Summary
The current paradigm for data use oversight of biomedical datasets is onerous, extending the timescale and resources needed to obtain access for secondary analyses, thus hindering scientific discovery. For a researcher to utilize a controlled-access dataset, a data access committee must review her research plans to determine whether they are consistent with the data use limitations (DULs) specified by the informed consent form. The newly created GA4GH data use ontology (DUO) holds the potential to streamline this process by making data use oversight computable. Here, we describe an open-source software platform, the Data Use Oversight System (DUOS), that connects with DUO terminology to enable automated data use oversight. We analyze dbGaP data acquired since 2006, finding an exponential increase in data access requests, which will not be sustainable with current manual oversight review. We perform an empirical evaluation of DUOS and DUO on selected datasets from the Broad Institute’s data repository. We were able to structure 118/123 of the evaluated DULs (96%) and 52/52 (100%) of research proposals using DUO terminology, and we find that DUOS’ automated data access adjudication in all cases agreed with the DAC manual review. This first empirical evaluation of the feasibility of automated data use oversight demonstrates comparable accuracy to human-based data access oversight in real-world data governance.
Keywords: Data Access Committee, Data Use, GA4GH, Passport
Graphical abstract
Highlights
Genomic data sharing is overly complex, resulting in significant delays that slow science
DUO is a new standardized vocabulary that structures data use restrictions
The DUOS platform uses DUO to expedite data access requests for researchers and reviewers
We provide empirical evidence that data governance can be automated in most cases
The GA4GH Data Use Ontology (DUO) is a structured approach to describing the secondary uses of data. Cabili et al. report the Data Use Oversight System (DUOS), an open-source software platform that leverages DUO and other data-sharing policy advancements as a first step toward automating data governance. They provide empirical evidence that DUOS performs comparable to a human data access committee in adjudication of data access requests, streamlining the process of gaining access to human biomedical data.
Introduction
The life sciences are in the midst of a data revolution. Inexpensive and accurate genome sequencing is a reality, advanced imaging is routine, and clinical data are increasingly stored in an electronic form.1 In principle, these advances have brought us to the threshold of a new era in medicine, one where the data sciences can propel our understanding and treatment of human diseases.
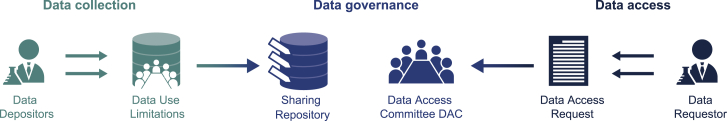
In practice, however, the analyses of these shared datasets are stymied by the operational challenges of managing access.2 The current prevailing data access framework for biomedical datasets shared in public repositories is schematized in Figure 1. (1) Researchers running studies collect data from research participants and deposit them in data repositories. Appropriate secondary uses of the data, as specified in the informed consent form, are listed as data use limitations (DULs) (e.g., “this dataset is available only for non-commercial breast cancer research”). (2) Researchers seeking to use this dataset for secondary research describe their research purpose via a data access request (DAR). (3) A data access committee (DAC) then determines whether the research purpose specified in the DAR is within the bounds of the DUL. This paradigm has been adopted by large-scale repositories for genomics and biomedical data around the world, including the Database of Genotypes and Phenotypes (dbGaP)3,4 in the United States and the European Genome-Phenome Archive (EGA)5 in Europe. Together, these are the two main public genomic data repositories in the world, jointly managing access to more than 4,000 datasets in total.
Figure 1.
Current workflow for data use oversight
In the most common current model for biomedical data sharing and data governance, researchers running studies involving human research participants establish data use limitations (DULs) and include them in the informed consent form prior to sample collection and data generation; the DULs specify how the data may be used going forward. The researchers then deposit the generated data in a data-sharing repository, which has an established data access committee (DAC) to govern what data other researchers may access. Researchers seeking to use the data for secondary research must submit a data access request (DAR) that describes their research purpose. The DAC then assesses the DUL and the DAR to see if the proposed research is within the bounds of the DULs.
However, this framework is not scalable, as both the number of datasets and the number of researchers seeking access to them are growing rapidly. It is becoming challenging for even the most efficient DACs to scale their review rate with the number of DARs, and long delays (e.g., multiple weeks) in processing data access requests are becoming increasingly common, thus slowing research. This calls for a fundamental rethinking of our overall paradigm for data use oversight, as both our team and colleagues in ELIXIR6 have called for.
The Global Alliance for Genomics and Health (GA4GH)1,7 was founded to address many of the challenges hindering rapid developments in genomics and health research. Through its multiple work streams, such as the Data Use and Researcher Identities (DURI), GA4GH identifies areas for standardization and mints community-led standards in response. Under the DURI work stream, standards such as the Data Use Ontology (DUO),8 GA4GH Passports,9 and Machine Readable Consent Guidance10 have been approved in an effort to facilitate data access and sharing.
