Abstract
Hirt et al.1 report an automated, high-throughput drug screening platform for organoid cultures to enable repurposing of previously approved drugs for pancreatic cancers harboring specific genetic alterations. The pancreatic cancer organoid biobank also represents a valuable tool to uncover new drug-gene interactions in pancreatic tumors.
Hirt et al. report an automated, high-throughput drug screening platform for organoid cultures to enable repurposing of previously approved drugs for pancreatic cancers harboring specific genetic alterations. The pancreatic cancer organoid biobank also represents a valuable tool to uncover new drug-gene interactions in pancreatic tumors.
Main text
Pancreatic cancer (PDAC) is one of the most lethal human malignancies, with a 5-year survival rate less than 10%, and where very few advances have been made in terms of novel therapies.2 Indeed, classic chemotherapy still represents the treatment of choice for the management of pancreatic cancer of all stages. Over the last few years, an improved understanding of the genomic landscape of PDAC has begun to emerge, raising hopes for the future of targeted therapies against this devastating malignancy.3 Activating mutations of KRAS have been found almost ubiquitously in PDAC, and also inactivation of TP53, SMAD4, and CDKN2A happens in more than 50% of cases. While the small GTPase KRAS could represent a very useful actionable target, it has unfortunately proven difficult to drug, despite decades of studies. Only recently, KRASG12C-specific inhibitors have entered clinical development, and this resulted in sotorasib as the first drug approved by the Food and Drug Administration (FDA) for the treatment of KRASG12C-driven non-small cell lung cancer.4 Unfortunately, KRASG12C mutants represent only 1% of KRAS mutations in PDAC,5 while for the most frequent amino acid substitutions G12D (41%), G12V (34%), and G12R (16%) targeted inhibitors have not yet reached the clinic. The identification of germline alterations in BRCA1/2 and PALB2 in a subset of PDAC patients, causing homologous repair deficiency (HRD), has recently led to the clinical approval of PARP inhibitors, representing the first targeted therapy for HRD-pancreatic cancer.6 However, the majority of alterations detected in PDAC are a heterogenous mix of lower frequency mutations. In light of this wide diversity, it becomes crucial to gain a deeper understanding of gene-drug interactions, in order to improve patient recruitment criteria and success rate for clinical trials for novel therapeutics.
Another obstacle to the discovery and clinical progression of new treatment options for pancreatic cancer is the limitation of the available experimental models. Genetically engineered mouse models and patient-derived xenografts have been very valuable for the study of the biology of pancreatic cancer, but they are not necessarily suitable for larger translational screening efforts. For this, in vitro models have proven more versatile and less time consuming. Traditionally, monolayer cell lines have been used to test therapeutic approaches. However, translation of these studies into clinical benefit has been extremely poor.7 Patient-derived organoids are 3D primary cultures that have the advantage to maintain the cell types and architecture of the organ of origin, and therefore constitute an attractive model for translational studies. In particular, it is not inconceivable that tumors can exert different dependencies, and therefore show different vulnerabilities, when cultured in tridimensional cultures, as opposed to monolayers. This has been shown already for MAPK dependency, which is a pathway virtually always altered in pancreatic cancer.8 Hence, there is a need to study PDAC in its tridimensional form.
