Abstract
The absorption, distribution, metabolism, excretion, and toxicity (ADMET) properties are important in drug discovery as they define efficacy and safety. In this work, we applied an ensemble of features, including fingerprints and descriptors, and a tree-based machine learning model, extreme gradient boosting, for accurate ADMET prediction. Our model performs well in the Therapeutics Data Commons ADMET benchmark group. For 22 tasks, our model is ranked first in 18 tasks and top 3 in 21 tasks. The trained machine learning models are integrated in ADMETboost, a web server that is publicly available at https://ai-druglab.smu.edu/admet.
Keywords: ADMET, Machine learning, XGBoost, Web server
1. Introduction
Properties such as absorption, distribution, metabolism, excretion, and toxicity (ADMET) are important in small molecule drug discovery and therapeutics. It was reported that many clinical trials fail due to the deficiencies in ADMET properties. Kola and Landis (2004); Kennedy (1997); M Honorio et al (2013); Waring et al (2015) While profiling ADMET in the early stage of drug discovery is desirable, experimental evaluation of ADMET properties is costly with limited available data. Moreover, computational studies of ADMET in the clinical trial stage can serve as an efficient design strategy that can allow researchers pay more attention to the most promising compounds. Göller et al (2020)
Recent developments in machine learning (ML) promote research in chemistry and biology Song et al (2020); Zhang et al (2020); Tian et al (2021b) and bring new opportunities for ADMET prediction. ADMETLab Dong et al (2018) provides 31 ADMET endpoints with six machine learning models, and further advanced to 53 endpoints using a multi-task graph attention network Xiong et al (2021). vNN Schyman et al (2017) is a web server that applies variable nearest neighborhood to predict 15 ADMET properties. admetSAR Cheng et al (2012) and admetSAR 2.0 Yang et al (2019) are also ML based web servers for drug discovery or environmental risk assessment with random forest, support vector machine, and k-nearest neighbors models. As a fingerprint-based random forest model, FP-ADMET Venkatraman (2021) evaluated over 50 ADMET and ADMET-related tasks. In these ML models, small molecules are provided in SMILES representations and further featurized using fingerprints, such as extended connectivity fingerprints Rogers and Hahn (2010) and Molecular ACCess System (MACCS) fingerprints Durant et al (2002). Beside these, there are many other fingerprints and descriptors that can be used for ADMET prediction, such as PubChem fingerprints and Mordred descriptors. Taking advantage of all possible features enables sufficient learning process for machine learning models.
One common issue is that many machine learning models in previous work are trained on different datasets, which leads to unfair comparison and evaluation of ML models. As a curated dataset, Therapeutics Data Commons (TDC) Huang et al (2021) unifies resources in therapeutics for systematic access and evaluation. There are 22 tasks in TDC ADMET benchmark group, each with small molecules SMILES representations and corresponding ADMET property values or labels.
Extreme gradient boosting (XGBoost) Chen and Guestrin (2016) is a powerful machine learning model and has been shown to be effective in regression and classification tasks in biology and chemistry Chen et al (2020); Tian et al (2020, 2021a); Deng et al (2021). In this work, we applied XGBoost to learn a feature ensemble, including multiple fingerprints and descriptors, for accurate ADMET prediction. Our model performs well in the TDC ADMET benchmark group with 11 tasks ranked first and 19 tasks ranked top 3.
2. Methods
Therapeutics Data Commons
Therapeutics Data Commons (v0.3.6) is a Python library with an open-science initiative. It holds many therapeutics tasks and datasets including target discovery, activity modeling, efficacy, safety, and manufacturing. TDC provides a unified and meaningful benchmark for fair comparison between different machine learning models. For each ADMET prediction task, TDC splits the dataset into the predefined 80% training set and 20% test set with scaffold split, which simulates the real-world application scenario. In practice, a well-trained machine learning model would be used to predict ADMET properties on unseen and structurally different drugs.
Fingerprints and Descriptors
Six featurizers from DeepChem Ramsundar et al (2019) were used to compute fingerprints and descriptors:
MACCS fingerprints are common structural keys that compute a binary string based on a molecule’s structural features.
Extended connectivity fingerprints compute a bit vector by breaking up a molecule into circular neighborhoods. They are widely used for structure-activity modeling.
Mol2Vec fingerprints Jaeger et al (2018) create vector representations of molecules based on an unsupervised machine learning approach.
