Abstract
Degradation of plant cell wall polysaccharides is of major importance in the food and feed, beverage, textile, and paper and pulp industries, as well as in several other industrial production processes. Enzymatic degradation of these polymers has received attention for many years and is becoming a more and more attractive alternative to chemical and mechanical processes. Over the past 15 years, much progress has been made in elucidating the structural characteristics of these polysaccharides and in characterizing the enzymes involved in their degradation and the genes of biotechnologically relevant microorganisms encoding these enzymes. The members of the fungal genus Aspergillus are commonly used for the production of polysaccharide-degrading enzymes. This genus produces a wide spectrum of cell wall-degrading enzymes, allowing not only complete degradation of the polysaccharides but also tailored modifications by using specific enzymes purified from these fungi. This review summarizes our current knowledge of the cell wall polysaccharide-degrading enzymes from aspergilli and the genes by which they are encoded.
This review summarizes our current knowledge on the different classes of enzymes involved in plant cell wall polysaccharide degradation produced by Aspergilli, the genes encoding these enzymes, and the regulation of these genes. The data from literature is presented in tables as much as possible to provide easy comparisons of the enzymes and genes reported so far. Only enzymes for which a detailed characterisation was published are presented this way, requiring at least a MW and one of three other characteristics (pI, pH optimum or T optimum). Enzymes that have been characterised in less detail are mentioned in the text when this provided additional information valuable to this review. The tables of genes list the assignment of the corresponding enzymes to the different glycosidase, polysaccharide lyase and carbohydrate esterase families (69, 145–147) as described by B. Henrissat at URL: http://afmb.cnrs-mrs.fr/∼pedro/CAZY/db.html.
Plant Cell Wall Polysaccharides
Plant cell wall polysaccharides are the most abundant organic compounds found in nature. They make up 90% of the plant cell wall and can be divided into three groups: cellulose, hemicellulose, and pectin (256). Cellulose represents the major constituent of cell wall polysaccharides and consists of a linear polymer of β-1,4-linked d-glucose residues. The cellulose polymers are present as ordered structures (fibers), and their main function is to ensure the rigidity of the plant cell wall.
Hemicelluloses are more heterogeneous polysaccharides and are the second most abundant organic structure in the plant cell wall. The major hemicellulose polymer in cereals and hardwood is xylan. Xylan consists of a β-1,4-linked d-xylose backbone and can be substituted by different side groups such as l-arabinose, d-galactose, acetyl, feruloyl, p-coumaroyl, and glucuronic acid residues (400). A second hemicellulose structure commonly found in soft- and hardwoods is (galacto)glucomannan (369), which consists of a backbone of β-1,4-linked d-mannose and d-glucose residues with d-galactose side groups (see “Structural features of galacto(gluco)mannan” below). Softwoods contain mainly galactoglucomannan, whereas in hardwoods glucomannan is the most common form. Xyloglucans are present in the cell walls of dicotyledonae and some monocotylodonae (e.g., onion). Xyloglucans consist of a β-1,4-linked d-glucose backbone substituted by d-xylose. l-Arabinose and d-galactose residues can be attached to the xylose residues, and l-fucose has been detected attached to galactose residues in xyloglucan. Xyloglucans interact with cellulose microfibrils by the formation of hydrogen bonds, thus contributing to the structural integrity of the cellulose network (56).
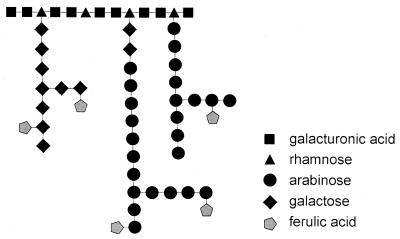
Pectins form another group of heteropolysaccharides and consist of a backbone of α-1,4-linked d-galacturonic acid residues (see “Structural features of pectin” below). In specific “hairy” regions the galacturonic acid backbone is interrupted by α-1,2-linked l-rhamnose residues. Long side chains consisting mainly of l-arabinose and d-galactose residues can be attached to these rhamnose residues. In pectins of certain origins (e.g., sugar beet and apple), ferulic acid can be present as terminal residues attached to O-5 of the arabinose residues or O-2 of the galactose residues.
The hemicellulose and pectin polysaccharides, as well as the aromatic polymer lignin, interact with the cellulose fibrils, creating a rigid structure strengthening the plant cell wall. They also form covalent cross-links, which are thought to be involved in limiting cell growth and reducing cell wall biodegradability. Two types of covalent cross-links have been identified between plant cell wall polysaccharides and lignin (117). The cross-link formed by diferulic acid bridges is studied in most detail. Diferulic acid bridges between polysaccharides and lignin have been identified in many plants. They have been shown to occur between arabinoxylans from bamboo shoot cell walls (162), between pectin polymers in sugar beet (275), and between lignin and xylan in wheat (22). A second type of cross-link is the ester linkage between lignin and glucuronic acid attached to xylan, which was identified in beech wood (160, 359). Recently, indications of a third type of cross-linking have been reported involving a protein- and pH-dependent binding of pectin and glucuronoarabinoxylan to xyloglucan (311). This not yet fully characterized binding is dependent on the presence of fucose on the xyloglucan.
Structural Features of Cellulose and Xyloglucan
Cellulose consists of linear β-1,4-linked d-glucopyranose chains that are condensed by hydrogen bonds into crystalline structures, called microfibrils (205). These microfibrils consist of up to 250 glucose chains and are linked by hemicelluloses (56). In addition to this crystalline structure, cellulose contains noncrystalline (amorphous) regions within the microfibrils. The relative amounts of crystalline and noncrystalline cellulose vary depending on the origin (232).
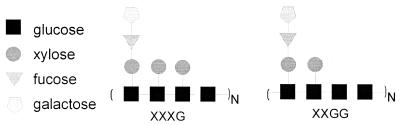
Two major types of xyloglucans have been identified in the plant cell wall (Fig. 1). Xyloglucan type XXXG consists of repeating units of three β-1,4-linked d-glucopyranose residues, substituted with d-xylopyranose via an α-1,6-linkage, which are separated by an unsubstituted glucose residue. In xyloglucan type XXGG, two xylose-substituted glucose residues are separated by two unsubstituted glucose residues. The structural features of these, as well as some other types of xyloglucans, have been discussed in detail by Vincken et al. (391). The xylose residues in xyloglucan can be substituted with α-1,2-l-fucopyranose-β-1,2-d-galactopyranose and α-1,2-l-galactopyranose-β-1,2-d-galactopyranose disaccharides (138, 391). l-Arabinofuranose has been detected α-1,2-linked to main-chain glucose residues or xylose side groups (151, 157, 308, 409). In addition, xyloglucans can contain O-linked acetyl groups (243, 339). Xyloglucans are strongly associated with cellulose and thus add to the structural integrity of the cell wall. They are thought to play an important role in regulating cell wall extension. The length of the xyloglucan polymers enables them to cross-link many cellulose microfibrils, creating a rigid structure (248).
FIG. 1.
Schematic presentation of the repeating units of the two major xyloglucan structures.
Structural Features of Xylan
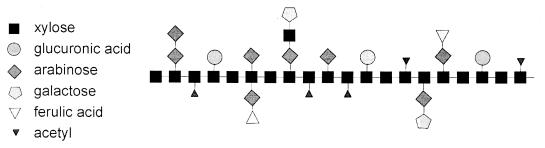
The structure of xylans found in cell walls of plants can differ greatly depending on their origin, but they always contain a β-1,4-linked d-xylose backbone (101, 399). The schematic representation of xylan (Fig. 2) also lists the different structures which can be attached to the xylan backbone and which result in the large variety of xylan structures found in plants. Although most xylans are branched structures, some linear polysaccharides have been isolated (102, 261). Cereal xylans contain large quantities of l-arabinose and are therefore often referred to as arabinoxylans, whereas hardwood xylans are often referred to as glucuronoxylans due to the large amount of d-glucuronic acid attached to the backbone.
FIG. 2.
Schematic presentation of xylan.
Arabinose is connected to the backbone of xylan via an α-1,2- or α-1,3-linkage either as single residues or as short side chains. These side chains can also contain xylose β-1,2-linked to arabinose, and galactose, which can be either β-1,5-linked to arabinose or β-1,4-linked to xylose. Acetyl residues are attached to O-2 or O-3 of xylose in the backbone of xylan, but the degree of acetylation differs greatly amongst xylans from different origin. Glucuronic acid and its 4-O-methyl ether are attached to the xylan backbone via an α-1,2-linkage, whereas aromatic (feruloyl and p-coumaroyl) residues have so far been found attached only to O-5 of terminal arabinose residues (324, 343, 397). As a consequence of all these features, the xylans form a very heterogeneous group of polysaccharides (27, 47, 156, 328).
Structural Features of Galacto(gluco)mannan
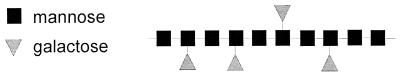
Galactomannans and galactoglucomannans form a second group of hemicellulolytic structures present in plant cell walls. They are the major hemicellulose fraction of gymnosperms (20), in which they represent 12 to 15% of the cell wall biomass. Galactomannans are most commonly found in the family of Leguminoseae, in which they represent 1 to 38% of seed dry weight, but have also been identified in species of other plants such as Ebenaceae and Palmae (75, 97). They consist of a backbone of β-1,4-linked d-mannose residues, which can be substituted by d-galactose residues via an α-1,6-linkage (Fig. 3). Depending on the source of the polysaccharide, mannose/galactose ratios can vary from 1.0 to 5.3 (75, 97).
FIG. 3.
Schematic presentation of galactomannan.
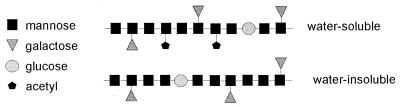
Galactoglucomannan is the major hemicellulolytic component of softwood. Two different structures can be identified within this group of polysaccharides (Fig. 4) (369). Both consist of a β-1,4-linked d-mannose backbone, which can be substituted by α-1,6-linked d-galactose. The galactoglucomannan backbone also contains β-1,4-linked d-glucose residues. Water-soluble galactoglucomannan has a higher galactose content than does water-insoluble galactoglucomannan and in addition contains acetyl residues attached to the main chain (369). Approximately 20 to 30% of the backbone glucose and/or mannose residues are esterified with acetyl groups at C-2 or C-3 (233). Recently, the structure of a galactoglucomannan from Nicotiana plumbaginifolia was analyzed (338). Apart from side chains consisting of single α-1,6-linked galactose residues, this polysaccharide also contained a disaccharide consisting of a galactose residue β-1,2-linked to a galactose residue that is α-1,6-linked to the main chain.
FIG. 4.
Schematic presentation of the two galactoglucomannan structures.
Structural Features of Pectin
Pectins are complex heteropolysaccharides which contain two different defined regions (85, 290). The “smooth” regions consist of a backbone of α-1,4-linked d-galacturonic acid residues, which can be acetylated at O-2 or O-3 or methylated at O-6. In the “hairy” regions, two different structures can be identified, a xylogalacturonan consisting of a d-xylose-substituted galacturonan backbone and rhamnogalacturonan I. In rhamnogalacturonan I (Fig. 5), the d-galacturonic acid residues in the backbone are interrupted by α-1,2-linked l-rhamnose residues, to which long arabinan and galactan chains can be attached at O-4. The arabinan chains consist of a main chain of α-1,5-linked l-arabinose residues that can be substituted by α-1,3-linked l-arabinose and by feruloyl residues attached terminally to O-2 of the arabinose residues (66, 134). The galactan side chains contain a main chain of β-1,4-linked d-galactose residues, which can be substituted by feruloyl residues at O-6 (66, 134). Approximately 20 to 30% of the feruloyl residues in sugar beet pectin are attached to arabinan side chains, whereas the other feruloyl residues are attached to galactan side chains (134). Rhamnogalacturonan I also contains acetyl groups ester-linked to O-2 or O-3 of galacturonic acid residues of the backbone (326, 327). Rhamnogalacturonan II is a polysaccharide of approximately 30 monosaccharide units with a backbone of galacturonic acid residues that is substituted by four side chains. The structures of these side chains have been determined and have been shown to contain several uncommon sugars such as 2-O-methyl-l-fucose and 3-deoxy-d-manno-2-octulosonic acid (247). The structural arrangement in which these side chains are attached to the backbone of rhamnogalacturonan II have also been determined and have demonstrated two possible arrangements for this oligosaccharide (148). Whether rhamnogalacturonan II is covalently linked to the pectin main chain is not known.
FIG. 5.
Schematic presentation of the hairy region of pectin.
Aromatic Residues in Plant Cell Wall Polysaccharides
Aromatic compounds are thought to play an important role in the structure and function of the plant cell wall. Ferulic acid can be linked to both the hemicellulose (343) and the pectin (314) fractions of plant cell walls and is able to cross-link these polysaccharides to each other as well as to the aromatic polymeric compound lignin (163, 230). This cross-linked structure results in an increased rigidity of the cell wall. An increase in ferulic acid cross-links during ageing of the plant cell suggests a function for these cross-links in limiting cell growth (118, 395). A role for these cross-links in preventing biodegradability of the plant cell wall by microorganisms has also been suggested. Indications for a limited enzymatic degradation of arabinoxylan due to ferulate cross-links have been obtained (104, 131). Additionally, the antimicrobial effects of these aromatic compounds (21) may contribute to to the plant defense mechanism against phytopathogenic microorganisms.
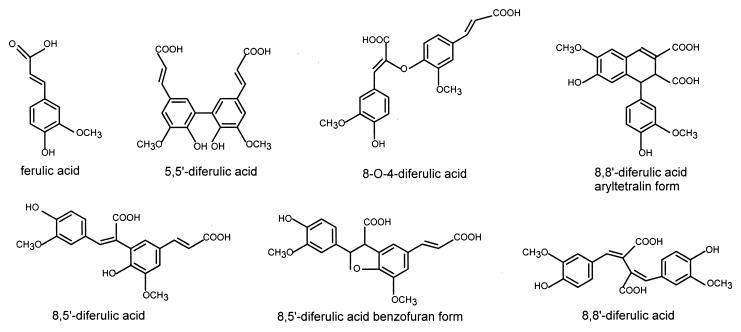
In cereals, cinnamic acids (mainly ferulic acid) are ester-linked to arabinose residues in arabinoxylan in the primary cell wall. Ferulic acid was detected both as terminal residues and as ferulate dimers linked in several ways (Fig. 6), such as 5,5′ or 5,8′ carbon-carbon bonds (163, 217).
FIG. 6.
Schematical presentation of ferulic acid and diferulic acid structures identified in plant cell walls.
BIODEGRADATION OF PLANT CELL WALL POLYSACCHARIDES
Aspergillus
The genus Aspergillus is group of filamentous fungi with a large number of species. The first record of this fungus can be found in Micheli's Nova Plantarum Genera (258), but a more detailed description of the aspergilli did not appear until the middle of the 19th century. In 1926 a first classification of these fungi was proposed describing 11 groups within the genus (366). A reexamination of the genus was published by Thom and Raper (367), identifying 14 distinct groups. Some of these groups consist of pathogenic fungi (e.g., A. fumigatus, A. flavus, and A. parasiticus), but most important for industrial applications are some members of the group of black aspergilli (A. niger and A. tubingensis). In addition to the morphological techniques traditionally applied, new molecular and biochemical techniques have been used in the reclassification of this group of aspergilli (137, 226, 257, 271, 283, 386). These analyses resulted in the clear distinction of eight groups of black aspergilli (A. niger, A. tubingensis, A. foetidus, A. carbonarius, A. japonicus, A. aculeatus, A. heteromorphus, and A. ellipticus) (283). Products of several of these species have obtained a GRAS (Generally Regarded As Safe) status, which allows them to be used in food and feed applications. The black aspergilli have a number of characteristics which make them ideal organisms for industrial applications, such as good fermentation capabilities and high levels of protein secretion. In particular, the wide range of enzymes produced by Aspergillus for the degradation of plant cell wall polysaccharides are of major importance to the food and feed industry. Recently, several Aspergillus spp. have received increased interest as hosts for heterologous protein production (74).
Degradation of Cellulose and the Xyloglucan Backbone
Four classes of enzymes are involved in the biodegradation of cellulose. Endoglucanases (EC 3.2.1.4) (Table 1) hydrolyze cellulose to glucooligosaccharides. Cellobiohydrolases (EC 3.2.1.91) release cellobiose from crystalline cellulose. β-Glucosidases (EC 3.2.1.21) (Table 1) degrade the oligosaccharides to glucose. Exoglucanases (Table 1) release glucose from cellulose and glucooligosaccharides. The distinction between exoglucanases and cellobiohydrolases is not always clear due to differences in the methods used to study these enzymes. All four classes of enzymes have been identified in aspergilli, although the number of isozymes produced by different species or even strains of the same species can differ. An analysis of the production of endoglucanases by 45 A. terreus isolates not only revealed different electrophoretic mobilities for the enzymes of the different isolates but also indicated the absence of endoglucanase I in a number of the isolates (341). Endoglucanases and β-glucosidases are also able to degrade the backbone of xyloglucan. From A. aculeatus an endoglucanase has been purified that is specific for the substituted xyloglucan backbone (287). This enzyme was not able to hydrolyze cellulose, and treatment of plant cell walls with the enzyme liberated only xyloglucan oligosaccharides.
TABLE 1.
Physical properties of endo- and exoglucanases and β-glucosidases from Aspergillus
| Species and enzyme type | Enzyme | Mol mass (kDa) | pHopt | Topt (°C) | pI | Reference(s) |
|---|---|---|---|---|---|---|
| Endoglucanases | ||||||
| A. aculeatus | XEG | 23.6 | 3.4 | 287 | ||
| A. aculeatus | FI-CMCase | 25 | 4.5 | 50 | 4.8 | 263 |
| A. aculeatus | FII-CMCase | 66 | 5.0 | 70 | 4.0 | 358 |
| A. aculeatus | FV-CMCase | 38 | 4.0 | 65 | 3.4 | 358 |
| A. aculeatus | hydrocellulase | 68 | 2.5 | 60 | 3.5 | 358 |
| A. aculeatus | FI-Avicelase | 109 | 5.5 | 65 | 4.7 | 358 |
| A. aculeatus | FIII-Avicelase | 112 | 2.5 | 65 | 4.0 | 358 |
| A. fumigatus | 12.5 | 4.8 | 60 | 7.1 | 286 | |
| A. nidulans | Endo-I | 25 | 6.0 | 65 | 24 | |
| A. nidulans | Endo-II | 32.5 | 5.0 | 50 | 24 | |
| A. nidulans | EG A | 35 | 6.5 | 50 | 58 | |
| A. niger | 26 | 3.8–4.0 | 45 | 158 | ||
| A. niger | A | 43 | 2–7 | 60 | 389 | |
| A. niger | B | 25 | 2–7 | 60 | 389 | |
| A. niger | 40 | 6.0–7.0 | 70 | 7 | ||
| A. niger | 31 | 4.0 | 45–50 | 3.67 | 273 | |
| A. oryzae | CelA | 31 | 5.0 | 55 | 193 | |
| A. oryzae | CelB | 53 | 4.0 | 45 | 193 | |
| Exoglucanases | ||||||
| A. nidulans | Exo-I | 29 | 5.5 | 50 | 24 | |
| A. niger | 52.5 | 5.5 | 50 | 340 | ||
| β-Glucosidases | ||||||
| A. aculeatus | β-Gluc1 | 133 | 4.5 | 55 | 4.7 | 358 |
| A. japonicus | >240 | 5.0 | 65 | 323 | ||
| A. nidulans | β-Gluco-I | 26 | 6.0 | 35 | 24 | |
| A. nidulans | β-Gluco-II | 14 | 5.0 | 65 | 24 | |
| A. nidulans | P-I | 125 | 5.0 | 50 | 4.4 | 227 |
| A. nidulans | P-II | 50 | 5.5 | 60 | 4.0 | 227 |
| A. niger | 325 | 4.6 | 60 | 408 | ||
| A. niger | I | 96 | 5.1 | 4.6 | 401 | |
| A. niger | II | 96 | 4.1 | 3.8 | 401 | |
| A. niger | 120 | 4.5 | 55–60 | 4.0 | 396, 407 | |
| A. niger | 137 | 3.8 | 373 | |||
| A. niger | 117–122 | 4.2 | 150 | |||
| A. niger | 100 | 4.8 | 65 | 120 | ||
| A. niger | β-Glu I | 49 | 5.0 | 55 | 3.2 | 406 |
| A. oryzae | BGI | 130 | 4.9 | 310 | ||
| A. oryzae | HGT-BG | 43 | 5.0 | 50 | 4.2 | 310 |
| A. terreus | 200 | 4.5 | 55–60 | 4.8 | 49, 313, 360 |
Three exoglucanases have been purified from A. nidulans (24) but only exoglucanase I (Exo-I) was studied in detail. Exo-I, Exo-II, and Exo-III differed significantly in their molecular mass (29, 72.5, and 138 kDa, respectively). Exo-II and Exo-III had a had a higher affinity for cellulose than did Exo-I (24). Two exoglucanases have been identified in A. terreus (170). Two cellobiohydrolases have been purified from A. ficuum (143) and A. terreus (170). The two enzymes from A. ficuum have very different molecular masses (128 and 50 kDa, respectively), whereas the molecular masses of the A. terreus cellobiohydrolases are nearly identical (28.5 and 29.5 kDa, respectively).
Production of cellulolytic enzymes by aspergilli has been observed using the following carbon sources: cellulose (164, 285, 301), sophorose and 2-O-β-d-glucopyranosyl-d-xylose (155), and cellobiose, glucose and xylose (8). However, other factors were important as well. Production of both endo- and exoglucanases in A. fumigatus was much higher when ammonia was used as a nitrogen source instead of nitrate (347), whereas the production of β-glucosidase in A. terreus was higher on nitrate than on ammonia (301). In A. nidulans, an endoglucanase was identified that was developmentally regulated and that was produced only during cleisthothecial development (25). For several β-glucosidases, transglycosylation activity has been observed using cellobiose (33, 396), cellotriose, methyl-β-glucoside, and ethyl-β-glucoside (407) as substrates.
Based on the derived amino acid sequences, the gene products have been assigned to different glycosidase families. Endoglucanases are assigned mainly to families 5 and 12 (Table 2), with the exception of CelB from A. oryzae. This enzyme was assigned to family 7, which also contains the Aspergillus cellobiohydrolases (Table 2). The only exoglucanase gene cloned so far was assigned to family 74 (Table 2). All β-glucosidases from Aspergillus have been assigned to family 3 of the glycosidases (Table 2). All cellulose-degrading enzymes have a retaining mechanism. The exoglucanase from A. aculeatus (family 74) is the only enzyme for which the catalytic mechanism has not yet been determined.
TABLE 2.
Genes encoding endoglucanases, exoglucanases, cellobiohydrolases, and β-glucosidases from Aspergillus and their assignment to the glycosidase families
| Species | Activitya | Gene | Glycosidase family | Database accession no. | Reference |
|---|---|---|---|---|---|
| A. aculeatus | EGL | cel1 | 5 | AF054512 | L. V. Kofod et al., unpublished data |
| A. aculeatus | EGL | cmc2 | 5 | AB015510 | M. Arai et al., unpublished data |
| A. aculeatus | EGL | xeg | 12 | AF043595 | 287 |
| A. aculeatus | EGL | 12 | D00546 | 274 | |
| A. kawachii | EGL | cekA | 12 | D12901 | 322 |
| A. nidulans | EGL | eglA | 5 | AB009402 | 58 |
| A. niger | EGL | eglA | 12 | AJ224451 | 383 |
| A. niger | EGL | eglB | 5 | AJ224452 | 383 |
| A. oryzae | EGL | celA | 12 | D83732 | 193 |
| A. oryzae | EGL | celB | 7 | D83731 | 193 |
| A. aculeatus | EXG | 74 | AB015511 | M. Arai et al., unpublished data | |
| A. aculeatus | CBH | cbhI | 7 | AB002821 | 357 |
| A. niger | CBH | cbhA, cbhB | 7 | AF156268, AF156269 | 124 |
| A. aculeatus | BGL | bgl1 | 3 | D64088 | 179 |
| A. kawachii | BGL | bglA | 3 | AB003470 | 171 |
| A. terreus | BGL | 3 | Z37722 | D. B. Mitchell, unpublished data | |
| A. wentii | BGL | bglA-3 | 3 | P29090 | 31 |
EGL, endoglucanase, EXG, exoglucanase; CBH, cellobiohydrolase; BGL, β-glucosidase.
Degradation of the Xylan Backbone
The biodegradation of the xylan backbone depends on two classes of enzymes. Endoxylanases (EC 3.2.1.8) are able to cleave the xylan backbone into smaller oligosaccharides, which can then be degraded further to xylose by β-xylosidases (EC 3.2.1.37). Both classes of enzymes, as well as their encoding genes, have been characterized from many organisms. Various endoxylanases have been identified in Aspergillus (Table 3). Although variation is detected in their molecular mass or pH optimum, the major difference between the enzymes is in their pI, which ranges from 3.5 (168) to 9.0 (119). Endoxylanases also differ in their specificity toward the xylan polymer. Some enzymes cut randomly between unsubstituted xylose residues, whereas the activity of other endoxylanases strongly depends on the substituents on the xylose residues neighboring the attacked residues.
TABLE 3.
Physical properties of Aspergillus endoxylanases and β-xylosidases
| Species and enzyme type | Enzyme | Mol mass (kDa) | pHopt | Topt (°C) | pI | Reference |
|---|---|---|---|---|---|---|
| Endoxylanases | ||||||
| A. aculeatus | FIa | 18 | 4.0 | 50 | 5.6 | 119 |
| A. aculeatus | FIb | 26 | 5.0 | 50 | 9.0 | 119 |
| A. aculeatus | FIII | 52 | 5.0 | 70 | 3.8 | 119 |
| A. awamori | I | 39 | 5.5–6.0 | 55 | 5.7–6.7 | 210 |
| A. awamori | II | 23 | 5.0 | 50 | 3.7 | 210 |
| A. awamori | III | 26 | 4.0 | 45–50 | 3.3–3.5 | 210 |
| A. flavipes | 45 | 5.0 | 55 | 334 | ||
| A. foetidus | I | 24 | 7.6 | 26 | ||
| A. foetidus | II | 26 | 4.6 | 26 | ||
| A. fumigatus | I | 22 | >9.4 | 26 | ||
| A. fumigatus | II | 10 | 5.7 | 26 | ||
| A. fumigatus | II | 19 | 5.5 | 55 | 337 | |
| A. kawachii | XylA | 35 | 5.5 | 6.7 | 168 | |
| A. kawachii | XylB | 26 | 4.5 | 4.4 | 168 | |
| A. kawachii | XylC | 29 | 2.0 | 3.5 | 168 | |
| A. nidulans | 34 | 6.0 | 56 | 3.4 | 108 | |
| A. niger | 33 | 4.0 | 50 | 4.2 | 130 | |
| A. niger | XYLI | 20.8 | 5 | 55 | 6.7 | 115 |
| A. niger | XYLII | 13 | 6 | 45 | 8.6 | 116 |
| A. niger | XYLIII | 13 | 5.5 | 45 | 9.0 | 116 |
| A. niger | XYLIV | 14 | 4.9 | 45 | 4.5 | 333 |
| A. niger | XYLV | 28 | 5.0 | 42 | 3.65 | 114 |
| A. oryzae | I | 28 | 5.0 | 7.0 | 26 | |
| A. oryzae | II | 26 | 5.0 | 4.9 | 26 | |
| A. oryzae | 46.5 | 5.0 | 55 | 3.6 | 128 | |
| A. sojae | X-I | 32.7 | 5.5 | 60 | 3.5 | 188 |
| A. sojae | X-II-B | 35.5 | 5.5 | 50 | 3.75 | 188 |
| A. sydowii | 30 | 5.5 | 60 | 123 | ||
| A. tubingensis | XlnA | 19 | 3.6 | 78 | ||
| β-Xylosidases | ||||||
| A. awamori | 110 | 6.5 | 70 | 4.2 | 210 | |
| A. foetidus | 83 | 4.4 | 26 | |||
| A. fumigatus | 90 | 4.5 | 75 | 5.4 | 199 | |
| A. fumigatus | 60 | 4.3 | 26 | |||
| A. nidulans | XlnD | 85 | 5.0 | 50 | 3.4 | 222 |
| A. niger | 5.0 | >75 | 370 | |||
| A. niger | 122 | 3.8–4.0 | 70 | 4.9 | 312 | |
| A. oryzae | 62 | 4.5 | 26 | |||
| A. oryzae | XylA | 110 | 4.0 | 60 | 198 | |
| A. pulverulentus | β-Xyl I | 65 | 2.5–3.5 | 60 | 4.7 | 350 |
| A. pulverulentus | β-Xyl II | 100 | 4.0–5.0 | 60 | 3.5 | 350 |
In several aspergilli, three different endoxylanases have been identified (119, 168, 210). The best-studied Aspergillus endoxylanases, with respect to substrate specificity, are the three enzymes from A. awamori (210). Counting from the reducing end, A. awamori endoxylanase I is unable to remove one unsubstituted xylose residue adjacent to singly substituted xylose residues or two unsubstituted xylose residues adjacent to doubly substituted xylose residues (206). A. awamori endoxylanase III was not able to remove two unsubstituted xylose residues adjacent to singly or doubly substituted xylose residues toward the reducing end (206).
