Abstract
Polar flagella of Vibrio species can rotate at speeds as high as 100,000 rpm and effectively propel the bacteria in liquid as fast as 60 μm/s. The sodium motive force powers rotation of the filament, which acts as a propeller. The filament is complex, composed of multiple subunits, and sheathed by an extension of the cell outer membrane. The regulatory circuitry controlling expression of the polar flagellar genes of members of the Vibrionaceae is different from the peritrichous system of enteric bacteria or the polar system of Caulobacter crescentus. The scheme of gene control is also pertinent to other members of the gamma purple bacteria, in particular to Pseudomonas species. This review uses the framework of the polar flagellar system of Vibrio parahaemolyticus to provide a synthesis of what is known about polar motility systems of the Vibrionaceae. In addition to its propulsive role, the single polar flagellum of V. parahaemolyticus is believed to act as a tactile sensor controlling surface-induced gene expression. Under conditions that impede rotation of the polar flagellum, an alternate, lateral flagellar motility system is induced that enables movement through viscous environments and over surfaces. Although the dual flagellar systems possess no shared structural components and although distinct type III secretion systems direct the simultaneous placement and assembly of polar and lateral organelles, movement is coordinated by shared chemotaxis machinery.
Flagella, which act as semirigid helical propellers, provide bacteria with a highly efficient means of locomotion. For example, many Vibrio and Pseudomonas species swim in liquid environments at speeds as fast as 60 μm/s (10, 11, 64, 189). The propellers are powered by reversible rotary motors embedded in the cell membrane, which can turn the flagellum at rates as high as 1,700 revolutions per s (rps) (115). Energy for rotation of the motor is derived from either the sodium or proton membrane potential (72, 117). The number and arrangement of the propellers can vary, but the mode of insertion is of two major types, i.e., polar or peritrichous. Flagella play other roles in addition to swimming in liquid (reviewed in reference 132). They can enable bacteria to move over and colonize surfaces, a process called swarming (63). They also participate in adhesion. Attachment of bacteria to surfaces is often first mediated by contact of the flagellum with the surface (127). As propulsive organelles, flagella seems to aid in overcoming negative electrostatic interactions and thus are believed to play a key role in the initial steps of adsorption of bacteria to surfaces, biofilm formation, and invasion of hosts (30, 39, 154). Studies using Vibrio alginolyticus have demonstrated that attachment to glass is directly proportional to swimming speed (90). Other studies have shown that by disabling the flagellar motor of the fish pathogen V. anguillarum, invasion into the fish host is severely reduced (142). In addition, some flagella are sheathed by a membrane that appears to be an extension of the outer cell membrane. The composition of this sheath (specifically, lipopolysaccharide and protein) may allow additional specific interactions between the bacterium and a surface (77, 163, 164).
Extensive structural and genetic analysis of the unsheathed, peritrichous flagella of Escherichia coli and Salmonella enterica serovar Typimurium has deciphered the complexity of the organelle, its assembly process, motor function, and the coordination of movement (i.e., chemotaxis) (reviewed in references 17, 21, 111, and 135). Many of these features are conserved in flagellar systems of other bacteria; however, novel permutations also exist. For example, the flagella of spirochetes are not external to the cell but are contained within the periplasmic space (103). These flagella also play a skeletal role in determining the spiral shape of the cell (133). Another example is the single flagellum of Caulobacter crescentus, which is assembled at one pole during asymmetric cell division and is later ejected and replaced by a stalk (reviewed in references 140 and 195). A number of bacteria are polarly flagellated, yet, aside from C. crescentus, comparatively little is known about polar flagellar systems. This review uses the polar flagellar system of V. parahaemolyticus as a focal point to build a framework for describing what is known about polar systems of the gamma purple bacteria, most particularly Vibrio but with some reference to Pseudomonas species. The reader is also referred to the excellent review by Yorimitsu and Homma, which focuses on flagellar motors of the Vibrionaceae family (202). Although our understanding of these polar motility systems is not comprehensive, it is hoped that this review will provide a general context for polar systems and points of contrast with what is known for the well-studied peritrichous systems of E. coli and S. enterica serovar Typhimurium.
V. parahaemolyticus is a common gram-negative bacterium in marine and estuarine environments. It is also a human pathogen and a worldwide cause of gastroenteritis. In areas of the world where seafood consumption is high, such as Southeast Asia, it is the primary cause of food poisoning (24, 78, 193). It is a serious emerging pathogen in North America, where it is the most common Vibrio species isolated from humans and the most frequent cause of Vibrio-associated gastroenteritis (37, 175). Infections are usually associated with consumption of raw or undercooked shellfish and result in acute gastroenteritis, but they can also result in wound infections and septicemia. As a member of the Vibrionaceae, V. parahaemolyticus is classified as a γ-proteobacterium within the enteric-vibrio branch of the γ-3 subgroup (192). In phylogenetic analyses, V. parahaemolyticus clusters most closely with V. harveyi, V. vulnificus, and V. alginolyticus. It is more distantly related to V. anguillarum, V. fischeri, and V. cholerae (188).
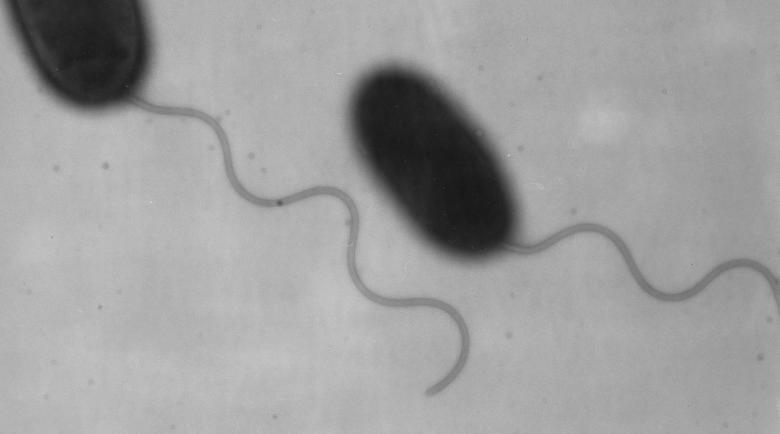
The flagellation patterns of these members of the Vibrionaceae family are presented in Table 1. V. parahaemolyticus and some other members of this family exhibit mixed flagellation, possessing polar and peritrichous flagella. When grown planktonically, the bacteria display polar flagella (Fig. 1). The flagellum is sheathed by what appears to be an extension of the cell outer membrane. When grown on solid medium or medium of high viscosity, e.g., medium supplemented with Ficoll, the organisms produce both polar and peritrichous (also called lateral) flagella (Fig. 2). As can be seen in Fig. 2, remarkable numbers of peritrichously arranged flagella are produced. These flagella are unsheathed and more fragile than the polar flagellum (2). Figure 2A shows plate-grown cells that have been stained with phosphotungstic acid. The polar flagellum, which is produced by liquid- and surface-grown bacteria, is distinguished from the lateral flagella by the increased thickness due to the sheath. Figure 2B emphasizes the striking difference in polar and lateral flagellar integrity: cells were stained with uranyl acetate, which causes deterioration of the lateral but not the polar flagellar structure. In liquid environments, the swimming speed of the marine vibrios is approximately 60 μm/s; however, as the viscosity increases, the polar flagellum is not an effective propulsive organelle and swimming slows (10, 82). In compensation, the lateral flagellar system is induced. These peritrichous flagella are quite functional in viscous environments and enable the bacterium to move over and colonize surfaces (162). The two motility systems are genetically distinct (121). Thus, in possessing dual flagellar systems suited for locomotion under different circumstances, V. parahaemolyticus seems highly adapted to survival in changing habitats, including life in planktonic environments and on surfaces or in biofilms.
TABLE 1.
Flagellation of Vibrio speciesa
| Organism | Sheathed polar flagellab | Unsheathed peritrichous flagellac |
|---|---|---|
| V. parahaemolyticus | Monotrichousd | +e |
| V. alginolyticus | Monotrichousd | +e |
| V. harveyi | Monotrichous | + |
| V. vulnificus | Monotrichous | − |
| V. anguillarum | Monotrichous | − |
| V. fischeri | Lophotrichous (2–8) | − |
| V. cholerae | Monotrichousd | − |
Determined when grown in liquid medium.
Determined when grown on solid medium. +, the majority of strains examined (>80%) possessed lateral flagella; −, none possessed lateral flagella.
Rotation has been demonstrated to be powered by the sodium motive force.
Rotation has been demonstrated to be powered by the proton motive force.
FIG. 1.
Sheathed polar flagellum of V. parahaemolyticus. An electron micrograph of cells grown in liquid and stained with 1% uranyl acetate is shown. Magnification, ×26,000.
FIG. 2.
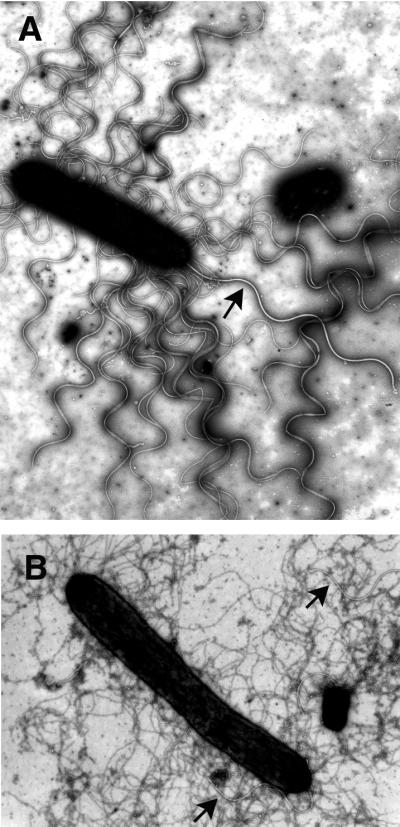
Polar and lateral flagella of surface-grown bacteria. The helical structure of lateral flagella, but not polar flagella, is destroyed by uranyl acetate. Electron micrographs of cells harvested from a plate and negatively stained with 0.5% phosphotungstic acid (A) or 1% uranyl acetate (B) are shown. Arrows indicate the sheathed polar flagellum. Magnification, approximately ×14,000.
POLAR FLAGELLAR STRUCTURE AND THE SHEATHED FLAGELLUM
Multiple Flagellin Genes
Flagellar filaments, which act as propellers, consist of self-assembling protein subunits (flagellin) arranged in a helix and forming a hollow tube (reviewed in reference 135). Subunits move down the hollow core and are polymerized at the tip of the flagellum. The V. parahaemolyticus polar organelle is a complex flagellum. There are six polar flagellin genes, organized in two loci (86, 122). The flagellins are similar to each other: FlaB and FlaA are 78% identical, FlaB and FlaC are 68% identical, FlaB and FlaD are 99% identical, FlaB and FlaF are 69% identical, and FlaB and FlaE are 50% identical. Despite the great protein similarity of FlaB and FlaD, the flagellins migrate differently on sodium dodecyl sulfate-polyacrylamide gel electrophoresis analysis, suggesting the possibility of posttranslational modification, e.g., glycosylation, which has been observed for flagellins of many bacteria including some spirochetes, Campylobacter species, and P. aeruginosa (27, 56, 171, 196), or phosphorylation, which has been detected for flagellins of P. aeruginosa (85). Analysis of the protein composition of purified flagella from wild-type strains and strains with mutations in flagellin genes suggests that all of the flagellins can be incorporated into the organelle and that FlaA, FlaB, and FlaD are the major subunits (121, 122; L. McCarter, unpublished data). Nothing is known with respect to their spatial arrangement in the flagellum. Loss of function of a single flagellin gene has little or no effect on swimming motility or flagellar structure (waveform or length). Thus, none of the six flagellin genes is essential for filament formation. Deletion of the flaFBA or the flaCD genes also has little effect on motility, but the deletion of both loci (ΔflaFBA ΔflaCD) completely abolishes motility (122).
Why are there six flagellin genes? It is not clear why the organism possesses such an extraordinary number of flagellins. The similarity of the gene products and the dispensability of the genes suggest that there are no special structural requirements, although the filament structure and function could be more complex and adapted to specific circumstances than our laboratory tests can reveal. Bacteria are known to modulate the antigenicity of their flagellar filaments by expression of different flagellin genes or by recombination and rearrangement of flagellin genes (reviewed in reference 190). Therefore, the capacity for immune system evasion in a host organism might account for some of the diversity. The multiplicity of flagellin genes suggests a significant reservoir for antigenic or phase variation. Although the sheath covers the filament and might be thought to provide a disguise, electron microscopy suggests that it may be fragile (2, 164). Thus, the sheath may not protect the filament against the immune response of a host. In some respects the endoflagella of the spirochetes are similar, for these flagella can also be viewed as being sheathed, polar organelles (reviewed in reference 103). The spirochete flagella are normally found in the periplasm, between the outer membrane and the cell cylinder and attached near each cell pole. Purification of periplasmic flagella demonstrated that these filaments are also complex, with generally two to four different flagellin proteins, encoded by distinct genes, as well as containing an accessory, nonflagellin protein.
Differential Flagellin Gene Expression
The six flagellin genes occur in five distinct transcriptional units; flaD and flaE appear to be cotranscribed (86, 122). The promoters for three of these operons (flaA, flaB, and flaDE) require a specialized sigma factor for transcription, ς28. Primer extension mapping has defined the consensus promoter. The fifth flagellin gene, flaF, is poorly expressed, although upstream sequences contain a potential ς28-recognition site. The sixth flagellin gene, flaC, is the most unusual with respect to transcription. Unlike flaA, flaB, and flaD, which are expressed in E. coli in a ς28-dependent manner, FlaC cannot be detected in E. coli unless expression of the gene is driven by an isopropyl-β-d-thiogalactoside (IPTG)-inducible tac promoter. The nucleotide sequence upstream of the start point of transcription of flaC appears unusual in that it does not contain sequences consistent with either the ς28-dependent promoter or the ς54-dependent consensus polar flagellar promoters. A major FlaC-encoding transcript can be detected in plate-grown cells but not in broth-grown cultures; therefore, flaC expression appears to be induced by growth on a surface. Furthermore, flaC expression requires an intact lateral flagellar genetic pathway. A defect in the lateral flagellar hook gene (lfgE) prevents the expression of many surface-dependent genes, including genes encoding lateral-specific flagellar ς28 and lateral flagellin (125). This defect also prevents surface-induced expression of flaC. Thus, flaC appears to be a unique flagellin gene with respect to transcription. What this means with respect to polar flagellar function and regulation, particularly in the context of growth on surfaces, remains to be investigated.
