Abstract
Aspergillus section Candidi encompasses white- or yellow-sporulating species mostly isolated from indoor and cave environments, food, feed, clinical material, soil and dung. Their identification is non-trivial due to largely uniform morphology. This study aims to re-evaluate the species boundaries in the section Candidi and present an overview of all existing species along with information on their ecology. For the analyses, we assembled a set of 113 strains with diverse origin. For the molecular analyses, we used DNA sequences of three house-keeping genes (benA, CaM and RPB2) and employed species delimitation methods based on a multispecies coalescent model. Classical phylogenetic methods and genealogical concordance phylogenetic species recognition (GCPSR) approaches were used for comparison. Phenotypic studies involved comparisons of macromorphology on four cultivation media, seven micromorphological characters and growth at temperatures ranging from 10 to 45 °C. Based on the integrative approach comprising four criteria (phylogenetic and phenotypic), all currently accepted species gained support, while two new species are proposed (A. magnus and A. tenebricus). In addition, we proposed the new name A. neotritici to replace an invalidly described A. tritici. The revised section Candidi now encompasses nine species, some of which manifest a high level of intraspecific genetic and/or phenotypic variability (e.g., A. subalbidus and A. campestris) while others are more uniform (e.g., A. candidus or A. pragensis). The growth rates on different media and at different temperatures, colony colours, production of soluble pigments, stipe dimensions and vesicle diameters contributed the most to the phenotypic species differentiation.
Taxonomic novelties: New species: Aspergillus magnus Glässnerová & Hubka; Aspergillus neotritici Glässnerová & Hubka; Aspergillus tenebricus Houbraken, Glässnerová & Hubka.
Citation: Glässnerová K, Sklenář F, Jurjević Ž, Houbraken J, Yaguchi T, Visagie CM, Gené J, Siqueira JPZ, Kubátová A, Kolařík M, Hubka V (2022). A monograph of Aspergillus section Candidi. Studies in Mycology 102: 1–51. doi: 10.3114/sim.2022.102.01
Keywords: Aspergillus candidus, Aspergillus tritici, genealogical concordance, integrative taxonomy, intraspecific variability, multispecies coalescent model
INTRODUCTION
Aspergillus is a large genus of filamentous fungi which is divided into 27 sections. Aspergillus section Candidi currently includes seven white- or yellow-sporulating species (Houbraken et al. 2020). Members of this section show a relatively uniform morphology and their identification to a species level is non-trivial when using only phenotypic characters. The first modern monograph of the section was published by Varga et al. (2007) who used a polyphasic approach for species delimitation (combination of morphology, physiology, exometabolite profiles and molecular data) and recognized A. candidus [MB 204868] and three other species: A. tritici [MB 309248] (Mehrotra & Basu 1976), A. campestris [MB 110495] (Christensen 1982) and A. taichungensis [MB 434473] (Yaguchi et al. 1995). Since then, another three species have been described, namely, A. subalbidus [MB 809190] (Visagie et al. 2014), A. pragensis [MB 800371] (Hubka et al. 2014) and A. dobrogensis [MB 821313] (Hubka et al. 2018b).
The studies of Varga et al. (2007), Visagie et al. (2014) and Hubka et al. (2014) used a combination of morphology, phylogeny and/or profiles of exometabolites for species description (polyphasic approach). Another widely used approach in fungal taxonomy, GCPSR (Genealogical Concordance Phylogenetic Species Recognition), has not been applied in section Candidi. This approach involves comparisons of topologies of single-locus phylogenetic trees and search for monophyletic genealogical groups that are concordantly well-supported (Taylor et al. 2000, Dettman et al. 2003a). More recently, new phylogenetic methods based on the multispecies coalescent model (MSC) have been implemented in taxonomy (Fujisawa et al. 2016). These methods take into account for example the incomplete lineage sorting, a most common reason for incongruences in tree topologies constructed on the basis of different loci (Edwards 2009, Mirarab et al. 2016). The application of these methods on section Candidi led to segregation of A. candidus into two species and description of A. dobrogensis (Hubka et al. 2018b).
The best known and the first described member of this section is A. candidus, a xerophilic species which is able to grow on substrates with low water activity like stored grains and seeds, where it can reduce the grains’ germinability (Thom & Raper 1945, Papavizas & Christensen 1960, Raper & Fennell 1965). It is also frequently found in the indoor environment, on food products (flour, cereals, spices, nuts), in soil or in the marine environment (Weidenbörner & Kunz 1994, Pitt & Hocking 1997, Klich 2002, Wei et al. 2007, Visagie et al. 2017). The section also encompasses several clinically relevant representatives which occasionally cause superficial (onychomycosis or otitis externa) or invasive mycoses, or contaminate clinical samples. Among these, A. tritici and A. candidus are confirmed etiological agents of some infections, while A. pragensis and A. dobrogensis have only been isolated from clinical samples and their clinical significance remains questionable (Moling et al. 2002, Hubka et al. 2012, Hubka et al. 2014, Becker et al. 2015, Nouripour-Sisakht et al. 2015, Gupta et al. 2016, Masih et al. 2016, Carballo et al. 2020, Kaur et al. 2021).
Members of the section Candidi produce many industrially important enzymes and secondary metabolites with a biotechnological potential. For example, they produce extracellular lipases, xylanases and cellulases which have potential to be used in the waste degradation processes (Milala et al. 2009, Garai & Kumar 2013, Farias et al. 2015) or metabolites with antioxidant activity which can be used in the process of maturing foods (Grazia et al. 1986, Pitt & Hocking 1997, Sunesen & Stahnke 2003). The production of bioactive metabolites with pharmacological potential has been identified in some species, for example chlorflavonin with antibiotic and antifungal effects (Munden et al. 1970, Marchelli & Vining 1973, Hubka et al. 2014), compounds with significant anti-oxidative activity such as candidusin B, 3-hydroxyterphenyllin or dihydroxymethyl pyranone (Yen et al. 2001, Elaasser et al. 2011), cytotoxic candidusin A, preussin or p-terphenyl derivatives (Kobayashi et al. 1982, Malhão et al. 2019, Lin et al. 2021) and many other recently discovered compounds with antitumor, antimicrobial, cytotoxic or antiviral activities such as ascandinines (Zhou et al. 2021), taichunines (El-Desoky et al. 2021), unguisins (Li et al. 2020) or other p-terphenyl derivatives (Han et al. 2020, Shan et al. 2020, Wang et al. 2020). The production of most of these secondary metabolites is usually attributed to A. candidus, but it is likely that the producer was misidentified in some cases with other species from section Candidi. For instance, production of p-terphenyl derivative 4″-deoxy-2′-methoxyterphenyllin attributed to A. candidus is in fact produced by A. subalbidus (Shan et al. 2020). Such re-identifications are, however, only possible when the isolate is accessioned and preserved in culture collections or DNA sequence data are available which is gradually becoming a standard in studies on compounds produced by microorganisms.
Previous studies focused on section Candidi included only a small number of strains and consequently, little is known about intraspecific genetic and phenotypic variability. Section Candidi members are widespread worldwide and occur on a wide range of substrates, but at low frequencies. During our past projects, a large collection of over a hundred strains belonging to the section Candidi has been gathered from various substrates and countries, and a high genetic and morphological variability has been observed within species which were previously considered to be relatively homogeneous. To determine if this variability reflects undescribed species diversity or a high intraspecific variability, we compared the data from phenotypic analyses, classical phylogenetic methods, GCPSR approach and delimitation methods based on coalescence theory. According to the resulting taxonomic conclusions, we revised the descriptions of all supported species in the section.
MATERIALS AND METHODS
Strains
This study builds upon previous taxonomic studies dealing with section Candidi (Varga et al. 2007, Hubka et al. 2014, Visagie et al. 2014, Hubka et al. 2018b). The majority of new and yet unpublished strains were isolated from indoor environments, caves and dung. The isolation techniques mostly followed the procedures previously described by Jurjević et al. (2015), Nováková et al. (2018), Guevara-Suarez et al. (2020) and Visagie et al. (2021). Remaining strains were obtained from culture collections and collaborators, and originated from diverse substrates and countries. In total, we gathered 113 strains and detailed information about their provenance is listed in Table 1. Dried holotype and isotype specimens of the newly described species were deposited into the herbarium of the Mycological Department, National Museum, Prague, Czech Republic (PRM).
Table 1.
List of strains from Aspergillus section Candidi examined in this study.
| Species | Strain No.a | Provenance: country, locality, substrate, year, collector/isolator |
GenBank/ENA/DDBJ accession Nos.
|
|||
|---|---|---|---|---|---|---|
| ITS | benA | CaM | RPB2 | |||
| A. campestris | CBS 348.81 T = IMI 259099T = NRRL 13001T = IBT 13382T = IBT 28561T = ATCC 44563T = IFM 50931T = CCF 5596T | USA, North Dakota, near Zap, storage pile of topsoil displaced from a native, mixed prairie (Agropyron spp., Bouteloua gracilis), 1979, R.M. Miller & S.M. Pippen | EF669577 | EU014091 | EF669535 | EF669619 |
| IBT 17867 = CCF 5700 | USA, Wyoming, Pillot Hill Road, Medicine Bow National Forest, near Laramie, soil, 1994, J.C. Frisvad | ON156368 | ON164544 | ON164594 | ON164491 | |
| CCF 5641 = IFM 66789 = EMSL 1816 | USA, Wyoming, Gillette, air of living room, 2012, Ž. Jurjević | ON156369 | ON164567 | ON164616 | ON164514 | |
| FMR 15224 = CBS 142984 = CCF 6075 = IFM 66790 | Spain, Castile and Leon, Palencia, Monte el Viejo, Deer Reserve Park, herbivore dung, 2016, J. Gené & J.Z. Siqueira | LT798902 | LT798915 | LT798916 | LT798917 | |
| FMR 15226 = CBS 142985 = CCF 6053 | Spain, Castile and Leon, Palencia, Monte el Viejo, Deer Reserve Park, herbivore dung, 2016, J. Gené & J.Z. Siqueira | LT798903 | LT798918 | LT798919 | LT798920 | |
| IBT 23172 = IMI 073462 = CCF 5699 = IFM 66791 | USA, California, rabbit (Oryctolagus cuniculus) dung, 1958, S. Stribling | ON156370 | ON164568 | ON164617 | ON164515 | |
| IMI 344489 | South Africa, Pretoria, mouse dung, deposited into IMI collection in 1991, R.Y. Anelich | ON156371 | ON164569 | ON164618 | ON164516 | |
| A. candidus | CBS 566.65T = IMI 091889T = NRRL 303T = IBT 28566T = ATCC 1002T = CCF 5594T | unknown, received by K.B. Raper and D.I. Fennell in 1909 from J. Westerdijk | EF669592 | EU014089 | EF669550 | EF669634 |
| CCF 488 = IBT 32273 | Czech Republic, tunnels of bark beetles, 1960, E. Kotýnková-Sychrová | FR733811 | LT626977 | LT626978 | LT626979 | |
| CCF 4029 = CMF ISB 1730 = IBT 32272 | Romania, Meziad Cave, bat droppings, 2009, A. Nováková | FR733813 | LT626980 | FR751424 | LT626981 | |
| CCF 3996 = CBS 134394 | Czech Republic, Prague, external auditory canal of 53-year-old man (otitis externa), 2010, P. Lysková | FR727137 | FR775325 | HE716843 | LT626982 | |
| CCF 5577 | Romania, Ungurului Cave, old bat guano, 2010, A. Nováková | LT626946 | LT626983 | LT626984 | LT626985 | |
| NRRL 58579 = CCF 5843 = IBT 33371 = EMSL 914 | USA, New York, indoor air of a home, 2008, Ž. Jurjević | LT626947 | LT626986 | LT626987 | LT626988 | |
| NRRL 58959 = CCF 5845 = EMSL 1252 | USA, Pennsylvania, indoor air of a home, 2009, Ž. Jurjević | LT626948 | LT626989 | LT626990 | LT626991 | |
| CCF 4912 = EMSL 2295 | USA, New Jersey, Cranbury, crawlspace metal duct (swab), 2014, Ž. Jurjević | LT626949 | LT626992 | LT626993 | LT626994 | |
| CCF 5172 | Czech Republic, Prague, mouse excrements in grain store, 2000, J. Hubert | LT626953 | LT627005 | LT627006 | LT627007 | |
| CCF 4713 | Romania, Ungurului Cave, bat guano, 2010, A. Nováková | LT626950 | LT626995 | LT626996 | LT626997 | |
| CCF 4659 | Czech Republic, Hustopeče, toenail of 52-year-old woman, 2012, S. Dobiášová | HG915889 | HG916672 | HG916681 | LT626998 | |
| CCF 4915 = EMSL 1285 | USA, Rhode Island, indoor air of living room, 2009, Ž. Jurjević | LT626951 | LT626999 | LT627000 | LT627001 | |
| NRRL 4646 = IMI 359076 = DTO 213-G2 = CBS 133061 | USA, barn litter, D.I. Fennell | EF669605 | EU014090 | EF669563 | EF669647 | |
| CCF 5675 = EMSL 2403 | USA, Pennsylvania, Feasterville, air in a basement of a house, 2014, Ž. Jurjević | LT626952 | LT627002 | LT627003 | LT627004 | |
| CCF 5576 = EMSL 2421 | USA, New York, Binghamton, air in bedroom, 2014, Ž. Jurjević | LT626954 | LT627008 | LT627009 | LT627010 | |
| DTO 223-E5 | The Netherlands, soil, 2012, M. Meijer | LT626955 | LT627011 | LT627012 | LT627013 | |
| CCF 6195 = EMSL 2721 | USA, Minnesota, Minneapolis, classroom dust, 2015, Ž. Jurjević | ON156372 | ON164545 | ON164634 | ON164492 | |
| CCF 6198 = EMSL 2863 | USA, Ohio, Westerville, air in basement (settle plates), 2015, Ž. Jurjević | ON156373 | ON164546 | ON164595 | ON164493 | |
| CCF 6200 = EMSL 2895 | USA, Massachusetts, Cohasset, air in bathroom, 2015, Ž. Jurjević | ON156374 | ON164547 | ON164596 | ON164494 | |
| CMW-IA 8 = CMW 58610 = CN138F7 = KAS 5864 | Canada, Nova Scotia, Wolfville, house dust, 2015, C.M. Visagie & A. Walker | — | ON164548 | ON164597 | ON164495 | |
| CMW-IA 9 = CMW 58611 = CN138F8 = KAS 6297 | Canada, Nova Scotia, Little Lepreau, house dust, 2015, C.M. Visagie & A. Walker | — | ON164549 | ON164598 | ON164496 | |
| CMW-IA 10 = CMW 58612 = CN138F9 = KAS 6134 | Canada, Nova Scotia, Wolfville, house dust, 2015, C.M. Visagie & A. Walker | — | ON164550 | ON164599 | ON164497 | |
| CMW-IA 11 = CMW 58613 = CN138G1 = KAS 6135 | Canada, Nova Scotia, Wolfville, house dust, 2015, C.M. Visagie & A. Walker | — | ON164551 | ON164600 | ON164498 | |
| A. dobrogensis | CCF 4651T = CCF 4655T = NRRL 62821T = IBT 32697T = CBS 143370T | Romania, Movile Cave, 2nd airbell, cave sediment, 2012, A. Nováková | LT626959 | LT627027 | LT558722 | LT627028 |
| IBT 29476 = CCF 5823 | Denmark, Høve Strand, mouse dung (in bed in summer house annex), 2007, J.C. Frisvad | LT907964 | LT907967 | LT907966 | LT907969 | |
| CCF 5567 | Romania, Meziad Cave, bat droppings, 2009, A. Nováková | LT626960 | LT627029 | LT627030 | LT627031 | |
| CCF 5568 | Romania, Liliecilor de la Guru Dobrogei Cave, Galeria Fossile, bat guano, 2010, A. Nováková | LT626961 | LT627032 | LT627033 | LT627034 | |
| CCF 4650 = CCF 4657 = NRRL 62820 = IBT 32699 | Romania, Meziad Cave, cave sediment, 2010, A. Nováková | LT626962 | LT627035 | LT627036 | LT627037 | |
| CCF 4649 = IBT 32700 | Romania, Liliecilor de la Guru Dobrogei Cave, Galeria Fossile, bat guano, 2010, A. Nováková | LT626963 | LT627038 | LT627039 | LT627040 | |
| CCF 5569 | Romania, Liliecilor de la Guru Dobrogei Cave, Galeria Fossile, bat guano, 2010, A. Nováková | LT626964 | LT627041 | LT627042 | LT627043 | |
| CCF 5570 | Romania, Liliecilor de la Guru Dobrogei Cave, Galeria Fossile, bat droppings, 2010, A. Nováková | LT626965 | LT627044 | LT627045 | LT627046 | |
| CCF 5573 | Romania, Liliecilor de la Guru Dobrogei Cave, cave air, 2011, A. Nováková | LT626966 | LT627047 | LT627048 | LT627049 | |
| CCF 5571 | Romania, Meziad Cave, hall in the upper floor, bat guano, 2010, A. Nováková | LT626967 | LT627050 | LT627051 | LT627052 | |
| CCF 5572 | Romania, Liliecilor de la Guru Dobrogei Cave, cave air, 2013, A. Nováková | LT626968 | LT627053 | LT627054 | LT627055 | |
| CCF 5574 | Romania, Liliecilor de la Guru Dobrogei cave, Galeria Fossile, bat droppings, 2014, A. Nováková | LT626969 | LT627056 | LT627057 | LT627058 | |
| CCF 5575 | Czech Republic, nails of 31-year-old woman, 2014, P. Lysková | LT626970 | LT627 059 | LT627060 | LT627061 | |
| CBS 225.80 = DTO 031-E6 | The Netherlands, human nail (contaminant), 1980, CBS | LT626971 | LT627062 | LT627063 | LT627064 | |
| DTO 025-I1 | Germany, carpet, 2006 | LT626972 | LT627077 | LT627078 | LT627079 | |
| DTO 013-C4 = IBT 28582 | The Netherlands, Maastricht, indoor air, 2006, J. Houbraken | LT626973 | LT627065 | LT627066 | LT627067 | |
| DTO 001-F9 = IBT 28576 | The Netherlands, surface of museum piece, 2004, J. Houbraken | LT626974 | LT627068 | LT627069 | LT627070 | |
| DTO 029-H2 | Germany, carpet, 2006 | LT626975 | LT627071 | LT627072 | LT627073 | |
| DTO 031-D9 = CBS 116945 = IBT 116945 = IBT 28573 | The Netherlands, Tiel, dust in museum, 2004, J. Houbraken | LT626976 | LT627074 | LT627075 | LT627076 | |
| CCF 5844 = EMSL No. 2810 | USA, New York, Lockport, indoor air in basement (settle plate), 2015, Ž. Jurjević | LT907963 | ON164552 | LT907965 | LT907968 | |
| FMR 15444 = CBS 142752 = CCF 6119 | Spain, Galicia, Lugo, Ribeira Sacra, herbivore dung, 2016, J. Guarro & M. Guevara-Suarez | LT798904 | LT798921 | LT798922 | LT798923 | |
| FMR 15601 = CCF 5846 | Spain, Galicia, Orense, Cortegada, herbivore dung, 2016, J. Guarro & M. Guevara-Suarez | ON156375 | LT962396 | ON164601 | ON164499 | |
| A. magnus | UAMH 1324T = IBT 14560T = CCF 6606T | Canada, Alberta, Edmonton, mouse collected on horsefarm, 1962, J.W. Carmichael | ON156376 | ON164570 | ON164619 | ON164517 |
| A. neotritici | CCF 3853T = IBT 32725T | Czech Republic, Prague, toenail of 62-year-old man, 2008, M. Skoøepová | FR727136 | FR775327 | HE661598 | LT627021 |
| CBS 266.81 = IBT 21956 = CCF 5842 | India, grains of Triticum aestivum, before 1976, B.S. Mehrotra | LT626958 | EU076293 | HG916678 | LT627017 | |
| CCF 4030 = CMF ISB 1300 = IBT 32729 | Czech Republic, Frýdek-Místek, vermicompost, 2001, A. Nováková | FR733814 | LT627018 | FR751425 | LT627019 | |
| CCF 4653 | Czech Republic, Prague, toenail of 58 -year-old woman, 2012, P. Lysková | HG915890 | HG916674 | HG916677 | LT627020 | |
| CCF 3314 | Czech Republic, Prague, outdoor air, 1996, A. Kubátová | FR733812 | LT627022 | FR751426 | LT627023 | |
| CCF 1649 | Czech Republic, Prague, flour, 1979, J. Svrčková | FR733810 | LT627024 | FR751427 | LT627025 | |
| NRRL 4847 = IMI 359077 = DTO 213-G3 = CBS 133055 = CCF 4809 | Japan (?), unknown, received by K.B. Raper and D.I. Fennell from CBS as Nakazawa′s strain of A. albus var. thermophilus | ON156395 | ON164587 | ON164628 | ON164537 | |
| CCF 4658 | Czech Republic, Prague, toenail of 69-year-old woman, 2008, M. Skoøepová | HG915891 | HG916675 | HG916676 | LT627026 | |
| DTO 201-D3 = CBS 129260 = RMF 7641 | USA, Nebraska, 1 mile north of Miller, loess soil, 6 feet deep, soil profile, 1984 | ON156397 | ON164591 | ON164632 | ON164541 | |
| DTO 201-G7 = RMF TC 215 = CBS 129307 | Unknown location, soil | ON156398 | ON164592 | ON164633 | ON164542 | |
| CCF 6202 = EMSL 3152 | USA, Florida, Key West, air in bedroom (settle plates), 2015, Ž. Jurjević | ON156396 | ON164588 | ON164629 | ON164538 | |
| CCF 6397 | Czech Republic, Havlíčkùv Brod, abdominal cavity of 62-year-old male with disseminated mycosis (probably caused by Rhizopus sp. and Trichosporon asahii), 2020, D. Lžičaøová | ON156394 | ON164589 | ON164630 | ON164539 | |
| CMW-IA 12 = CMW 58614 = CN117B9 | South Africa, Free State, Viljoenskroon, sunflower seed, 2020, C.M. Visagie | — | ON164590 | ON164631 | ON164540 | |
| IBT 12659 | USA, New Mexico, Seviletta National Wildlife Refuge, Socorro County, soil in a kangaroo rat burrow, 1989, L. Hawkins | ON156393 | ON164557 | ON164606 | ON164504 | |
| CCF 4914 = IFM 66793 = EMSL 2182 | USA, Arizona, Tucson, air in a hospital, 2013, Ž. Jurjević | ON156392 | ON164556 | ON164605 | ON164503 | |
| A. pragensis | CCF 3962T = CBS 135591T = IBT 32274T = NRRL 62491T | Czech Republic, Prague, toenail of 58-year-old man, 2007, M. Skoøepová | FR727138 | HE661604 | FR751452 | LN849445 |
| CCF 4654 = IBT 32701 | Czech Republic, Prague, toenail of 59-year-old man, 2013, P. Lysková | HG915888 | HG916673 | HG916680 | LT627014 | |
| CCF 5847 = EMSL 2216 | USA, Pennsylvania, Feasterville, carpet in bedroom, 2013, Ž. Jurjević | LT908112 | LT908040 | LT908041 | LT908042 | |
| NRRL 58614 = CCF 4911 = EMSL 1057 | USA, Pennsylvania, indoor air of a home, 2008, Ž. Jurjević | LT908111 | LT908037 | LT908038 | LT908039 | |
| CCF 5693 EMSL 2397 | USA, Maryland, Cohasset, outdoor air, 2014, Ž. Jurjević | LT908113 | LT908043 | LT908044 | LT908045 | |
| CMW-IA 13 = CMW 58615 = CN138F5 = KAS 6296 | Canada, Nova Scotia, Little Lepreau, house dust, 2015, C.M. Visagie & A. Walker | — | ON164553 | ON164602 | ON164500 | |
| CMW-IA 14 = CMW 58616 = CN138G6 = KAS 6138 | Canada, Nova Scotia, Little Lepreau, house dust, 2015, C.M. Visagie & A. Walker | — | ON164554 | ON164603 | ON164501 | |
| CMW-IA 15 = CMW 58617 = CN138G8 = KAS 6299 | Canada, Nova Scotia, Little Lepreau, house dust, 2015, C.M. Visagie & A. Walker | — | ON164555 | ON164604 | ON164502 | |
| A. subalbidus | CBS 567.65T = NRRL 312T = ATCC 16871T = IMI 230752T = CCF 5822T | Brazil, received by K.B. Raper and D.I. Fennell in 1939 from J. Reis (Instituto Biologica) | EF669593 | KP987050 | EF669551 | EF669635 |
| CBS 112449 = DTO 031-E3 = DTO 039-E7 | Germany, indoor environment | LT626956 | EU076294 | EU076307 | LT627015 | |
| NRRL 5214 = ATCC 26930 = IMI 16046 = CCF 4860 | Ghana, vegetable lard, International Mycological Institute, Egham, England, 1933, H.A. Dade (strain GC601) sent to IMI | LT908114 | LT908034 | LT908035 | LT908036 | |
| CCF 5696 =EMSL 2674 | USA, Georgia, Canton, settle plates in living room, 2015, Ž. Jurjević | ON156388 | ON164571 | ON164620 | ON164518 | |
| CCF 6199 = EMSL 2894 | USA, Florida, Bradenton, air in bathroom (settle plates), 2015, Ž. Jurjević | ON156386 | ON164572 | ON164621 | ON164519 | |
| CMW-IA 16 = CMW 58618 = CN162C8 = DN12 | Botswana, Okavango basin, Gcwihaba Caves, bat guano-contaminated soil, 2019, G. Modise, D. Nkwe & R. Mazebedi | — | ON164573 | MW480718 | ON164520 | |
| CMW-IA 17 = CMW 58619 = CN162C9 = DN13 | Botswana, Okavango basin, Gcwihaba Caves, bat guano-contaminated soil, 2019, G. Modise, D. Nkwe & R. Mazebedi | — | ON164574 | MW480719 | ON164521 | |
| CMW-IA 18 = CMW 58620 = CN162D1 = DN62 | Botswana, Okavango basin, Gcwihaba Caves, bat guano-contaminated soil, 2019, G. Modise, D. Nkwe & R. Mazebedi | — | ON164575 | MW480764 | ON164522 | |
| CMW-IA 19 = CMW 58621 = CN162D2 = DN67 | Botswana, Okavango basin, Gcwihaba Caves, bat guano-contaminated soil, 2019, G. Modise, D. Nkwe & R. Mazebedi | — | ON164576 | MW480769 | ON164523 | |
| CMW-IA 20 = CMW 58622 = CN162D3 = DN68 | Botswana, Okavango basin, Gcwihaba Caves, bat guano-contaminated soil, 2019, G. Modise, D. Nkwe & R. Mazebedi | — | ON164577 | MW480770 | ON164524 | |
| CN162D4 = DN69 | Botswana, Okavango basin, Gcwihaba Caves, bat guano-contaminated soil, 2019, G. Modise, D. Nkwe & R. Mazebedi | — | ON164578 | MW480771 | ON164525 | |
| CN162D5 = DN75 | Botswana, Okavango basin, Gcwihaba Caves, bat guano-contaminated soil, 2019, G. Modise, D. Nkwe & R. Mazebedi | — | ON164579 | MW480777 | ON164526 | |
| CN162D6 = DN80 | Botswana, Okavango basin, Gcwihaba Caves, bat guano-contaminated soil, 2019, G. Modise, D. Nkwe & R. Mazebedi | — | ON164580 | MW480781 | ON164527 | |
| CN162D7 = DN82 | Botswana, Okavango basin, Gcwihaba Caves, bat guano-contaminated soil, 2019, G. Modise, D. Nkwe & R. Mazebedi | — | ON164581 | MW480783 | ON164528 | |
| CN162D8 = DN85 | Botswana, Okavango basin, Gcwihaba Caves, bat guano-contaminated soil, 2019, G. Modise, D. Nkwe & R. Mazebedi | — | ON164582 | MW480785 | ON164529 | |
| CCF 5642 = IFM 66794 = EMSL 410 | USA, Connecticut, wall in a house, 2008, Ž. Jurjević | ON156377 | ON164558 | ON164607 | ON164505 | |
| CCF 6197 = IFM 66795 = EMSL 2731 | USA, Illinois, Mokena, air in crawl space (settle plates), 2015, Ž. Jurjević | ON156378 | ON164559 | ON164608 | ON164506 | |
| CCF 5643 = EMSL 2181 | USA, Maryland, Baltimore, carpet in living room, 2013, Ž. Jurjević | ON156380 | ON164561 | ON164610 | ON164508 | |
| NRRL 4809 = ATCC 11380 = IFO 4310 = DTO 213-F7 = CBS 133057 = CCF 5595 | Japan (?), chinese yeast cake, received by K.B. Raper and D.I. Fennell from the Institute for Fermentation, Osaka, Japan (No. 4319) as “A. albus var. 1”, M. Yamazaki, 1946 | EF669609 | EU014092 | EF669567 | EF669651 | |
| NRRL 58123 = CCF 6193 = IFM 66796 = EMSL 465 | USA, California, wall in a house, 2008, Ž. Jurjević | ON156382 | ON164565 | ON164614 | ON164512 | |
| CCF 5697 = IFM 66797 = EMSL 2180 | USA, New Jersey, Mont Clair, indoor air, 2013, Ž. Jurjević | ON156381 | ON164562 | ON164611 | ON164509 | |
| CCF 4913 = EMSL 2297 | USA, Maryland, Baltimore, house dust (living room), 2014, Ž. Jurjević | ON156379 | ON164560 | ON164609 | ON164507 | |
| CCF 5698 = EMSL 2369 | USA, Maryland, Baltimore, house dust (living room), 2014, Ž. Jurjević | ON156385 | ON164563 | ON164612 | ON164510 | |
| CCF 5848 = EMSL 2646 | USA, California, Rancho Mirage, settle plates in living room, 2014, Ž. Jurjević | ON156387 | ON164564 | ON164613 | ON164511 | |
| CCF 6205 = EMSL 3325 | USA, Texas, Harker Heights, air in basement (settle plates), 2015, Ž. Jurjević | ON156383 | ON164566 | ON164615 | ON164513 | |
| FMR 15733 = CBS 142983 = CCF 6052 = IFM 66798 | Spain, Canary Islands, Gran Canaria, Santa Brígida, herbivore dung, 2016, J. Gené & J.Z. Siqueira | LT798905 | LT798924 | LT798925 | LT798926 | |
| FMR 15736 = CBS 142982 = CCF 6074 | Spain, Canary Islands, Gran Canaria, Teror, herbivore dung, 2016, J. Gené & J.Z. Siqueira | LT798906 | LT798927 | LT798928 | LT798929 | |
| FMR 15877 = CBS 142667 = CCF 6058 | Spain, Canary Islands, Gran Canaria, North Coast, herbivore dung, 2016, J. Gené & J.Z. Siqueira | LT798907 | LT798930 | LT798931 | LT798932 | |
| DTO 196-E4 = CBS 126836 | Ecuador, Galapagos Islands, soil, 1965, D.P. Mahoney | ON156384 | ON164583 | ON164622 | ON164530 | |
| A. taichungensis | IBT 19404T = CCF 5597T= DTO 031-C6T | Taiwan, Taichung city, soil, 1994, T. Yaguchi | LT626957 | EU076297 | HG916679 | LT627016 |
| CMW-IA 21 = CMW 58623 = CN162D9 = DN07 | Botswana, Okavango basin, Gcwihaba Caves, bat guano-contaminated soil, 2019, G. Modise, D. Nkwe & R. Mazebedi | — | — | MW480714 | ON164531 | |
| DTO 266-G2 = CCF 5827 = IFM 66799 | Mexico, house dust, 2013, C.M. Visagie | KJ775572 | KJ866980 | ON164627 | ON164536 | |
| DTO 270-C9 = CCF 5826 | Mexico, house dust, 2013, C.M. Visagie | KJ775573 | KJ866981 | ON164626 | ON164535 | |
| A. tenebricus | DTO 337-H7T = CBS 147048T | South Africa, Robben island, soil, 2015, P.W. Crous & M. Meijer | ON156389 | ON164584 | ON164623 | ON164532 |
| DTO 440-E1 = CBS 147376 | Australia, Queensland, soil, 2009, P.W. Crous, N. Yilmaz & T. Hoogenhuijzen | ON156390 | ON164585 | ON164624 | ON164533 | |
| DTO 440-E2 | Australia, Queensland, soil, 2009, P.W. Crous & T. Hoogenhuijzen | ON156391 | ON164586 | ON164625 | ON164534 | |
| Aspergillus sp. | DTO 244-F1 | New Zealand, indoor environment, E. Whitfield, K. Mwange & T. Atkinson | ON156399 | ON164543 | ON164593 | ON164490 |
a Acronyms of culture collections in alphabetic order: ATCC, American Type Culture Collection, Manassas, Virginia; CBS, Westerdijk Fungal Biodiversity Institute (formerly Centraalbureau voor Schimmelcultures), Utrecht, the Netherlands; CCF, Culture Collection of Fungi, Department of Botany, Charles University, Prague, Czech Republic; CMF ISB, Collection of Microscopic Fungi of the Institute of Soil Biology, Academy of Sciences of the Czech Republic, České Budějovice, Czech Republic; CN, CMW & CMW-IA, working and formal culture collections housed at FABI (Forestry and Agricultural Biotechnology) Institute, University of Pretoria, South Africa; DN, working collection of David Nkwe, housed at the Department of Biological Sciences and Biotechnology, Botswana International University of Science and Technology, Botswana; DTO, Internal Culture Collection of The Department Applied and Industrial Mycology of the CBS-KNAW Fungal Biodiversity Centre, Utrecht, The Netherlands; EMSL, EMSL Analytical Inc., New Jersey, USA; FMR, Facultat de Medicina i Ciències de la Salut, Reus, Spain; FRR, Food Fungal Culture Collection, North Ryde, Australia; IBT, Culture Collection at Department of Biotechnology and Biomedicine, Lyngby, Denmark; IFM, Collection at the Medical Mycology Research Center, Chiba University, Japan; IFO, Institute for Fermentation, Osaka, Japan; IHEM (BCCM/IHEM), Belgian Coordinated Collections of Micro-organisms, Fungi Collection: Human and Animal Health, Sciensano, Brussels, Belgium; IMI, CABI’s collection of fungi and bacteria, Egham, UK; KAS, fungal collection of Keith A. Seifert, internal working culture collection at DAOMC (Culture collection of the National Mycological Collections, Agriculture & Agri-Food Canada), Ottawa, Canada; NRRL, Agricultural Research Service Culture Collection, Peoria, Illinois, USA; RMF, Rocky Mountain Fungi, Dept. of Botany, University of Wyoming, Laramie; UAMH, University of Alberta Microfungus collection and Herbarium, Edmonton, Alberta, Canada.