Hydrolysis of a glucuronoxylan by an endoxylanase from A. niger (130) resulted mainly in xylobiose, xylotriose, and xylose, but hydrolysis of an arabinoxylan by the same enzyme resulted mainly in oligosaccharides with a degree of polymerization of more than 3. This suggests that the action of this endoxylanase is reduced by the presence of arabinose residues on the xylan backbone. All xylanases that have been purified to date are produced when Aspergillus is grown on xylan. Most of these enzymes are also produced when xylose was used as a carbon source, but all at lower levels than on xylan. This is discussed in more detail below (see “Carbon catabolite repression”).
Several genes encoding endoxylanases from aspergilli have been cloned. The encoded enzymes have been assigned to glycosidase families 10 and 11 (Table 4), and they all work via a retaining mechanism. Based on the data of the A. kawachii endoxylanases, it would appear that the acidic endoxylanases belong to family 11 whereas the neutral endoxylanases belong to group 10. However, more data on other neutral and acidic endoxylanases are needed to verify this. Recently, a method was developed to experimentally determine whether an endoxylanase belongs to family 10 or 11 (272). This method is based on the irriversible inhibition of family 11 endoxylanases by epoxyl glycosides of d-xylose and xylooligosaccharides, whereas family 10 endoxylanases are unaffected (272).
TABLE 4.
Genes encoding Aspergillus endoxylanases and β-xylosidases and their assignment to the glycosidase families
| Species | Activitya | Gene | Glycosidase family | Database accession no. | Reference(s) |
|---|---|---|---|---|---|
| A. aculeatus | EXL | Fla | 10 | AB013110 | M. Arai et al., unpublished data |
| A. awamori | EXL | exlA | 11 | X78115 | 149 |
| A. kawachii | EXL | xynA | 10 | D14847 | 166 |
| A. kawachii | EXL | xynB, xynC | 11 | D38070, S45138 | 167; K. Ito, unpublished data |
| A. nidulans | EXL | xlnA, xlnB | 11 | Z49892, Z49893 | 291 |
| A. nidulans | EXL | xlnC | 10 | Z49894 | 238 |
| A. niger | EXL | 11 | A19535 | 215 | |
| A. niger | EXL | xynB | 11 | D38071 | 192 |
| A. niger | EXL | xyn4, xyn5 | 11 | U39785, U39784 | M. Luttig et al., unpublished data |
| A. oryzae | EXL | xynG1 | 11 | AB003085 | 191 |
| A. oryzae | EXL | F1 | 10 | AB011212 | 197 |
| A. tubingensis | EXL | xlnA | 11 | L26988 | 78 |
| A. nidulans | BXL | xlnD | 3 | Y13568 | 292 |
| A. niger | BXL | xlnD | 3 | Z84377 | 382 |
| A. oryzae | BXL | xylA | 3 | AB013851 | 198 |
| A. oryzae | BXL | xyl-1 | 3 | AB009972 | 141 |
EXL, endoxylanase; BXL, β-xylosidase.
β-Xylosidases have been identified in several aspergilli (Table 3). These enzymes are highly specific for small unsubstituted xylose oligosaccharides (degree of polymerization of up to 4), and their action results in the production of xylose. Although this activity is of major importance for the complete degradation of xylan, absence of the enzyme does not interfere with the induction of the xylanolytic system (382). The ability of an A. awamori β-xylosidase to release xylose from xylooligosaccharides was studied to determine its substrate specificity (206). This enzyme was able to release xylose from the nonreducing end of branched oligosaccharides only when two contiguous unsubstituted xylose residues were present adjacent to singly or doubly substituted xylose residues.
Based on the sequence of the corresponding genes, β-xylosidases from Aspergillus spp. have all been assigned to glycosidase family 3 (Table 4) and have a retaining mechanism.
For some β-xylosidases, transxylosylation activity has been detected (26, 200, 335, 350), allowing the production of novel xylose containing oligosaccharides using these enzymes. Production of xylooligosaccharides from xylose using β-xylosidase in a condensation reaction was also demonstrated (159), suggesting a possible application for these enzymes in the synthesis of specific oligosaccharides.
Degradation of the Galacto(gluco)mannan Backbone
The degradation of the galacto(gluco)mannan backbone depends on the action of β-endomannanases (EC 3.2.1.78) and β-mannosidases (EC 3.2.1.25), which are commonly produced by aspergilli (Table 5). β-Endomannanases, generally referred to as β-mannanases, hydrolyze the backbone of galacto(gluco)mannans, resulting in mannooligosaccharides. The ability of β-mannanases to degrade the mannan backbone depends on several factors, such as the number and distribution of the substituents on the backbone and the ratio of glucose to mannose (250). β-Mannanase is most active on galactomannans with a low substitution of the backbone (64). The presence of galactose residues on the mannan backbone significantly hinders the activity of β-mannanase (252), but this effect is small if the galactose residues in the vicinity of the cleavage point are all on the same side of the main chain (251). β-Mannanases release predominantly mannobiose and mannotriose from mannan, confirming that they are true endohydrolases (4, 64, 105, 306). It has been shown that A. niger β-mannanase binds to four mannose residues during catalysis (249).
TABLE 5.
Physical properties of Aspergillus β-d-mannanases and β-d-mannosidases
| Species and enzyme type | Enzyme | Mol mass (kDa) | pHopt | Topt (°C) | pI | Reference(s) |
|---|---|---|---|---|---|---|
| β-d-Mannanases | ||||||
| A. aculeatus | 45 | 5.0 | 60–70 | 4.5 | 59 | |
| A. aculeatus | FIIIa | 39 | 4.0 | 70 | 4.2 | 356 |
| A. niger | 40–45 | 3.0–3.8 | 65 | 3.7–3.95 | 4, 6, 105, 251 | |
| A. niger | 3.6 | >80 | 4.1 | 405 | ||
| A. niger | 56 | 3.0 | 50 | 4.9 | 307 | |
| A. oryzae | 110 | 6.0 | 40 | 3.5–4.5 | 307 | |
| A. tamarii | 53 | 4.5 | 64 | |||
| β-d-Mannosidases | ||||||
| A. aculeatus | 130 | 2.0 | 70 | 4.0 | 356 | |
| A. awamori | 96–100 | 3.8 | 66 | 4.55 | 270 | |
| A. niger | 120–130 | 3.5 | 55 | 4.7 | 46, 103 | |
| A. niger | 135 | 2.4–5.0 | 70 | 5.0 | 3 |
β-Mannosidases (EC 3.2.1.25) are exo-acting enzymes, which release mannose from the nonreducing end of mannooligosaccharides. The substrate specificity of A. niger β-mannosidase has recently been studied (3). The enzyme is able to completely release terminal mannose residues when one or more adjacent unsubstituted mannose residues are present. The presence of a galactose-substituted mannose residue adjacent to the terminal mannose residue reduces the activity of β-mannosidase to 18 to 43%, compared to unsubstituted substrates, depending on the size of the oligosaccharide (3). Both β-mannanase and β-mannosidase have transglycosylation activity (153, 169, 252) and can therefore be used for the synthesis of specific oligosaccharides.
Complete degradation of the galacto(gluco)mannan backbone to mannose by β-mannanase and β-mannosidase also depends on the action of β-glucosidase and α-galactosidase (see “α- and β-d-galactosidases” below). Galactomannan-degrading enzymes are produced when Aspergillus is grown on milled soybean (59), locust bean gum (4), galactomannan (64), and mannose (270). So far, only one β-mannanase-encoding gene (A. aculeatus man1 [accession no. L35487]) (59) and two β-mannosidase -encoding genes (A. aculeatus manB [accession no. AB015509] and A. niger mndA [accession no. AJ251874]) (1, 356) have been reported. Based on the sequences of these genes, the Aspergillus β-mannanase and β-mannosidases are assigned to glycosidase families 5 and 2, respectively. Both types of enzymes use a retaining mechanism for catalysis.
Degradation of the Pectin Backbone
The structural differences between the main chain of the hairy and smooth regions of pectin have implications for the enzymes involved in the degradation of these regions. The backbone of the smooth region can be hydrolyzed by pectin lyases (EC 4.2.2.10), pectate lyases (EC 4.2.2.2), and polygalacturonases (EC 3.2.1.15 and EC 3.2.1.67). In Aspergillus, families of genes encoding these types of enzymes have been identified (51, 140). Several classes of enzymes are involved in the degradation of the hairy-region backbone.
The pectin main-chain-degrading enzymes can be divided into hydrolases (Table 6) and lyases (Table 7). Six types of hydrolases have been identified in aspergilli. Several endopolygalacturonases are produced that all cleave within the pectin smooth region (10, 185, 284), whereas exopolygalacturonases cleave at the nonreducing terminal end of this region (35, 139, 182, 259). The A. aculeatus and A. tubingensis exopolygalacturonases were able to release galacturonic acid from polygalacturonic acid, sugar beet pectin, and xylogalacturonan (35, 180, 182). It also released the dimer β-Xyl-(1,3)-GalA from xylogalacturonan, indicating that the action of the enzyme is not hindered by the presence of xylose on the terminal galacturonic acid residue. The seven endopolygalacturonases produced by A. niger differ in their specific activity (varying from 25 to 4,000 U/mg), sensitivity to methylation of the substrate (36, 183, 281, 284), and mode of action. Four of the enzymes (endopolygalacturonases I, A, C, and D) show processive behavior, also known as multiple attack on a single chain (36, 281, 284), whereas the other three enzymes (endopolygalacturonases II, B, and E) work via a single-attack mechanism (36, 281, 284).
TABLE 6.
Physical properties of Aspergillus endo- and exopolygalacturonases and rhamnogalacturonan hydrolases
| Species and enzyme type | Enzyme | Mol mass (kDa) | pHopt | Topt (°C) | pI | Reference |
|---|---|---|---|---|---|---|
| Endopolygalacturonases | ||||||
| A. alliaceus | endoPG | 40 | 5.5 | 35 | 5.9 | 259 |
| A. carbonarius | PG-I | 61 | 4.0 | 55 | 10 | |
| A. carbonarius | PG-II | 42 | 4.1 | 50 | 10 | |
| A. carbonarius | PG-III | 47 | 4.3 | 55 | 10 | |
| A. niger | E1 | 35 | 4.1 | 68 | ||
| A. niger | E2 | 80 | 3.8 | 68 | ||
| A. niger | Endo-I | 55 | 4.9 | 3.2–3.5 | 185 | |
| A. niger | Endo-II | 38 | 4.8 | 4.6–5.9 | 185 | |
| A. niger | Endo-III A | 57 | 4.3 | 3.3 | 185 | |
| A. niger | Endo-III B | 57 | 4.5 | 3.3 | 185 | |
| A. niger | Endo-IV | 59 | 4.8 | 3.7 | 185 | |
| A. niger | PGA | 35 | 4.0 | 3.43 | 281 | |
| A. niger | PGB | 35 | 5.0 | 6.19 | 281 | |
| A. niger | PGD | 51 | 4.2 | 4.1 | 284 | |
| A. oryzae | PGI | 4.0 | 60 | 372 | ||
| A. oryzae | PGA | 41 | 5.0 | 45 | 195 | |
| A. oryzae | PGB | 39 | 5.0 | 55 | 195 | |
| Exopolygalacturonases | ||||||
| A. aculeatus | 42 | 4.3 | 35 | |||
| A. alliaceus | exoPG1 | 40 | 3.5 | 45–50 | 5.7 | 259 |
| A. alliaceus | exoPG2 | 40 | 6.0 | 30–35 | 6.3 | 259 |
| A. niger | exo-PG I | 66 | 3.8 | 60 | 5.6 | 139 |
| A. niger | exo-PG II | 63 | 4.5 | 60 | 5.8 | 139 |
| A. niger | Exo-I | 4.0 | 185 | |||
| Rhamnogalacturonan hydrolases | ||||||
| A. aculeatus | RhgA | 51 | 3–4 | 40–50 | 325 | |
| A. aculeatus | RGase A | 59 | 3.5 | 30–50 | 4.5 | 203 |
| A. aculeatus | RG-RH | 84 | 4 | 60 | 4.9–5.4 | 265 |
| A. aculeatus | RG-GH | 66 | 4 | 50 | 5.12 | 264 |
TABLE 7.
Physical properties of Aspergillus pectin, pectate, and rhamnogalacturonan lyases
| Species and enzyme type | Enzyme | Mol mass (kDa) | pHopt | Topt (°C) | pI | Reference |
|---|---|---|---|---|---|---|
| Pectin lyase | ||||||
| Aspergillus sp. strain CH-Y-1043 | 8.5–8.8 | 40–45 | 79 | |||
| A. japonicus | 32 | 6.0 | 55 | 7.7 | 161 | |
| A. niger | PL B | 40 | 8.5–9.0 | 5.9 | 184 | |
| A. niger | PLI | 37.5 | 3.65 | 380 | ||
| A. niger | PLII | 37.5 | 3.75 | 380 | ||
| A. oryzae | PL | 8.5 | 50–55 | 372 | ||
| Pectate lyase | ||||||
| A. nidulans | PL A | 40 | 4.2 | 76 | ||
| A. niger | PlyA | 43 | 7.5–8.5 | 38 | ||
| Rhamnogalacturonan lyase | ||||||
| A. aculeatus | RGase B | 55 | 6.0 | 50 | 5.2 | 203 |
Endorhamnogalacturonan hydrolases cleave within the main chain of rhamnogalacturonan and have been identified in several aspergilli (203, 325, 352). These enzymes are severely hindered in their activity by the presence of acetyl residues on the main chain and require the presence of rhamnogalacturonan acetyl esterase (see “Acetyl- and methylesterases” below) for efficient hydrolysis of the rhamnogalacturonan backbone (90). Also characterized are two exo-acting enzymes, rhamnogalacturonan rhamnohydrolase (265) and rhamnogalacturonan galacturonohydrolase (264), that further degrade the oligosaccharides from the nonreducing end. Recently the activity of an endo-acting xylogalacturonase has been characterized (377) that is specific for a xylose-substituted galacturonic acid backbone. The stereochemical course of hydrolysis of several enzymes involved in the degradation of the main chain of the pectin hairy regions was studied recently (39, 297). Enzymes acting on the hairy regions of pectin, exogalacturonase, rhamnogalacturonan hydrolase, rhamnogalacturonan rhamnohydrolase, and α-rhamnosidase (297), as well as enzymes acting on the smooth regions, endopolygalacturonase I and II (39), all hydrolyzed the substrate via an inverting mechanism.
Sequencing of the corresponding genes grouped all Aspergillus endo- and exopolygalacturonases and rhamnogalacturonases in the same glycosidase family, which has an inverting mechanism of hydrolysis (Table 8).
TABLE 8.
Genes encoding Aspergillus polygalacturonases, exopolygalacturonases, rhamnogalacturonases, endoxylogalacturonan hydrolases, pectin lyases, pectate lysases, and rhamnogalacturonan lyases and their assignment to the glycosidase families
| Species | Activitya | Gene(s) | Glycosidase family | Database accession no. | Reference(s) |
|---|---|---|---|---|---|
| A. aculeatus | PG | pgaI | 28 | AF054893 | S. Kauppinen et al., unpublished data |
| A. flavus | PG | pecA, pecB | 28 | U05015, U05020 | 398 |
| A. niger | PG | pgaI, pgaII | 28 | X58892, X58893 | 50, 52 |
| A. niger | PG | pgaA, pgaB | 28 | Y18804, Y18805 | 281 |
| A. niger | PG | pgaD, pgaE | 28 | Y18806, Y14386 | 282, 284 |
| A. niger | PG | pgaC | 28 | X64356 | 51 |
| A. oryzae | PG | pgaA, pgaB | 28 | D14282, AB007769 | 194, 195 |
| A. oryzae | PG | pecA | 28 | AF036848 | 122 |
| A. parasiticus | PG | pecA | 28 | L23523 | 57 |
| A. tubingensis | PG | pgaII | 28 | X58894, X54146 | 52, 321 |
| A. tubingensis | XPG | pgaX | 28 | X99795 | 182 |
| A. aculeatus | RHG | rhgA | 28 | L35499; X83525 | 203, 353 |
| A. niger | RHG | rhgA, rhgB | 28 | X94220, X94221 | 352 |
| A. tubingensis | XGH | xghA | 28 | AJ249460 | 377 |
| A. niger | PEL | pelA, pelB | 1 | X60724, X65552 | 224, 225 |
| A. niger | PEL | pelD | 1 | M55657 | 136 |
| A. nidulans | PLY | pelA | 1 | U05592 | 152 |
| A. niger | PLY | plyA | AJ276331 | 38 | |
| A. aculeatus | RGL | rhgB | 4 | L3550 | 203 |
PG, polygalacturonase; XPG, exopolygalacturonase; RHG, rhamnogalacturonase; XGH, endoxylogalacturonan hydrolase; PEL, pectin lyase; PLY, pectate lyase; RGL; rhamnogalacturonan lyase.
Pectin, pectate, and rhamnogalacturonan lyases cleave the pectin backbone by β-elimination, which results in the formation of a Δ4,5-unsaturated nonreducing end. Pectin lyases prefer substrates with a high degree of methylesterification, whereas pectate lyases prefer those with a low degree of esterification. A clearer distinction between these two types of enzymes can be made based on the absolute requirement of Ca2+ ions for catalysis by pectate lyases versus the lack of Ca2+ ion requirement by pectin lyases (173). Six pectin lyase genes have been identified in A. niger (37), but so far no indications have been obtained for the presence of more than one pectate lyase (38, 76). The A. niger pectin lyases characterized (A, B, and II) prefer substrates with a high degree of esterification.
Only one rhamnogalacturonan lyase has been identified in aspergilli (203, 266). This enzyme has a higher molecular mass than the pectin and pectate lyases and was positively influenced by Ca2+ but did not require Ca2+ ions for catalysis (266). The activity of the enzyme was positively affected by the presence of galactose side chains and negatively affected by the presence of arabinose side chains and acetyl residues (266).
Lyases working on the smooth regions (pectin and pectate lyases) and on the hairy regions (rhamnogalacturonan lyases) of pectin have been assigned to two different glycosidase families (Table 8), indicating that the differences in the structure of the substrates require a different enzyme structure as well. In this respect, the lyases are different from the galacturonan hydrolases, which all belong to the same glycosidase family.
Crystal structures have been obtained for endopolygalacturonase II (385) and pectin lyases A and B (246, 393) from A. niger and for rhamnogalacturonan hydrolase A from A. aculeatus (293). All enzymes have the same β-helical topology.
Accessory Enzymes Involved in the Degradation of Plant Cell Wall Polysaccharides
In contrast to the enzymes described in the previous section, which act on the main chain of plant cell wall polysaccharides, accessory enzymes act on the substituents or the side chains of these structures. Some of these enzymes act on linkages between a main-chain residue and a substituent, whereas other enzymes cleave internal or terminal linkages of side chains. This section deals with the different classes of accessory enzymes produced by aspergilli that act on plant cell wall polysaccharides.
α-d-Xylosidases.
α-d-Xylosidases can release α-linked xylose residues from xyloglucan. Only a limited number of α-xylosidases has been characterized from Aspergillus (Table 9). These enzymes are all highly specific for α-linked xylose residues (410, 411) but differ with respect to the type of glycoside they can hydrolyze. Both enzymes from A. niger were able to act on p-nitrophenyl-α-d-xylanopyranoside, isoprimeverose, and oligosaccharides derived from xyloglucan (244, 245). α-Xylosidase I from A. flavus is also able to act on all three types of substrates (410), but α-xylosidase II from this fungus is active only on p-nitrophenyl-α-d-xylanopyranoside and to a small extent on isoprimeverose (411). α-Xylosidase I from A. flavus is produced constitutively, whereas α-xylosidase II from this fungus is specifically induced by xylose (411).
TABLE 9.
Physical properties of Aspergillus α-d-xylosidases
| Species | Enzyme | Mol mass (kDa) | pHopt | Topt (°C) | pI | Reference |
|---|---|---|---|---|---|---|
| A. flavus | I | 100 | 4.5 | 45 | 410 | |
| A. flavus | II | 67 | 6.0 | 40 | 411 | |
| A. niger | I | 123 | 2.5–3.0 | 45 | 5.6 | 245 |
| A. niger | II | 800 | 2.5–3.0 | 40–45 | 244 |
α-l-Arabinofuranosidases and arabinoxylan arabinofuranohydrolases.
Arabinose residues can be removed by α-l-arabinofuranosidases (EC 3.2.1.55) and arabinoxylan arabinofuranohydrolases. These enzymes and their corresponding genes from many different microorganisms have been studied and have been shown to differ strongly in substrate specificity. Several arabinofuranosidases and arabinoxylan arabinofuranohydrolases have been purified from Aspergillus spp. (Table 10) and studied with respect to their activity on polymeric and oligomeric substrates. The A. niger arabinofuranosidase purified by Kaneko et al. (174) was able to release only terminal α-1,3-linked arabinose residues, whereas arabinofuranosidase B from A. niger was able to release terminal α-1,2-, α-1,3- and α-1,5-linked arabinose residues (34). Unlike some of the arabinofuranosidases, the arabinoxylan arabinofuranohydrolase (AXH) from A. awamori was not able to release arabinose from pectin or pectin-derived oligosaccharides but is highly specific for arabinose residues linked to xylan (209). Wood and McCrae (403) reported the ability of an A. awamori arabinofuranosidase to release feruloylated arabinose residues from wheat straw arabinoxylan. Large differences can be observed when the molecular mass and pI of the arabinofuranosidases characterized are compared (Table 10).
TABLE 10.
Physical properties of Aspergillus arabinofuranosidases, arabinoxylan arabinofuranohydrolases, and endo- and exoarbinanases
| Species and enzyme type | Enzyme | Mol mass (kDa) | pHopt | Topt (°C) | pI | Reference(s) |
|---|---|---|---|---|---|---|
| Arabinofuranosidases | ||||||
| A. aculeatus | B1 | 37 | 3.0–3.5 | 34 | ||
| A. aculeatus | B2 | 37 | 4.0–4.5 | 34 | ||
| A. awamori | 64 | 4.6 | 50 | 3.6, 3.2 | 403 | |
| A. nidulans | α-AFase | 36 | 5.5 | 55 | 4.3 | 108 |
| A. nidulans | AbfB | 65 | 4.0 | 65 | 3.3 | 303 |
| A. niger | AbfA | 83 | 3.4 | 46 | 3.3 | 376 |
| A. niger | AbfB | 61–67 | 3.8–4.0 | 56–60 | 3.5 | 135, 174, 376 |
| A. niger | A | 128 | 4.1 | 50 | 6–6.5 | 315 |
| A. niger | B | 60 | 3.7 | 60 | 5.5–6 | 315 |
| A. niger | 53 | 3.8 | 3.6 | 355 | ||
| A. niger var. Tieghem | α-L-AFS | 64 | 4.5 | 50 | 267 | |
| A. sojae | X-II-A | 34.3 | 5.0 | 50 | 3.9 | 188 |
| A. terreus | AbfA | 39 | 4.0 | 7.5 | 235 | |
| A. terreus | AbfB1 | 59 | 4.0 | 8.3 | 235 | |
| A. terreus | AbfB2 | 59 | 4.0 | 8.5 | 235 | |
| Arabinoxylan arabinofurano- hydrolases | ||||||
| A. awamori | Axh | 32 | 5.0 | 209 | ||
| A. tubingensis | AxhA | 32 | 3.6 | 126 | ||
| Endoarabinases | ||||||
| A. aculeatus | Endo-ara | 45 | 5.5 | 34 | ||
| A | ||||||
| A. nidulans | Abn | 40 | 5.5 | 68 | 3.25 | 303 |
| A. niger | 34.5 | 4.7 | 50–55 | 3.0 | 329 | |
| A. niger | 35 | 5.0 | 50 | 4.5–5.5 | 315 | |
| A. niger | AbnA | 43 | 4.6 | 51 | 3.0 | 376 |
| Exoarabinase | ||||||
| A. niger | 67 | 4.0 | 60 | 2.85 | M. Lahaye and J.-F. Thibault, unpublished results |
Production of arabinofuranosidases has been observed on arabinoxylan (174), sugar beet pulp (110, 376), and l-arabinose and l-arabitol (303, 375). Arabinoxylan arabinofuranohydrolases were produced when Aspergillus was grown on oat straw (209) and birchwood xylan (126). The induction of these enzymes is discussed in more detail below (see “Expression of specific genes responding to different inducers”).
The mode of action of AXH from A. awamori and two arabinofuranosidases (Arafurs A and B) from A. niger was studied on alkali-extractable wheat flour arabinoxylan (207). AXH specifically released α-1,2- and α-1,3-linked arabinose residues from singly substituted xylose residues. Whereas Arafur B was able to release arabinose only from terminal singly substituted residues, AXH and Arafur A were able to release arabinose from both terminal and nonterminal singly substituted xylose residues. AXH and Arafur B were able to release arabinose from the intact polysaccharide as well as from xylooligosaccharides, while Arafur A was able to release arabinose only from xylooligosaccharides. Additionally, AXH was not able to release arabinose from arabinan, sugar beet pulp, or pectin, whereas Arafur A and B were active on these substrates. Based on this information, it can be concluded that AXH is specifically involved in arabinoxylan degradation while Arafurs A and B are more general arabinose-releasing enzymes. Additional information about the substrate specificity of AXH was obtained from a study using a sorghum glucuronoarabinoxylan as a substrate (387, 388). It was demonstrated that AXH was not able to release arabinose from xylose residues adjacent to glucuronic acid-substituted xylose residues. The enzyme was also not able to remove arabinobiose side chains (387, 388).
