Abstract
Background and Aims
Hybridization is a common and important force in plant evolution. One of its outcomes is introgression – the transfer of small genomic regions from one taxon to another by hybridization and repeated backcrossing. This process is believed to be common in glacial refugia, where range expansions and contractions can lead to cycles of sympatry and isolation, creating conditions for extensive hybridization and introgression. Polyploidization is another genome-wide process with a major influence on plant evolution. Both hybridization and polyploidization can have complex effects on plant evolution. However, these effects are often difficult to understand in recently evolved species complexes.
Methods
We combined flow cytometry, analyses of transcriptomic sequences and pollen tube growth assays to investigate the consequences of polyploidization, hybridization and introgression on the recent evolution of several Erysimum (Brassicaceae) species from the South of the Iberian Peninsula, a well-known glacial refugium. This species complex differentiated in the last 2 million years, and its evolution has been hypothesized to be determined mainly by polyploidization, interspecific hybridization and introgression.
Key Results
Our results support a scenario of widespread hybridization involving both extant and ‘ghost’ taxa. Several taxa studied here, most notably those with purple corollas, are polyploids, probably of allopolyploid origin. Moreover, hybridization in this group might be an ongoing phenomenon, as pre-zygotic barriers appeared weak in many cases.
Conclusions
The evolution of Erysimum spp. has been determined by hybridization to a large extent. Species with purple (polyploids) and yellow flowers (mostly diploid) exhibit a strong signature of introgression in their genomes, indicating that hybridization occurred regardless of colour and across ploidy levels. Although the adaptive value of such genomic exchanges remains unclear, our results demonstrate the significance of hybridization for plant diversification, which should be taken into account when studying plant evolution.
Keywords: Hybridization, introgression, polyploidy, allopolyploidy, glacial refugium, Brassicaceae, Erysimum spp
INTRODUCTION
Hybridization is widespread across the tree of life, determining the branching and diversification patterns of many taxonomic groups (Rieseberg and Carney, 1998; Coyne and Orr, 2004; Abbott et al., 2013; Arnold, 2015). Because of its pervasiveness, hybridization has been a subject of research for a long time (Stebbins, 1959; Anderson, 1953; Arnold et al., 1999). However, it is only recently, with the advent of next-generation sequencing, that scientists have started to analyse the dynamics of hybridization at the scale of whole genomes, thus rekindling interest in the evolutionary relevance of this phenomenon. Although the patterns of hybridization remain unexplored for many groups, the renewed research efforts have undoubtedly increased our understanding of the role of hybridization in nature (Payseur and Rieseberg, 2016; Goulet et al., 2017; Taylor and Larson, 2019).
Hybridization is particularly relevant for plant evolution, with many plant species showing hybrid origins (Mallet, 2005; Soltis and Soltis, 2009). The evolutionary outcomes of hybridization may vary widely. Interspecific hybridization can hinder speciation and therefore diversification (Mayr, 1992; Schemske, 2000; Mallet, 2005; Saari and Faeth, 2012; Gómez et al. 2015a); however, in other cases, hybridization can actually foster the formation of new species (Rieseberg et al., 2003; Stelkens and Seehausen, 2009) or the introgression of novel genetic variation (by hybridization and repeated backcrossing; Anderson and Hubricht, 1938; Anderson, 1953; Rieseberg and Wendel, 1993). In addition, the fusion of genomes between two hybridizing species can lead to changes in ploidy levels (i.e. allopolyploidization; Soltis et al., 2014). There is evidence that introgression might even span ploidy levels (e.g. gene flow between diploid and tetraploid species of Senecio; Chapman and Abbott, 2010), which led to intriguing questions about the interplay of introgression and polyploidization. However, the specifics of how hybridization, introgression and polyploidization interact to affect the evolution of particular plant groups remain poorly understood. Advancements in genomic sequencing technology and analyses are now making the challenges of characterizing these processes far more feasible, even in recently diverged lineages and taxa.
Erysimum L. is one of the largest genera of the Brassicaceae, comprising >200 species (Polatschek, 1986), and has been described as a taxonomically complex genus with a reticulated evolutionary history in which polyploidization may have affected the evolution of some clades (Marhold and Lihová, 2006; Turner, 2006; Abdelaziz, 2013; Muñoz-Pajares, 2013). This genus is distributed mainly in Eurasia, with some species in North America and North Africa (Warwick et al., 2006). Notably, more than a hundred species have been described in the Mediterranean region (Greuter et al., 1986), with particular abundance in the Iberian Peninsula, where 21 (Polatschek, 1978, 2014) or 23 (Nieto-Feliner, 1993; Mateo et al., 1998) species have been described. Most Iberian Erysimum species have yellow flowers, but six have purple corollas (Nieto-Feliner, 1993; Gómez et al., 2015b). Interestingly, previous studies suggested that some purple species may have a recent, hybrid and allopolyploid origin (Nieto-Feliner, 1992, 1993; Abdelaziz et al., 2014; Gómez et al., 2014). A history of hybridization could further suggest the possibility that the purple flower colour has been transferred across the Iberian clade through hybridization and then maintained by natural selection. This scenario would indicate that introgression and polyploidization are intertwined in this group and might have contributed to the adaptive evolution of Erysimum spp.
Here we studied signals of hybridization across six species of Erysimum (E. mediohispanicum, E. nevadense, E. fitzii, E. popovii, E. baeticum and E. bastetanum) that inhabit the Baetic Mountains, an important and dynamic glacial refugium (Médail and Diadema, 2009). The evolution of several plant species has been hypothesized to have been affected by speciation and secondary contacts in this region (Médail and Diadema, 2009; Nieto-Feliner, 2011). The repeated expansion and contraction of ranges and the subsequent cycles of sympatry and isolation might have created conditions for extensive hybridization, introgression and allopolyploid formation. This species group appears to have differentiated relatively rapidly within the last 2 million years (Osuna-Mascaró et al., 2021). Previous authors have hypothesized that this rapid evolution has been strongly affected by polyploidization and hybridization, as this group spans several ploidy levels, and some species pairs have been reported to produce fertile hybrids (Abdelaziz et al., 2014, 2021). Species of this group show characteristics that may facilitate ongoing introgression, such as growing in sympatry in some locations and having a generalist pollination system that enables gene flow among different species possible.
The main goal of this study is to disentangle the history of hybridization for the Erysimum species complex in the Baetic Mountains. Specifically, we considered both whole-genome effects of hybridization (i.e. the interplay between hybridization and polyploidization) and local, potentially important, introgression of specific genomic regions. Moreover, we also quantified pre-zygotic barriers among extant taxa to estimate the likelihood of gene flow among them. We test the hypotheses that (1) genomes of this species complex must exhibit signals of multiple hybridization events; (2) some taxa might be allopolyploid; and (3) if purple corollas are the product of introgression, hybridization and gene flow should be detectable, and pre-zygotic barriers may be weak between (at least some) yellow and purple taxa.
MATERIALS AND METHODS
Plant samples
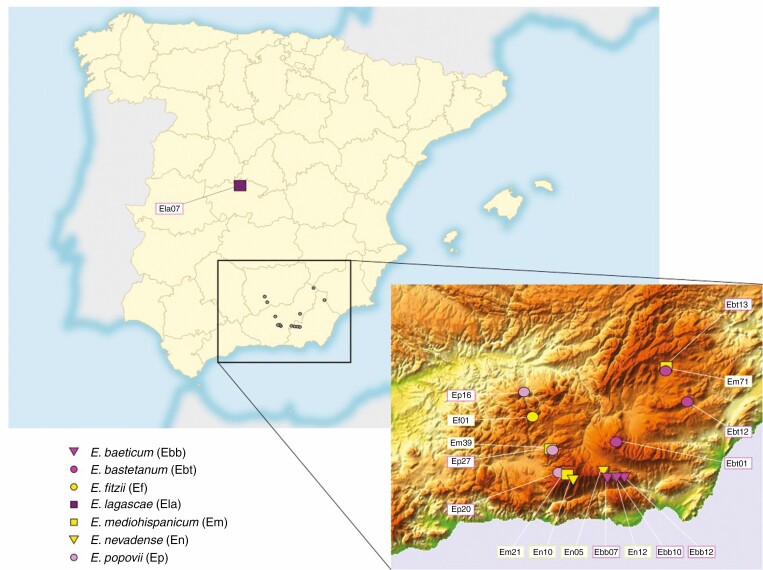
We studied six species in the genus Erysimum collected in the Baetic Mountains, South of Spain (Table 1; Fig. 1). Specifically, we sampled three different populations for E. mediohispanicum (yellow corollas; Em21, Em39 and Em71), E. nevadense (yellow corollas; En05, En10 and En12), E. popovii (purple corollas; Ep16, Ep20 and Ep27), E. bastetanum (purple corollas; Ebt01, Ebt12 and Ebt13) and E. baeticum (purple corollas; Ebb07, Ebb10 and Ebb12), and one population for E. fitzii (yellow corollas; Ef01). Some of these species appear in sympatry in some of the sampled localities (e.g. E. popovii, Ep20, and E. mediohispanicum, Em39; Table 1). Additionally, we sampled one population of E. lagascae (Ela07), an allopatric diploid species with purple corollas inhabiting Central Spain, posited as one potential parental species of the Baetic Mountain species studied here (Nieto-Feliner, 1993). We collected fully developed flower buds for transcriptomic analyses (five buds from an individual per population) and leaves for flow cytometry (6–10 individuals per population).
Table 1.
