum mostly relies on ABC-type transporters for solute uptake (155). The apparent absence of Na+/H+ antiporters suggests that the T. pallidum cell must tightly balance its H+ ion fluxes, i.e., the proton motive force-dependent solute uptake with proton motive force-generating efflux of fermentation products. Such a mechanism of proton motive force generation has been previously demonstrated in Streptococcus cremoris (112). Another possibility, of course, is that T. pallidum exclusively uses Na+ as a coupling ion. A survey of the secondary transporters encoded in the T. pallidum genome (155) appears to support this possibility. Most of them either are Na+ symporters or belong to the families of transporters that can be powered by either Na+ or H+ gradients. The former group includes predicted the Na+/alanine symporters TP0414 and TP0998, the Na+/branched-chain amino acid symporter TP0265, the Na+/phosphate symporter TP0771, and the Ca2+/Na+ antiporter TP1034 (155, 197), (Table 2). Most other T. pallidum permeases have unknown specificity and can be characterized only using transporter protein family assignment (170). This group includes TP0023, a member of the neurotransmitter/sodium symporter family (TC 2.A.22); TP0106, a member of the betaine/carnitine/choline transporter family (TC 2.A.15); TP0555 and TP0934, members of the dicarboxylate-amino acid/cation symporter family (TC 2.A.23); and TP0901, a member of the multiantimicrobial extrusion family (TC 2.A.66). While the exact substrate and coupling-ion specificities of the transporters in this second group are still obscure, some or all of them use Na+ gradient as the energy source.
The genome of T. denticola, a close relative of T. pallidum, is currently being sequenced at TIGR with support from the National Institute of Dental and Craniofacial Research (see http://www.nidr.nih.gov for more details). The currently sequenced part of T. denticola genome encodes an Na+ pump, but here it is NQR, not an oxaloacetate decarboxylase, as in T. pallidum (Table 2). This contrast between the two spirochetes could be due to a higher availability of oxygen in the oral cavity, which is the ecological niche of T. denticola. The absence of the NQR in T. pallidum then probably reflects the loss of nqr genes in the course of its adaptation to its own ecological niche. Indeed, it still encodes distant homologs of the NqrA and NqrB subunits in TP0152 and TP0151. In addition to NQR, T. denticola encodes NAD+/NADP+ transhydrogenase, a proton motive force generating-enzyme, an Na+/H+ antiporter, and several Na+- or H+-dependent transporters (Table 2). It is possible, of course, that NQR in T. denticola is functioning in the reverse direction and is used for Na+-dependent NAD+ reduction, as discussed above. Nevertheless, the fact that both Treponema spp. retain primary Na+ pumps supports the idea that they are dependent on the Na+ cycle for at least part of their membrane energetics.
Chlamydia trachomatis and C. pneumoniae
Both C. trachomatis and C. pneumoniae are extremely important pathogens. In addition to being the causative agent of trachoma, an eye infection that may lead to blindness, C. trachomatis is one of the most common pathogens of human genital tract (190). C. pneumoniae is a common cause of infections of the respiratory tract and can be found in many other organs; it appears that virtually every human is infected with C. pneumoniae at least once (69). Probably the most intriguing aspect of C. pneumoniae pathogenicity is its apparent involvement in the development of atherosclerosis (see references 22 and 69, and other reviews in the special issue of the Journal of Infectious Diseases, Vol. 181, Suppl. 3; June 2000). The enticing perspective of using antibiotics to prevent or treat coronary artery disease adds some urgency to the task of understanding the basics of chlamydial physiology. Recently, genome comparisons were used to identify the unusual DhnA-type fructose-1,6-bisphosphate aldolase as a potential target for a chlamydia-specific “magic bullet” (64). Genome analysis also shows that both C. trachomatis and C. pneumoniae encode a primary Na+ pump, NQR (190) (Table 1). They have another potential ion pump in cytochrome d-type terminal oxidase, but, as discussed above, the mechanism and energy yield of a cytochrome d complex remains unclear. Like Treponema spp. chlamydias have an H+- (or Na+)-transporting V-type ATPase, which turns out to be common in bacteria (66). Another similarity between these two phylogenetically very distant groups of bacteria is in the organization of their transport systems, which is probably due to their common reliance on the Na+ cycle. Like T. pallidum, each of the two chlamydias encodes two predicted Na+/alanine symporters (CT409 and CT735 in C. trachomatis, CPn0876 and CPn0536 in C. pneumoniae), an Na+/branched-chain amino acid symporter (CT554 and CPn0836), and an uncharacterized transporter of the neurotransmitter:sodium symporter family (CT231 and CPn0290) (155, 197). On the other hand, other chlamydial transporters, such as the phosphate permease PitA (CT692 and CPn0680), the glutamate transporter GltS (CT401 and CPn0528), ADP/ATP translocase (CT065, CT495, CPn0351, and CPn0614), amino acid-polyamine-organocation family (TC 2.A.3) transporters (CT374, CT216, CPn0282 and CPn1031), and several other predicted transporters are likely to be energized by an H+ rather than an Na+ gradient. Table 2 shows that generation of the proton motive force in chlamydiae could be accomplished through the action of either Na+/H+ antiporters of the NhaD type or cytochrome d-type terminal oxidases.
Porphyromonas gingivalis and Actinobacillus actinomycetemcomitans
P. gingivalis and A. actinomycetemcomitans, both of which are periodontitis-causing bacteria, encode both the primary Na+ pump NQR and NAD+/NADP+ transhydrogenase, an H+ pump (Table 2). The reason why these two residents of the oral cavity, where the pH almost never goes above 7.8 and is often much lower (117), would need a primary Na+ pump is not quite clear. One possible explanation is that an Na+ gradient might help in stabilizing the levels of the proton motive force in these bacteria, which could otherwise vary due to the huge swings of the pH in the oral cavity, caused by consumption of fluids of variable ion content. Such a “buffering” role of Na+ gradient has been experimentally demonstrated in several diverse bacterial species (19). Another reason for the existence of a primary Na+ pump in oral pathogens is that in combination with the Ca2+/Na+ antiporter, it could help in protecting the bacteria from excessive influx of Ca2+ ions from Ca2+-saturated saliva (117). Finally, the most convincing explanation of the possible role of NQR in P. gingivalis and A. actinomycetemcomitans relies on the fact that periodontitis caused by these bacteria is often accompanied by gum bleeding. As a result, the salt content in the ecological niche occupied by these microorganisms approaches that of the blood plasma, i.e., is characterized by a relatively high concentration of Na+ ions. Thus, NQR could be as important for such oral pathogens as T. denticola, P. gingivalis, and A. actinomycetemcomitans as it is for marine microorganisms. A possible consequence of this adaptation is that periodontal pathogens may be fit to survive in blood and cause bacteriemia. Indeed, P. gingivalis and A. actinomycetemcomitans have been recently detected in atherosclerotic plaques in the carotid artery (73).
Escherichia coli and Haemophilus influenzae
Early research of the membrane energetics of E. coli failed to demonstrate an Na+ pump in this organism (203). Later studies, however, provided ample evidence for primary active Na+ transport under conditions of low proton motive force (6, 7, 37). A ΔnhaA ΔnhaB mutant lacking two principal Na+/H+ antiporters retains the capacity to excrete Na+ ions when incubated in the presence of high concentrations of K+ (74). After the genome sequence of H. influenzae became available, it was found to contain an nqr operon very similar to the one in V. alginolyticus (Table 1). Soon thereafter, the presence of NQR in H. influenzae was demonstrated experimentally (82). Recent experiments with E. coli confirmed the presence of a primary Na+ pump in this organism (192). In spite of all these findings, the nqr genes in the E. coli genome sequence remained unidentified until now (Table 1), probably due to their unusual order in the operon and the likely nonorthologous displacement of the beta subunit.
Vibrio cholerae
While V. cholerae clearly has multiple H+ and Na+ pumps and a number of Na+/H+ antiporters (Table 2), it relies on Na+-dependent polar flagella for its motility (111). V. cholerae also provides the only well-documented case of the connection between transmembrane Na+ circulation and the expression of pathogenicity determinants (78). Dissipation of the sodium motive force by ionophores, nqr mutations, or NQR inhibitors in each case led to an increased expression of virulence-related genes encoding cholera toxin and toxin-coregulated pili (78). Changes in Na+ circulation appeared to affect a set of regulatory membrane proteins, TcpP and TcpH, which, in turn, are required for the expression of ToxT, a transcriptional activator of the major virulence factors in V. cholerae (77, 78). At high NaCl concentrations in the growth medium, the TcpP/TcpH-mediated activation of toxT was diminished (78). Thus, at least in V. cholerae, a functional linkage between the virulence factor expression machinery and the Na+ cycle has been shown experimentally. The reason for this connection is not clear, although it has been speculated that one of the functions of cholera toxin might be the generation of an Na+-rich environment in the intestinal lumen to boost the efficiency of the Na+ cycle in V. cholerae cells immersed in alkaline medium (9).
In many pathogens including V. cholerae, motility is considered a virulence factor (72, 150). However, the relationship between motility and virulence in V. cholerae is quite complicated, since the motility phenotype itself appears to affect the expression of virulence determinants. Indeed, some nonmotile mutants of V. cholerae showed increased toxT transcription and constitutive expression of cholera toxin and toxin-coregulated pili under alkaline conditions (67, 78). Deceleration of flagellar rotation by different means, such as high medium viscosity or inhibitory drugs, had a similar effect (78). On the other hand, cholera toxin and toxin-coregulated pilus expression was repressed in several spontaneous hypermotile mutants (67). A hypermotile phenotype was also observed in toxR mutants, which are defective in the regulatory protein ToxR, which also is required for expression of ToxT (67, 78). Thus, not only is the motility phenotype controlled by the ToxR regulon in this species, but also there appears to exist a specific signal transduction mechanism that monitors cell motility and conveys that information to the virulence regulatory cascade. However, the molecular mechanisms underlying these signal transduction events remain obscure. It could be noted in this regard that products of two ToxR-regulated genes, tcpI (VC0825) and acfB (VC0840), located on the pathogenicity island responsible for the expression of toxin-coregulated pili, are homologous to methyl-accepting chemotaxis receptors. Mutations in these two genes positively affect the motility of V. cholerae as assayed by swarm plate assays (57, 75). Further experiments are required to try to pinpoint the exact signals(s) that links the Na+ cycle and expression of pathogenicity factors in V. cholerae. The likely candidates for such a signal are changes in the total level of sodium motive force (78) or one of its components, e.g., the membrane potential (36). It would be also extremely interesting to determine which of the V. cholerae proteins acts as the primary “bioenergetic” sensor. In summary, V. cholerae seems to be a useful system for studying the possible involvement of Na+ cycle elements in the regulation of virulence.
NA+ PUMPS AS DRUG TARGETS
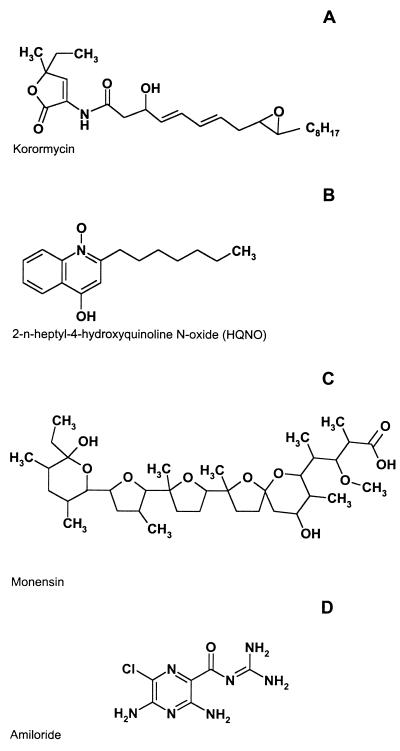
The structures of some of the inhibitors of the Na+ cycle discussed in this section are shown in Fig. 5.
