Abstract
Here we describe two new species of the genus Penicillium section Torulomyces with solitary phialides. Penicillium poederi sp. nov. was isolated from volcanic soils in Iceland. Penicillium tirolense sp. nov. was isolated from a sporocarp of Serpula lacrymans. Both species are characterised by slow growth rates and the production of a brown soluble pigment on CYA, conidiophores with solitary ampulliform phialides with smooth-walled stipes and warty, globose conidia and with connectives without visible rings. The spores of. P. poederi are 2.5 μm diam, while the spores of P. tirolense are 2.0 μm diam. In a multigene phylogeny based on the ITS, BenA, CaM and RPB2 gene regions P. tubakianum and P. wollemiicola are the closest relatives of P. poederi. This species differs from P. tubakianum and P. wollemiicola by its growth rates and by its pigmentation. The holotype of P. poederi is IB2017/0007, while SF014017 (CBS 147622) is a culture derived from the holotype. The closest relatives of P. tirolense are P. austricola and P. riverlandense. It differs from P. austricola by lower growth rates on all tested media and temperatures and by its larger spores. It differs from P. riverlandense by lower growth rates and the absence of growth at 37 °C. The holotype of P. tirolense is IBF2019/0162, while SF015108 (CBS 147625) is a culture derived from the holotype.
Citation: Kirchmair M, Embacher J, Heimdörfer D, Walch G, Neuhauser S (2022). Penicillium poederi and Penicillium tirolense, two new species of section Torulomyces. Fungal Systematics and Evolution 10: 91–101. doi: 10.3114/fuse.2022.10.03
Keywords: Austria, Eurotiomycetes, Iceland, new taxa, soil, taxonomy, Tyrol
INTRODUCTION
Newly created environments are found after the retreat of glaciers, after volcanic eruptions or on a newly created island like Surtsey. Studying such sites allows insight in the development of soils and organic matter. A well-studied volcanic succession site is Mt. Hekla, Iceland, where larger eruptions created a series of soil ecosystems of different age. Cutler et al. (2014) analysed the soil microbial communities in lava flows of different ages by amplicon-sequencing. In accordance with other studies on soil fungal communities in succession sites (Brown & Jumpponen 2014, Otaki et al. 2016, Delgado-Baquerizo et al. 2019), they found an increasing fungal diversity with older terrain age. We re-investigated the samples from Cutler et al. (2014) using culture-based approaches to link the sequences gathered by Cutler et al. (2014) with morphologically and physiologically well-characterised fungal cultures. We used a dilution-to-extinction approach to isolate also slow growing fungi which are easily overlooked or simply overgrown by other faster growing colonies (Unterseher & Schnittler 2009). Within this scope three strains of a hitherto unknown, very slow growing Penicillium with solitary phialides were isolated.
Recently the focus of research on the symbiosis and interaction of fungi and their environment started to study the fungal holobiont. The term “holobiont” dates back to Lynn Margulis describing the essential links between a fungus and algae in lichens (Margulis 1993). Currently the term holobiont is often used for every microbe and its associate microbes (Gordon et al. 2013), which is expanding the view of the holobiont beyond the original definition which was referring to close relationships between the partners rather than loose associations. Most studies on holobionts are from plants and animals (including humans) while fungal holobionts are still rare (Partida-Martínez 2017). Studies on fungal holobionts are usually focussing on the bacteriome (Deveau et al. 2018). Within the scope of a study on the Serpula lacrymans holobiont (Embacher et al. 2021) fungi associated with S. lacrymans tissue were isolated. Also in this case, a dilution-to-extinction approach was applied for isolating slow growing fungi. We isolated a Penicillium with solitary phialides which was morphologically different from the Mt. Hekla Penicillium and distinct from other known species of Penicillium.
It is the aim of this study to characterise and describe the two Penicillium species as new to science.
MATERIAL AND METHODS
Fungal strains
Soil samples were diluted 1:5 in a 0.9 % sodium chloride solution supplemented with 0.05 % Tween 80 and mixed for 2 h with an overhead shaker. The suspensions were plated onto potato dextrose agar (PDA) supplemented with 1 % Streptomycin, 0.05 % Tetracycline, and 0.01 % Dicloran. Agar plates were incubated at 25 °C for 14 d and screened daily for fungal growth. Colonies of interest were transferred onto PDA.
Parts of a sporocarp of S. lacrymans (0.1–1 g) were suspended in 20 mL 0.9 % sodium chloride solution supplemented with 0.05 % Tween 80 and mixed together with 10 sterilised glass beads (∅ 4 mm) for 10 min with an overhead shaker (Embacher et al. 2021). The suspension was plated onto malt extract agar (MEA; according to Pitt 1988) supplemented with 1 % Streptomycin, 0.05 % Tetracycline. Agar plates were incubated at 25 °C for 7 d and screened daily for fungal growth. Colonies of interest were transferred onto MEA.
A dilution to extinction approach was applied to find a maximum number of different fungal strains (Unterseher & Schnittler 2009). Ninety-six-well plates were prepared with PDA supplemented with antibiotics (see above). Each well contained 0.5 mL of PDA. Aliquots of 10 μL of the soil and S. lacrymans sporocarp suspension were pipetted into each well. The 96-well plates were incubated at 25 °C and screened daily for fungal growth. Agar plugs with a single visible fungal colony were transferred onto fresh MEA agar plates.
Cultures were deposited at the Jena Microbial Resource Collection (JMRC), at the culture collection (CBS) of the Westerdijk Fungal Biodiversity Institute (WI), Utrecht, the Netherlands, and at the Institute of Microbiology, University Innsbruck, Austria. Dried specimens were deposited at the National Science Collection of the Tiroler Landesmuseum (IBF).
Molecular characterisation
Fungal colonies were grown on PDA for 8–10 d, and DNA extraction performed using a CTAB-based extraction protocol (Neuhauser et al. 2009). The methods of Visagie et al. (2014) were followed for PCR amplification of the ITS, BenA, and CaM gene regions. For RPB2 a touch down PCR protocol was used (98 °C for 2 min, 4 cycles of 96 °C for 20 s, 60 °C for 45 s, 72 °C for 60 s, 4 cycles of 96 °C for 20 s, 58 °C for 45 s, 72 °C for 60 s, 28 cycles of 96 °C for 20 s, 55 °C for 45 s, 72 °C for 60 s and a final incubation step at 72°C for 7 min.
