Abstract
Exome and genome sequencing have become the tools of choice for rare disease diagnosis, leading to large amounts of data available for analyses. To identify causal variants in these datasets, powerful filtering and decision support tools that can be efficiently used by clinicians and researchers are required. To address this need, we developed seqr - an open source, web-based tool for family-based monogenic disease analysis that allows researchers to work collaboratively to search and annotate genomic callsets. To date, seqr is being used in several research pipelines and one clinical diagnostic lab. In our own experience through the Broad Institute Center for Mendelian Genomics, seqr has enabled analyses of over 10,000 families, supporting the diagnosis of more than 3,800 individuals with rare disease and discovery of over 300 novel disease genes. Here, we describe a framework for genomic analysis in rare disease that leverages seqr’s capabilities for variant filtration, annotation, and causal variant identification, as well as support for research collaboration and data sharing. The seqr platform is available as open-source software, allowing low-cost participation in rare disease research, and a community effort to support diagnosis and gene discovery in rare disease.
Keywords: data sharing, genomic analysis, novel gene discovery, rare disease diagnosis, research collaboration, variant filtration
1. Introduction
Approximately 1 in 20 people worldwide are affected by a rare genetic condition, but approximately 65% of cases go undiagnosed due to limitations in diagnostic technology used, a lack of understanding of human genomic variation, and insufficient delineation of the mechanisms underlying disease (Chong et al., 2015; Boycott et al., 2017). Many of these unsolved cases move into the research realm, where exome and genome sequencing have been shown to increase the diagnostic yield by identifying complex variants and novel causes of disease (Clark et al., 2018; Palmer et al., 2021). With the large-scale uptake of exome and genome sequencing by research programs, including the USA’s Centers for Mendelian Genomics (CMG) (Baxter et al., 2022) and Undiagnosed Disease Network (UDN) (Schoch et al., 2021), Canada’s Care4Rare program (Osmond et al., 2022), and the UK’s Deciphering of Developmental Disorders (DDD) project (Wright et al., 2015), and Genomics England 100,000 Genomes study (100,000 Genomes Project Pilot Investigators et al., 2021) among many others, vast and rapidly growing amounts of data are now available for analysis. To identify disease-causing variants in these large datasets, powerful filtering and decision support tools that can be easily accessed and used by researchers are needed.
Several existing tools have been developed in the academic sector for reviewing variants in defined gene lists (gene.iobio) (Di Sera et al., 2021) or for broader review and prioritization of variants in genomic analysis (Exomiser, GEMINI, PhenoDB, slivar) (Buske et al., 2013; Paila et al., 2013; Robinson et al., 2014; Sobreira et al., 2015). This is also an area of development in the commercial arena, with many fee-based platforms available, such as those listed on ClinGen’s Genomic Analysis Software Platforms list (https://clinicalgenome.org/tools/genomic-analysis-software-platform-list/). Each platform has its strengths, but none met all the needs of our team. Limitations included an inability to add specific features needed by our team for project management or analysis and inaccessibility to non-computationally experienced personnel. Additionally, proprietary software lacks features for broad data sharing and collaboration across the global rare disease research community, which is essential to uncover novel causes of rare genetic disease (Philippakis et al., 2015). Fee-based software can be cost-prohibitive for individual research groups, further limiting the utility of these platforms for rare disease research.
To meet the needs of the rare disease research community, our team developed seqr - an open-source, web-based platform for family-based analysis, data sharing, and project management. The National Institutes of Health (NIH) funded Broad Institute of MIT and Harvard Center for Mendelian Genomics (Broad CMG) has supported sequencing, analysis, and data sharing for an international collaborative network of rare disease researchers and clinicians since 2016 generating 23,166 exomes and 4,179 genomes from 16,803 families between 2016 and 2021. As the core analysis platform for the Broad CMG, seqr (Figure 1) has enabled analyses of over 10,000 unsolved families, facilitated identifying a diagnosis for more than 3,800 of these families, and supported the discovery of over 300 novel disease genes. Current applications of seqr are primarily research focused, but seqr has also been incorporated in a clinical diagnostic pipeline at the Victorian Clinical Genetic Services laboratory in Melbourne, Australia. In this review, we describe seqr’s capabilities for rare disease genomics, and the technology stack underlying seqr that has facilitated scaling the system to analyze tens of thousands of exome and genome samples. A demonstration of the features described in this paper can be viewed on the Broad CMG website tutorials (https://cmg.broadinstitute.org/using-seqr).
Figure 1:
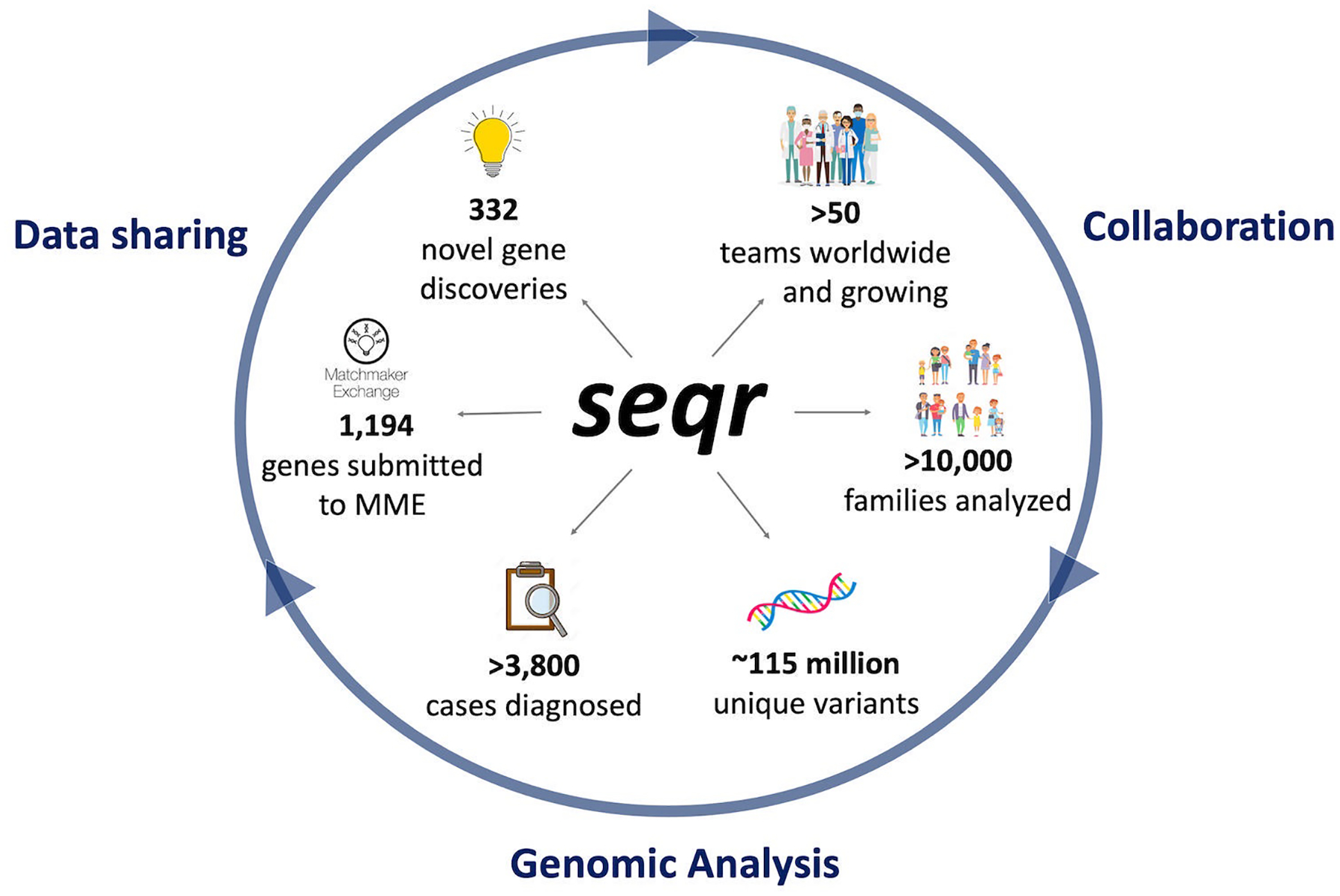
The seqr platform for family-based genomic analysis, research collaboration, and data sharing. MME, Matchmaker Exchange
2. Metadata storage, data upload, and project management functionality
An indispensable component of rare disease research programs is the need for affordable storage of large volumes of genomic datasets alongside easily accessible clinical information. A single platform with both functionalities streamlines the process of project management and analysis. The centralization of knowledge and data sharing across the teams that use the platform can also accelerate the pace of diagnosis and novel gene discovery.