Population code, location and details of sympatry status for all of the populations sampled
| Species | Population | Location | Elevation | Geographical co-ordinates | Flower colour | Sympatry with |
|---|---|---|---|---|---|---|
| E. baeticum | Ebb07 | Sierra Nevada, Almería, Spain | 2128 | 37°05′46″N, 3°01′01″W | Purple | |
| Ebb10 | Sierra Nevada, Almería, Spain | 2140 | 37°05′32″N, 3°00′40″W | Purple | En12 | |
| Ebb12 | Sierra Nevada, Almería, Spain | 2264 | 37°05′51″N, 2°58′06″W | Purple | ||
| E. bastetanum | Ebt01 | Sierra de Baza, Granada, Spain | 1990 | 37°22′52″N, 2°51′49″W | Purple | |
| Ebt12 | Sierra de María, Almería, Spain | 1528 | 37°41′03″N, 2°10′51″W | Purple | ||
| Ebt13 | Sierra Jureña, Granada, Spain | 1352 | 37°57′10″N, 2°29′24″W | Purple | Em71 | |
| E. fitzii | Ef01 | Sierra de la Pandera, Jaén, Spain | 1804 | 37°37′56″N, 3°46′46″W | Yellow | |
| E. lagascae | Ela07 | Sierra de San Vicente, Toledo, Spain | 516 | 44°05′49″N, 4°40′40″W | Purple | |
| E. mediohispanicum | Em21 | Sierra Nevada, Granada, Spain | 1723 | 37°08′04″N, 3°25′43″W | Yellow | |
| Em39 | Sierra de Huétor, Granada, Spain | 1272 | 37°19′08″N, 3°33′11″W | Yellow | Ep20 | |
| Em71 | Sierra Jureña, Granada, Spain | 1352 | 37°57′10″N, 2°29′24″W | Yellow | Ebt13 | |
| E. nevadense | En05 | Sierra Nevada, Granada, Spain | 2074 | 37°06′35″N, 3°01′32″W | Yellow | |
| En10 | Sierra Nevada, Granada, Spain | 2321 | 37°06′37″N, 3°24′18″W | Yellow | ||
| En12 | Sierra Nevada, Granada, Spain | 2255 | 37°05′37″N, 2°56′19″W | Yellow | Ebb10 | |
| E. popovii | Ep16 | Jabalcuz, Jaén, Spain | 796 | 37°45′26″N, 3°51′02″W | Purple | |
| Ep20 | Sierra de Huétor, Granada, Spain | 1272 | 37°19′08″N, 3°33′11″W | Purple | Em39 | |
| Ep27 | Llanos del Purche, Granada, Spain | 1470 | 37°07′46″N, 3°28′48″W | Purple |
Fig. 1.
Map of the Iberian Peninsula showing the location of the sampled populations. The insert shows a more detailed map of the Baetic Mountains. Purple species are represented with purple symbols and yellow species are represented with yellow symbols. The populations Ebt13–Em71, Ebb10–En12 and Em39–Ep27 represent population pairs located in sympatry.
Flow cytometry analyses
We used flow cytometry to assess genome size and estimate DNA ploidy levels. Nuclei were isolated from fresh leaf tissues by simultaneously chopping with a razor blade 0.5 cm2 of leaf and 0.5 cm2 of an internal reference standard (Galbraith et al., 1983). We used Solanum lycopersicum L. ‘Stupické’ with 2C = 1.96 pg or Raphanus sativus L. with 2C = 1.11 pg as internal reference standards (Doležel et al., 1992). The extraction of nuclei was carried out on a Petri dish containing 1 mL of WPB buffer (Loureiro et al., 2007). Then, the nuclear suspension was filtered using a 50 µm nylon mesh, and DNA was stained with 50 µg mL–1 of propidium iodide (PI; Fluka, Buchs, Switzerland). Additionally, 50 µg mL–1 of RNase (Fluka) was added to degrade double-stranded RNA (dsRNA). After a 5 min incubation, the samples were analysed in a Sysmex CyFlow Space flow cytometer (532 nm green solid-state laser, operating at 30 mW). Results were acquired using FloMax software v2.4d (Partec GmbH, Münster, Germany) in the form of four graphics: histogram of fluorescence pulse integral in linear scale (FL); forward light scatter (FS) vs. side light scatter (SS), both in logarithmic (log) scale; FL vs. time; and FL vs. SS in log scale. The FL histogram was gated using a polygonal region defined in the FL vs. SS histogram to avoid debris signals. At least 5000 particles were analysed per sample. Only coefficient of variation (CV) values of 2C peak of each sample below 5 % were accepted; otherwise, a new sample was prepared and analysed until quality standards were achieved (Greilhuber et al., 2007). In a few cases, samples produced histograms of poorer quality even after repetition due to the presence of cytosolic compounds. Thus, it was impossible to estimate ploidy level and/or genome size for some samples (Table 2).
Table 2.
Genome size estimates and DNA ploidy levels obtained in populations of Erysimum
| Species | Population | DNA ploidy level | Genome size (2C, pg) | ||||||
|---|---|---|---|---|---|---|---|---|---|
| 2n | N | Mean | s.d. | CV | Min | Max | N | ||
| E. baeticum | Ebb07 | 8x | 5 | 2.08 | 0.08 | 3.85 | 1.93 | 2.17 | 2 |
| Ebb10 | 8x | 6 | 2.07 | 0.09 | 4.35 | 1.93 | 2.17 | 5 | |
| Ebb12 | 8x | – | – | – | – | – | – | – | |
| E. bastetanum | Ebt01 | 4x | 4 | 1.06 | 0.06 | 5.66 | 0.97 | 1.10 | 4 |
| Ebt12 | 4x | 2 | 1.06 | 0.12 | 11.32 | 0.97 | 1.15 | 2 | |
| Ebt13 | 8x | 64 | 1.96 | 0.06 | 3.06 | 1.87 | 2.17 | 60 | |
| E. fitzii | Ef01 | 2x | 3 | 0.44 | 0.004 | 0.91 | 0.44 | 0.45 | 3 |
| E. lagascae | Ela07 | 2x | 10 | 0.46 | 0.02 | 4.35 | 0.44 | 0.50 | 10 |
| E. mediohispanicum | Em21 | 2x | 2 | 0.44 | 0.01 | 2.27 | 0.43 | 0.44 | 2 |
| Em39 | 2x | 21 | 0.46 | 0.02 | 4.35 | 0.43 | 0.49 | 19 | |
| Em71 | 4x | 59 | 0.98 | 0.04 | 4.08 | 0.93 | 1.13 | 59 | |
| E. nevadense | En05 | 2x | – | – | – | – | – | – | – |
| En10 | 2x | – | – | – | – | – | – | – | |
| En12 | 2x | 3 | 0.45 | 0.03 | 6.67 | 0.42 | 0.47 | 3 | |
| E. popovii | Ep16 | 4x | 3 | 0.98 | 0.02 | 2.041.86 | 0.95 | 1.00 | 3 |
| Ep20 | 10x | 15 | 2.49 | 0.06 | 2.416 | 2.40 | 2.60 | 10 | |
| Ep27 | 4x | 39 | 0.96 | 0.04 | 4.17 | 0.92 | 1.05 | 9 |
The following data are given for each population and ploidy level: mean, the standard deviation of the mean (s.d.), coefficient of variation (CV, %), minimum (Min) and maximum values (Max) of the holoploid genome size (2C, pg) followed by sample size for genome size estimates (N); DNA ploidy level (2n) and respective sample size (N) for ploidy estimates. DNA ploidy levels: 2x, diploid; 4x, tetraploid; 8x, octoploid; 10x, decaploid. For Ebb12, En05 and En10, samples it was not possible to estimate the ploidy levels, and we have used those described in Blanca et al. (1992).
We obtained the genome size in mass units (2C in pg; sensuGreilhuber et al., 2005) using the formula: sample 2C nuclear DNA content (pg) = (sample G1 peak mean/reference standard G1 peak mean) × genome size of the reference. The ploidy levels were inferred for each sample based on chromosome counts and genome size estimates available for the genus and species (Blanca et al., 1992).
RNA extraction and sequencing
Details of the sampling, RNA extraction and sequencing appear in Osuna-Mascaró et al. (2021). In summary, we stored collected flower buds of each individual in liquid nitrogen until RNA extraction. Floral buds were ground with a mortar and a pestle in liquid nitrogen. We used the Qiagen RNeasy Plant Mini Kit following the manufacturer’s protocol to isolate total RNA from 17 samples (one individual per population; three populations of E. baeticum, E. bastetanum, E. mediohispanicum, E. nevadense and E. popovii, and one population of E. fitzii and E. lagascae). Then, we checked the quality and quantity of the RNA using a NanoDrop 2000 spectrophotometer (Thermo Fisher Scientific, Wilmington, DE, USAs) and agarose gel electrophoresis. Library preparation and RNA sequencing were conducted at Macrogen Inc. (Seoul, Korea). Before sequencing, the quality of the RNA was analysed again with the Agilent 2100 Bioanalyzer system (Agilent Technologies Inc., Santa Clara, CA, USA), and an rRNA depletion procedure with Ribo-Zero (Illumina, San Diego, CA, USA) was used to enrich mRNA content and to avoid the sequencing of rRNA. Library preparation was performed using the TruSeq Stranded Total RNA LT Sample Preparation Kit (Plant). Sequencing of the 17 libraries (one per individual) was carried out using the Hiseq 3000–4000 sequencing protocol and TruSeq 3000–4000 SBS Kit v 3 reagent, following a paired-end 150 bp strategy on the Illumina HiSeq 4000 platform. A summary of sequencing statistics is shown in Supplementary data Table S1.
