Abstract
Tim molecules regulate T cell responses; however, the molecular basis of their ligand recognition remains largely unknown. In this issue of Immunity, Santiago et al. (2007) and Cao et al. (2007) report the crystal structures of several Tims and provide insights into the structure-function relationship of these molecules.
Main Text
The recently discovered Tim (T cell immunoglobulin mucin domain; also known as TIM) family has emerged as an important player in regulating T cell responses. The Tim family consists of eight members in mice (Havcr1 [Tim-1], Timd2 [Tim-2], Havcr2 [Tim-3], Timd4 [Tim-4], three genes that are predicted to encode Tims 5–7, and Dppa 1 [Tim-8]; the proteins are also referred to as mTIM-1, etc.) and three members in humans (HAVCR1 [TIM-1], HAVCR2 [TIM-3], and TIMD4 [TIM-4]). In mice, the genes encoding Tim proteins are encoded on mouse chromosome 11 in a genetic interval that has shown linkage to a number of autoimmune diseases, allergy, and atopy as well as asthma (Kuchroo et al., 2003). The syntenic region in humans, 5q33, has also been associated with asthma (McIntire et al., 2001). Furthermore, comparisons of the genes encoding Tim proteins in different strains of mice have revealed polymorphisms in those encoding Tim-1 and Tim-3, but not Tim-2 and Tim-4. Interestingly, Th1-prone strains (i.e., C57BL/6) and Th2-prone strains (i.e., Balb/c) express different Havcr1 (Tim-1) and Havcr2 (Tim-3) polymorphisms (Meyers et al., 2005). Indeed, mounting data support that the Tims are important regulators of effector T cells. Both mouse and human studies suggest a role for Tim-3 in regulating Th1 immunity and tolerance (Anderson and Anderson, 2006). Tim-1 has been shown to act as a costimulatory molecule and may have a specialized role in regulating Th2 responses (Meyers et al., 2005). Similarly, Tim-2 appears to be an important negative regulator of Th2 responses (Chakravarti et al., 2005). Tim-4 is exclusively expressed on antigen-presenting cells in the mouse and is a natural ligand for Tim-1 (Meyers et al., 2005). Thus, current data point to the Tim genes as important regulators of both Th1 and Th2 immunity and possibly as important determinants for susceptibility to both autoimmune and allergic diseases.
All of the Tim molecules share a common structural organization consisting of an N-terminal IgV domain followed by a mucin domain, a transmembrane domain, and a cytoplasmic tail. Of note are the four noncanonical cysteines in the IgV domain that are conserved in all the Tims in both mouse and man. In this issue of Immunity, Santiago et al. (2007) and Cao et al. (2007) report the crystal structures of mouse Tim-1, Tim-2, and Tim-3 and show that these four cysteines result in the formation of two disulfide bonds that reposition the classical loops formed between the F and G and C and C′ strands of the β sheet (FG and CC′ loops) to form a unique binding cleft (FG-CC′ cleft) not seen in the Ig domain of any other Ig superfamily members (Cao et al., 2007, Santiago et al., 2007).
Thus far, galectin-9 has been identified as a ligand for Tim-3 (Zhu et al., 2005), both Semaphorin 4a (Sema4a) and H-ferritin for Tim-2, and both hepatitis A virus (HAV) and Tim-4 for Tim-1 (Chen et al., 2005, Kuchroo et al., 2003, Meyers et al., 2005). Interestingly, Santiago et al. (2007) and Cao et al. (2007) demonstrate that the unique FG-CC′ cleft is responsible neither for galectin-9 binding to Tim-3 nor HAV or Tim-4 binding to Tim-1 ( Figure 1; Cao et al., 2007, Santiago et al., 2007). Thus, the presence of the FG-CC′ cleft opens the door for the possibility of as-yet-undiscovered Tim ligands. Indeed, Cao et al. (2007) show that unglycosylated Tim-3 IgV, which cannot bind to galectin-9, binds to several primary cell types and transformed cell lines from different species. Mutations in residues affecting the FG-CC′ cleft abolish this binding. Collectively, these findings support the existence of one or more evolutionarily conserved Tim-3 ligands that occupy the unique FG-CC′ cleft (Cao et al., 2007).