The seqr platform possesses capabilities for data storage, case tracking, and data sharing. It allows for bulk upload of individual phenotype metadata in tsv, csv, or json formats for handling larger cohorts. It stores thousands of sample IDs with family pedigrees, phenotype information using the human phenotype ontology (HPO) format, additional sample metadata, and quality metrics. Data loading in seqr is optimized for a joint-called variant call format (VCF) file from exome or genome data using the genomic analysis toolkit (GATK) pipeline, although joint called VCFs generated with other callers have also been used successfully. Copy number variant (CNV) calls from exome or genome data generated by tools such as GATK germline CNV (GATK-gCNV) or GATK-SV respectively can also be loaded (Collins et al., 2020; Fu et al., 2021). The user-friendly interface enables seqr to be used by a diverse team of variant analysts, researchers, clinicians, and project managers to manage sample data, review and prioritize variants, submit candidates to Matchmaker Exchange (MME), and generate custom reports.
In seqr, an overview of a cohort can be accessed via the Project Page (Figure 2). Summary data about the project includes information about the families and data loaded in the project, stats on analysis status and MME submissions, along with gene lists used in the project and sub-cohort analysis of groups of similar cases. This is followed by a collapsed list of families in the project showing the analysis status, who has analyzed the case, if and when data were loaded, a high-level case description, and any saved variants. Additional detailed information about a case, including phenotype stored as HPO terms and sections for notes about analysis progress, can be entered on the Family Page (Figure S1).
Figure 2:
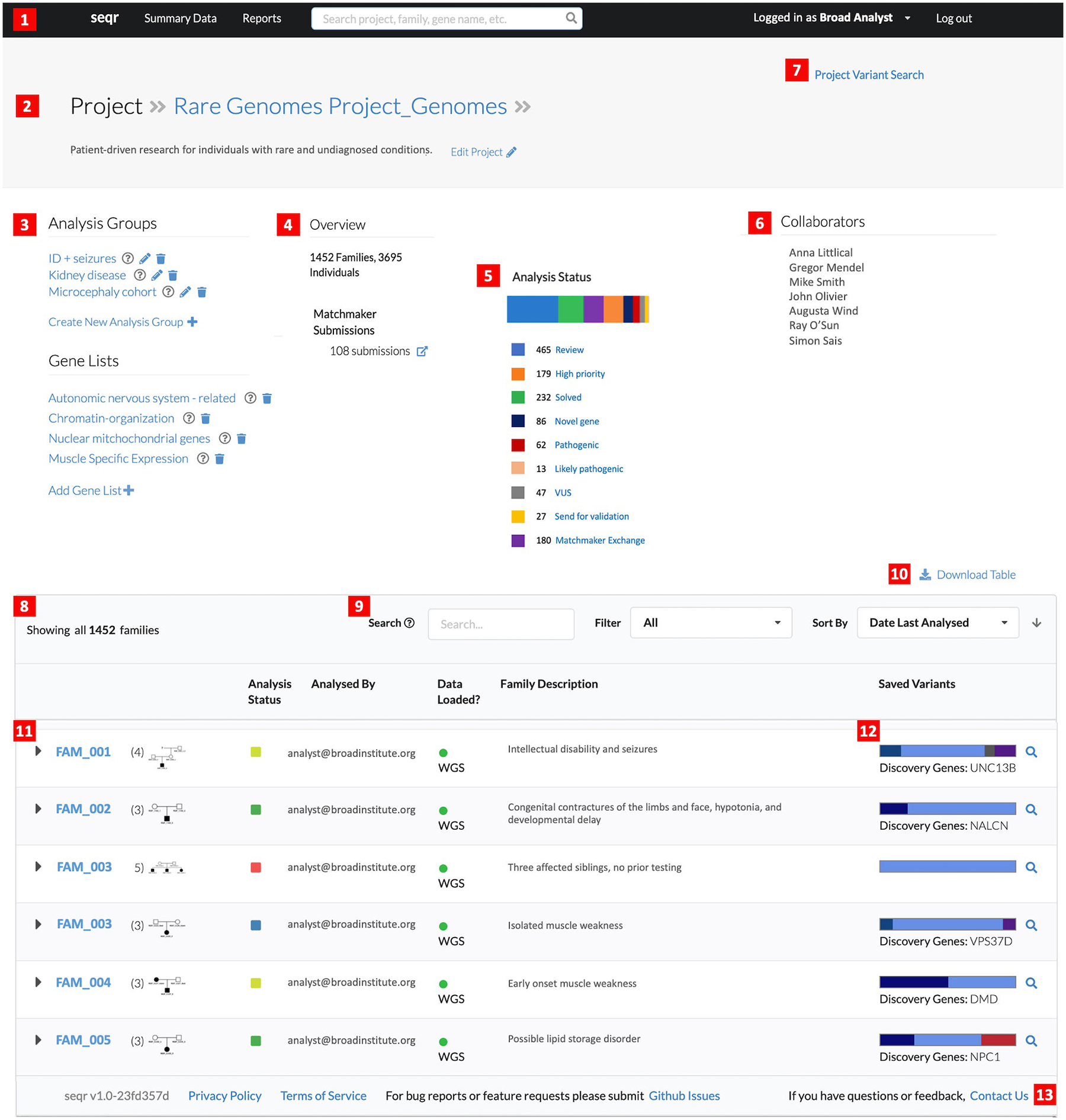
The Project Page in seqr, showing the Rare Genomes Project as an example. The top portion of the page contains (1) quick links to the main page and summary data; (2) project description details; (3) customizable analysis groups and gene lists; (4) project overview with high-level details of the number of families and Matchmaker submissions (5) variants tagged in the project; (6) users with access to the project; (7) project-wide variant search function. The lower portion of the page lists the following functions or summary data: (8) number of families in the project; (9) functions to search, filter, and sort through families; (10) ability to download the list of families with analysis details; (11) overview of each family with the pedigree, details of the analysis status, the user who has analyzed the case, date the data was loaded, description of the family, and overview of the saved variants. Selecting the family ID opens the Family Page; (12) colored box highlighting tagged variants and quick link to the variant search page; (13) link to the GitHub repository for bug reports or feature requests, and the option to contact the seqr team. VUS, variant of uncertain significance; WGS, whole genome sequencing
3. Search/Filtration functionality
Variant searches can yield tens to hundreds of candidates for manual review, depending on the search parameters, type of sequence data, and the availability of familial data. Current clinical filtration workflows are largely based on published gene-disease associations, potentially excluding novel causes of genetic disease. Additionally, separate analysis of different data types (e.g., single nucleotide variants (SNVs) and insertion deletion (indel) calls or SV calls) can limit the diagnostic rate due difficulties in recognizing compound heterozygous calls across variant classes.
