Abstract
The coagulase-negative bacterium Staphylococcus epidermidis is a member of the human skin microbiota. S. epidermidis is not merely a passive resident on skin but actively primes the cutaneous immune response, maintains skin homeostasis and prevents opportunistic pathogens from causing disease via colonization resistance. However, it is now appreciated that S. epidermidis and its interactions with the host exist on a spectrum of potential pathogenicity derived from its high strain-level heterogeneity. S. epidermidis is the most common cause of implant-associated infections and is a canonical opportunistic biofilm former. Additional emerging evidence suggests that some strains of S. epidermidis may contribute to the pathogenesis of common skin diseases. Here, we highlight new developments in our understanding of S. epidermidis strain diversity, skin colonization dynamics and its multifaceted interactions with the host and other members of the skin microbiota.
The human skin microbiota comprise a complex group of bacteria, viruses, fungi and archaea that mediate dynamic interactions with the host1. Considering the surface area of all skin niches, including hair follicles and sebaceous glands, the total area of inhabitable space for microorganisms is more than 30 m2, making it one of the largest epithelial surfaces for interactions with microorganisms2. Bacteria inhabit all layers of the skin from the exterior-facing stratum corneum to the interior dermal tissue3,4. Early life colonization with microorganisms shapes proper skin development5–7 and gene expression8. The microbiota also exclude pathogens from their skin niche via mechanisms collectively termed colonization resistance9. These mechanisms include competition for nutrients, direct inhibition by antimicrobial peptides (AMPs) and/or stimulation of the cutaneous immune response10. Taken together, microbial maintenance of skin barrier integrity is vital for homeostasis and disease prevention10,11.
The coagulase-negative bacterium Staphylococcus epidermidis is a ubiquitous human skin colonizer that is identified at most skin sites through traditional culture methods and metagenomics analyses1,12,13. Early phenotypic typing primarily distinguished between pathogenic Staphylococcus aureus and other staphylococci by the presence or absence of the coagulase enzyme, respectively13,14. Historically, S. epidermidis was used as a representative of all coagulase-negative staphylococci (CoNS), yet current evidence suggests that CoNS are far more genetically and functionally distinct from each other than previously appreciated10,13,15,16. Many studies regarded S. epidermidis as a benign or beneficial member of the skin microbiota that is involved in barrier development, maintenance of homeostasis and control of opportunistic pathogens, yet evidence suggests that S. epidermidis leads a far more nuanced lifestyle on skin than previously appreciated17. Previous literature also regarded S. epidermidis as an ‘accidental pathogen’, given that many S. epidermidis infections are derived from the same strains that inhabit the skin18–20. Indeed, S. epidermidis remains one of the most frequent causes of implant-associated infections in the United States, and these antibiotic-refractory, biofilm-associated infections are costly for patients and the healthcare system21. Further, the global spread of three strains of S. epidermidis nearly pan resistant to all classes of antibiotics is also cause for clinical concern22. Yet all of these descriptions and assumptions of S. epidermidis overly simplify a complex organism. The current question addressed in this Review is how we should appreciate the vast diversity of S. epidermidis isolates while reconciling the many benefits of this species with its serious propensity to cause disease.
In this Review, we discuss new developments in our understanding of S. epidermidis strain-level diversity, physiology and lifestyle on the skin. We compare positive and negative interactions between the host and S. epidermidis during skin colonization or infection. Finally, we discuss the many competitive interactions between S. epidermidis and other members of the skin microbiota. Together, a greater appreciation of this complicated organism is imperative for future translational applications, including growing interest in S. epidermidis as a candidate for ‘bacteriotherapies’23,24.
Commensal lifestyle
The skin is a harsh environment for microorganisms to colonize25. The outermost layer of the epidermis (the stratum corneum) is low in pH and nutrient availability but high in salinity, exposure and concentration of free fatty acids and AMPs11,25. By contrast, appendages such as the hair follicle are considerably more anaerobic, shielded from exposure and replete in lipids25. Skin can be categorized based on physiology into dry (volar forearm), moist (antecubital crease) or sebaceous or oily (face) sites1. Metagenomics analyses of healthy skin demonstrated that the microbial composition of these sites is unique in abundance and species composition26,27. The total composition and diversity of the skin microbiome remains stable for up to 2 years, which suggests that microorganisms maintain and reseed themselves in their preferred niche over time in the absence of a substantial disturbance28. Healthy human skin is colonized by an array of CoNS species, but S. epidermidis is the most abundant CoNS by either traditional culturing methods12 or metagenomics analyses27–29 and colonizes all types of skin sites, with a strong propensity for moist areas1 (FIG. 1). Additionally, skin sites are colonized by diverse strains of S. epidermidis that are genetically distinct within and between healthy humans27 (BOX 1).
Fig. 1 |. The commensal lifestyle of Staphylococcus epidermidis.
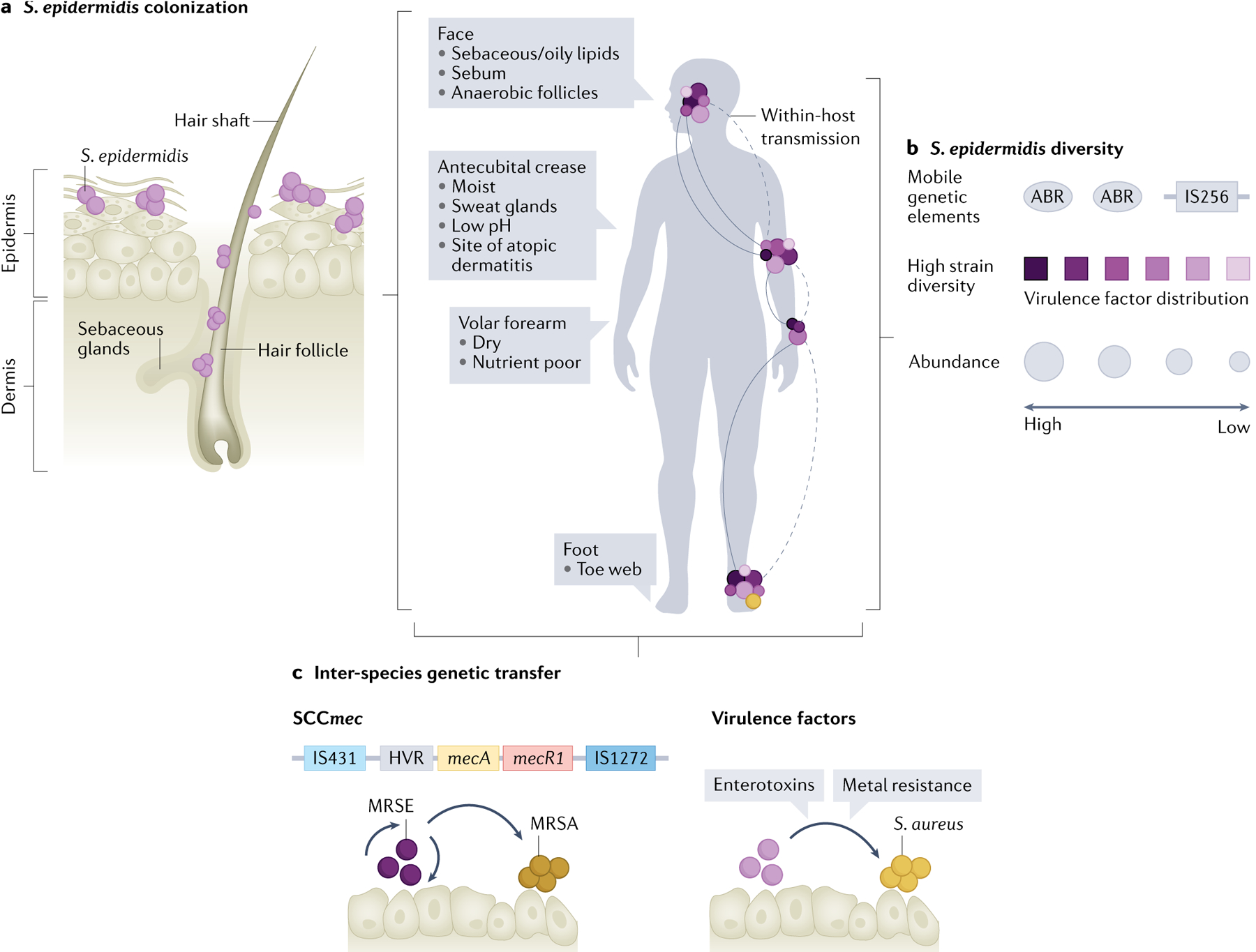
a | The skin can be subdivided into different ecological niches based on physio-chemical properties such as density of sebaceous and sweat glands, level of moisture and oxygen tension. Staphylococcus epidermidis colonizes all these environments at different densities, including the outermost stratum corneum as well as the interior of the hair follicle, and is a dominant colonizer of moist sites. b | Recent metagenomics studies revealed high strain-level heterogeneity of skin-colonizing S. epidermidis (represented by various colours) (BOX 1). These strains share genetic content, including virulence factors and antibiotic resistance factors (ABRs), via horizontal gene transfer of mobile genetic elements, including plasmids, phages and transposable elements (such as the insertion sequence IS256). Body sites are colonized by varying levels of S. epidermidis (indicated by circle size) with the highest density of S. epidermidis in moist or sebaceous sites. c | Horizontal gene transfer of the staphylococcal cassette chromosome mec (SCCmec), which encodes resistance to methicillin, between S. epidermidis and Staphylococcus aureus. Other virulence factors can also be exchanged between these organisms, including enterotoxins and genes for metal resistance. MRSA, methicillin-resistant S. aureus; MRSE, methicillin-resistant S. epidermidis.
Box 1 |. Staphylococcus epidermidis genetic flexibility.
Understanding of Staphylococcus epidermidis strain heterogeneity was previously limited by several factors, including the assumption that all S. epidermidis strains were genetically similar and, thus, behaved similarly and the focus on understanding host–pathogen relationships at the expense of commensals. Many initial efforts to genetically characterize S. epidermidis focused on multilocus sequence typing or pulse field gel electrophoresis to identify traits that could distinguish pathogenic strains from commensal strains170. These methods can reveal certain allelic relationships among strains, but they are inherently low resolution and do not provide substantial insight into the total genomic heterogeneity in a population. Recent advances in whole-metagenome shotgun sequencing and in silico modelling enabled higher taxonomic resolution and provided insight into microbial functionality in disparate environments, but a large gap remains in our understanding of intra-species diversity in the microbiome and its implications for skin health171,172.
A sequencing survey of healthy and nosocomial S. epidermidis isolates was the first to substantially improve the number of fully sequenced S. epidermidis genomes80. It revealed that the S. epidermidis genome is composed of approximately 80% core and 20% variable genes80. These variable genes indicated that S. epidermidis has an open pangenome, or high potential to acquire new genetic traits especially through mobile genetic elements (also called the mobilome), including phages and plasmids80,82. Indeed, the most enriched functions of the variable genome included replication, recombination and repair, as well as transcriptional regulators and defence mechamisms80. It is well established that mobile genetic elements profoundly affect S. epidermidis adaptation to various niches and lifestyles, and many mobile genetic elements have been associated with the propensity of certain S. epidermidis strains to cause infection77.
Underlying genetic diversity among strains and exchange of mobile genetic elements among strains or species can influence phenotypic behaviour and adaptation to various niches. S. epidermidis functional specialization during skin colonization is apparent in the variety and diversity of strains that colonize distinct ecological niches (dry, moist, sebaceous), especially those that are more isolated from intra-host transmission (toe web)95. The first longitudinal (two to four time points over 1 month), multibody site, multi-volunteer metagenomic interrogation of S. epidermidis strain-level diversity of healthy adult skin was published in 2020 (REF.95). Subjects from this study were colonized by multiple lineages of S. epidermidis with distinct genetic content, and the predicted functionality of this genetic content, including KEGG modules, lantibiotics and biosynthetic gene clusters, was dependent on the host and the skin site95. Further, there was strong evidence of horizontal gene transfer between skin sites, especially for antibiotic resistance and virulence-associated genes95. Together, S. epidermidis strain-level variation is likely to underlie niche specialization and colonization along with contextual potential for pathogenicity.
Genetic flexibility is also apparent during infection, as isolates of S. epidermidis from pacemaker-associated endocarditis were shown to undergo within-host adaptation to become stronger biofilm formers and more antibiotic tolerant173. Insertion elements are another driver of phenotypic diversity among S. epidermidis strains. The insertion sequence IS256 is essential for genetic plasticity as it mediates phase variation in biofilm formation and spontaneous chromosomal deletions, and can also function as a gene promoter170. Insertions in the exopolysaccharide-forming locus icaADBC and the quorum sensing accessory gene regulator (agr) system highlight some of the phenotypic variations driven by IS256 (REF.170). It has been reported that the presence of IS256 is associated with pathogenic lineages of S. epidermidis, and its presence or absence has been proposed as a ‘molecular marker’ to differentiate invasive strains from commensals77,107,174. However, there is at least one report of commensal S. epidermidis isolates that retain the IS256, although the relative prevalence of IS256-positive commensals was much lower than IS256-positive invasive strains80.
