Abstract
Background
Colorectal cancer is a leading cause of cancer deaths. Several screening tests, such as colonoscopy, can be used to find polyps or colorectal cancer. Colonoscopy reports are often written in unstructured narrative text. The information embedded in the reports can be used for various purposes, including colorectal cancer risk prediction, follow-up recommendation, and quality measurement. However, the availability and accessibility of unstructured text data are still insufficient despite the large amounts of accumulated data. We aimed to develop and apply deep learning-based natural language processing (NLP) methods to detect colonoscopic information.
Methods
This study applied several deep learning-based NLP models to colonoscopy reports. Approximately 280,668 colonoscopy reports were extracted from the clinical data warehouse of Samsung Medical Center. For 5,000 reports, procedural information and colonoscopic findings were manually annotated with 17 labels. We compared the long short-term memory (LSTM) and BioBERT model to select the one with the best performance for colonoscopy reports, which was the bidirectional LSTM with conditional random fields. Then, we applied pre-trained word embedding using large unlabeled data (280,668 reports) to the selected model.
Results
The NLP model with pre-trained word embedding performed better for most labels than the model with one-hot encoding. The F1 scores for colonoscopic findings were: 0.9564 for lesions, 0.9722 for locations, 0.9809 for shapes, 0.9720 for colors, 0.9862 for sizes, and 0.9717 for numbers.
Conclusions
This study applied deep learning-based clinical NLP models to extract meaningful information from colonoscopy reports. The method in this study achieved promising results that demonstrate it can be applied to various practical purposes.
Keywords: Natural language processing, Deep learning, Data processing, Information extraction, Colonoscopy
Background
Colorectal cancer is a leading cause of cancer deaths [1–3]. Cancer screening helps in early cancer detection before the appearance of symptoms and reduces cancer mortality. Several screening tests, such as fecal occult blood test, fecal immunochemical test, and colonoscopy, can be used to find polyps or colorectal cancer. The US Preventive Services Task Force recommends colorectal cancer screening in adults aged 50 to 75. [4]. In the United States, colonoscopy prevalence among adults aged 50 years and above tripled from 20% in 2000 to 61% in 2018, primarily due to the Medicare expansion of colonoscopy screening coverage from high-risk individuals to all beneficiaries in 2011 [3]. Since 1999, the National Cancer Screening Program (NCSP) has been implemented in Korea, which provides free screening services for the six most common cancers: stomach, breast, colorectal, cervix, lung, and liver cancer [5]. According to the NCSP protocol, adults aged 50 years and above are eligible to take colorectal screening tests. The participation rates for colonoscopies increased from 25.0% in 2005 to 64.4% in 2020 [6]. The prevalence of colonoscopy, the most reliable way to prevent and detect colorectal cancer, has been increasing.
With the rapid adoption of electronic health records (EHRs), hospitals have accumulated a large amount of unstructured text data in EHR systems, such as discharge summaries, radiology reports, operation notes, and pathology reports. The unstructured text data in EHR systems contain clinically significant information, which is vital for comprehensive care. Regarding colonoscopies, the information embedded in the report can be used for various purposes, including colorectal cancer risk prediction, follow-up recommendations, and quality measurement [7–13]. The information embedded in colonoscopy reports can be used for various purposes, including colorectal cancer risk prediction, follow-up recommendations, and quality measurement. The location, size, number, and appearance of target lesions such as polyps, ulcers, stenosis, and bleeding can determine the risk of colorectal cancer and the follow-up treatment. The colonoscopic findings and procedural information can be used for the assessment of quality indicators, such as adenoma detection rate [14]. However, the availability and accessibility of the unstructured data are still insufficient despite the large amounts of accumulated data.
Natural language processing (NLP) is a computer science subfield that uses computational techniques to learn, understand, and produce human language content [15]. With the impressive advances of deep learning in computer vision and pattern recognition, the recent research in NLP is increasingly emphasizing the use of deep learning methods to overcome the drawbacks of traditional NLP systems, which depend heavily on the time-consuming and often incomplete hand-crafted features [16]. Although clinical NLP research has been actively performed since the 1960s, its progress was slow and lagged behind the progress of NLP in the general domain [17]. Similar to other areas, deep learning-based NLP research in the medical field has repeatedly demonstrated its feasibility [18–20].
Research data integration is essential in cancer research, and there are many efforts to gather and utilize clinical data, such as OHDSI CDM [21]. Although the importance of data has been increasing, many portions of EHR remain unstructured. Clinical NLP is the key to unlocking the evidence buried in clinical narratives. Unfortunately, clinical NLP research still faces several challenges, such as insufficient datasets or the complexity of clinical narratives [22–24]. Although certain pioneering efforts have made clinical text data available for sharing, the number of training datasets are relatively small for practical application. The representative clinical text datasets are MIMIC-III [25] and NLP community challenges, such as n2c2 NLP Research Data Sets [26], ShARe/CLEF eHealth [27], and CEGS N-GRID [19]. Besides, most of the shared datasets emphasize a single type of clinical narrative, like discharge summary [19], which does not reflect the characteristics of various medical specialties, for example, the different types of anatomical structures and their pathologies.
Prior work
Clinical NLP research has recently emphasized the use of deep learning methods, and the publications are increasing yearly. Among deep learning models, recurrent neural network (RNN) has been widely employed in clinical NLP studies [18]. RNN [28] retains the memory of previous computations and uses it in current processing. Using this memory, RNN can capture the inherent sequential nature of language; therefore, it is suited for various NLP tasks such as named entity recognition (NER), machine translation, and speech recognition [16]. However, RNN suffers from the problem of vanishing and exploding gradients, which makes it challenging to learn and tune the parameters of the earlier layers in the networks. Its variants, such as long short-term memory (LSTM) [29] and gated recurrent unit (GRU) [30], have been proposed to overcome the limitation of RNN.
Clinical NER is an essential NLP task for extracting meaningful information from clinical narratives. Recently, numerous efforts have been made to combine RNN variants with other techniques, such as embedding techniques [31], attention mechanisms, and statistical modeling methods like CRFs [32, 33]. Among these techniques, word embedding (or distributed representation), such as Word2Vec [34], GloVe [35], and BERT [36], is a set of language modeling and feature learning techniques in NLP where words or phrases are mapped to a continuous vector space. Typically, word embedding is trained by optimizing an auxiliary objective in large unlabeled and semantic information [16]. Word embedding models trained by Word2Vec and GloVe assign the word to a certain vector, which means these models can only have context-independent representations [37]. BERT is one of the current state-of-the-art language models. Unlike traditional word embeddings such as Word2Vec and GloVe, BERT assign the word to embedding depending on the context, which means the word could have different representations in different contexts by utilizing a transformer network.
In previous studies, clinical standard terminologies such as UMLS or SNOMED CT have enriched word embedding using the semantic relations between clinical concepts [38, 39]. Although the embedding method using clinical standard terminologies is somewhat effective, it is unsuitable for dealing with various synonyms and abbreviated terms in colonoscopy reports. There are multiple expressions to describe colonoscopic findings; for example, "D-colon", "D colon", "Desc. colon", "DC", "mid-d-colon", and "proximal-d-colon" for descending colon; "T-ileum", "T ileum", "TI", and "T.I." for terminal ileum; and "H-flexure", "H flexure", "H-Fx", and "HF" for hepatic flexure. As mentioned in the result section, adding the pre-trained contextual information from the large unlabeled data to the embedding layer demonstrates a slightly better performance than merely learning with annotated data.
Several studies have been published on clinical NLP for colonoscopy, as shown in Table 1. The list of previous studies on clinical NLP for colonoscopies has been excerpted from Table 4 of Fevrier et al. [40], modified, and summarized in Table 1. Most studies have used statistical or rule-based NLP methods. Since there are no publicly available colonoscopy text data, all studies used data from each institution. Most of them focused on extracting information about polyps, such as presence, size, number, and type. Our study covered comprehensive endoscopic findings, such as stenosis, erosion, edema, ulcer, erythema, hyperemia, hemorrhage, and polyp. It isn't easy to directly compare the performance between this study and previous studies due to the difference in data sources and sizes.