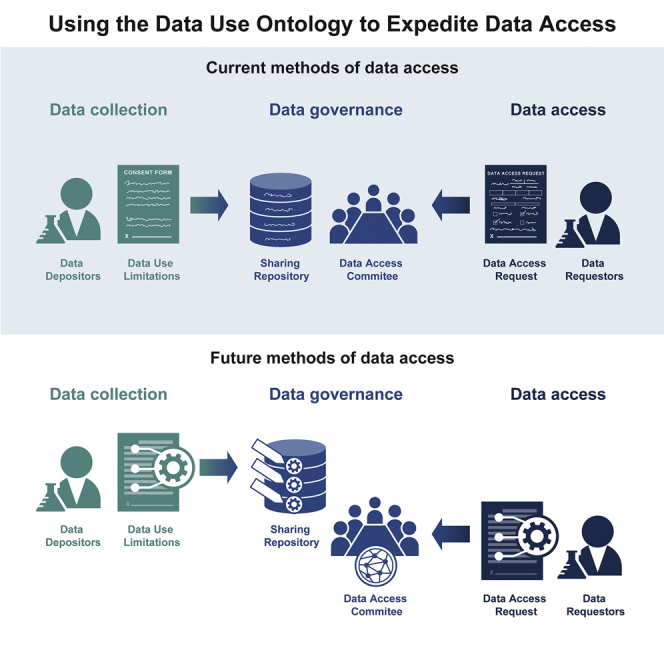
In this study, we evaluate the feasibility of more automated approaches to data use oversight in order to streamline managing access to biomedical datasets. Leveraging the new GA4GH DUO standard (see Lawson et al.8 in this issue), we have created a new open-source software platform called the Data Use Oversight System (DUOS) that streamlines many of the steps in data governance. We first analyze dbGaP datasets, finding an exponential increase in DARs, which will not be sustainable with current manual oversight review. We then perform an empirical evaluation of DUO on selected datasets from the Broad Institute’s data repository, including a direct comparison of human-based manual review by a DAC and our DUOS-enabled automated approach to data use oversight. We demonstrate that automated approaches are feasible and effective for real-world data use oversight and have comparable accuracy to human-based data access oversight. To our knowledge, this is the first empirical evaluation of the feasibility of automated data use oversight. We are extending this pilot study with additional testing of implementation across other programs and biomedical datasets.
Design
The current paradigm for data use oversight has three shortcomings. (1) It is not scalable—both the number of datasets and the number of researchers seeking to access them are growing, suggesting an exponential growth in DARs. (2) It is inconsistent—different DACs may reach different conclusions regarding appropriate secondary data uses. For example, it is not uncommon for a DUL to state that a dataset may only be reused for “diabetes and related conditions”; different DACs may have different interpretations of what constitutes a “related condition,” leading to inconsistent interpretation of access requests. (3) It is cumbersome—answering even straightforward questions such as “What samples can be used as controls for my study of autism?” require substantial human effort rather than a simple computational query.
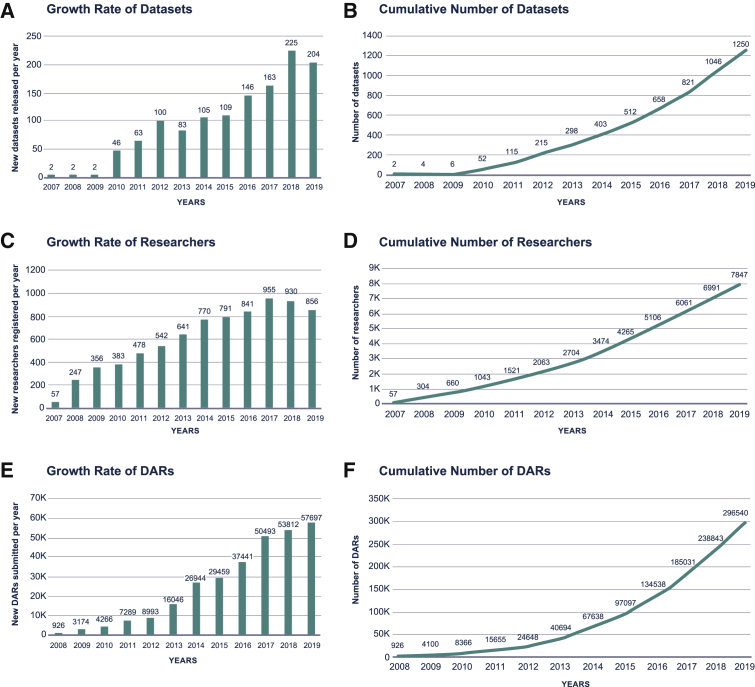
We first sought to better understand the landscape of current approaches to data use oversight by collecting quantitative data from an existing data-sharing repository, dbGaP, the National Institutes of Health’s (NIH) repository for archiving and sharing data from genotypic and phenotypic studies. Because the majority of NIH-sponsored genome-phenome datasets are deposited in dbGaP (with more than 1,200 datasets to date, Figure 2A), this repository provides deep and diverse data on the type of data use restrictions that are applied on genomics datasets as well as the type of data uses described in DARs. Here, we partnered closely with members of the dbGaP team to compile historic usage. We observed that the rate of growth in the number of datasets deposited in dbGaP was increasing (Figure 2A), implying exponential growth in the total number of datasets (Figure 2B). Similarly, we observed an increasing rate of growth in both the number of researchers seeking to access these datasets (Figures 2C and 2D) and the number of DARs (Figures 2E and 2F) they submitted. This results in exponential growth in the cumulative number of DARs. With the growing volumes of both datasets and researchers seeking to access dbGaP, we expect that the current processes for data access will not be able to scale to meet the volume of access requests projected in the next few years without significant investment in human resources to cover the exponential increase in tasks.