The seqr variant search (Figure 3) can efficiently filter SNVs, indels, and SVs (from exome or genome data) in tandem. The search functionality is based on six core parameters including: suspected inheritance, reported pathogenicity in ClinVar (Landrum et al., 2016), type of variant, frequency in population databases, in silico score (optional), chromosomal region or gene list location (optional), and variant call quality. Search results can be further sorted by position, protein consequence, allele frequency, gene constraint, pathogenicity per ClinVar, gene-disease association per Online Mendelian Inheritance in Man (OMIM) (Amberger et al., 2015), and in silico scores. To enable researchers of various skill sets to participate in the filtration process, seqr has predefined de novo/dominant and recessive searches (the latter includes homozygous, compound heterozygous, and X-linked recessive in one search) (Supporting Information Methods). If a suitable candidate has not been identified with these predefined searches, each search parameter can be customized, such as adjusting the inheritance filter to accommodate for incomplete penetrance in a family member, increasing the allele frequency to account for hypomorphic variants, or relaxing cutoffs for variant quality metrics to be more permissive. Customized searches can be saved for future use by a user.
Figure 3:

The Variant Search Page in seqr displaying settings for a de novo/dominant restrictive search from the (1) predefined searches with (2–7) options to customize each search parameter.
The platform’s advanced search functionalities allow the creation of sub-cohort analysis groups such as for joint analysis of families with similar clinical presentations. Users can also focus variant searches based on user-created gene lists. Gene lists are encouraged to be entered as public lists to promote use by other teams using seqr for analysis. Sharing candidate disease genes can help other researchers to identify additional cases for inclusion in a case series publication. Publicly available gene lists maintained by the seqr team include curated assessments of the clinical validity of gene-disease relationships from PanelApp (Martin et al., 2019) and novel gene candidates from across the CMG program.
4. Analysis functionality
Assessing a subset of filtered variants for potential clinical significance requires comprehensive variant and gene level information. For each variant returned in a search, seqr displays in a single view the genotype, gene and Ensembl ID, chromosomal location, Matched Annotation from NCBI and EMBL-EBI (MANE) transcript, disease association as per OMIM, missense and loss of function constraint metrics, variant quality scores, variant frequency in population databases and within the joint callset (shown here for the Broad Institute CMG callset, with case-level data only available to those with appropriate permissions), in silico scores, and ClinVar classification if available. While many systems include similar annotations, they are often displayed in a long series of columns that require more space and back-and-forth scrolling for review. By grouping and coloring data, using more vertical space per variant, and providing additional information by hover over or with links, seqr shows a large amount of data for each variant that is quickly interpretable by an analyst. Variant quality assessment is made easy through the Integrative Genomics Viewer (IGV) (Robinson et al., 2011) (Figure 4) for every variant returned in search, though use of this feature does require seqr to have the path and read access to the binary alignment map (BAM) or compressed columnar format (CRAM) files. Filtration of SNVs, indels, and SVs can be performed simultaneously, with results displayed together, to enable identification of compound heterozygous variation across SVs and short variants (Figure S2).