Transcriptome assembly and annotation
Details of the read quality control, trimming and de novo transcriptome assembly and annotation can be found in Osuna-Mascaró et al. (2021). Briefly, we used FastQC v0.11.5 (Andrews, 2010) to analyse the quality of each library’s raw reads. Then, we trimmed the adaptors in the raw reads using cutadapt v1.15 (Martin, 2011), and we quality-filtered the reads using Sickle v1.33 (Joshi and Fass, 2011). After trimming, we used FastQC v0.11.5 (Andrews, 2010) again to verify the trimming efficiency. To assemble the resulting high-quality, cleaned reads into contigs, we followed a de novo approach using Trinity v 2.8.4 (Grabherr et al. 2011). Before assembly, each library was normalized in silico to validate and reduce the number of reads using the ‘insilico_read_normalization.pl’ function in Trinity (Haas et al., 2013). Then we used the parameter ‘min_kmer_cov 2’ to eliminate single occurrence k-mers heavily enriched in sequencing errors, following Haas et al. (2013). Candidate open reading frames (ORFs) within transcript sequences were predicted and translated using TransDecoder v 5.2.0 (Haas et al., 2013). We performed functional annotation of Trinity transcripts with ORFs using Trinotate v 3.0.1 (Haas, 2015). Sequences were searched against UniProt (UniProt Consortium, 2014), using the SwissProt databases (Bairoch and Apweiler, 2000) (with BLASTX and BLASTP searching and an e-value cut-off of 10–5). We also used the Pfam database (Bateman et al., 2004) to annotate protein domains for each predicted protein sequence. Transcripts were filtered through the eggnog (Jensen et al., 2007), GO (Gene Ontology Consortium, 2004) and KEGG (Kanehisa and Goto, 2000) annotation databases.
Variant calling
We first ran a variant calling analysis, using the E. lagascae transcriptome as a reference. We indexed the E. lagascae transcriptome using BWA v 0.7.17 (Li and Durbin, 2009) to create a reference and then mapped all the trimmed raw reads to it using the BWA v 0.7.17 ‘mem’ option (see Supplementary data Table S1 for mapping details). We used SAMtools v 1.7 (Li et al., 2009) to convert and sort the alignment files. We then called single nucleotide polymorphisms (SNPs) using the SAMtools v 1.7 ‘mpileup’ command. Lastly, we used bcftools v 1.9 to filter the SNPs (Narasimhan et al., 2016), running the SAMtools v 1.7 Perl script ‘vcfutils.pl VarFilter’ with default parameters to filter down the candidate variants and to eliminate false positives.
Orthology inference
To reduce redundancy, we clustered the translated sequences using cd-hit v 4.6 (Li and Godzik, 2006), following the steps of the pipeline described in Yang and Smith (2014). For the inference of orthologues, we excluded untranslated regions (UTRs) and non-coding transcripts, using only coding DNA sequences (CDS) in order to avoid the inclusion of sequencing errors (Yang and Smith, 2014). We identified orthologue genes using the OrthoFinder v 2.3.3 pipeline (Emms and Kelly, 2015). In brief, this pipeline first made a BLASTP analysis with the protein sequences as input for searching the orthogroups (a set of potentially orthologous protein-coding genes derived from a single gene in the last common ancestor of all the species sampled), then clustered and aligned the orthologous sequences using MAFFT v 7.450 (Katoh and Standley, 2013) with default parameters. Finally, we obtained the maximum likelihood phylogenetic gene trees for all orthogroups using IQ-Tree v 1.6.1 (Nguyen et al., 2014). Then, each orthogroup that contained sequences from all sampled species was used to infer a species tree using STAG v 1.0.0 (Emms and Kelly, 2019). Then, we used DLCpar v 1.1 (Wu et al., 2014) to reconcile the species tree with the gene trees, considering gene duplication, losses and incomplete lineage sorting (ILS) as potential causes of discordance among trees.
Phylogenetic reconstruction
We obtained a coalescent species tree using ASTRAL v 5.6.3 (Mirarab et al., 2014) with default parameters. This method reconstructs a species tree from unrooted gene tree topologies. We used the gene trees previously obtained by maximum likelihood by using IQ-Tree v 1.6.1 as input. We used FigTree v 1.4.0 (Rambaut and Drummond, 2012) to visualize and edit the species tree. Then, we compared the alternative tree topologies with the phylogeny obtained from whole-chloroplast genome analyses for the same species (presented in Osuna-Mascaró et al., 2021) using the Shimodaira–Hasegawa test (SH test; Shimodaira and Hasegawa, 1999) from the R package phangorn v 2.5.5 (Schliep, 2011). Both phylogenies were also compared visually, plotting them as mirror images with the function cophyloplot, using the R package ape v 5.4 (Paradis et al., 2004).
Discriminant analysis of principal components
We conducted a discriminant analysis of principal components (DAPC; Jombart et al., 2010) of the SNP data to group the different genotypes avoiding any prior subjective bias using the R package adegenet v 2.1.3 (Jombart and Ahmed, 2011). DAPC is a multivariate method that identifies and describes clusters of genetically related individuals from large datasets, providing a measure of the optimal number of genetic clusters (K) across a range of K values by using the Bayesian information criterion (BIC). We set a range of K values from two to seven since K = 7 is the number of different species in our dataset. The existence of significant hybridization and introgression would result in K < 7. To identify the optimal number of K, we selected the model with the lowest BIC.
Phylogenetic inference of introgression
As a first step to detect introgression events between species pairs, we computed phylogenetic species networks. This approach provides a graphical extension of the phylogenetic tree model, representing the gene flow by edges connecting the operational taxonomic units (OTUs) that are likely to be linked by introgression. We used the software PhyloNet v 3.6.9 (Than et al., 2008; Wen et al., 2018), which implements a phylogenetic network method based on the frequencies of rooted trees accounting for ILS. To generate the input for PhyloNet, we first ultrametricized the trees obtained previously with IQ-Tree v 1.6.1, using the ‘nnls’ method in the ‘force.ultrametric’ function within the R package phytools v 0.6-99 (Revell, 2012). Due to computational limitations, we inferred the species networks using a maximum pseudo-likelihood (MPL) method (Yu and Nakhleh, 2015). We performed the search five times to avoid getting stuck at local optima. We estimated optimal networks among an optimal computational range of 0–15 introgression events, determining the most likely network based on Akaike’s information criterion (AIC; Bozdogan, 1987) with the generic function for AIC in R package stats v 3.6.1. As AIC may not provide precise values when using pseudo-likelihood phylogenetic networks (Cao et al., 2019), we also estimated the more optimal network by slope heuristic of log-likelihood values. The optimal network was then visualized with Dendroscope v 3.5.10 (Huson and Scornavacca, 2019).
ABBA–BABA statistic
To assess gene flow between species, we calculated D-statistics, also known as the ABBA–BABA statistic (Durand et al., 2011). To evaluate introgression among the seven species, we used the software Dsuite v 0.1 (Malinsky et al., 2021), which allows the assessment of gene flow across large datasets and directly from a variant call format (VCF) file. This algorithm computes the D statistic by considering multiple groups of four populations: P1, P2, P3 and O, grouped in asymmetric trees of the form {[(P1, P2), P3], O}. The site patterns are ordered such that the pattern BBAA refers to P1 and P2 sharing the derived allele (B-derived allele, A-ancestral allele), ABBA to P2 and P3 sharing the derived allele and BABA to P1 and P3 sharing the derived allele. The ABBA and BABA patterns are expected to occur with equal frequencies, assuming no gene flow (null hypothesis), while a significant deviation from that suggests possible introgression. To assess whether D is significantly different from zero, D-suite uses a standard block-jackknife procedure (Green et al., 2010; Durand et al., 2011), obtaining approximately normally distributed standard errors. As recommended by Malinsky et al. (2021), we used a conservative approach estimating the statistic Dmin, which gives the lowest D-statistic value in a given trio. We used the ruby script ‘plot_d.rb’ to plot into a heatmap the introgression among all the pairs of samples. To complement these analyses, we computed the Fbranch statistic implemented in Dsuite v 0.1 (Malinsky et al., 2018, 2021). The statistic allows the identification of gene flow events within specific internal branches of a phylogeny. Thus, evaluating the excess sharing of alleles between one species and the descendant or ancestral species helps to understand when the gene flow happened. We used the whole-chloroplast genomes phylogeny from Osuna-Mascaró et al. (2021) in Newick format to establish a reference phylogeny and specify which species could be more accurately treated as sister species (i.e. as P1 and P2) while always using E. lagascae as an outgroup.
Pollen tube growth
The existence of pre-zygotic barriers can fully impede interspecific hybridization. Therefore, the existence of such barriers may indicate that gene flow across a given set of species is highly unlikely, while the lack of such barriers may indicate plausible hybridization and introgression. To explore the existence of pre-zygotic barriers, we carried out a preliminary experiment on the growth of pollen tubes on a reduced set of co-occurring species (Table 1). We collected 20 individual plants each of E. mediohispanicum, E. bastetanum and E. popovii from natural populations. We grew the plants in a common garden (University of Granada facilities) and moved them into a greenhouse before flowering to exclude pollinators. When the flowers opened, we performed hand pollination experiments by tipping the anther with a small stick to remove the pollen and placing it on the stigma of a flower from different species previously emasculated (hybrid crosses) or of a flower from the same species but from different populations previously emasculated (intraspecific crosses). Moreover, we emasculated some flowers and hand-pollinated them with their own pollen (forced selfing crosses), and some flowers were not manipulated and left for spontaneous self-pollination (spontaneous selfing crosses).