Figure 2.
Growth in datasets and data access
(A and B) The number of datasets deposited in dbGaP (y axis) each year (x axis) demonstrates an increase in deposit rate (A), leading to an exponential growth in total number of datasets (B) (see STAR Methods, quantification and statistical analysis).
(C and D) The number of new researchers that become new users on dbGaP (y axis) each year (x axis) demonstrates an annual increase in dbGaP users (C), leading to exponential growth in the number of researchers (D) (see STAR Methods, quantification and statistical analysis).
(E and F) The number of DARs that are being submitted to dbGaP (y axis) every year (x axis) is increasing (E), leading to an exponential growth in the number of cumulative DARs (F). Note that, in (E) and (F), we include both new DARs and those submitted for renewal as these are all being reviewed following the same process (see STAR Methods, quantification and statistical analysis).
Given the above scalability challenges, we considered the possibility that these limitations could be addressed through automation.
Results
We took a three-part strategy to streamline the process of data use oversight. (1) The Data Use and Researcher Identity (DURI) Work Stream of the Global Alliance for Genomics and Health (GA4GH)7,8 developed an ontology to structure DULs (many authors of this paper were involved in that effort). This was aided by the prior efforts of dbGaP, as well as others in the GA4GH community.11,12 The resulting DUO is now a GA4GH standard.8 (2) We created a suite of interfaces that enable researchers to construct DARs that articulate their research purpose using this same ontology. (3) We developed an inference engine for automated checking of the compatibility between DULs and DARs expressed via DUO (e.g., a DAR for “melanoma” is compatible with a DUL for “cancer,” but not vice versa); this is described in the STAR Methods. This framework thus mimics the current framework for data use oversight but has the potential to streamline it by making many steps computable.
We developed a software package, DUOS, that encapsulates these three functionalities. Moreover, knowing that there will always be datasets whose DULs cannot be structured, DUOS also provides user interfaces to support the workflows of traditional DACs. The DUOS user interfaces substantially facilitate the work of a DAC, even in the non-automated setting (which is the vast majority of cases today), by: (1) clearly presenting the DULs and DARs to the DAC and simplifying how they track progress by compiling the number of DAC members that have voted and the results of their votes; (2) sending notices to DAC members when new DARs come in or when they are delayed in voting; (3) demonstrating to DAC members whether researchers have been approved by their institution to submit DARs; (4) allowing researchers to apply for access to many datasets from multiple DACs through a single DAR application form, thus lessening the number of DARs that need to be submitted; and (5) providing an application programming interface (API) that enumerates the “white-listed” researchers that have been approved to access various datasets, removing the need for communication between the DAC and the growing number of data repositories hosting these datasets. Support for DACs is an enormous unmet need that we hope systems like DUOS and related systems like REMS, a Resource Entitlement Management System developed by ELIXIR Finland, can fill.6
We sought to provide empirical support for the hypothesis that DUOS could serve as an automated alternative to traditional DACs (Figures 3A–3C). We chose to quantitatively validate three aspects of DUOS: (1) the fraction of DULs that could be structured with DUO, (2) the fraction of DARs that could be expressed with DUO, and (3) the concordance between DUOS and the adjudications of a traditional DAC. To do this rigorously, we convened a DAC comprised of individuals with expertise in data use oversight. This DAC both served as the source of truth for validation point 3 (concordance of DUOS with a DAC) and adjudicated whether DULs and DARs could be structured correctly (points 1 and 2). In all cases, the DAC was blinded to the results of DUOS (and to the adjudications of other DAC members) so that decisions would be unbiased.
Figure 3.