FIG. 5.
Structures of some inhibitors of the Na+ cycle. (A) Korormicin. (B) 2-n-Heptyl-4-hydroxyquinoline N-oxide (HQNO). (C) Monensin. (D) Amiloride.
Korormicin
The importance of the Na+ cycle in the energy metabolism of certain human pathogens suggests that primary Na+ pumps might hold promise as potential drug targets. Indeed, korormicin, a powerful inhibitor of NQR (223), was originally isolated as an antibiotic, secreted by a marine bacterium, Pseudoalteromonas sp. strain F-420 and demonstrating antibacterial activity against other marine bacteria (139, 224).
Korormicin is an extremely effective noncompetitive inhibitor (Ki ≈ 8 × 10−11 M) of the interaction of NQR with its quinone substrate. As a result, korormicin was approximately 103-fold more active in inhibiting purified NQR than was 2-heptyl-4-hydroxyquinoline-N-oxide (HQNO), a traditional and well-studied inhibitor of NQR (223). Korormicin proved to be a specific inhibitor of NQR, since it had no effect on Na+-independent NADH oxidase (NADH:menadione reductase) activity (223). At the cellular level, korormicin was active against a variety of gram-negative halophilic bacteria, including Vibrio alginolyticus, Shewanella putrefaciens, and Alteromonas macleodii (223). These observations show that inhibiting NQR is lethal for at least some marine bacteria and that perhaps the same approach could work against human pathogens that depend on the Na+ cycle. Other known inhibitors of the Na+ cycle in bacteria include Li+ and Ag+ions, amiloride, and monensin, an artificial electroneutral Na+/H+ antiporter.
Ag+
While antibacterial effects of silver salts were first noticed long ago (see reference 180 for a review), NQR has been recently recognized as one of the targets of Ag+ ions. In two independent studies, nanomolar concentrations of Ag+ ions were shown to inhibit energy-dependent Na+ transport in inside-out vesicles of alkalophilic Bacillus sp. strain FTU and to inhibit purified NQR of V. alginolyticus (81, 177). Later, Ag+ was shown to irreversibly bind to the beta subunit of NQR (NqrF or Nqr6), causing enzyme denaturation and the loss of its flavin adenine dirucleotide cofactor (139). Half-maximal inhibition of the enzyme activity was attained at concentrations between 0.5 and 2 nM Ag+, making NQR one of the most vulnerable targets of Ag+ ions.
Li+
A number of Na+-dependent bacterial permeases, including NhaA- and NhaB-type Na+/H+ antiporters, can use Li+ instead of Na+ (95, 146, 151). As a result, Li+ is effectively exported from the cell and thereby decreases its toxicity. Thus, growth inhibition of wild-type E. coli required as high as 700 mM Li+ in the medium, while a ΔnhaA ΔnhaB double mutant could not grow even in the presence of 30 mM Li+ (95). The growth of P. aeruginosa is also Li+ sensitive (96). NQR does not seem to use Li+ as substrate, and some data suggest that Li+ inhibits this enzyme (214). If so, Li+ might have potential as drug against Na+ cycle-dependent bacteria; however, it should be noted that Li+ is mildly toxic for humans. It is currently used in the treatment of bipolar affective disorder and is known to affect thyroid function, leading to hypothyroidism and goiter.
Monensin
An artificial electroneutral Na+/H+ antiporter, monensin has been traditionally used as a nutritional additive (growth promoter) in cattle (54, 213). Monensin addition reduces amino acid fermentation and, hence, ammonia production in the rumen by disrupting the Na+ cycle in ruminal Peptostreptococcus spp., which imports some amino acids in symport with Na+ ions (24, 25). An Na+-motive, biotin-dependent glutaconyl-CoA decarboxylase and an Na+-motive membrane ATPase are apparently operative in this bacterium (24). Well-established activity of monensin against many anaerobic bacteria including Clostridium perfringens, Streptococcus bovis, and others (21, 167) suggests that it holds promise as a prototype for new antibacterial drugs.
Amiloride
The diuretic drug amiloride and its 5-aminoalkylated derivatives are potent inhibitors of mammalian Na+/H+ antiporters of the NHE family (60, 145). Amiloride was found to be ineffective against the NhaA-type Na+/H+ antiporter in E. coli, but it inhibited the NhaB-type antiporter with K0.5 = 6.0 μM (159). Tenfold-higher concentrations of amiloride were reported to inhibit the NhaA antiporter of V. parahaemolyticus (116). However, high doses of amiloride should be used with caution, since at high concentrations it may act as a nonspecific uncoupler (32). Amiloride and some of its derivatives inhibit the Na+-dependent motility of V. parahaemolyticus, V. alginolyticus, and V. cholerae (4, 99), which indicates that it could be used in preventing colonization.
NQRA AS A VACCINE CANDIDATE
Because NQR is a membrane protein, whose phylogenetic distribution is apparently limited to marine bacteria and certain human and animal pathogens (Table 2), its subunits could make interesting vaccine candidates. A study of Actinobacillus pleuropneumoniae, the causative agent of pleuropneumonia in swine, showed that the NqrA protein (referred to as AopA by the authors) was immunogenic in infected pigs (31). Even though NqrA is a cytoplasmic membrane protein that was not detected in the outer membrane in any significant amounts, the serum of convalescent-phase pigs infected with A. pleuropneumoniae was found to contain anti-NqrA antibodies. It is tempting to speculate that switching off the Na+ pump of A. pleuropneumoniae might have helped those pigs to fight infection. The NqrA protein of Porphyromonas gingivalis has been patented in Australia as a “50 kD antigen PG1” (GenBank accession number AF144076), presumably due to its immunogenic properties. These observations, while still preliminary, suggest yet another direction of future studies of the role(s) of primary Na+ pumps in bacterial infection.
CONCLUSIONS AND PERSPECTIVES
Although analysis of the role of Na+ ions in bacterial virulence is still in its infancy, a few things are becoming increasingly clear. The presence of genes encoding primary Na+ pumps in the genomes of a number of phylogenetically diverse pathogenic bacteria (Table 1) indicates that generation of the Na+ gradient is an important part of their membrane energetics. It should be noted that such microorganisms as Mycoplasma genitalium, M. pneumoniae, B. burgdorferi, H. pylori, and Mycobacterium tuberculosis do not seem to encode any primary Na+ pumps (see Table 1) and may not depend on Na+ circulation. Most bacterial pathogens, however, encode both Na+ and H+ pumps and multiple Na+/H+ antiporters (Table 2) that ensure the maintenance of both Na+ and H+ gradients on their cytoplasmic membrane. It appears, therefore, that the sodium motive force supplements the proton motive force as an additional source of energy in these bacteria. In an extreme case, the syphilis spirochete T. pallidum appears to encode no primary H+ pumps; it thus might exclusively depend on the sodium motive force, generated by Na+-transporting oxaloacetate decarboxylase, for its energy metabolism. For several other important pathogens, including C. trachomatis, C. pneumoniae, and H. influenzae, NQR comprises the principal respiratory ionic pump; their genomes also encode pyrimidine nucleotide transhydrogenase and/or cytochrome bd-type terminal oxidase (Table 2). Several pathogens, including K. pneumoniae, V. cholerae, and S. enterica serovar Typhi, are capable of anaerobic citrate fermentation, which includes Na+ cycling across the cytoplasmic membrane.
One could think of several possible explanations for the widespread distribution of the elements of the Na+ cycle among pathogenic bacteria. First, Na+-based membrane energetics could improve the versatility of a pathogen by providing it with additional means of ATP synthesis, motility, and solute uptake. This would improve its chances for colonization of the host cells and survival in the host organisms, where defense mechanisms, such as generation of superoxide radicals impair the integrity of the bacterial membrane and decrease the levels of the proton motive force. Second, because Na+ concentrations in most natural environments are almost 106-fold higher than H+ concentrations, sodium motive force levels are unlikely to change as rapidly as proton motive force levels, making sodium motive force a much more reliable source of energy. Finally, the well-known similarity between the salt content of blood and seawater could create evolutionary pressure toward the development of similar adaptation mechanisms in human pathogens and marine microorganisms or, alternatively, acquisition of the corresponding genes through horizontal gene transfer.
The dualistic H+- and Na+-based character of membrane energetics in pathogenic bacteria propounds a number of intriguing questions. First, is the persistent appearance of elements of the Na+ cycle in very different pathogens just a consequence of the adaptive advantage of having more than one chemiosmotic coupling ion, or is there a more profound, mechanistic link between the presence of an Na+ cycle and virulence? And, if the latter is true, what is the exact mechanism linking Na+ energetics to the regulatory events responsible for the expression of pathogenicity determinants? In other words, does a change in Na+ homeostasis signal the pathogenic organism that it has reached its destination inside the host and that it is time to activate the expression of virulence factors? At least in the case of V. cholerae, the cells appear to respond to alterations in Na+ circulation by modulating the expression of the main virulence regulon (78). The exact nature of the potential sensor and the mechanism of this regulation are, unfortunately, still unknown.
From the practical point of view, the peculiar character of the Na+ cycle in bacterial pathogens makes its components attractive potential targets for the development of “smart” drugs and therapeutic strategies that would have minimal side effects at acceptable antimicrobial potency. A few examples considered in this review illustrate the potential of different chemicals acting as specific inhibitors of primary Na+ pumps (korormicin and Ag+), Na+/H+ antiporters (amiloride and its analogs), substrate analogs (Li+), and Na+-translocating ionophores (monensin). It should be stressed that for a large number of Na+ transporters, there are no known specific inhibitors. However, our analysis shows that a variety of new potential drug targets could be pinpointed by screening complete and partially complete bacterial genomes. The putative system of the Na+-dependent anaerobic fermentation of citrate in T. denticola, V. cholerae, S. enterica serovar Typhi, and some other pathogens may be mentioned as such a potential target. In addition, the immunogenic efficacy of NqrA demonstrates that Na+ pumps, residing in the cytoplasmic membrane of gram-negative bacteria, are much more appealing targets for the development of effective vaccines than it would have seemed from their “nonsurface” localization. Although drugs targeted against the components of Na+ cycle, such as NQR, would certainly have a limited antibacterial spectrum, they might be very helpful weapons against persistent infections caused by Treponema or Chlamydia species and potentially might even help in preventing coronary artery disease and atherosclerosis, to which chlamydias are now believed to contribute.
The presence of primary Na+ pumps in modern archaeal and bacterial hyperthermophiles suggests that the Na+ cycle was a primary mechanism of energy conservation in the common ancestor of these two branches of the Tree of Life (see references 124 and 220 for discussions). As is the case with free-living extremophiles (thermophiles, halophiles, and alkalophiles), human pathogens may rely on the Na+ cycle to survive and grow in the hostile environment created by host defense mechanisms. It seems reasonable to expect that elucidation of the precise role of Na+ circulation in pathogenic bacteria would open new avenues of research, which potentially could bring not only additional knowledge but also novel approaches to cure infectious diseases.
ACKNOWLEDGMENTS
This work was supported in part by Cancer Center Support grant CA 21765 and ALSAC (American Lebanese Syrian Associated Charities) to C.C.H. and by Manitoba Health Research Council operating grant 34021 to P.A.D.
We thank Floyd Dewhirst, Eugene Koonin, Peter Loewen, John Mekalanos, and Armen Mulkidjanian for helpful suggestions. Analysis of unfinished genome sequences has been made possible by generous submission to the public databases of preliminary sequence data by the Sanger Centre (C. difficile, S. enterica serovar Typhi, and S. pyogenes), The Institute for Genomic Research (P. gingivalis, T. denticola, and E. faecalis), Oklahoma University Advanced Center for Genome Technology (N. gonorrhoeae, A. actinomycetemcomitans, and S. pyogenes), University of Washington (C. difficile), and the Washington University Genome Sequencing Center (K. pneumoniae, and S. enterica serovar Paratyphi).