Transfer of mobile genetic elements among staphylococcal species was originally thought to be rare given the presence of ‘impenetrable’ restriction modification systems175 and CRISPR loci176 that degrade foreign DNA. However, more recent evidence suggests that the presence of CRISPR loci in coagulase-negative staphylococci (CoNs) is not as high as previously assumed177. additionally, there are multiple examples of inter-species genetic transfer between S. epidermidis and Staphylococcus aureus including exchange of enterotoxin-encoding genes93, metal toxicity resistance genes87 and the staphylococcal cassette chromosome mec (SCCmec), which encodes methicillin resistance84. In the context of patient care and antibiotic treatment, the importance of this genetic flexibility and exchange cannot be overstated. It suggests that genetic adaptations that are advantageous to one species can be transferred to another to promote the persistence of highly adapted, multidrug-resistant pathogenic lineages87. Together, the fundamental genetic flexibility of S. epidermidis enables it to be a highly adaptable specialist in various contexts from commensalism to invasive infection.
Adhesion and colonization
S. epidermidis must express various adhesins to colonize human skin30 (FIG. 2). One adhesin implicated in attachment to the stratum corneum is accumulation associated protein (Aap) (FIG. 2a). Aap is a large (140 kDa) rod-like fibril that is sortase-anchored to the cell wall and extends away from the cell body31. Aap is found in 85–95% of all S. epidermidis isolates, which suggests a broadly conserved mechanism for adhesion to corneocytes30,31. Many S. aureus strains express an orthologous giant surface protein, SasG, that also binds human skin components31. It was demonstrated in 2021 that Aap-mediated adhesion to corneocytes is dependent on the presence of the amino-terminal l-type lectin domain, although the specific host substrate that binds the lectin remains to be determined31,32.
Fig. 2 |. Staphylococcus epidermidis adhesion and biofilm formation.
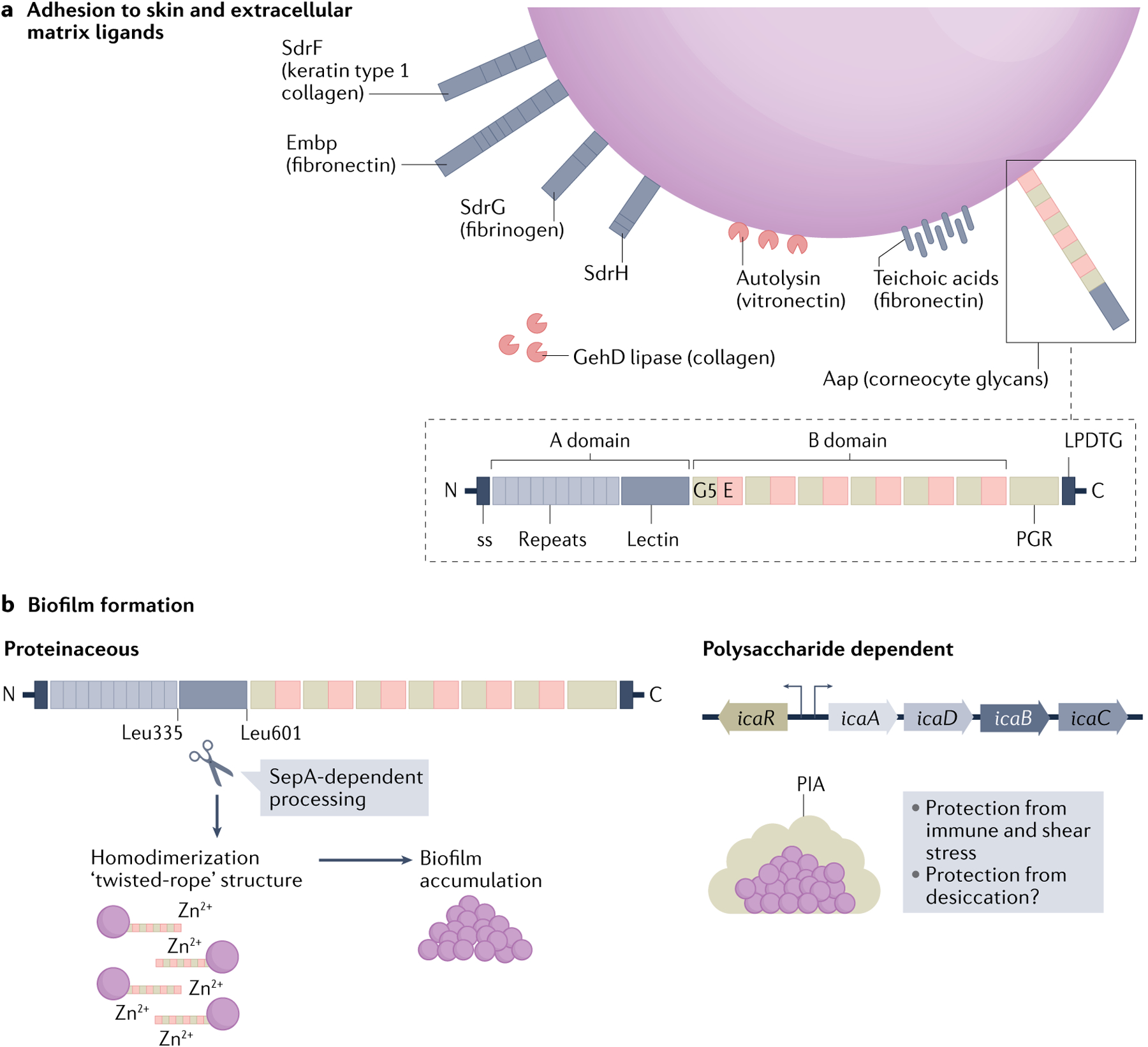
a | Staphylococcus epidermidis binds components of skin (ligands indicated, if known) and the extracellular matrix through expression of surface-anchored proteins, called adhesins. This family of microbial surface components recognizing adhesive matrix molecules (MSCRAMMs) includes several serine aspartate repeat containing (Sdr) proteins. Other non-MSCRAMM cell wall-associated proteins with roles in cellular adhesion include the giant Embp protein, the bifunctional autolysin and the bifunctional lipase GehD. Many S. epidermidis isolates encode the large (140 kDa), multi-domain accumulation associated protein (Aap) that is anchored to the cell wall by sortase via the LPDTG motif in the carboxy terminus. Aap export from the cell is facilitated by the amino-terminal secretion sequence (ss). The A domain is composed of short, 16 amino acid repeats followed by a lectin-like domain. The B domain contains a variable number (5–17) of 128 amino acid repeats of G5 domains (named for their conserved glycine residues) and E spacer regions. The C-terminal proline–glycine rich region (PGR) and the cell wall anchoring motif (LPDTG) are also indicted. Aap specifically mediates attachment to the host stratum corneum through the A-domain lectin, which leads to epithelial colonization. b | Aap also mediates proteinaceous biofilm attachment and accumulation. The metalloproteinase SepA cleaves the N-terminal A domain of Aap at specific amino acid residues (Leu335 and Leu601). Cleavage is followed by homodimerization of the B domain in a zinc-dependent manner. Many, but not all, S. epidermidis strains have the icaADBC operon, which encodes polysaccharide intracellular adhesin (PIA; also known as poly-N-acetylglucosamine (PNAG)), as well as the icaR gene, which encodes a regulatory protein. PIA is composed of repeating β-1,6-linked N-acetylglucosamine residues and its production promotes biofilm formation. Such polysaccharide-dependent biofilms may protect mature biofilms from immune, antibiotic or shear stress. In addition, the polysaccharide has been speculated to aid in protection against desiccation but further investigation is warranted105.
S. epidermidis also expresses various other cell wall-anchored proteins important for binding components of the skin matrix (FIG. 2a). One of the largest families of these proteins is the microbial surface components recognizing adhesive matrix molecules (MSCRAMMs), which are characterized by the presence of two adjacent IgG-like folded subdomains and their ability to bind substrates via a ‘dock lock and latch’ mechanism30,33. The S. epidermidis MSCRAMM repertoire is substantially smaller than that of S. aureus18. Members of this family include SdrF, which binds keratin and type I collagen18,34, and SdrG, which binds fibrinogen30. SdrH has also been identified, but its role in binding skin substrates remains to be determined35. In addition to the classic MSCRAMMs, other cell wall-associated proteins including the bifunctional autolysin Atl, the giant surface protein Embp and the bifunctional lipase GehD bind vitronectin, surface immobilized fibronectin and collagen, respectively18,30,36. Similar to Embp, S. epidermidis teichoic acid enhances binding to immobilized fibronectin37. Together, this suite of adhesins mediate specific interactions between S. epidermidis and the many layers of host skin tissue and the extracellular matrix.
Beneficial host interactions
It is now widely appreciated that the skin microbiota, similar to the gut microbiota, can profoundly influence human development and health7. In this section, we review how S. epidermidis colonization promotes skin barrier development, maintains homeostasis and controls opportunistic pathogens.
Immune priming in skin development and homeostasis.
There is a critical time window in neonatal skin development required for establishing tolerance to commensal microorganisms while maintaining a discrete response to pathogens5,38. Regulatory T cells (Treg cells) are important homeostatic mediators at barrier sites, including the gut and skin39. In humans, approximately 20% of tissue-resident CD4+ T cells are Treg cells, which is almost twice as many as are found in the colon39. Neonatal mice initiated an activated Treg cell-mediated response to commensal microorganism when they were colonized early with S. epidermidis that expressed a model T cell antigen5. This tolerogenic response was dependent on precise timing of S. epidermidis skin colonization and could not be restored later in life5,38. Activated Treg cells localize to the developing hair follicle, a primary site of immune–microbial crosstalk40 (FIG. 3a). Together with intrinsic hair follicle morphogenesis, S. epidermidis application on fetal skin tissue induced the chemokine CCL20 and subsequent Treg cell migration to the follicle40. Thus, early S. epidermidis skin colonization is one factor in proper development of this cutaneous barrier site. Yet, it is highly likely that many other organisms also contribute to this process given that application of another skin commensal, Corynebacterium accolens, similarly induced CCL20 signalling in fetal tissue40. Identification of other bacterial species or strains with similar properties will be informative for a global picture of microbial contributions to the development and maintenance of tolerance.
Fig. 3 |. Staphylococcus epidermidis mediates skin homeostasis, barrier repair after injury and colonization resistance.