The difference between arabinoxylan arabinofuranohydrolases and arabinofuranosidases is also apparent with respect to the assignment to the glycosidase families (Table 11). Arabinofuranosidases are assigned to families 51 and 54, which both have a retaining mechanism, whereas arabinoxylan arabinofuranohydrolases belong to family 62. Arabinofuranohydrolase from A. sojae was assigned to family 62 based on the amino acid sequence (189), but it has significantly different substrate specificity from that of AxhA from A. niger (126). The latter enzyme is active only on arabinoxylan, whereas arabinofuranohydrolase also releases arabinose from l-arabinan and arabinogalactan (189). For these enzymes, the hydrolysis mechanism has not yet been elucidated. AbfA from A. niger is assigned to a different family from AbfB from A. niger, which might reflect the differences in the substrate specificity of the enzymes. Both enzymes are able to release arabinose from arabinan and sugar beet pulp, but only AbfB is able to release arabinose from xylan.
TABLE 11.
Genes encoding Aspergillus arabinofuranosidases, AXH, and endoarabinanases and their assignment to the glycosidase families
| Organism | Activitya | Gene | Glycosidase family | Database accession no. | Reference |
|---|---|---|---|---|---|
| A. nidulans | ABF | abfB | 54 | Y13759 | 125 |
| A. niger | ABF | abfA | 51 | L29005 | 112 |
| A. niger | ABF | abfB | 54 | X74777 | 110 |
| A. niger | ABF | abf2 | 54 | U39942 | 71 |
| A. sojae | ABF | 62 | AB032289 | 189 | |
| A. niger | AXH | axhA | 62 | Z78011 | 126 |
| A. tubingensis | AXH | axhA | 62 | Z78010 | 126 |
| A. nigers | ABN | abnA | 43 | L23430 | 109 |
ABF, arabinofuranosidase; ABN, endoarabinanase.
Endo- and exoarabinases.
Endoarabinases (EC 3.2.1.99) hydrolyze the α-1,5-linkages of arabinan polysaccharides, which are present as side chains of pectin. Although some arabinofuranosidases are also able to hydrolyze polymeric arabinan (see the previous section), endoarabinases strongly enhance the efficiency of arabinan degradation and positively influence the action of arabinofuranosidases. So far, no indications have been obtained for the presence of more than one endoarabinase in any Aspergillus sp. (Table 10). The production of endoarabinases by Aspergillus spp. was observed on sugar beet pulp (376) and l-arabinose and l-arabitol (303, 375). In A. niger, induction of AbnA seems to occur simultaneously with the induction of AbfA and AbfB (111).
An analysis of the degradation patterns of linear (1–5)-α-l-arabino-oligosaccharides using A. niger endoarabinase demonstrated that the enzyme is not (or is hardly) able to release terminal residues but preferentially acts on internal linkages (100).
Only one endoarabinase-encoding gene has been found in Aspergillus spp. (Table 11). Based on the sequence of this gene, AbnA was assigned to family 43 of the glycosidases and has a inverting mechanism.
So far, only one exoarabinase has been purified from Aspergillus (228). This enzyme released mainly arabinobiose from sugar beet arabinan, although a small amount of arabinotriose was also liberated.
α- and β-d-galactosidases.
The removal of d-galactose residues from plant cell wall polysaccharides requires the action of α-galactosidases (EC 3.2.1.22) and β-galactosidases (EC 3.2.1.23) (Table 12). β-Galactosidases release terminal galactose residues from the galactan side chains of pectins. α-Galactosidases are involved in the degradation of galacto(gluco)mannan, removing galactose from the mannose residues of the backbone. The presence of terminal β-linked galactose residues in certain galactoglucomannans (338) suggest that both α- and β-galactosidases may play a role in the degradation of these polysaccharides. Studies addressing the activity of α- and β-galactosidases on xylan have not been reported. However, the production of α- and β-galactosidases on crude substrates containing xylan indicates a putative role for these enzymes in the degradation of xylan. Production of α-galactosidases has been reported on arabinoxylan (242), glucose (412), locust bean gum (81), wheat and rice bran (345), lactose and galactose (309), galactomannan (64), and guar flour (5). Aspergillus spp. produce β-galactosidase during growth on arabinoxylan (242), polygalacturonic acid (254), wheat bran (129), and lactose (305). Several different α-galactosidases have been purified from Aspergillus spp. (Table 12), but there are no indications for the production of more than one β-galactosidase by any Aspergillus sp. The differences in molecular mass observed for the purified β-galactosidases (Table 12) are most probably due to strain differences and differences in glycosylation of the enzymes.
TABLE 12.
Physical properties of Aspergillus α- and β-galactosidases and endo- and exogalactanases
| Species and enzyme type | Enzyme | Mol mass (kDa) | pHopt | Topt (°C) | pI | Reference |
|---|---|---|---|---|---|---|
| α-Galactosidases | ||||||
| A. ficuum | 70.8 | 6.0 | 60 | 412 | ||
| A. nidulans | 87 | 4–5 | 50 | 6.3 | 309 | |
| A. niger | AglA | 82 | 4.8 | 93 | ||
| A. niger | AglB | 54 | 4.5 | 50–55 | 4.2–4.6 | 242 |
| A. niger | 95 | 6.0 | 201 | |||
| A. niger | AglA | 82 | 4–4.5 | 50 | 3.73 | 330 |
| A. niger | α-gal I | 94 | 4.5 | 60 | 4.15 | 2 |
| A. niger | α-gal II-IV | 64 | 4.5 | 60 | 4.5–4.8 | 2 |
| A. niger | 45 | 4–4.5 | 5 | |||
| A. niger | 78, 69 | 5 | 50 | 345 | ||
| A. oryzae | 64 | 4.0 | 60 | 11 | ||
| A. tamarii | 88 | 4.2–4.3 | 63 | |||
| A. tamarii | 77.5 | 4.2–4.3 | 63 | |||
| A. tamarii | 56 | 4.8 | 64 | |||
| β-Galactosidases | ||||||
| A. fonsecaeus | 124 | 4.5 | 4.2 | 129 | ||
| A. niger | 93 | 4 | 60–65 | 4.6 | 242 | |
| A. niger | 117 | 4.9 | 133 | |||
| A. phoenicis | 4.0 | 70 | 305 | |||
| Endo-β-1,4-galactanases | ||||||
| A. aculeatus | 42 | 4.25 | 50 | 4–6 | 374 | |
| A. aculeatus | 38 | 3.5–4.0 | 50–55 | 2.8 | 229 | |
| A. aculeatus | 43 | 4.0–4.5 | 40–65 | <3.0 | 61 | |
| A. niger | 46 | 4–5 | 55 | 2.9 | 329 | |
| A. niger | 43 | 4.0 | 50–55 | 4–6 | 374 | |
| A. niger | 32 | 3.5 | 55 | 404 | ||
| A. niger var. Tieghem | 44 | 3.6 | 55 | 267 | ||
| A. sojae | 39.7 | 4.5 | 50 | 3.6 | 190 | |
| Endo-β-1,6-galactanase | ||||||
| A. niger | β-1,6- | 60 | 3.5 | 60 | 48 | |
| Exo-β-1,3-galactanase | ||||||
| A. niger | exo-β-1,3- | 66 | 4.5 | 40–50 | 288 | |
| Exo-β-1,4-galactanase | ||||||
| A. niger | exo-β-1,4- | 90–120 | 3.5 | 60 | 3.8–4.1 | 43 |
For one α-galactosidase from A. niger, α-galactosyltransferase activity has been detected (330). This enzyme transferred an α-galactosyl residue to the 4-position of a galactosyl receptor.
Several genes encoding α-galactosidases have been cloned and characterized from Aspergillus niger (2, 81, 94). Based on their sequence, AglA and AglB have been assigned to glycosidase family 27 (Table 13). AglC is highly homologous to Trichoderma reesei Agl2, a member of family 36, which consists mainly of bacterial α-galactosidases. Based on the sequence of the β-galactosidase-encoding gene (lacA) (223), this enzyme has been assigned to family 35 of the glycosidases. All Aspergillus galactosidases work via a retaining mechanism.
TABLE 13.
Genes encoding Aspergillus α-galactosidases, β-galactosidase, and β-1,4-endogalactanases and their assignment to the glycosidase families
| Species | Activitya | Gene | Glycosidase family | Database accession no. | Reference |
|---|---|---|---|---|---|
| A. niger | AGL | aglA | 27 | X63348 | 81 |
| A. niger | AGL | aglB | 27 | Y18586 | 94 |
| A. niger | AGL | aglC | 36 | AJ251873 | 1 |
| A. niger | LAC | lacA | 35 | L06037 | 223 |
| A. aculeatus | GAL | gal1 | 53 | L34599 | 61 |
| A. niger | GAL | galA | 53 | AJ305303 | R. P. de Vries et al., unpublished |
| A. tubingensis | GAL | galA | 53 | AJ012316 | 378 |
AGL, α-galactosidase; LAC, β-galactosidase; GAL, β-1,4-endogalactanase.
Endo- and exogalactanases.
The galactan side chains of pectin are hydrolyzed by endogalactanases (EC 3.2.1.89), exogalactanases, and β-galactosidases (see the previous section). Endogalactanases are able to hydrolyze the galactan polysaccharides, resulting in the liberation of galactobiose and galactose. Production of endogalactanases was observed on beet pulp (190), soybean (61), and locust bean gum (15). Differences between the enzymes exist with respect to their ability to hydrolyze β-1,3-, β-1,4- or β-1,6 linkages between galactose residues. Two types of arabinogalactans are present as side chains of pectins. Type I consists of a backbone of β-1,4-linked galactopyranose residues, while type II consists of a backbone of β-1,3-linked galactopyranose residues that can be branched by β-1,6-linked galactopyranose residues. For the complete degradation of these polysaccharides, all three types of endogalactanases would be required, but so far mainly β-1,4-endogalactanases have been reported (Table 12). Two exogalactanases have been purified from A. niger. The β-1,4-exogalactanase (43) was able to release galactose from galactooligosaccharides and potato galactan (44, 45). Additionally, this exogalactanase possessed galactose transferase activity (43–45), indicating a possible application for this enzyme in the production of specific galactooligosaccharides and a retaining mechanism of hydrolysis. The β-1,3-exogalactanase (288) was not active against native plant polysaccharides but had a high activity against a β-1,3-galactan obtained from gum arabic by partial acid hydrolysis and two Smith degradations. The enzyme was capable of releasing the β-1,6-side chains of type II arabinogalactans by hydrolyzing the β-1,3-linkages in the main chain adjacent to the branching point (288).
Genes encoding Aspergillus β-1,4-endogalactanase have been reported (Table 13). Based on these sequences, the enzymes were assigned to glycosidase family 53.
α-Glucuronidases.
Glucuronic acid residues and their 4-O-methyl ethers can be removed from the xylan backbone by α-glucuronidases (EC 3.2.1.131). The activity of this enzyme has been detected in a large number of fungal and bacterial culture filtrates, but α-glucuronidases have been purified from only a small number of organisms. α-Glucuronidases have been isolated from A. niger and A. tubingensis (Table 14). The enzyme is active mainly on small xylooligomers and therefore is dependent on the action of endoxylanases. α-Glucuronidases have the highest activity against oligosaccharides, whereas only low or no activity is observed against polymeric substrates (40, 93). Synergy between α-glucuronidases and endoxylanases and between α-glucuronidases and β-xylosidase has been reported (90, 93).
TABLE 14.
Physical properties of Aspergillus α-glucuronidases
| Species | Enzyme | Mol mass (kDa) | pHopt | Topt (°C) | pI | Reference |
|---|---|---|---|---|---|---|
| A. niger 5–16 | CM-I | 130 | 4.8 | 60 | 5.3 | 371 |
| A. niger 5–16 | CM-II | 150 | 4.8 | 60 | 5.3 | 371 |
| A. tubingensis | AguA | 107 | 4.5–6.0 | 70 | 5.2 | 93 |
Two genes encoding Aspergillus α-glucuronidases have been reported (A. niger aguA [accession no. AJ290451] and A. tubingensis aguA [accession no. Y15405]) (93). These genes show significant sequence identity to other fungal and bacterial α-glucuronidases and are assigned to glycosidase family 67. Recently it has been shown that this enzyme has an inverting mechanism (40).
Feruloyl and p-coumaroyl esterases.
Several types of feruloyl and p-coumaroyl esterases can be identified based on their physical properties as well as by substrate specificity (Table 15). All enzymes (except A. awamori p-coumaroyl esterase) are active on methylferulate, which is a synthetic substrate commonly used for feruloyl esterase assays. Studies of the activities of feruloyl esterases against natural substrates have focused mainly on xylan and xylan-derived oligosaccharides, from which most enzymes were able to release ferulic acid. Only two of these enzymes, FaeA (90, 92) and CinnAE (217), have been shown to release ferulic acid from pectin. A comparative study using A. niger FaeA and CinnAE (216) demonstrated a preference of FaeA for substrates with a methoxy group at position 3 of the aromatic ring, and an increase in activity was observed when the number of methoxy groups on the aromatic ring increased. The activity of CinnAE was low or absent on substrates containing a methoxy group at position 3 of the aromatic ring, whereas additional methoxy groups at other positions of the aromatic ring reduced CinnAE activity compared to unsubstituted compounds. Hydroxy substitutions on the aromatic ring increased the activity of CinnAE but reduced FaeA activity. These two enzymes were also studied with respect to their ability to release ferulic acid from oligosaccharides derived from sugar beet pulp and wheat bran (302). FaeA was able to release ferulic acid, which was linked to O-5 of arabinose (as present in wheat arabinoxylan). FaeA was not able to release ferulic acid linked to O-2 of arabinose (as present in sugar beet pectin) but did release ferulic acid linked to O-6 of galactose (also present in sugar beet pectin), suggesting a specificity for the linkage rather than the polymeric compound. CinnAE (FAE-I) was able to release ferulic acid from all oligosaccharides tested but was more active against arabinose-linked ferulic acid (302). These data suggest that the different feruloyl esterases from A. niger have complementary functions in the degradation of cell wall polysaccharides. Although this has not been studied in detail for other organisms, differences in substrate specificity have been identified for other feruloyl esterases. A. awamori produces a coumaroyl esterase, which is unable to hydrolyze feruloyl esters (253). A similar enzyme has not been reported for other organisms, but in nearly all purifications feruloyl esterase activity was monitored using methylferulate as a substrate. Coumaroyl esterase activity would therefore not be detected.
TABLE 15.
Physical properties of Aspergillus feruloyl, acetyl, and methyl esterases
| Species and enzyme type | Enzyme | Mol mass (kDa) | pHopt | Topt (°C) | pI | Reference |
|---|---|---|---|---|---|---|
| Feruloyl esterases | ||||||
| A. awamori | FE | 112 | 3.7 | 253 | ||
| A. awamori | CE | 75 | 4.2 | 253 | ||
| A. awamori | 35 | 5.0 | 45 | 3.8 | 212 | |
| A. niger | Fae-I | 63 | 3.0 | 107 | ||
| A. niger | Fae-II | 29 | 3.6 | 107 | ||
| A. niger | FaeA (FAE-III) | 36 | 5.0 | 60 | 3.3 | 92 |
| A. niger | CinnAE | 75.8 | 6.0 | 50 | 4.8 | 217 |
| A. niger | CE | 120 | 28 | |||
| A. oryzae | FAE | 30 | 4.5–6.0 | 3.6 | 363 | |
| A. tubingensis | FaeA | 36 | 5.0 | 60 | 3.4 | 92 |
| Acetylxylan esterase | ||||||
| A. awamori | ACEA | 31 | 7.0 | 40 | 213 | |
| A. niger | 30.5 | 5.5–6.0 | 50 | 3.0–3.2 | 208 | |
| A. niger | AXE | 35 | 7.0 | 35 | 234 | |
| Acetylgalactoglucomannan esterase | ||||||
| A. niger | 40 | 6 | 46 | 4.1 | 300 | |
| A. oryzae | AGME | 36 | 5.0–5.5 | 4.6 | 364 | |
| Rhamnogalacturonan acetylesterases | ||||||
| A. aculeatus | 32–34 | 6.0 | 40 | 178 | ||
| A. niger | RGAE | 42 | 5.5 | 50 | 4.5–6 | 315 |
| Pectin methylesterase | ||||||
| A. aculeatus | PME | 36.2 | 4.6 | 45 | 3.8 | 60 |
| A. niger | PE | 39 | 4.5 | 40 | 29 | |
| A. niger | 43 | 3.6 | 187 | |||
| Pectin acetyl esterase | ||||||
| A. niger | PAE | 60 | 5.5 | 50 | 4.1 | 332 |
To date, several genes encoding feruloyl esterases have been cloned from Aspergillus (Table 16). A region of the amino acid sequence of FaeA from A. niger and A. tubingensis has homology to the active site of lipases (92). In lipases the active site is a catalytic triad, which consists of a serine, an aspartic acid, and a histidine residue. The spacing between these residues in the amino acid sequences of lipases is conserved and is also present in FaeA, suggesting a similar active site for this enzyme. No lipase activity could be detected for FaeA (9).
TABLE 16.
Genes encoding Aspergillus feruloyl, acetyl- and methylesterases and their assignment to the carbohydrate esterase families.
| Species | Activity | Gene | Carbohydrate esterase family | Database accession no. | Reference |
|---|---|---|---|---|---|
| A. awamori | FE | ferA | AB032760 | T. Koseki et al., unpublished results | |
| A. niger | FE | faeA | Y09330 | 92 | |
| A. niger | FE | faeB | AJ309807 | R. P. de Vries et al., submitted | |
| A. tubingensis | FE | faeA | Y09331 | 92 | |
| A. awamori | AXE | aceA | 1 | D87681 | 213 |
| A. niger | AXE | axeA | 1 | A22880 | 90 |
| A. terreus | AXE | ORF1 | 3 | AF141924 | 187 |
| A. aculeatus | RGAE | rha1 | 12 | X89714 | 178 |
| A. niger | RGAE | rgaeA | 12 | AJ242854 | 90 |
| A. aculeatus | PME | pme1 | 8 | U49378 | 60 |
| A. niger | PME | pmeA | 8 | X54145 | 187 |
| A. oryzae | PME | pmeA | 8 | AB011211 | 196 |
FE, feruloyl esterase; AXE, acetylxylan esterase; PME, pectin methylesterase.
Acetyl- and methylesterases.
Acetylesterases and methylesterases release acetyl and methyl residues from the backbone of cell wall polysaccharides (Table 15). Acetylxylan esterases (EC 3.1.1.72) remove acetyl from O-2 or O-3 of xylose in the xylan-main chain. Although acetylxylan esterase activity has been detected in several aspergilli, such as A. niger, A. japonicus, and A. nidulans (41, 186, 342), only a limited number of acetylxylan esterases have been purified from Aspergillus spp. (208, 213, 351). Unlike most other accessory enzymes, acetylxylan esterases are highly active on the polymeric substrate and are thought to be important for efficient degradation of the xylan backbone by endoxylanases. The presence of the A. niger acetylxylan esterase enabled degradation of steamed birchwood xylan by three types of endoxylanase and a β-xylosidase, which could not degrade this substrate in the absence of the esterase (208), indicating the importance of this enzyme in xylan degradation.
Two acetylglucomannan esterases have been purified from Aspergillus (300, 364). The enzyme purified from A. niger (300) is highly specific for acetylated galacto(gluco)mannan, whereas the A. oryzae esterase is also active (to a lesser extent) on acetylated xylan (361, 364). Both esterases were active on polymeric and oligomeric substrates, and significantly influenced the activity of β-1,4-mannanase. However, the presence of β-1,4-mannanase had a greater influence on the activity of the A. oryzae esterase (364) than on the activity of the A. niger enzyme (300).
The acetyl and methyl residues in the smooth regions of pectins are removed by pectin acetylesterases (332) and pectin methylesterases (EC 3.1.1.11) (113, 187). Several pectin methylesterases have been purified from Aspergillus spp. (29, 60, 187). The ability of polygalacturonases and pectate lyases to degrade the pectin main chain depends on the activity of pectin methylesterase. Recently it has been shown that pectin methylesterase is unable to remove methyl residues from the nonreducing end of the pectin backbone and also cannot deesterify a methyl-esterified galacturonic acid dimer (181). Only one pectin acetyl esterase (PAE) from Aspergillus has been reported so far (332). 1H nuclear magnetic resonance spectroscopy experiments identified differences in the acetyl residues attached to the pectin backbone, and showed that the activity of A. niger PAE did depend on these differences (332). PAE works synergistically with pectin methylesterase and pectin lyase.
A rhamnogalacturonan acetylesterase (RGAE) has been purified from A. aculeatus (178) and from A. niger (90, 332). This enzyme was found to be essential for the action of rhamnogalacturonan hydrolases (90, 178). 1H nuclear magnetic resonance spectroscopy experiments also identified differences in the acetyl residues attached to the rhamnogalacturonan main chain, but RGAE was shown to randomly remove the different types of acetyl residues (332). Pectin acetylesterases can be easily distinguished from rhamnogalaturonan acetylesterases by their activity on triacetin, which cannot be hydrolyzed by the latter enzyme (332).
Only a limited number of genes encoding Aspergillus acetyl- or methylesterases have been reported so far (Table 16). Differences with respect to their substrate specificity are reflected by their assignment to the different carbohydrate esterase families (69).
Synergy between Polysaccharide-Degrading Enzymes
Efficient degradation of polysaccharides requires cooperative or synergistic interactions between the enzymes responsible for cleaving the different linkages. Synergy has been reported for many enzymes from Aspergillus involved in xylan degradation, usually between a main-chain-cleaving enzyme and one or more accessory enzymes. In this section, some examples will be given relating to different plant cell wall polysaccharides demonstrating that synergy is in fact a general phenomenon.
Synergistic action has been observed between endoxylanase, β-xylosidase, arabinoxylan arabinofuranohydrolase, and acetylxylan esterase in the degradation of different (glucuronoarabino)xylans (211). Synergy has also been observed between these enzymes and some of the other xylanolytic enzymes. The release of ferulic acid from xylan by a feruloyl esterase from A. niger was strongly enhanced by the addition of endoxylanases (30, 90, 92). Similarly, both endoxylanase and β-xylosidase positively influenced the release of 4-O-methylglucuronic acid from birchwood xylan by A. tubingensis α-glucuronidase (93). The latter enzyme enhanced the activity of endoxylanase and β-xylosidase on this substrate. Synergy has also been demonstrated between an endoxylanase (XylI) and AXH from A. awamori in the degradation of sorghum glucuronoarabinoxylan (388). A recent study revealed that synergistic interactions in the degradation of xylan not only are present between main-chain-cleaving enzymes and accessory enzymes but also occur among accessory enzymes and that nearly all accessory enzymes positively influence the activity of the main-chain-cleaving enzymes (82). A strong synergistic effect has been observed for the role of A. niger acetylxylan esterase in the hydrolysis of steamed birchwood xylan by three endoxylanases from A. niger (208). The addition of acetylxylan esterase resulted in an increase in the release of xylose and short xylooligosaccharides by a factor of 1.9 to 4.4, 6.8 to 14.7, and 2.5 to 16.3 for endoxylanase I, II, and III, respectively, depending on the incubation time (208).
Only a limited number of studies demonstrating synergy between pectinolytic enzymes from Aspergillus have been reported. Pectin methylesterase from A. aculeatus strongly enhanced the degradation and depolymerization of pectin by polygalacturonases (60). Similarly, RGAE from A. aculeatus had a positive effect on the hydrolysis of the backbone of pectic hairy regions by rhamnogalacturonase A and rhamnogalacturonan lyase from A. aculeatus (178). Although indications for inhibition of RGAE activity by the side chains of the hairy regions were obtained (178), pretreatment of pectin with arabinofuranosidase did not increase the acetyl release by RGAE (331). Pectin lyase positively influenced the release of ferulic acid from sugar beet pectin by a feruloyl esterase from A. niger, but only to a small extent (92). The release of ferulic acid from pectin by a second A. niger feruloyl esterase was positively affected by endoarabinase and arabinofuranosidase from A. niger (219), indicating that synergy also occurs amongst pectinolytic accessory enzymes. Recently, synergy in the degradation of hairy regions from sugar beet pectin was studied using six accessory enzymes and a main-chain-cleaving enzyme (90). The positive effect of RGAE on the degradation of the hairy-region backbone also positively affected the activity of feruloyl esterase A, β-galactosidase, and endogalactanase from A. niger. Additionally, synergistic effects among these three enzymes, an endoarabinase, and an arabinofuranosidase from A. niger were detected.
A similar effect was observed in the degradation of acetylgalactoglucomannan. The presence of galactose and acetyl residues on the backbone severely hindered the activity of β-mannanase (300). The presence of acetylmannanesterase and to a lesser extent α-galactosidase significantly increased the β-mannanase activity on this substrate. Additionally, the action of β-mannanase and α-galactosidase on acetylgalactoglucomannan was positively influenced by the removal of acetyl residues from the main chain by acetylgalactoglucomannan esterase (362, 364).
REGULATION OF GENE EXPRESSION
Coordinated Expression of Genes Encoding Xylanolytic and Cellulolytic Enzymes
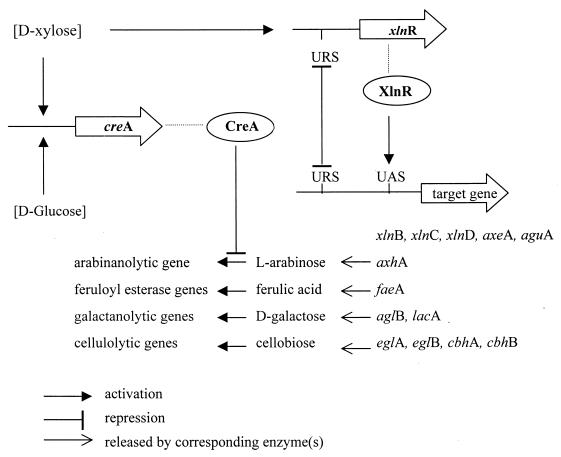
Xylanolytic enzymes from Aspergilli have all been found to be produced on xylose-, xylan-, and crude-xylan-containing substrates but not on other monomeric (e.g. glucose, galactose) or polymeric (e.g., cellulose and pectin) substrates. However, several cellulolytic enzymes are also produced in the presence of xylan and xylose. This suggests a general system of regulation of the genes encoding these enzymes. Several genes encoding xylanolytic enzymes have been studied with respect to regulation of expression, and all demonstrated expression in the presence of d-xylose, xylobiose, or xylan (78, 93, 108, 126, 222, 292, 382) but repression of expression in the presence of glucose (see “Carbon catabolite repression” below). Expression of some xylanolytic genes was also observed on l-arabinose (93, 95, 222). This might be due to the presence of small amounts of d-xylose in the l-arabinose preparations used, as detected in the l-arabinose preparation obtained from Sigma (R. P. de Vries, unpublished results). Additionally, induction of xylanolytic enzymes was observed on cellobiose and cellulose (154) and on a heterodisaccharide consisting of glucose and xylose (Glcβ1-2Xyl) (155) in A. terreus, which are compounds that also induce the cellulolytic system from this fungus. Sophorose and 2-O-β-d-xylopyranosyl-d-xylose specifically induced the synthesis of cellulolytic and xylanolytic enzymes, respectively, in this fungus (155). A xylose-induced, glucose-repressed, endoxylanase-encoding gene (xynG1) from A. oryzae was expressed in A. nidulans, resulting in expression of the gene on xylose as well as glucose (191). This indicates that regulation of the expression of xylanolytic genes is not identical in A. oryzae and A. nidulans, although identical regulation of xylanolytic genes has been reported for A. niger and A. tubingensis (78).