Chromosomal Organization of Flagellin Genes
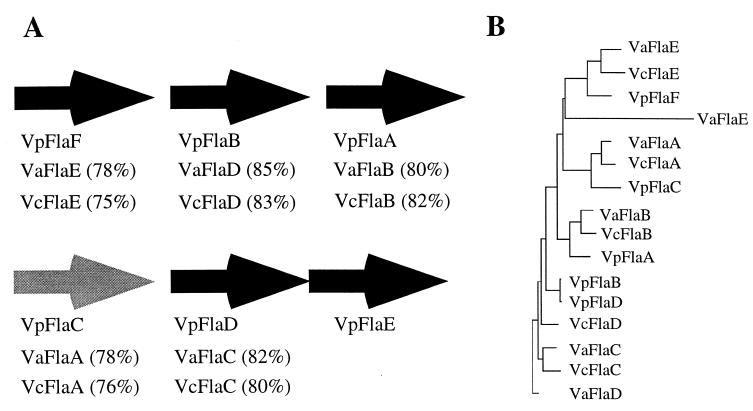
The multiple flagellin genes are found in two distinct locations on the chromosome, and the organization of these genes in V. parahaemolyticus is similar to that of the loci in V. anguillarum and V. cholerae (88, 126). However, these organisms possess only five flagellin genes and lack an equivalent of flaE. Figure 3A depicts the relationship of the flagellin genes with respect to chromosomal organization and protein homology. The phylogram (Fig. 3B) shows that each flagellin is more closely related to the predicted product of its spatially equivalent open reading frame (ORF) in the other organisms than to the other flagellins within the same organism. On the basis of nucleotide sequence, mutant analysis, and/or primer extension analysis, the open reading frames (ORFs) indicated by the black arrows are believed to be transcribed by ς28 in all organisms (86, 88, 126, 150). In each of the three species, one of the flagellin genes, indicated by the gray arrow, appears different from the other flagellin genes with respect to transcription and/or function. In V. anguillarum and V. cholerae, this gene, flaA, seems to be specifically required for motility and/or virulence. Disruption of V. anguillarum flaA significantly slows motility, and the gene is essential for infection of the fish (129), whereas mutations in any of the other flagellin genes have little or no effect on swimming or virulence. In V. cholerae, mutation of flaA completely abolishes motility (88). Expression of V. cholerae flaA is directly dependent on ς54, whereas the four other flagellin genes are dispensable and require ς28 (150). The critical flagellin genes of V. anguillarum and V. cholerae are most equivalent with respect to gene location and predicted protein sequence to V. parahaemolyticus flaC.
FIG. 3.
Comparison of multiple flagellin genes with respect to chromosomal location and predicted gene product similarity. (A) Chromosomal organization of flagellin genes. Genes are located in two loci: flaFBA and flaCDE in V. parahaemolyticus (Vp) and flaEDB and flaAC in V. anguillarum (Va) and V. cholerae (Vc). Arrowheads indicate the direction of transcription of each ORF. Evidence suggests that the genes indicated by the black arrows are transcribed by ς28. V. parahaemolyticus flaE is cotranscribed with flaD. V. anguillarum and V. cholerae lack the flaE equivalent. The gray arrow represents the ORF encoding a unique flagellin with respect to transcription and/or function. It encodes the major flagellin of V. anguillarum and V. cholerae and is transcribed by ς54. In V. parahaemolyticus, this gene is induced by growth on a surface and is not expressed when the organism is grown in liquid. The promoter sequences of the V. parahaemolyticus flaC gene do not resemble ς28- or ς54-dependent polar flagellar promoters. Below each gene, the percent identities of the V. anguillarum and V. cholerae gene products with V. parahaemolyticus flagellin are shown. (B) Grow/Tree phylogram of flagellins (produced by GCG Inc. program analysis).
The Basal Body
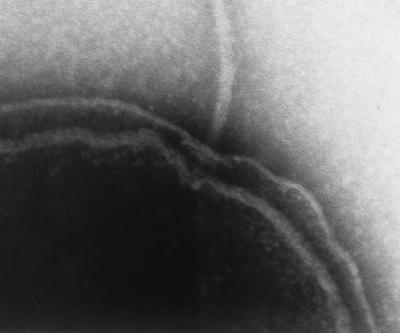
The flagellar filament is connected to a structure in the membrane known as the basal body. The basal body is composed of several rings and an axial structure. Some evidence exists that the polar basal-body structure differs from that of peritrichously inserted flagella. Two models for the basal organelle of polar flagella have been derived from electron microscopy studies of V. cholerae, Campylobacter fetus, Bdellovibrio bacteriovorus, and Wolinella succinogenes (44, 46, 180). In W. succinogenes, electron micrographs display a beautiful large basal disk described as an archimedian spiral (44). Such elements, which also have been described as concentric membrane rings, were found associated with the basal organelle of Aquaspirillum serpens, V. cholerae, and C. fetus (36, 46). Regardless of whether the flagellum is sheathed or unsheathed, all of the studies report the existence in the basal body complex of a large convex disk situated below the outer membrane, and thus the large disk does not seem to be a feature specific to sheath formation. It has been hypothesized that the disk provides reinforcement at the flagellar insertion site and disperses forces that are generated by the force of flagellar rotation (44). A large basal disk is visible in V. parahaemolyticus and is shown in Fig. 4. One model for the polar basal body suggests that the disk is the P-ring equivalent, acting as a bushing associated with peptidoglycan, and that for sheathed flagella the L ring, which is lipoplysaccharide or outer membrane associated, is not present. The second model places the large disk between the P and L rings. Genes for both the P and L rings exist in V. parahaemolyticus and V. cholerae. Whereas the P ring displays homology to enteric P rings over the entire molecule, the Vibrio L ring shows sequence divergence at the N terminus. Perhaps the nature of this protein is one key to differences in basal-body structure.
FIG. 4.
Electron micrograph of the polar basal body structure. Cells were adsorbed to a grid and treated with 1% Triton X-100 before being stained with 1% phosphotungstic acid. Magnification, ×90,000.
The Sheath
The flagellum is sheathed by an apparent extension of the cell membrane (2). The mechanism of how a sheathed flagellum rotates has not been elucidated. Potentially, the flagellar filament could rotate within the sheath or the two could rotate as a unit (50). Little is known about the composition, formation, or function of flagellar sheaths, which are found in many bacteria, including marine Vibrio species, V. cholerae, B. bacteriovorus, and Helicobacter pylori (reviewed in reference 164). Evidence from these organisms suggests that the sheath contains both lipopolysaccharide and proteins and that it may exist as a stable membrane domain distinct from the outer membrane (42, 51, 58, 69, 144). The lipid content of the sheath of B. bacteriovorus is distinct from that of the outer membrane, and the sheath appears to be a highly fluid, symmetric bilayer (179). How the sheath is formed remains essentially uninvestigated. It has been postulated that the sheath forms concomitantly with the elongation of the flagellar filament. However, it is provocative to note that “tubules” or structures that appear to be empty sheaths lacking filament have been observed, which suggests the interesting possibility of uncoupling of the flagellar core and the sheath assembly (2). One of three major sheath proteins of V. alginolyticus has been characterized. Genetic and biochemical evidence suggests that it is a lipoprotein (52). Another flagellar sheath protein that is a lipoprotein is HpaA of H. pylori (76, 144). There is some controversy about the role and cellular location of HpaA. Although HpaA has also been reported to be a cell surface adhesin (45), other groups have localized HpaA to the cytoplasm (144) or the flagellar sheath (76), and no adherence defect for hpaA mutants to eukaryotic cell lines has been demonstrated (76, 144). Experiments with V. anguillarum suggest the sheath is a virulence organelle. Mutants of V. anguillarum that lack a major flagellar sheath antigen are avirulent, even though the initial stages of infection are unaffected (137). Biochemical analysis indicates that this particular sheath antigen is lipopolysaccharide. Thus, the sheath may be important for specific interactions with the environment.
Implications of the Sheath for Filament Assembly: HAP Mutant Phenotypes
The sheath seems to provide some variation to the pathway of flagellar assembly. The hook–basal-body complex forms a channel through which proteins can be exported. In fact, not only are structural elements of the flagellum exported through this channel, but also regulatory molecules, e.g., the flagellar anti-ς factor FlgM, are secreted (70, 99, 139). For bacteria with unsheathed flagella, such as E. coli, mutants with defects in genes encoding three hook-associated proteins (HAPs) are nonmotile and secrete unpolymerized flagellin subunits (66). HAP1 and HAP3 are the connector proteins that join the filament to the hook. Without the ability to adapt flagellin subunits to the hook, the flagellins are secreted. HAP2 is also called the distal capping protein because its role is one of a cap or plug. Without this cap, flagellins are also secreted. Since purified flagellin subunits can assemble in vitro (9) and since an S. enterica serovar Typhimurium mutant lacking HAP2 can polymerize filaments if the concentration of flagellin in the external medium is high (67), the role of HAP2 has been viewed as capping the flagellar tip to retard subunit secretion sufficiently to increase the local concentration of flagellin and promote self-assembly. Recent work in Salmonella, analyzing cap-filament interactions by cryoelectron microscopy, suggests a model for the cap as being a flat, disklike pentameric structure that acts as a processive chaperone, preventing the loss of flagellin monomers and actively catalyzing folding and insertion into the filament (200).
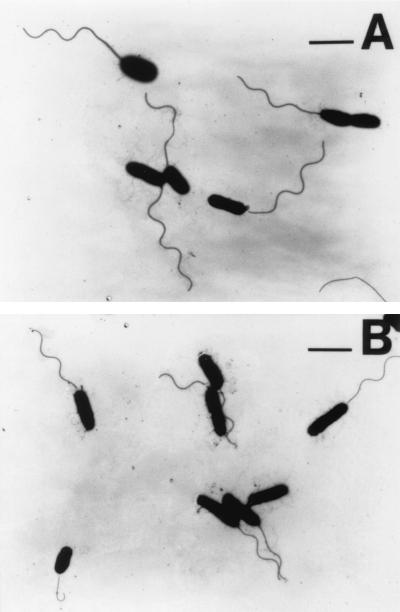
HAP mutants of V. parahaemolyticus display different phenotypes (122). The most striking difference is in the phenotypes of mutants with defects in the gene encoding HAP2. These mutants are competent for filament assembly and motile. Figure 5 compares the flagella of the wild type and mutants with defects in the gene that encodes HAP2, and the flagella seem mostly indistinguishable. This suggests that in the absence of HAP2 but in the presence of the flagellar sheath, the local concentrations of the flagellin monomers remain high enough to allow polymerization of subunits. HAP1 and HAP3 mutants of V. parahaemolyticus are nonmotile and nonflagellated; however, they produce detached, severely truncated filaments encased in a membrane (122). The sheath seems to act to retain flagellin monomers and allow subunit assembly. The polymerized flagellins cannot be connected to the hook and bleb off as abortive filaments surrounded by a membrane vesicle. Similar filamentless mutants that produce flagellin-containing membrane vesicles were isolated in V. alginolyticus, although the genetic lesions in these strains were not determined (136). Thus, the sheath itself appears to be able to substitute for the cap. The potential for the sheath to act in such a sealing capacity raises a question about the competence of the sheathed flagellum for secretion of the flagellar regulatory molecule FlgM. In E. coli, FlgM is sequestered inside the cell, where it acts as an anti-ς factor to prevent late flagellar gene expression, until the hook is completed and FlgM is exported (70). If the sheath acts as a passive barrier to export, the checkpoint for coordinating flagellar morphogenesis and gene expression may be somewhat different from the mechanism in E. coli.
FIG. 5.
The flagellar sheath can act as a cap. Electron micrographs of liquid-grown cells stained with 0.5% phosphotungstic acid are shown. (A) Wild-type strain. (B) HAP2-defective strain. Magnification, ×4,500.
ORGANIZATION AND REGULATION OF FLAGELLAR AND CHEMOTAXIS GENES
The V. parahaemolyticus polar flagellar gene system (Fla) comprises approximately 60 genes (86). It is the default motility system for the organism and is produced continuously; therefore, most of the polar flagellar genes have been named analogously to homologs in other bacteria. Genes in the lateral flagellar system (Laf) are expressed under particular conditions and have been assigned designations that are permutations of the fla nomenclature. Table 2 summarizes the organization, homology, and predicted function of the gene products. By comparison with E. coli, the full complement of genes encoding flagellar structural components and the export apparatus exist, and there are a few additions. Most of the genes are found in two loci that contain large, predicted flagellar operons. Precedence for large motility operons has been established in other bacteria, e.g., Borrelia burgdorferi (55). The closest homologs to many of the genes are found in V. cholerae, P. aeruginosa, and P. putida. The physical organization also seems highly conserved among organisms. The flagellar gene organization of V. cholerae is almost identical to that of V. parahaemolyticus, with the exception of a large insertion containing nonflagellar genes positioned between flhB and flhA in V. cholerae (65).
TABLE 2.
Polar flagellar and chemotaxis genes of V. parahaemolyticusa
 |
Genes Not Found in E. coli
One pair of genes found in V. parahaemolyticus and many other bacteria, including other Vibrio, Bacillus, Pseudomonas, Campylobacter, and Borrelia species, but not found in E. coli is flhF and flhG. FlhF shows homology to FtsY, which is a GTP-binding protein involved in the signal recognition particle targeting pathway (134). The flhF gene was first discovered in Bacillus subtilis, where it was demonstrated to be required for motility (29). A nonpolar, null mutation in flhF produced viable but nonmotile cells lacking flagella. Intriguingly, the V. parahaemolyticus and V. cholerae FlhF proteins contain an insertion of ∼170 amino acids that is not found in other FlhF sequences derived from organisms possessing proton-driven flagella. The inserted domain shows homology to a eukaryotic sodium channel. It should be quite interesting to probe the function of this domain. Immediately downstream of V. parahaemolyticus flhF is flhG, which encodes a protein that shows homology to MinD. MinD is a membrane ATPase involved in septum site determination (102, 118). The FlhF-FlhG pair is found in a number of polarly flagellated bacteria. Disruption of flhF in P. putida leads to random arrangement of flagellar insertion (147), and disruption of fleN, the flhG homolog in P. aeruginosa, leads to multiple polar flagella (38). Potentially, FlhF and FlhG could work as a pair to determine and/or control site selection of flagellar insertion as well as the number of flagella.
The gene products with the least homology to flagellar components of other bacteria are those that are candidates for flagellar chaperones, i.e., FlaI and FlgN. Perhaps this is not surprising. Flagella are assembled via a type III export pathway (reviewed in reference 110). It seems clear that classes of sequentially exported flagellar proteins exist (32, 130). No consensus flagellar export signal has been defined, although a number of models have been suggested. It has been proposed that the flagellar chaperones, e.g., FliT or FlgN, act to guide the secretion of substrates by preventing cytoplasmic assembly or degradation, by presenting substrates to the secretory apparatus, and by regulating translation to directly couple translation to secretion (48, 80). When grown on a surface, V. parahaemolyticus assembles two distinct types of flagella. The substrate signals and the export system for each flagellar system must be sufficiently divergent to exclude components from the second system. In some respects, the dual flagellar systems in V. parahaemolyticus seem ideal to test hypotheses with respect to type III secretion determinants and the specificity of export.
Chemotaxis Genes and Gene Organization
The complement of chemotaxis (che) genes and their organization are different from those in E. coli. One of these genes is cheV, which encodes a hybrid CheY/CheW. CheV has been found in a number of organisms, including Bacillus species, Campylobacter jejuni, H. pylori, P. aeruginosa, and V. cholerae. In B. subtilis, genetic analysis suggests that CheV and CheW are functionally redundant (155). Three unusual ORFs occur within the che gene cluster of region 2. ORF1 encodes a protein that resembles Soj of B. subtilis and other ATPase proteins involved in chromosome partitioning (152, 161). The other ORFs encode potential polypeptides that do not resemble proteins of known function. It seems curious that a Soj-like protein exists within a flagellar/chemotaxis operon, and this particular arrangement is conserved in other bacteria, e.g., P. aeruginosa, P. putida, and V. cholerae. Perhaps these novel ORFs will prove key for understanding the linkage between cell division and flagellation or development. It should be remarked that many of the V. parahaemolyticus che genes located in the flagellar clusters (Table 2) were discovered by mutant analysis; i.e., these genes produce defects in chemotaxis when mutated. The V. cholerae genome, as well as those of Pseudomonas species, indicates additional complexity with respect to a multiplicity of potential che genes.