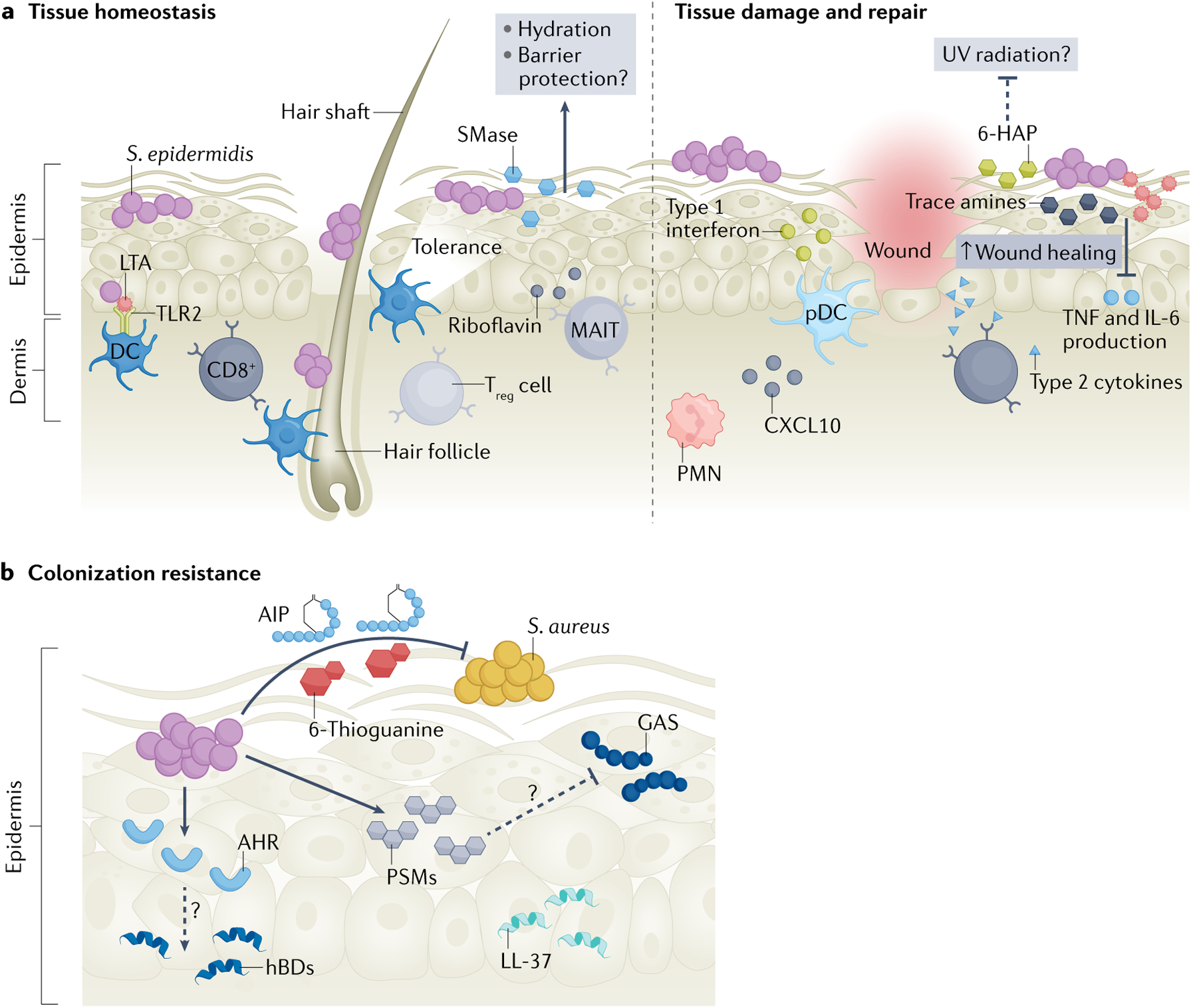
a | Homeostasis: early-life Staphylococcus epidermidis skin colonization in the hair follicle primes the cutaneous immune response to tolerate future commensal colonization via activation of regulatory T cells (Treg cells). S. epidermidis-derived riboflavin is sensed by unconventional mucosal-associated invariant T cells (MAIT cells), priming them to become a dominant type 17 effector subset. CD11B+ dendritic cells (DCs) sense S. epidermidis healthy adult skin colonization and stimulate CD8+ T cell migration. S. epidermidis lipoteichoic acid (LTA) is sensed by dendritic cells via Toll-like receptor 2 (TLR2) signalling. S. epidermidis produces a potent sphingomyelinase (SMase) on human skin that increases ceramide content, thus promoting skin hydration and, possibly, barrier integrity. Barrier repair: S. epidermidis and its molecular products including trace amines, lipopeptide 78 and LTA dampen the inflammatory response (TNF and IL-6 production) to promote wound healing. S. epidermidis-recruited polymorphonuclear leukocytes (PMNs) stimulate plasmacytoid dendritic cells (pDCs) via CXCL10 signalling to secret type I interferons in the wound site to aid healing. CD8+ T cells express type 2 cytokines to promote wound repair. 6-N-hydroxyaminopurine (6-HAP) may also protect skin from tumour development, although further studies are necessary to clarify the mechanism and the 6-HAP biosynthetic pathway (dashed line). b | Colonization resistance: S. epidermidis excludes Group A Streptococcus (GAS) and Staphylococcus aureus from skin. Autoinducing peptides (AIPs) block S. aureus quorum sensing and the purine analogue 6-thioguanine inhibits S. aureus purine biosynthesis. S. epidermidis phenol soluble modulins (PSMs) synergize with host antimicrobial peptides (AMPs) including LL-37 to augment killing of S. aureus and GAS, although the mechanism warrants further investigation (dashed line). Unidentified secreted factors from S. epidermidis signal via TLR2 to stimulate keratinocyte production of human β-defensin 2 (hBD2) and hBD3. Unidentified S. epidermidis factors activate the keratinocyte aryl hydrocarbon receptor (AHR) and increase hBD3 expression. Some strains of S. epidermidis produce AMPs that specifically kill certain pathogenic species.
S. epidermidis also promotes cutaneous immune priming through stimulation of mucosal-associated invariant T cells (MAIT cells), which are a predominant unconventional innate-like lymphoid T cell subset in barrier tissues that sense and respond to riboflavin-producing microorganisms41. MAIT cell priming depended on early exposure to S. epidermidis and other riboflavin-synthesizing bacteria41. Primed MAIT cells also mediated tissue repair after punch biopsy injury41. However, many skin microorganisms in addition to S. epidermidis also synthesize riboflavin and the cumulative contributions of the microbiota to MAIT cell priming remain to be elucidated.
Outside neonatal immune priming, S. epidermidis enhances adult skin barrier immunity through a coupled mechanism between dendritic cells and T cells42. S. epidermidis was detected by non-inflammatory skin-resident CD11B+ dendritic cells, which induced IL-17A+CD8+ T cell homing to the epidermis and enhanced barrier immunity against the opportunistic pathogen, Candida albicans42. The ability of S. epidermidis to induce CD8+ T cells was restricted to a specific clade (A20) that is enriched on skin from healthy individuals compared with skin from individuals with atopic dermatitis43. Finally, the major cell wall component lipoteichoic acid (LTA) of the common laboratory strain S. epidermidis O-47 stimulated a distinct ‘IL-10 balanced’ immune signature in dendritic cells via Toll-like receptor 2 (TLR2) signalling, whereas LTA from S. aureus did not recapitulate this phenotype44.
Maintenance of skin barrier integrity is vital for pre-serving homeostasis and preventing allergy or infection. Germ-free mice had altered skin barrier function that could be restored through application of a defined consortium45. However, few examples exist of specific microorganisms or microbial gene products that directly control skin barrier function. In 2022, it was revealed that S. epidermidis produces a potent sphingomyelinase on human skin that increased ceramide content and prevented skin dehydration in vivo46. The sphingomyelinase gene was highly conserved across a large cohort of S. epidermidis healthy skin isolates, which suggests a species-wide mechanism for skin barrier control46. Although this study did not reveal a role for other commensals, other microbial products that facilitate barrier integrity may be identified in follow-up studies.
S. epidermidis and the immune response to barrier injury.
S. epidermidis actively coordinates the skin response to injury (FIG. 3a). Commensal staphylococci, including S. epidermidis, induced neutrophil CXCL10 signalling in skin wounds that recruited type I interferon-producing plasmacytoid dendritic cells (pDCs) and drove T cell-independent wound repair47. S. epidermidis-primed CD8+ T cells have a dual purpose in cutaneous immunity and wound healing48. In addition to controlling skin barrier function through interactions with dendritic cells, these cells also maintained a poised type 2 state that could respond to injury and release type 2 cytokines to promote wound repair48. However, type 2 immunity is also implicated in the aetiology of various skin disorders49,50, which suggests that tightly controlled immune–microorganism crosstalk is necessary for optimal skin function and that breakdown of this crosstalk may contribute to immune dysregulation.
S. epidermidis-derived small molecules can have profound impacts on barrier response to insult or injury (FIG. 3a). Healthy skin is dominated by strains of S. epidermidis that convert aromatic amino acids into trace amines, and it was shown in 2020 that S. epidermidis-derived trace amines accelerated wound healing in a mouse model51. The S. epidermidis-produced small molecule 6-N-hydroxyaminopurine (6-HAP) selectively inhibited proliferation of tumour cell lines but not primary keratinocytes52. Exposure to ultraviolet B radiation is associated with chronic inflammation and DNA damage53, and application of 6-HAP-producing S. epidermidis on mouse skin significantly suppressed ultraviolet-induced tumour growth52. However, it remains to be determined whether 6-HAP synthesis is conserved in other skin-resident S. epidermidis strains, as this phenotype has only been observed in one strain and the genes encoding 6-HAP or its biosynthetic machinery remain elusive. Skin application of butyric acid, a major by-product of S. epidermidis glycerol fermentation, also dampened ultraviolet B-induced pro-inflammatory cytokines in vivo53. Finally, S. epidermidis-derived small molecules can act directly on innate immune sensors. The S. epidermidis lipopeptide 78 inhibited TLR3-induced skin inflammation to promote wound healing54. Further, LTA from some strains of S. epidermidis dampened TNF and IL-6 production after skin injury in a TLR3-dependent manner, which enabled efficient wound healing55. The LTA-mediated inhibition of TNF signalling is strain dependent, and LTA from strains isolated from healthy skin (54.3%), healthy conjunctiva (68.8%) and ocular infection (80.4%) inhibited TNF production in keratinocytes in vitro56. Additional novel small molecules or metabolic by-products from S. epidermidis or other CoNS may also protect skin from internal (that is, tumorous) or external (that is, wounding) injury but remain to be identified.
Colonization resistance.
S. aureus is the most common cause of skin and soft tissue infections in the United States and a frequent challenge to skin homeostasis57. Methicillin-resistant S. aureus (MRSA) is now pandemic and has been classified by the World Health Organization (WHO) as one of 12 pathogens that threaten human health58. S. epidermidis excludes pathogens, including S. aureus, from the skin through various direct and indirect mechanisms (FIG. 3b). S. epidermidis strains activated distinct innate immune responses and amplified the keratinocyte antimicrobial response to S. aureus infection via secretion of an unidentified factor or factors59. In the skin, neutrophil recruitment and netosis enhance S. aureus colonization and infection60. However, colonization with S. epidermidis reduced the number of migrating neutrophils to the site of S. aureus infection, although the mechanism of protection remains unclear60. An unidentified small molecule (<2 kDa) secreted by S. epidermidis activated the keratinocyte aryl hydrocarbon receptor (AHR) and downstream IL-1α and human β-defensin 3 (hBD3) expression to promote an innate immune defence response61. S. epidermidis-mediated skin protection is dependent on the integrity of the barrier itself; when mouse skin was tape stripped before challenge with S. aureus, the protective effects of S. epidermidis colonization were negated62.
S. epidermidis-derived AMPs also have protective functions on skin (FIG. 3b). The S. epidermidis phenol soluble modulins (PSMs) are a family of small, cytolytic amphipathic molecules18,63. Both PSMγ (also known as δ-toxin) and PSMδ had cooperative activity with the host AMP LL-37 and enhanced antimicrobial action against S. aureus and Group A Streptococcus (GAS)64. Further, S. epidermidis PSMγ was present in the epidermis and dermis of normal human skin and reduced GAS survival in pretreated mouse skin wounds65. These studies suggest that S. epidermidis-derived antimicrobials could be an additional layer of defence on healthy skin. As only synthetic peptides were used in either study, further investigation of isogenic S. epidermidis PSM mutants would be informative. In addition, S. epidermidis PSMs are immunomodulatory and share similarities to S. aureus PSMs, which can exacerbate skin dysfunction in atopic dermatitis18,66. Thus, the regulation of S. epidermidis PSMs and the context in which they are made may dictate their positive or negative effects on barrier integrity.
Pathogenic lifestyle
In addition to its role as a commensal, S. epidermidis often leads a dual lifestyle as an opportunistic pathogen17. Shifts in S. epidermidis abundance have been associated with skin diseases, including atopic dermatitis67, dandruff68,69, seborrhoeic dermatitis70,71 and rosacea72–74. These observations could suggest an underappreciated potential for S. epidermidis pathogenicity in common skin disorders, but the mechanism or mechanisms of interaction remain to be determined. Aside from observations of general dysbiosis, in this section we review specifics of S. epidermidis invasive infections, antibiotic resistance and virulence factor production, and the implications of this dual lifestyle for patient health.
Infections
One of the most concerning aspects of S. epidermidis physiology is its propensity to cause ‘accidental’ opportunistic infections, where the infectious strain is the same as a colonizing strain18–20. S. epidermidis is a leading cause of implant-associated infections, prosthetic valve endocarditis and cardiac pacemaker infections21,75. Implant infections are serious for the patient, costly for the healthcare system and often only resolved by device removal21. S. epidermidis is also responsible for 30–40% of nosocomial bloodstream infections, which are often the result of a prior biofilm-associated catheter infection that disseminated into the bloodstream75. Bloodstream infections are difficult to treat and can lead to serious complications including sepsis, septic shock and infective endocarditis21. S. epidermidis bloodstream infections are also a predominant cause of late-onset sepsis in the preterm neonatal population, with long-term complications from infection, including neurodevelopmental impairment and cerebral palsy76. Collectively, S. epidermidis remains a serious opportunistic pathogen particularly for patients who are immunocompromised.
Clinical prevalence
Although up to 60% of S. epidermidis infections are derived from diverse genetic backgrounds77,78, certain clonal lineages of S. epidermidis, particularly sequence type 2 (ST2), are over-represented in infections22,77,79. ST2 strains are almost exclusively nosocomial, hospital adapted and drug resistant22. Strains of this sequence type are robust biofilm formers and highly invasive18,80. Hospital-acquired S. epidermidis ST23 (HA-ST23) is now considered a globally important threat to patient health22. Strains of HA-ST23 had altered wall teichoic acid (WTA) profiles that augmented pathogenesis in a mouse sepsis model and enabled efficient phage transduction of genetic material between hospital-acquired methicillin-resistant S. epidermidis (MRSE) and MRSA, emphasizing the shared potential for virulence between these two organisms81. These S. epidermidis sequence types represent major pathogens of concern, but more work is needed to understand how they are maintained or spread in the community and why these sequence types, but not others, are associated with infectious disease.