We collected the pistils after 72 h and preserved them in ethanol at 4 °C until staining of pollen tubes, following the protocol of Mori et al. (2006) with minor modifications. In brief, each pistil was cleaned in 70 % ethanol (EtOH) for 10 min and then moved to 50 % EtOH, 30 % EtOH and finally distilled water. We softened the samples by placing them on a small Petri dish of 8 m NaOH for 1 h at room temperature (as recommended by Kearns and Inouye, 1993). Then, we transferred the pistils to distilled water for 10 min, and afterwards the stigmas were incubated with 0.1 % aniline blue in phosphate buffer (pH 8.3) for 2 h. The final slide preparations were examined under a fluorescence microscope with blue light (410 nm) to observe and measure pollen tube development.
RESULTS
Ploidy levels
Flow cytometry revealed a wide variation in genome size and, therefore, in DNA ploidy levels across but also within species (Table 2). We found that all samples of E. fitzii and E. nevadense were diploid. The other species with yellow corollas, E. mediohispanicum, also appeared to be predominantly diploid, although the Em71 population deviated from this pattern, being tetraploid. The genome size of E. lasgacae also corresponded to that of a diploid, while the other purple corolla species showed ploidy levels higher than diploidy (Table 2). Moreover, ploidy levels differed across populations in two of these species. Populations of E. bastetanum varied between 4x and 8x, while in E. popovii the range was even greater, from 4x to 10x. In three cases (Ebb12, En05 and En10; Table 2), it was not possible to establish the ploidy level of the samples, and we used those reported in Blanca et al. (1992).
Transcriptome assembly and orthology inference
The sequencing results and the corresponding summary statistics of the assembled transcriptomes can be found in Osuna-Mascaró et al. (2021). In summary, we obtained between 104 000 and 382 000 different Trinity transcripts, producing between 66 000 and 235 000 Trinity isogenes. The total assembled bases ranged from 92 Mbp (in the Em21 population of E. mediohispanicum) to 319 Mbp (in the En10 population of E. nevadense). The number of annotated unigenes ranged between 71 606 (E. nevadense, En12) and 197 069 (E. baeticum, Ebb10); mean value 146 314.35. The highest proportion of annotated unigenes was obtained using BLASTX to search against the SwissProt reference database. Details of the annotated unigenes using different protein databases can be found in Osuna-Mascaró et al. (2021). OrthoFinder assigned 1 519 064 protein gene sequences (96.4 % of the total) to 92 984 gene families (orthogroups) (Supplementary data Table S2). Among them, 16 941 orthogroups were shared by all species, and their corresponding gene trees were used for further analyses.
Phylogenetic trees and population clustering
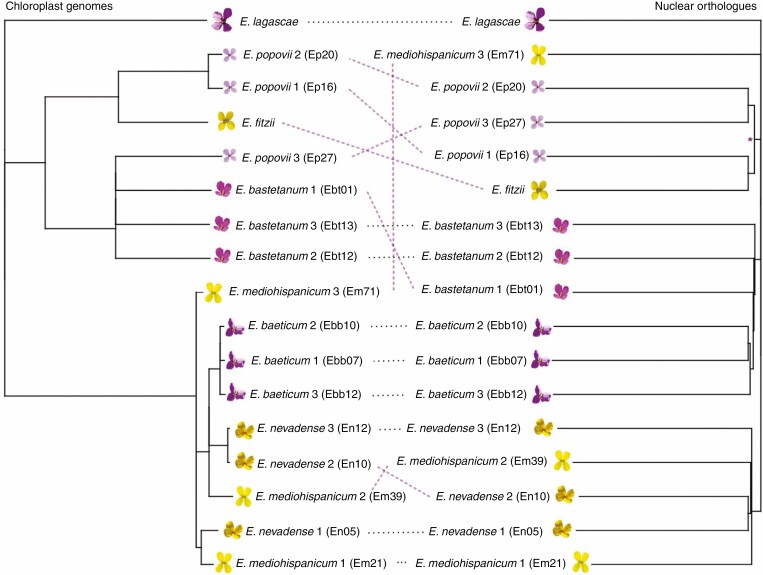
We inferred a coalescence tree using the 16 941 maximum likelihood gene trees obtained with IQ-Tree as input for ASTRAL (Fig. 2; Supplementary data Figs S1 and S2). This species tree was nearly fully resolved with high support, having only four nodes with low quartet scores results (posterior probabilities for these nodes: 0.78, 0.77, 0.70 and 0.53; see Supplementary data Fig. S1). In this tree, rooted with E. lagascae, the 4x population of E. mediohispanicum (yellow corollas; Em71) appeared as the first branching OTU. Three clades, although with low support, were evident. A clade was formed by E. bastetanum and E. baeticum, both species having purple corollas; another clade included E. fitzii (yellow corollas) and the three populations of E. popovii (purple corollas); and the last clade included the populations of E. nevadense and the 2x populations of E. mediohispanicum, both species with yellow corollas. Although there is some species clustering, not all species appear to be monophyletic, supporting a history of hybridization. Moreover, when comparing the species tree with the whole-chloroplast genomes phylogeny (Fig. 2), we find clear cytonuclear discordances resulting in a significant SH test result (Diff –ln L = 345 426.4, P-value < 0.01). This lack of congruence among both phylogenies also supports the hybridization hypothesis.
Fig. 2.
Cyto-nuclear discordance in Erysimum spp. The phylogeny on the left was obtained using whole plastid genomes in Osuna-Mascaró et al. (2021). The phylogeny on the right is a representation of nuclear genome evolution and was generated from the 16 941 maximum likelihood gene trees computed using the SNP data described in the present paper (see text for details).
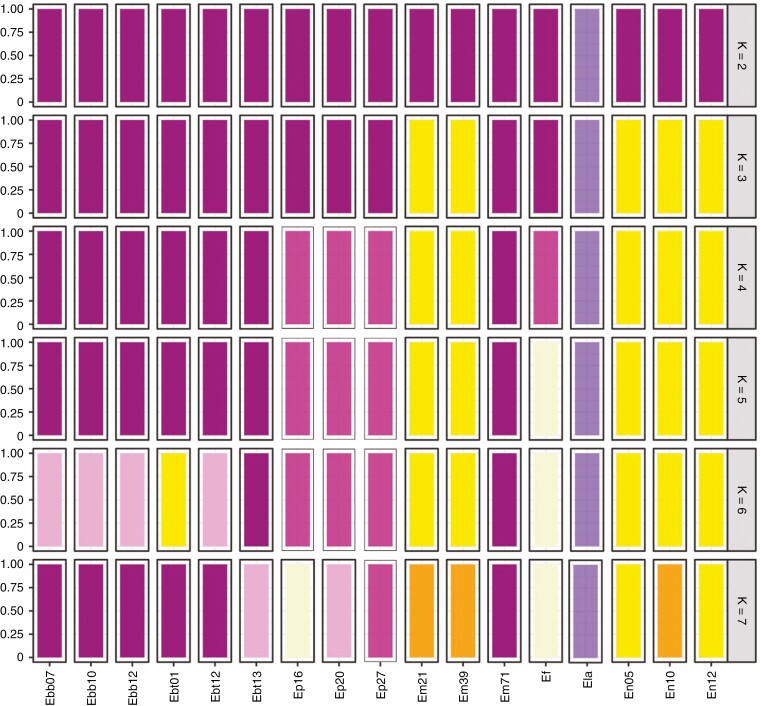
The discriminant analysis revealed K = 4 and K = 5 as the most likely number of genetic clusters (Fig. 3), both with very similar BIC values (K = 4, BIC = 189.99; K = 5, BIC = 188.99). The clusters corresponding to K = 4 produced the same clusters that appeared in the coalescence tree (Fig. 2). However, the clusters corresponding to K = 5 included three monospecific groups (for E. lagascae, purple corollas; E. fitzii, yellow corollas; and E. popovii, purple corollas), one for the diploid species with yellow corollas (the three populations of E. nevadense and the diploid populations of E. mediohispanicum) and the last including all the populations of E. baeticum (purple corollas), E. bastetanum (purple corollas), E. popovii (purple corollas) and Em71, the 4x population of E. mediohispanicum (yellow corollas).
Fig. 3.
Membership probability plot showing the DAPC results representing the populations grouped into pre-determined different clusters (ranging from K = 2 to K = 7, where each colour represents a cluster). The Bayesian analyses (BIC) revealed K = 5 as the most likely number of genetic clusters.
Analysis of introgression
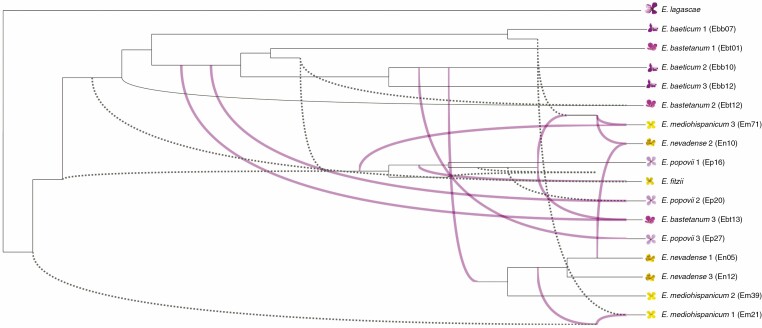
The network with 13 reticulation instances appeared as the most reliable based on the AIC values for the log-likelihood of the networks (Supplementary data Table S3). The estimates of the slope heuristic of log-likelihood values also supported the network with 13 reticulation instances as the most reliable network estimated. This network shows frequent hybridization events in the genealogy of these populations involving yellow and purple species (Fig. 4), as indicated by the edges connecting tree branches between different populations and species. Notably, this network includes edges connecting non-terminal branches (see Fig. 4), which indicates reticulations with past extinct taxa (i.e. ‘ghost species’) or incomplete sampled taxa.
Fig. 4.