Regulation of Gene Expression: a Potential Hierarchy of Gene Control
As can be seen in Table 2, a considerable number of genes are dedicated to the flagellar motility system; therefore, maintenance of flagellation is a sizable investment with respect to cellular economy. As a result, flagellar systems are highly regulated. In systems where it has been studied, the general scheme of gene control represents a hierarchical cascade of regulation that couples the sequential expression of specific classes of genes to the assembly of the organelle. Genes in each temporal class must be functional in order for expression of the subsequent class to occur. This also seems to be the case for V. parahaemolyticus, and a potential hierarchy is outlined in Table 3. The hierarchical scheme proposed for V. cholerae is very similar, with some variations that are discussed below (150).
TABLE 3.
Regulatory cascade for the Vibrio polar flagellar gene systema
| Early genes | Middle genes (ς54-dependent operons)b | Late genes (ς28-dependent operons)c |
|---|---|---|
| fliEFGHIHKLMNOPQR flhB (switch, export, and assembly proteins) | motAB (motor proteins) | |
| flaK (ς54-interacting regulator)d | flhAFG fliA cheYZAB orf1 orf2 cheW orf3 (assembly, ς28, and chemotaxis components) | motX (motor proteins) |
| flaLM (two-component sensor-response regulator pair) | flgBCDEFGHIJ (hook and basal-body parts) | flaAGHIJK (flagellin, chaperones, HAP2, and regulator K) |
| flgKL (HAP1 and HAP3) | flaB (flagellin) | |
| flaDE (flagellins) | ||
| motY (motor protein) | cheVR (chemotaxis proteins) | |
| flgMNc (anti-ς factor and chaperone) |
Promoters for the polar flagellar genes have been defined in V. parahaemolyticus (86) and V. cholerae (150).
The consensus ς54 recognition sequence derived from a comparison of V. parahaemolyticus promoters defined by primer extension analysis is TGGC N7 TTGC N11–13 + 1.
The consensus ς28-recognition sequence derived from comparison of V. parahaemolyticus promoters defined by primer extension analysis is CTAAG N14 G(C/T)CG(A/T)TAA N7 + 1.
In V. cholerae, the FlaK homolog is required to activate the transcription of the flaLM-like locus in a ς54-dependendent manner, and genes in the middle class are divided into FlaK- and FlaM-dependent subclasses (89, 150).
The flgMN genes are also transcribed from an upstream promoter; however, the start point of transcriptional initiation for the flgAMN operon has not been determined.
To summarize, at least three classes of transcription for the polar flagellar hierarchy of V. parahaemolyticus have been proposed (86). At an early level in the transcriptional hierarchy are master regulators that potentially interact with ς54, FlaK and FlaM. Flagellation in V. alginolyticus, V. cholerae, V. anguillarum, and P. aeruginosa requires the rpoN gene, which encodes ς54 (84, 89, 145, 184). It seems probable that ς54 will also direct the transcription of V. parahaemolyticus polar genes, although this has not yet been demonstrated. Genes in the middle class require ς54-type activation and are dedicated to assembly of the hook–basal-body structure. Additionally, one finds middle genes encoding HAP1 and HAP3, the motor gene motY, some chemotaxis genes, and fliA, which encodes ς28. In turn, this alternative ς factor is specific for the other large subset of flagellar promoters. The genes under this later level of expression encode additional motor parts (MotA, MotB, and MotX), additional chemotaxis proteins, the distal capping protein HAP2, the anti-ς28 factor FlgM, putative flagellar chaperones, and five flagellins.
Early Gene Expression: Master Regulatory Proteins That Interact with ς54
FlaK and FlaM appear to be master transcriptional regulators of polar genes. The genes are encoded by two linked operons, flaK and flaLM. Homologs of these regulators exist in V. cholerae and P. aeruginosa. The homologous gene clusters are shown in Table 4. Much is known about the role of the gene products in flagellar regulation in V. cholerae and P. aeruginosa (4, 35, 89, 153). Based on these studies, the following model has been proposed. FlaK homologs, acting as ς54-dependent transcription factors, activate transcription of the flaLM-like operons. FlaL homologs are histidine sensor kinases that activate the receiver domain of the response regulator FlaM homologs by phosphotransfer. Phosphorylated FlaM homologs are required for ς54-dependent transcription of genes in the middle class of flagellar genes. For V. cholerae and P. aeruginosa, the FlaK homologue is absolutely required for ς54-dependent activation of the flaLM-like operon and is essential for motility (4, 89). V. cholerae FlrB (the FlaL homolog) is an autokinase that can transfer phosphate to FlrC (the FlaM homolog), specifically to the aspartate that is equivalent to Asp57 of CheY. FlrC must be phosphorylated to activate flagellar gene transcription (35).
TABLE 4.
Polar flagellar regulatory genes
| Organism | Homologous gene clusters | |
|---|---|---|
| V. parahaemolyticus | flaK | flaLM |
| V. cholerae | flrA | flrBC |
| P. aeruginosa | fleQ | fleSR |
For V. cholerae, the middle genes have been further divided into two classes: those that require FlrA for activation (encoding the MS-ring-switch-export complex) and those that require FlrC for activation of transcription (encoding the remainder of the basal-body and hook structure) (150). For V. parahaemolyticus, like V. cholerae and P. aeruginosa, loss of function of flaM results in a completely nonmotile cell; however, disruption of the gene encoding FlaK (488 amino acids) by introduction of a chloramphenicol cassette at the position in the gene encoding amino acid 37 or 216 does not abolish motility (168; McCarter, unpublished). Mutants with flaK defects possess a slow-motility phenotype, which is significantly different from the completely nonmotile phenotype observed in the other organisms. One possible explanation for the difference is that readthrough transcription occurs in the V. parahaemolyticus flaK::Camr mutants, originating with the chloramphenicol cassette and continuing into flaLM, which is sufficient to circumvent the requirement for FlaK-mediated activation of flaLM. If true, this would suggest that FlaK is not required for transcription of other flagellar operons in V. parahaemolyticus and that the middle genes of the hierarchy are not subdivided as they are in V. cholerae. Clearly, more experiments are required.
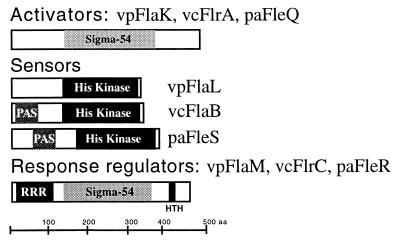
A schematic comparison of the functional domain of these regulators is shown in Fig. 6. The central domain of the FlaK homologs contains a potential ς54-interaction domain. The N-terminal domains of the FlaK homologs do not contain the hallmark residues in the receiver domain of response regulators; this suggests that they are ς54-dependent transcriptional regulatory proteins but that they do not participate in two-component phosphorelay signaling. The FlaM homologs resemble two-component response regulators, containing both ς54-interaction domains and response regulatory regions. The N-terminal response regulatory region of FlaM-like proteins contains the four residues that are highly conserved in a number of response regulators, i.e., residues that correspond to Asp12, Asp13, Asp57, and Lys109 of the paradigm response regulator CheY (33). The C terminus of the FlaM homologs contains a helix-turn-helix region. The three FlaL-like proteins each possess a histidine kinase domain; however, their N termini differ. V. cholerae FlrB and P. aeruginosa FleS possess N-terminal PAS domains, whereas V. parahaemolyticus FlaL does not contain a recognizable PAS domain. PAS modules have been identified in a number of signal transduction molecules and are thought to be signatures of proteins that play roles in detection and adaptation to environmental change, e.g., changes in light, redox potential, and oxygen (177). This suggests that the ways in which these molecules act as sensor kinases may be different, specifically with respect to input. Activation of the phosphorelay cascade may be responsive to different signals in the different organisms. Moreover, with respect to global control of flagellar gene expression, there are most probably other environmental signals that contribute to the regulation of flagellar gene expression, e.g., catabolite repression (199).
FIG. 6.
Comparison of the conserved domains in the master polar flagellar regulatory factors of V. parahaemolyticus (vp), V. cholerae (vc), and P. aeruginosa (pa). FlaK, FlrA, and FleQ contain a potential ς54-interaction domain (Sigma-54; pfam00158). FlaL, FlrB, and FleS are potential two-component sensors that contain the histidine kinase signaling domain (His kinase: pfam00512). FlrB and FleS also contain a PAS domain (Smart PAS domain or pfam00989), whereas FlaL does not. FlaM, FlrC, and FleR contain the ς54 domain, a response regulatory receiver domain (RRR; pfam00072), and a helix-turn-helix (HTH) region. Experiments performed with V. cholerae and P. aeruginosa (4, 89) support the following model for a transcriptional activation cascade: FlaK, FlrA, and FleQ activate transcription of the flrBC and fleSR operons in a ς54-dependent manner. FlaL, FlrB, and FleS act as sensor kinases to phosphorylate and activate the response regulators FlaM, FlrC, and FleR, which in turn control the transcription of other ς54-dependent flagellar promoters. Evidence from V. cholerae suggests that FlrA directly controls the transcription of other ς54-dependent flagellar genes (150).
THE SODIUM-DRIVEN MOTOR
The flagellar filament acts as a propeller that is turned by a reversible rotary motor, which is embedded in the membrane (reviewed in references 17, 19, 21, and 41). Energy to power flagellar rotation is derived from the transmembrane electrochemical potential of specific ions (101, 117, 128). Rotation appears to be tightly coupled to the flow of ions through the motor. Two kinds of motors, which are dependent on different coupling ions, have been described: H+ and Na+ motors (72, 112). In V. parahaemolyticus and V. alginolyticus, the sodium motive force drives polar flagellar rotation and the proton motive force powers rotation of the lateral flagella (11, 83). The polar flagellum of V. cholerae is also sodium driven (95).
Proton-type motors of E. coli and S. enterica serovar Typhimurium have been extensively characterized. Two cytoplasmic proteins, MotA and MotB, form the force-generating unit through which the protons are channeled (22, 23, 160, 169, 204, 206). Torque is transmitted from the MotA-MotB complex to the flagellar basal body. Thus, the torque generator, which is probably fastened to the cell wall via a peptidoglycan interaction domain found in the C terminus of MotB (34, 40), acts as a stator to transmit force to the rotor. Critical electrostatic interactions between MotA and FliG have been demonstrated (205). FliG is found at the base of the flagellar basal body. Below FliG is a cytoplasmic structure called the C-ring, containing FliM and FliN (47). The complex containing FliG, FliM, and FliN is also known as the switch complex and is essential for torque generation, flagellar assembly, and the control of the direction of flagellar rotation (73, 165, 173, 174, 183, 197, 198).
Sodium Channel-Blocking Drugs Specifically Interfere with Polar Flagellar Rotation
The pioneering bioenergetic studies by Imae and coworkers on Na+-driven motility were performed using alkalophilic Bacillus and marine Vibrio species (reviewed in reference 71). Imae demonstrated that sodium channel-blocking drugs, such as amiloride, specifically inhibited sodium-driven motility and could be used to probe motor function (170). Mutants were isolated that could swim in the presence of the sodium channel-blocking drug (92). Currently, an understanding of the architecture of the sodium-type flagellar motor is being developed by studying V. alginolyticus, V. cholerae, and V. parahaemolyticus (reviewed in reference 202). Rotation rates for the sodium-driven flagellum are quite remarkable; e.g., using laser dark-field microscopy, the polar flagellar rotation rate for V. alginolyticus was recorded to average 1,100 rps and to be as high as 1,700 rps (115). Four genes have been described that are required for sodium-dependent flagellar rotation is V. parahaemolyticus: motA, motB, motX, and motY (7, 25, 60, 123, 124, 141). In V. alginolyticus and V. cholerae, motA and motB are named pomA and pomB, respectively. Transposon insertions or deletions in these genes produce flagellated but paralyzed bacteria. Loss of function of any one of the four motor genes completely abolishes motility but does not prevent flagellar assembly.
The MotA-MotB Complex Translocates Sodium Ions
With respect to membrane topology and function, the Vibrio sodium-type proteins resemble MotA and MotB of the proton-type motor (8, 93). Many data suggest that these proteins form the Na+-conducting channel. Initial evidence implicating Vibrio MotA and MotB in Na+ translocation was provided by the isolation of mutants that could swim in the presence of the sodium channel inhibitor phenamil (74, 91). These mutants contained alterations in motA or motB. Some of the mutations conferring phenamil resistance altered the ion specificity of the motor, for example resulting in increased or decreased swimming rates in the presence of lithium ions (74). Some mutations, such as a substitution at residue Asp31 in motA, caused a slow-motility phenotype, suggesting that the negative charge of this residue contributes to optimal speed or efficiency of the motor (94). Reconstitution experiments with liposomes provided strong proof that purified V. alginolyticus MotA-MotB complexes catalyzed Na+ flux (157). Furthermore, these purification studies, as well as immunoprecipitation experiments, indicated that MotA and MotB physically interact and can be purified as a complex (157, 203). Interestingly, the molar ratio of MotA to MotB was calculated to be 2:1. Intermolecular cross-linking studies using cysteine substitutions within the periplasmic loop between the third and fourth transmembrane domains of MotA also suggested that adjacent MotA molecules were physically close within the motor (201). To further probe the stoichiometry of the torque generator, a chimeric construct was designed to produce a fused dimer of MotA molecules (158). Such a dimer was found to support motility; however, mutational inactivation of either half of the fusion dimer caused complete loss of swimming motility and loss of the ability to interact with MotB, suggesting that dual MotA components are essential for sodium-type motor function.
Chimeras Composed of Sodium and Proton Parts
Some intriguing experiments using chimeras of mixed sodium and proton motor parts have been performed. E. coli motor proteins can substitute to weakly power bacterial motility by using the proton motive force in a V. cholerae strain that has been gutted for the four sodium motor genes (60). Sodium-dependent motility could be restored to a MotA-deficient V. alginolyticus strain on provision of the proton-type MotA from Rhodobacter sphaeroides (5). Although R. sphaeroides MotB was not functional in V. alginolyticus, MotB chimeras that contained portions of the N-terminal transmembrane domain of R. sphaeroides and the C-terminal linker-peptidoglycan-binding domains of V. alginolyticus were able to reconstitute Na+-driven motility (6). These experiments suggest that the determinants of the specificity of the coupling ion do not reside exclusively within MotA and/or the transmembrane domain of MotB, and they raise the interesting question of how ion specificity is conferred.