Phenotypic studies
Macromorphological characters were studied on malt extract agar (MEA; malt extract from Oxoid Ltd., Basingstoke, UK), Czapek Yeast Extract Agar (CYA; yeast extract from Oxoid Ltd.), Czapek-Dox Agar (CZA), and Czapek Yeast Extract Agar with 20 % sucrose (CY20S). For the agar media composition, see Samson et al. (2014). The strains were inoculated in three points on growth media in 90 mm Petri dishes and colony dimensions were measured after seven and 14 d of incubation at 25 °C. After 14 d, the photographs of colonies were taken, and after 28 d, the colonies were checked for presence of sclerotia and soluble pigment production as these characters are frequently expressed with delay. To determine the cardinal temperatures, the strains were grown for 14 d on MEA at 10, 15, 20, 25, 30, 35, 37, 40, and 45 °C. The description and names of colours followed Kornerup & Wanscher (1967). Colony details were documented with a Canon EOS 500D camera and a Leica M205C stereo microscope with a Leica DMC 5400 digital camera.
Micromorphological characters (length and width of stipe, vesicle diameter, length of metulae and phialides, length and width of conidia) were photographed and measured from 7–21-d-old colonies on MEA incubated at 25 °C. Lactic acid (60%) was used as mounting medium. Every morphological character was recorded at least 35 times for each strain. Statistical differences, in particular phenotypic characters, were tested with one-way ANOVA followed by Tukey’s honest significant difference (HSD) test in R v. 4.1.2 (R Core Team 2021). Boxplots and violin graphs were created in R with package ggplot2 (Wickham 2016).
Microscopic photographs were taken using an Olympus BX51 microscope equipped with an Olympus DP72 camera. The photographic plates were made in CorelDRAW Graphic Suite 2021. Scanning electron microscopy (SEM) was performed using a JEOL-6380 LV microscope (JEOL Ltd. Tokyo, Japan) as described previously by Hubka et al. (2013b).
Molecular studies
Total genomic DNA was extracted from 7-d-old colonies growing on MEA using the ZR Fungal/ Bacterial DNA Kit™ (Zymo Research, Irvin, CA, USA). The quality of the isolated DNA was verified using a NanoDrop 1000 Spectrophotometer.
The PCR reaction volume of 20 μL contained 1 μL (50 ng mL-1) of DNA, 0.3 μL of both primers (25 pM mL-1), 0.2 μL of MyTaqTM DNA Polymerase (Bioline, GmbH, Germany) and 4 μl of 5 × MyTaq PCR buffer. The standard thermal cycle profile was 93 °C—2 min; 38 cycles of 93 °C—30 s, 55 °C—30 s, 72 °C—60 s; and a final extension of 72 °C—10 min. The internal transcribed spacer rDNA region (ITS) was amplified using forward primer ITS1 (White et al. 1990) and reverse primers NL4 (O’Donnell 1993) or ITS4 (White et al. 1990); the partial β-tubulin gene region (benA) was amplified using forward primers Bt2a (Glass & Donaldson 1995), T10 (O’Donnell & Cigelnik 1997) or Ben2f (Hubka & Kolařík 2012) and reverse primer Bt2b (Glass & Donaldson 1995); the partial calmodulin gene region (CaM) was amplified using forward primers CF1L, CF1M (Peterson 2008) or cmd5 (Hong et al. 2006) and reverse primers CF4 (Peterson 2008) or cmd6 (Hong et al. 2006); and the partial RNA polymerase II second largest subunit gene region (RPB2) using forward primers fRPB2-5F (Liu et al. 1999) or RPB2-F50-CanAre (Jurjević et al. 2015) and reverse primer fRPB2-7cR (Liu et al. 1999). The PCR products were separated by electrophoresis on 1 % agarose gel and subsequently purified using ExoSAP-IT™ (Thermo Fisher Scientific, Vilnius, Lithuania).
Sequences were inspected and assembled using BioEdit v. 7.2.5 (Hall 1999) and deposited into the GenBank database under accession numbers listed in Table 1.
Phylogenetic studies
Alignments of the benA, CaM and RPB2 loci were performed using the FFT-NS-i option implemented in the MAFFT online service (Katoh et al. 2019). ITS was excluded from analyses due to its low number of informative sites. The alignments were trimmed, concatenated and then analysed using maximum likelihood (ML), Bayesian inference (BI) and Maximum Parsimony (MP; this method was used only to construct single-gene phylogenies) methods. Suitable partitioning schemes and substitution models (Bayesian information criterion) for the analyses were selected using a greedy strategy implemented in PartitionFinder 2 (Lanfear et al. 2017) with settings allowing introns, exons and codon positions to be independent datasets. The optimal partitioning schemes for each analysed dataset along with basic alignment characteristics are listed in Table 2.
Table 2.
Characteristics of alignments, partition-merging results and best substitution model for each partition according to the Bayesian information criterion.
| Alignment | Length (bp) | Variable position | Parsimony informative sites | Phylogenetic method | Partitioning scheme (substitution model) |
|---|---|---|---|---|---|
| benA + CaM + RPB2 (Fig. 1) | 2069 | 455 | 353 | Maximum likelihood (ML) | Five partitions: 1st codon positions of benA, CaM & RPB2 & 2nd codon positions of CaM (TrN+I); 2nd codon positions of benA & RPB2 (JC); 3rd codon positions of benA & CaM (HKY+G); 3rd codon positions of RPB2 (K81uf+G); introns of benA & CaM (K80+G) |
| Bayesian inference (BI) | Five partitions: 1st codon positions of benA, CaM & RPB2 (HKY+I); 2nd codon positions of benA, CaM & RPB2 (F81+I); 3rd codon positions of benA & CaM (HKY+G); 3rd codon positions of RPB2 (HKY+G); introns of benA & CaM (K80+G) | ||||
| benA (Fig. 2) | 479 | 133 | 106 | ML | Three partitions: 1st & 2nd codon positions (JC); 3rd codon positions (HKY+G); introns (K80+G) |
| BI | Three partitions: 1st & 2nd codon positions (JC); 3rd codon positions (HKY+G); introns (K80+G) | ||||
| CaM (Fig. 2) | 576 | 120 | 88 | ML | Three partitions: 1st & 2nd codon positions (TrN+I); 3rd codon positions (HKY+G); introns (K80+G) |
| BI | Three partitions: 1st & 2nd codon positions (HKY+I); 3rd codon positions (HKY+G); introns (K80+G) | ||||
| RPB2 (Fig. 2) | 1014 | 202 | 159 | ML | Three partitions: 1st codon positions (HKY+I); 2nd codon positions (JC); 3rd codon positions (K81uf+G) |
| BI | Three partitions: 1st codon positions (HKY+I); 2nd codon positions (JC); 3rd codon positions (HKY+G) |
Maximum likelihood trees were constructed with IQ-TREE v. 1.4.4 (Nguyen et al. 2015) with nodal support determined by ultrafast bootstrapping (BS) with 100 000 replicates. Trees were rooted with the clade containing A. neotritici isolates. Bayesian posterior probabilities (PP) were calculated using MrBayes v. 3.2.6 (Ronquist et al. 2012). The analysis ran for 107 generations, two parallel runs with four chains each were used, every 1 000th tree was retained and the first 25 % of trees were discarded as burn-in. The convergence of the runs and effective sample sizes were checked in Tracer v. 1.6 (http://tree.bio.ed.ac.uk/software/tracer). Maximum parsimony (MP) trees were created using PAUP* v.4.0b10 (Swofford 2003). Analyses were performed using the heuristic search option with 100 random taxon additions; tree bisection-reconnection (TBR); maxtrees were set to 1 000. Branch support was assessed by bootstrapping with 500 replications.
The rules for the application of the GCPSR approach were adopted from Dettman et al. (2003a,b) and slightly modified to different design of this study (different number of loci and methods used). To recognize a clade as an evolutionary lineage, it had to satisfy either of two criteria: (a) genealogical concordance - the clade was present in the majority (2/3) of the single-locus genealogies; (b) genealogical nondiscordance - the clade was well supported in at least one single-locus genealogy, as judged by both ML and MP bootstrap proportions (≥70 %) and BI posterior probabilities (≥95 %), and was not contradicted in any other single-locus genealogy at the same level of support. When deciding which evolutionary lineages represent phylogenetic species, two additional criteria were applied and evaluated according to the combined phylogeny of three genes: (a) genetic differentiation - species had to be relatively distinct and well differentiated from other species to prevent minor tip clades from being recognized as a separate species; (b) all individuals had to be placed within a phylogenetic species, and no individuals were to be left unclassified.
In order to create hypotheses about species boundaries, we used one multi-locus multispecies coalescent (MSC) model-based method STACEY (Jones 2017) and four single-locus species MSC delimitation methods: (1) the general mixed Yule-coalescent method (GMYC) (Fujisawa & Barraclough 2013), (2) the Bayesian version of the general mixed yule-coalescent model (bGMYC) (Reid & Carstens 2012), (3) the Poisson tree processes model (PTP) and (4) the Bayesian Poisson tree processes model bPTP (Zhang et al. 2013).
For the single-locus species delimitation methods, we used the haplotype function from package pegas (Paradis 2010) in R to retain only unique sequences in alignments. The following nucleotide substitution models were selected by jModelTest v. 2.1.10 (Darriba et al. 2012) for benA, CaM and RPB2 loci according to the Bayesian information criterion: K80+I, TrNef+G and TrN+I. The GMYC analysis was performed in R with the package splits (Fujisawa & Barraclough 2013). The ultrametric input trees for the GMYC method were calculated in BEAST v. 2.6.6 (Bouckaert et al. 2014) with a chain length of 1 × 107 generations. As a model for creating input trees we set up Coalescent Constant Population prior and performed two tree reconstruction methods – one with Common Ancestor heights (CAh) setting and second with Median heights (Mh) setting. We only show delimitation results of both settings when they were different. The bGMYC analysis was performed with package bgmyc (Reid & Carstens 2012) in R v. 3.4.1. For this method, we firstly discarded the initial 25 % of the trees from the BEAST inference as burn-in and then we used R v. 4.1.2 the package ape (Paradis et al. 2004) in R to randomly select one hundred trees, which were then used as input. Two values (0.5 and 0.75) of bgmyc.point function were set up for all analyses. This function is crucial for the final division of strains into species. The authors of the software recommended value around 0.5, lower value delimits either the same number of species or less while a value >0.5 delimits the same number of species or more. For the PTP and bPTP method, 1 000 maximum likelihood standard bootstrap trees were calculated in IQ-TREE v. 1.6.12 (Nguyen et al. 2015) and used as input. The analysis was run in the Python v. 3 (van Rossum & Drake 2019) package ptp (Zhang et al. 2013).
The multi-locus species delimitation method STACEY was performed in BEAST v. 2.6.6 (Bouckaert et al. 2014) using the STACEY v. 1.2.5 add-on (Jones 2017). We set up the length of mcmc chain to 1 × 109 generations, the species tree prior was set to the Yule model, the molecular clock model was set to strict clock, growth rate prior was set to lognormal distribution (M = 5, S = 2), clock rate priors for all loci were set to lognormal distribution (M = 0, S = 1), PopPriorScale prior was set to lognormal distribution (M = -7, S = 2) and relativeDeathRate prior was set to beta distribution (α = 1, β = 1000). The output was processed with SpeciesDelimitationAnalyzer (Jones 2015). For the presentation of the results of STACEY, we firstly created a plot showing how the number of delimited species and the probability of the most probable scenarios change in relation to the value of collapseheight parameter, and then we created similarity matrices using code from Jones et al. (2015) with two different values (0.007, 0.01) of collapseheight chosen from the plot (see Results section - Species delimitation using STACEY). Phylogenetic trees generated during STACEY analysis were then used for the presentation of species delimitation results analysis. The graphical outputs were created in iTOL (Interactive Tree Of Life) (Letunic & Bork 2021).
We also employed the recently developed software DELINEATE (Sukumaran et al. 2021) to independently test species boundaries hypotheses. Firstly, the dataset was split into hypothetical populations with “A10” analysis in BPP v. 4.3 (Yang 2015). Then, the species tree for these populations was estimated in starBEAST (Heled & Drummond 2009) implemented in BEAST v. 2.6.6 (Bouckaert et al. 2014). Finally, the populations delimited by BPP were lumped into species based on the results of species delimitation methods and phenotypic characters, with several populations always left unassigned to be delimited by DELINEATE. In total, nine models of species boundaries were set up. The analysis was run in Python v. 3 (van Rossum & Drake 2019) package delineate (Sukumaran et al. 2021).
RESULTS
Integrative approach for determining species boundaries
For the species delimitation in section Candidi, three genetic loci (benA, CaM and RPB2) were examined across 113 strains, while phenotypic characters were measured and scored for 66 strains representing the genetic variability across the section. The results from the GCPSR approach were compared with MSC methods and phenotypic data to draw the final conclusions about species limits.
From the early beginning of this study, it was clear that creating initial hypotheses about species boundaries in section Candidi would be extremely difficult due to relatively uniform morphology and conflicting data from single-gene phylogenies. To overcome these difficulties and remain relatively consistent across the section, we decided to score support for delimitation of species and monitor four main criteria. In this integrative approach, we required that the delimited species meet at least three of the following four criteria: (1) no conflict in the assessment of species limits using the GCPSR approach, (2) support from the multi-locus MSC method STACEY (at least in one of the two most probable scenarios – see below), (3) support by the majority (10 from 18) of single-locus MSC methods and their settings (agreement on the delimitation of species in its exact form or delimitation of smaller entities within it but without any admixture with related species/populations) and (4) presence of phenotypic difference(s) from phylogenetically most closely related species. The resulting scoring is summarized in Table 3. Using this approach, delimitation of two novel species was supported, A. magnus and A. tenebricus (see below and section Taxonomy).
Table 3.
Support for delimitation of various species/populations using integrative approach consisting of four main components.
| Examined species/populations |
Support from four evaluated components
|
Overall support (3–4/4) | |||
|---|---|---|---|---|---|
| GCPSR | STACEY1 | Single-locus MSC methods2 | Morphology / physiology | ||
| A. campestris | NO | YES (2/2) | YES (12/18) | YES | YES |
| segregation of A. campestris into three species | NO3 | NO (0/2) | NO (6/18) | NO | NO |
| A. candidus | YES | YES (1/2) | NO (7/18) | N/A (trend) | ? |
| A. dobrogensis | YES | YES (1/2) | NO (7/18) | N/A (trend) | ? |
| Aspergillus sp. DTO 244-F1 | N/A | YES (2/2) | YES (11/18) | N/A | ? |
| A. magnus | N/A | YES (2/2) | YES (17/18) | YES | YES |
| A. neotritici | YES | YES (2/2) | YES (18/18) | YES | YES |
| segregation of CCF 4914 and IBT 12659 from A. neotritici | YES | NO (0/2) | NO (3/18) | YES | NO |
| A. pragensis | YES | YES (2/2) | YES (17/18) | YES | YES |
| A. subalbidus | YES | YES (1/2) | YES (17/18) | YES | YES |
| segregation of CCF 6199 and CCF 5642 (pop 6) from A. subalbidus | YES | YES (1/2) | NO (9/18) | NO | NO |
| A. taichungensis | YES | YES (2/2) | YES (13/18) | YES | YES |
| segregation of DTO 266-G2 from A. taichungensis | NO | NO (0/2) | YES (10/18) | NO | NO |
| A. tenebricus | YES | YES (2/2) | YES (13/18) | YES | YES |
MCS – multispecies coalescent model-based methods; N/A – data not available or analysis could not be performed (non-viable strain DTO 244-F1 could not be analyzed phenotypically; species/populations represented by one strains could not be evaluated using GCPSR; a part of A. dobrogensis and A. candidus isolates cannot be distinguished morphologically – see sections Results, Discussion and Taxonomy).
1Support in at least one of the two most probable scenarios with collapseheight parameters 0.007 and 0.01 – see Fig. 4B.
2Support by the majority (at least 10 out of 18) of single-locus methods and their settings, i.e., agreement on the delimitation of species in its exact form or delimitation of smaller entities within it but without any admixture with related species/populations – see Fig. 3.
Phylogenetic analysis and GCPSR approach
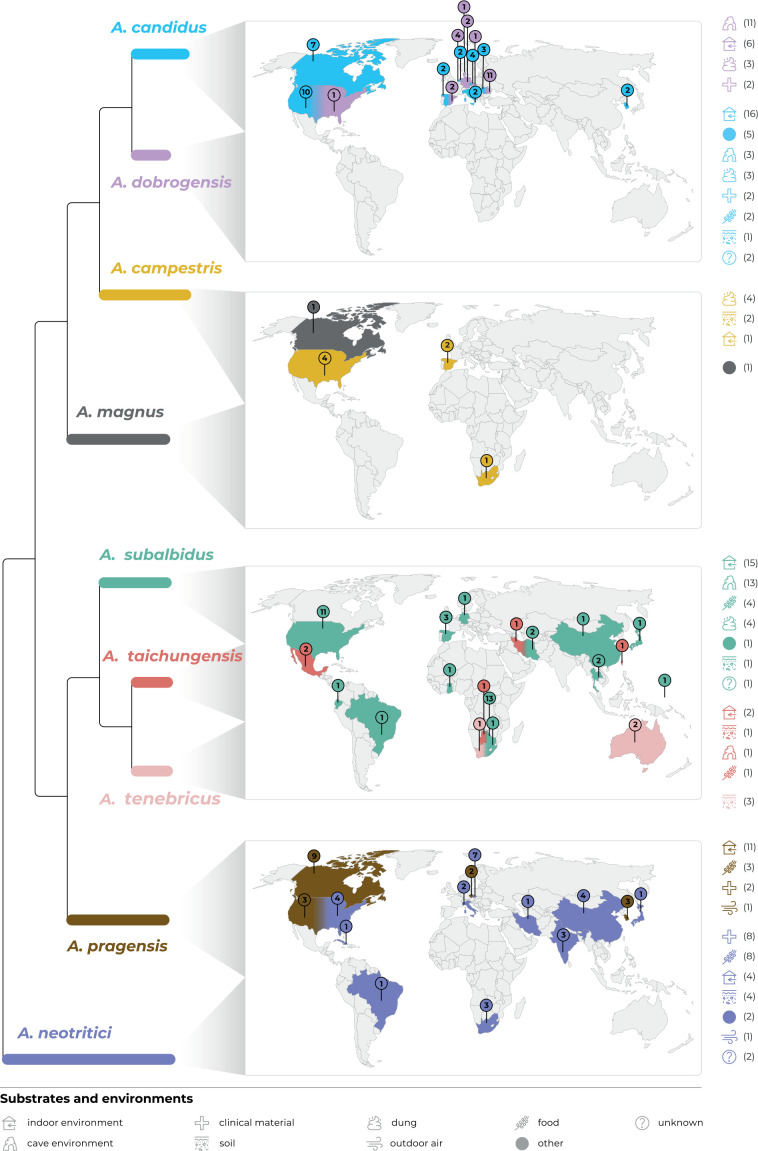
The best scoring ML tree based on the concatenated and partitioned alignment of 113 strains is shown in Fig. 1. The topology of the tree inferred by BI was almost identical and the posterior probabilities are appended to nodes together with bootstrap support values from ML analysis. Isolation source and geographical origin of strains are plotted on the tree (also listed in Table 1) together with the culture collection accession numbers. All alignments are available from the Dryad Digital Repository (https://doi.org/10.5061/dryad.3j9kd51mq) and basic alignment characteristics are listed in Table 2 together with partitioning scheme and substitution models used in the analyses.
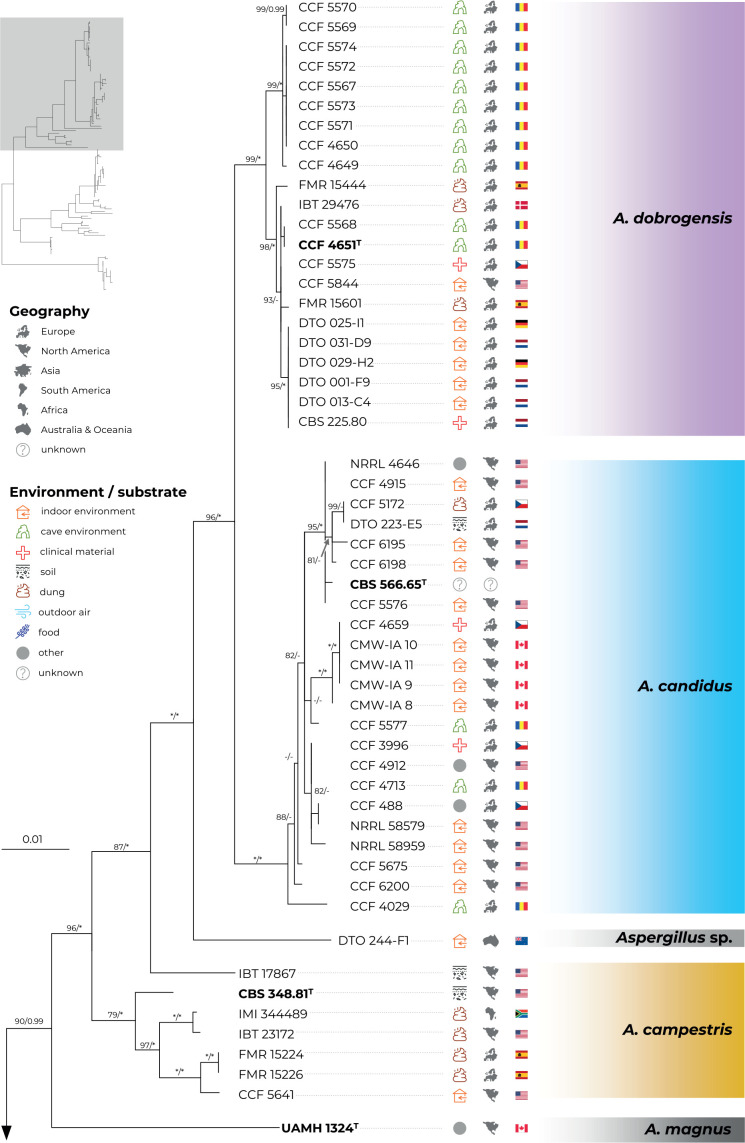
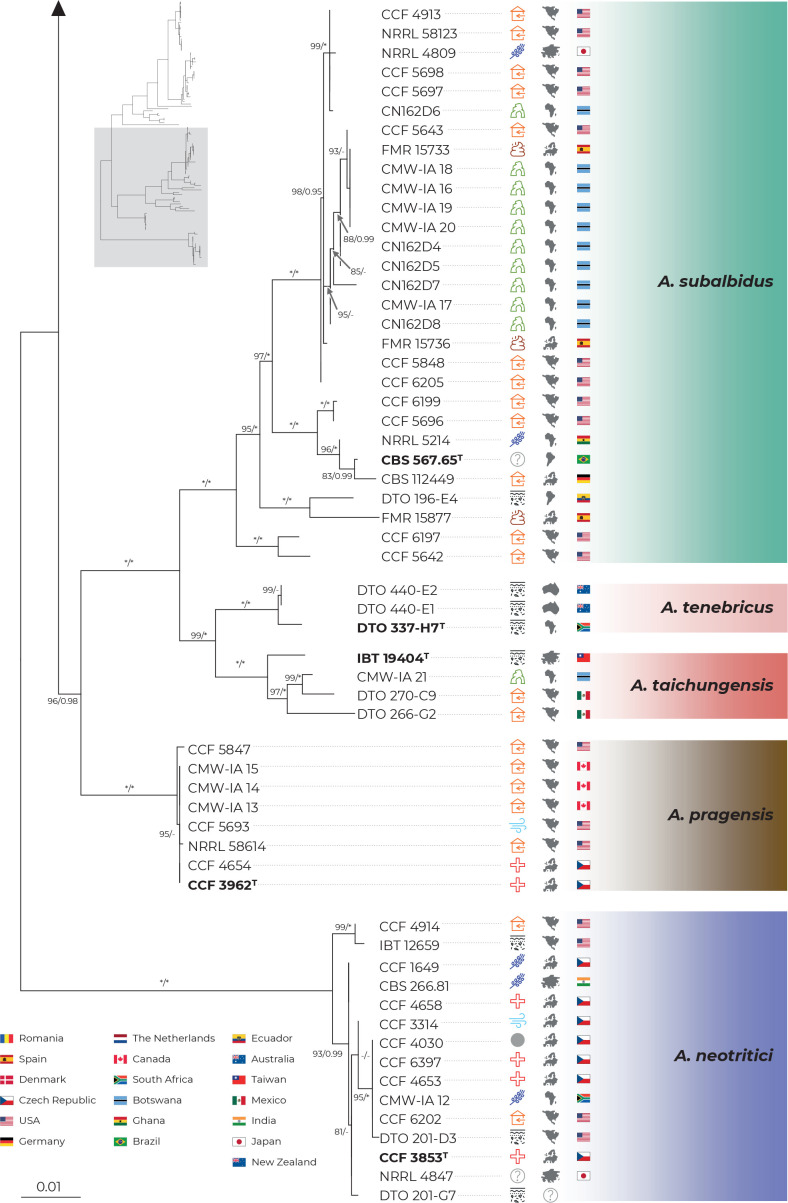
Fig. 1.