Table 1.
Previous studies on clinical NLP for colonoscopy reports
| Year | Author | NLP method (tool) | NLP category |
|---|---|---|---|
| Setting | Dataset | Performance | |
| Current study | Seong et al | Bi-LSTM-CRF, BioBERT | Deep learning-based NLP |
| Samsung Medical Center |
280,668 colonoscopy reports Training and Test: 1,000–5,000 Embedding: 280,668 |
F1 score: 0.9564–0.9862 | |
| 2022 | Bae et al.[13] | SmartTA |
Rule-based NLP (Commercial software) |
| Seoul National University Hospital |
54,562 colonoscopy reports and pathology reports Training: 2,000 Test: 1,000 |
Accuracy: 0.99–1.0 | |
| 2021 | Vadyala et al. [41] | Bio-Bi-LSTM-CRF | Deep learning-based NLP |
| Veterans Affair Medical Centers (VA) |
4,000 colonoscopy reports and pathology reports Training: 3,200 Test: 400 Validation: 400 |
F1 score: 0.85–0.964 | |
| 2020 | Fevrier et al. [40] | SAS PERL regular expression |
Rule-based NLP (Commercial software) |
| Kaiser Permanente Northern California (KPNC) |
401,566 colonoscopy reports and pathology reports Training: 1,000 Validation: 3,000 Test: 397,566 |
Cohen's κ: 0.93–0.99 | |
| 2020 | Karwa et al. [12] | Prolog |
Rule-based NLP (Logic program language) |
| Cleveland Clinic |
2,439 colonoscopy reports Validation: 263 |
Accuracy: 1.0 | |
| 2019 | Lee et al. [11] | Linguamatics I2E [42] |
Rule-based NLP (Commercial software) |
| Kaiser Permanente Northern California (KPNC) |
500 colonoscopy reports Validation: 300 |
Accuracy: 0.893–1.0 | |
| 2017 | Hong et al. [10] | SAS ECC [43] |
Rule-based NLP (Commercial software) |
| Samsung Medical Center (SMC) | 49,450 colonoscopy reports and pathology reports |
Precision: 0.9927 Recall: 0.9983 |
|
| 2017 | Carrell et al. [44] | HITEX [45] |
Statistical NLP (Clinical NLP framework) |
| University of Pittsburgh Medical Center (UPMC) |
3,178 colonoscopy reports and 1,799 pathology reports Training: 1,051 Validation: 2,127 |
F-measure: 0.57–0.99 | |
| 2015 | Raju et al. [46] | CAADRR | Rule-based NLP |
| MD Anderson |
12,748 colonoscopy reports and pathology reports Validation: 343 |
Positive predictive value: 0.913 | |
| 2014 | Gawron et al. [47] | UIMA [48] |
Statistical NLP (NLP framework) |
| Northwestern University |
34,998 colonoscopy reports and 10,186 pathology reports Validation: 200 |
F1 score: 0.81–0.95 | |
| 2013–2015 | Imler et al. [8, 9, 49] | cTAKES [50] |
Statistical NLP (Clinical NLP framework) |
| Veterans Administration medical center |
42,569 colonoscopy reports and pathology reports Training: 250 Test: 500 |
Accuracy: 0.87–0.998 | |
| 2011 | Harkema et al. [51] | GATE [52] |
Statistical NLP (NLP framework) |
| University of Pittsburgh Medical Center (UPMC) | 453 colonoscopy reports and 226 pathology reports |
Accuracy: 0.89 (0.62–1.0) F-measure: 0.74 (0.49–0.89) Cohen’s κ: 0.62 (0.09–0.86) |
Table 4.
Items of the colonoscopy report and assigned labels for annotation
| Items | Labels a |
|---|---|
| 1. Patient information | |
| 1.1 Brief history (disease, family, etc.) | |
| 1.2 Indication/reason for endoscopy | |
| 2. Procedures | |
| 2.1 Sedation and other drugs | |
| 2.1.1 Sedation | SEDATION |
| 2.1.1.1 Level of sedation | SEDATIONLEVEL |
| 2.1.1.2 Medication | MEDICATION |
| 2.1.1.3 Dosage | DOSAGE |
| 2.1.2 Antispasmodics | ANTISPASMODICS |
| 2.2 Equipment (endoscope) used | DEVICE |
| 2.2.1 Extent of examination | EXTENT |
| 2.3 Quality of cleansing/visualization | PREPARATION |
| 2.4 Procedural time | |
| 2.4.1 Time-to-cecum | |
| 2.4.2 Withdrawal time | |
| 2.5 Digital rectal examination | DRE |
| 3. Colonoscopic findings | |
| 3.1 Lesions and their attributes | |
| 3.1.1 Lesion | LESION, NEGATION b |
| 3.1.2 Anatomical site | LOCATION |
| 3.1.3 Shape | SHAPE |
| 3.1.4 Color | COLOR |
| 3.1.5 Size | SIZE |
| 3.1.6 Number | NUMBER |
| 3.2 Sampling (type of sample) | BIOPSY |
| 3.3 Adverse intraprocedural events | |
| 4. Conclusion | |
aA total of 17 labels are used in this study
b"NEGATION" is used to detect negated concepts
Objective
This study aimed to extract meaningful information from colonoscopy reports using deep learning approach. We applied pre-trained word embedding to a deep learning-based NER model using large unlabeled colonoscopy reports. We compared variants of the long short-term memory (LSTM) and BioBERT [53] model to select the one with the best performance for colonoscopy reports, which was the bidirectional LSTM with conditional random fields (CRF). Then we applied pre-trained word embedding using large unlabeled data to the selected model.
Methods
Data collection and text annotation
This study used colonoscopies performed at Samsung Medical Center from 2000 to 2015. Data for this study were extracted from DARWIN-C, the clinical data warehouse of Samsung Medical Center, launched in 2016. As shown in Table 2, the total number of extracted colonoscopy reports was 280,668, of which 5,000 reports from 2011 to 2015 were manually annotated using an open-source web-based text annotation tool named DOCCANO [54]. It provides annotation features for text classification, sequence labeling, and sequence-to-sequence tasks. We made the annotation based on the results of the DARWIN-C project. In the project, we performed text analysis to extract meaningful information from various clinical documents such as pathology, colonoscopy, gastro endoscopy, and radiology reports using a rule-based commercial software named SAS Enterprise Contents Categorization. The extracted results through the text analysis were reviewed and evaluated by clinicians of each department. In this study, two annotators performed the annotation using the tool DOCCANO, and then we manually reviewed all the annotations based on the results of the DARWIN-C project. All colonoscopy reports (280,668) were used for the pre-trained word annotation tool. All colonoscopy reports (280,668) were used for pre-trained word embedding, and the annotated reports (5,000) were used for training the NER model. Table 3 shows the statistics of data used for pre-trained word embedding and training and test.
Table 2.
The number of extracted colonoscopy reports and annotated reports by year
| Year | Colonoscopy reports | Annotated reports |
|---|---|---|
| 2000 | 2,620 | – |
| 2001 | 3,521 | – |
| 2002 | 4,196 | – |
| 2003 | 4,890 | – |
| 2004 | 5,299 | – |
| 2005 | 7,780 | – |
| 2006 | 9,525 | – |
| 2007 | 10,926 | – |
| 2008 | 17,108 | – |
| 2009 | 26,617 | – |
| 2010 | 30,387 | – |
| 2011 | 34,446 | 1,000 |
| 2012 | 32,441 | 1,000 |
| 2013 | 32,103 | 1,000 |
| 2014 | 34,156 | 1,000 |
| 2015 | 24,653 | 1,000 |
| Total | 280,668 | 5,000 |
Table 3.