REFERENCES
- 1.Altschul S F, Madden T L, Schaffer A A, Zhang J, Zheng Z, Miller W, Lipman D J. Gapped BLAST and PSI-BLAST—a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arigoni F, Talabot F, Peitsch M, Edgerton M D, Meldrum E, Allet E, Fish R, Jamotte T, Curchod M L, Loferer H. A genome-based approach for the identification of essential bacterial genes. Nat Biotechnol. 1998;16:851–856. doi: 10.1038/nbt0998-851. [DOI] [PubMed] [Google Scholar]
- 3.Asai Y, Kawagishi I, Sockett R E, Homma M. Hybrid motor with H+- and Na+-driven components can rotate Vibrio polar flagella by using sodium ions. J Bacteriol. 1999;181:6332–6338. doi: 10.1128/jb.181.20.6332-6338.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Atsumi T, Maekawa Y, Tokuda H, Imae Y. Amiloride at pH 7.0 inhibits the Na+-driven flagellar motors of Vibrio alginolyticus but allows cell growth. FEBS Lett. 1992;314:114–116. doi: 10.1016/0014-5793(92)80954-f. [DOI] [PubMed] [Google Scholar]
- 5.Atsumi T, McCarter L, Imae Y. Polar and lateral flagellar motors of marine Vibrio are driven by different ion-motive forces. Nature. 1992;355:182–184. doi: 10.1038/355182a0. [DOI] [PubMed] [Google Scholar]
- 6.Avetisyan A V, Bogachev A V, Murtasina R A, Skulachev V P. Involvement of a d-type oxidase in the Na+-motive respiratory chain of Escherichia coli growing under low ΔμH+ conditions. FEBS Lett. 1992;306:199–202. doi: 10.1016/0014-5793(92)80999-w. [DOI] [PubMed] [Google Scholar]
- 7.Avetisyan A V, Bogachev A V, Murtasina R A, Skulachev V P. ATP-driven Na+ transport and Na+-dependent ATP synthesis in Escherichia coli grown at low ΔμH+ FEBS Lett. 1993;317:267–270. doi: 10.1016/0014-5793(93)81290-g. [DOI] [PubMed] [Google Scholar]
- 8.Avetisyan A V, Dibrov P A, Semeykina A L, Skulachev V P, Sokolov M V. Adaptation of Bacillus FTU and Escherichia coli to alkaline conditions: the Na+-motive respiration. Biochim Biophys Acta. 1991;1098:95–104. [PubMed] [Google Scholar]
- 9.Bakeeva L E, Chumakov K M, Drachev A L, Metlina A L, Skulachev V P. The sodium cycle. III. Vibrio alginolyticus resembles Vibrio cholerae and some other vibriones by flagellar motor and ribosomal 5S-RNA structures. Biochim Biophys Acta. 1986;850:466–472. doi: 10.1016/0005-2728(86)90115-5. [DOI] [PubMed] [Google Scholar]
- 10.Beattle P, Tan K, Bourne R M, Leach D, Rich P R, Ward F B. Cloning and sequencing of four structural genes for the Na+-translocating NADH-ubiquinone oxidoreductase of Vibrio alginolyticus. FEBS Lett. 1994;356:333–338. doi: 10.1016/0014-5793(94)01275-x. [DOI] [PubMed] [Google Scholar]
- 11.Berg H C. Torque generation by the flagellar rotary motor. Biophys J. 1995;68:163S–166S. [PMC free article] [PubMed] [Google Scholar]
- 12.Berg M, Hilbi H, Dimroth P. Sequence of a gene cluster from Malonomonas rubra encoding components of the malonate decarboxylase Na+ pump and evidence for their function. Eur J Biochem. 1997;245:103–115. doi: 10.1111/j.1432-1033.1997.00103.x. [DOI] [PubMed] [Google Scholar]
- 13.Blattner F R, Plunkett G, 3rd, Bloch C A, Perna N T, Burland V, Riley M, Collado-Vides J, Glasner J D, Rode C K, Mayhew G F, Gregor J, Davis N W, Kirkpatrick H A, Goeden M A, Rose D J, Mau B, Shao Y. The complete genome sequence of Escherichia coli K-12. Science. 1997;277:1453–1474. doi: 10.1126/science.277.5331.1453. [DOI] [PubMed] [Google Scholar]
- 14.Bogachev A V, Murtasina R A, Shestopalov A I, Skulachev V P. The role of protonic and sodium potentials in the motility of E. coli and Bacillus FTU. Biochim Biophys Acta. 1993;1142:321–326. doi: 10.1016/0005-2728(93)90160-h. [DOI] [PubMed] [Google Scholar]
- 15.Bogachev A V, Murtazine R A, Shestopalov A I, Skulachev V P. Induction of the Escherichia coli cytochrome d by low ΔμH+ and by sodium ions. Eur J Biochem. 1995;232:304–308. doi: 10.1111/j.1432-1033.1995.tb20812.x. [DOI] [PubMed] [Google Scholar]
- 16.Bott M. Anaerobic citrate metabolism and its regulation in enterobacteria. Arch Microbiol. 1997;167:78–88. [PubMed] [Google Scholar]
- 17.Bott M, Pfister K, Burda P, Kalbermatter O, Woehike G, Dimroth P. Methylmalonyl-CoA decarboxylase from Propionigenium modestum—cloning and sequencing of the structural genes and purification of the enzyme complex. Eur J Biochem. 1997;250:590–599. doi: 10.1111/j.1432-1033.1997.0590a.x. [DOI] [PubMed] [Google Scholar]
- 18.Braune A, Bendrat K, Rospert S, Buckel W. The sodium ion translocating glutaconyl-CoA decarboxylase from Acidaminococcus fermentans: cloning and function of the genes forming a second operon. Mol Microbiol. 1999;31:473–487. doi: 10.1046/j.1365-2958.1999.01189.x. [DOI] [PubMed] [Google Scholar]
- 19.Brown I I, Galperin M Y, Glagolev A N, Skulachev V P. Utilization of energy stored in the form of Na+ and K+ ion gradients by bacterial cells. Eur J Biochem. 1983;134:345–349. doi: 10.1111/j.1432-1033.1983.tb07573.x. [DOI] [PubMed] [Google Scholar]
- 20.Brown M H, Paulsen I T, Skurray R A. The multidrug efflux protein NorM is a prototype of a new family of transporters. Mol Microbiol. 1999;31:394–395. doi: 10.1046/j.1365-2958.1999.01162.x. [DOI] [PubMed] [Google Scholar]
- 21.Callaway T R, Adams K A, Russell J B. The ability of “low G + C gram-positive” ruminal bacteria to resist monensin and counteract potassium depletion. Curr Microbiol. 1999;39:226–230. doi: 10.1007/s002849900449. [DOI] [PubMed] [Google Scholar]
- 22.Campbell L A, Rosenfeld M, Kuo C C. The role of Chlamydia pneumoniae in atherosclerosis—recent evidence from animal models. Trends Microbiol. 2000;8:255–257. doi: 10.1016/s0966-842x(00)01745-5. [DOI] [PubMed] [Google Scholar]
- 23.Chen C C, Tsuchiya T, Yamane Y, Wood J M, Wilson T H. Na+ (Li+)-proline cotransport in Escherichia coli. J Membr Biol. 1985;84:157–164. doi: 10.1007/BF01872213. [DOI] [PubMed] [Google Scholar]
- 24.Chen G, Russell J B. Transport and deamination of amino acids by a gram-positive, monensin-sensitive ruminal bacterium. Appl Environ Microbiol. 1990;56:2186–2192. doi: 10.1128/aem.56.7.2186-2192.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen G J, Russell J B. Sodium-dependent transport of branched-chain amino acids by a monensin-sensitive ruminal peptostreptococcus. Appl Environ Microbiol. 1989;55:2658–2663. doi: 10.1128/aem.55.10.2658-2663.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cheng J, Guffanti A A, Krulwich T A. A two-gene ABC-type transport system that extrudes Na+ in Bacillus subtilis is induced by ethanol or protonophore. Mol Microbiol. 1997;23:1107–1120. doi: 10.1046/j.1365-2958.1997.2951656.x. [DOI] [PubMed] [Google Scholar]
- 27.Chepuri V, Lemieux L, Au D C, Gennis R B. The sequence of the cyo operon indicates substantial structural similarities between the cytochrome o ubiquinol oxidase of Escherichia coli and the aa3-type family of cytochrome c oxidases. J Biol Chem. 1990;265:11185–11192. [PubMed] [Google Scholar]
- 28.Clarke D M, Loo T W, Gillam S, Bragg P D. Nucleotide sequence of the pntA and pntB genes encoding the pyridine nucleotide transhydrogenase of Escherichia coli. Eur J Biochem. 1986;158:647–653. doi: 10.1111/j.1432-1033.1986.tb09802.x. [DOI] [PubMed] [Google Scholar]
- 29.Claros M G, von Heijne G. TopPred II: an improved software for membrane protein structure predictions. Comput Appl Biosci. 1994;10:685–686. doi: 10.1093/bioinformatics/10.6.685. [DOI] [PubMed] [Google Scholar]
- 30.Cornelis G R, Van Gijsegem F. Assembly and function of type III secretory systems. Annu Rev Microbiol. 2000;54:735–774. doi: 10.1146/annurev.micro.54.1.735. [DOI] [PubMed] [Google Scholar]
- 31.Cruz W T, Nedialkov Y A, Thacker B J, Mulks M H. Molecular characterization of a common 48-kilodalton outer membrane protein of Actinobacillus pleuropneumoniae. Infect Immun. 1996;64:83–90. doi: 10.1128/iai.64.1.83-90.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Davies K, Solloz M. Assessment of uncoupling by amiloride analogs. Biochemistry. 1992;31:8055–8058. doi: 10.1021/bi00149a040. [DOI] [PubMed] [Google Scholar]
- 33.de Gier J W, Schepper M, Reijnders W N, van Dyck S J, Slotboom D J, Warne A, Saraste M, Krab K, Finel M, Stouthamer A H, van Spanning R J, van der Oost J. Structural and functional analysis of aa3-type and cbb3-type cytochrome c oxidases of Paracoccus denitrificans reveals significant differences in proton-pump design. Mol Microbiol. 1996;20:1247–1260. doi: 10.1111/j.1365-2958.1996.tb02644.x. [DOI] [PubMed] [Google Scholar]
- 34.Deguchi Y, Yamato I, Anraku Y. Nucleotide sequence of gltS, the Na+/glutamate symport carrier gene of Escherichia coli B. J Biol Chem. 1990;265:21704–21708. [PubMed] [Google Scholar]
- 35.Deppenmeier U, Lienard T, Gottschalk G. Novel reactions involved in energy conservation by methanogenic archaea. FEBS Lett. 1999;457:291–297. doi: 10.1016/s0014-5793(99)01026-1. [DOI] [PubMed] [Google Scholar]
- 36.Dibrov P. Lowering of the electric potential on the membrane as a possible signal modulating the expression of virulence factors in Vibrio cholerae. Mol Microbiol. 2000;35:473–475. doi: 10.1046/j.1365-2958.2000.01733.x. [DOI] [PubMed] [Google Scholar]
- 37.Dibrov P A. The role of sodium ion transport in Escherichia coli energetics. Biochim Biophys Acta. 1991;1056:209–224. doi: 10.1016/s0005-2728(05)80052-0. [DOI] [PubMed] [Google Scholar]
- 38.Dibrov P A, Lazarova R L, Skulachev V P, Verkhovskaya M L. The sodium cycle. II. Na+-coupled oxidative phosphorylation in Vibrio alginolyticus cells. Biochim Biophys Acta. 1986;850:458–465. doi: 10.1016/0005-2728(86)90114-3. [DOI] [PubMed] [Google Scholar]