The PCR products were purified using the Illustra GFX PCR DNA and Gel Band Purification Kit (GE Healthcare) and sequenced by GATC Biotech. Sequences were quality checked and aligned with sequences of known Torulomyces species (Table 1) using MAFFT v. 7.308 (Katoh & Stanley 2013) implemented in Geneious v. 9.1.7. To concatenate the multigene alignment the individual alignments were combined manually leaving a gap between the individual genes in the following order: ITS, BenA, CaM, RPB2. The datasets were analysed using Maximum Likelihood in MEGA X v. 10.1 (Kumar et al. 2018). The most suitable substitution model for each dataset (ITS, BenA, CaM, RPB2, and the concatenated alignment of all four genes) was selected using the model-test within MEGA, based on the lowest Bayesian information criterion (BIC) value (Table 2). An initial tree was calculated with the Bio-Neighbour-Joining (BioNJ) option, with the subsequent Heuristic search done with Nearest-Neighbour-Interchange (NNI). For calculating node support a bootstrap analysis with 1 000 replicates was performed. Alignments and raw trees have been deposited at Figshare (https://doi.org/10.6084/m9.figshare.20631792.v1).
Table 1.
Sequence data of Penicillum spp. used for phylogenetic analyses.
Table 2.
Length of datasets and models used for phylogenetic analyses.
| ITS | BenA | CaM | RPB2 | Concatenated | |
|---|---|---|---|---|---|
| Length | 587 bp | 532 bp | 597 bp | 887 bp | 2 621 bp |
| Model ML | T92+G | K2+G | T92+G+I | TN93+G+I | TN93+G+I |
Abbreviations: T92 = Tamura 3-parameter; K2 = Kimura 2-parameter; TN93 = Tamura-Nei; +G = Gamma distribution; +I = Invariant sites.
Morphology
Isolates were grown as described by Pitt (1988) and Visagie et al. (2014). Colony characteristics were recorded from strains grown on CREA (creatine sucrose agar), CYA (Czapek yeast autolysate agar), CYAS (CYA supplemented with 5 % NaCl), DG18 (dichloran 18 % glycerol agar), G25N (25 % glycerol nitrate agar), MEA, YES (yeast extract sucrose agar), OA (oatmeal agar), and SNA (synthetic nutrient-poor agar). All four isolates were grown on all media at 25 °C. Additionally all isolates were incubated at 10 °C, 30 °C, and 37 °C on CYA and MEA. Growth measurements are given in the form (minimum)-median-(maximum) after 7 and 14 d incubation. Colour names and codes used for descriptions refer to the Methuen Handbook of Colour (Kornerup & Wanscher 1967).
Microscopic characters were examined using a Nikon Optiphot compound microscope with Nomarski interference contrast, fitted with a Nikon DS-Fi3 microscope camera and pictures captured and analysed using Nikon NIS-elements D v. 3.0 software. Microscopic specimens were prepared from 7–14-d-old cultures grown on MEA, with 3 % KOH as mounting medium.
For scanning electron microscopy (SEM) culture discs were fixed with Roti®Histofix 4 % (Carl Roth) for 30 min, washed in PBS (phosphate buffered saline buffer: 0.02 mol/L sodium phosphate buffer with 0.15 mol/L sodium chloride, pH adjusted to 7.4), dehydrated in an ascending methanol series and critical point dried. The specimens were mounted on aluminium stubs and sputtered with gold. Micrographs were taken using a Zeiss DSM 950 scanning electron microscope (SEM).
Photo plates were prepared using Adobe® Photoshop® Creative Suite v. 5. Photomicrographs were modified for aesthetic purposes, without altering areas of scientific significance. All measurements are given in the form (minimum) mean ± standard deviation (maximum).
RESULTS
Taxonomy
Penicillium poederi Kirchm. & Neuh., sp. nov. MycoBank MB 845495. Figs 1–3.
Fig. 1.
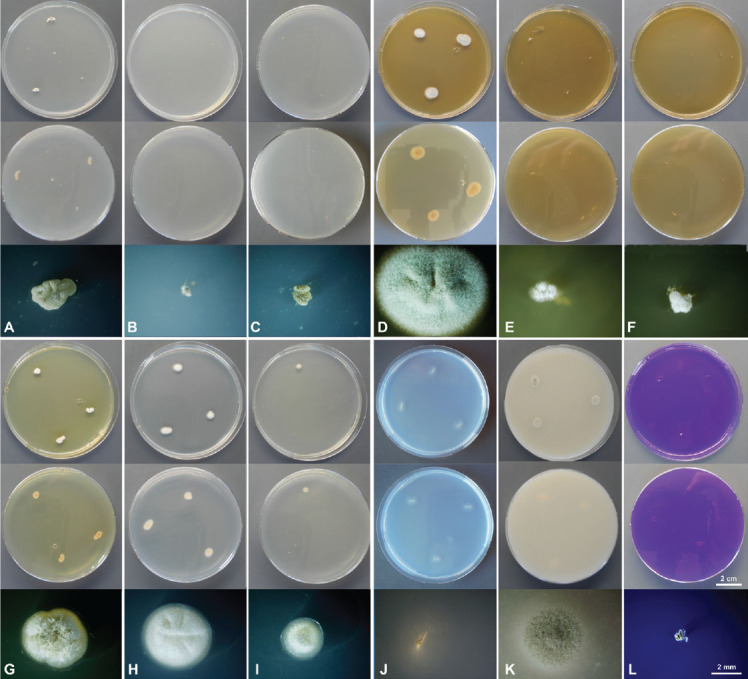
Culture characteristics of Penicillium poederi sp. nov. after 1 wk incubation. From top to bottom: obverse, reverse, single colony. A. CYA 25 °C. B. CYA 10 °C. C. CYA 30 °C. D. MEA 25 °C. E. MEA 10 °C. F. 30 °C. G. YES 25 °C. H. DG18 25 °C. I. G25N 25 °C. J. SNA 25 °C. K. OA 25°C. L. CREA 25 °C.
Fig. 3.