Data statistics
| Data | For pre-trained word embedding | For training and test |
|---|---|---|
| Year | 2000–2015 | 2011–2015 |
| Number of documents | 280,668 | 5,000 |
| Number of sentences | 4,193,814 | 81,666 |
| Number of types of words | 41,563 | 4,478 |
In general, a colonoscopy report includes various information, such as patient information (indication/reason), procedural information, colonoscopic findings, and assessment (interpretation, conclusion, and recommendation) [55, 56]. Among the several items that describe the result of colonoscopy, the items for colonoscopic findings are an essential part of the colonoscopy report. Table 4 lists the generally used items in a colonoscopy report; labels were assigned to the items to be extracted. A total of 17 labels were used in this study. As shown in Table 4, our study covered comprehensive endoscopic findings. For the lesion of the colonoscopic findings in Table 4, there are two labels; Lesion and Negation. The label "Lesion" presents the presence of lesions and abnormalities. The negation scope of this study is the absence of any lesions, abnormalities, or tumor recurrence. Finding the absence of lesions or tumor recurrence is crucial for determining cancer diagnosis. There are several patterns for negation clues to describe the absence in colonoscopy reports [57]. For example, "There was no evidence of tumor recurrence.", "There was no mucosal lesion.", "There was no other mucosal abnormality." and "There was no definite mass lesion.". But this study excluded a few items like family history, indication, and withdrawal time because most of our colonoscopy reports did not fully describe the information.
As shown in Textbox 1, the colonoscopy report can be divided into two parts: procedures and colonoscopic findings. Procedural information is written in semi-structured text (e.g., Level of sedation: Moderate). On the other hand, colonoscopic findings are often written in free text (e.g., On the distal descending colon, about 0.5 cm sized Is polyp was noticed.). Colonoscopic findings include lesions and their attributes to describe the lesions, such as their anatomical site, size, number, shape, and color. Colonoscopic findings are more complex than procedures, and the terms used for colonoscopic findings are often written using various expressions. For example, one of the anatomical sites, the ascending colon, is written in an unexpected form such as "a-colon", "a colon", "a. colon", "ac", "a. c.", and "a-c". As will be shown later, the accuracy of the extraction of procedural information was much better than that of colonoscopic findings. Our corpus contains a few discontinuous entities, though not many. We assigned the labels two ways for the discontinuous entities: grouping them together or separating them. For example, "on the ascending colon and descending colon" is divided into two entities: "ascending colon" and "descending colon". On the other hand, "on the ascending and descending colon" is assigned to an entity: "ascending and descending colon".
Textbox 1.
An example of the colonoscopy report with annotations. Italicized text indicates the target of annotation, and the terms in square brackets are labels for annotation
|
Clinical information Past (medical) Hx: AGC s/p STG B-II Antithrombotics: No Indication: Checkup Procedure Note Sedation: Yes [SEDATION]: midazolam [MEDICATION] 3 mg [DOSAGE] pethidine [MEDICATION] 50 mg [DOSAGE] Level of sedation: moderate [SEDATIONLEVEL] (paradoxical response: no) Antispasmodics (cimetropium 5 mg): Yes [ANTISPASMODICS] Digital rectal examination was normal [DRE] Bowel preparation was fair [PREPARATION] The CF 260AI [DEVICE] was inserted up to the terminal ileum [EXTENT] Colonoscopic finding On the terminal ileum [LOCATION], several [NUMBER] erosions [LESION] and shallow [SHAPE] ulcer [LESION] were noticed There were several [NUMBER] outpouching lesions [LESION] on the ascending colon [LOCATION]. On the distal descending colon [LOCATION], about 0.5 cm [SIZE] sized Is [SHAPE] polyp [LESION] was noticed. It was removed by cold biopsy. On the rectum [LOCATION], AV 10 cm [LOCATION] about 0.3 cm [SIZE] sized Is [SHAPE] polyp [LESION] was noticed. It was removed by cold biopsy biopsy + [BIOPSY] Conclusion 1. Colon polyp, removed 2. Rectal polyp, removed 3. A-colon diverticulum Comment No immediate complication |
An open-source web-based text annotation tool was used to create training and test datasets. As shown in Table 5, we made five different sizes of annotated datasets which were increased by 1,000, to compare the performance according to the amount of data. The datasets D1, D2, D3, and D4 were randomly generated from 5,000 annotated data. Table 6 shows the distribution of the assigned labels of the datasets. Most of the experiments were performed with dataset D1, except for the performance comparison according to the amount of data. We applied fivefold cross-validation to evaluate the model using all parts of the data. The output file of the text annotation tool was JSON formatted file. It was converted to IOB2 formatted data, where B refers to the beginning of the phrase, I the elements within the phrase, and O the elements outside the phrase [58]. Each token is classified using an IOB label. For example, "on the ascending colon" with the "LOCATION" label was tagged as "O O B-LOC I-LOC". We used partial matches to calculate the performance for the entity consisting of several tokens.
Table 5.
Training and test datasets
| Dataset a | D1 | D2 | D3 | D4 | D5 |
|---|---|---|---|---|---|
| Number of documents | 1,000 | 2,000 | 3,000 | 4,000 | 5,000 |
| Number of sentences | 16,417 | 32,821 | 49,048 | 65,279 | 81,668 |
| Number of words | 92,315 | 184,928 | 277,266 | 369,063 | 461,713 |
| Number of types of words | 2,001 | 2,771 | 3,410 | 3,922 | 4,478 |
aThe dataset sizes were increased by 1,000 to compare the performance according to the amount of data. For evaluation, fivefold cross-validation was applied
Table 6.
Training and test datasets
| Labels | D1 | D2 | D3 | D4 | D5 |
|---|---|---|---|---|---|
| PROCEDURE NOTE | |||||
| SEDATION | 860 | 1,735 | 2,586 | 3,443 | 4,312 |
| SEDATIONLEVEL | 679 | 1,361 | 2,027 | 2,706 | 3,404 |
| MEDICATION | 871 | 1,778 | 2,659 | 3,566 | 4,500 |
| DOSAGE | 872 | 1,781 | 2,663 | 3,576 | 4,515 |
| ANTISPASMODICS | 799 | 1,620 | 2,408 | 3,215 | 4,032 |
| DRE | 995 | 1,990 | 2,986 | 3,982 | 4,979 |
| PREPARATION | 996 | 1,993 | 2,985 | 3,977 | 4,971 |
| DEVICE | 999 | 2,000 | 2,997 | 3,994 | 4,992 |
| EXTENT | 1,000 | 2,000 | 2,998 | 3,995 | 4,992 |
| COLONOSCOPIC FINDINGS | |||||
| LESION | 1,043 | 2,053 | 3,201 | 4,237 | 5,336 |
| LOCATION | 1,118 | 2,269 | 3,481 | 4,599 | 5,757 |
| SHAPE | 719 | 1,513 | 2,296 | 3,024 | 3,795 |
| COLOR | 197 | 373 | 589 | 789 | 983 |
| SIZE | 726 | 1,530 | 2,318 | 3,037 | 3,831 |
| NUMBER | 219 | 416 | 639 | 853 | 1,052 |
| BIOPSY | 995 | 1,993 | 2,991 | 3,987 | 4,984 |
| NEGATION | 651 | 1,300 | 1,929 | 2,609 | 3,240 |
| Total | 13,739 | 27,705 | 41,753 | 55,589 | 69,675 |
Model
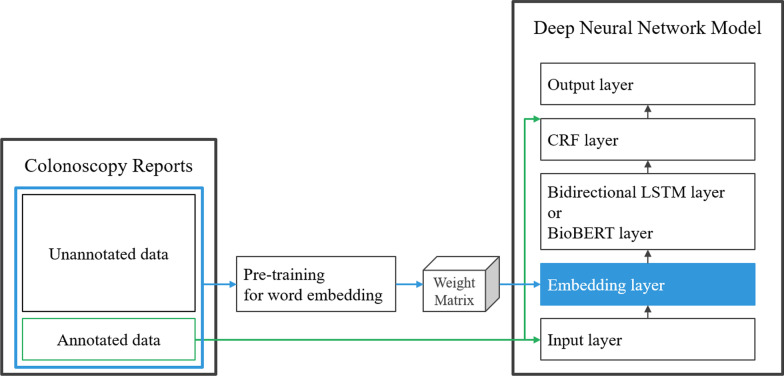
Figure 1 presents the overall architecture of the model. We selected the Bi-LSTM with a CRF layer (Bi-LSTM-CRF) and BioBERT as the model for the current study. It largely contained three layers: input and embedding layer, bidirectional LSTM layer or BioBERT layer, and CRF layer. Annotated data was composed of a set of words and labels used as input and output of the model. A pre-trained word embedding using unannotated data was applied to the embedding layer.