Interaction with the Switch Complex
On the basis of extensive mutational analysis (26, 53, 54, 73, 106, 107, 181, 205) and the determination of the crystal structure of the C-terminal domain of FliG, a structural model for the part of the rotor that interacts with the stator has been developed for the proton-type motor (108, 205). Residues known to be critical for torque generation in three switch components of the proton-type motor are conserved in the switch components of the V. parahaemolyticus and V. cholerae sodium-type motor. This suggests a common mechanism for energy transfer at the rotor-stator interface regardless of the driving force powering rotation. Such an idea is further supported by the ability to productively mix sodium and proton components to create functionally propulsive motors (6, 60). The sodium-type switch components FliM and FliN do possess unique charged domains not found in their proton-type homologs, and it will be of interest to determine whether these domains are important with respect to motor, switching, or assembly functions.
Unique Components
MotX and MotY proteins are the unique components of sodium-type flagellar systems, and their specific roles are not known. Loss of function of either motX or motY produces a paralyzed mutant completely defective for swimming but competent for flagellar assembly (123, 124, 141). Both proteins possess single membrane-spanning domains. The C terminus of MotY contains an extended domain that shows striking homology to a number of outer membrane proteins know to interact with peptidoglycan, e.g., OmpA and peptidoglycan-associated lipoproteins (124). The simplest hypothesis for the role of MotY is that the polar flagellar motor possesses two elements for anchoring the force generator. Perhaps extremely precise alignment of the stator with the rotor is required for a motor that spins as fast as 100,000 rpm. The role of MotX remains mysterious. It is known that MotX recruits MotY to the membrane when the proteins are coexpressed in E. coli (123). Furthermore, overexpression of MotX is lethal to E. coli in proportion to the external Na+ concentration, and lethality can be reversed by the presence of the sodium channel blocker amiloride. This suggests that the proteins may somehow participate in or modulate Na+ translocation. For example, MotX could act to modify or specify ion channel activity. Thus, it may be that all four proteins comprise and specify the sodium-type torque-generating unit. However, there is no existing evidence that places MotX and MotY in the physical context of MotA and MotB. It is possible that MotX and MotY may play a more distinct role in the generation of sodium-driven motility; e.g., they could participate in some other aspect of the sodium cycle.
CHEMOTAXIS
Bacterial chemotaxis has been most extensively studied in organisms with multiple, peritrichously arranged flagella, e.g., E. coli, S. enterica serovar Typhimurium, and B. subtilis (reviewed in reference 21). Bacteria respond to signals in the environment by modulating the direction of flagellar rotation. As viewed under the light microscope, the cells move in alternating periods of smooth, forward trajectories of swimming and tumbling. Forward translation, or running, occurs when the flagellar motor rotates in a direction that results in propagation of the semirigid helical wave of the flagellum, which acts like a propeller and exerts a pushing motion on the cell. Wave propagation proceeds from the cell-proximal to cell-distal end of the flagellum, and the multiple flagella coalesce to rotate synchronously and form a propulsive bundle (reviewed in reference 111). Tumbling is linked to changes in the quaternary structure of the flagellum (113). When the filaments are rotated in the opposite direction, structural changes are induced within the filament; the usual transformation is from a normal left-handed helix to semicoiled to a right-handed curly form. If most of the filaments change the direction of rotation at the same time, the bundle flies apart. However, not all of the filaments in the bundle need to reverse the direction of rotation in order to elicit tumbling. Fluorescence microscopy has revealed that changes in the direction of rotation of a single filament that result in transformation of the filament structure to the semicoiled form can elicit a change in the direction of movement of the cell body (185). The running mode allows positional translation, whereas the tumbling mode results in reorientation in three-dimensional space. In the absence of chemotaxis, these bacteria move in the alternating pattern of running and tumbling described as a random walk. In the presence of chemotactic stimuli, the time spent in one mode is biased, e.g., in the presence of a gradient of attractant, the probability of tumbling is decreased, thus prolonging the smooth period of swimming.
Although polarly flagellated V. parahaemolyticus cells do not tumble like peritrichously flagellated E. coli or S. enterica serovar Typhimurium, the motility pattern consists of alternating runs in a forward direction and changes of direction. In the light microscope, V. parahaemolyticus swims in smooth, slightly curved lines, punctuated by quick periods of directional change. Sometimes this change is a reversal of direction so that the cells back up, or it can be a rapid back-and-forth motion, but reversals are not usually exact and Brownian motion perturbs the cell's trajectory. Hence at other times, only an abrupt change of direction can be observed. Similar movement has been described for V. cholerae (formerly V. metschnikovii) (148), monoflagellated Pseudomonas citronellolis (176), and other monotrichous bacteria (reviewed in reference 16).
Very little is known about chemotaxis in the Vibrionaceae family. In contrast to the five E. coli sensory transducing proteins that mediate taxis (12), the V. cholerae genome contains 43 potential methyl-accepting chemotaxis proteins (MCPs) (65). Deciphering the roles of these potential MCPs will be a challenging task, particularly because there may be redundancy or overlap of signaling receptors. For example, P. aeruginosa possesses three MCPs with overlapping specificity for amino acids and two MCPs for phosphate (172, 194). V. alginolyticus and V parahaemolyticus are attracted to serine (68, 156). On addition of phenol, V. alginolyticus cells rapidly move back and forth (68). MCP localization has been performed in V. parahaemolyticus by using antibody directed against the E. coli chemoreceptor Trg (59). Consistent with observations in other bacteria, the MCP localizes to both cell poles (3, 62, 87, 109, 116, 166). In the elongated swarmer cell, MCPs are found at the poles and at intervals along the cell.
The chemotaxis system is one point of integration between the polar and lateral motility systems (156). Mutations in some of the central chemotaxis genes (i.e., defects in cheA or cheB) affect efficient translocation over surfaces on solidified swarming agar (1.5%) and through semisolid swimming-motility agar (0.3%). These mutations also reduce migration into capillary tubes in high- and low-viscosity media, i.e., optimal conditions under which the lateral system and polar system operate, respectively (156). Thus, it is clear that some of the cytoplasmic chemotaxis components are shared by the two motility systems; however the question remains to be investigated whether there are unique components dedicated to each system. For V. alginolyticus, the interesting observation has been made that the polar and lateral flagella show differences in adapting to repellent, which suggests that the chemoresponses of the two systems may not be identical (68).
THE POLAR FLAGELLUM AS A TACTILE SENSOR
The single polar flagellum efficiently propels the rod-shaped free-living bacterium in liquid environments; however, as viscosity increases, the swimming speed is reduced as polar rotation is impeded (10, 82, 114). In addition to its role as a propulsive organelle, the polar flagellum appears to act as a sensor. Growth on surfaces or in viscous environments induces differentiation to the swarmer cell. Cell division ceases, the cells become transiently elongated (∼30 μm), and the lateral flagellar system is induced (2, 15, 162). Polar flagellar function is coupled to expression of the swarmer cell gene system. The polar flagellum is produced constitutively, irrespective of liquid- or surface-associated growth. Physical conditions that impede flagellar rotation seem to act as a signal. All conditions that slow polar flagellar rotation lead to swarmer cell development. Such conditions include increasing viscosity or using antibodies to inhibit flagellar rotation (15, 121). Polar flagellar performance can be perturbed in other ways. If one uses sodium channel-blocking drugs to slow the motor, lateral flagellar gene induction increases proportionally to the decrease in flagellar rotation (82). Thus, the polar flagellum seems to act as a mechanosensor: interference with rotation somehow signals swarmer cell differentiation. The mechanism by which such signal transduction occurs remains a mystery.
Genetic interference with polar function also affects swarmer cell gene expression. All swimming-defective transposon mutants that have been isolated constitutively express swarmer cell genes when grown in liquid (121). These mutants include paralyzed mutants that cannot rotate the flagellum (25), which supports the hypothesis that sensing seems to require flagellar rotation. However, the phenotypes of some mutants are quite puzzling in the context of such a hypothesis. These include filamentless mutants, e.g., those with HAP1 or HAP3 defects or with deletions of the multiple flagellin genes. One would predict that for such mutants, motor rotation would be unaffected; however, these mutants produce lateral flagella in liquid. Clearly, these mutants need to be studied further.
THE LATERAL FLAGELLAR SYSTEM AND SWARMING MOTILITY
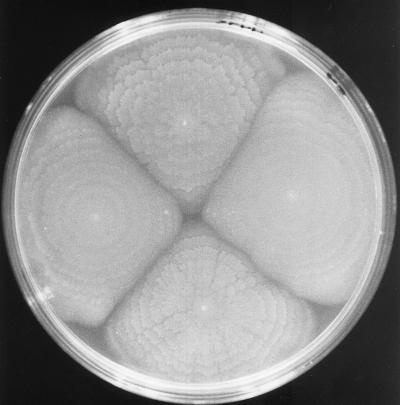
In response to growth on surfaces, the alternate, lateral-motility system is induced. The lateral flagella are polymerized from a single flagellin subunit, LafA. The lateral filaments of V. alginolyticus and V. parahaemolyticus detach easily and form giant coiled bundles that can be seen in the light microscope (186, 187). Other bacteria that swarm are known to produce extracellular surfactants, e.g., extracellular polysaccharide and surfactant-like polypeptides (61, 104, 119), that are essential for movement; however, agents acting as swarming facilitators have yet to be discovered for V. parahaemolyticus. Swarming enables bacteria to colonize surfaces, coordinate behavior, and form multicellular communities, which sometimes display the periodic architecture shown in Fig. 7.
FIG. 7.
Swarming colonies of V. parahaemolyticus on the surface of solidified medium.
The gene sets for the lateral and polar flagellar systems are entirely distinct. Mutants with defects in genes encoding polar structural or export components are competent for swarming, and swarm-defective mutants retain swimming motility. Not only do the lateral and polar flagella differ at the structural level, but also they are powered by different energy sources. Lateral flagella are driven by the flow of protons through the motor (11, 83). Although all of the lateral flagellar genes have not yet been identified, it seems that there are four classes of genes in the lateral hierarchy of expression (125). Class 4 contains the flagellin structural gene, and it is transcribed by a specialized ς28 that recognizes a unique lateral flagellar promoter. Genes identified in class 3 include those that encode motor parts, HAPs, and the lateral flagellar ς28. Class 2 contains the hook and rod genes. Class 1 genes have not yet been discovered, although class 1 seems likely to contain genes encoding master regulators similar to the flhDC operon of E. coli.
A number of bacteria are known to swarm. Some, like V. parahaemolyticus, show mixed flagellation with distinct polar and peritrichous organelles, e.g., Rhodospirillum centenum and Azospirillum species (75, 131), and others possess single peritrichous flagellar systems, e.g., Proteus mirabilis, Serratia species, E. coli, and S. enterica serovar Typhimurium (reviewed in references 49 and 63). In general, the extent of hyperflagellation of the swarmer cell correlates with the robustness of swarming. Little is known about how surface contact initiates swarmer cell development, although both chemotaxis and lipopolysaccharide have been implicated as playing important roles in swarming in the enteric organisms (14, 28, 61, 182). Paralyzed S. enterica serovar Typhimurium motor mutants show normal, surface-specific regulation of flagella (182). This is distinctly different from the phenotype of mot mutants of V. parahaemolyticus, which exhibit constitutive synthesis of lateral flagella (25). Results for E. coli also suggest that the chemotaxis pathway participates in surface signaling and is required to induce swarmer cell differentiation (28). In contrast, V. parahaemolyticus mutants with defects in the cytoplasmic chemotaxis components are able to fully induce swarmer cell genes (121). Thus, some evidence suggests that the mechanism of surface sensing and swarmer cell differentiation may differ among organisms.
SUMMARY AND PERSPECTIVES: COMPARISONS WITH FLAGELLAR SYSTEMS OF OTHER BACTERIA
A large variety of bacterial species are motile by means of flagellar propulsion. The flagella of V. parahaemolyticus are of particular interest because this organism possesses two flagellar systems. A single, sheathed polar flagellum propels the cell in liquid environments. Numerous unsheathed, lateral flagella move the cell over surfaces. Not only are flagella organelles of locomotion, but also they play important roles in attachment (79, 163), biofilm formation (143, 149), and pathogenesis (reviewed in reference 146). Powered by a rotary motor, the flagellum acts as semirigid helical propeller, which is attached via a flexible coupling known as the hook, to the basal body (reviewed in references 21, 41, and 111). The basal body consists of rings and rods that penetrate the membrane and peptidoglycan layers. Associating with the basal body and projecting into the cytoplasm is a structure termed the C-ring, which contains switch proteins and acts as the core, or rotating part, of the motor. There is coupling of the passage of protons or sodium through the flagellar motor with the generation of torque. A number of questions remain unanswered, with respect to both flagellar function in general and the polar organelle in particular. One is, how does the flagellar motor work? Although many models exist, the precise mechanism of how the transmembrane gradient of protons or sodium ions is converted to mechanical work is not known (18–20, 178). The sodium-type motor is remarkably fast and attractive for study because its architecture can be probed with sodium channel-blocking inhibitors and because the sodium motive force can be easily manipulated. More specific questions with regard to the sodium-type motors include the following: what is the architecture of the motor, how is it assembled, and how is ion specificity determined?
The polar flagellum is sheathed and produced continuously. What is the mechanism of sheath formation? Allen and Baumann have noted the regular appearance of tubule projections originating from the cell surface that resemble empty flagellar sheaths (2), suggesting that sheath formation can be an event independent of filament polymerization. In addition, very little is known about the role or function of flagellar sheaths. Does possession of a sheath alter the structure of the basal body? In fact, differences in basal-body structure have been observed between sheathed or unsheathed polar and peritrichous flagella. What are the determinants of site selection for polar placement of the flagellum? Two genes, flhF and flhG, that are not found in E. coli have recently been discovered that may play roles in site determination. Their products resemble components of the signal recognition particle-targeting pathway and cell division site selection pathway, respectively. Pseudomonas mutants with defects in these genes in show altered patterns of flagellation with respect to location and number (38, 147). Intriguingly, the deduced FlhF protein products from both V. parahaemolyticus and V. cholerae possess a unique domain, which has low similarity to a eukaryotic sodium channel.
Lateral flagella are unsheathed and expressed when the bacterium is on a surface or in viscous environments. Thus, under some conditions the bacterium simultaneously assembles two distinct flagellar organelles. Genetic analysis suggests that the gene systems are distinct and that no structural or assembly components are shared; therefore, the independent type III flagellar export systems must be sufficiently specific to discriminate polar from lateral parts. Thus, V. parahaemolyticus should be a good model organism for use in answering the question, what determines the specificity of type III flagellar export?
The chemotaxis sensory transduction system controls the direction of flagellar rotation to modulate behavior in response to environmental cues. The central cytoplasmic chemotaxis components are conserved in many bacteria, and paralogous systems exist for the control of other types of movement, e.g., twitching motility. Chemotaxis has been well studied using bacteria possessing multiple flagella that can form bundles. It controls the frequency of switching of the direction of flagellar rotation. Counterclockwise motor rotation results in smooth swimming, and clockwise rotation results in tumbling and reorientation of the cell. One reorientation solution for a uniflagellated bacterium is known: the single flagellum of Rhodobacter sphaeroides rotates unidirectionally, and rotation stops periodically to allow reorientation (159). However, the polar flagella of Vibrio species are reversible, and little is known about the mechanism or frequency of motor reversal. The genomes of V. cholerae and P aeruginosa are tantalizing with respect to chemotaxis, for they suggest additional complexity over that of E. coli due to a multiplicity of che-like genes. For example, five cheY-like genes can be identified in V. cholerae.