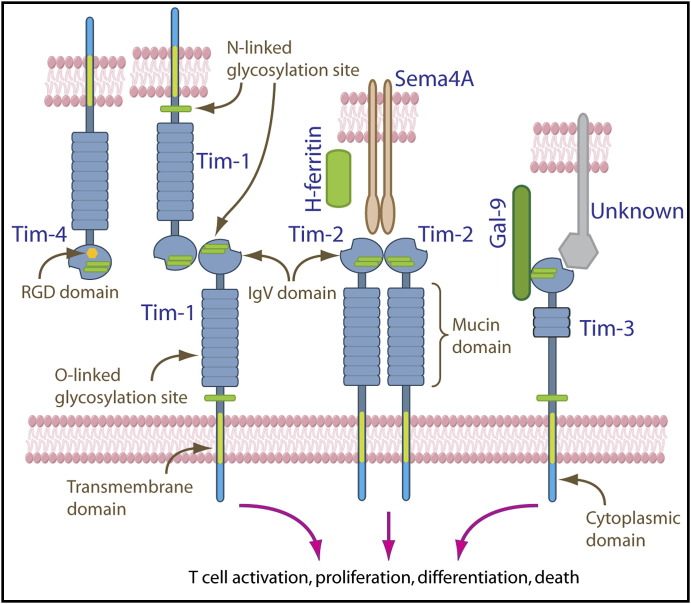
Figure 1.
Schematic Representation of the Mouse Tim Family Proteins and Their Ligands
Tim-4 can bind to Tim-1 to costimulate T cell activation and expansion. Tim-1 can also form homophilic interaction. Tim-2 proteins form a dimer that prevents homophilic binding. Semaphorin 4A (Sema4A) and H-ferritin have been identified as Tim-2 ligands. Galectin-9 binds to Tim-3 via carbohydrates present on the IgV domain. An unknown ligand(s) that is widely expressed in many cell types can interact with Tim-3 independent of Tim-3 glycosylation by involving the FG-CC′ cleft on the opposite side of the Tim-3 IgV domain. The cleft in Tim-3 is a feature shared by all Tim proteins. Glycosylation affects not only Tim protein structures but also their interaction with their ligands. The existence of multiple ligands for Tim proteins and the possibility of their own association suggest that each Tim protein might deliver multiple differential signals in regulating T cell responses such as activation, proliferation, differentiation, and death, depending on the receptor-ligand interaction.
That the FG-CC′ cleft is a common feature of all the Tims does not necessitate that they all recognize a common ligand. In fact, Santiago et al. (2007) show that the CC′ loop in Tim-2 has a markedly different conformation than that in Tim-1, which is more representative of the conformation seen in the rest of the Tims. This different conformation of the CC′ loop in Tim-2 likely mediates binding to different ligands via the FG-CC′ cleft and may in part explain the different functions of these two molecules in that Tim-1 appears to be costimulatory whereas Tim-2 is inhibitory. Furthermore, the structures of Tim-1 and Tim-2 reported by Santiago et al. (2007) demonstrate that Tim-2 forms a dimer and that homophilic Tim-1:Tim-1 interactions can take place on neighboring cells (Figure 1). The fact that this homophilic Tim-1 interaction is conserved in human TIM-1 suggests that it is biologically important. Lastly, Santiago et al. (2007) show that the carboyhydrate residues in the Tim-1 mucin domain also influence Tim-1 binding. Thus, although the Tims have common overall binding features such as the FG-CC′ cleft, unique sequence differences in the IgV and mucin domains will result in differential ligand binding (Santiago et al., 2007).