Fig. 1.
The architecture of bidirectional LSTM-CRF and BioBERT with pre-trained word embedding using unannotated data
Input and embedding layer
To prepare a sequence of tokens for the input layer, documents were split into sentences, and sentences were zero-padded to make the input equal in length. Special characters were removed within the criteria that do not alter the number and position of the words and sentences. For the input representation, we assigned each word with an integer value and then converted the unique integer value to a binary vector.
The weight matrix of the input word representation was created using Word2Vec [59], which uses a neural network model to learn word association from a large unlabeled corpus of text. Word2Vec utilizes continuous bag-of-words (CBOW) and skip-gram models. We applied the CBOW model, which learns the conditional probability of a target word given the context words surrounding it across a window. As shown in Table 3, 41,563 words (280,668 colonoscopy reports) were used for training the weight matrix. The embedding layer was seeded with the weight matrix, and the input words were mapped to word vectors.
Bidirectional LSTM layer
NER is a task for identifying meaningful words or phrases in a given text and classifying them into predefined semantic categories. Therefore, we focused on the principle of LSTM [29] to capture the context of the sentence and extract the meaning of each word from the sentence. LSTM is a variant of RNN composed of a cell, and three gates: input, forget, and output. The cell captures the long-term dependencies over any time interval, and the three gates regulate the flow of information into and out of the cell. This unique mechanism can effectively memorize the context of the entire input sentence and overcome vanilla RNN's vanishing and exploding gradient problem. Based on this principle, we constructed a bidirectional LSTM (Bi-LSTM) [60] layer to jointly capture past and future features to obtain a better text representation.
BioBERT layer
BERT utilizes a transformer network to pre-train a language model by jointly conditioning on both the left and right context in all layers. The transformer model introduces a multi-layer, multi-head self-attention mechanism that has demonstrated superiority over RNNs and LSTMs in exploiting GPU-based parallel computation and modeling long-range dependencies in a text [61]. The original BERT model was trained from general domain knowledge, such as Wikipedia and BookCorpus. According to the need for models that can perform better for each domain, domain-specific models such as BioBERT and ClinicalBERT have been developed [53, 62]. This study used an existing pre-trained contextualized word embedding, BiomedNLP-PubMedBERT, which was pre-trained using abstracts from PubMed and full-text articles from PubMedCentral.
CRF layer
The output of the Bi-LSTM was used as input to the CRF layer. CRFs [63], one of the often used statistical modeling methods in the field of NLP [64], are used for structured prediction. CRFs learn the dependencies between labels (i.e., IOB constraints) from training data. For example, I-tag does not appear in the first word of the phrase, and the O-I pattern does not exist in the IOB format. As will be shown later, the model with a CRF layer performs much better than learning without the layer.
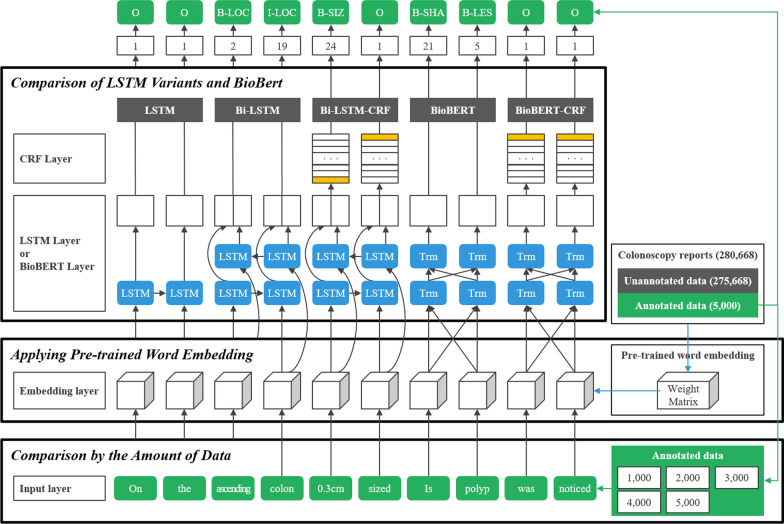
Experiment
This study conducted three experiments, as shown in Fig. 2. First, we compared variants of the LSTM and BioBERT model to the one with the best performance for colonoscopy reports. Then, we applied pre-trained word embedding using unannotated data to the selected model. Additionally, we compared the effect on performance as the training data increased.
Fig. 2.
Three experiments performed in this study
Comparison of LSTM and BioBERT variants
We compared LSTM, Bi-LSTM, Bi-LSTM-CRF, BioBERT, and BioBERT-CRF models with different loss functions and optimizers to select the appropriate model and parameters. For the loss function, Categorical Cross-Entropy (CCE), Kullback–Leibler (KL) divergence [65], and Poisson distribution were used. The CRF layer uses its loss function to learn a set of transition probabilities. For the optimizer, Adaptive Moment Estimation (ADAM) [66], Nesterov-accelerated Adaptive Moment Estimation (NADAM) [67], and Root Mean Square Propagation (RMSProp) [68] were used. For the BioBERT model, we used an existing pre-trained contextualized word embedding, BiomedNLP-PubMedBERT, which was pre-trained using abstracts from PubMed and full-text articles from PubMedCentral [61, 69]. The dataset D1 presented in Table 5 was used in this experiment, and one-hot encoding was used for the input word representation. The experimental parameters were 128 as the dimension of embedding, 256 as the dimension of the LSTM hidden layer, and ten as the epoch.
Applying pre-trained word embedding
As shown in Table 3, about 280,668 colonoscopy reports were trained using Word2Vec to demonstrate the effect of pre-trained word embedding. The CBOW training algorithm was used. For comparison, one-hot encoding and pre-trained word embedding were applied to the Bi-LSTM-CRF model with RMSProp in Table 7, respectively. The dataset D1 presented in Table 5 was used in this experiment. The experimental parameters were 128 as the dimension of word embedding, five as the size of the window, and three as the minimum count of words.
Table 7.