From A. niger a gene encoding a transcriptional activator has been isolated by complementation of a mutant unable to degrade xylan (384). Sequence analysis of this factor, XlnR, demonstrated that it is a member of the GAL4-like family of transcriptional activators. Characterization of XlnR showed that it was responsible for the expression of genes encoding endoxylanase and β-xylosidase. Analysis of the promoter region of these genes identified a putative XlnR binding site, GGCTAAA, of which the second G was determined to be essential for XlnR binding by band mobility shift assays and in vivo (384). A more detailed analysis of the role of XlnR in the regulation of genes involved in xylan, arabinan, and cellulose degradation indicated that this protein does not activate only the expression of xylanolytic genes (383). Genes encoding two endoxylanases (xlnB and xlnC), a β-xylosidase (xlnD), an arabinoxylan arabinofuranohydrolase (axhA), an acetylxylan esterase (axeA), an α-glucuronidase (aguA), a feruloyl esterase (faeA), and two endoglucanase (eglA and eglB) were found to be regulated by XlnR (383). However, genes encoding α-arabinofuranosidase (abfB) and β-glucosidase (bglA) were not regulated by this protein. This indicates that in addition to its role as a xylanolytic activator, XlnR also regulates the expression of some, but not all, genes encoding cellulolytic enzymes. A subsequent study demonstrated that XlnR is also involved in the regulation of α- and β-galactosidase genes (aglB and lacA respectively) (94), two cellobiohydrolase-encoding genes (cbhA and cbhB) (124), and a gene encoding xylose reductase (142). Analysis of the promoter regions of the genes that are regulated by XlnR demonstrated that the third A in the consensus for the binding site is variable, and the consensus sequence was therefore shortened to GGCTAA (383). However, the presence of a putative XlnR binding site does not automatically imply regulation by XlnR. A putative XlnR binding site was detected in the promoter of an endoglucanase from A. nidulans, but no expression of this gene was detected on xylose (58). Introduction of a large number of copies of a xylanolytic gene or the promoter of a xylanolytic gene results in a decrease in the expression of other XlnR-regulated genes (197, 381). This indicates that the production of XlnR is tightly balanced with the number of genes this protein regulates. Indications for a transcription activator from A. oryzae binding to a similar DNA region have been obtained (176).
Recently a model was suggested for the role of XlnR in the regulation of (hemi)cellulose degradation by A. niger (Fig. 7) (87). XlnR is activated during growth of A. niger in the presence of arabinoxylan by monomeric xylose which is already present in the substrate or released by endoxylanase B and β-xylosidase that are present at low constitutive levels. XlnR then activates the expression of (hemi)cellulolytic genes (94, 124, 383). However if the concentration of xylose is high, this causes CreA-mediated repression, resulting in reduced expression of these genes (see “Carbon catabolite repression” below). This effect is stronger in the presence of glucose, which prevents the expression of (hemi)cellulolytic genes to a large extent (78, 93). XlnR-mediated expression of (hemi)cellulolytic genes results in the release of arabinose, cellobiose, ferulic acid, and release galactose by the enzymes encoded by these genes. These compounds induce the expression of other genes (see “Expression of specific genes responding to different inducers” below).
FIG. 7.
Model for the role of XlnR and CreA in the regulation of the genes encoding (hemi)cellulose-degrading enzymes by A. niger. Other regulatory factors, such as the HAP complex and PacC, are not considered in this model. UAS, Upstream Activating Sequence; URS, Upstream Repressing Sequence. (Reprinted from reference 87 with permission of the publisher.)
Expression of Pectinolytic Genes
The production of pectin main-chain-cleaving enzymes (polygalacturonases, pectin lyases, rhamnogalacturonan hydrolases, and lyases) has been detected on sugar beet pectin (53), apple pectin (140, 352), polygalacturonic acid (320), a combination of rhamnose and galacturonic acid (354), and soybean flour (204). However, some pectinolytic enzymes have also been reported to be produced constitutively (259, 281). Expression of genes encoding pectinolytic main-chain-cleaving enzymes has been reported to occur on apple pectin (140, 224, 225), sugar beet pulp (140, 224, 225), galacturonic acid (182), and polygalacturonic acid (76). Strong differences in expression pattern are observed for the different polygalacturonase-encoding genes. Constitutively expressed polygalacturonase-encoding genes have been reported from A. flavus (398), A. parasiticus (57), and A. niger (281). In contrast, no expression could be detected on pectin, polygalacturonic acid, or galacturonic acid for the gene encoding endopolygalacturonase E from A. niger (282). Three other polygalacturonase-encoding genes from A. niger were induced in the presence of sugar beet pulp, and a promoter deletion analysis of one of these genes (pgaII) identified a region which was important for high-level gene expression (5′-TYATTGGTGGA-3′) (53, 392). A region of high similarity was detected in the promoter of pgaA from A. niger (281). This gene showed increased expression on d-galacturonic acid, which might be attributed to this region. The expression of pgaB, which does not contain this region, is not increased on galacturonic acid, although it has a similar expression profile to that of pgaA on sucrose and pectin. Comparison of this region to the promoters of the other genes (pgaI and pgaC) (392) identified a consensus sequence, which is similar to an upstream activating sequence of the Saccharomyces cerevisiae cycI gene, to which the HAP2/3/4-activating complex binds (see “Regulation by a HAP-like CCAAT binding complex” below). Additionally, a hexanucleotide sequence was detected in the promoters of several pectin lyase-encoding genes (CCCTGA) (37). However, the functions of these sequences have not yet been studied in detail.
Recently, additional evidence has been obtained for the role of galacturonic acid as a general inducer for pectinolytic genes in A. niger. Several genes encoding pectin main-chain-cleaving enzymes (pelA, plyA, pgaX, and rglA) and a gene encoding pectin methylesterase (pmeA) are expressed in the presence of galacturonic acid (280). Genes encoding arabinofuranosidases (abfA and abfB), endoarabinase (abnA), endogalactanase (galA), and β-galactosidase (lacA), all of which act on the pectin side chains, are also expressed on galacturonic acid (de Vries, unpublished). In addition, a gene encoding a xylanolytic function (α-glucuronidase, aguA) is expressed on galacturonic acid and glucuronic acid (de Vries, unpublished). This may indicate a general system activating expression in response to the presence of galacturonic (and glucuronic) acid.
Expression of Specific Genes Responding to Different Inducers
Apart from the transcription activation by XlnR (xylan degradation) and the induction on galacturonic acid (pectin degradation), other monomeric compounds also induce the expression of specific sets of genes. As mentioned above, arabinofuranosidases and endoarabinases are induced when Aspergillus is grown on sugar beet pulp whereas AXH are produced during growth on xylan (see “α-l-Arabinofranosidases and arabinoxylan arabinofuranohydrolases” above). Expression of the genes encoding these enzymes was also observed on monomeric carbon sources, such as l-arabinose and l-arabitol (111, 125, 126, 303, 375). The expression of two arabinofuranosidase-encoding genes (abfA and abfB) and one endoarabinase-encoding gene (abnA) has been studied in A. niger (111). Transformants containing additional copies of one of the genes showed reduced expression levels of the other two genes compared to a wild-type strain. This indicated that these genes are most probably under coordinated control of a common specific transcription factor. The reduced expression would then be caused by titration of the transcription factor by the additional copies of one of the genes. Indications that l-arabitol, and not l-arabinose, is the true inducing compound of this regulatory system were obtained from studies with an A. nidulans mutant defective in l-arabitol dehydrogenase activity (88). This mutant accumulated l-arabitol when grown on media containing glycerol and l-arabitol. Under these conditions, increased arabinofuranosidase, endoarabinase, and l-arabinose reductase activity was observed compared to a wild-type strain. These data also suggest coordinated induction of extracellular arabinose-releasing enzymes and enzymes involved in l-arabinose catabolism. This was confirmed in A. niger, where induction of arabinofuranosidases and endoarabinases occurred simultaneously with the induction of intracellular enzyme activities involved in l-arabinose catabolism (l-arabinose reductase and l-arabitol dehydrogenase) (375). In addition to the arabinases and the arabinose metabolic pathway functions, an endogalactanase-encoding gene (galA) and a β-galactosidase-encoding gene (lacA) are expressed in the presence of arabinose and arabitol (de Vries, unpublished), suggesting coordinated regulation of all pectin side-chain-cleaving functions in A. niger. Galactose and mannose are involved in the induction of α-galactosidases and β-mannosidases (1). However the expression pattern of α-galactosidases from A. niger varies significantly among the individual genes. den Herder et al. (81) demonstrated that the A. niger α-galactosidase A-encoding gene (aglA) was expressed on galactomannan. A recent study (94) compared the expression of this gene to two other α-galactosidase-encoding genes from A. niger (aglB and aglC) and the A. niger β-galactosidase-encoding gene (lacA). Of the three α-galactosidase -encoding genes, only aglA seemed to be specifically expressed on galactose and galactose-containing oligo- and polysaccharides. The aglB gene was constitutively expressed on all carbon sources tested but had increased expression on xylose and xylan. This has been attributed to regulation by the xylanolytic transcriptional activator XlnR (see “Coordinated expression of genes encoding xylanolytic and cellulolytic enzymes” above). Expression of aglC was detected only on glucose, which is surprising for an α-galactosidase-encoding gene. The expression of lacA was highest on xylose and xylan (regulated by XlnR [see above]), arabinose, and pectin (see “Expression of pectinolytic genes” above), whereas only low expression was observed on galactose. The expression on pectin is akin to the production of this enzyme by A. oryzae on polygalacturonic acid (254) and might indicate that this gene is coregulated with genes encoding pectin main-chain-degrading enzymes (see “Expression of pectinolytic genes” above).
The induction of faeA from A. niger by the product of the enzyme, ferulic acid, has recently been studied (95, 106). This gene is expressed during growth on xylan and xylose, which is mediated by XlnR (see “Coordinated expression of genes encoding xylanolytic and cellulolytic enzymes” above). The addition of ferulic acid to media containing xylan or xylose increased the expression of faeA, whereas other xylanolytic genes were unaffected, indicating a second regulatory system for the induction of faeA. Although the two systems positively influence each other with respect to the expression of faeA, ferulic acid-induced expression is not dependent on XlnR (95). In an XlnR-negative mutant, the level of expression of faeA was similar on ferulic acid and a combination of ferulic acid and xylose, while no expression of other xylanolytic genes was detected under these conditions.
Carbon Catabolite Repression
The major system responsible for carbon repression in Aspergillus is mediated by the carbon catabolite repressor protein CreA (99, 319). CreA is a zinc finger protein which binds to specific sites in the promoters (SYGGRG) of a wide range of target genes (221). In the presence of easily metabolizable substrates, such as glucose or fructose, CreA inhibits or decreases the expression of the target genes. Several A. nidulans and A. niger CreA mutants have been isolated which display (partially) derepressed phenotypes (19, 318, 336). Using these mutants, CreA-mediated repression of gene expression has been detected for the proline and alcohol gene clusters in A. nidulans (72, 278) and for several extracellular proteases in A. niger (172). Recently, the mechanism of CreA-mediated repression has been investigated in more detail (349). It appears that the creA gene is strongly expressed when monomeric repressing carbon sources are added to the culture, but CreA quickly down-regulates the expression of its own gene. This autoregulation has been demonstrated to be dependent on the formation of glucose-6-phosphate (349). Under carbon-derepressing conditions, a significantly higher creA expression was observed, but this did not result in the formation of the CreA-DNA complex. Formation of this complex was dependent on de novo protein synthesis (349). At this point it is not clear whether the change from active to inactive CreA and vice versa is caused by covalent modification of CreA or by protein degradation.
The influence of CreA on the plant cell wall-degrading enzyme systems from Aspergillus has been extensively studied. CreA-mediated repression in Aspergillus was detected for genes encoding arabinases and l-arabinose catabolic enzymes (318), several endoxylanases (75, 108, 292, 296, 318), and other xylanolytic functions such as β-xylosidase (222), AXH (126), and feruloyl esterase (95). Some of the pectinolytic genes from Aspergillus are also repressed in the presence of glucose (52, 178, 182, 241, 344).
Removal of four putative CreA binding sites from the promoter of the A. tubingensis xlnA gene resulted in an increased expression of this gene (78), indicating that at least one of these sites is involved in CreA binding. Gel mobility shift analysis of a fragment of the promoter of the A. nidulans pectate lyase-encoding gene (pelA) using a fusion protein containing the N-terminal part of CreA demonstrated binding of the fusion protein to this fragment (152) and a potential role for CreA in the regulation of (some) pectinolytic genes.
CreA-mediated repression is not restricted to glucose and fructose alone. Other monomeric carbon sources also result in (weaker) CreA-mediated repression of gene expression. Xylose is commonly regarded as the monomeric inducer of xylanolytic gene expression in Aspergillus (see “Coordinated expression of genes encoding xylanolytic and cellulolytic enzymes” above). Recently, the effect of different xylose concentrations on xylanolytic gene expression was studied (96), demonstrating that high concentrations of xylose result in CreA-mediated repression of the expression of xylanolytic genes. At lower xylose concentrations, higher expression levels were observed. A similar phenomenon was observed for the regulation of cellulase biosynthesis in A. terreus (8). At high concentrations of glucose, xylose, and cellobiose, a decrease in cellulase production was observed.
The ferulic acid-induced expression of the feruloyl esterase A-encoding gene (faeA) from A. niger was studied in combination with a number of different monomeric carbon sources (de Vries, unpublished). For nearly all combinations, higher faeA expression was detected in a CreA-derepressed mutant than in a wild-type strain, indicating that CreA-mediated repression occurred in the presence of all these carbon sources. Similarly, the addition of glycerol and glucose reduced the production of cellulolytic enzymes in A. nidulans (24) and A. japonicus (323), respectively. Addition of cyclic AMP relieved catabolite expression of cellulase production in glycerol-repressed A. nidulans, indicating a role for this compound in carbon catabolite repression (23).
pH-Dependent Expression
The major factor involved in pH-dependent expression in Aspergillus is the pH-regulatory protein PacC. At alkaline pH, PacC activates alkalin-expressed genes and represses acid-expressed genes; therefore, PacC has a dual function (368). The gene encoding this protein (pacC) has been cloned from A. nidulans (368) and was shown to complement A. nidulans pH-regulatory mutants, previously isolated as mutants defective in phosphatase expression (16, 18, 98). An A. niger homologue has also been cloned (240). PacC is activated under alkaline conditions by proteolytic modification of the C-terminal region, which enables it to bind to specific target sites (GCCARG, with R being A or G) in the promoter of target genes (276). Six genes (palA, palB, palC, palF, palH, and palI) are believed to be involved in the signal transduction pathway leading to the activation of PacC (17, 54, 368). A more detailed description of pH regulation of gene expression in fungi can be found in a recent review (82).
pH regulation of genes encoding cell wall-degrading enzymes has not been studied in detail in Aspergillus. However, indications for pH-dependent expression of xylanolytic and pectinolytic genes have been obtained. The production of different polygalacturonases was studied in A. kawachii using culture media with different pHs (204). From culture medium at pH 2, two polygalacturonases were purified, whereas only one polygalacturonase could be purified from culture medium at pH 5. The N-terminal amino acid sequences of these enzymes were different, demonstrating that the pH of the culture medium influences which polygalacturonase is produced by A. kawachii. Using PacC mutants, it was shown that in A. nidulans the expression of the major arabinofuranosidase-encoding gene (abfB) (125) and two endoxylanase-encoding genes (xlnA and xlnB) (239) is regulated by PacC. These two endoxylanase-encoding genes have opposite expression patterns with xlnA being expressed under alkaline conditions and xlnB being expressed under acidic conditions. Analysis of the promoter regions of xlnA and xlnB revealed the presence of two and one PacC consensus sites, respectively (239). Several putative PacC binding sites were also detected in the promoter of the A. nidulans β-xylosidase-encoding gene (xlnD), but no clear indications for PacC control of this gene were obtained (292).
The production of cellulases by A. fumigatus was reported to depend strongly on the pH (347). During growth on cellulose with ammonia as the nitrogen source, this species acidified the media, resulting in an increased cellulase production compared to cultures with nitrate as the nitrogen source.
Regulation by a HAP-Like CCAAT Binding Complex
The promoters of many Aspergillus genes, including genes encoding polysaccharide-degrading enzymes, contain CCAAT boxes. Recently, genes have been cloned from A. nidulans encoding homologues of the subunits of the HAP complex of S. cerevisiae. The hapC gene (279) has significant similarity to HAP3 of S. cerevisiae, whereas hapB and hapE contain regions of high similarity to S. cerevisiae HAP2 and HAP5, respectively. These three proteins have been produced in Escherichia coli and were shown to be necessary and sufficient for CCAAT binding in vitro (346). So far, no homologue of S. cerevisiae HAP4 has been detected in Aspergillus or other filamentous fungi. The function of a HAP-like complex has been studied in Aspergillus for the expression of amdS (acetamidase) (379), taaG2 (taka-amaylase) (269), and genes involved in penicillin biosynthesis (365), and the complex is involved in the enhancement of the expression levels of these genes. No studies of the role of HAP-like complexes in the regulation of genes encoding polysaccharide-degrading genes from Aspergillus have been reported, although CCAAT boxes have been identified in the promoters of many of the genes from Aspergillus encoding plant cell wall polysaccharide-degrading enzymes. However, the presence of CCAAT boxes and the involvement of HAP-like complexes in the regulation of xylanases (415) and cellulases (414) in Trichoderma reesei suggests that these systems might also be regulated by HAP-like complexes in Aspergillus.
INDUSTRIAL APPLICATIONS
The enzymes described in this review are involved in the degradation of complex plant cell wall polysaccharides. Plant cell walls are a major part of the crude biomass which is used in a wide variety of industrial processes. A first step in the industrial processing of biomass frequently involves (partial) degradation of the polymeric fraction. It is therefore obvious that enzymes capable of degrading the plant cell wall can be applied in many of these processes and provide a good alternative to chemical processing. In this section, examples of industrial applications of plant cell wall-degrading enzymes are given.
Applications of xylanolytic enzymes can be found in a variety of industrial processes. In the pulp and paper industry cellulase-free xylanolytic enzyme preparations can be of great value in the biobleaching of pulps (62, 390). Enzymatic degradation of the hemicellulose-lignin complexes present in pulps leaves the cellulose fibers intact and strongly reduces the amount of bleaching chemicals (e.g., chlorine) required. This not only results in a reduction in costs of chemicals but also reduces the environmental problems caused by the use of chlorine. The most important enzyme that is used in enzyme-aided bleaching is endoxylanase (62, 390), but the addition of other xylanolytic enzymes has also been shown to be effective (175). A second area in which the xylanolytic enzyme preparations are widely used is the bakery industry. In this context, their main effect is in solubilizing the arabinoxylan fraction of the dough, resulting in increased bread volume and an improved quality of the dough (237, 295, 298). Other applications in which xylanolytic enzymes are used include increasing the feed conversion efficiency of animal feed (32), clarifying juices (413), and producing xylose, xylobiose and xylooligomers (42, 289, 299). The oligosaccharides produced are used as functional food additives or alternative sweeteners with beneficial properties.
α-Galactosidases are used to improve the gelling capacity of galactomannans, which have applications in the food industry as well in the cosmetic and pharmaceutical industries (47, 70). Additionally, they reduce the concentration of raffinose and other oligosaccharides in soybean milk (262), cowpea meal (345), and sugar beet syrup (121).
Pectinases are of major importance in the beverage industry due to their ability to improve pressing and clarification of concentrated fruit juices (132). Pectin methylesterase and other pectinolytic enzymes are also used for the production of carrot puree (144). Whereas xylanolytic enzymes are used in the paper and pulp industry mainly for biobleaching, pectinolytic enzymes are used in enzymatic debarking (27, 304). Removal of the bark is the first step in all processing of wood and is traditionally a very energy-consuming process. A reduction in the amount of energy required can be obtained by using pectinolytic enzymes to aid in the process. Other applications for pectinolytic enzymes include increasing the yield of lemon peel oils (65) and the production of oligouronides (316).
The monosaccharides that are the building blocks of the pectin polymer all have different food and nonfood applications, and enzymatic release of these compounds is an important tool in industrial processes. Arabinose is a precursor of l-fructose and l-glucose, which can be used as sweeteners, and it can also be transformed to 5-deoxy-l-arabinose, a compound that has anti-Parkinson properties (394). Galacturonic acid can be enzymatically converted into l-ascorbic acid (220) or can be used to produce surface-active agents by esterification with various fatty acids (294). Chemical transformations of rhamnose result in aromas such as “furaneol,” which is used in caramel and fruit flavors (402).
All industrial applications to date utilize the enzymes during the processing of the crude plant material. Recently, in vivo modifications of plant cell wall polysaccharides using Aspergillus enzymes has received increasing interest. Transgenic plants have been obtained containing the A. aculeatus endogalactanase-encoding gene (277). The galactosyl content of rhamnogalacturonan I in these plants was reduced by 70%, indicating the potential applications of introducing Aspergillus genes in plants for in vivo polysaccharide modifications.
CONCLUDING REMARKS
We have presented an overview of the large number of enzymes and genes from Aspergilli involved in the degradation of plant cell wall polysaccharides. Although the enzymes reported cover most of the functions necessary for the complete degradation of plant cell wall polysaccharides, some functions remain to be isolated (e.g., enzymes removing fucose residues from xyloglucan). Also, the substrate specificity and mode of action of many of the enzymes have not been studied in detail. A good example of this is β-galactosidase. So far, only a single β-galactosidase has been found in any Aspergillus species, although β-linked galactose residues are found in xylan, xyloglucan, and pectin side chains. Whether this single enzyme is able to hydrolyze all these linkages or whether other β-galactosidases are produced by aspergilli is unclear at this point. The availability of genome sequences for several Aspergillus species will accelerate the discovery in the near future of new genes encoding polysaccharide-degrading enzymes.
Only recently, plant proteins have been identified that are able to inhibit the action of some fungal plant cell wal polysaccharide-degrading enzymes. The polygalacturonase inhibitor protein (PGIP) has been studied in the most detail and has been proposed to form a part of the plant “immune system” (80). Evidence for the production of PGIPs in plants as a response to wounding but not to fungal infection was obtained (84). Fungal pathogens produce a family of polygalacturonases, suggesting the neccessity of several PGIPs with different specificities. Recent studies confirmed this by demonstrating that inhibition depends on the type of PGIP and the type of endopolygalacturonase (67, 83, 348). A glycoprotein was identified in kiwi fruit that inhibits the action of a different pectin-degrading enzyme, pectin methylesterase (55, 127). In addition, several plant proteins have been isolated that inhibit the activity of endoxylanases (77, 255, 317). One of these studies also produced evidence for the production of an arabinofuranosidase inhibitor by wheat (317). It is likely that future studies will identify other plant proteins capable of inhibiting the action of fungal plant cell wall polysaccharide-degrading enzymes.
A second topic that still requires intensive study is the regulation of the genes encoding plant cell wall polysaccharide-degrading enzymes. So far only one negatively acting factor (CreA [see “Carbon catabolite repression” above]) and one positively acting factor (XlnR [see “Coordinated expression of genes encoding xylanolytic and cellulolytic enzymes” above]) have been studied in detail. However, evidence for the existence of positively acting factors responding to galacturonic and glucuronic acid (see “Expression of pectiolytic genes” above) arabinose and arabitol, galactose, and ferulic acid (see “Expression of specific genes responding to different inducers” above) has been obtained. It has also become clear that these factors are not specific for the genes involved in the degradation of one particular polysaccharide. Expression profiling and proteomics will be powerful tools to further elucidate the complexity of these systems.
A number of studies report the production of Aspergillus plant cell wall polysaccharide-degrading enzymes in other organisms, in particular S. cerevisiae (see e.g., references 73, 231, and 268). Most plant cell wall-degrading enzymes as produced by Aspergillus are highly glycosylated proteins. When these enzymes are produced by S. cerevisiae, they are usually overglycosylated and one should keep in mind that the properties might differ from those of the native enzyme.
REFERENCES
- 1.Ademark, P., R. P. de Vries, H. Stålbrand, and J. Visser. Cloning and characterisation of genes encoding a β-mannosidase and an α-galactosidase from Aspergillus niger involved in galactomannan degradation. Eur. J. Biochem. 268:2982–2990. [DOI] [PubMed]
- 2.Ademark P, Larsson M, Tjerneld F, Stålbrand H. Multiple α-galactosidases from Aspergillus niger: purification, characterization, and substrate specificities. Enzyme Microb Technol. 2001;29:441–448. [Google Scholar]
- 3.Ademark P, Lundqvist J, Hagglund P, Tenkanen M, Torto N, Tjerneld F, Stålbrand H. Hydrolytic properties of a β-mannosidase purified from Aspergillus niger. J Biotechnol. 1999;75:281–289. doi: 10.1016/s0168-1656(99)00172-8. [DOI] [PubMed] [Google Scholar]
- 4.Ademark P, Varga A, Medve J, Harjunpaa V, Drakenberg T, Tjerneld F, Stålbrand H. Softwood hemicellulose-degrading enzymes from Aspergillus niger: purification and properties of a β-mannanase. J Biotechnol. 1998;63:199–200. doi: 10.1016/s0168-1656(98)00086-8. [DOI] [PubMed] [Google Scholar]
- 5.Adya S, Elbein A D. Glycoprotein enzymes secreted by Aspergillus niger: purification and properties of a α-galactosidase. J Bacteriol. 1977;129:850–856. doi: 10.1128/jb.129.2.850-856.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ahlgren E, Eriksson K-E, Vesterberg O. Characterization of cellulases and related enzymes by isoelectric focusing, gel-filtration and zone electrophoresis. Acta Chem Scand. 1967;21:937–944. [Google Scholar]
- 7.Akiba S, Kimura Y, Yamamoto K, Kumagai H. Purification and characterization of a protease-resistant cellulase from Aspergillus niger. J Ferment Bioeng. 1995;79:125–130. [Google Scholar]
- 8.Ali S, Sayed A. Regulation of cellulase biosynthesis in Aspergillus terreus. World J Microbiol Biotechnol. 1992;8:73–75. doi: 10.1007/BF01200691. [DOI] [PubMed] [Google Scholar]
- 9.Aliwan F O, Kroon P A, Faulds C B, Pickersgill R, Williamson G. Ferulic acid esterase-III from Aspergillus niger does not exhibit lipase activity. J Sci Food Agric. 1999;79:457–459. [Google Scholar]
- 10.Anjana Devi N, Appu Rao A G. Fractionation, purification, and preliminary characterization of polygalacturonases produced by Aspergillus carbonarius. Enzyme Microb Technol. 1996;18:59–65. [Google Scholar]
- 11.Annunziato M E, Mahoney R R. Partial purification and characterization of α-galactosidase from Aspergillus oryzae. J Food Biochem. 1987;11:263–277. [Google Scholar]
- 12.Reference deleted.
- 13.Reference deleted.
- 14.Reference deleted.