At times, V. parahaemolyticus elaborates both polar and lateral flagella. How is the flow of sensory information channeled to produce a coordinated response in a cell with two propulsive systems? A partial answer can be given to this question, for it is known that some of the chemotaxis genes are shared between the two flagellar systems (156) and that mutations in these genes affect both swimming and swarming motility. It is not known whether all of the components are shared, and it is tempting to speculate that there might be separate swim- and swarm-specific components. For example, movement on surfaces might require the integration of a social component controlling behavior similar to systems found in Myxococcus xanthus (167). One could also imagine that there might be chemoattractants peculiar to life on surfaces. Another unexplored area is the number and nature of the chemosensory receptors. The V. cholerae genome (65) contains more than 40 predicted methyl-accepting chemotaxis proteins (in comparison to the 5 known receptors in E. coli) and suggests a tremendous capacity for chemotactic responses.
There is some link between polar flagellar performance and surface-induced gene expression. Inhibition of polar rotation leads to induction of expression of the genes required for swarming motility. How does the polar flagellum work as a tactile sensor? Does this mechanism of gene control extend to other bacteria? And what genes, in addition to those that encode the lateral flagellar system, does the polar flagellum regulate?
Flagellar systems are encoded by large gene sets that are highly regulated. A hierarchy of regulation has been elucidated for peritrichously flagellated E. coli and S. enterica serovar Typhimurium (96, 97, 100; reviewed in reference 31). This cascade of control couples the timing of gene expression to assembly of the organelle. The pyramid of expression possesses three tiers, or classes, of genes. Genes in each class must be functional for expression of the subsequent class to occur. Class 1 genes, flhD and flhC, encode the master transcriptional activators of class 2 flagellar gene expression. The flhDC operon is controlled by a ς70 promoter and a number of global regulatory factors (98). The majority of the class 2 flagellar genes encode components of the flagellar export system and the basal body. One class 2 gene encodes an alternative ς factor devoted to recognition of flagellar genes (138). Flagellar class 3 operons are positively controlled by the flagellar ς28 factor and negatively regulated by FlgM, an anti-ς factor (139). The anti-ς factor is retained within the cell until the flagellar basal body and hook are completed (70, 99). At this time, FlgM is exported, and ς28 becomes free to direct the expression of class 3 genes, which encode flagellin subunits and hook-associated, motor, and chemotaxis signal transduction proteins. There are additional intricacies to this cascade, e.g., transcriptional classes within classes, translational modulation coupled to basal-body assembly, and linkage between cell division and flagellar production (1, 81, 105, 151). The regulatory hierarchy established for E. coli and S. enterica serovar Typhimurium serves as the paradigm for peritrichous flagellar systems of many other bacteria.
The other well-characterized set of flagellar genes and scheme of flagellar control are those of C. crescentus (reviewed in references 140 and 195). In this organism, flagellation and cell division are strikingly coupled. On cell division, the daughter cell is motile and propelled by a polar flagellum whereas the mother cell is nonmotile and stalked. DNA replication is repressed in the motile cell until the point in the cell cycle when differentiation to a new stalked cell occurs. Many of the genes required for flagellar biosynthesis are homologs of E. coli and S. enterica serovar Typhimurium genes; however, the flagellar hierarchy differs between C. crescentus and the enteric bacteria. The flagellar genes of C. crescentus are organized in four levels of expression with two assembly checkpoints, completion of the MS-ring-switch export complex and completion of the basal body-hook structures. Genes at the lowest level of the hierarchy are transcriptionally regulated by ς54-dependent promoters and the two-component pair FlbE-FlbD (191). The master transcriptional regulator at the top of the hierarchy is also a member of the response regulator family of two-component signal transduction systems, and this regulator, CtrA, controls the initiation of DNA replication, DNA methylation, cell division, and flagellar biogenesis (43).
The polar flagellar hierarchy of members of the Vibrionaceae is different from the peritrichous system of the enteric bacteria or the polar system of C. crescentus. It also differs from what is known about the regulation of spirochete flagellar motility, for which transcription of an operon containing 26 flagellar genes is initiated by a ς70-type promoter (57). The system of gene control is relevant for a number of polarly flagellated bacteria including Pseudomonas species. Similar to E. coli, the flagellar genes at the lowest level of the Vibrio hierarchy are transcribed by a polar-flagellum-specific ς28. The gene encoding the ς28 factor is in an operon controlled by ς54-dependent promoter. Other genes in this middle level of control include those that encode the hook-basal body and export apparatus. The timing of the expression of certain motor and HAP components appears to be split among different levels of the hierarchy, which may reveal something about specific, ordered requirements in the morphogenesis pathway. Also, evidence obtained in experiments with V. cholerae suggests that the genes in the middle level may be divided into subclasses (150). Similar to C. crescentus, near the top of the hierarchy are genes encoding ς54-dependent regulatory factors including one member of a two-component, sensor-response regulatory pair. Thus, there are common themes in flagellar gene control and assembly, but there also appear to be variations among organisms. Some of the most interesting differences remain to be explored, e.g., the cellular and environmental factors that influence flagellar-gene expression and motile behavior.
ACKNOWLEDGMENTS
I thank those who have provided data and discussion, most particularly past and present members of my laboratory and Michael Silverman.
Work from my laboratory was supported by the funding of grant GM43196 from the National Institutes of Health.
REFERENCES
- 1.Aizawa S I, Kubori T. Bacterial flagellation and cell division. Genes Cells. 1998;3:625–634. doi: 10.1046/j.1365-2443.1998.00219.x. [DOI] [PubMed] [Google Scholar]
- 2.Allen R D, Baumann P. Structure and arrangement of flagella in species of the genus Beneckea and Photobacterium fischeri. J Bacteriol. 1971;107:295–302. doi: 10.1128/jb.107.1.295-302.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alley M R, Maddock J R, Shapiro L. Polar localization of a bacterial chemoreceptor. Genes Dev. 1992;6:825–836. doi: 10.1101/gad.6.5.825. [DOI] [PubMed] [Google Scholar]
- 4.Arora S W, Ritchings B W, Almira E C, Lory S, Ramphal R. A transcriptional activator FleQ, regulates mucin adhesion and flagellar gene expression in Pseudomonas aeruginosa in a cascade manner. J Bacteriol. 1997;179:5574–5581. doi: 10.1128/jb.179.17.5574-5581.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Asai Y, Kawagishi I, Sockett R, Homma M. Hybrid motor with H+- and Na+-driven components can rotate Vibrio polar flagella by using sodium ions. J Bacteriol. 1999;181:6332–6338. doi: 10.1128/jb.181.20.6332-6338.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Asai Y, Kawagishi I, Sockett R E, Homma M. Coupling ion specificity of chimeras between H(+)- and Na(+)-driven motor proteins, MotB and PomB, in Vibrio polar flagella. EMBO J. 2000;19:3639–3648. doi: 10.1093/emboj/19.14.3639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Asai Y, Kojima S, Kato H, Nishioka N, Kawagishi I, Homma M. Putative channel components for the fast-rotating sodium-driven flagellar motor of a marine bacterium. J Bacteriol. 1997;179:5104–5110. doi: 10.1128/jb.179.16.5104-5110.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Asai Y, Shoji T, Kawagishi I, Homma M. Cysteine-scanning mutagenesis of the periplasmic loop regions of PomA, a putative channel component of the sodium-driven flagellar motor in Vibrio alginolyticus. J Bacteriol. 2000;182:1001–1007. doi: 10.1128/jb.182.4.1001-1007.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Asakura S, Eguchi G, Iino T. Unidirectional growth of Salmonella flagella in vitro. J Mol Biol. 1968;35:227–236. doi: 10.1016/s0022-2836(68)80050-6. [DOI] [PubMed] [Google Scholar]
- 10.Atsumi T, Maekawa Y, Yamada T, Kawagishi I, Imae Y, Homma M. Effect of viscosity on swimming by the lateral and polar flagella of Vibrio alginolyticus. J Bacteriol. 1996;178:5024–5026. doi: 10.1128/jb.178.16.5024-5026.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Atsumi T, McCarter L, Imae Y. Polar and lateral flagellar motors of marine Vibrio are driven by different ion-motive forces. Nature. 1992;355:182–184. doi: 10.1038/355182a0. [DOI] [PubMed] [Google Scholar]
- 12.Barnakov A N, Barnakova L A, Hazelbauer G L. Comparison in vitro of a high- and a low-abundance chemoreceptor of Escherichia coli: similar kinase activation but different methyl-accepting activities. J Bacteriol. 1998;180:6713–6718. doi: 10.1128/jb.180.24.6713-6718.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Baumann P, Baumann L. The marine Gram-negative eubacteria: genera Photobacterium, Beneckea, Alteromonas, and Alcaligenes. In: Starr M, Stolp H, Trüper H, Balows A, Schlegel H, editors. The prokaryotes. New York, N.Y: Springer-Verlag; 1981. pp. 1302–1331. [Google Scholar]
- 14.Belas R, Goldman M, Ashliman K. Genetic analysis of Proteus mirabilis mutants defective in swarmer cell elongation. J Bacteriol. 1995;177:823–828. doi: 10.1128/jb.177.3.823-828.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Belas R, Simon M, Silverman M. Regulation of lateral flagella gene transcription in Vibrio parahaemolyticus. J Bacteriol. 1986;167:210–218. doi: 10.1128/jb.167.1.210-218.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Berg H C. Chemotaxis in bacteria. Annu Rev Biophys Bioeng. 1975;4:119–136. doi: 10.1146/annurev.bb.04.060175.001003. [DOI] [PubMed] [Google Scholar]
- 17.Berg H C. Constraints on models for the flagellar rotary motor. Philos Trans R Soc London Ser B. 2000;355:491–501. doi: 10.1098/rstb.2000.0590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Berg H C, Turner L. Torque generated by the flagellar motor of Escherichia coli. Biophys J. 1993;65:2201–2216. doi: 10.1016/S0006-3495(93)81278-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Berry R M. Theories of rotary motors. Philos Trans R Soc Lond Ser B. 2000;355:503–509. doi: 10.1098/rstb.2000.0591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Berry R M, Armitage J P. The bacterial flagellar motor. Adv Microb Physiol. 1999;41:291–337. doi: 10.1016/s0065-2911(08)60169-1. [DOI] [PubMed] [Google Scholar]
- 21.Blair D F. How bacteria sense and swim. Annu Rev Microbiol. 1995;49:489–522. doi: 10.1146/annurev.mi.49.100195.002421. [DOI] [PubMed] [Google Scholar]
- 22.Blair D F, Berg H C. The MotA protein of E. coli is a proton-conducting component of the flagellar motor. Cell. 1990;60:439–449. doi: 10.1016/0092-8674(90)90595-6. [DOI] [PubMed] [Google Scholar]
- 23.Blair D F, Berg H C. Mutations in the MotA protein of Escherichia coli reveal domains critical for proton conduction. J Mol Biol. 1991;221:1433–1442. doi: 10.1016/0022-2836(91)90943-z. [DOI] [PubMed] [Google Scholar]
- 24.Blake P A, Weaver R E, Hollis D G. Diseases of humans (other than cholera) caused by Vibrios. Annu Rev Microbiol. 1980;34:341–367. doi: 10.1146/annurev.mi.34.100180.002013. [DOI] [PubMed] [Google Scholar]
- 25.Boles B R, McCarter L L. Insertional inactivation of genes encoding components of the sodium-type flagellar motor and switch of Vibrio parahaemolyticus. J Bacteriol. 2000;182:1035–1045. doi: 10.1128/jb.182.4.1035-1045.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Braun T F, Poulson S, Gully J B, Empey J C, Van Way S, Putnam A, Blair D F. Function of proline residues of MotA in torque generation by the flagellar motor of Escherichia coli. J Bacteriol. 1999;181:3542–3551. doi: 10.1128/jb.181.11.3542-3551.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Brimer C D, Montie T C. Cloning and comparison of fliC genes and identification of glycosylation in the flagellin of Pseudomonas aeruginosa a-type strains. J Bacteriol. 1998;180:3209–3217. doi: 10.1128/jb.180.12.3209-3217.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Burkart M, Toguchi A, Harshey R M. The chemotaxis system, but not chemotaxis, is essential for swarming motility in Escherichia coli. Proc Natl Acad Sci USA. 1998;95:2568–2573. doi: 10.1073/pnas.95.5.2568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Carpenter P B, Hanlon D W, Ordal G W. flhF, a Bacillus subtilis flagellar gene that encodes a putative GTP-binding protein. Mol Microbiol. 1992;6:2705–2713. doi: 10.1111/j.1365-2958.1992.tb01447.x. [DOI] [PubMed] [Google Scholar]
- 30.Characklis W G. Biofilm processes. In: Characklis W G, Marshall K C, editors. Biofilms. New York, N.Y: John Wiley & Sons, Inc.; 1990. pp. 195–231. [Google Scholar]
- 31.Chilcott G S, Hughes K T. Coupling of flagellar gene expression to flagellar assembly in Salmonella enterica serovar Typhimurium and Escherichia coli. Microbiol Mol Biol Rev. 2000;64:694–708. doi: 10.1128/mmbr.64.4.694-708.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chilcott G S, Hughes K T. The type III secretion determinants of the flagellar anti-transcription factor, FlgM, extend from the amino-terminus into the anti-sigma28 domain. Mol Microbiol. 1998;30:1029–1040. doi: 10.1046/j.1365-2958.1998.01131.x. [DOI] [PubMed] [Google Scholar]
- 33.Cho H S, Lee S Y, Yan D, Pan X, Parkinson J S, Kustu S, Wemmer D E, Pelton J G. NMR structure of activated CheY. J Mol Biol. 2000;297:543–551. doi: 10.1006/jmbi.2000.3595. [DOI] [PubMed] [Google Scholar]