- 39.Dibrov P A, Lazarova R L, Skulachev V P, Verkhovskaya M L. A study on Na+-coupled oxidative phosphorylation: ATP formation supported by artificially imposed delta pNa and delta pK in Vibrio alginolyticus cells. J Bioenerg Biomembr. 1989;21:347–357. doi: 10.1007/BF00762726. [DOI] [PubMed] [Google Scholar]
- 40.Dibrov P A, Skulachev V P, Sokolov M V, Verkhovskaya M L. The ATP-driven primary Na+ pump in subcellular vesicles of Vibrio alginolyticus. FEBS Lett. 1988;233:355–358. doi: 10.1016/0014-5793(88)80459-9. [DOI] [PubMed] [Google Scholar]
- 41.Dimroth P. A new sodium-transport system energized by the decarboxylation of oxaloacetate. FEBS Lett. 1980;122:234–236. doi: 10.1016/0014-5793(80)80446-7. [DOI] [PubMed] [Google Scholar]
- 42.Dimroth P. Reconstitution of sodium transport from purified oxaloacetate decarboxylase and phospholipid vesicles. J Biol Chem. 1981;256:11974–11976. [PubMed] [Google Scholar]
- 43.Dimroth P. Decarboxylation and transport. Biosci Rep. 1982;2:849–860. doi: 10.1007/BF01114890. [DOI] [PubMed] [Google Scholar]
- 44.Dimroth P. Mechanisms of sodium transport in bacteria. Philos Trans R Soc London Ser B. 1990;326:465–477. doi: 10.1098/rstb.1990.0025. [DOI] [PubMed] [Google Scholar]
- 45.Dimroth P. Na+-coupled alternative to H+-coupled primary transport systems in bacteria. Bioessays. 1991;13:463–468. doi: 10.1002/bies.950130906. [DOI] [PubMed] [Google Scholar]
- 46.Dimroth P. The ATPases of Propionigenium modestum and Bacillus alcalophilus. Strategies for ATP synthesis under low energy conditions. Biochim Biophys Acta. 1992;1101:236–239. [PubMed] [Google Scholar]
- 47.Dimroth P. Bacterial sodium ion-coupled energetics. Antonie Leeuwenhoek. 1994;65:381–395. doi: 10.1007/BF00872221. [DOI] [PubMed] [Google Scholar]
- 48.Dimroth P. Primary sodium ion translocating enzymes. Biochim Biophys Acta. 1997;1318:11–51. doi: 10.1016/s0005-2728(96)00127-2. [DOI] [PubMed] [Google Scholar]
- 49.Dimroth P, Hilbi H. Enzymic and genetic basis for bacterial growth on malonate. Mol Microbiol. 1997;25:3–10. doi: 10.1046/j.1365-2958.1997.4611824.x. [DOI] [PubMed] [Google Scholar]
- 50.Dimroth P, Schink B. Energy conservation in the decarboxylation of dicarboxylic acids by fermenting bacteria. Arch Microbiol. 1998;170:69–77. doi: 10.1007/s002030050616. [DOI] [PubMed] [Google Scholar]
- 51.Dimroth P, Thomer A. Subunit composition of oxaloacetate decarboxylase and characterization of the alpha chain as carboxyltransferase. Eur J Biochem. 1983;137:107–112. doi: 10.1111/j.1432-1033.1983.tb07802.x. [DOI] [PubMed] [Google Scholar]
- 52.Dimroth P, Thomer A. Solubilization and reconstitution of the Na+-dependent citrate carrier of Klebsiella pneumoniae. J Biol Chem. 1990;265:7721–7724. [PubMed] [Google Scholar]
- 53.Dimroth P, Wang H, Grabe M, Oster G. Energy transduction in the sodium F-ATPase of Propionigenium modestum. Proc Natl Acad Sci USA. 1999;96:4924–4929. doi: 10.1073/pnas.96.9.4924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Dinius D A, Simpson M E, Marsh P B. Effect of monensin fed with forage on digestion and the ruminal ecosystem of steers. J Anim Sci. 1976;42:229–234. doi: 10.2527/jas1976.421229x. [DOI] [PubMed] [Google Scholar]
- 55.Efiok B J, Webster D A. A cytochrome that can pump sodium ion. Biochem Biophys Res Commun. 1990;173:370–375. doi: 10.1016/s0006-291x(05)81067-8. [DOI] [PubMed] [Google Scholar]
- 56.Efiok B J, Webster D A. Respiratory-driven Na+ electrical potential in the bacterium Vitreoscilla. Biochemistry. 1990;29:4734–4739. doi: 10.1021/bi00471a030. [DOI] [PubMed] [Google Scholar]
- 57.Everiss K D, Hughes K J, Kovach M E, Peterson K M. The Vibrio cholerae acfB colonization determinant encodes an inner membrane protein that is related to a family of signal-transducing proteins. Infect Immun. 1994;62:3289–3298. doi: 10.1128/iai.62.8.3289-3298.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Fan F, Macnab R M. Enzymatic characterization of Flil. An ATPase involved in flagellar assembly in Salmonella typhimurium. J Biol Chem. 1996;271:31981–31988. doi: 10.1074/jbc.271.50.31981. [DOI] [PubMed] [Google Scholar]
- 59.Fleischmann R D, Adams M D, White O, Clayton R A, Kirkness E F, Kerlavage A R, Bult C J, Tomb J-F, Dougherty B A, Merrick J M, McKenney K, Sutton G G, FitzHugh W, Fields C, Gocayne J D, Scott J, Shirley R, Liu L-I, Glodek A, Kelley J M, Weidman J F, Phillips C A, Spriggs T, Hedblom E, Cotton M D, Utterback T R, Hanna M C, Nguyen D, Saudek D M, Brandon R C, Fine L D, Frichtman J L, Fuhrmann J L, Geoghagen N S M, Gnehm C L, McDonald L A, Small K V, Fraser C M, Smith H O, Venter J C. Whole-genome random sequencing and assembly of Haemophilus influenzae Rd. Science. 1995;269:496–512. doi: 10.1126/science.7542800. [DOI] [PubMed] [Google Scholar]
- 60.Fliegel L, Dibrov P. Biochemistry and molecular biology of the Na+/H+ exchanger: an overview. In: Fliegel L, editor. The Na+/H+ exchanger. New York, N.Y: Chapman & Hall; 1996. pp. 1–20. [Google Scholar]
- 61.Fraser C M, Norris S J, Weinstock G M, White O, Sutton G G, Dodson R, Gwinn M, Hickey E K, Clayton R, Ketchum K A, Sodergren E, Hardham J M, McLeod M P, Salzberg S, Peterson J, Khalak H, Richardson D, Howell J K, Chidambaram M, Utterback T, McDonald L, Artiach P, Bowman C, Cotton M D, Fujii C, Garland S, Hatch B, Horst K, Roberts K, Sandusky M, Weidman J, Smith H O, Venter J C. Complete genome sequence of Treponema pallidum, the syphilis spirochete. Science. 1998;281:375–388. doi: 10.1126/science.281.5375.375. [DOI] [PubMed] [Google Scholar]
- 62.Galan J E, Collmer A. Type III secretion machines: bacterial devices for protein delivery into host cells. Science. 1999;284:1322–1328. doi: 10.1126/science.284.5418.1322. [DOI] [PubMed] [Google Scholar]
- 63.Galperin M, Dibrov P A, Glagolev A N. ΔμH+ is required for flagellar growth in Escherichia coli. FEBS Lett. 1982;143:319–322. doi: 10.1016/0014-5793(82)80125-7. [DOI] [PubMed] [Google Scholar]
- 64.Galperin M Y, Aravind L, Koonin E V. Aldolases of the DhnA family: a possible solution to the problem of pentose and hexose biosynthesis in archaea. FEMS Microbiol Lett. 2000;183:259–264. doi: 10.1111/j.1574-6968.2000.tb08968.x. [DOI] [PubMed] [Google Scholar]
- 65.Galperin M Y, Koonin E V. Searching for drug targets in microbial genomes. Curr Opin Biotechnol. 1999;10:571–578. doi: 10.1016/s0958-1669(99)00035-x. [DOI] [PubMed] [Google Scholar]
- 66.Galperin M Y, Tatusov R L, Koonin E V. Comparing microbial genomes: how the gene set determines the lifestyle. In: Charlebois R L, editor. Organization of the prokaryotic genome. Washington, D.C.: ASM Press; 1999. pp. 91–108. [Google Scholar]
- 67.Gardel C L, Mekalanos J J. Alterations in Vibrio cholerae motility phenotypes correlate with changes in virulence factor expression. Infect Immun. 1996;64:2246–2255. doi: 10.1128/iai.64.6.2246-2255.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Gosink K K, Häse C C. Requirements for conversion of the Na+-driven flagellar motor of Vibrio cholerae to the H+-driven motor of Escherichia coli. J Bacteriol. 2000;182:4234–4240. doi: 10.1128/jb.182.15.4234-4240.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Grayston J T. Background and current knowledge of Chlamydia pneumoniae and atheroscierosis. J Infect Dis. 2000;181:S402–S410. doi: 10.1086/315596. [DOI] [PubMed] [Google Scholar]
- 70.Green G N, Fang H, Lin R J, Newton G, Mather M, Georgiou C D, Gennis R B. The nucleotide sequence of the cyd locus encoding the two subunits of the cytochrome d terminal oxidase complex of Escherichia coli. J Biol Chem. 1988;263:13138–13143. [PubMed] [Google Scholar]
- 71.Grinkevich V A, Lysenko A M, Muntyan M S, Skripnikova E V, Afrikyan E K. Identification of Bacillus sp. FTU strain and the study of the caa3-type oxidase homology. Biochemistry (Moscow) 1997;62:718–724. [PubMed] [Google Scholar]
- 72.Guentzel M N, Berry L J. Motility as a virulence factor for Vibrio cholerae. Infect Immun. 1975;11:890–897. doi: 10.1128/iai.11.5.890-897.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Haraszthy V I, Zambon J J, Trevisan M, Zeid M, Genco R J. Identification of periodontal pathogens in atheromatous plaques. J Periodontol. 2000;71:1554–1560. doi: 10.1902/jop.2000.71.10.1554. [DOI] [PubMed] [Google Scholar]
- 74.Harel-Bronstein M, Dibrov P, Olami Y, Pinner E, Schuldiner S, Padan E. MH1, a second-site revertant of an Escherichia coli mutant lacking Na+/H+ antiporters (ΔnhaAΔnhaB), regains Na+ resistance and a capacity to excrete Na+ in a ΔμH+-independent fashion. J Biol Chem. 1995;270:3816–3822. doi: 10.1074/jbc.270.8.3816. [DOI] [PubMed] [Google Scholar]
- 75.Harkey C W, Everiss K D, Peterson K M. The Vibrio cholerae toxin-coregulated-pilus gene tcpI encodes a homolog of methyl-accepting chemotaxis proteins. Infect Immun. 1994;62:2669–2978. doi: 10.1128/iai.62.7.2669-2678.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Harold F M, Maloney P C. Energy transduction by ion currents. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C.: ASM Press; 1996. pp. 283–306. [Google Scholar]
- 77.Häse C C, Mekalanos J J. TcpP protein is a positive regulator of virulence gene expression in Vibrio cholerae. Proc Natl Acad Sci USA. 1998;95:730–734. doi: 10.1073/pnas.95.2.730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Häse C C, Mekalanos J J. Effects of changes in membrane sodium flux on virulence gene expression in Vibrio cholerae. Proc Natl Acad Sci USA. 1999;96:3183–3187. doi: 10.1073/pnas.96.6.3183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Hayashi M, Hirai K, Unemoto T. Cloning of the Na+-translocating NADH-quinone reductase gene from the marine bacterium Vibrio alginolyticus and the expression of the beta-subunit in Escherichia coli. FEBS Lett. 1994;356:330–332. doi: 10.1016/0014-5793(94)01274-1. [DOI] [PubMed] [Google Scholar]