- 15.Araujo A, Ward O P. Extracellular mannanases and galactanases from selected fungi. J Ind Microbiol. 1990;6:171–178. [Google Scholar]
- 16.Arst H N, Jr, Bailey C R, Penfold H A. A possible role for acid phosphatase in γ-amino-n-butyrate uptake in Aspergillus nidulans. Arch Microbiol. 1980;125:153–158. doi: 10.1007/BF00403213. [DOI] [PubMed] [Google Scholar]
- 17.Arst H N, Jr, Bignell E, Tilburn J. Two new genes involved in signalling ambient pH in Aspergillus nidulans. Mol Gen Genet. 1994;245:787–790. doi: 10.1007/BF00297286. [DOI] [PubMed] [Google Scholar]
- 18.Arst H N, Jr, Cove D J. Molybdate metabolism in Aspergillus nidulans. II. Mutations affecting phosphatase activity or galactose utilization. Mol Gen Genet. 1970;108:146–153. doi: 10.1007/BF02430520. [DOI] [PubMed] [Google Scholar]
- 19.Arst H N, Jr, Tollervey D, Dowzer C E A, Kelly J M. An inversion truncating the creA gene of Aspergillus nidulans results in carbon catabolite derepression. Mol Microbiol. 1990;4:851–854. doi: 10.1111/j.1365-2958.1990.tb00656.x. [DOI] [PubMed] [Google Scholar]
- 20.Aspinall G O. Chemistry of cell wall polysaccharides. In: Preiss J, editor. The biochemistry of plants. Vol. 3. New York, N.Y: Academic Press, Inc.; 1980. pp. 473–500. [Google Scholar]
- 21.Aziz N H, Farag S E, Mousa L A A, Abo-Zaid M A. Comparative antibacterial and antifungal effects of some phenolic compounds. Microbios. 1998;93:43–54. [PubMed] [Google Scholar]
- 22.Bach Tuyet Lam T, Iiyama K, Stone B A. Cinnamic acid bridges between cell wall polymers in wheat and phalaris internodes. Phytochemistry. 1992;31:1179–1183. [Google Scholar]
- 23.Bagga P S, Sandhu D K, Sharma S. Effect of exogenous cyclic AMP on catabolite repression of cellulase formation in Aspergillus nidulans. Acta Biotechnol. 1991;11:395–402. [Google Scholar]
- 24.Bagga P S, Sandhu D K, Sharma S. Purification and characterization of cellulolytic enzymes produced by Aspergillus nidulans. J Appl Bacteriol. 1990;68:61–68. doi: 10.1111/j.1365-2672.1990.tb02549.x. [DOI] [PubMed] [Google Scholar]
- 25.Bagga P S, Sharma S, Sandhu D K. Developmentally related changes in the production and expression of endo-β-1,4-glucanases in Aspergillus nidulans. Genome. 1988;32:288–292. doi: 10.1139/g89-442. [DOI] [PubMed] [Google Scholar]
- 26.Bailey M J, Puls J, Poutanen K. Purification and properties of two xylanases from Aspergillus oryzae. Biotechnol Appl Biochem. 1991;13:380–389. [Google Scholar]
- 27.Bajpai P. Microbial xylanolytic enzyme system: properties and applications. Adv Appl Microbiol. 1997;43:141–194. doi: 10.1016/s0065-2164(08)70225-9. [DOI] [PubMed] [Google Scholar]
- 28.Barbe C, Dubourdieu D. Characterisation and purification of a cinnamate esterase from Aspergillus niger industrial pectinase preparation. J Sci Food Agric. 1998;78:471–478. [Google Scholar]
- 29.Baron A, Rombouts F, Drilleau J F, Pilnik W. Purification et proprietes de la pectinesterase produite par Aspergillus niger. Lebensm-Wiss Technol. 1980;13:330–333. [Google Scholar]
- 30.Bartolome B, Faulds C B, Tuohy M, Hazlewood G P, Gilbert H J, Williamson G. Influence of different xylanases on the activity of ferulic acid esterase on wheat bran. Biotechnol Appl Biochem. 1995;22:65–73. [Google Scholar]
- 31.Bause E, Legler G. Isolation and structure of a tryptic glycopeptide from the active site of β-glucosidase A3 from Aspergillus wentii. Biochim Biophys Acta. 1980;626:459–465. doi: 10.1016/0005-2795(80)90142-7. [DOI] [PubMed] [Google Scholar]
- 32.Bedford M R, Classen H L. The influence of dietary xylanase on intestinal viscosity and molecular weight distribution of carbohydrates in rye-fed broiler chicks. In: Visser J, Beldman G, Kusters-van Someren M A, Voragen A G J, editors. Xylans and xylanases. Vol. 7. Amsterdam, The Netherlands: Elsevier Science; 1992. pp. 361–370. [Google Scholar]
- 33.Bekker E G, Gusakov A V, Sinitsyn A P. Transglycosylation reactions catalyzed by β-glucosidase from Aspergillus japonicus in the presence of lignin model compounds. Prikl Biokhim Mikrobiol. 1991;27:482–485. [Google Scholar]
- 34.Beldman G, M, Searle-van Leeuwen M J F, de Ruiter G A, Siliha H A, Voragen A G J. Degradation of arabinans by arabinases from Aspergillus aculeatus and Aspergillus niger. Carbohydr Polym. 1993;20:159–168. [Google Scholar]
- 35.Beldman G, van den Broek L A M, Schols H A, Searle-van Leeuwen M J F, van Laere K M J, Voragen A G J. An exogalacturonase from Aspergillus aculeatus able to degrade xylogalacturonan. Biotechnol Lett. 1996;18:707–712. [Google Scholar]
- 36.Benen J, Kester H, Visser J. Kinetic characterization of Aspergillus niger N400 endopolygalacturonases I, II, and C. Eur J Biochem. 1999;259:577–585. doi: 10.1046/j.1432-1327.1999.00080.x. [DOI] [PubMed] [Google Scholar]
- 37.Benen J, Parenicová L, Kusters-van Someren M, Kester H, Visser J. Molecular genetic and biochemical aspects of pectin degradation in Aspergillus. In: Visser J, Voragen A G J, editors. Pectins and pectinases. Vol. 14. Amsterdam, The Netherlands: Elsevier Science; 1996. pp. 331–346. [Google Scholar]
- 38.Benen J A E, Kester H C M, Parenicova L, Visser J. Characterization of Aspergillus niger pectate lyase A. Biochemistry. 2000;39:15563–15569. doi: 10.1021/bi000693w. [DOI] [PubMed] [Google Scholar]
- 39.Biely P, Benen J A E, Heinrichová K, Kester H C M, Visser J. Inversion of configuration during hydrolysis of alpha-1,4-galacturonidic linkage by three Aspergillus polygalacturonases. FEBS Lett. 1996;382:249–255. doi: 10.1016/0014-5793(96)00171-8. [DOI] [PubMed] [Google Scholar]
- 40.Biely P, de Vries R P, Vranská M, Visser J. Inverting character of α-glucuronidase A from Aspergillus tubingensis. Biochim Biophys Acta. 2000;1474:360–364. doi: 10.1016/s0304-4165(00)00029-5. [DOI] [PubMed] [Google Scholar]
- 41.Biely P, Puls J, Schneider H. Acetyl xylan esterases in fungal cellulolytic systems. FEBS Lett. 1985;186:80–84. [Google Scholar]
- 42.Biely P, Vrsanská M, Claeyssens M. The endo-1,4-beta-glucanase I from Trichoderma reesei—action on beta-1,4-oligomers and polymers derived from d-glucose and d-xylose. Eur J Biochem. 1991;200:157–163. doi: 10.1111/j.1432-1033.1991.tb21062.x. [DOI] [PubMed] [Google Scholar]
- 43.Bonnin E, Lahaye M, Vigoureux J, Thibault J-F. Preliminary characterization of a new exo-β-(1,4)-galactanase with transferase activity. Int J Biol Macromol. 1995;17:345–351. doi: 10.1016/0141-8130(96)81844-7. [DOI] [PubMed] [Google Scholar]
- 44.Bonnin E, Thibault J-F. Galactooligosaccharide production by transfer reaction of an exogalactanase. Enzyme Microb Technol. 1996;19:99–106. [Google Scholar]
- 45.Bonnin E, Vigoureux J, Thibault J-F. Kinetic parameters of hydrolysis and transglycosylation catalyzed by an exo-β-(1,4)-galactanase. Enzyme Microb Technol. 1997;20:516–522. [Google Scholar]
- 46.Bouquelet S, Spik G, Montreuil J. Properties of a beta-d-mannosidase from Aspergillus niger. Biochim Biophys Acta. 1978;522:521–530. doi: 10.1016/0005-2744(78)90084-0. [DOI] [PubMed] [Google Scholar]
- 47.Brillouet J-M, Joseleau J P. Investigation of the structure of a heteroxylan from the outer pericarp (beeswing bran) of wheat kernel. Carbohydr Res. 1987;159:109–126. [Google Scholar]
- 48.Brillouet J-M, Williams P, Moutounet M. Purification and some properties of a novel endo-β-(1-6)-d-galactanase from Aspergillus niger. Agric Biol Chem. 1991;55:1565–1571. [Google Scholar]
- 49.Buachidze T S, Tavobilov I M, Rodionova N A, Kvesitadze G I. Isolation and purification of β-d-glucosidase from the thermophilic mutant Aspergillus terreus. Appl Biochem Microbiol. 1987;23:187–196. [Google Scholar]
- 50.Bussink H J, Brouwer K B, de Graaff L H, Kester H C, Visser J. Identification and characterization of a second polygalacturonase gene of Aspergillus niger. Curr Genet. 1991;20:301–307. doi: 10.1007/BF00318519. [DOI] [PubMed] [Google Scholar]
- 51.Bussink H J D, Buxton F P, Fraaye B A, de Graaff L H, Visser J. The polygalacturonases of Aspergillus niger are encoded by a family of diverged genes. Eur J Biochem. 1992;208:83–90. doi: 10.1111/j.1432-1033.1992.tb17161.x. [DOI] [PubMed] [Google Scholar]
- 52.Bussink H J D, Buxton F P, Visser J. Expression and sequence comparison of the Aspergillus niger and Aspergillus tubingensis genes encoding polygalacturonase II. Curr Genet. 1991;19:467–474. doi: 10.1007/BF00312738. [DOI] [PubMed] [Google Scholar]
- 53.Bussink H J D, van den Hombergh J P T W, van den Ijssel P R L A, Visser J. Characterization of polygalacturonase-overproducing Aspergillus niger transformants. Appl Microbiol Biotechnol. 1992;37:324–329. [Google Scholar]
- 54.Caddick M X, Brownlee A G, Arst H N J. Regulation of gene expression by pH of the growth medium in Aspergillus nidulans. Mol Gen Genet. 1986;203:346–353. doi: 10.1007/BF00333978. [DOI] [PubMed] [Google Scholar]
- 55.Camardela L, Carratore V, Ciardello M A, Servillo L, Balestrieri C, Giovane A. Kiwi protein inhibitor of pectin methylesterase. Eur J Biochem. 2000;267:4561–4565. doi: 10.1046/j.1432-1327.2000.01510.x. [DOI] [PubMed] [Google Scholar]
- 56.Carpita N C, Gibeaut D M. Structural models of primary cell walls in flowering plants: consistency of molecular structure with the physical properties of the walls during growth. Plant J. 1993;3:1–30. doi: 10.1111/j.1365-313x.1993.tb00007.x. [DOI] [PubMed] [Google Scholar]
- 57.Cary J W, Brown R, Cleveland T E, Whitehead M, Dean R A. Cloning and characterization of a novel polygalacturonase-encoding gene from Aspergillus parasiticus. Gene. 1995;153:129–133. doi: 10.1016/0378-1119(94)00749-i. [DOI] [PubMed] [Google Scholar]
- 58.Chikamatsu G, Shirai K, Kato M, Kobayashi T, Tsukagoshi N. Structure and expression properties of the endo-β-1,4-glucanase A gene from the filamentous fungus Aspergillus nidulans. FEMS Microbiol Lett. 1999;175:239–245. doi: 10.1111/j.1574-6968.1999.tb13626.x. [DOI] [PubMed] [Google Scholar]
- 59.Christgau S, Kauppinen S, Vind J, Kofod L V, Dalboge H. Expression cloning, purification and characterization of β-1,4-mannanase from Aspergillus aculeatus. Biochem Mol Biol Int. 1994;33:917–925. [PubMed] [Google Scholar]
- 60.Christgau S, Kofod L V, Halkier T, Anderson L N, Hockauf M, Dorreich K, Dalboge H, Kauppinen S. Pectin methyl esterase from Aspergillus aculeatus: expression cloning in yeast and characterization of the recombinant enzyme. Biochem J. 1996;319:705–712. doi: 10.1042/bj3190705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Christgau S, Sandal T, Kofod L V, Dalboge H. Expression cloning, purification and characterization of a β-1,4-galactanase from Aspergillus aculeatus. Curr Genet. 1995;27:135–141. doi: 10.1007/BF00313427. [DOI] [PubMed] [Google Scholar]
- 62.Christov L P, Szakacs G, Balakrishnan H. Production, partial characterization and use of fungal cellulase-free xylanases in pulp bleaching. Process Biochem. 1999;34:511–517. [Google Scholar]
- 63.Civas A, Eberhard R, le Dizet P, Petek F. Glycosidases induced in Aspergillus tamarii. Mycelial α-d-galactosidases. Biochem J. 1984;219:849–855. doi: 10.1042/bj2190849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Civas A, Eberhard R, le Dizet P, Petek F. Glycosidases induced in Aspergillus tamarii. Secreted α-d-galactosidase and β-d-mannanase. Biochem J. 1984;219:857–863. doi: 10.1042/bj2190857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Coll L, Saura D, Ros J M, Moliner M, Laencina J. Enzymatic treatment in the extraction of cold-pressed lemon peel oils. In: Visser J, Voragen A G J, editors. Pectins and pectinases. Vol. 14. Amsterdam, The Netherlands: Elsevier Science; 1996. pp. 963–970. [Google Scholar]
- 66.Colquhoun I J, Ralet M-C, Thibault J-F, Faulds C B, Williamson G. Structure identification of feruloylated oligosaccharides from sugar beet pulp by NMR spectroscopy. Carbohydr Res. 1994;263:243–256. doi: 10.1016/0008-6215(94)00176-6. [DOI] [PubMed] [Google Scholar]
- 67.Cook B J, Clay R P, Bergmann C W, Albersheim P, Darvil A G. Fungal polygalacturonases exhibit different substrate degradation patterns and differ in their susceptibilities to polygalacturonase inhibiting proteins. Mol Plant-Microbe Interact. 1999;12:703–711. doi: 10.1094/MPMI.1999.12.8.703. [DOI] [PubMed] [Google Scholar]
- 68.Cooke R D, Ferber C E M, Kanagasabapathy L. Purification and characterisation of polygalacturonases from a commercial Aspergillus niger preparation. Biochim Biophys Acta. 1976;452:440–451. doi: 10.1016/0005-2744(76)90194-7. [DOI] [PubMed] [Google Scholar]
- 69.Coutinho P M, Henrissat B. Carbohydrate-active enzymes: an integrated approach. In: Gilbert H J, Davies G J, Henrissat B, Svensson B, editors. Recent advances in carbohydrate bioengineering. Cambridge, United Kingdom: The Royal Society of Chemistry; 1999. pp. 3–14. [Google Scholar]
- 70.Critchley P. Commercial aspects of biocatalysis in low-water systems. In: Laane C, Tramper J, Lilly M D, editors. Biocatalysis in organic media. Amsterdam, The Netherlands: Elsevier Science; 1987. pp. 173–183. [Google Scholar]
- 71.Crous J M, Pretorius I S, van Zyl W H. Cloning and characterization of the alpha-L-arabinofuranosidase gene (ABF2) of Aspergillus niger expressed in Saccharomyces cerevisiae. Appl Microbiol Biotechnol. 1995;46:256–260. doi: 10.1007/s002530050813. [DOI] [PubMed] [Google Scholar]
- 72.Cubero B, Scazzocchio C. Two different, adjacent and divergent zinc finger binding sites are necessary for CreA-mediated carbon catabolite repression in the proline gene cluster of Aspergillus nidulans. EMBO J. 1994;13:407–415. doi: 10.1002/j.1460-2075.1994.tb06275.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Dalboge H, Heldt-Hansen H P. A novel method for efficient expression cloning of fungal enzymes. Mol Gen Genet. 1994;243:253–260. doi: 10.1007/BF00301060. [DOI] [PubMed] [Google Scholar]
- 74.Davies R W. Heterologous gene expression and protein secretion in Aspergillus. Prog Ind Microbiol. 1994;29:527–560. [PubMed] [Google Scholar]
- 75.Dea I C M, Morrison A. Chemistry and interactions of seed galactomannans. Adv Carbohydr Chem Biochem. 1975;31:241–312. [Google Scholar]
- 76.Dean R A, Timberlake W A. Regulation of the Aspergillus nidulans pectate lyase gene (pelA) Plant Cell. 1989;1:275–284. doi: 10.1105/tpc.1.3.275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Debeyser W, Peumans W J, Van Damme E J M, Delcour J A. Triticum aestivum xylanase inhibitor (TAXI), a new class of enzyme inhibitor affecting breadmaking performance. J Cereal Sci. 1999;30:39–43. [Google Scholar]
- 78.de Graaff L H, van den Broeck H C, van Ooijen A J J, Visser J. Regulation of the xylanase-encoding xlnA gene of Aspergillus tubingensis. Mol Microbiol. 1994;12:479–490. doi: 10.1111/j.1365-2958.1994.tb01036.x. [DOI] [PubMed] [Google Scholar]
- 79.Delgado L, Trejo B A, Huitron C, Aguilar G. Pectin lyase from Aspergillus sp. CH-Y-1043. Appl Microbiol Biotechnol. 1992;39:515–519. [Google Scholar]
- 80.De Lorenzo G, Cervone F. Polygalacturonase-inhibiting proteins (PGIPs): their role in specificity and defense against pathogenic fungi. In: Stacy G, Keen N T, editors. Plant-microbe interactions. Vol. 3. New York, N.Y: Chapman & Hall; 1997. pp. 76–93. [Google Scholar]
- 81.den Herder I F, Mateo Rosell A M, van Zuilen C M, Punt P J, van den Hondel C A M J J. Cloning and expression of a member of the Aspergillus niger gene family encoding α-galactosidase. Mol Gen Genet. 1992;233:404–410. doi: 10.1007/BF00265437. [DOI] [PubMed] [Google Scholar]
- 82.Denison S H. pH regulation of gene expression in fungi. Fungal Genet Biol. 2000;29:61–71. doi: 10.1006/fgbi.2000.1188. [DOI] [PubMed] [Google Scholar]
- 83.Desiderio A, Aracri B, Leckie F, Mattei B, Salvi G, Tigelaar H, Van Roekel J S C, Baulcombe D C, Melchers L S, De Lorenzo G, Cervone F. Polygalacturonase-inhibiting proteins (PGIPs) with different specificities are expressed in Phaseolus vulgaris. Mol Plant-Microbe Interact. 1997;10:852–860. doi: 10.1094/MPMI.1997.10.7.852. [DOI] [PubMed] [Google Scholar]
- 84.Devoto A, Leckie F, Lupotto E, Cervone F, De Lorenzo G. The promotor of a gene encoding a polygalacturonase-inhibiting protein of Phaseolus vulgaris L. is activated by wounding but not by elicitors or pathogen infection. Planta. 1998;205:165–174. doi: 10.1007/s004250050308. [DOI] [PubMed] [Google Scholar]
- 85.de Vries J A, Rombouts F M, Voragen A G J, Pilnik W. Enzymic degradation of apple pectins. Carbohydr Polym. 1982;2:25–33. [Google Scholar]
- 86.Reference deleted.
- 87.de Vries, R. P., J. A. E. Benen, L. H. de Graaff, and J. Visser. Plant cell wall degrading enzymes produced by Aspergillus. In H. D. Osiewacz (ed.), Industrial applications, the mycota, vol. X. Springer-Verlag, Heidelberg, Germany, in press.
- 88.de Vries R P, Flipphi M J A, Witteveen C F B, Visser J. Characterisation of an Aspergillus nidulansl-arabitol dehydrogenase mutant. FEMS Microbiol Lett. 1994;123:83–90. doi: 10.1111/j.1574-6968.1994.tb07205.x. [DOI] [PubMed] [Google Scholar]
- 89.Reference deleted.
- 90.de Vries R P, Kester H C M, Poulsen C H, Benen J A E, Visser J. Synergy between accessory enzymes from Aspergillus in the degradation of plant cell wall polysaccharides. Carbohydr Res. 2000;327:401–410. doi: 10.1016/s0008-6215(00)00066-5. [DOI] [PubMed] [Google Scholar]
- 91.Reference deleted.
