Abstract
The discovery of regulatory domains has been limited to the investigation of transcription factors and homologous protein sequences. In this issue of Cell Genomics, motivated by an oncogenic protein fusion, Tak et al.1 direct the regulatory potential of a nontraditional effector domain to novel genomic loci with fusions to programmable DNA-binding domains.
The discovery of regulatory domains has been limited to the investigation of transcription factors and homologous protein sequences. In this issue of Cell Genomics, motivated by an oncogenic protein fusion, Tak et al.1 direct the regulatory potential of a nontraditional effector domain to novel genomic loci with fusions to programmable DNA-binding domains.
Main text
A translocation product in Ewing sarcoma results in a fusion of the transactivating region of the EWS protein and the DNA-binding domain (DBD) of FLI1, forming a novel EWS-FLI1 protein.2 This oncogenic fusion delivers the activation potential of the EWS protein to thousands of GGAA microsatellites across the genome because of the acquired GGAA-binding specificity of the FLI1 ETS domain. These new binding events can convert GGAA repeats into de novo enhancers that contribute to a Ewing sarcoma regulatory program. In this issue of Cell Genomics, Tak et al.1 used this oncogenic fusion as the inspiration for a series of synthetic proteins that fuse the EWS truncation product to the programmable DNA-binding proteins dCas9 or Cys2His2 zinc fingers (ZFs), both designed to bind GGAA repeats. Interestingly, the ZFs outperform dCas9 in this context, activating the UGT3A2 gene, a known EWS-FLI1 target, with similar activation levels to the EWS-FLI1 protein, while dCas9 lead to a modest increase in expression. In addition, the EWS- ZF array (ZFA) fusion was able to approximate Ewing-FLI1 activity across the genome as a large fraction of genes in proximity to GGAA repeats are similarly regulated with both the EWS-FLI1 and EWS-ZFA proteins. This work provides a proof of concept that fragment or domain swapping with programmable DBDs will allow the close investigation of protein function across the genome and generate new tools for synthetic biology.
The use of the EWS truncation for targeted activation is an interesting example of how activation domains can be mined from proteins that we don’t traditionally think of as having activating potential. A first assumption of gene regulation is that proteins with regulatory potential likely contain a DBD with the obvious exceptions of common cofactors and mediator components. However, the EWS fusion with the natural FLI1 ETS DBD—and in this manuscript with programmable domains—demonstrates that unanticipated proteins may have regulatory potential if targeted to genomic loci. This is not only interesting from a protein function point of view but also for the discovery of effector domains to be employed as synthetic tools. This is especially useful for activation because a limited number of activation domains are available to the community. The most common domains are derivatives of the viral VP16 domain.3 Although these domains produce a range of activities, they are limited in their therapeutic application because of their viral origin and limited in the insight they can provide for human regulatory mechanisms. Fortunately, with the EWS protein described here and multiple recent publications that have screened for human activating and repressing domains,4,5 we are beginning to address the paucity of human effector domains for the synthetic biology community to utilize.
The most common programmable tools used for directed gene regulation are based on various versions of dCas9.6 Interestingly, the authors demonstrated that the dCas9-EWS fusions worked extremely well to activate numerous genomic targets despite the fact that dCas9 activity was poor in comparison to FLI1 or the ZFAs for binding GGAA repeats. There are several potential explanations for the modest activity. For example, the size of dCas9 might interfere with the recruitment of additional transcription factors, or nucleosome effects near the GGAA repeats might have a greater steric influence on dCas9 than a ZFA. Expression levels may also differ because of the extreme difference in size. Conversely, the ZFA’s were able to closely mimic the activity of the EWS-FLI1 protein as a EWS-ZFA fusion suggesting the swap of common DBDs might provide a better approximation of natural protein function. In addition, these same ZFs expressed as a KRAB domain fusion led to repression of microsatellite-associated genes and toxicity in EWS precursor cell lines. In this way, the authors have demonstrated the ability to use ZF-human protein fusions to closely probe the functional activity of human proteins and juxtapose the influence of alternative domains when binding the same target sequences (Figure 1).
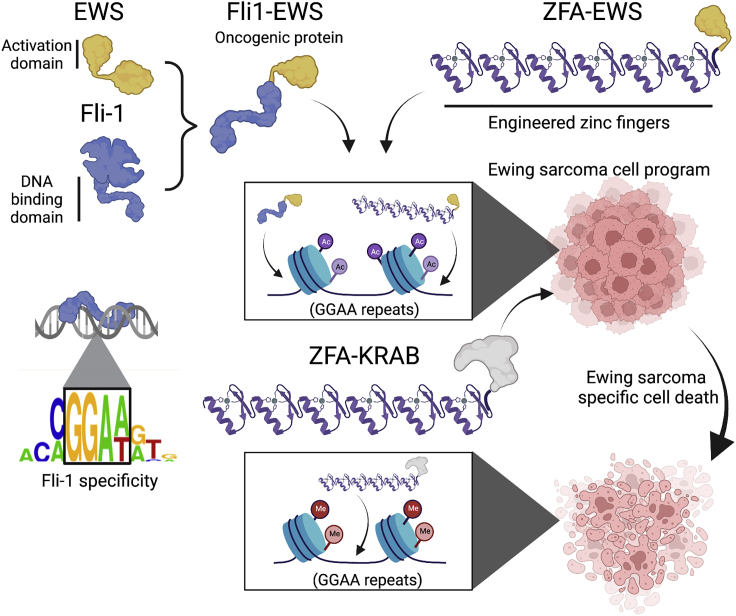
Figure 1.