Comparison of LSTM and BioBERT variations
| Model | Loss function a & optimizer b | Precision c | Recall c | F1 score c |
|---|---|---|---|---|
| LSTM | CCE + ADAM | 0.5267 | 0.5297 | 0.5282 |
| LSTM | CCE + NADAM | 0.5258 | 0.5285 | 0.5271 |
| LSTM | CCE + RMS | 0.5266 | 0.5297 | 0.5281 |
| LSTM | KL + ADAM | 0.5255 | 0.5286 | 0.5270 |
| LSTM | KL + NADAM | 0.5258 | 0.5287 | 0.5273 |
| LSTM | KL + RMS | 0.5260 | 0.5278 | 0.5269 |
| LSTM | POISSON + ADAM | 0.5255 | 0.5274 | 0.5264 |
| LSTM | POISSON + NADAM | 0.5245 | 0.5267 | 0.5256 |
| LSTM | POISSON + RMSProp | 0.5229 | 0.5258 | 0.5244 |
| Bi-LSTM | CCE + ADAM | 0.5880 | 0.6761 | 0.6290 |
| Bi-LSTM | CCE + NADAM | 0.5971 | 0.7056 | 0.6460 |
| Bi-LSTM | CCE + RMSProp | 0.5884 | 0.6763 | 0.6293 |
| Bi-LSTM | KL + ADAM | 0.5881 | 0.6768 | 0.6294 |
| Bi-LSTM | KL + NADAM | 0.5957 | 0.7039 | 0.6445 |
| Bi-LSTM | KL + RMSProp | 0.5884 | 0.6767 | 0.6295 |
| Bi-LSTM | POISSON + ADAM | 0.5873 | 0.6756 | 0.6284 |
| Bi-LSTM | POISSON + NADAM | 0.5949 | 0.7021 | 0.6433 |
| Bi-LSTM | POISSON + RMSProp | 0.5869 | 0.6758 | 0.6282 |
| Bi-LSTM-CRF | CRF + ADAM | 0.9828 | 0.9842 | 0.9835 |
| Bi-LSTM-CRF | CRF + NADAM | 0.9825 | 0.9851 | 0.9838 |
| Bi-LSTM-CRF | CRF + RMSProp | 0.9844 | 0.9853 | 0.9848 |
| BioBERT | CCE + ADAM | 0.9824 | 0.9821 | 0.9822 |
| BioBERT-CRF | CRF + ADAM | 0.9810 | 0.9815 | 0.9812 |
aLoss functions; CCE = categorical cross-entropy, KL = Kullback–Leibler divergence, POISSON = Poisson distribution
bOptimizers; ADAM = Adaptive Moment Estimation, NADAM = Nesterov-accelerated Adaptive Moment Estimation, RMSProp = Root Mean Square Propagation
cThe best results are marked in bold
Comparison by the amount of data
The datasets D1, D2, D3, D4, and D5 in Table 5 were used to compare the effect on performance as the amount of training data increased. The Bi-LSTM-CRF model with pre-trained word embedding in Table 8 was used in this experiment.
Table 8.
Comparison between one-hot encoding and pre-trained word embedding
| Labels | Bi-LSTM-CRF + one-hot encoding | Bi-LSTM-CRF + pre-trained word embedding | ||||
|---|---|---|---|---|---|---|
| Precision | Recall | F1 score | Precision | Recall | F1 score | |
| PROCEDURE NOTE a | ||||||
| SEDATION | 0.9881 | 0.9953 | 0.9916 | 0.9888 | 0.9950 | 0.9918 |
| SEDATIONLEVEL | 0.9987 | 0.9938 | 0.9962 | 0.9985 | 0.9958 | 0.9971 |
| MEDICATION | 0.9991 | 0.9954 | 0.9972 | 1 | 0.9959 | 0.9980 |
| DOSAGE | 0.9929 | 0.9897 | 0.9913 | 0.9959 | 0.9920 | 0.9939 |
| ANTISPASMODICS | 0.9962 | 1 | 0.9981 | 0.9978 | 1 | 0.9989 |
| DRE | 0.9967 | 0.9990 | 0.9978 | 0.9958 | 0.9989 | 0.9973 |
| PREPARATION | 0.9892 | 0.9914 | 0.9903 | 0.9879 | 0.9928 | 0.9904 |
| DEVICE | 0.9991 | 0.9991 | 0.9991 | 0.9980 | 0.9979 | 0.9979 |
| EXTENT | 0.9883 | 0.9951 | 0.9916 | 0.9960 | 0.9967 | 0.9963 |
| COLONOSCOPIC FINDINGS b | ||||||
| LESION | 0.9881 | 0.9953 | 0.9916 | 0.9888 | 0.9950 | 0.9918 |
| LOCATION | 0.9987 | 0.9938 | 0.9962 | 0.9985 | 0.9958 | 0.9971 |
| SHAPE | 0.9991 | 0.9954 | 0.9972 | 1 | 0.9959 | 0.9980 |
| COLOR | 0.9929 | 0.9897 | 0.9913 | 0.9959 | 0.9920 | 0.9939 |
| SIZE | 0.9962 | 1 | 0.9981 | 0.9978 | 1 | 0.9989 |
| NUMBER | 0.9967 | 0.9990 | 0.9978 | 0.9958 | 0.9989 | 0.9973 |
| BIOPSY | 0.9892 | 0.9914 | 0.9903 | 0.9879 | 0.9928 | 0.9904 |
| NEGATION | 0.9991 | 0.9991 | 0.9991 | 0.9980 | 0.9979 | 0.9979 |
| MICROAVG | 0.9883 | 0.9951 | 0.9916 | 0.9960 | 0.9967 | 0.9963 |
aProcedure note was written in semi-structured text. The best results are marked in bold
bColonoscopic findings were written in free text. The best results are marked in bold
Results
Comparison of LSTM and BioBERT variants
In Table 7, the experimental result shows the performance of LSTM variants with the combinations of loss functions and optimizers. As shown in Table 7, the Bi-LSTM-CRF model with RMSProp achieved the best performance with a precision of 0.9844, a recall of 0.9853, and an F1-score of 0.9848. The model with a CRF layer performed much better than the others. The CRF layer was a vital component in the NER problem. There was little difference in the performance depending on the loss functions and optimizers. We also compared GRU variants; there was no significant difference in the performance between LSTM and GRU models. All measures in Table 7 were micro-averaged to capture each label imbalance adequately.
Applying word embedding
About 280,668 colonoscopy reports were trained using Word2Vec to demonstrate the effect of pre-trained word embedding. Table 8 shows the performance of each label; the model with pre-trained word embedding performed better for most labels than the model with one-hot encoding. In the case of the labels for procedure notes, both one-hot encoding and pre-trained word embedding had F1 scores of more than 0.99 because the procedure note was written in semi-structured text. In the case of the labels for colonoscopic findings, adding pre-trained word embedding improved the performance at a certain rate. Figure 3 shows the effect of pre-trained word embedding for colonoscopic findings.
Fig. 3.
Performance of pre-trained word embedding
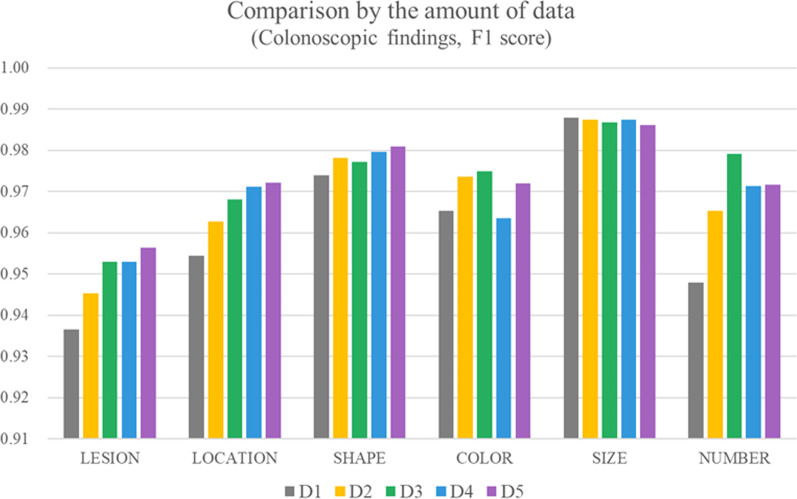
Comparison by the amount of data
As shown in Table 9, the performance slightly improved as data increased. As previously mentioned, F1 scores of the labels for procedure notes were more than 0.99 due to semi-structured patterns. As shown in Fig. 4, F1 scores of LESION, LOCATION, and SHAPE improved as the amount of data increased. COLOR and NUMBER had the best F1 scores in D3. SIZE had a similar performance for all data.
Table 9.