Figure 4:
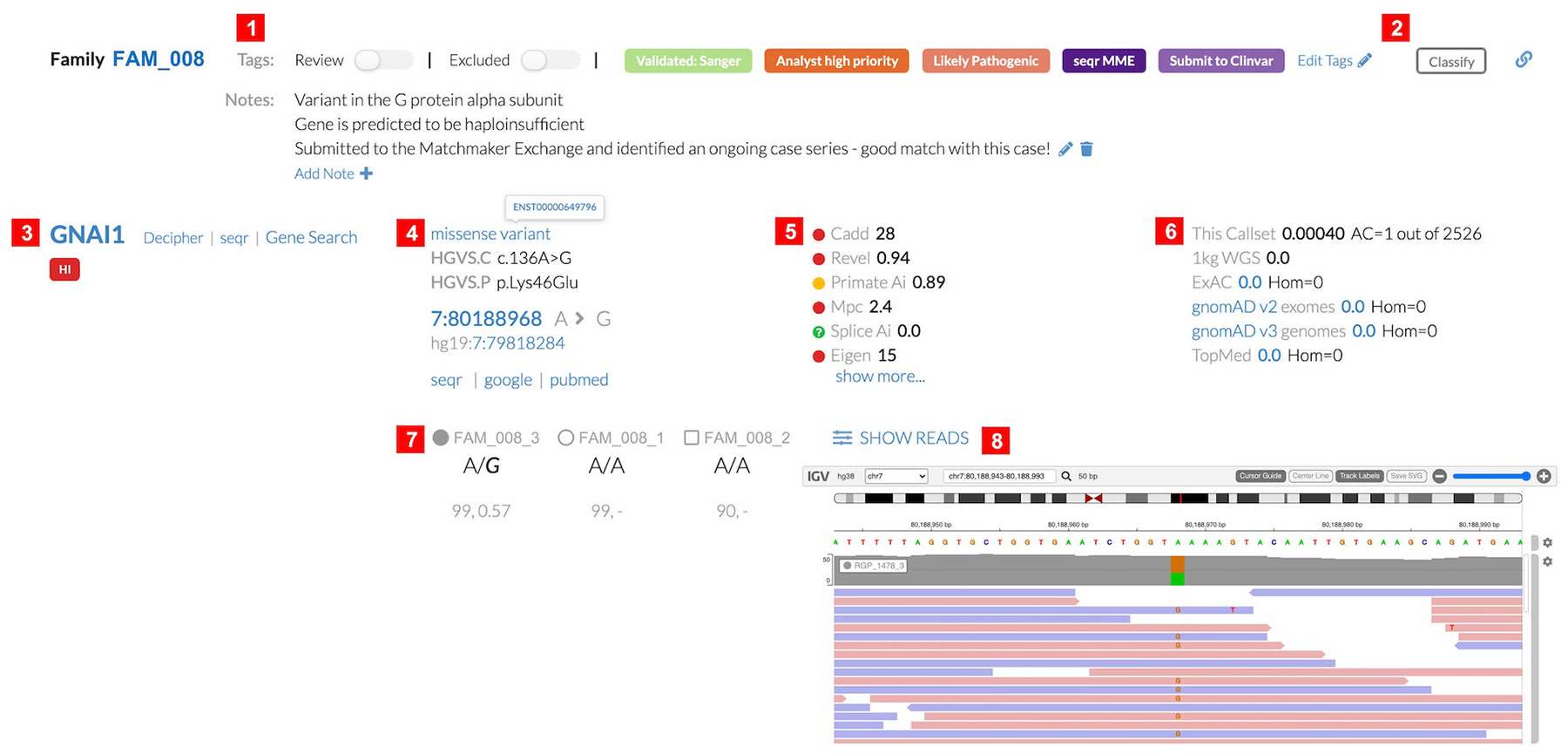
The seqr variant view showing a de novo missense variant in the gene GNAI1 as an example. (1) Tags have been used to mark the variant and additional notes have been added by an analyst; (2) link to the classification calculator to determine pathogenicity of a variant using ACMG/AMP criteria codes; variant information with (3) gene ID, link to the gene page in DECIPHER, seqr search function to display all previously saved variants in the gene across all of the user’s seqr projects, Gene Search for all variants in the gene present in any affected individual within this family; (4) variant HGVS nomenclature, with transcript ID, and annotations in other transcripts of the gene by clicking on “missense variant”, genomic location link to the UCSC browser, links to seqr search for this variant across all of the user’s seqr projects, Google and PubMed searches for the variant; (5) in silico scores; (6) variant allele frequency within the joint called VCF (“this callset”) and reference population databases with links to the gene page (e.g. gnomAD v2) or variant page (e.g. 0.0 which will link to a region page for variants absent from gnomAD); (7) genotype for each individual with the genotype quality score and allele balance; (8) IGV read data in seqr. ACMG, American College of Medical Genetics and Genomics; AMP, Association for Molecular Pathology; HGVS, Human Genome Variation Society; IGV, Integrative Genomics Viewer; VCF, variant call format; WGS, whole-genome sequencing
For genes with little or no evidence for disease, delineating the potential clinical significance of a variant is a laborious process of seeking gene-phenotype relationships by consulting numerous disease-associated databases. To aggregate available data for analysis, seqr displays links to external resources, including gene-related information (PubMed, Monarch, DECIPHER) (McMurry et al., 2016; Shefchek et al., 2020; Firth et al., 2009), transcript information (Genome Aggregation Database, Genotype-tissue Expression [GTEx]) (Cummings et al., 2020; GTEx Consortium, 2013), and functional animal model databases (Mouse Genome Informatics and International Mouse Phenotyping Consortium) (Bult et al., 2019; Dickinson et al., 2016) (Figure S3).
Tracking variants as they proceed through the analysis pipeline can be challenging in large, rare disease cohorts. In seqr, variant tags can be used to highlight variants of interest, marking them for further review, validation, or as the diagnosis. Users can filter for variants with a specific tag in a particular case or across a project such as those requiring orthogonal confirmation, classification of pathogenicity, or results ready for return to a participant. In addition to tags, the variant notes section can be used to record an analyst’s impressions and additional data gathered so that work is not duplicated across multiple searches on the same case. The public notes section is gene-specific and can be used to share information more broadly with all seqr users including relevant references, available functional studies, or case series in progress.
5. Data sharing through the Matchmaker Exchange
For many undiagnosed individuals with variants in genes with limited information, the presence of other cases with variants in the same gene can provide useful evidence for or against the role of the gene in disease. To enable data sharing among research labs and clinical centers across the globe, the Matchmaker Exchange (MME) (Philippakis et al., 2015) was developed to connect researchers and clinicians with cases that have variants in the same gene of interest. Through seqr, users can seamlessly submit a gene, variant, and HPO terms to MME. The seqr platform also makes contacting potential “matches” easy by autogenerating a customizable email (Figure S4) with relevant variant and phenotype information listed in seqr. Gene submissions and contacts are recorded for easy tracking, and users can list additional notes regarding the gene submission such as matches or progress made. To date, seqr has aided in submitting and tracking >6,500 match communications.
6. Reporting
One of the biggest hurdles to rapid and comprehensive data sharing is the lack of structured data storage. Details about gene/variant evidence, data sharing activities, or patient phenotype are commonly stored in free text fields, which can be impossible to parse, and thus are difficult to include in data exports or other analysis reports without substantial manual effort. We address this issue in seqr by storing all accompanying metadata in discrete fields that can be used for creating data sharing files. By allowing analysts and project managers to input this data into structured fields, rather than free text or attachments (e.g., PDFs), we can quickly meet the data model requirements for most genomic data sharing platforms.
In the case of the Broad CMG’s NIH progress reporting, we were able to substantially reduce the amount of time our team spent creating these semi-annual reports by transitioning from an entirely manual effort, which took weeks for our team to complete, to an automated report which takes minutes and can be generated at any time. Similarly, our CMG data is stored in the National Human Genome Research Institute’s Analysis Visualization and Informatics Lab-Space (NHGRI’s AnVIL; https://anvilproject.org) and our AnVIL metadata reports, which include over 60 values varying from family and individual level data to sample and discovery details, can be generated by the project management staff on demand with minimal to no manual manipulation. The design of additional report formats takes development and software engineering time but allows for reproducible and expedient report generation. This infrastructure has allowed us to scale our data sharing activities without adding extra burden on our analysts and project managers.
7. Under the hood
The seqr platform is constructed as an open-source project, designed to balance the needs of a small development team with those of a broad and ever growing user base. The underlying technologies were chosen for their maintainability and flexibility to allow their continued usage and evolution and also for their ease at being bundled and connected to streamline open-source installations. Additionally, databases were selected and designed to both support seqr’s robust searching needs and ensure data integrity and stability.