Multi-locus phylogeny of Aspergillus section Candidi based on three loci (benA, CaM, RPB2) and comprising 113 isolates. Best scoring Maximum Likelihood tree inferred in the IQ-TREE is shown; Maximum likelihood bootstrap values and Bayesian posterior probabilities are appended to nodes; only support values higher than 70 % and 0.95, respectively, are shown. The ex-type strains are designated with a superscripted T and bold print. Alignment characteristics, partitioning scheme and substitution models are listed in Table 2. The information on geographic origin and isolation source was plotted on the tree – see legend.
Phylogenetic relationships between section Candidi members are well resolved in the combined phylogeny and the species clustered into three main monophyletic clades. The first clade included A. candidus, A. dobrogensis, Aspergillus sp. DTO 244-F1, A. campestris and A. magnus. Although it is highly probable that strain DTO 244-F1 represents an undescribed species according to this phylogeny and molecular analyses mentioned below, it is no longer viable and thus only molecular data could be analysed. The second clade comprised A. subalbidus, A. taichungensis, A. tenebricus and A. pragensis. Aspergillus neotritici formed a single-species lineage, relatively distant from the other species. This topology was almost identical to the trees generated in STACEY analysis and starBEAST with one notable exception. In the ML and BI trees, A. campestris was resolved as polyphyletic due to the position of the strain IBT 17867. This strain formed a single-strain lineage clustering with Aspergillus sp. DTO 244-F1, A. candidus and A. dobrogensis. This is caused by its atypical RPB2 sequences influencing the topology not only of the RPB2 tree (Fig. 2) but also of the whole multi-locus phylogeny (Fig. 1). The RPB2 sequence of A. campestris IBT 17867 shares many variable positions with A. candidus and A. dobrogensis and probably represents a phenomenon of ancestral polymorphism/incomplete lineage sorting or could be caused by past recombination/hybridization. By contrast, in the tree from the multi-locus MSC method STACEY (Fig. 3) and starBEAST (see Delineate analysis), all seven strains of A. campestris formed a monophyletic clade. Other sequences and phenotypic characters of IBT 17867 were typical of A. campestris.
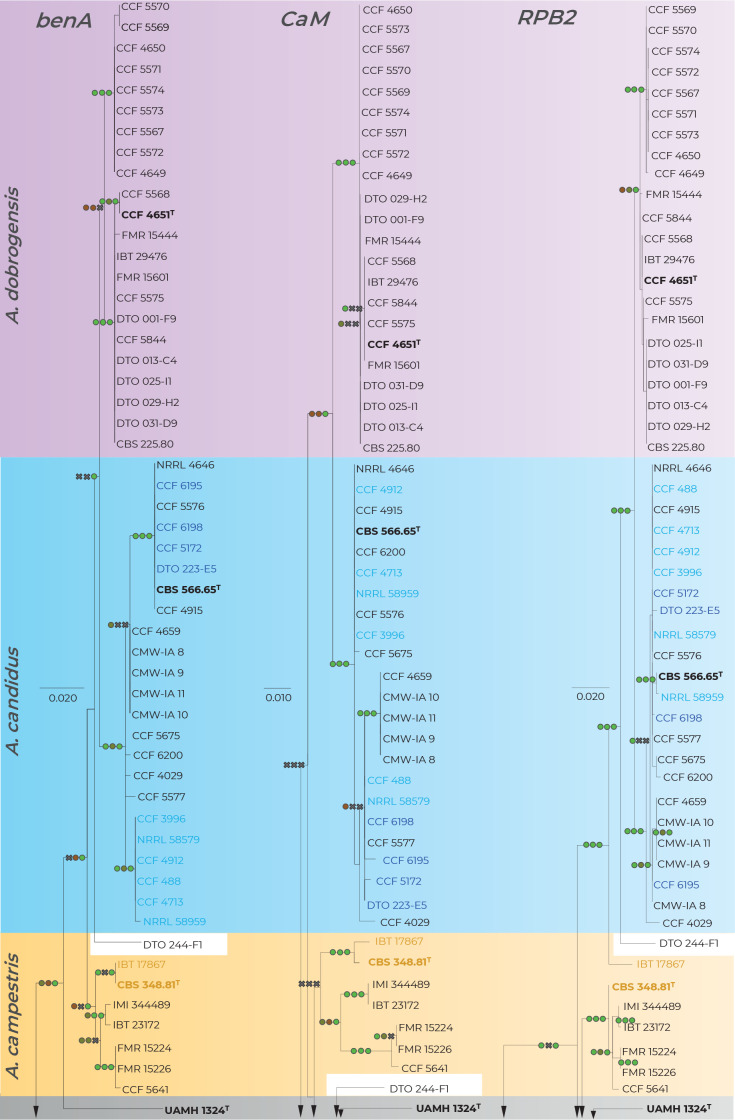
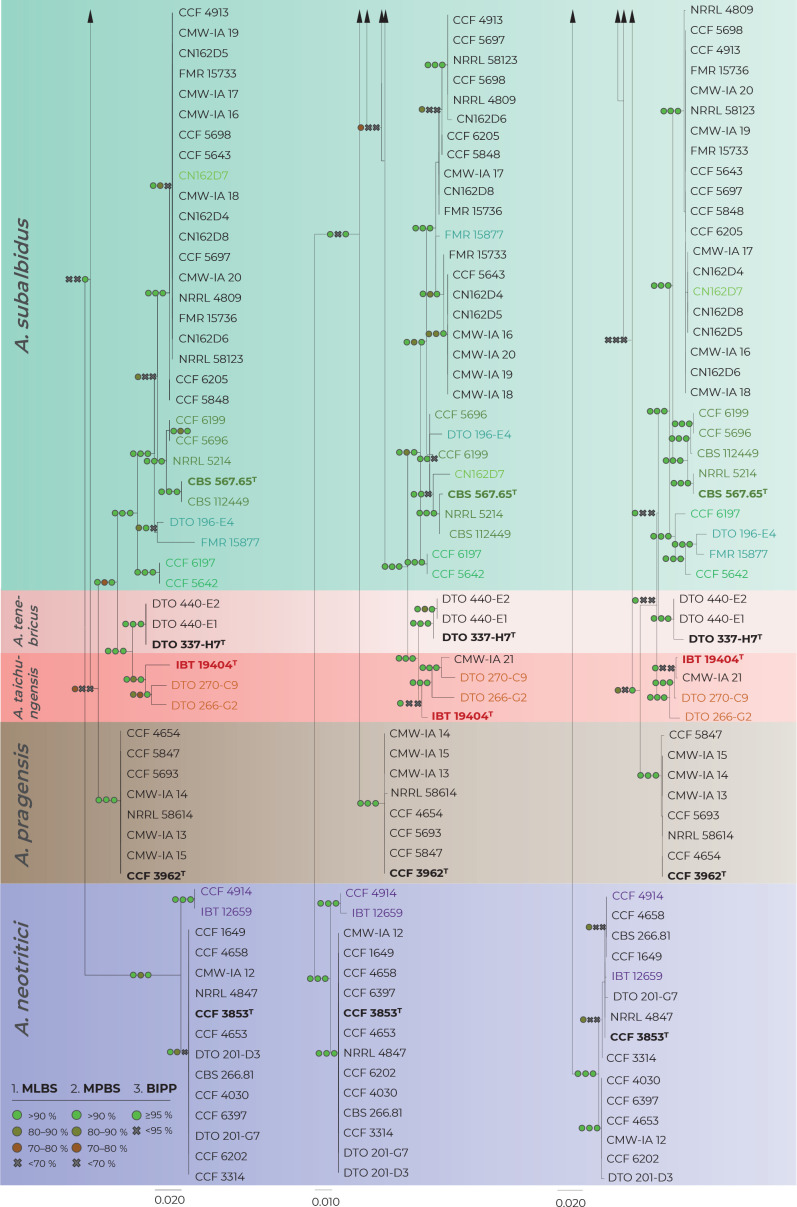
Fig. 2.
Comparison of single-gene genealogies based on the alignments of benA, CaM and RPB2 loci and created by three different phylogenetic methods. Single-locus maximum likelihood trees are shown; maximum likelihood bootstrap supports (MLBS), maximum parsimony bootstrap supports (MPBS) and Bayesian inference posterior probabilities (BIPP) are appended to nodes. Only MLBS and MPBS values ≥70 % and BIPP ≥0.95, respectively, are shown. A cross indicates lower statistical support for a specific node or the absence of a node in the MP and BI phylogenies. Selected strains or strain groups causing incongruences across single-gene phylogenies because of their unstable position are colour highlighted. The ex-type strains are designated with a superscripted T and bold print.
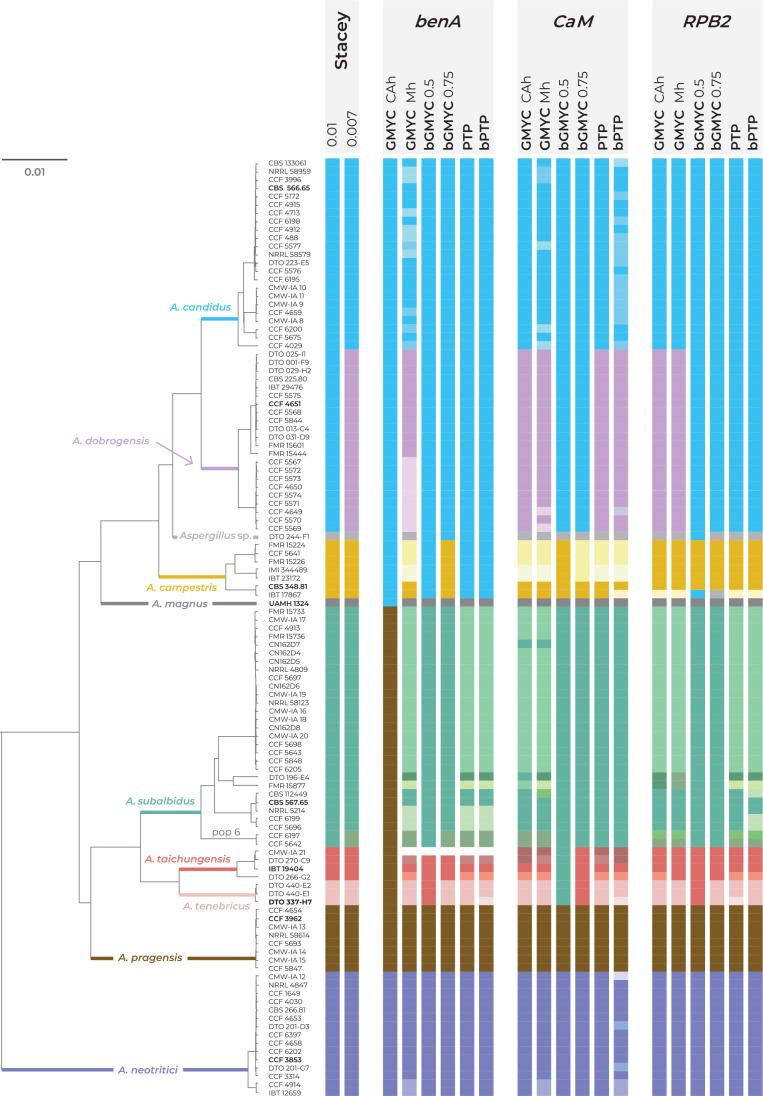
Fig. 3.
Schematic representation of results of species delimitation methods in the section Candidi. One multi-locus method (STACEY) and five single-locus methods (GMYC, bGMYC, PTP, bPTP) were applied on dataset of three loci (benA, CaM, RPB2). The results are depicted by coloured bars with different colours or shades indicating tentative species delimited by specific method and setting. Ex-type isolates of accepted species are highlighted with bold print. STACEY results with two values of collapseheight parameter, 0.01 and 0.007, are shown. For the GMYC method, the Coalescent Constant Population tree model was used as an input with both common ancestor heights (CAh) and Median heights (Mh) settings. For the bGMYC method, values 0.5 and 0.75 were used for the bgmyc.point function. The phylogenetic tree was calculated in starBEAST analysis and it is used solely for the comprehensive presentation of the results from different methods.
When comparing topologies of single-gene trees and applying rules of the GCPSR concept (Fig. 2), no conflicts were found between A. candidus and A. dobrogensis supporting definition of these species in their known limits (Hubka et al. 2018b). There was some exchange of isolates between clades within the A. candidus lineage and we thus consider this species a single phylogenetic species (PS). Three evolutionary lineages are supported among seven examined strains of A. campestris: IBT 17867 + CBS 348.81 (1), IMI 344489 + IBT 23172 (2) and FMR 15224 + FMR 15226 + CCF 5641 (3). Except for lineage 1, these lineages are present in all three single-gene trees without conflict. However, as mentioned above, the deviating RPB2 sequence and phylogenetic position of IBT 17867 in the RPB2 tree contradicts support of lineages 1 in the RPB2 tree. It is difficult to decide if there is a support for three PS using GCPSR because lineage 1 does not occur in the combined phylogeny reconstructed using ML and BI (Fig. 1) but at the same time, it is present in the species trees from other multigene analyses as mentioned above.
Aspergillus pragensis received clear support because this lineage contained little intra-species variation and it is well separated from other species in all phylogenies. The GCPSR approach also clearly supported delimitation of A. tenebricus and its sister species A. taichungensis. There are some incongruences within the A. taichungensis lineage not allowing this species to be defined other than in the form of four strains (IBT 19404, DTO 266-G2, DTO 270-C9 and genetically invariable CMW-IA 21). Significant incongruences can be found in the robust and structured lineage of A. subalbidus. GCPSR approach supports segregation of basal clade with strains CCF 5642 and CCF 6197 (referred to as population “pop 6” according to the DELINEATE analysis – see below) from A. subalbidus. Speciafically, segregation of this PS is supported by benA, CaM and combined phylogenies but not by RPB2 phylogeny (Figs 1–2).
All species belonging to the section Candidi are biseriate, however, within the lineage of A. neotritici, there is a subclade with two uniseriate strains CCF 4914 and IBT 12659 (no biseriate conidiophores observed) which is supported by benA, CaM and combined phylogenies but not by RPB2 (Figs 1–2). Although GCPSR supported segregation of this subclade from A. neotritici, MSC methods preferred a broad concept of A. neotritici (see below).
Singleton lineages with unstable phylogenetic position, namely, Aspergillus sp. DTO 244-F1 and UAMH 1324 (A. magnus), were excluded from evaluation using GCPSR because it is not possible to evaluate potential discordant positions of similar isolates across genealogies. On the other hand, both strains formed their own singleton lineages in all single-gene and multigene trees.
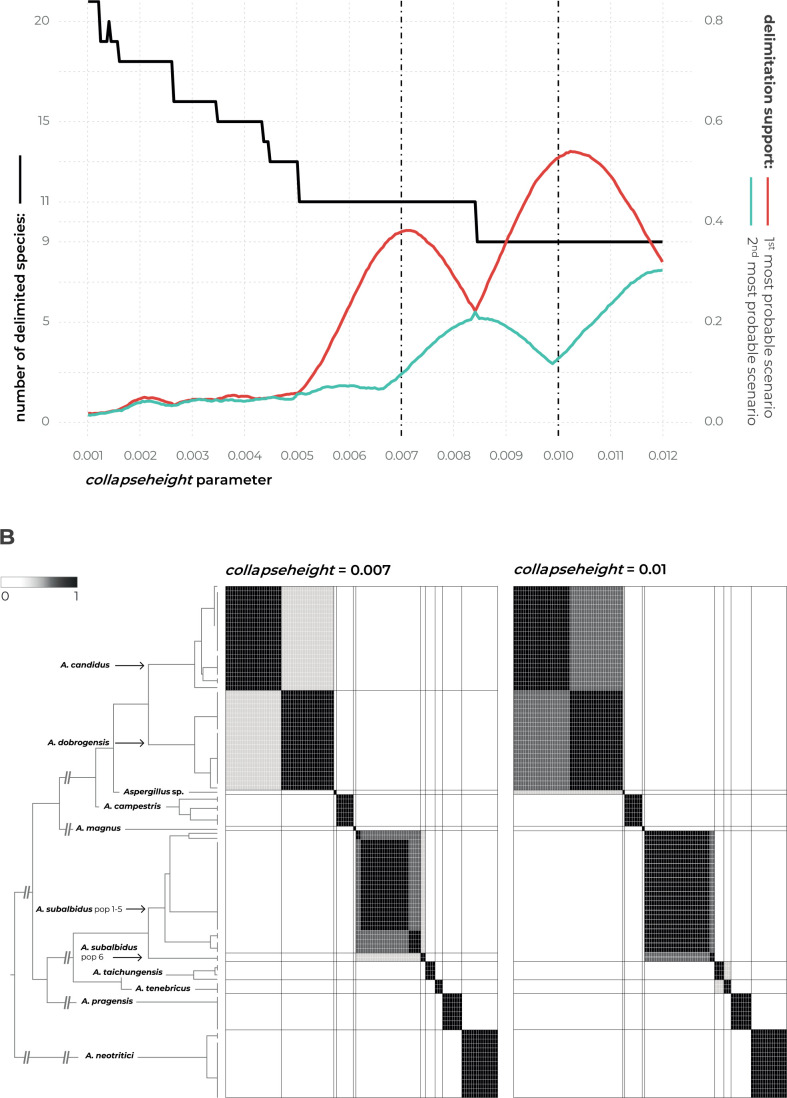
Species delimitation using STACEY
Detailed results of the multi-locus method STACEY are shown in Fig. 4 where subfigure A illustrates the effect of the collapseheight parameter value on the number of delimited species. This collapseheight parameter is plotted on the x-axis while on the left y-axis, there is a number of delimited species with the given collapseheight value (black line). The support for the most probable scenario (red line), and the second most probable scenario (turquoise line) are shown; other less supported scenarios are omitted. The changing value o the collapseheight parameter has consequences especially for the delimitation of A. dobrogensis, A. candidus, and species in the clade containing A. subalbidus, A. taichungensis and A. tenebricus. There are two main scenarios which gained reasonable support, i.e., with 9 and 11 delimited species (Fig. 4A). The vertical dashed lines in subfigure A represent these scenarios illustrated in detail in subfigures B (Fig. 4B) in the form of similarity matrices showing posterior probabilities of each pair of isolates being included in the same species.
Fig. 4.
The results of species delimitation by using STACEY method. A. Dependence of delimitation results on collapseheight parameter. The black solid line represents the number of delimited species (left y-axis) depending on the changing value of collapseheight parameter (x-axis). The red line represents the probability (range from 0 to 1, right y-axis) of the most probable scenario at specific collapseheight value. The turquoise line represents the probability of the second most probable scenario at specific collapseheight value. Dashed vertical lines mark two values (0.007, 0.01) of collapseheight parameter whose results are shown in detail by similarity matrices (subfigure B). B. The similarity matrices give the posterior probability of every two isolates belonging to the same multispecies coalescent cluster (tentative species). The darkest black shade corresponds to a posterior probability of 1, while the white colour is equal to 0. Thicker horizontal and vertical lines in the similarity matrices delimit species or their populations that gained delimitation support in some scenarios.
The scenario with 11 species reached support of approximately 0.4 (y-axis on the right side; maximum is 1) when the collapseheight parameter value is around 0.007. In this scenario, A. dobrogensis, A. candidus, A. taichungensis and A. tenebricus are delimited as separate species. Strains of A. subalbidus are divided into two tentative species – status of the separate species is supported for the clade “pop 6” designated according to the DELINEATE analysis (see below).
In the scenario with 9 species and collapseheight parameter value of approximately 0.01, A. subalbidus is delimited as a single broad species, while A. dobrogensis is merged with A. candidus. Both A. taichungensis and A. tenebricus are still delimited but with lower support. The delimitation support of the other species was stable and with high support in both scenarios (Aspergillus sp. DTO 244-F1, A. campestris, A. magnus, A. pragensis and A. neotritici).
Delimitation using single-locus MSC methods
There was a high overall agreement across single-locus MSC methods and their different settings on the delimitation of A. magnus, A. pragensis and A. neotritici while for the other species, different methods generated a plethora of possible arrangements (Fig. 3). Aspergillus pragensis and A. magnus have been consistently defined from other species by all mentioned methods except the GMYC method with Common Ancestor node heights settings (GMYC CAh) based on benA. In this setting, A. magnus was lumped with A. candidus, A. dobrogensis, A. campestris and Aspergillus sp. DTO 244-F1, while A. pragensis was lumped with A. subalbidus, A. taichungensis and A. tenebricus. Only three analyses delimited one or more additional species within A. neotritici lineage, namely GMYC method with Median node heights setting (GMYC Mh) based on benA and CaM loci, and bPTP method based on CaM locus. All these three analyses agreed on the separation of clade containing uniseriate isolates CCF 4914 and IBT 12659 from A. neotritici. Most of the analyses (12/18) also supported delimitation of singleton species Aspergillus sp. DTO 244-F1 which is related to A. candidus, A. dobrogensis and A. campestris.
The agreement of the methods on the delimitation and arrangement of the remaining species was much lower. The majority of analyses did not support the delimitation of A. dobrogensis from A. candidus (Fig. 3). Only seven analyses distinguished A. candidus and A. dobrogensis or delimited a couple of additional species within these species. Delimitation of A. campestris in its broad concept (seven isolates) was only supported by bGMYC with value 0.5 for the bgmyc.point function (bGMYC 0.5) based on CaM locus and with value 0.75 (bGMYC 0.75) based on benA locus. This broad concept is in agreement with results of STACEY with both collapseheight parameters. Some analyses based on benA locus (PTP, bPTP, GMYC CAh and bGMYC 0.5 methods) lumped A. campestris with A. candidus and A. dobrogensis. All methods based on CaM locus except bGMYC 0.5 and also GMYC Mh based on benA locus delimited 2–4 species within A. campestris. Because the RPB2 sequence of strain IBT 17867 is atypical and relatively dissimilar from other A. campestris isolates, all single-locus methods based on RPB2 failed to connect this strain with A. campestris. This strain was either delimited as a singleton species or it was lumped with Aspergillus sp. DTO 244-F1 (bGMYC 0.75) or with the clade containing A. candidus and A. dobrogensis (bGMYC 0.5).
Aspergillus taichungensis was delimited as a species with four strains (IBT 19404, DTO 270-C9, DTO 266-G2 and CMW-IA 21) by STACEY with both values of the collapseheight parameter. This arrangement was only supported by three single-locus methods, namely, bGMYC 0.75 based on benA and RPB2 and GMYC Mh based on RPB2. The majority of analyses (10/18) delimited 2–4 species within A. taichungensis (Fig. 3). Three analyses merged A. taichungensis with A. tenebricus and two analyses (bGMYC 0.5 based on CaM and GMYC Cah based on benA) even merged these species with A. subalbidus and/or A. pragensis. Similarly to STACEY, most of the analyses (10/18) delimited A. tenebricus as a species comprising three strains: DTO 337-H7, DTO 440-E1 and DTO 440-E2. Three analyses supported segregation into two species and the remaining five analyses merged A. tenebricus with related species as mentioned above. Aspergillus subalbidus was represented by a high number of strains (n = 29) which were structured into several clades and frequently delimited as separate species by single-locus MSC methods. STACEY proposed a broad concept of A. subalbidus with 29 or 27 strains, only supporting segregation of “pop 6” (see DELINEATE analysis below) in the setting with low collapseheight parameter (0.007). This broad concept of A. subalbidus was supported by six out of 18 single-locus MSC analyses and the remaining analyses usually segregated “pop 6” (nine out of 18) and/or delimited several additional species with variable arrangements. Two analyses lumped A. subalbidus with neighbouring species (Fig. 3).
Phenotypic analysis - macromorphology
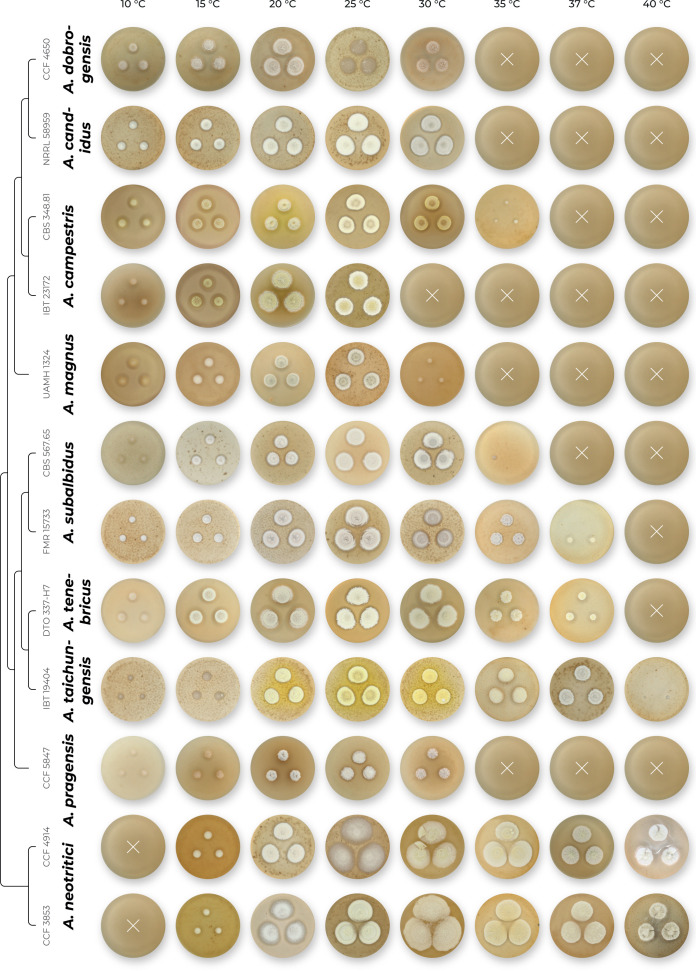
All species were able to grow on the four cultivation media (MEA, CYA, CZA, CY20S) at 25 °C. Colony diameters on these media and selected macromorphological characters relevant for the species identification are summarized in Table 4. Almost all species have the largest colony diameter on CY20S compared to the other media with a lower sugar content. Aspergillus magnus and A. pragensis had the smallest colony diameters and their restricted growth was even more accentuated on CZA.
Table 4.