Comparison by the amount of data (F1 score)
| Labels | D1 | D2 | D3 | D4 | D5 |
|---|---|---|---|---|---|
| COLONOSCOPIC FINDINGS a | |||||
| LESION | 0.9366 | 0.9453 | 0.9530 | 0.9530 | 0.9564 |
| LOCATION | 0.9545 | 0.9627 | 0.9681 | 0.9711 | 0.9722 |
| SHAPE | 0.9739 | 0.9782 | 0.9772 | 0.9797 | 0.9809 |
| COLOR | 0.9653 | 0.9736 | 0.9749 | 0.9636 | 0.9720 |
| SIZE | 0.9879 | 0.9874 | 0.9867 | 0.9875 | 0.9862 |
| NUMBER | 0.9480 | 0.9653 | 0.9791 | 0.9713 | 0.9717 |
| BIOPSY | 0.9975 | 0.9985 | 0.9988 | 0.9989 | 0.9992 |
| NEGATION | 0.9772 | 0.9784 | 0.9845 | 0.9815 | 0.9858 |
| MICROAVG | 0.9892 | 0.9912 | 0.9921 | 0.9921 | 0.9924 |
aThe best results are marked in bold
Fig. 4.
Comparison by the amount of data
As an analysis of errors of this model, the errors came from various reasons, such as vocabulary: synonyms, acronyms, and typos (i.e., easy touch bleeingd), grammatical mistakes (i.e., from cecum ~ t colon), and incorrect extraction of size and location (i.e., on the 80 cm from anal verge).
Discussion
This study has shown the feasibility of deep learning-based NLP methods to extract meaningful information from colonoscopy reports. Since there are no publicly available colonoscopy text data, 5,000 out of 280,668 colonoscopy reports were manually annotated using an open-source web-based text annotation tool. In the initial planning phase of this study, we assumed that 275,668 unannotated data could improve performance because it contained real-world synonyms and acronyms used in our institution. We compared LSTM, Bi-LSTM, Bi-LSTM-CRF, BioBERT, and BioBERT-CRF models to select the one with the best performance for colonoscopy reports. Although BioBERT performed much better than Bi-LSTM without a CRF layer, the performance of Bi-LSTM with a CRF layer was slightly higher than others. We applied pre-trained word embedding using large unlabeled data to the Bi-LSTM-CRF model. Therefore, the NER model with pre-trained word embedding performed better for most labels than the model without pre-trained word embedding. Although the deep learning-based NLP method performs much better than the traditional NLP method, the currently available public data is insufficient to cover the characteristics of various medical specialties. The method in this study could be effective in the absence of a shared colonoscopy dataset.
This study has the following limitations. First, the study was conducted in a single institution, so it is possible that the model could not handle colonoscopy reports from other institutions. They can differ in many aspects, such as writing patterns, templates, and vocabulary. Although our model may not apply to other colonoscopy reports directly, our approach can be used in others. Second, there are no available colonoscopy datasets to compare the performance of our model. Evaluating the performance of the model is not possible, but we believe that the performance level of our model is sufficient for clinical applications. Third, we need to consider synonyms, acronyms, and typos and be able to process them. There are various synonyms and acronyms to describe colonoscopic findings and anatomical sites; for example, "D-colon", "D colon", "Desc. colon", "DC", "mid-d-colon", and "proximal-d-colon" for descending colon; "T-ileum", "T ileum", "TI", and "T.I." for terminal ileum; and "H-flexure", "H flexure", "H-Fx", and "HF" for hepatic flexure.
Conclusions
Realizing the full potential of precision medicine begins with identifying better ways to collect, share, and make decisions based on data [70]. Although the importance of data has been increasing, clinical data is still challenging to use as many portions of EHR remain unstructured. Clinical NLP is the key to unlocking the evidence buried in clinical narratives. With the advancements in deep learning techniques, NLP research has achieved remarkable performance in the clinical domain. However, obtaining sufficient public data for training is difficult. The currently available public data is insufficient to cover the characteristics of various medical specialties.
This study addresses the first deep learning-based NLP method for NER in colonoscopy reports. It is an important problem for clinical utility, such as cancer risk prediction, follow-up recommendation, and quality measurement. We applied a deep learning-based clinical NER with pre-trained word embedding. The method in this study achieves promising results that demonstrate it can be helpful for various practical purposes, including clinical document summarization and automated structured report generation.
Acknowledgements
Not applicable
Abbreviations
- ADAM
Adaptive moment estimation
- Bi-LSTM
Bidirectional LSTM
- Bi-LSTM-CRF
Bidirectional LSTM with CRF layer
- CBOW
Continuous bag-of-words
- CCE
Categorical cross-entropy
- CRF
Conditional random fields
- EHR
Electronic health record
- GRU
Gated recurrent unit
- KL
Kullback–Leibler divergence
- LSTM
Long short-term memory
- NADAM
Nesterov-accelerated adaptive moment estimation
- NCSP
National cancer screening program
- NER
Named entity recognition
- NLP
Natural language processing
- RMSProp
Root mean square propagation
- RNN
Recurrent neural network
Author contributions
All of the authors contributed to the study. DS wrote the first draft of the manuscript. DS and BY conceived the idea for this manuscript. DS and SS contributed to this project's data collection and text annotation. DS and YC contributed to the preliminary design of the method and implementation of the project. SS and BY provided critical comments and revised the manuscript. All of the authors read and approved the final manuscript.
Funding
This research was supported by a grant of the Korea Health Technology R&D Project through the Korea Health Industry Development Institute (KHIDI), funded by the Ministry of Health & Welfare, Republic of Korea (Grant Number: HI19C1328).
Availability of data and materials
The datasets generated and analyzed during the current study are not publicly available because of privacy issues but are available from the corresponding author upon reasonable request. The code implementation of this study is publicly accessible from GitHub, https://github.com/dhseong/Deep-Learning-based-NLP-for-Colonoscopy [71].
Declarations
Ethics approval and consent to participate
This study was approved by the Institutional Review Board of Samsung Medical Center (IRB No. 2019–08-022) and was conducted in full concordance with the Declaration of Helsinki. The requirement for informed patient consent was waived by the Institutional Review Board of Samsung Medical Center due to its retrospective design.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, Bray F. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71(3):209–249. doi: 10.3322/caac.21660. [DOI] [PubMed] [Google Scholar]
- 2.Kang MJ, Won Y-J, Lee JJ, Jung K-W, Kim H-J, Kong H-J, Im J-S, Seo HG. Cancer statistics in Korea: incidence, mortality, survival, and prevalence in 2019. Cancer Res Treat. 2022;54(2):330–344. doi: 10.4143/crt.2022.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Siegel RL, Miller KD, Goding Sauer A, Fedewa SA, Butterly LF, Anderson JC, Cercek A, Smith RA, Jemal A. Colorectal cancer statistics, 2020. CA Cancer J Clin. 2020;70(3):145–164. doi: 10.3322/caac.21601. [DOI] [PubMed] [Google Scholar]
- 4.US Preventive Services Task Force Screening for colorectal cancer: us preventive services task force recommendation statement. JAMA. 2021;325(19):1965–1977. doi: 10.1001/jama.2021.6238. [DOI] [PubMed] [Google Scholar]
- 5.Korea National Cancer Center. National Cancer Control Programs. https://www.ncc.re.kr/main.ncc?uri=english/sub04_ControlPrograms. Accessed 20 Jan 2023.
