Abstract
The Aspergillus series Nigri contains biotechnologically and medically important species. They can produce hazardous mycotoxins, which is relevant due to the frequent occurrence of these species on foodstuffs and in the indoor environment. The taxonomy of the series has undergone numerous rearrangements, and currently, there are 14 species accepted in the series, most of which are considered cryptic. Species-level identifications are, however, problematic or impossible for many isolates even when using DNA sequencing or MALDI-TOF mass spectrometry, indicating a possible problem in the definition of species limits or the presence of undescribed species diversity. To re-examine the species boundaries, we collected DNA sequences from three phylogenetic markers (benA, CaM and RPB2) for 276 strains from series Nigri and generated 18 new whole-genome sequences. With the three-gene dataset, we employed phylogenetic methods based on the multispecies coalescence model, including four single-locus methods (GMYC, bGMYC, PTP and bPTP) and one multilocus method (STACEY). From a total of 15 methods and their various settings, 11 supported the recognition of only three species corresponding to the three main phylogenetic lineages: A. niger, A. tubingensis and A. brasiliensis. Similarly, recognition of these three species was supported by the GCPSR approach (Genealogical Concordance Phylogenetic Species Recognition) and analysis in DELINEATE software. We also showed that the phylogeny based on benA, CaM and RPB2 is suboptimal and displays significant differences from a phylogeny constructed using 5 752 single-copy orthologous proteins; therefore, the results of the delimitation methods may be subject to a higher than usual level of uncertainty. To overcome this, we randomly selected 200 genes from these genomes and performed ten independent STACEY analyses, each with 20 genes. All analyses supported the recognition of only one species in the A. niger and A. brasiliensis lineages, while one to four species were inconsistently delimited in the A. tubingensis lineage. After considering all of these results and their practical implications, we propose that the revised series Nigri includes six species: A. brasiliensis, A. eucalypticola, A. luchuensis (syn. A. piperis), A. niger (syn. A. vinaceus and A. welwitschiae), A. tubingensis (syn. A. chiangmaiensis, A. costaricensis, A. neoniger and A. pseudopiperis) and A. vadensis. We also showed that the intraspecific genetic variability in the redefined A. niger and A. tubingensis does not deviate from that commonly found in other aspergilli. We supplemented the study with a list of accepted species, synonyms and unresolved names, some of which may threaten the stability of the current taxonomy.
Citation: Bian C, Kusuya Y, Sklenář F, D’hooge E, Yaguchi T, Ban S, Visagie CM, Houbraken J, Takahashi H, Hubka V (2022). Reducing the number of accepted species in Aspergillus series Nigri. Studies in Mycology 102: 95–132. doi: 10.3114/sim.2022.102.03
Keywords: Aspergillus luchuensis; Aspergillus niger; Aspergillus tubingensis; clinical fungi; indoor fungi; infraspecific variability; multigene phylogeny, multispecies coalescence model; ochratoxin A; species delimitation
INTRODUCTION
Aspergillus niger and its relatives play important roles in food mycology (Taniwaki et al. 2018), biotechnology (Schuster et al. 2002, Yang et al. 2017), the fermentation industry (Hong et al. 2014) and medical mycology (Howard et al. 2011). Consequently, A. niger is the most frequently cited species name in the genus Aspergillus (Samson et al. 2017). Since, on most occasions, species in section Nigri have been classified as GRAS (generally regarded as safe) by the Food and Drug Administration of the US government, they are suitable organisms for genetic manipulation and thus are used by biotechnology industries to produce hydrolytic enzymes (such as amylases and lipases) or organic acids (such as citric acid and gluconic acid) (Ward 1989, Bennett & Klich 1992, Andersen et al. 2011). On the other hand, Aspergillus section Nigri species (including some A. niger strains) cause food spoilage or contaminate a wide variety of food products (Samson et al. 2019) and may produce mycotoxins such as ochratoxin A, fumonisins and oxalic acid (Frisvad et al. 2011, 2018). Aspergillus niger, A. tubingensis and their lesser known cryptic species (phylogenetically supported but phenotypically nearly identical or undistinguishable) can also act as opportunistic pathogens that cause invasive mycoses in immunocompromised patients and non-invasive mycoses (aspergilloma, allergic aspergillosis, fungal otitis externa and keratomycosis) in otherwise healthy patients (Hubka et al. 2012, D’hooge et al. 2019, Hashimoto et al. 2017, Salah et al. 2019, Vidal-Acuña et al. 2019, Gits-Muselli et al. 2021, Nargesi et al. 2022). Some previous studies also demonstrated that different antifungal susceptibility patterns to azole derivates are present between A. niger and A. tubingensis complexes (Alcazar-Fuoli et al. 2009, Hendrickx et al. 2012, Szigeti et al. 2012).
The infrageneric classification of Aspergillus has a long history with Thom & Church (1926), Thom & Raper (1945) and Raper & Fennell (1965) recognizing that its species could be classified into “groups” based on their morphological similarities. This infrageneric classification was formalised when Gams et al. (1986) introduced names as subgenera and sections in Aspergillus, e.g., all black aspergilli were classified in section Nigri in the subgenus Circumdati. Recently, Houbraken et al. (2020) reviewed this infrageneric classification with the help of multigene phylogenies, and showed that it mostly agreed with the infrageneric classification proposed by Gams et al. (1986). They introduced series rank and subdivided section Nigri into five series. The most economically important section Nigri species, including A. niger, A. tubingensis and A. luchuensis belong to the series Nigri.
The taxonomy of section Nigri at the species level has been turbulent over time. Originally, Mosseray (1934) proposed 35 species. This number was reduced by Raper & Fennell (1965), who recognized 12 species and two varieties, and provided detailed morphological characteristics to distinguish among their accepted species. Al-Musallam (1980) revised the species limits using cluster analysis based on morphological and cultural parameters, suggesting at least seven species, including A. carbonarius, A. ellipticus, A. helicothrix, A. heteromorphus, A. japonicus, A. foetidus and A. niger. In this classification, A. niger was subdivided into six varieties and two forms. Kozakiewicz (1989), who drew her taxonomic conclusions from conidial ornamentation under scanning electron microscopy, distinguished 16 taxa, among which three belong to the current series Nigri, namely, A. acidus, A. niger with six varieties and A. citricus with two varieties. Early molecular genetic studies indicated that A. niger consisted of at least two cryptic species, A. niger and A. tubingensis (Kusters-van Someren et al. 1991, Varga et al. 1994, Peterson 2000), which are morphologically undistinguishable (Pitt & Hocking 2009, Crous et al. 2009). Using a polyphasic approach combining morphological features, extrolite profiling and β-tubulin DNA sequences, Samson et al. (2004) accepted 15 species in the section, comprising eight species in the clade currently known as series Nigri, i.e., A. brasiliensis, A. costaricensis, A. foetidus, A. lacticoffeatus, A. niger, A. piperis, A. tubingensis and A. vadensis. Subsequent phylogenetic studies supported the recognition of several cryptic phylogenetic species in the series Nigri, such as A. awamori (Perrone et al. 2011), later synonymized with another cryptic species, A. welwitschiae (Hong et al. 2013). Similarly, Varga et al. (2011) recognized A. acidus, which was later synonymized with the resurrected species A. luchuensis (Hong et al. 2013). Another two cryptic species related to A. tubingensis, i.e., A. eucalypticola and A. neoniger, were described by Varga et al. (2011), who also synonymized A. foetidus and A. lacticoffeatus with A. niger. As a result, the last overview of accepted Aspergillus species (Houbraken et al. 2020) assigned ten species to the series Nigri: A. brasiliensis, A. costaricensis, A. eucalypticola, A. luchuensis, A. neoniger, A. niger, A. piperis, A. tubingensis, A. vadensis and A. welwitschiae. Four novel cryptic species have been proposed since. Silva et al. (2020) introduced A. vinaceus as a close relative of A. niger and Khuna et al. (2021) introduced A. chiangmaiensis, A. pseudopiperis and A. pseudotubingensis as close relatives of A. tubingensis.
Vesth et al. (2018) and de Vries et al. (2017) de novo sequenced the genomes of the majority of accepted section Nigri species and performed phenotypic and genomic comparative analyses. The whole genomes of the ex-neotype strain of A. niger (CBS 554.65) and 24 A. niger sensu stricto strains were subsequently sequenced, and the mating-type distribution was analysed (Ellena et al. 2021, Seekles et al. 2022). Isolates with both MAT1-2-1 and MAT1-1-1 mating-type gene idiomorphs were found among genome-sequenced strains of A. niger, in agreement with previous PCR-based detection studies (Varga et al. 2014, Mageswari et al. 2016). This fact indicates that the species might have a cryptic sexual cycle, as previously observed in A. tubingenis (Horn et al. 2013). Additionally, recent phylogenomic studies indicated that section Nigri might belong to the subgenus Nidulantes (de Vries et al. 2017, Steenwyk et al. 2019) and not subgenus Circumdati (Jurjević et al. 2015, Kocsubé et al. 2016).
The reliable identification of section Nigri species is of great importance, as evidenced by their diverse positive and negative significances for humans. However, there is an increasing number of isolates that cannot be satisfactorily classified into the currently recognized species despite using multilocus sequence data (Howard et al. 2011, Negri et al. 2014, D’hooge et al. 2019). This fact also resulted in the recent description of several cryptic species related to A. niger and A. tubingensis (Silva et al. 2020, Khuna et al. 2021). The narrow species definition in the series Nigri is also associated with unsatisfactory identification results of MALDI-TOF MS (matrix-assisted laser desorption ionization time-of-flight mass spectrometry), a widely used identification tool in diagnostic laboratories and applied spheres (Gautier et al. 2016, D’hooge et al. 2019, Ban et al. 2021). All of the abovementioned problems may indicate that the species limits are not defined correctly and that the species definitions applied in the series are too narrow. This situation prompted the initiation of this study, where we are concerned with the verification of species boundaries at the molecular level based on a large number of strains.
A common requirement of species delimitation methods is proper sampling (a high number of strains of all studied species ideally isolated from a wide range of substrates and localities) and the presence of intraspecific variability (Carstens et al. 2013). This condition is, however, frequently difficult to fulfil in fungal taxonomy (Ahrens et al. 2016). From this point of view, series Nigri represents a perfect model group for studying species limits on a large scale due to the frequent occurrence of its species in many habitats and a high representation of molecular data in public databases. To do so, we have gathered extensive sequence data and applied a wide range of phylogenetic methods. The synthesis of the resulting data and the consideration of its practical taxonomic implications have led to a significant reduction in the number of species, as detailed below.
METHODS
Molecular studies
The DNA sequences of three loci, β-tubulin (benA), calmodulin (CaM) and the RNA polymerase II second largest subunit (RPB2), were gathered for a total of 276 strains from series Nigri. For this process, we removed short sequences or those that contained obvious errors. Additional species from other series of section Nigri were selected as outgroups or reference species depending on the analysis. The sequences were either downloaded from the GenBank database and mostly originated from previous studies, or they were taken from genes amplified and sequenced in this study. Sequences downloaded from the GenBank database were mostly from studies by D’hooge et al. (2019), Peterson (2008), Jurjević et al. (2012), Fungaro et al. (2017) and Hashimoto et al. (2017). Sequence alignments from the Silva et al. (2020) study were kindly provided by the authors. Information about the provenance of all strains is listed in Table 1.
Table 1.
List of Aspergillus series Nigri strains included in the phylogenetic analyses.
| Species | Strain No.1 | Country | Substrate |
GenBank/ENA/DDBJ accession Nos.2
|
||||||
|---|---|---|---|---|---|---|---|---|---|---|
| benA | CaM | RPB2 | ITS | |||||||
| A. brasiliensis | CBS 101740 = IMI 381727 = ATCC MYA 4553 = IBT 21946T | Brazil | soil | genome2 | genome | EF661063 | MH862749 | |||
|
| ||||||||||
| PPRI 26017 = CMV 007D1 | South Africa | onion | MK451119 | MK451325 | MK450774 | MK450631 | ||||
| PPRI 3388 = CMV 010B8 | South Africa | garlic (Allium sativum) | MK451152 | MK451326 | MK450775 | MK450632 | ||||
| PPRI 3328 = CMV 004I8 | South Africa | mangrove leaves | MK451015 | MK451324 | MK450773 | MK450630 | ||||
| NRRL 26651 | USA | unknown | EF661094 | EF661160 | EF661064 | KC796389 | ||||
| NRRL 26650 | USA | unknown | EF661079 | EF661159 | EF661062 | EF661196 | ||||
| NRRL 35542 | USA | peanut seed | EF661096 | EF661162 | EU021641 | EF661199 | ||||
| NRRL 26652 | USA | unknown | EF661095 | EF661161 | EF661063 | EF661198 | ||||
| CBS 121619 = DTO 24-D5 = ITEM 6139 | Portugal | grapes | AM295185 | AM295176 | OP081976 | AM295181 | ||||
| CBS 121618 = DTO 24-D2 = ITEM 4539 | Portugal | grapes (Tinta barroca) | AM295182 | AM295179 | OP081975 | — | ||||
| NBRC 9455 = ATCC 16404 = CBS 733.88 = DSM 1387 = DSM 1988 = KCTC 6317 = MUCL 29039 = MUCL 30113 = CECT 2574 = IFO 9455 = NCPF 2275 = IHEM 3766 | USA | blueberry (Vaccinium sp.) | genome | genome | genome | — | ||||
| NBRC 105650 = ATCC 9642 = CBS 246.65 = JCM 16265 = NRRL 3536 = ATHUM 2856 = CECT 2700 = DSM 63263 = IFO 6342 = IMI 091855 = MUCL 19001 = IHEM 3797 | Australia | wireless set | genome | genome | genome | — | ||||
| IFM 66950 = CCF 3991 | Spain | cave air | genome | genome | genome | — | ||||
| IFM 66951 = CCF 4962 | Romania | microbial mat in cave | genome | genome | genome | — | ||||
| IHEM 23046 | Belgium | unknown | MH614413 | MH644886 | OP081977 | MH613107 | ||||
| IHEM 23047 | Belgium | unknown | MH614444 | MH644887 | OP081978 | MH613108 | ||||
| IHEM 5185 = NRRL 2276 | Unknown | unknown | MH614565 | MH644892 | OP081982 | MH613109 | ||||
| IHEM 23049 | Belgium | unknown | MH614414 | MH644889 | OP081979 | MH613081 | ||||
| IHEM 3766 | USA | fruit, Vaccinium subg. Cyanococcus | MH614415 | MH644891 | OP081980 | MH613079 | ||||
| IHEM 3797 | Australia | radio set | MH614416 | MH644890 | OP081981 | MH613110 | ||||
| A. niger | CBS 554.65 = ATCC 16888 = IFO 33023 = IHEM 3415 = JCM 10254 = NRRL 326 = CCFC 222006 = IMI 050566 = NBRC 33023T | USA | tannin-gallic acid fermentation | EF661089 | EF661154 | EF661058 | FJ629337 | |||
| CBS 101883 = CECT 20581 = IBT 22031T1 | Indonesia | coffee bean | genome | genome | genome | — | ||||
| PPRI 8682 = CMV 005B2 | South Africa | mopane debris | MK451027 | MK451462 | MK450792 | — | ||||
| PPRI 8734 = CMV 005B7 | South Africa | mopane debris | MK451032 | MK451464 | MK450793 | — | ||||
| PPRI 26011 = CMV 005G9 | South Africa | soil | MK451060 | MK451468 | MK450794 | — | ||||
| PPRI 18719 = CMV 004I3 | South Africa | frass of Busseola fusca feeding inside maize stems | MK451010 | MK451458 | MK450789 | — | ||||
| PPRI 3640 = CMV 004I6 | South Africa | old photo paper | MK451013 | MK451459 | MK450790 | — | ||||
| IFM 63326 | Japan | human sputum | MK854742 | OP081973 | genome | — | ||||
| IFM 63604 | Japan | human sputum | MK854748 | OP081974 | genome | — | ||||
| IFM 59636 | Japan | human abdominal drain | OP081896 | OP081969 | genome | — | ||||
| 104 | Brazil | coffee bean | OP081838 | OP081913 | OP082051 | — | ||||
| 1400 | Brazil | brazil nuts | OP081842 | OP081917 | OP082055 | — | ||||
| 1551 | Brazil | brazil nuts | OP081848 | OP081923 | OP082061 | — | ||||
| 2334 | Brazil | brazil nuts | OP081860 | OP081935 | OP082073 | — | ||||
| 2504 | Brazil | brazil nuts | OP081861 | OP081936 | OP082074 | — | ||||
| 4.17 | Brazil | grapes | OP081863 | OP081938 | OP082076 | — | ||||
| 4.18 | Brazil | grapes | OP081864 | OP081939 | OP082077 | — | ||||
| 643 | Brazil | coffee bean | OP081878 | OP081953 | OP082091 | — | ||||
| 6504 | Brazil | brazil nuts | OP081879 | OP081954 | OP082092 | — | ||||
| 7061 | Brazil | brazil nuts | OP081884 | OP081959 | OP082097 | — | ||||
| 8219 | Brazil | coffee bean | OP081888 | OP081963 | OP082101 | — | ||||
| 902 | Brazil | brazil nuts | OP081890 | OP081965 | OP082103 | — | ||||
| 1.2 | Brazil | grapes | OP081836 | OP081911 | OP082049 | — | ||||
| 1357 | Brazil | brazil nuts | OP081841 | OP081916 | OP082054 | — | ||||
| 23.115 | Brazil | onions | OP081859 | OP081934 | OP082072 | — | ||||
| CBS 117.80 | Unknown | unknown | OP081891 | GU195632 | OP082104 | — | ||||
| IHEM 18069 | French Guiana, France | soil under palm tree | MH614489 | MH645000 | OP082110 | MH613155 | ||||
| IHEM 2312 | Belgium | cosmetics | MH614521 | MH645010 | OP082127 | MH613218 | ||||
| IHEM 17892 | Belgium | chronic sinusitus | MH614520 | MH644877 | OP082108 | KP131598 | ||||
| IHEM 23979 | Belgium | otitis | MH614500 | MH645005 | OP082139 | MH613182 | ||||
| IHEM 25482 | France | human bronchoalveolar lavage | MH614474 | MH644860 | OP082150 | MH613113 | ||||
| IHEM 23907 | Belgium | human ear | MH614492 | MH645003 | OP082136 | MH613168 | ||||
| IHEM 24459 | India | otitis | MH614502 | MH645006 | OP082144 | MH613185 | ||||
| IHEM 5788 | Unknown | Chinese gall | MH614437 | MH645007 | OP082169 | MH613213 | ||||
| IHEM 21604 | Cuba | outdoor air | MH614478 | MH644996 | OP082117 | MH613124 | ||||
| IHEM 5844 | Unknown | unknown | MH614434 | MH645002 | OP082170 | MH613157 | ||||
| IHEM 5296 | Unknown | leather | MH614519 | MH645009 | OP082167 | MH613217 | ||||
| IHEM 6126 | Belgium | human sputum | MH614453 | MH644998 | OP082171 | MH613135 | ||||
| IHEM 5622 | Unknown | unknown | MH614451 | MH645001 | OP082168 | MH613156 | ||||
| IHEM 4023 | Belgium | mycotic otitis externa | MH614496 | MH645004 | OP082162 | KP131606 | ||||
| IHEM 9673 | France | dwelling environment dust | MH614523 | MH645012 | OP082176 | MH613219 | ||||
| IHEM 25481 | France | human bronchoalveolar lavage | MH614473 | MH644859 | OP082149 | MH613112 | ||||
| IHEM 21605 | Cuba | nickel deposit | MH614479 | MH644997 | OP082118 | MH613125 | ||||
| IHEM 6147 | Belgium | human nose | MH614439 | MH645008 | OP082172 | MH613215 | ||||
| IHEM 9709 | France | cement | MH614522 | MH645011 | OP082177 | MH613084 | ||||
| IFM 56816 | Japan | human ear | OP081895 | OP081968 | genome | — | ||||
| 2.6 | Brazil | grapes | OP081856 | OP081931 | OP082069 | — | ||||
| 2.8 | Brazil | grapes | OP081858 | OP081933 | OP082071 | — | ||||
| 102.2706 | Brazil | grapes | OP081837 | OP081912 | OP082050 | — | ||||
| 13.09 | Brazil | coffee bean | OP081840 | OP081915 | OP082053 | — | ||||
| 148.727 | Brazil | onions | OP081846 | OP081921 | OP082059 | — | ||||
| ITAL 47.456 = IBT 35556T2 | Brazil | surface of grape berries | MN583579 | MN583580 | MN583581 | MN575692 | ||||
| ITAL 49.577 | Brazil | grapes | OP081869 | OP081944 | OP082082 | — | ||||
| ITAL 57.1243 | Brazil | grapes | OP081875 | OP081950 | OP082088 | — | ||||
| ITAL 62.1583 | Brazil | grapes | OP081877 | OP081952 | OP082090 | — | ||||
| ITAL 66.1929 | Brazil | grapes | OP081881 | OP081956 | OP082094 | — | ||||
| ITAL 48.545 | Brazil | grapes | OP081867 | OP081942 | OP082080 | — | ||||
| 47.514 | Brazil | grapes | OP081865 | OP081940 | OP082078 | — | ||||
| 48.544 | Brazil | grapes | OP081866 | OP081941 | OP082079 | — | ||||
| 49.580 | Brazil | grapes | OP081870 | OP081945 | OP082083 | — | ||||
| 55.1175 | Brazil | grapes | OP081874 | OP081949 | OP082087 | — | ||||
| 58.1292 | Brazil | grapes | OP081876 | OP081951 | OP082089 | — | ||||
| CBS 139.54T3 | Namibia | Welwitschia mirabilis | MN969369 | genome | MN969100 | MH857271 | ||||
| PPRI 23385 = CMV 004I5 | South Africa | animal feed | MK451012 | MK451548 | MK450816 | MK450663 | ||||
| PPRI 4966 = CMV 004I1 | South Africa | Chrysomelidae beetle | MK451008 | MK451546 | MK450814 | MK450661 | ||||
| PPRI 13255 = CMV 004I2 | South Africa | chicken house bedding, mostly sunflower husks | MK451009 | MK451547 | MK450815 | MK450662 | ||||
| PPRI 26014 = CMV 006G2 | South Africa | soil | MK451097 | MK451562 | MK450819 | MK450669 | ||||
| PPRI 9362 = CMV 005D1 | South Africa | twigs and leaves from Colophospermum mopane | MK451039 | MK451556 | MK450818 | MK450666 | ||||
| IFM 50267 | Japan | human sputum | OP081894 | OP081967 | genome | — | ||||
| IFM 63309 | Japan | human, bronchoalveolar lavage fluid | MK854741 | OP081972 | genome | — | ||||
| IFM 49718 | Japan | human sputum | OP081893 | OP081966 | genome | — | ||||
| IFM 62618 | Japan | human, bronchoalveolar lavage fluid | MK854725 | OP081971 | genome | — | ||||
| IFM 60653 | Japan | human sputum | OP081897 | OP081970 | genome | — | ||||
| 15477 | Brazil | onions | OP081847 | OP081922 | OP082060 | — | ||||
| 15580 | Brazil | onions | OP081849 | OP081924 | OP082062 | — | ||||
| 15646 | Brazil | onions | OP081850 | OP081925 | OP082063 | — | ||||
| 16158 | Brazil | onions | OP081851 | OP081926 | OP082064 | — | ||||
| 16285 | Brazil | onions | OP081852 | OP081927 | OP082065 | — | ||||
| 171804 | Brazil | onions | OP081854 | OP081929 | OP082067 | — | ||||
| 181832 | Brazil | onions | OP081855 | OP081930 | OP082068 | — | ||||
| 2.7 | Brazil | grapes | OP081857 | OP081932 | OP082070 | — | ||||
| 48282 | Brazil | onions | OP081868 | OP081943 | OP082081 | — | ||||
| 53295 | Brazil | onions | OP081871 | OP081946 | OP082084 | — | ||||
| 53a | Brazil | yerba mate | OP081872 | OP081947 | OP082085 | — | ||||
| 54298 | Brazil | onions | OP081873 | OP081948 | OP082086 | — | ||||
| 6573 | Brazil | brazil nuts | OP081880 | OP081955 | OP082093 | — | ||||
| 69363 | Brazil | onions | OP081883 | OP081958 | OP082096 | — | ||||
| 714 | Brazil | brazil nuts | OP081885 | OP081960 | OP082098 | — | ||||
| 77390 | Brazil | onions | OP081887 | OP081962 | OP082100 | — | ||||
| 88430 | Brazil | onions | OP081889 | OP081964 | OP082102 | — | ||||