PubChem fingerprints consist of 881 structural keys that cover a wide range of substructures and features. It is used by PubChem for similarity searching.
Mordred descriptors Moriwaki et al (2018) calculate a set of chemical descriptors such as the count of aromatic atoms or all halogen atoms.
RDKit descriptors calculate a set of chemical descriptors such as molecular weight and the number of radical electrons.
Extreme Gradient Boosting
Extreme gradient boosting is a powerful machine learning model. It boosts model performance through ensemble that includes decision tree models trained in sequence.
Let represents a training set with m features and n labels. The j-th decision tree in XGBoost model makes a prediction for sample (xi, yi) by gj(xi) = wq(xi) where wq is the leaf weights. The final prediction of XGBoost model is the sum of all M decision tree predictions with .
The objective function consists of a loss function l and a regularization term Ω to reduce overfitting:
| (1) |
where . T represents the number of leaves while γ, λ are parameters for regularization.
During training, XGBoost iteratively trains a new decision tree based on the output of the previous tree. The prediction of the t-th iteration . The objective function of the t-th iteration is:
| (2) |
XGBoost introduces first and second derivatives of this objective function, which can be expressed as follows by applying Taylor expansion at second order:
| (3) |
A total of seven parameters are being fine-tuned with selected value options and are listed in Table 1. Default values are used for other parameters.
Table 1.
Fine-tuned XGBoost Parameters
| Name and Description | Values |
|---|---|
| n_estimators: Number of gradient boosted trees. | [50, 100, 200, 500, 1000] |
| max_depth: Maximum tree depth. | [3, 4, 5, 6, 7] |
| learning_rate: Boosting learning rate. | [0.01, 0.05, 0.1, 0.2, 0.3] |
| subsample: Subsample ratio of instances. | [0.5, 0.6, 0.7, 0.8, 0.9, 1.0] |
| colsample_bytree: Subsample ratio of columns. | [0.5, 0.6, 0.7, 0.8, 0.9, 1.0] |
| reg_alpha: L1 regularization weights. | [0, 0.1, 1, 5, 10] |
| reg_lambda: L2 regularization weights. | [0, 0.1, 1, 5, 10] |
Performance Criteria
For regression tasks, mean absolute error (MAE) and Spearman’s correlation coefficient are considered to evaluate model performance:
MAE is used to measure the deviation between predictions yi and real values xi in n sample size.
| (4) |
Spearman’s correlation coefficient ρ measures the correlation strength between two ranked variables. Where di represents the difference in paired ranks,
| (5) |
For binary classification tasks, area under curve (AUC) is calculated with receiver operating characteristic (ROC) and precision-recall curve (PRC). For both metrics, a higher value indicates a more powerful model.
AUROC is the area under the curve where x-axis is false positive rate and y-axis is true positive rate.
AUPRC is the area under the curve where x-axis is recall and y-axis is precision.
All metrics are calculated with evaluation functions provided by the TDC APIs.
3. Results and Discussion
Model Performance
We first used a random seed to split the overall dataset into a training set (80%) and a test set (20%). XGBoost model was trained with the training set using 5-fold cross validation (CV). A randomized grid search CV was applied to optimize hyperparameters. The parameter set with the highest CV score is used, and the model performance is evaluated on the test set. We repeat this process five times with varying random seeds from zero to four following the TDC guideline. The evaluation results are listed on Table 2. In each task, there are at least seven other models or featurization methods being compared with, including DeepPurpose Huang et al (2020), AttentiveFP Xiong et al (2019), ContextPred Hu et al (2019), NeuralFP Lee et al (2021), AttrMasking Hu et al (2019) and graph convolutional network Kipf and Welling (2016). For all 22 tasks, XGBoost is ranked first for 18 and top 3 for 21 out of 22 tasks, demonstrating the success of XGBoost model in predicting ADMET tasks.
Table 2.