Results of the automated data use oversight trial
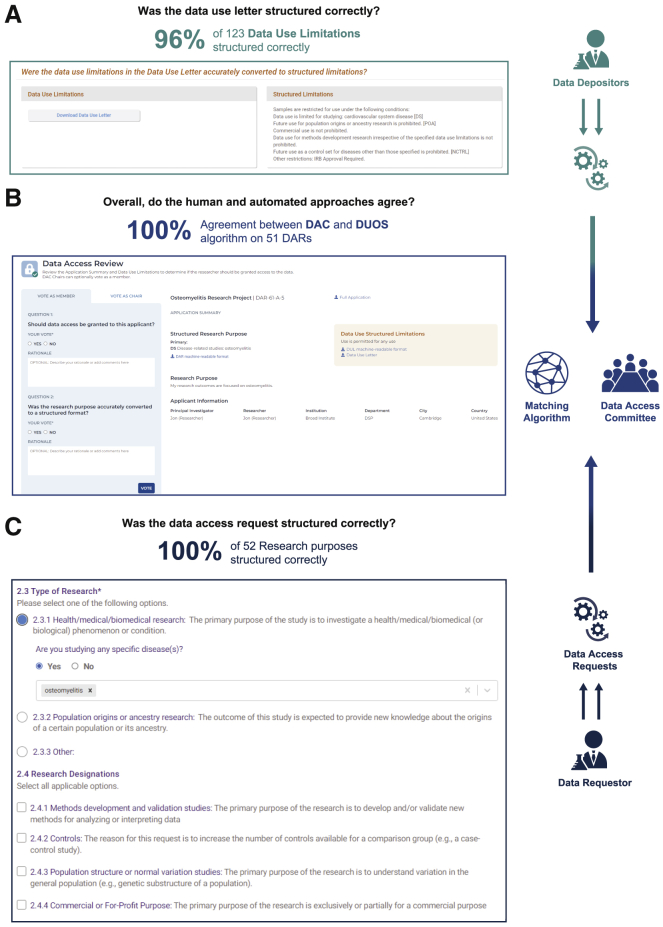
An illustration summarizing the results of the DUOS trial and the interfaces used to collect data in each phase of the trial.
(A) A screenshot of the DUOS interface used by the DAC to evaluate whether the data use restriction of a specific dataset can be structured using DUO. The trial found that 96% of the datasets DULs could be structured with DUO.
(B) A screenshot where DAC members evaluated the overall percentage of DUL-DAR pairs for which the DUOS automated approach and the DAC agreed and found it to be 100% (n = 51).
(C) A screenshot of the DUOS interface used by researchers to log a DUR, which is, in turn, structured in the backend into DUO codes. The DUOS trial found that 100% (n = 52) of DARs that were included in the trial could be structured.
The results of this empirical validation of DUOS are shown in Figure 3D. We observed that out of 123 evaluated DULs, 118 (96%) of them could be structured using DUO. The five that could not be structured are listed in Table S1; in each case, the data use limitations fell outside of our current DUO. Similarly, we observed that 100% of research proposals (52 out of 52) could be correctly structured using the DUO. Finally, we note that, in all cases (51 out of 51) that were reviewed by the DAC and DUOS, the DAC and DUOS were in agreement (1 of the 52 DARs was classified by DUOS for manual review and thus the automated process was not applied).
Discussion
In this study, we have empirically evaluated the feasibility of using the new DUO standard in real-world data governance. To the best of our knowledge, this is the first such trial of automated data use oversight. In it, we provide initial evidence that (1) DULs can be structured using the DUO standard, (2) DARs can be structured using this same ontology, and (3) the work of a DAC to align data access requests with data use limitations can, in most cases, be done using a more automated approach. Moreover, we describe the creation of DUOS, a production-grade software platform that automates data use oversight and simplifies the DAC workflow even when traditional non-automated approaches are used by structuring both research proposals and permitted data uses using DUO terminology.
Certainly, there will be DULs that cannot be structured using the DUO standard; for example, in this study, we observed that 5 of 123 DULs could not be cleanly mapped onto the DUO (these 5 DULs are listed in Table S1). In each of these cases, we recognized during our manual review, before conducting these analyses, that it would not be possible to map these more complex consent clauses to DUO terms. For example, one of the DULs stated that “general research use of aggregate level is prohibited,” and this specific terminology has not been included in DUO. For the datasets for which DUOS is not able to map DULs onto DUO, researchers would identify this upon registering datasets in DUOS and would alternatively be able to describe their datasets’ DULs using free text. Such cases would necessitate that subsequent access requests follow the current protocol of human-based DAC review but would not be more onerous than the current state. The GA4GH DURI workstream understands that not all consent forms will map to DUO as described above yet aspires to have principal investigators (PIs) and institutional review boards (IRBs) that seek to enable data sharing leverage existing DUO terms or provide suggestions for new terms that numerous community members support adopting into the DUO standard. Indeed, DUO has established a process to allow community members to update the ontology (see Lawson et al.8 for details).
Similarly, many datasets have DULs that are somewhat ambiguous (e.g., “this dataset is only available for research into diabetes and related conditions”). In these situations, we interpreted the DUL in a more conservative way (i.e.., “this dataset is only available for research into diabetes”), on the grounds that we consider it better to allow for potentially reducing access to the dataset rather than to violate the terms specified within the informed consent form. In addition, the researcher will be informed by email whether their request was declined for not meeting the DULs, and if they feel their research is compliant with the data use restrictions specified by the informed consent form, they can apply directly to the DAC with an explanation as to why the DUO term is overly conservative. Thus, in the vast majority of cases, automated data use oversight would appear to be feasible, and in the rare cases where it is not, we can default to the current human-based DAC review processes with confidence in the same level of compliancy or effectiveness.