Drug resistance
S. epidermidis strains encode various antibiotic resistance genes often on mobile genetic elements82. Staphylococci acquire resistance to methicillin, previously the drug of choice for infections83, primarily through acquisition of the mecA gene18 encoded on a mobile genomic island referred to as the staphylococcal cassette chromosome mec (SCCmec)84. Methicillin resistance in S. epidermidis nosocomial isolates has been reported at 70% or higher, although many studies have not distinguished between S. epidermidis and other contaminating CoNS isolates22,79,85. Resistance to methicillin is frequently correlated with resistance to other classes of antibiotics, and resistance to all classes of antibiotics has been reported in S. epidermidis22. In addition to nosocomial infections, MRSE is also frequently isolated from normal skin in adult86,87 and neonatal88,89 populations. It has been widely speculated that S. epidermidis or other CoNS are the primary reservoir for SCCmec acquisition in MRSA and there are now several examples of inter-species transfer of this mobile genetic element, as well as others such as the arginine catabolic mobile element (ACME), between S. aureus and S. epidermidis84,87,90–92. The high levels of shared antibiotic resistance and pathogenicity genes between MRSA and MRSE are a continued threat to patient care and infection outcomes87.
Virulence factors
The canonical S. epidermidis virulence factor arsenal is small compared with S. aureus and composed of factors that may have a dual role in commensalism and infection, including proteases, lipases and the PSMs18. S. epidermidis strains generally do not encode many toxins or exoenzymes, although enterotoxigenic genes were identified in nine strains of S. epidermidis in 2019 (REF.93). Whole-genome sequencing of more commensal and nosocomial S. epidermidis isolates has revealed a larger virulence gene repertoire (that is, genes for biofilm formation, adhesins, haemolysins and exoenzymes) than previously appreciated, which is most likely due to the genetic flexibility of this organism94 (BOX 1).
Current evidence suggests that the pathogenic or commensal behaviour of S. epidermidis often depends on the context95. Pathogenic traits are not typically restricted to a single S. epidermidis clonal lineage96, unlike certain S. aureus clonal lineages, which have evolved to be more pathogenic than others97. A 2020 metagenomics analysis of healthy skin isolates revealed that S. epidermidis virulence genes were variably distributed across subjects and skin sites95. Further, the probability of horizontal gene transfer of mobile genetic elements on skin was found to be quite high, with many putative plasmid and phage sequences identified in the genome scaffolds of these healthy skin isolates95. Many plasmid scaffolds contained multiple antibiotic resistance genes and the presence of these plasmids was distributed across S. epidermidis isolates from multiple body sites, which suggests dynamic horizontal gene transfer among isolates during colonization and represents a potential reservoir of strains with contextually important virulence factors or antibiotic resistance even on healthy skin95.
The most compelling mechanistic studies of S. epidermidis virulence factor production on skin come from 2020 (REF.98) and 2021 (REF.99) work on the extracellular cysteine protease EcpA. The EcpA protease cleaved integral skin components including desmogelin 1, a key component of corneodesmosomes, and the AMP LL-37 (REF.99). EcpA transcript levels were positively correlated with the abundance of S. epidermidis in severe atopic dermatitis lesional sites, tying observational data of S. epidermidis abundance in atopic dermatitis to the mechanism of action99.
EcpA-mediated barrier degradation extends beyond atopic dermatitis lesions, and also has deleterious effects in Netherton syndrome lesions98. Patients with Netherton syndrome have a single gene mutation in the serine protease inhibitor Kazal type 5 (SPINK5) that leads to aberrant epidermal serine protease activity and results in skin barrier disruption and inflammation98. Similar to atopic dermatitis, there was a strong positive correlation between the abundance of S. epidermidis and the transcription of ecpA in lesional Netherton syndrome sites of human volunteers98. Further, application of a high EcpA-producing S. epidermidis strain on mouse skin recapitulated many of the same phenotypes as application of a staphopain-producing S. aureus strain98. These data suggest that S. epidermidis EcpA is an important virulence factor in atopic dermatitis and Netherton syndrome. However, it remains unclear why all S. epidermidis strains retain the ecpA gene, but not all strains express EcpA at the same level99.
Biofilm formation and role in atopic dermatitis
S. epidermidis biofilm formation and regulation have been extensively reviewed, and we refer the reader to the outstanding articles in REFS.75,100,101. In brief, many S. epidermidis strains mediate biofilm accumulation through the production of polysaccharide intracellular adhesin (PIA; also known as poly-N-acetylglucosamine or PNAG), which is encoded in the icaADBC operon102 (FIG. 2b). PIA is composed of repeating β-1,6-linked N-acetylglucosamine residues and its production has been linked to protection of the mature biofilm from immune, antibiotic or shear stress75,103,104. Production of PIA could also potentially trap water and protect cells from desiccation on the skin, although this is currently only speculation and remains to be investigated105. The presence of the ica locus has been proposed as a potential determinant of invasive versus commensal S. epidermidis isolates with varying success106,107. Infection isolates of S. epidermidis, including those isolated from high shear stress environments (that is, catheters), may encode the icaADBC locus and produce variable amounts of polysaccharide103. However, the same study also found that more than 50% of high shear isolates did not encode the ica locus yet could mediate biofilm formation through ica-independent mechanisms103. Other studies demonstrated that ica-positive S. epidermidis switches between polysaccharide-dependent and protein-dependent biofilm formation103. Strikingly, many S. epidermidis isolates from healthy skin do not encode the icaADBC locus108. Further, icaADBC carriage was associated with a fitness defect during human skin colonization and competition with an isogenic icaADBC mutant108.
As an alternative to polysaccharide-based biofilms, S. epidermidis also forms proteinaceous biofilms through interactions of surface-associated proteins, including Aap. Initial attachment to surfaces is mediated by the A domain102,109. Subsequent biofilm accumulation is dependent on two steps: the metalloproteinase SepA-mediated processing of the A domain to reveal the homodimeric B domain109,110; and a ‘zinc zipper’ mechanism, whereby the B-domain G5 repeats homodimerize in a zinc-dependent manner to form a biofilm111 (FIG. 2b). The orthologous S. aureus SasG protein also forms proteinaceous biofilms using a similar zinc-dependent mechanism112. Importantly for patient care, S. epidermidis and S. aureus can form heterogeneous biofilm complexes through SasG and Aap interactions, which suggests a potential role for mixed biofilms during infection, although a role for mixed biofilms on the skin remains unclear112.
Infection with S. aureus is associated with worse disease outcomes for patients with atopic dermatitis; however, some patients experience blooms of S. epidermidis rather than S. aureus67,113,114. Staphylococcal biofilms have been identified in atopic dermatitis eccrine ducts, but not on other body sites unaffected by atopic dermatitis lesions115. Further, S. epidermidis isolates from lesional atopic dermatitis sites were strong biofilm producers115. S. epidermidis biofilms in atopic dermatitis lesions may also occlude sweat ducts and exacerbate disease through the induction of itch, although more investigation of the mechanisms of itch induction is warranted115,116.
In 2021, the biofilm-forming properties of S. aureus and S. epidermidis were assessed in 400 patients enrolled in the Mechanisms of Progression of Atopic Dermatitis to Asthma in Children (MPAACH) cohort, which is the first US-based longitudinal study of a paediatric population and the potential progression from atopic dermatitis to other atopic diseases117. Corneocyte-associated staphylococcal biofilms were identified through scanning electron microscopy analysis of tape-stripped skin in most samples collected117. Nineteen per cent of patients were co-colonized with S. aureus and S. epidermidis, and co-isolated strains of these organisms tended to act cooperatively during in vitro biofilm growth117. Although more work is needed to fully describe mechanisms of biofilm cooperativity in atopic dermatitis, the synergistic interactions between S. aureus and S. epidermidis may be an important mediator of disease outcomes and impair treatment efficacy for patients.
Notably, S. epidermidis most likely does not exist only in a biofilm state in atopic dermatitis as the final step in the biofilm life cycle is dispersal75. In staphylococci, biofilm dispersal is mediated through upregulation of the accessory gene regulator (agr) quorum sensing system118,119. Thus, there is likely to be a balance in the S. epidermidis atopic dermatitis life cycle, whereby cells expressing agr seed eccrine ducts or other niches, turn down agr to form biofilms and then reseed these areas with sequential induction of agr. Future work may determine the consequences of these regulatory steps for disease outcomes in atopic dermatitis or other skin conditions.
Microbial competition
S. epidermidis competes with opportunistic pathogens and endogenous members of the normal skin flora for niche dominance. In this section, we review some of these competitive interactions and the implications of these interactions for skin health or disease.
Quorum sensing interference
Among the CoNS, one mechanism of competition is through the agr quorum sensing system (BOX 2). This system is conserved across staphylococci120–122 but best characterized in S. aureus, in which it is a master regulator of virulence factors and required for productive skin infection123,124. Most studies have focused on direct interactions among CoNS including S. epidermidis and S. aureus as a means of dampening S. aureus virulence factor production and protecting the skin barrier from damage (inter-species competition)66,125–128. S. epidermidis autoinducing peptide I (AIP-I) was the first identified inter-species inhibitor of S. aureus agr quorum sensing, and it has been postulated that this crosstalk could be one mechanism that enables S. epidermidis domination on healthy skin while excluding S. aureus125.
Box 2 |. The Staphylococcus epidermidis accessory gene regulator quorum sensing system.
The Staphylococcus epidermidis accessory gene regulator (agr) is a conserved two-component quorum sensing system that senses and responds to changes in bacterial population density120,178. Upon reaching sufficient external concentration (a quorum), the signal autoinducing peptide (AIP) binds to the membrane-bound histidine kinase receptor AgrC, which induces receptor dimerization and autophosphorylation followed by subsequent phosphorylation of the response regulator, AgrA120,122 (see the figure, left panel). AgrA binds between chromosomal promoters P2 and P3 to induce transcription of the agrBDCA operon and the major effector transcript RNAIII, which post-transcriptionally regulates a small suite of factors involved in colonization and pathogenesis of the organism120,122. AgrA also directly regulates expression of the immunomodulatory, cytolytic phenol soluble modulins (PSMs)63.
A hyper-variable region spanning agrBDC determines the agr class, or allelic variant, of every S. epidermidis strain (see the figure, right panel). There are four described allelic variants of S. epidermidis, and each senses and responds to its own cognate AIP (denoted type I–type IV)95,129. The structures of S. epidermidis AIP-I, AIP-II and AIP-III (grey) were confirmed by mass spectrometric analysis129, whereas AIP-IV is the most recently discovered AIP and the structure has not yet been confirmed (white)95. Certain S. epidermidis agr classes cross-inhibit non-cognate agr systems via intra-species crosstalk. AIP-I inhibited agr-II and agr-III systems (and vice versa), whereas AIP-II and AIP-III do not cross inhibit, which is likely to be due to similarities in their structures129. Although a functional role for S. epidermidis agr heterogeneity remains to be determined, it is widely speculated that it may enable kin selection via intra-species competition and/or functional advantages in one niche over another95,125.
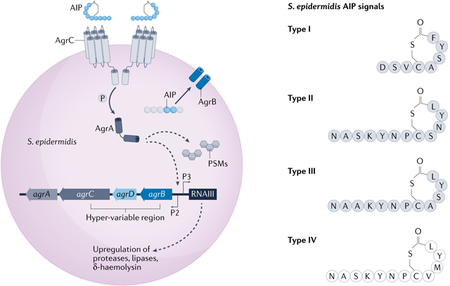
However, just as the agr system is a master virulence factor regulator in S. aureus, it also regulates a smaller but substantial suite of virulence factors in S. epidermidis18. The extracellular cysteine protease EcpA (discussed above) is directly controlled by S. epidermidis agr signalling129. Conversely, agr also regulates potentially important mediators of skin colonization including the glycerol ester hydrolase (lipase geh), PSMs and likely the SepA and Esp proteases129. Functional S. epidermidis agr signalling was also necessary for colonization of porcine skin ex vivo129. Accordingly, agr may have a dual purpose for S. epidermidis colonization and potential for pathogenicity.
Although fewer agr-mediated competitive interactions have been studied between CoNS and S. epidermidis, a 2021 study revealed that Staphylococcus hominis, another ubiquitous skin commensal, produced an AIP that inhibited the S. epidermidis agr quorum sensing system and dampened EcpA production99. This type of competitive quorum sensing interaction, or lack thereof, may be insightful in understanding why S. epidermidis can bloom and exacerbate atopic dermatitis lesions where the diversity of other CoNS and inhibitory AIP molecules is quite low67,99.
Antimicrobial peptides and small molecules
Healthy skin is dominated by many CoNS strains that produce antimicrobials (including bacteriocins), and loss of antimicrobial-producing strains correlates with development of opportunistic S. aureus infections130. Although there is evidence that antimicrobial production is a substantial metabolic burden for the producer131, this conserved mechanism of bacterial competition is an important component of inter-bacterial warfare on skin130,132. Bacteriocins often have a narrow target range, but a few broad-spectrum bacteriocins have been identified in S. epidermidis and other CoNS133–135. Some skin isolates of S. epidermidis produce novel bacteriocins such as epidermicin, which had potent antimicrobial activity against strains of S. aureus and vancomycin-resistant enterococci136. Other strains of S. epidermidis produce lantibiotics (which are bacteriocins that contain a thioether lanthionine ring)132 such as nukacin IVK45 (REF.137) and epilancin 15X (REF.138) that may modulate both the skin and nasal microbiome composition, although more work is needed to understand the distribution and relative abundance of these strains on healthy skin.