| 127a | Brazil | yerba mate | OP081839 | OP081914 | OP082052 | — | ||||
| 145723 | Brazil | onions | OP081843 | OP081918 | OP082056 | — | ||||
| 146.724 | Brazil | onions | OP081844 | OP081919 | OP082057 | — | ||||
| 146725 | Brazil | onions | OP081845 | OP081920 | OP082058 | — | ||||
| 361 | Brazil | brazil nuts | OP081862 | OP081937 | OP082075 | — | ||||
| 670 | Brazil | brazil nuts | OP081882 | OP081957 | OP082095 | — | ||||
| 716 | Brazil | brazil nuts | OP081886 | OP081961 | OP082099 | — | ||||
| 17.179 | Brazil | grapes | OP081853 | OP081928 | OP082066 | — | ||||
| DTO 267-I3 | Thailand | house dust | OP081892 | KP330149 | OP082105 | — | ||||
| IHEM 22373 | India | otomycosis | MH614468 | MH644935 | OP082121 | KP131600 | ||||
| IHEM 17902 | Belgium | chronic sinusitus | MH614508 | MH644965 | OP082109 | KP131599 | ||||
| IHEM 25483 | France | human sputum | MH614447 | MH644929 | OP082151 | MH613114 | ||||
| IHEM 3095 | Mauritius | soil | MH614488 | MH644942 | OP082160 | MH613151 | ||||
| IHEM 3090 | Mauritius | soil | MH614487 | MH644884 | OP082159 | MH613150 | ||||
| IHEM 20620 | Cuba | soil | MH614516 | MH644973 | OP082115 | MH613208 | ||||
| IHEM 23644 | India | otitis | MH614435 | MH644948 | OP082129 | MH613171 | ||||
| IHEM 6727 | France | bronchopulmonary cancer | MH614450 | MH644972 | OP082174 | KP131609 | ||||
| IHEM 21970 | Mauritius | soil | MH614484 | MH644938 | OP082120 | MH613111 | ||||
| IHEM 24434 | India | otitis | MH614498 | MH644952 | OP082141 | MH613179 | ||||
| IHEM 18351 | French Guiana, France | bark kapok tree | MH614481 | MH644934 | OP082112 | MH613128 | ||||
| IHEM 23822 | India | otitis | MH614493 | MH644947 | OP082132 | MH613169 | ||||
| IHEM 23651 | India | otitis | MH614491 | MH644946 | OP082131 | MH613167 | ||||
| IHEM 24707 | Belgium | otitis | MH614503 | MH644957 | OP082145 | MH613189 | ||||
| IHEM 25101 | Belgium | Unknown | MH614441 | MH644961 | OP082147 | MH613194 | ||||
| IHEM 21697 | Belgium | onion | MH614448 | MH644939 | OP082119 | MH613137 | ||||
| IHEM 23892 | Belgium | human | MH614570 | MH644944 | OP082134 | MH613161 | ||||
| IHEM 23965 | France | otitis | MH614497 | MH644951 | OP082138 | MH613178 | ||||
| IHEM 15980 | Belgium | ball from Pakistan | MH614518 | MH644976 | OP082107 | MH613210 | ||||
| IHEM 20621 | Cuba | soil | MH614480 | MH644933 | OP082116 | MH613126 | ||||
| IHEM 18080 | French Guiana, France | soil of riverside | MH614513 | MH644969 | OP082111 | MH613077 | ||||
| IHEM 25485 | France | human bronchoaspiration | MH614477 | MH644932 | OP082153 | MH613116 | ||||
| IHEM 5077 | Belgium | human ear | MH614515 | MH644971 | OP082166 | MH613078 | ||||
| IHEM 25340 | Belgium | human toenail | MH614506 | MH644962 | OP082148 | MH613195 | ||||
| IHEM 18676 | Peru | guano | MH614446 | MH644928 | OP082113 | MH613082 | ||||
| IHEM 24328 | Unknown | horse (crust) - clinical sample | MH614499 | MH644953 | OP082140 | MH613180 | ||||
| IHEM 2951 | Belgium | mycotic otitis externa | MH614569 | MH644963 | OP082156 | KP131605 | ||||
| IHEM 2969 | India | soil | MH614449 | MH644941 | OP082157 | MH613149 | ||||
| IHEM 22432 | Somalia | otitis | MH614509 | MH644966 | OP082126 | KP131604 | ||||
| IHEM 26623 | Belgium | human ear | MH614486 | MH644940 | OP082155 | MH613146 | ||||
| IHEM 26214 | Belgium | otitis | MH614505 | MH644960 | OP082154 | MH613192 | ||||
| IHEM 23648 | India | otitis | MH614495 | MH644950 | OP082130 | MH613174 | ||||
| IHEM 24450 | India | otitis | MH614469 | MH644955 | OP082142 | MH613184 | ||||
| IHEM 25484 | France | human bronchoaspiration | MH614476 | MH644931 | OP082152 | MH613115 | ||||
| IHEM 22377 | Belgium | conjunctivitis - human eye | MH614483 | MH644937 | OP082123 | KP131602 | ||||
| IHEM 22378 | Somalia | human auditory canal | MH614510 | MH644967 | OP082124 | MH613076 | ||||
| IHEM 23913 | Belgium | human ear | MH614501 | MH644954 | OP082137 | MH613183 | ||||
| IHEM 4781 | Belgium | mycotic otitis externa | MH614454 | MH644974 | OP082165 | KP131608 | ||||
| IHEM 22374 | India | otomycosis | MH614482 | MH644936 | OP082122 | KP131601 | ||||
| IHEM 23642 | India | otitis | MH614452 | MH644861 | OP082128 | MH613170 | ||||
| IHEM 4063 | Bahamas | sea water | MH614440 | MH644995 | OP082163 | MH613152 | ||||
| IHEM 23895 | Belgium | human ear secretions | MH614584 | MH644945 | OP082135 | MH613163 | ||||
| IHEM 14389 | Belgium | chronic obstructive bronchopneumopathy (bronchoalveolar lavage) | MH614517 | MH644975 | OP082106 | KP131597 | ||||
| IHEM 18678 | Peru | guano | MH614475 | MH644930 | OP082114 | MH613074 | ||||
| IHEM 3710 | Unknown | unknown | MH614490 | MH644943 | OP082161 | MH613158 | ||||
| IHEM 24767 | India | otitis | MH614507 | MH644964 | OP082146 | MH613198 | ||||
| IHEM 9388 | France | air | MH614438 | MH644979 | OP082175 | MH613220 | ||||
| IHEM 3019 | Zaire | soil from palm grove (Elaeis guineensis) | MH614514 | MH644970 | OP082158 | MH613206 | ||||
| IHEM 24454 | India | otitis | MH614470 | MH644956 | OP082143 | MH613186 | ||||
| IHEM 22379 | India | otomycosis | MH614511 | MH644968 | OP082125 | KP131603 | ||||
| IHEM 4461 | Belgium | mycotic otitis externa | MH614585 | MH644978 | OP082164 | KP131607 | ||||
| IHEM 23888 | Belgium | otitis | MH614494 | MH644949 | OP082133 | MH613173 | ||||
| IHEM 6350 | Belgium | dried food for fish | MH614471 | MH644977 | OP082173 | MH613214 | ||||
| A. eucalypticola | CBS 122712 = DTO 53-A2 = IBT 29274T | Australia | leaves of Eucalyptus sp. | genome | genome | genome | OL711732 | |||
| A. luchuensis | CBS 205.80 = IFM 47726 = NBRC 4281 = IFO 428 = KACC 46772 = RIB 2642T | Unknown | awamori-koji | JX500062 | JX500071 | MN969081 | JX500081 | |||
| NRRL 4750 = CBS 128.52 = KACC 47005T4 | USA | unknown | EF661087 | EF661152 | EF661052 | MH856956 | ||||
| RIB 2601 = IFO 4033 = NBRC 111188 = ATCC 38854 = IFM 46994 | Unknown | unknown | genome | genome | genome | LC573600 | ||||
| IFO 4308 = NBRC 4308 | Unknown | unknown | genome | genome | genome | AB573884 | ||||
| IHEM 26285 | France | human broncho-aspiration | MH614561 | MH644869 | OP082038 | MH613141 | ||||
| IHEM 25486 | France | human trimming liquid | MH614563 | MH644862 | OP082030 | MH613118 | ||||
| CBS 112811 = CECT 20582 = IBT 26239T5 | Denmark | black pepper | genome | genome | genome | OL711715 | ||||
| IHEM 23904 | India | otitis | MH614551 | MH644990 | OP082022 | MH613164 | ||||
| PPRI 13091 = CMV 010I3 | South Africa | soil | MK451175 | MK451492 | MK450797 | — | ||||
| IHEM 5316 | Belgium | human ear | MH614562 | MH644893 | OP082046 | MH613216 | ||||
| PPRI 9197 = CMV 005C9 | South Africa | soil | MK451038 | MK451491 | MK450796 | MK450641 | ||||
| PPRI 13230 = CMV 011A9 | South Africa | maize roots | MK451187 | MK451493 | MK450798 | — | ||||
| PPRI 8983 = CMV 005C2 | South Africa | twigs and leaves from Colophospermum mopane | MK451035 | MK451490 | MK450795 | MK450640 | ||||
| A. pseudotubingensis | SDBR CMUO2T | Thailand | rhizosphere soil | MK457206 | MK457205 | MK457208 | — | |||
| SDBR CMUO8 | Thailand | rhizosphere soil | MW602908 | MW602907 | MW602909 | — | ||||
| SDBR CMU20 | Thailand | rhizosphere soil | MW602913 | MW602912 | MW602914 | — | ||||
| A. tubingensis | CBS 133056 = DTO 213-F6 = NRRL 4875T | Unknown | unknown | EF661086 | EF661151 | EF661055 | EF661193 | |||
| PPRI 8683 = CMV 005B3 | South Africa | soil | MK451028 | MK451452 | MK450786 | — | ||||
| PPRI 8733 = CMV 005B6 | South Africa | plant debris from Colophospermum mopane | MK451031 | MK451453 | MK450787 | — | ||||
| PPRI 21344 = CMV 004I4 | South Africa | seed of soybean (Glycine max) | MK451011 | MK451448 | MK450782 | — | ||||
| IFM 57258 | Nagasaki, Japan | soil | OP081902 | OP081909 | genome | — | ||||
| IHEM 18329 | French Guiana, France | rotten mango | MH614467 | MH644992 | OP082002 | MH613202 | ||||
| IHEM 18205 | French Guiana, France | soil | MH614419 | MH644865 | OP082001 | MH613127 | ||||
| IHEM 21599 | Cuba | rail feather | MH614458 | MH644985 | OP082008 | MH613073 | ||||
| IHEM 18097 | French Guiana, France | rotten mango | MH614417 | MH644981 | OP081998 | MH613119 | ||||
| IHEM 21607 | Cuba | indoor environment | MH614464 | MH644988 | OP082012 | MH613133 | ||||
| IHEM 18042 | French Guiana, France | soil under mango tree | MH614462 | MH644980 | OP081997 | MH613117 | ||||
| IHEM 21597 | Cuba | indoor air | MH614457 | MH644984 | OP082007 | MH613122 | ||||
| IHEM 2463 | Belgium | nutmeg (Myristica fragrans) | MH614465 | MH644989 | OP082028 | MH613147 | ||||
| IHEM 21593 | Cuba | cotton cloth | MH614460 | MH644987 | OP082006 | MH613131 | ||||
| IHEM 18136 | French Guiana, France | soil under palm tree | MH614456 | MH644983 | OP082000 | MH613121 | ||||
| IHEM 21592 | Cuba | human ear | MH614420 | MH644866 | OP082005 | MH613130 | ||||
| IHEM 21606 | Cuba | indoor environment | MH614466 | MH644991 | OP082011 | MH613199 | ||||
| IHEM 21590 | Cuba | indoor air | MH614418 | MH644986 | OP082004 | MH613129 | ||||
| IHEM 22375 | India | otomycosis | MH614461 | MH644867 | OP082015 | KP131633 | ||||
| IHEM 22376 | India | otomycosis | MH614463 | MH644868 | OP082016 | KP131634 | ||||
| IHEM 18106 | French Guiana, France | leaf of mango tree | MH614455 | MH644982 | OP081999 | MH613083 | ||||
| IHEM 21602 | Argentina | clinical material | MH614459 | MH644864 | OP082010 | MH613123 | ||||
| PPRI 21345 = CMV 005A8 | South Africa | seed of soybean (Glycine max) | MK451024 | MK451450 | MK450784 | — | ||||
| PPRI 21347 = CMV 005A9 | South Africa | seed of soybean (Glycine max) | MK451025 | MK451451 | MK450785 | — | ||||
| IHEM 21971 | Mauritius | soil | MH614546 | MH644993 | OP082013 | MH613134 | ||||
| IHEM 25327 | Belgium | laboratory contaminant | MH614421 | MH644994 | OP082029 | MH613139 | ||||
| IHEM 21601 | Argentina | clinical material | MH614564 | MH644917 | OP082009 | — | ||||
| PPRI 7393 = CMV 005A5 | South Africa | unknown | MK451021 | MK451542 | MK450813 | MK450660 | ||||
| NRRL 4851 = ATCC 16879 = CBS 115.48 = CBS 558.65 | Wisconsin, USA | unknown | EF661085 | EF661150 | EF661054 | — | ||||
| CBS 134.48 = ITEM 7040 | Unknown | unknown | AY820007 | AJ964876 | EF661055 | FJ629354 | ||||
| IHEM 5802 | Unknown | unknown | MH614558 | MH644919 | OP082048 | MH613204 | ||||
| IHEM 22772 | Belgium | human ear | MH614545 | MH644871 | OP082018 | MH613132 | ||||
| PPRI 25991 = CMV 001B7 | South Africa | walnut kernels (Juglans regia) | MK450891 | MK451540 | MK450811 | MK450658 | ||||
| PW3161 | Hong Kong | human nail | LC000547 | LC000560 | LC000573 | AB987902 | ||||
| PPRI 6720 = CMV 005A3 | South Africa | beetle (Aspidimorpha areata) | MK451019 | MK451541 | MK450812 | MK450659 | ||||
| IFM 54640 | Japan | human sputum | OP081898 | OP081905 | genome | — | ||||
| IFM 55763 | Japan | vineyard | OP081899 | OP081906 | genome | — | ||||
| IFM 57143 | Japan | human | OP081901 | OP081908 | genome | — | ||||
| IFM 56815 | Japan | human ear | OP081900 | OP081907 | genome | — | ||||
| IFM 61612 | Japan | human, bronchoalveolar lavage fluid | OP081903 | OP081910 | genome | — | ||||
| IHEM 22370 | Belgium | otitis | MH614525 | MH644894 | OP082014 | KP131632 | ||||
| IHEM 1941 | Belgium | dust from mattress | MH614550 | MH644900 | OP082003 | MH613159 | ||||
| IHEM 17170 | Belgium | chronic sinusitus | MH614537 | MH644923 | OP081989 | KP131625 | ||||
| IHEM 25487 | France | environment | MH614524 | MH644879 | OP082031 | MH613120 | ||||
| IHEM 23971 | Belgium | mycotic otitis externa | MH614555 | MH645013 | OP082025 | MH613181 | ||||
| IHEM 17440 | Belgium | chronic sinusitus | MH614540 | MH644924 | OP081991 | KP131627 | ||||
| IHEM 23890 | Belgium | human ear | MH614530 | MH644905 | OP082020 | MH613172 | ||||
| IHEM 16879 | Morocco | otitis human ear | MH614559 | MH644921 | OP081986 | MH613211 | ||||
| IHEM 26291 | Belgium | unknown | MH614552 | MH644903 | OP082039 | MH613165 | ||||
| IHEM 17439 | Belgium | chronic sinusitus | MH614539 | MH644876 | OP081990 | KP131626 | ||||
| IHEM 13662 | Belgium | hospital environment | MH614553 | MH644898 | OP081985 | MH613153 | ||||
| IHEM 17033 | Belgium | mycotic otomycosis externa | MH614536 | MH644922 | OP081987 | KP131623 | ||||
| IHEM 23911 | Belgium | otitis | MH614566 | MH644908 | OP082024 | MH613177 | ||||
| IHEM 17904 | Belgium | chronic sinusitus | MH614538 | MH644875 | OP081996 | MH613212 | ||||
| IHEM 17893 | Belgium | chronic sinusitus | MH614542 | MH644926 | OP081994 | KP131630 | ||||
| IHEM 25949 | France | human sputum | MH614528 | MH644873 | OP082034 | MH613144 | ||||
| IHEM 26200 | Belgium | otitis | MH614568 | MH644901 | OP082036 | MH613160 | ||||
| IHEM 26645 | Belgium | human ear secretions | MH614549 | MH644897 | OP082043 | MH613148 | ||||
| IHEM 26163 | Belgium | otitis externa | MH614527 | MH644872 | OP082035 | MH613140 | ||||
| IHEM 23900 | Belgium | otitis | MH614529 | MH644904 | OP082021 | MH613166 | ||||
| IHEM 26661 | Belgium | onychomycosis human toe nail | MH614556 | MH644911 | OP082044 | MH613193 | ||||
| IHEM 17903 | Belgium | chronic sinusitus | MH614543 | MH644927 | OP081995 | KP131631 | ||||
| IHEM 17441 | Belgium | chronic sinusitus | MH614541 | MH644925 | OP081992 | KP131628 | ||||
| IHEM 26350 | France | air | MH614548 | MH644883 | OP082040 | MH613143 | ||||
| IHEM 17168 | Belgium | chronic sinusitus | MH614534 | MH644920 | OP081988 | KP131624 | ||||
| IHEM 26622 | Belgium | human ear | MH614567 | MH644896 | OP082042 | MH613145 | ||||
| IHEM 25777 | Belgium | human wound | MH614423 | MH644913 | OP082033 | MH613197 | ||||
| IHEM 25773 | Belgium | otitis externa | MH614472 | MH644912 | OP082032 | MH613196 | ||||
| IHEM 4379 | unknown | unknown | MH614554 | MH644899 | OP082045 | MH613154 | ||||
| IHEM 23985 | Belgium | otitis | MH614531 | MH644907 | OP082026 | MH613176 | ||||
| IHEM 26215 | Belgium | otomycosis | MH614433 | MH644914 | OP082037 | MH613200 | ||||
| IHEM 17442 | Belgium | chronic sinusitus | MH614533 | MH644915 | OP081993 | KP131629 | ||||
| IHEM 10349 | China | grains | MH614535 | MH644918 | OP081984 | MH613207 | ||||
| IHEM 23886 | Belgium | otitis | MH614424 | MH644910 | OP082019 | MH613188 | ||||
| IHEM 5615 | Unknown | unknown | MH614425 | MH644916 | OP082047 | MH613201 | ||||
| IHEM 24447 | India | otitis | MH614532 | MH644909 | OP082027 | MH613187 | ||||
| CBS 115574 = IBT 23401 = ITEM 7555 = CECT 20579T6 | Costa Rica | soil | genome | genome | HE984361 | MH862988 | ||||
| CBS 553.65 = NRRL 5121 = ATCC 16880 = IMI 23599 | Costa Rica | soil | FJ629278 | OP081904 | OP081983 | FJ629327 | ||||
| CBS 115656 = IBT 20973 = NRRL 62634T7 | Venezuela | mangrove water | KC796361 | KC796377 | genome | OL711719 | ||||
| IHEM 23909 | India | otitis | MH614422 | MH644906 | OP082023 | MH613175 | ||||
| IHEM 22394 | India | conjunctivitis - human eye | MH614544 | MH644870 | OP082017 | KP131635 | ||||
| SDBR CMUI4T8 | Thailand | rhizosphere soil | MK457200 | MK457199 | MK457202 | — | ||||
| SDBR CMU15 | Thailand | rhizosphere soil | MW602898 | MW602897 | MW602899 | — | ||||
| SDBR CMUI1T9 | Thailand | rhizosphere soil | MK457194 | MK457193 | MK457196 | — | ||||
| SDBR CMUI7 | Thailand | rhizosphere soil | MW602903 | MW602902 | MW602904 | — | ||||
| A. vadensis | CBS 113365 = IMI 142717 = CECT 20584 = IBT 24658T | Egypt | air | AY585531 | genome | HE984371 | — | |||
| IHEM 26351 | France | human sputum | MH614547 | MH644878 | OP082041 | MH613142 | ||||
| A. carbonarius | CBS 111.26 = ITEM 4503 = NRRL 369 = ATCC 1025 = ATHUM 2854 = CBS 556.65 = IMI 016136 = MUCL 13583 = NCTC 1325 = NRRL 1987 = NRRL 369 = ITEM 5010T | Unknown | paper | EF661099 | EF661167 | EF661068 | EF661204 | |||
| NRRL 67 = NBRC 5864 = ATCC 8740 = CBS 420.64 = DSM 872 = IFO 5864 = IMI 41875 = MUCL 30479 = NRRL 1737 = NRRL 605 | Unknown | unknown | EF661097 | EF661165 | EF661066 | EF661202 | ||||
| NRRL 4849 = CBS 114.29 | USA | unknown | EF661100 | EF661168 | EF661069 | EF661205 | ||||
| NRRL 346 = ATCC 6277 = CBS 146284 | Honduras | unknown | EF661098 | EF661166 | EF661067 | EF661203 | ||||
| Additional strains used only in DELINEATE and STACEY analyses | ||||||||||
| A. carbonarius | IHEM 661 | France | indoor air in bakery | MH614442 | MH645014 | — | — | |||
| IHEM 1931 | Belgium | dust from mattress | MH61458 | MH644880 | — | — | ||||
| IHEM 25902 | France | human sputum | MH614575 | MH645015 | — | — | ||||
| DTO 179-F4 | South Africa | house dust | KP329846 | KJ775280 | — | — | ||||
| DTO 179-C6 | South Africa | house dust | KP329845 | KJ775278 | — | — | ||||
| CCF 3388 | Czech Republic | toenails, human clinical material | HE577803 | HE649500 | — | — | ||||
| 144-3-K2 | Unknown | unknown | KJ599604 | KJ599576 | — | — | ||||
| A-1759 | Israel | grape | KC520549 | KC520552 | — | — | ||||
| G187 | Slovakia | dried vine fruits | MT166308 | MK046876 | — | — | ||||
| A-2160 | Spain | grape | KC520550 | KC520553 | — | — | ||||
| A. ibericus | NRRL 35644 = CBS 121593 = IMI 391429 = DTO 24-F1 = ITEM 4776 = IHEM 23498T | Portugal | wine grapes | EF661102 | EF661163 | XM_025715438 | — | |||
| NRRL 35645 = ITEM 6601 = IMI 391430 | Portugal | wine grapes | EF661101 | EF661164 | EF661065 | — | ||||
| A. ellipticus | CBS 707.79 = IMI 172283T | Unknown | unknown | AY585530 | EF661170 | EF661051 | — | |||
| CBS 677.79 = IMI 278383 = ITEM 4499T10 | Costa Rica | unknown | AY819993 | AM117810 | HE984363 | — | ||||
| A. heteromorphus | NRRL 4747 = CBS 117.55 = ATCC 12064 = IMI 172288 = IHEM 5801 = ITEM 7045T | Brazil | culture contaminant | EF661103 | EF661169 | EF661050 | — | |||
| IHEM 18645 | France, Martinique | indoor wall | MH614573 | MH645030 | — | — | ||||
| A. sclerotiicarbonarius | CBS 121057 = IBT 121057T | Unknown | culture contaminant | EU159229 | EU159235 | MN969091 | — | |||
| A. sclerotioniger | CBS 115572 = IBT 22905 = ITEM 7560T | Unknown | unknown | FJ629304 | FN594557 | HE984369 | — | |||
| A. japonicus | CBS 114.51 = ITEM 7034T | Unknown | unknown | HE577804 | AJ964875 | MN969079 | — | |||
| PPRI 4286 = CMV 005H7 | South Africa | soil | MK451062 | MK451430 | MK450779 | — | ||||
| ITEM 14787 | USA | indoor air | HE984413 | HE984430 | HE984377 | — | ||||
| ITEM 14805 | USA | indoor air | HE984416 | — | HE984378 | — | ||||
| CBS 123.27 = NRRL 360 = ATCC 1042 = IFO 4106 = IMI 358697 | Puerto Rico | soil | HE577805 | EF661141 | EF661047 | — | ||||
| NRRL 1782 = ATCC 16873 = CBS 568.65 = IMI 211387 | Panama | soil | — | EF661144 | EF661048 | — | ||||
| NRRL 35494 = ITEM 15926 | Unknown | unknown | — | EU021690 | EU021639 | — | ||||
| NRRL 35541 | USA | peanut field soil | EF661104 | EF661143 | EU021640 | — | ||||
| NRRL 4839 = IHEM 5627 = NCTC 3792 = MUCL 13578 = IMI 312983 = CBS 113.48 | Unknown | unknown | MH614586 | EF661142 | EF661049 | — | ||||
| IHEM 26043 | France | human sputum | MH614587 | MH645026 | — | — | ||||
| CBS 115571 | Bahamas | marine environment | EU482434 | EU482432 | — | — | ||||
| AJP01 | Unknown | unknown | JX103558 | JX103559 | — | — | ||||
| CCF 4079 | Unknown | unknown | HE577811 | FR751423 | — | — | ||||
| ITEM 14789 | USA | indoor air | HE9844174 | HE984432 | — | — | ||||
| NBRC 4408 | Unknown | soil | LC573650 | LC573708 | — | — | ||||
| NBRC 32856 | Japan | living leaf | LC573651 | LC573709 | — | — | ||||
| A. aculeatus | CBS 172.66 = IHEM 5796 = NRRL 5094 = IMI 211388 = CCRC 32190 = ATCC 16872 = ITEM 7046T | Unknown | tropical soil | HE577806 | EF661148 | XM020198603 | — | |||
| PPRI 7513 = CMV 005A6 | Unknown | unknown | MK451022 | MK451291 | MK450752 | — | ||||
| PPRI 26016 = CMV 007C9 | South Africa | soil | MK451118 | MK451296 | MK450756 | — | ||||
| F-719 | Unknown | unknown | HE577810 | HE578093 | — | — | ||||
| PPRI 4070 = CMV 005F1 | South Africa | soil | MK451044 | MK451292 | MK450753 | — | ||||
| ITEM 14807 | USA | indoor air | HE984409 | HE984424 | HE984372 | — | ||||
1Acronyms of culture collections in alphabetic order: ATCC, American Type Culture Collection, Manassas, Virginia; ATHUM, Athens Collection of Fungi, University of Athens, Athens, Greece; CBS, Westerdijk Fungal Biodiversity Institute (formerly Centraalbureau voor Schimmelcultures), Utrecht, the Netherlands; CCF, Culture Collection of Fungi, Department of Botany, Charles University, Prague, Czech Republic; CCFC,Canadian Collection of Fungal Cultures, living fungal collection associated with the DAOM herbarium (Agriculture and Agri-Food Canada), Ottawa, Canada; CCM (F-), Czech Collection of Microorganisms, Brno, Czech Republic; CCRC (=BCRC), Bioresources Collection and Research Center, Food Industry Research and Development Institute, Hsinchu, Taiwan; CECT, Colección Española de Cultivos Tipo, Universidad de València, Edificio de Investigación, Burjassot, Spain; CMV, working and formal culture collections housed at FABI (Forestry and Agricultural Biotechnology), Innovation Africa, University of Pretoria, South Africa; DSM, Leibniz Institute DSMZ-German Collection of Microorganisms and Cell Cultures, Braunschweig, Germany; DTO, internal culture collection of the Department Applied and Industrial Mycology of the Westerdijk Fungal Biodiversity Institute, Utrecht, The Netherlands; IBT, Culture Collection at the Department of Biotechnology and Biomedicine, Lyngby, Denmark; IFM, Culture Collections for Pathogenic Fungi and Actinomycetes, Medical Mycology Research Center, Chiba University, Chiba, Japan; IFO, Institute for Fermentation, Osaka, Japan (IFO strains were transferred to the NBRC NITE collection); IHEM (BCCM/IHEM), Belgian Coordinated Collections of Micro-organisms, Fungi Collection: Human and Animal Health, Sciensano, Brussels, Belgium; IMI, CABI’s collection of fungi and bacteria, Wallingford, UK; JCM, Japan Collection of Microorganisms, Tsukuba, Japan; KACC, Korean Agricultural Culture Collection, Wanju, South Korea; KCTC, Korean Collection for Type Cultures, Korea Research Institute of Bioscience and Biotechnology, Jeongeup, South Korea; MUCL (BCCM/MUCL), Agro-food & Environmental Fungal Collection, Louvain-la-Neuve, Belgium; NBRC (NITE), National Institute of Technology and Evaluation, Biological Resource Center, Department of Biotechnology, Kisarazu, Chiba, Japan; NCTC, National Collection of Type Cultures, Central Public Laboratory Service, London, UK; ITEM, Agri-Food Toxigenic Fungi Culture Collection, Institute of Sciences of Food Production, Bari, Italy; NRRL, Agricultural Research Service Culture Collection, Peoria, Illinois, USA; PPRI, culture collection of the National Collections of Fungi, housed at the Agricultural Research Council - Plant Health and Protection, Roodeplaat, South Africa; RIB, National Research Institute of Brewing, Tax Administration Agency, Higashihiroshima, Hiroshima, Japan; SDBR, Sustainable Development of Biological Resources Laboratory, Faculty of Science, Chiang Mai University, Chiang Mai Province, Thailand. Other acronyms represent personal strain numbers (without permanent preservation).