- 92.de Vries R P, Michelsen B, Poulsen C H, Kroon P A, van den Heuvel R H H, Faulds C B, Williamson G, van den Hombergh J P T W, Visser J. The faeA genes from Aspergillus niger and Aspergillus tubigensis encode ferulic acid esterases involved in the degradation of complex cell wall polysaccharides. Appl Environ Microbiol. 1997;63:4638–4644. doi: 10.1128/aem.63.12.4638-4644.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.de Vries R P, Poulsen C H, Madrid S, Visser J. aguA, the gene encoding an extracellular α-glucuronidase from Aspergillus tubingensis, is specifically induced on xylose and not on glucuronic acid. J Bacteriol. 1998;180:243–249. doi: 10.1128/jb.180.2.243-249.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.de Vries R P, van den Broeck H C, Dekkers E, Manzanares P, de Graaff L H, Visser J. Differential expression of three α-galactosidase genes and a single β-galactosidase gene from Apergillus niger. Appl Environ Microbiol. 1999;65:2453–2460. doi: 10.1128/aem.65.6.2453-2460.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.de Vries R P, Visser J. Regulation of the feruloyl esterase (faeA) gene from Aspergillus niger. Appl Environ Microbiol. 1999;65:5500–5503. doi: 10.1128/aem.65.12.5500-5503.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.de Vries R P, Visser J, de Graaff L H. CreA modulates the XlnR induced expression on xylose of Aspergillus niger genes involved in xylan degradation. Res Microbiol. 1999;150:281–285. doi: 10.1016/s0923-2508(99)80053-9. [DOI] [PubMed] [Google Scholar]
- 97.Dey P M. Biochemistry of plant galactomannans. Adv Carbohydr Chem Biochem. 1978;35:3411–376. [Google Scholar]
- 98.Dorn G. Phosphatase mutants in Aspergillus nidulans. Science. 1965;150:1183–1184. doi: 10.1126/science.150.3700.1183. [DOI] [PubMed] [Google Scholar]
- 99.Dowzer C E A, Kelly J M. Analysis of the creA gene, a regulator of carbon catabolite repression in Aspergillus nidulans. Mol Cell Biol. 1991;11:5701–5709. doi: 10.1128/mcb.11.11.5701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Dunkel M P H, Amado R. Analysis of endo-(1–5)-α-l-arabinanase degradation patterns of linear (1–5)-α-l-arabino-oligosaccharides by high-performance anion-exchange chromatography with pulsed amperometric detection. Carbohydr Res. 1995;268:151–158. doi: 10.1016/0008-6215(94)00305-y. [DOI] [PubMed] [Google Scholar]
- 101.Ebringerová A, Heinze T. Xylan and xylan derivatives—biopolymers with valuable properties. 1. Naturally occurring xylan structures, isolation procedures and properties. Macromol Rapid Commun. 2000;21:542–556. [Google Scholar]
- 102.Eda S, Ohnishi A, Kato K. Xylan isolated from the stalk of Nicotiana tabacum. Agric Biol Chem. 1976;40:359–364. [Google Scholar]
- 103.Elbein A D, Adya S, Lee Y C. Purification and properties of a β-mannosidase from Aspergillus niger. J Biol Chem. 1977;252:2026–2031. [PubMed] [Google Scholar]
- 104.Eraso F, Hartley R D. Monomeric and dimeric phenolic constituents of plant cell walls—possible factors influencing wall biodegradability. J Sci Food Agric. 1990;51:163–170. [Google Scholar]
- 105.Eriksson K-W, Winell M. Purification and characterization of a fungal β-mannanase. Acta Chem Scand. 1968;22:1924–1934. [Google Scholar]
- 106.Faulds C B, de Vries R P, Kroon P A, Visser J, Williamson G. Influence of ferulic acid on the production of feruloyl esterases by Aspergillus niger. FEMS Microbiol Lett. 1997;157:239–244. doi: 10.1111/j.1574-6968.1997.tb12779.x. [DOI] [PubMed] [Google Scholar]
- 107.Faulds C B, Williamson G. Ferulic acid esterase from Aspergillus niger: purification and partial characterization of two forms from a commercial source of pectinase. Biotechnol Appl Biochem. 1993;17:349–359. [PubMed] [Google Scholar]
- 108.Fernandez-Espinar M, Pinaga F, de Graaff L, Visser J, Ramon D, Valles S. Purification, characterization, and regulation of the synthesis of an Aspergillus nidulans acidic xylanase. Appl Microbiol Biotechnol. 1994;42:555–562. [Google Scholar]
- 109.Flipphi M J A, Panneman H, van der Veen P, Visser J, de Graaff L H. Molecular cloning, expression and structure of the endo-1,5-α-l-arabinase gene of Aspergillus niger. Appl Microbiol Biotechnol. 1993;40:318–326. doi: 10.1007/BF00170387. [DOI] [PubMed] [Google Scholar]
- 110.Flipphi M J A, van Heuvel M, van der Veen P, Visser J, de Graaff L H. Cloning and characterisation of the abfB gene coding for the major α-l-arabinofuranosidase (AbfB) of Aspergillus niger. Curr Genet. 1993;24:525–532. doi: 10.1007/BF00351717. [DOI] [PubMed] [Google Scholar]
- 111.Flipphi M J A, Visser J, van der Veen P, de Graaff L H. Arabinase gene expression in Aspergillus niger: indications for co-ordinated gene expression. Microbiology. 1994;140:2673–2682. doi: 10.1099/00221287-140-10-2673. [DOI] [PubMed] [Google Scholar]
- 112.Flipphi M J A, Visser J, van der Veen P, de Graaff L H. Cloning of the Aspergillus niger gene encoding α-L-arabinofuranosidase A. Appl Microbiol Biotechnol. 1993;39:335–340. doi: 10.1007/BF00192088. [DOI] [PubMed] [Google Scholar]
- 113.Forster H. Pectin esterase from Phytophtera infestans. Methods Enzymol. 1988;161:355–361. [Google Scholar]
- 114.Fournier R A, Frederick M M, Frederick J R, Reilly P J. Purification and characterization of endo-xylanases from Aspergillus niger. III. An enzyme of pI 3.65. Biotechnol Bioeng. 1985;27:539–546. doi: 10.1002/bit.260270422. [DOI] [PubMed] [Google Scholar]
- 115.Frederick M M, Frederick J R, Frayzke A R, Reilly P J. Purification and characterization of a xylobiose- and xylose-producing endo-xylanase from Aspergillus niger. Carbohydr Res. 1981;97:87–103. [Google Scholar]
- 116.Frederick M M, Kang S-H, Frederick J R, Reilly P J. Purification and characterization of endo-xylanases from Aspergillus niger. I. Two isozymes active on xylan backbones near branch points. Biotechnol Bioeng. 1985;27:525–532. doi: 10.1002/bit.260270420. [DOI] [PubMed] [Google Scholar]
- 117.Fry S C. Cross-linking of matrix polymers in the growing cell walls of angiosperms. Annu Rev Plant Physiol. 1986;37:165–186. [Google Scholar]
- 118.Fry S C. Phenolic components of the primary cell wall and their possible role in the hormonal regulation of growth. Planta. 1979;146:343–351. doi: 10.1007/BF00387807. [DOI] [PubMed] [Google Scholar]
- 119.Fujimoto H, Ooi T, Wang S-L, Takiwaza T, Hidaka H, Murao S, Arai M. Purification and properties of three xylanases from Aspergillus aculeatus. Biosci Biotechnol Biochem. 1995;59:538–540. [Google Scholar]
- 120.Galas E, Romanowska I. Purification and some properties of β-glucosidase from Aspergillus niger IBT-90. Acta Microbiol Pol. 1997;46:241–252. [PubMed] [Google Scholar]
- 121.Ganter C, Bock A, Buckel P, Mattes R. Production of thermostable, recombinant α-galactosidase suitable for raffinose elimination from sugar beet syrup. J Biotechnol. 1988;8:301–310. [Google Scholar]
- 122.Geiser D M, Pitt J I, Taylor J W. Cryptic speciation and recombination in the aflatoxin-producing fungus Aspergillus flavus. Proc Natl Acad Sci USA. 1998;95:388–393. doi: 10.1073/pnas.95.1.388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Ghosh M, Nanda G. Purification and some properties of a xylanase from Aspergillus sydowii MG49. Appl Environ Microbiol. 1994;60:4620–4623. doi: 10.1128/aem.60.12.4620-4623.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Gielkens M M C, Dekkers E, Visser J, de Graaff L H. Two cellobiohydrolase-encoding genes from Aspergillus niger require d-xylose and the xylanolytic transcriptional activator XlnR for their expression. Appl Environ Microbiol. 1999;65:4340–4345. doi: 10.1128/aem.65.10.4340-4345.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Gielkens M M C, Gonzales-Candelas L, Sanchez-Torres P, van de Vondervoort P J I, de Graaf L H, Visser J. The abfB gene encoding the major α-l-arabinofuranosidase of Aspergillus nidulans: nucleotide sequence, regulation and construction of a disrupted strain. Microbiology. 1999;145:735–741. doi: 10.1099/13500872-145-3-735. [DOI] [PubMed] [Google Scholar]
- 126.Gielkens M M C, Visser J, de Graaff L H. Arabinoxylan degradation by fungi: characterisation of the arabinoxylan arabinofuranohydrolase encoding genes from Aspergillus niger and Aspergillus tubingensis. Curr Genet. 1997;31:22–29. doi: 10.1007/s002940050172. [DOI] [PubMed] [Google Scholar]
- 127.Giovane A, Balestrieri C, Quagliuolo L, Castaldo D, Servillo L. A glycoprotein inhibitor of pectin methylesterase in kiwi fruit. Eur J Biochem. 1995;233:926–929. doi: 10.1111/j.1432-1033.1995.926_3.x. [DOI] [PubMed] [Google Scholar]
- 128.Golubev A M, Ibatullin F H, Kilimnik A Y, Rodionova N A, Neustroev K N. Isolation and properties of endoxylanase and β-xylosidase from Aspergillus oryzae. Biochemistry. 1993;58:565–570. [Google Scholar]
- 129.Gonzalez R R, Monsan P. Purification and some characteristics of β-galactosidase from Aspergillus fonsecaeus. Enzyme Microb Technol. 1991;13:349–352. [Google Scholar]
- 130.Gorbacheva I V, Rodionova N A. Studies on xylan degrading enzymes. I. Purification and characterization of endo-1,4-β-xylanase from Aspergillus niger str. 14. Biochim Biophys Acta. 1977;484:79–93. doi: 10.1016/0005-2744(77)90114-0. [DOI] [PubMed] [Google Scholar]
- 131.Grabber J H, Ralph J, Hatfield R D. Ferulate cross-links limit the enzymatic degradation of synthetically lignified primary walls of maize. J Agric Food Chem. 1998;46:2609–2614. [Google Scholar]
- 132.Grassin C, Fauquembergue P. Applications of pectinases in beverages. In: Visser J, Voragen A G J, editors. Pectin and pectinases. Vol. 14. Amsterdam, The Netherlands: Elsevier Science; 1996. pp. 453–462. [Google Scholar]
- 133.Greenberg N A, Mahoney R R. Rapid purification of β-galactosidase (Aspergillus niger) from a commercial preparation. J Food Sci. 1981;46:684–687. [Google Scholar]
- 134.Guillon F, Thibault J-F. Enzymic hydrolysis of the “hairy” fragments of sugarbeet pectins. Carbohydr Res. 1989;190:97–108. [Google Scholar]
- 135.Gunata Z, Brillouet J-M, Voirin S, Baumes R, Cordonnier R. Purification and some properties of an α-L-arabinofuranosidase from Aspergillus niger. Action on grape monoterpenyl arabinofuranosylglucosides. J Agric Food Chem. 1990;38:772–776. [Google Scholar]
- 136.Gysler C, Harmsen J A M, Kester H C M, Visser J, Heim J. Isolation and structure of the pectin lyase D-encoding gene from Aspergillus niger. Gene. 1990;89:101–108. doi: 10.1016/0378-1119(90)90211-9. [DOI] [PubMed] [Google Scholar]
- 137.Hamari Z, Kevei F, Kovacs E, Varga J, Kozakiewicz Z, Croft J H. Molecular and phenotypic characterization of Aspergillus japonicus and Aspergillus aculeatus strains with special regard to their mitochondrial DNA polymorphism. Antonie Leeuwenhoek. 1997;72:337–347. doi: 10.1023/a:1000578913759. [DOI] [PubMed] [Google Scholar]
- 138.Hantus S, Pauly M, Darvill A G, Albersheim P, York W S. Structural characterization of novel l-galactose-containing oligosaccharide subunits of jojoba seed xyloglucans. Carbohydr Res. 1997;304:11–20. doi: 10.1016/s0008-6215(97)00200-0. [DOI] [PubMed] [Google Scholar]
- 139.Hara T, Lim J Y, Fujio Y, Ueda S. Purification and some properties of exo-polygalacturonase from Aspergillus niger cultured in the medium containing Satsuma mandarin peel. Nippon Shokuhin Kogyo Gakkaishi. 1984;31:581–586. [Google Scholar]
- 140.Harmsen J A M, Kusters-van Someren M A, Visser J. Cloning and expression of a second Aspergillus niger pectin lyase gene (pelA): indications of a pectin lyase gene family in A. niger. Curr Genet. 1990;18:161–166. doi: 10.1007/BF00312604. [DOI] [PubMed] [Google Scholar]
- 141.Hashimoto T, Morishita M, Nakata Y, Tsuji Y, Ito K. Cloning of β-xylosidase gene of Aspergillus oryzae and expression in heterologous microorganisms. In: Ohmiya K, Sakka K, Karita S, Hayashi H, Kobayashi Y, Kimura T, editors. Genetics, biochemistry and ecology of cellulose degradation. Tokyo, Japan: Uni Publishers Co., Ltd.; 1999. pp. 397–404. [Google Scholar]
- 142.Hasper A A, Visser J, de Graaff L H. The Aspergillus niger transcriptional activator XlnR, which is involved in the degradation of the polysaccharides xylan and cellulose, also regulates d-xylose reductase gene expression. Mol Microbiol. 2000;36:193–200. doi: 10.1046/j.1365-2958.2000.01843.x. [DOI] [PubMed] [Google Scholar]
- 143.Hayashida S, Mo K, Hosoda A. Production and characteristics of avicel-digesting and non-avicel-digesting cellobiohydrolases from Aspergillus ficuum. Appl Environ Microbiol. 1988;54:1523–1529. doi: 10.1128/aem.54.6.1523-1529.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Heldt-Hansen H P, Kofod L V, Budolfsen G, Nielsen P M, S. H, Bladt T. Application of tailor made pectinases. In: Visser J, Voragen A G J, editors. Pectin and pectinases. Vol. 14. Amsterdam, The Netherlands: Elsevier Science; 1996. pp. 463–474. [Google Scholar]
- 145.Henrissat B. A classification of glycosidases based on amino-acid sequence similarities. Biochem J. 1991;280:309–316. doi: 10.1042/bj2800309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Henrissat B, Bairoch A. New families in the classification of glycosidases based on amino acid sequence similarities. Biochem J. 1993;293:781–788. doi: 10.1042/bj2930781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Henrissat B, Bairoch A. Updating the sequence based classification of glycosidases. Biochem J. 1996;316:695–696. doi: 10.1042/bj3160695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Herve du Penhoat C, Gey C, Pellerin P, Perez S. An NMR solution study of the mega-oligosaccharide, rhamnogalacturonan II. J Biomol NMR. 1999;14:253–271. doi: 10.1023/a:1008312423877. [DOI] [PubMed] [Google Scholar]
- 149.Hessing J G M, van Rotterdam C, Verbakel J M, Roza M, Maat J, van Gorcom R F, van den Hondel C A M J J. Isolation and characterization of a 1,4-beta-endoxylanase gene of A. awamori. Curr Genet. 1994;26:228–232. doi: 10.1007/BF00309552. [DOI] [PubMed] [Google Scholar]
- 150.Himmel M E, Adney W S, Fox J W, Mitchell D J, Baker J O. Isolation and characterization of two forms of β-D-glucosidase from Aspergillus niger. Appl Biochem Biotechnol. 1993;39/40:213–225. doi: 10.1007/BF02918991. [DOI] [PubMed] [Google Scholar]
- 151.Hisamatsu M, York W S, Darvill A G, Albersheim P. Characterization of seven xyloglucan oligosaccharides containing from seventeen to twenty glycosyl residues. Carbohydr Res. 1992;227:45–71. doi: 10.1016/0008-6215(92)85060-d. [DOI] [PubMed] [Google Scholar]
- 152.Ho M-C, Whitehead M P, Cleveland T E, Dean R A. Sequence analysis of the Aspergillus nidulans pectate lyase pelA gene and evidence for binding of promoter regions to CREA, a regulator of carbon catabolite repression. Curr Genet. 1995;27:142–149. doi: 10.1007/BF00313428. [DOI] [PubMed] [Google Scholar]
- 153.Holazo A, Shinoyama H, Kamiyama Y, Yasui T. Screening for β-mannosidases with transmannosidation capacity. Biosci Biotechnol Biochem. 1992;56:822–824. doi: 10.1271/bbb.56.822. [DOI] [PubMed] [Google Scholar]
- 154.Hrmová M, Biely P, Vrsanská M. Cellulose- and xylan-degrading enzymes of Aspergillus terreus and Aspergillus niger. Enzyme Microb Technol. 1989;11:610–616. [Google Scholar]
- 155.Hrmová M, Petráková E, Biely P. Induction of cellulose- and xylan-degrading enzyme systems in Aspergillus terreus by homo- and heterodisaccharides composed of glucose and xylose. J Gen Microbiol. 1991;137:541–547. doi: 10.1099/00221287-137-3-541. [DOI] [PubMed] [Google Scholar]
- 156.Huisman M M H, Schols H A, Voragen A G J. Glucuronoarabinoxylans from maize kernel cell walls are more complex than those from sorghum kernel cell walls. Carbohydr Pol. 2000;43:269–279. [Google Scholar]
- 157.Huisman M M H, Weel K G C, Schols H A, Voragen A G J. Xyloglucan from soybean (Glycine max) meal is composed of XXXG-type building units. Carbohydr Pol. 2000;42:185–191. [Google Scholar]
- 158.Hurst P L, Nielsen J, Sullivan P A, Shepherd M G. Purification and properties of a celullase from Aspergillus niger. Biochem J. 1977;165:33–41. doi: 10.1042/bj1650033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Iizuka Y, Shinoyama H, Kamiyama Y, Yasui T. The condensation reaction of Aspergillus niger crude β-xylosidase using xylose. Biosci Biotechnol Biochem. 1992;56:331–332. [Google Scholar]
- 160.Imamura T, Watanabe T, M. K, Koshijima T. Ester linkages between lignin and glucuronic acid in lignin-carbohydrate complexes from Fagus crenata. Phytochemistry. 1994;37:1165–1173. doi: 10.1016/s0031-9422(00)89551-5. [DOI] [PubMed] [Google Scholar]
- 161.Ishii S, Yokotsuka T. Purification and properties of pectin lyase from Aspergillus japonicus. Agric Biol Chem. 1975;39:313–321. [Google Scholar]
- 162.Ishii T. Isolation and characterization of a diferuloyl arabinoxylan hexasaccharide from bamboo shoot cell-walls. Carbohydr Res. 1991;219:15–22. doi: 10.1016/0008-6215(91)89039-i. [DOI] [PubMed] [Google Scholar]
- 163.Ishii T. Structure and functions of feruloylated polysaccharides. Plant Sci. 1997;127:111–127. [Google Scholar]
- 164.Ismail A M, Abdel-Naby M A, Abdel-Fattah A F. Utilization of water hyacinth cellulose for the production of cellobiase-rich preparation by Aspergillus niger 1. Microbios. 1995;83:191–198. [PubMed] [Google Scholar]
- 165.Reference deleted.
- 166.Ito K, Ikemasu T, Ishikawa T. Cloning and sequencing of the xynA gene encoding xylanase A of Aspergillus kawachii. Biosci Biotechnol Biochem. 1992;56:906–912. doi: 10.1271/bbb.56.906. [DOI] [PubMed] [Google Scholar]
- 167.Ito K, Iwashita K, Iwano K. Cloning and sequencing of the xynC gene encoding xylanase C of Aspergillus kawachii. Biosci Biotechnol Biochem. 1992;56:1338–1340. doi: 10.1271/bbb.56.1338. [DOI] [PubMed] [Google Scholar]
- 168.Ito K, Ogasawara H, Sugimoto T, Ishikawa T. Purification and properties of acid stable xylanases from Aspergillus kawachii. Biosci Biotechnol Biochem. 1992;56:547–550. doi: 10.1271/bbb.56.547. [DOI] [PubMed] [Google Scholar]
- 169.Itoh H, Kamiyama Y. Synthesis of alkyl β-mannosides from mannobiose by Aspergillus niger β-mannosidase. J Ferment Bioeng. 1995;80:510–512. [Google Scholar]
- 170.Ivanova G S, Beletskaya O P, Okunev O N, Golovlev E L, Kulaev I S. Fractionation of cellulase complex of the fungus Aspergillus terreus. Appl Biochem Microbiol. 1983;19:275–281. [PubMed] [Google Scholar]
- 171.Iwashita K, Nagahara T, Kimura H, Takano M, Shimoi H, Ito K. The bglA gene from Aspergillus kawachii encodes both extracellular and cell wall bound β-glucosidases. Appl Environ Microbiol. 1999;65:5546–5553. doi: 10.1128/aem.65.12.5546-5553.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Jarai G, Buxton F. Nitrogen, carbon, and pH regulation of extracellular acidic proteases of Aspergillus niger. Curr Genet. 1994;26:238–244. doi: 10.1007/BF00309554. [DOI] [PubMed] [Google Scholar]
- 173.Jurnak F, Kita N, Garrett M, Heffron S E, Scavetta R, Boyd C, Keen N. Functional implications of the three-dimensional structures of pectate lyases. In: Visser J, Voragen A G J, editors. Pectin and pectinases. Vol. 14. Amsterdam, The Netherlands: Elsevier Science; 1996. pp. 295–308. [Google Scholar]
- 174.Kaneko S, Shimasaki T, Kusakabe I. Purification and some properties of intracellular α-L-arabinofuranosidase from Aspergillus niger 5–16. Biosci Biotechnol Biochem. 1993;57:1161–1165. doi: 10.1271/bbb.57.1161. [DOI] [PubMed] [Google Scholar]
- 175.Kantelinen A, Ratto M, Sundquist J, Ranua M, Viikari L, Linko M. Hemicelluloses and their potential role in bleaching. 1988. pp. 1–9. . 1988 Int. Pulp Bleaching Conf. [Google Scholar]
- 176.Kato M, Mimura S, Rao U, Tanaka A, Kitamoto N, Yoshino S, Tsukagoshi N. Regulation of the xynF1 gene encoding the major family F xylanase of Aspergillus oryzae. In: Ohmiya K, Hayashi K, Sakka K, Koyabashi Y, Karita S, Kimura T, editors. Genetics, biochemistry and ecology of cellulose degradation. Tokyo, Japan: Uni Publishers Co., Ltd.; 1999. pp. 331–335. [Google Scholar]
- 177.Reference deleted.
- 178.Kauppinen S, Christgau S, Kofod L V, Halkier T, Dorreich K, Dalboge H. Molecular cloning and characterization of a rhamnogalacturonan acetylesterase from Aspergillus aculeatus. Synergism between rhamnogalacturonan degrading enzymes. J Biol Chem. 1995;270:27172–27178. doi: 10.1074/jbc.270.45.27172. [DOI] [PubMed] [Google Scholar]
- 179.Kawaguchi T, Enoki T, Tsurumaki S, Sumitani J, Ueda M, Ooi T, Arai M. Cloning and sequencing of the cDNA encoding β-glucosidase 1 from Aspergillus aculeatus. Gene. 1996;173:287–288. doi: 10.1016/0378-1119(96)00179-5. [DOI] [PubMed] [Google Scholar]
- 180.Kester H C M, Benen J A E, Visser J. The exopolygalacturonase from Aspergillus tubingensis is also active on xylogalacturonan. Biotechnol Appl Biochem. 1999;30:53–57. [PubMed] [Google Scholar]
- 181.Kester H C M, Esteban Warren M, Orlando R, Benen J A E, Bergmann C, Visser J. Tandem mass spectrometric analysis of Aspergillus niger pectin methyl esterase; mode of action on fully methylesterified oligogalacturonates. Biochem J. 2000;346:469–474. [PMC free article] [PubMed] [Google Scholar]
- 182.Kester H C M, Kusters-van Someren M A, Muller Y, Visser J. Primary structure and characterization of an exopolygalacturonase from Aspergillus tubingensis. Eur J Biochem. 1996;240:738–746. doi: 10.1111/j.1432-1033.1996.0738h.x. [DOI] [PubMed] [Google Scholar]
- 183.Kester H C M, Magaud D, Roy C, Anker D, Doutheau A, Shevchik V, Hugouvieux-Cotte-Pattat N, Benen J A E, Visser J. Performance of selected microbial pectinases on synthetic monomethyl-esterified di- and trigalacturonates. J Biol Chem. 1999;274:37053–37059. doi: 10.1074/jbc.274.52.37053. [DOI] [PubMed] [Google Scholar]
- 184.Kester H C M, Visser J. Purification and characterization of pectin lyase B, a novel pectinolytic enzyme from Aspergillus niger. FEMS Microbiol Lett. 1994;120:63–68. [Google Scholar]
- 185.Kester H C M, Visser J. Purification and characterization of polygalacturonases produced by the hyphal fungus Aspergillus niger. Biotechnol Appl Biochem. 1990;12:150–160. [PubMed] [Google Scholar]
- 186.Khan A W, Lamb K A, Overend R P. Comparison of natural hemicellulose and chemically acetylated xylan as substrates for the determination of acetyl-xylan esterase activity in Aspergilli. Enzyme Microb Technol. 1990;12:127–130. [Google Scholar]
- 187.Khanh N Q, Ruttkowski E, Leidinger K, H. A, Gottschalk M. Characterisation and expression of a genomic pectin methyl esterase-encoding gene in Aspergillus niger. Gene. 1991;106:71–77. doi: 10.1016/0378-1119(91)90567-u. [DOI] [PubMed] [Google Scholar]
- 188.Kimura I, Sasahara H, Tajima S. Purification and characterization of two xylanases and an arabinofuranosidase from Aspergillus sojae. J Ferment Bioeng. 1995;80:334–339. [Google Scholar]
- 189.Kimura I, Yoshioka N, Kimura Y, Tajima S. Cloning, sequencing and expression of an α-l-arabinofuranosidase from Aspergillus sojae. J Biosci Bioeng. 2000;89:262–266. doi: 10.1016/s1389-1723(00)88830-1. [DOI] [PubMed] [Google Scholar]
- 190.Kimura I, Yoshioka N, Tajima S. Purification and characterization of an endo-1,4-β-d-galactanase from Aspergillus sojae. J Ferment Bioeng. 1998;85:48–52. [Google Scholar]
- 191.Kimura T, Kitamoto N, Kito Y, Karita S, Sakka K, Ohmiya K. Molecular cloning of xylanase gene xynG1 from Aspergillus oryzae KBN 616, a Shoyu Koji mold, and analysis of its expression. J Ferment Bioeng. 1998;85:10–16. [Google Scholar]
- 192.Kinoshita K, Takano M, Koseki T, Ito K, Iwano K. Cloning of the xynNB gene encoding xylanase B from Aspergillus niger and its expression in Aspergillus kawachii. J Ferment Bioeng. 1994;79:422–428. [Google Scholar]
- 193.Kitamoto N, Go M, Shibayama T, Kimura T, Kito Y, Ohmiya K, Tsukagoshi N. Molecular cloning, purification and characterization of two endo-1,4-β-glucanases from Aspergillus oryzae KBN616. Appl Microbiol Biotechnol. 1996;46:538–544. doi: 10.1007/s002530050857. [DOI] [PubMed] [Google Scholar]
- 194.Kitamoto N, Kimura T, Kito Y, Ohmiya K, Tsukagoshi N. Structural features of a polygalacturonase gene cloned from Aspergillus oryzae KBN616. FEMS Microbiol Lett. 1993;111:37–41. doi: 10.1111/j.1574-6968.1993.tb06358.x. [DOI] [PubMed] [Google Scholar]
- 195.Kitamoto N, Matsui J, Kawai Y, Kato A, Yoshino S, Ohmiya K, Tsukagoshi N. Utilization of the TEF1-alpha gene (TEF1) promoter for expression of polygalacturonase genes, pgaA and pgaB, in Aspergillus oryzae. Appl Microbiol Biotechnol. 1998;50:85–92. doi: 10.1007/s002530051260. [DOI] [PubMed] [Google Scholar]
- 196.Kitamoto N, Okada H, Yoshino S, Ohmiya K, Tsukagoshi N. Pectin methylesterase gene (pmeA) from Aspergillus oryzae KBN616: its sequence analysis and overexpression, and characterization of the gene product. Biosci Biotechnol Biochem. 1999;63:120–124. doi: 10.1271/bbb.63.120. [DOI] [PubMed] [Google Scholar]
- 197.Kitamoto N, Yoshino S, Ito M, Kimura T, Ohmiya K, Tsukagoshi N. Repression of the expression of genes encoding xylanolytic enzymes in Aspergillus oryzae by introduction of multiple copies of the xynF1 promoter. Appl Microbiol Biotechnol. 1998;50:558–563. doi: 10.1007/s002530051334. [DOI] [PubMed] [Google Scholar]
- 198.Kitamoto N, Yoshino S, Ohmiya K, Tsukagoshi N. Sequence analysis, overexpression, and antisense inhibition of a β-xylosidase gene, xylA, from Aspergillus oryzae KBN616. Appl Environ Microbiol. 1999;65:20–24. doi: 10.1128/aem.65.1.20-24.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Kitpreechavanich V, Hayashi M, Nagai S. Purification and characterisation of extracellular β-xylosidase and β-glucosidase from Aspergillus fumigatus. Agric Biol Chem. 1986;50:1703–1711. [Google Scholar]
- 200.Kizawa H, Shinoyama H, Yasui T. The synthesis of new xylosyloligosaccharides by transxylosylation with Aspergillus niger β-xylosidase. Agric Biol Chem. 1991;55:671–678. [PubMed] [Google Scholar]
- 201.Knap, I. H., M. Carsten, T. Halkier, and L. V. Kofod. Oct. 1994. An α-galactosidase enzyme. International patent WO 94/23022.
- 202.Reference deleted.
- 203.Kofod L V, Kauppinen S, Christgau S, Andersen L N, Heldt-Hansen H P, Dorreich K, Dalboge H. Cloning and characterization of two structurally and functionally divergent rhamnogalacturonases from Aspergillus aculeatus. J Biol Chem. 1994;269:29182–29189. [PubMed] [Google Scholar]
- 204.Kojima Y, Sakamoto T, Kishida M, Sakai T, Kawasaki H. Acidic condition-inducible polygalacturonase of Aspergillus kawachii. J Mol Catal Ser B. 1999;6:351–357. [Google Scholar]
- 205.Kolpak F J, Blackwell J. Determination of the structure of cellulose II. Macromolecules. 1976;9:273–278. doi: 10.1021/ma60050a019. [DOI] [PubMed] [Google Scholar]
- 206.Kormelink F J M, Gruppen H, Vietor R J, Voragen A G J. Mode of action of the xylan-degrading enzymes from Aspergillus awamori on alkali-extractable cereal arabinoxylans. Carbohydr Res. 1993;249:355–367. doi: 10.1016/0008-6215(93)84100-k. [DOI] [PubMed] [Google Scholar]
- 207.Kormelink F J M, Gruppen H, Voragen A G J. Mode of action of (1,4)-β-d-arabinoxylan arabinofuranohydrolase (AXH) and α-l-arabinofuranosidases on alkali-extractable wheat-flour arabinoxylan. Carbohydr Res. 1993;249:345–353. doi: 10.1016/0008-6215(93)84099-r. [DOI] [PubMed] [Google Scholar]
- 208.Kormelink F J M, Lefebvre B, Strozyk F, Voragen A G J. The purification and characterisation of an acetyl xylan esterase from Aspergillus niger. J Biotechnol. 1993;27:267–282. [Google Scholar]
- 209.Kormelink F J M, Searle-van Leeuwen M J F, Wood T M, Voragen A G J. Purification and characterization of a (1,4)-β-d-arabinoxylan arabinofuranohydrolase from Aspergillus awamori. Appl Microbiol Biotechnol. 1991;35:753–758. [Google Scholar]
- 210.Kormelink F J M, Searle-van Leeuwen M J F, Wood T M, Voragen A G J. Purification and characterization of three endo-(1,4)-β-xylanases and one β-xylosidase from Aspergillus awamori. J Biotechnol. 1993;27:249–265. [Google Scholar]
- 211.Kormelink F J M, Voragen A G J. Degradation of different [(glucurono)arabino]xylans by combination of purified xylan-degrading enzymes. Appl Microbiol Biotechnol. 1993;38:688–695. [Google Scholar]
- 212.Koseki T, Furuse S, Iwano K, Matsuzawa H. Purification and characterization of a feruloylesterase from Aspergillus awamori. Biosci Biotechnol Biochem. 1998;62:2032–2034. doi: 10.1271/bbb.62.2032. [DOI] [PubMed] [Google Scholar]
- 213.Koseki T, Furuse S, Iwano K, Sakai H, Matsuzawa H. An Aspergillus awamori acetylesterase: purification of the enzyme, and cloning and sequencing of the gene. Biochem J. 1997;326:485–490. doi: 10.1042/bj3260485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Reference deleted.