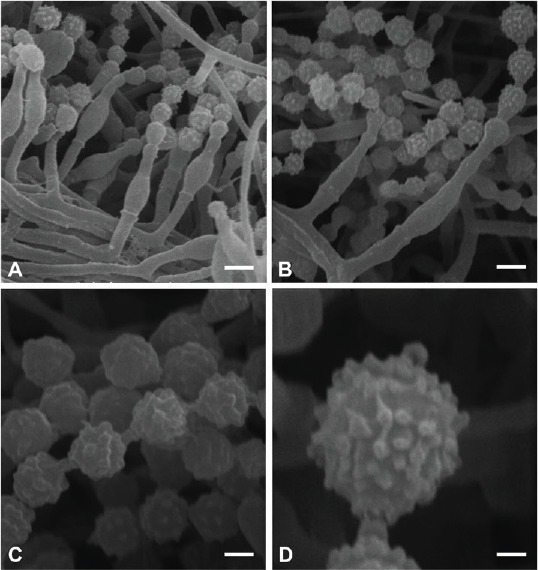
Micromorphological characters of Penicillium poederi sp. nov. (SEM). A, B. Solitary phialides, conidial chains. C. Warty conidia, connectives without visible rings. D. Conidium with tubercles. Scale bars: A, B = 2 μm, C = 1 μm, D = 0.5 μm.
Etymology: Dedicated to our teacher, mentor, and friend, the Austrian mycologist Reinhold “Bodo” Pöder, who died on Aug. 20th 2015 at the age of 66.
Typus: Iceland, from soil on the lava flow from 1158 (19°52’W, 63°59’N) at Mt Hekla, 24 Nov. 2016, D. Heimdörfer & M. Kirchmair (holotype IBF2017/0007, preserved as dried specimen, ex-type culture SF014017 = CBS 147622 = Bq214).
Soil characteristics: pH = 5.3–6.6; SOM = 10.8 ± 0.9 %, Mean grain size: coarse sand 24.7 % ± 1.2 %, fine sand 70.8 % ± 1.2 %, silt 4.5 % ± 0.2 %, clay 0.0 %, total N 0.1 ± 0.01, total PM (mg/L) 13.4 ± 0.3, total PO (mg/L) 7.6± 1.5, total KM (mg/L) 1.1 ± 0.1, total KO (mg/L) 0.4 ± 0.1. (Cutler et al. 2014).
ITS Barcode: MF611757 (alternative markers: BenA = MF611760; CaM = MF611763; RPB2 = MF611766).
Additional cultures examined: Iceland, from soil at Mt Hekla, D. Heimdörfer & M. Kirchmair, culture IB2017/0006 = SF014018 = CBS 147623 = Ad249; ibid., culture IB2017/0008 = SF014019 = CBS 147624 = Ar233.
Colony diam, 7 d (in mm): CYA 25 °C (1–)2(–3); CYA 10 °C microcolonies (< 1); CYA 30 °C microcolonies (< 1); CYA 37 °C no germination; MEA 25 °C (9–)10(–12); MEA 10 °C 1(–3); MEA 30 °C microcolonies (< 1); MEA 37 °C no germination; G25N 25 °C (2–)3(–3); YES 25 °C (3–)4(–6); OA 25°C (6–)7(–9); DG18 25°C (4–)6(–7); CYAS 25 °C no germination; SNA 25 °C (6–)7(–10); CREA 25 °C: 1.
Colony characters, 7 d (Fig. 1): CYA 25 °C: crateriform to irregularly sulcate, mycelia white, sporulation sparse, soluble pigments absent, obverse and reverse yellowish white (4A2); CYA 10 °C: floccose, no sporulation, soluble pigment absent, colours not determinable (colonies too small); CYA 30 °C: floccose, no sporulation, soluble pigment absent, colours not determinable (colonies too small); MEA 25 °C: velvety, sulcation only indicated, mycelia white, sporulation moderately dense, conidia en masse greenish grey (28E2), soluble pigments absent, reverse light brown (5C3); MEA 10 °C: floccose, mycelia white, no sporulation, soluble pigment absent, colours not determinable (colonies too small); MEA 30 °C: floccose, mycelia white, no sporulation, soluble pigment absent, colours not determinable (colonies too small); YES 25 °C: crateriform to slightly sulcate, fasciculate at the centre, mycelia white, sporulation sparse, soluble pigment light brown, reverse yellowish white (4A3); DG18 25 °C: slightly sulcate, mycelia white, sporulation sparse, soluble pigments absent, obverse and reverse white; G25N 25 °C: velvety, mycelia white, sporulation sparse, soluble pigments absent, obverse and reverse white; SNA 25 °C: low, mycelia white, sporulation sparse, soluble pigments absent, obverse and reverse white (4A1); OA 25 °C: velvety, mycelia white sporulation moderately dense, conidia en masse greenish grey (28E2), soluble pigments absent; CREA 25 °C: acid not produced.
Colony diam, 14 d (in mm): CYA 25 °C: (4–)4(–5); CYA 10 °C 1; CYA 30 °C: (microcolonies) 1; CYA 37 °C: no germination; MEA 25 °C: (17–)20(–23); MEA 10 °C: (3–)4(–6); MEA 30 °C: (1–)1(–2); MEA 37 °C: no germination; YES 25 °C: (5–)7(–10); DG18 25 °C: (13–) 14(–16); G25N 25 °C: (5–)7(–9); CYAS 25 °C: no germination; SNA 25 °C: (10–)16(–20); OA 25 °C: (15–)17(–20); CREA 25 °C: 1.
Colony characters, 14 d (supplementary Fig. S1): CYA 25 °C: irregularly sulcate, mycelia white, sporulation sparse, soluble pigment brown, obverse yellowish white (4A3), reverse olive brown (4E5); CYA 10 °C: dense with a floccose overlay, mycelia white, sporulation sparse, soluble pigment absent, obverse and reverse white; CYA 30 °C: dense, waxy, mycelia white, no sporulation, soluble pigment absent, obverse and reverse white; CYA 37 °C: no germination; MEA 25 °C: radially sulcate, low with concentrical rings near the margin, mycelia white, sporulation moderately to dense, soluble pigment absent, obverse greenish grey (28E2) when sporulating, in non or sparsely sporulating cultures white, reverse brown to dark brown (5F4–5F5); MEA 10 °C: velvety to slightly fasciculate; mycelia white, sporulation sparse, soluble pigment absent, obverse and revers white; MEA 30 °C: velvety, mycelia white, sporulation sparse, soluble pigment absent, obverse and revers white; MEA 37 °C: no germination; YES 25 °C: dense, irregularly sulcate, occasionally crateriform, velvety to slightly fasciculate at the centre; mycelia white, sporulation sparse, soluble pigment brown, obverse white, reverse yellowish brown (5E5); DG18 25 °C: plane irregularly to radially sulcate, velvety, mycelia white, sporulation sparse, soluble pigment absent, obverse olive brown (4D3) when sporulating, in non or sparsely sporulating cultures yellowish grey (4B2), reverse olive brown (4F5) when sporulating, in non or sparsely sporulating cultures greyish yellow (4B4); G25N 25 °C: dense, irregularly sulcate, occasionally crateriform, velvety to slightly fasciculate, mycelia white, sporulation sparse, soluble pigment absent, obverse greenish grey (28E2), revers greyish green (28E5); CYAS 25 °C: no germination; SNA 25 °C: plane with irregular margins, mycelia white, sporulation sparse, soluble pigment absent, obverse greenish grey (26B2), reverse white to light greyish. OA 25 °C: plane, velutinous, mycelia white, sporulation moderate, soluble pigment absent, conidia en masse greenish grey (28E2); CREA 25 °C: acid not produced.