2Accession numbers to genomic sequences analysed in this study are listed in Supplementary Table S3.
Tex-type strain. T1 = A. lacticoffeatus; T2 = A. vinaceus; T3 = A. welwitschiae; T4 = A. nakazawae; T5 = A. piperis; T6 = A. costaricensis; T7 = A. neoniger; T8 = A. chiangmaiensis; T9 = A. pseudopiperis; T10 = A. helicothrix.
Total genomic DNA was isolated from 5-d-old cultures using the Zymo Research Fungal/Bacterial DNA Kit™ (Zymo Research, Irvin, CA, USA) or the Invisorb Spin Plant Mini Kit (Invitek, Berlin, Germany). PCR amplification of CaM was performed using the primer pairs cmd5 and cmd6 (Hong et al. 2006) or CF1L and CF4 (Peterson 2008); benA with Bt2a and Bt2b (Donaldson et al. 1995) or Ben2f (Hubka & Kolarik 2012) and Bt2b; and RPB2 with fRPB2-5F and fRPB2-7CR (Liu et al. 1999). Samples were amplified using the following cycling parameters: one initial step of 10 min at 95 °C followed by 35 cycles of 1 min at 95 °C, 1 min at 55 °C, and 1 min at 72 °C and a single extension step of 10 min at 72 °C. The PCRs were performed in 25 μL volume containing 2 μL of DNA (2 pM), 2 μL of forward primer, 2 μL of reverse primer, 19 μL of H2O, and PuReTaq Ready-To-Go PCR beads (GE Healthcare UK, Little Chalfont, UK). The PCR products were sequenced using the BigDye Terminator cycle sequencing ready reaction kit (Applied Biosystems, Foster City, CA, USA) using the same primers used for PCR and analysed on an ABI Prism 3130 genetic analyser (Applied Biosystems) according to the manufacturer’s instructions.
Newly obtained DNA sequences were inspected and assembled in Geneious Prime v. 2020.2.4 (Biomatters Ltd.). Sequences were deposited into GenBank with the accession numbers listed in Table 1.
Phylogenetic analyses and species delimitation
Alignments of benA, CaM and RPB2 were performed using the FFT-NS-i option implemented in the MAFFT online service (Katoh et al. 2019). The alignments were trimmed, concatenated, and then analysed using maximum likelihood (ML), maximum parsimony (MP) and Bayesian inference (BI) methods. The final alignments are available from the DRYAD digital repository (https://doi.org/10.5061/dryad.866t1g1td).
Suitable partitioning schemes and substitution models for the analyses were selected based on the Bayesian information criterion using a greedy strategy implemented in PartitionFinder 2 (Lanfear et al. 2017) with settings allowing introns, exons and codon positions to be independent datasets (Supplementary Table S1). ML trees were constructed with IQ-TREE v. 1.4.4 (Nguyen et al. 2015) with nodal support determined by ultrafast bootstrapping (BS) with 105 replicates. Bayesian posterior probabilities were calculated using MrBayes v. 3.2.6 (Ronquist et al. 2012). The analysis ran for 107 generations, two parallel runs with four chains each were used, every 1 000th tree was retained, and the first 25 % of trees were discarded as burn-in. The convergence of the runs and effective sample sizes were checked in Tracer v. 1.6 (http://tree.bio.ed.ac.uk/software/tracer). Maximum parsimony (MP) trees were calculated using PAUP* v. 4.0b10 (Swofford 2003). Analyses were performed using the heuristic search option with 100 random taxon additions; tree bisection-reconnection (TBR); maxtrees were set to 1 000. Branch support was assessed by bootstrapping with 500 replications.
The rules for the application of the GCPSR approach were adopted from Dettman et al. (2003a, b) and slightly modified for the different design of this study (different numbers of loci and methods used). To recognize a clade as an “evolutionary lineage”, it had to satisfy one of two criteria: (a) genealogical concordance - the clade was present in the majority (2/3) of the single-locus genealogies; (b) genealogical nondiscordance - the clade was well supported in at least one single-locus genealogy, as judged by ML ultrafast bootstrap proportions ≥95 %, MP bootstrap proportions ≥70 % and BI posterior probabilities ≥95 %, and was not contradicted in any other single-locus genealogy at the same level of support. When deciding which evolutionary lineages represent “phylogenetic species”, two additional criteria were applied and evaluated according to the combined phylogeny of three genes: (a) genetic differentiation - species had to be relatively distinct and well differentiated from other species to prevent minor tip clades from being recognized as a separate species; (b) all individuals had to be placed within a phylogenetic species, and no individuals were to be left unclassified.
Species delimitation analyses based on the multispecies coalescent model were performed as described previously (Sklenář et al. 2021). In brief, the haplotype function from R v. 4.0.2 (R Core Team 2016) package PEGAS (Paradis 2010) was used to retain only unique sequences in the alignments. The best fitting models were obtained in jModelTest v. 2.1.7. Parameters and the other settings of the STACEY, GMYC, bGMYC, PTP, and bPTP methods were consistent with previous studies (Sklenář et al. 2021, Glässnerová et al. 2022). Two different settings were used for the GMYC method. The ultrametric input trees for this method were calculated in BEAST v. 2.6.6 (Bouckaert et al. 2014) with a chain length of 107 generations. As a model for creating input trees, we set up the coalescent constant population prior and performed two tree reconstruction methods: one with the common ancestor height (CAh) setting and a second with the median height (Mh) setting. We only show the delimitation results of both settings when they were different. The results of all analyses are graphically summarised in iTOL v. 6 (Interactive Tree of Life) (Letunic & Bork 2021).
The multilocus species delimitation method STACEY was performed in BEAST (Bouckaert et al. 2014) using the STACEY v. 1.2.5 add-on (Jones 2017). For this and the DELINEATE analysis (see below), we added several additional species from other series of section Nigri (Table 1). We set up the length of MCMC chain to 109 generations, the species tree prior was set to the Yule model, the molecular clock model was set to a strict clock, the growth rate prior was set to a lognormal distribution (M = 5, S = 2), the clock rate priors for all loci were set to a lognormal distribution (M = 0, S = 1), the PopPriorScale prior was set to a lognormal distribution (M = -7, S = 2) and the relative DeathRate prior was set to a beta distribution (α = 1, β = 1 000). The substitution models were as follows: benA - K80+I; CaM - TrNef+G; RPB2 - TrNef+G. The output was processed with SpeciesDelimitationAnalyzer (Jones et al. 2015). For the presentation of the STACEY results, we first created a plot showing how the number of delimited species and the probability of the most likely scenarios changed in relation to the value of the collapseheight parameter, and then we created similarity matrices using code from Jones et al. (2015) with different values of collapseheight chosen from the plot (see the Results section).
Independent testing of various species boundary hypotheses was performed in the DELINEATE software (Sukumaran et al. 2021). After splitting the dataset into hypothetical populations (Supplementary Table S2) using the “A10” analysis in BPP v. 4.4 (Yang 2015), the species tree for these populations was estimated in starBEAST (Heled & Drummond 2010) implemented in BEAST v. 2.6.7 (Bouckaert et al. 2014). The analysis was run in the Python (van Rossum & Drake Jr FL 2014) package DELINEATE (Sukumaran et al. 2021). In every simulated model, some populations delimited by BPP were assigned to tentative (fixedly defined) species based on the species delimitation results from previous analyses or were based on the common species definitions from previous studies. Other populations were left free to be delimited (unassigned to any species). As a result, the algorithm implemented in DELINEATE can either recognize the unassigned population as a separate species, define a wider monophyletic species composed of several unassigned populations, or merge unassigned populations with some of the predefined species.
Whole genome sequencing and assembly
Whole-genome 100 bp paired-end (PE) sequencing was performed in 18 strains with the aid of HiSeq 1500 (Illumina, San Diego, CA) using the HiSeq reagent kit v. 1, according to the manufacturer’s instructions. A 300 bp PE sequencing was performed on a MiSeq (Illumina) using the MiSeq Reagent Kit v. 3, according to the manufacturer’s instructions. Long read sequencing of IFM 61612 was performed on the MinION platform (ONT; Oxford Nanopore Technologies, Cambridge, UK). A DNA library for ONT sequencing was prepared using a short-read eliminator kit (Nippon Genetics, Tokyo, Japan) and a ligation sequencing kit (SQK-LSK109), and sequencing was performed with a FLO-MIN106D flow cell (R9.4.1) for 48 h, according to the manufacturer’s instructions. A total of 83 443 ONT reads were generated with a mean length of 12 011 bp using ONT Guppy v. 2.3.5.
Adapters and low-quality bases of Illumina reads were trimmed by Trim Galore v. 0.6.4 (Krueger 2015) with the default settings selected (http://www.bioinformatics.babraham.ac.uk/projects/trim_galore). The mitochondrial genomes were assembled using GetOrganelle v. 1.6.4 (Jin et al. 2020) with trimmed reads. To filter the mitochondrial reads, the trimmed reads were aligned against the mitochondrial genomes by BWA v. 0.7.17-r1188 (Li & Durbin 2009), and the mapped reads were filtered by SAMtools v. 1.9 (Li et al. 2009) and SeqKit (Shen et al. 2016). For eight isolates sequenced by MiSeq, the filtered reads were used to assemble the nuclear genomes using SPAdes v. 3.14.0 (Bankevich et al. 2012) with the options ‘--cov-cutoff auto’ and ‘--careful’. For six isolates sequenced by HiSeq 1500, the assembly of nuclear genomes was carried out as follows: (1) the filtered reads were used to assemble the contigs using VelvetOptimiser v. 2.2.6 (Zerbino & Birney 2008); (2) the contigs were used to generate in silico long mate-pair reads with inserts of 3 000 ± 300 bp using Wgsim v. 0.3.1-r13 (https://github.com/lh3/wgsim) with the options ‘-e 0 -1 100 -2 100 -r 0 -R 0 -X 0 -d 3000 -s 300 -N 4000000’; (3) Illumina PE and simulated mate-pair (MP) reads were assembled by ALLPATHS-LG v. 52488 (Gnerre et al. 2011). The genome of IFM 61612 was assembled using NECAT v. 0.0.1_update20200803 (Chen et al. 2021), and polished with the ONT reads by using Racon v. 1.4.20 (Vaser et al. 2017) and Medaka v. 1.4.4 (https://github.com/nanoporetech/medaka) and with Illumina PE reads by using HyPo v. 1 (Kundu et al. 2019). Most tools were obtained through Bioconda (Grüning et al. 2018).
Genome annotation
The annotation of assembled genomes was performed using the Funannotate pipeline v. 1.7.4 (https://funannotate.readthedocs.io/en/latest/). Following the identification of repeat sequences by RepeatModeler v. 1.0.11 (http://www.repeatmasker.org/RepeatModeler.html) and RepeatMasker v. 4.0.7 (https://www.repeatmasker.org), Funannotate ab initio prediction was performed with the option ‘--busco_seed_species=aspergillus_oryzae’ by Augustus v. 3.3.3 (Stanke et al. 2006), GeneMark-ES v. 4.38 (Ter-Hovhannisyan et al. 2008), GlimmerHMM v. 3.0.4 (Majoros et al. 2004), and SNAP v. 2006-07-28 (Korf 2004) using exon hints from the proteins of A. niger CBS 513.88 and A. oryzae RIB40 downloaded from the Aspergillus Genome Database (Bairoch & Apweiler 2000, Cerqueira et al. 2014). The functional annotations of predicted genes were performed using the Swiss-Prot, InterPro v. 5.42-78.0 (Jones et al. 2014), eggNOG v. 4.5.1 (Buchfink et al. 2015, Huerta-Cepas et al. 2016), MEROPS v. 12.0 (Rawlings et al. 2018, Mitchell et al. 2019), and the dbCAN v. 8.0 (Yin et al. 2012) databases. Genome annotations of the reference strains were obtained from the Joint Genome Institute (http://genome.jgi.doe.gov/) (Supplementary Table S3). The completeness of the draft genomes and predicted proteins were evaluated using BUSCO v. 4.0.6 (Seppey et al. 2019) with the database “eurotiales_odb10”.
Phylogenetic analyses based on the genomic sequences
Orthologous genes among the 31 strains (18 newly sequenced and 13 available from previous studies) were identified by OrthoFinder v. 2.3.12 (Emms & Kelly 2019). Then, the amino acid sequences of 5 752 single-copy orthologous proteins were aligned using the G-INS-i option in MAFFT v. 7.471 and concatenated into one long protein sequence. A phylogenetic tree was constructed using multithreaded RAxML (Stamatakis 2014), selecting the PROTGAMMAWAG model and using 100 bootstrap replicates to determine node support.
The predicted protein sets of 31 strains were evaluated with BUSCO protein mode. A total of 2 754 genes present at a single copy were identified among all strains. From these, 200 genes were randomly selected for analyses using the STACEY method and randomly distributed into ten independent analyses, each containing 20 genes (Supplementary Table S4). Coding regions (exons) of the corresponding 200 nucleotide sequences were extracted for each strain from the de novo assemblies. Each sequence matrix was aligned using MAFFT, and well-aligned regions were extracted using Gblocks v. 0.91b (Talavera & Castresana 2007) with settings allowing no gap positions.
RESULTS
Combined phylogeny based on three loci
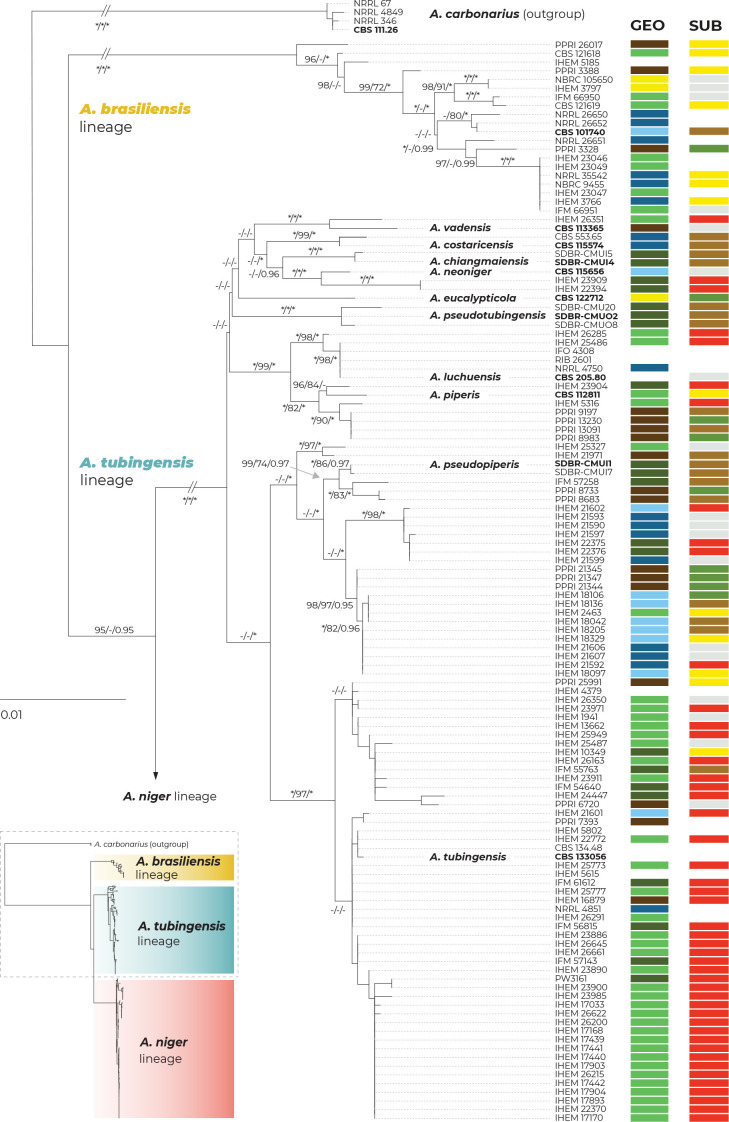
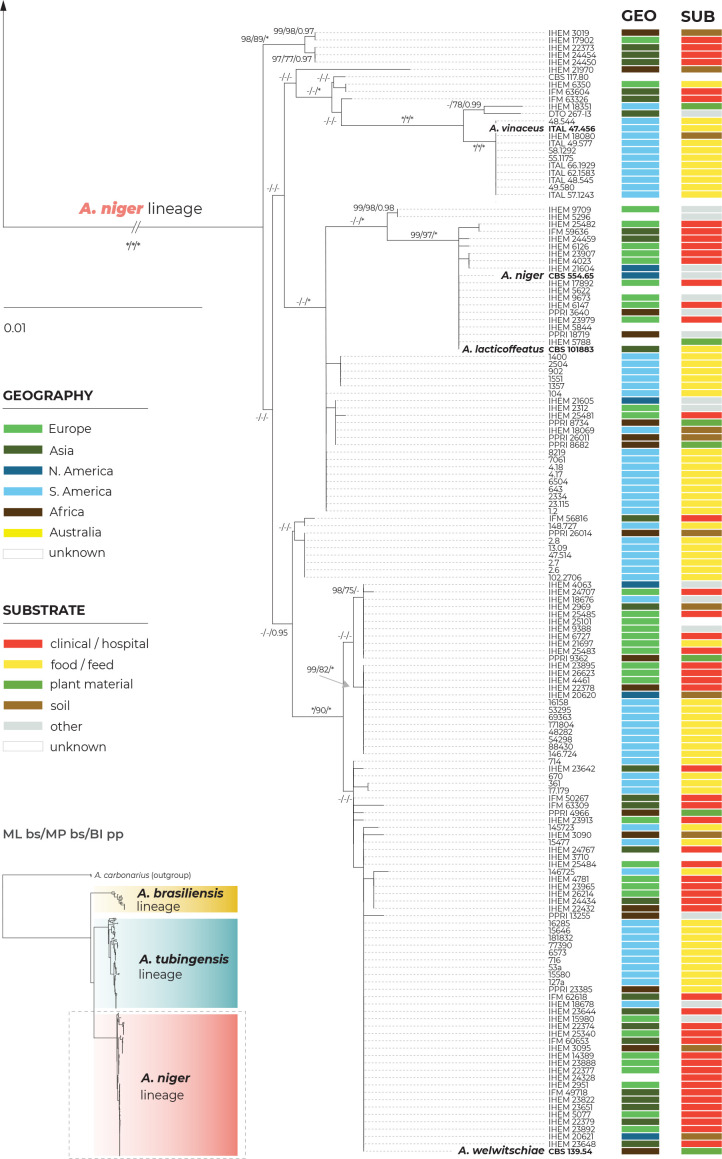
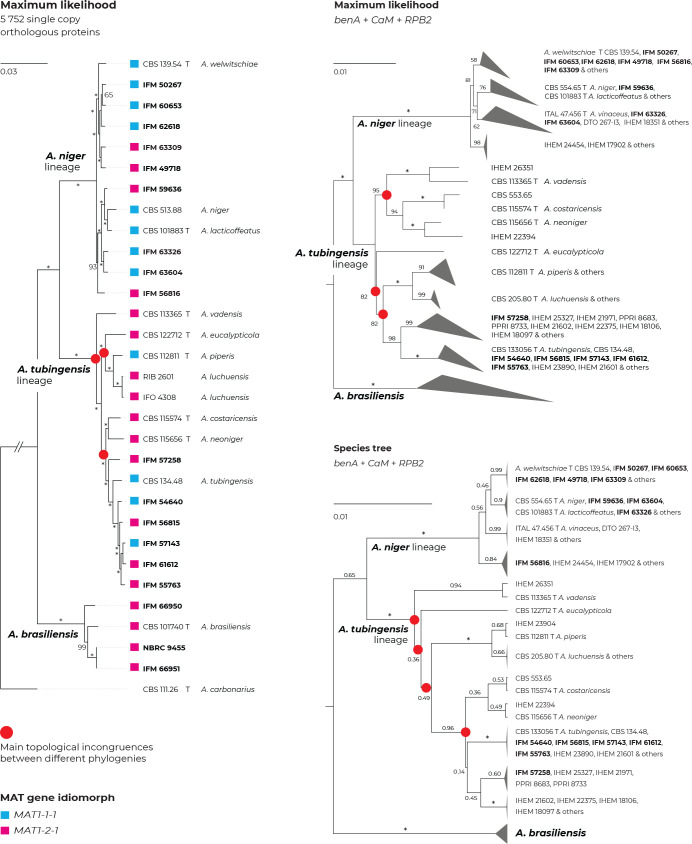
A maximum likelihood (ML) phylogenetic tree based on three loci in 280 strains (276 belonging to the series Nigri and four A. carbonarius strains used as outgroups) is presented in Fig. 1, with the geographic origin and substrate plotted on the tree in the form of colour strips. The topology of the trees inferred by the maximum parsimony (MP) and Bayesian inference (BI) methods was similar to ML and the bootstrap support values and posterior probabilities, respectively, were appended to the nodes.
Fig. 1.
Multilocus phylogeny of Aspergillus series Nigri based on three loci (benA, CaM, RPB2) and 276 isolates (and four A. carbonarius isolates as an outgroup). The best-scoring maximum likelihood (ML) tree inferred in the IQ-TREE is shown; ultrafast bootstrap support values (ML bs) are appended to nodes along with maximum parsimony bootstrap support values (MP bs) and Bayesian inference posterior probabilities (BI pp); only support values ≥95 %, ≥70 % and ≥0.95, respectively, are shown; a dash indicates lower statistical support for a specific node or the absence of a node in the phylogeny while an asterisk indicates full support; the ex-type strains are designated with a bold print; the information on geographic origin and isolation source is plotted on the tree (see legend). Alignment characteristics, partitioning schemes and substitution models are listed in Supplementary Table S1.
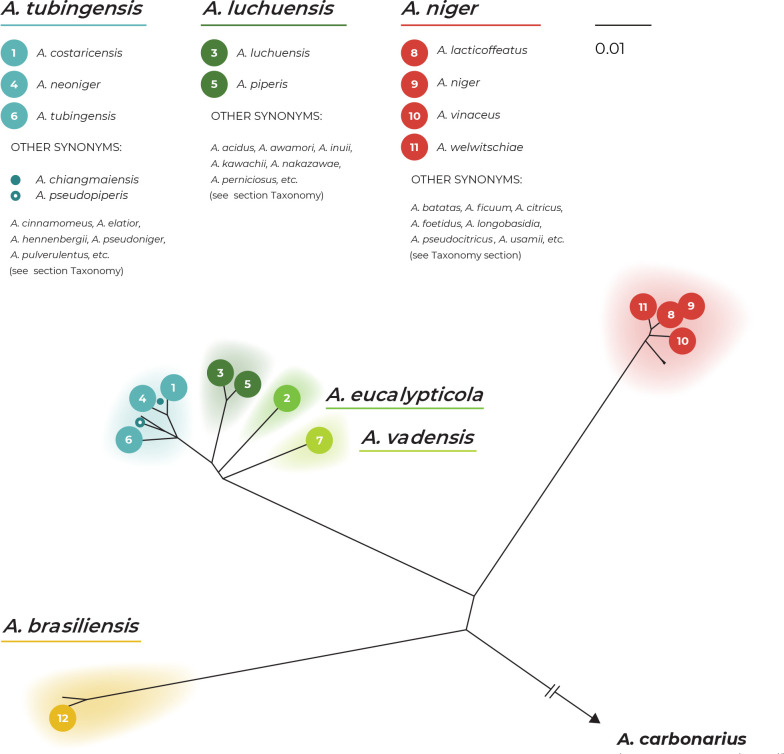
There are three main lineages in the tree and for practical reasons, we named them based on the priority rules. The A. brasiliensis lineage (ABL) only contains the ex-type isolate of A. brasiliensis, while the A. niger lineage (ANL) contains the ex-type isolates of A. lacticoffeatus, A. niger, A. vinaceus and A. welwitschiae (the old species names, which are usually considered synonyms, are omitted). The Aspegillus tubingensis lineage (ATL) contains ex-types of ten species: A. chiangmaiensis, A. costaricensis, A. eucalypticola, A. luchuensis, A. neoniger, A. piperis, A. pseudopiperis, A. pseudotubingensis, A. tubingensis and A. vadensis. The sequences of the three recently described species, A. chiangmaiensis, A. pseudopiperis and A. pseudotubingensis (Khuna et al. 2021), were of poor quality and contained a large number of errors (indels) in the coding regions and probably also in the noncoding regions of all three genes. Due to this fact and the absence of genomic sequences, we excluded these species from most analyses. However, we manually corrected some apparent errors in the exonic regions (partially revised sequences can be found in the alignments deposited in the Dryad digital repository: https://doi.org/10.5061/dryad.866t1g1td) and added these species to the classical phylogenetic trees based on the ML, MP and BI methods. In these phylogenies, the positions of A. chiangmaiensis and A. pseudotubingensis differ significantly from the tree presented by Khuna et al. (2021). The combined phylogeny without these species is shown in Supplementary Fig. S1.
The bootstrap support values in the combined trees (Fig. 1 and Supplementary Fig. S1) are high on many terminal branches in ATL, and the majority of the currently accepted species form their own well-supported terminal clades. On the other hand, some deeper nodes are poorly supported and the relationships among the species in ATL are mostly unresolved. If only the monophyly and statistical support of branches in these combined phylogenetic trees were considered, all currently recognized species in ATL could be accepted. Bootstrap values in the trees based on concatenated datasets are, however, often falsely high and are not a suitable criterion for deciding about species limits (Kubatko & Degnan 2007, Seo 2008). The boundaries between currently accepted species in ANL are much less clear, as there are several poorly supported clades in the proximity of A. vinaceus, A. niger and A. welwitschiae whose species identification is unclear.