Model Evaluation on the TDC ADMET Leaderboarda
| TDC | Current Top 1 | XGBoost | |||
|---|---|---|---|---|---|
| Task | Metric | Method | Score | Score | Rank |
| Absorption | |||||
| Caco2 | MAE | RDKit2D + MLP | 0.393 ± 0.024 | 0.288 ± 0.011 | 1st |
| HIA | AUROC | AttrMasking | 0.978 ± 0.006 | 0.987 ± 0.002 | 1st |
| Pgp | AUROC | AttrMasking | 0.929 ± 0.006 | 0.911 ± 0.002 | 4th |
| Bioav | AUROC | RDKit2D + MLP | 0.672 ± 0.021 | 0.700 ± 0.010 | 1st |
| Lipo | MAE | ContextPred | 0.535 ± 0.012 | 0.533 ± 0.005 | 1st |
| AqSol | MAE | AttentiveFP | 0.776 ± 0.008 | 0.727 ± 0.004 | 1st |
| Distribution | |||||
| BBB | AUROC | ContextPred | 0.897 ± 0.004 | 0.905 ± 0.001 | 1st |
| PPBR | MAE | NeuralFP | 9.292 ± 0.384 | 8.251 ± 0.115 | 1st |
| VDss | Spearman | RDKit2D + MLP | 0.561 ± 0.025 | 0.612 ± 0.018 | 1st |
| Metabolism | |||||
| CYP2C9 Inhibition | AUPRC | AttentiveFP | 0.749 ± 0.004 | 0.794 ± 0.004 | 1st |
| CYP2D6 Inhibition | AUPRC | AttentiveFP | 0.646 ± 0.014 | 0.721 ± 0.003 | 1st |
| CYP3A4 Inhibition | AUPRC | AttentiveFP | 0.851 ± 0.006 | 0.877 ± 0.002 | 1st |
| CYP2C9 Substrate | AUPRC | Morgan + MLP | 0.380 ± 0.015 | 0.387 ± 0.018 | 1st |
| CYP2D6 Substrate | AUPRC | RDKit2D + MLP | 0.677 ± 0.047 | 0.648 ± 0.023 | 3rd |
| CYP3A4 Substrate | AUPRC | CNN | 0.662 ± 0.031 | 0.680 ± 0.005 | 1st |
| Excretion | |||||
| Half Life | Spearman | Morgan + MLP | 0.329 ± 0.083 | 0.396 ± 0.027 | 1st |
| CL-Hepa | Spearman | ContextPred | 0.439 ± 0.026 | 0.420 ± 0.011 | 2nd |
| CL-Micro | Spearman | RDKit2D + MLP | 0.586 ± 0.014 | 0.587 ± 0.006 | 1st |
| Toxicity | |||||
| LD50 | MAE | Morgan + MLP | 0.649 ± 0.019 | 0.602 ± 0.006 | 1st |
| hERG | AUROC | RDKit2D + MLP | 0.841 ± 0.020 | 0.806 ± 0.005 | 3rd |
| Ames | AUROC | AttrMasking | 0.842 ± 0.008 | 0.859 ± 0.002 | 1st |
| DILI | AUROC | AttrMasking | 0.919 ± 0.008 | 0.933 ± 0.011 | 1st |
Only models that have been evaluated by most of the tasks are considered.
The superior prediction results of XGBoost are explainable. As shown in Table 2, previously, there are 13 tasks which the top models are trained using descriptors (RDKit 2D + MLP model) or fingerprints (Morgan + MLP model and AttentiveFP). Inspired by this, XGBoost was trained using a combination of fingerprints and descriptors. These featurization methods cover both structural features (MACCS, extended connectivity, Mol2Vec, and PubChem fingerprints) to chemical descriptors (Mordred and RDKit descriptors) for each given SMILES representation. For a specific property prediction task, XGBoost can take the consideration of all possible molecular features, select the best set of them for prediction, while avoiding over-fitting by controlling tree complexity. Together, these would boost the prediction performance of XGBoost to be superior to other models.