The seqr web platform is built using a Python web framework to handle server-side functionality such as URL routing and database querying (see Supporting Information Methods for details about the framework and databases). While seqr is designed to be platform-agnostic, it is currently hosted on Google Cloud Platform (GCP) and Kubernetes is used to manage deployment and resource management within GCP. Data storage in seqr is separated into two different databases: a read-only database which includes all annotated variant information, and a read/write database which includes all project and user metadata, including user-generated data like variant tags and notes. Additionally, seqr stores external reference data in a separate database. Such reference data includes gene level information from sources such as GENCODE (Frankish et al., 2019), OMIM, and dbNSFP (Liu et al., 2020), as well as structured phenotype data from HPO (Köhler et al., 2021). This reference data is updated periodically to ensure accuracy but is not editable by seqr application users. Databases are backed up at regular intervals, conforming to best practices and Federal Information Security Management Act (FISMA) requirements. Due to its intended use as a research system, seqr does not currently have data fields for entry of personally identifiable information or protected health information.
To be more broadly available to researchers, seqr has been made available as a connected application in the NHGRI’s AnVIL. AnVIL is an application powered by Terra - a scalable and secure cloud-based platform for biological research. As a connected application, seqr legally resides within Terra’s security boundary. This means that users can be assured that the security controls around their data in Terra extend to seqr as well. Terra and AnVIL users can bring their own joint called VCFs, or create one using supported Terra workflows, and then load that data to their own private seqr projects for analysis. There are no restrictions for who can create an AnVIL account and then store data in their own private GCP bucket, which empowers researchers from a variety of backgrounds and technical abilities to access the seqr platform. A guide to launching seqr through Terra can be accessed on Terra’s support page (https://support.terra.bio/hc/en-us/articles/4402431367949-Launching-seqr-through-Terra). The use of seqr through Terra is currently free to all users, though in the future the costs of data storage and loading may be passed on to users. Additionally, to request data be loaded in seqr, data must be temporarily stored in a Google Cloud Platform bucket on Terra, and operations such as running workflows, accessing, and storing data, can incur charges. Detailed information on understanding and controlling costs are described on Terra’s support page (https://support.terra.bio/hc/en-us/articles/360048632271).
In addition to being available to all AnVIL users, the seqr platform is available as an open-source project. Institutions or laboratories that prefer to operate their own seqr installation and have the technical resources to support it are invited to do so. GitHub is used to host the seqr code base as a public repository (https://github.com/broadinstitute/seqr) and is the same repository used by the Broad’s seqr deployment. Copyleft protection for seqr is licensed under the GNU Affero General Public License v3.0. Instructions for deployment are available in GitHub (see Supporting Information Methods).
GitHub is also used for managing seqr technical requests. Bug reports and feature requests can be submitted via the repository’s “Issues” tab. For questions and general seqr discussion, users are invited to join the seqr user forum (https://github.com/broadinstitute/seqr/discussions). In addition, seqr accepts code contribution for features and bug fixes from external groups via pull requests to the repository. We recommend users to first submit an issue to discuss such contributions with the seqr development team before starting on implementation.
8. Collaboration
Collaboration is a key feature of seqr. For example, our Broad CMG analysts work closely with over 50 collaborating research teams worldwide to diagnose cases by enabling groups to access a common workspace, tag candidate variants for group discussion, and jointly contribute to analyses. In addition, Broad CMG analysts search for candidate genes across multiple projects for potential matches and connect the respective research groups. This collaborative approach has empowered numerous diagnoses and novel gene discoveries across the more than 10,000 families we have studied to date (Coppens et al., 2021; Donkervoort et al., 2019; Mohassel et al., 2021). Due to joint calling, the allele count across the joint callset is displayed for all variants in the search results and aggregation of cases with common etiologies is easily enabled. Data loaded through AnVIL requests is not visible to our analysis team or other seqr users. Using the callset allele count, analysts with project-specific access to case-level data at Broad Institute identified three other cases with the same homozygous splice variant in TRAPPC4, which led to its association as a relatively common cause of early-infantile neurodegenerative syndrome (Figure S5) (Ghosh et al., 2021).
Both the Broad and University of Washington centers participating in the recently launched NHGRI Genomic Research to Elucidate the Genetics of Rare Diseases (GREGoR) consortium are using seqr as their primary analysis and project management platform. Some of the sites using seqr outside of GREGoR include the Harvard International Center for Genetic disease (iCGD) (Boston, MA, USA), Garvan Institute of Medical Research (Sydney, Australia), Murdoch Children’s Research Institute (MCRI; Melbourne, Australia), National Institute for Allergy and Infectious Disease (Bethesda, MD, USA), Yale University (New Haven, CT, USA), University of Tartu (Tartu, Estonia), and University of Southampton (Southampton, UK). While most local seqr installations are deployed on Google Cloud, Boston Children’s Hospital (Boston, MA, USA) has seqr running on Amazon Web Services cloud computing.
New feature ideas and feedback on existing features often come from groups that use seqr through CMG collaborations. They submit feedback through seqr’s GitHub page which has led to many improvements in seqr’s analysis capabilities. In other cases, outside developers have added features needed by a local group using seqr and we have integrated these into the primary version of seqr so these features can be accessed by all users. An example of this is the recent integration of code for Genomics England’s and Australia’s PanelApp tool, making >300 curated disease panels now selectable in seqr. Another example is the addition of filtration by in silico scores and the integration of a module to allow application of the American College of Medical Genetics and Genomics/Association for Molecular Pathology (ACMG/AMP) criteria codes (Richards et al., 2015) for classifying variants according to pathogenicity. From regular discussions between analysts and developers, the variant search interface undergoes continuous iterative enhancements to improve the variant search experience.
9. Discussion
The seqr platform was designed to be a resource for the global rare disease research community, enabling efficient, high-quality analysis and collaboration. The main feature driving its utility is the user-friendly interface which allows quick onboarding to the platform by researchers and clinicians who are familiar with disease architecture and patient phenotypes but lack a computational background. The recommendation to use prepopulated standardized searches allows for consistency and reproducibility in the variant filtering and prioritization process across teams, while also allowing adjustment as needed. Recent development efforts have focused on incorporating additional data types (e.g., SVs from the GATK-SV pipeline) and providing support and guidance materials for data loading from different file formats from external users. With the addition of CNV calls from exome data with the gCNV caller, we have been able to make dozens of diagnoses (Cloney et al., 2021) of de novo CNVs, homozygous CNVs with recessive inheritance, and compound heterozygous variants, most often with a CNV in trans with an SNV or indel. It is in these cases where analysis is needed across the CNV and short variant data that seqr is particularly powerful for analysis. A popular feature of seqr is the ability to directly view the raw read data supporting a specific variant through a web-IGV integration. Current feature development includes visualization integrations of RNA sequencing data to interrogate changes in expression and splicing with integrated searching for rare variants in the DNA sequence data. Future development may include the ability to use seqr for Variant Matching to query for variants of interest (where appropriate permissions are in place).