The geography and substrate of isolation showed no clear patterns that could be associated with particular species, although clinical isolates from European countries (mostly strains from the study of D’hooge et al. (2019)) were overrepresented in the clade containing the A. tubingensis ex-type strain CBS 133056, while isolates from food and South America were more common throughout ANL (strains predominantly from Silva et al. (2020)).
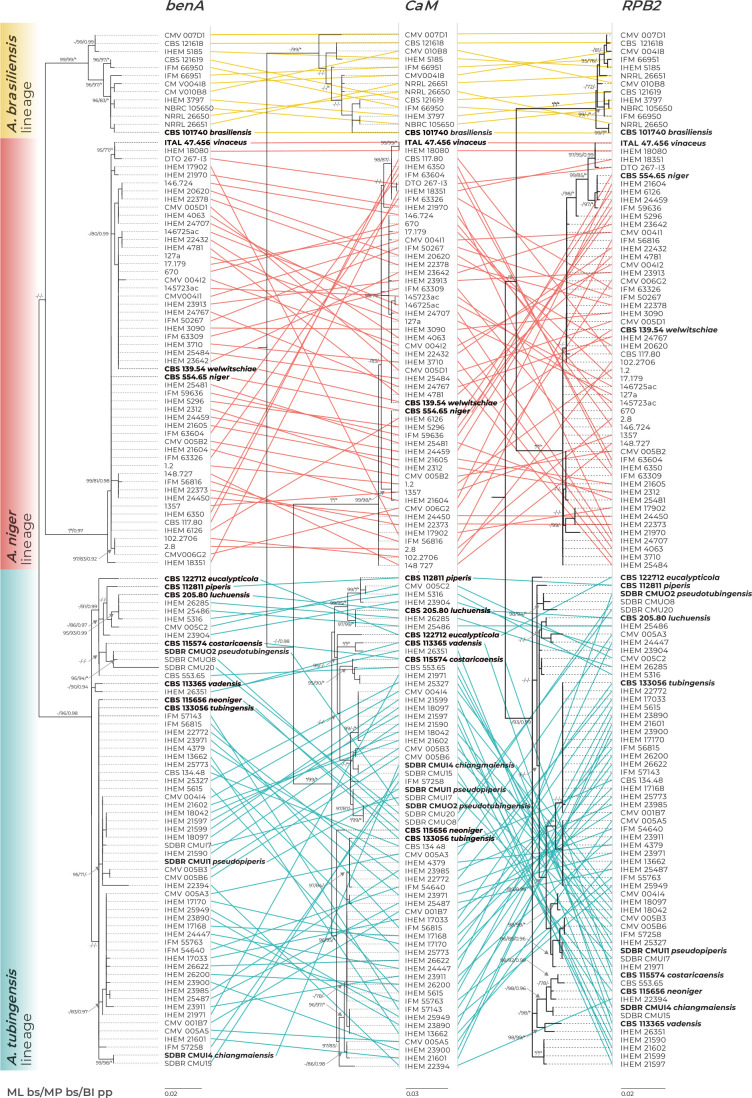
Incongruences between single gene trees: consequences for the GCPSR approach and BLAST similarity searches
Single-gene trees based on benA, CaM and RPB2 loci are shown in Fig. 2 and Supplementary Fig. S2 (only one isolate per unique multilocus haplotype was included). The coloured connecting lines show changes in the positions of isolates between single-gene trees (Fig. 2); the branches were rotated so that the trees maximally correspond to each other; the trees without connecting lines are shown in Supplementary Fig. S2. There was no conflict in topology among the three main lineages, i.e., there was no isolate changing position between ABL, ANL and ATL in different single-gene trees. When applying the rules of the GCPSR approach in ABL and ANL, only one phylogenetic species (PS) can be recognized in each lineage because the composition of their subclades is variable in terms of the isolates resolved in them, while the statistical support of the subclades is low or unstable between phylogenies. A more complicated situation is present in the ATL, where some clades can be recognized as evolutionary lineages, as they are well supported in at least two phylogenetic trees. For example, the clade containing the two strains of A. vadensis (CBS 113365 and IHEM 26351) was present and supported in all phylogenies, while the clade containing seven strains attributed to A. luchuensis/A. piperis was present and supported in the benA and CaM trees. The singleton lineage containing the ex-type of A. eucalypticola (CBS 122712) was also present in all trees, but it cannot be evaluated by the GCPSR criteria as there are no isolates clustering with this strain. Although the abovementioned evolutionary lineages were recognized by the grouping criteria, they cannot be recognized as PS because doing so would have resulted in a situation where other strains belonging to the ATL and forming several nonmonophyletic clades in the combined tree (Fig. 1) could not be assigned to any phylogenetic species and were left unclassified. As a result, applying GCPSR criteria in the ATL would also lead to the recognition of only one phylogenetic species.
Fig. 2.
Comparison of single-gene genealogies based on the benA, CaM and RPB2 loci and created by three different phylogenetic methods (only one isolate per unique multilocus haplotype is included in each phylogeny). The coloured connecting lines show changes in the positions of isolates between single-gene trees (the branches were rotated so that the trees maximally correspond to each other). Best-scoring single-locus maximum likelihood (ML) trees are shown; ML ultrafast bootstrap support values (ML bs), maximum parsimony bootstrap support values (MP bs) and Bayesian inference posterior probabilities (BI pp) are appended to nodes. Only support values ≥95 %, ≥70 % and ≥0.95, respectively, are shown. A dash indicates lower statistical support for a specific node, or the absence of a node in the phylogeny, while an asterisk indicates full support. The ex-type strains are designated with a bold print. Alignment characteristics, partitioning schemes and substitution models are listed in Supplementary Table S1.
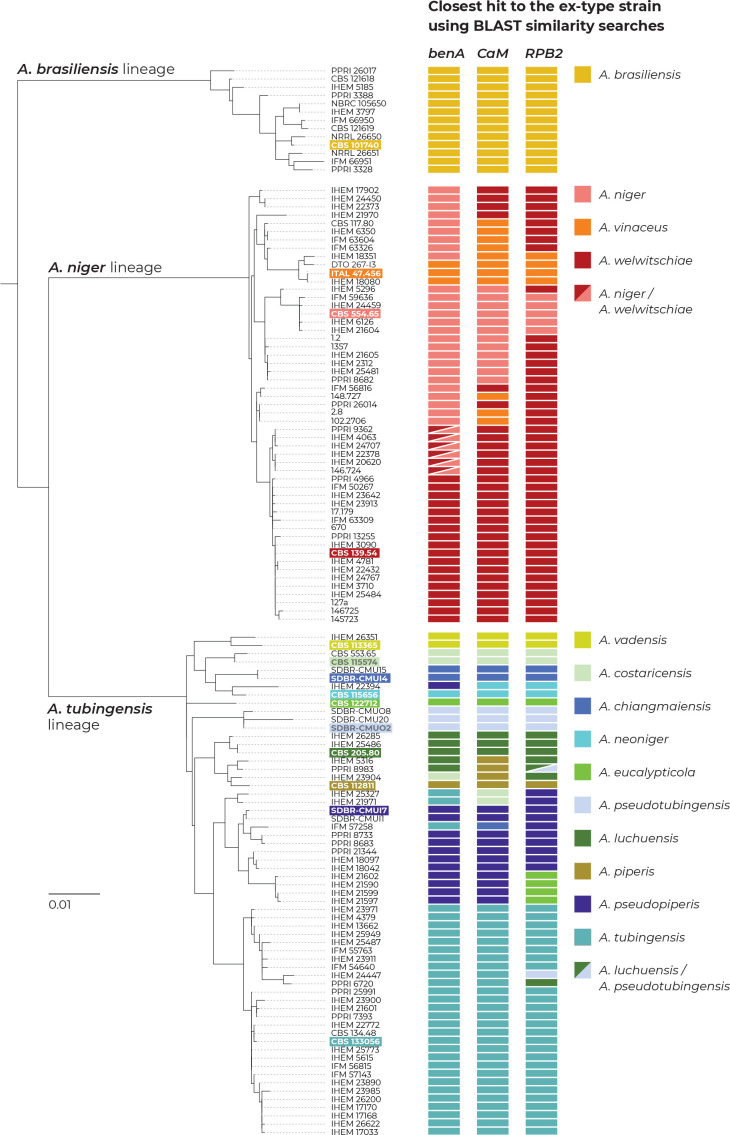
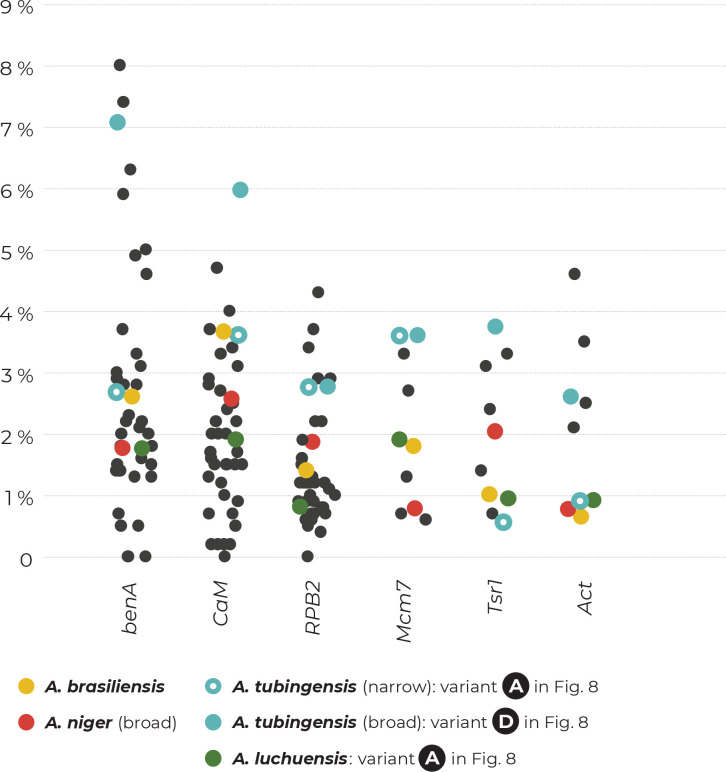
To show the practical consequences of the incongruences between single-gene datasets, we created a local BLAST database containing sequences of benA, CaM and RPB2 of the ex-type strains belonging to the series Nigri. Then, we performed BLAST searches for all sequences of these genes against this database (only one isolate per unique multilocus haplotype was used) in Geneious PRIME v. 2020.2.4 (https://www.geneious.com). The results of the BLAST searches and the closest similarities to the ex-type strains are presented in Fig. 3. It is apparent that sequences of different genes from the same strain frequently resulted in the closest sequence similarities with ex-type strains of different species and thus resulted in variable species identifications. This phenomenon does not involve A. brasiliensis.
Fig. 3.
The results of BLAST similarity searches of three loci (benA, CaM and RPB2) derived from strains with unique multilocus haplotypes across the genetic diversity of series Nigri. Coloured rectangles represent the closest hits to one of the 14 ex-type strains (every species has its unique colour; the ex-type of A. lacticoffeatus was omitted because it has an identical genotype to the ex-type of A. niger). If there was an identical similarity to two ex-type strains, the rectangles were diagonally divided. Ex-type isolates are marked with bold font and a coloured background. The phylogenetic tree was calculated in IQ-TREE using partitioned analysis and 105 ultrafast bootstrap replicates.
In the ANL, different species identifications commonly resulted for different loci, which was observed in 21 out of 53 haplotypes (39.6 %). In seven haplotypes (13 %; CBS 117.80, IFM 63326, IFM 63604, IHEM 6350, 148.727, 2.8 and 102.2706), the three BLAST searches resulted in three different identifications, i.e., A. niger, A. vinaceus and A. welwitschiae. This demonstrates that there are phylogenetic conflicts throughout the entire clade and that there is no barcode gap for these species at any of these loci. These incongruences are also visible in the single-gene phylogenetic trees shown in (Fig. 2, Supplementary Fig. S2).
In ATL, unstable identification based on different genes was less prevalent than in ANL, but was still present in 14 out of 54 haplotypes (24 %). It occurred mostly between A. tubingensis, A. costaricensis, A. neoniger, A. pseudopiperis, A. luchuensis and A. piperis. Four haplotypes (7 %; IHEM 23904, IHEM 25327, IHEM 21971 and IFM 57258) were identified as a different species for each gene region. This phenomenon also has its basis in the single-gene tree incongruences (Fig. 2, Supplementary Fig. S2).
Phylogenomic tree
In Fig. 4, the topology of the phylogenomic tree was compared to the topology of the three-locus ML tree (alignment reduced to unique multilocus haplotypes; A. chiangmaiensis, A. pseudopiperis and A. pseudotubingensis were excluded; list of haplotypes in Supplementary Table S5) and to the topology of the species tree constructed in starBEAST. The phylogenomic tree was based on 5 752 protein orthologues extracted from 30 whole-genome sequences of series Nigri and one sequence of A. carbonarius as an outgroup. Nine out of 18 newly sequenced genomes (Supplementary Table S3; designated by bold print in Fig. 4) belonged to ANL, six to ATL and three to ABL. Almost all clades in the phylogenomic tree gained full statistical support, in contrast to the trees based on the benA, CaM and RPB2 loci. The topologies among the different analyses were mostly congruent, but several incongruences were observed (most of them were present at unresolved nodes with low statistical support). For example, A. vadensis was resolved as a separate branch sister to all other species in the ATL in the phylogenomic and species tree but clustered with A. costaricensis and A. neoniger with a high support of 95 % in the ML tree. In the phylogenomic and species tree, A. neoniger and A. costaricensis clustered with A. tubingensis isolates, in contrast to the ML tree, where they clustered distantly from the latter. Last, A. eucalypticola clusters with A. luchuensis and A. piperis in the phylogenomic tree, while in ML and species trees, it formed a long separate branch.
Fig. 4.
Comparison of topologies of phylogenetic trees constructed on the basis of different methods and data. Incongruences between phylogenies are designated with red circles. Maximum likelihood (ML) tree constructed based on the 5 752 orthologous proteins extracted from the 31 whole genome sequences (WGS; Supplementary Table S3) (left side); ML tree based on benA, CaM and RPB2 loci estimated in IQ-TREE (top right); species tree based on benA, CaM and RPB2 loci estimated in starBEAST (bottom right); the last two mentioned trees were constructed from combined three-gene alignments reduced to unique multilocus haplotypes (Table S5), and A. chiangmaiensis, A. pseudopiperis and A. pseudotubingensis were excluded. The most significant differences can be observed in the positions of A. neoniger, A. costaricensis and A. eucalypticola. Bootstrap support values or posterior probabilities are appended to the nodes, the support values equal to 100 % and 1.00, respectively, are designated with an asterisk; the ex-type strains are designated with the letter “T”. Isolates for which whole genomic sequences were generated in this study are designated with a bold print; mating-type gene idiomorphs are plotted on the tree based on the WGS data.
Both MAT1-2-1 and MAT1-1-1 mating-type gene idiomorphs were found in the genomes of strains belonging to the ANL and ATL, while only the MAT1-2-1 idiomorph was found in the A. brasiliensis strains (Fig. 4).
Multispecies coalescent model-based (MSC) methods
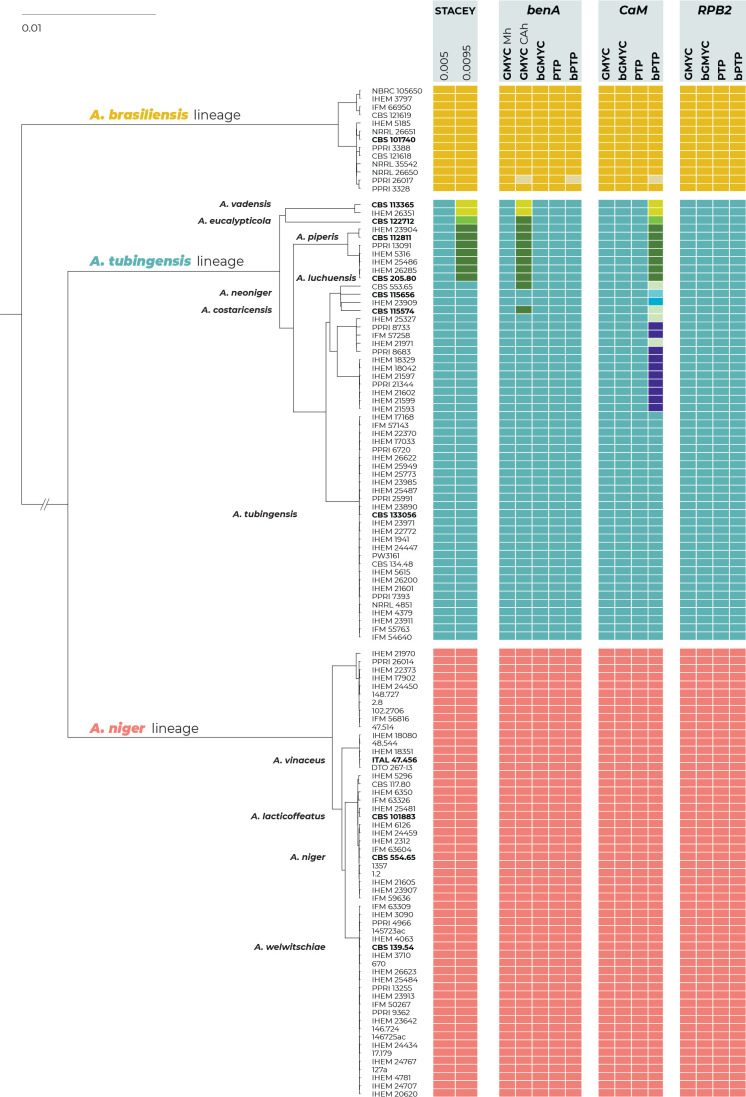
The results of the species delimitation using four single-locus species delimitation methods (GMYC, bGMYC, PTP and bPTP) and one multilocus method, STACEY, are summarised in Fig. 5. For the GMYC and STACEY methods, we showed the results of two different settings, as they differed in the number of delimited species.
Fig. 5.
Schematic representation of the results of species delimitation methods in the series Nigri. One multilocus method (STACEY) and four single-locus methods (GMYC, bGMYC, PTP, bPTP) were applied to a dataset of three loci (benA, CaM, RPB2). The results are depicted by coloured bars with different colours or shades indicating species delimited by specific methods and settings. Ex-type isolates are highlighted with a bold print. The STACEY results with two collapseheight parameter values, 0.005 and 0.0095, are shown. For the GMYC method, the coalescent constant population tree model was used as an input with both common ancestor heights (CAh) and median height (Mh) settings (both settings are only shown when they produced different delimitation results). The phylogenetic tree was calculated by STACEY analysis and is used solely for the comprehensive presentation of the results from different methods.
All methods and their settings delimited the entire ANL as a single species without any exception. The method STACEY and all single-locus methods except for three delimited only one species in the A. brasiliensis lineage. Three methods (GMYC with a common ancestor height (CAh) setting; bPTP based on benA, and bPTP based on CaM) recognized isolate PPRI 26017 as a singleton species separated from A. brasiliensis. All single-locus methods except for two unequivocally proposed that ATL should be considered a single broad species. The first exception was the GMYC method based on benA with a CAh setting that divided the ATL into three species: A. vadensis and another two species that are not monophyletic in any multigene phylogeny that are shown in Fig. 1 and 3. The second was the bPTP method based on CaM, which divided ATL into eight species corresponding to A. vadensis, A. eucalypticola, A. tubingensis, a species comprising both A. piperis and A. luchuensis, and four additional species whose delimitation has no support in the multigene phylogeny as they are not monophyletic (Figs 1 and 3).
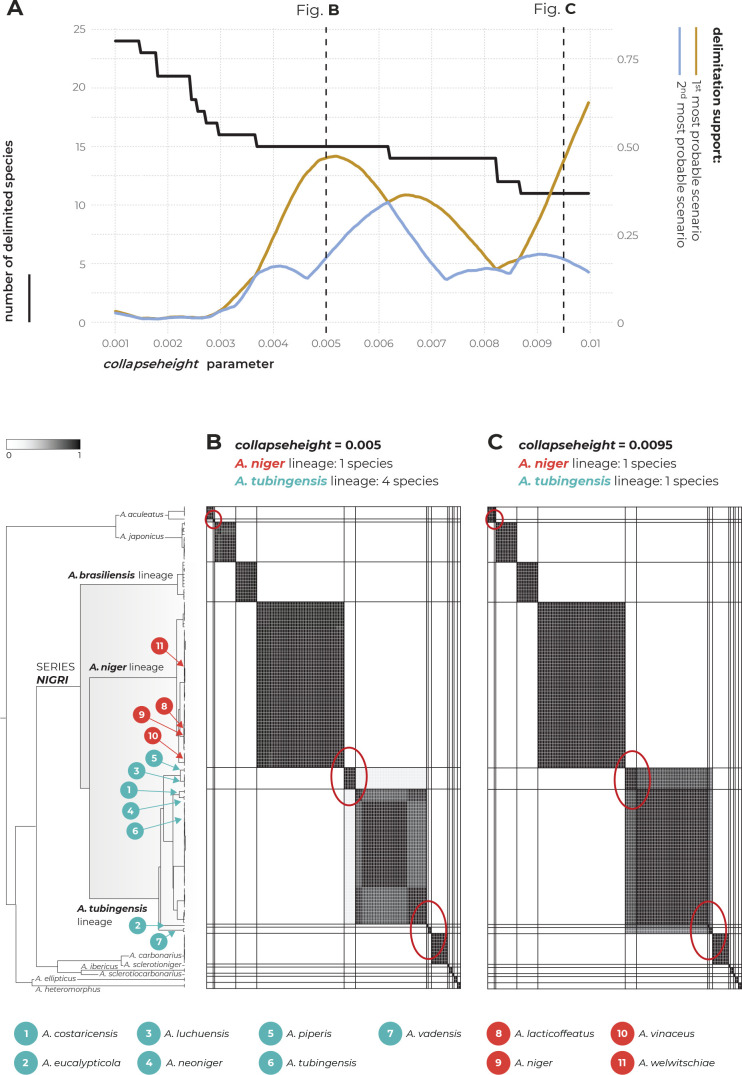
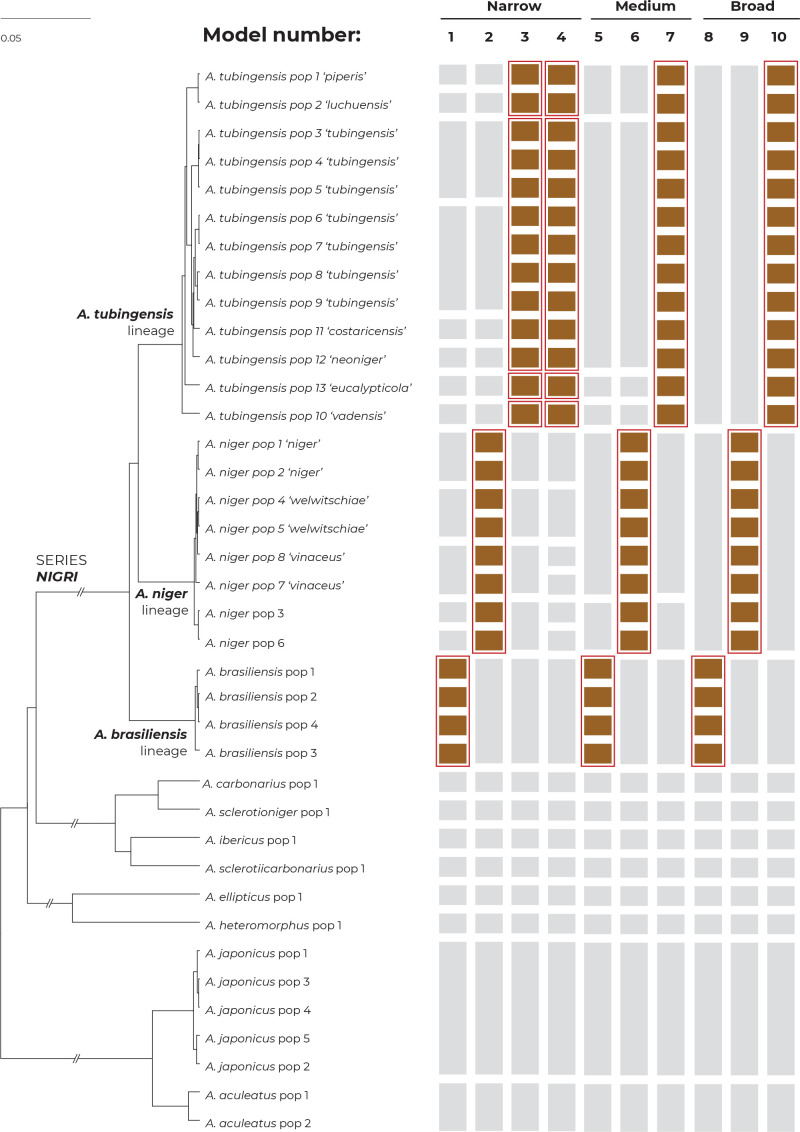
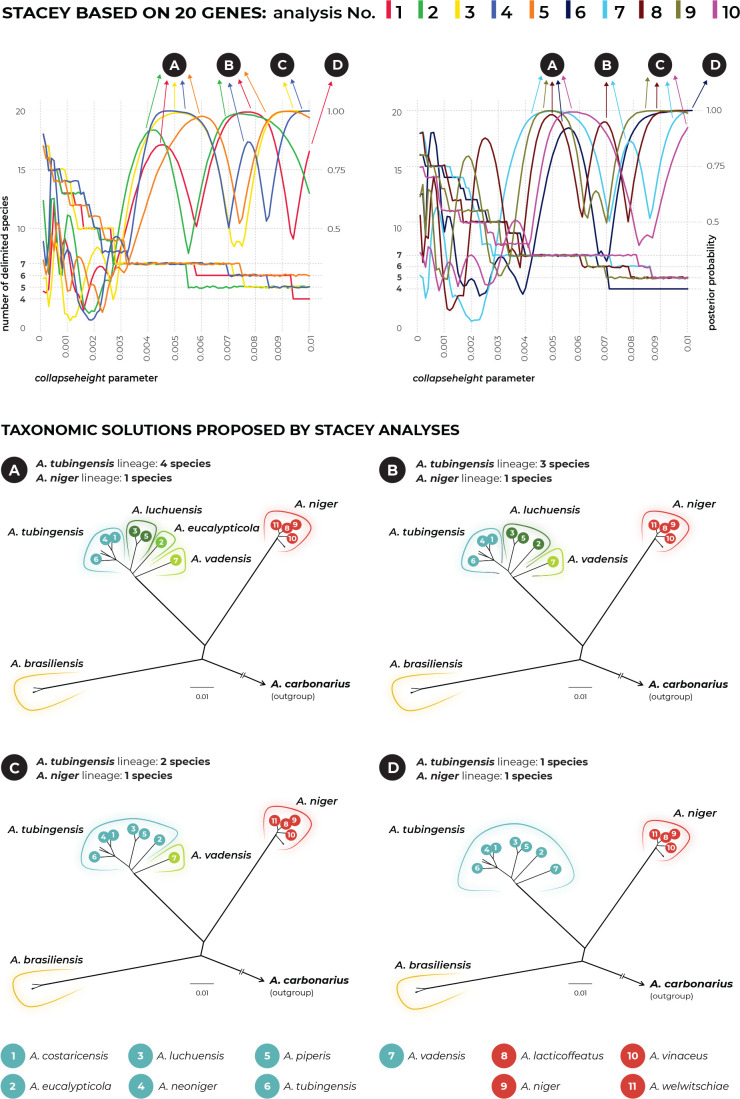
The multilocus method STACEY was performed using two different datasets. The first dataset contained unique multilocus haplotypes from the Nigri series and the second dataset was supplemented with eight additional species outside the Nigri series. The results of both analyses were identical and supported the delimitation of one or four species in the ATL depending on the value of the collapseheight parameter. Detailed results of STACEY based on the second dataset are presented in Fig. 6, where subfigure A illustrates the effect of the collapseheight parameter value on the number of delimited species. On the y-axis on the left side, there is the number of delimited species with the given collapseheight value (black line). The support for the most likely scenario (yellow line), and the support for the second most likely scenario (blue line) are shown; other less supported scenarios were omitted. The vertical dashed lines in subfigure A represent the scenarios illustrated in detail in Fig. 6B and Fig. 6C in the form of similarity matrices showing posterior probabilities of each pair of isolates being included in the same species. There are three main scenarios that gained reasonable support and which delimited 15 (collapseheight = ~0.004–0.006), 14 (collapseheight = ~0.006–0.008) and 11 species (collapseheight = ~0.009 and higher) (Fig. 6), corresponding to 3 or 7 species in the series Nigri. In all three scenarios, the ANL and ABL were always resolved as single species. At collapseheight of ~0.005, four species were delimited in the ATL: (1) A. costaricensis + A. neoniger + A. tubingensis; (2) A. luchuensis + A. piperis; (3) A. eucalypticola; and (4) A. vadensis. When the collapseheight increases and reaches values in the range 0.006–0.008, the support for the delimitation of multiple species in the ATL decreases, but four species are still supported and not lumped together. The only difference in the scenario with 14 species compared to that with 15 species is outside series Nigri, namely, A. aculeatus is delimited as one or two species, respectively. In the scenario with 11 species and a collapseheight of approximately 0.009 and higher, the whole A. tubingensis lineage is delimited as a single species.