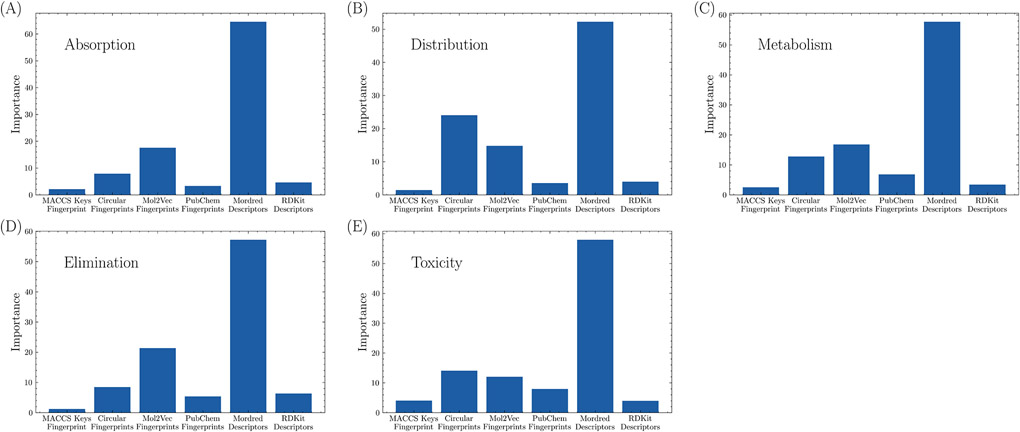
To further understand the importance of each fingerprint and descriptor, for each ADMET task, averaged feature importance is calculated for each feature set and is plotted in Figure 1. It is shown that Mordred descriptors are consistently the most important feature in all tasks, followed by Mol2Vec and Circular fingerprints. MACCS Keys fingerprint set is the least important among the five groups of features. As Mordred descriptors are considered significantly more important than other features, we retrained the models in each task using only this feature set. The results are listed in Table 3. XGBoost with only Mordred outperformed the base model in three tasks (HIA, Aqsol and PPBR). However, metabolism and excretion predictions were not improved using XGBoost with Mordered alone but were comparable in other tasks.
Fig. 1.
Average feature importance of fingerprints and descriptors in (A) absorption, (B) distribution, (C) metabolism, (D) elimination, and (E) toxicity tasks.
Table 3.
Performance comparison of XGBoost models trained with all features and with only Mordred.
| TDC | XGBoost with all features | XGBoost with Mordred | |
|---|---|---|---|
| Task | Metric | Score | Score |
| Absorption | |||
| Caco2 | MAE | 0.288 ± 0.011 | 0.301 ± 0.008 |
| HIA | AUROC | 0.987 ± 0.002 | 0.990 ± 0.002 |
| Pgp | AUROC | 0.911 ± 0.002 | 0.909 ± 0.005 |
| Bioav | AUROC | 0.700 ± 0.010 | 0.692 ± 0.016 |
| Lipo | MAE | 0.533 ± 0.005 | 0.538 ± 0.003 |
| AqSol | MAE | 0.727 ± 0.004 | 0.720 ± 0.003 |
| Distribution | |||
| BBB | AUROC | 0.905 ± 0.001 | 0.900 ± 0.001 |
| PPBR | MAE | 8.251 ± 0.115 | 7.897 ± 0.061 |
| VDss | Spearman | 0.612 ± 0.018 | 0.610 ± 0.005 |
| Metabolism | |||
| CYP2C9 Inhibition | AUPRC | 0.794 ± 0.004 | 0.781 ± 0.002 |
| CYP2D6 Inhibition | AUPRC | 0.721 ± 0.003 | 0.694 ± 0.005 |
| CYP3A4 Inhibition | AUPRC | 0.877 ± 0.002 | 0.862 ± 0.002 |
| CYP2C9 Substrate | AUPRC | 0.387 ± 0.018 | 0.334 ± 0.004 |
| CYP2D6 Substrate | AUPRC | 0.648 ± 0.023 | 0.594 ± 0.034 |
| CYP3A4 Substrate | AUPRC | 0.680 ± 0.005 | 0.649 ± 0.013 |
| Excretion | |||
| Half Life | Spearman | 0.396 ± 0.027 | 0.373 ± 0.008 |
| CL-Hepa | Spearman | 0.420 ± 0.011 | 0.378 ± 0.020 |
| CL-Micro | Spearman | 0.587 ± 0.006 | 0.576 ± 0.010 |
| Toxicity | |||
| LD50 | MAE | 0.602 ± 0.006 | 0.602 ± 0.006 |
| hERG | AUROC | 0.806 ± 0.005 | 0.763 ± 0.007 |
| Ames | AUROC | 0.859 ± 0.002 | 0.856 ± 0.002 |
| DILI | AUROC | 0.933 ± 0.011 | 0.928 ± 0.003 |
Searching for the parameter set with best validation performance is necessary. However, there are over 100,000 parameter combinations in the current search space, and it could growth exponentially with additional features being considered. It is challenging to iterate over all possible parameter set to find the best parameter set. In the current study, a randomized grid search CV was used. It should be noted that the randomized grid search CV does not necessarily lead to the global optimum parameter set due to the randomness nature. In recent decades, Bayesian optimization has been developed to search in the hyperparameter space, such as hyperopt Bergstra et al (2013), which might be promising under such tasks.