Based on this pilot study, we are now initiating a larger-scale effort to evaluate DUOS as an automated approach to data use oversight in a real-world setting. Specifically, we have engaged seven institutes and centers within NIH that collectively utilize four DACs: the National Human Genome Research Institute (NHGRI), National Heart Lung and Blood Institute (NHLBI), National Institute for Allergy and Infectious Diseases (NIAID), and the Joint Addiction Aging and Mental Health (JAAMH) DAC, which represents the National Institute on Drug Abuse (NIDA), National Institute on Alcohol Abuse and Alcoholism, National Institute on Aging, and National Institute of Mental Health, to conduct a trial of DUOS as a system of automated data use oversight. The first phase of this pilot commenced in June 2019 with test datasets, DULs, and access request content with a single DAC. Since then, the pilots have concentrically expanded to a second phase that includes testing of legitimate real-time DARs for actual NIH datasets and DULs, conducted by the growing number of DACs described above. The second phase of the pilot is ongoing and already approaching scale (more than 600 DULs mapped to DUO and more than 65 DARs processed in the first round of testing). We expect this second phase to be carried out over the next 2 years, with testing expanding to additional NIH DACs, datasets, and data types.
Limitations of the study
While this study is the first to demonstrate the feasibility of automating data governance, this was an initial pilot study intended to validate the use of the GA4GH DUO to describe real DULs and the ability to compare the decisions of a DUO-backed algorithm with human manual review, although many practical obstacles remain before the vision of routine automated data governance is a reality.
First, there is the task of retrospectively coding the many existing DULs to the DUO standard. Here, we are encouraged by our initial experiences with the large-scale NIH trial mentioned above. There, a single individual knowledgeable in the DUO standard was able to codify more than 600 NIH datasets in a matter of weeks, working far from full time. It is our experience that DACs are often highly motivated to code their DULs, as it can result in many fewer requests for access to data where the research purpose is obviously at odds with the DUL (certainly, this is true in the dbGaP experience), lessening unproductive work of the DAC. Moreover, the DAC at our institution now requests data depositors to send the DUL in a template form that asks simple questions like “Are there restrictions on commercial use?” and “Are there any disease-specific restrictions?” that correspond to DUO fields. These are easy for an investigator or signing official to fill out and greatly simplifies the task of mapping the DUL to DUO. We believe that it is feasible for other DACs to start adopting a similar approach to lightweight formatting of the DULs they receive.
Second, there is the task of changing the language used in drafting informed consent forms. Here, researchers will be greatly aided by the GA4GH Machine Readable Consent Guidance,10 which ensures that consent forms include data use language in verbatim DUO terms and definitions through the template language provided by the Machine Readable Consent Guidance. A significant task will be encouraging uptake of this guidance by IRBs and researchers developing consent forms. Achieving this will require extensive outreach to raise awareness of the empirical evidence demonstrating the efficacy and benefit of DUO for preserving research participant protections and its validated capacity to promote responsible data use. We hope that this work represents a first step toward providing such an evidence base.
Moreover, our group and others within the DURI workstream of GA4GH are beginning to address two other barriers to automated data use oversight. First, there is the challenge of “identity proofing”—that is, verifying that a data requester is both a bona fide researcher and that they are giving their true identity rather than a pseudonym.13 Related to this is the task of validating that they have an institution legally standing behind them in the event of misuse. The GA4GH DURI work stream has recently approved the GA4GH Passport12 that addresses these challenges (see Voisin et al.9 in this issue). Here, the Passport is a new GA4GH standard to encode machine-readable access permissions that have been approved by DACs. After proofing identity, organizational affiliation, and bona fide researcher status, the GA4GH Passport allows these to be embedded in encrypted strings (a.k.a., “JSON Web Tokens” or JWTs) that are then consumed by web services to allow researchers to access various datasets. The net result is to greatly streamline the communication of the access rights of the user within appropriate data systems.
Lastly, there is the challenge of institutional signoff, such that a signing official at every institution must review every DAR rather than giving a blanket approval to researchers. Our experience is that signing officials are overwhelmed by DARs and generally do not review them in detail. By instead moving to a model where bona fide researchers are pre-approved by their signing official to submit DARs (which we define as providing the researchers with a “Library Card”), this further removes a manual impediment to data access. This pre-approval can be implemented in practice by a digital object similar to a GA4GH Passport that represents the “Library Card” and captures this pre-authorization.13 Our group is initiating pilots with the NIH and in concert with the GA4GH DURI workstream on the feasibility of this approach. Moreover, we have established a DAC at our own institution for non-NIH datasets that we govern, and this uses such a “Library Card” approach in which the requesting institutions’ signing officials pre-authorize their researchers to submit data access requests to our DAC (15 institutions requesting access to data are already using this process for more than 50 researchers).