Other S. epidermidis molecular products mediate distinct competitive interactions with S. aureus. Some strains of S. epidermidis synthesize and export the purine analogue 6-thioguanine, which inhibited S. aureus growth, de novo purine biosynthesis and virulence factor production and protected skin from dermonecrotic damage139 (FIG. 3b). However, far fewer strains of S. epidermidis contained the biosynthetic pathway for 6-thioguanine compared with other CoNS, which suggests that this may not be a primary mechanism of pathogen exclusion for S. epidermidis139.
Competition with cutibacterium acnes
There is substantial interest in understanding the interactions between Cutibacterium acnes (formerly Propionibacterium acnes) and S. epidermidis given their coexistence in the hair follicle and inconclusive contributions to the pathophysiology of acne vulgaris140–142. C. acnes engages in various strain-dependent antagonistic interactions with S. epidermidis through the production of antimicrobial compounds143 (FIG. 4). C. acnes strains also produce short-chain fatty acids that inhibited S. epidermidis polysaccharide-dependent biofilm formation and sensitized S. epidermidis to antibiotic treatment144. It was discovered in 2020 that the novel C. acnes thiopeptide antibiotic, cutimycin, directly controlled staphylococcal colonization of the hair follicle145. The cutimycin locus was present in the C. acnes population in the majority of a cohort of healthy volunteers and its presence was stable on a specific individual over time145. Sampling of hair follicle content revealed significantly greater abundances of C. acnes compared with S. epidermidis when the locus for cutimycin was present, which suggests a specific role for C. acnes in controlling S. epidermidis growth and niche colonization in the hair follicle.
Fig. 4 |. Staphylococcus epidermidis competes with Cutibacterium acnes in the hair follicle.
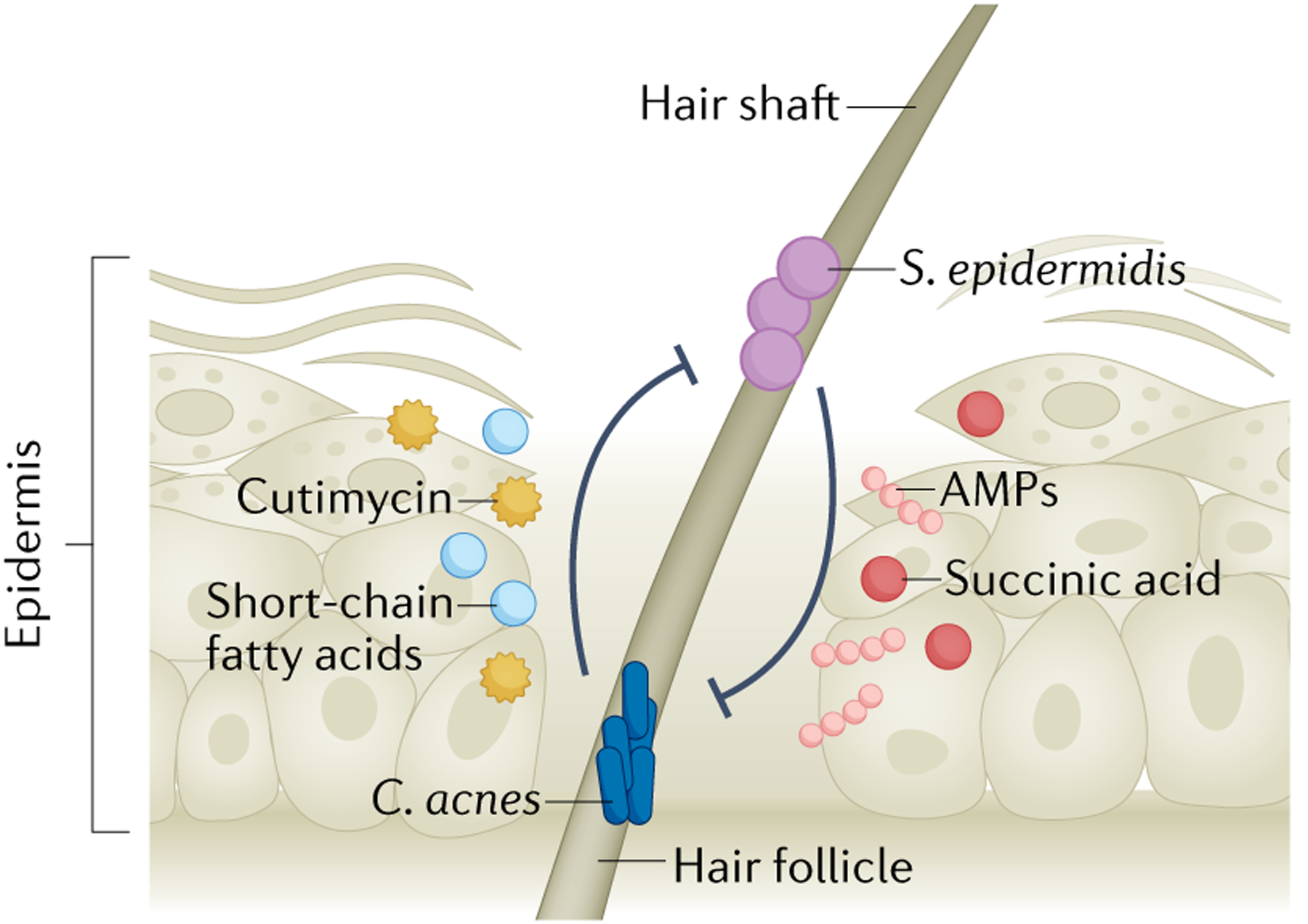
Cutibacterium acnes is a dominant hair follicle colonizer, and its expansion is correlated with progression of the common skin disease acne vulgaris. C. acnes encounters and competes with follicle-resident Staphylococcus epidermidis through production of the antimicrobial peptide (AMP) cutimycin. C. acnes inhibits S. epidermidis biofilm formation and sensitizes S. epidermidis to antibiotic killing through production of several short-chain fatty acids. C. acnes strains may produce other AMPs but this remains unclear. S. epidermidis counters this competition through production of AMPs and fermentation of follicle-available glycerol to multiple short-chain fatty acids, including acetic acid, butyric acid, lactic acid and succinic acid, to suppress C. acnes overgrowth. S. epidermidis strains may also utilize an ESAT6 secretion system to compete with C. acnes (not indicated), but this remains to be experimentally determined.
S. epidermidis–C. acnes competition is not unidirectional, and S. epidermidis directly competes with C. acnes through the fermentation of skin-available glycerol146. S. epidermidis ferments glycerol to short-chain fatty acids, including acetic acid, butyric acid, lactic acid and succinic acid146. However, succinic acid was the most effective inhibitor of C. acnes growth in vitro and in vivo146. As both S. epidermidis and C. acnes can ferment glycerol, sucrose has shown promise as a S. epidermidis-specific fermentation initiator to boost its antimicrobial production against C. acnes147. Supporting a potential anti-inflammatory role in acne, S. epidermidis LTA dampened C. acnes-induced skin inflammation in vivo via TLR2 induction of the keratinocyte small RNA miR-143 (REF.148). In another potential mechanism of competition, some strains of S. epidermidis encode an ESAT6 secretion system, similar to the type seven secretion systems identified in Mycobacterium tuberculosis and S. aureus143. This system might be used to engage in inter-bacterial competition with C. acnes, although an isogenic mutant in the S. epidermidis ESAT6 system is needed to confirm these genotypic findings. Additionally, it is unclear whether this system is only important for competition with C. acnes or whether it could also be used for competition with other organisms143. Together, S. epidermidis may have a beneficial role in containing C. acnes pathogenesis in certain contexts, but skewing conditions to favour S. epidermidis outgrowth in the follicle could also have negative consequences for the host given the intrinsic ability of S. epidermidis to cause accidental or opportunistic infections.
Competition with S. aureus in the nasal cavity
S. aureus is considered a poor or transient healthy skin colonizer and more often asymptomatically colonizes the anterior nares149–151. Prior nasal colonization with S. aureus is strongly associated with autoinfection of distal skin sites149,150,152. However, high nasal colonization with S. epidermidis is associated with lower abundance of opportunistic pathogens153. Much like skin colonization resistance, S. epidermidis actively excludes S. aureus from the nasal environment. Some isolates of S. epidermidis secrete the extracellular serine protease Esp that degrades S. aureus biofilms154. Esp cleaved many S. aureus cell wall-associated proteins in an established biofilm matrix, including the fibronectin binding protein A (FnbA), the fibrinogen-binding protein Efb and the IgG-binding protein (Spa)155. However, S. aureus also encodes the homologous V8 serine protease156. It remains unclear if or how these strains may respond to Esp-mediated clearance and a role for the V8 protease was not addressed in either Esp study. Some strains of S. epidermidis may also compete with S. aureus for binding sites on desquamated nasal epithelial cells (that is, squames)157. The S. epidermidis Aap and S. aureus SasG are homologous, cell surface-associated proteins that individually mediate attachment to nasal squames157. However, a clear role for competition between these two proteins in the nose remains to be determined. Many S. epidermidis strains also induce expression of the AMPs LL-37 and human hBD3 in nasal keratinocytes to kill S. aureus153.
Therapeutic applications
There is growing interest in the usefulness of S. epidermidis as a biotherapeutic, whereby its beneficial attributes could be harnessed to treat dysbiotic skin disorders such as atopic dermatitis, psoriasis or others23,133,141 (TABLE 1). There are several approaches to formulating biotherapeutics, including application of live probiotics, supplementation with prebiotics to support the growth of select organisms or administration of inanimate microbial products via postbiotic therapy158. Clinically, it will be imperative to select the correct strain of S. epidermidis so that the benefits of its anti-inflammatory or immunomodulatory properties outweigh the risks of its potential to harbour or acquire virulence factors. In a small trial, application of lyophilized S. epidermidis on the face of human volunteers significantly boosted facial lipid concentration and moisture retention compared with controls159. Probiotic application of an Esp protease-producing strain of S. epidermidis154 could represent a viable alternative to standard mupirocin treatment160 to clear S. aureus from the nares as long as the growth or potential for pathogenicity of the strain could be managed. One approach for controlling growth of a live biotherapeutic is through nutrient auxotrophy, whereby a specific substrate must be provided in trans for an organism to grow. An alanine auxotroph engineered strain of S. epidermidis recently showed promise for use as a live biotherapeutic whereby bacterial growth and colonization levels could be modulated without the need for antibiotic marker insertion or antibiotic application161.
Table 1 |.
Staphylococcus epidermidis and its molecular products in therapeutic applications
| Application | Treatment | Description | Refs. |
|---|---|---|---|
| Prebiotic | Sucrose | Increased Staphylococcus epidermidis fermentation and production of short-chain fatty acids, which inhibit Cutibacterium acnes | 147 |
| Poly(ethylene glycol) dimethacrylate | Increased S. epidermidis fermentation and production of short-chain fatty acids, which inhibit MRSA | 167 | |
| Colloidal oata | Increased S. epidermidis growth, lactic acid production Increased lactic acid levels on moderate to severe dry skin of human volunteers |
168 | |
| Lactobacillus brevis a | Topically applied cream extract promoted S. epidermidis growth and improved skin barrier function in human volunteers | 163 | |
| Probiotic | Alanine auxotroph | Colonization of human skin explants maintained by addition of alanine Proposed for future clinical studies given safety and no requirement for antibiotics |
161 |
| AMP stimulator | |||
| High dependency on which S. epidermidis strain is applied | Nose153 | ||
| Co-applicationa | Staphylococcus hominis and S. epidermidis lantibiotic strains in formulated lotion decreased S. aureus colonization in atopic dermatitis lesions of human volunteers | 130 | |
| Esp producera | Nasal S. aureus decolonization in human volunteers | 154,155 | |
| 6-HAP producer | Strain-dependent expression, reduced tumour incidence in mice exposed to repeated ultraviolet B radiation | 52 | |
| Postbiotic | Lyophilized bacteriaa | Strains isolated from patients, lyophilized and reapplied in cream Increased skin hydration and lipid content in human volunteers |
159 |
| Bacteriocins and lantibiotics | Selective host range to kill pathogens, but not clinically validated | 132,136,169 | |
| Proteases | |||
| Sphingomyelinase increased skin barrier hydration, and expression was detected on arms/faces of human volunteers | Sphingomyelinase46 | ||
| Small molecules | |||
| 6-Thioguanine inhibited S. aureus purine biosynthesis | 6-Thioguanine139 |
6-HAP, 6-N-hydroxyaminopurine; AIP, autoinducing peptide; AMP, antimicrobial peptide; MRSA, methicillin-resistant Staphylococcus aureus.
Treatment has been performed on human volunteers.