Micromorphology (Fig. 2): Conidiophores as solitary phialides; stipes smooth-walled, (4.0–)6.5 ± 1.4(–9.3) × (1.3–)1.7 ± 0.2(–2.1) μm (n = 25); phialides ampulliform, (4.9–)6.1 ± 0.9(–9.0) × (2.1–) 2.6 ± 0.3(–3.2) μm (n = 37); conidia warty, globose, (2.1–)2.5 ± 0.2(–2.9) μm (n = 38), average width/length quotient = 1; sclerotia not produced.
Fig. 2.
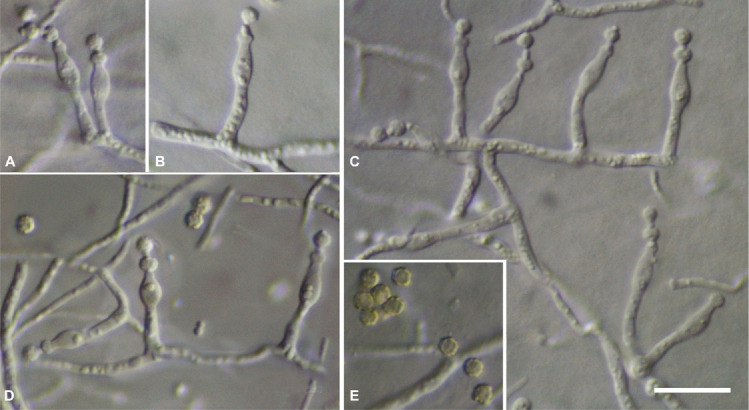
Micromorphological characters of Penicillium poederi sp. nov. (light microscopy). A–D. Solitary phialides. E. Mature conidia. Scale bar = 10 μm.
SEM observations (Fig. 3): Conidia warty, tubercles (0.26–)0.36 ± 0.04(–0.43) μm diam, (0.14–)0.19 ± 0.03(–0.21) high, connectives long without visible rings.
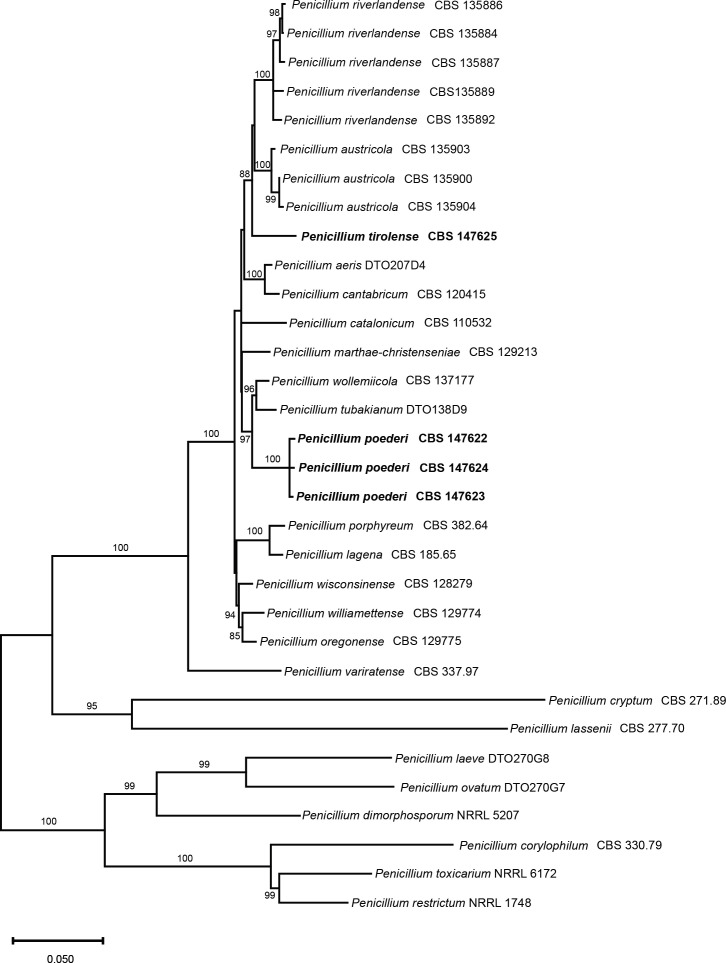
Notes: In ML phylogenies of the RBP2, CaM, BenA as well as in the combined dataset (Fig. 7); Supplementary Figs S3–S6, P. poederi forms one clade together with P. tubakianum and P. wollemiicola, and branches consequently as a separate and distinct clade.
Fig. 7.
Multigene phylogeny (maximum likelihood) for a combined ITS, BenA, CaM & RPB2 dataset of Penicillium sect. Torulomyces. Bootstrap values higher than 80 % are indicated above branches. The new species are highlighted in bold font.
Penicillium poederi differs from P. tubakianum and P. wollemiicola by its lower growth rates on CYA when incubated on 25 °C and 30 °C, lower growth rates on YES and DG18 incubated at 25 °C, and higher growth rates on MEA at 25 °C. The obverse on MEA 25 °C is P. poederi is greenish grey when sporulating or white in non-sporulating cultures, while in P tubakianum the obverse is orange-white. In contrast to P. tubakianum, a brown soluble pigment is produced on CYA and YES at 25 °C by P. poederi (Table 3).
Table 3.