Future directions
Our hope is that, through a combination of both technological innovation and empirical validation of these innovations, we will be able to demonstrate the feasibility of truly automated approaches to data use oversight for all types of biomedical research data. We envision a world where researchers log in to data repositories, such as dbGaP, with a digital Library Card that was digitally pre-signed by a signing official and that allows them to perform DARs. The researcher will then describe their research purpose using the DUO standard using a DUOS-like system that, in turn, will automatically grant access to the majority of datasets where the codified data use restrictions are consistent with the stated research purpose.
If these goals are achieved, it will enable a world where qualified researchers, who are performing investigations that are consistent with the terms of the informed consent, have instantaneous access to biomedical data. Given that the process of gaining access to data can currently take months, such a change would accelerate biomedical and health research tremendously. This trial represents a first step toward that goal.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Deposited data | ||
| dbGaP use statistics | NCBI | https://duos.broadinstitute.org/dataset_catalog; Dataset ID: DUOS-000137 |
| Software and algorithms | ||
| Data Use Ontology | Lawson et al.8 | https://github.com/EBISPOT/DUO |
| Data Use Oversight System | This manuscript | https://github.com/DataBiosphere/duos-ui; https://duos.broadinstitute.org/home |
Resource availability
Lead contact
Further information and requests for resources should be directed to and will be fulfilled by the lead contact, Anthony Philippakis (aphilipp@broadinstitute.org).
Materials availability
This study did not generate new unique reagents.
Method details
The Data Use Ontology Matching Algorithm
DUOS uses DUO terms to structure both DULs and DARs and automate pattern matching between them, checking if the DUO terms associated with a DAR are a subset of those associated with the DUL. This information is presented to Data Access Committees so that they can make an informed decision about whether to approve access to a Dataset from a Researcher’s DAR.
The DUO defines terms for secondary data use, such as general research use, commercial use, populations research (ethnicities and gender), methods development, aggregate statistics as well as disease-specific research which relies on terms imported from the Human Disease Ontology14,15. DUO provides a cross-product of sub-ontologies, one for each of the above fields. The details of DUO are described in the companion manuscript.9
To determine if a researcher’s DAR should be approved for a dataset, DUOS uses business rules that evaluate if the researcher’s stated purpose is within the bounds of the datasets’ DULs. More technically, DUOS checks to see if the purpose of the DAR is a hierarchical descendant of the DULs (for example, a purpose of ‘Cancer-specific research’ for a dataset with ‘General Research Use’ DULs would be approved as ‘Cancer-specific research’ would be lower on the ontological hierarchy), and checks to see if other boolean operators are triggered (described further in Figure S3 and Methods S1). In order to lower latency, the ontologies are indexed and pre-calculated for each ontology term. Further details on the algorithm are described in Methods S1.
As noted in the text, the quantitative validation of the Data Use Ontology Matching Algorithm by the DAC in DUOS established (1) the fraction of DULs that could be structured with our ontology, (2) the fraction of DARs that could be expressed with this ontology, and (3) the concordance between DUOS and the adjudications of a traditional DAC. The dataset for this exercise consists of the 123 datasets in the Broad Institutes’ data repository for which both consent forms and proposed codified versions of those consent forms’s DULs were provided by each datasets’ IRB of record. The DARs and their research proposals were sent by researchers requesting access based on their research needs to the same selection of datasets starting in March 2018 through January 2020.
For point 1, the DAC received a PDF copy of either the original consent form or the IRB’s approved DULs represented in the consent form juxtaposed with a proposed codified version of those DULs in DUO terms. The DAC voted individually to confirm if they believed this was an acceptable translation or not. If not, further codified iterations would be passed to the DAC until either agreement was reached or it was determined that the DULs were unable to be codified in DUO terms. See Figure S1 and Table S2.
For point 2, in the DAR review page the DAC was presented with the researcher’s narrative description of their research purpose, as well as a codified version of the research purpose in DUO terms - self-selected by the researcher. The DAC then voted on if the researcher’s codifying of the narrative research purpose in DUO terms was accurate or not. See Figure S2.
For point 3, the DAC was presented with the inputs mentioned above: the codified DULs in DUO terms, the narrative research purpose, and the research purpose in DUO terms. With these inputs, the DAC was then asked to vote on whether or not the research purpose was within the bounds of the DULs. Simultaneously, the Data Use Ontology Matching Algorithm evaluated the same question using the research purpose and DULs in DUO terms and saved it’s result to the system alongside the DAC’s final vote. See Table S3.
Quantification and statistical analysis
We worked with NIH/NCBI’s dbGaP team to review and evaluate the system’s statistics with a primary focus on the 1) number of datasets registered, 2) number of researchers, and 3) numbers of DARs submitted over time. Our quantification of each of these categories looked primarily at unique new data for each category on a yearly basis, as well as a cumulative quantification demonstrated with quarter-based data points.
In Figure 2 we display quantitative results of an analysis of datasets, researchers, and data access requests in dbGaP since its inception in 2006 through 2019. The left column with graphs A, C, and E show the unique new numbers of each in the dbGaP per year throughout this time period. The right most images display the cumulative count of these additions over time for each of the three categories. There are a few important caveats on this data:
First, the datasets count is a cumulative total of unique datasets released by NIH in dbGaP, and does not include re-releases and/or updated versions of the datasets which would increase the total number of datasets from 1,250 to 1,745.