- 80.Hayashi M, Hirai K, Unemoto T. Sequencing and the alignment of structural genes in the nqr operon encoding the Na+-translocating NADH-quinone reductase from Vibrio alginolyticus. FEBS Lett. 1995;363:75–77. doi: 10.1016/0014-5793(95)00283-f. [DOI] [PubMed] [Google Scholar]
- 81.Hayashi M, Miyoshi T, Sato M, Unemoto T. Properties of respiratory chain-linked Na+-independent NADH-quinone reductase in a marine Vibrio alginolyticus. Biochim Biophys Acta. 1992;1099:145–151. [PubMed] [Google Scholar]
- 82.Hayashi M, Nakayama Y, Unemoto T. Existence of Na+-translocating NADH-quinone reductase in Haemophilus influenzae. FEBS Lett. 1996;381:174–176. doi: 10.1016/0014-5793(96)00114-7. [DOI] [PubMed] [Google Scholar]
- 83.Heidelberg J F, Eisen J A, Nelson W C, Clayton R A, Gwinn M L, Dodson R J, Haft D H, Hickey E K, Peterson J D, Umayam L, Gill S R, Nelson K E, Read T D, Tettelin H, Richardson D, Ermolaeva M D, Vamathevan J, Bass S, Qin H, Dragoi I, Sellers P, McDonald L, Utterback T, Fleishmann R D, Nierman W C, White O, Salzberg S L, Smith H O, Colwell R R, Mekalanos J J, Venter J C, Fraser C M. DNA sequence of both chromosomes of the cholera pathogen Vibrio cholerae. Nature. 2000;406:477–483. doi: 10.1038/35020000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Heyne R I, de Vrij W, Crielaard W, Konings W N. Sodium ion-dependent amino acid transport in membrane vesicles of Bacillus stearothermophilus. J Bacteriol. 1991;173:791–800. doi: 10.1128/jb.173.2.791-800.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Hilpert W, Schink B, Dimroth P. Life by a new decarboxylation-dependent energy conservation mechanism with sodium as coupling ion. EMBO J. 1984;3:1665–1670. doi: 10.1002/j.1460-2075.1984.tb02030.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Hindersson P, Thomas D, Stamm L, Penn C, Norris S, Joens L A. Interaction of spirochetes with the host. Res Microbiol. 1992;143:629–639. doi: 10.1016/0923-2508(92)90121-4. [DOI] [PubMed] [Google Scholar]
- 87.Hiramatsu T, Kodama K, Kuroda T, Mizushima T, Tsuchiya T. A putative multisubunit Na+/H+ antiporter from Staphylococcus aureus. J Bacteriol. 1998;180:6642–6648. doi: 10.1128/jb.180.24.6642-6648.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Hoenke S, Schmid M, Dimroth P. Sequence of a gene cluster from Klebsiella pneumoniae encoding malonate decarboxylase and expression of the enzyme in Escherichia coli. Eur J Biochem. 1997;246:530–538. doi: 10.1111/j.1432-1033.1997.00530.x. [DOI] [PubMed] [Google Scholar]
- 89.Honer zu Bentrup K, Ubbink-Kok T, Lolkema J S, Konings W N. An Na+-pumping V1V0-ATPase complex in the thermophilic bacterium Clostridium fervidus. J Bacteriol. 1997;179:1274–1279. doi: 10.1128/jb.179.4.1274-1279.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Hoshino T, Kose K, Uratani Y. Cloning and nucleotide sequence of the gene braB coding for the sodium-coupled branched-chain amino acid carrier in Pseudomonas aeruginosa PAO. Mol Gen Genet. 1990;220:461–467. doi: 10.1007/BF00391754. [DOI] [PubMed] [Google Scholar]
- 91.Huder J B, Dimroth P. Sequence of the sodium ion pump methylmalonyl-CoA decarboxylase from Veillonella parvula. J Biol Chem. 1993;268:24564–24571. [PubMed] [Google Scholar]
- 92.Huder J B, Dimroth P. Expression of the sodium ion pump methylmalonyl-coenzyme A-decarboxylase from Veillonella parvula and of mutated enzyme specimens in Escherichia coli. J Bacteriol. 1995;177:3623–3630. doi: 10.1128/jb.177.13.3623-3630.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Hueck C J. Type III protein secretion systems in bacterial pathogens of animals and plants. Microbiol Mol Biol Rev. 1998;62:379–433. doi: 10.1128/mmbr.62.2.379-433.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Ikegami M, Kawano M, Takase K, Yamato I, Igarashi K, Kakinuma Y. Enterococcus hirae vacuolar ATPase is expressed in response to pH as well as sodium. FEBS Lett. 1999;454:67–70. doi: 10.1016/s0014-5793(99)00776-0. [DOI] [PubMed] [Google Scholar]
- 95.Inaba K, Kuroda T, Shimamoto T, Kayahara T, Tsuda M, Tsuchiya T. Lithium toxicity and Na+(Li+)/H+ antiporter in Escherichia coli. Biol Pharm Bull. 1994;17:395–398. doi: 10.1248/bpb.17.395. [DOI] [PubMed] [Google Scholar]
- 96.Inaba K, Utsugi J, Kuroda T, Tsuda M, Tsuchiya T. Na+(Li+)/H+ antiporter in Pseudomonas aeruginosa and effect of Li+ on cell growth. Biol Pharm Bull. 1997;20:621–624. doi: 10.1248/bpb.20.621. [DOI] [PubMed] [Google Scholar]
- 97.Ivey D M, Guffanti A A, Bossewitch J S, Padan E, Krulwich T A. Molecular cloning and sequencing of a gene from alkaliphilic Bacillus firmus OF4 that functionally complements an Escherichia coli strain carrying a deletion in the nhaA Na+/H+ antiporter gene. J Biol Chem. 1991;266:23483–23489. [PubMed] [Google Scholar]
- 98.Iwao E, Hirayama F, Takagi S, Yokoyama Y, Ikeda Y. Virulence factors of Helicobacter pylori affecting its gastric colonization in Mongolian gerbils. J Gastroenterol. 1999;34:47–54. [PubMed] [Google Scholar]
- 99.Jaques S, Kim Y K, McCarter L L. Mutations conferring resistance to phenamil and amiloride, inhibitors of sodium-driven motility of Vibrio parahaemolyticus. Proc Natl Acad Sci USA. 1999;96:5740–5745. doi: 10.1073/pnas.96.10.5740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Kaim G, Dimroth P. A double mutation in subunit c of the Na+-specific F1F0-ATPase of Propionigenium modestum results in a switch from Na+ to H+-coupled ATP synthesis in the Escherichia coli host cells. J Mol Biol. 1995;253:726–738. doi: 10.1006/jmbi.1995.0586. [DOI] [PubMed] [Google Scholar]
- 101.Kaim G, Dimroth P. A triple mutation in the a subunit of the Escherichia coli/Propionigenium modestum F1F0 ATPase hybrid causes a switch from Na+ stimulation to Na+ inhibition. Biochemistry. 1998;37:4626–4634. doi: 10.1021/bi973022f. [DOI] [PubMed] [Google Scholar]
- 102.Kakinuma Y, Igarashi K. Electrogenic Na+ transport by Enterococcus hirae Na+-ATPase. FEBS Lett. 1995;359:255–258. doi: 10.1016/0014-5793(95)00056-f. [DOI] [PubMed] [Google Scholar]
- 103.Kakinuma Y, Yamato I, Murata T. Structure and function of vacuolar Na+-translocating ATPase in Enterococcus hirae. J Bioenerg Biomembr. 1999;31:7–14. doi: 10.1023/a:1005499126939. [DOI] [PubMed] [Google Scholar]
- 104.Kalman S, Mitchell W, Marathe R, Lammel C, Fan J, Hyman R W, Olinger L, Grimwood J, Davis R W, Stephens R S. Comparative genomes of Chlamydia pneumoniae and C. trachomatis. Nat Genet. 1999;21:385–389. doi: 10.1038/7716. [DOI] [PubMed] [Google Scholar]
- 105.Kamata H, Akiyama S, Morosawa H, Ohta T, Hamamoto T, Kambe T, Kagawa Y, Hirata H. Primary structure of the alanine carrier protein of thermophilic bacterium PS3. J Biol Chem. 1992;267:21650–21655. [PubMed] [Google Scholar]
- 106.Karpel R, Alon T, Glaser G, Schuldiner S, Padan E. Expression of a sodium proton antiporter (NhaA) in Escherichia coli is induced by Na+ and Li+ ions. J Biol Chem. 1991;266:21753–21759. [PubMed] [Google Scholar]
- 107.Kelly D J. The physiology and metabolism of the human gastric pathogen Helicobacter pylori. Adv Microb Physiol. 1998;40:137–189. doi: 10.1016/s0065-2911(08)60131-9. [DOI] [PubMed] [Google Scholar]
- 108.Kim K J, Chi P Y, Hwang K W, Stark B C, Webster D A. Study of cytochrome bo function in Vitreoscilla using a cyo− knockout mutant. J Biochem (Tokyo) 2000;128:49–55. doi: 10.1093/oxfordjournals.jbchem.a022729. [DOI] [PubMed] [Google Scholar]
- 109.Kogure K. Bioenergetics of marine bacteria. Curr Opin Biotechnol. 1998;9:278–282. doi: 10.1016/s0958-1669(98)80059-1. [DOI] [PubMed] [Google Scholar]
- 110.Kogure K, Tokuda H. Respiration-dependent primary Na+ pump in halophilic marine bacterium Alcaligenes strain 201. FEBS Lett. 1989;256:147–149. [Google Scholar]
- 111.Kojima S, Yamamoto K, Kawagishi I, Homma M. The polar flagellar motor of Vibrio cholerae is driven by an Na+ motive force. J Bacteriol. 1999;181:1927–1930. doi: 10.1128/jb.181.6.1927-1930.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Konings W N, Otto R. Energy transduction and solute transport in streptococci. Antonie Leeuwenhoek. 1983;49:247–257. doi: 10.1007/BF00399501. [DOI] [PubMed] [Google Scholar]
- 113.Koonin E V, Galperin M Y. Prokaryotic genomes: the emerging paradigm of genome-based microbiology. Curr Opin Genet Dev. 1997;7:757–763. doi: 10.1016/s0959-437x(97)80037-8. [DOI] [PubMed] [Google Scholar]
- 114.Koonin E V, Tatusov R L, Galperin M Y. Beyond the complete genomes: from sequences to structure and function. Curr Opin Struct Biol. 1998;8:355–363. doi: 10.1016/s0959-440x(98)80070-5. [DOI] [PubMed] [Google Scholar]
- 115.Kostyrko V A, Semeykina A L, Skulachev V P, Smirnova I A, Vaghina M L, Verkhovskaya M L. The H+-motive and Na+-motive respiratory chains in Bacillus FTU subcellular vesicles. Eur J Biochem. 1991;198:527–534. doi: 10.1111/j.1432-1033.1991.tb16046.x. [DOI] [PubMed] [Google Scholar]
- 116.Kuroda T, Shimamoto T, Mizushima T, Tsuchiya T. Mutational analysis of amiloride sensitivity of the NhaA Na+/H+ antiporter from Vibrio parahaemolyticus. J Bacteriol. 1997;179:7600–7602. doi: 10.1128/jb.179.23.7600-7602.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Lagerlof F, Oliveby A. Caries-protective factors in saliva. Adv Dent Res. 1994;8:229–238. doi: 10.1177/08959374940080021601. [DOI] [PubMed] [Google Scholar]
- 118.Laubinger W, Dimroth P. Characterization of the Na+-stimulated ATPase of Propionigenium modestum as an enzyme of the F1F0 type. Eur J Biochem. 1987;168:475–480. doi: 10.1111/j.1432-1033.1987.tb13441.x. [DOI] [PubMed] [Google Scholar]