Project management features have given users the ability to efficiently track the status of samples and cohort analyzed in seqr, and facilitated sharing of metadata in standardized formats for AnVIL as well as submitting candidate genes through the Matchmaker Exchange. And by enabling several variant types in seqr, users can have all project results in a single location. In cases where a causal variant is identified through an independent method, but is missing or inaccurate in the short-read variant calls stored in seqr, such as many short tandem repeat (STR) expansions, there is also an option for users to manually add the variant to seqr to ensure these diagnoses are recorded.
Seamless real-time integration with the MME platform, and detailed support for communication, has been a critical addition to seqr, given that most novel gene discoveries made by the Broad CMG have benefited from matches identified through MME. We continue to expand our interactions as new nodes are added to the federated MME platform.
Initially, seqr was designed to meet the project management, collaborative analysis, and data sharing needs of the Broad CMG. Local instances can be deployed but require computational expertise for installation and maintenance. To enable more widespread access to users without software engineering skills, seqr is now available on the AnVIL platform. Anyone can upload a joint called VCF file into a private workspace in Terra or generate a joint called VCF from data available in AnVIL and then request the joint VCF be loaded in seqr. To further empower the global rare disease community, we have made written and video-based materials and training workshops materials available (https://cmg.broadinstitute.org/using-seqr).
New projects and new users will want additional features to be implemented in seqr, and we look forward to continuing to work with developers to improve and expand the platform. The recent Panel App gene list integration from the team at MCRI is a successful example of how features built to support a local use case can benefit all seqr users.
In summary, many platforms currently exist to support rare disease case analysis; however, most are expensive commercial platforms or proprietary and confined in use to a single laboratory. Access to a free and publicly accessible platform that enables collaborative analysis and communal code contribution is critical to enabling widespread equitable involvement in rare disease diagnosis and gene discovery as well as harnessing the latest methodological advances in genome analysis. Engaging the larger rare disease community to contribute to developing seqr and increasing the rare disease genomic data that can be shared within seqr or easily exported to other platforms will advance rare disease science as well as patient diagnosis and treatment.
Supplementary Material
Acknowledgements
Funding for platform development was supported by the National Human Genome Research Institute (NHGRI), the National Eye Institute, and the National Heart, Lung and Blood Institute Grant UM1HG008900, and NHGRI Grants R01HG009141, U24HG0- 10262, and U01HG011755. The authors would like to thank Simon Sadedin, David Ma, and Tommy Li from the Victoria Clinical Genetics Service for their contributions to integrating PanelApp into seqr. They would also like to thank Alireza Haghighi, Jasmin Bakalovic, and Yvonne Chekaluk from Harvard International Center for Genetic Disease, along with input from the Laboratory for Molecular Medicine, for integration of a variant classifier tool based on ACMG/AMP criteria and the addition of filtration by in silico scores into seqr. We would also like to thank Thomas Mullen, Monkol Lek, Elise Valkanas, Stacey Hall, Kayla Delano, Katie Larkin, Jaime Chang, Grace Tiao, Kristen Larrichia, Julia Goodrich, and Steve Jahl for their contributions in developing the platform, and Mark Daly and Brett Thomas for their contributions to the development of an early precursor to the seqr platform.
Funding information:
NIH, Grant/Award Numbers: R01HG009141, U01HG011755, U24HG010262, UM1HG008900
Conflict of Interest Statement
Daniel G. MacArthur is a founder with equity in Goldfinch Bio and is an advisor to Insitro, Variant Bio, GSK, and Foresite Labs. Heidi L. Rehm receives funding from Illumina, Inc to support rare disease gene discovery and diagnosis. Anne O’Donnell-Luria is on the Scientific Advisory Board for Congenica. While work was performed at the Broad Institute, Alysia Lovgren is now an employee of Invitae Corporation, Stacy Mano an employee at X4 Pharmaceuticals, and Harindra Arachchi an employee at Foundation Medicine. Other authors have no disclosures relevant to the manuscript.
Data Availability Statement
The seqr platform code is available at https://github.com/broadinstitute/seqr. Code contribution guidelines are available at https://github.com/broadinstitute/seqr#contributing-to-seqr. Candidate genes can be shared through seqr to Matchmaker Exchange, as submitted by users of the platform. Training materials are available through the Broad CMG website (https://cmg.broadinstitute.org/using-seqr).
References
- 100,000 Genomes Project Pilot Investigators, Smedley D, Smith KR, Martin A, Thomas EA, McDonagh EM, Cipriani V, Ellingford JM, Arno G, Tucci A, Vandrovcova J, Chan G, Williams HJ, Ratnaike T, Wei W, Stirrups K, Ibanez K, Moutsianas L, Wielscher M, … Caulfield M (2021). 100,000 Genomes Pilot on Rare-Disease Diagnosis in Health Care - Preliminary Report. The New England Journal of Medicine, 385(20), 1868–1880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amberger JS, Bocchini CA, Schiettecatte F, Scott AF, & Hamosh A (2015). OMIM.org: Online Mendelian Inheritance in Man (OMIM®), an online catalog of human genes and genetic disorders. Nucleic Acids Research, 43(Database issue), D789–D798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baxter SM, Posey JE, Lake NJ, Sobreira N, Chong JX, Buyske S, Blue EE, Chadwick LH, Coban-Akdemir ZH, Doheny KF, Davis CP, Lek M, Wellington C, Jhangiani SN, Gerstein M, Gibbs RA, Lifton RP, MacArthur DG, Matise TC, … O’Donnell-Luria A (2022). Centers for Mendelian Genomics: A decade of facilitating gene discovery. Genetics in Medicine: Official Journal of the American College of Medical Genetics. 10.1016/j.gim.2021.12.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boycott KM, Rath A, Chong JX, Hartley T, Alkuraya FS, Baynam G, Brookes AJ, Brudno M, Carracedo A, den Dunnen JT, Dyke SOM, Estivill X, Goldblatt J, Gonthier C, Groft SC, Gut I, Hamosh A, Hieter P, Höhn S, … Lochmüller H (2017). International Cooperation to Enable the Diagnosis of All Rare Genetic Diseases. American Journal of Human Genetics, 100(5), 695–705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bult CJ, Blake JA, Smith CL, Kadin JA, Richardson JE, & Mouse Genome Database Group. (2019). Mouse Genome Database (MGD) 2019. Nucleic Acids Research, 47(D1), D801–D806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buske OJ, Manickaraj A, Mital S, Ray PN, & Brudno M (2013). Identification of deleterious synonymous variants in human genomes. Bioinformatics, 29(15), 1843–1850. [DOI] [PubMed] [Google Scholar]