Overview of selected macromorphological characters and growth parameters at 25 °C.
| Species (no. of examined strains) |
Colony diameter after 7 d in mm (mean) |
Colony diameter after 14 d in mm (mean) |
Colony colours (CYA and MEA) | Soluble pigment (present : absent)1 | Sclerotia (present : absent)1 | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| MEA | CYA | CZA | CY20S | MEA | CYA | CZA | CY20S | ||||
| A. campestris (7) | 11–17 (15) | 14–20 (17) | 6–14 (9) | 10–21 (18) | 16–28 (24) | 22–36 (28) | 10–20 (16) | 25–34 (30) | yellow, sulphur yellow, yellowish white | 7 : 0 (CZA > CYA > CY20S) | 4 : 3 (CZA, CYA) |
| A. candidus (9) | 12–17 (15) | 15–21 (18) | 9–15 (11) | 16–25 (22) | 20–27 (24) | 20–32 (27) | 16–25 (21) | 25–45 (36) | white, white with a yellowish tinge | 5 : 4 (CZA > CYA) | 5 : 4 (CZA) |
| A. dobrogensis (10) | 17–20 (19) | 20–24 (21) | 9–14 (10) | 18–28 (24) | 22–35 (29) | 26–39 (32) | 17–22 (19) | 32–48 (41) | white, white with a yellowish tinge | 0 : 10 | 2 : 8 (CZA > MEA, CYA, CY20S ) |
| A. magnus (1) | 12–14 (13) | 12–14 (13) | 4–6 (5) | 13–15 (14) | 20–22 (21) | 16–17 (17) | 11–13 (12) | 19–20 (20) | pale yellow, yellowish gray | 1 : 0 (CYA) | 0 : 1 |
| A. neotritici (11) | 10–25 (18) | 14–28 (22) | 5–19 (12) | 19–31 (27) | 20–47 (31) | 21–50 (38) | 14–39 (25) | 28–60 (51) | white, yellowish white | 6 : 5 (CZA) | 4 : 7 (CZA) |
| A. pragensis (5) | 8–10 (9) | 9–14 (12) | 3–6 (5) | 13–18 (15) | 15–18 (16) | 16–22 (20) | 11–15 (12) | 24–30 (27) | white | 5 : 0 (CZA, CYA) | 2 : 3 (CYA > MEA) |
| A. subalbidus (19) | 11–17 (14) | 14–24 (20) | 8–18 (13) | 18–27 (22) | 18–30 (24) | 24–42 (32) | 16–26 (21) | 24–45 (33) | white, yellowish white | 12 : 7 (CYA > CZA) | 7 : 12 (CZA > CYA > MEA > CY20S) |
| A. taichungensis (3) | 16–18 (17) | 22–28 (25) | 10–12 (11) | 28–31 (30) | 25–29 (27) | 33–43 (38) | 19–21 (20) | 40–51 (46) | pastel yellow | 2 : 1 (CYA, MEA) | 2 : 1 (CZA, CY20S) |
| A. tenebricus (3) | 19–21 (20) | 19–23 (21) | 12–14 (13) | 28–30 (29) | 26–33 (30) | 29–34 (31) | 22–24 (23) | 48–52 (50) | white, yellowish white | 3 : 0 (CYA, CZA) | 2 : 1 (CZA) |
1Production was scored after 4 wk of cultivation on MEA, CYA, CZA and CY20S; the ratio shows the number of strains producing and not producing pigment/sclerotia; media on which pigment or slerotia were produced, are listed in the parentheses and sorted by frequency. See section Taxonomy for more details concerning colours of soluble pigments and sclerotia, and strain numbers.
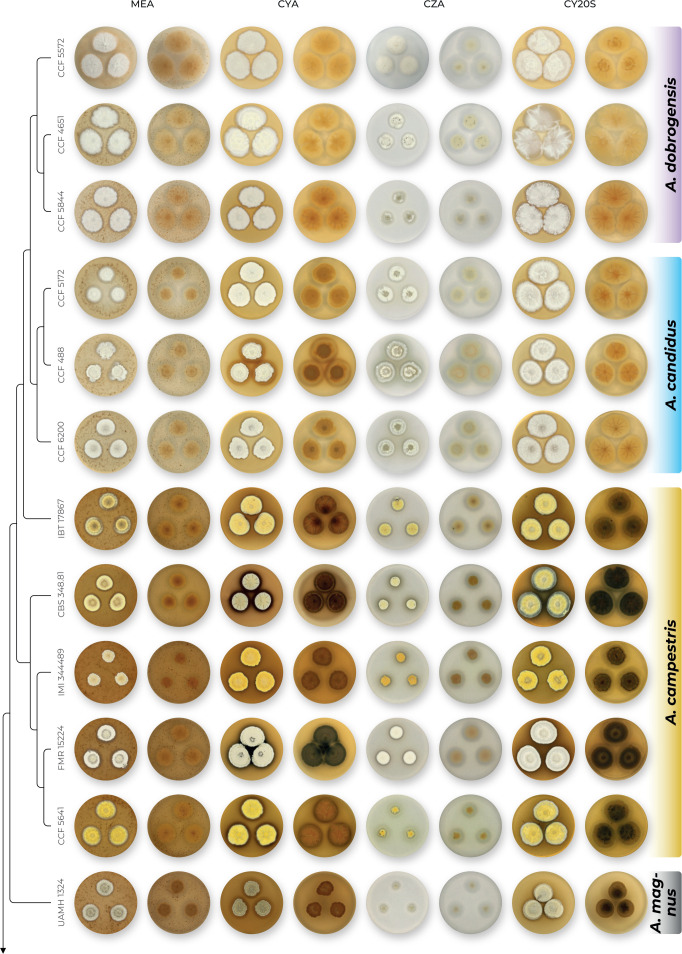
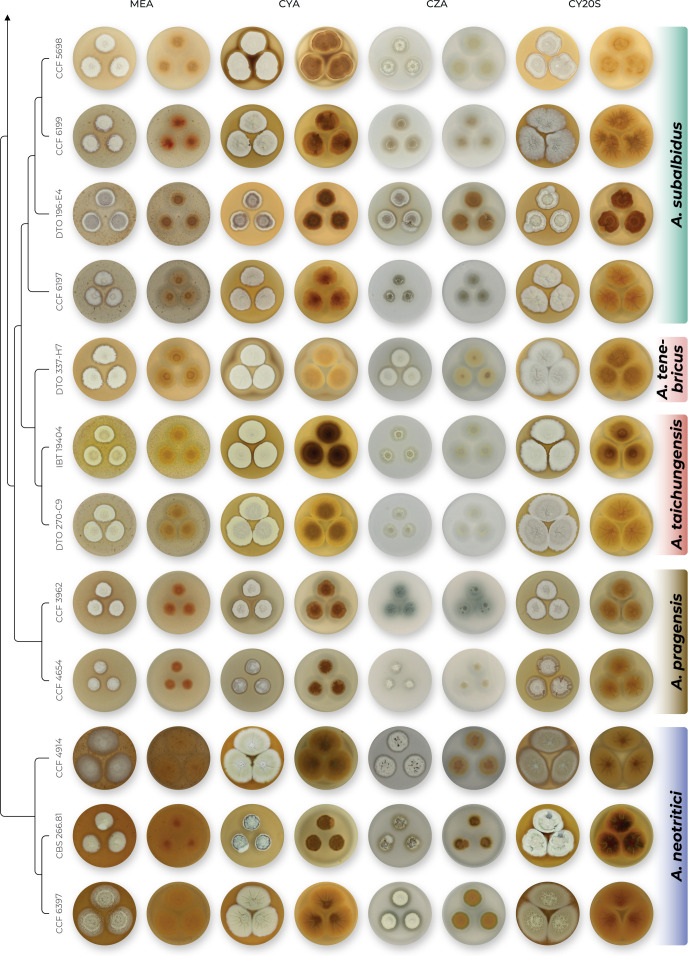
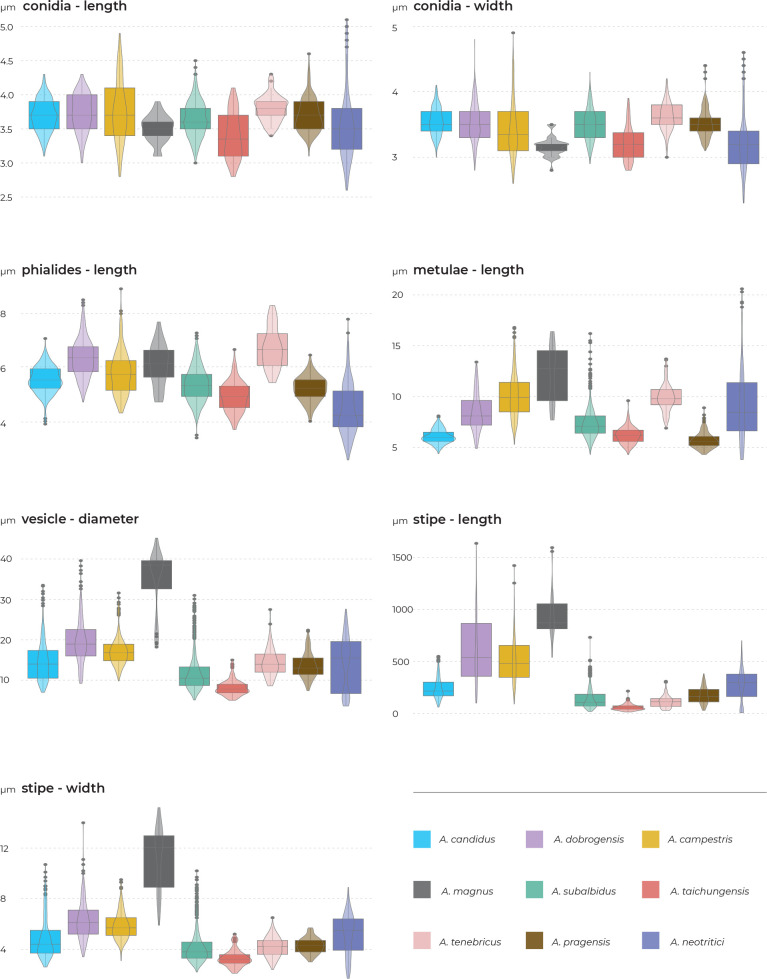
The overview of macromorphology of all species and their various morphotypes are shown in Fig. 5. Colony morphology of A. candidus is very similar to A. dobrogensis, but colonies of A. dobrogensis strains have a larger diameter on MEA, CYA and CY20S (Table 4). The colony colour of most species in the section Candidi is white or yellowish white, but shades of yellow dominate in A. campestris (usually yellow to sulphur yellow), A. magnus (greyish yellow on MEA, CYA and CY20S) and A. taichungensis (pastel yellow on CYA and MEA). Intraspecific variability in colony colour and dimensions is sometimes high. For instance, colonies of A. campestris, usually ascribed as yellow, sulphur yellow or bright yellow, were rather white or yellowish-white in FMR 15224. In addition, there were also significant differences in the shape and profile of colonies, production of soluble pigment and reverse colour among strains (Fig. 5), but without any phylogenetic pattern. Another phenotypically variable species was A. subalbidus with strains showing significant differences in their colony surfaces, dimensions and colours (Fig. 5). Two strains of A. subalbidus (DTO 196-E4 and FMR 15877) differed from other strains by their greyish beige colonies, but otherwise they did not display any other unique characteristics. Neither the other morphological variability observed in A. subalbidus displayed any phylogenetic pattern that could lead to considerations of segregation into multiple monophyletic species. Two strains of A. neotritici (CCF 4914 and IBT 12659), which formed their own subclade (Fig. 1, Fig. 2, Fig. 3), differed from other strains by more velvety colony surface and significantly deviated in micromorphology (see below).
Fig. 5.
Overview of macromorphological patterns (obverse and reverse) within section Candidi on four cultivation media (MEA, CYA, CZ, CY20S) grown for 14 d at 25 °C. Macromorphological characters were scored in detail for 68 isolates and only unique phenotypic patterns are shown for every species.
Production of sclerotia was observed in all species (except A. magnus), mostly on CZA, CYA and MEA (Table 4). Except A. dobrogensis, at least some strains of every species produced soluble pigments after 4 wk of cultivation, most often on CZA and CYA (Table 4). In general, the production of sclerotia and soluble pigments was mostly strain-specific rather than species-specific. But in several species represented by more strains, we observed some trends, and in few cases, production of pigments contributed to the species differentiation. For instance, a dark brown soluble pigment was produced by A. tenebricus compared to yellow soluble pigment produced by some strains of A. taichungensis. Dark soluble pigments were also produced by all strains of A. campestris and A. pragensis but with variable intensity and location. In contrast, soluble pigments were observed to be absent in A. dobrogensis strains while in the closely related A. candidus, five out of nine examined strains produced pigments.
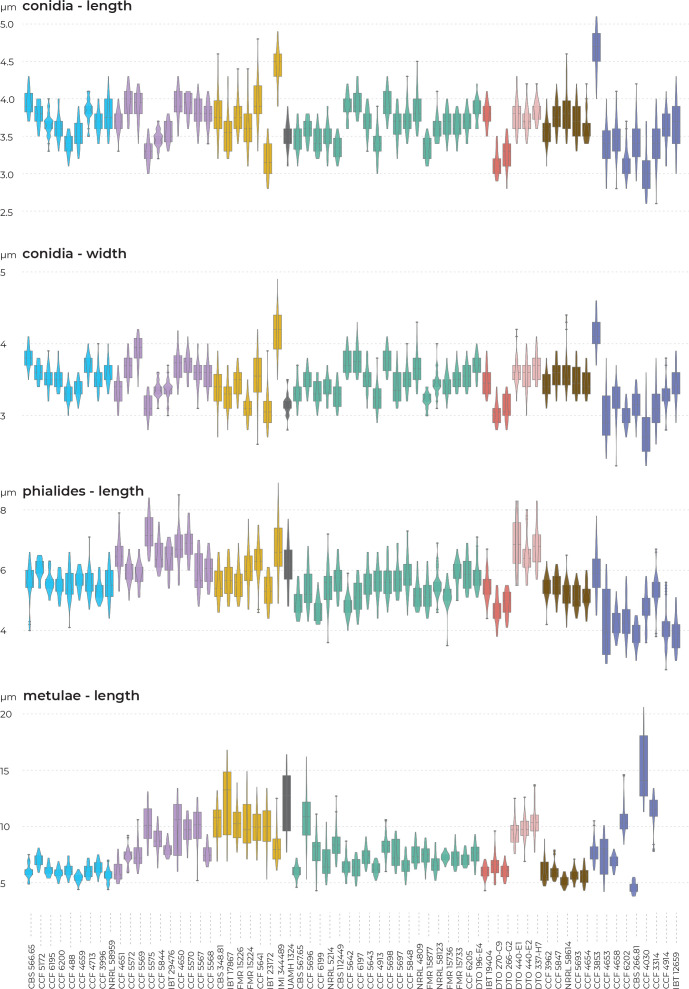
Phenotypic analysis – micromorphology
Dimensions of micromorphological characters are summarized in Table 5 and statistical significances of differences in these characters between species are detailed in Supplementary Table S1. Interesting phenomenon can be seen in A. neotritici because some strains of this species produce atypically short and uniseriate conidiophores only (CCF 4914 and IBT 12659) in contrast to other A. neotritici isolates and other section Candidi species. These strains formed a separate clade in benA and CaM phylogenies and were delimited as separate species by three single-locus MSC methods. The strain CBS 266.81, a reference strain of invalidly described A. tritici, produces atypically short, distorted and septate conidiophores. Colonies of this strain also have a smaller diameter than other A. neotritici isolates on all media and the strain produces abundant sclerotia.
Table 5.
| Species (no. of examined strains) |
Stipe
|
Metulae length; mean ± sd | Phialides length; mean ± sd | Vesicle diam; mean ± sd | Conidia (longer dimension); mean ± sd | |
|---|---|---|---|---|---|---|
| Length; mean ± sd | Width; mean ± sd | |||||
| A. campestris (7) | (90–)250–850(–1 300); 507 ± 217.5 | (4–)5–7.5(–9); 5.9 ± 1.1 | (7–)8–14(–17); 10.1 ± 2.3 | (4.5–)5–7(–9); 5.9 ± 0.8 | (10.5–)14–22(–31.5); 17.1 ± 3.7 | (2.8–)3–4.5(–4.9); 3.8 ± 0.5 |
| A. candidus (9) | (70–)125–400(–520); 242 ± 101.5 | (2.5–)3.5–8.5(–9.5); 4.9 ± 1.6 | (4.5–)5.5–7.5(–8); 6 ± 0.7 | (4.5–)5–6.5(–7); 5.6 ±0.5 | (8–)10–23(–33); 14.7 ± 5.0 | (3.1–)3.3–4(–4.3); 3.7 ± 0.2 |
| A. dobrogensis (10) | (100–)150–1 150(–2 000); 609 ± 330.2 | (3–)5–7(–14); 6.2 ± 1.5 | (5–)5.5–12(–14); 8.4 ± 1.9 | (5–)5.5–7.5(–8.5); 6.4 ± 0.7 | (9–)10–30(–39.5); 19.6 ± 5.6 | (3–)3.4–4.1(–4.3); 3.7 ± 0.3 |
| A. magnus (1) | (540–)810–1 150(–1 600); 945 ± 225.8 | (6–)9–13(–16); 11.2 ± 2.4 | (7.5–)9–14(–17); 12.1 ± 2.8 | (4.5–)5.5–7(–8); 6.1 ± 0.8 | (18–)33–49(–45); 34.8 ± 8.0 | (3.2–)3.4–3.6(–3.8); 3.5 ± 0.2 |
| A. neotritici (9) | (140–)250–500(–700); 363 ± 109.73 | (3.5–)4–8(–9); 6.1 ± 1.03 | (4–)7–17(–21); 9.9 ± 3.33 | (2.5–)3.5–6(–8); 4.5 ± 0.9 | (11–)14–24(–28); 18.4 ± 3.73 | (2.6–)3–4.6(–5.1); 3.5 ± 0.5 |
| A. pragensis (5) | (40–)110–260(–380); 175 ± 82.1 | (3–)3.5–5(–6); 4.3 ± 0.6 | (4–)5–6.5(–8); 5.7 ± 0.8 | (4–)5–6(–6.5); 5.3 ± 0.4 | (7–)10–17(–23); 13.6 ± 3.1 | (3.1–)3.5–4(–4.6); 3.7 ± 0.3 |
| A. subalbidus (19) | (20–)50–330(–730); 136 ± 91.0 | (2.5–)3–8(–10); 4.2 ± 1.3 | (4–)6–11(–16.5); 7.4 ± 1.5 | (3.5–)4.5–6(–7.5); 5.4 ± 0.6 | (5–)7–23(–31); 11.8 ± 4.7 | (3–)3.2–4.2(–4.5); 3.7 ± 0.3 |
| A. taichungensis (3) | (15–)40–90(–215); 60 ± 31.4 | (2–)3–4(–5); 3.3 ± 0.7 | (4–)5.5–7(–9.5); 6.2 ± 0.8 | (3.5–)4.5–6.5(–7.5); 5.0 ± 0.5 | (5–)7–11(–15); 8.2 ± 2.0 | (2.8–)3–3.8(–4.1); 3.4 ± 0.3 |
| A. tenebricus (3) | (30–)80–140(–310); 114 ± 57.9 | (2.5–)3.5–5(–6.5); 4.2 ± 0.8 | (7–)8–12(–14); 9.9 ± 1.3 | (5.5–)6–7(–8.5); 6.7 ± 0.7 | (8.5–)11–18(–27); 14.3 ± 3.4 | (3.4–)3.6–3.9(–4.3); 3.8 ± 0.2 |
1Measured after 1–3 wk of cultivation on MEA at 25 °C.
2Statistical comparison of micromorphological characters between species is listed in Supplementary Table 1.
3Strains CCF 4914, IBT 12659 and CBS 266.81 were excluded because they produced short, uniseriate or atypical conidiophores.
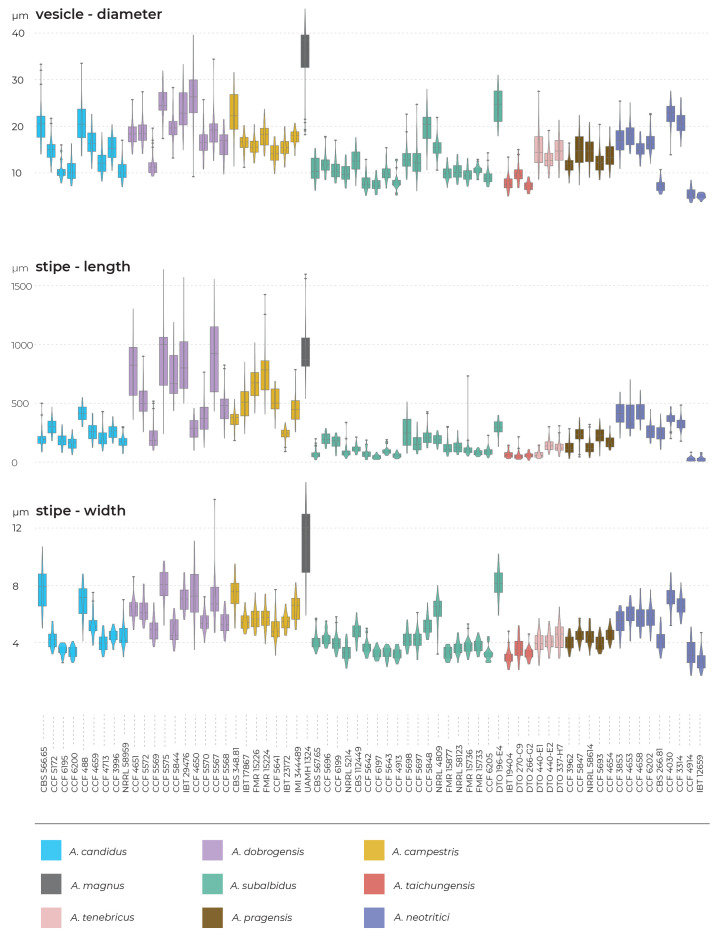
Results of the micromorphological analysis show that the most important characters for distinguishing species are the length and width of stipe, vesicle diameter and the length of metulae. In contrast, conidial dimensions are characters that do not contribute to species differentiation (Fig. 6). Aspergillus magnus is easily distinguishable from other species. It has the longest and widest stipes, the largest vesicle and the longest metulae. Aspergillus dobrogensis has larger dimensions of stipes, vesicles, metulae and phialides compared to the closely related A. candidus (Fig. 6). This statement is however valid only for the species as a whole as there are several individual strains not differing from A. candidus (Fig. 7). The possibilities of micromorphological differentiation between related species A. subalbidus, A. taichungensis, A. tenebricus and A. pragensis are limited. Among these species, A. tenebricus typically has longer phialides and metulae. It also has a larger vesicle than A. taichungensis. Some phylogenetic methods supported segregation of clade “pop 6” from A. subalbidus or segregation of DTO 266-G2 from A. taichungensis. We did not observe any specific feature connecting these strains and differentiating them from other A. subalbidus strains. The strain DTO 266-G2 also did not show unique characters compared to remaining A. taichungensis strains except much weaker production of yellow soluble pigment.
Fig. 6.
Summary overview of seven micromorphological characters across species of section Candidi. Boxplots show median, interquartile range, values within ± 1.5 of interquartile range (whiskers) and outliers.
Fig. 7.
A detailed overview of seven micromorphological characters at a strain level. Boxplots show median, interquartile range, values within ± 1.5 of interquartile range (whiskers) and outliers.
In some species, we observed remarkable differences in individual microscopic characters between strains. For example, in the A. campestris lineage, strain IMI 344489 produced larger conidia while strain CBS 348.81 had larger vesicles (Fig. 7). In the A. subalbidus lineage, strain DTO 196-E4 produced significantly larger stipes and vesicles, and among A. neotritici strains, CCF 4030 had longer metulae and CCF 3853 produced larger conidia (Fig. 7). These phenotypically exceptional strains did not form any well-defined phylogenetic units.
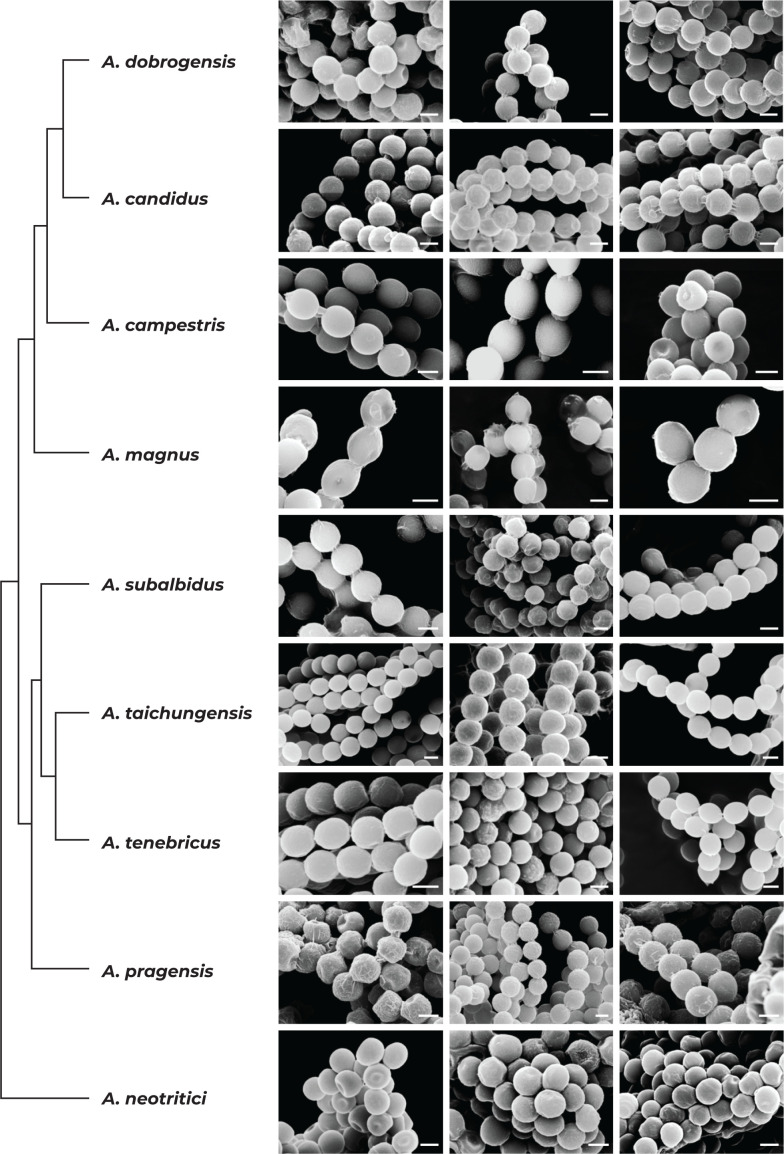
We did not observe any differences in surface ornamentation of conidia between species when using SEM (Fig. 8). Conidia of all species were smooth-walled or occasionally finely roughened.
Fig. 8.
Conidia under scanning electron microscopy (SEM). Examined strains from left to right: A. dobrogensis - CCF 4649, IBT 29476, IBT 29476; A. candidus - CCF 3996, CBS 566.65, CCF 3996; A. campestris - FMR 15224, FMR 15226, IBT 23172; A. magnus - UAMH 1324 only; A. subalbidus - FMR 15877, CBS 122449, CCF 5643; A. taichungensis - DTO 266-G2 only; A. tenebricus - DTO 440-E1, DTO 337-H7, DTO 337-H7; A. pragensis - CCF 4654, CCF 3962, CCF 4654; A. neotritici - CCF 4653, CBS 266.81, IBT 12659. Scale bars = 2 μm.
Physiology
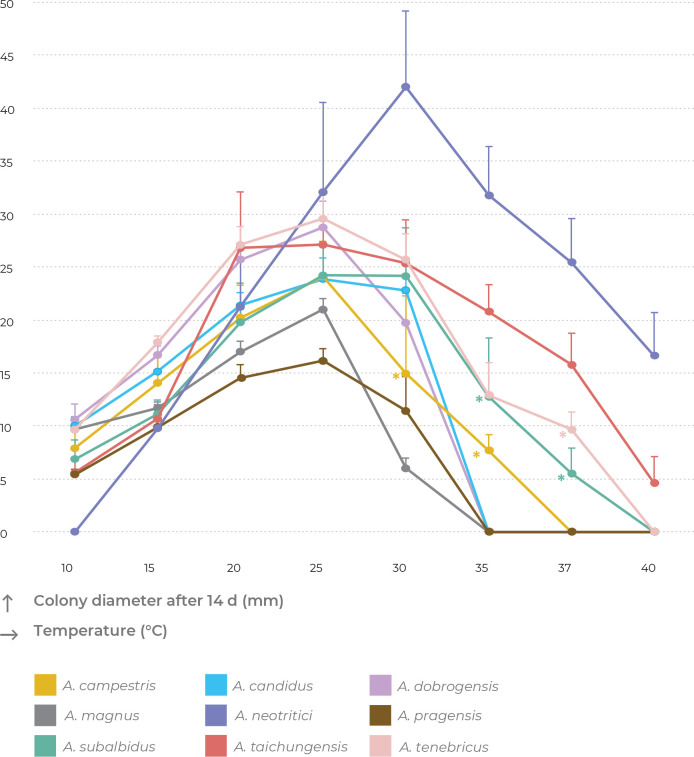
Cardinal temperatures were assessed on MEA at nine different temperatures ranging from 10 to 45 °C. The most common growth patterns are shown in Fig. 9. Aspergillus neotritici is the only species which is not able of growing or at least germinating at 10 °C and its optimal growth temperature is around 30 °C, while all other species have optima around 25 °C or do not grow faster at 30 °C compared to 25 °C. At least some strains of four species can grow at 37 °C, namely, A. neotritici, A. subalbidus, A. taichungensis and A. tenebricus. All strains of A. taichungensis and A. neotritici grow or germinate at 40 °C (Fig. 10). An unusually wide variability was observed in the temperature maximum of A. campestris. Some strains of this species do not grow at 30 °C (IBT 17867, IBT 23172), while all others grow well at this temperature, and the ex-type strain (CBS 348.81) grows up to 35 °C. Similarly, only some strains (CBS 567.65, FMR 15736, NRRL 58123, FMR 15733, CCF 5643, NRRL 4809, CCF 5697, CCF 6197) of A. subalbidus were able to grow at 35 °C and only some strains of A. subalbidus (CCF 5643, CCF 5697, FMR 15733) and A. tenebricus (DTO 337-H7, DTO 440-E2) could grow at 37 °C (Table 6, Fig. 9, Fig. 10).
Fig. 9.
Temperature growth profiles of Aspergillus section Candidi members after 14 d on MEA at temperatures ranging from 10 °C to 40 °C. Physiological characters were scored in detail for 52 isolates and only predominant patterns are shown for every species. For details, see Table 6 and section Taxonomy.