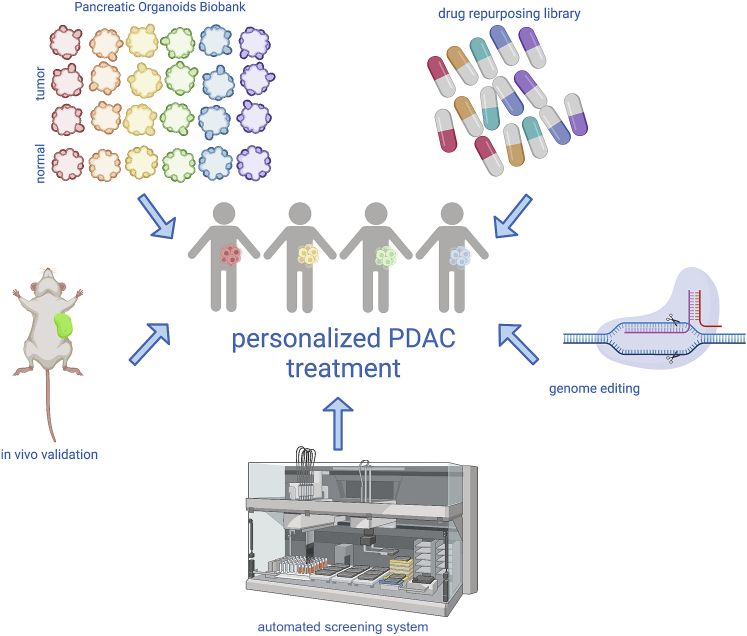
In this issue of Cell Genomics, Hirt et al. describe a newly derived pancreatic organoids biobank, comprising 31 tumor and 9 healthy 3D cell lines, and use it to investigate novel drug-gene interactions with potential translational value. Importantly for their objective, this organoid collection proved to recapitulate the frequency of driver genes mutations previously reported by sequencing studies in patients. Nevertheless, the question always remains whether organoids can predict drug response better as compared to traditional monolayer cultures. To this point, the authors show that the in vivo response of PDAC models to the EGFR inhibitor erlotinib is better recapitulated in the 3D in vitro cultures as compared to 2D cultures.
The authors also perform a remarkable drug screening effort in two of their organoids to search for compounds with an anti-tumor effect in 3D. High-throughput screenings in organoids have been notoriously difficult for reasons including the laborious handling of cells in Matrigel, the wide range of proliferation rates, the instability of the organoid composition that may affect drug response, and limitations in the readouts that can be applied to tridimensional cultures. Therefore, the effort described by Hirt and colleagues appears notable in scale, as for the first time 1,172 FDA-approved compounds were tested in multiple organoids. Through this strategy, the authors identify 22 drugs with anti-cancer effects that could be grouped into 8 common mechanisms of action, and they proceed to validate one compound per group in vivo. Remarkably, 6 out of the 8 compounds showed some anti-tumor activity in PDX models. Interestingly, at least two of the hits from the screening could not be validated in isogenic monolayer cultures, again emphasizing the need for similar screening platforms that address 3D-specific dependencies.
It is worth noting that none of the targets was more effective than the standard of care gemcitabine. This suggests that new target-discovery efforts are still needed for pancreatic cancer. Nevertheless, it is not uncommon that targeted therapies need to be administered in combination in order to achieve a significant anti-cancer effect.9,10 Thus, drug-repurposing efforts such as the one described in this paper could still hold value to uncover novel compounds for combinatorial use, as well as second-line treatments effective in pre-treated chemo-resistant specimens.
The search for novel drug-gene interactions in pancreatic cancer also led the authors to some unexpected observations. First, that BRCA2 mutations don’t confer increased sensitivity to PARP inhibitors in PDAC organoids. This is surprising giving that PARP inhibitors have been approved for BRCA mutant HDR pancreatic cancer6 and raises the question of whether organoid-specific culture conditions might still be too artificial and masking some of the actual tumor dependencies. It would be important for future studies to validate how those BRCA2 mutant organoids respond to PARP inhibitors when grown in mice under more physiological conditions. Second, the authors find that, surprisingly, ARID1A missense, but not null mutants, are specifically sensitive to ATR inhibitors. This observation has potential translational implications, in particular for the selection of patients that could benefit from ATR inhibitors treatment and should motivate further exploration of the molecular mechanisms underlying this finding. It is important to point out that creating a gene knockout in a cancer that never acquired the loss of that gene during its natural history may not yield the same phenotype as a cancer that mutated that gene during its development.11 Therefore, the results obtained with cells with engineered mutations must be interpreted with caution.
In conclusion, the paper by Hirt et al. highlights the value of improved technologies for 3D-culture-based, high-throughput drug screening. When applied to such compelling clinical needs as the search for new treatments for pancreatic cancer, and in combination with thorough in vivo validation (Figure 1), this technology has the potential to speed up the bridging between bench and bedside, for the benefit of PDAC patients.
Figure 1.
Graphical summary
Schematic overview of the techniques integrated by Hirt et al.,1 with the objective of improving personalized medicine for pancreatic cancer. Created with BioRender.com.