- 34.Chun S Y, Parkinson J S. Bacterial motility: membrane topology of the Escherichia coli MotB protein. Science. 1988;230:276–277. doi: 10.1126/science.2447650. [DOI] [PubMed] [Google Scholar]
- 35.Correa N E, Lauriano C M, McGee R, Klose K E. Phosphorylation of the flagellar regulatory protein FlrC is necessary for Vibrio cholerae motility and enhanced colonization. Mol Microbiol. 2000;35:743–755. doi: 10.1046/j.1365-2958.2000.01745.x. [DOI] [PubMed] [Google Scholar]
- 36.Coulton J W, Murray R G E. Cell envelope associations of Aquaspirillum serpens flagella. J Bacteriol. 1978;136:1037–1049. doi: 10.1128/jb.136.3.1037-1049.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Daniels N A, MacKinnon L, Bishop R, Altekruse S, Ray B, Hammond R M, Thompson S, Wilson S, Bean N H, Griffin P M, Slutsker L. Vibrio parahaemolyticus infections in the United States, 1973–1998. J Infect Dis. 2000;181:1661–1666. doi: 10.1086/315459. [DOI] [PubMed] [Google Scholar]
- 38.Dasgupta N, Arora S K, Ramphal R. fleN, a gene that regulates flagellar number in Pseudomonas aeruginosa. J Bacteriol. 2000;182:357–364. doi: 10.1128/jb.182.2.357-364.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Davey M, O'Toole G. Microbial biofilms: from ecology to molecular genetics. Microbiol Mol Biol Rev. 2000;64:847–867. doi: 10.1128/mmbr.64.4.847-867.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.De Mot R, Vanderleyden J. The C-terminal sequence conservation between OmpA-related outer membrane proteins and MotB suggests a common function in both gram-positive and gram-negative bacteria, possibly in the interaction of these domains with peptidoglycan. Mol Microbiol. 1994;12:333–334. doi: 10.1111/j.1365-2958.1994.tb01021.x. [DOI] [PubMed] [Google Scholar]
- 41.DeRosier D J. The turn of the screw the bacterial flagellar motor. Cell. 1998;93:17–20. doi: 10.1016/s0092-8674(00)81141-1. [DOI] [PubMed] [Google Scholar]
- 42.Doig P, Trust T J. Identification of surface-exposed outer membrane antigens of Helicobacter pylori. Infect Immun. 1994;62:4526–4533. doi: 10.1128/iai.62.10.4526-4533.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Domian I J, Reisenauer A, Shapiro L. Feedback control of a master bacterial cell-cycle regulator. Proc Natl Acad Sci USA. 1999;96:6648–6653. doi: 10.1073/pnas.96.12.6648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Engelhardt H, Schuster S C, Baeuerlein E. An archimedian spiral: the basal disk of the Wolinella flagellar motor. Science. 1993;262:1046–1048. doi: 10.1126/science.8235620. [DOI] [PubMed] [Google Scholar]
- 45.Evans D G, Karjalainen T K, Evans D J, Jr, Graham D Y, Lee C H. Cloning, nucleotide sequence, and expression of a gene encoding an adhesin subunit protein of Helicobacter pylori. J Bacteriol. 1993;175:674–683. doi: 10.1128/jb.175.3.674-683.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ferris F G, Beveridge T J, Marceau-Day M L, Larson A D. Structure and cell envelope associations of flagellar basal complexes of Vibrio cholerae and Campylobacter fetus. Can J Microbiol. 1984;30:322–333. doi: 10.1139/m84-048. [DOI] [PubMed] [Google Scholar]
- 47.Francis N R, Sosinsky G E, Thomas D, DeRosier D J. Isolation, characterization and structure of bacterial flagellar motors containing the switch complex. J Mol Biol. 1994;235:1261–1270. doi: 10.1006/jmbi.1994.1079. [DOI] [PubMed] [Google Scholar]
- 48.Fraser G M, Bennett J C, Hughes C. Substrate-specific binding of hook-associated proteins by FlgN and FliT, putative chaperones for flagellum assembly. Mol Microbiol. 1999;32:569–580. doi: 10.1046/j.1365-2958.1999.01372.x. [DOI] [PubMed] [Google Scholar]
- 49.Fraser G M, Hughes C. Swarming motility. Curr Opin Microbiol. 1999;2:630–635. doi: 10.1016/s1369-5274(99)00033-8. [DOI] [PubMed] [Google Scholar]
- 50.Fuerst J A. Bacterial sheathed flagella and the rotary motor model for the mechanism of bacterial motility. J Theor Biol. 1980;4:761–774. doi: 10.1016/s0022-5193(80)80032-4. [DOI] [PubMed] [Google Scholar]
- 51.Fuerst J A, Perry J W. Demonstration of lipopolysaccharide on sheathed flagella of Vibrio cholerae O:1 by protein A-gold immunoelectron microscopy. J Bacteriol. 1988;170:1488–1494. doi: 10.1128/jb.170.4.1488-1494.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Furuno M, Sato K, Kawagishi I, Homma M. Characterization of a flagellar sheath component, PF60, and its structural gene in marine Vibrio. J Biochem (Tokyo) 2000;127:29–36. doi: 10.1093/oxfordjournals.jbchem.a022580. [DOI] [PubMed] [Google Scholar]
- 53.Garza A G, Biran R, Wohlschlegel J A, Manson M D. Mutations in motB suppressible by changes in stator or rotor components of the bacterial flagellar motor. J Mol Biol. 1996;258:270–285. doi: 10.1006/jmbi.1996.0249. [DOI] [PubMed] [Google Scholar]
- 54.Garza A G, Harris-Haller L W, Stoebner R A, Manson M D. Motility protein interactions in the bacterial flagellar motor. Proc Natl Acad Sci USA. 1995;92:1970–1974. doi: 10.1073/pnas.92.6.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ge Y, Charon N W. Identification of a large motility operon in Borrelia burgdorferi by semi-random PCR chromosome walking. Gene. 1997;189:195–201. doi: 10.1016/s0378-1119(96)00848-7. [DOI] [PubMed] [Google Scholar]
- 56.Ge Y, Li C, Corum L, Slaughter C A, Charon N W. Structure and expression of the FlaA periplasmic flagellar protein of Borrelia burgdorferi. J Bacteriol. 1998;180:2418–2425. doi: 10.1128/jb.180.9.2418-2425.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ge Y, Old I G, Saint Girons I, Charon N W. Molecular characterization of a large Borrelia burgdorferi motility operon which is initiated by a consensus sigma70 promoter. J Bacteriol. 1997;179:2289–2299. doi: 10.1128/jb.179.7.2289-2299.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Geis G, Suerbaum S, Forsthoff B, Leying H, Opferkuch W. Ultrastructure and biochemical studies of the flagellar sheath of Helicobacter pylori. J Med Microbiol. 1993;38:371–377. doi: 10.1099/00222615-38-5-371. [DOI] [PubMed] [Google Scholar]
- 59.Gestwicki J E, Lamanna A C, Harshey R M, McCarter L L, Kiessling L L, Adler J. Evolutionary conservation of methyl-accepting chemotaxis protein location in Bacteria and Archaea. J Bacteriol. 2000;182:6499–6502. doi: 10.1128/jb.182.22.6499-6502.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gosink K K, Hase C C. Requirements for conversion of the Na+-driven flagellar motor of Vibrio cholerae to the H+-driven motor of Escherichia coli. J Bacteriol. 2000;182:4234–4240. doi: 10.1128/jb.182.15.4234-4240.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gygi D, Rahman M M, Lai H C, Carlson R, Guard-Petter J, Hughes C. A cell-surface polysaccharide that facilitates rapid population migration by differentiated swarm cells of Proteus mirabilis. Mol Microbiol. 1995;17:1167–1175. doi: 10.1111/j.1365-2958.1995.mmi_17061167.x. [DOI] [PubMed] [Google Scholar]
- 62.Harrison D M, Skidmore J, Armitage J P, Maddock J R. Localization and environmental regulation of MCP-like proteins in Rhodobacter sphaeroides. Mol Microbiol. 1999;31:885–892. doi: 10.1046/j.1365-2958.1999.01226.x. [DOI] [PubMed] [Google Scholar]
- 63.Harshey R M. Bees aren't the only ones: swarming in Gram-negative bacteria. Mol Microbiol. 1994;13:389–394. doi: 10.1111/j.1365-2958.1994.tb00433.x. [DOI] [PubMed] [Google Scholar]
- 64.Harwood C, Fosnaugh K, Dispensa M. Flagellation of Pseudomonas putida and analysis of its motile behavior. J Bacteriol. 1989;171:4063–4066. doi: 10.1128/jb.171.7.4063-4066.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Heidelberg J F, Eisen J A, Nelson W C, Clayton R A, Gwinn M L, Dodson R J, Haft D H, Hickey E K, Peterson J D, Umayam L, Gill S R, Nelson K E, Read T D, Tettelin H, Richardson D, Ermolaeva M D, Vamathevan J, Bass S, Qin H, Dragoi I, Sellers P, McDonald L, Utterback T, Fleishmann R D, Nierman W C, White O. DNA sequence of both chromosomes of the cholera pathogen Vibrio cholerae. Nature. 2000;406:477–483. doi: 10.1038/35020000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Homma M, Fujita H, Yamaguchi S, Iino T. Excretion of unassembled flagellin by Salmonella typhimurium mutants deficient in hook-associated proteins. J Bacteriol. 1984;159:1056–1059. doi: 10.1128/jb.159.3.1056-1059.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Homma M, Iino T, Kutsukake K, Yamaguchi S. In vitro reconstitution of flagellar filaments onto hooks of filamentless mutants of Salmonella typhimurium by addition of hook-associated proteins. Proc Natl Acad Sci USA. 1986;83:6169–6173. doi: 10.1073/pnas.83.16.6169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Homma M, Oota H, Kojima S, Kawagishi I, Imae Y. Chemotactic responses to an attractant and a repellent by the polar and lateral flagellar systems of Vibrio alginolyticus. Microbiology. 1996;142:2777–2783. doi: 10.1099/13500872-142-10-2777. [DOI] [PubMed] [Google Scholar]
- 69.Hranitzky K W, Mulholland A, Larson A D, Eubanks E R, Hart L T. Characterization of a flagellar sheath protein of Vibrio cholerae. Infect Immun. 1980;27:597–603. doi: 10.1128/iai.27.2.597-603.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hughes K T, Gillen K L, Semon M J, Karlinsey J E. Sensing structural intermediates in bacterial flagellar assembly by export of a negative regulator. Science. 1993;262:1277–1280. doi: 10.1126/science.8235660. [DOI] [PubMed] [Google Scholar]
- 71.Imae Y, Atsumi T. Na+-driven bacterial flagellar motors. J Bioenerg Biomembr. 1989;21:705–716. doi: 10.1007/BF00762688. [DOI] [PubMed] [Google Scholar]
- 72.Imae Y, Matsukura H, Kobayashi S. Sodium-driven flagellar motors motors of alkalophilic Bacillus. Methods Enzymol. 1986;125:582–592. doi: 10.1016/s0076-6879(86)25047-8. [DOI] [PubMed] [Google Scholar]
- 73.Irikura V M, Kihara M, Yamaguchi S, Sockett H, Macnab R M. Salmonella typhimurium fliG and fliN mutations causing defects in assembly, rotation, and switching of the flagellar motor. J Bacteriol. 1993;175:802–810. doi: 10.1128/jb.175.3.802-810.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Jaques S, Kim Y K, McCarter L L. Mutations conferring resistance to phenamil and amiloride, inhibitors of sodium-driven motility of Vibrio parahaemolyticus. Proc Natl Acad Sci USA. 1999;96:5740–5745. doi: 10.1073/pnas.96.10.5740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Jiang Z Y, Rushing B G, Bai H, Gest H, Bauer C E. Isolation of Rhodospirillum centenum mutants defective in phototactic colony motility by transposon mutagenesis. J Bacteriol. 1998;180:1248–1255. doi: 10.1128/jb.180.5.1248-1255.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Jones A C, Logan R P, Foynes S, Cockayne A, Wren B W, Penn C W. A flagellar sheath protein of Helicobacter pylori is identical to HpaA, a putative N-acetylneuraminyllactose-binding hemagglutinin, but is not an adhesin for AGS cells. J Bacteriol. 1997;179:5643–5647. doi: 10.1128/jb.179.17.5643-5647.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Jones G W, Freter R. Adhesive properties of Vibrio cholerae: nature of the interaction with isolated rabbit brush border membranes and human erythrocytes. Infect Immun. 1976;14:240–245. doi: 10.1128/iai.14.1.240-245.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Joseph S W, Colwell R R, Kaper J B. Vibrio parahaemolyticus and related halophilic Vibrios. Crit Rev Microbiol. 1982;10:77–124. doi: 10.3109/10408418209113506. [DOI] [PubMed] [Google Scholar]
- 79.Kaneko T, Colwell R R. Adsorption of V. parahaemolyticus onto chitin and copepods. Appl Microbiol. 1975;29:269–274. doi: 10.1128/am.29.2.269-274.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Karlinsey J, Lonner J, Brown K, Hughes K. Translation/secretion coupling by type III secretion systems. Cell. 2000;102:487–497. doi: 10.1016/s0092-8674(00)00053-2. [DOI] [PubMed] [Google Scholar]
- 81.Karlinsey J E, Tsui H C, Winkler M E, Hughes K T. Flk couples flgM translation to flagellar ring assembly in Salmonella typhimurium. J Bacteriol. 1998;180:5384–5397. doi: 10.1128/jb.180.20.5384-5397.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kawagishi I, Imagawa M, Imae Y, McCarter L, Homma M. The sodium-driven polar flagellar motor of marine Vibrio as the mechanosensor that regulates lateral flagellar expression. Mol Microbiol. 1996;20:693–699. doi: 10.1111/j.1365-2958.1996.tb02509.x. [DOI] [PubMed] [Google Scholar]
- 83.Kawagishi I, Maekawa Y, Atsumi T, Homma M, Imae Y. Isolation of the polar and lateral flagellum-defective mutants in Vibrio alginolyticus and identification of their flagellar driving energy sources. J Bacteriol. 1995;177:5158–5160. doi: 10.1128/jb.177.17.5158-5160.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Kawagishi I, Nakada M, Nishioka N, Homma M. Cloning of a Vibrio alginolyticus rpoN gene that is required for polar flagellar formation. J Bacteriol. 1997;179:6851–6854. doi: 10.1128/jb.179.21.6851-6854.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Kelly-Wintenberg K, South S L, Montie T C. Tyrosine phosphate in a- and b-type flagellins of Pseudomonas aeruginosa. J Bacteriol. 1993;175:2458–2461. doi: 10.1128/jb.175.8.2458-2461.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Kim Y K, McCarter L L. Analysis of the polar flagellar gene system of Vibrio parahaemolyticus. J Bacteriol. 2000;182:3693–3704. doi: 10.1128/jb.182.13.3693-3704.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Kirby J R, Niewold T B, Maloy S, Ordal G W. CheB is required for behavioural responses to negative stimuli during chemotaxis in Bacillus subtilis. Mol Microbiol. 2000;35:44–57. doi: 10.1046/j.1365-2958.2000.01676.x. [DOI] [PubMed] [Google Scholar]
- 88.Klose K E, Mekalanos J J. Differential regulation of multiple flagellins in Vibrio cholerae. J Bacteriol. 1998;180:303–316. doi: 10.1128/jb.180.2.303-316.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Klose K E, Mekalanos J J. Distinct roles of an alternative sigma factor during both free-swimming and colonizing phases of the Vibrio cholerae pathogenic cycle. Mol Microbiol. 1998;28:501–520. doi: 10.1046/j.1365-2958.1998.00809.x. [DOI] [PubMed] [Google Scholar]