Combinatorial bacteriotherapy is another potential option for patients. A cream mixture of S. epidermidis and S. hominis with antimicrobial activity significantly reduced the S. aureus burden on the skin of five patients with atopic dermatitis after 24 h130. Another group found that combinatorial therapy with S. epidermidis and a skin commensal strain of Staphylococcus cohnii significantly decreased the disease activity index score in germ-free mice that were topically infected with S. aureus162. In the context of re-establishing a disrupted microbiota (that is, prebiotic therapy), a commensal strain of Lactobacillus brevis promoted S. epidermidis recolonization and improved barrier function in a group of patients with severe dry skin163. However, L. brevis is not considered a member of the normal skin flora and the mechanism of this interaction remains unclear. These early studies suggest promising applications for S. epidermidis in the treatment of various skin diseases. Nevertheless, there remain significant challenges to implementation of bacteriotherapy. More work is needed to fully appreciate potential S. epidermidis pathogenicity, especially possession or acquisition of virulence factors as well as broad-spectrum resistance to methicillin and other antibiotics.
Conclusions and future perspectives
S. epidermidis is a complex skin colonizer that mediates positive and negative interactions with the host. Given the rise of antibiotic-resistant infections and the link between many skin diseases and microbial dysbiosis, there is intense interest in harnessing commensals and their molecular products for bacteriotherapies23,158 to treat conditions such as acne141,147, atopic dermatitis23,164–166 or psoriasis158. S. epidermidis is easy to isolate1,12, often stimulates positive immune responses41,52,59 and produces various novel antimicrobials65,134,138. However, S. epidermidis retains considerable potential for pathogenicity through strain-level variation95, genetic flexibility77,84 and production of an array of virulence factors that have been implicated in worsening the pathologies of atopic dermatitis99 and Netherton syndrome98. Aside from S. epidermidis, other members of the CoNS normal community, including S. hominis, have also shown great promise as a source of novel antimicrobials130,133 and for live bacteriotherapy application in atopic dermatitis lesions24. It is time to develop novel therapeutics or probiotics, but we must proceed with caution and respect the unintended consequences of reshaping microorganisms such as S. epidermidis while expecting them to behave predictably.
Acknowledgements
M.M.S. was supported by the National Institute of Allergy and Infectious Diseases (NIAID) Ruth L. Kirschstein National Research Award Predoctoral Fellowship AI157052. A.R.H. was supported by the NIAID grants AI153185 and AI162964 and the US Department of Veteran Affairs grant BX002711.
Glossary
- Stratum corneum
The topmost layer of the epidermis, composed of dead keratinocytes linked together in a ‘brick and mortar’ structure by extracellular lipids.
- Coagulase
A polypeptide secreted by Staphylococcus aureus that promotes blood clotting and was historically used to distinguish this organism from less pathogenic coagulase-negative staphylococci (CoNS) for clinical identification.
- Sortase
An enzyme that links surface proteins to the gram-positive cell wall.
- Corneocytes
Dead keratinocytes that compose the topmost layer of the epidermis.
- Extracellular matrix
A structural support system within the dermal skin layer composed of various proteins including collagen and elastin.
- Sphingomyelinase
A bacterial esterase, often implicated in virulence, that cleaves sphingomyelin.
- Pangenome
The entire set of genes within a species, including conserved, core genes found in every strain and variable genes not conserved in every strain.
- KEGG modules
Families of genes that are linked to specific cellular functions.
- Phase variation
The reversible, heterogeneous switch in gene expression profiles within a clonal population of bacterial cells.
- Plasmacytoid dendritic cells
(pDCs). A specialized subpopulation of dendritic cells involved in immunosurveillance and antigen recognition.
- Corneodesmosomes
The major intercellular adhesives within the stratum corneum.
- Netherton syndrome
A rare genetic disorder characterized by a mutation in a serine protease inhibitor that leads to severe, often life-threatening, skin abnormalities.
- Bacteriocins
Ribosomally synthesized peptides made by many types of bacteria with varying mechanisms of antimicrobial activity.
- Kin selection
The theory that genetic relatedness of a population of cells should lead to more cooperativity and increased fitness of that specific population.
- Probiotics
Live microorganisms applied to re-regulate a dysbiotic community or actively exclude an opportunistic pathogen.
- Prebiotics
Nutrients or other substrates added to alter species composition that favour select microbial growth and exclude opportunistic pathogens.
- Postbiotic
Dead or otherwise inanimate bacteria or a bacterial product applied to promote skin barrier function.
Footnotes
Competing interests
The authors declare no competing interests.
References
- 1.Byrd AL, Belkaid Y & Segre JA The human skin microbiome. Nat. Rev. Microbiol 16, 143–155 (2018). [DOI] [PubMed] [Google Scholar]
- 2.Gallo RL Human skin is the largest epithelial surface for interaction with microbes. J. Invest. Dermatol 137, 1213–1214 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]; This perspective highlights the previously underappreciated large surface area of skin.
- 3.Bay L et al. Universal dermal microbiome in human skin. mBio 11, e02945–19 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nakatsuji T et al. The microbiome extends to subepidermal compartments of normal skin. Nat. Commun 4, 1431 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Scharschmidt TC et al. A wave of regulatory T cells into neonatal skin mediates tolerance to commensal microbes. Immunity 43, 1011–1021 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Naik S et al. The microbiome in patients with atopic dermatitis. J. Allergy Clin. Immunol 143, 26–35 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Naik S et al. Compartmentalized control of skin immunity by resident commensals. Science 337, 1115–1119 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Meisel JS et al. Commensal microbiota modulate gene expression in the skin. Microbiome 6, 20 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Leshem A, Liwinski T & Elinav E Immune–microbiota interplay and colonization resistance in infection. Mol. Cell 78, 597–613 (2020). [DOI] [PubMed] [Google Scholar]
- 10.Parlet CP, Brown MM & Horswill AR Commensal staphylococci influence Staphylococcus aureus skin colonization and disease. Trends Microbiol. 27, 497–507 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Del Rosso J, Zeichner J, Alexis A, Cohen D & Berson D Understanding the epidermal barrier in healthy and compromised skin: clinically relevant information for the dermatology practitioner. J. Clin. Aesthet. Dermatol 9, 2–8 (2011). [PMC free article] [PubMed] [Google Scholar]
- 12.Kloos WE & Schleifer KH Isolation and characterization of staphylococci from human skin. Descriptions of four new species: Staphylococcus warneri, Staphylococcus capitis, Staphylococcus hominis, and Staphylococcus simulans. Int. J. Syst. Bacteriol 25, 62–79 (1975). [Google Scholar]
- 13.Becker K, Heilmann C & Peters G Coagulase-negative staphylococci. Clin. Microbiol. Rev 27, 870–926 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fleer A & Verhoef J New aspects of staphylococcal infections: emergence of coagulase-negative staphylococci as pathogens. Antonie van Leeuwenhoek 50, 72–744 (1984). [DOI] [PubMed] [Google Scholar]
- 15.Rendboe AK et al. The epidome — a species-specific approach to assess the population structure and heterogeneity of Staphylococcus epidermidis colonization and infection. BMC Microbiol. 20, 362 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lamers RP et al. Phylogenetic relationships among Staphylococcus species and refinement of cluster groups based on multilocus data. BMC Evol. Biol 12, 171 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brown MM & Horswill AR Staphylococcus epidermidis — skin friend or foe? PloS Pathog. 16, e1009026 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Otto M Staphylococcus epidermidis — the ‘accidental’ pathogen. Nat. Rev. Microbiol 7, 555–567 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Milisavljevic V et al. Genetic relatedness of Staphylococcus epidermidis from infected infants and staff in the neonatal intensive care unit. Am. J. Infect. Control 33, 341–347 (2005). [DOI] [PubMed] [Google Scholar]
- 20.Vuong C & Otto M Staphylococcus epidermidis infections. Microbes Infect. 4, 481–489 (2002). [DOI] [PubMed] [Google Scholar]
- 21.Oliveira WF et al. Staphylococcus aureus and Staphylococcus epidermidis infections on implants. J. Hosp. Infect 98, 111–117 (2018). [DOI] [PubMed] [Google Scholar]
- 22.Lee JYH et al. Global spread of three multidrug-resistant lineages of Staphylococcus epidermidis. Nat. Microbiol 3, 1175–1185 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nakatsuji T, Cheng JY & Gallo RL Mechanisms for control of skin immune function by the microbiome. Curr. Opin. Immunol 72, 324–330 (2021). [DOI] [PubMed] [Google Scholar]
- 24.Nakatsuji T et al. Development of a human skin commensal microbe for bacteriotherapy of atopic dermatitis and use in a phase 1 randomized clinical trial. Nat. Med 27, 700–709 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]; This study demonstrates the utility of commensal staphylococci as ‘biotherapeutics’ for the common skin ailment atopic dermatitis.
- 25.Chen YE, Fischbach MA & Belkaid Y Skin microbiota–host interactions. Nature 533, 427–436 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Grice EA et al. Topographical and temporal diversity of the human skin microbiome. Science 324, 1190–1192 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Oh J et al. Biogeography and individuality shape function in the human skin metagenome. Nature 514, 59–64 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Oh J et al. Temporal stability of the human skin microbiome. Cell 165, 854–866 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kong HH et al. Temporal shifts in the skin microbiome associated with disease flares and treatment in children with atopic dermatitis. Genome Res. 22, 850–859 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Foster TJ Surface proteins of Staphylococcus epidermidis. Front. Microbiol 11, 1829 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Roy P, Horswill AR & Fey PD Glycan-dependent corneocyte adherence of Staphylococcus epidermidis mediated by the lectin subdomain of Aap. mBio 12, e02908–e02920 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]; This study identifies the specific proteinaceous interaction between S. epidermidis Aap and human corneocytes necessary for colonization of the stratum corneum.
- 32.Macintosh RL et al. The terminal A domain of the fibrillar accumulation-associated protein (Aap) of Staphylococcus epidermidis mediates adhesion to human corneocytes. J. Bacteriol 191, 7007–7016 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Foster TJ The MSCRAMM family of cell-wall-anchored surface proteins of Gram-positive cocci. Trends Microbiol. 27, 927–941 (2019). [DOI] [PubMed] [Google Scholar]
- 34.Trivedi S et al. The surface protein SdrF mediates Staphylococcus epidermidis adherence to keratin. J. Infect. Dis 215, 1846–1854 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mccrea KW et al. The serine-aspartate repeat (Sdr) protein family in Staphylococcus epidermidis. Microbiology 146, 1535–1546 (2000). [DOI] [PubMed] [Google Scholar]
- 36.Büttner H et al. A giant extracellular matrix binding protein of Staphylococcus epidermidis binds surface-immobilized fibronectin via a novel mechanism. mBio 11, e01612–e01620 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hussain M, Heilmann C, Peters G & Herrmann M Teichoic acid enhances adhesion of Staphylococcus epidermidis to immobilized fibronectin. Microb. Pathog 31, 261–270 (2001). [DOI] [PubMed] [Google Scholar]
- 38.Leech JM et al. Toxin-triggered interleukin-1 receptor signaling enables early-life discrimination of pathogenic versus commensal skin bacteria. Cell Host Microbe 26, 1–15 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ali N & Rosenblum MD Regulatory T cells in skin. Immunology 152, 372–381 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Scharschmidt TC et al. Commensal microbes and hair follicle morphogenesis coordinately drive Treg migration into neonatal skin. Cell Host Microbe 21, 467–477.e5 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Constantinides MG et al. MAIT cells are imprinted by the microbiota in early life and promote tissue repair. Science 366, eaax6624 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]; This study identifies that crosstalk between non-classical immune cells and the microbiota is important for tissue development and wound healing.
- 42.Naik S et al. Commensal–dendritic-cell interaction specifies a unique protective skin immune signature. Nature 520, 104–108 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Linehan JL et al. Non-classical immunity controls microbiota impact on skin immunity and tissue repair. Cell 172, 784–796.e18 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Volz T et al. Induction of IL-10-balanced immune profiles following exposure to LTA from Staphylococcus epidermidis. Exp. Dermatol 27, 318–326 (2018). [DOI] [PubMed] [Google Scholar]
- 45.Uberoi A et al. Commensal microbiota regulates skin barrier function and repair via signaling through the aryl hydrocarbon receptor. Cell Host Microbe 29, 1235–1248 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]; This study demonstrates that a defined microbial consortia was sufficient to regulate skin barrier integrity and repair.
- 46.Zheng Y et al. Commensal Staphylococcus epidermidis contributes to skin barrier homeostasis by generating protective ceramides. Cell Host Microbe 30, 1–13 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]; This study identifies a specific S. epidermidis molecular product that protects skin from dehydration.