- 215.Krengel U, Dijkstra B W. Three-dimensional structure of Endo-1,4-beta-xylanase I from Aspergillus niger: molecular basis for its low pH optimum. J Mol Biol. 1996;263:70–78. doi: 10.1006/jmbi.1996.0556. [DOI] [PubMed] [Google Scholar]
- 216.Kroon P A, Faulds C B, Brezillon C, Williamson G. Methyl phenylalkanoates as substrates to probe the active sites of esterases. Eur J Biochem. 1997;248:245–251. doi: 10.1111/j.1432-1033.1997.00245.x. [DOI] [PubMed] [Google Scholar]
- 217.Kroon P A, Faulds C B, Williamson G. Purification and characterisation of a novel esterase induced by growth of Aspergillus niger on sugar-beet pulp. Biotechnol Appl Biochem. 1996;23:255–262. [PubMed] [Google Scholar]
- 218.Kroon P A, Garcia-Conesa M T, Fillingham I J, Hazlewood G P, Williamson G. Release of ferulic acid dehydrodimers from plant cell walls by feruloyl esterases. J Sci Food Agric. 1999;79:428–434. [Google Scholar]
- 219.Kroon P A, Williamson G. Release of ferulic acid from sugar-beet pulp by using arabinase, arabinofuranosidase and an esterase from Aspergillus niger. Biotechnol Appl Biochem. 1996;23:263–267. [PubMed] [Google Scholar]
- 220.Kulbe K D, Czarnetzki H, Giray J, Schmidt H, Miemietz G, Novalic S, Mattes R. Transformation of pectin to l-ascorbic acid by enzyme. Proc. 4th Int. 1997. Workshop Carbohydr. Org. Raw Mater. [Google Scholar]
- 221.Kulmburg P, Mathieu M, Dowzer C, Kelly J, Felenbok B. Specific binding sites in the alcR and alcA promoters of the ethanol regulon for the CreA repressor mediating carbon catabolite repression in Aspergillus nidulans. Mol Microbiol. 1993;7:847–857. doi: 10.1111/j.1365-2958.1993.tb01175.x. [DOI] [PubMed] [Google Scholar]
- 222.Kumar S, Ramon D. Purification and regulation of the synthesis of a β-xylosidase from Aspergillus nidulans. FEMS Microb Lett. 1996;135:287–293. [Google Scholar]
- 223.Kumar V, Ramakrishnan S, Teeri T T, Knowles J K C, Hartley B S. Saccharomyces cerevisiae cells secreting an Aspergillus niger β-galactosidase grown on whey permeate. Bio/Technology. 1992;10:82–85. doi: 10.1038/nbt0192-82. [DOI] [PubMed] [Google Scholar]
- 224.Kusters-van Someren M, Flipphi M, de Graaff L H, van den Broeck H, Kester H, Hinnen A, Visser J. Characterisation of the Aspergillus niger pelB gene: structure and regulation of expression. Mol Gen Genet. 1992;234:113–120. doi: 10.1007/BF00272352. [DOI] [PubMed] [Google Scholar]
- 225.Kusters-van Someren M A, Harmsen J A M, Kester H C M, Visser J. The structure of the Aspergillus niger pelA gene and its expression in Aspergillus niger and Aspergillus nidulans. Curr Genet. 1991;20:293–299. doi: 10.1007/BF00318518. [DOI] [PubMed] [Google Scholar]
- 226.Kusters-van Someren M A, Samson R A, Visser J. The use of RFLP analysis in classification of the black Aspergilli — reinterpretation of the Aspergillus niger aggregate. Curr Genet. 1991;19:21–26. [Google Scholar]
- 227.Kwon K S, Kang H G, Hah Y C. Purification and characterization of two extracellular β-glucosidases from Aspergillus nidulans. FEMS Microbiol Lett. 1992;76:149–153. doi: 10.1016/0378-1097(92)90378-2. [DOI] [PubMed] [Google Scholar]
- 228.Reference deleted.
- 229.Lahaye M, Vigouroux J, Thibault J-F. Endo-β-1,4-d-galactanase from Aspergillus niger var. aculeatus: purification and some properties. Carbohydr Polyme. 1991;15:431–444. [Google Scholar]
- 230.Lam T B T, Iiyama K, Stone B A. An approach to the estimation of ferulic acid bridges in unfractioned cell walls of wheat internodes. Phytochemistry. 1994;37:327–333. [Google Scholar]
- 231.Lang C, Looman A C. Efficient expression and secretion of Aspergillus niger RH5344 polygalacturonase in Saccharomyces cerevisiae. Appl Microbiol Biotechnol. 1995;44:147–156. doi: 10.1007/BF00164494. [DOI] [PubMed] [Google Scholar]
- 232.Lin J S, Tang M-Y, Fellers J F. Fractal analysis of cotton cellulose as characterized by small-angle X-ray scattering. ACS Symp Ser. 1987;340:233–254. [Google Scholar]
- 233.Lindberg B, Rosell K-G, Svensson S. Positions of the O-acetyl groups in pine glucomannan. Svensk Papperstidn. 1973;76:383–384. [Google Scholar]
- 234.Linden J, Samara M, Decker S, Johnson E, Boyer M, Pecs M, Adney W, Himmel M. Purification and characterisation of an acetyl esterase from Aspergillus niger. Appl Biochem Biotechnol. 1994;45/46:383–393. doi: 10.1007/BF02941813. [DOI] [PubMed] [Google Scholar]
- 235.Luonteri E, Siika-aho M, Tenkanen M, Viikari L. Purification and characterization of three alpha-arabinofuranosidases from Aspergillus terreus. J Biotechnol. 1995;38:279–291. [Google Scholar]
- 236.Reference deleted.
- 237.Maat J, Roza M, Verbakel J, Stam H, Santos da Silva M, Borrel M, Egmond M R, Hagemans M L D, van Gorcom R F M, Hessing J G M, van den Hondel C A M J J, van Rotterdam C. Xylanases and their applications in bakery. In: Visser G B J, Kusters-van Someren M A, Voragen A G J, editors. Xylans and xylanases. Vol. 7. Amsterdam, The Netherlands: Elsevier Science; 1992. pp. 349–360. [Google Scholar]
- 238.MacCabe A P, Fernandez-Espinar M T, de Graaff L H, Visser J, Ramon D. Identification, isolation and sequence of the Aspergillus nidulans xlnC gene encoding the 34-kDa xylanase. Gene. 1996;175:29–33. doi: 10.1016/0378-1119(96)00116-3. [DOI] [PubMed] [Google Scholar]
- 239.MacCabe A P, Orejas M, Perez-Gonzalez J A, Ramon D. Opposite patterns of expression of two Aspergillus nidulans xylanase genes with respect to ambient pH. J Bacteriol. 1998;180:1331–1333. doi: 10.1128/jb.180.5.1331-1333.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.MacCabe A P, van den Hombergh J P T W, Tilburn J, Arst H N J, Visser J. Identification, cloning and analysis of the Aspergillus niger gene pacC, a wide domain regulatory gene responsive to ambient pH. Mol Gen Genet. 1996;250:367–374. doi: 10.1007/BF02174395. [DOI] [PubMed] [Google Scholar]
- 241.Maldonado M C, Strasser de Saad A M, Callieri D. Catabolite repression of the synthesis of inducible polygalacturonase and pectinesterase by Aspergillus sp. Curr Microbiol. 1989;18:303–306. [Google Scholar]
- 242.Manzanares P, de Graaff L H, Visser J. Characterization of galactosidases from Aspergillus niger: purification of a novel α-galactosidase activity. Enzyme Microb Technol. 1998;22:383–390. doi: 10.1016/s0141-0229(97)00207-x. [DOI] [PubMed] [Google Scholar]
- 243.Maruyama K, Goto C, Numata M, Suzuki T, Nakagawa Y, Hoshino T, Uchiyama T. o-Acetylated xyloglucan in extracellular polysaccharides from cell-suspension cultures of Mentha. Phytochemistry. 1996;41:1309–1314. doi: 10.1016/0031-9422(95)00739-3. [DOI] [PubMed] [Google Scholar]
- 244.Matsushita J, Kato Y, Matsuda K. Characterization of α-d-xylosidase II from Aspergillus niger. Agric Biol Chem. 1987;51:2015–2016. [Google Scholar]
- 245.Matsushita J, Kato Y, Matsuda K. Purification and properties of an α-d-xylosidase from Aspergillus niger. J Biochem. 1985;98:825–832. doi: 10.1093/oxfordjournals.jbchem.a135341. [DOI] [PubMed] [Google Scholar]
- 246.Mayans O, Scott M, Connerton I, Gravesen T, Benen J, Visser J, Pickersgill R, Jenkins J. Two crystal structures of pectin lyase A from Aspergillus reveal a pH driven conformational change and striking divergences in the substrate-binding clefts of pectin and pectate lyases. Structure. 1997;5:677–689. doi: 10.1016/s0969-2126(97)00222-0. [DOI] [PubMed] [Google Scholar]
- 247.Mazeau K, Perez S. The preferred conformations of the four oligomeric fragments of rhamnogalacturonan II. Carbohydr Res. 1998;311:203–217. doi: 10.1016/s0008-6215(98)00190-6. [DOI] [PubMed] [Google Scholar]
- 248.McCann M C, Roberts K. Architecture of the primary cell wall. In: Lloyd C W, editor. The cytoskeletal basis of plant growth and form. New York, N.Y: Academic Press, Inc.; 1991. pp. 109–129. [Google Scholar]
- 249.McCleary B V. β-d-Mannanase. Methods Enzymol. 1988;160:596–610. [Google Scholar]
- 250.McCleary B V. Comparison of endolytic hydrolases that depolymerize 1,4-β-d-mannan, 1,5-α-l-arabinan, and 1,4-β-d-galactan. In: Leatham G F, Himmel M E, editors. Enzymes in biomass conversion. ACS Symposium Series. Vol. 460. Washington, D.C.: American Chemical Society; 1991. pp. 437–449. [Google Scholar]
- 251.McCleary B V. Modes of action of β-mannanase enzymes of diverse origin on legume seed galactomannans. Phytochemistry. 1979;18:757–763. [Google Scholar]
- 252.McCleary B V, Mathesen N K. Action patterns and substrate-binding requirements of β-d-mannanase with mannosaccharides and mannan-type polysaccharides. Carbohydr Res. 1983;119:191–219. [Google Scholar]
- 253.McCrae S I, Leith K M, H. G A, Wood T M. Xylan degrading enzyme system produced by the fungus Aspergillus awamori: isolation and characterization of a feruloyl esterase and a p-coumaroyl esterase. Enzyme Microbi Technol. 1994;16:826–834. [Google Scholar]
- 254.McKay A M. Extracellular β-galactosidase production during growth of filamentous fungi on polygalacturonic acid. Lett Appl Microbiol. 1991;12:75–77. [Google Scholar]
- 255.McLauchlan W R, Garcia-Conesa M T, Williamson G, Roza M, Ravestein P, Maat A novel class of protein from wheat which inhibits xylanases. Biochem J. 1999;338:441–446. [PMC free article] [PubMed] [Google Scholar]
- 256.McNeill M, Darvill A G, Fry S C, Albersheim P. Structure and function of the primary cell walls of plants. Annu Rev Biochem. 1984;53:625–663. doi: 10.1146/annurev.bi.53.070184.003205. [DOI] [PubMed] [Google Scholar]
- 257.Megnegneau B, Debets F, Hoekstra R F. Genetic variability and relatedness in the complex group of black Aspergilli based on random amplification of polymorphic DNA. Curr Genet. 1993;23:323–329. doi: 10.1007/BF00310893. [DOI] [PubMed] [Google Scholar]
- 258.Micheli P A. Nova Plantarum Genera. 1729. Florentiae. [Google Scholar]
- 259.Mikhailova R V, Sapunova L I, Lobanok A G. Three polygalacturonases constitutively synthesized by Aspergillus alliaceus. World J Microbiol Biotechnol. 1995;11:330–332. doi: 10.1007/BF00367111. [DOI] [PubMed] [Google Scholar]
- 260.Reference deleted.
- 261.Montgomery R, Smith F, Srivastava H. Structure of corn hull hemicellulose. I. Partial hydrolysis and identification of 2-O-(α-glucopyranosuluronic acid)-D-xylopyranose. J Am Chem Soc. 1956;78:2837–2839. [Google Scholar]
- 262.Mulimani V H, Ramalingam R. Enzymic hydrolysis of raffinose and stachiose in soymilk by alpha-galactosidase from Gibberella fujikuroi. Biochem Mol Biol Int. 1995;36:897–905. [PubMed] [Google Scholar]
- 263.Murao S, Sakamoto S, Arai M. Cellulases of Aspergillus aculeatus. Methods Enzymol. 1988;160:275–299. [Google Scholar]
- 264.Mutter M, Beldman G, Pitson S M, Schols H A, Voragen A G J. Rhamnogalacturonan α-d-galactopyranosyluronohydrolase. An enzyme that specifically removes the terminal nonreducing galacturonosyl residue in rhamnogalacturonan regions of pectin. Plant Physiol. 1994;117:153–163. doi: 10.1104/pp.117.1.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 265.Mutter M, Beldman G, Schols H A, Voragen A G J. Rhamnogalacturonan α-l-rhamnopyranosylhydrolase. A novel enzyme specific for the terminal nonreducing rhamnosyl unit in rhamnogalacturonan regions of pectin. Plant Physiol. 1994;106:241–250. doi: 10.1104/pp.106.1.241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266.Mutter M, Colquhoun I J, Schols H A, Beldman G, Voragen A G J. Rhamnogalacturonase B from Aspergillus aculeatus is a rhamnogalacturonan α-l-rhamnopyranosyl -(1,4)-α-d-galactopyranosyluronide lyase. Plant Physiol. 1996;110:73–77. doi: 10.1104/pp.110.1.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267.Muzakhar K, Hayashi H, Kawaguchi T, Sumitani J, Arai M. Purification and properties of α-l-arabinofuranosidase and endo-β-d-1,4-galactanase from Aspergillus niger v. Tieghem KF-267 which liquified the Okara. In: Ohmiya K, Sakka K, Karita S, Hayashi H, Kobayashi Y, Kimura T, editors. Genetics, biochemistry and ecology of cellulose degradation. Tokyo, Japan: Uni Publishers Co., Ltd.; 1999. pp. 134–143. [Google Scholar]
- 268.Nagai M, Ozawa A, Katsuragi T, Kawasaki H, Sakai T. Cloning and heterologous expression of gene encoding a polygalacturonase from Aspergillus awamori. Biosci Biotechnol Biochem. 2000;64:1580–1587. doi: 10.1271/bbb.64.1580. [DOI] [PubMed] [Google Scholar]
- 269.Nagata O, Takashima T, Tanaka M, Tsukagoshi N. Aspergillus nidulans nuclear proteins bind to a CCAAT element and the adjacent upstream sequence in the promoter region of the starch-inducible taka-amylase A gene. Mol Gen Genet. 1993;237:251–260. doi: 10.1007/BF00282807. [DOI] [PubMed] [Google Scholar]
- 270.Neustroev K N, Krylov A S, Firsov L M, Abroskina O L, Khorlin A Y. Isolation and properties of β-mannosidase from Aspergillus awamori. Biokhimiya. 1991;56:1406–1412. [Google Scholar]
- 271.Nikkuni S, Kosaka N, Suzuki C, Mori K. Comparitive sequence analysis on the 18S rRNA gene of Apergillus oryzae, A. sojae, A. flavus, A. parasiticus, A. niger, A. awamori and A. tamari. J Gen Appl Microbiol. 1996;42:181–187. [Google Scholar]
- 272.Ntarima P, Nerinckx W, Klarslov K, Devreese B, Bhat M K, Van Beeumen J, Claeyssens M. Epoxyalkyl glycosides of d-xylose and xylo-oligosaccharides are active-site markers of xylanases from glycoside family 11, not from family 10. Biochem J. 2000;347:865–873. [PMC free article] [PubMed] [Google Scholar]
- 273.Okada G. Purification and properties of a cellulase from Aspergillus niger. Agric Biol Chem. 1985;49:1257–1265. [Google Scholar]
- 274.Ooi T, Shinmyo A, Okada H, Murao S, Kawaguchi T, Arai M. Complete nucleotide sequence of a gene coding for Aspergillus aculeatus cellulase (FI-CMCase) Nucleic Acids Res. 1990;18:5884. doi: 10.1093/nar/18.19.5884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 275.Oosterveld, A., J. H. Grabber, G. Beldman, R. J., and A. G. J. Voragen. 1997. Formation of ferulic acid dehydrodimers through oxidative cross-linking of sugar beet pectin. Carbohydr. Res. 300:179–189.
- 276.Orejas M, Espeso E A, Tilburn J, Sarkar S, Arst H N J, Penalva M A. Activation of the Aspergillus PacC transcription factor in response to alkaline ambient pH requires proteolysis of the carboxy-terminal moiety. Genes Dev. 1995;9:1622–1632. doi: 10.1101/gad.9.13.1622. [DOI] [PubMed] [Google Scholar]
- 277.Oxenboll Sorensen S, Pauly M, Bush M, Skjot M, McCann M C, Borkhardt B, Ulvskov P. Pectin engineering: modification of potato pectin by in vivo expression of an endo-1,4-β-d-galactanase. Proc Natl Acad Sci USA. 2000;97:7639–7644. doi: 10.1073/pnas.130568297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 278.Panozzo C, Cornillot E, Felenbok B. The CreA repressor is the sole DNA-binding protein responsible for carbon catabolite repression of the alcA gene in Aspergillus nidulans via its binding to a couple of specific sites. J Biol Chem. 1998;273:6367–6372. doi: 10.1074/jbc.273.11.6367. [DOI] [PubMed] [Google Scholar]
- 279.Papagiannopoulos P, Adrianopoulos A, Sharp J A, Davis M A, Hynes M J. The hapC gene of Aspergillus nidulans is involved in the expression of CCAAT-containing promoters. Mol Gen Genet. 1996;251:412–421. doi: 10.1007/BF02172369. [DOI] [PubMed] [Google Scholar]
- 280.Parenicova L. Pectinases of Aspergillus niger: a molecular and biochemical characterisation. Ph.D. thesis. Wageningen, The Netherlands: Wageningen University; 2000. [Google Scholar]
- 281.Parenicová L, Benen J A E, Kester H C M, Visser J. pgaA and pgaB encode two constitutively expressed endopolygalacturonases of Aspergillus niger. Biochem J. 2000;345:637–644. [PMC free article] [PubMed] [Google Scholar]
- 282.Parenicová L, Benen J A E, Kester H C M, Visser J. pgaE encodes a fourth member of the endopolygalacturonase gene family from Aspergillus niger. Eur J Biochem. 1998;251:72–80. doi: 10.1046/j.1432-1327.1998.2510072.x. [DOI] [PubMed] [Google Scholar]
- 283.Parenicová L, Benen J A E, Samson R A, Visser J. Evaluation of RFLP analysis of the classification of selected black aspergilli. Mycol Res. 1997;101:810–814. [Google Scholar]
- 284.Parenicová L, Kester H C M, Benen J A E, Visser J. Characterization of a novel endopolygalacturonase from Aspergillus niger with unique kinetic properties. FEBS Lett. 2000;467:333–336. doi: 10.1016/s0014-5793(00)01173-x. [DOI] [PubMed] [Google Scholar]
- 285.Paroda S, Mishra M M. Growth and enzyme production by Aspergillus terreus in holocellulose. Ann Microbiol. 1984;135A:397–302. doi: 10.1016/s0769-2609(84)80012-5. [DOI] [PubMed] [Google Scholar]
- 286.Parry J B, Stewart J C, Heptinstall J. Purification of the major endoglucanase from Aspergillus fumigatus Fresenius. Biochem J. 1983;312:437–444. doi: 10.1042/bj2130437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 287.Pauly M, Andersen L N, Kaupinnen S, Kofod L V, York W S, Albersheim P, Darvill A. A xyloglucan-specific endo-β-1,4-glucanase from Aspergillus aculeatus: expression cloning in yeast, purification and characterization of the recombinant enzyme. Glycobiology. 1999;9:93–100. doi: 10.1093/glycob/9.1.93. [DOI] [PubMed] [Google Scholar]
- 288.Pellerin P, Brillouet J-M. Purification and properties of an exo-(1→3)-β-d-galactanase from Aspergillus niger. Carbohydr Res. 1994;264:281–291. doi: 10.1016/s0008-6215(05)80012-6. [DOI] [PubMed] [Google Scholar]
- 289.Pellerin P, Gosselin M, Lepoutre J-P, Samain E, Debeire P. Enzymic production of oligosaccharides from corncob xylan. Enzyme Microb Technol. 1991;13:617–621. [Google Scholar]
- 290.Perez S, Mazeau K, Herve du Penhoat C. The three-dimensional structures of the pectic polysaccharides. Plant Physiol Biochem. 2000;38:37–55. [Google Scholar]
- 291.Perez-Gonzalez J A, de Graaff L H, Visser J, Ramon D. Molecular cloning and expression in Saccharomyces cerevisiae of two Aspergillus nidulans xylanase genes. Appl Environ Microbiol. 1996;62:2179–2182. doi: 10.1128/aem.62.6.2179-2182.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 292.Perez-Gonzalez J A, van Peij N N M E, Bezoen A, MacCabe A P, Ramon D, de Graaff L H. Molecular cloning and transcriptional regulation of the Aspergillus nidulans xlnD gene encoding β-xylosidase. Appl Environ Microbiol. 1998;64:1412–1419. doi: 10.1128/aem.64.4.1412-1419.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 293.Petersen R, Kauppinen S, Larsen S. The crystal structure of rhamnogalacturonase A from Aspergillus aculeatus: a right-handed β helix. Structure. 1997;5:533–544. doi: 10.1016/s0969-2126(97)00209-8. [DOI] [PubMed] [Google Scholar]
- 294.Petit, S., R. Ralainirina, S. Favre, and R. de Baynast. February 1993. International Patent 93/02092.