Fig. 6.
The results of species delimitation by using the STACEY method based on the three loci: benA, CaM and RPB2. (A) Dependence of the delimitation results on the collapseheight parameter. The black solid line represents the number of delimited species (left y-axis) depending on the changing value of the collapseheight parameter (x-axis). The yellow line represents the probability (range from 0 to 1, right y-axis) of the most likely scenario at specific collapseheight value. The blue line represents the probability of the second most probable scenario at a specific collapseheight value; curves representing other less probable scenarios are omitted. Dashed vertical lines mark two values of the collapseheight parameter (0.005 and 0.0095) whose results are shown in detail by similarity matrices in subfigures B and C. The similarity matrices give the posterior probability of every two isolates belonging to the same multispecies coalescent cluster (tentative species). The darkest black shade corresponds to a posterior probability of 1, while the white colour is equal to 0. Thicker horizontal and vertical lines in the similarity matrices delimit species or their populations that gained delimitation support in some scenarios.
Species delimitation using DELINEATE
The species hypotheses were independently tested in DELINEATE software, where we set up 10 different models. These models were divided into three categories based on the broadness of the fixed species predefined within the ANL and ATL: narrow (Models 1–4), medium (Models 5–7) and broad (Models 8–10). The results are summarised in Fig. 7, where every column represents one model with individual populations either assigned into species (grey bars = predefined species), or left unassigned and free to be delimited (brown bars). The red frames show the resulting solution proposed by DELINEATE for every model, i.e., an unassigned population either recognized as a separate species, merged with other unassigned population(s), or merged with some predefined species.
Fig. 7.
Species delimitation using DELINEATE software. In total, ten models of species boundaries were set up and tested. The brown bars represent unassigned populations left free to be delimited, while the grey bars represent the predefined species. The resulting solutions suggested by DELINEATE are depicted by red frames around the bars. The populations were delimited by using BPP software; isolates belonging to each population are listed in Supplementary Table S2. The displayed tree was calculated in starBEAST based on the benA, CaM and RPB2 loci and is only used for the comprehensive presentation of the results from different models.
Models 1, 5 and 8 focused on delimitation within the ABL, where four populations were recognized by BPP software. In all three models, these populations were left unassigned, and they were always lumped to form a single broad species regardless of whether the species in the ANL and ATL were predefined as narrow or broad.
Models 2, 6 and 9 focused on the ANL, in which all populations were left free to be delimited, while the populations of the ATL were segregated into eight narrow species (Model 2), four broader species (Model 6) and one broad species (Model 10). In all of these settings, the ANL was always recognized as a single broad species regardless of the species number and species limits in the ATL.
More variable delimitation results were obtained in the ATL. Models 3 and 4 simulated situations where A. niger populations (pop1 to pop8) were segregated into four and six species, respectively. In these cases, the ATL populations (pop1 to pop13) were arranged into four species as follows: (1) A. piperis + A. luchuensis; (2) A. tubingensis + A. costaricensis + A. neoniger; (3) A. eucalypticola; and (4) A. vadensis. In Models 7 and 10, the populations of the ANL were predefined into two and single species, respectively. In both cases, the ATL was delimited as a single species.
STACEY analysis using genomic data
To analyse species limits using MSC methods on the genome scale, we randomly selected 200 single-copy orthologous genes from the genomes. The alignments of these genes were randomly distributed into ten datasets each containing 20 genes that were analysed separately (Supplementary Table S4). The results are summarized in Fig. 8. The upper part of the figure shows the course of each analysis using different colours and the dependence of the delimitation results on the changing value of the collapseheight parameter. For every analysis, there are two curves, the first representing the number of delimited species (left y-axis) and the second representing the probability of the most likely species delimitation scenario at a specific collapseheight value (ranging from 0 to 1, right y-axis).
Fig. 8.
Species delimitation results from ten independent STACEY analyses, each based on 20 genes randomly selected from genomes of series Nigri species (n = 30; plus one genome of A. carbonarius as the outgroup). The upper part of the figure shows the dependence of the delimitation results on the changing value of the collapseheight parameter; each analysis is designated with a different colour. There are two curves for each analysis, the first representing the number of delimited species (left y-axis) and the second representing the probability of the most likely species delimitation scenario at a specific collapseheight value (ranging from 0 to 1, right y-axis). Every analysis proposed two to three taxonomic solutions having similar probabilities. In total, there were four possible scenarios (A, B, C, D) differing in the number of delimited species within the A. tubingensis lineage (one to four species). These delimitation results are schematically shown in the lower part of the figure. The list of genes used in every analysis can be found in Supplementary Table S4.
Surprisingly, every analysis proposed two to three taxonomic solutions having high probabilities (two to three major peaks). There were four possible scenarios (A, B, C, D) resulting in 3–6 delimited species in the Nigri series. However, none of these analyses supported the division of the ANL or ABL into more species. Thus, all of these scenarios only differed in the number of delimited species in the ATL, ranging from one to four delimited species. These delimitation results are schematically shown in the lower part of Fig. 8. All ten analyses supported the scenario with four species: (1) A. costaricensis + A. neoniger + A. tubingensis; (2) A. luchuensis + A. piperis; (3) A. eucalypticola; and (4) A. vadensis. In addition, six analyses supported scenarios with three or two species in the ATL, and only two analyses lumped all species into one at high collapseheight values (Fig. 8).
Intraspecific genetic diversity in Aspergillus and series Nigri
To determine whether the intraspecific genetic variability in the redefined species in the series Nigri falls within the range commonly found in Aspergillus, we made an intraspecific diversity comparison across Aspergillus (basic data in Supplementary Table S6). We downloaded sequences of the benA, CaM and RPB2 genes from 34 species outside section Nigri (fewer common markers, such as Mcm7, Tsr1 and Act, were available for only some species). The criteria for inclusion were as follows: (1) species boundaries were re-examined using MSC methods, and (2) the strains of the species were obtained from at least three countries (sufficient sampling).
The results of the comparison (Fig. 9) showed that dissimilarites in the common taxonomic markers were usually lower than 4 % except in benA, where a higher degree of dissimilarity was found occasionally. This was mainly caused by long intronic regions in benA. However, the two highest dissimilarity values in benA belong to A. vitricola (sect. Restricti) and A. subalbidus (sect. Candidi), which could each be a complex of two species because the species delimitation results were ambiguous (Sklenář et al. 2017, Glässnerová et al. 2022). Thus, the real maximum sequence dissimilarity in benA may be lower than that displayed in Fig. 9.
Fig. 9.
Jitter plot showing maximum sequence dissimilarity between isolates of the same Aspergillus species whose species limits have been delimited using methods based on a multispecies coalescent model. Only species represented by isolates from at least three countries were included to ensure relevant sampling and thus representative intraspecific genetic variability. In total, 34 species were included that were classified outside section Nigri. The data from benA, CaM and RPB2 were collected from all 34 species, while less common markers (minichromosome maintenance factor 7 - Mcm7, ribosome biogenesis protein - Tsr1 and actin - Act) were available for only a limited number of species. Species belonging to the series Nigri are designated with coloured circles. The basic data used for construction of the jitter plot are listed in the Supplementary Table S6.
The intraspecific variability in the broadly defined A. niger (corresponding to the whole ANL) and in A. brasiliensis falls comfortably within the range frequently found in Aspergillus. The intraspecific genetic diversity of the broadly defined A. tubingensis in benA and CaM is higher than that commonly found in Aspergillus, while the variability within the narrowly defined A. tubingensis (excluding A. luchuensis/A. piperis, A. eucalypticola and A. vadensis) is within the range common for Aspergillus (Fig. 9).
TAXONOMY
The following list contains the names of accepted species in the series Nigri and their synonyms. Illegitimate and invalid names are also included as some of them are still used in the applied sphere. This list builds on previously published lists of accepted species and their synonyms in section Nigri (Thom & Church 1926, Thom & Raper 1945, Raper & Fennell 1965, Al-Musallam 1980, Kozakiewicz 1989). Compared to previous lists, this list is primarily based on available molecular data. If this data is missing, the names are included in the sub-list of unresolved or doubtful names.
One of the main goals of this list is to point out that the taxonomy of series Nigri is still unresolved and to stimulate further research to resolve names for which original material is available in the form of cultures or herbarium specimens. Some of these names may still threaten the stability of the taxonomy of series Nigri. Most importantly, some of the synonyms or unresolved names compete for priority with economically significant species such as A. tubingensis, A. luchuensis or even A. niger. Althoug A. niger Tiegh. 1867 was conserved against Ustilago ficuum Reichard 1867 and Ustilago phoenicis Corda 1840 (Kozakiewicz et al. 1992), it can still be predated by A. phaeocephalus Durieu & Mont. 1848 (no specimens available), A. nigrescens C.P. Robin 1853 (no specimens available) and A. nanus Mont. 1856 whose type is preserved in the Muséum National d’Histoire Naturelle in France but no DNA data are available. The situation is even more complicated with A. tubingensis Mosseray 1934. This species is conspecific with several older species names, such as A. pulverulentus (basionym Sterigmatocystis pulverulenta McAlpine 1896), A. schiemanniae Thom 1916, A. elatior Mosseray 1934, A. pseudoniger Mosseray 1934 and possibly some other species listed among unresolved species. Aspergillus tubingensis is the most frequently used name compared to other competing names and members of the International Commission of Penicillium and Aspergillus (ICPA) are thus currently in discussions about its possible conservation.
Accepted species and their synonyms in Aspergillus section Nigri series Nigri
1. Aspergillus brasiliensis Varga, Frisvad & Samson, Int. J. Syst. Evol. Microbiol. 57: 1929. 2007. MycoBank MB 510581. — Type: CBS 101740. Ex-type: CBS 101740 = IMI 381727 = IBT 101740. DNA barcodes: ITS = FJ629321; benA = FJ629272; CaM = FN594543; RPB2 = EF661063.
2. Aspergillus eucalypticola Varga, Frisvad & Samson, Stud. Mycol. 69: 9. 2011. MycoBank MB 560387. — Type: CBS H-20627. Ex-type: CBS 122712 = IBT 29274. DNA barcodes: ITS = EU482439; benA = EU482435; CaM = EU482433; RPB2 = MN969070.
3. Aspergillus luchuensis Inui, J. Coll. Sci. Imp. Univ. Tokyo 15: 469. 1901. MycoBank MB 151291. — Type: CBS H-24280. Ex-type: CBS 205.80 = NBRC 4281 = KACC 46772 = IFM 47726 = RIB 2642. DNA barcodes: ITS = JX500081; benA = JX500062; CaM = JX500071; RPB2 = MN969081.
Synonym: Aspergillus perniciosus Inui, J. Coll. Sci. Imp. Univ. Tokyo 15: 473. 1901. [no MycoBank record]. Synonymized by Hong et al. (2013). No living culture available.
Synonym: Aspergillus awamori Nakaz., Rep. Govt. Res. Inst. Formosa: 1. 1907. [MycoBank MB 119955]. Synonymized by Hong et al. (2014). Culture ex-type: JCM 2261 = IAM 2112 = KACC 47234.
Synonym: Aspergillus velutinus Mosseray, La Cellule 43: 252. 1934. [MycoBank MB 255342]. The benA sequence (FJ629283) derived from the ex-type strain CBS 139.48 (= NRRL 4877 = CCM F-286 = VKM F-2084 = WB 4877) is identical to the ex-type of A. luchuensis.
Synonym: Aspergillus aureus var. acidus Nakaz., Simo & A. Watan., Bull. Agric. Chem. Soc. Japan 12: 960. 1936. [MycoBank MB 493809]. Not validly published [Art. 39.1; Turland et al. (2018)]. Synonymized by Hong et al. (2013). Culture ex-type: NBRC 4121 = NRRL 4796 = CBS 564.65 = KACC 45131.
Synonym: Aspergillus aureus var. pallidus Nakaz., Simo & A. Watan., Bull. Agric. Chem. Soc. Japan 12: 961. 1936. [MycoBank MB 372388]. Not validly published [Art. 39.1; Turland et al. (2018)]. Synonymized by Hong et al. (2014). Culture ex-type: NBRC 4123 = NRRL 4797 = CBS 565.65 = KACC 47004.
Synonym: Aspergillus awamori var. piceus Nakaz., Simo & A. Watan., Bull. Agric. Chem. Soc. Japan 12: 956. 1936. [MycoBank MB 250923]. Not validly published [Art. 39.1; Turland et al. (2018)]. Synonymized by Hong et al. (2013). Representative strain: CBS 119.52 = NRRL 4846.
Synonym: Aspergillus kawachii Kitahara & Yoshida. J. Ferment. Technol. 27. 1949. [No MycoBank record]. Not validly published [Art. 39.1; Turland et al. (2018)]. Synonymized by Hong et al. (2013). Culture ex-type: NBRC 4308 = CBS 117.80 = NRRL 4886 = KACC 46771. Hong et al. (2013) and Ban et al. (2021) examined phylogenetic position of A. kawachii based on strains KACC 46771 and NBRC 4308, respectively. Their sequences are identical with the ex-type of A. kawachii in contrast to the sequences from strain CBS 117.90 that belong to the ANL (Table 1). Our conclusion is that the strain CBS 117.90 is contaminated with A. niger.
Synonym: Aspergillus citricus var. pallidus (Nakaz., Simo & A. Watan.) Kozak., Mycol. Pap. 161: 114. 1989. [MycoBank MB 127748]. Not validly published – basionym A. aureus var. pallidus Nakaz., Simo & A. Watan. is not a validly published [Art. 41.5; Turland et al. (2018)].
Synonym: Aspergillus inuii Sakag., Iizuka & M. Yamaz., J. Agric. Chem. Soc. Japan 3: 97. 1950. [Mycobank MB 309227]. Not validly published [Art. 39.1; Turland et al. (2018)]. Synonymized by Hong et al. (2014). Culture ex-type: JCM 22302 = IAM 2255 = CBS 125.52 = KACC 47235.
Synonym: Aspergillus nakazawae [as nakazawai] Sakag., Iizuka & M. Yamaz., J. Agric. Chem. Soc. Japan 4: 3. 1950. [Mycobank MB 309232]. Not validly published [Art. 39.1; Turland et al. (2018)]. Synonymized by Hong et al. (2014). Culture ex-type: NRRL 4750 = CBS 128.52 = KACC 47005.
Synonym: Aspergillus foetidus var. acidus (Nakaz., Simo & A. Watan.) Raper & Fennell, The Genus Aspergillus: 326. 1965. [MycoBank MB 353273]. Synonymized by Hong et al. (2013). Not validly published – basionym A. aureus var. acidus Nakaz., Simo & A. Watan. is not a validly published name [Art. 41.5; Turland et al. (2018)].
Synonym: Aspergillus foetidus var. pallidus (Nakaz., Simo & A. Watan.) Raper & Fennell, The Genus Aspergillus: 325. 1965. [MycoBank MB 353274]. Synonymized by Hong et al. (2014). Not validly published – basionym A. aureus var. pallidus Nakaz., Simo & A. Watan. is not a validly published name [Art. 41.5; Turland et al. (2018)].
Synonym: Aspergillus niger var. awamori (Nakaz.) Al-Musallam, Revision of the black Aspergillus species: 60. 1980. Doctoral diss., Rijksuniversiteit Utrecht, Netherlands. [MycoBank MB 118405]. Hong et al. (2014) showed that the ex-type strain NRRL 4948 (= CBS 557.65 = KACC 46996 = ATCC 16877) is closely related to the ex-type of A. welwitschiae (ANL) and is synonymized here with A. niger.
Synonym: Aspergillus acidus Kozak., Mycol. Pap. 161: 110. 1989. [MycoBank MB 127765]. Synonymized by Hong et al. (2013). Culture ex-type: KACC 45131 = CBS 564.65 = ATCC 16874 = NBRC 4121 = IMI 104688 = NRRL 4796.
Synonym: Aspergillus coreanus Yu T.S. et al., J. Microbiol. Biotechnol. 14:182. 2004. [No Mycobank record]. Illegitimate name [Art. 54.1; Turland et al. (2018)] and nomen nudum. Synonymized by Hong et al. (2013). Authentic strain: KACC 41731 = CBS 119384.
Synonym: Aspergillus piperis Samson & Frisvad, Stud. Mycol. 50: 57. 2004. [MycoBank MB 500009]. Synonymized in this study.
Synonym: Aspergillus luchuensis mut. kawachii (Kitahara & Yoshida) S.B. Hong, O. Yamada & R.A. Samson, Appl. Microbiol. Biotechnol. 98: 560. 2014. [No Mycobank record]. Not validly published: basionym was not validly published [Art. 41.5; Turland et al. (2018)] and protologue does not include citation of the identifier issued for the name by a recognized electronic repository, e.g. MycoBank [Art. F.5.1; Turland et al. (2018)].
4. Aspergillus niger Tiegh., Ann. Sci. Nat., Bot. ser. 5, 8: 240. 1867. MycoBank MB 284309. Nomen conservandum taking precedence over Ustilago phoenicis Corda and Ustilago ficuum Reichard which were published earlier (Kozakiewicz et al. (1992). — Type: CBS 554.65. Ex-type: CBS 554.65 = NRRL 326 = ATCC 16888 = IFO 33023 = IHEM 3415 = IMI 050566ii = IMI 50566 = JCM 10254 = QM 9270 = QM 9946 = Thom 2766 = WB 326. DNA barcodes: ITS = EF661186; BenA = EF661089; CaM = EF661154; RPB2 = EF661058.
Synonym: Ustilago ficuum Reichardt, Verh. Zool.-Bot. Ges. Wien 17: 335. 1867. [MycoBank MB 187740]. Nomen rejiciendum, rejected name in favour of A. niger Tiegh. – see Kozakiewicz et al. (1992). The ex-type strain IHEM 3710 (= WB 364 = NRRL 364 = IMI 91881 = CBS 555.65= ATCC 16882) derived from the type IMI 91881 (Kozakiewicz et al. 1992) is located in the ANL (Fig. 1).
Synonym: Eurotium nigrum (Tiegh.) de Bary, Beiträge zur Morphologie und Physiologie der Pilze: 21. 1870. [No MycoBank record]. Basionym: A. niger Tiegh.
Synonym: Sterigmatocystis nigra (Tiegh.) Tiegh., Bull. Soc. Bot. France 24: 102. 1877. [MycoBank MB 231646]. Basionym: A. niger Tiegh.
Synonym: Ustilago welwitschiae Bres. in Sacc, Bol. Soc. Brot. 11: 68. 1893. [MycoBank MB 176748]. Synonymized in this study.
Sterigmatocystis ficuum (Reichardt) Henn., Hedwigia 34: 86. 1895. [MycoBank MB 100573]. See Ustilago ficuum Reichardt (basionym).
Synonym: Sterigmatocystis welwitschiae (Bres.) Henn. in H. Baum, Kunene-Zambesi Expedit., Berlin: 168. 1903. [MycoBank MB 233714]. See Ustilago welwitschiae Bres. in Sacc. (basionym).
Synonym: Aspergillus batatas [as batatae] Saito, Zentralbl. Bakteriol. Parasitenkn. Abt. II, 18: 34. 1907. [MycoBank MB 214829]. Synonymized by Hong et al. (2014). Culture ex-type: NRRL 4785 = CBS 115.34 = NBRC 4034 = KACC 46995.
Synonym: Aspergillus welwitschiae (Bres.) Henn. in Wehmer, Zentrbl. Bakteriol. ParasitK. Abt. II, 18: 294. 1907. [MycoBank MB 490584]. See Ustilago welwitschiae Bres. in Sacc. (basionym).
Synonym: Aspergillopsis nigra (Tiegh.) Speg., Anales Mus. Nac. Hist. Nat. Buenos Aires, ser. 3, 13: 435. 1911. [MycoBank MB 210007]. Basionym: A. niger Tiegh.
Synonym: Rhopalocystis nigra (Tiegh.) Grove, J. Econ. Biol. 6: 41. 1911. [MycoBank MB 432029]. Basionym: A. niger Tiegh.
Synonym: Aspergillus niger var. altipes E. Schiemann, Z. Indukt. Abstamm. Vererbungsl. 8: 1–35. 1912. [Mycobank MB 493817]. This variety belongs to the ANL based on the ITS sequence (MH854603) derived from the ex-type strain CBS 102.12 (= ATCC 10549 = IBT 19348 = IBT 26343 = IFO 4067 = MUCL 13608 = NRRL 4863 = WB 4863).
Synonym: Sterigmatocystis batatas [as batatae] (Saito) Sacc. Syll. Fung. 22: 1261. 1913. [MycoBank MB 209678]. See A. batatas Saito (basionym).
Synonym: Aspergillus aureus Nakazawa, Rep. Govt. Res. Inst. Formosa 4: 215–222. 1915. [No MycoBank record]. Synonymized by Hong et al. (2014). Culture ex-type: CBS 121.28 = NRRL 4784 = NBRC 4031 = RIB 2014 = KACC 47003 = IFO 4031 = IMI 104687 = WB 4784. Illegitimate name [Art. 54.1; Turland et al. (2018)]. Not A. aureus Berk. 1836 [MycoBank MB 214770].
Synonym: Aspergillus ficuum (Reichardt) Thom & Currie, J. Agric. Res. 7: 12. 1916. [MycoBank MB 101909]. See Ustilago ficuum Reichardt (basionym).
Synonym: Aspergillus citricus Mosseray, Ann. Univ. Sci. Budap. Rolando Eotvos Nominatae Biol. 43: 262. 1934. [MycoBank MB 251457]. Synonymized by Frisvad et al. (2011) and Houbraken et al. (2014). The genotype of the ex-type strain IHEM 5622 (= CBS 126.48 = WB 337 = NRRL 337 = NCTC 1692 = MUCL 28130 = IMI 15954 = IFO 6428 = DSM 734 = CCRC 30206 = ATCC 10254) is identical to the ex-type strains of A. niger (Fig. 1).
Synonym: Aspergillus bainieri Mosseray, Ann. Soc. Sci. Brux. 54: 79. 1934. [No MycoBank record]. The description is based on the same strain like A. longobasidia Bainier ex Mosseray (CBS 121.48).
Synonym: Aspergillus longobasidia Bainier ex Mosseray, La Cellule 43: 227. 1934. [MycoBank MB 253086]. The ex-type strain CBS 121.48 (= NRRL 4857) belongs to the ANL (Hong et al. 2013).
Synonym: Aspergillus pseudocitricus Mosseray, La Cellule 43: 228. 1934. [Mycobank MB 254148]. The ex-type strain CBS 127.48 (= NRRL 4869) can be identified as A. niger based on benA (Hong et al. 2013).
Synonym: Aspergillus niger mut. fusca Blochwitz, Ann. Mycol. 32: 87. [No MycoBank record]. The ex-type strain CBS 113.33 (= NRRL 4864 = ATCC 10548 = WB 4864) can be identified as A. niger based on benA (Hong et al. 2013).
Synonym: Aspergillus hennebergii Blochwitz, Ann. Mycol. 33: 238. 1935. [MycoBank MB 266969]. Not validly published [Art. 39.1; Turland et al. (2018)]. The ex-type strain CBS 118.35 (= NRRL 4883 = CECT 2801 = QM 9704 = WB 4883 = VKM F-4392) can be identified as A. niger based on benA (Hong et al. 2013).
Synonym: Aspergillus aureus var. brevior [as brevius] Nakaz., Simo & A. Watan. Bull. Agric. Chem. Soc. Japan 12: 962. 1936. [MycoBank MB 493810]. Not validly published [Art. 39.1; Turland et al. (2018)]. The representative strain CBS 113.52(= WB 4841) can be identified as A. niger based on benA (Hong et al. 2013).
Synonym: Aspergillus awamori var. fuscus Nakaz., Simo & A. Watan., Bull. Agric. Chem. Soc. Japan 12: 959. 1936. [MycoBank MB 250921]. Not validly published [Art. 39.1; Turland et al. (2018)]. Synonymized by Hong et al. (2013, 2014). Representative strain: CBS 117.52 = NRRL 4844 = WB 4844.