- Clark MM, Stark Z, Farnaes L, Tan TY, White SM, Dimmock D, & Kingsmore SF (2018). Meta-analysis of the diagnostic and clinical utility of genome and exome sequencing and chromosomal microarray in children with suspected genetic diseases. NPJ Genomic Medicine, 3, 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cloney T, Gallacher L, Pais LS, Tan NB, Yeung A, Stark Z, Brown NJ, McGillivray G, Delatycki MB, de Silva MG, Downie L, Stutterd CA, Elliott J, Compton AG, Lovgren A, Oertel R, Francis D, Bell KM, Sadedin S, … Tan TY (2021). Lessons learnt from multifaceted diagnostic approaches to the first 150 families in Victoria’s Undiagnosed Diseases Program. Journal of Medical Genetics. 10.1136/jmedgenet-2021-107902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins RL, Brand H, Karczewski KJ, Zhao X, Alföldi J, Francioli LC, Khera AV, Lowther C, Gauthier LD, Wang H, Watts NA, Solomonson M, O’Donnell-Luria A, Baumann A, Munshi R, Walker M, Whelan CW, Huang Y, Brookings T, … Talkowski ME (2020). A structural variation reference for medical and population genetics. Nature, 581(7809), 444–451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coppens S, Barnard AM, Puusepp S, Pajusalu S, Õunap K, Vargas-Franco D, Bruels CC, Donkervoort S, Pais L, Chao KR, Goodrich JK, England EM, Weisburd B, Ganesh VS, Gudmundsson S, O’Donnell-Luria A, Nigul M, Ilves P, Mohassel P, … Kang PB (2021). A form of muscular dystrophy associated with pathogenic variants in JAG2. American Journal of Human Genetics, 108(6), 1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cummings BB, Karczewski KJ, Kosmicki JA, Seaby EG, Watts NA, Singer-Berk M, Mudge JM, Karjalainen J, Satterstrom FK, O’Donnell-Luria AH, Poterba T, Seed C, Solomonson M, Alföldi J, Genome Aggregation Database Production Team, Genome Aggregation Database Consortium, Daly MJ, & MacArthur DG (2020). Transcript expression-aware annotation improves rare variant interpretation. Nature, 581(7809), 452–458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickinson ME, Flenniken AM, Ji X, Teboul L, Wong MD, White JK, Meehan TF, Weninger WJ, Westerberg H, Adissu H, Baker CN, Bower L, Brown JM, Caddle LB, Chiani F, Clary D, Cleak J, Daly MJ, Denegre JM, … Murray SA (2016). High-throughput discovery of novel developmental phenotypes. Nature, 537(7621), 508–514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Sera T, Velinder M, Ward A, Qiao Y, Georges S, Miller C, Pitman A, Richards W, Ekawade A, Viskochil D, Carey JC, Pace L, Bale J, Clardy SL, Andrews A, Botto L, & Marth G (2020). gene.iobio: an interactive web tool for versatile, clinically-driven variant interrogation and prioritization. medRxiv : The Preprint Server for Health Sciences. 10.1101/2020.11.05.20224865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donkervoort S, Sabouny R, Yun P, Gauquelin L, Chao KR, Hu Y, Al Khatib I, Töpf A, Mohassel P, Cummings BB, Kaur R, Saade D, Moore SA, Waddell LB, Farrar MA, Goodrich JK, Uapinyoying P, Chan SHS, Javed A, … Shutt TE (2019). MSTO1 mutations cause mtDNA depletion, manifesting as muscular dystrophy with cerebellar involvement. Acta Neuropathologica, 138(6), 1013–1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Firth HV, Richards SM, Bevan AP, Clayton S, Corpas M, Rajan D, Van Vooren S, Moreau Y, Pettett RM, & Carter NP (2009). DECIPHER: Database of Chromosomal Imbalance and Phenotype in Humans Using Ensembl Resources. American Journal of Human Genetics, 84(4), 524–533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frankish A, Diekhans M, Ferreira A-M, Johnson R, Jungreis I, Loveland J, Mudge JM, Sisu C, Wright J, Armstrong J, Barnes I, Berry A, Bignell A, Carbonell Sala S, Chrast J, Cunningham F, Di Domenico T, Donaldson S, Fiddes IT, … Flicek P (2019). GENCODE reference annotation for the human and mouse genomes. Nucleic Acids Research, 47(D1), D766–D773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu JM, Satterstrom FK, Peng M, Brand H, Collins RL, Dong S, Klei L, Stevens CR, Cusick C, Babadi M, Banks E, Collins B, Dodge S, Gabriel SB, Gauthier L, Lee SK, Liang L, Ljungdahl A, Mahjani B, … iPSYCH-BROAD Consortium. (2021). Rare coding variation illuminates the allelic architecture, risk genes, cellular expression patterns, and phenotypic context of autism. In bioRxiv. 10.1101/2021.12.20.21267194 [DOI] [Google Scholar]
- Ghosh SG, Scala M, Beetz C, Helman G, Stanley V, Yang X, Breuss MW, Mazaheri N, Selim L, Hadipour F, Pais L, Stutterd CA, Karageorgou V, Begtrup A, Crunk A, Juusola J, Willaert R, Flore LA, Kennelly K, … Gleeson JG (2021). A relatively common homozygous TRAPPC4 splicing variant is associated with an early-infantile neurodegenerative syndrome. European Journal of Human Genetics: EJHG, 29(2), 271–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GTEx Consortium. (2013). The Genotype-Tissue Expression (GTEx) project. Nature Genetics, 45(6), 580–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Köhler S, Gargano M, Matentzoglu N, Carmody LC, Lewis-Smith D, Vasilevsky NA, Danis D, Balagura G, Baynam G, Brower AM, Callahan TJ, Chute CG, Est JL, Galer PD, Ganesan S, Griese M, Haimel M, Pazmandi J, Hanauer M, … Robinson PN (2021). The Human Phenotype Ontology in 2021. Nucleic Acids Research, 49(D1), D1207–D1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landrum MJ, Lee JM, Benson M, Brown G, Chao C, Chitipiralla S, Gu B, Hart J, Hoffman D, Hoover J, Jang W, Katz K, Ovetsky M, Riley G, Sethi A, Tully R, Villamarin-Salomon R, Rubinstein W, & Maglott DR (2016). ClinVar: public archive of interpretations of clinically relevant variants. Nucleic Acids Research, 44(D1), D862–D868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Li C, Mou C, Dong Y, & Tu Y (2020). dbNSFP v4: a comprehensive database of transcript-specific functional predictions and annotations for human nonsynonymous and splice-site SNVs. Genome Medicine, 12(1), 103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin AR, Williams E, Foulger RE, Leigh S, Daugherty LC, Niblock O, Leong IUS, Smith KR, Gerasimenko O, Haraldsdottir E, Thomas E, Scott RH, Baple E, Tucci A, Brittain H, de Burca A, Ibañez K, Kasperaviciute D, Smedley D, … McDonagh EM (2019). PanelApp crowdsources expert knowledge to establish consensus diagnostic gene panels. Nature Genetics, 51(11), 1560–1565. [DOI] [PubMed] [Google Scholar]