- 47.Di Domizio J et al. The commensal skin microbiota triggers type I IFN-dependent innate repair responses in injured skin. Nat. Immunol 21, 1034–1045 (2020). [DOI] [PubMed] [Google Scholar]
- 48.Harrison OJ et al. Commensal-specific T cell plasticity promotes rapid tissue adaptation to injury. Science 363, eaat6280 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Belmesk L et al. Prominent role of type 2 immunity in skin diseases — beyond atopic dermatitis. J. Cutan. Med. Surg 26, 33–49 (2021). [DOI] [PubMed] [Google Scholar]
- 50.Kim J, Kim BE & Leung DYM Pathophysiology of atopic dermatitis: clinical implications. Allergy Asthma Proc. 40, 84–92 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Luqman A et al. Trace amines produced by skin bacteria accelerate wound healing in mice. Commun. Biol 3, 277 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nakatsuji T et al. A commensal strain of Staphylococcus epidermidis protects against skin neoplasia. Sci. Adv 4, eaao4502 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Keshari S et al. Butyric acid from probiotic Staphylococcus epidermidis in the skin microbiome down-regulates the ultraviolet-induced pro-inflammatory IL-6 cytokine via short-chain fatty acid receptor. Int. J. Mol. Sci 20, 4477 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Li D et al. Lipopeptide 78 from Staphylococcus epidermidis activates β-catenin to inhibit skin inflammation. J. Immunol 202, 1219–1228 (2019). [DOI] [PubMed] [Google Scholar]
- 55.Lai Y et al. Commensal bacteria regulate Toll-like receptor 3-dependent inflammation after skin injury. Nat. Med 12, 1377–1382 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.García-Gómez E et al. Staphylococcus epidermidis lipoteichoic acid: exocellular release and ltaS gene expression in clinical and commensal isolates. J. Med. Microbiol 66, 864–873 (2017). [DOI] [PubMed] [Google Scholar]
- 57.Hersh AL, Chambers HF, Maselli JH & Gonzales R National trends in ambulatory visits and antibiotic prescribing for skin and soft-tissue infections. Arch. Intern. Med 168, 1585–1591 (2008). [DOI] [PubMed] [Google Scholar]
- 58.Craft KM, Nguyen JM, Berg LJ & Townsend SD Methicillin-resistant Staphylococcus aureus (MRSA): antibiotic-resistance and the biofilm phenotype. Medchemcomm 10, 1231–1241 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wanke I et al. Skin commensals amplify the innate immune response to pathogens by activation of distinct signaling pathways. J. Invest. Dermatol 131, 382–390 (2011). [DOI] [PubMed] [Google Scholar]
- 60.Bitschar K et al. Staphylococcus aureus skin colonization is enhanced by the interaction of neutrophil extracellular traps with keratinocytes. J. Invest. Dermatol 140, 1054–1065.e4 (2020). [DOI] [PubMed] [Google Scholar]
- 61.Rademacher F et al. Staphylococcus epidermidis activates aryl hydrocarbon receptor signaling in human keratinocytes: implications for cutaneous defense. J. Innate Immun 11, 125–135 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Burian M, Bitschar K, Dylus B, Peschel A & Schittek B The protective effect of microbiota on S. aureus skin colonization depends on the integrity of the epithelial barrier. J. Invest. Dermatol 137, 976–979 (2017). [DOI] [PubMed] [Google Scholar]; This short report demonstrates the dual importance of S. epidermidis colonization and barrier integrity in resisting S. aureus skin colonization.
- 63.Peschel A & Otto M Phenol-soluble modulins and staphylococcal infection. Nat. Rev. Microbiol 11, 667–673 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Cogen AL et al. Selective antimicrobial action is provided by phenol-soluble modulins derived from staphylococcus epidermidis, a normal resident of the skin. J. Invest. Dermatol 130, 192–200 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Cogen AL et al. Staphylococcus epidermidis antimicrobial δ-toxin (phenol-soluble modulin-γ) cooperates with host antimicrobial peptides to kill group A Streptococcus. PloS ONE 5, e8557 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Williams MR et al. Quorum sensing between bacterial species on the skin protects against epidermal injury in atopic dermatitis. Sci. Transl Med 11, eaat8329 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Byrd AL et al. Staphylococcus aureus and Staphylococcus epidermidis strain diversity underlying pediatric atopic dermatitis. Sci. Transl Med 9, eaal4651 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]; This study is one of the first to identify that S. epidermidis, akin to S. aureus, can expand in some lesional atopic dermatitis sites.
- 68.Saxena R et al. Comparison of healthy and dandruff scalp microbiome reveals the role of commensals in scalp health. Front. Cell. Infect. Microbiol 8, 364 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Xu Z et al. Dandruff is associated with the conjoined interactions between host and microorganisms. Sci. Rep 6, 24877 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sanders MGH, Nijsten T, Verlouw J, Kraaij R & Pardo LM Composition of cutaneous bacterial microbiome in seborrheic dermatitis patients: a cross-sectional study. PloS ONE 16, e0251136 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Tanaka A et al. Comprehensive pyrosequencing analysis of the bacterial microbiota of the skin of patients with seborrheic dermatitis. Microbiol. Immunol 60, 521–526 (2016). [DOI] [PubMed] [Google Scholar]
- 72.Woo YR, Lee SH, Cho SH, Lee JD & Kim HS Characterization and analysis of the skin microbiota in rosacea: impact of systemic antibiotics. J. Clin. Med 9, 185 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Holmes AD Potential role of microorganisms in the pathogenesis of rosacea. J. Am. Acad. Dermatol 69, 1025–1032 (2013). [DOI] [PubMed] [Google Scholar]
- 74.Kim HS Microbiota in rosacea. Am. J. Clin. Dermatol 21, 25–35 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Schilcher K & Horswill AR Staphylococcal biofilm development: structure, regulation, and treatment strategies. Microbiol. Mol. Biol. Rev 84, e00026–19 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Joubert IA, Otto M, Strunk T & Currie AJ Look who’s talking: host and pathogen drivers of Staphylococcus epidermidis virulence in neonatal sepsis. Int. J. Mol. Sci 23, 860 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Both A et al. Distinct clonal lineages and within-host diversification shape invasive Staphylococcus epidermidis populations. PloS Pathog. 17, e1009304 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]; This study utilizes patient-matched isolates to demonstrate S. epidermidis genetic flexibility and adaptation to various environments.
- 78.Miragaia M, Thomas JC, Couto I, Enright MC & De Lencastre H Inferring a population structure for Staphylococcus epidermidis from multilocus sequence typing data. J. Bacteriol 189, 2540–2552 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Mendes RE, Deshpande LM, Costello AJ & Farrell DJ Molecular epidemiology of Staphylococcus epidermidis clinical isolates from US hospitals. Antimicrob. Agents Chemother 56, 4656–4661 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Conlan S et al. Staphylococcus epidermidis pan-genome sequence analysis reveals diversity of skin commensal and hospital infection-associated isolates. Genome Biol. 13, R64 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]; This study increases the number of publicly available S. epidermidis draft genomes and highlights genetic differences between commensal and nosocomial isolates.
- 81.Du X et al. Staphylococcus epidermidis clones express Staphylococcus aureus-type wall teichoic acid to shift from a commensal to pathogen lifestyle. Nat. Microbiol 6, 757–768 (2021). [DOI] [PubMed] [Google Scholar]
- 82.Fišarová L et al. Staphylococcus epidermidis phages transduce antimicrobial resistance plasmids and mobilize chromosomal islands. mSphere 6, e00223–21 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Thakuria B & Lahon K The β-lactam antibiotics as an empirical therapy in a developing country: an update on their current status and recommendations to counter the resistance against them. J. Clin. Diagn. Res 7, 1207–1214 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ray MD, Boundy S & Archer GL Transfer of the methicillin resistance genomic island among staphylococci by conjugation. Mol. Microbiol 100, 675–685 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Krediet TG et al. Molecular epidemiology of coagulase-negative staphylococci causing sepsis in a neonatal intensive care unit over an 11-year period. J. Clin. Microbiol 42, 992–995 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Månsson E et al. Methicillin-resistant Staphylococcus epidermidis lineages in the nasal and skin microbiota of patients planned for arthroplasty surgery. Microorganisms 9, 1–14 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Mé Ric G et al. Ecological overlap and horizontal gene transfer in Staphylococcus aureus and Staphylococcus epidermidis. Genome Biol. Evol 7, 1313–1328 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Datta MS et al. Rapid methicillin resistance diversification in Staphylococcus epidermidis colonizing human neonates. Nat. Commun 12, 6062 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Salgueiro VC et al. High rate of neonates colonized by methicillin-resistant Staphylococcus species in an intensive care unit. J. Infect. Dev. Ctries 13, 810–816 (2019). [DOI] [PubMed] [Google Scholar]
- 90.Barbier F et al. Methicillin-resistant coagulase-negative staphylococci in the community: high homology of SCCmec Iva between Staphylococcus epidermidis and major clones of methicillin-resistant Staphylococcus aureus. J. Infect. Dis 202, 270–281 (2010). [DOI] [PubMed] [Google Scholar]
- 91.Miragaia M et al. Genetic diversity of arginine catabolic mobile element in Staphylococcus epidermidis. PloS ONE 4, e7722 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Hung WC et al. Skin commensal staphylococci may act as reservoir for fusidic acid resistance genes. PloS ONE 10, e0143106 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Banaszkiewicz S et al. Genetic diversity of composite enterotoxigenic Staphylococcus epidermidis pathogenicity islands. Genome Biol. Evol 11, 3498–3509 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Argemi X, Hansmann Y, Prola K & Prévost G Coagulase-negative staphylococci pathogenomics. Int. J. Mol. Sci 20, 1215 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Zhou W et al. Host-specific evolutionary and transmission dynamics shape the functional diversification of Staphylococcus epidermidis in human skin. Cell 180, 454–470.e18 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]; This study is the first longitudinal, multi-patient assessment of S. epidermidis strain-level diversity on healthy skin.
- 96.Méric G et al. Disease-associated genotypes of the commensal skin bacterium Staphylococcus epidermidis. Nat. Commun 9, 5034 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Harris S et al. Evolution of MRSA during hospital transmission and intercontinental spread. Science 327, 466–469 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Williams MR et al. Interplay of staphylococcal and host proteases promotes skin barrier disruption in Netherton syndrome. Cell Rep. 30, 2923–2933.e7 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Cau L et al. Staphylococcus epidermidis protease EcpA can be a deleterious component of the skin microbiome in atopic dermatitis. J. Allergy Clin. Immunol 147, 955–966.e16 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]; This study implicates the production of the S. epidermidis EcpA protease in the disruption and exacerbation of atopic skin lesions.
- 100.Otto M Staphylococcal biofilms. Curr. Top. Microbiol. Immunol 322, 207–228 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Paharik AE & Horswill AR The staphylococcal biofilm: adhesins, regulation, and host response. Microbiol. Spectr 4, 529–566 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Schaeffer CR et al. Accumulation-associated protein enhances Staphylococcus epidermidis biofilm formation under dynamic conditions and is required for infection in a rat catheter model. Infect. Immun 83, 214–226 (2015).103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Schaeffer CR et al. Versatility of biofilm matrix molecules in Staphylococcus epidermidis clinical isolates and importance of polysaccharide intercellular adhesin expression during high shear stress. mSphere 1, e00165–16 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Fey PD & Olson ME Current concepts in biofilm formation of Staphylococcus epidermidis. Fut. Microbiol 5, 917–933 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.O’Gara JP Into the storm: chasing the opportunistic pathogen Staphylococcus aureus from skin colonisation to life-threatening infections. Environ. Microbiol 19, 3823–3833 (2017). [DOI] [PubMed] [Google Scholar]
- 106.Kozitskaya S et al. Clonal analysis of Staphylococcus epidermidis isolates carrying or lacking biofilm-mediating genes by multilocus sequence typing. J. Clin. Microbiol 43, 4751–4757 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Kozitskaya S et al. The bacterial insertion sequence element IS256 occurs preferentially in nosocomial Staphylococcus epidermidis isolates: association with biofilm formation and resistance to aminoglycosides. Infect. Immun 72, 1210–1215 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Rogers KL, Rupp ME & Fey PD The presence of icaADBC is detrimental to the colonization of human skin by Staphylococcus epidermidis. Appl. Environ. Microbiol 74, 6155–6157 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Paharik AE et al. The metalloprotease SepA governs processing of accumulation-associated protein and shapes intercellular adhesive surface properties in Staphylococcus epidermidis. Mol. Microbiol 103, 860–874 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Rohde H et al. Induction of Staphylococcus epidermidis biofilm formation via proteolytic processing of the accumulation-associated protein by staphylococcal and host proteases. Mol. Microbiol 55, 1883–1895 (2005). [DOI] [PubMed] [Google Scholar]
- 111.Conrady DG et al. A zinc-dependent adhesion module is responsible for intercellular adhesion in staphylococcal biofilms. Proc. Natl Acad. Sci. USA 105, 19456–19461 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Formosa-Dague C, Speziale P, Foster TJ, Geoghegan JA & Dufrêne YF Zinc-dependent mechanical properties of Staphylococcus aureus biofilm-forming surface protein SasG. Proc. Natl Acad. Sci. USA 113, 410–415 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Salava A & Lauerma A Role of the skin microbiome in atopic dermatitis. Clin. Transl Allergy 4, 1–6 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Meylan P et al. Skin colonization by Staphylococcus aureus precedes the clinical diagnosis of atopic dermatitis in infancy. J. Invest. Dermatol 137, 2497–2504 (2017). [DOI] [PubMed] [Google Scholar]
- 115.Allen HB et al. The presence and impact of biofilm-producing staphylococci in atopic dermatitis. JAMA Dermatol. 150, 260–265 (2014). [DOI] [PubMed] [Google Scholar]
- 116.Allen HB, Mueller JL & Herbert Allen CB A novel finding in atopic dermatitis: film-producing Staphylococcus epidermidis as an etiology. Int. J. Dermatol 50, 992–993 (2011). [DOI] [PubMed] [Google Scholar]
- 117.Gonzalez T et al. Biofilm propensity of Staphylococcus aureus skin isolates is associated with increased atopic dermatitis severity and barrier dysfunction in the MPAACH pediatric cohort. Allergy 76, 302–313 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]; This work presents a clinically robust and mechanistic assessment of paediatric atopic dermatitis development and progression in a US cohort.