Summary of the most important morphological characters. Conspicuous differences between the new species (bold) and their closest relatives are printed in bold.
| P. tirolense | P. austricola* | P. riverlandense* | P. poederi | P. tubakianum* | P. wollemiicola* | |||
|---|---|---|---|---|---|---|---|---|
| growth rates (mm) | CYA 37 | n.g. | n.g. | 4–7 | n.g. | n.g. | n.g. | |
| CYA 30 | 2–5 | 10–11 | 9–14 | < 1 | 5–6 | 6–7 | ||
| CYAS | 1–3 | 8–10 | 8–9 | n.g. | 2–4 | 4–5 | ||
| CREA | 1–2 | 4 | 3–4 | 1 | n.g. | n.g. | ||
| CYA | 3–5 | 10–13 | 10–14 | 1–3 | 8–9 | 7–8 | ||
| OA | 2–5 | 9–10 | 10–14 | 6–9 | 8–9 | 9–10 | ||
| MEA | 4–6 | 4–8 | 6–8 | 9–12 | 4–5 | 4–5 | ||
| YES | 3–6 | 10–11 | 10–15 | 3–6 | 7–8 | 8–9 | ||
| SNA | 2–6 | 9–11 | 10–12 | 6–10 | 5–7 | 7–9 | ||
| DG18 | 4–7 | 9–11 | 10–12 | 4–7 | 9–10 | 9–11 | ||
| brown soluble pigment | present | present | present | present | absent | present | ||
| acid on CREA | absent | absent | absent | absent | absent | absent | ||
| Conidia | size [μm] | 2.0 ± 0.1 | 2.1 ± 0.1 | 1.8 ± 0.1 | 2.5 ± 0.2 | 2.1 ± 0.1 | 2.3 ± 0.1 | |
| shape | globose | globose | globose | globose | globose | globose | ||
| tubercle size [μm] | 0.23–0.34 | 0.19–0.30 | 0.19–0.35 | 0.26–0.46 | 0.17–0.28 | 0.22–0.41 | ||
| connectives | present, lack visible rings | long, lack visible rings | long, lack visible rings | long, lack visible rings | present, lack visible rings | long, lack visible rings |
*Data from Visagie et al. (2016).
Penicillium tirolense Kirchm., Embacher & Neuh., sp. nov. MycoBank MB 845496. Figs 4–6.
Fig. 4.
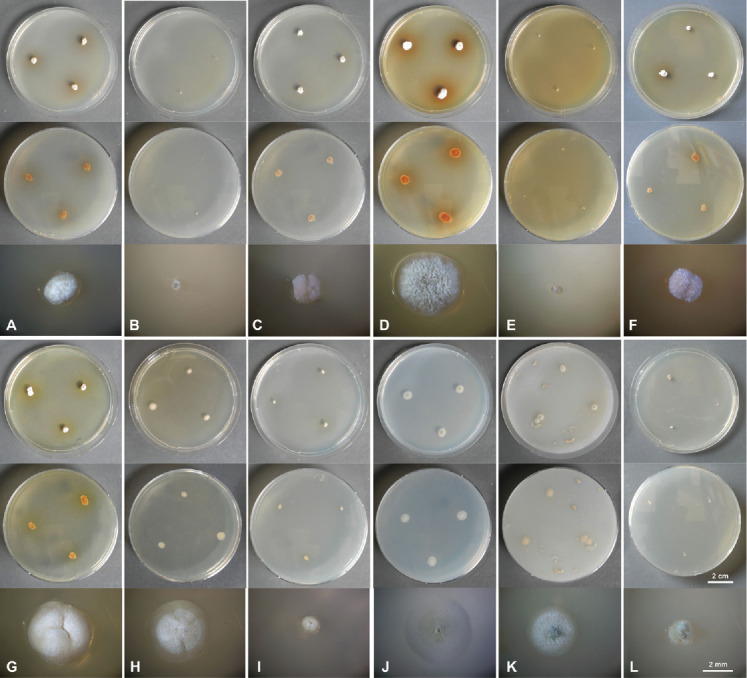
Culture characteristics of Penicillium tirolense sp. nov. after 1 wk incubation. From top to bottom: obverse, reverse, single colony. A. CYA 25 °C. B. CYA 10 °C. C. CYA 30 °C. D. MEA 25 °C. E. MEA 10 °C. F. 30 °C. G. YES 25 °C. H. DG18 25 °C. I. G25N 25 °C. J. SNA 25 °C. K. OA 25 °C. L. CYAS 25 °C.
Fig. 6.
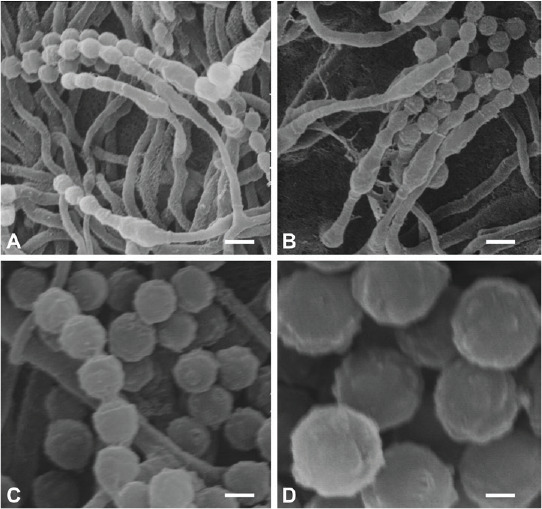
Micromorphological characters of Penicillium tirolense sp. nov. (SEM). A, B. Solitary phialides, conidial chains. C. Warty conidia, connectives without visible rings. D. Conidia with tubercles. Scale bars: A, B = 2 μm, C = 1 μm, D = 0.5 μm.
Etymology: Named after Tyrol, a province in Austria from where the new species was isolated.
Typus: Austria, Tyrol, Matrei am Brenner (11°7’W, 47°27’N), from a sporocarp of Serpula lacrymans, 14 Oct. 2019, J. Embacher & M. Kirchmair (holotype IBF2019/0162, preserved as dried specimen, culture ex-type SF014017 = CBS 147625).
ITS Barcode: MW145398 (alternative markers: BenA = MW143069; CaM = MW143068; RPB2 = MW143067).
Colony diam, 7 d (in mm): CYA 25 °C: (3–)4(–5); CYA 10 °C: microcolonies (< 1); CYA 30 °C (2–)3(–5); CYA 37 °C: no germination; MEA 25 °C: (4–)5(–6); MEA 10 °C: microcolonies (< 1); MEA 30 °C: (2–)3(–5); MEA 37 °C : no germination; G25N 25 °C: (1–)2(–3); YES 25 °C: (3–)5(–6); OA (2–)4(–5); DG18 25 °C: (4–) 5(–7); CYAS: (1–)2(–3); SNA 25 °C: (2–)3(–6); CREA 25 °C: 1(–2).