Synonym: Aspergillus awamori var. minimus Nakaz., Simo & A. Watan., Bull. Agric. Chem. Soc. Japan 12: 955. 1936. [MycoBank MB 250922]. Not validly published [Art. 39.1; Turland et al. (2018)]. Synonymized by Hong et al. (2013). Representative strain: CBS 118.52 [the whole-genome sequence was generated by Seekles et al. (2022)].
Synonym: Aspergillus miyakoensis Nakaz., Simo & A. Watan., Bull. Agric. Chem. Soc. Japan 12: 963. 1936. [MycoBank MB 253392]. Not validly published [Art. 39.1; Turland et al. (2018)]. Synonymized by Hong et al. (2014). Culture ex-type: NRRL 4859 = CBS 117.51 = KACC 46998.
Synonym: Aspergillus pyri W.H. English, Res. Stud. State Coll. Wash. 8: 127. 1940. [MycoBank MB 453796]. Nomen nudum. Subsequent work by English led to recognition of the name as a synonym of A. niger and withdrawal of the name (Thom & Raper 1945, Raper & Fennell 1945).
Synonym: Aspergillus foetidus Thom & Raper, A manual of the Aspergilli: 219. 1945. [Mycobank MB 284300]. Synonymized by Varga et al. (2011). Culture ex-type: CBS 121.28 = NRRL 4784 = NBRC 4031 = RIB 2014 = KACC 47003 = IFO 4031 = IMI 104687 = WB 4784.
Synonym: Aspergillus usamii Sakag., Iizuka & M. Yamaz., J. Appl. Mycol. Japan 4: 1. 1950. [No MycoBank record]. Not validly published [Art. 39.1; Turland et al. (2018)]. Validly published later – see A. usamii Sakag., Iizuka & M. Yamaz. ex Iizuka & K. Sugiy.
Synonym: Aspergillus usamii mut. shirousamii Iizuka & Yamaguchi. J. Gen. Appl. Microbiol. 1: 194–200. 1954. [No MycoBank record]. Not validly described. Synonymized by Hong et al. (2014). Culture ex-type: NRRL 4889 = KACC 47001.
Synonym: Aspergillus usamii Sakag., Iizuka & M. Yamaz. ex Iizuka & K. Sugiy., J. Jap. Bot.: 232. 1965. [MycoBank MB 326664]. Synonymized by Yamada et al. (2011) and Hong et al. (2014). Culture ex-type: CBS 139.52 = NRRL 4760 = ATCC 11364 = ATCC 14331 = NBRC 4388 = RIB 2602 = IAM 2185 = KACC 47000 = IFO 4388 = QM 8164 = WB 4760.
Synonym: Aspergillus niger f. hennebergii Blochwitz ex Al-Musallam, Revision of the black Aspergillus species: 68. 1980. Doctoral diss., Rijksuniversiteit Utrecht, Netherlands. [MycoBank MB 118404]. See A. hennebergii Blochwitz (basionym). Synonym: Aspergillus niger var. usamii (Sakag., Iizuka & M. Yamaz. ex Iizuka & K. Sugiy.) Al-Musallam, Revision of the black Aspergillus species: 64. 1980. Doctoral diss., Rijksuniversiteit Utrecht, Netherlands. [MycoBank MB 118400]. See A. usamii Sakag., Iizuka & M. Yamaz. ex Iizuka & K. Sugiy. (basionym).
Synonym: Aspergillus niger var. ficuum (Reichardt) Kozak., Mycol. Pap. 161: 112. 1989. [MycoBank MB 127745]. See Ustilago ficuum Reichardt (basionym).
Synonym: Aspergillus lacticoffeatus Frisvad & Samson, Stud. Mycol. 50: 52. 2004. [Mycobank MB 500008]. Synonymized by Varga et al. (2011) and confirmed in this study.
Synonym: Aspergillus vinaceus Ferranti et al., J. Fungi 6: 371 - p14. 2020. [MycoBank MB 833399]. Synonymized in this study.
5. Aspergillus tubingensis Mosseray, La Cellule 43: 245. 1934. [MycoBank MB 255209]. — Type: CBS H-24288. Ex-type: NRRL 4875 = CBS 133056 = QM 8904 = WB 4875. DNA barcodes: ITS = EF661193; benA = EF661086; CaM = EF661151; RPB2 = EF661055.
Synonym: Sterigmatocystis pulverulenta McAlpine, Agric. Gaz. N.S.W., Sydney 7: 302. 1896 (1897). [MycoBank MB 194998]. Synonymized by Houbraken et al. (2014). The ex-type strain IHEM 5615 (= CBS 558.65 = CBS 115.48 = MUCL 13592 = MUCL 15630 = NRRL 4851 = WB 4851 = IMI 211396 = ATCC 16879) is close to the ex-type of A. tubingensis (Fig. 1).
Synonym: Aspergillus pulverulentus (McAlpine) Wehmer, Zentralbl. Bakteriol. Parasitenkd., Abt. II, 18: 394. 1907. [MycoBank MB 121243]. See Sterigmatocystis pulverulenta McAlpine (basionym).
Synonym: Aspergillus cinnamomeus E. Schiemann, Z. Indukt. Abstamm. Vererbungsl. 8: 1–35. 1912. [MycoBank MB 122195]. The benA sequence (FJ629307) derived from the ex-type strain CBS 103.12 = ATCC 1027 = IBT 16906 = IBT 28139 = IBT 29892 = IFO 4043 = IMI 016148 = LSBH Ac42 = MZKI A-76 = NCTC 3774 = NRRL 348 = QM 326 = Thom 3534B = WB 348) is identical with the ex-type of A. tubingensis.
? Synonym: Aspergillus fuscus E. Schiem., Z. Indukt. Abstamm. Vererbungsl. 8: 1–35. 1912. [MycoBank MB 450741]. Illegitimate name [Art. 54.1; Turland et al. (2018)]. Not A. fuscus Bonord. 1861 [MycoBank MB 206602]. Illegitimate name A. fuscus was replaced by nomen novum A. schiemanniae Thom 1916 (see A. schiemanniae).
? Synonym: Aspergillus proteus E. Schiem., Z. Indukt. Abstamm. Vererbungsl. 8: 1–35. 1912. [No MycoBank record]. Illegitimate name – not accepted by the author itself in the original publication [Art. 36.1; Turland et al. (2018)].
Synonym: Aspergillus pulverulentus (McAlpine) Thom, J. Agric. Res. 7: 11. 1916. [No MycoBank record]. Illegitimate name [Art. 54.1; Turland et al. (2018)] - a later homonym of A. pulverulentus (McAlpine) Wehmer 1907.
? Synonym: Aspergillus schiemanniae Thom [as schiemanni], J. Agric. Res. 7: 11. 1916. [MycoBank MB 102113]. The ITS sequence (MH854950) derived from the ex-type strain CBS 122.28 (= ATCC 1040 = NRRL 361 = IFO 4091 = IMI 091895 = IMI 16269 = LSHB AC.37 = NCTC 3781 = QM 327 = Thom 3534C = WB 361) belongs to the ATL.
Synonym: Aspergillus elatior Mosseray, Ann. Univ. Sci. Budap. Rolando Eotvos Nominatae Biol. 43: 253. 1934. [MycoBank MB 251950]. The ex-type strain IHEM 5615 (= CBS 558.65 = CBS 115.48 = MUCL 13592 = MUCL 15630 = NRRL 4851 = WB 4851 = IMI 211396 = ATCC 16879) is similar to the ex-type of A. tubingensis (Fig. 1).
Synonym: Aspergillus pseudoniger Mosseray, La Cellule 43: 256. 1934. [MycoBank MB 472168]. The ex-type strain IHEM 4379 (= CBS 128.48 = IMI 313491 = WB 4870 = MUCL 31312) is similar to A. tubingensis (Fig. 1).
Synonym: Aspergillus niger mut. cinnamomeus (E. Schiemann) Thom & Raper, A manual of the Aspergilli: 223. 1945. [MycoBank MB 440825]. See A. cinnamomeus E. Schiemann (basionym).
? Synonym: Aspergillus niger mut. schiemanniae (Thom) Thom & Raper [as schiemanni], A manual of the Aspergilli: 224. 1945. [MycoBank MB 123940]. See A. schiemanniae Thom (basionym).
Synonym: Aspergillus saitoi Sakag., Iizuka & M. Yamaz., J. Appl. Mycol. Japan 3: 68. 1950. [No MycoBank record]. Not validly published [Art. 39.1; Turland et al. (2018)]. Validly published later – see A. saitoi Sakag., Iizuka & M. Yamaz. ex Iizuka & K. Sugiy.
Synonym: Aspergillus saitoi var. kagoshimaensis Sakag., Iizuka & M. Yamaz., J. Appl. Mycol. Japan 4: 3. 1950. [No MycoBank record]. Not validly published [Art. 39.1; Turland et al. (2018)]. Validly published later – see A. saitoi var. kagoshimaensis Sakag., Iizuka & M. Yamaz. ex Iizuka & K. Sugiy.
Synonym: Aspergillus awamori var. hominis Bat. & Maia, An. Soc. Biol. Pernambuco 15: 186. 1957. [MycoBank MB 351895]. The benA sequence (FJ629318) derived from the ex-type strain CBS 107.55 (= NRRL 4740 = ATCC 12074 = WB 4740) is almost identical (419/420 bp) to the ex-type of A. tubingensis.
Synonym: Aspergillus saitoi Sakag., Iizuka & M. Yamaz. ex Iizuka & K. Sugiy., J. Jap. Bot. 40: 230. 1965. [Mycobank MB 326655]. Synonymized by Hong et al. (2014). Culture ex-type: CBS 136.52 = CBS 552.65 = NRRL 4757 = IAM 2209 = ATCC 11362 = IMI 211395 = KACC 46993 = WB 4757.
Synonym: Aspergillus saitoi var. kagoshimaensis Sakag., Iizuka & M. Yamaz. ex Iizuka & K. Sugiy., J. Jap. Bot. 40: 231. 1965. [MycoBank MB 349043]. Synonymized by Hong et al. (2014). Culture ex-type: CBS 137.52 = NRRL 4758 = IMI 214827 = ATCC 11363 = KACC 46994 = QM 8162 = IAM 2190 = WB 4758.
Synonym: Aspergillus niger f. pulverulentus (McAlpine) Al-Musallam, Revision of the black Aspergillus species: 58. 1980. Doctoral diss., Rijksuniversiteit Utrecht, Netherlands. [MycoBank MB 118403]. See Sterigmatocystis pulverulenta McAlpine (basionym).
Synonym: Aspergillus niger var. pulverulentus (McAlpine) Kozak., Mycol. Pap. 161: 113. 1989. [MycoBank MB 127747]. See Sterigmatocystis pulverulenta McAlpine (basionym).
Synonym: Aspergillus niger var. tubingensis (Mosseray) Kozak., Mycol. Pap. 161: 112. 1989. [MycoBank MB 127746]. Basionym: A. tubingensis Mosseray.
Synonym: Aspergillus costaricensis [as costaricaensis] Samson & Frisvad, Stud. Mycol. 50: 52. 2004. [MycoBank MB 369151]. Synonymized in this study.
Synonym: Aspergillus neoniger Varga, Frisvad & Samson, Stud. Mycol. 69: 16. 2011. [MycoBank MB 560390]. Synonymized in this study.
Synonym: Aspergillus chiangmaiensis S. Khuna, N. Suwannarach & S. Lumyong, Front. Microbiol. 12: 705896 - p6. 2021. [Mycobank MB 830887]. Synonymized in this study.
Synonym: Aspergillus pseudopiperis S. Khuna, N. Suwannarach & S. Lumyong, Front. Microbiol. 12: 705896 - p6. 2021. [MycoBank MB 830888]. Synonymized in this study.
6. Aspergillus vadensis Samson, de Vries, Frisvad & Visser, Antonie van Leeuwenhoek 87: 201. 2005. [MycoBank MB 340234]. — Type: CBS 113365. Ex-type: CBS 113365 = CECT 20584 = IMI 313493 = IBT 24658. DNA barcodes: ITS = AY585549; benA = AY585531; CaM = FN594560; RPB2 = HE984371.
Synonym: Aspergillus vadensis de Vries et al., Appl. Environ. Microbiol. 70: 3954. 2004. [MycoBank MB 560390]. Not validly described (nomen nudum).
Unresolved or doubtful names most probably belonging to the series Nigri
Aspergillus phaeocephalus Durieu & Mont., Exploration scientifique de l’Algérie 1: 342. 1848. [MycoBank MB 179669]. No specimens are available.
Synonym: Sterigmatocystis phaeocephala (Durieu & Mont.) Sacc., Syll. Fung. 4: 76. 1886. [MycoBank MB 233071].
Aspergillus nigrescens C.P. Robin, Histoire naturelle des végétaux parasites: 518. 1853. [MycoBank MB 182212]. Considered either A. fumigatus or A. niger by various authors (Wilhelm 1877, Wehmer 1901, Raper & Fennell 1965), no material is available (species described from pathological specimens only).
Aspergillus nanus Mont., Syll. Gen. Sp. Crypt. (Paris): 300. 1856. [MycoBank MB 193617]. Typus in Muséum National d’Histoire Naturelle, France, Paris (PC); no DNA sequence available. The strains of A. niger var. nanus examined by Al-Musallam (1980) belong either to the ANL or ATL, e.g., CBS 105.47 (MH856174; ATL), CBS 106.47 (FJ629281; ATL), CBS 110.30 (MH855092; ANL), CBS 115.50 (MH856565; ANL), CBS 131.52 (the whole-genome sequence was generated by Seekles et al. (2022); ANL).
Synonym: Aspergillus niger var. nanus (Mont.) Al-Musallam, Revision of the black Aspergillus species: 62. 1980. Doctoral diss., Rijksuniversiteit Utrecht, Netherlands. [MycoBank MB 118406].
Sterigmatocystis antacustica C.E. Cramer, Vierteljahrsschr. Naturforsch. Ges. Zürich 4: 325. 1859. [MycoBank MB 209963].
Synonym: Rhopalocystis antacustica (C.E. Cramer) Grove, J. Econ. Biol.: 41. 1911. [MycoBank MB 503610].
Aspergillus nigricans Wreden, C. R. Hebd. Seances Acad. Sci. 65: 368. 1867. [No MycoBank record]. No specimen is available.
Aspergillus fuliginosus Peck, Bull. Buffalo Soc. Nat. Sci. 1: 69. 1873. [MycoBank MB 208679]. Typus in New York State Museum, USA (NYSf 1265); no DNA sequence available.
Synonym: Sterigmatocystis fuliginosa Bainer, Bull. Soc. Bot. France 28: 78. 1881. [No MycoBank record].
Aspergillus nigricans Cooke, Grevillea 6: 127. 1878. [MycoBank MB 182128]. Illegitimate name. Not A. nigricans Wreden 1867.
Alliospora sapucaya [as sapucayae] Pim, J. Bot. London 21: 234. 1883. [Mycobank MB 215122].
Aspergillus cookei Sacc., Syll. Fung. 4: 71. 1886. [MycoBank MB 206435]. A replaced synonym for illegitimate name A. mucoroides Cooke.
Basionym: Aspergillus mucoroideus Cooke, Grevillea 12: 9. 1883. [MycoBank MB 187861]. Illegitimate name [Art. 54.1; Turland et al. (2018)]. Not A. mucoroides Corda 1838 [MycoBank MB 187771].
Aspergillus subfuscus Johan-Olsen, Nordiskt Med. Ark. 18: 14. 1886. [MycoBank MB 184212].
Basionym: Sterigmatocystis subfusca (Johan-Olsen) Sacc., Syll. Fung. 10: 526. 1892. [MycoBank MB 226258].
Aspergillus nigriceps Berk. & Curt., Grevillea 17: 21. 1888. [No MycoBank record]. A slide from material in the Harvard Collection (Curtis collection, Wright no. 927) showed species inseparable from A. niger (Raper & Fennell 1965).
Aspergillus ustilago Beck in H. Wawra, Itinera Principum S. Coburgi 2: 148. 1888. [MycoBank MB 161832].
Synonym: Sterigmatocystis ustilago (Beck) Sacc., Syll. Fung. 10: 526. 1892. [MycoBank MB 198044].
Sterigmatocystis castanea F. Patt., Bull. Torrey Bot. Club 27: 284. 1900. [MycoBank MB 195664]. Typus in BPI Herbarium (BPI 410863).
Aspergillus gallomyces Calmette, Germ. Pat. Abstr. 129: 164. 1902. [No MycoBank record].
Aspergillus strychni Lindau, Hedwigia 43: 306. 1904. [MycoBank MB 183848].
Synonym: Sterigmatocystis strychni (Lindau) Sacc. & D. Sacc., Syll. Fung. 18: 516. 1906. [MycoBank MB 225824].
Sterigmatocystis pseudonigra Costantin & Lucet, Bull. Soc. Mycol. France 19: 33. 1903. [MycoBank MB 195028]. No specimen available.
Synonym: Aspergillus pseudoniger (Costantin & Lucet) Saincl., Centralbl. Gesammte Forstwesen: 103. 1949. [MycoBank MB 292856]. Illegitimate name. Not A. pseudoniger Mosseray 1934. [MycoBank MB 472168].
Sterigmatocystis luteonigra M.L. Lutz, Bull. Soc. Bot. France 53: 50. 1907. [MycoBank MB 228634].
Synonym: Aspergillus luteoniger (M.L. Lutz) Thom & Church, The Aspergilli: 166. 1926. [MycoBank MB 269972].
Aspergillopsis intermedia Speg., Anales Mus. Nac. Hist. Nat. Buenos Aires, ser. 3, 13: 435. 1911. [MycoBank MB 209760]. Typus deposited in the Herbarium of Museo de La Plata, Argentiva (LPS 12691); no DNA sequence is available. The strains considered A. niger var. intermedius by Al-Musallam (1980) belong either to the ANL or ATL, e.g., CBS 117.32 (MH855233; ATL), CBS 115.29 (MH855018; ATL), CBS 117.52 (MH856951; ANL), CBS 130.52 (FJ629359, FJ629310; ATL), CBS 107.55 (FJ629367, FJ629318; ATL).
Synonym: Aspergillus niger var. intermedius (Speg.) Al-Musallam, Revision of the black Aspergillus species: 66. 1980. Doctoral diss., Rijksuniversiteit Utrecht, Netherlands. [MycoBank MB 118402].
Aspergillus phoenicis (Corda) Thom, J. Agric. Res. 7: 14. 1916. [MycoBank MB 101952].
Basionym: Ustilago phoenicis Corda, Icones fungorum hucusque cognitorum 4: 9. 1840. [MycoBank MB 169060]. Nomen rejiciendum, rejected name in favour of A. niger Tiegh. (Kozakoewict et. al. 1992). Type located in Corda′s herbarium (PRM, Czech Republic) but no sequence derived from the original material is available. Representative strains NRRL 363, NRRL 365 and NRRL 1956 do either belong to ANL or ATL (Houbraken et al. 2014).
Synonym: Sterigmatocystis phoenicis (Corda) Pat. & Delacr., Bull. Soc. Mycol. France 7: 119. 1891. [MycoBank MB 232882].
Synonym: Aspergillus niger var. phoenicis (Corda) Al-Musallam, Revision of the black Aspergillus species: 56. 1980. Doctoral diss., Rijksuniversiteit Utrecht, Netherlands. [MycoBank MB 118401].
Aspergillopsis tropicalis Speg., Bol. Acad. Nac. Cienc. Córdoba 23: 588. 1918. [MycoBank MB 209817].
Aspergillus fumaricus Wehmer ex Thom & Church, The Aspergilli: 181. 1926. [MycoBank MB 122538].
Sterigmatocystis gigantea Mattlet, Ann. Soc. Belge Méd. Trop. 6: 31. 1926. [MycoBank MB 252349].
Synonym: Aspergillus giganteus (Mattlet) C.W. Dodge, Medical Mycology: 629. 1935. [No MycoBank record]. Illegitimate name. Not A. giganteus Wehmer 1901 [MycoBank MB 206765].
Synonym: Aspergillus mattletii Hendr., Publ. Inst. Nat. Étude Agron. Congo Belge 35: 7. 1948. [MycoBank MB 284307].
Aspergillus atropurpureus Blochwitz, Ann. Mycol. 32: 86. 1934. [No MycoBank record]. Illegitimate name. Not A. atropurpureus Zimm. 1902 [MycoBank MB 214593].
Aspergillus biourgei Mosseray, La Cellule 43: 241. 1934. [MycoBank MB 251009].
Aspergillus buntingii Mosseray, La Cellule 43: 236. 1934. [MycoBank MB 251145].
Aspergillus churchii Mosseray, La Cellule 43: 242. 1934. [MycoBank MB 251413].
Aspergillus densus Mosseray, La Cellule 43: 232. 1934. [MycoBank MB 251792].
Aspergillus granulatus Mosseray, La Cellule 43: 249. 1934. [MycoBank MB 252446]. Culture ex-type: CBS 120.48 = NRRL 4855 = WB 4855 (no DNA sequence available).
Aspergillus guttifer Mosseray, La Cellule 43: 235. 1934. [MycoBank MB 252493].
Aspergillus microcephalus Mosseray, La Cellule 43: 225. 1934. [MycoBank MB 253336]. The strain CBS 122.48 is considered the ex-type strain (Al-Musallam 1980) but no DNA sequence is available.
Aspergillus niger var. fermentarius Nakaz., Simo & A. Watan., J. Agric. Chem. Soc. Japan 10: 171. 1934. [No MycoBank record].
Aspergillus olivaceofuscus Mosseray, La Cellule 43: 258. 1934. [MycoBank MB 253675].
Aspergillus praecox Mosseray, Ann. Soc. Sci. Brux. 54: 79. 1934. [No MycoBank record].
Aspergillus pseudoelatior Mosseray, La Cellule 43: 255. 1934. [MycoBank MB 254152].
Aspergillus rutilans Mosseray La Cellule 43: 234. 1934. [MycoBank MB 254522].
Aspergillus sclerotifer Mosseray, Ann. Soc. Sci. Brux. 54: 79. 1934. [No MycoBank record].
Aspergillus variegatus Mosseray La Cellule 43: 238. 1934. [MycoBank MB 255332].
Aspergillus macfiei C.W. Dodge, Medical Mycology: 629. 1935. [MycoBank MB 253139]. Not validly described [Art. 39.1, 40.1; Turland et al. (2018)].
Aspergillus aureus var. minor Nakaz., Simo & A. Watan., Bull. Agric. Chem. Soc. Japan 12: 958. 1936. [MycoBank MB 534300]. Not validly published [Art. 39.1; Turland et al. (2018)].
Aspergillus aureus var. murinus Nakaz., Simo & A. Watan., Bull. Agric. Chem. Soc. Japan 12: 959. 1936. [MycoBank MB 534301]. Not validly published [Art. 39.1; Turland et al. (2018)].
Aspergillus awamori var. ferrugineus Nakaz., Simo & A. Watan., Bull. Agric. Chem. Soc. Japan 12: 957. 1936. [Mycobank MB 250919]. Not validly published [Art. 39.1; Turland et al. (2018)].
Aspergillus awamori var. fumeus Nakaz., Simo & A. Watan., Bull. Agric. Chem. Soc. Japan 12: 961. 1936. [MycoBank MB 250920]. Not validly published [Art. 39.1; Turland et al. (2018)]. Representative strain: CBS 116.52 = NRRL 4843 = WB 4843 = IBT 16908 (no DNA sequence available).
Aspergillus luchuensis var. rubeolus Y.K. Shih, Lingnan Sci. J. 15: 374. 1936. [Mycobank MB 253092]. The ITS sequence (DQ196202) derived from the representative strain CBS 114.37 (= IAM 2185 = IBT 4944 = IBT 29898 = IMI 313493 = NRRL 4856 = WB 4856) belongs to the ATL.
Cladosarum olivaceum E. Yuill & J.L. Yuill, Trans. Brit. Mycol. Soc. 22: 199. 1938. [MycoBank MB 272878]. Culture ex-type: CBS 147.38 (no DNA sequence available).
Aspergillus niger var. arecae Lal & Ram Chandra, J. Scient. Res. Banaras Hindu Univ. 3: 123. 1953. [MycoBank MB 349040]. Not validly published [Art. 39.1; Turland et al. (2018)].
Aspergillus niger var. taxi D.P. Zhou, K. Zhao & Ping, J. Appl. Microbiol. 107: 1206. 2009. [MycoBank MB 542212]. Not validly described [Art. 39.1, 40.1; Turland et al. (2018)]. The ITS sequence (EU853157) derived from the original strain belongs to the ATL.
Aspergillus pseudotubingensis S. Khuna, N. Suwannarach & S. Lumyong, Front. Microbiol. 12: 705896 - p9. 2021. [MycoBank MB 830889]. See comments in sections Results and Discussion.
DISCUSSION
Taxonomic studies on A. niger, A. tubingensis and their related species have been the subject of many taxonomic controversies, rearrangements, name resurrections and repeated synonymizations. Currently, the species number in series Nigri is at one of its historical peaks and involves 14 accepted species (Houbraken et al. 2020, Silva et al. 2020, Khuna et al. 2021). Distinguishing among these species using morphology is impossible, and DNA sequencing is the gold standard for reliable classification and species identification (Howard et al. 2011, Varga et al. 2011). Therefore, these species can be considered cryptic in the truest sense. However, even when using molecular methods, some isolates cannot be identified satisfactorily at the species level (Howard et al. 2011, Negri et al. 2014, D’hooge et al. 2019). Contradictory species identifications in significant part of isolates using BLAST similarity searches with different genes (Fig. 3) can serve as evidence of taxonomic issues and may indicate the need for a taxonomic reclassification. To verify this assumption, we gathered a large dataset of a series Nigri sequences from three common phylogenetic markers and de novo sequenced 18 genomes and employed various species delimitation methods. All of these methods unequivocally suggested species number reduction.
Phylogenetic support of species reduction
We demonstrated that MSC delimitation methods using three genes broadly agreed on the distinction of only three species in the series Nigri (A. brasiliensis, A. niger and A. tubingensis). Only four analyses from among the 15 depicted in Fig. 5 proposed the delimitation of more species. These conclusive results, especially in the A. niger lineage (ANL) and A. brasiliensis lineage (ABL), are supported by the collection of a large number of strains well representing the intraspecific genetic variability.