Optimal species network. The graph represents a maximum pseudo-likelihood (MPL) tree with 13 reticulations computed using PhyloNet. These events are represented by edges connecting the tree branches between different individuals and indicate likely hybridization between different taxa. Note that in some instances, introgression appears to involve ancestral or extinct taxa (i.e. ghost species, dotted lines).
The ABBA–BABA analyses support this scenario of frequent hybridization, even using a conservative approach (D-min). We summarized the tested topologies and the inferred D-statistics with corrected P-values for all triplet combinations in Supplementary data Table S4 and Fig. S3. The highest signal of introgression occurred between E. fitzii (yellow corollas) and E. baeticum (purple corollas; populations Ebb12 and Ebb10) and E. popovii (purple corollas; Ep16); and between E. popovii (purple corollas; Ep16) and E. bastetanum (purple corollas; Ebt12) and E. baeticum (purple corollas; Ebb07, Ebb10 and Ebb12). There was also evidence of interspecific gene flow as manifested by the fbranch statistic (Supplementary data Fig. S4) that identifies gene flow events into specific internal branches of the phylogeny while accounting for potential false-positive results due to correlated introgression signatures among closely related species. Specifically, we found the highest signal of gene flow between E. bastetanum (purple corollas; Ebt12) and E. mediohispanicum (yellow corollas; Em71 and Em21), E. nevadense (yellow corollas; En12 and En05) and E. baeticum (purple corollas; Ebb07 and Ebb10); between E. bastetanum (purple corollas; Ebt13) and E. mediohispanicum (yellow corollas; Em71) and E. popovii (purple corollas; Ep20); and between E. bastetanum (purple corollas; Ebt01) and E. mediohispanicum (yellow corollas; Em21 and Em39) and E. nevadense (yellow corollas; En12). In addition, we have detected other gene flow events with ancestral or non-sampled taxa (Supplementary data Fig. S4).
Pollen tube growth
A total of 103 preparations of Erysimum pistils were examined: 52 from hybrid crosses, 24 from forced selfing crosses, 16 from spontaneous selfing crosses and 11 from intraspecific crosses (Supplementary data Table S5). Our results showed full growth of pollen tubes (i.e. reaching the ovary) in 63.33 % of intraspecific crosses, 51.92 % of hybrid crosses (χ 2 = 0.50, P-value = 0.513 compared with intraspecific crosses), only in 29.16 % of forced selfing crosses (χ 2 = 3.73, P-value = 0.074) and in 25.16 % of spontaneous selfing crosses (χ 2 = 4.03, P-value = 0.057). Although these last values were not significant, when selfing classes were pooled, a significant reduction in the growth of pollen tubes was shown (χ 2 = 4.93, P-value = 0.039) (Supplementary data Fig. S5). Cases in which pollen tubes grew but did not reach the ovary were treated as non-growing. In these cases, we could not estimate whether tube growth had completely stopped or if it was ongoing but developed too slowly to reach the ovary during the duration of the experiment.
Discussion
Our results suggest that the Erysimum species studied here have a strong signature of hybridization and introgression in their genomes. This result is supported by the pollen tube growth experiments that showed that pollen tubes could grow all the way to the ovary in some hybrid crosses, indicating very weak or non-existent pre-zygotic barriers. Moreover, we found that species with purple flowers are polyploid and have a strong signature of introgression, suggesting an allopolyploid origin. We also found a hybridization signature in the (mostly diploid) yellow species, indicating that hybridization occurred across both colours and ploidy levels.
Several phylogenetic reconstructions have been performed for western Mediterranean Erysimum species (e.g. Abdelaziz et al. 2014; Gómez et al., 2014, 2015b; Züst et al., 2020) that have used different strategies (several populations per species or only one representative per species; several nuclear and cytoplasmic sequences; or next-generation sequencing transcriptomic data). Abdelaziz et al. (2014) found that populations of E. mediohispanicum and E. bastetanum, species analysed here, did not appear as monophyletic (with some populations placed within other branches of the phylogeny), which was indicative that introgression probably produced important reticulation at the population level. Our analyses support this hypothesis. The reticulate nature of these phylogenies imposes some caution in interpreting phylogenies based on only a few nuclear or cytoplasmic sequences, as suggested by Chan and Levin (2005). In these cases, major divisions may reflect the reality of some old phylogenetic splits. However, it will be challenging for more recent speciation events to provide a clear picture of the phylogeny without interrogating complete genomes or transcriptomes.
Overall, our results support a hybrid origin for the purple polyploid Eysimum Iberian species, as suggested in previous studies (Nieto-Feliner, 1993; Abdelaziz et al., 2014; Gómez et al., 2015b; Osuna-Mascaró, 2020). In particular, we found support for E. popovii (purple corollas and polyploid) and E. fitzii (yellow corollas and diploid) as sister species. Also, the genome of E. popovii exhibited signatures of a hybridization process in which E. fitzii may have been involved. The possible hybrid origin of E. popovii with E. fitzii as a potential parental taxon was previously proposed by Nieto-Feliner (1993) based on morphology. Similarly, a hybrid origin of E. baeticum (purple corollas and polyploid) had been previously suggested, in this case with E. nevadense (yellow corollas and diploid) implicated as a potential parent (Nieto-Feliner, 1992). Our results showed that these two species appear to be closely related, and E. baeticum may have had an introgression signature of E. nevadense. Moreover, our results also suggested a complex scenario for E. bastetanum (purple corollas and polyploid), which appears closely related to E. baeticum (purple corollas and polyploid). In fact, E. bastetanum has been considered a subspecies of E. baeticum until recently (Lorite et al., 2015). Therefore, the general pattern that emerged from our results is that these purple species are polyploids of hybrid origin, descending from crosses between an unidentified parent and some diploid, often yellow taxon.
However, our results also suggested a complex evolutionary history for the mostly diploid yellow species. The contributing lineages also often involve unidentified taxa. This might be attributable to insufficient sampling, as we did not include some Erysimum species (E. rondae and E. myriophyllum, yellow corollas; and E. cazorlense, purple corollas) that also inhabit the Baetic Mountains, although with a limited distribution (Nieto-Feliner, 1993). At this point, it is impossible to establish whether these taxa may have acted as a source of introgression. In any case, our results show that hybridization and introgression are major contributors to the evolutionary history of this species complex, deserving further research.
Interestingly, we did not find a consistent, predictable pattern of hybridization for most species. Populations of the same species showed differences in their hybridization history, as demonstrated by the ABBA–BABA test (which detected multiple and diverse introgression events) and the PhyloNet reconstructions (which yielded a tree with 13 reticulations as the most optimal network). In the same vein, the DAPC results did not support a scenario with populations clustered by species. Our results are similar to those of previous studies describing asymmetric hybridization patterns as a consequence of differences in ecological pressures across populations and geographical areas (Payton et al., 2019; Sujii et al., 2019; Wang et al., 2019). At this stage, we cannot unambiguously identify any ecological factor behind the asymmetries we detected. However, we did observe variation in pollinators’ preferences and flowering time across populations, which might lead to local differences in gene flow patterns (unpublished data). Thus, to fully understand this asymmetry in hybridization and why some populations have more introgression signatures than others, future studies considering different ecological pressures for these species and including pollinator censures of wild populations are required. Furthermore, it would also be interesting to conduct a functional analysis of the introgressed regions to establish clearly the metabolic routes affected by them. This information, coupled with detailed ecological studies, could throw light on the adaptive processes that foster genetic exchanges in this group.
Evidence of hybridization between at least some of these species has been reported previously (Abdelaziz et al., 2014). Thus, E. mediohispanicum and E. nevadense show a hybrid zone in a sector of the Spanish Sierra Nevada (Abdelaziz et al., 2021). Pollinators do not appear to constitute strong pre-pollinating barriers since all of these species are extreme generalists and share most pollinators (Gómez et al., 2015b). Moreover, we have found that pre-zygotic, post-pollination barriers may not be effective since pollen tubes are often growing in hybrid crosses. Contemporary gene flow between different cytotypes of E. mediohispanicum seems negligible, as evidenced by an almost complete absence of triploids and other minority cytotypes in the contact zone between tetraploid and diploid populations of this species (Muñoz-Pajares et al., 2018). Historical dynamics of genetic isolation and sympatry might have also played a role (Albaladejo and Aparicio, 2007; Rifkin et al., 2019; Zieliński et al., 2019). These Erysimum species are located in a well-known glacial refugium (Médail and Diadema, 2009; Hughes and Woodward, 2017), and thus the isolation and then re-establishment of gene flow (i.e. secondary contact zones) among populations of different species may have favoured locally specific hybridization patterns (Coyne and Orr, 2004; Harrison and Larson, 2014; Arnold, 2015). A better knowledge of the historical dynamics of species and populations and overlap of past ranges is required to fully understand the genomic pattern of divergence between closely related species. For instance, combining macroecological methods with niche models and phylogenetic approaches could clarify the opportunity for hybridization through evolutionary time (Folk et al., 2018; Aguirre-Liguori et al., 2021).
Furthermore, we detected signatures of ghost introgression, implying that ancestral species have influenced the hybridization history of these Erysimum species. This result was first evidenced by cytonuclear discordance, which might be due to past organellar introgression from extinct species (Huang et al., 2014; Folk et al., 2017; Lee‐Yaw et al., 2019). We also found a clear signature of ancestral introgression in the phylogenetic species network, in which some of the reticulations appeared from introgression involving ‘ghost’ taxa. Similarly, the fbranch statistic identified gene flow events in internal branches that concurred with introgression with ghost species. Specifically, we observed that some ancestral form of E. popovii (purple corollas and polyploid) could have been related to E. fitzii (yellow corollas and polyploid). Also, we detected evidence of gene flow between an ancestor of E. mediohispanicum (yellow corollas and diploid; Em21), E. bastetanum (purple corollas and polyploid) and E. baeticum (purple corollas and polyploid). Moreover, the results showed that many past gene flow events could have occurred between E. baeticum (purple corollas and polyploid), E. nevadense (yellow corollas and diploid) and E. bastetanum (purple corollas and polyploid). In light of these results, it seems that some unidentified ancestral species played a role as introgression sources for both the purple and yellow species. However, as previously noted, we include only a sub-set of the Iberian Erysimum species in this study; accordingly, we may be mistaking the signal of the unsampled species for that of ancestral taxa. Further research about the ghost introgression’s influence on Erysimum evolution, including all the Iberian species and high-quality genome assemblies, would be required to thoroughly understand the hybridization history.