Second, the researcher figures display counts of the individual PIs submitting DARs and do not include counts of any internal collaborator PIs or lab staff who are often included in the research and access of data, and denoted in DAC applications. Thus the actual number of researchers accessing data is conservative.
The DAR counts factor in annual renewals/extensions of the DAR which require re-review and approval by the DAC under the same process as the initial review, and if not completed, data access for the researcher would be shut off.
Acknowledgments
The authors were generously funded by awards from EMBL-EBI Core Funds, Wellcome Trust GA4GH award number 201535/Z/16/Z, CINECA project (H2020 No 825775), and the “The AnVIL Data Ecosystem,” NHGRI- U24HG010262. The authors would like to acknowledge contribution from the developers of the Data Use Oversight System (DUOS), notably David An, Andrea Haessly, Justin Thomas, Linda Zhang, Richard Fricke, Rae Johanek, Diego Gil, Veronica Vicario, Nadya Lopez Zalba, Leo Forconesi, Tadeo Riveros, Walter Lo Forte, and Marianie Simeon. Thanks to Mike Feolo and colleagues at NIH/NCBI dbGaP for providing reference data for this publication. Thanks to Yvonne Blanco for developing the graphics and design for this manuscript and to Lou Strano and Nancy Lao for their work on the DUOS user interface. We are also grateful for the ongoing collaborations and support from the NHGRI AnVIL Data Access Working Group, NIH Office of Science Policy, NIH Genomic Data Submission and Management Taskforce, the GA4GH Data Use & Researcher Identities Workstream, and GA4GH leadership. The views expressed by L.L.R. are her own and not necessarily representative of those of her organization.
Author contributions
Conceptualization, M.N.C., J.L., A.S., T.N., M.C., S.D., and A.A.P.; methodology, M.N.C., J.L., A.S., G.R., T.N., M.C., S.D., and A.A.P.; formal analysis, M.N.C. and J.L.; investigation, M.N.C., J.L., A.S., P.O., J.W., L.L.R., S.D., and A.A.P.; data curation, M.N.C. and J.L.; validation, P.O., J.W., L.L.R., S.D., and A.A.P.; writing, M.N.C., J.L., T.N., M.C., S.D., and A.A.P.; project administration, A.S.; software, G.R.; supervision, S.D. and A.A.P.; funding acquisition, A.A.P.
Declaration of interests
A.A.P. is a venture partner at GV, a venture capital group within Alphabet. In that capacity, he makes investments in companies in both the life sciences and data sciences. He has received funding from Alphabet, Microsoft, Intel, IBM, Rakuten, Bayer, Novartis, and Pfizer. M.N.C. is an employee and equity holder of Foundation Medicine. J.W. is an employee and equity holder of Biogen.
Published: November 10, 2021
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.xgen.2021.100031.
Contributor Information
Stacey Donnelly, Email: sdonnell@broadinstitute.org.
Anthony A. Philippakis, Email: aphilipp@broadinstitute.org.
Supplemental information
Data and code availability
The code for DUOS is distributed under the three-clause BSD open source license, which allows for redistribution and use in source and forms, with or without modifications under certain conditions, and is in the “DataBiosphere” GitHub repositories located here:
- •
- •
- •
- •
The “DUOS-UI” is the front end user interface for the application. “Consent” is the application component where all user, data access requests, and dataset access decisions are stored. “Consent Ontology” is the application component that can make ontologically valid decisions about whether a dataset and a data access request are compatible using the GA4GH (DUO) and Human Disease Ontology (DOID) ontologies. “Consent Data Use” captures all the ontologies we referenced in the article for efficient access by the DUOS service.
In addition, we have made DUOS available as a publicly accessible web-service for registering, requesting, and reviewing access requests for datasets at https://duos.broadinstitute.org. Individuals and organizations unable to initiate and maintain the DUOS code are welcome to inquire with the corresponding author on how to use DUOS as a web service.