- 119.Laublnger W, Dimroth P. The sodium ion translocating adenosinetriphosphatase of Propionigenium modestum pumps protons at low sodium ion concentrations. Biochemistry. 1989;28:7194–7198. doi: 10.1021/bi00444a010. [DOI] [PubMed] [Google Scholar]
- 120.Laussermair E, Schwarz E, Oesterhelt D, Reinke H, Beyreuther K, Dimroth P. The sodium ion translocating oxaloacetate decarboxylase of Klebsiella pneumoniae. Sequence of the integral membrane-bound subunits beta and gamma. J Biol Chem. 1989;264:14710–14715. [PubMed] [Google Scholar]
- 121.Lee A, Mao W, Warren M S, Mistry A, Hoshino K, Okumura R, Ishida H, Lomovskaya O. Interplay between efflux pumps may provide either additive or multiplicative effects on drug resistance. J Bacteriol. 2000;182:3142–3150. doi: 10.1128/jb.182.11.3142-3150.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Lewis K. Multidrug resistance pumps in bacteria: variations on a theme. Trends Biochem Sci. 1994;19:119–123. doi: 10.1016/0968-0004(94)90204-6. [DOI] [PubMed] [Google Scholar]
- 123.Lewis K, Hooper D C, Ouellette M. Multidrug resistance pumps provide broad defense. ASM News. 1997;63:605–610. [Google Scholar]
- 124.Lolkema J S, Speelmans G, Konings W N. Na+-coupled versus H+-coupled energy transduction in bacteria. Biochim Biophys Acta. 1994;1187:211–215. doi: 10.1016/0005-2728(94)90113-9. [DOI] [PubMed] [Google Scholar]
- 125.Lombard M, Touati D, Fontecave M, Niviere V. Superoxide reductase as a unique defense system against superoxide stress in the microaerophile Treponema pallidum. J Biol Chem. 2000;275:27021–27026. doi: 10.1074/jbc.M004201200. [DOI] [PubMed] [Google Scholar]
- 126.Macnab R M. Flagella and motility. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C.: ASM Press; 1996. pp. 123–145. [Google Scholar]
- 127.Magagnin S, Werner A, Markovich D, Sorribas V, Stange G, Biber J, Murer H. Expression cloning of human and rat renal cortex Na/Pi cotransport. Proc Natl Acad Sci USA. 1993;90:5979–5983. doi: 10.1073/pnas.90.13.5979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Maloney P C, Wilson T H. Ion-coupled transport and transporters. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C.: ASM Press; 1996. pp. 1130–1148. [Google Scholar]
- 129.Marchal K, Sun J, Keijers V, Haaker H, Vanderleyden J. A cytochrome cbb3 (cytochrome c) terminal oxidase in Azospirillum brasilense Sp7 supports microaerobic growth. J Bacteriol. 1998;180:5689–5696. doi: 10.1128/jb.180.21.5689-5696.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Marty-Teysset C, Lolkema J S, Schmitt P, Divies C, Konings W N. Membrane potential-generating transport of citrate and malate catalyzed by CitP of Leuconostoc mesenteroides. J Biol Chem. 1995;270:25370–25376. doi: 10.1074/jbc.270.43.25370. [DOI] [PubMed] [Google Scholar]
- 131.May B J, Zhang Q, Li L L, Paustian M L, Whittam T S, Kapur V. Complete genomic sequence of Pasteurella multocida Pm70. Proc Natl Acad Sci USA. 2001;98:3460–3465. doi: 10.1073/pnas.051634598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.McGee D J, Mobley H L. Mechanisms of Helicobacter pylori infection: bacterial factors. Curr Top Microbiol Immunol. 1999;241:155–180. doi: 10.1007/978-3-642-60013-5_9. [DOI] [PubMed] [Google Scholar]
- 133.Morita Y, Kataoka A, Shiota S, Mizushima T, Tsuchiya T. NorM of Vibrio parahaemolyticus is an Na+-driven multidrug efflux pump. J Bacteriol. 2000;182:6694–6697. doi: 10.1128/jb.182.23.6694-6697.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Morita Y, Kodama K, Shiota S, Mine T, Kataoka A, Mizushima T, Tsuchiya T. NorM, a putative multidrug efflux protein, of Vibrio parahaemolyticus and its homolog in Escherichia coli. Antimicrob Agents Chemother. 1998;42:1778–1782. doi: 10.1128/aac.42.7.1778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Muntyan M S, Bloch D A, Drachev L A, Skulachev V P. Kinetics of CO binding to putative Na+-motive oxidases of the o-type from Bacillus FTU and of the d-type from Escherichia coli. FEBS Lett. 1993;327:347–350. doi: 10.1016/0014-5793(93)81018-u. [DOI] [PubMed] [Google Scholar]
- 136.Murata T, Yamato I, Igarashi K, Kakinuma Y. Intracellular Na+ regulates transcription of the ntp operon encoding a vacuolar-type Na+-translocating ATPase in Enterococcus hirae. J Biol Chem. 1996;271:23661–23666. doi: 10.1074/jbc.271.39.23661. [DOI] [PubMed] [Google Scholar]
- 137.Nakao T, Yamato I, Anraku Y. Nucleotide sequence of putP, the proline carrier gene of Escherichia coli K12. Mol Gen Genet. 1987;208:70–75. doi: 10.1007/BF00330424. [DOI] [PubMed] [Google Scholar]
- 138.Nakayama Y, Hayashi M, Unemoto T. Identification of six subunits constituting Na+-translocating NADH-quinone reductase from the marine Vibrio alginolyticus. FEBS Lett. 1998;422:240–242. doi: 10.1016/s0014-5793(98)00016-7. [DOI] [PubMed] [Google Scholar]
- 139.Nakayama Y, Hayashi M, Yoshikawa K, Mochida K, Unemoto T. Inhibitor studies of a new antibiotic, korormicin, 2-n-heptyl-4-hydroxyquinoline N-oxide and Ag+ toward the Na+-translocating NADH-quinone reductase from the marine Vibrio alginolyticus. Biol Pharm Bull. 1999;22:1064–1067. doi: 10.1248/bpb.22.1064. [DOI] [PubMed] [Google Scholar]
- 140.Natale D A, Galperin M Y, Tatusov R L, Koonin E V. Using the COG database to improve gene recognition in complete genomes. Genetica. 2000;108:9–17. doi: 10.1023/a:1004031323748. [DOI] [PubMed] [Google Scholar]
- 141.Nelson K E, Clayton R A, Gill S R, Gwinn M L, Dodson R J, Haft D H, Hickey E K, Peterson J D, Nelson W C, Ketchum K A, McDonald L, Utterback T R, Malek J A, Linher K D, Garrett M M, Stewart A M, Cotton M D, Pratt M S, Phillips C A, Richardson D, Heidelberg J, Sutton G G, Fleischmann R D, Eisen J A, Fraser C M. Evidence for lateral gene transfer between Archaea and bacteria from genome sequence of Thermotoga maritima. Nature. 1999;399:323–329. doi: 10.1038/20601. [DOI] [PubMed] [Google Scholar]
- 142.Neumann S, Matthey U, Kaim G, Dimroth P. Purification and properties of the F1F0 ATPase of Ilyobacter tartaricus, a sodium ion pump. J Bacteriol. 1998;180:3312–3316. doi: 10.1128/jb.180.13.3312-3316.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Nguyen L, Paulsen I T, Tchieu J, Hueck C J, Saier M H., Jr Phylogenetic analyses of the constituents of Type III protein secretion systems. J Mol Microbiol Biotechnol. 2000;2:125–144. [PubMed] [Google Scholar]
- 144.Nielsen H, Engelbrecht J, Brunak S, von Heijne G. Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng. 1997;10:1–6. doi: 10.1093/protein/10.1.1. [DOI] [PubMed] [Google Scholar]
- 145.Noel J, Pouyssegur J. Hormonal regulation, pharmacology, and membrane sorting of vertebrate Na+/H+ exchanger isoforms. Am J Physiol. 1995;268:C283–C296. doi: 10.1152/ajpcell.1995.268.2.C283. [DOI] [PubMed] [Google Scholar]
- 146.Nozaki K, Kuroda T, Mizushima T, Tsuchiya T. A new Na+/H+ antiporter, NhaD, of Vibrio parahaemolyticus. Biochim Biophys Acta. 1998;1369:213–220. doi: 10.1016/s0005-2736(97)00223-x. [DOI] [PubMed] [Google Scholar]
- 147.Ogawa W, Kim Y M, Mizushima T, Tsuchiya T. Cloning and expression of the gene for the Na+-coupled serine transporter from Escherichia coli and characteristics of the transporter. J Bacteriol. 1998;180:6749–6752. doi: 10.1128/jb.180.24.6749-6752.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Oh S, Kogure K, Ohwada K, Simidu U. Correlation between possession of a respiration-dependent Na+ pump and Na+ requirement for growth of marine bacteria of marine bacteria. Appl Environ Microbiol. 1991;57:1844–1846. doi: 10.1128/aem.57.6.1844-1846.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Oosawa K, Ueno T, Aizawa S. Overproduction of the bacterial flagellar switch proteins and their interactions with the MS ring complex in vitro. J Bacteriol. 1994;176:3683–3691. doi: 10.1128/jb.176.12.3683-3691.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Ottemann K M, Miller J F. Roles for motility in bacterial-host interactions. Mol Microbiol. 1997;24:1109–1117. doi: 10.1046/j.1365-2958.1997.4281787.x. [DOI] [PubMed] [Google Scholar]
- 151.Padan E, Schuldiner S. Molecular physiology of Na+/H+ antiporters, key transporters in circulation of Na+ and H+ in cells. Biochim Biophys Acta. 1994;1185:129–151. doi: 10.1016/0005-2728(94)90204-6. [DOI] [PubMed] [Google Scholar]
- 152.Park C, Moon J Y, Cokic P, Webster D A. Na+-translocating cytochrome bo terminal oxidase from Vitreoscilla: some parameters of its Na+ pumping and orientation in synthetic vesicles. Biochemistry. 1996;35:11895–11900. doi: 10.1021/bi9530503. [DOI] [PubMed] [Google Scholar]
- 153.Parkhill J, Achtman M, James K D, Bentley S D, Churcher C, Klee S R, Morelli G, Basham D, Brown D, Chillingworth T, Davies R M, Davis P, Devlin K, Feltwell T, Hamlin N, Holroyd S, Jagels K, Leather S, Moule S, Mungall K, Quail M A, Rajandream M A, Rutherford K M, Simmonds M, Skelton J, Whitehead S, Spratt B G, Barrell B G. Complete DNA sequence of a serogroup A strain of Neisseria meningitidis Z2491. Nature. 2000;404:502–506. doi: 10.1038/35006655. [DOI] [PubMed] [Google Scholar]
- 154.Paulsen I T, Brown M H, Skurray R A. Proton-dependent multidrug efflux systems. Microbiol Rev. 1996;60:575–608. doi: 10.1128/mr.60.4.575-608.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Paulsen I T, Nguyen L, Sliwinski M K, Rabus R, Saier M H., Jr Microbial genome analyses: comparative transport capabilities in eighteen prokaryotes. J Mol Biol. 2000;301:75–100. doi: 10.1006/jmbi.2000.3961. [DOI] [PubMed] [Google Scholar]