In the A. tubingensis lineage (ATL), the multilocus method STACEY gave similar support to the delimitation of one or four species. This dilemma has not been satisfactorily resolved even at the genome level because ten STACEY analyses based on 20 genes each proposed several solutions ranging from one to four species in the ATL (Fig. 8), with four species being most preferred. The datasets containing 20 genes (and not higher) were selected due to the relatively high computational requirements of the method and because 20 genes are usually enough to resolve the phylogeny with a similar level of accuracy to the genome-wide data (Rokas et al. 2003, Yang & Rannala 2010, 2014). The uncertainty regarding the number of delimited species can probably be attributed to the underrepresentation of isolates related to A. vadensis and A. eucalypticola and, to a lesser extent, other species in the ATL, with the exception of A. tubingensis itself. Generally, it is thought that when a dataset contains a few isolates with large genetic distances, the delimitation methods may be prone to over delimitation, but with a large set of isolates representing genetic variability of every species, the probability of methods finding real species boundaries increases (Pante et al. 2015, Chambers & Hillis 2020). Another reason for the conflicting results can be the widely present incongruences between single-gene genealogies as typically present in recently speciating species or in populations that have not completed their speciation process. In recently diverged species, it is typically caused by retaining ancestral polymorphisms (incomplete lineage sorting), or it can be caused by past hybridization events (Hubka et al. 2018, Steenkamp et al. 2018, Matute & Sepúlveda 2019).
At this point, it is appropriate to mention that the single-gene phylogenies based on benA, CaM and RPB2 loci were highly incongruent (Fig. 2, Supplementary Fig. S2) and that the topology of the resulting multigene phylogeny shows several conflicts or poorly resolved clades compared to the whole-genome phylogeny, as we demonstrated in Fig. 4. Suboptimal phylogenetic signals contained in these three loci can further contribute to the discrepancy between the results of the MSC methods (support of a single species in the ATL) and those of the STACEY analyses based on 20 loci randomly selected from genomes (predominant support of four species in the ATL) (Fig. 8). When species limits are evaluated using the GCPSR approach (Taylor et al. 2000, Dettman et al. 2003a) using the benA, CaM and RPB2 loci only, the obvious conclusion is the recognition of only three species in the series Nigri: A. brasiliensis, A. niger and A. tubingensis. The recently proposed A. vinaceus (Silva et al. 2020) from the ANL was also described based on the GCPSR approach, but the phylogeny lacked some isolates with intermediate genotypes included here. The presence of these strains in the phylogenies contradicts the delimitation of A. vinaceus as a separate phylogenetic species.
Another independent testing of species hypotheses was performed in the recently released software DELINEATE (Sukumaran et al. 2021). The software needs some a priori defined species that are correctly delimited in the dataset. This requirement was fulfilled by the inclusion of some species belonging to the other series of section Nigri. In addition, A. brasiliensis is well defined and can serve as a “reference species” when dealing with delimitation in A. niger and A. tubingensis lineages. The results of the various models were relatively stable, always supporting the broad concept of A. niger (comprising A. niger, A. welwitschiae and A. vinaceus) and A. brasiliensis (Fig. 7). In the ATL a broad species definition (comprising all species in the lineage) gained support in Models 7 and 10 when A. niger was defined as a broad species as well. In models where the ANL was divided into four and six species, the unassigned ATL populations were segregated into four tentative species. This scenario is, however, improbable because the ANL was never divided into several species in Models 2, 6 and 9. Model 6 shows the situation where populations of the ATL are predefined into four species identical to those delimited by Models 3 and 4, while populations of the ANL are unassigned. In this situation, A. niger was also delimited as a single broad species. In summary, DELINEATE analysis convincingly supported that the series Nigri contains only three species based on benA, CaM and RPB2.
Phenotype and sexual reproduction in series Nigri
Species in the series Nigri are all biseriate and present colonies with brown, dark brown to black sporulating areas and white to yellowish white mycelial areas. Generally, the species exhibit largely overlapping characteristics in terms of both macromorphology (colony diameter and colour) and micromorphology (size, shape and ornamentation of conidia, diameter and shape of vesicle, and length and width of stipes) (Table 2). Not only particular species but also the main lineages of A. niger and A. tubingensis are indistinguishable from each other using morphology. Additionally, species identification is sometimes complicated by the occurrence of atypical strains exhibiting unusual morphological characteristics. For example, A. lacticoffeatus was described based on its different colony colour and a specific extrolite pattern (Samson et al. 2004). However, it was later synonymized with A. niger, because it was phylogenetically inseparable (Varga et al. 2011). Within the ATL, A. vadensis stands out among the others for its slow growth and short stipes (Table 2), but the original description is based on only one isolate (Vries et al. 2005).
Table 2.
Overview of macro- and micromorphological features of species belonging to the series Nigri.
| Species |
CYA, 7 d (mm)
|
MEA, 7 d, 25 °C (mm) | Prevailing colony colour(s) on CYA | Vesicle diam (μm), shape |
Stipe (μm)
|
Conidia: diam (μm), shape, surface | Sclerotia (mm), colour | References 1 | ||
|---|---|---|---|---|---|---|---|---|---|---|
| 25 °C | 37 °C | Length | Width | |||||||
| A. brasiliensis | 71–76 | 71–76 | 52–70 | cream-coloured to light brown | 30–45, nearly globose | 700–1 700 | 8–13 | 3.5–4.8, subglobose, echinulate | 1–1.5, white | Varga et al. (2007) |
| 70–85 | 70–85 | n/a | black | 45–80, globose | 1 500–3 000 | 15–20 | 3.5–5.5, globose, finely to distinctly roughened | 0.5–0.7, cream | Crous et al. (2009), Samson et al. (2010), Frisvad et al. (2014) | |
| A. lacticoffeatus | 71–76 | 59–75 | 52–70 | cream to light brown | 40–65, nearly globose | (200–)300–1 200 | (7–)10–15(–18) | 3.5–4.1 × 3.4–3.9, subglobose, usually smooth to very finely roughened | not observed | Samson et al. (2004) |
| A. vinaceus | 64–66 | 60–63.5 | 35–39 | black | 63–75, globose to subglobose | 1 300–1 800 | 16–21 | 3–5.5, globose to subglobose, finely roughened to echinulate | 0.9–1.5, white to cream | Silva et al. (2020) |
| A. welwitschiae | similar to A. niger | similar to A. niger | similar to A. niger | black | 45–85, globose | similar to A. niger | n/a | 3.5–5.5, globose, finely to distinctly roughened | not observed | Hong et al. (2013), Silva et al. (2020) |
|
| ||||||||||
| A. chiangmaiensis | 62–63 | 57–66 | 68–70 | dark brown to black | 35–68, globose to ellipsoidal | 370–1 430 | 10–15 | 3–4.5, globose to subglobose, echinulate | 0.4–1.4, white | Khuna et al. (2021) |
| A. costaricensis | 63–78 | 58–62 | 26–62 | white with sparse black-sporulating areas | 40–90, globose | (800–)1 000–1 700(–1 900) | (12–)13–20(–22) | 3.1–4.5, globose to subglobose, smooth to distinctly roughened | 1.2–1.8, pink to yellow | Samson et al. (2004) |
| A. eucalypticola | 68–72 | 30–50 | 46–51 | beige to cream yellow | 30–55, globose | n/a | 8–14 | 2.5–3.5, globose, smooth to coarsely roughened | not observed | Varga et al. (2011) |
| A. luchuensis 2 | 37–80 | 30–67 | 43–68 | white, gray, brown, black | 15–90, subglobose to globose | 1500 | 10–22 | 3–4.5, globose, smooth | 0.8–1.3, cream | Samson et al. (2010), Varga et al. (2011), Hong et al. (2013), Frisvad et al. (2014) |
| A. neoniger | 72–80 | 37–67 | 54–61 | gray to black | 30–50, globose | n/a | 8–12 | 3.5–5, globose, coarsely roughened to echinulate | observed (without description) | Varga et al. (2011), Frisvad et al. (2014) |
| A. piperis | 60–75 | 64–82 | 59–78 | cream-coloured with sparse black-sporulating areas | 40–55, subglobose | (300–)400–3 000 | (7–)12–15(–20) | 2.8–3.6, subglobose to broadly ellipsoidal, distinctly roughened | 1–1.7, yellow to pink brown | Samson et al. (2004) |
| A. pseudopiperis | 63–65 | 61–64 | 55–60 | greenish brown to dark brown | 15–39, globose to ellipsoidal | 200–2 125 | 10–18 | 3–5, globose to subglobose, roughened to spinulose | 0.2–1 × 0.2–0.8, light yellow to pinkish orange | Khuna et al. (2021) |
| A. pseudotubingensis | 51–58 | 55–68 | 62–63 | grayish brown to dark brown | 20–70, globose to ellipsoidal | 740–3 060 | 10–18 | 3–6, globose to subglobose, fine spiny surface | not observed | Khuna et al. (2021) |
| A. tubingensis | n/a | n/a | 40–65 | beige, grayish brown, brown to black | 27–80, globose | 550–2 300(–6 000) | 10–35 | 3–5, globose, coarsely roughened | 0.4–1.8, cream to pinkish tan | Samson et al. (2007), Varga et al. (2007), Hong et al. (2013), Horn et al. (2013), Ismail (2017) |
| A. vadensis | 25 | n/a | 30 | light brown to olive green brown | 25–35, globose | up to 150 | 6–15 | 3–4, globose, rough-walled to finely echinulate | not observed | de Vries et al. (2005) |
n/a - exact data not available.
1if there are multiple studies in the column “references”, the description and dimensions are shown as a summary from all these sources.
2the dimensions shown here represent summary from description of A. luchuensis and its synononym A. acidus as they appeared in the publications of Varga et al. (2011) and Hong et al. (2013).
Series Nigri species are known for producing a wide range of extrolites, and their spectra were summarised by Samson et al. (2007), Nielsen et al. (2009) and Frisvad et al. (2018). It is often asserted that species in series Nigri could be identified by different secondary metabolite patterns. However, the extrolite profiles can vary among strains, which makes its use to distinguish between species complicated. Additionally, the usability of secondary metabolite production for taxonomy and evolutionary biology in general might be limited due to the potential for horizontal gene transfer of secondary metabolism gene clusters between species of the same genus or even between fungal genera (Richards 2011, Slot & Rokas 2011, Szöllősi et al. 2015). The high frequency of this phenomenon is also suggested from the genomes of series Nigri species (Vesth et al. 2018). Considering our proposed reclassification of series Nigri, the extrolite profiles of the six species we accepted need to be reassessed to determine if they support the delineation proposed here based on a phylogenetic species concept.
Sexual reproduction in heterothallic fungi is governed by mating-type (MAT) genes (Dyer & O’Gorman 2011). In section Nigri, the development of asci and ascospores occurs in the stroma of sclerotia (Horn et al. 2013). Therefore, sclerotia formation is considered a prerequisite for sexual reproduction. In the ATL, sclerotia were reported in most species except for A. eucalypticola, A. pseudotubingensis and A. vadensis. In the ANL, sclerotia were formed in some strains of A. vinaceus and A. niger (Frisvad et al. 2014, Silva et al. 2020). Sclerotia in the ATL were mostly pink to yellow, while those in the ANL were white to cream (Table 2). Despite the heterogeneity in sclerotia production among series Nigri species, most of them were proven to be heterothallic or protoheterothallic, which means that mating-type gene idiomorph(s) associated with heterothallism have been detected among the examined strains of these species (Houbraken et al. 2020). Currently, A. tubingensis is the only species in series Nigri with an experimentally proven sexual cycle (Horn et al. 2013). In this study, we detected both MAT1-1-1 and MAT1-2-1 idiomorphs among isolates of ANL and ATL (Fig. 4), in agreement with previous studies (Horn et al. 2013, Varga et al. 2014, Mageswari et al. 2016, Seekles et al. 2022). An interesting observation was made by Horn et al. (2013), who sequenced five genes in the parental isolates of A. tubingensis and their progeny resulting from sexual reproduction. The authors found that A. neoniger cannot be separated from A. tubingensis in the benA or tsr1 phylogenies, while in the other phylogenies (CaM, Mcm7 and RPB2), it is resolved outside the least inclusive clade containing all A. tubingensis strains and its ex-type. In our opinion, this fact demonstrates gene flow between A. tubingensis and A. neoniger and supports the synonymization made in this study. Aspergillus costaricensis was not included by Horn et al. (2013), but it is a close relative of A. neoniger and must be synonymized together to retain the monophyly of the redefined A. tubingensis (Fig. 5).
Updated taxonomy of series Nigri and species identification
We showed based on various independent species delimitation analyses that the number of accepted species in the Nigri series is too high and needs to be reduced. The methods broadly agreed that the ABL and ANL contain only one species each. The only uncertainty was about the species number in the ATL, where single-gene MSC methods and the GCPSR approach based on the commonly used phylogenetic markers benA, CaM and RPB2 supported only one species, while STACEY analyses supported up to four species: A. tubingensis, A. luchuensis, A. eucalypticola and A. vadensis. After considering all results and practical consequences, we decided on a conservative solution by retaining four species in the ATL. This newly proposed taxonomic treatment of series Nigri with six accepted species overall is shown in Fig. 10.
Fig. 10.
Taxonomic rearrangement of series Nigri into six species with marked synonymizations. The proposed taxonomy is schematically shown in the form of a radial tree (species tree based on benA, CaM and RPB2 calculated in starBEAST). The positions of A. chiangmaiensis and A. pseudopiperis are approximated based on the tree shown in Figs 1, 3 as they were not included in this phylogenetic analysis.
The following findings played a major role in our decision-making process. Aspergillus luchuensis is industrially extremely important (Hong et al. 2013, 2014), and its imprudent synonymization without further confirmation could cause unnecessary taxonomic instability. The phylogenetic signal from the dataset containing only benA, CaM and RPB2 loci may be suboptimal when compared to the predominant signal obtained from whole genome data (Fig. 4); thus, the results should be considered with caution before data from more genes/genomes are evaluated. We also showed that the intraspecific genetic diversity of the broadly defined A. tubingensis would be higher than that commonly found for Aspergillus species, while the variability within A. tubingensis, excluding A. luchuensis, A. eucalypticola and A. vadensis comfortably falls within this range (Fig. 9).
We believe that the user community will benefit from the simplified taxonomy of series Nigri with a lower number of species. This new classification will facilitate species identification that is currently complicated by inconsistent identification results when using sequence data of different genes (Fig. 3) and the impossibility of finding species-specific mass spectra for some narrow species when using the MALDI-TOF method (Gautier et al. 2016, D’hooge et al. 2019, Ban et al. 2021). DNA sequence identification of these newly defined species is possible by using benA, CaM and RPB2 markers, although in a minority of cases, there can be conflicting identification in the ATL (Fig. 3). Because of this, we recommend that if identification to a species level is required in the ATL, it should be done through DNA sequencing of all three markers and performing phylogenetic analysis. The ITS barcoding sequence can only distinguish among ABL, ANL and ATL (Supplementary Fig. S3).
As mentioned above, we excluded three newly described species, A. chiangmaiensis, A. pseudopiperis and A. pseudotubingensis (Khuna et al. 2021), from the analyses based on the MSC model because of the poor quality of their available sequences. However, we propose that A. chiangmaiensis and A. pseudopiperis are synonyms of A. tubingensis, as their phylogenetic position in the combined ML, MP and BI trees is within the redefined A. tubingensis: the A. pseudopiperis ex-type strain (SDBR-CMUI1) is located in a clade sister to the clade containing the A. tubingensis ex-type (CBS 133056), and A. chiangmaiensis is closely related to A. costaricensis and A. neoniger (Figs 1, 3), which are also synonymized with A. tubingensis. The status of A. pseudotubingensis remains uncertain and needs to be revised in the future as it forms a relatively separate lineage in the combined phylogeny based on three loci similar to A. eucalypticola (Figs 1, 3). The phylogeny presented by Khuna et al. (2021) is affected by numerous errors in the DNA sequences of the newly described species and it shows clear signs of the long branch attraction phenomenon (Kück et al. 2012). We believe that a more reliable position is shown in our phylogenies constructed based on partially corrected sequences (Figs 1, 3).
The species reduction proposed in this study is in line with some other studies using MSC model-based methods, genomic data or more complex approaches for species delimitation in fungi. For example, species number was significantly reduced in the plant pathogens from the Diaporthe eres complex (Hilário et al. 2021) and Alternaria section Alternaria (Woudenberg et al. 2015), the edible mushroom Flammulina (Wang et al. 2018), the lichen-forming fungus Bryoria (Boluda et al. 2019), the indoor fungi from Aspergillus series Versicolores (Sklenář et al. 2022) and the opportunistic human and animal pathogens from Aspergillus series Viridinutantes (Hubka et al. 2018). Numerous examples can also be found outside the Fungi kingdom (Li et al. 2019, Feng et al. 2021, Parker et al. 2021). These studies demonstrate that species over splitting similar to that observed in series Nigri is relatively common in extensively studied groups and species number reduction can become an increasingly common trend in taxonomy.
Future prospects
Further reduction of species number in the series Nigri cannot be ruled out in the future after collecting more strains and more whole genome sequences from underrepresented taxa such as A. eucalypticola and A. vadensis. Inclusion of more variability from these rare taxa should resolve persistent ambiguities in the definition of species limits and increase the probability of methods to delimit species more conclusively. As mentioned above, the position of A. pseudotubingensis should be re-examined, and higher quality DNA sequences should be obtained, ideally whole genome sequences. For now, we recommend avoiding using this name until the abovementioned issues are resolved.
Advanced phylogenetic methods spanning the whole genome will allow for a systematic and comparable metric of species differentiation. For example, reciprocal monophyly and a high level of concordance between thousands of markers on the genome scale should allow better differentiation between processes such as recombination, incomplete lineage sorting and speciation in most cases (Kobmoo et al. 2019, Matute & Sepúlveda 2019, Mavengere et al. 2020). A dense population-level representation in genome-scale phylogenetic studies is important for accurate identification of species boundaries and for resolving evolutionary relationships between species and higher-level taxonomic ranks in Aspergillus (Steenwyck et al. 2022). Our efforts in the following years will be directed towards achieving this goal.
Acknowledgments
Vit Hubka was supported by the Czech Ministry of Health (grant NU21-05-00681), the Charles University Research Centre program no. 204069 and the Czech Academy of Sciences Long-term Research Development Project (RVO: 61388971). František Sklenář was supported by the project of Charles University Grant Agency (GAUK 380821). This work has been partly supported by JST SPRING awarded to Cai Bian (JPMJSP2109). CMV acknowledges the Plant Health and Protection - Agricultural Research Council (ARC-PHP), where research on the PPRI strains included in this study was initiated and financially supported by a grant (nr 110441; reference FBIS170406226088) received from the Foundational Biodiversity Information Programme (FBIP) of the National Research Foundation of South Africa. VH is grateful for the support from the Japan Society for the Promotion of Science - Postdoctoral Fellowships for Research in Japan (Standard) and Grant-in-aid for a JSPS research fellow (No. 20F20772). We are grateful to Jan Karhan for the help with graphical adjustments of analysis outputs. We thank Kateřina Glässnerová for her assistance in the laboratory and Osamu Yamada for providing copies of some old taxonomic studies.
DECLARATION ON CONFLICT OF INTEREST
The authors declare that there is no conflict of interest.
Supplementary Material: https://studiesinmycology.org/
Multilocus phylogeny of Aspergillus series Nigri based on three loci (benA, CaM and RPB2) without species A. chiangmaiensis, A. pseudopiperis and A. pseudotubingensis. Best-scoring Maximum Likelihood (ML) tree inferred in the IQ-TREE is shown; ultrafast bootstrap support values (ML bs) are appended to nodes; only support values ≥95 % are shown; the ex-type strains are designated with a bold print; the information on geographic origin and isolation source is plotted on the tree (see the legend).
Comparison of single-gene genealogies based on the benA, CaM and RPB2 loci and created by three different phylogenetic methods (only one isolate per unique multilocus haplotype is included in each phylogeny). Best-scoring single-locus maximum likelihood (ML) trees are shown; ML ultrafast bootstrap support values (ML bs), maximum parsimony bootstrap support values (MP bs) and Bayesian inference posterior probabilities (BI pp) are appended to nodes. Only support values ≥95 %, ≥70 % and ≥0.95, respectively, are shown. A dash indicates lower statistical support for a specific node, or the absence of a node in the phylogeny, while an asterisk indicates full support. The ex-type strains are designated with a bold print. Alignment characteristics, partitioning schemes and substitution models are listed in Supplementary Table S1.
Best scoring maximum likelihood tree inferred in IQ-TREE based on ITS rDNA region sequences of series Nigri members (Table 1). It is apparent from the tree that a limited variability present in this locus can only differentiate the three main lineages, and more detailed differentiation is not possible. The ITS sequence of the A. vadensis ex-type was excluded from the analysis because it contains numerous errors and unresolved positions (accession number AY585549). Alignment characteristics, partitioning schemes and substitution models are listed in Supplementary Table S1.
Characteristics of alignments, partition-merging results and best substitution model for each partition according to the Bayesian information criterion.
Assignment of strains into populations for analysis in DELINEATE based on BPP v. 4.3.
Aspergillus series Nigri genomes analyzed in this study and their basic characteristics.
Distribution of 200 orthologous genes into ten separate STACEY analyses.
List of unique multilocus haplotypes.
Maximum sequence dissimilarity between isolates of the same Aspergillus species whose species limits have been delimited using methods based on a multispecies coalescent model; only species represented by isolates from at least three countries were included.