Fig. 10.
Growth of section Candidi members after 14 d on MEA at temperatures ranging from 10 °C to 40 °C. The points on the curves show the mean values and standard deviations. When isolates of the same species differed in their maximum growth temperatures, zero values were omitted for calculation of mean values (these points are designated by asterisks).
Table 6.
Overview of growth parameters at different temperatures.
| Species (no. of examined strains) |
Colony diameter after 14 d of cultivation on MEA in mm (mean value)
|
|||||||
|---|---|---|---|---|---|---|---|---|
| 10 °C | 15 °C | 20 °C | 25 °C | 30 °C | 35 °C | 37 °C | 40 °C | |
| A. campestris (7) | 4–10 (8) | 10–17 (14) | 15–26 (20) | 16–28 (24) | 0–22 (18)1 | 0–4 (3)1 | — | — |
| A. candidus (5) | 8–11 (10) | 12–17 (15) | 20–23 (21) | 20–27 (24) | 18–25 (23) | — | — | — |
| A. dobrogensis (6) | 9–13 (11) | 15–18 (17) | 24–27 (26) | 23–32 (29) | 17–24 (20) | — | — | — |
| A. magnus (1) | 9–10 (10) | 11–12 (12) | 16–18 (17) | 20–22 (21) | 5–7 (6) | — | — | — |
| A. neotritici (6) | — | 4–13 (10) | 11–26 (21) | 23–47 (32) | 32–51 (42) | 30–42 (32) | 18–33 (25) | 4–22 (17) |
| A. pragensis (5) | 5–6 (5) | 9–11 (10) | 13–17 (15) | 15–18 (16) | 3–14 (11) | — | — | — |
| A. subalbidus (16) | 3–9 (7) | 9–14 (11) | 15–30 (20) | 18–30 (24) | 9–29 (24) | 0–22 (13)1 | 0–9 (6)1 | — |
| A. taichungensis (3) | 4–7 (6) | 9–12 (11) | 20–32 (27) | 25–29 (27) | 20–29 (25) | 18–25 (21) | 12–20 (16) | 1–7 (5) |
| A. tenebricus (3) | 9–11 (10) | 17–19 (18) | 25–29 (27) | 26–33 (30) | 22–29 (26) | 8–16 (13) | 0–12 (10)1 | — |
1The mean was calculated from non-zero values only.
— no growth.
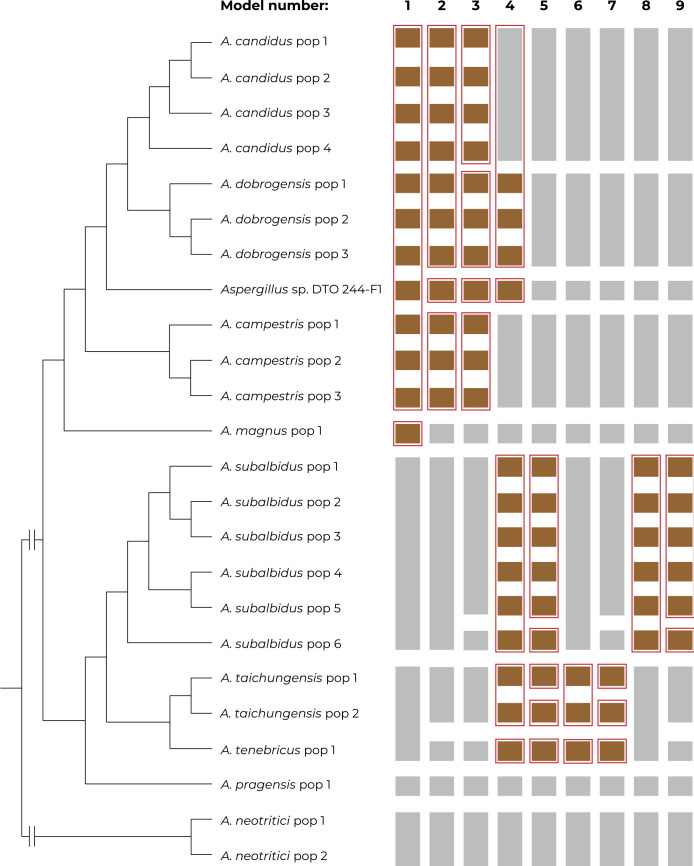
Species delimitation using DELINEATE software
The species hypotheses were independently tested in DELINEATE software where we set up nine different models. The results are summarized in Fig. 11. Individual populations were either assigned into species according to the previous results of species delimitation (Supplementary Table S2) - grey coloured bars, or they were left unassigned and free to be delimited - brown coloured bars. The red frames show the resulting solution proposed by DELINEATE for every model.
Fig. 11.
The results of species validation using the software DELINEATE. Nine models of species boundaries were set up and tested. The brown bars represent unassigned populations left free to be delimited, while the grey bars represent the predefined species. The resulting solutions suggested by DELINEATE are depicted by red frames around bars. The populations of each species were delimited by BPP software (Supplementary Table S2) and the displayed tree was calculated in starBEAST.
The first model left all populations of A. candidus, A. dobrogensis, A. campestris, A. magnus and Aspergillus sp. DTO 244-F1 unassigned and the other species were defined as follows: all populations of A. subalbidus formed one species (including “pop 6”), A. taichungensis was merged with A. tenebricus. In this setting, unassigned populations were divided into two species: A. magnus and a broad species comprising all other species (A. candidus + A. dobrogensis + A. campestris + Aspergillus sp. DTO 244-F1). The second model differed from the first by predefined species status of A. magnus, and also A. tenebricus was separated from A. taichungensis. In this setting, A. campestris and Aspergillus sp. DTO 244-F1 were recognized as a separate species, while A. candidus and A. dobrogensis remained lumped together. The third model differed from the second by predefined species status for subpopulation “pop 6” of A. subalbidus. When “pop 6” was separated, A. candidus and A. dobrogensis were supported as separate species in contrast to model 2. In the fourth model, populations of A. dobrogensis, Aspergillus sp. DTO 244-F1, A. subalbidus, A. taichungensis and A. tenebricus were left unassigned and other species were determined within their usual boundaries. This model resulted in recognizing A. subalbidus as a single species, A. tenebricus was divided from A. taichungensis, and A. dobrogensis was lumped together with A. candidus.
The remaining models focused on the clade containing A. subalbidus, A. taichungensis and A. tenebricus. In all models, there was a predefined species status for A. dobrogensis and all other species in their usual boundaries. In the fifth model, all populations of A. subalbidus, A. taichungensis and A. tenebricus were left free to be delimited. In this setting A. subalbidus “pop 6” was segregated from A. subalbidus, A. tenebricus was segregated from A. taichungensis, and A. taichungensis was divided into two species. The populations of A. taichungensis and A. tenebricus were the only unassigned populations in the sixth and seventh model. Aspergillus subalbidus was predefined as one (model 6) or two species (model 7) with “pop 6” treated as separate species. In both cases, A. tenebricus was recognized as a separate species, while A. taichungensis was divided into two species in the seventh model. In the last two models, only populations of A. subalbidus were left free to be delimited. In the eighth model, A. subalbidus was recognized as a single species in the situation when A. taichungensis and A. tenebricus were predefined as one species. By contrast, when A. taichungensis and A. tenebricus were predefined as separate species, population “A. subalbidus pop 6” was segregated from A. subalbidus.
Ecology
To analyse distribution and substrate preferences of section Candidi members, we downloaded all sequences of benA, CaM and RPB2 and associated data from GenBank (accessed on 20 February 2022) (Supplementary Table S3) and analysed them together with sequences listed in Table 1. Then we constructed a combined tree (Supplementary Fig. S1), determined probable species limits and visualized ecological data (Fig. 12). Although there are many other records on the occurrence of section Candidi members in literature, we restricted our focus to data with identification verified by DNA sequencing of variable loci.
Fig. 12.
Geographic distribution of Aspergillus section Candidi members based on available sequence data from benA, CaM and RPB2 genes generated in this study (accession numbers are listed in Table 1) and available in the NCBI database (accession numbers in Supplementary Table S3), accessed on February 20, 2022. The numbers in the location pointers correspond to a total number of reliably identified strains reported from a specific country; the numbers following icons of substrates represent a total number of reliably identified strains reported from this substrate.
Eight from nine species (except A. tenebricus) have been found in North America from where we gathered 52 records (Supplementary Table S3), followed by Europe with 51 records representing seven species (except A. magnus and A. tenebricus). Aspergillus neotritici and A. subalbidus were the most geographically widespread species in our dataset occurring on five continents each.
The most common and diverse habitat for section Candidi is the indoor environment, where seven out of nine species were found (55 records), and only A. magnus and A. tenebricus were missing. Four species, A. dobrogensis, A. candidus, A. subalbidus and A. taichungensis have been isolated from caves (28 records). Food-borne species (19 records) comprised A. neotritici, A. candidus, A. subalbidus, A. pragensis and A. taichungensis. Four species are known as coprophilous (14 records), namely, A. campestris, A. candidus, A. dobrogensis and A. subalbidus. Clinical isolates (14 records) mostly belonged to A. neotritici (eight records) and the remaining strains were A. pragensis, A. dobrogensis and A. candidus. Five species (12 records) have been found in soil, i.e., A. candidus, A. campestris, A. subalbidus, A. neotritici, A. taichungensis and A. tenebricus. Strains from outdoor air were poorly represented in our dataset and were restricted to A. neotritici and A. pragensis.
Our set of strains included many from less usual substrates (scored as “other”) like vermicompost, tunnels of bark beetles, metal duct, egg-mass, barn litter and the newly described species, A. magnus, that was isolated from a mouse.
Based on the number of strains included in this study and number of sequences deposited in GenBank, A. candidus and A. subalbidus seem to be the most encountered species. None of the species which were represented by a high number of strains seem to be substrate-specific or geographically restricted. On the other hand, there were apparently preferred substrates/environments of occurrence in some species, for instance dung in A. campestris.
Species identification in practice using molecular and phenotypic characters
All nine species recognized in this study can be identified by using DNA sequencing of any of three phylogenetic markers employed, i.e., benA, CaM and RPB2. The ITS region which was excluded from the phylogenetic analyses due to the low number of variable positions, can only be used for the identification of A. campestris, A. taichungensis and A. neotritici. The ITS rDNA sequences of these species include the following diagnostic positions which are mostly found in the ITS1 region (the numbering is according to the ITS alignment deposited in the DRYAD digital repository: https://doi.org/10.5061/dryad.3j9kd51mq): A. campestris position 95 T/C; A. taichungensis 109 T/C; and A. neotritici 89 A/G, 97 C/T and 498 C/T. Remaining species are indistinguishable from each other by ITS region.
As detailed above, the phenotypic species identification is non-trivial due to the relatively high intraspecific variability in morphological and physiological features detected in this work. Additionally, we do not have information about the degree of variability in species represented by one or few strains. For this reason, any identification key is burdened with a significant inaccuracy. For approximate phenotypic identification in practice, we have created the following synoptic key.
Synoptic key to accepted species in the Aspergillus section Candidi
A. campestris M. Chr. ...................................................................................................................................................................................... 1
A. candidus Link ............................................................................................................................................................................................ 2
A. dobrogensis A. Nováková et al. ................................................................................................................................................................ 3
A. magnus Glässnerová & Hubka ................................................................................................................................................................... 4
A. neotritici Glässnerová & Hubka .................................................................................................................................................................. 5
A. pragensis Hubka et al. ................................................................................................................................................................................ 6
A. subalbidus Visagie, Hirooka & Samson .................................................................................................................................................... 7
A. taichungensis Yaguchi, Someya & Udagawa ........................................................................................................................................... 8
A. tenebricus Houbraken, Glässnerová & Hubka .......................................................................................................................................... 9
I. Macromorphology
Isolates must be grown on the following media at 25 °C for 14 d and 28 d for the presence of soluble pigment.
Colony colour on MEA and CYA
white .............................................................................................................................................................................................. 2, 3, 5, 6, 7, 9
yellowish white/pale yellow/pastel yellow ...................................................................................................................................... 1, 4, 5, 7, 8, 9
yellowish grey ........................................................................................................................................................................................... 1, 4, 7
sulphur yellow ................................................................................................................................................................................................... 1
Growth on MEA
≤25 mm .............................................................................................................................................................................. 1, 2, 3, 4, 5, 6, 7, 8
26–34 mm ................................................................................................................................................................................ 1, 2, 3, 5, 7, 8, 9
≥35 mm ....................................................................................................................................................................................................... 3, 5
Growth on CYA
≤25 mm ....................................................................................................................................................................................................... 1, 2, 4, 5, 6, 7
> 25 mm ....................................................................................................................................................................................................... 1, 2, 3, 5, 7, 8, 9
Growth on CZA
≤15 mm ....................................................................................................................................................................................................... 1, 4, 5, 6, 7, 8
> 15 mm ....................................................................................................................................................................................................... 1, 2, 3, 5, 7, 8, 9
Growth on CY20S
≤30 mm ....................................................................................................................................................................................................... 1, 2, 4, 5, 6, 7
31–45 mm ....................................................................................................................................................................................................... 1, 2, 3, 5, 7, 8, 9
> 45 mm ....................................................................................................................................................................................................... 3, 5, 8, 9
Soluble pigment on CZA
brown, dark brown, black ....................................................................................................................................................................................................... 1, 2, 5, 7, 9
green, dark green, greyish green ....................................................................................................................................................................................................... 1, 5, 6, 7
yellow, yellowish brown ....................................................................................................................................................................................................... 2, 5
no soluble pigment ....................................................................................................................................................................................................... 1, 2, 3, 4, 5, 7, 8
Soluble pigment on CYA
brown, dark brown ....................................................................................................................................................................................................... 1, 2, 4, 6, 7, 9
yellow ....................................................................................................................................................................................................... 8
no soluble pigment ....................................................................................................................................................................................................... 1, 2, 3, 5, 6, 7, 8
II. Physiology
Isolates must be grown on MEA at specified temperatures for 14 d.
Growth at 10 °C
no growth ....................................................................................................................................................................................................... 5
< 8 mm ....................................................................................................................................................................................................... 1, 6, 7, 8
≥8 mm ....................................................................................................................................................................................................... 1, 2, 3, 4, 9
Growth at 30 °C
≤25 mm ....................................................................................................................................................................................................... 1, 2, 3, 4, 6, 7, 8, 9
26–30 mm ....................................................................................................................................................................................................... 7, 8, 9
> 30 mm ....................................................................................................................................................................................................... 5
Growth at 35 °C
no growth ....................................................................................................................................................................................................... 1, 2, 3, 4, 6, 7
growth ....................................................................................................................................................................................................... 1, 5, 7, 8, 9
Growth at 37 °C
no growth ....................................................................................................................................................................................................... 1, 2, 3, 4, 6, 7, 9
growth ....................................................................................................................................................................................................... 5, 7, 8, 9
Growth at 40 °C
no growth ....................................................................................................................................................................................................... 1, 2, 3, 4, 6, 7
growth ....................................................................................................................................................................................................... 5, 8
III. Micromorphology
Isolates must be grown on MEA at 25 °C for 7–21 d.
Length of stipes
< 500 μm ....................................................................................................................................................................................................... 1, 2, 3, 5, 6, 7, 8, 9
500–800 μm ....................................................................................................................................................................................................... 1, 3, 4, 5
> 800 μm ....................................................................................................................................................................................................... 1, 3, 4
Width of stipe
≤6.5 μm ....................................................................................................................................................................................................... 1, 2, 3, 4, 5, 6, 7, 8, 9
7–9.5 μm ....................................................................................................................................................................................................... 1, 2, 3, 4, 5, 7
≥10 μm ....................................................................................................................................................................................................... 7
Vesicle diameter
≤17 μm ....................................................................................................................................................................................................... 1, 2, 3, 5, 6, 7, 8, 9
17–30 μm ....................................................................................................................................................................................................... 1, 2, 3, 5, 6, 7, 9
> 30 μm ....................................................................................................................................................................................................... ………… 4
Length of metulae
≤8 μm ....................................................................................................................................................................................................... …………………………………………………………………………… 1, 2, 3, 5, 6, 7, 8
≥10 μm ....................................................................................................................................................................................................... 1, 3, 4, 5, 7, 9
TAXONOMY
Aspergillus section Candidi W. Gams et al., Adv. Pen. Asp. Syst.: 61. 1986 [1985]. MB832512. Emended description.
Typus: Aspergillus candidus Link, Mag. Ges. Naturf. Freunde Berlin 3: 16. 1809.
Section Candidi encompasses nine species with white- or yellow-coloured colonies. Conidiophores biseriate (exceptionally uniseriate in some A. neotritici strains), stipes hyaline, smooth-walled, rarely finely roughened, non-septate or septate, vesicles usually globose or subglobose, typically reaching diameter of 10–30 μm (Table 5), conidia subglobose to globose, rarely elliptical, smooth-walled or finely roughened (smooth to microtuberculate in SEM). The species are moderately xerophilic and grow more rapidly on media with higher content of osmotically active substances (e.g. CY20S) compared to conventional media such as MEA and CYA. The majority of species produce soluble pigments and/or sclerotia after 4 wk of cultivation (Table 4). Sclerotia are purple, brown to black. Sexual state has not been observed. All species (except for A. neotritici) are able to grow at 10 °C and some species (A. neotritici, A. subalbidus, A. taichungensis and A. tenebricus) are able to grow at 37 °C or even higher temperatures. The members of sect. Candidi are worldwide distributed across all climate zones and are mostly isolated from indoor and cave environments, stored food/feed, soil and clinical samples (Fig. 12).
The majority of species produce the secondary metabolites chloroflavonins, terphenyllins, candidusins and xanthoascins (Rahbæk et al. 2000, Varga et al. 2007, Hubka et al. 2014, Hubka et al. 2018b), in addition to taichunins (Kato et al. 2019) and taichunamides (Kagiyama et al. 2016) which are found in some species.
The members of the phylogenetically related section Petersoniorum (Jurjević et al. 2015) are not xerophilic and their occurrence is restricted to the tropical region. They produce conidia that are grey-green or blue-green on masse and have tuberculate surface. Their vesicles do not exceed 20 μm and sclerotia are yellow to brown. The secondary metabolites of section Petersoniorum are poorly explored but similarly to section Candidi, they produce terphenyl-type of secondary metabolites, arenarins (Oh et al. 1998).
Aspergillus campestris M. Chr., Mycologia 74: 212. 1982. MycoBank MB110495. Fig. 13.
Fig. 13.
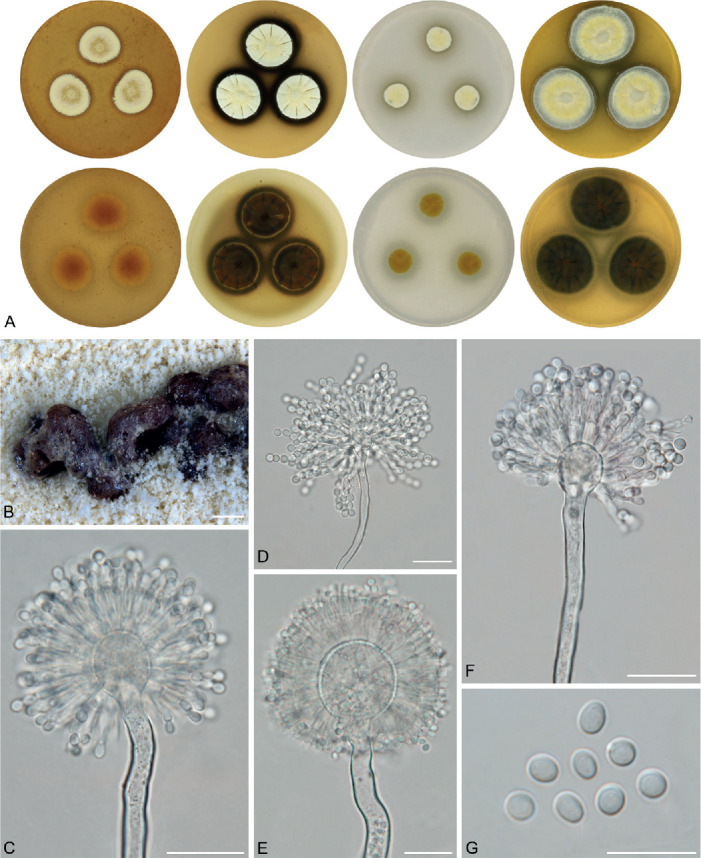
Macromorphology and micromorphology of Aspergillus campestris. A. Colonies after 14 d at 25 °C, left to right: MEA, CYA, CZA, CY20S. B. Sclerotia produced on CYA (photograph taken after 28 d of cultivation). C–F. Conidiophores. G. Conidia. Scale bars: B = 500 μm; C–F = 20 μm, G = 10 μm.
Holotype: NY ST 2–3–1 (catalogue number 00936735). Culture ex-type: CBS 348.81 = NRRL 13001 = IBT 13382 = IBT 28561 = IMI 259009 = ATCC 44563 = IFM 50931 = CCF 5596.
Colony diam, 25 °C, 7 d / 14 d (mm): MEA: 11–17 / 16–28; CYA: 14–20 / 22–36; CZA: 6–14 / 10–20; CY20S: 10–21 / 25–34.
Cardinal temperatures (on MEA, after 2 wk): Aspergillus campestris is able to grow at 10 °C (4–10 mm) and its optimum growth temperature is 25 °C (16–28 mm). Some strains did not grow or only germinate at 30 °C (IBT 17867, IBT 23172), while all other strains grew well at this temperature (12–22 mm). The only strain capable of growing at 35 °C was CBS 348.81T (3–4 mm). No growth at 37 °C.
Culture characteristics (at 25 °C after 2 wk): Colonies on MEA pale yellow (2A3), light yellow (3A5), yellowish white (2A2) or greyish yellow (4B4) with pale yellow margins (2A3), flat, texture granular or floccose, sporulation abundant, margins entire, exudate absent, soluble pigment absent, reverse yellow (3A6), light yellow (4A5) or greyish yellow (4B5). Colonies on CYA yellowish white (3A2), pale yellow (3A3), light yellow (3A5) or yellowish grey (4B2) with pale yellow margins (2A3), flat or centrally raised, usually radially wrinkled, texture granular or floccose, sporulation abundant, margins entire or delicately undulate, reverse yellowish brown (5D5), greyish yellow (4B5), brownish orange (5C5) or dark blond (5D4), exudate absent, soluble pigment dark brown after 4 wk in some strains with varying intensity or location: 4–5 mm large circle around colonies in CBS 348.81T and CCF 5641; 1–3 mm large circle around colonies in FMR 15226 and IMI 344489; pigment in central area between colonies in FMR 15224. Colonies on CZA yellowish white (3A2), yellow (3A6) or light yellow (2A5), flat, texture granular or floccose, sporulation abundant, margins entire or irregular, reverse pale yellow (3A3), light yellow (3A5) or greyish yellow (3B5), exudate absent, soluble pigment present in some strains after 4 wk: dark brown to black in CBS 348.81T and IMI 344489 (3–5 mm large circle around colonies), greenish grey in FMR 15226 (6–7 mm around colonies) and brown in IBT 17867, FMR 15224 and IBT 23172 (2 mm large circle around colonies). Colonies on CY20S yellowish white (3A2), pale yellow (3A3) or pastel yellow (3A4) with bluish grey margins (23D2), flat, centrally raised or umbonate, texture floccose, granular or downy, sporulation abundant, margins entire, less frequently undulate or irregular, reverse olive (2E5), olive yellow (3C8) or yellowish brown (5E4) with greyish yellow margins (3B5, 4B4), exudate absent, soluble pigment present in some strains after 4 wk: dark green to black in CBS 348.81T (3 mm large circle around colonies), dark brown to black in FMR 15224 (5 mm around colonies), and brown in IBT 17867 and CCF 5641 (only central area between colonies). Sclerotia produced superficially after 4 wk of cultivation, in IBT 17867, CCF 5641, FMR 15226 and FMR 15224. Sclerotia absent in CBS 348.81T, IBT 23172 and IMI 344489. Dark grey or black sclerotia were observed on CZA (IBT 17867, CCF 5641) and CYA (FMR 15224), dark brown sclerotia were present on CYA in strain FMR 15226. No sclerotia were observed in any strain on MEA and CY20S.
Micromorphology: Conidial heads radiate, biseriate. Diminutive conidiophores occasionally present. Stipes (excluding diminutive) hyaline, smooth-walled, occasionally finely roughened, undulate in CBS 348.81T, usually non-septate, (90–)250–850(–1 300) × (4–)5–7.5(–9) μm, vesicles globose or subglobose, (10.5–)14–22(–31.5) μm diam, metulae cylindrical or wedge-shaped (V-shaped), (7–)8–14(–17) μm long, covering three quarters to entire surface of vesicle; phialides ampulliform, (4.5–)5–7(–9) μm long. Conidia subglobose to globose, ovate or limoniform, (2.5–)3–4.5(–5) (3.8 ± 0.5 μm) × (2.5–)3–4.5(–5) (3.4 ± 0.4 μm), smooth-walled, rarely finely roughened. In IMI 344489 conidia wider and broader, subglobose or ovate, (3.5–)4–4.5(–5) (4.4 ± 0.3 μm) × (3–)4–4.5(–5) (4.2 ± 0.3 μm).
Diagnosis: Aspergillus campestris is most closely related to A. candidus, A. dobrogensis and A. magnus. Strains of A. campestris are usually easily differentiated from other members of section Candidi by their bright yellow colonies. Conidiophores of A. magnus have longer and wider stipes and broader vesicles compared to A. campestris, while the stipes of A. candidus are shorter. In comparison with A. dobrogensis, A. campestris isolates produce dark soluble pigments and grow more restricted on all cultivation media. The remaining species in sect. Candidi, i.e., A. neotritici, A. pragensis, A. taichungensis, A. tenebricus and A. subalbidus have shorter stipes than A. campestris. These species, except for A. neotritici, have also narrower stipes and smaller vesicles (Table 5). Aspergillus campestris colonies have a smaller diameter than A. taichungensis, A. tenebricus and A. neotritici on all media, but larger than A. pragensis (Table 4). In addition, the majority of strains of A. subalbidus, A. taichungensis, A. tenebricus and A. neotritici grow at 35 °C or higher temperatures (Table 6) in contrast to A. campestris where only one strain germinated at 35 °C (CBS 348.81T).
Ecology (only records verified by DNA sequencing): The species is known on dung from herbivorous animals, indoor air and soil. It has been isolated in South Africa, Spain and the USA (Fig. 12, Table 1).
Aspergillus candidus Link, Mag. Gesell. Naturf. Freunde, Berlin 3(1-2): 16. 1809. MycoBank MB 204868. Fig. 14.
Fig. 14.
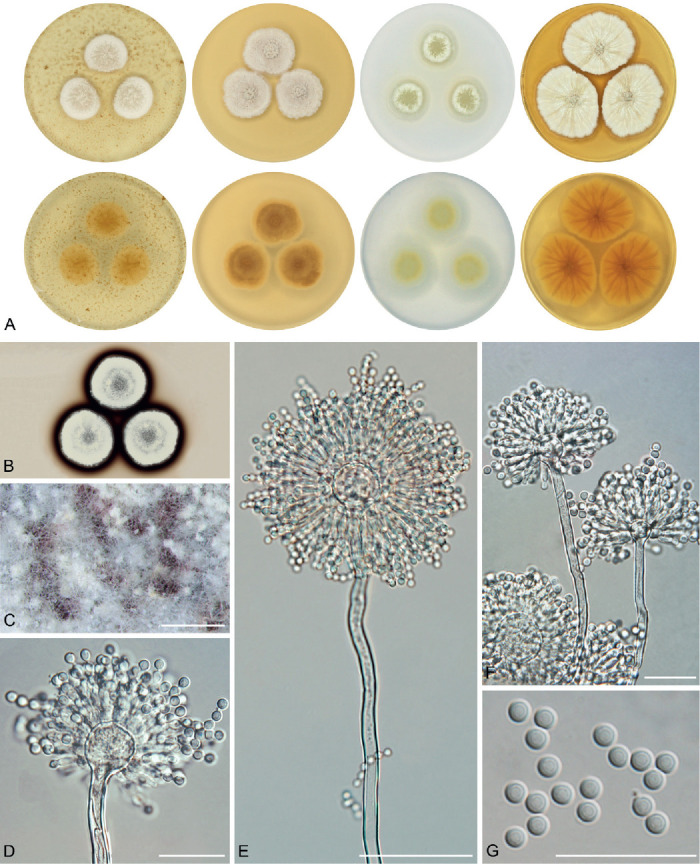
Macromorphology and micromorphology of Aspergillus candidus. A. Colonies after 14 d at 25 °C, left to right: MEA, CYA, CZA, CY20S. B. Soluble pigment on CZA (photograph taken after 28 d of cultivation). C. Sclerotia produced on CZA (photograph taken after 28 d of cultivation). D–F. Conidiophores. G. Conidia. Scale bars: C = 1 mm; D, F, G = 20 μm; E = 50 μm.
Neotype (designated by Samson & Gams 1985): CBS 566.65. Culture ex-type: CBS 566.65 = NRRL 303 = IMI 091889 = ATCC 1002 = IBT 28566 = CCF 5594.
Colony diam, 25 °C, 7 d / 14 d (mm): MEA: 12–17 / 20–27; CYA: 15–21 / 20–32; CZA: 9–15 / 16–25; CY20S: 16–25 / 25–45.