Data reported in this paper will be shared by the lead contact upon request.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- 1.Rehm H.L., Page A.J.H., Smith L., Adams J.B., Alterovitz G., Babb L.J., Barkley M.P., Baudis M., Beauvais M.J.S., Beck T., et al. GA4GH: international policies and standards for data sharing across genomic research and healthcare. Cell Genomics. 2021;1 doi: 10.1016/j.xgen.2021.100029. 100029-1–100029-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Powell K. The broken promise that undermines human genome research. Nature. 2021;590:198–201. doi: 10.1038/d41586-021-00331-5. [DOI] [PubMed] [Google Scholar]
- 3.Mailman M.D., Feolo M., Jin Y., Kimura M., Tryka K., Bagoutdinov R., Hao L., Kiang A., Paschall J., Phan L., et al. The NCBI dbGaP database of genotypes and phenotypes. Nat. Genet. 2007;39:1181–1186. doi: 10.1038/ng1007-1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Paltoo D.N., Rodriguez L.L., Feolo M., Gillanders E., Ramos E.M., Rutter J.L., Sherry S., Wang V.O., Bailey A., Baker R., et al. National Institutes of Health Genomic Data Sharing Governance Committees Data use under the NIH GWAS data sharing policy and future directions. Nat. Genet. 2014;46:934–938. doi: 10.1038/ng.3062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lappalainen I., Almeida-King J., Kumanduri V., Senf A., Spalding J.D., Ur-Rehman S., Saunders G., Kandasamy J., Caccamo M., Leinonen R., et al. The European Genome-phenome Archive of human data consented for biomedical research. Nat. Genet. 2015;47:692–695. doi: 10.1038/ng.3312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Linden M., Procházka M., Lappalainen I., Bucik D., Vyskocil P., Kuba M., Silén S., Belmann P., Sczyrba A., Newhouse S., et al. Common ELIXIR Service for Researcher Authentication and Authorisation. F1000Res. 2018;7:1199. doi: 10.12688/f1000research.15161.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Global Alliance for Genomics and Health GENOMICS. A federated ecosystem for sharing genomic, clinical data. Science. 2016;352:1278–1280. doi: 10.1126/science.aaf6162. [DOI] [PubMed] [Google Scholar]
- 8.Lawson J., Cabili M.N., Kerry G., Boughtwood T., Thorogood A., Alper P., Bowers S.R., Boyles R.R., Brookes A.J., Brush M., et al. The Data Use Ontology to streamline responsible access to human biomedical datasets. Cell Genomics. 2021;1 doi: 10.1016/j.xgen.2021.100028. 100028-1–100028-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Voisin C., Linden M., Dyke S.O.M., Bowers S.R., Reinold K., Lawson J., Li S., Ota Wang V., Barkley M.P., Bernick D., et al. GA4GH Passport standard for digital identity and access permissions. Cell Genomics. 2021;1 doi: 10.1016/j.xgen.2021.100030. 100030-1–100030-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.The Global Alliance for Genomics and Health (2021). Genomics Data Toolkit, Machine Readable Consent Guidance. https://www.ga4gh.org/genomic-data-toolkit/. August 21st, 2021.
- 11.Woolley J.P., Kirby E., Leslie J., Jeanson F., Cabili M.N., Rushton G., Hazard J.G., Ladas V., Veal C.D., Gibson S.J., et al. Responsible sharing of biomedical data and biospecimens via the “Automatable Discovery and Access Matrix” (ADA-M) NPJ Genom. Med. 2018;3:17. doi: 10.1038/s41525-018-0057-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dyke S.O.M., Philippakis A.A., Rambla De Argila J., Paltoo D.N., Luetkemeier E.S., Knoppers B.M., Brookes A.J., Spalding J.D., Thompson M., Roos M., et al. Consent Codes: Upholding Standard Data Use Conditions. PLoS Genet. 2016;12:e1005772. doi: 10.1371/journal.pgen.1005772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cabili M.N., Carey K., Dyke S.O.M., Brookes A.J., Fiume M., Jeanson F., Kerry G., Lash A., Sofia H., Spalding D., et al. Simplifying research access to genomics and health data with Library Cards. Sci. Data. 2018;5:180039. doi: 10.1038/sdata.2018.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schriml L.M., Mitraka E., Munro J., Tauber B., Schor M., Nickle L., Felix V., Jeng L., Bearer C., Lichenstein R., et al. Human Disease Ontology 2018 update: classification, content and workflow expansion. Nucleic Acids Res. 2019;47(D1):D955–D962. doi: 10.1093/nar/gky1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shefchek K.A., Harris N.L., Gargano M., Matentzoglu N., Unni D., Brush M., Keith D., Conlin T., Vasilevsky N., Zhang X.A., et al. The Monarch Initiative in 2019: an integrative data and analytic platform connecting phenotypes to genotypes across species. Nucleic Acids Res. 2020;48(D1):D704–D715. doi: 10.1093/nar/gkz997. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The code for DUOS is distributed under the three-clause BSD open source license, which allows for redistribution and use in source and forms, with or without modifications under certain conditions, and is in the “DataBiosphere” GitHub repositories located here:
- •
- •
- •
- •
The “DUOS-UI” is the front end user interface for the application. “Consent” is the application component where all user, data access requests, and dataset access decisions are stored. “Consent Ontology” is the application component that can make ontologically valid decisions about whether a dataset and a data access request are compatible using the GA4GH (DUO) and Human Disease Ontology (DOID) ontologies. “Consent Data Use” captures all the ontologies we referenced in the article for efficient access by the DUOS service.
In addition, we have made DUOS available as a publicly accessible web-service for registering, requesting, and reviewing access requests for datasets at https://duos.broadinstitute.org. Individuals and organizations unable to initiate and maintain the DUOS code are welcome to inquire with the corresponding author on how to use DUOS as a web service.
Data reported in this paper will be shared by the lead contact upon request.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.