It should be noted that, while TDCommons provides a benchmark dataset useful to evaluate and compare different machine learning models, there are some limitations when applying this dataset. First, in the ADMET prediction scenario, all classification and regression predictions are single-instance: we only predict one value for each task. Thus, multitask learning is not feasible under the current framework. Moreover, the dataset has not been constantly updated. The dataset version should be mentioned when reporting related results.
Web Server
The trained machine learning models are hosted on the SMU high computing center at https://ai-druglab.smu.edu/admet. A SMILES representation is required for ADMET predictions. On the result page, molecule structures in both 2D and 3D are displayed using Open Babel O’Boyle et al (2011). A table is present to summary prediction results under 22 tasks. The optimal levels are referenced from Drug-Like Soft Rule and empirical ranges from ADMETLab 2.0 Xiong et al (2021). For each prediction, green, yellow, and red colors are used to indicate whether the prediction lies in optimal, medium, or poor ranges, suggested in ADMETLab 2.0. The web server has been tested rigorously to respond within seconds.
4. Conclusion
In this study, we applied XGBoost for ADMET prediction. XGBoost can effectively learn molecule features ranging from fingerprints to descriptors. For the 22 tasks on TDC benchmark, our model is ranked first in 11 tasks with all tasks ranked in top 5. The web server, ADMETboost, can be freely accessed at https://ai-druglab.smu.edu/admet.
Acknowledgments
Research reported in this paper was supported by the National Institute of General Medical Sciences of the National Institutes of Health under Award No. R15GM122013. Computational time was generously provided by Southern Methodist University’s Center for Research Computing. The preprint version of this work is available on arXiv with DOI number 2204.07532 under CC BY-NC-ND 4.0 license.
Footnotes
Competing interests: The authors declare no competing financial interest.
Availability of data and materials: The data used in this study is publicly available in TDC ADMET benchmark group https://tdcommons.ai/benchmark/admet_group/overview/. The dataset can be downloaded through the TDC Python package (v0.3.6). The default training and testing data were used for model training. We shared the related codes, model parameters for each task, and the ready-to-use featurization results on GitHub at https://github.com/smu-tao-group/ADMET_XGBoost. The web server can be accessed at https://ai-druglab.smu.edu/admet. .
References
- Bergstra J, Yamins D, Cox D (2013) Making a science of model search: Hyperparameter optimization in hundreds of dimensions for vision architectures. In: Int. Conf. Mach. Learn, PMLR, pp 115–123 [Google Scholar]
- Chen C, Zhang Q, Yu B, et al. (2020) Improving protein-protein interactions prediction accuracy using xgboost feature selection and stacked ensemble classifier. Comput Biol Med 123:103,899. [DOI] [PubMed] [Google Scholar]
- Chen T, Guestrin C (2016) Xgboost: A scalable tree boosting system. In: Proceedings of the 22nd acm sigkdd international conference on knowledge discovery and data mining, pp 785–794 [Google Scholar]
- Cheng F, Li W, Zhou Y, et al. (2012) admetsar: a comprehensive source and free tool for assessment of chemical admet properties. J Chem Inf Model 52(11):3099–3105 [DOI] [PubMed] [Google Scholar]
- Deng D, Chen X, Zhang R, et al. (2021) Xgraphboost: Extracting graph neural network-based features for a better prediction of molecular properties. J Chem Inf Model 61(6):2697–2705 [DOI] [PubMed] [Google Scholar]
- Dong J, Wang NN, Yao ZJ, et al. (2018) Admetlab: a platform for systematic admet evaluation based on a comprehensively collected admet database. J Cheminf 10(1):1–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durant JL, Leland BA, Henry DR, et al. (2002) Reoptimization of mdl keys for use in drug discovery. J Chem Inf Comput Sci 42(6):1273–1280 [DOI] [PubMed] [Google Scholar]
- Göller AH, Kuhnke L, Montanari F, et al. (2020) Bayer’s in silico admet platform: A journey of machine learning over the past two decades. Drug Discovery Today 25(9):1702–1709 [DOI] [PubMed] [Google Scholar]