- 295.Petit-Benvegnen M-D, Saulnier L, Rouau X. Solubilization of arabinoxylans from isolated water-unextractable pentosans and wheat flour doughs by cell wall degrading enzymes. Cereal Chem. 1998;75:551–556. [Google Scholar]
- 296.Pinaga F, Fernandez-Espinar M T, Valles S, Ramon D. Xylanase production in Aspergillus nidulans: Induction and carbon catabolite repression. FEMS Microbiol Lett. 1994;115:319–324. doi: 10.1111/j.1574-6968.1994.tb06622.x. [DOI] [PubMed] [Google Scholar]
- 297.Pitson S M, Mutter M, van den Broek L A M, Voragen A G J, Beldman G. Stereochemical course of hydrolysis catalysed by α-l-rhamnosyl and α-d-galacturonosyl hydrolases from Aspergillus aculeatus. Biochem Biophys Res Commun. 1998;242:552–559. doi: 10.1006/bbrc.1997.8009. [DOI] [PubMed] [Google Scholar]
- 298.Poutanen K-J. Enzymes. An important tool in the improvement of the quality of cereal foods. Trends Food Sci Technol. 1997;8:300–306. [Google Scholar]
- 299.Puls J, Borneman A, Gottschalk D, Wiegel J. Xylobiose and xylooligomers. Methods Enzymol. 1988;160:528–536. [Google Scholar]
- 300.Puls J, Schorn B, Schuseil J. Acetylmannanesterase: a new component in the arsenal of wood mannan degrading enzymes. In: Kuwahara M, Shimada M, editors. Biotechnology in pulp and paper industry. Tokyo, Japan: Uni Publishers Co., Ltd.; 1992. pp. 357–363. [Google Scholar]
- 301.Pushalkar S, Rao K K, Menon K. Production of β-glucosidase by Aspergillus terreus. Curr Microbiol. 1995;30:255–258. doi: 10.1007/BF00295497. [DOI] [PubMed] [Google Scholar]
- 302.Ralet M-C, Faulds C B, Williamson G, Thibault J-F. Degradation of feruloylated oligosaccharides from sugar beet pulp and wheat bran by ferulic acid esterases from Aspergillus niger. Carbohydr Res. 1994;263:257–269. doi: 10.1016/0008-6215(94)00177-4. [DOI] [PubMed] [Google Scholar]
- 303.Ramon D, van der Veen P, Visser J. Arabinan degrading enzymes from Aspergillus nidulans: induction and purification. FEMS Microbiol Lett. 1993;113:15–22. doi: 10.1111/j.1574-6968.1993.tb06481.x. [DOI] [PubMed] [Google Scholar]
- 304.Rättö M, Viikari L. Pectinases in wood debarking. In: Visser J, Voragen A G J, editors. Pectins and pectinases. Vol. 14. Amsterdam, The Netherlands: Elsevier Science; 1996. pp. 979–982. [Google Scholar]
- 305.Reczey K, Stalbrand H, Hahn-Hagerdal B, Tjerneld F. Mycelia-associated β-galactosidase activity in microbial pellets of Aspergillus and Penicillium strains. Appl Microbiol Biotechnol. 1992;38:393–397. [Google Scholar]
- 306.Reese E T, Shibata Y. β-Mannanases of fungi. Can J Microbiol. 1965;11:167–183. doi: 10.1139/m65-023. [DOI] [PubMed] [Google Scholar]
- 307.Regalado C, Garcia-Almendarez B E, Venegas-Barrera L M, Tellez-Jurado A, Rodriguez-Serrano G, Huerta-Ochoa S, Whitaker J R. Production, partial purification and properties of β-mannanases obtained by solid substrate fermentation of spent soluble coffee wastes and copra paste using Aspergillus oryzae and Aspergillus niger. J Sci Food Agric. 2000;80:1343–1350. [Google Scholar]
- 308.Ring S G, Selvendran R R. An arabinogalactoxyloglucan from the cell wall of Solanum tuberosum. Phytochemistry. 1981;20:2511–2519. [Google Scholar]
- 309.Rios S, Pedregosa A M, Fernandez Monistrol I, Laborda F. Purification and molecular properties of an α-galactosidase synthesized and secreted by Aspergillus nidulans. FEMS Microbiol Lett. 1993;112:35–42. doi: 10.1111/j.1574-6968.1993.tb06420.x. [DOI] [PubMed] [Google Scholar]
- 310.Riou C, Salmon J-M, Vallier M-J, Gunata Z, Barre P. Purification, characterization, and substrate specificity of a novel high glucose-tolerant β-glucosidase from Aspergillus oryzae. Appl Environ Microbiol. 1998;64:3607–3614. doi: 10.1128/aem.64.10.3607-3614.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 311.Rizk S E, Abdel-Massih R M, Baydoun E A-H, Brett C T. Protein- and pH-dependent binding of nascent pectin and glucuronoarabinoxylan to xyloglucan in pea. Planta. 2000;211:423–429. doi: 10.1007/s004250000303. [DOI] [PubMed] [Google Scholar]
- 312.Rodionova N A, Tavobilov I M, Bezborodov M. β-Xylosidase from Aspergillus niger 15: purification and properties. J Appl Biochem. 1983;5:300–312. [PubMed] [Google Scholar]
- 313.Rodionova N A, Tavobilov I M, Martinovich L I, Buachidze T S, Kvesitadze G I, Bezborodov A M. β-Glucosidases from cellulolytic fungi Aspergillus terreus, Geotrichum candidum, and Trichoderma longibrachiatum as typical glycosidases. Biotechnol Appl Biochem. 1987;9:239–250. doi: 10.1111/j.1470-8744.1987.tb00475.x. [DOI] [PubMed] [Google Scholar]
- 314.Rombouts F M, Thibault J-F. Feruloylated pectic substances from sugar-beet pulp. Carbohydr Res. 1986;154:177–187. [Google Scholar]
- 315.Rombouts F M, Voragen A G J, Searle-van Leeuwen M F, Geraeds C C J M, Schols H A, Pilnik W. The arabinases of Aspergillus niger: purification and characterization of two α-l-arabinofuranosidases and an endo-1,5-α-l-arabinase. Carbohydr Polym. 1988;9:25–47. [Google Scholar]
- 316.Ross J M, Saura D, Coll L, M. M, Laencina J. Oligouronides of pectins in membrane reactor by enzymatic degradation of pectins from citrus peel. A preliminary study. In: Visser J, Voragen A G J, editors. Pectins and pectinases. Vol. 14. Amsterdam, The Netherlands: Elsevier Science; 1996. pp. 983–990. [Google Scholar]
- 317.Rouau X, Surget A. Evidence for the presence of a pentosanase inhibitor in wheat. J Cereal Sci. 1998;28:63–70. [Google Scholar]
- 318.Ruijter G J G, Vanhanen S I, Gielkens M M C, van de Vondervoort P J I, Visser J. Isolation of Aspergillus niger creA mutants: effects on expression of arabinases and l-arabinose catabolic enzymes. Microbiology. 1997;143:2991–2998. doi: 10.1099/00221287-143-9-2991. [DOI] [PubMed] [Google Scholar]
- 319.Ruijter G J G, Visser J. Carbon repression in aspergilli. FEMS Microbiol Lett. 1997;151:103–114. doi: 10.1111/j.1574-6968.1997.tb12557.x. [DOI] [PubMed] [Google Scholar]
- 320.Ruttkowski E, Khanh N Q, Wientjes F-J, Gottschalk M. Characterisation of a polygalacturonase gene of Aspergillus niger RH5344. Mol Microbiol. 1991;5:1353–1361. doi: 10.1111/j.1365-2958.1991.tb00782.x. [DOI] [PubMed] [Google Scholar]
- 321.Ruttkowski E, Labitzke R, Khanh N Q, Loffler F, Gottschalk M, Jany K D. Cloning and DNA sequence analysis of a polygalacturonase cDNA from Aspergillus niger RH5344. Biochim Biophys Acta. 1990;1087:104–106. doi: 10.1016/0167-4781(90)90130-t. [DOI] [PubMed] [Google Scholar]
- 322.Sakamoto S, Tamura G, Ito K, Ishikawa T, Iwano K, Nishiya N. Cloning and sequencing of cellulase cDNA from Aspergillus kawachii and its expression in Saccharomyces cerevisiae. Curr Genet. 1995;27:435–439. doi: 10.1007/BF00311212. [DOI] [PubMed] [Google Scholar]
- 323.Sanyal A, Kundu R K, Dube S, Dube D K. Extracellular cellulolytic enzyme system of Aspergillus japonicus. 2. Purification and characterization of an inducible extracellular β-glucosidase. Enzyme Microb Technol. 1988;10:91–99. [Google Scholar]
- 324.Saulnier L, Vigouroux J, Thibault J-F. Isolation and partial characterization of feruloylated oligosaccharides from maize bran. Carbohydr Res. 1995;272:241–253. doi: 10.1016/0008-6215(95)00053-v. [DOI] [PubMed] [Google Scholar]
- 325.Schols H A, Geraeds C J M, Searle-van Leeuwen M F, Kormelink F J M, Voragen A G J. Rhamnogalacturonase: a novel enzyme that degrades the hairy regions of pectins. Carbohydr Res. 1990;206:104–115. [Google Scholar]
- 326.Schols H A, Voragen A G J. Complex pectins: structure elucidation using enzymes. In: Visser J, Voragen A G J, editors. Pectin and pectinases. Vol. 14. Amsterdam, The Netherlands: Elsevier Science; 1996. pp. 793–798. [Google Scholar]
- 327.Schols H A, Voragen A G J. Occurrence of pectic hairy regions in various plant cell wall materials and their degradability by rhamnogalacturonase. Carbohydr Res. 1994;256:83–95. doi: 10.1016/0008-6215(94)84230-2. [DOI] [PubMed] [Google Scholar]
- 328.Schooneveld-Bergmans M E F, Hopman A M C P, Beldman G, Voragen A G J. Extraction and partial characterization of feruloylated glucuronoarabinoxylans from wheat bran. Carbohydr Polym. 1998;35:39–47. [Google Scholar]
- 329.Schopplein E, Dietrich H. Charakterisierung polysaccharidspaltender Begleitaktivitaten in einem technischen Pektinasepreparat. Dtsche Lebensm-Rundsch. 1991;87:212–219. [Google Scholar]
- 330.Scigelova M, Crout D H G. Purification of α-galactosidase from Aspergillus niger for application in the synthesis of complex oligosaccharides. J Mol Catal. 2000;8:175–181. [Google Scholar]
- 331.Searle-van Leeuwen M J F, van den Broek L A M, Schols H A, Beldman G, Voragen A G J. Rhamnogalacturonan acetylesterase: a novel enzyme from Aspergillus aculeatus, specific for the deacetylation of hairy (ramified) regions of pectins. Appl Microbiol Biotechnol. 1992;38:347–349. [Google Scholar]
- 332.Searle-van Leeuwen M J F, Vincken J-P, Schipper D, Voragen A G J, Beldman G. Acetyl esterases of Aspergillus niger: purification and mode of action on pectins. In: Visser J, Voragen A G J, editors. Pectins and pectinases. Vol. 14. Amsterdam, The Netherlands: Elsevier Science; 1996. pp. 793–798. [Google Scholar]
- 333.Shei J C, Fratzke A R, Frederick M M, Frederick J R, Reilly P J. Purification and characterization of endo-xylanases from Aspergillus niger. II. An enzyme of pI 4.5. Biotechnol Bioeng. 1985;27:533–538. doi: 10.1002/bit.260270421. [DOI] [PubMed] [Google Scholar]
- 334.Sherief A A. Separation and some properties of an endo-1,4-β-d-xylanase from Aspergillus flavipes. Acta Microbiol Hung. 1990;37:301–306. [PubMed] [Google Scholar]
- 335.Shinoyama H, Ando A, Fujii T, Yasui T. The possibility of enzymatic synthesis of a variety of β-xylosides using the transfer reaction of Aspergillus niger β-xylosidase. Agric Biol Chem. 1991;55:849–850. [Google Scholar]
- 336.Shroff R A, Lockington R A, Kelly J M. Analysis of mutations in the creA gene involved in carbon catabolite repression in Aspergillus nidulans. Can J Microbiol. 1996;42:950–959. doi: 10.1139/m96-122. [DOI] [PubMed] [Google Scholar]
- 337.Silva C H C, Puls J, Valle de Sousa M, Ferreira Filho E X. Purification and characterization of low molecular weight xylanase from solid-state cultures of Aspergillus fumigatus Fresenius. Rev Microbiol. 1999;30:114–119. [Google Scholar]
- 338.Sims I, Craik D J, Bacic A. Structural characterisation of galactoglucomannan secreted by suspension-cultured cells of Nicotiana plumbaginifolia. Carbohydr Res. 1997;303:79–92. doi: 10.1016/s0008-6215(97)00144-4. [DOI] [PubMed] [Google Scholar]
- 339.Sims I, Munro S L A, Currie G, Craik D, Bacic A. Structural characterisation of xyloglucan secreted by suspension-cultured cells of Nicotiana plumbaginifolia. Carbohydr Res. 1996;293:147–172. doi: 10.1016/0008-6215(96)00142-5. [DOI] [PubMed] [Google Scholar]
- 340.Singh A, Agrawal K A, Abidi A B, Darmwal N S. Properties of exoglucanase from Aspergillus niger. J Gen Appl Microbiol. 1990;36:245–253. [Google Scholar]
- 341.Singh S, Brar K, Sandhu D K, Kaur A. Isozyme polymorphism of cellulases in Aspergillus terreus. J Basic Microbiol. 1996;36:289–296. doi: 10.1002/jobm.3620360412. [DOI] [PubMed] [Google Scholar]
- 342.Smith D C, Bhat K M, Wood T M. Xylan-hydrolizing enzymes from thermophilic and mesophilic fungi. World J Microbiol Biotechnol. 1991;7:475–484. doi: 10.1007/BF00303373. [DOI] [PubMed] [Google Scholar]
- 343.Smith M M, Hartley R D. Occurrence and nature of ferulic acid substitution of cell wall polysaccharides in graminaceous plants. Carbohydr Res. 1983;118:65–80. [Google Scholar]
- 344.Solis-Pereira S, Favela-Torres E, Viniegra-Gonzalez G, Gutierrez-Rojas M. Effects of different carbon sources on the synthesis of pectinase by Aspergillus niger in submerged and solid state fermentations. Appl Microbiol Biotechnol. 1993;39:36–41. [Google Scholar]
- 345.Somiari R I, Balogh E. Properties of an extracellular glycosidase of Aspergillus niger suitable for removal of oligosaccharides from cowpea meal. Enzyme Microb Technol. 1995;17:311–316. [Google Scholar]
- 346.Steidl S, Papagiannopoulus P, Litzka O, Adrianopoulos A, Davis M A, Brakhage A A, Hynes M J. AnCF, the CCAAT binding complex of Aspergillus nidulans contains products of the hapB, hapC, and hapE genes and is required for activation by the pathway-secific regulatory gene amdR. Mol Cell Biol. 1999;19:99–106. doi: 10.1128/mcb.19.1.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 347.Stewart J C, Parry J B. Factors influencing the production of celulase by Aspergillus fumigatus (Fresenius) J Gen Microbiol. 1981;125:33–39. doi: 10.1099/00221287-125-1-33. [DOI] [PubMed] [Google Scholar]
- 348.Stotz H U, Bishop J G, Bergmann C W, Koch M, Albersheim P, Darvill A G, Labavitch J M. Identification of target amino acids that affect interactions of fungal polygalacturonases and their plant inhibitors. Physiol Mol Plant Pathol. 2000;56:117–130. [Google Scholar]
- 349.Strauss J, Horvath H K, Abdallah B M, Kindermann J, Mach R L, Kubicek C P. The function of CreA, the carbon catabolite repressor of Aspergillus nidulans, is regulated at the transcriptional and post-transcriptional level. Mol Microbiol. 1999;32:169–178. doi: 10.1046/j.1365-2958.1999.01341.x. [DOI] [PubMed] [Google Scholar]
- 350.Sulistyo J, Kamiyama Y, Yasui T. Purification and some properties of Aspergillus pulverulentus β-xylosidase with transxylosylation capacity. J Ferment Bioeng. 1995;79:17–22. [Google Scholar]
- 351.Sundberg M, Poutanen K, Markkanen P, Linko M. An extracellular esterase of Aspergillus awamori. Biotechnol Appl Biochem. 1990;12:670–680. [Google Scholar]
- 352.Suykerbuyk M E G, Kester H C M, Schaap P J, Stam H, Musters W, Visser J. Cloning and characterization of two rhamnogalacturonan hydrolase genes from Aspergillus niger. Appl Environ Microbiol. 1997;63:2507–2515. doi: 10.1128/aem.63.7.2507-2515.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 353.Suykerbuyk M E G, Schaap P J, Stam H, Musters W, Visser J. Cloning, sequence and expression of the gene for rhamnogalacturonan hydrolase of Aspergillus aculeatus; a novel pectinolytic enzyme. Appl Microbiol Biotechnol. 1995;43:861–870. doi: 10.1007/BF02431920. [DOI] [PubMed] [Google Scholar]
- 354.Suykerbuyk M E G, van de Vondervoort P J I, Schaap P J, Visser J. Identification of regulatory mutants of Aspergillus aculeatus affected in rhamnogalacturonan hydrolase expression. Curr Genet. 1996;30:439–446. doi: 10.1007/s002940050154. [DOI] [PubMed] [Google Scholar]
- 355.Tagawa K, Kaji A. α-l-Arabinofuranosidase from Aspergillus niger. Methods Enzymol. 1988;160:707–712. [Google Scholar]
- 356.Takada G, Kawaguchi T, Kaga T, Sumitani J-I, Arai M. Cloning and sequencing of β-mannosidase gene from Aspergillus aculeatus No.F-50. Biosci Biotechnol Biochem. 1999;63:206–209. doi: 10.1271/bbb.63.206. [DOI] [PubMed] [Google Scholar]
- 357.Takada G, Kawaguchi T, Sumitani J-I, Arai M. Cloning, nucleotide sequence, and transcriptional analysis of Aspergillus aculeatus No.F-50 cellobiohydrolase I (cbhI) gene. J Ferment Bioeng. 1998;85:1–9. [Google Scholar]
- 358.Takada G, Kawaguchi T, Yoneda T, Kawasaki M, Sumitani J-I, Arai M. Molecular cloning and expression of the cellulolytic system from Aspergillus aculeatus. In: Ohmiya K S K, Karita S, Hayashi H, Kobayashi Y, Kimura T, editors. Genetics, biochemistry and ecology of cellulose degradation. Tokyo, Japan: Uni Publishers Co., Ltd.; 1999. pp. 364–373. [Google Scholar]
- 359.Takahashi N, Koshijima T. Ester linkages between lignin and glucuronoxylan in a lignin-carbohydrate complex from beech (Fagus crenata) wood. Wood Sci Technol. 1988;22:231–241. [Google Scholar]
- 360.Tavolibov I M, Buachidze T S, Rodionova N A, Kvesitadze G I, Bezborodov A M. Substrate specificity and certain catalytic characteristics of β-glucosidase from Aspergillus terreus. Biochemistry. 1988;49:847–853. [Google Scholar]
- 361.Tenkanen M. Action of Tricoderma reesei and Aspergillus oryzae esterases in the deacetylation of hemicelluloses. Biotechnol Appl Biochem. 1998;27:19–24. doi: 10.1111/j.1470-8744.1998.tb01370.x. [DOI] [PubMed] [Google Scholar]
- 362.Tenkanen M, Puls J, Ratto M, Viikari L. Enzymatic deacetylation of galactoglucomannans. Appl Microbiol Biotechnol. 1993;39:159–165. [Google Scholar]
- 363.Tenkanen M, Schuseil J, Puls J, Poutanen K. Production, purification and characterisation of an esterase liberating phenolic acids from lignocellulosics. J Biotechnol. 1991;18:69–84. [Google Scholar]
- 364.Tenkanen M, Thornton J, Viikari L. An acetylglucomannan esterase of Aspergillus oryzae; purification, characterization and role in the hydrolysis of O-acetyl-galactoglucomannan. J Biotechnol. 1995;42:197–206. doi: 10.1016/0168-1656(95)00080-a. [DOI] [PubMed] [Google Scholar]
- 365.Then Bergh K, Litzka O, Brakhage A A. Identification of a major cis-acting DNA element controlling the bidirectionally transcribed penicillium biosynthesis genes acvA (pcbAB) and ipnA (pcbC) of Aspergillus nidulans. J Bacteriol. 1996;178:3908–3916. doi: 10.1128/jb.178.13.3908-3916.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 366.Thom C, Church M B. The aspergilli. Baltimore, Md: The Williams & Wilkins Co.; 1926. [Google Scholar]
- 367.Thom C, Raper K B. A manual of the aspergilli. Baltimore, Md: The Williams & Wilkins Co.; 1945. [Google Scholar]
- 368.Tilburn J, Sarkar S, Widdick D A, Espeso E A, Orejas M, Mungroo J, Penalva M A, Arst H A., Jr The Aspergillus PacC zinc finger transcription factor mediates regulation of both acidic and alkaline expressed genes by ambient pH. EMBO J. 1995;14:779–790. doi: 10.1002/j.1460-2075.1995.tb07056.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 369.Timell T E. Recent progress in the chemistry of wood hemicelluloses. Wood Sci Technol. 1967;1:45–70. [Google Scholar]
- 370.Uchida H, Kusakabe I, Kawabata Y, Ono T, Murakami K. Production of xylose from xylan with intracellular enzyme system of Aspergillus niger 5-16. J Ferment Bioeng. 1992;74:153–158. [Google Scholar]
- 371.Uchida H, Nanri T, Kawabata Y, Kusakabe I, Murakami K. Purification and characterization of intracellular α-glucuronidase from Aspergillus niger 5-16. Biosci Biotechnol Biochem. 1992;56:1608–1615. [Google Scholar]
- 372.Ueda S, Fujio Y, Lim J Y. Production and some properties of pectic enzymes from Aspergillus oryzae A-3. J Appl Biochem. 1982;4:524–532. [Google Scholar]
- 373.Unno T, Ide K, Yazaki T, Tanaka Y, Nakakuki T, Okada G. High recovery purification and some properties of a β-glucosidase from Aspergillus niger. Biosci Biotechnol Biochem. 1993;57:2172–2173. [Google Scholar]
- 374.van de Vis J W, Searle-van Leeuwen M J F, Siliha H A, Kormelink F J M, Voragen A G J. Purification and characterization of endo-1,4-β-d-galactanases from Aspergillus niger and Aspergillus aculeatus: use in combination with arabinases from Aspergillus niger in enzymic conversion of potato arabinogalactan. Carbohydr Polym. 1991;16:167–187. [Google Scholar]
- 375.van der Veen P, Flipphi M J A, Voragen A G J, Visser J. Induction of extracellular arabinases on monomeric substrates in Aspergillus niger. Arch Microbiol. 1993;159:66–71. doi: 10.1007/BF00244266. [DOI] [PubMed] [Google Scholar]
- 376.van der Veen P, Flipphi M J A, Voragen A G J, Visser J. Induction, purification and characterisation of arabinases produced by Aspergillus niger. Arch Microbiol. 1991;157:23–28. doi: 10.1007/BF00245330. [DOI] [PubMed] [Google Scholar]
- 377.van der Vlugt-Bergmans C J B, Meeuwsen P J A, Voragen A G J, van Ooyen A J J. Endo-xylogalacturonan hydrolase, a novel pectinolytic enzyme. Appl Environ Microbiol. 2000;66:36–41. doi: 10.1128/aem.66.1.36-41.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 378.van der Vlugt-Bergmans C J B, van Ooyen A J J. Expression cloning in Kluyveromyces lactis. BioTechniques. 1999;13:87–92. [Google Scholar]
- 379.van Heeswijck R, Hynes M J. The amdR product and a CAATT-binding factor bind to adjacent, possibly overlapping DNA sequences in the promoter region of the Aspergillus nidulans amdS gene. Nucleic Acids Res. 1991;19:2655–2660. doi: 10.1093/nar/19.10.2655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 380.van Houdenhoven F E A. Studies on pectin lyase. Ph.D. thesis. Wageningen, The Netherlands: Wageningen Agricultural University; 1975. [Google Scholar]
- 381.van Peij N N M E. Transcriptional regulation of the xylanolytic enzyme system of Aspergillus. Ph.D. thesis. Wageningen, The Netherlands: Wageningen University; 1999. [Google Scholar]
- 382.van Peij N N M E, Brinkmann J, Vrsanska M, Visser J, de Graaff L H. β-Xylosidase activity, encoded by xlnD, is essential for complete hydrolysis of xylan by Aspergillus niger but not for induction of the xylanolytic enzyme spectrum. Eur J Biochem. 1997;245:164–173. doi: 10.1111/j.1432-1033.1997.00164.x. [DOI] [PubMed] [Google Scholar]
- 383.van Peij N N M E, Gielkens M M C, de Vries R P, Visser J, de Graaff L H. The transcriptional activator xlnR regulates both xylanolytic and endoglucanase expression in Aspergillus niger. Appl Environ Microbiol. 1998;64:3615–3619. doi: 10.1128/aem.64.10.3615-3619.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 384.van Peij N N M E, Visser J, de Graaff L H. Isolation and analysis of xlnR, encoding a transcriptional activator coordinating xylanolytic expression in Aspergillus niger. Mol Microbiol. 1998;27:131–142. doi: 10.1046/j.1365-2958.1998.00666.x. [DOI] [PubMed] [Google Scholar]
- 385.van Santen Y, Benen J A E, Schröter K-H, Kalk K H, Armand S, Visser J, Dijkstra B W. 1.68-Å crystal structure of endopolygalacturonase II from Aspergillus niger and identification of active site residues by site-directed mutagenesis. J Biol Chem. 1999;274:30474–30480. doi: 10.1074/jbc.274.43.30474. [DOI] [PubMed] [Google Scholar]
- 386.Varga J, Kevei F, Vriesema A, Debets F, Kozakiewicz Z, Croft J H. Mitochondrial DNA restriction fragment length polymorphisms in field isolates of the Aspergillus niger aggregate. Can J Microbiol. 1994;40:612–621. doi: 10.1139/m94-098. [DOI] [PubMed] [Google Scholar]
- 387.Verbruggen M A, Beldman G, Voragen A G J. Enzymic degradation of sorghum glucuronoarabinoxylans leading to tentative structures. Carbohydr Res. 1998;306:275–282. doi: 10.1016/s0008-6215(97)10065-9. [DOI] [PubMed] [Google Scholar]
- 388.Verbruggen M A, Beldman G, Voragen A G J. Structures of enzymically derived oligosaccharides from sorghum glucuronoarabinoxylan. Carbohydr Res. 1998;306:265–274. doi: 10.1016/s0008-6215(97)10064-7. [DOI] [PubMed] [Google Scholar]
- 389.Vidmar S, Turk V, Kregar I. Cellulolytic complex of Aspergillus niger under conditions for citric acid production. Isolation and characterization of two β-(1→4)-glucan hydrolases. Appl Microbiol Biotechnol. 1984;20:326–330. [Google Scholar]
- 390.Viikari L, Kantelinen A, Sundquist J, Linko M. Xylanases in bleaching — from an idea to the industry. FEMS Microbiol Rev. 1994;13:335–350. [Google Scholar]
- 391.Vincken J-P, York W S, Beldman G, Voragen A G J. Two general branching patterns of xyloglucan, XXXG and XXGG. Plant Physiol. 1997;114:9–13. doi: 10.1104/pp.114.1.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 392.Visser J, Bussink H-J, Witteveen C. Gene expression in filamentous fungi. In: Smith A, editor. Gene expression in recombinant microorganisms. New York, N.Y: Marcel Dekker, Inc; 1994. pp. 241–308. [Google Scholar]
- 393.Vitali J, Schick B, Kester H C M, Visser J, Jurnak F. The three-dimensional structure of Aspergilus niger pectin lyase B at 1.7-Å resolution. Plant Physiol. 1998;116:69–80. doi: 10.1104/pp.116.1.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 394.Vogel M. Alternative utilization of sugar beet pulp. Zuckerindustrie. 1991;116:265–270. [Google Scholar]
- 395.Wakabayashi K, Hososn T, Kamisaka S. Osmotic stress suppresses cell wall stiffening and the increase in cell wall-bound ferulic and diferulic acids in wheat coleoptiles. Plant Physiol. 1997;113:407–411. doi: 10.1104/pp.113.3.967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 396.Watanabe T, Sato T, Yoshioka S, Koshijima T, Kuwahara M. Purification and properties of Aspergillus niger β-glucosidase. Eur J Biochem. 1992;209:651–659. doi: 10.1111/j.1432-1033.1992.tb17332.x. [DOI] [PubMed] [Google Scholar]
- 397.Wende G, Fry S C. O-feruloylated, O-acetylated oligosaccharides as side-chains of grass xylans. Phytochemistry. 1997;44:1011–1018. doi: 10.1016/s0031-9422(96)00648-6. [DOI] [PubMed] [Google Scholar]
- 398.Whitehead M P, Shieh M T, Cleveland T E, Cary J W, Dean R A. Isolation and characterization of polygalacturonase genes (pecA and pecB) from Aspergillus flavus. Appl Environ Microbiol. 1995;61:3316–3322. doi: 10.1128/aem.61.9.3316-3322.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 399.Wilkie K C B. The hemicelluloses of grasses and cereals. Adv Carbohydr Chem Biochem. 1979;36:215–264. [Google Scholar]
- 400.Wilkie K C B, Woo S-L. A heteroxylan and hemicellulosic materials from bamboo leaves, and a reconsideration of the general nature of commonly occurring xylans and other hemicelluloses. Carbohydr Res. 1977;57:145–162. [Google Scholar]
- 401.Witte K, Wartenberg A. Purification and properties of two β-glucosidases isolated from Aspergillus niger. Acta Biotechnol. 1989;9:179–190. [Google Scholar]
- 402.Wong C-H, Mazenod F P, Whiteside G M. Chemical and enzymatic synthese of 6-deoxyhexoses. Cobversion to 2,5-dimethyl-4-hydroxy-2,3-dihydrofuran-3-one (furaneol) and analogues. J Org Chem. 1983;48:3493–3497. [Google Scholar]
- 403.Wood T M, McCrae S I. Arabinoxylan-degrading enzyme system of the fungus Aspergillus awamori: purification and properties of an α-l-arabinofuranosidase. Appl Microbiol Biotechnol. 1996;45:538–545. doi: 10.1007/BF00578468. [DOI] [PubMed] [Google Scholar]
- 404.Yamaguchi F, Inoue S, Hatanaka C. Purification and properties of endo-β-1,4-galactanase from Aspergillus niger. Biosci Biotech Biochem. 1995;59:1742–1744. doi: 10.1271/bbb.59.1742. [DOI] [PubMed] [Google Scholar]
- 405.Yamazaki N, Sinner M, Dietrichs H H. Isolation and properties of β-1,4-mannanase from Aspergillus niger. Holzforschung. 1976;30:101–109. [Google Scholar]
- 406.Yan T-R, Lin C-L. Purification and characterization of a glucose-tolerant β-glucosidase from Aspergillus niger CCRC 31494. Biosci Biotechnol Biochem. 1997;61:965–970. doi: 10.1271/bbb.61.965. [DOI] [PubMed] [Google Scholar]
- 407.Yan T-R, Lin Y-H, Lin C-L. Purification and characterization of an extracellular β-glucosidase II with high hydrolysis and transglucosylation activities from Aspergillus niger. J Agric Food Chem. 1998;46:431–437. doi: 10.1021/jf9702499. [DOI] [PubMed] [Google Scholar]
- 408.Yeoh H H, Tan T K, Chua S L, Lim G. Properties of β-glucosidase purified from Aspergillus niger. Mircen J Appl Microbiol Biotechnol. 1988;4:425–430. [Google Scholar]
- 409.York W S, Kumar Kolli V S, Orlando R, Albersheim P, Darvill A G. The structures of arabinoxyloglucans produced by solanaceous plants. Carbohydr Res. 1996;285:99–128. doi: 10.1016/s0008-6215(96)90176-7. [DOI] [PubMed] [Google Scholar]
- 410.Yoshikawa K, Yamamoto K, Okada S. Isolation of Aspergillus flavus MO-5 producing two types of intracellular α-d-xylosidases: purification and characterization of α-d-xylosidase I. Biosci Biotechnol Biochem. 1993;57:1275–1280. doi: 10.1271/bbb.57.1275. [DOI] [PubMed] [Google Scholar]
- 411.Yoshikawa K, Yamamoto K, Okada S. Purification and characterization of an intracellular α-d-xylosidase II from Aspergillus flavus MO-5. Biosci Biotechnol Biochem. 1993;57:1281–1285. doi: 10.1271/bbb.57.1281. [DOI] [PubMed] [Google Scholar]
- 412.Zapater I G, Ullah A H J, Wodzinski R J. Extracellular α-galactosidase (EC 3.2.1.22) from Aspergillus ficuum NRRL 3135: purification and characterisation. Prep Biochem. 1990;20:263–296. doi: 10.1080/00327489008050201. [DOI] [PubMed] [Google Scholar]
- 413.Zeikus J G, Lee C, Lee Y E, Saha B C. Thermostable saccharidases. New sources, uses and biodesign. ACS Symp Ser. 1991;460:36–51. [Google Scholar]
- 414.Zeilinger S, Mach R, Kubicek C P. Two adjacent protein binding motifs in the cbh2 (cellobiohydrolase-II encoding) promoter of the fungus Hypocrea jecorina (Trichoderma reesei) cooperate in the induction by cellulose. J Biol Chem. 1998;273:34463–34471. doi: 10.1074/jbc.273.51.34463. [DOI] [PubMed] [Google Scholar]
- 415.Zeilinger S, Mach R, Schindler M, Herzog P, Kubicek C P. Different inducibility of expression of the two xylanase genes xyn1 and xyn2 in Trichoderma reesei. J Biol Chem. 1996;271:25624–25629. doi: 10.1074/jbc.271.41.25624. [DOI] [PubMed] [Google Scholar]