- 90.Kogure K, Ikemoto E, Morisaki H. Attachment of Vibrio alginolyticus to glass surfaces is dependent on swimming speed. J Bacteriol. 1998;180:932–937. doi: 10.1128/jb.180.4.932-937.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Kojima S, Asai Y, Atsumi T, Kawagishi I, Homma M. Na+-driven flagellar motor resistant to phenamil, an amiloride analog, caused by mutations in putative channel components. J Mol Biol. 1999;285:1537–1547. doi: 10.1006/jmbi.1998.2377. [DOI] [PubMed] [Google Scholar]
- 92.Kojima S, Atsumi T, Muramoto K, Kudo S, Kawagishi I, Homma M. Vibrio alginolyticus mutants resistant to phenamil, a specific inhibitor of the sodium-driven flagellar motor. J Mol Biol. 1997;265:310–318. doi: 10.1006/jmbi.1996.0732. [DOI] [PubMed] [Google Scholar]
- 93.Kojima S, Kuroda M, Kawagishi I, Homma M. Random mutagenesis of the pomA gene encoding a putative channel component of the Na(+)-driven polar flagellar motor of Vibrio alginolyticus. Microbiology. 1999;145:1759–1767. doi: 10.1099/13500872-145-7-1759. [DOI] [PubMed] [Google Scholar]
- 94.Kojima S, Shoji T, Asai Y, Kawagishi I, Homma M. A slow-motility phenotype caused by substitutions at residue Asp31 in the PomA channel component of a sodium-driven flagellar motor. J Bacteriol. 2000;182:3314–3318. doi: 10.1128/jb.182.11.3314-3318.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Kojima S, Yamamoto K, Kawagishi I, Homma M. The polar flagellar motor of Vibrio cholerae is driven by an Na+ motive force. J Bacteriol. 1999;181:1927–1930. doi: 10.1128/jb.181.6.1927-1930.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Komeda Y. Fusions of flagellar operons to lactose genes on a Mu lac bacteriophage. J Bacteriol. 1982;150:16–26. doi: 10.1128/jb.150.1.16-26.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Komeda Y. Transcriptional control of flagellar genes in Escherichia coli K-12. J Bacteriol. 1986;168:1315–1318. doi: 10.1128/jb.168.3.1315-1318.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Kutsukake K. Autogenous and global control of the flagellar master operon, flhDC, in Salmonella typhimurium. Mol Gen Genet. 1997;24:440–448. doi: 10.1007/s004380050437. [DOI] [PubMed] [Google Scholar]
- 99.Kutsukake K, Iino T. Role of the FliA-FlgM regulatory system on the transcriptional control of the flagellar regulon and flagellar formation in Salmonella typhimurium. J Bacteriol. 1994;176:3598–3605. doi: 10.1128/jb.176.12.3598-3605.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Kutsukake K, Ohya Y, Iino T. Transcriptional analysis of the flagellar regulon of Salmonella typhimurium. J Bacteriol. 1990;172:741–747. doi: 10.1128/jb.172.2.741-747.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Larsen S H, Adler J, Gargus J J, Hogg R W. Chemomechanical coupling without ATP the source of energy for motility and chemotaxis in bacteria. Proc Natl Acad Sci USA. 1974;71:1239–1243. doi: 10.1073/pnas.71.4.1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Levin P A, Shim J J, Grossman A D. Effect of minCD on FtsZ ring position and polar septation in Bacillus subtilis. J Bacteriol. 1998;180:6048–6051. doi: 10.1128/jb.180.22.6048-6051.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Li C, Motaleb A, Sal M, Goldstein S F, Charon N W. Spirochete periplasmic flagella and motility. J Mol Microbiol Biotechnol. 2000;2:345–354. [PubMed] [Google Scholar]
- 104.Lindum P, Anthoni W U, Christophersen C, Eberl L, Molin S, Givskov M. N-Acyl-l-homoserine lactone autoinducers control production of an extracellular lipopeptide biosurfactant required for swarming motility of Serratia liquefaciens MG1. J Bacteriol. 1998;180:6384–6388. doi: 10.1128/jb.180.23.6384-6388.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Liu X, Matsumura P. Differential regulation of multiple overlapping promoters in flagellar class II operons in Escherichia coli. Mol Microbiol. 1996;21:613–620. doi: 10.1111/j.1365-2958.1996.tb02569.x. [DOI] [PubMed] [Google Scholar]
- 106.Lloyd S A, Blair D F. Charged residues of the rotor protein FliG essential for torque generation in the flagellar motor of Escherichia coli. J Mol Biol. 1997;266:733–744. doi: 10.1006/jmbi.1996.0836. [DOI] [PubMed] [Google Scholar]
- 107.Lloyd S A, Tang H, Wang X, Billings S, Blair D F. Torque generation in the flagellar motor of Escherichia coli: evidence of a direct role for FliG but not for FliM or FliN. J Bacteriol. 1996;178:223–231. doi: 10.1128/jb.178.1.223-231.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Lloyd S A, Whitby F G, Blair D F, Hill C P. Structure of the C-terminal domain of FliG, a component of the rotor in the bacterial flagellar motor. Nature. 1999;400:472–475. doi: 10.1038/22794. [DOI] [PubMed] [Google Scholar]
- 109.Lybarger S R, Maddock J R. Differences in the polar clustering of the high- and low-abundance chemoreceptors of Escherichia coli. Proc Natl Acad Sci USA. 2000;97:8057–8062. doi: 10.1073/pnas.130195397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Macnab R M. The bacterial flagellum: reversible rotary propellor and type III export apparatus. J Bacteriol. 1999;181:7149–7153. doi: 10.1128/jb.181.23.7149-7153.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Macnab R M. Flagella and motility. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B Jr, Magasanik B, Reznikoff W, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C.: ASM Press; 1996. pp. 123–146. [Google Scholar]
- 112.Macnab R M. Proton-driven bacterial flagellar motor. Methods Enzymol. 1986;125:563–579. doi: 10.1016/s0076-6879(86)25046-6. [DOI] [PubMed] [Google Scholar]
- 113.Macnab R M, Ornston M K. Normal-to-curly flagellar transitions and their role in bacterial tumbling. Stabilization of an alternative quaternary structure by mechanical force. J Mol Biol. 1977;112:1–30. doi: 10.1016/s0022-2836(77)80153-8. [DOI] [PubMed] [Google Scholar]
- 114.Magariyama Y, Sugiyama S, Muramoto K, Kawagishi I, Imae Y, Kudo S. Simultaneous measurement of bacterial flagellar rotation rate and swimming speed. Biophys J. 1995;69:2154–2162. doi: 10.1016/S0006-3495(95)80089-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Magariyama Y, Sugiyama S, Muramoto K, Maekawa Y, Kawagishi I, Imae Y, Kudo S. Very fast flagellar rotation. Nature. 1994;371:752. doi: 10.1038/371752b0. [DOI] [PubMed] [Google Scholar]
- 116.Maki N, Gestwicki J E, Lake E M, Kiessling L L, Adler J. Motility and chemotaxis of filamentous cells of Escherichia coli. J Bacteriol. 2000;182:4337–4342. doi: 10.1128/jb.182.15.4337-4342.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Manson M D, Tedesco P, Berg H C, Harold F M, Van der Drift C. A protonmotive force drives bacterial flagella. Proc Natl Acad Sci USA. 1977;74:3060–3064. doi: 10.1073/pnas.74.7.3060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Marston A L, Thomaides H B, Edwards D H, Sharpe M E, Errington J. Polar localization of the MinD protein of Bacillus subtilis and its role in selection of the mid-cell division site. Genes Dev. 1998;12:3419–3430. doi: 10.1101/gad.12.21.3419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Matsuyama T, Kaneda K, Nakagawa Y, Isa K, Hara-Hotta H, Yano I. A novel extracellular cyclic lipopeptide which promotes flagellum-dependent and -independent spreading growth of Serratia marcescens. J Bacteriol. 1992;174:1769–1776. doi: 10.1128/jb.174.6.1769-1776.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.McCarter L. The multiple identities of Vibrio parahaemolyticus. J Mol Microbiol Biotechnol. 1999;1:51–57. [PubMed] [Google Scholar]
- 121.McCarter L, Hilmen M, Silverman M. Flagellar dynamometer controls swarmer cell differentiation of V. parahaemolyticus. Cell. 1988;54:345–351. doi: 10.1016/0092-8674(88)90197-3. [DOI] [PubMed] [Google Scholar]
- 122.McCarter L L. Genetic and molecular characterization of the polar flagellum of Vibrio parahaemolyticus. J Bacteriol. 1995;177:1595–1609. doi: 10.1128/jb.177.6.1595-1609.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.McCarter L L. MotX, the channel component of the sodium-type flagellar motor. J Bacteriol. 1994;176:5988–5998. doi: 10.1128/jb.176.19.5988-5998.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.McCarter L L. MotY, a component of the sodium-type flagellar motor. J Bacteriol. 1994;176:4219–4225. doi: 10.1128/jb.176.14.4219-4225.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.McCarter L L, Wright M E. Identification of genes encoding components of the swarmer cell flagellar motor and propeller and a sigma factor controlling differentiation of Vibrio parahaemolyticus. J Bacteriol. 1993;175:3361–3371. doi: 10.1128/jb.175.11.3361-3371.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.McGee K, Horstedt P, Milton D L. Identification and characterization of of additional flagellin genes from Vibrio anguillarum. J Bacteriol. 1996;178:5188–5198. doi: 10.1128/jb.178.17.5188-5198.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Meadows P S. The atachment of bacteria to solid surfaces. Arch Mikrobiol. 1971;75:374–381. doi: 10.1007/BF00407699. [DOI] [PubMed] [Google Scholar]
- 128.Meister M, Lowe G, Berg H C. The proton flux through the bacterial flagellar motor. Cell. 1987;49:643–650. doi: 10.1016/0092-8674(87)90540-x. [DOI] [PubMed] [Google Scholar]
- 129.Milton D, O'Toole R, Horstedt P, Wolf-Watz H. Flagellin A is essential for virulence of Vibrio anguillarum. J Bacteriol. 1996;178:1310–1319. doi: 10.1128/jb.178.5.1310-1319.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Minamino T, Doi H, Kutsukake K. Substrate specificity switching of the flagellum-specific export apparatus during flagellar morphogenesis in Salmonella typhimurium. Biosci Biotechnol Biochem. 1999;63:1301–1303. doi: 10.1271/bbb.63.1301. [DOI] [PubMed] [Google Scholar]
- 131.Moens S, Schloter M, Vanderleyden J. Expression of the structural gene, laf1, encoding the flagellin of the lateral flagella in Azospirillum brasilense Sp7. J Bacteriol. 1996;178:5017–5019. doi: 10.1128/jb.178.16.5017-5019.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Moens S, Vanderleyden J. Functions of bacterial flagella. Crit Rev Microbiol. 1996;22:67–100. doi: 10.3109/10408419609106456. [DOI] [PubMed] [Google Scholar]
- 133.Motaleb M A, Corum L, Bono J L, Elias A F, Rosa P, Samuels D S, Charon N W. Borrelia burgdorferi periplasmic flagella have both skeletal and motility functions. Proc Natl Acad Sci USA. 2000;97:10899–10904. doi: 10.1073/pnas.200221797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Muller M, Koch H G, Beck K, Schafer U. Protein traffic in bacteria multiple routes from the ribosome to and across the membrane. Prog Nucleic Acid Res Mol Biol. 2000;66:107–157. doi: 10.1016/s0079-6603(00)66028-2. [DOI] [PubMed] [Google Scholar]
- 135.Namba K, Vonderviszt F. Molecular architecture of bacterial flagellum. Q Rev Biophys. 1997;30:1–65. doi: 10.1017/s0033583596003319. [DOI] [PubMed] [Google Scholar]
- 136.Nishioka N, Furuno M, Kawagishi I, Homma M. Flagellin-containing membrane vesicles excreted from Vibrio alginolyticus mutants lacking a polar-flagellar filament. J Biochem (Tokyo) 1998;123:1169–1173. doi: 10.1093/oxfordjournals.jbchem.a022057. [DOI] [PubMed] [Google Scholar]
- 137.Norqvist A, Wolf-Watz H. Characterization of a novel chromosomal virulence locus involved in expression of a major surface flagellar sheath antigen of the fish pathogen Vibrio anguillarum. Infect Immun. 1993;61:2434–2444. doi: 10.1128/iai.61.6.2434-2444.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Ohnishi I, Kutsukake K, Suzuki H, Iino T. Gene fliA encodes an alternative sigma factor specific for flagellar operons in Salmonella typhimurium. Mol Gen Genet. 1990;221:1139–1147. doi: 10.1007/BF00261713. [DOI] [PubMed] [Google Scholar]
- 139.Ohnishi I, Kutsukake K, Suzuki H, Iino T. A novel transcriptional regulation mechanism in the flagellar regulon of Salmonella typhimurium: an antisigma factor inhibits the activity of the flagellum-specific sigma factor, sigma F. Mol Microbiol. 1992;6:3149–3157. doi: 10.1111/j.1365-2958.1992.tb01771.x. [DOI] [PubMed] [Google Scholar]
- 140.Ohta M, Newton A. Signal transduction in the cell cycle regulation of Caulobacter differentiation. Trends Microbiol. 1996;8:326–332. doi: 10.1016/0966-842x(96)10050-0. [DOI] [PubMed] [Google Scholar]
- 141.Okunishi I, Kawagishi I, Homma M. Cloning and characterization of motY, a gene coding for a component of the sodium-driven flagellar motor in Vibrio alginolyticus. J Bacteriol. 1996;178:2409–2415. doi: 10.1128/jb.178.8.2409-2415.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Ormonde P, Horstedt P, O'Toole R, Milton D L. Role of motility in adherence to and invasion of a fish cell line by Vibrio anguillarum. J Bacteriol. 2000;182:2326–2328. doi: 10.1128/jb.182.8.2326-2328.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.O'Toole G A, Kolter R. Flagellar and twitching motility are necessary for Pseudomonas aeruginosa biofilm development. Mol Microbiol. 1998;30:295–304. doi: 10.1046/j.1365-2958.1998.01062.x. [DOI] [PubMed] [Google Scholar]
- 144.O'Toole P W, Janzon L, Doig P, Huang J, Kostrzynska M, Trust T J. The putative neuraminyllactose-binding hemagglutinin HpaA of Helicobacter pylori CCUG 17874 is a lipoprotein. J Bacteriol. 1995;177:6049–6057. doi: 10.1128/jb.177.21.6049-6057.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.O'Toole R, Milton D L, Horstedt P, Wolf-Watz H. RpoN of the fish pathogen Vibrio (Listonella) anguillarum is essential for flagellum production and virulence by the water-borne but not intraperitoneal route of inoculation. Microbiology. 1997;143:3849–3859. doi: 10.1099/00221287-143-12-3849. [DOI] [PubMed] [Google Scholar]
- 146.Ottemann K M, Miller J F. Roles for motility in bacterial-host interactions. Mol Microbiol. 1997;24:1109–1117. doi: 10.1046/j.1365-2958.1997.4281787.x. [DOI] [PubMed] [Google Scholar]
- 147.Pandza S, Baetens M, Park C H, Au T, Keyhan M, Matin A. The G-protein FlhF as a role in polar flagellar placement and general stress response induction in Pseudomonas putida. Mol Microbiol. 2000;36:414–423. doi: 10.1046/j.1365-2958.2000.01859.x. [DOI] [PubMed] [Google Scholar]
- 148.Pijper A, Nunn A J. Flagella and motility of Vibrio metschnikovii. J R Microsc Soc. 1949;69:138–142. doi: 10.1111/j.1365-2818.1949.tb00972.x. [DOI] [PubMed] [Google Scholar]