- McMurry JA, Köhler S, Washington NL, Balhoff JP, Borromeo C, Brush M, Carbon S, Conlin T, Dunn N, Engelstad M, Foster E, Gourdine J-P, Jacobsen JOB, Keith D, Laraway B, Xuan JN, Shefchek K, Vasilevsky NA, Yuan Z, … Haendel MA (2016). Navigating the Phenotype Frontier: The Monarch Initiative. Genetics, 203(4), 1491–1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohassel P, Donkervoort S, Lone MA, Nalls M, Gable K, Gupta SD, Foley AR, Hu Y, Saute JAM, Moreira AL, Kok F, Introna A, Logroscino G, Grunseich C, Nickolls AR, Pourshafie N, Neuhaus SB, Saade D, Gangfuß A, … Bönnemann CG (2021). Childhood amyotrophic lateral sclerosis caused by excess sphingolipid synthesis. Nature Medicine, 27(7), 1197–1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osmond M, Hartley T, Dyment DA, Kernohan KD, Brudno M, Buske OJ, Innes AM, Boycott KM, & Care4Rare Canada Consortium. (2022). Outcome of over 1500 matches through the Matchmaker Exchange for rare disease gene discovery: The 2-year experience of Care4Rare Canada. Genetics in Medicine: Official Journal of the American College of Medical Genetics, 24(1), 100–108. [DOI] [PubMed] [Google Scholar]
- Paila U, Chapman BA, Kirchner R, & Quinlan AR (2013). GEMINI: integrative exploration of genetic variation and genome annotations. PLoS Computational Biology, 9(7), e1003153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer EE, Sachdev R, Macintosh R, Melo US, Mundlos S, Righetti S, Kandula T, Minoche AE, Puttick C, Gayevskiy V, Hesson L, Idrisoglu S, Shoubridge C, Thai MHN, Davis RL, Drew AP, Sampaio H, Andrews PI, Lawson J, … Kirk E (2021). Diagnostic Yield of Whole Genome Sequencing After Nondiagnostic Exome Sequencing or Gene Panel in Developmental and Epileptic Encephalopathies. Neurology, 96(13), e1770–e1782. [DOI] [PubMed] [Google Scholar]
- Philippakis AA, Azzariti DR, Beltran S, Brookes AJ, Brownstein CA, Brudno M, Brunner HG, Buske OJ, Carey K, Doll C, Dumitriu S, Dyke SOM, den Dunnen JT, Firth HV, Gibbs RA, Girdea M, Gonzalez M, Haendel MA, Hamosh A, … Rehm HL (2015). The Matchmaker Exchange: a platform for rare disease gene discovery. Human Mutation, 36(10), 915–921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, Grody WW, Hegde M, Lyon E, Spector E, Voelkerding K, Rehm HL, & ACMG Laboratory Quality Assurance Committee. (2015). Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genetics in Medicine: Official Journal of the American College of Medical Genetics, 17(5), 405–424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson JT, Thorvaldsdóttir H, Winckler W, Guttman M, Lander ES, Getz G, & Mesirov JP (2011). Integrative genomics viewer. Nature Biotechnology, 29(1), 24–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson PN, Köhler S, Oellrich A, Sanger Mouse Genetics Project, Wang K, Mungall CJ, Lewis SE, Washington N, Bauer S, Seelow D, Krawitz P, Gilissen C, Haendel M, & Smedley D (2014). Improved exome prioritization of disease genes through cross-species phenotype comparison. Genome Research, 24(2), 340–348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoch K, Esteves C, Bican A, Spillmann R, Cope H, McConkie-Rosell A, Walley N, Fernandez L, Kohler JN, Bonner D, Reuter C, Stong N, Mulvihill JJ, Novacic D, Wolfe L, Abdelbaki A, Toro C, Tifft C, Malicdan M, … Shashi V (2021). Clinical sites of the Undiagnosed Diseases Network: unique contributions to genomic medicine and science. Genetics in Medicine: Official Journal of the American College of Medical Genetics, 23(2), 259–271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shefchek KA, Harris NL, Gargano M, Matentzoglu N, Unni D, Brush M, Keith D, Conlin T, Vasilevsky N, Zhang XA, Balhoff JP, Babb L, Bello SM, Blau H, Bradford Y, Carbon S, Carmody L, Chan LE, Cipriani V, … Osumi-Sutherland D (2020). The Monarch Initiative in 2019: an integrative data and analytic platform connecting phenotypes to genotypes across species. Nucleic Acids Research, 48(D1), D704–D715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobreira N, Schiettecatte F, Boehm C, Valle D, & Hamosh A (2015). New tools for Mendelian disease gene identification: PhenoDB variant analysis module; and GeneMatcher, a web-based tool for linking investigators with an interest in the same gene. Human Mutation, 36(4), 425–431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright CF, Fitzgerald TW, Jones WD, Clayton S, McRae JF, van Kogelenberg M, King DA, Ambridge K, Barrett DM, Bayzetinova T, Bevan AP, Bragin E, Chatzimichali EA, Gribble S, Jones P, Krishnappa N, Mason LE, Miller R, Morley KI, … DDD study. (2015). Genetic diagnosis of developmental disorders in the DDD study: a scalable analysis of genome-wide research data. The Lancet, 385(9975), 1305–1314. [DOI] [PMC free article] [PubMed] [Google Scholar]
Web Resources
- 1. https://anvilproject.org/
- 2. http://care4rare.ca/
- 3. http://compbio.cs.toronto.edu/silva/
- 4. http://mendelian.org/
- 5. http://www.informatics.jax.org/
- 6. https://clinicalgenome.org/
- 7. https://gemini.readthedocs.io/en/latest/
- 8. https://genome.ucsc.edu/
- 9. https://gnomad.broadinstitute.org/
- 10. https://gtexportal.org/home/
- 11. https://igv.org/
- 12. https://iobio.io/
- 13. https://monarchinitiative.org/
- 14. https://omim.org/
- 15. https://panelapp.genomicsengland.co.uk/
- 16. https://phenodb.org/
- 17. https://terra.bio/
- 18. https://www.deciphergenomics.org/
- 19. https://www.matchmakerexchange.org/
- 20. https://www.mousephenotype.org/
- 21. https://www.ncbi.nlm.nih.gov/clinvar/
- 22. https://www.sanger.ac.uk/tool/exomiser/
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The seqr platform code is available at https://github.com/broadinstitute/seqr. Code contribution guidelines are available at https://github.com/broadinstitute/seqr#contributing-to-seqr. Candidate genes can be shared through seqr to Matchmaker Exchange, as submitted by users of the platform. Training materials are available through the Broad CMG website (https://cmg.broadinstitute.org/using-seqr).