Conclusions
Our results indicate that complex evolutionary dynamics have shaped present-day Iberian Erysimum diversity. The genomes of extant taxa are the product of multiple polyploidizations, hybridization and introgression events. Understanding these multifaceted processes and their interplay is crucial to characterize the evolution of Erysimum spp. and probably of angiosperms in general. Although the evolution of the Iberian Erysimum might have been particularly dynamic, this group could be representative of the evolutionary response of multispecies complexes to drastic environmental fluctuations. Further research that incorporates a wider taxonomic sample, whole-genome sequences and complex demographic and evolutionary statistical methods is needed to precisely characterize the patterns described here.
SUPPLEMENTARY DATA
Supplementary data are available online at https://academic.oup.com/aob and consist of the following.
Figure S1: phylogeny inferred after a species tree analysis of 16 941 gene trees. Figure S2: densitree plot based on 16 941 gene trees. Figure S3: heatmap depicting the introgression found by Dsuite. Figure S4: heatmap depicting the gene flow among phylogeny branches estimated with the fbranch statistic. Figure S5: pollen tube growth as a result of hand pollination crosses. Table S1: summary of sequencing and mapping statistics. Table S2: summary of OrthoFinder results. Table S3: log likelihood and AIC for all the estimated phylogenetic species network. Table S4: D-statistics with corrected P-values for all triplet combinations. Table S5: Erysimum pistil preparations.
ACKNOWLEDGEMENTS
The authors thank Modesto Berbel Cascales, Tatiana López Pérez, Mercedes Sánchez Cabrera, Raquel Sánchez Fernández, Javier Valverde and Mohamed Abdelaziz for their help in the lab and fieldwork. Thanks to Pamela Soltis and Douglas Soltis for their help during the first steps of this work. The authors thank the Sierra Nevada National Park headquarters for providing the permits to work in the National Park. C.O.M., R.R., J.M.G. and F.P. conceived and designed the study. C.O.M. analysed the data with the help of F.P., J.L. and R.H. J.L. and S.C. performed the flow cytometry analyses. C.O.M. wrote the first draft. The final version of the manuscript was redacted with the contribution of all the authors.
Contributor Information
Carolina Osuna-Mascaró, Departamento de Genética, Universidad de Granada, Granada, Spain; Research Unit Modeling Nature, Universidad de Granada, Granada, Spain; Department of Biology, University of Nevada, Reno, 1664 North Virginia Street, Reno, NV 89557-0314, USA.
Rafael Rubio de Casas, Research Unit Modeling Nature, Universidad de Granada, Granada, Spain; Departamento de Ecología, Universidad de Granada, Granada, Spain.
José M Gómez, Research Unit Modeling Nature, Universidad de Granada, Granada, Spain; Departamento de Ecología Funcional y Evolutiva, Estación Experimental de Zonas Áridas (EEZA‐CSIC), Almería, Spain.
João Loureiro, Centre for Functional Ecology, Department of Life Sciences, University of Coimbra, Coimbra, Portugal.
Silvia Castro, Centre for Functional Ecology, Department of Life Sciences, University of Coimbra, Coimbra, Portugal.
Jacob B Landis, BTI Computational Biology Center, Boyce Thompson Institute, Ithaca, NY 14853, USA; School of Integrative Plant Science, Section of Plant Biology and the L.H. Bailey Hortorium, Cornell University, Ithaca, NY, USA.
Robin Hopkins, Department of Organismic and Evolutionary Biology, Harvard University, Cambridge, MA, USA; The Arnold Arboretum, 1300 Centre Street, Boston, MA, USA.
Francisco Perfectti, Departamento de Genética, Universidad de Granada, Granada, Spain; Research Unit Modeling Nature, Universidad de Granada, Granada, Spain.
Data Availability
Data are available in the NCBI Sequence Read Archive BioProject PRJNA607615 under the following accession numbers: E. popovii: Ep27 (SRX7756239), Ep20 (SRX7756238), Ep16 (SRX7756237); E. lagascae: Ela07 (SRX7756236); E. fitzii: Ef01 (SRX7756235); and E. bastetanum: Ebt13 (SRX7756233), Ebt12 (SRX7756232), Ebt01 (SRX7756231); and BioProject PRJNA473238 under the following accession numbers: E. baeticum: Ebb12 (SRX4130243), Ebb10 (SRX4130242), Ebb07 (SRX4130235); E. mediohispanicum: Em39 (SRX4130241), Em71 (SRX4130240), Em21 (SRX4130233); and E. nevadense: En12 (SRX4130237), En10 (SRX4130236), En05 (SRX4130234).
Funding
This research is supported by grants from FEDER/Junta de Andalucía-Consejería de Economía y Conocimiento A-RNM-505-UGR18 and P18-FR-3641. This research was also funded by the Spanish Ministry of Science and Innovation (CGL2016-79950-R and CGL2017-86626-C2-2-P), including EU FEDER funds. C.O.M. was supported by the Ministry of Economy and Competitiveness (BES-2014-069022).
LITERATURE CITED
- Abbott R, Albach D, Ansell S, et al. 2013. Hybridization and speciation. Journal of Evolutionary Biology 26: 229–246. doi: 10.1111/j.1420-9101.2012.02599.x. [DOI] [PubMed] [Google Scholar]
- Abdelaziz M. 2013. How species are evolutionarily maintained? Pollinator-mediated divergence and hybridization in Erysimum mediohispanicum and Erysimum nevadense. PhD Thesis, Universidad de Granada, Spain. [Google Scholar]
- Abdelaziz M, Muñoz-Pajares AJ, Lorite J, Herrador MB, Perfectti F, Gómez Reyes JM. 2014. Phylogenetic relationships of Erysimum (Brassicaceae) from the Baetic Mountains (SE Iberian peninsula). Anales del Jardín Botánico de Madrid 71: e005. [Google Scholar]
- Abdelaziz M, Muñoz-Pajares AJ, Berbel M, García-Muñoz A, Gómez JM, Perfectti F. 2021. Asymmetric reproductive barriers and gene flow promote the rise of a stable hybrid zone in the Mediterranean high mountain. Frontiers in Plant Science 12: 687094. doi: 10.3389/fpls.2021.687094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aguirre-Liguori JA, Ramírez-Barahona S, Gaut BS. 2021. The evolutionary genomics of species’responses to climate change. Nature Ecology & Evolution 5: 1–11. [DOI] [PubMed] [Google Scholar]
- Albaladejo RG, Aparicio A. 2007. Population genetic structure and hybridization patterns in the Mediterranean endemics Phlomis lychnitis and P. crinita (Lamiaceae). Annals of Botany 100: 735–746. doi: 10.1093/aob/mcm154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson E. 1953. Introgressive hybridization. Biological Reviews 28: 280–307. doi: 10.1111/j.1469-185x.1953.tb01379.x. [DOI] [Google Scholar]
- Anderson E, Hubricht L. 1938. Hybridization in Tradescantia. III. The evidence for introgressive hybridization. American Journal of Botany: 396–402. [Google Scholar]
- Andrews S. 2010. FastQC: a quality control tool for high throughput sequence data. http://www.bioinformatics.babraham.ac.uk/projects/fastqc/ [Google Scholar]
- Arnold ML. 2015. Divergence with genetic exchange. Oxford: Oxford University Press. [Google Scholar]
- Arnold ML, Bulger MR, Burke JM, Hempel AL, Williams JH. 1999. Natural hybridization: how low can you go and still be important? Ecology 80: 371–381. doi: 10.1890/0012-9658(1999)080[0371:nhhlcy]2.0.co;2. [DOI] [Google Scholar]
- Bairoch A, Apweiler R. 2000. The SWISS-PROT protein sequence database and its supplement TrEMBL in 2000. Nucleic Acids Research 28: 45–48. doi: 10.1093/nar/28.1.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bateman A, Coin L, Durbin R, et al. 2004. The Pfam protein families database. Nucleic Acids Research 32: D138–D141. doi: 10.1093/nar/gkh121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanca G, Torres MCM, Rejón MR. 1992. El género ‘Erysimum’ L.(‘Cruciferae’) en Andalucía (España). Anales del Jardín Botánico de Madrid 49: 201–214. [Google Scholar]
- Bozdogan H. 1987. Model selection and Akaike’s information criterion (AIC): The general theory and its analytical extensions. Psychometrika 52: 345–370. doi: 10.1007/bf02294361. [DOI] [Google Scholar]
- Cao Z, Liu X, Ogilvie HA, Yan Z, Nakhleh L. 2019. Practical aspects of phylogenetic network analysis using phylonet. BioRxiv. doi: 10.1101/746362. [Preprint]. [DOI] [Google Scholar]
- Chan KM, Levin SA. 2005. Leaky prezygotic isolation and porous genomes: rapid introgression of maternally inherited DNA. Evolution 59: 720–729. [PubMed] [Google Scholar]
- Chapman MA, Abbott RJ. 2010. Introgression of fitness genes across a ploidy barrier. New Phytologist 186: 63–71. [DOI] [PubMed] [Google Scholar]
- Coyne JA, Orr HA. 2004. Speciation . Sunderland, MA: Sinauer Associates. [Google Scholar]
- Doležel J, Sgorbati S, Lucretti S. 1992. Comparison of three DNA fluorochromes for flow cytometric estimation of nuclear DNA content in plants. Physiologia Plantarum 85: 625–631. [Google Scholar]