Colony characters, 7 d (Fig. 4): CYA 25 °C: dense, somewhat fasciculate, mycelia white, sporulation sparse, soluble pigment brown, obverse white (4A1), reverse brown (5F4); CYA 10 °C: dense, mycelia white, no sporulation, soluble pigment absent, obverse and reverse white (4A1); CYA 30 °C: dense, muriform, irregularly sulcate, mycelium white, sporulation sparse, soluble pigment absent, obverse white (4A1), revers greyish beige to golden grey (4C2); CYA 37 °C: no germination. MEA 25 °C: crateriform, irregularly sulcate, velvety to slightly fasciculate, mycelia white, sporulation moderate, soluble pigment brown, obverse greenish grey (26C2–26D2, 25C2–25D2, 27D2), reverse brown to reddish brown (7B8 and 8B8). MEA 10 °C: waxy; mycelia white, sporulation sparse, soluble pigment absent, obverse and reverse white (4A2); MEA 30 °C: dense, crateriform, mycelia white, sporulation sparse, soluble pigment absent, obverse white (4A1), revers yellowish brown to brown (5E5 and 6E4). MEA 37 °C: no germination. YES 25 °C: dense, crateriform, irregularly sulcate; mycelia white, sporulation sparse, soluble pigment brown, obverse white (4A1), reverse yellowish brown (5E5). DG18 25 °C: plane to crateriform, velvety, mycelia white, sporulation sparse, soluble pigment brown (very weak), obverse yellowish grey (4B2), reverse greyish yellow (4B4) , G25N 25 °C: dense, crateriform, velvety to slightly fasciculate, mycelia white, sporulation sparse, soluble pigment brown (very weak), obverse white (4A1), revers yellowish white (4A2); SNA 25 °C: plane with inconspicuous concentric rings, mycelia white, sporulation moderate, soluble pigment absent, obverse white to olive green (4A1, 2E3), reverse white (4A1). OA 25 °C: plane, velutinous, mycelia white, sporulation moderate, soluble pigment brown (very weak), obverse greenish grey (27D3), reverse white (4A1). CYAS 25°C: dense, somewhat fasciculate, mycelia white, sporulation sparse, soluble pigment absent, obverse white to grey (8A1 to 8B1, 5C1), revers white (4A1). CREA 25 °C: acid not produced.
Colony diam, 14 d (in mm): CYA 25 °C: (8–)9(–10); CYA 10 °C: (1–)2(–3); CYA 30 °C: (7–)8(–9); CYA 37 °C: no germination; MEA 25 °C: (15–)16(–17); MEA 10 °C: (2–)3(–4); MEA 30 °C: (9–)10(–11); MEA 37 °C: no germination. YES 25 °C: (12–)14(–15); DG18 25 °C: (9–)12(–13); G25N 25 °C: (5–)6(–7); SNA 25 °C: (11–)14(–16); OA 25 °C: (7–)8(–10); CYAS 25 °C: (3–)4(–6); CREA 25 °C: (3–)4(–5).
Colony characters, 14 d (supplementary Fig. S2): CYA 25 °C: dense, crateriform, irregularly sulcate, mycelia white, sporulation sparse, soluble pigment brown, obverse grey (28C1 and 29B1–29C1), reverse dark brown (7F7–7F8 and 8F8); CYA 10 °C: dense, mycelia white, no sporulation, obverse and reverse white (4A1), soluble pigment absent. CYA 30 °C: dense, moriform, irregularly sulcate, mycelium white, sporulation sparse, soluble pigment absent, obverse white (4A1), revers greyish beige to golden grey (4C2). CYA 37 °C: no germination. MEA 25 °C: irregularly to radially sulcate, velvety to slightly fasciculate, mycelia white, sporulation moderate, soluble pigment brown, obverse greenish grey (26C2–26D2, 25C2–25D2, 27D2), reverse brown to reddish brown (7B8 and 8B8). MEA 10 °C: velvety to slightly fasciculate; mycelia white, sporulation sparse, soluble pigment absent, obverse greyish yellow (4B3), revers white to greenish grey (26A1 and 26C2). MEA 30 °C: dense, crateriform, irregularly sulcate, mycelia white, sporulation sparse, soluble pigment absent, obverse greenish grey (28C2 to 28B2), revers greyish brown to brown (7E3 and 6E4). MEA 37 °C: no germination. YES 25 °C: dense, crateriform, irregularly sulcate; mycelia white, sporulation moderate, soluble pigment brown, obverse greyish orange to dull yellow (5B3 and 4A2), reverse greyish brown (5B3 and 8D3). DG18 25 °C: plane radially sulcate, velvety, mycelia white, sporulation sparse to moderate, soluble pigment brown present, obverse olive brown (4D3) when sporulating, in non or sparsely sporulating cultures yellowish grey (4B2), reverse olive brown (4F5) when sporulating; in non or sparsely sporulating cultures greyish yellow (4B4) , G25N 25 °C: dense, irregularly sulcate, velvety to slightly fasciculate, mycelia white, sporulation sparse, soluble pigment brown, obverse white (3A1), revers yellowish white to pale yellow (4A2 to 4A3). SNA 25 °C: plane with inconspicuous concentric rings, mycelia white, sporulation moderate, soluble pigment absent, obverse olive green to dull green (2E3, 2D2, 26E3), reverse white to greenish grey (2A1 to 2B2). OA 25 °C: plane, velutinous, mycelia white, sporulation moderate, soluble pigment brown (very weak), obverse greenish grey to dull green (27D3, 27C2), reverse olive to dull green (30D4, 3D3). CYAS 25 °C: dense, slightly sulcate, mycelia white, sporulation sparse, soluble pigment absent, obverse white to grey (8A1 to 8B1, 5C1), revers brownish grey (5C2 to 5D2). CREA 25 °C: acid not produced.
Micromorphology (Fig. 5): Conidiophores as solitary phialides; stipes smooth-walled; phialides ampulliform, (4.6–)6.2 ± 1.0 (–8.5) × (1.9–)2.6 ± 0.3(–3.2) μm (n = 32); conidia smooth to slightly rough, globose, (1.8–)2.0 ± 0.1(–2.2) μm (n = 41), average width/length quotient = 1; sclerotia not produced.