References
- 1.Hirt C.K., Booij T.H., Grob L., Simmler P., Toussaint N.C., Keller D., Taube D., Ludwig V., Goryachkin A., Pauli C., et al. Drug screening and genome editing in human pancreatic cancer organoids identifies drug-gene interactions and candidates for off-label treatment. Cell Genomics. 2022;2 doi: 10.1016/j.xgen.2022.100095. this issue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mizrahi J.D., Surana R., Valle J.W., Shroff R.T. Pancreatic cancer. Lancet. 2020;395:2008–2020. doi: 10.1016/S0140-6736(20)30974-0. [DOI] [PubMed] [Google Scholar]
- 3.Pishvaian M.J., Bender R.J., Halverson D., Rahib L., Hendifar A.E., Mikhail S., Chung V., Picozzi V.J., Sohal D., Blais E.M., et al. Molecular Profiling of Patients with Pancreatic Cancer: Initial Results from the Know Your Tumor Initiative. Clin. Cancer Res. 2018;24:5018–5027. doi: 10.1158/1078-0432.CCR-18-0531. [DOI] [PubMed] [Google Scholar]
- 4.Hong D.S., Fakih M.G., Strickler J.H., Desai J., Durm G.A., Shapiro G.I., Falchook G.S., Price T.J., Sacher A., Denlinger C.S., et al. KRASG12C Inhibition with Sotorasib in Advanced Solid Tumors. N. Engl. J. Med. 2020;383:1207–1217. doi: 10.1056/NEJMoa1917239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Witkiewicz A.K., McMillan E.A., Balaji U., Baek G., Lin W.C., Mansour J., Mollaee M., Wagner K.U., Koduru P., Yopp A., et al. Whole-exome sequencing of pancreatic cancer defines genetic diversity and therapeutic targets. Nat. Commun. 2015;6:6744. doi: 10.1038/ncomms7744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Golan T., Hammel P., Reni M., Van Cutsem E., Macarulla T., Hall M.J., Park J.O., Hochhauser D., Arnold D., Oh D.Y., et al. Maintenance Olaparib for Germline BRCA-Mutated Metastatic Pancreatic Cancer. N. Engl. J. Med. 2019;381:317–327. doi: 10.1056/NEJMoa1903387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ben-David U., Siranosian B., Ha G., Tang H., Oren Y., Hinohara K., Strathdee C.A., Dempster J., Lyons N.J., Burns R., et al. Genetic and transcriptional evolution alters cancer cell line drug response. Nature. 2018;560:325–330. doi: 10.1038/s41586-018-0409-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Riedl A., Schlederer M., Pudelko K., Stadler M., Walter S., Unterleuthner D., Unger C., Kramer N., Hengstschläger M., Kenner L., et al. Comparison of cancer cells in 2D vs 3D culture reveals differences in AKT-mTOR-S6K signaling and drug responses. J. Cell Sci. 2017;130:203–218. doi: 10.1242/jcs.188102. [DOI] [PubMed] [Google Scholar]
- 9.Kopetz S., Grothey A., Yaeger R., Van Cutsem E., Desai J., Yoshino T., Wasan H., Ciardiello F., Loupakis F., Hong Y.S., et al. Encorafenib, Binimetinib, and Cetuximab in BRAF V600E-Mutated Colorectal Cancer. N. Engl. J. Med. 2019;381:1632–1643. doi: 10.1056/NEJMoa1908075. [DOI] [PubMed] [Google Scholar]
- 10.Long G.V., Stroyakovskiy D., Gogas H., Levchenko E., de Braud F., Larkin J., Garbe C., Jouary T., Hauschild A., Grob J.J., et al. Combined BRAF and MEK inhibition versus BRAF inhibition alone in melanoma. N. Engl. J. Med. 2014;371:1877–1888. doi: 10.1056/NEJMoa1406037. [DOI] [PubMed] [Google Scholar]
- 11.Beijersbergen R.L., Wessels L.F.A., Bernards R. Synthetic Lethality in Cancer Therapeutics. Annual Review of Cancer Biology. 2017;1:141–161. [Google Scholar]