Domain swapping for directed gene regulation
The oncogenic Fli1-EWS protein is the result of a translocation in Ewing sarcoma. This fusion attaches the regulatory poteintial of the EWS protein to the Fli1 DNA-binding domain that binds GGAA-like sequences. As a result, the Fli1-EWS protein binds GGAA microsatellites across the genome. Inspired by this oncogenic fusion, Tak et al. demonstrate that ZFAs engineered to bind GGAA repeats can mimic the Fli1-EWS activation program. In addition, the same ZFAs are toxic in Ewing sarcoma cell lines when fused to a KRAB domain, suggesting that repression of the same regulatory program can counter the oncogenic program.
This successful application of the ZFAs may suggest a unique and powerful advantage of using designed ZFAs to probe transcription factor function, but ZF engineering has been quite challenging for the field. This is not due to any failing of the domain itself, as multiple examples have shown that ZFs are able to interact with any sequence of DNA.7,8 The challenge is understanding array assembly. Adjacent ZFs within an array can influence one another as their contacts with the DNA are made in close proximity and, in some cases, the target sequences’ overlap.9 Therefore, understanding ZF compatibility is as important as understanding specificity. As a result, the field has been left with a small set of resources that are mostly limited to binding G-rich targets because of their high affinity that will likely overcome any incompatibility between adjacent fingers in the array.10 Nevertheless, investigations such as those detailed by Tak et al.1 will hopefully provide the motivation for a renewed interest in understanding this common but complex domain.
Finally, the use of ZFs and human effector domains have interesting therapeutic potential. As most disease-associated SNPs are found in the non-coding regions of the genome, the misregulation of genes appears to be a common mechanism of disease. Tools that allow us to correct misregulation could be applied across a wide range of disease targets. However, if the transgene is expressed for long periods of time within a patient, this could produce a harmful immune response. This is particularly relevant for activation as continuous expression of the activating protein is likely necessary, suggesting that TAL domains or dCas9 could be problematic because of their prokaryotic origin. ZFs, on the other hand, represent the most common DBD in human, utilized by roughly 50% of our transcription factors. Therefore, deployment of artificial proteins that use a human activating sequence with a modified human ZF scaffold, as described by Tak et al., significantly reduces the chance of an immune response. Advancement of ZF design and engineering platforms would enable a therapeutic regime that will prove challenging for the commonly used prokaryotic platforms.
Acknowledgments
Declaration of interests
Dr. Noyes is a cofounder of Navega Therapeutics. He is also the inventor on multiple zinc finger engineering patents.
References
- 1.Tak E., Boulay G., Lee L., Iyer S., Perry N., Schultz H., Garcia S., Broye L., Horng J., Rengarajan S., et al. Genome-wide functional perturbation of human microsatellite repeats using engineered zinc finger transcription factors. Cell Genomics. 2022;2:100119-1–100119-10. doi: 10.1016/j.xgen.2022.100119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Delattre O., Zucman J., Plougastel B., Desmaze C., Melot T., Peter M., Kovar H., Joubert I., de Jong P., Rouleau G., et al. Gene fusion with an ETS DNA-binding domain caused by chromosome translocation in human tumours. Nature. 1992;359:162–165. doi: 10.1038/359162a0. [DOI] [PubMed] [Google Scholar]
- 3.Sadowski I., Ma J., Triezenberg S., Ptashne M. GAL4-VP16 is an unusually potent transcriptional activator. Nature. 1988;335:563–564. doi: 10.1038/335563a0. [DOI] [PubMed] [Google Scholar]
- 4.Alerasool N., Leng H., Lin Z.Y., Gingras A.C., Taipale M. Identification and functional characterization of transcriptional activators in human cells. Mol. Cell. 2022;82:677–695.e7. doi: 10.1016/j.molcel.2021.12.008. [DOI] [PubMed] [Google Scholar]
- 5.Tycko J., DelRosso N., Hess G.T., Aradhana, Banerjee A., Mukund A., Van M.V., Ego B.K., Yao D., Spees K., et al. High-throughput discovery and characterization of human transcriptional effectors. Cell. 2020;183:2020–2035.e16. doi: 10.1016/j.cell.2020.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gilbert L., Horlbeck M., Adamson B., Villalta J., Chen Y., Whitehead E., Guimaraes C., Panning B., Ploegh H., Bassik M., et al. Genome-scale CRISPR-Mediated Control of gene repression and activation. Cell. 2014;159:647–661. doi: 10.1016/j.cell.2014.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Persikov A.V., Wetzel J.L., Rowland E.F., Oakes B.L., Xu D.J., Singh M., Noyes M.B. A systematic survey of the Cys2His2 zinc finger DNA-binding landscape. Nucleic Acids Res. 2015;43:1965–1984. doi: 10.1093/nar/gku1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Najafabadi H.S., Mnaimneh S., Schmitges F.W., Garton M., Lam K.N., Yang A., Albu M., Weirauch M.T., Radovani E., Kim P.M., et al. C2H2 zinc finger proteins greatly expand the human regulatory lexicon. Nat. Biotechnol. 2015;33:555–562. doi: 10.1038/nbt.3128. [DOI] [PubMed] [Google Scholar]
- 9.Wolfe S.A., Nekludova L., Pabo C.O. DNA recognition by Cys2His2 zinc finger proteins. Annu. Rev. Biophys. Biomol. Struct. 2000;29:183–212. doi: 10.1146/annurev.biophys.29.1.183. [DOI] [PubMed] [Google Scholar]
- 10.Sander J.D., Dahlborg E.J., Goodwin M.J., Cade L., Zhang F., Cifuentes D., Curtin S.J., Blackburn J.S., Thibodeau-Beganny S., Qi Y., et al. Selection-free zinc-finger-nuclease engineering by context-dependent assembly (CoDA) Nat. Methods. 2011;8:67–69. doi: 10.1038/nmeth.1542. [DOI] [PMC free article] [PubMed] [Google Scholar]