- 156.Paulsen I T, Sliwinski M K, Saier M H., Jr Microbial genome analyses: global comparisons of transport capabilities based on phylogenies, bioenergetics and substrate specificities. J Mol Biol. 1998;277:573–592. doi: 10.1006/jmbi.1998.1609. [DOI] [PubMed] [Google Scholar]
- 157.Pfenninger-Li X D, Dimroth P. NADH formation by Na+-coupled reversed electron transfer in Klebsiella pneumoniae. Mol Microbiol. 1992;6:1943–1948. doi: 10.1111/j.1365-2958.1992.tb01367.x. [DOI] [PubMed] [Google Scholar]
- 158.Pinner E, Padan E, Schuldiner S. Cloning, sequencing, and expression of the nhaB gene, encoding a Na+/H+ antiporter in Escherichia coli. J Biol Chem. 1992;267:11064–11068. [PubMed] [Google Scholar]
- 159.Pinner E, Padan E, Schuldiner S. Amiloride and harmaline are potent inhibitors of NhaB, a Na+/H+ antiporter from Escherichia coli. FEBS Lett. 1995;365:18–22. doi: 10.1016/0014-5793(95)00364-f. [DOI] [PubMed] [Google Scholar]
- 160.Pos K M, Dimroth P. Functional properties of the purified Na+-dependent citrate carrier of Klebsiella pneumoniae: evidence for asymmetric orientation of the carrier protein in proteoliposomes. Biochemistry. 1996;35:1018–1026. doi: 10.1021/bi951609t. [DOI] [PubMed] [Google Scholar]
- 161.Pourcher T, Bassilana M, Sarkar H K, Kaback H R, Leblanc G. The melibiose/Na+ symporter of Escherichia coli: kinetic and molecular properties. Philos Trans R Soc London Ser B. 1990;326:411–423. doi: 10.1098/rstb.1990.0021. [DOI] [PubMed] [Google Scholar]
- 162.Puustinen A, Finel M, Haltia T, Gennis R B, Wikstrom M. Properties of the two terminal oxidases of Escherichia coli. Biochemistry. 1991;30:3936–3942. doi: 10.1021/bi00230a019. [DOI] [PubMed] [Google Scholar]
- 163.Puustinen A, Finel M, Virkki M, Wikstrom M. Cytochrome o (bo) is a proton pump in Paracoccus denitrificans and Escherichia coli. FEBS Lett. 1989;249:163–167. doi: 10.1016/0014-5793(89)80616-7. [DOI] [PubMed] [Google Scholar]
- 164.Raitio M, Wikstrom M. An alternative cytochrome oxidase of Paracoccus denitrificans functions as a proton pump. Biochim Biophys Acta. 1994;1186:100–106. [Google Scholar]
- 165.Reizer J, Reizer A, Saier M H., Jr The Na+/pantothenate symporter (PanF) of Escherichia coli is homologous to the Na+/proline symporter (PutP) of E. coli and the Na+/glucose symporters of mammals. Res Microbiol. 1990;141:1069–1072. doi: 10.1016/0923-2508(90)90080-a. [DOI] [PubMed] [Google Scholar]
- 166.Reizer J, Reizer A, Saier M H., Jr A functional superfamily of sodium/solute symporters. Biochim Biophys Acta. 1994;1197:133–166. doi: 10.1016/0304-4157(94)90003-5. [DOI] [PubMed] [Google Scholar]
- 167.Russell J B, Strobel H J. Effect of ionophores on ruminal fermentation. Appl Environ Microbiol. 1989;55:1–6. doi: 10.1128/aem.55.1.1-6.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Saez L P, Garcia P, Martinez-Luque M, Klipp W, Blasco R, Castillo F. Role for draTG and mf genes in reduction of 2,4-dinitrophenol by Rhodobacter capsulatus. J Bacteriol. 2001;183:1780–1783. doi: 10.1128/JB.183.5.1780-1783.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Saier M H., Jr Families of transmembrane transporters selective for amino acids and their derivatives. Microbiology. 2000;146:1775–1795. doi: 10.1099/00221287-146-8-1775. [DOI] [PubMed] [Google Scholar]
- 170.Saier M H., Jr A functional-phylogenetic classification system for transmembrane solute transporters. Microbiol Mol Biol Rev. 2000;64:354–411. doi: 10.1128/mmbr.64.2.354-411.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Saier M H, Jr, Beatty J T, Goffeau A, Harley K T, Heijne W H, Huang S C, Jack D L, Jahn P S, Lew K, Liu J, Pao S S, Paulsen I T, Tseng T T, Virk P S. The major facilitator superfamily. J Mol Microbiol Biotechnol. 1999;1:257–279. [PubMed] [Google Scholar]
- 172.Sanchez L B, Galperin M Y, Muller M. Acetyl-CoA synthetase from the amitochondriate eukaryote Giardia lamblia belongs to the newly recognized superfamily of acyl-CoA synthetases (nucleoside diphosphate-forming) J Biol Chem. 2000;275:5794–5803. doi: 10.1074/jbc.275.8.5794. [DOI] [PubMed] [Google Scholar]
- 173.Schmehl M, Jahn A, Meyer zu Vilsendorf A, Hennecke S, Masepohl B, Schuppler M, Marxer M, Oelze J, Klipp W. Identification of a new class of nitrogen fixation genes in Rhodobacter capsulatus: a putative membrane complex involved in electron transport to nitrogenase. Mol Gen Genet. 1993;241:602–615. doi: 10.1007/BF00279903. [DOI] [PubMed] [Google Scholar]
- 174.Schrader M, Fendler K, Bamberg E, Gassel M, Epstein W, Altendorf K, Drose S. Replacement of glycine 232 by aspartic acid in the KdpA subunit broadens the ion specificity of the K+-translocating KdpFABC complex. Biophys J. 2000;79:602–613. doi: 10.1016/S0006-3495(00)76337-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Schultz J, Milpetz F, Bork P, Ponting C P. SMART, a simple modular architecture research tool: identification of signaling domains. Proc Natl Acad Sci USA. 1998;95:5857–5864. doi: 10.1073/pnas.95.11.5857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Schwarz E, Oesterhelt D, Reinke H, Beyreuther K, Dimroth P. The sodium ion translocating oxalacetate decarboxylase of Klebsiella pneumoniae. Sequence of the biotin-containing alpha-subunit and relationship to other biotin-containing enzymes. J Biol Chem. 1988;263:9640–9645. [PubMed] [Google Scholar]
- 177.Semeykina A L, Skulachev V P. Submicromolar Ag+ increases passive Na+ permeability and inhibits the respiration-supported formation of Na+ gradient in Bacillus FTU vesicles. FEBS Lett. 1990;269:69–72. doi: 10.1016/0014-5793(90)81120-d. [DOI] [PubMed] [Google Scholar]
- 178.Semeykina A L, Skulachev V P, Verkhovskaya M L, Bulygina E S, Chumakov K M. The Na+-motive terminal oxidase activity in an alkalo- and halo-tolerant Bacillus. Eur J Biochem. 1989;183:671–678. doi: 10.1111/j.1432-1033.1989.tb21097.x. [DOI] [PubMed] [Google Scholar]
- 179.Shigenobu S, Watanabe H, Hattori M, Sakaki Y, Ishikawa H. Genome sequence of the endocellular bacterial symbiont of aphids Buchnera sp. APS Nature. 2000;407:81–86. doi: 10.1038/35024074. [DOI] [PubMed] [Google Scholar]
- 180.Silver S, Phung L T. Bacterial heavy metal resistance: new surprises. Annu Rev Microbiol. 1996;50:753–789. doi: 10.1146/annurev.micro.50.1.753. [DOI] [PubMed] [Google Scholar]
- 181.Singh A E, Romanowski B. Syphilis: review with emphasis on clinical, epidemiologic, and some biologic features. Clin Microbiol Rev. 1999;12:187–209. doi: 10.1128/cmr.12.2.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Skulachev V P. Membrane-linked energy buffering as the biological function of Na+/K+ gradient. FEBS Lett. 1978;87:171–179. doi: 10.1016/0014-5793(78)80326-3. [DOI] [PubMed] [Google Scholar]
- 183.Skulachev V P. Membrane-linked energy transductions. Bioenergetic functions of sodium: H+ is not unique as a coupling ion. Eur J Biochem. 1985;151:199–208. doi: 10.1111/j.1432-1033.1985.tb09088.x. [DOI] [PubMed] [Google Scholar]
- 184.Skulachev V P. The sodium cycle: a novel type of bacterial energetics. J Bioenerg Biomembr. 1989;21:635–647. doi: 10.1007/BF00762683. [DOI] [PubMed] [Google Scholar]
- 185.Skulachev V P. Chemiosmotic systems in bioenergetics: H+-cycles and Na+-cycles. Biosci Rep. 1991;11:387–441. doi: 10.1007/BF01130214. [DOI] [PubMed] [Google Scholar]
- 186.Skulachev V P. The laws of cell energetics. Eur J Biochem. 1992;208:203–209. doi: 10.1111/j.1432-1033.1992.tb17175.x. [DOI] [PubMed] [Google Scholar]
- 187.Smith M A, Finel M, Korolik V, Mendz G L. Characteristics of the aerobic respiratory chains of the microaerophiles Campylobacter jejuni and Helicobacter pylori. Arch Microbiol. 2000;174:1–10. doi: 10.1007/s002030000174. [DOI] [PubMed] [Google Scholar]
- 188.Speelmans G, Poolman B, Abee T, Konings W N. Energy transduction in the thermophilic anaerobic bacterium Clostridium fervidus is exclusively coupled to sodium ions. Proc Natl Acad Sci USA. 1993;90:7975–7979. doi: 10.1073/pnas.90.17.7975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Speelmans G, Poolman B, Konings W N. Amino acid transport in the thermophilic anaerobe Clostridium fervidus is driven by an electrochemical sodium gradient. J Bacteriol. 1993;175:2060–2066. doi: 10.1128/jb.175.7.2060-2066.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Stephens R S, Kalman S, Lammel C, Fan J, Marathe R, Aravind L, Mitchell W, Olinger L, Tatusov R L, Zhao Q, Koonin E V, Davis R W. Genome sequence of an obligate intracellular pathogen of humans: Chlamydia trachomatis. Science. 1998;282:754–759. doi: 10.1126/science.282.5389.754. [DOI] [PubMed] [Google Scholar]
- 191.Stermitz F R, Lorenz P, Tawara J N, Zenewicz L A, Lewis K. Synergy in a medicinal plant: antimicrobial action of berberine potentiated by 5′-methoxyhydnocarpin, a multidrug pump inhibitor. Proc Natl Acad Sci USA. 2000;97:1433–1437. doi: 10.1073/pnas.030540597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Steuber J, Schmid C, Rufibach M, Dimroth P. Na+ translocation by complex I (NADH:quinone oxidoreductase) of Escherichia coli. Mol Microbiol. 2000;35:428–434. doi: 10.1046/j.1365-2958.2000.01712.x. [DOI] [PubMed] [Google Scholar]
- 193.Stock A M, Martinez-Hackert E, Rasmussen B F, West A H, Stock J B, Ringe D, Petsko G A. Structure of the Mg2+-bound form of CheY and mechanism of phosphoryl transfer in bacterial chemotaxis. Biochemistry. 1993;32:13375–13380. doi: 10.1021/bi00212a001. [DOI] [PubMed] [Google Scholar]