Cardinal temperatures (on MEA, after 2 wk): Aspergillus candidus is able to grow at 10 °C (8−11 mm), the optimum growth temperature is 25 °C (20−27 mm). The maximum growth temperature is 30 °C (18−25 mm). No growth at 35 °C.
Culture characteristics (at 25 °C after 2 wk): Colonies on MEA white (3A1) with a tint of yellowish white (4A2), flat, texture granular to floccose, sporulation abundant, margins entire, exudate absent, reverse light yellow (4A4) or pale yellow (4A3). Colonies on CYA white (3A1) with a tint of yellowish white (4A2) or orange white (5A2), flat or slightly centrally raised, texture granular to floccose, sporulation abundant, margins entire or slightly undulate, exudate absent, reverse light yellow (4A5) or brownish orange (5C3). Colonies on CZA white (3A1) to pale yellow (1A3, 3A3), flat, texture waxy to granular, sporulation weak, margins entire or irregular, exudate absent, reverse pale yellow (3A3) or light yellow (4A4). Colonies on CY20S white (3A1), flat or slightly centrally raised, texture floccose, granular or velvety, radially or irregularly folded, sporulation abundant, margins entire to diffuse or slightly erose, exudate absent, reverse light yellow (4A4, 4A5). Soluble pigment production after 4 wk was examined in nine strains: five of them (CCF 488, CCF 5172, CCF 6200, CCF 3996, NRRL 58959) were able to produce it and four (CBS 566.65T, CCF 4659, CCF 4713, CCF 6195) not. Dark brown to black soluble pigment (3−4 mm around colonies) was observed on CZA in strains CCF 488 and CCF 6200. Brown soluble pigment was observed on CZA in strains CCF 5172 and CCF 3996 (2−4 mm around colonies) and on CYA in strain CCF 3996. Brownish yellow soluble pigment was observed on CZA in strain NRRL 58959. No soluble pigment was observed on MEA and CY20S in any strain. Sclerotia production after 4 wk was examined in nine strains. Five of them (CCF 488, CCF 5172, CCF 6200, CCF 3996, NRRL 58959) were able to produce sclerotia, while no sclerotia were observed in the remaining strains (CBS 566.65T, CCF 4659, CCF 4713, CCF 6195). Brown to black sclerotia covered by mycelium were present on CZA in five mentioned strains. No sclerotia were observed on MEA, CYA and CY20S in any examined strain.
Micromorphology: Conidial heads radiate, biseriate. Diminutive conidiophores occasionally present. Stipes (excluding diminutive) hyaline, smooth-walled, usually non-septate, occasionally with septa, (70–)125–400(–520) × (2.5–)3.5–8.5(–9.5) μm, vesicles predominantly globose, (8–)10–23(–33) μm diam, metulae cylindrical or wedge-shaped (V-shaped), (4.5–)5.5–7.5(–8) μm long, usually covering entire surface of vesicle; phialides ampulliform, (4–)5–6.5(–7) μm long. Conidia globose to subglobose, (3–)3.5–4(–4.5) (3.7 ± 0.2) μm × (3–)3.5–4(–4.5) (3.5 ± 0.2 μm), smooth-walled, rarely finely roughened.
Diagnosis: Aspergillus candidus is sister to A. dobrogensis and closely related to A. campestris and A. magnus. In general, A. candidus has smaller vesicles, shorter and narrower stipes and also shorter phialides and metulae compared to A. dobrogensis (Table 5). Aspergillus dobrogensis colonies also have a larger diameter than A. candidus on MEA, CYA and CY20S (Table 4). However, most of these differences can be interpreted as a trend across the whole species, while some isolates show identical dimensions to A. dobrogensis strains (Fig. 7). Thus, distinguishing between them can be problematic in practice. Aspergillus magnus colonies have a smaller diameter and it has wider and longer stipes, vesicles and metulae than A. candidus. Aspergillus campestris has usually bright yellow colonies and produces stipes, vesicles and metulae which are longer/wider than in A. candidus. Aspergillus pragensis grows more restricted on all media while A. neotritici, A. tenebricus and A. taichungensis colonies have a larger diameter on MEA, CYA and CY20S. The latter three species grow at 35 °C or higher temperatures (Table 6). Aspergillus taichungensis and A. tenebricus produce shorter stipes and A. tenebricus has longer phialides and metulae compared to A. candidus.
Ecology (only records verified by DNA sequencing): The species is known from indoor environments (air, dust), caves, soil, dung, clinical samples, food, metal duct, tunnels of bark beetles, the red flour beetle (Tribolium castaneum) and egg-mass of Arctoscopus japonicus. Aspergillus candidus has been isolated in Canada, Czech Republic, Italy, the Netherlands, Romania, South Korea, Spain and the USA (Fig. 12, Table 1, Supplementary Table S3).
Aspergillus dobrogensis A. Nováková, Jurjević, F. Sklenar, Frisvad, Houbraken & Hubka, Int. J. Syst. Evol. Microbiol. 68: 1004. 2018. MycoBank MB 821313. Fig. 15.
Fig. 15.
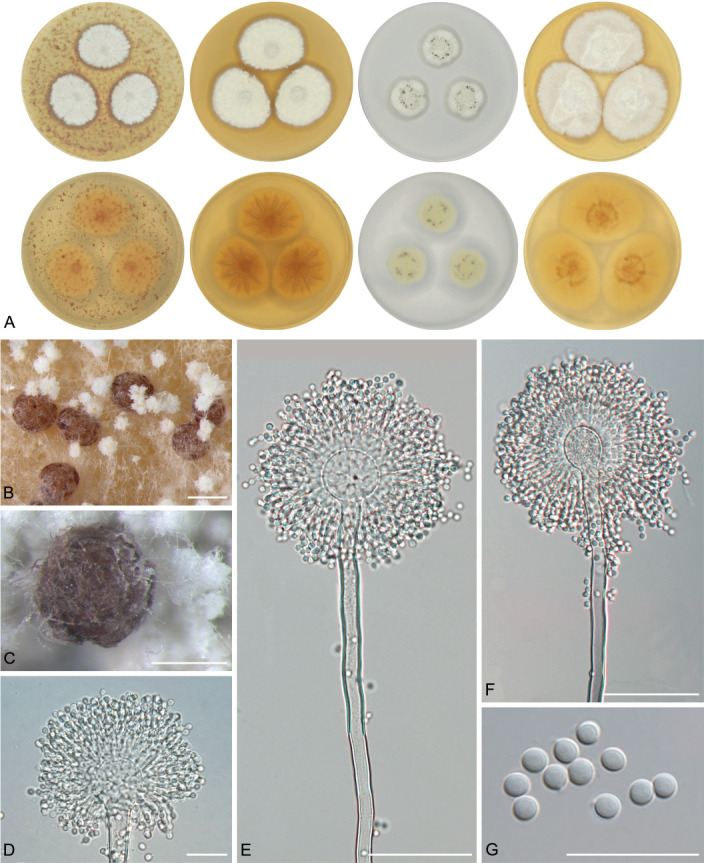
Macromorphology and micromorphology of Aspergillus dobrogensis. A. Colonies after 14 d at 25 °C, left to right: MEA, CYA, CZA, CY20S. B–C. Sclerotia produced on CYA (B) and CZA (C), photographs taken after 28 d of cultivation. D–F. Conidiophores. G. Conidia. Scale bars: B = 750 μm; C = 500 μm; D, G = 20 μm; E, F = 50 μm.
Holotype: PRM 935751. Culture ex-type: CCF 4651 = CCF 4655 = CBS 143370 = NRRL 62821 = IBT 32697.
Colony diameters (at 25 °C, 7 d / 14 d, mm): MEA: 17–20 / 22–34; CYA: 20–24 / 26–39; CZA: 9–15 / 16–25; CY20S: 18–28 / 32–48.
Cardinal temperatures (on MEA, after 2 wk): Aspergillus dobrogensis is able to grow at 10 °C (9–13 mm) and the optimum growth temperature is 25 °C (23–32 mm). The maximum growth temperature is 30 °C (17–24 mm). No growth at 35 °C.
Culture characteristics (at 25 °C after 2 wk): Colonies on MEA white (4A1), sometimes with a tint of yellowish white (4A2), flat or slightly centrally raised, texture floccose, downy or granular, sporulation abundant, margins entire to slightly erose, exudate absent, soluble pigment absent, reverse pale yellow (4A3) or light yellow (4A4). Colonies on CYA white (4A1), sometimes with a tint of yellowish white (4A2), flat, centrally raised or umbonate, texture floccose, downy or granular, sporulation abundant, margins entire, exudate absent, soluble pigment absent, reverse light yellow (4A4, 4A5). Colonies on CZA white (3A1) to yellowish white (3A2, 4A2), flat, texture waxy to granular, sporulation weak and located in a small area in the colony, margins entire or irregular, exudate absent, soluble pigment absent, reverse yellowish white (4A2, 3A2) or pale yellow (3A3, 4A3). Colonies on CY20S white (3A1), flat, centrally raised or umbonate, texture floccose, granular or downy, velvety margins in some strains, sporulation abundant, margins entire, exudate absent, soluble pigment absent, reverse light yellow (4A4, 4A5). Sclerotia production after 4 wk was examined in 10 strains, two of them (CCF 4651T, CCF 5570) were able to produce sclerotia and eight of them (CCF 5567, CCF 5568, CCF 4650, CCF 5844, IBT 29476, CCF 5575, CCF 5569 and CCF 5572) not. They were produced superficially, less commonly covered by felt of mycelium. Strain CCF 4651T was able to produce sclerotia on all four media; dark brown sclerotia on MEA, CZA and CY20S and brown sclerotia on CYA. Dark brown sclerotia covered by felt of mycelium were present on CZA in CCF 5570. No sclerotia were observed on MEA, CYA and CY20S in remaining examined strains.
Micromorphology: Conidial heads radiate, biseriate. Diminutive conidiophores occasionally present. Stipes (excluding diminutive) hyaline, smooth-walled, occasionally finely roughened, usually non-septate, rarely septate, (100–)150–1 150(–2 000) × (3.5–)5–7(–14) μm, vesicles predominantly globose, sometimes subglobose or elongated, (9–)10–30(–39.5) μm diam, metulae cylindrical or wedge-shaped (V-shaped), (5–)5.5–12(–14) μm long, usually covering the entire surface of vesicle; phialides ampulliform, (5–) 5.5–7.5(–8.5) μm long. Conidia globose to subglobose, (3–)3.5–4(–4.5) (3.7 ± 0.3) μm × (2.5–)3–4(–4.5) (3.5 ± 0.3 μm), smooth-walled, rarely finely roughened.
Diagnosis: See section “Diagnosis” of A. candidus for more details. Aspergillus dobrogensis differs from A. candidus, A. pragensis, A. taichungensis, A. tenebricus and A. subalbidus by its longer and broader stipes and larger vesicles (Table 5, Fig. 6). Aspergillus campestris can be differentiated by its yellow colonies. Aspergillus dobrogensis colonies have a larger diameter than A. campestris, A. candidus, A. pragensis, A. magnus and A. subalbidus on MEA, CYA and CY20S, while A. neotritici and A. taichungensis colonies have a larger diameter than A. dobrogensis on these media (Table 4). Aspergillus subalbidus, A. taichungensis, A. tenebricus and A. neotritici can be differentiated by their ability to grow at 35 °C or even higher temperatures (Table 6).
Ecology (only records verified by DNA sequencing): The species is known from caves (bat droppings and guano, sediment, air), indoor environments (air, dust, carpet), mouse dung, herbivore dung and clinical material. It has been isolated in the Czech Republic, Denmark, Germany, the Netherlands, Romania, Spain and the USA (Fig. 12, Table 1).
Aspergillus magnus Glässnerová & Hubka, sp. nov. MycoBank MB 844202. Fig. 16.
Fig. 16.
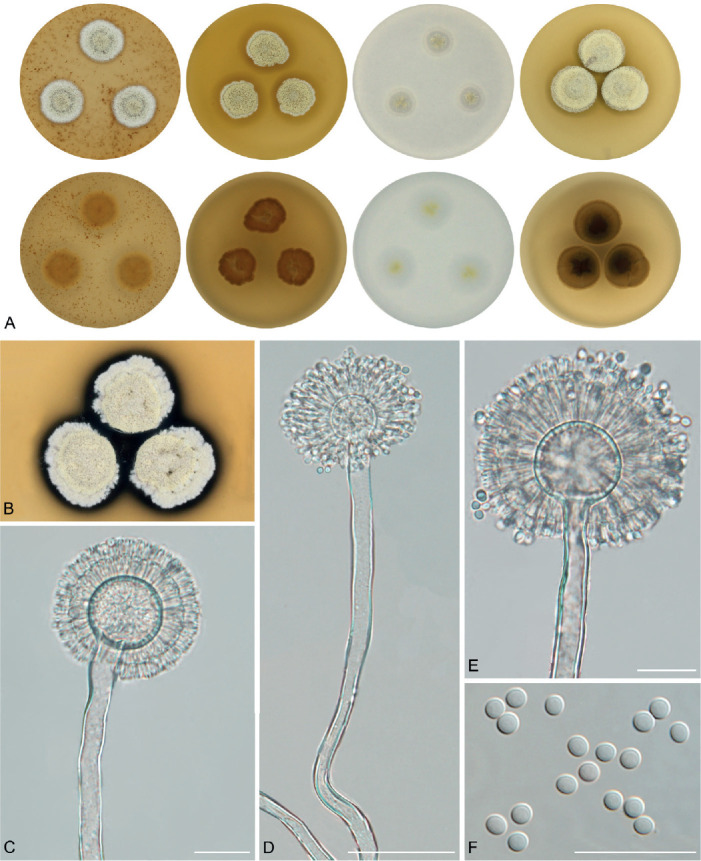
Macromorphology and micromorphology of Aspergillus magnus. A. Colonies after 14 d at 25 °C, left to right: MEA, CYA, CZA, CY20S. B. Soluble pigment produced on CYA (photograph taken after 28 d of cultivation). C–E. Conidiophores. F. Conidia. Scale bars: C, E, F = 20 μm; D = 50 μm.
Etymology: Latin adj. magnus -a -um, referring to its larger conidiophores (stipe length and width, metulae length and vesicle diameter) compared to related species.
Typus: Canada, Alberta, Edmonton, mouse collected on horse farm, 1962, J.W. Carmichael (holotype PRM 956934 (dried culture, metabolically inactive), isotypes PRM 956935–956937 (dried culture, metabolically inactive), culture ex-type UAMH 1324 = IBT 14560).
Colony diam, 25 °C, 7 d / 14 d (mm): MEA: 12–14 / 20–22; CYA: 12–14 / 16–17; CZA: 4–6 / 11–13; CY20S: 13–15 / 19–21.
Cardinal temperatures (on MEA, after 2 wk): Aspergillus magnus is able to grow at 10 °C (9–10 mm) and the optimum growth temperature is 25 °C (20–22 mm). The maximum growth temperature is 30 °C (5–7 mm). No growth at 35 °C.
Culture characteristics (at 25 °C after 2 wk): Colonies on MEA yellowish grey to pale yellow (3C2–3A3) in the centre, yellowish white (3A2) on the margins, flat, texture floccose, sporulation abundant, margins entire, exudate absent, soluble pigment absent, reverse light yellow (4A4). Colonies on CYA pale yellow (3A3) in the centre, yellowish grey (4B2) on the margins, flat, texture floccose, sporulation abundant, margins entire, exudate absent, soluble pigment dark brown (3 mm around colonies after 4 wk), reverse greyish yellow to dark blond (4B4-5D4). Colonies on CZA pale yellow (3A3), flat, texture floccose, sporulation abundant, margins entire, exudate absent, soluble pigment absent, reverse pale yellow (3A3). Colonies on CY20S pale yellow (3A3), centrally raised, texture floccose, sporulation abundant, margins entire, exudate absent, soluble pigment absent, reverse greyish yellow to dark blond (4B4–5D4). Sclerotia production after 4 wk was not observed on any medium.
Micromorphology: Conidial heads radiate, biseriate. Diminutive conidiophores usually present. Stipes (excluding diminutive) hyaline, smooth-walled, occasionally finely roughened, non-septate or occasionally septate, (540–)810–1 150(–1 600) × (6–)9–13(–16) μm, vesicles globose to subglobose, (18–)33–39(–45) μm diam, metulae cylindrical or wedge-shaped (V-shaped), (7.5–)9–14(–17) μm long, covering the entire surface of vesicle; phialides ampulliform, (4.5–)5.5–7(–8) μm long. Conidia subglobose to globose, (3–)3.4–3.6(–4) (3.5 ± 0.2 μm) × (2.5–)3.1–3.3(–3.5) (3.2 ± 0.2 μm), occasionally broadly ellipsoidal, smooth-walled, rarely finely roughened.
Diagnosis: Aspergillus magnus differs from other section Candidi members by its larger vesicles, longer and wider stipes and longer metulae (Table 5, Fig. 6). Some strains of A. dobrogensis reach similar stipe lengths but they differ from A. magnus by other characters, especially its white colony colour. Aspergillus magnus grows more restricted than other species on all media except for A. pragensis (Table 4). Aspergillus neotritici, A. tenebricus, A. taichungensis and A. subalbidus can be differentiated by their ability to grow at 35 °C or even higher temperatures.
Ecology (only records verified by DNA sequencing): The species is known only from a single strain, isolated from a mouse collected on a horse farm in Canada.
Aspergillus neotritici Glässnerová & Hubka, sp. nov. MycoBank MB 844204. Fig. 17.
Fig. 17.
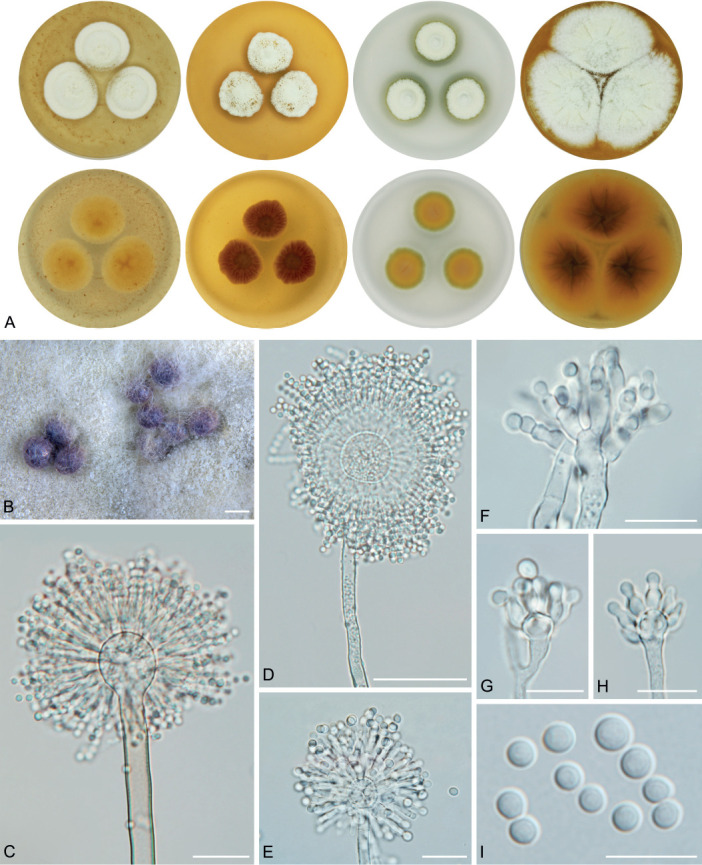
Macromorphology and micromorphology of Aspergillus neotritici. A. Colonies after 14 d at 25 °C, left to right: MEA, CYA, CZA, CY20S. B. Sclerotia produced on CZA (photograph taken after 28 d of cultivation). C–E. Biseriate conidiophores (typical). F. Atypical biseriate conidiophores produced by strain CBS 266.81. G–H. Atypical uniseriate conidiophores produced by strains CCF 4914 and IBT 12659. I. Conidia. Scale bars: B = 500 μm; C, E, F = 20 μm; D = 50 μm; G–I = 10 μm.
= Aspergillus tritici [as triticus] B.S. Mehrotra & M. Basu, Nova Hedwigia 27 (3-4): 599. 1976. MycoBank MB309248. Not validly published [Art. 8.1, Art. 8.4, Art 40.1, Art. 40.4 (Turland et al. 2018)].
Etymology: Latin preposition neo- referring to a new description of previously invalidly described species A. tritici.
Typus: Czech Republic, Prague, toenail of 62-year-old man, 2008, isolated by M. Skořepová (holotype PRM 956940 (dried culture, metabolically inactive), isotypes PRM 956938, PRM 956939 and PRM 956941 (metabolically inactive), culture ex-type CCF 3853 = IBT 32725).
Colony diam, 25 °C, 7 d / 14 d (mm): MEA: 10–25 / 20–47; CYA: 14–28 / 21–50; CZA: 5–19 / 14–39; CY20S: 19–31 / 28-60.
Cardinal temperatures (on MEA, after 2 wk): Aspergillus neotritici does not grow at 10 °C, and its minimum growth temperature is 15 °C (4–13 mm). The optimum growth temperature is 30 °C (32–51 mm) and the maximum growth temperature is 40 °C (4–22 mm). No growth at 45 °C.
Culture characteristics (at 25 °C after 2 wk): Colonies on MEA white (3A1) to yellowish white (2A2, 3A2), flat, texture floccose to granular, less commonly velvety (in CCF 4914 and IBT 12659), sporulation abundant, margins entire, exudate absent, soluble pigment absent, reverse light yellow (4A4), greyish yellow to golden yellow (4B5–5B7). Colonies on CYA white (3A1) to yellowish white (3A2), flat to centrally raised, usually radially wrinkled, texture floccose, velvety or floccose in CCF 4914 and IBT 12659, margins entire or diffuse, sporulation abundant, exudate usually absent, minute yellow droplets present in CCF 6202, soluble pigment absent, reverse greyish yellow (4B5), reddish yellow (4A6) or brownish orange (5B4) in the centre, light yellow (4A4) on margins. Colonies on CZA white (3A1) to yellowish white (3A2), flat to centrally raised, texture granular, floccose or downy, sporulation abundant, margins entire or irregular, exudate usually absent, minute yellow droplets present in CCF 6202, soluble pigment present in some strains after 4 wk, dark brown soluble pigment (1–2 mm around colonies) present in strains CCF 4030 and CCF 6397, dark green soluble pigment present in strain CCF 4653 (2 mm around colonies), yellow soluble pigment present in strains CCF 3314, CCF 3853T and CCF 4914 (1–5 mm around or between colonies), reverse greyish yellow to pale orange (4B5–5A3). Colonies on CY20S white to yellowish white (1A1–2A2) or pale yellow in CCF 4914 and IBT 12659 (2A2–2A3), flat or centrally raised, radially wrinkled in some strains (CBS 266.81, CCF 4914 and IBT 12659), texture floccose, downy to velvety in CCF 4914 and IBT 12659, sporulation abundant, margins entire or diffuse, exudate absent, soluble pigment absent, reverse yellow to reddish yellow (3A6–4A6) or greyish yellow (4C5). Sporulation was abundant in all strains on all tested media except for strains NRRL 4847 (ex-type of A. albus var. thermophilus) and CCF 1649 which were sterile. Sclerotia were present on the colony surface after 4 wk of cultivation in strains CBS 266.81, CCF 3314, CCF 4914 and IBT 12659, absent in CCF 4030, CCF 4653, CCF 3853T, CCF 4658, CCF 6202, CCF 1649 and CCF 6397. Purple sclerotia were present only on CZA in all mentioned strains, no sclerotia were observed on MEA, CYA and CY20S. Purple sclerotia were present in both strains on CZA only.
Micromorphology: Conidial heads radiate, predominantly biseriate, uniseriate in CCF 4914 and IBT 12659 (dimensions of micromorphological features are given separately in the next paragraph). Diminutive conidiophores occasionally present. Stipes (excluding diminutive) hyaline, smooth-walled or finely roughened, usually non-septate, occasionally with septa (extensively septate in CBS 266.81), (140–)200–500(–700) × (3.5–)4–8(–9) μm, vesicles globose, subglobose or elongated, (11–)14–24(–28) μm diam, (5–) 6–8(–11) μm diam in CBS 266.81, metulae cylindrical or wedge-shaped (V-shaped), (4–)7–17(–21) μm long, (3.5–)4–5(–5.5) μm long in CBS 266.81, usually covering the entire surface of vesicle; phialides ampulliform, (2.5–)3.5–6(–8) μm long. Conidia (including CCF 4914 and IBT 12659) globose to subglobose or ellipsoidal, (2.5–)3–4.5(–5) (3.5 ± 0.5) μm × (2.5–)3–4(–4.5) (3.2 ± 0.5) μm, smooth-walled, rarely finely roughened.
Conidiophores in CCF 4914 and IBT 12659 short, uniseriate, stipes (6–)18–45(–85) × (2–)2.5–4(–5) μm, vesicles (3.5–)4–7(–8.5) μm, phialides (3–)3.5–4.5(–6) μm long, conidia had similar dimension to other strain, (3–)3.5–4(–4.5) (3.6 ± 0.3) μm × (3–) 3–3.5(–4) (3.4 ± 0.3) μm.
Diagnosis: Aspergillus neotritici colonies have a larger diameter than A. campestris, A. candidus, A. magnus and A. pragensis on all media at 25 °C (Table 4). It grows better than any other species at 30–40 °C and its optimum growth temperature is 30 °C, while in other species, except for A. subalbidus, it is 25 °C (Table 6). It is also the only species which is not able to grow or at least germinate at 10 °C and can grow at 40 °C. Aspergillus pragensis, A. dobrogensis, A. candidus, A. campestris and A. magnus can be differentiated from A. neotritici by their inability to grow at 37 °C. The stipe length and vesicle diameter of A. neotritici is generally longer when compared to A. subalbidus, A. pragensis, A. tenebricus and A. taichungensis (Fig. 7), noting that this omits strains with atypical or uniseriate conidiophores.
Ecology (only records verified by DNA sequencing): The species is known from clinical samples, food (flour, corn, sunflower seed, cave cheese rind, grains of Triticum sativum), soil, indoor environments, outdoor air, vermicompost, and deep litter for pig raising. It has been reported in Brazil, China, Cuba, Czech Republic, India, Iran, Italy, South Africa, and the USA (Fig. 12, Table 1, Supplementary Table S3).
Aspergillus pragensis Hubka, Frisvad & M. Kolařík, Med. Mycol. 52: 570. 2014. MycoBank MB 800371. Fig. 18.
Fig. 18.
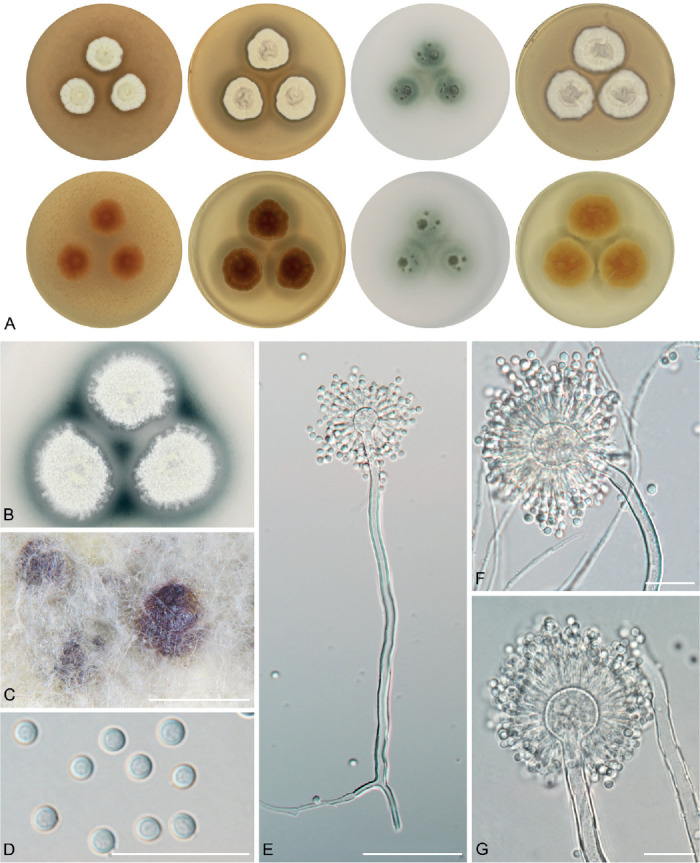
Macromorphology and micromorphology of Aspergillus pragensis. A. Colonies after 14 d at 25 °C, left to right: MEA, CYA, CZA, CY20S. B. Soluble pigment produced on CZA (photograph taken after 28 d of cultivation). C. Sclerotia produced on CYA (photograph taken after 28 d of cultivation). D. Conidia. E–G. Conidiophores. Scale bars: C = 500 μm; D, F, G = 20 μm; E = 50 μm.
Holotype: PRM 922702. Culture ex-type: CCF 3962 = CBS 135591 = NRRL 62491 = IBT 32274.
Colony diam, 25 °C, 7 d / 14 d (mm): MEA: 8–10 / 15–18; CYA: 9–14 / 16–22; CZA: 3–6 / 11–15; CY20S: 13–18 / 24–30.