REFERENCES
- Ahrens D, Fujisawa T, Krammer H-J, et al. (2016). Rarity and incomplete sampling in DNA-based species delimitation. Systematic Biology 65: 478–494. [DOI] [PubMed] [Google Scholar]
- Alcazar-Fuoli L, Mellado E, Alastruey-Izquierdo A, et al. (2009). Species identification and antifungal susceptibility patterns of species belonging to Aspergillus section Nigri. Antimicrobial Agents and Chemotherapy 53: 4514–4517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Musallam (1980). A revision of the black Aspergillus species. Ph.D. dissertation. Rijksuniversiteit; Utrecht, Netherlands. [Google Scholar]
- Andersen MR, Salazar MP, Schaap PJ, et al. (2011). Comparative genomics of citric-acid-producing Aspergillus niger ATCC 1015 versus enzyme-producing CBS 513.88. Genome Research 21: 885–897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bairoch A, Apweiler R. (2000). The SWISS-PROT protein sequence database and its supplement TrEMBL in 2000. Nucleic Acids Research 28: 45–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ban S, Kasaishi R, Kamijo T, et al. (2021). An exploratory MALDI-TOF MS library based on SARAMIS superspectra for rapid identification of Aspergillus section Nigri. Mycoscience 62: 224–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bankevich A, Nurk S, Antipov D, et al. (2012). SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. Journal of Computational Biology 19: 455–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett JW, Klich MA. (1992). Aspergillus: biology and industrial applications. Butterworth-Heinemann, USA. [Google Scholar]
- Boluda CG, Rico VJ, Divakar PK, et al. (2019). Evaluating methodologies for species delimitation: the mismatch between phenotypes and genotypes in lichenized fungi (Bryoria sect. Implexae, Parmeliaceae). Persoonia 42: 75–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouckaert R, Heled J, Kühnert D, et al. (2014). BEAST 2: a software platform for Bayesian evolutionary analysis. PLoS Computational Biology 10: e1003537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchfink B, Xie C, Huson DH. (2015). Fast and sensitive protein alignment using DIAMOND. Nature Methods 12: 59–60. [DOI] [PubMed] [Google Scholar]
- Carstens BC, Pelletier TA, Reid NM, Satler JD. (2013). How to fail at species delimitation. Molecular Ecology 22: 4369–4383. [DOI] [PubMed] [Google Scholar]
- Cerqueira GC, Arnaud MB, Inglis DO, et al. (2014). The Aspergillus Genome Database: multispecies curation and incorporation of RNA-Seq data to improve structural gene annotations. Nucleic Acids Research 42: D705–710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambers EA, Hillis DM. (2020). The multispecies coalescent over-splits species in the case of geographically widespread taxa. Systematic Biology 69: 184–193. [DOI] [PubMed] [Google Scholar]
- Chen Y, Nie F, Xie S-Q, et al. (2021). Efficient assembly of nanopore reads via highly accurate and intact error correction. Nature Communications 12: 60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crous PW, Verkley GJM, Groenewald JZ, et al. (eds) (2009). Fungal Biodiversity. CBS Laboratory Manual Series No. 1. Centraalbureau voor Schimmelcultures, Utrecht, The Netherlands. [Google Scholar]
- de Vries RP, Riley R, Wiebenga A, et al. (2017). Comparative genomics reveals high biological diversity and specific adaptations in the industrially and medically important fungal genus Aspergillus. Genome Biology 18: 28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dettman JR, Jacobson DJ, Taylor JW. (2003a). A multilocus genealogical approach to phylogenetic species recognition in the model eukaryote Neurospora. Evolution 57: 2703–2720. [DOI] [PubMed] [Google Scholar]
- Dettman JR, Jacobson DJ, Turner E, et al. (2003b). Reproductive isolation and phylogenetic divergence in Neurospora: comparing methods of species recognition in a model eukaryote. Evolution 57: 2721–2741. [DOI] [PubMed] [Google Scholar]
- D’hooge E, Becker P, Stubbe D, et al. (2019). Black aspergilli: A remaining challenge in fungal taxonomy? Medical Mycology 57: 773–780. [DOI] [PubMed] [Google Scholar]
- Donaldson GC, Ball LA, Axelrood PE, et al. (1995). Primer sets developed to amplify conserved genes from filamentous ascomycetes are useful in differentiating Fusarium species associated with conifers. Applied and Environmental Microbiology 61: 1331–1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyer PS, O’Gorman CM. (2011). A fungal sexual revolution: Aspergillus and Penicillium show the way. Current Opinion in Microbiology 14: 649–654. [DOI] [PubMed] [Google Scholar]
- Ellena V, Seekles SJ, Vignolle GA, et al. (2021). Genome sequencing of the neotype strain CBS 554.65 reveals the MAT1–2 locus of Aspergillus niger. BMC Genomics 22: 679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emms DM, Kelly S. (2019). OrthoFinder: phylogenetic orthology inference for comparative genomics. Genome Biology 20: 238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng X, Wang X, Chiang Y, et al. (2021). Species delimitation with distinct methods based on molecular data to elucidate species boundaries in the Cycas taiwaniana complex (Cycadaceae). Taxon 70: 477–491. [Google Scholar]
- Frisvad JC, Larsen TO, Thrane U, et al. (2011). Fumonisin and ochratoxin production in industrial Aspergillus niger strains. PLoS ONE 6: e23496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frisvad JC, Møller LLH, Larsen TO, et al. (2018). Safety of the fungal workhorses of industrial biotechnology: update on the mycotoxin and secondary metabolite potential of Aspergillus niger, Aspergillus oryzae, and Trichoderma reesei. Applied Microbiology and Biotechnology 102: 9481–9515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frisvad JC, Petersen LM, Lyhne EK, et al. (2014). Formation of sclerotia and production of indoloterpenes by Aspergillus niger and other species in section Nigri. PLoS ONE 9: e94857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fungaro MHP, Ferranti LS, Massi FP, et al. (2017). Aspergillus labruscus sp. nov., a new species of Aspergillus section Nigri discovered in Brazil. Scientific Reports 7: 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gams W, Christensen M, Onions AH, et al. (1986). Infrageneric Taxa of Aspergillus. In: Advances in Penicillium and Aspergillus Systematics. Springer, US: 55–62. [Google Scholar]
- Gautier M, Normand A-C, Ranque S. (2016). Previously unknown species of Aspergillus. Clinical Microbiology and Infection 22: 662–669. [DOI] [PubMed] [Google Scholar]
- Gits-Muselli M, Hamane S, Verillaud B, et al. (2021). Different repartition of the cryptic species of black aspergilli according to the anatomical sites in human infections, in a French University hospital. Medical Mycology 59: 985–992. [DOI] [PubMed] [Google Scholar]
- Glässnerová K, Sklenář F, Jurjević Ž, et al. (2022). A monograph of Aspergillus section Candidi. Studies in Mycology 102: 1–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gnerre S, MacCallum I, Przybylski D, et al. (2011). High-quality draft assemblies of mammalian genomes from massively parallel sequence data. Proceedings of the National Academy of Sciences of the United States of America 108: 1513–1518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grüning B, Dale R, Sjödin A, et al. (2018). Bioconda: sustainable and comprehensive software distribution for the life sciences. Nature Methods 15: 475–476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto A, Hagiwara D, Watanabe A, et al. (2017). Drug sensitivity and resistance mechanism in Aspergillus section Nigri strains from Japan. Antimicrobial Agents and Chemotherapy 61: e02583-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heled J, Drummond AJ. (2010). Bayesian inference of species trees from multilocus data. Molecular Biology and Evolution 27: 570–580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendrickx M, Beguin H, Detandt M. (2012). Genetic re-identification and antifungal susceptibility testing of Aspergillus section Nigri strains of the BCCM/IHEM collection. Mycoses 55: 148–155. [DOI] [PubMed] [Google Scholar]
- Hilário S, Gonçalves MFM, Alves A. (2021). Using genealogical concordance and coalescent-based species delimitation to assess species boundaries in the Diaporthe eres complex. Journal of Fungi 7: 507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong S-B, Cho HS, Shin HD, et al. (2006). Novel Neosartorya species isolated from soil in Korea. International Journal of Systematic and Evolutionary Microbiology 56: 477–486. [DOI] [PubMed] [Google Scholar]
- Hong S-B, Lee M, Kim DH, et al. (2013). Aspergillus luchuensis, an industrially important black Aspergillus in east Asia. PLoS ONE 8: e63769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong S-B, Yamada O, Samson RA. (2014). Taxonomic re-evaluation of black koji molds. Applied Microbiology and Biotechnology 98: 555–561. [DOI] [PubMed] [Google Scholar]
- Horn BW, Olarte RA, Peterson SW, et al. (2013). Sexual reproduction in Aspergillus tubingensis from section Nigri. Mycologia 105: 1153–1163. [DOI] [PubMed] [Google Scholar]
- Houbraken J, de Vries RP, Samson RA. (2014). Modern taxonomy of biotechnologically important Aspergillus and Penicillium species. Advances in Applied Microbiology 86: 199–249. [DOI] [PubMed] [Google Scholar]
- Houbraken J, Kocsubé S, Visagie CM, et al. (2020). Classification of Aspergillus, Penicillium, Talaromyces and related genera (Eurotiales): an overview of families, genera, subgenera, sections, series and species. Studies in Mycology 95: 5–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howard SJ, Harrison E, Bowyer P, et al. (2011). Cryptic species and azole resistance in the Aspergillus niger complex. Antimicrobial Agents and Chemotherapy 55: 4802–4809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubka V, Barrs V, Dudová Z, et al. (2018). Unravelling species boundaries in the Aspergillus viridinutans complex (section Fumigati): opportunistic human and animal pathogens capable of interspecific hybridization. Persoonia 41: 142–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubka V, Kolarik M. (2012). β-tubulin paralogue tubC is frequently misidentified as the benA gene in Aspergillus section Nigri taxonomy: primer specificity testing and taxonomic consequences. Persoonia 29: 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubka V, Kubatova A, Mallatova N, et al. (2012). Rare and new etiological agents revealed among 178 clinical Aspergillus strains obtained from Czech patients and characterized by molecular sequencing. Medical Mycology 50: 601–610. [DOI] [PubMed] [Google Scholar]
- Huerta-Cepas J, Szklarczyk D, Forslund K, et al. (2016). eggNOG 4.5: a hierarchical orthology framework with improved functional annotations for eukaryotic, prokaryotic and viral sequences. Nucleic Acids Research 44: D286–D293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ismail MA. (2017). Incidence and significance of black aspergilli in agricultural commodities: a review, with a key to all species accepted to-date. European Journal of Biological Research 7: 207–222. [Google Scholar]
- Jin JJ, Yu W bin, Yang JB, et al. (2020). GetOrganelle: a fast and versatile toolkit for accurate de novo assembly of organelle genomes. Genome Biology 21: 241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones G. (2017). Algorithmic improvements to species delimitation and phylogeny estimation under the multispecies coalescent. Journal of Mathematical Biology 74: 447–467. [DOI] [PubMed] [Google Scholar]
- Jones G, Aydin Z, Oxelman B. (2015). DISSECT: an assignment-free Bayesian discovery method for species delimitation under the multispecies coalescent. Bioinformatics 31: 991–998. [DOI] [PubMed] [Google Scholar]
- Jones P, Binns D, Chang HY, et al. (2014). InterProScan 5: genome-scale protein function classification. Bioinformatics 30: 1236–1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurjević Ž, Peterson SW, Stea G, et al. (2012). Two novel species of Aspergillus section Nigri from indoor air. IMA Fungus 3: 159–173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurjević Ž, Kubátová A, Kolařík M, Hubka V. (2015). Taxonomy of Aspergillus section Petersonii sect. nov. encompassing indoor and soil-borne species with predominant tropical distribution. Plant Systematics and Evolution 301: 2441–2462. [Google Scholar]
- Katoh K, Rozewicki J, Yamada KD. (2019). MAFFT online service: multiple sequence alignment, interactive sequence choice and visualization. Briefings in Bioinformatics 20: 1160–1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khuna S, Suwannarach N, Kumla J, et al. (2021). Growth enhancement of Arabidopsis (Arabidopsis thaliana) and onion (Allium cepa) with inoculation of three newly identified mineral-solubilizing fungi in the genus Aspergillus section Nigri. Frontiers in Microbiology 12: 705896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kocsubé S, Perrone G, Magistà D, et al. (2016). Aspergillus is monophyletic: evidence from multiple gene phylogenies and extrolites profiles. Studies in Mycology 85: 91–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korf I. (2004). Gene finding in novel genomes. BMC Bioinformatics 5: 59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozakiewicz Z. (1989). Aspergillus species on stored products. Mycological Papers 161: 1–188. [Google Scholar]
- Kozakiewicz Z, Frisvad JC, Hawksworth DL, Pitt JI, Samson RA, Stolk AC. (1992). Proposal for nomina specifica conservanda and rejicienda in Aspergillus and Penicillium (Fungi). Taxon 41: 109–113. [Google Scholar]
- Krueger F. (2015). Trim Galore!: a wrapper tool around Cutadapt and FastQC to consistently apply quality and adapter trimming to FastQ files. Babraham Institute https://www.bioinformatics.babraham.ac.uk/projects. [Google Scholar]
- Kubatko LS, Degnan JH. (2007). Inconsistency of phylogenetic estimates from concatenated data under coalescence. Systematic Biology 56: 17–24. [DOI] [PubMed] [Google Scholar]
- Kück P, Mayer C, Wägele J-W, et al. (2012). Long branch effects distort maximum likelihood phylogenies in simulations despite selection of the correct model. PLoS ONE 7: e36593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kundu R, Casey J, Sung WK. (2019). HyPo: super fast & accurate polisher for long read genome assemblies. bioRxiv doi: 10.1101/2019.12.19.882506. [Google Scholar]
- Kusters-van Someren MA, Samson RA, Visser J. (1991). The use of RFLP analysis in classification of the black Aspergilli: reinterpretation of the Aspergillus niger aggregate. Current Genetics 19: 21–26. [Google Scholar]
- Lanfear R, Frandsen PB, Wright AM, et al. (2017). PartitionFinder 2: new methods for selecting partitioned models of evolution for molecular and morphological phylogenetic analyses. Molecular Biology and Evolution 34: 772–773. [DOI] [PubMed] [Google Scholar]
- Letunic I, Bork P. (2021). Interactive Tree Of Life (iTOL) v5: an online tool for phylogenetic tree display and annotation. Nucleic Acids Research 49: W293–W296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Durbin R. (2009). Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25: 1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, et al. (2009). 1000 genome project data processing subgroup. The sequence alignment/map format and Samtools. Bioinformatics 25: 2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Wen J, Ren Y, et al. (2019). From seven to three: integrative species delimitation supports major reduction in species number in Rhodiola section Trifida (Crassulaceae) on the Qinghai-Tibetan Plateau. Taxon 68: 268–279. [Google Scholar]
- Liu YJ, Whelen S, Hall BD. (1999). Phylogenetic relationships among ascomycetes: evidence from an RNA polymerse II subunit. Molecular Biology and Evolution 16: 1799–1808. [DOI] [PubMed] [Google Scholar]
- Mageswari A, Kim J, Cheon K-H, et al. (2016). Analysis of the MAT1-1 and MAT1-2 gene ratio in black Koji molds isolated from Meju. Mycobiology 44: 269–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majoros WH, Pertea M, Salzberg SL. (2004). TigrScan and GlimmerHMM: two open source ab initio eukaryotic gene-finders. Bioinformatics 20: 2878–2879. [DOI] [PubMed] [Google Scholar]
- Matute DR, Sepúlveda VE. (2019). Fungal species boundaries in the genomics era. Fungal Genetics and Biology 131: 103249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell AL, Attwood TK, Babbitt PC, et al. (2019). InterPro in 2019: improving coverage, classification and access to protein sequence annotations. Nucleic Acids Research 47: D351–D360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosseray R. (1934). Les Aspergillus de la section” Niger” Thom & Church. La Cellule 43: 203–285. [Google Scholar]
- Nargesi S, Jafarzadeh J, Najafzadeh MJ, et al. (2022). Molecular identification and antifungal susceptibility of clinically relevant and cryptic species of Aspergillus sections Flavi and Nigri. Journal of Medical Microbiology 71: 001480. [DOI] [PubMed] [Google Scholar]
- Negri CE, Gonçalves SS, Xafranski H, et al. (2014). Cryptic and rare Aspergillus species in Brazil: prevalence in clinical samples and in vitro susceptibility to triazoles. Journal of Clinical Microbiology 52: 3633–3640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen L-T, Schmidt HA, von Haeseler A, et al. (2015). IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Molecular Biology and Evolution 32: 268–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen KF, Mogensen JM, Johansen M, et al. (2009). Review of secondary metabolites and mycotoxins from the Aspergillus niger group. Analytical and Bioanalytical Chemistry 395: 1225–1242. [DOI] [PubMed] [Google Scholar]
- Noonim P, Mahakarnchanakul W, Nielsen KF, et al. (2008). Isolation, identification and toxigenic potential of ochratoxin A-producing Aspergillus species from coffee beans grown in two regions of Thailand. International Journal of Food Microbiology 128: 197–202. [DOI] [PubMed] [Google Scholar]
- Pante E, Puillandre N, Viricel A, et al. (2015). Species are hypotheses: avoid connectivity assessments based on pillars of sand. Molecular Ecology 24: 525–544. [DOI] [PubMed] [Google Scholar]
- Paradis E. (2010). pegas: an R package for population genetics with an integrated-modular approach. Bioinformatics 26: 419–420. [DOI] [PubMed] [Google Scholar]
- Parker E, Dornburg A, Struthers CD, et al. (2021). Phylogenomic species delimitation dramatically reduces species diversity in an Antarctic adaptive radiation. Systematic Biology 71: 58–77. [DOI] [PubMed] [Google Scholar]
- Perrone G, Stea G, Epifani F, et al. (2011). Aspergillus niger contains the cryptic phylogenetic species A. awamori. Fungal Biology 115: 1138–1150. [DOI] [PubMed] [Google Scholar]
- Peterson SW. (2000). Phylogenetic relationships in Aspergillus based on rDNA sequence analysis. In: Integration of Modern Taxonomic Methods for Penicillium and Aspergillus Classification (Samson RA, Pitt JI, eds). Harwood Academic Publishers, UK: 323–355. [Google Scholar]
- Peterson SW. (2008). Phylogenetic analysis of Aspergillus species using DNA sequences from four loci. Mycologia 100: 205–226. [DOI] [PubMed] [Google Scholar]
- Pitt JI, Hocking AD. (2009). Aspergillus and related teleomorphs. In: Fungi and Food Spoilage. Springer, USA: 275–337. [Google Scholar]
- R Core Team (2016). R: a language and environment for statistical computing. R foundation for statistical computing, Vienna, Austria. [Google Scholar]
- Raper KB, Fennell DI. (1965). The genus Aspergillus. Williams & Wilkins, USA. [Google Scholar]
- Rawlings ND, Barrett AJ, Thomas PD, et al. (2018). The MEROPS database of proteolytic enzymes, their substrates and inhibitors in 2017 and a comparison with peptidases in the PANTHER database. Nucleic Acids Research 46: D624–D632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards TA. (2011). Genome evolution: horizontal movements in the fungi. Current Biology 21: R166–R168. [DOI] [PubMed] [Google Scholar]
- Rokas A, Williams BL, King N, et al. (2003). Genome-scale approaches to resolving incongruence in molecular phylogenies. Nature 425: 798–804. [DOI] [PubMed] [Google Scholar]
- Ronquist F, Teslenko M, van der Mark P, et al. (2012). MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Systematic Biology 61: 539–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salah H, Lackner M, Houbraken J, et al. (2019). The emergence of rare clinical Aspergillus species in Qatar: molecular characterization and antifungal susceptibility profiles. Frontiers in Microbiology 10: 1677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samson RA, Houbraken J, Thrane U, et al. (2010). Food and indoor fungi. CBS Laboratory Manual Series 2. CBS-KNAW Fungal Biodiversity Centre, The Netherlands. [Google Scholar]
- Samson RA, Houbraken J, Thrane U, et al. (2019). Food and Indoor Fungi. Westerdijk Fungal Biodiversity Institute, The Netherlands. [Google Scholar]
- Samson RA, Houbraken JAMP, Kuijpers AFA, et al. (2004). New ochratoxin A or sclerotium producing species in Aspergillus section Nigri. Studies in Mycology 50: 45–61. [Google Scholar]
- Samson RA, Hubka V, Varga J, et al. (2017). Response to Pitt & Taylor 2016: conservation of Aspergillus with A. niger as the conserved type is unnecessary and potentially disruptive. Taxon 66: 1439–1446. [Google Scholar]
- Samson RA, Noonim P, Meijer M, et al. (2007). Diagnostic tools to identify black aspergilli. Studies in Mycology 59: 129–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuster E, Dunn-Coleman N, Frisvad J, et al. (2002). On the safety of Aspergillus niger - a review. Applied Microbiology and Biotechnology 59: 426–435. [DOI] [PubMed] [Google Scholar]
- Seekles SJ, Punt M, Savelkoel N, et al. (2022). Genome sequences of 24 Aspergillus niger sensu stricto strains to study strain diversity, heterokaryon compatibility, and sexual reproduction. G3 (Bethesda) 12: jkac124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo T-K. (2008). Calculating bootstrap probabilities of phylogeny using multilocus sequence data. Molecular Biology and Evolution 25: 960–971. [DOI] [PubMed] [Google Scholar]
- Seppey M, Manni M, Zdobnov EM. (2019). BUSCO: assessing genome assembly and annotation completeness. Methods in Molecular Biology 1962: 227–245. [DOI] [PubMed] [Google Scholar]
- Shen W, Le S, Li Y, et al. (2016). SeqKit: a cross-platform and ultrafast toolkit for FASTA/Q file manipulation. PLoS ONE 11: e0163962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva da JJ, Iamanaka BT, Ferranti LS, et al. (2020). Diversity within Aspergillus niger clade and description of a new species: Aspergillus vinaceus sp. nov. Journal of Fungi 6: 371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sklenář F, Glässnerová K, Jurjević Ž, et al. (2022). Taxonomy of Aspergillus series Versicolores: species reduction and lessons learned about intraspecific variability. Studies in Mycology 102: 53–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sklenář F, Jurjević Ž, Houbraken J, et al. (2021). Re-examination of species limits in Aspergillus section Flavipedes using advanced species delimitation methods and description of four new species. Studies in Mycology 99: 100120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sklenář F, Jurjević Ž, Zalar P, et al. (2017). Phylogeny of xerophilic aspergilli (subgenus Aspergillus) and taxonomic revision of section Restricti. Studies in Mycology 88: 161–236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slot JC, Rokas A. (2011). Horizontal transfer of a large and highly toxic secondary metabolic gene cluster between fungi. Current Biology 21: 134–139. [DOI] [PubMed] [Google Scholar]
- Stamatakis A. (2014). RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30: 1312–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanke M, Schöffmann O, Morgenstern B, et al. (2006). Gene prediction in eukaryotes with a generalized hidden Markov model that uses hints from external sources. BMC Bioinformatics 7: 62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steenkamp ET, Wingfield MJ, McTaggart AR, et al. (2018). Fungal species and their boundaries matter – definitions, mechanisms and practical implications. Fungal Biology Reviews 32: 104–116. [Google Scholar]
- Steenwyk JL, Shen X-X, Lind AL, et al. (2019). A robust phylogenomic time tree for biotechnologically and medically important fungi in the genera Aspergillus and Penicillium. mBio 10: e00925-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steenwyk JL, Balamurugan C, Raja HA. et al. (2022). Phylogenomics reveals extensive misidentification of fungal strains from the genus Aspergillus. bioRxiv doi: 10.1101/2022.11.22.517304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sukumaran J, Holder MT, Knowles LL. (2021). Incorporating the speciation process into species delimitation. PLoS Computational Biology 17: e1008924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swofford DL. (2003). PAUP* Phylogenetic analysis using parsimony, (*and other methods); version 4.0 b10. Sinauer Associates, USA. [Google Scholar]
- Szigeti G, Sedaghati E, Mahmoudabadi AZ, et al. (2012). Species assignment and antifungal susceptibilities of black aspergilli recovered from otomycosis cases in Iran. Mycoses 55: 333–338. [DOI] [PubMed] [Google Scholar]
- Szöllősi GJ, Davín AA, Tannier E, et al. (2015). Genome-scale phylogenetic analysis finds extensive gene transfer among fungi. Philosophical Transactions of the Royal Society B: Biological Sciences 370: 20140335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talavera G, Castresana J. (2007). Improvement of phylogenies after removing divergent and ambiguously aligned blocks from protein sequence alignments. Systematic Biology 56: 564–577. [DOI] [PubMed] [Google Scholar]
- Taniwaki MH, Pitt JI, Magan N. (2018). Aspergillus species and mycotoxins: occurrence and importance in major food commodities. Current Opinion in Food Science 23: 38–43. [Google Scholar]
- Taylor JW, Jacobson DJ, Kroken S, et al. (2000). Phylogenetic species recognition and species concepts in fungi. Fungal Genetics and Biology 31: 21–32. [DOI] [PubMed] [Google Scholar]
- Ter-Hovhannisyan V, Lomsadze A, Chernoff YO, et al. (2008). Gene prediction in novel fungal genomes using an ab initio algorithm with unsupervised training. Genome Research 18: 1979–1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thom C. (1916). Aspergillus niger group. Journal of Agricultural Research 8: 1–15. [Google Scholar]
- Thom C, Church MB. (1926). The Aspergilli. Baltimore: Williams & Wilkins. [Google Scholar]
- Thom C, Raper KB. (1945). A Manual of the Aspergilli. Baltimore: Williams & Wilkins. [Google Scholar]
- Turland NJ, Wiersema JH, Barrie FR, et al. (2018). International Code of Nomenclature for algae, fungi, and plants (Shenzhen Code) adopted by the Nineteenth International Botanical Congress Shenzhen, China, July 2017. Regnum Vegetabile 159. Glashütten: Koeltz Botanical Books. [Google Scholar]
- van Rossum G, Drake FL., Jr (2014). The python language reference. Python software foundation. [Google Scholar]
- Varga J, Szigeti G, Baranyi N, et al. (2014). Aspergillus: sex and recombination. Mycopathologia 178: 349–362. [DOI] [PubMed] [Google Scholar]
- Varga J, Frisvad JC, Kocsubé S, et al. (2011). New and revisited species in Aspergillus section Nigri. Studies in Mycology 69: 1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varga J, Kevei F, Vriesema A, et al. (1994). Mitochondrial DNA restriction fragment length polymorphisms in field isolates of the Aspergillus niger aggregate. Canadian Journal of Microbiology 40: 612–621. [DOI] [PubMed] [Google Scholar]
- Vaser R, Sović I, Nagarajan N, et al. (2017). Fast and accurate de novo genome assembly from long uncorrected reads. Genome Research 27: 737–746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vesth TC, Nybo JL, Theobald S, et al. (2018). Investigation of inter- and infraspecies variation through genome sequencing of Aspergillus section Nigri. Nature Genetics 50: 1688–1695. [DOI] [PubMed] [Google Scholar]
- Vidal-Acuña MR, Ruiz M, Torres MJ, et al. (2019). Prevalence and in vitro antifungal susceptibility of cryptic species of the genus Aspergillus isolated in clinical samples. Enfermedades Infecciosas y Microbiologia Clinica (English ed.) 37: 296–300. [DOI] [PubMed] [Google Scholar]
- Wang PM, Liu X bin, Dai YC, et al. (2018). Phylogeny and species delimitation of Flammulina: taxonomic status of winter mushroom in East Asia and a new European species identified using an integrated approach. Mycological Progress 17: 1013–1030. [Google Scholar]
- Ward OP. (1989). Fermentation Biotechnology. Prentice Hall, Englewood Cliffs, New York, USA. [Google Scholar]
- Wehmer C. (1907). Zur Kenntnis einiger Aspergillus-Arten. Centralblatt für Bakteriologie und Parasitenkunde, Abt. II 18: 385–395. [Google Scholar]
- Wilhelm KA. (1877). Beiträge zur Kenntnis des Pilzgattung Aspergillus. Doctoral Dissertation, Strasburg, Germany. [Google Scholar]
- Woudenberg JHC, Seidl MF, Groenewald JZ, et al. (2015). Alternaria section Alternaria: species, formae speciales or pathotypes? Studies in Mycology 82: 1–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada O, Takara R, Hamada R, et al. (2011). Molecular biological researches of Kuro-Koji molds, their classification and safety. Journal of Bioscience and Bioengineering 112: 233–237. [DOI] [PubMed] [Google Scholar]
- Yang L, Lübeck M, Lübeck PS. (2017). Aspergillus as a versatile cell factory for organic acid production. Fungal Biology Reviews 31: 33–49. [Google Scholar]
- Yang Z. (2015). The BPP program for species tree estimation and species delimitation. Current Zoology 61: 854–865. [Google Scholar]
- Yang Z, Rannala B. (2010). Bayesian species delimitation using multilocus sequence data. Proceedings of the National Academy of Sciences 107: 9264–9269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Rannala B. (2014). Unguided species delimitation using DNA sequence data from multiple loci. Molecular Biology and Evolution 31: 3125–3135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin Y, Mao X, Yang J, et al. (2012). dbCAN: a web resource for automated carbohydrate-active enzyme annotation. Nucleic Acids Research 40: W445–451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu T-S, Yeo S-H, Kim H-S. (2004). A new species of hyphomycetes, Aspergillus coreanus sp. nov., isolated from traditional Korean nuruk. Journal of Microbiology and Biotechnology 14: 182–187. [Google Scholar]
- Zerbino DR, Birney E. (2008). Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Research 18: 821–829. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Multilocus phylogeny of Aspergillus series Nigri based on three loci (benA, CaM and RPB2) without species A. chiangmaiensis, A. pseudopiperis and A. pseudotubingensis. Best-scoring Maximum Likelihood (ML) tree inferred in the IQ-TREE is shown; ultrafast bootstrap support values (ML bs) are appended to nodes; only support values ≥95 % are shown; the ex-type strains are designated with a bold print; the information on geographic origin and isolation source is plotted on the tree (see the legend).
Comparison of single-gene genealogies based on the benA, CaM and RPB2 loci and created by three different phylogenetic methods (only one isolate per unique multilocus haplotype is included in each phylogeny). Best-scoring single-locus maximum likelihood (ML) trees are shown; ML ultrafast bootstrap support values (ML bs), maximum parsimony bootstrap support values (MP bs) and Bayesian inference posterior probabilities (BI pp) are appended to nodes. Only support values ≥95 %, ≥70 % and ≥0.95, respectively, are shown. A dash indicates lower statistical support for a specific node, or the absence of a node in the phylogeny, while an asterisk indicates full support. The ex-type strains are designated with a bold print. Alignment characteristics, partitioning schemes and substitution models are listed in Supplementary Table S1.
Best scoring maximum likelihood tree inferred in IQ-TREE based on ITS rDNA region sequences of series Nigri members (Table 1). It is apparent from the tree that a limited variability present in this locus can only differentiate the three main lineages, and more detailed differentiation is not possible. The ITS sequence of the A. vadensis ex-type was excluded from the analysis because it contains numerous errors and unresolved positions (accession number AY585549). Alignment characteristics, partitioning schemes and substitution models are listed in Supplementary Table S1.
Characteristics of alignments, partition-merging results and best substitution model for each partition according to the Bayesian information criterion.
Assignment of strains into populations for analysis in DELINEATE based on BPP v. 4.3.
Aspergillus series Nigri genomes analyzed in this study and their basic characteristics.
Distribution of 200 orthologous genes into ten separate STACEY analyses.
List of unique multilocus haplotypes.
Maximum sequence dissimilarity between isolates of the same Aspergillus species whose species limits have been delimited using methods based on a multispecies coalescent model; only species represented by isolates from at least three countries were included.