All of these structural data have numerous implications for the biochemical signaling pathways triggered by the Tims. Indeed, Santiago et al. (2007) observe that homophilic Tim-1 binding is responsible for Tim-1 clustering at the intercellular junction of transfected cells. This clustering could presumably facilitate aggregation and phosphorylation of signaling mediators. Indeed, this could underlie the observed phosphorylation of Tim-1 and activation of NFAT and AP-1 in T cells and T cell lines overexpressing Tim-1 (de Souza et al., 2005, Santiago et al., 2007). Similarly, the dimerization of Tim-2 may facilitate aggregation of signaling mediators and may provide an explanation for the ligand-independent repression of NFAT and AP-1 that has been reported in Tim-2 transfectants (Knickelbein et al., 2006). The existence of multiple binding modes could translate into triggering of multiple biochemical signaling modes and different functional outcomes depending on which ligand(s) are binding to the Tims. Lastly, whether signaling pathways will synergize or antagonize if two ligands bind to two different faces of the Tim molecules remains to be seen.
Thus, the accumulating biological data that underscore the importance of the Tim molecules in immunity together with the structural data discussed above highlight the need for understanding the complex interaction of Tim molecules with their ligands. How can this all take place? One can envision that the regulation of complex ligand:receptor interactions could take place at different levels such as the anatomic distribution of different ligands, differential affinity for different ligands, and the modulation of the expression of the Tims and different Tim ligands by environmental triggers such as inflammation. The identification of novel Tim ligands, their expression pattern and measurements of the affinity of different Tim:ligand pairs, and the elucidation of the pattern and regulation of Tim and Tim ligand expression will likely fuel investigation in this field for many years to come.
References
- Anderson A.C., Anderson D.E. Curr. Opin. Immunol. 2006;18:665–669. doi: 10.1016/j.coi.2006.09.009. [DOI] [PubMed] [Google Scholar]
- Cao E., Zang X., Ramagopal U.A., Mukhopadhaya A., Fedorov A., Fedorov E., Zencheck W.D., Lary J.W., Cole J.L., Deng H., et al. Immunity. 2007;26:311–321. doi: 10.1016/j.immuni.2007.01.016. this issue. [DOI] [PubMed] [Google Scholar]
- Chakravarti S., Sabatos C.A., Xiao S., Illes Z., Cha E.K., Sobel R.A., Zheng X.X., Strom T.B., Kuchroo V.K. J. Exp. Med. 2005;202:437–444. doi: 10.1084/jem.20050308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen T.T., Li L., Chung D.H., Allen C.D., Torti S.V., Torti F.M., Cyster J.G., Chen C.Y., Brodsky F.M., Niemi E.C., et al. J. Exp. Med. 2005;202:955–965. doi: 10.1084/jem.20042433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Souza A.J., Oriss T.B., O'Malley K.J., Ray A., Kane L.P. Proc. Natl. Acad. Sci. USA. 2005;102:17113–17118. doi: 10.1073/pnas.0508643102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knickelbein J.E., de Souza A.J., Tosti R., Narayan P., Kane L.P. J. Immunol. 2006;177:4966–4970. doi: 10.4049/jimmunol.177.8.4966. [DOI] [PubMed] [Google Scholar]
- Kuchroo V.K., Umetsu D.T., DeKruyff R.H., Freeman G.J. Nat. Rev. Immunol. 2003;3:454–462. doi: 10.1038/nri1111. [DOI] [PubMed] [Google Scholar]
- McIntire J.J., Umetsu S.E., Akbari O., Potter M., Kuchroo V.K., Barsh G.S., Freeman G.J., Umetsu D.T., DeKruyff R.H. Nat. Immunol. 2001;2:1109–1116. doi: 10.1038/ni739. [DOI] [PubMed] [Google Scholar]
- Meyers J.H., Sabatos C.A., Chakravarti S., Kuchroo V.K. Trends Mol. Med. 2005;11:362–369. doi: 10.1016/j.molmed.2005.06.008. [DOI] [PubMed] [Google Scholar]
- Santiago C., Ballesteros A., Tami C., Martínez-Muñoz L., Kaplan G.G., Casasnovas J.M. Immunity. 2007;26:299–310. doi: 10.1016/j.immuni.2007.01.014. this issue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu C., Anderson A.C., Schubart A., Xiong H., Imitola J., Khoury S.J., Zheng X.X., Strom T.B., Kuchroo V.K. Nat. Immunol. 2005;6:1245–1252. doi: 10.1038/ni1271. [DOI] [PubMed] [Google Scholar]