Fig. 5.
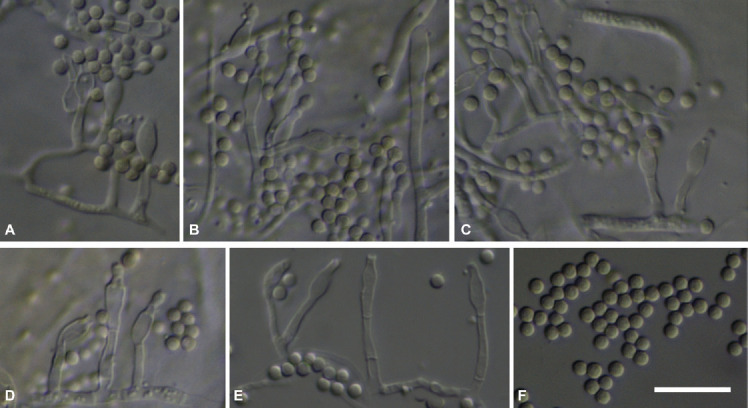
Micromorphological characters of Penicillium tirolense sp. nov. (light microscopy). A–E. Solitary phialides. F. Mature conidia. Scale bar = 10 μm.
SEM observations (Fig. 6): Conidia warty, tubercles (0.23–) 0.29 ± 0.04(–0.34) μm diam, (0.11–)0.13 ± 0.02(–0.16) high, connectives short without visible rings.
Notes: In ML phylogenies of the RBP2, CaM, BenA as well as in the combined datasets (Fig. 7; Supplementary Figs S3–S6), P. tirolense forms one branch together with P. austricola and P. riverlandense. From those species, P. tirolense differs in its lower growth rates on all tested media and temperatures. The obverse and reverse on DG18 are olive brown in sporulating cultures while in P. austricola the obverse is greenish white to pale green and the revers dull green. In P. riverlandense the obverse is white to pale yellow and the reverse yellowish white to dull yellow.
DISCUSSION
The genus Torulomyces (type species: T. lagena) was morphologically characterised by solitary phialides and dry, basipetal conidial chains (Delitsch 1943). Torulomyces viscosus was described at the same time, but the description is patchy and type material is lacking. Therefore, the species is considered as doubtful (Stolk & Samson 1983). Torulomyces lagena was recognised as asexual morph of Eupenicillium limoneum and the asexual morph was combined to Penicillium lagena (Stolk & Samson 1983). Pitt & Hocking (1985) argued that P. lagena should not be considered a Penicillium because of its solitary phialides. Ando et al. (1998) followed this argument and concluded that T. lagena and T. brunneus (≡ Monocillium humicola var. brunneum) are two distinct species. These authors expanded the genus by three further species, namely T. parviverrucosus, T. ovatus, and T. laevis. Houbraken & Samson (2011) investigated the phylogeny of the genus Prenicillium based on partial RPB1, RPB2, Tsr1 (putative ribosome biogenesis protein) and Cct8 (putative chaperonin complex component TCP-1) gene sequences and transferred Torulomyces as section to the genus Penicillium. Visagie et al. (2016) described 12 new species within the section Torulomyces. To date, 14 species are currently accepted in section Torulomyces (Houbraken et al. 2020).
Originally shape, surface structure (tubercle size), and size of conidia were considered to be the main distinguishing characteristics (Ando et al. 1998). With an increasing number of species being recognised in the Sect. Torulomyces, a morphological separation did prove to be more difficult (Visagie et al. 2016), as morphologically identical or at least very similar conidia are shared by several species. Growth rates on different media and temperatures allowed a separation into smaller phenotypic groups which in combination with molecular data allows to discriminate between taxonomic entities. The species newly described here have a slower growth compared to their closest relatives, which is notable as a slow growth rate is generally observed in all Torulomyces species. Specifically, P. tirolense has a remarkably slow growth on all media. For example, its growth rate on CYA is 3–5 mm per week – which is less than half of the growth rate of its closest relatives P. austricola and P. riverlandense. Morphologically P. tirolense differs from P. austricola and P. riverlandense by brown colours (obverse and reverse) on DG18. Penicillium austricola and P. riverlandense were isolated from the South African fynbos soils (Visagie et al. 2016) while P. tirolense was isolated from a sporocarp of S. lacrymans from Tyrol.
Penicillium poederi is also slow growing, but has higher growth rates on MEA and SNA than its closest relatives P. tubakianum and P. wollemiicola. In contrast to P. poederi, P. tubakianum does not produce a soluble pigment. In cultures of P. wollemiicola colourless sclerotia are abundant. Penicillium tubakianaum was originally isolated from bark from Cyathea in New Zealand while P. wollemiciicola was isolated from Wollemi pine litter. The three isolates of P. poederi on the other hand were isolated from soil samples from a lava flow in Iceland, possibly partly explaining the very restricted growth at 30 °C and 37 °C. Slow growing species of fungi are often overlooked in isolation-based approaches because they are outgrown and/or outcompeted by other fungi. Here the isolates were found by using a dilution to extinction approach. This highlights the need of specific sampling approaches to describe and sample the biodiversity of slow growing fungi.
Acknowledgments
SN was funded by the Austrian Science Fund (FWF) project Y801-B16. We are indebted to Nick Cutler for providing us with soil samples from Iceland. We would like to thank Bettina Schneidhofer for assistance in the laboratory.
Footnotes
Conflict of interest: The authors declare that there is no conflict of interest.
Supplementary Material: http://fuse-journal.org/
Culture characteristics of Penicillium poederi sp. nov. after 2 wk incubation. From top to bottom: obverse, reverse, single colony. A. CYA 25 °C. B. CYA 10 °C. C. CYA 30 °C. D. MEA 25 °C. E. MEA 10 °C. F. 30 °C. G. YES 25 °C. H. DG18 25 °C. I. G25N 25 °C. J. SNA 25 °C. K. OA 25°C. L. CREA 25 °C.
Culture characteristics of Penicillium tirolense sp. nov. after 2 wk incubation. From top to bottom: obverse, reverse, single colony. A. CYA 25 °C. B. CYA 10 °C. C. CYA 30 °C. D. MEA 25 °C. E. MEA 10 °C. F. 30 °C. G. YES 25 °C. H. DG18 25 °C. I. G25N 25 °C. J. SNA 25 °C. K. OA 25 °C. L. CYAS 25 °C.