Cardinal temperatures (on MEA, after 2 wk): Aspergillus pragensis grows restrictedly at 10 °C (5–6 mm, colonies having character of minimally sporulating waxy drops) and the optimum growth temperature is 25 °C (15–18 mm). The maximum growth temperature is 30 °C (3–14 mm). No growth at 35 °C.
Culture characteristics (at 25 °C after 2 wk): Colonies on MEA white (3A1), slightly raised, crateriform in the centre, radially sulcate, texture velvety to downy, sporulation abundant, margins entire to slightly undulate, exudate absent, soluble pigment absent, reverse brownish yellow (5C6) in the centre, orange white (5A2) on the margins. Colonies on CYA white (3A1), centrally raised to umbonate, texture floccose to granular, sporulation abundant, margins entire or slightly undulate, exudate absent, soluble pigment dark brown in CCF 3962T, CCF 4654, CCF 5693, CCF 5847 after 4 wk (4–6 mm wide circle around colonies or in the area between colonies), reverse light brown to brownish orange (5D5–5C4). Colonies on CZA white (3A1) or brownish grey (4D2), flat, texture waxy to floccose, sporulation weak and located only in the colony centre, margins irregular or submerged, exudate absent, soluble pigment intense dark green to greyish blue in all strains after 4 wk (3–6 mm around colonies), reverse yellowish grey (3D2). Colonies on CY20S white (3A1) with a tint of yellowish white (3A2), flat to slightly raised, texture downy, granular or floccose, sporulation abundant, margins entire, exudate absent, soluble pigment absent, reverse light yellow (4A5). Sclerotia were present after 4 wk of cultivation in NRRL 58614 and CCF 5847, absent in CCF 3962T, CCF 4654 and CCF 5693. Dark brown to purple sclerotia covered by felt of mycelium were observed on CYA in NRRL 58614 and CCF 5847, and on MEA in CCF 5847. No sclerotia were observed on CZA and CY20S.
Micromorphology: Conidial heads radiate, biseriate. Diminutive conidiophores occasionally present. Stipes (excluding diminutive) hyaline, smooth-walled, occasionally finely roughened, usually non-septate, occasionally with septa, (40–)110–260(–380) × (3–)3.5–5(–6) μm, vesicles predominantly globose, sometimes subglobose, (7–)10–17(–23) μm diam, metulae cylindrical or wedge-shaped (V-shaped), (4–)5–6.5(–8) μm long, covering three quarters to entire surface of vesicle; phialides ampulliform, (4–)5–6(–6.5) μm long. Conidia globose to subglobose, (3–)3.5–4(–4.5) (3.7.± 0.3 μm) × (3–)3.5–4(–4.5) (3.5 ± 0.2 μm), smooth-walled, rarely finely roughened.
Diagnosis: Aspergillus pragensis is phylogenetically basal to A. subalbidus, A. tenebricus and A. taichungensis. Together with A. magnus, it is unique by its small colony diameters on all media, especially on CZA. Aspergillus pragensis and A. magnus can be easily distinguished by colony colour and strikingly different micromorphology (Fig. 5, Fig. 6). Aspergillus subalbidus, A. taichungensis, A. tenebricus and A. neotritici can be differentiated from A. pragensis by their ability to grow at 35 °C or higher temperatures (Table 6). Aspergillus pragensis differs from A. dobrogensis, A. magnus and A. campestris by its shorter and narrower stipes, shorter metulae and smaller vesicles (Table 5).
Ecology (only records verified by DNA sequencing): The species is known from indoor environments (indoor air, carpet, dust), food (rice bran), clinical samples and outdoor air. It has been isolated in Canada, Czech Republic, South Korea and the USA (Fig. 12, Table 1, Supplementary Table S3).
Aspergillus subalbidus Visagie, Hirooka & Samson, Stud. Mycol. 78: 101. 2014. MycoBank MB 809190. Fig. 19.
Fig. 19.
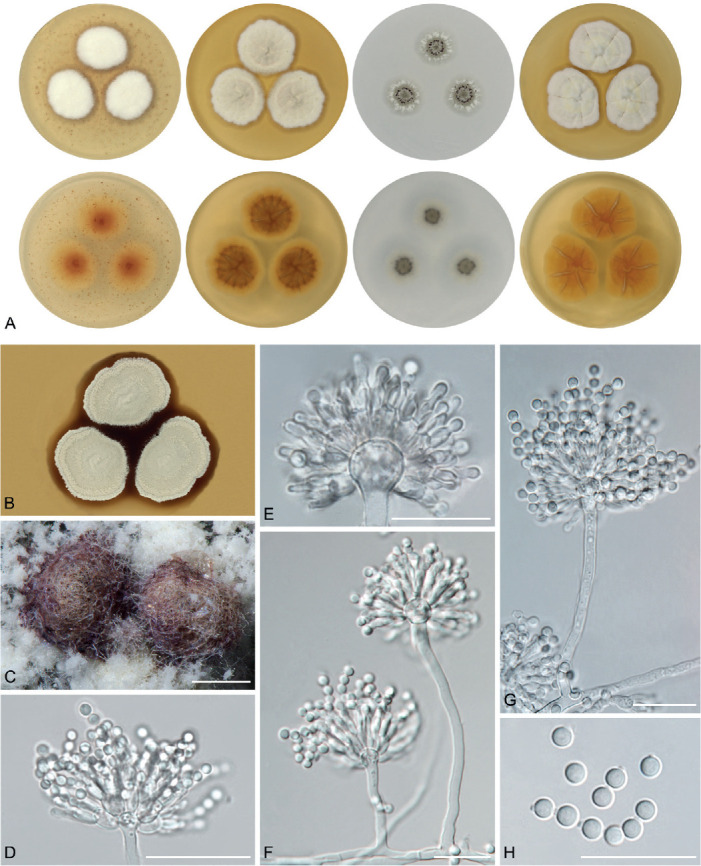
Macromorphology and micromorphology of Aspergillus subalbidus. A. Colonies after 14 d at 25 °C, left to right: MEA, CYA, CZA, CY20S. B. Soluble pigment produced on CYA (photograph taken after 28 d of cultivation). C. Sclerotia produced on CZA (photograph taken after 28 d of cultivation). D–G. Conidiophores. H. Conidia. Scale bars: C = 250 μm; D–H = 20 μm.
Holotype: CBS H-21807. Culture ex-type: CBS 567.65T = NRRL 312 = IMI 230752 = ATCC 16871 = CCF 5822.
Colony diam, 25 °C, 7 d / 14 d (mm): MEA: 11–17 / 18–30; CYA: 14–24 / 24–42; CZA: 8–18 / 16–25; CY20S: 18–27 / 25–45.
Cardinal temperatures (on MEA, after 2 wk): Aspergillus subalbidus germinates or grows restrictedly at 10 °C (3–9 mm). The optimum growth temperature is between 25 °C (18–30 mm) and 30 °C (9–29 mm). Only eight out of 12 tested strains (CBS 567.65T, FMR 15736, NRRL 58123, FMR 15733, CCF 5643, NRRL 4809, CCF 5697, CCF 6197) were able to grow at 35 °C (2–22 mm) and only three out of 12 tested strains (CCF 5643, CCF 5697 and FMR 15733) were able to grow at 37 °C (3–9 mm). No growth at 40 °C.
Culture characteristics (at 25 °C after 2 wk): Colonies on MEA white (4A1) to yellowish white (2A2, 3A2, 4A2), greyish yellow (4B3) in CBS 112449 and orange grey (5B2) in FMR 15877, flat, slightly raised to crateriform, texture floccose to granular, downy or fluffy, sporulation abundant, margins entire, less frequently slightly irregular or erose, exudate absent, soluble pigment absent, reverse greyish yellow (4B5), light yellow (4A5) or reddish orange (7B8) in the centre, pale yellow (4A3) or light orange (5A4) on margins. Colonies on CYA white (4A1), yellowish white (2A2, 3A2, 4A2), yellowish grey (4B2), brownish grey (8C2) or greyish brown (8E3) with white (8A1) margins, flat or centrally raised, texture granular, floccose, velvety or downy, sporulation abundant, margins entire, less frequently delicately undulate or erose, reverse brownish orange (5C4, 5C5), greyish yellow (4B4, 4B5, 4B6) or dark blond (5D4) in the centre, light yellow (4A4, 4A5) or golden brown (5D7) on margins, exudate absent, soluble pigment brown in some strains with varying intensity or location after 4 wk: central area between colonies in CCF 4913, CBS 112449, CCF 5697 and CBS 567.65T; 3–4 mm large circle around colonies in NRRL 58123 and CCF 5698; 5–6 mm large circle around colonies in CCF 5643; slightly pigmented areas around sclerotia in strain CCF 6199; rusty brown soluble pigment was present in strain CCF 6197 (6–7 mm around colonies). Colonies on CZA white (4A1), yellowish white (1A2, 2A2, 3A2), greyish yellow (4B3) or reddish grey (7B2) in strain DTO 196-E4, flat or submerged, floccose, waxy or granular, sporulation usually abundant, weak in CCF 5696, CCF 6199, CCF 5642 and CCF 6197, margins entire, less frequently delicately undulate or erose, reverse pale yellow (3A3, 4A3), greyish brown (5E3, 5F3) or greyish yellow (4B3), exudate absent, soluble pigment present in some strains after 4 wk: dark green to black in FMR 15736, NRRL 58123, CCF 5643 (all over the Petri dish), in the central area between colonies in CBS 567.65T, weak greyish green soluble pigment was present in the central area between colonies and slightly around colonies in NRRL 5214, and dark brown soluble pigment was present in DTO 196-E4 (3 mm around colonies). Colonies on CY20S white (4A1), yellowish white (2A2, 3A2, 4A2) or yellowish grey (4B2), flat or centrally raised, irregularly folded, texture floccose, less frequently granular, downy or fluffy, sporulation abundant, margins entire, less frequently erose or undulate, exudate absent, soluble pigment absent, reverse brownish orange (5C5, 5C6), light yellow (4A5), greyish yellow (4B4, 4B5, 4B6) or reddish yellow (4A6) in the centre, light yellow (4A4, 4A5) on margins. Sclerotia were produced superficially, less commonly covered by felt of mycelium (NRRL 4809 and CCF 6199 on CZA). After 4 wk of cultivation, they were observed in CBS 567.65T, NRRL 4809, FMR 15877, CCF 6197, CCF 6911, NRRL 5214 and CBS 112449, absent in CCF 5642, CCF 5696, DTO 196-E4, FMR 15733, CCF 4913, CCF 5848, CCF 5697, CCF 6205, CCF 5643, CCF 5698, NRRL 58123 and FMR 15736. Dark brown sclerotia were observed on MEA in FMR 15877, CCF 6197 and CCF 6199, on CYA in CBS 567.65T, CCF 6197, CCF 6199, NRRL 5214 and CBS 112449, on CZA in CCF 6197 and on CY20S in CCF 6197. Brown sclerotia on CZA were observed in CBS 112449, on CY20S in FMR 15877, and sclerotia in various shades of brown were observed on CZA in FMR 15877.
Micromorphology: Conidial heads radiate, biseriate. Diminutive conidiophores occasionally present. Stipes (excluding diminutive) hyaline, smooth-walled, occasionally finely roughened, usually non-septate, (20–)50–330(–730) × (2.5–)3–8(–10) μm, vesicles globose or subglobose, (5–)7–23(–31) μm diam, metulae cylindrical or wedge-shaped (V-shaped), (4–)6–11(–16.5) μm long, usually covering the entire surface of vesicle; phialides ampulliform, (3.5–) 4.5–6(–7.5) μm long. Conidia globose to subglobose, (3–)3.5–4(–4.5) (3.7 ± 0.3 μm) × (3–)3.5–4(–4.5) (3.5 ± 0.3 μm), smooth-walled, rarely finely roughened.
Diagnosis: Aspergillus subalbidus is closely related to A. taichungensis and A. tenebricus. Aspergillus taichungensis differs from A. subalbidus by its larger colony diameters on MEA, CYA, CY20S, and by its pastel yellow colonies and yellow soluble pigment produced by some strains on CYA and MEA (Table 4). Aspergillus tenebricus has longer phialides and metulae than A. subalbidus. Aspergillus subalbidus differs from A. dobrogensis, A. magnus, A. neotritici, A. candidus and A. campestris by its shorter and narrower stipes and smaller vesicles (Table 5, Fig. 6). Aspergillus pragensis and A. magnus colonies have a smaller diameter compared to A. subalbidus. Aspergillus taichungensis and A. neotritici can be differentiated by their larger colony diameters at 37 °C and by their ability to grow at 40 °C (Table 6).
Ecology (only records verified by DNA sequencing): The species is known from indoor environments (indoor air, walls, dust, carpet, settle plates), caves, food (Chinese yeast cake, vegetable lard, agriculture products), dung, soil, and from the Dubia roach (Blaptica dubia). It has been isolated in Botswana, Brazil, China, Federated States of Micronesia, Germany, Ghana, South Africa, Spain, Thailand and the USA (Fig. 12, Table 1, Supplementary Table S3).
Aspergillus taichungensis Yaguchi, Someya & Udagawa, Mycoscience 36: 421. 1995. MycoBank MB 434473. Fig. 20.
Fig. 20.
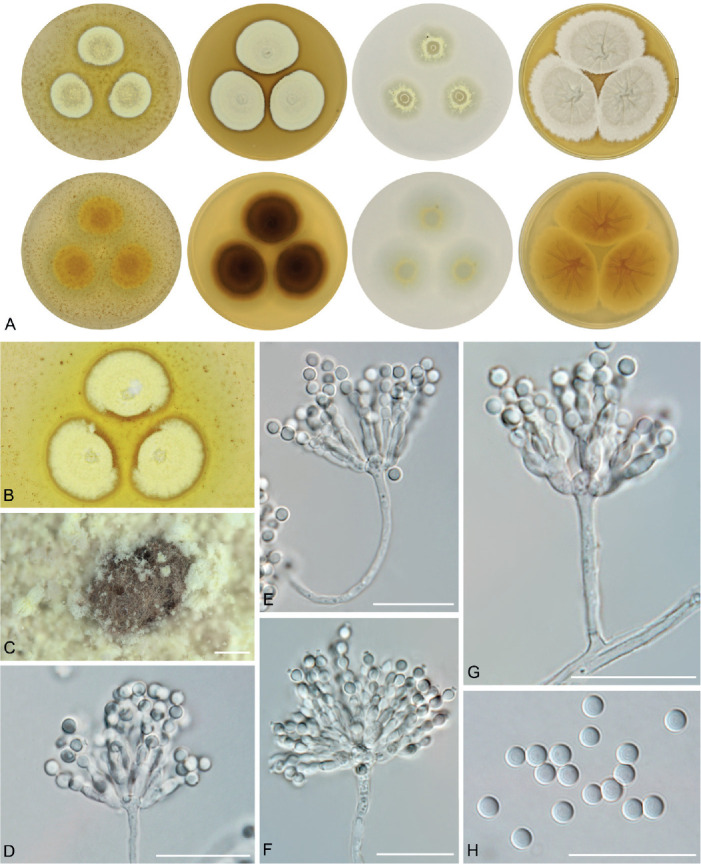
Macromorphology and micromorphology of Aspergillus taichungensis. A. Colonies after 14 d at 25 °C, left to right: MEA, CYA, CZA, CY20S. B. Soluble pigment produced on CZA (photograph taken after 28 d of cultivation). C. Sclerotia produced on CY20S (photograph taken after 28 d of cultivation). D–G. Conidiophores. H. Conidia. Scale bars: C = 1.4 mm; D–H = 20 μm.
Holotype: PF1167. Culture ex-type: IBT 19404 = CCF 5597 = DTO 031-C6.
Colony diam, 25 °C, 7 d / 14 d (mm): MEA: 16–18 / 25–29; CYA: 22–28 / 33–43; CZA: 11–12 / 19–21; CY20S: 28–31 / 36–51.
Cardinal temperatures (on MEA, after 2 wk): Aspergillus taichungensis germinates or grows restrictedly at 10 °C (4–7 mm). The optimum growth temperature is between 20 °C (20–32 mm) and 25 °C (25–29 mm). Aspergillus taichungensis is able to grow at 37 °C (12–20 mm) and the maximum growth temperature is 40 °C (1–7 mm), but the strain IBT 19404T only germinates at this temperature (1–2 mm).
Culture characteristics (at 25 °C after 2 wk): Colonies on MEA pastel yellow (1A4), flat, texture floccose to granular or velvety, sporulation abundant, margins entire, exudate absent, soluble pigment yellow (all over the Petri dish, predominantly around colonies) present in DTO 266-G2 and IBT 19404T, reverse yellow (2A8, 3A6) or light yellow (2A5). Colonies on CYA pastel yellow (1A4) or pale yellow (1A3), flat, texture granular, sporulation abundant, margins entire to slightly undulate, exudate absent, soluble pigment yellow (all over the Petri dish) present in IBT 19404T and DTO 266-G2, reverse yellow (3B8) or deep yellow (4B8) with vivid yellow margins (3A8) or yellowish brown (5D5) with reddish yellow (4A6) margins. Colonies on CZA pastel yellow (1A4), flat, texture granular or waxy, sporulation weak and located in irregular zones, margins irregular, exudate absent, soluble pigment absent, reverse pastel yellow (1A4, 2A4). Colonies on CY20S white (2A1) with a tint of yellowish white (2A2), flat to centrally raised, texture granular, downy or floccose, irregularly folded, sporulation abundant, margins entire to slightly erose, exudate absent, soluble pigment absent, reverse light yellow (4A5) or reddish yellow (4A6). Sclerotia produced superficially, less commonly covered by felt of mycelium (DTO 270-C9 on CY20S). After 4 wk of cultivation, they were observed in strains DTO 270-C9 and IBT 19404T, absent in DTO 266-G2. Brown sclerotia were observed on CZA in strain IBT 19404T, and dark brown to black sclerotia on CY20S in strain DTO 270-C9. No sclerotia were observed on MEA and CYA.
Micromorphology: Conidial heads radiate, biseriate. Diminutive conidiophores present. Stipes (excluding diminutive) hyaline, smooth-walled, occasionally finely roughened, usually non-septate, occasionally with septa, (15–)40–90(–215) × (2–)3–4(–5) μm, vesicles hemispherical, elongated, subglobose or globose, (5–)7–11(–15) μm diam, metulae cylindrical or wedge-shaped (V-shaped), (4–)5.5–7(–9.5) μm long, covering three quarters to entire surface of vesicle; phialides ampulliform, (3.5–)4.5–5.5(–6.5) μm long. Conidia globose to subglobose, (2.5–)3–3.5(–4) (3.4 ± 0.3 μm) × (2.5–)3–3.5(–4) (3.2 ± 0.3 μm), smooth-walled, rarely finely verrucose.
Diagnosis: Aspergillus taichungensis has the shortest and narrowest stipes and smallest vesicles in section Candidi (Table 5, Fig. 6). It is micromorphologically most similar to A. tenebricus and A. subalbidus, but differs from these by its pastel yellow colonies, production of yellow soluble pigment by some strains on CYA and MEA, and by its shorter stipes, metulae and phialides, and smaller vesicles. In addition, colonies of A. taichungensis have a larger diameter on CYA and CY20S compared to A. tenebricus and A. subalbidus (Table 4). Aspergillus candidus, A. dobrogensis, A. magnus and A. pragensis can be differentiated by their inability to grow at 37 °C. Colonies of A. neotritici have a larger diameter than A. taichungensis at 37 °C and 40 °C (Table 6).
Ecology (only records verified by DNA sequencing): The species is known from soil, agriculture products, caves and house dust. It has been isolated in Iran, Mexico and Taiwan (Fig. 12, Table 1, Supplementary Table S3).
Aspergillus tenebricus Houbraken, Glässnerová & Hubka, sp. nov. MycoBank MB 844203. Fig. 21.
Fig. 21.
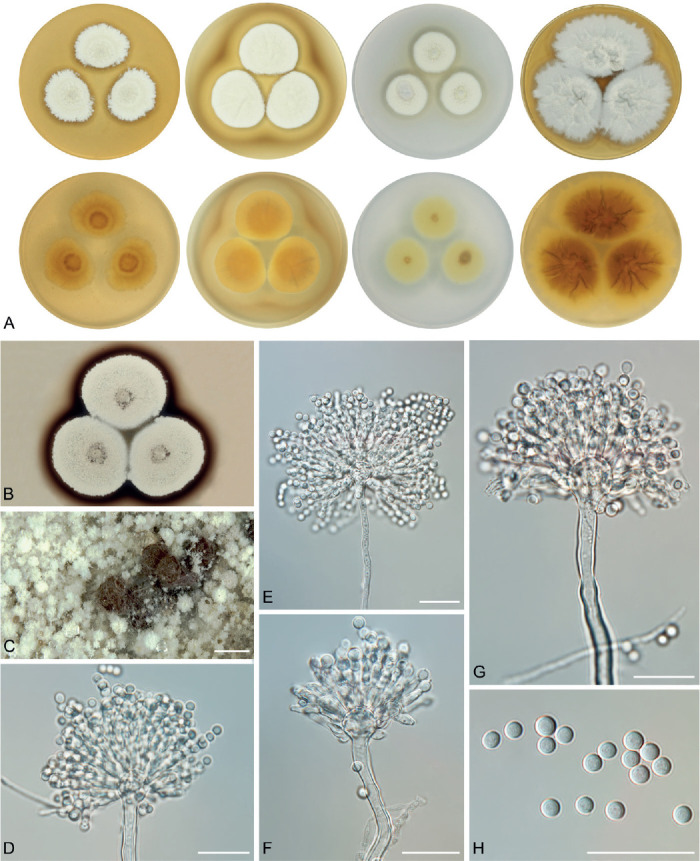
Macromorphology and micromorphology of Aspergillus tenebricus. A. Colonies after 14 d at 25 °C, left to right: MEA, CYA, CZA, CY20S. B. Soluble pigment produced on CZA (photograph taken after 28 d of cultivation). C. Sclerotia produced on CZA (photograph taken after 28 d of cultivation). D–G. Conidiophores. H. Conidia. Scale bars: C = 1 mm; D–H = 20 μm.
Etymology: Latin adj. tenebricus -a -um, referring to dark soluble pigment which is produced on CYA and CZA unlike its close relative A. taichungensis.
Typus: South Africa, Robben Island, soil, 2015, collected by P.W. Crous and isolated by M. Meijer (holotype PRM 957108 (dried culture, metabolically inactive), isotypes PRM 957109–957110, culture ex-type DTO 337-H7 = CBS 147048).
Colony diam, 25 °C, 7 d / 14 d (mm): MEA: 19–21 / 26–33; CYA: 19–23 / 29–34; CZA: 12–14 / 22–24; CY20S: 28–30 / 48–52.
Cardinal temperatures (on MEA, after 2 wk): Aspergillus tenebricus is able to grow at 10 °C (9–11 mm). The optimum growth temperature is 25 °C (26–33 mm). Strains DTO 337-H7T and DTO 440-E2 are able to grow at 37 °C (7–12 mm) in contrast to strain DTO 440-E1. No growth at 40 °C.
Culture characteristics (at 25 °C after 2 wk): Colonies on MEA white (4A1) to yellowish white (4A2, 2A2), flat, texture floccose, sporulation abundant, margins slightly erose, exudate absent, soluble pigment absent, reverse greyish yellow (4B4). Colonies on CYA yellowish white (4A2), flat with slight radial folding, texture granular, sporulation abundant, margins entire, exudate absent, soluble pigment dark brown after 4 wk (3–6 mm large circle around colonies), reverse light yellow (4A5, 4A4) or dark blond (5D4). Colonies on CZA white (2A1) to yellowish white (2A2), flat, texture granular, sporulation abundant, margins entire, exudate absent, soluble pigment dark brown after 4 wk (3–5 mm large circle around colonies), reverse pale yellow (3A3). Colonies on CY20S white (2A1), flat, texture granular to floccose, irregularly folded, sporulation abundant, margins slightly erose or entire, exudate absent, soluble pigment absent, reverse greyish yellow (4C6) with pastel yellow (3A4) margins. Sclerotia were produced superficially and only on CZA after 4 wk of cultivation. Brown sclerotia were observed in strains DTO 440-E1 and DTO 337-H7T, and absent in DTO 440-E2. No sclerotia were observed on MEA, CYA and CY20S in any of the examined isolates.
Micromorphology: Conidial heads radiate, biseriate. Diminutive conidiophores present. Stipes (excluding diminutive) hyaline, smooth-walled, occasionally finely roughened, usually non-septate, occasionally with septa, (30–)80–140(–310) × (2.5–)3.5–5(–6.5) μm, vesicles subglobose to globose, (8.5–)11–18(–27) μm diam, metulae cylindrical or wedge-shaped (V-shaped), (7–)8–12(–14) μm long, covering three quarters to entire surface of vesicle; phialides ampulliform, (5.5–)6–7(–8.5) μm long. Conidia subglobose to globose, (3–)3.5–4(–4.5) (3.8 ± 0.2 μm) × (3–)3.5–4(–4.5) (3.6 ± 0.2 μm), smooth-walled, rarely finely roughened.
Diagnosis: Aspergillus tenebricus is closely related to A. taichungensis and A. subalbidus. It has larger vesicles and longer phialides and metulae than A. taichungensis. Aspergillus tenebricus also produces a dark brown soluble pigment on CYA and CZA after 4 wk compared to A. taichungensis, which can produce a yellow soluble pigment on CYA and MEA. The length of phialides and metulae of A. tenebricus is comparable to A. dobrogensis, A. campestris and A. magnus but longer than other species (Table 5, Fig. 6). The colonies of A. taichungensis have a larger diameter than A. tenebricus on CYA and CY20S (Table 4). Aspergillus candidus, A. dobrogensis, A. magnus and A. pragensis can be differentiated by their inability to grow at 35–37 °C while colonies of A. neotritici and A. taichungensis have a larger diameter than A. tenebricus at 37 °C (Table 6).
Ecology (only records verified by DNA sequencing): The species is known only from soil. It has been isolated in Australia and South Africa (Fig. 12, Table 1).
DISCUSSION
A polyphasic approach (or consilient concept of species) incorporating phylogeny, morphology and/or extrolite data is currently a standard for species delimitations in Aspergillus (Samson et al. 2014). A key part of this approach is represented by phylogenetic methods, which are implemented in practice by different methodologies and approaches. To revise species limits and explore the intraspecific variability, we gathered a hitherto largest collection of section Candidi members (n = 113). To deal with conflicting or variable results from various methods and to maintain some consistency in the decision-making process about species limits, we defined an integrative approach consisting of four main components, three of which should be met to support the species (see Results section; Table 3). Two out of four criteria in this approach were devoted to MSC methods. We considered the results of STACEY to some extent superior to other MSC methods as it is based on several genes and thus it can better accommodate incomplete lineage sorting and similar phenomena. The criterion based on single-locus MSC methods required that the species is supported by the majority of single-locus MSC analyses in its exactly defined form or split into smaller entities, but without any admixture with neighbouring species/populations. Variability of results from single-gene MSC methods is relatively common between loci and settings, especially within genetically highly variable species. Oversplitting and conflicting delimitation results within specific lineages usually indicate ongoing gene flow and recombination within this lineage as previously observed for instance in A. iizukae (Sklenář et al. 2021), A. restrictus, A. penicillioides (Sklenář et al. 2017) and A. udagawae (Hubka et al. 2018a). Thus, splitting of species into multiple smaller lineages by one or several single-gene MSC methods is not a contradiction of a broader species definition. But the delimitation of “species” that involves some strains/populations from neighbouring species delimited by the majority of other methods is a clear conflict.
Among existing species, the least support was observed for A. dobrogensis. Its delimitation was clearly supported by GCPSR and by one scenario in STACEY, but support from other MSC methods was low (Fig. 3). Its delimitation was only supported by some models used in DELINEATE (Fig. 11) and morphological support was also ambiguous. The dimensions of the majority of micromorphological characters were larger in A. dobrogensis than in A. candidus (Fig. 6) and the species also has a slightly larger diameter of colonies at 25 °C than A. candidus (Table 4) in agreement with observations of Hubka et al. (2018b). However, we identified some isolates which were phenotypically indistinguishable from A. candidus (Fig. 7). We can conclude that we observe a clear trend towards the formation of larger colonies and conidiophores in A. dobrogensis compared to A. candidus. Because we did not observe any conflict between single-gene phylogenies, we decided not to synonymize these species. Recently, we made a similar decision regarding A. polyporicola and A. spelaeus in section Flavipedes (Sklenář et al. 2021). These two species were proposed by Hubka et al. (2015) based on phylogeny and the differences in their ecology and production of accessory conidia. After obtaining additional strains, it was shown that mentioned species are not distinguishable as previously thought, but they were retained as cryptic species (morphologically indistinguishable species which form a separate phylogenetic lineages and can be distinguished by molecular and phylogenetic methods) supported by GCPSR, STACEY and some other MSC methods.
Previous studies dealing with species boundaries in section Candidi contained only the ex-type strain of A. campestris (CBS 348.81) in the phylogenetic analyses (Varga et al. 2007, Hubka et al. 2014, Visagie et al. 2014, Hubka et al. 2018b). Among seven strains of this species included in the present study, we identified relatively high intraspecific variability. There were several subclades in the trees without any specific morphological features that would trace the phylogenetic pattern. All strains of A. campestris were also lumped together by STACEY and DELINEATE analyses further supporting the broad concept of this species. It is apparent from the example of deviating the RPB2 sequence of IBT 17867 that the multi-locus MSC method STACEY can better deal with incongruent phylogenies and incomplete lineage sorting. This method correctly lumped this strain with A. campestris and resolved the whole species as monophyletic (Fig. 3) in contrast to polyphyletic resolution in ML and BI trees (Fig. 1).