- Durand EY, Patterson N, Reich D, Slatkin M. 2011. Testing for ancient admixture between closely related populations. Molecular Biology and Evolution 28: 2239–2252. doi: 10.1093/molbev/msr048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emms DM, Kelly S. 2015. OrthoFinder: solving fundamental biases in whole genome comparisons dramatically improves orthogroup inference accuracy. Genome Biology 16: 157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emms DM, Kelly S. 2019. OrthoFinder: phylogenetic orthology inference for comparative genomics. Genome Biology 20: 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folk RA, Mandel JR, Freudenstein JV. 2017. Ancestral gene flow and parallel organellar genome capture result in extreme phylogenomic discord in a lineage of angiosperms. Systematic Biology 66: 320–337. doi: 10.1093/sysbio/syw083. [DOI] [PubMed] [Google Scholar]
- Folk RA, Visger CJ, Soltis PS, Soltis DE, Guralnick RP. 2018. Geographic range dynamics drove ancient hybridization in a lineage of angiosperms. The American Naturalist 192: 171–187. doi: 10.1086/698120. [DOI] [PubMed] [Google Scholar]
- Galbraith DW, Harkins KR, Maddox JM, et al. 1983. Rapid flow cytometric analysis of the cell cycle in intact plant tissues. Science 220: 1049–1051. doi: 10.1126/science.220.4601.1049. [DOI] [PubMed] [Google Scholar]
- Gene Ontology Consortium. 2004. The Gene Ontology (GO) database an informatics resource. Nucleic Acids Research 32: D258–D261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gómez JM, Perfectti F, Klingenberg CP. 2014. The role of pollinator diversity in the evolution of corolla-shape integration in a pollination-generalist plant clade. Philosophical Transactions of the Royal Society B: Biological Sciences 369: 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gómez JM, González-Mejias A, Lorite J, Abdelaziz M, Perfectti F. 2015a. The silent extinction: climate change and the potential for hybridization-mediated extinction of endemic high-mountain plants. Biodiversity and Conservation 24: 1843–1857. [Google Scholar]
- Gómez JM, Perfectti F, Lorite J. 2015b. The role of pollinators in floral diversification in a clade of generalist flowers. Evolution 69: 863–878. doi: 10.1111/evo.12632. [DOI] [PubMed] [Google Scholar]
- Goulet BE, Roda F, Hopkins R. 2017. Hybridization in plants: old ideas, new techniques. Plant Physiology 173: 65–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grabherr MG, Haas BJ, Yassour M, et al. 2011. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nature Biotechnology 29: 644–652. doi: 10.1038/nbt.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green RE, Krause J, Briggs AW, et al. 2010. A draft sequence of the Neandertal genome. Science 328: 710–722. doi: 10.1126/science.1188021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greilhuber J, Doležel J, Lysak MA, Bennett MD. 2005. The origin, evolution and proposed stabilization of the terms ‘genome size’and ‘C-value’ to describe nuclear DNA contents. Annals of Botany 95: 255–260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greilhuber J, Temsch EM, Loureiro JC. 2007. Nuclear DNA content measurement. In: Dolezel J, Greilhuber J, Suda J, eds. Flow cytometry with plant cells: analysis of genes, chromosomes and genomes. Weinheim, Germany: Wiley, 67–101. [Google Scholar]
- Greuter W, Burdet HM, Long G. 1986. Dicotyledones (Convolvulaceae-Labiatae). Med-Checklist 3: 106–116. [Google Scholar]
- Haas BJ. 2015. Trinotate: transcriptome functional annotation and analysis. https://trinotate.github.io/ [Google Scholar]
- Haas BJ, Papanicolaou A, Yassour D, et al. 2013. De novo transcript sequence reconstruction from RNA-Seq using the Trinity platform for reference generation and analysis. Nature Protocols 8: 1494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison RG, Larson EL. 2014. Hybridization, introgression, and the nature of species boundaries. Journal of Heredity 105: 795–809. doi: 10.1093/jhered/esu033. [DOI] [PubMed] [Google Scholar]
- Huang DI, Hefer CA, Kolosova N, Douglas CJ, Cronk QC. 2014. Whole plastome sequencing reveals deep plastid divergence and cytonuclear discordance between closely related balsam poplars, Populus balsamifera and P. trichocarpa (Salicaceae). New Phytologist 204: 693–703. [DOI] [PubMed] [Google Scholar]
- Hughes PD, Woodward JC. 2017. Quaternary glaciation in the Mediterranean mountains: a new synthesis. Geological Society London Special Publications 433: 1–23. [Google Scholar]
- Huson DH, Scornavacca C. 2019. User Manual for Dendroscope V 3.6.2. https://software-ab.informatik.uni-tuebingen.de/download/dendroscope/manual.pdf [Google Scholar]
- Jensen LJ, Julien P, Kuhn M, et al. 2007. eggNOG: automated construction and annotation of orthologous groups of genes. Nucleic Acids Research 36: D250–D254. doi: 10.1093/nar/gkm796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jombart T, Ahmed I. 2011. adegenet 1.3-1: new tools for the analysis of genome-wide SNP data. Bioinformatics 27: 3070–3071. doi: 10.1093/bioinformatics/btr521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jombart T, Devillard S, Balloux F. 2010. Discriminant analysis of principal components: a new method for the analysis of genetically structured populations. BMC Genetics 11: 94. doi: 10.1186/1471-2156-11-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi NA, Fass JN. 2011. Sickle: a sliding-window, adaptive, quality-based trimming tool for FastQ files (Version1.33) [Software]. https://github.com/najoshi/sickle [Google Scholar]
- Kanehisa M, Goto S. 2000. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Research 28: 27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Molecular Biology and Evolution 30: 772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kearns CA, Inouye DW. 1993. Techniques for pollination biologists. Niwot: University Press of Colorado. [Google Scholar]
- Lee‐Yaw JA, Grassa CJ, Joly S, Andrew RL, Rieseberg LH. 2019. An evaluation of alternative explanations for widespread cytonuclear discordance in annual sunflowers (Helianthus). New Phytologist 221: 515–526. [DOI] [PubMed] [Google Scholar]
- Li H, Durbin R. 2009. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics 25: 1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, et al. 2009. The sequence alignment/map format and SAMtools. Bioinformatics 25: 2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Godzik A. 2006. Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics 22: 1658–1659. doi: 10.1093/bioinformatics/btl158. [DOI] [PubMed] [Google Scholar]
- Lorite J, Perfectti F, Gómez JM. 2015. A new combination in Erysimum (Brassicaceae) for Baetic mountains (South-eastern Spain). Phytotaxa 201: 103–105. doi: 10.11646/phytotaxa.201.1.10. [DOI] [Google Scholar]
- Loureiro J, Rodriguez E, Doležel J, Santos C. 2007. Two new nuclear isolation buffers for plant DNA flow cytometry: a test with 37 species. Annals of Botany 100: 875–888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malinsky M, Svardal H, Tyers AM, et al. 2018. Whole-genome sequences of Malawi cichlids reveal multiple radiations interconnected by gene flow. Nature Ecology & Evolution 2: 1940–1919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malinsky M, Matschiner M, Svardal H. 2021. Dsuite-Fast D-statistics and related admixture evidence from VCF files. Molecular Ecology Resources 21: 584–595. doi: 10.1111/1755-0998.13265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallet J. 2005. Hybridization as an invasion of the genome. Trends in Ecology & Evolution 20: 229–237. doi: 10.1016/j.tree.2005.02.010. [DOI] [PubMed] [Google Scholar]
- Marhold K, Lihová J. 2006. Polyploidy, hybridization and reticulate evolution: lessons from the Brassicaceae. Plant Systematics and Evolution 259: 143–174. doi: 10.1007/s00606-006-0417-x. [DOI] [Google Scholar]
- Martin M. 2011. Cutadapt removes adapter sequences from high throughput sequencing reads. EMBnetJ 17: 10–12. [Google Scholar]
- Mateo G, Villalba MBC, Udias SL. 1998. Acerca del orófito minusvalorado de la Sierra de Javalambre (Teruel). Flora Montiberica 9: 41–45. [Google Scholar]
- Mayr E. 1992. A local flora and the biological species concept. American Journal of Botany 79: 222–238. doi: 10.1002/j.1537-2197.1992.tb13641.x. [DOI] [Google Scholar]
- Médail F, Diadema K. 2009. Glacial refugia influence plant diversity patterns in the Mediterranean Basin. Journal of Biogeography 36: 1333–1345. doi: 10.1111/j.1365-2699.2008.02051.x. [DOI] [Google Scholar]
- Mirarab S, Reaz R, Bayzid MS, Zimmermann T, Swenson MS, Warnow T. 2014. ASTRAL:genome-scale coalescent-based species tree estimation. Bioinformatics 30: i541–i548. doi: 10.1093/bioinformatics/btu462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori T, Kuroiwa H, Higashiyama T, Kuroiwa T. 2006. Generative Cell Specific 1 is essential for angiosperm fertilization. Nature Cell Biology 8: 64–71. doi: 10.1038/ncb1345. [DOI] [PubMed] [Google Scholar]