ITS1-5.8S-ITS2 phylogeny (maximum likelihood) of Penicillium sect. Torulomyces. Bootstrap values higher than 80 % are indicated above branches. The new species are highlighted in bold font.
BenA phylogeny (maximum likelihood) of Penicillium sect. Torulomyces. Bootstrap values higher than 80 % are indicated above branches. The new species are highlighted in bold font.
CaM phylogeny (maximum likelihood) of Penicillium sect. Torulomyces. Bootstrap values higher than 80 % are indicated above branches. The new species are highlighted in bold font.
RBP2 phylogeny (maximum likelihood) of Penicillium sect. Torulomyces. Bootstrap values higher than 80 % are indicated above branches. The new species are highlighted in bold font.
REFERENCES
- Ando A, Nawawi A, Manoch L, et al. (1998). Three new species and a new combination in the genus Torulomyces from soil. Mycoscience 39: 313–318. [Google Scholar]
- Bastida F, Bastidal F, Garcia C, et al. (2019). Global ecological predictors of the soil priming effect. Nature Communications 10: 3481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown SP, Jumpponen A. (2014). Contrasting primary successional trajectories of fungi and bacteria in retreating glacier soils. Molecular Ecology 23: 481–497. [DOI] [PubMed] [Google Scholar]
- Cutler NA, Chaput DL, van der Gast CJ. (2014). Long-term changes in soil microbial communities during primary succession. Soil Biology & Biochemistry 69: 359–370. [Google Scholar]
- Delitsch H. (1943). Systematik der Schimmelpilze. J. Neumann, Neudamm, Germany. [Google Scholar]
- Deveau A, Bonito G, Uehling J, et al. (2018). Bacterial-fungal interactions: ecolopgy, mechanisms and challenges. FEMS Microbiology Reviews 42: 335–352. [DOI] [PubMed] [Google Scholar]
- Embacher J, Neuhauser S, Zeilinger S, et al. (2021). Microbiota associated with different developmental stages of the dry rot fungus Serpula lacrymans. Journal of Fungi 7: 354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon J, Knowlton N, Relman DA, et al. (2013). Superorganisms and holobionts. Microbe 8: 152–153. [Google Scholar]
- Guindon S, Dufayard JF, Lefort V, et al. (2010). New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Systematic Biology 59: 307–321. [DOI] [PubMed] [Google Scholar]
- Huelsenbeck JP, Ronquist F. (2001). MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics 17: 754–755. [DOI] [PubMed] [Google Scholar]
- Katoh K, Standley DM. (2013). MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Molecular Biology and Evolution 30: 772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornerup A, Wanscher JH. (1978). Methuen Handbook of Colour, 3rd edn. Methuen & Co., London, UK. [Google Scholar]
- Kumar S, Stecher G, Li M. (2018). MEGA X: molecular evolutionary genetics analysis across computing platforms. Molecular Biology and Evolution 35: 1547–1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margulis L. (1993). Symbiosis in Cell Evolution 2nd edn. Freeman, New York, NY. [Google Scholar]
- Neuhauser S, Huber L, Kirchmair M. (2009). A DNA based detection method for Roesleria subterranea in grapevine roots and soil samples. Phytopathologia Mediterranea 48: 59–72. [PMC free article] [PubMed] [Google Scholar]
- Partida-Martínez LP. (2017). The fungal holobiont: Evidence from early diverging fungi. Environmental Biology 19: 2919–2923. [DOI] [PubMed] [Google Scholar]
- Pitt JI. (1988). A laboratory guide to Common Penicillium species. 2nd edn. Commonwealth Scientific and Industrial Research Organization, Division of Food Processing, Australia. [Google Scholar]
- Pitt JI, Hocking AD. (1985). Interfaces among genera related to Aspergillus and Penicillium. Mycologia 77: 810–824. [Google Scholar]
- Stamatakis A. (2006). RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22: 2688–2690. [DOI] [PubMed] [Google Scholar]
- Stolk AC, Samson RA. (1983). The ascomycete genus Eupenicillium and related Penicillium anamorphs. Studies in Mycology 23: 1–149. [Google Scholar]
- Unterseher M, Schnittler M. (2009). Dilution-to-extinction cultivation of leaf-inhabiting endophytic fungi in beech (Fagus sylvatica L.) – different cultivation techniques influence fungal biodiversity assessment. Mycological Research 113: 645–654. [DOI] [PubMed] [Google Scholar]
- Visagie CM, Houbraken J, Frisvad JC, et al. (2014). Identification and nomenclature of the genus Penicillium. Studies in Mycology 78: 343–371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visagie CM, Houbraken J, Dijksterhuis J, et al. (2016). A taxonomic review of Penicillium species producing conidiophores with solitary phialides, classified in section Torulomyces. Persoonia 36: 134–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Culture characteristics of Penicillium poederi sp. nov. after 2 wk incubation. From top to bottom: obverse, reverse, single colony. A. CYA 25 °C. B. CYA 10 °C. C. CYA 30 °C. D. MEA 25 °C. E. MEA 10 °C. F. 30 °C. G. YES 25 °C. H. DG18 25 °C. I. G25N 25 °C. J. SNA 25 °C. K. OA 25°C. L. CREA 25 °C.
Culture characteristics of Penicillium tirolense sp. nov. after 2 wk incubation. From top to bottom: obverse, reverse, single colony. A. CYA 25 °C. B. CYA 10 °C. C. CYA 30 °C. D. MEA 25 °C. E. MEA 10 °C. F. 30 °C. G. YES 25 °C. H. DG18 25 °C. I. G25N 25 °C. J. SNA 25 °C. K. OA 25 °C. L. CYAS 25 °C.
ITS1-5.8S-ITS2 phylogeny (maximum likelihood) of Penicillium sect. Torulomyces. Bootstrap values higher than 80 % are indicated above branches. The new species are highlighted in bold font.
BenA phylogeny (maximum likelihood) of Penicillium sect. Torulomyces. Bootstrap values higher than 80 % are indicated above branches. The new species are highlighted in bold font.
CaM phylogeny (maximum likelihood) of Penicillium sect. Torulomyces. Bootstrap values higher than 80 % are indicated above branches. The new species are highlighted in bold font.
RBP2 phylogeny (maximum likelihood) of Penicillium sect. Torulomyces. Bootstrap values higher than 80 % are indicated above branches. The new species are highlighted in bold font.