- 194.Stover C K, Pham X-Q T, Erwin A L, Mizoguchi S D, Warrener P, Hickey M J, Brinkman F S L, Hufnagle W O, Kowalik D J, Lagrou M, Garber R L, Goltry L, Tolentino E, Westbrook-Wadman S, Yuan Y, Brody L L, Coulter S N, Folger K R, Kas A, Larbig K, Lim R M, Smith K A, Spencer D H, Wong G K-S, Wu Z, Paulsen I T, Reizer J, Saier M H, Hancock R E W, Lory S, Olson M V. Complete genome sequence of Pseudomonas aeruginosa PA01, an opportunistic pathogen. Nature. 2000;406:959–964. doi: 10.1038/35023079. [DOI] [PubMed] [Google Scholar]
- 195.Stucky K, Hagting A, Klein J R, Matern H, Henrich B, Konings W N, Plapp R. Cloning and characterization of brnQ, a gene encoding a low-affinity, branched-chain amino acid carrier in Lactobacillus delbruckii subsp. lactis DSM7290. Mol Gen Genet. 1995;249:682–690. doi: 10.1007/BF00418038. [DOI] [PubMed] [Google Scholar]
- 196.Subramanian G, Koonin E V, Aravind L. Comparative genome analysis of the pathogenic spirochetes Borrelia burgdorferi and Treponema pallidum. Infect Immun. 2000;68:1633–1648. doi: 10.1128/iai.68.3.1633-1648.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Tatusov R L, Galperin M Y, Natale D A, Koonin E V. The COG database: a tool for genome-scale analysis of protein functions and evolution. Nucleic Acids Res. 2000;28:33–36. doi: 10.1093/nar/28.1.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Taylor B L, Zhulin I B. PAS domains: internal sensors of oxygen, redox potential, and light. Microbiol Mol Biol Rev. 1999;63:479–506. doi: 10.1128/mmbr.63.2.479-506.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Tettelin H, Saunders N J, Heidelberg J, Jeffries A C, Nelson K E, Eisen J A, Ketchum K A, Hood D W, Peden J F, Dodson R J, Nelson W C, Gwinn M L, DeBoy R, Peterson J D, Hickey E K, Haft D H, Salzberg S L, White O, Fleischmann R D, Dougherty B A, Mason T, Ciecko A, Parksey D S, Blair E, Cittone H, Clark E B, Cotton M D, Utterback T R, Khouri H, Qin H, Vamathevan J, Gill J, Scarlato V, Masignani V, Pizza M, Grandi G, Sun L, Smith H O, Fraser C M, Moxon E R, Rappuoli R, Venter J C. Complete genome sequence of Neisseria meningitidis serogroup B strain MC58. Science. 2000;287:1809–1815. doi: 10.1126/science.287.5459.1809. [DOI] [PubMed] [Google Scholar]
- 200.Thomas D R, Morgan D G, DeRosier D J. Rotational symmetry of the C ring and a mechanism for the flagellar rotary motor. Proc Natl Acad Sci USA. 1999;96:10134–10139. doi: 10.1073/pnas.96.18.10134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Thony-Meyer L, Beck C, Preisig O, Hennecke H. The ccoNOQP gene cluster codes for a cb-type cytochrome oxidase that functions in aerobic respiration of Rhodobacter capsulatus. Mol Microbiol. 1994;14:705–716. doi: 10.1111/j.1365-2958.1994.tb01308.x. [DOI] [PubMed] [Google Scholar]
- 202.Tokuda H, Unemoto T. A respiration-dependent primary sodium extrusion system functioning at alkaline pH in the marine bacterium Vibrio alginolyticus. Biochem Biophys Res Commun. 1981;102:265–271. doi: 10.1016/0006-291x(81)91516-3. [DOI] [PubMed] [Google Scholar]
- 203.Tokuda H, Unemoto T. Growth of a marine Vibrio alginolyticus and moderately halophilic V. costicola becomes uncoupler resistant when the respiration-dependent Na+ pump functions. J Bacteriol. 1983;156:636–643. doi: 10.1128/jb.156.2.636-643.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Tokuda H, Unemoto T. Na+ is translocated at NADH:quinone oxidoreductase segment in the respiratory chain of Vibrio alginolyticus. J Biol Chem. 1984;259:7785–7790. [PubMed] [Google Scholar]
- 205.Toledo-Cuevas M, Barquera B, Gennis R B, Wikstrom M, Garcia-Horsman J A. The cbb3-type cytochrome c oxidase from Rhodobacter sphaeroides, a proton-pumping heme-copper oxidase. Biochim Biophys Acta. 1998;1365:421–434. doi: 10.1016/s0005-2728(98)00095-4. [DOI] [PubMed] [Google Scholar]
- 206.Udagawa T, Unemoto T, Tokuda H. Generation of Na+ electrochemical potential by the Na+-motive NADH oxidase and Na+/H+ antiport system of a moderately halophilic Vibrio costicola. J Biol Chem. 1986;261:2616–2622. [PubMed] [Google Scholar]
- 207.Ueno S, Kaieda N, Koyama N. Characterization of a P-type Na+-ATPase of a facultatively anaerobic alkaliphile, Exiguobacterium aurantiacum. J Biol Chem. 2000;275:14537–14540. doi: 10.1074/jbc.275.19.14537. [DOI] [PubMed] [Google Scholar]
- 208.Ueno T, Oosawa K, Aizawa S. M ring, S ring and proximal rod of the flagellar basal body of Salmonella typhimurium are composed of subunits of a single protein, FliF. J Mol Biol. 1992;227:672–627. doi: 10.1016/0022-2836(92)90216-7. [DOI] [PubMed] [Google Scholar]
- 209.Unemoto T, Akagawa A, Mizugaki M, Hayashi M. Distribution of Na+-dependent respiration and a respiration-driven electrogenic Na+ pump in moderately halophilic bacteria. J Gen Microbiol. 1992;138:1999–2005. [Google Scholar]
- 210.Utsugi J, Inaba K, Kuroda T, Tsuda M, Tsuchiya T. Cloning and sequencing of a novel Na+/H+ antiporter gene from Pseudomonas aeruginosa. Biochim Biophys Acta. 1998;1398:330–334. doi: 10.1016/s0167-4781(98)00058-x. [DOI] [PubMed] [Google Scholar]
- 211.van der Rest M E, Siewe R M, Abee T, Schwarz E, Oesterhelt D, Konings W N. Nucleotide sequence and functional properties of a sodium-dependent citrate transport system from Klebsiella pneumoniae. J Biol Chem. 1992;267:8971–8976. [PubMed] [Google Scholar]
- 212.van Geest M, Lolkema J S. Membrane topology of the sodium ion-dependent citrate carrier of Klebsiella pneumoniae. Evidence for a new structural class of secondary transporters. J Biol Chem. 1996;271:25582–25589. doi: 10.1074/jbc.271.41.25582. [DOI] [PubMed] [Google Scholar]
- 213.Van Nevel C J, Demeyer D I. Effect of monensin on rumen metabolism in vitro. Appl Environ Microbiol. 1977;34:251–257. doi: 10.1128/aem.34.3.251-257.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Verkhovskaya M L, Verkhovsky M I, Wikstrom M. The respiration-driven active sodium transport system in E. coli does not function with lithium. FEBS Lett. 1996;388:217–218. doi: 10.1016/0014-5793(96)00531-5. [DOI] [PubMed] [Google Scholar]
- 215.Wei Y, Guffanti A A, Krulwich T A. Sequence analysis and functional studies of a chromosomal region of alkaliphilic Bacillus firmus OF4 encoding an ABC-type transporter with similarity of sequence and Na+ exclusion capacity to the Bacillus subtilis NatAB transporter. Extremophiles. 1999;3:113–120. doi: 10.1007/s007920050106. [DOI] [PubMed] [Google Scholar]
- 216.Weidner U, Geier S, Ptock A, Friedrich T, Leif H, Weiss H. The gene locus of the proton-translocating NADH:ubiquinone oxidoreductase in Escherichia coli. Organization of the 14 genes and relationship between the derived proteins and subunits of mitochondrial complex I. J Mol Biol. 1993;233:109–122. doi: 10.1006/jmbi.1993.1488. [DOI] [PubMed] [Google Scholar]
- 217.Weinstock G M, Hardham J M, McLeod M P, Sodergren E J, Norris S J. The genome of Treponema pallidum: new light on the agent of syphilis. FEMS Microbiol Rev. 1998;22:323–332. doi: 10.1111/j.1574-6976.1998.tb00373.x. [DOI] [PubMed] [Google Scholar]
- 218.Wheeler D L, Chappey C, Lash A E, Leipe D D, Madden T L, Schuler G D, Tatusova T A, Rapp B A. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2000;28:10–14. doi: 10.1093/nar/28.1.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Wilson D M, Wilson T H. Cation specificity for sugar substrates of the melibiose carrier in Escherichia coli. Biochim Biophys Acta. 1987;904:191–200. doi: 10.1016/0005-2736(87)90368-3. [DOI] [PubMed] [Google Scholar]
- 220.Wilson T H, Lin E C. Evolution of membrane bioenergetics. J Supramol Struct. 1980;13:421–446. doi: 10.1002/jss.400130403. [DOI] [PubMed] [Google Scholar]
- 221.Woehlke G, Wifling K, Dimroth P. Sequence of the sodium ion pump oxaloacetate decarboxylase from Salmonella typhimurium. J Biol Chem. 1992;267:22798–22803. [PubMed] [Google Scholar]
- 222.Wooldridge K G, Ketley J M. Campylobacter-host cell interactions. Trends Microbiol. 1997;5:96–102. doi: 10.1016/S0966-842X(97)01004-4. [DOI] [PubMed] [Google Scholar]
- 223.Yoshikawa K, Nakayama Y, Hayashi M, Unemoto T, Mochida K. Korormicin, an antibiotic specific for gram-negative marine bacteria, strongly inhibits the respiratory chain-linked Na+-translocating NADH:quinone reductase from the marine Vibrio alginolyticus. J Antibiot (Tokyo) 1999;52:182–185. doi: 10.7164/antibiotics.52.182. [DOI] [PubMed] [Google Scholar]
- 224.Yoshikawa K, Takadera T, Adachi K, Nishijima M, Sano H. Korormicin, a novel antibiotic specifically active against marine gram-negative bacteria, produced by a marine bacterium. J Antibiot (Tokyo) 1997;50:949–953. doi: 10.7164/antibiotics.50.949. [DOI] [PubMed] [Google Scholar]
- 225.Young G M, Badger J L, Miller V L. Motility is required to initiate host cell invasion by Yersinia enterocolitica. Infect Immun. 2000;68:4323–4326. doi: 10.1128/iai.68.7.4323-4326.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Young G M, Schmiel D H, Miller V L. A new pathway for the secretion of virulence factors by bacteria: the flagellar export apparatus functions as a protein-secretion system. Proc Natl Acad Sci USA. 1999;96:6456–6461. doi: 10.1073/pnas.96.11.6456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Zhang Y, Fillingame R H. Changing the ion binding specificity of the Escherichia coli H+-transporting ATP synthase by directed mutagenesis of subunit c. J Biol Chem. 1995;270:87–93. doi: 10.1074/jbc.270.1.87. [DOI] [PubMed] [Google Scholar]
- 228.Zhou W, Bertsova Y V, Feng B, Tsatsos P, Verkhovskaya M L, Gennis R B, Bogachev A V, Barquera B. Sequencing and preliminary characterization of the Na+-translocating NADH:ubiquinone oxidoreductase from Vibrio harveyi. Biochemistry. 1999;38:16246–16252. doi: 10.1021/bi991664s. [DOI] [PubMed] [Google Scholar]