- Muñoz-Pajares AJ. 2013. Erysimum mediohispanicum at the evolutionary crossroad: phylogrography, phenotype, and pollinators. PhD Thesis, Universidad de Granada, Spain. [Google Scholar]
- Muñoz‐Pajares AJ, Perfectti F, Loureiro J, et al. 2018. Niche differences may explain the geographic distribution of cytotypes in Erysimum mediohispanicum. Plant Biology 20: 139–147. [DOI] [PubMed] [Google Scholar]
- Narasimhan V, Danecek P, Scally A, Xue Y, Tyler-Smith C, Durbin R. 2016. BCFtools/RoH: a hidden Markov model approach for detecting autozygosity from next-generation sequencing data. Bioinformatics 32: 1749–1751. doi: 10.1093/bioinformatics/btw044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen LT, Schmidt HA, von Haeseler A, Minh BQ. 2014. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Molecular Biology and Evolution 32: 268–274. doi: 10.1093/molbev/msu300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieto-Feliner G. 1992. Los ‘Erysimum’ orófilos nevadenses de flor amarilla y purpúreo-violácea: ¿son coespecíficos? Anales del Jardín Botánico de Madrid 50: 272–274. [Google Scholar]
- Nieto-Feliner G. 1993. Erysimum L. In: Flora iberica. Vol. 4. Cruciferae-Monotropaceae. Madrid: Real Jardín Botánico, 48–76. [Google Scholar]
- Nieto-Feliner G. 2011. Southern European glacial refugia: a tale of tales. Taxon 60: 365–372. [Google Scholar]
- Osuna Mascaró C. 2020. Hybridization as an evolutionary driver for speciation: a case in the Southern European Erysimum species. PhD Thesis, Universidad de Granada, Spain. [Google Scholar]
- Osuna-Mascaró C, Rubio de Casas R, Landis JB, Perfectti F. 2021. Genomic resources for Erysimum spp. (Brassicaceae): transcriptome and chloroplast genomes. Frontiers in Ecology and Evolution 9: 206. [Google Scholar]
- Paradis E, Claude J, Strimmer K. 2004. APE: analyses of phylogenetics and evolution in R language. Bioinformatics 20: 289–290. doi: 10.1093/bioinformatics/btg412. [DOI] [PubMed] [Google Scholar]
- Payseur BA, Rieseberg LH. 2016. A genomic perspective on hybridization and speciation. Molecular Ecology 25: 2337–2360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payton AC, Naranjo AA, Judd W, Gitzendanner M, Soltis PS, Soltis DE. 2019. Population genetics, speciation, and hybridization in Dicerandra (Lamiaceae), a North American Coastal Plain endemic, and implications for conservation. Conservation Genetics 20: 531–543. doi: 10.1007/s10592-019-01154-8. [DOI] [Google Scholar]
- Polatschek A. 1978. Die arten der gattung Erysimum auf der Iberischen Halbinsel. Annalen des Naturhistorischen Museums in Wien 325: 362. [Google Scholar]
- Polatschek A. 1986. Erysimum. In: Strid A, ed. Mountain flora of Greece. Cambridge:Cambridge University Press, 239–247. [Google Scholar]
- Polatschek A. 2014. Revision der gattung Erysimum (Cruciferae): Nachträge zu den bearbeitungen derIberischen. Halbinsel und Makaronesiens. Annalen des Naturhistorischen Museums in Wien. Serie B für Botanik und Zoologie 87: 105. [Google Scholar]
- Rambaut A, Drummond AJ. 2012. FigTree version 1.4.0. https://github.com/rambaut/figtree/ [Google Scholar]
- Revell LJ. 2012. phytools: an R package for phylogenetic comparative biology (and other things). Methods in Ecology and Evolution 3: 217–223. [Google Scholar]
- Rieseberg LH, Carney SE. 1998. Plant hybridization. New Phytologist 140: 599–624. doi: 10.1046/j.1469-8137.1998.00315.x. [DOI] [PubMed] [Google Scholar]
- Rieseberg LH, Wendel JF. 1993. Introgression and its consequences in plants. Hybrid Zones and the Evolutionary Process 70: 109. [Google Scholar]
- Rieseberg LH, Raymond O, Rosenthal DM, et al. 2003. Major ecological transitions in wild sunflowers facilitated by hybridization. Science 301: 1211–1216. doi: 10.1126/science.1086949. [DOI] [PubMed] [Google Scholar]
- Rifkin JL, Castillo AS, Liao IT, Rausher MD. 2019. Gene flow, divergent selection and resistance to introgression in two species of morning glories (Ipomoea). Molecular Ecology 28: 1709–1729. doi: 10.1111/mec.14945. [DOI] [PubMed] [Google Scholar]
- Saari S, Faeth SH. 2012. Hybridization of Neotyphodium endophytes enhances competitive ability of the host grass. New Phytologist 195: 231–236. doi: 10.1111/j.1469-8137.2012.04140.x. [DOI] [PubMed] [Google Scholar]
- Schemske DW. 2000. Understanding the Origin of Species 1. Evolution 54: 1069–1073. doi: 10.1554/0014-3820(2000)054[1069:utoos]2.3.co;2. [DOI] [Google Scholar]
- Schliep KP. 2011. phangorn: phylogenetic analysis in R. Bioinformatics 27: 592–593. doi: 10.1093/bioinformatics/btq706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimodaira H, Hasegawa M. 1999. Multiple comparisons of log-likelihoods with applications to phylogenetic inference. Molecular Biology and Evolution 16: 1114–1116. doi: 10.1093/oxfordjournals.molbev.a026201. [DOI] [Google Scholar]
- Soltis DE, Visger CJ, Soltis PS. 2014. The polyploidy revolution then and now: Stebbins revisited. American Journal of Botany 101: 1057–1078. doi: 10.3732/ajb.1400178. [DOI] [PubMed] [Google Scholar]
- Soltis PS, Soltis DE. 2009. The role of hybridization in plant speciation. Annual Review of Plant Biology 60: 561–588. [DOI] [PubMed] [Google Scholar]
- Stebbins GL. 1959. The role of hybridization in evolution. Proceedings of the American Philosophical Society 103: 231–251. [Google Scholar]
- Stelkens R, Seehausen O. 2009. Genetic distance between species predicts novel trait expression in their hybrids. Evolution 63: 884–897. doi: 10.1111/j.1558-5646.2008.00599.x. [DOI] [PubMed] [Google Scholar]
- Sujii PS, Cozzolino S, Pinheiro F. 2019. Hybridization and geographic distribution shapes the spatial genetic structure of two co-occurring orchid species. Heredity 123: 458–469. doi: 10.1038/s41437-019-0254-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor SA, Larson EL. 2019. Insights from genomes into the evolutionary importance and prevalence of hybridization in nature. Nature Ecology & Evolution 3: 170–177. doi: 10.1038/s41559-018-0777-y. [DOI] [PubMed] [Google Scholar]
- Than C, Ruths D, Nakhleh L. 2008. PhyloNet: a software package for analyzing and reconstructing reticulate evolutionary relationships. BMC Bioinformatics 9: 322. doi: 10.1186/1471-2105-9-322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner BL. 2006. Taxonomy and nomenclature of the Erysimum asperum–E. capitatum complex (Brassicaceae). Phytologia 88: 279–287. doi: 10.5962/bhl.part.10454. [DOI] [Google Scholar]
- UniProt Consortium. 2014. UniProt: a hub for protein information. Nucleic Acids Research 43: D204–D212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D, Wang Z, Kang X, Zhang J. 2019. Genetic analysis of admixture and hybrid patterns of Populus hopeiensis and P. tomentosa. Scientific Reports 9: 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warwick SI, Francis A, Al-Shehba IA. 2006. Brassicaceae: species checklist and database on CD-Rom. Plant Systematics and Evolution 259: 249–258. [Google Scholar]
- Wen D, Yu Y, Zhu J, Nakhleh L. 2018. Inferring phylogenetic networks using PhyloNet. Systematic Biology 67: 735–740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu YC, Rasmussen MD, Bansal MS, Kellis M. 2014. Most parsimonious reconciliation in the presence of gene duplication, loss, and deep coalescence using labeled coalescent trees. Genome Research 24: 475–486. doi: 10.1101/gr.161968.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Smith SA. 2014. Orthology inference in nonmodel organisms using transcriptomes and low-coverage genomes: improving accuracy and matrix occupancy for phylogenomics. Molecular Biology and Evolution 31: 3081–3092. doi: 10.1093/molbev/msu245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Y, Nakhleh L. 2015. A maximum pseudo-likelihood approach for phylogenetic networks. BMC Genomics 16: S10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zieliński P, Dudek K, Arntzen JW, et al. 2019. Differential introgression across newt hybrid zones: evidence from replicated transects. Molecular Ecology 28: 4811–4824. doi: 10.1111/mec.15251. [DOI] [PubMed] [Google Scholar]
- Züst T, Strickler SR, Powell AF, et al. 2020. Independent evolution of ancestral and novel defenses in a genus of toxic plants (Erysimum, Brassicaceae). eLife 9: e51712. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data are available in the NCBI Sequence Read Archive BioProject PRJNA607615 under the following accession numbers: E. popovii: Ep27 (SRX7756239), Ep20 (SRX7756238), Ep16 (SRX7756237); E. lagascae: Ela07 (SRX7756236); E. fitzii: Ef01 (SRX7756235); and E. bastetanum: Ebt13 (SRX7756233), Ebt12 (SRX7756232), Ebt01 (SRX7756231); and BioProject PRJNA473238 under the following accession numbers: E. baeticum: Ebb12 (SRX4130243), Ebb10 (SRX4130242), Ebb07 (SRX4130235); E. mediohispanicum: Em39 (SRX4130241), Em71 (SRX4130240), Em21 (SRX4130233); and E. nevadense: En12 (SRX4130237), En10 (SRX4130236), En05 (SRX4130234).