- 149.Pratt L A, Kolter R. Genetic analysis of Escherichia coli biofilm formation roles of flagella, motility, chemotaxis and type I pili. Mol Microbiol. 1998;30:285–293. doi: 10.1046/j.1365-2958.1998.01061.x. [DOI] [PubMed] [Google Scholar]
- 150.Prouty M, Correa N, Klose K. The novel ς54- and ς28- dependent flagellar gene transcription hierarchy of Vibrio cholerae. Mol Microbiol. 2001;39:1595–1609. doi: 10.1046/j.1365-2958.2001.02348.x. [DOI] [PubMed] [Google Scholar]
- 151.Prüss B M, Matsumura P. Cell cycle regulation of flagellar genes. J Bacteriol. 1997;179:5602–5604. doi: 10.1128/jb.179.17.5602-5604.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Quisel J D, Grossman A D. Control of sporulation gene expression in Bacillus subtilis by the chromosome partitioning proteins Soj (ParA) and Spo0J (ParB) J Bacteriol. 2000;182:3446–3451. doi: 10.1128/jb.182.12.3446-3451.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Ritchings B W, Almira E C, Lory S, Ramphal R. Cloning and phenotypic characterization of fleS and fleR, new response regulators of Pseudomonas aeruginosa which regulate motility and adhesion to mucin. Infect Immun. 1995;63:4868–4876. doi: 10.1128/iai.63.12.4868-4876.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Rodgers H J. Adhesion of microroganisms to surfaces: some general considerations of the role of the envelope. In: Ellwood D C, Melling J, Rutter P, editors. Adhesion of microroganisms to surfaces. New York. N.Y: Academic Press, Inc.; 1979. pp. 28–55. [Google Scholar]
- 155.Rosario M M L, Fredrick K L, Ordal G, Helmann J. Chemotaxis in Bacillus subtilis requires either of two functionally redundant CheW homologs. J Bacteriol. 1994;176:2736–2739. doi: 10.1128/jb.176.9.2736-2739.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Sar N, McCarter L, Simon M, Silverman M. Chemotactic control of the two flagellar systems of Vibrio parahaemolyticus. J Bacteriol. 1990;172:334–341. doi: 10.1128/jb.172.1.334-341.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Sato K, Homma M. Functional reconstitution of the Na(+)-driven polar flagellar motor component of Vibrio alginolyticus. J Biol Chem. 2000;275:5718–5722. doi: 10.1074/jbc.275.8.5718. [DOI] [PubMed] [Google Scholar]
- 158.Sato K, Homma M. Multimeric structure of PomA, a component of the Na+-driven polar flagellar motor of Vibrio alginolyticus. J Biol Chem. 2000;275:20223–20228. doi: 10.1074/jbc.M002236200. [DOI] [PubMed] [Google Scholar]
- 159.Shah D S H, Porter S L, Martin A C, Hamblin P A, Armitage J P. Fine tuning bacterial chemotaxis analysis of Rhodobacter sphaeroides behaviour under aerobic and anaerobic conditions by mutation of the major chemotaxis operons and cheY genes. EMBO J. 2000;19:4602–4613. doi: 10.1093/emboj/19.17.4601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Sharp L L, Zhou J, Blair D F. Features of MotA proton channel structure revealed by tryptophan-scanning mutagenesis. Proc Natl Acad Sci USA. 1995;92:7946–7950. doi: 10.1073/pnas.92.17.7946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Sharpe M E, Errington J. The Bacillus subtilis soj-spo0J locus is required for a centromere-like function involved in prespore chromosome partitioning. Mol Microbiol. 1996;21:501–509. doi: 10.1111/j.1365-2958.1996.tb02559.x. [DOI] [PubMed] [Google Scholar]
- 162.Shinoda S, Okamoto K. Formation and function of Vibrio parahaemolyticus lateral flagella. J Bacteriol. 1977;129:1266–1271. doi: 10.1128/jb.129.3.1266-1271.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Sjoblad R D, Doetsch R N. Adsorption of polarly flagellated bacteria to surfaces. Curr Microbiol. 1982;7:191–194. [Google Scholar]
- 164.Sjoblad R D, Emala C W, Doetsch R N. Bacterial sheaths: structures in search of function. Cell Motil. 1983;3:93–103. doi: 10.1002/cm.970030108. [DOI] [PubMed] [Google Scholar]
- 165.Sockett H, Yamaguchi S, Kihara M M, Irikura V M, Macnab R M. Molecular analysis of the flagellar switch protein FliM of Salmonella typhimurium. J Bacteriol. 1992;174:793–806. doi: 10.1128/jb.174.3.793-806.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Sourjik V, Berg H C. Localization of components of the chemotaxis machinery of Escherichia coli using fluorescent protein fusions. Mol Microbiol. 2000;37:740–751. doi: 10.1046/j.1365-2958.2000.02044.x. [DOI] [PubMed] [Google Scholar]
- 167.Spormann A M. Gliding motility in bacteria: insights from studies of Myxococcus xanthus. Microbiol Mol Biol Rev. 1999;63:621–641. doi: 10.1128/mmbr.63.3.621-641.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Stewart B J, McCarter L L. Vibrio parahaemolyticus FlaJ, a homologue of FliS, is required for production of a flagellin. Mol Microbiol. 1996;20:137–149. doi: 10.1111/j.1365-2958.1996.tb02496.x. [DOI] [PubMed] [Google Scholar]
- 169.Stolz B, Berg H C. Evidence for interactions between MotA and MotB, torque-generating elements of the flagellar motor of Escherichia coli. J Bacteriol. 1991;173:7033–7037. doi: 10.1128/jb.173.21.7033-7037.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Sugiyama S J, Cragoe E J, Imae Y. Amiloride, a specific inhibitor for the Na+-driven flagellar motors of alkalophilic Bacillus. J Biol Chem. 1988;263:8215–8219. [PubMed] [Google Scholar]
- 171.Szymanski C M, Yao R, Ewing C P, Trust T J, Guerry P. Evidence for a system of general protein glycosylation in Campylobacter jejuni. Mol Microbiol. 1999;32:1022–1030. doi: 10.1046/j.1365-2958.1999.01415.x. [DOI] [PubMed] [Google Scholar]
- 172.Taguchi K, Fukutomi H, Kuroda A, Kato J, Ohtake H. Genetic identification of chemotactic transducers for amino acids in Pseudomonas aeruginosa. Microbiology. 1997;143:3223–3229. doi: 10.1099/00221287-143-10-3223. [DOI] [PubMed] [Google Scholar]
- 173.Tang H, Billings S, Wang X, Sharp L, Blair D F. Regulated underexpression and overexpression of the FliN protein of Escherichia coli and evidence for an interaction between FliN and FliM in the flagellar motor. J Bacteriol. 1995;177:3496–3503. doi: 10.1128/jb.177.12.3496-3503.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Tang H, Blair D F. Regulated underexpression of the FliM protein of Escherichia coli and evidence for a location in the flagellar motor distinct from the MotA/MotB torque generators. J Bacteriol. 1995;177:3485–3495. doi: 10.1128/jb.177.12.3485-3495.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Tauxe R V. Emerging foodborne diseases an evolving public health challenge. Emerg Infect Dis. 1997;3:425–434. doi: 10.3201/eid0304.970403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Taylor B L, Koshland D E., Jr Reversal of flagellar rotation in monotrichous and peritrichous bacteria: generation of changes in direction. J Bacteriol. 1974;119:640–642. doi: 10.1128/jb.119.2.640-642.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Taylor B L, Zhulin I B. PAS domains: internal sensors of oxygen, redox potential, and light. Microbiol Mol Biol Rev. 1999;63:479–506. doi: 10.1128/mmbr.63.2.479-506.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Thomas D R, Morgan D G, DeRosier D J. Rotational symmetry of the C ring and a mechanism for the flagellar rotary motor. Proc Natl Acad Sci USA. 1999;96:10134–10139. doi: 10.1073/pnas.96.18.10134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Thomashow L S, Rittenberg S C. Isolation and composition of sheathed flagella from Bdellovibrio bacteriovorus 109J. J Bacteriol. 1985;163:1047–1054. doi: 10.1128/jb.163.3.1047-1054.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Thomashow L S, Rittenberg S C. Waveform analysis and structure of flagella and basal complexes from Bdellovibrio bacteriovorus 109J. J Bacteriol. 1985;163:1038–1046. doi: 10.1128/jb.163.3.1038-1046.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Togashi F, Yamaguchi S, Kihara M, Aizawa S-I, Macnab R M. An extreme clockwise switch bias mutation in fliG of Salmonella typhimurium and its suppression by slow-motile mutations in motA and motB. J Bacteriol. 1997;179:2994–3003. doi: 10.1128/jb.179.9.2994-3003.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Toguchi A, Siano M, Burkart M, Harshey R M. Genetics of swarming motility in Salmonella enterica serovar Typhimurium: critical role for lipopolysaccharide. J Bacteriol. 2000;182:6308–6321. doi: 10.1128/jb.182.22.6308-6321.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Toker A S, Macnab R M. Distinct regions of bacterial flagellar switch protein FliM interact with FliG, FliN, and Che Y. J Mol Biol. 1997;273:623–634. doi: 10.1006/jmbi.1997.1335. [DOI] [PubMed] [Google Scholar]
- 184.Totten P A, Lara J C, Lory S. The rpoN gene product of Pseudomonas aeruginosa is required for expression of diverse genes, including the flagellin gene. J Bacteriol. 1990;172:389–396. doi: 10.1128/jb.172.1.389-396.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Turner L, Ryu W S, Berg H C. Real-time imaging of fluorescent flagellar filaments. J Bacteriol. 2000;182:2793–2801. doi: 10.1128/jb.182.10.2793-2801.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Ulitzur S. Induction of swarming in Vibrio parahaemolyticus. Arch Microbiol. 1974;101:357–363. doi: 10.1007/BF00455952. [DOI] [PubMed] [Google Scholar]
- 187.Ulitzur S, Kessel M. Giant flagellar bundles of Vibrio alginolyticus (NCMB1803) Arch Mikrobiol. 1973;94:331–339. doi: 10.1007/BF00769028. [DOI] [PubMed] [Google Scholar]
- 188.Urakawa H, Kita-Tsukamoto K, Ohwada K. 16s rDNA genotyping using PCR/RFLP (restriction fragment length polymorphism) analysis among the family Vibrionaceae. FEMS Microbiol Lett. 1997;152:125–132. doi: 10.1111/j.1574-6968.1997.tb10418.x. [DOI] [PubMed] [Google Scholar]
- 189.Vaituzis Z, Doetsch R. Motility tracks: technique for quantitative study of bacterial movement. Appl Microbiol. 1969;17:584–588. doi: 10.1128/am.17.4.584-588.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Wilson D R, Beveridge T J. Bacterial flagellar filaments and their component flagellins. Can J Microbiol. 1993;39:451–472. doi: 10.1139/m93-066. [DOI] [PubMed] [Google Scholar]
- 191.Wingrove J A, Gober J W. Identification of an asymmetrically localized sensor histidine kinase responsible for temporally and spatially regulated transcription. Science. 1996;274:597–601. doi: 10.1126/science.274.5287.597. [DOI] [PubMed] [Google Scholar]
- 192.Woese C R, Weisburg W G, Hahn C M, Paster C M, Zablen L B, Lewis B J, Macke T J, Ludwig W, Stackebrandt E. The phylogeny of purple bacteria of the gamma subdivision. Syst Appl Microbiol. 1985;6:25–33. [Google Scholar]
- 193.Wong H C, Liu C C, Yu C M, Lee Y S. Utilization of iron sources and its possible roles in the pathogenesis of V. parahaemolyticus. Microbiol Immunol. 1996;40:791–798. doi: 10.1111/j.1348-0421.1996.tb01144.x. [DOI] [PubMed] [Google Scholar]
- 194.Wu H, Kato J, Kuroda A, Ikeda T, Takiguchi N, Ohtake H. Identification and characterization of two chemotactic transducers for inorganic phosphate in Pseudomonas aeruginosa. J Bacteriol. 2000;182:3400–3404. doi: 10.1128/jb.182.12.3400-3404.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Wu J, Newton A. Regulation of the Caulobacter flagellar gene hierarchy: not just for motility. Mol Microbiol. 1997;24:233–239. doi: 10.1046/j.1365-2958.1997.3281691.x. [DOI] [PubMed] [Google Scholar]
- 196.Wyss C. Flagellins, but not endoflagellar sheath proteins, of Treponema pallidum and of pathogen-related oral spirochetes are glycosylated. Infect Immun. 1998;66:5751–5754. doi: 10.1128/iai.66.12.5751-5754.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Yamaguchi S, Aizawa S-I, Kihara M, Isomura M, Jones C J, Macnab R M. Genetic evidence for a switching and energy-transducing complex in the flagellar motor of Salmonella typhimurium. J Bacteriol. 1986;168:1172–1179. doi: 10.1128/jb.168.3.1172-1179.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Yamaguchi S, Fujita H, Ishihara A, Aizawa S-I, Macnab R M. Subdivision of flagellar genes of Salmonella typhimurium into regions responsible for assembly, rotation, and switching. J Bacteriol. 1986;166:187–193. doi: 10.1128/jb.166.1.187-193.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Yokota T, Kuwahara S. Adenosine 3′,5′-cyclic monophosphate-deficient mutants of Vibrio cholerae. J Bacteriol. 1974;120:106–113. doi: 10.1128/jb.120.1.106-113.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Yonekura K, Maki S, Morgan D G, DeRosier D J, Vonderviszt F, Imada K, Namba K. The bacterial flagellar cap as the rotary promoter of flagellin self-assembly. Science. 2000;290:2148–2152. doi: 10.1126/science.290.5499.2148. [DOI] [PubMed] [Google Scholar]
- 201.Yorimitsu T, Asai Y, Sato K, Homma M. Intermolecular cross-linking between the periplasmic loop3–4 regions of PomA, a component of the Na+-driven flagellar motor of Vibrio alginolyticus. J Biol Chem. 2000;275:31387–31391. doi: 10.1074/jbc.M000848200. [DOI] [PubMed] [Google Scholar]
- 202.Yorimitsu T, Homma M. Na+-driven flagellar motor of Vibrio. Biochim Biophys Acta. 2001;1505:82–93. doi: 10.1016/s0005-2728(00)00279-6. [DOI] [PubMed] [Google Scholar]
- 203.Yorimitsu T, Sato K, Asai Y, Kawagishi I, Homma M. Functional interaction between PomA and PomB, the Na+-driven flagellar motor components of Vibrio alginolyticus. J Bacteriol. 1999;181:5103–5106. doi: 10.1128/jb.181.16.5103-5106.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Zhou J, Blair D F. Residues of the cytoplasmic domain of MotA essential for torque generation in the bacterial flagellar motor. J Mol Biol. 1997;273:428–439. doi: 10.1006/jmbi.1997.1316. [DOI] [PubMed] [Google Scholar]
- 205.Zhou J, Lloyd S A, Blair D F. Electrostatic interactions between rotor and stator in the bacterial flagellar motor. Proc Natl Acad Sci USA. 1998;95:6436–6441. doi: 10.1073/pnas.95.11.6436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Zhou J, Sharp L L, Tang H L, Lloyd S A, Billings S, Braun T F, Blair D F. Function of protonatable residues in the flagellar motor of Escherichia coli: a critical role for Asp 32 of MotB. J Bacteriol. 1998;180:2729–2735. doi: 10.1128/jb.180.10.2729-2735.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]