- 6.Park B, Lee YY, Song SY, Shin HY, Suh M, Choi KS, Jun JK. Trends of colorectal cancer screening rates in Korea: Korean national cancer screening survey 2005–2020. Gut Liver. 2022. [DOI] [PMC free article] [PubMed]
- 7.Hou JK, Chang M, Nguyen T, Kramer JR, Richardson P, Sansgiry S, D'Avolio LW, El-Serag HB. Automated identification of surveillance colonoscopy in inflammatory bowel disease using natural language processing. Dig Dis Sci. 2013;58(4):936–941. doi: 10.1007/s10620-012-2433-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Imler TD, Morea J, Imperiale TF. Clinical decision support with natural language processing facilitates determination of colonoscopy surveillance intervals. Clin Gastroenterol Hepatol. 2014;12(7):1130–1136. doi: 10.1016/j.cgh.2013.11.025. [DOI] [PubMed] [Google Scholar]
- 9.Imler TD, Morea J, Kahi C, Cardwell J, Johnson CS, Xu H, Ahnen D, Antaki F, Ashley C, Baffy G, et al. Multi-center colonoscopy quality measurement utilizing natural language processing. Am J Gastroenterol. 2015;110(4):543–552. doi: 10.1038/ajg.2015.51. [DOI] [PubMed] [Google Scholar]
- 10.Hong SN, Son HJ, Choi SK, Chang DK, Kim YH, Jung SH, Rhee PL. A prediction model for advanced colorectal neoplasia in an asymptomatic screening population. PLoS ONE. 2017;12(8):e0181040. doi: 10.1371/journal.pone.0181040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee JK, Jensen CD, Levin TR, Zauber AG, Doubeni CA, Zhao WK, Corley DA. Accurate identification of colonoscopy quality and polyp findings using natural language processing. J Clin Gastroenterol. 2019;53(1):e25–e30. doi: 10.1097/MCG.0000000000000929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Karwa A, Patell R, Parthasarathy G, Lopez R, McMichael J, Burke CA. Development of an automated algorithm to generate guideline-based recommendations for follow-up colonoscopy. Clin Gastroenterol Hepatol. 2020;18(9):2038–2045. doi: 10.1016/j.cgh.2019.10.013. [DOI] [PubMed] [Google Scholar]
- 13.Bae JH, Han HW, Yang SY, Song G, Sa S, Chung GE, Seo JY, Jin EH, Kim H, An D. Natural language processing for assessing quality indicators in free-text colonoscopy and pathology reports: development and usability study. JMIR Med Inform. 2022;10(4):e35257. doi: 10.2196/35257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rex DK, Schoenfeld PS, Cohen J, Pike IM, Adler DG, Fennerty MB, Lieb JG, Park WG, Rizk MK, Sawhney MS, et al. Quality indicators for colonoscopy. Gastrointest Endosc. 2015;81(1):31–53. doi: 10.1016/j.gie.2014.07.058. [DOI] [PubMed] [Google Scholar]
- 15.Hirschberg J, Manning CD. Advances in natural language processing. Science. 2015;349(6245):261–266. doi: 10.1126/science.aaa8685. [DOI] [PubMed] [Google Scholar]
- 16.Young T, Hazarika D, Poria S, Cambria E. Recent trends in deep learning based natural language processing. IEEE Comput Intell Mag. 2018;13(3):55–75. [Google Scholar]
- 17.Sfakianaki P, Koumakis L, Sfakianakis S, Iatraki G, Zacharioudakis G, Graf N, Marias K, Tsiknakis M. Semantic biomedical resource discovery: a Natural Language Processing framework. BMC Med Inform Decis Mak. 2015;15(77). [DOI] [PMC free article] [PubMed]
- 18.Wu S, Roberts K, Datta S, Du J, Ji Z, Si Y, Soni S, Wang Q, Wei Q, Xiang Y, et al. Deep learning in clinical natural language processing: a methodical review. J Am Med Inform Assoc. 2020;27(3):457–470. doi: 10.1093/jamia/ocz200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Spasic I, Nenadic G. Clinical Text Data in Machine Learning: Systematic Review. JMIR Med Inform. 2020;8(3):e17984. doi: 10.2196/17984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Spasic I, Uzuner O, Zhou L. Emerging clinical applications of text analytics. Int J Med Inform. 2020;134(103974). [DOI] [PubMed]
- 21.Ryu B, Yoon E, Kim S, Lee S, Baek H, Yi S, Na HY, Kim J-W, Baek R-M, Hwang H, et al. Transformation of pathology reports into the common data model with oncology module: use case for colon cancer. J Med Internet Res. 2020;22(12):e18526. doi: 10.2196/18526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wen A, Fu S, Moon S, El Wazir M, Rosenbaum A, Kaggal VC, Liu S, Sohn S, Liu H, Fan J. Desiderata for delivering NLP to accelerate healthcare AI advancement and a Mayo Clinic NLP-as-a-service implementation. NPJ Digit Med. 2019;2:130. doi: 10.1038/s41746-019-0208-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chapman WW, Nadkarni PM, Hirschman L, D'Avolio LW, Savova GK, Uzuner O. Overcoming barriers to NLP for clinical text: the role of shared tasks and the need for additional creative solutions. J Am Med Inform Assoc. 2011;18(5):540–543. doi: 10.1136/amiajnl-2011-000465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Leaman R, Khare R, Lu Z. Challenges in clinical natural language processing for automated disorder normalization. J Biomed Inform. 2015;57:28–37. doi: 10.1016/j.jbi.2015.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Johnson AE, Pollard TJ, Shen L, Lehman LW, Feng M, Ghassemi M, Moody B, Szolovits P, Celi LA, Mark RG. MIMIC-III, a freely accessible critical care database. Sci Data. 2016;3:160035. doi: 10.1038/sdata.2016.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.n2c2 NLP Research Data Sets. https://portal.dbmi.hms.harvard.edu/projects/n2c2-nlp/. Accessed 20 Jan 2023.
- 27.Suominen H, Salanterä S, Velupillai S, Chapman WW, Savova G, Elhadad N, Pradhan S, South BR, Mowery DL, Jones GJF et al. Overview of the ShARe/CLEF eHealth Evaluation Lab 2013. In: Lecture Notes in Computer Science. Springer Berlin Heidelberg; 2013: 212–231.
- 28.Elman JL. Finding structure in time. Cogn Sci. 1990;14(2):179–211. [Google Scholar]
- 29.Hochreiter S, Schmidhuber J. Long short-term memory. Neural Comput. 1997;9(8):1735–1780. doi: 10.1162/neco.1997.9.8.1735. [DOI] [PubMed] [Google Scholar]
- 30.Cho K, Van Merriënboer B, Gulcehre C, Bahdanau D, Bougares F, Schwenk H, Bengio Y. Learning phrase representations using RNN encoder-decoder for statistical machine translation. 2014. arXiv preprint arXiv:14061078.
- 31.Peters ME, Neumann M, Iyyer M, Gardner M, Clark C, Lee K, Zettlemoyer L. Deep contextualized word representations. 2018.
- 32.Xu K, Zhou Z, Hao T, Liu W. A Bidirectional LSTM and Conditional Random Fields Approach to Medical Named Entity Recognition. In: Proceedings of the International Conference on Advanced Intelligent Systems and Informatics 2017: Springer International Publishing; 2018:355–365.
- 33.Huang Z, Xu W, Yu K. Bidirectional LSTM-CRF Models for Sequence Tagging. 2015.
- 34.Mikolov T, Sutskever I, Chen K, Corrado G, Dean J. Distributed representations of words and phrases and their compositionality. 2013.
- 35.Pennington J, Socher R, Manning C. Glove: global vectors for word representation. In: Proceedings of the 2014 Conference on Empirical Methods in Natural Language Processing (EMNLP): Association for Computational Linguistics; 2014.
- 36.Devlin J, Chang M-W, Lee K, Toutanova K. BERT: Pre-training of deep bidirectional transformers for language understanding. 2018.
- 37.Sun C, Yang Z, Wang L, Zhang Y, Lin H, Wang J. Biomedical named entity recognition using BERT in the machine reading comprehension framework. J Biomed Inform. 2021;118:103799. doi: 10.1016/j.jbi.2021.103799. [DOI] [PubMed] [Google Scholar]
- 38.Alawad M, Hasan SMS, Christian JB, Tourassi G. Retrofitting word embeddings with the UMLS metathesaurus for clinical information extraction. In: 2018 IEEE International Conference on Big Data; 2018:2838–2846.
- 39.Soriano IM, Peña JLC, Breis JTF, Román IS, Barriuso AA, Baraza DG. Snomed2Vec: Representation of SNOMED CT terms with Word2Vec. In: 2019 IEEE 32nd International Symposium on Computer-Based Medical Systems (CBMS); 2019:678–683.