- 118.Yarwood JM, Bartels DJ, Volper EM & Greenberg EP Quorum sensing in Staphylococcus aureus biofilms. J. Bacteriol 186, 1838–1850 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Kavanaugh JS & Horswill AR Impact of environmental cues on staphylococcal quorum sensing and biofilm development. J. Biol. Chem 291, 12556–12564 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Thoendel M, Kavanaugh JS, Flack CE & Horswill AR Peptide signaling in the staphylococci. Chem. Rev 111, 117–151 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Novick RP & Geisinger E Quorum sensing in staphylococci. Annu. Rev. Genet 42, 541–564 (2008). [DOI] [PubMed] [Google Scholar]
- 122.Yang T, Tal-Gan Y, Paharik AE, Horswill AR & Blackwell HE Structure–function analyses of a Staphylococcus epidermidis autoinducing peptide reveals motifs critical for AgrC-type receptor modulation. ACS Chem. Biol 11, 1982–1991 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Wright JS, Jin R & Novick RP Transient interference with staphylococcal quorum sensing blocks abscess formation. Proc. Natl Acad. Sci. USA 102, 1691–1696 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Nakamura Y et al. Staphylococcus Agr virulence is critical for epidermal colonization and associates with atopic dermatitis development. Sci. Transl Med 12, eaay4068 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Otto M, Echner H, Voelter W & Götz F Pheromone cross-inhibition between Staphylococcus aureus and Staphylococcus epidermidis. Infect. Immun 69, 1957–1960 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Brown MM et al. Novel peptide from commensal Staphylococcus simulans blocks methicillin-resistant Staphylococcus aureus quorum sensing and protects host skin from damage. Antimicrob. Agents Chemother 64, e00172–20 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Khan BA, Yeh AJ, Cheung GY & Otto M Investigational therapies targeting quorum-sensing for the treatment of Staphylococcus aureus infections. Expert Opin. Investig. Drugs 24, 689–704 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Harraghy N, Kerdudou S & Herrmann M Quorum-sensing systems in staphylococci as therapeutic targets. Anal. Bioanal. Chem 387, 437–444 (2007). [DOI] [PubMed] [Google Scholar]
- 129.Olson ME et al. Staphylococcus epidermidis agr quorum-sensing system: signal identification, cross talk, and importance in colonization. J. Bacteriol 196, 3482–3493 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Nakatsuji T et al. Antimicrobials from human skin commensal bacteria protect against Staphylococcus aureus and are deficient in atopic dermatitis. Sci. Transl Med 9, eaah4680 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Ebner P et al. Lantibiotic production is a burden for the producing staphylococci. Sci. Rep 8, 7471 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Heilbronner S, Krismer B, Brötz-Oesterhelt H & Peschel A The microbiome-shaping roles of bacteriocins. Nat. Rev. Microbiol 19, 726–739 (2021). [DOI] [PubMed] [Google Scholar]
- 133.Liu Y et al. Skin microbiota analysis-inspired development of novel anti-infectives. Microbiome 8, 85 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.O’Sullivan JN, Rea MC, O’Connor PM, Hill C & Ross RP Human skin microbiota is a rich source of bacteriocin-producing staphylococci that kill human pathogens. FEMS Microbiol. Ecol 95, fiy241 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Hibbing ME, Fuqua C, Parsek MR & Peterson SB Bacterial competition: surviving and thriving in the microbial jungle. Nat. Rev. Microbiol 8, 15–25 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Sandiford S & Upton M Identification, characterization, and recombinant expression of epidermicin NI01, a novel unmodified bacteriocin produced by Staphylococcus epidermidis that displays potent activity against staphylococci. Antimicrob. Agents Chemother 56, 1539–1547 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Janek D, Zipperer A, Kulik A, Krismer B & Peschel A High frequency and diversity of antimicrobial activities produced by nasal Staphylococcus strains against bacterial competitors. PloS Pathog. 12, e1005812 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Ekkelenkamp MB et al. Isolation and structural characterization of epilancin 15X, a novel lantibiotic from a clinical strain of Staphylococcus epidermidis. FEBS Lett. 579, 1917–1922 (2005). [DOI] [PubMed] [Google Scholar]
- 139.Chin D et al. Coagulase-negative staphylococci release a purine analog that inhibits Staphylococcus aureus virulence. Nat. Commun 12, 1887 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Claudel JP et al. Staphylococcus epidermidis: a potential new player in the physiopathology of acne? Dermatology 235, 287–294 (2019). [DOI] [PubMed] [Google Scholar]
- 141.Woo TE & Sibley CD The emerging utility of the cutaneous microbiome in the treatment of acne and atopic dermatitis. J. Am. Acad. Dermatol 82, 222–228 (2020). [DOI] [PubMed] [Google Scholar]
- 142.Fitz-Gibbon S et al. Propionibacterium acnes strain populations in the human skin microbiome associated with acne. J. Invest. Dermatol 133, 2152–2160 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Christensen GJM et al. Antagonism between Staphylococcus epidermidis and Propionibacterium acnes and its genomic basis. BMC Genomics 17, 152 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Nakamura K et al. Short chain fatty acids produced by Cutibacterium acnes inhibit biofilm formation by Staphylococcus epidermidis. Sci. Rep 10, 21237 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Claesen J et al. A Cutibacterium acnes antibiotic modulates human skin microbiota composition in hair follicles. Sci. Transl Med 12, 5445 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]; This study identifies a novel antimicrobial made by C. acnes in the hair follicle to facilitate inter-bacterial competition.
- 146.Wang Y et al. Staphylococcus epidermidis in the human skin microbiome mediates fermentation to inhibit the growth of Propionibacterium acnes: implications of probiotics in acne vulgaris. Appl. Microbiol. Biotechnol 98, 411–424 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Wang Y et al. A precision microbiome approach using sucrose for selective augmentation of Staphylococcus epidermidis fermentation against Propionibacterium acnes. Int. J. Mol. Sci 17, 1870 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Xia X et al. Staphylococcal LTA-induced miR-143 inhibits Propionibacterium acnes-mediated inflammatory response in skin. J. Invest. Dermatol 136, 621–630 (2016). [DOI] [PubMed] [Google Scholar]
- 149.Sakr A, Brégeon F, Mège JL, Rolain JM & Blin O Staphylococcus aureus nasal colonization: an update on mechanisms, epidemiology, risk factors, and subsequent infections. Front. Microbiol 9, 2419 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Totté JEE et al. Prevalence and odds of Staphylococcus aureus carriage in atopic dermatitis: a systematic review and meta-analysis. Br. J. Dermatol 175, 687–695 (2016). [DOI] [PubMed] [Google Scholar]
- 151.Krismer B, Weidenmaier C, Zipperer A & Peschel A The commensal lifestyle of Staphylococcus aureus and its interactions with the nasal microbiota. Nat. Rev. Microbiol 15, 675–687 (2017). [DOI] [PubMed] [Google Scholar]
- 152.Oh J, Conlan S, Polley EC, Segre JA & Kong HH Shifts in human skin and nares microbiota of healthy children and adults. Genome Med. 4, 1–11 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Liu Q et al. Staphylococcus epidermidis contributes to healthy maturation of the nasal microbiome by stimulating antimicrobial peptide production. Cell Host Microbe 27, 1–11 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Iwase T et al. Staphylococcus epidermidis Esp inhibits Staphylococcus aureus biofilm formation and nasal colonization. Nature 465, 346–349 (2010). [DOI] [PubMed] [Google Scholar]
- 155.Sugimoto S et al. Staphylococcus epidermidis Esp degrades specific proteins associated with Staphylococcus aureus biofilm formation and host–pathogen interaction. J. Bacteriol 195, 1645–1655 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Chen C et al. Secreted proteases control autolysin-mediated biofilm growth of Staphylococcus aureus. J. Biol. Chem 288, 29440–29452 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Roche FM, Meehan M & Foster TJ The Staphylococcus aureus surface protein SasG and its homologues promote bacterial adherence to human desquamated nasal epithelial cells. Microbiology (Reading) 149, 2759–2767 (2003). [DOI] [PubMed] [Google Scholar]
- 158.McLoughlin IJ, Wright EM, Tagg JR, Jain R & Hale JDF Skin microbiome — the next frontier for probiotic intervention. Probiotics Antimicrob. Proteins 10.1007/s12602-021-09824-1 (2021). [DOI] [PubMed] [Google Scholar]
- 159.Nodake Y et al. Pilot study on novel skin care method by augmentation with Staphylococcus epidermidis, an autologous skin microbe — a blinded randomized clinical trial. J. Dermatol. Sci 79, 119–126 (2015). [DOI] [PubMed] [Google Scholar]
- 160.Sakr A, Brégeon F, Rolain JM & Blin O Staphylococcus aureus nasal decolonization strategies: a review. Expert Rev. Anti Infect. Ther 17, 327–340 (2019). [DOI] [PubMed] [Google Scholar]
- 161.Dodds D et al. Controlling the growth of the skin commensal Staphylococcus epidermidis using d-alanine auxotrophy. mSphere 5, e00360–20 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Ito Y et al. Staphylococcus cohnii is a potentially biotherapeutic skin commensal alleviating skin inflammation. Cell Rep. 35, 109052 (2021). [DOI] [PubMed] [Google Scholar]
- 163.Holz C et al. Novel bioactive from Lactobacillus brevis DSM17250 to stimulate the growth of Staphylococcus epidermidis: a pilot study. Benef. Microbes 8, 121–131 (2017). [DOI] [PubMed] [Google Scholar]
- 164.Williams MR & Gallo RL Evidence that human skin microbiome dysbiosis promotes atopic dermatitis. J. Invest. Dermatol 137, 2460–2461 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Myles IA et al. First-in-human topical microbiome transplantation with Roseomonas mucosa for atopic dermatitis. JCI Insight 3, e120608 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Volz T et al. Nonpathogenic bacteria alleviating atopic dermatitis inflammation induce IL-10-producing dendritic cells and regulatory Tr1 cells. J. Invest. Dermatol 134, 96–104 (2014). [DOI] [PubMed] [Google Scholar]
- 167.Kao MS et al. Microbiome precision editing: using PEG as a selective fermentation initiator against methicillin-resistant Staphylococcus aureus. Biotechnol. J 12, 1600399 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Liu-Walsh F et al. Prebiotic colloidal oat supports the growth of cutaneous commensal bacteria including S. epidermidis and enhances the production of lactic acid. Clin. Cosmet. Investig. Dermatol 14, 73–82 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Götz F, Perconti S, Popella P, Werner R & Schlag M Epidermin and gallidermin: staphylococcal lantibiotics. Int. J. Med. Microbiol 304, 63–71 (2014). [DOI] [PubMed] [Google Scholar]
- 170.Schoenfelder SMK et al. Success through diversity — how Staphylococcus epidermidis establishes as a nosocomial pathogen. Int. J. Med. Microbiol 300, 380–386 (2010). [DOI] [PubMed] [Google Scholar]
- 171.Ferretti P et al. Experimental metagenomics and ribosomal profiling of the human skin microbiome. Exp. Dermatol 26, 211–219 (2017). [DOI] [PubMed] [Google Scholar]
- 172.Van Rossum T, Ferretti P, Maistrenko OM & Bork P Diversity within species: interpreting strains in microbiomes. Nat. Rev. Microbiol 18, 491–506 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Dengler Haunreiter V et al. In-host evolution of Staphylococcus epidermidis in a pacemaker-associated endocarditis resulting in increased antibiotic tolerance. Nat. Commun 10, 1149 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Gu J et al. Bacterial insertion sequence IS256 as a potential molecular marker to discriminate invasive strains from commensal strains of Staphylococcus epidermidis. J. Hosp. Infect 61, 342–348 (2005). [DOI] [PubMed] [Google Scholar]
- 175.Lee JYH et al. Mining the methylome reveals extensive diversity in Staphylococcus epidermidis restriction modification. mBio 10, e02451–19 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Marraffini L & Sontheimer E CRISPR interference limits horizontal gene transfer in staphylococci by targeting DNA. Science 322, 1843–1845 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Rossi CC, Souza-Silva T, Araújo-Alves AV & Giambiagi-deMarval M CRISPR–Cas systems features and the gene-reservoir role of coagulase-negative staphylococci. Front. Microbiol 8, 1545 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Lyon GJ & Novick RP Peptide signaling in Staphylococcus aureus and other Gram-positive bacteria. Peptides 25, 1389–1403 (2004). [DOI] [PubMed] [Google Scholar]