- Hu W, Liu B, Gomes J, et al. (2019) Strategies for pre-training graph neural networks. arXiv preprint arXiv:190512265 [Google Scholar]
- Huang K, Fu T, Glass LM, et al. (2020) Deeppurpose: a deep learning library for drug–target interaction prediction. Bioinformatics 36(22-23):5545–5547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang K, Fu T, Gao W, et al. (2021) Therapeutics data commons: Machine learning datasets and tasks for drug discovery and development. Proceedings of Neural Information Processing Systems, NeurIPS Datasets and Benchmarks [Google Scholar]
- Jaeger S, Fulle S, Turk S (2018) Mol2vec: unsupervised machine learning approach with chemical intuition. J Chem Inf Model 58(1):27–35 [DOI] [PubMed] [Google Scholar]
- Kennedy T (1997) Managing the drug discovery/development interface. Drug discovery today 2(10):436–444 [Google Scholar]
- Kipf TN, Welling M (2016) Semi-supervised classification with graph convolutional networks. arXiv preprint arXiv:160902907 [Google Scholar]
- Kola I, Landis J (2004) Can the pharmaceutical industry reduce attrition rates? Nat Rev Drug Discovery 3(8):711–716 [DOI] [PubMed] [Google Scholar]
- Lee WH, Millman S, Desai N, et al. (2021) Neuralfp: Out-of-distribution detection using fingerprints of neural networks. In: 2020 25th International Conference on Pattern Recognition (ICPR), IEEE, pp 9561–9568 [Google Scholar]
- Honorio MK, Moda LT, Andricopulo DA (2013) Pharmacokinetic properties and in silico adme modeling in drug discovery. Med Chem 9(2):163–176 [DOI] [PubMed] [Google Scholar]
- Moriwaki H, Tian YS, Kawashita N, et al. (2018) Mordred: a molecular descriptor calculator. J Cheminf 10(1):1–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Boyle NM, Banck M, James CA, et al. (2011) Open babel: An open chemical toolbox. J Cheminf 3(1):1–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramsundar B, Eastman P, Walters P, et al. (2019) Deep learning for the life sciences: applying deep learning to genomics, microscopy, drug discovery, and more. O’Reilly Media [Google Scholar]
- Rogers D, Hahn M (2010) Extended-connectivity fingerprints. J Chem Inf Model 50(5):742–754 [DOI] [PubMed] [Google Scholar]
- Schyman P, Liu R, Desai V, et al. (2017) vnn web server for admet predictions. Front Pharmacol 8:889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Z, Zhou H, Tian H, et al. (2020) Unraveling the energetic significance of chemical events in enzyme catalysis via machine-learning based regression approach. Commun Chem 3(1):1–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian H, Trozzi F, Zoltowski BD, et al. (2020) Deciphering the allosteric process of the phaeodactylum tricornutum aureochrome 1a lov domain. J Phys Chem B 124(41):8960–8972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian H, Jiang X, Tao P (2021a) Passer: prediction of allosteric sites server. Mach Learn: Sci Technol 2(3):035,015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian H, Jiang X, Trozzi F, et al. (2021b) Explore protein conformational space with variational autoencoder. Front Mol Biosci 8:781,635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkatraman V (2021) Fp-admet: a compendium of fingerprint-based admet prediction models. J Cheminf 13(1):1–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waring MJ, Arrowsmith J, Leach AR, et al. (2015) An analysis of the attrition of drug candidates from four major pharmaceutical companies. Nat Rev Drug Discovery 14(7):475–486 [DOI] [PubMed] [Google Scholar]
- Xiong G, Wu Z, Yi J, et al. (2021) Admetlab 2.0: an integrated online platform for accurate and comprehensive predictions of admet properties. Nucleic Acids Res 49(W1):W5–W14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong Z, Wang D, Liu X, et al. (2019) Pushing the boundaries of molecular representation for drug discovery with the graph attention mechanism. J Med Chem 63(16):8749–8760 [DOI] [PubMed] [Google Scholar]
- Yang H, Lou C, Sun L, et al. (2019) admetsar 2.0: web-service for prediction and optimization of chemical admet properties. Bioinformatics 35(6):1067–1069 [DOI] [PubMed] [Google Scholar]
- Zhang Q, Heldermon CD, Toler-Franklin C (2020) Multiscale detection of cancerous tissue in high resolution slide scans. In: Int. Symp. Vis. Comput, Springer, pp 139–153 [Google Scholar]