In the remaining species, the species delimitation results were either very clear and consistent (A. magnus, A. neotritici and A. pragensis) or species limits were identified by a combination of approaches. Incongruences between single-gene phylogenies basically did not give much space for a different solution in A. tenebricus and A. taichungensis than that proposed here (Figs 1–3). The most controversial was the inclusion of the basal clade “pop 6” in A. subalbidus. This clade was delimited as separate species using GCPSR, and one half of MSC methods including one STACEY scenario. Interestingly, in models simulated in DELINEATE, A. dobrogensis has never been delimited from A. candidus when this subpopulation was treated as an integral part of A. subalbidus. If this population was segregated from A. subalbidus, DELINEATE also supported segregation of DTO 266-G2 (A. taichungensis population “pop 2”, Fig. 11) from A. taichungensis. In the case of “pop 6” where delimitation results were very ambiguous, we followed the results of phenotypic studies which did not support delimitation of “pop 6” from A. subalbidus in contrast to a pair A. dobrogensis and A. candidus. We also believe that ambiguous support of “pop 6” may be further compromised in future with the addition of further isolates from other localities. These are the main reasons why we decided to be taxonomically conservative and not describe this potential species.
In general, the studies of intraspecific variability in Aspergillus are relatively rare and limited to a few extensively studied species or species complexes (Peronne et al. 2006, Drott et al. 2021, Lofgren et al. 2021, Bian et al. 2022, Sklenář et al. 2022). In this aspect, the present study is unique in showing both genetic and phenotypic variability in multiple markers and characters across strains of several related species. We showed that dimensions of morphological features, morphology of colonies and cardinal temperatures display significant variability within some species. Gathering of a considerable number of strains is needed to capture the full spectrum of variability and to identify taxonomically informative features. We also observed that atypical strains, significantly deviating from values typical for the whole species, are relatively common. This fact also contributed to the proposal of A. tritici which was described based on a single strain showing numerous atypical characters. Before the molecular era, such strains were frequently a cause of erroneous taxonomic conclusions and descriptions of superfluous species. For instance, we can find many examples where a single atypical isolate was proposed as a new species based on its inability to develop normal ascomata, or production of atypical ascospores, conidia and other characters. Such cases were very common especially in sections Aspergillus (Hubka et al. 2013a, Chen et al. 2017) and Nidulantes (Chen et al. 2016, Hubka et al. 2016) and most of these species were synonymized in the molecular era. Exceptionally, these species names, proposed on the basis of incorrect assumptions about their uniqueness, provide a name for the whole species thanks to the priority rules. In these cases, the type is morphologically dissimilar from other representatives of the same species (e.g. A. proliferans). This would be also the case of A. tritici, but this species name was not validly published. We redescribed this species in the present study as A. neotritici for which we selected a holotype better representing typical species morphology (see section Taxonomy).
We believe that collecting more strains or sequencing more genes in future will help to better understand speciation processes in section Candidi and to resolve persistent ambiguities in the definition of species limits in some species. It is possible that the markers used in this study are suboptimal for section Candidi, and that they do not reflect the predominant phylogenetic signal that would be detected when analysing more markers or the entire genomes. It is believed that the phylogenetic methods spanning the whole genome will allow for a systematic and comparable metric of species differentiation in fungi. Reciprocal monophyly and high level of concordance between thousands of markers on the genome scale should allow differentiation between processes such as recombination, incomplete lineage sorting and speciation in most cases (Kobmoo et al. 2019, Matute & Sepúlveda 2019, Mavengere et al. 2020). The differentiation of these processes is non-trivial in practice, especially if a low number of markers and strains per species are available, as this study has shown.
Acknowledgments
The project was supported by Czech Ministry of Health (grant NU21-05-00681), the Charles University Research Centre program no. 204069 and Czech Academy of Sciences Long-term Research Development Project (RVO: 61388971). Kateřina Glässnerová was supported by the project of Charles University Grant Agency (GAUK140520). We are grateful to Radek Zmítko and Jan Karhan for the help with graphical adjustments of some analysis outputs. We thank Milada Chudíčková and Lenka Zídková for their invaluable assistance in the laboratory. We thank Miroslav Hyliš and Jana Nebesářová for technical assistance with scanning electron microscopy. Vit Hubka is grateful for the support from the Japan Society for the Promotion of Science Postdoctoral Fellowships for Research in Japan (Standard). This study was partially supported by the Grant-in-aid for JSPS research fellow (No. 20F20772). Cobus M Visagie was financially supported by the Future Leaders - African Independent Research fellowship programme (FLAIR, FLR\R1\201831). The FLAIR Fellowship Programme is a partnership between the African Academy of Sciences and the Royal Society funded by the UK Government’s Global Challenges Research Fund. We would like to acknowledge Keith A Seifert, David Nkwe and Stephen W. Peterson who provided some strains used in this study.
DECLARATION ON CONFLICT OF INTEREST
The authors declare that there is no conflict of interest.
Supplementary Material: https://studiesinmycology.org/
Multi-locus phylogenetic tree (benA, CaM and RPB2 loci) of Aspergillus section Candidi comprising isolates from GenBank (Supplementary Table S3) and those included in Table 1. Best scoring Maximum Likelihood tree inferred in the IQ-Tree is shown, bootstrap values are appended to nodes, only support values higher than 70 % are shown. The ex-type strains are designated with a superscripted T and bold print.
Comparison of micromorphological characters between species of Aspergillus section Candidi and statistical significances.
Delimitation of isolates into populations by BPP v. 4.3.
Aspergillus isolates from the section Candidi used in the analysis of ecology together with strains listed in Table 1.
REFERENCES
- Becker A, Sifaoui F, Gagneux M, et al. (2015). Drug interactions between voriconazole, darunavir/ritonavir and tenofovir/emtricitabine in an HIV-infected patient treated for Aspergillus candidus lung abscess. International Journal of STD & AIDS 26: 672–675. [DOI] [PubMed] [Google Scholar]
- Bian C, Kusuya Y, Sklenář F, et al. (2022). Reducing the number of accepted species in Aspergillus series Nigri. Studies in Mycology 102: under review. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouckaert R, Heled J, Kühnert D, et al. (2014). BEAST 2: a software platform for Bayesian evolutionary analysis. PLoS Computational Biology 10: 1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carballo GM, Miranda JA, Arechavala A, et al. (2020). Onicomicosis en paciente inmunocompetente por Aspergillus sección Candidi. Ars Medica 45: 42–46. [Google Scholar]
- Chen A, Frisvad JC, Sun B, et al. (2016). Aspergillus section Nidulantes (formerly Emericella): Polyphasic taxonomy, chemistry and biology. Studies in Mycology 84: 1–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen A, Hubka V, Frisvad JC, et al. (2017). Polyphasic taxonomy of Aspergillus section Aspergillus (formerly Eurotium), and its occurrence in indoor environments and food. Studies in Mycology 88: 37–135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen M. (1982). The Aspergillus ochraceus group: two new species from western soils and a synoptic key. Mycologia 74: 210–225. [Google Scholar]
- Darriba D, Taboada GL, Doallo R, et al. (2012). jModelTest 2: more models, new heuristics and parallel computing. Nature Methods 9: 772–772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dettman JR, Jacobson DJ, Taylor JW. (2003a). A multilocus genealogical approach to phylogenetic species recognition in the model eukaryote Neurospora. Evolution 57: 2703–2720. [DOI] [PubMed] [Google Scholar]
- Dettman JR, Jacobson DJ, Turner E, Pringle A, Taylor JW. (2003b) Reproductive isolation and phylogenetic divergence in Neurospora: comparing methods of species recognition in a model eukaryote. Evolution 57: 2721–2741. [DOI] [PubMed] [Google Scholar]
- Drott MT, Rush TA, Satterlee TR, et al. (2021). Microevolution in the pansecondary metabolome of Aspergillus flavus and its potential macroevolutionary implications for filamentous fungi. Proceedings of the National Academy of Sciences 118: e2021683118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards SV. (2009). Is a new and general theory of molecular systematics emerging? Evolution: International Journal of Organic Evolution 63: 1–19. [DOI] [PubMed] [Google Scholar]
- El-Desoky AH, Inada N, Maeyama Y, et al. (2021). Taichunins E–T, Isopimarane Diterpenes and a 20-nor-Isopimarane, from Aspergillus taichungensis (IBT 19404): Structures and inhibitory effects on RANKL-induced formation of multinuclear osteoclasts. Journal of Natural Products 84: 2475–2485. [DOI] [PubMed] [Google Scholar]
- Elaasser MM, Abdel-Aziz MM, El-Kassas RA. (2011). Antioxidant, antimicrobial, antiviral and antitumor activities of pyranone derivative obtained from Aspergillus candidus. Journal of Microbiology and Biotechnology 1: 5–17. [Google Scholar]
- Farias CM, De Souza OC, Sousa MA, et al. (2015). High-level lipase production by Aspergillus candidus URM 5611 under solid state fermentation (SSF) using waste from Siagrus coronata (Martius) Becari. African Journal of Biotechnology 14: 820–828. [Google Scholar]
- Fujisawa T, Barraclough TG. (2013). Delimiting species using single-locus data and the Generalized Mixed Yule Coalescent approach: a revised method and evaluation on simulated data sets. Systematic Biology 62: 707–724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujisawa T, Aswad A, Barraclough TG. (2016). A rapid and scalable method for multilocus species delimitation using Bayesian model comparison and rooted triplets. Systematic Biology 65: 759–771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garai D, Kumar V. (2013). Response surface optimization for xylanase with high volumetric productivity by indigenous alkali tolerant Aspergillus candidus under submerged cultivation. 3 Biotech 3: 127–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glass NL, Donaldson GC. (1995). Development of primer sets designed for use with the PCR to amplify conserved genes from filamentous ascomycetes. Applied and Environmental Microbiology 61: 1323–1330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grazia L, Romano P, Bagni A, et al. (1986). The role of moulds in the ripening process of salami. Food Microbiology 3: 19–25. [Google Scholar]
- Guevara-Suarez M, García D, Cano-Lira J, et al. (2020). Species diversity in Penicillium and Talaromyces from herbivore dung, and the proposal of two new genera of penicillium-like fungi in Aspergillaceae. Fungal Systematics and Evolution 5: 39–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta A, Gupta G, Jain H, et al. (2016). The prevalence of unsuspected onychomycosis and its causative organisms in a multicentre Canadian sample of 30 000 patients visiting physicians’ offices. Journal of the European Academy of Dermatology and Venereology 30: 1567–1572. [DOI] [PubMed] [Google Scholar]
- Hall T. (1999). BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symposium Series: 95–98.10780396 [Google Scholar]
- Han J, Lu F, Bao L, et al. (2020). Terphenyl derivatives and terpenoids from a wheat-born mold Aspergillus candidus. The Journal of Antibiotics 73: 189–193. [DOI] [PubMed] [Google Scholar]
- Heled J, Drummond AJ. (2009). Bayesian inference of species trees from multilocus data. Molecular Biology and Evolution 27: 570–580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong S-B, Cho H-S, Shin H-D, et al. (2006). Novel Neosartorya species isolated from soil in Korea. International Journal of Systematic and Evolutionary Microbiology 56: 477–486. [DOI] [PubMed] [Google Scholar]
- Houbraken J, Kocsubé S, Visagie C, et al. (2020). Classification of Aspergillus, Penicillium, Talaromyces and related genera (Eurotiales): An overview of families, genera, subgenera, sections, series and species. Studies in Mycology 95: 5–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubka V, Kolařík M. (2012). β-tubulin paralogue tubC is frequently misidentified as the benA gene in Aspergillus section Nigri taxonomy: primer specificity testing and taxonomic consequences. Persoonia 29: 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubka V, Kubatova A, Mallatova N, et al. (2012). Rare and new etiological agents revealed among 178 clinical Aspergillus strains obtained from Czech patients and characterized by molecular sequencing. Medical Mycology 50: 601–610. [DOI] [PubMed] [Google Scholar]
- Hubka V, Kolařík M, Kubátová A, et al. (2013a). Taxonomic revision of Eurotium and transfer of species to Aspergillus. Mycologia 105: 912–937. [DOI] [PubMed] [Google Scholar]
- Hubka V, Peterson SW, Frisvad JC, et al. (2013b). Aspergillus waksmanii sp. nov. and Aspergillus marvanovae sp. nov., two closely related species in section Fumigati. International Journal of Systematic and Evolutionary Microbiology 63: 783–789. [DOI] [PubMed] [Google Scholar]
- Hubka V, Lyskova P, Frisvad JC, et al. (2014). Aspergillus pragensis sp. nov. discovered during molecular reidentification of clinical isolates belonging to Aspergillus section Candidi. Sabouraudia 52: 565–576. [DOI] [PubMed] [Google Scholar]
- Hubka V, Nováková A, Kolařík M, et al. (2015). Revision of Aspergillus section Flavipedes: seven new species and proposal of section Jani sect. nov. Mycologia 107: 169–208. [DOI] [PubMed] [Google Scholar]
- Hubka V, Nováková A, Peterson SW, et al. (2016). A reappraisal of Aspergillus section Nidulantes with descriptions of two new sterigmatocystin-producing species. Plant Systematics and Evolution 302: 1267–1299. [Google Scholar]
- Hubka V, Barrs V, Dudová Z, et al. (2018a). Unravelling species boundaries in the Aspergillus viridinutans complex (section Fumigati): opportunistic human and animal pathogens capable of interspecific hybridization. Persoonia 41: 142–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubka V, Nováková A, Jurjević Ž, et al. (2018b). Polyphasic data support the splitting of Aspergillus candidus into two species; proposal of Aspergillus dobrogensis sp. nov. International Journal of Systematic and Evolutionary Microbiology 68: 995–1011. [DOI] [PubMed] [Google Scholar]
- Jones G, Aydin Z, Oxelman B. (2015). DISSECT: an assignment-free Bayesian discovery method for species delimitation under the multispecies coalescent. Bioinformatics 31: 991–998. [DOI] [PubMed] [Google Scholar]
- Jones G. (2017). Algorithmic improvements to species delimitation and phylogeny estimation under the multispecies coalescent. Journal of Mathematical Biology 74: 447–467. [DOI] [PubMed] [Google Scholar]
- Jurjević Ž, Kubátová A, Kolařík M, et al. (2015). Taxonomy of Aspergillus section Petersonii sect. nov. encompassing indoor and soil-borne species with predominant tropical distribution. Plant Systematics and Evolution 301: 2441–2462. [Google Scholar]
- Kagiyama I, Kato H, Nehira T, et al. (2016). Taichunamides: prenylated indole alkaloids from Aspergillus taichungensis (IBT 19404). Angewandte Chemie International Edition 55: 1128–1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Rozewicki J, Yamada KD. (2019). MAFFT online service: multiple sequence alignment, interactive sequence choice and visualization. Briefings in Bioinformatics 20: 1160–1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaur M, Singla N, Bhalla M, et al. (2021). Aspergillus candidus eumycetoma with review of literature. Journal of Medical Mycology 31: 1–4. [DOI] [PubMed] [Google Scholar]
- Klich MA. (2002). Biogeography of Aspergillus species in soil and litter. Mycologia 94: 21–27. [PubMed] [Google Scholar]
- Kato H, Sebe M, Nagaki M, et al. (2019). Taichunins A–D, norditerpenes from Aspergillus taichungensis (IBT 19404). Journal of Natural Products 82: 1377-1381. [DOI] [PubMed] [Google Scholar]
- Kobayashi A, Takemura A, Koshimizu K, et al. (1982). Candidusin A and B: new p-terphenyls with cytotoxic effects on sea urchin embryos. Agricultural and Biological Chemistry 46: 585–589. [Google Scholar]
- Kobmoo N, Mongkolsamrit S, Arnamnart N, et al. (2019). Population genomics revealed cryptic species within host-specific zombie-ant fungi (Ophiocordyceps unilateralis). Molecular Phylogenetics and Evolution 140: 106580. [DOI] [PubMed] [Google Scholar]
- Kornerup A, Wanscher JH. (1978). Methuen handbook of colour (ten nazev kurzivou). 3rd ed. Eyre Methuen, London. [Google Scholar]
- Lanfear R, Frandsen PB, Wright AM, et al. (2017). PartitionFinder 2: new methods for selecting partitioned models of evolution for molecular and morphological phylogenetic analyses. Molecular Biology and Evolution 34: 772–773. [DOI] [PubMed] [Google Scholar]
- Letunic I, Bork P. (2021). Interactive Tree Of Life (iTOL) v5: an online tool for phylogenetic tree display and annotation. Nucleic Acids Research 49: W293–W296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Jiao F-W, Wang J-Q, et al. (2020). Unguisin G, a new kynurenine-containing cyclic heptapeptide from the sponge-associated fungus Aspergillus candidus NF2412. Tetrahedron Letters 61: 1–5. [Google Scholar]
- Lin Y-K, Xie C-L, Xing C-P, et al. (2021). Cytotoxic p-terphenyls from the deep-sea-derived Aspergillus candidus. Natural Product Research 35: 1627–1631. [DOI] [PubMed] [Google Scholar]
- Liu YJ, Whelen S, Hall BD. (1999). Phylogenetic relationships among ascomycetes: evidence from an RNA polymerse II subunit. Molecular Biology and Evolution 16: 1799–1808. [DOI] [PubMed] [Google Scholar]
- Lofgren LA, Ross BS, Cramer RA, Stajich JE. (2021) Combined pan-, population-, and phylo-genomic analysis of Aspergillus fumigatus reveals population structure and lineage-specific diversity. bioRxiv 2021.12.12.472145; doi: 10.1101/2021.12.12.472145. [DOI] [Google Scholar]
- Malhão F, Ramos AA, Buttachon S, et al. (2019). Cytotoxic and antiproliferative effects of Preussin, a hydroxypyrrolidine derivative from the marine sponge-associated fungus Aspergillus candidus KUFA 0062, in a panel of breast cancer cell lines and using 2D and 3D cultures. Marine Drugs 17: 1–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchelli R, Vining L. (1973). The biosyethetic origin of chlorflavonin, a flavonoid antibiotic from Aspergillus candidus. Canadian Journal of Biochemistry 51: 1624–1629. [DOI] [PubMed] [Google Scholar]
- Masih A, Singh PK, Kathuria S, et al. (2016). Identification by molecular methods and matrix-assisted laser desorption ionization–time of flight mass spectrometry and antifungal susceptibility profiles of clinically significant rare Aspergillus species in a referral chest hospital in Delhi, India. Journal of Clinical Microbiology 54: 2354–2364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matute DR, Sepúlveda VE. (2019). Fungal species boundaries in the genomics era. Fungal Genetics and Biology 131: 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mavengere H, Mattox K, Teixeira MM, et al. (2020). Paracoccidioides genomes reflect high levels of species divergence and little interspecific gene flow. MBio 11: 1–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehrotra B, Basu M. (1976). Some interesting new isolates of Aspergillus from stored wheat and flour. Nova Hedwigia 27: 597–607. [Google Scholar]
- Milala M, Shehu B, Zanna H, et al. (2009). Degradation of agro-waste by cellulase from Aspergillus candidus. Asian Journal of Biotechnology 1: 51–56. [Google Scholar]
- Mirarab S, Bayzid MS, Warnow T. (2016). Evaluating summary methods for multilocus species tree estimation in the presence of incomplete lineage sorting. Systematic Biology 65: 366–380. [DOI] [PubMed] [Google Scholar]
- Moling O, Lass-Floerl C, Verweij P, et al. (2002). Chronic and acute Aspergillus meningitis. Mycoses 45: 504–511. [DOI] [PubMed] [Google Scholar]
- Munden J, Butterworth D, Hanscomb G, et al. (1970). Production of chlorflavonin, an antifungal metabolite of Aspergillus candidus. Applied Microbiology 19: 718–720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen L-T, Schmidt HA, Von Haeseler A, et al. (2015). IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Molecular Biology and Evolution 32: 268–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nouripour-Sisakht S, Mirhendi H, Shidfar M, et al. (2015). Aspergillus species as emerging causative agents of onychomycosis. Journal de Mycologie Medicale 25: 101–107. [DOI] [PubMed] [Google Scholar]
- Nováková A, Hubka V, Valinová Š, et al. (2018). Cultivable microscopic fungi from an underground chemosynthesis-based ecosystem: a preliminary study. Folia Microbiologica 63: 43–55. [DOI] [PubMed] [Google Scholar]
- O’Donnell K. (1993). Fusarium and its near relatives. In: The Fungal Holomorph: Mitotic, Meiotic and Pleomorphic Speciation in Fungal Systematics (Reynolds DR, Taylor JW, eds). CAB International, Wallingford: 225–233. [Google Scholar]
- O’Donnell K, Cigelnik E. (1997). Two divergent intragenomic rDNA ITS2 types within a monophyletic lineage of the fungus Fusarium are nonorthologous. Molecular Phylogenetics and Evolution 7: 103–116. [DOI] [PubMed] [Google Scholar]
- Oh H, Gloer JB, Wicklow DT, Dowd PF. (1998). Arenarins A−C: new cytotoxic fungal metabolites from the sclerotia of Aspergillus arenarius. Journal of Natural Products 61: 702–705. [DOI] [PubMed] [Google Scholar]
- Papavizas G, Christensen C. (1960). Grain storage studies. XXIX. Effect of invasion by individual species and mixtures of species of Aspergillus upon germination and development of discolored germs in wheat. Cereal Chemistry 37: 197–203. [Google Scholar]
- Paradis E, Claude J, Strimmer K. (2004). APE: analyses of phylogenetics and evolution in R language. Bioinformatics 20: 289–290. [DOI] [PubMed] [Google Scholar]
- Paradis E. (2010). pegas: an R package for population genetics with an integrated–modular approach. Bioinformatics 26: 419–420. [DOI] [PubMed] [Google Scholar]
- Perrone G, Susca A, Epifani F, et al. (2006). AFLP characterization of Southern Europe population of Aspergillus section Nigri from grapes. International Journal of Food Microbiology 111: S22–S27. [DOI] [PubMed] [Google Scholar]
- Peterson SW. (2008). Phylogenetic analysis of Aspergillus species using DNA sequences from four loci. Mycologia 100: 205–226. [DOI] [PubMed] [Google Scholar]
- Pitt JI, Hocking AD. (1997). Aspergillus and related teleomorphs. In: Fungi and Food Spoilage (Pitt JI, Hocking AD, eds). Springer, London: 339–416. [Google Scholar]
- R Core Team (2021). R: A language and environment for statistical computing, Vienna, Austria. [Google Scholar]
- Rahbæk L, Frisvad JC, Christophersen C. (2000) An amendment of Aspergillus section Candidi based on chemotaxonomical evidence. Phytochemistry 53: 581–586. [DOI] [PubMed] [Google Scholar]
- Raper KB, Fennell DI. (1965). The genus Aspergillus. Baltimore, Williams & Wilkins. [Google Scholar]
- Reid NM, Carstens BC. (2012). Phylogenetic estimation error can decrease the accuracy of species delimitation: a Bayesian implementation of the general mixed Yule-coalescent model. BMC Evolutionary Biology 12: 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronquist F, Teslenko M, Van Der Mark P, et al. (2012). MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Systematic Biology 61: 539–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samson RA, Gams W. (1985). Typification of the species of Aspergillus and associated teleomorphs. In: Advances in Penicillium and Aspergillus systematics (Samson RA, Pitt JI, eds). Plenum Press, New York: 31–54. [Google Scholar]
- Samson RA, Visagie CM, Houbraken J, et al. (2014). Phylogeny, identification and nomenclature of the genus Aspergillus. Studies in Mycology 78: 141–173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shan T, Wang Y, Wang S, et al. (2020). A new p-terphenyl derivative from the insect-derived fungus Aspergillus candidus Bdf-2 and the synergistic effects of terphenyllin. PeerJ 8: e8221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sklenář F, Jurjević Ž, Zalar P, et al. (2017). Phylogeny of xerophilic aspergilli (subgenus Aspergillus) and taxonomic revision of section Restricti. Studies in Mycology 88: 161–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sklenář F, Jurjević Ž, Houbraken J, et al. (2021). Re-examination of species limits in Aspergillus section Flavipedes using advanced species delimitation methods and description of four new species. Studies in Mycology 99: 1–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sklenář F, Glässnerová K, Jurjević Ž, et al. (2022). Taxonomy of Aspergillus series Versicolores: species reduction and lesson learned about intraspecific variability. Studies in Mycology 102: under revision. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sukumaran J, Holder MT, Knowles LL. (2021). Incorporating the speciation process into species delimitation. PLoS Computational Biology 17: 1–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunesen L, Stahnke L. (2003). Mould starter cultures for dry sausages-selection, application and effects. Meat Science 65: 935–948. [DOI] [PubMed] [Google Scholar]
- Swofford DL. (2003) PAUP* Phylogenetic analysis using parsimony, (*and other methods); version 4.0 b10; Sunderland, Sinauer Associates. [Google Scholar]
- Taylor JW, Jacobson DJ, Kroken S, et al. (2000). Phylogenetic species recognition and species concepts in fungi. Fungal Genetics and Biology 31: 21–32. [DOI] [PubMed] [Google Scholar]
- Thom C, Raper K. (1945). A Manual of the Aspergilli. Baltimore, Williams & Wilkins. [Google Scholar]
- Turland NJ, Wiersema JH, Barrie FR, et al. (2018). International Code of Nomenclature for algae, fungi, and plants (Shenzhen Code) adopted by the Nineteenth International Botanical Congress Shenzhen, China, July 2017. Koeltz Botanical Books. [Google Scholar]
- Van Rossum G, Drake F. (2019). Python language reference, version 3. Python Software Foundation. [Google Scholar]
- Varga J, Frisvad JC, Samson RA. (2007). Polyphasic taxonomy of Aspergillus section Candidi based on molecular, morphological and physiological data. Studies in Mycology 59: 75–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visagie CM, Goodwell M, Nkwe D. (2021). Aspergillus diversity from the Gcwihaba Cave in Botswana and description of one new species. Fungal Systematics and Evolution 8: 81–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visagie CM, Hirooka Y, Tanney JB, et al. (2014). Aspergillus, Penicillium and Talaromyces isolated from house dust samples collected around the world. Studies in Mycology 78: 63–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visagie CM, Yilmaz N, Renaud JB, et al. (2017). A survey of xerophilic Aspergillus from indoor environment, including descriptions of two new section Aspergillus species producing eurotium-like sexual states. MycoKeys 19: 1–30. [Google Scholar]
- Wang D, Qu P, Zhou J, et al. (2020). p-Terphenyl alcohols from a marine sponge-derived fungus, Aspergillus candidus OUCMDZ-1051. Marine Life Science & Technology 2: 262–267. [Google Scholar]
- Wei H, Inada H, Hayashi A, et al. (2007). Prenylterphenyllin and its dehydroxyl analogs, new cytotoxic substances from a marine-derived fungus Aspergillus candidus IF10. The Journal of Antibiotics 60: 586–590. [DOI] [PubMed] [Google Scholar]
- Weidenbörner M, Kunz B. (1994). Contamination of different muesli components by fungi. Mycological Research 98: 583–586. [Google Scholar]
- White TJ, Bruns T, Lee S, et al. (1990). Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. PCR Protocols: a Guide to Methods and Applications 18: 315–322. [Google Scholar]
- Wickham H. (2016). ggplot2: elegant graphics for data analysis. Verlag, New York, Springer. [Google Scholar]
- Yaguchi T, Someya A, Udagawa S-I. (1995). Aspergillus taichungensis, a new species from Taiwan. Mycoscience 36: 421–424. [Google Scholar]
- Yang Z. (2015). The BPP program for species tree estimation and species delimitation. Current Zoology 61: 854–865. [Google Scholar]
- Yen G-C, Chang Y-C, Sheu F, et al. (2001). Isolation and characterization of antioxidant compounds from Aspergillus candidus broth filtrate. Journal of Agricultural and Food Chemistry 49: 1426–1431. [DOI] [PubMed] [Google Scholar]
- Zhang J, Kapli P, Pavlidis P, et al. (2013). A general species delimitation method with applications to phylogenetic placements. Bioinformatics 29: 2869–2876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou G, Sun C, Hou X, et al. (2021). Ascandinines A–D, indole diterpenoids, from the sponge-derived fungus Aspergillus candidus HDN15-152. The Journal of Organic Chemistry 86: 2431–2436. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Multi-locus phylogenetic tree (benA, CaM and RPB2 loci) of Aspergillus section Candidi comprising isolates from GenBank (Supplementary Table S3) and those included in Table 1. Best scoring Maximum Likelihood tree inferred in the IQ-Tree is shown, bootstrap values are appended to nodes, only support values higher than 70 % are shown. The ex-type strains are designated with a superscripted T and bold print.
Comparison of micromorphological characters between species of Aspergillus section Candidi and statistical significances.
Delimitation of isolates into populations by BPP v. 4.3.
Aspergillus isolates from the section Candidi used in the analysis of ecology together with strains listed in Table 1.