- 40.Fevrier HB, Liu L, Herrinton LJ, Li D. A transparent and adaptable method to extract colonoscopy and pathology data using natural language processing. J Med Syst. 2020;44(9):151. doi: 10.1007/s10916-020-01604-8. [DOI] [PubMed] [Google Scholar]
- 41.Vadyala SR, Sherer EA. Natural language processing accurately categorizes indications, findings and pathology reports from multicenter colonoscopy. ArXiv. 2021;abs/2108.11034.
- 42.Hartmann J, Van Keuren L. Text mining for clinical support. J Med Libr Assoc. 2019;107(4):603–605. doi: 10.5195/jmla.2019.758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Strauss JA, Chao CR, Kwan ML, Ahmed SA, Schottinger JE, Quinn VP. Identifying primary and recurrent cancers using a SAS-based natural language processing algorithm. J Am Med Inform Assoc. 2013;20(2):349–355. doi: 10.1136/amiajnl-2012-000928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Carrell DS, Schoen RE, Leffler DA, Morris M, Rose S, Baer A, Crockett SD, Gourevitch RA, Dean KM, Mehrotra A. Challenges in adapting existing clinical natural language processing systems to multiple, diverse health care settings. J Am Med Inform Assoc. 2017;24(5):986–991. doi: 10.1093/jamia/ocx039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zeng QT, Goryachev S, Weiss S, Sordo M, Murphy SN, Lazarus R. Extracting principal diagnosis, co-morbidity and smoking status for asthma research: evaluation of a natural language processing system. BMC Med Inform Decis Mak. 2006;6:30. doi: 10.1186/1472-6947-6-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Raju GS, Lum PJ, Slack RS, Thirumurthi S, Lynch PM, Miller E, Weston BR, Davila ML, Bhutani MS, Shafi MA, et al. Natural language processing as an alternative to manual reporting of colonoscopy quality metrics. Gastrointest Endosc. 2015;82(3):512–519. doi: 10.1016/j.gie.2015.01.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gawron AJ, Thompson WK, Keswani RN, Rasmussen LV, Kho AN. Anatomic and advanced adenoma detection rates as quality metrics determined via natural language processing. Am J Gastroenterol. 2014;109(12):1844–1849. doi: 10.1038/ajg.2014.147. [DOI] [PubMed] [Google Scholar]
- 48.Ferrucci D, Lally A. UIMA: an architectural approach to unstructured information processing in the corporate research environment. Nat Lang Eng. 2004;10(3–4):327–348. [Google Scholar]
- 49.Imler TD, Morea J, Kahi C, Imperiale TF. Natural language processing accurately categorizes findings from colonoscopy and pathology reports. Clin Gastroenterol Hepatol. 2013;11(6):689–694. doi: 10.1016/j.cgh.2012.11.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Savova GK, Masanz JJ, Ogren PV, Zheng J, Sohn S, Kipper-Schuler KC, Chute CG. Mayo clinical text analysis and knowledge extraction system (cTAKES): architecture, component evaluation and applications. J Am Med Inform Assoc. 2010;17(5):507–513. doi: 10.1136/jamia.2009.001560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Harkema H, Chapman WW, Saul M, Dellon ES, Schoen RE, Mehrotra A. Developing a natural language processing application for measuring the quality of colonoscopy procedures. J Am Med Inform Assoc. 2011;18(Suppl 1):i150–156. doi: 10.1136/amiajnl-2011-000431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cunningham H, Tablan V, Roberts A, Bontcheva K. Getting more out of biomedical documents with GATE's full lifecycle open source text analytics. PLoS Comput Biol. 2013;9(2):e1002854. doi: 10.1371/journal.pcbi.1002854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lee J, Yoon W, Kim S, Kim D, Kim S, So CH, Kang J. BioBERT: a pre-trained biomedical language representation model for biomedical text mining. Bioinformatics. 2019. [DOI] [PMC free article] [PubMed]
- 54.DOCCANO, open-source text annotation tool. https://github.com/doccano/doccano. Accessed 20 January 2023
- 55.Aabakken L, Barkun AN, Cotton PB, Fedorov E, Fujino MA, Ivanova E, Kudo SE, Kuznetzov K, de Lange T, Matsuda K, et al. Standardized endoscopic reporting. J Gastroenterol Hepatol. 2014;29(2):234–240. doi: 10.1111/jgh.12489. [DOI] [PubMed] [Google Scholar]
- 56.Aabakken L, Rembacken B, LeMoine O, Kuznetsov K, Rey JF, Rosch T, Eisen G, Cotton P, Fujino M. Minimal standard terminology for gastrointestinal endoscopy–MST 3.0. Endoscopy. 2009;41(8):727–728. doi: 10.1055/s-0029-1214949. [DOI] [PubMed] [Google Scholar]
- 57.Solarte Pabón O, Montenegro O, Torrente M, Rodríguez González A, Provencio M, Menasalvas E. Negation and uncertainty detection in clinical texts written in Spanish: a deep learning-based approach. Peer J Comput Sci. 2022;8:e913. doi: 10.7717/peerj-cs.913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kudo T, Matsumoto Y. Chunking with support vector machines. In: Second meeting of the north american chapter of the association for computational linguistics; 2001.
- 59.Mikolov T, Sutskever I, Chen K, Corrado GS, Dean J. Distributed representations of words and phrases and their compositionality. In: NIPS Proceedings. 2013:3111–3119.
- 60.Graves A, Schmidhuber J. Framewise phoneme classification with bidirectional LSTM networks. In: Proceedings 2005 IEEE International Joint Conference on Neural Networks; 2005:2047–2052. [DOI] [PubMed]
- 61.Gu Y, Tinn R, Cheng H, Lucas M, Usuyama N, Liu X, Naumann T, Gao J, Poon H. Domain-specific language model pretraining for biomedical natural language processing. ACM Trans Comput Healthc. 2022;3(1):1–23. [Google Scholar]
- 62.Alsentzer E, Murphy JR, Boag W, Weng W-H, Jin D, Naumann T, McDermott MBA. Publicly Available Clinical BERT Embeddings. ArXiv. 2019;abs/1904.03323.
- 63.Lafferty J, McCallum A, Pereira FC. Conditional random fields: Probabilistic models for segmenting and labeling sequence data. In: ICML '01: Proceedings of the Eighteenth International Conference on Machine Learning; 2001:282–289.
- 64.Uzuner O, South BR, Shen S, DuVall SL. 2010 i2b2/VA challenge on concepts, assertions, and relations in clinical text. J Am Med Inform Assoc. 2011;18(5):552–556. doi: 10.1136/amiajnl-2011-000203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kullback S, Leibler RA. On information and sufficiency. Ann Math Stat. 1951;22(1):79–86. [Google Scholar]
- 66.Kingma DP, Ba J. Adam: a method for stochastic optimization. 2014. arXiv preprint arXiv:14126980.
- 67.Dozat T. Incorporating nesterov momentum into adam. In: ICLR; 2016.
- 68.Tieleman T, Hinton G. Lecture 6.5-rmsprop: Divide the gradient by a running average of its recent magnitude. COURSERA: Neural networks for machine learning. 2012;4(2):26–31.
- 69.BiomedNLP-PubMedBERT-base-uncased-abstract-fulltext. https://huggingface.co/microsoft/BiomedNLP-PubMedBERT-base-uncased-abstract-fulltext/tree/main. Accessed 20 January 2023
- 70.Dzau VJ, Ginsburg GS. Realizing the full potential of precision medicine in health and health care. JAMA. 2016;316(16):1659–1660. doi: 10.1001/jama.2016.14117. [DOI] [PubMed] [Google Scholar]
- 71.GitHub. Deep learning-based natural language processing for colonoscopy reports. https://github.com/dhseong/Deep-Learning-based-NLP-for-Colonoscopy. Accessed 20 January 2023
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets generated and analyzed during the current study are not publicly available because of privacy issues but are available from the corresponding author upon reasonable request. The code implementation of this study is publicly accessible from GitHub, https://github.com/dhseong/Deep-Learning-based-NLP-for-Colonoscopy [71].